Abstract
The role of gut microbes in health and disease has often been surmised from stool, which is easily sampled and rich in microbial diversity, density, and abundance. Microbial analyses of stool have been accepted as measures to determine the relationship of gut microbiomes with host health and disease, based on the belief it represents all microbial populations throughout the gut. However, functional heterogeneity of each gastrointestinal tract (GIT) tract segment gives rise to regional differences in gut microbial populations. Herein, we summarize the literature regarding the microbial landscape along the rostral to caudal, i.e. horizontal mouth to anus, axis of the GIT. We aim to identify gaps in the literature, particularly regarding small intestinal microbiota abundance and diversity, highlight the importance of regional microbiota on host health and disease, as well as discuss opportunities to advance this line of research.
In Brief
Microbes inhabit the length of the gastrointestinal tract but differ in type and abundance in a regional fashion (mouth to colon). In this review, Martinez-Guryn, Leone, et al. examine host and environmental pressures that drive regional heterogeneity and how the regional gut microbiota influences physiological host processes and disease development.
Exploring new microbial territories
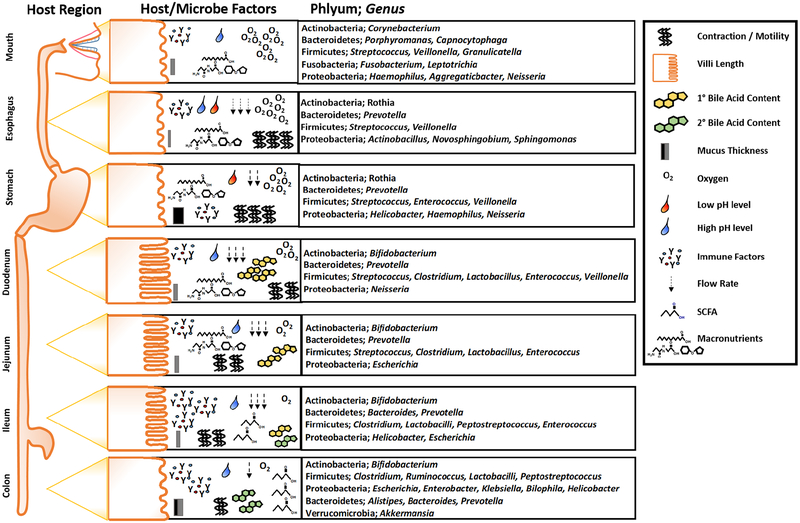
The gastrointestinal tract (GIT) is a multi-organ system with great regional diversity, housing extensive gut microbes and providing diverse functions. Each region is geographically specialized in gene expression and function, regulating complex and diverse digestive, immunological, metabolic, and endocrinological processes. Whether it is the acidity of gastric juices, the entry point of bile acids (BAs) and pancreatic secretions, the digestive and absorptive surfaces of the small intestine (SI), or water extraction and stool formation in the colon, these properties create critical environmental conditions that determine microbial community assemblage, fitness, and functions that mutually benefit host and microbes (Figure 1). Acquisition of regional microbiota is not random but arises from careful selection customized to the specific host needs through dynamic mutually reinforcing interactions.
Figure 1. Representative host and microbial factors and community membership along the rostral to caudal (horizontal) axis of the GI tract.
Different regions of the GI tract perform unique functions in regards to macro- and micronutrient digestion and absorption. These unique features along with a number of host derived factors, such as epithelial cell types and surfaces, mucus thickness, motility and contractility, pH, oxygen tension, and flow rate drive diversity and abundance of gut microbes from mouth to anus. Additionally, each area of the intestine secretes unique immune factors that can interact with the intestinal microbiota, shaping the community membership in a region-specific manner. Several host factors secreted into the intestine, such as primary bile acids (BAs), are important for digestion, but can also elicit direct antimicrobial effects i.e., primary vs. secondary BAs. Other regions of the intestine, due to pH and oxygen tension support microbial fermentation of complex fiber sources, resulting in an increasing abundance of short chain fatty acids in the distal portions of the GI tract that not only impact local epithelial cells, but further influence the regional microbial communities and distal tissue sites important for health and disease.
This review examines the extensive heterogeneity of gut microbes along the rostral to caudal axis of the GIT. We bring together current knowledge of regional gut microbiota composition and function, outlining how disturbances created by the host, environment, or diet can have detrimental consequences. Many challenges currently hinder studies of regional gut microbiota, especially in humans. This being said, emerging technologies and model systems will enable major advances in the future.
Exploring and mapping the rostral to caudal axis of GIT microbiomes
The abundance and diversity of microbes along the entire GIT generally increases from the proximal to distal intestine, influenced by host features as well as microbial community dynamics (Tropini et al., 2017). Regional oxygen level, nutrient bioavailability, pH, BAs, GI transit time, mucus, and immune factors are all important determinants of microbial selection (Figure 1, Friedman et al., 2018). A deep understanding of the microbial side has been evasive, in that most studies are limited to description of regional membership via 16S rRNA sequencing, with few functional insights.
Mouth
Digestion begins in the mouth with mastication of food and enzymatic digestion of macronutrients facilitated by coordinated action between teeth, tongue and salivary secretions. Multiple factors influence oral cavity microbial assemblage, including differences in surface structure at the micron scale, function and shedding of those surfaces, as well as nutrient flow and oxygen level. Despite ecological differences relative to the distal GIT, the oral microbial community is complex and diverse, containing nearly 20 billion microbes representing over 700 identified bacterial species (Dewhirst et al., 2010).
The microbiota of oral tissues such as the teeth, gingiva, tongue, hard and soft palatal mucosa, and tonsils are distinct and highly structured. For example, the microbiota of supragingival plaques are spatially organized with a “hedgehog” arrangement of Corynebacterium setting the foundation of plaque-forming filaments that extend outward, while other taxa take on distinct niches around and within this structure. This confers a strong structural base that resists physical disruption, i.e. teeth brushing (Mark Welch et al., 2016). Non-shedding surfaces such as the teeth, dentures, or implants also serve as a foundation for biofilm formation in the mouth (Lynge Pedersen and Belstrøm, 2019).
Functional features of the mouth include breathing, sensory receptor expression, secretion of saliva for mastication and enzymatic digestion of food, as well as serving as an immunological barrier between the host and environment. These features determine assemblage and elicit bidirectional relationships with resident microbes. For example, direct exposure to environmental oxygen favors aerobes and facultative anaerobes. Sensory perception of fat is affected by gut microbes, as germ-free (GF) mice display decreased expression of the long chain fatty acid translocase CD36, a lipid sensor on the tongue (Duca et al., 2012). Salivary production impacts microbial composition in various ways such as 1) flushing food and bacteria from the mouth, 2) liberating nutrients via digestive enzymes for bacterial metabolism, 3) releasing mucins that promote bacterial aggregation, and 4) delivering various immune factors, including antimicrobial compounds. Notably, decreased salivary flow can alter pH to affect the oral microbiome (Lynge Pedersen and Belstrøm, 2019).
Commensal bacteria in the oral cavity enhance immune function and confer protection from pathogenic microbes involved in forming supragingival and dental biofilms that promote dental caries. Commensal microbes such as Veillonella, Streptococcus, and Granulicatella gingiva increase anti-microbial peptide (AMP) production and inflammatory cytokine secretion, leading to increased epithelial barrier function and mucosal thickness in 3D-reconstructed human gingiva (Figure 1, Shang et al., 2018).
In normal conditions, oral microbes are continuously transmitted to the distal GI in numbers greater than expected by ingestion alone (Schmidt et al., 2019). However, transmission of certain oral microbes can potentially promote disease in the distal gut in genetically susceptible hosts. Orally-derived Klebsiella was found to colonize the colon, which promoted mucosal inflammation through TH1 cell activation and inflammation (Atarashi et al. 2017). Similarly, Klebsiella strains isolated from Crohn’s Disease (CD) and ulcerative colitis (UC) patients induced a Th1 immune response in GF mice compared to several other isolates of different genera. Notably, these Klebsiella strains were antibiotic resistant, making them well-suited to establish a colonic niche and mount an inflammatory response in genetically susceptible hosts, i.e., IL-10−/− mice.
Esophagus
As a conduit for partially digested food into the stomach, the esophagus houses a complex environment of microorganisms and host immune factors. The esophageal microbiota composition is similar to the oral microbiome with major phyla including Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Fusobacteria, and Saccharibacteria and genera Prevotella, Veillonella, and Streptococcus (Figure 1, reviewed in May and Abrams, 2018).
Much like other GIT regions, the esophageal microbiota is highly influenced by diet. For example, an obesogenic diet alters esophageal microbiota composition with increased representation of the genus Clostridium sensu strico (Kaakoush et al., 2017). Others found that increased dietary fiber intake and low fat diet (LFD) promote greater abundance of Firmicutes and reduced abundance of Proteobacteria and gram-negative bacteria, whereas low fiber intake was associated with greater abundance of Prevotella, Neisseria, and Eikenella. Whether these changes are associated with esophageal disease was not investigated (Nobel et al., 2018).
Stomach
The stomach continues digestion through peristaltic action and breakdown of food components within its acidic environment, playing an important role in regulating food intake through its endocrine function. It contains fewer total microbes than in distal regions of the GIT, ~101 to 103 CFU/ml (O’Hara and Shanahan, 2006). Despite gastric pH, mucosal thickness, and peristalsis that limit growth of microbes, the gastric microbiota is diverse, including major phyla such as Firmicutes, Bacteroidetes, Fusobacteria, Actinobacteria, and Proteobacteria, and at the genus level Prevotella, Streptococcus, Veillonella, Rothia and Haemophilus (Figure 1, Nardone and Compare, 2015).
The presence of gut microbes may contribute important host functions of the stomach, and may even contribute to systemic diseases. For example, GF mice have significantly elevated levels of the GI hormone ghrelin, which stimulates appetite, food intake, gastric motility and adiposity (Slade et al., 2018) as compared to conventionally-raised mice, suggesting a microbial role in its regulation (Martinez-Guryn et al., 2018). As G cells in the stomach produce ghrelin, gastric microbes may affect host endocrine signaling and metabolism.
Small intestine (SI)
The single layer of polarized intestinal epithelial cells (IECs) of the SI includes diverse cell types, such as absorptive, goblet, Paneth, Tuft, enteroendocrine, and other cell types that are differentially distributed along the length of the SI and impart region-specific assembly rules that determine microbial abundance, diversity, and function. One possible reason why the microbial load is relatively low in the SI compared to more distal regions is that it is purposefully designed for macro- and micronutrient absorption and immune regulation. Its motility pattern, for instance, is primarily determined by a gradient of non-propulsive annular contractions that maximizes mixing of digestive juices and content and produces transit times ranging 3 to 5 hrs (Szarka and Camilleri, 2012). In comparison, the colonic transit time is much longer (>30 hrs). This discrepancy may account for lower microbiota diversity and abundance merely due to less time to colonize and establish stability in the SI. Consistent with this notion, when SI stasis occurs, the luminal microbiota converts to a colonic-like microbiota that exhibit greater diversity, richness, and abundance (Ward et al., 2016).
1. Duodenum
Despite its shorter length and rapid transit of gastric chyme, the duodenum is a major site for digestion and absorption where biliary and pancreatic secretions facilitate macronutrient breakdown. Primary bile acids (BAs) made by the liver from cholesterol are conjugated with glycine (humans), taurine (mice), or sulfate prior to delivery into the duodenum (Russel and Setchell, 1992). BAs are amphipathic detergents that not only emulsify fat for digestion, but also favor bile-tolerant microbiota, and signal through nuclear receptors to affect host gene expression. The absorption of digested proteins, carbohydrates, and lipids by IECs begins here, facilitated by a thinner overlying mucus barrier in comparison to that seen in distal gut regions (Ermund et al., 2013).
The duodenum contains roughly 101 to 103 CFU/ml (O’Hara and Shanahan, 2006). Decreased bacterial abundance in the upper SI is attributed to reduced transit time, higher oxygen levels, antimicrobial compounds such as BAs, elevated pH levels, and presence of digestive enzymes. Oxygen gradients created by swallowed air, delivery from host tissues, and oxygenation through pancreatic and biliary secretions (Friedman et al., 2018) can influence microbial abundance and type in the upper SI, which consist mostly of Actinobacteria and Proteobacteria in mice. In turn, significant shifts in duodenal gut microbiota membership and function may have important implications to the host. Analysis of duodenal metagenomes in obese and lean participants (Angelakis et al., 2015) showed microbial genes involved in carbohydrate metabolism were decreased, whereas an increase in genes related to lipid metabolism were observed among obese individuals. Thus, duodenal microbiota may affect bioavailability of dietary nutrients to the host.
2. Jejunum
The jejunum is a major site of nutrient absorption, although not easily accessible. Endoscopic samples are the major sources of information, which limits understanding of the healthy upper GI microbiome because these individuals usually have some underlying condition. Thus, the majority of our knowledge of these regions has been gleaned from animal studies, where both mucosal and luminal samples can be readily obtained. Factors that influence the jejunal microbiota include diet, oxygen levels, bile acids, transit time, and a thinner overlying mucus. The jejunal microbiota is estimated to consist of 104 to 107 CFU/ml, primarily represented by Firmicutes, but also containing Proteobacteria, Actinobacteria, and Bacteroidetes (El Aidy et al., 2013; O’Hara and Shanahan, 2006). Similar taxa are found in the jejunum of human subjects that markedly differs from fecal microbiota (Seekatz et al., 2019).
Fecal microbial transplantation into GF mice causes functional changes in the jejunal transcriptome as early as one day (El Aidy et al., 2013), whereby metabolic genes (e.g. fat metabolism) were significantly altered compared to GF counterparts. This acute transcriptional response was specific to the jejunum and not observed elsewhere in the GIT. However, a limitation of this study was the use of fecal microbiota which differs from SI microbiota. Our studies, for instance, have shown that high fat diets (HFD) affect membership and function of the murine SI microbiota to a much greater extent than that of cecal and stool microbiota, with increased Firmicutes, particularly the family Clostridiaceae (Martinez-Guryn et al., 2018). When HFD-induced jejunal microbiota were transferred into GF mice, lipid absorption was increased compared to GF mice receiving LFD-induced jejunal microbiota. Thus, HFD-induced microbiota differentially primes the naïve GF gut compared to LFD microbiota, enhancing fat absorption.
3. Ileum
Digestion and absorption in the ileum is unique from the other regions, and not surprisingly its microbial community membership and functions differ. The ileum has an estimated microbial load of 103–108 CFU/mL, mostly comprised of facultative and obligate anaerobes. From limited studies in humans, the ileal microbiota appear to consist mainly of Bacteroides, Clostridium, Enterobacteria, Enterococcus, Lactobacillus, and Veillonella (Li et al., 2017; Zoetendal et al., 2012). Collectively, ileal IECs function as the primary absorption site for nutrients such as B vitamins, residual nutrients not absorbed more proximally, as well as BA reuptake and reentry into enterohepatic circulation, the latter influenced greatly by gut microbes.
The distal ileum expresses nuclear receptors involved in BA signaling, such as the farnesoid x receptor (FXR) and the G-protein coupled receptor TGR5, that elicit profound effects on downstream metabolic pathways (Ridlon et al., 2016a). GF mice have elevated levels of total BAs as well as abundance of SI muricholic acid compared to conventionally-raised animals (Sayin et al., 2013). The distal ileum of conventional mice expressed increased levels of basolateral BA transporters as well as FXR target genes, specifically Shp and Fgf15, important mediators of the BA enterohepatic pool. The microbially-mediated upregulation of Fgf15 decreased hepatic BA synthesis mediated by decreased expression of hepatic Cyp7a1, a rate limiting BA synthesis enzyme. Conversely, levels of these genes could be restored to those observed in GF mice by treating conventionally-raised animals with antibiotics (Sayin et al., 2013).
BAs themselves can in turn directly and indirectly impact the regional gut microbiota. Oral administration of certain conjugated BAs inhibits bacterial overgrowth and restores barrier integrity (Hegade et al., 2016). Recent work revealed activation of FXRα-upregulated mucosal immune factors, specifically in the ileum, which protected against bile duct ligation-induced overgrowth of both anaerobic and aerobic bacteria (Inagaki et al., 2006). Thus, BAs and gut microbiota can profoundly affect enterohepatic circulation to impact host metabolism and immunity.
Specific microbial community members can alter the genetic landscape of the ileum in isolation. Monoassociation of adult GF mice with Bacteroides thetaiotamicron significantly influenced expression of genes related to micro- and macronutrient uptake (Hooper et al., 2001). These findings partially explain why GF mice consume more calories daily to maintain the same amount of body weight compared to conventionally-raised counterparts (Bäckhed et al., 2007). In addition, B. thetaiotamicron dramatically increases colipase expression of ileal crypts and many genes associated with intestinal barrier function, immune responses, and host xenobiotic metabolism. IgA-producing B cells and Polymeric Immunoglobulin Receptor (PIgR) involved in IgA transcytosis were also elevated following colonization, indicating that both innate and adaptive immune function in the ileum are dramatically impacted by gut microbes (Hooper et al., 2001).
The terminal ileum is an important site for mucosal immunity where the mucus is thinner and Paneth cells produce a wide array of innate immune factors such as AMPs, including cathelicidins, C-type lectins, and defensins. Various microbe-associated molecular patterns (MAMPs) including LPS, peptidoglycan, flagella, bacterial DNA/RNA, fungal cell walls, and Lipid A can induce AMP expression and other mucosal adaptive immune components, such as IgA. Conventionalization of GF mice increases transcriptional and translational levels of AMPs (Natividad et al., 2013), particularly REGIIIγ, a C-type lectin made by both Paneth cells and absorptive enterocytes that exhibits bacteriocidal activity against Gram-positive organisms (Hooper and Macpherson, 2010). Commensal gut microbiota increase REGIIIγ expression (Cash et al., 2006), while HFD-induced microbial communities in conventionally-raised mice decreases expression of AMPs, including RegIIIγ, in the ileum (Everard et al., 2014), suggesting an important mechanism of dietary modulation of regional gut immune function.
Microbes are essential to drive both innate and adaptive mucosal immunity of the ileum as well as systemic immune function. For example, CD4+ RORγt+ T helper 17 (Th17) cells, which produce Interleukin (IL)-17A/IL-17F, IL-21, and IL-22, are dependent upon the presence of Segmented Filamentous Bacteria (SFB) (Ivanov et al., 2009). Indeed, GF animals, as well as newborns exhibit very few, if any, Th17 cells within the lamina propria. However, SFB adheres tightly to enterocytes and Peyer’s patches in the terminal ileum, a requirement for its immune-mediated stimulation of Th17 cells (Atarashi et al., 2015). Moreover, IL17a expressing RORγt+ T cells were found to be localized to the ileum in response to SFB (Sano et al., 2015). SFB can deter enteric pathogens like Citrobacter rodentium, Salmonella Typhimurium, or the nematode Nippostrongylus brasiliensis by enhancing Th17-mediate immunity (Stockinger and Omenetti, 2017). The presence of SFB may also mitigate the development of diabetes in the non-obese diabetic mouse model (Kriegel et al., 2011), although it does not appear to affect obesity outcomes following exposure to HF diet (Harley and Karp, 2012).
Circadian rhythmicity aids in maintaining metabolic function and immunity, that can be disrupted based on meal timing or type of diet. Several studies recently revealed a link between the host circadian system and gut microbiota. Gut microbiota, for example, drive rhythmic signaling events downstream of innate immune toll-like receptors (TLRs) within the ileal IEC compartment to regulate molecular circadian clock function (Mukherji et al., 2013). Gut microbiota also exhibit diurnal patterns in community membership and function that provide important inputs into host circadian rhythms and downstream metabolic functions (Leone et al., 2015). Both dietary and host factors are major determinants of microbial membership and their oscillatory properties. The distal gut microbiota (cecum, colon, and feces) also exhibit the diurnal variation that can be affected by circadian disruption by genetic manipulation, induction of jet lag, as well as to time of food intake (Thaiss et al., 2014; Zarrinpar et al., 2014). In the SI, circadian transcription nuclear factor interleukin 3 (NFIL3 or E4BP4) drives immune and metabolic function in a microbe-dependent manner (Wang et al., 2017). Here, ileal IECs from GF mice exhibit reduced expression of Nfil3 relative to conventionally-raised counterparts. IEC-specific Nfil3 knockout mice remain lean relative to WT littermates even on HFD, similar to that observed in GF animals. NFIL3 was ultimately shown to contribute to diet-induced obesity via affecting lipid absorption, as Nfil3 knockout mice had decreased epithelial lipid levels and increased lipids in the stool, overall demonstrating an important microbiota, host circadian, and metabolic interaction in the ileum.
Colon
The colon harbors much greater microbial abundance and diversity relative to the SI, through its unique functions, storage capacity, and physiological role. Despite its shorter length, the colon has a transit time that is considerably longer (>30hr) than the SI (Szarka and Camilleri, 2012). The thicker inner and outer mucus layers are essential barriers against the estimated ~1010 to 1012 CFUs/ml of (O’Hara and Shanahan, 2006) bacterial phyla such as Firmicutes and Bacteroidetes and families like Lachnospiraceae, Bacteroidaceae, and Prevotellaceae. The colon has functionally distinct regions, where the cecum and proximal regions of the colon are the major sites of fermentation, and the distal colon primarily extracts fluid and electrolytes (~ 1.3L/day) (O’Hara and Shanahan, 2006). Regional heterogeneity in metagenomic profiles of colonic mucosa-associated microbiota can also be appreciated when unprepped colonoscopy is performed (Wang et al., 2010). Here, proximal and distal colonic microbiota differed in representation by functional subsystems including short chain fatty acid (SCFA) production, primary to secondary BA conversion, and regulation of GI motility.
Strict colonic anaerobes such as Clostridia, Eubacteria, and Roseburia ferment complex carbohydrates and fiber (Koh et al., 2016), producing SCFAs like acetate, propionate, butyrate, and valerate (Sommer and Backhed, 2016) that are mildly acidic 2 to 5 carbon molecules and primary fuel sources for colonocytes. They are actively and passively transported by epithelial cells, but also activate G-protein coupled receptors (GPCRs) in the colon and peripheral tissues. Both butyrate and propionate modulate histone deacetylases (HDACs), which are implicated in immune function and cancer (Koh et al., 2016). SCFAs also enhance anti-inflammatory properties of adaptive immune cells (Tao et al., 2007) and influence a range of physiological processes (MacFabe et al., 2011; Weitkunat et al., 2016).
The cecum and colon are predominant sites for conversion of primary to over 20 types of secondary BAs via a broad range of microbial processes (Ridlon et al., 2016a). While oxidation and epimerization of primary BAs via hydroxysteroid dehydrogenase enzymes are major pathways, conversion of primary glycine or taurine-conjugated BAs to secondary BAs occurs by deconjugation via bile salt hydrolase (BSH). A broad range of bacterial genera harbored in the colon, including Clostridium, Lactobacillus, Bifidobacterium, Bacteroides, and enteric pathogens such as Listeria encode BSH and are able to perform this function (Gerard, 2014). Depending on the nature of the microbe, BSH activity detoxifies BAs. Colonic BSH deconjugation is followed by 7α/β-dehydroxylation resulting in the formation of deoxycholic (DCA) and lithocholic acids (LCA) predominantly. The BA-inducible gene network that encodes dehydroxylation enzymes were identified in Clostridium scindens and C. hyelomonae (Kang et al., 2008; Ridlon et al., 2016b). Short-term human consumption of an animal-based diet led to increased DCA abundance in stool that corresponded with increased BSH expression compared to baseline or after plant-based diet consumption (David et al., 2014). Both DCA and LCA have been linked to colon cancer. While DCA can be reconjugated with taurine or glycine in the liver reducing its toxicity, LCA must undergo amino acid conjugation and sulfation, after which it is mainly eliminated from the body in feces (Ridlon et al., 2016b).
Conversion of primary to secondary BAs involves formation of esters via microbially-derived esterification enzymes as well as sulfatase activity. Rodents fed high saturated fat diets derived from milk promoted an expansion of cecal and colonic Bilophila wadsworthia, known for its ability to utilize the taurine-derived sulfite as a terminal electron acceptor (Devkota et al., 2012). This expansion corresponded with increased Th1 interferon-gamma producing CD4+ T cells, leading to increased severity of colitis in genetically-susceptible IL-10−/− mice (Devkota et al., 2012). This finding was supported by the aforementioned human study in which consumption of animal-based products increased bile-tolerant Bilophila, Alistipes, and Bacteroides abundance along with increased gene levels of the bacterial sulfite reductase enzyme (dsrA) that reduces sulfite to H2S (David et al., 2014).
GI motility is impacted by types and functions of microbes harbored in the colon. GF mice have reduced GI motility and increased transit time vs conventional mice. Ex vivo stimulation of colonic rings from GF and conventionally-raised counterparts revealed GF colons are more sensitive to carbachol (a cholinergic agonist that stimulates muscle contraction) and KCl, indicating hyper-reactivity (Touw et al., 2017). Constipation induced by Loperamide, which slows intestinal transit, alters gut microbiota composition and functional capacity, which directly affected colonic contraction (Kashyap et al., 2013; Touw et al., 2017). Microbes impact GI motility via SCFA-induced peristalsis, as SCFA treatment of ex vivo rat middle to distal colons induced peristalsis via the release of 5-hydroxytryptamine (5-HT, serotonin) (Grider and Piland, 2007). Furthermore, tryptophan production by spore-forming gut microbes associated with a healthy gut can increase 5-HT production by colonic enterochromaffin cells, to stimulate GI motility (Yano et al., 2015). Together, these results highlight the bi-directional communication between host and microbe in a region-specific manner to regulate motility.
Region-specific gut microbiota and disease
The regional tropism of GIT microbiota can be a determinant of specific diseases. For instance, gastric Helicobacter pylori, can promote peptic ulcer diseases and gastric cancer in humans, whereas different species like Helicobacter hepaticus can cause colitis in susceptible mice (Fox et al., 2011). Similarly, severe perturbations in the colonic microbiota and niche conditions (e.g. increased primary bile acids) caused by broad spectrum antibiotics favor the germination and bloom of Clostridium difficile, and development of colitis.
1. Diseases of the Mouth, Esophagus, and Stomach
Dental caries and periodontal disease are influenced by periodontal pathogens including Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, and Actinomyces israelii (Shim et al., 2018). Saccharibacteria which is plentiful in dental and supragingival plaque formation may also contribute to periodontal disease (Liu et al., 2012).
Esophageal microbiota were previously overlooked as having a role in the development of esophageal diseases. However, their role in disorders such as Barrett’s esophagus, eosinophilic esophagitis, esophageal adenocarcinoma, and esophageal squamous cell carcinoma are now being revisited (reviewed in May and Abrams, 2018). Fecal microbiota from HFD-fed L2-IL1B mice , a mouse model of Barrett’s esophagus, increased tumor development when transferred into LFD-fed GF mice, suggesting a causal role for microbes in esophageal tumor development (Münch et al., 2019). However, since fecal microbiota were used in this study, the precise role of esophageal microbes in this disease remains unclear.
Gastric acid creates a unique niche for H. pylori, which was one of the most prevalent human indigenous microbes due to its ability to thrive under such conditions. Prior to improved hygiene, many individuals benefited from H. pylori through enhanced induction of gastric acid and mucosal immune mechanisms capable of deterring many pathogens. However, as hygiene improved and life span lengthened across societies, the negative consequences of indigenous H. pylori became apparent, e.g. peptic diseases and cancer (Nardone and Compare, 2015). In addition, immunological tolerance to H. pylori-induced T cell responses has been shown to increase bacterial levels by ten-fold in murine neonates (Fung et al., 2019), potentially affecting digestion and absorption and susceptibility to other pathogens.
2. The Small Intestine: Small Intestine Bacterial Overgrowth (SIBO), Bariatric Surgery, Undernutrition, and Celiac Disease
SIBO caused by decreased small bowel motility, impaired host immune functions, and reduced gastric acid barrier, is characterized by a bacterial load of 105 or 106 CFU/ml as opposed to normal levels of 103/ml (Dukowicz et al., 2007). In addition to maldigestion and malabsorption, SIBO can contribute to several other conditions including irritable bowel syndrome, celiac disease, Crohn’s disease, nonalcoholic steatohepatitis, and obesity. The stasis associated with SIBO creates conditions that favor colonization by microbes may not be indigenous to the region (e.g. colonic-like), potentially resulting in overgrowth of microbes that compete for nutrients, perturb critical commensal microbe-host interactions, and/or promote mucosal inflammation (reviewed in Bohm et al., 2013).
Restrictive and bypass bariatric surgery, e.g. roux-en-y gastric bypass (RYGB), sleeve gastrectomy, gastric banding and biliopancreatic diversion with duodenal switch (BPD/DS) promote weight loss by altering small bowel anatomy and function, but also likely set into play other mechanisms that change the state of host metabolism. For example, these procedures alter small bowel microbiota which can directly prevent diet-induced obesity (Liou et al., 2013). Similarly, dramatic changes in small bowel microbiota were observed following biliary diversion to the jejunum or ileum were associated with prevention of diet-induced obesity in mice (Pierre et al., 2016). These effects are due to the direct impact of bile acids on the host (e.g. through FXR activation) or through bile-mediated alterations in the gut microbiota.
Undernourished children in Bangladesh present with a condition known as environmental enteropathy (Petri et al., 2014) characterized by small intestinal villus blunting and inflammation with an associated immature microbiota. GF mice receiving fecal microbiota transplant (FMT) from growth-stunted vs healthy infants fed a Malawian diet displayed impaired growth, while cohousing with mice given healthy donor microbiota prevented growth impairments, implicating a causal role for microbiota in undernutrition (Blanton et al., 2016). Supplementing with ready-to-use therapeutic food only partially restored metabolic disruptions (Smith et al., 2013). Similarly, microbe accessible carbohydrate (MAC)-deficient diets led to the generational loss of commensal microorganisms. However, replenishment of MAC did not restore the abundance of depleted microorganisms, which were only restored by conventionalization (Sonnenburg et al., 2016). Notably, both groups examined stool microbiota, which begs the question whether “missing” regional microbiota might be better in restoring SI digestive and absorptive functions.
Celiac disease (CD) is a chronic enteropathy of genetically-prone patients expressing the MHC molecules HLA-DQ2 or DQ8, which is triggered by the dietary component gluten found in wheat, rye, and barley (Kim and Jabri, 2015). Chronic mucosal inflammation can cause long-term damage to the gut epithelium leading to maldigestive and malabsorptive issues. While HLA-DQ haplotype is a requirement, additional factors likely play a role because only 2 to 5% of HLA-DQ2/DQ8 individuals develop CD. Among these, changes in small bowel microbiota, resulting from altered immune states associated with HLA-DQ haplotype, have been implicated. While some studies suggest an expansion of proinflammatory SI microbiota, others have posited that the loss of key microbes important for proper immune development plays a role (reviewed in Verdu et al., 2015). Regional biopsy samples from duodenum and jejunum have revealed changes in the local gut microbiota of patients with active CD. For instance, rod vs. cocciform bacteria were identified adhered to the mucosa in distal duodenum/proximal jejunal biopsies collected from children with untreated CD, treated CD, oral gluten-challenged CD and controls. Mucosally-adhered rod-shaped bacteria increased in untreated and oral gluten-challenged CD patient biopsies relative to controls and treated CD subjects (Forsberg et al., 2004). Several studies have also shown increases in duodenal Proteobacteria in CD subjects relative to controls (Sánchez et al., 2013; Wacklin et al., 2014), while others have shown that SCFAs are dramatically decreased in active CD patients, which can be restored following a year’s long adherence to a gluten-free diet (Tjellström et al., 2013).
Interleukin-15 (IL15), a cytokine made by IECs, is often upregulated in CD (Abadie and Jabri, 2014). In mice that overexpress IEC IL15 (IL15tg), gut microbiota are affected throughout the intestine, with the most pronounced differences observed in the ileum (Meisel et al., 2017). Relative abundance of butyrate-producing bacteria and butyrate levels were significantly decreased. While the IL15Tg mice did not develop overt CD, these findings showed that altered IL-15 expression can significantly alter microbial community membership and function, possibly contributing to CD progression.
3. Colonic Diseases: Inflammatory Bowel Diseases and C. difficile-induced colitis
Inflammatory bowel diseases (IBD) are complex immune disorders that arise from convergence of host genetic, gut microbial, and environmental factors. Regionality and topical nature of lesions are hallmarks of the diseases and suggest that local factors (microbe and microbial products) are germane to their development. Ulcerative colitis (UC), one of the clinical IBD phenotypes, only involves the colon, usually starting at the rectum and proceeding proximally in a confluent mucosal inflammatory front, suggestive of propagative processes. In individuals with left-sided UC, a clear line of delineation between diseased and non-diseased regions is often seen. Here the differences in mucosa-associated microbiota are considered an important underlying factor causing or contributing to this phenomenon. Similarly, Crohn’s disease, another clinical phenotype of IBD, can involve any part of the GI tract, from mouth to anus. The typical starting lesion, an apthous ulcer, starts at the mucosal surface and as the disease progresses, the lesions widen and deepen, with the latter capable of penetrating the bowel wall. Yet, tissue surrounding the Crohn’s mucosal lesion is endoscopically and histologically normal. A few studies (Hirano et al., 2018; Walker et al., 2011) have attempted to map differences in taxonomical profiles of involved and non-involved mucosal regions in IBD patients based on 16S rRNA amplicons. While differences have been reported, it remains unknown whether they are a cause or an effect and what their functional significance might be. Crohn’s involving the terminal ileum is often associated with a perturbed bacteria populations and increased fungal load (Liguori et al., 2016). With regard to the latter, many patients exhibit serum levels of anti-Saccharomyces cerevisiae antibody (ASCA), which is directed against the oligo-mannin component of the cell wall of polymorphic fungi such as Candida albicans. Antibody levels to other microorganisms including Ecsherichia coli and Pseduomonas fluorescens can also be seen (Mitsuyama et al., 2016).
The obligate spore-forming Gram-positive anaerobe, Clostridium difficile, is the major cause of recurrent antibiotic-induced toxin-mediated colitis (reviewed in Libertucci and Young, 2019). While C. difficile has been observed at low levels in healthy humans, particularly infants, antibiotic-induced disruption of the indigenous colonic microbiota and resulting increases in colonic primary bile acid concentrations promote its outgrowth, toxin production, and invasion (Seekatz and Young, 2014). The latter is important for germination of C. difficile spores (Theriot et al., 2016), while production of specific secondary BAs can inhibit this process (Libertucci and Young, 2019). Commensal microbes, such as C. scindens and C. sordellii, which have the capacity to perform 7α-dehydroxylation play a role in inhibiting outgrowth of pathogenic C. difficile through secretion of antibiotic-like molecules derived from tryptophan metabolism (Kang et al., 2019). The actions of these molecules are further enhanced by the presence of secondary BAs, DCA and LCA. First line treatment of C. difficile colitis continues to be with antibiotics such as metronidazole or vancomycin, but FMT from a healthy donor is becoming increasingly used for recurrent C. difficile infection. Following therapeutic FMT, increased levels of DCA, LCA and SCFAs, even after 6 months, are observed and attributed to the expansion of families like Lachnospiraceae, Ruminococcaceae, and Clostridiales (Seekatz et al., 2018).
On the horizon: Next Generation technologies and approaches to navigate the unknown
The human gut microbiota from stomach to distal ileum (regions difficult to sample) remain largely unexplored. What can be inferred from animal studies has been useful to gain conceptual insights, but human microbiota in these regions are likely very different in speciation, composition, and function. The technical challenges and confounders associated with human studies of this domain are formidable.
Sampling of the upper GI is invasive and limited to disease settings where endoscopic examination is indicated. Thus, information of “healthy” upper GIT microbiomes is very limited. One possible non-invasive approach is to sample patients with ileostomies, but even then, their underlying disease could be a confounding factor. Non-invasive approaches are being developed such as the esophageal string test, inflatable balloons covered in cotton mesh and attached to a rubber tube, or the Cytosponge, which is a tethered capsule that dissolves in the stomach to release a 3cm mesh screen that is then pulled back through the mouth (described in May and Abrams et al. 2018). The limitation to these methods is potential contamination with oral microbes upon retrieval. Additionally, swallowed bio-sampling capsules programmed or triggered to sample luminal content during passage through the GIT are under development (Nakamura et al., 2017), although they have currently no capacity to sample the more stable mucosa-associated regional microbiota.
Other challenges to consider are those generic to the study of microbial communities in general. The field has relied heavily on 16S rRNA amplicon sequencing which provide limited, if any, functional information. Metagenomics and metatranscriptomics on regionally-collected samples could provide more information, but are often difficult to perform because of limited biomass and heavy contamination by human DNA and transcripts. Resolution of these data to identify genomes and functions of specific microbial strains or consortia is a major hurdle, although headway is being made through newer metagenome-assemble genome approaches (Eren et al., 2015). Matrix-assisted laser desorption ionization time-of-flight mass-spectrometry (MALDI-TOF) coupled to culturomics and sequencing based approaches has also been successfully used to isolate and identify single strains in human fecal samples in the clinical microbiology setting (Seng et al., 2013; Stevenson et al., 2010). Development of microfluidic cultivation systems (Liu and Walther-Antonio, 2017) has allowed for more rapid isolation and characterization of previously non-cultivable strains of microbes (Ma et al., 2014). Others have since coupled high-throughput microfluidic droplet platforms that allow for single cell assessments to gain information about functional parameters of individual microbial community members (Terekhov et al., 2018). These approaches applied to regional samples may aid to identify and characterize novel strains that are important to critical host-microbe interactions.
There is also the need to understand organizational structure of regional microbial communities of the gut. Confocal microscopy combined with immunofluorescence can provide information that complements more standard approaches using FISH (Tropini et al., 2017). The Sonnenburg group showed that fluorescently-tagged Bacteroides strains were useful to gain insight into the dynamic interactions between mucosally-associated bacteria and the host in different intestinal regions (Whitaker et al., 2017). Engineering bacterial sensors through synthetic biology has allowed for both detection and tracking of specific events within the intestine (Kotula et al., 2014). Similar approaches have been used to engineer bacteria, particularly strains of E. coli, that can sense and/or modulate the environment in a potentially therapeutic manner (Kurtz et al., 2019; Tscherner et al., 2019), a few of which have already passed Phase I trials in humans. Refinement of these tools and identification of novel targets could be accomplished using swallowable devices that can be made to detect important host or microbial factors.
To identify and probe mechanisms behind region-specific host-microbe interactions, an expansion on ex vivo and in vitro model systems is required. For instance, organoids (intestinal epithelial organoids) can be derived from differentiated cells obtained via biopsy (from human) or at the time of tissue harvest (mice) from varying regions of the gut, or they can be made from human intestinal organoids (HIO) (Bartfeld and Clevers, 2017) derived from iPS cells/embryonic stem cells. Both 2- and 3-dimensional organoids can be treated with either conditioned media (Martinez-Guryn et al., 2018; Wang et al., 2017) from individual organisms or micro-injected with live organisms to examine how they impact regional specific epithelial-based pathways involved in macro-nutrient uptake or inflammation (Leslie and Young, 2016). These approaches, combined with additional tools, such as gut-on-a-chip (Kim and Ingber, 2013), co-culture experiments with immune cell populations procured from regional mucosa, and/or bioreactors could provide novel insight into regional specific host-microbe interactions in health and disease.
The future: Things are only impossible until they are not
Much remains to be discovered, among them how to define healthy versus unhealthy microbiomes of the gut, which is likely to be determined by host, microbial, and environmental factors. An important step towards this goal is to gain a better understanding of the regional organization and functions of gut microbial communities along the GI tract and their relevance to conditions of health and disease. While there are many challenges on the horizon, particularly in studies of human subjects, great promise lies ahead to advance the field through new approaches and next generation technologies002E
Acknowledgements
This work was supported by the NIH NIDDK under grants P30DK42086 for the Digestive Diseases Research Center Core (DDRCC), T32 DK07074, DK115221, and K01 DK111785 (V.L.). We have no conflicts of interest to disclose.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Abadie V, and Jabri B (2014). IL-15 : a central regulator of celiac disease immunopathology. Immunol. Rev 260, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Aidy S, Derrien M, Merrifield CA, Levenez F, Doré J, Boekschoten MV, Dekker J, Holmes E, Zoetendal EG, Van Baarlen P, et al. (2013). Gut bacteria-host metabolic interplay during conventionalisation of the mouse germfree colon. ISME J 7, 743–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelakis E, Armougom F, Carri F, Bachar D, Henrissat B, and Raoult D (2015). A Metagenomic Investigation of the Duodenal Microbiota Reveals Links with Obesity. PLoS One 10, e0137784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, et al. (2015). Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 163, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Manchester JK, Semenkovich CF, and Gordon JI (2007). Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci 104, 979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartfeld S, and Clevers H (2017). Stem cell-derived organoids and their application for medical research and patient treatment. J. Mol. Med 95, 729–738. [DOI] [PubMed] [Google Scholar]
- Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Ilkaveya O, Subramanian S, Manary MJ, Trehan I, Jorgensen JM, Fan Y, et al. (2016). Gut bacteria that rescue growth impairments transmitted by immature microbiota from undernourished children. Science (80–.) 351, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm M, Siwiec RM, and Wo JM (2013). Diagnosis and Management of Small Intestinal Bacterial Overgrowth. Nutr. Clin. Pract 28, 289–299. [DOI] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, and Hooper LV (2006). Symbiotic Bacteria Direct Expression of an Intestinal Bactericidal Lectin. Science (80-.) 313, 1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AVA, Devlin S, Varma Y, Fischbach MA, et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-peach H, Nadimpalli A, Antonopoulos DA, Jabri B, and Chang EB (2012). Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 487, 104–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, Yu W-H, Lakshmanan A, and Wade WG (2010). The human oral microbiome. J. Bacteriol 192, 5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca FA, Swartz TD, Sakar Y, and Covasa M (2012). Increased oral detection, but decreased intestinal signaling for fats in mice lacking gut microbiota. PLoS One 7, e39748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukowicz AC, Lacy BE, and Levine GM (2007). Small Intestinal Bacterial Overgrowth: A Comprehensive Review. Gastroenterol. Hepatol. (N. Y) 3, 112–122. [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Esen C, Quince C, Vineis JH, Morrison HG, Sogin ML, and Delmont TO (2015). Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3, e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermund A, Schütte A, Johansson MEV, Gustafsson JK, and Hansson GC (2013). Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer ‘ s patches. Am J Physiol Gastrointest Liver Physiol 305, 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Lazarevic V, Gaı N, Johansson M, Backhed F, Delzenne NM, Schrenzel J, Franc P, and Cani PD (2014). Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J 8, 2116–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg G, Fahlgre A, Hörstedt P, and Hammarström Sten Hammarström, Olle Hernell M-L (2004). Presence of Bacteria and Innate Immunity of Intestinal Epithelium in Childhood Celiac Disease. Am. J. Gastroenterol 99, 894–904. [DOI] [PubMed] [Google Scholar]
- Fox J, Ge Z, Whary M, Erdman S, and Horwitz B (2011). Helicobacter hepaticus infection in mice: Models for understanding lower bowel inflammation and cancer JG. Mucosal Immunol 4, 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman ES, Bittinger K, Esipova TV, Hou L, Chau L, Jiang J, Mesaros C, Lund PJ, Liang X, Fitzgerald GA, et al. (2018). Microbes vs. chemistry in the origin of the anaerobic gut lumen. Proc. Natl. Acad. Sci 115, 4170–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung C, Tan S, Nakajima M, Skoog EC, Camarillo-Guerrero LF, Klein JA, Lawley TD, Solnick JV, Fukami T, and Amieva MR (2019). High-resolution mapping reveals that microniches in the gastric glands control Helicobacter pylori colonization of the stomach. PLOS Biol 17, e3000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard P (2014). Metabolism of Cholesterol and Bile Acids by the Gut Microbiota. Pathogens 3, 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grider JR, and Piland BE (2007). The peristaltic reflex induced by short-chain fatty acids is mediated by sequential release of 5-HT and neuronal CGRP but not BDNF. Am J Physiol Gastrointest Liver Physiol 292, 429–437. [DOI] [PubMed] [Google Scholar]
- Harley ITW, and Karp CL (2012). Obesity and the gut microbiome: Striving for causality. Mol. Metab 1, 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegade VS, Speight RA, Etherington RE, and Jones DEJ (2016). Novel bile acid therapeutics for the treatment of chronic liver diseases. Therap. Adv. Gastroenterol 9, 376–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano A, Umeno J, Okamoto Y, Shibata H, Ogura Y, Moriyama T, Torisu T, Fujioka S, Fuyuno Y, Kawarabayasi Y, et al. (2018). Comparison of the microbial community structure between inflamed and non-inflamed sites in patients with ulcerative colitis. J. Gastroenterol. Hepatol 33, 1590–1597. [DOI] [PubMed] [Google Scholar]
- Hooper L, and Macpherson A (2010). Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol 10, 159–169. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, and Gordon JI (2001). Molecular Analysis of Commensal Host-Microbial Relationships in the Intestine. Science (80−.) 291, 881–885. [DOI] [PubMed] [Google Scholar]
- Inagaki T, Moschetta A, Lee Y, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, et al. (2006). Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci 103, 3920–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush NO, Lecomte V, Maloney CA, and Morris MJ (2017). Cross-talk among metabolic parameters, esophageal microbiota, and host gene expression following chronic exposure to an obesogenic diet. Sci. Rep 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Ridlon JM, Ray D, Ii M, Barnes S, and Hylemon PB (2008). Clostridium scindens baiCD and baiH genes encode stereo-specific. Biochim. Biophys. Acta 1781, 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JD, Myers CJ, Harris SC, Bajaj JS, Zhou H, and Hylemon PB (2019). Bile Acid 7α-Dehydroxylating Gut Bacteria Secrete Antibiotics that Inhibit Clostridium difficile: Role of Secondary Bile Acids. Cell Chem. Biol 26, 27–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap PC, Marcobal A, Ursell LK, Larauche M, Duboc H, Earle KA, Sonnenburg ED, Ferreyra JA, Higginbottom SK, Million M, et al. (2013). Complex Interactions Among Diet, Gastrointestinal Transit, and Gut Microbiota in Humanized Mice. Gastroenterology 144, 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, and Ingber DE (2013). Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol 5, 1130–1140. [DOI] [PubMed] [Google Scholar]
- Kim SM, and Jabri B (2015). Best Practice & Research Clinical Gastroenterology Innate immunity : Actuating the gears of celiac disease pathogenesis 29, 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A, De Vadder F, Kovatcheva-Datchary P, and Bäckhed F (2016). From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345. [DOI] [PubMed] [Google Scholar]
- Kotula JW, Kerns SJ, Shaket LA, Siraj L, Collins JJ, and Way JC (2014). Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl. Acad. Sci 111, 4838–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel MA, Sefik E, Hill JA, Wu H-J, Benoist C, and Mathis D (2011). Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl. Acad. Sci 108, 11548–11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz CB, Millet YA, Puurunen Marja K. Perreault M, Charbonneau, Isabella Mark R., Kotula Vincent M., Jonathan W Antipov E, Dagon Y, Denney WS, Wagner DA, West KA, Degar AJ, Brennan AM, et al. (2019). An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci Transl Med 11, 7975. [DOI] [PubMed] [Google Scholar]
- Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, Pierre JF, Heneghan AF, Nadimpalli A, Hubert N, et al. (2015). Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe 17, 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie J, and Young VB (2016). A Whole New Ball Game: Stem Cell-Derived Epithelia in the Study of Host-Microbe Interactions. Anaerobe 37, 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Chen H, Mao B, Yang Q, Zhao J, Gu Z, Zhang H, Yang Q, Chen YQ, and Chen W (2017). Microbial Biogeography and Core Microbiota of the Rat Digestive Tract. Sci. Rep 7, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libertucci J, and Young VB (2019). The role of the microbiota in infectious diseases. Nat. Microbiol 4, 35–45. [DOI] [PubMed] [Google Scholar]
- Liguori G, Lamas B, Richard ML, Brandi G, Hoffmann TW, Pierluigi M, Simone D, Calabrese C, Poggioli G, Langella P, et al. (2016). Fungal Dysbiosis in Mucosa-associated Microbiota of Crohn’s Disease Patients. J. Crohn’s Colitis 10, 296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou A, Paziuk M, Luevano J-M Jr, Machineni S, Turnbaugh PJ, and Kaplan LM (2013). Conserved Shifts in the Gut Microbiota Due to Gastric Bypass Reduce Host Weight and Adiposity. Sci. Transl. Med 5, 178ra41–178ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, and Walther-Antonio M (2017). Microfluidics: A new tool for microbial single cell analyses in human microbiome studies. Biomicrofluidics 11, 061501. [Google Scholar]
- Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Pop M, and Amar S (2012). Deep Sequencing of the Oral Microbiome Reveals Signatures of Periodontal Disease. PLoS One 7, e37919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynge Pedersen AM, and Belstrøm D (2019). The role of natural salivary defences in maintaining a healthy oral microbiota. J. Dent 80, S3–S12. [DOI] [PubMed] [Google Scholar]
- Ma L, Kim J, Hatzenpichler R, Karymov MA, Hubert N, Hanan IM, Chang EB, and Ismagilov RF (2014). Gene-targeted microfluidic cultivation validated by isolation of a gut bacterium listed in Human Microbiome Project ‘ s Most Wanted taxa 111, 9768–9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFabe DF, Nathan CE, Boon F, Ossenkopp K-P, and Cain DP (2011). Effects of the enteric bacterial metabolic product propionic acid on object-directed behavior, social behavior, cognition, and neuroinflammation in adolescent rats: Relevance to autism spectrum disorder. Behav. Brain Res 217, 47–54. [DOI] [PubMed] [Google Scholar]
- Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, and Borisy GG (2016). Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci 113, 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Guryn K, Hubert N, Frazier K, Urlass S, Musch MW, Ojeda P, Pierre JF, Miyoshi J, Sontag TJ, Cham CM, et al. (2018). Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe 23, 458–469.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May M, and Abrams JA (2018). Emerging Insights into the Esophageal Microbiome. Curr. Treat. Options Gastroenterol 16, 72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel M, Mayassi T, Fehlner-peach H, Koval JC, Brien SLO, Hinterleitner R, Lesko K, Kim S, Bouziat R, Chen L, et al. (2017). Interleukin-15 promotes intestinal dysbiosis with butyrate deficiency associated with increased susceptibility to colitis. ISME J 11, 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuyama K, Niwa M, Takedatsu H, Yamasaki H, Kuwaki K, Yoshioka S, Yamauchi R, Fukunaga S, and Torimura T (2016). Antibody markers in the diagnosis of inflammatory bowel disease. World J. Gastroenterol 22, 1304–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji A, Kobiita A, Ye T, and Chambon P (2013). Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 153, 812–827. [DOI] [PubMed] [Google Scholar]
- Münch NS, Fang H-Y, Ingermann J, Maurer HC, Anand A, Kellner V, Sahm V, Wiethaler M, Baumeister T, Wein F, et al. (2019). High-fat Diet Accelerates Carcinogenesis in a Mouse Model of Barrett’s Esophagus via IL8 and Alterations to the Gut Microbiome. Gastroenterology epub ahead [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura R, Izumi S, Kawaguchi H, H O, and Yoshimoto M (2017). A swallowable sensing device platform with wireless power feeding and chemical reaction actuator 39th Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 3040–3043. [DOI] [PubMed] [Google Scholar]
- Nardone G, and Compare D (2015). The human gastric microbiota : Is it time to rethink the pathogenesis of stomach diseases? United Eur. Gastroenterol J 3, 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natividad JMM, Hayes CL, Motta J-P, Jury J, Galipeau HJ, Philip V, Garcia-Rodenas CL, Kiyama H, Bercik P, and Verdu EF (2013). Differential Induction of Antimicrobial REGIII by the Intestinal Microbiota and Bifidobacterium breve NCC2950. Appl. Environ. Microbiol 79, 7745–7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobel Y, Snider E, Compres G, Freedber D, Khiabanian H, Lightdale C, Toussaint N, and Abrams J (2018). Increasing dietary fiber intake is associated with a distinct esophageal microbiome. Clin Transl Gastroenterol 9, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara AM, and Shanahan F (2006). The gut flora as a forgotten organ. EMBO Rep 7, 688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri W, Naylor C, and Haque R (2014). Environmental enteropathy and malnutrition: Do we know enough to intervene? BMC Med 12, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre JF, Martinez KB, Ye H, Nadimpalli A, Morton TC, Yang J, Wang Q, Patno N, Chang EB, and Yin DP (2016). Activation of bile acid signaling improves metabolic phenotypes in high-fat diet-induced obese mice. Am. J. Physiol. Liver Physiol 311, G286–G304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Harris SC, Bhowmik S, Kang D, Hylemon B, Ridlon JM, Harris SC, Bhowmik S, and Kang D (2016a). Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7, 22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Wolf PG, and Gaskins HR (2016b). Taurocholic acid metabolism by gut microbes and colon cancer 7, 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel D, and Setchell K (1992). Bile Acid Biosynthesis. Biochemistry 31, 4737–4749. [DOI] [PubMed] [Google Scholar]
- Sánchez E, Donat E, Ribes-koninckx C, and Fernández-murga ML (2013). Duodenal-Mucosal Bacteria Associated with Celiac Disease in Children. Appl. Environ. Microbiol 79, 5472–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, Lee JY, Ziel JW, Miraldi ER, Domingos AI, et al. (2015). An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell 163, 381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, and Bäckhed F (2013). Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17, 225–235. [DOI] [PubMed] [Google Scholar]
- Schmidt TS, Hayward MR, Coelho LP, Li SS, Costea PI, Voigt AY, Wirbel J, Maistrenko OM, Alves RJ, Bergsten E, et al. (2019). Extensive transmission of microbes along the gastrointestinal tract. Elife 8, 42693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seekatz AM, and Young VB (2014). Clostridium difficile and the microbiota. J Clin Invest 124, 4182–4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seekatz AM, Theriot CM, Rao K, Chang Y-M, Freeman AE, Kao JY, and Young VB (2018). Restoration of short chain fatty acid and bile acid metabolism following fecal microbiota transplantation in patients with recurrent Clostridium difficile infection. Anaerobe 53, 64–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seekatz AM, Schnizlein MK, Koenigsknecht MJ, Baker JR, Hasler WL, Bleske BE, Young VB, and Sun D (2019). Spatial and Temporal Analysis of the Stomach and Small-Intestinal Microbiota in Fasted Healthy Humans. MSphere 4, e00126–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seng P, Abat C, Rolain M, Colson P, Lagier J, and Gouriet F (2013). Laboratory : Impact of Matrix-Assisted Laser Desorption Ionization – Time of Flight Mass Spectrometry 51, 2182–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang L, Deng D, Buskermolen JK, Janus MM, Krom BP, Roffel S, Waaijman T, Loveren C Van, Crielaard W, and Gibbs S (2018). Multi-species oral biofilm promotes reconstructed human gingiva epithelial barrier function. Sci. Rep 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JS, Park DS, Baek DH, Jha N, Park SI, Yun HJ, Kim WJ, and Ryu JJ (2018). Antimicrobial activity of NO-releasing compounds against periodontal pathogens. PLoS One 13, e0199998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade E, Williams L, and Gagnon J (2018). Hydrogen sulfide suppresses ghrelin secretion in vitro and delays postprandial ghrelin secretion while reducing appetite in mice. Physiol. Rep 6, 13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, et al. (2013). Gut Microbiomes of Malawian Twin. Science (80–.) 339, 548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, and Backhed F (2016). Know your neighbor: Microbiota and host epithelial cells interact locally to control intestinal function and physiology. BioEssays 38, 455–464. [DOI] [PubMed] [Google Scholar]
- Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, and Sonnenburg JL (2016). Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson LG, Drake SK, Shea YR, Zelazny AM, and Murray PR (2010). Evaluation of Matrix-Assisted Laser Desorption Ionization – Time of Flight Mass Spectrometry for Identification of Clinically Important Yeast Species ▪. J Clin Microbiol 48, 3482–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger B, and Omenetti S (2017). The dichotomous nature of T helper 17 cells. Nat. Rev. Immunol 17, 535–544. [DOI] [PubMed] [Google Scholar]
- Szarka L, and Camilleri M (2012). Methods for the Assessment of Small Bowel and Colonic Transit. Semin Nucl Med 42, 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, de Zoeten E, Ozkaynak E, Chen C, Wang L, Porrett P, Li B, Turka L, Olson E, Greene M, et al. (2007). Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med 13, 1299–1307. [DOI] [PubMed] [Google Scholar]
- Terekhov SS, Smirnov IV, Malakhova MV, Samoilov AE, and Manolov AI (2018). Ultrahigh-throughput functional profiling of microbiota communities 115, 9551–9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss CA, Zeevi D, Levy M, Zilberman-schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, et al. (2014). Article Transkingdom Control of Microbiota Diurnal Oscillations Promotes Metabolic Homeostasis. Cell 159, 514–529. [DOI] [PubMed] [Google Scholar]
- Theriot CM, Bowman AA, and Young B (2016). Antibiotic-Induced Alterations of the Gut Microbiota Alter Secondary Bile Acid Production and Allow for Clostridium difficile Spore Germination and Outgrowth in the Large Intestine. Am. Soc. Microbiol 1, e00045–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjellström B, Högberg L, Stenhammar L, Fälth-Magnusson K, Magnusson K-E, Norin E, Sundqvist T, and Midtvedt T (2013). Faecal short-chain fatty acid pattern in childhood coeliac disease is normalised after more than one year’s gluten-free diet. Microb Ecol Heal. Dis 1, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touw K, Ringus DL, Hubert N, Wang Y, Leone VA, Nadimpalli A, Theriault BR, Huang YE, Tune JD, Herring PB, et al. (2017). Mutual reinforcement of pathophysiological host-microbe interactions in intestinal stasis models. Physiol. Rep 5, e13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropini C, Earle KA, Huang KC, and Sonnenburg JL (2017). Review The Gut Microbiome : Connecting Spatial Organization to Function. Cell Host Microbe 21, 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherner M, Giessen TW, Markey L, Kumamoto CA, and Silver PA (2019). A Synthetic System That Senses Candida albicans and Inhibits Virulence Factors. ACS Synth Biol 8, 434–444. [DOI] [PubMed] [Google Scholar]
- Verdu EF, Galipeau HJ, and Jabri B (2015). Novel players in coeliac disease pathogenesis: role of the gut microbiota. Nat. Rev. Gastroenterol. Hepatol 12, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacklin P, Laurikka P, Lindfors K, Collin P, Salmi T, Lähdeaho M-L, Saavalainen P, Mäki M, Mättö J, Kurppa K, et al. (2014). Altered Duodenal Microbiota Composition in Celiac Disease Patients Suffering From Persistent Symptoms on a Long-Term Gluten-Free Diet. Am. J. Gastroenterol 109, 1933–1941. [DOI] [PubMed] [Google Scholar]
- Walker AW, Sanderson JD, Churcher C, Parkes GC, Hudspith BN, Rayment N, Brostoff J, Parkhill J, Dougan G, and Petrovska L (2011). High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol 11, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Devkota S, Musch MW, Jabri B, Nagler C, Antonopoulos DA, Chervonsky A, and Chang EB (2010). Regional mucosa-associated microbiota determine physiological expression of TLR2 and TLR4 in murine colon. PLoS One 5, e13607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kuang Z, Yu X, Ruhn KA, Kubo M, and Hooper LV (2017). The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science (80–.) 357, 912–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MA, Pierre JF, Leal RF, Huang Y, Shogan B, Dalal SR, Weber CR, Leone VA, Musch MW, An GC, et al. (2016). Insights into the pathogenesis of ulcerative colitis from a murine model of stasis-induced dysbiosis, colonic metaplasia, and genetic susceptibility. Am. J. Physiol. Liver Physiol 310, G973–G988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitkunat K, Schumann S, Nickel D, Kappo KA, Kipp AP, Blaut M, and Klaus S (2016). Importance of propionate for the repression of hepatic lipogenesis and improvement of insulin sensitivity in high-fat diet-induced obesity. Mol Nutr Food Res 60, 2611–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker WR, Shepherd ES, Sonnenburg JL, Whitaker WR, Shepherd ES, and Sonnenburg JL (2017). Tunable Expression Tools Enable Single-Cell Strain Distinction in the Gut Microbiome Resource Tunable Expression Tools Enable Single-Cell Strain Distinction in the Gut Microbiome. Cell 169, 538–538.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano JM, Yu K, Mazmanian SK, Hsiao EY, Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al. (2015). Indigenous Bacteria from the Gut Microbiota Regulate Host Serotonin Biosynthesis Article Indigenous Bacteria from the Gut Microbiota Regulate Host Serotonin Biosynthesis. Cell 161, 264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar A, Chaix A, Yooseph S, and Panda S (2014). Article Diet and Feeding Pattern Affect the Diurnal Dynamics of the Gut Microbiome. Cell Metab 20, 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Raes J, Bogert B Van Den, Booijink CCGM, Troost FJ, Bork P, and Wels M (2012). The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J 6, 1415–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]