Abstract
Background
Inflammatory breast cancer (IBC) is an aggressive form of breast cancer. The triple-negative subtype of IBC (TN-IBC) is particularly aggressive. Identification of molecular differences between TN-IBC and TN-non-IBC may help clarify the unique clinical behaviors of TN-IBC. However, our previous study comparing gene expression between TN-IBC and TN-non-IBC did not identify any TN-IBC-specific molecular signature. Lehmann et al recently reported that the mesenchymal stem-like (MSL) TNBC subtype consisted of infiltrating tumor-associated stromal cells but not cancer cells. Therefore, we compared the gene expression profiles between TN-IBC and TN-non-IBC patient samples not of the MSL subtype.
Methods
We classified 88 TNBC samples from the World IBC Consortium into subtypes according to the Vanderbilt classification and Insight TNBCtype, removed samples of MSL and unstable subtype, and compared gene expression profiles between the remaining TN-IBC and TN-non-IBC samples.
Results
In the Vanderbilt analysis, we identified 75 genes significantly differentially expressed between TN-IBC and TN-non-IBC at an FDR of 0.2. In the Insight TNBCtype analysis, we identified 81 genes significantly differentially expressed between TN-IBC and TN-non-IBC at an FDR of 0.4. In both analyses, the top canonical pathway was “Fc Receptor-mediated Phagocytosis in Macrophages and Monocytes”, and the top 10 differentially regulated genes included PADI3 and MCTP1, which were up-regulated, and CDC42EP3, SSR1, RSBN1, and ZC3H13, which were downregulated.
Conclusions
Our data suggest that the activity of macrophages might be enhanced in TN-IBC compared with TN-non-IBC. Further clinical and preclinical studies are needed to determine the cross-talk between macrophages and IBC cells.
Introduction
Inflammatory breast cancer (IBC) is a highly aggressive form of breast cancer and is associated with higher rates of recurrence and metastasis and a lower survival rate than non-IBC [1]. Several molecular changes have been found to contribute to the aggressiveness of IBC, including loss of WISP3 and overexpression of RhoC GTPase [2], E-cadherin [3], translation initiation factor eIF4GI [4], and tazarotene-induced gene 1 [5]. Our research group also reported that EGFR signaling promoted inflammation and cancer stem-like cell activity in IBC [6], and the EGFR pathway is a promising therapeutic target for patients with triple-negative IBC (TN-IBC) [7, 8]. Recent studies suggested that immune cells in the tumor microenvironment, especially macrophages, play a major role in regulating the malignant phenotype of IBC [9, 10]. Although these findings have improved our understanding of the molecular mechanisms underlying the aggressive behavior of IBC, unique genes that contribute to the aggressiveness of IBC have not yet been identified. Identification of such genes is critical to facilitate development of US Food and Drug Administration-approved targeted therapies for this disease.
Triple-negative breast cancer (TNBC) is characterized by lack of expression of estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2. Patients with TNBC have a worse prognosis than other breast cancer patients. Generally, 10% to 20% of patients with non-IBC have TNBC (TN-non-IBC), whereas 20% to 40% of patients with IBC have TN-IBC [11–13]. It has been speculated that the high percentage of TNBC among patients with IBC may be associated with the more aggressive clinical course and decreased overall and breast cancer-specific survival of patients with IBC [14].
A research group at Vanderbilt University reported that TNBC can be classified into 7 molecular subtypes on the basis of differential gene expression and gene ontologies, including basal-like 1 (BL1), basal-like 2 (BL2), immunomodulatory (IM), mesenchymal (M), mesenchymal stem-like (MSL), luminal androgen receptor (LAR), and unstable (UNS) [15]. In a previous study, our research group identified these 7 TNBC subtypes in patients with TN-IBC [16]. Because IBC is more aggressive than non-IBC, we hypothesized that the distribution of the 7 TNBC subtypes differs between TN-IBC and TN-non-IBC. However, our findings did not support this hypothesis: we found no significant difference in the distribution between TN-IBC and TN-non-IBC. Furthermore, comparison of gene expression profiles between patients with TN-IBC and TN-non-IBC did not identify any promising molecular signatures specific to the TN-IBC group. This study led us to conclude that not only tumor cells but also their microenvironment and other factors, such as inflammation, immune pathways, and mutations, may contribute to the specific biology of TN-IBC.
Recently, the Vanderbilt research group reported that the MSL gene expression signature was contributed from infiltrating tumor-associated stromal cells but not breast cancer cells [17]. Because surgical specimens may contain large amounts of stromal cells and normal cells, TNBC specimens of MSL subtype would provide inappropriate information for analysis. Therefore, in a new attempt to identify cancer-specific genes that contribute to the aggressiveness of TN-IBC, we decided to compare the gene expression profiles between TN-IBC and TN-non-IBC patient samples not of the MSL subtype.
Material and methods
Patients
This study was approved by the Institutional Review Board of The University of Texas MD Anderson Cancer Center (protocol number PA15-0954). We retrospectively analyzed gene expression profiles and clinical data of all 88 patients with TNBC with known IBC status (39 patients with IBC and 49 with non-IBC) from the World IBC Consortium dataset, contributed by MD Anderson Cancer Center, Houston, TX; General Hospital Sint-Augustinus, Antwerp, Wilrijk, Belgium; and Institut Paoli-Calmettes, Marseille, France [18]. Patients at each site gave informed consent for voluntary participation, and the study was approved by the institutional review boards of the 3 participating centers. IBC was identified according to the consensus diagnostic criteria [12]. TNBC was diagnosed according to gene expression profiling as reported in our previous paper [16].
TNBC was divided into 7 molecular subtypes (BL1, BL2, M, MSL, IM, LAR, and UNS) according to the Vanderbilt classification [15]. TNBC was also divided into 6 molecular subtypes (BL1, BL2, M, MSL, LAR, and UNS) according to Insight TNBCtype (Insight Genetics, Inc., Nashville, TN, USA), a new assay for TNBC subtyping that reduces the number of genes from the original 2188 genes described by Lehmann et al to 101 genes, including control housekeeping genes [19, 20]. IM subtype was removed in Insight TNBCtype because IM subtype likely reflects infiltrating lymphocytes within tumor. After subtyping was complete, the samples classified as MSL and UNS were excluded. Samples of MSL and UNS subtype were not included in the statistical analyses.
Statistical analysis
The data processing and statistical analyses were performed in R version 3.3.0 [21]. Differences between IBC and non-IBC at the gene expression level were examined using feature-by-feature 2-sample t tests followed by a beta-uniform mixture model to adjust for multiple comparisons [22]. Genes with significantly different expression between TN-IBC and TN-non-IBC were counted at different false discovery rates (FDRs).
To identify pathways that may contribute to the aggressiveness of TN-IBC, we performed Ingenuity Pathway Analysis of genes differentially expressed between non-MSL TN-IBC and non-MSL TN-non-IBC in both the Vanderbilt and Insight TNBCtype analyses.
To study the association of genes of interest with overall survival, Kaplan-Meier plots were generated for patients with TNBC in The Cancer Genome Atlas (TCGA) (N = 115) with high and low gene expression (http://cancergenome.nih.gov/). For each specific gene, a log-rank test was performed to compare the survival functions between patients with gene expression above and below the median level (“high” and “low” gene expression groups, respectively).
Western blotting
For Western blotting, cells were lysed in lysis buffer [50 mM Tris (pH 7.5), 150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate acid, 0.1% sodium dodecyl sulfate, protease inhibitor cocktail (Bimake.com, Houston, TX), and phosphatase inhibitor cocktail (Bimake.com)]. Proteins were fractioned by SDS-PAGE on 4% to 12% NuPAGE Bis-Tris gels (Thermo Fisher Scientific, Waltham, MA) and transferred to Immun-Blot PVDF membrane (Bio-Rad, Hercules, CA). Proteins of interest were probed using anti-PADI3 antibody (rabbit polyclonal, Sigma-Aldrich, St. Louis, MO) and anti-β-actin antibody (Sigma-Aldrich).
siRNA transfection
Using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s instructions, 4 × 105 SUM149 cells were transfected with ON-TARGETplus non-targeting siRNA (Dharmacon, Lafayette, CO) or SMARTpool: ON-TARGETplus PADI3 siRNA (a mixture of 4 designed siRNAs targeting PADI3; Dharmacon) at a final siRNA concentration of 16.7 μM.
Cell proliferation assay
Seventy-two hours after siRNA transfection, 1 × 105 cells were seeded in complete medium in a 12-well plate and incubated for 72 h. Viable cells were counted using the trypan blue dye exclusion method by Vi-CELL XR (Beckman Coulter, Brea, CA).
Anchorage-independent growth
Seventy-two hours after siRNA transfection, 8 × 103 cells were resuspended in 0.38% agarose medium and then plated in 12-well plates coated with solidified 0.75% agarose medium. Three weeks later, colonies greater than 50 μm in diameter were counted using the Gel Count system (Oxford Optronix Ltd., Milton Park, Abingdon, UK).
Results
Genes differentially expressed between non-MSL TN-IBC and non-MSL TN-non-IBC
The characteristics of the 88 patients with TNBC were described in our previous publication [23].
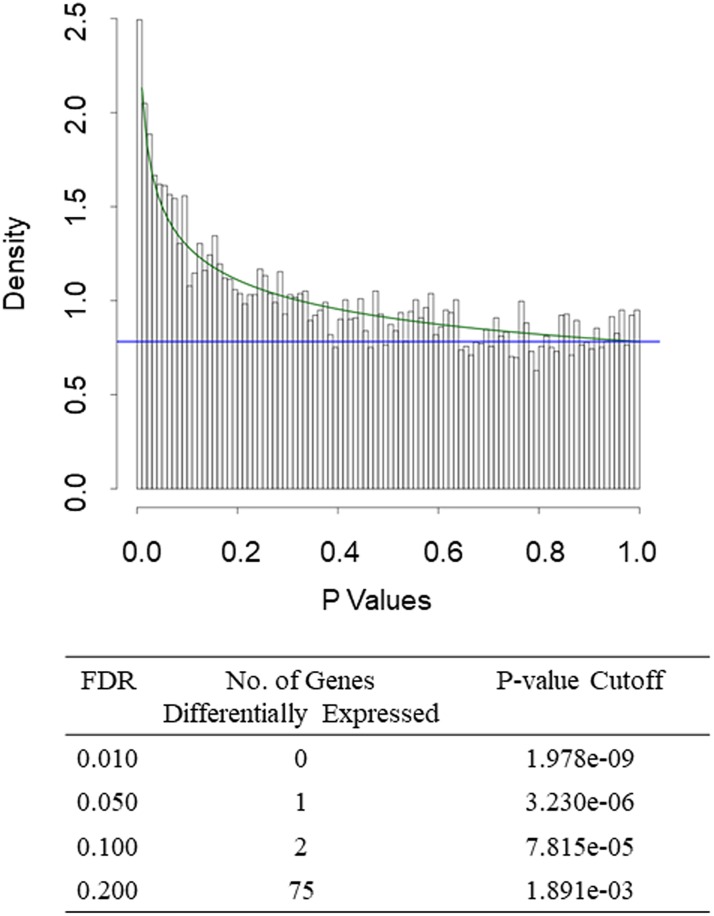
Classification of samples from the 88 patients with TNBC (39 with IBC and 49 with non-IBC) according to the Vanderbilt classification [15] revealed 11 samples (8 TN-IBC and 3 TN-non-IBC) with MSL subtype and 7 samples (2 TN-IBC and 5 TN-non-IBC) with UNS subtype. After excluding these samples, we had 29 TN-IBC samples and 41 TN-non-IBC samples. Comparison of the gene expression profiles between these non-MSL TN-IBC and TN-non-IBC samples revealed 75 genes differentially expressed at an FDR of 0.2 (Fig 1 and S1 Table).
Fig 1. Gene expression in IBC versus non-IBC in 70 patients with non-MSL TNBC according to the Vanderbilt classification.
Top, Histogram of P values from 2-sample t tests for gene expression in TN-IBC vs. TN-non-IBC. The overlaid curve is the fitted BUM model. Bottom, Counts of differentially expressed genes with various FDR cutoffs.
Classification of samples from the 88 patients with TNBC according to Insight TNBCtype revealed 31 samples (16 TN-IBC and 15 TN-non-IBC) with MSL subtype and 5 samples (1 TN-IBC and 4 TN-non-IBC) with UNS subtype. After excluding these samples, we had 22 TN-IBC samples and 30 TN-non-IBC samples. Comparison of the gene expression profiles between these non-MSL TN-IBC and TN-non-IBC samples revealed 81 genes differentially expressed at an FDR of 0.4 (Fig 2A and S2 Table). We also compared the gene expression profiles between the MSL TN-IBC and MSL TN-non-IBC samples and did not identify any significantly differentially expressed genes (Fig 2B).
Fig 2. Gene expression in IBC versus non-IBC in (A) 52 patients with non-MSL TNBC and (B) 31 patients with MSL TNBC according to Insight TNBCtype.
Top of each panel, Histogram of P values from 2-sample t test for gene expression in TN-IBC versus TN-non-IBC. The overlaid curves are the fitted BUM models. Bottom of each panel, Counts of differentially expressed genes with various FDR cutoffs.
Genes and signaling pathways that may contribute to the aggressiveness of TN-IBC
Ingenuity Pathway Analysis showed that the top canonical pathway in both analyses was “Fc Receptor-mediated Phagocytosis in Macrophages and Monocytes” (Table 1).
Table 1. Top 5 canonical pathways derived from Ingenuity Pathway Analysis gene ontology algorithmsa.
| Canonical pathway | P value | Overlap%b |
|---|---|---|
| Vanderbilt | ||
| Fc Receptor-mediated Phagocytosis in Macrophages and Monocytes | 1.72E-04 | 4.3 (4/93) |
| Systemic Lupus Erythematosus Signaling | 4.58E-03 | 1.8 (4/225) |
| Paxillin Signaling | 4.69E-03 | 2.7 (3/113) |
| Molecular Mechanisms of Cancer | 5.15E-03 | 1.3 (5/374) |
| Ethanol Degradation II | 5.46E-03 | 5.4 (2/37) |
| TNBCtype | ||
| Fc Receptor-mediated Phagocytosis in Macrophages and Monocytes | 3.81E-03 | 3.2 (3/93) |
| Heme Biosynthesis from Uroporphyrinogen-III I | 1.34E-02 | 25.0 (1/4) |
| Protein Citrullination | 1.67E-02 | 20.0 (1/5) |
| RAR Activation | 2.62E-02 | 1.6 (3/190) |
| VDR/RXR Activation | 2.84E-02 | 2.6 (2/78) |
a Results are based on 75 differentially expressed genes identified in the analysis based on the Vanderbilt classification and 81 differentially expressed genes identified in the analysis based on Insight TNBCtype.
b The overlap in Canonical pathways represents the ratio of analysis ready dataset molecules over the total molecules present in the particular Canonical Pathway.
We further identified the top 10 genes differentially expressed between non-MSL TN-IBC and non-MSL TN-non-IBC (Table 2). In both the Vanderbilt and Insight TNBCtype analyses, the top 10 differentially regulated genes included PADI3 and MCTP1, which were up-regulated, and CDC42EP3, SSR1, RSBN1, and ZC3H13, which were down-regulated. The functions of these 6 genes are summarized in Table 3. Analysis of the association between the expression of these genes and overall survival in 115 patients with TNBC from the TCGA database did not identify any significant associations (S1 Fig).
Table 2. Top 10 up-regulated and down-regulated genes in non-MSL TN-IBC versus non-MSL TN-non-IBCa.
| Up-regulated in TN-IBC | Down-regulated in TN-IBC | ||
|---|---|---|---|
| Molecule | Log ratio | Molecule | Log ratio |
| Vanderbilt | |||
| *PADI3 | 0.604 | SERPINE2 | -0.941 |
| ARF6 | 0.574 | CNN3 | -0.836 |
| *MCTP1 | 0.551 | *CDC42EP3 | -0.622 |
| TFPI | 0.471 | NME7 | -0.535 |
| ZFP36 | 0.399 | *SSR1 | -0.528 |
| CLEC1A | 0.351 | SSR3 | -0.527 |
| PAK4 | 0.344 | *RSBN1 | -0.466 |
| NOTCH4 | 0.309 | SNRNP40 | -0.461 |
| EFCC1 | 0.298 | CRK | -0.443 |
| PLXND1 | 0.282 | *ZC3H13 | -0.442 |
| TNBCtype | |||
| *PADI3 | 0.750 | *CDC42EP3 | -0.752 |
| *MCTP1 | 0.675 | LOC730101 | -0.682 |
| TCF4 | 0.671 | *SSR1 | -0.652 |
| RPL37A | 0.670 | TARDBP | -0.609 |
| COA1 | 0.647 | *ZC3H13 | -0.609 |
| MICAL2 | 0.558 | PRKD3 | -0.584 |
| FPR1 | 0.538 | KDM3B | -0.559 |
| LPIN2 | 0.496 | COPG1 | -0.551 |
| MERTK | 0.439 | *RSBN1 | -0.548 |
| MAPKAPK3 | 0.364 | DOPEY1 | -0.510 |
a Results are based on 75 differentially expressed genes identified in the analysis based on the Vanderbilt classification and 81 differentially expressed genes identified in the analysis based on Insight TNBCtype.
* Genes that were differentially regulated in both the Vanderbilt and Insight TNBCtype analyses.
Table 3. Main functions of genes differentially expressed between non-MSL TN-IBC and non-MSL TN-non-IBC in both the Vanderbilt and Insight TNBCtype analyses.
| Gene symbol | Gene name | Main functions |
|---|---|---|
| PADI3 | Peptidyl arginine deiminase 3 | Protein citrullination in hair follicles, keratinocytes, and macrophages |
| MCTP1 | Multiple C2 and transmembrane domain containing 1 | Unknown |
| CDC42EP3 | CDC42 effector protein 3 | Organization of actin cytoskeleton; regulation of tumor-associated fibroblasts |
| SSR1 | Signal sequence receptor subunit 1 | Part of a glycosylated endoplasmic reticulum membrane receptor |
| RSBN1 | Round spermatid basic protein 1 | Unknown |
| ZC3H13 | Zinc finger CCCH-type containing 13 | Regulation of mRNA degradation |
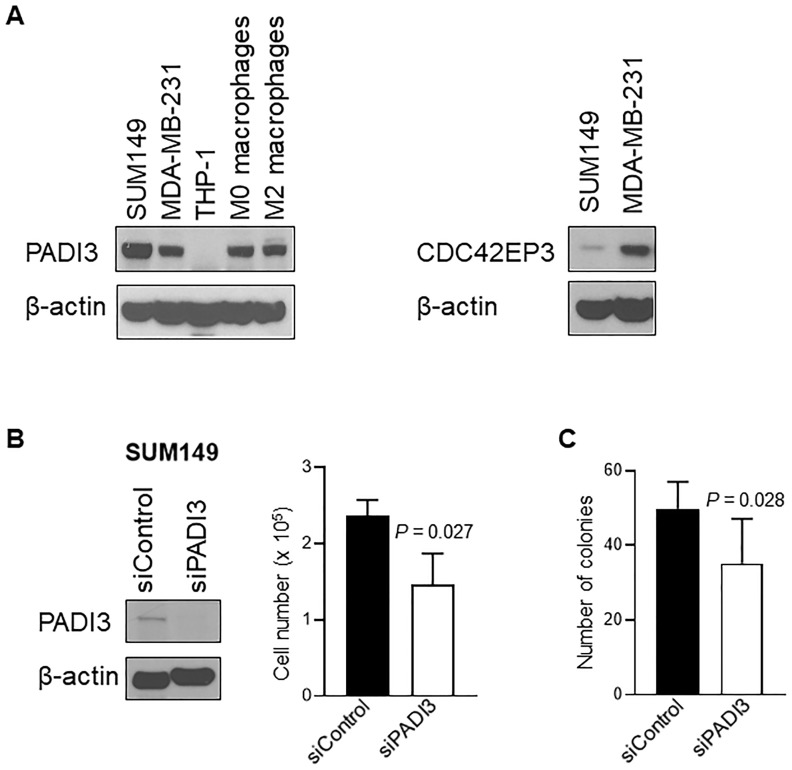
Next we examined the expression of PADI3 and CDC42EP3 in TN-IBC SUM149 and TN-non-IBC MDA-MB-231 cells using Western blotting. As shown in Fig 3A, PADI3 protein expression was higher in SUM149 cells than in MDA-MB-231 cells. In contrast, CDC42EP3 expression was lower in SUM149 cells than in MDA-MB-231 cells. These results validated the findings regarding differential gene expression of PADI3 and CDC42EP3 in TN-IBC and TN-non-IBC (Table 2). It has been reported that PADI3 is related to macrophages [24], so we examined the expression level of PADI3 in THP-1-derived M0 and M2 macrophages. Our data showed that both M0 and M2 macrophages expressed PADI3 protein (Fig 3A).
Fig 3. Analysis of PADI3 and CDC42EP3.
(A) The expression of PADI3 and CDC42EP3 in breast cancer cell lines and macrophages. Expressions of PADI3 in SUM149 (TN-IBC), MDA-MB-231 (TN-non-IBC) cells, THP-1 (monocytes), M0 macrophages (THP-1-derived immature macrophages) and M2 macrophages (THP-1–derived M2 macrophages) were analyzed with Western blotting. Expressions of CDC42EP3 in SUM149 and MDA-MB231 were analyzed with Western blotting. (B) PADI3 knockdown suppresses cell growth in SUM149. Left panel: The expression of PADI3 in SUM149 cells was depleted with siRNA and the expression of PADI3 in siControl and siPADI3 was analyzed with western blot. Right panel: Proliferation of SUM149 cells transfected with siControl and siPADI3 was measured by Trypan blue exclusion assay. Bars, SD. (C) PADI3 knockdown suppresses anchorage-independent growth of SUM149 cells. Bars, SD.
To further understand the function of PADI3 in TN-IBC, we depleted the expression of PADI3 in TN-IBC SUM149 cells using siRNA and measured cell proliferation. As shown in Fig 3B, compared to siControl, siPADI3 reduced the proliferation of SUM149 cells by 38% (P = 0.027). We also examined whether depletion of PADI3 affects anchorage-independent growth of SUM149 cells. The number of colonies formed by siPADI3 was less than that of siControl (Fig 3C, P = 0.028). These results suggested that PADI3 may play a role in the growth of TN-IBC cells.
Discussion
Compared with our previous study, our present study identified more genes that were differentially expressed in TN-IBC versus TN-non-IBC: 75 genes in the analysis based on the Vanderbilt classification (vs. 0 in our previous study) at an FDR of 0.2 [16] and 81 genes in the analysis based on Insight TNBCtype (vs. 38 in our previous study) at an FDR of 0.4 [16]. We therefore decided to analyze the overlapping genes between the 2 sets of results. We recognize that these genes were identified at a high FDR, which limited statistical power. However, this alternative approach allowed us to identify molecules and signaling pathways that may contribute to the aggressiveness of IBC. Indeed, we were able to validate the higher expression of PADI3 in TN-IBC SUM149 cells than in TN-non-IBC MDA-MB-231 cells (Fig 3A).
Analysis of the 31 MSL samples (16 TN-IBC and 15 TN-non-IBC) classified according to Insight TNBCtype did not detect any gene differentially expressed between TN-IBC and TN-non-IBC even at an FDR of 0.5 (Fig 2B).
The functions of the 6 genes that were among the top 10 differentially regulated genes in both the Vanderbilt and Insight TNBCtype analyses are summarized in Table 3. Among these genes, PADI3 (peptidyl arginase deiminase 3) and CDC42EP3 (CDC42 effector protein 3) are of particular interest because of their roles in macrophages and tumor biology. PADI3 is responsible for the citrullination of proteins in a calcium-dependent manner [25]. It is known to be distributed in hair follicles and keratinocytes, and a recent report indicated that PADI3 was strongly stained in macrophages (CD68-positive cells) in rheumatoid nodules and synovial tissue [24]. Consistent with that report, we found expression of PADI3 protein in THP-1 polarized M0 and M2 macrophages (Fig 3A). Furthermore, the higher expression of PADI3 in TN-IBC SUM149 cells than in TN-non-IBC MDA-MB-231 cells (Fig 3A) and the finding depletion of PADI3 reduced the proliferation of SUM149 cells (Fig 3B) indicated that PADI3 may play an important role in promoting the aggressiveness of IBC. CDC42EP3 is a downstream molecule of CDC42 and is involved in organization of the actin cytoskeleton [26, 27]. A recent study suggested that CDC42EP3 is a key regulator of cancer-associated fibroblasts. The expression of CDC42EP3 potentiates cellular responses to mechanical stimulation, leading to signaling and transcriptional adaptations required for activation of the cancer-associated fibroblast phenotype [28]. However, in our study, CDC42EP3 gene expression was downregulated in TN-IBC samples, which suggests that CDC42EP3-regulated cancer-associated fibroblasts may not contribute to the aggressiveness of TN-IBC. Consistent with our gene analysis data, we found that protein expression of CDC42EP3 was lower in TN-IBC SUM149 cells than in TN-non-IBC MDA-MB-231 cells (Fig 3A). The functions of the other genes that were differentially expressed between non-MSL TN-IBC and non-MSL TN-non-IBC in both the Vanderbilt and Insight TNBCtype analyses (MCTP1, SSR1, RSBN1, and ZC3H13) are still poorly understood and need to be further studied.
Our findings that Fc Receptor-mediated Phagocytosis in Macrophages and Monocytes was the top canonical pathway in both datasets and PADI3 was the top up-regulated gene suggest that the activity of macrophages may contribute to the specific biology of TN-IBC. Recent studies have indicated that the tumor microenvironment is a critical driver of the IBC clinical phenotype. It has been reported that macrophages enhance the migration of IBC cells through RhoC GTP signaling [9], and macrophages isolated from IBC patient samples secrete chemotactic cytokines that may increase IBC tumor cell dissemination and metastasis [18]. Woodward’s group showed that macrophage-educated mesenchymal stem cells promote the invasion, mammosphere formation, and IL-6 secretion of IBC cells [10]. We also previously found that high expression of CD163, an M2 macrophage marker, correlated with short disease-free survival of patients with IBC (unpublished data). Our current study further supports the critical role of the tumor microenvironment in promoting IBC progression. Further study is needed to determine the role of the differentially expressed genes that we identified in the aggressiveness of TN-IBC.
In conclusion, our study identified molecules and signaling pathways related to the activity of macrophages that may contribute to the unique biology of TN-IBC. These candidates need to be further validated in preclinical and clinical studies.
Supporting information
(TIF)
(DOCX)
(DOCX)
(XLSX)
Acknowledgments
The authors thank Ms. Stephanie P. Deming (Scientific Publications, Research Medical Library, The University of Texas MD Anderson Cancer Center) for her expert editorial assistance.
Data Availability
We have provided an Excel file of the 88 patients’ clinical data as a minimal anonymized data set.
Funding Statement
This work was supported by National Institutes of Health grant 1R01CA205043-01A1 (Dr. Ueno), Breast Cancer Research Foundation grant BCRF-18-164 (Dr. Ueno), the Morgan Welch Inflammatory Breast Cancer Research Program, a State of Texas Rare and Aggressive Breast Cancer Research Program grant (Dr. Ueno), and National Cancer Institute award P30CA016672, which supports the MD Anderson Cancer Center Clinical Trials Support Resource. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hance KW, Anderson WF, Devesa SS, Young HA, Levine PH. Trends in inflammatory breast carcinoma incidence and survival: the surveillance, epidemiology, and end results program at the National Cancer Institute. J Natl Cancer Inst. 2005;97(13):966–75. Epub 2005/07/07. 10.1093/jnci/dji172 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Golen KL, Davies S, Wu ZF, Wang Y, Bucana CD, Root H, et al. A novel putative low-affinity insulin-like growth factor-binding protein, LIBC (lost in inflammatory breast cancer), and RhoC GTPase correlate with the inflammatory breast cancer phenotype. Clin Cancer Res. 1999;5(9):2511–9. Epub 1999/09/28. . [PubMed] [Google Scholar]
- 3.Alpaugh ML, Tomlinson JS, Ye Y, Barsky SH. Relationship of sialyl-Lewis(x/a) underexpression and E-cadherin overexpression in the lymphovascular embolus of inflammatory breast carcinoma. Am J Pathol. 2002;161(2):619–28. Epub 2002/08/07. 10.1016/S0002-9440(10)64217-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silvera D, Arju R, Darvishian F, Levine PH, Zolfaghari L, Goldberg J, et al. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat Cell Biol. 2009;11(7):903–8. Epub 2009/06/16. 10.1038/ncb1900 . [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Saso H, Iwamoto T, Xia W, Gong Y, Pusztai L, et al. TIG1 promotes the development and progression of inflammatory breast cancer through activation of Axl kinase. Cancer Res. 2013;73(21):6516–25. Epub 2013/09/10. 10.1158/0008-5472.CAN-13-0967 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Reyes ME, Zhang D, Funakoshi Y, Trape AP, Gong Y, et al. EGFR signaling promotes inflammation and cancer stem-like activity in inflammatory breast cancer. Oncotarget. 2017;8(40):67904–17. Epub 2017/10/06. 10.18632/oncotarget.18958 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuda N, Lim B, Wang X, Ueno NT. Early clinical development of epidermal growth factor receptor targeted therapy in breast cancer. Expert opinion on investigational drugs. 2017;26(4):463–79. 10.1080/13543784.2017.1299707 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuda N, Wang X, Lim B, Krishnamurthy S, Alvarez RH, Willey JS, et al. Safety and Efficacy of Panitumumab Plus Neoadjuvant Chemotherapy in Patients With Primary HER2-Negative Inflammatory Breast Cancer. JAMA oncology. 2018;4(9):1207–13. 10.1001/jamaoncol.2018.1436 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen SG, Chen YC, Madden JM, Fournier CL, Altemus MA, Hiziroglu AB, et al. Macrophages Enhance Migration in Inflammatory Breast Cancer Cells via RhoC GTPase Signaling. Scientific reports. 2016;6:39190 Epub 2016/12/20. 10.1038/srep39190 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfe AR, Trenton NJ, Debeb BG, Larson R, Ruffell B, Chu K, et al. Mesenchymal stem cells and macrophages interact through IL-6 to promote inflammatory breast cancer in pre-clinical models. Oncotarget. 2016;7(50):82482–92. Epub 2016/10/21. 10.18632/oncotarget.12694 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaher N, Arias-Pulido H, Terki N, Qualls C, Bouzid K, Verschraegen C, et al. Molecular and epidemiological characteristics of inflammatory breast cancer in Algerian patients. Breast Cancer Res Treat. 2012;131(2):437–44. Epub 2011/03/02. 10.1007/s10549-011-1422-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawood S, Ueno NT, Valero V, Woodward WA, Buchholz TA, Hortobagyi GN, et al. Differences in survival among women with stage III inflammatory and noninflammatory locally advanced breast cancer appear early: a large population-based study. Cancer. 2011;117(9):1819–26. Epub 2011/04/22. 10.1002/cncr.25682 . [DOI] [PubMed] [Google Scholar]
- 13.Li J, Gonzalez-Angulo AM, Allen PK, Yu TK, Woodward WA, Ueno NT, et al. Triple-negative subtype predicts poor overall survival and high locoregional relapse in inflammatory breast cancer. Oncologist. 2011;16(12):1675–83. Epub 2011/12/08. 10.1634/theoncologist.2011-0196 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zell JA, Tsang WY, Taylor TH, Mehta RS, Anton-Culver H. Prognostic impact of human epidermal growth factor-like receptor 2 and hormone receptor status in inflammatory breast cancer (IBC): analysis of 2,014 IBC patient cases from the California Cancer Registry. Breast Cancer Res. 2009;11(1):R9 Epub 2009/02/21. 10.1186/bcr2225 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–67. Epub 2011/06/03. 10.1172/JCI45014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masuda H, Baggerly KA, Wang Y, Iwamoto T, Brewer T, Pusztai L, et al. Comparison of molecular subtype distribution in triple-negative inflammatory and non-inflammatory breast cancers. Breast Cancer Res. 2013;15(6):R112 Epub 2013/11/28. 10.1186/bcr3579 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehmann BD, Jovanovic B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS One. 2016;11(6):e0157368 Epub 2016/06/17. 10.1371/journal.pone.0157368 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Laere SJ, Ueno NT, Finetti P, Vermeulen P, Lucci A, Robertson FM, et al. Uncovering the molecular secrets of inflammatory breast cancer biology: an integrated analysis of three distinct affymetrix gene expression datasets. Clin Cancer Res. 2013;19(17):4685–96. Epub 2013/02/12. 10.1158/1078-0432.CCR-12-2549 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Li J, Gray WH, Lehmann BD, Bauer JA, Shyr Y, et al. TNBCtype: A Subtyping Tool for Triple-Negative Breast Cancer. Cancer Inform. 2012;11:147–56. Epub 2012/08/09. 10.4137/CIN.S9983 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ring BZ, Hout DR, Morris SW, Lawrence K, Schweitzer BL, Bailey DB, et al. Generation of an algorithm based on minimal gene sets to clinically subtype triple negative breast cancer patients. BMC Cancer. 2016;16:143 Epub 2016/02/26. 10.1186/s12885-016-2198-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://wwwR-projectorg/. 2016.
- 22.Pounds S, Morris SW. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics. 2003;19(10):1236–42. Epub 2003/07/02. 10.1093/bioinformatics/btg148 . [DOI] [PubMed] [Google Scholar]
- 23.Harano K, Wang Y, Lim B, Seitz RS, Morris SW, Bailey DB, et al. Rates of immune cell infiltration in patients with triple-negative breast cancer by molecular subtype. PLoS One. 2018;13(10):e0204513 Epub 2018/10/13. 10.1371/journal.pone.0204513 authors RSS, SWM, DBB, DRH, RLS, and BZR. This does not alter our adherence to PLOS ONE policies on sharing data and materials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turunen S, Huhtakangas J, Nousiainen T, Valkealahti M, Melkko J, Risteli J, et al. Rheumatoid arthritis antigens homocitrulline and citrulline are generated by local myeloperoxidase and peptidyl arginine deiminases 2, 3 and 4 in rheumatoid nodule and synovial tissue. Arthritis Res Ther. 2016;18(1):239 Epub 2016/10/22. 10.1186/s13075-016-1140-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanno T, Kawada A, Yamanouchi J, Yosida-Noro C, Yoshiki A, Shiraiwa M, et al. Human peptidylarginine deiminase type III: molecular cloning and nucleotide sequence of the cDNA, properties of the recombinant enzyme, and immunohistochemical localization in human skin. J Invest Dermatol. 2000;115(5):813–23. Epub 2000/11/09. 10.1046/j.1523-1747.2000.00131.x . [DOI] [PubMed] [Google Scholar]
- 26.Hirsch DS, Pirone DM, Burbelo PD. A new family of Cdc42 effector proteins, CEPs, function in fibroblast and epithelial cell shape changes. J Biol Chem. 2001;276(2):875–83. Epub 2000/10/18. 10.1074/jbc.M007039200 . [DOI] [PubMed] [Google Scholar]
- 27.Joberty G, Perlungher RR, Macara IG. The Borgs, a new family of Cdc42 and TC10 GTPase-interacting proteins. Mol Cell Biol. 1999;19(10):6585–97. Epub 1999/09/22. 10.1128/mcb.19.10.6585 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farrugia AJ, Calvo F. Cdc42 regulates Cdc42EP3 function in cancer-associated fibroblasts. Small GTPases. 2017;8(1):49–57. Epub 2016/06/02. 10.1080/21541248.2016.1194952 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(DOCX)
(DOCX)
(XLSX)
Data Availability Statement
We have provided an Excel file of the 88 patients’ clinical data as a minimal anonymized data set.