SUMMARY
Cooperative binding of transcription factors (TFs) to chromatin orchestrates gene expression programming and cell fate specification. However, the biophysical principles of TF cooperativity remain incompletely understood. Here we use single-molecule fluorescence microscopy to study the partnership between Sox2 and Oct4, two core members of the pluripotency gene regulatory network. We find that the ability of Sox2 to target DNA inside nucleosomes is strongly affected by the translational and rotational positioning of its binding motif. In contrast, Oct4 can access nucleosomal sites with equal capacities. Furthermore, the Sox2-Oct4 pair displays nonreciprocal cooperativity, with Oct4 modulating interaction of Sox2 with the nucleosome but not vice versa. Such cooperativity is conditional upon the composite motif’s residing at specific nucleosomal locations. These results reveal that pioneer factors possess distinct chromatin-binding properties and suggest that the same set of TFs can differentially regulate gene activities on the basis of their motif positions in the nucleosomal context.
In Brief
Using single-molecule fluorescence imaging, Li et al. investigate the pioneer activities of pluripotency factors Sox2 and Oct4 and find that they exhibit distinct nucleosome binding preferences as well as context-dependent cooperativity, which potentially allows gene-specific transcriptional regulation.
Graphical Abstract
INTRODUCTION
Transcription factors (TFs) access and interpret the genome by recognizing specific DNA sequences and regulating the transcriptional activity of selected sets of genes (Lambert et al., 2018; Ptashne and Gann, 2002). In eukaryotic nuclei, genomic DNA is wrapped around histone octamers, forming nucleosome building blocks and higher order chromatin structures (Luger et al., 2012; McGinty and Tan, 2015). The compaction of DNA into chromatin often occludes TFs from their cognate binding motifs, thereby constituting an important regulatory layer of gene expression control and cell identity determination (Li et al., 2007; Segal and Widom, 2009). A subset of TFs, known as pioneer factors (PFs), possess the ability to access nucleosomal DNA and closed chromatin, which further recruits other chromatin-binding proteins and the transcription machinery to their target sites and initiate transcriptional reprogramming and cell fate transitions (Meers et al., 2019; Zaret and Mango, 2016).
TF pairs can exhibit cooperative binding behavior; that is, binding of one factor to DNA facilitates targeting of the other (Morgunova and Taipale, 2017). Such cooperativity can be mediated by direct TF-TF interaction or through the DNA substrate (Jolma et al., 2015; Kim et al., 2013). Alternatively, because of the competition between TFs and histones for DNA binding, TF-TF cooperativity can be manifested indirectly and often nonspecifically in the context of chromatin (Adams and Workman, 1995; Mirny, 2010; Polach and Widom, 1996; Vashee et al., 1998). In this scenario, PFs are the first to engage and open up closed chromatin, making it more accessible to other TFs (Sartorelli and Puri, 2018; Zaret and Carroll, 2011).
One of the most prominent examples of TFs shaping gene expression pattern is the Yamanaka factors (Sox2, Oct4, Klf4, and c-Myc), which can convert mammalian somatic cells into induced pluripotent stem cells (Takahashi and Yamanaka, 2006). It was shown that Sox2, Oct4, and Klf4, but not c-Myc, can function as PFs by binding to nucleosomes in vitro and silent, DNase-resistant chromatin in vivo (Soufi et al., 2012, 2015). During reprogramming toward pluripotency, these factors cooperatively target selected enhancers and activate or repress the expression of distinct genes in a stage-specific manner (Chronis et al., 2017; Soufi et al., 2012).
Among these pluripotency TFs, Sox2, and Oct4 are also core members of the transcriptional regulatory network that governs embryogenesis and the maintenance of embryonic stem cells (Li and Belmonte, 2017; Rizzino and Wuebben, 2016). Sox2 contains an HMG domain that binds to the minor groove of the DNA helix, whereas Oct4 harbors a bipartite POU domain that interacts with the DNA major groove, allowing the formation of Sox2-Oct4-DNA ternary complexes with the TF pair binding to adjacent motifs (Reményi et al., 2003; Williams et al., 2004). Chromatin immunoprecipitation sequencing (ChIP-seq) experiments revealed that Sox2 and Oct4 co-occupy the cis-regulatory elements of a large number of target genes (Boyer et al., 2005; Chen et al., 2008; Whyte et al., 2013), suggesting that this TF pair works synergistically to regulate gene expression. Indeed, juxtaposed HMG:POU composite motifs are found upstream of many pluripotency-associated genes (Ambrosetti et al., 1997; Nishimoto et al., 1999; Okumura-Nakanishi et al., 2005; Rodda et al., 2005; Tomioka et al., 2002).
Despite extensive research, the capacity of and mutual relationship between Sox2 and Oct4, and PFs in general, in targeting nucleosomal DNA remains a matter of debate (Biddle et al., 2019; Chronis et al., 2017; Franco et al., 2015; Iwafuchi-Doi et al., 2016; Soufi et al., 2015; Swinstead et al., 2016). Recent data suggested that the pioneer activity of a given TF may be conditional and dependent on the local chromatin environment (King and Klose, 2017; Liu and Kraus, 2017; Soufi et al., 2015; Swinstead et al., 2016; Yu and Buck, 2019). Popular methods of choice for studying TF-chromatin interaction, such as genome-wide binding and bulk biochemical assays, lack sufficient temporal resolution to inform the time order of binding events (usually occurring on the order of seconds) by multiple TFs. Single-particle tracking in living cells can capture the dynamic nature of TF binding (Chen et al., 2014; Lam et al., 2012; White et al., 2016), but the underlying DNA sequence and chromatin state in these assays are usually not well defined. By contrast, in vitro single-molecule measurements allow precise control of the substrates and have been used to provide quantitative information on the binding/dissociation kinetics of TFs and chromatin regulators (Choi et al., 2017; Donovan et al., 2019; Gibson et al., 2017; Kilic et al., 2015; Luo et al., 2014).
In this work, we used single-molecule fluorescence microscopy to measure the binding dynamics of Sox2 and Oct4—both individually and in combination—on a variety of DNA and nucleosome substrates. We found that although both classified as PFs, Sox2 and Oct4 exhibit markedly distinct properties of nucleosome targeting. Sox2 strongly favors nucleosome dyad-positioned sites over end-positioned ones, whereas Oct4 indiscriminately binds to targets at all positions. Oct4 and Sox2 are hierarchically recruited to composite nucleosomal motifs, with Oct4 predominantly engaging first. We further demonstrated that the Sox2-Oct4 pair displays nonreciprocal cooperativity and that such cooperativity is dependent on the location of the composite motif with respect to the nucleosome. Consistent with these in vitro results based on a strong nucleosome positioning sequence (NPS), our analyses of the genomic data showed that the DNA binding sites of Sox2, but not Oct4, are preferentially located toward the center of the nucleosome. This study helps clarify the biophysical rules governing the Sox2-Oct4 partnership and suggests that genes may be differentially regulated by the same set of TFs on the basis of their motif positioning in the nucleosomal context.
RESULTS
Single-Molecule Analysis of Sox2 Binding to DNA
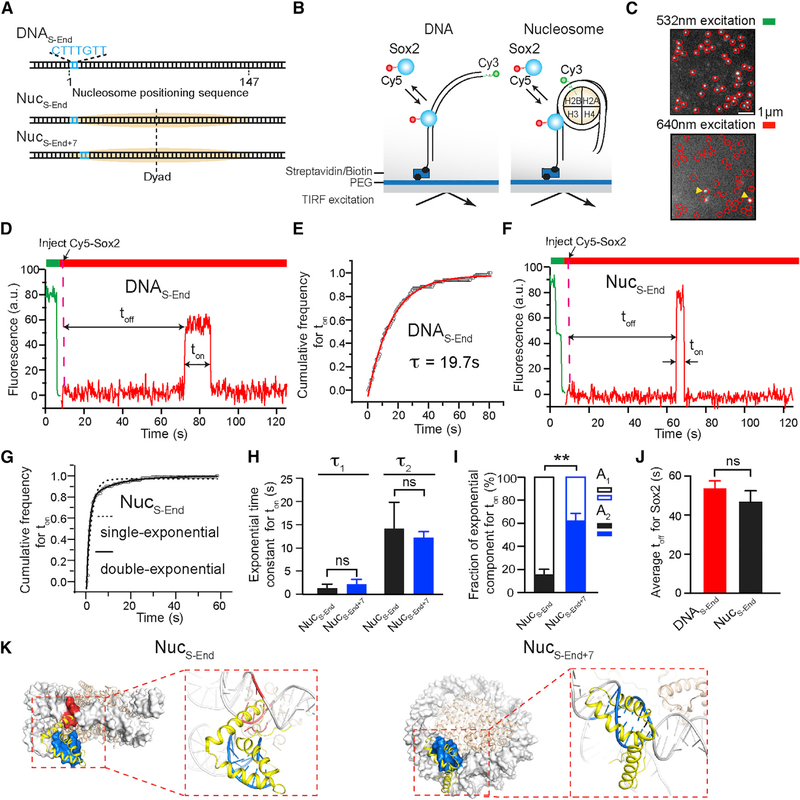
We labeled the purified full-length human Sox2 with a Cy5 fluorophore near its C terminus (Figure S1). We also constructed a DNA template containing the 601 NPS (Lowary and Widom, 1998) with a canonical Sox2 binding motif (CTTTGTT) located at its end (nucleotides 1–7), which was termed DNAS-End (Figure 1A). Cy3-labeled DNA templates were immobilized on a glass coverslip and their locations visualized using total internal reflection fluorescence (TIRF) microscopy (Figures 1B and 1C). Cy5-Sox2 was injected into the flow chamber at a concentration of ~2 nM. Binding and dissociation of individual Sox2 molecules at the DNA loci were monitored in real time (Figure 1C). Single-molecule fluorescence trajectories allowed us to measure the waiting time (toff) before the first binding event occurred, as well as the residence time (ton) of Sox2 on DNA (Figure 1D). The distribution of ton built from many binding events can be well fit by a single-exponential function (Figures 1E and S2). After correcting for dye photobleaching (Figure S1C), we determined the characteristic lifetime (τ) of Sox2 binding to DNAS-End, governed by the corresponding dissociation rate constant, to be 19.7 ± 2.3 s.
Figure 1. Sox2 Displays Differential Binding Kinetics on DNA and Nucleosome Substrates.
(A) Diagram of DNA and nucleosome substrates containing a Sox2 binding motif (blue) located near the end of a 601 NPS (orange).
(B) Schematic of the single-molecule TF binding assay using a total internal reflection fluorescence microscope.
(C) A representative field of view under green and red laser excitation showing immobilized substrates (marked by circles) and bound TFs (marked by arrowheads), respectively.
(D) A representative fluorescence-time trajectory showing Cy5-labeled Sox2 binding to a Cy3-labeled DNAS-End substrate. A 532 nm laser was first turned on to locate the surface-immobilized substrates. Then a 640 nm laser was switched on to monitor Sox2 binding and dissociation. Sox2 was injected at the time point indicated by the dashed line. The waiting time before the first binding event occurred (toff) and the lifetime of binding events (ton) were measured.
(E) Cumulative distribution (open circles) of the Sox2 residence time (ton) on DNAS-End and its fit (red curve) to a single-exponential function, y(t) = A × exp(−t/τ) + y0.
(F) A representative fluorescence-time trajectory showing Cy5-Sox2 binding to a Cy3-labeled NucS-End substrate. Two photobleaching steps under green laser excitation confirm the existence of two Cy3-labeled H2B, suggesting an intact nucleosome.
(G) Cumulative distribution (open circles) of the Sox2 residence time (ton) on NucS-End and its fit (solid curve) to a double-exponential function, y(t) = A1 × exp(−t/τ1) + A2 × exp(−t/τ2) + yo. The dashed curve displays a poor fit to a single-exponential function.
(H) Time constants for the two exponential components (τ1 and τ2) from the double-exponential fit shown in (G) (black bars for NucS-End, blue bars for NucS-End+7).
(I) Relative weights of the fast (A1) and slow (A2) exponential components for NucS-End (black) and NucS-End+7 (blue).
(J) Average waiting time (toff) before Sox2 binding to DNAS-End or NucS-End.
(K) The Sox2hmg:DNA structure (PDB: 1GT0; yellow) superimposed on the 601 nucleosome structure (PDB: 3LZ0; gray) aligned by the DNA motif (blue), which spans nucleotides 1–7 for NucS-End (left) or nucleotides 8–14 for NucS-End+7 (right). Steric clash between Sox2 and the nucleosome is highlighted in red.
Data are represented as mean ± SD. See also Figures S1, S2, and S3.
Sox2 Fails to Stably Bind to End-Positioned Nucleosomal DNA
Sox2 is considered a PF, capable of targeting DNA sites within nucleosome-occupied genomic regions (Zaret and Mango, 2016). To directly observe the behavior of Sox2 on nucleosomal DNA, we created a mononucleosome substrate, termed NucS-End, which was assembled with DNAS-End and Cy3-labeled histone H2B as well as unlabeled H2A, H3, and H4 (Figure 1A). Fluorescence signal from Cy3-H2B was used to localize individual nucleosome substrates on the surface (Figure 1F). We used DNase I digestion followed by sequencing to show that NucS-End shares an identical digestion pattern with nucleosomes assembled with the unmodified 601 NPS (Figure S3), suggesting that nucleosome positioning is not perturbed by the engineered Sox2 binding motif.
Interestingly, we found that the average residence time of Sox2 on NucS-End (4.6 ± 1.7 s) is 5-fold shorter than that on DNAS-End. The ton distribution on NucS-End cannot be described by a single-exponential model but is well fit by two exponential components: a predominant, shorter lived population (τ1 = 1.3 s, fraction A1 = 85%) and a rare, longer lived one (τ2 = 14.1 s, A2 = 15%) (Figures 1G–1I). The fast component likely corresponds to the nonspecific sampling of Sox2 on chromatin substrates as reported before (Chen et al., 2014). The slow component, on the other hand, likely represents the specific interaction between Sox2 and its cognate DNA sequence. We also compared the average toff between DNAS-End and NucS-End and found no significant difference (Figure 1J), suggesting similar association rates for Sox2 binding to DNA and nucleosome substrates.
Notably, the specific binding mode of Sox2 on NucS-End constitutes only a minor fraction of all binding events, indicating that nucleosome packing makes the DNA motif less accessible to Sox2. Indeed, when we superimposed the Sox2-DNA structure with the nucleosome structure, we observed substantial steric clash of the Sox2 HMG domain against the histone H3 and the neighboring DNA gyre (Figure 1K; NucS-End). Then we moved the Sox2 binding motif inward (toward the nucleosome dyad) by 7 bp and created a new nucleosome substrate, NucS-End+7. This change shifts the phasing of the minor groove face of the DNA motif, which is recognized by Sox2, relative to the histone octamer, such that the steric hindrance between Sox2 and the nucleosome is much reduced (Figure 1K; NucS-End+7). Accordingly, we observed a significantly larger fraction of long-lived binding events compared with NucS-End (A2 = 62% for NucS-End+7 versus 15% for NucS-End; Figure 1I), supporting the notion that these events correspond to the specific Sox2-nucleosome interaction mode.
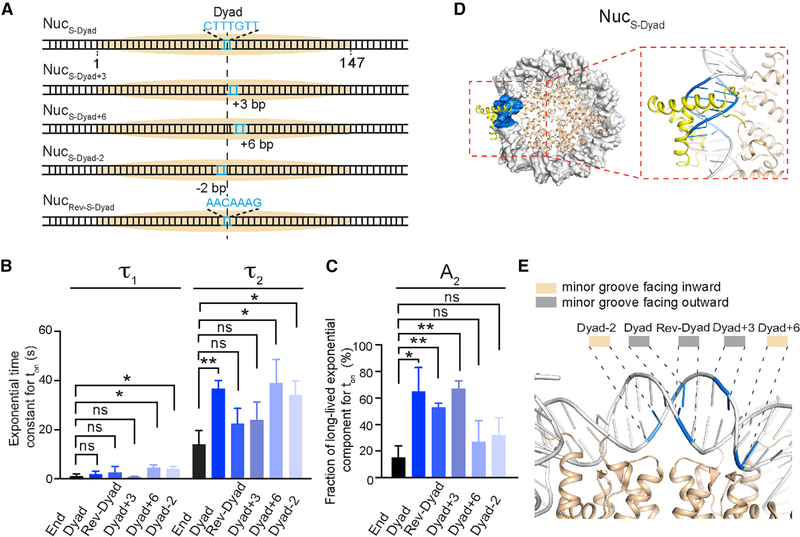
Sox2 Stably Engages with Binding Sites near the Nucleosome Dyad
The data above show that nucleosome wrapping can significantly inhibit Sox2 binding to an end-positioned nucleosomal DNA motif. To test whether this inhibitory effect is universal among all nucleosomal DNA sites, we placed the Sox2 binding motif at the center of the 601 NPS (nucleotides 72–78) and assembled a nucleosome substrate, NucS-Dyad (Figure 2A). In stark contrast to its behavior on NucS-End, Sox2 exhibited prolonged binding to NucS-Dyad (an average ton of 22.2 s for NucS-Dyad versus 4.6 s for NucS-End). The residence time distribution for Sox2 on NucS-Dyad is also characterized by a double-exponential function (τ1 = 2.1 s, τ2 = 36.8 s). Importantly, specific Sox2 binding events on NucS-Dyad are both longer lived (τ2) and more prevalent (A2) than those on NucS-End (Figures 2B and 2C, compare End and Dyad columns), indicating that the dyad position presents an optimal environment for Sox2 engagement. This interpretation is supported by structural superposition that shows minimal interference imposed by histones and nucleosomal DNA on Sox2 (Figure 2D). This is also in accordance with a recent study reporting that Sox family TFs exhibit preferred binding around the nucleosome dyad region (Zhu et al., 2018).
Figure 2. The Pioneer Activity of Sox2 Is Regulated by the Rotational Phasing of Its Binding Motif.
(A) Diagram of nucleosome substrates harboring a Sox2 binding motif around the nucleosome dyad axis. In the NucRev-S-Dyad construct, the Sox2 motif is placed in the opposite direction to those in the other constructs.
(B) Time constants for the fast and slow exponential components (τ1 and τ2) that describe the residence time of Sox2 on different nucleosome substrates.
(C) Fraction of long-lived, specific Sox2 binding events (A2) for the different nucleosome substrates.
(D) Structural superposition illustrating the putative binding configuration of Sox2 on the NucS-Dyad substrate. The Sox2 HMG domain and the DNA motif are shown in yellow and blue, respectively.
(E) Zoomed-in view of the nucleosome dyad region displaying the orientation of the DNA minor groove. The midpoints of the Sox2 binding motif placed at different positions (Dyad-2, Dyad, Rev-Dyad, Dyad+3, and Dyad+6) are indicated in blue.
Data are represented as mean ± SD. See also Figures S2, S3, and S4.
To evaluate whether the extended dyad region is in general favored by Sox2, we generated three more nucleosome substrates by shifting the Sox2 motif away from the dyad axis, either to the right by 3 or 6 bp or to the left by 2 bp (NucS-Dyad+3, NucS-Dyad+6, and NucS-Dyad-2, respectively; Figure 2A). We found that all dyad-positioned sites can better accommodate specific Sox2 binding than the end-positioned site, but to different degrees (Figures 2B and 2C). NucS-Dyad and NucS-Dyad+3, in which the minor groove of the Sox2 binding motif is mostly outward facing, have higher fractions of long-lived binding. On the other hand, the minor groove is mostly inward facing for NucS-Dyad+6 and NucS-Dyad-2, which also feature lower fractions of specific Sox2 interaction (Figures 2C–2E and S4A–S4C). Thus, even though Sox2 generally prefers the dyad region, the likelihood and lifetime of its specific nucleosome-binding mode are nonetheless influenced by the rotational phasing of the DNA motif.
Next we asked whether the direction of Sox2 on DNA plays any role in its nucleosome-targeting activity. To this end, we engineered a reverse Sox2 motif at the dyad axis by placing the canonical CTTTGTT sequence in the opposite strand and reconstituted another nucleosome substrate, termed NucRev-S-Dyad (Figure 2A). This change flips the direction of the bound Sox2 on DNA but keeps the minor groove outward facing (Figure S4D). We found that the lifetime of the specific binding events (τ2) on NucRev-S-Dyad is reduced compared with NucS-Dyad (Figure 2B), which may be attributed to increased steric hindrance or altered local sequence context. The relative weight of the specific binding mode (A2) on NucRev-S-Dyad still remains high (Figure 2C), which can be explained by the outward-facing minor groove (Figure 2E). Collectively, these results demonstrate that the pioneer activity of Sox2 is sensitively modulated by the position, orientation, and polarity of its binding motif within the nucleosome.
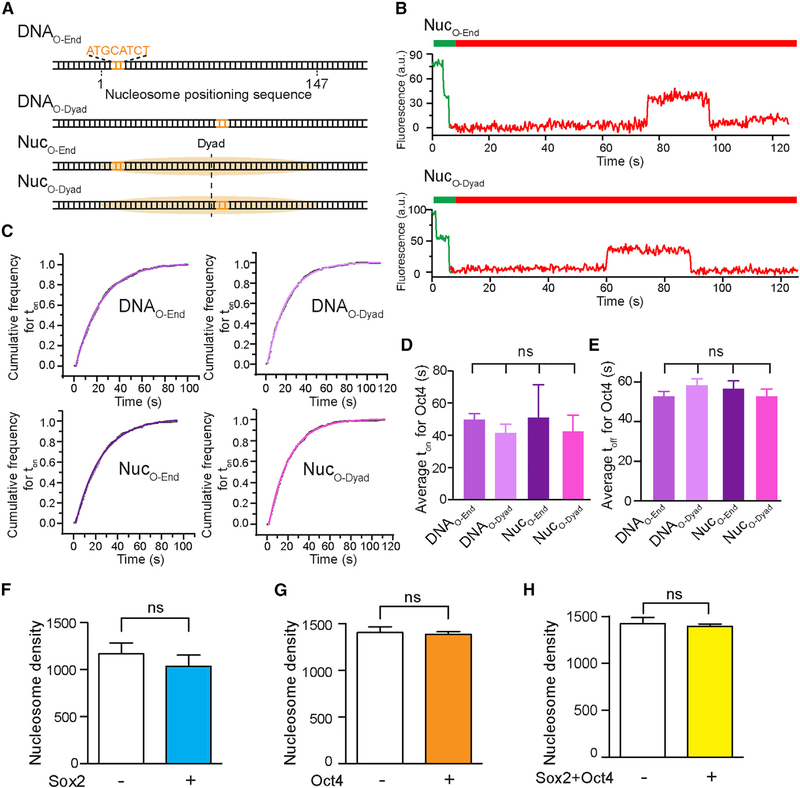
Oct4 Binds Equally to End- and Dyad-Positioned Nucleosomal DNA Motifs
Next we examined the behavior of the other core pluripotency TF, Oct4, on DNA and nucleosome substrates. We purified and fluorescently labeled full-length human Oct4 with Cy5 (Figure S1), incorporated an 8-bp-long Oct4 binding motif into the 601 NPS at either the end or the dyad position (DNAO-End and DNAO-Dyad) and assembled nucleosomes with these DNA templates (NucO-End and NucO-Dyad; Figure 3A). We then conducted single-molecule TIRF experiments to measure the interaction between Oct4 and these substrates (Figure 3B). The ton distribution of Oct4 on each substrate can be well described by single-exponential kinetics (Figure 3C). Interestingly, unlike Sox2, the residence times (ton) of Oct4 on these substrates are statistically identical among each other (Figure 3D), as are the average toff values that describe the association rates of Oct4 (Figure 3E). These results suggest that Oct4 displays no discrimination between bare DNA and nucleosome substrates or between end-positioned and dyad-positioned nucleosomal DNA motifs (Figures S4E and S4F). In addition, reversing the direction of the nucleosomal DNA motif does not significantly change the binding behavior of Oct4 either (Figures S4G and S4H).
Figure 3. The Nucleosome-Targeting Activity of Oct4 Is Insensitive to Its Motif Position.
(A) Diagram of DNA and nucleosome substrates containing an Oct4 binding motif located near the end or dyad of the 601 NPS.
(B) Representative single-molecule fluorescence trajectories showing Cy5-labeled Oct4 binding to nucleosome substrates labeled with Cy3.
(C) Cumulative distributions (open circles) of the Oct4 residence time on different DNA and nucleosome substrates and their respective single-exponential fits (curves).
(D) Average Oct4 residence times (ton) on different substrates.
(E) Average waiting time (toff) before Oct4 binding to a DNA or nucleosome substrate, which reports on the corresponding association rate.
(F) Average number of surface-immobilized and fluorescently labeled nucleosomes per field of view before (white bar) and 10 min after (blue bar) the addition of 2 nM Sox2.
(G) Surface density of fluorescent nucleosomes before (white bar) and after (orange bar) the addition of 2 nM Oct4.
(H) Surface density of fluorescent nucleosomes before (white bar) and after (yellow bar) the addition of both Sox2 and Oct4. The results in (F)-(H) suggest that Sox2 and Oct4 binding does not cause significant nucleosome disassembly.
Data are represented as mean ± SD. See also Figures S1, S2, and S4.
We note that in all of the Sox2/Oct4 binding experiments described above, the density of fluorescent nucleosomes on the surface did not appreciably decrease after addition of the TFs (Figures 3F–3H), suggesting that Sox2/Oct4 binding does not evict histones from the DNA.
Nonreciprocal Regulation between Sox2 and Oct4 in Nucleosome Binding
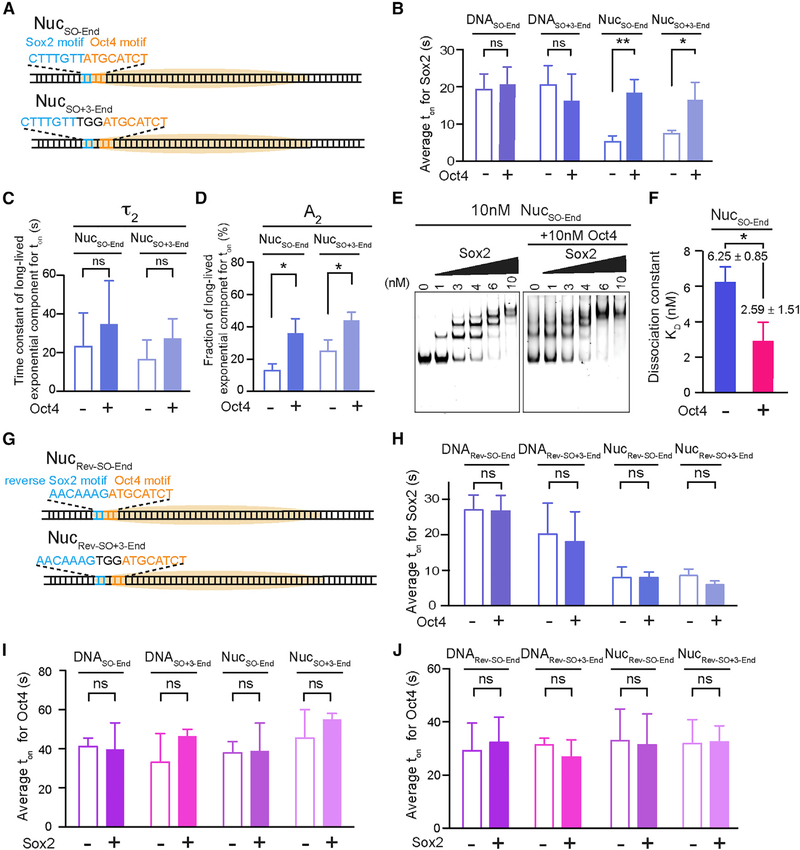
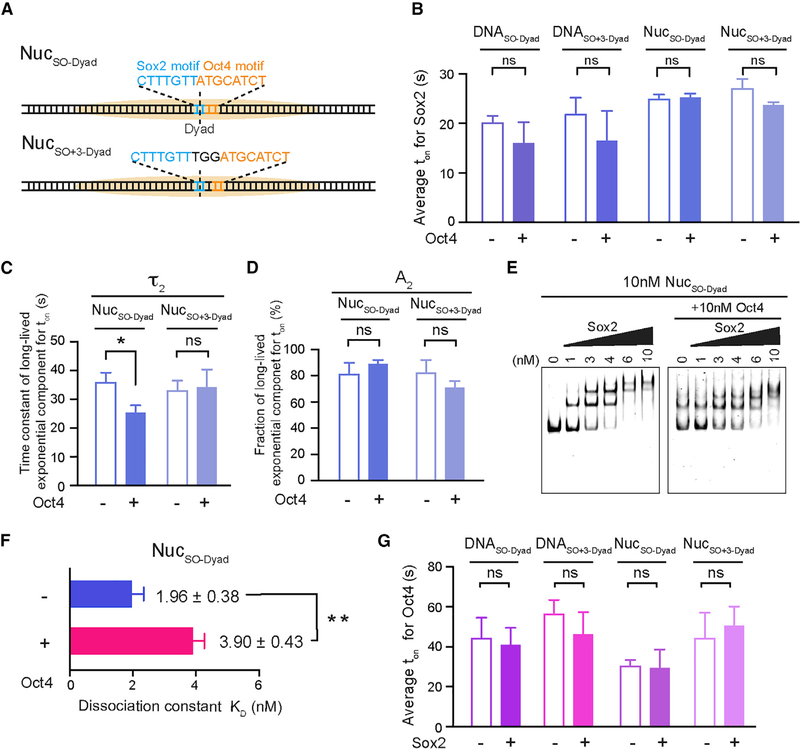
Having characterized the individual behaviors of Sox2 and Oct4, next we set out to interrogate the cooperativity between this TF pair in nucleosome targeting. First we engineered a composite Sox2:Oct4 binding motif into the 601 NPS at the end position (nucleotides 1–15) and assembled the nucleosome substrate, termed NucSO-End (Figure 4A). We then examined the effect of Oct4 on Sox2 binding by complementing Cy5-Sox2 with unlabeled Oct4 in the single-molecule experiments. Remarkably, we found that Oct4 prolonged the average residence time of Sox2 on NucSO-End by 3-fold (Figure 4B). A detailed kinetic analysis revealed that the lengthened dwell time is attributed mainly to increased weight of the specific Sox2 binding mode (A2 = 13% without Oct4 versus 36% with Oct4; Figures 4C and 4D). On the other hand, Oct4 had little impact on the association kinetics of Sox2 (Figure S5A). Therefore, the net effect is an enhanced affinity of Sox2 to an end-positioned nucleosomal target in the presence of Oct4. This conclusion was corroborated by the bulk electrophoretic mobility shift assay (EMSA), which showed that the dissociation constant (KD) for Sox2-NucSO-End interaction is smaller in the presence of Oct4 than in its absence (Figures 4E, 4F, and S5C). Notably, the stimulatory effect of Oct4 on Sox2 is restricted to the nucleosome targets, as Oct4 exerts no influence on Sox2 interaction with the bare DNA substrate DNASO-End (Figures 4B and S5A).
Figure 4. Oct4 and Sox2 Exhibit Nonreciprocal Cooperativity.
(A) Diagram of nucleosome substrates containing an end-positioned Sox2:Oct4 composite motif, either with no gap (NucSO-End) or with a 3 bp gap between the Sox2 and Oct4 binding motifs (Nucso+3-End).
(B) Average residence times (ton) of Sox2 on different DNA and nucleosome substrates shown in (A) in the absence or presence of Oct4.
(C) Time constants for the long-lived, specific Sox2 binding mode (τ2) on NucSO-End and NucSO+3-End in the absence or presence of Oct4.
(D) Relative populations of specific Sox2 binding events (A2) on NucSO-End and NucSO+3-End in the absence or presence of Oct4.
(E) A representative EMSA gel showing the formation of Sox2-NucSO-End complexes, or the formation of Sox2-NucSO-End-Oct4 ternary complexes when Oct4 is present, at different Sox2 concentrations. Cy5-labeled Sox2 and unlabeled Sox2 exhibited virtually identical binding patterns (not shown).
(F) Dissociation constant (KD) for the Sox2-NucSO-End interaction in the absence or presence of Oct4 determined from the EMSA results. n = 3 experimental replicates.
(G) Diagram of nucleosome substrates containing an end-positioned composite motif in which Sox2 and Oct4 sites are oriented in opposite directions, either with no gap (NucRev-SO-End) or with a 3 bp gap (NucRev-SO+3-End).
(H) Average residence times (ton) of Sox2 on different DNA and nucleosome substrates shown in (G) in the absence or presence of Oct4.
(I) Average residence times (ton) of Oct4 on different DNA and nucleosome substrates containing an end-positioned composite motif in the absence or presence of Sox2.
(J) Same as (I), except that the substrates contain a composite motif in which Sox2 and Oct4 sites are oriented in opposite directions.
Data are represented as mean ± SD. See also Figures S5 and S6.
Besides the canonical composite motif in which Sox2 and Oct4 binding sites are immediately juxtaposed with each other, a variant motif composed of Sox2 and Oct4 sites separated by 3 bp is also found in some cis-regulatory elements, such as the Fgf4 enhancer (Ambrosetti et al., 1997). Thus we generated a nucleosome substrate that contains such a gapped composite motif at its end position (NucSO+3-End; Figure 4A) and found that Oct4 exerts a similar, albeit somewhat weaker, positive effect on Sox2 engagement (compare NucSO+3-End with NucSO-End, Figures 4B–4D).
To assess how the relative direction of Sox2 and Oct4 binding in a composite site might influence their nucleosome-mediated cooperativity, we juxtaposed a reverse Sox2 motif and a forward Oct4 motif at the nucleosome end position, either with no gap or with a 3 bp gap (NucRev-SO-End and NucRev-SO+3-End; Figure 4G). In contrast to the results for NucSO-End and NucSO+3-End, Oct4 did not stabilize Sox2 binding in the oppositely oriented configuration (Figure 4H).
We then conducted the converse experiments by using Cy5-Oct4 and unlabeled Sox2 to check the influence of Sox2 on the interaction of Oct4 with DNA and nucleosome substrates. Neither ton nor toff of Oct4 on any of these substrates was significantly affected by Sox2 (Figures 4I, 4J, and S5B). Therefore, despite both being classified as PFs, Oct4 can stabilize the engagement of Sox2 with certain nucleosomal DNA sites but not vice versa.
Hierarchically Ordered Targeting of Oct4 and Sox2 to Nucleosomes
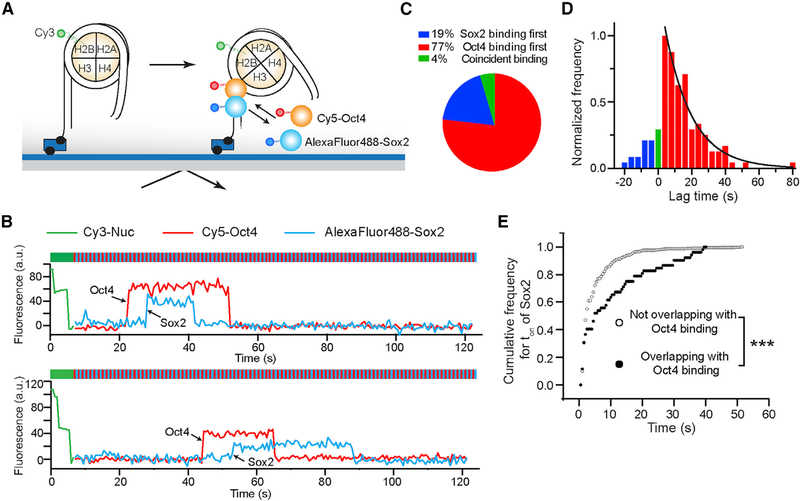
To directly follow the order of binding by Sox2 and Oct4 to the same nucleosome target, we labeled the TF pair with distinct fluorophores—Oct4 with Cy5 and Sox2 with AlexaFluor488—and used an alternating laser excitation scheme to simultaneously monitor their behaviors (Figure 5A). We found that the vast majority of overlapping Sox2/Oct4 binding events featured Oct4 binding first followed by Sox2 arrival (Figures 5B–5D). This result, together with the single-color residence time measurements described above, suggests that Oct4 facilitates the stable interaction of Sox2 with target sites located inside the nucleosome. Consistently, in the dual-color experiment, the Sox2 binding events that overlapped with an Oct4 binding event were significantly longer lived than those that did not overlap (Figure 5E).
Figure 5. Hierarchical Engagement of Oct4 and Sox2 in Nucleosome Targeting.
(A) Schematic of the three-color TIRF assay that simultaneously monitors Sox2 and Oct4 binding. Histone H2B, Oct4, and Sox2 are labeled with Cy3, Cy5, and AlexaFluor488, respectively.
(B) Representative fluorescence-time trajectories showing overlapping Sox2 and Oct4 binding events on NucSO-End, which reveal the order of TF engagement.
(C) Pie chart showing the distribution of different scenarios regarding the order of Sox2/Oct4 binding to nucleosome substrates. n = 134 (number of overlapping Sox2 and Oct4 binding events analyzed).
(D) Histogram of the lag time between Oct4 and Sox2 binding. The positive part of the histogram (red) corresponds to Oct4-first events and is fit to a single-exponential function (black curve).
(E) Cumulative distributions of the Sox2 residence time for the binding events that overlapped with an Oct4 binding event (filled circles) and for those that did not overlap (open circles) (p = 3.3 × 10−11, two-sided Kolmogorov-Smirnov test).
See also Figures S5 and S6.
Effect of Oct4 on the Nucleosome Binding Activity of Sox2 Is Position Dependent
We then asked whether the enhanced binding of Sox2 in the presence of Oct4 could be observed at other nucleosomal positions besides the end. In particular, we were curious as to whether binding to the nucleosome dyad, which is already favored by Sox2, could be further stimulated by Oct4. To answer this question, we placed a canonical Sox2:Oct4 composite motif at the dyad position (nucleotides 72–86) of the 601 NPS (NucSO-Dyad; Figure 6A). Surprisingly, this construct yielded a markedly different picture than NucSO-End: Oct4 had a negligible impact on ton of Sox2 on NucSO-Dyad (Figure 6B); it moderately reduced the lifetime of specific Sox2 binding (Figure 6C), but did not affect its relative population (Figure 6D). Oct4 also did not cause a noticeable change in toff of Sox2 on NucSO-Dyad (Figure S5A). Moreover, EMSA results showed that Kd for the Sox2-NucSO-Dyad interaction was increased by the presence of Oct4 (Figures 6E, 6F, and S5D). These results together illustrate that instead of promoting Sox2 binding as observed at end-positioned sites, Oct4 weakly diminishes the affinity of Sox2 to the nucleosome dyad. We speculated that this mild inhibition might be caused by the geometrical interference between the two TFs. Indeed, when a 3 bp gap was inserted between Sox2 and Oct4 binding sites at the dyad position (NucSO+3-Dyad; Figure 6A), the negative effect of Oct4 on the specific Sox2 binding mode was attenuated (Figure 6C). On the other hand, Sox2 exerts minimal influence on Oct4 binding to dyad-positioned sites (Figures 6G and S5B), similar to the results obtained with end-positioned sites.
Figure 6. Oct4 Negatively Influences Sox2 Binding to the Nucleosome Dyad.
(A) Diagram of nucleosome substrates containing a dyad-positioned Sox2:Oct4 composite motif, either with no gap (NucSO-Dyad) or with a 3 bp gap between the Sox2 and Oct4 motifs (NucSO+3-Dyad).
(B) Average residence times (ton) of Sox2 on different DNA and nucleosome substrates shown in (A) in the absence or presence of Oct4.
(C) Time constants for the specific Sox2 binding mode (τ2) on NucSO-Dyad and NucSO+3-Dyad in the absence or presence of Oct4.
(D) Relative populations of specific Sox2 binding events (A2) in the absence or presence of Oct4.
(E) A representative EMSA gel showing the formation of Sox2-NucSO-Dyad complexes at different Sox2 concentrations in the absence or presence of Oct4.
(F) Dissociation constant (KD) for the Sox2-NucSO-Dyad interaction in the absence or presence of Oct4 determined from the EMSA results. n = 3 experimental replicates.
(G) Average residence times (ton) of Oct4 on different DNA and nucleosome substrates containing a dyad-positioned composite motif in the absence or presence of Sox2.
Data are represented as mean ± SD. See also Figures S5 and S6.
Overall, our data show that the effect of Oct4 on the nucleosome binding activity of Sox2 could be positive, negative, or neutral depending on the position and configuration of the composite motif in the nucleosome context. Moreover, it is worth mentioning that the nonreciprocal and conditional cooperativity between Oct4 and Sox2 can be observed at different salt concentrations (Figures S5E and S5F), indicating that it is a generalizable phenomenon independent of the particular state of nucleosomal DNA accessibility.
Pioneer Activities of Pluripotency TFs at a Native Genomic Locus
Next we explored the binding behavior of Sox2 and Oct4 at a natural genomic site. We chose the human LIN28B locus, which encodes a key protein regulating cell pluripotency and reprogramming (Shyh-Chang and Daley, 2013). We cloned a 162-bp-long DNA segment from this region, which is occupied by a well-positioned nucleosome and targeted by both Sox2 and Oct4 (Soufi et al., 2015). We then used this DNA template to reconstitute nucleosomes, termed NucLIN28B. The predicted cognate Sox2 and Oct4 binding motifs are both located between the end and center of NucLIN28B (Figure S6A) (Soufi et al., 2015). We found that the average residence time of Sox2 on NucLIN28B is longer than that on NucS-End but shorter than that on NucS-Dyad (Figure S6B). In contrast, ton for Oct4 on NucLIN28B is indistinguishable from those on NucO-End and NucO-Dyad (Figure S6C). These results lend further support to our conclusion that Sox2’s pioneer activity is position dependent, while Oct4’s is not. We did not observe clear cooperativity between Sox2 and Oct4 on NucLIN28B (Figures S6D and S6E). This can be rationalized by the fact that the Sox2 and Oct4 binding motifs within NucLIN28B are oriented in opposite directions and separated by 3 bp, consistent with the results for NucRev-SO-End and NucRev-SO+3-End (Figures 4G and 4H).
The LIN28B locus is also bound by c-Myc, another Yamanaka TF that, unlike Oct4 and Sox2, is thought to lack the pioneer activity and preferentially bind to open chromatin (Soufi et al., 2015). We purified and fluorescently labeled c-Myc together with its heterodimeric partner Max (Figure S1) and tested its ability to bind DNA and nucleosome substrates at the single-molecule level (Figure S6F). We found that c-Myc bound to DNALIN28B and NucLIN28B much more transiently compared with Sox2 and Oct4 (Figure S6G). Thus, c-Myc possesses an inherent, albeit weak, ability to target nucleosomes.
Genome-wide Binding Preference of Sox2 and Oct4 with Respect to Nucleosome Positioning
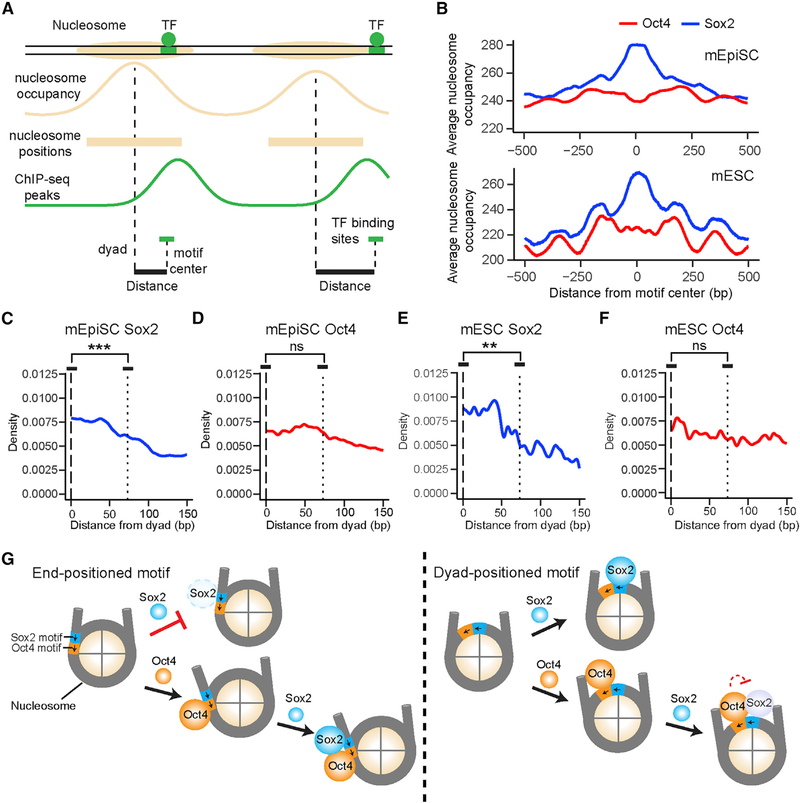
The in vitro single-molecule data described above revealed the differential positional preference of Sox2 and Oct4 for nucleosomal DNA. To investigate whether such a difference can be recapitulated on a genomic scale, we mined published nucleosome mapping and Sox2/Oct4 ChIP-seq data and interrogated the distributions of Sox2 and Oct4 binding sites relative to nucleosome locations (Figure 7A). We first calculated the aggregate nucleosome-positioning scores surrounding Sox2/Oct4 binding sites in mouse epiblast stem cells (mEpiSCs) (Matsuda et al., 2017) and mouse embryonic stem cells (mESCs) (Teif et al., 2012). In both cell types, the average nucleosome occupancy across all Sox2 binding sites is greater than that for all Oct4 binding sites (Figure 7B), indicating that Sox2 binding sites are more enriched in nucleosome-occupied regions.
Figure 7. Genome-wide Analysis of the Positional Preference of Sox2 and Oct4 Binding Relative to Nucleosome Positioning.
(A) Diagram illustrating nucleosome occupancy and TF binding sites in the genome. Nucleosome positions were derived from MNase-seq data. TF binding sites were identified by searching for a cognate sequence motif near a ChIP-seq peak for the given TF.
(B) Nucleosome occupancy scores within a 1,000 bp window surrounding a Sox2 (blue) or Oct4 (red) binding site averaged over all sites identified in mouse epiblast stem cells (top) and embryonic stem cells (bottom). Position 0 corresponds to the center of an identified TF binding site.
(C) Distribution of the distance between the center of a Sox2 binding site and the nearest nucleosome dyad in mEpiSCs (n = 14,101; n denotes the number of binding sites analyzed). Position 0 (dashed line) corresponds to the dyad; the dotted line approximates the edges of the nucleosome. Displayed significance is from t test conducted between a 13 bp window centered at the dyad and a 13 bp window inside the nucleosome edge (p = 3.6 × 10−7).
(D) Same as (C), except for analyzing the distribution of Oct4 binding sites with respect to the nearest nucleosome (n = 21,050, p = 0.47).
(E) Same as (C), except for analyzing the distribution of Sox2 binding sites in mESCs (n = 1,438, p = 0.0027).
(F) Same as (C), except for analyzing the distribution of Oct4 binding sites in mESCs (n = 2,437, p = 0.073).
(G) Schematic model illustrating the differential pioneer activity of Sox2 and Oct4, as well as their position-dependent cooperativity in the nucleosome context. The short arrows indicate the relative orientation of Sox2 and Oct4 binding motifs.
See also Figure S7.
We then plotted the distribution of Sox2/Oct4 binding sites in mEpiSCs averaged over all nucleosome-bound regions aligned by their dyad positions. Within the 147 bp window corresponding to the nucleosome, Sox2 binding sites exhibited a strong preference for the dyad region over the edges of the nucleosome (Figure 7C). In contrast, the distribution of Oct4 binding sites within the nucleosome showed no significant difference between nucleosome dyad and ends (Figure 7D). Similar patterns were observed in mESCs (Figures 7E and 7F) and human ESCs (Figure S7). Intriguingly, the distributions of noncanonical Sox2 and Oct4 motifs in the human genome, which are enriched in nucleosome-occupied regions as previously reported (Soufi et al., 2015), showed distinct patterns from those of canonical motifs (Figure S7), implying that the genome encodes subtle and diverse regulatory information that instructs TF targeting.
DISCUSSION
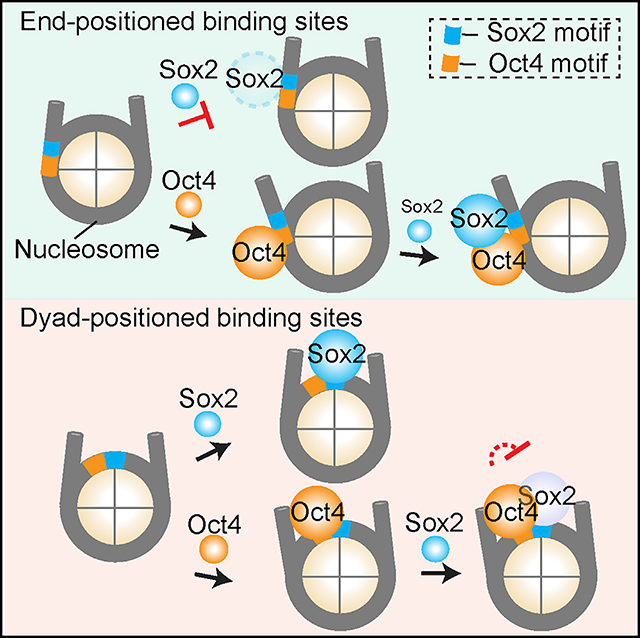
TF binding positions are usually classified into nucleosome- depleted and nucleosome-enriched regions. In this work, we went beyond such general localization analysis by dissecting the dynamic binding pattern of a TF within a nucleosome. Compared with bulk biochemical and genome-wide binding assays, our single-molecule platform affords higher temporal resolution and unique kinetic information, which enabled us to quantitatively determine the cooperativity between pluripotency TFs in the nucleosome context and discover an unexpectedly intricate Sox2-Oct4 partnership (Figure 7G). These results are corroborated by analysis of the in vivo genomic data, suggesting that the biophysical principles revealed by the in vitro reconstituted system may indeed be exploited by the cell to perform gene regulation.
Distinct Nucleosome-Targeting Properties of PFs
TFs generally prefer binding to nucleosome-depleted regions in the genome (Wang et al., 2012), with the exception of PFs that are able to access closed chromatin (Slattery et al., 2014; Zaret and Mango, 2016). The ensuing question then is, does a PF target all nucleosomal DNA sites with equivalent kinetics? We show that, at least for some PFs, the answer is no. In accordance with earlier bulk studies (Liu and Kraus, 2017; Zhu et al., 2018), our single-molecule results demonstrate that the pioneer activity of Sox2 is sensitive to both translational and rotational positioning of its cognate motif within the nucleosome. Dyad-positioned sites support prolonged Sox2 binding compared with end-positioned sites. This may be because the dyad region, where only one DNA gyre is wound, can better accommodate DNA bending and minor groove widening that are essential for recognition by the HMG domain of Sox2 (Scaffidi and Bianchi, 2001). The specific Sox2 binding mode at the dyad region is even more stable than Sox2 interaction with bare DNA, implying additional contacts between the histones and Sox2. As to the rotational setting of the Sox2 binding motif, a more accessible minor groove (facing away from the histone octamer) generally corresponds to a higher affinity. A preference for specific motif positioning in the nucleosome context has also been suggested for other TFs, such as p53, which favors exposed sites (Cui and Zhurkin, 2014) and nucleosome edges (Yu and Buck, 2019).
On the contrary, we found no positional preference for Oct4, which indiscriminately targets nucleosomal DNA at different locations. This could be rationalized by the fact that Oct4 contains two DNA-binding domains (POUS and POUHD), each recognizing a 4 bp half-motif locating at opposite sides of the DNA helix (Esch et al., 2013). Regardless of the nucleosomal DNA position, one of these half-motifs is solvent exposed, which is apparently sufficient for mediating stable Oct4-nucleosome interaction. In support of this idea, partial motifs are prevalently found in Oct4 target sites located within nucleosome-enriched genomic regions (Soufi et al., 2015). It was recently reported that POU family TFs exhibit a preference toward nucleosome edges (Zhu et al., 2018). However, this conclusion was drawn from the behaviors of the TF DNA-binding domains, while we used full-length Oct4 in the current study.
Therefore, although both regarded as PFs, Sox2 and Oct4 display drastically different nucleosome binding profiles. We note that our interpretations are based on the superposition between the available structures of TF-DNA complexes and unbound nucleosomes. It is conceivable that the nucleosome structure may be remodeled upon TF engagement.
We found that c-Myc, previously thought not to have an intrinsic nucleosome-targeting capability, can nonetheless bind to nucleosomal DNA, albeit transiently. With more sensitive methods such as single-molecule imaging being deployed to interrogate TF-chromatin interaction, the list of nucleosome-binding TFs is expected to continue to grow. Our data further suggest that the pioneer activity of these TFs, governed by the structural characteristics of their respective DNA-binding domains, is not a binary property but rather falls on a continuous spectrum.
Nonreciprocal and Conditional TF-TF Cooperativity
Clustered binding of TFs is a hallmark of cis-regulatory elements, such as promoters and enhancers, which integrate multiple TF inputs to direct gene expression. High-throughput methods have been developed to systematically determine the binding patterns of TF pairs on DNA (Chang et al., 2017; Jolma et al., 2015; Siggers et al., 2011; Slattery et al., 2011). However, the biophysical basis for cooperative TF binding in the nucleosome context remains underexplored. In particular, the relationship of chromatin targeting between a PF and a non-pioneer factor, and between a pair of PFs, is still under debate. Using the Sox2-Oct4 TF pair and 601 NPS as a model system, our study sheds new light on this issue. First of all, cooperativity can be unilateral. Oct4 stabilizes the binding of Sox2 to an end-positioned motif, perhaps by opening up a stretch of nucleosomal DNA and generating a local environment permissive to Sox2 binding (Figure 7G). Conversely, Sox2 has no effect on the behavior of Oct4 on the nucleosome. Using multi-color imaging, we directly followed the order of TF engagement with the nucleosome and found that Oct4 preceding Sox2 is the predominant scenario. This is notably different from live-cell results (Chen et al., 2014), which concluded that Sox2 is the lead TF guiding Oct4 to its target sites. This conclusion was based on single-color imaging as well as mathematical modeling and was later challenged by theoretical re-analysis (Biddle et al., 2019). Such discrepancy could be due to the heterogeneous chromatin states inside the cell, which may complicate data interpretation. In any case, the pioneer activity of a TF appears to be hierarchical, reinforcing the aforementioned notion that it should not be considered as an all-or-none trait.
Second, we showed that the Sox2-Oct4 cooperativity is strongly dependent on the geometry of the composite motif. Contrary to the positive effect exerted at nucleosomal-end positions, Oct4 has a negative impact on Sox2’s access to the dyad region, where Sox2 exhibits a robust pioneer activity by itself. We speculate that without the benefit of creating extra free DNA surface for Sox2, steric hindrance between the two proteins may become the deciding factor around the dyad region. Therefore, when two TFs are invading the same nucleosome, multiple mechanisms can contribute to their interplay, yielding synergistic or antagonistic binding depending on the specific motif arrangement. The determinants of Sox2-Oct4 cooperativity have been studied in depth with DNA substrates (Jauch et al., 2011; Merino et al., 2014; Reményi et al., 2003; Tapia et al., 2015). Future structural work on the Sox2-Oct4-nucleosome ternary complexes will help illuminate the mechanism by which the nonreciprocal and conditional cooperativity is accomplished.
The notion of mixed cooperativity across distinct genomic loci was also recently proposed in a theoretical study to explain in vivo Sox2 and Oct4 binding data (Biddle et al., 2019). Whether this phenomenon is applicable to other TF pairs warrants further investigation. Additionally, in this work we mainly used the 601 NPS, which has an exceptionally strong affinity to histone octamers (Vasudevan et al., 2010). It is worthwhile to evaluate to what extent the TF binding patterns hold using natural nucleosomal DNA sequences with higher accessibility (Takizawa et al., 2018).
Perspective
Combinatorial control of gene expression by specific TF circuits underlies the operation of diverse gene regulatory networks (Thompson et al., 2015). Our results point to an intriguing scenario in which the same group of TFs can activate one set of genes while repressing another set on the basis of their differential cooperativity in specific chromatin contexts. Importantly, TFs need not directly interact with one another in order to achieve nucleosome-mediated cooperativity, which greatly broadens the potential scope of this model in gene regulation. Future research seeking direct evidence for this paradigm will help us better understand how TF-chromatin association and its variation underpin normal cell physiology and disease (Deplancke et al., 2016) and how dynamic and stochastic molecular interactions lead to deterministic and precise gene expression programs.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and request for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shixin Liu (shixinliu@rockefeller.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains and growth conditions
E. coli Rosetta (DE3) pLysS cells were cultured in a Luria-Bertani (LB) medium containing 50 μg/ml kanamycin and 34 μg/ml chloramphenicol. E. coli BL21 (DE3) cells were cultured in an LB medium containing 100 μg/ml ampicillin.
METHOD DETAILS
Expression, purification, and fluorescent labeling of TFs
The human Sox2 and Oct4 genes were purchased from Addgene and the human c-Myc and Max genes were amplified from HeLa cell cDNA (US Biological T5595–0449). Each gene was cloned into the bacterial vector pET28B with a hexahistidine tag at its N terminus. All proteins were expressed in Rosetta (DE3) plyS cells (Novagen #70956–3) in an LB medium. Cell were grown at 37°C until OD600 reached 0.6, and then induced with 0.5 mM IPTG at37°Cfor4 hforOct4 or 2 hforSox2, at 30°C overnight for c-Myc, or at 25°C overnight for Max. ForSox2, Oct4, and c-Myc, cells were harvested and lysed by sonication in a denaturing buffer, followed by centrifugation at 15,000 rpmfor40 min. Proteins were first purified on a Ni-NTA affinity column. The eluted Sox2 and Oct4 were refolded by dialyzing to 2 M urea and then to 0 urea using a desalting column (GE healthcare). Further purification was carried out by gel filtration using a Superdex 200 10/300 GL column (GE Healthcare). Ni-NTA-purified c-Myc was mixed with Max at a molar ratio of 1:2.5 as described previously (Farina et al., 2004), and then purified by gel filtration as described above.
The only cysteine in Sox2 located at amino acid #265 near the flexible C terminus was labeled with Cy5 maleimide (GE healthcare) or AlexaFluor488 C5 maleimide (Thermo Fisher) at a mixing ratio of 1:1.5 after desalting column purification. An Sfp tag was introduced to the C terminus of Oct4 and c-Myc (Yin et al., 2005). After desalting column purification, the Sfp-tagged proteins were incubated with the Sfp synthase (purified in-house) and CoA-Cy5 (synthesized and purified in-house) at a molar ratio of 1:1.5:2.5 at 4°C overnight. To synthesize CoA-Cy5, the coenzyme A trilithium salt (Sigma-Aldrich) was conjugated with Cy5 maleimide at room temperature for 2 h and purified with a C18 250× 4.6mm column (Agilent) as previously described (Yin et al., 2006). Free dye molecules and the Sfp synthase were removed from the labeled proteins by gel filtration.
Preparation of histones and DNA templates and assembly of nucleosomes
Xenopus laevis histones were recombinantly expressed in BL21 (DE3) cells. H2B T49C mutant was generated by site-directed mutagenesis. The mutant histone was purified and labeled with Cy3 maleimide (GE Healthcare) under denaturing conditions (Harada et al., 2016). Histone octamers were reconstituted with equal ratio of each histone and purified by gel filtration as described previously (Luger et al., 1999). DNA templates were made by PCR using biotinylated primers and a plasmid containing a 601 NPS (Li et al., 2010) that was modified such that a Sox2 motif (CTTTGTT), a reverse Sox2 motif (AACAAAG), an Oct4 motif (ATGCATCT), a reverse Oct4 motif (AGATGCAT), a composite Sox2:Oct4 motif (CTTTGTTATGCATCT), a composite motif containing a reverse Sox2 motif (AACAAAGATGCATCT), a composite Sox2:Oct4 motif with a 3-bp spacer (CTTTGTTTGGATGCATCT), or a composite motif containing a reverse Sox2 motif and a 3-bp spacer (AACAAGTGGATGCATCT) was placed at indicated positions. The LIN28B genomic DNA fragment was purchased from IDT. The DNA products were purified by ion-exchange chromatography on a Mono Q column (GE Healthcare) and stored in TE buffer (10 mM Tris-HCl, pH 8.0, 0.1 mM EDTA). For fluorescent labeling, DNA modified with a primary amine group was mixed with Cy3 NHS ester (GE Healthcare) at room temperature for 2 h. Free dyes were subsequently removed by a Sephadex G-25 column (GE Healthcare). Nucleosomes were assembled by salt gradient dialysis as previously described (Lee and Narlikar, 2001). The assembly efficiency was optimized by titrating the DNA:octamer molar ratio. The purity of the products was evaluated on a 5% native TBE-PAGE gel.
Single-molecule experiments
Glass slides and coverslips were cleaned by sonication in acetone and 1 M KOH, followed by treatment with Nanostrip (VWR). They were then subjected to argon plasma cleaning (Harrick Plasma) followed by silanization with 2% 3-aminopropyltriethoxysilane in acetone. Cleaned slides and coverslips were passivated with a mixture of polyethylene glycol (PEG) and biotin-PEG (Laysan Bio) for 2 h, followed by a second round of PEGylation with 4 mM MS4-PEG (Thermo Fisher Scientific) for 1 h. The assembled flow chamber was infused with 40 μL of 0.2 mg/ml streptavidin (Thermo Fisher Scientific), incubated for 5 min, and washed with 100 μL of T50 buffer (10 mM Tris-HCl, pH 7.5, 50 mM NaCl). Biotinylated DNA or nucleosome substrates were injected into the chamber and immobilized through streptavidin-biotin linkage. Single-molecule imaging was conducted on a total-internal-reflection fluorescence microscope (Olympus IX83 cellTIRF) equipped with an EMCCD camera (Andor iXon Ultra897). Unless noted otherwise, the imaging buffer contained 40 mM Tris-HCl, 12 mM HEPES, pH 7.5, 60 mM KCl, 3 mM MgCl2, 10% (v/v) glycerol, 0.02% (v/v) Igepal CA-630, 0.32 mM EDTA, 0.5 mg/ml BSA, and an oxygen scavenging system [1% (w/v) glucose, 1 mg/ml glucose oxidase, 0.04 mg/ml catalase, 2 mM trolox (Sigma-Aldrich)]. Movies were recorded at room temperature with a frame rate of 300 ms. Positions of the immobilized substrates were determined during the initial 20 frames using a 532-nm laser. TF binding dynamics were then observed using a 640-nm laser. To simultaneously monitor two fluorescently labeled TF species, an alternating excitation scheme was adopted in which a 640-nm laser and a 488-nm laser were each switched on for one frame in an interlaced fashion. Labeled and unlabeled TFs were typically used at a concentration of 2 nM and 20 nM, respectively. Single-molecule fluorescence-time trajectories were extracted and analyzed with the SPARTAN software (Juette et al., 2016). TF binding events were identified using a fluorescence intensity threshold. Histogram building and curve fitting were performed with the Origin software (OriginLab).
Electrophoretic mobility shift assay (EMSA)
10 nM of DNA or nucleosome substrates were incubated with indicated TFs in a binding buffer (10 mM Tris-HCl, pH 7.5, 1 mM MgCl2, 1 mM DTT, 10 mM KCl, 0.5 mg/ml BSA, and 5% glycerol) at room temperature for 10 min. The reaction mixture was loaded on a 5% non-denaturing polyacrylamide gel, which was run in 0.5 × Tris-Borate-EDTA at 4°C at 80 V, stained by SYBR Gold (Invitrogen), and scanned by a Typhoon FLA 7000 gel imager (GE Healthcare). Band intensities were extracted by ImageQuant (GE healthcare). The fraction of substrates bound by Sox2 is defined as: /Sox2:substrate/(/Sox2:substrate + /free substrate). The fraction of substrates bound by Sox2 in the presence of Oct4 is defined as: (/TF:substrate − /Oct4:substrate)/(/TF:substrate + /free substrate). /TF:substrate denotes the intensity for substrates bound by any TF; /Oct4:substrate denotes the intensity for substrates bound by Oct4 alone.
DNase footprinting
200 ng of bare DNA or reconstituted nucleosome substrates (in 50 μL of Buffer 1:10 mM Tris-HCl, pH 7.5,1 mM MgCl2,10 μM ZnCl2, 0.2 mM DTT, 10 mM KCl, 0.5 mg/ml BSA, and 5% glycerol) were treated with 0.008 units of DNase I (Invitrogen; in 50 μL of Buffer II: 10 mM MgCl2 and 5 mM CaCl2) at 25°C for 3 min. The digestion reaction was stopped by the addition of 90 μL of Buffer III (20 mM Tris-HCl, pH 7.5, 50 mM EDTA, 2% SDS, and 0.2 mg/ml proteinase K) and chilled on ice for 10 min. The digested DNA was cleaned by TE-buffer-saturated phenol:chloroform:isoamyl alcohol (25:24:1, v/v) and then prepared for Illumina sequencing with the NEBNext Ultra II DNA Library Prep Kit (New England BioLabs). The adaptor-ligated DNA was amplified for 10 cycles following the manufacturer’s protocol. Sequencing was performed on a MiSeq platform. The paired-end reads were aligned to the correspondent DNA sequence. The read counts at each nucleotide position across the template sequence were extracted with custom Perl scripts.
Structure alignment
The structures of DNA in complex with Oct4POU and Sox2HMG (PDB: 1GT0) and the 601 nucleosome (PDB: 3LZ0) were obtained from the RCSB protein data bank (Reményi et al., 2003; Vasudevan et al., 2010). The DNA bound to Sox2HMG or Oct4POU was superimposed on specific positions of the nucleosomal DNA using the “align” command in PyMOL for aligning the 3′-, 4’-, and 5′-carbon atoms in the ribose sugar.
Nucleosome positioning
Raw sequencing paired-end reads were downloaded from the NCBI Sequence Read Archive (SRA) for MNase-seq nucleosome mapping data from human ESCs (SRP028172) (Yazdi et al., 2015) and MNase-seq data from mouse ESCs (SRX187610) (Teif et al., 2012). Using bowtie1 (v. 1.2.2) (Langmead et al., 2009) in “-v” alignment mode, raw sequencing reads with a maximum of 2 mismatches were aligned to the appropriate reference assembly (GRCh38 for human data and mm9 for mouse data), excluding nonstandard chromosomes. Only reads mapping to unique locations were kept for further analysis. DANPOS2 in “dpos” mode (v. 2.2.2) (Chen et al., 2013) was used to infer nucleosome positioning from mapped paired-end reads pooled across runs. Positions were represented as both genomic intervals defining peak locations as well as quantitative nucleosome position scores for 10-bp non-overlapping windows along the genome. Peaks were filtered using a custom script (https://github.com/LiZhaoLab/Oct4Sox2_nuclpos/tree/master/scripts/danpos_xls_process.py) to retain only those with a summit height of over 1.5 times the genomic mean. Nucleosome dyads are assumed to correspond to the summit positions.
TF binding sites
For mouse ESCs, ChIP-seq data were downloaded as raw single-end reads from the NCBI SRA (Oct4, SRR713340; Sox2, SRR713341; control, SRR713343) (Whyte et al., 2013). Fastq data were aligned using hisat2 (v. 2.0.5) (Kim et al., 2015) with default parameters, except with splice-awareness disabled, to the UCSC mm9 assembly of the mouse genome, excluding unmapped and random chromosomes. ChIP-seq peak locations were then called using MACS2 (v. 2.1.2) (Zhang et al., 2008) using the same settings as previously described (defaults except genome size 1.87 × 109, P-value threshold 10−9, and “-keep-dup” set to “auto”) (Whyte et al., 2013). For mouse EpiSCs, ChIP-seq data were downloaded as BED intervals from the NCBI GEO (Oct4, GSM1924747; Sox2, GSM1924746) (Matsuda et al., 2017). We defined TF binding sites as locations conforming to the canonical binding motif within 200 bp of the respective ChIP-seq peak. Genomic sequences around the ChIP-seq peaks from each cell type were then checked for TF binding motifs using FIMO (v. 4.11.2) (Grant et al., 2011) with default parameters and motifs from JASPAR (Oct4, MA1115.1; Sox2, MA0143.3). When more than one genomic location for a TF motif were found within a given ChIP-seq peak, all locations were analyzed. When no good match was found within a given ChIP-seq peak, those genomic regions and the nucleosomes therein were no longer considered in the subsequent analysis. For human ESCs, ChIP-seq peaks were downloaded from NCBI GEO (Oct4, GSM896985; Sox2, GSM896986) (Soufi et al., 2012) and converted from hg18 to GRCh38 via UCSC liftOver (http://hgdownload.cse.ucsc.edu/downloads.html) (Hinrichs et al., 2006). The peaks were then checked for canonical TF binding motifs using FIMO as described above. To identify noncanonical binding sites, these same windows were also checked for the noncanonical motifs identified by (Soufi et al., 2015), with a P-value threshold of 0.001.
Genomic data analysis
Nucleosome positions and motif centers described above were integrated using custom scripts. To obtain the aggregate nucleo- some occupancy, we calculated the average of the nucleosome positioning scores of all 1-kb regions centered on each motif center. To calculate the distribution of distances between nucleosome centers and TF binding sites, we aligned nucleosome positions with TF binding sites and extracted the distance between the center of the nucleosome and the center of the motif location.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical significance was determined by unpaired two-tailed Student’s t tests using GraphPad Prism. Welch’s two-sample t tests were conducted using the base implementation in the “stats” package of R (v. 3.4.3) on the count values for the central 13 bp and the 13 bp just inside the nucleosome boundary. The difference between two groups was considered statistically significant when the p value is less than 0.05 (*: p < 0.05; **: p < 0.01; ***: p < 0.001; ns: not significant).
DATA AND CODE AVAILABILITY
Raw data are available upon reasonable request. For the genomic data analysis, full conda, R/bioconductor, and python environment details, as well as custom scripts and Jupyter notebooks for data analysis and figure generation, are available at the project repository (https://www.github.com/LiZhaoLab/Oct4Sox2_nuclpos/).
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Escherichia coli Rosetta (DE3) pLysS | Novagen | Cat#70956-3 |
| Escherichia coli BL21 (DE3) | Invitrogen | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cy3 NHS Ester mono-reactive dye | GE Healthcare | Cat# PA23001 |
| Cy3 maleimide mono-reactive dye | GE Healthcare | Cat# PA23031 |
| Cy5 maleimide mono-reactive dye | GE Healthcare | Cat# PA15131 |
| Alexa Fluor 488 C5 maleimide | Invitrogen | Cat# A10254 |
| Coenzyme A trilithium salt | Sigma-Aldrich | Cat# C3019 |
| (3-Aminopropyl)triethoxysilane | Vector Laboratories | Cat# SP-1800 |
| Trolox | Sigma-Aldrich | Cat# 238813 |
| Glucose oxidase | Sigma-Aldrich | Cat# G2133 |
| Dextrose monohydrate | Sigma-Aldrich | Cat# D9559 |
| Catalase | Sigma-Aldrich | Cat# C100 |
| Bio-PEG-SVA, MW 5,000 | Laysan Bio | Cat# 143-117 |
| mPEG-SVA, MW 5,000 | Laysan Bio | Cat# 144-136 |
| MS(PEG)4 methyl-PEG4-NHS Ester | Thermo Fisher Scientific | Cat# 22341 |
| Streptavidin | Thermo Fisher Scientific | Cat# 434302 |
| Nanostrip | VWR | Cat# 10135-756 |
| Igepal CA-630 | Sigma-Aldrich | Cat# I8896 |
| DNase I | Invitrogen | Cat# 18068015 |
| NEBNext Ultra II DNA Library Prep Kit | New England Biolabs | Cat# E7645 |
| SYBR Gold | Invitrogen | Cat# S11494 |
| Oligonucleotides | ||
| See Table S1 | This paper | N/A |
| Recombinant DNA | ||
| pEP4 E02S CK2M EN2L | Addgene | Cat# 20924 |
| human Sox2 in pET28B vector | This paper | N/A |
| human Oct4 with C-terminal Sfp tag in pET28B vector | This paper | N/A |
| human c-Myc with C-terminal Sfp tag in pET28B vector | This paper | N/A |
| human Max in pET28B vector | This paper | N/A |
| Sfp enzyme in pET29B vector | Addgene | Cat# 75015 |
| Xenopus laevis histones | This paper | N/A |
| 601 plasmid in pBluescript II SK vector | (Lietal., 2010) | N/A |
| mutant 601 plasmids | This paper | N/A |
| Software and Algorithms | ||
| SPARTAN | (Juette et al., 2016) | https://www.scottcblanchardlab.com/software |
| ORIGIN | OriginLab | https://www.originlab.com |
| MATLAB | MathWorks | https://www.mathworks.com/products/matlab.html |
| PRISM | GraphPad | https://www.graphpad.com/scientific-software/prism |
| PYMOL | Schrödinger, LLC | http://www.pymol.org |
| Custom analysis scripts | This paper | https://www.github.com/LiZhaoLab/Oct4Sox2_nuclpos/ |
Highlights.
Nucleosome engagement by Sox2 is sensitive to the position of its binding motif
Oct4 stabilizes Sox2 binding to nucleosome ends but mildly inhibits dyad binding
Sox2 has little effect on Oct4 binding, thus their cooperativity is not reciprocal
Pioneer activities of transcription factors are multimodal and context dependent
ACKNOWLEDGMENTS
We thank Bryan Harada and Rachel Leicher for help with sample preparation, Michael Wasserman for single-molecule data analysis, Xiangwu Ju for the DNase footprinting assay, and other members of the Liu laboratory for discussions. We also thank Abdenour Soufi for sharing the human Sox2/Oct4 ChIP-seq dataset. S. Li was supported by a Tri-Institutional Starr Stem Cell Scholars Fellowship. E.B.Z. was supported by a Medical Scientist Training Program grant from the NIH (T32GM007739) to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program. L.Z. was supported by the Robertson Foundation, a Monique Weill-Caulier Career Scientist Award, and an Alfred P. Sloan Research Fellowship (FG-2018–10627). S. Liu was supported by the Robertson Foundation, the Quadrivium Foundation, a Monique Weill-Caulier Career Scientist Award, a March of Dimes Basil O’Connor Starter Scholar Award (5-FY17–61), a Kimmel Scholar Award, a Sinsheimer Scholar Award, an NIH Pathway to Independence Award (R00GM107365), and an NIH Director’s New Innovator Award (DP2HG010510).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.07.103.
REFERENCES
- Adams CC, and Workman JL (1995). Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol. Cell. Biol 15, 1405–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosetti DC, Basilico C, and Dailey L (1997). Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol 17, 6321–6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddle JW, Nguyen M, and Gunawardena J (2019). Negative reciprocity, not ordered assembly, underlies the interaction of Sox2 and Oct4 on DNA. eLife 8, e41017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YK, Srivastava Y, Hu C, Joyce A, Yang X, Zuo Z, Havranek JJ, Stormo GD, and Jauch R (2017). Quantitative profiling of selective Sox/ POU pairing on hundreds of sequences in parallel by Coop-seq. Nucleic Acids Res. 45, 832–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. (2008). Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117. [DOI] [PubMed] [Google Scholar]
- Chen K, Xi Y, Pan X, Li Z, Kaestner K, Tyler J, Dent S, He X, and Li W (2013). DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res. 23, 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. (2014). Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156, 1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Bachmann AL, Tauscher K, Benda C, Fierz B, and Muller J (2017). DNA binding by PHF1 prolongs PRC2 residence time on chromatin and thereby promotes H3K27 methylation. Nat. Struct. Mol. Biol 24, 10391047. [DOI] [PubMed] [Google Scholar]
- Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, Ernst J, and Plath K (2017). Cooperative binding of transcription factors orchestrates reprogramming. Cell 168, 442–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F, and Zhurkin VB (2014). Rotational positioning of nucleosomes facilitates selective binding of p53 to response elements associated with cell cycle arrest. Nucleic Acids Res. 42, 836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Alpern D, and Gardeux V (2016). The genetics of transcription factor DNA binding variation. Cell 166, 538–554. [DOI] [PubMed] [Google Scholar]
- Donovan BT, Chen H, Jipa C, Bai L, and Poirier MG (2019). Dissociation rate compensation mechanism for budding yeast pioneer transcription factors. eLife 8, e43008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch D, Vahokoski J, Groves MR, Pogenberg V, Cojocaru V, Vom Bruch H, Han D, Drexler HC, Araiuzo-Bravo MJ, Ng CK, et al. (2013). A unique Oct4 interface is crucial for reprogramming to pluripotency. Nat. Cell Biol 15, 295–301. [DOI] [PubMed] [Google Scholar]
- Farina A, Faiola F, and Martinez E (2004). Reconstitution of an E box-binding Myc:Max complex with recombinant full-length proteins expressed in Escherichia coli. Protein Expr. Purif 34, 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco HL, Nagari A, and Kraus WL (2015). TNFα signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol. Cell 58, 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson MD, Gatchalian J, Slater A, Kutateladze TG, and Poirier MG (2017). PHF1 Tudor and N-terminal domains synergistically target partially unwrapped nucleosomes to increase DNA accessibility. Nucleic Acids Res. 45, 3767–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, and Noble WS (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada BT, Hwang WL, Deindl S, Chatterjee N, Bartholomew B, and Zhuang X (2016). Stepwise nucleosome translocation by RSC remodeling complexes. eLife 5, e10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al. (2006). The UCSC Genome Browser database: update 2006. Nucleic Acids Res. 34, D590–D598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, and Zaret KS (2016). The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol. Cell 62, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauch R, Aksoy I, Hutchins AP, Ng CK, Tian XF, Chen J, Palasingam P, Robson P, Stanton LW, and Kolatkar PR (2011). Conversion of Sox17 into a pluripotency reprogramming factor by reengineering its association with Oct4 on DNA. Stem Cells 29, 940–951. [DOI] [PubMed] [Google Scholar]
- Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, and Taipale J (2015). DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527, 384–388. [DOI] [PubMed] [Google Scholar]
- Juette MF, Terry DS, Wasserman MR, Altman RB, Zhou Z, Zhao H, and Blanchard SC (2016). Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat. Methods 13, 341–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic S, Bachmann AL, Bryan LC, and Fierz B (2015). Multivalency governs HP1α association dynamics with the silent chromatin state. Nat. Commun 6, 7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Broströmer E, Xing D, Jin J, Chong S, Ge H, Wang S, Gu C, Yang L, Gao YQ, et al. (2013). Probing allostery through DNA. Science 339, 816–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King HW, and Klose RJ (2017). The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. eLife 6, e22631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam CS, Mistri TK, Foo YH, Sudhaharan T, Gan HT, Rodda D, Lim LH, Chou C, Robson P, Wohland T, and Ahmed S (2012). DNA-dependent Oct4-Sox2 interaction and diffusion properties characteristic of the pluripotent cell state revealed by fluorescence spectroscopy. Biochem. J 448, 21–33. [DOI] [PubMed] [Google Scholar]
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, and Weirauch MT (2018). The human transcription factors. Cell 172, 650–665. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, and Narlikar G (2001). Assembly of nucleosomal templates by salt dialysis. Curr. Protoc. Mol. Biol. 54, 21.6.1–21.6.16. [DOI] [PubMed] [Google Scholar]
- Li M, and Belmonte JC (2017). Ground rules of the pluripotency gene regulatory network. Nat. Rev. Genet 18, 180–191. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, and Workman JL (2007). The role of chromatin during transcription. Cell 128, 707–719. [DOI] [PubMed] [Google Scholar]
- Li G, Margueron R, Hu G, Stokes D, Wang YH, and Reinberg D (2010). Highly compacted chromatin formed in vitro reflects the dynamics of transcription activation in vivo. Mol. Cell 38, 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, and Kraus WL (2017). Catalytic-independent functions of PARP-1 determine Sox2 pioneer activity at intractable genomic loci. Mol. Cell 65, 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, and Widom J (1998). New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol 276, 19–42. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, and Richmond TJ (1999). Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304, 3–19. [DOI] [PubMed] [Google Scholar]
- Luger K, Dechassa ML, and Tremethick DJ (2012). New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol 13, 436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, North JA, Rose SD, and Poirier MG (2014). Nucleosomes accelerate transcription factor dissociation. Nucleic Acids Res. 42, 3017–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Mikami T, Oki S, lida H, Andrabi M, Boss JM, Yamaguchi K, Shigenobu S, and Kondoh H (2017). ChIP-seq analysis of genomic binding regions of five major transcription factors highlights a central role for ZIC2 in the mouse epiblast stem cell gene regulatory network. Development 144, 1948–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty RK, and Tan S (2015). Nucleosome structure and function. Chem. Rev 115, 2255–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meers MP, Janssens DH, and Henikoff S (2019). Pioneer factor-nucleosome binding events during differentiation are motif encoded. Mol. Cell 75, 562–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino F, Ng CKL, Veerapandian V, Schöler HR, Jauch R, and Cojocaru V (2014). Structural basis for the SOX-dependent genomic redistribution of OCT4 in stem cell differentiation. Structure 22, 1274–1286. [DOI] [PubMed] [Google Scholar]
- Mirny LA (2010). Nucleosome-mediated cooperativity between transcription factors. Proc. Natl. Acad. Sci. USA 107, 22534–22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgunova E, and Taipale J (2017). Structural perspective of cooperative transcription factor binding. Curr. Opin. Struct. Biol 47, 1–8. [DOI] [PubMed] [Google Scholar]
- Nishimoto M, Fukushima A, Okuda A, and Muramatsu M (1999). The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol 19, 5453–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura-Nakanishi S, Saito M, Niwa H, and Ishikawa F (2005). Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J. Biol. Chem. 280, 5307–5317. [DOI] [PubMed] [Google Scholar]
- Polach KJ, and Widom J (1996). A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J. Mol. Biol 258, 800–812. [DOI] [PubMed] [Google Scholar]
- Ptashne M, and Gann A (2002). Genes and Signals (Cold Spring Harbor Lab Press; ). [Google Scholar]
- Reményi A, Lins K, Nissen LJ, Reinbold R, Schöler HR, and Wilmanns M (2003). Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 17, 2048–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzino A, and Wuebben EL (2016). Sox2/Oct4: a delicately balanced partnership in pluripotent stem cells and embryogenesis. Biochim. Biophys. Acta 1859, 780–791. [DOI] [PubMed] [Google Scholar]
- Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, and Robson P (2005). Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem 280, 24731–24737. [DOI] [PubMed] [Google Scholar]
- Sartorelli V, and Puri PL (2018). Shaping gene expression by landscaping chromatin architecture: lessons from a master. Mol. Cell 71, 375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, and Bianchi ME (2001). Spatially precise DNA bending is an essential activity of the sox2 transcription factor. J. Biol. Chem 276, 47296–47302. [DOI] [PubMed] [Google Scholar]
- Segal E, and Widom J (2009). From DNA sequence to transcriptional behaviour: a quantitative approach. Nat. Rev. Genet. 10, 443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, and Daley GQ (2013). Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell 12, 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggers T, Chang AB, Teixeira A, Wong D, Williams KJ, Ahmed B, Ragoussis J, Udalova IA, Smale ST, and Bulyk ML (2011). Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-κB family DNA binding. Nat. Immunol 13, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, Zhou T, Rohs R, Honig B, Bussemaker HJ, and Mann RS (2011). Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147, 1270–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Zhou T, Yang L, Dantas Machado AC, Gordân R, and Rohs R (2014). Absence of a simple code: how transcription factors read the genome. Trends Biochem. Sci 39, 381–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, and Zaret KS (2012). Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151, 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, and Zaret KS (2015). Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinstead EE, Miranda TB, Paakinaho V, Baek S, Goldstein I, Hawkins M, Karpova TS, Ball D, Mazza D, Lavis LD, et al. (2016). Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell 165, 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Takizawa Y, Tanaka H, Machida S, Koyama M, Maehara K, Ohkawa Y, Wade PA, Wolf M, and Kurumizaka H (2018). Cryo-EM structure of the nucleosome containing the ALB1 enhancer DNA sequence. Open Biol. 8, 170255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia N, MacCarthy C, Esch D, Gabriele Marthaler A, Tiemann U, Araúzo-Bravo MJ, Jauch R, Cojocaru V, and Schöler HR (2015). Dissecting the role of distinct OCT4-SOX2 heterodimer configurations in pluripotency. Sci. Rep 5, 13533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teif VB, Vainshtein Y, Caudron-Herger M, Mallm JP, Marth C, Heifer T, and Rippe K (2012). Genome-wide nucleosome positioning during embryonic stem cell development. Nat. Struct. Mol. Biol 19, 1185–1192. [DOI] [PubMed] [Google Scholar]
- Thompson D, Regev A, and Roy S (2015). Comparative analysis of gene regulatory networks: from network reconstruction to evolution. Annu. Rev. Cell Dev. Biol 31, 399–428. [DOI] [PubMed] [Google Scholar]
- Tomioka M, Nishimoto M, Miyagi S, Katayanagi T, Fukui N, Niwa H, Muramatsu M, and Okuda A (2002). Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 30, 3202–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee S, Melcher K, Ding WV, Johnston SA, and Kodadek T (1998). Evidence for two modes of cooperative DNA binding in vivo that do not involve direct protein-protein interactions. Curr. Biol. 8, 452–458. [DOI] [PubMed] [Google Scholar]
- Vasudevan D, Chua EYD, and Davey CA (2010). Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J. Mol. Biol 403, 1–10. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. (2012). Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 22, 1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MD, Angiolini JF, Alvarez YD, Kaur G, Zhao ZW, Mocskos E, Bruno L, Bissiere S, Levi V, and Plachta N (2016). Long-lived binding of Sox2 to DNA predicts cell fate in the four-cell mouse embryo. Cell 165, 75–87. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, and Young RA (2013). Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DC Jr., Cai M, and Clore GM (2004). Molecular basis for synergistic transcriptional activation by Oct1 and Sox2 revealed from the solution structure of the 42-kDa Oct1.Sox2.Hoxb1-DNA ternary transcription factor complex. J. Biol. Chem 279, 1449–1457. [DOI] [PubMed] [Google Scholar]
- Yazdi PG, Pedersen BA, Taylor JF, Khattab OS, Chen YH, Chen Y, Jacobsen SE, and Wang PH (2015). Nucleosome organization in human embryonic stem cells. PLoS ONE 10, e0136314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Straight PD, McLoughlin SM, Zhou Z, Lin AJ, Golan DE, Kelleher NL, Kolter R, and Walsh CT (2005). Genetically encoded short peptide tag for versatile protein labeling by Sfp phosphopantetheinyl transferase. Proc. Natl. Acad. Sci. USA 102, 15815–15820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Lin AJ, Golan DE, and Walsh CT (2006). Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat. Protoc 1, 280–285. [DOI] [PubMed] [Google Scholar]
- Yu X, and Buck MJ (2019). Defining TP53 pioneering capabilities with competitive nucleosome binding assays. Genome Res. 29, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, and Carroll JS (2011). Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, and Mango SE (2016). Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr. Opin. Genet. Dev 37, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Farnung L, Kaasinen E, Sahu B, Yin Y, Wei B, Dodonova SO, Nitta KR, Morgunova E, Taipale M, et al. (2018). The interaction landscape between transcription factors and the nucleosome. Nature 562, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data are available upon reasonable request. For the genomic data analysis, full conda, R/bioconductor, and python environment details, as well as custom scripts and Jupyter notebooks for data analysis and figure generation, are available at the project repository (https://www.github.com/LiZhaoLab/Oct4Sox2_nuclpos/).