Abstract
Introduction
The Shewanella putrefaciens group are ubiquitous microorganisms recently isolated from different freshwater fish species and causing serious health disorders. The purpose of the study was to characterise isolates of the S. putrefaciens group with special emphasis on elucidating serological diversity and determining putative virulence factors.
Material and Methods
Isolates collected from freshwater fish (n = 44) and reference strains were used. The identification of bacteria was carried out using biochemical kits and 16S rRNA sequencing. Polyclonal antibodies were prepared against the S. putrefaciens group. The bacterium’s susceptibility to antimicrobial agents, its enzymatic properties, and its adhesion ability to fish cell lines were also tested. Finally, selected isolates were used in challenge experiments in common carp and rainbow trout.
Results
Excluding six isolates undeterminable for species, the bacteria were classified to three species: S. putrefaciens, S. xiamenensis, and S. oneidensis, and showed some phenotypic diversity. Fourteen serological variants of the S. putrefaciens group were determined with the newly developed serotyping scheme.
Conclusion
Serodiversity may play an important role in the virulence of particular isolates. Further, S. putrefaciens group members adhere to epithelial cells and produce enzymes which may contribute to their virulence. Challenge tests confirmed the pathogenicity of the S. putrefaciens group for fish.
Keywords: bacterial fish diseases, serotyping, virulence, adhesion
Introduction
Shewanella (S.) putrefaciens, a gammaproteobacterium belonging to the Alteromonadaceae family, is an important agent of food spoilage, mainly spoilage of fish stored at low temperatures (3, 12). This heterogeneous group of microorganisms, known to be halophilic, currently occurs in various environments including marine and freshwater ecosystems (4, 10). S. putrefaciens is considered to be the causative agent of shewanellosis, a disease causing health disorders in different freshwater fish species. Its clinical symptoms include lethargy and darkening and ulceration of the skin. Internally, haemorrhages in the kidneys and spleen are noticed (8, 10, 13, 15, 19, 22, 23, 24).
Bacteria belonging to the S. putrefaciens group were previously characterised according to their phenotypic properties (10, 15, 17, 22); however, the serological diversity of these microorganisms is still unknown. The serodiversity of bacteria plays an important role in the pathogenicity of many of them, for example Aeromonas spp. and Yersinia ruckeri (9, 20, 27). In case of S. putrefaciens, some serological differences have been observed; however, they concerned only a few strains isolated from food (11). Because of that, it is difficult to correlate the serodiversity results with the potential pathogenicity of S. putrefaciens for fish.
The mechanism by which shewanellosis infection develops is still not entirely clear. Many virulence factors might contribute to the pathogenicity of the S. putrefaciens group, and among them, the contribution of enzymes seems to be very important. In the case of S. putrefaciens, high enzymatic activity such as the ability to produce proteases, lipases, and exoenzymes such as haemolysins was confirmed (8, 17, 22, 28). However, the relationship between the pathogenicity of the S. putrefaciens group and production of enzymes has not been definitively explained. Interactions between bacteria and host cells also play an important role in fish diseases. The adhesion ability to epithelial cells of a few isolates of S. putrefaciens collected from humans and the environment has previously been studied; however, no correlation between pathogenicity and adhesion ability was found (17).
The aim of the present studies was to characterise collected isolates of the S. putrefaciens group with special emphasis on identification of antigenic properties with the newly developed serotyping scheme and determination of putative virulence factors.
Material and Methods
Bacterial isolates. A total of 44 isolates of S. putrefaciens group were collected from 2007 to 2016. Bacteria were isolated from freshwater fish according to methods described previously (22). Most of the isolates (n = 29) were obtained from cultured fish such as carp (Cyprinus carpio L.), rainbow trout (Oncorhynchus mykiss, Walbaum, 1792), Siberian sturgeon (Acipenser baerii), and European eel (Anguilla anguilla L.) (Table 1). Nine isolates were collected from wild fish: grass carp (Ctenopharyngodon idella), brown trout (Salmo trutta m. trutta), silver carp (Hypophthalmichthys molitrix), European whitefish (Coregonus lavaretus), ide (Leuciscus idus), and common roach (Rutilus rutilus). S. putrefaciens group specimens (n = 6) were also isolated from ornamental fish imported to Poland from Israel and Cameroon: zebrafish (Brachydanio rerio), slender krib (Pelvicachromis taeniatus), and koi carp (Cyprinus carpio L.). Isolates were collected both from apparently healthy fish as well as from individuals exhibiting clinical disorders. Characteristic disease symptoms observed in fish were ulceration and necrotic skin lesions as well as necrosis of the gills. Bacteria were also collected from the kidneys if swollen, the spleen if abnormally enlarged, and from internal organs generally if they showed disorders like haemorrhaging.
Table 1.
S. putrefaciens group isolates collected from freshwater fish
| Molecular identification | Symbol of isolate | APINE profile | Fish species | Symptoms of diseases/mortality |
|---|---|---|---|---|
| S. xiamenensis | K736 | 1451354 | Common carp | Yes/Yes |
| S. xiamenensis | K733 | Common carp | No/Yes | |
| S. xiamenensis | KK3 | Koi carp | Yes/Yes | |
| S. xiamenensis | KOI2 | Koi carp | Yes/Yes | |
| S. xiamenensis | W24 | European eel | Yes/Yes | |
| S. xiamenensis | W28 | European eel | Yes/Yes | |
| Shewanella sp. | Jes15 | Siberian sturgeon | No/No | |
| S. xiamenensis | St16 | Slender krib | Yes/Yes | |
| S. xiamenensis | St8 | Slender krib | Yes/Yes | |
| Shewanella sp. | Am6 | Grass carp | No/No | |
| S. putrefaciens | K677 | 1410344 | Common carp | No/No |
| S. putrefaciens | K678 | Common carp | No/No | |
| Shewanella sp. | K769 | Common carp | Yes/No | |
| S. oneidensis | Pt400 | Rainbow trout | nd/Yes | |
| S. putrefaciens | Pt411 | Rainbow trout | Yes/Yes | |
| S. putrefaciens | K313 | 1450754 | Common carp | Yes/Yes |
| S. putrefaciens | K530 | Common carp | Yes/Yes | |
| S. putrefaciens | K604 | Common carp | Yes/Yes | |
| S. putrefaciens | K679 | Common carp | No/No | |
| S. putrefaciens | K680 | Common carp | No/No | |
| Shewanella sp. | K575 | 1450744 | Common carp | Yes/Yes |
| S. putrefaciens | GIII41(3) | Brown trout | Yes/Yes | |
| S. putrefaciens | St4 | Rainbow trout | Yes/Yes | |
| S. putrefaciens | Tp5 | 1410354 | Silver carp | Yes/Yes |
| S. oneidensis | Plo1 | Common roach | No/No | |
| S. oneidensis | Jes34 | Siberian sturgeon | No/No | |
| S. putrefaciens | Tsk20 | 1410754 | Brown trout | Yes/Yes |
| S. putrefaciens | St5 | European whitefish | Yes/Yes | |
| S. putrefaciens | E6 | Rainbow trout | No/No | |
| S. putrefaciens | Sh1 | 1411754 | Common carp | Yes/Yes |
| S. putrefaciens | Sh2 | Common carp | Yes/Yes | |
| Shewanella sp. | Jes1 | 1451754 | Siberian sturgeon | No/No |
| S. putrefaciens | Sh4 | Common carp | Yes/Yes | |
| S. putrefaciens | Jaz4 | Ide | No/No | |
| S. putrefaciens | TIII | 1411744 | Brown trout | Yes/Yes |
| S. putrefaciens | K869 | Common carp | Yes/Yes | |
| S. oneidensis | K696 | 1450344 | Common carp | Yes/Yes |
| S. putrefaciens | E9 | 1010344 | Rainbow trout | No/No |
| S. putrefaciens | E3 | 1011754 | Rainbow trout | No/No |
| S. xiamenensis | 16/c/29 | 1051344 | Zebrafish | Yes/Yes |
| S. putrefaciens | W9 | 1400354 | European eel | Yes/Yes |
| Shewanella sp. | St15 | 1411344 | Slender krib | Yes/Yes |
| S. putrefaciens | K751 | 1450354 | Common carp | No/No |
| S. putrefaciens | GIII46(3) | 1451344 | Brown trout | Yes/Yes |
nd – no data
Reference strains. The following reference strains of Shewanella spp. were used: S. putrefaciens ATCC BAA-1097, ATCC 8071 and ATCC 8073, S. xiamenensis JCM16212T, and S. oneidensis ATCC700550. In addition, four reference strains of other fish pathogens were used for serodiversity assays: Aeromonas hydrophila ATCC 7966, A. salmonicida subsp. salmonicida ATCC 14174, Pseudomonas fluorescens ATCC 53794, and Yersinia ruckeri NCTC 12266.
Preliminary identification of the isolates. Isolates of the S. putrefaciens group (n = 44) and reference strains were cultured on blood agar (BA) and identified with API 20E and API 20NE kits (BioMérieux, France) according to the manufacturer’s instructions with the exception of the incubation temperature, which was 27 ± 2°C. The growth ability of the bacteria in the presence of NaCl was tested for 48 h at 27 ± 2°C on tryptic soy agar (TSA) supplemented with 0.0%, 0.5%, 2.0%, 3.0%, 4.0%, 5.0%, or 6.0% of NaCl.
Molecular identification of the isolates. DNA extraction and 16S rRNA gene sequence analysis were performed as described previously (21). Sequences were deposited in the GenBank database under accession numbers KC607503, KC607506–KC607522, KC607524, KC607525, KC607527–KC607530, KY630559, KY817247, KY817248–KY817252, KY817255, KY817257, KY817258, MF958276, MF958964, MF959701, MF962592, MF962594– MF962596, MF962598, MG930082, and MG930083 and compared with other sequences from the database. The sequence similarity examination was performed using MegaBLAST (highly similar sequences). The criteria for identification to each species were estimated based on a similarity level of ≥ 99.5% (14).
Shewanella serodiversity assay. Preparation of antisera: New Zealand white rabbits (Oryctolagus cuniculus) were immunised with selected isolates (n = 20) of the S. putrefaciens group and three reference strains (ATCC BAA-1097, ATCC 8071, and ATCC 8073) in order to obtain specific antisera (designated in the following as the strain or isolate number preceded by “S-”), as described previously (26), with our modifications. Two rabbits were immunised with sterile phosphate buffered saline (PBS) as a control group. The selection of isolates used for rabbit immunisation was based on their properties (Table 1). A total of 20 field and 3 reference strains of S. putrefaciens were cultured on tryptic soy broth (TSB) for 24 h at 27 ± 2°C and washed twice in PBS with 0.4% formalin. Inactivated bacteria were estimated to the concentration of 5 × 109 cells/mL and used for rabbit immunisation. Each rabbit was immunised by intramuscular injection of an inactivated bacterial suspension of a given strain at intervals of six days, with increasing antigen doses starting from 0.5 mL. After the fourth immunisation a blood sample was taken in order to check antibody titres. If the antibody level was satisfactory (≥ 320), the rabbits were bled from the cervical vein. The blood was incubated for 2 h at room temperature and next at 4 ± 2°C for 24 h. After incubation, the serum was separated by centrifugation (15 min, 1,000 g, 4°C) and stored for future analysis at −80°C.
Preparation of antigens: A total of 44 isolates of the S. putrefaciens group and reference strains of S. putrefaciens were used to prepare antigens (designated in the following as the strain or isolate number preceded by “A-”). Bacteria were cultured on TSB for 24 h at 27 ± 2°C. The preparation of the antigens was carried out in two ways. In the first method, after incubation the cells were centrifuged (15 min, 1,000 g, 4°C), and then washed in sterile PBS with 0.4% formalin. In the second approach, after washing in PBS, antigens were heated at 100°C for 30 and 60 min. Pellets were dissolved in PBS with 0.4% formalin to the final concentration of 2 × 109 cells/mL. All obtained antigens were stored at 4 ± 2°C until analysis.
Agglutination assay: The test was performed using 96-well plates as described by Davies (7). All plates were incubated at 37 ± 2°C for 24 h. In order to determine the reference antibody level, antisera were tested with homologous antigens. Then all antigens were examined with serum in concentrations decreasing from 1:20 to 1:2,560. Serum from control rabbits was used as a negative control.
Cross-reactivity: Cross-reactivity between all the obtained anti-Shewanella sera and antigens of reference strains such as ATCC 7966, ATCC 14174, ATCC 53794, and NCTC 12266 was also established experimentally. Antigen production and agglutination assays were carried out as described above.
Susceptibility to antimicrobial agents. The antimicrobial susceptibility of the S. putrefaciens group (n = 44) was assayed by the disc diffusion method according to the recommendations of the Clinical and Laboratory Standards Institute, USA (5). The inoculum of bacteria with 0.5 McFarland turbidity was prepared in sterile PBS and cultured on Mueller–Hinton agar for 24 h at 28 ± 2°C. After the incubation, the inhibition zones were measured. Nine compounds were used representing the antimicrobials frequently used in aquaculture belonging to the folate-path inhibitor, quinolone, tetracycline, and phenicol classes (Table 4). Since there are no interpretation criteria for Shewanella, for better understanding of the susceptibility results some criteria for Aeromonas salmonicida were used (6).
Table 4.
Distribution of inhibition zone diameter values of different antimicrobial agents for the Shewanella isolates (n = 44)
| Inhibition zone diameter (mm) | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antimicrobial (concentration) | Bacteria species | 7 | 11 | 14 | 30 | |||||||||||||||||
| 6 | - | - | - | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | - | >35 | Criteria* | %NWT | ||
| Shewanella | 10 | 13 | 16 | 35 | ||||||||||||||||||
| sulphonamides (300μg) |
putrefaciens
(n = 25) |
24 | 1 | n.a. | n.d. | |||||||||||||||||
|
xiamenensis
(n = 9) |
9 | n.d. | ||||||||||||||||||||
|
oneidensis
(n = 4) |
4 | n.d. | ||||||||||||||||||||
| sp.** (n = 6) |
6 | n.d. | ||||||||||||||||||||
| sulphamethoxazole/ trimethoprim (23.75/1.25μg) |
putrefaciens
(n = 25) |
1 | 2 | 3 | 1 | 5 | 3 | 2 | 4 | 4 | NWT≤19 mm | 4 | ||||||||||
|
xiamenensis
(n = 9) |
3 | 1 | 2 | 1 | 1 | 1 | 33 | |||||||||||||||
|
oneidensis
(n = 4) |
1 | 1 | 1 | 1 | 0 | |||||||||||||||||
| sp.** (n = 6) |
1 | 1 | 1 | 1 | 2 | 0 | ||||||||||||||||
| oxolic acid (2μg) |
putrefaciens
(n = 25) |
2 | 3 | 5 | 1 | 1 | 7 | 3 | 1 | 2 | NWT ≤29 mm | 100 | ||||||||||
|
xiamenensis
(n = 9) |
6 | 1 | 1 | 1 | 100 | |||||||||||||||||
|
oneidensis
(n = 4) |
1 | 1 | 1 | 1 | 75 | |||||||||||||||||
| sp.** (n = 6) |
1 | 1 | 4 | 33 | ||||||||||||||||||
| flumequine (30μg) |
putrefaciens
(n = 25) |
1 | 1 | 1 | 5 | 16 | 1 | n.a. | n.d. | |||||||||||||
|
xiamenensis
(n = 9) |
5 | 1 | 1 | 1 | 1 | n.d. | ||||||||||||||||
|
oneidensis
(n = 4) |
1 | 3 | n.d. | |||||||||||||||||||
| sp.** (n = 6) |
1 | 4 | 1 | n.d. | ||||||||||||||||||
| enrofloxacin (5μg) |
putrefaciens
(n = 25) |
1 | 1 | 2 | 1 | 1 | 1 | 2 | 3 | 12 | 1 | n.a. | n.d. | |||||||||
|
xiamenensis
(n = 9) |
3 | 2 | 1 | 3 | n.d. | |||||||||||||||||
|
oneidensis
(n = 4) |
1 | 3 | n.d. | |||||||||||||||||||
| sp.** (n = 6) |
1 | 5 | n.d. | |||||||||||||||||||
| norfloxacine (10μg) |
putrefaciens
(n = 25) |
2 | 3 | 3 | 1 | 3 | 2 | 1 | 3 | 7 | n.a. | n.d. | ||||||||||
|
xiamenensis
(n = 9) |
4 | 1 | 1 | 1 | 2 | n.d. | ||||||||||||||||
|
oneidensis
(n = 4) |
1 | 1 | 1 | 1 | n.d. | |||||||||||||||||
| sp.** (n = 6) |
1 | 1 | 4 | n.d. | ||||||||||||||||||
| oxytetracycline (30μg) |
putrefaciens
(n = 25) |
1 | 1 | 4 | 1 | 7 | 4 | 4 | 1 | 2 | NWT ≤27 mm | 100 | ||||||||||
|
xiamenensis
(n = 9) |
6 | 2 | 1 | 100 | ||||||||||||||||||
|
oneidensis
(n = 4) |
2 | 1 | 1 | 100 | ||||||||||||||||||
| sp.** (n = 6) |
1 | 1 | 2 | 1 | 1 | 100 | ||||||||||||||||
| doxycycline (30μg) |
putrefaciens
(n = 25) |
2 | 2 | 6 | 3 | 2 | 2 | 1 | 1 | 5 | 1 | n.a. | n.d. | |||||||||
|
xiamenensis
(n = 9) |
2 | 1 | 5 | 1 | n.d. | |||||||||||||||||
|
oneidensis
(n = 4) |
1 | 1 | 1 | 1 | n.d. | |||||||||||||||||
| sp.** (n = 6) |
1 | 1 | 1 | 1 | 1 | 1 | n.d. | |||||||||||||||
| florphenicol (30μg) |
putrefaciens
(n = 25) |
5 | 16 | 4 | NWT ≤26 mm | 0 | ||||||||||||||||
|
xiamenensis
(n = 9) |
8 | 1 | 0 | |||||||||||||||||||
|
oneidensis
(n = 4) |
4 | 0 | ||||||||||||||||||||
| sp.** (n = 6) |
2 | 1 | 3 | 0 | ||||||||||||||||||
*NWT criteria for Aeromonas salmonicida according to CLSI, ** isolates not classified to species level, NWT – non-wild-type,
%NWT – the number (%) of isolates potentially resistant to antimicrobial agents, n.a. – not available, n.d. – no data
Enzymatic properties. Enzyme secretion of the S. putrefaciens group (n = 44) and reference strains was detected using the API ZYM system (BioMérieux) as well as classical methods. For exoenzyme detection bacteria were cultured on agar supplemented with milk (caseinase activity), starch (amylolytic activity), or Tween 80 (lipolytic activity) as described previously (16, 18). Culture incubations were performed at 27 ± 2°C for 24 h. Haemolytic activity was tested on BA (agar supplemented with 5% horse blood) at 6 ± 2°C and 27 ± 2°C after 24 h incubation. Haemolysin production was investigated by lysis of erythrocytes on blood agar: β-haemolysis (complete lysis) or α-haemolysis (partial lysis).
Adhesion ability. The adhesion ability of the S. putrefaciens group isolates (n = 21) to cell cultures was tested according to Wang et al. (29) with a few modifications. Four cell lines were used: epithelioma papulosum cyprini (EPC), bluegill fry (BF-2), chinook salmon embryo (CHSE), and rainbow trout gonad (RTG-2). The density of each cell line was estimated in a Burker’s chamber, giving 6.3 ± 0.6 × 107/1 mL for BF-2, 6.6 ± 0.7 × 107/1 mL for EPC, 3.4 ± 0.1 × 107/1 mL for CHSE, and 5.5 ± 0.1 × 107/1 mL for RTG-2. All cells grew in 24-well plates at 24 ± 2°C.
S. putrefaciens group isolates were cultured on TSB (24 ± 2°C, 18 h) and washed three times in PBS without Ca++ and Mg++ by centrifugation (2,000 g, 15 min, 4°C). Each isolate was individually inoculated to each cell line culture (in duplicate), to a final concentration of 106 bacterial cells per well. In order to attach the bacteria with the EPC monolayer, the plate was centrifuged at low speed (800 g, 5 min, 4°C) and incubated at 24 ± 2°C for 2 h. After that, each well was washed three times in PBS to remove non-adherent bacteria. Lysis of EPC cells was carried out with 1% Triton X-100 for 10 min. The lysate was centrifuged twice (2,000 g, 15 min, 4°C), washed each time, and finally the obtained pellet was suspended in 1 mL of PBS. The number of adherent cells was determined according to the Koch method using serial dilutions (10−1 to 10−5) in PBS. A 100 μL volume of each dilution was inoculated into TSA in duplicate. The number of colony-forming units (CFU) was determined after 48 h incubation at 27 ± 2°C and the data were analysed using a one-way ANOVA test. Values of p < 0.05 were considered statistically significant.
Fish pathogenicity experiments. Common carp and rainbow trout weighing 100 g were acclimatised to laboratory conditions for 14 days before experimental infection. Animals were maintained in 300 L tanks with aerated fresh water at 12 ± 2°C. During adaptation, the fish were observed for behaviour and feed intake. A classical ichthyopathological examination series including parasitological, bacteriological, and anatomopathological examinations were also performed before experiments. Each fish species was divided into eight separate experimental groups and one control group containing 20 and 35 fish, respectively.
Eight isolates of the S. putrefaciens group were cultured on TSB at 27 ± 2°C. After 24 h incubation, the bacteria were washed three times in PBS and adjusted spectrophotometrically to a final concentration of 108 cells/mL. The fish were anaesthetised using MS-222 (75 and 150 μg/L for rainbow trout and common carp, respectively) and injected intraperitoneally with a single isolate of the S. putrefaciens group at a dose of 5 × 107 CFU per animal, while the control group received only PBS. Common carp and rainbow trout were challenged with the isolates Tp5, St5, K575, KK3, K696 K604, Jaz4, and Sh1. Clinical signs and mortality were recorded through a 28-day observation period. All dead and moribund fish were immediately examined and affected tissues and internal organs were taken for bacteriological tests. Samples were diluted 1:1 (v/v) in PBS and after homogenisation were cultured on TSA for 48 h. Pure colonies were re-isolated and identified with an API 20NE kit (BioMérieux). All bacteriological culture and biochemical tests were performed at 27 ± 2°C.
Results
Identification and characterisation of S. putrefaciens group isolates. All isolates produced cytochrome oxidase and gelatine hydrolase, reduced NO3 to NO2, and assimilated N-acetyl-glucosamine and maltose. Negative reactions were observed for ortho-nitrophenyl-β-galactoside (ONPG), acid production from glucose, mannitol, inositol, sorbitol, rhamnose, melibiose, and amygdalin fermentation, urease hydrolysis, tryptophan deaminase, indole production, and Voges–Proskauer response. Bacteria were also incapable of assimilating mannose, adipic acid, or phenylacetic acid.
Capacities for ornithine decarboxylation, citrate utilisation, aesculin hydrolysis, and assimilation of glucose, malic acid or capric acid were dependent on bacterial isolates (Table 2). Therefore, different biochemical profiles were determined. In the API20E system (BioMérieux), six biochemical profiles were observed in tested isolates. The bacteria showed the following profile numbers: 0502004 (34% of isolates), 0702004 (23% of isolates), 0402004 (23% of isolates), 0402006 (10% of isolates), and 0502006 (5% of isolates), and isolates representing these codes were classified to the S. putrefaciens group at excellent or very good match level (99.9–99.3%). An acceptable match for identification (89%) was observed in 5% of isolates (profile 0400004). In the API20NE system, the bacteria showed 17 different numeric codes (Table 1). More than half of the isolates were classified to the S. putrefaciens group with excellent, very good or good identification level (profiles 1451354, 1410344, 1410354, 1411754, 1010344, 1011754, 1051344, 1400354, 1411344, 1451344, and 1450354). About 30% of the tested bacteria showed doubtful or acceptable levels of identification (profiles 1410754, 1411744, 1450344, 1450754, and 1451754). The remaining isolates (7%) were identified as Aeromonas salmonicida subsp. salmonicida (profile 1450744) at good match level. All collected isolates were able to grow in the presence of 0.0–2.0% sodium chloride and almost all in 3.0%, whereas none of the isolates showed this ability in the presence of 6.0% NaCl (Table 2).
Table 2.
Biochemical properties and growth condition of S. putrefaciens group isolates (n = 44) and reference strains
| S. putrefaciens isolates | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Biochemical properties |
S. xiamenensis
(n = 9) |
S. oneidensis
(n = 4) |
S. putrefaciens
(n = 25) |
Shewanella sp. (n = 6) |
ATCC BAA-1097 | ATCC 8071 | ATCC 8073 | ATCC 700550 | JCM 16212T |
| Ornithine decarboxylase | 6 | 1 | 14 | 5 | - | - | − | + | + |
| Citrate utilisation | 2 | 0 | 4 | 2 | +w | +w | − | + | + |
| Acid from arabinose | 9 | 0 | 3 | 1 | + | + | + | − | + |
| Aesculin hydrolysis | 9 | 4 | 23 | 6 | +w | +w | + | + | + |
| Assimilation: | |||||||||
| Glucose | 8 | 2 | 12 | 0 | – | − | + | − | + |
| Arabinose | 9 | 0 | 8 | 1 | − | + | + | − | + |
| Potassium gluconate | 0 | 0 | 18 | 1 | − | − | − | − | − |
| Capric acid | 7 | 1 | 16 | 0 | + | + | − | − | + |
| Malic acid | 9 | 4 | 25 | 5 | + | + | − | − | + |
| Growth condition NaCl range (%) | |||||||||
| 0.0–2.0 | 9 | 4 | 25 | 6 | + | + | + | + | + |
| 3.0 | 9 | 3 | 25 | 5 | + | + | + | + | + |
| 4.0 | 5 | 0 | 25 | 1 | + | + | +w | + | + |
| 5.0 | 3 | 0 | 24 | 1 | +w | − | +w | +w | +w |
| 6.0 | 0 | 0 | 0 | 0 | − | − | − | − | − |
w – weak reaction; ATCC BAA-1097, ATCC 8071, ATCC 8073 0150 – S. putrefaciens; ATCC 700550 – S. oneidensis; JCM 16212T – S. xiamenensis
Analysis of the 16S rRNA gene classified the isolates into three species: S. putrefaciens (n = 25), S. xiamenensis (n = 9), and S. oneidensis (n = 4) (Table 1). The highest similarity among our sequences was observed to the three GenBank reference strain sequences of S. putrefaciens (acc. no. X82133), S. xiamenensis (acc. no. NR116732), and S. oneidensis (acc. no. AF005251). Therefore, these sequences were used in further analysis. Six isolates: K769, St15, Jes15, Am6, Jes1, and K575 could not be classified by their Shewanella species, because the similarity level between these bacteria and reference strains was <99.5%.
Shewanella serological variants. The antigens inactivated by heating did not react with any of the antisera, therefore differentiation of the S. putrefaciens group was achieved using antigens prepared with 0.4% formalin without heating. Titre values are shown according to homologous agglutinations between serum and antigen obtained with the same S. putrefaciens group isolate (Table 3). For identification of potential serological variants, the reactions were arranged according to the following criteria: 1) high (≥640) homologous titre, 2) medium (160–320) homologous titre, and 3) no titre (<160) in heterogeneous assays. Fourteen homologous sera-antigen combinations were adopted as the “type assay” for 14 presumptive serological variants arbitrarily numbered from I to XIV. Within this set of variants only two heterogeneous agglutinations were observed (titre 160). The remaining homologous titres (n = 9) were rejected due to multiple heterologous reactions.
Table 3.
Matrix showing homologous titres for serological variant definition (upper part) and Shewanella distribution in defined variants (lower part)
| Antisera* | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Titre | S-K604 | S-K575 | S-St5 | S-St16 | S-K530 | S-ATCC BAA 1097 | S-K696 | S-KK3 | S-Am6 | S-TIII | S-ATCC 8071 | S-Sh1 | S-K678 | S-St8 | |
| antigens | A-K604 | 2,560 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A-K575 | 40 | 2,560 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| A-St5 | 0 | 0 | 2,560 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| A-St16 | 0 | 0 | 0 | 2,560 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| A-K530 | 0 | 0 | 0 | 0 | 2,560 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| A- ATCC BAA 1097 | 0 | 0 | 0 | 0 | 0 | 2,560 | 0 | 0 | 0 | 0 | 0 | 160 | 0 | 0 | |
| A-K696 | 0 | 0 | 0 | 0 | 0 | 0 | 1,280 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| A-KK3 | 0 | 0 | 0 | 0 | 0 | 160 | 0 | 1,280 | 0 | 0 | 0 | 0 | 0 | 0 | |
| A-Am6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1,280 | 0 | 0 | 0 | 0 | 0 | |
| A-TIII | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1,280 | 0 | 0 | 0 | 0 | |
| A-ATCC 8071 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1,280 | 0 | 0 | 0 | |
| A-Sh1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 640 | 0 | 0 | |
| A-K678 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 320 | 0 | |
| A-St8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 160 | |
| Variant | I | II | III | IV | V | VI | VII | VIII | IX | X | XI | XII | XIII | XIV | |
| isolates | ID | K604 ATCC 8073 Tsk20 |
K575 E9, Tp5 K313 K677 |
St5 W24 W28 |
St16 | K530 | ATCC BAA 1097 |
K696 Jes1 K733 K736 |
KK3 St15 Pt400 Plo1 |
Am6 Jes15 |
TIII E6 E3 Jaz4 |
ATCC 8071 |
Sh1 KOI2 Sh4 |
K678 | St8 |
| n** | 3 | 5 | 3 | 1 | 1 | 1 | 4 | 4 | 2 | 4 | 1 | 3 | 1 | 1 | |
| % | 6% | 11% | 6% | 2% | 2% | 2% | 9% | 9% | 4% | 9% | 2% | 6% | 2% | 2% |
* only antisera used for serological variant identification are listed; sera showing multiple heterologous reaction were omitted (n = 9; S-GEIII46(3), S-Pt411, S-St4, S-St15, S-K679, S-K751, S-K869, S-ATCC 8073, S-KOI2); ** 13 (28%) isolates remained unrecognised due to reactions of their antigens with multiple sera: autoagglutination/rough antigen structure (n = 8; GEIII46(3), Pt411, St4, K869, K769, 16/c/29, GEIII41(3), W9) or no reaction with relevant sera (n = 5; K679, K751, Jes34, Sh2, K680)
Following serological variant definition, all assays were reassessed to classify S. putrefaciens group isolates according to their serological features. Titre ≥80 was considered positive. If more than one antiserum gave a positive reaction, the highest titre was used for serological variant assignment. The reference strain ATCC8073 isolated from food was classified to serological variant I with two isolates collected from fish, whereas ATCC BAA-1097 isolated from sediment and ATCC 8071 collected from food products were classified to separate serological variants (VI and XI, respectively) (Table 3). Over 70% of tested isolates were classified to one of the serological variants. The largest serological variant (II) contained 11% of the isolates, whereas variants VII, VIII, and X laid claim to 9% of the isolates each. Variants IV, V, XIII, and XIV were represented by single isolates. No cross reactivity between antigens of the other reference strains ATCC 7966, ATCC 14174, ATCC 53794, and NCTC 12266 and S. putrefaciens group antisera was observed.
Antimicrobial susceptibility. The range of disk diffusion diameters of the S. putrefaciens group is presented in Table 4. A no-inhibition zone (6 mm), indicating possible resistance, was noted in sulphonamides. Phenicols caused inhibition of ≥30 mm for most of isolates. Sulphamethoxazole/trimethoprim, quinolones, and tetracyclines produced a wide range of inhibition zones depending on the isolate. All isolates (100% NWT) were resistant to oxytetracycline and susceptible to florphenicol, whereas various sensitivities to sulphamethoxazole/trimethoprim and oxalic acid were observed (Table 4).
Enzymatic properties. High enzymatic activity was observed in all isolates of the S. putrefaciens group. They produced alkaline phosphatase and N-acetyl β-glucosaminidase, but not lipase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, α-mannosidase, or α-fucosidase. Some differentiation was observed between isolates for secretion of esterase, esterase lipase, leucin arylamidase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, caseinase, amylase and Tween 80 hydrolase. Two types of haemolysis (α and β) were observed in Shewanella isolates. Haemolysis of α type was observed in almost all isolates cultured at 27°C, with the exception of three collected from Siberian sturgeon (Jes1, Jes34, Jes15). Bacteria incubation at 6°C showed both types of haemolysis: β in 25% of isolates and α in 75%. Nonetheless, β-haemolysis was observed only in isolates of S. putrefaciens.
Adhesion ability. Evaluation of adhesion ability was carried out for 21 representatives of each species: S. putrefaciens (n = 11), S. xiamenensis (n = 4), S. oneidensis (n = 2) and Shewanella sp. (n = 4) (Table 1). Most isolates adhered to tested cell lines; however, greater adherence was observed to EPC cell monolayers. The adhesion ability of isolates of the S. putrefaciens group to EPC cell monolayers ranged from 1.1 × 102 to 2.5 × 103 bacterial cells (CFU) (Fig. 1), but no statistical difference between the adhesion ability to the EPC line of different bacteria species was observed.
Fig. 1.
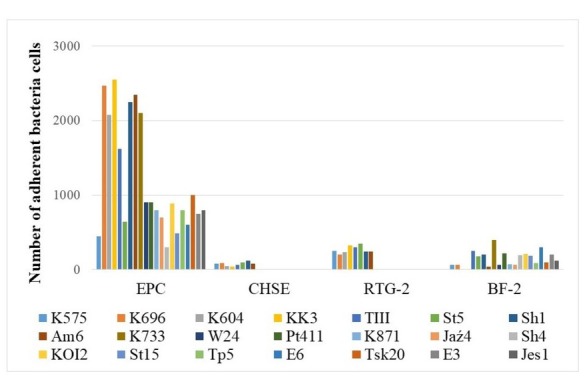
Adhesion ability of S. putrefaciens group to different fish cell lines
The average number of adherent bacteria (CFU) from two repetitions is shown. The standard errors of means were less than 10% of mean values in each case. Cell lines used: EPC – epithelioma papulosum cyprini, BF-2 – bluegill fry, CHSE – chinook salmon embryo, RTG-2 – rainbow trout gonad
Experimental infection. Fish challenge tests were carried out for eight selected isolates. Representative examples of each species were used: Tp5, K604, Sh1, St5, and Jaz4 for S. putrefaciens, KK3 for S. xiamenensis, K696 for S. oneidensis, and K575 for Shewanella sp. (Table 1). These bacteria were collected from fish with clinical signs of diseases (seven isolates) as well as from apparently healthy fish (one isolate).
During fish acclimatisation no health disorders were observed. Bacteriological and parasitological examinations were negative.
Common carp challenge: regardless of the isolate, infected common carp showed skin ulceration. The first lesions in carp infected with Tp5, Jaz4, St5 (S. putrefaciens), and K575 (Shewanella sp.) isolates were observed seven days post injection. The fish showed ulcers on the skin (Fig. 2) and mortality reached 40% of the challenge group. The first disorders in fish infected with KK3 (S. xiamenensis), K696 (S. oneidensis), K604 and Sh1 (S. putrefaciens) bacteria were observed on 10 day after injection. Small ulcers were observed in about 40% of infected fish, the mortality among which was 11%. Congestion of internal organs was revealed in most infected fish (Fig. 3). S. putrefaciens group specimens were reisolated in monoculture from skin samples while from the kidneys the bacteria were isolated along with Aeromonas sobria.
Fig. 2.
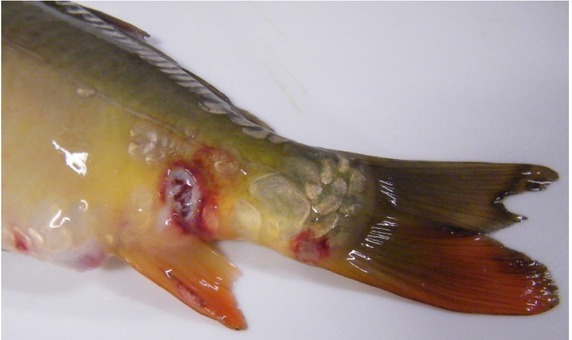
Ulcers on the skin in common carp
Fig. 3.
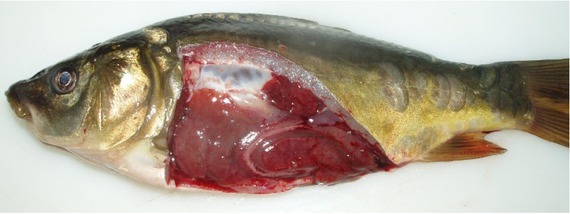
Congestion of internal organs in common carp
Rainbow trout challenge: the first clinical signs occurred between 7 and 16 days after injection with Tp5, K604, Sh1 (S. putrefaciens), and K575 (Shewanella sp.) isolates. Mortality rates of 10%, 35%, 50%, and 100% were recorded in rainbow trout infected with the Sh1, K575, K604, and Tp5 isolates, respectively. All moribund fish showed darkness of skin, exophthalmia (mono or bilateral), and lesions on the gill covers (Fig. 4). Small ulcers on the skin and around the fins were observed. The anatomopathological examination showed a pale liver and spleen enlargement. S. putrefaciens group specimens were re-isolated in monoculture from eyes, kidneys, and gills. No mortality or disease symptoms were observed in fish infected with St5, Jaz4 (S. putrefaciens), K696 (S. oneidensis), or KK3 (S. xiamenensis) isolates.
Fig. 4.
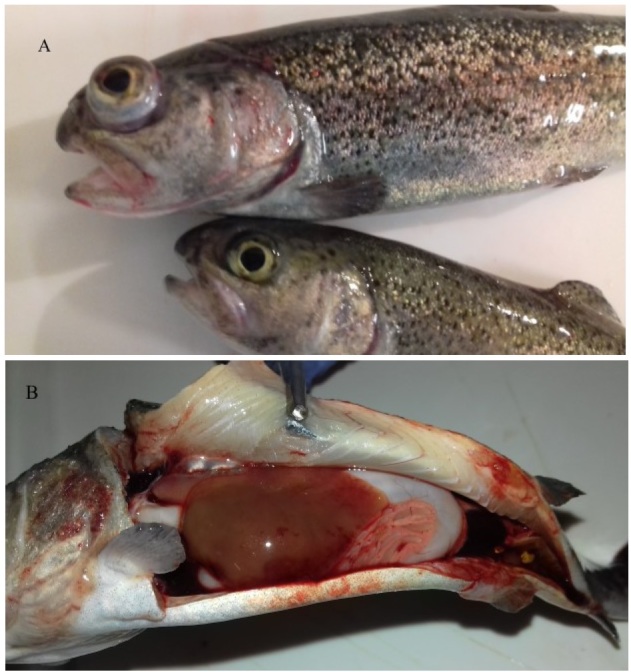
Exophthalmia, lesions on gill cover (A) and pale liver (B) in rainbow trout
Discussion
Our study contributes to the identification and characterisation of Shewanella group isolates and pathogenicity of the bacterium for fish. Among the isolates collected from freshwater fish, three species were identified: the predominant S. putrefaciens, and S. xiamenensis and S. oneidensis. These species as well as S. seohaensis, S. baltica, and S. algae were previously isolated from fish with health disorders (8, 10, 13, 15, 19, 22, 23, 24, 25). Interestingly, some of our isolates were not classified to these Shewanella species, which indicates the isolation of other, perhaps yet undescribed species within the genus Shewanella.
Despite the genotype diversity, based on biochemical properties the isolates of the S. putrefaciens group formed a homogeneous group, which has been observed previously (15, 22). However, some properties such as production of acid from arabinose and assimilation of arabinose and potassium gluconate were not possessed by every bacterial species (Table 2), and also for this phenomenon similar results had been obtained previously (15). Assimilation and acidification of arabinose were typical for S. xiamenensis but not for S. oneidensis, neither for field nor reference strains. Therefore, these properties may be used for differentiation of these two Shewanella species. Assimilation of potassium gluconate was observed only in isolates of S. putrefaciens collected from fish, which indicates diversity between our isolates and reference strains obtained from food (ATCC 8071 and ATCC8073) or sediment (ATCC BAA-1097). In concurrence with the report from Hu et al. (13), the tested S. putrefaciens and S. xiamenensis isolates grew at higher concentrations of NaCl than those of S. oneidensis. None of the isolates collected from freshwater fish had the ability to grow in the presence of 6% sodium chloride, which justifies consideration of S. putrefaciens as a halophytic microorganism and confirms adaptation of the bacteria to environmental condition.
The tested isolates of the S. putrefaciens group showed quite some diversity in susceptibility to antimicrobial agents, but further studies to develop interpretation criteria and a definition of possible resistance mechanisms are still necessary. The antimicrobial susceptibility of these bacteria for clinical purposes should also be investigated at 8°C because clinical signs in fish are usually observed at this temperature.
Fonnesbech et al. (11) used polyclonal and monoclonal antibodies to show low serological diversity of S. putrefaciens collected from food. To our knowledge, serotyping of fish isolates is novel and has not yet been reported in the literature. Although we used polyclonal antibodies and a limited number of isolates, up to 14 serological variants of the S. putrefaciens group (I–XIV) were defined. The serotyping scheme developed here embraced not only fish isolates, but also reference strains of food origin. Interestingly, the clustering of ATCC 8073 strain (from food) into the same serological variant as two isolates from fish exhibiting health disorders suggests common serological properties. The other two reference strains were scattered rather far from any fish isolates. Contrary to the case with Shewanella of food origin, our collection of freshwater fish isolates appears to be a serologically diverse group of bacteria. Although 14 serological variants were identified, several isolates remained unclassified. This could result from unspecific heterogeneous reactions due to the roughness of particular isolates, but might also suggest the presence of still unrecognised serological variants of the S. putrefaciens group.
The enzymatic activity of the S. putrefaciens group isolated from freshwater fish was found to be related to clinical signs of shewanellosis. Our study showed that haemolytic activity depended on bacterial species and temperature of incubation. The finding of α-haemolytic activity of almost all tested Shewanella species at higher (27°C) temperature and the shift to β-haemolysis exclusively in S. putrefaciens at low (6°C) temperature is of great value in the context of clinical signs observed in fish. This finding correlates well with the fact that S. putrefaciens infections are usually observed in spring when water temperature is approximately 8°C. Therefore, haemolytic activity seems to contribute to the pathogenicity of S. putrefaciens at lower temperatures. In addition, the clinical signs of shewanellosis such as ulceration of the skin may be correlated with proteolytic and lipolytic activity.
The pathogenicity of bacteria might be also linked to their adhesion to host cells. In one of the few reports on the topic, Bagge et al. (1) related adhesion to metal reduction by Shewanella, which use iron as a terminal electron acceptor for anaerobic respiration and therefore adhere to iron-containing compounds. Another study showed a limited degree of S. putrefaciens adhesion to human epithelial cells (17). These authors described adherence in the relative degree, therefore it was not possible to compare their results accurately with ours. The higher affiliation of the S. putrefaciens group to epithelial cells compared to other tested cell lines might indicate that these bacteria have affinity for epithelial tissue, which was observed in the clinical signs of the disease (ulceration). The fish’s body is covered by a mucous layer which is the first barrier protecting the fish from pathogens (2) and this invests the possible tissue affinity of S. putrefaciens with significance. In future research, adhesion of the S. putrefaciens group to mucus should be also considered as a virulence factor.
Most of the above findings of clinical relevance were produced by clinical experiments. The pathogenicity of S. putrefaciens and S. xiamenensis for fish had been confirmed previously (10, 19, 22) and in our study Koch’s postulates for disease development were also fulfilled.
The results of the challenge tests indicate that the clinical signs and mortality rate in infected fish depend on either fish species or bacterial species and the specific isolate. Noteworthily, symptoms observed in experimental infection of common carp and rainbow trout are also seen in naturally infected fish (8, 15, 22). The only exception seems to be exophthalmia, which was typical in the experimental model, but observed only once in a confirmed case of Shewanella in rainbow trout.
It is also noteworthy that the pathogenicity of the particular isolates seems to be correlated with their serological variant. Bacteria classified to serological variants I and II were usually collected from fish with health disorders and challenge tests confirmed their pathogenicity for fish. Furthermore, isolates belonging to these serological variants showed adhesion ability to epithelial cells and produced enzymes important for pathogenicity such as haemolysin, caseinase, and leucine arylamidase. Meanwhile, isolates classified to serological variants III, VII, VIII, and X did not cause any disorders in rainbow trout after experimental infection, in contrast to their nocuous effects on common carp. Future studies should clarify the role of particular serological variants in the pathogenicity of Shewanella spp. for different fish species.
In conclusion, the current study highlights the diversity of the S. putrefaciens group responsible for freshwater fish health disorders. This group of bacteria has been classified to several species and some of them still require definite identification. The large serodiversity determined within the studied group might be relevant for the virulence of particular isolates. However unclear disease course development may still be, the adhesion to the epithelial cells seems to be the first important stage initiating the infection. Then the process leads to skin ulcerations and tissue penetration, probably due to proteolytic, lipolytic, and haemolytic activity of the bacterium. Since the S. putrefaciens group is part of the normal microbiota of the freshwater environment and cannot be eliminated, further studies are needed to elucidate the complex interactions between the bacterium, animal host, and environmental conditions.
Acknowledgements
The authors are grateful to A. Kozińska, PhD, ScD. Special thanks are expressed to A. Sandomierska for preparation of cell lines and to members of the technical team of the laboratories.
Footnotes
Conflict of Interest
Conflict of Interests Statement: The authors declare that there is no conflict of interests regarding the publication of this article.
Financial Disclosure Statement: This work was supported by the National Science Centre, Poland as a project titled “Studies on genotypic characterisation of Shewanella putrefaciens group isolates from freshwater fish in Poland” (grant no. 2015/19/N/NZ7/01687).
Animal Rights Statement: The experiments on animals were conducted in accordance with the local Ethical Committee laws and regulations as regards care and use of experimental animals. Local Ethics Commission No. II in Lublin (resolutions nos 49/2014 and 47/2017) approved all procedures on animals.
References
- 1.Bagge D., Hjelm M., Johansen C., Huber I., Gram L.. Shewanella putrefaciens adhesion and biofilm formation on food processing surfaces. Appl Environ Microbiol. 2001;67:2319–2325. doi: 10.1128/AEM.67.5.2319-2325.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benhamed S., Guardiola F.A., Mars M., Esteban M.A.. Pathogen bacteria adhesion to skin mucus of fishes. Vet Microbiol. 2014;171:1–12. doi: 10.1016/j.vetmic.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Borch E., Kant-Muermans M.L., Blixt Y.. Bacterial spoilage of meat and cured meat products. Int J Food Microbiol. 1996;33:103–120. doi: 10.1016/0168-1605(96)01135-x. [DOI] [PubMed] [Google Scholar]
- 4.Buller N.B. Bacteria and fungi from fish and other aquatic animals: a practical identification manual. CABI Publishing; Wallingford: 2014. pp. 168–176. [Google Scholar]
- 5.Clinical and Laboratory Standards Institute (CLSI): VET03-A Methods for antimicrobial disk susceptibility testing of bacteria isolated from aquatic animals;Approved Guideline. 23. Vol. 26. CLSI; Wayne: 2006. [Google Scholar]
- 6.Clinical and Laboratory Standards Institute (CLSI): VET03/VET04-S2 Performance standards for antimicrobial susceptibility testing of bacteria isolated from aquatic animals;Second Informational Supplement. 15. Vol. 34. CLSI; Wayne: 2014. [Google Scholar]
- 7.Davies R.L.. O-serotyping of Yersinia ruckeri with special emphasis on European isolates. Vet Microbiol. 1990;22:299–307. doi: 10.1016/0378-1135(90)90016-o. [DOI] [PubMed] [Google Scholar]
- 8.El-Barbary M.I.. First recording of Shewanella putrefaciens in cultured Oreochromis niloticus and its identification by 16S rRNA in Egypt. Egypt J Aquat Res. 2017;43:101–107. doi: 10.1016/j.ejar.2017.01.002. [DOI] [Google Scholar]
- 9.Esteve C., Amaro C., Toranzo A.E.. O-serogrouping and surface components of Aeromonas hydrophila and Aeromonas jandaei pathogenic for eels. FEMS Microbiol Lett. 1994;117:85–90. doi: 10.1111/j.1574-6968.1994.tb06746.x. [DOI] [PubMed] [Google Scholar]
- 10.Esteve C., Merchán R., Alcaide E.. An outbreak of Shewanella putrefaciens group in wild eels Anguilla anguilla L. favoured by hypoxic aquatic environments. J Fish Dis. 2017;40:929–939. doi: 10.1111/jfd.12574.. [DOI] [PubMed] [Google Scholar]
- 11.Fonnesbech B., Frøkiaer H., Gram L., Jespersen C.M.. Production and specificity of poly- and monoclonal antibodies raised against Shewanella putrefaciens. J Appl Bacteriol. 1993;74:444–451. doi: 10.1111/j.1365-2672.1993.tb05152.x. [DOI] [PubMed] [Google Scholar]
- 12.Gram L., Huss H.H.. Microbiological spoilage of fish and fish products. Int J Food Microbiol. 1996;33:121–137. doi: 10.1016/0168-1605(96)01134-8. [DOI] [PubMed] [Google Scholar]
- 13.Hu X., Li X., Lü A., Sun J., Shi H., Chen C., Li L., Xianghui K.. Characterization of Shewanella sp. isolated from cultured loach Misgurnus anguillicaudatus. IJA. 2016;68:1–9. [Google Scholar]
- 14.Janda J.M., Abbott S.L.. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 2007;45:2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung-Schroers V., Jung A., Ryll M., Bauer J., Teitge F., Steinhagen D.. Methods for identification and differentiation of different Shewanella spp. isolates for diagnostic use. J Fish Dis. 2018;41:689–714. doi: 10.1111/jfd.12772.. [DOI] [PubMed] [Google Scholar]
- 16.Kędzia W. Microbiological diagnostics in medicine. Państwowy Zakład Wydawnictw Lekarskich. Toruń: 1990. p. 196. [Google Scholar]
- 17.Khashe S., Janda J.M.. Biochemical and pathogenic properties of Shewanella alga and Shewanella putrefaciens. J Clin Microbiol. 1998;36:783–787. doi: 10.1128/jcm.36.3.783-787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozińska A.. Dominant pathogenic species of mesophilic aeromonads isolated from diseased and healthy fish cultured in Poland. J Fish Dis. 2007;30:293–301. doi: 10.1111/j.1365-2761.2007.00813.x.. [DOI] [PubMed] [Google Scholar]
- 19.Kozińska A., Pękala A.. First isolation of Shewanella putrefaciens from freshwater fish – a potential new pathogen of the fish. Bull Eur Assoc Fish Pathol. 2004;24:199–203. [Google Scholar]
- 20.Kozińska A., Pękala A.. Serotyping of Aeromonas species isolated from polish fish farms in relation to species and virulence phenotype of the bacteria. Bull Vet Inst Pulawy. 2010;54:315–320. [Google Scholar]
- 21.Paździor E., Pękala-Safińska A., Wasyl D.. Genotypic diversity among Shewanella spp. collected from freshwater fish. J Fish Dis. 2019;42:677–684. doi: 10.1111/jfd.12971.. [DOI] [PubMed] [Google Scholar]
- 22.Pękala A., Kozińska A., Paździor E., Głowacka H.. Phenotypical and genotypical characterization of Shewanella putrefaciens strains isolated from diseased freshwater fish. J Fish Dis. 2015;38:283–293. doi: 10.1111/jfd.12231.. [DOI] [PubMed] [Google Scholar]
- 23.Qin L., Zhu M., Xu L.. First report of Shewanella sp. and Listonella sp. infection in freshwater cultured loach, Misgurnus anguillicaudatus. Aquacult Res. 2012;45:602–608. doi: 10.1111/j.1365-2109.2012.03260.x.. [DOI] [Google Scholar]
- 24.Rusev V., Rusenova N., Simeonov R., Stratev D.. Staphylococcus warneri and Shewanella putrefaciens co-infection in Siberian sturgeon Acipenser baerii and Hybrid sturgeon Huso huso x Acipenser baerii. J Microbiol Exp. 2016;3:1–4. doi: 10.15406/jmen.2016.03.00078. [DOI] [Google Scholar]
- 25.Saeed M.O., Alamoudi M.M., Al-Harbi A.H.. A Pseudomonas associated with disease in cultured rabbitfish Siganus rivulatus in the Red Sea. Dis Aquat Organ. 1987;3:177–180. [Google Scholar]
- 26.Sakazaki R., Shimada T.. O-serogrouping scheme for mesophilic Aeromonas strains. Jpn J Med Sci Biol. 1984;37:247–255. doi: 10.7883/yoken1952.37.247. [DOI] [PubMed] [Google Scholar]
- 27.Tobback E., Decostere A., Hermans K., Haesebrouck F., Chiers K.. Yersinia ruckeri infections in salmonid fish. J Fish Dis. 2007;30:257–268. doi: 10.1111/j.1365-2761.2007.00816.x.. [DOI] [PubMed] [Google Scholar]
- 28.Vogel B.F., Jorgensen K., Christensen H., Olsen J.E., Gram L.. Differentiation of Shewanella putrefaciens and Shewanella alga on the basis of whole-cell protein profiles, ribotyping, phenotypic characterization, and 16S rRNA gene sequence analysis. Appl Environ Microbiol. 1997;63:2189–2199. doi: 10.1128/aem.63.6.2189-2199.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X.H., Oon H.L., Ho G.W., Wong W.S., Lim T.M., Leung K.Y.. Internalization and cytotoxicity are important virulence mechanisms in Vibrio-fish epithelial cell interactions. Microbiology. 1998;144:2987–3002. doi: 10.1099/00221287-144-11-2987. [DOI] [PubMed] [Google Scholar]


