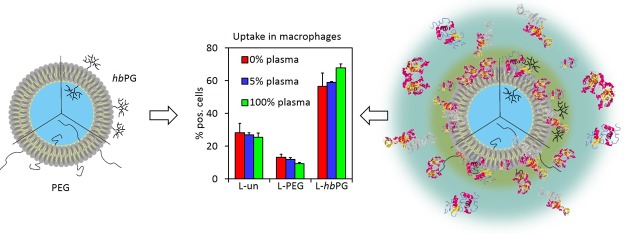
Abstract
Liposomes are established drug carriers that are employed to transport and deliver hydrophilic drugs in the body. To minimize unspecific cellular uptake, nanocarriers are commonly modified with poly(ethylene glycol) (PEG), which is known to minimize unspecific protein adsorption. However, to date, it has not been studied whether this is an intrinsic and specific property of PEG or if it can be transferred to hyperbranched polyglycerol (hbPG) as well. Additionally, it remains unclear if the reduction of unspecific cell uptake is independent of the “basic” carrier at which a surface functionalization with polymers is usually applied. Therefore, we studied the protein corona of differently functionalized liposomes (unfunctionalized vs PEG or hbPG-functionalized) using PEGylated and PGylated lipids. Their cellular uptake in macrophages was compared. For all three liposomal samples, rather similar protein corona compositions were found, and also—more importantly—the total amount of proteins adsorbed was very low compared to other nanoparticles. Interestingly, the cellular uptake was then significantly changed by the surface functionalization itself, despite the adsorption of a small amount of proteins: although the PEGylation of liposomes resulted in the abovementioned decreased cell uptake, functionalization with hbPG lead to enhanced macrophage interaction—both in the media with and without proteins. In comparison to other nanocarrier systems, this seems to be a liposome-specific effect related to the low amount of adsorbed proteins.
Introduction
Drug delivery plays an increasingly important role in modern nanomedicine, where nanocarriers are designed to transport a drug to a specific target location in the body. The advantages of such a delivery system include in particular the protection of the active component from degradation by the metabolism, as well as the decrease of systemic side effects.1−3
Among the different delivery systems, liposomes are a valuable class of nanocarriers for several reasons. They are spherical vesicles composed of one or multiple phospholipid bilayers. Because of the amphiphilic character of the phospholipids, the membrane bilayer displays an aqueous core and a hydrophilic outer surface as well as a lipophilic membrane interspace. Therefore, depending on their polarity, a high variety of lipophilic and hydrophilic drugs can be encapsulated, either in the membrane or in the core, respectively. Additionally, by choice of natural or synthetic (phospho)lipids as components, liposomes may be designed to be biocompatible and even biodegradable. These factors render liposomes promising drug carriers.4−6 Moreover, they already found their way into the market, for example, as the formulation Doxil, which contains the chemotherapeutic drug doxorubicin encapsulated in a poly(ethylene glycol) (PEG)ylated liposome,7,8 as well as in a non-PEGylated liposome formulation called Myocet9 and others.10
To predict the behavior of potential nanocarriers in a biological system, it is necessary to consider and analyze the protein adsorption on the surface of the nanomaterial.11,12 Liposomes are no exception here, as they also adsorb proteins on their surface, as shown by Caracciolo.13 This adsorption process occurs as soon as the nanomaterial comes into contact with body fluids, for example, via intravenous injection into the bloodstream. The drug carrier is confronted with a high amount of proteins and other biomolecules, which adsorb onto the material’s surface to form the so-called protein corona.14−18 Thereby, the adsorbed proteins cover the liposomal surface, thus contributing to its biological identity. For many other colloidal nanomaterials, it has been shown that this biological identity determines the organism’s response toward the nanocarrier, influencing cell uptake, clearance, and body distribution.19
Depending on the binding affinity, the protein corona can be divided into the so-called hard and soft corona, according to the respective experimental accessibility. The hard corona consists of proteins that exhibit a high binding affinity and that are directly adsorbed onto the nanomaterial’s surface. These proteins are tightly bound and show long residence times, which allows to easily isolate and analyze them, for example, by centrifugation.11,12 Soft corona proteins on the other hand are more loosely bound and show high exchange rates in a biological medium. Because of the low binding affinities and highly dynamic behavior, the isolation of proteins from the soft corona is experimentally challenging.12,14,20 To access the loosely bound proteins, we already successfully implemented asymmetric flow field-flow fractionation (AF4) in our previous work.21 AF4 is a chromatography-like separation technique, whose separation principle is based on diffusion coefficients and thereby separates the sample according to size. Because of the lack of a stationary phase, only minimum shear stress is applied to the sample. Consequently, it is a very mild technique, which can also separate free proteins from fragile analytes like protein–particle complexes including strongly and weakly bound proteins or other self-assembled systems like liposomes or micelles, which in some cases (depending on their molecular composition) are too fragile to be centrifuged.22,23
Even liposomes formed from natural phospholipids are still recognized as foreign objects by the body and are therefore rapidly cleared from the body by the mononuclear phagocytic system.4 To prevent this rapid clearance, hydrophilic long-chain polymers can be attached to the liposomal surface, with the most prominent being PEG.6 The attachment of these polymers potentially decreases the unspecific protein adsorption and forms “stealth” liposomes, which show a longer blood circulation time and decreased clearance by the immune system.24−28
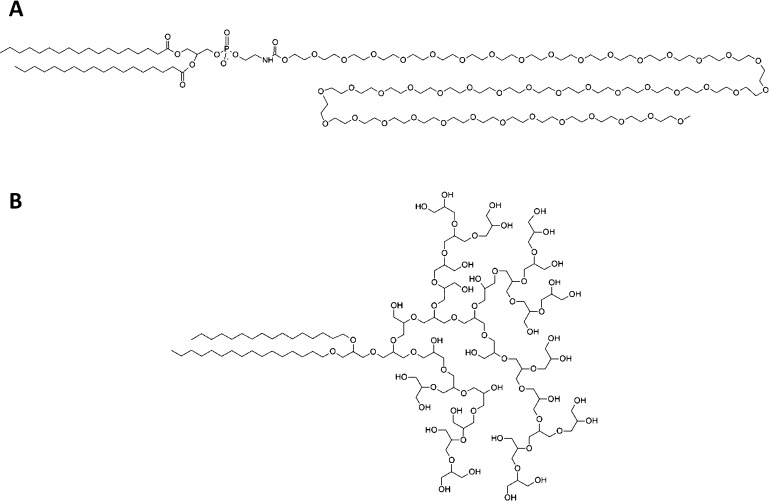
Caracciolo et al. investigated the protein corona of PEGylated liposomes and lipoplexes as well as the ability of different PEG chain lengths to decrease unspecific cell uptake.13,29−31 However, the biological behavior of liposomes functionalized with polymers exhibiting a different structure such as hyperbranched polyglycerol (hbPG) has not been analyzed with regard to the protein corona to this point. This polymer with its tree-like structure provides a hydroxyl group on each branch end, leading to a high hydrophilicity comparable to PEG, but with more possible functionalization sites for targeted delivery (Figure 1).32−34
Figure 1.
Structure of (A) linear PEG and (B) hbPG. Repeating units are displayed individually to allow for a better structural comparison between the two polymers.
So far, PEG- and hbPG-functionalized liposomal systems were already analyzed with respect to cellular uptake,35 and a comparison by Wagener et al. with respect to their in vivo fate by positron emission tomography imaging found a comparable biodistribution in mice but differences in splenic uptake depending on the functionalization.36 However, the protein corona, as well as possible functionalization effects on a cellular level, have not been studied to date.
In this paper, we analyzed the influence of the abovementioned liposome surface functionalizations (unfunctionalized, PEG, and hbPG) on the protein corona. We evaluated the separation by AF4 compared to the usually applied centrifugation to isolate the protein corona.12,21 By combining these two techniques, the influence of the surface functionalization of liposomes on the soft and the hard protein corona was investigated. Moreover, we evaluated the influence of the different surface functionalities in the presence and absence of proteins on the biological response by executing cell uptake experiments in macrophages.
We show that despite the striking similarities in the protein corona, the uptake behavior varies significantly between the linear and hyperbranched stealth-type structures, which correlates with a generally low adsorbed protein mass. We conclude that the intrinsic properties of the “stealth” functionalization itself can have a strong and sometimes superseding impact on cellular uptake and accordingly on the biological identity of liposomal formulations, depending on the overall amount of adsorbed proteins.
Experimental Section
Materials
Phosphate-buffered saline (PBS) and Sephacryl S500-HR were procured from Sigma-Aldrich (USA), whereas ethanol (99.5%) and cholesterol (Chol) were acquired from Carl Roth, Germany. A Milli Q device (Merck Millipore, Germany) was used to obtain demineralized water. Egg phosphatidyl choline (EPC) and mPEG-DSPE (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-3000], both from Lipoid, Germany) were used for liposome formulation. The dialkyl-based hbPG amphiphiles were synthesized as described in the Supporting Information,33 where also the polymer characterization can be found. The membrane dye DiI (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate) was purchased from Thermo Fisher Scientific.
Human Blood Plasma
In total, 10 healthy participants donated their blood plasma at the Transfusion Center of the University Clinic of Mainz, Germany, according to standard guidelines, which was then combined in a pooled batch. The blood plasma was stored at −20 °C and centrifuged before use (20 000 g and 4 °C for 1 h, Sigma 3-30K, Germany) to eliminate protein precipitates and cell fragments.
Liposome Formulation by Dual Centrifugation
All lipids and amphiphilic polymers were dissolved in ethanol and stored at −20 °C. After thawing at room temperature, the stock solutions of Chol (20 mg mL–1), EPC (50 mg mL–1), and amphiphilic polymer (20 mg mL–1) were combined in a polymerase chain reaction (PCR) tube (Kisker Biotech, Steinfurt, Germany) to yield the intended compositions. The unfunctionalized bare liposomes (L-un) consisted of 55:45 mol % of EPC–Chol. For liposomes functionalized with PEG or hbPG, 5 mol % of EPC was substituted with either mPEG-DSPE (2750 g mol–1) or amphiphilic dialkyl-based hbPG polymer (2750 g mol–1), resulting in a composition of 55:40:5 mol % of EPC–Chol–PEG/hbPG. A 0.2 mol % of the membrane dye DiI was added to each composition. The combined lipid solutions with a total lipid mass of 5 mg were dried in a SpeedVac vacuum centrifuge (Eppendorf, Hamburg, Germany) at 30 °C for at least 6 h and then in a lyophilization unit (Alpha 2-4 LD Christ, Osterode am Harz, Germany) for at least 24 h. PBS (9.3 μL) was added to the dry lipids and incubated for 10 min at room temperature. After adding 71 mg of ceramic beads (SiLiBeads ZY, 0.3–0.4 mm, Sigmund Lindner, Warmensteinach, Germany), the PCR tube was subjected to a dual centrifuge (Rotanta 400 with a prototype dc-rotor, Hettich, Tuttlingen, Germany) in 3D-printed insets for PCR tubes (in-house Helm Group, Johannes Gutenberg University, Mainz, Germany) for 20 min at 2500 rpm. The obtained vesicular phospholipid gel was then diluted with 28.5 μL of PBS and subjected again to dual centrifugation for 2 × 2 min at 2500 rpm while turning the reaction tube by 180° in between. The highly concentrated resulting liposome suspension was stored at 4 °C until usage.
Liposome Purification
Preparative size exclusion chromatography was performed via an Agilent 1100 System (Agilent, Germany) to remove the nonencapsulated cargo and free lipids from the nanocarrier solution. A volume of 60 μL of the liposome suspension as obtained after dual centrifugation was injected into the system running with PBS at a flow rate of 1 mL min–1. A BioRad UNO Q1 column (BioRad, Munich, Germany) filled with Sephacryl S500-HR was used for separation. A multiwavelength detector (G1365A Agilent 1100 Series, Germany) was used for the detection of the absorption of DiI-labeled liposomes at 550 nm. An automated fraction collector collected the resulting purified liposome solution with a volume of 600 μL.
Light Scattering
The presented light scattering (LS) experiments were performed with an ALV spectrometer (ALV-GmbH, Germany). The setup consists of a goniometer and an ALV/LSE-5004 multiple-tau full-digital correlator with 320 channels. As a light source, a He–Ne laser was used at a wavelength of 632.8 nm. The samples were filtered through Millex LCR 0.45 μm syringe filters (Merck, Germany) into cylindrical quartz cuvettes (18 mm diameter, Hellma, Germany). To remove the dust from the cuvettes, they were cleaned in an acetone fountain prior to usage. Liposome samples were prepared in a concentration of 0.001 mg mL–1 in PBS. The shown experiments were performed in triplet, and the CONTIN algorithm37,38 was used for data analysis.
Zeta Potential
A Malvern Zetasizer Nano series (UK) with disposable folded capillary cells was used to determine the zeta-potential (ζ-potential) of the liposome samples. The liposome solutions were diluted to a concentration of 0.1 mg mL–1 in 1 mM KCl.
Protein Corona Preparation
The liposomes were diluted in PBS containing 5% human blood plasma to give a final concentration of 2 mg mL–1. Under constant agitation, the dispersion was incubated for 1 h at 37 °C.
The obtained liposome–protein complexes (see below) were resuspended in the corresponding solvent. To quantify the protein concentration, the Pierce 660 nm Protein Assay (Thermo Scientific, Germany) was used according to the manufacturer’s instruction.
Separation of Liposomes with Protein Corona by Centrifugation
Centrifugation at 20 000 g at 4 °C for 1 h (Sigma 3-30K, Germany) removed unbound proteins. The resulting pellet consisted of liposomes with adsorbed proteins and was resuspended in 1 mL of PBS and subsequently washed by three centrifugation steps at 20 000 g and 4 °C for 1 h. After the last centrifugation step, the sample was resuspended in 200 μL of PBS. The samples were dried by a SpeedVac Concentrator (Savant DNA120, Thermo Scientific, USA) and resuspended again to align the sample preparation to the AF4 procedure.
Separation of Liposomes with Protein Corona by Asymmetric Flow Field-Flow Fractionation
The used Postnova AF2000 system was equipped with an autosampler, the tip and focus pumps, a degasser, a smart stream splitter, and a fraction collector. A stainless steel frit covered with a regenerated cellulose membrane with a molecular cutoff of 10 kDa and a 500 μm spacer formed the separation channel. As detectors, a fluorescence detector (1260 Infinity, Agilent Technologies, USA) at 549/565 nm and a UV detector (SPD-20A, Postnova, Germany) at 280 nm were used. Data evaluation was executed with an AF2000Control 2.0.8.0 (Postnova, Germany).
The mixture of liposomes and proteins was separated under the following flow conditions: the channel flow was split to result in a detector flow of 0.2 mL min–1. The initial crossflow was 1 mL min–1 and kept constant for 7.2 min. Then it was decreased exponentially over 20 min to a crossflow of 0.05 mL min–1, which was kept constant for another 7 min. A second exponential decrease was used over 20 min to lower the crossflow to 0 mL min–1. The separation was continued without the crossflow for another 30 min. PBS was used as a carrier liquid.
The samples were prepared as described above, and a sample volume of 50 μL was injected. After the the fractions were collected by the fraction collector, they were dried in a SpeedVac and then resuspended in 350 μL of H2O for further analysis.
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis
A protein mass of 1 μg in a volume of 16.25 μL was combined with 2.5 μL of the NuPAGE sample reducing agent and 6.25 μL of the NuPAGE lithium dodecyl sulfate sample buffer. After an incubation step at 70 °C for 10 min, the mixture was applied on a NuPAGE 10% Bis–Tris Protein Gel. SeeBlue Plus2 Pre-Stained Standard (all Invitrogen, Thermo Fisher Scientific, USA) served as a molecular ladder. Electrophoresis was carried out at 100 V for 1.5 h in the NuPAGE MES SDS running buffer.
The SilverQuest Silver Staining Kit (Thermo Fisher Scientific, USA) was used according to manufacturer’s instructions to stain the gel.
Transmission Electron Microscopy
A volume of 2 μL of the corresponding liposome sample, embedded in 1% trehalose with 4% uranyl acetate, was placed on a lacey grid, and measurements were executed on a FEI Tecnai F20 transmission electron microscope with a working voltage of 200 kV. An Ultrascan 1000 (Gatan, USA) charge-coupled device camera was used to obtain the electron micrographs, and the images were collected with the Digital Micrograph software (Gatan, USA).
Detection of Corona Proteins (IgG) on the Surface of Liposomes by Flow Cytometry
Liposomes (1 μg) were incubated for 30 min at room temperature with 5 μL of a protein-labeling reagent. Zenon Alexa Fluor 647 human immunoglobulin G (IgG) labeling reagent (200 μg mL–1 as provided by Thermo Fisher, USA) served for this purpose. The solution was filled up to 1 mL with PBS. An Attune NxT flow cytometer (Thermo Fisher, USA) was used for measurement. A dot plot (SSC vs YL1in logarithmical scale) displayed the liposomes, whereas the negative control was defined as liposomes without corona and set to 1% of Alexa Fluor 647-positive liposomes.
Cellular Uptake
RAW264.7 cells (obtained from ATCC TIB-71) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA). The medium was supplemented with 2 mM glutamine, 100 mg mL–1 of streptomycin, 100 U mL–1 of penicillin, and 10% fetal bovine serum (all Invitrogen, Germany).
At 80% confluency, the cells were split, and after detaching, the cells were seeded out in a cell culture medium in 24-well plates (100 000 cells/well). The medium was changed to a serum-free medium after 24 h. Liposomes were added to the cell culture medium to achieve a final concentration of 7.5 or 75 μg mL–1, and the mixture was incubated with cells for 2 or 24 h at 37 °C. For flow cytometry analysis on an Attune NxT flow cytometer (Thermo Fisher, USA), the cells were detached with 2.5% trypsin (Gibco, Germany).
Confocal Laser Scanning Microscopy
For confocal laser scanning microscopy (CLSM) experiments, 50 × 104 cells (RAW264.7) were seeded in Ibidi iTreat μ-dishes (IBIDI, Germany) for 24 h, washed with PBS, and kept in DMEM without additional proteins for 2 h. In the following step, liposomes with and without corona were added to cells for either 2 or 24 h at concentrations of 7.5 or 75 μg mL–1. Subsequently, the cells were washed with PBS and then fixed for 15 min with Roti-Histofix 4% (Carl Roth GmbH, Germany). Staining of the cell membrane was achieved by CellMask Deep Red (dilution of 1:5000 in PBS, Thermo Fisher, USA). Images were taken on a Leica TCS SP5 II microscope with an HC PL APO CS 63×/1.4 oil objective using the LAS AF 3000 software, and detection took place in a serial scan mode. Liposomes are pseudocolored in green and the cell membrane is pseudocolored in red.
Statistical Analysis
For the comparison of two experimental groups, an unpaired Student’s t test assuming equal variances was performed. The calculated p-values were defined as follows: *p < 0.05, **p < 0.01, ***p < 0.001. Not significant differences are marked as n.s.
Results and Discussion
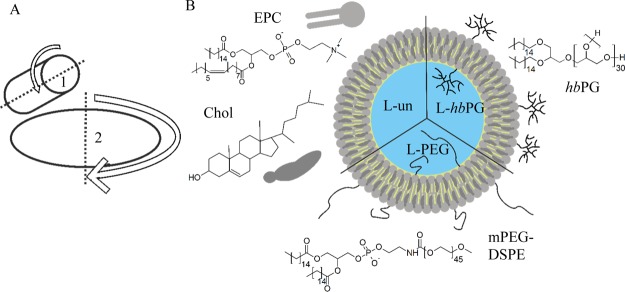
Liposomes were formulated by dual centrifugation, a rather new and easy-to-use “in-vial” homogenization technique for fast and reproducible liposome formulation within the single-digit milligram range of the total lipids (for the principle, see Figure 2A).39,40 This method allows aseptic formulation of up to 90 individual liposome samples within 30 min, even for experimental materials with limited availability. For the following experiments, three different liposome formulations were chosen. Besides investigating a conventional, unfunctionalized liposome sample (L-un) with the same lipid composition as that of Myocet, two liposome samples with potential stealth functionalizations were also analyzed: liposomes exhibiting PEG chains (L-PEG) as in Doxil linked to the headgroup of DSPE (Figure 2B) as well as liposomes with hbPG (L-hbPG) on the surface attached to the hydroxyl group of a 1,2-bis-n-hexadecyl glyceryl ether. For these systems, we consider the initial liposome components forming the lipid double layer (such as EPC, Chol, and DSPE) as the “base material”, whereas PEG and hbPG represent additional surface functionalizations. In case of a liposomal system, the base material is very similar to cell membranes, so that the base carrier could be considered as a “body-own” material. The liposome structure, individual components, composition, and physicochemical characteristics of all liposome samples are shown in Figure 2B and Table 1.
Figure 2.
(A) Principle of dual centrifugation. The sample is subjected to the sample holder 1, which rotates contrary to the main axis 2. (B) Schematic structure of each liposomal composition together with the chemical structures of the used components, EPC, Chol, hbPG, and mPEG-DSPE.
Table 1. Composition and Physicochemical Characteristics of All Used Liposomes.
| sample | compositiona | Rh/nm | Rg/nm | ρ ratiobRg/Rh | PD.I.c | ζ-potential/mV |
|---|---|---|---|---|---|---|
| L-un | 55 mol % EPC, 45 mol % Chol | 86 ± 9 | 81 ± 8 | 0.94 | 0.08 ± 0.03 | –20 ± 1 |
| L-PEG | 50 mol % EPC, 45 mol % Chol, 5 mol % mPEG-DSPE | 61 ± 6 | 68 ± 7 | 1.15 | 0.10 ± 0.03 | –26 ± 1 |
| L-hbPG | 50 mol % EPC, 45 mol % Chol, 5 mol % hbPG | 91 ± 9 | 94 ± 9 | 1.03 | 0.13 ± 0.06 | –21 ± 1 |
At low grafting densities, in the so-called “mushroom regime”, PEG chains can freely move without direct interaction. With increasing grafting density and size of the polymer, the PEG chains begin to sterically impede each other and therefore extend like brushes, increasing the liposome membrane thickness. According to Lee and Larson,41 the surface of liposomes functionalized with 5 mol % of PEG3000 should be completely covered with the polymers in the brush regime. We tentatively suggest that the rather compact architecture of the hbPG structure may lead to dense surface coverage of the liposomes already at lower concentration of polymer lipids than the linear PEG chains that can be present in a rather extended conformation. Accordingly, we assume for both L-PEG and L-hbPG that the highest possible functionalization density was reached.
The liposomes were incubated with 5 vol % citrate plasma to form the protein corona. The 5 vol % plasma was chosen to not overload the AF4 channel with proteins but still provide an excess of free proteins. The newly formed liposome–protein corona complexes were then separated from the medium by AF4 or by centrifugation, respectively. The former preparation was used for the analysis of the whole complex including most of the soft protein corona, whereas the latter represented the standard analysis of the hard protein corona. AF4 was used because of its particular suitability for fragile and self-assembled systems (e.g., liposomes or micelles), which often cannot be separated from free proteins by other means. In our case, the liposome samples were sufficiently stable to be centrifuged because of their high Chol content. Hence, we were able to compare the protein corona after AF4 with the one obtained after centrifugation.
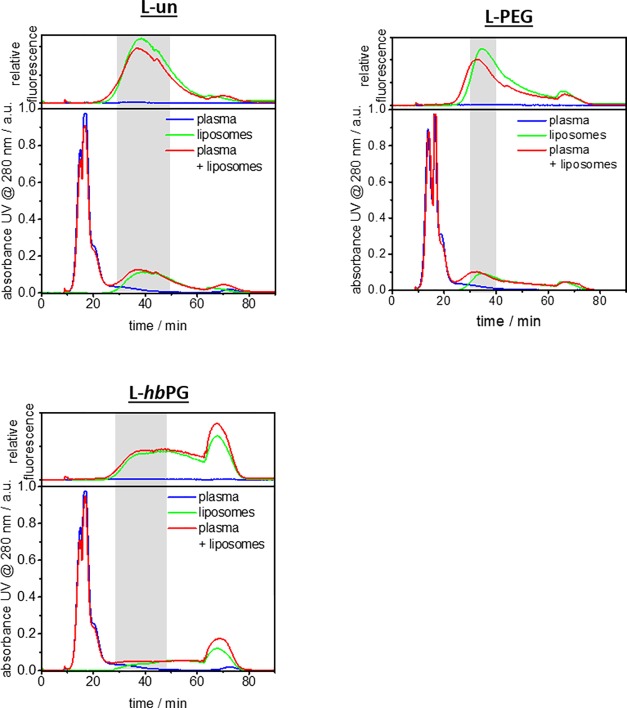
The AF4 elugrams of all liposomes are displayed in Figure 3. The green line represents the elution of the pristine liposomes. Noticeably, all liposomes show a different elution profile depending on their surface functionalization. In theory, interactions between the sample and the AF4 membrane would be expected to be minimal or nonexistent. However, in practice, for some surface functionalizations, the liposomes seem to interact preferably with the regenerated cellulose membrane. L-un and L-PEG demonstrate defined elution peaks, even though the L-PEG peak shows slightly more peak tailing, whereas L-hbPG interacts strongly with the membrane, which retards the elution and results in a constant elution of the sample over the remaining separation time. The high number of hydroxyl end groups at hbPG most probably causes the strong interaction with the membrane. Interestingly, also after plasma incubation (red line), the strong interactions with the membrane still occur. This indicates that the proteins do not cover the surface functionalization completely and a significant amount of the hydroxyl end groups are still accessible for membrane interactions.
Figure 3.
AF4 elugrams of all liposomes incubated with 5% plasma are shown in red. The individual elugrams are given as a reference, pure plasma in blue and pristine liposomes in green. The lower graph always represents the UV detector signal at 280 nm, whereas the upper one shows the fluorescence intensity. The fractions that were collected for further analysis are indicated by the gray box.
The shoulder in all elugrams at 62 min corresponds to the time at which the crossflow is stopped. Subsequently, no retention force acts on the sample anymore, and the remaining species that are left in the channel at that moment elute at once. Thus, no further analysis was performed on this last fraction. Free proteins are expected to elute first from the separation channel, as, according to the AF4 principle, the smallest components in a sample have the shortest retention time. In agreement with this, the plasma protein elution takes place in the first 25 min as seen in the reference elugram of pure plasma (blue line, Figure 3). The red line represents the incubated mixture of pristine liposomes and blood plasma. The signal of the mixed sample nicely correlates with the pure components. The successful separation of incubated liposomes from free plasma proteins was confirmed by the distinct elution time. Indeed, free plasma proteins present in the mixture were eluting until 25 min, whereas the liposome signal just starts at that time. The origin of the signal cannot be identified by the UV detector alone; hence, a fluorescence signal was recorded to verify that the second peak was generated by the fluorescently labeled liposomes and not by protein artifacts or something else. Thereby, the second peak between 25 and 50 min was identified as that of the corresponding liposomes—presumably with proteins bound. The AF4 results indicate that an interaction between the proteins and the liposomes must have taken place, as the peak shape of the liposomes slightly changed in all cases after incubation with the plasma. For further analysis of the liposome–protein complexes, fractions were collected as indicated by the gray boxes in each elugram.
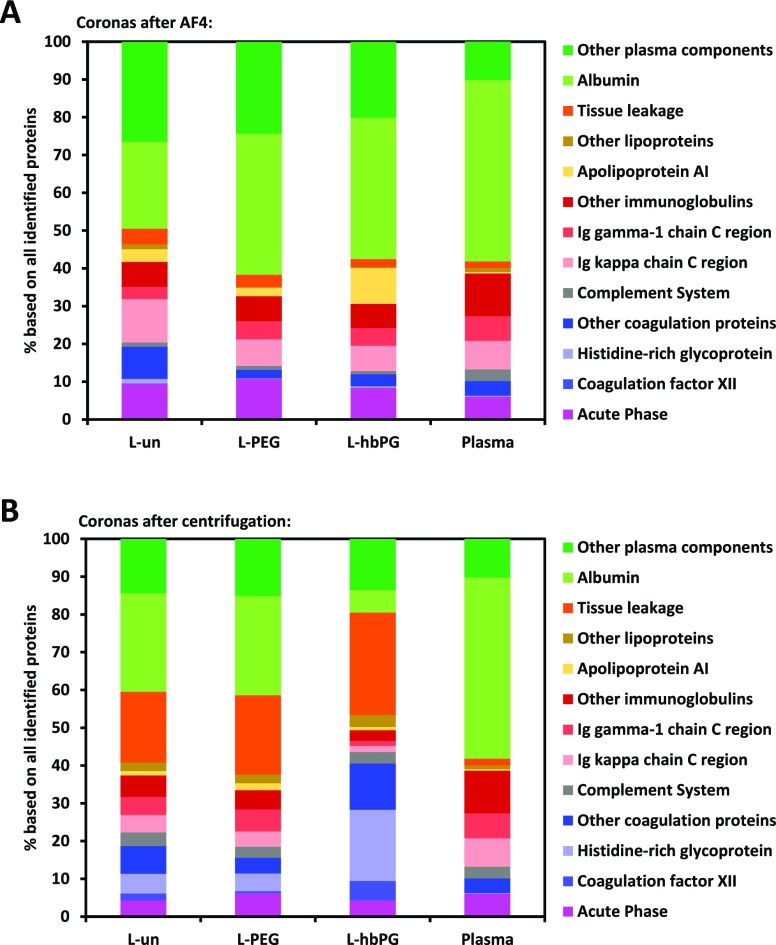
To compare the protein corona after AF4, the sample was also separated via centrifugation. Therefore, the sample was centrifuged at 4 °C for 1 h at 20 000g to pellet the liposome–protein complex and remove the free proteins in the supernatant. Subsequently, the sample was washed for another three times with PBS and resuspended. In the last step, the proteins were detached and afterward quantified via a Pierce protein assay. For all three samples, the protein-containing solution was then concentrated to the highest possible extent (see Experimental Section). Still, the amount of protein detected in the assay was low compared to other more hydrophobic non-PEGylated nanoparticle systems, where amounts in the range of 3–4 mg m–2 are typical.12,42 More specifically, the obtained values were above the limit of detection (LOD = 0.075), but below the limit of quantification (LOQ = 0.095) (for these samples corresponding to 0.3 and 0.7 mg m–2, respectively), for all three liposomal formulations (for the details of calculation, see Supporting Information). Accordingly, the detected protein amounts for all formulations cannot be reliably compared to each other, but they certainly were below 0.7 mg m–2, which is in the range of other hydrophilic nanocarriers.12 The protein compositions of the coronas obtained after AF4 and centrifugation were determined by liquid chromatography–mass spectrometry (LC–MS) in addition to a plasma reference, as shown in Figure 4. The identified proteins were grouped according to their function to represent the most abundant protein types. Proteins that showed the most significant differences are displayed individually. A detailed list with all identified proteins can be found in the Supporting Information.
Figure 4.
MS data of the protein corona. Panel (A) shows the coronas of the different liposomes after AF4, compared to plasma and (B) shows the same for the coronas after centrifugation. Significant proteins are displayed individually. Experiments were performed with two biological replicates containing three technical replicates each.
Interestingly, when comparing the protein corona patterns between the differently functionalized liposomes, the identified proteins varied only to some extent. Compared to pure plasma, the relative amount of albumin was lower in all cases but still enough to constitute the main protein in the corona. This decrease of albumin was the most dominant for L-hbPG after centrifugation and for L-un after AF4. Also, the amount of apolipoprotein AI (Apo AI) slightly increased for all samples and especially for L-hbPG after AF4. Depending on the preparation of the corona, however, more significant differences were obtained concerning the protein fractions. Most prominent are the changes regarding coagulation and tissue leakage proteins, which were significantly increased after centrifugation compared to AF4. Two proteins which stand out are the histidine-rich glycoprotein and coagulation factor XII which were significantly enriched after centrifugation for L-hbPG. On the other hand, immunoglobulins, albumin, and acute phase proteins were more abundant after AF4. In general, immunoglobulins and albumin are mostly known as proteins with lower binding affinities and, therefore, part of the soft protein corona, which is supported by our data. Previously, for different kinds of PEGylated nanomaterials, an enrichment of apolipoproteins was reported,42,43 which surprisingly was not the case for the PEGylated liposome sample. Also, the functionalization with hbPG did not result in a strong attraction of apolipoproteins besides Apo AI, although the monomer building blocks are in principle similar to PEG. It seems that for this liposomal system the different surface functionalizations—with the exception of slight changes possibly induced by hbPG—did not significantly change the properties of the surface with regard to the protein interaction, as already observed for other nanocarriers. Instead, the hydrophilic base material underneath the hydrophilic polymers (PEG and hbPG) forming the liposomal membranes (phospholipids and Chol) apparently was mainly responsible for determining the protein pattern.
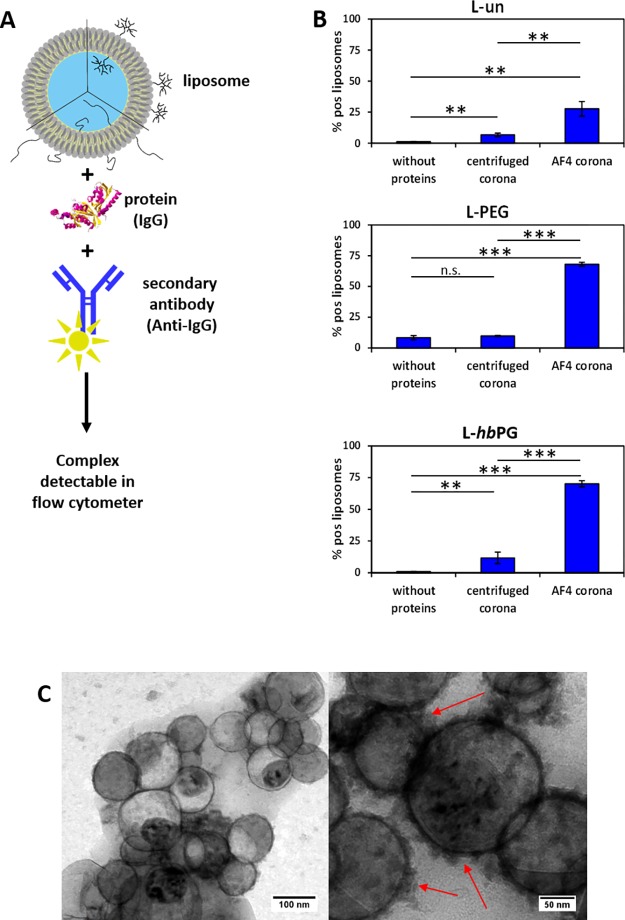
To make sure that the corona obtained after AF4 can indeed be compared with the one obtained after centrifugation, it is important to exclude the possibility that the observed corona proteins are just co-eluting with the liposomes. In this respect, an assay was conducted, in which a fluorescently labeled Fab fragment (antigen-binding fragment) selective for the Fc region (fragment-crystallizable region) of all IgG types was incubated with the liposome–protein mixtures after AF4 and after centrifugation, to form complexes with a specific protein of the protein corona. Only if the selected proteins are actually attached on the liposomal surface, the formed complex of liposome, protein, and labeling agent is large enough to be detected by flow cytometry (Figure 5A). For this test, IgG was chosen as the protein to be tested, as it was found as a highly abundant component in all coronas. The results are displayed in Figure 5B. The fluorescence signal of the fluorescently labeled anti-IgG antibody fragment is directly proportional to the number of liposomes with IgG in the corona. From the results, it can be confirmed that the investigated proteins were constituents of the corona after centrifugation and AF4 and also that the amount of the immunoglobulins after AF4 was significantly higher. This is in good agreement with the results of the LC–MS experiment (Figure 4). Therefore, we conclude that IgG was indeed part of the liposomal protein corona for all samples and that the obtained protein pattern is not a result of a co-elution effect.
Figure 5.
(A) Schematic illustration of the experimental design. (B) Liposomes were incubated with human plasma for 1 h. Protein-coated liposomes were either centrifuged to remove unbound proteins or applied to AF4. Secondary fluorescently labeled antihuman IgG antibodies were incubated with liposomes for 30 min in the dark. The fluorescence intensity of secondary fluorescently labeled antibodies, which were bound to liposomes, was detected by flow cytometry. *p < 0.05, **p < 0.01, ***p < 0.001. (C) TEM micrograph with two magnifications of liposomes incubated with plasma after centrifugation. The arrows indicate protein corona formation.
Additionally, the corona formation around the liposomes was visualized44 with transmission electron microscopy (TEM, see Figure 5C). The spherical liposome structure was preserved during the centrifugation process, which is not universally valid for all liposomes. The liposomes were comparably stable because of the high amount of Chol that was incorporated into the lipid membrane. When zoomed in on the sample, even parts of the protein corona itself could be observed as indicated by the red arrows.
As the protein coronas were quite similar for all liposome samples, with only L-hbPG showing slight deviations with regard to their corona composition, the influence of the different surface functionalizations on the cellular uptake by macrophages (RAW264.7) was investigated. A decreased uptake of a nanocarrier by macrophages is known to lead to reduced clearance by the immune system and, therefore, to a potentially longer blood circulation time with a correspondingly higher probability for the nanocarrier to reach its target location in the body.45,46
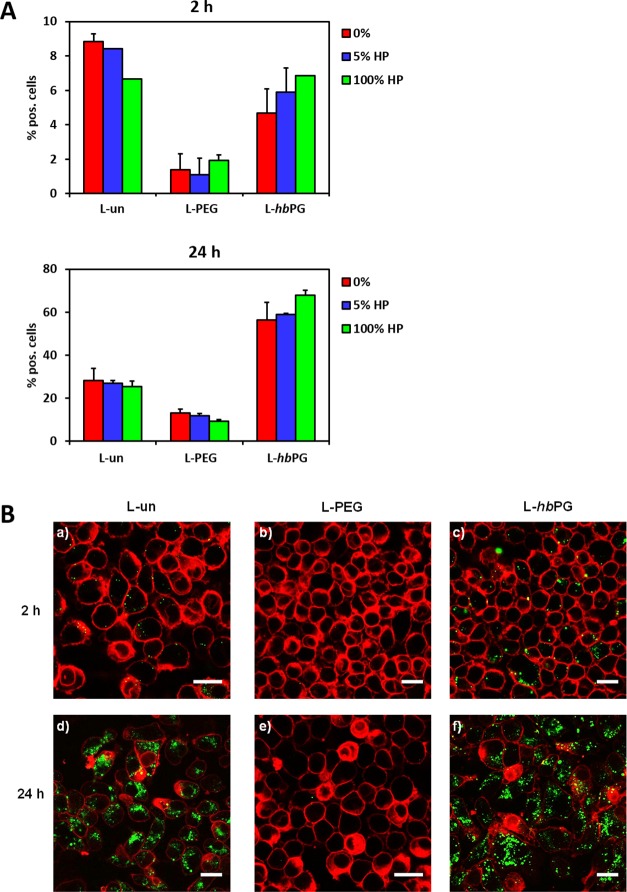
In Figure 6A, the cell uptake results obtained from flow cytometry measurements after 2 and 24 h of incubation for all liposome samples (7.5 μg mL–1) are represented. For the formation of the protein corona, different plasma concentrations (0, 5, and 100%) were chosen to detect any concentration dependency, as observed by Monopoli et al. for different systems.47 Interestingly, when first comparing the liposome uptake behavior with and without protein corona, there was no significant change visible. After 2 h incubation time, the number of positive cells was generally very low (<10%). Liposomes functionalized with PEG chains (L-PEG) showed an even lower cellular uptake than the other two samples, even though their protein corona composition only differed very slightly from the unfunctionalized liposomes (L-un). This reduced cellular uptake of L-PEG could be caused by an inhibition of the scavenger receptor-mediated cellular uptake. These scavenger receptors can be found on macrophages and were reported to recognize PEG, especially in combination with albumin.48,49 The PEG–albumin complexes could potentially block the scavenger pathway and consequently decrease the cellular uptake of the liposomes employed. Additionally, small amounts of proteins released from cells during the cell uptake experiments could also cause this effect, although no proteins were added to the culture medium, and might then lead to the reduced uptake observed.
Figure 6.
(A) Influence of protein corona formation on the cellular uptake behavior of liposomes. Liposomes were either directly incubated with RAW 264.7 cells (referred to as 0%) or preincubated with human plasma (5 or 100%) and further added to cells at a concentration of 7.5 μg mL–1. Cellular interaction was analyzed by flow cytometry after 2 and 24 h. The amount of fluorescence-positive cells (%) is shown. (B) Representative CLSM images. Liposomes were treated with 100% human plasma and incubated with RAW264.7 cells for 2 or 24 h at a concentration of 75 μg mL–1. The cell membrane was stained with CellMask Deep Red and is pseudocolored in red. Liposomes are pseudocolored in green. Scale bar: 20 μm.
After 24 h, the uptake increased for all samples but most prominently for L-hbPG. Additional cell experiments using a higher liposome concentration (75 μg mL–1, Figure S3) confirmed the presented results (Figure 6A), and the CLSM images verified the intracellular localization of the liposomes (Figure 6B). As the fluorescent marker used to stain the liposomes was located in the liposome membranes, the images indicate that the liposomes were taken up completely via the endolysosomal pathway including their protein corona. In the case of fusion with the cell membrane, which could also be proposed as an interaction pathway with cells, the cell membrane would also be stained with the liposome dye. Because of the self-assembled and “soft” nature of liposomes, it is not fully clear how their membrane fluidity and mechanical characteristics influence their uptake. However, as the liposomes used in this study contained a very high Chol content (45 mol %), their membrane supposedly was rather stable—leading to the fact that they retain their morphology during centrifugation. In this case, their uptake mechanism could be similar to the one of solid nanomaterials.
To sum up, hbPG did not lead to reduced cell uptake, although the attached chains are very hydrophilic. In contrast to the PEG functionalization with methoxy groups at the chain termini, the hbPG chains exhibit multiple hydroxyl groups at their ends, which in this system appear to mediate a stronger cellular internalization into macrophages. The observed reduction in cellular uptake for L-PEG is generally in line with other reports for reduced unspecific uptake of PEGylated materials but is in this case clearly not a result of the protein corona composition. The same applies to the increase in the cellular uptake for hbPG-functionalized liposomes, which was also independent of the adsorbed proteins. However, it is still possible that the differences observed in the corona of hbPG-functionalized liposomes lead to, for example, different endocytosis pathways in the macrophages or variations in other cell lines. The fact that the cellular uptake of L-hbPG differs from the one of L-PEG is interesting. Previously, a comparable in vivo distribution of both liposomes was observed with slightly higher amounts of hbPG liposomes in the spleen compared to the PEGylated ones.36 As the spleen contains over half of the monocytes in the body,50 the different biodistribution could be a consequence of different macrophage uptake. Of course, it has to be considered that the in vivo situation is much more complex and the interplay of different cell types as well as kinetic effects play a role. Generally, the cellular uptake behavior of some carrier systems containing hbPG was already reported.51,52 However, the results were not compared to the unfunctionalized or PEGylated equivalents; hence, it is unclear how the introduced hydroxyl groups affected the interaction with cells. Similarly, the effect of adsorbed proteins in these studies was not determined.
We conclude that even though protein adsorption takes place on the liposomes, the liposomal material itself, including surface functionalization with hydrophilic polymers, also plays a crucial role in the biological response. This is probably a result of the generally low amount of adsorbed proteins as described above, so that the polymer chains used for functionalization were still accessible. Also, the general binding affinity of adsorbed proteins might be relatively low, so that the protein corona could be a rather dynamic structure. We explain this low protein adsorption tendency and the associated effect of the surface functionalization with the fact that the liposomal material underneath the polymer functionalization itself is already very hydrophilic and similar to the composition of cellular membranes.
As the tested liposomes showed very low protein adsorption, and the protein corona was not the key factor for the cellular uptake, this presents an advantage for the successful application in patients. It was already shown that the plasma composition is subject to individual deviations and additionally differs between healthy donors and diseased patients, leading to a “personalized protein corona”.53−56 This means that by reducing the overall influence of the protein corona, the complications introduced by personalization effects could also be potentially minimized.
Conclusions
The investigation of the protein corona and its influence on cellular interactions is important for the application of drug carriers in nanomedicine, as it is commonly accepted that adsorbed proteins are responsible for the associated biological behavior. In this study, we investigated the protein corona of differently functionalized liposomes (unfunctionalized, PEG, and hbPG) and subsequently evaluated the effect of functionalization and protein adsorption on cellular uptake in macrophages. It was found that the protein corona of all liposome samples was surprisingly similar with small deviations found for the hbPG-functionalized system. Additionally, the overall adsorbed protein amount was very low compared to other, for example, polymeric nanomaterials. Interestingly, the macrophage uptake was then found to not primarily correlate with the protein adsorption. More specifically, the amount of liposomes internalized into cells remained almost unchanged regardless of the presence or absence of a protein corona. Instead, the different surface functionalizations significantly impacted the macrophage uptake: although the internalization of PEGylated liposomes was reduced compared to unfunctionalized ones, hbPG functionalization resulted in the opposite effect. We tentatively attribute the minor influence of protein corona to the fact that the hydrophilic liposomal membrane material resulted in a generally low protein adsorption. Thus, a significant fraction of the attached PEG and hbPG polymer chains was probably still accessible and, therefore, influenced cellular interaction. In consequence, it becomes clear that the properties of PEG and hbPG are not universally comparable but depend on the nature of the nanocarrier they are attached to. As such, it might be advisable to take more advantage of hydrophilic, “body-similar” nanomaterials like liposomes for future nanocarrier design, eventually reducing the disease-specific protein corona effects and patient variations.
Acknowledgments
We thank Ulrike Kemmer-Jonas and Dr. Matthias Worm for help with the synthesis of hbPG. We acknowledge DFG SFB1066 for financial support.
Glossary
Abbreviations
- PEG
poly(ethylene glycol)
- AF4
asymmetric flow field-flow fractionation
- PET
positron emission tomography
- EPC
egg phosphatidyl choline
- Chol
cholesterol
- hbPG
hyperbranched polyglycerol
- mPEG-DSPE
1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-3000]
- DTT
dithiothreitol
- IAA
indole-3-acetic acid
- Apo AI
apolipoprotein A1
- IgG
immunoglobulin G
- L-un
unfunctionalized liposomes
- L-PEG
PEGylated liposomes
- L-hbPB
liposomes functionalized with hyperbranched polyglycerol
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biomac.9b00539.
Detailed description of the synthesis and characterization of the dialkyl-based hbPG amphiphiles, as well as details on the protein quantification; experimental procedure of LC-MS measurements; TEM micrographs of the liposomes after centrifugation; and results of additional cell experiments at higher liposome concentration for CLSM experiments (PDF)
List of all identified proteins (XLSX)
Author Contributions
C.W. and M.V. contributed equally. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Allen T. M.; Cullis P. R. Drug Delivery Systems: Entering the Mainstream. Science 2004, 303, 1818–1822. 10.1126/science.1095833. [DOI] [PubMed] [Google Scholar]
- Havel H.; Finch G.; Strode P.; Wolfgang M.; Zale S.; Bobe I.; Youssoufian H.; Peterson M.; Liu M. Nanomedicines: From Bench to Bedside and Beyond. AAPS J. 2016, 18, 1373–1378. 10.1208/s12248-016-9961-7. [DOI] [PubMed] [Google Scholar]
- van Elk M.; Murphy B. P.; Eufrásio-da-Silva T.; O’Reilly D. P.; Vermonden T.; Hennink W. E.; Duffy G. P.; Ruiz-Hernández E. Nanomedicines for advanced cancer treatments: Transitioning towards responsive systems. Int. J. Pharm. 2016, 515, 132–164. 10.1016/j.ijpharm.2016.10.013. [DOI] [PubMed] [Google Scholar]
- Akbarzadeh A.; Rezaei-Sadabady R.; Davaran S.; Woo Joo S.; Zarghami N.; Hanifehpour Y.; Samiei M.; Kouhi M.; Nejati-Koshki K. Liposome: classification, preparation, and applications. Nanoscale Res. Lett. 2013, 8, 102. 10.1186/1556-276x-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian T.; Ho R. J. Y. Trends and developments in liposome drug delivery systems. J. Pharm. Sci. 2001, 90, 667–680. 10.1002/jps.1023. [DOI] [PubMed] [Google Scholar]
- Sharma A.; Sharma U. S. Liposomes in drug delivery: Progress and limitations. Int. J. Pharm. 1997, 154, 123–140. 10.1016/s0378-5173(97)00135-x. [DOI] [Google Scholar]
- Gabizon A.; Catane R.; Uziely B.; Kaufman B.; Safra T.; Cohen R.; Martin F.; Huang A.; Barenholz Y. Prolonged Circulation Time and Enhanced Accumulation in Malignant Exudates of Doxorubicin Encapsulated in Polyethylene-glycol Coated Liposomes. Cancer Res. 1994, 54, 987–992. [PubMed] [Google Scholar]
- Gabizon A. A. Pegylated Liposomal Doxorubicin: Metamorphosis of an Old Drug into a New Form of Chemotherapy. Cancer Invest. 2001, 19, 424–436. 10.1081/cnv-100103136. [DOI] [PubMed] [Google Scholar]
- Swenson C. E.; Perkins W. R.; Roberts P.; Janoff A. S. Liposome technology and the development of MyocetTM; (liposomal doxorubicin citrate). Breast 2001, 10, 1–7. 10.1016/s0960-9776(01)80001-1.14965549 [DOI] [Google Scholar]
- Bobo D.; Robinson K. J.; Islam J.; Thurecht K. J.; Corrie S. R. Nanoparticle-Based Medicines: A Review of FDA-Approved Materials and Clinical Trials to Date. Pharm. Res. 2016, 33, 2373–2387. 10.1007/s11095-016-1958-5. [DOI] [PubMed] [Google Scholar]
- Lundqvist M.; Stigler J.; Elia G.; Lynch I.; Cedervall T.; Dawson K. A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 14265–14270. 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzen S.; Schoettler S.; Baier G.; Rosenauer C.; Mailaender V.; Landfester K.; Mohr K. Complementary analysis of the hard and soft protein corona: sample preparation critically effects corona composition. Nanoscale 2015, 7, 2992–3001. 10.1039/c4nr05982d. [DOI] [PubMed] [Google Scholar]
- Caracciolo G. Liposome-protein corona in a physiological environment: challenges and opportunities for targeted delivery of nanomedicines. Nanomedicine 2015, 11, 543–557. 10.1016/j.nano.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Walkey C. D.; Chan W. C. W. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 2012, 41, 2780–2799. 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- Kharazian B.; Hadipour N. L.; Ejtehadi M. R. Understanding the nanoparticle-protein corona complexes using computational and experimental methods. Int. J. Biochem. Cell Biol. 2016, 75, 162–174. 10.1016/j.biocel.2016.02.008. [DOI] [PubMed] [Google Scholar]
- Aggarwal P.; Hall J. B.; McLeland C. B.; Dobrovolskaia M. A.; McNeil S. E. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv. Drug Delivery Rev. 2009, 61, 428–437. 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monopoli M. P.; Åberg C.; Salvati A.; Dawson K. A. Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 2012, 7, 779–786. 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- Pearson R. M.; Juettner V. V.; Hong S. Biomolecular corona on nanoparticles: a survey of recent literature and its implications in targeted drug delivery. Front. Chem. 2014, 2, 108. 10.3389/fchem.2014.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczyk D.; Bombelli F. B.; Monopoli M. P.; Lynch I.; Dawson K. A. What the Cell “Sees” in Bionanoscience. J. Am. Chem. Soc. 2010, 132, 5761–5768. 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]
- Milani S.; Baldelli Bombelli F.; Pitek A. S.; Dawson K. A.; Rädler J. Reversible versus Irreversible Binding of Transferrin to Polystyrene Nanoparticles: Soft and Hard Corona. ACS Nano 2012, 6, 2532–2541. 10.1021/nn204951s. [DOI] [PubMed] [Google Scholar]
- Weber C.; Simon J.; Mailänder V.; Morsbach S.; Landfester K. Preservation of the soft protein corona in distinct flow allows identification of weakly bound proteins. Acta Biomater. 2018, 76, 217–224. 10.1016/j.actbio.2018.05.057. [DOI] [PubMed] [Google Scholar]
- Messaud F. A.; Sanderson R. D.; Runyon J. R.; Otte T.; Pasch H.; Williams S. K. R. An overview on field-flow fractionation techniques and their applications in the separation and characterization of polymers. Prog. Polym. Sci. 2009, 34, 351–368. 10.1016/j.progpolymsci.2008.11.001. [DOI] [Google Scholar]
- Malik M. I.; Pasch H. Field-flow fractionation: New and exciting perspectives in polymer analysis. Prog. Polym. Sci. 2016, 63, 42–85. 10.1016/j.progpolymsci.2016.03.004. [DOI] [Google Scholar]
- Shehata T.; Ogawara K.-i.; Higaki K.; Kimura T. Prolongation of residence time of liposome by surface-modification with mixture of hydrophilic polymers. Int. J. Pharm. 2008, 359, 272–279. 10.1016/j.ijpharm.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Müller L. K.; Landfester K. Natural liposomes and synthetic polymeric structures for biomedical applications. Biochem. Biophys. Res. Commun. 2015, 468, 411–418. 10.1016/j.bbrc.2015.08.088. [DOI] [PubMed] [Google Scholar]
- Woodle M. C.; Matthay K. K.; Newman M. S.; Hidayat J. E.; Collins L. R.; Redemann C.; Martin F. J.; Papahadjopoulos D. Versatility in lipid compositions showing prolonged circulation with sterically stabilized liposomes. Biochim. Biophys. Acta, Biomembr. 1992, 1105, 193–200. 10.1016/0005-2736(92)90194-q. [DOI] [PubMed] [Google Scholar]
- Siegers C.; Biesalski M.; Haag R. Self-Assembled Monolayers of Dendritic Polyglycerol Derivatives on Gold That Resist the Adsorption of Proteins. Chem.—Eur. J. 2004, 10, 2831–2838. 10.1002/chem.200306073. [DOI] [PubMed] [Google Scholar]
- Papahadjopoulos D.; Allen T. M.; Gabizon A.; Mayhew E.; Matthay K.; Huang S. K.; Lee K. D.; Woodle M. C.; Lasic D. D.; Redemann C. Sterically stabilized liposomes: improvements in pharmacokinetics and antitumor therapeutic efficacy. Proc. Natl. Acad. Sci. U.S.A. 1991, 88, 11460–11464. 10.1073/pnas.88.24.11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigdeli A.; Palchetti S.; Pozzi D.; Hormozi-Nezhad M. R.; Baldelli Bombelli F.; Caracciolo G.; Mahmoudi M. Exploring Cellular Interactions of Liposomes Using Protein Corona Fingerprints and Physicochemical Properties. ACS Nano 2016, 10, 3723–3737. 10.1021/acsnano.6b00261. [DOI] [PubMed] [Google Scholar]
- Caracciolo G.; Pozzi D.; Capriotti A. L.; Cavaliere C.; Piovesana S.; La Barbera G.; Amici A.; Laganà A. The liposome–protein corona in mice and humans and its implications for in vivo delivery. J. Mater. Chem. B 2014, 2, 7419–7428. 10.1039/c4tb01316f. [DOI] [PubMed] [Google Scholar]
- Pozzi D.; Colapicchioni V.; Caracciolo G.; Piovesana S.; Capriotti A. L.; Palchetti S.; De Grossi S.; Riccioli A.; Amenitsch H.; Laganà A. Effect of polyethyleneglycol (PEG) chain length on the bio-nano-interactions between PEGylated lipid nanoparticles and biological fluids: from nanostructure to uptake in cancer cells. Nanoscale 2014, 6, 2782–2792. 10.1039/c3nr05559k. [DOI] [PubMed] [Google Scholar]
- Fritz T.; Hirsch M.; Richter F. C.; Müller S. S.; Hofmann A. M.; Rusitzka K. A. K.; Markl J.; Massing U.; Frey H.; Helm M. Click Modification of Multifunctional Liposomes Bearing Hyperbranched Polyether Chains. Biomacromolecules 2014, 15, 2440–2448. 10.1021/bm5003027. [DOI] [PubMed] [Google Scholar]
- Hofmann A. M.; Wurm F.; Hühn E.; Nawroth T.; Langguth P.; Frey H. Hyperbranched Polyglycerol-Based Lipids via Oxyanionic Polymerization: Toward Multifunctional Stealth Liposomes. Biomacromolecules 2010, 11, 568–574. 10.1021/bm901123j. [DOI] [PubMed] [Google Scholar]
- Hofmann A. M.; Wurm F.; Frey H. Rapid Access to Polyfunctional Lipids with Complex Architecture via Oxyanionic Ring-Opening Polymerization. Macromolecules 2011, 44, 4648–4657. 10.1021/ma200367c. [DOI] [Google Scholar]
- Fritz T.; Voigt M.; Worm M.; Negwer I.; Müller S. S.; Kettenbach K.; Ross T. L.; Roesch F.; Koynov K.; Frey H.; Helm M. Orthogonal Click Conjugation to the Liposomal Surface Reveals the Stability of the Lipid Anchorage as Crucial for Targeting. Chem.—Eur. J. 2016, 22, 11578–11582. 10.1002/chem.201602758. [DOI] [PubMed] [Google Scholar]
- Wagener K.; Worm M.; Pektor S.; Schinnerer M.; Thiermann R.; Miederer M.; Frey H.; Rösch F. Comparison of Linear and Hyperbranched Polyether Lipids for Liposome Shielding by 18F-Radiolabeling and Positron Emission Tomography. Biomacromolecules 2018, 19, 2506–2516. 10.1021/acs.biomac.8b00115. [DOI] [PubMed] [Google Scholar]
- Provencher S. W. CONTIN: A general purpose constrained regularization program for inverting noisy linear algebraic and integral equations. Comput. Phys. Commun. 1982, 27, 229–242. 10.1016/0010-4655(82)90174-6. [DOI] [Google Scholar]
- Provencher S. W. Inverse problems in polymer characterization: Direct analysis of polydispersity with photon correlation spectroscopy. Die Makromolekulare Chem. 1979, 180, 201–209. 10.1002/macp.1979.021800119. [DOI] [Google Scholar]
- Massing U.; Cicko S.; Ziroli V. Dual asymmetric centrifugation (DAC)--a new technique for liposome preparation. J. Controlled Release 2008, 125, 16–24. 10.1016/j.jconrel.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Massing U.; Ingebrigtsen S. G.; Skalko-Basnet N.; Holsoeter A. M.. Dual Centrifugation—A Novel “In-Vial” Liposome Processing Technique in Liposomes; Catala A., Ed.; IntechOpen, 2017; pp. 3–28. [Google Scholar]
- Lee H.; Larson R. G. Adsorption of Plasma Proteins onto PEGylated Lipid Bilayers: The Effect of PEG Size and Grafting Density. Biomacromolecules 2016, 17, 1757–1765. 10.1021/acs.biomac.6b00146. [DOI] [PubMed] [Google Scholar]
- Schöttler S.; Becker G.; Winzen S.; Steinbach T.; Mohr K.; Landfester K.; Mailänder V.; Wurm F. R. Protein adsorption is required for stealth effect of poly(ethylene glycol)- and poly(phosphoester)-coated nanocarriers. Nat. Nanotechnol. 2016, 11, 372–377. 10.1038/nnano.2015.330. [DOI] [PubMed] [Google Scholar]
- Gref R.; Lück M.; Quellec P.; Marchand M.; Dellacherie E.; Harnisch S.; Blunk T.; Müller R. H. “Stealth” corona-core nanoparticles surface modified by polyethylene glycol (PEG): influences of the corona (PEG chain length and surface density) and of the core composition on phagocytic uptake and plasma protein adsorption. Colloids Surf., B 2000, 18, 301–313. 10.1016/s0927-7765(99)00156-3. [DOI] [PubMed] [Google Scholar]
- Kokkinopoulou M.; Simon J.; Landfester K.; Mailänder V.; Lieberwirth I. Visualization of the protein corona: towards a biomolecular understanding of nanoparticle-cell-interactions. Nanoscale 2017, 9, 8858–8870. 10.1039/c7nr02977b. [DOI] [PubMed] [Google Scholar]
- Immordino M. L.; Dosio F.; Cattel L. Stealth liposomes: review of the basic science, rationale, and clinical applications, existing and potential. Int. J. Nanomed. 2006, 1, 297–315. [PMC free article] [PubMed] [Google Scholar]
- Suk J. S.; Xu Q.; Kim N.; Hanes J.; Ensign L. M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Delivery Rev. 2016, 99, 28–51. 10.1016/j.addr.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monopoli M. P.; Walczyk D.; Campbell A.; Elia G.; Lynch I.; Baldelli Bombelli F.; Dawson K. A. Physical-chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534. 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- Allen R. J.; Mathew B.; Rice K. G. PEG-Peptide Inhibition of Scavenger Receptor Uptake of Nanoparticles by the Liver. Mol. Pharmaceutics 2018, 15, 3881–3891. 10.1021/acs.molpharmaceut.8b00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumhover N. J.; Duskey J. T.; Khargharia S.; White C. W.; Crowley S. T.; Allen R. J.; Rice K. G. Structure–Activity Relationship of PEGylated Polylysine Peptides as Scavenger Receptor Inhibitors for Non-Viral Gene Delivery. Mol. Pharmaceutics 2015, 12, 4321–4328. 10.1021/acs.molpharmaceut.5b00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swirski F. K.; Nahrendorf M.; Etzrodt M.; Wildgruber M.; Cortez-Retamozo V.; Panizzi P.; Figueiredo J.-L.; Kohler R. H.; Chudnovskiy A.; Waterman P.; Aikawa E.; Mempel T. R.; Libby P.; Weissleder R.; Pittet M. J. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science 2009, 325, 612–616. 10.1126/science.1175202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L.; Xu Y.-H.; Akasaka T.; Abe S.; Komatsu N.; Watari F.; Chen X. Polyglycerol-coated nanodiamond as a macrophage-evading platform for selective drug delivery in cancer cells. Biomaterials 2014, 35, 5393–5406. 10.1016/j.biomaterials.2014.03.041. [DOI] [PubMed] [Google Scholar]
- Wang L.; Neoh K. G.; Kang E.-T.; Shuter B. Multifunctional polyglycerol-grafted Fe3O4@SiO2 nanoparticles for targeting ovarian cancer cells. Biomaterials 2011, 32, 2166–2173. 10.1016/j.biomaterials.2010.11.042. [DOI] [PubMed] [Google Scholar]
- Corbo C.; Molinaro R.; Tabatabaei M.; Farokhzad O. C.; Mahmoudi M. Personalized protein corona on nanoparticles and its clinical implications. Biomater. Sci. 2017, 5, 378–387. 10.1039/c6bm00921b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajipour M. J.; Laurent S.; Aghaie A.; Rezaee F.; Mahmoudi M. Personalized protein coronas: a “key” factor at the nanobiointerface. Biomater. Sci. 2014, 2, 1210–1221. 10.1039/c4bm00131a. [DOI] [PubMed] [Google Scholar]
- Colapicchioni V.; Tilio M.; Digiacomo L.; Gambini V.; Palchetti S.; Marchini C.; Pozzi D.; Occhipinti S.; Amici A.; Caracciolo G. Personalized liposome–protein corona in the blood of breast, gastric and pancreatic cancer patients. Int. J. Biochem. Cell Biol. 2016, 75, 180–187. 10.1016/j.biocel.2015.09.002. [DOI] [PubMed] [Google Scholar]
- Prozeller D.; Pereira J.; Simon J.; Mailänder V.; Morsbach S.; Landfester K. Prevention of Dominant IgG Adsorption on Nanocarriers in IgG-Enriched Blood Plasma by Clusterin Precoating. Adv. Sci. 2019, 6, 1802199. 10.1002/advs.201802199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.