Abstract
Nuclear-import receptors (NIRs) bind nuclear-localization signals (NLSs) of protein cargo in the cytoplasm and transport them into the nucleus. Here, we review advances establishing that NIRs also function in the cytoplasm to prevent and reverse functional and aberrant phase transitions of their cargo, including neurodegenerative disease-linked RNA-binding proteins (RBPs) with prion-like domains, such as TDP-43, FUS, hnRNPA1, and hnRNPA2. NIRs selectively extract cargo from condensed liquid phases thereby regulating functional phase separation. Consequently, NIRs sculpt cytoplasmic membraneless organelles and regulate cellular organization beyond their canonical role in nuclear import. Elevating NIR expression dissolves cytoplasmic RBP aggregates, restores functional RBPs to the nucleus, and rescues disease-linked RBP toxicity. Thus, NIRs could be leveraged therapeutically to restore RBP homeostasis and mitigate neurodegeneration.
Keywords: nuclear-import receptor, phase transition, neurodegeneration, Karyopherin-β2, FUS, ALS
Nuclear-import receptors (NIRs) import specific protein cargo into the nucleus
The passage of diverse macromolecules between the cytoplasm and nucleus occurs via transport across the nuclear pore complex (NPC; see glossary) [1]. The NPC permits passive transit of small molecules, but proteins larger than ~20–40kDa must typically engage nuclear-import receptors (NIRs) for transport across the NPC [1]. Thus, NIRs bind nuclear-localization signals (NLSs) of protein cargo in the cytoplasm and transport them across the central channel of the NPC into the nucleus (Figure 1) [1]. Nuclear import of cargo by NIRs is orchestrated by a small GTPase, Ran, which regulates interactions between NIRs and cargo (Figure 1, Box 1) [1]. In this way, Ran-regulated NIRs enable nuclear localization of many proteins, which would otherwise be unable to cross the permeability barrier imposed by the NPC [1].
Figure 1. Mechanisms of nuclear import.
Kapβ2 has low affinity for Ran-GDP (see Box 1) in the cytoplasm and can bind cargo bearing a PY-NLS. Kapβ2 then transports cargo into the nucleus, where upon binding to Ran-GTP with high affinity, Kapβ2 undergoes a conformational change and unloads its cargo. This mechanism of nuclear import is distinct from that used by Kapβ1, which must form a heterodimer with a member of the Impα family to exert its function. Impαs recognize cNLSs, but only when in complex with Kapβ1. When this ternary complex enters the nucleus, Ran-GTP binds Kapβ1, causing it to release Impα. Once Impα is separated from Kapβ1 in the nucleus, its associated cargo is released.
Box 1. The Ran Cycle and Nuclear Transport.
Ran is a small GTPase, which can hydrolyze GTP and switch between a GTP- and GDP-bound state [19]. The nucleotide state of Ran is regulated by the activity of guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). GEFs stimulate the exchange of GDP for GTP and the Ran-GEF, Rcc1, is enriched in the nucleus [19]. GAPs, on the other hand, work to accelerate GTPase activity. RanGAP is enriched in the cytoplasm where it promotes the conversion of RanGTP to RanGDP [19]. By differentially localizing RanGAP and Rcc1, the cell creates a concentration gradient where Ran-GTP is concentrated in the nucleus and Ran-GDP is concentrated in the cytoplasm. The Ran concentration gradient is leveraged to facilitate directional nuclear transport. Kapβ1 and Kapβ2 both have a high affinity for Ran in its GTP-bound state [18, 126]. Thus, in the cytoplasm where Ran-GDP is the dominant species, NIRs exist in a conformation that allows cargo binding. Upon entering the nucleus, where Ran-GTP is abundant, NIRs undergo a conformational rearrangement that results in the loaded NLS-bearing cargo being displaced (Figure 1).
Importantly, NIRs drive the nuclear localization of several notorious RNA-binding proteins (RBPs) with prion-like domains (PrLDs) that are connected with neurodegenerative disease via pathology and genetics [1–6]. In disease, these predominantly nuclear RBPs become mislocalized to cytoplasmic aggregates in degenerating cells [4]. Here, we review NIR structure and function in light of advances establishing that NIRs prevent and reverse functional and aberrant phase separation [7–9] of their cargo, including neurodegenerative disease-linked RBPs with PrLDs, such as TDP-43, FUS, TAF15, EWSR1, hnRNPA1, and hnRNPA2 [10–13].
Structure of NIRs
Typically, NIRs are members of the Karyopherin-β (Kapβ) family of proteins, which drive the majority of nuclear-cytoplasmic transport in eukaryotic cells [1]. For example, Karyopherin-β2 (Kapβ2, also known as Transportin-1 or Importin-β2) is a NIR that shuttles cargo bearing a proline-tyrosine (PY)-NLS into the nucleus [2]. Similar to other NIRs, Kapβ2 is predominantly comprised of HEAT repeats, which are structural motifs composed of two antiparallel helices A and B [1]. The HEAT repeats coil to form a stacked superhelix in which the B helices form the inner hydrophobic core and the A helices form the outer convex surface (Figure 2A) [3, 14–16]. The two arches of the superhelix are structurally similar, but differ in their binding partners. The N-terminal arch (HEAT repeats 1–8) binds to Ran, which regulates interactions with cargo (Figure 1; Box 1), while the C-terminal arch (HEAT repeats 12–20) binds to PY-NLS-bearing cargo (Figure 2A) [1–3, 17].
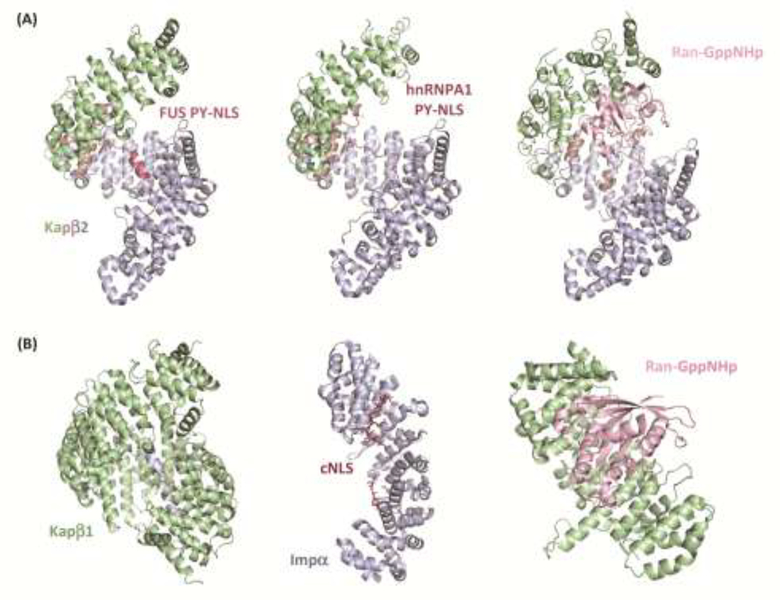
Figure 2. Structures of nuclear-import receptors.
(A) Structural architecture of Kapβ2. Kapβ2 acts as a single molecule to transport proteins bearing a PY-NLS into the nucleus. Kapβ2 is comprised of 20 HEAT repeats. HEAT repeats 1–7 are shown in green, HEAT repeat 8 in taupe, and HEAT repeats 9–20 in blue. Kapβ2 cargos are varied, and can have structured regions, as with the PY-NLS (dark pink) of FUS (Left, PDB: 4FDD), or completely unstructured regions, as with the PY-NLS (dark pink) of hnRNPA1 (Middle, PDB: 2H4M). To release cargo in the nucleus, the N-terminal arch of Kapβ2 (HEAT repeats 1–7, green, and HEAT repeat 8, taupe) binds to Ran-GTP (light pink), causing an extended acidic loop in HEAT repeat 8 to displace the cargo loaded into the C-terminal arch (HEAT repeats 12–20) (Right, PDB: 1QBK).
(B) Structural architecture of Kapβ1 (green) and Impα (blue). Cargo bearing a cNLS are transported into the nucleus by a heterodimer of Kapβ1 (left, PDB: 1QGK) and Impα (middle, PDB: 5E6Q). Impα binds to a cNLS (shown in dark pink) only when associated with Kapβ1. The N-terminal importin-beta-binding (IBB) domain of Impα binds Kapβ1. Upon entering the nucleus, Ran-GTP (light pink) binds to Kapβ1, thus displacing Impα (right, PDB: 1IBR). As a result of this dissociation, Impα releases its cargo.
Communication between the N- and C-terminal arches of Kapβ2 is mediated by an extended acidic loop between the A and B helices of HEAT repeat 8 [2, 18]. This loop is critical for Ran-GTP-dependent cargo release in the nucleus (Figure 1 and 2A; Box 1) [18]. Kapβ2 has a low affinity for Ran-GDP, which is enriched in the cytoplasm due to an asymmetric distribution of the Ran GTPase activating protein (GAP) in the cytoplasm and the Ran Guanine nucleotide Exchange Factor (GEF) in the nucleus (Box 1) [1]. Thus, in the cytoplasm Kapβ2 can engage PY-NLS bearing cargo (Figure 1). Once Kapβ2 enters the nucleus with its cargo, there must be a specific mechanism for cargo release. Here is where the acidic loop functions: upon binding to Ran-GTP with high affinity, Kapβ2 undergoes a conformational change and the acidic loop is moved to the cargo-binding site, unloading the cargo (Figure 1 and 2A) [2, 18, 19]. Indeed, when this acidic loop is truncated, Kapβ2 maintains its ability to engage cargo in the cytoplasm, but is unable to release cargo in the nucleus in a Ran-GTP-dependent manner [16, 18].
This mechanism of nuclear import differs from that used by another Karyopherin, Kapβ1 (also known as Importin-β1) (Figure 2B). Kapβ1 forms a functional heterodimer with a member of the Importin-α (Impα) family (Figure 1 and 2B) [2, 20–22]. Impα recognizes the canonical, lysine-rich NLS (cNLS), but only when in complex with Kapβ1 [21–23]. When this ternary complex enters the nucleus, Ran-GTP binds Kapβ1, causing it to release Impα (Figure 1 and 2B) [2, 24, 25]. Once Impα is separated from Kapβ1 in the nucleus, its associated cargo is released (Figure 1) [24, 26]. Separating the cargo-binding function from the nuclear-import activity of the Kapβ1/Impα system provides an additional level of regulation, a safeguard to ensure proper nuclear transport of over 50% of nuclear proteins [19, 24–27].
The “all-in-one” design of Kapβ2 makes it an attractive NIR to study. Kapβ2 recognizes cargo and navigates through the gel-like phase of the nuclear pore on its own [2, 3, 18, 28]. This dual functionality is likely important for newly realized functions ascribed to Kapβ2, including preventing and reversing functional and aberrant phase transition of cargo in the cytoplasm [10–13, 29].
Cargo Recognition by NIRs
Kapβ2 cargo generally harbor a PY-NLS (Figure 3), which can be ~15–100 amino acids in length [1, 2]. Unlike the much larger class of lysine-rich cNLSs recognized by Impα/β, PY-NLSs have three important epitopes: (1) an N-terminal hydrophobic or basic epitope, (2) the arginine of the R-X2–5P-Y/Φ (Φ is a hydrophobic residue) motif, and (3) the PY or homologous PΦ motif of the C-terminal R-X(2–5)-PY/Φ motif [2, 3, 20, 30]. The list of cargo recognized by Kapβ2 is still growing and includes several disease-linked RBPs with PrLDs (Figure 3) [1, 3]. RBPs with PrLDs have garnered attention due to their involvement in liquid-liquid phase separation (LLPS) and membraneless organelle biogenesis [7–9], as well as for their mislocalization to cytoplasmic aggregates in neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) [4, 5, 8, 9, 31–38]. Thus, understanding how Kapβ2 ferries these RBPs between the nucleus and the cytoplasm is an important area of research.
Figure 3. Domain map of NIR cargo.
Domain architecture of FUS, TAF15, EWSR1, hnRNPA1, hnRNPA2, and TDP-43. Domains are indicated: prion-like domain (PrLD, green), Gly-rich domain (purple), RNA-Recognition Motif (RRM, orange), RGG domain (yellow), Zinc finger (ZNF, blue), and NLS (red).
Proteins that have a PY-NLS are the most well studied class of cargo recognized by Kapβ2, but we are only beginning to appreciate the diverse array of cargo that Kapβ2 is capable of engaging [1]. Identifying proteins harboring a PY-NLS has been a predictive heuristic for identifying Kapβ2 cargo, but this sequence is surprisingly not necessary, and Kapβ2 can engage and transport proteins to the nucleus that vary dramatically in composition. For example, Kapβ2 recognizes the histone H3 tail, which lacks the titular PY residues of the PY-NLS [30]. Instead, Kapβ2 binds the H3 tail through electrostatic and entropic interactions [30]. Despite lacking a PY epitope, the H3 tail binds to Kapβ2 with high affinity, with a Kd similar to other characterized Kapβ2-cargo interactions [30]. Kapβ2 also interacts with several other proteins that lack a PY-NLS, including RNA helicase DDX3, molecular chaperones Hsp70 and Hsp90, FOXO transcription factors, and HuR [3, 18, 39–41]. In some cases, these proteins may interact with other cargo with a canonical PY-NLS and thus ride “piggy-back” into the nucleus, but in other cases direct interaction with Kapβ2 may be important [1, 2]. The diversity observed in the repertoire of Kapβ2 binding partners further supports the hypothesis that cargo recognition by Kapβ2 is sensitive to a combination of features, each of which has its own range of allowable properties [2, 18].
Epitope sensitivity can be understood by comparing how Kapβ2 binds to different domains within each cargo. The distribution of binding energy has been probed by systematically mutating residues of the well-characterized PY-NLS of hnRNPA1 to alanine and assessing the effect of these mutations on binding affinity [2]. Surprisingly, single mutations to either the Pro or Tyr of the PY-NLS resulted in only a modest decrease in binding affinity, and single mutations elsewhere along the binding interface had little effect [2]. Furthermore, when combined, alanine mutations have a non-additive effect, suggesting that Kapβ2 binding is cooperative [2]. Remarkably, once bound to a PY-NLS, Kapβ2 can make secondary contacts with the cargo [10, 11, 13]. For example, Kapβ2 binding to FUS depends on the PY-NLS, but upon engaging the PY-NLS, Kapβ2 can make secondary low-affinity interactions with other domains of FUS, including the RGG domains (a domain enriched in repeats of Arg-Gly-Gly often interspersed with hydrophobic residues), RNA-recognition motif (RRM), and PrLD [11, 13]. Interestingly, these interactions cause FUS to eject bound RNA, thereby enabling an apoform of the RBP to be transported back to the nucleus where it can bind new RNAs [11, 13].
Cargo and Stress Granules
During stress, cells must conserve and redirect their limited energy to avoid severe damage. To this end, cells will pause translation of most mRNAs except for those encoding various heat-shock proteins, and store transcripts and RBPs in cytoplasmic membraneless organelles called stress granules (SGs) [9, 42–44]. Many NIR cargos and NIRs themselves become sequestered in SGs [10, 45]. The formation of SGs is a normal protective process, but it can become deleterious if SGs persist for too long in the cytoplasm [10, 37, 42–44, 46–48]. Notably, mutations that promote SG assembly or prevent SG disassembly in RBPs with PrLDs have been linked to the development of neurodegenerative disorders, including ALS, FTD, and multisystem proteinopathy (MSP) [10, 32, 35, 43, 49–57]. Within SGs, there is a high local concentration of aggregation-prone proteins, and improper temporal maintenance promotes intra- and inter-molecular interactions, thereby facilitating protein aggregation [4, 5, 10, 31–34, 43, 46, 58, 59]. Kapβ2 plays an important role in SG regulation. Many of the RBPs found in SGs are cargo recognized by Kapβ2 [2, 3, 10]. Indeed, Kapβ2 can selectively extract cargo from SGs without affecting SG assembly per se [10]. Nonetheless, complete resolution of SGs is in part dependent on Kapβ2 activity [10, 11]. This dependence becomes more apparent in cases where cargo harbor mutations that reduce the affinity of Kapβ2 binding [10, 11, 13, 17, 60].
Cargo and Disease
Many NIR cargos are RBPs with PrLDs that are connected with ALS and FTD, including TDP-43, FUS, EWSR1, TAF15, hnRNPA1, and hnRNPA2 (Figure 3) [4–6]. These RBPs mislocalize to cytoplasmic inclusions in degenerating neurons [4–6, 32, 61, 62]. For example, TDP-43 is mislocalized to cytoplasmic aggregates in degenerating neurons of ~97% of ALS cases and ~45% of FTD cases [4]. By contrast, FUS is mislocalized to cytoplasmic aggregates in degenerating neurons of ~1% of ALS cases and ~9% of FTD cases [4]. TDP-43 and FUS pathology are mutually exclusive [4]. Cytoplasmic EWSR1 and TAF15 aggregates are found in all FTD cases with FUS pathology and have been observed in sporadic ALS [4, 61, 62]. Cytoplasmic hnRNPA1 and hnRNPA2 inclusions are found in degenerating tissues of patients with multisystem proteinopathy (MSP), an inherited pleiotropic degenerative disorder that can affect muscle, bone, and the nervous system [4, 32, 63]. MSP is very rare and has been diagnosed in several hundred people worldwide [63]. Operationally, MSP is defined as a combination of two or more of inclusion body myopathy, Paget’s disease of bone, and ALS/FTD [63].
In addition to pathology, mutations in the genes that encode these RBPs are linked to sporadic and familial forms of disease [4–6, 32, 61, 62]. Mutations in the gene encoding TDP-43 are found in ~1% of ALS cases [64]. Likewise, mutations in the gene encoding FUS are also found in ~1% of ALS cases [64]. Mutations in the genes encoding TAF15, EWSR1, and hnRNPA1 are very rare in ALS [32, 61, 62, 65], whereas mutations in the genes encoding hnRNPA1 and hnRNPA2 occur in MSP [32, 63]. Often, these mutations accelerate RBP fibrillization, aberrant phase transitions, or promote cytoplasmic mislocalization [4].
Several disease-linked mutations have been found in the PrLDs of RBPs [4]. PrLDs possess a distinctive low-complexity composition enriched in uncharged polar amino acids and glycine [4–6]. PrLDs are similar to prion domains, which enable various yeast proteins such as Sup35 and Rnq1 to form prions in yeast [4–6]. In the context of human RBPs, PrLDs are important protein-protein interaction domains that enable functionality, and promote LLPS and fibrillization [4–6, 35, 43, 66–68]. Additional intrinsically-disordered domains can also contribute to LLPS [8]. For example, FUS also has two intrinsically-disordered RGG domains (Figure 3) that contribute to LLPS [4, 11, 13, 69]. Cation-π or π-π interactions between the arginine residues in RGG domains and tyrosine residues in the PrLD of FUS promote FUS LLPS [11–13, 69, 70]. However, if FUS dwells in a dense liquid state for too long then it converts to a gel or solid phase, which is comprised of self-templating fibrils as the PrLD switches from an intrinsically disordered state to a cross-β polymeric state [10, 71, 72]. This transition from liquid to gel or solid is termed an aberrant phase transition, and can be accelerated by disease-linked mutations in PrLDs and other domains [35, 71, 73].
Disease-linked mutations in FUS and hnRNPA1 can also fall in the PY-NLS recognized by Kapβ2 [4, 10, 17]. Importantly, the degree to which ALS-associated mutations affect Kapβ2 binding correlates with the degree of impaired nucleocytoplasmic transport, as well as the severity of disease [17, 74]. For example, a mutation to the PY-NLS of FUS underlies particularly aggressive form of juvenile ALS [10, 17, 60, 74]. In this mutation, P525L, the Pro of the PY-NLS is mutated to Leu, resulting in a nine-fold decrease in affinity for Kapβ2 [17]. Consequently, cells expressing the P525L mutant exhibit cytoplasmic mislocalization of FUS, and the severity of this mislocalization is inversely correlated with age of disease onset [74]. Similar mutations to the critical P of the PY-NLS of hnRNPA1 have also been linked to ALS [51, 75]. Because RBPs tend to self-associate, a mutation in just one RBP can have dominant, far-reaching effects on nucleocytoplasmic homeostasis [4]. When one RBP is cytoplasmically mislocalized, it will sequester additional copies of the RBP, even if their NLS is wild type (WT) [4].
Nucleocytoplasmic transport dysfunction in disease
Accumulating evidence suggests that nucleocytoplasmic transport defects are a shared mechanism contributing to the disease initiation and progression of ALS and FTD, even in cases where no apparent mutations in RBP NLSs are observed [76, 77]. For example, in FTD, WT FUS mislocalization is seen in ~9% of cases [78, 79]. Additionally, the RBP TDP-43 is cytoplasmically mislocalized in ~97% of ALS patient and ~45% of FTD patients [80]. In most cases, WT TDP-43 accumulates in cytoplasmic aggregates, and only ~1% of ALS cases present with TDP-43 mutations, which are usually located in the TDP-43 PrLD [4, 64, 80]. Moreover, cytoplasmic TDP-43 aggregates sequester NIRs, leading to dysfunction of nucleocytoplasmic transport [81]. The involvement of NIRs in ALS/FTD pathogenesis is suggested by studies where post-mortem tissue from ALS and FTD patients revealed reduced NIR expression, NIR mislocalization, or both in the brain and spinal cord [79].
Several studies also connect C9orf72 hexanucleotide repeat expansion (HRE), the most common genetic cause of ALS and FTD, to aberrant nucleocytoplasmic transport [82–86]. C9orf72 HRE can induce toxicity via both the repeat-containing RNA itself and the polydipeptide repeat products (DPRs) that are produced by RAN (Repeat-associated non-AUG) translation from the sense or antisense transcript of the HRE [87–90]. Additionally, the C9orf72 HRE can reduce C9orf72 expression, which may result in haploinsufficiency phenotypes [90]. Genetic models of ALS that express the HRE transcript, DPRs, or both, have been used to identify modifiers of HRE toxicity [82–86]. The modifiers identified include Kapβ family members, Ran, RanGAP, Ran-GEF, and NPC components [82–86]. Specifically, knockdown of the fly homologs of Kapβ1 or Kapβ2 acts as an enhancer of toxicity in fly models of C9-ALS [82, 85]. Furthermore, overexpression of the yeast homolog of Kapβ2 (Kap104) in DPRexpressing yeast acts as a potent suppressor of toxicity, and overexpression of Kapβ1 in mouse primary neurons expressing the poly-PR DPR suppresses toxicity [77, 83].
In addition, nucleocytoplasmic transport defects are also observed in other neurodegenerative diseases. For example, Impα1 is found in inclusions in neurons from patients with Alzheimer’s diseases [91, 92]. General defects in nucleocytoplasmic transport have also been demonstrated in a variety of models for Huntington’s disease that express polyQ-expanded huntingtin protein or the RAN-translation products of the CAG repeat expansion [93, 94]. A common risk factor for these neurodegenerative diseases is aging. Interestingly, NIRs are downregulated and can be mislocalized upon aging [95–97]. Moreover, nucleocytoplasmic transport defects can be caused by the accumulation of misfolded proteins in the cytoplasm [98]. Thus, nucleocytoplasmic transport dysfunction may contribute to aging phenotypes and age-related neurodegenerative diseases.
The connection between nucleocytoplasmic transport dysfunction and neurodegenerative disease is further supported findings that nuclear-export inhibitors are neuroprotective agents in C9-ALS, TDP-43-ALS, and Huntington’s disease [81, 94]. For example, two small-molecule inhibitors of nuclear export, KPT-335 and KPT-276, rescued primary mouse cortical neurons transfected with ALS-linked TDP-43Q331K and reduced the number of cells with abnormal nuclear morphology [81]. However, the mechanism by which KPT-335 and KPT-276 rescue toxicity remains uncertain and is unlikely to be due to a direct effect on TDP-43 export. Indeed, TDP-43 diffuses passively out of the nucleus and is not exported by exportin-1 (CRM1/XPO1), the target of KPT-335 and KPT-276 [99, 100]. Additionally, Thiamet-G, an O-GlcNAcase inhibitor that results in increased glycosylation events involved in NPC homeostasis, or KPT-350, an inhibitor of canonical nuclear export, can restore nucleocytoplasmic transport function and rescue cell death in primary neurons transfected with full-length Huntingtin with a pathogenic polyQ expansion and in a Drosophila HD model [94].
Although the link between protein aggregation, nucleocytoplasmic transport, and neurodegeneration is established, the temporal relationship of these events is not clear. For cases where a disease-linked RBP bears a mutation in its NLS, RBP mislocalization likely precedes aggregation. However, nucleocytoplasmic transport defects may also be an early event in ALS/FTD pathogenesis. For example, expression of C9orf72 HRE results in defects in the NPC and in nucleocytoplasmic transport, which elicit TDP-43 mislocalization and aggregation [84]. Moreover, upon stress, NIRs are recruited into SGs, which might reduce the effective NIR concentration and trigger protein aggregation [10, 45]. Alternatively, TDP-43 aggregates could sequester components of the NPC and nucleocytoplasmic transport machinery and cause nucleocytoplasmic transport dysfunction [81]. A positive feed-forward loop is also proposed, in which protein aggregates or DPRs obstruct the nuclear pore, which, in turn, causes TDP-43 and other aggregation-prone proteins to accumulate in the cytoplasm, resulting in further nuclear pore damage and prolonged transport defects [81, 101]. Irrespective of the precise mechanism, agents that simultaneously reverse RBP aggregation and mislocalization would rescue toxicity associated with dysfunctional nucleocytoplasmic transport [10, 102–104].
New NIR functions in preventing and reversing aberrant phase separation
Several groups have reported new functions for NIRs [10–13]. We reported that NIRs potently inhibit and reverse LLPS, aberrant phase transition, and fibrillization of several RBPs with PrLDs implicated in ALS, FTD, and MSP [10]. While NIRs have been found to function as chaperones before [105–107], the ability to rapidly disaggregate cargo that has converted to stable cross-β polymers, and to prevent and reverse aberrant phase transitions was unanticipated [10–13]. For example, we established that Kapβ2 prevents the fibrillization and LLPS of PY-NLS bearing RBPs including FUS, EWSR1, TAF15, hnRNPA1, and hnRNPA2 [10]. Importantly, we demonstrated that Kapβ2 also disassembles preformed fibrils, liquid droplets, and hydrogels composed of these disease proteins [10]. Kapβ2 does so by engaging the PY-NLS of these RBPs [10, 13]. Similarly, we established that Impα and Kapβ1 engage the cNLS to prevent and reverse TDP-43 fibrillization [10].
We discovered that Kapβ2 rapidly dissolves FUS, EWSR1, and TAF15 fibrils [10]. After binding the exposed PY-NLS on FUS, EWSR1, and TAF15 fibrils, Kapβ2 might extract individual RBPs from the fibril via entropic pulling or ‘collision pressure’ upon binding the exposed PY-NLS on FUS, EWSR1, and TAF15 fibrils [108, 109]. However, unlike Kapβ2, an antibody that binds to the FUS PY-NLS is unable to dissolve FUS fibrils, which argues against this model [10]. Instead, binding to the PY-NLS enables Kapβ2 to engage secondary binding sites in the PrLD and rapidly and forcibly disrupt intermolecular contacts that maintain fibril integrity [10]. On the other hand, we found that Kapβ2 only slowly disassembles hnRNPA1 and hnRNPA2 fibrils [10]. Unlike FUS, the PY-NLS of hnRNPA1 or hnRNPA2 is not at the extreme C-terminal end of the protein (Figure 3). Thus, accessing the PY-NLS in hnRNPA1 or hnRNPA2 fibrils is likely more difficult since it is sequestered from solvent in the cross-β fibril core [110]. Nonetheless, thermal fluctuations may enable Kapβ2 to engage the PY-NLS to dissolve hnRNPA1 and hnRNPA2 fibrils [10]. Moreover, Kapβ2 dissolved hnRNPA2 fibrils more effectively that MSP-linked hnRNPA2D290V fibrils [10], which may be due to increased stability of hnRNPA2D290V fibrils compared to WT hnRNPA2 fibrils [111].
The chaperone and disaggregation activity of Kapβ2 is regulated by the small GTPase, Ran (Box 1) [10, 13]. Indeed, we established that Ran-GDP allows Kapβ2 chaperone and disaggregation activity, whereas Ran-GTP inhibits Kapβ2 chaperone and disaggregation activity by preventing Kapβ2 interactions with the PY-NLS of cargo [10]. This regulation by Ran implies that the chaperone and disaggregation activity of Kapβ2 is likely restricted to the cytoplasm [10, 13]. Once the Kapβ2:cargo complex reaches the nucleus it is dissociated by Ran-GTP. Thus, cargo is now free to perform nuclear functions, and Kapβ2 can be recycled to the cytoplasm for further rounds of chaperone or disaggregation activity. In this way, Ran-GTP completes Kapβ2-mediated disaggregation and makes the nuclear-import system as a whole a catalytic disaggregase [10].
These findings change our view of NIRs. They are not only involved in nuclear import. Rather, they chaperone and disaggregate cargo in the cytoplasm [10, 13]. These findings also change our view of the NLS. It is not merely a signal for nuclear localization. Rather, by recruiting NIRs the NLS is an anti-aggregation and disaggregation signal in the cytoplasm [10, 13]. Moreover, we have established that the TDP-43 cNLS contains a poly(ADP-ribose) (PAR)-binding motif, which enables PAR to stimulate TDP-43 LLPS and recruit TDP-43 to PAR-rich SGs [46]. Thus, NLSs serve multiple functions that are distinct from their canonical role in nuclear import.
In diverse cellular models, Kapβ2 expression prevents and reverses FUS accumulation in SGs and increases cell viability (Figure 4A) [10, 11]. The activity of Kapβ2 to reduce FUS association with SGs is independent of its nuclear-import activity [10, 11]. We found that FUS fibril dissolution by Kapβ2 yields soluble Kapβ2:FUS complexes, which are competent for nuclear transport (Figure 4A) [10]. Indeed, we discovered that Kapβ2 disassembled cytoplasmic FUS foci and restored FUS nuclear localization in yeast [10]. In ALS-patient fibroblasts bearing the FUSR521H mutation in the PY-NLS, FUS activity in pre-mRNA splicing is reduced [10]. However, upon increased Kapβ2 expression, FUS activity in pre-mRNA splicing is increased in these patient cell lines, indicating that Kapβ2 restores both FUS localization and function [10]. Finally, elevated Kapβ2 expression suppressed neurodegeneration and increased lifespan in a Drosophila model where FUSR521H is expressed only in motor neurons [10]. Likewise, Kapβ2 suppressed muscle degeneration elicited by MSP-linked hnRNPA2D290V [10]. Thus, we suggest that increasing NIR expression could have therapeutic utility in ALS, FTD, and MSP [10].
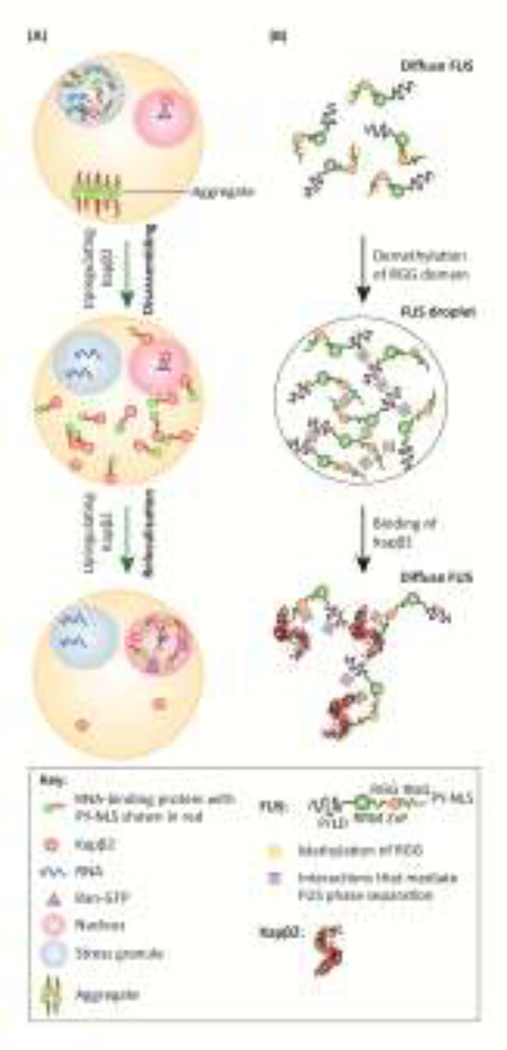
Figure 4. New NIR functions in preventing and reversing aberrant phase separation.
(A) In ALS/FTD, mislocalized nuclear RBPs with PrLDs are recruited into SGs upon stress. Prolonged stress leads to an aberrant phase transition and formation of pathological fibrils (aggregate). Upregulating Kapβ2 reverses recruitment of RBPs into SGs without disassembling the SGs. Upregulating Kapβ2 also solubilizes aggregated RBPs. Once solubilized, Kapβ2 transports RBPs back to the nucleus where Ran-GTP dissociates Kapβ2:RBP complexes enabling RBPs to perform nuclear functions and Kapβ2 to be recycled to catalyze further rounds of disaggregation.
(B) Phase separation of FUS is mediated by the intrinsically disordered PrLDs, as well as cation-π and π-π interactions between tyrosines in the PrLD and arginines in RGG domains. These interactions are modulated by post-translational arginine methylation, where methylation of RGG domains inhibits FUS LLPS. Kapβ2 inhibits FUS LLPS by binding tightly to the PY-NLS and by weakly binding to multiple regions across FUS thereby disrupting FUS-FUS interactions. Direct binding of Kapβ2 to arginines in RGG domains contributes to its chaperoning mechanism by disrupting cation-π and π-π interactions.
Independently, Qamar and colleagues showed Kapβ2 reduces phase separation and gelation of methylated and hypomethylated FUS [12]. In ex vivo Xenopus retinal neurons, FUS condensation into stable cross-β hydrogels disrupts RNP granule function at nerve terminals and impairs new protein synthesis [12]. Like FUS, Kapβ2 is a resident of these RNP granules [12]. Remarkably, modestly increasing Kapβ2 expression fluidizes the granule phase imposed by aberrant FUS gelation, thereby restoring granule function and local translation in axon terminals [12]. Thus, Kapβ2 can suppress FUS toxicity in diverse settings [10, 12]. Moreover, these findings establish that NIRs can rescue RBP toxicity by reversing aberrant phase transitions, in addition to their canonical function in nuclear import [10, 12]. Collectively, these activities enable NIRs to shape the contents and architecture of cytoplasmic membraneless organelles [10–13, 29].
How does Kapβ2 inhibit FUS LLPS? NMR analyses suggest that Kapβ2 binds to the FUS PY-NLS with high affinity, tethering the proteins together [11, 13]. Once firmly bound, Kapβ2 disrupts FUS self-association and blocks LLPS by establishing multiple weak intermolecular contacts distributed across FUS domains that mediate LLPS, including the PrLD and the RGG domains (Figure 4B) [11, 13, 69, 112, 113]. Kapβ2 also binds to RGG regions and the ZnF and RRM domains that were previously shown to facilitate FUS binding to RNA [11, 13]. Indeed, Kapβ2 competes with RNAs for FUS binding [11, 13]. Because RNA concentration can influence FUS aggregation and LLPS [114], these data suggest that Kapβ2 may also modulate LLPS of FUS by mimicking interactions with RNA. Interestingly, the phase separation of a chimeric FUS protein where the PY-NLS of FUS is replaced with a nuclear-export sequence (NES), was not inhibited by its cognate Kapβ protein, exportin-1 (CRM1/XPO1), even though the two proteins bound to each other tightly [13]. Thus, chaperone activity may be restricted to NIRs, although whether exportins regulate phase transitions of their endogenous cargo rather than a chimeric protein remains unclear. Although FUS and TDP-43 are predicted to have well-defined NESs, their export from the nucleus does not appear to depend on exportins [99, 100, 115, 116]. Notably, we found that 75% of human PrLD-containing proteins harbor a cNLS or PY-NLS [10]. Thus, NIRs likely function broadly to antagonize aberrant phase transitions of proteins with PrLDs. Moreover, even when the PY-NLS of FUS was deleted, high concentrations of Kapβ2 could disrupt FUS LLPS by making many weak contacts, indicating the chaperone function of NIRs could extend beyond NLS-containing proteins, and that NIRs could help regulate an extended catalog of membraneless organelles [13]. Indeed, Kapβ2 could employ the same weak and highly dynamic interactions that disrupt FUS self-assembly to pass through hydrogels formed by phenylalanine-glycine (FG) repeats in various nucleoporins to traverse the NPC [28, 117].
The nuclear-import activity of Kapβ2 is affected by several factors. Mutations in the PY-NLS can reduce binding affinity between Kapβ2 and cargo, disrupting nuclear import [17, 74]. We discovered that Kapβ2 chaperone and disaggregation activity is also impaired by mutations in the cargo PY-NLS [10]. Therefore, other factors affecting Kapβ2-PY-NLS binding affinity might also regulate Kapβ2 chaperone activity. One factor of great interest is Arg-methylation in RGG domains, a modification that is lost in FTD-FUS patients [118]. Indeed, hypomethylation promotes nuclear import of ALS-linked FUS variants with mutations in the PY-NLS [118]. Furthermore, Arg-methylation of FUS also suppresses FUS LLPS and SG partitioning [11, 12] (Figure 4B). Arg-methylation inhibits FUS LLPS by dismantling cation-π or π-π interactions between arginine residues in the C-terminal RGG domains and tyrosine residues in the N-terminal PrLD, which drive FUS LLPS [11, 12, 70]. Direct binding of Kapβ2 to arginine residues in the RGG domains of FUS contributes to its chaperoning mechanism by disrupting cation-π or π-π interactions [11]. Interestingly, Kapβ2 has higher affinity for hypomethylated FUS, and its disaggregation activity is not impaired when FUS is hypomethylated [11, 12]. Nonetheless, in FTD-FUS, Kapβ2 is mislocalized to cytoplasmic FUS inclusions indicating that the chaperone or disaggregation activity of Kapβ2 is somehow overwhelmed or dysregulated [119]. It will be interesting to study the effect of other cargo post-translational modifications (PTMs) on the activity of Kapβ2.
Concluding remarks
We have established that NIRs inhibit spontaneous and seeded fibrillization of cargo RBPs with PrLDs, which could prevent pathological spreading by prion-like conformers in disease [10]. Moreover, NIRs dissolve fibrils and reverse aberrant phase transitions of cargo RBPs with PrLDs, which we suggest could also be neuroprotective [10]. Indeed, we found that increasing NIR concentrations antagonized neurodegeneration and toxicity caused by cargo RBPs with PrLDs [10]. Moreover, increased expression of NIRs can buffer toxicity caused by toxic DPRs in several model systems [82–84]. Thus, small molecules that increase the expression of NIRs or enhance their chaperone or disaggregation activity could be effective therapeutics for several presently intractable neurodegenerative disorders, including ALS, FTD, and MSP. Likewise, delivery of NIRs via adeno-associated viruses (AAVs) to afflicted neurons might represent another therapeutic opportunity.
In neurodegenerative disease, NIRs likely become overwhelmed and fail to counter excessive aggregation by cargo RBPs such as TDP-43, FUS, or hnRNPA1. Indeed, expression of some NIRs appears to be reduced in the brain of FTD patients [120]. Moreover, Kapβ2 is sequestered in cytoplasmic inclusions in FTD-FUS, which may limit Kapβ2 activity [121]. PTMs may also reduce interactions between NIRs and RBP cargo, and might also contribute to reduced NIR activity in disease [11, 12, 118]. Likewise, specific ALS-linked mutations such as FUSP525L and FUSR495X reduce or eliminate Kapβ2 binding [17, 74]. Key future goals in precision medicine will focus on engineering NIRs with enhanced disaggregation activity against specific substrates, such as FUSP525L, which display reduced affinity for Kapβ2 [17]. Likewise, isolation of small-molecule drugs that increase the affinity of Kapβ2 for FUSP525L could also lead to therapeutics for severe forms of juvenile ALS caused by this mutation [60, 122].
We have now established that the NLS can function as an anti-aggregation and disaggregation signal in the cytoplasm [10]. We suggest that NIRs function broadly in preventing and reversing the aggregation of their cargo in the cytoplasm. This possibility raises the question of whether other short linear motifs engaged by transport factors in various locales may serve as signals that enable transport-factor-mediated disaggregation. Importantly, signal-recognition particles display disaggregase activity specifically against their signal-bearing clients [123–125]. Thus, it will be important to determine whether signal-dependent disaggregation activity extends to other soluble transport factors such as peroxisomal import factors, Guided Entry of Tail-anchored protein system component Get3, and nuclear-export factors. These agents may be critical in preventing and reversing aberrant phase transitions in multiple settings.
Highlights.
Nuclear-localization sequences (NLSs) function as anti-aggregation and disaggregation signals in the cytoplasm
Nuclear-import receptors (NIRs) prevent and reverse functional and pathological phase separation of NLS-bearing cargo, including RNA-binding proteins (RBPs) with prion-like domains connected to neurodegenerative disease
NIRs shape cytoplasmic membraneless organelles and regulate cellular organization beyond their canonical role in nuclear import.
Elevating NIR expression dissolves cytoplasmic RBP aggregates, restores functional RBPs to the nucleus, and rescues disease-linked RBP toxicity
NIRs could be leveraged therapeutically to restore RBP homeostasis and mitigate neurodegeneration
Outstanding Questions.
How are NIRs regulated? Kapβ2 and other NIRs can function to prevent and reverse phase separation. However, NIRs also colocalize with pathological aggregates in disease. Thus, it is possible that dysregulation of NIRs is another way by which neurodegeneration manifests. One common mechanism of regulation is altering expression levels. Are there specific transcription factors associated with NIR expression? Do NIR levels change under stress? Do NIR levels vary in specific neuronal populations? Do NIR levels explain selective neuronal vulnerability in ALS and FTD? Can we uncover small-molecule drugs that selectively increase NIR expression? Answering these questions will allow us to design more nuanced therapeutic strategies that leverage beneficial NIR functions.
What is the minimal functional unit of NIRs that confers chaperone or disaggregation activity? Given their repetitive structure, we may be able to design small peptides that replicate specific actions of the entire NIR. For example, a small series of HEAT repeats could be engineered to bind to specific domains or recognize PTMs to combat aberrant phases.
Can NIRs be evolved or engineered to recognize any aggregation-prone protein? NIRs have some degree of inherent promiscuity with respect to their binding partners. Therefore, it may be possible to apply engineering or directed evolution to uncover NIRs that can specifically disaggregate proteins such as α-synuclein, Aβ, tau, or polyglutamine.
Acknowledgements
We thank Bo Lim (Linda) Lee and Hana Odeh for comments on the manuscript. This work was funded by the Ellison Medical Foundation (LG), American Federation for Aging Research (LG), Alzheimer’s Association (LG), Life Extension Foundation (JS), ALS Association (JS), Department of Biochemistry and Biophysics Pilot Grant (JS), Target ALS (JS and LG), the Robert Packard Center for ALS Research at Johns Hopkins (JS), the Office of the Assistant Secretary of Defense for Health Affairs, through the Amyotrophic Lateral Sclerosis Research Program under award no. W81XWH-17–1-0237 (JS), and NIH grants T32GM008275 (CMF), R01GM099836 (JS), and R21NS090205 (JS).
Glossary
- Aberrant phase transitions
The transition of the material state from a liquid to a solid-like state. Aberrant phase transition leads to a less dynamic material state and the formation of protein hydrogel, aggregates, and fibrils. Aberrant phase transition is often accelerated by disease mutations in the PrLD and other intrinsically-disordered domains
- Adeno-associated virus (AAV)
a small virus that infects humans, but elicits only a very mild immune response. AAVs have been adapted for gene-therapy purposes where they can express a gene from an extrachromosomal state without integrating into the host genome. Gene delivery by AAVs is now a FDA-approved therapy for congential blindness [127]
- Disaggregase
An agent that catalytically drives the disassembly of protein aggregates
- Gel
a material network formed by crosslinks between the component macromolecules. The crosslinks can be covalent (i.e. a chemical gel) or non-covalent (i.e. a physical gel). Gels vary widely in their material properties, and are determined as a function of the extent, pattern, and longevity of crosslinking
- Membraneless organelle
A membrane-bound organelle is a cellular compartment that is delimited by a lipid bilayer. In contrast, a membraneless organelle is non-membrane-bound cellular compartment that is formed by LLPS and is typically a condensate of proteins and nucleic acid. Examples of membraneless organelles include the nucleolus in the nucleus and stress granules in the cytoplasm
- Molecular chaperones
Proteins that assist protein folding, unfolding, and the assembly or disassembly of other macromolecular structures
- Nuclear pore complex (NPC)
The nuclear pore complex regulates transport to and from the nucleus. Embedded throughout the nuclear envelope, each nuclear pore is a ~125 MDa structure with three major regions: cytoplasmic-facing filaments, a central pore, and a nuclear basket. Interestingly, NIRs interact with a subset of NPC proteins that have regions enriched in phenylalanine and glycine residues. These FG-rich nucleoporins (FG-Nups) are essential for cell viability, have no clear secondary structure, and can form gels under certain conditions. Thus, it is hypothesized that NIRs make multiple weak and transient contacts with FG-Nups, which enable them to traverse the nuclear pore
- Phase separation
Phase separation, also known as liquid-liquid phase separation or liquidliquid demixing, occurs when a single-phase fluid converts into at least two distinct, immiscible liquid phases
- Prion
Prions are infectious proteins capable of conformational replication, which occurs as the prion conformer templates the folding of other copies of the protein to the prion state
- Prion-like domain (PrLD)
A specific type of low-complexity domain enriched in uncharged polar amino acids and glycine. A PrLD resembles in amino-acid composition the prion domain that enables specific yeast proteins, such as Rnq1 and Sup35, to form prions
- Stress Granules (SGs)
SGs are reversible cytoplasmic membraneless organelles that form in response to cellular stress. SGs are dense structures composed of proteins, non-translating mRNAs, and stalled translation-initiation complexes. When stress passes, SGs disassemble
- RAN translation
Repeat-associated non-AUG (RAN) translation is a form of non-canonical translation, which is initiated by repetitive sequences in the absence of an AUG start codon. RAN translation can proceed in multiple reading frames and produces multiple homopolymeric or dipeptide repeat-containing polypeptides
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Soniat M and Chook YM (2015) Nuclear localization signals for four distinct karyopherin-beta nuclear import systems. Biochem J 468 (3), 353–62. [DOI] [PubMed] [Google Scholar]
- 2.Lee BJ et al. (2006) Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 126 (3), 543–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Twyffels L et al. (2014) Transportin-1 and Transportin-2: protein nuclear import and beyond. FEBS Lett 588 (10), 1857–68. [DOI] [PubMed] [Google Scholar]
- 4.Harrison AF and Shorter J (2017) RNA-binding proteins with prion-like domains in health and disease. Biochem J 474 (8), 1417–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King OD et al. (2012) The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res 1462, 61–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.March ZM et al. (2016) Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res 1647, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin Y and Brangwynne CP (2017) Liquid phase condensation in cell physiology and disease. Science 357 (6357). [DOI] [PubMed] [Google Scholar]
- 8.Gomes E and Shorter J (2018) The molecular language of membraneless organelles. J BiolChem [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boeynaems S et al. (2018) Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol 28 (6), 420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo L et al. (2018) Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains. Cell 173 (3), 677–692 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofweber M et al. (2018) Phase Separation of FUS Is Suppressed by Its Nuclear Import Receptor and Arginine Methylation. Cell 173 (3), 706–719 e13. [DOI] [PubMed] [Google Scholar]
- 12.Qamar S et al. (2018) FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-pi Interactions. Cell 173 (3), 720–734 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshizawa T et al. (2018) Nuclear Import Receptor Inhibits Phase Separation of FUS through Binding to Multiple Sites. Cell 173 (3), 693–705 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook A et al. (2007) Structural biology of nucleocytoplasmic transport. Annu Rev Biochem 76, 647–71. [DOI] [PubMed] [Google Scholar]
- 15.Kim YH et al. (2017) The molecular mechanism for nuclear transport and its application. Anat Cell Biol 50 (2), 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chook YM and Blobel G (1999) Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature 399 (6733), 230–7. [DOI] [PubMed] [Google Scholar]
- 17.Zhang ZC and Chook YM (2012) Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the Fused in Sarcoma protein (FUS). Proc Natl Acad Sci U S A 109 (30), 12017–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chook YM et al. (2002) Uncoupling Kapbeta2 substrate dissociation and ran binding. Biochemistry 41 (22), 6955–66. [DOI] [PubMed] [Google Scholar]
- 19.Gorlich D et al. (1996) Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J 15 (20), 5584–94. [PMC free article] [PubMed] [Google Scholar]
- 20.Kosugi S et al. (2009) Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J Biol Chem 284 (1), 478–85. [DOI] [PubMed] [Google Scholar]
- 21.Gorlich D et al. (1996) A 41 amino acid motif in importin-alpha confers binding toimportin-beta and hence transit into the nucleus. EMBO J 15 (8), 1810–7. [PMC free article] [PubMed] [Google Scholar]
- 22.Weis K et al. (1996) The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO J 15 (8), 1818–25. [PMC free article] [PubMed] [Google Scholar]
- 23.Kobe B (1999) Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat Struct Biol 6 (4), 388–97. [DOI] [PubMed] [Google Scholar]
- 24.Gilchrist D and Rexach M (2003) Molecular basis for the rapid dissociation of nuclear localization signals from karyopherin alpha in the nucleoplasm. J Biol Chem 278 (51), 51937–49. [DOI] [PubMed] [Google Scholar]
- 25.Rexach M and Blobel G (1995) Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83 (5), 683–92. [DOI] [PubMed] [Google Scholar]
- 26.Freitas N and Cunha C (2009) Mechanisms and signals for the nuclear import of proteins. Curr Genomics 10 (8), 550–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lange A et al. (2007) Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem 282 (8), 5101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt HB and Gorlich D (2016) Transport Selectivity of Nuclear Pores, Phase Separation, and Membraneless Organelles. Trends Biochem Sci 41 (1), 46–61. [DOI] [PubMed] [Google Scholar]
- 29.Mikhaleva S and Lemke EA (2018) Beyond the Transport Function of Import Receptors: What’s All the FUS about? Cell 173 (3), 549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soniat M and Chook YM (2016) Karyopherin-beta2 Recognition of a PY-NLS Variant that Lacks the Proline-Tyrosine Motif. Structure 24 (10), 1802–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keller BA et al. (2012) Co-aggregation of RNA binding proteins in ALS spinal motor neurons: evidence of a common pathogenic mechanism. Acta Neuropathol 124 (5), 733–47. [DOI] [PubMed] [Google Scholar]
- 32.Kim HJ et al. (2013) Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495 (7442), 467–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Y et al. (2015) Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell 60 (2), 208–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mateju D et al. (2017) An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J 36 (12), 1669–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molliex A et al. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163 (1), 123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saha S and Hyman AA (2017) RNA gets in phase. J Cell Biol 216 (8), 2235–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shorter J and Taylor JP (2013) Disease mutations in the prion-like domains of hnRNPA1 and hnRNPA2/B1 introduce potent steric zippers that drive excess RNP granule assembly. Rare Dis 1, e25200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blokhuis AM et al. (2013) Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol 125 (6), 777–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guttinger S et al. (2004) Transportin2 functions as importin and mediates nuclear import of HuR. Proc Natl Acad Sci U S A 101 (9), 2918–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Putker M et al. (2013) Redox-dependent control of FOXO/DAF-16 by transportin-1. Mol Cell 49 (4), 730–42. [DOI] [PubMed] [Google Scholar]
- 41.Rebane A et al. (2004) Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA 10 (4), 590–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchan JR and Parker R (2009) Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36 (6), 932–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li YR et al. (2013) Stress granules as crucibles of ALS pathogenesis. J Cell Biol 201 (3), 361–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Protter DS and Parker R (2016) Principles and Properties of Stress Granules. Trends Cell Biol 26 (9), 668–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang K et al. (2018) Stress Granule Assembly Disrupts Nucleocytoplasmic Transport. Cell 173 (4), 958–971 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGurk L et al. (2018) Poly(ADP-Ribose) Prevents Pathological Phase Separation of TDP-43 by Promoting Liquid Demixing and Stress Granule Localization. Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sahoo PK et al. (2018) Axonal G3BP1 stress granule protein limits axonal mRNA translation and nerve regeneration. Nature Communications 9 (1), 3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riback JA et al. (2017) Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell 168 (6), 1028–1040 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boeynaems S et al. (2017) Phase Separation of C9orf72 Dipeptide Repeats Perturbs Stress Granule Dynamics. Mol Cell 65 (6), 1044–1055 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen L and Liu B (2017) Relationships between Stress Granules, Oxidative Stress, and Neurodegenerative Diseases. Oxid Med Cell Longev 2017, 1809592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naruse H et al. (2018) Molecular epidemiological study of familial amyotrophic lateral sclerosis in Japanese population by whole-exome sequencing and identification of novel HNRNPA1 mutation. Neurobiol Aging 61, 255 e9–255 e16. [DOI] [PubMed] [Google Scholar]
- 52.Prpar Mihevc S et al. (2017) Nuclear trafficking in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Brain 140 (1), 13–26. [DOI] [PubMed] [Google Scholar]
- 53.Shorter J (2017) Liquidizing FUS via prion-like domain phosphorylation. EMBO J 36 (20), 2925–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolozin B (2012) Regulated protein aggregation: stress granules and neurodegeneration. Mol Neurodegener 7, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchan JR et al. (2013) Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153 (7), 1461–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee Y et al. (2018) TIA1 variant drives myodegeneration in multisystem proteinopathy with SQSTM1 mutations. J Clin Invest 128 (3), 1164–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mackenzie IR et al. (2017) TIA1 Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Promote Phase Separation and Alter Stress Granule Dynamics. Neuron 95 (4), 808–816 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kapeli K et al. (2017) Genetic mutations in RNA-binding proteins and their roles in ALS. Hum Genet 136 (9), 1193–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pak CW et al. (2016) Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein. Mol Cell 63 (1), 72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conte A et al. (2012) P525L FUS mutation is consistently associated with a severe form of juvenile amyotrophic lateral sclerosis. Neuromuscul Disord 22 (1), 73–5. [DOI] [PubMed] [Google Scholar]
- 61.Couthouis J et al. (2012) Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet 21 (13), 2899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Couthouis J et al. (2011) A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci U S A 108 (52), 20881–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor JP (2015) Multisystem proteinopathy: intersecting genetics in muscle, bone, and brain degeneration. Neurology 85 (8), 658–60. [DOI] [PubMed] [Google Scholar]
- 64.Renton AE et al. (2014) State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci 17 (1), 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Couthouis J et al. (2014) Targeted exon capture and sequencing in sporadic amyotrophic lateral sclerosis. PLoS Genet 10 (10), e1004704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson BS et al. (2009) TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem 284 (30), 20329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun Z et al. (2011) Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol 9 (4), e1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kato M et al. (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149 (4), 753–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J et al. (2018) A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 174 (3), 688–699 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bogaert E et al. (2018) Molecular Dissection of FUS Points at Synergistic Effect of Low-Complexity Domains in Toxicity. Cell Reports 24 (3), 529–537.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patel A et al. (2015) A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162 (5), 1066–77. [DOI] [PubMed] [Google Scholar]
- 72.Murray DT et al. (2017) Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains. Cell 171 (3), 615–627 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Murakami T et al. (2015) ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron 88 (4), 678–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dormann D et al. (2010) ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J 29 (16), 2841–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Q et al. (2016) Whole-exome sequencing identifies a missense mutation in hnRNPA1 in a family with flail arm ALS. Neurology 87 (17), 1763–1769. [DOI] [PubMed] [Google Scholar]
- 76.Kim HJ and Taylor JP (2017) Lost in Transportation: Nucleocytoplasmic Transport Defects in ALS and Other Neurodegenerative Diseases. Neuron 96 (2), 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jovicic A et al. (2016) Nuclear transport dysfunction: a common theme in amyotrophic lateral sclerosis and frontotemporal dementia. J Neurochem 138 Suppl 1, 134–44. [DOI] [PubMed] [Google Scholar]
- 78.Neumann M et al. (2009) A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain 132 (Pt 11), 2922–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mackenzie IR and Neumann M (2012) FET proteins in frontotemporal dementia and amyotrophic lateral sclerosis. Brain Res 1462, 40–3. [DOI] [PubMed] [Google Scholar]
- 80.Neumann M et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314 (5796), 130–3. [DOI] [PubMed] [Google Scholar]
- 81.Chou CC et al. (2018) TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat Neurosci 21 (2), 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Freibaum BD et al. (2015) GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525 (7567), 129–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jovicic A et al. (2015) Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18 (9), 1226–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang K et al. (2015) The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525 (7567), 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boeynaems S et al. (2016) Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci Rep 6, 20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boeynaems S et al. (2016) Inside out: the role of nucleocytoplasmic transport in ALS and FTLD. Acta Neuropathol 132 (2), 159–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.DeJesus-Hernandez M et al. (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72 (2), 245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lagier-Tourenne C et al. (2013) Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A 110 (47), E4530–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zu T et al. (2011) Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A 108 (1), 260–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gitler AD and Tsuiji H (2016) There has been an awakening: Emerging mechanisms of C9orf72 mutations in FTD/ALS. Brain Res 1647, 19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee HG et al. (2006) Aberrant localization of importin alpha1 in hippocampal neurons in Alzheimer disease. Brain Res 1124 (1), 1–4. [DOI] [PubMed] [Google Scholar]
- 92.Sheffield LG et al. (2006) Nuclear pore complex proteins in Alzheimer disease. J Neuropathol Exp Neurol 65 (1), 45–54. [DOI] [PubMed] [Google Scholar]
- 93.Gasset-Rosa F et al. (2017) Polyglutamine-Expanded Huntingtin Exacerbates Age-Related Disruption of Nuclear Integrity and Nucleocytoplasmic Transport. Neuron 94 (1), 48–57 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grima JC et al. (2017) Mutant Huntingtin Disrupts the Nuclear Pore Complex. Neuron 94 (1), 93–107 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu T et al. (2004) Gene regulation and DNA damage in the ageing human brain. Nature 429 (6994), 883–91. [DOI] [PubMed] [Google Scholar]
- 96.Mertens J et al. (2015) Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell 17 (6), 705–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pujol G et al. (2002) Age-associated reduction of nuclear protein import in human fibroblasts. Biochem Biophys Res Commun 294 (2), 354–8. [DOI] [PubMed] [Google Scholar]
- 98.Woerner AC et al. (2016) Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science 351 (6269), 173–6. [DOI] [PubMed] [Google Scholar]
- 99.Ederle H et al. (2018) Nuclear egress of TDP-43 and FUS occurs independently of Exportin-1/CRM1. Sci Rep 8 (1), 7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pinarbasi ES et al. (2018) Active nuclear import and passive nuclear export are the primary determinants of TDP-43 localization. Sci Rep 8 (1), 7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shi KY et al. (2017) Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc Natl Acad Sci U S A 114 (7), E1111–E1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shorter J (2016) Engineering therapeutic protein disaggregases. Mol Biol Cell 27 (10), 1556–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shorter J (2017) Designer protein disaggregases to counter neurodegenerative disease. Curr Opin Genet Dev 44, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jackrel ME et al. (2014) Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell 156 (1–2), 170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jackel S et al. (2015) Nuclear import factor transportin and arginine methyltransferase 1 modify FUS neurotoxicity in Drosophila. Neurobiol Dis 74, 76–88. [DOI] [PubMed] [Google Scholar]
- 106.Milles S et al. (2013) Facilitated aggregation of FG nucleoporins under molecular crowding conditions. EMBO Rep 14 (2), 178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Odeh HM et al. (2018) The SUMO-specific isopeptidase SENP2 is targeted to intracellular membranes via a predicted N-terminal amphipathic alpha-helix. Mol Biol Cell 29 (15), 1878–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sousa R et al. (2016) Clathrin-coat disassembly illuminates the mechanisms of Hsp70 force generation. Nat Struct Mol Biol 23 (9), 821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.De Los Rios P and Goloubinoff P (2016) Hsp70 chaperones use ATP to remodel native protein oligomers and stable aggregates by entropic pulling. Nat Struct Mol Biol 23 (9), 766–9. [DOI] [PubMed] [Google Scholar]
- 110.Xiang S et al. (2015) The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei. Cell 163 (4), 829–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Murray DT et al. (2018) Structural characterization of the D290V mutation site in hnRNPA2 low-complexity-domain polymers. Proc Natl Acad Sci U S A 115 (42), E9782–E9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Monahan Z et al. (2017) Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J 36 (20), 2951–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Burke KA et al. (2015) Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol Cell 60 (2), 231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Maharana S et al. (2018) RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360 (6391), 918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hock EM et al. (2018) Hypertonic Stress Causes Cytoplasmic Translocation of Neuronal, but Not Astrocytic, FUS due to Impaired Transportin Function. Cell Rep 24 (4), 987–1000 e7. [DOI] [PubMed] [Google Scholar]
- 116.Archbold HC et al. (2018) TDP43 nuclear export and neurodegeneration in models of amyotrophic lateral sclerosis and frontotemporal dementia. Sci Rep 8 (1), 4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hough LE et al. (2015) The molecular mechanism of nuclear transport revealed by atomic-scale measurements. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dormann D et al. (2012) Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J 31 (22), 4258–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Neumann M et al. (2012) Transportin 1 accumulates specifically with FET proteins but no other transportin cargos in FTLD-FUS and is absent in FUS inclusions in ALS with FUS mutations. Acta Neuropathol 124 (5), 705–16. [DOI] [PubMed] [Google Scholar]
- 120.Nishimura AL et al. (2010) Nuclear import impairment causes cytoplasmic transactivation response DNA-binding protein accumulation and is associated with frontotemporal lobar degeneration. Brain 133 (Pt 6), 1763–71. [DOI] [PubMed] [Google Scholar]
- 121.Troakes C et al. (2013) Transportin 1 colocalization with FUS inclusions is not characteristic for ALS-FUS confirming disrupted nuclear import of mutant FUS and distinguishing it from FTLD with FUS inclusions. Neuropathol Appl Neurobiol 39 (5), 553–61. [DOI] [PubMed] [Google Scholar]
- 122.Zou ZY et al. (2013) De novo FUS gene mutations are associated with juvenile-onset sporadic amyotrophic lateral sclerosis in China. Neurobiol Aging 34 (4), 1312 e1–8. [DOI] [PubMed] [Google Scholar]
- 123.Jaru-Ampornpan P et al. (2013) Mechanism of an ATP-independent protein disaggregase: II. distinct molecular interactions drive multiple steps during aggregate disassembly. J Biol Chem 288 (19), 13431–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jaru-Ampornpan P et al. (2010) ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP subunit. Nat Struct Mol Biol 17 (6), 696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nguyen TX et al. (2013) Mechanism of an ATP-independent protein disaggregase: I. structure of a membrane protein aggregate reveals a mechanism of recognition by its chaperone. J Biol Chem 288 (19), 13420–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Izaurralde E et al. (1997) The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J 16 (21), 6535–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Apte RS (2018) Gene Therapy for Retinal Degeneration. Cell 173 (1), 5. [DOI] [PubMed] [Google Scholar]