The genus Enterobacter is a member of the ESKAPE group, which contains the major resistant bacterial pathogens. First described in 1960, this group member has proven to be more complex as a result of the exponential evolution of phenotypic and genotypic methods. Today, 22 species belong to the Enterobacter genus. These species are described in the environment and have been reported as opportunistic pathogens in plants, animals, and humans.
KEYWORDS: β-lactamases, Enterobacter spp., clinical aspects, diagnosis, efflux, epidemiology, impermeability, multidrug resistance, pathogenicity
SUMMARY
The genus Enterobacter is a member of the ESKAPE group, which contains the major resistant bacterial pathogens. First described in 1960, this group member has proven to be more complex as a result of the exponential evolution of phenotypic and genotypic methods. Today, 22 species belong to the Enterobacter genus. These species are described in the environment and have been reported as opportunistic pathogens in plants, animals, and humans. The pathogenicity/virulence of this bacterium remains rather unclear due to the limited amount of work performed to date in this field. In contrast, its resistance against antibacterial agents has been extensively studied. In the face of antibiotic treatment, it is able to manage different mechanisms of resistance via various local and global regulator genes and the modulation of the expression of different proteins, including enzymes (β-lactamases, etc.) or membrane transporters, such as porins and efflux pumps. During various hospital outbreaks, the Enterobacter aerogenes and E. cloacae complex exhibited a multidrug-resistant phenotype, which has stimulated questions about the role of cascade regulation in the emergence of these well-adapted clones.
INTRODUCTION
The genus Enterobacter includes facultative anaerobic Gram-negative bacilli that are 2 mm long, are motile by means of peritrichous flagella, and belong to the family Enterobacteriaceae. It was first described in 1960, but changes in taxonomy have occurred in the last 50 years (1). For example, E. sakazakii has been reassigned to a new genus, Cronobacter (2).
To date, 22 species have been found in the genus Enterobacter: E. aerogenes, E. amnigenus, E. arachidis, E. asburiae, E. carcinogenus, E. cloacae, E. cowanii, E. dissolvans, E. gergoviae, E. helveticus, E. hormaechei, E. kobei, E. ludwigii, E. mori, E. nimipressuralis, E. oryzae, E. pulveris, E. pyrinus, E. radicincitans, E. soli, E. taylorae, and E. turicensis. Among these species, seven are grouped within the Enterobacter cloacae complex group: E. cloacae, E. asburiae, E. hormaechei, E. kobei, E. ludwigii, E. mori, and E. nimipressuralis. This nomenclature is based on the sharing of phenotypic and especially genotypic characteristics, determined by whole-genome DNA-DNA hybridizations. Indeed, E. cloacae shares at least 60% similarity in its genome with the other six members of this group (3).
The genus Enterobacter is associated with a variety of environmental habitats. These bacteria are recovered from soil and water and are endophytic or considered phytopathogens for various plant species (4). Some species are frequently associated with bioprocessing and metabolic engineering approaches (5, 6). Moreover, Enterobacter spp. are also natural commensals of the animal and human gut microbiota. Among these bacteria, only certain subspecies/species have been associated with hospital-acquired infections and outbreaks (7–12). Enterobacter species are members of the ESKAPE group (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species), which are described as the leading cause of resistant nosocomial infections (7, 10, 11, 13–20). Enterobacter aerogenes, E. cloacae, and E. hormaechei represent the most frequently isolated species described in clinical infections, especially in immunocompromised patients and those hospitalized in an intensive care unit (ICU), due to the adaptation of these species to antimicrobial agents and their behavior as opportunistic pathogens. Several hospital outbreaks have been reported in Europe since the mid-1990s, and the wide use of extensive broad-spectrum antibiotics has stimulated the spread of resistant clones (21–23). These pathogens are frequently associated with a multidrug resistance (MDR) phenotype, mainly due to their adaptation to the hospital environment and their ability to easily acquire numerous genetic mobile elements containing resistance and virulence genes. These species have an intrinsic resistance to ampicillin, amoxicillin, first-generation cephalosporins, and cefoxitin due to the expression of a constitutive AmpC β-lactamase. Moreover, the production of extended-spectrum β-lactamases has been reported in these bacteria, which make their treatment difficult (24, 25). Antibiotic resistance, regulation of resistance genes and the clinical implications of these situations have been extensively studied (26–31).
The accurate identification of species and subspecies remains a challenge. The development of genome sequencing has rapidly modified the phylogeny of the genus, particularly that of the E. cloacae complex (32–34). Due to use of modern molecular techniques, the genus has undergone modifications in classification, and several species have been transferred to and from this genus. For example, four species first identified as Enterobacter have been reclassified to the genus Kosakonia (E. cowanii, E. arachidis, E. oryzae, and E. radicinintans), E. intermedium has been reclassified to the genus Kluyvera, and E. sakazakii has been reclassified to the genus Cronobacter (2, 35, 36). Moreover, the current taxonomic position of E. aerogenes is still discussed. Since 1971, the proposition of reclassification in the genus Klebsiella as K. aerogenes, K. mobilis, or K. aeromobilis because of its motility remains unsettled (37). The phenotypic differences between E. aerogenes and the genus Klebsiella include motility and the presence of ornithine decarboxylase and the lack of urease activity in E. aerogenes. However, results from full-genome sequence analysis confirmed that the closest species to E. aerogenes was K. pneumoniae (37). Considering the taxonomic requirements, the epithet mobilis was discussed (38). Recently, both CLSI and EUCAST (https://clsi.org/media/1974/ast_news_update_jan18.pdf, http://www.eucast.org/eucast_news/news_singleview/?tx_ttnews%5Btt_news%5D=316&cHash=000ba6c5b8c2516c98467ef8fbd8dccc) have included this modification in their documents, which now mention Enterobacter aerogenes as Klebsiella aerogenes. However, until now, scientific reports only occasionally have used Klebsiella aerogenes and usually have used E. aerogenes. Multilocus sequence analysis (MLSA) of housekeeping genes, in part, and sequencing of the 16S rRNA have recently allowed the characterization of new Enterobacter species isolated from human infections or from plants (39, 40). The total genome sequences of the various Enterobacter spp. have allowed reevaluation of the genus phylogeny and better evaluation of the importance of those cases where some species were misidentified as other species by routine identification techniques, as was the case of E. hormaechei in the E. cloacae complex (41).
TAXONOMY OF THE GENUS
Enterobacter amnigenus
The species E. amnigenus, described in 1981 by Izard et al., is a rarely isolated bacterium (42). It was suggested by Brady et al. that it be reclassified in the genus Lelliottia based on multilocus sequence analysis; however, that change has never been validly published, and E. amnigenus still remains the official nomenclature (35). It comprises two genotypically and phenotypically different groups that have been called biogroup 1 and biogroup 2 by the CDC. The strains are generally ornithine decarboxylase (ODC) positive and lysine decarboxylase (LDC) and urease negative and ferment melibiose. Strains of E. amnigenus biogroup 1 ferment sucrose and raffinose but not d-sorbitol. They are arginine dihydrolase (ADH) negative. Strains of the E. amnigenus biogroup 2 ferment d-sorbitol but not sucrose and raffinose. Additional identifying characteristics are presented in Table 1. Strains of E. amnigenus are generally susceptible to piperacillin, imipenem, gentamicin, tobramycin, amikacin, nalidixic acid, norfloxacin, ciprofloxacin, and colistin and are resistant to ampicillin, amoxicillin-clavulanic acid, ticarcillin, and cephalothin. Resistance is variable across strains for second- and third-generation cephalosporins, moxalactam, doxycycline, chloramphenicol, and co-trimoxazole (43–45).
TABLE 1.
Biochemical characteristics for the identification of species of the genus Enterobactera
| Organism | Yellow pigment | LDC | ADH | URE | ESC | Fermentation |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| INO | SOR | SAC | MEL | RAF | AMG | DUL | ADO | ARL | ||||||
| E. amnigenus biogroup 1 | − | − | − | − | + | − | − | + | + | + | V | − | − | − |
| E. amnigenus biogroup 2 | − | − | V | − | + | − | + | − | + | − | + | − | − | − |
| E. cancerogenus | − | − | + | − | + | − | − | − | − | − | − | − | − | − |
| E. asburiae | − | − | (−) | +/− | + | − | + | + | V | (+) | + | − | − | − |
| E. cloacae subsp. cloacae | − | − | + | − | − | (−) | + | + | + | (+) | (+) | (−) | (−) | (−) |
| E. cloacae subsp. dissolvens | − | − | + | +w | + | V | + | + | + | + | + | − | − | − |
| E. hormaechei subsp. hormaechei | − | − | (+) | (+) | − | − | − | + | − | − | + | + | − | − |
| E. kobei | − | − | + | + | − | + | + | + | + | + | + | V | − | − |
| E. ludwigii | − | (−) | + | − | − | + | + | + | + | + | + | − | − | |
| E. nimipressuralis | − | − | − | − | + | − | + | − | + | − | + | − | − | |
| E. aerogenes | − | + | − | − | + | + | + | + | + | + | + | − | + | |
| E. gergoviae | − | (+) | − | +w | + | − | − | + | + | + | − | − | − | + |
| E. mori | + | + | ND | ND | + | ND | + | ND | + | ND | ND | ND | ND | + |
| E. bugandensis | − | − | + | − | +w | + | + | ND | + | + | ND | + | − | ND |
Data are from references 4, 5, 12, 48, 55, 75, 82, 86, and 120. Abbreviations: LDC, lysine decarboxylase; ADH, arginine dihydrolase; URE, urease; ESC, esculin; INO, inositol; SOR, sorbitol; SAC, saccharose; MEL, melibiose; RAF, raffinose; AMG α-methyl-d-glucoside; ADO, adonitol; ARL, d-arabitol. +, positive reaction (>,90% of strains); (+), generally positive reaction; +w, weakly positive reaction; V, variable reaction; (−), generally negative reaction; −, negative reaction (<90% of strains); +/−, variable depending on the method used; ND, not determined.
Enterobacter cancerogenus
Formerly known as Erwinia cancerogena, this species was transferred in 1988 to the genus Enterobacter after DNA-DNA hybridizations were studied for the three strains (46). Then, in 1989, Grimont and Ageron observed the synonymy between E. cancerogenus and Enterobacter taylorae (46). Currently, both denominations are still valid. Nevertheless, the name E. cancerogenus should be retained because it was established earlier. These strains are ODC and ADH positive and LDC and urease negative. They do not ferment d-sorbitol, sucrose, melibiose, dulcitol, and raffinose, and they are not gelatinolytic. Additional identification characteristics are presented in Table 1. Studied E. cancerogenus strains are generally susceptible to third-generation cephalosporins, moxalactam, imipenem, gentamicin, kanamycin, norfloxacin, ciprofloxacin, and colistin, and often they are susceptible to carbenicillin, ticarcillin, azlocillin, mezlocillin, and piperacillin. They are naturally resistant to aminopenicillins, some cephalosporins, and co-trimoxazole (47). They have an inducible chromosomal AmpC β-lactamase (45, 48–50).
Enterobacter cloacae Complex
The Enterobacter cloacae complex includes the following species: Enterobacter asburiae, Enterobacter carcinogenus, Enterobacter cloacae, Enterobacter hormaechei, Enterobacter kobei, Enterobacter nimipressuralis, and Enterobacter mori. All these species are genotypically very close, with more than 60% DNA-DNA homology. A phylogenetic study by Hofmann and Roggenkamp, based on sequences of four housekeeping genes, confirmed the genetic diversity of the Enterobacter cloacae complex, of which all affiliated species, although genetically related, form distinct clusters (3). E. cloacae and E. hormaechei are the most frequently isolated in human clinical specimens.
Enterobacter asburiae.
E. asburiae was described in 1986 by Brenner et al. (5) from strains of the enteric group 17, of which atypical bacterial strains of the genus Citrobacter or of the genus Enterobacter had been collected since 1978. It is sometimes described as Enterobacter muelleri. These strains are sometimes immobile, often having a Voges-Proskauer (VP)-negative reaction. They are indole negative, they ferment d-sorbitol and sucrose, and they generally do not ferment melibiose. When they are VP positive, it is necessary to differentiate them from E. cloacae (E. asburiae does not have ADH and does not ferment l-rhamnose) and other VP-positive species (E. asburiae does not possess LDC, Tween 80 esterase, or DNase, characteristics possessed by Serratia marcescens and Serratia liquefaciens). Additional identifying characteristics are shown in Table 1. E. asburiae has occasionally been described as a bacterium with clinical significance, mainly in blood cultures. However, de Florio et al. observed its gradual increase in 2017 (51). The complete genome of a clinical E. asburiae isolate from a bone marrow transplant patient has been sequenced (52). The strains studied by Brenner et al. (5) were still susceptible to gentamicin and sulfadiazine and generally to carbenicillin, kanamycin, streptomycin, chloramphenicol, nalidixic acid, and colistin. They were all resistant to ampicillin, cefalotine, and tetracycline. Environmental strains isolated from watercourses in the United States have been shown to be resistant to imipenem by producing a plasmid-derived IMI-2 carbapenemase, thus confirming the hypothesis of an environmental reservoir of this type of resistance gene (53, 54).
Enterobacter cloacae.
E. cloacae is the type species of the genus Enterobacter. It is now divided into two subspecies: E. cloacae subsp. cloacae, for which the esculin test is negative, and E. cloacae subsp. dissolvens, with a positive esculin reaction (55). The strains are ODC and ADH positive and LDC negative (among the bacteria of the genus Enterobacter, only the species E. cloacae, E. taylorae, E. kobei, and possibly E. amnigenus have this profile of decarboxylases). They ferment d-sorbitol, sucrose, and melibiose. Additional characteristics are presented in Table 1. Some pathogenicity factors have been identified as hemolytic and leukotoxic membrane cell cytotoxins (56). With regard to epidemiological dissemination, several studies have confirmed that E. cloacae colonizations/infections correspond to dissemination of several clusters corresponding to known major multilocus sequences types and that no relationship exists with the geographical source. The various clones have a widespread dissemination and are continuously arising and expanding. Clinical isolates come from various sources and reservoirs, representing sampling from the diversity of the species in the population, and patients are potentially colonized in different ways before entering the hospital (57). E. cloacae is naturally resistant to ampicillin, amoxicillin-clavulanic acid, cephalothin, and cefoxitin by the low-level production of the Bush group 1 inducible natural cephalosporinase (class C). Ureidopenicillins and carboxypenicillins are active on at least half of the strains (12). In AmpC chromosomal cephalosporinase, derepression and constitutive production by mutation can lead to resistance to a large number of β-lactams, particularly third-generation cephalosporins, except for cefepime (12, 58, 59). This AmpC-type resistance accounts for 50% of β-lactam resistance in clinical strains and coexists frequently with that due to extended-spectrum β-lactamase (ESBL) expression.
In 1989, the first cases of nosocomial infections due to strains possessing an ESBL were identified (18). Since then, various ESBLs, such as TEM, SHV, and CTX-M, have been characterized in E. cloacae, including inhibitor-resistant TEMs (IRP) (60–63). E. cloacae, along with Escherichia coli and K. pneumoniae, is one of the most common Enterobacteriaceae resistant to third-generation cephalosporins. Nevertheless, in recent years, clinical isolates that are resistant through the production of carbapenemases have been identified (64–67). In particular, in Asia, strains harboring IMP, NDM, GIM, or KPC enzymes have been described (68–70). Lee et al. found an imipenem resistance rate of 0.4% in E. cloacae (71). For aminoglycosides, the percentages of resistant strains range from 0% to 51% for gentamicin and from 0% to 34% for amikacin, whereas ciprofloxacin is active in 64% to 100% of cases (12). A recent study showed that 77% of clinical strains in China are plasmid positive with determinants of aminoglycoside resistance (70). Regarding quinolones, Enterobacter cloacae is one of the enterobacteria, along with Escherichia coli and Klebsiella pneumoniae, in which resistance of plasmid origin due to the QnrA protein was initially observed (70, 72–74). These determinants of plasmid resistance to fluoroquinolones are found in more than 60% of the strains (70).
Enterobacter hormaechei.
E. hormaechei was designated by O’Hara et al. in 1989 to describe previously assembled strains in enteric group 75 (75). These are LDC- and gelatinase-negative strains that are generally ODC, ADH, and urease positive, and they ferment sucrose and l-rhamnose but not d-sorbitol or melibiose. These characteristics generally make it possible to differentiate this entity from phenotypically close species. Additional identifying characteristics are shown in Table 1. Hoffmann et al. have subdivided E. hormaechei into three subspecies based on the differential biochemical characteristics for d-adonitol, d-arabitol, d-sorbitol, d-melibiose, and dulcitol: E. hormaechei subsp. oharae, which ferments only melibiose; E. hormaechei subsp. hormaechei, which ferments only dulcitol; and E. hormaechei subsp. steigerwaltii, which ferments all of these compounds except for dulcitol (76). Two additional subspecies have now been characterized by whole-genome comparisons and average nucleotide identity from complete genome sequencing: E. hormaechei subsp. xiangfangensis and E. hormaechei subsp. hoffmannii (41).
Most strains described by O’Hara et al. have been isolated from samples of human origin. These isolates were sensitive or moderately sensitive to azlocillin, mezlocillin, piperacillin, cefotaxime, ceftriaxone, ceftazidime, moxalactam, imipenem, gentamicin, tobramycin, amikacin, chloramphenicol, co-trimoxazole, and trimethoprim (75). Rare strains harbored ESBLs and hyperproduced AmpC cephalosporinase, thereby having resistance to third-generation cephalosporins (77). Strains producing carbapenemases have been recently identified (78). Davin-Regli et al. reported a nosocomial outbreak involving 21 isolates of E. hormaechei subsp. oharae which were resistant to fluoroquinolones, and their isolation was always preceded by treatment of patients with a fluoroquinolone (17).
Enterobacter kobei.
The name Enterobacter kobei was proposed by Kosako et al. to group strains belonging to the NIH 21 group, which was previously included in E. cloacae (79). These strains differ from E. cloacae by a negative VP reaction. However, recently, a new VP-positive biotype causing urinary tract infection (UTI) has been characterized (77). The clinical significance of these strains is often uncertain. However, the species is now also of concern because of ESBL production (80).
Enterobacter ludwigii.
The new species Enterobacter ludwigii has been described on the basis of genotypic and phenotypic characteristics of 16 strains with a clinical origin (81). It is genetically close to E. hormaechei, and a Biotype 100 Gallery API system was used to distinguish it from other Enterobacter spp. by its ability to use myo-inositol and methyl-d-glucopyranose. Whole-genome sequencing of the type strain was conducted (82). The strains are ADH and ODC positive and LDC and urease negative. All strains are naturally resistant to ampicillin, amoxicillin-clavulanic acid, and cefoxitin. Some strains of clinical origin hyperproduced AmpC cephalosporinase but were sensitive to co-trimoxazole, gentamicin, imipenem, and ciprofloxacin. Moreover, recently a nosocomial bloodstream infection outbreak in neonates caused by Enterobacter ludwigii coharboring CTX-M-8, SHV-12, and TEM-15 (83) and a clinical isolate responsible for a postoperative bone infection coharboring NDM-1 and OXA-48 carbapenemases in India (84) were described.
Enterobacter mori.
Enterobacter mori was first described as a phytopathogenic bacterium (85). It was isolated from diseased mulberry roots. The species could be differentiated from closely related species by the presence of lysine decarboxylase activity and the ability to use d-arabitol. The type strain R18-2T (= CGMCC 1.10322T = LMG 25706T) was sequenced. The 16S rRNA gene sequence and MLSA indicated that E. mori is closed related to E. asburiae (E. muelleri) (4). Sixty-six genes potentially involved in the secretion system have been identified, explaining the phytopathogenic nature of this species (39). However, recently it has been identified as being responsible for human infection in Austria, and the isolates carried an IMI-2 carbapenemase (39).
Enterobacter nimipressuralis.
Enterobacter nimipressuralis was first considered an Erwinia sp. before being reclassified as Enterobacter in 1986 by Brenner et al. (5, 86) on the basis of DNA-DNA hybridizations. Strains of E. nimipressuralis are not pigmented and are generally ADH positive and urease negative. They ferment d-sorbitol and melibiose but not sucrose. These characteristics most often make it possible to differentiate them from species that are phenotypically similar to the genus Enterobacter. Additional identifying characteristics are shown in Table 1. No isolate of human origin has been described except from a pseudobacteremia (87). However, Brenner et al. believed that this species could be involved in clinical practice because of the phenotypic proximity of these two species to E. cloacae (5); strains of clinical origin could be reported under that name. Hoffmann cluster X is E. nimipressuralis, and it has been suggested that it be reclassified as Lelliottia nimipressuralis by MLSA (3).
Enterobacter aerogenes
Enterobacter aerogenes phenotypically and genotypically resembles a mobile Klebsiella pneumoniae strain because of its peritrichous, ODC-positive, urease-negative, and indole-negative flagella (Table 1). Izard et al. proposed to reclassify E. aerogenes as Klebsiella under the name Klebsiella mobilis (88). While it is taxonomically justified (by DNA-DNA hybridization), this proposal has not been accepted by medical microbiologists, who maintain the name E. aerogenes. From genome sequencing performed on a resistant clinical isolate, Diene et al. (37) have recently proposed the shift of the E. aerogenes species to the genus Klebsiella as K. aeromobilis. The successive acquisition of additional genes from genetic mobile elements and other species, which are efficiently integrated and translated, contributes to its notable phenotype (37). Interestingly, several genes involved in the bacterial mobility could have been borrowed from the Serratia genus, and the conjugative plasmid also could have been constructed from various transposons or genetic mobile elements (37).
E. aerogenes has been regularly involved in nosocomial infections since 1992, particularly in Western Europe (9, 13, 16, 89–92). In 2012, in France, E. aerogenes represented the fifth enterobacterium responsible for nosocomial infections and the seventh Gram-negative bacillus. Its prevalence has fallen sharply since 2000 (21). Although Enterobacter cloacae is now the Enterobacter sp. most frequently isolated in clinical settings and the species expressing the widest panel of new β-lactamases or carbapenemases, E. aerogenes more easily leads to septic shock in infected patients, is associated with higher mortality (39% of patients), and shows greater virulence (93, 94).
Strains of E. aerogenes have a broad ability to acquire antibiotic resistance mechanisms (95). They possess a low-level natural AmpC chromosomal cephalosporinase (Bush group 1) that induces resistance to first-generation cephalosporins. The hyperproduction of chromosomal AmpC in clinical strains leads, after induction by the third-generation cephalosporins or after mutation, to resistance to all β-lactams except cefepime, cefpirome, and carbapenems (96). The plasmid-borne AmpC cephalosporinase gene (blaCMY-10) gives the same phenotypes (97). In 1993, the first cases of nosocomial infections caused by ESBL-producing strains were observed. In 1998, Pitout et al. isolated ESBL-producing strains resistant to gentamicin and co-trimoxazole (98). Various ESBLs were identified as being in the TEM, SHV, and CTX β-lactamases families, but TEM-24 remains associated with the preferential conjugative plasmid of this species (60, 99–104). In these producer strains, the sensitivity to carbapenems is generally preserved. In parallel, a number of imipenem-resistant clinical strains have been described (61, 93, 95, 99). In these isolates, the lack of antibiotic penetration is associated mainly with a modification of the porin expression: an alteration of the Omp35/Omp36 balance is detected and then followed by a total defect of the porins in the strains collected during the treatment (61, 93, 105–108). Interestingly, an original mechanism of impermeability has been reported, with the presence of a mutation in Omp36 that strongly alters the channel properties (106, 107). Finally, since 2008, carbapenemases of the IPM, NDM, or KPC type have been described as responsible for carbapenem resistance (95). Moreover, approximately 40% of multidrug-resistant (MDR) clinical strains have active efflux (108). Resistance to quinolones is due to modification of the target or due to plasmid-borne resistance (qnrS or qepA, encoding an efflux pump) transmitted by other species. Finally, total resistance is not an exceptional phenotype in E. aerogenes, since a strain resistant to all antibiotics, including colistin by mutation of pmrA, has been isolated and studied (37, 109).
Enterobacter gergoviae
Enterobacter gergoviae was described for the first time by Richard et al. in 1976 using a multidrug-resistant hospital strain isolated in Gergòvia near Clermont-Ferrand, France (110). Its classification was confirmed in 1980 by Brenner et al. after a DNA-DNA hybridization study (5, 111). Recently, a suggestion was made to include this species in the genus Pluribacter as P. gergoviae, based on the sequence analysis of four genes according to MLSA (35). The strains are generally LDC and ODC positive and gelatinase negative. They are urease positive and do not ferment inositol, d-sorbitol, and mucate, characteristics that differentiate them from E. aerogenes. Unlike other Enterobacter bacteria, E. gergoviae does not grow in potassium cyanide broth. Additional characteristics are presented in Table 1. Enterobacter gergoviae is rarely clinically isolated and has been exceptionally resistant to antibiotics (5, 35, 45, 112). Recently, however, ESBL type SHV and carbapenemase type (IMP or KPC) producers were described in this species (113). With regard to biocides, due to membrane modifications, esterase production, and the modulation of enzymes involved in oxidative detoxification, this species has a natural resistance to the parabens, triclosan, and methylisothiazolinone-chloromethylisothiazolinone (MIT-CMIT), which are preservatives used in this type of product (114, 115). Such results explain the ability of this species to contaminate cosmetics from a source probably of unknown plant origin (116, 117).
Recent Species Descriptions
Enterobacter bugandensis, E. timonensis, E. massiliensis, E. chengduensis, E. sichuanensis, and E. roggenkampii were recently described based on a computational analysis of sequenced Enterobacter genomes or MLSA of housekeeping genes (20, 40, 41, 118, 119). E. bugandensis was responsible for a 3-month outbreak of septicemia in a neonatal ward in Tanzania and was also identified from the environment of the International Space Station and studied for its MDR phenotype (4, 118). On the basis of whole-genome sequencing, this species was found to be phylogenetically close to E. hormaechei. It is capsule-forming and motile, and its biochemical properties are presented in Table 1. All strains harbored a blaCTX-M-15 gene and were resistant to quinolones, tetracycline, and sulfamides.
E. timonensis and E. massiliensis (characterized in the Timone Hospital laboratory, Marseille, France) were described on the basis of mass spectrometry (MS) and 16S rRNA DNA-DNA hybridization among strains isolated from the gut microflora of patients from Africa (40). These isolates were not associated with human infections. Enterobacter chengduensis and E. sichuanensis were isolated in China from a human blood sample and from urine, respectively, and considered to be new species due to particular phenotypic characteristics and by phylogenetic analysis using MLSA (4, 119). E. roggenkampii (rog.gen.kampʹi.i. N.L. gen. m. Roggenkamp) was named in honor of Andreas Roggenkamp, a German bacteriologist who helped with the understanding of the phylogenetic structure of the E. cloacae complex. This creates a novel clade of the E. cloacae complex, on the basis of cluster determination within the E. cloacae complex using hsp60 genes as marker genes (41). Enterobacter roggenkampii sp. nov. is the type strain for Hoffmann cluster IV.
EPIDEMIOLOGY AND GLOBAL SPREAD
Environmental Sources
Members of the genus Enterobacter are environmental organisms and opportunistic pathogens of plants and humans. They are commonly found in water, sewage, soil, plants, or animal feces (120). E. amnigenus biogroups 1 and 2 have been isolated from drinking water, surface water, and wild soils. E. asburiae has been isolated from water, soils, plants, food, hospital environments, and health care staff equipment, such as probes, catheters, etc. (121). E. cloacae has been isolated from food, especially from samples of formula containing plants, raw vegetables, and roots, as well as from drinking water (45, 122). Dugleux et al. described an outbreak of E. cloacae septicemia in a hospital due to the contamination of parenteral nutrition preparations stored in a refrigerator (123). Similarly, outbreaks due to human albumin flasks (124), humidifiers and respiratory therapy equipment (125), and hydrotherapy water in a burn unit (126) have been described.
E. gergoviae has been isolated from the environment, from fruits and vegetables, from various sterility controls, and in batches of various types of cosmetic products (48, 110, 116, 117). The reservoir of E. gergoviae is unknown but could be associated with a plant biotope. Numerous Enterobacter species are endophytic bacteria and are present in plant rhizospheres.
Human Reservoirs and Hospital-Acquired Infections
E. amnigenus has been isolated from respiratory tract samples, wounds, and stool, as well as from a catheter and a series of blood cultures in a patient undergoing cardiac transplantation (43). E. cancerogenus has been found in specimens of cerebrospinal fluid, blood, osteomyelitis, bile, tracheal secretions, and urine, for which its clinical significance has been proven. It has also been isolated from skin specimens, without evidence that E. cancerogenus was responsible for the infection (49, 50, 127–129). In 1997, Abbott and Janda, reported 5 cases of E. cancerogenus infections as bacteremia in patients with significant injuries or trauma (129). In one of them, E. cancerogenus was isolated for 2 months in the liquid drainage.
E. asburiae has been isolated from urine, respiratory samples, blood, stool, wounds, skin, and gallbladder (52). E. cloacae is present in the normal flora of the human gastrointestinal tract. This species is very often isolated in samples of clinical origin: urine, sputum, and blood culture (130, 131). Currently, the bacterium is found frequently during systematic sampling of neonates who have been colonized early (132). It is involved in 10% of postsurgical peritonitis cases, 5% of nosocomial sepsis and pneumonia cases, and 4% of urinary tract infections (3). Fata et al. reported a fatal case of myositis in a neutropenic patient (133). E. cloacae has been implicated in cases of endophthalmitis (134), brain abscess (135), meningitis (136), spondylodiscitis (137), and endocarditis (138).
E. hormaechei has been isolated from wounds, sputum, and blood cultures. In 1997, Wenger et al. (19) reported an outbreak in 1993 in an ICU involving 10 premature infants and including 5 cases of bacteremia.
Clinical strains of E. kobei have been isolated from various clinical samples: blood, sputum, urine, and especially intra-abdominal samples. Recently, a nosocomial bloodstream infection outbreak occurred in a neonatal ICU in a Venezuelan hospital and was caused by Enterobacter ludwigii coharboring three different ESBLs (84). E. aerogenes is quite frequently isolated in human samples (respiratory, urinary, blood, abscess, gastrointestinal, or cutaneous samples) or from materials such as ureteric stents. The species is isolated particularly from patients hospitalized in ICUs (15, 22, 23, 109, 139). The spread from patient to patient due to inadequate attention to infection control measures, especially handwashing, represents the main risk factor (16). Particular infections were described as endocarditis, endophthalmitis, and postneurosurgical meningitis (140).
Finally, E. gergoviae was isolated from respiratory samples, wounds, blood cultures, stools, and urine. Except for its involvement in a nosocomial epidemic of urinary tract infections in France in 1976 and an epidemic of bacteremia in newborns following the contamination of a parenteral glucose solution, sporadic cases have been described (141), such as pulmonary pneumopathy (142).
CLINICAL ASPECTS
Pathogenicity
Little is known about the pathogenicity and virulence factors of Enterobacter spp. due to the paucity of studies in this area. Like other enterobacteria, they possess a flagellum. In addition to facilitating motility, flagella possess several other functions: biofilm formation, protein export, and adhesion (143). Enterobacter spp. also possesses different endotoxins (12). Barnes et al. observed that Enterobacter species strains secreted in vitro enterotoxins, alpha-hemolysins, and cytotoxins similar to Shiga-like toxins II, “thiol-activated pore-forming cytotoxins” (144, 145). In Gram-negative bacteria, the type III secretion system (TTSS) is recognized as a pathogenicity factor. One study showed that 27% of E. cloacae isolates from clinical infections possessed this factor (10). The E. cloacae complex strains may also induce apoptosis of Hep2 cells (146). The acquisition of the plasmid pQC described by Paauw et al., containing virulence-encoding (ter and sea) and resistance-encoding (blaCTX-M-9, qnrA1, aadB, aadA2, sukK, and sat) genes, contributes to the virulence and adaptation of E. cloacae clade 1 (10). Additionally, E. hormaechei has been reported to be more virulent than other species due to the presence of a high-pathogenicity island (HPI) that is frequently detected on its chromosome.
The ability of bacteria to assimilate iron through chelators is important for bacterial metabolism and for the establishment of infection. The siderophore-encoding genes are generally observed in HPIs, especially in Yersinia spp. Among these genes, the irp2 gene has been identified in Enterobacter spp. (147). Finally, E. cloacae complex strains can harbor curli-encoding genes involving host cell adhesion and invasion. A recent study showed that 78% of the clinical strains studied (n = 11) had the csgBA operon (which encodes curli). The authors observed a significant correlation between biofilm formation by these strains and gene expression of csgA (coding for the main subunit of curli, curlin) and csgD (coding for an activator of the operon) (148).
In the genus Enterobacter, some differences in pathogenicity could be noted between E. aerogenes and E. cloacae (149). Azevedo et al. reported the presence of virulence-encoding genes in E. aerogenes that have been identified in Klebsiella pneumoniae (149). For instance, the fimH and mrkD genes, encoding adhesins of type 1 and type 3 fimbriae, and ycfM are detected, and they play a key role in bacterial adhesion and in biofilm formation, which are important aspects of bacterial virulence (150). Regarding iron transport, kfu, entB, and ybtS, genes that are involved in the production of siderophores, are identified in E. aerogenes (151). In this regard, it is important to note that Kfu is often detected in hypervirulent K. pneumoniae strains, and the allS gene, which is involved in allantoin metabolism, is closely associated with K. pneumoniae isolates detected in liver abscesses. Finally, the virulence of TEM-24-producing E. aerogenes was evaluated in the Caenorhabditis elegans model (93). A significant reduction of E. aerogenes virulence was observed in resistant strains that have modifications of membrane permeability involved in drug resistance. This difference is noticeable even though this species exhibits a moderate virulence in this model, although the studied strains harbored the HPI virulence factor-encoding genes. The alteration of outer membrane (OM) permeability, e.g., a lack of porins that are a prominent entry pathway for nutrients, has an important impact on bacterial fitness. The antibiotic pressure promotes the emergence of resistant strains having porin deficiency and lipopolysaccharide (LPS) modifications that generate a nonphysiological membrane state (29). This causes an unfavorable fitness cost that consequently alters the infection/colonization capability (93).
Characteristics of Affected Patients
Enterobacter spp. are involved in nosocomial infections and especially in ICUs, where they affect immunocompromised patients, such as neonates, premature infants, patients with diabetes mellitus, burned or multiply traumatized patients, and patients with leukemia or who are undergoing immunosuppressive therapy. Invasive procedures, such as catheterization and intubation, which are frequently found in an ICU, represent a main source of infection (99, 152–155). The patients also harbored numerous comorbidities with a high Charlson score. Among them, diabetes mellitus and its main complications (chronic vascular and renal diseases) represent a risk factor for Enterobacter infection (152). Finally, Enterobacter spp. preferentially affect patients with a long median duration of hospitalization. This time increases the digestive carriage, which represents a high-risk factor for transmission (156, 157). A persistence of digestive carriage over at least a 5-year period was demonstrated (158).
Frequently, the acquisition of Enterobacter spp. concerned MDR strains. Nosocomial acquisition and the median number of antibiotics used represent risk factors for these bacteria (159). An ICU stay of >14 days, presence of a tracheostomy, prior central venous catheter use, prior receipt of mechanical ventilation, and previous exposure to broad-spectrum antibiotics or any antibiotic during the 30 days before the infection were also associated with acquisition of this MDR (160, 161). Due to the immune context of the patients and the high rate of multidrug resistance, the presence of Enterobacter spp. in the bloodstream represents a high risk of mortality (152, 154, 162).
Clinical Manifestations
Among Enterobacter spp., E. aerogenes and the E. cloacae complex have been described in various nosocomial outbreaks that correspond to more than 5% of the bacteremia acquired in the hospital, 5% of pneumonia, 4% of UTIs, and 10% of postsurgical peritonitis cases (12). Enterobacter spp. are involved in numerous infections, including cerebral abscess, pneumonia, meningitis, septicemia, and wound, urinary tract (particularly catheter related), and abdominal cavity/intestinal infections (24, 163). These species are especially described in ICUs, as previously mentioned, and have also been involved in sepsis occurring in neonatology (164, 165). Moreover, E. hormaechei has also been identified in intravascular device-related infections, in surgical site infections (primarily postoperative in orthopedic trauma or related to devices), and, notably, after organ transplants (160, 166–171).
Within the E. cloacae complex, the most isolated species are E. hormaechei (clusters VIII and VI), with 40% to 48% of strains isolated, followed by E. cloacae cluster III, with 25% to 42% of strains being isolated (11, 34, 172). In 2012, Kremer and Hoffmann were interested in the types of infections caused by the different species of the group (172). They studied 196 strains which had been isolated from various samples: blood cultures; catheters; pleural fluids; and respiratory, urogenital, digestive, and cutaneous samples. In blood cultures, E. hormaechei subsp oharae (cluster VI) was significantly more prevalent, whereas E. asburiae was not represented at all. On the other hand, E. asburiae was more frequently isolated in respiratory samples than in samples from other sites. E. hormaechei subsp. steigerwaltii (cluster VIII) was overrepresented in skin injury swabs, particularly in burns. No clonality relationship was identified between the strains. Paauw et al. studied 156 strains and similarly showed that clade 1 was significantly more involved than clade 2 in infections, suggesting that this clade had greater pathogenicity (34). This clade is more common in the nosocomial environment, and its implication in infections could be the result of a better adaptation in this environment rather than a higher pathogenicity. This hypothesis is supported by the detection of the pQC plasmid in clade 1 but not in clade 2 species (10). Reports of several outbreaks of sepsis in neonatal ICUs in Brazil and the United States have been reported, with E. hormaechei being implicated (165). In 2016, Akbari et al. studied 50 Enterobacter strains isolated from UTIs (7). Twenty-five were part of cluster VI, 9 part of cluster III, and 6 part of cluster VIII. Clusters IV, X, XII, and XIII were absent (7). In 2009, the first study on the involvement of E. cloacae specifically in infected orthopedic implants was published (11). Fifteen strains (71%) belonged to E. hormaechei (5 of cluster VI and 10 of cluster VIII), 2 (9%) to E. cloacae cluster III, and 2 (9%) to E. ludwigii (cluster V). E. cloacae subsp. cloacae and E. asburiae were identified only once, and other species were not observed. The authors found a significant difference between the prevalence of cluster III in this type of infection and that of cluster III in the other samples. Cluster III was less commonly present in infected orthopedic implants compared to their overall distribution. In addition, in hip prosthesis samples, only E. hormaechei was isolated (9 out of 9). The authors hypothesized that different species would be implicated in different infections. Finally, E. cloacae was one of the most prevalent species isolated from diabetic foot infections using a culturomics approach (173). This result confirms its role in wounds and bone infections.
First-Line Antibiotics and Treatment
As infections due to Enterobacter are mainly nosocomial, most isolates present a broad resistance to third-generation cephalosporins, penicillins, and quinolones due to previous treatment of infected patients located in the same or next hospital ward. Some antibiotics remain effective for treatment: for instance, among the beta-lactams, the fourth-generation cephalosporins and carbapenems are the most attractive options, even if limiting carbapenem use should be encouraged, and the aminoglycosides have a good activity.
The use of third-generation cephalosporins and the monobactams (e.g., aztreonam) represents an important risk of in vivo derepression of AmpC β-lactamases, which can be due to a mutation in the repressor, during the treatment inducing high-level resistance to many β-lactam antibiotics. The interest in the concomitant use of aminoglycoside to prevent this type of resistance is mixed (9, 174). The use of fourth-generation cephalosporins (e.g., cefepime and cefpirome) seems to be more effective, mostly due to their activity against AmpC-hyperproducing Enterobacter strains (175). Compared to older cephalosporins, these molecules present (i) an efficient diffusion across the outer membrane porins, (ii) a significant stability against chromosomal β-lactamases, and (iii) an enhanced affinity for key penicillin-binding proteins located in the Enterobacter periplasm (176, 177). Many publications have demonstrated interest in them (178, 179).
Carbapenems are very efficient against a wide variety of enterobacteria (180). AmpC-overproducing Enterobacter spp. typically remains susceptible to carbapenems. However, the use of carbapenems could induce a loss of porin production and an impermeability of the bacteria (see “Membrane-associated mechanisms” in the section Antimicrobial Resistance) (152). This resistance remains rare to date.
Recently, the use of the piperacillin-tazobactam combination has been observed to be a valuable treatment option for bloodstream infections due to Enterobacter spp. (178). Different new antibiotics have been tested against Enterobacter spp. The novel siderophore cephalosporin cefiderocol shows excellent results against these bacteria (181). Different combinations of cephalosporins and β-lactamase inhibitors (cefepime-zidebactam, cefepime-tazobactam, ceftolozane-tazobactam, ceftazidime-avibactam, etc.) also show high antibacterial efficacy against these pathogens (182–184). However, their use is not encouraged as a first approach because of the aim to limit the emergence of bacteria resistant to these new antibiotic solutions.
Finally, aminoglycosides, and particularly amikacin, remain active in more than 95% of Enterobacter spp. These rates have been stable over time. In Enterobacter, the aminoglycoside resistance is usually due to the presence of a plasmid coding for aminoglycoside-modifying enzymes (185).
ANTIMICROBIAL RESISTANCE
Development of Antimicrobial Resistance
Among Enterobacter spp., E. cloacae and E. aerogenes are mainly affected by development of antimicrobial resistance (14, 186). Regarding E. aerogenes, β-lactam uptake is closely associated with the presence of general porins, such as Omp35 and Omp36, which are homologous to the OmpC and OmpF porins that are the archetypes of the general nonspecific enterobacterial porins (187–190).
Several publications have described a modification of the porin pattern present in antibiotic-resistant isolates: resistant E. aerogenes can exhibit a shift in the type of porin expressed (Omp35 to Omp36), a reduction in the production level, or the synthesis of a porin exhibiting mutations in the porin structure that alter channel functions (for reviews, see references 29 and 188). These interplays of membrane impermeability and enzymatic barriers were first mentioned in H. Nikaido 's model (191, 192).
Consequently, the clinical isolates collected in the patient during antibiotherapy show a serious loss in susceptibility to cephalosporins and carbapenems (193, 194). This alteration of porin profiles is also often reported with a concomitant synthesis of degradative enzymes such as β-lactamases, cephalosporinases, or carbapenemases, which generate a worrying level of β-lactam resistance (193, 195–197).
Moreover, the dissemination of resistance genes via genetic mobile elements is an important aspect of the antibiotic resistance in Gram-negative bacteria in the ESKAPE group (14, 198).
Molecular Mechanisms of Resistance
A summary of the varieties of acquired resistance described in E. cloacae complex bacteria is given in Table 2.
TABLE 2.
Acquired resistances described in Enterobacter cloacae complex bacteria
| Antibiotic group | Mechanism of resistance | Gene(s) | Species | Reference(s) |
|---|---|---|---|---|
| β-Lactams | Enzymatic β-lactamases | |||
| Class A | blaTEM, blaSHV, blaCTX-M | E. cloacae | 63, 315 | |
| E. hormaechei | 270 | |||
| blaVEB, blaGES/IBC, blaKPC, blaFRI | E. cloacae | 210, 316 | ||
| E. hormaechei | 317 | |||
| blaNMCA, blaIMI | E. cloacae | 318 | ||
| E. asburiae | 54 | |||
| Class B | blaVIM, blaGIM | E. cloacae | 69, 319, 320 | |
| blaNDM | E. ludwigii | 84 | ||
| blaIMP, blaNDM | E. cloacae, E. hormaechei | 209, 321 | ||
| Class C | ampC | E. cloacae | 288, 295 | |
| E. asburiae | 3 | |||
| E. hormaechei | 213 | |||
| E. kobei, E. ludwigii, E. nimipressuralis | 163 | |||
| Class D | blaOXA-48 | E. cloacae | 320 | |
| Impermeability/efflux | acrAB-tolC, ompC, ompF, ompX | E. cloacae | 25, 31, 240, 241, 263, 322 | |
| Fluoroquinolones | Target mutation | gyrA, gyrB, parC, parE | E. cloacae | 291 |
| Enzymatic (acetyltransferase) | aac(6′)-Ib-cr | E. cloacae | 148 | |
| Protective mechanism target | qnr (A, B, S, C, D) | E. cloacae, E. hormaechei | 323 | |
| Efflux | qepA, acrAB-tolC, oqxAB, sugE, emmdR | E. cloacae | 258, 259, 263 | |
| Aminoglycosides | Enzymatic (acetyl, phospho, nucleotidyltransferase) | aac, aph, ant | E. cloacae | 323 |
| Methylase | armA, rmtB | E. cloacae | 324 | |
| Cyclines | Efflux | acrAB-tolC | E. cloacae, E. hormaechei | 325 |
Enzymatic barriers and epidemiology.
In most Enterobacter spp., the production of β-lactamases is the prominent mechanism responsible for β-lactam resistance, and E. aerogenes and E. cloacae have a broad ability to modulate these mechanisms of resistance. Importantly, these bacteria are able to produce a low level of a chromosomal AmpC β-lactamase-type cephalosporinase that generates a resistance to first-generation cephalosporins (24, 196). The chromosomally acquired resistance promotes the overproduction of this AmpC cephalosporinase, for instance, during incubation with a subinhibitory concentration of carbapenem (199). Following the inactivation of AmpR or the acquisition of a plasmid-borne ampC, an overproduction of AmpC β-lactamase contributes to the resistance toward the third-generation cephalosporins (24, 200–202). E. aerogenes is also able to integrate a large plasmid (130 kb) that contains the ampC gene of chromosomal origin (blaCMY-10). In the absence of antibiotic pressure, this genetic transmission can contribute to a systematic spreading of the resistance mechanism (203). This AmpC-related resistance, described in 50% of clinical isolates, is frequently associated with the expression of ESBLs (24).
The first hospital-acquired infections caused by these strains that exhibit resistance to common β-lactams due to the expression of ESBLs were reported in 1993 (98). The TEM-24 enzyme was associated with E. aerogenes clonal dissemination in hospitals in France (8, 23, 102, 103). Other TEM types or CTX-M types (e.g., CTX-M-2) are also often reported, but TEM-24 remains associated with preferential conjugative Enterobacter plasmids (10, 24, 73, 99, 100). These enzymes contribute to a global resistance toward all β-lactams except carbapenems (62). In Enterobacteriaceae, E. cloacae is now identified as the third most common bacterium resistant to third-generation cephalosporins, after enteric E. coli and K. pneumoniae (204). Different enzymes (ESBLs) belonging to the TEM, SHV, and CTX-M classes have been characterized in E. cloacae, and these also include resistant TEM inhibitors or inhibitor-resistant TEM (IRT) enzymes. Notably, some variants that exhibit CTX-M production have been identified, and others, such as TEM or SHV, have been described from epidemic episodes (24). A transfer of a genomic resistance island is also possible in Enterobacter spp. For instance, a variant of AGI1 that belongs to the Salmonella genomic island/Proteus genomic island/Acinetobacter genomic island family has been recently described in E. cloacae (205). This isolate was resistant to all the antibiotics tested except imipenem and amikacin.
Imipenem is the most effective antibiotic for the treatment of E. cloacae infections (24). Carbapenemases that belong to the NDM and VIM types have been identified in E. aerogenes and E. cloacae. KPC or class D β-lactamases possessing carbapenemase properties, such as the OXA-48 type, have been identified in Europe, Asia, and America (24, 206, 207). In 2010, the CDC reported the first carriage of NDM-1-producing E. cloacae in patients previously treated in India (208, 209). Recently, metallo-β-lactamases that comprise IMP-type enzymes and NDM-, GIM-, VIM-, and serine-carbapenemase-type KPC and FRI have been characterized (24, 210, 211). OXA-48-type serine carbapenemase seems to be the most prevalent (206, 212). In E. cloacae, an increase in the rate of imipenem resistance has been observed. Moreover, an epidemic survey of E. cloacae blood infection reported the presence of metallo-β-lactamases (24). E. cloacae is the third Enterobacteriaceae in the production of carbapenemase, and several strains have been described to simultaneously express two carbapenemases (213). A report mentioned that the KPC enzyme was the most frequently identified in the SMART global surveillance program from 2008 to 2014 (214). This KPC prevalence was also reported in another study, indicating a limited emergence of NDM-1 (215). In contrast, a longitudinal study (2013 to 2017) performed in China indicated that an NDM producer is predominant in E. cloacae (216). Moreover, the carbapenem resistance in the E. cloacae complex had noticeably increased in a recent study of carbapenem susceptibility performed by the U.S. Veterans Health Administration from 2006 to 2015 (217). A recent publication based on genomic epidemiology of carbapenemase-producing Enterobacter spp., comprising predominantly E. xiangfangensis and E. hormaechei isolates, reports that the most common enzyme identified is VIM, followed by NDM, KPC, OXA-48, and IMP (218). Finally, an association of carbapenemase production with a loss of porin expression has been demonstrated in strains resistant to combinations of β-lactamase inhibitors (relactam) plus carbapenems (219).
Regarding aminoglycoside resistance, aminoglycoside-modifying enzymes are distributed among acetyltransferases (aminoglycoside-N-acetyltransferases [AACs]), phosphotransferases (aminoglycoside-O-phosphotransferases [APHs]), adenylyltransferases (aminoglycoside adenylyltransferases [AADs] or aminoglycoside-O-nucleotidyltransferases [ANTs]), and 16S rRNA methyltransferases, such as ArmA and RmtB (70). They are often plasmid encoded or associated with transposable elements, which facilitate the acquisition of resistance phenotypes (163).
The aminoglycoside-modifying genes, e.g., aac(3)-IIa, aac(6')-Ib, and ant(2″)-Ia, are involved in aminoglycoside resistance affecting, at different levels, tobramycin, gentamicin, and amikacin. The clinical strains frequently contain more than one enzyme (70, 185, 220). The enzymatic resistance to fluoroquinolones has been characterized as a two-point mutation allele of aac(6′)-Ib [named aac(6′)-Ib-cr], the aminoglycoside resistance enzymatic determinant, which became able to acetylate ciprofloxacin and norfloxacin (70, 221). The association with the blaOXA-1 gene in various genetic mobile elements contributes to a rapid spreading of this new mechanism (24).
Membrane-associated mechanisms.
Numerous imipenem-resistant clinical strains have been described, and these present a severe alteration of porin expression associated or not with the overexpression of efflux pumps that occurs during antibiotic therapy (24, 29).
Membrane-associated mechanisms of resistance, including porin defects and increased levels of efflux pumps, are now recognized to strongly participate in the MDR phenotype by controlling the internal concentration of antibiotics (29). These “concentration barriers” can also induce the emergence/induction of other mechanisms, such as target mutations (e.g., mutated gyrase) or the expression of detoxifying enzymes, including β-lactamases (193). Interestingly, the alteration of LPS also has been described in many resistant isolates (222, 223).
(i) Omps, porins, and OM permeability.
OmpA was first reported and characterized in 1983, and the Tsx channel involved in nucleoside uptake was reported in 1997 in E. aerogenes (224, 225). Today, three general (nonspecific) porins, Omp35, Omp36, and Omp37, have been identified in Enterobacter spp., and two additional specific porins, LamB and PhoE, have been identified and exhibit some similarities with E. coli OM proteins that have been largely studied (for reviews see references 187–189, and 226). Importantly, due to their specific trimeric organization in the membrane, these OM proteins need important posttranslational steps that (i) perform the maturation of precursor forms, (ii) correctly insert the protein into the OM, and (iii) manage the rate and dynamics of the final trimeric assembly of newly synthesized proteins (227–229). Like the archetypes OmpC and OmpF, Omp35 and Omp36 are assembled in stable trimers, and each subunit contains a hydrophilic channel presenting a typical β-barrel structure organizing an internal eyelet that constricts the lumen and creates a strong transverse electric field guiding the diffusion of charged molecules. Recently, the three-dimensional (3D) structures of E. aerogenes and E. cloacae porins have been solved, and the trimeric structures have been published. Their structures exhibit a high sequence identity, and their channel properties, e.g., conductance and selectivity, which are determined by their planar lipid bilayers, are very similar; for instance, the transversal electric field located in the lumen of the channel is well conserved in the OmpF group, which is more permeable to anionic compounds (230). Regarding the OmpC group, which includes Omp36, OmpE36, and OmpK36 for E. aerogenes, E. cloacae, and K. pneumoniae, a smaller lumen of the pore, a lower conductance (approximately 3 nS), and a higher cation selectivity (with a PK+/PCl− ratio of approximately 2.1 to 2.2) are obtained compared to those obtained from the OmpF orthologs (230). Importantly, the channel is able to promote antibiotic travel across the OM and ensure accumulation inside the periplasmic space, as demonstrated with purified Omp36 or by using intact cells and labeled antibiotics (231, 232). Moreover, immunological and functional comparisons of E. aerogenes and E. coli porins have reported some conserved and variable features in the antigenic profile and in the reception/translocation functions for bacteriocins (233). This illustrates an adaptive evolution of specific exposed domains when the pore activity is preserved (230) (Table 3).
TABLE 3.
Some examples of the membrane proteins identified in Enterobacter spp.
| Bacterium | Protein | Function | Characteristics | Reference(s) |
|---|---|---|---|---|
| E. cloacae | OmpE35 | General porin | Trimer | 230, 233 |
| OmpE36 | General porin | Trimer | 230, 233 | |
| OmpE37 | Quiescent porin | Trimer | ||
| OmpX | Monomer | 240 | ||
| OmpA | OM architecture | NDa | ||
| LamB | 326 | |||
| PhoE | 242 | |||
| EmmdR | IM transporter | 258, 259 | ||
| SugE | IM transporter | 258, 259 | ||
| TolC | OM channel | Trimer | 254 | |
| AcrA | Adapter | ND | 254 | |
| AcrB | IM transporter | Trimer | 254 | |
| E. aerogenes | Omp35 | General porin | Trimer | 230, 243 |
| Omp36 | General porin | Trimer | 230, 243 | |
| Omp37 | Quiescent porin | Trimer | C. Bornet, unpublished data | |
| OmpX | Small channel | ND | 239 | |
| OmpA | OM architecture | Monomer | 199, 224 | |
| LamB | Maltoporin | Trimer | 238 | |
| PhoE | Phosphoporin | ND | ND | |
| Tsx | Nucleoside uptake | ND | 225 | |
| TolC | OM channel | Trimer | 253 | |
| AcrA | Adapter | ND | 253 | |
| AcrB | IM transporter | Trimer | 253 | |
| EefC | OM channel | Trimer | 327, 328 | |
| EefA | Adapter | ND | 327, 328 | |
| EefB | IM transporter | Trimer | 327, 328 |
ND, not determined.
Similarly, to the case for E. coli porin expression (28, 234, 235), the regulation of these OM general channels is sophisticated in Enterobacter spp., and several models have been proposed to integrate not only the Mar and Ram regulation cascades but also the two-component system (TCS) regulator pathways that are directly involved in the sensing, transmission, and control of porin transcription, translation, and assembly (24, 29, 193). These different means of regulation are involved in the Enterobacter response and adaptation to the presence of antibiotics; they represent the first barrier and the main lever for controlling the penetration flux and accumulation level of antimicrobials [see “(iii) Efflux pumps and antibiotic activity” below].
(ii) Porins and antibiotic activity.
Interestingly, a pioneer study has determined that the porin-deficient phenotype is present in approximately 6 to 7% of the β-lactam-resistant E. aerogenes isolates collected during a 1-year period (104, 236). During antibiotic treatment of patients, a sequential alteration in the balance between Omp35 and Omp36 has been reported: from the expression of Omp36 and 35 in the carbapenem-susceptible isolate, to the Omp36-producing strain lacking Omp35 and exhibiting intermediate susceptibility, and finally to a carbapenem-resistant isolate having no porins (93, 237). Furthermore, LamB porin can be expressed in place of Omp35 and Omp36, generating a low susceptibility to β-lactams (238). In some isolates, OmpX, a small OM protein, is involved in the downregulation of porin expression that is associated with a decrease of antibiotic susceptibility (239–241). An E. cloacae PhoE porin has been purified and characterized; however, no information has been obtained regarding its role in clinical strains (242). Resistant isolates collected from patients who received antibiotherapy were extensively studied with regard to their levels of porin expression and pore activity (106, 107, 199, 238, 242). The loss of porins has been reported in many studies carried out on E. cloacae and E. aerogenes clinical isolates, and due to space limitations, only a limited part of the published studies are indicated here (10, 200, 219, 243–248).
Importantly, a key mutation has been identified in a resistant isolate: this mutation, causing a Gly-to-Asp change located in the eyelet region of the Omp36 lumen, induced a strong modification inside the channel conformation, causing alteration of both conductance and selectivity. Consequently, the mutated porin promotes a noticeable resistance to β-lactams but preserves a limited nutrient permeation through the porin (106, 107). This selected “porin strategy” maintains a minimal cost fitness for the bacterial cell associated with a solid decrease of antibiotic diffusion contributing to the resistance (249). Interestingly, as previously mentioned, the porin loss has an impact on the pathogenicity of E. aerogenes isolates, which become less virulent in a Caenorhabditis elegans model (93). This adverse effect of resistance mechanisms on K. pneumoniae virulence has been extensively analyzed in the case of OmpK36 and OmpK35 in a recent review (250, 251). Moreover, similar observations have been reported for E. coli (252), and we may hypothesize that the porin expression is necessary for some important steps involved in virulence or contributes to the envelope stability during colonization or virulence.
In addition to general porins, TolC, the outer membrane channel involved in the efflux activity and secretion (RND), has been described and characterized in E. aerogenes and E. cloacae (251–253). In addition, E. aerogenes TolC and EefC have been documented and present different channel properties determined by using electrophysiology assays (254).
(iii) Efflux pumps and antibiotic activity.
Multidrug efflux pumps present on the Enterobacter genomes belong to the ABC, MF, SMR, MATE, PACE, and RND superfamilies described previously (255, 256); for a classification of membrane transporters, see reference 257 and the Paulsen site (http://www.membranetransport.org/transportDB2/index.html) (Table 3). Inner membrane (IM) transporters correspond to a single membrane protein located in the inner bacterial membrane, and they function as monomers or as dimers (255). These IM transporters pump out the drugs from the cytoplasm (or the inner leaflet of the IM) to the periplasmic space, such as reported for EmmdR or SugE in E. cloacae (258, 259). Interestingly, these IM transporters belonging to the SMR or MATE families can cooperate with the RND family in order to efficiently expel antibacterial compounds outside the bacterial cell (260). In the bacterial envelope, an RND complex, the tripartite efflux system that comprises an inner membrane transporter, a periplasmic adapter protein, and an outer membrane channel, recognizes and translocates the drugs across the OM to the external medium (255, 256, 261). These RND efflux pumps contribute to the removal of a large number of chemically diverse compounds, such as antibiotics, detergents, biocides, preservatives, etc., that are present in the bacterial volume and can be deleterious to the bacteria (255, 262).
With regard to the AcrAB-TolC pump, the complex has been identified and described in E. cloacae and E. aerogenes (253, 254, 263). Purification and biochemical characterization have been performed for TolC and EefC of E. aerogenes, and their channel properties have been documented (264, 265). Regarding the involvement of this efflux pump in the resistance of clinical strains, various publications have reported the expression of AcrAB-TolC and its contribution in the reduced susceptibility of the isolates produced (199, 237, 238, 253, 266). Finally, the protein AcrZ, which has been described in the E. coli AcrZ-AcrAB-TolC efflux pump, has also been characterized in the Enterobacter/Klebsiella genome (267, 268).
Recently, the OqxAB operon has been identified in E. cloacae and E. aerogenes strains, and this efflux pump contributes to a decreased susceptibility to quinolones in Enterobacteriaceae (269). In addition, selective efflux pumps also have been identified and described to play a role in heavy-metal resistance/tolerance in E. hormaechei and E. asburiae isolates (270).
An important point is the relevance and the prevalence of AcrAB-TolC in clinical isolates. In a study published in 2008, the evaluation of efflux activity, measured by using an efflux pump inhibitor (PAβN) in clinical isolates collected within an 8-year period (1995 to 2003), indicated a noticeable increase of efflux expression during this interval (108). Moreover, this study clearly pinpoints the importance of evaluating the prevalence of membrane barriers, e.g., impermeability due to porin loss or/and efflux expression, in clinical strains submitted to antibiotherapy treatment, as recently mentioned (265). This aspect is important when taking into account the role of AcrAB-TolC in the susceptibility of E. aerogenes to macrolides (271).
A main concern is the correlation that exists between efflux activity and intracellular accumulation of antibiotics (209). A series of publications focused on fluoroquinolone accumulation in E. aerogenes and E. coli strains expressing or not the AcrAB-TolC pump has clearly illustrated the impact of the efflux expression on the accumulation rate inside the bacterial cell (272–275). The expression of the AcrAB-TolC complex is able to maintain the internal concentration of antibiotics under the threshold required for triggering the bacterial killing (263, 276). Moreover, by using a microspectrofluorimetry method, the authors found that the fluorescence drug signal varied among the individual bacteria in a uniformly treated population (272, 273). These important data illustrate the heterogeneity of the intrabacterial accumulation of an antibiotic during early incubation times. This heterogeneity may reflect different levels of resistant phenotypes coexisting in the isogenic population due to different growth phase or division steps (272, 277). This may pave the way for identifying the bacterial adaptation and persister formation inside a bacterial population subjected to antibiotic stress (272, 273).
In addition, these RND pumps are involved in the bacterial pathogenicity and in the acquisition of additional mechanisms of resistance in Enterobacteriaceae (195, 262, 265). Using a mouse model for measuring the competitiveness and virulence of AcrAB-TolC parental or deletion E. cloacae strains, G. Bou’s group has clearly demonstrated the involvement of this pump in the bacterial physiology (278).
(iv) LPS modification and polymyxin susceptibility.
Various alterations of the OM structure are also associated with the LPS modifications in Enterobacter clinical isolates that induce some changes in polymyxin susceptibility (109, 279, 280). In some cases, the plasmid-mediated colistin resistance mcr-1 has been identified in resistant E. aerogenes and E. cloacae strains (280–283). A study reports that the overall prevalence of colistin resistance corresponds to 0.67% of the total enterobacterial isolates collected during a 4-year period. The colistin resistance was higher in E. cloacae (4.2%) than in E. coli and K. pneumoniae (0.5% and 0.4%, respectively). Although the authors reported that this resistance was not associated with the mcr genes, unfortunately, the molecular and genetic characterization of this resistance is lacking (284). In a recent study, Guérin et al. analyzed a collection of 124 strains of the E. cloacae complex and concluded that the PhoP/PhoQ TCS would play a role in colistin resistance regulation (285). This observation is similar to work performed on E. aerogenes clinical isolates collected from patients receiving imipenem (237). In a polymyxin-resistant strain, the authors identified mutations located on phoQ and pmrB, which are part of the well-described TCS controlling the LPS-modifying enzymes (237). The genome of the colistin-resistant strain identified in 2005 has been sequenced and analyzed, and a mutation in pmrA has been identified as the cause of the alteration of LPS biosynthesis that had been previously observed (37, 109). This type of chromosomal mutation that efficiently alters the OM structure and generates a noticeable decrease of polymyxin activity may be involved in the emergence of resistant strains devoid of mcr-1 and mcr-2 (247). Moreover, the dynamic of colistin resistance in E. cloacae during selective decontamination of the digestive tract in intensive care units has been recently reported, suggesting a possible clonal transmission (286).
Mutations in antibiotic targets.
Regarding β-lactam resistance, the target mutation occurs rarely in Enterobacter spp. However, the diverse β-lactamases reported today are the result of a series of mutations that have successively appeared in the original β-lactamase genes (287). Moreover, mutations affecting the ampR gene have been described in strains where AmpC cephalosporinase is derepressed (200, 288, 289). In MDR Enterobacter isolates, multiple point sequence alterations can be found in ampC but are not in regions corresponding to the serine active site or the β-lactam-binding site and have no correlation with the resistance phenotype (246). However, amino acid deletions in the Ω loop of E. cloacae AmpC involved in enzyme competitiveness and point mutations suspected to affect the enzymatic activity were found in strains selected with ceftaroline-avibactam (290).
Regarding the mutations that contribute to antibiotic resistance, the best characterized are those that affect the quinolone target and, as found more recently, those involved in polymyxin resistance (see the previous section). In Enterobacter spp., mutations located in the quinolone resistance-determining regions (QRDRs) of targeted enzymes, e.g., gyrase or topoisomerase, have been described to confer high-level resistance (24, 237, 291–293). This is the common resistance mechanism identified in clinical isolates with recent descriptions of plasmid-mediated quinolone resistance (70, 72, 73, 294, 295). Recent studies in South Africa reported that qnr genes were commonly detected in resistant Enterobacter isolates collected in a hospital (296, 297). Interestingly, recently the qnrE1 gene was reported as probably originating from the Enterobacter chromosome (298). This “target-protective mechanism” confers low-level resistance to first-generation quinolones when present alone (72–74). Importantly, these mechanisms exhibit a noticeable spread and have been reported in approximately 60% of clinical isolates due to the presence of various genes coding for ESBLs or AmpC-type β-lactamases on the same plasmid (70, 289, 295).
Lastly, MDR has recently been described in Enterobacter isolates (E. cloacae and E. aerogenes) and in MDR-associated porin alteration, target mutation β-lactamase production, and efflux overexpression that are accumulated during antibiotic treatment (24, 237). Some mechanisms are intertwined and controlled by regulators in a complex genetic cascade.
MDR and genetic regulation.
Recently, several chemical inducers that are able to modulate the expression of Enterobacter membrane transporters, including porins and/or efflux pumps, have been described, e.g., salicylate, chloramphenicol, etc. (24, 193, 239, 299). Interestingly, the regulation of porin expression is a fast event, occurring shortly after the addition of chemicals in the culture medium or with the addition of subinhibitory concentrations of antibiotics. During incubation with low imipenem concentrations, an increase in the efflux pump expression that is mediated by the overproduction of MarA has been observed (104, 243, 299).
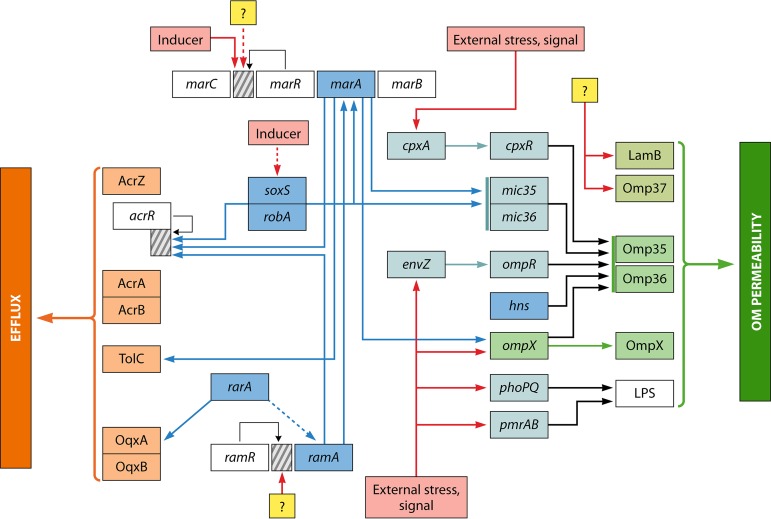
A schematic representation of the regulation pathways that control the expression of porins and efflux pumps and their interconnection in Enterobacter spp. is shown in Fig. 1.
FIG 1.
Schematic representation of the regulation pathways that control the expression of porins and efflux pumps and their interconnection in Enterobacter spp. The multiple regulation cascades that can modulate the outer membrane permeability (porins and LPS) and the expression of major efflux pumps (AcrAB and OqxAB) are summarized. Blue arrows represent transcriptional activation/repression of different genes, such as acrR and ompX, by the global regulators (e.g., Mar, Ram, and Sox). Red arrows symbolize the external stress signals that can activate/repress some gene expression. Black arrows indicate the negative regulation of gene expression (thin line directly on promoter/operator region). Yellow squares illustrate some unknown regulation that can modulate gene expression on promoter/operator regions (represented by dashed boxes) of key loci. Dashed lines represent hypothesized regulation.
(i) MarA, RamA, SoxS, and RobA.
Regarding global regulators involved in the control of antibiotic resistance, importantly, the RamA regulator has been characterized in Enterobacter, Salmonella, and Klebsiella, but it has not been detected in Escherichia, in contrast to the case for the Mar regulon (24, 26).
RamA has been detected in E. aerogenes and E. cloacae and has generated a noticeable resistance to various antibiotics (chloramphenicol, tetracycline, tigecycline, fluoroquinolones, trimethoprim, etc.), in conjunction with a decreased expression of Omp35 and an active efflux in E. aerogenes (24, 271). RamA seems to be a “superregulator” of the membrane permeability, acting directly or via MarA and controlling the influx and the efflux of antibacterial agents in Enterobacter (24, 30). In addition, rarA may also contribute to the combined regulation of the RamA-MarA cascade during the emergence of antibiotic resistance (300, 301). RarA, which belongs to the AraC-type transcriptional regulators, is overproduced when the negative regulator OqxR is inactivated (25, 302). SoxS and Rob can also play a role by sharing some information detected via other signaling systems in Enterobacter (25, 278). Regarding environmental stress, H-NS (histone-like structuring nucleoid protein) modulates the level and balance (e.g., the Omp35/Omp36 ratio) of porins in the outer membrane in response to osmotic stress (24, 25).
(ii) Other regulators.
With these global regulators, several other partners play a key role in monitoring the expression of porins: OmpX, a small OM protein, and different small noncoding RNAs (sRNAs), such as Mic35 and Mic36 (239–241). In addition, several TCSs, such as EnvZ-OmpR, and CpxA-CpxR, can regulate the expression level of porins. In parallel, other TCSs, PmrA-PmrB and PhoQ-PhoR, involved in the synthesis of LPS, which is involved in the last step of porin assembly in the OM, may also modulate the porin level in the OM. Importantly, some mutations have been identified in TCSs, such as PmrAB or CpxAB, indicating that under antibiotic treatment, clinical strains are able to select mutations that can modify the membrane permeability in order to acquire low susceptibility against antibiotics that are used (237).
Several local regulators, repressors such as acrR or rarR, play a role by controlling the expression of efflux pumps; a more detailed description of the regulators involved in the control of drug membrane transporters in Enterobacter and Klebsiella has recently been published (25), and an illustration of this complex network is presented in Fig. 1 This illustration has been constructed by using the publications on this subject and descriptions of the genes and proteins in the data bank.
(iii) Inducers and chemical effectors.
Some inducers are described as binding directly to the repressor MarR, such as salicylate and tetracycline, thereby impairing the repressor action (25). The transcriptional activators respond to a variety of chemically unrelated compounds, including antibiotics, biocides, carbonyl cyanide m-chlorophenylhydrazone, cyclohexane, salicylate, acetylsalicylate (aspirin), acetaminophen, sodium benzoate, paraquat, and phenolic rings (24). However, the exact mode of action of this induction is not yet elucidated, and further investigations are necessary to understand the targeted step in the regulation cascade (Fig. 1).
DIAGNOSIS OF SPECIES
The routine identification of Enterobacter spp. has classically been performed by an evaluation of their phenotypic characteristics by using commercialized systems: the Api 20E gallery or Vitek 2 (bioMérieux). Although these systems are adapted to differentiate E. aerogenes and E. cloacae, they are not able to differentiate the species in the E. cloacae complex except for E. cloacae and E. asburiae.
Mass spectrometry, which is increasingly used today, can identify the E. cloacae complex but fails to differentiate the species within the group. The two most commonly used devices have failures with this identification. E. nimipressuralis is identified correctly, while, on the other hand, E. hormaechei, E. cloacae, E. asburiae, E. kobei, and E. ludwigii are not discriminated (51, 303).
Molecular biology techniques are needed to identify the species precisely. Sequencing of 16S rRNA is widely used to identify Enterobacter spp., but this technology cannot differentiate two closely related species; thus, it is not applicable to the case of the E. cloacae complex (40). The technique of microarray comparative genomic hybridization (microarray-CGH) is also a powerful identification method, but this method is time-consuming, expensive, and therefore difficult to implement routinely. It showed the existence of two distinct, genetically different clades within the complex. Possibly, this method could be coupled with MLSA. The first clade is then divided into two clusters, with the second being more heterogeneous and containing five clusters. Most strains associated with infections belong to the first clade (34). Recently, MLSA employing sequencing of four to seven housekeeping genes, such as atpD, fusA, infB, gyrB, leuS, dnaA, pyrG, rplB, rpoB, and hsp60, appeared to be the most valuable tool to identify of E. cloacae complex species and the most recent Enterobacter species (4, 118). The MLSA results were strongly validated by single-nucleotide polymorphism analysis by whole-genome sequence (4). By sequencing the hsp60 gene, Hoffman and Roggenkamp discriminated 13 clusters (clusters I to XII and cluster XIII, which corresponds to an unstable sequence crowd) within the group (3, 55). More recently, Beyrouthy et al., using the same technique, confirmed the characterization of the two new subspecies of E. hormaechei obtained by complete genome sequencing and helped to complete the Enterobacter cloacae complex clusters (41, 208) (Table 4). Twelve clusters were attributed to existing species and more recently described species such as E. roggenkampii, E. bugandensis, and E. hormaechei subsp. hoffmanii (Table 4). The name E. hormaechei is sometimes given as a generic name for strains belonging to different clusters.
TABLE 4.
Distribution of clusters and species of the Enterobacter cloacae complex using the hsp60 sequencing technique (163, 208)
| Cluster | Species |
|---|---|
| I | E. asburiae |
| II | E. kobei |
| III | E. hormaechei subsp. hoffmannii |
| IV | E. roggenkampii |
| V | E. ludwigii |
| VI | E. hormaechei subsp. oharae, E. hormaechei subsp. xiangfangensis |
| VII | E. hormaechei subsp. hormaechei |
| VIII | E. hormaechei subsp. steigerwaltii |
| IX | E. bugandensis |
| X | E. nimipressuralis |
| XI | E. cloacae subsp. cloacae |
| XII | E. cloacae subsp. dissolvens |
| XIII | E. cloacae sequence crowd |
Recently, a German team implemented a combination of two techniques to identify the species within the E. cloacae complex: matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MS coupled with real-time PCR (303). Since mass spectrometry does not allow the species to be distinguished from each other, this group requires associations to be established using real-time PCR that amplifies the dnaJ gene. This gene, identified by Pham et al., encodes a chaperone protein of the Hsp40 family and is specific for the E. cloacae species (304). Precise identification of the species in the E. cloacae complex is important in the biological diagnosis of infections because species are differently implicated in human pathology (11). Phenotypic techniques can lead to misidentifications between very different species harboring different virulence potentials, such as E. hormaechei and Cronobacter sakazakii. However, a species identification error has no impact on antibiotic therapy, as the species of the E. cloacae complex have the same antibiotic resistance profiles.
CONCLUSION
Until now, only few virulence factors/genes have been described in Enterobacter. In addition, the precise regulation mechanisms and the molecular and functional properties involved in bacterial adaptation to various environmental stresses/conditions have not yet been precisely determined (10, 56, 145, 305–309).
A main biological behavior reported in the Enterobacteriaceae seems to be associated with its ability to evade the activity of a huge collection of antibacterial agents, including antibiotics, disinfectants, biocides, etc. For instance, Enterobacter possesses a versatile and sophisticated system for regulating the envelope permeability, as characterized above in clinical strains. It must be noted that the RamA operon, which is not present in Escherichia spp., plays a key role both directly and indirectly via the Mar operon: it can control the expression of porins and RND efflux complexes, and it can complement or link its proper regulation cascade to other global regulators such as SoxS or Rob. The advantage of this additional global regulator associated with the Mar regulon remains to be clarified in E. aerogenes and E. cloacae, where it can play the role of an enhancer ring or accelerate the bacterial adaptation to environmental stress and contribute to the prevalence of these Gram-negative bacteria in human infectious diseases.
The complete genome sequences of E. aerogenes and E. cloacae strains have illuminated the presence of resistance genes that contribute to the MDR phenotype, virulence, quorum sensing, and competition against other microorganisms (37, 120, 237, 310–313). Moreover, the whole-genome sequencing of clinical isolates collected during patient antibiotherapy in association with proteomic and functional studies has strongly illuminated the strategies developed by these important Gram-negative pathogens (237, 310). In this regard, the genus-level analysis and comparison of Enterobacter spp. versus K. pneumoniae indicates high genetic variation (average nucleotide diversity of 0.16 compared to 0.02) (314). This difference suggests that these individual species may have different mechanisms and evolutionary pressures that govern them. The regulation cascades involved during colonization, virulence, and resistance support physiological changes that ensure rapid and appropriate responses to environmental stresses, and this remarkable adaptability explains the presence of Enterobacter in the ESKAPE group.
The reemergence of Enterobacter as a worrying resistant pathogen is an important health concern, especially when the scarcity of new antibiotics active against Gram-negative bacteria is considered. Consequently, much effort is necessary to identify and dissect the molecular and genetic events that manage the Enterobacter adaptation and to clarify the remaining unknown aspects in the regulation cascade.
ACKNOWLEDGMENTS
The research leading to the discussions presented here was conducted as part of the TRANSLOCATION consortium and has received support from the Innovative Medicines Initiatives Joint Undertaking under Grant Agreement no. 115525, resources which are composed of financial contributions from the European Union’s seventh framework program (FP7/2007-2013) and EFPIA company in-kind contribution. This work was also supported by Aix-Marseille University, INSERM, and the Service de Santé des Armées.
J.-P. Lavigne belongs to the FHU InCh (Aviesan, University Montpellier, University Hospitals Montpellier and Nîmes).
Biographies

Anne Davin-Regli is an associate professor of the Faculty of Pharmacy on the Medical Campus at Aix-Marseille University and a member of the team UMR_MD1 U1261 INSERM “Membranes and Therapeutic Targets.” She is a hospital practitioner and head of the unit of health and safety at the Aubagne hospital. Her research interests focus on epidemiology of hospital-acquired bacteria, antibiotic and biocide resistance of Gram-negative bacteria, and genetic regulation of multiresistance in Enterobacteriaceae. She has over 50 international scientific papers. Among them, 27 publications concern the genus Enterobacter.

Jean-Philippe Lavigne has been Medical Doctor and Professor of Microbiology at University Hospital of Nîmes and in the Faculty of Medicine Montpellier-Nîmes, France, since 2011. He is the head of a team in INSERM U1047, “Bacterial Virulence and Infectious Diseases,” at the University of Montpellier. He is the co-Director of the University Hospital Federation on Chronic Infection (Aviesan) and of the French National Reference Centre of Brucellosis. He studies the relationships between the different mechanisms of resistance developed by bacteria and their virulence, the cooperation between these bacteria, and the link between commensalism and pathogenicity. His work focuses on the modelling of chronic infections, particularly diabetic foot ulcers. He showed the adaptation of pathogenic bacteria in the stressful environment encountered in these chronic wounds and the influence of efflux pumps and outer membrane porins in this adaptation. By metagenomics, culturomics, and metabolomics approaches, he proposed new concepts to differentiate colonization and infection. He has published 233 papers in international journals, including Diabetes, Diabetes Care, Advanced Functional Materials, and Scientific Reports.

Jean-Marie Pagès has been Emeritus Research Director at INSERM (Institut National de la Santé et de la Recherche Médicale) and a member of the research unit “Membranes and Therapeutic Targets” at the Medical Campus at Aix-Marseille University, Marseille, France, since 2017. He was previously Chair of the UMR-MD1 “Membrane Transporters, Chemoresistance and Drug Design” (Aix-Marseille University-Health Department of the French Army) from 2008 to 2017. He currently studies bacterial envelope permeability, including genetic regulation, biological activities, and clinical aspects. His work focuses on outer membrane porins, drug efflux pumps, and their key role in bacterial adaptation. Recently, as chair of a working group in the IMI-Translocation consortium, he developed new methods to dissect drug translocation across membranes of Gram-negative bacteria and proposed new concepts to understand the molecular bases of antibiotic resistance. As a member of the “Scientific Committee on Emerging and Newly Identified Health Risks,” EC-DG SANCO, Brussels (2004 to 2010), he has contributed to several published scientific opinions, and he is member of several scientific committees of European institutes. He has published over 300 papers and abstracts in international journals, including Nature Microbiology, Proceedings of the National Academy of Sciences of the United States of America, Nature Reviews Microbiology, and The EMBO Journal, and has presented his work and concepts during the main top international meetings (Gordon Research Conferences, Nature Conferences, ECCMID, ICAAC, etc.).
REFERENCES
- 1.Hormaeche E, Edwards P. 1960. A proposed genus Enterobacter. Int Bull Bacteriol Nomencl Taxon 10:71–74. doi: 10.1099/0096266X-10-2-71. [DOI] [Google Scholar]
- 2.Iversen C, Mullane N, McCardell B, Tall BD, Lehner A, Fanning S, Stephan R, Joosten H. 2008. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int J Syst Evol Microbiol 58:1442–1447. doi: 10.1099/ijs.0.65577-0. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann H, Roggenkamp A. 2003. Population genetics of the Nomenspecies Enterobacter cloacae. Appl Environ Microbiol 69:5306–5318. doi: 10.1128/AEM.69.9.5306-5318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh NK, Bezdan D, Checinska Sielaff A, Wheeler K, Mason CE, Venkateswaran K. 2018. Multi-drug resistant Enterobacter bugandensis species isolated from the International Space Station and comparative genomic analyses with human pathogenic strains. BMC Microbiol 18:175. doi: 10.1186/s12866-018-1325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenner DJ, McWorther AC, Kai A, Steigerwalt AG, Farmer JJ. 1986. Enterobacter asburiae sp nov, a new species found in clinical specimens, and reassignment of Erwinia dissolvens and Erwinia nimipressuralis to the genus Enterobacter as Enterobacter dissolvens comb. nov. and Enterobacter nimipressuralis comb. nov. J Clin Microbiol 23:1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhuang L, Zhou S, Yuan Y, Liu T, Wu Z, Cheng J. 2011. Development of Enterobacter aerogenes fuel cells: from in situ biohydrogen oxidization to direct electroactive biofilm. Bioresour Technol 102:284–289. doi: 10.1016/j.biortech.2010.06.038. [DOI] [PubMed] [Google Scholar]
- 7.Akbari M, Bakhshi B, Najar Peerayeh S. 2016. Particular distribution of Enterobacter cloacae strains isolated from urinary tract infection within clonal complexes. Iran Biomed J 20:49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertrand X, Hocquet D, Boisson K, Siebor E, Plésiat P, Talon D. 2003. Molecular epidemiology of Enterobacteriaceae producing extended-spectrum β-lactamase in a French university-affiliated hospital. Int J Antimicrob Agents 22:128–133. doi: 10.1016/S0924-8579(03)00098-0. [DOI] [PubMed] [Google Scholar]
- 9.Chow JW, Fine MJ, Shlaes DM, Quinn JP, Hooper DC, Johnson MP, Ramphal R, Wagener MM, Miyashiro DK, Yu VL. 1991. Enterobacter bacteremia: clinical features and emergence of antibiotic resistance during therapy. Ann Intern Med 115:585–590. doi: 10.7326/0003-4819-115-8-585. [DOI] [PubMed] [Google Scholar]
- 10.Paauw A, Caspers MP, Leverstein-van Hall MA, Schuren FH, Montijn RC, Verhoef J, Fluit AC. 2009. Identification of resistance and virulence factors in an epidemic Enterobacter hormaechei outbreak strain. Microbiology 155:1478–1488. doi: 10.1099/mic.0.024828-0. [DOI] [PubMed] [Google Scholar]
- 11.Morand PC, Billoet A, Rottman M, Sivadon-Tardy V, Eyrolle L, Jeanne L, Tazi A, Anract P, Courpied JP, Poyart C, Dumaine V. 2009. Specific distribution within the Enterobacter cloacae complex of strains isolated from infected orthopedic implants. J Clin Microbiol 47:2489–2495. doi: 10.1128/JCM.00290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanders WE, Sanders CC. 1997. Enterobacter spp.: pathogens poised to flourish at the turn of the century. Clin Microbiol Rev 10:220–241. doi: 10.1128/CMR.10.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allerberger F, Koeuth T, Lass-Florl C, Dierich MP, Putensen C, Schmutzhard E, Mohsenipour I, Grundmann H, Hartung D, Bauernfeind A, Eberlein E, Lupski JR. 1996. Epidemiology of infections due to multiresistant Enterobacter aerogenes in a university hospital. Eur J Clin Microbiol Infect Dis 15:517–521. doi: 10.1007/BF01691323. [DOI] [PubMed] [Google Scholar]
- 14.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 15.Canton R, Oliver A, Coque TM, Varela M. d C, Perez-Diaz JC, Baquero F. 2002. Epidemiology of extended-spectrum β-lactamase producing Enterobacter isolates in a Spanish hospital during a 12-years period. J Clin Microbiol 40:1237–1243. doi: 10.1128/JCM.40.4.1237-1243.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davin-Regli A, Monnet D, Saux P, Bosi C, Charrel RN, Barthelemy A, Bollet C. 1996. Molecular epidemiology of Enterobacter aerogenes acquisition: one-year prospective study in two intensive care units. J Clin Microbiol 34:1474–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davin-Regli A, Bosi C, Charrel RN, Ageron E, Papazian L, Grimont PAD, Cremieux A, Bollet C. 1997. A nosocomial outbreak due to Enterobacter cloacae strains with the E. hormachei genotype in patients treated with fluoroquinolone treatement. J Clin Microbiol 35:1008–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Champs C, Sauvant MP, Chanal C, Sirot D, Gazuy N, Malhuret R, Baguet JC, Sirot J. 1989. Prospective survey of colonization and infection caused by expanded spectrum-beta-lactamase-producing members of the family Enterobacteriaceae in an intensive care unit. J Antimicrob Chemother 27:2887–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wenger PN, Tokars JI, Brennan P, Samel C, Bland L, Miller M, Carson L, Arduino M, Edelstein P, Aguero S, Riddle C, O'Hara C, Jarvis W. 1997. An outbreak of Enterobacter hormachei infection and colonization in an intensive care nursery. Clin Infect Dis 24:1243–1244. doi: 10.1086/513650. [DOI] [PubMed] [Google Scholar]
- 20.Wu W, Feng Y, Zong Z. 2018. Enterobacter sichuanensis sp. nov., recovered from human urine. Int J Syst Evol Microbiol 68:3922–3927. doi: 10.1099/ijsem.0.003089. [DOI] [PubMed] [Google Scholar]
- 21.Anastay M, Lagier E, Blanc V, Chardon H. 2013. Epidémiologie des bêtalactamases à spectre étendu (BLSE) chez les entérobactéries dans un hôpital du sud de la France, 1997–2007. Path Biol 61:38–43. doi: 10.1016/j.patbio.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Arpin C, Coze C, Rogues AM, Gachie JP, Bebear C, Quentin C. 1996. Epidemiological study of an outbreak due to multidrug-resistant Enterobacter aerogenes in a medical intensive care unit. J Clin Microbiol 34:2163–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bosi C, Davin-Regli A, Bornet C, Mallea M, Pages JM, Bollet C. 1999. Most Enterobacter aerogenes strains in France belong to a prevalent clone. J Clin Microbiol 37:2165–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davin-Regli A, Pagès JM. 2015. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front Microbiol 6:392. doi: 10.3389/fmicb.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davin-Regli A, Masi M, Bialek S, Nicolas-Chanoine MH, Pagès JM. 2016. Antimicrobial resistance and drug efflux pumps in Enterobacter and Klebsiella, p 281–306. In Li X-Z, Elkins CA, Zgurskaya HI (ed), Efflux-mediated drug resistance in bacteria: mechanisms, regulation and clinical implications. Springer International Publishing, Basel, Switzerland. [Google Scholar]
- 26.Chollet R, Bollet C, Chevalier J, Malléa M, Pagès JM, Davin-Regli A. 2002. mar operon involved in multidrug resistance of Enterobacter aerogenes. Antimicrob Agents Chemother 46:1093–1097. doi: 10.1128/aac.46.4.1093-1097.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chollet R, Chevalier J, Bryskier A, Pagès JM. 2004. The AcrAB-TolC pump is involved in macrolide resistance but not in telithromycin efflux in Enterobacter aerogenes and Escherichia coli. Antimicrob Agents Chemother 48:3621–3624. doi: 10.1128/AAC.48.9.3621-3624.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dam S, Pagès JM, Masi M. 2018. Stress responses, outer membrane permeability control and antimicrobial resistance in Enterobacteriaceae. Microbiology 164:260–267. doi: 10.1099/mic.0.000613. [DOI] [PubMed] [Google Scholar]
- 29.Masi M, Réfregiers M, Pos KM, Pagès JM. 2017. Mechanisms of envelope permeability and antibiotic influx and efflux in Gram-negative bacteria. Nat Microbiol 2:17001. doi: 10.1038/nmicrobiol.2017.1. [DOI] [PubMed] [Google Scholar]
- 30.Molitor A, James CE, Fanning S, Pagès JM, Davin-Regli A. 2018. Ram locus is a key regulator to trigger multidrug resistance in Enterobacter aerogenes. J Med Microbiol 67:148–159. doi: 10.1099/jmm.0.000667. [DOI] [PubMed] [Google Scholar]
- 31.Pérez A, Poza M, Fernández A, del Carmen Fernández M, Mallo S, Merino M, Rumbo-Feal S, Cabral MP, Bou G. 2012. Involvement of the AcrAB-TolC efflux pump in the resistance, fitness, and virulence of Enterobacter cloacae. Antimicrob Agents Chemother 56:2084–2090. doi: 10.1128/AAC.05509-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izdebski R, Baraniak A, Herda M, Fiett J, Bonten MJM, Carmeli Y, Goossens H, Hryniewicz W, Brun-Buisson C, Gniadkowski M, Grabowska A, Nikonorow E, Derde LPG, Dautzenberg MJ, Adler A, Kazma M, Navon-Venezia S, Malhotra-Kumar S, Lammens C, Dumpis U, Giamarellou H, Muzlovic I, Nardi G, Petrikkos GL, Stammet P, Salomon J, Lawrence C, Legrand P, Rossini A, Salvia A, Samso JV, Fierro J, Paul M, Lerman Y, MOSAR WP2, WP3 and WP5 Study Groups. 2015. MLST reveals potentially high-risk international clones of Enterobacter cloacae. J Antimicrob Chemother 70:48–56. doi: 10.1093/jac/dku359. [DOI] [PubMed] [Google Scholar]
- 33.Liu WY, Wong CF, Chung KM, Jiang JW, Leung FC. 2013. Comparative genome analysis of Enterobacter cloacae. PLoS One 8:e74487. doi: 10.1371/journal.pone.0074487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paauw A, Caspers MPM, Schuren FHJ, Hall MAL, Delétoile A, Montijn RC, Verhoef J, Fluit AC. 2008. Genomic diversity within the Enterobacter cloacae complex. PLoS One 3:e3018. doi: 10.1371/journal.pone.0003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brady C, Cleenwerck I, Venter S, Coutinho T, De Vos P. 2013. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst Appl Microbiol 36:309–319. doi: 10.1016/j.syapm.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 36.Izard D, Gavini F, Leclerc H. 1980. Polynucleotide sequence relatedness and genome size among Enterobacter intermedium sp nov and the species Enterobacter cloacae and Klebsiella pneumoniae. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig Reihe C 1:51–60. doi: 10.1016/S0172-5564(80)80016-9. [DOI] [Google Scholar]
- 37.Diene SM, Merhej V, Henry M, El Filali A, Roux V, Robert C, Azza S, Gavory F, Barbe V, La Scola B, Raoult D, Rolain JM. 2013. The rhizome of the multidrug-resistant Enterobacter aerogenes genome reveals how new “killer bugs” are created because of a sympatric lifestyle. Mol Biol Evol 30:369–383. doi: 10.1093/molbev/mss236. [DOI] [PubMed] [Google Scholar]
- 38.Tindall BJ, Sutton G, Garrity GM. 2017. Enterobacter aerogenes Hormaeche and Edwards 1960 (Approved Lists 1980) and Klebsiella mobilis Bascomb et al. 1971 (Approved Lists 1980) share the same nomenclatural type (ATCC 13048) on the Approved Lists and are homotypic synonyms, with consequences for the name Klebsiella mobilis Bascomb et al. 1971 (Approved Lists 1980). Int J Syst Evol Microbiol 67:502–504. doi: 10.1099/ijsem.0.001572. [DOI] [PubMed] [Google Scholar]
- 39.Hartl R, Kerschner H, Gattringer R, Lepuschitz S, Allerberger F, Sorschag S, Ruppitsch W, Apfalter P. 2018. Whole-genome analysis of a human Enterobacter mori isolate carrying a blaIMI-2 carbapenemase in Austria. Microb Drug Resist doi: 10.1089/mdr.2018.0098. [DOI] [PubMed] [Google Scholar]
- 40.Tidjani Alou M, Cadoret F, Brah S, Diallo A, Sokhna C, Mehrej V, Lagier JC, Fournier PE, Raoult D. 2017. 'Khelaifiella massiliensis', 'Niameybacter massiliensis', 'Brachybacterium massiliense', 'Enterobacter timonensis', 'Massilibacillus massiliensis', new bacterial species and genera isolated from the gut microbiota of healthy infants. New Microbes New Infect 19:1–7. doi: 10.1016/j.nmni.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sutton GG, Brinkac LM, Clarke TH, Fouts DE. 2018. Enterobacter hormaechei subsp. hoffmannii subsp. nov., Enterobacter hormaechei subsp. xiangfangensis comb. nov., Enterobacter roggenkampii sp. nov., and Enterobacter muelleri is a later heterotypic synonym of Enterobacter asburiae based on computational analysis of sequenced Enterobacter genomes. F1000 Res 7:521. doi: 10.12688/f1000research.14566.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Izard D, Gavini F, Trinel PA, Leclerc H. 1981. Desoxyribonucleic acid relatedness between Enterobacter cloacae and Enterobacter amnigenus sp nov. Int J Syst Bacteriol 31:35–42. doi: 10.1099/00207713-31-1-35. [DOI] [Google Scholar]
- 43.Bollet C, Elkouby A, Pietri P, Micco P. 1991. Enterobacter amnigenus isolated from a heart transplant recipient. Eur J Clin Microbiol Infect Dis 10:1071–1073. doi: 10.1007/BF01984933. [DOI] [PubMed] [Google Scholar]
- 44.Freney J, Husson MO, Gavini F, Madier S, Martra A, Izard D, Leclerc H, Fleurette J. 1988. Susceptibility to antibiotics and antiseptics of new species of the family Enterobacteriaceae. Antimicrob Agents Chemother 32:873–876. doi: 10.1128/aac.32.6.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muytjens HL, van der Ros-van de Repe J. 1986. Comparative in vitro susceptibilities of eight Enterobacter species, with special reference to Enterobacter sakazakii. Antimicrob Agents Chemother 29:367–370. doi: 10.1128/aac.29.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grimont PAD, Ageron E. 1989. Enterobacter cancerogenus (Urosevic, 1966) Dickey and Zumoff 1988, a senior subjective synonym of Enterobacter taylorae Farmer et al (1985). Res Microbiol 140:459–465. doi: 10.1016/0923-2508(89)90067-3. [DOI] [PubMed] [Google Scholar]
- 47.Stock I, Wiedemann B. 2002. Natural antibiotic susceptibility of Enterobacter amnigenus, Enterobacter cancerogenus, Enterobacter gergoviae and Enterobacter sakasakii. Clin Microbiol Infect 8:564–578. doi: 10.1046/j.1469-0691.2002.00413.x. [DOI] [PubMed] [Google Scholar]
- 48.Farmer JJ III, Fanning GR, Davis BR, O’Hara CM, Riddle C, Hickman-Brenner FW, Asbury MA, Lowery VA III, Brenner DJ. 1985. Escherichia fergusonii and Enterobacter taylorae, two new species of Enterobactriaceae isolated from clinical specimens. J Clin Microbiol 21:77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freney J, Gavini F, Ploton C, Leclerc H, Fleurette J. 1986. Isolement d’une souche d’Enterobacter taylorae chez un brûlé. Méd Mal Infect 5:320–321. doi: 10.1016/S0399-077X(86)80248-7. [DOI] [Google Scholar]
- 50.Westblom TU, Coggins ME. 1987. Osteomyelitis caused by Enterobacter taylorae, formerly enteric group 19. J Clin Microbiol 25:2432–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Florio L, Riva E, Giona A, Dedej E, Fogolari M, Cella E, Spoto S, Lai A, Zehender G, Ciccozzi M, Angeletti S. 2018. MALDI-TOF MS identification and clustering applied to Enterobacter species in nosocomial setting. Front Microbiol 9:1885. doi: 10.3389/fmicb.2018.01885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu B, Wang S, Li O, Hussain A, Hussain A, Shen J, Ibrahim M. 2017. High-quality genome sequence of human pathogen Enterobacter asburiae type strain 1497-78T. J Glob Antimicrob Resist 8:104–105. doi: 10.1016/j.jgar.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 53.Aubron C, Poirel L, Ash RJ, Nordmann P. 2005. Carbapenemase-producing Enterobacteriaceae, US rivers. Emerg Infect Dis 11:260–264. doi: 10.3201/eid1102.030684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rotova V, Papagiannitsis CC, Chudejova K, Medvecky M, Skalova A, Adamkova V, Hrabak J. 2017. First description of the emergence of Enterobacter asburiae producing IMI-2 carbapenemase in the Czech Republic. J Glob Antimicrob Resist 11:98–99. doi: 10.1016/j.jgar.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 55.Hoffmann H, Stindl S, Ludwig W, Stumpf A, Mehlen A, Heesemann J, Monget D, Schleifer KH, Roggenkamp A. 2005. Reassignment of Enterobacter dissolvens to Enterobacter cloacae as E. cloacae subspecies dissolvens comb. nov. and emended description of Enterobacter asburiae and Enterobacter kobei. Syst Appl Microbiol 28:196–205. doi: 10.1016/j.syapm.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 56.Paraje MG, Barnes AI, Albesa I. 2005. An Enterobacter cloacae toxin able to generate oxidative stress and to provoke dose-dependent lysis of leukocytes. Int J Med Microbiol 295:109–116. doi: 10.1016/j.ijmm.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 57.Moradigaravand D, Reuter S, Martin V, Peacock SJ, Parkhill J. 2016. The dissemination of multidrug-resistant Enterobacter cloacae throughout the UK and Ireland. Nat Microbiol 1:16173. doi: 10.1038/nmicrobiol.2016.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ito A, Nishikawa T, Ota M, Ito-Horiyama T, Ishibashi N, Sato T, Tsuji M, Yamano Y. 2018. Stability and low induction propensity of cefiderocol against chromosomal AmpC β-lactamases of Pseudomonas aeruginosa and Enterobacter cloacae. J Antimicrob Chemother 73:3049–3052. doi: 10.1093/jac/dky317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zaher A, Cimolai N. 1998. ERIC-PCR typing profiles of Enterobacter cloacae are stable after development of advanced cephalosporin resistance. Int J Antimicrobial Agents 9:165–167. doi: 10.1016/S0924-8579(97)00046-0. [DOI] [PubMed] [Google Scholar]
- 60.Arpin C, Labia R, Dubois V, Noury P, Souquet M, Quentin C. 2002. TEM-80, a novel inhibitor-resistant β-lactamase in a clinical isolate of Enterobacter cloacae. Antimicrob Agents Chemother 46:1183–1189. doi: 10.1128/AAC.46.5.1183-1189.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bornet C, Davin-Regli A, Bosi C, Pages JM, Bollet C. 2000. Imipenem resistance of Enterobacter aerogenes mediated by outer membrane permeability. J Clin Microbiol 38:1048–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pitout JDD, Moland ES, Sanders CC, Thomson KS, Fitzsimmons SR. 1997. β-Lactamases and detection of β-lactam resistance in Enterobacter spp. Antimicrob Agents Chemother 41:35–39. doi: 10.1128/AAC.41.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szabo D, Melan MAA, Hujer AM, Bonomo RA, Hujer KM, Bethel CR, Kristof K, Paterson DL. 2005. Molecular analysis of the simultaneous production of two SHV-type extended-spectrum beta-lactamases in a clinical isolate of Enterobacter cloacae by using single-nucleotide polymorphism genotyping. Antimicrob Agents Chemother 49:4716–4720. doi: 10.1128/AAC.49.11.4716-4720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Galani I, Souli M, Chryssouli Z, Orlandou K, Giamarellou H. 2005. Characterization of a new integron containing, bla (VIM-1) and aac(6')-IIc in an Enterobacter cloacae clinical isolate from Greece. J Antimicrob Chemother 55:634–638. doi: 10.1093/jac/dki073. [DOI] [PubMed] [Google Scholar]
- 65.Jin C, Zhang J, Wang Q, Chen H, Wang X, Zhang Y, Wang H. 2018. Molecular characterization of carbapenem-resistant Enterobacter cloacae in 11 Chinese cities. Front Microbiol 9:1597. doi: 10.3389/fmicb.2018.01597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nordmann P, Mariotte S, Naas T, Labia R, Nicolas MH. 1993. Biochemical properties of a carbapenem-hydrolyzing beta-lactamase from Enterobacter cloacae and cloning of the gene into Escherichia coli. Antimicrob Agents Chemother 37:939–946. doi: 10.1128/aac.37.5.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raimondi A, Traverso A, Nikaido H. 1991. Imipenem- and meropenem-resistant mutants of Enterobacter cloacae and Proteus rettgeri lack porins. Antimicrob Agents Chemother 35:1174–1180. doi: 10.1128/aac.35.6.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dai W, Sun S, Yang P, Huang S, Zhang X, Zhang L. 2013. Characterization of carbapenemases, extended spectrum β-lactamases and molecular epidemiology of carbapenem-non-susceptible Enterobacter cloacae in a Chinese hospital in Chongqing. Infect Genet Evol 14:1–7. doi: 10.1016/j.meegid.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 69.Hamprecht A, Poirel L, Gottig S, Seifert H, Kaase M, Nordmann P. 2013. Detection of the carbapenemase GIM-1 in Enterobacter cloacae in Germany. J Antimicrob Chemother 68:558–561. doi: 10.1093/jac/dks447. [DOI] [PubMed] [Google Scholar]
- 70.Huang S, Dai W, Sun S, Zhang X, Zhang L. 2012. Prevalence of plasmid-mediated quinolone resistance and aminoglycoside resistance determinants among carbapeneme non-susceptible Enterobacter cloacae. PLoS One 7:e47636. doi: 10.1371/journal.pone.0047636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee HK, Park YJ, Kim JY, Chang E, Cho SG, Chae HS, Kang CS. 2005. Prevalence of decreased susceptibility to carbapenems among Serratia marcescens, Enterobacter cloacae, and Citrobacter freundii and investigation of carbapenemases. Diagn Microbiol Infect Dis 52:331–336. doi: 10.1016/j.diagmicrobio.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 72.Corkill JE, Anson JJ, Hart CA. 2005. High prevalence of the plasmid-mediated quinolone resistance determinant qnrA in multidrug-resistant Enterobacteriaceae from the blood cultures in Liverpool, UK. J Antimicrob Chemother 56:1115–1117. doi: 10.1093/jac/dki388. [DOI] [PubMed] [Google Scholar]
- 73.Kanamori H, Yano H, Hirakata Y, Hirotani A, Arai K, Endo S, Ichimura S, Ogawa M, Shimojima M, Aoyagi T, Hatta M, Yamada M, Gu Y, Tokuda K, Kunishima H, Kitagawa M, Kaku M. 2012. Molecular characteristics of extended-spectrum β-lactamases and qnr determinants in Enterobacter species from Japan. PLoS One 7:e37967. doi: 10.1371/journal.pone.0037967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Poirel L, Mammeri H, Nordmann P. 2004. TEM-121, a novel complex mutant of TEM-type beta-lactamase from Enterobacter aerogenes. Antimicrob Agents Chemother 48:4528–4531. doi: 10.1128/AAC.48.12.4528-4531.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Hara CM, Steigerwalt AG, Hill BC, Farmer JJ III, Fanning GR, Brenner DJ. 1989. Enterobacter hormaechei, a new species of the family Enterobacteriaceae formerly known as enteric group 75. J Clin Microbiol 27:2046–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoffmann H, Stindl S, Ludwig W, Stumpf A, Mehlen A, Monget D, Pierard D, Ziesing S, Heesemann J, Roggenkamp A, Schleifer KH. 2005. Enterobacter hormaechei subsp. oharae subsp. nov., E. hormaechei subsp. hormaechei comb. nov., and E. hormaechei subsp. steigerwaltii subsp. nov., three new subspecies of clinical importance. J Clin Microbiol 43:3297–3303. doi: 10.1128/JCM.43.7.3297-3303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoffmann H, Schmoldt S, Trulzsch K, Stumpf A, Bengsch S, Blankenstein T, Heesemann J, Roggenkamp A. 2005. Nosocomial urosepsis caused by Enterobacter kobei with aberrant phenotype. Diagn Microbiol Infect Dis 53:143–147. doi: 10.1016/j.diagmicrobio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 78.Wendel AF, Meyer S, Deenen R, Köhrer K, Kolbe-Busch S, Pfeffer K, Willmann M, Kaasch AJ, MacKenzie CR. 2018. Long-term, low-frequency cluster of a German-imipenemase-1-producing Enterobacter hormaechei ssp. steigerwaltii ST89 in a tertiary care hospital in Germany. Microb Drug Resist doi: 10.1089/mdr.2017.0433. [DOI] [PubMed] [Google Scholar]
- 79.Kosako YK, Tamura K, Sakazaki R, Miki K. 1996. Enterobacter kobei sp. nov., a new species of the family Enterobacteriaceae resembling Enterobacter cloacae. Curr Microbiol 33:261–265. doi: 10.1007/s002849900110. [DOI] [PubMed] [Google Scholar]
- 80.Zhou K, Yu W, Bonnet R, Cattoir V, Shen P, Wang B, Rossen JW, Xiao Y. 2017. Emergence of a novel Enterobacter kobei clone carrying chromosomal-encoded CTX-M-12 with diversified pathogenicity in northeast China. New Microbes New Infect 17:7–10. doi: 10.1016/j.nmni.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hoffmann H, Stindl S, Stumpf A, Mehlen A, Monget D, Heesemann J, Schleifer KH, Roggenkamp A. 2005. Description of Enterobacter ludwigii sp. nov., a novel Enterobacter species of clinical relevance. Syst Appl Microbiol 28:206–212. doi: 10.1016/j.syapm.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 82.Li G, Hu Z, Zeng P, Zhu B, Wu L. 2015. Whole genome sequence of Enterobacter ludwigii type strain EN-119T, isolated from clinical specimens. FEMS Microbiol Lett 362:fnv033. doi: 10.1093/femsle/fnv033. [DOI] [PubMed] [Google Scholar]
- 83.Flores-Carrero A, Labrador I, Paniz-Mondolfi A, Peaper DR, Towle D, Araque M. 2016. Nosocomial outbreak of extended-spectrum β-lactamase-producing Enterobacter ludwigii co-harbouring CTX-M-8, SHV-12 and TEM-15 in a neonatal intensive care unit in Venezuela. J Glob Antimicrob Resist 7:114–118. doi: 10.1016/j.jgar.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 84.Khajuria A, Praharaj AK, Grover N, Kumar M. 2013. First report of an Enterobacter ludwigii isolate coharboring NDM-1 and OXA-48 carbapenemases. Antimicrob Agents Chemother 57:5189–5190. doi: 10.1128/AAC.00789-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu B, Lou MM, Xie GL, Wang GF, Zhou Q, Wang F, Fang Y, Su T, Li B, Duan YP. 2011. Enterobacter mori sp. nov., associated with bacterial wilt on Morus alba L. Int J Syst Evol Microbiol 61:2769–2774. doi: 10.1099/ijs.0.028613-0. [DOI] [PubMed] [Google Scholar]
- 86.Steigerwalt AG, Fanning GR, Fife-Asbury MA, Brenner DJ. 1976. DNA relatedness among species of Enterobacter and Serratia. Can J Microbiol 22:121–137. doi: 10.1139/m76-018. [DOI] [PubMed] [Google Scholar]
- 87.Kim DM, Jang SJ, Neupane GP, Jang MS, Kwon SH, Kim SW, Kim WY. 2010. Enterobacter nimipressuralis as a cause of pseudobacteremia. BMC Infect Dis 10:315. doi: 10.1186/1471-2334-10-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Izard D, Gavini F, Trinel PA, Krubwa F, Leclerc H. 1980. Contribution to DNA-DNA hybridization to the transfer of Enterobacter aerogenes to the genus Klebsiella as K. mobilis. Zentralbl Bakteriol Hyg 1:257–263. doi: 10.1016/S0172-5564(80)80006-6. [DOI] [Google Scholar]
- 89.De Gheldre Y, Maes N, Rost F, De Ryck R, Clevenbergh P, Vincent JL, Struelens MJ. 1997. Molecular epidemiology of an outbreak of multidrug-resistant Enterobacter aerogenes infections and in vivo emergence of imipenem resistance. J Clin Microbiol 35:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Georghiou PR, Hamill RJ, Wright CE, Versalovic J, Koeuth T, Watson DA, Lupski JR. 1995. Molecular epidemiology of infections due to Enterobacter aerogenes: identification of hospital outbreak-associated strains by molecular techniques. Clin Infect Dis 20:84–94. doi: 10.1093/clinids/20.1.84. [DOI] [PubMed] [Google Scholar]
- 91.Grattard F, Pozzetto B, Tabard L, Petit M, Ros A, Gaudin OG. 1995. Characterization of nosocomial strains of Enterobacter aerogenes by arbitrarily primed PCR analysis and ribotyping. Infect Control Hosp Epidemiol 16:224–230. doi: 10.2307/30140982. [DOI] [PubMed] [Google Scholar]
- 92.Jalaluddin S, Devaster JM, Scheen R, Gerard M, Butzler JP. 1998. Molecular epidemiological study of nosocomial Enterobacter aerogenes isolates in a Belgian hospital. J Clin Microbiol 36:1846–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lavigne JP, Sotto A, Nicolas-Chanoine MH, Bouziges N, Bourg G, Davin-Regli A, Pagès JM. 2012. Membrane permeability, a pivotal function involved in antibiotic resistance and virulence in Enterobacter aerogenes clinical isolates. Clin Microbiol Infect 18:539–545. doi: 10.1111/j.1469-0691.2011.03607.x. [DOI] [PubMed] [Google Scholar]
- 94.Song E-H, Park K-H, Jang E-Y, Lee E-J, Chong Y-P, Cho O-H, Kim S_H, Lee S-O, Sung H, Kim MN, Jeong JY, Kim YS, Woo JH, Choi SH. 2010. Comparison of the clinical and microbiologic characteristics of patients with Enterobacter cloacae and Enterobacter aerogenes bacteremia: a prospective observation study. Diagn Microbiol Infect Dis 66:436–440. doi: 10.1016/j.diagmicrobio.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 95.Miro E, Alonso C, Navarro F, Mirelis B, Prats G. 1995. Resistencia al imipenem en Enterobacter aerogenes. Enferm Infecc Microbiol Clin 13:278–282. [PubMed] [Google Scholar]
- 96.Preston KE, Radomski CCA, Venezia RA. 2000. Nucleotide sequence of the chromosomal ampC gene of Enterobacter aerogenes. Antimicrob Agents Chemother 44:3158–3162. doi: 10.1128/AAC.44.11.3158-3162.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee SH, Jeong SH, Park YM. 2003. Characterization of blaCMY-10 a novel, plasmid-encoded AmpC-type β-lactamase gene in a clinical isolate of Enterobacter aerogenes. J Appl Microbiol 95:744–752. doi: 10.1046/j.1365-2672.2003.02040.x. [DOI] [PubMed] [Google Scholar]
- 98.Pitout JDD, Thomson KS, Hanson ND, Ehrhardt AF, Coudron P, Sanders CC. 1998. Plasmid-mediated resistance to expanded-spectrum cephalosporins among Enterobacter aerogenes strains. Antimicrob Agents Chemother 42:596–600. doi: 10.1128/AAC.42.3.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Biendo M, Canarelli B, Thomas D, Rousseau F, Hamdad F, Adjide C, Laurans G, Eb F. 2008. Successive emergence of extended-spectrum beta-lactamase-producing and carbapenemase-producing Enterobacter aerogenes isolates in a university hospital. J Clin Microbiol 46:1037–1044. doi: 10.1128/JCM.00197-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dumarche P, De Champs C, Sirot D, Chanal C, Bonnet R, Sirot J. 2002. TEM derivative-producing Enterobacter aerogenes strains: dissemination of a prevalent clone. Antimicrob Agents Chemother 46:1128–1131. doi: 10.1128/aac.46.4.1128-1131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bousquet A, van der Mee-Marquet N, Dubost C, Bigaillon C, Larréché S, Bugier S, Surcouf C, Mérat S, Blanchard H, Mérens A. 2017. Outbreak of CTX-M-15-producing Enterobacter cloacae associated with therapeutic beds and syphons in an intensive care unit. Am J Infect Control 45:1160–1164. doi: 10.1016/j.ajic.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 102.Lavigne JP, Bouziges N, Chanal C, Mahamat A, Michaux-Charachon S, Sotto A. 2004. Molecular epidemiology of Enterobacteriaceae isolates producing extended-spectrum beta-lactamases in a French hospital. J Clin Microbiol 42:3805–3808. doi: 10.1128/JCM.42.8.3805-3808.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Neuwirth C, Siebor E, Lopez J, Pechinot A, Kazmierczak A. 1996. Outbreak of TEM-24-producing Enterobacter aerogenes in an intensive care unit and dissemination of the extended-spectrum β-lactamase to other members of the family Enterobacteriacae. J Clin Microbiol 34:76–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Charrel RN, Pagès JM, De Micco P, Mallea M. 1996. Prevalence of outer membrane porin alteration in beta-lactam-antibiotic-resistant Enterobacter aerogenes. Antimicrob Agents Chemother 40:2854–2858. doi: 10.1128/AAC.40.12.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fernández-Cuenca F, Rodríguez-Martínez JM, Martínez-Martínez L, Pascual A. 2006. In vivo selection of Enterobacter aerogenes with reduced susceptibility to cefepime and carbapenems associated with decreased expression of a 40 kDa outer membrane protein and hyperproduction of AmpC β–lactamase. Int J Antimicrob Agents 27:549–552. doi: 10.1016/j.ijantimicag.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 106.Thiolas A, Bornet C, Davin-Régli A, Pagès JM, Bollet C. 2004. Resistance to imipenem, cefepime, and cefpirome associated with mutation in Omp36 osmoporin of Enterobacter aerogenes. Biochem Biophys Res Commun 317:851–856. doi: 10.1016/j.bbrc.2004.03.130. [DOI] [PubMed] [Google Scholar]
- 107.Dé E, Baslé A, Jaquinod M, Saint N, Malléa M, Molle G, Pagès JM. 2001. A new mechanism of antibiotic resistance in Enterobacteriaceae induced by a structural modification of the major porin. Mol Microbiol 41:189–198. doi: 10.1046/j.1365-2958.2001.02501.x. [DOI] [PubMed] [Google Scholar]
- 108.Chevalier J, Mulfinger C, Garnotel E, Nicolas P, Davin-Régli A, Pagès JM. 2008. Identification and evolution of drug efflux pump in clinical Enterobacter aerogenes strains isolated in 1995 and 2003. PLoS One 3:e3203. doi: 10.1371/journal.pone.0003203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Thiolas A, Bollet C, La Scola B, Raoult D, Pages J-M. 2005. Successive emergence of Enterobacter aerogenes strains resistant to imipenem and colistin in a patient. Antimicrob Agents Chemother 49:1354–1358. doi: 10.1128/AAC.49.4.1354-1358.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Richard C, Joly B, Sirot J, Stoleru GH, Popoff M. 1976. Étude de souches d’Enterobacter appartenant à un groupe particulier proche de E. aerogenes. Ann Microbiol Inst Pasteur 127A:545–548. [PubMed] [Google Scholar]
- 111.Brenner DJ, Richard C, Steigerwalt AG, Asbury MA, Mandel M. 1980. Enterobacter gergoviae sp. nov.: a new species of Enterobacteriaceae found in clinical specimens and the environment. Int J Syst Bacteriol 30:1–6. doi: 10.1099/00207713-30-1-1. [DOI] [Google Scholar]
- 112.Cheng Y, Chen M. 1994. Extended-spectrum beta-lactamases in clinical isolates of Enterobacter gergoviae and Escherichia coli in China. Antimicrob Agents Chemother 38:2838–2842. doi: 10.1128/aac.38.12.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Freire MP, de Oliveira Garcia D, Cury AP, Spadão F, Di Gioia TS, Francisco GR, Bueno MF, Tomaz M, de Paula FJ, de Faro LB, Piovesan AC, Rossi F, Levin AS, David Neto E, Nahas WC, Pierrotti LC. 2016. Outbreak of IMP-producing carbapenem-resistant Enterobacter gergoviae among kidney transplant recipients. J Antimicrob Chemother 71:2577–2585. doi: 10.1093/jac/dkw165. [DOI] [PubMed] [Google Scholar]
- 114.Périamé M, Pagès JM, Davin-Regli A. 2015. Enterobacter gergoviae membrane modifications are involved in the adaptive response to preservatives used in cosmetic industry. J Appl Microbiol 118:49–61. doi: 10.1111/jam.12676. [DOI] [PubMed] [Google Scholar]
- 115.Périamé M, Philippe N, Condell O, Fanning S, Pagès JM, Davin-Regli A. 2015. Phenotypic changes contributing to Enterobacter gergoviae biocide resistance. Lett Appl Microbiol 61:121–129. doi: 10.1111/lam.12435. [DOI] [PubMed] [Google Scholar]
- 116.Davin-Regli A, Chollet R, Bredin J, Chevalier J, Lepine F, Pagès JM. 2006. Enterobacter gergoviae and the prevalence of efflux in parabens resistance. J Antimicrob Chemother 57:757–760. doi: 10.1093/jac/dkl023. [DOI] [PubMed] [Google Scholar]
- 117.Périamé M, Pagès JM, Davin-Regli A. 2014. Enterobacter gergoviae adaptation to preservatives commonly used in cosmetic industry. Int J Cosmet Sci 36:386–395. doi: 10.1111/ics.12140. [DOI] [PubMed] [Google Scholar]
- 118.Doijad S, Imirzalioglu C, Yao Y, Pati NB, Falgenhauer L, Hain T, Foesel BU, Abt B, Overmann J, Mirambo MM, Mshana SE, Chakraborty T. 2016. Enterobacter bugandensis sp. nov., isolated from neonatal blood. Int J Syst Evol Microbiol 66:968–974. doi: 10.1099/ijsem.0.000821. [DOI] [PubMed] [Google Scholar]
- 119.Wu W, Feng Y, Zong Z. 2018. Characterization of a strain representing a new Enterobacter species, Enterobacter chengduensis sp. nov. 2018. Antonie Van Leeuwenhoek doi: 10.1007/s10482-018-1180-z. [DOI] [PubMed] [Google Scholar]
- 120.Humann JL, Wildung M, Cheng CH, Lee T, Stewart JE, Drew JC, Triplett EW, Main D, Schroeder BK. 2011. Complete genome of the onion pathogen Enterobacter cloacae EcWSU1. Stand Genomic Sci 5:279–286. doi: 10.4056/sigs.2174950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Richard C. 1989. Nouvelles Enterobacteriaceae rencontrées en bactériologie médicale: Moellerella wisconsensis, Koserella trabulsii, Leclercia adercarboxylata, Escherichia fergusonii, Enterobacter asburiae, Rahnella aquatilis. Ann Biol Clin 47:231–236. [PubMed] [Google Scholar]
- 122.Cordoba MA, Roccia IL, De Luca MM, Pezzani BC, Basualdo JA. 2001. Resistance to antibiotics in injured coliforms isolated from drinking water. Microbiol Immunol 45:383–386. doi: 10.1111/j.1348-0421.2001.tb02634.x. [DOI] [PubMed] [Google Scholar]
- 123.Dugleux G, Le Coutour X, Hecquard C, Oblin I. 1991. Septicemia caused by contaminated parenteral nutrition pouches: the refrigerator as an unusual cause. J Parenter Enteral Nutr 15:474–475. doi: 10.1177/0148607191015004474. [DOI] [PubMed] [Google Scholar]
- 124.Wang SA, Tokars JI, Bianchine PJ, Carson LA, Arduino MJ, Smith AL, Hansen NC, Fitzgerald EA, Epstein JS, Jarvis WR. 2000. Enterobacter cloacae bloodstream infections traced to contamined human albumine. Clin Infect Dis 30:35–40. doi: 10.1086/313585. [DOI] [PubMed] [Google Scholar]
- 125.Wang CC, Chu ML, Ho LJ, Hwang RC. 1991. Analysis of plasmid pattern in paediatric intensive care unit outbreaks of nosocomial infection due to Enterobacter cloacae. J Hosp Infect 19:33–40. doi: 10.1016/0195-6701(91)90126-S. [DOI] [PubMed] [Google Scholar]
- 126.Mayhall CG, Lamb VA, Gayle WE Jr, Haynes BW Jr. 1979. Enterobacter cloacae septicemia in a burn center: epidemiology and control of an outbreak. J Infect Dis 139:166–171. doi: 10.1093/infdis/139.2.166. [DOI] [PubMed] [Google Scholar]
- 127.Garazzino S, Aprato A, Maiello A, Massé A, Biasibetti A, De Rosa FG, Di Perri G. 2005. Osteomyelitis caused by Enterobacter cancerogenus infection following a traumatic injury: case report and review of the literature. J Clin Microbiol 43:1459–1461. doi: 10.1128/JCM.43.3.1459-1461.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rubistein EM, Klevjer-Anderson P, Smith CA, Drouin MT, Evans Patterson J. 1993. Enterobacter taylorae, a new opportunistic pathogen: report of four cases. J Clin Microbiol 31:249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Abbott S, Janda M. 1997. Enterobacter cancerogenus (“Enterobacter taylorae”) infections associated with severe trauma or crush injuries. Am J Clin Pathol 107:359–361. doi: 10.1093/ajcp/107.3.359. [DOI] [PubMed] [Google Scholar]
- 130.Fernández-Baca V, Ballesteros F, Hervás JA, Villalón P, Domínguez MA, Benedí VJ, Albertí S. 2001. Molecular epidemiological typing of Enterobacter cloacae isolates from a neonatal intensive care unit: three-year prospective study. J Hosp Infect 49:173–182. doi: 10.1053/jhin.2001.1053. [DOI] [PubMed] [Google Scholar]
- 131.Fluit AC, Schmitz FJ, Verhoef J. 2001. Multi-resistance to antimicrobial agents for the ten most frequently isolated bacterial pathogens. Int J Antimicrob Agents 18:147–160. doi: 10.1016/S0924-8579(01)00357-0. [DOI] [PubMed] [Google Scholar]
- 132.Talon D, Menget P, Thouverez M, Thiriez G, Gbaguidi Haore H, Fromentin C, Muller A, Bertrand X. 2004. Emergence of Enterobacter cloacae as a common pathogen in neonatal units: pulse-field gel electrophoresis analysis. J Hosp Infect 57:119–125. doi: 10.1016/j.jhin.2004.01.023. [DOI] [PubMed] [Google Scholar]
- 133.Fata F, Chittivelu S, Tessler S, Kupfer Y. 1996. Gas gangrene of the arm due to Enterobacter cloacae in a neutropenic patient. South Med J 89:1095–1096. doi: 10.1097/00007611-199611000-00014. [DOI] [PubMed] [Google Scholar]
- 134.Okhravi N, Ficker L, Matheson MM, Lightman S. 1998. Enterobacter cloacae endophtalmitis: report of four cases. J Clin Microbiol 36:48–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Saini AG, Rathore V, Ahuja CK, Chhabra R, Vaidya PC, Singhi P. 2017. Multiple brain abscesses due to Enterobacter cloacae in an immune-competent child. J Infect Public Health 10:674–677. doi: 10.1016/j.jiph.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 136.Chauhan S, Noor J, Yegneswaran B, Kodali H. 2016. Enterobacter meningitis and challenges in treatment. J Clin Diagn Res 10:OD10–OD11. doi: 10.7860/JCDR/2016/20759.9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jayaweera JA, Kothalawala M, Devakanthan B, Arunan S, Galgamuwa D, Rathnayake M. 2016. Spondylodiscitis caused by Enterobacter agglomerans. Case Rep Infect Dis 2016:8491571. doi: 10.1155/2016/8491571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Karasahin O, Yildiz Z, Unal O, Arslan U. 2017. A rare cause of healthcare-associated infective endocarditis: Enterobacter cloacae. Front Microbiol 8:2343. doi: 10.3389/fmicb.2017.02343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Arpin C, Dubois V, Coulange L, André C, Fischer I, Noury P, Grobost F, Brochet JP, Jullin J, Dutilh B, Larribet G, Lagrange I, Quentin C. 2003. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in community and private health care centers. Antimicrob Agents Chemother 47:3506–3514. doi: 10.1128/aac.47.11.3506-3514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bouraoui H, Trimeche B, Kaabia N, Zaaraoui J, Mahdhaoui A, Ernez-Hajri S, Jeridi G, Ammar H. 2005. Endocardite infectieuse à Enterobacter aerogenes. Rev Méd Intern 26:71–80. doi: 10.1016/j.revmed.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 141.Ganeswire R, Thong KL, Puthucheary SD. 2003. Nosocomial outbreak of Enterobacter gergoviae bacteraemia in a neonatal intensive care unit. J Hosp Infect 53:292–296. doi: 10.1053/jhin.2002.1371. [DOI] [PubMed] [Google Scholar]
- 142.Kesieme EB, Kesieme CN, Akpede GO, Okonta KE, Dongo AE, Gbolagade AM, Eluenhike SU. 2012. Tension pneumatocele due to Enterobacter gergoviae pneumonia: a case report. Case Report Med 2012:808630. doi: 10.1155/2012/808630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Haiko J, Westerlund-Wikström B. 2013. The role of the bacterial flagellum in adhesion and virulence. Biology (Basel) 2:1242–1267. doi: 10.3390/biology2041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Barnes AI, Ortiz C, Paraje MG, Balanzino LE, Albesa I. 1997. Purification and characterization of a cytotoxin from Enterobacter cloacae. Can J Microbiol 43:729–733. doi: 10.1139/m97-105. [DOI] [PubMed] [Google Scholar]
- 145.Krzymińska S, Mokracka J, Koczura R, Kaznowski A. 2009. Cytotoxic activity of Enterobacter cloacae human isolates. FEMS Immunol Med Microbiol 56:248–252. doi: 10.1111/j.1574-695X.2009.00572.x. [DOI] [PubMed] [Google Scholar]
- 146.Krzymińska S, Koczura R, Mokracka J, Puton T, Kaznowski A. 2010. Isolates of the Enterobacter cloacae complex induce apoptosis of human intestinal epithelial cells. Microb Pathog 49:83–89. doi: 10.1016/j.micpath.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 147.Souza Lopes AC, Rodrigues JF, Cabral AB, da Silva ME, Leal NC, da Silveira VM, de Morais Júnior MA. 2016. Occurrence and analysis of irp2 virulence gene in isolates of Klebsiella pneumoniae and Enterobacter spp. from microbiota and hospital and community-acquired infections. Microb Pathog 96:15–19. doi: 10.1016/j.micpath.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 148.Kim S-M, Lee H-W, Choi Y-W, Kim S-H, Lee J-C, Lee Y-C, Seol S-Y, Cho D-T, Kim J. 2012. Involvement of curli fimbriae in the biofilm formation of Enterobacter cloacae. J Microbiol 50:175–178. doi: 10.1007/s12275-012-2044-2. [DOI] [PubMed] [Google Scholar]
- 149.Azevedo PAA, Furlan JPR, Oliveira-Silva M, Nakamura-Silva R, Gomes CN, Costa KRC, Stehling EG, Pitondo-Silva A. 2018. Detection of virulence and β-lactamase encoding genes in Enterobacter aerogenes and Enterobacter cloacae clinical isolates from Brazil. Braz J Microbiol 49(Suppl 1):224–228. doi: 10.1016/j.bjm.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.El Fertas-Aissani R, Messai Y, Alouache S, Bakour R. 2013. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol Biol (Paris) 61:209–216. doi: 10.1016/j.patbio.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 151.Compain F, Babosan A, Brisse S, Genel N, Audo J, Ailloud F, Kassis-Chikhani N, Arlet G, Decre D. 2014. Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J Clin Microbiol 52:4377–4380. doi: 10.1128/JCM.02316-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lavigne JP, Sotto A, Nicolas-Chanoine MH, Bouziges N, Pagès JM, Davin-Regli A. 2013. An adaptive response of Enterobacter aerogenes to imipenem: regulation of porin balance in clinical isolates. Int J Antimicrob Agents 41:130–136. doi: 10.1016/j.ijantimicag.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 153.Rostad CA, Wehrheim K, Kirklin JK, Naftel D, Pruitt E, Hoffman TM, L’Ecuyer T, Berkowitz K, Mahle WT, Scheel JN. 2017. Bacterial infections after pediatric heart transplantation: epidemiology, risk factors and outcomes. J Heart Lung Transplant 36:996–1003. doi: 10.1016/j.healun.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 154.Huh K, Kang CI, Kim J, Cho SY, Ha YE, Joo EJ, Chung DR, Lee NY, Peck KR, Song JH. 2014. Risk factors and treatment outcomes of bloodstream infection caused by extended-spectrum cephalosporin-resistant Enterobacter species in adults with cancer. Diagn Microbiol Infect Dis 78:172–177. doi: doi: 10.1016/j.diagmicrobio.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 155.Gaynes RP, Edwards JR, Jarvis WR, Culver DH, Tolson JS, Martone WJ. 1996. Nosocomial infections among neonates in high-risk nurseries in the United States. Pediatrics 98:357–361. [PubMed] [Google Scholar]
- 156.Vidal-Navarro L, Pfeiffer C, Bouziges N, Sotto A, Lavigne JP. 2010. Faecal carriage of multidrug-resistant Gram-negative bacilli during a non-outbreak situation in a French university hospital. J Antimicrob Chemother 65:2455–2458. doi: 10.1093/jac/dkq333. [DOI] [PubMed] [Google Scholar]
- 157.Boutet-Dubois A, Pantel A, Prère MF, Bellon O, Brieu-Roche N, Lecaillon E, Le Coustumier A, Davin-Regli A, Villeneuve L, Bouziges N, Gleize E, Lamarca R, Dunyach-Remy C, Sotto A, Lavigne JP. 2013. Faecal carriage of oxyiminocephalosporin-resistant Enterobacteriaceae among paediatric units in different hospitals in the south of France. Eur J Clin Microbiol Infect Dis 32:1063–1068. doi: 10.1007/s10096-013-1851-7. [DOI] [PubMed] [Google Scholar]
- 158.Arpin C, Dubois V, Maugein J, Jullin J, Dutilh B, Brochet JP, Larribet G, Fischer I, Quentin C. 2005. Clinical and molecular analysis of extended-spectrum β-lactamase-producing enterobacteria in the community setting. J Clin Microbiol 43:5048–5054. doi: 10.1128/JCM.43.10.5048-5054.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lee EH, Collatz E, Trias J, Gutmann L. 1992. Diffusion of beta-lactam antibiotics into proteoliposomes reconstituted with outer membranes of isogenic imipenem-susceptible and -resistant strains of Enterobacter cloacae. J Gen Microbiol 138:2347–2351. doi: 10.1099/00221287-138-11-2347. [DOI] [PubMed] [Google Scholar]
- 160.Tebano G, Geneve C, Tanaka S, Grall N, Atchade E, Augustin P, Thabut G, Castier Y, Montravers P, Desmard M. 2016. Epidemiology and risk factors of multidrug-resistant bacteria in respiratory samples after lung transplantation. Transpl Infect Dis 18:22–30. doi: 10.1111/tid.12471. [DOI] [PubMed] [Google Scholar]
- 161.Hyle EP, Ferraro MJ, Silver M, Lee H, Hooper DC. 2010. Ertapenem-resistant Enterobacteriaceae: risk factors for acquisition and outcomes. Infect Control Hosp Epidemiol 31:1242–1249. doi: 10.1086/657138. [DOI] [PubMed] [Google Scholar]
- 162.Ivády B, Kenesei É, Tóth-Heyn P, Kertész G, Tárkányi K, Kassa C, Ujhelyi E, Mikos B, Sápi E, Varga-Heier K, Guóth G, Szabó D. 2016. Factors influencing antimicrobial resistance and outcome of Gram-negative bloodstream infections in children. Infection 44:309–321. doi: 10.1007/s15010-015-0857-8. [DOI] [PubMed] [Google Scholar]
- 163.Mezzatesta ML, Gona F, Stefani S. 2012. Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol 7:887–902. doi: 10.2217/fmb.12.61. [DOI] [PubMed] [Google Scholar]
- 164.Dalben M, Varkulja G, Basso M, Krebs VLJ, Gibelli MA, van der Heijden I, Rossi F, Duboc G, Levin AS, Costa SF. 2008. Investigation of an outbreak of Enterobacter cloacae in a neonatal unit and review of the literature. J Hosp Infect 70:7–14. doi: 10.1016/j.jhin.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 165.Campos LC, Lobianco LF, Seki LM, Santos RMR, Asensi MD. 2007. Outbreak of Enterobacter hormaechei septicaemia in newborns caused by contaminated parenteral nutrition in Brazil. J Hosp Infect 66:95–97. doi: 10.1016/j.jhin.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 166.Tuon FF, Cieslinski J, Ono AFM, Goto FL, Machinski JM, Mantovani LK, Kosop LR, Namba MS, Rocha JL. 2018. Microbiological profile and susceptibility pattern of surgical site infections related to orthopaedic trauma. Int Orthop doi: 10.1007/s00264-018-4076-7. [DOI] [PubMed] [Google Scholar]
- 167.Kalakouti E, Simillis C, Pellino G, Mughal N, Warren O, Mills S, Tan E, Kontovounisios C, Tekkis PP. 2017. Characteristics of surgical site infection following colorectal surgery in a tertiary center: extended-spectrum β-lactamase-producing bacteria culprits in disease. Wounds 30:108–113. [PubMed] [Google Scholar]
- 168.Alexiou K, Drikos I, Terzopoulou M, Sikalias N, Ioannidis A, Economou N. 2017. A prospective randomised trial of isolated pathogens of surgical site infections (SSI). Ann Med Surg (Lond) 21:25–29. doi: 10.1016/j.amsu.2017.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Scheufele F, Aichinger L, Jäger C, Demir IE, Schorn S, Sargut M, Erkan M, Kleeff J, Friess H, Ceyhan GO. 2017. Effect of preoperative biliary drainage on bacterial flora in bile of patients with periampullary cancer. Br J Surg 104:e182–e188. doi: 10.1002/bjs.10450. [DOI] [PubMed] [Google Scholar]
- 170.Chen H, Zhang Y, Chen YG, Yu YS, Zheng SS, Li LJ. 2009. Sepsis resulting from Enterobacter aerogenes resistant to carbapenems after liver transplantation. Hepatobiliary Pancreat Dis Int 8:320–322. [PubMed] [Google Scholar]
- 171.Hadano Y, Tamagawa K, Ohkusu K. 2018. Trauma wound related infection caused by Enterobacter cancerogenus and Aeromonas hydrophilia. Intern Med 57:131–133. doi: 10.2169/internalmedicine.9171-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kremer A, Hoffmann H. 2012. Prevalences of the Enterobacter cloacae complex and its phylogenetic derivatives in the nosocomial environment. Eur J Clin Microbiol Infect Dis 31:2951–2955. doi: 10.1007/s10096-012-1646-2. [DOI] [PubMed] [Google Scholar]
- 173.Jneid J, Cassir N, Schuldiner S, Jourdan N, Sotto A, Lavigne JP, La Scola B. 2018. Exploring the microbiota of diabetic foot infections with culturomics. Front Cell Infect Microbiol 8:282. doi: 10.3389/fcimb.2018.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jacobson KL, Cohen SH, Inciardi JF, King JH, Lippert WE, Iglesias T, VanCouwenberghe CJ. 1995. The relationship between antecedent antibiotic use and resistance to extended-spectrum cephalosporins in group I b-lactamase-producing organisms. Clin Infect Dis 21:1107–1113. doi: 10.1093/clinids/21.5.1107. [DOI] [PubMed] [Google Scholar]
- 175.Segreti J, Levin S. 1996. Bacteriologic and clinical applications of a new extended-spectrum parental cephalosporin. Am J Med 100:976–982. doi: 10.1016/S0002-9343(96)00107-6. [DOI] [PubMed] [Google Scholar]
- 176.Fung-Tomc J, Dougherty TJ, DeOrio FJ, Simich-Jacobson V, Kessler RE. 1989. Activity of cefepime against ceftazidime- and cefotaxime-resistant gram-negative bacteria and its relationship to beta-lactamase levels. Antimicrob Agents Chemother 33:498–502. doi: 10.1128/aac.33.4.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.McKamey L, Venugopalan V, Cherabuddi K, Borgert S, Voils S, Shah K, Klinker KP. 2018. Assessing antimicrobial stewardship initiatives: clinical evaluation of cefepime or piperacillin/tazobactam in patients with bloodstream infections secondary to AmpC-producing organisms. Int J Antimicrob Agents 52:719–723. doi: 10.1016/j.ijantimicag.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 178.Cheng L, Nelson BC, Mehta M, Seval N, Park S, Giddins MJ, Shi Q, Whittier S, Gomez-Simmonds A, Uhlemann AC. 2017. Piperacillin-tazobactam versus other antibacterial agents for treatment of bloodstream infections due to AmpC β-lactamase-producing Enterobacteriaceae. Antimicrob Agents Chemother 61:e00276-17. doi: 10.1128/AAC.00276-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sanders WE, Tenney JH, Kessler RE. 1996. Efficacy of cefepime in the treatment of infections due to multiply resistant Enterobacter species. Clin Infect Dis 23:454–461. doi: 10.1093/clinids/23.3.454. [DOI] [PubMed] [Google Scholar]
- 180.Harris PNA, Wei JY, Shen AW, Abdile AA, Paynter S, Huxley RR, Pandeya N, Doi Y, Huh K, O'Neal CS, Talbot TR, Paterson DL. 2016. Carbapenems versus alternative antibiotics for the treatment of bloodstream infections caused by Enterobacter, Citrobacter or Serratia species: a systematic review with meta-analysis. J Antimicrob Chemother 71:296–306. doi: 10.1093/jac/dkv346. [DOI] [PubMed] [Google Scholar]
- 181.Dobias J, Dénervaud-Tendon V, Poirel L, Nordmann P. 2017. Activity of the novel siderophore cephalosporin cefiderocol against multidrug-resistant Gram-negative pathogens. Eur J Clin Microbiol Infect Dis 36:2319–2327. doi: 10.1007/s10096-017-3063-z. [DOI] [PubMed] [Google Scholar]
- 182.Livermore DM, Mushtaq S, Warner M, Vickers A, Woodford N. 2017. In vitro activity of cefepime/zidebactam (WCK 5222) against Gram-negative bacteria. J Antimicrob Chemother 72:1373–1385. doi: 10.1093/jac/dkw593. [DOI] [PubMed] [Google Scholar]
- 183.Sader HS, Castanheira M, Mendes RE, Flamm RK, Jones RN. 2017. Antimicrobial activity of high-proportion cefepime-tazobactam (WCK 4282) against a large number of Gram-negative isolates collected worldwide in 2014. Antimicrob Agents Chemother 61:e02409-16. doi: 10.1128/AAC.02409-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mischnik A, Baumert P, Hamprecht A, Rohde A, Peter S, Feihl S, Knobloch J, Gölz H, Kola A, Obermann B, Querbach C, Willmann M, Gebhardt F, Tacconelli E, Gastmeier P, Seifert H, Kern WV, DZIF-ATHOS Study Group. 2017. Susceptibility to cephalosporin combinations and aztreonam/avibactam among third-generation cephalosporin-resistant Enterobacteriaceae recovered on hospital admission. Int J Antimicrob Agents 49:239–242. doi: 10.1016/j.ijantimicag.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 185.Haidar G, Alkroud A, Cheng S, Churilla TM, Churilla BM, Shields RK, Doi Y, Clancy CJ, Nguyen MH. 2016. Association between the presence of aminoglycoside-modifying enzymes and in vitro activity of gentamicin, tobramycin, amikacin, and plazomicin against Klebsiella pneumoniae carbapenemase- and extended-spectrum-β-lactamase-producing Enterobacter species. Antimicrob Agents Chemother 60:5208–5214. doi: 10.1128/AAC.00869-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Boucher HW, Talbot GH, Benjamin DK Jr, Bradley J, Guidos RJ, Jones RN, Murray BE, Bonomo RA, Gilbert D, Infectious Diseases Society of America. 2013. 10 x '20 Progress–development of new drugs active against Gram-negative bacilli: an update from the Infectious Diseases Society of America. Clin Infect Dis 56:1685–1694. doi: 10.1093/cid/cit152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/mmbr.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pagès JM, James CE, Winterhalter M. 2008. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol 6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 189.Delcour AH. 2009. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta 1794:808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zgurskaya HI, Löpez CA, Gnanakaran S. 2015. Permeability barrier of Gram-negative cell envelopes and approaches to bypass it. ACS Infect Dis 1:512–522. doi: 10.1021/acsinfecdis.5b00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Nikaido H. 1985. Role of permeability barriers in resistance to ß-lactams antibiotics. Pharmacol Ther 27:197–231. doi: 10.1016/0163-7258(85)90069-5. [DOI] [PubMed] [Google Scholar]
- 192.Nikaido H. 1989. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother 33:1831–1836. doi: 10.1128/aac.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Valade E, Davin-Regli A, Bolla JM, Pagès JM. 2014. Bacterial membrane, a key for controlling drug influx and efflux, p 217–240. In Gualerzi CO, Brandi L, Fabbretti A, Pon CL (ed), Antibiotics—targets, mechanims and resistance. Wiley-VCH Verlag GmbH & Co. KGaA Publications, Weinheim, Germany. [Google Scholar]
- 194.Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. 2015. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 195.Bush K. 2018. Past and present perspectives on β-lactamases. Antimicrob Agents Chemother 62:e01076-18. doi: 10.1128/AAC.01076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Carter MQ, Pham A, Huynh S, He X. 2017. Complete genome sequence of a Shiga toxin-producing Enterobacter cloacae clinical isolate. Genome Announc 5:e00883-17. doi: 10.1128/genomeA.00883-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Martínez-Martínez L. 2008. Extended-spectrum beta-lactamases and the permeability barrier. Clin Microbiol Infect 14:82–89. doi: 10.1111/j.1469-0691.2007.01860.x. [DOI] [PubMed] [Google Scholar]
- 198.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Malléa M, Chevalier J, Bornet C, Eyraud A, Davin-Regli A, Bollet C, Pagès JM. 1998. Porin alteration and active efflux: two in vivo drug resistance strategies used by Enterobacter aerogenes. Microbiology 144:3003–3009. doi: 10.1099/00221287-144-11-3003. [DOI] [PubMed] [Google Scholar]
- 200.Babouee Flury B, Ellington MJ, Hopkins KL, Turton JF, Doumith M, Woodford N. 2016. The differential importance of mutations within AmpD in cephalosporin resistance of Enterobacter aerogenes and Enterobacter cloacae. Int J Antimicrob Agents 48:555–558. doi: 10.1016/j.ijantimicag.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 201.Jean SS, Hsueh PR, SMART Asia-Pacific Group. 2017. Distribution of ESBLs, AmpC β-lactamases and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal and urinary tract infections in the Asia-Pacific region during 2008-14: results from the Study for Monitoring Antimicrobial Resistance Trends (SMART). J Antimicrob Chemother 72:166–171. doi: 10.1093/jac/dkw398. [DOI] [PubMed] [Google Scholar]
- 202.Zhou K, Yu W, Cao X, Shen P, Lu H, Luo Q, Rossen JWA, Xiao Y. 2018. Characterization of the population structure, drug resistance mechanisms and plasmids of the community-associated Enterobacter cloacae complex in China. J Antimicrob Chemother 73:66–76. doi: 10.1093/jac/dkx361. [DOI] [PubMed] [Google Scholar]
- 203.Lee SO, Kim YS, Kim BN, Kim MN, Woo JH, Ryu J. 2002. Impact of previous use of antibiotics on development of resistance to extended-spectrum cephalosporins in patients with Enterobacter bacteremia. Eur J Clin Microbiol Infect Dis 21:577–581. doi: 10.1007/s10096-002-0772-7. [DOI] [PubMed] [Google Scholar]
- 204.Jarlier V, INVS. 2014. Surveillance of multidrug resistant bacteria in French healthcare facilities BMR-Raisin network Données 2012 Saint-Maurice: Institut de veille sanitaire. http://www.invs.sante.fr.
- 205.Siebor E, de Curraize C, Neuwirth C. 2018. Identification of AGI1-A, a variant of Acinetobacter genomic island 1 (AGI1), in a French clinical isolate belonging to the Enterobacter cloacae complex. J Antimicrob Chemother doi: 10.1093/jac/dky442. [DOI] [PubMed] [Google Scholar]
- 206.Torres E, López-Cerero L, Del Toro MD, Pascual A. 2014. First detection and characterization of an OXA-48-producing Enterobacter aerogenes isolate. Enferm Infecc Microbiol Clin 32:469–470. doi: 10.1016/j.eimc.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 207.Khajuria A, Praharaj AK, Kumar M, Grover N. 2014. Carbapenem resistance among Enterobacter species in a tertiary care hospital in central India. Chemother Res Pract 2014:972646. doi: 10.1155/2014/972646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Beyrouthy R, Barets M, Marion E, Dananché C, Dauwalder O, Robin F, Gauthier L, Jousset A, Dortet L, Guérin F, Bénet T, Cassier P, Vanhems P, Bonnet R. 2018. Novel Enterobacter lineage as leading cause of nosocomial outbreak involving carbapenemase-Producing Strains. Emerg Infect Dis 24:1505–1515. doi: 10.3201/eid2408.180151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Boyd DA, Mataseje LF, Davidson R, Delport JA, Fuller J, Hoang L, Lefebvre B, Levett PN, Roscoe DL, Willey BM, Mulvey MR. 2017. Enterobacter cloacae complex isolates harboring blaNMC-A or blaIMI-type class A carbapenemase genes on novel chromosomal integrative elements and plasmids. Antimicrob Agents Chemother 61:e02578-16. doi: 10.1128/AAC.02578-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kubota H, Uwamino Y, Matsui M, Sekizuka T, Suzuki Y, Okuno R, Uchitani Y, Ariyoshi T, Aoki W, Suzuki S, Kuroda M, Shinkai T, Yokoyama K, Sadamasu K, Funakoshi T, Murata M, Hasegawa N, Iwata S. 2018. FRI-4 carbapenemase-producing Enterobacter cloacae complex isolated in Tokyo, Japan. J Antimicrob Chemother doi: 10.1093/jac/dky291. [DOI] [PubMed] [Google Scholar]
- 211.Meunier D, Findlay J, Doumith M, Godoy D, Perry C, Pike R, Gronthoud F, Shryane T, Poirel L, Welfare W, Woodford N, Hopkins KL. 2017. FRI-2 carbapenemase-producing Enterobacter cloacae complex in the UK. J Antimicrob Chemother 72:2478–2482. doi: 10.1093/jac/dkx173. [DOI] [PubMed] [Google Scholar]
- 212.Potron A, Poirel L, Rondinaud E, Nordmann P. 2013. Intercontinental spread of OXA-48 β-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill 18:20549. doi: 10.2807/1560-7917.ES2013.18.31.20549. [DOI] [PubMed] [Google Scholar]
- 213.Izdebski R, Baraniak A, Zabicka D, Sekowska A, Gospodarek-Komkowska E, Hryniewicz W, Gniadkowski M. 2018. VIM/IMP carbapenemase-producing Enterobacteriaceae in Poland: epidemic Enterobacter hormaechei and Klebsiella oxytoca lineages. J Antimicrob Chemother doi: 10.1093/jac/dky257. [DOI] [PubMed] [Google Scholar]
- 214.Karlowsky JA, Lob SH, Kazmierczak KM, Badal RE, Young K, Motyl MR, Sahm DF. 2017. In vitro activity of imipenem against carbapenemase-positive Enterobacteriaceae isolates collected by the SMART Global Surveillance Program from 2008 to 2014. J Clin Microbiol 55:1638–1649. doi: 10.1128/JCM.02316-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chavda KD, Chen L, Fouts DE, Sutton G, Brinkac L, Jenkins SG, Bonomo RA, Adams MD, Kreiswirth BN. 2016. Comprehensive genome analysis of carbapenemase-producing Enterobacter spp.: new insights into phylogeny, population structure, and resistance mechanisms. mBio 7:e02093-16. doi: 10.1128/mBio.02093-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wang Q, Wang X, Wang J, Ouyang P, Jin C, Wang R, Zhang Y, Jin L, Chen H, Wang Z, Zhang F, Cao B, Xie L, Liao K, Gu B, Yang C, Liu Z, Ma X, Jin L, Zhang X, Man S, Li W, Pei F, Xu X, Jin Y, Ji P, Wang H. 2018. Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012-2016). Clin Infect Dis 67Suppl 2):S196–S205. doi: 10.1093/cid/ciy660. [DOI] [PubMed] [Google Scholar]
- 217.Wilson BM, El Chakhtoura NG, Patel S, Saade E, Donskey CJ, Bonomo RA, Perez F. 2017. Carbapenem-resistant Enterobacter cloacae in patients from the US Veterans Health Administration, 2006-2015. Emerg Infect Dis 23:878–880. doi: 10.3201/eid2305.162034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Peirano G, Matsumura Y, Adams MD, Bradford P, Motyl M, Chen L, Kreiswirth BN, Pitout J. 2018. Genomic epidemiology of global carbapenemase-producing Enterobacter spp, 2008-2014. Emerg Infect Dis 24:1010–1019. doi: 10.3201/eid2406.171648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Gomez-Simmonds A, Stump S, Giddins MJ, Annavajhala MK, Uhlemann AC. 2018. Clonal background, resistance gene profile, and porin gene mutations modulate in vitro susceptibility to imipenem-relebactam in diverse Enterobacteriaceae. Antimicrob Agents Chemother 62:e00573-18. doi: 10.1128/AAC.00573-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Miró E, Grünbaum F, Gómez L, Rivera A, Mirelis B, Coll P, Navarro F. 2013. Characterization of aminoglycoside-modifying enzymes in Enterobacteriaceae clinical strains and characterization of the plasmids implicated in their diffusion. Microb Drug Resist 19:94–99. doi: 10.1089/mdr.2012.0125. [DOI] [PubMed] [Google Scholar]
- 221.Khennouchi NC, Loucif L, Boutefnouchet N, Allag H, Rolain JM. 2015. MALDI-TOF MS as a tool to detect a nosocomial outbreak of extended-spectrum-β-lactamase- and ArmA methyltransferase-producing Enterobacter cloacae clinical isolates in Algeria. Antimicrob Agents Chemother 59:6477–6483. doi: 10.1128/AAC.00615-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bakthavatchalam YD, Pragasam AK, Biswas I, Veeraraghavan B. 2018. Polymyxin susceptibility testing, interpretative breakpoints and resistance mechanisms: an update. J Glob Antimicrob Resist 12:124–136. doi: 10.1016/j.jgar.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 223.Ebbensgaard A, Mordhorst H, Aarestrup FM, Hansen EB. 2018. The role of outer membrane proteins and lipopolysaccharides for the sensitivity of Escherichia coli to antimicrobial peptides. Front Microbiol 9:2153. doi: 10.3389/fmicb.2018.02153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Braun G, Cole ST. 1983. Molecular characterization of the gene coding for major outer membrane protein OmpA from Enterobacter aerogenes. Eur J Biochem 137:495–500. doi: 10.1111/j.1432-1033.1983.tb07853.x. [DOI] [PubMed] [Google Scholar]
- 225.Nieweg A, Bremer E. 1997. The nucleoside-specific Tsx channel from the outer membrane of Salmonella typhimurium, Klebsiella pneumoniae and Enterobacter aerogenes: functional characterization and DNA sequence analysis of the tsx genes. Microbiology 143:603–615. doi: 10.1099/00221287-143-2-603. [DOI] [PubMed] [Google Scholar]
- 226.Schulz GE. 2002. The structure of bacterial outer membrane proteins. Biochim Biophys Acta 1565:308–317. doi: 10.1016/S0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- 227.Fairman JW, Noinaj N, Buchanan SK. 2011. The structural biology of β-barrel membrane proteins: a summary of recent reports. Curr Opin Struct Biol 21:523–531. doi: 10.1016/j.sbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Selkrig J, Leyton DL, Webb CT, Lithgow T. 2014. Assembly of β-barrel proteins into bacterial outer membranes. Biochim Biophys Acta 1843:1542–1550. doi: 10.1016/j.bbamcr.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 229.Sperandeo P, Martorana AM, Polissi A. 2017. Lipopolysaccharide biogenesis and transport at the outer membrane of Gram-negative bacteria. Biochim Biophys Acta Mol Cell Biol Lipids 1862:1451–1460. doi: 10.1016/j.bbalip.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 230.Acosta-Gutiérrez S, Ferrara L, Pathania M, Masi M, Wang J, Bodrenko I, Zahn M, Winterhalter M, Stavenger RA, Pagès JM, Naismith JH, van den Berg B, Page MGP, Ceccarelli M. 2018. Getting drugs into Gram-negative bacteria: rational rules for permeation through general porins. ACS Infect Dis 4:1487–1498. doi: 10.1021/acsinfecdis.8b00108. [DOI] [PubMed] [Google Scholar]
- 231.James CE, Mahendran KR, Molitor A, Bolla JM, Bessonov AN, Winterhalter M, Pagès JM. 2009. How beta-lactam antibiotics enter bacteria: a dialogue with the porins. PLoS One 4:e5453. doi: 10.1371/journal.pone.0005453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Chevalier J, Malléa M, Pagès JM. 2000. Comparative aspects of the diffusion of norfloxacin, cefepime and spermine through the F porin channel of Enterobacter cloacae. Biochem J 348:223–227. doi: 10.1042/bj3480223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Malléa M, Simonet V, Lee EH, Gervier R, Collatz E, Gutmann L, Pagès JM. 1995. Biological and immunological comparisons of Enterobacter cloacae and Escherichia coli porins. FEMS Microbiol Lett 129:273–279. doi: 10.1111/j.1574-6968.1995.tb07592.x. [DOI] [PubMed] [Google Scholar]
- 234.De la Cruz MA, Calva E. 2010. The complexities of porin genetic regulation. J Mol Microbiol Biotechnol 18:24–36. doi: 10.1159/000274309. [DOI] [PubMed] [Google Scholar]
- 235.Masi M, Winterhalter M, Pagès JM. 2019. Outer membrane porins, p 1–54. In Kuhn A. (ed), Bacterial cell walls and membranes. Springer Nature Publications, Basel, Switzerland. [Google Scholar]
- 236.Bornet C, Chollet R, Malléa M, Chevalier J, Davin-Regli A, Pagès JM, Bollet C. 2003. Imipenem and expression of multidrug efflux pump in Enterobacter aerogenes. Biochem Biophys Res Commun 301:985–990. doi: 10.1016/S0006-291X(03)00074-3. [DOI] [PubMed] [Google Scholar]
- 237.Philippe N, Maigre L, Santini S, Pinet E, Claverie JM, Davin-Régli AV, Pagès JM, Masi M. 2015. In vivo evolution of bacterial resistance in two cases of Enterobacter aerogenes infections during treatment with imipenem. PLoS One 10:e0138828. doi: 10.1371/journal.pone.0138828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Gayet S, Chollet R, Molle G, Pagès JM, Chevalier J. 2003. Modification of outer membrane protein profile and evidence suggesting an active drug pump in Enterobacter aerogenes clinical strains. Antimicrob Agents Chemother 47:1555–1559. doi: 10.1128/aac.47.5.1555-1559.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Dupont M, James CE, Chevalier J, Pagès JM. 2007. An early response to environmental stress involves regulation of OmpX and OmpF, two enterobacterial outer membrane pore-forming proteins. Antimicrob Agents Chemother 51:3190–3198. doi: 10.1128/AAC.01481-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Stoorvogel J, van Bussel MJ, Tommassen J, van de Klundert JA. 1991. Molecular characterization of an Enterobacter cloacae outer membrane protein (OmpX). J Bacteriol 173:156–160. doi: 10.1128/jb.173.1.156-160.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Stoorvogel J, van Bussel MJ, van de Klundert JA. 1991. Biological characterization of an Enterobacter cloacae outer membrane protein (OmpX). J Bacteriol 173:161–167. doi: 10.1128/jb.173.1.161-167.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Bauer K, Schmid A, Boos W, Benz R, Tommassen J. 1988. Pore formation by pho-controlled outer-membrane proteins of various Enterobacteriaceae in lipid bilayers. Eur J Biochem 174:199–205. doi: 10.1111/j.1432-1033.1988.tb14082.x. [DOI] [PubMed] [Google Scholar]
- 243.Bornet C, Saint N, Fetnaci L, Dupont M, Davin-Régli A, Bollet C, Pagès JM. 2004. Omp35, a new Enterobacter aerogenes porin involved in selective susceptibility to cephalosporins. Antimicrob Agents Chemother 48:2153–2158. doi: 10.1128/AAC.48.6.2153-2158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Doumith M, Ellington MJ, Livermore DM, Woodford N. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother 63:659–667. doi: 10.1093/jac/dkp029. [DOI] [PubMed] [Google Scholar]
- 245.Dupont H, Gaillot O, Goetgheluck AS, Plassart C, Emond JP, Lecuru M, Gaillard N, Derdouri S, Lemaire B, Girard de Courtilles M, Cattoir V, Mammeri H. 2016. Molecular characterization of carbapenem-nonsusceptible enterobacterial isolates collected during a prospective interregional survey in France and susceptibility to the novel ceftazidime-avibactam and aztreonam-avibactam combinations. Antimicrob Agents Chemother 60:215–221. doi: 10.1128/AAC.01559-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Babouee Flury B, Ellington MJ, Hopkins KL, Turton JF, Doumith M, Loy R, Staves P, Hinic V, Frei R, Woodford N. 2016. Association of novel nonsynonymous single sucleotide polymorphisms in AmpD with cephalosporin resistance and phylogenetic variations in AmpC, AmpR, OmpF, and OmpC in Enterobacter cloacae isolates that are highly resistant to carbapenems. Antimicrob Agents Chemother 60:2383–2390. doi: 10.1128/AAC.02835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Bedenić B, Vranić-Ladavac M, Venditti C, Tambić-Andrašević A, Barišić N, Gužvinec M, Karčić N, Petrosillo N, Ladavac R, di Caro A. 2018. Emergence of colistin resistance in Enterobacter aerogenes from Croatia. J Chemother 30:120–123. doi: 10.1080/1120009X.2017.1382121. [DOI] [PubMed] [Google Scholar]
- 248.Hao M, Ye M, Shen Z, Hu F, Yang Y, Wu S, Xu X, Zhu S, Qin X, Wang M. 2018. Porin deficiency in carbapenem-resistant Enterobacter aerogenes strains. Microb Drug Resist 24:1277–1283. doi: 10.1089/mdr.2017.0379. [DOI] [PubMed] [Google Scholar]
- 249.Phan K, Ferenci T. 2017. The fitness costs and trade-off shapes associated with the exclusion of nine antibiotics by OmpF porin channels. ISME J 11:1472–1482. doi: 10.1038/ismej.2016.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Hennequin C, Robin F. 2016. Correlation between antimicrobial resistance and virulence in Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis 35:333–341. doi: 10.1007/s10096-015-2559-7. [DOI] [PubMed] [Google Scholar]
- 251.Ferreira RL, da Silva BCM, Rezende GS, Nakamura-Silva R, Pitondo-Silva A, Campanini EB, Brito MCA, da Silva EML, Freire CCM, da Cunha AF, Pranchevicius MDS. 2019. High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and β-lactamase encoding genes in a Brazilian intensive care unit. Front Microbiol 9:3198. doi: 10.3389/fmicb.2018.03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Pantel A, Dunyach-Remy C, Ngba Essebe C, Mesureur J, Sotto A, Pagès JM, Nicolas-Chanoine MH, Lavigne JP. 2016. Modulation of membrane influx and efflux in Escherichia coli sequence type 131 has an impact on bacterial motility, biofilm formation, and virulence in a Caenorhabditis elegans model. Antimicrob Agents Chemother 60:2901–2911. doi: 10.1128/AAC.02872-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Pradel E, Pagès JM. 2002. The AcrAB-TolC efflux pump contributes to multidrug resistance in the nosocomial pathogen Enterobacter aerogenes. Antimicrob Agents Chemother 46:2640–2643. doi: 10.1128/aac.46.8.2640-2643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Pérez A, Canle D, Latasa C, Poza M, Beceiro A, Tomás Mdel M, Fernández A, Mallo S, Pérez S, Molina F, Villanueva R, Lasa I, Bou G. 2007. Cloning, nucleotide sequencing, and analysis of the AcrAB-TolC efflux pump of Enterobacter cloacae and determination of its involvement in antibiotic resistance in a clinical isolate. Antimicrob Agents Chemother 51:3247–3253. doi: 10.1128/AAC.00072-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Li XZ, Plésiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Du D, Wang-Kan X, Neuberger A, van Veen HW, Pos KM, Piddock LJV, Luisi BF. 2018. Multidrug efflux pumps: structure, function and regulation. Nat Rev Microbiol 16:523–539. doi: 10.1038/s41579-018-0048-6. [DOI] [PubMed] [Google Scholar]
- 257.Saier MH Jr, Reddy VS, Tamang DG, Västermark A. 2014. The transporter classification database. Nucleic Acids Res 42:D251–D258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.He GX, Thorpe C, Walsh D, Crow R, Chen H, Kumar S, Varela MF. 2011. EmmdR, a new member of the MATE family of multidrug transporters, extrudes quinolones from Enterobacter cloacae. Arch Microbiol 193:759–765. doi: 10.1007/s00203-011-0738-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.He GX, Zhang C, Crow RR, Thorpe C, Chen H, Kumar S, Tsuchiya T, Varela MF. 2011. SugE, a new member of the SMR family of transporters, contributes to antimicrobial resistance in Enterobacter cloacae. Antimicrob Agents Chemother 55:3954–3957. doi: 10.1128/AAC.00094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Schuldiner S. 2018. The Escherichia coli effluxome. Res Microbiol 169:357–362. doi: 10.1016/j.resmic.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 261.Neuberger A, Du D, Luisi BF. 2018. Structure and mechanism of bacterial tripartite efflux pumps. Res Microbiol 169:401–413. doi: 10.1016/j.resmic.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 262.Nikaido H, Pagès JM. 2012. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol Rev 36:340–363. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Guérin F, Isnard C, Sinel C, Morand P, Dhalluin A, Cattoir V, Giard JC. 2016. Cluster-dependent colistin hetero-resistance in Enterobacter cloacae complex. J Antimicrob Chemother 71:3058–3061. doi: 10.1093/jac/dkw260. [DOI] [PubMed] [Google Scholar]
- 264.Masi M, Pagès JM, Pradel E. 2003. Overexpression and purification of the three components of the Enterobacter aerogenes AcrA-AcrB-TolC multidrug efflux pump. J Chromatogr B Anal Technol Biomed Life Sci 786:197–205. doi: 10.1016/S1570-0232(02)00746-8. [DOI] [PubMed] [Google Scholar]
- 265.Masi M, Saint N, Molle G, Pagès JM. 2007. The Enterobacter aerogenes outer membrane efflux proteins TolC and EefC have different channel properties. Biochim Biophys Acta 1768:2559–2567. doi: 10.1016/j.bbamem.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 266.Tran QT, Dupont M, Lavigne JP, Chevalier J, Pagès JM, Sotto A, Davin-Regli A. 2009. Occurrence of efflux mechanism and cephalosporinase variant in a population of Enterobacter aerogenes and Klebsiella pneumoniae isolates producing extended-spectrum beta-lactamases. Antimicrob Agents Chemother 53:1652–1656. doi: 10.1128/AAC.00822-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF. 2014. Structure of the AcrAB-TolC multidrug efflux pump. Nature 509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Hobbs EC, Yin X, Paul BJ, Astarita JL, Storz G. 2012. Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proc Natl Acad Sci U S A 109:16696–16701. doi: 10.1073/pnas.1210093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wong MH, Chan EW, Chen S. 2015. Evolution and dissemination of OqxAB-like efflux pumps, an emerging quinolone resistance determinant among members of Enterobacteriaceae. Antimicrob Agents Chemother 59:3290–3297. doi: 10.1128/AAC.00310-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Andrade LN, Siqueira TES, Martinez R, Darini A. 2018. Multidrug-Resistant CTX-M-(15, 9, 2) and KPC-2-Producing Enterobacter hormaechei and Enterobacter asburiae isolates possessed a set of acquired heavy metal tolerance genes including a chromosomal sil operon (for acquired silver resistance). Front Microbiol 9:539. doi: 10.3389/fmicb.2018.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Chollet R, Chevalier J, Bollet C, Pagès JM, Davin-Regli A. 2004. RamA is an alternate activator of the multidrug resistance cascade in Enterobacter aerogenes. Antimicrob Agents Chemother 48:2518–2523. doi: 10.1128/AAC.48.7.2518-2523.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Cinquin B, Maigre L, Pinet E, Chevalier J, Stavenger RA, Mills S, Réfrégiers M, Pagès JM. 2015. Microspectrometric insights on the uptake of antibiotics at the single bacterial cell level. Sci Rep 5:17968. doi: 10.1038/srep17968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Kaščáková S, Maigre L, Chevalier J, Réfrégiers M, Pagès JM. 2012. Antibiotic transport in resistant bacteria: synchrotron UV fluorescence microscopy to determine antibiotic accumulation with single cell resolution. PLoS One 7:e38624. doi: 10.1371/journal.pone.0038624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Vergalli J, Dumont E, Cinquin B, Maigre L, Pajovic J, Bacqué E, Mourez M, Réfrégiers M, Pagès JM. 2017. Fluoroquinolone structure and translocation flux across bacterial membrane. Sci Rep 7:9821. doi: 10.1038/s41598-017-08775-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Vergalli J, Dumont E, Pajović J, Cinquin B, Maigre L, Masi M, Réfrégiers M, Pagés JM. 2018. Spectrofluorimetric quantification of antibiotic drug concentration in bacterial cells for the characterization of translocation across bacterial membranes. Nat Protoc 13:1348–1361. doi: 10.1038/nprot.2018.036. [DOI] [PubMed] [Google Scholar]
- 276.Ghisalberti D, Masi M, Pagès JM, Chevalier J. 2005. Chloramphenicol and expression of multidrug efflux pump in Enterobacter aerogenes. Biochem Biophys Res Commun 328:1113–1118. doi: 10.1016/j.bbrc.2005.01.069. [DOI] [PubMed] [Google Scholar]
- 277.Fisher RA, Gollan B, Helaine S. 2017. Persistent bacterial infections and persister cells. Nat Rev Microbiol 15:453–464. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 278.Pérez A, Poza M, Aranda J, Latasa C, Medrano FJ, Tomás M, Romero A, Lasa I, Bou G. 2012. Effect of transcriptional activators SoxS, RobA, and RamA on expression of multidrug efflux pump AcrAB-TolC in Enterobacter cloacae. Antimicrob Agents Chemother 56:6256–6266. doi: 10.1128/AAC.01085-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Tzouvelekis LS, Tzelepi E, Kaufmann ME, Mentis AF. 1994. Consecutive mutations leading to the emergence in vivo of imipenem resistance in a clinical strain of Enterobacter aerogenes. J Med Microbiol 40:403–407. doi: 10.1099/00222615-40-6-403. [DOI] [PubMed] [Google Scholar]
- 280.Baron S, Bardet L, Dubourg G, Fichaux M, Rolain JM. 2017. mcr-1 plasmid-mediated colistin resistance gene detection in an Enterobacter cloacae clinical isolate in France. J Glob Antimicrob Resist 10:35–36. doi: 10.1016/j.jgar.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 281.Uechi K, Tada T, Shimada K, Nakasone I, Kirikae T, Fujita J. 2019. Emergence of a carbapenem-resistant and colistin-heteroresistant Enterobacter cloacae clinical isolate in Japan. J Infect Chemother 25:285–288. doi: 10.1016/j.jiac.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 282.Telke AA, Olaitan AO, Morand S, Rolain JM. 2017. SoxRS induces colistin hetero-resistance in Enterobacter asburiae and Enterobacter cloacae by regulating the acrAB-tolC efflux pump. J Antimicrob Chemother 72:2715–2721. doi: 10.1093/jac/dkx215. [DOI] [PubMed] [Google Scholar]
- 283.Zeng K-J, Doi Y, Patil S, Huang X, Tian G-B. 2016. Emergence of the plasmid-mediated mcr-1 gene in colistin-resistant Enterobacter aerogenes and Enterobacter cloacae. Antimicrob Agents Chemother 60:3862–3863. doi: 10.1128/AAC.00345-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Prim N, Turbau M, Rivera A, Rodríguez-Navarro J, Coll P, Mirelis B. 2017. Prevalence of colistin resistance in clinical isolates of Enterobacteriaceae: a four-year cross-sectional study. J Infect 75:493–498. doi: 10.1016/j.jinf.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 285.Guérin F, Lallement C, Isnard C, Dhalluin A, Cattoir V, Giard JC. 2016. Landscape of resistance-nodulation-cell division (rnd)-type efflux pumps in Enterobacter cloacae complex. Antimicrob Agents Chemother 60:2373–2382. doi: 10.1128/AAC.02840-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Dautzenberg MJD, Bayjanov JR, Leverstein-van Hall MA, Muller AE, Gelinck LBS, Jansen CL, Leyten EMS, Ruys T, Scharringa J, van der Starre RE, Fluit AC, Bonten M. 2018. Dynamics of colistin and tobramycin resistance among Enterobacter cloacae during prolonged use of selective decontamination of the digestive tract. Antimicrob Resist Infect Control 7:67. doi: 10.1186/s13756-018-0356-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Kohlmann R, Bähr T, Gatermann SG. 2018. Species-specific mutation rates for ampC derepression in Enterobacterales with chromosomally encoded inducible AmpC β-lactamase. J Antimicrob Chemother 73:1530–1536. doi: 10.1093/jac/dky084. [DOI] [PubMed] [Google Scholar]
- 288.Guérin F, Isnard C, Cattoir V, Giard JC. 2015. Complex regulation pathways of AmpC-mediated β-lactam resistance in Enterobacter cloacae complex. Antimicrob Agents Chemother 59:7753–7761. doi: 10.1128/AAC.01729-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Islam MA, Islam M, Hasan R, Hossain MI, Nabi A, Rahman M, Goessens WHF, Endtz HP, Boehm AB, Faruque SM. 2017. Environmental spread of New Delhi metallo-β-lactamase-1-producing multidrug-resistant bacteria in Dhaka, Bangladesh. Appl Environ Microbiol 83:e00793-17. doi: 10.1128/AEM.00793-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Livermore DM, Mushtaq S, Barker K, Hope R, Warner M, Woodford N. 2012. Characterization of β-lactamase and porin mutants of Enterobacteriaceae selected with ceftaroline + avibactam (NXL104). J Antimicrob Chemother 67:1354–1358. doi: 10.1093/jac/dks079. [DOI] [PubMed] [Google Scholar]
- 291.Deguchi T, Yasuda M, Nakano M, Ozeki S, Kanematsu E, Nishino Y, Ishihara S, Kawada Y. 1997. Detection of mutations in the gyrA and parC genes in quinolone-resistant clinical isolates of Enterobacter cloacae. J Antimicrob Chemother 40:543–549. doi: 10.1093/jac/40.4.543. [DOI] [PubMed] [Google Scholar]
- 292.De Jong A, Muggeo A, El Garch F, Moyaert H, de Champs C, Guillard T. 2018. Characterization of quinolone resistance mechanisms in Enterobacteriaceae isolated from companion animals in Europe (ComPath II study). Vet Microbiol 216:159–167. doi: 10.1016/j.vetmic.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 293.Linde HJ, Notka F, Irtenkauf C, Decker J, Wild J, Niller HH, Heisig P, Lehn N. 2002. Increase in MICs of ciprofloxacin in vivo in two closely related clinical isolates of Enterobacter cloacae. J Antimicrob Chemother 49:625–630. doi: 10.1093/jac/49.4.625. [DOI] [PubMed] [Google Scholar]
- 294.Nordmann P, Poirel L. 2005. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother 56:463–469. doi: 10.1093/jac/dki245. [DOI] [PubMed] [Google Scholar]
- 295.Park YJ, Yu JK, Kim SI, Lee K, Arakawa Y. 2009. Accumulation of plasmid-mediated fluoroquinolone resistance genes, qepA and qnrS1, in Enterobacter aerogenes co-producing RmtB and class A beta-lactamase LAP-1. Ann Clin Lab Sci Winter 39:55–59. [PubMed] [Google Scholar]
- 296.Jlili NE-H, Rejiba S, Smaoui H, Guillard T, Chau F, Kechrid A, Cambau E. 2014. Trend of plasmid-mediated quinolone resistance genes at the Children's Hospital in Tunisia. J Med Microbiol 63:195–202. doi: 10.1099/jmm.0.062216-0. [DOI] [PubMed] [Google Scholar]
- 297.Osei Sekyere J, Amoako DG. 2017. Genomic and phenotypic characterisation of fluoroquinolone resistance mechanisms in Enterobacteriaceae in Durban, South Africa. PLoS One 12:e0178888. doi: 10.1371/journal.pone.0178888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Albornoz E, Tijet N, De Belder D, Gomez S, Martino F, Corso A, Melano RG, Petroni A. 2017. qnrE1, a member of a new family of plasmid-located quinolone resistance genes, originated from the chromosome of Enterobacter species. Antimicrob Agents Chemother 61:e02555-16. doi: 10.1128/AAC.02555-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Davin-Regli A, Bolla J-M, James CE, Lavigne J-P, Chevalier J, Garnotel E, Molitor A, Pagès JM. 2008. Membrane permeability and regulation of drug “influx and efflux” in enterobacterial pathogens. Curr Drug Targets 9:750–759. doi: 10.2174/138945008785747824. [DOI] [PubMed] [Google Scholar]
- 300.Cha MK, Kang CI, Park GE, Kim SH, Chung DR, Peck KR, Song JH. 2018. Genetic characterisation of tigecycline-resistant Enterobacter spp. in blood isolates causing bacteraemia. J Glob Antimicrob Resist 13:115–118. doi: 10.1016/j.jgar.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 301.Veleba M, Higgins PG, Gonzalez G, Seifert H, Schneiders T. 2012. Characterization of RarA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae. Antimicrob Agents Chemother 56:4450–4458. doi: 10.1128/AAC.00456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Veleba M, De Majumdar S, Hornsey M, Woodford N, Schneiders T. 2013. Genetic characterization of tigecycline resistance in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J Antimicrob Chemother 68:1011–1018. doi: 10.1093/jac/dks530. [DOI] [PubMed] [Google Scholar]
- 303.Pavlovic M, Konrad R, Iwobi AN, Sing A, Busch U, Huber I. 2012. A dual approach employing MALDI-TOF MS and real-time PCR for fast species identification within the Enterobacter cloacae complex. FEMS Microbiol Lett 328:46–53. doi: 10.1111/j.1574-6968.2011.02479.x. [DOI] [PubMed] [Google Scholar]
- 304.Pham HN, Ohkusu K, Mishima N, Noda M, Monir Shah M, Sun X, Hayashi M, Ezaki T. 2007. Phylogeny and species identification of the family Enterobacteriaceae based on dnaJ sequences. Diagn Microbiol Infect Dis 58:153–161. doi: 10.1016/j.diagmicrobio.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 305.Mishra M, Patole S, Mohapatra H. 2017. Draft genome sequences of nonclinical and clinical Enterobacter cloacae isolates exhibiting multiple antibiotic resistance and virulence factors. Genome Announc 5:e01218-17. doi: 10.1128/genomeA.01218-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Muresu R, Maddau G, Delogu G, Cappuccinelli P, Squartini A. 2010. Bacteria colonizing root nodules of wild legumes exhibit virulence-associated properties of mammalian pathogens. Antonie Van Leeuwenhoek 97:143–153. doi: 10.1007/s10482-009-9396-6. [DOI] [PubMed] [Google Scholar]
- 307.Paton AW, Paton JC. 1996. Enterobacter cloacae producing a Shiga-like toxin II-related cytotoxin associated with a case of hemolytic-uremic syndrome. J Clin Microbiol 34:463–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Probert WS, McQuaid C, Schrader K. 2014. Isolation and identification of an Enterobacter cloacae strain producing a novel subtype of Shiga toxin type 1. J Clin Microbiol 52:2346–2351. doi: 10.1128/JCM.00338-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Simi S, Carbonell GV, Falcón RM, Gatti MS, Joazeiro PP, Darini AL, Yano T. 2003. A low molecular weight enterotoxic hemolysin from clinical Enterobacter cloacae. Can J Microbiol 49:479–482. doi: 10.1139/w03-060. [DOI] [PubMed] [Google Scholar]
- 310.Grazziotin AL, Vidal NM, Palmeiro JK, Dalla-Costa LM, Venancio TM. 2016. Genome sequencing of four multidrug-resistant Enterobacter aerogenes isolates from hospitalized patients in Brazil. Front Microbiol 7:1649. doi: 10.3389/fmicb.2016.01649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Lau YY, Yin WF, Chan KG. 2014. Enterobacter asburiae strain L1: complete genome and whole genome optical mapping analysis of a quorum sensing bacterium. Sensors (Basel) 14:13913–13924. doi: 10.3390/s140813913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Liu F, Yang J, Xiao Y, Li L, Yang F, Jin Q. 2016. Complete genome sequence of a clinical isolate of Enterobacter asburiae. Genome Announc 4:e00523-16. doi: 10.1128/genomeA.00523-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Ren Y, Ren Y, Zhou Z, Guo X, Li Y, Feng L, Wang L. 2010. Complete genome sequence of Enterobacter cloacae subsp. cloacae type strain ATCC 13047. J Bacteriol 192:2463–2464. doi: 10.1128/JB.00067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Brooks LE, Ul-Hasan S, Chan BK, Sistrom MJ. 2018. Quantifying the evolutionary conservation of genes encoding multidrug efflux pumps in the ESKAPE pathogens to identify antimicrobial drug targets. mSystems 3:e00024-18. doi: 10.1128/mSystems.00024-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Ho PL, Shek RH, Chow KH, Duan RS, Mak GC, Lai EL, Yam WC, Tsang KW, Lai WM. 2005. Detection and characterization of extended-spectrum beta-lactamases among bloodstream isolates of Enterobacter spp. in Hong Kong, 2000-2002. J Antimicrob Chemother 55:326–332. doi: 10.1093/jac/dki010. [DOI] [PubMed] [Google Scholar]
- 316.Bratu S, Landman D, Alam M, Tolentino E, Quale J. 2005. Detection of KPC carbapenem-hydrolyzing enzymes in Enterobacter spp. from Brooklyn, New York. Antimicrob Agents Chemother 49:776–778. doi: 10.1128/AAC.49.2.776-778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Yang B, Feng Y, McNally A, Zong Z. 2018. Occurrence of Enterobacter hormaechei carrying blaNDM-1 and blaKPC-2 in China. Diagn Microbiol Infect Dis 90:139–142. doi: 10.1016/j.diagmicrobio.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 318.Naas T, Cattoen C, Bernusset S, Cuzon G, Nordmann P. 2012. First identification of blaIMI-1 in an Enterobacter cloacae clinical isolate from France. Antimicrob Agents Chemother 56:1664–1665. doi: 10.1128/AAC.06328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Falcone M, Mezzatesta ML, Perilli M, Forcella C, Giordano A, Cafiso V, Amicosante G, Stefani S, Venditti M. 2009. Infections with VIM-1 metallo-β-lactamase-producing Enterobacter cloacae and their correlation with clinical outcome. J Clin Microbiol 47:3514–3519. doi: 10.1128/JCM.01193-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Guillard T, Cholley P, Limelette A, Hocquet D, Matton L, Guyeux C, Lebreil AL, Bajolet O, Brasme L, Madoux J, Vernet-Garnier V, Barbe C, Bertrand X, de Champs O, CarbaFrEst Group. 2015. Fluoroquinolone resistance mechanisms and population structure of Enterobacter cloacae non-susceptible to ertapenem in north-eastern France. Front Microbiol 23:1186. doi: 10.3389/fmicb.2015.01186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Bogaerts P, Bouchahrouf W, de Castro RR, Deplano A, Berhin C, Piérard D, Denis O, Glupczynski Y. 2011. Emergence of NDM-1-producing Enterobacteriaceae in Belgium. Antimicrob Agents Chemother 55:3036–3038. doi: 10.1128/AAC.00049-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Ghosh R, Aggeler R. 1987. Effect of lipid fluidity upon the activity and structure of the 39 kDa porin from Enterobacter cloacae 908S. FEBS Lett 222:154–158. doi: 10.1016/0014-5793(87)80210-7. [DOI] [PubMed] [Google Scholar]
- 323.Gibson JS, Cobbold RN, Heisig P, Sidjabat HE, Kyaw-Tanner MT, Trott DJ. 2010. Identification of Qnr and AAC(6′)-1b-cr plasmid-mediated fluoroquinolone resistance determinants in multidrug-resistant Enterobacter spp. isolated from extraintestinal infections in companion animals. Vet Microbiol 143:329–336. doi: 10.1016/j.vetmic.2009.11.031. [DOI] [PubMed] [Google Scholar]
- 324.Amin M, Mehdipour G, Navidifar T. 2019. High distribution of 16S rRNA methylase genes rmtB and armA among Enterobacter cloacae strains isolated from an Ahvaz teaching hospital, Iran. Acta Microbiol Immunol Hung doi: 10.1556/030.66.2019.009. [DOI] [PubMed] [Google Scholar]
- 325.Keeney D, Ruzin A, Bradford PA. 2007. RamA, a transcriptional regulator, and AcrAB, an RND-type efflux pump, are associated with decreased susceptibility to tigecycline in Enterobacter cloacae. Microb Drug Resist 13:1–6. doi: 10.1089/mdr.2006.9990. [DOI] [PubMed] [Google Scholar]
- 326.Werts C, O'Callaghan D, Hofnung M, Charbit A. 1993. Immunological relatedness of the LamB proteins among members of Enterobacteriaceae. J Gen Microbiol 139:881–887. doi: 10.1099/00221287-139-4-881. [DOI] [PubMed] [Google Scholar]
- 327.Masi M, Pagès JM, Villard C, Pradel E. 2005. The eefABC multidrug efflux pump operon is repressed by H-NS in Enterobacter aerogenes. J Bacteriol 187:3894–3897. doi: 10.1128/JB.187.11.3894-3897.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Masi M, Pagès JM, Pradel E. 2006. Production of the cryptic EefABC efflux pump in Enterobacter aerogenes chloramphenicol-resistant mutants. J Antimicrob Chemother 57:1223–1226. doi: 10.1093/jac/dkl139. [DOI] [PubMed] [Google Scholar]