Abstract
The transfer of protein-encoding genetic information from DNA to RNA to protein, a process formalized as the “Central Dogma of Molecular Biology”, has undergone a significant evolution since its inception. It was amended to account for the information flow from RNA to DNA, the reverse transcription, and for the information transfer from RNA to RNA, the RNA-dependent RNA synthesis. These processes, both potentially leading to protein production, were initially described only in viral systems, and although RNA-dependent RNA polymerase activity was shown to be present, and RNA-dependent RNA synthesis found to occur, in mammalian cells, its function was presumed to be restricted to regulatory. However, recent results, obtained with multiple mRNA species in several mammalian systems, strongly indicate the occurrence of protein-encoding RNA to RNA information transfer in mammalian cells. It can result in the rapid production of the extraordinary quantities of specific proteins as was seen in cases of terminal cellular differentiation and during cellular deposition of extracellular matrix molecules. A malfunction of this process may be involved in pathologies associated either with the deficiency of a protein normally produced by this mechanism or with the abnormal abundance of a protein or of its C-terminal fragment. It seems to be responsible for some types of familial thalassemia and may underlie the overproduction of beta amyloid in sporadic Alzheimer’s disease. The aim of the present article is to systematize the current knowledge and understanding of this pathway. The outlined framework introduces unexpected features of the mRNA amplification such as its ability to generate polypeptides non-contiguously encoded in the genome, its second Tier, a physiologically occurring intracellular polymerase chain reaction, iPCR, a “Two-Tier Paradox” and RNA “Dark Matter”. RNA-dependent mRNA amplification represents a new mode of genomic protein-encoding information transfer in mammalian cells. Its potential physiological impact is substantial, it appears relevant to multiple pathologies and its understanding opens new venues of therapeutic interference, it suggests powerful novel bioengineering approaches and its further rigorous investigations are highly warranted.
Keywords: RNA-dependent amplification of mammalian mRNA, Physiologically occurring intracellular PCR, iPCR, RNA-dependent RNA polymerase, RdRp, Chimeric RNA, Sense-strand RNA, Antisense-strand RNA
1. Introduction
RNA-dependent RNA synthesis and the corresponding enzymatic activity, RNA-dependent RNA polymerase (RdRp), were discovered in studies of mengovirus and polio virus [1–6]. Eventually, with the discovery of the RNA-dependent DNA synthesis and the enzyme associated with this process [7,8], it became clear that among RNA viruses, retroviruses utilize reverse transcriptase for their genomic information transfer whereas almost all other RNA viruses encode and utilize RdRp for this purpose [9,10]. The exceptions are viruses that do not encode their own RNA-dependent RNA polymerase but rely on RdRp activity of either their partners (helper viruses within the same cells) or their hosts. The latter category includes plant viroids and hepatitis delta virus, HDV [11]. These viruses do not encode an RdRp yet they undergo a robust RNA replication once inside the host cells [9–11]. These findings imply that both groups utilize cellular RNA-dependent RNA polymerase activities: viroids in plant cells and HDV in mammalian cells. The presence of RdRp in plant cells is well documented [12–14]. The ability of RdRp-deficient HDV to vigorously replicate inside its mammalian hosts establishes the occurrence and functionality of RdRp activity in mammalian cells. Indeed, it became clear that RdRp activity, apparently in a non-conventional form, is constitutively present in most, if not in all, mammalian cells [15–21]; (see more in section 9 below). Because non-conventional mammalian RdRp activity was shown to produce short transcripts, because of its apparent involvement in RNA interference phenomena, and because double-stranded RNA is known to trigger cellular responses leading to its degradation, it was generally assumed that the function of RdRp activity in mammalian cells is restricted to regulatory. However, at the same time, an RdRp activity [18] capable of generating complete antisense RNA complements of mRNAs, as well as its products [22], were discovered in mammalian cells undergoing terminal differentiation. Moreover, observations of widespread synthesis of antisense RNA initiating at the 3’poly (A) of mRNAs in human cells [17] suggested an extensive cellular utilization of mammalian RdRp activity. These results led to the development of a model of RdRp-facilitated and antisense RNA-mediated amplification of mRNA in mammalian cells [22–25]. A major prediction of this model is generation of a chimeric RNA composed of covalently joined antisense and sense strands of the same molecule and uniquely defined by self-priming of the antisense molecule and the extension of its 3’terminus into the sense, mRNA, strand. Recently, this prediction was borne out by the detection of such chimeric RNA molecules in terminally differentiating cells and in cells overproducing the components of the extracellular matrix [23,24]. Moreover, a putative end product of mRNA amplification, which translates into polypeptides indistinguishable from the translation product of genome-originated mRNA, has been identified at levels that are unprecedented for conventional mRNA transcripts [23]. Below, we discuss in depth the current understanding of RNA-dependent mammalian mRNA amplification processes, their potentially multiple outcomes, possible regulation, and prospective significance. We also describe in detail novel experimental bioengineering designs suggested by the understanding of the amplification processes, and consider future directions of investigations into mechanisms underlying this powerful pathway of mammalian gene expression.
2. mRNA Amplification Process May Preserve the Protein-Encoding Information Content of a Conventional mRNA
The model for RdRp-facilitated, antisense RNA-mediated amplification of mammalian mRNA described below was developed in studies of the extraordinarily massive production of alpha and beta globin chains in murine cells/tissue undergoing erythroid differentiation [22,23]. This model is diagrammed in Figure 1 and can be summarized as follows. The amplification process starts with transcription of the antisense complement from a conventional mature spliced mRNA template, initiated at its 3’poly (A), possibly with the help of a uridylated protein as in some viral [26] systems (Figure 1, Step 1). Such complete antisense transcripts containing 5’poly (U) and a full complement of mature spliced murine globin mRNA were indeed detected, cloned and sequenced [22]. Generation of a complete antisense transcript requires the presence of an eligible RNA template and a compatible polymerase activity. The enzymatic activity central to the mRNA amplification process is RdRp. In mammalian cells, it was first detected in anemic rabbit reticulocytes [18]. It is now apparent that it operates in most, if not all, mammalian cells [15–21]. However, the outcomes are qualitatively different in different types of cells. In cells undergoing erythroid differentiation, full-length antisense transcripts of mature globin mRNA are produced [22], whereas in other “normal” cell types only short antisense transcripts are generated [16,17]. It appears that under regular circumstances mammalian RdRp activity lacks a processivity component, and that in special circumstances requiring a substantial overproduction of specific proteins (i.e. hemoglobin in erythroid differentiation, extracellular matrix proteins during connective tissue deposition) or in some pathologies, what is induced is not RdRp activity, but rather its processivity conferring co-factor. A proof of concept for the notion of such RdRp co-factor that allows production of complete antisense transcripts is discussed in section 9 below.
Figure 1: Projected steps in RdRp-facilitated and antisense RNA-mediated amplification of mammalian mRNA with chimeric RNA end product retaining the intact coding capacity of conventional mRNA.
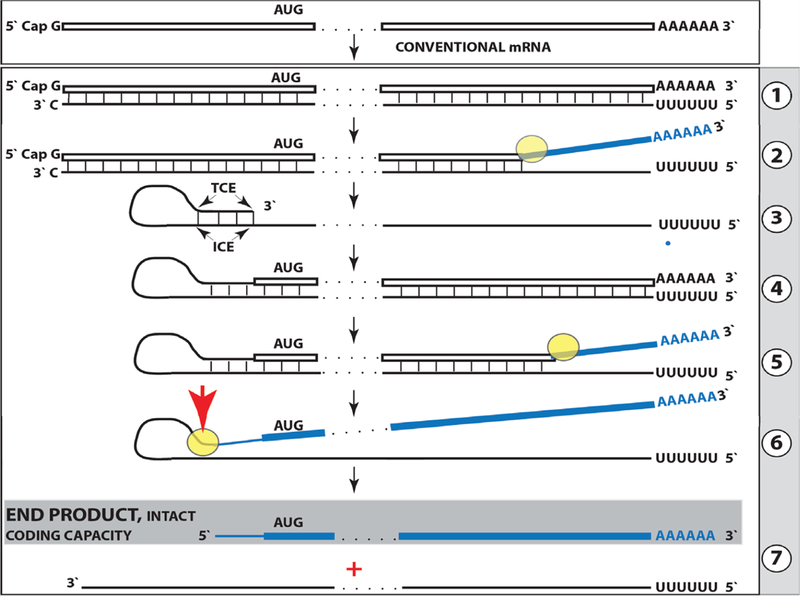
Top panel: conventional, genome-originated mRNA molecule. Bottom panel: projected stages of antisense RNA-mediated mRNA amplification. Boxed line – sense strand RNA. Single line – antisense strand RNA. “AUG” – functional translation initiation codon (could be other than “AUG”). “TCE”– 3’-terminal complementary element; “ICE”– internal complementary element, both on the antisense RNA strand. Yellow circle – helicase/modifying activity complex. Blue lines (both single and boxed) – RNA strand modified and separated from its complement by a helicase complex. Red arrowhead – position of cleavage of the chimeric intermediate. Step 1 : synthesis of antisense strand; step 2: strand separation; step 3: folding of antisense strand into self-priming configuration; step 4: extension of self-primed antisense RNA; step 5: strand separation; step 6: cleavage of the chimeric intermediate; stage 7: end-products of amplification. Note that chimeric RNA end product retains the intact coding capacity of conventional mRNA.
The resulting double-stranded sense/antisense structure is then separated (Figure 1, Step 2) into single-stranded molecules by a helicase activity that appears to mount the poly(A) segment of the 3’poly(A)-containing strand (the sense-oriented strand) of the double helical structure [23]. As helicase activity proceeds in 3’ to 5’ direction along this strand, it modifies some of its nucleotides. On average, every fifth nucleotide is modified in the process and it appears that only adenosine and guanosine, purines but not pyrimidines, are modified ([23]; Figure 1, Step 2). The nature, possible functions, and consequences of these modifications are discussed in section 8 below. The 5’ poly (U)-containing antisense strand remains unmodified during and after the separation [23], this being essential for the production of a new sense strand since, as discussed in section 8 below, modifications could interfere with complementary interactions required in this process. It could be argued that the occurrence of double-stranded RNA may trigger a dsRNA cellular response resulting in its degradation but this apparently does not happen. This issue, along with other regulatory aspects, is discussed in section 9 below.
As for the template eligibility, the only major prerequisite for a potential RNA template appears to be the presence of the poly (A) segment at its 3’ terminus [17,22,23]. The vast majority of mammalian mRNA species contains 3’-terminal poly (A) segments. The notion that many, or possibly most, of them could be eligible templates for RdRp was suggested in earlier studies [22]. Subsequent observations by Kapranov et al. [17] showed a widespread synthesis of antisense RNA initiating, apparently indiscriminately, at the 3’ poly (A) of diverse mRNAs in human cells. This, apparently undiscerning, RdRp template eligibility of the bulk of mammalian mRNA species raises questions with regard to mechanisms underlying the manifestly stringent specificity of the mRNA amplification process as seen, for example, in erythropoietic differentiation [22,23]. The specificity of the amplification process appears to be determined at the 3’ terminus of an antisense transcript by its ability or inability to support production of a complementary sense strand molecule [22–25].
The generation of a sense strand on an antisense template occurs via extension of the 3’ terminus of a self-primed antisense template and requires the presence within the antisense transcript of two spatially independent complementary elements whose occurrence in antisense globin molecules was found to be evolutionary conserved across mammalian species [27]. One of these is the strictly 3’-Terminal Complementary Element (TCE), the other is the Internal Complementary Element (ICE). These elements (Figure 1, Step 3) must be complementary to an extent sufficient to form a priming structure but may contain both mismatches and unconventional G/U pairings. Generation of a sense strand also requires the thermodynamic feasibility, enhanced/enabled by the occurrence of these two complementary and topologically compatible elements, of the antisense strand folding into a self-priming configuration. The requirement for terminal localization of the TCE appears to be stringent; an overhang of even a single nucleotide diminishes self-priming [27].
Provided that a self-priming structure is formed, the 3′ end of the folded antisense strand is extended by RdRp into a sense-orientation molecule terminating with the poly (A) at the 3′end (Figure 1, Step 4), thus generating a hairpin-structured chimeric intermediate consisting of covalently joined sense and antisense strands. The double-stranded portion of the resulting structure is separated by a helicase activity invoked above, which mounts the 3′poly(A) of a newly synthesized sense strand component of the chimeric intermediate and proceeds along this strand in the 5’ direction modifying the molecule as it advances (Figure 1, Step 5). When the helicase activity reaches a single stranded portion of the hairpin structure, it, or an associated activity, cleaves the molecule either within the TCE, at a TCE/ICE mismatch, or immediately upstream of the TCE; a cleavage was shown to occur between the 5’ hydroxyl group and the 3’ phosphate ([22,23]; red arrow, Figure 1, Step 6).
Strand separation, in conjunction with the cleavage, produces two single-stranded molecules (Figure 1, Step 7) one of which is a chimeric mRNA, the functional mRNA end product of the amplification and the basis for defining this pathway as the “chimeric”. The chimeric nature of this end product is due to the presence at its 5’ end of a 3’-terminal segment of the antisense strand consisting, depending on the site of cleavage of the chimeric intermediate, of either the entire TCE or a portion thereof covalently attached, in a 5’ to 3’ orientation, to the 5’-truncated sense strand. This chimeric molecule is modified and 3’polyadenylated; it cannot be further amplified, because its antisense complement would be lacking the TCE, but can be translated into a conventional mRNA-encoded polypeptide [23]. Such RNA end product of amplification was putatively identified for murine globin-encoding mRNA and is described in detail and discussed in section 8 below. Figure 1 illustrates the situation whereby the ICE of the antisense strand is located within its segment corresponding to the 5’-untranslated region (5’UTR) of a genome-encoded mRNA. Consequently, the chimeric end product contains the entire protein coding region of a conventional mRNA and can be translated into the original, conventional mRNA-encoded, polypeptide [23]. Another single-stranded product of step 7, Figure 1 is the antisense RNA molecule. It is truncated at the 3’terminus and lacks either a portion of or the entire TCE, depending on the position of the cleavage site; its suitability for further amplification is discussed in sections 3 and 9 below. Such antisense beta-globin RNA, cleaved at the three-nucleotide TCE/ICE mismatch (Figure 4, bottom panel, D), was detected, cloned and sequenced and found to be a predominant variant of β globin antisense RNA in murine erythroid cells [22].
The occurrence of RNA-dependent amplification of mammalian mRNA was initially shown for globin-encoding mRNA at rather extreme circumstances of terminal erythroid differentiation [22,23], in cells destined to die in just a few days. For this reason, it could be argued that the amplification mechanism might be in restricted use and not relevant to more conventional tissue formation and functions. Recently, however, the evidence for RNA-dependent amplification was obtained for mRNAs encoding all three constituent chains of laminin 111 in a tissue producing extraordinarily large amounts of extracellular matrix proteins (Figure 2), indicating that such an amplification process may operate also during normal development when the production of large quantities of certain polypeptides is required [24].
Figure 2: Detection of chimeric junction sequences containing antisense and sense segments of mRNA encoding α1, β1 and γ1 chains of laminin and the projected pathways of their generation.
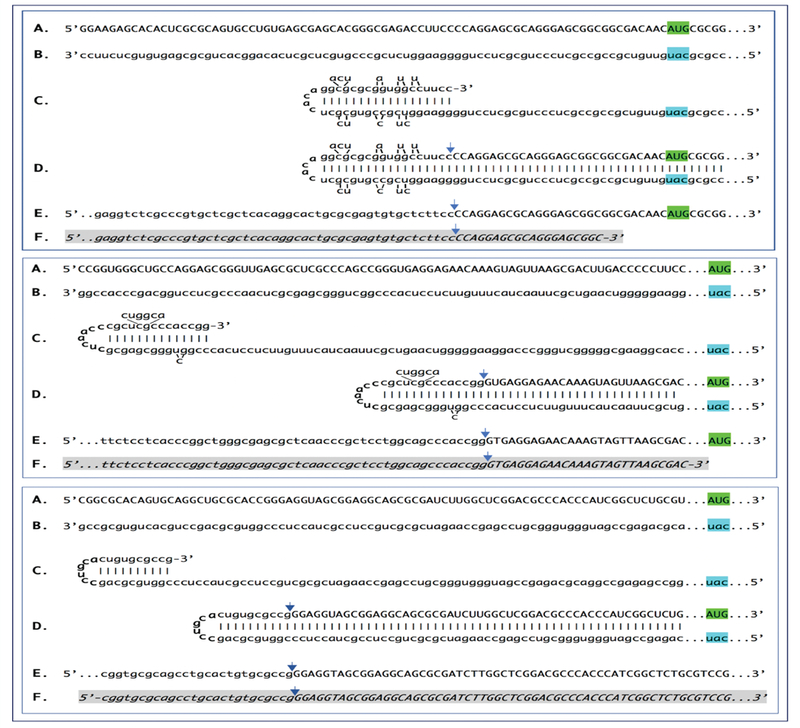
Chimeric junction sequences for mRNAs molecules encoding α1, β1, and γ1 chains of laminin were detected in cells of the extracellular matrix-generating Engelbreth-Holm-Swarm (EHS) mouse tumor, best known for producing extraordinarily large amounts of “Matrigel”. Cytoplasmic RNA was isolated from tumor cells harvested ten days after tumor implantation, and, following depletion of ribosomal RNA, used to generate sequencing libraries. Libraries were sequenced on an Illumina Next500 instrument. Sequencing data were converted into blast databases and analyzed by blasting with chimeric reference sequences. Sequences of interest were extracted from raw data and analyzed. Top panel: α1 laminin sequences; middle panel: β1 laminin sequences; bottom panel: γ1 laminin sequences. Uppercase letters – nucleotide sequence of the sense strand; lowercase letters – nucleotide sequence of the antisense strand. Highlighted in green – “AUG” translation initiation codon on the sense strand; highlighted in blue – “uac” complement of translation initiation codon on the antisense strand. In italics and highlighted in grey – detected chimeric fragments. Blue arrows: position of antisense/sense junctions. A: 5’ terminus of conventional laminin mRNA. B: antisense complement of the 5’ terminus of conventional laminin mRNA. C: folding of the antisense strand into self-priming configuration. D: extension of self-primed antisense strand into sense-oriented sequence. E: projected chimeric junction sequence. F: detected chimeric junction sequence. Note that the priming occurs within the segment of antisense strand corresponding to the 5’UTR of mRNA, thus preserving the coding capacity of amplified mRNA.
Among RNA molecules described above, the best, if not the only, definitive identifier of the occurrence of mRNA amplification is a chimeric junction between sense and antisense components. Although the chimeric end product of amplification would be the most abundant source of the junction sequences, it may not provide the best, or possibly even any, evidence for the occurrence of the amplification. This is because the antisense component could be too short for a definitive identification or even indistinguishable from a sense-oriented sequence. Indeed, if there are no G/U pairings, if a strand-separating activity would cleave at the first mismatch within the TCE or if the TCE would be perfectly complementary to the ICE, the sequence of an antisense component of a chimeric RNA molecule would be identical to and indistinguishable from the sequence of the regular mRNA in the region of interest, and its only characteristic feature would be a 5’ truncation that is insufficient for a definitive identification. The most suitable identifying feature, therefore, appears to be a nucleotide sequence of a yet unmodified portion of non-cleaved chimeric intermediate, containing sufficiently long and unmistakably identifiable sense and antisense junction components. Such chimeric junctions, detected for RNA encoding all three constituent chains of murine laminin 111, and the projected pathways of their generation are presented in Figure 2 above.
3. iPCR: Physiologically Occurring Intracellular Polymerase Chain Reaction, the Second Tier of RNA-dependent mRNA Amplification
In addition to chimeric sequences of antisense RNA extended into a sense RNA, described above, the analysis of globin RNA molecules in murine erythroid cells identified sense globin RNA sequences with non-conventionally templated 5’-terminal poly (U) added to the 5’UTR truncated at positions corresponding to potential cleavage sites within the antisense component of the chimeric intermediate [23]. What could be the origin of such sequences? Provided the presence of 3’poly (A) on an RNA molecule is necessary and sufficient for initiation of RNA-dependent RNA synthesis, another mRNA amplification paradigm, constituting the second Tier of RNA-dependent mRNA amplification, may be considered. If an antisense transcript is polyadenylated at the 3’ end by a known or a novel cellular poly (A) polymerase, it would become a valid template for RdRp. Since the antisense strand has, by virtue of initiation within the poly (A) of conventional mRNA, a poly (U) stretch at the 5’ end, its transcription by RdRp would result in a sense strand with poly (U) at the 5’ end and poly (A) at the 3’ end, also a legitimate RdRp template. Since strand separation mechanisms are in place and the described sequence of events can occur repeatedly, the process will amount to an intracellular polymerase chain reaction, iPCR. The obvious question regarding such a process is its specificity. If 3’ polyadenylation of an antisense molecule was coupled with the cleavage of the chimeric intermediate, the specificity of iPCR would be equal to that of the initial chimeric amplification round.
Such a process is diagrammatically presented in Figure 3. Up to the step 6, all processes occur exactly as described and shown above in Figure 1. In steps 6/7, the cleavage at a TCE/ICE mismatch or at the 5’ end of the TCE (red arrow) is coupled with polyadenylation of a newly created 3’terminus of the antisense strand. This event marks a conclusion of the chimeric cycle of amplification. One of its end products, a chimeric RNA marked in Figure 3 as “END PRODUCT, TIER ONE”, is identical to the chimeric end product of Figure 1. The other end product is different. It is 3’-truncated in the same position as depicted in Figure 1, but in addition to the 5’-terminal poly (U), it also contains the 3’-terminal poly (A) (Figure 3, step 7). This molecule constitutes the initial template of a polymerase chain reaction. Indeed, just as in step 1, RdRp activity initiates transcription at the 3’poly (A) and generates the sense strand containing 5’poly (U) and 3’poly (A) (Figure 3, step 8). After strand separation (Figure 3, step 9) there are now two templates, each containing 3’poly (A) and 5’poly (U), and the iPCR is under way. Strand separation and associated nucleotide modification probably commences at 3’poly(A) as soon as it becomes double-stranded (see sections 8 and 9 for more discussion). Therefore, at steady state there would be many more non-modified poly (U)-containing than poly (A)-containing ends and because of problems with analyzing modified RNA (discussed in section 8), their detection would be much more likely.
Figure 3: RNA-dependent mammalian mRNA amplification as a Two-Tier process.
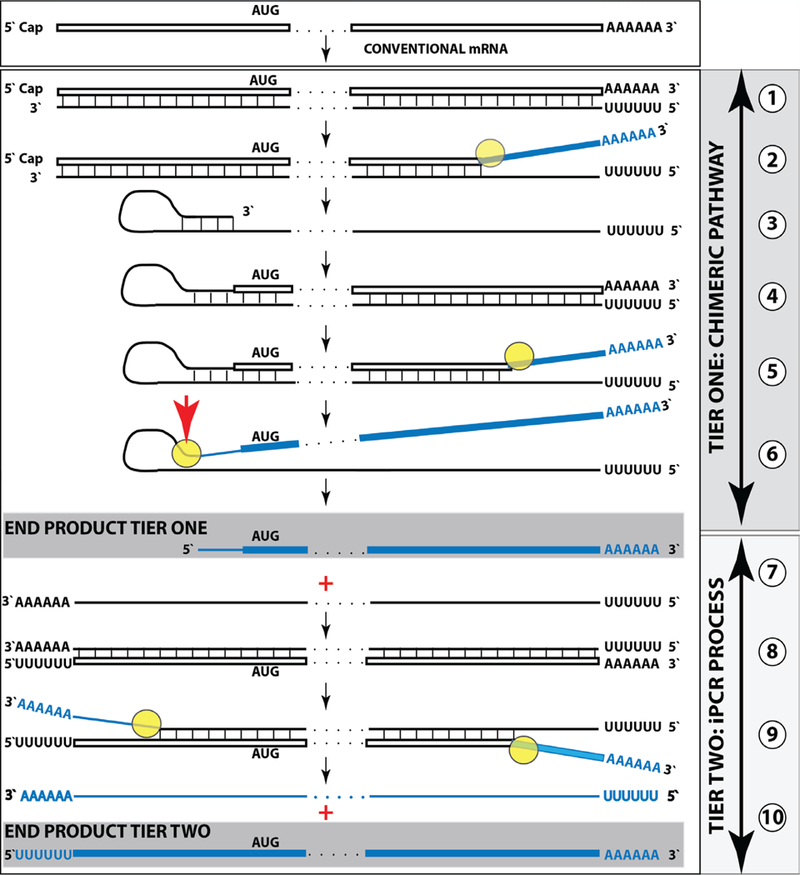
Top panel: conventional, genome-originated mRNA molecule. Bottom panel: Tier One, the chimeric pathway merging with Tier Two, the iPCR process. Boxed line – sense strand RNA. Single line – antisense strand RNA. “AUG” – functional translation initiation codon (could be other than “AUG”). Yellow circle – helicase/modifying activity complex. Blue lines (both single and boxed) – RNA strand modified and separated from its complement by a helicase complex. Red arrowhead – position of cleavage of the chimeric intermediate. Step 1: synthesis of antisense strand; step 2: strand separation; step 3: folding of antisense strand into self-priming configuration; step 4: extension of self-primed antisense RNA into sense RNA; step 5: strand separation; step 6: cleavage of the chimeric intermediate coupled with 3’polyadenylation of the antisense RNA; step 7: end-products of amplification; step 8: RdRp-mediated synthesis of the sense strand initiated at the 3’poly (A) of antisense RNA; step 9: strand separation. Note that each strand constitutes an iPCR template; step 10: iPCR products. The antisense can be further amplified whereas the sense strand can be used either for amplification or for translation. Note that the iPCR-amplified sense strand retains the intact coding content of conventional mRNA. For details see main text.
Conceptually, amplification of a nucleic acid molecule by a polymerase chain reaction necessitates, beside the presence of the building blocks, the occurrence of a template, the priming arrangement for the initial nucleic acid strand and for its complement, a polymerase, and the arrangement for strand separation that doubles the number of template molecules in each cycle. In a conventional PCR reaction, a single-stranded DNA molecule serves as a template, oligonucleotides complementary to the initial DNA strand and to its complement in desired and appropriate positions function as primers (the “forward” and the “return” primers), DNA polymerase of choice extends the 3’ end of a primer, generating a double-stranded molecule, and a thermal treatment separates the strands to enable the next cycle of the chain reaction.
In the cellular iPCR process, an eligible single-stranded RNA molecule acts as a template, the process is driven by an RdRp that generates a complementary strand, the priming arrangements are reflected in the template eligibility requirements that are satisfied by the occurrence of poly (A) segment at the 3’ termini of an RNA template as well as of its transcript/complement, and strand separation is carried out by a helicase activity. When a full-length 5’ poly (U)-containing antisense strand is generated (transcribed by RdRp from a conventional mRNA molecule) and separated from its template (steps 1 and 2, Tier One, Figure 1), it is not an iPCR-eligible template because it lacks the 3’-terminal poly (A) segment. Instead, provided that it contains the 3’-terminal and internal complementary elements, it self-primes its extension into a sense strand molecule; thus generating an intermediate in the chimeric pathway of mRNA amplification. It is the processing of this intermediate that has the potential to produce, in addition to chimeric RNA end product, an iPCR-eligible template. This requires cleavage-coupled 3’ polyadenylation of the antisense strand, which already contains 5’-terminal poly (U) transcribed from the poly (A) of a conventional mRNA progenitor molecule. The presence of the 3’-terminal poly (A) segment would allow the RdRp to initiate and to proceed with the synthesis of a sense strand complement that commences with the 5’-terminal poly(U) and concludes with the 3’-terminal poly (A) segments, also an iPCR-eligible template. The following separation of strands by a helicase activity would enable the next cycle of a polymerase chain reaction. Thus, the key feature underlying the feasibility of iPCR is that both the initial cleavage/polyadenylation-released antisense molecule and its transcript are eligible RdRp templates. Potentially and purely hypothetically, if an antisense RNA transcribed from a conventional mRNA were polyadenylated at the 3’end, it would become an eligible iPCR template. This, however, is highly unlikely because such a process would completely lack specificity. In the Two-Tier amplification process, because the generation of the initial iPCR template is coupled with and enabled by the concluding step of the previous Tier, the specificity of the iPCR process is as stringent as that of the preceding chimeric pathway, which, in turn, is defined by the occurrence within an RdRp transcript of the TCE and the ICE features and by the ability of the initial antisense transcript to self-prime its extension. On the other hand, whether or not the second Tier of amplification, iPCR, occurs does not affect in any way the first Tier, a chimeric pathway. The regulatory aspects of the iPCR amplification pathway are further discussed in section 9 below.
Figure 4 shows detected chimeric junction sequences and 5’-truncated polyurydilated mRNA sequences encoding α and β globin chains and the projected Two-Tier pathways of their generation. It should be noted that 5’-truncated and polyurydilated sense RNA, the product of Tier Two of amplification process, retains the entire coding information content of conventional mRNA. Such RNA can either serve as a template for further iPCR amplification or be utilized for translation of conventional polypeptide.
Figure 4: Detection of chimeric junction sequences and of 5’-truncated polyurydilated mRNA sequences encoding α and β globin chains and the projected Two-Tier pathways of their generation.
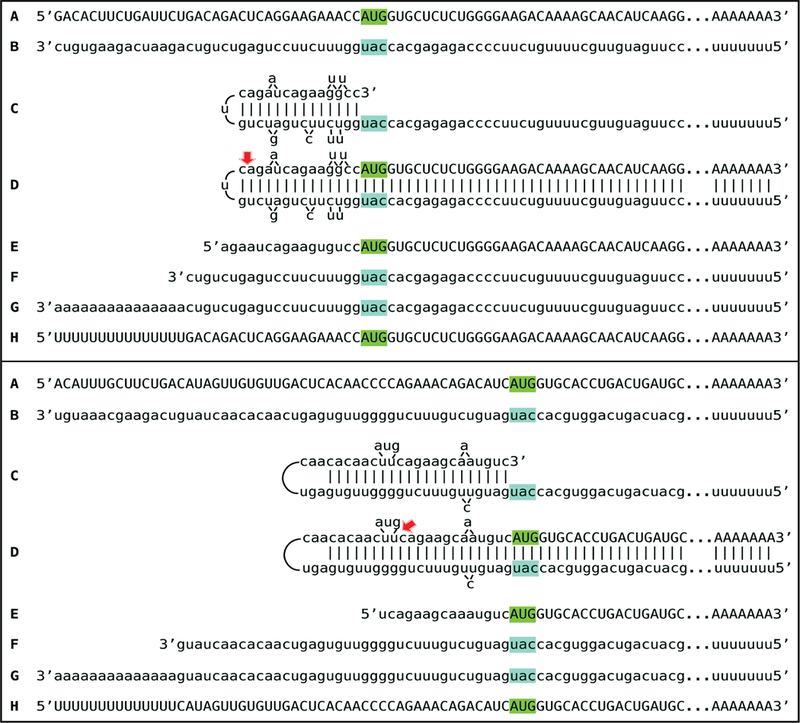
Chimeric RNA sequences and terminal 5’-truncated and polyurydilated mRNA fragments were detected in spleen cells of anemic mice. Hemolytic anemia was induced by daily intraperitoneal injections of 0.1ml of 0.8% neutralized phenylhydrazine. Cytoplasmic RNA from spleen cells collected after seven days of hemolytic treatment was used, after rRNA depletion, for construction of sequencing libraries. Libraries were sequenced on Illumina MiSeq instrument. Top panel: RNA–dependent amplification of alpha globin mRNA; bottom panel: RNA-dependent amplification of beta globin mRNA. Uppercase letters – nucleotide sequence of the sense strand; lowercase letters – nucleotide sequence of the antisense strand. Highlighted in green – “AUG” translation initiation codon on the sense strand; highlighted in blue – “uac” complement of translation initiation codon on the antisense strand. Red arrows: position of cleavage of the chimeric intermediate coupled with the 3’polyadenylation of the antisense RNA. A: conventional genome-originated mRNA. B: antisense complement of conventional mRNA. C: folding of the antisense strand into self-priming configuration; 3’terminal “c” is a transcript of the 5’capG of mRNA. D: extension of self-primed antisense strand into sense-oriented sequence. E: projected chimeric RNA end product. F: cleavage-generated 3’-truncated antisense RNA; G: projected 3’polyadenylated antisense RNA, the initial template of the iPCR process. H: 5’poly(U)-containing sense strand RNA transcript of the antisense RNA initiated at its 3’poly(A). It can either serve as a template for further iPCR amplification or utilized for translation. Note that it retains the intact coding content of conventional mRNA.
The translationally functional end product of the iPCR amplification pathway is a non-chimeric sense strand RNA identical to a conventional genome-encoded mRNA in informational content and all other aspects except three – it contains modified nucleotides, it is 5’polyuridylated, and it is 5’truncated. The truncations are no larger than the TCE of a corresponding antisense RNA, and since TCEs appear to be relatively short, so would the 5’truncations be. Mechanisms underlying the iPCR Tier of amplification could explain the not yet elucidated observations in human cells of a class of unconventional mRNA molecules that differ from their conventional counterparts only in two aspects: (a) their genome-encoded portions are truncated at the 5’ end, typically by 14 to 18 nucleotides, not affecting their protein-encoding capacity, and (b) they contain poly (U) segments appended to their truncated 5’ termini [17]. In terms of the Two-Tier mRNA amplification mechanisms, such molecules could be the products of the second stage of the mRNA amplification process, and the extent of 5’ truncations in their genome-encoded portions could reflect the average size of an antisense 3’ truncation at the conclusion of the chimeric pathway, i.e. the distance between the cleavage site within a chimeric intermediate and the 3’ end of an antisense strand.
4. mRNA Amplification Process May Reduce or Change the Protein-Encoding Information Content of a Conventional mRNA. Connection to Sporadic Alzheimer’s Disease
Previous sections discussed a situation where both complementary elements required for an appropriate folding and self-priming of the antisense strand, TCE and ICE, are located within its segment corresponding to the 5’UTR of a conventional genome-encoded mRNA. In such a situation, depicted in steps 3 trough 7 of Figure 5, the chimeric RNA end product contains the entire protein coding region of a conventional mRNA and can be translated into the original, conventional mRNA-encoded, polypeptide. In the chimeric mRNA amplification pathway, the position of the TCE within the antisense molecule is fixed; it is strictly 3’-terminal. In contrast, the intramolecular location of the ICE is variable, and potentially it can be positioned within a segment of the antisense strand corresponding to the coding portion of an mRNA, a scenario diagrammed in steps 3’ trough 7’ of Figure 5. In this scenario, the chimeric RNA end product would consist of a 3’-terminal segment of the antisense strand (the TCE or its fraction) and a 3’ portion of a conventional mRNA progenitor with a 5’-truncated coding region. In such a case, the translational outcome would be decided by the position of the first functional (capable of initiation of translation) AUG or other initiation-competent codon. If it were in-frame with the protein-encoding information content of conventional mRNA, translation would result in the C-terminal fragment, CTF, of a conventionally encoded polypeptide.
Figure 5: RNA-dependent mRNA amplification can result in a 5’-truncated molecule encoding C-terminal fragment of a conventionally encoded polypeptide.
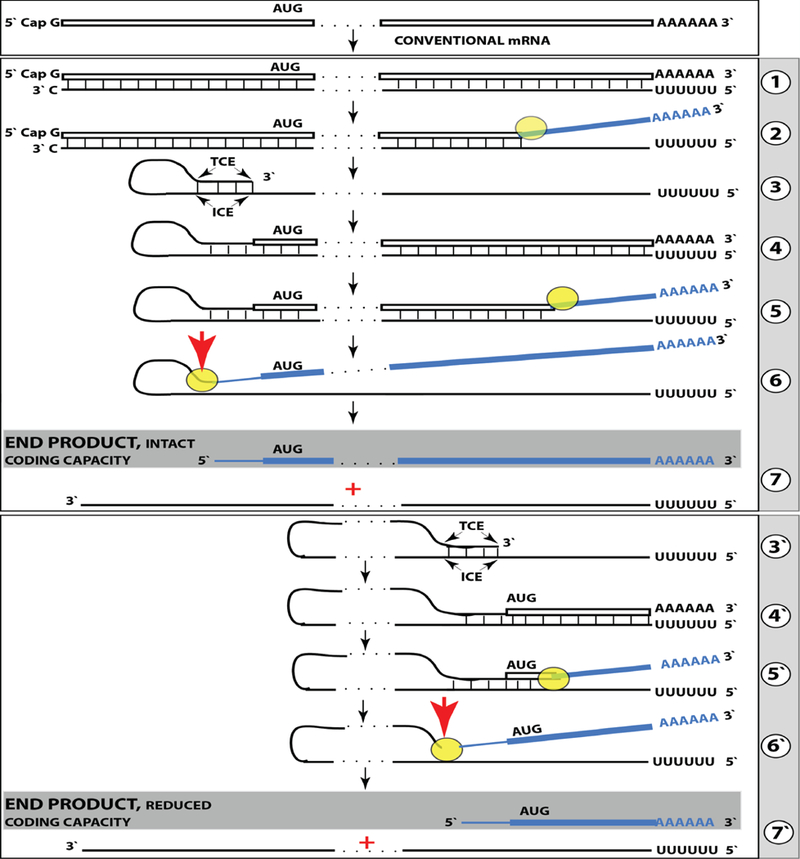
Boxed line-sense strand RNA. Single line-antisense strand RNA. “AUG”-functional translation initiation codon (could be other than AUG). “TCE”- 3’-terminal complementary element; “ICE”- internal complementary element, both on the antisense RNA strand. Yellow circle – helicase/modifying activity complex. Blue lines (both single and boxed) – RNA strand modified and separated from its complement by a helicase complex. Red arrow – position of cleavage of the chimeric intermediate. Step 1: synthesis of antisense strand; step 2: strand separation; step 3: folding of antisense strand into self-priming configuration; step 4: extension of self-primed antisense RNA; step 5: strand separation; step 6: cleavage of the chimeric intermediate; stage 7: end-products of RNA amplification. Steps 3’−7’ correspond to steps 3-7. Top panel: Conventional, genome-transcribed mRNA molecule. Middle panel: projected stages of RNA-dependent mRNA amplification. “ICE” is located within a segment of antisense RNA corresponding to the 5’UTR of conventional mRNA; the chimeric RNA end product contains the entire coding content of conventional mRNA. Bottom panel: “ICE” is located within a segment of antisense RNA corresponding to the coding region of conventional mRNA. The amplified chimeric end product contains a 5’-truncated coding region of conventional mRNA. The translational outcome is decided by position of the first functional translation initiation codon; if in-frame, a CTF of conventional polypeptide is produced.
The scenario resulting in a CTF of conventionally encoded polypeptide as the translational outcome of mRNA amplification can be illustrated with the case of beta-amyloid overproduction in sporadic Alzheimer’s disease. Beta amyloid, Aβ, the peptide associated with and, when overproduced, widely believed to have a pivotal early role in the etiology of Alzheimer’s Disease (AD), is normally generated by proteolytic cleavages of a much larger molecule, β amyloid precursor protein, βAPP [28–36]. Two sequential cleavages of βAPP are involved in the production of Aβ. The first is a cleavage of βAPP by the β secretase enzyme. It occurs between residues 671 and 672 of the βAPP molecule, generating the N-terminus of Aβ. The second cleavage, by γ secretase activity, generates the C-terminus of Aβ [29–31]. In inherited familial cases, constituting about 1% of the AD burden (the rest are sporadic cases) it was shown that the overproduction of Aβ is due to mutations around cleavage sites resulting in an abnormal proteolysis [37,38]. Since the pathological lesions and symptoms in the non-hereditary form of the disease are analogous to those seen in the familial forms, it has been assumed that abnormal proteolytic processing of βAPP also underlies the pathogenesis of sporadic AD [33–35]. It was further assumed, therefore, that the suppression of APP proteolysis might alleviate the disease [39–41]. And, indeed, inhibition of beta secretase rescued functional impairments and actually reversed the disease in animal models bioengineered to imitate familial AD [42–45]. However, in several large human clinical trials such inhibition was completely inefficient in sporadic AD [25]. This suggested that, in sporadic AD, beta-amyloid could be produced in the amyloid precursor protein-independent manner. But how?
In βAPP mRNA, the Aβ-coding segment is preceded immediately, contiguously and in-frame by the AUG codon normally encoding methionine in position 671 of the APP. If translation were initiated at this position, it would produce Aβ-containing CTF independently of βAPP. Interestingly, the AUG in question is situated within a nucleotide context optimal for the initiation of translation (an “A” in position −3 and a “G” in position +4 relative to the “A” of the AUG codon). In fact, of the twenty AUG codons encoding methionine residues in the βAPP mRNA, only the AUG encoding Met671, not even Met1, is located within an optimal translation initiation context. Such favorable positioning of the AUG encoding Met671 was the basis for an early proposal that in AD, Aβ may be generated independently from βAPP by the internal initiation of translation of the intact βAPP mRNA [46]. The possibility of the internal initiation of translation has been subsequently ruled out by the elegant experiments of Citron and co-investigators [47]. There is, however, another possibility of utilization of the AUG in question as a translation initiation codon, namely the generation of a severely 5’-truncated βAPP mRNA in which the AUG encoding Met671 in the intact mRNA becomes the first AUG codon.
To determine if an mRNA species of interest can potentially be a subject of RNA-dependent mRNA amplification, one needs to assess whether its antisense complement contains both TCE and ICE and is capable of folding into a self-priming configuration. If it were, the position of the ICE would indicate the possible translational outcome. Such an assessment can be conducted in a model experiment where an mRNA of interest serves as a template for synthesis of cDNA and is subsequently removed by RNAse H activity present in a preparation of reverse transcriptase used. If an mRNA were fully transcribed, if complementary elements were present within the antisense strand (cDNA), if one of them were 3’-terminal, and if they were topologically compatible, self-priming and the extension synthesis of a segment of the sense strand would occur. The junction between the antisense and sense components would define the site of self-priming and facilitate identification of the TCE and ICE. Just such an experiment was inadvertently carried out with human βAPP mRNA [48]. The results of this experiment, misinterpreted and eventually dismissed by the authors as an artifact [49], indicated the occurrence of topologically compatible TCE and ICE within the antisense strand of βAPP mRNA and defined their sequence as well as the position of self-priming. Based on these results, the TCE/ICE-guided folding of the antisense strand of βAPP mRNA [25,50–52] can be depicted as shown in Figure 6.
Figure 6: Projected topology of RNA-dependent generation of 5’-truncated mRNA encoding beta amyloid-containing C-terminal fragment of human amyloid precursor protein.
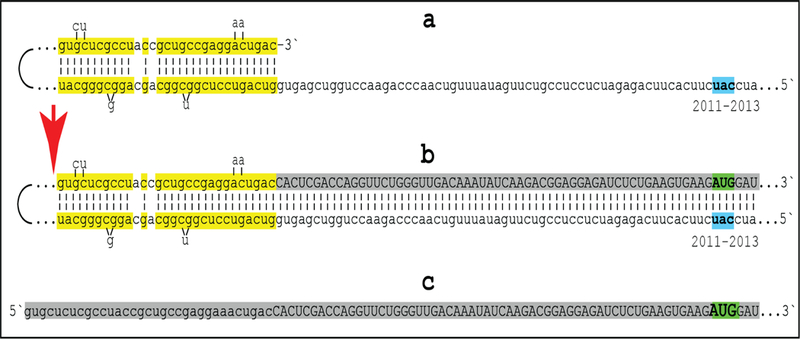
Lowercase letters-nucleotide sequence of the antisense RNA; uppercase letters-nucleotide sequence of the sense RNA. Double-stranded portions highlighted in yellow: TCE (top) and ICE (bottom) elements of the antisense RNA. Note that the TCE and the ICE are separated by about 2000 nucleotides. “2011–2013”: nucleotide positions on the antisense RNA (starting from the complement of the AUG encoding Met1 of the APP) of the “uac” (highlighted in blue) corresponding to the “AUG” (highlighted in green) encoding Met671 in the APP mRNA. a: TCE/ICE-guided folding of the antisense APP RNA; 3’-terminal “c” corresponds to one of multiple transcription start sites of APP mRNA 149 nucleotides upstream from its AUG initiation codon [25]. b: extension of self-primed antisense RNA into sense RNA and cleavage (red arrow; may also occur at one of the TCE/ICE mismatches), after strand separation, of the chimeric intermediate. c: chimeric RNA end product contains 5’terminal antisense segment extending into severely 5’-truncated APP mRNA. Its translation initiates from the “AUG” (highlighted in green and encoding Met671 in conventional APP mRNA) immediately preceding the beta amyloid-encoding region.
Approximately a 30 nucleotide-long 3’-terminal segment of the antisense strand of βAPP mRNA constitutes the TCE. Its counterpart, the ICE, is separated by nearly 2000 nucleotides, yet these elements are topologically compatible and the folding of the antisense molecule results in a self-priming configuration (Figure 6a). The TCE serves as a primer and is extended; thus, generating the sense strand as shown in the Figure 6b. Strands are then separated as illustrated in Steps 5’ and 6’ of Figure 5, and cleavage occurs either at the mismatches within the TCE or immediately upstream from it as indicated by the arrow in Figure 6b. The resulting chimeric RNA end product, shown in Figure 6c, consists of an antisense segment (TCE or its portion) continued into a sense-orientation molecule. The translational outcome is decided by the first, 5’-most, initiation-competent AUG codon. As shown in Figure 6b–c, the first AUG codon is located 58 nucleotides downstream from the TCE portion of the chimeric RNA end-product and it is, in fact, the AUG encoding Met671 in the intact βAPP mRNA! Translation from this position would produce beta amyloid-containing CTF in the APP-independent manner. The major prediction of such a mechanism is a complete inefficiency of beta secretase inhibition in sporadic Alzheimer’s disease. This prediction was, in fact, borne out in several massive stage III clinical trials [25].
The generation of a CTF of a conventionally encoded polypeptide, described above, requires that the first functional translation initiation codon in the chimeric RNA end product were in-frame with the conventional coding sequence. If it is out-of-frame, but is followed by an open reading frame, ORF, a polypeptide unrelated to a conventionally encoded protein can be produced. If it is out-of-frame and is not followed by an ORF, an abortive translation would result. If there were no functional initiation codon, no translation would occur. An abortive translation or a translational incompetence may appear to be a functional failure of the amplification process, but, as discussed in the next section, even such an apparent malfunction, in terms of the mRNA amplification translational outcome, can potentially be rescued.
5. Two-Tier Paradox: Asymmetry of Outcomes
Section 3 described a Two-Tier scenario where the ICE of the antisense strand is located within its segment corresponding to the 5’UTR of the mRNA, and the RNA end product resulting from the chimeric Tier of amplification contains the intact protein-encoding information of a conventional mRNA. The result of the following iPCR Tier of amplification is the same, namely amplified sense strand has protein coding information content identical to that of conventional mRNA. In a section 4 scenario above, the ICE is located within a segment of the antisense strand corresponding to the coding region of mRNA and the chimeric amplification pathway Tier results in an RNA molecule encoding a CTF of a conventional polypeptide. In this scenario, as in the previous one (section 3), the chimeric pathway may merge into the second, iPCR Tier of amplification, and one may intuitively think that the iPCR-amplified sense strand would also encode the same CTF as the chimeric RNA end product. This, however, is not the case because of the phenomenon of “Two-Tier Paradox”.
Two Tier Paradox is defined as follows: Regardless of the translational outcome of the chimeric pathway Tier of mRNA amplification – a conventional polypeptide, a CTF, an unrelated polypeptide, and even an abortive translation or translational incompetence – the sense RNA strand produced in the iPCR pathway Tier would always retain the entire coding information of a genome-encoded mRNA and would direct the translation of a polypeptide identical to its conventionally encoded counterpart.
As was mentioned in section 3 above, whether or not the second Tier of amplification, iPCR, occurs, does not affect in any way the first Tier, a chimeric pathway. In turn, the translational outcome of the first, chimeric, Tier does not affect in any way the translational outcome of the second, iPCR, Tier. This is because, in sharp contrast to and irrespective of any of the potentially multiple translational outcomes of the chimeric pathway, there is only one possible translational outcome of the iPCR stage of mRNA amplification - an intact conventional polypeptide. This disparity reflects the asymmetry in the truncation of sense and antisense components of the chimeric intermediate during its cleavage at the conclusion of the Tier One. The source of the asymmetry is that the truncation of molecules in question is determined by two different, spatially independent, elements. The truncation of the sense strand component of the chimeric RNA end product is defined by the position of the ICE whereas the TCE position determines the truncation of the antisense strand. Because the ICE can be anywhere in the antisense RNA molecule, so can the sense strand’s 5’ truncation be, and because the TCE, by definition, can be only 3’-terminal, the antisense always loses only a short segment at its 3’end. This phenomenon is demonstrated in Figure 5. Whereas the sizes of chimeric RNA end products are drastically different in steps 7 and 7’, the sizes of antisense RNA end products in the same steps are identical. The 3’-truncated antisense RNA, if polyadenylated at the conclusion of Tier One, gives rise in Tier Two to poly (U)-containing sense strand truncated at the 5’end by exactly the same extent as the 3’ antisense truncation, which in most, if not all, instances is substantially shorter than a typical 5’UTR of a conventional mammalian mRNA [53]. Consequently, because the 5’ poly (U)-containing sense RNA molecule, derived from the 3’ poly(A)-containing antisense protagonist by the iPCR process, retains the bulk of its 5’UTR and the intact protein-encoding portion of a conventional mRNA, regardless of the translational outcome of the Tier One, even if it is an abortive translation, in the Tier Two the translational outcome would always be the intact polypeptide, identical to that attained with a conventional progenitor mRNA. The translational outcome of the Tier Two would match that of the Tier One only when the ICE is positioned within the antisense RNA segment corresponding to the 5’UTR of conventional mRNA (Figures 1 and 3), as seen in the cases of RNA encoding both globin chains (Figure 4) and all three chains of laminin (Figure 2). In other cases, the situation can be quite remarkable. It is conceivable, for example, that the outcome of Tier One is an abortive translation or translational incompetence, yet Tier Two would produce an intact, conventional polypeptide. The same can be said for cases where Tier One results in a CTF of a conventional protein whereas the Tier Two would produce, in parallel, the intact conventionally encoded polypeptide.
The possible beta APP-independent generation of beta-amyloid in sporadic Alzheimer’s disease discussed above provides a good illustration of such a phenomenon. If the antisense beta APP RNA strand, resulting from the cleavage of the chimeric intermediate and lacking only its TCE (or a fraction thereof), were 3’-polyadenylated in a cleavage-coupled manner and utilized as an iPCR template in the second Tier of the mRNA amplification process, it would give rise to a 5’-truncated and poly (U)-containing sense strand retaining the bulk of the 5’UTR and the complete coding region of the conventional beta APP mRNA. Following its translation, an intact beta APP polypeptide would be generated alongside its beta amyloid-carrying CTF, produced from the 5’-truncated sense strand in the proteolysis-independent manner following the first, chimeric pathway Tier of the mRNA amplification process.
6. mRNA Amplification Process May Enhance Protein-Coding Information Content of Conventional mRNA and Generate Polypeptides Non-Contiguously Encoded in the Genome
The previous sections considered RNA-dependent mRNA amplification scenarios where protein coding information is contained only in the sense RNA strand. In these scenarios, the chimeric RNA end product either retains the entire coding information of a conventional mRNA and is translated from the original translation initiation codon (sections 2 and 3) or loses the 5’-portion of the coding region, and its translation depends on the availability of a functional initiation codon within the remaining portion of the coding region (section 4). There is, however, another possibility. If the TCE of an antisense RNA contains a functional translation initiation codon and if, after the cleavage of the chimeric intermediate, it was retained in the chimeric RNA end product, translation would be initiated from the initiation codon within the antisense portion of the chimeric RNA end product. In such a case, if the folding/self-priming of the antisense RNA occurs within its segment corresponding to the 5’UTR of conventional mRNA and if the initiation codon within the antisense portion of the chimeric RNA end product is in-frame with the mRNA coding sequence, translation will result in a chimeric protein. It will contain, in its entirety, the polypeptide encoded by a conventional mRNA, enhanced at its N-end by additional amino acids encoded by the antisense portion of the chimeric RNA end product and by a segment of the 5’UTR of conventional mRNA. Thus, interestingly, the protein-encoding information content of the chimeric RNA end product of amplification would be non-contiguously encoded in the genome.
If, however, the folding/self priming of the antisense strand occurs within the segment corresponding to the coding region of conventional mRNA, and if the initiation codon within the antisense portion of the chimeric RNA end product is in-frame with the mRNA coding sequence, the translational outcome would be a CTF of the conventional polypeptide, enhanced at its N-end by additional amino acids encoded by the antisense portion of the chimeric RNA end product, also a chimeric protein non-contiguously encoded in the genome. Both variants described above are diagrammatically illustrated in Figure 7.
Figure 7: RNA-dependent mRNA amplification process may enhance protein-encoding information content of a conventional mRNA and generate polypeptides non-contiguously encoded in the genome.
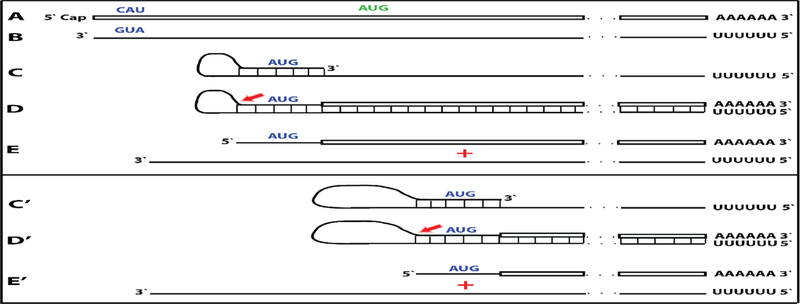
Boxed line: sense strand RNA. Single line: antisense strand RNA. “AUG”: functional translation initiation codon (could be other than AUG). 5’-CAU-3’ on the sense RNA: complement of 5’-AUG-3’ (both in blue) on the antisense RNA. Red arrow: position of cleavage of the chimeric intermediate. A: conventional mRNA with translation initiation “AUG” in green. B: antisense complement of conventional mRNA; 3’-GUA-5’ (in blue) is the 5’-AUG-3’. C: folding of the antisense strand into self-priming configuration. D: extension of self-primed antisense strand into sense-oriented sequence followed by strand separation and cleavage of the chimeric intermediate. E: chimeric RNA and 3’-truncated antisense RNA end products of RNA-dependent mRNA amplification. Note that translation of the chimeric end product starts from the antisense RNA-originated initiation codon (“AUG”, in blue) and produces a chimeric polypeptide non-contiguously encoded in the genome. Steps C’, D’, and E’ correspond to C, D, and E. Top panel: antisense folding/ self priming occurs within its segment corresponding to the 5’UTR of conventional mRNA. Bottom panel: antisense folding/self priming occurs within its segment corresponding to the coding region of conventional mRNA.
In this scenario of antisense RNA contributing a functional translation initiation codon to a chimeric RNA end product, if the chimeric pathway was to merge into the iPCR process, the rules of the Two-Tier Paradox described above would fully apply. Regardless of translational outcome of the chimeric Tier of amplification – enhanced conventional polypeptide, enhanced CTF of a conventionally encoded protein, or any other outcome, translational outcome of the second, iPCR Tier of amplification would always be a conventional genome-encoded polypeptide.
7. mRNA Amplification May Activate Dormant Protein-Encoding Information with Resulting Polypeptides Non-Contiguously Encoded in the Genome
This section considers a hypothetical yet plausible scenario where an mRNA is translated only under conditions, for example a particular stress, that activate the RNA-dependent mRNA amplification process. It should be emphasized that regardless of the extent of its physiological occurrence, the understanding of such a scenario is important because it could, and actually does as described in section 10 below, suggest very useful experimental and bioengineering designs. In this scenario, RNA encodes a polypeptide but lacks a functional translation initiation codon in-frame with the encoded information. Since its potential to produce a protein is present but unrealized under regular circumstances, it can be considered a silent, or “dormant” mRNA. The potential to direct synthesis of a polypeptide can be realized, and a dormant mRNA activated, by the mRNA amplification process. In the chimeric pathway of mRNA amplification, the chimeric RNA end product contains a 5’-terminal segment contributed by the antisense RNA and not present in the conventional genome-encoded RNA molecule. If this additional segment contains a functional translation initiation codon and if this codon were in-frame with the encoded information, the protein-encoding content of a conventional “dormant” mRNA would be activated. The resulting translational outcome would be a chimeric protein consisting of a conventionally encoded polypeptide with the N-end encoded by the antisense RNA. Such a chimeric protein would be, therefore, non-contiguously encoded in the genome. This scenario is diagrammed in Figure 8.
Figure 8: RNA-dependent mRNA amplification process may activate a dormant protein-encoding information and generate polypeptides non-contiguously encoded in the genome.
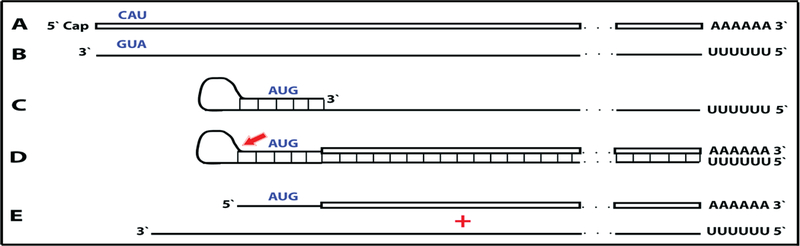
Boxed line: sense strand RNA. Single line: antisense strand RNA. “AUG” (in blue): functional translation initiation codon (could be other than “AUG”). 5’-CAU-3’ (in blue) on the sense RNA: complement of 5’-AUG-3’ on the antisense RNA. Red arrow: position of cleavage of the chimeric intermediate. A: genome-originated “dormant” mRNA containing protein-encoding information but lacking in-frame translation initiation codon. B: antisense complement of conventional mRNA. Note that 3’-GUA-5’(in blue) is the 5’-AUG-3’. C: folding of the antisense strand into self-priming configuration. D: extension of self-primed antisense strand into sense-oriented sequence followed by strand separation and cleavage of the chimeric intermediate. E: end products of RNA-dependent mRNA amplification. Note that translation of the chimeric RNA end product starts from the antisense RNA-originated initiation codon (“AUG”, in ‘‘blue) and produces a chimeric polypeptide non-contiguously encoded in the genome.
In this scenario, if the chimeric pathway were to merge into the iPCR process, the rules of the Two-Tier Paradox, namely that the translational outcome of the iPCR Tier of mRNA amplification is always identical to the outcome of the conventional gene expression, would still remain in force – the resulting sense strand RNA would be translationally dormant, just as the conventional genome-encoded RNA.
An interesting variant of this scenario is one where in the folded/self-primed antisense RNA strand, the functional translation initiation codon (TIC) is 3′-terminal and immediately precedes a conventional coding region of the resulting chimeric RNA or, if in the antisense the TIC is followed by a nucleotide sequence, this sequence is identical to the 5′ portion of a conventional coding region and aligns with it. In this case, the antisense-encoded portion of the resulting polypeptide may consist solely of the amino acid encoded by TIC and, if it is methionine, it could be removed by N-terminal methionyl aminopeptidase thus producing a completely conventional polypeptide product of the otherwise “dormant” mRNA and relieving it of its “chimeric” attribute but not of its “chimeric” origin.
8. The RNA End Product of Mammalian mRNA Amplification is a “Dark Matter”
The understanding of the RNA end product of the RNA-dependent mRNA amplification process is derived largely from a study of globin mRNA amplification during erythroid differentiation [23]. Deficiency of red blood cells in mammals, due to blood loss or induced chemically, by selective degradation of circulating red blood cells with phenylhydrazine injections, stimulates conversion of the spleen into an erythropoietic organ [23,54–56]. This splenic reaction is particularly dramatic in rodents; as shown in Figure 9 (top panel), at seven days post-induction of hemolytic anemia in mice the spleen mass increases nearly 20 fold, and it consists almost entirely of basophilic erythroblasts [23,56]. Electrophoresis of total cytoplasmic RNA from such spleens on denaturing methylmercury hydroxide/agarose gels shows the presence of an increasingly pronounced band that is slightly smaller than globin mRNA (Figure 9, bottom panel) and whose emergence is associated with the duration of hemolytic treatment and with hemoglobin accumulation [23]. Based on the size of this RNA, its kinetics and abundance, and in light of results described below, it was designated pepRNA (putative end product of globin mRNA amplification). Eventually, erythroid cells mature into reticulocytes and are released into the bloodstream; during this erythroblast/reticulocyte transition the cellular pepRNA levels drop sharply [23]. The pepRNA band produced no signal upon stringent hybridization of Northern blots with globin-specific probes [22,23]. Since nucleotide modifications can interfere with hybridization properties [57], the nucleoside composition of pepRNA was analyzed [23]. In addition to the nucleoside peaks seen in digests of control RNA, two new peaks were evident in the profile of pepRNA [23]. Mass-spectrometry produced mass numbers of 304 and 320 for the new peaks and standard numbers for the four conventional peaks (267, 283, 244 and 243 for A, G, U and C, respectively). Together, the new peaks comprised 18%, nearly one fifth, of all nucleoside residues [23]. Multiple considerations [23] indicated that the modified nucleotides are adenosine and guanosine with the same size modifying group with a mass of 37 appended to both. Considering that methylmercury interacts with the NH and allowing for the mass requirement, one possible candidate for the modifying group is LiCH2NH2.
Figure 9: During induced anemia mouse spleen converts to an erythropoietic organ and produces new type of RNA.
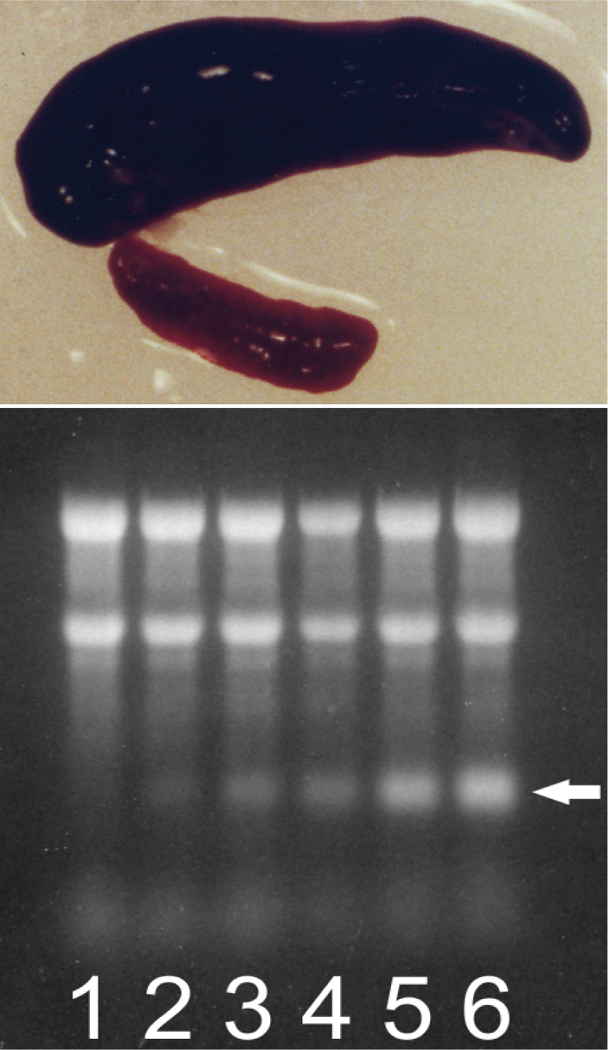
Hemolytic anemia was induced by daily intra-peritoneal injections of 0.1 ml of 0.8% neutralized phenylhydrazine hydrochloride. Spleen cells were lysed with non-ionic detergent, nuclei were removed, cytoplasmic fraction was treated with proteinase K in the presence of SDS, and RNA was extracted with phenol/chloroform and treated with DNase. Note that Trizol extraction of whole cell cannot be used because pepRNA separates with the DNA/protein fraction during such extraction [23]. Cytoplasmic RNA samples from spleen cells collected at different stages of hemolytic treatment were resolved on denaturing agarose/ methylmercury hydroxide gels and visualized by ethidium bromide staining. Top panel: Conversion of spleen into an erythropoietic organ. At seven days post-induction of hemolytic anemia the spleen mass increases almost 20 fold and nearly 90% of spleen cells are erythroid with characteristics of basophilic erythroblasts [22]. Upper image: spleen at seven days post-induction. Lower image: spleen from untreated animal. Bottom panel: Proportion of pepRNA (putative end product of mRNA amplification) increases with the duration of hemolytic treatment. Lanes 1–6: number of daily phenylhydrazine injections. Spleens were collected and processed 24 hours after final injection. Two prominent upper bands in each lane: 28S and 18S ribosomal RNA. Horizontal arrow: pepRNA. Qualitatively similar results were obtained in mice with induced hemorrhagic anemia [22].
With some, but not all, oligonucleotide globin-specific probes, at low stringency hybridization, the pepRNA band produced a signal. However all attempted extension reactions with pepRNA, but not with conventional globin mRNA, terminated after incorporation of only few nucleotides [23]. When a set of random primers was used with pepRNA, but not with conventional mRNA, only very short labeled fragments were produced [23]. It was concluded that modifications interfere both with hybridization and with the primer extension. When oligonucleotide probes were used in RNase H digest mapping, nearly identical maps were obtained for globin mRNA and pepRNA [23]. The main differences were that instead of a diffuse poly (A) band with conventional globin RNA, a uniformly short poly (A) was seen at the 3’ end with pepRNA whereas at the 5’ end fragments of pepRNA were uniformly shorter than corresponding 5’UTR fragments of conventional globin RNA; the truncations did not affect the coding regions. When two, one alpha-and another beta-globin specific probes were used together in digests, a complete cleavage of pepRNA by RNase H was observed, attesting to its homogeneity [23]. The end-labeling experiments indicated that at both 5’ and 3’ ends pepRNA terminates with the (OH) group (23). Finally, the notion that pepRNA is a variant of globin mRNA was confirmed by cell-free translation of pepRNA. The resulting polypeptides were indistinguishable from the translation product of conventional globin mRNA [23].
There is very little doubt that pepRNA is a variant of globin mRNA. But is it the projected chimeric RNA end product of the amplification process? Multiple considerations strongly support an affirmative answer. First, the 5’(OH) group of pepRNA indicates that its 5’ terminus is generated by cleavage, just as projected for the chimeric RNA end product. Second, 5’ truncations of pepRNA seen with both alpha-globin probe (about 20–25 nucleotides) and beta-globin probe (about 30–35 nucleotides) match very closely with the sizes projected for 5’ truncations of the chimeric RNA end product (20 nucleotides for chimeric alpha-globin RNA and 36 nucleotides for chimeric beta-globin RNA). Third, the massive cellular levels of pepRNA are unprecedented for and vastly exceed the observed levels of even most abundant conventional mRNA transcripts. In differentiating mammalian erythroid cells, the mass of conventional globin mRNA is about 0.01% of that of ribosomal RNA [58,59]. At its peak, pepRNA constitutes about 15% of ribosomal RNA ([23]; Figure 9), a 1500-fold increase and a clear indicator that an amplification process is at work.
If the “visualization” of a nucleic acid species is defined as the determination of its nucleotide sequence, the modified RNA end products of both Tiers of mRNA amplification can be categorized as a “Dark Matter” because they are invisible to the methods of detection, i.e. nucleotide sequencing, based on the reverse transcription of RNA of interest. As was mentioned above, the RNA end product of amplification contains a large proportion of modified nucleotides, presumably “A”s and “G”s, both appended apparently by the same group with a mass of 37 [23]. The modifications are apparently introduced on the 3’poly (A)-containing strand during strand-separation process [23]; in the case of globin mRNA, on average, about every fifth nucleotide of the amplified end product is modified [23]. Apparently, the modified nucleotides are either impassable or create a structure that is impassable for a reverse transcriptase; in related studies, both sequence-specific and random primers were ineffective or extended only by few nucleotides with the modified end product of mRNA amplification as a template, in sharp contrast to their considerable extension with the conventional mRNA template [22,23]. Modified pepRNA showed strong affinity to ribosomal RNA [23]. It appears that pepRNA forms complexes with ribosomal RNA where modifying groups are sequestered, enabling a codon/anticodon interaction [23]. Modified pepRNA appears to be “sticky” and can be lost in storage. The possible regulatory functions of nucleotide modifications in pepRNA are discussed in section 9 below.
In the chimeric pathway of mRNA amplification, the cleavage of the chimeric intermediate following the strand separation and the associated modification of its poly (A)-containing component of the double-stranded structure is the ultimate act in the generation of the chimeric mRNA end product [23]. Consequently, it is formed already modified [23] and, unlike its hairpin chimeric intermediate/precursor, is never present in the unmodified form and therefore cannot be detected by sequencing methods dependent on the lack of modifications. On the other hand, the chimeric hairpin intermediate remains unmodified until the extension of a self-primed antisense strand into the sense-orientation molecule is concluded by the synthesis of the 3’ poly (A) segment transcribed from the 5’ poly (U) of the antisense strand and required for the commencement of strand separation/modification. Moreover, following the completion of the extension and the commencement of separation, the sense/antisense chimeric junction is not modified for the duration of the strand separation process downstream from the junction thus generating a certain steady-state level and enabling the detection of the yet unmodified junction segment by the conventional reverse transcription-based methods.
In the iPCR stage of mRNA amplification, because the helicase/modifying activity requires the presence of the 3’-terminal poly (A) in a double-stranded configuration to commence strand separation, the 5’ poly (U)-containing mRNA end product remains unmodified until its synthesis is completed. Even after the sense strand RNA product of the iPCR acquires its 3’ poly(A) segment, transcribed from the 5’poly (U) of the antisense strand, its 5’ portion remains unmodified for the limited duration of the strand-separating/modifying activity traversing the length of the molecule. These two processes, the completion of the synthesis and the strand-separation/modification downstream from the region of interest, provide a temporal “window of opportunity” that, apparently, enabled the detection of the yet unmodified 5’-terminal poly (U)-containing segment of the mRNA end product of the iPCR Tier of the mRNA amplification process (Figure 4).
A temporal “window of opportunity” is much smaller with respect to the detection of yet unmodified 3’-terminal poly (A)-containing segment of the antisense strand. At the conclusion of the chimeric pathway, it remains unmodified for probably only a short duration between the cleavage/3’ polyadenylation of the chimeric intermediate and its separation from a probably still nascent 5’poly (U)-containing sense strand being transcribed from it. During the iPCR process this duration could be even shorter, comprised only of the period between completion of the 3’ polyadenylated antisense synthesis and the commencement of the modification starting at its newly added poly (A) component as soon as poly (U) is transcribed. In such a case the modifying activity could be lagging closely behind the polymerase complex. It should be emphasized that the detected, not yet modified, 5’-terminal poly(U)-containing segment of the sense strand product of Tier Two represents probably a very minor subpopulation, with a substantial populations of modified molecules remaining for now, alongside the probably highly abundant end product of the chimeric pathway, a “Dark Matter”.
9. Regulatory Aspects of Mammalian RNA-dependent mRNA Amplification Process
The ubiquitous presence of conventional RdRp was shown in numerous eukaryotic organisms but not in mammals [9]. However, the ability of viruses not encoding the RdRp, such as HDV, to robustly replicate in mammalian cells attested to the cellular presence and functionality of this activity [9,10]. The RdRp activity in mammalian cells appears to be non-conventional; two possible candidates for this role are the RNA polymerase II complex or its components [19,20] and RdRp activity of the TERT complex [21], both ubiquitously present in all cells. Under regular circumstances the RdRp activity in mammalian cells produces only short antisense RNA transcripts. For example, a widespread synthesis of diverse short antisense RNA transcripts initiating at the 3’poly (A) of mRNA was observed in human cells [17]. On the other hand, RdRp activity isolated from rabbit reticulocytes [18] was able to produce, in assays, long antisense RNA transcripts. Subsequent studies identified full-length antisense transcripts of globin mRNA in erythroid cells [22]. It could be argued that the component responsible for the production of long antisense transcripts in mammalian cells is a processivity conferring co-factor of RdRp activity that is induced under special circumstances when over-production of specific proteins is required [23]. The notion of a processivity co-factor is strongly supported by studies of HDV replication in “normal” (i.e. apparently lacking processivity co-factor) mammalian cells. Within the framework of the above considerations, the ability of RdRp-deficient viruses to use RdRp activity of mammalian cell for their replication implies that they should encode a processivity co-factor of cellular RdRp. In case of HDV, it appears to be hepatitis delta antigen HDAg, the only protein encoded by HDV. HDAg is essential [71] both for production of long transcripts by cellular RdRp, and for viral replication. In its absence only short transcripts are generated [71]. These observations provide a proof of concept for the notion of RdRp processivity co-factor, central for our understanding of mammalian mRNA amplification. Identification of a cellular homolog of HDAg, DIPA [72,73] suggests directions for a search for the cellular RdRp processivity co-factor.
Since HDV apparently needs only HDAg, presumably a processivity co-factor, to enable its replication within a mammalian host cell by cellular RdRp activity, it could be argued that all that is required for activation of mammalian RNA-dependent mRNA amplification is the induction of cellular RdRp processivity component, or its (or HDAg) expression from an exogeneous vector. This, however, is probably not the case. HDV replication is initiated by primer extension and occurs via rolling circle mechanism [71]. Accordingly, strands are separated by the advancing polymerase complex, while cleavages of newly synthesized RNA into individual units are effected by ribozyme activities present in HDV RNA [71]. On the other hand, whereas the nature of initiation of antisense RNA synthesis within 3’poly (A) of conventional mRNA remains to be elucidated, mammalian mRNA amplification involves strand separating/nucleotide modifying activity as well as a chimeric RNA cleaving activity. To understand what is required for the activation of mRNA amplification in mammalian cells, we have to know which of the relevant activities are constitutive and which are inducible. Our current understanding indicates that mammalian RdRp activity is constitutive and its processivity co-factor is inducible; it doesn’t inform us on the status of other activities involved. Elucidation of the nature of all activities involved in RNA-dependent mRNA amplification would, when attained, enable complex and powerful bioengineering designs.
The observed uniformly short 3’poly(A) region of the pepRNA, transcribed from 5’poly(U) of the antisense strand [23] indicates that synthesis of the antisense strand starts within the 3’poly(A) region of the sense strand, in a relatively narrow distance range from the 3’UTR of a conventional mRNA. The manner of antisense RNA initiation remains to be determined. Interestingly, at the 3’terminus of the antisense strand, RdRp appears to be capable of copying the cap”G” of conventional mRNA despite its inverted orientation. The rationale for this conclusion is based on the detected antisense/sense junction sequences of chimeric alpha-globin RNA amplification intermediate shown in Figure 4; it is summarized in Figure 10. If the cap “G” is not transcribed, the antisense strand would terminate with the 3’ “c” corresponding to the transcription start site of mRNA (panel A; highlighted in blue). In such a case, antisense RNA folding/self-priming configuration would be as shown in panel A, and following the extension of self-primed antisense RNA, the antisense/sense junction would consist of the “c/A” (highlighted in blue) as depicted in panel B. The experimental results presented in Figure 4 are different. They show that the sequence of the antisense/sense junction is, in fact, the “cc/A” (panel C; highlighted in green). Since the genomic sequence upstream of the transcription start site cannot account for the additional 3’-terminal “c” (panel D; highlighted in green) in the antisense strand [23], the remaining possibility is that the “c” in question is a transcript of the cap “G” of the sense strand and that the antisense folding into a self-priming configuration occurs as shown in panel D. It should be mentioned that conceptually similar results were obtained with chimeric RNA intermediate of murine alpha 1 laminin mRNA amplification [24].
Figure 10: RNA-dependent RNA polymerase can transcribe the cap “G” of mRNA.
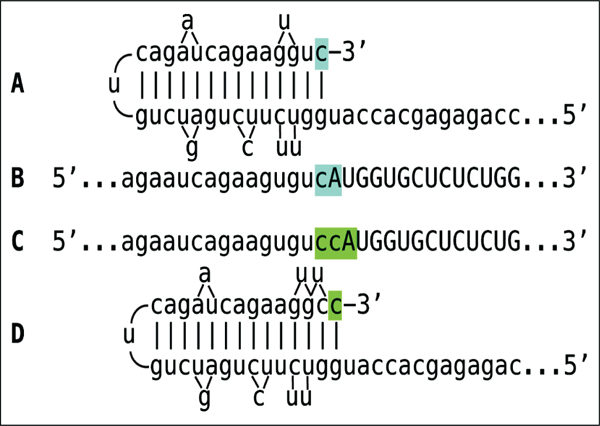
Data shown is adopted from Figure 4, top panel. Uppercase letters – nucleotide sequence of the sense strand; lowercase letters – nucleotide sequence of the antisense strand. “c” highlighted in blue – 3′-terminal nucleotide of the antisense strand corresponding to the transcription start site of mRNA; “cA” highlighted in blue –the projected junction structure in the absence of the cap “G” transcription. “c” highlighted in green – transcript of the cap ”G”; “ccA” highlighted in green – the resulting junction structure when cap “G” is transcribed. A: projected self-priming configuration of the antisense strand in the absence of the cap “G” transcription. B. Projected nucleotide sequence of the sense/antisense junction in the absence of the cap “G” transcription. C. Detected nucleotide sequence of the sense/antisense junction. D. Self-priming configuration of the antisense strand as defined by experimental results. Note that the genomic sequence upstream of the TSS cannot account for the additional 3’-terminal “C” in the antisense strand [23].
Transcription of conventional mRNA by RdRp activity results in a double-stranded molecule. Separation of strands and associated modifications of one of the strands starts at the poly(A) that is in double-stranded configuration with its poly (U) counterpart. As discussed below, the separation appears to be very rapid. It is possible that a separating/modifying activity mounts the poly (A) segment as soon as the poly (U) is transcribed and lags closely behind the polymerase complex as it progresses. Importantly, during separation of the antisense strand from its conventional RNA template, only the latter is modified. This enables the TCE/ICE interaction and subsequent priming/extension that would be impaired if the antisense strands were modified.
Globin genes contain the “TATA” regulatory element and a fixed transcription start site (TSS). Globin antisense RNA contains both the TCE and the ICE and is eligible for RdRp-mediated amplification. On the other hand, with mRNA transcripts of TATA-less genes where transcription can be initiated from multiple positions, the eligibility for RdRp-mediated amplification appears to be regulated by a shift in the TSS position. For example, genes for all three chains of laminin 111 are TATA-less and have multiple TSS positions [61,62], most of which are inconsistent with the eventual generation of antisense molecules capable of self-priming within their segments corresponding to the 5’UTRs of mRNA because neither one of the complementary elements on the antisense strand is 3’-terminal. The same is true also for the APP gene discussed in section 4 above. However, a shift in TSS position can make one of the elements 3’-terminal and thus define its eligibility. The concepts of such a regulation are summarized in Figure 11. If the 3’-distant complementary element of the antisense strand is terminal (Figure 11, panel A), it can form a self-priming structure. If, however, both elements are internal (Figure 11, panel B), the downstream shift of the TSS position can make one of them a 3’-terminal and thus enable self-priming. When, on the other hand, the 3’ segment of the antisense strand has no topologically compatible complementary sequences, an upstream shift of the TSS position (Figure 11, panel C) can generate such elements and make the transcript eligible for amplification. The events diagrammed in panels B and C of Figure 11 can also occur in reverse, making previously eligible mRNAs ineligible for the amplification process.
Figure 11: TSS shift as potential regulator of the eligibility of an mRNA for the amplification process.
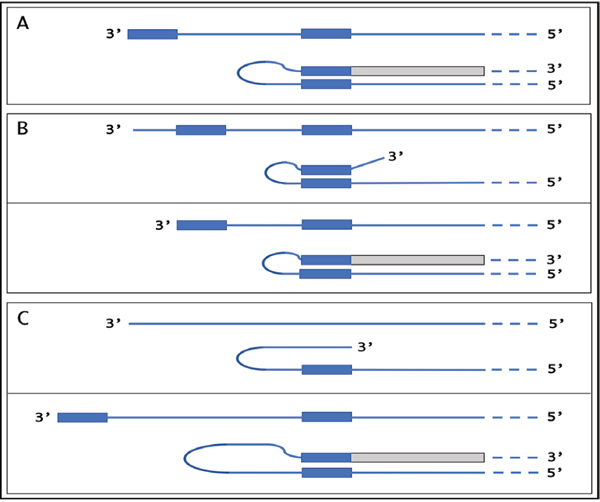
Single line: 3’terminus of the antisense strand. Filled grey boxes: sense strand. Filled blue boxes: topologically compatible complementary elements on the antisense strand. A: one of the complementary elements is 3’-terminal; folding results in a self-priming structure that is extended into the sense strand. B: both complementary elements are internal, no self-priming is possible; TSS shift in the downstream direction makes one of the elements 3’-terminal and allows self-priming and extension into the sense strand. C: there are no complementary elements/no self-priming; TSS shift in the upstream direction generates 3’-terminal complementary element and thus enables self-priming and extension. Note that processes depicted in panels B and C can occur in reverse, resulting in a loss, rather than the acquisition, of the eligibility.
Separation of strands of the chimeric hairpin intermediate occurs as described above for the antisense/conventional mRNA complex. At the conclusion of strand separation, the chimeric intermediate is cleaved either at or near a TCE/ICE mismatch or at or near the 5’end of the TCE. The cleavage releases the chimeric RNA end product and a 3’-truncated antisense RNA. If cleavage occurs close to the 3’ end of the TCE and a substantial portion of the TCE were retained after the cleavage, the self-priming structure could remain stable and be re-used, apparently after 3’-P to 3’-OH phosphatase-mediated conversion, to generate another chimeric intermediate. In such a case, the site of antisense/sense RNA junction would shift upstream from the previous one by the size of the antisense truncation resulting from the initial cleavage. If such a process occurs more than once, multiple antisense truncations are generated and every time the site of the antisense/sense RNA junction is shifted. In fact, such a process was observed with chimeric intermediates of globin mRNA amplification. As can be seen in Figure 1 of [23], one shift of the antisense/sense junction was observed with α-globin RNA and two shifts were seen with β-globin RNA. Eventually, the substantial portion of or the entire TCE would be cleaved off the antisense RNA and it could not be re-used. A possible antisense transcript of the chimeric RNA end product would also lack the TCE. The only molecule that can be productively and repeatedly re-used as a template in the chimeric Tier of amplification is a conventional mRNA. Therefore, for the chimeric amplification pathway to operate, it has to be presumed that conventional mRNA, once utilized and thus modified, can be transcribed by RdRp. Similarly, the iPCR pathway starts with unmodified 3’polyadenylated antisense RNA but apparently utilizes modified RNA templates in the process.
Whereas the occurrence and regulation of the chimeric pathway is entirely independent from the iPCR process, the occurrence of the latter depends on the completion of the former. The occurrence of the iPCR also requires that the last step of the chimeric pathway, a cleavage of the chimeric intermediate, is coupled with the 3’polyadenylation of the truncated antisense RNA strand. Arguably, the iPCR process may vary in a regulated manner in accordance with the needs for the production of a protein encoded by an mRNA being amplified. For example, although 5’-truncated polyuridylated RNA, the identifier of the iPCR process, was detected in the globin system [23], no significant amount of this product was seen in the RNase H digest analysis [23]. Both sequencing analysis and RNase H digest studies were conducted at the very late stage of splenic erythropoiesis. It is conceivable that at this stage the chimeric pathway is sufficient to maintain hemoglobin production and that the iPCR process occurred primarily at early stages of splenic erythropoiesis and only at low rate at late stages. Considering that the presence of the 3’-terminal poly (A) segment is a presumably single major template eligibility requirement for RNA-dependent RNA synthesis [17,22,23], and allowing for the operational presence of the RdRp, a key component of both the chimeric pathway and the iPCR process, it is reasonable to suggest that the principal regulatory checkpoint for the entry into the Tier Two of mRNA amplification could be the rate of the chimeric intermediate cleavage-coupled 3’ polyadenylation of the antisense RNA strand. In the extreme case, if the polyadenylation (but not the cleavage) was completely suppressed, so would the Tier Two be, but neither the outcome nor the efficiency of the Tier One process would be affected.
In the iPCR process, if, as indicated above, RdRp initiates transcription within the poly (A) (rather than at its 3’ end), the poly (A)/poly (U) shortening will occur in successive iPCR rounds until template eligibility would be lost unless poly(A) elongation takes place simultaneously with its shortening. This is in contrast to the chimeric pathway where repeated use of conventional mRNA as a template for antisense RNA synthesis apparently has no impact on the length of its 3’poly (A). Additional augmentation of the iPCR’s efficiency could occur at the level of the iPCR template availability, which is comprised of two components: stability of both sense and antisense strands produced in the reaction and the utilization of the end product of the second Tier. The iPCR is, as any chain reaction, potentially an exponential process provided that the entire output of every cycle is utilized in the next cycle of a chain. However, if all sense strand products in every cycle were withdrawn because of recruitment for protein synthesis, this Tier of amplification would be linear, akin to the chimeric pathway. In all probability, some sense iPCR-produced molecules are removed for translation and some are utilized as templates for further amplification, making the process faster than linear but slower than exponential. As for the stability of the iPCR templates, it could be regulated, at least in part, by nucleotide modifications present in both RNA strands. Indeed, when cells undergoing erythroid differentiation progress from the stage of rapid hemoglobin accumulation to the stage of the maintenance of steady hemoglobin levels (erythroblast/reticulocyte transition), modified RdRp-amplified globin mRNA accrues to extremely high levels during the former but rapidly disappears during the latter stage. This suggests that the amplification-associated RNA modification could mark the molecule for degradation when it is no longer needed.
Considering that the RNA-dependent mRNA amplification process proceeds trough the double-stranded RNA stages, an obvious question is how it evades the dsRNA response? One possibility is that the amplification process is compartmentalized and the components of the dsRNA response have no access to it. Another possibility is the involvement of factors protecting from the dsRNA response. What is known experimentally, is that double-stranded RNA components associated with RNA-dependent mRNA amplification appear to be extremely short-lived as reflected in their steady-state levels [23,24]. Thus, the levels of reads for chimeric RNA intermediates for each of three chains of laminin 111 are only one per 3×107 [24], whereas levels of reads for chimeric intermediates for both chains of globin [23] are one in 105 (in both systems, after ribosomal RNA depletion). This means that strand separation occurs very rapidly and double stranded molecules have very limited exposure. Moreover, provided that one of functions of modifications is strand separation, when already modified RNA is used as a template for RdRp, as in the re-use of conventional mRNA as a template or in the iPCR process, strand separation would occur concurrently with the progression of the polymerase complex.
The functions of nucleotide modifications occurring during mRNA amplification remain to be elucidated. They could include the facilitation of RNA strand separation, stimulation of the cap-independent translation of the amplification end product, and regulation of its stability. The latter is suggested by a drastic decrease in levels of pep RNA [23], but not of conventional globin mRNA [56], during the erythroblast/reticulocyte transition. There may also be a connection between nucleotide modifications of amplified mRNA and the activation of the mRNA amplification process. The amplified and heavily modified [23] mRNA could behave in ways that are different spatially, qualitatively and quantitatively from those of conventional mRNA. For example, the amplification of mRNA species encoding secreted proteins, such as laminin [24], could overwhelm the ER, cause the ER stress and trigger cell death. One cellular response to ER stress appears to be a shift of translation and secretion outside the ER [62]. It could be suggested, therefore, that nucleotide modifications of amplified mRNA may direct its translation and secretion of the resulting protein via pathways bypassing the ER, despite the presence of a signal peptide sequence. The initial ER stress resulting from increased transcription and subsequent translation of conventional mRNA encoding a secreted protein could be one of potentially multiple cellular events, probably certain types of stresses, which may trigger mRNA amplification processes. In this case, one can envision that conventional overproduction of a secreted protein induces ER stress and activates multiple transcription factors [63,64], which, in turn, activate mRNA amplification pathway thus facilitating overproduction and relieving the ER stress, since in the mRNA amplification process, a portion of conventionally produced mRNA molecules, used as templates for the production of antisense RNA, is apparently modified during strands separation (Figure 1, step 2) and would be translationally processed outside the ER, alongside the modified RNA end product of mRNA amplification.
10. A Definitive Proof of the Occurrence of Mammalian mRNA Amplification: Novel Experimental Designs
The detection of a full-size antisense RNA initiated at the 3’poly (A) of its conventional mRNA counterpart [22], and the identification of the putative end product that accumulates to massive cellular levels unprecedented for conventionally expressed mRNA and matches the expected properties of a projected end product [23] strongly support the mechanism for RdRp-mediated amplification of mRNA described above. The strongest evidence in support of the occurrence of this process is the detection, in two different experimental systems and for five different mammalian mRNA species, of uniquely defined chimeric RNA intermediates containing antisense and sense sequences covalently joined in a rigorously predicted manner [23,24]. Experimental approaches used for the detection of these sequences included multiple procedures that were safeguarded by numerous checkpoints. Yet, very strong arguments for the cellular origin of these chimeric RNA molecules notwithstanding, their possible inadvertent generation during experimental manipulations could not be ruled out with absolute certainty. A definitive proof for the occurrence of RNA-dependent amplification of mammalian mRNA, it appears, requires a direct measurement, without any outside experimental manipulations whatever, of a key cellular product of this process.
A novel experimental strategy that would enable such direct measurement is suggested by a scenario for RNA-dependent mRNA amplification discussed in section 7 (Figure 8 above). In fact, the experimental designs described below actually re-create it. In this scenario, a latent mRNA contains ‘‘dormant” protein-coding information that cannot be translated for the lack of a functional in-frame initiation codon. It can, however, be activated by the RNA-dependent mRNA amplification if the antisense portion of the chimeric RNA end product generated in the amplification process contains a functional translation initiation codon in-frame with the mRNA-encoded content. In such a case, not only would a “dormant” mRNA be translated, but a distinctively defined chimeric polypeptide non-contiguously encoded in the genome would also be produced, with the N-end portion encoded by the antisense RNA. The detection, by direct measurement in cell lysate, of such a molecule, the ultimate end product, would constitute a definitive proof for the occurrence of RNA-dependent mRNA amplification.
Two experimental models and associated experimental designs, both based on experimental results described above, exemplify this novel approach. The first experimental model is the in vitro amplification of mRNA encoding the murine alpha1 chain of laminin. The experimental design, outlined in Figure 12, starts with the elucidated pathway shown in the top panel of Figure 2 and modifies it, by editing the alpha1 laminin gene, to achieve the desired outcome. In the top panel of Figure 12, line A shows the 5’terminal portion of conventional mRNA and line B shows the 5’terminal portion of the projected chimeric RNA end product of amplification (capital letters-sense RNA; lower case letters-antisense RNA); it should be noted that both share a portion of the 5’UTR and contain the same coding regions. Line C shows the polypeptide (highlighted in blue) resulting from translation of either conventional or amplified chimeric laminin mRNA. In the middle panel, line A shows the 5’terminal portion of edited mRNA and line B shows its antisense counterpart. “CAU” highlighted in blue (line A) is transcribed into 5’-aug-3’ highlighted in green (shown as 3’-gua-5’) in the antisense strand (line B).
Figure 12: Novel experimental design 1: in vitro generation of a chimeric polypeptide containing murine alpha 1 laminin chain, initiated from the antisense RNA and non-contiguously encoded in the genome.
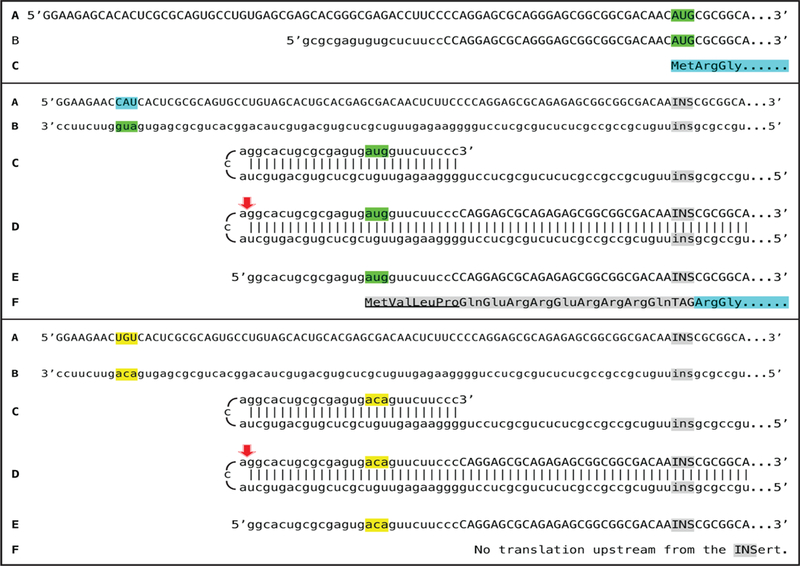
Uppercase letters: sense strand RNA. Lowercase letters: antisense strand RNA. Highlighted in green: AUG translation initiation codon. 5’-CAU-3’ (highlighted in blue) on the sense RNA: complement of 5’-aug-3’ (highlighted in green) on the antisense RNA. “INS” highlighted in grey: insert encoding a Tag peptide and lacking initiation codon; “TAG” highlighted in grey: peptide encoded by “INS”. “ins” highlighted in grey: “INS” complement on the antisense RNA. Amino acid sequence highlighted in blue: polypeptide encoded by conventional mRNA or by the amplified mRNA. Amino acid sequence highlighted in grey: the N-end extension of conventional polypeptide; underlined portion: amino acids encoded by the antisense RNA. Highlighted in yellow: editing changes resulting in replacement of the “aug” by the “aca” on the antisense RNA. Red arrowhead: position of cleavage of the chimeric intermediate. Top panel: A-conventional mRNA encoding alpha 1 chain of laminin. B-chimeric RNA end product of amplification (adopted from Figure 2) encoding the same polypeptide as “A”. C-amino acid sequence encoded by either “A” or “B”. Middle panel: A-projected edited “dormant” mRNA originated from edited alpha 1 laminin gene. B-antisense complement of edited alpha 1 laminin mRNA. C-folding of the antisense strand into self-priming configuration; 3’terminal “c” is a transcript of the 5’capG of mRNA. D-extension of self-primed antisense strand into sense-oriented sequence followed by strand separation and cleavage of the chimeric intermediate. E-chimeric RNA end product of RNA-dependent amplification of edited alpha 1 laminin mRNA. F-projected translational outcome. Bottom panel: same as the middle panel with the exception of editing changes resulting in replacement of the “aug” by the “aca” (highlighted in yellow) on the antisense RNA.
The major objectives of the editing are the following: (1) To delete translation initiation AUG codon of the conventional mRNA; (2) To replace it with an insert (marked “INS” in Figure 12 and 13) lacking translation initiation codon and encoding a Tag peptide so as to enable detection and isolation of the resulting polypeptide; (3) To eliminate all potential translation initiation codons [65,66] upstream from and in-frame with the “INS”/protein-encoding sequence in the edited mRNA; (4) To introduce within the TCE of the projected antisense strand an AUG codon in the 5’ to 3’ orientation, in the optimal translation initiation context, and in-frame with the “INS”/protein-encoding sequence in the projected chimeric RNA end product of amplification; (5) To minimize TCE/ICE mismatches so as to assure that the cleavage of a chimeric intermediate does not occur downstream from the “AUG” and to maximize the length of the 5’UTR in the projected chimeric RNA end product; (6) To ascertain that the “AUG” of the antisense RNA is followed by a codon encoding Val so as to confer to the resulting protein the maximum half-life, in accordance with the N-end rule pathway of protein degradation [67], if the N-terminal Met is removed by the N-terminal methionyl aminopeptidase [68]; (7) To minimally interfere with the 5’terminus of the sense strand transcript so as to preserve the position of the TSS.
Stages C and D of the middle panel of Figure 12 show the folding of the antisense RNA (stage C) and its extension into chimeric intermediate followed by strand separation and cleavage (red arrow, stage D). Line E shows the projected edited chimeric RNA end product. In its antisense portion it contains an AUG codon (“aug” highlighted in green) in an optimal translation initiation context (purine in position-3 and “G” in position +4 relative to the “A” of the AUG) and in-frame with the INS/laminin coding sequence. Translation of the edited chimeric RNA end product would result in a chimeric polypeptide shown in line F of the middle panel and comprised of a laminin portion (highlighted in blue and identical to that shown in top panel, line C), a Tag peptide, a portion (highlighted in grey) encoded by the sense-strand RNA sequence upstream of the “INS” (an analog of a portion of the 5’UTR of conventional mRNA) and the N-end portion encoded by the antisense RNA component of the chimeric RNA end product (highlighted in grey and underlined); it would be non-contiguously encoded in the genome. The detection of a Tag peptide would indicate that a chimeric polypeptide has been produced and the isolation and sequencing of this polypeptide would provide a definitive and irrefutable proof of the occurrence of the RNA-dependent mRNA amplification process.
Identification/sequencing of the projected chimeric polypeptide would be sufficient to make a determination of the occurrence of RNA-dependent mRNA amplification, but the above experimental design allows a number of additional controls. One of these is illustrated in the bottom panel of Figure 12. The AUG codon of the antisense strand can be mutated into a codon (ACA) not known to initiate translation, without changing the complementary relationship between the TCE and the ICE components of the antisense RNA. In such a case, the amplification would proceed similarly to that depicted in the middle panel, but the resulting chimeric RNA end product (bottom panel, line E) would lack a functional in-frame initiation codon and no translation of INS/laminin sequence would occur. Other possible controls are frame-shifts of the edited RNA shown in the middle panel of Figure 12. Effecting a frame-shift of one, two but not three nucleotides in the region between the AUG of the antisense portion and the “INS” of the chimeric RNA end product (middle panel, line E) should abolish translation of the INS/laminin whereas a frame-shift of either one or two or three nucleotides upstream from the AUG of the antisense RNA segment within the same molecule should have no such effect.
In the framework of the above experimental design, the edited gene would produce translationally “silent” mRNA. The protein-coding content could be activated in a rigorously predicted “enhanced” manner only when mRNA amplification process is induced. Such an experiment can be carried out in a murine astrocyte cell line known to elevate the expression of laminin under hypoxic conditions [69]. There are good reasons to believe that severe chronic hypoxia would induce the mRNA amplification process in these cells and enable the testing of the above experimental design. It would be of great interest to employ this experimental design in an animal model. To do it with laminin RNA, however, would be problematic because laminin is expressed diffusely and in a shifting manner throughout the organism, complicating the analysis. The second experimental model, that of in vivo murine phenylhydrazine-driven/hemolytic anemia-induced splenic erythropoiesis, doesn’t have these shortcomings. It takes place in a large, nearly homogeneous, organ, and its features and dynamics are well understood both in basic terms and in terms of RNA-dependent globin mRNA amplification [22,23]. β-globin mRNA is much better suited for such an experiment than α-globin RNA because it has a longer 5’UTR and its antisense strand contains a conveniently positioned AUG codon (Figure 4, bottom panel, stages C and D) that can be incorporated in experimental design. The major objectives of the editing are the same as defined above. Only one gene copy should be edited making mice “heterogeneous” so as to interfere minimally with the physiological function of erythroid cells. Circulating spleen-originated reticulocytes (over 90% of circulating red blood cells in this model system are reticulocytes) can be repeatedly collected from the same animals for time-points analyses prior to the collection and analysis of spleen cells. The experimental design for in vivo testing of the occurrence of RNA-dependent amplification of murine beta-globin mRNA is presented in Figure 13.
Figure 13: Novel experimental design 2: in vivo generation of a chimeric polypeptide containing murine beta globin chain, initiated from the antisense RNA and non-contiguously encoded in the genome.
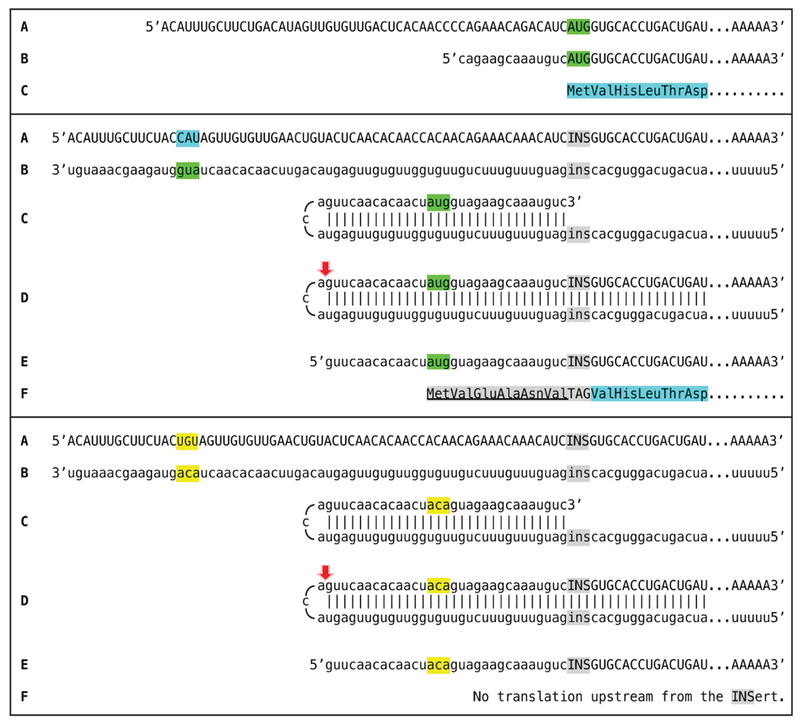
Uppercase letters: sense strand RNA. Lowercase letters: antisense strand RNA. Highlighted in green: AUG translation initiation codon. 5’-CAU-3’ (highlighted in blue) on the sense RNA: complement of 5’-aug-3’ (highlighted in green) on the antisense RNA. “INS” highlighted in grey: insert encoding a Tag peptide and lacking initiation codon; “TAG” highlighted in grey: peptide encoded by “INS”. “ins” highlighted in grey: “INS” complement in the antisense RNA. Amino acid sequence highlighted in blue: polypeptide encoded by conventional mRNA or by the amplified mRNA. Amino acid sequence highlighted in grey and underlined: the N-end extension encoded by the antisense RNA. Highlighted in yellow: editing changes resulting in replacement of the “aug” by the “aca” in the antisense RNA. Red arrow: position of cleavage of the chimeric inter mediate. Top panel: A-conventional mRNA encoding beta globin chain. B-chimeric RNA end product of amplification (adopted from Figure 4) encoding the same polypeptide as “A”. C-amino acid sequence encoded by either “A” or “B”. Middle panel: A-projected edited “dormant” mRNA originated from edited beta globin gene. B-antisense complement of edited beta globin mRNA. C-folding of the antisense strand into self-priming configuration; 3’terminal “c” is a transcript of the 5’capG of mRNA. D-extension of self-primed antisense strand into sense-oriented sequence followed by strand separation and cleavage of the chimeric intermediate. E-chimeric RNA end product of RNA-dependent amplification of edited beta globin mRNA. F-projected translational outcome. Bottom panel: same as the middle panel with the exception of editing changes resulting in replacement of the “aug” by the “aca” (highlighted in yellow) on the antisense RNA.
It starts with the elucidated pathway shown in the bottom panel of Figure 4 and modifies it, by editing the beta globin gene, to achieve the desired outcome. In the top panel of Figure 13, line A shows the 5’terminal portion of conventional mRNA and line B shows the 5’terminal portion of the chimeric RNA end product of amplification (capital letters - sense RNA; lower case letters - antisense RNA). Conventional (A) and chimeric (B) mRNAs differ only in their 5’UTRs and contain identical coding regions. Line C shows the polypeptide (highlighted in blue) resulting from translation of either conventional or amplified chimeric beta globin mRNA.
In the middle panel of Figure 13, line A shows the 5’terminal portion of edited beta globin mRNA and line B shows its antisense counterpart. Note that “CAU” highlighted in blue (line A) is transcribed into 5’-aug-3’ highlighted in green (shown as 3’-gua-5’) in the antisense strand (line B). Stages C and D of the middle panel of Figure 13 show the folding of the antisense RNA (stage C) and its extension into chimeric intermediate followed by strand separation and cleavage (red arrow, stage D). Line E shows the projected edited chimeric RNA end product. It contains, in its antisense RNA portion, an AUG codon (“aug” highlighted in green), in an optimal translation initiation context and in-frame with the INS/globin coding sequence. Translation of the edited chimeric RNA end product would result in a chimeric polypeptide shown in line F of the middle panel in Figure 13 and comprised of a globin portion (highlighted in blue and identical to that shown in top panel, line C), a Tag peptide (highlighted in grey), and the N-end portion encoded by the antisense RNA component of the chimeric RNA end product (highlighted in grey and underlined); it would be non-contiguously encoded in the genome. The detection of a Tag peptide would indicate that a chimeric polypeptide has been produced and the isolation and sequencing of this polypeptide would provide a definitive and irrefutable proof of the occurrence of the RNA-dependent mRNA amplification process.
The bottom panel of Figure 13 details one of numerous possible controls. In it, the AUG codon of the antisense strand is mutated into a codon, ACA, not known to initiate translation, without changing the complementary relationship between the TCE and the ICE components of the antisense RNA. In such a control, the amplification would proceed similarly to that depicted in the middle panel, but the resulting chimeric RNA end product (bottom panel, line E) would lack a functional in-frame initiation codon and no translation of INS/globin nucleotide sequence would occur. Other possible controls are frame-shifts of the edited RNA shown in the middle panel of Figure 13. Effecting a frame-shift of one, two but not three nucleotides in the region between the AUG of the antisense portion and the “INS” of the chimeric RNA end product (middle panel, line E) should abolish translation of the INS/globin whereas a frame-shift of either one or two or three nucleotides upstream from the AUG of the antisense RNA segment within the same molecule should have no such effect.
Finally, a possibility of non-specific side effects resulting from the administration of phenylhydrazine can be addressed by parallel measurements in “edited” animals following the induction of hemorrhagic anemia shown to trigger, similarly to hemolytic anemia, globin mRNA amplification processes as described previously [22]. Moreover, whereas induced anemia-driven splenic erythropoiesis is indispensible for the analysis of RNA components and enzymatic activities involved in the RNA-dependent mRNA amplification process as described in section 11 below, the projected chimeric polypeptide non-contiguously encoded by the edited beta-globin gene as discussed above (Figure 13) should be detectable, albeit at lower abundance, in erythroid cells of “edited” animals under physiologically normal, i.e. no induced anemia, conditions.
11. Significance and Directions of Future Investigations
The potential physiological, therapeutic and bioengineering significance of RNA-dependent mammalian mRNA amplification, with every genome-originated mRNA molecule acting as a possible template, a process analogous in its effect to a massive gene amplification, can hardly be overstated. Malfunctions of this process may be involved in pathologies associated either with the deficiency of a protein normally produced by this mechanism or with the overproduction of a protein normally not involved in such a process. One of the experimental approaches to assess its involvement in such pathologies would be to interfere in vivo with the extent of complementarity between the two elements involved in the antisense RNA self-priming. In fact, multiple experiments of this sort have been carried out by nature. Thus, at least four different types of familial beta thalassemia, characterized by the reduced production of beta-globin chains, are associated with different point mutations in the 5’UTR of human beta-globin mRNA that clearly impede the amplification-associated complementary TCE/ICE relationship within the antisense beta globin RNA and appear to solely account for the disease [70]. A possible example of an opposite impact, the mRNA amplification-driven pathology that is due to the abnormal overproduction of a polypeptide, is the increased generation of beta amyloid in sporadic Alzheimer’s disease. As discussed in section 4 above, this can be a consequence of RNA-dependent amplification of a 3’-portion of the beta APP mRNA encoding a CTF of the conventional protein that would enable amyloid precursor protein-independent generation of beta-amyloid peptide [25,50–52]. The increased understanding of components and mechanisms involved in RNA-dependent mammalian mRNA amplification could open up new ways and approaches not only for therapeutic interference in multiple pathologies but also for novel and powerful forms of bioengineering.
Multiple directions of investigation would be instrumental in bringing about this increased understanding. Virtually every step of RNA-dependent mRNA amplification discussed above, from the nature of RdRp activity and its postulated processivity conferring co-factor to strand-separating/modifying activity and the nature of nucleotide modifications, to chimeric intermediate-cleaving activity and the 3’polyadenylating activity coupled with it, needs to be further elucidated. Importantly, both genetic/molecular biology research and biochemical studies should be involved in these investigations. In the first category, a well targeted DNA editing and the assessment of its consequences could be crucial in eliciting additional information. In the second category, the choice of the starting material could be decisive in studying enzymatic activities involved in the amplification process and their networking and regulation. The first identification of mammalian RdRp activity capable of generating long antisense transcripts was made in and the activity was isolated from circulating reticulocytes produced during the hemolytic anemia-induced splenic erythropoiesis [18]. But now we understand that it was only a residual, remnant, activity carried over from the previous differentiation stage. We know this from the kinetics of the accumulation and subsequent loss of the putative chimeric RNA end product of globin mRNA amplification; its levels increase massively through the erythroblast stage, while still in spleen, but decline drastically when cells reach the reticulocyte stage and are released into circulation [23]. Therefore, cells undergoing anemia-induced splenic erythropoiesis, in a nearly homogeneous large organ with about 90% of cells differentiating into erythroblasts, appear to be by far the best choice of starting material. Moreover, different stages of splenic erythropoiesis could be best suited to address diverse specific aspects of RNA-dependent mRNA amplification. For example, RdRp activity and its components could be best evaluated during later stages of splenic erythropoiesis whereas certain activities, such as 3’polyadenylating activity coupled with the cleavage of the chimeric intermediate and essential for the iPCR process, should be, for the reasons discussed in section 9 above, investigated at the earlier stages of splenic erythroid differentiation.
12. Conclusion
RNA-dependent mRNA amplification represents a new mode of genomic protein-encoding information transfer in mammalian cells. Its potential physiological impact is substantial, it appears relevant to multiple pathologies and its understanding opens new venues of therapeutic interference, it suggests powerful novel bioengineering approaches and its further rigorous investigations are highly warranted.
Acknowledgements
Great thanks to Sophia Rits for many fruitful discussions and to Bjorn Olsen material support and for critical reading of the manuscript and helpful suggestions.
Funding
NIH RO1AR036819.
References
- 1.Kitamura N, Semler BL, Rothberg PG, Larsen GR, Adler CJ, Dorner AJ, et al. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981;291(5816):547–53 [DOI] [PubMed] [Google Scholar]
- 2.Dunnebacke TH, Levinthal JD, Williams RC. Entry and release of poliovirus as observed by electron microscopy of cultured cells. J Virol. 1969;4(4):505–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter J, Saunders V. In: Virology: Principles and applications. Wiley and Sons; 2007. [Google Scholar]
- 4.Baltimore D Structure of the poliovirus replicative intermediate RNA. J Mol Biol. 1968;32(2):359–68. [DOI] [PubMed] [Google Scholar]
- 5.Lundquist RE, Maizel JV Jr. In vivo regulation of the poliovirus RNA polymerase. Virology. 1978;89(2):484–93. [DOI] [PubMed] [Google Scholar]
- 6.Traub A, Duskin B, Rosenberg H, Kalmar E. Isolation and properties of the replicase of encephalomyocarditis virus. J Virol. 1976;18(2):375–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Temin HM, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226(5252):1211–3. [DOI] [PubMed] [Google Scholar]
- 8.Baltimore D RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226(5252):1209–11. [DOI] [PubMed] [Google Scholar]
- 9.Lai MM 1. RNA replication without RNA-dependent RNA polymerase: surprises from hepatitis delta virus. J Virol. 2005;79(13):7951–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor J Replication of human hepatitis delta virus: recent developments. Trends Microbiol. 2003;11:185–90. [DOI] [PubMed] [Google Scholar]
- 11.Rackwitz H, Rohde W, Sanger H. DNA-dependent RNA polymerase II of plant origin transcribes viroid RNA into full-length copies. Nature. 1981;291:297–301. [DOI] [PubMed] [Google Scholar]
- 12.Astier-Manifacier S Cornuet P. RNA-dependent RNA polymerase in Chinese cabbage. Biochim Biophys Acta. 1971;232(3):484–93. [DOI] [PubMed] [Google Scholar]
- 13.Duda C, Zaitlin M, Siegel, A. In vitro synthesis of double-stranded RNA by an enzyme system isolated from tobacco leaves. Biochim Biophys Acta. 1973;319(1):62–71. [DOI] [PubMed] [Google Scholar]
- 14.Fraenkel-Conrat H RNA-dependent RNA polymerases of plants. Proc Natl Acad Sci U S A. 1983;80(2):422–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahlquist P RNA-dependent RNA polymerases, viruses, and RNA silencing. Science. 2002;296(5571):1270–3. [DOI] [PubMed] [Google Scholar]
- 16.Iyer L, Koonin E, Aravind L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct Biol. 2003;3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapranov P, Ozsolak F, Kim SW, Foissac S, Lipson D, Hart C, et al. New class of gene-termini-associated human RNAs suggests a novel RNA copying mechanism. Nature. 2010;466(7306):642–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Downey KM, Byrnes JJ, Jurmark BS, So AG. Reticulocyte RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A. 1973;70(12):3400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehmann E, Brueckner F, Cramer P. Molecular basis of RNA-dependent RNA polymerase II activity. Nature. 2007;450(7168):445–9. [DOI] [PubMed] [Google Scholar]
- 20.Wagner SD, Yakovchuk P, Gilman B, Ponicsan SL, Drullinger LF, Kugel JF, et al. RNA polymerase II acts as an RNA-dependent RNA polymerase to extend and destabilize a non-coding RNA. EMBO J. 2013;32(6):781–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maida Y, Yasukawa M, Masutomi K. De Novo RNA Synthesis by RNA-Dependent RNA Polymerase Activity of Telomerase Reverse Transcriptase. Mol Cell Biol. 2016;36(8):1248–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volloch V, Schweitzer B, Rits S. Antisense globin RNA in mouse erythroid tissues: Structure, origin, and possible function. Proc Natl Acad Sci USA. 1996;93(9):2476–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rits S, Olsen B, Volloch V. RNA-dependent synthesis of mammalian mRNA: Identification of chimeric intermediate and putative end-product. Bio Rxiv, 2016;071266. [Google Scholar]
- 24.Volloch V, Rits S, Olsen B. RNA-dependent amplification of mammalian mRNA encoding extracellular matrix components: identification of chimeric RNA intermediates for a1, ß1, and γ1 chains of laminin. Bio Rxiv. 2018;376293. [PMC free article] [PubMed] [Google Scholar]
- 25.Volloch V, Rits S. Results of beta secretase-inhibitor clinical trials support amyloid precursor protein-independent generation of beta amyloid in sporadic Alzheimer’s disease. Med Sci (Basel). 2018;6(2).pii: E45,6020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richards OC, Ehrenfeld E. Poliovirus RNA replication. Curr Top Microbiol Immunol. 1990;161:89–119. [DOI] [PubMed] [Google Scholar]
- 27.Volloch V, Schweitzer B, Rits S. Evolutionary conserved elements in the 5’ untranslated region of beta globin mRNA mediate site-specific priming of a unique hairpin structure during cDNA synthesis. Nucl. Acids Res. 1994;22:5302–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iizuka T, Shoji M, Kawarabayashi T, Sato M, Kobayashi T, Tada N, et al. Intracellular generation of amyloid ß-protein from amyloid ß-protein precursor fragment by direct cleavage with ß- and γ -secretase. Biochem Biophys Res Commun. 1996;218(1):238–42. [DOI] [PubMed] [Google Scholar]
- 29.DeStrooper B, Annaert W. Proteolytic processing and cell biological functions of the amyloid precursor protein. J Cell Sci. 2000;113(11):1857–70. [DOI] [PubMed] [Google Scholar]
- 30.Barber RC. The genetics of Alzheimer’s disease. Scientifica (Cairo). 2012;2012:246210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beyreuther K, Masters CL. Amyloid precursor protein (APP) and beta A4 amyloid in the etiology of Alzheimer’s disease: precursor-product relationships in the derangement of neuronal function. Brain Pathol. 1991;1(4):241–51. [DOI] [PubMed] [Google Scholar]
- 32.Hardy J, Allsop D. Amyloid deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol Sci. 1991;12(10):383–8. [DOI] [PubMed] [Google Scholar]
- 33.Selkoe DJ. The molecular pathology of Alzheimer’s disease. Neuron. 1991;6(4):487–98. [DOI] [PubMed] [Google Scholar]
- 34.Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256(5054):184–5. [DOI] [PubMed] [Google Scholar]
- 35.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–6. [DOI] [PubMed] [Google Scholar]
- 36.Dyrks T, Dyrks E, Monning U, Urmoneit B, Turner J, Beyreuther K. Generation of beta A4 from the amyloid protein precursor and fragments thereof. FEBS Lett. 1993;335(1):89–93. [DOI] [PubMed] [Google Scholar]
- 37.Haass C, Lemere CA, Capell A, Citron M, Seubert P, Schenk D, et al. The Swedish mutation causes early-onset Alzheimer’s disease by beta-secretase cleavage within the secretory pathway. Nat Med. 1995;1(12):1291–6. [DOI] [PubMed] [Google Scholar]
- 38.Hunter S, Brayne C. Understanding the roles of mutations in the amyloid precursor protein in Alzheimer disease. Mol Psychiatry. 2018;23(1):81–93. [DOI] [PubMed] [Google Scholar]
- 39.Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, et al. Betasecretase cleavage of Alheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286(5440):735–41. [DOI] [PubMed] [Google Scholar]
- 40.Hussain I, Powell D, Howlett DR, Tew DG, Meek TD, Chapman C, et al. Identification of a novel aspartic protease (Asp 2) as beta-secretase. Mol Cell Neurosci. 1999;14(6):419–27. [DOI] [PubMed] [Google Scholar]
- 41.Vassar R BACE inhibitor drugs in clinical trials for Alzheimer’s disease. Alzheimers Res Ther. 2014;6(9):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stamford AW, Scott JD, Li SW, Babu S, Tadesse D, Hunter R, et al. Discovery of an Orally Available, Brain Penetrant BACE1 Inhibitor that Affords Robust CNS Aβ Reduction. ACS Med Chem Lett. 2012;3(11):897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keskin AD, KekuAj M, Adelsberger H, Neumann U, Shimshek DR, Song B, et al. BACE inhibition-dependent repair of Alzheimer’s pathophysiology. Proc Natl Acad Sci U S A. 2017;114(32):8631–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neumann U, Rueeger H, Machauer R, Veenstra SJ, Lueoend RM, Tintelnot-Blomley M. A novel BACE inhibitor NB-360 shows a superior pharmacological profile and robust reduction of amyloid-ft and neuroinflammation in APP transgenic mice. Mol Neurodegener. 2015;10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu X, Das B, Hou H, He W, Yan R. BACE1 deletion in the adult mouse reverses preformed amyloid deposition and improves cognitive functions. J Exp Med. 2018;215:927–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Breimer LH, Denny P. Alzheimer amyloid aspects. Nature. 1987;326(6115):749–50. [DOI] [PubMed] [Google Scholar]
- 47.Citron M, Haass C, Selkoe DJ. Production of amyloid-beta-peptide by cultured cells: no evidence for internal initiation of translation at Met596. Neurobiol Aging. 1993;14(6):571–3. [DOI] [PubMed] [Google Scholar]
- 48.Mita S, Sadlock J, Herbert J, Schon EA. A cDNA specifying the human amyloid beta precursor protein encodes a 95-kDa polypeptide. Nucleic Acids Res. 1988;16(19):9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mita S, Sadlock J, Herbert J, Schon E. A cDNA specifying the human amyloid beta precursor protein encodes a 95-kDa polypeptide. Nucl Acids Res. 1988;16:11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volloch V A mechanism for β-amyloid overproduction in Alzheimer’s disease: Precursor-independent generation of β-amyloid via antisense RNA-primed mRNA synthesis. FEBS Lett. 1996;390(2):124–8. [DOI] [PubMed] [Google Scholar]
- 51.Volloch V Mechanism for β-amyloid overproduction in Alzheimer’s Disease: Possible antisense RNA-mediated generation of a 5’-truncated βAPP mRNA encoding 12 kDa C-terminal fragment of βAPP, the immediate precursor of Aβ In Molecular Mechanisms of Dementia; Wasco W, Tanzi R, Eds.; Humana Press; 1997. [Google Scholar]
- 52.Volloch V Possible mechanism for resistance to Alzheimer’s disease (AD) in mice suggests new approach to generate a mouse model for sporadic AD and may explain familial resistance to AD in man. Exp Neurol. 1997;144(1):214–8. [DOI] [PubMed] [Google Scholar]
- 53.Lodish H, Berk A, Zipursk L. In: Molecular Cell Biology. New York, Freeman and Co; 2010. [Google Scholar]
- 54.Jain SK, Subrahmanyam D. On the mechanism of phenylhydrazine-induced hemolytic anemia. Biochem Biophys Res Commun. 1978;82(4):1320–4. [DOI] [PubMed] [Google Scholar]
- 55.Conkie D, Kleiman L, Harrison PR, Paul J. Increase in the accumulation of globin mRNA in immature erythroblasts in response to erythropoietin in vivo or in vitro. Exp Cell Res. 1975;93(2):315–24. [DOI] [PubMed] [Google Scholar]
- 56.Bastos RN, Volloch Z, Aviv H. Messenger RNA population analysis during erythroid differentiation: a kinetical approach. J Mol Biol. 1977;110(2):191–203. [DOI] [PubMed] [Google Scholar]
- 57.Hundley HA, Bass BL. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem Sci. 2010;35(7):377–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clissold PM, Arnstein HR, Chesterton CJ. Quantitation of globin mRNA levels during erythroid differentiation in the rabbit. Cell. 1977;11(2):353–61. [DOI] [PubMed] [Google Scholar]
- 59.British Society for Cell Biology. https://bscb.org/learning-resources/softcell-e-learning/ribosome/
- 60.Piccinni SA, Bolcato-Bellemin AL, Klein A, Yang VW, Kedinger M, Simon-Assmann P, et al. Kruppel-like factors regulate the Lama1 gene encoding the laminin alpha1 chain. J Biol Chem. 2004;279(10):9103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okano R, Mita T, Matsui T. Characterization of a novel promoter structure and its transcriptional regulation of the murine laminin B1 gene. Biochim Biophys Acta. 1992;1132(1):49–57. [DOI] [PubMed] [Google Scholar]
- 62.Rabouille C Pathways of Unconventional Protein Secretion. Trends Cell Biol. 2017;27(3):230–40. [DOI] [PubMed] [Google Scholar]
- 63.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER Ssress to produce a highly active transcription factor. Cell. 2001;107(7):881–91. [DOI] [PubMed] [Google Scholar]
- 64.Jiang S, Zhang A, Zhang R, Xiangiang L. Altered activity patterns of transcription factors induced by endoplasmic reticulum stress. BMC Biochem. 2016;17:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Juba A, Chaput J, Wellenslek B. Exploring the Role of AUG Triplets in Human Cap Independent Translation Enhancing Elements. Biochem. 2018;57;6308–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kearse MG, Wilusz JE. Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev. 2017;31(17):1717–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Varshavsky A The N-end rule pathway of protein degradation. Genes to Cells. 1997;2(1):13–28. [DOI] [PubMed] [Google Scholar]
- 68.Giglione C, Boularot A, Meinnel T. Protein N-terminal methionine excision. Cell Mol Life Sci. 2004;61(12):1455–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Halder S, Kant R, Milner R. Chronic mild hypoxia increases expression of laminins 111 and 411 and the laminin receptor a6ß1 integrin at the blood-brain barrier. Brain Res. 2018;1700:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cao A, Galanello R. Beta-thalassemia. Genet Med. 2010;12(2):61–76. [DOI] [PubMed] [Google Scholar]
- 71.Tseng CH, Lai MM. Hepatitis delta virus RNA replication. Viruses. 2009;1(3):818–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brazas R, Ganem D. A cellular homolog of hepatitis delta antigen: implications for viral replication and evolution. Science. 1996;274(5284):90–4. [DOI] [PubMed] [Google Scholar]
- 73.Huang CR, Lo SJ. Evolution and diversity of the human hepatitis d virus genome. Adv Bioinformatics. 2010, ID 323654. [DOI] [PMC free article] [PubMed] [Google Scholar]


