Abstract
This study aims to screen differentially expressed host miRNAs that could be used as diagnostic markers for liver alveolar echinococcosis (LAE).
Differentially expressed miRNAs were first screened by miRNA microarray in liver tissues from2 LAE patients and normal liver tissues from 3 LAE patients, followed by qRT-PCR validation in 15 LAE tissues and 15 normal tissues. Target genes of differentially expressed miRNAs were predicted using Targetscan, PITA and microRNAorg database, and the overlapped predicted target genes were analyzed by GO and KEGG.
The hsa-miR-1237-3p, hsa-miR-33b-3p, and hsa-miR-483-3p were up-regulated whereas the hsa-miR-4306 was down-regulated in LAE tissues compared with normal controls (P < .05). The expression change of miR-483-3p was further confirmed in both liver tissues and plasma. Several predicted targets of miR-1237-3p, miR-4306, and miR-483-3p were related to DNA-dependent transcriptional regulation, developmental regulation of multicellular organisms, and biological functions such as cellular immune responses (T cell proliferation). The overlapped predicted target genes of the 4 differentially expressed miRNAs were enriched in mRNA surveillance, cancer signaling pathway, intestinal immune network, and other signal pathways.
Our results indicate that miR-483-3p is a potential marker for the diagnosis of LAE, and targets of this miRNA could be the focus of further studies.
Keywords: Echinococcus multilocularis, GO and KEGG analysis, liver alveolar echinococcosis, miR-483-3p
1. Introduction
Liver alveolar echinococcosis (LAE) is a disease caused by larvae stage of Echinococcus multilocularis (E multilocularis) in humans. This disease is invasive and has tumor-like characteristics, which is called “bug cancer”.[1,2] LAE has the characteristics of long incubation period, no obvious symptoms at the beginning of infection, and easy transfer in the later stage.[3] In China, it is mainly reported in Qinghai, Tibet, Xinjiang and other animal husbandry areas, of which Sanjiangyuan area in Qinghai Province is one of the most reported area.[2]
At present, the diagnosis of LAE mainly relies on traditional diagnostic methods such as clinical diagnosis, imaging diagnosis, and immunological diagnosis. However, these diagnostic methods have certain deficiencies, such as strong subjectivity and inconsistency among patients for clinical diagnosis,[4] and low sensitivity and specificity for imaging and immunological diagnosis.[5] The development of molecular technology in recent years greatly improved the early diagnosis of patients with LAE.[6]
MicroRNAs (miRNAs) are a class of endogenous non-coding RNA with the length of 22nt, which could regulate gene expression at post-transcriptional level by targeted cleavage of mRNA or repression of translation and participate in the regulation of several physiology and pathology processes.[7,8] For example, it is found that miR211 can promote cell proliferation, migration, and differentiation by targeting ZFPM2 gene in human osteoblasts.[9] Several differentially expressed miRNAs are found during the growth of Hela cells.[10] Deep sequencing and miRNA microarray technology reveal that the expression of miRNAs in host cells or tissues are regulated during parasite infection and play an important role in host response to pathogens [Xu, 2013 #4118;Yadong, 2013 #4121]. Jin et al found that infection with E multilocularis disrupted the expression of 40% of genes involved in miRNA synthesis in mouse liver, including AGO1 and AGO2, and thus 46 differentially expressed miRNAs were found by deep sequencing analysis.[11] In the process of E multilocularis infection in humans, miRNAs also play important roles,[12] and some of these miRNAs participate in the immune and inflammatory responses to parasite infection.[13] Therefore, detection of miRNAs involved in pathogen-infection process could improve the sensitivity and specificity of early diagnosis for parasite.[14]
Circulating miRNAs are a class of molecule that are stably present in body fluids such as serum and plasma, and are stably detected in blood or body fluids of humans and animals infected with worms.[15–17] Circulating miRNAs have been used as potential biomarkers in various tumors, heart diseases, liver damage, and other diseases for early diagnosis, classification, and prognosis.[18–22] Circulating miRNAs have also been reported as biomarkers in parasitic infections.[23–25] For example, serum miR-146a and miR-223 were reported to be used as biomarkers for the diagnosis of sepsis, with high specificity and sensitivity.[26,27]
In this study, we used miRNA microarray to detect miRNA expression in patients with LAE. The significantly dysregulated miRNAs between LAE patients and normal controls were selected for further validation in the liver tissues and plasma of a larger cohort of patients. Our results will provide candidate for circulating miRNAs that could be used as biomarkers for LAE diagnosis.
2. Materials and methods
2.1. Patients
Two cases with LAE (1 male and 1 female with average age of 34.75) who were diagnosed between Jun, 2016 and May, 2018 at the Affiliated Hospital of Qinghai University, and 3 healthy controls (1 male and 2 female with average age of 41.5) were enrolled for microRNA microarray. A total of 15 cases of LAE patients (7 male and 8 female with average age of 46.1) who were diagnosed between, 2016 and May, 2018 at the Affiliated Hospital of Qinghai University, and 15 healthy controls (7 male and 8 female with average age of 44.6) were enrolled for validation. Prior written and informed consent were obtained from every patient and the study was approved by the ethics review board of the Hospital affiliated to Qinghai University.
2.2. Inclusion and exclusion criteria
All patients with LAE were first diagnosed and confirmed according to the diagnostic criteria by Expert consensus on diagnosis and treatment of liver echinococcosis (2015 edition). The patients were at early stage of LAE when the tissue samples were collected. Patients or normal controls with hepatitis B or tuberculosis or other liver diseases were excluded from this study.
2.3. Sample collection
A total of 0.5 to 250 mg normal or infected liver tissues were collected by resection, and stored in liquid nitrogen until analysis. EDTA-K2 treated whole blood was collected from patients and healthy controls and plasma was isolated by centrifugation for 5 minutes at 1000 rpm at 4°C. The plasma was stored at −80°C until further analysis.
2.4. RNA extraction and microRNA microarray
miRNAs were isolated from liver tissues using mirVanaTM RNA Isolation Kit (AM1561, Thermo Fisher Scientific) according to the manufacturer's instructions. The miRNA was quantified using NanoDrop ND-2000 (Thermo Fisher Scientific) and the integrity of RNA was determined using Agilent 2100 Bioanalyzer (Agilent Technologies).
Agilent Human miRNA (8∗60K, Design ID: 070156) microarrays were used. The RNA labeling and array hybridization were conducted using the miRNA Complete Labeling and Hyb Kit (Agilent technologies, 5190–0456). Slides were scanned by Agilent Scanner (#G2505C Agilent technologies).
Raw data were obtained by Agilent Feature Extraction software (version 10.7.1.1; Agilent Technologies), and the quantile algorithm was performed using Genespring software (version 12.5; Agilent Technologies) with default settings.[20] The total gene signal between groups was normalize to the 75th percentile signal intensity. After acquiring the raw data, the differently expressed miRNAs were calculated using the t test. Those with fold change ≥ 2 and P value < .05 were regarded as significantly different. Volcano plots and heat maps were generated in R software.
2.5. Target gene prediction and analysis
Targets of miRNAs were predicted using Targetscan (http://www.targetscan.org), miroRNAorg (http://www.microrna.org/) and PITA (https://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html). The function analysis was conducted using the GO analysis (http://www.geneontology.org). The pathway analysis was conducted using the KEGG (https://www.genome.jp/kegg/).
2.6. Quantitative real-time PCR
Total RNAs were isolated from liver tissues using Trizol Reagent (Thermo Fisher Scientific). Plasma miRNA was extracted using serum/plasma miRNA extraction and separation kit (#DP503, Tiangen Biotech, China). RNA was reverse transcribed using miRNA First Strand cDNA Synthesis (Tailing Reaction) into cDNA (Sangon Biotech Co. Ltd., China). Quantitative real-time PCR was performed using miRNA Fluorescence Quantitative PCR Kit (Dye Method) (Sangon Biotech Co. Ltd., China) on an ABI 7500 Real-time PCR system (Thermo Fisher Scientific). Primers used were as follows: hsa-miR-1237–3p forward 5’- agtccttctgctccgtc -3’; hsa-miR-483-3p forward 5’- gcagtcactcctctcct -3’; hsa-miR-4306 forward 5’- cgcagtggagagaaagg -3’. Reverse primer was universal primer from Sangon (Shanghai, China). Primers for cel-miR-39 were purchased from Tiangen Biotech (Beijing) Co. Ltd. The relative expression levels were evaluated using the 2−ΔΔCt method. The expression of cel-miR-39 was used as internal control.
2.7. Statistical analysis
Data analysis was carried out using the SPSS 19.0 (IBM Corp, Chicago, IL, USA) and expressed as mean ± SD. Differences between groups were evaluated for significance using independent sample T test. P < .05 was considered as statistically significant.
3. Results
3.1. Screening for differentially expressed miRNAs
To identify miRNAs that may play important roles in LAE, we investigated the differentially expressed miRNAs in LAE liver tissues of 2 patients with LAE and normal liver tissues from 3 controls. As shown in Table 1 and Figure 1A, there were 9 miRNAs that were differently expressed between the 2 groups (|Fold change|>1.3, P < .05 & coefficient of variation%<12%) among the 2549 investigated miRNAs. In these 9 miRNAs, we further focused on miRNAs with |log2(Fold change)|>1, which were deemed as differentially expressed miRNA. As a result, we detected the upregulation of hsa-miR-1237-3p (log2 (fold change) = 3.4), hsa-miR-33b-3p (log2 (fold change) = 3.03) and hsa-miR-483-3p (log2 (fold change) = 3.13), and the downregulation of hsa-miR-4306 (log2 (fold change) = −5.73) in LAE tissues compared with normal tissues (Fig. 1A). Unsupervised clustering of these 4 miRNAs showed the obvious differences of their expression between LAE tissues and controls (Fig. 1B). These results suggest that these 4 miRNAs of hsa-miR-1237-3p, hsa-miR-33b-3p, hsa-miR-483-3p, and hsa-miR-4306 were significantly differentially expressed between LAE and control tissues and that they may play a role in the development of LAE.
Table 1.
MiRNAs with significantly different expression (P < .05) in livers between 2 cases of LAE and 3 healthy controls.
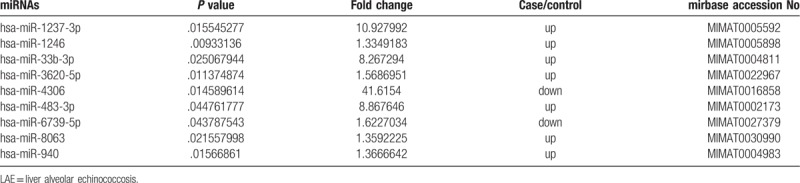
Figure 1.
Differential expression of miRNAs in patients with LAE and healthy controls. Microarray of normal liver tissues from 3 healthy controls or E. multilocularis infected liver tissues from 2 cases of LAE. (A) Volcanic plot of the differentially expressed miRNAs. Gray dots, P ≥ .05; green dots, |log2(FC)|<2 and P < .05; red dots, |log2(FC)| ≥2 and P < .05. (B) Unsupervised clustering of the 4 significantly differenced miRNAs between cases and controls.
3.2. Validation of miRNA expression
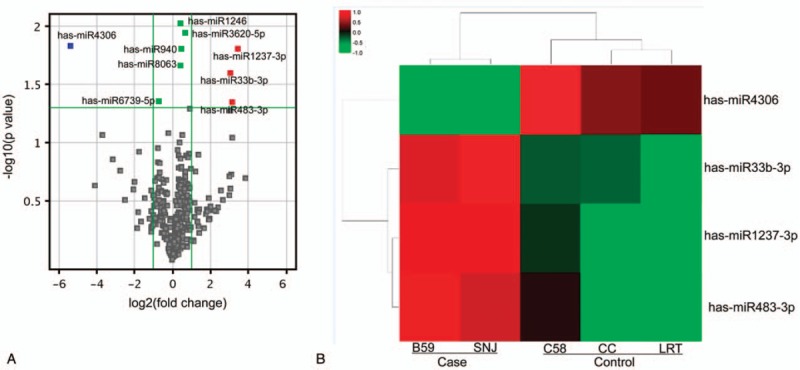
To confirm the expression changes of these miRNAs, 3 miRNAs with higher fold changes were selected for validation by qRT-PCR in a larger set of samples. Compared with normal tissues, only miR-483-3p showed significant upregulation in LAE tissues (P < .01), whereas no significant changes were found for miR-1237-3p and miR-4306 (Fig. 2A). Expression of miR-483-3p was further investigated in plasma, in which similar results were obtained (P < .01; Fig. 2B). These data indicate that miR-483-3p may be a potential diagnostic marker for the early detection of LAE.
Figure 2.
Real time PCR analysis of miRNAs. Real time PCR was used to validate miRNA expression in liver tissues and plasma. (A) Expression of miR-1237-3p, miR-483-3p, and miR-4306 in normal liver tissues from 60 healthy controls and from 47 cases of LAE (case). (B) Expression of miR-483-3p in plasma from 60 healthy controls (control) and 47 cases of LAE (case). ∗∗P < .01, independent sample t test.
3.3. Target gene prediction for miRNAs
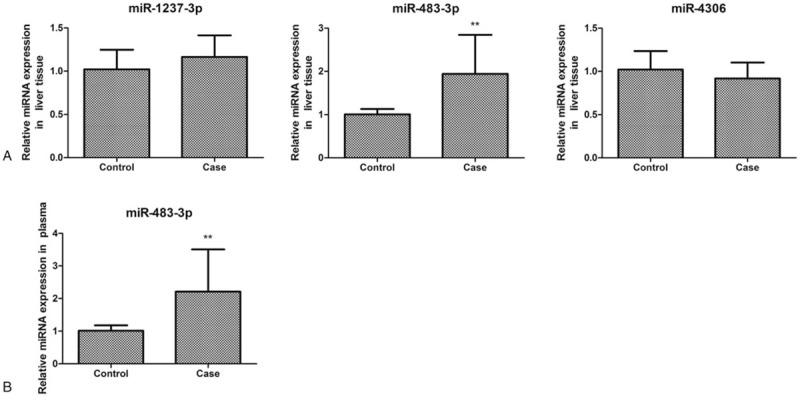
We first predicted the target genes of the differentially expressed miRNAs using 3 database of Targetscan (http://www.targetscan.org), miroRNAorg (http://www.microrna.org/), and PITA (https://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html). Different numbers of predicted target genes were obtained (Table 2). A total of 137 common target genes were obtained by overlapping the results from the 3 databases (Fig. 3). The target genes of miR-1237-3p, miR-4306, and miR-483-3p included genes related to immune responses and liver lesion.[28–30]
Table 2.
Target gene prediction for the 4 differentially expressed miRNAs.
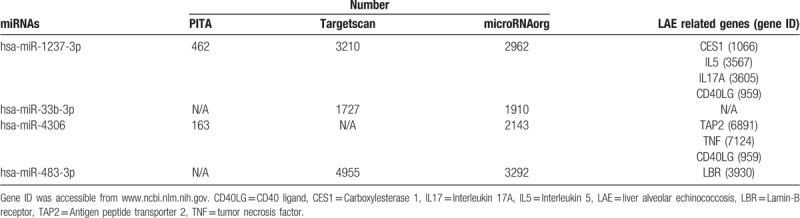
Figure 3.
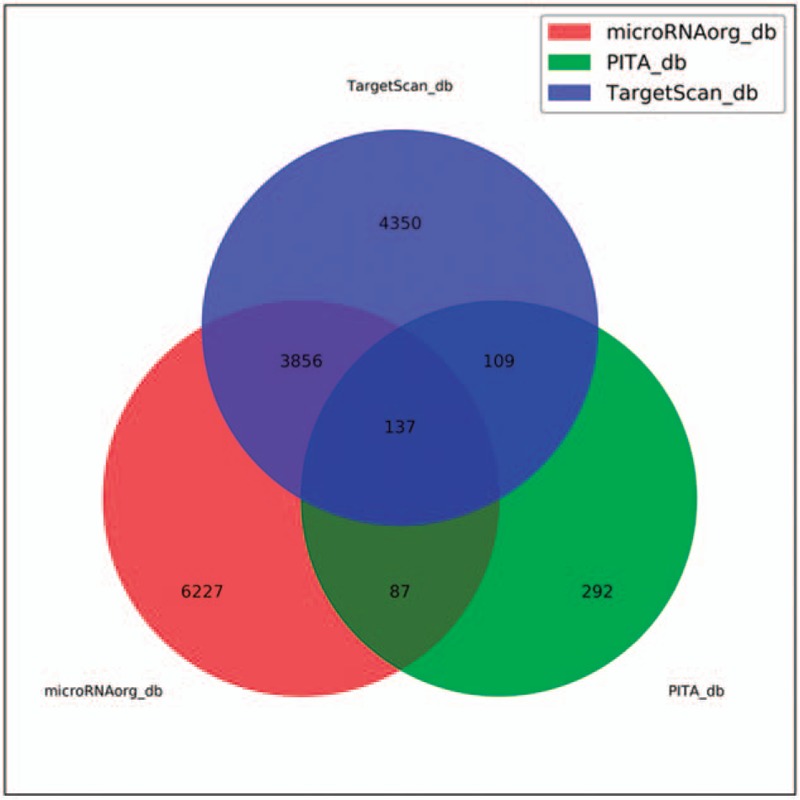
Venn graph showing the prediction results of miR-1237-3p, miR-483-3p, miR-4306, and miR-33b-3p targets in PITA, Targetscan, and microRNAorg softwares.
3.4. GO analysis of predicted target genes
We next performed GO analysis to analyze the functions of predicted target genes. As shown in Figure 4, the target genes of differentially expressed miRNAs were mainly involved in biological processes such as DNA-dependent transcriptional regulation, developmental regulation of multicellular organisms, and cellular immune responses (T cell proliferation). Their molecular function was enriched in proteins such as sequence -specific DNA binding protein, ubiquitin-binding enzyme, and reverse transporters. They also enriched in cell components related to mRNA cleavage factor complex and vesicle membrane. The results of GO analysis provided a set of genes for subsequent studies on LAE.
Figure 4.
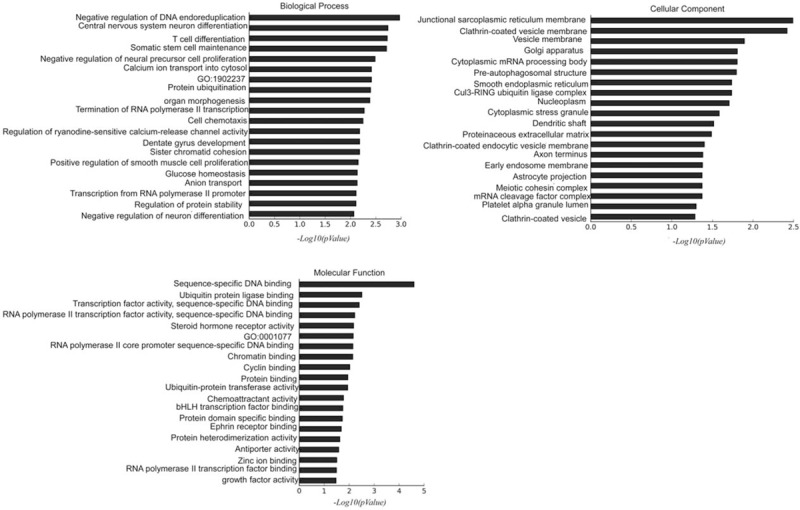
GO analysis of the predicted target genes for miR-1237-3p, miR-483-3p, miR-4306, and miR-33b-3p.
3.5. KEGG analysis of predicted target genes
The pathway analysis of the predicted target gene was performed using the KEGG database. In the KEGG mapping of the differentially expressed miRNA target genes, the top 20 entries were displayed. These target genes were enriched in signaling pathways such as mRNA surveillance pathways, cancer signaling pathways, and intestinal immune networks (Fig. 5).
Figure 5.
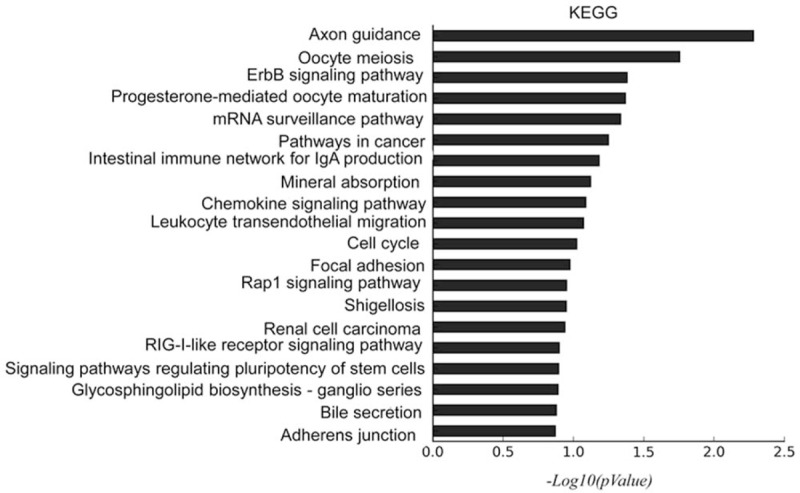
KEGG pathway analysis of the predicted target genes for miR-1237-3p, miR-483-3p, miR-4306, and miR-33b-3p. The top 20 terms were shown.
4. Discussion
In this study, we compared the host miRNA expression in the infected liver tissues and normal tissues from LAE patients using miRNA microarray. Among the 2549 detected miRNAs, a total of 4 miRNAs were differentially expressed, of which 3 (miR-1237-3p, miR-33b-3p, and miR-483-3p) were significantly upregulated and 1 (miR-4306) were significantly downregulated in LAE tissues. Consistent with the microarray data, qRT-PCR results showed that the expression level of miR-483-3p was significantly elevated in LAE tissues.
All of the 4 differentially expressed miRNAs were reported to be related with human diseases. For example, decreased expression of miR-1237-3p is associated with poor survival in patients with chordoma.[31] It is predicted that miR-33b-3p can regulate metabolic processes in arthritic patients and serum miR-33b-3p expression could be used as a potential biomarker for assessing the risk and progression of arthritis.[32] In hepatic carcinoma cells, miR-145-5p, and miR-483-3p have dual effect on cell proliferation dependent on the concentration of glucose.[33] Hsa-miR-4306 is an important marker of disease recurrence and survival in human Papillomavirus (HPV)-negative head and neck squamous cell carcinoma (HNSCC) patients.[34] Here, to the best of our knowledge, this is the first study on host miRNA expression changes upon E multilocularis infection in Qinghai, China. The differential expressions of these 4 miRNAs indicate that these miRNAs may play important roles in the process of E multilocularis infection.
Circulating miRNAs are a class of molecules that are stably present in body fluids such as serum and plasma, which are involved in the transmission of information among cells.[35,36] Cell-free circulating miRNAs are usually present in ribonucleoprotein complexes or high-density lipoproteins, or released from liposomes, exosomes, or apoptotic bodies.[36] Therefore, circulating miRNAs can reflect the body's homeostasis and disease progression. Since circulating miRNAs are stable and resistant to endogenous RNase digestion, they are appropriate biomarkers for the diagnosis and prognosis of diseases and clinical applications of circulating miRNAs are present in areas such as cancer, diabetes, and neurological disorders.[27,37–39] However, studies on circulation miRNAs in LAE progression is just emerging, which prompts us to find a circulating miRNA as possible diagnostic marker for LAE. We found significantly increased expression of miR-483-3p in the plasma of LAE patients, indicating that miR-483-3p may be used as a diagnostic marker for echinococcosis.
Roles of hsa-miR-483-3p have been reported in wound healing and cancer progression.[40–42] It is an important regulator of endothelial integrity in patients with type 2 diabetes, and is a potential treatment target for rescue endothelial regeneration.[40–43] Hsa-miR-483-3p also plays a negative regulatory role in diabetes-related heart disease.[44] In summary, expression of miR-483-3p is dysregulated in several diseases and plays a role in regulating disease progression.
A previous study in E multilocularis-infected mice found that several targets of E multilocularis-responsive miRNAs were associated with the host immune responses.[45] In this study, 137 predicted target genes were found for the 4 differentially expressed miRNAs when predicted by TargetScan, PITA and microRNAorg database. The predicted targets of miR-1237-3p, miR-4306, and miR-483-3p are involved in host immune responses and hepatic echinococcosis. For instance, CD40LG (IgM) and antigen peptide transporter 2 (TAP2), the predicted targets of miR-4306, are correlated with hepatic echinococcosis infection.[46,47] The mean concentrations of IgG, IgA, and IgM in patients with alveolar hydatidosis and the mean levels of IgG and IgM in patients with cystic hydatid disease were significantly higher than those in the normal control group,[47] while the polymorphism of the TAP2 gene is related to cystic hydatidosis.[46] IL17A and IL5 were predicted targets of miR-1237-3p, and these 2 genes were reported to be downregulated after cystic echinococcosis infection, leading to significantly reduced severity of OVA-induced airway inflammation.[48] As the predicted target of miR-483-3p, lamin B receptor is a multi-transmembrane protein of nuclear membrane that is often used as a “reporter” of nuclear membrane dynamics.[49] Lamin B receptor is associated with primary biliary cirrhosis.[50]
GO analysis of the overlapped target genes showed that these genes are related to biological processes such as cellular immune response, which in line with the finding that infection of E multilocularis can elicit immune responses in patients.[47] KEGG analysis indicated that these genes were enriched in signaling pathways such as mRNA surveillance pathways, cancer signaling pathways, and intestinal immune networks. These signaling pathways may have implications for the prevention and treatment of LAE in the future.
This study has some limitations. First, the sample size was relatively small. Second, only 1 miRNA was identified as the potential marker for the diagnosis of LAE and thus the number was limited. Further studies are warranted.
To sum up, based on the differentially expressed miRNAs identified by liver tissue miRNA microarray, we found that plasma miR-483-3p is significantly differentially expressed between normal and LAE liver tissues, which indicates that miR-483-3p is a potential circulating marker for LAE. Our study thus provides new approach for the clinical diagnosis of LAE.
Author contributions
Data curation: Haining Fan.
Formal analysis: Bin Ren, Li Ren, Cairang Yangdan, Haining Fan.
Funding acquisition: Yi Lv.
Methodology: Ying Zhou, Haining Fan.
Project administration: Yi Lv.
Resources: Haijiu Wang, Cairang Yangdan.
Software: Ying Zhou.
Writing – original draft: Bin Ren, Haijiu Wang, Li Ren, Cairang Yangdan.
Writing – review & editing: Yi Lv.
Footnotes
How to cite this article: Ren B, Wang H, Ren L, Yangdan C, Zhou Y, Fan H, Lv Y. miRNAs in liver alveolar echinococcosis. Medicine. 2019;98:37(e17156).
Abbreviations: HNSCC = HPV-negative head and neck squamous cell carcinoma, HPV = human Papillomavirus, LAE = liver alveolar echinococcosis.
This work was supported by the Science and Technology Project of Qinghai (2015-ZJ-753).
The authors report no conflicts of interest.
References
- [1].Bauder B, Auer H, Schilcher F, et al. Experimental investigations on the B and T cell immune response in primary alveolar echinococcosis. Parasite Immunol 1999;21:409–21. [DOI] [PubMed] [Google Scholar]
- [2].Wang TP, Cao ZG. Progress in prevention and control of Chinese hydatid disease and its existing problems. Chin J Parasitol Parasit Dis 2018;36:291–6. [Google Scholar]
- [3].Moss JE, Chen X, Li T, et al. Reinfection studies of canine echinococcosis and role of dogs in transmission of Echinococcus multilocularis in Tibetan communities, Sichuan, China. Parasitology 2013;140:1685–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yan CB, Mulati H. Study on the feature retrieval method based on gray histogram of CT images of endemic high-incidence liver echinococcosis in Xinjiang province. J Biomed Eng 2013;29:76–9. [Google Scholar]
- [5].Huang SB, Mi Yy, Liu AQ. Diagnosis and progress of liver hydatid disease. Modern BiomedProgr 2016;16:797–9. [Google Scholar]
- [6].Kumar MJ, Toe K, Banerjee RD. Hydatid cyst of liver. Postgrad Med J 2003;79:113–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ouyang XY, Jiang X, Gu D, et al. Dysregulated serum MiRNA profile and promising biomarkers in dengue-infected patients. Int J Med Sci 2016;13:195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sakthivel S, Subramanian Tamil S, Govindaraju A, et al. MicroRNAs -the next generation therapeutic targets in human diseases. Theranostics 2013;3:930–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zheng X, Dai J, Zhang H, et al. MicroRNA-221 promotes cell proliferation, migration, and differentiation by regulation of ZFPM2 in osteoblasts. Braz J Med Biol Res 2018;51:e7574. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [10].Cheng AM, Byrom MW, Shelton J, et al. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 2005;33:1290–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jin X, Guo X, Zhu D, et al. miRNA profiling in the mice in response to Echinococcus multilocularis infection. Acta Trop 2017;166:39–44. [DOI] [PubMed] [Google Scholar]
- [12].Zheng YD, Cai XP, Bradley JE. microRNAs in parasites and parasite infection. RNA Biol 2013;10:371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Judice CC, Bourgard C, Kayano AC, et al. MicroRNAs in the host-apicomplexan parasites interactions: a review of immunopathological aspects. Front Cell Infect Microbiol 2016;6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Manzano-Romã nR, Siles-Lucas M. MicroRNAs in parasitic diseases: potential for diagnosis and targeting. Mol Biochem Parasitol 2012;186:81–6. [DOI] [PubMed] [Google Scholar]
- [15].Cheng G, Luo R, Hu C, et al. Deep sequencing-based identification of pathogen-specific microRNAs in the plasma of rabbits infected with Schistosoma japonicum. Parasitology 2013;140:1751–61. [DOI] [PubMed] [Google Scholar]
- [16].Hoy AM, Lundie RJ, Ivens A, et al. Parasite-derived microRNAs in host serum as novel biomarkers of helminth infection. PLoS Negl Trop Dis 2014;8:e2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xu MJ, Zhou DH, Nisbet AJ, et al. Characterization of mouse brain microRNAs after infection with cyst-forming Toxoplasma gondii. Parasit Vectors 2013;6:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Brase JC, Wuttig D, Kuner R, et al. Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer 2010;9:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Grasedieck S, Sorrentino A, Langer C, et al. Circulating microRNAs in hematological diseases: principles, challenges, and perspectives. Blood 2013;121:4977–84. [DOI] [PubMed] [Google Scholar]
- [20].Kinet V, Halkein J, Dirkx E, et al. Cardiovascular extracellular microRNAs: emerging diagnostic markers and mechanisms of cell-to-cell RNA communication. Front Genet 2013;4:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Trebicka J, Anadol E, Elfimova N, et al. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J Hepatol 2013;58:234–9. [DOI] [PubMed] [Google Scholar]
- [22].Zheng H, Liu JY, Song FJ, et al. Advances in circulating microRNAs as diagnostic and prognostic markers for ovarian cancer. Cancer Biol Med 2013;10:123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Benz F, Roy S, Trautwein C, et al. Circulating MicroRNAs as biomarkers for sepsis. Int J Mol Sci 2016;17(1.): [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chamnanchanunt S, Fucharoen S, Umemura T. Circulating microRNAs in malaria infection: bench to bedside. Malar J 2017;16:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Moran J, Ramirez-Martinez G, Jimenez-Alvarez L, et al. Circulating levels of miR-150 are associated with poorer outcomes of A/H1N1 infection. Exp Mol Pathol 2015;99:253–61. [DOI] [PubMed] [Google Scholar]
- [26].He X, Sai X, Chen C, et al. Host serum miR-223 is a potential new biomarker for Schistosoma japonicum infection and the response to chemotherapy. Parasit Vectors 2013;6:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Khoo SK, Petillo D, Kang UJ, et al. Plasma-based circulating MicroRNA biomarkers for Parkinson's disease. J Parkinsons Dis 2012;2:321–31. [DOI] [PubMed] [Google Scholar]
- [28].Cicalese MP, Gerosa J, Baronio M, et al. Circulating follicular helper and follicular regulatory T cells are severely compromised in human CD40 deficiency: a case report. Front Immunol 2018;9:1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lin F, Noyer CM, Ye Q, et al. Autoantibodies from patients with primary biliary cirrhosis recognize a region within the nucleoplasmic domain of inner nuclear membrane protein LBR. Hepatology 1996;23:57–61. [DOI] [PubMed] [Google Scholar]
- [30].Meng J, Li W, Zhang M, et al. An update meta-analysis and systematic review of TAP polymorphisms as potential biomarkers for judging cancer risk. Pathol Res Pract 2018;214:1556–63. [DOI] [PubMed] [Google Scholar]
- [31].Zou MX, Huang W, Wang XB, et al. Reduced expression of miRNA-1237-3p associated with poor survival of spinal chordoma patients. Eur Spine J 2015;24:1738–46. [DOI] [PubMed] [Google Scholar]
- [32].Ntoumou E, Tzetis M, Braoudaki M, et al. Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes. Clin Epigenetics 2017;9:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lupini L, Pepe F, Ferracin M, et al. Over-expression of the miR-483-3p overcomes the miR-145/TP53 pro-apoptotic loop in hepatocellular carcinoma. Oncotarget 2016;7:31361–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hess J, Unger K, Maihoefer C, et al. A five-microrna signature predicts survival and disease control of patients with head and neck cancer negative for HPV infection. Infection Clin Cancer Res 2019;25:1505–16. [DOI] [PubMed] [Google Scholar]
- [35].Giannopoulou L, Zavridou M, Kasimir-Bauer S, et al. Liquid biopsy in ovarian cancer: the potential of circulating miRNAs and exosomes. Transl Res 2019;205:77–91. [DOI] [PubMed] [Google Scholar]
- [36].Hamam R, Hamam D, Alsaleh KA, et al. Circulating microRNAs in breast cancer: novel diagnostic and prognostic biomarkers. Cell Death Dis 2017;8:e3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chin LJ, Slack FJ. A truth serum for cancer--microRNAs have major potential as cancer biomarkers. Cell Res 2008;18:983–4. [DOI] [PubMed] [Google Scholar]
- [38].Gandhi R, Healy B, Gholipour T, et al. Circulating microRNAs as biomarkers for disease staging in multiple sclerosis. Ann Neurol 2013;73:729–40. [DOI] [PubMed] [Google Scholar]
- [39].Guay C, Regazzi R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat Rev Endocrinol 2013;9:513–21. [DOI] [PubMed] [Google Scholar]
- [40].Abue M, Yokoyama M, Shibuya R, et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int J Oncol 2015;46:539–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bertero T, Gastaldi C, Bourget-Ponzio I, et al. miR-483-3p controls proliferation in wounded epithelial cells. FASEB J 2011;25:3092–105. [DOI] [PubMed] [Google Scholar]
- [42].Pepe F, Visone R, Veronese A. The glucose-regulated miR-483-3p influences key signaling pathways in cancer. Cancers (Basel) 2018;10:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kuschnerus K, Straessler ET, Muller MF, et al. Increased expression of miR-483-3p impairs the vascular response to injury in type 2 diabetes. Diabetes 2019;68:349–60. [DOI] [PubMed] [Google Scholar]
- [44].Qiao Y, Zhao Y, Liu Y, et al. miR-483-3p regulates hyperglycaemia-induced cardiomyocyte apoptosis in transgenic mice. Biochem Biophys Res Commun 2016;477:541–7. [DOI] [PubMed] [Google Scholar]
- [45].Guo X, Zheng Y. Expression profiling of circulating miRNAs in mouse serum in response to Echinococcus multilocularis infection. Parasitology 2017;144:1079–87. [DOI] [PubMed] [Google Scholar]
- [46].Kiper N, Gerceker F, Utine E, et al. TAP1 and TAP2 gene polymorphisms in childhood cystic echinococcosis. Parasitol Int 2010;59:283–5. [DOI] [PubMed] [Google Scholar]
- [47].Zhang J, Liu B, Li Y, et al. Comparison of serum immunoglobulin levels in patients with alveolar and cystic echinococcosis. Chin J Parasitol Parasit Dis 1990;8:38–40. [PubMed] [Google Scholar]
- [48].Wang H, Li J, Pu H, et al. Echinococcus granulosus infection reduces airway inflammation of mice likely through enhancing IL-10 and down-regulation of IL-5 and IL-17A. Parasit Vectors 2014;7:522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Giannios I, Chatzantonaki E, Georgatos S. Dynamics and structure-function relationships of the Lamin B Receptor (LBR). PLoS One 2017;12:e0169626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Invernizzi P, Crosignani A, Battezzati PM, et al. Comparison of the clinical features and clinical course of antimitochondrial antibody-positive and -negative primary biliary cirrhosis. Hepatology 1997;25:1090–5. [DOI] [PubMed] [Google Scholar]


