Abstract
The aminotransferase gene family in the model plant Arabidopsis thaliana consists of 44 genes, eight of which are suggested to be alanine aminotransferases. One of the putative alanine aminotransferases genes, At3g08860, was attributed the function of alanine:glyoxylate aminotransferase/β‐alanine:pyruvate aminotransferase based on the analysis of gene expression networks and homology to other β‐alanine aminotransferases in plants. It was earlier demonstrated that At3g08860 is specifically upregulated in response to osmotic stress, but not other stresses (β‐alanine is an osmoprotectant in plants). Furthermore, it was shown that the expression of At3g08860 is highly coordinated with the genes of the uracil degradation pathway leading to the non‐proteinogenic amino acid β‐alanine. These evidence were suggestive of the involvement of At3g08860 in β‐alanine metabolism. However, direct experimental evidence for the function of At3g08860 was lacking, and therefore, the goal of this study was to elucidate the function of the uncharacterized aminotransferase annotated by the locus tag At3g08860. The cDNA of At3g08860 was demonstrated to functionally complement two E. coli mutants auxotrophic for the amino acids, L‐alanine (proteinogenic) and β‐alanine (non‐proteinogenic). Enzyme activity using purified recombinant At3g08860 further demonstrated that the enzyme is endowed with L‐alanine:glyoxylate aminotransferase activity.
Keywords: aminotransferase, Arabidopsis thaliana, At3g08860, L‐alanine aminotransferase, transaminase, β‐alanine aminotransferase
1. INTRODUCTION
Aminotransferases or transaminases [EC:2.6.1.X] are pyridoxal‐5′‐phosphate (PLP) dependent enzymes that catalyze reversible reactions between amino acids and α‐keto (2‐oxo) acids. Aminotransferase enzymes function via a bimolecular double displacement ping‐pong mechanism where an amino acid usually serves as the amino donor and an α‐keto acid serves as the amino acceptor (Nelson & Cox, 2000). Aminotransferases are ubiquitous in the three domains of life and are involved in a variety of metabolic pathways including amino acid metabolism, nitrogen assimilation, gluconeogenesis, responses to a number of biotic/abiotic stresses, and among other pathways (Liepman & Olsen, 2004; McAllister, Facette, Holt, & Good, 2013; Rocha et al., 2010; de Sousa & Sodek, 2003).
Alanine aminotransferase usually refers to enzymes which transfer an amino group from L‐alanine to 2‐oxoglutarate, producing pyruvate and L‐glutamate [EC:2.6.1.2] (Figure 1a). However, the term alanine aminotransferase is also used in the literature to describe enzymes which transfer an amino group from L‐alanine to other substrates, for example, L‐alanine‐pyruvate aminotransferase or L‐alanine:glyoxylate aminotransferase. β‐Alanine:pyruvate transaminase or β‐alanine aminotransferase [EC:2.6.1.18] refers to enzymes involved in the degradation of β‐alanine. This enzyme transfers the amino group from β‐alanine to pyruvate to yield L‐alanine and malonate semialdehyde.
Figure 1.
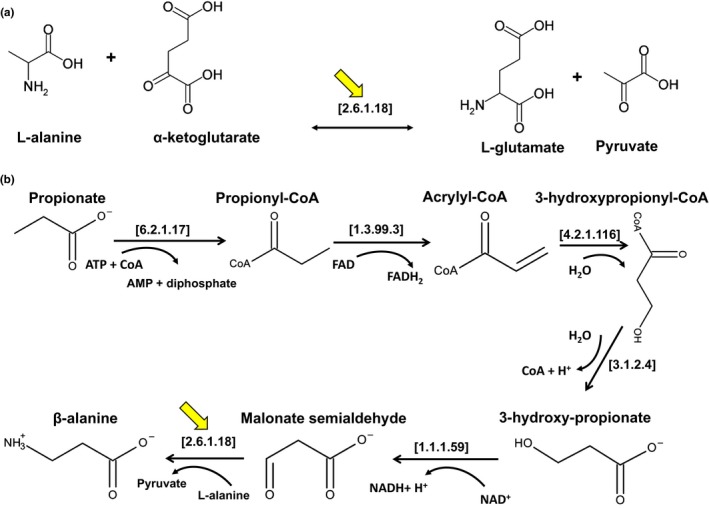
Alanine‐ and β‐alanine aminotransferase. (a) Catalysis of alanine aminotransferase (indicated by the yellow arrow). The enzyme utilizes alanine as the amino donor and α‐ketoglutarate as the amino acceptor to synthesize L‐glutamate and pyruvate in one direction. The enzyme utilizes glutamate and pyruvate as the cognate pair in the reverse direction. (b) β‐alanine synthesis from the propionate pathway. β‐Alanine aminotransferase (indicated by the yellow arrow) catalyzes the synthesis of β‐alanine and pyruvate using L‐alanine as the amino donor and malonate semialdehyde as the amino acceptor. The EC numbers of the enzymes shown in brackets correspond to the following enzymes: [6.2.1.17] = propionyl‐CoA synthetase, [1.3.99.3] = acyl‐CoA dehydrogenase, [4.2.1.116] = 3‐hydroxypropionyl‐CoA dehydratase, [3.2.1.4] = 3‐hydroxyisobutyryl‐CoA hydrolase, [1.1.1.59] = 3‐hydroxypropionate dehydrogenase, [2.6.1.18] = β‐alanine:pyruvate aminotransferase, and [2.6.1.2] = alanine aminotransferase. The yellow arrows indicate the reactions catalyzed by β‐alanine and L‐alanine aminotransferase
The genome of the model plant Arabidopsis thaliana contains 44 annotated genes as part of the aminotransferase gene family (Liepman & Olsen, 2004). In this family, 8 of the 44 genes are annotated as putative alanine aminotransferases with the following loci tags: At2g13360, At4g39660, At2g38400, At1g23310, At1g70580, At1g17290, At1g72330, and At3g08860 (Liepman & Olsen, 2004; Niessen et al., 2012). The literature mostly supports the idea that L‐alanine accumulates during hypoxia with an increase in alanine aminotransferase activity as plants return to normoxia (de Sousa & Sodek, 2003). This is perhaps a mechanism for maintenance via an increase of the nitrogen pool/skeletons since the assimilation of inorganic nitrogen affects anaerobic tolerance (Miyashita, Dolferus, Ismond, & Good, 2007). During hypoxia/anoxia in plant tissues, fermentative products including acetaldehyde, ethanol, and lactate can accumulate, whereby the regeneration of NAD+ by lactate dehydrogenase and alcohol dehydrogenase enhances seedling survival (Ismond, Dolferus, Pauw, Dennis, & Good, 2003). Pyruvate is an immediate precursor of lactate and in A. thaliana it was reported that pyruvate decarboxylase was specifically induced during oxygen limitation, but not other stresses (Kürsteiner, Dupuis, & Kuhlemeier, 2003).
An alternative way to counter hypoxia would be through alanine aminotransferase, which could reduce the flux of carbon through lactate by consuming pyruvate (Ricoult, Echeverria, Cliquet, & Limami, 2006). Pyruvate is not only a known activator of the alternative oxidase (Vanlerberghe, Yip, & Parsons, 1999), but has also recently been shown to interfere with the hypoxia‐induced inhibition of respiration (Gupta, Zabalza, & Dongen, 2009; Zabalza et al., 2009). The phenotypes of various alanine aminotransferases over‐expressed in the A. thaliana and in the alanine aminotransferase At1g17290 and At1g72330 knockouts suggests that nitrogen use efficiency (NUE) could be improved in plants by the overexpression of alanine aminotransferases (McAllister & Good, 2015). Gene regulation studies of alanine aminotransferases in response to low‐oxygen stress, light and nitrogen have been studied in many plants and it was shown that hypoxia induces the expression of two distinct alanine aminotransferase genes, namely At1g17290 and At1g72330 in A. thaliana (Miyashita et al., 2007; Niessen et al., 2007).
Moreover, in plants the metabolism of L‐alanine and the non‐proteinogenic amino acid β‐alanine are linked. β‐Alanine biosynthesis can be initiated from four different precursors: (a) the polyamines spermine and spermidine (Galston & Sawhney, 1990; Rastogi & Davies, 1989; Terano & Suzuki, 1978), (b) the nucleotide base uracil (Barnes & Naylor, 1962; Campbell, 1960; Traut & Loechel, 1984), (c) propionate (Hayaishi, Nishizuka, Tatibana, Takeshita, & Kuno, 1961; Stinson & Spencer, 1969a, 1969b), and (d) L‐aspartate (Ottenhof et al., 2004; Shi, Blundell, & Mizuguchi, 2001). L‐alanine enters the propionate pathway of β‐alanine synthesis via the reverse reaction of the β‐alanine‐pyruvate aminotransferase [EC:2.6.1.18], exchanging an amino group with malonate semialdehyde, and generating pyruvate and β‐alanine (Figure 1b).
Aminotransferases use a range of amino donors and acceptors, suggesting possible overlapping roles in metabolism (Liepman & Olsen, 2003). Zrenner et al. (2009) postulated that the locus tag At3g08860 could be a β‐alanine:pyruvate aminotransferase, as its expression is highly coordinated with the genes of the uracil degradation pathway leading to β‐alanine (Zrenner et al., 2009). Other work based on gene expression networks in A. thaliana suggested that alanine:glyoxylate aminotransferase/β‐alanine:pyruvate aminotransferase could be putative functions for the locus tag At3g08860 (Obayashi, Hayashi, Saeki, Ohta, & Kinoshita, 2009; Obayashi & Kinoshita, 2010). The locus tag At4g39660 was linked to β‐alanine metabolism since knockout lines in A. thaliana accumulate more β‐alanine compared with wild‐type (Wu et al., 2016). At4g39660 is therefore considered a candidate for a β‐alanine:pyruvate aminotransferase and is homologous to At2g38400, At3g08860, and Zm01g05170, a maize gene linked to β‐alanine metabolism (Wu et al., 2016). Although genome‐wide analysis and knockout studies link all four genes into a large gene family of β‐alanine aminotransferases conserved both in monocot and in dicot plants, At3g08860 clusters in a separate branch from other dicots (Wu et al., 2016). Notwithstanding these predictions for β‐alanine aminotransferase activity of the gene product of At3g08860, its function in A. thaliana has not been directly elucidated. Here, we present data to show that the locus tag e At3g08860 encodes an L‐alanine:glyoxylate aminotransferase using in vivo functional complementation assays in addition to in vitro biochemical assays. Although the cDNA was shown to complement an E. coli mutant auxotrophic for β‐alanine, β‐alanine:pyruvate aminotransferase activity was not detected using in vitro experiments.
2. MATERIALS AND METHODS
2.1. Plant growth and conditions
Arabidopsis thaliana Col 7 from the Arabidopsis Biological Resource Center (ABRC) was grown on Murashige and Skoog (MS) medium with a 16‐hr light (light intensity was approximately 120 Em−2 s−1) and an 8‐hr dark period, with temperatures of 24°C during the light period and 20°C during the dark period.
2.2. RNA isolation
Total RNA was isolated from 7‐day‐old Col 7 A. thaliana seedlings grown on MS medium using TriZol reagent (Life Technologies). One hundred milligrams of seedlings was ground in liquid nitrogen and homogenized in 1 ml of TriZol, followed by incubation at room temperature for two minutes. Total RNA was precipitated using 1 ml of 100% (v/v) isopropanol. The RNA pellet was washed three times with 1 ml of 75% (v/v) ethanol. The air‐dried RNA pellet was resuspended in 30 µl of Diethyl Pyro Carbonate (DEPC)‐treated water and quantified using a NanoDrop spectrophotometer.
2.3. cDNA synthesis
The Reverse Transcription System Kit (Promega) was used to synthesize a cDNA library following the manufacturer's protocol. One microgram of total RNA from 7‐day‐old seedlings was used to synthesize cDNA. The reaction contained 1µl oligo‐dT primer, 1 µg total RNA, 1 µl of 10 mM dNTP mix, and DEPC‐treated water up to 13 µl. The mixture was incubated at 65ºC for 5 min followed by an incubation on ice for 5 min.
2.4. Amplification and cloning of the At3g08860 cDNA
The full‐length protein encoded by At3g08860 is predicted to be 481 amino acids in length. The protein was predicted to be localized to the mitochondria using the TargetP and SUBA subcellular localization prediction tools (Emanuelsson, Brunak, Heijne, & Nielsen, 2007; Heazlewood, Verboom, Tonti‐Filippini, Small, & Millar, 2007). The first 93 nucleotides of the full‐length cDNA were predicted to encode the signal sequence that denote localization to the mitochondria. As such, the first 93 nucleotides were excluded when cloning the cDNA. The At3g08860 cDNA was amplified via PCR. The PCR (50 µl) reaction contained 1 µl (12 p.m./µl) each of the forward primer 5′‐CACCATGTCCTCCGTCCGCGAGACCGAGACCGAA‐3′ and the reverse primer 5′‐CTGCAGTCACATCTTGGACATGGCGTGATCCATCAC‐3′, 1 mM MgSO4, 0.4 mM of each of the four deoxynucleotide triphosphates, 2 µl of cDNA library, and 1 unit of platinum Pfx DNA polymerase (Invitrogen Corporation). The following PCR conditions were used: 1 cycle at 94ºC for 3 min, followed by 35 cycles of 94ºC for 30 s, 60ºC for 30 s, and 72ºC for 2 min and an indefinite soak of 4ºC. The cDNA amplicon was ligated into the pET100/D‐TOPO vector (Invitrogen Corporation, Carlsbad, CA, USA). The fidelity of the pET100D::At3g08860 construct was confirmed via Sanger nucleotide sequencing using T7 promoter (5′‐TAATACGACTCACTATAGGG‐3′) and the T7 terminator (5′‐TATGCTAGTTATTGCTCAG‐3′) primers located on the pET100/D‐TOPO plasmid backbone.
2.5. Construction of the plasmid for functional complementation
The plasmid pBAD33::At3g08860 used for functional complementation experiments of the E. coli mutants auxotrophic for β‐alanine (panD) and L‐alanine (HYE032) was constructed by sub‐cloning facilitated by the Xba1 and Pst1 restriction endonuclease sites from the pET100D::At3g08860 construct into pBAD33 (Guzman, Belin, Carson, & Beckwith, 1995).
2.6. Functional complementation assay
Auxotrophic E. coli mutants for L‐alanine (HYE032) (avtA::GM, yfbQ::KM, yfdZ::FRT, Ala −) were obtained from Dr. Hiroshi Yoneyama from Tohoku University (Yoneyama et al., 2011). The E. coli mutant auxotrophic for β‐alanine (panD) (F‐, Δ(araD‐araB)567, ΔpanD748::kan, ΔlacZ4787c (::rrnB‐3), λ −, rph‐1, Δ(rhaD‐rhaB)568, hsdR514) was obtained from the Coli Genetic Stock Center (CGSC #8404) (http://cgsc2.biology.yale.edu/) (Baba et al., 2006). The auxotrophic strains were transformed with pBAD33 or pBAD33::At3g08860. Transformants were selected on LB media supplemented with chloramphenicol (34 μg/ml). Colonies were then replica plated on M9 agar plates containing M9 salts (1X), 2 mM MgSO4, 0.1 mM CaCl2, and 0.1% glycerol (w/v), ±0.2% glucose or arabinose, and ±β‐alanine/L‐alanine (10 µg/µl). In testing of the panD (β‐alanine auxotroph), uracil was also required (10 µg/µl).
2.7. Enzyme expression and purification
The plasmid pET100D::At3g08860 was transformed into E. coli BL21‐CodonPlus‐RIPL strain (Stratagene). For protein expression and purification, the strain was grown in 1.0 L LB broth containing 50 μg/ml ampicillin and 34 μg/ml chloramphenicol at 37°C to an OD600 nm of 0.6. Protein expression was induced by adding isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) to a final concentration of 1 mM with continued growth at 18°C for 18 hr. For purification, the cells were lysed by sonication in a solution of 50 mM sodium phosphate and 300 mM NaCl (pH 8.0). The soluble protein extract was incubated with 0.5 ml bed volume of Talon Metal Affinity Resin (Clontech) for 30 min at 4°C. The resin was washed with 300 ml of sonication buffer containing 10 mM imidazole (pH 8.0), and the enzyme was eluted using 50 ml of sonication buffer containing 250 mM imidazole (pH 8.0). The purified enzyme was concentrated using an Amicon Ultra 10,000 molecular weight cutoff filter device, replacing the elution buffer with 100 mM HEPES‐NaOH containing 1 mM DTT and 1 mM EDTA (pH 8.0). For long‐term storage, the enzyme was stored in 50% (v/v) glycerol. Protein concentration was measured using the Bradford assay with bovine serum albumin as the standard (Bradford, 1976).
2.8. Enzyme assays
The assay measured the production of amino acceptors facilitated by the removal of the amino group from the donor. The assay for L‐alanine aminotransferase consisted of varying amounts of L‐alanine, 10 mM glyoxylate, and 0.66 µg of pure recombinant At3g08860, 1.4 µg of pyruvate dehydrogenase, 0.25 mM CoA, and 0.2 mM NADH in 100 mM HEPES‐NaOH (pH 8.0) to a final volume of 0.5 ml. The reverse assay consisted of varying amounts of glyoxylate, 10 mM L‐alanine, 1.4 µg pyruvate dehydrogenase, 0.25 mM CoA, and 0.2 mM NAD in 100 mM HEPES‐NaOH (pH 8.0) to a final volume of 0.5 ml. The assay to detect β‐alanine aminotransferase activity was similar with only difference being the amino donor (β‐alanine) and glycine dehydrogenase as the coupling enzyme since glycine and malonate semialdehyde are the products of the reaction. The production of NADH was measured at A 340. Both the forward and the reverse assays were incubated at 30°C. All kinetic constants were calculated using GraphPad Prism V5 software by using the nonlinear regression analysis algorithm feature. A Beckman DU640 Spectrophotometer was used to monitor enzyme activity.
2.9. Bioinformatic analyses
To identify sequences from related species, the At3g08860 amino acid sequence was used as a query against the protein database using the Basic Local Alignment Search Tool (BLAST) protein program (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) employing the standard parameters. Sequences were obtained from the taxonomy report, representing the first fifteen identified species. The multiple sequence alignment of the At3g08860 amino acid sequence with other plant aminotransferase sequences was constructed via ClustalW algorithm in MEGA7 (Kumar, Stecher, & Tamura, 2016). The GONNET amino acid substitution matrix was used (Gonnet, Cohen, & Benner, 1992). ClustalW alignment of top five best hits was used to create the images with BOXSHADE (https://embnet.vital-it.ch/software/BOX_form.html). Three‐dimensional homology model for the At3g08860 protein was constructed via SWISS‐MODEL (Schwede, Kopp, Guex, & Peitsch, 2003) after alignment to the Arthrobacter aurescens TC1 phospholyase template, a class III transaminase homolog (PDB ID: 5G4I) (Cuetos et al., 2016). The template was chosen as the crystal structure with the best sequence identity (35.82%) to the enzyme based on a search with HHBlits against the SWISS‐MODEL template library (Remmert, Biegert, Hauser, & Söding, 2012). The model was built based on the alignment between reference and query protein sequence. Conserved coordinates were directly copied while insertions and deletions were modeled from the SWISS‐MODEL fragment library.
3. RESULTS
Functional complementation assays showed that the plasmid harboring the At3g08860 cDNA was able to rescue both the L‐alanine and β‐alanine auxotrophic mutants. The assays show that both E. coli mutants were able to grow on β‐alanine and L‐alanine free media compared with the vector only control (Figure 2a,b). Interestingly, in the panD background, the cDNA rescued growth under repressible conditions (with glucose) and not under inducible conditions (with arabinose) (Figure 2a) whereas the opposite is true in the HYE032 background (Figure 2b). This result suggests that the enzyme is probably involved in L‐alanine catabolism. As a control to test enzyme promiscuity, a complementation assay was also performed by expressing At3g08860 in the E. coli mutant DL39. DL39 harbors a mutation in the tyrosine aminotransferase (tyrB) gene which leads to auxotrophy for L‐tyrosine and L‐phenylalanine. While the At5g36160 cDNA was able to complement DL39, At3g08860 was not able to (data not shown) (Prabhu and Hudson, 2010). The enzyme was purified to homogeneity using metal affinity chromatography with an apparent monomeric size of approximately 50 kDa (Figure 3a) and showed the characteristic spectrum of a PLP‐containing protein (Figure 3b). In vitro aminotransferase, enzymatic assays with the purified recombinant At3g08860 enzyme showed activity with L‐alanine as the amino donor and glyoxylate as the amino acceptor. The Michaelis‐Menten (KM) constants for the substrates were 0.63 and 0.56 mM for L‐alanine and glyoxylate respectively with a specific enzyme activity of 0.63 µmol min−1 mg−1 (Figure 3c,d). Enzymatic activity was not detected using β‐alanine as the amino donor with either pyruvate or glyoxylate as amino acceptors.
Figure 2.
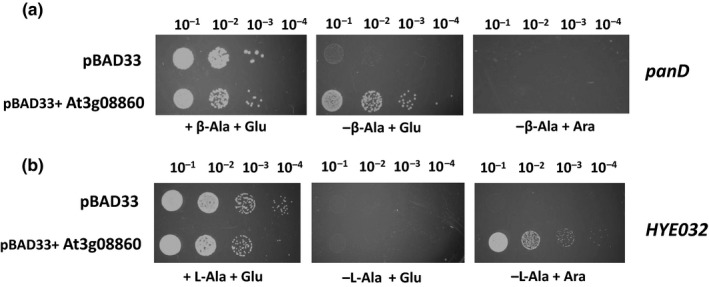
Functional complementation assay. (a) Functional complementation of the panD E. coli mutant, which is auxotrophic for β‐alanine. (b) Functional complementation of the E. coli HYE032 mutant, which is auxotrophic for L‐alanine. The plasmids used were pBAD33 and pBAD33::At3g08860. Transformants harboring pBAD33 or pBAD33::At3g08860 were grown to an OD of 1.0 measured at 600 nm. The strains were serially diluted to 10–1, 10–2, 10–3, and 10–4 using 0.85% (w/v) saline. Five µl was replica plated on M9 medium with or without β‐alanine or L‐alanine supplemented with 0.2% (w/v) arabinose or glucose
Figure 3.
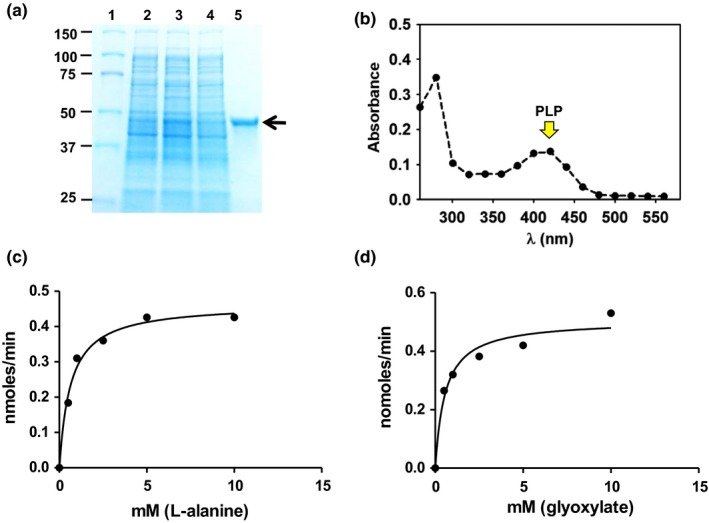
Protein purification and biochemical characterization. (a) Purification of the At3g08860 protein to apparent homogeneity by affinity chromatography followed by SDS‐PAGE. Along with the molecular mass markers (kDa) (lane 1), the gel shows the profile of 10 μg of uninduced soluble proteins (lane 2), 10 μg of induced soluble proteins (lane 3) and 0.5 μg of the purified recombinant enzyme (black arrow) (lane 4). (b) A wavelength scan of the protein shows a peak around 420 nm (indicated by the yellow arrow), which is characteristic of PLP‐containing proteins
Multiple sequence alignment and comparison of At3g08860 with orthologs from Cannabis sativa, Capsella rubella, Arabidopsis lyrata, and Brassica rapa show that the protein is conserved including the PLP‐binding motif with the conserved lysine residue in the active site involved in catalysis (Figure 4). A three‐dimensional homology model of At3g08860 using a phospholyase transaminase (PDB ID 5G4I) template show a predicted dimeric structure containing two v‐shaped active sites (Figure 5a). The model quality, assessed with z‐scores for the QMEAN function (−2.04), all atom pairwise energy (−0.16), CBeta interaction energy (−2.11), solvation energy (0.65), and torsion angle energy (−1.77), indicates an acceptable structure prediction (Figure 5b). The local quality estimate across the protein sequence indicates good local quality scores for areas of determined secondary structure (alpha helices or beta strands) above 0.70. Areas lacking secondary structural elements (i.e., disordered regions or loops) were areas with lower local quality scores (Figure 5c). The dimeric structural nature of At3g08860 is similar to that of other aminotransferases that have been solved by X‐ray crystallography including L,L‐diaminopimelate aminotransferase (DapL) and N‐acetylornithine aminotransferase (Rajaram, Ratna Prasuna, Savithri, & Murthy, 2008; Triassi et al., 2014).
Figure 4.

Multiple amino acid sequence alignment. Multiple amino acid sequence alignment comparing the At3g08860 with orthologs from a variety of plant species. The line designates the conserved PLP‐binding motif, and the asterisk denotes the conserved lysine involved in PLP binding
Figure 5.
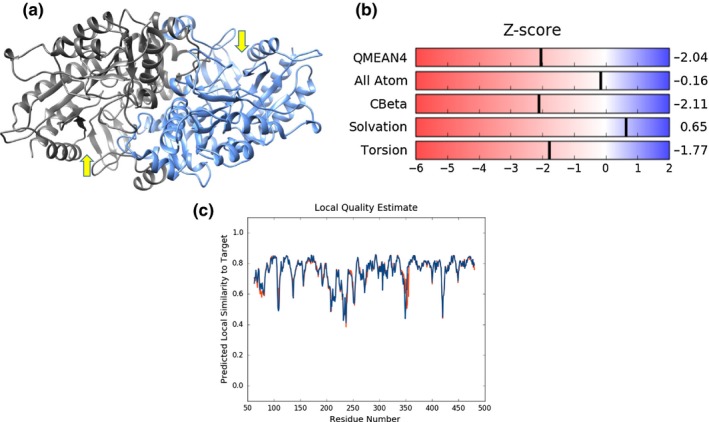
Structural analysis of At3g08860. (a) Homology model of the At3g08860. The model was constructed via SWISS‐MODEL and aligned to the phospholyase transaminase homolog template PDB ID: 5G4I. The two active sites are highlighted with the black arrows. 35.82% sequence identity, GMQE 0.66 and QMEAN −2.04; (b) gives a local quality estimate for different regions of the proteins and (c) summarizes the Z‐score data generated via SWISS‐MODEL when related to high quality X‐ray crystal structures for proteins of similar size
4. DISCUSSION
The physiological role of At3g08860 according to earlier studies (Obayashi & Kinoshita, 2010; winter et al., 2007; Wu et al., 2016; Zrenner et al., 2009) was suggested to be β‐alanine metabolism. β‐Alanine is a known osmoprotectant in plants. It was recently demonstrated that in the A. thaliana At3g08860 is upregulated in response to osmotic stress (300 mM mannitol or 150 mM sodium chloride), but not cold, drought, UV radiation, oxidative stress, or wounding, suggesting a role in osmotic balance, possibly via β‐alanine metabolism (Winter et al., 2007), based on visualizing gene expression in plants using the BAR tool (http://bar.utoronto.ca/).
Even though the cDNA was able to rescue both L‐alanine and β‐alanine auxotrophs, we were not able to corroborate the in vivo complementation results using in vitro assays using β‐alanine as a substrate with two different acceptors (pyruvate and α‐ketoglutarate). This could be due to a low‐level promiscuous activity for β‐alanine in vivo, which is not detectable in vitro. The co‐expression analysis of At3g08860 shown in Figure 6 suggests an involvement in multiple processes in the plant life cycle. Studies investigating genes involved in a wide array of metabolic/cellular processes have identified the At3g08860 locus as responsive to changes in light, which could indirectly affect carbon/oxygen availability/concentration (Thum et al., 2008). Another study demonstrated that the highest levels of expression of At3g08860 occur in the silique and roots of A. thaliana (Schmid et al., 2005).
Figure 6.
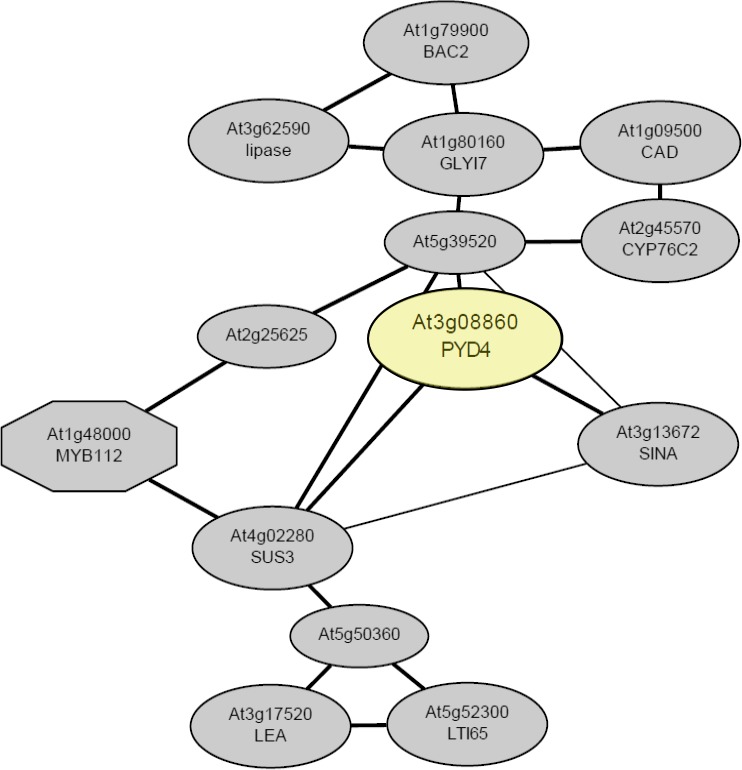
Gene expression network of At3g08860. The predicted network of genes co‐expressed with At3g08860, generated based on the ATTED‐II tool (Obayashi et al., 2009; Obayashi & Kinoshita, 2010). The gene loci and their corresponding protein functions are as follows: At3g08860 = Pyd4 = Alanine:glyoxylate aminotransferase, putative/ β‐Alanine‐pyruvate aminotransferase, putative/AGT, putative; At5g52300 = LTI65 (low‐temperature induced 65); At5g50360 = Unknown protein; At3g17520 = Late embryogenesis abundant (LEA) domain‐containing protein; At1g79900 = BAC‐2: L‐Ornithine transmembrane transporter/binding/ carnitine‐acylcarnitine antiporter; At3gt3672 = Seven in absentia (SINA) family protein; At1g80160 = Lactoyl‐glutathione lyase family protein/ glyoxylase I family protein; At2g25625 = Unknown protein; At4g02280 = Sucrose synthase (SUS)‐3, UDP‐glycosyltransferase/ sucrose synthase/ transferase, glycosyl transfer; At3g62590 = Lipase class 3 family protein; At5g39520 = Unknown protein; At2g45570 = CYP76C2: electron carrier, heme binding/ iron ion binding/ oxygen binding/ monooxygenase; At1g09500 = Cinnamyl alcohol dehydrogenase (CAD) family, and At1g48000 = MYB domain protein112 (MYB112), DNA binding/ transcription factor
Regarding the subcellular distribution of the At3g08860 protein, localization to either the mitochondria or the peroxisome has been suggested (Niessen et al., 2012). It was demonstrated that alanine:glyoxylate aminotransferase activity was the only aminotransferase activity detected within the mitochondria (Niessen et al., 2012). They also demonstrated that alanine degradation resulted in increased CO2 release following the addition of alanine to mitochondrial extracts, implying that alanine degradation increased photorespiratory activity. The enzyme activity was measured at pH 8.0 in this study, which is similar to the mitochondrial pH of 7.8, but much higher than the pH cytosol or the peroxisome (pH 7.0). The visualization of cellular localization using GFP‐tagged proteins was unsuccessful. The question of whether the protein At3g08860 is localized in the peroxisome or the mitochondria remains unanswered.
5. CONCLUSIONS
This study identified the activity of At3g08860 using an in vivo functional complementation assay showing that the enzyme can complement L‐alanine and β‐alanine auxotrophy of E. coli mutants. In vitro analysis using purified recombinant enzyme corroborated the in vivo results using L‐alanine and glyoxlyate as substrates. Although the enzyme was able to complement the β‐alanine auxotrophy in E. coli, we were not able to identify this activity using in vitro assays. The enzyme has orthologs in higher plants, is related to other alanine aminotransferases in A. thaliana and contains the PLP‐binding motif with the conserved active site lysine residue. The dimeric structure that is predicted for the enzyme is consistent with other structurally characterized aminotransferases. In summary, the physiological role of the enzyme At3g08860 is suggested to be L‐alanine catabolism in addition to response to osmotic stress based on expression studies.
CONFLICT OF INTEREST
The authors declare no conflict of interest associated with the work described in this manuscript.
AUTHOR CONTRIBUTIONS
AOH developed the overall strategy, designed experiments, and coordinated the project. AP, LEA, and FCS performed experiments. AP, LEA, and AOH wrote the manuscript.
ACKNOWLEDGMENTS
This research was supported by a United States National Science Foundation (NSF) award (MCB‐#1120541) to AOH. We thank Dr. Hiroshi Yoneyama from Tohoku University for providing the E. coli HYE032 mutant. We would like to acknowledge the College of Science and the Thomas H. Gosnell School of Life Sciences at the Rochester Institute of Technology for ongoing support.
Parthasarathy A, Adams LE, Savka FC, Hudson AO. The Arabidopsis thaliana gene annotated by the locus tag At3g08860 encodes alanine aminotransferase. Plant Direct. 2019;3:1–10. 10.1002/pld3.171
This manuscript was previously deposited as a preprint at https://doi.org/10.1101/576041
REFERENCES
- Baba, T. , Ara, T. , Hasegawa, M. , Takai, Y. , Okumura, Y. , Baba, M. , … Mori, H. (2006). Construction of Escherichia coli K‐12 in‐frame, single‐gene knockout mutants: The Keio collection. Molecular Systems Biology, 2(1), 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, R. L. , & Naylor, A. W. (1962). Formation of beta‐alanine by pine tissues supplied with intermediates in uracil & orotic acid metabolism. Plant Physiology, 37(2), 171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Analytical Biochemistry, 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Campbell, L. L. (1960). Reductive degradation of pyrimidines. 5. Enzymatic conversion of N‐carbamyl‐beta‐alanine to beta‐alanine, carbon dioxide, and ammonia. Journal of Biological Chemistry, 235, 2375–2378. [PubMed] [Google Scholar]
- Cuetos, A. , Seffen‐Munsberg, F. , Mangas‐Sanchez, J. , Frese, A. , Bornscheuer, U. T. , Hohne, M. , & Grogan, G. (2016). Structural basis for phospholyase activity of a class III transaminase homologue. ChemBioChem, 17, 2308–2311. [DOI] [PubMed] [Google Scholar]
- de Sousa, C. A. F. , & Sodek, L. (2003). Alanine metabolism and alanine aminotransferase activity in soybean (Glycine max) during hypoxia of the root system and subsequent return to normoxia. Environmental and Experimental Botany, 50, 1–8. [Google Scholar]
- Emanuelsson, O. , Brunak, S. , von Heijne, G. , & Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols, 2(4), 953–971. [DOI] [PubMed] [Google Scholar]
- Galston, A. W. , & Sawhney, R. K. (1990). Polyamines in plant physiology. Plant Physiology, 94(2), 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonnet, G. H. , Cohen, M. A. , & Benner, S. A. (1992). Exhaustive matching of the entire protein sequence database. Science, 256, 1433–1445. 10.1126/science.1604319 [DOI] [PubMed] [Google Scholar]
- Gupta, K. J. , Zabalza, A. , & van Dongen, J. T. (2009). Regulation of respiration when the oxygen availability changes. Physiologia Plantarum, 137(4), 383–391. [DOI] [PubMed] [Google Scholar]
- Guzman, L. M. , Belin, D. , Carson, M. J. , & Beckwith, J. (1995). Tight regulation, modulation, and high‐level expression by vectors containing the arabinose PBAD promoter. Journal of Bacteriology, 177(14), 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayaishi, O. , Nishizuka, Y. , Tatibana, M. , Takeshita, M. , & Kuno, S. (1961). Enzymatic studies on the metabolism of beta‐alanine. Journal of Biological Chemistry, 236, 781–790. [PubMed] [Google Scholar]
- Heazlewood, J. L. , Verboom, R. E. , Tonti‐Filippini, J. , Small, I. , & Millar, A. H. (2007). SUBA: The Arabidopsis subcellular database. Nucleic Acids Research, 5, D213–D218. 10.1093/nar/gkl863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismond, K. P. , Dolferus, R. , de Pauw, M. , Dennis, E. S. , & Good, A. G. (2003). Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiology, 132(3), 1292–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , & Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kürsteiner, O. , Dupuis, I. , & Kuhlemeier, C. (2003). The pyruvate decarboxylase1 gene of Arabidopsis is required during anoxia but not other environmental stresses. Plant Physiology, 132(2), 968–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman, A. H. , & Olsen, L. J. (2003). Alanine aminotransferase homologs catalyze the glutatmate:Glyoxylate aminotransferase reaction in peroxisomes of Arabidopsis . Plant Physiology, 131(1), 215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman, A. H. , & Olsen, L. J. (2004). Genomic analysis of aminotransferases in Arabidopsis thaliana . Critical Reviews in Plant Sciences, 23(1), 73–89. [Google Scholar]
- McAllister, C. H. , Facette, M. , Holt, A. , & Good, A. G. (2013). Analysis of the enzymatic properties of a broad family of alanine aminotransferases. PLoS ONE, 8(2), e55032 10.1371/journal.pone.0055032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister, C. H. , & Good, A. G. (2015). Alanine aminotransferase variants conferring diverse NUE phenotypes in Arabidopsis thaliana . PLoS ONE, 10(4), e0121830 10.1371/journal.pone.0121830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita, Y. , Dolferus, R. , Ismond, K. P. , & Good, A. G. (2007). Alanine aminotransferase catalyses the breakdown of alanine after hypoxia in Arabidopsis thaliana . The Plant Journal, 49(6), 1108–1121. [DOI] [PubMed] [Google Scholar]
- Nelson, D. , & Cox, M. (2000). Lehninger, principles of biochemistry. New York, NY: Worth Publishing. [Google Scholar]
- Niessen, M. , Krause, K. , Horst, I. , Staebler, N. , Klaus, S. , Gaertner, S. , … Peterhansel, C. (2012). Two alanine aminotranferases link mitochondrial glycolate oxidation to the major photorespiratory pathway in Arabidopsis and rice. Journal of Experimental Botany, 63(7), 2705–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen, M. , Thiruveedhi, K. , Rosenkranz, R. , Kebeish, R. , Hirsch, H. J. , Kreuzaler, F. , & Peterhansel, C. (2007). Mitochondrial glycolate oxidation contributes to photorespiration in higher plants. Journal of Experimental Botany, 58(10), 2709–2715. [DOI] [PubMed] [Google Scholar]
- Obayashi, T. , Hayashi, S. , Saeki, M. , Ohta, H. , & Kinoshita, K. (2009). ATTED‐II provides coexpressed gene networks for Arabidopsis . Nucleic Acids Research, 37, D987–D991. 10.1093/nar/gkn807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi, T. , & Kinoshita, K. (2010). Coexpression landscape in ATTED‐II: Usage of gene list and gene network for various types of pathways. Journal of Plant Research, 123, 311–319. 10.1007/s10265-010-0333-6 [DOI] [PubMed] [Google Scholar]
- Ottenhof, H. H. , Ashurst, J. , Whitney, H. , Saldanha, S. , Schmitzberger, F. , Gweon, H. , … Smith, A. G. (2004). Organisation of the pantothenate (vitamin B5) biosynthesis pathway in higher plants. The Plant Journal, 37(1), 61–72. [DOI] [PubMed] [Google Scholar]
- Prabhu, P. , & Hudson, A. O. (2010). Identification and partial characterization of an L-Tyrosine aminotransferase from Arabidopsis thaliana . Biochemistry Research International, 549572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaram, V. , Ratna Prasuna, P. , Savithri, H. S. , & Murthy, M. R. (2008). Structure of biosynthetic N‐acetylornithine aminotransferase from Salmonella typhimurium: Studies on substrate specificity and inhibitor binding. Proteins, 70(2), 429–441. [DOI] [PubMed] [Google Scholar]
- Rastogi, R. , & Davies, P. (1989). Polyamine metabolism in ripening tomato fruit. Plant Physiology, 94, 1449–1455. 10.1104/pp.94.3.1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmert, M. , Biegert, A. , Hauser, A. , & Söding, J. (2012). HHblits: Lightning‐fast iterative protein sequence searching by HMM‐HMM alignment. Nature Methods, 9, 173–175. 10.1038/nmeth.1818 [DOI] [PubMed] [Google Scholar]
- Ricoult, C. , Echeverria, L. O. , Cliquet, J. B. , & Limami, A. M. (2006). Characterization of alanine aminotransferase (AlaAT) multigene family and hypoxic response in young seedlings of the model legume Medicago truncatula . Journal of Experimental Botany, 57(12), 3079–3089. [DOI] [PubMed] [Google Scholar]
- Rocha, M. , Licausi, F. , Araújo, W. L. , Nunes‐Nesi, A. , Sodek, L. , Fernie, A. R. , & van Dongen, J. T. (2010). Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus . Plant Physiology, 152(3), 1501–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M. , Davison, T. S. , Henz, S. R. , Pape, U. J. , Demar, M. , Vingron, M. , … Lohmann, J. U. (2005). A gene expression map of Arabidopsis thaliana development. Nature Genetics, 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Schwede, T. , Kopp, J. , Guex, N. , & Peitsch, M. C. (2003). SWISS‐MODEL: An automated protein homology‐modeling server. Nucleic Acid Research, 31(13), 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Blundell, T. L. , & Mizuguchi, K. (2001). FUGUE: Sequence‐structure homology recognition using environment‐specific substitution tables and structure‐dependent gap penalties. Journal of Molecular Biology, 310(1), 243–257. [DOI] [PubMed] [Google Scholar]
- Stinson, R. A. , & Spencer, M. S. (1969a). Beta‐alanine as an ethylene precursor. investigations towards preparation, and properties, of a soluble enzyme system from a subcellular particulate fraction of bean cotyledons. Plant Physiology, 44(9), 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinson, R. A. , & Spencer, M. S. (1969b). Beta alanine aminotransferase(s) from a plant source. Biochemical and Biophysical Research Communications, 34(1), 120–127. [DOI] [PubMed] [Google Scholar]
- Terano, S. , & Suzuki, Y. (1978). Formation of β‐alanine from spermine and spermidine in maize shoots. Phytochemistry, 17, 148–149. 10.1016/S0031-9422(00)89700-9 [DOI] [Google Scholar]
- Thum, K. E. , Shin, M. J. , Gutiérrez, R. A. , Mukherjee, I. , Katari, M. S. , Nero, D. , … Coruzzi, G. M. (2008). An integrated genetic, genomic and systems approach defines gene networks regulated by the interaction of light and carbon signaling pathways in Arabidopsis. BMC Systems Biology, 2, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut, T. W. , & Loechel, S. (1984). Pyrimidine catabolism: Individual characterization of the three sequential enzymes with a new assay. Biochemistry, 23(11), 2533–2539. [DOI] [PubMed] [Google Scholar]
- Triassi, A. J. , Wheatley, M. S. , Savka, M. A. , Gan, H. M. , Dobson, R. C. J. , & Hudson, A. O. (2014). L, L‐Diaminopimelate aminotransferase (DapL): A putative target for the development of narrow‐spectrum antibacterial compounds. Frontiers in Microbiology, 5, 509 10.3389/fmicb.2014.00509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe, G. C. , Yip, J. Y. , & Parsons, H. L. (1999). In organello and in vivo evidence of the importance of the regulatory sulfhydryl/disulfide system and pyruvate for alternative oxidase activity in tobacco. Plant Physiology, 121(3), 793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, D. , Vinegar, B. , Nahal, H. , Ammar, R. , Wilson, G. V. , & Provart, N. J. (2007). An “Electronic Fluorescent Pictograph” Browser for Exploring and Analyzing Large‐Scale Biological Data Sets. PLoS ONE, 2(8), e718 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S. I. , Alseekh, S. , Cuadros‐Inostroza, Á. , Fusari, C. M. , Mutwil, M. , Kooke, R. , … Brotman, Y. (2016). Combined use of genome‐wide association data and correlation networks unravels key regulators of primary metabolism in Arabidopsis thaliana . PLoS Genetics, 12(10), e1006363 10.1371/journal.pgen.1006363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama, H. , Hori, H. , Lim, S. J. , Murata, T. , Ando, T. , Isogai, E. , & Katsumata, R. (2011). Isolation of a mutant auxotrophic for L‐alanine and identification of three major aminotransferases that synthesize L‐alanine in Escherichia coli . Bioscience, Biotechnology, and Biochemistry, 75(5), 930–938. [DOI] [PubMed] [Google Scholar]
- Zabalza, A. , van Dongen, J. T. , Froehlich, A. , Oliver, S. N. , Faix, B. , Gupta, K. J. , … Geigenberger, P. (2009). Regulation of respiration and fermentation to control the plant internal oxygen concentration. Plant Physiology, 149(2), 1087–1098. 10.1104/pp.108.129288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner, R. , Riegler, H. , Marquard, C. R. , Lange, P. R. , Geserick, C. , Bartosz, C. E. , … Slocum, R. D. (2009). A functional analysis of the pyrimidine catabolic pathway in Arabidopsis . New Phytologist., 183, 117–132. 10.1111/j.1469-8137.2009.02843.x [DOI] [PMC free article] [PubMed] [Google Scholar]


