Abstract
The centerpiece of the sample cell assembly in analytical ultracentrifugation holds the sample solution between windows, sealed against high vacuum, and is shaped such that macromolecular migration in centrifugal fields exceeding 200 000g can proceed undisturbed by walls or convection while concentration profiles are imaged with optical detection systems aligned perpendicular to the plane of rotation. We have recently shown that 3D printing using various materials allows inexpensive and rapid manufacturing of centerpieces. In the present work, we expand this endeavor to examine the accuracy of the measured sedimentation process, as well as short-term durability of the centerpieces. We find that 3D-printed centerpieces can be used many times and can provide data equivalent in quality to commonly used commercial epoxy resin centerpieces. Furthermore, 3D printing enables novel designs adapted to particular experimental objectives because they offer unique opportunities, for example, to create well-defined curved surfaces, narrow channels, and embossed features. We present examples of centerpiece designs exploiting these capabilities for improved AUC experiments. This includes narrow sector centerpieces that substantially reduce the required sample volume while maintaining the standard optical path length; thin centerpieces with integrated window holders to provide very short optical pathlengths that reduce optical aberrations at high macromolecular concentrations; long-column centerpieces that increase the observable distance of macromolecular migration for higher-precision sedimentation coefficients; and three-sector centerpieces that allow doubling the number of samples in a single run while reducing the sample volumes. We find each of these designs allows unimpeded macromolecular sedimentation and can provide high-quality sedimentation data.
Graphical Abstract
Analytical ultracentrifugation (AUC) is a traditional technique of physical chemistry to characterize molecules and particles free in solution through observation of their collective behavior after application of a centrifugal field.1,2 The foundations of this technique were laid by The Svedberg in the 1920s,3 leading to the discovery of biological macromolecules and the birth of cell biology.4,5 AUC allows the measurement of a variety of properties including size, shape, polydispersity, and macromolecular interactions with KD values ranging from pM to mM,6–8 and it is still growing in applications in many fields of molecular sciences including biochemistry, cell biology, immunology, food sciences, biotechnology, polymer chemistry, colloid chemistry, and nanoparticles.9–14
The present work is focused on the AUC sample holder that sandwiches the macromolecular solutions between optical windows and seals against the high vacuum in the centrifugal chamber while spinning in the rotor at 200 000 to 300 000g. For sedimentation velocity (SV) experiments that aim at interpreting the time-course of sedimentation, they are sector-shaped to ensure that particles can sediment radially unimpeded by the walls, thereby preventing macroscopic convective flows originating in lateral density gradients.2,15 Besides the basic design featuring a ≈14 mm long sector of ≈2.5° in single sector or side-by-side double sector configuration,16,17 the early decades of AUC have also seen considerable experimentation with sample holders specialized for measuring particle hydrodynamic volumes,18 achieving preparative separation,19 overlaying solutions,20,21 and for short and ultrashort solution columns for sedimentation equilibrium.22 Strikingly, however, the basic design of the most widely used modern commercial AUC centerpiece is virtually identical to that originally developed nearly a century ago by Svedberg and colleagues. Such centerpieces are currently commercially available in an Epon resin (Beckman Coulter and Spin Analytics), aluminum (Beckman Coulter), and titanium (Nanolytics). Besides variation in optical path length (3 mm and 12 mm for Epon centerpieces; 1.5 to 20 mm for titanium centerpieces) for SV experiments, they offer little or no flexibility in terms of sample number, sample volume, and radial observation range.
A significant impediment in the development of AUC centerpieces has been the manufacturing process, which until recently was prohibitive in terms of expertise for the vast majority of laboratories and limited the choice to commercially available designs. However, open-source 3D printing has revolutionized manufacturing of laboratory instrumentation,23–27 enabling laboratories to create cost-effective and optimized custom instrumentation. Exploiting these advances in manufacturing, we recently discovered that 3D printing allows for the creation of AUC centerpieces from a variety of materials including acrylic, carboxylate, polypropylene carbonate, and ABS-like resins28 in a quality sufficient to withstand the extreme mechanical stress up to 200 000–300 000g while sealing samples in high vacuum. We have shown that these can allow macromolecular sedimentation to proceed in a way that seems indistinguishable from commercial Epon centerpieces.28 The cost of 3D-printed centerpieces is a small fraction of commercial Epon centerpieces. Therefore, an obvious first application is in training, and a second application arises in work with samples that benefit from disposable centerpieces.28 However, 3D printing opens new possibilities: for example, we have previously presented centerpiece designs with elevated 3 mm optical path length sectors optimized for use with the confocal fluorescence detection system,28,29 and more recently, we have created 1.5 mm and 1.75 mm optical path length centerpieces for the purpose of working with very high sample concentrations.7
The purpose of the present communication is 2-fold: First, we study the durability of 3D-printed centerpieces in repeated SV experiments and report on the accuracy and statistical precision of the main features of the resulting sedimentation coefficient distributions of a test sample of bovine serum albumin (BSA). Furthermore, we demonstrate that the sedimentation process of a monodisperse sample follows the ideal sedimentation model with precision equal to or better than commercial Epon centerpieces. This verifies the utility of 3D-printed centerpieces for the study of macromolecular sedimentation processes. Next, we exploit a capability intrinsic to additive manufacturing that allows the creation of curved surfaces and channels that have not been possible in previous centerpieces. These features lead to a variety of novel designs that should enhance precision and can substantially increase throughput of SV experiments.
EXPERIMENTAL SECTION
Fabrication of Centerpieces.
3D designs were created in OpenSCAD,30 a freeware programming language for parametric script-based design of 3D objects. The code for the centerpieces was structured to utilize predefined variables (such as optical path length and sector angle top and bottom) and parametrized subroutines to allow easier customization. The code can be downloaded along with compiled stereo-lithography (stl) files from the NIH 3D Print Exchange31 (3DPX-010758).
Unless mentioned otherwise, centerpieces were printed from an epoxy-based photopolymer “MicroFine Green”, using microstereo-lithography in 0.001” layers with a modified 3DSystems Viper printer at protolabs.com (ProtoLabs, Inc., Maple Plain, MN). No postprocessing was applied other than an isopropyl alcohol rinse to remove left-over resin and UV curing. The printing layers were oriented in the plane of rotation (stacking layers in z-direction perpendicular to sedimentation) to avoid stair-casing artifacts on the sector side walls in the direction of sedimentation, and to maximize smoothness of top and bottom surfaces facing windows. A microscopic view of a sector side-wall is shown in SI Figure S1, from which layers in z-direction can be faintly observed, but not in the x–y-plane.
Centerpiece prototypes were also printed in-house by fused deposition modeling in ABS (Stratasys part no. 340–21201) on a Fortus 250mc, and by polyjet printing in acrylic-like VeroBlackPlus (glossy and matte) and VeroClear, or in polypropylene-like Rigur on an Eden260vs For the cell housing lids, the build was in “RenShape SL 7820” normal resolution (0.004 in. layers) stereolithography, also from ProtoLabs, Inc.
Analytical Ultracentrifugation.
SV experiments were carried out in a ProteomeLab XL-A/I (Beckman Coulter, Indianapolis IN) following standard techniques15 unless mentioned otherwise. For details on SV and protein samples, see SI Materials and Methods. Calibration factors for radial magnification, scan time, and actual temperature were measured as described.32,33 All data analyses were carried out using c(s) analysis or single species Lamm equation models in SEDFIT (sedfitsedphat.nibib.nih.gov),34 extended with buffer mismatch modeling35 where necessary, and results were plotted using GUSSI.36 Statistical analysis was based on 95% confidence intervals using the error projection method and F-statistics.37 From histograms of residuals the H-value was used to quantify deviations from Gaussian distribution as described.38
RESULTS AND DISCUSSION
General Features.
In our previous report introducing 3D-printed centerpieces for AUC, we have demonstrated that they can be fabricated from a wide range of different materials, from different sources, and using different printing technologies, such that they withstand centrifugation up to 60 000 rpm, while holding liquid samples in the high vacuum of the ultracentrifuge chamber with sufficient precision for SV experiments.28 In the present work, we show how 3D printing permits improved centerpiece designs that enhance AUC experiments. For this we have focused on builds with an epoxy-based photopolymer termed “MicroFine Green” printed by stereolithography.
As we have reported previously, the vacuum seal of 3D-printed centerpieces can be enhanced by creating a narrow, embossed ridge surrounding the sample sectors28 (Figure 1A). Extending the previous design, such an embossed seal can be added on both sides of the centerpiece. Applying a torque of ≈100–120 in.-pound (11.3–13.5 N m) with the conventional AUC torque stand, a vacuum seal is created that persists during centrifugation, without any extra gaskets. We have also gained more experience in the dimensions of this seal and consider an optimal height to be 0.15–0.2 mm with a width of at least 0.25 mm. Shallower seals have not consistently printed, and we have observed narrower seals to separate from the body of the centerpiece after repeated runs. More sophisticated gaskets with optimized shape, such as triangular or half-spherical cross sections, have been conceived but not tested.
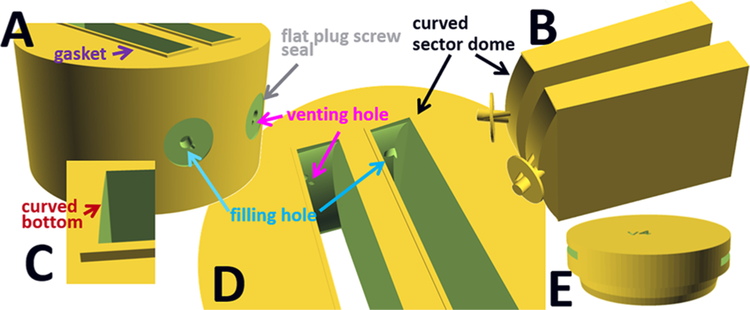
Figure 1.
Design features of 3D-printed analytical ultracentrifugation centerpieces. All pictures are from the design of a 12 mm optical path length centerpiece with 11 mm radius. In (A) a side-view is shown highlighting the compressible gasket and holes exiting the plug screw flat. (B) highlights the negative of domed sectors with filling and venting holes and plug screw flat. Close-up views of the curved bottom (C) and top of the domed sectors (D). (E) shows a cell housing plug. Pictures of the fabricated centerpieces in this design are shown in SI Figure S1.
Another aspect of 3D printing convenient for manufacturing AUC centerpieces is that small channels can easily be created. This allows the filling hole to be included into the design. Thus, unless otherwise noted, all centerpieces include a filling hole with 0.5 mm radius generally from the center of the plug screw opening in the cell housing to the top of the narrow end of the solution sectors (top with respect to the centrifugal force) (Figure 1). This dimension is suitable for the long gel loading tips that we prefer for sample loading but may be easily adjusted to fit syringes or tubing. Further, the ≈4 mm diameter circular flat areas of the centerpiece against which the filling hole plug screws seal (Figure 1A) can accommodate additional channels. Thus, we have routinely included venting holes of 0.2 mm radius that originate next to the filling hole and lead to an entry point in the sector distant from the filling hole. We have found the presence of a secondary hole not to be of concern for creating the vacuum seal with the plug screw (Figure 1). This simplifies loading of the cells by minimizing the formation of air locks in the filling hole, which can be a problem with solutions of low surface tension (such as protein solution at high concentrations or containing detergent) and/or when attempting to maximize the solution column (see below).15
Both the filling and the venting hole can easily be curved and tapered. However, even though the minimum layer thickness is 0.001 in. using this material, the minimum feature thickness is 0.008 in. or 0.203 mm, respectively, which poses some constraints on the path for the channels and the minimum centerpiece thickness (see below). Another constraint is the need for channels to be accessible for rinsing to remove unreacted polymer as the final step of fabrication.
To ensure that these capabilities are not unique to the particular material used, as proof of principle, we have also fabricated centerpieces in different materials in the long-channel design described below. We observed similar vacuum seal in acrylic-like and polypropylene-like centerpieces. These also allowed SV experiments at 50 000 rpm, with best results in acrylic-like materials (SI Figure S2); this was not further pursued in the present work. We note that no vacuum seal of the liquid sample could be achieved in ABS centerpieces—even when using with separate gaskets—which we attribute to porosity of the material when 3D-printed using fused deposition modeling. We note that seals would likely have to be omitted when printing in metals; in that case, separate gaskets can be cut from polyethylene, Teflon, or other materials; such gaskets are routinely used in conjunction with commercial aluminum and titanium centerpieces and commercially available. However, such gaskets can also be cut, for example, from inexpensive 0.005 in. Teflon sheet using standard electronic cutting machines (Dr. Jeffrey Fagan, personal communication). Generally, builds from material other than MicroFine Green may require minor adjustments in the centerpiece radius to fit smoothly and snugly into the standard barrels of commercial cell assemblies.
An example of useful ancillary 3D-printed equipment for AUC are housing plugs (Figure 1E) that snap into the aluminum housing. Printed out of black plastic, they prevent light from entering the filled solution columns during setup, thus protecting light-sensitive chromophores from bleaching. This is particularly useful in conjunction with fluorescence optical detection.39
Repeatability and Durability of 3D-Printed Center-pieces in Standard Design.
3D-printed MicroFine Green centerpieces with embossed seals and venting holes but otherwise conforming to the standard dimensions of a commercial 12 mm Epon centerpiece were tested for durability and reproducibility of the measured sedimentation process. To this end, three 3D-printed centerpieces (that had previously been used twice) were subjected to 11 consecutive rounds of filling with 400 μL solutions of 0.5 mg/mL BSA in PBS in the sample sector and an equivalent volume of PBS in the reference sector, temperature equilibration, standard SV centrifugation at 50 000 rpm with absorbance and Rayleigh interference data acquisition, and stopping of the run, followed by disassembly and cleaning of the centerpieces (except for the first two replicates, which were just mixed in situ). Three conventional 12 mm Epon centerpieces containing the same sample were run alongside as controls.
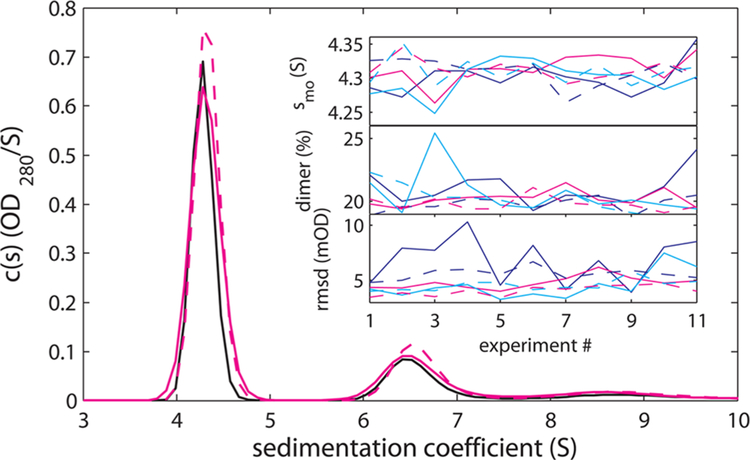
For all data from 3D-printed centerpieces, fits with excellent quality to the c(s) sedimentation model were achieved (see SI Figure S3 for an example data set from last run #11), with randomly distributed residuals close to the noise of data acquisition, and with the sedimentation coefficient distribution exhibiting the expected peak structure for BSA monomer and oligomers (Figure 2). More quantitatively, after application of calibration factors, we obtain an average monomer s-value at (4.30 ± 0.02) S from interference data and (4.33 ± 0.02) S from absorbance data. These results are in excellent agreement with the published value from the multilaboratory benchmark study (4.33 ± 0.03) S.40 Furthermore, there was no trend visible in the performance of any of the centerpieces, as judged by either monomer s-value, dimer percentage, or root-mean-square-deviation (rmsd) (Figure 2; for a detailed table of results, see SI Table 1). The dimer percentage may exhibit greater variability, but larger statistics would be required to establish this. In summary, this demonstrates the accuracy of the data from 3D-printed centerpieces and that MicroFine Green centerpieces can be reused many times. Other materials may show different repeatability; this has not been tested.
Figure 2.
Durability and accuracy of 3D-printed centerpieces in replicate SV experiment series with BSA. The main panel shows the c(s) distributions of centerpiece #2 from run #1 (black solid line) and run #11 (magenta solid line). For comparison, the sedimentation coefficient distribution of a conventional Epon centerpiece in run # 11 is shown as magenta dotted line. The insets shows results as a function of replicate number for 3D-printed centerpiece #1 (black), #2 (magenta), and #3 (cyan) for 3D-printed centerpieces (solid lines) and conventional Epon centerpieces for the monomer s-value (top inset), dimer percentage (middle inset), and rmsd of the c(s) fit (bottom inset). All data are based on absorbance data, except the signal fraction of the dimer, which is based on interference data as the absorbance based c(s) does not resolve the peaks well for all absorbance data.
Long-Column Sedimentation Velocity Centerpiece.
Since sedimentation velocity in essence measures the distance of migration with time, the precision of sedimentation coefficients will directly scale with the length of the solution column across which macromolecular migration can be observed. Conventional commercial centerpieces span a radial range from ≈5.75 cm to ≈7.2 cm (measured from the center of rotation), but a significantly shorter range is experimentally usable: (1) The maximal filling volumes are limited by the need to accommodate an air bubble above the solution so that the menisci are visible, and, in practice, by air locks that prevent complete filling of the sector. (2) Near the bottom of the solution column, a radial region is dominated by back-diffusion, which limits the observation of sedimentation boundaries.
We have relaxed the first limitation by creating a circular dome-shaped ceiling along the direction of the optical path, which leaves the sector edge at 5.75 cm at both sides but reduces the distance to the filling hole in the center by half (Figure 1B). This provides an additional 46 μL to accommodate a virtually unavoidable air bubble and allows filling the centerpiece higher. On the other end of the sector, we found that the bottom can be positioned further out to7.275 cm while still allowing a reliable seal against the windows. This moves the back-diffusion region to a radial range that is obscured from imaging, but in most cases, back-diffusion is excluded from the data analysis and therefore acquisition of this data is usually not required.34
Creating well-defined curved surfaces is trivial with 3D printing technology. Therefore, the bottom of the sectors can be fitted with a radial shape relative to the center of rotation (Figure 1C), as opposed to a flat bottom in the initial designs (and presumably present in commercial centerpieces). Since sedimentation is strictly radial, the curved bottom allows the process to conform to the rotational symmetry assumed in the one-dimensional sedimentation models. Even though the curvature at a distance of 72 mm from the center of rotation is small, the difference in radii between flat and curved bottom surface amounts to ≈0.02 mm in standard designs, which exceeds the nominal resolution of optical detection systems.
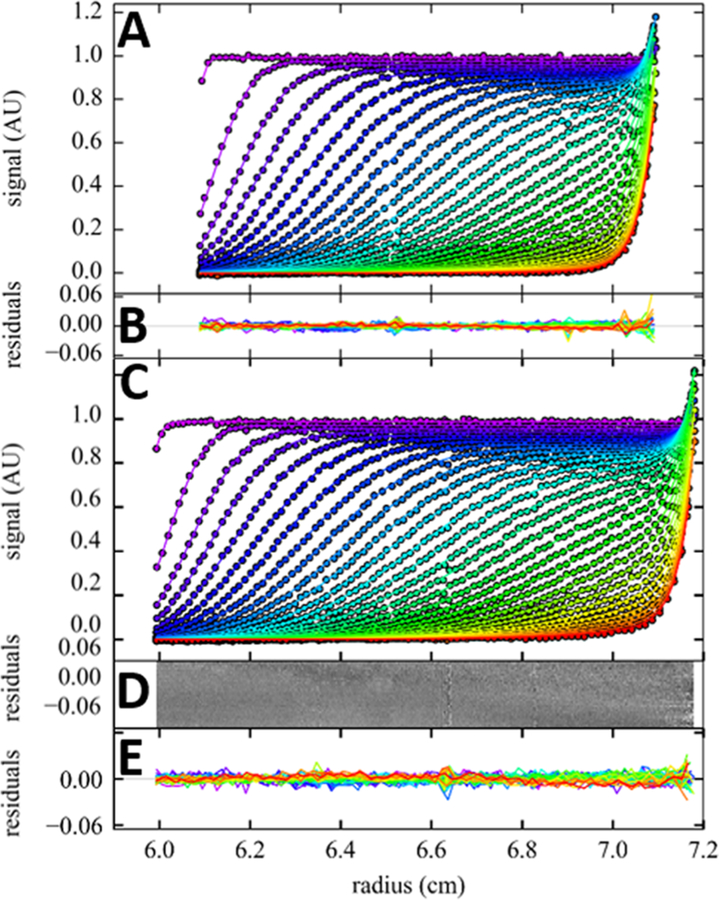
Figure 3 shows data from the long-column SV centerpiece in comparison with a commercial Epon centerpiece from the side-by-side sedimentation of the same lysozyme sample at 50 000 rpm. (Data demonstrating mechanical stability of this cell design at 60 000 rpm are shown in SI Figure S4.) The gain in solution column height from both ends can be easily discerned. Lysozyme, when dissolved in 10 mM acetate pH 4.6 with moderate ionic strength (100 mM NaCl), possesses repulsive charges that suppress self-association.41 Therefore, as a stringent test for both the purity of the protein as well as the quality of the centerpiece to allow unimpeded sedimentation, we fit the data with a single-species Lamm equation model. As shown in Figure 3, this results in an excellent fit with residuals that are close to normally distributed (H = 1.2%) and exhibit an rmsd of 0.0051 OD in the 3D-printed centerpiece, which compares favorably to the residuals with rmsd of 0.0053 OD with H = 3.7% in the commercial Epon centerpiece (for residuals histograms see SI Figure S5). Best-fit sedimentation coefficients are 1.906 (1.900–1.925) S in the 3D-printed centerpiece and 1.895 (1.882–1.912) S in the standard centerpiece. This highlights the fact that 3D-printed center-pieces are of sufficiently high quality for quantitative sedimentation studies. By design, the longer solution column offers ≈20% more data points, and the analysis exhibits a ≈17% reduced error for s-values.
Figure 3.
Comparison of SV data of lysozyme with single species fit for conventional commercial Epon and 3D-printed long-column SV centerpieces. Shown are absorbance optical data acquired at 50 000 rpm for lysozyme in a standard commercial 12 mm path length Epon centerpiece using 400 μL of sample (A,B) and in a long-column SV 3D-printed centerpiece using 500 μL sample (C,D,E). For clarity, only every 5th data point is shown, out of a total of 11 707 and 13 975 data points for the standard and long-column centerpiece, respectively. The solid lines are the best fit from a single species Lamm equation solution, with residuals overlays and bitmaps shown in panels B and D/E, respectively. For the standard centerpiece, the best-fit parameters are a sedimentation coefficient of 1.895 (1.882–1.912) S and apparent molar mass of 14.1 (13.7–14.6) kDa with rmsd of 0.0053 OD; for the long-column 3D-printed centerpiece, the bestfit parameters are a sedimentation coefficient of 1.906 (1.900–1.925) S and an apparent molar mass of 14.3 (13.8–14.7) kDa, with an rmsd of 0.0051 OD. The best-fit bottom position was at 7.214 cm for the conventional centerpiece and 7.303 cm for the 3D-printed centerpiece.
Narrow-Sector Centerpieces.
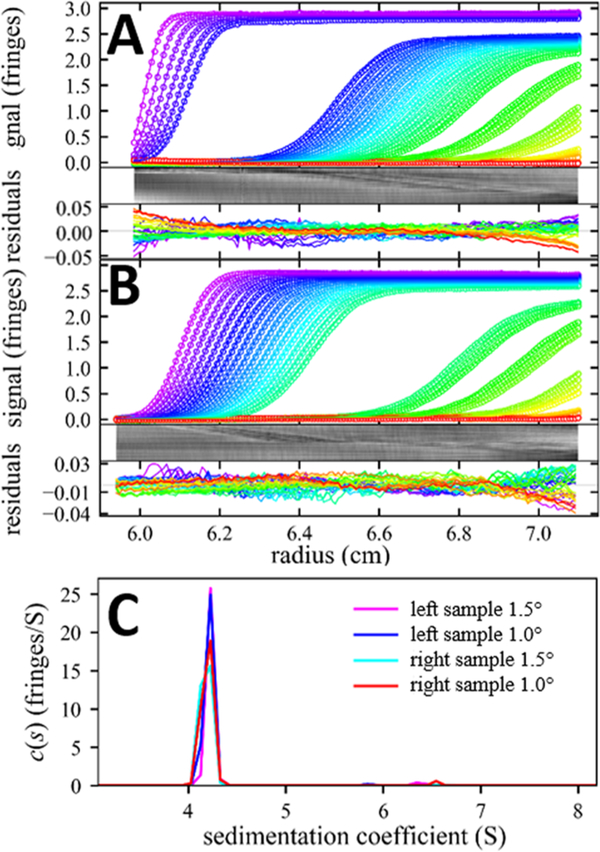
Another objective that can be pursued by 3D printing is the reduction of the sample volumes. This can be achieved in a straightforward way—without compromising solution column lengths and precision—through the reduction of the sector angle. Unfortunately, absorbance optical data acquisition in current commercial Beckman Coulter AUCs requires standard center-piece angles for reliable “delay calibration” (i.e., synchronization of rotor rotation and data acquisition timing) and is not compatible with narrow centerpieces. However, data acquisition angles in the Rayleigh interference optical system can be manually adjusted in the most widely used ProteomeLab and XLA/I ultracentrifuges. For the Rayleigh interference system, an essential requirement is that parallel beams can simultaneously pass through sample and reference solution. In order to enhance fringe contrast, narrow beams are desirable, which prior to the availability of pulsed lasers was achieved with slits in the lower window holder.42 Thus, narrow sectors are naturally suitable for the interference optical system. The ≈4 mm distance between the parallel beams poses an ultimate design constraint because the beams are not radial and must be accommodated through the entire length of the solution column. Both the 1.5° and the 1.0° designs shown in Figure 4A satisfy this requirement with the laser beam simultaneously centered in both sectors. In practice, the 1.0° centerpiece is the narrowest sector possible, considering that exposure angles can be adjusted only in 0.1° increments and allowing for the finite beam width.
Figure 4.
Other specialized designs of 3D-printed analytical ultracentrifugation centerpieces. Each row shows different centerpiece designs and a negative of the solution sectors, filling holes, and venting holes. (A) Narrow centerpieces with 1.5° (left) and 1.0° (middle) sectors, with a negative of the 1.0° design (right) featuring curved filling hole channels. (B) Thin cup centerpiece with 1.0 mm optical path length with integrated (half-height) window holders: centerpiece (left), negative of the sectors and channels (middle), and counterpart serving as half-height window holder and spacer. (C) Three-sector centerpiece for SV experiments with doubled capacity featuring two sample sectors on the left and right and a joint buffer reference in the middle: centerpiece (left), negative of sectors and channels (middle), and schematics of parallel beams of Rayleigh interference optics for detection of left sample (a) and right sample(b) both utilizing the same solvent reference.
A naïve implementation of this idea would in practice not allow the centerpiece to be filled with solution at the desired volumes because of trapped air bubbles. Therefore, the addition of venting channels is essential. Further, because of the mismatch in position between the fill ports in the conventional cell housing and the top of the narrow sectors, we created curved filling channels that allow bendable tubing to reach the bottom of the sectors. This prevents air locks that would form with straight filling holes that would confine the loading tip to the top of the sector and instead allows the solution to be dispensed from the bottom of the sector.
The narrow centerpieces with 1.5° and 1.0° angle reduce sample volumes to ≈55% or ≈40% of the volumes of conventional centerpieces. From the sedimentation of a monomeric BSA sample at 50 000 rpm, we obtained data that are indistinguishable from those of conventional center-pieces. As shown in the SI Figure S6, the fringe data could be fit with 0.26% rmsd relative to loading signal and close to normal residuals distribution (H = 1.5%), leading to an s-value of 4.23 S in the 1.5° centerpiece, rmsd of 0.25% and H = 1.0% with a 4.24 S peak in the 1.0° centerpiece, as compared with the conventional centerpiece with rmsd of 1.8% and slightly more non-normal residual distribution with H = 4.3% and an s-value of 4.20 S. Consistent with these excellent fits, the sedimentation coefficient distributions c(s) virtually super-impose (SI Figure S6).
Thin Centerpiece.
When working with highly concentrated solutions, as required for studying weak macromolecular interactions,43,44 it is imperative with any optical detection system to use short optical path length centerpieces because optical aberrations from refractive index gradients increase with the third power of the path length.45 In addition, the absorbance optical system has a limited linear range for which Lambert–Beer’s law is valid, and the Rayleigh optical interference system is limited in the strongest gradients that can be recorded without integral fringe offsets between neighboring radial points.46 For equilibrium experiments, very short pathlengths have been achieved previously by utilizing gaskets made of thin films to serve as centerpiece,47,48 but we found the loading process to be very problematic and were unable to create long solution columns suitable for SV with this approach.
A different strategy is based on the standard 3D printing design for SV centerpieces, for which the path length can be easily reduced. We have previously described the application of such centerpieces with optical pathlengths of 1.75 and 1.5 mm,7 but a further reduction would be desirable to extend the concentration range that can be studied. We found the filling holes can be reduced to a designed diameter of 0.6 mm when using 30 or 32 gauge needles (0.23–0.31 mm OD) instead of the wider gel loading tips. Further slight reduction is possible, in principle, but we found the compression of the material after torqueing can hinder passage of needles for filling. With the minimum feature size of 0.2 mm on either side of the filling hole, an optical path length of 1 mm was found to be practical. In practice, the embossed seals slightly increase the optical path length by 0.1–0.2 mm dependent on the torque.
It is obvious that such reduced path length centerpieces are thin discs that can break easily while mounting the cell assembly. This is exacerbated by the problem that the seal against the plug screw is limited to a small area and that too much pressure from the plug screw may also cause breakage. Also, since the plug screw diameter exceeds the centerpiece thickness, it may rest on the window holder rather than sealing the centerpiece, dependent on slight differences in window holder and centerpiece diameter. To avoid this problem, improve the stability, and achieve reliable seals, we integrated the centerpiece into two-sided, cup-shaped window holders (Figure 4B, left). These accommodate half of the window thickness, while the other half of the windows are held in place by 3D-printed counterparts that resemble shallow window holders integrated into suitable spacers (Figure 4B, right). In this design, the added thickness of the centerpiece provides stability and normal seal of the filling ports by the plug screws.
Designed for samples at high concentration, we have equipped these centerpieces with long solution columns as described above, to increase the solution column length where back-diffusion-free sedimentation can be observed. As with the narrow centerpieces, air vents facilitate filling with well-defined solution volumes. For highest quality data, we note that a residual small air pocket is necessary to create a well-defined meniscus.
Using these centerpieces, we have been able to carry out SV experiments with concentrated protein solutions in excess of 100 mg/mL (manuscript in preparation). As a test for its performance, we carried out a SV experiment with the same lysozyme preparation as used above. Because of the short path length, the concentration was raised to 10 mg/mL, and the single-species analysis was extended to include nonideality parameters.49 As can be discerned from Figure 5, an excellent fit was achieved with residuals amounting to only 0.23% of the loading signal. The best-fit sedimentation coefficient of (1.96 ± 0.02) S and the best-fit apparent molar mass of 14.3 kDa are close to the value measured in 12 mm path length cells (above), and they compare well with those measured side-by-side with the same sample in a control experiment in standard 3 mm Epon centerpieces (1.88 S and 14.7 kDa, respectively).
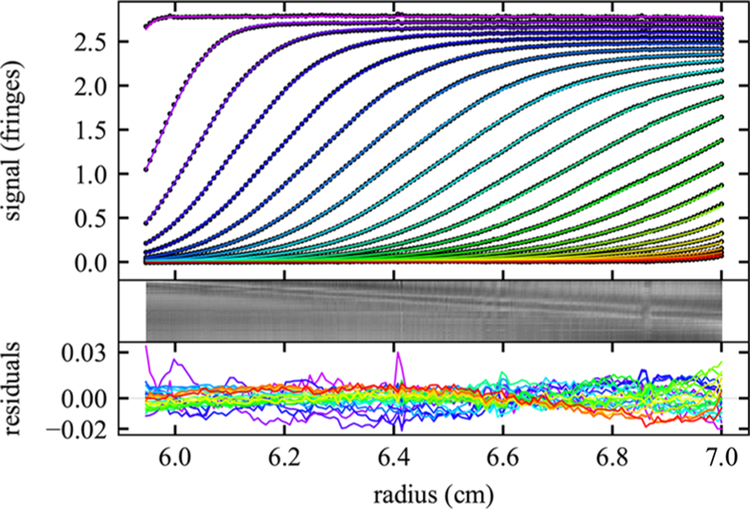
Figure 5.
SV data from 3D-printed 1 mm path length ceneterpiece for 10 mg/mL lysozyme with single nonideal species fit. Rayleigh interferometric SV data at 50 000 rpm, 20 °C (circles) are shown for every 20th data point of each 2nd scan. The solid lines are the best fit from a single nonideal species Lamm equation solution, resulting in a sedimentation coefficient (at infinite dilution) of 1.96 S, and an apparent molar mass of 14.3 kDa. Residuals are shown as bitmap and overlay; they have an rmsd of 0.0065 fringes (0.23% of the loading signal) and are nearly Gaussian (H = 0.9%).
Three-Sector Centerpiece.
As mentioned above, the Rayleigh interference optical detection allows the use of narrow sectors. As a consequence, the centerpiece can easily accommodate another sector (Figure 4C). With a third sector, two samples can be studied if they share the same solvent reference. In the present design, the bottom of both outer sectors will be slightly obscured by the window holder, which could be remedied by a shift of the bottom to a smaller radius by ≈1 mm or by modification of the window holder. However, this would not be desirable in practice for most samples exhibiting back-diffusion, which is customarily excluded from the analysis and therefore does not need to be recorded, as illustrated for the long-column SV centerpiece above.
The basic idea for data acquisition is depicted on the right of Figure 4C: In configuration (b), light passes through the right sample sector and the middle solvent sector as in a standard centerpiece. In configuration (a), the orientation of sample and reference is reversed, but this is of no further consequence for Rayleigh interferometry, other than a negative of the macromolecular concentrations is reported, which can be remedied trivially by multiplying all data by –1. Naturally, the exposure angle for the configurations (a) and (b) will be slightly different, but the difference will be constant and equal for all cells. Even though the most recent model of Beckman AUCs does not allow user control of exposure angles, the most commonly used ProteomeLab and older instruments do. Therefore, the extra channels can be recorded simply by periodically switching between the known two sets of angles. The particular time points of switching are irrelevant, and intermittent lack of scan data is without consequence, as long as data in both configurations span the entire duration of the experiment.34 Suitable software for automatic switching will be described elsewhere.
We have built centerpieces in this design with either 1.5° sectors or 1.0° sectors. Figure 6A,B shows data from BSA sedimenting at 50 000 rpm in such a three-channel cell with1.5° sectors. In this particular experiment, the angles were switched manually 11 times during the experiment. The fits to the recorded sedimentation processes are of excellent quality (Figure 6C), similar in both sectors and similar to those obtained in three channel centerpieces with 1.0° sectors.
Figure 6.
Sedimentation of BSA monomer in three-channel 3D-printed centerpieces with 1.5° sectors. Raw data and c(s) fit of BSA monomer sedimenting at 50 000 rpm (only every 5th scan and 10th data point shown). (A) Configuration with left sample shown as (a) in the schematics of Figure 4C, leading to an rmsd of 0.0105 fringes and an H-value of 1.4%; (B) Configuration with right sample as shown (b) in the schematics of Figure 4C, leading to an rmsd of0.0090 fringes and an H-value 0.2%. (C) Sedimentation coefficient distribution c(s) obtained from the data shown in A and B, as well as analogous experiments in three-channel 3D-printed centerpieces with1.0° sectors run side-by-side in the same rotor.
CONCLUSIONS
Open-source 3D printing has the potential to revolutionize scientific instrumentation as illustrated in areas ranging from labware,31 to optical tools,50 microfluidics,51 and devices in analytical chemistry.27,52,53 Beyond resource reallocation due to inexpensive manufacturing, a significant additional benefit is rapid prototyping of customized equipment that is optimized for specific applications. Additive manufacturing can implement designs that are hard or impossible to achieve through traditional molding or machining. Current freeware design software such as OpenSCAD,30 parametrized open-source designs,50 and open-source printers and commercial 3D printing services allow rapid turnaround from conception to application.
In our present work on AUC centerpieces, we demonstrate how 3D printing creates new experimental opportunities for various advanced applications. The recent discovery that centerpieces 3D printed in various materials can withstand the mechanical stresses at 200 000–300 000g while providing a seal with the sapphire or quartz windows for the sample solutions against high vacuum in the rotor chamber was unexpected.28 After gaining more experience with such centerpieces, as described in the present work, we found such centerpieces can be durable for many experiments and confirmed that they allow macromolecular sedimentation to be observed at a quality on par with commercial standard centerpieces. Therefore, we believe 3D printing of AUC centerpieces will offer similar advantages as 3D printing of other scientific tools and usher in a renewed period of experimentation in experimental configurations in AUC. Recent interest has arisen already, for example, in centerpieces capable of layering solutions,54 and in very short path length centerpieces to study highly concentrated solutions.7
One limitation of the present study is that, for practical reasons, we did not determine at which number of experiments they ultimately brake. This may be revealed in future long-term studies. 3D-printed centerpieces may still be far less durable than commercial Epon centerpieces, but their inexpensive fabrication cost may compensate for this, while the new experimental opportunities can offer significant advantages. Another limitation of the present study is the focus on one particular proprietary epoxy-like material that we found attractive due to the ease of forming seals and small features. However, as shown by successful prototypes in the present and previous work,28 similar centerpieces could be fabricated using different printing technologies and from other plastics or even 3D-printed metal (notably a 14 K gold centerpiece would now be at a cost less than twice the commercial epoxy resin centerpiece).
In the present communication, we have presented four novel centerpiece designs optimized for different objectives. Three designs optimize along three spatial dimensions, and features may be combined as needed on the basis of experimental constraints. First, along the radial dimension, the long-column SV centerpiece exploits the observation that back-diffusion from the bottom of the sector is usually at least partially excluded from data analysis34,55 and therefore may be moved to higher radii into a location partially obscured from detection without detrimental effects on the information content of the experiment. This is combined with the elevation of the top of the sector, including a new dome shape ceiling and venting hole that facilitate loading. This allows unimpeded sedimentation to be monitored for longer distances, which directly translates into higher precision and higher resolution for SV analysis. Second, along the axis of optical detection, we have presented a new design with 1 mm optical path length, which we found needs to include part of the window holder in order to adapt to the cell housing with its plug screw for sealing the solution columns. Such short optical pathlengths are a key strategy for extending SV to highly concentrated solutions because optical aberrations from refractive index gradients scale with the third power of the optical path length.7,45 Third, in the dimension of the rotation angle, we have demonstrated that significant savings in the sample volumes can be achieved with narrow centerpieces. With current Beckman-Coulter XLA/I and ProteomeLab ultracentrifuges, narrower center-pieces are not compatible with required angular tolerances of absorbance optical detection, but they can be seamlessly used with Rayleigh interference optical detection. In fact, the design is reminiscent of the window holders with narrow slits that were in use to enhance fringe contrast prior to the advent of pulsed lasers. Since higher concentrated samples often require interference optical detection, narrow centerpieces have significant practical potential. Combination of all features into a long, narrow, and thin centerpiece would reduce the sample volume to ≈17 μL.
The fourth design doubles the capacity of SV experiments when using interferometric detection. It is based on the observation that multiple narrow sectors can be easily accommodated in the standard centerpiece dimensions. The current data provide a proof of principle for this approach. We have previously introduced a strategy to double the sample capacity for absorbance optical detection using intensity data and accommodating position-dependent variation in photo-multiplier response using time-invariant noise models.56 In principle, intensity data acquisition from each sector could triple AUC capacity. Unfortunately, both methods cannot currently be combined due to the required angular tolerances for the absorbance system. However, there is no fundamental reason this could not be accomplished in future instruments.
In conclusion, we believe the present work shows that 3D-printed centerpiece designs can substantially expand the limits of current AUC, in areas of throughput, detection limits, and precision, in a way that is synergistic to the development of new instrumentation, detection systems, theoretical sedimentation models, and data analysis.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Shi-Chia Tso for the purification of the monomeric BSA sample. This work was supported by the Intramural Research Programs of the National Institute of Biomedical Imaging and Bioengineering National Institutes of Health.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.9b00202.
Detailed analytical ultracentrifugation methods; detailed results from replicate series, raw data and fits from 3D-printed long-column and narrow centerpieces (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Svedberg T; Pedersen KO The Ultracentrifuge; Oxford University Press: London, 1940. [Google Scholar]
- (2).Schachman HK Ultracentrifugation in Biochemistry; Academic Press: New York, 1959. [Google Scholar]
- (3).Svedberg T; Rinde HJ Am. Chem. Soc 1923, 45 (4), 943–954. [Google Scholar]
- (4).Van Holde KE; Hansen JC Chemtracts Biochem. Mol. Biol 1998, 11, 933–943. [Google Scholar]
- (5).de Duve C Nobel Lecture; Presented at Université Catholique de Louvain, Belgium and The Rockefeller University, New York, NY, December 12, 1974; pp 152–170. [Google Scholar]
- (6).Zhao H; Mayer ML; Schuck P Anal. Chem 2014, 86 (6), 3181–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Chaturvedi SK; Ma J; Brown PH; Zhao H; Schuck P Nat. Commun 2018, 9 (1), 4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Schuck P; Zhao H Sedimentation Velocity Analytical Ultra-centrifugation: Interacting Systems; CRC Press: Boca Raton, FL, 2017. [Google Scholar]
- (9).Harding SE; Rowe AJ Biochem. Soc. Trans 2010, 38 (4), 901–907. [DOI] [PubMed] [Google Scholar]
- (10).Sousa AA; Hassan SA; Knittel LL; Balbo A; Aronova MA; Brown PH; Schuck P; Leapman R D. Nanoscale 2016, 8 (12), 6577–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Berkowitz SA; Philo JS In Biophysical Characterization of Proteins in Developing Biopharmaceuticals; Houde DJ, Berkowitz SA, Eds.; Elsevier: Amsterdam, 2015; pp 211–260. [Google Scholar]
- (12).Krayukhina E; Uchiyama S; Nojima K; Okada Y; Hamaguchi I; Fukui KJ Biosci. Bioeng 2013, 115, 104–110. [DOI] [PubMed] [Google Scholar]
- (13).Cölfen H; Tirosh S; Zaban A Langmuir 2003, 19 (26), 10654–10659. [Google Scholar]
- (14).Mehn D; Rio-Echevarria IM; Gilliland D; Kaiser M; Vilsmeier K; Schuck P; Wohlleben W NanoImpact 2018, 10, 87–96. [Google Scholar]
- (15).Schuck P; Zhao H; Brautigam CA; Ghirlando R Basic Principles of Analytical Ultracentrifugation; CRC Press: Boca Raton, FL, 2015. [Google Scholar]
- (16).Pickels EG Rev. Sci. Instrum 1942, 13 (1942), 426. [Google Scholar]
- (17).Boestad G; Pedersen KO; Svedberg T Rev. Sci. Instrum 1938, 9 (11), 346–353. [Google Scholar]
- (18).Bendet IJ; Smith CE; Lauffer MA Arch. Biochem. Biophys 1960, 88 (2), 280–286. [Google Scholar]
- (19).Yphantis DA; Waugh DF J. Phys. Chem 1956, 60, 630–635. [Google Scholar]
- (20).Hersh RT; Schachman HK J. Am. Chem. Soc 1955, 77 (20), 5228–5234. [Google Scholar]
- (21).Vinograd J; Bruner R; Kent R; Weigle J Proc. Natl. Acad. Sci. U. S. A 1963, 49 (6), 902–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Yphantis DA Ann. N. Y. Acad. Sci 1960, 88 (3), 586–601. [DOI] [PubMed] [Google Scholar]
- (23).Pearce JM Science 2012, 337 (6100), 1303–1304. [DOI] [PubMed] [Google Scholar]
- (24).Hietanen I; Heikkinen ITS; Savin H; Pearce JM HardwareX 2018, 4, No. e00042. [Google Scholar]
- (25).Hoang T; Moskwa N; Halvorsen K PLoS One 2018, 13 (4), No. e0195907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Winters BJ; Shepler D HardwareX 2018, 3, 62–81. [Google Scholar]
- (27).Fichou D; Morlock GE Anal. Chem 2017, 89 (3), 2116–2122. [DOI] [PubMed] [Google Scholar]
- (28).Desai A; Krynitsky J; Pohida TJ; Zhao H; Schuck P PLoS One 2016, 11 (8), No. e0155201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).MacGregor IK; Anderson AL; Laue TM Biophys. Chem 2004, 108 (1–3), 165–185. [DOI] [PubMed] [Google Scholar]
- (30).Kintel M. OpenSCAD. http://www.openscad.org/.
- (31).National Institutes of Health 3-D print exchange. http://3dprint.nih.gov/.
- (32).Ghirlando R; Balbo A; Piszczek G; Brown PH; Lewis MS; Brautigam CA; Schuck P; Zhao H Anal. Biochem 2013, 440 (1), 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zhao H; Ghirlando R; Piszczek G; Curth U; Brautigam CA; Schuck P Anal. Biochem 2013, 437 (1), 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Schuck P Sedimentation Velocity Analytical Ultracentrifugation: Discrete Species and Size-Distributions of Macromolecules and Particles; CRC Press: Boca Raton, FL, 2016. [Google Scholar]
- (35).Zhao H; Brown PH; Balbo A; Fernandez-Alonso MC; Polishchuck N; Chaudhry C; Mayer ML; Ghirlando R; Schuck P; et al. Macromol. Biosci 2010, 10 (7), 736–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Brautigam CA Methods Enzymol. 2015, 562, 109–133. [DOI] [PubMed] [Google Scholar]
- (37).Johnson ML Anal. Biochem 1992, 206, 215–225. [DOI] [PubMed] [Google Scholar]
- (38).Ma J; Zhao H; Schuck P Anal. Biochem 2015, 483 (1), 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Chaturvedi SK; Ma J; Zhao H; Schuck P Nat. Protoc 2017, 12 (9), 1777–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Zhao H; Ghirlando R; Alfonso C; Arisaka F; Attali I; Bain DL; Bakhtina MM; Becker DF; Bedwell GJ; Bekdemir A; et al. PLoS One 2015, 10 (5), No. e0126420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Kuehner DE; Heyer C; Ramsch C; Fornefeld UM; Blanch HW; Prausnitz JM Biophys. J 1997, 73 (6), 3211–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Richards EG; Schachman HK J. Phys. Chem 1959, 63 (10), 1578–1591. [Google Scholar]
- (43).Chaturvedi SK; Schuck P AAPS J. 2019, 21 (3), 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Chaturvedi SK; Sagar V; Zhao H; Wistow G; Schuck PJ Am. Chem. Soc 2019, 141, 2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Forsberg R; Svensson H Opt. Acta 1954, 1 (2), 90–93. [Google Scholar]
- (46).Schilling K; Krause F PLoS One 2015, 10 (3), No. e0120820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Braswell EH J. Phys. Chem 1972, 76 (26), 4026–4030. [Google Scholar]
- (48).Minton AP; Lewis MS Biophys. Chem 1981, 14 (4), 317–324. [DOI] [PubMed] [Google Scholar]
- (49).Solovyova A; Schuck P; Costenaro L; Ebel C Biophys. J 2001, 81 (4), 1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Zhang C; Anzalone NC; Faria RP; Pearce JM PLoS One 2013, 8 (3), No. e59840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Bhattacharjee N; Urrios A; Kang S; Folch A Lab Chip 2016, 16 (10), 1720–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Lambert A; Valiulis S; Cheng Q ACS Sensors 2018, 3 (12), 2475–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Cardoso RM; Mendonça DMH; Silva WP; Silva MNT; Nossol E; da Silva RAB; Richter EM; Muñoz RAA Anal. Chim. Acta 2018, 1033, 4 9–57. [DOI] [PubMed] [Google Scholar]
- (54).Schneider CM; Haffke D; Cölfen H Anal. Chem 2018, 90, 10659. [DOI] [PubMed] [Google Scholar]
- (55).Schuck P Eur. Biophys. J 2010, 39 (8), 1261–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Kar SR; Kingsbury JS; Lewis MS; Laue TM; Schuck P Anal. Biochem 2000, 285 (1), 135–142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.