Abstract
Prader–Willi syndrome (PWS) is a neurodevelopmental disorder that arises from lack of expression of paternally inherited genes known to be imprinted and located in the chromosome 15q11-q13 region. PWS is considered the most common syndromal cause of life-threatening obesity and is estimated at 1 in 10 000 to 20 000 individuals. A de novo paternally derived chromosome 15q11-q13 deletion is the cause of PWS in about 70% of cases, and maternal disomy 15 accounts for about 25% of cases. The remaining cases of PWS result either from genomic imprinting defects (microdeletions or epimutations) of the imprinting centre in the 15q11-q13 region or from chromosome 15 translocations. Here, we describe the clinical presentation of PWS, review the current understanding of causative cytogenetic and molecular genetic mechanisms, and discuss future directions for research.
Prader–Willi syndrome (PWS) is a neurogenetic disorder characterised by neurodevelopmental features including hyperphagia (increased ingestion of food) and early childhood obesity that is extremely severe in some cases (Refs 1, 2, 3); indeed, PWS is considered the most common genetic syndrome leading to life-threatening obesity. It is estimated to occur at a frequency of 1 in 10 000 to 20 000 individuals (Ref. 1), with 350 000–400 000 people affected worldwide, and presents in all ethnic groups but is reported disproportionately more in Caucasians (Refs 2, 4). Most cases are sporadic and the chance of recurrence is less than 1% (Ref. 3); however, the risk might be much higher (e.g. 50%) in PWS families in which the father carries imprinting deletions in the chromosome 15q11-q13 region.
The PWS disorder arises from lack of expression of paternally inherited genes known to be imprinted and located in the chromosome 15q11-q13 region. Genomic imprinting, a process first described in plant genetics and not known in human genetics until its discovery in PWS, is an epigenetic phenomenon whereby phenotype is modified depending on the sex of the parent contributing that allele (Refs 5, 6). It arises from epigenetic changes (such as methylation) to genes during gametogenesis that result in gene expression dependent on the parent of origin of the gene. This process results in a reversible specific marking of a fraction of the genome that is parent specific and produces mono-allelic gene expression of either the maternal or paternal allele of a particular imprinted locus. Approximately 70% of PWS cases are caused by a non-inherited (i.e. de novo) deletion in the paternally derived chromosome 15q11-q13 region; approximately 25% of cases result from maternal disomy 15 (i.e. two maternal chromosome 15s and no paternal chromosome 15); and the remaining cases arise either from genomic imprinting defects (microdeletions or epimutations) of the imprinting centre located in the 15q11-q13 region or balanced chromosome 15 translocations (Refs 2, 3, 7, 8, 9, 10, 11). The typical 15q11-q13 deletion has been classified into two types, type I and type II, depending on the size and chromosome breakpoint position.
Clinical description, natural history and treatment
The cardinal features of PWS include infantile hypotonia (diminished muscle tone) and feeding difficulties, hypogonadism and hypogenitalism, hyperphagia and onset of obesity in early childhood, small hands and feet, mild mental deficiency (average IQ of 65), behavioural problems (skin picking, temper tantrums, stubbornness) and a characteristic facial appearance (small upturned nose, narrow bifrontal diameter, dolichocephaly, strabismus, down-turned corners of the mouth and almond-shaped eyes) (Refs 1, 2, 3). Other findings include short stature, scoliosis (lateral curvature of the spine), dental problems, endocrine disturbances such as growth hormone (GH) deficiency and diabetes mellitus, sleep apnea and comorbidities relating to obesity. Approximately 50% of children with PWS develop temper tantrums and stubbornness between 3–5 years of age and may display depression during adolescence and adulthood; behavioural problems may be precipitated by withholding food. Nonetheless, children with PWS can be affectionate and caring. Intolerance to changes in routine can be a severe problem for many adolescents and adults with PWS, as well as poor peer interaction, immaturity and inappropriate social behaviour.
PWS can be divided into two distinct clinical stages. Stage 1 of the clinical course is first recognised in neonates and infants, with a delay in reaching developmental milestones, feeding difficulties, profound hypotonia and the presence of distinctive facial and body features. Stage 2 of the clinical course appears with the onset of hyperphagia and associated obesity in early childhood. Food foraging ensues and behavioural difficulties arise, often initiated by withholding food. The appearance of PWS clinical findings with age is summarised in Table 1.
Table 1.
Clinical characteristics of Prader–Willi syndromea
| Stage 1 | Stage 2 | ||
|---|---|---|---|
| Pregnancy and delivery | Neonatal period and infancy | Childhood | Adolescence and adulthood |
| Reduced fetal activity | Narrow forehead | Short stature | Short stature |
| Breech delivery | Undescended testicles | Small hands and feet | Small hands and feet |
| Pre-term or post-term delivery | Small genitals and testicles | Light skin and eye colour | Scoliosis |
| Poor muscle tone (hypotonia) | Almond-shaped eyes | Osteoporosis | |
| Strabismus | Delayed puberty | ||
| Feeding problems | Myopia | Diabetes mellitus | |
| Poor suck | Skin picking | Depression | |
| Sticky saliva | Dental caries | Excessive sleepiness | |
| Weak cry | Excessive appetite/food foraging | ||
| Temperature instability | |||
| Developmental delay | Obesity | ||
| Intellectual disability | |||
| Behavioural problems | |||
| Temper tantrums | |||
| Stubbornness | |||
| Obsessive-compulsive behaviour | |||
As a result of better awareness and advances in genetic testing, the diagnosis of PWS is made earlier than in the past. Early diagnosis is important since nutritional intervention is required. Therapy with GH is frequently prescribed during early childhood to improve stature and body composition, energy level and metabolism (Refs 12, 13, 14, 15, 16). Physical strength and activity also increases during GH treatment (Refs 12, 17, 18, 19). No specific medication has universal benefit in treating the abnormal behavioural problems seen in PWS; however, specific serotonin re-uptake inhibitors have been useful in controlling behaviour such as skin picking (Refs 2, 20). If weight is adequately controlled, life expectancy should be similar to that of other mildly retarded individuals. For example, a person with PWS who died at age 71 years was described in 1994 (Ref. 21) and a second PWS individual at 68 years of age was reported in 2000 (Ref. 22). Caloric diet restriction (e.g. approximately 60% of normal) throughout life is important to control the obesity and co-morbidity. A review of causes of death in 25 individuals with PWS indicated that obesity and related complications led to the death of 14 subjects (Ref. 23), with an average age at the time of death of 23 years. Additional causes of death in children with PWS, including possible complications associated with GH therapy, have recently been reviewed and interventions recommended (Refs 16, 24, 25, 26).
Cytogenetics
The chromosome 15q deletion was first reported in PWS by Ledbetter et al. in 1981 as a result of high-resolution chromosome analysis (Ref. 27) but was found not to be present in all individuals with PWS. In 1983, Butler and Palmer reported that chromosome 15 was normal in each of the parents of children with PWS and demonstrated, using staining patterns of chromosome 15 polymorphisms, that the de novo deletion always occurred on the paternally derived chromosome 15 (Ref. 28). This suggested that the parent of origin of a chromosomal aberration influenced the phenotype. The analysis of cases of PWS in which no chromosome 15 deletion was present, using cloned DNA markers specific for the 15q11-q13 region, showed that some individuals had inherited two chromosome 15s from their mother; thus, the 15q11-q13 region must be inherited from each parent for normal development (Ref. 29). Furthermore, cytogenetic analysis of Angelman syndrome also showed a deletion in the 15q11-q13 region; however, in Angelman syndrome, the maternally derived chromosome is affected (Ref. 8). Angelman syndrome has a very different phenotype to PWS, characterised by: severe mental retardation, usually with no speech; seizures; unusual behaviour including a happy demeanor, frequent laughter, and easy excitability; and delayed head growth with microcephaly by age 2 years. PWS and Angelman syndrome are classic examples of imprinting disorders resulting from the failure of proper epigenetic regulation of gene expression.
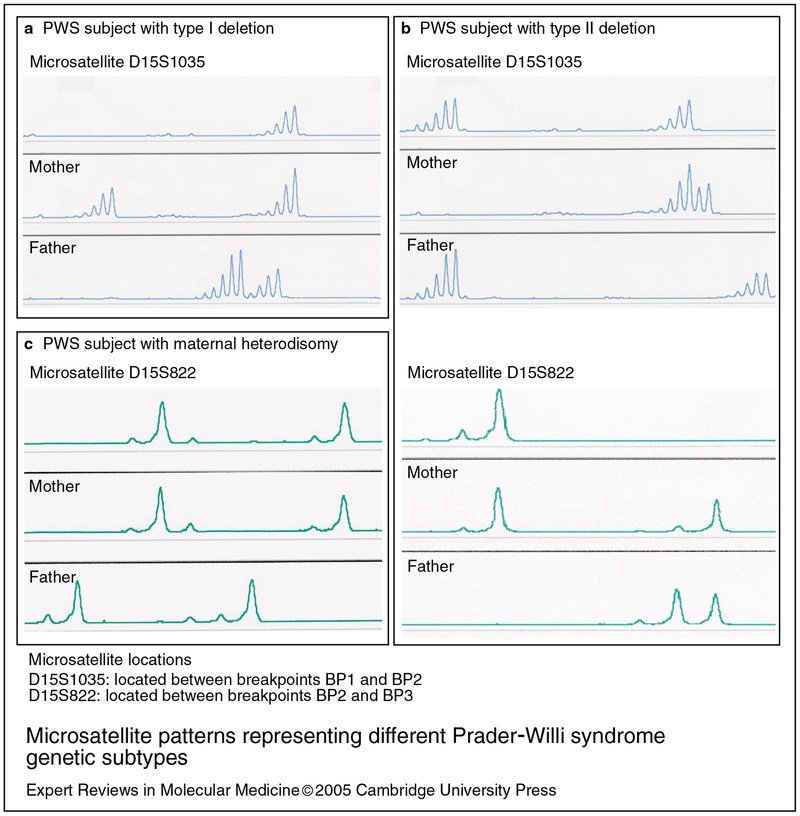
The PWS critical region (PWSCR) located in the chromosome 15q11-q13 region is shown in Figure 1. The typical deletion of the 15q11-q13 region is the most common cause of PWS, presumably due to unequal crossing over in meiosis at repeated transcribed DNA sequences (i.e. HERC2 genes) located at the proximal and distal ends of the 15q11-q13 region (Refs 30, 31). However, it should be noted that overall chromosome breakage in the 15q11-q13 region has not been reported to be increased in PWS subjects compared with controls (Ref. 32). The typical deletion is of two classes – longer type I and shorter type II – and involves a distal breakpoint (BP3) at the end of the 15q11-q13 region and either of two proximally positioned breakpoints (BP1 or BP2) (Ref. 33) (Fig. 1). The type I deletion, involving breakpoints BP1 and BP3, is approximately 5 Mb in size; the type II deletion, involving breakpoints BP2 and BP3, is about 500 kb smaller (Ref. 33). Microsatellite DNA analysis of chromosome 15q11-q13 loci using genomic DNA from subjects with PWS and type I or type II deletions are shown in Figure 2a and 2b, respectively.
Figure 1. Ideogram of chromosome 15, showing genes located in the typical deletion region of Prader–Willi syndrome.
The locations of genes in this region, 15q11-q13, and their imprinting statuses are shown. The gene order is based on the UCSC Genome Bioinformatics website (http://genome.ucsc.edu). Approximately 40% of subjects with the typical deletion have the type I deletion, and approximately 60% have the type II deletion. Abbreviations: Cen, centromere; Tel, telomere; BP, breakpoint; IC, imprinting centre; snoRNA, small nucleolar RNA.
Figure 2. Microsatellite patterns representing different Prader–Willi syndrome genetic subtypes.
Microsatellite patterns are generated by PCR amplification of specific highly polymorphic regions of the genome. The amplified fragments contain a fluorescent tag that is detected after capillary electrophoresis to separate the fragments based on size. The microsatellite patterns are from three different Prader–Willi syndrome (PWS) families. (a) Individuals with type I deletions have only one DNA peak with genotyping analysis using the proximally placed microsatellite marker D15S1035 located between breakpoints BP1 and BP2, indicating the chromosome break occurred at BP1. (b) Individuals with type II deletions have two DNA peaks for D15S1035 but only one peak for a second DNA marker, D15S822, located in the middle of the PWS critical region, indicating the deletion breakpoints at BP2 and BP3. (c) Individuals with maternal heterodisomy have two maternally derived DNA peaks for microsatellite markers from the 15q11-q13 region (e.g. D15S822) and no paternally derived peaks since the paternally derived chromosome is absent.
The maternal disomy 15 that is found in some individuals with PWS is thought to occur from nondisjunction during maternal meiosis, resulting in a trisomy 15 fetus (Refs 7, 29). Through trisomy rescue of the fetus and loss of the father’s chromosome 15, the pregnancy is salvaged and not spontaneously aborted. The fetus is delivered at term having PWS with normal cytogenetic findings but with maternal disomy 15. The microsatellite pattern from a subject with maternal disomy is shown in Figure 2c. Maternal disomy 15 or uniparental disomy (UPD) is of two types: heterodisomy or isodisomy. Maternal heterodisomy occurs when the baby inherits each of the mother’s chromosome 15s but no chromosome 15 from the father (Fig. 2c). Maternal isodisomy results when two identical chromosome 15s are inherited from the mother as a result of nondisjunction in meiosis II or from nondisjunction in meiosis I with crossing over occurring in the proximal long arm of chromosome 15. Maternal isodisomy presents the possibility of additional genetic disorders in the PWS patient if the mother is a carrier of an autosomal recessive gene mutation on chromosome 15, such as the Bloom syndrome gene (BLM) located at 15q26.1 (Ref. 34).
Fluorescence in situ hybridisation (FISH) studies have confirmed that approximately 70% of cases of PWS have the typical chromosome 15q11-q13 deletion. The remaining cases have no obvious defect revealed by FISH and approximately 25% of these cases result from maternal disomy 15. About 3% of cases have biparental or normal inheritance of chromosome 15 and PWS results from defects (microdeletions or epimutations) of the imprinting centre or, rarely, other chromosome 15q11-q13 rearrangements such as translocations, inversions or marker chromosomes (Refs 10, 11, 35).
Molecular biology: candidate genes for PWS
PWS arises from loss of expression of paternally derived genes from the chromosome 15q11-q13 region that are imprinted. Several genes or transcripts have been mapped to the 15q11-q13 region that are imprinted, with most having only paternal expression, including SNURF–SNRPN, small nucleolar RNAs (snoRNAs), necdin, MKRN3 and MAGEL2 (Fig. 1). Candidate genes for causing PWS should be paternally expressed and maternally silenced, located within the chromosome 15q11-q13 region and involved directly or indirectly in brain development and function. To date, the most extensively studied paternally expressed gene that is active in brain tissue is the bicistronic gene SNURF–SNRPN (Refs 36, 37). Exons 4–10 (termed SNRPN, for small nuclear ribonucleoprotein N) encode a core spliceosomal protein (SmN) involved in mRNA splicing in the brain, whereas exons 1–3 (termed SNURF, for ‘SNRPN upstream reading frame’) encode a 71 amino acid protein enriched in arginine residues without known function as yet. The promoter and first exon of SNURF–SNRPN are an integral component of the imprinting centre that controls the regulation of imprinting or gene activity throughout the chromosome 15q11-q13 region. A disruption (i.e. paternal deletion, maternal disomy 15, imprinting mutations/defects or chromosome translocations) of this complex locus will cause loss of function of paternally expressed genes in this region, leading to PWS (Refs 11, 36).
Necdin (NDN), a member of the melanoma-associated antigen gene (MAGE) family of proteins, is expressed only from the paternal allele in the 15q11-q13 region. The function of the MAGE family of proteins remains unclear but they might play a role in cell-cycle regulation and apoptosis (Ref. 38). Necdin is detected in all developing neurons of the embryonic mouse, in both the central and peripheral nervous system, with the highest expression levels in the diencephalon and the hindbrain. After embryonic day 13, necdin has been shown to be essential for axonal outgrowth (Ref. 39) and is expressed in specific structures of the nervous system such as the hypothalamus, thalamus and pons, suggesting a developmental role (Ref. 40). Mice deficient for necdin showed hypothalamic deficiency, neonatal lethality and behavioural changes similar to changes observed in PWS. Necdin might also be responsible for at least some of the other clinical features of PWS (Refs 41, 42), including respiratory abnormalities (Refs 43, 44).
The three breakpoint sites within the 15q11-q13 region are 450–500 kb sequences containing a large transcribed gene (HERC2) and many partially duplicated copies, some of which are transcribed (Refs 30, 45). Other, uncharacterised transcribed sequences are contained within these segments. The functional HERC2 gene located at BP3 encodes a highly conserved giant protein that is distantly related to HERC1, a guanine nucleotide exchange factor (GEF) implicated in vesicular trafficking (Refs 46, 47). The mouse genome contains a single Herc2 locus, located in the jdf2 (juvenile development and fertility-2) interval of chromosome 7C. Mutations in Herc2 lead to defects in neuromuscular secretory vesicles and sperm acrosomes, other developmental abnormalities and juvenile lethality of jdf2 mice. These findings suggest that HERC2 is an important gene encoding a GEF involved in protein trafficking and degradation pathways in the cell (Refs 46, 47). HERC2 repeated sequences have also been implicated in unequal crossing over in meiosis and, as described above, participate in the cytogenetic 15q11-q13 deletion of two sizes (type I and type II) (Refs 30, 36, 46). In addition, an individual with PWS with the typical deletion has been reported also to exhibit a duplication of the proximal end of the 15q11-q13 region inherited from her father (Ref. 48).
Other genes implicated in PWS include IPW (for ‘imprinted in Prader–Willi’), MAGEL2, and a series of highly repeated sequences encoding snoRNAs (Refs 8, 37, 49, 50, 51, 52, 53, 54, 55, 56). IPW is a component of the large SNURF–SNRPN transcript and is spliced and polyadenylated but apparently does not encode a protein; however, it may be a functional RNA, similar to H19 and XIST (Ref. 54). Also encoded within the SNURF–SNRPN transcript are multiple snoRNAs. Two snoRNAs–HBII-52 and HBII-85 – are encoded in a tandemly repeated array of 47 or 24 units, respectively (Ref. 57). These snoRNAs were absent from the brain cortex of an individual with PWS and from a PWS mouse model, demonstrating paternal expression and a possible causal role in PWS. However, the HBII-52 snoRNAs have recently been excluded from having a significant role in PWS because a small deletion encompassing the HBII-52 segment had no obvious effect (Ref. 58). MAGEL2 is expressed predominantly in brain from the paternal allele, with lack of expression in the central nervous system of individuals with PWS. The orthologous mouse gene (Magel2) is paternally expressed predominantly in late developmental stages and adult brain (Ref. 59). In humans, the loss of expression of MAGEL2 might account for brain abnormalities and possibly the dysmorphic features seen in PWS.
Comparing overlapping deletion regions among cases of PWS has allowed the identification of a minimal critical region for PWS (Ref. 57). This critical region includes an approximately 121 kb segment within the >460 kb SNRPN locus. This region contains only the PWCR1/HBII-85 cluster of snoRNAs and the single HBII-438A snoRNA, which further implicates the snoRNAs in playing a significant role in the PWS phenotype (Ref. 57). The function of the snoRNAs in the 15q11-q13 region is unknown but the sequence complementarity of some of the snoRNAs (i.e. HBII-52) to critical regions of other functional gene sequences suggests a role in the processing of specific mRNAs (Ref. 60).
Thus, although PWS is considered a contiguous gene syndrome, the number of causative genes or how the loss of expression of these genes ultimately leads to the PWS phenotype remains unclear. In addition, very little consideration has been given to the potential contribution to the PWS phenotype by altered gene expression of nonimprinted genes. Recently, the analysis of gene expression by microarray technology suggested that several genes/transcripts within or nearby the PWSCR thought to have biallelic expression have expression reduced by more than 50% in those with 15q11-q13 deletions. Furthermore, cells with maternal disomy also had reduced expression of these genes even though two alleles were present (Refs 61, 62). Several genes appeared to have a paternal bias in expression pattern (i.e. greater expression from the paternal allele), most notably the inhibitory neurotransmitter gamma aminobutyric acid (GABA) receptor genes (GABRA5 and GABRB3; discussed below) located in the 15q11.2-q13 region. Thus, the loss of the paternal allele results in a reduction of gene expression greater than 50% since the paternal allele accounts for a larger percentage of gene expression. GABAergic mechanisms have been implicated in a number of symptoms associated with PWS, including hunger (Refs 63, 64, 65), obsessive–compulsive disorder (Refs 66, 67), metabolism (Ref. 68), and visual perception and memory (Refs 69, 70). Presumably, even a modest decrease in the synthesis of GABA receptor proteins could have a significant effect on receptor formation and brain development, with permanent consequences for central nervous system function.
Physiology of eating behaviour
Abnormal levels of multiple neuropeptides and hormones involved in eating behaviour and neurological function, such as ghrelin, growth and sex hormones, and GABA, have been reported in PWS (Ref. 71). The molecular mechanisms responsible for these imbalances and clinical features are not understood. However, data from molecular studies, neuropathology and microarray gene expression analyses of somatic and brain tissues are beginning to illustrate gene(s) interaction patterns and pathways within and outside of the 15q11-q13 region, shedding light on the role of molecular mechanisms in causing features recognised in PWS (Fig. 3).
Figure 3. Speculative diagram illustrating possible interconnected and interactive mechanisms leading to Prader–Willi syndrome.
Chromosome 15q11-q13 abnormalities in Prader–WIlli syndrome (PWS) lead to altered or lack of expression of genes in the region grouped into three categories (imprinted, biased or biallelic). In addition, expression of an interactive network of downstream genes is affected as a result of trans effects (for example due to loss of expression of 15q11-q13 genes involved in RNA processing) and cis effects (for example due to loss of or reduced expression of SGNE1, which might lead to poor transport and processing of proteins such as vasopressin). Many of these perturbations of gene expression might impact on RNA and/or protein processing/trafficking of neuroregulators/hormones. This in turn might lead to misregulation of neuronal development and endocrine dysfunction. The disruption of the expression of other genes, such as those encoding NDN (a cell cycle regulator), the GABA receptors (GABRB3, GABRA5) and olfactory receptors (OR4N4), might have more direct effects on neuronal function, and P (OCA2) disruption directly affects pigmentation. Representative examples of genes grouped according to their expression pattern and their presumed role in the clinical outcome are shown in the figure. Abbreviations: GABR, gamma aminobutyric acid receptor; Gpr15, G-protein-coupled receptor 15; HERC2, hect domain and RLD (renal cell carcinoma-like domain) 2; IPW, imprinted in Prader–Willi; MAGEL2, Mage-like 2; NDN, Necdin; NIPA1, not imprinted in PWS A1; OR4N4, olfactory receptor, family 4, subfamily N, member 4; P (OCA2), oculocutaneous albinism II; POMC, pro-opiomelanocortin; SGNE1, secretory granule neuroendocrine protein 1; snoRNAs, small nucleolar RNAs; SNRPN, small nuclear ribonucleoprotein N; SNURF, SNRPN upstream reading frame; Tcerg1, transcription elongation regulator 1.
The peptide ghrelin is produced and secreted by the stomach into the circulation but is synthesised in several tissues, suggesting both endocrine and paracrine effects. These include increasing appetite, stimulation of GH, prolactin and adenocorticotropic hormone secretion, and regulation of energy homeostasis (Refs 72, 73). Infants, adolescents and adults with PWS have been reported to have significantly higher ghrelin levels than either lean or obese comparison subjects (Refs 74, 75, 76, 77). Ghrelin stimulates eating whereas peptide YY released by the intestine inhibits eating (Refs 78, 79). To characterise these peptides further in PWS, preliminary expression studies have been carried out for ghrelin and peptide YY genes and their receptors using gene microarray technology and quantitative reverse transcription PCR from several regions of the brain and from lymphoblastoid cell lines (Ref. 80). It was found that these peptide genes are active in all regions of the brain studied in both PWS and control subjects (Ref. 80).
There is a 30% reduction in the GH-releasing hormone (GHRH) neurons in the arcuate nucleus in the brain in PWS (Refs 81, 82) and this might explain why GH responsiveness to GHRH is impaired in PWS (Refs 83, 84). Furthermore, PWS has been suggested as a genetic model of starvation owing to abnormal hypothalamic function that results in the body interpreting the absence of satiation as a state of starvation (Ref. 85).
The SGNE1 protein (secretory granule, neuroedocrine protein 1; also known as 7B2 peptide) is detected by antibodies against SGNE1 in the supraoptic and paraventricular nucleus of the hypothalamus in control subjects, but there was no reaction in the majority of individuals with PWS studied (Ref. 86). SGNE1 is a neuroendocrine chaperone protein that interacts with prohormone convertase 2 (PC2) and is involved in the regulation of secretory pathways in the brain, including the release of hormones. The gene encoding SGNE1 is located on chromosome 15, close to the 15q11-q13 region.
GABA is widely distributed throughout the central nervous system; it is estimated that up to 40% of neurons in the brain and spinal cord utilise GABA as their neurotransmitter, making it, quantitatively at least, the most important inhibitory brain neurotransmitter (Ref. 87). Plasma GABA levels are increased three- to fourfold in individuals with PWS compared with control subjects (Ref. 88). Furthermore, three members of the large gene family encoding subunits for GABA-A receptors – GABRA5, GABRB4 and GABRG3 – are located in the 15q11-q13 region (expression changes discussed above). A recent study using positron emission tomography suggested a reduction in GABA receptors in the cingulate, frontal and temporal neocortices and insula in six adult PWS patients as measured by reduced benzodiazepine binding (Ref. 89). We postulate that an increase in GABA level is a direct response to the reduction of A5, B3 and G3 subunit expression in PWS subjects. Abnormal function of the genetic material on chromosome 15 might alter synthesis, release, metabolism, binding, intrinsic activity, or reuptake of specific neurotransmitters, or alter the receptor numbers and/or distribution involved in modulating appetite. Although a mechanistic explanation has not yet been demonstrated, the evidence strongly implicates a significant role of altered GABAergic pathways in the PWS phenotype (Ref. 20).
As mentioned, most PWS patients have reduced GH-secretory capacity and hypogonadotropic hypogonadism, suggesting hypothalamic–pituitary dysfunction. Replacement of GH and/or sex hormones has been shown to be beneficial in several clinical trials in children with PWS (Refs 14, 81, 90, 91, 92). Characteristics such as decreased growth velocity despite the onset of obesity, reduced lean body mass in the presence of adiposity, small hands and feet, and relatively low insulin-like growth factor 1 (IGF-1) and insulin levels are probably the direct result of the hypothalamic GH deficiency in PWS (Refs 14, 17, 91, 93).
Leptin is a peptide produced by adipose tissue and is involved in the regulation of appetite and fat storage (Refs 94, 95, 96, 97). An association has been investigated between genetic variants of the human obesity gene (OB; encoding leptin) and body mass index (BMI) or weight in a group of individuals with PWS and age- and gender-matched lean and obese subjects without PWS (Ref. 98). There was no evidence for reduced leptin in PWS as leptin levels are consistent with increased adiposity associated with PWS or with simple obesity (Ref. 94). In addition, no differences in leptin levels were seen in PWS individuals with maternal disomy or 15q deletions (Ref. 94). Furthermore, the functional leptin receptor (OBRb) has been reported to be present in PWS lymphoblasts (Ref. 95); however, there have been no reported OBRb gene studies from the human hypothalamus. Reduction of leptin levels after GH treatment (Ref. 99) and associated reduction of body fat in PWS patients further suggest that leptin is not misregulated in PWS. There is no evidence of abnormalities of leptin receptors, neuropeptide Y or agouti-related protein in subjects with PWS.
Adiponectin is another peptide produced by adipose tissue and plays a role in regulating adiposity. Recently, serum adiponectin levels in PWS subjects were found to be significantly lower compared with lean subjects and significantly higher compared with obese controls (Ref. 100; L. Kennedy et al., Children’s Mercy Hospitals and Clinics, unpublished). In addition, there were no differences in adiponectin levels between PWS subjects with 15q deletions compared with those with maternal disomy 15. No correlation was found between adiponectin and anthropometrical parameters or measures of insulin sensitivity or between adiponectin and IGF-binding protein 1 or IGF-1 levels in PWS. Furthermore, adiponectin levels did not change during GH treatment in PWS (Ref. 100). Recent evidence suggests individuals with PWS are less likely to develop diabetes than comparison subjects without PWS but with comparable BMI (Ref. 76). This may be a consequence of altered fat distribution in subjects with PWS, resulting in greater adiponectin production than comparison subjects.
Levels of oxytocin, another important neuropeptide produced by the hypothalamus and involved with behaviour, have been reported to be significantly higher in the cerebral spinal fluid of individuals with PWS relative to controls (Ref. 101). However, circulating oxytocin levels were abnormally low in relationship to obesity after GH treatment of PWS (Refs 93, 102).
Diabetes mellitus is not a diagnostic criterion for PWS but is often present (Refs 103, 104). The aetiology for diabetes mellitus in PWS might be related to the morbid obesity and consequent insulin resistance. A decrease of oxytocin neurons and leptin resistance in PWS might also induce hyperphagia and obesity. However, treatment with GH is beneficial for the majority of GH-deficient PWS children, resulting in a relative decrease in fat mass and an increase in fat-free mass, hence decreasing obesity and concomitant insulin resistance.
Genetic subclass comparison in PWS
Individuals with PWS with the typical chromosome 15 deletion are more homogeneous in their clinical presentation and have hypopigmentation (due to deletion of the pigmentation gene P (OCA2) located in the 15q11-q13 region) compared with PWS individuals with nondeleted chromosomes or maternal disomy (Refs 1, 105, 106, 107, 108). Individuals with PWS as a result of maternal disomy have fewer typical facial features and are less likely to have certain behavioural findings such as skin picking, unusual skill with jigsaw puzzles, a high pain threshold and articulation problems. The diagnosis of PWS is reported to occur later in PWS individuals with maternal disomy compared with the 15q deletion, reflecting a milder phenotype in the maternal disomy subjects (Refs 107, 109, 110). Only recently have detailed studies been conducted to identify specific clinical and behavioural differences using gene expression analysis or further delineation of the deletion subtype. The characteristics of genetic subtypes are summarised in Table 2.
Table 2.
Comparison of genetic subtypes in Prader–Willi syndromea
| Genetic defect | Characteristics |
|---|---|
| Typical 15q11-q13 deletion | Hypopigmentation, homogeneous clinical findings (more-typical facial appearance), lower birth weight, greater birth length (males), more self-injurious behaviour (skin picking), higher pain threshold, greater jigsaw puzzle skills relative to maternal disomy |
| Type I deletion | Increased maladaptive and compulsive behaviour relative to type II and maternal disomy; poorer academic performance relative to type II and maternal disomy |
| Type II deletion | Better adaptive behaviour relative to type I; better social skills relative to type I or maternal disomy |
| Maternal disomy | Shorter course of gavage feeding (females), higher verbal IQ scores, greater numeric calculation skills, superior visual memory, poorer object assembly and visual perceptual skills, increased psychosis relative to typical deletion |
Behavioural problems including obsessive–compulsive and self-injurious behaviour are often reported in PWS. Self-injurious behaviour in individuals with intellectual impairment, autism and developmental disabilities without PWS ranges from 5–60% (Ref. 111). However, self-injurious behaviour is reported in 69% of PWS adolescents (Ref. 111) and 81% of adults (Ref. 112). Self-injurious behaviour has been analysed in a cohort of PWS individuals and skin picking was found to be the most common form of self-injury, particularly in those PWS individuals with the typical 15q11-q13 deletion (Ref. 113). Additionally, PWS individuals hoard and arrange items excessively. Compulsive symptoms have been found in about 60% of PWS patients (Ref. 114). Those with the deletion recorded higher scores on the Compulsive Behaviour Checklist, indicating a greater number of compulsive symptoms and more symptom-related distress than those with maternal disomy 15 (Ref. 114). It was also reported that PWS children with the deletion showed relative strengths on standardised visual–spatial tasks such as object assembly compared with ageand IQ-matched control subjects with mixed mental retardation (Ref. 115). Interestingly, the PWS group outperformed normal peers in proficiency at jigsaw puzzle placement.
In a relatively large study of genetic, clinical and behaviour assessments performed at Vanderbilt University (TN, USA), PWS individuals with the typical 15q11-q13 deletions exhibited significantly more self-injury, particularly skin picking, than found in both the obese comparison group and the maternal disomy PWS subgroup (Ref. 116). Furthermore, the typical deletion subgroup displayed higher compulsivity scores than the obese comparison group and spent more time engaging in compulsive behaviour. They were also less able to control their compulsive behaviour. Both PWS subgroups showed significantly greater global severity of compulsive behaviours than found in an obese comparison group. The typical deletion group showed the most severe symptoms of obsessive–compulsive behaviour. In addition, acute psychosis has been reported in PWS, particularly adults with maternal disomy 15 (Ref. 117) and possibly more seizures found in the typical deletion PWS subjects (Ref. 118).
Measures of intelligence and academic achievement in PWS have shown that those with maternal disomy had significantly higher verbal IQ scores than those with the typical deletion (Ref. 105). However, PWS deletion subjects scored higher than the maternal disomy subjects on object-assembly subtests, supporting specific visual perceptual skills as being relative strengths for the deletion group. Additionally, discrimination of shape and motion testing showed that the kinetic form visual performance of the maternal disomy group was significantly worse than the comparison or typical deletion PWS groups (Refs 119, 120). The reason for poorer performance in the maternal disomy group could be due to retinal function and visual processing differences related to excessive GABA levels and abnormal GABA subunit receptors. However, with respect to visual recognition and memory tasks, PWS individuals with maternal disomy performed better than either typical deletion or comparison subjects (Ref. 121). The superior visual recognition memory in the maternal disomy subjects further supports the possible role of two active alleles of maternally expressed genes from chromosome 15, requiring further studies.
In summary, although all individuals with PWS share many common characteristics, comparison of the effects of the deletion (loss of 4–5 Mb) with maternal disomy (two maternal chromosome 15s) reveals several distinctions between the genetic subtypes. In general, individuals with deletions have poorer outcomes compared with individuals with maternal disomy. Paternally expressed genes will be inactive in both subtypes. Therefore, the increased severity associated with the deletion must be due to reduced expression of hemizygous genes, which are compensated when two maternal copies are present, or disruption of interactive genes on other chromosomes by the altered expression of chromosome 15 genes. In addition, some of the differences might be due to changes in expression of genes distal to the PWSCR that could be affected by the deletion process, repositioning of the genes on chromosome 15 or maternal disomy. Currently, several paternally expressed genes in the 15q11-q13 region are candidates for clinical features seen in PWS (e.g. snoRNAs, NDN) and in genotype–phenotype studies.
Genetic research in progress in PWS
Clinical findings associated with type I versus type II typical deletions
Recently, clinical findings associated with type I versus type II typical deletions have been compared (Table 2). Clinical, anthropometric and behavioural data were analysed in 12 PWS individuals shown to have type I deletions and 14 PWS subjects with type II deletions, as determined by the presence or absence of DNA markers located between breakpoints BP1 and BP2 (Ref. 122). Within this chromosome region (between BP1 and BP2) are four recently recognised genes including NIPA1, a gene widely expressed in the central nervous system and brain. Abnormalities of NIPA1 have been reported to occur in familial spastic paraplegia (Ref. 123). Progressive spastic paraplegia appears to result from mutations in NIPA1 as haploinsufficiency does not result in spastic paraplegia in PWS individuals with the type I deletion (Ref. 123). However, PWS individuals with type I deletions scored significantly worse in certain self-injurious and maladaptive behaviour assessments in our study compared with the PWS subjects with type II deletions. Obsessive–compulsive behaviour was also more evident in PWS subjects with type I deletions along with more impairment of visual perception (Ref. 122).
Interestingly, individuals with type II deletions exhibited significantly more self-injury than those with maternal disomy but less than those with type I deletions. The disassociation of compulsivity and skin picking is consistent with our previous factor analytic study revealing that skin picking does not factor with compulsivity using data from the Compulsive Behaviour Checklist (Ref. 124). Psychobehavioural phenotypic characteristics of individuals with PWS who had type I or longer deletions (i.e. involving BP1) were similar in several aspects to individuals with uncharacterised typical deletions (Refs 125, 126, 127, 128) but differed from those with type II or shorter deletions (i.e. involving BP2), the latter group resembling PWS individuals with maternal disomy.
Several academic achievement scores differed between those with shorter or longer typical deletions, which might reflect a difference in intellectual functioning as well as differences in visual perception affecting reading ability. Therefore, loss of genetic material between BP1 and BP2 apparently increases the severity of behavioural and psychological problems seen in this syndrome. The observation of behavioural differences between type I and type II deletions will require replication and further studies using gene expression analysis to understand the role of the genes involved in features seen in PWS.
Gene expression microarrays
The loss of function of genes from the 15q11-q13 region does not directly explain the phenotype of PWS: with the exception of necdin, loss of expression of these genes has not been shown to cause a phenotype observed in PWS. Because it is likely that multiple interacting networks of genes are disrupted by the 15q11-q13 defect – including genes on chromosome 15 (cis effects) as well as genes on other chromosomes (trans effects) – gene expression microarrays have been used to examine expression patterns linked with the PWS phenotype.
Custom-made microarrays were used to examine expression of 73 genes/transcripts to identify higher-level regulatory mechanisms perturbed in PWS and the 15q11-q13 region (Refs 61, 62). Several genes/transcripts (e.g. GABRA5, GABRB3) showed increased expression in PWS cell lines established from individuals with maternal disomy compared with those with the deletion. However, these genes showed less expression in PWS individuals with maternal disomy than in control individuals. Therefore, two maternal copies of the genes produced less RNA than one maternal and one paternal copy of the gene, indicating paternal bias of expression (Ref. 61). In addition, several transcripts outside the PWSCR also had increased expression in individuals with typical deletions relative to controls or PWS individuals with maternal disomy. These observations were confirmed using similar methodology in Angelman syndrome (Ref. 62). This expression pattern is presumably a consequence of repositioning of these genes on the proximal long arm region of the deleted chromosome 15. These results warrant further investigations since they indicate that interconnected mechanisms can produce subtle and unexpected changes in gene expression that may relate to phenotypic differences observed among the genetic subtypes of PWS.
A recent report of microarray analysis of a transgenic mouse model of PWS indicated that genes located in a region on mouse chromosome 18 are modestly upregulated (1.5-fold) by the loss of gene expression from the PWSCR, suggesting a trans-acting regulatory mechanism (Ref. 129). The genes that are upregulated include Pp2ab, Tcerg1, Lars, Pou4f3, Rbm27, Gpr151, C330008R14Rik and Sh3rf2, which all map to a single chromosomal domain. The authors suggest that a gene(s) from the PWSCR interacts with a regulatory element within this domain to reduce expression from this group of genes under normal conditions. Tcerg1 is associated with the RNA polymerase II holoenzyme and is involved in the regulation of transcription elongation, and Gpr151 is a G-protein-coupled receptor. Both genes are widely expressed in the central nervous system and could affect neuronal development. The subtle transactivational control of these genes by genes within the PWSCR suggest that more dramatic changes remain to be discovered at either the translational level or perhaps at the protein level to explain the dramatic phenotypic effects of the loss of expression of the genes in 15q11-q13. Additional studies to examine global regulatory mechanisms at both the transcription and translation level are needed to further our understanding of the complex nature of multigenic disorders such as PWS.
Brain imaging
Preliminary brain imaging studies using functional magnetic resonance imaging (MRI) from a cohort of individuals with PWS and controls revealed post-prandial hyperfunction in the limbic and paralimbic cortical regions of the brain in PWS (Ref. 130). Conversely, brain imaging of individuals with healthy weights indicated increased activity of the limbic and paralimbic regions prior to consumption of a meal (Ref. 131). After eating, these regions of the brain were normalised and neuronal activity decreased substantially. In contrast to healthy-weight individuals, PWS individuals failed to normalise these regions of the brain after meal consumption and instead developed a dramatic increase in neurological activity (Ref. 130). Thus, the failure of satiation following a meal appears to have a neurological basis in PWS. These preliminary results need confirmation in obese individuals without PWS using a wider variety of stimuli to further understand eating behaviour and its regulation in PWS.
Interesting future questions
PWS individuals exhibit reduced selectivity in choosing food items for consumption (i.e. they may consume inappropriate food items). There is a known correlation between smell and taste and/or selection of food for consumption. Interestingly, two olfactory receptor genes (OR4M2, OR4N4) are located proximally at the boundary of the PWSCR on chromosome 15 (UCSC Genome Browser, http://genome.ucsc.edu/). If these genes are misregulated or perturbed in PWS owing to genetic rearrangements, they might contribute to reduced olfactory sensitivity or normal olfaction. This disturbance could lead to failure to discriminate between appropriate and inappropriate food items for consumption. Therefore, the expression of the olfactory receptor genes might be affected more in the larger typical type I deletion. One proposed study would investigate gene expression patterns in PWS to identify gene expression differences that correlate with food-seeking and food-preference behaviour (Ref. 132). The application of proteomic technology in brain and peripheral tissues might also be helpful in elucidating the role of deleted or perturbed genes in neurobehavioural outcomes in PWS.
There is growing evidence for the loss of expression of SNRPN and snoRNAs in causing some or all features recognised in PWS, although there is no evidence that these genes are directly causal. As described before, it appears that these genes are involved in the correct processing/splicing of other genes, particularly in the hypothalamus where they are known to be paternally expressed. There are a number of nonimprinted genes acting in the hypothalamus that are involved in appetite and behaviour regulation with complex processing and splicing required prior to translation (e.g. ghrelin, ghrelin receptor, vasopressin receptor). It is possible that faulty production of mature mRNA of these and/or other downstream genes as a result of the loss of SNRPN function and/or the snoRNAs could produce PWS. This requires further investigation.
Like SNRPN and the snoRNAs, SGNE1 is expressed in the hypothalamus (Ref. 86). SGNE1 produces a chaperone protein (peptide 7B2) implicated in the proper transport and processing of proteins, such as vasopressin in the paraventricular nucleus. Vasopressin reduction in brain specimens of PWS individuals has been reported (Refs 81, 82) and immunohistochemical studies have provided evidence that SGNE1 is downregulated in PWS, resulting in reduced production of mature vasopressin (Refs 86, 133). Perhaps other genes that require complex post-translational processing (e.g. pro-opiomelanocortin (POMC)] might be affected by the misregulation of SGNE1. Genomic and proteomic tools will undoubtedly identify more interactive genes outside the 15q11-q13 region that are affected by incorrect post-transcriptional or post-translational processing.
Concluding remarks
A great deal has been learned about PWS in the past 25 years. Understanding of the basic defect and careful characterisation of the phenotype have dramatically improved the management and therapeutic intervention for individuals with PWS. However, the genes directly responsible for the features of PWS remain obscure. Understanding the interconnection of candidate genes for PWS, their tissue specificity and their level of expression in PWS, along with other interactive genes involved as a result of altered transcription and translation, will improve our knowledge of causation of PWS and hopefully lead to improved therapies.
Acknowledgements and funding
The authors acknowledge the anonymous peer referees for their helpful comments. Funding was supplied by the Hall Foundation of Kansas City and the National Institute of Health (NICHD RO1 41672).
Contributor Information
Douglas C. Bittel, Section of Medical Genetics and Molecular Medicine, Children’s Mercy Hospitals and Clinics; and Assistant Professor, University of Missouri-Kansas City School of Medicine, 2401 Gillham Rd, Kansas City, MO 64108, USA..
Merlin G. Butler, Section of Medical Genetics and Molecular Medicine, Children’s Mercy Hospitals and Clinics; and Professor of Pediatrics, University of Missouri-Kansas City School of Medicine, 2401 Gillham Rd, Kansas City, MO 64108, USA..
References
- 1.Butler MG (1990) Prader-Willi syndrome: current understanding of cause and diagnosis. Am J Med Genet 35, 319–332, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler MG and Thompson T (2000) Prader-Willi syndrome: clinical and genetic findings. The Endocrinol 10, 35–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cassidy SB (1997) Prader-Willi syndrome. J Med Genet 34, 917–923, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler MG, Weaver DD and Meaney FJ (1982) Prader-Willi syndrome: are there population differences? Clin Genet 22, 292–294, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanel ML and Wevrick R (2001) The role of genomic imprinting in human developmental disorders: lessons from Prader-Willi syndrome. Clin Genet 59, 156–164, [DOI] [PubMed] [Google Scholar]
- 6.Walter J and Paulsen M (2003) Imprinting and disease. Semin Cell Dev Biol 14, 101–110, [DOI] [PubMed] [Google Scholar]
- 7.Mascari MJ et al. (1992) The frequency of uniparental disomy in Prader-Willi syndrome. Implications for molecular diagnosis. N Engl J Med 326, 1599–1607, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glenn CC et al. (1997) Genomic imprinting: potential function and mechanisms revealed by the Prader-Willi and Angelman syndromes. Mol Hum Reprod 3, 321–332, [DOI] [PubMed] [Google Scholar]
- 9.Robinson WP et al. (1998) Maternal meiosis I non-disjunction of chromosome 15: dependence of the maternal age effect on level of recombination. Hum Mol Genet 7, 1011–1019, [DOI] [PubMed] [Google Scholar]
- 10.Ohta T et al. (1999) Imprinting-mutation mechanisms in Prader-Willi syndrome. Am J Hum Genet 64, 397–413, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buiting K et al. (2003) Epimutations in Prader-Willi and Angelman syndromes: a molecular study of 136 patients with an imprinting defect. Am J Hum Genet 72, 571–577, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paterson WF and Donaldson MD (2003) Growth hormone therapy in the Prader-Willi syndrome. Arch Dis Child 88, 283–285, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrel AL and Allen DB (2000) Effects of growth hormone on body composition and bone metabolism. Endocrine 12, 163–172, [DOI] [PubMed] [Google Scholar]
- 14.Dattani M and Preece M (2004) Growth hormone deficiency and related disorders: insights into causation, diagnosis, and treatment. Lancet 363, 1977–1987, [DOI] [PubMed] [Google Scholar]
- 15.Harris M, Hofman PL and Cutfield WS (2004) Growth hormone treatment in children: review of safety and efficacy. Paediatr Drugs 6, 93–106, [DOI] [PubMed] [Google Scholar]
- 16.Van Vliet G et al. (2004) Sudden death in growth hormone-treated children with Prader-Willi syndrome. J Pediatr 144, 129–131, [DOI] [PubMed] [Google Scholar]
- 17.Carrel AL and Allen DB (2001) Prader-Willi syndrome: how does growth hormone affect body composition and physical function? J Pediatr Endocrinol Metab 14 Suppl 6, 1445–1451, [PubMed] [Google Scholar]
- 18.Carrel AL et al. (1999) Growth hormone improves body composition, fat utilization, physical strength and agility, and growth in Prader-Willi syndrome: A controlled study. J Pediatr 134, 215–221, [DOI] [PubMed] [Google Scholar]
- 19.Carrel AL et al. (2002) Benefits of long-term GH therapy in Prader-Willi syndrome: a 4-year study. J Clin Endocrinol Metab 87, 1581–1585, [DOI] [PubMed] [Google Scholar]
- 20.Dimitropoulos A et al. (2000) Appetitive behavior, compulsivity, and neurochemistry in Prader-Willi syndrome. Ment Retard Dev Disabil Res Rev 6, 125–130, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carpenter PK (1994) Prader-Willi syndrome in old age. J Intellect Disabil Res 38 (Pt 5), 529–531, [DOI] [PubMed] [Google Scholar]
- 22.Butler MG (2000) A 68-year-old white female with Prader-Willi syndrome. Clin Dysmorphol 9, 65–67, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanchett JM et al. (1996) Age and causes of death in Prader-Willi syndrome patients. Am J Med Genet 62, 211 [Google Scholar]
- 24.Eiholzer U (2005) Deaths in children with Prader-Willi syndrome. A contribution to the debate about the safety of growth hormone treatment in children with PWS. Horm Res 63, 33–39, [DOI] [PubMed] [Google Scholar]
- 25.Schrander-Stumpel CT et al. (2004) Prader-Willi syndrome: causes of death in an international series of 27 cases. Am J Med Genet A 124, 333–338, [DOI] [PubMed] [Google Scholar]
- 26.Stevenson DA et al. (2004) Unexpected death and critical illness in Prader-Willi syndrome: report of ten individuals. Am J Med Genet A 124, 158–164, [DOI] [PubMed] [Google Scholar]
- 27.Ledbetter DH et al. (1981) Deletions of chromosome 15 as a cause of the Prader-Willi syndrome. N Engl J Med 304, 325–329, [DOI] [PubMed] [Google Scholar]
- 28.Butler MG and Palmer CG (1983) Parental origin of chromosome 15 deletion in Prader-Willi syndrome. Lancet 1, 1285–1286, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholls RD et al. (1989) Genetic imprinting suggested by maternal heterodisomy in nondeletion Prader-Willi syndrome. Nature 342, 281–285, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amos-Landgraf JM et al. (1999) Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet 65, 370–386, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christian SL et al. (1999) Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13). Hum Mol Genet 8, 1025–1037, [DOI] [PubMed] [Google Scholar]
- 32.Butler MG and Jenkins BB (1989) Analysis of chromosome breakage in the Prader-Labhart-Willi syndrome. Am J Med Genet 32, 514–519, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christian SL et al. (1995) Molecular characterization of two proximal deletion breakpoint regions in both Prader-Willi and Angelman syndrome patients. Am J Hum Genet 57, 40–48, [PMC free article] [PubMed] [Google Scholar]
- 34.Woodage T et al. (1994) Bloom syndrome and maternal uniparental disomy for chromosome 15. Am J Hum Genet 55, 74–80, [PMC free article] [PubMed] [Google Scholar]
- 35.Buiting K et al. (2000) Imprinting centre deletions in two PWS families: implications for diagnostic testing and genetic counseling. Clin Genet 58, 284–290, [DOI] [PubMed] [Google Scholar]
- 36.Nicholls RD and Knepper JL (2001) Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet 2, 153–175, [DOI] [PubMed] [Google Scholar]
- 37.Lee S, Walker CL and Wevrick R (2003) Prader-Willi syndrome transcripts are expressed in phenotypically significant regions of the developing mouse brain. Gene Expr Patterns 3, 599–609, [DOI] [PubMed] [Google Scholar]
- 38.Barker PA and Salehi A (2002) The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J Neurosci Res 67, 705–712, [DOI] [PubMed] [Google Scholar]
- 39.Lee S et al. (2005) Essential role for the Prader-Willi syndrome protein necdin in axonal outgrowth. Hum Mol Genet 14, 627–637, [DOI] [PubMed] [Google Scholar]
- 40.Andrieu D et al. (2003) Expression of the Prader-Willi gene Necdin during mouse nervous system development correlates with neuronal differentiation and p75NTR expression. Gene Expr Patterns 3, 761–765, [DOI] [PubMed] [Google Scholar]
- 41.Gerard M et al. (1999) Disruption of the mouse necdin gene results in early post-natal lethality. Nat Genet 23, 199–202, [DOI] [PubMed] [Google Scholar]
- 42.Muscatelli F et al. (2000) Disruption of the mouse Necdin gene results in hypothalamic and behavioral alterations reminiscent of the human Prader-Willi syndrome. Hum Mol Genet 9, 3101–3110, [DOI] [PubMed] [Google Scholar]
- 43.Butler JV et al. (2002) Prevalence of, and risk factors for, physical ill-health in people with Prader-Willi syndrome: a population-based study. Dev Med Child Neurol 44, 248–255, [DOI] [PubMed] [Google Scholar]
- 44.DiMario FJ Jr. et al. (1996) Respiratory sinus arrhythmia in patients with Prader-Willi syndrome. J Child Neurol 11, 121–125, [DOI] [PubMed] [Google Scholar]
- 45.Buiting K et al. (1998) Expressed copies of the MN7 (D15F37) gene family map close to the common deletion breakpoints in the Prader-Willi/Angelman syndromes. Cytogenet Cell Genet 81, 247–253, [DOI] [PubMed] [Google Scholar]
- 46.Ji Y et al. (2000) Structure of the highly conserved HERC2 gene and of multiple partially duplicated paralogs in human. Genome Res 10, 319–329, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ji Y et al. (1999) The ancestral gene for transcribed, low-copy repeats in the Prader-Willi/Angelman region encodes a large protein implicated in protein trafficking, which is deficient in mice with neuromuscular and spermiogenic abnormalities. Hum Mol Genet 8, 533–542, [DOI] [PubMed] [Google Scholar]
- 48.Butler MG, Bittel D and Talebizadeh Z (2002) Prader-Willi syndrome and a deletion/duplication within the 15q11-q13 region. J Med Genet 39, 202–204, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Francke U (1998) Imprinted genes in the Prader-Willi deletion. Novartis Found Symp 214, 264–275; discussion 275–269, [DOI] [PubMed] [Google Scholar]
- 50.Meguro M et al. (2001) Large-scale evaluation of imprinting status in the Prader-Willi syndrome region: an imprinted direct repeat cluster resembling small nucleolar RNA genes. Hum Mol Genet 10, 383–394, [DOI] [PubMed] [Google Scholar]
- 51.Muralidhar B, Marney A and Butler MG (1999) Analysis of imprinted genes in subjects with Prader-Willi syndrome and chromosome 15 abnormalities. Genet Med 1, 141–145, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Runte M et al. (2001) The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet 10, 2687–2700, [DOI] [PubMed] [Google Scholar]
- 53.Wevrick R and Francke U (1997) An imprinted mouse transcript homologous to the human imprinted in Prader-Willi syndrome (IPW) gene. Hum Mol Genet 6, 325–332, [DOI] [PubMed] [Google Scholar]
- 54.Wevrick R, Kerns JA and Francke U (1994) Identification of a novel paternally expressed gene in the Prader-Willi syndrome region. Hum Mol Genet 3, 1877–1882, [DOI] [PubMed] [Google Scholar]
- 55.Wevrick R, Kerns JA and Francke U (1996) The IPW gene is imprinted and is not expressed in the Prader-Willi syndrome. Acta Genet Med Gemellol (Roma) 45, 191–197, [DOI] [PubMed] [Google Scholar]
- 56.Saitoh S et al. (1996) Minimal definition of the imprinting center and fixation of chromosome 15q11-q13 epigenotype by imprinting mutations. Proc Natl Acad Sci U S A 93, 7811–7815, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gallagher RC et al. (2002) Evidence for the role of PWCR1/HBII-85 C/D box small nucleolar RNAs in Prader-Willi syndrome. Am J Hum Genet 71, 669–678, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Runte M et al. (2005) Exclusion of the C/D box snoRNA gene cluster HBII-52 from a major role in Prader-Willi syndrome. Hum Genet 116, 228–230, [DOI] [PubMed] [Google Scholar]
- 59.Lee S et al. (2000) Expression and imprinting of MAGEL2 suggest a role in Prader-willi syndrome and the homologous murine imprinting phenotype. Hum Mol Genet 9, 1813–1819, [DOI] [PubMed] [Google Scholar]
- 60.Cavaille J et al. (2000) Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci U S A 97, 14311–14316, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bittel DC et al. (2003) Microarray analysis of gene/transcript expression in Prader-Willi syndrome: deletion versus UPD. J Med Genet 40, 568–574, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bittel DC et al. (2005) Microarray analysis of gene/transcript expression in Angelman syndrome: deletion versus UPD. Genomics 85, 85–91, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horvath TL et al. (1997) Heterogeneity in the neuropeptide Y-containing neurons of the rat arcuate nucleus: GABAergic and non-GABAergic subpopulations. Brain Res 756, 283–286, [DOI] [PubMed] [Google Scholar]
- 64.Kalra SP et al. (1999) Interacting appetite-regulating pathways in the hypothalamic regulation of body weight. Endocr Rev 20, 68–100, [DOI] [PubMed] [Google Scholar]
- 65.Dahir GA and Butler MG (1991) Is GABA-A receptor B3 subunit abnormality responsible for obesity in persons with Prader-Willi syndrome? Dysmorphol Clin Genet 5, 112–113 [Google Scholar]
- 66.Baxter LR Jr. et al. (1996) Brain Mediation of Obsessive-Compulsive Disorder Symptoms: Evidence From Functional Brain Imaging Studies in the Human and Nonhuman Primate. Semin Clin Neuropsychiatry 1, 32–47, [DOI] [PubMed] [Google Scholar]
- 67.Rosenberg DR and Keshavan MS (1998) A.E. Bennett Research Award. Toward a neurodevelopmental model of of obsessive—compulsive disorder. Biol Psychiatry 43, 623–640, [DOI] [PubMed] [Google Scholar]
- 68.Lee HS et al. (2001) Activation of metabotropic glutamate receptors inhibits GABAergic transmission in the rat subfornical organ. Neuroscience 102, 401–411, [DOI] [PubMed] [Google Scholar]
- 69.Berninger B et al. (1995) GABAergic stimulation switches from enhancing to repressing BDNF expression in rat hippocampal neurons during maturation in vitro. Development 121, 2327–2335, [DOI] [PubMed] [Google Scholar]
- 70.Huang B, Mitchell CK and Redburn-Johnson DA (2000) GABA and GABA(A) receptor antagonists alter developing cone photoreceptor development in neonatal rabbit retina. Vis Neurosci 17, 925–935, [DOI] [PubMed] [Google Scholar]
- 71.Butler MG et al. (2005) Genetics and obesity: Prader-Willi syndrome, an illustrative example In Focus on Obesity Research (Ling PR, ed.), pp. 51–88, Nova Science Publishers, Inc., Hauppage, NY, USA [Google Scholar]
- 72.Korbonits M and Grossman AB (2004) Ghrelin: update on a novel hormonal system. Eur J Endocrinol 151 Suppl 1, S67–70, [DOI] [PubMed] [Google Scholar]
- 73.Meier U and Gressner AM (2004) Endocrine regulation of energy metabolism: review of pathobiochemical and clinical chemical aspects of leptin, ghrelin, adiponectin, and resistin. Clin Chem 50, 1511–1525, [DOI] [PubMed] [Google Scholar]
- 74.Butler MG, Bittel DC and Talebizadeh Z (2004) Plasma peptide YY and ghrelin levels in infants and children with Prader-Willi syndrome. J Pediatr Endocrinol Metab 17, 1177–1184, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cummings DE et al. (2002) Elevated plasma ghrelin levels in Prader-Willi syndrome. Nat Med 8, 643–644, [DOI] [PubMed] [Google Scholar]
- 76.Goldstone AP et al. (2004) Elevated fasting plasma ghrelin in Prader-Willi syndrome adults is not solely explained by their reduced visceral adiposity and insulin resistance. J Clin Endocrinol Metab 89, 1718–1726, [DOI] [PubMed] [Google Scholar]
- 77.Haqq AM et al. (2003) Serum ghrelin levels are inversely correlated with body mass index, age, and insulin concentrations in normal children and are markedly increased in Prader-Willi syndrome. J Clin Endocrinol Metab 88, 174–178, [DOI] [PubMed] [Google Scholar]
- 78.Halford JC, Cooper GD and Dovey TM (2004) The pharmacology of human appetite expression. Curr Drug Targets 5, 221–240, [DOI] [PubMed] [Google Scholar]
- 79.Small CJ and Bloom SR (2004) Gut hormones and the control of appetite. Trends Endocrinol Metab 15, 259–263, [DOI] [PubMed] [Google Scholar]
- 80.Talebizadeh Z et al. (2005) Ghrelin, peptide YY and their receptors: gene expression in brain from subjects with and without Prader-Willi syndrome. Int J Mol Med 15, 707–711, [PMC free article] [PubMed] [Google Scholar]
- 81.Swaab DF (1997) Prader-Willi syndrome and the hypothalamus. Acta Paediatr Suppl 423, 50–54, [DOI] [PubMed] [Google Scholar]
- 82.Swaab DF (2004) Neuropeptides in hypothalamic neuronal disorders. Int Rev Cytol 240, 305–375, [DOI] [PubMed] [Google Scholar]
- 83.Grugni G, Guzzaloni G and Morabito F (2001) Impairment of GH responsiveness to GH-releasing hexapeptide (GHRP-6) in Prader-Willi syndrome. J Endocrinol Invest 24, 340–348, [DOI] [PubMed] [Google Scholar]
- 84.Grugni G et al. (1998) Reduced growth hormone (GH) responsiveness to combined GH-releasing hormone and pyridostigmine administration in the Prader-Willi syndrome. Clin Endocrinol (Oxf) 48, 769–775, [DOI] [PubMed] [Google Scholar]
- 85.Holland A, Whittington J and Hinton E (2003) The paradox of Prader-Willi syndrome: a genetic model of starvation. Lancet 362, 989–991, [DOI] [PubMed] [Google Scholar]
- 86.Gabreels BA et al. (1994) Differential expression of the neuroendocrine polypeptide 7B2 in hypothalami of Prader-(Labhart)-Willi syndrome patients. Brain Res 657, 281–293, [DOI] [PubMed] [Google Scholar]
- 87.Bloom FE and Iversen LL (1971) Localizing 3H-GABA in nerve terminals of rat cerebral cortex by electron microscopic autoradiography. Nature 229, 628–630, [DOI] [PubMed] [Google Scholar]
- 88.Ebert MH et al. (1997) Elevated plasma gamma-aminobutyric acid (GABA) levels in individuals with either Prader-Willi syndrome or Angelman syndrome. J Neuropsychiatry Clin Neurosci 9, 75–80, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lucignani G et al. (2004) GABA A receptor abnormalities in Prader-Willi syndrome assessed with positron emission tomography and [11C]flumazenil. Neuroimage 22, 22–28, [DOI] [PubMed] [Google Scholar]
- 90.Burman P, Ritzen EM and Lindgren AC (2001) Endocrine dysfunction in Prader-Willi syndrome: a review with special reference to GH. Endocr Rev 22, 787–799, [DOI] [PubMed] [Google Scholar]
- 91.Eiholzer U, Bachmann S and l’Allemand D (2000) Is there growth hormone deficiency in prader-willi Syndrome? Six arguments to support the presence of hypothalamic growth hormone deficiency in Prader-Willi syndrome. Horm Res 53 Suppl 3, 44–52, [DOI] [PubMed] [Google Scholar]
- 92.Rittinger O (2001) [Clinical aspects and genetics of Prader-Willi syndrome]. Klin Padiatr 213, 91–98, [DOI] [PubMed] [Google Scholar]
- 93.Hoybye C (2004) Endocrine and metabolic aspects of adult Prader-Willi syndrome with special emphasis on the effect of growth hormone treatment. Growth Horm IGF Res 14, 1–15, [DOI] [PubMed] [Google Scholar]
- 94.Butler MG et al. (1998) Comparison of leptin protein levels in Prader-Willi syndrome and control individuals. Am J Med Genet 75, 7–12, [PMC free article] [PubMed] [Google Scholar]
- 95.Goldstone AP et al. (2002) Resting metabolic rate, plasma leptin concentrations, leptin receptor expression, and adipose tissue measured by whole-body magnetic resonance imaging in women with Prader-Willi syndrome. Am J Clin Nutr 75, 468–475, [DOI] [PubMed] [Google Scholar]
- 96.Ostlund RE Jr. et al. (1996) Relation between plasma leptin concentration and body fat, gender, diet, age, and metabolic covariates. J Clin Endocrinol Metab 81, 3909–3913, [DOI] [PubMed] [Google Scholar]
- 97.Pijl H, Toornvliet AC and Meinders AE (1996) Serum leptin in normal-weight and obese humans. N Engl J Med 334, 1544, [DOI] [PubMed] [Google Scholar]
- 98.Butler MG et al. (1998) Genetic variants of the human obesity (OB) gene in subjects with and without Prader-Willi syndrome: comparison with body mass index and weight. Clin Genet 54, 385–393, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zadik Z et al. (2001) Interrelationship between insulin, leptin and growth hormone in growth hormone-treated children. Int J Obes Relat Metab Disord 25, 538–542, [DOI] [PubMed] [Google Scholar]
- 100.Hoybye C et al. (2004) Serum adiponectin levels in adults with Prader-Willi syndrome are independent of anthropometrical parameters and do not change with GH treatment. Eur J Endocrinol 151, 457–461, [DOI] [PubMed] [Google Scholar]
- 101.Martin A et al. (1998) Cerebrospinal fluid levels of oxytocin in Prader-Willi syndrome: a preliminary report. Biol Psychiatry 44, 1349–1352, [DOI] [PubMed] [Google Scholar]
- 102.Hoybye C et al. (2003) Peptides associated with hyperphagia in adults with Prader-Willi syndrome before and during GH treatment. Growth Horm IGF Res 13, 322–327, [DOI] [PubMed] [Google Scholar]
- 103.Ristow M (2004) Neurodegenerative disorders associated with diabetes mellitus. J Mol Med 82, 510–529, [DOI] [PubMed] [Google Scholar]
- 104.Nagai T and Mori M (1999) Prader-Willi syndrome, diabetes mellitus and hypogonadism. Biomed Pharmacother 53, 452–454, [DOI] [PubMed] [Google Scholar]
- 105.Roof E et al. (2000) Intellectual characteristics of Prader-Willi syndrome: comparison of genetic subtypes. J Intellect Disabil Res 44 (Pt 1), 25–30, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cassidy SB et al. (1997) Comparison of phenotype between patients with Prader-Willi syndrome due to deletion 15q and uniparental disomy 15. Am J Med Genet 68, 433–440, [PubMed] [Google Scholar]
- 107.Gunay-Aygun M et al. (1997) Delayed diagnosis in patients with Prader-Willi syndrome due to maternal uniparental disomy 15. Am J Med Genet 71, 106–110, [PubMed] [Google Scholar]
- 108.Veltman MW et al. (2004) Prader-Willi syndrome—a study comparing deletion and uniparental disomy cases with reference to autism spectrum disorders. Eur Child Adolesc Psychiatry 13, 42–50, [DOI] [PubMed] [Google Scholar]
- 109.Gillessen-Kaesbach G et al. (1995) Genotype-phenotype correlation in a series of 167 deletion and non-deletion patients with Prader-Willi syndrome. Hum Genet 96, 638–643, [DOI] [PubMed] [Google Scholar]
- 110.Mitchell J et al. (1996) Comparison of phenotype in uniparental disomy and deletion Prader-Willi syndrome: sex specific differences. Am J Med Genet 65, 133–136, [DOI] [PubMed] [Google Scholar]
- 111.Thompson T and Gray DB, eds (1994) Destructive Behavior in Developmental Disabilities: Diagnosis and Treatment, Sage Publishers, Thousand Oaks, CA, USA [Google Scholar]
- 112.Whitman BY and Accardo P (1987) Emotional symptoms in Prader-Willi syndrome adolescents. Am J Med Genet 28, 897–905, [DOI] [PubMed] [Google Scholar]
- 113.Symons FJ et al. (1999) Self-injurious behavior and Prader-Willi syndrome: behavioral forms and body locations. Am J Ment Retard 104, 260–269, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dykens EM, Leckman JF and Cassidy SB (1996) Obsessions and compulsions in Prader-Willi syndrome. J Child Psychol Psychiatry 37, 995–1002, [DOI] [PubMed] [Google Scholar]
- 115.Dykens EM (2002) Are jigsaw puzzle skills ‘spared’ in persons with Prader-Willi syndrome? J Child Psychol Psychiatry 43, 343–352, [DOI] [PubMed] [Google Scholar]
- 116.Thompson T and Butler MG (2003) Prader-Willi syndrome: clinical, behavioral and genetic findings In Disorders of Development and Learning (3rd edn) (Wolraich ML, ed.), pp. 276–281, B.C. Decker, Inc., Hamilton, Ontario, Canada [Google Scholar]
- 117.Vogels A et al. (2003) Chromosome 15 maternal uniparental disomy and psychosis in Prader-Willi syndrome. J Med Genet 40, 72–73, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Varela MC et al. (2005) Impact of molecular mechanisms, including deletion size, on Prader-Willi syndrome phenotype: study of 75 patients. Clin Genet 67, 47–52, [DOI] [PubMed] [Google Scholar]
- 119.Fox R et al. (2001) Kinetic form discrimination in Prader-Willi syndrome. J Intellect Disabil Res 45, 317–325, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fox R et al. (1999) Visual capacity and Prader-Willi syndrome. J Pediatr Ophthalmol Strabismus 36, 331–336, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Joseph B et al. (2001) Possible dosage effect of maternally expressed genes on visual recognition memory in Prader-Willi syndrome. Am J Med Genet 105, 71–75, [PubMed] [Google Scholar]
- 122.Butler MG et al. (2004) Behavioral differences among subjects with Prader-Willi syndrome and type I or type II deletion and maternal disomy. Pediatrics 113, 565–573, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rainier S et al. (2003) NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6). Am J Hum Genet 73, 967–971, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Feurer ID et al. (1998) The latent variable structure of the Compulsive Behaviour Checklist in people with Prader-Willi syndrome. J Intellect Disabil Res 42 (Pt 6), 472–480, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Butler MG, Meaney FJ and Palmer CG (1986) Clinical and cytogenetic survey of 39 individuals with Prader-Labhart-Willi syndrome. Am J Med Genet 23, 793–809, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Magenis RE et al. (1990) Comparison of the 15q deletions in Prader-Willi and Angelman syndromes: specific regions, extent of deletions, parental origin, and clinical consequences. Am J Med Genet 35, 333–349, [DOI] [PubMed] [Google Scholar]
- 127.Robinson WP et al. (1991) Molecular, cytogenetic, and clinical investigations of Prader-Willi syndrome patients. Am J Hum Genet 49, 1219–1234, [PMC free article] [PubMed] [Google Scholar]
- 128.Zori R et al. (1990) Parental origin of del(15)(q11-q13) in Angelman and Prader-Willi syndromes. Am J Med Genet 37, 294–295, [DOI] [PubMed] [Google Scholar]
- 129.Stefan M et al. (2005) A nonimprinted Prader-Willi Syndrome (PWS)-region gene regulates a different chromosomal domain in trans but the imprinted pws loci do not alter genome-wide mRNA levels. Genomics 85, 630–640, [DOI] [PubMed] [Google Scholar]
- 130.Holsen LM et al. Neural mechanisms underlying food motivation in children and adolescents. Neuro Image, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shapira NA et al. (2005) Satiety dysfunction in Prader-Willi syndrome demonstrated by fMRI. J Neurol Neurosurg Psychiatry 76, 260–262, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Young J et al. Measures of food seeking in Prader-Willi syndrome. Journal of Intellectual Disability Research (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gabreels BA et al. (1998) Attenuation of the polypeptide 7B2, prohormone convertase PC2, and vasopressin in the hypothalamus of some Prader-Willi patients: indications for a processing defect. J Clin Endocrinol Metab 83, 591–599, [DOI] [PubMed] [Google Scholar]
- 134.Butler MG, Lee PDK and Whitman BY, eds (2005) Management of Prader-Willi Syndrome (3rd edn), Springer-Verlag, New York, NY, USA [Google Scholar]