Cryo-EM structure of PTCH1-ShhNC24II complex reveals multiple sterol binding sites and a possible sterol translocation pathway.
Abstract
Hedgehog signaling is central in embryonic development and tissue regeneration. Disruption of the pathway is linked to genetic diseases and cancer. Binding of the secreted ligand, Sonic hedgehog (ShhN) to its receptor Patched (PTCH1) activates the signaling pathway. Here, we describe a 3.4-Å cryo-EM structure of the human PTCH1 bound to ShhNC24II, a modified hedgehog ligand mimicking its palmitoylated form. The membrane-embedded part of PTCH1 is surrounded by 10 sterol molecules at the inner and outer lipid bilayer portion of the protein. The annular sterols interact at multiple sites with both the sterol-sensing domain (SSD) and the SSD-like domain (SSDL), which are located on opposite sides of PTCH1. The structure reveals a possible route for sterol translocation across the lipid bilayer by PTCH1 and homologous transporters.
INTRODUCTION
The hedgehog pathway is one of the key mechanisms of the developmental patterning in mammalian embryonic development and is essential in tissue regeneration and homeostasis (1, 2). Disrupting mutations in the components of the hedgehog pathway are linked to diseases such as holoprosencephaly (3), Curry-Jones syndrome (4), and Gorlin syndrome (5). Furthermore, abnormal hedgehog pathway activation is associated with different forms of cancer, including medulloblastoma and basal cell carcinoma (6, 7).
The hedgehog morphogens are small secreted proteins. Three homologous hedgehog ligands have been identified in mammals: Sonic (Shh), Desert (Dhh), and Indian (Ihh) hedgehog (8). Shh is the best studied of the three and is presumed to play the key role in the hedgehog pathway (9). The processed form of Shh is posttranslationally palmitoylated at its N-terminal residue C24 (10). Furthermore, the protein undergoes autoproteolysis, leading to production of the mature ShhN fragment that contains a C-terminal cholesteryl moiety (11). The dually lipid-modified ShhN is released from the hedgehog-producing cells and can elicit short- and long-range hedgehog pathway activation in the developing embryo (12). The hedgehog pathway is activated in the hedgehog-receiving cells upon binding of the hedgehog ligand to its receptor, protein patched homolog 1 (PTCH1), along with several recently identified coreceptors (13, 14). In the absence of ShhN, PTCH1 suppresses the activity of smoothened homolog (SMO), a G protein–coupled receptor (15). Upon binding of ShhN to PTCH1, SMO translocates to the Ellis-van Creveld (EvC) zone of the primary cilium (16) and activates the GLI family of transcription factors, the hallmark of the hedgehog signaling pathway activation (17).
PTCH1 is a member of the resistance-nodulation-division (RND) transporter family (18). It is a 12–transmembrane (TM) domain membrane protein, featuring two extended ectodomains that are involved in ligand recognition. The TM2–6 bundle of PTCH1 is annotated as the sterol-sensing domain (SSD) based on sequence similarity with similar domains in proteins involved in cholesterol homeostasis (18, 19). The SSD is presumed to be involved in the interaction with the hedgehog ligand, as suggested by recent structural and biochemical studies. Several recent reports described the interaction of ShhN with human PTCH1 in detail. Gong et al. (20) obtained a high-resolution cryogenic electron microscopy (cryo-EM) structure of PTCH1 bound to a bacterially expressed ShhN fragment, including residues 39 to 190 of human ShhN. The structure featured the ligand bound to the two ectodomains, with extreme N- and C-terminal portions of ShhN unresolved. Separately, Qi et al. (21) solved a cryo-EM structure of PTCH1 bound to a native ShhN fragment that includes the palmitoylated N terminus, including residues 24 to 188. Contrary to the structure reported by Gong et al. (20), this reconstruction showed that the N terminus of the ShhN ligand is buried deep within the receptor, with palmitoyl moiety wedged within the “neck” region between the ectodomain and the membrane-embedded part of the receptor. The density of the hedgehog ligand in this reconstruction was inconsistent with that reported by Gong et al. (20). This inconsistency was settled by the same authors who followed their study with a structure of a native ShhN-bound PTCH1 captured in a state, where two PTCH1 molecules A and B were bound to ShhN via a ShhN-core interface (PTCH1B) and via the palmitoylated N terminus of ShhN (PTCH1A) (22). This remarkable structure explained the seemingly contradictory results obtained previously with the monomeric PTCH1 molecules trapping the bound ligand in opposite orientations. The structure also provided structural evidence supporting the previously reported ability of an acylated 22-residue N-terminal peptide of ShhN to activate the hedgehog pathway by direct interaction with PTCH1 (23). All three of the published structures of PTCH1 featured two extra densities corresponding to sterols bound at the lipid-protein interface of PTCH1 (20–22). Recently, a ligand-free PTCH1 structure was reported, featuring a dimeric arrangement of the mouse PTCH1 homolog. The ligand-free mouse PTCH1 displayed several lipid-like density elements along a duct connecting the outer leaflet SSD region and the lipid site in the extracellular domain (ECD). Supported by biochemical evidence, the structure suggested a possible role of PTCH1 in cholesterol transport as a mechanism of SMO regulation (24).
Previous studies identified several ShhN variants that can be used for hedgehog pathway activation, with differing efficacies (25). The dually lipid-modified ShhN showed the highest potency of activation, whereas the unmodified ShhN fragment was the least effective in vitro. Substitution of the C24 residue of the nonlipidated ShhN with two Ile residues (C24II-ShhN, here referred to as ShhNC24II) resulted in a substantial increase in the activity of the ShhN fragment. Since the discovery of this effect of the C24II substitution on ShhN, the ShhNC24II has become an indispensable tool in a wide range of studies of the hedgehog pathway. On the basis of the close to native activity of the ShhNC24II ligand, structural characterization of the PTCH1-ShhNC24II complex should accurately describe the recognition of the native hedgehog ligand by its receptor, PTCH1.
RESULTS AND DISCUSSION
To gain insights into the recognition of the modified ShhNC24II hedgehog ligand by PTCH1, we solved the cryo-EM structure of the human PTCH1 in a complex with ShhNC24II. The protein construct PTCH1Δ combining the C-terminal truncation (residues 1 to 1188) and the Y645A mutation, devoid of the two PPXY motifs recognized by the HECT E3 ligases (26, 27), had excellent biochemical properties and was used for structure determination (Fig. 1A and fig. S1, C to E). The ligand, ShhNC24II, was generated by the expression in Escherichia coli as an N-terminally SUMO (small ubiquitin-like modifier)–tagged fusion (fig. S1, A and B). The purified ShhNC24II protein was biologically active, with an apparent median effective concentration (EC50) an order of magnitude lower than that of the recombinant human ShhN (Fig. 1B). The protein was mixed with the purified PTCH1Δ at a 2:1 molar ratio and subjected to cryo-EM analysis (figs. S2 and S3). The cryo-EM map obtained by image processing of the PTCH1Δ-ShhNC24II dataset was refined to 3.4-Å resolution and was used for building a complete model of the complex (Fig. 1, C and D, and fig. S3). The atomic model of PTCH1Δ-ShhNC24II covers the sequence of PTCH1 residues D46-P1186, with an unresolved loop region comprising residues F614-T728. The model features six well-resolved glycosylation sites, the extended N terminus of the protein, and a part of the intracellular loop 3 (IL3), which were missing in the previous reconstructions of PTCH1 (Figs. 2 and 3, A and B, and fig. S4).
Fig. 1. Structure of the PTCH1Δ-ShhNC24II complex.
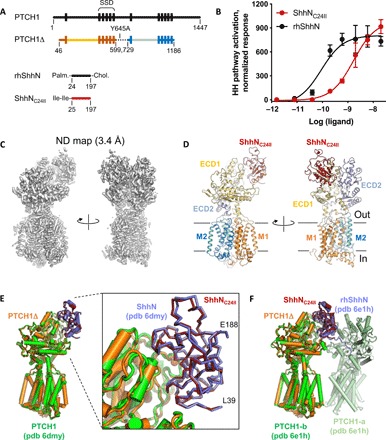
(A) A schematic representation of the constructs used in this study. For comparison, the wild-type full-length proteins (PTCH1 and ShhN) are shown. (B) The hedgehog pathway activation assay confirms the functionality of the ShhNC24II preparation. NIH 3T3 cells were treated with the indicated concentrations of the ligands, and hedgehog pathway activity was evaluated analyzing Gli1 mRNA levels by qPCR; error bars indicate SD (n = 3). (C) Density map of the reconstituted PTCH1Δ-ShhNC24II complex. (D) The views of the modeled complex corresponding to the views shown in (C). The key components of the complex are labeled, including the TM1–6 (“M1”), TM7–12 (“M2”), and the extracellular domains (ECD1–2). (E and F) Comparison of the structure to the previously solved PTCH1-ShhN (E) and PTCH1:rhShhN 2:1 complex (F). The bound ShhNC24II perfectly aligns to the previously observed complexes mediated by the metal-binding site interface of the ligand.
Fig. 2. Bound sterol molecules revealed by the PTCH1Δ-ShhNC24II structure.
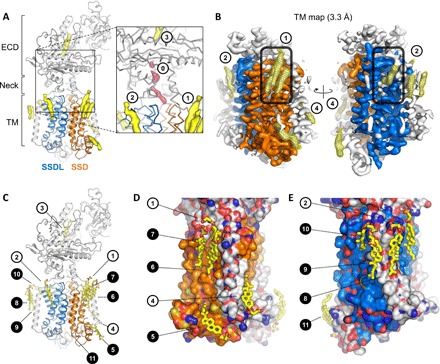
(A) The 3D reconstruction features several well-resolved density elements in the ECD region and surrounding the TM domains of PTCH1Δ (isolated density are colored yellow). The SSD (TM2–6) and the SSDL (TM8–12) are colored orange and blue. Inset: Previously observed sterol sites are indicated as sites 1, 2, and 3; additional partial density is present in the neck region of PTCH1Δ (site 0, pink), suggesting partial occupancy of the site. (B) Focused density map of the TM region (TM map) indicates the well-resolved sites 1, 2, and 4, as well as a number of novel sites, indicated with a mesh; the pocket region within the SSD and the corresponding SSDL is indicated by a rounded rectangle. (C) The CHS molecules were modeled into the well-defined density map regions (B) using 10σ cutoff of the unsharpened refined density map. The closed circles correspond to the novel sterol sites revealed by the PTCH1Δ-ShhNC24II reconstruction. (D and E) Views of the sterols surrounding the SSD and SSDL domains. The protein model is shown as a surface, colored according to atom type; carbon atoms in the SSD and SSDL are colored orange and blue, respectively.
Fig. 3. The inner leaflet SSD sterol site points at a functional role in PTCH1.
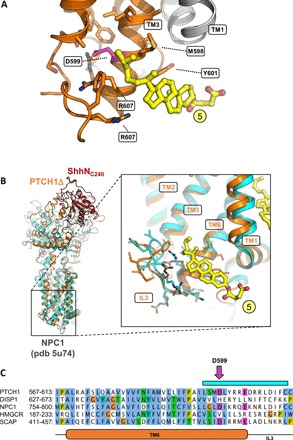
(A) The site 5 sterol (as defined in Fig. 2B; colored yellow) is within contact of the residue D599, previously shown to be functionally critical in the SSDs of SCAP and Drosophila Patched. (B) A similar site, unoccupied by a sterol molecule, has been previously observed in the structure of the cholesterol transporter NPC1, homologous to PTCH1. The PTCH1Δ-ShhNC24II structure was aligned to the NPC1 X-ray structure (cyan). Inset: Alignment of the two structures using the residues of TM6 shows a great degree of similarity in the region surrounding the site 5 sterol. (C) Sequence alignment of key SSD-containing proteins, PTCH1, DISP1, NPC1, HMG-CoA reductase, and SCAP, shows the similarity between the elements involved in inner leaflet sterol binding site. The pink arrow indicates the residue D599 [shown in (A)]. The cyan bar corresponds to residues shown with side chains in (B). TM6 and IL3 of PTCH1 are indicated below the sequence alignment.
Comparisons of PTCH1Δ-ShhNC24II and the two previously published reconstructions revealed that the unmodified ShhN binds in a near-identical manner (Fig. 1, E and F). The N terminus of ShhNC24II, similar to that of ShhN, projects outside of the PTCH1 tunnel (Fig. 1E); the divalent metal-binding site of the hedgehog ligand is close to the PTCH1 binding interface. Comparison to the rhShhN (recombinant human ShhN)–bound PTCH1 structure shows that PTCH1Δ bound to the ShhNC24II matches the B chain of the dimer (Fig. 1F).
Similar to the previously published reconstructions, our cryo-EM map contains four strong density elements likely corresponding to the bound sterol molecules (Fig. 2), here denoted as site 1 (at the outer leaflet portion of the SSD; Figs. 2 and 4, A and B), site 2 (at the binding site formed by TM9, TM11, and TM12; Figs. 2 and 4, A and B), site 3 (buried within the ECD region; Fig. 2, A and C), and site 4 (at the inner leaflet of the SSD; Fig. 2, B to D). The neck region of the protein features a partial density element (site 0; Fig. 1A, inset), indicative of partial occupancy by an added sterol or a copurified lipid-like molecule.
Fig. 4. Sterol binding site overlap observed at the SSD and SSDL domains of PTCH1.
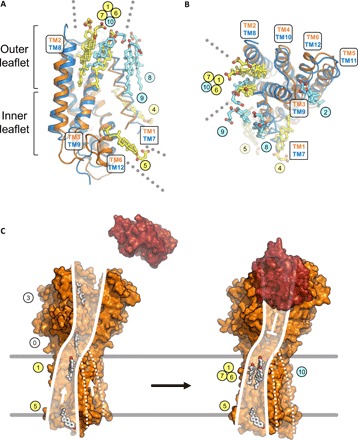
(A and B) The M1 and M2 regions of PTCH1Δ were aligned using the SSD (orange) and SSDL domains (blue). The SSD and SSDL regions are represented as cartoons; the TM helices TM1 and TM7 are shown as ribbons. Sterols bound at the SSD (yellow) and SSDL regions (light blue) are labeled using corresponding colored circles. The outer leaflet sites 1, 6, and 7 at the SSD and site 10 at the SSDL overlap. The dotted lines in (A) and (B) indicate access of sterols via the inner leaflet to site 5 (SSD) and via the outer leaflet to pocket sites 1, 6, and 7 (SSD) and site 10 (SSDL). (C) Putative model of sterol translocation via PTCH1. Left: In the absence of hedgehog ligand (red), the protein (illustrated using PDB ID: 6D4H) moves the inner leaflet sterols along the path formed by the SSD. Right: Binding of the ligand negatively regulates this activity; the sterols accumulate at the SSD pocket region, evidenced by our 3D reconstruction. Presence of sterols at the SSDL, along with its similarity with SSD, may suggest a similar functional role (marked by the dashed lines).
The structure of PTCH1Δ-ShhNC24II revealed a previously unidentified feature: the presence of eight prominent density elements at the protein-lipid interface, additionally to sites 1, 2, and 4 (Figs. 2, A to E, and 4, A and B). These novel positions of the bound sterols became apparent with the improvement of the density map corresponding to PTCH1Δ. The focused refinement of the TM domain of PTCH1Δ led to a density map at 3.3-Å resolution, allowing us to describe the membrane-embedded portion of the protein with an unprecedented level of detail (Fig. 3B, fig. S3, and movie S1). The positions of the membrane-embedded sterols surround the PTCH1Δ at sites within SSD (TM2–6), at the SSD-like region of the protein corresponding to TM8–12 (here referred to as “SSDL”), and at several sites not in direct contact with either SSD or SSDL (Fig. 2, C to E).
Sites 1, 6, and 7 occupy the outer leaflet “pocket” of the SSD (Fig. 2, B to D). Site 5 is located at the inner leaflet portion of the SSD adjacent to the pocket (Figs. 2, C and D, and 3). Site 4 sterol at the SSD is sterically separated from site 5 by the TM1 (Figs. 2, C and D, and 4, A and B). The outer leaflet SSDL region is decorated with sterols at sites 2 and 10. Sites 8 and 9 are adjacent to the TM7. The site 11 sterol is located apart from other sterol molecules, bound to the N-terminal α helix that precedes TM1 and lies parallel to the lipid bilayer plane (Fig. 2, C and E).
The presence of the well-resolved density of the sterol site 5 explains a previous observation of functional coupling between residues in this region and the activity of the SSD domain–containing proteins. It has been shown previously that D443N mutation in the SSD of SCAP (sterol regulatory element-binding protein cleavage-activating protein), an endoplasmic reticulum (ER)–resident SSD-containing protein, disrupts the cholesterol sensitivity of the protein (28). A substitution of the equivalent residue D584 to an asparagine in Drosophila melanogaster Patched affected the ability of the protein to repress smoothened (29). It is noteworthy that a similar mutation, D585N, in a mouse PTCH1 homolog appeared to have little or no effect on the function of the protein under cell culture conditions, indicating that the effect of this SSD mutation may be species or cellular context specific (30). Our reconstruction shows that the sterol bound at site 5 is in immediate proximity to D599 (Figs. 2 and 3A), corresponding to D443 of the yeast SCAP or D584 of the Drosophila Patched. Comparison of PTCH1Δ to the x-ray structure of NPC1, a cholesterol transporter homologous to PTCH1, showed that a very similar structural motif is present in both proteins, likely forming a structurally conserved sterol binding site (Fig. 3B). Sequence alignment of PTCH1 and SSD domain–containing proteins further confirmed the conservation of the residues in the region proximal to the site 5 sterol in PTCH1 and NPC1, two RND proteins presumed to be involved in cholesterol transport (Fig. 3C).
Structural alignment of the two pseudosymmetric intramembrane halves of PTCH1Δ, using the SSD and SSDL regions for alignment, showed that most of the bound sterol molecules appear to be unique to the observed binding sites (Fig. 4, A and B). However, there is a clear overlap between the pocket sterols: sites 1, 6, and 7 in the SSD and site 10 in the SSDL. This points to a possibility of a conserved role of both domains of the protein in the interaction with the sterols, in line with the previously proposed model where cholesterol may access the inner tunnel at the neck region within PTCH1 from both sides (22). This would also be consistent with the presence of disease-linked mutations on both sides of PTCH1 (fig. S5A). The mutations at the SSD side, more densely populated with bound sterol molecules, coincide with the residues directly involved in sterol binding (fig. S5B). The mutations linked to holoprosencephaly 7, disrupting hedgehog signaling (3, 31), and the mutations linked to the basal cell nevus syndrome, inducing aberrant hedgehog signaling (32), have been mapped to the SSD and the SSDL regions of PTCH1 (fig. S5, B and C), suggesting a likely functional conservation of the two domains.
The PTCH1Δ-ShhNC24II structure provides the missing clues for linking the putative sterol translocation pathway from the inner to the outer leaflet of the SSD (Fig. 4, A to C). The existence of this pathway was proposed by the recent reports (24), and our structure suggests the plausible routes for the sterol movement along the SSD surface from the inner to the outer leaflet of the lipid bilayer (Fig. 4C). The SSD domain of PTCH1 is capable of accumulating multiple sterol molecules in the pocket as well as at the inner leaflet of the lipid bilayer. It is possible that the presence of ShhNC24II, a ligand that blocks the activity of PTCH1, establishes the conformation of the PTCH1 conducive to the accumulation of the sterols at the sites along the translocation pathway. The observed sterol sites are likely functionally coupled, as a number of mutations in this region abolish the activity of the protein, as described above. Furthermore, the presence of sterols at equivalent positions in the pocket region of the SSDL region of PTCH1 indicates that this domain may play a similar role as that of the SSD. Although the SSD regions have been defined on the basis of sequence similarity to the SSD regions in 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase and SCAP (33), it has been noted previously that the distinction between the SSD and the regions corresponding to the SSDL domain in the RND family proteins may be somewhat arbitrary (18). The structural comparison of SSD and SSDL in PTCH1Δ confirms this suggestion and points at the functional similarity of these domains in sterol recognition (Fig. 4, A and B).
To test whether disruption of sterol sites in PTCH1 SSD and SSDL may have a direct effect on the hedgehog signaling, we designed several mutations in the corresponding TM regions of PTCH1 (fig. S6). Mutations to a bulky residue (Trp) were chosen to disrupt the sterol sites 1 and 6 (L116W; fig. S6A), site 5 (M598W; fig. S6B) in the SSD, and site 10 (T768W; fig. S6C) in the SSDL. The rationale for the Trp mutations was to sterically hinder potential movement of the substrates at these sites. The plasmids encoding the wild-type and the mutated PTCH1 and PTCH1Δ were transfected into the PTCH1−/− cells together with a luciferase hedgehog signaling reporter, and the pathway activation was assessed (fig. S6D). The mock-transfected cells showed constitutive pathway activation, as expected for the PTCH1−/− mouse embryo fibroblasts (MEFs). In contrast, all of the constructs, including the wild-type PTCH1 and PTCH1Δ controls and the mutants, were capable of inhibiting the pathway activation (fig. S6D). The same was true for the double mutants featuring simultaneously mutated SSD and SSDL outer leaflet sites. This experiment does not rule out the involvement of the observed sterol sites in substrate transport by PTCH1. However, it is clear that mutating the sterol sites in PTCH1 may not suffice to shut down the downstream pathway activation. It is possible that the mutations do not fully obstruct the sterol sites to prevent sterol translocation by PTCH1. It is also possible that the mutations have a direct effect on sterol transport by PTCH1, but the residual activity of the protein is sufficient to block the downstream pathway activation (i.e., the assay is likely not sensitive to different levels of PTCH1 activity). A reduction of cholesterol transport activity of any PTCH1 mutant could be compensated by the superphysiological expression of the constructs. The development of molecular tools to directly track cholesterol movement facilitated by PTCH1 will be necessary to address this point experimentally.
Several well-resolved sterol densities are present in our three-dimensional (3D) reconstruction at sites distinct from the SSD and SSDL (sites 8, 9, and 11). The sterols bound at these sites may play an important structural role, stabilizing the membrane-embedded parts of the protein. These sites may also have a functional role, either directly participating in the substrate flux mediated by PTCH1 or by modulating its activity in various lipidic environments within or outside of the cilia membrane. The role of each lipid interaction will require careful investigation, and our structure provides the framework for the future in-depth analysis of the protein-lipid interactions in PTCH1 and other SSD domain–containing membrane proteins.
Although our reconstruction features a monomeric state of the protein, the previously published biochemical evidence suggested that the hedgehog pathway RND proteins D. melanogaster Ptch (34) and Disp1 (35) may exist as trimers, similarly to bacterial RND transporters. Recent structural studies showed that Shh-mediated dimerization (22) and tetramerization of PTCH1 may also be possible (36). One or more of these oligomeric states, or combinations thereof, may be required for PTCH1 to act as a sterol transporter. In this case, the sterol translocation pathway may be formed at the interface between the adjacent PTCH1 molecules. It is likely that PTCH1-mediated sterol transport may involve several distinct stages, including interactions in the plane of the membrane followed by movement of the sterol to the extracellular space. Future investigations will address how the interplay of the sterol sites, the ion transport, and the oligomerization of PTCH1 contribute to its role as a sterol transporter and a master regulator of the hedgehog signaling pathway.
MATERIALS AND METHODS
PTCH1 expression and purification
The synthetic DNA fragment (Genewiz) encoding the human wild-type full-length PTCH1 protein (UniProt ID: Q13635) was cloned into pcDNA3.1 vector modified to contain a C-terminal green fluorescent protein (GFP)–10xHis tag fusion protein. PTCH1 was previously shown to be recognized by the HECT E3 ubiquitin ligases, via two PPXY motifs in the C terminus (residues 1313 to 1316) and in the cytosolic loop (residues 642 to 645) (26, 27). Disruption of these motifs was shown to reduce PTCH1 degradation (27). Thus, we designed the PTCH1 expression construct to maximize the protein yield by removing the two PPXY motifs; the C-terminally truncated version of the protein, PTCH1-C (residues 1 to 1188), and the construct PTCH1Δ (comprising the residues 1 to 1188, with a mutation Y645A) were generated by polymerase chain reaction (PCR). The latter was subcloned into a modified pACMV plasmid (37), resulting in an expression cassette containing the PTCH1∆ construct fused to the C-terminal 3C-YFP-twinStrep tag. This was used to generate a stable tetracycline-inducible human embryonic kidney (HEK) 293-GnTi− cell line expressing PTCH1Δ.
Small-scale expression tests were performed using HEK293F cells. Expression plasmids encoding GFP-tagged wild-type full-length PTCH1 and derivative constructs were transfected into the HEK293F cells grown in six-well plates using a polyethylenimine (PEI)–based method in six wells. Cells were harvested 48 hours later by centrifugation, resuspended in buffer A [50 mM tris-HCl (pH 8.0), 200 mM NaCl, 10% glycerol], and sonicated and solubilized using 1% dodecyl maltoside (DDM). The lysates were centrifuged for 30 min at 25,000g (Eppendorf), and supernatants were analyzed by SDS–polyacrylamide gel electrophoresis and in-gel fluorescence using Amersham Imager 600 (GE: 29083461).
For large-scale expression, the PTCH1Δ cell line cultures were expanded to 5 liters in 2-liter Erlenmeyer flasks at 37°C in protein expression medium (PEM) supplemented with 5% fetal calf serum (FCS), penicillin/streptomycin, GlutaMAX, and nonessential amino acids until cell density of 2 × 106 cells/ml was reached, and the expression of the protein was induced by adding 2 μg/ml tetracycline and 5 mM sodium butyrate. Cells were grown for an additional 48 hours at 37°C and were subsequently collected by centrifugation at 500g, frozen, and stored at −80°C until the day of the experiment.
For protein purification, the frozen pellets of 5 liters of HEK293-GnTi− cells expressing PTCH1 were resuspended in 150 ml of buffer A supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and one tablet per 50 ml of Roche complete protease inhibitor cocktail (EDTA free). Cells were homogenized using a Dounce homogenizer and were solubilized directly by the addition of 1% DDM and 0.2% cholesteryl semisuccinate (CHS) at 4°C for 1 hour. The lysates were clarified by centrifugation (Ti45 rotor, 35,000 revolutions per minute, 30 min). The supernatants were added to 3 ml of cyanogen bromide (CNBr)–sepharose resin coupled with a purified anti-GFP nanobody (38). Following 1 hour of incubation with slow rotation at 4°C, the resin was collected in a gravity column (Bio-Rad), washed with 30 column volumes of buffer A containing DDM/CHS mixture (0.025% DDM/0.005% CHS) and with 10 column volumes of buffer A containing 0.1% digitonin. The protein was eluted from the resin by cleavage with 3C protease overnight at 4°C. The eluted fractions were passed through 1 ml of NiNTA resin to remove the 3C protease. The protein was further purified using size exclusion chromatography using Superose 6 Increase 10/300 GL column equilibrated in buffer B [50 mM tris-HCl (pH 8.0), 100 mM NaCl, 0.1% digitonin]. The eluted fractions corresponding to PTCH1 were concentrated to 5 mg/ml (~38 μM) using an Amicon Ultra-4 concentrator (100 kDa cutoff, Millipore) and used for cryo-EM grid freezing immediately.
ShhNC24II expression and purification
The synthetic DNA fragment (Genewiz) encoding the human ShhNC24II protein (residues 25 to 197, modified at the N terminus with a Cys24-Ile-Ile substitution; UniProt ID: Q15465), fused with the N-terminal 6xHis-SUMO tag, was cloned into pET28a plasmid. The plasmid was transformed into BL21(DE3) RIPL E. coli cells. The transformed cultures were grown in a shaking incubator at 37°C until OD600 of 0.8, at which point the temperature was switched to 30°C and the expression was induced with 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG). After 3 hours, cells were collected by centrifugation, frozen, and stored at −80°C until the day of the experiment.
ShhNC24II-expressing E. coli pellets (0.5-liter culture) were thawed, resuspended in 10 ml of buffer C [50 mM tris-HCl (pH 7.5), 200 mM NaCl] containing 25 mM imidazole, disrupted by sonication, and cleared using centrifugation with a benchtop Eppendorf centrifuge for 30 min at 25,000g. The supernatant was added to 0.5 ml of NiNTA resin, incubated with rotation for 1 hour. The resin was collected on a gravity column (Bio-Rad), washed with 30 column volumes of buffer C containing 25 mM imidazole, followed by 10 column volumes of buffer C containing 50 mM imidazole. The protein was eluted with buffer C containing 250 mM imidazole. The eluted fractions were pooled, diluted eightfold to reduce the concentration of imidazole, and incubated with 250 μg of purified SUMO protease Ulp1 overnight at 4°C. The mixture was passed through 1 ml of immobilized NiNTA resin to remove uncleaved protein, excess cleaved SUMO tag, and Ulp1, and the flow-through was concentrated to 1 ml and applied to a Superdex 200 Increase 10/300 GL column. The fractions corresponding to ShhNC24II were pooled, supplemented with 10% glycerol, aliquoted into 100-μl batches, flash frozen in liquid nitrogen, and stored at −80°C. Prior to cryo-EM grid freezing, the protein was thawed, desalted into buffer B, and concentrated to a final concentration of ~76 μM using an Amicon Ultra-4 concentrator (10-kDa cutoff, Millipore).
Hedgehog reporter assays
To evaluate hedgehog pathway activation in NIH 3T3 cells, quantitative PCR (qPCR) assays were performed. Confluent cultures of NIH 3T3 cells were starved overnight in Dulbecco’s modified Eagle’s medium (DMEM) and treated for 24 hours with different concentrations of either rhShhN or ShhNC24II. Following incubation, RNA was extracted using the RNeasy Mini Kit (Qiagen) and reverse transcribed using the QuantiTect Reverse Transcription Kit (Qiagen). Transcription of the mouse Gli1 gene was measured by qPCR using KAPA SYBR FAST (Sigma) on the LightCycler 480 System (Roche). mRNA expression of the SDHA gene was used for normalization. The data (as shown in Fig. 1B) were presented as fold enrichment with respect to the untreated sample. The sequences for the gene-specific primers were as follows: Gli1, GAATTCGTGTGCCATTGGGG (forward) and GGACTTCCGACAGCCTTCAA (reverse); SDHA, TTCCGTGTGGGGAGTGTATTGC (forward) and AGGTCTGTGTTCCAAACCATTCC (reverse). All qPCR experiments were performed in triplicate starting from independent cell cultures.
The luciferase assays were performed in PTCH1−/− MEFs (the cells were provided by M. P. Scott, Stanford University School of Medicine). In brief, PTCH1−/− MEFs were transfected using the TransIT-LT1 Transfection Reagent (catalog no. MIR2304). For transfection, cells were plated with the transfection mixture in 96-well plates. A mixture containing the plasmids encoding 8xGli-BS firefly luciferase (39) and cytomegalovirus (CMV)–Renilla luciferase (Promega), the latter constituting of 8xGli-BS firefly luciferase plasmid, was added to the plasmids encoding the GFP, PTCH1, and PTCH1Δ constructs (1:1 with the luciferase plasmid mixture). At confluence, cells cultured in DMEM in the presence of 10 to 15% FCS were shifted to DMEM with 0.5% FCS and incubated for 48 hours. Cell lysates were prepared following the Dual-Glo Luciferase Assay System (Promega) protocol, and luciferase activity was measured using the GloMax Explorer Multimode Microplate Reader. For each sample, firefly luciferase values were normalized to the corresponding Renilla luciferase values to account for the differences in transfection efficiency between samples.
Electron microscopy data acquisition
To prepare cryo-EM grids containing the PTCH1Δ-ShhNC24II complex, the purified components, PTCH1Δ and ShhNC24II, were mixed at a 1:2 molar ratio, with a final concentration of PTCH1 of 2.5 mg/ml (20 μM). The complex was incubated for 10 min at room temperature, followed by 30 min of incubation on ice. Aliquots of the protein mixture (3.5 μl) were applied to the glow-discharged UltrAuFoil 1.2/1.3 Au 300-mesh grids. The grids were blotted for 3 s, plunge frozen in liquid ethane using Vitrobot Mark IV (Thermo Fisher Scientific), and stored in liquid nitrogen until the day of high-resolution data collection.
Cryo-EM data collection was performed using a Titan Krios electron microscope (Thermo Fisher Scientific) equipped with a K2 Summit direct electron detector (Gatan) and a GIF Quantum energy filter (slit width of 20 eV) at EMBL Heidelberg. The micrographs were recorded in counting mode with pixel size of 0.814 Å per pixel using SerialEM. The defocus range was set from −0.6 to −2.5 μm. Each micrograph was dose fractionated to 40 frames with a total exposure time of 8 s, resulting in a total dose of ~44.7 e−/Å2.
The cryo-EM dataset for PTCH1Δ-apo (ligand free) was collected at BioEM Lab, C-Cina (University of Basel), using a similar setup (Titan Krios, K2 Camera, Quantum GIF) and methodology. The pixel size was 0.831 Å per pixel, and the defocus range was −0.8 to −3 μm; each micrograph was dose fractionated to 40 frames with a total exposure time of 10 s, resulting in a total dose of 75 e−/Å2.
Cryo-EM image analysis
A total of 7628 cryo-EM movies were motion corrected using MotionCor2 (40). Contrast transfer function (CTF) was estimated using Gctf on non–dose-weighted aligned images (41); micrographs with estimated resolution exceeding 4 Å were discarded, leaving 3260 micrographs for downstream processing. The PTCH1Δ-ShhNC24II particles were autopicked in relion-2.1.0 using templates generated by 2D classification of 1306 manually picked particles (42), resulting in a selection of 707,620 particles. After several rounds of 2D classification, 430,811 particles were selected for 3D classification. The procedure for 3D classification with four classes was performed in relion-2.1.0, using a 3D model of PTCH1Δ-apo low pass filtered to 30 Å. The initial model was generated de novo in cisTEM using a dataset composed of 165,533 PTCH1Δ-apo particles selected after 2D classification in relion-2.0. The best 3D class of the PTCH1Δ-ShhNC24II complex consisted of 200,679 particles, showed clear secondary structure elements (α helices), and featured a bound molecule of the hedgehog ligand. This class was selected for 3D refinement. Refinement of this class gave a map with a resolution of 3.6 Å [based on Fourier shell correlation (FSC) at 0.143]. Reprocessing of this class using relion-3.0, including motion correction, CTF refinement, and Bayesian particle polishing (43), resulted in a new map with a resolution of 3.5 Å (“F”). Further refinement was performed using the mask that excluded the detergent micelle, continuing from the last iteration. This resulted in a map at 3.4-Å resolution. The same procedure was repeated substituting the mask with a detergent and ectodomain-free mask, resulting in a 3.3-Å resolution map after postprocessing. Postprocessing was performed in relion-3.0 using a B factor of −50 Å2. The processing steps and map improvements are illustrated in figs. S2 and S3.
Model building
Model building was performed in COOT (44) using the previously solved structures of PTCH1 [Protein Data Bank (PDB) ID: 6DMY] and ShhN (PDB ID: 3N1R) for guidance. Bound sterol molecules were modeled as CHS (PDB code: Y01). The manually built model was refined using phenix.real_space_refine (45) implemented in PHENIX (46). For model validation, the atom positions of the refined model were randomly displaced by a maximum of 0.5 Å using the PDB tools in PHENIX. The derived perturbed model was subjected to real-space refinement against one of the refined half maps (half-map1). Map versus model FSC comparison was made for the model against the corresponding half-map1 used in the refinement job, and for the same model versus the half-map2 that was not used during refinement (47). The model geometry was validated using MolProbity (48). Figures featuring the models and density maps were prepared using PyMOL (49) and UCSF Chimera (50).
Supplementary Material
Acknowledgments
We thank the Electron Microscopy Facility at PSI, Villigen (E. Mueller-Gubler and T. Ishikawa) for support. We thank the BioEM Lab service facility at the University of Basel (M. Chami, K. Goldie, and L. Kovacik) for the support in cryo-EM data collection. We also thank the Electron Microscopy Core Facility at EMBL Heidelberg (F. Weis) for the support and expertise in high-resolution cryo-EM data collection. Funding: This study has been supported by the Swiss National Science Foundation (SNF Professorship, 150665) and the ETH grants (ETH-29 15-1) to V.M.K. Author contributions: C.Q. performed the experiments, analyzed the data, and wrote the manuscript; G.D.M. designed and performed the experiments, analyzed the data, and wrote the manuscript; I.V. designed and performed the experiments; A.W. analyzed the data and wrote the manuscript; and V.M.K. designed and performed the experiments, analyzed the data, and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The cryo-EM maps have been deposited in the Electron Microscopy Data Bank with the accession numbers EMD-4936 and EMD-4939. The atomic coordinates have been deposited in the Protein Data Bank, with the PDB ID 6RMG. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/9/eaaw6490/DC1
Fig. S1. Purification of the hedgehog ligand and PTCH1.
Fig. S2. Cryo-EM and single-particle analysis workflow.
Fig. S3. FSC plots and 3D reconstruction details.
Fig. S4. Features of the map and the atomic model of the protein.
Fig. S5. Disease-linked mutations in PTCH1 are present within the SSD and SSDL regions of the protein.
Fig. S6. Mutations designed to disrupt sterol binding sites in PTCH1 fail to suppress hedgehog pathway activation.
Movie S1. The density elements corresponding to bound sterols in the PTCH1 TM-focused map.
Table S1. Cryo-EM data collection, single-particle analysis, and model building statistics.
REFERENCES AND NOTES
- 1.Briscoe J., Thérond P. P., The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 416–429 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Ingham P. W., Nakano Y., Seger C., Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 12, 393–406 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Ming J. E., Kaupas M. E., Roessler E., Brunner H. G., Golabi M., Tekin M., Stratton R. F., Sujansky E., Bale S. J., Muenke M., Mutations in PATCHED-1, the receptor for SONIC HEDGEHOG, are associated with holoprosencephaly. Hum. Genet. 110, 297–301 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Twigg S. R. F., Hufnagel R. B., Miller K. A., Zhou Y., McGowan S. J., Taylor J., Craft J., Taylor J. C., Santoro S. L., Huang T., Hopkin R. J., Brady A. F., Clayton-Smith J., Clericuzio C. L., Grange D. K., Groesser L., Hafner C., Horn D., Temple I. K., Dobyns W. B., Curry C. J., Jones M. C., Wilkie A. O. M., A recurrent mosaic mutation in SMO, encoding the hedgehog signal transducer smoothened, is the major cause of Curry-Jones syndrome. Am. J. Hum. Genet. 98, 1256–1265 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foulkes W. D., Kamihara J., Evans D. G. R., Brugières L., Bourdeaut F., Molenaar J. J., Walsh M. F., Brodeur G. M., Diller L., Cancer surveillance in Gorlin syndrome and Rhabdoid tumor predisposition syndrome. Clin. Cancer Res. 23, e62–e67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pak E., Segal R. A., Hedgehog signal transduction: Key players, oncogenic drivers, and cancer therapy. Dev. Cell 38, 333–344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasca di Magliano M., Hebrok M., Hedgehog signalling in cancer formation and maintenance. Nat. Rev. Cancer 3, 903–911 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Ingham P. W., McMahon A. P., Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 15, 3059–3087 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Xavier G. M., Seppala M., Barrell W., Birjandi A. A., Geoghegan F., Cobourne M. T., Hedgehog receptor function during craniofacial development. Dev. Biol. 415, 198–215 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Pepinsky R. B., Zeng C., Wen D., Rayhorn P., Baker D. P., Williams K. P., Bixler S. A., Ambrose C. M., Garber E. A., Miatkowski K., Taylor F. R., Wang E. A., Galdes A., Identification of a palmitic acid-modified form of human Sonic hedgehog. J. Biol. Chem. 273, 14037–14045 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Porter J. A., Ekker S. C., Park W.-J., von Kessler D. P., Young K. E., Chen C.-H., Ma Y., Woods A. S., Cotter R. J., Koonin E. V., Beachy P. A., Hedgehog patterning activity: Role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell 86, 21–34 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Gritli-Linde A., Lewis P., McMahon A. P., Linde A., The whereabouts of a morphogen: Direct evidence for short- and graded long-range activity of Hedgehog signaling peptides. Dev. Biol. 236, 364–386 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Allen B. L., Tenzen T., McMahon A. P., The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 21, 1244–1257 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada A., Charron F., Morin S., Shin D. S., Wong K., Fabre P. J., Tessier-Lavigne M., McConnell S. K., Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nature 444, 369–373 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Rohatgi R., Milenkovic L., Scott M. P., Patched1 regulates Hedgehog signaling at the primary cilium. Science 317, 372–376 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Dorn K. V., Hughes C. E., Rohatgi R., A Smoothened-Evc2 complex transduces the Hedgehog signal at primary cilia. Dev. Cell 23, 823–835 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haycraft C. J., Banizs B., Aydin-Son Y., Zhang Q., Michaud E. J., Yoder B. K., Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLOS Genet. 1, e53 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hausmann G., von Mering C., Basler K., The hedgehog signaling pathway: Where did it come from? PLOS Biol. 7, e1000146 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuwabara P. E., Labouesse M., The sterol-sensing domain: Multiple families, a unique role? Trends Genet. 18, 193–201 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Gong X., Qian H., Cao P., Zhao X., Zhou Q., Lei J., Yan N., Structural basis for the recognition of Sonic Hedgehog by human Patched1. Science 361, eaas8935 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Qi X., Schmiege P., Coutavas E., Wang J., Li X., Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature 560, 128–132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi X., Schmiege P., Coutavas E., Li X., Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex. Science 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tukachinsky H., Petrov K., Watanabe M., Salic A., Mechanism of inhibition of the tumor suppressor Patched by Sonic Hedgehog. Proc. Natl. Acad. Sci. U.S.A. 113, E5866–E5875 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Bulkley D. P., Xin Y., Roberts K. J., Asarnow D. E., Sharma A., Myers B. R., Cho W., Cheng Y., Beachy P. A., Structural basis for cholesterol transport-like activity of the Hedgehog receptor Patched. Cell 175, 1352–1364.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor F. R., Wen D., Garber E. A., Carmillo A. N., Baker D. P., Arduini R. M., Williams K. P., Weinreb P. H., Rayhorn P., Hronowski X., Whitty A., Day E. S., Boriack-Sjodin A., Shapiro R. I., Galdes A., Pepinsky R. B., Enhanced potency of human Sonic hedgehog by hydrophobic modification. Biochemistry 40, 4359–4371 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Kim J., C. Hsia E. Y., Brigui A., Plessis A., Beachy P. A., Zheng X., The role of ciliary trafficking in Hedgehog receptor signaling. Sci. Signal. 8, ra55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yue S., Tang L.-Y., Tang Y., Tang Y., Shen Q.-H., Ding J., Chen Y., Zhang Z., Yu T.-T., Zhang Y. E., Cheng S. Y., Requirement of Smurf-mediated endocytosis of Patched1 in sonic hedgehog signal reception. eLife 3, e02555 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hua X., Nohturfft A., Goldstein J. L., Brown M. S., Sterol resistance in CHO cells traced to point mutation in SREBP cleavage–activating protein. Cell 87, 415–426 (1996). [DOI] [PubMed] [Google Scholar]
- 29.Strutt H., Thomas C., Nakano Y., Stark D., Neave B., Taylor A. M., Ingham P. W., Mutations in the sterol-sensing domain of Patched suggest a role for vesicular trafficking in Smoothened regulation. Curr. Biol. 11, 608–613 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Johnson R. L., Zhou L., Bailey E. C., Distinct consequences of sterol sensor mutations in Drosophila and mouse patched homologs. Dev. Biol. 242, 224–235 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Ribeiro L. A., Murray J. C., Richieri-Costa A., PTCH mutations in four Brazilian patients with holoprosencephaly and in one with holoprosencephaly-like features and normal MRI. Am. J. Med. Genet. A 140, 2584–2586 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Johnson R. L., Rothman A. L., Xie J., Goodrich L. V., Bare J. W., Bonifas J. M., Quinn A. G., Myers R. M., Cox D. R., Epstein E. H. Jr., Scott M. P., Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272, 1668–1671 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Brown M. S., Goldstein J. L., A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. U.S.A. 96, 11041–11048 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu X., Liu S., Kornberg T. B., The C-terminal tail of the Hedgehog receptor Patched regulates both localization and turnover. Genes Dev. 20, 2539–2551 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Etheridge L. A., Crawford T. Q., Zhang S., Roelink H., Evidence for a role of vertebrate Disp1 in long-range Shh signaling. Development 137, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian H., Cao P., Hu M., Gao S., Yan N., Gong X., Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm. Nat. Commun. 10, 2320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaudhary S., Pak J. E., Gruswitz F., Sharma V., Stroud R. M., Overexpressing human membrane proteins in stably transfected and clonal human embryonic kidney 293S cells. Nat. Protoc. 7, 453–466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kubala M. H., Kovtun O., Alexandrov K., Collins B. M., Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. 19, 2389–2401 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sasaki H., Hui C., Nakafuku M., Kondoh H., A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development 124, 1313–1322 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J. H., Lindahl E., Scheres S. H. W., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Afonine P. V., Headd J. J., Terwelliger T. C., Adams P. D., New tool: Phenix. real-space-refine. Comput. Crystallogr. Newslett. 4, 43–44 (2013). [Google Scholar]
- 46.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amunts A., Brown A., Bai X. C., Llacer J. L., Hussain T., Emsley P., Long F., Murshudov G., Scheres S. H. W., Ramakrishnan V., Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen V. B., Arendall W. B. III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anonymous (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
- 50.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/9/eaaw6490/DC1
Fig. S1. Purification of the hedgehog ligand and PTCH1.
Fig. S2. Cryo-EM and single-particle analysis workflow.
Fig. S3. FSC plots and 3D reconstruction details.
Fig. S4. Features of the map and the atomic model of the protein.
Fig. S5. Disease-linked mutations in PTCH1 are present within the SSD and SSDL regions of the protein.
Fig. S6. Mutations designed to disrupt sterol binding sites in PTCH1 fail to suppress hedgehog pathway activation.
Movie S1. The density elements corresponding to bound sterols in the PTCH1 TM-focused map.
Table S1. Cryo-EM data collection, single-particle analysis, and model building statistics.


