Protozoan Plasmodium parasites are the causative agents of malaria, a deadly disease that continues to afflict hundreds of millions of people every year. Infections with malaria parasites can be asymptomatic, with mild or severe symptoms, or fatal, depending on many factors such as parasite virulence and host immune status. Malaria can be treated with various drugs, with artemisinin-based combination therapies (ACTs) being the first-line choice.
KEYWORDS: association studies, genetic mapping, genome diversity, population structure, evolutionary selection
SUMMARY
Protozoan Plasmodium parasites are the causative agents of malaria, a deadly disease that continues to afflict hundreds of millions of people every year. Infections with malaria parasites can be asymptomatic, with mild or severe symptoms, or fatal, depending on many factors such as parasite virulence and host immune status. Malaria can be treated with various drugs, with artemisinin-based combination therapies (ACTs) being the first-line choice. Recent advances in genetics and genomics of malaria parasites have contributed greatly to our understanding of parasite population dynamics, transmission, drug responses, and pathogenesis. However, knowledge gaps in parasite biology and host-parasite interactions still remain. Parasites resistant to multiple antimalarial drugs have emerged, while advanced clinical trials have shown partial efficacy for one available vaccine. Here we discuss genetic and genomic studies of Plasmodium biology, host-parasite interactions, population structures, mosquito infectivity, antigenic variation, and targets for treatment and immunization. Knowledge from these studies will advance our understanding of malaria pathogenesis, epidemiology, and evolution and will support work to discover and develop new medicines and vaccines.
INTRODUCTION
Malaria is a deadly disease caused by Plasmodium species, a large and diverse taxonomic group that includes parasites of birds, reptiles, rodents, monkeys, apes, and humans. Hundreds of millions of people are infected annually across tropical and subtropical regions, largely Africa, South and Central America, India, Southeast Asia, and Oceania (1–3). In addition to studies of the major species infecting humans (Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, Plasmodium ovale, and Plasmodium knowlesi), malaria research employs models including infections of nonhuman primates (e.g., Plasmodium reichenowi [infecting chimpanzees] and Plasmodium cynomolgi and P. knowlesi [infecting Macaca fascicularis]), rodents (e.g., Plasmodium berghei, Plasmodium chabaudi, and Plasmodium yoelii [infecting Mus musculus]), and birds (e.g., Plasmodium gallinaceum [infecting Gallus gallus domesticus]) (4–10).
Malaria parasites are transmitted by mosquitoes. The sporozoite forms inoculated by mosquito feeding produce blood stages that cause a wide range of clinical outcomes, ranging from no symptom at all to severe malaria and death. The onset of malaria often manifests by constitutional symptoms similar to those of “the flu,” such as fever, chills, headache, dizziness, back pain, myalgia, joint and bone pains, cough, chest pain, weakness, prostration, nausea, vomiting, and diarrhea. Progression to severe malaria happens in a small fraction of patients, with life-threatening or outright lethal developments of coma (cerebral malaria), pulmonary edema, acute renal failure, severe anemia, acidosis, hypoglycemia, and/or bleeding (11). Interestingly, many adults in regions of endemicity in African countries often carry parasites in the bloodstream, including gametocytes that are infectious to mosquitoes, but do not have clinical symptoms and are thus considered “asymptomatic.” However, some adults can develop recurrent episodes of symptomatic parasitemia, chronic anemia, maternal morbidity, coinfection with invasive bacterial disease, and cognitive impairment (12).
Sporozoites inoculated into the skin by mosquito feeding transit through the dermis and enter the bloodstream, where they are carried to the liver (13). The parasites then enter hepatocytes and complete a cycle of schizogony, growing and dividing into exoerythrocytic merozoites. In P. vivax and P. ovale infections, some parasites become dormant forms, called “hypnozoites,” after invasion of their host hepatocytes (14), where they can remain quiescent for months or years before entering schizogony (15, 16); P. falciparum, P. malariae, and P. knowlesi parasites do not produce such hypnozoites. After schizogony, merozoites are released from the hepatocytes into bloodstream, where they invade erythrocytes. There they start new cycles of schizogony, with each cycle progressing from morphologically characteristic ring-stage forms to trophozoite and then schizont stages. Mature schizonts have a segmented appearance from mitotic divisions that produce many daughter merozoites, each for invasion of a new erythrocyte upon its release from the consumed and destroyed host cell. The duration of each erythrocytic cycle is approximately 48 h for P. falciparum, P. vivax, and P. ovale, 24 h for P. knowlesi, and 72 h for P. malariae.
After several erythrocytic cycles and under likely stimulations from host immune responses, fever, and even antimalarial therapy, some of the ring forms develop into male and female gametocytes that are infective to mosquitoes (schizogenic replication is asexual). When mosquitoes take blood meals from the infected patient, male and female gametes differentiated from gametocytes fertilize in the mosquito midgut to produce zygotes that develop into motile ookinetes. Ookinetes penetrate through the mosquito midgut wall and develop into oocysts. Approximately 2 weeks later (depending on the parasite species), thousands of sporozoites result from schizogony in each oocyst. The mature sporozoites then migrate via hemolymph circulation and penetrate the mosquito salivary gland, where they await injection into another person when the mosquito takes a blood meal again (17).
Various drugs are used for malaria prevention and cure. These include quinine (QN), chloroquine (CQ), amodiaquine, piperaquine (PPQ), mefloquine (MQ), lumefantrine (LUM), pyrimethamine (PYR), proguanil, sulfadoxine, atovaquone, primaquine, and artemisinin and its derivatives (ART) (18). Artemisinin-based combination therapies (ACTs) are now recommended worldwide for treatment of P. falciparum infections (19). Primaquine and the recently approved tafenoquine are the only antimalarials available against liver-stage parasites and hypnozoites (20, 21). Unfortunately, resistance (or treatment failure) has been reported from nearly all malarious regions (22). Efforts are being made to discover new antimalarial drugs and to understand the molecular mechanisms of drug resistances (23).
THE PLASMODIUM GENOMES
Genome Sequences and Characteristics of Important Plasmodium Species
Since completion of the first draft sequence of the P. falciparum 3D7 genome in 2002 (5), genomic research on malaria parasites has advanced rapidly in step with next-generation sequencing (NGS) technologies and reduction in costs (24). In addition to the sequences of important species that infect humans (4, 6, 8, 25–27), public databases now provide the genome information of primate, rodent, and avian parasites, including those of widely used disease models such as P. berghei, P. chabaudi, P. yoelii, P. reichenowi, P. cynomolgi, P. relictum, and P. gallinaceum (Table 1) (7, 28–33). The evolutionary relationships of Plasmodium spp. have been extensively investigated using various DNA sequences, with clustering of parasite species largely depending on their host origins (34–36). Data from large numbers of laboratory lines and field isolates of P. falciparum and P. vivax have been deposited from studies of genome diversity, parasite evolution, population genetics, and drug resistances (37–44), leading to generation of an updated reference P. falciparum genome with greatly improved annotation (45). More recently, single-cell sequencing techniques have begun to yield exciting information for mixed infections, genetic recombination, and parasite differentiation (46–48).
TABLE 1.
Summary of genome sequence statistics for important human and animal Plasmodium speciesa
| Host | Parasite species or strain | No. of PIRb genes | Genome size (Mb) | GC content (%) | No. of scaffolds | No. of predicted genesc | No. of predicted proteinsc | Data release date (mo/day/yr) | Data update date (mo/day/yr) | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|---|
| Humans | P. falciparum 3D7 | 189 | 23.2 | 19.3 | 14 | 5,712 | 5,460 | 10/2/02 | 3/31/16 | 27, 29 |
| P. vivax | 1,212 | 29.1 | 39.7 | 374 | 6,830 | 6,677 | 10/6/16 | 10/6/16 | 8, 285 | |
| P. ovale | >2,100 | 33.5 | 29.4 | 779 | 6,986 | 6,228 | 8/3/16 | 9/15/16 | 33 | |
| P. ovale subsp. wallikeri | 1,375 | 33.5 | 28.9 | 1,914 | 8,582 | 8,421 | 6/16/16 | 6/16/16 | 8 | |
| P. ovale subsp. curtisi | 1,949 | 33.5 | 28.4 | 4,025 | 7,280 | 7,162 | 6/16/16 | 6/16/16 | 8 | |
| P. malariae | 255 | 33.6 | 24.7 | 63 | 6,709 | 6,573 | 9/23/16 | 9/23/16 | 8 | |
| P. knowlesi | 71 | 24.4 | 38.7 | 28 | 5,483 | 5,323 | 5/15/17 | 5/15/17 | 6, 8 | |
| P. inui San Antonio | 27.4 | 42.4 | 323 | 5,879 | 5,832 | 1/31/14 | 4/31/14 | https://plasmodb.org/plasmo/app/record/organism/NCBITAXON_1237626 | ||
| Chimpanzees | P. reichenowi | 351 | 24.0 | 19.3 | 261 | 5,909 | 5,741 | 3/25/18 | 3/25/18 | 29 |
| Old World monkeys | P. cynomolgi B | 265 | 26.2 | 40.4 | 1,663 | 5,776 | 5,716 | 5/31/12 | 9/16/15 | 31 |
| P. coatneyi Hackeri | 771 | 27.7 | 39.7 | 14 | 5,575 | 5,516 | 7/6/16 | 2/2/17 | https://plasmodb.org/plasmo/app/record/organism/TMPTX_pcoaHackeri | |
| Rodents | P. yoelii 17X | 980 | 22.8 | 21.1 | 154 | 6,257 | 6,091 | 8/27/14 | 10/26/17 | 28, 286 |
| P. berghei ANKA | 217 | 18.5 | 22.1 | 100 | 5,245 | 4,928 | 8/27/14 | 10/24/17 | 28, 286 | |
| P. chabaudi AS | 201 | 18.9 | 23.6 | 39 | 5,364 | 5,217 | 8/27/14 | 5/13/16 | 28, 286 | |
| P. vinckei | 18.2 | 23.4 | 49 | 5,009 | 4,954 | 6/16/14 | 7/30/14 | 287, 288 | ||
| Birds | P. relictum SGS1 | 4 | 22.6 | 18.4 | 498 | 5,306 | 5,138 | 11/17/16 | 1/9/17 | 30 |
| P. gallinaceum 8A | 20 | 23.8 | 17.8 | 152 | 5,439 | 5,280 | 1/9/17 | 30, 289, 290 |
All information was obtained through PubMed searches. More sequences from different strains of each species may be available.
PIR, Plasmodium interspersed repeat.
Includes updated information from PlasmoDB.
Except for a short diploid phase after fertilization in the mosquito midgut, Plasmodium parasites are haploid throughout their life cycle. The genomes of different species are roughly 2 to 3 times larger than that of brewer’s yeast (Saccharomyces cerevisiae), ranging from 20 to 35 megabases (Mb) and containing 14 chromosomes, a circular plastid genome of ∼35 kb, and multiple copies of a 6-kb mitochondrial DNA (Table 1). Comparison of genomes from different species showed that homologous genes are often found in syntenic blocks arranged in different orders among different chromosomes (49, 50). The adenine-thymine (AT) contents of Plasmodium spp. can also be very different: e.g., ∼80% AT in P. falciparum, P. reichenowi, and P. gallinaceum; ∼75% AT in rodent malaria parasites; and ∼60% AT in P. vivax, P. knowlesi, and P. cynomolgi (Table 1). AT content is often higher in introns and intergenic noncoding regions than in protein-coding exons, with an average of 80.6% AT for the whole P. falciparum genome versus 86.5% for noncoding sequences (27). The high AT content of P. falciparum reflects large numbers of low-complexity regions, simple sequence repeats, and microsatellites, as well as a highly skewed codon usage bias (51–54). Polymorphisms of AT-rich repeats provide abundant markers for linkage mapping of drug resistance genes (55–58) and for tracing the evolution and structure of parasite populations (59–61).
Gene Families Playing Important Roles in Parasite Development, Virulence, and Transmission
Malaria parasite genomes carry multigene families that serve important roles in parasite interactions with their hosts, including antigenic variation, signaling, protein trafficking, and adhesion (27, 62, 63). Several polymorphic families are found among species of the Laverania subgenus (P. falciparum [infecting humans], P. reichenowi, P. gaboni, and P. billcollinsi [infecting chimpanzees], and P. praefalciparum, P. adleri, and P. blacklocki [infecting gorillas]) (64). Among the gene families, the genes encoding P. falciparum erythrocyte membrane protein 1 (PfEMP1) (65, 66) have been studied most extensively. Each individual P. falciparum parasite carries a unique set of 50 to 150 copies of the var genes in its genome, where switches of gene expression produce antigenic variation (67–69). PfEMP1 plays an important role in the pathogenesis of clinical developments such as in cerebral and placental malaria, in which it mediates the cytoadherence of infected red blood cells (iRBCs; infected erythrocytes) in the deep tissues (70–72). Different PfEMP1 molecules bind to various host molecules, including α2-macroglobulin, CD36, chondroitin sulfate A (CSA), complement 1q, CR1, E-selectins and P-selectins, endothelial protein C receptor (EPCR), heparan sulfate, ICAM1, IgM, IgG, PECAM1, thrombospondin (TSP), and VCAM1 (62). Such binding leads to activation of various host inflammatory responses. Hemoglobinopathies, including the hemoglobin C and hemoglobin S trait conditions, interfere with PfEMP1 display in knob structures of the iRBCs. This poor display of PfEMP1 on the host cell surface offers protection against malaria by reducing the cytoadherence and activation of inflammatory processes that promote the development of severe disease (73–75).
A second group of genes receiving recent attention is the large Plasmodium interspersed repeat (pir) multigene family (63). Members of the pir family are named differently by parasite species: yir in P. yoelii, bir in P. berghei, vir in P. vivax, and so on. Several P. falciparum gene families (stevor, rif, and PfMC-2TM) are classified with pir by their similar gene structures (63), which characteristically include a short first exon, a long second exon, and a third exon encoding a transmembrane domain. In a recent study, the pir genes from P. chabaudi (cir) were shown to be expressed in different cellular locations, within and on the surface of iRBCs, and in merozoites (76). Additionally, a subset of recombinant CIR (P. chabaudi) proteins bound to mouse red blood cells (RBCs), suggesting a role for CIR in rosette formation and/or invasion. In another study, sequences of the P. berghei fam-a, fam-b, and bir multigene families were compared with those of P. yoelii and P. chabaudi; expressions of mRNA and selected proteins were analyzed (77). The majority of fluorescently tagged proteins were transported into the iRBC cytoplasm and into the parasitophorous vacuole of the liver stage, suggesting potential functions in parasite development and/or in manipulating the host immune response. Interestingly, Fam-A proteins carrying a steroidogenic acute regulatory-related lipid transfer (START) domain were found to transfer phosphatidylcholine in vitro, suggesting that these proteins transport host phosphatidylcholine for parasite membrane synthesis. Using the P. chabaudi AS parasite and C57BL/6 mice, Brugat et al. (78) showed that chronic infections were characterized by expression of distinctive clusters of cir genes, independent of adaptive immunity, and that the initial composition of parasite population dictated chronicity and virulence of infection. These observations suggest involvement of some cir genes in regulating the establishment of chronic infections and virulence. In an earlier study, vector transmission was shown to influence subsequent P. chabaudi asexual blood-stage infection (79). Transmission through the mosquito was also associated with a reduction in the severity of the resulting infection and altered the mammalian immune response as evidenced by decreased levels of circulating proinflammatory chemokines and cytokines during the acute stage (79). Overall, the expression of the cir multigene family was increased following mosquito transmission, as opposed to the hierarchical expression pattern observed in non-vector-transmitted parasites (80). Malaria parasites devote large portions of their genomes to gene families that ensure evasion of host immune defenses and protection of molecular processes essential to infection. These families emphasize the importance of research on their roles in parasite-host interactions and virulence, despite the difficulties inherent to their investigation.
Another polymorphic gene family worth mentioning is a group of 14 genes encoding proteins with six cysteines (6-Cys) (81). These proteins often localize on the parasite surface interacting with host proteins, are likely under various selection pressures such as immunity, and are promising vaccine candidates (82–85). These genes are expressed at different parasite developmental stages: pf230, pf48/45, pf230p, pf47, and pfPSOP12 are expressed in P. falciparum sexual stages; pf52, pf36, pfLISP2, and pfB9 are expressed in the preerythrocytic stages; and pf12, pf12p, pf41, pf38, and pf92 are expressed in asexual erythrocytic stages (81). The proteins have diverse functions. Pf48/45, Pf230, and Pf47 share unique disulfide-bonded structures and have been shown to play an essential role in parasite fertilization (86). Both Pf48/45 and Pf47 were shown to have high inbreeding coefficients, suggesting that these two molecules may play a significant role in mating interaction (87). Indeed, the pf47 gene was shown to play a critical role in the evasion of the Anopheles gambiae innate immune response, and replacement of the pf47 haplotypes in a P. falciparum isolate changed its compatibility to a different mosquito species (88, 89). PfP52 and PfP36 mediate sporozoite invasion of hepatocytes (90, 91). The proteins expressed in asexual stages are generally polymorphic and/or under selection, suggesting that they could be targets of the host immune response; however, their functions in parasite development remain largely unknown (81).
GENOME POLYMORPHISMS AND LINKAGE ANALYSIS
Early Work on Restriction Fragment Length Polymorphism
The polymorphic nature of Plasmodium genomes was recognized before large-scale genome sequencing was available. Early studies of the smaller P. falciparum chromosomes by pulsed-field gel electrophoresis (PFGE) showed size polymorphisms involving tens to hundreds of kilobases (92–94). Successful separation of the larger chromosomes by PFGE subsequently defined the nuclear content of 14 chromosomes (95) and, by analysis with rare-cutting restriction enzymes, demonstrated that the chromosome structure in P. falciparum is largely conserved in central regions but extensively polymorphic in both length and sequence near the telomeres (96–98). Much of this subtelomeric variation was explained by recombination within blocks of repetitive sequences and families of genes (99, 100). At a time when whole-genome sequencing was not available, restriction fragment length polymorphism (RFLP) analysis was employed to map the linkage groups of chromosomes in different parasite strains (101). These advances supported the localization of genes in P. falciparum crosses governing the heritable traits of PYR and CQ resistance (CQR) (102, 103).
Simple Sequence Repeats (Microsatellites)
The frequency of simple sequence repeats (microsatellites) in P. falciparum is estimated to be approximately one polymorphic microsatellite per kb DNA, a high rate that may involve the AT-rich nature of the genome (52, 53). This abundance of microsatellites facilitated the generation of high-density genetic linkage maps (53, 104) and expedited PCR-based mapping of the CQR locus in P. falciparum (57). Additionally, the rich wealth of genome variations supported development of rapid genotyping methods that have greatly facilitated parasite characterization and studies of P. falciparum epidemiology, population structure, and transmission (105, 106). Hundreds of microsatellites have also been identified from the P. yoelii genome and have been employed to type various strains and map genes contributing to virulence (104, 107, 108). Microsatellites seems to be less frequent in other Plasmodium species that have genomes with lower AT contents (104, 107–109).
Single Nucleotide Polymorphisms
Advances in DNA sequencing technologies have also facilitated the identification of large numbers of single nucleotide polymorphisms (SNPs). Early work compared amplified DNA segments from 204 P. falciparum genes on chromosome 3 (110). A total of 403 polymorphic sites (238 SNPs and 165 microsatellites) were identified from five parasite clones, establishing a chromosome-wide map with one polymorphic marker per 2.3 kb. Similarly, 191 SNPs and 44 size polymorphisms were identified from five P. vivax isolates after amplification and sequencing a DNA segment of ∼100 kb of DNA syntenic to a segment of P. falciparum chromosome 3 (109). Later, three independent laboratories comprehensively sequenced additional P. falciparum isolates, establishing high-density SNP maps for the parasite (111–113). Using these SNP maps, signatures of selection and drug-selective sweeps were identified from field parasite populations.
Copy Number Variation
Another important type of genomic variation in Plasmodium is copy number variation (CNV). CNVs have been found to affect important traits of drug resistance (114–119), erythrocyte invasion (120, 121), cytoadherence (122), and transcriptional regulation (123, 124). Increased gene copy numbers and levels of expression of P. falciparum multiple-drug resistance gene 1 (pfmdr1) were associated with decreased susceptibility to MQ and halofantrine (114). Amplification of the P. falciparum GTP-cyclohydrolase I (pfgch1) gene, associated with resistance to antifolate drugs, has also been described (44, 117, 125). Genomic breakpoints are frequently located in AT-rich regions or near homopolymeric tracks of poly(A) or poly(T) nucleotides (126, 127). Gene amplification is often unstable and can revert to single copy without pressure. However, DNA duplications flanked by distant A/T tracks could be a first step for desirable evolutionary events under selection pressure (127). Duplication between chromosome subtelomeric regions followed by sequence divergence was a likely mechanism for the generation of two histidine-rich genes of P. falciparum, PfHRP-II and PfHRP-III (95, 128).
GENETICS, GENOMICS, AND DRUG RESISTANCE
Candidate Gene Association Studies
Genetics and genomics information has been widely applied to identify genes contributing to various parasite traits, particularly those of drug resistance. Such studies have included strategies of linkage mapping using genetic crosses, genome-wide association studies (GWAS) of parasite field populations, and in vitro selection followed by gene expression analysis and/or genome-wide sequence surveys for mutations. Candidate genes may also be suggested based on known determinants of biological processes in different species. For example, some ATP-binding cassette (ABC) transporters are known to confer drug resistance in cancer treatment by efflux of drugs out of cancer cells. One of the ABC transporters in the P. falciparum parasite, P. falciparum multiple-drug resistance 1 (PfMDR1, the product of the pfmdr1 gene), was thought to confer CQR by pumping the drug out of the parasite food vacuole (129, 130). An association between CQR and several alleles of pfmdr1 was reported (131); however, analysis of a genetic cross between the CQ-sensitive (CQS) HB3 and the CQR Dd2 parasites found a lack of linkage between the CQR phenotype and inheritance of pfmdr1 (102) (see “Genetic Crosses and Quantitative Trait Analysis of Parasite and Host Phenotypes,” below). Interestingly, pfmdr1 amplification was associated with higher half-maximal inhibitory concentrations (IC50) for MQ in P. falciparum, P. vivax, and P. chabaudi (114, 132, 133); in other work, parasites with amplified pfmdr1 genes became more sensitive to CQ (134). A large number of studies using candidate gene association have been published; space is not available to discuss and cite all of the work here (for a recent review, see reference 22).
Genetic Crosses and Quantitative Trait Analysis of Parasite and Host Phenotypes
Experimental crosses of malaria parasites provide a powerful tool for the identification of genes underlying parasite traits (Fig. 1). To this end, four P. falciparum crosses have been performed through chimpanzee hosts and additional crosses through humanized mice (102, 135–138). These crosses have been employed to study the determinants for a variety of important phenotypes, including drug resistance, parasite invasion of RBCs, nutrient transport, infectivity to mosquitoes, hemoglobin catabolism, and gene expression (56, 88, 123, 136, 139–144). The first P. falciparum cross was generated using HB3 and 3D7 parasites and was used to map a phenotype of PYR resistance to the parasite dihydrofolate reductase-thymidylate synthase gene (pfdhfr-ts) (103, 135). The second genetic cross was performed using the CQR Dd2 and CQS HB3 parasites and linked CQR to a determinant on P. falciparum chromosome 7 (57, 102); the gene for the P. falciparum CQ resistance transporter (pfcrt) was identified after further fine-mapping and parasite transformation experiments (145). The third cross, between 7G8 and GB4 P. falciparum clones, identified a novel erythrocyte-binding protein, PfRH5, that mediates species-specific erythrocyte invasion and virulence of P. falciparum infection to Aotus nancymaae monkeys (136). PfRH5 was independently shown to bind human basigin, a key erythrocyte receptor used by P. falciparum merozoites to invade RBCs (146), and is a promising vaccine candidate against parasite invasion of RBC (147). The HB3 × Dd2 and 7G8 × GB4 crosses also supported discovery of the clag3 genes, which are responsible for channel-mediated nutrient uptake by iRBC (141), pfs47 alleles, which mediate evasion of the mosquito immune system (88), and modulation of parasite responses to amodiaquine and CQ by pfcrt and pfmdr1 (148). The fourth P. falciparum cross was performed between Cambodian 803 and Ghanaian GB4 parasites to investigate ART response phenotypes in vitro and in vivo and to evaluate phenotypes attributed to the PfK13 C580Y Kelch propeller mutation (137). In monkeys receiving three daily doses of intravenous artesunate (AS) after infection with the progenies, recrudescences were not more frequent in propeller mutant than in wild-type PfK13 infections, nor were clearances of the mutant parasites substantially slower in vivo, even though a greater in vitro ring-stage survival rate was linked to the PfK13 C580Y mutation in progenies of the cross after an ART pulse. Additional laboratory crosses of human malaria parasites are being performed through human-liver chimeric mice now that research use of chimpanzees is restricted (138).
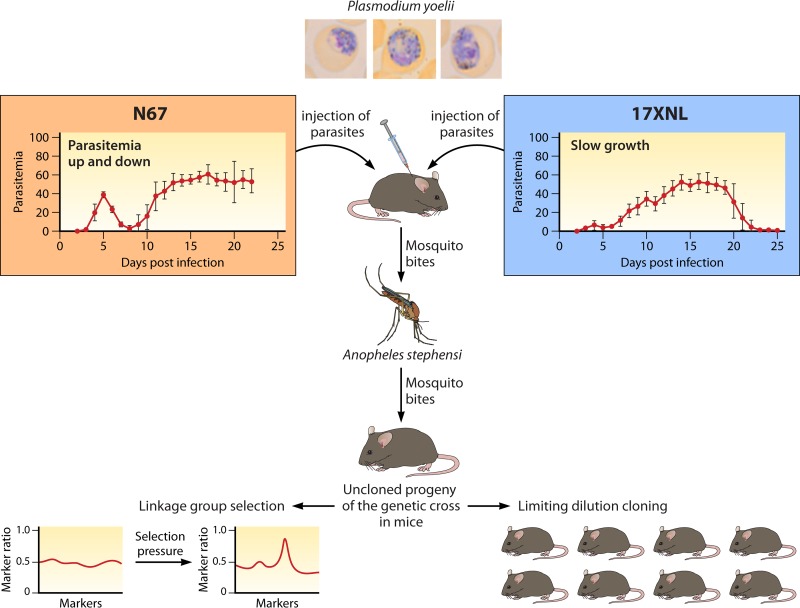
FIG 1.
Diagram illustrating the principle of a malaria genetic cross of rodent malaria parasites. A cross starts with an intravenous or intraperitoneal injection of blood samples containing gametocytes from two parasite strains (in this case, Plasmodium yoelii subsp. nigeriensis N67 and P. yoelii subsp. yoelii 17XNL, that have different growth characteristics and virulence in mice). Mice infected with mixtures of gametocytes are anesthetized and fed to Anopheles stephensi mosquitoes. Approximately 15 to 17 days after feeding, the infected mosquitoes with salivary gland sporozoites are allowed to feed on new mice. Daily blood smears are made to monitor parasitemia. The resulting parasites are cloned through limiting dilution by injecting a single parasite into a mouse or are frozen in liquid nitrogen for future studies. The parasite mixtures can be also used for linkage group selection (LGS) after applying selection pressure such as drugs.
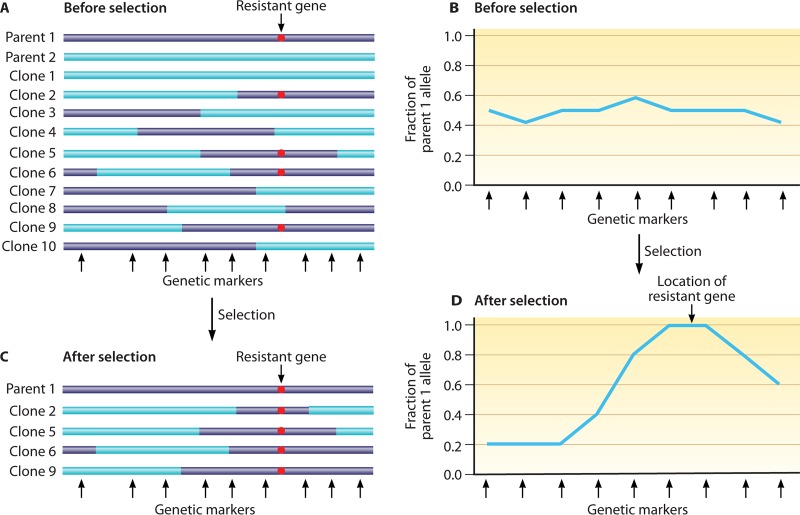
Genetic crosses of rodent malaria parasites have also been performed to investigate various phenotypes such as drug resistance, disease severity, or parasite development (104, 149–155). To optimize recombination in these crosses, the parasitemias of the parents are first tested in different ratios for gametocytemias that give maximum cross-fertilization (156). For characterization of inherited traits and linkage relationships, progeny clones may be obtained by injecting a large number of mice with inocula containing individual parasites (usually obtained by limiting dilution), a process that is more difficult and expensive than the cloning of P. falciparum progenies from in vitro cultures. Analysis of the crosses is sometimes possible by an alternative strategy of linkage group selection (LGS) (132, 149–152, 157–159) (Fig. 2). In LGS, samples of uncloned progeny populations are subjected to a specific pressure that selects an inherited trait (151). The survivors of selection and the original mixtures of unselected progenies are then compared in genome-wide searches for evidence of reduced marker diversity (“selection valleys”) in a chromosome region. This strategy has been used to map the genes conferring resistances to PYR (dihydrofolate reductase or pcdhfr) and ART (a gene encoding a deubiquitinating enzyme) and strain-specific immunity in P. chabaudi (150–152). LGS avoids the labor-intensive processes of parasite cloning and evaluations of their individual phenotypes.
FIG 2.
Diagram illustrating the principle of genetic recombination and linkage group selection. (A) Genetic recombination between parental lines (purple and blue) results in progenies harboring various combinations of the parental chromosomal segments. The bars represent chromosomal segments with a putative resistance marker from parent 1 (red dot) distributed among the progenies. Black arrows at the bottom denote positions of polymorphic genetic markers between the two parents. (B) Ratios of resistant alleles from parent 1, showing approximately 50% of allele ratios of the parents before selection. (C) After selection, only the parasites carrying the resistant allele (red dot) survive. (D) A plot of the ratios of resistant alleles shows increased frequency (to 100%) at one locus of the chromosome segment, suggesting at least one genetic determinant contributing to parasite survival or resistance to selection pressure in the locus. Fine-mapping with additional recombinant progenies and genetic markers may identify the gene(s) conferring the resistance.
Genetic crosses of rodent malaria parasites are a valuable resource for studies of phenotypes that are affected by host environment and the interplay of infections from different parasite strains. A number of P. yoelii crosses have been performed to map genes involved in host-parasite interaction, the host immune response, parasite growth, and virulence (104, 153, 160–162). In a recent example, a genome-wide quantitative trait locus (QTL) scan of 43 progenies from a P. yoelii cross linked two major loci on chromosomes 1 and 7 to phenotypes of parasite growth and host mortality, respectively (153). Fine-mapping of the chromosome 7 locus identified a gene encoding a HECT-like E3 ubiquitin ligase (pyheul), which was confirmed by allelic exchange experiments and modification of gene expression to modulate both parasite growth and host mortality.
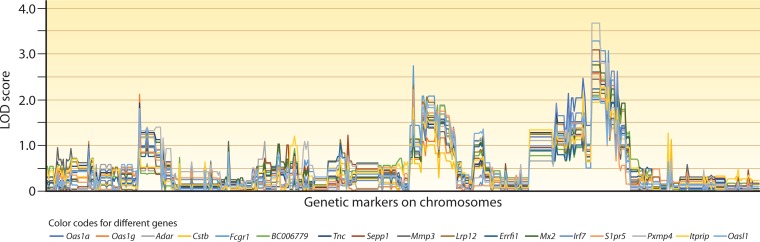
Progenies of Plasmodium crosses in mice are also a powerful resource for the identification of host genes that respond to parasite infections. More than 1,000 host genes were significantly linked to many parasite genetic loci after transspecies expression QTL (ts-eQTL) analysis (161). In one example, the host transcriptome responses to infections of 24 progenies from a P. yoelii genetic cross were obtained as phenotypes (by microarray analysis) and were analyzed against the parasite genotypes of hundreds of parasite microsatellites. Genome-wide patterns of logarithm-of-odds (LOD) scores (GPLS) were used to cluster host genes that likely function in related pathways of host responses, identifying a number of regulators of the type I interferon (IFN-I) response (Fig. 3). Functional assays have confirmed the roles of individual candidate regulators in IFN-I responses (163).
FIG 3.
Plots of host genes based on genome-wide patterns of LOD scores. This figure is based on data published in reference 161. In the study, mice were individually infected with 24 progenies from a genetic cross of Plasmodium yoelii subsp. yoelii 17XNL and Plasmodium yoelii subsp. nigeriensis N67 parasites. mRNA samples from infected spleens were extracted at day 4 postinfection and were hybridized to a microarray representing ∼19,100 unique mouse genes. Transspecies expression quantitative trait locus (ts-eQTL) analysis was used to analyze genome-wide transcription data from the infections against 479 microsatellite (MS) markers typed on the corresponding progenies. This analysis provided a genome-wide pattern of LOD scores (GPLS) for each host gene. Genes with expression levels linked to at least one MS marker (out of 479) with a LOD score of ≥2.0 were chosen, and their GPLSs were clustered based on pattern similarity. This figure shows a group of genes with similar GPLSs including a major peak of LOD score on one end of the parasite chromosome 13. Genes in an individual cluster are often related through roles in the same host response pathways; examples shown in this figure include type I interferon (IFN-I)-stimulated genes (isg) or genes that likely regulate IFN-I responses (Oas1a, Oas1g, Adar, Mx2, Irf7, S1pr5, or Oasl1). The names of the genes in the cluster are indicated under the plot.
Genome-Wide Association Studies for Genes Contributing to Parasite Drug Responses
The development of a relatively high-density linkage map with hundreds of microsatellites (53) made it possible to conduct GWAS using parasite isolates adapted to in vitro culture. In an early study, 87 culture-adapted worldwide P. falciparum isolates were typed with 342 highly polymorphic microsatellite markers (59). Extensive linkage disequilibrium (LD) of markers near pfcrt with decay of LD suggested that strong CQ directional selective sweeps originated from multiple CQR founder events (59). Publication of the first draft of the P. falciparum genome sequence (27) provided a further resource for the identification and characterization of large numbers of SNPs and microsatellites. Sequencing of additional P. falciparum parasites and their use in GWAS identified a large number of SNPs and expanded the availability of these high-density polymorphic markers (111, 112, 164). In one of these GWAS, the responses to seven antimalarial drugs in 189 culture-adapted P. falciparum parasites were analyzed with a custom-built array containing approximately 3,000 SNPs, averaging ∼1 SNP per 7 kb (165). In addition to identifying candidate genes significantly associated with parasite responses to various drugs, the results of the study provided information about parasite population structure, recombination rate, and loci under recent positive selection. Another GWAS, performed with a higher-density array of over 17,000 SNPs (∼0.7 SNP/kb) and 57 culture-adapted P. falciparum parasites, suggested an association of the Duffy binding-like merozoite surface protein 2 gene (pfdblmsp2) (gene ID, PF3D7_1036300 or PF10_355) with parasite resistance to halofantrine, MQ, and LUM (166); genetic knockout of this gene was subsequently found to increase parasite sensitivity to these three drugs (167). In a GWAS of 22 antimalarial drug responses among 35 Kenyan P. falciparum isolates (168), signals were detected from previously unreported genes as well as genes known to be involved in drug resistance (pfdhfr, pfdhps, pfmdr1, pfnhe, and pfcrt).
Genetic Analysis of P. falciparum Responses to Artemisinin Derivatives and Artemisinin-Based Combination Therapy
Artemisinin and its derivatives, collectively termed ART, are now the critical components of first-line antimalarial therapies recommended worldwide (169). In these therapies, the ART component serves as a powerful, short-acting drug for rapid parasitemia reduction, and a second, longer-acting partner drug is provided to eliminate parasites that can survive ART and cause recrudescence. The requirement for a partner drug in ART treatment was recognized in early clinical studies by Li et al. (170), who demonstrated that 3 days of ART monotherapy was followed within 28 days by recrudescence in more than 40% of P. falciparum-infected patients. The importance of ACTs for definitive cures was further highlighted by findings of frequent recrudescence after 3 to 7 days of ART treatment alone, including results from a trial in Vietnam where patients received artemisinin doses of 10 mg/kg on day 1 followed by 5 mg/kg/day on days 2 to 7 (170–172).
The failures of ART monotherapy to cure P. falciparum infections were recognized by Li et al. (170) as a phenotype of RI resistance, defined by the World Health Organization (WHO) as microscopic clearance followed by recrudescence within 28 days (173). This category is one of the four WHO classifications of drug response (173): (i) S, drug sensitive by clearance of asexual parasitemia within 7 days of the first day of treatment without recrudescence; (ii) RI, resistance characterized by microscopic clearance of asexual parasitemia, followed by recrudescence within 28 days; (iii) RII, resistance defined by reduction of asexual parasitemia but no clearance observed by microscopy; and (iv) RIII, high-level resistance in which there is no substantial reduction of asexual parasitemia. Resistances to CQ, sulfadoxine-PYR (SP), and certain other drugs have been reported at all levels of RI, RII, and RIII. Infections that fail CQ or SP treatment yield isolates that have half-maximum inhibitory concentrations (IC50s) that are significantly increased over those of drug-sensitive isolates in conventional 72-h dose-response in vitro assays. IC50 increases of 10× to 1,000× are linked to mutations in the pfcrt and pfdhfr-ts genes, respectively (145, 174). These mutations are useful molecular markers for drug resistance in population surveys (175).
ACT failures in Southeast Asia have raised concerns about increasing ART or partner drug resistance or both (176–178). In these areas of Southeast Asia, however, parasite isolates were not found to have substantially higher ART IC50 values than elsewhere (165, 179–182). Explorations for ART “resistance” therefore turned to alternative phenotypes such as delayed parasite clearance (DPC) or clearance half-life (t1/2) (179, 183); however, in patients treated with monotherapy, these measures were not associated with increased recrudescence or fever clearance time (180, 184–187). The t1/2s of erythrocyte ring stages were instead associated with improved survival of these forms (but not later trophozoite and schizont stages) after a short pulse of high-concentration ART in vitro (ring survival assay [RSA]) (188).
Using data from a number of field studies, GWAS have been conducted to identify genes associated with these various phenotypes. An early study examined dihydroartemisinin (DHA) IC50 values from culture-adapted parasites to search for SNPs associated with ART responses (165); however, the small differences between IC50 values precluded their definitive associations with candidate resistant genes. Cheeseman et al. (189) analyzed 6,969 SNPs in 91 parasites collected from Cambodia, Thailand, and Laos and identified 33 genome regions under strong selection; further study of 715 isolates from Thailand with SNPs using microsatellites revealed a signature of selective sweep on chromosome 13 that was significantly associated with DPC. Takala-Harrison et al. (190) used a P. falciparum array of 8,079 SNPs to survey 342 parasite samples from artemisinin-treated patients in Bangladesh, in northwestern Thailand near the Myanmar border, and at two sites in western Cambodia. One SNP on chromosome 10, two on chromosome 13, and one on chromosome 14 were significantly associated with DPC, of which the two SNPs on chromosome 13 were near those identified by Cheeseman et al. (189). Whole-genome sequencing of clinical isolates from Cambodia and an in vitro line selected for ART resistance then identified a gene with Kelch propeller domain mutations (pfk13; gene ID, PF3D7_1343700 or PF13_0238) from this chromosome segment. Correlation of these mutations with in vivo parasite clearance t1/2s and RSA survivals provided evidence for selection of pfk13 alleles under ACT pressure in Cambodia (182). More recently, a large multicenter GWAS of parasite samples from 15 locations in Southeast Asia found that at least 20 mutations in the pfk13 gene were associated with a low parasite clearance rate after treatment with ART (191). Multiple additional candidate gene association studies have been recently reviewed (192).
In view of the frequent recrudescences recognized ever since the original clinical studies of ART (170), the absence of ART IC50 signals from isolates after treatment failure, and the studies showing no significant association of clinical recrudescences with DPC or clearance t1/2s after 3 days of monotherapy (180, 184–187), the nature of ART “resistance” and whether clinical levels of resistance to the drug are now substantially greater have been controversial (183, 193–197). The debate is compounded by observations of greater gametocytogenesis with pfk13 mutant strains (198, 199). Because mature gametocytes are poorly cleared by ART, the mutant parasites may have better survival rates than those with wild-type pfk13, and P. falciparum transmission to mosquitoes may be protected as a survival mechanism. Additionally, parasites from infections clearing more slowly after treatment (i.e., those with a t1/2 of >5 h) (200) may have IC50 values as low as or lower than those for parasites from shorter clearance infections (t1/2s of <3 h). Clearance t1/2s can be affected by host genetic background and factors including immunity, fever, and anemia as well as parasitemia, and differences of 2-fold or less may be difficult to assess in infected patients (196, 201).
RSA is an in vitro parasite survival assay that was developed based on the observation that the ring stage of some parasite strains could survive better under a short period of ART treatment (202, 203). For P. falciparum, this surrogate measure of ART resistance has been set at a threshold ring-stage survival rate of ≥1% after exposure to 700 nM DHA for 6 h (181, 188). This measure ignores the full life cycle, during which ring-stage parasites mature through trophozoite and schizont stages that show little or no K13-mediated difference in survival after the DHA pulse, which is approximately 1% regardless of their pfk13 status (188). In the continuing presence of drug, a ring-stage parasite that survives 6 h of exposure will be subsequently killed as it becomes a more mature trophozoite stage, so that RSA cannot reliably reflect or predict failure to cure infection.
In a recently reported cross of P. falciparum parasites (803 × GB4), parasite clearances and recrudescence in Aotus monkeys treated with three daily doses of intravenous AS were not significantly associated with the inheritance of different parental pfk13 alleles (137), although the C580Y codon in pfk13 was linked to increased RSA values. Moreover, a C580 wild-type clone, produced by allelic exchange of a C580Y mutant-type progeny, yielded a monkey infection that recrudesced every time after 13 individual 3-day artesunate treatments over a period of 500 days. These results reinforce the fact that ART-treated P. falciparum infections frequently recrudesce with or without K13 mutations and highlight the need, as emphasized by Li et al. (170), for effective ACT partner drugs to completely cure parasites that can persist after exposure to ART. Unfortunately, ACT treatment failures from ineffective partner drugs have been increasingly reported from western Cambodia. For example, DHA plus PPQ, an ACT with a historically good track record, now fails frequently in regions of Cambodia (204, 205). PPQ resistance from new mutations in pfcrt is a major contributor to these failures (206), necessitating switches to alternative ACTs such as MQ plus AS (200, 207). Recent findings of plasmepsin gene amplifications, modified pfmdr1 copy numbers, and selection of different pfmdr1 genotypes (208–210) in Cambodian parasites have led to suggestions that rotations among different ACTs or triple ACT drug combinations may help reduce difficulties with drug resistance (211, 212). Multicopy plasmepsin 2 was proposed as a surrogate molecular marker to track PPQ resistance; however, the roles of plasmepsin 2 require further investigation relative to the H97Y, F145I, M343L, or G353V mutation of PfCRT in parasite resistance to PPQ (206). Readers who are interested in additional information on antimalarial drug development and mechanisms of drug resistance can consult the several excellent reviews on these subjects (22, 23, 213–218).
Mapping Genes Using Signatures of Selection and Selective Sweeps
Malaria parasites are under continual evolutionary selection from pressures including host immunity and antimalarial drugs. Because the blood-stage parasites have a haploid genome, signatures of selection may be comparatively easy to identify in the neighborhood (valley or peak) of relevant genes. Such signatures can be followed across regions where malaria is endemic by analyzing the temporal and spatial patterns of genetic markers in parasite populations. Signatures of selective sweeps have thus been reported for CQ, SP, and ART (59, 61, 219–223). For discovery of the selective-sweep signals of reduced genetic diversity or LD decay in gene neighborhoods, microsatellites have been particularly useful because of their relatively high mutation rates (52). Microsatellites were employed in the first demonstration of sweeps from multiple founder events of CQ-resistant parasites, including a major sweep from southeast Asia into Africa (59). In addition to mutations directly responsible for drug resistance, selective sweeps can identify signals of positive versus negative (purifying) selections as well as balancing selection. For example, genome-wide analysis of sequence variation was performed on more than 150 P. falciparum clinical isolates from a region of the Republic of Guinea with high endemicity; signatures of selection by antimalarial drugs and host immunity were detected (224). The citations here have included just a few of many reports from selective studies.
CHEMICAL GENOMICS: SELECTION, SCREENING, AND TARGET IDENTIFICATION
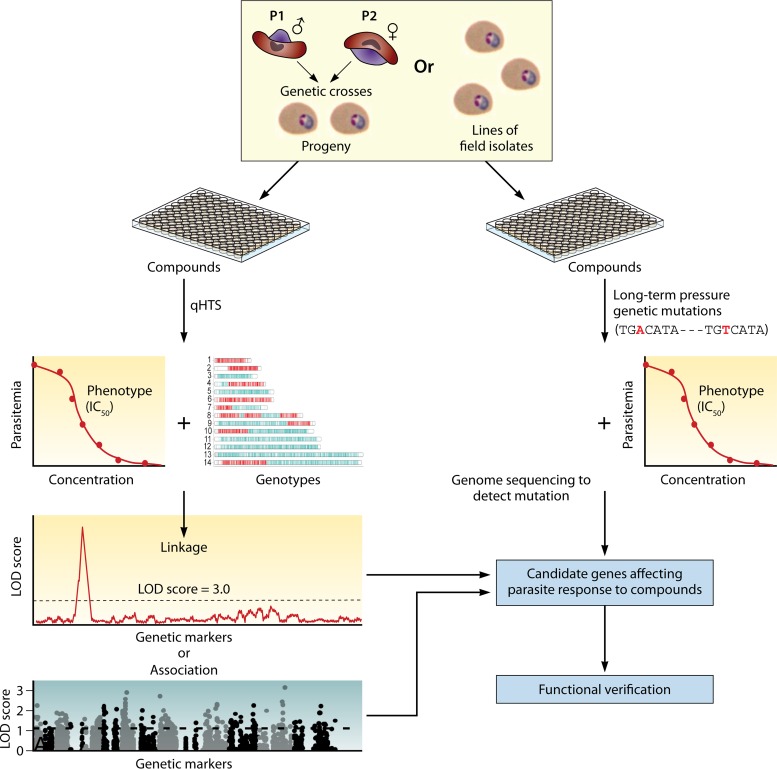
As data and tools to study genome-wide variations grow in availability and high-throughput chemical screening platforms expand, innovative combinations of these powerful approaches will drive new discoveries of gene functions and gene interactions. Analysis of differential responses to small molecules (SMs) and various biologics will unveil targets for exploration in programs to discover new drugs and malaria interventions. For large-scale studies, parasites exposed to libraries of SMs can generate large numbers of mutants with phenotypic variations (Fig. 4). These may involve mutations in direct targets or in other molecules (e.g., transporters) that affect SM susceptibility.
FIG 4.
Chemical genomics approaches to identify interactions of small molecules (SMs) and parasite genes. Malaria parasites, either progenies from genetic crosses or field isolates, can be screened for response (half-maximal inhibitory concentration [IC50]) against libraries of SMs using quantitative high-throughput screening (qHTS). Parasites can also be placed under long-term SM pressure for mutations that may play a role in parasite survival. The parasites are genotyped with a large number of genetic markers or are genome sequenced to detect polymorphism or mutations under SM pressure. Linkage analysis, including LGS, or genome-wide association analysis (GWAS) can be performed to link SM and parasite genes playing roles in the SM responses. Genetic mutations and changes in gene expression can be detected after genome sequencing or microarray/RNA-seq from parasites before and after SM pressure. Candidate genes can be further verified using CRISPR/Cas9-based gene knockout, knock-in, or regulation of gene expression to confirm the potential SM target or genes in SM transport.
Mutations underlying the phenotypic changes such as responses to SMs can be identified using various genetic and genomic approaches, while changes in gene expression can be evaluated by microarray or RNA sequencing (RNA-seq) methods (225). Variations in proteins or metabolite levels can be detected using spectrometry-based methods such as matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS) or gas chromatography-MS (GC-MS) (226, 227), chromatin modifications can be revealed by chromatin immunoprecipitation (ChIP) with microarray technology (ChIP-chip) or ChIP-sequencing (ChIP-seq) (228), and DNA polymorphisms can be mapped using genetic linkage analysis or GWAS (229–231). Mass spectrometry has been employed to characterize the metabolomic profiles of P. falciparum-infected RBCs (232) or parasite development throughout its 48-h intraerythrocytic developmental cycle (233). In a recent example of the use of this approach, Lewis et al. (144) performed QTL analysis of metabolites as phenotypes and identified elevated levels of hemoglobin-derived peptides in CQR parasites of the HB3 × Dd2 genetic cross.
Differential Chemical Phenotypes, Selection of Mutations with Small Molecules, and Identification of Target Genes
The power of using SMs to detect differences between malaria parasites was demonstrated in a quantitative high-throughput screening (qHTS) study of seven P. falciparum strains exposed to the LOPAC 1280 library (Sigma) (Fig. 4) (229). More than 600 differential chemical phenotypes (DCPs), defined as pairwise IC50 differences of 5-fold or more between parasite lines, were detected. Particular SMs (e.g., dihydroergotamine methanesulfonate, trimethoprim, and triamterene) that produced large differences in DCP values were further tested on 32 progenies from a GB4 × 7G8 cross (136). QTL analysis of the DCPs with the genotypes of 285 microsatellite markers identified a major locus on chromosome 5 (LOD score = 16.4) and a second locus on chromosome 12 (LOD score = 3.0), both of which were linked to the dihydroergotamine methanesulfonate response. A candidate gene on chromosome 5, pfmdr1, was confirmed by allelic substitutions to have an important role in this response (229).
Another powerful genomic approach for identifying drug targets is to subject a parasite to prolonged pressure of drugs or SMs (Fig. 4). Under drug pressure, the parasite may increase expression of genes that can help its survival by changing gene copy number or level of transcription. Nucleotide substitutions (or SNPs) in target genes may also occur, leading to parasites that are more resistant to a drug. Large-scale detection of these changes has been employed to identify a number of mutated genes after selection with SMs (214, 234–237). In one study, CNV of the P. falciparum 1-deoxy-d-xylulose 5-phosphate reductoisomerase gene (pfdxr) was detected using a tiling microarray after in vitro selection with fosmidomycin (235). Studies using ART/DHA-selected P. falciparum parasites, genome sequencing, and transcriptome analysis have also reported changes in CNV, gene expression, and/or nucleotide substitutions in genes such as pfmdr1, pfk13, and pfcoronin and genes in the antioxidant defense network (182, 238, 239). In a recent work, 262 P. falciparum parasites were selected to become resistant to 37 diverse compounds. Subsequent genome sequence analysis revealed 159 gene amplifications and 148 nonsynonymous changes in 83 genes that could be associated with resistance to the compounds (240). In another study, selection of P. falciparum with putative inhibitors of P. falciparum NADH dehydrogenase 2 (PfNDH2) led to mutations in the more likely target: the parasite cytochrome B gene expressed by the mitochondrial genome (237). Consistent with these findings, deletion of the pfndh2 gene has been shown to be dispensable in asexual blood stages, and further, its deletion does not alter the parasite’s susceptibility to multiple mitochondrial electron transport chain (mtETC) inhibitors (241).
Genome-Wide Association Studies of Genes Playing a Role in Parasite Responses to Small Molecules
GWAS is also useful for insights into the responses of P. falciparum isolates to SMs (230). In a qHTS screen of parasite responses to serial dilutions of 2,816 individual compounds, Yuan et al. (230) used 3,354 SNPs and 61 P. falciparum parasite lines to identify 32 highly active compounds and several chromosomal loci significantly associated with 49 DCPs (230). These loci were further confirmed by linkage analysis and the production and testing of parasites with genetically modified candidate genes. The results of this study identified not only SMs that were more effective against parasites expressing wild-type genes but also compounds that had better efficacy against parasites with mutant forms of these genes, suggesting the potential for drug combinations to treat both wild-type and mutant forms of molecules such as PfCRT and PfMDR1.
VACCINE TARGET IDENTIFICATION, MOLECULAR EVOLUTION, AND EPIDEMIOLOGY
In addition to its usefulness for insights into drug resistance and important parasite traits, information on the genome diversity and variation of malaria parasites is valuable for research on vaccine development, disease epidemiology, and parasite evolution. A large amount of work has been done in these subject areas. Here we draw attention to just some examples.
Genome Surveys for Selection Signatures of Vaccine Targets
Vaccine candidates such as PfMSP1, PfAMA1, PfCSP, or PfEMP1 have highly polymorphic regions where these molecules are under host immune pressure (242, 243). Vaccines developed from these polymorphic antigens may be confounded by allelic restriction of the host immune response, and parasites expressing alternative epitopes can escape host immune clearance. A vaccine that does not provide complete protection may select parasite variants having elevated virulence and thereby be “imperfect” (244). Based on the observation of a polymorphic nature of immune targets, potential antigens can be identified through genome-wide analysis of genetic diversity (111). Population genetic studies may uncover signatures of balancing selection in the polymorphisms of genes encoding immune targets (245). In a genome-wide survey of P. falciparum parasites isolated from 65 Gambian clinical patients, candidate genes with signatures of balancing selection were identified for studies of immune mechanisms and potential vaccine development (246). The gene with the highest values of Tajima’s D was Pfdblmsp2 (PF3D7_1036300 or PF10_0355), a gene previously associated with parasite responses to halofantrine, MQ, and LUM by GWAS and gene expression analysis (166). Interestingly, indirect immunofluorescence assay (IFA) analysis showed that the PfDBLMSP2 protein was expressed in only a small fraction of mature schizonts (246). In another study, a survey of 3,539 predicted P. falciparum genes from four cloned isolates (Dd2, HB3, D10, and 7G8) for elevated diversity detected 56 highly polymorphic genes (with θ values 2 standard deviations higher than the mean θ value for 1,920 genes). In vitro protein expression and Western blotting using pooled infected human sera confirmed the antigenicity of 11 proteins, including seven previously unknown antigens (111). Using samples collected from patients, Conway et al. (247) measured fixation index (FST) values for multiple microsatellite loci and polymorphic sites in block 2 of PfMSP1 and showed that the block 2 was under immune selection or an immune target. These studies demonstrate that survey of genome diversity is an effective approach for identifying genes under immune selection for vaccine candidates.
Applications to Studies of Parasite Molecular Evolution
Genome diversity data support inferences about parasite origin and evolutionary history, including estimates of time to the most recent common ancestor (TMRCA). Genetic polymorphism information can be used to date historical events based on assumptions of a molecular clock and the absence of selection. Under such assumptions, the rate of molecular evolution for the average protein is taken to be approximately constant over time, although estimates of rates can be time dependent and influenced by factors of natural selection, calibration errors, model misspecification, and other artifacts (248). Thus, special care should be taken to collect data on “neutral” genetic sequences for evolutionary analysis. Such care helps to avoid the use of antigen genes that can inflate overall estimates of genome diversity and mutation rate. On the other hand, the effects of antimalarial drugs can reduce genome diversity of parasite populations by removing drug-sensitive parasites and overrepresenting parasites that carry drug resistance genes. A single haploid mutant parasite can multiply, produce male and female gametocytes, infect mosquitoes, and quickly generate a genetically homogeneous population (clonal expansion outbreak). Parasite samples from such a malaria outbreak or from parasite populations massively treated with antimalarial drugs may show greatly reduced genome diversity, potentially affecting an estimate of TMRCA.
Studies of P. falciparum populations and their evolutionary origin have led to different estimated TMCRAs and proposed “malaria’s eve” hypotheses (249). Some studies, finding poor correlation between the strength of LD and nucleotide distances in the circumsporozoite (csp) gene, as well as no synonymous substitutions in 10 genes, have suggested that all extant world populations of the P. falciparum parasite are from an ancestral strain (or a small number of strains) that was subjected to an ancestral sweep a few thousand years ago (250, 251). Additional data from SNPs in 25 introns of 10 housekeeping genes and the 6-kb mitochondrial genome were also consistent with a relatively recent origin of P. falciparum populations (252, 253). However, a much longer TMRCA and large effective population size have also been proposed (109, 110, 254, 255). Recent findings from DNA sequences in the fecal samples of wild-living apes suggest that a bottleneck or eve event likely occurred from an ancient transmission of a great ape parasite to a human in Africa (36, 256). Such an event would be consistent with close genomic relationships between P. falciparum and other Plasmodium species of the Laverania subgenus, which infect gorillas (P. praefalciparum, P. adleri, and P. blacklocki) and chimpanzees (P. reichenowi, P. gaboni, and P. billcollinsi) (257). Indeed, recent analysis suggests that P. falciparum began to emerge in humans from P. praefalciparum around 40,000 to 60,000 years ago, followed by a population bottleneck around 4,000 to 6,000 years ago (64).
Population Structure and Geographic Origin of Parasites
Population structures of malaria parasites have been investigated using polymorphic genes and genome-wide distributions of sequence variations. Using 342 highly polymorphic microsatellite markers from a genetic map, a survey of a worldwide collection of P. falciparum isolates demonstrated geographical and continental clustering (59). Additional insight into the structure of P. falciparum populations was gained from genotypes of ∼3,000 SNPs, which suggested an evolutionary separation of Cambodian parasites from those of Thailand (165). More recently, analysis of genome variation in 825 P. falciparum samples from Asia and Africa identified unique divisions in the parasite population structure in western Cambodia, suggesting three different subpopulations associated with ACT responses (41). Differences among these subpopulations were not associated with regional gene flow or high rates of transmission (258). In other studies, clustering of polymorphic P. falciparum apical membrane antigen 1 (PfAMA1) sequences from 97 parasite clones from around the world and 61 isolates from Mali showed patterns of polymorphisms that were independent of geographic location (259), which is likely due to multiple instances of diversifying selection on the gene (260).
Knowledge of population structure can be useful for studies of epidemiological developments and changes in patterns of disease. Initiatives such as the Malaria Genomic Epidemiology Network (MalariaGEN; https://www.malariagen.net) and the PlasmoDB resource (http://plasmodb.org/plasmo/app/search/dataset/AllDatasets/result) provide genomic sequences, omics data sets, disease information, genetic resources, and analysis capabilities for multiple Plasmodium species and infections from regions where malaria is endemic worldwide. Approximately 3,500 P. falciparum parasite isolates from 23 countries and 228 P. vivax samples from 13 countries have been sequenced by MalariaGEN. Sequence data from the project have been used to investigate drug resistance, parasite population structure, diversity, epidemiology, and transmission (41, 43, 191, 258, 261, 262). A recent study examined genome sequences from 1,492 P falciparum samples from 11 locations across southeast Asia, including 464 samples from western Cambodia, and found that an amino acid substitution within the product of a gene encoding an exonuclease (E415G) and plasmepsin 2/3 amplification are frequently associated with piperaquine resistance and dihydroartemisinin-piperaquine failures in Cambodia (261).
GENOMICS AND APPLICATIONS ON POPULATION DIVERSITY, PARASITE ORIGIN, AND RELAPSE OF P. VIVAX INFECTION
Genome Diversity of P. vivax and Genes under Selection
P. vivax is another malaria parasite of major public health burden and was responsible for ∼7.5 million clinical cases in 2018 (263). P. vivax and other non-P. falciparum human malaria species have attracted more attention recently. A number of reports now provide genetic and genomic analyses on parasite population diversity, signatures of selection, and molecular evolution (25, 33, 39, 49, 262, 264–266). One of the early attempts to characterize P. vivax genome diversity was the sequencing of a 100-kb contiguous chromosome segment from five isolates. The data showed P. vivax to be a genetically diverse species with an abundance of SNPs but a relatively lower level of polymorphic tandem repeats than that in P. falciparum, possibly due to its more balanced guanine-cytosine content (109). Whole-genome sequencing of a Peruvian P. vivax isolate obtained ex vivo without further propagation identified a large number of SNPs and genes under selection, including the homolog of pfcrt (pvcrt), genes encoding reticulocyte-binding proteins, and AMA-1 (265). More recently, genome sequencing of global collections of P. vivax parasite isolates confirmed higher genomic diversity in P. vivax than in the more virulent P. falciparum parasite and revealed signals of recent evolutionary selection on drug resistance genes (39, 262, 267). The higher levels of SNPs also led to estimates of TMRCA that were longer than that of P. falciparum (267–269).
Origin of P. vivax Parasites
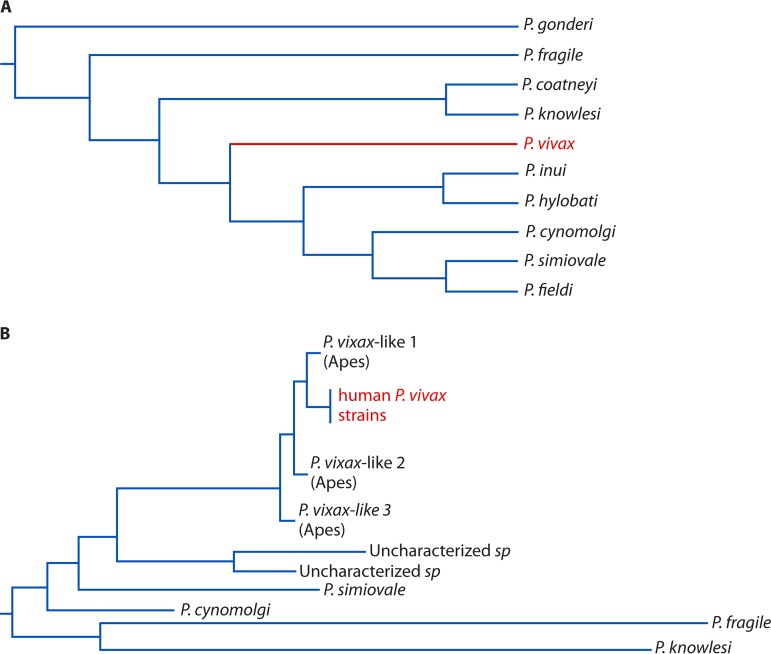
The P. vivax parasite is genetically closely related to several parasites infecting nonhuman primates, such as P. cynomolgi, P. knowlesi, and P. vivax-like parasites (264, 270, 271). Early phylogenetic analyses using mitochondrial DNA or genes from the plastid and nuclear genome placed P. vivax among the Asian primate malaria parasites (Fig. 5A) (35, 269). The near-perfect genetic identity between P. vivax and P. simium, a New World primate malaria species, also suggested the possibility of transfer between humans and the New World monkeys (272–274). Indeed, studies using the complete mitochondrial genome sequence or genes from the plastid and nuclear genome indicated an Asian origin via a host switch from macaque monkeys (268, 269). On the other hand, a relative protection against P. vivax in western and central Africa by the highly prevalent Duffy-negative blood type suggests that P. vivax might have a long history in Africans (275–277). More recent identification and sequencing of genomes of P. vivax-like parasites support an evolutionary transfer of P. vivax-like parasites from African great apes to humans (Fig. 5B) (36, 264, 278). Additionally, analyses of the apicoplast genome from 18 Plasmodium species have shown that P. vivax is positioned next to the divergence of the African guenon parasite, P. gonderi, before the common ancestor of Asian primate parasites, supporting an African origin (279). Much about the relationships and evolutionary history of P. vivax, P. vivax-like, P. simium, P. cynomolgi, and P. knowlesi parasites remains to be discovered.
FIG 5.
Molecular phylogenetic trees of Plasmodium vivax and other related nonhuman primate parasites. (A) A phylogenetic tree of P. vivax and related parasites based on sequences from two nuclear genes (β-tubulin and cell division cycle 2) and a plastid gene (the elongation factor Tu). The tree is adapted from Fig. 1B of reference 269 with permission of the National Academy of Sciences. (B) A phylogenetic tree of P. vivax and P. vivax-like parasites from nonhuman primates, including apes. The tree is adapted (simplified) from Fig. 2 of reference 278 with permission of Springer Nature/Macmillan Publishers Limited.
P. vivax Relapses versus Recrudescences and Reinfections
An important characteristic of P. vivax infection is malaria relapse arising from dormant liver-stage forms called hypnozoites (16). One of the main challenges in studying relapses in regions of endemicity is the difficulty in distinguishing parasites of relapse from those of reinfection or recrudescence of drug-resistant parasites. Genetic typing and more recently genome sequencing have been employed to distinguish parasites of relapse from reinfection and/or recrudescence of drug-resistant parasites (280–282). A study on Australian soldiers experiencing relapses of P. vivax malaria after exposure in East Timor used the polymorphic regions of three P. vivax genes to demonstrate the predominance of a single allelic type in relapsing parasites. However, the second relapse was often from a different allelic type, indicating a different strain (283). Therefore, multiple relapses in an individual may arise from the activation of hypnozoites of different parasite strains. Another study showed that relapsing parasites are often polyclonal, but with some degrees of relatedness to the parasites in a mixed initial infection (282). Different parasite clones might emerge during the course of the relapsing infections, which is consistent with the study of Australian soldiers. Genome sequencing studies including single-cell analysis of relapsing parasites can provide high resolution with a large number of polymorphic sites; however, adequate studies of the low-level parasite populations characteristic of P. vivax will remain challenging. The recent advances in genomics will certainly contribute greatly to the study and resolution of relapses in P. vivax infection (284).
CONCLUSIONS
Knowledge of genetic variations and genome diversity of malaria parasite populations has greatly improved our understanding of the biology, gene function, drug resistance, population dynamics, transmission, and molecular evolution of malaria parasites. Information gained from these studies is particularly important when ethical considerations, resources limitations, or confounding factors limit direct experimental infections of human hosts. In addition, investigations of disease phenotypes in rodent or other animal models of malaria under controlled laboratory conditions, combined with parasite genetics and genomics, can provide helpful insights into pathogenesis and new information on disease control and management in humans.
ACKNOWLEDGMENTS
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), USA.
We thank Brian Brown, NIH Library Writing Center, for manuscript editing assistance.
We declare no competing interests.
Biographies

Xin-zhuan Su, Ph.D., is a senior investigator and the Chief of the Malaria Functional Genomics Section, National Institutes of Health, USA. Dr. Su obtained his bachelor’s and master’s degrees in parasitology from Xiamen University, China, and his Ph.D. in parasitology from the University of Georgia. His research interests include mechanisms of drug resistance, drug screening, genetic mapping, genomics, molecular evolution, population genetics, host-parasite interaction, and molecular signaling of the host immune response. His major contributions to malaria research include mapping of the gene encoding the Plasmodium falciparum chloroquine resistance transporter (pfcrt), discovery of P. falciparum var genes, development of the first P. falciparum linkage map, and detection of worldwide selective sweeps of pfcrt. Dr. Su is a board member for several scientific journals and has received various awards, including the Bailey K. Ashford Award from the American Society of Tropical Medicine and Hygiene and the NIH Director’s Award. He is a Fellow of the American Academy of Microbiology.

Kristin D. Lane, Ph.D., M.S., is a research associate in the Division of Intramural Research, National Institute of Allergy and Infectious Diseases. She holds a Ph.D. in microbiology and immunology with concentrations in molecular biology and microbial pathogenesis from Virginia Commonwealth University and a master’s of science in biology from Old Dominion University. Her interests encompass mechanisms of molecular pathogenesis and drug resistance in the malaria parasite Plasmodium falciparum, transcriptional regulation, molecular piracy, and viral pathogenesis. Dr. Lane’s major contributions to malaria research include drug target identification and resistance mechanisms in P. falciparum, particularly those involving cytochrome B (PfCytB) and the P. falciparum chloroquine resistance transporter (PfCRT).

Lu Xia is a Ph.D. student majoring in medical genetics at the State Key Laboratory of Medical Genetics of the Central South University, China. In 2017, she joined the Laboratory of Malaria and Vector Research, NIAID, as a student trainee supported by the China Scholarship Council. Her research focuses mainly on genetics and bioinformatics of human inherited diseases and Plasmodium genetics and genomics. Her expertise includes genome-wide expression and association analyses to study molecular mechanisms of neurodevelopmental disorders and immune pathways in response to malaria infections. She has published 12 peer-reviewed papers in the fields of malaria research and human genetic diseases.

Juliana M. Sá, Ph.D., received a bachelor’s degree in biological sciences from the Universidade Estadual Paulista, Brazil. She obtained a master’s degree in biochemistry and a Ph.D. in parasitology from the Universidade de São Paulo, Brazil. Dr. Sá pursued postdoctoral training at the National Institute of Allergy and Infectious Diseases (NIAID), USA, where she holds a Staff Scientist position investigating antimalarial resistance. Her interest in genetics of drug resistance results from a passion for basic science and applicability. Dr. Sá has been in the field of malaria drug resistance for 18 years.

Thomas E. Wellems, M.D., Ph.D., is an NIH Distinguished Investigator in the Division of Intramural Research, National Institute of Allergy and Infectious Diseases. His interests embrace the biology, pathogenesis, and drug responses of the various species of malaria parasites. Major reports from his work include the descriptions of the Plasmodium falciparum chloroquine resistance transporter (PfCRT), the P. falciparum var gene family (responsible for antigenic variation and immune evasion), a molecular mechanism for malaria protection by sickle-cell trait and hemoglobin C, the PfHRP-II protein used in malaria rapid diagnostic tests (RDTs), and development of P. falciparum transformation methods. Dr. Wellems is a member of the U.S. National Academy of Sciences and the National Academy of Medicine and a former president of the American Society of Tropical Medicine and Hygiene, and he has served on a number of advisory committees for foundations and public-private partnerships, including the Medicines for Malaria Venture.
REFERENCES
- 1.Feachem RG, Phillips AA, Hwang J, Cotter C, Wielgosz B, Greenwood BM, Sabot O, Rodriguez MH, Abeyasinghe RR, Ghebreyesus TA, Snow RW. 2010. Shrinking the malaria map: progress and prospects. Lancet 376:1566–1578. doi: 10.1016/S0140-6736(10)61270-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snow RW, Amratia P, Kabaria CW, Noor AM, Marsh K. 2012. The changing limits and incidence of malaria in Africa: 1939-2009. Adv Parasitol 78:169–262. doi: 10.1016/B978-0-12-394303-3.00010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noor AM, Kinyoki DK, Mundia CW, Kabaria CW, Mutua JW, Alegana VA, Fall IS, Snow RW. 2014. The changing risk of Plasmodium falciparum malaria infection in Africa: 2000-10: a spatial and temporal analysis of transmission intensity. Lancet 383:1739–1747. doi: 10.1016/S0140-6736(13)62566-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bright AT, Tewhey R, Abeles S, Chuquiyauri R, Llanos-Cuentas A, Ferreira MU, Schork NJ, Vinetz JM, Winzeler EA. 2012. Whole genome sequencing analysis of Plasmodium vivax using whole genome capture. BMC Genomics 13:262. doi: 10.1186/1471-2164-13-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlton JM, Angiuoli SV, Suh BB, Kooij TW, Pertea M, Silva JC, Ermolaeva MD, Allen JE, Selengut JD, Koo HL, Peterson JD, Pop M, Kosack DS, Shumway MF, Bidwell SL, Shallom SJ, van Aken SE, Riedmuller SB, Feldblyum TV, Cho JK, Quackenbush J, Sedegah M, Shoaibi A, Cummings LM, Florens L, Yates JR, Raine JD, Sinden RE, Harris MA, Cunningham DA, Preiser PR, Bergman LW, Vaidya AB, van Lin LH, Janse CJ, Waters AP, Smith HO, White OR, Salzberg SL, Venter JC, Fraser CM, Hoffman SL, Gardner MJ, Carucci DJ. 2002. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 419:512–519. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- 6.Pain A, Böhme U, Berry AE, Mungall K, Finn RD, Jackson AP, Mourier T, Mistry J, Pasini EM, Aslett MA, Balasubrammaniam S, Borgwardt K, Brooks K, Carret C, Carver TJ, Cherevach I, Chillingworth T, Clark TG, Galinski MR, Hall N, Harper D, Harris D, Hauser H, Ivens A, Janssen CS, Keane T, Larke N, Lapp S, Marti M, Moule S, Meyer IM, Ormond D, Peters N, Sanders M, Sanders S, Sargeant TJ, Simmonds M, Smith F, Squares R, Thurston S, Tivey AR, Walker D, White B, Zuiderwijk E, Churcher C, Quail MA, Cowman AF, Turner CMR, Rajandream MA, Kocken CHM, Thomas AW, Newbold CI, Barrell BG, Berriman M. 2008. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature 455:799–803. doi: 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tachibana S, Sullivan SA, Kawai S, Nakamura S, Kim HR, Goto N, Arisue N, Palacpac NM, Honma H, Yagi M, Tougan T, Katakai Y, Kaneko O, Mita T, Kita K, Yasutomi Y, Sutton PL, Shakhbatyan R, Horii T, Yasunaga T, Barnwell JW, Escalante AA, Carlton JM, Tanabe K. 2012. Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat Genet 44:1051–1055. doi: 10.1038/ng.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rutledge GG, Bohme U, Sanders M, Reid AJ, Cotton JA, Maiga-Ascofare O, Djimde AA, Apinjoh TO, Amenga-Etego L, Manske M, Barnwell JW, Renaud F, Ollomo B, Prugnolle F, Anstey NM, Auburn S, Price RN, McCarthy JS, Kwiatkowski DP, Newbold CI, Berriman M, Otto TD. 2017. Plasmodium malariae and P. ovale genomes provide insights into malaria parasite evolution. Nature 542:101–104. doi: 10.1038/nature21038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinelli A, Culleton R. 2018. Non-human primate malaria parasites: out of the forest and into the laboratory. Parasitology 145:41–54. doi: 10.1017/S0031182016001335. [DOI] [PubMed] [Google Scholar]
- 10.Valkiūnas G, Iezhova TA. 2018. Keys to the avian malaria parasites. Malar J 17:212. doi: 10.1186/s12936-018-2359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trampuz A, Jereb M, Muzlovic I, Prabhu RM. 2003. Clinical review: severe malaria. Crit Care 7:315–323. doi: 10.1186/cc2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen I, Clarke SE, Gosling R, Hamainza B, Killeen G, Magill A, O’Meara W, Price RN, Riley EM. 2016. “Asymptomatic” malaria: a chronic and debilitating infection that should be treated. PLoS Med 13:e1001942. doi: 10.1371/journal.pmed.1001942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinnis P, Zavala F. 2012. The skin: where malaria infection and the host immune response begin. Semin Immunopathol 34:787–792. doi: 10.1007/s00281-012-0345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams JH, Mueller I. 2017. The biology of Plasmodium vivax. Cold Spring Harb Perspect Med 7:a025585. doi: 10.1101/cshperspect.a025585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shanks GD, White NJ. 2013. The activation of vivax malaria hypnozoites by infectious diseases. Lancet Infect Dis 13:900–906. doi: 10.1016/S1473-3099(13)70095-1. [DOI] [PubMed] [Google Scholar]
- 16.White NJ, Imwong M. 2012. Relapse. Adv Parasitol 80:113–150. doi: 10.1016/B978-0-12-397900-1.00002-5. [DOI] [PubMed] [Google Scholar]
- 17.Smith RC, Vega-Rodríguez J, Jacobs-Lorena M. 2014. The Plasmodium bottleneck: malaria parasite losses in the mosquito vector. Mem Inst Oswaldo Cruz 109:644–661. doi: 10.1590/0074-0276130597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorobets NY, Sedash YV, Singh BK, Poonam, Rathi B. 2017. An overview of currently available antimalarials. Curr Top Med Chem 17:2143–2157. doi: 10.2174/1568026617666170130123520. [DOI] [PubMed] [Google Scholar]
- 19.Phillips MA, Burrows JN, Manyando C, van Huijsduijnen RH, Van Voorhis WC, Wells T. 2017. Malaria. Nat Rev Dis Primers 3:17050. doi: 10.1038/nrdp.2017.50. [DOI] [PubMed] [Google Scholar]
- 20.Thriemer K, Ley B, Bobogare A, Dysoley L, Alam MS, Pasaribu AP, Sattabongkot J, Jambert E, Domingo GJ, Commons R, Auburn S, Marfurt J, Devine A, Aktaruzzaman MM, Sohel N, Namgay R, Drukpa T, Sharma SN, Sarawati E, Samad I, Theodora M, Nambanya S, Ounekham S, Mudin RNB, Da Thakur G, Makita LS, Deray R, Lee S-E, Boaz L, Danansuriya MN, Mudiyanselage SD, Chinanonwait N, Kitchakarn S, Nausien J, Naket E, Duc TN, Do Manh H, Hong YS, Cheng Q, Richards JS, Kusriastuti R, Satyagraha A, Noviyanti R, Ding XC, Khan WA, Swe Phru C, Guoding Z, Qi G, Kaneko A, Miotto O, Nguitragool W, Roobsoong W, Battle K, Howes RE, Roca-Feltrer A, Duparc S, Bhowmick IP, Kenangalem E, Bibit J-A, Barry A, Sintasath D, Abeyasinghe R, Sibley CH, McCarthy J, von Seidlein L, Baird JK, Price RN. 2017. Challenges for achieving safe and effective radical cure of Plasmodium vivax: a round table discussion of the APMEN Vivax Working Group. Malar J 16:141. doi: 10.1186/s12936-017-1784-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baird JK. 2007. Neglect of Plasmodium vivax malaria. Trends Parasitol 23:533–539. doi: 10.1016/j.pt.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 22.Blasco B, Leroy D, Fidock DA. 2017. Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat Med 23:917–928. doi: 10.1038/nm.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wells TN, Hooft van Huijsduijnen R, Van Voorhis WC. 2015. Malaria medicines: a glass half full? Nat Rev Drug Discov 14:424–442. doi: 10.1038/nrd4573. [DOI] [PubMed] [Google Scholar]
- 24.Le Roch KG, Chung DW, Ponts N. 2012. Genomics and integrated systems biology in Plasmodium falciparum: a path to malaria control and eradication. Parasite Immunol 34:50–60. doi: 10.1111/j.1365-3024.2011.01340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlton J. 2003. The Plasmodium vivax genome sequencing project. Trends Parasitol 19:227–231. doi: 10.1016/S1471-4922(03)00066-7. [DOI] [PubMed] [Google Scholar]
- 26.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RMR, Crabb BS, del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kang’a S, Kooij TWA, Korsinczky M, Meyer EV-S, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. 2008. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otto TD, Bohme U, Jackson AP, Hunt M, Franke-Fayard B, Hoeijmakers WA, Religa AA, Robertson L, Sanders M, Ogun SA, Cunningham D, Erhart A, Billker O, Khan SM, Stunnenberg HG, Langhorne J, Holder AA, Waters AP, Newbold CI, Pain A, Berriman M, Janse CJ. 2014. A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol 12:86. doi: 10.1186/s12915-014-0086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otto TD, Rayner JC, Bohme U, Pain A, Spottiswoode N, Sanders M, Quail M, Ollomo B, Renaud F, Thomas AW, Prugnolle F, Conway DJ, Newbold C, Berriman M. 2014. Genome sequencing of chimpanzee malaria parasites reveals possible pathways of adaptation to human hosts. Nat Commun 5:4754. doi: 10.1038/ncomms5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bohme U, Otto TD, Cotton JA, Steinbiss S, Sanders M, Oyola SO, Nicot A, Gandon S, Patra KP, Herd C, Bushell E, Modrzynska KK, Billker O, Vinetz JM, Rivero A, Newbold CI, Berriman M. 2018. Complete avian malaria parasite genomes reveal features associated with lineage-specific evolution in birds and mammals. Genome Res 28:547–560. doi: 10.1101/gr.218123.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasini EM, Bohme U, Rutledge GG, Voorberg-Van der Wel A, Sanders M, Berriman M, Kocken CH, Otto TD. 2017. An improved Plasmodium cynomolgi genome assembly reveals an unexpected methyltransferase gene expansion. Wellcome Open Res 2:42. doi: 10.12688/wellcomeopenres.11864.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auburn S, Bohme U, Steinbiss S, Trimarsanto H, Hostetler J, Sanders M, Gao Q, Nosten F, Newbold CI, Berriman M, Price RN, Otto TD. 2016. A new Plasmodium vivax reference sequence with improved assembly of the subtelomeres reveals an abundance of pir genes. Wellcome Open Res 1:4. doi: 10.12688/wellcomeopenres.9876.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ansari HR, Templeton TJ, Subudhi AK, Ramaprasad A, Tang J, Lu F, Naeem R, Hashish Y, Oguike MC, Benavente ED, Clark TG, Sutherland CJ, Barnwell JW, Culleton R, Cao J, Pain A. 2016. Genome-scale comparison of expanded gene families in Plasmodium ovale wallikeri and Plasmodium ovale curtisi with Plasmodium malariae and with other Plasmodium species. Int J Parasitol 46:685–696. doi: 10.1016/j.ijpara.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Escalante AA, Ayala FJ. 1994. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc Natl Acad Sci U S A 91:11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perkins SL, Schall JJ. 2002. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J Parasitol 88:972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 36.Loy DE, Liu W, Li Y, Learn GH, Plenderleith LJ, Sundararaman SA, Sharp PM, Hahn BH. 2017. Out of Africa: origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax. Int J Parasitol 47:87–97. doi: 10.1016/j.ijpara.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talundzic E, Ravishankar S, Kelley J, Patel D, Plucinski M, Schmedes S, Ljolje D, Clemons B, Madison-Antenucci S, Arguin PM, Lucchi NW, Vannberg F, Udhayakumar V. 2018. Next-generation sequencing and bioinformatics protocol for malaria drug resistance marker surveillance. Antimicrob Agents Chemother 62:e02474-17. doi: 10.1128/AAC.02474-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Oliveira TC, Rodrigues PT, Menezes MJ, Goncalves-Lopes RM, Bastos MS, Lima NF, Barbosa S, Gerber AL, Loss de Morais G, Berna L, Phelan J, Robello C, de Vasconcelos ATR, Alves JMP, Ferreira MU. 2017. Genome-wide diversity and differentiation in New World populations of the human malaria parasite Plasmodium vivax. PLoS Negl Trop Dis 11:e0005824. doi: 10.1371/journal.pntd.0005824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hupalo DN, Luo Z, Melnikov A, Sutton PL, Rogov P, Escalante A, Vallejo AF, Herrera S, Arévalo-Herrera M, Fan Q, Wang Y, Cui L, Lucas CM, Durand S, Sanchez JF, Baldeviano GC, Lescano AG, Laman M, Barnadas C, Barry A, Mueller I, Kazura JW, Eapen A, Kanagaraj D, Valecha N, Ferreira MU, Roobsoong W, Nguitragool W, Sattabonkot J, Gamboa D, Kosek M, Vinetz JM, González-Cerón L, Birren BW, Neafsey DE, Carlton JM. 2016. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat Genet 48:953–958. doi: 10.1038/ng.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ocholla H, Preston MD, Mipando M, Jensen AT, Campino S, MacInnis B, Alcock D, Terlouw A, Zongo I, Oudraogo JB, Djimde AA, Assefa S, Doumbo OK, Borrmann S, Nzila A, Marsh K, Fairhurst RM, Nosten F, Anderson TJ, Kwiatkowski DP, Craig A, Clark TG, Montgomery J. 2014. Whole-genome scans provide evidence of adaptive evolution in Malawian Plasmodium falciparum isolates. J Infect Dis 210:1991–2000. doi: 10.1093/infdis/jiu349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miotto O, Almagro-Garcia J, Manske M, Macinnis B, Campino S, Rockett KA, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Duong S, Nguon C, Chuor CM, Saunders D, Se Y, Lon C, Fukuda MM, Amenga-Etego L, Hodgson AVO, Asoala V, Imwong M, Takala-Harrison S, Nosten F, Su X-Z, Ringwald P, Ariey F, Dolecek C, Hien TT, Boni MF, Thai CQ, Amambua-Ngwa A, Conway DJ, Djimdé AA, Doumbo OK, Zongo I, Ouedraogo J-B, Alcock D, Drury E, Auburn S, Koch O, Sanders M, Hubbart C, Maslen G, Ruano-Rubio V, Jyothi D, Miles A, O’Brien J, Gamble C, Oyola SO, Rayner JC, Newbold CI, Berriman M, Spencer CCA, McVean G, Day NP, White NJ, Bethell D, Dondorp AM, Plowe CV, Fairhurst RM, Kwiatkowski DP. 2013. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet 45:648–655. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan ER, Barnwell JW, Zimmerman PA, Serre D. 2015. Comparative analysis of field-isolate and monkey-adapted Plasmodium vivax genomes. PLoS Negl Trop Dis 9:e0003566. doi: 10.1371/journal.pntd.0003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manske M, Miotto O, Campino S, Auburn S, Almagro-Garcia J, Maslen G, O’Brien J, Djimde A, Doumbo O, Zongo I, Ouedraogo J-B, Michon P, Mueller I, Siba P, Nzila A, Borrmann S, Kiara SM, Marsh K, Jiang H, Su X-Z, Amaratunga C, Fairhurst R, Socheat D, Nosten F, Imwong M, White NJ, Sanders M, Anastasi E, Alcock D, Drury E, Oyola S, Quail MA, Turner DJ, Ruano-Rubio V, Jyothi D, Amenga-Etego L, Hubbart C, Jeffreys A, Rowlands K, Sutherland C, Roper C, Mangano V, Modiano D, Tan JC, Ferdig MT, Amambua-Ngwa A, Conway DJ, Takala-Harrison S, Plowe CV, Rayner JC, Rockett KA, Clark TG, Newbold CI, Berriman M, MacInnis B, Kwiatkowski DP. 2012. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 487:375–379. doi: 10.1038/nature11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miles A, Iqbal Z, Vauterin P, Pearson R, Campino S, Theron M, Gould K, Mead D, Drury E, O’Brien J, Ruano Rubio V, MacInnis B, Mwangi J, Samarakoon U, Ranford-Cartwright L, Ferdig M, Hayton K, Su X-Z, Wellems T, Rayner J, McVean G, Kwiatkowski D. 2016. Indels, structural variation, and recombination drive genomic diversity in Plasmodium falciparum. Genome Res 26:1288–1299. doi: 10.1101/gr.203711.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Böhme U, Otto T, Sanders M, Newbold C, Berriman M. 2019. Progression of the canonical reference malaria parasite genome from 2002–2019 [version 2; peer review: 3 approved]. Wellcome Open Res 4:58. doi: 10.12688/wellcomeopenres.15194.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trevino SG, Nkhoma SC, Nair S, Daniel BJ, Moncada K, Khoswe S, Banda RL, Nosten F, Cheeseman IH. 2017. High-resolution single-cell sequencing of malaria parasites. Genome Biol Evol 9:3373–3383. doi: 10.1093/gbe/evx256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nair S, Nkhoma SC, Serre D, Zimmerman PA, Gorena K, Daniel BJ, Nosten F, Anderson TJ, Cheeseman IH. 2014. Single-cell genomics for dissection of complex malaria infections. Genome Res 24:1028–1038. doi: 10.1101/gr.168286.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poran A, Notzel C, Aly O, Mencia-Trinchant N, Harris CT, Guzman ML, Hassane DC, Elemento O, Kafsack B. 2017. Single-cell RNA sequencing reveals a signature of sexual commitment in malaria parasites. Nature 551:95–99. doi: 10.1038/nature24280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carlton J, Silva J, Hall N. 2005. The genome of model malaria parasites, and comparative genomics. Curr Issues Mol Biol 7:23–37. [PubMed] [Google Scholar]
- 50.Kooij TW, Carlton JM, Bidwell SL, Hall N, Ramesar J, Janse CJ, Waters AP. 2005. A Plasmodium whole-genome synteny map: indels and synteny breakpoints as foci for species-specific genes. PLoS Pathog 1:e44. doi: 10.1371/journal.ppat.0010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DePristo MA, Zilversmit MM, Hartl DL. 2006. On the abundance, amino acid composition, and evolutionary dynamics of low-complexity regions in proteins. Gene 378:19–30. doi: 10.1016/j.gene.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 52.Su X, Wellems TE. 1996. Toward a high-resolution Plasmodium falciparum linkage map: polymorphic markers from hundreds of simple sequence repeats. Genomics 33:430–444. doi: 10.1006/geno.1996.0218. [DOI] [PubMed] [Google Scholar]
- 53.Su X, Ferdig MT, Huang Y, Huynh CQ, Liu A, You J, Wootton JC, Wellems TE. 1999. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science 286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- 54.Aravind L, Iyer LM, Wellems TE, Miller LH. 2003. Plasmodium biology: genomic gleanings. Cell 115:771–785. doi: 10.1016/S0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- 55.Su X, Hayton K, Wellems TE. 2007. Genetic linkage and association analyses for trait mapping in Plasmodium falciparum. Nat Rev Genet 8:497–506. doi: 10.1038/nrg2126. [DOI] [PubMed] [Google Scholar]
- 56.Ferdig MT, Cooper RA, Mu J, Deng B, Joy DA, Su X-Z, Wellems TE. 2004. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol Microbiol 52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- 57.Su X, Kirkman LA, Fujioka H, Wellems TE. 1997. Complex polymorphisms in an approximately 330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell 91:593–603. doi: 10.1016/S0092-8674(00)80447-X. [DOI] [PubMed] [Google Scholar]
- 58.Figan CE, Sa JM, Mu J, Melendez-Muniz VA, Liu CH, Wellems TE. 2018. A set of microsatellite markers to differentiate Plasmodium falciparum progeny of four genetic crosses. Malar J 17:60. doi: 10.1186/s12936-018-2210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, Su XZ. 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- 60.Roper C, Pearce R, Bredenkamp B, Gumede J, Drakeley C, Mosha F, Chandramohan D, Sharp B. 2003. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet 361:1174–1181. doi: 10.1016/S0140-6736(03)12951-0. [DOI] [PubMed] [Google Scholar]
- 61.Anderson TJ, Roper C. 2005. The origins and spread of antimalarial drug resistance: lessons for policy makers. Acta Trop 94:269–280. doi: 10.1016/j.actatropica.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 62.Wahlgren M, Goel S, Akhouri RR. 2017. Variant surface antigens of Plasmodium falciparum and their roles in severe malaria. Nat Rev Microbiol 15:479–491. doi: 10.1038/nrmicro.2017.47. [DOI] [PubMed] [Google Scholar]
- 63.Cunningham D, Lawton J, Jarra W, Preiser P, Langhorne J. 2010. The pir multigene family of Plasmodium: antigenic variation and beyond. Mol Biochem Parasitol 170:65–73. doi: 10.1016/j.molbiopara.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 64.Otto TD, Gilabert A, Crellen T, Bohme U, Arnathau C, Sanders M, Oyola SO, Okouga AP, Boundenga L, Willaume E, Ngoubangoye B, Moukodoum ND, Paupy C, Durand P, Rougeron V, Ollomo B, Renaud F, Newbold C, Berriman M, Prugnolle F. 2018. Genomes of all known members of a Plasmodium subgenus reveal paths to virulent human malaria. Nat Microbiol 3:687–697. doi: 10.1038/s41564-018-0162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aley SB, Sherwood JA, Howard RJ. 1984. Knob-positive and knob-negative Plasmodium falciparum differ in expression of a strain-specific malarial antigen on the surface of infected erythrocytes. J Exp Med 160:1585–1590. doi: 10.1084/jem.160.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leech JH, Barnwell JW, Miller LH, Howard RJ. 1984. Identification of a strain-specific malarial antigen exposed on the surface of Plasmodium falciparum-infected erythrocytes. J Exp Med 159:1567–1575. doi: 10.1084/jem.159.6.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. 1995. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 68.Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. 1995. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 69.Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, Miller LH. 1995. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82:101–110. doi: 10.1016/0092-8674(95)90056-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pasternak ND, Dzikowski R. 2009. PfEMP1: an antigen that plays a key role in the pathogenicity and immune evasion of the malaria parasite Plasmodium falciparum. Int J Biochem Cell Biol 41:1463–1466. doi: 10.1016/j.biocel.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 71.Tuikue Ndam N, Moussiliou A, Lavstsen T, Kamaliddin C, Jensen ATR, Mama A, Tahar R, Wang CW, Jespersen JS, Alao JM, Gamain B, Theander TG, Deloron P. 2017. Parasites causing cerebral falciparum malaria bind multiple endothelial receptors and express EPCR and ICAM-1-binding PfEMP1. J Infect Dis 215:1918–1925. doi: 10.1093/infdis/jix230. [DOI] [PubMed] [Google Scholar]
- 72.Fried M, Duffy PE. 2017. Malaria during pregnancy. Cold Spring Harb Perspect Med 7:a025551. doi: 10.1101/cshperspect.a025551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fairhurst RM, Baruch DI, Brittain NJ, Ostera GR, Wallach JS, Hoang HL, Hayton K, Guindo A, Makobongo MO, Schwartz OM, Tounkara A, Doumbo OK, Diallo DA, Fujioka H, Ho M, Wellems TE. 2005. Abnormal display of PfEMP-1 on erythrocytes carrying haemoglobin C may protect against malaria. Nature 435:1117–1121. doi: 10.1038/nature03631. [DOI] [PubMed] [Google Scholar]
- 74.Cholera R, Brittain NJ, Gillrie MR, Lopera-Mesa TM, Diakite SA, Arie T, Krause MA, Guindo A, Tubman A, Fujioka H, Diallo DA, Doumbo OK, Ho M, Wellems TE, Fairhurst RM. 2008. Impaired cytoadherence of Plasmodium falciparum-infected erythrocytes containing sickle hemoglobin. Proc Natl Acad Sci U S A 105:991–996. doi: 10.1073/pnas.0711401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cyrklaff M, Srismith S, Nyboer B, Burda K, Hoffmann A, Lasitschka F, Adjalley S, Bisseye C, Simpore J, Mueller AK, Sanchez CP, Frischknecht F, Lanzer M. 2016. Oxidative insult can induce malaria-protective trait of sickle and fetal erythrocytes. Nat Commun 7:13401. doi: 10.1038/ncomms13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yam XY, Brugat T, Siau A, Lawton J, Wong DS, Farah A, Twang JS, Gao X, Langhorne J, Preiser PR. 2016. Characterization of the Plasmodium interspersed repeats (PIR) proteins of Plasmodium chabaudi indicates functional diversity. Sci Rep 6:23449. doi: 10.1038/srep23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fougere A, Jackson AP, Bechtsi DP, Braks JA, Annoura T, Fonager J, Spaccapelo R, Ramesar J, Chevalley-Maurel S, Klop O, van der Laan AM, Tanke HJ, Kocken CH, Pasini EM, Khan SM, Bohme U, van Ooij C, Otto TD, Janse CJ, Franke-Fayard B. 2016. Variant exported blood-stage proteins encoded by Plasmodium multigene families are expressed in liver stages where they are exported into the parasitophorous vacuole. PLoS Pathog 12:e1005917. doi: 10.1371/journal.ppat.1005917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brugat T, Reid AJ, Lin J, Cunningham D, Tumwine I, Kushinga G, McLaughlin S, Spence P, Bohme U, Sanders M, Conteh S, Bushell E, Metcalf T, Billker O, Duffy PE, Newbold C, Berriman M, Langhorne J. 2017. Antibody-independent mechanisms regulate the establishment of chronic Plasmodium infection. Nat Microbiol 2:16276. doi: 10.1038/nmicrobiol.2016.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spence PJ, Jarra W, Levy P, Reid AJ, Chappell L, Brugat T, Sanders M, Berriman M, Langhorne J. 2013. Vector transmission regulates immune control of Plasmodium virulence. Nature 498:228–231. doi: 10.1038/nature12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Llinas M, Bozdech Z, Wong ED, Adai AT, DeRisi JL. 2006. Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res 34:1166–1173. doi: 10.1093/nar/gkj517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arredondo SA, Kappe S. 2017. The s48/45 six-cysteine proteins: mediators of interaction throughout the Plasmodium life cycle. Int J Parasitol 47:409–423. doi: 10.1016/j.ijpara.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Healer J, McGuinness D, Hopcroft P, Haley S, Carter R, Riley E. 1997. Complement-mediated lysis of Plasmodium falciparum gametes by malaria-immune human sera is associated with antibodies to the gamete surface antigen Pfs230. Infect Immun 65:3017–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Canepa GE, Molina-Cruz A, Yenkoidiok-Douti L, Calvo E, Williams AE, Burkhardt M, Peng F, Narum D, Boulanger MJ, Valenzuela JG, Barillas-Mury C. 2018. Antibody targeting of a specific region of Pfs47 blocks Plasmodium falciparum malaria transmission. NPJ Vaccines 3:26. doi: 10.1038/s41541-018-0065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee SM, Plieskatt J, Krishnan S, Raina M, Harishchandra R, King CR. 2019. Expression and purification optimization of an N-terminal Pfs230 transmission-blocking vaccine candidate. Protein Expr Purif 160:56–65. doi: 10.1016/j.pep.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Williamson KC. 2003. Pfs230: from malaria transmission-blocking vaccine candidate toward function. Parasite Immunol 25:351–359. doi: 10.1046/j.1365-3024.2003.00643.x. [DOI] [PubMed] [Google Scholar]
- 86.van Dijk MR, van Schaijk BC, Khan SM, van Dooren MW, Ramesar J, Kaczanowski S, van Gemert GJ, Kroeze H, Stunnenberg HG, Eling WM, Sauerwein RW, Waters AP, Janse CJ. 2010. Three members of the 6-cys protein family of Plasmodium play a role in gamete fertility. PLoS Pathog 6:e1000853. doi: 10.1371/journal.ppat.1000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anthony TG, Polley SD, Vogler AP, Conway DJ. 2007. Evidence of non-neutral polymorphism in Plasmodium falciparum gamete surface protein genes Pfs47 and Pfs48/45. Mol Biochem Parasitol 156:117–123. doi: 10.1016/j.molbiopara.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 88.Molina-Cruz A, Garver LS, Alabaster A, Bangiolo L, Haile A, Winikor J, Ortega C, van Schaijk BC, Sauerwein RW, Taylor-Salmon E, Barillas-Mury C. 2013. The human malaria parasite Pfs47 gene mediates evasion of the mosquito immune system. Science 340:984–987. doi: 10.1126/science.1235264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Molina-Cruz A, Canepa GE, Kamath N, Pavlovic NV, Mu J, Ramphul UN, Ramirez JL, Barillas-Mury C. 2015. Plasmodium evasion of mosquito immunity and global malaria transmission: The lock-and-key theory. Proc Natl Acad Sci U S A 112:15178–15183. doi: 10.1073/pnas.1520426112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ishino T, Chinzei Y, Yuda M. 2005. Two proteins with 6-cys motifs are required for malarial parasites to commit to infection of the hepatocyte. Mol Microbiol 58:1264–1275. doi: 10.1111/j.1365-2958.2005.04801.x. [DOI] [PubMed] [Google Scholar]
- 91.VanBuskirk KM, O’Neill MT, De La Vega P, Maier AG, Krzych U, Williams J, Dowler MG, Sacci JB Jr, Kangwanrangsan N, Tsuboi T, Kneteman NM, Heppner DG Jr, Murdock BA, Mikolajczak SA, Aly AS, Cowman AF, Kappe SH. 2009. Preerythrocytic, live-attenuated Plasmodium falciparum vaccine candidates by design. Proc Natl Acad Sci U S A 106:13004–13009. doi: 10.1073/pnas.0906387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Van der Ploeg LH, Smits M, Ponnudurai T, Vermeulen A, Meuwissen JH, Langsley G. 1985. Chromosome-sized DNA molecules of Plasmodium falciparum. Science 229:658–661. doi: 10.1126/science.3895435. [DOI] [PubMed] [Google Scholar]
- 93.Kemp DJ, Corcoran LM, Coppel RL, Stahl HD, Bianco AE, Brown GV, Anders RF. 1985. Size variation in chromosomes from independent cultured isolates of Plasmodium falciparum. Nature 315:347–350. doi: 10.1038/315347a0. [DOI] [PubMed] [Google Scholar]
- 94.Corcoran LM, Forsyth KP, Bianco AE, Brown GV, Kemp DJ. 1986. Chromosome size polymorphisms in Plasmodium falciparum can involve deletions and are frequent in natural parasite populations. Cell 44:87–95. doi: 10.1016/0092-8674(86)90487-3. [DOI] [PubMed] [Google Scholar]
- 95.Wellems TE, Walliker D, Smith CL, do Rosario VE, Maloy WL, Howard RJ, Carter R, McCutchan TF. 1987. A histidine-rich protein gene marks a linkage group favored strongly in a genetic cross of Plasmodium falciparum. Cell 49:633–642. doi: 10.1016/0092-8674(87)90539-3. [DOI] [PubMed] [Google Scholar]
- 96.Ellis J, Irving DO, Wellems TE, Howard RJ, Cross GA. 1987. Structure and expression of the knob-associated histidine-rich protein of Plasmodium falciparum. Mol Biochem Parasitol 26:203–214. doi: 10.1016/0166-6851(87)90144-7. [DOI] [PubMed] [Google Scholar]
- 97.Wellems T, Oduola AMJ, Fenton B, Desjardins R, Panton LJ, DoRosario VE. 1988. Chromosome size variation occurs in cloned Plasmodium falciparum on in vitro cultivation. Rev Brazilian Genetics 11:813–825. [Google Scholar]
- 98.Sinnis P, Wellems TE. 1988. Long-range restriction maps of Plasmodium falciparum chromosomes: crossingover and size variation among geographically distant isolates. Genomics 3:287–295. doi: 10.1016/0888-7543(88)90117-6. [DOI] [PubMed] [Google Scholar]
- 99.Corcoran LM, Thompson JK, Walliker D, Kemp DJ. 1988. Homologous recombination within subtelomeric repeat sequences generates chromosome size polymorphisms in P. falciparum. Cell 53:807–813. doi: 10.1016/0092-8674(88)90097-9. [DOI] [PubMed] [Google Scholar]
- 100.Freitas-Junior LH, Bottius E, Pirrit LA, Deitsch KW, Scheidig C, Guinet F, Nehrbass U, Wellems TE, Scherf A. 2000. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature 407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 101.Walker-Jonah A, Dolan SA, Gwadz RW, Panton LJ, Wellems TE. 1992. An RFLP map of the Plasmodium falciparum genome, recombination rates and favored linkage groups in a genetic cross. Mol Biochem Parasitol 51:313–320. doi: 10.1016/0166-6851(92)90081-T. [DOI] [PubMed] [Google Scholar]
- 102.Wellems TE, Panton LJ, Gluzman IY, do Rosario VE, Gwadz RW, Walker-Jonah A, Krogstad DJ. 1990. Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature 345:253–255. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- 103.Peterson DS, Walliker D, Wellems TE. 1988. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci U S A 85:9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li J, Pattaradilokrat S, Zhu F, Jiang H, Liu S, Hong L, Fu Y, Koo L, Xu W, Pan W, Carlton JM, Kaneko O, Carter R, Wootton JC, Su XZ. 2011. Linkage maps from multiple genetic crosses and loci linked to growth-related virulent phenotype in Plasmodium yoelii. Proc Natl Acad Sci U S A 108:E374–E82. doi: 10.1073/pnas.1102261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Su X-Z, Wellems T. 1998. Genome discovery amd malaria research: current status and promise, p 253–266. In Sherman IW. (ed), Malaria: parasite biology, pathogenesis, and protection. American Society for Microbiology, Washington, DC. [Google Scholar]
- 106.Su X-Z, Wellems TE. 1997. Plasmodium falciparum: a rapid DNA fingerprinting method using microsatellite sequences within var clusters. Exp Parasitol 86:235–236. doi: 10.1006/expr.1997.4174. [DOI] [PubMed] [Google Scholar]
- 107.Li J, Zhang Y, Sullivan M, Hong L, Huang L, Lu F, McCutchan TF, Su XZ. 2007. Typing Plasmodium yoelii microsatellites using a simple and affordable fluorescent labeling method. Mol Biochem Parasitol 155:94–102. doi: 10.1016/j.molbiopara.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li J, Zhang Y, Liu S, Hong L, Sullivan M, McCutchan TF, Carlton JM, Su XZ. 2009. Hundreds of microsatellites for genotyping Plasmodium yoelii parasites. Mol Biochem Parasitol 166:153–158. doi: 10.1016/j.molbiopara.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Feng X, Carlton JM, Joy DA, Mu J, Furuya T, Suh BB, Wang Y, Barnwell JW, Su XZ. 2003. Single-nucleotide polymorphisms and genome diversity in Plasmodium vivax. Proc Natl Acad Sci U S A 100:8502–8507. doi: 10.1073/pnas.1232502100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mu J, Duan J, Makova KD, Joy DA, Huynh CQ, Branch OH, Li WH, Su XZ. 2002. Chromosome-wide SNPs reveal an ancient origin for Plasmodium falciparum. Nature 418:323–326. doi: 10.1038/nature00836. [DOI] [PubMed] [Google Scholar]
- 111.Mu J, Awadalla P, Duan J, McGee KM, Keebler J, Seydel K, McVean GA, Su X-Z. 2007. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat Genet 39:126–130. doi: 10.1038/ng1924. [DOI] [PubMed] [Google Scholar]
- 112.Jeffares DC, Pain A, Berry A, Cox AV, Stalker J, Ingle CE, Thomas A, Quail MA, Siebenthall K, Uhlemann AC, Kyes S, Krishna S, Newbold C, Dermitzakis ET, Berriman M. 2007. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat Genet 39:120–125. doi: 10.1038/ng1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, Milner DA Jr, Daily JP, Sarr O, Ndiaye D, Ndir O, Mboup S, Duraisingh MT, Lukens A, Derr A, Stange-Thomann N, Waggoner S, Onofrio R, Ziaugra L, Mauceli E, Gnerre S, Jaffe DB, Zainoun J, Wiegand RC, Birren BW, Hartl DL, Galagan JE, Lander ES, Wirth DF. 2007. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet 39:113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- 114.Wilson CM, Volkman SK, Thaithong S, Martin RK, Kyle DE, Milhous WK, Wirth DF. 1993. Amplification of pfmdr1 associated with mefloquine and halofantrine resistance in Plasmodium falciparum from Thailand. Mol Biochem Parasitol 57:151–160. doi: 10.1016/0166-6851(93)90252-S. [DOI] [PubMed] [Google Scholar]
- 115.Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Suwanarusk R, Chavchich M, Russell B, Jaidee A, Chalfein F, Barends M, Prasetyorini B, Kenangalem E, Piera KA, Lek-Uthai U, Anstey NM, Tjitra E, Nosten F, Cheng Q, Price RN. 2008. Amplification of pvmdr1 associated with multidrug-resistant Plasmodium vivax. J Infect Dis 198:1558–1564. doi: 10.1086/592451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nair S, Miller B, Barends M, Jaidee A, Patel J, Mayxay M, Newton P, Nosten F, Ferdig MT, Anderson TJ. 2008. Adaptive copy number evolution in malaria parasites. PLoS Genet 4:e1000243. doi: 10.1371/journal.pgen.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Anderson TJ, Patel J, Ferdig MT. 2009. Gene copy number and malaria biology. Trends Parasitol 25:336–343. doi: 10.1016/j.pt.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Eastman RT, Dharia NV, Winzeler EA, Fidock DA. 2011. Piperaquine resistance is associated with a copy number variation on chromosome 5 in drug-pressured Plasmodium falciparum parasites. Antimicrob Agents Chemother 55:3908–3916. doi: 10.1128/AAC.01793-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Triglia T, Duraisingh MT, Good RT, Cowman AF. 2005. Reticulocyte-binding protein homologue 1 is required for sialic acid-dependent invasion into human erythrocytes by Plasmodium falciparum. Mol Microbiol 55:162–174. doi: 10.1111/j.1365-2958.2004.04388.x. [DOI] [PubMed] [Google Scholar]
- 121.Hostetler JB, Lo E, Kanjee U, Amaratunga C, Suon S, Sreng S, Mao S, Yewhalaw D, Mascarenhas A, Kwiatkowski DP, Ferreira MU, Rathod PK, Yan G, Fairhurst RM, Duraisingh MT, Rayner JC. 2016. Independent origin and global distribution of distinct Plasmodium vivax Duffy binding protein gene duplications. PLoS Negl Trop Dis 10:e0005091. doi: 10.1371/journal.pntd.0005091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Biggs BA, Kemp DJ, Brown GV. 1989. Subtelomeric chromosome deletions in field isolates of Plasmodium falciparum and their relationship to loss of cytoadherence in vitro. Proc Natl Acad Sci U S A 86:2428–2432. doi: 10.1073/pnas.86.7.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gonzales JM, Patel JJ, Ponmee N, Jiang L, Tan A, Maher SP, Wuchty S, Rathod PK, Ferdig MT. 2008. Regulatory hotspots in the malaria parasite genome dictate transcriptional variation. PLoS Biol 6:e238. doi: 10.1371/journal.pbio.0060238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mackinnon MJ, Li J, Mok S, Kortok MM, Marsh K, Preiser PR, Bozdech Z. 2009. Comparative transcriptional and genomic analysis of Plasmodium falciparum field isolates. PLoS Pathog 5:e1000644. doi: 10.1371/journal.ppat.1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Samarakoon U, Gonzales JM, Patel JJ, Tan A, Checkley L, Ferdig MT. 2011. The landscape of inherited and de novo copy number variants in a Plasmodium falciparum genetic cross. BMC Genomics 12:457. doi: 10.1186/1471-2164-12-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nair S, Nash D, Sudimack D, Jaidee A, Barends M, Uhlemann AC, Krishna S, Nosten F, Anderson TJ. 2007. Recurrent gene amplification and soft selective sweeps during evolution of multidrug resistance in malaria parasites. Mol Biol Evol 24:562–573. doi: 10.1093/molbev/msl185. [DOI] [PubMed] [Google Scholar]
- 127.Guler JL, Freeman DL, Ahyong V, Patrapuvich R, White J, Gujjar R, Phillips MA, DeRisi J, Rathod PK. 2013. Asexual populations of the human malaria parasite, Plasmodium falciparum, use a two-step genomic strategy to acquire accurate, beneficial DNA amplifications. PLoS Pathog 9:e1003375. doi: 10.1371/journal.ppat.1003375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wellems TE, Howard RJ. 1986. Homologous genes encode two distinct histidine-rich proteins in a cloned isolate of Plasmodium falciparum. Proc Natl Acad Sci U S A 83:6065–6069. doi: 10.1073/pnas.83.16.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Krogstad DJ, Schlesinger PH, Gluzman IY. 1989. Chloroquine and acid vesicle function. Prog Clin Biol Res 313:53–59. [PubMed] [Google Scholar]
- 130.Foote SJ, Thompson JK, Cowman AF, Kemp DJ. 1989. Amplification of the multidrug resistance gene in some chloroquine- resistant isolates of P. falciparum. Cell 57:921–930. doi: 10.1016/0092-8674(89)90330-9. [DOI] [PubMed] [Google Scholar]
- 131.Foote SJ, Kyle DE, Martin RK, Oduola AM, Forsyth K, Kemp DJ, Cowman AF. 1990. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature 345:255–258. doi: 10.1038/345255a0. [DOI] [PubMed] [Google Scholar]
- 132.Cravo PV, Carlton JM, Hunt P, Bisoni L, Padua RA, Walliker D. 2003. Genetics of mefloquine resistance in the rodent malaria parasite Plasmodium chabaudi. Antimicrob Agents Chemother 47:709–718. doi: 10.1128/aac.47.2.709-718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Imwong M, Pukrittayakamee S, Pongtavornpinyo W, Nakeesathit S, Nair S, Newton P, Nosten F, Anderson TJ, Dondorp A, Day NP, White NJ. 2008. Gene amplification of the multidrug resistance 1 gene of Plasmodium vivax isolates from Thailand, Laos, and Myanmar. Antimicrob Agents Chemother 52:2657–2659. doi: 10.1128/AAC.01459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Barnes DA, Foote SJ, Galatis D, Kemp DJ, Cowman AF. 1992. Selection for high-level chloroquine resistance results in deamplification of the pfmdr1 gene and increased sensitivity to mefloquine in Plasmodium falciparum. EMBO J 11:3067–3075. doi: 10.1002/j.1460-2075.1992.tb05378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Walliker D, Quakyi IA, Wellems TE, McCutchan TF, Szarfman A, London WT, Corcoran LM, Burkot TR, Carter R. 1987. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science 236:1661–1666. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- 136.Hayton K, Gaur D, Liu A, Takahashi J, Henschen B, Singh S, Lambert L, Furuya T, Bouttenot R, Doll M, Nawaz F, Mu J, Jiang L, Miller LH, Wellems TE. 2008. Erythrocyte binding protein PfRH5 polymorphisms determine species-specific pathways of Plasmodium falciparum invasion. Cell Host Microbe 4:40–51. doi: 10.1016/j.chom.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sa JM, Kaslow SR, Krause MA, Melendez-Muniz VA, Salzman RE, Kite WA, Zhang M, Moraes Barros RR, Mu J, Han PK, Mershon JP, Figan CE, Caleon RL, Rahman RS, Gibson TJ, Amaratunga C, Nishiguchi EP, Breglio KF, Engels TM, Velmurugan S, Ricklefs S, Straimer J, Gnadig NF, Deng B, Liu A, Diouf A, Miura K, Tullo GS, Eastman RT, Chakravarty S, James ER, Udenze K, Li S, Sturdevant DE, Gwadz RW, Porcella SF, Long CA, Fidock DA, Thomas ML, Fay MP, Sim BKL, Hoffman SL, Adams JH, Fairhurst RM, Su XZ, Wellems TE. 2018. Artemisinin resistance phenotypes and K13 inheritance in a Plasmodium falciparum cross and Aotus model. Proc Natl Acad Sci U S A 115:12513–12518. doi: 10.1073/pnas.1813386115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vaughan AM, Pinapati RS, Cheeseman IH, Camargo N, Fishbaugher M, Checkley LA, Nair S, Hutyra CA, Nosten FH, Anderson TJ, Ferdig MT, Kappe SH. 2015. Plasmodium falciparum genetic crosses in a humanized mouse model. Nat Methods 12:631–633. doi: 10.1038/nmeth.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Vaidya AB, Morrisey J, Plowe CV, Kaslow DC, Wellems TE. 1993. Unidirectional dominance of cytoplasmic inheritance in two genetic crosses of Plasmodium falciparum. Mol Cell Biol 13:7349–7357. doi: 10.1128/mcb.13.12.7349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Vaidya AB, Muratova O, Guinet F, Keister D, Wellems TE, Kaslow DC. 1995. A genetic locus on Plasmodium falciparum chromosome 12 linked to a defect in mosquito-infectivity and male gametogenesis. Mol Biochem Parasitol 69:65–71. doi: 10.1016/0166-6851(94)00199-W. [DOI] [PubMed] [Google Scholar]
- 141.Nguitragool W, Bokhari AA, Pillai AD, Rayavara K, Sharma P, Turpin B, Aravind L, Desai SA. 2011. Malaria parasite clag3 genes determine channel-mediated nutrient uptake by infected red blood cells. Cell 145:665–677. doi: 10.1016/j.cell.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Reilly Ayala HB, Wacker MA, Siwo G, Ferdig MT. 2010. Quantitative trait loci mapping reveals candidate pathways regulating cell cycle duration in Plasmodium falciparum. BMC Genomics 11:577. doi: 10.1186/1471-2164-11-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sanchez CP, Liu CH, Mayer S, Nurhasanah A, Cyrklaff M, Mu J, Ferdig MT, Stein WD, Lanzer M. 2014. A HECT ubiquitin-protein ligase as a novel candidate gene for altered quinine and quinidine responses in Plasmodium falciparum. PLoS Genet 10:e1004382. doi: 10.1371/journal.pgen.1004382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lewis IA, Wacker M, Olszewski KL, Cobbold SA, Baska KS, Tan A, Ferdig MT, Llinas M. 2014. Metabolic QTL analysis links chloroquine resistance in Plasmodium falciparum to impaired hemoglobin catabolism. PLoS Genet 10:e1004085. doi: 10.1371/journal.pgen.1004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naude B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell 6:861–871. doi: 10.1016/S1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Crosnier C, Bustamante LY, Bartholdson SJ, Bei AK, Theron M, Uchikawa M, Mboup S, Ndir O, Kwiatkowski DP, Duraisingh MT, Rayner JC, Wright GJ. 2011. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature 480:534–537. doi: 10.1038/nature10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Douglas AD, Williams AR, Knuepfer E, Illingworth JJ, Furze JM, Crosnier C, Choudhary P, Bustamante LY, Zakutansky SE, Awuah DK, Alanine DG, Theron M, Worth A, Shimkets R, Rayner JC, Holder AA, Wright GJ, Draper SJ. 2014. Neutralization of Plasmodium falciparum merozoites by antibodies against PfRH5. J Immunol 192:245–258. doi: 10.4049/jimmunol.1302045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sa JM, Twu O, Hayton K, Reyes S, Fay MP, Ringwald P, Wellems TE. 2009. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci U S A 106:18883–18889. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Walliker D, Carter R, Sanderson A. 1975. Genetic studies on Plasmodium chabaudi: recombination between enzyme markers. Parasitology 70:19–24. doi: 10.1017/S0031182000048824. [DOI] [PubMed] [Google Scholar]
- 150.Hunt P, Afonso A, Creasey A, Culleton R, Sidhu AB, Logan J, Valderramos SG, McNae I, Cheesman S, do Rosario V, Carter R, Fidock DA, Cravo P. 2007. Gene encoding a deubiquitinating enzyme is mutated in artesunate- and chloroquine-resistant rodent malaria parasites. Mol Microbiol 65:27–40. doi: 10.1111/j.1365-2958.2007.05753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Culleton R, Martinelli A, Hunt P, Carter R. 2005. Linkage group selection: rapid gene discovery in malaria parasites. Genome Res 15:92–97. doi: 10.1101/gr.2866205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Martinelli A, Cheesman S, Hunt P, Culleton R, Raza A, Mackinnon M, Carter R. 2005. A genetic approach to the de novo identification of targets of strain-specific immunity in malaria parasites. Proc Natl Acad Sci U S A 102:814–819. doi: 10.1073/pnas.0405097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Nair SC, Xu R, Pattaradilokrat S, Wu J, Qi Y, Zilversmit M, Ganesan S, Nagarajan V, Eastman RT, Orandle MS, Tan JC, Myers TG, Liu S, Long CA, Li J, Su XZ. 2017. A Plasmodium yoelii HECT-like E3 ubiquitin ligase regulates parasite growth and virulence. Nat Commun 8:223. doi: 10.1038/s41467-017-00267-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bopp SE, Rodrigo E, Gonzalez-Paez GE, Frazer M, Barnes SW, Valim C, Watson J, Walker JR, Schmedt C, Winzeler EA. 2013. Identification of the Plasmodium berghei resistance locus 9 linked to survival on chromosome 9. Malar J 12:316. doi: 10.1186/1475-2875-12-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Raine JD, Ecker A, Mendoza J, Tewari R, Stanway RR, Sinden RE. 2007. Female inheritance of malarial lap genes is essential for mosquito transmission. PLoS Pathog 3:e30. doi: 10.1371/journal.ppat.0030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Qi Y, Zhu F, Li J, Fu Y, Pattaradilokrat S, Hong L, Liu S, Huang F, Xu W, Su XZ. 2013. Optimized protocols for improving the likelihood of cloning recombinant progeny from Plasmodium yoelii genetic crosses. Exp Parasitol 133:44–50. doi: 10.1016/j.exppara.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Carlton J, Mackinnon M, Walliker D. 1998. A chloroquine resistance locus in the rodent malaria parasite Plasmodium chabaudi. Mol Biochem Parasitol 93:57–72. doi: 10.1016/S0166-6851(98)00021-8. [DOI] [PubMed] [Google Scholar]
- 158.Cheesman S, O’Mahony E, Pattaradilokrat S, Degnan K, Knott S, Carter R. 2010. A single parasite gene determines strain-specific protective immunity against malaria: the role of the merozoite surface protein I. Int J Parasitol 40:951–961. doi: 10.1016/j.ijpara.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 159.Pattaradilokrat S, Culleton RL, Cheesman SJ, Carter R. 2009. Gene encoding erythrocyte binding ligand linked to blood stage multiplication rate phenotype in Plasmodium yoelii yoelii. Proc Natl Acad Sci U S A 106:7161–7166. doi: 10.1073/pnas.0811430106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pattaradilokrat S, Li J, Wu J, Qi Y, Eastman RT, Zilversmit M, Nair SC, Huaman MC, Quinones M, Jiang H, Li N, Zhu J, Zhao K, Kaneko O, Long CA, Su XZ. 2014. Plasmodium genetic loci linked to host cytokine and chemokine responses. Genes Immun 15:145–152. doi: 10.1038/gene.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wu J, Cai B, Sun W, Huang R, Liu X, Lin M, Pattaradilokrat S, Martin S, Qi Y, Nair SC, Bolland S, Cohen JI, Austin CP, Long CA, Myers TG, Wang RF, Su XZ. 2015. Genome-wide analysis of host-Plasmodium yoelii interactions reveals regulators of the type I interferon response. Cell Rep 12:661–672. doi: 10.1016/j.celrep.2015.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wu J, Tian L, Yu X, Pattaradilokrat S, Li J, Wang M, Yu W, Qi Y, Zeituni AE, Nair SC, Crampton SP, Orandle MS, Bolland SM, Qi CF, Long CA, Myers TG, Coligan JE, Wang R, Su XZ. 2014. Strain-specific innate immune signaling pathways determine malaria parasitemia dynamics and host mortality. Proc Natl Acad Sci U S A 111:E511–E20. doi: 10.1073/pnas.1316467111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cai B, Wu J, Yu X, Su XZ, Wang RF. 2017. FOSL1 inhibits type I interferon responses to malaria and viral infections by blocking TBK1 and TRAF3/TRIF interactions. mBio 8:e02161-16. doi: 10.1128/mBio.02161-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Neafsey DE, Schaffner SF, Volkman SK, Park D, Montgomery P, Milner DA Jr, Lukens A, Rosen D, Daniels R, Houde N, Cortese JF, Tyndall E, Gates C, Stange-Thomann N, Sarr O, Ndiaye D, Ndir O, Mboup S, Ferreira MU, Moraes SD, Dash AP, Chitnis CE, Wiegand RC, Hartl DL, Birren BW, Lander ES, Sabeti PC, Wirth DF. 2008. Genome-wide SNP genotyping highlights the role of natural selection in Plasmodium falciparum population divergence. Genome Biol 9:R171. doi: 10.1186/gb-2008-9-12-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mu J, Myers RA, Jiang H, Liu S, Ricklefs S, Waisberg M, Chotivanich K, Wilairatana P, Krudsood S, White NJ, Udomsangpetch R, Cui L, Ho M, Ou F, Li H, Song J, Li G, Wang X, Seila S, Sokunthea S, Socheat D, Sturdevant DE, Porcella SF, Fairhurst RM, Wellems TE, Awadalla P, Su XZ. 2010. Plasmodium falciparum genome-wide scans for positive selection, recombination hot spots and resistance to antimalarial drugs. Nat Genet 42:268–271. doi: 10.1038/ng.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Van Tyne D, Park DJ, Schaffner SF, Neafsey DE, Angelino E, Cortese JF, Barnes KG, Rosen DM, Lukens AK, Daniels RF, Milner DA Jr, Johnson CA, Shlyakhter I, Grossman SR, Becker JS, Yamins D, Karlsson EK, Ndiaye D, Sarr O, Mboup S, Happi C, Furlotte NA, Eskin E, Kang HM, Hartl DL, Birren BW, Wiegand RC, Lander ES, Wirth DF, Volkman SK, Sabeti PC. 2011. Identification and functional validation of the novel antimalarial resistance locus PF10_0355 in Plasmodium falciparum. PLoS Genet 7:e1001383. doi: 10.1371/journal.pgen.1001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Van Tyne D, Uboldi AD, Healer J, Cowman AF, Wirth DF. 2013. Modulation of PF10_0355 (MSPDBL2) alters Plasmodium falciparum response to antimalarial drugs. Antimicrob Agents Chemother 57:2937–2941. doi: 10.1128/AAC.02574-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wendler JP, Okombo J, Amato R, Miotto O, Kiara SM, Mwai L, Pole L, O’Brien J, Manske M, Alcock D, Drury E, Sanders M, Oyola SO, Malangone C, Jyothi D, Miles A, Rockett KA, MacInnis BL, Marsh K, Bejon P, Nzila A, Kwiatkowski DP. 2014. A genome wide association study of Plasmodium falciparum susceptibility to 22 antimalarial drugs in Kenya. PLoS One 9:e96486. doi: 10.1371/journal.pone.0096486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Institute of Medicine. 2004. Saving lives, buying time: economics of malaria drugs in an age of resistance. Institute of Medicine, Washington, DC. [PubMed] [Google Scholar]
- 170.Li GQ, Arnold K, Guo XB, Jian HX, Fu LC. 1984. Randomised comparative study of mefloquine, qinghaosu, and pyrimethamine-sulfadoxine in patients with falciparum malaria. Lancet 2:1360–1361. doi: 10.1016/s0140-6736(84)92057-9. [DOI] [PubMed] [Google Scholar]
- 171.Looareesuwan S, Kyle DE, Viravan C, Vanijanonta S, Wilairatana P, Charoenlarp P, Canfield CJ, Webster HK. 1992. Treatment of patients with recrudescent falciparum malaria with a sequential combination of artesunate and mefloquine. Am J Trop Med Hyg 47:794–799. doi: 10.4269/ajtmh.1992.47.794. [DOI] [PubMed] [Google Scholar]
- 172.Nguyen TA. 1993. Malaria in Vietnam. Environment, prevention and treatment. Bull Soc Pathol Exot 86:494–499. [PubMed] [Google Scholar]
- 173.World Health Organization. 1967. Chemotherapy of malaria: report of a WHO scientific group. Technical report series no. 375. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 174.Peterson DS, Milhous WK, Wellems TE. 1990. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc Natl Acad Sci U S A 87:3018–3022. doi: 10.1073/pnas.87.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV, Coulibaly D. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med 344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 176.Fairhurst RM, Dondorp AM. 2016. Artemisinin-resistant Plasmodium falciparum malaria. Microbiol Spectr 4. doi: 10.1128/microbiolspec.EI10-0013-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dondorp AM. 2017. New genetic marker for piperaquine resistance in Plasmodium falciparum. Lancet Infect Dis 17:119–121. doi: 10.1016/S1473-3099(16)30414-5. [DOI] [PubMed] [Google Scholar]
- 178.Ouji M, Augereau JM, Paloque L, Benoit-Vical F. 2018. Plasmodium falciparum resistance to artemisinin-based combination therapies: a sword of Damocles in the path toward malaria elimination. Parasite 25:24. doi: 10.1051/parasite/2018021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM, Artemisinin R, In Cambodia 1, Study C . 2008. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med 359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 180.Bethell D, Se Y, Lon C, Tyner S, Saunders D, Sriwichai S, Darapiseth S, Teja-Isavadharm P, Khemawoot P, Schaecher K, Ruttvisutinunt W, Lin J, Kuntawungin W, Gosi P, Timmermans A, Smith B, Socheat D, Fukuda MM. 2011. Artesunate dose escalation for the treatment of uncomplicated malaria in a region of reported artemisinin resistance: a randomized clinical trial. PLoS One 6:e19283. doi: 10.1371/journal.pone.0019283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Witkowski B, Khim N, Chim P, Kim S, Ke S, Kloeung N, Chy S, Duong S, Leang R, Ringwald P, Dondorp AM, Tripura R, Benoit-Vical F, Berry A, Gorgette O, Ariey F, Barale JC, Mercereau-Puijalon O, Menard D. 2013. Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in western Cambodia. Antimicrob Agents Chemother 57:914–923. doi: 10.1128/AAC.01868-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois A-C, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale J-C, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Ménard D. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Taylor SM, Juliano JJ, Meshnick SR. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361:1807. (Author reply, 361:1808.) [DOI] [PubMed] [Google Scholar]
- 184.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Saunders D, Khemawoot P, Vanachayangkul P, Siripokasupkul R, Bethell D, Tyner S, Se Y, Rutvisuttinunt W, Sriwichai S, Chanthap L, Lin J, Timmermans A, Socheat D, Ringwald P, Noedl H, Smith B, Fukuda M, Teja-Isavadharm P. 2012. Pharmacokinetics and pharmacodynamics of oral artesunate monotherapy in patients with uncomplicated Plasmodium falciparum malaria in western Cambodia. Antimicrob Agents Chemother 56:5484–5493. doi: 10.1128/AAC.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Das D, Tripura R, Phyo AP, Lwin KM, Tarning J, Lee SJ, Hanpithakpong W, Stepniewska K, Menard D, Ringwald P, Silamut K, Imwong M, Chotivanich K, Yi P, Day NP, Lindegardh N, Socheat D, Nguon C, White NJ, Nosten F, Dondorp AM. 2013. Effect of high-dose or split-dose artesunate on parasite clearance in artemisinin-resistant falciparum malaria. Clin Infect Dis 56:e48–e58. doi: 10.1093/cid/cis958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kyaw MP, Nyunt MH, Chit K, Aye MM, Aye KH, Aye MM, Lindegardh N, Tarning J, Imwong M, Jacob CG, Rasmussen C, Perin J, Ringwald P, Nyunt MM. 2013. Reduced susceptibility of Plasmodium falciparum to artesunate in southern Myanmar. PLoS One 8:e57689. doi: 10.1371/journal.pone.0057689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, Lim P, Mao S, Sopha C, Sam B, Anderson JM, Duong S, Chuor CM, Taylor WR, Suon S, Mercereau-Puijalon O, Fairhurst RM, Menard D. 2013. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis 13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cheeseman IH, Miller BA, Nair S, Nkhoma S, Tan A, Tan JC, Al Saai S, Phyo AP, Moo CL, Lwin KM, McGready R, Ashley E, Imwong M, Stepniewska K, Yi P, Dondorp AM, Mayxay M, Newton PN, White NJ, Nosten F, Ferdig MT, Anderson TJ. 2012. A major genome region underlying artemisinin resistance in malaria. Science 336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Takala-Harrison S, Clark TG, Jacob CG, Cummings MP, Miotto O, Dondorp AM, Fukuda MM, Nosten F, Noedl H, Imwong M, Bethell D, Se Y, Lon C, Tyner SD, Saunders DL, Socheat D, Ariey F, Phyo AP, Starzengruber P, Fuehrer HP, Swoboda P, Stepniewska K, Flegg J, Arze C, Cerqueira GC, Silva JC, Ricklefs SM, Porcella SF, Stephens RM, Adams M, Kenefic LJ, Campino S, Auburn S, Macinnis B, Kwiatkowski DP, Su XZ, White NJ, Ringwald P, Plowe CV. 2013. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc Natl Acad Sci U S A 110:240–245. doi: 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Miotto O, Amato R, Ashley EA, MacInnis B, Almagro-Garcia J, Amaratunga C, Lim P, Mead D, Oyola SO, Dhorda M, Imwong M, Woodrow C, Manske M, Stalker J, Drury E, Campino S, Amenga-Etego L, Thanh TN, Tran HT, Ringwald P, Bethell D, Nosten F, Phyo AP, Pukrittayakamee S, Chotivanich K, Chuor CM, Nguon C, Suon S, Sreng S, Newton PN, Mayxay M, Khanthavong M, Hongvanthong B, Htut Y, Han KT, Kyaw MP, Faiz MA, Fanello CI, Onyamboko M, Mokuolu OA, Jacob CG, Takala-Harrison S, Plowe CV, Day NP, Dondorp AM, Spencer CC, McVean G, Fairhurst RM, White NJ, Kwiatkowski DP. 2015. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet 47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tilley L, Straimer J, Gnadig NF, Ralph SA, Fidock DA. 2016. Artemisinin action and resistance in Plasmodium falciparum. Trends Parasitol 32:682–696. doi: 10.1016/j.pt.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Meshnick S. 2012. Perspective: artemisinin-resistant malaria and the wolf. Am J Trop Med Hyg 87:783–784. doi: 10.4269/ajtmh.2012.12-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Krishna S, Kremsner PG. 2013. Antidogmatic approaches to artemisinin resistance: reappraisal as treatment failure with artemisinin combination therapy. Trends Parasitol 29:313–317. doi: 10.1016/j.pt.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 195.Ferreira PE, Culleton R, Gil JP, Meshnick SR. 2013. Artemisinin resistance in Plasmodium falciparum: what is it really? Trends Parasitol 29:318–320. doi: 10.1016/j.pt.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 196.Hastings IM, Kay K, Hodel EM. 2015. How robust are malaria parasite clearance rates as indicators of drug effectiveness and resistance? Antimicrob Agents Chemother 59:6428–6436. doi: 10.1128/AAC.00481-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Phyo AP, Ashley EA, Anderson TJ, Carrara VI, Woodrow CJ, White NJ, Nosten F. 2016. Reply to Meshnick and Hastings et al. Clin Infect Dis 63:1528–1529. doi: 10.1093/cid/ciw584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han K-T, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, Rahman MR, Hasan MM, Islam A, Miotto O, Amato R, MacInnis B, Stalker J, Kwiatkowski DP, Bozdech Z, Jeeyapant A, Cheah PY, Sakulthaew T, Chalk J, Intharabut B, Silamut K, Lee SJ, Vihokhern B, Kunasol C, Imwong M, Tarning J, Taylor WJ, Yeung S, Woodrow CJ, Flegg JA, Das D, Smith J, Venkatesan M, Plowe CV, Stepniewska K, Guerin PJ, Dondorp AM, Day NP, White NJ. 2014. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lozano S, Gamallo P, Gonzalez-Cortes C, Presa Matilla JL, Fairhurst RM, Herreros E, Amaratunga C, Rodrigues J. 2018. Gametocytes from K13 propeller mutant Plasmodium falciparum clinical isolates demonstrate reduced susceptibility to dihydroartemisinin in the male gamete exflagellation inhibition assay. Antimicrob Agents Chemother 62:e01426-18. doi: 10.1128/AAC.01426-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.World Health Organization Global Malaria Programme. 2017. Status report on artemisinin and artemisinin-based combination therapy resistance. WHO, Geneva, Switzerland. [Google Scholar]
- 201.Abdulla S, Ashley EA, Bassat Q, Bethell D, Björkman A, Borrmann S, D’Alessandro U, Dahal P, Day NP, Diakite M, Djimde AA, Dondorp AM, Duong S, Edstein MD, Fairhurst RM, Faiz MA, Falade C, Flegg JA, Fogg C, Gonzalez R, Greenwood B, Guérin PJ, Guthmann J-P, Hamed K, Hien TT, Htut Y, Juma E, Lim P, Mårtensson A, Mayxay M, Mokuolu OA, Moreira C, Newton P, Noedl H, Nosten F, Ogutu BR, Onyamboko MA, Owusu-Agyei S, Phyo AP, Premji Z, Price RN, Pukrittayakamee S, Ramharter M, Sagara I, Se Y, Suon S, Stepniewska K, Ward SA, White NJ, Winstanley PA. 2015. Baseline data of parasite clearance in patients with falciparum malaria treated with an artemisinin derivative: an individual patient data meta-analysis. Malar J 14:359. doi: 10.1186/s12936-015-0874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Amaratunga C, Neal AT, Fairhurst RM. 2014. Flow cytometry-based analysis of artemisinin-resistant Plasmodium falciparum in the ring-stage survival assay. Antimicrob Agents Chemother 58:4938–4940. doi: 10.1128/AAC.02902-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Amaratunga C, Witkowski B, Dek D, Try V, Khim N, Miotto O, Menard D, Fairhurst RM. 2014. Plasmodium falciparum founder populations in western Cambodia have reduced artemisinin sensitivity in vitro. Antimicrob Agents Chemother 58:4935–4937. doi: 10.1128/AAC.03055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Spring MD, Lin JT, Manning JE, Vanachayangkul P, Somethy S, Bun R, Se Y, Chann S, Ittiverakul M, Sia-Ngam P, Kuntawunginn W, Arsanok M, Buathong N, Chaorattanakawee S, Gosi P, Ta-Aksorn W, Chanarat N, Sundrakes S, Kong N, Heng TK, Nou S, Teja-Isavadharm P, Pichyangkul S, Phann ST, Balasubramanian S, Juliano JJ, Meshnick SR, Chour CM, Prom S, Lanteri CA, Lon C, Saunders DL. 2015. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: an observational cohort study. Lancet Infect Dis 15:683–691. doi: 10.1016/S1473-3099(15)70049-6. [DOI] [PubMed] [Google Scholar]
- 205.Leang R, Barrette A, Bouth DM, Menard D, Abdur R, Duong S, Ringwald P. 2013. Efficacy of dihydroartemisinin-piperaquine for treatment of uncomplicated Plasmodium falciparum and Plasmodium vivax in Cambodia, 2008 to 2010. Antimicrob Agents Chemother 57:818–826. doi: 10.1128/AAC.00686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ross LS, Dhingra SK, Mok S, Yeo T, Wicht KJ, Kumpornsin K, Takala-Harrison S, Witkowski B, Fairhurst RM, Ariey F, Menard D, Fidock DA. 2018. Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat Commun 9:3314. doi: 10.1038/s41467-018-05652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.U.S. AID. 2018. President’s malaria initiative, Cambodia. Malaria operational plan FY 2018. U.S. AID, Washington, DC. [Google Scholar]
- 208.Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, Almagro-Garcia J, Neal AT, Sreng S, Suon S, Drury E, Jyothi D, Stalker J, Kwiatkowski DP, Fairhurst RM. 2017. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis 17:164–173. doi: 10.1016/S1473-3099(16)30409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, Sam B, Dek D, Try V, Amato R, Blessborn D, Song L, Tullo GS, Fay MP, Anderson JM, Tarning J, Fairhurst RM. 2016. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis 16:357–365. doi: 10.1016/S1473-3099(15)00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Witkowski B, Duru V, Khim N, Ross LS, Saintpierre B, Beghain J, Chy S, Kim S, Ke S, Kloeung N, Eam R, Khean C, Ken M, Loch K, Bouillon A, Domergue A, Ma L, Bouchier C, Leang R, Huy R, Nuel G, Barale J-C, Legrand E, Ringwald P, Fidock DA, Mercereau-Puijalon O, Ariey F, Ménard D. 2017. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype-genotype association study. Lancet Infect Dis 17:174–183. doi: 10.1016/S1473-3099(16)30415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lim P, Dek D, Try V, Sreng S, Suon S, Fairhurst RM. 2015. Decreasing pfmdr1 copy number suggests that Plasmodium falciparum in Western Cambodia is regaining in vitro susceptibility to mefloquine. Antimicrob Agents Chemother 59:2934–2937. doi: 10.1128/AAC.05163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Taylor AR, Flegg JA, Holmes CC, Guerin PJ, Sibley CH, Conrad MD, Dorsey G, Rosenthal PJ. 2017. Artemether-lumefantrine and dihydroartemisinin-piperaquine exert inverse selective pressure on Plasmodium falciparum drug sensitivity-associated haplotypes in Uganda. Open Forum Infect Dis 4:ofw229. doi: 10.1093/ofid/ofw229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ekland EH, Fidock DA. 2007. Advances in understanding the genetic basis of antimalarial drug resistance. Curr Opin Microbiol 10:363–370. doi: 10.1016/j.mib.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Flannery EL, Fidock DA, Winzeler EA. 2013. Using genetic methods to define the targets of compounds with antimalarial activity. J Med Chem 56:7761–7771. doi: 10.1021/jm400325j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tse EG, Korsik M, Todd MH. 2019. The past, present and future of anti-malarial medicines. Malar J 18:93. doi: 10.1186/s12936-019-2724-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Martin RE, Shafik SH, Richards SN. 2018. Mechanisms of resistance to the partner drugs of artemisinin in the malaria parasite. Curr Opin Pharmacol 42:71–80. doi: 10.1016/j.coph.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 217.Kumar S, Bhardwaj TR, Prasad DN, Singh RK. 2018. Drug targets for resistant malaria: historic to future perspectives. Biomed Pharmacother 104:8–27. doi: 10.1016/j.biopha.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 218.Baird JK. 2009. Resistance to therapies for infection by Plasmodium vivax. Clin Microbiol Rev 22:508–534. doi: 10.1128/CMR.00008-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T. 2004. Intercontinental spread of pyrimethamine-resistant malaria. Science 305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- 220.Nair S, Williams JT, Brockman A, Paiphun L, Mayxay M, Newton PN, Guthmann JP, Smithuis FM, Hien TT, White NJ, Nosten F, Anderson TJ. 2003. A selective sweep driven by pyrimethamine treatment in southeast Asian malaria parasites. Mol Biol Evol 20:1526–1536. doi: 10.1093/molbev/msg162. [DOI] [PubMed] [Google Scholar]
- 221.Imwong M, Nair S, Pukrittayakamee S, Sudimack D, Williams JT, Mayxay M, Newton PN, Kim JR, Nandy A, Osorio L, Carlton JM, White NJ, Day NP, Anderson TJ. 2007. Contrasting genetic structure in Plasmodium vivax populations from Asia and South America. Int J Parasitol 37:1013–1022. doi: 10.1016/j.ijpara.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 222.McCollum AM, Basco LK, Tahar R, Udhayakumar V, Escalante AA. 2008. Hitchhiking and selective sweeps of Plasmodium falciparum sulfadoxine and pyrimethamine resistance alleles in a population from central Africa. Antimicrob Agents Chemother 52:4089–4097. doi: 10.1128/AAC.00623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Vinayak S, Alam MT, Mixson-Hayden T, McCollum AM, Sem R, Shah NK, Lim P, Muth S, Rogers WO, Fandeur T, Barnwell JW, Escalante AA, Wongsrichanalai C, Ariey F, Meshnick SR, Udhayakumar V. 2010. Origin and evolution of sulfadoxine resistant Plasmodium falciparum. PLoS Pathog 6:e1000830. doi: 10.1371/journal.ppat.1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Mobegi VA, Duffy CW, Amambua-Ngwa A, Loua KM, Laman E, Nwakanma DC, MacInnis B, Aspeling-Jones H, Murray L, Clark TG, Kwiatkowski DP, Conway DJ. 2014. Genome-wide analysis of selection on the malaria parasite Plasmodium falciparum in West African populations of differing infection endemicity. Mol Biol Evol 31:1490–1499. doi: 10.1093/molbev/msu106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109–126. doi: 10.1016/S0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 226.Rix U, Superti-Furga G. 2009. Target profiling of small molecules by chemical proteomics. Nat Chem Biol 5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 227.Urbaniak MD, Mathieson T, Bantscheff M, Eberhard D, Grimaldi R, Miranda-Saavedra D, Wyatt P, Ferguson MA, Frearson J, Drewes G. 2012. Chemical proteomic analysis reveals the drugability of the kinome of Trypanosoma brucei. ACS Chem Biol 7:1858–1865. doi: 10.1021/cb300326z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Huebert DJ, Kamal M, O’Donovan A, Bernstein BE. 2006. Genome-wide analysis of histone modifications by ChIP-on-chip. Methods 40:365–369. doi: 10.1016/j.ymeth.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 229.Yuan J, Johnson RL, Huang R, Wichterman J, Jiang H, Hayton K, Fidock DA, Wellems TE, Inglese J, Austin CP, Su XZ. 2009. Genetic mapping of targets mediating differential chemical phenotypes in Plasmodium falciparum. Nat Chem Biol 5:765–771. doi: 10.1038/nchembio.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Yuan J, Cheng KC, Johnson RL, Huang R, Pattaradilokrat S, Liu A, Guha R, Fidock DA, Inglese J, Wellems TE, Austin CP, Su XZ. 2011. Chemical genomic profiling for antimalarial therapies, response signatures, and molecular targets. Science 333:724–729. doi: 10.1126/science.1205216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Li J, Yuan J, Cheng KC, Inglese J, Su XZ. 2013. Chemical genomics for studying parasite gene function and interaction. Trends Parasitol 29:603–611. doi: 10.1016/j.pt.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sana TR, Gordon DB, Fischer SM, Tichy SE, Kitagawa N, Lai C, Gosnell WL, Chang SP. 2013. Global mass spectrometry based metabolomics profiling of erythrocytes infected with Plasmodium falciparum. PLoS One 8:e60840. doi: 10.1371/journal.pone.0060840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Olszewski KL, Morrisey JM, Wilinski D, Burns JM, Vaidya AB, Rabinowitz JD, Llinas M. 2009. Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell Host Microbe 5:191–199. doi: 10.1016/j.chom.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Rottmann M, McNamara C, Yeung BK, Lee MC, Zou B, Russell B, Seitz P, Plouffe DM, Dharia NV, Tan J, Cohen SB, Spencer KR, Gonzalez-Paez GE, Lakshminarayana SB, Goh A, Suwanarusk R, Jegla T, Schmitt EK, Beck HP, Brun R, Nosten F, Renia L, Dartois V, Keller TH, Fidock DA, Winzeler EA, Diagana TT. 2010. Spiroindolones, a potent compound class for the treatment of malaria. Science 329:1175–1180. doi: 10.1126/science.1193225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Dharia NV, Sidhu AB, Cassera MB, Westenberger SJ, Bopp SE, Eastman RT, Plouffe D, Batalov S, Park DJ, Volkman SK, Wirth DF, Zhou Y, Fidock DA, Winzeler EA. 2009. Use of high-density tiling microarrays to identify mutations globally and elucidate mechanisms of drug resistance in Plasmodium falciparum. Genome Biol 10:R21. doi: 10.1186/gb-2009-10-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Hoepfner D, McNamara CW, Lim CS, Studer C, Riedl R, Aust T, McCormack SL, Plouffe DM, Meister S, Schuierer S, Plikat U, Hartmann N, Staedtler F, Cotesta S, Schmitt EK, Petersen F, Supek F, Glynne RJ, Tallarico JA, Porter JA, Fishman MC, Bodenreider C, Diagana TT, Movva NR, Winzeler EA. 2012. Selective and specific inhibition of the Plasmodium falciparum lysyl-tRNA synthetase by the fungal secondary metabolite cladosporin. Cell Host Microbe 11:654–663. doi: 10.1016/j.chom.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lane KD, Mu J, Lu J, Windle ST, Liu A, Sun PD, Wellems TE. 2018. Selection of Plasmodium falciparum cytochrome B mutants by putative PfNDH2 inhibitors. Proc Natl Acad Sci U S A 115:6285–6290. doi: 10.1073/pnas.1804492115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Cui L, Wang Z, Miao J, Miao M, Chandra R, Jiang H, Su XZ, Cui L. 2012. Mechanisms of in vitro resistance to dihydroartemisinin in Plasmodium falciparum. Mol Microbiol 86:111–128. doi: 10.1111/j.1365-2958.2012.08180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Demas AR, Sharma AI, Wong W, Early AM, Redmond S, Bopp S, Neafsey DE, Volkman SK, Hartl DL, Wirth DF. 2018. Mutations in Plasmodium falciparum actin-binding protein coronin confer reduced artemisinin susceptibility. Proc Natl Acad Sci U S A 155:12799–12804. doi: 10.1073/pnas.1812317115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Cowell AN, Istvan ES, Lukens AK, Gomez-Lorenzo MG, Vanaerschot M, Sakata-Kato T, Flannery EL, Magistrado P, Owen E, Abraham M, LaMonte G, Painter HJ, Williams RM, Franco V, Linares M, Arriaga I, Bopp S, Corey VC, Gnadig NF, Coburn-Flynn O, Reimer C, Gupta P, Murithi JM, Moura PA, Fuchs O, Sasaki E, Kim SW, Teng CH, Wang LT, Akidil A, Adjalley S, Willis PA, Siegel D, Tanaseichuk O, Zhong Y, Zhou Y, Llinas M, Ottilie S, Gamo FJ, Lee MCS, Goldberg DE, Fidock DA, Wirth DF, Winzeler EA. 2018. Mapping the malaria parasite druggable genome by using in vitro evolution and chemogenomics. Science 359:191–199. doi: 10.1126/science.aan4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ke H, Ganesan SM, Dass S, Morrisey JM, Pou S, Nilsen A, Riscoe MK, Mather MW, Vaidya AB. 2019. Mitochondrial type II NADH dehydrogenase of Plasmodium falciparum (PfNDH2) is dispensable in the asexual blood stages. PLoS One 14:e0214023. doi: 10.1371/journal.pone.0214023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Tanabe K, Mita T, Palacpac NM, Arisue N, Tougan T, Kawai S, Jombart T, Kobayashi F, Horii T. 2013. Within-population genetic diversity of Plasmodium falciparum vaccine candidate antigens reveals geographic distance from a Central sub-Saharan African origin. Vaccine 31:1334–1339. doi: 10.1016/j.vaccine.2012.12.039. [DOI] [PubMed] [Google Scholar]
- 243.Chan JA, Fowkes FJ, Beeson JG. 2014. Surface antigens of Plasmodium falciparum-infected erythrocytes as immune targets and malaria vaccine candidates. Cell Mol Life Sci 71:3633–3657. doi: 10.1007/s00018-014-1614-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Gandon S, Mackinnon MJ, Nee S, Read AF. 2001. Imperfect vaccines and the evolution of pathogen virulence. Nature 414:751–756. doi: 10.1038/414751a. [DOI] [PubMed] [Google Scholar]
- 245.Weedall GD, Conway DJ. 2010. Detecting signatures of balancing selection to identify targets of anti-parasite immunity. Trends Parasitol 26:363–369. doi: 10.1016/j.pt.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 246.Amambua-Ngwa A, Tetteh KK, Manske M, Gomez-Escobar N, Stewart LB, Deerhake ME, Cheeseman IH, Newbold CI, Holder AA, Knuepfer E, Janha O, Jallow M, Campino S, Macinnis B, Kwiatkowski DP, Conway DJ. 2012. Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites. PLoS Genet 8:e1002992. doi: 10.1371/journal.pgen.1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Conway DJ, Cavanagh DR, Tanabe K, Roper C, Mikes ZS, Sakihama N, Bojang KA, Oduola AM, Kremsner PG, Arnot DE, Greenwood BM, McBride JS. 2000. A principal target of human immunity to malaria identified by molecular population genetic and immunological analyses. Nat Med 6:689–692. doi: 10.1038/76272. [DOI] [PubMed] [Google Scholar]
- 248.Ho SY, Lanfear R, Bromham L, Phillips MJ, Soubrier J, Rodrigo AG, Cooper A. 2011. Time-dependent rates of molecular evolution. Mol Ecol 20:3087–3101. doi: 10.1111/j.1365-294X.2011.05178.x. [DOI] [PubMed] [Google Scholar]
- 249.Su X-Z, Mu J, Joy DA. 2003. The “malaria’s eve” hypothesis and the debate concerning the origin of the human malaria parasite Plasmodium falciparum. Microbes Infect 5:891–896. doi: 10.1016/S1286-4579(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 250.Rich SM, Hudson RR, Ayala FJ. 1997. Plasmodium falciparum antigenic diversity: evidence of clonal population structure. Proc Natl Acad Sci U S A 94:13040–13045. doi: 10.1073/pnas.94.24.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Rich SM, Licht MC, Hudson RR, Ayala FJ. 1998. Malaria’s eve: evidence of a recent population bottleneck throughout the world populations of Plasmodium falciparum. Proc Natl Acad Sci U S A 95:4425–4430. doi: 10.1073/pnas.95.8.4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Conway DJ, Fanello C, Lloyd JM, Al-Joubori BM, Baloch AH, Somanath SD, Roper C, Oduola AM, Mulder B, Povoa MM, Singh B, Thomas AW. 2000. Origin of Plasmodium falciparum malaria is traced by mitochondrial DNA. Mol Biochem Parasitol 111:163–171. doi: 10.1016/S0166-6851(00)00313-3. [DOI] [PubMed] [Google Scholar]
- 253.Volkman SK, Barry AE, Lyons EJ, Nielsen KM, Thomas SM, Choi M, Thakore SS, Day KP, Wirth DF, Hartl DL. 2001. Recent origin of Plasmodium falciparum from a single progenitor. Science 293:482–484. doi: 10.1126/science.1059878. [DOI] [PubMed] [Google Scholar]
- 254.Hughes AL, Verra F. 1998. Ancient polymorphism and the hypothesis of a recent bottleneck in the malaria parasite Plasmodium falciparum. Genetics 150:511–513. (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Hughes AL, Vierra F. 2001. Very large long-term effective population size in the virulent human malaria parasite Plasmodium falciparum. Proc R Soc Lond B Biol Sci 268:1855–1860. doi: 10.1098/rspb.2001.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Liu W, Li Y, Learn GH, Rudicell RS, Robertson JD, Keele BF, Ndjango JB, Sanz CM, Morgan DB, Locatelli S, Gonder MK, Kranzusch PJ, Walsh PD, Delaporte E, Mpoudi-Ngole E, Georgiev AV, Muller MN, Shaw GM, Peeters M, Sharp PM, Rayner JC, Hahn BH. 2010. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature 467:420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Liu W, Sundararaman SA, Loy DE, Learn GH, Li Y, Plenderleith LJ, Ndjango JB, Speede S, Atencia R, Cox D, Shaw GM, Ayouba A, Peeters M, Rayner JC, Hahn BH, Sharp PM. 2016. Multigenomic delineation of Plasmodium species of the Laverania subgenus infecting wild-living chimpanzees and gorillas. Genome Biol Evol 8:1929–1939. doi: 10.1093/gbe/evw128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Duffy CW, Ba H, Assefa S, Ahouidi AD, Deh YB, Tandia A, Kirsebom FCM, Kwiatkowski DP, Conway DJ. 2017. Population genetic structure and adaptation of malaria parasites on the edge of endemic distribution. Mol Ecol 26:2880–2894. doi: 10.1111/mec.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Duan J, Mu J, Thera MA, Joy D, Kosakovsky Pond SL, Diemert D, Long C, Zhou H, Miura K, Ouattara A, Dolo A, Doumbo O, Su XZ, Miller L. 2008. Population structure of the genes encoding the polymorphic Plasmodium falciparum apical membrane antigen 1: implications for vaccine design. Proc Natl Acad Sci U S A 105:7857–7862. doi: 10.1073/pnas.0802328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Polley SD, Conway DJ. 2001. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics 158:1505–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Amato R, Pearson RD, Almagro-Garcia J, Amaratunga C, Lim P, Suon S, Sreng S, Drury E, Stalker J, Miotto O, Fairhurst RM, Kwiatkowski DP. 2018. Origins of the current outbreak of multidrug-resistant malaria in southeast Asia: a retrospective genetic study. Lancet Infect Dis 18:337–345. doi: 10.1016/S1473-3099(18)30068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Pearson RD, Amato R, Auburn S, Miotto O, Almagro-Garcia J, Amaratunga C, Suon S, Mao S, Noviyanti R, Trimarsanto H, Marfurt J, Anstey NM, William T, Boni MF, Dolecek C, Hien TT, White NJ, Michon P, Siba P, Tavul L, Harrison G, Barry A, Mueller I, Ferreira MU, Karunaweera N, Randrianarivelojosia M, Gao Q, Hubbart C, Hart L, Jeffery B, Drury E, Mead D, Kekre M, Campino S, Manske M, Cornelius VJ, MacInnis B, Rockett KA, Miles A, Rayner JC, Fairhurst RM, Nosten F, Price RN, Kwiatkowski DP. 2016. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat Genet 48:959–964. doi: 10.1038/ng.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.WHO. 2018. World malaria report. https://apps.who.int/iris/bitstream/handle/10665/275867/9789241565653-eng.pdf?ua=1.
- 264.Gilabert A, Otto TD, Rutledge GG, Franzon B, Ollomo B, Arnathau C, Durand P, Moukodoum ND, Okouga AP, Ngoubangoye B, Makanga B, Boundenga L, Paupy C, Renaud F, Prugnolle F, Rougeron V. 2018. Plasmodium vivax-like genome sequences shed new insights into Plasmodium vivax biology and evolution. PLoS Biol 16:e2006035. doi: 10.1371/journal.pbio.2006035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Dharia NV, Bright AT, Westenberger SJ, Barnes SW, Batalov S, Kuhen K, Borboa R, Federe GC, McClean CM, Vinetz JM, Neyra V, Llanos-Cuentas A, Barnwell JW, Walker JR, Winzeler EA. 2010. Whole-genome sequencing and microarray analysis of ex vivo Plasmodium vivax reveal selective pressure on putative drug resistance genes. Proc Natl Acad Sci U S A 107:20045–20050. doi: 10.1073/pnas.1003776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Daniels RF, Rice BL, Daniels NM, Volkman SK, Hartl DL. 2015. The utility of genomic data for Plasmodium vivax population surveillance. Pathog Glob Health 109:153–161. doi: 10.1179/2047773215Y.0000000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Neafsey DE, Galinsky K, Jiang RH, Young L, Sykes SM, Saif S, Gujja S, Goldberg JM, Young S, Zeng Q, Chapman SB, Dash AP, Anvikar AR, Sutton PL, Birren BW, Escalante AA, Barnwell JW, Carlton JM. 2012. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat Genet 44:1046–1050. doi: 10.1038/ng.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Mu J, Joy DA, Duan J, Huang Y, Carlton J, Walker J, Barnwell J, Beerli P, Charleston MA, Pybus OG, Su XZ. 2005. Host switch leads to emergence of Plasmodium vivax malaria in humans. Mol Biol Evol 22:1686–1693. doi: 10.1093/molbev/msi160. [DOI] [PubMed] [Google Scholar]
- 269.Escalante AA, Cornejo OE, Freeland DE, Poe AC, Durrego E, Collins WE, Lal AA. 2005. A monkey’s tale: the origin of Plasmodium vivax as a human malaria parasite. Proc Natl Acad Sci U S A 102:1980–1985. doi: 10.1073/pnas.0409652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, Thomas A, Conway DJ. 2004. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet 363:1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 271.Imwong M, Madmanee W, Suwannasin K, Kunasol C, Peto TJ, Tripura R, von Seidlein L, Nguon C, Davoeung C, Day NPJ, Dondorp AM, White NJ. 2019. Asymptomatic natural human infections with the simian malaria parasites Plasmodium cynomolgi and Plasmodium knowlesi. J Infect Dis 219:695–702. doi: 10.1093/infdis/jiy519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Ayala FJ, Escalante AA, Rich SM. 1999. Evolution of Plasmodium and the recent origin of the world populations of Plasmodium falciparum. Parassitologia 41:55–68. [PubMed] [Google Scholar]
- 273.de Alvarenga DAM, de Pina-Costa A, de Sousa TN, Pissinatti A, Zalis MG, Suaréz-Mutis MC, Lourenço-de-Oliveira R, Brasil P, Daniel-Ribeiro CT, de Brito CFA. 2015. Simian malaria in the Brazilian Atlantic forest: first description of natural infection of capuchin monkeys (Cebinae subfamily) by Plasmodium simium. Malar J 14:81. doi: 10.1186/s12936-015-0606-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Brasil P, Zalis MG, de Pina-Costa A, Siqueira AM, Júnior CB, Silva S, Areas ALL, Pelajo-Machado M, de Alvarenga DAM, da Silva Santelli ACF, Albuquerque HG, Cravo P, Santos de Abreu FV, Peterka CL, Zanini GM, Suárez Mutis MC, Pissinatti A, Lourenço-de-Oliveira R, de Brito CFA, de Fátima Ferreira-da-Cruz M, Culleton R, Daniel-Ribeiro CT. 2017. Outbreak of human malaria caused by Plasmodium simium in the Atlantic Forest in Rio de Janeiro: a molecular epidemiological investigation. Lancet Glob Health 5:e1038–e1046. doi: 10.1016/S2214-109X(17)30333-9. [DOI] [PubMed] [Google Scholar]
- 275.Carter R. 2003. Speculations on the origins of Plasmodium vivax malaria. Trends Parasitol 19:214–219. doi: 10.1016/S1471-4922(03)00070-9. [DOI] [PubMed] [Google Scholar]
- 276.Zimmerman PA. 2017. Plasmodium vivax infection in Duffy-negative people in Africa. Am J Trop Med Hyg 97:636–638. doi: 10.4269/ajtmh.17-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Gunalan K, Niangaly A, Thera MA, Doumbo OK, Miller LH. 2018. Plasmodium vivax infections of Duffy-negative erythrocytes: historically undetected or a recent adaptation? Trends Parasitol 34:420–429. doi: 10.1016/j.pt.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Liu W, Li Y, Shaw KS, Learn GH, Plenderleith LJ, Malenke JA, Sundararaman SA, Ramirez MA, Crystal PA, Smith AG, Bibollet-Ruche F, Ayouba A, Locatelli S, Esteban A, Mouacha F, Guichet E, Butel C, Ahuka-Mundeke S, Inogwabini B-I, Ndjango J-BN, Speede S, Sanz CM, Morgan DB, Gonder MK, Kranzusch PJ, Walsh PD, Georgiev AV, Muller MN, Piel AK, Stewart FA, Wilson ML, Pusey AE, Cui L, Wang Z, Färnert A, Sutherland CJ, Nolder D, Hart JA, Hart TB, Bertolani P, Gillis A, LeBreton M, Tafon B, Kiyang J, Djoko CF, Schneider BS, Wolfe ND, Mpoudi-Ngole E, Delaporte E, Carter R, Culleton RL, Shaw GM, Rayner JC, Peeters M, Hahn BH, Sharp PM. 2014. African origin of the malaria parasite Plasmodium vivax. Nat Commun 5:3346. doi: 10.1038/ncomms4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Arisue N, Hashimoto T, Kawai S, Honma H, Kume K, Horii T. 2019. Apicoplast phylogeny reveals the position of Plasmodium vivax basal to the Asian primate malaria parasite clade. Sci Rep 9:7274. doi: 10.1038/s41598-019-43831-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Craig AA, Kain KC. 1996. Molecular analysis of strains of Plasmodium vivax from paired primary and relapse infections. J Infect Dis 174:373–379. doi: 10.1093/infdis/174.2.373. [DOI] [PubMed] [Google Scholar]
- 281.Bright AT, Manary MJ, Tewhey R, Arango EM, Wang T, Schork NJ, Yanow SK, Winzeler EA. 2014. A high resolution case study of a patient with recurrent Plasmodium vivax infections shows that relapses were caused by meiotic siblings. PLoS Negl Trop Dis 8:e2882. doi: 10.1371/journal.pntd.0002882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Popovici J, Friedrich LR, Kim S, Bin S, Run V, Lek D, Cannon MV, Menard D, Serre D. 2018. Genomic analyses reveal the common occurrence and complexity of Plasmodium vivax relapses in Cambodia. mBio 9:e01888-17. doi: 10.1128/mBio.01888-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Chen N, Auliff A, Rieckmann K, Gatton M, Cheng Q. 2007. Relapses of Plasmodium vivax infection result from clonal hypnozoites activated at predetermined intervals. J Infect Dis 195:934–941. doi: 10.1086/512242. [DOI] [PubMed] [Google Scholar]
- 284.Ablack JN, Metz PJ, Chang JT, Cantor JM, Ginsberg MH. 2015. Ubiquitylation of CD98 limits cell proliferation and clonal expansion. J Cell Sci 128:4273–4278. doi: 10.1242/jcs.178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Pinney JW, Papp B, Hyland C, Wambua L, Westhead DR, McConkey GA. 2007. Metabolic reconstruction and analysis for parasite genomes. Trends Parasitol 23:548–554. doi: 10.1016/j.pt.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 286.Stanton A, Harris LM, Graham G, Merrick CJ. 2016. Recombination events among virulence genes in malaria parasites are associated with G-quadruplex-forming DNA motifs. BMC Genomics 17:859. doi: 10.1186/s12864-016-3183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Liew KJ, Hu G, Bozdech Z, Peter PR. 2010. Defining species specific genome differences in malaria parasites. BMC Genomics 11:128. doi: 10.1186/1471-2164-11-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Shutler D, Reece SE, Mullie A, Billingsley PF, Read AF. 2005. Rodent malaria parasites Plasmodium chabaudi and P. vinckei do not increase their rates of gametocytogenesis in response to mosquito probing. Proc Biol Sci 272:2397–2402. doi: 10.1098/rspb.2005.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Lauron EJ, Aw Yeang HX, Taffner SM, Sehgal RN. 2015. De novo assembly and transcriptome analysis of Plasmodium gallinaceum identifies the Rh5 interacting protein (ripr), and reveals a lack of EBL and RH gene family diversification. Malar J 14:296. doi: 10.1186/s12936-015-0814-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Omori S, Sato Y, Isobe T, Yukawa M, Murata K. 2007. Complete nucleotide sequences of the mitochondrial genomes of two avian malaria protozoa, Plasmodium gallinaceum and Plasmodium juxtanucleare. Parasitol Res 100:661–664. doi: 10.1007/s00436-006-0333-6. [DOI] [PubMed] [Google Scholar]