i. Summary
Immunohistochemistry (IHC) is a powerful technique that exploits the specific binding between an antibody and antigen to detect and localize specific antigens in cells and tissue, most commonly detected and examined with the light microscope. A standard tool in many fields in the research setting, IHC has become an essential ancillary technique in clinical diagnostics in anatomic pathology (1) with the advent of antigen retrieval methods allowing it to be performed conveniently on formalin fixed paraffin embedded (FFPE) tissue (2, 3) and automated methods for high volume processing with reproducibility (4). IHC is frequently utilized to assist in the classification of neoplasms, determination of a metastatic tumor’s site of origin and detection of tiny foci of tumor cells inconspicuous on routine hematoxylin and eosin (H&E) staining. Furthermore, it is increasingly being used to provide predictive and prognostic information, such as in testing for HER2 amplification in breast cancer (5) in addition to serving as markers for molecular alterations in neoplasms, including IDH1 and ATRX mutations in brain tumors (6). In this section we describe the basic methods of immunohistochemical staining which has become an essential tool in the daily practice of anatomic pathology worldwide.
Keywords: Immunohistochemistry, Antibodies, Antigens, Antigen retrieval, Light microscopy
1. Introduction
Immunohistochemistry (IHC) is a widely used ancillary testing method in anatomic surgical pathology for cell classification and diagnosis and utilizes antibodies targeted against certain antigens in specific tissues and cells to facilitate determination of cell type and organ of origin. The method is most commonly performed on formalin fixed paraffin embedded (FFPE) tissue which has the advantage of being amenable to easy storage, although it was first developed on frozen sections and can also be done on plastic embedded tissue (7, 8). The use of IHC has recently further expanded to assess predictive and prognostic biomarkers in many malignancies including those of the breast, gastrointestinal tract, lung, hematolymphoid and central nervous systems (9, 10). Guidelines for the standardization and analytic validation of immunohistochemical tests have been established by the College of American Pathologists (1, 11).
The sequential steps in IHC can be summarized as follows: antigen retrieval (AR), addition of primary antibody, application of a secondary antibody that binds the primary antibody, and addition of a detection reagent to localize the primary antibody (Fig 1). The first step in IHC is usually antigen retrieval (AR), which involves the pretreatment of tissue to retrieve antigens masked by fixation and make them more accessible to antibody binding (12). This technique, first described by Shi et al, has significantly increased the sensitivity of IHC and consequently greatly expanded its application (3, 13). There are various methods of antigen retrieval depending on the specific target antigen and antibody (Table 1), but most generally involve the breaking of protein cross-links caused by fixation, such as formalin, through chemical or physical means. Physical treatments include heat and ultrasound while chemical methods include enzyme digestion and denaturant treatment, although many utilize both, such as chemical treatment with heat. Currently, the most popular method is heat induced antigen retrieval (HIAR) using microwave ovens most commonly, as well as pressure cookers, autoclaves and water baths (14).
Fig 1:
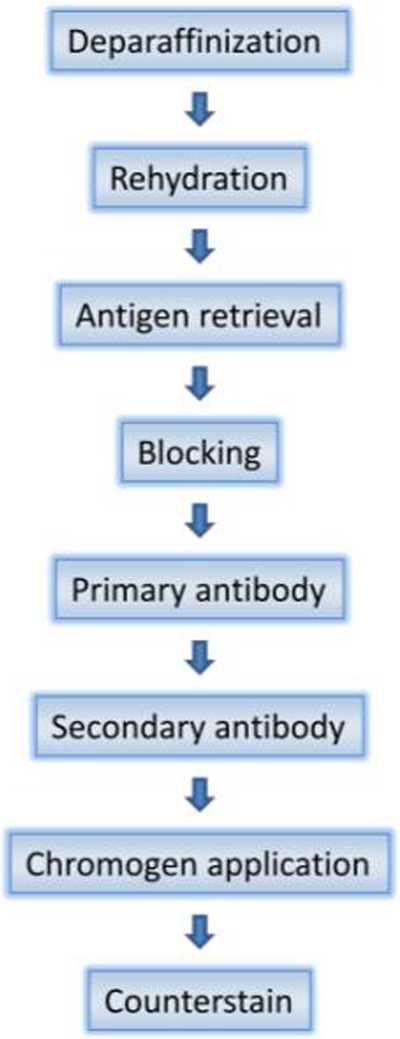
Immunohistochemistry flow chart
Table 1: Methods used to retrieve antigens.
(Adapted from D’Amico et al. (14))
| Method | Type | Uses |
|---|---|---|
| Heat (microwave, pressure cooker, autoclave, water bath) | Physical | Most commonly used, provides good tissue morphology |
| Ultrasound | Physical | Minimize protein changes caused by fixation |
| Enzyme digestion | Chemical | For epitopes which may lose antigenicity with heat, may destroy epitopes and tissue morphology |
| Denaturant (formic acid, urea) | Chemical | Formic acid for prion and neurofilament protein |
| Detergent (Triton X-100, SDS) | Chemical | Minimize contamination of sections |
| Oxidizing (hydrogen peroxide) | Chemical | For some drugs such as bleomycin, daunomycin and pepleomycin |
The primary antibody, which can be monoclonal or polyclonal, is titrated to optimize contrast between positively staining tissue and nonspecific background staining, with the highest primary antibody dilution to prevent waste (1, 7). In general, monoclonal antibodies, which target a single epitope, tend to be more specific while polyclonal antibodies, which can bind many different epitopes, tend be more sensitive (2). Usually starting with the dilution recommended by the manufacturer or published in the literature for the tissue of interest, a more concentrated and less concentrated dilution flanking the recommended dilution is tested on a series of tissues with the appropriate positive control. This may be combined with various combinations of dilutions of the secondary antibody in the setting of the particular AR method and chromogen to produce optimum staining (7). For the initial titration, an antibody concentration of 1 to 5 μg/mL is usually recommended (1).
To visualize the antigen antibody interaction under light microscopy, either the primary antibody or secondary antibody, which is targeted against the immunoglobulin of the species in which the primary antibody was produced, must be labeled (2). In the direct method, the primary antibody is labeled and applied to the tissue in a quick one step process; however, this method is not commonly used due to lack of signal amplification and thus the requirement for a higher concentration of antibody as well as the need to label each primary antibody. In the indirect method, the secondary antibody is labeled, allowing for signal amplification and use with many different primary antibodies. There are various labels that can be used, such as fluorescent molecules and enzymes such as horseradish peroxidase or alkaline phosphatase which produce a colored product after incubation with a chromogenic substrate such as diaminobenzidine (DAB) (2, 7). Immunofluorescence techniques using fluorescent compounds are also available but require a fluorescence microscope (2). The avidin-biotin-peroxidase method suffers from high background staining due to binding of endogenous biotin, and the method is now largely obsolete. Polymer based methods utilize many peroxidase molecules and secondary antibodies which are attached to a dextran polymer backbone and allows for increased sensitivity (7).
Background staining may be due to nonspecific antibody binding, more common in polyclonal antibodies, and endogenous peroxidase activity, more problematic in tissues with abundant hematopoietic elements such as bone marrow. Nonspecific antibody binding can be decreased by preincubation with normal serum from the same species as the secondary antibody or with a commercially available universal blocking agent. Endogenous enzyme activity can be inhibited by pretreating the tissue with solutions containing hydrogen peroxide prior to application of the antibody (2).
Quality control is critical, and a positive and negative control should be performed with each run. Positive controls are tissues that contain an antigen known to stain with a certain antibody, and ideally should be run on the same slide as the tissue of interest so that the control tissue undergoes the same reaction conditions as the sample tissue. To eliminate the possibility of nonspecific antibody binding with the secondary antibody, negative controls consist of the sample tissue that undergoes the identical staining conditions minus the primary antibody or with a non-immune immunoglobulin from the same species (7). False positives and negatives can be due to the immunohistochemical method itself but also to a myriad of other factors including preparation and fixation (see Notes section).
2. Materials
2.1. Solutions
Deionized and distilled water
Xylene
Ethanol, anhydrous denatured, histological grade (100, 95, 80, and 70%)
Hematoxylin solution
Tacha’s Bluing solution: for bluing hematoxylin stained nuclei
Wash buffers: follow vendor recommendations.
Antigen retrieval buffer, depends on specific antigen retrieval method: for HIAR e.g. 10X Antigen Decloaker (Biocare, Pacheco, CA, USA) diluted 1:10 with deionized water
0.1% TBS-Tween
3% hydrogen peroxide (for blocking endogenous peroxidase); can also use a ready-to-use peroxidase blocking solution available from various manufacturers
Blocking reagent to decrease nonspecific background staining e.g. Background Sniper (Biocare, Pacheco, CA, USA)
Charged or adhesion slides to promote tissue retention onto the slide
2.2. Antibodies
Primary antibodies: labeled or unlabeled (if unlabeled, secondary antibodies are needed); many available from numerous vendors- in general, follow their recommendations.
Secondary antibodies: labeled such as with an enzyme (e.g. horseradish peroxidase), enhanced sensitivity due to signal amplification, choice depends on species and immunoglobulin isotype (class) of primary antibody.
2.3. Detection
Detection system: such as horseradish peroxidase or alkaline phosphatase, but polymer based systems are preferred (7)
Chromogens: e.g. DAB for peroxidase, NovaRed for alkaline phosphatase
3. Methods
3.1. Slide preparation
Cool blocks on ice or in a refrigerator at 4 °C and section at a thickness of 4–7 μm (thickness will vary depending on type of tissue, e.g. brain tissue is usually cut thicker compared to other types of tissue) and mount on adhesion treated slides.
Allow freshly cut paraffin sections to dry overnight or for at least several hours.
Place slides in a 60°C oven for at least 2 hours (or overnight ideally).
3.2. Deparaffinization and rehydration using standard methods (sample protocol below)
Immerse slides in 3 washes of xylene, each for 10 min for a total of 30 minutes (change solutions after they have been used for approximately 30 slides).
Dip slides (approximately 15 dips each) in graded alcohols sequentially from 100%, 100%, 80%, to 70%.
Immerse in two changes of deionized water.
Let sit in deionized water for 5 min (slides can remain in deionized water for up to 30 minutes).
3.3. Antigen retrieval
Antigen retrieval methods include HIAR and enzymatic (protease-induced) which is not further discussed here. HIAR using either a microwave oven or a pressure cooker is a common approach as they typically have a higher success rate for restoring antigenicity than an enzymatic method. We provide sample protocols below of HIAR with a microwave oven or a pressure cooker (Decloaking chamber, (Biocare Medical, Pacheco, CA, USA)). Other pressure cookers may use different protocols and manufacturer instructions should be followed. It is emphasized that only one HIAR method need be utilized- not both microwave and pressure cooker.
3.3.1. Microwave oven (3,14)
Place deparaffinized and rehydrated slides in 3% hydrogen peroxide for 5 minutes.
Wash slides with deionized water for 5 minutes.
Place slides in retrieval buffer (such as 10 mM citrate) and heat in the microwave at 100 °C for 5 to 10 minutes. Make sure the buffer level is adequate throughout the heating process.
Cool slides for 15 minutes at room temperature.
Wash slides with deionized water twice.
Wash slides in phosphate buffered saline for 5 minutes.
3.3.2. Pressure cooker
Place removable pan in body of Decloaking chamber.
Add 500mL deionized water to pressure cooker and submerge heat shield, positioning in center of pan.
Ensure gasket properly positioned on the removable pan.
Prepare antigen retrieval buffer (10X Biocare Antigen Decloaker diluted 1:10 with deionized water).
Fill polypropylene staining dish(es) with prepared antigen retrieval buffer (100mL in each dish).
Place full basket(s) into dish(es) (inserting blank slides into empty slots) and transfer dish(es) into decloaking chamber.
Input BioCare Medical Decloaking Chamber settings to Set point (SP)1 at 95°C for 30 minutes, SP2 at 90°C for 10 seconds and with a SP limit of 10°C for a maximum of 10 minutes.
Secure lid on pressure cooker tightly.
After completion, open lid and remove dish(es), and leaving the basket in dish with the retrieval buffer, set the dish on the bench to cool for 20–30 minutes.
Immerse in 2 changes of deionized water and let sit in deionized water for 5 minutes.
Immediately transfer slides to 0.1% TBS-Tween for 5 minutes (can remain in 0.1% TBS-Tween for up to 1 hr).
3.4. Blocking endogenous peroxidase
Immerse slides in 3% hydrogen peroxide solution in Coplin jar for 5 minutes (can also use a ready-to-use peroxidase blocking solution).
Rinse in 3 changes of 0.1% TBS-Tween.
3.5. Blocking with serum or universal blocking buffer (e.g. Biocare Background Sniper) to decrease background staining
Prepare diluted solutions of appropriate antibodies (Note: should spin down the antibodies for approximately 3 seconds in small centrifuge prior to diluting)
Vortex the prepared antibodies for 2 seconds. Place in 4°C until application.
Dry slide around the tissue using a kimwipe and using a hydrophobic pen (e.g. Dako Pap Pen) draw a barrier around the tissue leaving some space between the tissue and barrier.
Each slide should get one quick dip in 0.1% TBS-Tween immediately after pap-penning.
Apply 3–5 drops of blocking buffer to each slide for 15 minutes in trays containing deionized water (close tray as soon as it is applied).
Rinse off blocking buffer with 0.1% TBS-Tween.
3.6. Antibody application
The concentration of antibody that will provide the strongest staining of the target antigen and lowest background staining must be determined by serial dilutions of a concentrated antibody. Generally, it is easiest to start with the dilution recommended by the manufacturer with one dilution above and one dilution below it. For example, if the recommended dilution is 1:400, then testing serial dilutions of 1:200, 1:400, and 1:800 is likely to show optimal signal to background staining on one of the dilutions.
Apply 200μl of diluted primary antibody to slides and incubate at room temperature for 80 minutes (or time optimized for specific antibody) (Note: can also incubate primary antibody overnight at 4°C and resume at next step).
Wash in 3 changes of 0.1% TBS-Tween using a different staining dish for each wash (leave in each dish for 3 minutes for a total of 9 minutes washing).
Apply species specific secondary antibody onto slides according to manufacturer specifications [e.g. if using Mach-4 secondary kit, apply a few drops of Yellow Probe (enough to cover sample) and let sit for 15 minutes, then wash with 0.1% TBS-Tween, add a few drops of orange horseradish peroxidase and let sit for 15 minutes].
Wash in 3 changes of 0.1% TBS-Tween using a different staining dish for each wash (leave in each dish for 5 minutes for a total of 15 minutes washing).
3.7. Chromogen application
Prepare NovaRED Chromogen (Vector Laboratories) or other chromogen depending on detection system according to manufacturer specifications.
Remove slides from 0.1% TBS-Tween and quickly pipette 3–5 drops NovaRED Chromogen onto slide to entirely cover the tissue.
Incubate 6.5 minutes, using shorter incubations times if background staining is too high or longer incubation times of up to 2 minutes longer depending on the age of the tissue, antibody, or chromogen.
Immerse in 3 changes of deionized water or until water is clear, then let sit in deionized water for 3 minutes.
3.8. Counterstaining, dehydration, clearing, and mounting
Place slides in hematoxylin for 30 seconds.
Rinse in multiple changes of distilled water until clear and then let sit in distilled water for 2 minutes.
Incubate in Tacha’s Bluing solution for 1.5 minutes.
Rinse in multiple changes of distilled water until clear and then let sit in distilled water for 2 minutes.
Oven dry for 10–15 minutes.
Immerse in 3 washes of Xylene (for 5 minutes each).
Mount in permount and coverslip.
4. Notes
Proper fixation is essential in ensuring optimum staining as both under- (less than 12 hours) and over-fixation (more than 48 hours) can lead to false negative results or weak staining. Furthermore, necrotic regions may stain indiscriminately or not at all so it is best to avoid sampling of these areas (7). A decalcification procedure can also lead to false negative results with certain antigens (1).
Drying out of tissue, air bubbles in the reagents, or incomplete dehydration during the processing step can lead to false negative results. Expired reagents and reagents not stored properly will also cause false negativity. Ensure that reagents are fresh and check for compatibility with other reagents when used together (7).
Incorporating a blocking step using nonimmune serum from the same animal species as the secondary antibody will help reduce false positive staining caused by nonspecific protein binding (7).
A too high concentration or too long incubation time of the antibody or chromogen may also result in false positive or excessive background staining. Conversely, a too dilute concentration or too short of an incubation time may cause false negatives.
Too thick tissue sections may cause an artifactual increase in staining or even a false positive; it is recommended to cut sections at 4 μm for most tissues (7).
False negative staining is more common than false positive staining, and it is crucial to examine both positive and negative controls, preferably internal (1).
False negative staining may also be due to the improper AR method for a specific antigen-antibody combination and some recommend testing with different AR protocols (1). For example, excessive enzyme digestion may destroy epitopes and cause false negative staining (14).
Acknowledgement
This work was supported in part by NIH:NCI P50-CA211015, NIH:NIMH U24 MH100929, the Art of the Brain Foundation, and the Henry E. Singleton Brain Cancer Research Program.
References
- 1.Lin F, Chen Z (2014) Standardization of diagnostic immunohistochemistry: literature review and Geisinger experience. Arch Pathol Lab Med 138:1564–1577 [DOI] [PubMed] [Google Scholar]
- 2.Taylor CR, Shi S-R, Barr NJ (2010) Techniques of immunohistochemistry: principles, pitfalls, and standardization In: Dabbs DJ (ed) Diagnostic immunohistochemistry: theranostic and genomic applications, 3rd edn. Saunders, Philadelphia [Google Scholar]
- 3.Shi SR, Key ME, Kalra KL (1991) Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J Histochem Cytochem 39:741–748 [DOI] [PubMed] [Google Scholar]
- 4.Prichard J, Hicks D, Hammond E (2015) Automated immunohistochemistry overview In: Fan L, Jeffrey P (eds) Handbook of practical immunohistochemistry: frequently asked questions, 2nd edn. Springer, New York [Google Scholar]
- 5.Wolff AC, Hammond MEH, Hicks DG et al. (2014) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med 138:241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Appin CL, Brat DJ (2015) Biomarker-driven diagnosis of diffuse gliomas. Mol Aspects Med doi: 10.1016/j.mam.2015.05.002 [DOI] [PubMed] [Google Scholar]
- 7.Taylor CR (2014) Immunohistochemistry in surgical pathology: Principles and practice In: Day CE (ed) Histopathology: methods and protocols. Methods in molecular biology; Springer, New York, pp 81–110 [DOI] [PubMed] [Google Scholar]
- 8.Coons AH, Creech HJ, Jones RN (1941) Immunological Properties of an antibody containing a fluorescent group. Proc Soc Exp Biol Med 47:200–202 [Google Scholar]
- 9.Chen ZE, Lin F (2015) Overview of predictive biomarkers and integration of IHC into molecular pathology In: Fan L, Jeffrey P (eds) Handbook of practical immunohistochemistry: frequently asked questions, 2nd edn. Springer, New York [Google Scholar]
- 10.Yong WH, Dry SM, Shabihkhani M (2014) A practical approach to clinical and research biobanking. Methods Mol Biol 1180:137–162 [DOI] [PubMed] [Google Scholar]
- 11.Fitzgibbons PL, Bradley LA, Fatheree LA et al. (2014) Principles of analytic validation of immunohistochemical assays: guideline from the college of american pathologists pathology and laboratory quality center. Arch Pathol Lab Med 138:1432–1443 [DOI] [PubMed] [Google Scholar]
- 12.Cregger M, Berger AJ, Rimm DL (2006) Immunohistochemistry and quantitative analysis of protein expression. Arch Pathol Lab Med 130:1026–1030 [DOI] [PubMed] [Google Scholar]
- 13.Shi S-R, Shi Y, Taylor CR (2011) Antigen retrieval immunohistochemistry: review and future prospects in research and diagnosis over two decades. J Histochem Cytochem 59:13–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Amico F, Skarmoutsou E, Stivala F (2009) State of the art in antigen retrieval for immunohistochemistry. J Immunol Methods 341:1–18 [DOI] [PubMed] [Google Scholar]


