Abstract
Novel genes provide important genetic resource for organism innovation. However, the evidence from genetic experiment is limited. In plants, γ-aminobutyric acid (GABA) transporters (GATs) primarily transport GABA and further involve in plant growth, development, and response to various stresses. In this study, we have identified the GATs family in Populus species and characterized their functional evolution and divergence in a desert poplar species (Populus euphratica). We found that the GATs underwent genus-specific expansion via multiple whole-genome duplications in Populus species. The purifying selection were identified across those GATs evolution and divergence in poplar diversity, except two paralogous PeuGAT2 and PeuGAT3 from P. euphratica. The both genes arose from a tandem duplication event about 49 million years ago and have experienced strong positive selection, suggesting that the divergence in PeuGAT3 protein function/structure might define gene function better than in expression pattern. Both PeuGAT genes were functionally characterized in Arabidopsis and poplar, respectively. The overexpression of PeuGAT3 increased the thickness of xylem cells walls in both Arabidopsis and poplar and enhanced the lignin content of xylem tissues and the proline accumulation in poplar leaves, all of which may improve tolerance of salt/drought stress in desert poplars. Our findings help clarify the genetic mechanisms underpinning high tolerance in desert poplars and suggest that PeuGAT3 could be an attractive candidate gene for engineering trees with improved brown-rot resistance.
Keywords: γ-aminobutyric acid transporters, novel gene, functional divergence, poplars, adaptive evolution
Introduction
Novel genes provide important genetic resource for organism innovation. The novel genes may originate from various models, such as gene duplication, exon shuffling, and retroposition, which all may play important roles in plant evolution (Long et al., 1999). The novel gene often evolves rapidly and diversify in function, including expression pattern, location after translation, and biochemical function (Tautz and Domazet-Lošo, 2011), which all contribute to the formation of their specific traits in species or lineage, such as tolerance to stresses (Force et al., 1999; Shukla et al., 2014). The divergence of the novel genes in function may occur in multiple levels (Force et al., 1999; Hittinger and Carroll, 2007; Gagnon-Arsenault et al., 2013; Nan et al., 2017). In general, the primary mechanisms responsible for functional divergence include site-specific regulatory modification (Bergström et al., 2014) and changes in enzymatic activity and protein specificity (Force et al., 1999; Voordeckers et al., 2012). Of them, motif loss could potentially lead to changes in protein structure, which could in turn alter the function of protein.
Amino acid transporters (AATs) are membrane-integrated proteins that mediate the transport of amino acids across cellular membranes in flowering plants and play indispensable roles in various processes of plant growth and development (Wipf et al., 2002; Tegeder, 2012). There are two major families of plant AATs: the amino acid/auxin permease (AAAP) family and the amino acid–polyamine–choline transporter family (APC) (Ortiz-Lopez et al., 2000; Saier et al., 2008). The AAAP family has at least six subfamilies, including amino acid permeases (AAPs), lysine and histidine transporters (LHTs), proline transporters (ProTs), γ-aminobutyric acid transporters (GATs), auxin transporters (AUXs), and aromatic and neutral amino acid transporters (ANTs) (Okumoto and Pilot, 2011). Those subfamilies, refer to Arabidopsis, define with the type of the transporting amino acids/or metabolite molecular and diversify in the number of motifs of coding region and spatial–temporal expression, thus involve widely in various processes of plant growth and development (Schwacke et al., 1999). In AAAP family, GATs mainly transport the ubiquitous amino acid γ-aminobutyric acid (GABA) (Narayan and Nair, 1990), which act as a signal molecule that affects C/N balance, pH regulation, nitrogen storage, and defense mechanisms (Shelp et al., 1999; Bouche and Fromm, 2004). For example, GABA accumulation can modulate the activity of plant-specific anion transporters, cause changes in root growth, and improved salt stress tolerance (Bouche and Fromm, 2004). Only one GAT1 transporter has been identified in Arabidopsis; it cannot transport proline but has a high affinity for GABA (Schwacke et al., 1999), and are suggested to mediate interactions between carbon and nitrogen (Batushansky et al., 2015). Interestingly, in Populus, more putative GAT copies were identified in the genome of black cottonwood (Tuskan et al., 2006), Populus euphratica (Ma et al., 2013) and Populus alba (Ma et al., 2019). It seems clear that a genus-specific expansion of GAT genes occurred at some point in the evolutionary history of the poplars and that the duplicated GATs diversified, thereby avoiding functional redundancy and leading to behaviors that improve the salt tolerance of woody plants and increase their xylem tissue growth.
P. euphratica Oliv. is mainly distributed in central Asia and prefers dry and central deserts with deep underground water sources (Ottow et al., 2005). As an important tree with high resistance to salt and drought, P. euphratica has been studied extensively in efforts to understand stress resistance mechanisms in woody plants. Its leaves are phenotypically polymorphic: they become succulent under salt stress, which increases their ability to tolerate highly saline conditions (Ottow et al., 2005). P. euphratica also has a well-developed root system with a high capacity for hydrotropism, increasing its access to ground water and its likelihood of surviving in desert ecosystems. Finally, its wood is rigid and dense, helping to prevent excessive evaporation of water. The publication of genomic data for P. euphratica led to the identification of several gene families associated with responses to various stresses that have expanded via gene duplication events followed by functional diversification, such as HKT (Ma et al., 2013), and WRKY (Ma et al., 2016) families. These studies show that many of the genes responsible for the high stress resistance of P. euphratica arose via gene duplication and subsequent functional divergence, which may confer an adaptive advantage for desert poplars in arid and saline environments (Perry et al., 2007). Therefore, more research is needed to identify genetic resources that could be exploited in molecular tree breeding.
Here, we present a phylogenetic analysis of a set of GAT-like proteins in Populus. By inferring from phylogeny trees, we show that the GAT genes probably underwent specific-genus expansion in Populus species. To better characterize the function among GATs in poplar, a novel gene arose by gene duplication, and its paralogs from P. euphratica have been overexpressed in P. alba var. pyramidalis and Arabidopsis thaliana (col-0), respectively. We then compared the effect on plant growth, plant wood properties, wood structure, and proline accumulation under salt stress. Our results provide highlight insight into the role of GAT novel gene in the response to environment stress P. euphratica.
Materials and Methods
Plant Materials and Salt Treatments
Two-year-old seedlings of P. euphratica were obtained by germinating seeds collected from Akesu in Xinjiang province, China. Two-year-old saplings of P. alba var. pyramidalis were obtained from cuttings. All seedlings and saplings were cultivated in pots with loam soil and grown under greenhouse conditions with a constant temperature of 25°C, a 16-h (16/8 h) photoperiod with a photon flux of 80 µmol m−2 s−1 and 60% relative air humidity. Saplings and seedlings from two poplars with similar growth statuses were treated with 0, 150, and 300 mM NaCl solutions for a week under the same growth conditions (150 and 300 mM, representing moderate and severe stresses, respectively). All samples were collected and examined to determine the treatments’ physiological effects.
Phylogenetic Analyses of GAT Genes in Plants
GAT genes were identified by TBLASTN searches of the five genome databases P. euphratica (Ma et al., 2013), P. alba (Ma et al., 2018), P. trichocarpa (Tuskan et al., 2006), P. pruinosa (Ma et al., 2016), and willow (Dai et al., 2014) using 22 full-length AAAP sequences from Arabidopsis (Arabidopsis thaliana; Dixon and Edwards, 2010a) with an e-value cut-off 1-E-30. These candidate full-length AAAPs were primarily analyzed using the NCBI (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and MEME (http://meme-suite.org/doc/overview.html) conserved domain search to identify typical AAAP Aa_trans domain in their protein structures. When an AAAP candidate was identified to contain typical AAAP Aa_trans domain in its protein structure, it was considered as an AAAP gene. A total of 188 AAAPs (Table S3) represent six clades (GAT, ProT, LHT, AAP, AUX, and ANT) defined by the NCBI conserved domain database (Marchler et al., 2005) and MEME methods (Bailey et al., 2006). Subsequently, these Aa_trans domain sequences of candidate genes were aligned using CLUSTALW (Tamura et al., 2007). Phylogenetic analyses were conducted using the neighbor-joining (NJ) method as implemented in the MEGA version 4.1 software package, using the pairwise deletion option to handle alignment gaps and the Poisson correction model for distance computation. Bootstrap tests were performed using 1,000 replicates. The branch lengths in the resulting phylogenetic tree are proportional to phylogenetic distances. The rate of nonsynonymous substitution rate (Ka) and synonymous substitution (Ks) of paralogs were calculated using DNAsp version 6 (Rozas et al., 2017). Transmembrane domain analysis was performed using the TMHMM server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/), and protein tertiary structure prediction was performed using the SWISS-MODEL (https://swissmodel.expasy.org/).
PeuGAT2 and PeuGAT3 Overexpress in Arabidopsis and Poplars
PeuGAT2 (CCG029175.1) and PeuGAT3 (CCG029176.1) coding regions were amplified using specific primers (Table S1), subcloned into the PENTR/D TOPO vector (Invitrogen, Carlsbad, CA, USA), and then transferred into the GATAWAY destination pk7WG2D.1 vector (Karimi et al., 2002). In the latter vector, the CaMV 35S promoter drove the expression of the gene of interest (PeuGAT2 and PeuGAT3, respectively) and the reporter gene green fluorescent protein (GFP) in an LR Clonase reaction (Invitrogen, Carlsbad, CA, USA). The 35S::PeuGAT2 and 35S::PeuGAT3 constructs were introduced into the Agrobacterium tumefaciens strain GV3101 and then transformed into wild-type (WT) Populus species (P. alba) using the leaf disk method (Horsch et al., 1985) with some modifications (Ma et al., 2019). Putative transgenic plants were selected on Murashige and Skoog (MS) agar (0.8%, w/v) medium containing 100 mg/l kanamycin, and successful transformation was verified by semiquantitative reverse transcription PCR (RT-PCR) using PeuGAT2- and PeuGAT3-specific primers. The transcript levels were normalized to endogenous ACTIN transcripts amplified with primers Actin F/R (Table S1). Each set of experiments was repeated three times, and the relative quantification method (2−ΔΔCT) was used to evaluate quantitative variation (Livak and Schmittgen, 2001). We obtained eight independent transgenic lines. The putative transgenic poplar plantlets were incubated in a growth chamber at 25 ± 1°C with a 16-h-light/8-h-dark photoperiod and a relative humidity (RH) of 50–60%.
Transgenic Arabidopsis lines were obtained by transforming WTA. thaliana (ecotype Columbia-0) using floral dipping methods as described by Clough and Bent (1998). The T1 transgenic lines were screened on 1/2 MS medium containing 50 mg/L kanamycin. The selection of transgenic lines were continued until T3 generations to obtain transgenic homozygous lines with a single T-DNA locus (e.g.,35S::PeuGAT2 and 35S::PeuGAT3). The T-DNA insertion lines were identified using the SIGnAL T-DNA Express Arabidopsis Gene Mapping Tool (http://signal.salk.edu/cgi-bin/tdnaexpress), and the PeuGAT2- and PeuGAT3-specific primers (Table S1) were used as the screening primers. The selected transgenic seedlings were grown on MS agar (0.8%, w/v) medium for 5–7 days. A total of 16 independent transgenic Arabidopsis lines (nine 35S::PeuGAT2 lines and seven 35S::PeuGAT3 lines, respectively) (Figures S2A, B) were obtained. We found that the phenotypes of these transgenic lines were correlated with the expression levels. Therefore, 35S::PeuGAT2L9 and 35S::PeuGAT3L6 lines were selected for the subsequent experiments because of their high and similar expression levels. Similarly, the 35S::PeuGAT2L9 and 35S::PeuGAT3L4 from the confirmed transgenic poplar saplings (1035S::PeuGAT2 and 835S::PeuGAT3, respectively; Figures S2C, D) were selected for the subsequent experiments. The selected transgenic poplar saplings were propagated by cutting, grown for 3 weeks in culture vessels, and then transplanted to soil under a photoperiod of 16-h light/8-h dark with a relative humidity of 50–60% for 3–4 weeks at 25°C.
Abiotic Stress Treatments of Arabidopsis
WT and transgenic Arabidopsis seeds were vernalized at 4°C for 2 days after water absorption. After disinfection with mercury bichloride solution, the seeds were planted on MS, 50mM and 100mM NaCl MS medium for germination, respectively. Five days later, the germinated seeds were separately transferred to common medium and several stress-conditioned MS medium (75 mM NaCl and 150 mM NaCl for salt treatments; 5% PEG6000 and 10% PEG6000 for drought treatments) to continue cultivate for 1 week, and then, the phenotypes of wide-type and transgenic Arabidopsis were determined.
Anatomical Observations in the Transgenic Arabidopsis and Populus
For histological observations, rooted WT and transgenic plants (at least seven individuals for each line) were transplanted into soil and maintained in the greenhouse, respectively. Fresh stems from the base internode of 6-week-old Arabidopsis (L9 for 35S::PeuGAT2 and L6 for 35S::PeuGAT3) and 3-month-old WT (P. alba) and transgenic poplars (L9 for 35S::PeuGAT2 and L4 for 35S::PeuGAT3) (at least 2 cm in diameter) were fixed with 2% formaldehyde and passed through a graded ethanol series. The stems were then embedded in paraffin and 8-µm-thick sections were cut with a rotary microtome. After removing residual paraffin, the sections were stained with 0.05% toluidine blue and examined with a light microscope. Images were captured under bright field using an ECLIPSE 80i microscope (Nikon, Tokyo, Japan). The radial widths of the phloem, cambium, and xylem were measured (at 20× and 40× magnification, respectively) using the Image Tool software package (UTHSCSA, San Antonio, TX, USA).
The Examination of Chemical Compositions in Transgenic Poplars and Proline Level Assay
Two-year-old WT and transgenic P. alba saplings were grown in soil with a 16-h-light/8-h-dark cycle at 25°C and 50% relative humidity under uniform illumination. Plants with stem diameters above 2 cm were selected for further experimentation. The wood composition of the selected saplings was studied using a Bruker Tensor 27 Fourier transform infrared (FTIR) spectrometer (Bruker, Germany) as described by Rodrigues et al. (1998). Five major parameters, I1505/I1425 (the relative content of lignin), I1505/I895 (the ratio of lignin to cellulose), I1505/I1738 (the ratio of lignin to hemicellulose), and I1505/I1382 and I1505/I1158 (the ratio of lignin to total cellulose) were measured to characterize the saplings’ wood composition, while cell wall thickness was measured using a dissecting microscope to characterize their anatomical structure. Leaf materials detached from WT and transgenic P. alba saplings were measured to determine their Pro contents using previously reported protocols (Li et al., 2017). These experiments were independently replicated three times under identical conditions.
Statistical Analysis
All statistical analyses were performed using the SPSS 17.0 software package. All quoted p values were derived using Student’s t-test based on one-way analysis of variance; p values below 0.05 and 0.01 are denoted by one asterisk (*) and two asterisks (**), respectively. Significance was evaluated by one- and two-way analyses of variance. The data were normalized, and all samples were normally distributed with homogeneity of variance.
Results
GATs Evolution Among Populus Species
GAT has been clustered into the AAAP family and sister with other five groups corresponding to the AAP, LHT, AUX, ANT, and ProT subfamilies (Ortiz-Lopez et al., 2000). The ProT and GAT subfamilies separated recently and clustered together. In four poplar species, a total of 18 GAT genes have been retrieved. Among them, two salt-resistive poplars (P. euphratica and P. pruinosa) have four GAT members, while five members were found in two salt-sensitive poplars (P. trichocarpa and P. alba) (Table 1). In Arabidopsis, only one GAT member has been found, whereas three ProT members have been retained (Figure 1). Phylogenetic analysis showed that GAT and ProT subfamilies separated from each other due to one whole-genome duplication. Then, GAT expanded via gene duplications with genus-specific in Populus. These genes have conserved protein domains similar to those found in various plant amino acid transporters, and sequence alignments revealed high homology with glutamate derived γ-aminobutyric acid transporters (GATs) from Arabidopsis (Batushansky et al., 2015).
Table 1.
Ks-dating and Ka/Ks ratios of GAT paralogs from different poplar species.
| Duplicated genes | Ka/Ks ratio | Duplicated genes | Ka/Ks ratio |
|---|---|---|---|
| PeuGAT1/PeuGAT2 | 0.1438 | PalGAT3/PalGAT4 | 0.0106 |
| PeuGAT1/PeuGAT3 | 1 | PtrGAT1/PtrGAT2-1 | 0.0842 |
| PeuGAT1/PeuGAT4 | 0.1463 | PtrGAT1/PtrGAT2-2 | 0.3959 |
| PeuGAT2/PeuGAT3 | 1.1747 | PtrGAT1/PtrGAT3 | 0.5931 |
| PeuGAT2/PeuGAT4 | 0.1750 | PtrGAT1/PtrGAT4 | 0.5637 |
| PeuGAT3/PeuGAT4 | 0.2023 | PtrGAT2-1/PtrGAT2-2 | 0.1630 |
| PprGAT1/PprGAT2-1 | 0.2508 | PtrGAT2-1/PtrGAT3 | 0.0746 |
| PprGAT1/PprGAT2-2 | 0.2961 | PtrGAT2-1/PtrGAT4 | 0.2272 |
| PprGAT1/PprGAT3 | 0.1367 | PtrGAT2-2/PtrGAT3 | 0.2206 |
| PprGAT2-1/PprGAT2-2 | 0.1637 | PtrGAT2-2/PtrGAT4 | 0.2131 |
| PprGAT2-1/PprGAT3 | 0.2032 | PtrGAT3/PtrGAT4 | 0.4209 |
| PprGAT2-2/PprGAT3 | 0.5273 | WiGAT1/WiGAT2-1 | 0.2081 |
| PalGAT1/PalGAT2-1 | 0.2406 | WiGAT1/WiGAT2-2 | 0.2419 |
| PalGAT1/PalGAT2-2 | 0.8139 | WiGAT1/WiGAT3 | 0.1471 |
| PalGAT1/PalGAT3 | 0.1487 | WiGAT1/WiGAT4 | 0.1457 |
| PalGAT1/PalGAT4 | 0.1333 | WiGAT2-1/WiGAT2-2 | 0.1196 |
| PalGAT2-1/PalGAT2-2 | 0.1762 | WiGAT2-1/WiGAT3 | 0.8407 |
| PalGAT2-1/PalGAT3 | 0.8042 | WiGAT2-1/WiGAT4 | 0.3160 |
| PalGAT2-1/PalGAT4 | 0.1428 | WiGAT2-2/WiGAT3 | 0.8323 |
| PalGAT2-2/PalGAT3 | 0.7459 | WiGAT2-2/WiGAT4 | 0.2661 |
| PalGAT2-2/PalGAT4 | 0.2841 | WiGAT3/WiGAT4 | 0.1263 |
Ka, nonsynonymous substitution rate; Ks, synonymous substitution rate; Ptr, P. trichocarpa; Ppr, P. pruinosa; Peu, P. euphratica; willow, Salix integra.
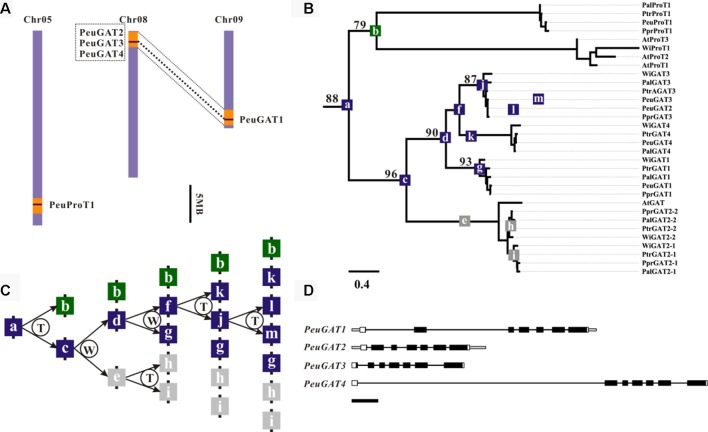
Figure 1.
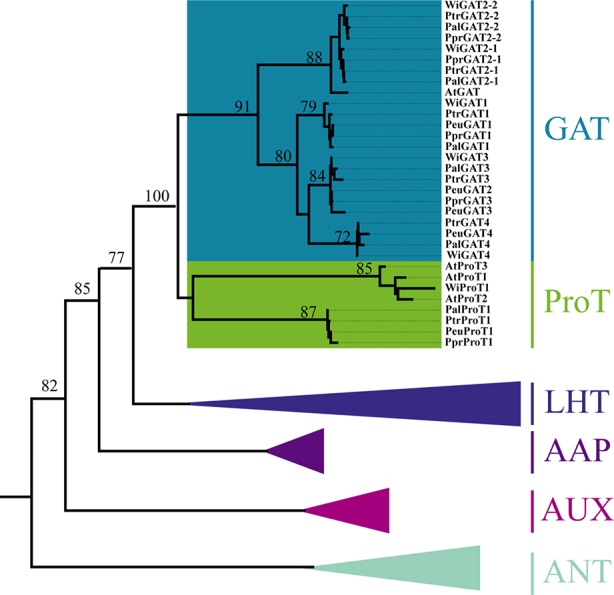
Phylogenetic position of AAAP isoforms among other transporters from Populus species and Arabidopsis thaliana. All transporter proteins were clustered into six clades, classified as AAP, LHT, AUX, ANT, ProT, and GAT transporters and marked with different background colors. Plant species are as follows: At, Arabidopsis thaliana; Ptr, Populus trichocarpa; Ppr, P. pruinosa; Peu, P. euphratica; willow, Salix integra.
Identification of Novel Gene
From the phylogenetic analysis as above, two P. euphratica genes PeuGAT2 and PeuGAT3 have clustered into one group, while in other species, especially for P. pruninose, a closely related poplar of P. euphratica, there was only one copy in that group; thus, PeuGAT3 are lineage specific (Figure 2A). Gene colinearity analysis found that the PeuGAT2 and PeuGAT3 located on eighth chromosome and are separated by an interval of ∼900 bp (Figure 2A); this suggested that the two paralogs arose from a tandem duplication event since the recent whole-genome duplication (Figure 2C). Protein structure analysis showed that PeuGAT3 was partially deleted at the N-terminus compared to PeuGAT2 (Figures S1A, B), and this deletion resulted in the lack a transmembrane region in PeuGAT3 compared to PeuGAT2 (Figure S1C). The estimation of divergence times among PeuGAT genes using nucleotide substitution rate of 1.2 × 10−8 (Ingvarsson, 2008) found that PeuGAT2 and PeuGAT3 diversified about 49 million years ago (Mya) (Table S2), indicating the two genes duplicated after the recent WGD of Populus. Finally, the Ka/Ks ratio was used to identify positive selection for all Populus GAT paralogs. Most of GATs of Ka/Ks ratio were lower than 1, suggesting that the GATs suffered from strong purifying selection, while Ka/Ks ratio between PeuGAT2 and PeuGAT3 was higher than 1, indicating that both genes experienced positive selection.
Figure 2.
Genomic localization, phylogenetic relationships, and hypothetical duplication histories of the PeuGAT genes. (A) Regions that are assumed to correspond to homologous genome blocks are shaded in orange and connected with lines. (B) Phylogenetic relationships were reconstructed using the WAG + I + G model. Numbers on branches indicate the bootstrap percentage values calculated from 1,000 bootstrap replicates. (C) The letters T and W in the schematic diagram showing the hypothetical origins of PeuGAT genes indicate tandem duplication and whole-genome duplication, respectively. Gray boxes represent the presumed pseudogenes or lost genes. Blue and green boxes represent PeuGAT and PeuProT genes, respectively. (D) The Aa_trans-coding domain is colored black, and the remaining exon sequences are shown as white boxes. Gray boxes represent untranslated regions, while lines represent introns. Scale bar: 500 bp.
Expression Pattern of PeuGAT2 and PeuGAT3 in P. Euphratica During Salt Stress
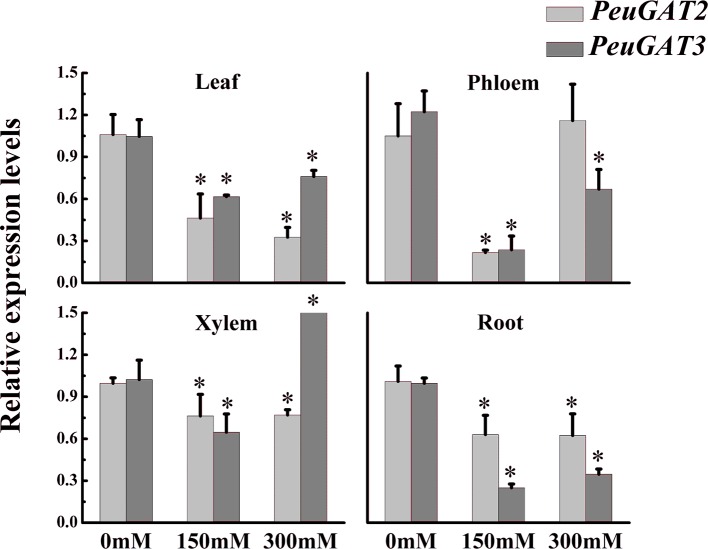
To identify potential differences in expression patterns between PeuGAT2 and PeuGAT3, we quantified their expression in the stems, roots, and leaves of P. euphratica seedlings. Both genes were expressed simultaneously in all studied organs (Figure 3). However, salt stress induced by treatment with different NaCl concentrations reduced the expression of both genes in leaves and roots. Furthermore, compared with PeuGAT3, the overall expression of PeuGAT2 in leaves were more severely suppressed, while in roots, it exhibited the opposite result. In xylems, the two genes exhibited similar patterns under moderate salt stress, while under severe stress, PeuGAT3 exhibited significant higher expression level compared with PeuGAT2. On the contrary, PeuGAT2 could maintain relatively high expression in phloem under severe stress. Those results show that the two genes have different expression patterns and that these differences become more pronounced in response to salt stress.
Figure 3.
The expression pattern of PeuGAT2 and PeuGAT3 in P. euphratica seedlings treated with different concentrations of NaCl for 2 weeks. RNA was extracted from leaves, roots, phloem, and xylem, and the relative expression of the two genes was quantified by real-time PCR; 18S rRNA was used as a reference. Three replicates of each sample were analyzed. All values are means ± SE (n = 3). Values labeled with different letters differ significantly (*p < 0.05).
Overexpression PeuGAT3 Enhances Abiotic Stress Tolerance in Arabidopsis and Improves Lignin Accumulation in Arabidopsis and Poplar
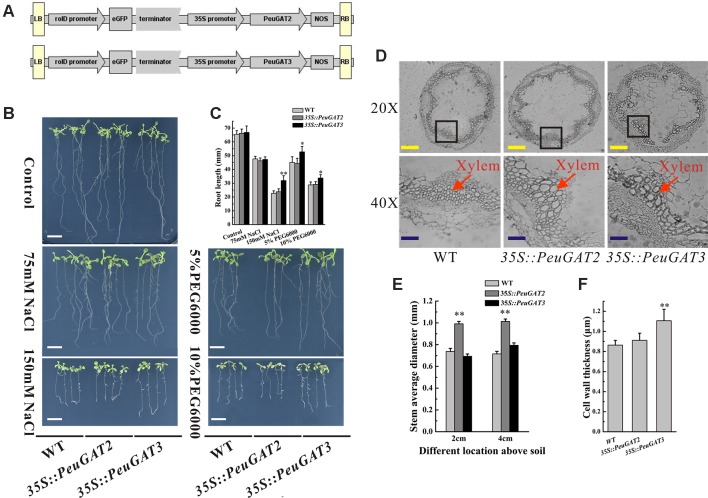
To investigate the differences between the function of PeuGAT2 and PeuGAT3 in plants, they were overexpressed in Arabidopsis (Figure 4A) and Populus (Figure 5A) species through transgenic methods. Under normal condition, the WT and two transgenic Arabidopsis lines showed similar phenotype. In the cases of abiotic stress treatment, except PeuGAT3 overexpression lines, the root growths of the remaining Arabidopsis lines were significantly inhibited (Figures 4B, C). However, in the process of germination, there was no significant difference between wild type and two overexpressing Arabidopsis lines, whether in normal condition or salt stress conditions (Figure S3). Furthermore, in vegetative growth phase, the 35S::PeuGAT2 line exhibited significantly (p < 0.05) thicker stems as determined by measuring the stem diameter at heights of 2 and 4 cm above the soil (Figure 4E), while 35S::PeuGAT3 line exhibited a more thicker cell wall (Figures 4D, F). In transgenic Populus lines, similar phenotypes were observed but the difference were less pronounced. Lines overexpressing PeuGAT3 produced ligneous tissues with thicker cell walls than were seen in lines overexpressing PeuGAT2 (Figures 5B, D), indicating that the neo-functionalization of PeuGAT3 may confer plasticity that allows trees to limit water losses.
Figure 4.
Morphology phenotype of transgenic Arabidopsis. (A) Constructs of vectors used in transgenic Arabidopsis and poplar. (B) Phenotypes of wild-type and transgenic Arabidopsis under salt and drought stresses. (D) Stem anatomical structures of wild-type and transgenic Arabidopsis. The root length (C) diameter of stem (E) and cell wall thickness (F) of wild-type and transgenic Arabidopsis. Each sample was analyzed in triplicate. All values are means ± SE (n = 20). Values labeled with different letters differ significantly (*p < 0.05, **p < 0.01). Bars: white, 10 mm; yellow, 20 µm; blue, 40 µm.
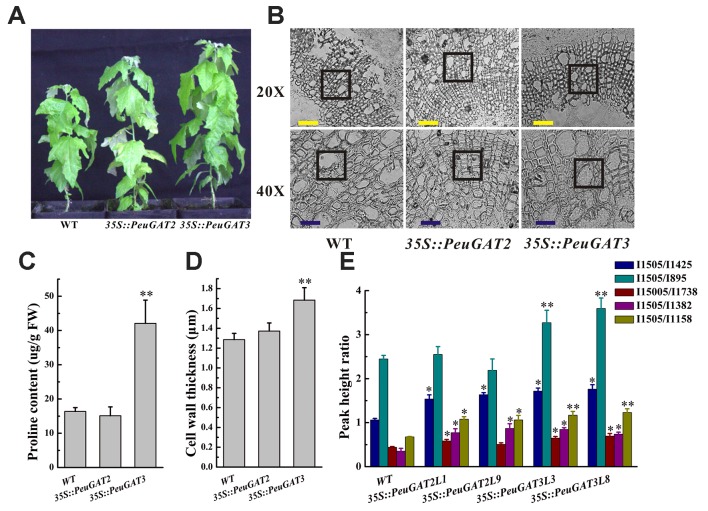
Figure 5.
The phenotype (A) and stem anatomy (B) of wild and transgenic poplars overexpressing PeuGAT2 and PeuGAT3. (C) Proline content in wild-type and transgenic poplar leaves. (D)The cell wall thickness of wild and transgenic poplars overexpressing PeuGAT2 and PeuGAT3. (E)The wood composition of wild and transgenic poplars overexpressing PeuGAT2 and PeuGAT3. I1505/I1382 and I1505/I1158 represent the ratio of lignin to total cellulose, while I1505/I1425, I1505/I895, and I1505/I1738 represent the relative content of lignin, the lignin to cellulose ratio, and the lignin to hemicellulose ratio, respectively. Values labeled with different letters differ significantly (*p < 0.05 and **p < 0.01). Bars: yellow, 20 µm; blue, 40 µm.
To characterize the morphological and anatomical differences between the transgenic Populus and Arabidopsis lines, their wood composition was studied by FTIR (BRUKER TENSOR 27, Germany), and their anatomical structure was studied using a dissecting microscope. 35S::PeuGAT2 overexpression in poplars increased the size of xylem cells in the stem, whereas overexpression of 35S::PeuGAT3 caused the cell walls of stem xylem cells to thicken significantly (Figures 5B, D). Similar results were obtained in the transgenic Arabidopsis lines: lines overexpressing 35S::PeuGAT2 produced larger xylem cells, whereas those overexpressing 35S::PeuGAT3 produced thicker xylem cell walls (Figures 4D, F). In addition, five key IR spectroscopic ratios were determined to characterize the wood composition of the transgenic and WT Populus plants (Figure 5E). Measurements of the I1505/I1425 ratio (i.e., the ratio of lignin to cellulose) indicated that transgenic plants had higher lignin contents than did their WT counterparts. The I1505/I895 ratio (i.e., the ratio of lignin to hemicellulose) was significantly higher in lines overexpressing 35S::PeuGAT3 than those overexpressing 35S::PeuGAT2, but the I1505/I1738; I1505/I1382 and I1505/I1158 ratios (representing the ratio of lignin to total cellulose) were similar for both transgenic lines.
The proline content of the leaves typically reflects plants’ tolerance for various biotic and abiotic stresses. We therefore measured the proline levels in the leaves of the WT and transgenic poplars. As shown in Figure 5C, the 35S::PeuGAT3 poplars had significantly higher levels of proline than WT poplars, but the 35S::PeuGAT2 poplars had similar leaf proline levels to WT poplars.
Discussion
The GAT genes in Populus Expanded in Specific Genus
The novel gene arose through gene duplication and divergence provide important genetic resources that drive evolution (Kondrashov et al., 2002). In the immediate aftermath of a duplication event, the duplicated genes will typically exhibit almost complete functional redundancy. Copies that acquire detrimental mutations are likely to be eliminated by purifying selection, but those that diverge functionally may be retained (Force et al., 1999), potentially leading to the emergence of novel and evolutionarily beneficial functions (Long and Langley, 1993; Long et al., 1999; Katju and Lynch, 2006; Nan et al., 2017). Some of these duplicated genes may be fixed by selection because they are adaptive, as may be the case if they increase stress tolerance and growth flexibility (Kondrashov et al., 2002). In AAAP family of plants, subfamilies GAT and ProT diversified recently from a common ancestral gene. The GAT gene family then expanded with specific genus in Populus but not in Arabidopsis, while in the ProT family, the opposite is true. In Arabidopsis, only one AtGAT gene has been identified and has a high affinity for GABA but cannot transport proline (Schwacke et al., 1999), suggesting that GAT and ProT subfamilies have diversified completely in function. Whereas in poplar, the genus-specific expansion of GATs would improve poplar species adaptation or contribute the formation of polymorphic traits, such as wood property and tolerance to stress. Of them, PeuGAT3, sister gene of PeuGAT2, underwent rapid evolution and have suffered from positive selection. Furthermore, no PeuGAT3 orthologs have been found in the closely related poplars, indicating PeuGAT3 arose lineage specific through a duplication event (Tautz and Domazet-Lošo, 2011). Therefore, PeuGAT3 has been identified as a novel gene that created through gene duplication or divergence event. It is clear that the genus-specific expansion of GAT in Populus and subsequent diversification provide important genetic resource in the formation of specific adaptive traits (Shukla et al., 2014), while the divergence of novel PeuGAT3 in protein function/structure might define its function better than in expression pattern.
Functional Divergence Between PeuGAT2 and PeuGAT3 Occurred in Multiple Levels
The functional divergence among paralogous genes (including novel genes) plays an important role in plant adaptation (Lynch et al., 2001; He and Zhang, 2005). The proteins encoded by duplicated genes have diversified in their expression patterns and/or biochemical functions, leading to changes in gene structure and the properties of the corresponding proteins (Force et al., 1999; Nan et al., 2017), site-specific regulatory modification of proteins, variation of splicing sites among isoforms (Bergström et al., 2014), and protein binding specificity (Hittinger and Carroll, 2007; Gagnon-Arsenault et al., 2013). In the desert poplars species, two paralogous PeuGAT2 and PeuGAT3 have diversified not only in expression pattern during salt treatment but also in gene structure and function. First, PeuGAT3 express in phloem and leaves, especially in salt treatment, while PeuGAT2 express in lignin. The novel genes express in tissue specific and involve in response to environmental stresses and in formation of lineage-specific traits (Arendsee and Li, 2014). Second, PeuGAT3 lacks one motif found in PeuGAT2, which lead to lack of a transmembrane domain in PeuGAT3. Finally, the overexpression of PeuGAT2 improved the Arabidopsis rapid growth of the aerial parts by keep the size of cells in xylem, while the overexpression of PeuGAT3 in Arabidopsis and poplar enhanced the accumulation of proline in leaves and lignin content in the xylem tissues, which enable cells to better regulate their osmotic potential and further accelerate proline transport (Yancey, 2005; Verslues et al., 2006). Meanwhile, the higher expression of PeuGAT3 than PeuGAT2 in leaves could improve ion homeostasis between leaves and roots (Foyer and Noctor, 2005). Obviously, in poplar, PeuGAT3 maybe have more affinity to proline than PeuGAT2.
PeuGAT3 Remains the Proline Transport That May Improve the Stress Tolerance in P. euphratica
Proline is a beneficial solute whose accumulation allows plant cells to increase their osmolarity and thereby avoid damage caused by osmotic stress (Yancey, 2005; Lehmann et al., 2010; Szabados and Savoure, 2010). In Arabidopsis, three ProTs copies involved in proline transporting are known to exhibit differential expression in different tissues under various stresses (Grallath et al., 2005). It might be expected that a proline transporter would be induced by stress when proline levels are also increasing. In Populus, only one ProT protein was responsible for proline transport. However, overexpression PeuGAT3, a closely related gene of PeuProTs, can increase proline content in poplar and improve tolerance to salt and drought in Arabidopsis. Therefore, this novel gene PeuGAT3 would be responsible for partial proline transport, and this greatly contributes to the adaptation of P. euphratica to arid and high salt environments. Additionally, overexpression PeuGAT3 increased the thickness of cell walls in the ligneous tissues. This may be an adaptive anatomical feature that helps plants such as P. euphratica to reduce the risk of cavitation in arid climates (Tyree and Sperry, 1989; Sperry et al., 2008).It is well-known that P. euphratica do not fall down and even rot when it is dead because of its hard tree trunk. PeuGAT3 overexpression increased the lignin content in the xylem tissues of transgenic poplar lines to a greater degree than was observed for PeuGAT2 overexpression. High lignin levels in the xylem of transgenic poplars could enhance their resistance to the erosion by harsh environment, indicating that this activity of novel gene PeuGAT3 in P. euphratica may partly explain why the trunks of dead P. euphratica trees often persist without rotting for extended periods of time. However, the transgenic poplars examined in this work were too young to study their resistance. It would certainly be desirable to investigate this in future.
Data Availability
All datasets for this study are included in the manuscript and the Supplementary Files.
Author Contributions
DW designed the experiments and wrote the manuscript. XB, JX, XS, and ZN performed the experiments. WL and CG contributed to the data analyses.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The research was supported by the National Natural Science Foundation of China (No. 31870580) and the Fundamental Research Funds for the Central Universities (lzujbky-2017-k14). Core Facility of School of Life Sciences, Lanzhou University provided us with qRT-PCR facilities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01083/full#supplementary-material
References
- Arendsee Z. W., Li L. (2014). Wurtele ES. Coming of age: orphan genes in plants. Trends Plant Sci 19 (11), 698–708. 10.1016/j.tplants.2014.07.003 [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Williams N., Misleh C., Li W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. 10.1093/nar/gkl198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batushansky A., Kirma M., Grillich N., Pham P. A., Rentsch D., Galili G., et al. (2015). The transporter GAT1 plays an important role in GABA-mediated carbon–nitrogen interactions in Arabidopsis. Front. Plant Sci. 6, 785. 10.3389/fpls.2015.00785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström A., Simpson J. T., Salinas F., Barré B., Parts L., Zia A., et al. (2014). A high-definition view of functional genetic variation from natural yeast genomes. Mol. Biol. Evol. 31 (4), 872–888. 10.1093/molbev/msu037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche N., Fromm H. (2004). GABA in plants: just a metabolite? Trends Plant Sci. 9 (3), 110–115. 10.1016/j.tplants.2004.01.006 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 (6), 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Dixon D. P., Edwards R. (2010. a). Glutathione transferases. The Arabidopsis Book/American Society of Plant Biologists, 8. 10.1199/tab.0131 [DOI] [PMC free article] [PubMed]
- Dai X., Hu Q., Cai Q., Feng K., Ye N., Tuskan G. A., et al. (2014). The willow genome and divergent evolution from poplar after the common genome duplication. Cell Res. 24 (10), 1274. 10.1038/cr.2014.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., Postlethwait J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C. H., Noctor G. (2005). Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17 (7), 1866–1875. 10.1105/tpc.105.033589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon-Arsenault I., Blanchet F. C. M., Rochette S., Diss G., Dubé A. K., Landry C. R. (2013). Transcriptional divergence plays a role in the rewiring of protein interaction networks after gene duplication. J. Proteomics 81, 112–125. 10.1016/j.jprot.2012.09.038 [DOI] [PubMed] [Google Scholar]
- Grallath S., Weimar T., Meyer A., Gumy C., Suter-Grotemeyer M., Neuhaus J. M., et al. (2005). The AtProT family. Compatible solute transporters with similar substrate specificity but differential expression patterns. Plant Physiol. 137, 117–126. 10.1104/pp.104.055079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X. L., Zhang J. Z. (2005). Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169, 1157–1164. 10.1534/genetics.104.037051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger C. T., Carroll S. B. (2007). Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449, 677–681. 10.1038/nature06151 [DOI] [PubMed] [Google Scholar]
- Horsch R., Fry J., Hoffmann N., Wallroth M. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. 10.1126/science.227.4691.1229 [DOI] [PubMed] [Google Scholar]
- Ingvarsson P. K. (2008). Molecular evolution of synonymous codon usage in Populus. BMC Evol. Biol. 8 (1), 307. 10.1186/1471-2148-8-307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 75, 193–195. 10.1016/S1360-1385(02)02251-3 [DOI] [PubMed] [Google Scholar]
- Katju V., Lynch M. (2006). On the formation of novel genes by duplication in the Caenorhabditis elegans genome. Mol. Biol. Evol. 23, 1056–1067. 10.1093/molbev/msj114 [DOI] [PubMed] [Google Scholar]
- Koch M. A., Haubold B., Mitchell-Olds T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17 (10), 1483–1498. 10.1093/oxfordjournals.molbev.a026248 [DOI] [PubMed] [Google Scholar]
- Kondrashov F. A., Rogozin I. B., Wolf Y. I., Koonin E. V. (2002). Selection in the evolution of gene duplications. Genome Biol. 3, research0008. 10.1186/gb-2002-3-2-research0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann S., Funck D., Szabados L., Rentsch D. (2010). Proline metabolism and transport in plant development. Amino Acids 39, 949–962. 10.1007/s00726-010-0525-3 [DOI] [PubMed] [Google Scholar]
- Li J., Shi S., Srivastava S. P., Kitada M., Nagai T., Nitta K., et al. (2017). FGFR1 is critical for the anti-endothelial mesenchymal transition effect of N-acetyl-seryl-aspartyl-lysyl-proline via induction of the MAP4K4 pathway. Cell Death Dis. 8 (8), e2965. 10.1038/cddis.2017.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–DDCT method. Methods 25 (4), 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Long M. Y., Langley C. H. (1993). Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science 260, 91–95. 10.1126/science.7682012 [DOI] [PubMed] [Google Scholar]
- Long M. Y., Wang W., Zhang J. (1999). Origin of new genes and source for N-terminal domain of the chimerical gene, jingwei, in Drosophila. Gene 238, 135–141. 10.1016/S0378-1119(99)00229-2 [DOI] [PubMed] [Google Scholar]
- Lynch M., O’Hely M., Walsh B., Force A. (2001). The probability of preservation of a newly arisen gene duplicate. Genetics 159, 1789–1804. 10.1017/S0016672301005365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., He X., Bai X., Niu Z., Duan B., Chen N., et al. (2016). Genome-wide survey reveals transcriptional differences underlying the contrasting trichome phenotypes of two sister desert poplars. Genes 7, 111. 10.3390/genes7120111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Wan D., Duan B., Bai X., Bai Q., Chen N., et al. (2019). Genome sequence and genetic transformation of a widely distributed and cultivated poplar. Plant Biotechnol. J. 17 (2) 451–460. 10.1111/pbi.12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Wang J., Zhou G., Yue Z., Hu Q., Chen Y., et al. (2013). Genomic insights into salt adaptation in a desert poplar. Nat. Comm. 4, 2797. 10.1038/ncomms3797 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Anderson J. B., Cherukuri P. F., Weese-Scott C., Geer L. Y., Gwadz M., et al. (2005). CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33, D192–D196. 10.1093/nar/gki069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan Q., Qian D., Niu Y., He Y., Tong S., Niu Z., et al. (2017). Plant actin-depolymerizing factors possess opposing biochemical properties arising from key amino acid changes throughout evolution. Plant Cell 29, 395–408. 10.1105/tpc.16.00690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan V. S., Nair P. M. (1990). Metabolism, enzymology and possible roles of 4-aminobutyrate in higher plants. Phytochemistry 29 (2), 367–375. 10.1016/0031-9422(90)85081-P [DOI] [Google Scholar]
- Okumoto S., Pilot G. (2011). Amino acid export in plants: a missing link in nitrogen cycling. Mol. Plant 4, 453–463. 10.1093/mp/ssr003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Lopez A., Chang H. C., Bush D. R. (2000). Amino acid transporters in plants. Biochim. Biophys. Acta Biomembr. 1465, 275–280. 10.1016/S0005-2736(00)00144-9 [DOI] [PubMed] [Google Scholar]
- Ottow E. A., Brinker M., Teichmann T., Fritz E., Kaiser W., Brosché M., et al. (2005). Populus euphratica displays apoplastic sodium accumulation, osmotic adjustment by decreases in calcium and soluble carbohydrates, and develops leaf succulence under salt stress. Plant Physiol. 139 (4), 1762–1772. 10.1104/pp.105.069971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G. H., Dominy N. J., Claw K. G. (2007). Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39, 1256–1260. 10.1038/ng2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues J., Faix O., Pereira H. (1998). Determination of lignin content of Eucalyptus globulus wood using FTIR spectroscopy. Holzforschung 52 (1), 46–50. 10.1515/hfsg.1998.52.1.46 [DOI] [Google Scholar]
- Rozas J., Ferrer-Mata A., Sanchez-DelBarrio J. C., Guirao-Rico S., Librado P., Ramos-Onsins S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. 10.1093/molbev/msx248 [DOI] [PubMed] [Google Scholar]
- Saier M. H., Yen M. R., Noto K., Tamang D. G., Elkan C. (2008). The transporter classification database: recent advances. Nucleic Acids Res. 37, 274–278. 10.1093/nar/gkn862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R., Grallath S., Breitkreuz K. E., Stransky E., Stransky H., Frommer W. B., et al. (1999). LeProT1, a transporter for proline, glycine betaine, and γ-amino butyric acid in tomato pollen. Plant Cell 11 (3), 377–391. 10.1105/tpc.11.3.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelp B. J., Bown A. W., McLean M. D. (1999). Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci. 4 (11), 446–452. 10.1016/S1360-1385(99)01486-7 [DOI] [PubMed] [Google Scholar]
- Shukla V., Habib F., Kulkarni A., Ratnaparkhi G. S. (2014). Gene duplication, lineage-specific expansion, and subfunctionalization in the MADF-BESS family patterns the Drosophila wing hinge. Genetics 196 (2), 481–496. 10.1534/genetics.113.160531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry J. S., Meinzer F. C., McCulloh K. A. (2008). Safety and efficiency conflicts in hydraulic architecture: scaling from tissues to trees. Plant Cell Environ. 31, 632–645. 10.1111/j.1365-3040.2007.01765.x [DOI] [PubMed] [Google Scholar]
- Szabados L., Savoure A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15 89–97. 10.1016/j.tplants.2009.11.009 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 (8), 1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Tautz D., Domazet-Lošo T. (2011). The evolutionary origin of orphan genes. Nat. Rev. Genet. 12 (10), 692–702. 10.1038/nrg3053 [DOI] [PubMed] [Google Scholar]
- Tegeder M. (2012). Transporters for amino acids in plant cells: some functions and many unknowns. Curr. Opin. Plant Biol. 15, 1–7. 10.1016/j.pbi.2012.02.001 [DOI] [PubMed] [Google Scholar]
- Tuskan G. A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. 10.1126/science.1128691 [DOI] [PubMed] [Google Scholar]
- Tyree M. T., Sperry J. S. (1989). Vulnerability of xylem to cavitation and embolism. Annu. Rev. Plant Biol. 40, 19–38. 10.1146/annurev.pp.40.060189.000315 [DOI] [Google Scholar]
- Verslues P. E., Agarwal M., Katiyar-Agarwal S., Zhu J., Zhu J. K. (2006). Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 45, 523–539. 10.1111/j.1365-313X.2005.02593.x [DOI] [PubMed] [Google Scholar]
- Voordeckers K., Brown C. A., Vanneste K., van der Zande E., Voet A., Maere S., et al. (2012). Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biol. 10 (12), e1001446. 10.1371/journal.pbio.1001446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipf D., Ludewig U., Tegeder M., Rentsch D., Koch W., Frommer W. B. (2002). Conservation of amino acid transporters in fungi, plants and animals. Trends Biochem. Sci. 27, 139–147. 10.1016/S0968-0004(01)02054-0 [DOI] [PubMed] [Google Scholar]
- Yancey P. H. (2005). Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 208, 2819–2830. 10.1242/jeb.01730 [DOI] [PubMed] [Google Scholar]
- Zhang C. H., Ma T., Luo W. C., Xu J. M., Liu J. Q., Wan D. S. (2015). Identification of 4CL genes in desert poplars and their changes in expression in response to salt stress. Genes 6 (3), 901–917. 10.3390/genes6030901 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets for this study are included in the manuscript and the Supplementary Files.