Abstract
Evaluation of microRNAs (miRNAs) could allow characterization of the obstructive sleep apnea (OSA) and help diagnose it more accurately. We aimed to examine circulating miRNA profiles to establish the differences between non-OSA and OSA patients. Additionally, we aimed to analyse the effect of continuous positive airway pressure (CPAP) treatment on the miRNA profile. This observational, longitudinal study included 230 subjects referred to the Sleep Unit due to suspected OSA. Expression profiling of 188 miRNAs in plasma was performed in 27 subjects by TaqMan-Low-Density-Array. OSA-related miRNAs were selected for validation by RT-qPCR in 203 patients. Prediction models were built to discriminate between non-OSA and OSA: 1) NoSAS-score, 2) differentially expressed miRNAs, and 3) combination of NoSAS-score plus miRNAs. The differentially expressed miRNAs were measured after 6 months of follow-up. From the 14 miRNAs selected for validation, 6 were confirmed to be differentially expressed. The areas under the curve were 0.73 for the NoSAS-score, 0.81 for the miRNAs and 0.86 for the combination. After 6 months of CPAP treatment, miRNA levels in the OSA group seem to approximate to non-OSA levels. A cluster of miRNAs was identified to differentiate between non-OSA and OSA patients. CPAP treatment was associated with changes in the circulating miRNA profile.
Subject terms: Diagnostic markers, Respiratory tract diseases
Introduction
Obstructive sleep apnea (OSA) is a prevalent disease that affects approximately 10–17% of the adult population1,2. OSA is characterized by repetitive episodes of upper airway collapse during sleep leading to arousal, nocturnal hypoxemia and changes in intrathoracic pressure. Altogether, these events link OSA to the risk of cardiovascular diseases, metabolic disturbances, and cancer and to higher overall mortality mechanisms3–6. This association is of special relevance in the young population, in which OSA could have a higher damaging effect, as the observed cancer incidence is increased in young patients7 and the impact of OSA on cardiovascular diseases may not be reversible when treatment is delayed8. Thus, new methods of early diagnosis of the disease are needed.
Up to 80% of individuals with moderate-to-severe OSA remain undiagnosed9. Diagnosis requires overnight recordings, including time- and resource-consuming procedures9. High-throughput diagnostic systems may improve the underdiagnoses of the disease and could reduce unnecessary procedures, decreasing the use of healthcare resources.
Currently, the NoSAS score is one of the best-validated tools for screening OSA populations (area under the curve (AUC) 0.74). The NoSAS score performed significantly better than did the STOP-Bang and Berlin scores10. NoSAS is based on a questionnaire and does not include any molecular variables that could help with diagnosis. The search for biomarkers that can help physicians not only diagnose OSA but also understand the physiopathology of the disease is a research priority in the field9,11. In this context, microRNAs (miRNAs) have emerged as an opportunity in the era of precision medicine for the screening, diagnosis and management of several diseases12–16. miRNAs are a class of small non-coding RNAs that negatively regulate gene expression post-transcriptionally by binding to target messenger RNA (mRNA), leading to either degradation or translational repression and protein synthesis17. They play a pivotal role in several biological processes, such as stress response, apoptosis, proliferation and differentiation, and many studies suggest that they are deregulated in several diseases, including cardiovascular diseases, metabolic disturbances and cancer18. miRNAs are present in body fluids, resistant to degradation and easily and rapidly (8–10 hours) measurable, fulfilling the criteria of an ideal biomarker in an era of evolving precision medicine19.
We aimed to examine circulating miRNA profiles to establish the differences between non-OSA and OSA patients and to explore their clinical significance and contribution to disease diagnosis. Moreover, we aimed to evaluate changes in miRNA patterns after 6 months of continuous positive airway pressure (CPAP) treatment.
Methods
Study cohort and sample collection
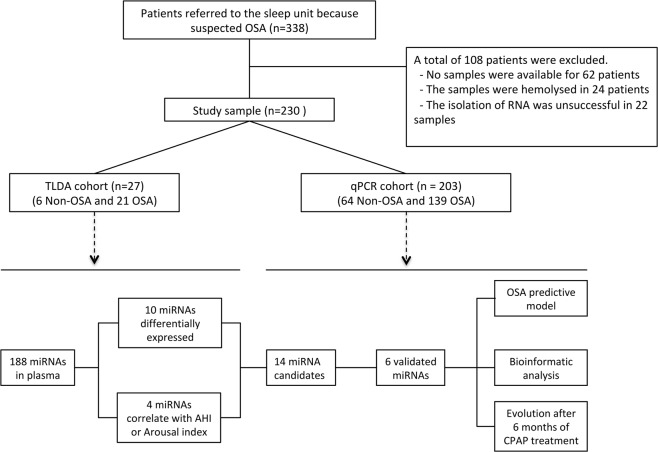
A total of 230 consecutive subjects who were aged between 18 and 60 years, were referred because of suspected OSA and had undergone full polysomnography were enrolled at the Sleep Unit of the Arnau de Vilanova-Santa Maria University Hospital of Lleida (NCT03513926). Subjects were grouped according to their Apnea-Hypopnea Index (AHI) gain into the following: 1) non-OSA group (subjects without moderate-to-severe OSA: AHI <15 events/h), 2) OSA group (moderate to severe OSA: AHI ≥15 events/h). Patients in the TaqMan low-density array (TLDA) cohort were carefully selected. Only patients taking medications related to the most habitual OSA associated pathologies (e.g., hypertension, dyslipidaemia, and cardiovascular events) were not excluded. No patients were excluded from the validation cohort, except those with previous CPAP use or any condition that, in the opinion of the responsible physician investigator, made the person unsuitable for the study (e.g., pregnancy, drug or alcohol consumption, or less than one year of life expectancy). On the other hand, patients without samples or with a haemolysed sample were excluded from the analyses. Additionally, samples with a recovery of RNA less than 1% were excluded (Fig. 1). All recruited patients signed an informed consent form, and the ethics committee of the centre (Clinical Research Ethics Committee of the Arnau de Vilanova-Santa Maria Hospital University Hospital) approved the study. All methods were performed in accordance with current clinical practice guidelines and regulations.
Figure 1.
Flowchart of the study. Patients who were referred because of suspected OSA were divided into the TLDA cohort and the qPCR cohort and further divided for study on the basis of non-OSA and OSA. Six non-OSA and 21 OSA patients were used to perform a general screening of 188 miRNAs. Differentially expressed miRNAs were validated in an independent cohort. The analyses in the validation cohort were stratified by sex.
A venous fasting blood sample was obtained from each patient at baseline in the morning immediately after the sleep study between 08:00 and 09:00 a.m. An additional fasting blood sample was obtained at the 6-month follow-up between 08:00 and 09:00 a.m. The blood samples were centrifuged to separate plasma, and all specimens were immediately aliquoted, frozen, and stored in a dedicated −80 °C freezer. No freeze-thaw cycles were performed during the experiment.
Clinical measurements
All patients underwent full polysomnography at baseline. Apnea was defined as an interruption or reduction in oronasal airflow ≥90% that lasted at least 10 seconds. Hypopnoea was defined as a 30% to 90% reduction in oronasal airflow for at least 10 seconds associated with oxygen desaturation of at least 3% or an arousal on the electroencephalogram. The AHI was defined as the number of apnea and hypopnoea events per hour of sleep. The NoSAS score for screening sleep-disordered breathing was evaluated. The NoSAS score ranges from 0–17 based on neck circumference, body mass index (BMI), age, and gender10. All patients were evaluated at baseline and after 6 months of follow-up. Following the National Clinical Guidelines20, patients with OSA were treated with CPAP.
Circulating RNA extraction and purification
RNA was extracted from 300 μL of plasma using a mirVana PARIS isolation kit (Applied Biosystems, Vilnius, Lithuania) according to the manufacturer’s instructions. Non-human cel-miR-39 was spiked into the plasma immediately before extraction. RNA isolation efficiency was examined by RT-qPCR quantification of cel-miR-39 to guarantee homogeneous retrotranscription and cDNA synthesis.
Circulating miRNA profiling with taqman low density array
To identify plasma miRNA profiles that could help to diagnose OSA, we performed a general screening of 188 circulating miRNAs that have been reported as the most consistent and reliable miRNAs in human plasma samples21,22. This circulating profile was evaluated in a cohort of 27 patients (TLDA cohort) that included 6 non-OSA and 21 OSA patients matched by nearest neighbor matching (a procedure to match each OSA patient to a non-OSA subject with the most similar value of the estimated propensity score)23 for age and body mass index (Fig. 1). At this point, due to sample size, only male subjects were evaluated. Multiple real-time (RT)-PCRs were performed using a customized TaqMan Low Density Array (TLDA, Life Technologies, Foster City, CA, USA). Briefly, a fixed volume of 3 μL of RNA solution from the 40 μL of RNA isolation eluate was used as the input for the retrotranscription, using a TaqMan MicroRNA Reverse Transcription Kit and TaqMan MicroRNA Multiplex RT Assays, which are customized to run TLDAs. Preamplification was performed using TaqMan PreAmp Master Mix and Megaplex PreAmp Primers for our selected miRNAs. RT-PCR was carried out using an Applied Biosystems QuantStudio™ 7 Flex Real-Time PCR System. Data were processed with the Relative Quantification tool (powered by Thermo Fisher cloud), with a minimal threshold above the baseline (ΔRn = 0.012) and less than 35 thermal cycles (Ct). The results were normalized using a mean-centre normalization method, the gold-standard method when screening a large number of miRNAs24.
Analysis of individual miRNAs
The most reliable and consistent candidates for endogenous control (i.e., hsa-miR-106a and hsa-miR-186) were identified in TLDA analyses and were used together with spike-in (cel-miR-39) for normalization25. Then, individual TaqMan hydrolysis probes (Applied Biosystems) were applied to analyse the expression of the set of differentially expressed miRNAs in the qPCR cohort (Fig. 1). Additionally, to assess the evolution of miRNAs after CPAP treatment, we evaluated the changes in miRNA expression of the validated candidates after 6 months of CPAP treatment, comparing non-OSA subjects with OSA patients treated with CPAP.
Enrichment analysis
The web-based computational tools miRWalk, DIANA-mirPath, TargetScan, DAVID and miRanda were used to predict the target genes and altered pathways of the differentially expressed miRNAs.
Statistical analysis
To prevent biases due to differential miRNA pattern expression, all analyses were stratified by gender. Comparability between non-OSA and OSA patients was assessed using the Mann-Whitney U test for quantitative characteristics and Fisher’s exact test for qualitative variables. The differences in miRNA expression between groups were evaluated by relative quantification using linear models for arrays (LIMMA)26. Spearman’s rank correlation coefficient was used to evaluate the association between miRNA levels and polysomnography parameters. False discovery rate-adjusted p-values were calculated to adjust for the performance of multiple paired comparisons.
Three logistic models were fitted for OSA risk modelling: 1) miRNAs showing differential expression in the validation phase, which were dichotomized by selecting the cut-off point as the point on the receiver operating characteristic (ROC) curve closest to the upper left corner of the unit square and were included in the model; 2) NoSAS score as a continuous variable; and 3) The combination of NoSAS and differentially expressed miRNAs. The Hosmer-Lemeshow test was used to test model calibration. A nonparametric test comparing the AUC of ROC curves27 was evaluated. The AUC of the combined model (NoSAS and miRNA) was evaluated in a 10-fold cross-validation. The effect of CPAP on the changes in miRNA expression at the six-month follow-up was estimated by the difference in the mean change between treated OSA patients and non-OSA patients. R software (R Project for Statistical Computing, Vienna, Austria) was used for statistical analysis.
Results
Patient characteristics
Of the 338 patients evaluated, 230 were ultimately included (Fig. 1). Patients were mainly middle aged, overweight-obese and male, especially in the OSA group (Table 1). Significant differences were identified in baseline characteristics between non-OSA and OSA groups in age (median 47 vs. 51 years old, p-value = 0.002), BMI (median 27.5 vs. 31.7 kg/m2, p-value = 0.015) and sex (68.6% vs. 83.8% male, p-value < 0.001).
Table 1.
Baseline characteristics of the patients.
| All | Non-OSA (AHI < 15) | OSA (AHI ≥ 15) | p-value | |
|---|---|---|---|---|
| N = 230 | N = 70 | N = 160 | ||
| Demographic and clinical variables | ||||
| Age (years) -median [IQR]- | 49.0 [44.0;55.0] | 47.0 [40.0;52.0] | 51.0 [45.0;55.0] | 0.002 |
| Sex (men), n (%) | 182 (79.1%) | 48 (68.6%) | 134 (83.8%) | 0.015 |
| BMI (kg/m2) -median [IQR]- | 30.9 [26.5;34.7] | 27.5 [25.4;31.9] | 31.7 [28.1;35.2] | <0.001 |
| Systolic blood pressure (mmHg) -median [IQR]- | 134 [123;146] | 127 [116;137] | 136 [127;148] | <0.001 |
| Diastolic blood pressure (mmHg) -median [IQR]- | 86.0 [79.5;94.5] | 81.8 [75.2;87.8] | 88.5 [82.5;95.0] | <0.001 |
| Smoking status: -n(%)- | 0.564 | |||
| Non-smoker | 81 (35.5%) | 28 (40.0%) | 53 (33.5%) | |
| Former smoker | 79 (34.6%) | 24 (34.3%) | 55 (34.8%) | |
| Smoker | 68 (29.8%) | 18 (25.7%) | 50 (31.6%) | |
| Respiratory parameters | ||||
| AHI (events/h) -median [IQR]- | 29.5 [12.1;51.0] | 8.44 [5.00;11.4] | 43.8 [27.4;63.9] | <0.001 |
| TSat90 (%) -median [IQR]- | 2.40 [0.21;12.7] | 0.05 [0.00;0.58] | 5.15 [2.00;20.6] | <0.001 |
| Arousal index (events/h) -median [IQR]- | 33.8 [21.1;53.6] | 18.9 [13.7;25.9] | 43.3 [32.1;60.7] | <0.001 |
| Minimum SaO2 (%) -median [IQR]- | 82.0 [73.0;87.0] | 89.0 [86.0;91.0] | 79.0 [71.0;83.0] | <0.001 |
| Mean SaO2 (%) -median [IQR]- | 93.0 [91.0;94.0] | 94.0 [93.0;95.0] | 93.0 [91.0;94.0] | <0.001 |
| ESS (0–24) -median [IQR]- | 10.0 [7.00;13.0] | 10.0 [7.00;14.0] | 10.0 [7.00;13.0] | 0.469 |
Abbreviations: BMI = Body Mass Index; AHI = Apnea-Hypoapnea Index; TSat90 = Night Time with Oxygen Saturation Less Than 90%; ESS = Epworth Sleepiness Scale.
Identification of plasma miRNAs relevant to OSA
The miRNA profiles were used to identify miRNAs related to OSA in the TLDA cohort (a cohort of men paired by BMI and age). After the analysis, a subset of 14 miRNAs was selected based on an individual p-value < 0.05 when comparing non-OSA and OSA groups and/or a high correlation (r > 0.4) with the AHI or arousal index (Table 2). miR-451, miR-486-3p and miR-133a were selected based on their p-values. On the other hand, miR-181a, hsa-let-7d, miR-199a and miR-199b were selected because of their high correlations with the sleep parameters. Finally, miR-181a-2, miR-495, miR-486, miR-660, miR-345, miR-340 and miR-107 were selected because they met both criteria. From the 14 miRNAs selected, 8 seemed to have less expression in the OSA patients than in the non-OSA patients.
Table 2.
miRNA candidates that were differentially expressed between non-OSA and OSA male subjects. Fourteen miRNAs were selected based on their fold change and their correlation with the AHI and arousal index.
| miRNA | Fold change | p-value | AHI correlation | Arousal index correlation |
|---|---|---|---|---|
| hsa-miR-181a-2 | 3.72 | 0.001 | −0.51 | −0.42 |
| hsa-miR-495 | 2.22 | 0.004 | −0.58 | −0.79 |
| hsa-miR-451 | 0.52 | 0.006 | 0.18 | −0.06 |
| hsa-miR-486 | 0.52 | 0.007 | 0.54 | 0.28 |
| hsa-miR-660 | 0.65 | 0.015 | 0.42 | 0.22 |
| hsa-miR-345 | 0.69 | 0.021 | 0.47 | 0.41 |
| hsa-miR-340 | 1.51 | 0.026 | −0.61 | −0.49 |
| hsa-miR-107 | 1.62 | 0.029 | −0.63 | −0.45 |
| hsa-miR-486-3p | 0.55 | 0.03 | 0.38 | 0.31 |
| hsa-miR-133a | 0.37 | 0.049 | −0.13 | −0.06 |
| hsa-miR-181a | 0.73 | 0.066 | 0.47 | 0.5 |
| hsa-let-7d | 1.35 | 0.1 | −0.52 | −0.67 |
| hsa-miR-199a | 1.3 | 0.157 | −0.28 | −0.41 |
| hsa-miR-199b | 0.81 | 0.332 | 0.42 | 0.36 |
Validation of plasma miRNAs relevant to OSA
All potential OSA-related miRNAs selected from TLDA were validated in the qPCR cohort. Among men, the analysis confirmed that the circulating concentrations of hsa-miR-181a, hsa-miR-199b, hsa-miR-345, hsa-miR-133a, hsa-miR-340 and hsa-miR-486-3p were decreased in the OSA group after adjustment for BMI and age (Table 3 and Fig. 2). Of these, miR-181a seemed to be correlated with the AHI (rho = 0.29) and with the arousal index (rho = 0.25) and miR-345 with the AHI (rho = 0.21). To further explore the results, mild OSA patients were removed in order to confidently report the results between non-OSA (AHI <5 events/h) and OSA (AHI ≥15 events/h) subjects. This analysis reported increased differences between non-OSA and OSA subjects (except for miR-199b and miR-486-3p) (see e-Table 3).
Table 3.
Validation of miRNA candidates. Six miRNAs were found to be differentially expressed between non-OSA and OSA patients. P-values adjusted for age and BMI.
| miRNA | Fold change | p-value | FDR correction |
|---|---|---|---|
| hsa-miR-181a | 0.59 | 0.001 | 0.013 |
| hsa-miR-199b | 0.42 | 0.008 | 0.056 |
| hsa-miR-345 | 0.71 | 0.014 | 0.056 |
| hsa-miR-133a | 0.47 | 0.016 | 0.056 |
| hsa-miR-340 | 0.67 | 0.029 | 0.072 |
| hsa-miR-486-3p | 0.64 | 0.031 | 0.072 |
| hsa-miR-181a2 | 0.51 | 0.066 | 0.133 |
| hsa-miR-199a | 0.54 | 0.184 | 0.318 |
| hsa-miR-660 | 0.79 | 0.214 | 0.318 |
| hsa-miR-451 | 0.82 | 0.227 | 0.318 |
| hsa-let-7d | 0.93 | 0.591 | 0.752 |
| hsa-miR-486 | 1.06 | 0.665 | 0.776 |
| hsa-miR-107 | 1.07 | 0.817 | 0.832 |
| hsa-miR-495 | 0.93 | 0.832 | 0.832 |
FDR <0.1 was considered statistically significant.
Figure 2.
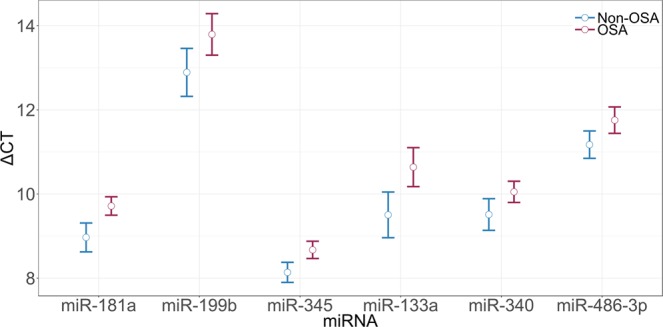
Delta Ct plot means of validated miRNAs (FDR < 0.1).
Among women, only hsa-miR-451 (fold change = 1.6 and p-value = 0.057) seemed to be differentially expressed after adjustment for BMI and age, but this significance is lost after FDR correction (corrected p-value = 0.54) (see e-Table 4).
Prediction model
Logistic regression models were generated for male patients as a tool to differentiate between non-OSA and OSA patients. Three multivariate models were fitted to classify the patients: A) the NoSAS score; B) the validated miRNAs and C) a combination of NoSAS score and validated miRNAs (Fig. 3). All the models exhibited good calibration and discrimination. The AUCs were 0.73 for the NoSAS score, 0.81 for the miRNAs and 0.86 for the combination. A comparison of the models based on AUC showed that the combination of NoSAS score and miRNAs was the best model for discriminating OSA patients (Table 4). The AUC of the combined model showed similar results in cross-validation (AUC = 0.83).
Figure 3.
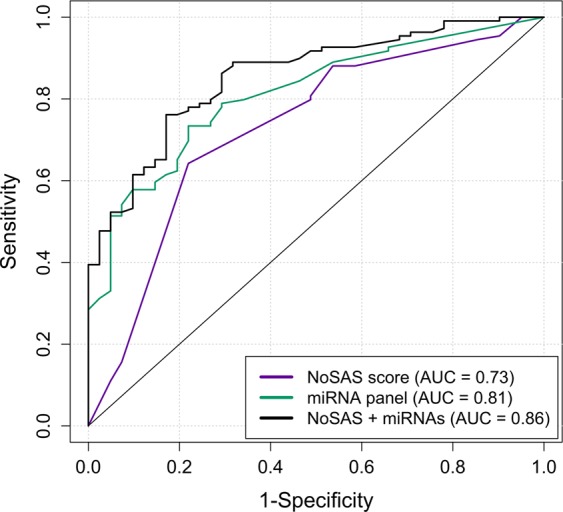
Discriminatory ROC plot of the models for screening OSA using (A) NoSAS; (B) 6 miRNAs; and (C) a combination of NoSAS and miRNAs.
Table 4.
Comparison of ROC models. A) NoSAS scores, B) miRNA model, C) combination.
| Comparison | AUC difference | p-value | Correlation |
|---|---|---|---|
| A vs. B | −0.08 | 0.16 | 0.01 |
| A vs. C | −0.13 | 0.002 | 0.54 |
| B vs. C | −0.04 | 0.065 | 0.76 |
Changes in validated plasma miRNAs after CPAP treatment
We evaluated the miRNAs that were differentially expressed and validated, at baseline and after 6 months of follow-up. We found that in OSA CPAP-treated patients (median adherence 5.24 h/night), miRNA levels seem to increase, except for miR-486-3p, compared to those in non-OSA patients. Despite this observation, only changes in miR-345 reached statistical significance (Table 5).
Table 5.
Post-pre mean differences in differentially expressed miRNAs between non-OSA and OSA patients after 6 months of CPAP treatment.
| miRNA | Estimate difference | Lower bound | Upper bound | p-value |
|---|---|---|---|---|
| hsa-miR-181a | −0.514 | −1.524 | 0.495 | 0.314 |
| hsa-miR-199b | −0.372 | −1.714 | 0.971 | 0.581 |
| hsa-miR-345 | −1.419 | −2.416 | −0.423 | 0.006 |
| hsa-miR-133a | −0.216 | −1.443 | 1.012 | 0.728 |
| hsa-miR-340 | −0.418 | −1.477 | 0.642 | 0.435 |
| hsa-miR-486-3p | 0.550 | −0.567 | 1.668 | 0.330 |
Bioinformatic analysis
The enrichment analysis, including the 6 miRNAs confirmed as differentially expressed between non-OSA and OSA patients at baseline, revealed 69 dysregulated pathways. A total of 1,743 genes were identified to be deregulated; 352 genes were targeted by miR-133a, 188 genes by miR-181, 167 genes by miR-199b, 47 genes by miR-340, 185 genes by miR-345, and 804 genes by miR-486-3p. Of the 1,743 predicted targeted genes, only 626 were unique genes, and IGF1R, KRAS, RUNX1T1 and SLC9A8 were regulated by 3 miRNAs.The most frequently enriched pathways were categorized under biological processes connected with cancer (renal cell carcinoma, melanoma, etc.), cardiovascular diseases (dilated cardiomyopathy, hypertrophic cardiomyopathy, etc.), and signalling (sphingolipid signalling, adrenergic signalling of cardiomyocytes, etc.) (see e-Table 6 and e-Fig. 6).
Discussion
A singular male-specific cluster of miRNAs functionally associated with sleep, cardiovascular diseases, metabolic disorders and cancer significantly contributes to the discrimination between non-OSA and OSA patients in the sleep unit. The present study also showed that CPAP use is associated with changes in the miRNA profile that could influence the overall risk of suffering OSA-related diseases.
Circulating concentrations of 6 miRNAs were significantly reduced in OSA subjects compared with those in non-OSA subjects. We also investigated the discriminatory ability of the miRNAs. We compared the NoSAS score, miRNAs and a combination of both. The NoSAS score AUC was consistent with previous results, with an AUC of 0.73, but lower than that obtained with miRNAs (AUC = 0.81)10,28. The correlation between both AUCs is very low (0.01), which could be argued because NoSAS and miRNAs could explain different components of OSA, resulting in a higher AUC of 0.86 for the combination of both, which could represent a very powerful tool that combines anthropometric and molecular variables to explain OSA variability. Finally, the longitudinal study showed that after CPAP treatment, miRNAs seem to increase their circulating concentration, at least partially recovering the non-OSA phenotype. Decreased blood pressure could have contributed to the miRNA changes detected after CPAP treatment. In fact, as previously reported by our group, CPAP treatment changes the miRNA profile, and these changes correlate with blood pressure changes that occur following CPAP treatment. Additionally, these changes are associated with the response to treatment of the patient14.
This study identifies the utility of miRNAs as biomarkers that could contribute to the personalized management of OSA in the sleep unit. Recent studies have demonstrated the importance of miRNAs in the control of many processes in health and disease14,19,21. To the best of our knowledge, this is one of the first studies to use miRNAs to identify a signature and characterize OSA in patients who are referred to a sleep unit. A similar study was conducted29, in which the researchers identified that in a preliminary phase, 104 miRNAs were differentially expressed in serum between OSA and non-OSA subjects. Of these miRNAs, only 4 were validated by qPCR. We also attempted to validate miR-107 and miR-199 in our study. In both studies, miR-199 was downregulated when comparing OSA patients vs. non-OSA subjects; however, in our study, miR-107 was not confirmed to be differentially expressed. These possible discrepancies could be due to the different sample types utilized (plasma vs. serum) and our different normalization methods (endogenous control + spike-in vs. spike-in). Additionally, the data reported in this previous study did not seem to be adjusted by confounding variables.
A total of 6 miRNAs have been identified to be differentially expressed between non-OSA and OSA patients. This set of miRNAs seems to have the ability to differentiate between non-OSA and OSA patients, which is effective and practical for clinical use and enables the identification of patients with OSA. Recently, a seminal study developed by our group identified miRNAs as a useful tool in the management of patients with OSA and resistant hypertension14. In this study, we demonstrated the utility of miRNAs by identifying a singular cluster of miRNAs (HIPARCO-Score) that identifies those patients who will have a favourable blood pressure response to CPAP treatment. The present study corroborates the clinical usefulness of miRNAs for the diagnosis of OSA at sleep unit level.
The set of 6 miRNAs that are differentially expressed between non-OSA and OSA patients is related to relevant canonical pathways. First, we identified that miRNAs are misregulating pathways related to cardiovascular diseases, specifically several cardiomyopathies, in concordance with the hypothesis of a difference in cardiovascular risk between non-OSA and OSA patients. Second, identifying the molecular pathways related to both cancer and OSA is one of the hallmarks of OSA research. Additionally, an early diagnosis of OSA could be especially important because the association of OSA with cancer incidence is limited to young people7. Several studies have shown the association of OSA with several types of cancer, especially in young patients30. In the present study, we identified several cancer-related pathways that could be altered in patients with OSA, and some of the altered pathways are related to melanoma. These results are in accordance with previous literature in which the impact of OSA on melanoma and some of the intermediate pathways have been addressed31,32. Similar results were found in a previous work25, where bioinformatic analyses showed that OSA could alter cardiovascular and cancer-related pathways.
Specifically, these miRNAs play an important role in some pathways related to common diseases associated with OSA. OSA is well known to be related to deregulated nutrient sensing, especially glucose. Patients with OSA are more likely to develop insulin resistance due to oxidative stress and a lower concentration of oxygen in pancreatic ß-cells33. Recently, miR-486 has been associated with the HOMA index and fasting levels of glucose and insulin34. Moreover, miR-340 seems to deregulate IGF1R gene expression, as shown in bioinformatic analyses. Previous studies observed associations between OSA and melanoma. Additionally, miR-340 is known to regulate MITF gene expression, which is a gene that plays a key role in melanocyte development. Recently, one study found that miR-340 regulates RAS–RAF–mitogen activated protein kinase (MAPK) signalling by modulating the expression of multiple components of this pathway35, and this association could explain, at least in part, the relationship between OSA and melanoma. Several genes regulated by these miRNAs are involved in cardiovascular diseases. miR-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart36 and is dysregulated in human myocardial infarction37. Finally, miR-199b is involved in the response to the hypoxia-repressing HIF1A gene38. HIF1A is a key gene in the response to hypoxic stress, and its expression is deregulated in OSA patients39. Moreover, IGF1R, KRAS, RUNX1T1 and SLC9A8 are regulated by 3 miRNAs. All these genes are related to melanoma pathways and play an important role in the transformation of melanocytes. Recently, one study assessed the molecular relationship between OSA and melanoma, concluding that OSA increased circulating VCAM-1 levels in melanoma patients31. This relationship could be an indirect consequence of IGF1R, KRAS, RUNX1T1 and SLC9A8 regulation.
Notably, all miRNAs validated in the present study were downregulated in patients with OSA. As previously reported, chronic hypoxia impairs Dicer (DICER1) expression and activity, resulting in global downregulation of miRNA expression40.
Diagnostic and personalized therapeutic decision-making tools are needed to manage OSA, especially in the young population9,10,41. Together, these results suggest that the use of miRNAs in OSA management could be a powerful tool for the diagnosis of OSA and to establish the benefits of the treatment for this complex chronic disease.
The present study has several limitations that deserve comment. First, only patients between 18 and 60 years old were studied, and larger studies should be performed to determine the validity of this tool in patients of other ages. The validation showed poor results in women, reflecting the different physiopathology of OSA between women and men. Recent studies have shown that miRNAs have sex-dimorphic expression patterns42. Because of the ages of the subjects, oestrogens could play a very important role in these expression patterns. As previously reported, sex-related differences may cause abnormal miRNA expression, and for this reason, stratified analyses are recommended. Further studies are needed to evaluate the whole profile of miRNAs in women with OSA. Despite, RNA isolation efficiency was measured with spike-in cel-miR-39 to guarantee homogeneous retrotranscription and cDNA synthesis; no data about RNA integrity or quantity are available. Although we performed a quantification of a large number of miRNAs (a total of 188 miRNAs) that were previously identified as the most stable miRNAs in plasma and were quantified in at least 70% of the subjects21,22, we did not perform whole miRNA quantification. This approximation permits us to use gold standard technology and enables an accurate quantification in a short period of time, with the highest dynamic range43. By last, this tool is not able to distinguish mild OSA patients from non-OSA subjects; further studies including a higher number of patients with AHI <5 events/h is required. The strengths of this study reside in a large number of patients who were analysed in two different sets for the TLDA and qPCR cohorts, which permitted us to detect high-magnitude associations. Additionally, some of the basal associations were modified after CPAP use, confirming the importance of these miRNAs in OSA. Finally, all patients were recruited from the sleep unit, performing gold-standard methods for OSA diagnosis (polysomnography) and for miRNA measurement (RT-qPCR).
A singular male-specific cluster of miRNAs was identified that specifically differentiates between non-OSA and OSA patients. This miRNA model, which improves the discriminatory ability, is based in anthropometric and clinical variables. Finally, the use of CPAP treatment was associated with changes in the circulating miRNA profile that partly recovered the non-OSA phenotype.
Supplementary information
Acknowledgements
The founding sources have no role in the writing, data collection, analysis or interpretation of the study. The project is supported by PI 14/01266 and PI 18/00449 from the Instituto de Salud Carlos III (ISCIII), Fondo Europeo de Desarrollo Regional (FEDER), Sociedad Española de Neumología y Cirugía Torácica (SEPAR) and Societat Catalana de Pneumologia (SOCAP). The work is supported by IRBLleida Biobank (B.0000682) and PLATAFORMA BIOBANCOS PT17/0015/0027”. F.S.-M. is the recipient of a predoctoral fellowship from “AGAUR-University of Lleida” and Convocatòria d’Ajuts 2018 de Promoció a la Recerca en Salut.
Author Contributions
M.S.T. is the guarantor of the study. F.S.M., I.B., J.M.F.R., F.B., F.O. and M.S.T. contributed to study design F.S.M., A.Z., C.G., L.P., L.P., A.C., M.D., F.O., M.S.T. contributed to data acquisition F.S.M., I.B., F.O., M.S.T. contributed to data analysis and interpretation F.S.M., I.B., A.Z., C.G., L.P., L.P., A.C., M.D., J.M.F.R., F.B., F.O. and M.S.T. contributed to drafting of the manuscript F.S.M., I.B., A.Z., C.G., L.P., L.P., A.C., M.D., J.M.F.R., F.B., F.O. and M.S.T. contributed to revision of the manuscript for intellectual content and approval of the final version.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-49940-1.
References
- 1.Peppard P. E. et al. Increased prevalence of sleep-disordered breathing in adults. Am. J. Epidemiol. 177 (2013). [DOI] [PMC free article] [PubMed]
- 2.Heinzer R, et al. Prevalence of sleep-disordered breathing in the general population: the HypnoLaus study. Lancet Respir. Med. 2015;3:310–318. doi: 10.1016/S2213-2600(15)00043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sánchez-de-la-Torre M, Campos-Rodriguez F, Barbé F. Obstructive sleep apnoea and cardiovascular disease. Lancet Respir. Med. 2013;1:61–72. doi: 10.1016/S2213-2600(12)70051-6. [DOI] [PubMed] [Google Scholar]
- 4.Bradley TD, Floras JS. Obstructive sleep apnoea and its cardiovascular consequences. Lancet. 2009;373:82–93. doi: 10.1016/S0140-6736(08)61622-0. [DOI] [PubMed] [Google Scholar]
- 5.Santamaria-Martos F, Sánchez-de-la-Torre M, Martínez-García MAS. and Cancer: Clinical Studies and Opportunities for Personalized Medicine. Curr. Sleep Med. Reports Current Sleep Medicine Reports. 2017;3:11–21. doi: 10.1007/s40675-017-0063-6. [DOI] [Google Scholar]
- 6.Jennum P, Tønnesen P, Ibsen R, Kjellberg J. All-cause mortality from obstructive sleep apnea in male and female patients with and without continuous positive airway pressure treatment: a registry study with 10 years of follow-up. Nat. Sci. Sleep. 2015;7:43–50. doi: 10.2147/NSS.S75166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campos-Rodriguez F. et al. Association between obstructive sleep apnea and cancer incidence in a large multicenter spanish cohort. Am. J. Respir. Crit. Care Med. 187 (2013). [DOI] [PubMed]
- 8.Arias MA, et al. Obstructive sleep apnea syndrome affects left ventricular diastolic function: Effects of nasal continuous positive airway pressure in men. Circulation. 2005;112:375–383. doi: 10.1161/CIRCULATIONAHA.104.501841. [DOI] [PubMed] [Google Scholar]
- 9.Fleming WE, et al. Use of blood biomarkers to screen for obstructive sleep apnea. Nat. Sci. Sleep. 2018;10:159–167. doi: 10.2147/NSS.S164488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marti-Soler H, et al. The NoSAS score for screening of sleep-disordered breathing: a derivation and validation study. Lancet Respir. Med. 2016;4:742–748. doi: 10.1016/S2213-2600(16)30075-3. [DOI] [PubMed] [Google Scholar]
- 11.Sánchez-de-la-Torre M, Gozal D. Obstructive sleep apnea: in search of precision. Expert Rev. Precis. Med. Drug Dev. Taylor & Francis. 2017;2:217–228. doi: 10.1080/23808993.2017.1361319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGuire A, Brown JAL, Kerin MJ. Metastatic breast cancer: the potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev. 2015;34:145–155. doi: 10.1007/s10555-015-9551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schultz NA, et al. MicroRNA Biomarkers in Whole Blood for Detection of Pancreatic Cancer. JAMA. 2014;311:392. doi: 10.1001/jama.2013.284664. [DOI] [PubMed] [Google Scholar]
- 14.Sánchez-de-la-Torre M, et al. Spanish Sleep Network. Precision Medicine in Patients With Resistant Hypertension and Obstructive Sleep Apnea. J. Am. Coll. Cardiol. 2015;66:1023–1032. doi: 10.1016/j.jacc.2015.06.1315. [DOI] [PubMed] [Google Scholar]
- 15.Alipoor SD, et al. The roles of miRNAs as potential biomarkers in lung diseases. Eur. J. Pharmacol. 2016;791:395–404. doi: 10.1016/j.ejphar.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sayed ASM, Xia K, Salma U, Yang T, Peng J. Diagnosis, Prognosis and Therapeutic Role of Circulating miRNAs in Cardiovascular Diseases. Hear. Lung Circ. 2014;23:503–510. doi: 10.1016/j.hlc.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Latronico MVG, Condorelli G. MicroRNAs and cardiac pathology. Nat. Rev. Cardiol. 2009;6:418–429. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- 18.Khalyfa A, et al. Circulating plasma extracellular microvesicle MicroRNA cargo and endothelial dysfunction in children with obstructive sleep apnea. Am. J. Respir. Crit. Care Med. 2016;194:1116–1126. doi: 10.1164/rccm.201602-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anfossi S., Babayan A., Pantel K., Calin G. A. Clinical utility of circulating non-coding RNAs — an update. Nat. Rev. Clin. Oncol. (2018). [DOI] [PubMed]
- 20.Lloberes P, et al. Diagnosis and treatment of sleep apnea-hypopnea syndrome. Spanish Society of Pulmonology and Thoracic Surgery. Arch. Bronconeumol. 2011;47:143–156. doi: 10.1016/j.arbres.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Ortega FJ, et al. Targeting the Circulating MicroRNA Signature of Obesity. Clin. Chem. 2013;59:781–792. doi: 10.1373/clinchem.2012.195776. [DOI] [PubMed] [Google Scholar]
- 22.Latorre J, et al. Decreased lipid metabolism but increased FA biosynthesis are coupled with changes in liver microRNAs in obese subjects with NAFLD. Int. J. Obes. 2017;41:620–630. doi: 10.1038/ijo.2017.21. [DOI] [PubMed] [Google Scholar]
- 23.Ho DE, et al. Matching as Nonparametric Preprocessing for Reducing Model Dependence in Parametric Causal Inference. Polit. Anal. 2007;15:199–236. doi: 10.1093/pan/mpl013. [DOI] [Google Scholar]
- 24.Mestdagh P, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santamaria-Martos F, et al. Identification and validation of circulating miRNAs as endogenous controls in obstructive sleep apnea. Taguchi Y, editor. PLoS One. 2019;14:e0213622. doi: 10.1371/journal.pone.0213622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 28.Lebkuchen A, et al. Metabolomic and lipidomic profile in men with obstructive sleep apnoea: implications for diagnosis and biomarkers of cardiovascular risk. Sci. Rep. 2018;8:11270. doi: 10.1038/s41598-018-29727-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li K, Wei P, Qin Y, Wei Y. MicroRNA expression profiling and bioinformatics analysis of dysregulated microRNAs in obstructive sleep apnea patients. Medicine (Baltimore). 2017;96:e7917. doi: 10.1097/MD.0000000000007917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martínez-García Miguel A., Campos-Rodriguez Francisco, Durán-Cantolla Joaquín, de la Peña Mónica, Masdeu María J., González Mónica, del Campo Félix, Serra Pablo Catalán, Valero-Sánchez Irene, Ferrer M.J. Selma, Marín José M., Barbé Ferrán, Martínez M., Farré Ramón, Montserrat José M. Obstructive sleep apnea is associated with cancer mortality in younger patients. Sleep Medicine. 2014;15(7):742–748. doi: 10.1016/j.sleep.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 31.Santamaria-Martos F, et al. Biomarkers of carcinogenesis and tumour growth in patients with cutaneous melanoma and obstructive sleep apnoea. Eur. Respir. J. 2018;51:1701885. doi: 10.1183/13993003.01885-2017. [DOI] [PubMed] [Google Scholar]
- 32.Almendros Isaac, Montserrat Josep M., Torres Marta, Dalmases Mireia, Cabañas Maria L., Campos-Rodríguez Francisco, Navajas Daniel, Farré Ramon. Intermittent hypoxia increases melanoma metastasis to the lung in a mouse model of sleep apnea. Respiratory Physiology & Neurobiology. 2013;186(3):303–307. doi: 10.1016/j.resp.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Morgenstern Michael, Wang Janice, Beatty Norman, Batemarco Tom, Sica Anthony L., Greenberg Harly. Obstructive Sleep Apnea. Endocrinology and Metabolism Clinics of North America. 2014;43(1):187–204. doi: 10.1016/j.ecl.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Santamaria-Martos F. et al. Comparative and functional analysis of plasma membrane-derived extracellular vesicles from obese vs. nonobese women. Clin. Nutr. European Society for Clinical Nutrition and Metabolism; (2019). [DOI] [PubMed]
- 35.Poenitzsch Strong AM, Spiegelman VS. microRNA-340 as a modulator of RAS–RAF–MAPK signaling in melanoma. Arch. Biochem. Biophys. Academic Press. 2014;563:118–124. doi: 10.1016/j.abb.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boštjančič E, Zidar N, Štajer D, Glavač D. MicroRNAs miR-1, miR-133a, miR-133b and miR-208 Are Dysregulated in Human Myocardial Infarction. Cardiology Karger Publishers. 2010;115:163–169. doi: 10.1159/000268088. [DOI] [PubMed] [Google Scholar]
- 37.Liu N, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. Cold Spring Harbor Laboratory Press. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shang W, et al. MiR199b Suppresses Expression of Hypoxia-Inducible Factor 1α (HIF-1α) in Prostate Cancer Cells. Int. J. Mol. Sci. Multidisciplinary Digital Publishing Institute. 2013;14:8422–8436. doi: 10.3390/ijms14048422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lavie L. Obstructive sleep apnoea syndrome - An oxidative stress disorder. Sleep Med. Rev. 2003;7:35–51. doi: 10.1053/smrv.2002.0261. [DOI] [PubMed] [Google Scholar]
- 40.Ho JJD, et al. Functional importance of Dicer protein in the adaptive cellular response to hypoxia. J. Biol. Chem. 2012;287:29003–29020. doi: 10.1074/jbc.M112.373365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pack AI. Application of Personalized, Predictive, Preventative, and Participatory (P4) Medicine to Obstructive Sleep Apnea. A Roadmap for Improving Care? Ann. Am. Thorac. Soc. 2016;13:1456–1467. doi: 10.1513/AnnalsATS.201604-235PS. [DOI] [PubMed] [Google Scholar]
- 42.Guo L, Zhang Q, Ma X, Wang J, Liang T. miRNA and mRNA expression analysis reveals potential sex-biased miRNA expression. Sci. Rep. 2017;7:39812. doi: 10.1038/srep39812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat. Rev. Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.