Abstract
Nuclear receptors (NRs) are ligand-activated transcription factors that are expressed in a wide variety of cells and play a major role in lipid signaling. NRs are key regulators of immune and metabolic functions in macrophages and are linked to macrophage responses to microbial pathogens. Pathogens are also known to induce the expression of specific NRs to promote their own survival. In this review, we focus on the NRs recently shown to influence macrophage responses to Mycobacterium tuberculosis (M.tb), a significant cause of morbidity and mortality worldwide. We provide an overview of NR-controlled transcriptional activity and regulation of macrophage activation. We also discuss in detail the contribution of specific NRs to macrophage responses to M.tb, including influence on macrophage phenotype, cell signaling, and cellular metabolism. We pay particular attention to PPARγ since it is required for differentiation of alveolar macrophages, an important niche for M.tb, and its role during M.tb infection is becoming increasingly appreciated. Research into NRs and M.tb is still in its early stages, therefore continuing to advance our understanding of the complex interactions between M.tb and macrophage NRs may reveal the potential of NRs as pharmacological targets for the treatment of tuberculosis.
Keywords: Macrophage, Nuclear Receptors, Mycobacterium tuberculosis, PPARγ
1. Introduction
Mycobacterium tuberculosis (M.tb), the etiological agent of tuberculosis (TB), is arguably the oldest known human bacterial pathogen. TB is currently the ninth leading cause of death worldwide and the leading cause from a single infectious agent, surpassing deaths caused by HIV/AIDS [1]. In 2016, there were 10.4 million cases of TB reported [1], demonstrating an urgent need for new therapies (targeting the bacterium and the host) to halt infection and progression to active TB. Drug-resistant TB is an ongoing threat with 600,000 new cases of M.tb resistant to the most effective first-line drug, rifampicin, and 490,000 cases of multi-drug resistant TB [1]. According to the World Health Organization (WHO), as of 2017 there are 17 drugs in clinical trials and various new combination regimens and several repurposed drugs [1].
A promising host-directed target for anti-TB treatment are members of a superfamily of intracellular transcription factors referred to as nuclear receptors (NRs). Immune cells such as macrophages utilize NRs to sense their local environment and shape the immune response. NRs are key players in homeostasis, metabolism (especially lipid and the lipid-based eicosanoids), and transcriptional regulation [2–8]. Approximately 13% of drugs approved for sale in the United States target NRs, representing $27.5 billion in sales revenue in 2009 [9]. As nuclear receptors are increasingly appreciated in the context of M.tb pathogenesis [10–18], targeting NRs may provide a new, largely unexplored area in TB drug development. In this review, we discuss NR regulation of transcription and macrophage responses. We focus on NRs that have been shown to play a role in M.tb infection and consider their anti-TB therapeutic potential.
2. Nuclear Receptors
2.1. Structure
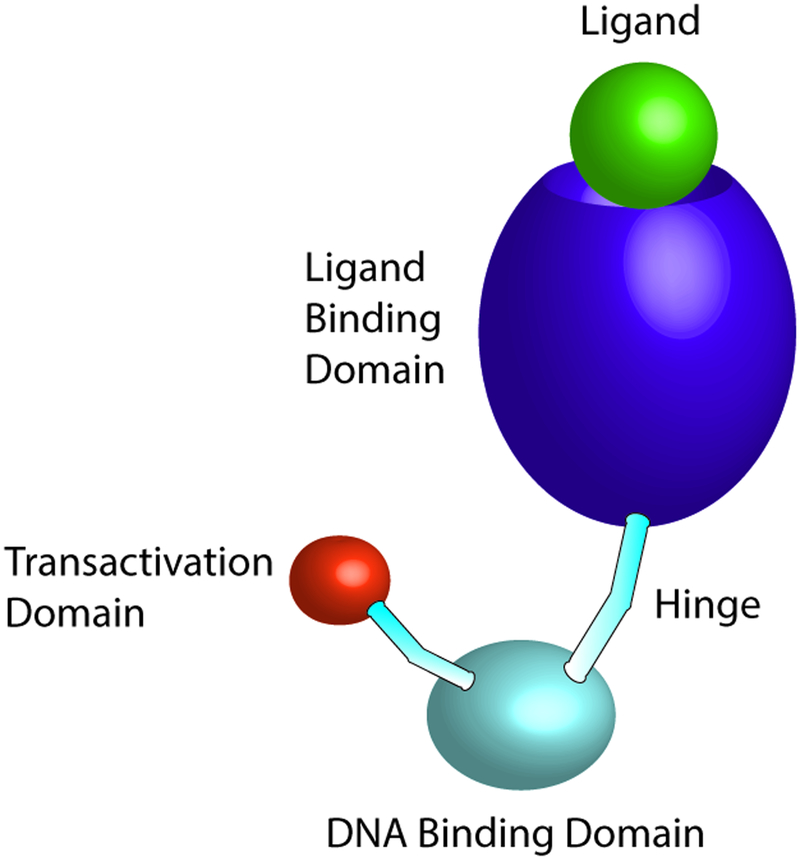
NRs are ligand-dependent and nearly all have a common architecture with a highly conserved DNA binding domain (DBD) and carboxy-terminal ligand-binding domain (LBD) (Fig 1) [19]. There are 48 NRs in the human genome [20] and 49 in the rodent genome, of which 28 are associated with macrophages [21]. NRs are typically activated by lipid-soluble, membrane-permeable ligands. The two zinc-finger motifs of the DBD target specific DNA sequences known as hormone response elements. The LBD has a high specificity for its ligand. After interacting with the NR’s respective ligand, the NR undergoes a conformational change which can then lead to recruitment of co-activator complexes as well as association with and stabilization of co-repressors that alter the transcriptional regulatory function of the receptor [22]. Ligand binding, along with other factors in vivo, can also lead to dissociation of co-repressor complexes such as nuclear co-repressor (NCoR) and the related silencing mediator of retinoic acid and thyroid hormone receptors (SMRT) [23]. NRs have a variable hinge region that links the DBD and LBD, permitting the structural flexibility of the receptor [24]. Members of this superfamily of receptors have historically been categorized into three classes: conventional steroid/thyroid hormone receptors (i.e. estrogen receptor, progesterone receptor), orphan receptors for which the ligand has either not been identified or that appear to function without a ligand, and adopted orphan receptors for which a ligand has been discovered [i.e. liver X receptors (LXRs), peroxisome proliferator-activated receptors (PPARs), and retinoid X receptors (RXRs)] [25]. The transcriptional activity and protein stabilization of NRs can also be regulated via post-translational modifications including phosphorylation, acetylation, sumoylation and ubiquitination [26]. For example, the influences of phosphorylation, acetylation and sumoylation of PPARγ can increase or decrease this transcription factor’s activity, depending on the site and type of modification [26].
Figure 1. Nuclear receptor domain structure.
Nuclear receptors consist of a DNA binding domain, ligand binding domain and a flexible hinge region which allows for conformational changes following ligand binding. The transactivation domain interacts at the promoter with coactivators to induce gene transcription.
2.2. Transcriptional Regulation
A primary and critical role of NRs is the regulation of transcription via activation, repression, or trans-repression [4–6]. NRs positively regulate transcription by binding to specific response elements of the target gene as homodimers or heterodimers. PPARs and LXRs constitutively bind to DNA as heterodimers with RXRs and can do so with or without a ligand [5, 27]. Without a ligand, these heterodimers often function as transcriptional repressors and interact with co-repressor complexes containing NCoR and SMRT [28–30]. NRs, including PPARs and LXRs, often regulate transcription through indirect targeting of target genes, a process referred to as trans-repression, rather than direct binding and inhibition of other transcription factors like NF-kB, AP1, and STATs [25, 31].
RXR forms heterodimers with one third of known human NRs, most of which require RXR as an obligatory partner for DNA binding and transcriptional regulation [32]. RXR heterodimers are classified as either permissive or non-permissive. Permissive heterodimers such as RXR and its partners (i.e. PPAR/RXR, LXR/RXR) can be activated by the ligands of either partner. However, heterodimers of RXR and a non-permissive partner (i.e. retinoic acid receptor (RAR)/RXR and VDR/RXR) can only be activated by the agonist of the dominant partner receptor [32].
2.3. Macrophage Activation
Macrophages are capable of various activities which are dependent on the local cytokine milieu [33–35]. In general, macrophages stimulated with the cytokine interferon-γ (IFN-γ) activate to a classical or M1 polarization state that is largely pro-inflammatory and anti-microbial [35, 36]. Conversely, macrophages stimulated with the cytokines interleukin-4 (IL-4) and/or IL-13 are activated to an alternative or M2 polarization state that promotes anti-inflammatory and wound healing responses and are more permissive to M.tb infection [36, 37]. Alveolar macrophages (AMs), which are unable to efficiently clear M.tb, are classically thought of as M2, but it must be noted that the M1/M2 paradigm does not fully describe the spectrum of macrophage activation states, with many cells displaying a mixed phenotype dependent on numerous factors [36, 38, 39].
Macrophage activation is often only characterized by responses to polarizing cytokines, however, NRs also play a significant role in macrophage responses. For example, PPARγ expression is augmented by the Th2-associated cytokine IL-4, which induces the generation of PPARγ ligands, and contributes to the maturation of M2 macrophages [40–43]. PPARγ also aids in the induction of Th2 polarization in murine T cells in vitro and is essential for IL-33 production [44], another cytokine that plays a role in M2 activation [45, 46].
Numerous other NRs have been shown to play significant roles in macrophage activation responses. For example, agonists of LXR inhibited inducible nitric oxide (iNOS), COX-2, and IL-6 in response to LPS and E. coli in vitro [31]. In fact, many genes inhibited by LXR agonists were targets of NF-κB [31], indicating an inhibitory effect on M1 responses. REV-ERBα, a constitutive repressor, is more highly expressed in M1 activated compared to M2 activated human monocyte-derived macrophages (hMDMs) [14]. REV-ERBα negatively regulates TNF-α and macrophage chemotactic protein-1 (MCP-1) in hMDMs stimulated with LXR agonists [47]. These data demonstrate that macrophage activation phenotype is shaped by signaling of NRs, signifying the importance of these receptors in macrophage responses to pathogens. In this review, we focus on NRs shown to influence macrophage responses to M.tb, which can result in a more permissive or anti-bacterial phenotype of these phagocytes.
3. NRs and TB
M.tb can affect the expression of various NRs and a growing number of these have been implicated in macrophage responses to M.tb [2, 17, 48]. NRs play vital roles in disease pathogenesis and in macrophage-mediated host defense. The following sections focus on the specific NR-dependent responses of macrophages to mycobacterial infection.
3.1. PPARs
PPARs are ligand activated transcription factors that control fatty acid metabolism, including transport, synthesis, mobilization, activation, and oxidation of fatty acids [3]. There are three PPAR subtypes in mammals: PPARα, PPARγ, and PPARβ/δ (also referred to as NR1C1, NR1C3, and NR1C2, respectively) which exhibit different expression patterns and functions. PPARα and PPARβ/δ are ubiquitously expressed, and PPARγ is expressed in immune cells and aids in storage of fatty acids. PPARγ also plays an important role in macrophage anti-inflammatory responses [49, 50]. PPARs can be activated by a diverse group of ligands due to their large ligand-binding pocket. PPAR ligands include endogenous native and modified fatty acids as well as synthetic ligands such as PPARγ agonists thiazolidinediones (TZDs) rosiglitazone and pioglitazone, used most commonly to treat diabetes [PPAR ligands are comprehensively reviewed in [51]].
3.1.1. PPARγ
PPARγ is important for the generation of alveolar macrophages which are permissive to M.tb intra-macrophage growth [52]. Inhibition or knockdown of PPARγ reduces mycobacteria growth in human and murine macrophages in vitro and in mice (Table 1) [10, 11, 53, 54], while activation of PPARγ with rosiglitazone increases M.tb growth in human macrophages [10]. Multiple macrophage model systems have revealed that infection with M.tb or M. bovis Bacillus Calmette-Guérin (BCG) and stimulation with certain M.tb cell wall components [i.e. mannose-capped lipoarabinomannan (ManLAM) or P19 (an M.tb cell wall lipoprotein)] are capable of up-regulating expression and activity of PPARγ, as observed in PBMCs from TB patients [10, 11, 53, 55–57]. In contrast to M.tb, M. smegmatis does not increase PPARγ expression [11, 53]. The inability of M. smegmatis to up-regulate PPARγ could be partly responsible for its less virulent nature. Similarly, M. bovis BCG does not induce PPARγ to the same extent as M.tb in hMDMs [11] and actually appears to repress its expression in murine AMs in vivo [58]. This suggests that more virulent mycobacteria have evolved to induce PPARγ during infection to alter the environment to be more permissive to M.tb growth.
Table 1:
Role of NRs on Macrophage Responses and Mycobacterial Infection
PPARγ contributes to dampening iNOS expression and nitric oxide secretion in macrophages [59]. PPARγ also plays an inhibitory role in the secretion of M1 macrophage effector molecules TNF-α and IL-6 and increases IL-8 and IL-10 in isolated macrophages as well as murine lungs (Fig 2) [10, 11, 53, 54]. It is interesting to note that M.tb and M. bovis BCG use contrasting signaling pathways to up-regulate IL-8. M.tb induces IL-8 through an NF-κB-independent (but mannose receptor [MR]- and PPARγ-dependent) pathway in human macrophages, while M. bovis BCG uses an NF-κB-dependent, and PPARγ-independent, pathway [11]. These data suggest that the use of disparate host signaling pathways could be an indicator of M.tb’s immune evasion strategy.
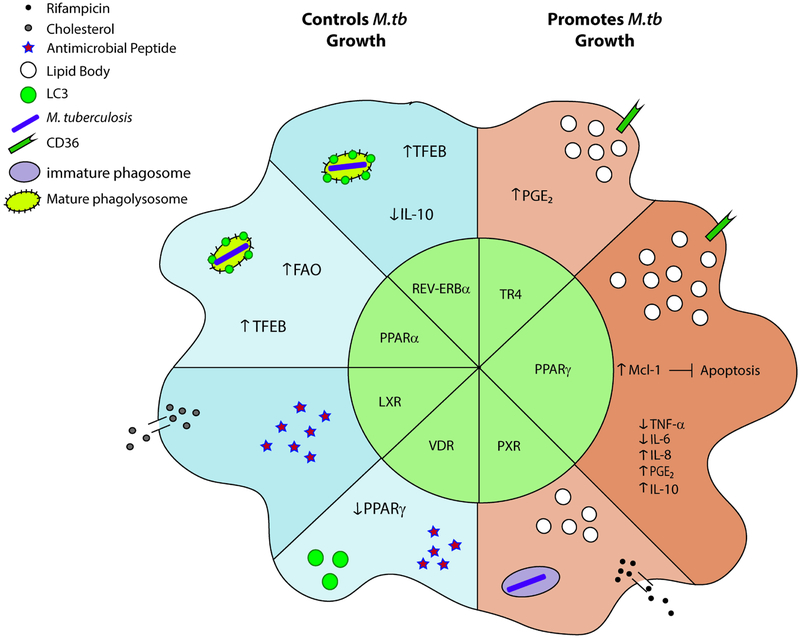
Figure 2. Nuclear receptor-regulated macrophage responses to M. tuberculosis.
Nuclear receptors play a major role in regulating macrophage responses following infection with M.tb which can be divided into responses that aid in controlling M.tb growth or that result in increased bacterial burden. The receptors TR4, PPARγ, and PXR are associated with increased susceptibility to M.tb and lipid body formation whereas PPARα, REV-ERBα, LXRs and VDR are associated with resistance to M.tb. Upon infection, TR4 activation can result in increased PGE2 and CD36 expression. PPARγ is also associated with CD36 expression as well as regulation of cytokine production, PGE2 production, and decreased apoptosis via increased Mcl-1 expression. PXRs are largely responsible for drug efflux, negating the effects of antibacterial rifampicin and also decrease phagolysosomal maturation, shown here as a phagosome devoid of mature endosome markers. VDR and LXRs are associated with antimicrobial peptide production following M.tb infection. VDR is also associated with decreased expression of PPARγ and increased LC3 positive phagosomes. LXRs play a role in cholesterol efflux and decreased in lipid body formation. PPARα and REV-ERBα stimulation is linked to lysosomal maturation, formation of autophagosomes, and increased activity of TFEB. PPARα is also associated with increased FAO. REV-ERBα represses IL-10 production during M.tb infection, aiding in antimicrobial activities of the macrophages. Abbreviations: PGE2, prostaglandin E2; TFEB, transcription factor EB; FAO, fatty acid oxidation; LAMP, lysosomal membrane protein
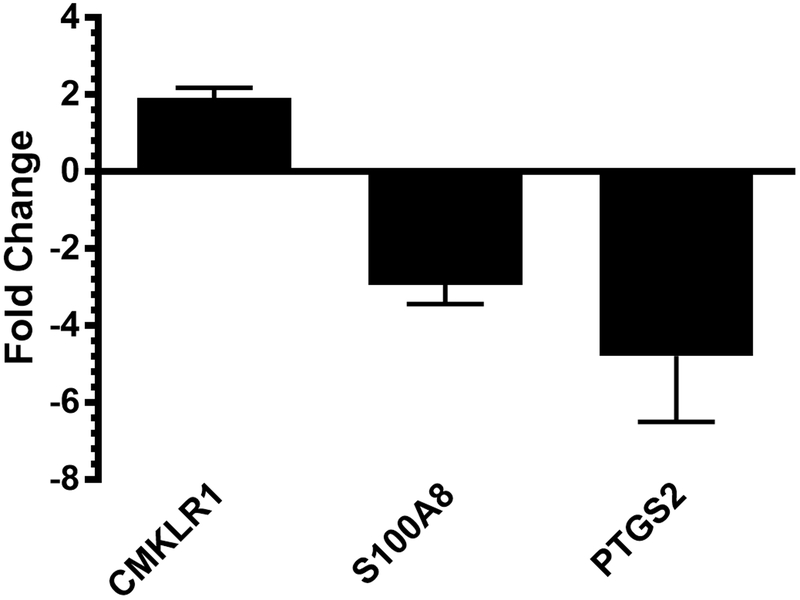
M.tb induced PPARγ appears to be mediated, in part, by distinct pattern recognition receptors (PRRs) that detect mycobacteria. M. bovis BCG and M.tb P19 as well as ManLAM up-regulate PPARγ in a Toll-like receptor 2 (TLR2) dependent manner in mouse macrophages [53, 55]. In human and mouse macrophages, M.tb and ManLAM induce PPARγ following recognition by MR [11, 59], a hallmark surface marker of M2 macrophages. PPARγ activity in these cells also requires cytosolic phospholipase A2 (cPLA2) and 15-lipoxygenase (15-LOX) [11], which are required for production of the eicosanoids 13-hydroxyoctadecadienoic acid (13-HODE) and 15-hydroxyeicosatetraenoic acid (15-HETE), identifying these products as endogenous ligands during M.tb infection. NanoString analysis recently undertaken by our lab identified many genes important for host immune responses as being regulated by PPARγ during M.tb infection of human macrophages [60]. Of note, genes whose expression is affected by PPARγ include those involved in eicosanoid and resolvin signaling including PTGS2, S100A8, and CMKLR1 (Fig 3; data adapted from [60]). These data suggest that PPARγ regulates expression of lipid mediators of inflammation during M.tb infection.
Figure 3. Genes significantly altered with PPARγ knockdown in human macrophages.
Macrophages were transfected with scrambled and PPARγ specific siRNA with Mirus X2, then infected with M.tb at MOI 5. After 24h, total RNA was isolated and subjected to NanoString analysis. Shown are selected genes that displayed at least a 1.5x fold change after PPARγ knockdown, N=3. Results adapted from: [60].
There is recent evidence that in M.tb-infected THP-1 macrophages, PPARγ is also capable of increasing CD36 expression [10], a major receptor for the uptake of low density lipoproteins which contributes to the generation of foamy macrophages. CD36 interacts with surfactant lipids (found throughout the lungs) and can enhance M.tb growth in human macrophages in vitro [61]. In M. bovis BCG-infected macrophages, CD36 directly interacts with TLR2 as evidenced by co-immunoprecipitation of the two receptors [56]. Neutralization of CD36 subsequently decreased PPARγ expression, as well as lipid body formation and PGE2 secretion [56]. These data demonstrate a critical role for CD36 in inducing PPARγ-mediated macrophage responses to Mycobacteria species and may be an effective target for pharmacological intervention against TB.
Apoptosis has been linked to mycobacterial virulence, since more virulent mycobacteria induce less apoptosis during infection of macrophages, and this mode of cell death can limit M.tb growth [62, 63]. Our laboratory recently confirmed that PPARγ regulates apoptosis during M.tb infection through the induction of anti-apoptotic Mcl-1 [60]. Inhibition of PPARγ, 15-LOX (which is required for PPARγ activity, mentioned above), or Mcl-1 all led to significant increases in human macrophage apoptosis. This work further identified Mcl-1 and 15-LOX as promising targets for host directed therapy during TB, since inhibition of either of these molecules significantly reduced M.tb growth in macrophages. Excitingly, inhibition of Mcl-1 also limited M.tb growth in an in vitro granuloma model [60, 64].
Altogether, these data further support the idea that M.tb has evolved to modulate macrophage signaling processes to promote its own survival. Considering that PPARγ is critical for promoting anti-inflammatory activities, it may be beneficial to block PPARγ early in infection to enhance host defense, and, in contrast, promote its anti-inflammatory activities with active TB to limit tissue inflammation. Intriguingly, pyrazinamide treatment up-regulates PPARγ expression and reduces release of pro-inflammatory cytokines in mice during M.tb infection [65], supporting the notion that temporal control of PPARγ could be critical to control M.tb infection and disease. An alternative therapy to targeting PPARγ could involve inhibition of the MR or other molecules upstream of PPARγ activation [11, 59]. Recently elucidated MR signaling during M.tb infection revealed the importance of this receptor for M.tb uptake, inhibition of phagolysosomal fusion, and intracellular M.tb survival [66]. The role of PPARγ in progression of TB has not been thoroughly established. Further elucidation of the signaling pathways in which PPARγ plays a role will be advantageous to our understanding of macrophage-M.tb interactions and should help identify additional pathways that can specifically be targeted to limit M.tb growth.
3.1.2. PPARα
Compared to PPARγ, much less is known about the role of PPARα and M.tb pathogenesis. PPARα is generally a negative regulator of inflammatory responses and tends to antagonize the activities of NF-kB and activator protein-1 (AP-1) families through trans-repression [67]. PPARα also regulates lipid transport, gluconeogenesis, and fatty acid oxidation (FAO) [51]. Endogenous ligands include conjugated linoleic acid, 1-palmitoyl-2-oleoyl-sn-glycerol-3-phoshocholine, and the eicosanoid leukotriene B4 [51]. Despite its known activities identified above, a recent study by Kim, et al. revealed that PPARα is essential for anti-mycobacterial responses. PPARα deficiency in mice led to increased bacterial burden and inflammatory responses in the lungs and spleen. BMDMs from PPARa−/− mice had decreased activation of transcription factor EB (TFEB, a critical regulator of autophagy) and increased formation of lipid bodies following infection with M.tb or M. bovis BCG [68]. Addition of PPARα agonists increased autophagy, lysosomal biogenesis, phagosomal maturation, and anti-mycobacterial defenses in BMDMs [68]. PPARα agonist treatment also increased the mitochondrial respiration rates and FAO, which were decreased in BMDMs from PPARα−/− mice [68]. All together, these data indicate that PPARα aids in mediating anti-mycobacterial responses through the activation of TFEB, autophagy, lipid catabolism, and FAO although more work needs to be done.
It is interesting to note that PPARγ and PPARα, both members of the same NR subfamily, have such contrasting roles in macrophage responses to M.tb. Since PPARγ supports M.tb growth, while PPARα’s role appears to support anti-mycobacterial activity, further investigation into PPARα’s anti-TB activity is necessary to understand the mechanism(s) underlying these contradictory phenotypes.
3.2. TR4
Testicular receptor 4 (TR4, NR2C2) is an NR found widely throughout the body and important for roles such as cerebellar development, gluconeogenesis, lipogenesis, and bone and muscle development [69]. TR4 can bind to response elements targeted by other NRs, including VDR, RAR, RXR, and PPAR, thus competing with these NRs for their downstream targets [69]. Interestingly, TR4 can repress activation of VDR and PPARα targets, but enhances PPARγ targets [69]. Molecules known to trans-activate TR4 include ligands associated with PPARγ, including eicosanoid intermediates 15-HETE, 13-HODE, and the TZD family of drugs [70]. The M.tb lipid keto-mycolic acid was recently shown to stably bind TR4 in a non-canonical fashion, leading to the induction of foamy macrophages and granuloma formation both in vitro (PBMC granuloma model) and in vivo (murine lung granulomas) (Table 1)[15]. Similar to PPARγ, TR4 is important for M.tb growth in macrophages since it promotes an M2-like macrophage phenotype and decreases reactive oxygen species production [10]. There is evidence that TR4 and PPARγ augment each other, as knockdown of both receptors has an additive effect compared to knockdown of the individual receptor in control of M.tb growth [10]. TR4 binds to a response element in the CD36 promoter, thus increasing expression of CD36, a major receptor for the uptake of low density lipoproteins which contributes to the generation of foamy macrophages [70] and TB pathogenesis [61]. Knockdown of TR4 marginally reduced the bacterial burden of macrophages infected with the attenuated M.tb strain H37Ra in vitro and also resulted in decreased PGE2 production [10]. In addition, a knockdown of 50–60% of TR4 in alveolar macrophages in vivo corresponded with reduced survival of M.tb H37Rv [15]. It is interesting to note, however, that TR4 gene expression is not changed in TB-infected patients compared to healthy controls [10].
These data indicate that activation of TR4 plays an important role in the survival of M.tb in mice and in human cell lines in vitro. However, very little is known about TR4 and downstream effects during M.tb infection. Further research into the translational aspects of TR4 regulation of conditions conducive to M.tb survival in human primary macrophages is necessary to delineate if targeting this NR or its signaling pathways is a feasible approach to anti-TB therapy.
3.3. LXRs
Liver X receptors (LXRs) are regulated by oxidized forms of cholesterol (oxysterols) and intermediate products of cholesterol biosynthetic pathways [71, 72], aiding in tight regulation of lipid homeostasis and transport. LXRs have two identified isoforms, LXRα (NR1H3) and LXRβ (NR1H2). LXRs form obligate heterodimers with RXR [27] and are known to play a role in macrophage survival, preventing bacterial-induced apoptosis [73, 74].
In both mouse models and in vitro macrophage assays, LXRs have shown a propensity for anti-mycobacterial activity (Fig 2; Table 1) [10, 16]. LXRα, but not LXRβ, is up-regulated in response to M.tb infection in macrophages [75] and knockdown of LXRα results in increased bacterial burden [10]. In M.tb-infected macrophages, LXRα was shown to bind to Alu/DR4 elements which are associated with multiple genes implicated in lipid metabolism [75]. THP-1 macrophages infected with M.tb H37Ra contain an increased number of lipid bodies with decreased gene expression for ABCA1 and ABCG1, genes implicated in cholesterol efflux [10]. Treatment with the LXRα agonist TO901317 increased the expression of ABCA1 and ABCG1 in THP-1 macrophages, which was further enhanced during H37Ra infection and resulted in decreased lipid body formation [75]. Thus, activation of cholesterol efflux through the NR LXRa could prove a viable target to enhance anti-M.tb macrophage activity.
LXRs and LXR target genes are up-regulated in CD11c+ cells in the lung and bronchoalveolar lavage fluid as well as in BMDMs following M.tb infection [16, 17]. Mice deficient in both LXRα and LXRβ were more susceptible to M.tb demonstrating increased bacterial burden and granulomatous lesions as well as a decreased Th1/Th17 immune response, though only the LXRα single knockout mouse recapitulated these results [16]. Addition of LXR agonists to WT mice both prophylactically and therapeutically resulted in decreased bacterial burden and increased Th1/Th17 function in the lungs [16]. A recent study showed that M.tb-induced IL-36 production increased the generation of the LXR ligand oxysterol and subsequently inhibited M.tb growth in macrophages [76]. The IL-36/LXR axis was also responsible for antimicrobial peptide production [76], partially explaining the anti-mycobacterial effects of IL-36 and LXRs.
Analysis of LXR single nucleotide polymorphisms (SNPs) was performed on TB patients in the Chinese Han population. Eight common variants in the LXR genes were identified, of which two were associated with an increased risk of developing TB [12]. The other six SNPs appeared to be protective against TB, with three showing significant protection. Altogether, these data indicate that LXRs play a fundamental role in genetic susceptibility to TB.
LXRs help shape the macrophage response to M.tb and mediate lipid metabolism and decreased lipid body formation which is conducive to M.tb eradication. Interestingly, use of cholesterol reducing statins aids in TB treatment in animal models and clinical trials [77–79]. This is suggested to occur through reduction of LXR activity. LXR agonist treatment therapeutically aided in fighting M.tb infection in mice [16] and additional research is necessary to verify if anti-TB treatments targeting LXRs would translate to humans. A recent study examined the effects of LXR agonists in human hypercholesterolemic patients treated with statins. They noted a reverse in cholesterol transport pathways, however murine and NHP models did not show the increased LDL cholesterol and decreased circulating neutrophils observed in statin-treated and non-treated hypercholesterolemic patients [80]. It is possible that the activity of statins is redundant with LXR-mediated signaling, thus the decreased LXR activity may be due to a reduction in needed cholesterol efflux. Further evaluation of statins for TB treatment and the roles of LXRs is required to definitively determine how statins alter LXR activity and how this intervention impacts TB treatment.
3.4. REV-ERBα
REV-ERBα is a unique member of the NR superfamily. It has an atypical LBD lacking the carboxy-terminal activation function 2 (AF2) region [81], which is responsible for transcriptional activation. Thus, REV-ERBα is a constitutive transcriptional repressor with constitutive binding of co-repressors such as NCoR1 [82]. REV-ERBα competes for response elements with NRs known to have transcription activation activity, including PPARs and LXRs [47, 83, 84]. REV-ERBα is responsible for regulation of the circadian rhythm, cellular metabolism, and immune function [81]. REV-ERBα was referred to as an orphan receptor for quite a while until its ligand, the porphyrin heme, was identified in 2007 [85, 86]. REV-ERBα is encoded by the gene NR1D1, which is the opposite strand, or reverse, of the ERBA oncogene [87], hence the name REV-ERBα.
Very little is known about REV-ERBα and its activities during M.tb infection. REV-ERBα appears to play a role in antimicrobial immune responses in macrophages, positively regulating autophagy and lysosome biogenesis, two mechanisms used to combat M.tb infection. The promotor region of the immunoregulatory cytokine IL-10 contains a REV-ERBα binding site in humans and nonhuman primates, but not in mice [14], demonstrating a species disparity in model systems. Over-expression of REV-ERBα induced repression of IL-10 which led to increased anti-M.tb activity in macrophages due, in part, to increased phagolysosome maturation (Fig 2) [14]. Knockdown of REV-ERBα with siRNA results in decreased lysosomal-associated membrane protein 1 (LAMP-1) expression, a marker of phagolysosome maturation, as well as decreased expression of TFEB [13], indicating that REV-ERBα plays a role in lysosome biogenesis. Treatment of THP-1 macrophages with the REV-ERBα agonist GSK4112 resulted in an increased number of autophagosomes and lysosomes and the levels of MAP1LC3-II, a hallmark molecule of autophagy progression, leading to enhanced M.tb clearance [13].
As REV-ERBα is a constitutive repressor, these data indicate an indirect and complex mechanism of action used by this NR which involves NCoR and histone deacetylase 3 [14, 82]. It is unclear if REV-ERBα’s repressive activity on IL-10 transcription is altered by M.tb, which would likely aid in bacterial survival. Furthermore, how REV-ERBα promotes autophagy and lysosomal biogenesis is uncertain. Additional studies designed to fully elucidate the cellular pathway(s) used by REV-ERBα as well as its downstream targets in macrophages during M.tb infection is required. However, work concerning the immune response and cytokine balance will be limited by the inability to study these interactions in mice.
3.5. PXR
The human xenobiotic nuclear receptor pregnane X receptor (PXR) is an adopted orphan nuclear receptor. It is expressed in immune cells such as monocytes/macrophages and lymphocytes, but is predominantly expressed in the liver and intestine [88]. The major role of PXRs is drug metabolism. Activation of PXRs up-regulates genes important for lipid uptake and lipogenic pathways [89]. PXRs can also inhibit both innate and adaptive immune responses [90]. In hMDMs, PXR has been shown to augment M.tb H37Ra survival and promote foamy macrophage formation as well as decrease phagolysosomal fusion, inflammatory responses, and apoptosis (Table 1) [91]. Some of these findings were confirmed in the humanized PXR mouse model, resulting in increased M.tb survival in vivo [91]. The study also showed that M.tb cell wall lipids, namely mycolic acid, were able to crosstalk with the human PXR via interaction with its promiscuous LBD [91].
In a subsequent study, the same research group showed that PXR can modulate macrophage drug-efflux transporter expression and activity, compromising the effect of rifampicin in vitro in hMDMs (Fig 2)[92]. Previous studies showed that rifampicin is a potent PXR activator that can induce expression of important metabolizing enzymes [93]. In mice infected with M.tb, the PXR antagonist ketoconazole rescued the activity of rifampicin [92]. Other rifamycin derivatives such as rifapentine and rifabutin do not stimulate PXR regulation of metabolizing enzymes to the same extent as rifampicin [93], and could potentially be used as an alternative to combat PXR-mediated drug non-responsiveness. Further, rifalazil does not induce metabolizing enzymes and no effect on PXR has been observed in animal models [94]. PXRs have also been implicated in the toxicity effects of certain TB drugs. Co-treatment of rifampicin and isoniazid in PXR-humanized mice disrupted the heme biosynthesis pathway resulting in liver injury [95]. These findings were not recapitulated with isoniazid metabolites, illustrating a mechanism for rifampicin and isoniazid-induced hepatotoxicity that is dependent on PXR signaling pathways yet independent of isoniazid metabolism [95].
To date, very few studies have examined the role of PXRs in the modulation of infectious disease, thus knowledge of PXR pathway regulation during infection is virtually nonexistent. PXRs have been documented to play a role in CD36 expression and activity, with PXR deficiency resulting in decreased lipid uptake [89], which would be beneficial for an M.tb-infected host. In terms of TB treatment, it would appear that the critical role of PXRs is efflux of the powerful anti-M.tb drug rifampicin as well as contribution to liver toxicity. Additional research into blocking PXR activity in order to increase rifampicin’s effectiveness and curb side effects could prove useful in TB treatment.
3.6. VDR
Vitamin D (cholecalciferol, vitamin D3, or 1,25-dihydroxyvitamn D3) has long been studied as an anti-TB therapy and administering vitamin D along with standard anti-TB drug regimens has improved clinical outcomes in some studies [96]. The vitamin D receptor (VDR) is a ligand dependent transcription factor and part of the NR superfamily which heterodimerizes with RXR and is constitutively expressed in macrophages [18, 97]. Polymorphisms in the VDR gene are well-studied due to their association with increased susceptibility to TB [98, 99]. Numerous studies have identified VDR polymorphisms associated with increased risk of developing TB, including Fokl, Taql, Msml, and Apal, however meta-analysis data have shown inconsistent results [99–104]. Larger studies with increased diversity of TB patients and controls are required for more definitive conclusions.
On the cellular level, M.tb and certain M.tb proteins can activate the VDR. Stimulation of monocytes with M.tb or the M.tb lipoprotein LpqH can induce nuclear translocation of VDR, where it can activate certain signaling pathways, without the addition of exogenous vitamin D [105]. Knockdown of VDR reduced control of M.tb strain H37Ra [10]. Ligation of VDR with its ligand, vitamin D, leads to the induction of the antimicrobial peptides cathelicidin and human beta-defensin 2 (HBD2), which can kill intracellular M.tb [106, 107]. The promoter region for hCAP-18, the only human cathelicidin, has multiple VDR response elements [18], showing a strong correlation between VDR ligand binding and up-regulation of this anti-mycobacterial protein, whereas the HBD2 promoter contains fewer VDR response elements and is also regulated by NF-κB. Stimulation of monocytes with LpqH also activated antibacterial autophagy in a cathelicidin-dependent manner [105], while stimulation with the prostaglandin PGE2 reduced VDR expression and abrogated vitamin D-mediated increases in cathelicidin expression and autophagy, and M.tb control [108]. These results demonstrate a link between VDR and autophagy as a method to control intra-macrophage M.tb growth.
In human leukocytes and the THP-1 macrophage cell line, transcriptome analysis of M.tb-infected cells showed an increase in VDR-regulated gene expression and revealed a correlation between VDR and lipid metabolism [109]. Interestingly, the addition of vitamin D decreased the number of lipid droplets in M.tb-infected THP-1 macrophages to that of uninfected cells by down-regulating PPARγ [109]. Addition of PPARγ agonists restored the lipid droplet formation, as well as negated the anti-M.tb effects of the VDR [109]. Thus, these data demonstrate that vitamin D regulates both VDR and PPARγ, and that VDR plays a role in lipid metabolism during M.tb infection.
One of the longest standing, somewhat effective TB therapies involved convalescence in the open air or in mountainous locations where patients would likely increase their vitamin D production and subsequently stimulate VDR signaling. It is interesting to note that while the VDR plays a role in combating TB through production of cathelicidin and at least partial regulation of lipid metabolism, treatment of exogenous vitamin D has had limited effectivity on its own. Since treatment with PPARγ agonists was shown to negate the effects of VDR signaling, perhaps the pathways regulated by PPARγ are dominant to those initiated by VDR, thus resulting in conditions permissive to M.tb growth. Numerous studies have linked polymorphisms of the VDR gene and increased susceptibility to TB. In addition, vitamin D supplementation has resulted in improved clinical outcomes when administered with standard anti-TB drug regimens [96]. Thus, in the context of TB, the importance of this NR cannot be refuted.
4. Conclusion
Targeting NRs as novel approaches for TB treatment appears to be a viable option considering that these transcription factors play a pivotal role in macrophage lipid metabolism, cholesterol efflux, phagosome maturation, and production of antimicrobial byproducts. The NRs PPARγ, LXR, and VDR have been the most studied in terms of M.tb infection, however there is still much to learn about the signaling pathways these NRs help regulate. Other NRs, including PXRs, REV-ERBα, TR4, and PPARα have been only recently implicated in progression or resistance to TB and it is mostly unclear how these NRs interact with each other, in addition to how these NRs are regulated during M.tb infection. Since this receptor superfamily consists of 48 identified NRs in humans [20], it is likely that more NRs will be associated with TB in the near future. In addition, the existing use of pharmacological interventions targeting NRs strongly suggests that following this line of research will be feasible for the discovery of novel methods to combat TB. Future NR interventions will need to be more specific given the current off target effects of NR modulators on the market today. This is an exciting time in NR and TB research with the potential for an effective treatment just around the corner.
| Nuclear Receptor | Model System | Mycobacterial species, strain, or molecule | Host and Macrophage Response | Mycobacterial Effects | Ref |
|---|---|---|---|---|---|
| PPARα | Mouse | H37Rv | controls bacterial burden in lung, spleen, and liver, inhibits inflammatory cytokine production | aids in anti-mycobacterial activity | [68] |
| Mouse | M. bovis BCG | controls pulmonary bacterial burden, prevents neutrophilic inflammation and COX2 expression in lung tissue, inhibits inflammatory cytokine production | aids in anti-mycobacterial activity | [68] | |
| BMDMs | H37Rv | aids in transcription factor EB activation, lipid body formation, autophagy, lysosomal biogenesis, phagosomal maturation, mitochondrial respiratory function | aids in anti-mycobacterial activity | [68] | |
| PPARγ | hMDMs | H37Rv and ManLam | Induces PPARγ expression in MR-dependent manner, upregulates IL-8 and COX2 independent of TLR-2 and NF-kB, PPARγ activity requires cPLA2 | supports Mycobacterial persistence | [11] |
| hMDMs | M. bovis BCG | Induces PPARγ to a lesser extent than M.tb or ManLam, induces IL-8 in an NF-kB-dependent, PPARγ-independent manner | [11] | ||
| hMDMs | M. smegmatis | Does not induce PPARγ | [11] | ||
| hMDMs | H37Rv | PPARγ knockdown results in differential gene expression of numerous immunology-related genes during M.tb infection including apoptosis related genes Bax and Mcl-1, 15-lipoxygenase is needed for PPARγ-mediated Mcl-1 production which limits apoptosis and aids in M.tb survival | supports Mycobacterial persistence | [60] | |
| Mouse | M. bovis BCG | PPARγ is downregulated in M. bovis BCG infected alveolar macrophages, addition of PPARγ agonist decreased BCG-induced PGE2 production | [58] | ||
| Murine peritoneal macrophage | M. bovis BCG | increased NF-kB activation and PPARγ expression in a TLR-2 dependent manner, neutralization of CD36 decreased PPARγ expression, lipid body formation and PGE2 production | [56] | ||
| WBC 264–9C macrophage cell line | H37Rv and P19 | increases PPARγ expression, p38 phosphorylation and IL-6 and TNF-α production, all effects were dependent on TLR2 | [55] | ||
| THP-1 | H37Ra and H37Rv | Aids in macrophage lipidation, PGE2 production, promoted an M2 macrophage phenotype, decreased ROS production, crosstalks with H37Ra lipids | supports Mycobacterial persistence | [10] | |
| THP-1 | H37Rv | increased PPARγ expression and lipid biogenesis, decreased lipolysis, increased surface levels of GLUT proteins dependent on PPARγ and AKT | [57] | ||
| TR4 | Mouse | H37Rv | TR4 knockdown results in reduced size of follicular granulomas and increased clearance of bacteria | supports Mycobacterial persistence | [15] |
| Mouse | Ketomycolic acid + M. smegmatis | multiple well-formed granulomas compared to M. smegmatis alone, TR4 knockdown mice had fewer granulomas | supports Mycobacterial persistence | [15] | |
| hMDMs | M. smegmatis and H37Rv | addition of ketomycolic acid increases bacterial survival which was abrogated in hMDMs with TR4 knocked down | supports Mycobacterial persistence | [15] | |
| THP-1 | H37Ra and H37Rv | aids in macrophage lipidation, PGE2 production, promotes an M2 macrophage phenotype, decreases ROS production, shown to crosstalk with M.tb lipids | supports Mycobacterial persistence | [10] | |
| LXRα/LXRβ | hMDMs and THP-1 | H37Rv | IL-36 upregulates LXR ligands to activate LXRs and induces the production of cathelicidin and defensins | aids in anti-mycobacterial activity | [76] |
| hMDMs | H37Rv | upregulated in response to M.tb | [75] | ||
| THP-1 | H37Ra | upregulated in response to M.tb, LXRα binds to Alu/DR4 elements which are associated with lipid metabolism, treatment with LXRα agonist increased expression of ABCA1 andABCGI and lead to decreased lipid body formation | [75] | ||
| Mouse | H37Rv | are upregulated in response to M.tb, increased Th1/Th17 responses, restricted lipid loading of macrophages | aids in anti-mycobacterial activity | [16] | |
| Mouse | H37Rv | transcript ionally regulates AIM, a macrophage apoptosis inhibitor that is upregulated in the serum of M.tb infected mice | aids in anti-mycobacterial activity | [110] | |
| THP-1 | H37Ra | prevents lipidation of macrophages, crosstalks with H37Ra lipids | aids in anti-mycobacterial activity | [10] | |
| REV-ERBα | hMDMs | H37Ra and H37Rv | represses IL-10 production, allows for phagolysosome maturation | aids in anti-mycobacterial activity | [14] |
| THP-1 | H37Ra and H37Rv | enhances autophagy progression, positively regulates LAMP1 andTFEB | aids in anti-mycobacterial activity | [13] | |
| PXR | hMDMs | H37Rv, Rifampicin-resistant M. tuberculosis (Zopf) | Modulates drug-efflux transporter expression, expedites drug efflux | supports Mycobacterial persistence | [92] |
| hMDMs | H37Ra and H37Rv | promotes foamy macrophage formation, decreases phagolysosomal fusion, inflammatory responses, and apoptosis | supports Mycobacterial persistence | [91] | |
| humanized mice overexpressing PXR | H37Rv | allows pulmonary M.tb growth in the presence of rifampicin, phenotype is rescued by addition of ketoconazole or during treatment with rifabutin | allows M.tb rifampicin resistance and increased bacterial survival | [92] | |
| humanized mice overexpressing PXR | H37Rv | decreased bacterial clearance from the lungs | supports Mycobacterial persistence | [91] | |
| VDR | human monocytes | LpqH (an M.tb lipoprotein) | initiates antibacterial autophagy and cathelicidin production | aids in anti-mycobacterial activity | [105] |
| THP-1 | H37Rv | transcriptome analysis revealed a correlation between VDR and lipid metabolism, decreased the number of lipid droplets, downregulated PPARγ | aids in anti-mycobacterial activity | [109] | |
| THP-1 | H37Ra | knockdown of VDR resulted in increased bacterial growth | aids in anti-mycobacterial activity | [10] |
Abbreviations: BMDMs, bone marrow derived macrophages; BCG, Bacillus Calmette-Guerin; cPLA2, cytosolic phospholipase A2; COX2, cyclooxygenase 2; hMDMs, human monocyte derived macrophages; LAMP1, lysosomal-associated membrane protein 1; LXR, liver X receptor; ManLam, mannosylated lipoarabinomannan; MR, mannose receptor; NF-kB, nuclear factor-kappa B; PXR, pregnane X receptor; PPAR, peroxisome proliferator-activated receptor; PGE2, prostaglandin E2; TLR2, toll-like receptor 2; TR4, testicular receptor 4; TFEB, transcription factor EB; VDR, vitamin D receptor
Funding:
This work was supported by the Texas Biomedical Research Institute Cowles Postdoctoral Fellowship (to CMLW), National Institutes of Health grants AI059636 and AI052458 and Texas Biomedical Research Institute Funds (to LSS). The funders had no role in the decision to publish or preparation of the manuscript. This work was presented in part at the Texas Tuberculosis Research Symposium (TTRS) 2018, El Paso, Texas, USA, sponsored by the Texas Tech University Health Sciences Center El Paso.
Each article needs to have a financial disclosure line, usually within the author bio section. Publication of this supplement was supported by The University of Texas Health Science Center at Houston.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: The authors declare that they have no competing interests.
References
- 1.Global tuberculosis report 2017. Geneva: World Health Organization, 2017 CC BY-NC-SA 3.0 IGO. [Google Scholar]
- 2.Mahajan S, et al. , Frienemies of infection: A chronic case of host nuclear receptors acting as cohorts or combatants of infection. Crit Rev Microbiol, 2016. 42(4): p. 526–34. [DOI] [PubMed] [Google Scholar]
- 3.Chawla A, Control of macrophage activation and function by PPARs. Circ Res, 2010. 106(10): p. 1559–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagy L, et al. , Nuclear hormone receptors enable macrophages and dendritic cells to sense their lipid environment and shape their immune response. Physiol Rev, 2012. 92(2): p. 739–89. [DOI] [PubMed] [Google Scholar]
- 5.Glass CK and Saijo K, Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol, 2010. 10(5): p. 365–76. [DOI] [PubMed] [Google Scholar]
- 6.Kiss M, Czimmerer Z, and Nagy L, The role of lipid-activated nuclear receptors in shaping macrophage and dendritic cell function: From physiology to pathology. J Allergy Clin Immunol, 2013. 132(2): p. 264–86. [DOI] [PubMed] [Google Scholar]
- 7.Rigamonti E, Chinetti-Gbaguidi G, and Staels B, Regulation of macrophage functions by PPAR-alpha, PPAR-gamma, and LXRs in mice and men. Arterioscler Thromb Vasc Biol, 2008. 28(6): p. 1050–9. [DOI] [PubMed] [Google Scholar]
- 8.Chawla A, et al. , Nuclear receptors and lipid physiology: opening the X-files. Science, 2001. 294(5548): p. 1866–70. [DOI] [PubMed] [Google Scholar]
- 9.Via M, Nuclear Receptors: The Pipeline Outlook. 2010. [Google Scholar]
- 10.Mahajan S, et al. , Mycobacterium tuberculosis modulates macrophage lipid-sensing nuclear receptors PPAR gamma and TR4 for survival. J Immunol, 2012. 188(11): p. 5593–603. [DOI] [PubMed] [Google Scholar]
- 11.Rajaram MV, et al. , Mycobacterium tuberculosis activates human macrophage peroxisome proliferator-activated receptor gamma linking mannose receptor recognition to regulation of immune responses. J Immunol, 2010. 185(2): p. 929–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han M, et al. , Liver X receptor gene polymorphisms in tuberculosis: effect on susceptibility. PLoS One, 2014. 9(5): p. e95954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandra V, et al. , NR1D1 ameliorates Mycobacterium tuberculosis clearance through regulation of autophagy. Autophagy, 2015. 11(11): p. 1987–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandra V, et al. , Human IL10 gene repression by Rev-erbalpha ameliorates Mycobacterium tuberculosis clearance. J Biol Chem, 2013. 288(15): p. 10692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dkhar HK, et al. , Mycobacterium tuberculosis keto-mycolic acid and macrophage nuclear receptor TR4 modulate foamy biogenesis in granulomas: a case of a heterologous and noncanonical ligand-receptor pair. J Immunol, 2014. 193(1): p. 295–305. [DOI] [PubMed] [Google Scholar]
- 16.Korf H, et al. , Liver X receptors contribute to the protective immune response against Mycobacterium tuberculosis in mice. J Clin Invest, 2009. 119(6): p. 1626–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saini A, et al. , An Accord of Nuclear Receptor Expression in M. tuberculosis Infected Macrophages and Dendritic Cells. Sci Rep, 2018. 8(1): p. 2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selvaraj P, Vitamin D, vitamin D receptor, and cathelicidin in the treatment of tuberculosis. Vitam Horm, 2011. 86: p. 307–25. [DOI] [PubMed] [Google Scholar]
- 19.Mangelsdorf DJ, et al. , The nuclear receptor superfamily: the second decade. Cell, 1995. 83(6): p. 835–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maglich JM, et al. , Comparison of complete nuclear receptor sets from the human, Caenorhabditis elegans and Drosophila genomes. Genome Biol, 2001. 2(8): p. RESEARCH0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barish GD, et al. , A Nuclear Receptor Atlas: macrophage activation. Mol Endocrinol, 2005. 19(10): p. 2466–77. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez-Martinez R, et al. , Vitamin D-dependent recruitment of corepressors to vitamin D/retinoid X receptor heterodimers. Mol Cell Biol, 2008. 28(11): p. 3817–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perissi V, et al. , A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell, 2004. 116(4): p. 511–26. [DOI] [PubMed] [Google Scholar]
- 24.Aranda A and Pascual A, Nuclear hormone receptors and gene expression. Physiol Rev, 2001. 81(3): p. 1269–304. [DOI] [PubMed] [Google Scholar]
- 25.Glass CK and Ogawa S, Combinatorial roles of nuclear receptors in inflammation and immunity. Nat Rev Immunol, 2006. 6(1): p. 44–55. [DOI] [PubMed] [Google Scholar]
- 26.Ahmadian M, et al. , PPAR gamma signaling and metabolism: the good, the bad and the future. Nat Med, 2013. 19(5): p. 557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bourguet W, et al. , Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell, 2000. 5(2): p. 289–98. [DOI] [PubMed] [Google Scholar]
- 28.Rosenfeld MG, Lunyak VV, and Glass CK, Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev, 2006. 20(11): p. 1405–28. [DOI] [PubMed] [Google Scholar]
- 29.Wagner BL, et al. , Promoter-specific roles for liver X receptor/corepressor complexes in the regulation of ABCA1 and SREBP1 gene expression. Mol Cell Biol, 2003. 23(16): p. 5780–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu X, et al. , Liver X receptors interact with corepressors to regulate gene expression. Mol Endocrinol, 2003. 17(6): p. 1019–26. [DOI] [PubMed] [Google Scholar]
- 31.Joseph SB, et al. , Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med, 2003. 9(2): p. 213–9. [DOI] [PubMed] [Google Scholar]
- 32.Germain P, et al. , International Union of Pharmacology. LXIII. Retinoid X receptors. Pharmacol Rev, 2006. 58(4): p. 760–72. [DOI] [PubMed] [Google Scholar]
- 33.Leopold Wager CM and Wormley FL, Classical versus alternative macrophage activation: the Ying and the Yang in host defense against pulmonary fungal infections. Mucosal Immunol, 2014. 7(5): p. 1023–1035. [DOI] [PubMed] [Google Scholar]
- 34.Mantovani A, et al. , The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol, 2004. 25(12): p. 677–86. [DOI] [PubMed] [Google Scholar]
- 35.Mosser DM and Edwards JP, Exploring the full spectrum of macrophage activation. Nat Rev Immunol, 2008. 8(12): p. 958–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajaram MV, et al. , Macrophage immunoregulatory pathways in tuberculosis. Semin Immunol, 2014. 26(6): p. 471–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez FO, Helming L, and Gordon S, Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol, 2009. 27: p. 451–83. [DOI] [PubMed] [Google Scholar]
- 38.Martinez FO and Gordon S, The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep, 2014. 6: p. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torrelles JB and Schlesinger LS, Integrating Lung Physiology, Immunology, and Tuberculosis. Trends Microbiol, 2017. 25(8): p. 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang JT, et al. , Interleukin-4-dependent production of PPAR-gamma ligands in macrophages by 12/15-lipoxygenase. Nature, 1999. 400(6742): p. 378–82. [DOI] [PubMed] [Google Scholar]
- 41.Odegaard JI, et al. , Macrophage-specific PPAR gamma controls alternative activation and improves insulin resistance. Nature, 2007. 447(7148): p. 1116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szanto A, et al. , STAT6 transcription factor is a facilitator of the nuclear receptor PPAR gamma-regulated gene expression in macrophages and dendritic cells. Immunity, 2010. 33(5): p. 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Czimmerer Z, et al. , Identification of novel markers of alternative activation and potential endogenous PPAR gamma ligand production mechanisms in human IL-4 stimulated differentiating macrophages. Immunobiology, 2012. 217(12): p. 1301–14. [DOI] [PubMed] [Google Scholar]
- 44.Nobs SP, et al. , PPAR gamma in dendritic cells and T cells drives pathogenic type-2 effector responses in lung inflammation. J Exp Med, 2017. 214(10): p. 3015–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitz J, et al. , IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity, 2005. 23(5): p. 479–90. [DOI] [PubMed] [Google Scholar]
- 46.Kurowska-Stolarska M, et al. , IL-33 amplifies the polarization of alternatively activated macrophages that contribute to airway inflammation. J Immunol, 2009. 183(10): p. 6469–77. [DOI] [PubMed] [Google Scholar]
- 47.Fontaine C, et al. , The nuclear receptor Rev-erbalpha is a liver X receptor (LXR) target gene driving a negative feedback loop on select LXR-induced pathways in human macrophages. Mol Endocrinol, 2008. 22(8): p. 1797–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Almeida PE, et al. , PPAR gamma Expression and Function in Mycobacterial Infection: Roles in Lipid Metabolism, Immunity, and Bacterial Killing. PPAR Res, 2012. 2012: p. 383829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ricote M, et al. , The peroxisome proliferator-activated receptor-gamma is a negative regulator of macrophage activation. Nature, 1998. 391(6662): p. 79–82. [DOI] [PubMed] [Google Scholar]
- 50.Jiang C, Ting AT, and Seed B, PPAR-gamma agonists inhibit production of monocyte inflammatory cytokines. Nature, 1998. 391(6662): p. 82–6. [DOI] [PubMed] [Google Scholar]
- 51.Harmon GS, Lam MT, and Glass CK, PPARs and lipid ligands in inflammation and metabolism. Chem Rev, 2011. 111(10): p. 6321–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider C, et al. , Induction of the nuclear receptor PPAR-gamma by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat Immunol, 2014. 15(11): p. 1026–37. [DOI] [PubMed] [Google Scholar]
- 53.Almeida PE, et al. , Mycobacterium bovis bacillus Calmette-Guerin infection induces TLR2-dependent peroxisome proliferator-activated receptor gamma expression and activation: functions in inflammation, lipid metabolism, and pathogenesis. J Immunol, 2009. 183(2): p. 1337–45. [DOI] [PubMed] [Google Scholar]
- 54.Guirado E, et al. , Deletion of PPAR-gamma in lung macrophages provides an immunoprotective response against M. tuberculosis infection in mice. Tuberculosis (Edinb), 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu L, et al. , Mycobacterium tuberculosis 19-kDa lipoprotein induces Toll-like receptor 2-dependent peroxisome proliferator-activated receptor gamma expression and promotes inflammatory responses in human macrophages. Mol Med Rep, 2015. 11(4): p. 2921–6. [DOI] [PubMed] [Google Scholar]
- 56.Almeida PE, et al. , Differential TLR2 downstream signaling regulates lipid metabolism and cytokine production triggered by Mycobacterium bovis BCG infection. Biochim Biophys Acta, 2014. 1841(1): p. 97–107. [DOI] [PubMed] [Google Scholar]
- 57.Dasgupta S and Rai RC, PPAR-gamma and Akt regulate GLUT1 and GLUT3 surface localization during Mycobacterium tuberculosis infection. Mol Cell Biochem, 2018. 440(1–2): p. 127–138. [DOI] [PubMed] [Google Scholar]
- 58.Kogiso M, et al. , Role of PPAR gamma in COX-2 activation in mycobacterial pulmonary inflammation. Inflammation, 2012. 35(5): p. 1685–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan Q, et al. , A single-stranded DNA aptamer against mannose-capped lipoarabinomannan enhances anti-tuberculosis activity of macrophages through downregulation of lipid-sensing nuclear receptor peroxisome proliferator-activated receptor gamma expression. Microbiol Immunol, 2017. 61(2): p. 92–102. [DOI] [PubMed] [Google Scholar]
- 60.Arnett E, et al. , PPARγ is critical for Mycobacterium tuberculosis induction of Mcl-1 and limitation of human macrophage apoptosis. PLoS Pathog, 2018. 14(6): p. e1007100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dodd CE, et al. , CD36-Mediated Uptake of Surfactant Lipids by Human Macrophages Promotes Intracellular Growth of Mycobacterium tuberculosis. J Immunol, 2016. 197(12): p. 4727–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lamkanfi M and Dixit VM, Manipulation of host cell death pathways during microbial infections. Cell Host Microbe, 2010. 8(1): p. 44–54. [DOI] [PubMed] [Google Scholar]
- 63.Behar SM, et al. , Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol, 2011. 4(3): p. 279–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guirado E, et al. , Characterization of host and microbial determinants in individuals with latent tuberculosis infection using a human granuloma model. MBio, 2015. 6(1): p. e02537–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manca C, et al. , Host targeted activity of pyrazinamide in Mycobacterium tuberculosis infection. PLoS One, 2013. 8(8): p. e74082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rajaram MVS, et al. , M. tuberculosis-Initiated Human Mannose Receptor Signaling Regulates Macrophage Recognition and Vesicle Trafficking by FcRgamma-Chain, Grb2, and SHP-1. Cell Rep, 2017. 21(1): p. 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pontis S, et al. , Macrophage-derived lipid agonists of PPAR-alpha as intrinsic controllers of inflammation. Crit Rev Biochem Mol Biol, 2016. 51(1): p. 7–14. [DOI] [PubMed] [Google Scholar]
- 68.Kim YS, et al. , PPAR-alpha Activation Mediates Innate Host Defense through Induction of TFEB and Lipid Catabolism. J Immunol, 2017. 198(8): p. 3283–3295. [DOI] [PubMed] [Google Scholar]
- 69.Lin SJ, et al. , Minireview: Pathophysiological roles of the TR4 nuclear receptor: lessons learned from mice lacking TR4. Mol Endocrinol, 2014. 28(6): p. 805–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie S, et al. , TR4 nuclear receptor functions as a fatty acid sensor to modulate CD36 expression and foam cell formation. Proc Natl Acad Sci U S A, 2009. 106(32): p. 13353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Janowski BA, et al. , An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature, 1996. 383(6602): p. 728–31. [DOI] [PubMed] [Google Scholar]
- 72.Janowski BA, et al. , Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc Natl Acad Sci U S A, 1999. 96(1): p. 266–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Joseph SB, et al. , LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell, 2004. 119(2): p. 299–309. [DOI] [PubMed] [Google Scholar]
- 74.Valledor AF, et al. , Activation of liver X receptors and retinoid X receptors prevents bacterial-induced macrophage apoptosis. Proc Natl Acad Sci U S A, 2004. 101(51): p. 17813–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bouttier M, et al. , Alu repeats as transcriptional regulatory platforms in macrophage responses to M. tuberculosis infection. Nucleic Acids Res, 2016. 44(22): p. 10571–10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahsan F, et al. , IL-36/LXR axis modulates cholesterol metabolism and immune defense to Mycobacterium tuberculosis. Sci Rep, 2018. 8(1): p. 1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bah SY, et al. , Immune oxysterols: Role in mycobacterial infection and inflammation. J Steroid Biochem Mol Biol, 2017. 169: p. 152–163. [DOI] [PubMed] [Google Scholar]
- 78.Su VY, et al. , Statin Use Is Associated With a Lower Risk of TB. Chest, 2017. 152(3): p. 598–606. [DOI] [PubMed] [Google Scholar]
- 79.Dutta NK, et al. , Statin adjunctive therapy shortens the duration of TB treatment in mice. J Antimicrob Chemother, 2016. 71(6): p. 1570–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kirchgessner TG, et al. , Beneficial and Adverse Effects of an LXR Agonist on Human Lipid and Lipoprotein Metabolism and Circulating Neutrophils. Cell Metab, 2016. 24(2): p. 223–33. [DOI] [PubMed] [Google Scholar]
- 81.Kojetin DJ and Burris TP, REV-ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov, 2014. 13(3): p. 197–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yin L and Lazar MA, The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol, 2005. 19(6): p. 1452–9. [DOI] [PubMed] [Google Scholar]
- 83.Guillaumond F, et al. , Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms, 2005. 20(5): p. 391–403. [DOI] [PubMed] [Google Scholar]
- 84.Gervois P, et al. , Fibrates increase human REV-ERBalpha expression in liver via a novel peroxisome proliferator-activated receptor response element. Mol Endocrinol, 1999. 13(3): p. 400–9. [DOI] [PubMed] [Google Scholar]
- 85.Raghuram S, et al. , Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol, 2007. 14(12): p. 1207–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Forman BM, et al. , Cross-talk among ROR alpha 1 and the Rev-erb family of orphan nuclear receptors. Mol Endocrinol, 1994. 8(9): p. 1253–61. [DOI] [PubMed] [Google Scholar]
- 87.Miyajima N, et al. , Identification of two novel members of erbA superfamily by molecular cloning: the gene products of the two are highly related to each other. Nucleic Acids Res, 1988. 16(23): p. 11057–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qiao E, et al. , Expression of the PXR gene in various types of cancer and drug resistance. Oncol Lett, 2013. 5(4): p. 1093–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sui Y, et al. , Deficiency of PXR decreases atherosclerosis in apoE-deficient mice. J Lipid Res, 2011. 52(9): p. 1652–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dubrac S, et al. , Modulation of T lymphocyte function by the pregnane X receptor. J Immunol, 2010. 184(6): p. 2949–57. [DOI] [PubMed] [Google Scholar]
- 91.Bhagyaraj E, et al. , Human Xenobiotic Nuclear Receptor PXR Augments Mycobacterium tuberculosis Survival. J Immunol, 2016. 197(1): p. 244–55. [DOI] [PubMed] [Google Scholar]
- 92.Bhagyaraj E, et al. , A human xenobiotic nuclear receptor contributes to nonresponsiveness of Mycobacterium tuberculosis to the antituberculosis drug rifampicin. J Biol Chem, 2018. 293(10): p. 3747–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shehu AI, et al. , The pregnane X receptor in tuberculosis therapeutics. Expert Opin Drug Metab Toxicol, 2016. 12(1): p. 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mae T, et al. , Effect of a new rifamycin derivative, rifalazil, on liver microsomal enzyme induction in rat and dog. Xenobiotica, 1998. 28(8): p. 759–66. [DOI] [PubMed] [Google Scholar]
- 95.Li F, et al. , Human PXR modulates hepatotoxicity associated with rifampicin and isoniazid co-therapy. Nat Med, 2013. 19(4): p. 418–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sutaria N, Liu CT, and Chen TC, Vitamin D Status, Receptor Gene Polymorphisms, and Supplementation on Tuberculosis: A Systematic Review of Case-Control Studies and Randomized Controlled Trials. J Clin Transl Endocrinol, 2014. 1(4): p. 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pinette KV, et al. , Vitamin D receptor as a drug discovery target. Mini Rev Med Chem, 2003. 3(3): p. 193–204. [DOI] [PubMed] [Google Scholar]
- 98.Azad AK, Sadee W, and Schlesinger LS, Innate immune gene polymorphisms in tuberculosis. Infect Immun, 2012. 80(10): p. 3343–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gao L, et al. , Vitamin D receptor genetic polymorphisms and tuberculosis: updated systematic review and meta-analysis. Int J Tuberc Lung Dis, 2010. 14(1): p. 15–23. [PubMed] [Google Scholar]
- 100.Areeshi MY, et al. , A reappraised meta-analysis of the genetic association between vitamin D receptor BsmI (rs1544410) polymorphism and pulmonary tuberculosis risk. Biosci Rep, 2017. 37(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fol M, et al. , Immune response gene polymorphisms in tuberculosis. Acta Biochim Pol, 2015. 62(4): p. 633–40. [DOI] [PubMed] [Google Scholar]
- 102.Huang L, et al. , Vitamin D Receptor Gene FokI Polymorphism Contributes to Increasing the Risk of Tuberculosis: An Update Meta-Analysis. Medicine (Baltimore), 2015. 94(51): p. e2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cao Y, et al. , Vitamin D receptor gene FokI polymorphisms and tuberculosis susceptibility: a meta-analysis. Arch Med Sci, 2016. 12(5): p. 1118–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen C, et al. , Vitamin D receptor gene polymorphisms on the risk of tuberculosis, a meta-analysis of 29 case-control studies. PLoS One, 2013. 8(12): p. e83843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shin DM, et al. , Mycobacterial lipoprotein activates autophagy via TLR2/1/CD14 and a functional vitamin D receptor signalling. Cell Microbiol, 2010. 12(11): p. 1648–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu PT, et al. , Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science, 2006. 311(5768): p. 1770–1773. [DOI] [PubMed] [Google Scholar]
- 107.Liu PT, et al. , Convergence of IL-1beta and VDR activation pathways in human TLR2/1-induced antimicrobial responses. PLoS One, 2009. 4(6): p. e5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wan M, et al. , Prostaglandin E2 suppresses hCAP18/LL-37 expression in human macrophages via EP2/EP4: implications for treatment of Mycobacterium tuberculosis infection. FASEB J, 2018: p. fj201701308. [DOI] [PubMed] [Google Scholar]
- 109.Salamon H, et al. , Cutting edge: Vitamin D regulates lipid metabolism in Mycobacterium tuberculosis infection. J Immunol, 2014. 193(1): p. 30–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sanjurjo L, et al. , The scavenger protein apoptosis inhibitor of macrophages (AIM) potentiates the antimicrobial response against Mycobacterium tuberculosis by enhancing autophagy. PLoS One, 2013. 8(11): p. e79670. [DOI] [PMC free article] [PubMed] [Google Scholar]