Abstract
We recently isolated Candida auris from a blood culture and cutaneous swabs of a patient in her mid-70s. Our routine phenotypic methods failed to identify the microorganism, but it was identified by molecular tests and MALDI-TOF MS analysis. Our report, the first from Italy, further underlines the geographically wide distribution of C. auris and the need to confirm species identification of any suspicious colony as soon as possible to stop its spread.
Keywords: Candida, antifungal resistance, fungal infection, epidemic clone, MDR, epidemiology
Candida auris is a rapidly spreading, multidrug-resistant, healthcare-associated pathogen [1].
Since the first description of this novel Candida species, isolated from a patient in a Japanese hospital in 2009, the emergence of C. auris has been documented on all continents [2]. Invasive infections have been associated with high rates of treatment failure and mortality ranging from 30% to 72% [3]. C. auris is thus considered to pose a serious global public health threat.
C. auris can colonise patients for a long time, as well as persist on surfaces in healthcare environments [4]. Furthermore, C. auris can be misidentified in the diagnostic laboratory when traditional phenotypic methods are used. These features of C. auris contribute to its spread in healthcare facilities.
Here, we report a case of C. auris being isolated from a female in her mid-70s suffering from vascular disease.
Case report
On 16 June 2019 (day 1), the patient who had a history of hypertension and dyslipidaemia, was admitted to hospital for endovascular repair of an abdominal aortic aneurysm and left renal artery stenting. Because of surgical complications she was transferred to the intensive care unit (ICU). The postoperative course was further complicated and required left subclavian artery stenting on day 14. The patient first developed a fever on day 17, the same day the patient was transferred to vascular surgery department from the ICU. A computed tomography performed on day 26 for worsening condition and respiratory symptoms showed bilateral ground glass opacities. The patient was then retransferred to the ICU on day 26. Blood cultures were repeatedly collected during intermittent fever episodes that were unresponsive to antimicrobial treatment with meropenem and linezolid. Serum (1,3)-beta-D-glucan tests were performed repeatedly during fever episodes and were constantly negative. Ultimately, C. auris grew from blood cultures collected on day 31. The patient was immediately transferred to a single room in the infectious disease unit. C. auris also grew from axillary and ear swabs collected on day 41 and day 47 respectively. Treatment with caspofungin was started and all subsequent blood cultures and swabs were negative. On day 39, the patient’s improved clinical condition led to being discharged from the ICU and being admitted to the infectious diseases ward for completion of the antifungal treatment, i.e. until 14 days after the first negative blood culture. At the time of this communication, the patient is still hospitalised in the infectious diseases ward and in stable condition.
Identification of Candida auris and phylogenetic analysis
The pathogen was not identified by VITEK2 Advanced Expert System software version 8.01 (bioMérieux, Macy-l'Étoile, France), reporting a low discrimination between C. guillermondi (50%) and Cryptococcus laurentii (50%). It was also not identified by MALDI-TOF mass spectrometry (MALDI-TOF MS) analysis using VITEK MS software version 3 (bioMérieux). The microorganism was identified as C. auris by a species-specific PCR for GPI protein-encoding genes [5]. MALDI-TOF MS analysis using BD MALDI Biotyper System (Bruker Daltonics, Bremen, Germany) identified the yeast as C. auris (score 2.7). This result was further confirmed by D1/D2 region and internal transcribed spacer (ITS) sequencing [6,7]. The BLAST tool at the National Center for Biotechnology Information (NCBI) database was used to perform sequences similarity searches. Our sequences (GenBank accession numbers MN275234 and MN294701) showed > 99% homology with C. auris.
The D1/D2 sequences were aligned with the ClustalW programme [8].
A neighbour-joining tree based on 26S rRNA gene D1/D2 domains sequences was generated using MEGA software version X [9].
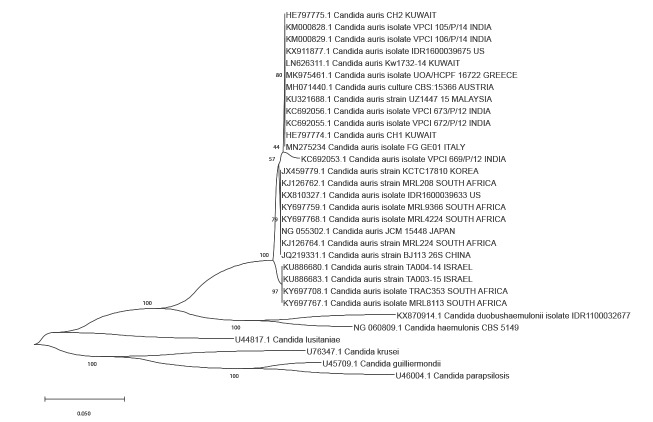
Phylogenetic analyses showed that our isolate, C. auris FG_GE01, clustered with the southern Asian strains (Figure).
Figure.
Dendrogram of Candida auris with other related Candida species, July 2019
MEGAX software was used to align 26S rRNA gene D1/D2 domains sequences and to draw the bootstrap (550 replicates) unweighted pair group method. The tree is drawn to scale with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The Kimura 2-parameter method was used to compute the evolutionary distances.
Antifungal susceptibility
Antifungal susceptibility was determined using the Clinical and Laboratory Standards Institute (CLSI) microdilution method [10] and Sensititre YeastOne (Thermo Scientific, Waltham, Massachusetts, United States (US)) The following minimum inhibitory concentration (MIC) values were observed: > 256 mg/L (fluconazole), 2mg/L (amphotericin B), 0.5 mg/L (flucytosine), 4 mg/L (voriconazole), 0.25mg/L (posaconazole), 0.5 mg/L (itraconazole), 0.12 mg/L (micafungin), 0.25 mg/L (anidulafungin) and 0.12 mg/L (caspofungin).
Epidemiological and environmental investigations
Preliminary epidemiological and environmental investigations have not yet revealed the source of the infection. The patient had no history of recent travel abroad or hospital admission and to date, C. auris has not been isolated from any of the case’s close contacts. A close contact was defined as a patient who was hospitalised in the same room during the same period (at least 12 hours) of the case or a patient who occupied the same bed as the case immediately after. No contacts outside the hospital, e.g. family members, were swabbed.
In August 2019, screening was performed of the environment and medical devices of the vascular ward, ICU and operating theatre where the patient stayed during the course of hospitalisation. The 36 environmental samples tested (dynamic mattresses, n = 5; bedrails, n = 10; trolley, n = 5; ventilators, n = 5; suction apparatus, n = 5; floor, n = 2; washbasin, n = 2; bed bell, n = 2) were negative. Additional cleaning using hydrogen peroxide and hypochlorite was implemented across the hospital.
Discussion
To our knowledge, this is the first isolation of C. auris in Italy. However, this is not particularly surprising given the widespread distribution of this pathogen and its previous detection in several nearby European countries, including France, Spain and Greece [11].
We were able to correctly identify the microorganism by using MALDI-TOF as well as molecular techniques, approaches that are still not available in many diagnostic laboratories. The first finding of C. auris in Italy should encourage a careful approach to yeast identification when non-albicans Candida strains are detected. Given the present scenario, confirmation of species identification of any suspicious colony is clearly essential.
Four major phylogenetically distinct clades of C. auris have been described: clade I (South Asian), clade II (East Asian), clade III (African) and clade IV (South American) [12]. A potential fifth clade has recently been reported in Iran [13].
The isolate in this study clusters with the southern Asian strains. Clade I has been already described in European hospitals and in hospitals in the US, and linked to outbreaks with invasive infections [14].
Since the potential source of the infection has not been identified, we currently have no basis for any speculation about the origin of C. auris in the hospital. The prompt isolation of the patient seems to have stopped the spread. Although specific breakpoints for C. auris have not been defined by international committees, susceptibility data published to date suggests that this pathogen exhibits resistance to fluconazole (MICs > 32 mg/L) and different level of susceptibility to the other azoles, echinocandins and amphotericin B. A considerable percentage of C. auris strains investigated had high MICs for voriconazole and amphotericin B (MICs > 1). In general, our susceptibility results are in agreement with previous observations [15]. Adopting CLSI breakpoints for closely related species (C.guillermondi and C. parapsilosis), C. auris FG_GE01 was categorised as susceptible to echinocandins. This finding is supported by clinical data: caspofungin treatment was effective and blood samples collected 7 days after starting the treatment were negative. However, development of resistance to echinocandins has also been described in C. auris and other Candida species [16,17].
This isolation of C. auris is further confirmation its intercontinental distribution, and that judicious use of antifungals coupled with strengthened infection control measures are needed to prevent and control the spread of C. auris.
Acknowledgements
We thank C Perfumo from the Laboratory Unit of the Ospedali Galliera Hospital, Genoa, Italy, for her support with the Bruker MALDI-TOF analysis. We also thank C Viscoli, G Icardi and G Mazzarello from the Policlinico San Martino, Genoa, Italy for critical discussion about the clinical case.
We are grateful to the Infection Control Team from Policlinico San Martino, Genoa, Italy (A Battistini, AM Di Bella, B Guglielmi, D Bellina, A Talamini, V Daturi and R Ziferro) for their support in the management of environmental investigations.
Conflict of interest: None declared.
Authors’ contributions: Francesca Crea: analysis and interpretation of patient's primary cultures; isolation of C. auris; analysis and interpretation of conventional identification and susceptibility tests; critical revision of the manuscript.
Giulia Codda: analysis and interpretation of molecular data; GenBank deposit; contribution to critical revision of manuscript.
Anna Marchese: acquisition, analysis and interpretation of data; integration of information submitted by all contributors; drafting and revising the manuscript.
Andrea Orsi, Alberto Battaglini: epidemiological follow up of patient; revising the epidemiological aspects of the manuscript.
Daniele Roberto Giacobbe, Emanuele Delfino, Riccardo Ungaro: administration of antimicrobial therapy; follow-up of patient; contribution to the acquisition of data; and drafting and revising the clinical aspects of the manuscript.
References
- 1. Jeffery-Smith A, Taori SK, Schelenz S, Jeffery K, Johnson EM, Borman A, et al. Candida auris: a Review of the Literature. Clin Microbiol Rev. 2017;31(1):1-18. 10.1128/CMR.00029-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rhodes J, Fisher MC. Global epidemiology of emerging Candida auris. Curr Opin Microbiol. 2019;52:84-9. 10.1016/j.mib.2019.05.008 [DOI] [PubMed] [Google Scholar]
- 3. Cortegiani A, Misseri G, Fasciana T, Giammanco A, Giarratano A, Chowdhary A. Epidemiology, clinical characteristics, resistance, and treatment of infections by Candida auris. J Intensive Care. 2018;6(1):69. 10.1186/s40560-018-0342-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC). Candida auris: A Drug-resistant Germ That Spreads in Healthcare Facilities. Atlanta: CDC; 21 Dec 2018. Available from: https://www.cdc.gov/fungal/candida-auris/c-auris-drug-resistant.html
- 5. Ruiz-Gaitán AC, Fernández-Pereira J, Valentin E, Tormo-Mas MA, Eraso E, Pemán J, et al. Molecular identification of Candida auris by PCR amplification of species-specific GPI protein-encoding genes. Int J Med Microbiol. 2018;308(7):812-8. 10.1016/j.ijmm.2018.06.014 [DOI] [PubMed] [Google Scholar]
- 6. Kurtzman CP, Robnett CJ. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol. 1997;35(5):1216-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim MN, Shin JH, Sung H, Lee K, Kim EC, Ryoo N, et al. Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis. 2009;48(6):e57-61. 10.1086/597108 [DOI] [PubMed] [Google Scholar]
- 8. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673-80. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35(6):1547-9. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute (CLSI). M27-S4 Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, 4th edition. Wayne, Pennsylvania: CLSI; 2012. Available from: https://clsi.org/standards/products/microbiology/documents/m27/
- 11. Kohlenberg A, Struelens MJ, Monnet DL, Plachouras D, The Candida Auris Survey Collaborative Group Candida auris: epidemiological situation, laboratory capacity and preparedness in European Union and European Economic Area countries, 2013 to 2017. Euro Surveill. 2018;23(13):1800136. 10.2807/1560-7917.ES.2018.23.13.18-00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Welsh RM, Sexton DJ, Forsberg K, Vallabhaneni S, Litvintseva A. Insights into the Unique Nature of the East Asian Clade of the Emerging Pathogenic Yeast Candida auris. J Clin Microbiol. 2019;57(4):e00007-00019. 10.1128/JCM.00007-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chow NA, de Groot T, Badali H, Abastabar M, Chiller TM, Meis JF. Potential Fifth Clade of Candida auris, Iran, 2018. Emerg Infect Dis. 2019;25(9):1780-1. 10.3201/eid2509.190686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spivak ES, Hanson KE. Candida auris: an Emerging Fungal Pathogen. J Clin Microbiol. 2018;56(2):e01588-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arendrup MC, Prakash A, Meletiadis J, Sharma C, Chowdhary A. Comparison of EUCAST and CLSI Reference Microdilution MICs of Eight Antifungal Compounds for Candida auris and Associated Tentative Epidemiological Cutoff Values. Antimicrob Agents Chemother. 2017;61(6):e00485-17. 10.1128/AAC.00485-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64(2):134-40. 10.1093/cid/ciw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lortholary O, Desnos-Ollivier M, Sitbon K, Fontanet A, Bretagne S, Dromer F, French Mycosis Study Group Recent exposure to caspofungin or fluconazole influences the epidemiology of candidemia: a prospective multicenter study involving 2,441 patients. Antimicrob Agents Chemother. 2011;55(2):532-8. 10.1128/AAC.01128-10 [DOI] [PMC free article] [PubMed] [Google Scholar]