Abstract
Recent advances in our understanding of the genetic architecture of schizophrenia have shed light on the schizophrenia etiology. While common variation is one of the major genetic contributors, the majority of common variation reside in non-coding genome, posing a significant challenge in understanding the functional impact of this class of genetic variation. Functional genomic datasets that range from expression quantitative trait loci (eQTL) to chromatin interactions are critical to identify the potential target genes and functional consequences of non-coding variation. In this review, we discuss how three-dimensional chromatin landscape, identified by a technique called Hi-C, has facilitated the identification of potential target genes impacting schizophrenia risk. We outline key steps for Hi-C driven gene mapping, and compare Hi-C defined schizophrenia risk genes defined across developmental epochs and cell types, which offer rich insights into the temporal window and cellular etiology of schizophrenia. In contrast with a neurodevelopmental hypothesis in schizophrenia, Hi-C defined schizophrenia risk genes are postnatally enriched, suggesting that postnatal development is also important for schizophrenia pathogenesis. Moreover, Hi-C defined schizophrenia risk genes are highly expressed in excitatory neurons, highlighting excitatory neurons as a central cell type for schizophrenia. Further characterization of Hi-C defined schizophrenia risk genes demonstrated enrichment for genes that harbor loss-of-function variation in neurodevelopmental disorders, suggesting a shared genetic etiology between schizophrenia and neurodevelopmental disorders. Collectively, moving the search space from risk variants to the target genes lays a foundation to understand the neurobiological basis of schizophrenia.
1. Introduction
Schizophrenia is a highly heritable and polygenic neuropsychiatric condition characterized by psychosis, emotional withdrawal, and cognitive deficits. Heritability estimates of schizophrenia are ~80%, indicating a strong genetic inherited contribution to the disorder (Hilker et al., 2018; Wray and Visscher, 2010). While the genetic contribution is substantial, each individual variant confers only a small portion of risk, making it difficult to understand the coherent mechanism.
Three distinct classes of genetic variation have been reported to be associated with schizophrenia risk, including rare loss-of-function (LoF) variation, copy number variation (CNV), and common variation. While elevated burden of rare LoF variants in schizophrenia affected individuals has been reported, these were less often present than in individuals with autism spectrum disorder (ASD) or developmental disorder (DD), suggesting a significant but small contribution of rare disruptive variation to schizophrenia risk (Singh et al., 2016). In contrast, excess CNV burden in schizophrenia affected individuals was much stronger. Eight loci were recurrently detected as schizophrenia-associated CNVs, reaching genome-wide significance (Singh et al., 2017). These eight CNVs can contribute to 0.85% of the total population variance in liability (a theoretical trait normally distributed in the population that represents the risk to develop a certain condition; individuals with liability greater than a specific threshold are thought to develop the condition) to develop schizophrenia. However, the biggest genetic contributor to schizophrenia etiology is common variation. Genome-wide association studies (GWAS) of schizophrenia have identified more than a hundred genome-wide significant (GWS) loci, which explain a significant proportion of risk for schizophrenia (15.6% of single nucleotide polymorphism (SNP) heritability and >3% of variance in liability) (Pardinas et al., 2018; Schizophrenia Working Group of the Psychiatric Genomics Consortium et al., 2014). Unlike rare LoF variation and CNV, the majority of common variants conferring risk of schizophrenia reside in non-coding regions, making it a substantial challenge to understand the functional impact of GWS loci (Edwards et al., 2013; Schizophrenia Working Group of the Psychiatric Genomics Consortium et al., 2014).
In this review, we outline strategies to link GWS loci to genes with a focus of leveraging physical chromatin interactions obtained by Hi-C, a genome-wide chromosome conformation capture technique. Recently, high-resolution Hi-C datasets from the human brain have enabled the identification of schizophrenia risk genes across brain development and in different brain cell types. Since a direct comparison between schizophrenia risk genes identified in different studies has not been conducted, we compare them and generate a set of high-confidence schizophrenia risk genes defined by multiple Hi-C datasets. This gene set lays a foundation for deciphering the molecular mechanisms underpinning schizophrenia pathogenesis, by providing rich insights into the spatiotemporal resolutions, key cell types, and relationships with other psychiatric disorders.
2. Linking non-coding variation to genes
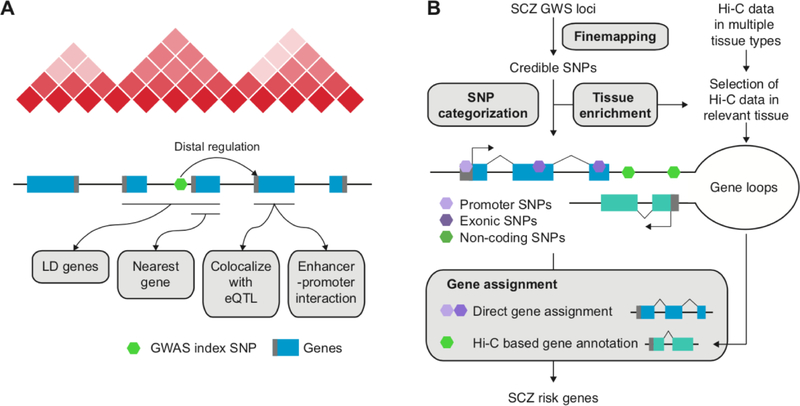
The common practice for nominating target genes for non-coding variants is to assign them to the nearest gene (Figure 1A). However, previous findings have shown that this approach gives correct target genes for only 20–30% of instances (Gusev et al., 2016; Won et al., 2016). Another commonly used method is to select all genes within the region that are in linkage disequilibrium (LD) with the index SNP (a SNP showing the strongest association with a trait within a GWS locus, Figure 1A). Given that LD structures do not reflect chromatin organization or regulatory relationships (Whalen and Pollard, 2018), this approach can miss actual target genes and can also lead to many false positive hits within the LD block.
Figure 1. Defining putative target genes of genome-wide significant (GWS) loci for schizophrenia (SCZ).
A. Four widely used methods to identify target genes for a GWS locus. The nearest gene approach is to assign an index SNP to its nearest gene. The LD gene approach is to expand the search space to the LD region to include all genes located within the LD. Two methods leverage distal regulatory information such as eQTL and enhancer-promoter interactions. Enhancer-promoter interactions can be experimentally validated by Hi-C or computationally predicted. B. GWS loci for schizophrenia are finemapped to credible SNPs, which are subsequently categorized into promoter/exonic SNPs and non-coding SNPs. Promoter/exonic SNPs are directly assigned to their target genes, while non-coding SNPs are assigned to their target genes based on chromatin interaction profiles (Hi-C). The enrichment patterns of credible SNPs in regulatory regions of different tissue and cell types can inform us about which Hi-C dataset should be used to identify correct target genes.
Currently, two types of functional genomic datasets are most frequently used to identify the target genes for GWAS loci. One approach is to leverage expression quantitative trait loci (eQTLs), and the other approach is to leverage chromatin interactions (Figure 1A). A number of frameworks to integrate GWAS and eQTLs have been developed, including a (1) colocalization-based GWS loci mapping strategy such as coloc (Giambartolomei et al., 2014), SMR (Zhu et al., 2016), and eCAVIAR (Hormozdiari et al., 2016), and (2) genome-wide mapping strategy that leverages sub-threshold SNPs such as TWAS (Gusev et al., 2016) and PrediXcan (Gamazon et al., 2015). While eQTL-based GWAS mapping is a powerful tool, obtaining enough sample size is often a challenge, especially for tissues with limited access such as brains. The PsychENCODE consortium has recently compiled more than a thousand samples to obtain eQTLs for ~33k genes in the prefrontal cortex (PFC, Wang et al., 2018). While this provides an unprecedented genomic resource, most of the eQTL studies have been conducted in the cortex, and building such resources for other brain regions and/or in different cell types is challenging, given the difficulties in obtaining such samples from hundreds of individuals.
The other complementary approach is to map GWS loci based on chromatin interaction profiles (Figure 1). While most of the GWS loci reside in non-coding regions of the genome, they are enriched in regulatory elements (Finucane et al., 2015), suggesting that they may have a regulatory function via enhancer-promoter interactions. Experimentally validated interactions, obtained from chromosome conformation capture-derived techniques, are widely used, but machine learning-based predictors of enhancer-promoter interactions such as TargetFinder (Whalen et al., 2016) and GEME (Cao et al., 2017) can be also used. These algorithms computationally predict enhancer-promoter interactions from independent datasets, such as epigenetic marks and gene expression data. While they may be useful to predict enhancer-promoter interactions in cell- or tissue-types that are difficult to obtain, a recent study has shown that the accuracy of these predictors can be inflated due to overfitting (Xi and Beer, 2018). Therefore, it is desired to use actual Hi-C datasets when available.
A key assumption for the Hi-C based gene mapping strategy is that non-coding risk variants exert their regulatory effects by physical interactions with their target genes. Indeed, a comparison between Hi-C and eQTLs suggests that ~30% of eQTLs can be explained by chromatin interactions (Wang et al., 2018). However, SNPs may affect gene regulation by changing the local or global structure of the genome. Moreover, the mechanism of action for distal regulation – whether SNPs exert their effects by changing the loop structure or by changing the transcription factor (TF) binding property while loops still remain – is unclear. Therefore, the impact of genetic variation in hierarchical structures of the genome needs to be further explored.
Due to their nature, Hi-C based mapping has some limitations. Since Hi-C contact frequency by default is exponentially decreased upon the genomic distance (Lieberman-Aiden et al., 2009), it is difficult to distinguish between genomic and physical distance for adjacent regions. Therefore, when chromatin contact matrices at 5–10kb resolution are used to detect gene loops, proximal interactions within a 10–20kb window are difficult to capture. The resolution can be increased up to 1kb by either (1) using a 4 cutter restriction enzyme and increasing the sequencing depth (Rao et al., 2014), or (2) enriching the interactions of interest, such as promoter and enhancer interactions (Mifsud et al., 2015; Mumbach et al., 2017).
In addition, chromatin interactions do not give the direction of the regulatory effect on a gene – whether risk alleles increase or decrease expression. Both active and inactive regions are engaged in chromatin interactions in a way that physically interacting regions share similar epigenetic states (Lieberman-Aiden et al., 2009; Mifsud et al., 2015; Won et al., 2016). Therefore, risk alleles mapped to the target genes may either downregulate or upregulate the target gene expression. In order to determine directionality, a combination of functional genomics approaches can be utilized, including reporter assays and CRISPR/Cas9-mediated genome engineering to identify a causal relationship between risk variants, the direction of the effect, and target genes. For example, our previous work employed Hi-C in the fetal brain, luciferase assays, and CRISPR/Cas9-mediated genome engineering and identified a causal relationship between a regulatory SNP (rs1191551) and the target gene (FOXG1, Won et al., 2016).
An important advantage of Hi-C based gene mapping is that high-resolution contact profiles can be achieved by deep sequencing without a large number of samples. Compared with eQTL studies which require hundreds of samples to get robust signals, less than a dozen samples are typically used for Hi-C. This allows flexibility to construct chromatin contact profiles for multiple brain regions, across developmental epochs, and even in different cell types. Indeed, chromatin architecture is dynamically regulated in different cell- and tissue-types and across brain development (Dixon et al., 2015; Schmitt et al., 2016; Wang et al., 2018), emphasizing the importance of building a comprehensive Hi-C map in the human brain that encompasses multiple regions, developmental epochs, and cell types. Moreover, it has been recently reported that disease genes are connected to larger regulatory domains, while depleted of eQTLs, raising a possibility that eQTL-based gene mapping requires a much larger sample size to identify eQTLs for disease genes (Wang and Goldstein, 2018). Therefore, Hi-C can complement the eQTL approach when the sample availability is limited and target genes are depleted of eQTLs.
3. Chromatin interactions identify putative target genes for schizophrenia GWS loci.
Hi-C data from brain tissues and brain cell types have successfully identified putative target genes for schizophrenia GWS loci (Table 1). In this section, we outline how Hi-C based gene mapping has been applied to schizophrenia GWAS to allow a deeper understanding of the schizophrenia etiology. Hi-C based gene mapping is a multi-step process, which includes finemapping, SNP categorization, and gene assignment (Figure 1B).
Table 1.
Hi-C data in human brain identify schizophrenia (SCZ) risk genes. NPC, neural progenitor cells; hiPSC, human induced pluripotent stem cells; res, resolution. Only the number of protein-coding genes was reported except (Giusti-Rodriguez and Sullivan, 2018).
| SCZ risk genes defined by Hi-C | Hi-C data obtained from | res | SCZ GWAS |
|---|---|---|---|
| CP: 314 / GZ: 310 (Won et al., 2012) | Two cortical laminae of the developing cortex: CP, cortical plates; GZ, germinal zone. | 10kb | (Schizophrenia Working Group of the Psychiatric Genomics Consortium et al., 2014) |
| CP: 285 / GZ: 293 (Pardinas et al., 2018) | (Pardinas et al., 2018) | ||
| Adult brain: 414 (Wang et al., 2018) | Adult dorsolateral prefrontal cortex. | 10kb | |
| NPC: 345 / Neuron: 331 / Glia: 170 (Rajarajan et al., 2018) | hiPSC-derived NPC, Neurons, Glia. | 10kb | |
| Adult brain: 1,038 (GiustiRodriguez and Sullivan, 2018) | Fetal cortex, Adult anterior temporal cortex. | 10kb |
3–1. Finemapping
An index SNP is not necessarily the causal variant, because hundreds of variants are often in high linkage disequilibrium (LD) with the index SNP, hindering the identification of causal variants at a risk locus. To address this issue, multiple computational algorithms, including CAVIAR (Hormozdiari et al., 2014) and FINEMAP (Benner et al., 2016), have been developed to identify a set of SNPs that are likely to contain causal SNPs, hereby referred to as “credible SNPs”. CAVIAR and FINEMAP use similar statistical frameworks and allow multiple causal variants. The main difference is that CAVIAR uses a greedy algorithm, while FINEMAP uses a shotgun stochastic algorithm. Since the combination of causal configurations exponentially grows upon the increased number of causal variants, CAVIAR can be computationally extensive when there are more than five causal variants. FINEMAP solves this issue by performing a stochastic search rather than evaluating all potential combinations. As a result, FINEMAP runs quicker than CAVIAR, but may miss some causal configurations by chance. Indeed, different finemapping algorithms do not necessarily give the same set of credible SNPs.
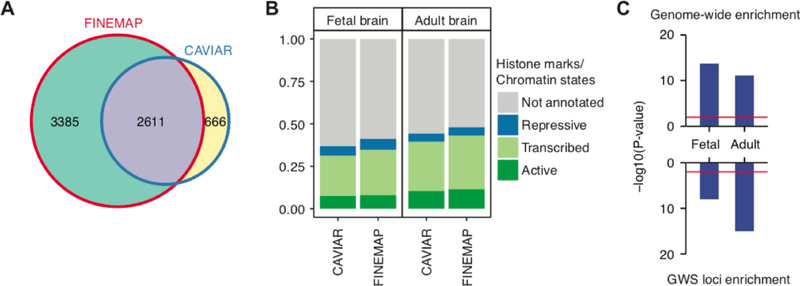
We compared credible SNPs for schizophrenia GWS loci using CAVIAR and FINEMAP (Figure 2A). While the majority of credible SNPs overlapped, FINEMAP identified 2,719 more SNPs than CAVIAR. A higher proportion of FINEMAP credible SNPs were located in active or transcribed regulatory elements in the human brain compared with CAVIAR credible SNPs (Figure 2B). Since all downstream analyses are dependent on the credible SNP selection, evaluating different finemapping algorithms is essential for Hi-C based gene mapping. We decided to use FINEMAP to define schizophrenia risk variants, because FINEMAP credible SNPs were more enriched in the brain regulatory elements than CAVIAR credible SNPs (Figure 2B).
Figure 2. Finemapping of GWS loci.
A. Different finemapping algorithms (FINEMAP and CAVIAR) give different sets of credible SNPs for schizophrenia GWS loci. B. A larger proportion of FINEMAP credible SNPs can be functionally annotated compared with CAVIAR credible SNPs. C. Credible SNPs for schizophrenia are more strongly enriched in enhancers in the adult brain than in the fetal brain.
3–2. Categorization of credible SNPs
Credible SNPs can be divided into three categories: (1) exonic SNPs, (2) promoter SNPs, and (3) non-coding SNPs (Figure 1B). SNPs that are located in the gene body and gene promoters are referred as exonic SNPs and promoter SNPs, respectively. Exonic SNPs can be further refined into functional SNPs, which cause nonsense mediated decay (NMD), missense variation, or protein-truncating variation. Promoter SNPs are subject to change based on the definition of promoter regions. We typically define promoters as 2kb upstream of transcription start site (TSS). The remaining intergenic and intronic SNPs are defined as non-coding SNPs or unannotated SNPs. The majority of credible SNPs for schizophrenia (76.3%) fall onto this category, highlighting the importance of functional annotation of non-coding SNPs. Both promoter and non-coding SNPs can be further grouped into those that reside in regulatory elements and/or those with higher evolutionary conservation.
3–3. Assign risk variants to target genes
Exonic SNPs and promoter SNPs can be directly assigned to their target genes based on their genomic coordinates, while non-coding SNPs can be mapped to their target genes via Hi-C based gene mapping (Figure 1B).
Hi-C datasets across two major developmental epochs (fetal vs. adult) and three brain cell types (neural progenitor cells, neurons, and glia) have been used to map schizophrenia GWS loci (Table 1). Within the fetal brain, Hi-C datasets are available for two cortical layers (Won et al., 2016), cortical plates (CP) and germinative zones (GZ). GZ is mainly comprised of neural progenitors, while CP is mainly comprised of post-mitotic neurons, adding an extra layer to dissect chromatin architecture during neurogenesis and neural differentiation, respectively. Adult brain Hi-C datasets are available for three brain regions, including the dorsolateral prefrontal cortex (DLPFC), anterior temporal cortex, and hippocampus (Giusti-Rodriguez and Sullivan, 2018; Schmitt et al., 2016; Wang et al., 2018).
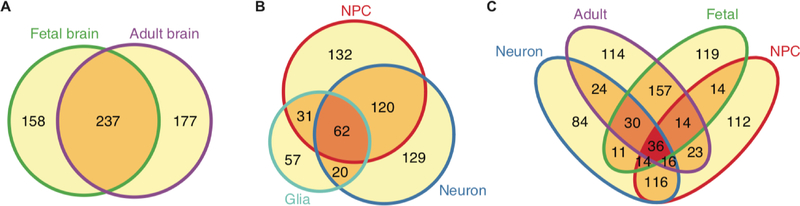
Developmental stages and cell types are key drivers of chromatin reorganization (GiustiRodriguez and Sullivan, 2018; Prashanth et al., 2018; D Wang et al., 2018). Different brain regions in the adult brain share similar chromatin architecture, which is in contrast to the substantial difference in chromatin architecture between fetal and adult brains (Giusti-Rodriguez and Sullivan, 2018; Wang et al., 2018). In addition, human induced pluripotent stem cell (hiPSC)-derived neurons and glia showed a remarked difference in chromatin architecture, including a striking difference in the number of gene loops. Neurons showed smaller number of gene loops compared with neural progenitor cells (NPCs) and glia (Rajarajan et al., 2018). Notably, even across the development, cell-type specificity appears to be the key mediator of chromatin reorganization, as changes in chromatin architecture between fetal and adult brains largely reflected changes in cell type composition (Wang et al., 2018). As expected from these results, there was a substantial difference in putative target genes for schizophrenia GWS loci upon different developmental epochs and cell types (Figure 3), which allows a new lens to understand developmental and cell type specificity of schizophrenia.
Figure 3. Comparisons of schizophrenia risk genes defined by Hi-C in different developmental stages and cell types.
Fetal brain (Won et al., 2016); Adult brain (Wang et al., 2018); NPC, Neuron, Glia (Rajarajan et al., 2018).
One important question in schizophrenia research is whether schizophrenia is a neurodevelopmental disorder (Owen et al., 2011). The role of brain development in schizophrenia etiology has recently gained attention, supported by co-expression network (Gulsuner et al., 2013) and heritability enrichment (de la Torre-Ubieta et al., 2018; Finucane et al., 2015). However, schizophrenia GWS loci are more strongly enriched in regulatory elements in the adult brain (Figure 2C), suggesting that postnatal development is also critical to schizophrenia etiology. Notably, ~40% of genes mapped to schizophrenia GWS loci differ between fetal and adult brains (Figure 3A). While the significance of this discrepancy has not been well studied, comparing these genes identified in fetal versus adult brains may provide a complementary perspective about the developmental stages implicated in schizophrenia (Figure 3A).
The cellular etiology of schizophrenia formulates another important question. Glutamate and dopamine hypotheses have been two major hypotheses in schizophrenia (Coyle et al., 2012; Snyder, 1976), suggesting that excitatory and/or dopaminergic neurons are the central cell types for schizophrenia etiology. However, it has not been well understood whether schizophrenia risk is mediated by a central cell type or a constellation of different cell types. A recent elaborate study highlighted excitatory neurons in the cortex and medium spiny neurons in the striatum as central cell types by integrating single cell transcriptomic data with schizophrenia GWAS (Skene et al., 2018). Moreover, Hi-C datasets in hiPSC-derived NPC, neurons, and glia allowed cell-type specific mapping of schizophrenia risk variants (Figure 3B, Rajarajan et al., 2018). Notably, interactions anchored to schizophrenia risk loci were enriched in NPCs and neurons, but not glia, highlighting excitatory neurons as primary cell types central to schizophrenia. Once Hi-C datasets in more refined cell types become available, cell-type specific regulatory landscape will help prioritize the cell types which can be further used to guide targeted therapeutic strategies.
4. Characteristics of schizophrenia risk genes
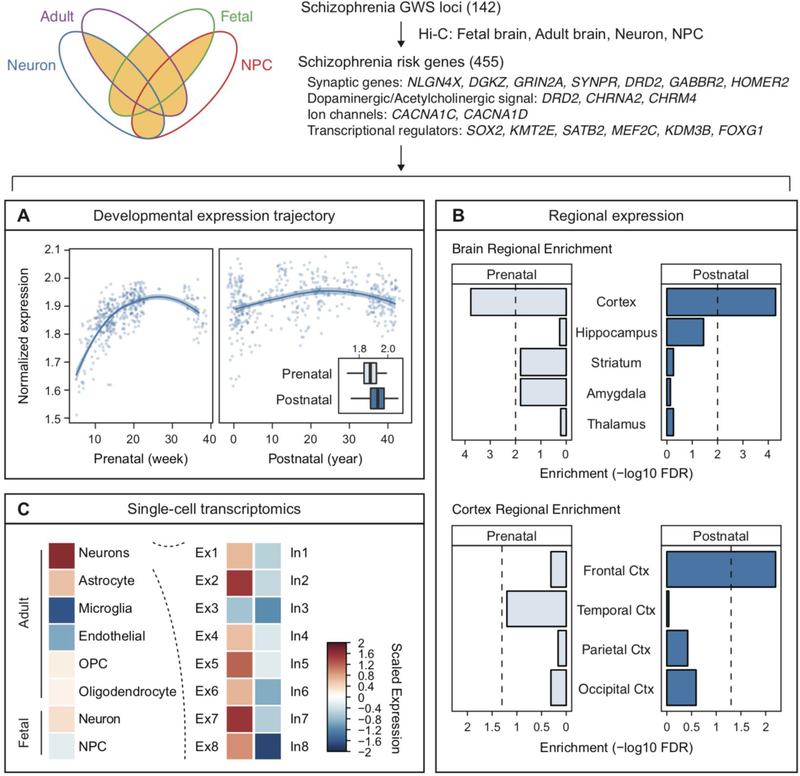
Since schizophrenia putative target genes have been defined by multiple Hi-C datasets (Table 1), we will define schizophrenia risk genes as a set of genes that have more than two evidence sources from the Hi-C datasets in the fetal cortex (Won et al., 2016), adult DLPFC (Wang et al., 2018), NPCs and neurons (Rajarajan et al., 2018). We excluded genes defined from glial Hi-C datasets, as glial interactions were not enriched for schizophrenia risk loci. To focus on risk genes assigned by Hi-C based mapping, we excluded genes mapped to promoter/exonic SNPs. In total, we got 455 Hi-C defined schizophrenia risk genes (Figure 3C, Supplementary Table 1).
Moving the search space from risk variants to targetable genes facilitates the understanding of molecular mechanisms underpinning schizophrenia. Gene ontology analysis demonstrated that Hi-C defined schizophrenia risk genes were enriched for cell adhesion molecules, chromatin remodelers, and synaptic genes (Figure 4). Moreover, interrogating the cell-type specific expression signatures and developmental/regional expression profiles of schizophrenia risk genes may refine cellulo-spatio-temporal resolution of schizophrenia (Darmanis et al., 2015; Kang et al., 2010; Lake et al., 2016). For example, schizophrenia risk genes are postnatally enriched (Figure 4A), which is in line with the enrichment of credible SNPs for regulatory elements in the adult brain (Figure 2C). They are enriched in the cortex for both prenatal and postnatal stages. Within the cortex, they show selective expression in the frontal cortex (Figure 4B). Given that the PFC is one of the key brain regions implicated in schizophrenia pathophysiology (Selemon and Zecevic, 2015), it is intriguing that the brain region impacted by genetic variants also converges onto the frontal cortex. In addition, schizophrenia risk genes are highly expressed in the excitatory neurons than in inhibitory neurons or glia (Figure 4C). This is consistent with enrichment of schizophrenia heritability in genes highly expressed in glutamatergic neurons (Skene et al., 2018).
Figure 4. Cellulo-spatio-temporal resolution of Hi-C defined schizophrenia risk genes.
Schizophrenia risk genes were defined by having more than two sources of Hi-C evidence from adult brains, fetal brains, neural progenitor cells (NPC), and neurons. A. Developmental expression trajectory of Hi-C defined schizophrenia risk genes suggests midgestation and postnatal periods as critical windows. B. Regional enrichment of schizophrenia risk genes in the frontal cortex. C. Single-cell expression values of schizophrenia risk genes highlight excitatory neurons as central cell types. Ctx, cortex; Ex, excitatory neurons; In, inhibitory neurons; OPC, oligodendrocyte precursor cells.
Common variation in schizophrenia has been found to impact LoF mutation-intolerant genes (Pardinas et al., 2018). This finding was recapitulated by another set of schizophrenia risk genes defined by multiple functional genomic resources in the adult human brain (D Wang et al., 2018), indicating that the enrichment for LoF-intolerant genes is an important feature of schizophrenia risk genes. Another interesting characteristic of schizophrenia risk genes is that they are co-regulated both at the level of transcription and protein interactions (Prashanth et al., 2018), which resembles ASD risk genes that converge onto a small number of co-expression networks (Parikshak et al., 2013). This result suggests that despite the genetic heterogeneity, risk genes for psychiatric disorders may lay onto a smaller number of convergent biological pathways. Therefore, genetics can provide an alternative lens to identify the molecular mechanisms, developmental window, brain circuits, and cell types that are impacted by the risk variants.
5. Crosstalk between common and rare variation
Genetic variation in schizophrenia range from common variation to rare LoF variation and CNV. Therefore, one important question is whether common variation and rare variation converge onto the same set of genes. Schizophrenia risk genes defined by a Hi-C driven approach (Figure 4) significantly overlap with genes that reside in recurrent CNV in schizophrenia, but not with genes that harbor rare de novo LoF variation in schizophrenia (Figure 5A, Marshall et al., 2017; Singh et al., 2016). Since the schizophrenia exome study was not yet powered to find genes at genome-wide significance (Singh et al., 2016), it is yet unclear whether rare and common variation in schizophrenia impact different genes and biological pathways. However, the significant overlap between genes impacted by common variation and CNV hints potential crosstalk between common and rare variation in schizophrenia.
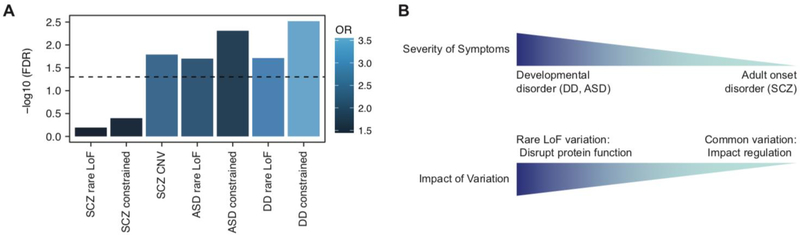
Figure 5.
A. Schizophrenia risk genes are enriched for genes that harbor rare LoF variation in neurodevelopmental disorders, such as ASD and DD. OR, odds ratio; rare LoF, genes that harbor rare loss-of-function variation in each disorder; constrained; LoF-intolerant genes that harbor rare variation in each disorder. B. Psychiatric disorders display widespread pleiotropy that the same set of genes can be impacted in multiple psychiatric disorders. This widespread pleiotropy can be partly explained by the mode of genetic variation: e.g. schizophrenia risk genes impacted by common variation may also cause neurodevelopmental disorders when impacted by LoF rare variation. However, cautions need to be exercised as this does not necessarily suggest that schizophrenia is simply a less severe form of neurodevelopmental disorders. Instead, this result suggests that identifying the mode of variation (common vs. rare, non-coding vs. protein-disrupting) is as important as variation discovery to allow comprehensive understanding of pleiotropy among psychiatric disorders.
Notably, schizophrenia risk genes significantly overlap with genes that harbor de novo LoF variation in DD and ASD, indicating pleiotropy between schizophrenia and neurodevelopmental disorders (Figure 5A, Deciphering Developmental Disorders, 2017; Sanders et al., 2015). Indeed, it has been shown that many psychiatric disorders display shared genetic etiology (Brainstorm Consortium et al., 2018). Understanding how mutations in the same gene lead to a range of clinical manifestations is therefore a critical step to explain widespread pleiotropy in psychiatric disorders.
Here we propose that different types of mutations may have a different impact on the gene function, which leads to different phenotypic severity. For example, LoF variation causes a disruption of the protein function, which leads to the earlier onset of clinical manifestations. In contrast, common variation affects gene regulation, the impact of which may appear later in life (Figure 5B). This is similar to what has been observed in Rett syndrome: mutations in MECP2 can cause a wide range of phenotypes that are dependent on the X chromosome inactivation and types of mutations (Amir et al., 2000; Zoghbi, 2003). In this model, understanding the impact of mutations on gene function is as important as gene prioritization. To achieve this goal, elucidation of comprehensive genetic architecture of psychiatric disorders that encompasses both the realm of rare and common variation is critical.
6. Discussion
Schizophrenia is a highly polygenic disorder with more than a hundred risk loci the functional consequence of which have not been fully addressed. Mapping risk loci to their target genes is often the first step to decode the functional consequence of risk loci. Hi-C datasets have become one of the key functional genomic resources to map risk loci to target genes. Recent studies fueled by the PsychENCODE consortium have provided wealth of Hi-C datasets in the human brain that span multiple developmental epochs and brain cell types. Notably, target genes for schizophrenia GWS loci differ across brain development and in different cell types, emphasizing the importance of building a comprehensive Hi-C resource that encompasses multiple brain regions, developmental epochs, and cell types. While single cell Hi-C has not been widely used to map cell-type specific chromatin landscape, we expect that it will provide a new avenue to deconvolve the complexity of chromatin architecture of the human brain. Collectively, these resources will provide an alternative perspective to refine a cellular and spatiotemporal resolution of schizophrenia etiology.
While Hi-C is an invaluable tool, it becomes much more powerful when combined with gene expression profiles, regulatory elements, eQTLs, and single cell genomics. Therefore, building a large corpus of functional genomic datasets in the human brain, as exemplified by the recent efforts of the CommonMind consortium (Fromer et al., 2016) and the PsychENCODE consortium (Akbarian et al., 2015), will greatly improve our toolbox to understand underlying mechanisms of psychiatric disorders. In addition, systemic experimental validation of regulatory elements via massively parallel reporter assay (MPRA; Melnikov et al., 2012), self-transcribing active regulatory region sequencing (STARR-seq; Arnold et al., 2013), and CRISPR-screening (Gasperini et al., 2019) will provide a complementary platform to decipher the functional impact of risk variants.
Finally, recent advancement in sequencing technology and consortia-level efforts have enabled a deep dive into the genetic architecture of not only schizophrenia but a range of psychiatric disorders. These studies have revealed that the genetic architecture of psychiatric disorders is much more complex than initially hypothesized, and both rare and common variation play a role. For example, while schizophrenia was thought to be primarily mediated by common variation, a recent exome study discovered small but significant elevated burden of rare protein disrupting variants (Singh et al., 2016). On the contrary, developmental disorders were considered as monogenic, while the first large scale GWAS in developmental disorders discovered striking burden of inherited common variants (Niemi et al., 2018). Therefore, it is crucial to identify risk variants across the allele frequency spectrum. Then, the functional genomic toolbox can be used to identify putative target genes impacted by different types of variation. If they converge onto specific biological pathways, this indicates the core mechanism for a given disorder and offers therapeutic avenues. If they point to divergent pathways, this may inform strategies to stratify genetically heterogeneous samples. Once expanded to multiple psychiatric disorders, we envision that this framework will greatly advance our understanding of the neurobiological mechanisms underlying shared genetic basis of psychiatric disorders.
Supplementary Material
Acknowledgements
This work was supported by the NIH grant R00MH113823 and NARSAD Young Investigator Award (H.W.) and Helen Lyng White Fellowship (W.M.).
Funding Source
NIH/NIMH R00MH113823 (H.W.)
NARSAD Young Investigator Award (H.W.)
Helen Lyng White Fellowship (W.M.)
Footnotes
Conflict of Interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, Jaffe AE, Pinto D, Dracheva S, Geschwind DH, Mill J, Nairn AC, Abyzov A, Pochareddy S, Prabhakar S, Weissman S, Sullivan PF, State MW, Weng Z, Peters MA, White KP, Gerstein MB, Amiri A, Armoskus C, Ashley-Koch AE, Bae T, Beckel-Mitchener A, Berman BP, Coetzee GA, Coppola G, Francoeur N, Fromer M, Gao R, Grennan K, Herstein J, Kavanagh DH, Ivanov NA, Jiang Y, Kitchen RR, Kozlenkov A, Kundakovic M, Li M, Li Z, Liu S, Mangravite LM, Mattei E, Markenscoff-Papadimitriou E, Navarro FCP, North N, Omberg L, Panchision D, Parikshak N, Poschmann J, Price AJ, Purcaro M, Reddy TE, Roussos P, Schreiner S, Scuderi S, Sebra R, Shibata M, Shieh AW, Skarica M, Sun W, Swarup V, Thomas A, Tsuji J, Van Bakel H, Wang D, Wang Y, Wang K, Werling DM, Willsey AJ, Witt H, Won H, Wong CCY, Wray GA, Wu EY, Xu X, Yao L, Senthil G, Lehner T, Sklar P, Sestan N, 2015. The PsychENCODE project. Nat. Neurosci 18 10.1038/nn.4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van Den Veyver IB, Schultz R, Malicki DM, Tran CQ, Dahle EJ, Philippi A, Timar L, Percy AK, Motil KJ, Lichtarge O, O’Brian Smith E, Glaze DG, Zoghbi HY, 2000. Influence of mutation type and X chromosome inactivation on Rett syndrome phenotypes. Ann. Neurol 47, 670–679. [DOI] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Stelzer C, Boryń ŁM, Rath M, Stark A, 2013. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339, 1074–7. 10.1126/science.1232542 [DOI] [PubMed] [Google Scholar]
- Benner C, Spencer CC, Havulinna AS, Salomaa V, Ripatti S, Pirinen M, 2016. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–1501. 10.1093/bioinformatics/btw018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brainstorm Consortium, T.B., Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, Duncan L, Escott-Price V, Falcone GJ, Gormley P, Malik R, Patsopoulos NA, Ripke S, Wei Z, Yu D, Lee PH, Turley P, Grenier-Boley B, Chouraki V, Kamatani Y, Berr C, Letenneur L, Hannequin D, Amouyel P, Boland A, Deleuze J-F, Duron E, Vardarajan BN, Reitz C, Goate AM, Huentelman MJ, Kamboh MI, Larson EB, Rogaeva E, St George-Hyslop P, Hakonarson H, Kukull WA, Farrer LA, Barnes LL, Beach TG, Demirci FY, Head E, Hulette CM, Jicha GA, Kauwe JSK, Kaye JA, Leverenz JB, Levey AI, Lieberman AP, Pankratz VS, Poon WW, Quinn JF, Saykin AJ, Schneider LS, Smith AG, Sonnen JA, Stern RA, Van Deerlin VM, Van Eldik LJ, Harold D, Russo G, Rubinsztein DC, Bayer A, Tsolaki M, Proitsi P, Fox NC, Hampel H, Owen MJ, Mead S, Passmore P, Morgan K, Nöthen MM, Rossor M, Lupton MK, Hoffmann P, Kornhuber J, Lawlor B, McQuillin A, Al-Chalabi A, Bis JC, Ruiz A, Boada M, Seshadri S, Beiser A, Rice K, van der Lee SJ, De Jager PL, Geschwind DH, Riemenschneider M, Riedel-Heller S, Rotter JI, Ransmayr G, Hyman BT, Cruchaga C, Alegret M, Winsvold B, Palta P, Farh K-H, Cuenca-Leon E, Furlotte N, Kurth T, Ligthart L, Terwindt GM, Freilinger T, Ran C, Gordon SD, Borck G, Adams HHH, Lehtimäki T, Wedenoja J, Buring JE, Schürks M, Hrafnsdottir M, Hottenga J-J, Penninx B, Artto V, Kaunisto M, Vepsäläinen S, Martin NG, Montgomery GW, Kurki MI, Hämäläinen E, Huang H, Huang J, Sandor C, Webber C, Muller-Myhsok B, Schreiber S, Salomaa V, Loehrer E, Göbel H, Macaya A, Pozo-Rosich P, Hansen T, Werge T, Kaprio J, Metspalu A, Kubisch C, Ferrari MD, Belin AC, van den Maagdenberg AMJM, Zwart J-A, Boomsma D, Eriksson N, Olesen J, Chasman DI, Nyholt DR, Avbersek A, Baum L, Berkovic S, Bradfield J, Buono R, Catarino CB, Cossette P, De Jonghe P, Depondt C, Dlugos D, Ferraro TN, French J, Hjalgrim H, Jamnadas-Khoda J, Kälviäinen R, Kunz WS, Lerche H, Leu C, Lindhout D, Lo W, Lowenstein D, McCormack M, Møller RS, Molloy A, Ng P-W, Oliver K, Privitera M, Radtke R, Ruppert A-K, Sander T, Schachter S, Schankin C, Scheffer I, Schoch S, Sisodiya SM, Smith P, Sperling M, Striano P, Surges R, Thomas GN, Visscher F, Whelan CD, Zara F, Heinzen EL, Marson A, Becker F, Stroink H, Zimprich F, Gasser T, Gibbs R, Heutink P, Martinez M, Morris HR, Sharma M, Ryten M, Mok KY, Pulit S, Bevan S, Holliday E, Attia J, Battey T, Boncoraglio G, Thijs V, Chen W-M, Mitchell B, Rothwell P, Sharma P, Sudlow C, Vicente A, Markus H, Kourkoulis C, Pera J, Raffeld M, Silliman S, Boraska Perica V, Thornton LM, Huckins LM, William Rayner N, Lewis CM, Gratacos M, Rybakowski F, Keski-Rahkonen A, Raevuori A, Hudson JI, Reichborn-Kjennerud T, Monteleone P, Karwautz A, Mannik K, Baker JH, O’Toole JK, Trace SE, Davis OSP, Helder SG, Ehrlich S, Herpertz-Dahlmann B, Danner UN, van Elburg AA, Clementi M, Forzan M, Docampo E, Lissowska J, Hauser J, Tortorella A, Maj M, Gonidakis F, Tziouvas K, Papezova H, Yilmaz Z, Wagner G, Cohen-Woods S, Herms S, Julià A, Rabionet R, Dick DM, Ripatti S, Andreassen OA, Espeseth T, Lundervold AJ, Steen VM, Pinto D, Scherer SW, Aschauer H, Schosser A, Alfredsson L, Padyukov L, Halmi KA, Mitchell J, Strober M, Bergen AW, Kaye W, Szatkiewicz JP, Cormand B, Ramos-Quiroga JA, Sánchez-Mora C, Ribasés M, Casas M, Hervas A, Arranz MJ, Haavik J, Zayats T, Johansson S, Williams N, Dempfle A, Rothenberger A, Kuntsi J, Oades RD, Banaschewski T, Franke B, Buitelaar JK, Arias Vasquez A, Doyle AE, Reif A, Lesch K-P, Freitag C, Rivero O, Palmason H, Romanos M, Langley K, Rietschel M, Witt SH, Dalsgaard S, Børglum AD, Waldman I, Wilmot B, Molly N, Bau CHD, Crosbie J, Schachar R, Loo SK, McGough JJ, Grevet EH, Medland SE, Robinson E, Weiss LA, Bacchelli E, Bailey A, Bal V, Battaglia A, Betancur C, Bolton P, Cantor R, Celestino-Soper P, Dawson G, De Rubeis S, Duque F, Green A, Klauck SM, Leboyer M, Levitt P, Maestrini E, Mane S, De-Luca DM, Parr J, Regan R, Reichenberg A, Sandin S, Vorstman J, Wassink T, Wijsman E, Cook E, Santangelo S, Delorme R, Rogé B, Magalhaes T, Arking D, Schulze TG, Thompson RC, Strohmaier J, Matthews K, Melle I, Morris D, Blackwood D, McIntosh A, Bergen SE, Schalling M, Jamain S, Maaser A, Fischer SB, Reinbold CS, Fullerton JM, Guzman-Parra J, Mayoral F, Schofield PR, Cichon S, Mühleisen TW, Degenhardt F, Schumacher J, Bauer M, Mitchell PB, Gershon ES, Rice J, Potash JB, Zandi PP, Craddock N, Ferrier IN, Alda M, Rouleau GA, Turecki G, Ophoff R, Pato C, Anjorin A, Stahl E, Leber M, Czerski PM, Cruceanu C, Jones IR, Posthuma D, Andlauer TFM, Forstner AJ, Streit F, Baune BT, Air T, Sinnamon G, Wray NR, MacIntyre DJ, Porteous D, Homuth G, Rivera M, Grove J, Middeldorp CM, Hickie I, Pergadia M, Mehta D, Smit JH, Jansen R, de Geus E, Dunn E, Li QS, Nauck M, Schoevers RA, Beekman AT, Knowles JA, Viktorin A, Arnold P, Barr CL, Bedoya-Berrio G, Bienvenu OJ, Brentani H, Burton C, Camarena B, Cappi C, Cath D, Cavallini M, Cusi D, Darrow S, Denys D, Derks EM, Dietrich A, Fernandez T, Figee M, Freimer N, Gerber G, Grados M, Greenberg E, Hanna GL, Hartmann A, Hirschtritt ME, Hoekstra PJ, Huang A, Huyser C, Illmann C, Jenike M, Kuperman S, Leventhal B, Lochner C, Lyon GJ, Macciardi F, Madruga-Garrido M, Malaty IA, Maras A, McGrath L, Miguel EC, Mir P, Nestadt G, Nicolini H, Okun MS, Pakstis A, Paschou P, Piacentini J, Pittenger C, Plessen K, Ramensky V, Ramos EM, Reus V, Richter MA, Riddle MA, Robertson MM, Roessner V, Rosário M, Samuels JF, Sandor P, Stein DJ, Tsetsos F, Van Nieuwerburgh F, Weatherall S, Wendland JR, Wolanczyk T, Worbe Y, Zai G, Goes FS, McLaughlin N, Nestadt PS, Grabe H-J, Depienne C, Konkashbaev A, Lanzagorta N, Valencia-Duarte A, Bramon E, Buccola N, Cahn W, Cairns M, Chong SA, Cohen D, Crespo-Facorro B, Crowley J, Davidson M, DeLisi L, Dinan T, Donohoe G, Drapeau E, Duan J, Haan L, Hougaard D, Karachanak-Yankova S, Khrunin A, Klovins J, Kučinskas V, Lee Chee Keong J, Limborska S, Loughland C, Lönnqvist J, Maher B, Mattheisen M, McDonald C, Murphy KC, Nenadic I, van Os J, Pantelis C, Pato M, Petryshen T, Quested D, Roussos P, Sanders AR, Schall U, Schwab SG, Sim K, So H-C, Stögmann E, Subramaniam M, Toncheva D, Waddington J, Walters J, Weiser M, Cheng W, Cloninger R, Curtis D, Gejman PV, Henskens F, Mattingsdal M, Oh S-Y, Scott R, Webb B, Breen G, Churchhouse C, Bulik CM, Daly M, Dichgans M, Faraone SV, Guerreiro R, Holmans P, Kendler KS, Koeleman B, Mathews CA, Price A, Scharf J, Sklar P, Williams J, Wood NW, Cotsapas C, Palotie A, Smoller JW, Sullivan P, Rosand J, Corvin A, Neale BM, Schott JM, Anney R, Elia J, Grigoroiu-Serbanescu M, Edenberg HJ, Murray R, 2018. Analysis of shared heritability in common disorders of the brain. Science 360, eaap8757 10.1126/science.aap8757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Anyansi C, Hu X, Xu L, Xiong L, Tang W, Mok MTS, Cheng C, Fan X, Gerstein M, Cheng ASL, Yip KY, 2017. Reconstruction of enhancer-target networks in 935 samples of human primary cells, tissues and cell lines. Nat Genet 49, 1428–1436. 10.1038/ng.3950 [DOI] [PubMed] [Google Scholar]
- Coyle JT, Basu A, Benneyworth M, Balu D, Konopaske G, 2012. Glutamatergic synaptic dysregulation in schizophrenia: therapeutic implications. Handb Exp Pharmacol 267–295. 10.1007/978-3-642-25758-2_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, Quake SR, 2015. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A 112, 7285–7290. 10.1073/pnas.1507125112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Stein JL, Won H, Opland CK, Liang D, Lu D, Geschwind DH, 2018. The Dynamic Landscape of Open Chromatin during Human Cortical Neurogenesis. Cell. 10.1016/j.cell.2017.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deciphering Developmental Disorders, S., 2017. Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438. 10.1038/nature21062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, Diao Y, Liang J, Zhao H, Lobanenkov VV, Ecker JR, Thomson JA, Ren B, 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336. 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SL, Beesley J, French JD, Dunning AM, 2013. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet 93, 779–797. 10.1016/j.ajhg.2013.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, Anttila V, Xu H, Zang C, Farh K, Ripke S, Day FR, ReproGen C, Schizophrenia Working Group of the Psychiatric Genomics, C., Consortium, R., Purcell S, Stahl E, Lindstrom S, Perry JR, Okada Y, Raychaudhuri S, Daly MJ, Patterson N, Neale BM, Price AL, 2015. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47, 1228–1235. 10.1038/ng.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, Ruderfer DM, Oh EC, Topol A, Shah HR, Klei LL, Kramer R, Pinto D, Gumus ZH, Cicek AE, Dang KK, Browne A, Lu C, Xie L, Readhead B, Stahl EA, Xiao J, Parvizi M, Hamamsy T, Fullard JF, Wang YC, Mahajan MC, Derry JM, Dudley JT, Hemby SE, Logsdon BA, Talbot K, Raj T, Bennett DA, De Jager PL, Zhu J, Zhang B, Sullivan PF, Chess A, Purcell SM, Shinobu LA, Mangravite LM, Toyoshiba H, Gur RE, Hahn CG, Lewis DA, Haroutunian V, Peters MA, Lipska BK, Buxbaum JD, Schadt EE, Hirai K, Roeder K, Brennand KJ, Katsanis N, Domenici E, Devlin B, Sklar P, 2016. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci 19, 1442–1453. 10.1038/nn.4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, Eyler AE, Denny JC, Consortium, G.Te., Nicolae DL, Cox NJ, Im HK, 2015. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 47, 1091–1098. 10.1038/ng.3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini M, Hill AJ, McFaline-Figueroa JL, Martin B, Kim S, Zhang MD, Jackson D, Leith A, Schreiber J, Noble WS, Trapnell C, Ahituv N, Shendure J, 2019. A Genome-wide Framework for Mapping Gene Regulation via Cellular Genetic Screens. Cell 176, 377–390.e19. 10.1016/j.cell.2018.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, Plagnol V, 2014. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 10, e1004383 10.1371/journal.pgen.1004383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusti-Rodriguez PMD, Sullivan PF, 2018. Schizophrenia and a high-resolution map of the three-dimensional chromatin interactome of adult and fetal cortex. bioRxiv. [Google Scholar]
- Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW, Jansen R, de Geus EJ, Boomsma DI, Wright FA, Sullivan PF, Nikkola E, Alvarez M, Civelek M, Lusis AJ, Lehtimaki T, Raitoharju E, Kahonen M, Seppala I, Raitakari OT, Kuusisto J, Laakso M, Price AL, Pajukanta P, Pasaniuc B, 2016. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 48, 245–252. 10.1038/ng.3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilker R, Helenius D, Fagerlund B, Skytthe A, Christensen K, Werge TM, Nordentoft M, Glenthoj B, 2018. Heritability of Schizophrenia and Schizophrenia Spectrum Based on the Nationwide Danish Twin Register. Biol Psychiatry 83, 492–498. 10.1016/j.biopsych.2017.08.017 [DOI] [PubMed] [Google Scholar]
- Hormozdiari F, Kostem E, Kang EY, Pasaniuc B, Eskin E, 2014. Identifying causal variants at loci with multiple signals of association. Genetics 198, 497–508. 10.1534/genetics.114.167908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, van de Bunt M, Segre AV, Li X, Joo JWJ, Bilow M, Sul JH, Sankararaman S, Pasaniuc B, Eskin E, 2016. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am J Hum Genet 99, 1245–1260. 10.1016/j.ajhg.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E, 2010. Variance component model to account for sample structure in genome-wide association studies. Nat Genet 42, 348–354. 10.1038/ng.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung HL, Chen S, Vijayaraghavan R, Wong J, Chen A, Sheng X, Kaper F, Shen R, Ronaghi M, Fan JB, Wang W, Chun J, Zhang K, 2016. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science (80-. ). 352, 1586–1590. 10.1126/science.aaf1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J, 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science (80-. ). 326, 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, Antaki D, Shetty A, Holmans PA, Pinto D, Gujral M, Brandler WM, Malhotra D, Wang Z, Fajarado KVF, Maile MS, Ripke S, Agartz I, Albus M, Alexander M, Amin F, Atkins J, Bacanu SA, Belliveau RA Jr., Bergen SE, Bertalan M, Bevilacqua E, Bigdeli TB, Black DW, Bruggeman R, Buccola NG, Buckner RL, Bulik-Sullivan B, Byerley W, Cahn W, Cai G, Cairns MJ, Campion D, Cantor RM, Carr VJ, Carrera N, Catts SV, Chambert KD, Cheng W, Cloninger CR, Cohen D, Cormican P, Craddock N, Crespo-Facorro B, Crowley JJ, Curtis D, Davidson M, Davis KL, Degenhardt F, Del Favero J, DeLisi LE, Dikeos D, Dinan T, Djurovic S, Donohoe G, Drapeau E, Duan J, Dudbridge F, Eichhammer P, Eriksson J, Escott-Price V, Essioux L, Fanous AH, Farh KH, Farrell MS, Frank J, Franke L, Freedman R, Freimer NB, Friedman JI, Forstner AJ, Fromer M, Genovese G, Georgieva L, Gershon ES, Giegling I, Giusti-Rodriguez P, Godard S, Goldstein JI, Gratten J, de Haan L, Hamshere ML, Hansen M, Hansen T, Haroutunian V, Hartmann AM, Henskens FA, Herms S, Hirschhorn JN, Hoffmann P, Hofman A, Huang H, Ikeda M, Joa I, Kahler AK, Kahn RS, Kalaydjieva L, Karjalainen J, Kavanagh D, Keller MC, Kelly BJ, Kennedy JL, Kim Y, Knowles JA, Konte B, Laurent C, Lee P, Lee SH, Legge SE, Lerer B, Levy DL, Liang KY, Lieberman J, Lonnqvist J, Loughland CM, Magnusson PKE, Maher BS, Maier W, Mallet J, Mattheisen M, Mattingsdal M, McCarley RW, McDonald C, McIntosh AM, Meier S, Meijer CJ, Melle I, Mesholam-Gately RI, Metspalu A, Michie PT, Milani L, Milanova V, Mokrab Y, Morris DW, Muller-Myhsok B, Murphy KC, Murray RM, Myin-Germeys I, Nenadic I, Nertney DA, Nestadt G, Nicodemus KK, Nisenbaum L, Nordin A, O’Callaghan E, O’Dushlaine C, Oh SY, Olincy A, Olsen L, O’Neill FA, Van Os J, Pantelis C, Papadimitriou GN, Parkhomenko E, Pato MT, Paunio T, Psychosis Endophenotypes International, C., Perkins DO, Pers TH, Pietilainen O, Pimm J, Pocklington AJ, Powell J, Price A, Pulver AE, Purcell SM, Quested D, Rasmussen HB, Reichenberg A, Reimers MA, Richards AL, Roffman JL, Roussos P, Ruderfer DM, Salomaa V, Sanders AR, Savitz A, Schall U, Schulze TG, Schwab SG, Scolnick EM, Scott RJ, Seidman LJ, Shi J, Silverman JM, Smoller JW, Soderman E, Spencer CCA, Stahl EA, Strengman E, Strohmaier J, Stroup TS, Suvisaari J, Svrakic DM, Szatkiewicz JP, Thirumalai S, Tooney PA, Veijola J, Visscher PM, Waddington J, Walsh D, Webb BT, Weiser M, Wildenauer DB, Williams NM, Williams S, Witt SH, Wolen AR, Wormley BK, Wray NR, Wu JQ, Zai CC, Adolfsson R, Andreassen OA, Blackwood DHR, Bramon E, Buxbaum JD, Cichon S, Collier DA, Corvin A, Daly MJ, Darvasi A, Domenici E, Esko T, Gejman PV, Gill M, Gurling H, Hultman CM, Iwata N, Jablensky AV, Jonsson EG, Kendler KS, Kirov G, Knight J, Levinson DF, Li QS, McCarroll SA, McQuillin A, Moran JL, Mowry BJ, Nothen MM, Ophoff RA, Owen MJ, Palotie A, Pato CN, Petryshen TL, Posthuma D, Rietschel M, Riley BP, Rujescu D, Sklar P, St Clair D, Walters JTR, Werge T, Sullivan PF, O’Donovan MC, Scherer SW, Neale BM, Sebat J, Cnv, Schizophrenia Working Groups of the Psychiatric Genomics, C., 2017. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet 49, 27–35. 10.1038/ng.3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, Callan CG, Kinney JB, Kellis M, Lander ES, Mikkelsen TS, 2012. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol 30, 271–277. 10.1038/nbt.2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, Wingett SW, Andrews S, Grey W, Ewels PA, Herman B, Happe S, Higgs A, LeProust E, Follows GA, Fraser P, Luscombe NM, Osborne CS, 2015. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet 47, 598–606. 10.1038/ng.3286 [DOI] [PubMed] [Google Scholar]
- Mumbach MR, Satpathy AT, Boyle EA, Dai C, Gowen BG, Cho SW, Nguyen ML, Rubin AJ, Granja JM, Kazane KR, Wei Y, Nguyen T, Greenside PG, Corces MR, Tycko J, Simeonov DR, Suliman N, Li R, Xu J, Flynn RA, Kundaje A, Khavari PA, Marson A, Corn JE, Quertermous T, Greenleaf WJ, Chang HY, 2017. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat Genet 49, 1602–1612. 10.1038/ng.3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi MEK, Martin HC, Rice DL, Gallone G, Gordon S, Kelemen M, McAloney K, McRae J, Radford EJ, Yu S, Gecz J, Martin NG, Wright CF, Fitzpatrick DR, Firth HV, Hurles ME, Barrett JC, 2018. Common genetic variants contribute to risk of rare severe neurodevelopmental disorders. Nature 562, 268–271. 10.1038/s41586-018-0566-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen MJ, O’Donovan MC, Thapar A, Craddock N, 2011. Neurodevelopmental hypothesis of schizophrenia. Br J Psychiatry 198, 173–175. 10.1192/bjp.bp.110.084384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardinas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, Legge SE, Bishop S, Cameron D, Hamshere ML, Han J, Hubbard L, Lynham A, Mantripragada K, Rees E, MacCabe JH, McCarroll SA, Baune BT, Breen G, Byrne EM, Dannlowski U, Eley TC, Hayward C, Martin NG, McIntosh AM, Plomin R, Porteous DJ, Wray NR, Caballero A, Geschwind DH, Huckins LM, Ruderfer DM, Santiago E, Sklar P, Stahl EA, Won H, Agerbo E, Als TD, Andreassen OA, Baekvad-Hansen M, Mortensen PB, Pedersen CB, Borglum AD, Bybjerg-Grauholm J, Djurovic S, Durmishi N, Pedersen MG, Golimbet V, Grove J, Hougaard DM, Mattheisen M, Molden E, Mors O, Nordentoft M, Pejovic-Milovancevic M, Sigurdsson E, Silagadze T, Hansen CS, Stefansson K, Stefansson H, Steinberg S, Tosato S, Werge T, Consortium G, Consortium C, Collier DA, Rujescu D, Kirov G, Owen MJ, O’Donovan MC, Walters JTR, Consortium, G., Consortium, C., Consortium, G., Consortium, C., 2018. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet 50, 381–389. 10.1038/s41588-018-0059-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH, 2013. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021. 10.1016/j.cell.2013.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prashanth R, Borrman T, Liao W, Schrode N, Flaherty E, Casiño C, Powell S, Yashaswini C, LaMarca E, Kassim B, Javidfar B, Espeso-Gil S, Li A, Won H, Geschwind DH, Ho S, MacDonald M, Hoffman GE, Roussos P, Zhang B, Hahn CG, Weng Z, Brennand KJ, Akbarian S, 2018. Neuron-specific Signatures in the Chromosomal Connectome Are Associated with Schizophrenia Risk. Science (80-. ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajarajan P, Borrman T, Liao W, Schrode N, Flaherty E, Casiño C, Powell S, Yashaswini C, LaMarca EA, Kassim B, Javidfar B, Espeso-Gil S, Li A, Won H, Geschwind DH, Ho S-M, MacDonald M, Hoffman GE, Roussos P, Zhang B, Hahn C-G, Weng Z, Brennand KJ, Akbarian S, 2018. Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science 362, eaat4311 10.1126/science.aat4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL, 2014. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 159, 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong S, Goldberg AP, Jinlu C, Keaney JF, Klei L, Mandell JD, Moreno-De-Luca D, Poultney CS, Robinson EB, Smith L, Solli-Nowlan T, Su MY, Teran NA, Walker MF, Werling DM, Beaudet AL, Cantor RM, Fombonne E, Geschwind DH, Grice DE, Lord C, Lowe JK, Mane SM, Martin DM, Morrow EM, Talkowski ME, Sutcliffe JS, Walsh CA, Yu TW, Autism Sequencing Consortium, D.H., Ledbetter DH, Martin CL, Cook EH, Buxbaum JD, Daly MJ, Devlin B, Roeder K, State MW, 2015. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 87, 1215–1233. 10.1016/j.neuron.2015.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium, Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, Holmans PA, Lee P, Bulik-Sullivan B, Collier DA, Huang H, Pers TH, Agartz I, Agerbo E, Albus M, Alexander M, Amin F, Bacanu SA, Begemann M, Jr RAB, Bene J, Bergen SE, Bevilacqua E, Bigdeli TB, Black DW, Bruggeman R, Buccola NG, Buckner RL, Byerley W, Cahn W, Cai G, Campion D, Cantor RM, Carr VJ, Carrera N, Catts SV, Chambert KD, Chan RCK, Chen RYL, Chen EYH, Cheng W, Cheung EFC, Chong SA, Cloninger CR, Cohen D, Cohen N, Cormican P, Craddock N, Crowley JJ, Curtis D, Davidson M, Davis KL, Degenhardt F, Del Favero J, Demontis D, Dikeos D, Dinan T, Djurovic S, Donohoe G, Drapeau E, Duan J, Dudbridge F, Durmishi N, Eichhammer P, Eriksson J, Escott-Price V, Essioux L, Fanous AH, Farrell MS, Frank J, Franke L, Freedman R, Freimer NB, Friedl M, Friedman JI, Fromer M, Genovese G, Georgieva L, Giegling I, Giusti-Rodríguez P, Godard S, Goldstein JI, Golimbet V, Gopal S, Gratten J, Haan L. de, Hammer C, Hamshere ML, Hansen M, Hansen T, Haroutunian V, Hartmann AM, Henskens FA, Herms S, Hirschhorn N, Hoffmann P, Hofman A, Hollegaard MV, Hougaard DM, Ikeda M, Joa I, Julià A, Kahn RS, Kalaydjieva L, Karachanak-Yankova S, Karjalainen J, Kavanagh D, Keller MC, Kennedy JL, Khrunin A, Kim Y, Klovins J, Knowles JA, Konte B, Kucinskas V, Kucinskiene ZA, Kuzelova-Ptackova H, Kähler AK, Laurent C, Keong JLC, Lee SH, Legge SE, Lerer B, Li M, Li T, Liang K-Y, Lieberman J, Limborska S, Loughland CM, Lubinski J, Lönnqvist J, Jr MM, Magnusson PKE, Maher BS, Maier W, Mallet J, Marsal S, Mattheisen M, Mattingsdal M, McCarley RW, McDonald C, McIntosh AM, Meier S, Meijer CJ, Melegh B, Melle I, Mesholam-Gately RI, Metspalu A, Michie PT, Milani L, Milanova V, Mokrab Y, Morris DW, Mors O, Murphy KC, Murray RM, Myin-Germeys I, Müller-Myhsok B, Nelis M, Nenadic I, Nertney DA, Nestadt G, Nicodemus KK, Nikitina-Zake L, Nisenbaum L, Nordin A, O’Callaghan E, O’Dushlaine C, O’Neill FA, Oh S-Y, Olincy A, Olsen L, Van Os J, Pantelis C, Papadimitriou GN, Papiol S, Parkhomenko E, Pato MT, Paunio T, Pejovic-Milovancevic M, Perkins DO, Pietiläinen O, Pimm J, Pocklington AJ, Powell J, Price A, Pulver AE, Purcell SM, Quested D, Rasmussen HB, Reichenberg A, Reimers MA, Richards AL, Roffman JL, Roussos P, Ruderfer DM, Salomaa V, Sanders AR, Schall U, Schubert CR, Schulze TG, Schwab SG, Scolnick EM, Scott RJ, Seidman LJ, Shi J, Sigurdsson E, Silagadze T, Silverman JM, Sim K, Slominsky P, Smoller JW, So H-C, Spencer CA, Stahl EA, Stefansson H, Steinberg S, Stogmann E, Straub RE, Strengman E, Strohmaier J, Stroup TS, Subramaniam M, Suvisaari J, Svrakic DM, Szatkiewicz JP, Söderman E, Thirumalai S, Toncheva D, Tosato S, Veijola J, Waddington J, Walsh D, Wang D, Wang Q, Webb BT, Weiser M, Wildenauer DB, Williams NM, Williams S, Witt SH, Wolen AR, Wong EHM, Wormley BK, Xi HS, Zai CC, Zheng X, Zimprich F, Wray NR, Stefansson K, Visscher PM, Consortium WTC-C, Adolfsson R, Andreassen OA, Blackwood DHR, Bramon E, Buxbaum JD, Børglum AD, Cichon S, Darvasi A, Domenici E, Ehrenreich H, Esko T, Gejman PV, Gill M, Gurling H, Hultman CM, Iwata N, Jablensky AV, Jönsson EG, Kendler KS, Kirov G, Knight J, Lencz T, Levinson DF, Li QS, Liu J, Malhotra AK, McCarroll SA, McQuillin A, Moran JL, Mortensen PB, Mowry BJ, Nöthen MM, Ophoff RA, Owen MJ, Palotie A, Pato CN, Petryshen TL, Posthuma D, Rietschel M, Riley BP, Rujescu D, Sham PC, Sklar P, Clair DS, Weinberger DR, Wendland JR, Werge T, Daly MJ, Sullivan PF, O’Donovan MC, 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, Tan CL, Li Y, Lin S, Lin Y, Barr CL, Ren B, 2016. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Rep 17, 2042–2059. 10.1016/j.celrep.2016.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selemon LD, Zecevic N, 2015. Schizophrenia: a tale of two critical periods for prefrontal cortical development. Transl. Psychiatry 5, e623–e623. 10.1038/tp.2015.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T, Kurki MI, Curtis D, Purcell SM, Crooks L, McRae J, Suvisaari J, Chheda H, Blackwood D, Breen G, Pietilainen O, Gerety SS, Ayub M, Blyth M, Cole T, Collier D, Coomber EL, Craddock N, Daly MJ, Danesh J, DiForti M, Foster A, Freimer NB, Geschwind D, Johnstone M, Joss S, Kirov G, Korkko J, Kuismin O, Holmans P, Hultman CM, Iyegbe C, Lonnqvist J, Mannikko M, McCarroll SA, McGuffin P, McIntosh AM, McQuillin A, Moilanen JS, Moore C, Murray RM, Newbury-Ecob R, Ouwehand W, Paunio T, Prigmore E, Rees E, Roberts D, Sambrook J, Sklar P, St Clair D, Veijola J, Walters JT, Williams H, Swedish Schizophrenia S, Study I, Study DDD, Consortium UK, Sullivan PF, Hurles ME, O’Donovan MC, Palotie A, Owen MJ, Barrett JC, 2016. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci 19, 571–577. 10.1038/nn.4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T, Walters JTR, Johnstone M, Curtis D, Suvisaari J, Torniainen M, Rees E, Iyegbe C, Blackwood D, McIntosh AM, Kirov G, Geschwind D, Murray RM, Di Forti M, Bramon E, Gandal M, Hultman CM, Sklar P, Study I, Consortium UK, Palotie A, Sullivan PF, O’Donovan MC, Owen MJ, Barrett JC, 2017. The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat Genet 49, 1167–1173. 10.1038/ng.3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene NG, Bryois J, Bakken TE, Breen G, Crowley JJ, Gaspar HA, Giusti-Rodriguez P, Hodge RD, Miller JA, Munoz-Manchado AB, O’Donovan MC, Owen MJ, Pardinas AF, Ryge J, Walters JTR, Linnarsson S, Lein ES, Major Depressive Disorder Working Group of the Psychiatric Genomics, C., Sullivan PF, Hjerling-Leffler J, 2018. Genetic identification of brain cell types underlying schizophrenia. Nat Genet 50, 825–833. 10.1038/s41588-018-0129-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder SH, 1976. The dopamine hypothesis of schizophrenia: focus on the dopamine receptor. Am J Psychiatry 133, 197–202. 10.1176/ajp.133.2.197 [DOI] [PubMed] [Google Scholar]
- Wang D, Liu S, Warrell J, Won H, Shi X, Navarro F, Clarke D, Gu M, Emani P, Xu M, Yang YT, Park JJ, Rhie SK, Manakongtreecheep K, Zhou H, Nathan A, Zhang J, Peters M, Mattei E, Fitzgerald D, Brunetti T, Moore J, Consortium P, Sestan N, Jaffe AE, White K, Weng Z, Geschwind DH, Knowles J, Gerstein M, 2018. Comprehensive functional genomic resource and integrative model for the adult brain. Science (80-. ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, Clarke D, Gu M, Emani P, Yang YT, Xu M, Gandal MJ, Lou S, Zhang J, Park JJ, Yan C, Rhie SK, Manakongtreecheep K, Zhou H, Nathan A, Peters M, Mattei E, Fitzgerald D, Brunetti T, Moore J, Jiang Y, Girdhar K, Hoffman GE, Kalayci S, Gümüş ZH, Crawford GE, PsychENCODE Consortium, P., Roussos P, Akbarian S, Jaffe AE, White KP, Weng Z, Sestan N, Geschwind DH, Knowles JA, Gerstein MB, 2018. Comprehensive functional genomic resource and integrative model for the human brain. Science 362, eaat8464 10.1126/science.aat8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Goldstein DB, 2018. Enhancer redundancy predicts gene pathogenicity and informs complex disease gene discovery. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen S, Pollard KS, 2018. Most regulatory interactions are not in linkage disequilibrium. bioRxiv. 10.1101/272245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen S, Truty RM, Pollard KS, 2016. Enhancer-promoter interactions are encoded by complex genomic signatures on looping chromatin. Nat Genet 48, 488–496. 10.1038/ng.3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, Gandal MJ, Sutton GJ, Hormozdiari F, Lu D, Lee C, Eskin E, Voineagu I, Ernst J, Geschwind DH, 2016. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527. 10.1038/nature19847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J, Ha S, Chung C, Jung ES, Cho YS, Park SG, Lee JS, Lee K, Kim D, Bae YC, Kaang BK, Lee MG, Kim E, 2012. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature 486, 261–265. 10.1038/nature11208 [DOI] [PubMed] [Google Scholar]
- Wray NR, Visscher PM, 2010. Narrowing the boundaries of the genetic architecture of schizophrenia. Schizophr Bull 36, 14–23. 10.1093/schbul/sbp137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi W, Beer MA, 2018. Local epigenomic state cannot discriminate interacting and non-interacting enhancer-promoter pairs with high accuracy. bioRxiv. 10.1101/420372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, Montgomery GW, Goddard ME, Wray NR, Visscher PM, Yang J, 2016. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 48, 481–487. 10.1038/ng.3538 [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, 2003. Postnatal neurodevelopmental disorders: meeting at the synapse? Science 302, 826–30. 10.1126/science.1089071 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.