Abstract
Background
The molecular characterization of carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-hvKP) isolates is not well studied. Our goal was to investigate the molecular epidemiology of CR-hvKP strains that were isolated from a Chinese hospital.
Results
All clinical carbapenem-resistant K. pneumoniae (CR-KP) isolates were collected and identified from patient samples between 2014 and 2017 from a Chinese hospital. The samples were subjected to screening for CR-hvKP by string test and the detection of the aerobactin gene. CR-hvKP isolates were further confirmed through neutrophil phagocytosis and a mice lethality assay. The CR-hvKP isolates were investigated for their capsular genotyping, virulence gene profiles, and the expression of carbapenemase genes by PCR and DNA sequencing. Multilocus sequence type (MLST) and pulsed-field gel electrophoresis (PFGE) were performed to exclude the homology of these isolates. Twenty strains were identified as CR-hvKP. These strains were resistant to imipenem and several other antibiotics, however, most were susceptible to amikacin. Notably, two isolates were not susceptible to tigecycline. Capsular polysaccharide synthesis genotyping revealed that 17 of the 20 CR-hvKP strains belonged to the K2 serotype, while the others belonged to serotypes other than K1, K2, K5, K20, and K57. The strains were found to be positive for 10 types of virulence genes and a variety of these genes coexisted in the same strain. Two carbapenemase genes were identified: blaKPC-2 (13/20) and blaNDM-1 (1/20). PFGE typing revealed eight clusters comprising isolates that belonged to MLST types ST25, ST11 and ST375, respectively. PFGE cluster A was identified as the main cluster, which included 11 isolates that belong to ST25 and mainly from ICU department.
Conclusions
Our findings suggest that hospital-acquired infections may contribute in part to the CR-hvKP strains identified in this study. It also suggests that ST25 CR-hvKP strain has a clonal distribution in our hospital. Therefore, effective surveillance and strict infection control strategies should be implemented to prevent outbreak by CR-hvKP strains in hospitals setting.
Keywords: Carbapenem-resistant, Carbapenemase, Hypervirulent, Klebsiella pneumoniae, Serotype
Background
Hypervirulent Klebsiella pneumoniae (hvKP) is a variant of K. pneumoniae, and was first reported in Taiwan in 1986 [1]. Compared with classic K. pneumoniae (cKP), hvKP causes life-threatening, community-acquired infection, especially in younger, healthy global population [2].
In previous studies, the resistance of hvKP to commonly used antimicrobial agents has rarely been reported, except for an intrinsic resistance to ampicillin [3]. However, there has been an increased occurrence of multiple-resistant hvKP strains, including extended spectrum β-lactamases-producing isolates and third-generation cephalosporin-resistant strains [3–5]. Recently, carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKP) isolates have also been described in some case reports [6–9]. Furthermore, several studies have shown that Klebsiella pneumoniae carbapenemase (KPC)-like genes have the potential for dissemination among hvKP isolates [8, 9]. Carbapenem-resistant K. pneumoniae (CR-KP) isolates have the potential to be converted into CR-hvKP through the acquisition of virulence related plasmids, such as pLVPK [9, 10]. The confluence of multidrug resistance and enhanced virulence may cause serious outbreaks and public-health problems.
A variety of factors lead to carbapenem resistance, including the production of carbapenemases, decreased expression of outer membrane proteins and overexpression of efflux pumps [11]. In particular, K. pneumoniae have acquired carbapenemases, which are enzymes capable of breaking down most β-lactams, including carbapenems, and thus conferring resistance to these drugs. Carbapenemases can be divided into metallo-carbapenemases (zinc-dependent class B) and non-metallo-carbapenemases (zinc-independent classes A, C, and D), according to their dependency on divalent cations for enzyme activation. The primary carbapenemase that was detected in K. pneumoniae was of the class A, such as the KPC enzymes [12]. However, there have only been a few reports of KPC production in hvKP isolates [6, 7, 13, 14].
Multilocus sequence type (MLST) has been used as a powerful technique to genotype and characterize bacterial strains. MLST 258 (ST258) is an important CR-KP strain that is responsible for the extensive global spread of KPC-producing K. pneumoniae, while ST11, which is closely related to ST258, is a prevalent clone associated with the spread of KPC-producing K. pneumoniae in Asia (particularly in China and Taiwan) [15–17]. Additional sequence types of CR-hvKP isolates have also been reported, including ST23, ST11, ST25, ST65, ST86, etc. [18].
Studies have found that the K1 serotype of hvKP is mainly related to ST23, whereas the K2 serotype of hvKP is related to a more diverse number of sequence types, including ST25, ST65, ST66, and ST86 [18–21]. Notably, ST23 hvKP clones of the K1 serotype are associated with pyogenic liver abscesses, whereas ST65 hvKP clones of the K2 serotype are correlated with various invasive infections [21].
Although many studies have reported hvKP infections, especially involving pyogenic liver abscesses, there is limited literature regarding the prevalence, and molecular characteristics of CR-hvKP isolates. In this study, we report 20 CR-hvKP isolates that were identified from a hospital in Hunan, mid-south China. We further investigated the molecular characteristics of these CR-hvKP isolates. Our study provides novel insight into the prevalence of CR-hvKP and may provide important information regarding the treatment and prevention CR-hvKP infections.
Results
Determination and clinical characteristics of CR-hvKP
A total of 562 CR-KP isolates were collected. The results of the string test show that 26 CR-KP isolates were positive, with a rate of 4.6%. Twenty-three CR-KP isolates were positive for the aerobactin gene. The combination of the results of the string test and the detection of the aerobactin gene resulted in 20 CR-hvKP isolates being filtered out (Table 1). The 20 CR-hvKP isolates were from different samples of 15 patients, 13 hospitalized patients and 2 outpatients. All of these patients were males, which is consistent with the findings of a previous study [22]. The main specimens were sputum samples (65.0%, 13/20; Table 1). It is worth noting that seven of the CR-hvKP isolates were detected in two patients (P1 and P3) during multiple hospitalizations. P1 had three consecutive visits to two different wards of the hospital and a total of four CR-hvKP isolates (CS11, CS12, CS70 and CS129) were detected. P3 had two consecutive visits to the same ward of the hospital and three CR-hvKP isolates (CS15, CS48 and CS57) were identified.
Table 1.
Patient information and clinical features of the CR-hvKP isolates
| Strain Number | Patient Number | Age-range | Department | Collection Date | Source | Outcome | Resistance Determinants Genes | PFGE type | MLST |
|---|---|---|---|---|---|---|---|---|---|
| CS1 | P2 | 21–30 | ICU | 2016/12/27 | csf | Death | blaKPC-2, blaSHV-1, blaTEM-1, blaCTX-M-3 | A | 25 |
| CS11 | P1a | 41–50 | ICU | 2016/11/28 | sp | Survived | blaKPC-2, blaSHV-1, blaTEM-1, blaCTX-M-3 | A | 25 |
| CS12 | P1 | 41–50 | ICU | 2016/12/6 | sp | Survived | blaKPC-2, blaSHV-1, blaTEM-1, blaCTX-M-3 | A | 25 |
| CS15 | P3a | 51–60 | IMD | 2016/12/21 | sp | Survived | blaKPC-2, blaSHV-1, blaTEM-1, blaCTX-M-3 | A | 25 |
| CS17 | P4 | 71–80 | ICU | 2016/12/3 | sp | Giving up Treatmentb | blaKPC-2, blaSHV-1, blaTEM-1, blaCTX-M-3 | A | 25 |
| CS45 | P5 | 41–50 | ICU | 2016/11/15 | sp | Giving up treatment | blaKPC-2, blaTEM-1, blaCTX-M-3 | A | 25 |
| CS47 | P6 | 71–80 | ICU | 2016/12/28 | blood | Giving up treatment | blaKPC-2, blaTEM-1, blaCTX-M-3 | A | 25 |
| CS48 | P3 | 51–60 | IMD | 2016/12/31 | sp | Survived | blaKPC-2, blaSHV-1, blaTEM-1, blaCTX-M-3 | A | 25 |
| CS57 | P3 | 51–60 | IMD | 2017/3/11 | sp | Survived | blaKPC-2, blaSHV-1, blaTEM-1, blaCTX-M-3 | A | 25 |
| CS60 | P14 | 61–70 | CSD | 2017/6/16 | puncture fluid | Giving up treatment | bla SHV-1 | H | 25 |
| CS61 | P15 | 31–40 | ICU | 2017/6/4 | drainage liquid | Death | bla SHV-1 | H | 25 |
| CS62 | P8 | 51–60 | ICU | 2017/6/16 | sp | Survived | bla SHV-1 | C | 25 |
| CS70 | P1 | 41–50 | RD | 2017/6/22 | sp | Survived | blaTEM-1, blaCTX-M-3 | A | 25 |
| CS80 | P13 | 61–70 | Burn Unit | 2017/7/2 | traumatic secretion | Death | blaKPC-2, blaSHV-1, blaTEM-1, blaCTX-M-65 | G | 11 |
| CS90 | P9 | 51–60 | Outpatient | 2017/3/27 | drainage liquid | Survived | blaSHV-1, blaCTX-M-65 | D | 375 |
| CS103 | P11 | 41–50 | ICU | 2017/5/22 | sp | Giving up treatment | blaKPC-2, blaSHV-1 | F | 11 |
| CS118 | P12 | 71–80 | ICU | 2017/3/18 | sp | Giving up treatment | blaKPC-2, blaSHV-1, blaTEM-1, blaCTX-M-65 | F | 11 |
| CS127 | P10 | 41–50 | Outpatient | 2017/5/11 | blood | Survived | bla SHV-1 | E | 25 |
| CS129 | P1 | 41–50 | ICU | 2017/3/16 | sp | Survived | blaKPC-2, blaSHV-1, blaCTX-M-3 | A | 25 |
| CS59497 | P7 | 51–60 | ICU | 2014/10/5 | sp | Survived | blaNDM-1, blaSHV-1, blaTEM-1 | B | 25 |
Department: indicates the department at the Xiangya Hospital, where the samples were collected; ICU Intensive care unitm, IMD Integrative Medicine Department, CSD Cerebrovascular Surgery Department, RD Rehabilitation Department, mCIM Modified Carbapenem Inactivation Method, PFGE Pulsed Field Gel Electrophoresis, MLST Multilocus sequence type
“CSF” Cerebrospinal fluid, “Sp” Sputum
aPatients P1 and P3 are immunocompromised patients. Repeated infections occurred in these two patients during 2016 and 2017
bThe patient’s condition deteriorated so the patient stopped any further treatment
The 20 CR-hvKP isolates had high drug resistance rates for most of the antibacterial drugs that were tested, except for tigecycline and amikacin. Two isolates were not susceptible to tigecycline (CS45 and CS129, with MICs of 1.5 μg/mL and 4 μg/mL, respectively) (shown in Table 2).
Table 2.
Resistance to antimicrobial agents of the CR-hvKP isolates
| Antimicrobial Agents | R (%) | I (%) | S (%) |
|---|---|---|---|
| Ceftazidime | 65.0 | 15.0 | 20.0 |
| Ceftriaxone | 85.0 | 0.0 | 15.0 |
| Cefepime | 45.0 | 30.0 | 25.0 |
| Aztreonam | 80.0 | 0.0 | 20.0 |
| Piperacillin/Tazobactam | 60.0 | 20.0 | 20.0 |
| Amikacin | 15.0 | 0.0 | 85.0 |
| Gentamycin | 55.0 | 5.0 | 40.0 |
| Ciprofloxacin | 80.0 | 0.0 | 20.0 |
| Levofloxacin | 45.0 | 5.0 | 50.0 |
| Trimethoprim-Sulfamethoxazole | 65.0 | 0.0 | 35.0 |
| Imipenem | 100.0 | 0.0 | 0.0 |
| Tigecycline | 5.0 | 5.0 | 90.0 |
R Resistant, I Intermediary, S Susceptible
Four CR-hvKP isolates were randomly selected for neutrophil phagocytosis and mice lethality assay. The results of the neutrophil phagocytosis show that the survival rates of the four tested CR-hvKP strains and NTUH-K2044 (a hypervirulent wild strain) were significantly higher than that of K. pneumoniae ATCC 700603 (a standard strain for low virulence and low resistance; P < 0.05; Fig. 1). The mice lethality assay results show that the survival time of mice that were infected with the CR-hvKP isolates or NTUH-K2044 was 5–6 d, while those infected with K. pneumoniae ATCC 700603 all survived the monitoring period (10 d; Fig. 2).
Fig. 1.
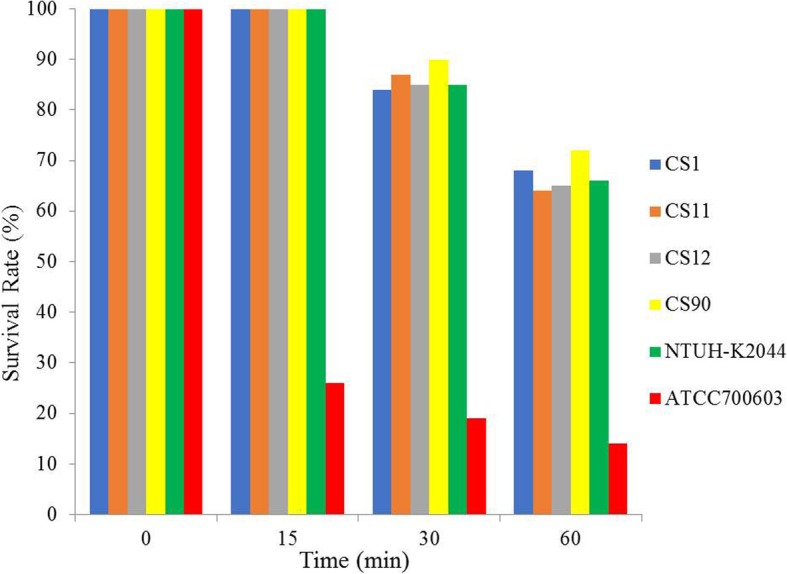
Neutrophil phagocytosis analysis of four randomly selected CR-hvKP isolates (CS1, CS11, CS12 and CS90). NTUH-K2044: a hypervirulent wild strain of K. pneumoniae; ATCC 700603: a standard strain of K. pneumoniae with low virulence and low resistance
Fig. 2.
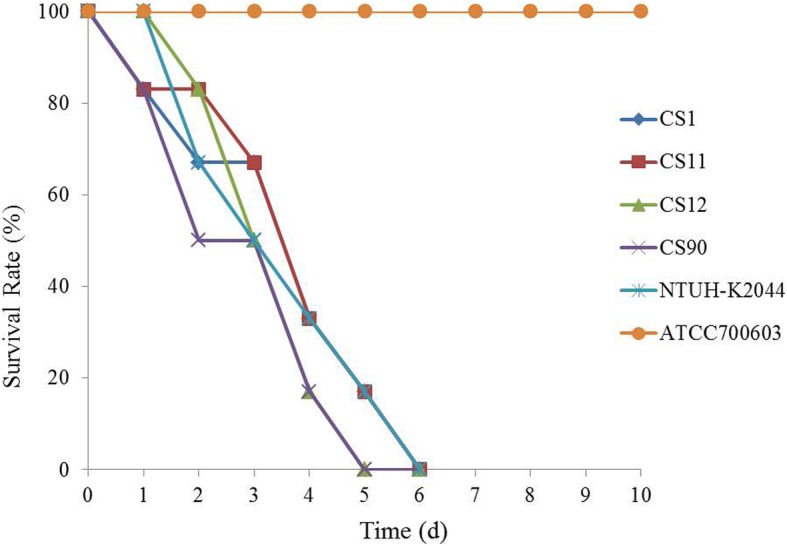
Mice lethality assay using four randomly selected CR-hvKP isolates (CS1, CS11, CS12 and CS90). NTUH-K2044: a hypervirulent wild strain of K. pneumoniae; ATCC 700603: a standard strain of K. pneumoniae with low virulence and low resistance
Capsular serotyping and virulence genes
Among the 20 CR-hvKP isolates, 17 were identified as the K2 serotype, and 3 (CS80, CS103, and CS118) were serotypes other than K1, K2, K5, K20, and K57 (Table 3). We also found that isolates were positive for each of the 10 virulence genes tested, but the proportion of isolates positive for each virulence gene varied, from 10 to 100% (Table 3). Some virulence genes had a low positive rate, such as allS (10%) and fimH (30%). It suggests that these genes might be less important for high virulence in these strains.
Table 3.
Serotypes and the presence of virulence genes in the CR-hvKP isolates
| Strain Number | Serotyping | Virulence Gene | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rmpA | iucB | iroB | uge | wabG | ureA | allS | ybtS | fimH | entB | ||
| CS1 | K2 | – | + | + | + | + | + | – | + | + | + |
| CS11 | K2 | + | + | + | + | + | + | – | + | – | + |
| CS12 | K2 | + | + | + | + | + | + | – | + | + | + |
| CS15 | K2 | + | + | + | + | + | + | – | + | + | + |
| CS17 | K2 | + | + | + | + | + | + | – | – | – | + |
| CS45 | K2 | + | + | + | + | + | + | – | – | – | + |
| CS47 | K2 | + | + | + | + | + | + | – | – | – | + |
| CS48 | K2 | + | + | + | + | + | + | – | + | – | + |
| CS57 | K2 | + | + | + | + | + | + | – | + | – | + |
| CS60 | K2 | + | + | – | + | + | + | + | – | – | + |
| CS61 | K2 | + | + | – | + | + | + | + | – | – | + |
| CS62 | K2 | + | – | + | + | + | + | – | – | – | + |
| CS70 | K2 | + | – | + | + | + | + | – | – | – | + |
| CS80 | NA | – | + | – | + | + | + | – | + | + | + |
| CS90 | K2 | + | + | – | + | + | + | – | – | – | + |
| CS103 | NA | – | + | – | + | + | + | – | + | + | + |
| CS118 | NA | – | + | – | + | + | + | – | – | – | + |
| CS127 | K2 | + | + | – | + | + | + | – | – | – | + |
| CS129 | K2 | + | + | + | + | + | + | – | – | + | + |
| CS59497 | K2 | + | + | + | + | + | + | – | + | – | – |
| Positive rate of genes (%) | 80 | 90 | 65 | 100 | 100 | 100 | 10 | 45 | 30 | 95 | |
Carbapenemase phenotype and carbapenemase gene detection
To further confirm the presence of carbapenemase, carbapenemase genes were tested and two families of carbapenemase genes were identified: blaKPC-2 (13/20) and blaNDM-1 (1/20; Table 1). All strains were found to be negative for the other carbapenemase genes tested, including blaVIM, blaIMP and blaOXA-48. As shown in Table 1, two isolates (CS80 and CS103) that were negative for mCIM were blaKPC-positive. In addition, other β-lactamase genes, including blaTEM-1, blaSHV-1 and blaCTX-M-3, were frequently detected in these CR-hvKP isolates. Notably, three stains carried blaCTX-M-65 (Table 1).
CR-hvKP fingerprinting
PFGE analysis revealed that the 20 CR-hvKP isolates showed 8 PFGE types (type A to type H), with type A as the dominating type, being expressed in 55.0% of the isolates (11/20). The majority of the type A isolates came from the intensive care unit (ICU; Fig. 3; Table 1). MLST detected three sequence types, including ST25, ST11 and ST375. ST25 was the main sequence type, accounting for 80.0% of the isolates. Among them, PFGE types A, B, C, E and H belong to ST25, type F and type G are ST11, while type D belongs to ST375 (Table 1). Interestingly, three isolates (CS11, CS12 and CS129) were all from samples that were collected from patients in the ICU that were collected during the same time period, and had the same carbapenemase phenotypes and PFGE profile, suggesting that they may be hospital-acquired infections.
Fig. 3.
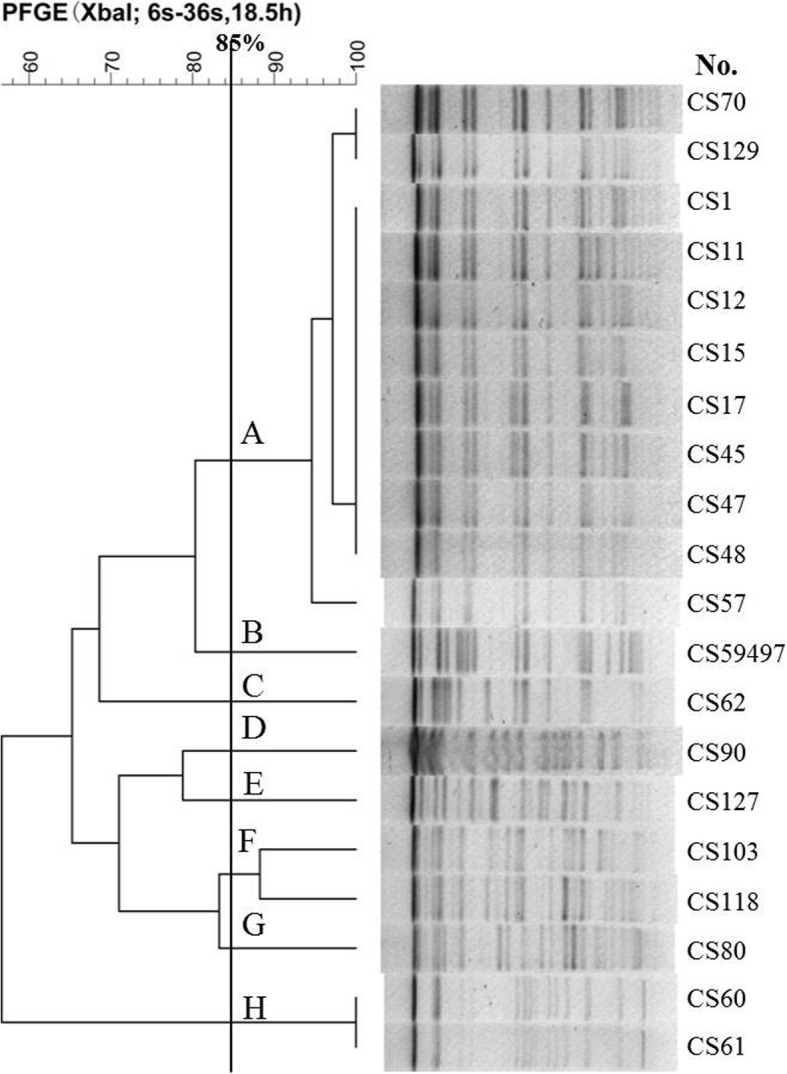
Relationships of the 20 CR-hvKP isolates based on Pulsed Field Gel Electrophoresis (PFGE). The 20 CR-hvKP isolates were analyzed by PFGE using XbaI. The interpretation of the PFGE patterns was performed with BioNumerics software using the Dice similarity coefficient. The tree indicates relative genetic similarity and was constructed on the basis of the unweighted pair group method of averages (UPGMA). A PFGE pattern with more than 85% DNA bands that are different from the others is taken to be a unique PFGE pattern
Discussion
Increasing rates of hvKP infection have been reported worldwide over the past few decades. However, until the last several years, only a few cases of CR-hvKP infections have been reported [2, 6–9]. In this study, 20 CR-hvKP isolates were identified from a single hospital in mid-south China. Several of the isolates were identified from the same patients. Additionally, several of these isolates were sampled from the ICU and share some common phenotypes, suggesting that they might be a hospital-acquired infection. Notably, hvKP is considered to be the predominant cause of pyogenic liver abscess [18, 23, 24]. However, we found that the isolates were mostly identified from sputum samples taken from patients that did not have symptoms of a pyogenic liver abscess, suggesting that CR-hvKP can also cause infection or colonize in other parts of the human body.
Serotypes K1 and K2 are the most common and virulent strains of hvKP [25, 26]. Similarly, in this study we found 17 isolates that belonged to serotype K2 and the remaining 3 were nontypable. Serotype K2 has been identified in many countries, including China, Germany, Spain and Singapore [10, 19, 24, 27, 28]. Serotype K2 generally belongs to a diverse sequence of types, such as ST25, ST65, ST66 and ST86 [18]. In our current study, we found that the serotype K2 CR-hvKP isolates all belonged to ST25, except for one CR-hvKP strain (ST375).
mCIM is a novel method that was recommended by the CLSI in 2017 for the detection of carbapenemase phenotypes and is considered to have high sensitivity and specificity [29, 30]. Our results show that 60% (12/20) of the CR-hvKP strains were mCIM positive. We then tested the strains for carbapenemase genes and 14 of the 20 CR-hvKP strains were positive. All of the 12 mCIM-positive strains were positive for the carbapenemase gene, indicating that the specificity of mCIM is as high as 100.0%. We also found that 2 mCIM-negative CR-hvKP strains were positive for the blaKPC-2 gene. We previously investigated 259 clinical K. pneumoniae strains [75 CR-KP and 184 carbapenem-susceptible K. pneumoniae (CS-KP)] by mCIM and PCR assay. Our results showed that 69 of the tested 75 CR-KP strains were mCIM positive, in accordance with the results of PCR assay, while all of the CS-KP isolates were mCIM negative. Among the six negative CR-KP isolates, three were positive for carbapenem genes (blaKPC/blaVIM) with a high expression of AmpC (Jun Li, Ziyan Huang, Xiaoyan Tao, etc., unpublished data). We speculate that the carbapenem gene may be partially deleted. For example, a truncated transposon Tn4401 variant Tn4401d is not able to express the blaKPC gene, causing a loss of carbapenemase activity, subsequently leading to the negative results of mCIM [31].
The epidemiology of KPC-producing K. pneumoniae has been mainly associated with the global expansion of cKP resistance to carbapenems. However, there are limited reports of CR-hvKP strains that carry the blaKPC gene [6, 7, 13, 14]. In this study, we found that the KPC-producing CR-hvKP strains were predominant, suggesting that KPC was the main mechanism of carbapenem resistance in hvKP strains. In addition, it has been reported that the blaKPC gene can be transferred to hvKP by plasmid conjugation [10], suggesting that drug resistance genes can be acquired by hvKP from the environment, producing a hypervirulent strain with high drug resistance. Previous studies showed that most of KPC-producing K. pneumoniae isolates belonged to ST11, which is a single-locus variant of the pandemic ST258 clone and is considered to be a high-risk epidemic clone owing to its ability to spread rapidly and disseminate carbapenemases. Several studies also found a few KPC-producing K. pneumoniae isolates belonging to ST11 and clone dissemination in hospitals. For example, one report showed that five patients died of severe pneumonia in a hospital due to infection of KPC-producing hvKP isolates belonging to ST11 [9]. In our study, 3 patients were infected with KPC-producing hvKP isolates belonging to ST11 in the hospital and one died. It was rarely reported that KPC-producing hvKP isolates belong to ST25 [27]. However, we found 10 KPC-producing hvKP isolates all belonging to ST25, suggesting that ST25 may play an important role in promoting the dissemination of blaKPC-carried hvKP isolates in our hospital. It is worth noting that one CR-hvKP isolate was positive for the blaNDM-1 gene, which was first identified in 2010 in a extensively drug-resistant (XDR) K. pneumoniae clinical isolate [32].
PFGE is a molecular typing technique that is broadly used in bacterial epidemiological studies. PFGE analysis of the 20 CR-hvKP strains identified eight genotypes from type A to type H. Type A was the main genotype, accounting for 55.0% of the strains. Interestingly, this genotype was mainly isolated from patients in the ICU, suggesting that the CR-hvKP strains that were isolated from this hospital could possibly have clone transmission.
Conclusions
In conclusion, our study revealed that a small percentage (20 out of 562) CR-KP are hypervirulent and that these CR-hvKP strains are highly resistant to most of the commonly used antibacterial drugs, which could bring great challenges to clinical anti-infection treatment. We believe that our findings may provide new insights into the treatment and prevention of CR-hvKP infections, especially in China. One limitation of our current study is the possible inconsistency of sample collection, due to the difficulty and limitation of working in a clinical setting. A more comprehensive and consistent sample collection (such as collection from the same sample source) could greatly improve the outcome.
Methods
Ethics statement and study subjects
This study was carried out in accordance with the recommendations of the Ethics Committee of Central South University (Changsha, Hunan Province, China) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The protocol was approved by the Ethics Committee of Central South University (Changsha, Hunan Province, China). All of the participants provided written consent prior to the study.
Collection, identification and antimicrobial susceptibility testing of K. pneumoniae clinical isolates
K. pneumoniae isolates, which were identified by Matrix Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS; Bruker Daltonics GmbH, Germany) and examined by VITEK-2 automated microbiology analyzer (bioMérieux, Marcy l’Etoile, France), were collected from patients at the Xiangya Hospital Central South University between September 1st 2014 and September 31st 2017. This hospital has 3500 beds and is located in Changsha, Hunan Province, China. To avoid duplicate samples, the majority of isolates were collected from different types of specimen from each patient. In some cases, isolates were collected from the same type of specimen. In these cases, the interval between the collection of the two samples was at least 1 week. Escherichia coli ATCC 25922 and K. pneumoniae ATCC 700603 (American Type Culture Collection, Manassas, VA) were used as controls for the species identification.
The minimal inhibitory concentrations (MICs) of imipenem for imipenem-resistant K. pneumoniae were verified by E-test method according to the guidelines recommended by the Clinical and Laboratory Standards Institute [29]. The MICs of tigecycline were performed by E-test method and interpretation of the results was based on break points of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org/clinical_breakpoints). E. coli ATCC 25922 was used as a control.
Identification of CR-hvKP isolates
To identify CR-hvKP isolates, the CR-KP isolates were screened by string test and the detection of the aerobactin gene, as has been described previously (Zhang et al., 2016b). The primers used for PCR were as follows: aerobactin forward, 5′-GCATAGGCGGATACGAACAT-3′; aerobactin reverse, 5′-CACAGGGCAATTGCTTACCT-3′. The reaction mixture was kept at 95 °C for 5 min, followed by 30 cycles of 95 °C for 1 min, 50 °C for 1 min, 72 °C for 1 min, and 72 °C for 10 min. The PCR products were visualized and analyzed by agarose gel electrophoresis and sequencing.
Neutrophil phagocytosis
Neutrophil phagocytosis was performed as described previously [6, 19]. Briefly, four CR-hvKP isolates were selected for analysis. The 4 CR-hvKP isolates include all three sequence types (2 ST25 and 1 ST11 were randomly selected and 1 ST375 was selected) (Table 1). K. pneumoniae ATCC 700603 (a standard strain for low virulence and low resistance) and K. pneumoniae NTUH-K2044 (a hypervirulent wild strain) were used as the controls. Neutrophil cells were isolated from the venous blood of healthy volunteers. A solution of 200 μL 1 × 106 neutrophil cells (or 200 μL sterilized saline in the control group), 200 μL 1 × 106 CR-hvKP suspension and 600 μL RPMI 1640 medium (Biosun, Shanghai, China) was mixed in a cell culture plate, and incubated on a rocking bed at 37 °C with constant shaking. Ten microliters of 0.1% saponin were added to each plate at 0, 15, 30 and 60 min incubation time. The plates were then placed on ice for 15 min and the CR-hvKP isolates were inoculated on Luria-Bertani solid medium overnight. The colony formation unit (CFU) was counted. The survival rate was determined using the following equation: (CFU in the presence of neutrophils)/(CFU with sterilized saline) × 100. The experiment was repeated three times.
Mouse lethality assay
The mouse lethality assay was performed as described previously [6, 19]. Briefly, four CR-hvKP isolates were randomly selected for the experiment and K. pneumoniae ATCC 700603 and K. pneumoniae NTUH-K2044 were used as the controls. CR-hvKP suspensions of 1 × 106 cfu/mL were injected into the abdominal cavity of BALB/C mice (that were obtained from the Animal Center of the Central South University). The mortality rates were observed over 10 days. Three mice were used in each CR-hvKP isolate condition and the experiment was repeated three times. After the experiments, mice were all euthanized using 100% CO2 at a flow rate of ~ 2 l per minute.
Modified carbapenem inactivation method (mCIM)
To identify the carbapenemase phenotypes, mCIM was performed with meropenem for all of the CR-hvKP isolates, according to the operation standard of CLSI [29]. The zone diameter around the meropenem was measured. The result was considered to be positive when the diameter was < 16 mm, while the result was considered to be negative when the diameter was > 19 mm. The result was considered to be intermediary when the inhibition zone diameter was between 16 and 18 mm. K. pneumoniae ATCC BAA-1705 and K. pneumoniae ATCC BAA-1706 were used as the positive and negative controls, respectively [29].
Capsular serotyping, detection of resistance genes and virulence genes
PCR was used to detect the serotype genes (including K1, K2, K5, K20, and K57), virulence genes (rmpA, iucB, ironB, uge, wabG, ureA, alls, ybtS, fimH, and entB), carbapenemase genes (blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA-48), and other β-lactamase genes (blaCTX − M, blaTEM, blaSHV, blaDHA, and blaCMY). PCR primers and conditions have been described elsewhere [33, 34]. All the positive products were sequenced and analyzed using the BLAST website (https://blast.ncbi.nlm.nih.gov/blast.cgi).
Molecular epidemiology
Genetic relatedness among the CR-hvKP isolates was determined by pulsed-field gel electrophoresis (PFGE). In brief, genomic DNA was prepared by embedding K. pneumoniae cells in agarose plugs (ThermoFisher, China), followed by XbaI (Promega, USA) digestion for 18.5 h at 37 °C. Electrophoresis was performed at 14 °C for 20 h using the Bio-Rad CHEF III system (120° angle, 6 V/cm, switch times of 6 and 36 s). Cluster analysis was performed with BioNumerics software Version 5.1 (Applied Maths, Austin, TX) using the Dice Similarity Coefficient. Isolates with pattern similarities > 85% were considered to be from the same PFGE cluster [35]. A subset of isolates that represented the different PFGE clusters were further studied by multilocus sequence typing (MLST), according to the Institut Pasteur scheme [36].
Statistical analysis
Statistical analysis was conducted using SPSS19.0 software (SPSS Inc., USA). For the mouse modality test, survival curves were assessed using Kaplan-Meier analysis and the log-rank test. A value of P < 0.05 was considered to be statistically significant.
Acknowledgements
We are thankful to the staff in the Laboratory of the Xiangya Hospital of Central South University for their help in the collection and identification of the clinical CR-KP isolates. We would also like to thank Haijian Zhou, a teacher at the Center for Disease Control (CDC) in China for her help with the PFGE experiments.
Abbreviations
- BLAST
Basic Local Alignment Search Tool
- CFU
Colony Formation Unit
- cKP
classic K. pneumoniae
- CLSI
Clinical and LaboratoryStandards Institute
- CR-hvKP
Carbapenem-resistant hypervirulent Klebsiella pneumoniae
- CR-KP
Carbapenem-resistant Klebsiella pneumoniae
- EUCAST
European Committee on Antimicrobial Susceptibility Testing
- hvKP
hypervirulent Klebsiella pneumoniae
- K. pneumoniae
Klebsiella pneumoniae
- KPC
Klebsiella pneumoniae carbapenemase
- mCIM
modified Carbapenem Inactivation Method
- MICs
Minimal Inhibitory Concentrations
- MLST
Multilocus sequence type
- PCR
Polymerase Chain Reaction
- PFGE
Pulsed-field gel electrophoresis
- SPSS
Statistical Package for the Social Sciences
- ST
Sequence Type
- XDR
Extensively Drug-resistant
Authors’ contributions
Study design: JL and ZYH. Study conduct: ZYH and XYT. Data collection: TY. Data analysis: MXZ. Data interpretation: HCW and YMH. Drafting manuscript: ZYH and XYT. Revising manuscript content: JL and MXZ. Approving final version of manuscript: MXZ and JL. JL take responsibility for the integrity of the data analysis. All authors read and approved the final manuscript.
Funding
This study was supported by the Hunan Provincial Natural Science Foundation (Grant 2017JJ3478) and the National Natural Science Foundation of China (81702068). The authors declare that they have no financial relationship with the organization that sponsored the research, and the funding body was not involved in study design, data collection, analysis and writing of the study.
Availability of data and materials
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Central South University (Changsha, Hunan Province, China). All procedures performed in studies involving human participants were in accordance with the ethics standards of the institutional and national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethics standards. Written informed consent was obtained from all individual participants included in this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jun Li, Email: lijun198412@126.com.
Zi-Yan Huang, Email: 312991657@qq.com.
Ting Yu, Email: tingyu96@126.com.
Xiao-Yan Tao, Email: 1124443724@qq.com.
Yong-Mei Hu, Email: 626973094@qq.com.
Hai-Chen Wang, Email: 337228840@qq.com.
Ming-Xiang Zou, Phone: +8613907496278, Email: zoumingxiang@csu.edu.cn.
References
- 1.Liu YC, Cheng DL, Lin CL. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch Intern Med. 1986;146:1913–1916. doi: 10.1001/archinte.1986.00360220057011. [DOI] [PubMed] [Google Scholar]
- 2.Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, et al. Antimicrobial resistance of Hypervirulent Klebsiella pneumoniae: epidemiology, Hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;7:483. doi: 10.3389/fcimb.2017.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, Zhao C, Wang Q, Wang X, Chen H, Li H, et al. High prevalence of Hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. 2016;60:6115–6120. doi: 10.1128/AAC.01127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Surgers L, Boyd A, Girard PM, Arlet G, Decre D. ESBL-producing strain of Hypervirulent Klebsiella pneumoniae K2, France. Emerg Infect Dis. 2016;22:1687–1688. doi: 10.3201/eid2209.160681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie Y, Tian L, Li G, Qu H, Sun J, Liang W, et al. Emergence of the third-generation cephalosporin-resistant hypervirulent Klebsiella pneumoniae due to the acquisition of a self-transferable blaDHA-1-carrying plasmid by an ST23 strain. Virulence. 2018;9:838–844. doi: 10.1080/21505594.2018.1456229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Zeng J, Liu W, Zhao F, Hu Z, Zhao C, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Inf Secur. 2015;71:553–560. doi: 10.1016/j.jinf.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. Emergence of Carbapenem-resistant serotype K1 Hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother. 2016;60:709–711. doi: 10.1128/AAC.02173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Y, Lu Y, Yao Z, Zong Z. Carbapenem-resistant Hypervirulent Klebsiella pneumoniae of sequence type 36. Antimicrob Agents Chemother. 2018;62:1–6. [DOI] [PMC free article] [PubMed]
- 9.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 10.Siu LK, Huang DB, Chiang T. Plasmid transferability of KPC into a virulent K2 serotype Klebsiella pneumoniae. BMC Infect Dis. 2014;14:176. doi: 10.1186/1471-2334-14-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JY, Ko KS. OprD mutations and inactivation, expression of efflux pumps and AmpC, and metallo-beta-lactamases in carbapenem-resistant Pseudomonas aeruginosa isolates from South Korea. Int J Antimicrob Agents. 2012;40:168–172. doi: 10.1016/j.ijantimicag.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei DD, Wan LG, Deng Q, Liu Y. Emergence of KPC-producing Klebsiella pneumoniae hypervirulent clone of capsular serotype K1 that belongs to sequence type 11 in mainland China. Diagn Microbiol Infect Dis. 2016;85:192–194. doi: 10.1016/j.diagmicrobio.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Wei DD, Wan LG, Liu Y. Draft genome sequence of an NDM-1- and KPC-2-coproducing Hypervirulent Carbapenem-resistant Klebsiella pneumoniae strain isolated from burn wound infections. Genome Announc. 2018;6:1–2. [DOI] [PMC free article] [PubMed]
- 15.Chiu SK, Wu TL, Chuang YC, Lin JC, Fung CP, Lu PL, et al. National surveillance study on carbapenem non-susceptible Klebsiella pneumoniae in Taiwan: the emergence and rapid dissemination of KPC-2 carbapenemase. PLoS One. 2013;8:e69428. doi: 10.1371/journal.pone.0069428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adler A, Hussein O, Ben-David D, Masarwa S, Navon-Venezia S, Schwaber MJ, et al. Persistence of Klebsiella pneumoniae ST258 as the predominant clone of carbapenemase-producing Enterobacteriaceae in post-acute-care hospitals in Israel, 2008–13. J Antimicrob Chemother. 2015;70:89–92. doi: 10.1093/jac/dku333. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Yu J, Chen F, Yu J, Simner P, Tamma P, et al. Emergence and establishment of KPC-2-producing ST11 Klebsiella pneumoniae in a general hospital in Shanghai, China. Eur J Clin Microbiol Infect Dis. 2018;37:293–299. doi: 10.1007/s10096-017-3131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siu LK, Fung CP, Chang FY, Lee N, Yeh KM, Koh TH, et al. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J Clin Microbiol. 2011;49:3761–3765. doi: 10.1128/JCM.00977-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin JC, Koh TH, Lee N, Fung CP, Chang FY, Tsai YK, et al. Genotypes and virulence in serotype K2 Klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut Pathog. 2014;6:21. doi: 10.1186/1757-4749-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu TT, Zhou JC, Jiang Y, Shi KR, Li B, Shen P, et al. Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in East China. BMC Infect Dis. 2015;15:161. doi: 10.1186/s12879-015-0899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Y, Wang S, Zhan L, Jin Y, Duan J, Hao Z, et al. Microbiological and clinical characteristics of Hypermucoviscous Klebsiella pneumoniae isolates associated with invasive infections in China. Front Cell Infect Microbiol. 2017;7:24. doi: 10.3389/fcimb.2017.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu YM, Li BB, Zhang YY, Zhang W, Shen H, Li H, et al. Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland China. Antimicrob Agents Chemother. 2014;58:5379–5385. doi: 10.1128/AAC.02523-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye M, Tu J, Jiang J, Bi Y, You W, Zhang Y, et al. Clinical and genomic analysis of liver abscess-causing Klebsiella pneumoniae identifies new liver abscess-associated virulence genes. Front Cell Infect Microbiol. 2016;6:165. doi: 10.3389/fcimb.2016.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossi B, Gasperini ML, Leflon-Guibout V, Gioanni A, de Lastours V, Rossi G, et al. Hypervirulent Klebsiella pneumoniae in cryptogenic liver abscesses, Paris, France. Emerg Infect Dis. 2018;24:221–229. doi: 10.3201/eid2402.170957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fung CP, Chang FY, Lin JC, Ho DM, Chen CT, Chen JH, et al. Immune response and pathophysiological features of Klebsiella pneumoniae liver abscesses in an animal model. Lab Investig. 2011;91:1029–1039. doi: 10.1038/labinvest.2011.52. [DOI] [PubMed] [Google Scholar]
- 26.Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis. 2015;37:107–112. doi: 10.1016/j.ijid.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 27.Cubero M, Grau I, Tubau F, Pallares R, Dominguez MA, Linares J, et al. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007-2013) Clin Microbiol Infect. 2016;22:154–160. doi: 10.1016/j.cmi.2015.09.025. [DOI] [PubMed] [Google Scholar]
- 28.Pichler C, Buchsel M, Rossen JW, Vavra M, Reuter S, Kern WV, et al. First report of invasive liver abscess syndrome with endophthalmitis caused by a K2 serotype ST2398 hypervirulent Klebsiella pneumoniae in Germany, 2016. New Microbes New Infect. 2017;17:77–80. doi: 10.1016/j.nmni.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clinical and Laboratory Standards Institute (CLSI) The performance stands for antimicrobial susceptibility testing. 2017. [Google Scholar]
- 30.Pierce VM, Simner PJ, Lonsway DR, Roe-Carpenter DE, Johnson JK, Brasso WB, et al. Modified Carbapenem inactivation method for phenotypic detection of Carbapenemase production among Enterobacteriaceae. J Clin Microbiol. 2017;55:2321–2333. doi: 10.1128/JCM.00193-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L, Chavda KD, Mediavilla JR, Jacobs MR, Levi MH, Bonomo RA, et al. Partial excision of blaKPC from Tn4401 in carbapenem-resistant Klebsiella pneumoniae. Antimicrob Agents Chemother. 2012;56:1635–1638. doi: 10.1128/AAC.06182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C, Xu X, Pu S, Huang S, Sun J, Yang S, et al. Characterization of carbapenemases, extended spectrum beta-lactamases, quinolone resistance and aminoglycoside resistance determinants in carbapenem-non-susceptible Escherichia coli from a teaching hospital in Chongqing, Southwest China. Infect Genet Evol. 2014;27:271–276. doi: 10.1016/j.meegid.2014.07.031. [DOI] [PubMed] [Google Scholar]
- 34.Yan Q, Zhou M, Zou M, Liu WE. Hypervirulent Klebsiella pneumoniae induced ventilator-associated pneumonia in mechanically ventilated patients in China. Eur J Clin Microbiol Infect Dis. 2016;35:387–396. doi: 10.1007/s10096-015-2551-2. [DOI] [PubMed] [Google Scholar]
- 35.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.


