SUMMARY
Cellular heterogeneity is frequently observed in cancer, but the biological significance of heterogeneous tumor clones is not well defined. Using multicolor reporters and CRISPR-Cas9 barcoding, we trace clonal dynamics in a mouse model of sarcoma. We show that primary tumor growth is associated with a reduction in clonal heterogeneity. Local recurrence of tumors following surgery or radiation therapy is driven by multiple clones. In contrast, advanced metastasis to the lungs is driven by clonal selection of a single metastatic clone (MC). Using RNA sequencing (RNA-seq) and in vivo assays, we identify candidate suppressors of metastasis, namely, Rasd1, Reck, and Aldh1a2. These genes are downregulated in MCs of the primary tumors prior to the formation of metastases. Overexpression of these suppressors of metastasis impair the ability of sarcoma cells to colonize the lungs. Overall, this study reveals clonal dynamics during each step of tumor progression, from initiation to growth, recurrence, and distant metastasis.
In Brief
Tang et al. demonstrate that distinct clonal architecture underlies different stages of tumor progression, from tumor initiation, local recurrence after therapy, to advanced metastasis.
Graphical Abstract
INTRODUCTION
Most cancers are composed of heterogeneous populations of cells. During tumor progression, the genomic instability of cancer cells and differences in the tumor microenvironment can confer tumor clones with different genetic and epigenetic alterations (Marusyk et al., 2012; McGranahan and Swanton, 2017; Swanton, 2012). These alterations may endow phenotypic differences in tumor clones, which are subjected to competition and selection. Cancer progression can be viewed as an evolutionary process where tumor clones compete and are selected upon over the course of the disease (Greaves and Maley, 2012). Although the phenomenon of clonal heterogeneity at the morphological, histological, and genetic levels are well characterized, the functional impact of different tumor clones during tumor evolution is not well understood, especially in mesenchymal cancers (de Bruin et al., 2014; Gerlinger et al., 2012; Gulati et al., 2014; Makohon-Moore et al., 2017). In particular, how different tumor clones contribute to primary tumor growth, recurrence, and metastasis is not fully defined. During primary tumor progression, it remains unclear whether growth is driven by the continuous expansion of a dominant clone or the maintenance of the overall clonal architecture. Furthermore, the clonal composition of distant metastasis is poorly understood. Specifically, it is not known whether all tumor clones are equally capable of forming distant metastases and whether spatially distinct metastatic lesions in the receptive tissue are clonally related. Insights into the clonal composition and evolution of tumors throughout the major stages of disease progression are essential to understanding the biological consequences of intratumoral heterogeneity and developing new therapy to impede cancer progression.
Our understanding of tumor clonal dynamics is largely derived from next-generation sequencing of tumor genomes (Gundem et al., 2015; Hao et al., 2016; Wang et al., 2014; Yates et al., 2015). Based on shared somatic mutations, the phylogenetic relationship of tumor clones and their life history can be retrospectively constructed (Schwartz and Schäffer, 2017). These studies captured snapshots of tumor heterogeneity and clearly demonstrated that most tumors are composed of multiple clones. Although genomic analyses of clonal dynamics revealed important insights into tumor evolution, such as the frequency of mutations and the order in which specific somatic mutations may arise, their ability to prospectively track tumor evolution in an unperturbed manner is limited (Landau et al., 2013; Nik-Zainal et al., 2012; Welch, 2014). This is especially the case for many solid cancers, where longitudinal sampling is difficult and patient-matched primary tumors and metastases samples are scarce. Moreover, it is difficult to ascribe the genetic variations used to identify tumor clones to cellular phenotypes. To functionally investigate the impact of various tumor clones during different stages of tumor progression, in vivo models of lineage tracing are required.
In this study, we traced tumor evolution in a mouse model of undifferentiated pleomorphic sarcoma (UPS), one of the most common and lethal soft tissue sarcomas in human adults (Burningham et al., 2012; Kirsch et al., 2007). Using the R26R-Confetti lineage tracing system and endogenous DNA barcoding with CRISPR-Cas9, we investigated clonal dynamics during tumor initiation, local recurrence after therapy, and metastasis.
RESULTS
Modeling Clonal Evolution in Sarcoma
To track tumor evolution in sarcoma, we utilized two complementary lineage tracing mouse models. First, we crossed the homozygous R26R-Confetti system with a spatially and temporally restricted model of UPS, carrying a Cre-inducible oncogenic KrasLSL-G12D allele and homozygous p53f/f alleles (KrasLSL-G12D; p53f/f; Confetti/Confetti; KPCC mice in Figure S1A) (Snippert et al., 2010). Ras/mitogen-activated protein kinase (MAPK) signaling is commonly activated in human soft tissue sarcomas, including UPS, and inactivating p53 mutations or silencing of the p53 pathway is one of the most common mutations in UPS (Cancer Genome Atlas Research Network, 2017; Pérot et al., 2010; Serrano et al., 2016; Yoo et al., 1999). Tumors formed by the activation of oncogenic KrasG12D and deletion of p53 were similar to human UPS in histology and molecular profile (Blum et al., 2013; Kirsch et al., 2007; Mito et al., 2009; Sato et al., 2016). In the KrasLSL-G12D; p53f/f; Confetti/Confetti (KPCC) animals, following injection of Cre-expressing adenovirus into the gastrocnemius muscle, primary sarcomas expressing different fluorescent reporters were formed at the injection site between 9 and 12 weeks, with 100% penetrance, similar to previous reports (Figure S1B) (Kirsch et al., 2007; Mito et al., 2009). We observed up to 8 distinct fluorescent protein combinations, labeling on average ~60% of cells in tumors (Figure S1C). The expression of the R26R-Confetti allele did not affect tumor histology (Figures S1B). Consistent with other reports using the R26R-Confetti allele (Baggiolini et al., 2015), fewer cells recombined to express GFP compared to the other fluorescent reporters (Figures S1C). Long-term culture experiments showed that the expression of fluorescent proteins was permanent and stable (Figure S1D). Distinct tumor clones can be identified by the expression of different fluorescent reporters, and these clones shared founder mutations and grew in close proximity within the tumor (Figure S1E). Intermixing of tumor clones is commonly observed in KPCC tumors (Figure S1F).
Because the R26R-Confetti alleles randomly labeled tumor cells with 6–8 colors, and a given tumor may theoretically have more than 8 clones, it is possible that different clones may be labeled by the same color. To complement the KPCC model, we utilized another KP sarcoma model induced by CRISPR-Cas9-mediated genome editing (Huang et al., 2017). The CRISPR-Cas9 system can produce a vast array of indels and, thus, is not limited to the maximum number of colors observed in the R26R-Confetti alleles (McKenna et al., 2016; Shin et al., 2017). In this model, the Cre-inducible KrasG12D allele is retained, and the Rosa26 locus expressed Cas9 endonuclease. Sarcomas were initiated by injecting adenovirus expressing Cre and single guide RNA (sgRNA) targeting p53 (K-sgP53-Cas9 in Figure S1A). The K-sgP53-Cas9 tumors were histologically similar to KP and KPCC tumors (Figure S1B). The genetic edits generated by CRISPR-Cas9 at the targeted region of p53 can serve as barcodes marking independent tumor clones.
Primary Tumor Progression Is Associated with a Loss of Clonal Heterogeneity
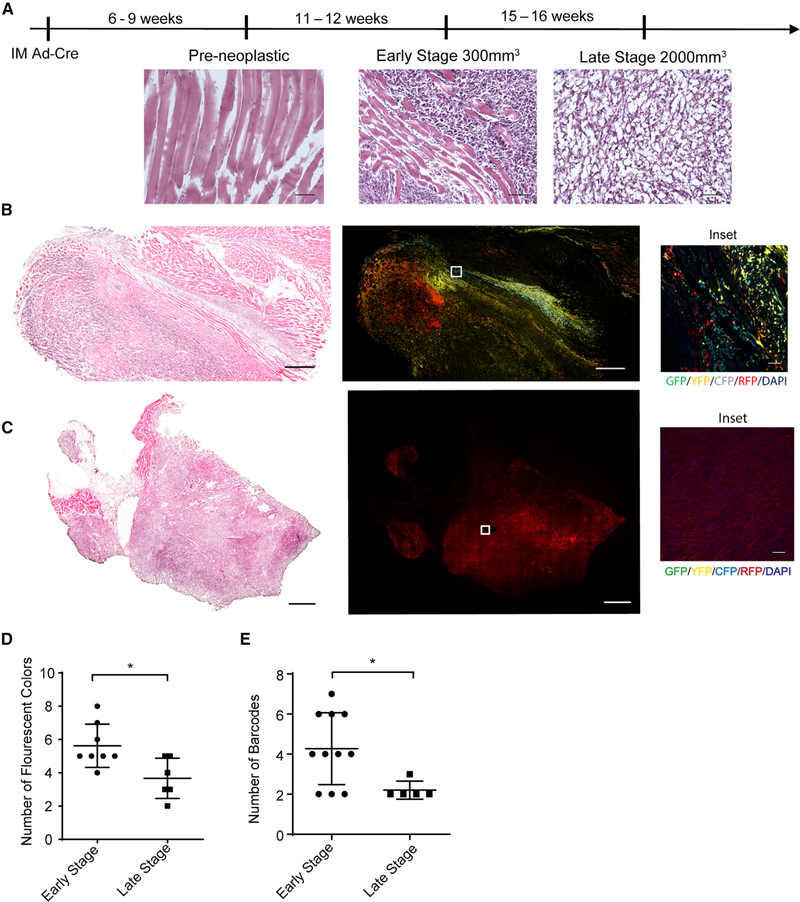
Using the KPCC and K-sgP53-Cas9 models, we first examined how clonality changed during primary tumor progression. Tumors were harvested between 250mm3 and 300mm3 to determine the clonality at a relatively early stage of sarcomagenesis and were then compared to late stage tumors that were approximately 2,000 mm3 in volume (Figure 1A). The cytology observed in early-stage tumors consisted of pleomorphic spindle cells with nuclear atypia, which had a similar cytological appearance to late stage tumors (Figure 1A). We observed significantly more fluorescent colors in early-stage tumors compared to late stage tumors (Figures 1B–1D). Furthermore, we scored each image from tumor sections as monochromatic, consisting of mostly 1 fluorescent color; as dichromatic, consisting of mostly 2 fluorescent colors; or as polychromatic, consisting of 3 or more fluorescent colors. The early-stage tumors had significantly higher polychromatic scores compared to late stage tumors (Figures S2A and S2B).
Figure 1. Primary Tumor Progression Is Associated with a Loss of Clonal Heterogeneity.
(A) Tissue and tumor histology from H&E staining at different time points after injection of Ad-Cre virus into the gastrocnemius muscle (scale, 100 μm).
(B) H&E staining (scale, 1 mm) and confocal microscopy images of a representative early-stage tumor. GFP (green), yellow fluorescent protein (YFP; yellow), RFP (red), CFP (cyan), 4′,6-diamidino-2-phenylindole (DAPI; blue) (scale, 1 mm; inset scale, 50 μm).
(C) H&E staining (scale, 1 mm) and confocal microscopy images of a representative late stage tumor (scale, 1 mm; inset scale, 50 μm).
(D) Graph comparing the number of fluorescent reporters observed in early and late tumors of KPCC model. Data are represented as mean ± SD (*p < 0.05, Student’s t test).
(E) Graph comparing the number of barcodes from the K-sgP53-Cas9 mice between early- and late-stage tumors. The barcodes are generated by deep sequencing of the p53 allele. Data are represented as mean ± SD (*p < 0.05, Student’s t test).
For the K-sgP53-Cas9 model, we harvested tumors at similar early and late stages and performed deep sequencing of the p53 alleles by using genomic DNA extracted from tumor masses. Early-stage tumors had significantly more barcodes compared to late stage tumors (Figures 1E and S3A). This is consistent with our findings in the KPCC model, indicating that primary sarcoma progression is associated with the sweeping growth of a dominant clone.
The KPCC and K-sgP53-Cas9 models require separate animals to study clonal evolution. To trace tumor clones in the same animal over time, we developed an intravital microscopy model in the mouse pinnae. Differently labeled tumor cells were injected into the pinnae of nude mice. These transplanted tumors maintained the histological features of primary sarcoma (Figure S2C). Consistent with the autochthonous models, the growth of transplanted tumors is associated with a reduction in clonal heterogeneity (Figure S2D).
Multiple Tumor Clones Give Rise to Local Recurrence
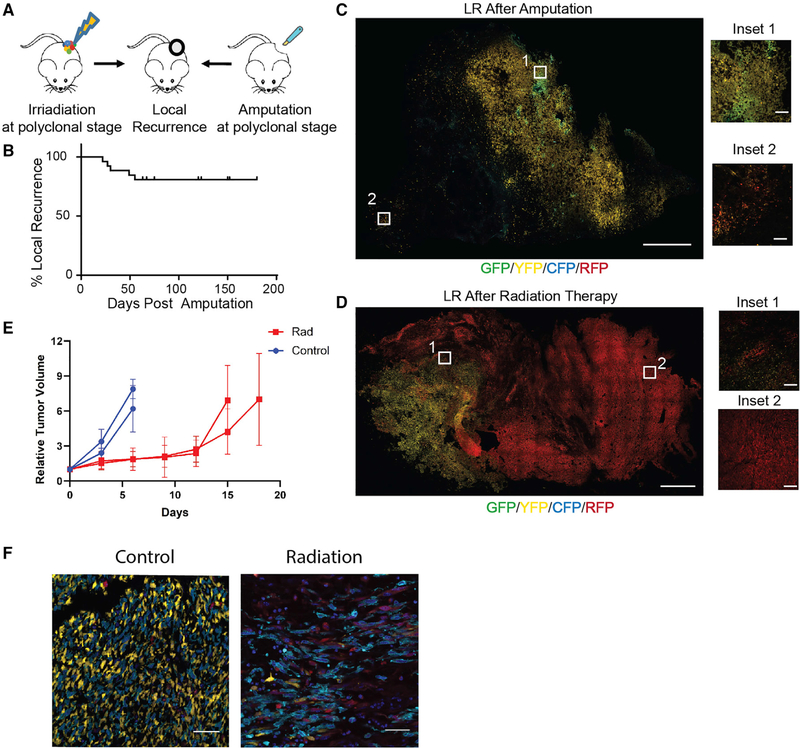
Local recurrence following therapy causes substantial morbidity and is an important prognostic risk factor for survival in many cancers, including sarcoma (Daigeler et al., 2014). To investigate whether local recurrence is derived from a single or multiple clones (Figure 2A), we treated early-stage polyclonal KPCC tumors by either surgically amputating the tumor-bearing leg (Sachdeva et al., 2014) or by delivering focal image-guided radiation (Moding et al., 2015). Amputation of the tumor-bearing legs was performed at the femoral head, and the entire leg was removed. In the amputated mice, 5/26 (19%) developed locally recurrent tumors near the pelvic bone at the surgical margins, with a median of 30 days (Figure 2B). We analyzed the recurrent disease between approximately 300mm3 and 800mm3 and found that the recurrent tumors consisted of multiple clones (Figure 2C). Recurrence following radiation therapy was defined by the continued growth of the tumor following irradiation and the tumors were harvested between 800mm3 and 1500mm3. Similar to local recurrence following surgery, the recurrent tumors (n = 6) following radiation therapy consisted of multiple clones (Figure 2D). To assess the effect of radiation on recurrence for the same tumor, we transplanted 2 polyclonal tumors into nude mice and gave half of the mice a single dose of radiation therapy. By imaging the control tumors and irradiated tumors, we found that similar to the autochthonous model, recurrence following irradiation in the transplant model was derived from multiple tumor clones (Figure 2E and 2F). Together, these data demonstrate that multiple clones gave rise to local recurrence after surgery or radiation therapy.
Figure 2. Local Recurrence in Sarcoma Is Driven by Multiple Clones.
(A) Schematic of experiments for local recurrence (LR). Tumor-bearing KPCC mice were either amputated or focally irradiated.
(B) Kaplan-Meier curve representing the percentage of mice and time to local recurrence following surgery.
(C) Confocal microscopy images of locally recurrent tumor after surgery. GFP (green), YFP (yellow), RFP (red), CFP (cyan) (scale, 1 mm; inset scale, 100 μm).
(D) Confocal microscopy images of recurrent tumor after irradiation (scale, 1 mm; inset scale, 100 μm).
(E) Two polyclonal tumors were transplanted into multiple nude mice and stereotactic irradiation delayed tumor growth. The graph shows the effect of irradiation on tumor growth over time. Data represented as mean ± SD of relative tumor volume to starting tumor volume.
(F) Confocal microscopy images of recurrent and control tumors after radiation therapy showed recurrence was driven by multiple clones. GFP(green), YFP (yellow), RFP (red), CFP (cyan), DAPI (blue) (scale, 50 μm).
Advanced Distant Metastases Are Clonal in Origin
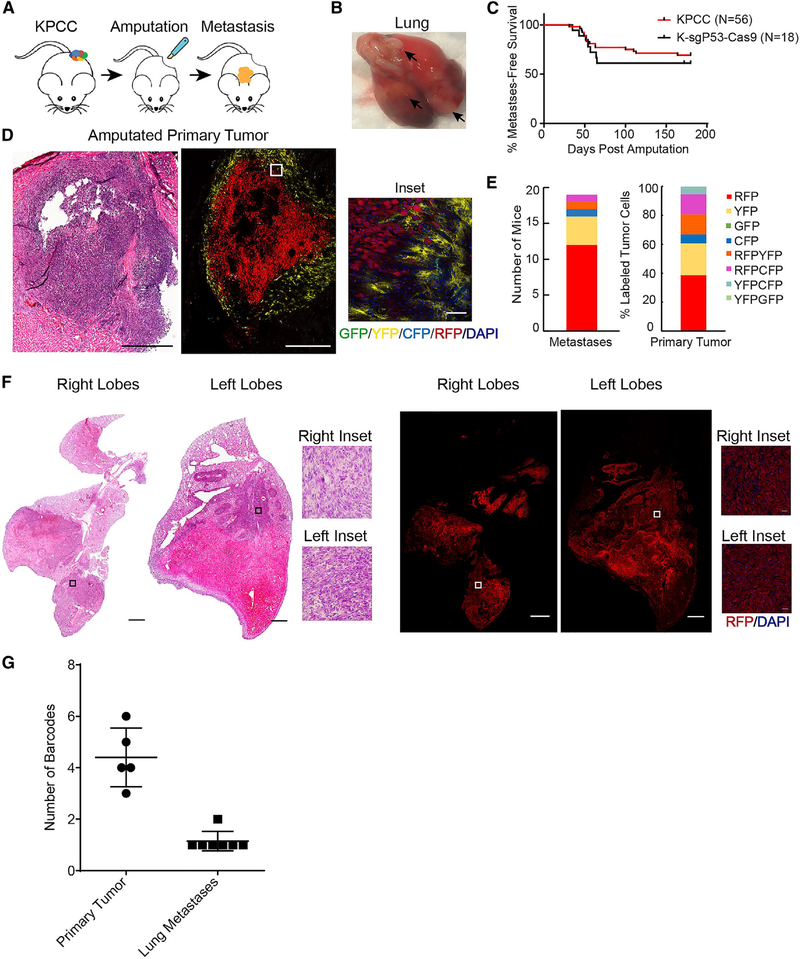
Metastasis accounts for the vast majority of cancer-related mortality. To investigate the clonality of metastases, we amputated the tumor-bearing limb and monitored the animals for up to 6 months (Figure 3A). In the KPCC mice, approximately 40% of mice developed metastasis after amputation, with a median survival of 54 days post-surgery (Figures 3B and 3C). This is similar to the frequency of metastasis in human patients (Digesu et al., 2016). The primary tumors were resected at the early stage (250mm3–350mm3) when they were polyclonal (Figure 3D). The majority of the metastases we observed were to the lungs (n = 18/19), which is the most common site of metastasis in human patients; 1/19 animal developed metastasis to the kidney (Figure S4A). Upon examination of the lung metastases, every lung lesion within a mouse with multiple lesions, ranging from 3–52 lesions per mouse, expressed the same fluorescent reporter (Figures 3E and 3F). We examined the metastases from KPCC animals by confocal microscopy and found that the percentages of different fluorescent reporters expressed in the metastases across animals were similar to the percentages of different fluorescent reporters observed in the primary tumors (Figure 3E). Two animals also developed liver metastases in addition to their lung metastases. The tumor cells in the liver expressed the same reporter as tumor cells in the lungs (Figure S4B).
Figure 3. Advanced Lung Metastases Are Driven by a Single Clone.
(A) Schematic of amputation experiments and development of lung metastases.
(B) Gross examination of harvested lungs with arrows indicating metastases.
(C) Kaplan-Meier curve representing the percent metastasis-free survival of amputated KPCC and K-sgP53-Cas9 mice.
(D) H&E staining (scale, 500 μm) and confocal microscopy images of a representative tumor after amputation. GFP (green), YFP (yellow), RFP (red), CFP (cyan), DAPI (blue) (scale, 500 μm; inset scale, 50 μm).
(E) Stacked bar graph of the number of KPCC mice that developed metastases with the corresponding color, and stacked bar graph of the percentage of differently labeled cells in the primary tumor.
(F) H&E staining (scale, 1.2 mm) and confocal microscopy images of representative lung metastases from a KPCC animal. RFP (red), DAPI (blue) (scale, 1 mm; inset scale, 25 μm).
(G) Graph showing the number of p53 barcodes in K-sgP53-Cas9 primary tumor and metastases.
Because labeling by the R26R-Confetti allele is random, it is possible for distinct clones to be labeled by the same fluorescent reporter. We statistically determined the probability of this occurring over multiple animals with metastases. We reasoned that the likelihood of two or more fluorescently indistinguishable tumor clones forming metastases would decrease with an increasing number of animals that had metastases comprised of cells expressing the same reporter (Methods S1, Section I). In other words, if metastasis is polyclonal in origin, the probability of not observing multi-colored lesions would decrease with an increasing number of mice. For our sample size (n = 19), the probability that two or more clones expressing the same reporter contributing to advanced lung metastasis is less than 9.71e-15. Therefore, advanced metastasis from sarcoma is most likely derived from a single clone in the primary tumor.
To further establish the clonal contribution of metastases experimentally without ambiguity, we used the K-sgP53-Cas9 sarcoma model. We amputated the extremity of these animals when the tumors were at early stage (approximately 350 mm3). The amputated mice developed metastases at a similar latency and frequency as the KPCC model (n = 7/18) (Figure 3C). Deep sequencing of the p53 alleles in matched bulk tumors and multiple metastases showed that although the primary sarcomas were polyclonal containing an average of 4–5 barcodes, the metastases from each mouse showed only 1 (n = 6 mice) or 2 barcodes (n = 1 mouse) that matched the barcodes from the original primary tumor (Figure 3G; Figure S3B). The wild-type (WT) p53 reads observed in the primary tumors and metastases were likely derived from normal stromal or immune cells in the primary tumors and at the metastatic site. In the metastases with 2 barcodes, the percentages of reads for each barcode were similar, suggesting that different edits were likely introduced to each p53 allele of the same clone (Figure S3B). The combined strength of lineage-tracing data from two independent and complementary mouse models indicate that advanced lung metastases originated from a single metastatic clone (MC) in primary sarcoma. Moreover, in these models, distinct metastatic lesions in the same animal are clonally related.
Clonal Metastasis Is a Deterministic Process
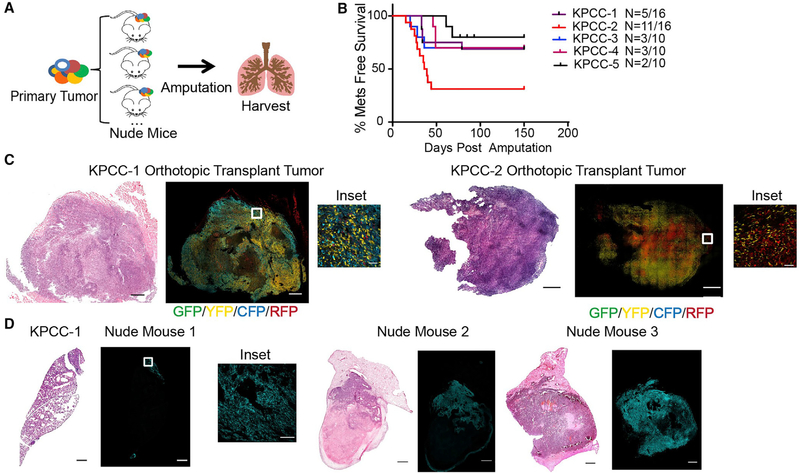
The emergence of a dominant clone in tissues can occur through either stochastic neutral drift or deterministic clonal selection (Lopez-Garcia et al., 2010; Morrissy et al., 2016; Snippert et al., 2010). To determine whether the cellular mechanism of clonal origin of advanced metastases is due to neutral drift where a single clone overtakes the lungs by chance or selection of a tumor clone that is most capable of metastatic colonization, we orthotopically transplanted polyclonal primary KPCC tumor cells into the left gastrocnemius muscle of multiple nude mice (Figure 4A). We hypothesized that if metastases were due to neutral drift, then the metastases in each nude mouse engrafted with the same KPCC tumor would be formed by different clones, which can be observed by the expression of different fluorescent reporters. On the other hand, if the clonality of metastases was due to the biological selection of a clone with enhanced metastatic capacity, then the same clone would form metastases in all the nude mice transplanted with the same KPCC tumor.
Figure 4. Monoclonal Origin of Metastases Is Driven by Selection.
(A) Schematic of orthotopic transplantation and amputation experiment. Independent primary KPCC tumors were injected into the left gastrocnemius muscle of multiple nude mice, and the tumors were amputated.
(B) Kaplan-Meier curve representing the percentage of metastasis-free survival for each transplanted KPCC tumor.
(C) H&E (scale, 1.2 mm) and confocal microscopy images of representative transplanted tumors at the time of amputation. GFP (green), YFP (yellow), RFP (red), CFP (cyan) (scale, 1 mm; inset scale, 50 μm).
(D) H&E (scale, 1 mm) and confocal microscopy images of lungs from nude mice transplanted with tumor KPCC-1. GFP (green), YFP (yellow), RFP (red), CFP (cyan) (scale, 1 mm; inset scale, 100 μm).
We transplanted 5 primary KPCC tumors into the gastrocnemius muscle of 10–16 nude mice per KPCC tumor (Figure 4A). After the orthotopic transplants, we amputated the tumor-bearing legs from the nude mice at the early stage (300 mm3–350 mm3) and followed the mice for the development of metastases (Figure 4B). We confirmed the transplanted tumors were polyclonal, indicating that the differently labeled clones were tumorigenic, and the transplanted tumors recapitulated the clonal heterogeneity of the primary tumor (Figure 4C). In mice that were transplanted with the same KPCC tumor, metastases in each nude mouse expressed the same fluorescent reporter, indicating they were derived from the same clone (Figure 4D). This result was consistent across 5 groups of animals transplanted with different primary tumors (Figure 4B; Figure S4C). Statistically, we considered the possibility of a single clone forming metastases in all the nude mice transplanted with the same tumor by random chance. We used an intracluster correlation coefficient approach to determine the probability of chance giving rise to our results (Kerry and Bland, 1998). Based on our sample size, the probability that all of the metastases in different nude mice randomly expressed the same fluorescent reporter is less than 3% (Methods S1, Section II). Therefore, the consistent presence of the same fluorescent reporter found in the metastases of nude mice indicates that metastasis is likely driven by a deterministic process by clonal selection.
Because late-stage primary tumor and advanced metastasis were dominated by a single clone, we sought to determine whether the MCs were derived from the dominant clones in each primary tumor. Because amputation of late-stage tumors resulted in overwhelming mortality and rapid onset of local recurrence, we orthotopically transplanted 3 independent polyclonal KPCC tumors, in which the identity of the MC cells was known, into nude mice and allowed these tumors to grow to late stage. We analyzed the transplanted tumors by confocal microscopy and fluorescence-activated cell sorting (FACS) to determine the abundance of each tumor clone. The results showed that in KPCC-1, the MC was present as a minor clone when the transplanted tumor reached late stage (Figure S5A). In the other 2 transplanted KPCC tumors, the MCs were dominant in the late-stage primary tumors (Figures S5B and S5C). However, in KPCC-4, the dominance of MC in the late-stage transplanted tumor was not consistent in every nude mouse, wherein one of the transplanted tumors, the MC was not the dominant population (Figure S5C). Taken together, these results suggest that, in our model, MCs can be either the dominant or minor clone in late-stage tumors and a growth advantage in the primary location is not always associated with enhanced metastatic ability.
Clonality of Early Metastatic Process
Next, we sought to determine the clonality of early metastatic seeding. Advanced lesions in distant organs are the result of a multi-step cascade that involves intravasation into the bloodstream, survival in circulation, extravasation, engraftment, and, ultimately, metastatic colonization (Lambert et al., 2017). Selective pressure can act on each step to influence clonal dynamics (Naxerova and Jain, 2015). We examined the intravasation step by determining the clonal composition of circulating tumor cells (CTCs). Blood from KPCC mice (n = 5) with primary sarcomas (approximately 300 mm3) was collected by cardiac puncture and stained with hematopoietic lineage marker CD45 and analyzed by flow cytometry. CTCs were identified as cells in the blood expressing R26R-Confetti reporters. We found CTCs in 4/5 mice. On average, 1.34% (±0.61% SEM) of CD45− cells expressed fluorescent reporters, and in all 4 animals, multiple reporters were present (Figures S6A–S6D). Nonetheless, in 2/4 animals, a single reporter was more dominant in the CTCs (Figure S6B), whereas in the other 2/4 animals, multiple fluorescent cells were represented (Figure S6C).
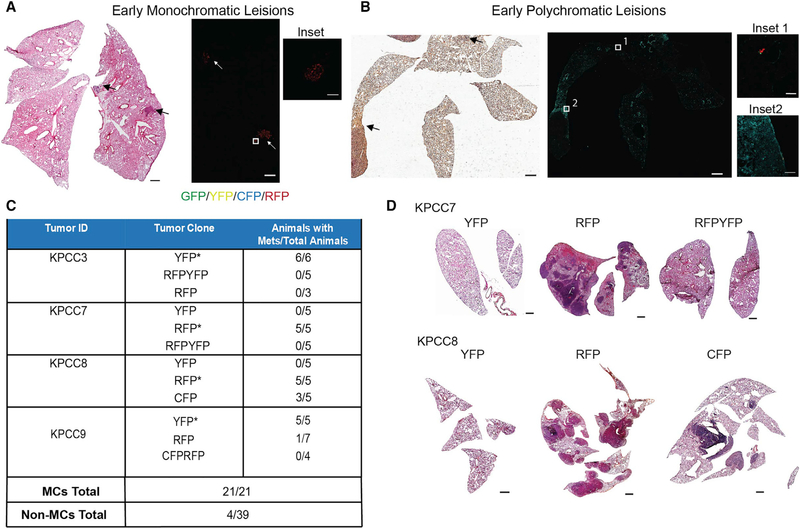
We looked further in the metastatic cascade by examining the lungs of KPCC mice before the onset of weight loss or labored breathing. In contrast to earlier experiments where the lungs were harvested when animals died from metastases or displayed clear symptoms of metastasis (ranging 54–164 days post-amputation), lungs were harvested from mice 35–45 days post-amputation. We examined the lungs histologically for the presence of early metastatic lesions. The early metastatic lesions were defined as clusters of less than 500 cells and were found in 5 of 15 amputated mice. In 3/5 mice, the early lesions were monoclonal (Figure 5A), but in 2 mice, the lungs contained polyclonal cell clusters, with 3 colors in each mouse (Figure 5B). Furthermore, in the polyclonal animals, the relatively larger early metastatic lesions, which were defined as a cluster of approximately 30 to 500 cells, were mostly monoclonal (Figure 5B). In an additional 2 animals, we observed single KPCC cells in the lungs that expressed the same reporter (Figure S6E). Taken together, these data suggest that early metastatic seeding into the lung can be polyclonal or monoclonal.
Figure 5. Clonality of Early Metastatic Process.
(A) H&E (scale, 1 mm) and confocal images of monoclonal early metastatic lesions from a KPCC mouse (scale, 1 mm; inset scale, 100 μm).
(B) H&E (scale, 1 mm) and confocal images of polyclonal early metastatic lesions from a KPCC mouse (scale, 1 mm; inset 1 scale, 50 μm; inset 2 scale, 100 μm).
(C) Table indicating the number of tail-vein-injected mice that developed metastases for each sorted population and their totals (*p < 0.05, chi-square test comparing the number of mice with and without metastases between different clones).
(D) H&E images of representative lung metastases from each sorted population (scale, 1 mm).
To determine the ability of each individual clone to colonize the lungs, we FACS-sorted different clones from 4 primary KPCC tumors. The individual clones were injected into separate groups of 3–5 nude mice by the tail vein. We followed the animals for 3–6 weeks and compared the metastatic ability of the different tumor clones. The MCs that formed metastases in the amputated primary KPCC mouse formed metastases in 21/21 nude mice. Most of the other primary tumor clones did not form metastases. Overall, 4/39 nude mice developed advanced metastases after tail vein injection of the other clones, which we termed Non-MCs (Figure 5C and 5D). These results suggest that in the polyclonal tumor, most tumor clones have a limited capacity to form overt metastases, consistent with the notion that advanced metastasis is largely driven by a single MC.
Transcriptome of MCs and Non-MCs
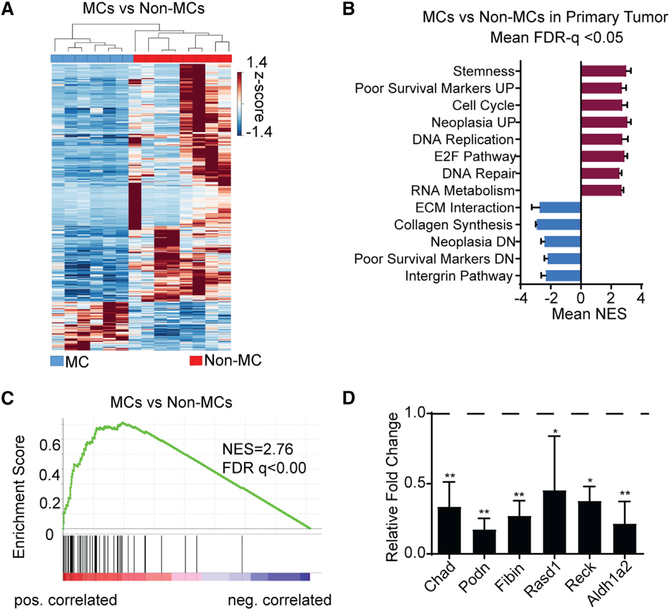
To gain insight into the molecular mechanism that distinguishes MCs, which are capable of forming advanced metastatic growth, from Non-MCs, we performed RNA sequencing on MCs and Non-MCs from the same primary tumors. When the primary sarcomas were amputated in the autochthonous model, the mice were followed for the development of lung metastases. By matching the color of the fluorescent reporter in the lung metastases to the clones in the primary tumor, we were able to define primary tumor clones as MCs or Non-MCs. We performed RNA sequencing on MCs, Non-MCs and matched lung metastases (Lung-Mets). Clustering of samples by gene expression largely distinguished MCs from Non-MCs (Figure 6A). The expression of 853 genes was differentially expressed in the MCs relative to Non-MCs. Gene set enrichment analysis (GSEA) of MCs versus Non-MCs showed positive enrichment of processes involved in stemness, cancer aggressiveness, cell cycle, and DNA repair (Subramanian et al., 2005). Cellular processes involved in extracellular matrix interaction, collagen synthesis, and integrin were negatively enriched (Figure 6B; Table S1). Several gene signatures associated with poor cancer outcome were positively enriched in MCs. For example, CINSARC (Complexity Index in Sarcomas), a well-established gene set prognostic of poor survival in human sarcomas (Chibon et al., 2010), was enriched in MCs (Figure 6C). Because the majority of (74%) of the differentially regulated genes were suppressed in MCs compared to Non-MCs, we sought to investigate potential candidate suppressors of metastasis. Because the top negatively enriched processes in our GSEA involved cell-matrix interaction, integrin, and cell adhesion, we focused on genes that were associated with these processes. We took a candidate approach and selected six top differentially suppressed genes in MCs relative to Non-MCs that were not previously known to play a role in sarcoma metastasis, namely, Chad, Podn, Fibin, Rasd1, Reck, and Aldh1a2. The downregulation of these candidate genes in MCs was confirmed by qRT-PCR (Figure 6D).
Figure 6. Transcriptome Analysis of Tumor Clones from Primary KPCC Sarcomas.
(A) Hierarchical clustering of gene expression heatmap between MCs (n = 6) and Non-MCs (n = 8).
(B) GSEA analysis of molecular pathways enriched in MCs versus Non-MCs.
(C) CINSARC signature enrichment in MCs versus Non-MCs.
(D) qRT-PCR validation 6 candidate downregulated genes in MCs in RNA-seq. Data represented as mean ± SEM (*p < 0.05, **p < 0.01, one-way ANOVA).
A comparison of the overall gene expression patterns between Lung-Mets, MCs, and Non-MCs showed that the Lung-Mets were distinct from clones in the primary tumor (Figure S7A and S7B). The gene expression differences between Lung-Mets and Non-MCs were greater compared to the differences between MCs and Non-MCs, with 5,162 genes differentially expressed. GSEA between Lung-Mets and Non-MCs showed pathways specifically enriched in the Lung-Mets (Figure S7A; Table S2). Notably, processes involved in hypoxia, cell metabolism, and Wnt signaling were positively enriched in metastases, whereas, transforming growth factor α (TNF-α), epidermal growth factor (EGF), and immune response were negatively enriched. In addition, we compared the gene expression profile of Lung-Mets versus MCs, and the expression of 3,519 genes was significantly different (Figure S7B). Many of these genes were associated with mitochondrial metabolism, DNA damage response, and protein degradation (Figure S7B; Table S3). These differences likely reflect additional molecular changes in the MCs in response to the lung microenvironment as they grew and colonized the tissue over time. Although the CINSARC gene signature was enriched in Lung-Mets compared to Non-MCs, the enrichment was not statistically significant between Lung-Mets and MCs (Figure S7C). This suggests that the CINSARC enrichment may be specific for differences between metastatic and non-metastatic tumor cells.
Overexpression of Suppressed MC Genes Inhibits Metastasis
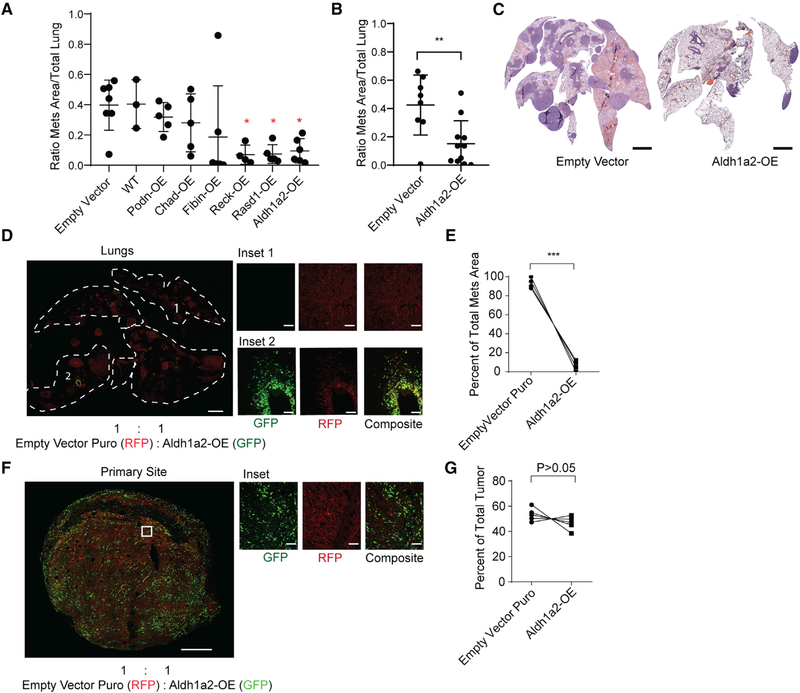
To investigate the potential for the six candidate metastasis suppressor genes (Figure 6D) to inhibit metastasis in vivo, we overexpressed each gene in metastatic KPCC cells (Figure S7D). Cells overexpressing each candidate gene or empty vector control cells were injected into nude mice (n = 3–5 for each construct) via the tail vein. Metastatic ability was then assessed by averaging the relative total area of the metastatic lesions across the lungs and by enumerating the number of metastatic lesions. Three of the six genes (Rasd1, Reck, and Aldh1a2) showed a 70%–75% reduction in metastatic burden (Figure 7A; Figure S7E).
Figure 7. Identification of In Vivo Metastasis Suppressors.
(A) Effect of stable overexpression of each candidate gene on the metastatic ability of KPCC cells after tail-vein injection. Each point represents the relative metastatic area in the lungs of an individual nude mouse. Data represented as mean ± SD (*p < 0.05, one-way ANOVA).
(B) Metastatic burden following tail-vein injection of control or Aldh1a2 overexpressing cells in an independent set of animals quantified by relative area of metastatic lesions. Data represented as mean ± SD (**p < 0.01, Student’s t test).
(C) Representative H&E images of lungs from mice injected with Aldh1a2-OE and empty-vector control cells (scale, 1.5 mm).
(D) Confocal lung metastasis after competition between Aldh1a2-OE (labeled green) and empty-vector control cells (labeled red) injected at 1:1 ratio into the tail vein of nude mice (scale, 1.5 mm; inset scale, 100 μm).
(E) Quantification of metastases area. Data represented as mean ± SD (***p < 0.001, Student’s t test).
(F) Confocal microscopy of orthotopic competition between Aldh1a2-OE cells (labeled green) and control cells (labeled red) injected at 1:1 ratio into the muscle of the extremity (scale, 700 μm; inset scale, 100 μm).
(G) Quantification of tumor tissue area for each fluorescent reporter in tumor sections (Student’s t test).
From the 3 candidate suppressors of metastasis, we focused on Aldh1a2, one of the least characterized members of the aldehyde dehydrogenase family that functions in retinoic acid biosynthesis. MCs were enriched for the downregulated gene signature associated with the knock out of retinoic acid receptors, the downstream transcription factors of retinoic acid signaling (Al Tanoury et al., 2014) (Figure S7F). In addition, the loss of ALDH1A2 copy number in human soft tissue sarcoma was correlated with decreased survival (Figure S7G). We further validated the overexpression of Aldh1a2 in transduced cells by western blot (Figure S7H) and performed tail vein injection in an independent cohort of nude mice. The results showed a consistent decrease in lung-mets (Figures 7B, 7C, and S7I). Because MCs have increased competitive capacity to establish colonies in the lungs compared to Non-MCs, we hypothesized that overexpression of Aldh1a2 might reduce their competitiveness. To test this, we performed a competition assay by co-injecting GFP+ Aldh1a2-OE cells with red fluorescent protein (RFP)+ control cells at a 1:1 ratio through the tail vein of nude mice. In these animals, the resulting metastases were dominated by control cells, where less than 10% of the metastases included Aldh1a2-OE cells (Figures 7D and 7E). We then tested the competitiveness between Aldh1a2-OE cells and the empty vector control cells in the primary tumor site. An equal number of Aldh1a2-OE cells and control cells were orthotopically engrafted into the gastrocnemius muscle of nude mice. The resulting tumors showed relatively similar numbers of Aldh1a2-OE cells and control cells (Figures 7F and 7G), suggesting that overexpression of Aldh1a2 preferentially impairs competitive fitness in the lungs.
DISCUSSION
Competition and selection of tumor clones are essential elements of cancer evolution (Marusyk and Polyak, 2010). In this study, we prospectively traced tumor clones throughout disease progression, from sarcoma initiation, growth, and local recurrence to metastasis. Using models of multicolor fluorescent reporters, intravital microscopy, and CRISPR-Cas9-based barcoding, we demonstrated the functional roles of different tumor clones throughout the major stages of sarcoma progression. These models closely recapitulated important aspects of tumor evolution. First, in our models, tumor clones were initiated with the same founder mutations, KrasG12D, and homozygous deletion of p53. Second, contrary to many other autochthonous tumor models, such as non-small-cell lung cancer, where the individual tumor clones are spatially separated throughout the organ with limited contact (Chuang et al., 2017), the tumor clones in our models grow in proximity as part of a single tumor mass where clonal intermixing is common. Third, our model captured the stepwise progression observed in human cancers, where tumor clones from one primary location grow, disseminate, and metastasize to form multiple lesions in distant organs. Taken together, the models in this study mimic tumor evolution where distinct tumor clones occupy overlapping tissue habitats and progress by competition and selection (Greaves and Maley, 2012).
The K-sgP53-Cas9 model generated different indels targeted by the sgRNA within the p53 gene, which served as barcodes for different tumor clones. Although the majority of the indels induced are frameshift mutations or gene truncation (Pinello et al., 2016; Ran et al., 2013), it is still possible that different indels may confer distinct functional consequences on the gene. In our previous work, we tested the expression of p53 in cells generated from sarcomas in the KP-sgP53-Cas9 model and found that p53 expression is lost as a result of the various indels (Huang et al., 2017). Furthermore, because our results with the KPsgP53-Cas9 model mirror the results from the KPCC model, where p53 alleles were deleted by Cre-Lox recombination, we concluded that the sarcomas in the K-sgP53-Cas9 model had lost p53 function. Based on the fluorescent reporter model and DNA barcoding, we showed that primary tumor progression is associated with the expansion of a dominant clone. Moreover, we demonstrated that local recurrence and metastasis are driven by distinct clonal dynamics, where local recurrences are polyclonal and advanced distant metastases are clonal in origin. Importantly, to investigate the clonal origin of metastasis, the tumors were amputated at an early stage, prior to the establishment of a dominant clone in the primary tumor, indicating metastatic ability is gained before clonal sweep in the primary tumor. Amputation of late-stage tumors where clonal dominance has occurred could not be performed effectively because of mortality during surgery and rapid onset of local recurrence in the survivors, which prevented the study of gross metastases. By transplanting polyclonal tumors that were metastatic into nude mice and allowing the tumors to reach late stage, we showed MCs could be either the minor or the dominant clones in late-stage primary tumors. This is consistent with the notion that metastatic ability can be independent of primary tumor dominance and is in agreement with other reports of sequencing-matched tumors and metastases in human patients where metastatic seeding can derive from rare or truncal clones (Bao et al., 2018; Fidler, 2003; Gundem et al., 2015; Makohon-Moore et al., 2017).
Through transplantation of the same polyclonal tumor into multiple animals, we demonstrated that the MC arises through a deterministic selection process instead of neutral population drift. The statistical methods we used to exclude the probability of multiple clones being labeled with the same reporter by chance will be valuable for future investigations of clonal dynamics. By tracing the early steps of the metastatic cascade, our data demonstrated polyclonal CTCs and polyclonal early metastatic lesions in some animals. This suggests that metastatic seeding can be polyclonal. As such, the critical selective bottleneck leading to the monoclonality of clinically relevant, advanced metastases may be in the adaptation and colonization of the lungs. Although we cannot rule out the possibility that a few cells from other clones in the early polyclonal metastatic seeds may be residing below the threshold of detection in advanced stage metastases, in these models the vast majority of animals with detectable, clinically relevant metastases are clonal. Our data also suggest that selective pressure and competition between clones can act at each successive step of the metastatic cascade. Furthermore, it is conceivable that in certain types of cancer, a primary tumor could harbor more than one clone each with the MC phenotype and clonally derived metastases may be generated by multiple tumor clones. Nonetheless, in our models, when the clones were transplanted individually, the inability of most clones to form metastases or at a reduced rate to form metastases suggests that the MC cells likely can outcompete the other clones over time, leading to the monoclonality of advanced metastases.
Previous lineage tracing studies for metastases were in epithelial cancers and found that metastases can derive from multiple clones (Cheung et al., 2016; Maddipati and Stanger, 2015; Reeves et al., 2018). In the breast cancer study, the labeled tumor cells were generated from single-cell-derived tumor organoids. The progenies of these organoids may not faithfully represent the divergent properties that exist between clones of an autochthonous tumor developed over the course of the disease (Cheung et al., 2016). Reeves et al. (2018) labeled tumor cells in clonally derived skin squamous cell carcinoma after the tumors had progressed and found polyclonal metastases. The difference in the clonality of sarcoma metastases in our model may be related to the biology of mesenchymal tumors, or more likely, in the method in which tumor clones are defined. In some of the previous studies, tumor cells are randomly labeled late during tumor progression (Cheung et al., 2016; Reeves et al., 2018). In these studies, it is unclear whether distinctly labeled tumor cells are different tumor clones that exhibit phenotypic diversity or if they are progenies of the same clonal population with similar biological properties but are labeled differently by chance. In our model, tumor clones were distinguishable at tumor initiation, so that the competition and selection of different clones can be traced throughout different stages of diseases progression. In the pancreatic cancer model, where tumor clones are traced from initiation, even though regional metastases were polyclonal, the large distant metastases to the lungs were frequently monoclonal, consistent with our observations (Maddipati and Stanger, 2015).
Genes that are associated with metastasis are usually identified by comparing matched pairs of established metastases and primary tumor (Lambert et al., 2017). The differential expression of genes that underlie the metastatic properties within phenotypically different primary tumor clones has not been studied and may shed light on the early metastatic process. By focusing on genes altered in MCs, we identified potential suppressors of metastasis, namely, Aldh1a2, Reck, and Rasd1. The expression of these genes is altered in the primary tumor preceding cells reaching the distant organ and may represent the early gene expression changes that contribute to metastasis initiation. Aldh1a2 is a member of the aldehyde dehydrogenase family of enzymes that catalyze the oxidation of aldehydes to carboxylic acids and participates in the biosynthesis of retinoic acid (RA) (Niederreither and Dollé, 2008). Its role in cancer metastasis has not been described previously. During tissue development, members of the Aldh1a family, including Aldh1a2, are known to play pivotal roles. The loss of Aldh1a2 disrupts mesodermal patterning and formation of the forelimb bud (Niederreither and Dollé, 2008). More recently, genetic knock out of RA receptors, the transcriptional factors downstream of RA signaling, is reported to inhibit cell adhesion to matrix substrates (Al Tanoury et al., 2014). In our study, Aldh1a2 expression was suppressed in MCs, and genes associated with RA signaling were negatively enriched. Pathways involved in cell adhesion and cell-matrix interaction were among the top cellular processes negatively enriched in MCs. Rasd1 encodes a member of the Ras superfamily of GTPases and may play a role in cell matrix interactions and cell adhesion (Tu and Wu, 1999). The RECK protein is a membrane-bound glycoprotein that functions in extracellular matrix remodeling by inhibiting the activity of metalloproteinases (Walsh et al., 2015). The expression of Reck is downregulated in breast cancer by hypermethylation and contributes to tumor invasion and angiogenesis (Walsh et al., 2015). Future studies into the molecular mechanisms in which these candidate genes regulate metastasis may reveal novel pathways and potential interactions between metastasis suppressor genes that contribute to the metastatic process.
Overall, by modeling clonal competition and selection, we describe in detail the clonal dynamics of sarcoma progression during each step, from tumor initiation, local recurrence after therapy, and to advanced metastasis. The clonal origin of advanced metastases offers the possibility of targeting specific clonal populations within a heterogeneous tumor to impede disease progression. The model we used here traced clonal evolution from the beginning of the tumorigenic process; however, during tumor evolution, each clone may give rise to distinct subclones, creating additional heterogeneity. In future studies, approaches with single-cell resolution such as single-cell RNA sequencing (scRNA-seq) may help identify additional transcriptional heterogeneity within individual clonal populations. Furthermore, the metastatic process involves a complex interplay between the tumor intrinsic alterations, such as changes in the genome and epigenome, with extrinsic alterations such as the tumor microenvironment. Systematic investigation of the tumor genome and microenvironmental interactions at the clonal level may reveal new insights into the selective forces that enable certain clones to gain the metastatic advantage.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVALIABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by lead contact, Benjamin Alman (ben.alman@duke.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse Models
Animal studies were performed in accordance with approved protocols from the Duke University Institutional Animal Care and Use Committee. Males and female animals are used for autochthonous mouse models. The R26R-Confetti mice were obtained from Jackson Laboratories (Stock No: 013731). These mice were crossed with previously described KRASLSL-G12D; p53f/f mice (Kirsch et al., 2007) to generate the KRASLSL-G12D; p53f/f; R26R-Confetti/Confetti (KPCC) mice. The KRASLSL-G12D mice were provided by Dr. Tyler Jacks (MIT, MA) and the p53f/f mice were provided by Anton Berns (NKI, Netherlands). To induce tumors, adenovirus expressing Cre recombinase were mixed with 2M CaCl2 in DMEM and were directly injected into the hind gastrocnemius muscle of 7–10-week-old mice (Kirsch et al., 2007). Tumor volume was calculated using the formula V = (π × L × W × H)/6, with L, W, and H representing the length, width, and height of the tumor in mm. All mice were on a mixed genetic background. The K-sgP53-Cas9 mice were generated by crossing KrasLSL-G12D mice with Rosa26-LSL-Cas9 (Stock No: 024857, Jackson Laboratory) mice or Rosa26-Loxp-Cas9 (Huang et al., 2017). Tumor initiation in K-sgP53-Cas9 mice was as described previously (Huang et al., 2017). For tumor transplantation experiments, 6 to 7-week-old male Foxn1nu/nu were purchased from Duke University Division of Laboratory Animals and Resources. Tumor cells were diluted in 1XPBS for injection. The cells were injected into the hind gastrocnemius at 500,000 cells per mouse or the pinnae at 100,000 cells per mouse.
METHOD DETAILS
Amputation Surgery, Orthotopic Transplantation, Tail-Vein and Pinna Injections
For amputation surgery, primary tumors in KPCC and K-sgP53-Cas9 mice were amputated when their size reached an average of 250–350mm3. The tumor-bearing limb was amputated under 2% isoflurane induced anesthesia. Subcutaneous injection of 0.1mg/kg buprenorphine and 5mg/kg ketoprofen and 0.25% bupivacaine were administered to provide analgesia. The mice were followed for the development of lung metastases. The amputated tumors were digested into single cells using a mixture of collagenase, dispase, and trypsin (Wei et al., 2015). Mice were examined every 2 days following amputation until they developed signs of systemic illness, such as hunched posture, lethargy, or significant weight loss. When clear symptoms were observed, the mice were euthanized, and the lungs were harvested and analyzed for the presence of metastases. To observe advanced metastasis, usually complete local control of the tumor is required. The animals that developed local recurrence following surgery were euthanized and the recurrent tumors were harvested for histology and microscopy. For orthotopic transplants, the tumors were amputated at early stage. The nude mice were monitored using the same protocol as the autochthonous model. Tail-vein injection experiments were performed by injecting 250,000 to 500,000 tumor cells diluted in 1XPBS into the tail-vein of nude mice. The mice were euthanized when they developed clear symptoms of metastases, and the lungs were harvested. Pinna injections were performed by injecting single cell suspensions of primary KPCC tumor cells with a 27G needle into the pinna of 6–7-week-old nude mice. Successful injections were confirmed by swelling immediately after injection. The pinnae were imaged using a two-photon microscope (FluoView FV1000, Olympus, Central Valley, PA).
Isolation of Circulating Tumor Cells
Blood from KPCC mice was harvested via cardiac puncture and the animals were euthanized. The blood was then centrifuged in Ficoll-Paque PLUS (GE-Health Care, PA) and the nucleated cells were isolated from the buffy coat. The nucleated cells were immediately stained with CD45-PECy7 anti-mouse antibody (561868, RRID:AB_10893599, BD Biosciences, NJ) and analyzed using MoFlo Astrios Flow cytometer (Beckman Coulter) for RFP, YFP, CFP, and GFP expression. Propidium Iodine (ThermoFisher, MA) was used as a live/dead indicator.
In Vivo Competition Assay
The empty vector puro cells were generated by transducing parental cells with lentivirus carrying the pCDH-CMV-MCS-EF1α-Puro vector (System Biosciences, CA). For competition in metastasis, 250,000 cells with the overexpression vector were mixed with 250,000 empty vector cells in 1XPBS. The mixture was injected into nude mice via the tail-vein. For competition in orthotopic tumors, 150,000 cells from each group were mixed in 1XPBS, and injected into the left gastrocnemius muscle of nude mice. Quantification of the metastases area and tumor area was done using FIJI (Version 2.0) by calculating the area of each fluorescent reporter with intensity threshold.
Radiation Treatment
Sarcoma irradiation was performed using the X-RAD 225Cx small animal image-guided irradiator (Precision X-Ray) (Newton et al., 2011). The tumors were irradiated at an average of 350mm3. The radiation field was centered on the target by fluoroscopy with 40-kilovolt peak (kVp), 2.5 mA X-rays using a 2-mm aluminum filter. Radiation therapy was performed with parallel-opposed anterior and posterior fields at an average dose rate of 300 cGy/min prescribed to midplane with 225 kVp, 13 mA X-rays using a 0.3-mm copper filter and a collimator with a 40 × 40 mm2 radiation field at treatment isocenter (Moding et al., 2013). The mice were placed under anesthesia with continuous 2% isoflurane with 2L of oxygen per minute. The mice were also treated with 0.1mg/kg buprenorphine and 5mg/kg of ketoprofen, and local injection of 0.25% bupivacaine for analgesia. For autochthonous sarcomas, an average of 35 Gy was delivered in a single fraction, and for transplanted tumors in nude mice, the total dose was reduced to 25 Gy in a single fraction because of increased normal tissue injury observed in Foxn1nu/nu mice.
Histology
For frozen sections, the lung samples were inflated with 4% PFA through the trachea. The tumor and lung samples were fixed in 4% paraformaldehyde at 4°C for 1 hour and 3 hours respectively, followed by incubation in 15% and 30% w/v sucrose for 24 hours. The tissues were then embedded in Optimum Cutting Temperature medium (OCT, Tissue-Tek) and frozen on dry ice. 10–12μm sections were cut using a Cryostat (Leica CM1950). Sections were taken 50–80μm apart across 1–2mm of tissue to limit sampling bias. The tissue sections were mounted with Prolong Diamond Antifade Mountant with or without DAPI (ThermoFisher). For formalin-fixed and paraffin-embedded sections, samples were fixed in formalin overnight, washed with PBS and ethanol, followed by embedding in paraffin. The tissues were cut at 5 μm. At least 1–2 mm of tissue from each sample were analyzed. H&E images were taken with Zeiss Axio Imager Z2 (Zeiss, Germany) at either 5X, 10X, or 20X, and the entire sections were scanned.
Confocal and Intravital Microscopy
Fluorescent images of primary and transplanted tumors were taken using Leica SP8 confocal microscope with a 20X and 40X objective. The argon laser 488 nm line was used to excite GFP, 514 nm line for YFP, 561nm red diode laser for RFP, 458 nm laser line for CFP and UV laser line for DAPI. The collection wavelengths for GFP, YFP, RFP, CFP, and DAPI were: 493–510 nm, 537–565 nm, 612–650 nm, 468–498 nm, and 417–497 nm, respectively. Airy unit 1 was used for all images. The images were taken using Leica Application Suite X. Intravital imaging of sarcoma ear allograft was performed using a two-photon microscope (FluoView FV1000, Olympus, Central Valley, PA). This microscope is equipped with a 680–1050 nm tunable ultrafast laser (Spetra-Physics, CA). To image transplanted tumors in the ear pinna, tumor-bearing mice were anesthetized with 2% isoflurane and the ear pinna was fixed to the stage using double-sided tape. YFP was excited by 860–980 nm laser and detected using a 495/540 nm bandpass filter. RFP was excited by 980 nm laser and detected using a 575/630 nm bandpass filter. CFP was excited by 830–850nm laser with a 460–500 nm bandpass filter. Collagen was imaged by second harmonic generation using 860–920 nm excitation and 420/460 nm detection. Image processed with Imaris Microscopy Image Analysis Software (v 9.30).
Tumor Digestion, Cell Sorting, and Cell Culture
Primary sarcoma tumors from KPCC mice were mechanically and enzymatically dissociated into single cell suspensions as previously described (Wei et al., 2015). Briefly, the tumors were mechanically dissociated into small pieces and enzymatically digested with a mixture of 10 mg/mL of collagenase IV (Worthington), 2.4U/ml of Dispase (Stem Cell Technologies, BC), and 0.05% trypsin (ThermoFisher). Red blood cells were excluded with ACK lysis buffer (ThermoFisher). FACS for isolating the R26R-Confetti fluorescent proteins was performed using Moflo Astrios Sorter (Beckman Coulter, CA) at the Duke Cancer Institute Flow Cytometry Core (Duke University, NC). CFP was detected by 405 nm violate laser with a 488/59 bandpass filter, and RFP was detected by 561 nm red laser with a 579/16 bandpass filter. GFP and YFP were detected by 488 nm argon laser with a 510/20 bandpass filter for GFP and a 546/20 bandpass filter for YFP. Propidium iodide was used to remove dead cells. The sorted cells were collected in PBS and cultured in DMEM (ThermoFisher) supplemented with 10%FBS.
Cell Lines
Sorted cells for tail-vein injections and metastatic cells used for overexpression experiments were from digested primary KPCC animals. The purity of the different sorted populations was checked by FACS after the cell lines were passaged once in vitro. All cell lines were confirmed to be negative for mycoplasma using the MycoAlert Detection Kit (Lonza, Brazil).
DNA Isolation, PCR and Targeted Deep Sequencing
Multiple pieces of the primary tumor masses and multiple lesions from the lung metastases of K-sgP53-Cas9 mice were harvested. The bulk DNA from the harvested tissues were isolated using DNeasy Blood and Tissue Kit (QIAGEN) following manufacturers’ protocol and then sequenced. To sequence the CRISPR-Cas9 induced barcodes, the 200bp region near the sgRNA target site was PCR amplified using proof-reading AccuPrime Taq Polymerase (ThermoFisher). The forward and reverse primer sequence are 5′CAGAAGCTGGGGAAGAAACA3′, and 3′GTAGGGAGCGACTTCACCTG5′ respectively. The PCR products were sequenced by Massachusetts General Hospital DNA Core (Boston, MA). Sequencing depth was > 5,000X. Sequences representing 1% of total reads for each sample was used as a cut-off. PCR to check the recombination of LSL-KrasG12D and p53 deletion were performed as described previously (DuPage et al., 2009; Marino et al., 2000). DNA was extracted from sorted cells of primary KPCC tumors. The primers for LSL-KrasG12D are 5′ GTC TTT CCC CAG CAC AGT GC 3′, 5′ CTC TTG CCT ACG CCA CCA GCT C 3′, 5′ AGC TAG CCA CCA TGG CTT GAG TAA GTC TGC A 3′. The primers for p53 are 5′ CAC AAA AAC AGG TTA AAC CCA G 3′, 5′ GAA GAC AGA AAA GGG GAG GG 3′, 5′ AAG GGG TAT GAG GGA CAA GG 3′. Tail genomic DNA is used as the negative control.
RNA Isolation and RNA-Sequencing
Primary KPCC tumor cells were FACS sorted into MCs and Non-MCs, and the RNA was extracted using Norgen RNA Purification Kit (Norgen, ON). RNA from bulk metastatic lesions were isolated using Directzol RNA kit (Zymogen, CA). Standard manufacturer’s protocol is used for both kits. The total RNA for RNA-sequencing was performed at the Duke Center for Genomic and Computational Biology (Duke University, NC). The RNA was prepared using KAPA stranded mRNA-seq kit (Sopachem, Belgium). The first 50 bases of these libraries were sequenced on a HiSeq 4000 system (Illumina, CA). Over 50 million reads were generated per sample.
Molecular Cloning, Lentivirus Generation and Transduction
To generate viral vectors overexpressing the six-candidate metastasis suppressor genes (Chad, Podn, Fibin, Rasd1, Reck, and Aldh1a2), cDNA constructs were ordered from Origene (TrueORF, Origene, MD). The cDNA for each gene was cloned into EcoRI and FseI sites of pLenti-MCS2-CMV-GFP-2A-Puro or pCDH-CMV-MCS-EF1α-Puro vector (System Biosciences, CA). Successful cloning was confirmed via Sanger sequencing. Lentivirus was generated using the Delta8.2 and VSV-G packaging plasmids. To generate stable overexpression cells, virus carrying the cDNA constructs and empty vectors were transduced into the cells with 8 μg/mL of polybrene. After two days, the cells were selected with puromycin, and overexpression was validated by qPCR. Overexpression of Aldh1a2 was also validated with Western Blot.
RT-qPCR
Reverse-transcription of RNA to generate cDNA was done using iScript gDNA Clear cDNA Synthesis Kit following the manufacturer’s instructions (Bio-Rad, CA). Quantitative PCR (qPCR) was performed using TaqMan Fast Advanced Master Mix (ThermoFisher, MA) with QuantStudio Flex 6 (ThermoFisher, MA). The denaturation step was carried out at 95°C for 20 s; the annealing/extension step was carried out at 60°C for 20 s, and the total cycle is 40. Relative gene expression was determined using ΔΔCt with beta-Actin as the internal control gene.
Western Blot
Whole-cell protein extracts from cells were denatured, separated on SDS–PAGE gels, and dry-transferred to nitrocellulose membranes. After blocking in 5% milk in Tris-buffered saline–Tween, membranes were probed overnight at 4°C with either vinculin (1: 5,000, Cell Signaling Technology) or Aldh1a2 (1:500; ProteinTech, IL). After incubation, the secondary antibody was added and incubated for 1 hour in room temperature. Results were detected using SuperSignal West Femto Maximum Sensitivity Substrate, according to the manufacturer’s instructions (ThermoFisher, MA).
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantitative Image Analysis
Quantification of images was performed by manually counting the fluorescent reporter positive cells and total DAPI+ cells using FIJI (version 2.0 with cell counter plugin). For each tissue section, we either scanned the entirety of the slide or examined approximately 20 areas of view. For categorization of the images, each picture was categorized as monochromatic if ≥ 90% of fluorescent cells were of a single color, as dichromatic if the two colors represented ≥ 90% of total fluorescent cells, and as polychromatic if at least 3 colors were present, each representing 10%–20% of total fluorescent cells. Quantification of metastases in the lungs was done by measuring the number of metastatic lesions and the total area of the metastatic lesions using FIJI (version 2.0). For both measurements, approximately 5 sections were randomly selected across 1–2 mm of lung tissues and the average number of metastases and the average area of metastatic lesions per animal was used for statistical comparison. For in vivo competition assays, approximately 5 sections from across 1–2 mm of lung tissues or tumor tissues were randomly examined. The area of each fluorescent reporter was calculated by color threshold using FIJI (version 2.0) and divided by the total tissue area of the sections.
RNA-Seq Analysis
RNA-seq analysis was performed on MCs (6 biological replicates with 5 technical replicates), Non-MCs (8 biological replicates with 1 technical replicate), and Lung-Mets (5 biological replicates and 1 technical replicate). The RNA sequencing reads were processed using the Trimmomatic (v0.36) (Bolger et al., 2014) to trim low-quality bases and Illumina sequencing adapters from the 3′ end. Only reads that were 20nt or longer after trimming were kept for further analysis. Reads were mapped to the mouse transcriptome using the STAR aligner (v2.5.2b) (Dobin et al., 2013). Gene counts were compiled using the HTSeq tool implemented in STAR program (Anders et al., 2015). The reference sequence and GTF file were obtained from the UCSC GRCm38 bundle available from the iGenomes collection. Normalization and differential expression were carried out using the DESeq2 (v1.16.1) (Anders and Huber, 2010). Gene differential expression was performed within the framework of a negative binomial model using R (v3.4.4)(R Core Team, 2016). Statistical analyses were adjusted for multiple testing within the framework of control of the false-discovery rate (Bass et al., 2015; Benjamini and Hochberg, 1995). Statistical analyses were mainly scripted using the R statistical environment (R Core Team, 2016) along with its extension packages from the comprehensive R archive network (CRAN; https://cran.r-project.org/) and the Bioconductor project. (Gentleman et al., 2004). We considered genes with p < 0.005 for hypothesis generation. Gene set enrichment analysis (GSEA) was performed to identify differentially regulated pathways and gene ontology (Subramanian et al., 2005). Statistically significant gene sets are defined as Nom p-val < 0.05 and FDR q-val < 0.05. Gene sets describing similar processes were grouped together, and the average NES and FDR q-val were determined for each group. The genes list for CINSARC and Rar-KO enrichment were obtained from Chibon et al. (2010) and Al Tanoury et al., 2014. For selection of genes in the Rar-KO list, genes that are differentially expressed between Rar-KO and WT cells in RNA-seq were selected (Padj < 0.01 was used as a cutoff). Copy number information for ALDH1A2 and corresponding survival analysis from The Cancer Genome Atlas (TCGA) was accessed using UCSC Xena (http://biorxiv.org/lookup/doi/10.1101/326470).
Statistical Analyses
Graphs and statistics for most biological experiments were generated using the GraphPad Prism software (Prism 6) unless indicated otherwise. Statistical significance was calculated using the two-tailed unpaired Student’s t test for comparison between two groups or one-way ANOVA for comparison between multiple groups unless described otherwise. Holm-Sidak method was used post hoc correction of multiple comparisons in ANOVA, adjusted p < 0.05 is considered significant. For details on the statistics used to determine the likelihood of monoclonality of metastasis through biological selection, see Supplemental Items Methods S1.
DATA AND CODE AVAILABILITY
The RNA-seq data reported in this paper is uploaded to the NCBI GEO database with accession number GSE118489 shown in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Aldh1a2, Rabbit Polyclonal | ProteinTech | Cat#: 13951–1-AP; RRID:AB_2224033 |
| Vinculin | Cell Signaling | Cat#: 4650S; RRID:AB_10559207 |
| cDNA Clones and Virus Strains | ||
| Ad5-CMV-Cre | Viral Vector Core Facility University of Iowa Health Care | N/A |
| Ad-P-Cre | Huang et al., 2017 | N/A |
| pLenti-MCS2-CMV-GFP-2A-Puro | This study | N/A |
| pCDH-CMV-MCS-EF1 a-Puro | System Biociences | CD510B-1 |
| Aldh1a2-ORF | Origene | Cat#: MR208298 |
| Chad-ORF | Origene | Cat#: MR205471 |
| Fibin-ORF | Origene | Cat#: MR202431 |
| Podn-ORF | Origene | Cat#: MR209397 |
| Rasd1-ORF | Origene | Cat#: MR203784 |
| Reck-ORF | Origene | Cat#: MR227392 |
| Critical Commercial Assays | ||
| AccuPrime Taq DNA Polymerase, high fidelity | ThermoFisher | Cat#: 12346094 |
| Taqman Fast Advanced Master Mix | ThermoFisher | Cat#:4444556 |
| Taqman primer for Aldh1a2 | ThermoFisher | Cat#: Mm00501306_m1 |
| Taqman primer for Beta-Actin | ThermoFisher | Cat#: Mm02619580_g1 |
| Taqman primer for Gapdh | ThermoFisher | Cat#: Mm99999915_g1 |
| Taqman primer for Chad | ThermoFisher | Cat#: Mm00483284_m1 |
| Taqman primer for Fibin | ThermoFisher | Cat#: Mm00471003_s1 |
| Taqman primer for Podn | ThermoFisher | Cat#: Mm01263602_m1 |
| Taqman primer for Rasd1 | ThermoFisher | Cat#: Mm00842185_g1 |
| Taqman primer for Reck | ThermoFisher | Cat#: Mm01299530_m1 |
| Deposited Data | ||
| RNA-seq Data | This Study | NCBI GEO Database Accession number: GSE118489 |
| Experimental Models: Organisms/Strains | ||
| LSL-KrasG12D; p53flox/flox; R26R-Confetti/ Confetti (KPCC) | This Study | N/A |
| LSL-KrasG12D; R26-Cas9 (K-sgp53-Cas9) | Huang et al., 2017 | N/A |
| Nude mice | Duke University Rodent Breeding Core | N/A |
| Software and Algorithms | ||
| FIJI v2.0 | N/A | https://fiji.sc/ |
| Trimmomatic v0.36 | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| STAR Aligner v2.5.2b | Dobin et al., 2013 | N/A |
| STAR Program | Anders et al., 2015 | N/A |
| Deseq2 | Anders and Huber, 2010 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| R v3.4.4 | R Core Team, 2016 | https://cran.r-project.org/ |
| Gene Set Enrichment Analysis (GSEA) | Subramanian et al., 2005 | http://software.broadinstitute.org/gsea/index.jsp |
| Prism GraphPad | GraphPad Software | https://www.graphpad.com/ |
| Imaris v9.30 | N/A | https://imaris.oxinst.com |
| USCS Xena | http://biorxiv.org/lookup/doi/10.1101/326470 | https://xena.ucsc.edu/ |
| Others | ||
| High Glucose DMEM | ThermoFisher | Cat#: 11995065 |
| Collagenase Type IV | Worthington | Cat#: LS004188 |
| Leica SP8 Upright Confocal Microscope | Leica | N/A |
| FV1000 Two-Photon Microscope | Olympus | N/A |
| Hisea2000 | Illumina | N/A |
Supplementary Material
Highlights.
Primary tumor progression is associated with loss of clonal heterogeneity
Local recurrence following therapy is polyclonal
Advanced metastasis is driven by a single metastatic clone by biological selection
Overexpression of Aldh1a2 is sufficient to impair sarcoma metastasis to the lungs
ACKNOWLEDGMENTS
This study was supported by a grant from the National Cancer Institute of the NIH (R01CA183811 to B.A.A.). The authors thank Ms. Consuelo Mendoza for helping with image analysis; Dr. Michael Cook and Lynn Martinek from the Duke University Flow Cytometry Core for flow cytometry support; Dr. David Corcoran from the Duke Center for Genomic and Computational Biology for assistance with analysis of RNA-seq and performing GSEA analysis; and Dr. Kouros Owzar and team from the Duke Cancer Institute for input on bioinformatics.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.08.029.
REFERENCES
- Al Tanoury Z, Piskunov A, Andriamoratsiresy D, Gaouar S, Lutzing R, Ye T, Jost B, Keime C, and Rochette-Egly C (2014). Genes involved in cell adhesion and signaling: a new repertoire of retinoic acid receptor target genes in mouse embryonic fibroblasts. J. Cell Sci 127, 521–533. [DOI] [PubMed] [Google Scholar]
- Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggiolini A, Varum S, Mateos JM, Bettosini D, John N, Bonalli M, Ziegler U, Dimou L, Clevers H, Furrer R, and Sommer L (2015). Premigratory and migratory neural crest cells are multipotent in vivo. Cell Stem Cell 16, 314–322. [DOI] [PubMed] [Google Scholar]
- Bao L, Qian Z, Lyng MB, Wang L, Yu Y, Wang T, Zhang X, Yang H, Brünner N, Wang J, and Ditzel HJ (2018). Coexisting genomic aberrations associated with lymph node metastasis in breast cancer. J. Clin. Invest 128, 2310–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J, Swcf A, Dabney A, and Robinson D (2015). qvalue: Q-value estimation for false discovery rate control. R package version 280 https://bioconductor.org/packages/release/bioc/html/qvalue.html. [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B 57, 289–300. [Google Scholar]
- Blum JM, Añó L, Li Z, Van Mater D, Bennett BD, Sachdeva M, Lagutina I, Zhang M, Mito JK, Dodd LG, et al. (2013). Distinct and overlapping sarcoma subtypes initiated from muscle stem and progenitor cells. Cell Rep. 5, 933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burningham Z, Hashibe M, Spector L, and Schiffman JD (2012). The epidemiology of sarcoma. Clin. Sarcoma Res. 2, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2017). Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell 171, 950–965.e928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KJ, Padmanaban V, Silvestri V, Schipper K, Cohen JD, Fairchild AN, Gorin MA, Verdone JE, Pienta KJ, Bader JS, and Ewald AJ (2016). Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl. Acad. Sci. USA 113, E854–E863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibon F, Lagarde P, Salas S, Pérot G, Brouste V, Tirode F, Lucchesi C, de Reynies A, Kauffmann A, Bui B, et al. (2010). Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Nat. Med 16, 781–787. [DOI] [PubMed] [Google Scholar]
- Chuang CH, Greenside PG, Rogers ZN, Brady JJ, Yang D, Ma RK, Caswell DR, Chiou SH, Winters AF, Grüner BM, et al. (2017). Molecular definition of a metastatic lung cancer state reveals a targetable CD109-Janus kinase-Stat axis. Nat. Med 23, 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigeler A, Zmarsly I, Hirsch T, Goertz O, Steinau HU, Lehnhardt M, and Harati K (2014). Long-term outcome after local recurrence of soft tissue sarcoma: a retrospective analysis of factors predictive of survival in 135 patients with locally recurrent soft tissue sarcoma. Br. J. Cancer 110, 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, Jamal-Hanjani M, Shafi S, Murugaesu N, Rowan AJ, et al. (2014). Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digesu CS, Wiesel O, Vaporciyan AA, and Colson YL (2016). Management of Sarcoma Metastases to the Lung. Surg. Oncol. Clin. N. Am 25, 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M, Dooley AL, and Jacks T (2009). Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 4, 1064–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler IJ (2003). The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer 3, 453–458. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, et al. (2012). Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med 366, 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves M, and Maley CC (2012). Clonal evolution in cancer. Nature 481, 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati S, Martinez P, Joshi T, Birkbak NJ, Santos CR, Rowan AJ, Pickering L, Gore M, Larkin J, Szallasi Z, et al. (2014). Systematic evaluation of the prognostic impact and intratumour heterogeneity of clear cell renal cell carcinoma biomarkers. Eur. Urol 66, 936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JMC, Papaemmanuil E, Brewer DS, Kallio HML, Högnäs G, Annala M, et al. ; ICGC Prostate Group (2015). The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao JJ, Lin DC, Dinh HQ, Mayakonda A, Jiang YY, Chang C, Jiang Y, Lu CC, Shi ZZ, Xu X, et al. (2016). Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat. Genet. 48, 1500–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Chen M, Whitley MJ, Kuo HC, Xu ES, Walens A, Mowery YM, Van Mater D, Eward WC, Cardona DM, et al. (2017). Generation and comparison of CRISPR-Cas9 and Cre-mediated genetically engineered mouse models of sarcoma. Nat. Commun 8, 15999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerry SM, and Bland JM (1998). The intracluster correlation coefficient in cluster randomisation. BMJ 316, 1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch DG, Dinulescu DM, Miller JB, Grimm J, Santiago PM, Young NP, Nielsen GP, Quade BJ, Chaber CJ, Schultz CP, et al. (2007). A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat. Med 13, 992–997. [DOI] [PubMed] [Google Scholar]
- Lambert AW, Pattabiraman DR, and Weinberg RA (2017). Emerging Biological Principles of Metastasis. Cell 168, 670–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, et al. (2013). Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152, 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia C, Klein AM, Simons BD, and Winton DJ (2010). Intestinal stem cell replacement follows a pattern of neutral drift. Science 330, 822–825. [DOI] [PubMed] [Google Scholar]
- Maddipati R, and Stanger BZ (2015). Pancreatic Cancer Metastases Harbor Evidence of Polyclonality. Cancer Discov. 5, 1086–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makohon-Moore AP, Zhang M, Reiter JG, Bozic I, Allen B, Kundu D, Chatterjee K, Wong F, Jiao Y, Kohutek ZA, et al. (2017). Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat. Genet 49, 358–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S, Vooijs M, van Der Gulden H, Jonkers J, and Berns A (2000). Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14, 994–1004. [PMC free article] [PubMed] [Google Scholar]
- Marusyk A, and Polyak K (2010). Tumor heterogeneity: causes and consequences. Biochim. Biophys. Acta 1805, 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A, Almendro V, and Polyak K (2012). Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer 12, 323–334. [DOI] [PubMed] [Google Scholar]
- McGranahan N, and Swanton C (2017). Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 168, 613–628. [DOI] [PubMed] [Google Scholar]
- McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, and Shendure J (2016). Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito JK, Riedel RF, Dodd L, Lahat G, Lazar AJ, Dodd RD, Stangenberg L, Eward WC, Hornicek FJ, Yoon SS, et al. (2009). Cross species genomic analysis identifies a mouse model as undifferentiated pleomorphic sarcoma/malignant fibrous histiocytoma. PLoS One 4, e8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moding EJ, Clark DP, Qi Y, Li Y, Ma Y, Ghaghada K, Johnson GA, Kirsch DG, and Badea CT (2013). Dual-energy micro-computed tomography imaging of radiation-induced vascular changes in primary mouse sarcomas. Int. J. Radiat. Oncol. Biol. Phys 85, 1353–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moding EJ, Castle KD, Perez BA, Oh P, Min HD, Norris H, Ma Y, Cardona DM, Lee CL, and Kirsch DG (2015). Tumor cells, but not endothelial cells, mediate eradication of primary sarcomas by stereotactic body radiation therapy. Sci. Transl. Med 7, 278ra34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissy AS, Garzia L, Shih DJ, Zuyderduyn S, Huang X, Skowron P, Remke M, Cavalli FM, Ramaswamy V, Lindsay PE, et al. (2016). Divergent clonal selection dominates medulloblastoma at recurrence. Nature 529, 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naxerova K, and Jain RK (2015). Using tumour phylogenetics to identify the roots of metastasis in humans. Nat. Rev. Clin. Oncol 12, 258–272. [DOI] [PubMed] [Google Scholar]
- Newton J, Oldham M, Thomas A, Li Y, Adamovics J, Kirsch DG, and Das S (2011). Commissioning a small-field biological irradiator using point, 2D, and 3D dosimetry techniques. Med. Phys 38, 6754–6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither K, and Dollé P (2008). Retinoic acid in development: towards an integrated view. Nat. Rev. Genet 9, 541–553. [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. ; Breast Cancer Working Group of the International Cancer Genome Consortium (2012). Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérot G, Chibon F, Montero A, Lagarde P, de Thé H, Terrier P, Guillou L, Ranchère D, Coindre JM, and Aurias A (2010). Constant p53 pathway inactivation in a large series of soft tissue sarcomas with complex genetics. Am. J. Pathol 177, 2080–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinello L, Canver MC, Hoban MD, Orkin SH, Kohn DB, Bauer DE, and Yuan GC (2016). Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 34, 695–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2016). R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing; ). [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves MQ, Kandyba E, Harris S, Del Rosario R, and Balmain A (2018). Multicolour lineage tracing reveals clonal dynamics of squamous carcinoma evolution from initiation to metastasis. Nat. Cell Biol 20, 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M, Mito JK, Lee CL, Zhang M, Li Z, Dodd RD, Cason D, Luo L, Ma Y, Van Mater D, et al. (2014). MicroRNA-182 drives metastasis of primary sarcomas by targeting multiple genes. J. Clin. Invest 124, 4305–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Tang YJ, Wei Q, Hirata M, Weng A, Han I, Okawa A, Takeda S, Whetstone H, Nadesan P, et al. (2016). Mesenchymal Tumors Can Derive from Ng2/Cspg4-Expressing Pericytes with β-Catenin Modulating the Neoplastic Phenotype. Cell Rep. 16, 917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R, and Schäffer AA (2017). The evolution of tumour phylogenetics: principles and practice. Nat. Rev. Genet 18, 213–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano C, Romagosa C, Hernández-Losa J, Simonetti S, Valverde C, Moliné T, Somoza R, Pérez M, Vélez R, Vergés R, et al. (2016). RAS/MAPK pathway hyperactivation determines poor prognosis in undifferentiated pleomorphic sarcomas. Cancer 122, 99–107. [DOI] [PubMed] [Google Scholar]
- Shin HY, Wang C, Lee HK, Yoo KH, Zeng X, Kuhns T, Yang CM, Mohr T, Liu C, and Hennighausen L (2017). CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat. Commun 8, 15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, and Clevers H (2010). Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton C (2012). Intratumor heterogeneity: evolution through space and time. Cancer Res. 72, 4875–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y, and Wu C (1999). Cloning, expression and characterization of a novel human Ras-related protein that is regulated by glucocorticoid hormone. Biochim. Biophys. Acta 1489, 452–456. [DOI] [PubMed] [Google Scholar]
- Walsh LA, Roy DM, Reyngold M, Giri D, Snyder A, Turcan S, Badwe CR, Lyman J, Bromberg J, King TA, and Chan TA (2015). RECK controls breast cancer metastasis by modulating a convergent, STAT3-dependent neoangiogenic switch. Oncogene 34, 2189–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, Chen K, Scheet P, Vattathil S, Liang H, et al. (2014). Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512, 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Tang YJ, Voisin V, Sato S, Hirata M, Whetstone H, Han I, Ailles L, Bader GD, Wunder J, and Alman BA (2015). Identification of CD146 as a marker enriched for tumor-propagating capacity reveals targetable pathways in primary human sarcoma. Oncotarget 6, 40283–40294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JS (2014). Mutation position within evolutionary subclonal architecture in AML. Semin. Hematol 51, 273–281. [DOI] [PubMed] [Google Scholar]
- Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, Aas T, Alexandrov LB, Larsimont D, Davies H, et al. (2015). Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med 21, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J, Robinson RA, and Lee JY (1999). H-ras and K-ras gene mutations in primary human soft tissue sarcoma: concomitant mutations of the ras genes. Mod. Pathol 12, 775–780. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data reported in this paper is uploaded to the NCBI GEO database with accession number GSE118489 shown in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Aldh1a2, Rabbit Polyclonal | ProteinTech | Cat#: 13951–1-AP; RRID:AB_2224033 |
| Vinculin | Cell Signaling | Cat#: 4650S; RRID:AB_10559207 |
| cDNA Clones and Virus Strains | ||
| Ad5-CMV-Cre | Viral Vector Core Facility University of Iowa Health Care | N/A |
| Ad-P-Cre | Huang et al., 2017 | N/A |
| pLenti-MCS2-CMV-GFP-2A-Puro | This study | N/A |
| pCDH-CMV-MCS-EF1 a-Puro | System Biociences | CD510B-1 |
| Aldh1a2-ORF | Origene | Cat#: MR208298 |
| Chad-ORF | Origene | Cat#: MR205471 |
| Fibin-ORF | Origene | Cat#: MR202431 |
| Podn-ORF | Origene | Cat#: MR209397 |
| Rasd1-ORF | Origene | Cat#: MR203784 |
| Reck-ORF | Origene | Cat#: MR227392 |
| Critical Commercial Assays | ||
| AccuPrime Taq DNA Polymerase, high fidelity | ThermoFisher | Cat#: 12346094 |
| Taqman Fast Advanced Master Mix | ThermoFisher | Cat#:4444556 |
| Taqman primer for Aldh1a2 | ThermoFisher | Cat#: Mm00501306_m1 |
| Taqman primer for Beta-Actin | ThermoFisher | Cat#: Mm02619580_g1 |
| Taqman primer for Gapdh | ThermoFisher | Cat#: Mm99999915_g1 |
| Taqman primer for Chad | ThermoFisher | Cat#: Mm00483284_m1 |
| Taqman primer for Fibin | ThermoFisher | Cat#: Mm00471003_s1 |
| Taqman primer for Podn | ThermoFisher | Cat#: Mm01263602_m1 |
| Taqman primer for Rasd1 | ThermoFisher | Cat#: Mm00842185_g1 |
| Taqman primer for Reck | ThermoFisher | Cat#: Mm01299530_m1 |
| Deposited Data | ||
| RNA-seq Data | This Study | NCBI GEO Database Accession number: GSE118489 |
| Experimental Models: Organisms/Strains | ||
| LSL-KrasG12D; p53flox/flox; R26R-Confetti/ Confetti (KPCC) | This Study | N/A |
| LSL-KrasG12D; R26-Cas9 (K-sgp53-Cas9) | Huang et al., 2017 | N/A |
| Nude mice | Duke University Rodent Breeding Core | N/A |
| Software and Algorithms | ||
| FIJI v2.0 | N/A | https://fiji.sc/ |
| Trimmomatic v0.36 | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| STAR Aligner v2.5.2b | Dobin et al., 2013 | N/A |
| STAR Program | Anders et al., 2015 | N/A |
| Deseq2 | Anders and Huber, 2010 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| R v3.4.4 | R Core Team, 2016 | https://cran.r-project.org/ |
| Gene Set Enrichment Analysis (GSEA) | Subramanian et al., 2005 | http://software.broadinstitute.org/gsea/index.jsp |
| Prism GraphPad | GraphPad Software | https://www.graphpad.com/ |
| Imaris v9.30 | N/A | https://imaris.oxinst.com |
| USCS Xena | http://biorxiv.org/lookup/doi/10.1101/326470 | https://xena.ucsc.edu/ |
| Others | ||
| High Glucose DMEM | ThermoFisher | Cat#: 11995065 |
| Collagenase Type IV | Worthington | Cat#: LS004188 |
| Leica SP8 Upright Confocal Microscope | Leica | N/A |
| FV1000 Two-Photon Microscope | Olympus | N/A |
| Hisea2000 | Illumina | N/A |