Abstract
Although integrin α9 (ITGA9) is known to be involved in cell adhesion and motility, its expression in cancer and its role in tumor growth and metastasis remain largely unknown. This study was designed to investigate the role of ITGA9 in triple negative breast cancer (TNBC). ITGA9 expression in TNBC cells were knocked out (KO) using CRISPR/Cas9 technology. Four orthotopic mouse mammary xenograft tumor models coupled with cell culture studies were performed to determine the effect of ITGA9 depletion on TNBC tumor growth and metastasis and the underlying mechanism. Bioinformatics analysis showed that ITGA9 level is significantly higher in TNBC than other breast cancer subtypes; and higher ITGA9 level is associated with significantly worse distant metastasis free survival and recurrence free survival in TNBC patients. Experimentally, ITGA9 KO significantly reduced TNBC cell cancer stem cell (CSC)-like property, tumor angiogenesis, tumor growth and metastasis by promoting β-catenin degradation. Further mechanistic studies revealed that ITGA9 KO causes integrin-linked kinase (ILK) relocation from the membrane region to the cytoplasm, where it interacts with protein kinase A (PKA) and inhibits PKA activity leading to increased activity of glycogen synthase kinase 3 (GSK3) and subsequent β-catenin degradation. Overexpressing β-catenin in ITGA9 KO cells reversed the inhibitory effect of ITGA9 KO on tumor growth and metastasis. Furthermore, ITGA9 down-regulation in TNBC tumors by nanoparticle-mediated delivery of ITGA9 siRNA drastically decreased tumor angiogenesis, tumor growth and metastasis. These findings indicate that ITGA9 depletion suppresses TNBC tumor growth and metastasis by promoting β-catenin degradation through the ILK/PKA/GSK3 pathway.
Keywords: integrin α9 (ITGA9), triple negative breast cancer (TNBC), integrin-linked kinase (ILK), protein kinase A (PKA), β-catenin, metastasis
Introduction
Integrins are a large family of heterodimeric transmembrane receptors and each integrin consists of an α and β subunit. Integrins are key mediators of cell adhesion and most members of the integrin family are found to play critical roles in cell migration, invasion, and tumor metastasis.1–3 Human integrin subunit α9 (ITGA9), known to partner with the subunit β1 to form integrin α9β1, is one of the least studied integrins.4,5 While studies showed that ITGA9 has important roles in regulating cell adhesion, motility, lymphangenesis and angiogenesis, its expression pattern in cancer and its role in tumor growth and metastasis remain largely unknown.4
Triple negative breast cancer (TNBC) is a breast cancer subtype that does not express estrogen and progesterone receptor and lacks the amplification of human epidermal growth factor receptor 2, accounting for 10–20% of newly diagnosed breast cancer cases.6–8 TNBC overlaps largely with basal-like breast cancer that is one of breast cancer subtypes classified by microarray-based gene expression profiling studies.9,10 Clinically, TNBCs usually show worse metastasis and poorer patient prognosis than other breast cancer subtypes.11–13 It was reported that the worst survival among all breast cancer subtypes having metastatic disease was seen in TNBC with metastasis cases with a median survival of about 7 months.11,12 However, the mechanism of TNBC displaying more aggressive metastatic behavior is still poorly understood. A previous study of 141 invasive breast carcinomas and 10 normal breast tissues showed strong associations between the expression of integrin α9β1 and the expression of basal cytokeratins, the triple negative status of breast carcinoma and poorer patient survival,14 suggesting a potentially important role of integrin α9β1 in TNBC. Nonetheless, whether ITGA9 plays a role in TNBC metastasis and the underlying mechanism are currently unknown.
β-Catenin is a critical signaling molecule in the canonical Wnt/β-Catenin pathway, which plays key roles in normal development and cancer.15 Importantly, the Wnt/β-catenin pathway is critically involved in the generation and maintenance of cancer stem cells (CSCs) that play pivotal roles in cancer initiation and metastasis.16,17 Interestingly, studies showed that the Wnt/β-catenin pathway is activated and enriched in TNBC.18–20 However, how the Wnt/β-catenin pathway is activated and enriched in TNBC is still poorly understood and it remains to be determined whether ITGA9 plays a role in regulating the β-catenin pathway in TNBC.
In this study, we found that ITGA9 expression level is significantly higher in TNBC cells and tumors; and higher ITGA9 expression level is associated with significantly worse distant metastasis free survival (DMFS) and recurrence free survival (RFS) in TNBC patients. ITGA9 depletion in TNBC cells or tumor tissues significantly reduced TNBC cell stemness, tumor angiogenesis, tumor growth and metastasis by promoting β-catenin degradation. Further mechanistic studies revealed that ITGA9 depletion promote β-catenin degradation through the ILK/PKA/GSK3 pathway.
Materials and Methods
Cell lines and cell culture
MCF-7, T-47D, BT-474, MDA-MB-453, Hs578T and BT-549 cells were purchased from ATCC (Manassas, VA). MDA-MB-231 cells were obtained from Dr. Suyun Huang (MD Anderson Cancer). MDA-MB-231-LM2 (LM2) cells were provided by Dr. Joan Massagué Lab (Memorial Sloan-Kettering Cancer Center). The LM2 cells, a derivative of MDA-MB-231 cells, were selected for its strong ability to metastasize to the lung.21 All above cells were cultured following instructions from ATCC. SUM-159 cells were obtained from Dr. Stephen Ethier (Wayne State University) and cultured in F-12 supplemented with 5% FBS, 1% penicillin/streptomycin at 37 °C in a humidified 5% CO2 atmosphere.
Generation of ITGA9 CRISPR/Cas9 knockout (KO) cells
ITGA9 CRISPR/Cas9 KO TNBC LM2 and SUM-159 cells were generated following the procedures described in our recent study.22 Briefly, ITGA9-specific gRNA oligos were cloned into the pX459 two-in-one CRISPR targeting vector (Addgene, Cambridge, MA). Two target sequences for ITGA9 KO are GGTGCTGGCGCTGGTGGTCGCGG and CCTCGACCCGCAGCGCCCCGTGC. After successful cloning, the ITGA9 CRISPR/Cas9 targeting vector was transfected into LM2 and SUM-159 cells and selected with puromycin for generating ITGA KO cells, which was confirmed by DNA sequencing and Western blot analysis. Two confirmed ITGA9 KO clones from each cell line were pooled together and used in this study.
ITGA9, β-catenin and PKA catalytic subunit α (PKA-Cα) stable overexpression in ITGA9 KO LM2 and SUM-159 cells
Full length human ITGA9, β-catenin and PKA-Cα cDNAs were obtained from OriGene Technologies (Rockville, MD) and Addgene and cloned into lentiviral expression vector pLenti6.3⁄V5-DEST™ (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. Vector control, ITGA9-, β-catenin-, and PKA-Cα-expressing lentiviral particles were packaged as described in our recent publications.22,23 To stably overexpress ITGA9, β-catenin or PKA-Cα in ITGA9 KO cells, ITGA9 KO LM2 and SUM-159 cells were transduced with vector control (pLenti6.3), ITGA9-expressing (pLenti6.3-ITGA9), β-catenin-expressing (pLenti6.3-β-catenin) or PKA-Cα-expressing (pLenti6.3-PKA-Cα) lentiviral particles and selected with blasticidin.
Orthotopic mouse mammary xenograft tumor model studies and IVIS bioluminescent imaging analysis
All following animal study protocols were reviewed and approved by the University of Kentucky IACUC. Seven-week-old female nude mice (Nu/Nu; Charles River Laboratories, Wilmington, MA) were used and the orthotopic mouse mammary xenograft tumors were established as described in our recent studies.22,24 Four orthotopic mammary xenograft tumor model studies were carried out: (i) To study the effect of ITGA9 KO on mammary tumor growth and metastasis, mouse mammary fat pad was injected with 1 × 106 of LM2 parental cells (n=6 mice) or LM2 ITGA9 KO cells (n=5 mice). Mice injected with LM2 parent cells or LM2 ITGA9 KO cells were euthanized 6 and 10 weeks after the injection, respectively. (ii) To study the rescue effect of ITGA9 re-expressing in ITGA9 KO cells on the inhibitory effect of ITGA9 KO on mammary tumor growth and metastasis, mouse mammary fat pad was injected with 1 × 106 of LM2 ITGA9 KO vector control (LM2-ITGA9KO-pLenti6.3) (n=5 mice) or LM2 ITGA9 KO with ITGA9 re-expressing (LM2-ITGA9KO-pLenti6.3-ITGA9) (n=5 mice) cells. All mice were euthanized 7 weeks after the injection. (iii) To study the effect of siRNA knocking down IGTA9 expression in mouse mammary xenograft tumor tissues on tumor growth and metastasis, 1 × 106 of parental LM2 cells were injected into mouse mammary fat pad to produce orthotopic mammary xenograft tumors. When mammary tumor volumes reached about 200 mm3 two weeks after injection, tumor bearing mice were randomly divided into two groups (n=5 mice in each group) and the average tumor volumes between two groups were not significantly different. One group of mice were treated with control siRNA oligoes and the other group of mice were treated with ITGA9 siRNA oligoes. Both control and ITGA9 ON-TARGETplus SMARTpool siRNA oligoes (a mixture of 4 different siRNAs provided as a single reagent) were purchased from Dharmacon (Lafayette, CO), packaged into hybrid nanoparticles (HNP) that were developed recently in our laboratory,25 and given to mice via intratumoral injection at a dose of 12.5 μg/mouse, three times per week for three weeks. All mice were euthanized one week after the last intratumoral injection. (iv) To study the effect of β-catenin overexpression in ITGA9 KO cells on the inhibitory effect of ITGA9 KO on mammary tumor growth and metastasis, mouse mammary fat pad was injected with 1 × 106 of β-catenin overexpressing LM2 ITGA9 KO cells (LM2-ITGA9KO-pLenti6.3-β-catenin) (n=5 mice). Mice were euthanized 6 weeks after injection. All mammary tumors were collected and fixed in 10% formalin solution for H&E and vascular marker CD31 immunofluorescence (IF) staining as described in our recent studies.22,26 LM2 cells also stably express the luciferase 2 reporter, so the lungs of mice injected with LM2 cells were collected and imaged with the IVIS bioluminescent imaging system to determine lung metastasis as described in our recent studies.22
Suspension serum-free culture sphere formation assay
The suspension serum-free culture sphere formation assay was carried out to determine the presence of cancer stem cell (CSC)-like cells as previously described.27,28 Briefly, cells were plated into the ultra-low adhesion 24-well cell culture plates (2500 cells per well) and cultured for 10 days for sphere formation. At the end of culture, spheres with a diameter bigger than 100 μm were photographed and counted.
Flow cytometry analysis of CD44+/CD24- cells
The parental LM2 and SUM-159 cells and the ITGA9 KO LM2 and SUM-159 cells (1 × 106 cells) were harvested, washed with PBS and stained with anti-CD44-PE (Thermo Fisher # 12–0441-82) and/or anti-CD24-APC (Thermo Fisher # 17–0247-42) on ice for 30 minutes following the manufacturer’s instructions. At the end of antibody staining, cells were washed and stained with DAPI and processed for BD FACS (LSR II) analysis. The results were analyzed using BD FlowJo software (BD Biosciences).
Quantitative PCR (Q-PCR) analysis
The TRIzol Reagent was used to extract cellular total RNA. Gene mRNA expression levels were measured by Q-PCR analysis carried out in ABI QuantStudio 3 Q-PCR System using ABI TaqMan gene expression assay and β-actin was used to normalize gene mRNA expression levels as described in our previous study.23
Western blot, cell fractionation and Co-immunoprecipitation (Co-IP) analysis
Western blot, cell fractionation and Co-IP analysis were carried out as described in our previous publications.23,26,29 The following primary antibodies were used: anti-ITGA9, anti-CD31 (Abcam, Cambridge, MA); anti-β-catenin, anti-non-phospho-β-catenin, phospho-β-catenin (Ser33/37/Thr41, Thr41/Ser45, Ser675), phospho-GSK3α (Ser21), phospho-GSK3β (Ser9), total GSK3α, total GSK3β, phosphor-CREB (Ser133), total CREB, integrin-linked kinase (ILK), PKA-Cα, and PARP-1 (Cell Signaling Technology, Beverly, MA); anti-PKA RIIα regulatory unit (Santa Cruz Biotechnology, Dallas, TX); anti-β-actin (Sigma, St. Louis, MO).
Immunofluorescence (IF) staining of cultured cells and mouse tumor tissue sections
Cellular β-catenin, ILK, and PKA RIIα regulatory subunit IF staining were performed following our previous protocols.22,26 Mouse mammary tumor section β-catenin and CD31 staining was performed as described in our previous studies.26,30 The CD31 staining was quantified by counting CD31 positive staining structures in 10 randomly-selected fields (magnification: × 200) from each mouse mammary tumor section and sections from three mouse tumors in each group were stained and counted; and presented as the number of CD31 positive-stained structures per field of view (FOV). Mouse tumor tissue β-catenin IF images were taken using a confocal microscope (Nikon Ti2). The presented images for cultured cells and mouse tumor tissues are overlaid images of a specific protein positive staining with nuclear DNA 4’6-diamidino-2-phenylindole (DAPI) staining.
Statistical analysis
The statistical analyses for the significance in the presented numerical data (as mean ± SD) were performed by testing for different treatment effects by one-way analysis of variance (ANOVA). The differences between treatment groups were determined using two-tailed student t-test. A p value of <0.05 was considered statistically significant.
Results
ITGA9 expression level is significantly higher in basal-like breast tumors and basal mesenchymal-like TNBC cells than other subtype breast cancers and higher ITGA9 expression level is associated with significantly worse DMFS and RFS in TNBC patients
The expression level of ITGA9 among breast cancer subtypes is not well-known. To determine the potential role of ITGA9 in breast cancer, we first compared ITGA9 expression levels among different subtypes of breast cancer by performing bioinformatics analysis of 942 breast cancer patient gene expression data from TCGA PanCancer Atlas Breast Invasive Carcinoma dataset (http://www.cbioportal.org/index.do). As shown in Fig. 1a, the expression level of ITGA9 is significantly higher in basal-like breast cancers than other breast cancer subtypes. Western blot analysis showed that ITGA9 protein level is significantly higher in strongly migratory and invasive basal mesenchymal-like TNBC cells than other breast cancer cells (Fig. 1b). Since basal-like breast cancer overlaps largely with TNBC, these findings suggest that ITGA9 level is significantly higher in TNBC tumor tissues and cell lines.
Fig. 1.
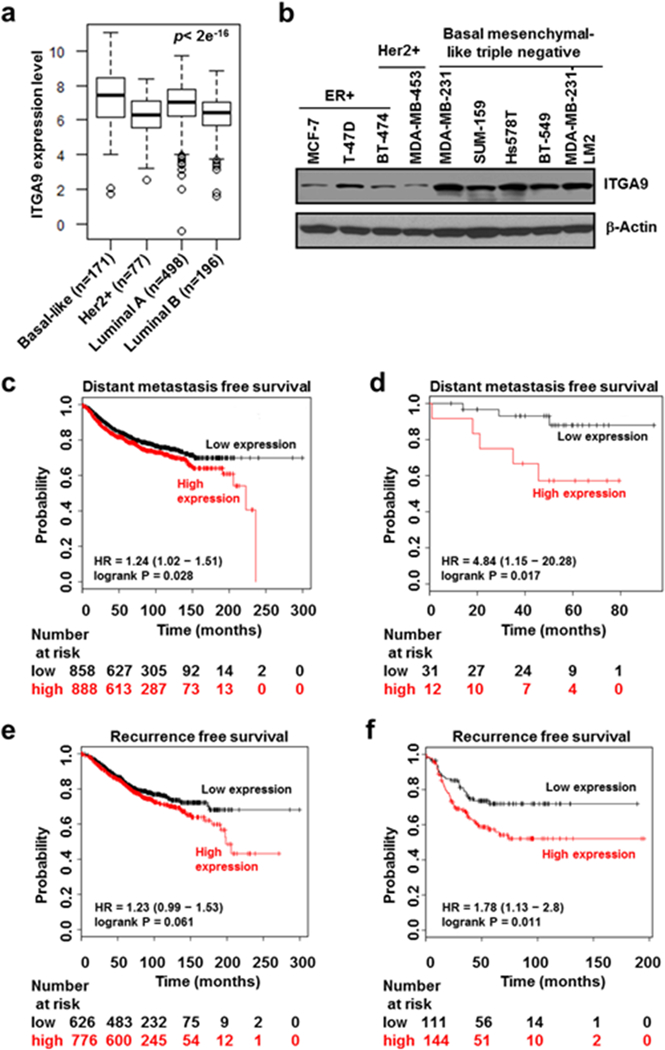
ITGA9 expression level is significantly higher in TNBC than other breast cancer subtypes and high ITGA9 expression level is associated with significantly worse DMFS and RFS in TNBC patients. a ITGA9 expression level analysis among different breast cancer subtypes. The patient gene expression data was retrieved from TCGA PanCancer Atlas Breast Invasive Carcinoma dataset (http://www.cbioportal.org/index.do) and log2 transformed. The differences among groups were determined by one way ANOVA. b Representative images of Western blot analysis of ITGA9 protein level among different type of breast cancer cells. c, d DMFS analysis. The KM plotter (http://kmplot.com/analysis/) was used to analyze the relationship between ITGA9 expression level and DMFS for all breast cancer subtypes (c) or TNBC subtype (d). e, f RFS analysis. The KM plotter was used to analyze the relationship between ITGA9 expression level and RFS for all breast cancer subtypes (e) or TNBC subtype (f).
Moreover, by analyzing a large number of breast cancer patients’ ITGA9 expression and survival data from KM plotter (http://kmplot.com), we found that a higher ITGA9 expression level is associated with significantly worse distant metastasis free survival (DMFS) in patients (Fig. 1c). Importantly, this reverse association is only significant in the TNBC subtype group after stratifying different breast cancer subtypes including ER+, PR+, Her2+ and triple negative (Fig. 1d). Similarly, a higher ITGA9 expression level is also associated with worse recurrence free survival (RFS) in breast cancer patients (p=0.061) (Fig. 1e); and this reverse association became statistically significant (p=0.011) after stratifying the TNBC subtype (Fig. 1f). These findings suggest that higher ITGA9 expression may contribute significantly to breast cancer metastasis and recurrence, particularly to TNBC metastasis and recurrence.
ITGA9 depletion by CRISPR/Cas9 knockout (KO) suppresses TNBC cell cancer stem cell (CSC)-like property, tumor angiogenesis, tumor growth and metastasis
We next used CRISPR/Cas9 technology to knock out ITGA9 expression in two TNBC cell lines (LM2 and SUM-159) for determining the role of ITGA9 in TNBC. ITGA9 depletion in LM2 and SUM-159 cells was confirmed by Western blot (Fig. 2a). To ensure that the effects observed below in ITGA9 KO cells were indeed caused by ITGA9 down-regulation, we also stably re-expressed ITGA9 in ITGA9 KO cells for performing rescue experiments (Fig. 2a).
Fig. 2.
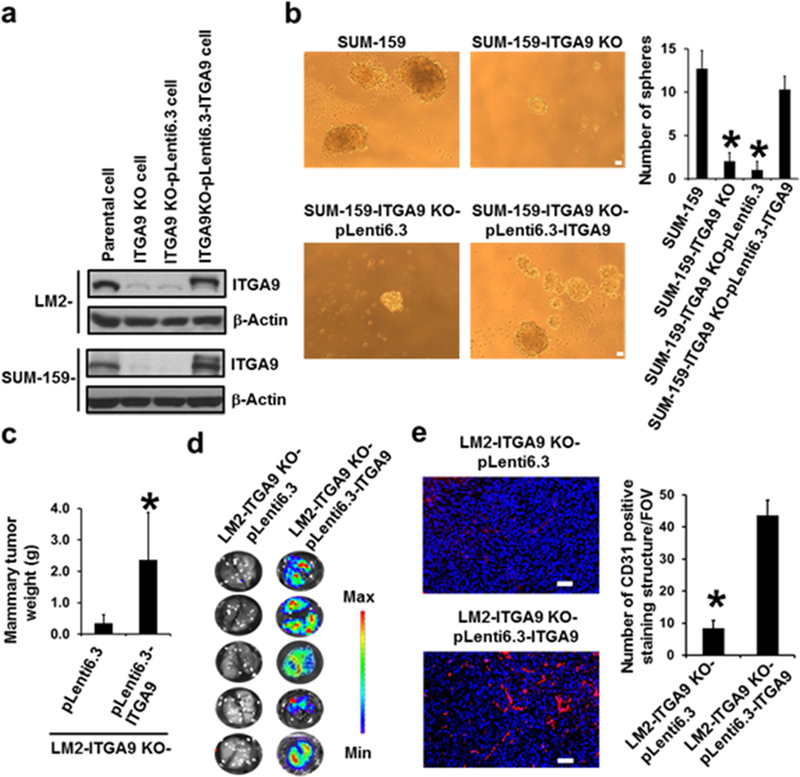
ITGA9 KO by CRISPR/Cas9 significantly reduces TNBC cell CSC-like property, tumor angiogenesis, tumor growth and metastasis. a Representative images of Western blot analysis of ITGA9 level in parental, ITGA9 KO and ITGA9 re-expressing TNBC cells. b Representative images of suspension mammary spheres and quantifications. The numbers of spheres formed in each group were counted and presented as mean ± SD (n=3). *p< 0.05, compared to parental SUM-159 cells or ITGA9 re-expressing ITGA9 KO cells. Scale bar, 50 μm. c Mouse mammary xenograft tumor weight (mean ± SD, n=5). d Images of mouse lung ex vivo IVIS bioluminescence imaging analysis. e Representative overlaid images of IF staining of CD31 (red) and nuclear DNA DAPI (blue) and quantifications of CD31 positive staining in mouse mammary xenograft tumors. The CD31 positive staining were counted and presented as numbers of CD31 positive staining structures per field of view (FOV) (mean ± SD, n=30). *p<0.05. Scale bar, 50 μm.
CSCs play key roles in cancer metastasis and recurrence.31,32 We thus first determined the effect of ITGA9 KO on TNBC cell CSC-like property using a well-established serum-free suspension culture mammary sphere formation assay. It was found that ITGA9 KO significantly reduces the number of mammary spheres formed by the TNBC SUM-159 cells (Fig. 2b). Stably re-expressing ITGA9 in ITGA9 KO cells significantly recovered the capability of ITGA9 KO cells forming mammary spheres (Fig. 2b). In addition, we also performed flow cytometry analysis to determine the effect of ITGA9 KO on the number of cells with CD44+/CD24-, a mark used for identifying breast CSC. It was found that ITGA9 KO significantly reduces the number of cells with CD44+/CD24- in both LM2 and SUM-159 cells (Fig. S1a-b). Together, these findings suggest that ITGA9 KO is capable of reducing TNBC cell CSC-like property.
Our recent studies showed that nude mouse mammary fat pad injection of LM2 cells produced rapidly growing and strongly metastatic mammary tumors.22,28 To determine the effect of ITGA9 KO on TNBC tumor growth and metastasis, we produced our first nude mouse orthotopic mammary xenograft tumor model by mammary fat pad injection of LM2 parental cells or LM2 ITGA9 KO cells. It was found that tumors produced by LM2 parental cells reached allowed size limit 6–7 weeks post injection; in contrast, tumors produced by LM2 ITGA9 KO cells still did not reach the allowed size limit 10 weeks post injection (Fig. S1c). The LM2 cells also stably express luciferase 2 reporter facilitating the determination of lung metastasis using IVIS Spectrum imaging system. Bioluminescence imaging analysis showed that all mice injected with LM2 parental cells developed lung metastasis as evidenced by the appearance of strong bioluminescent signals in the lungs detected by the IVIS imaging (reported in our recent publication, Ref. 22); however, none of mice injected with LM2 ITGA9 KO cells developed lung metastasis as evidenced by the absence of bioluminescent signals in the lungs (Fig. S1d). To determine that the aforementioned inhibitory effect on tumor growth and metastasis was indeed caused by ITGA9 KO, we produced our second nude mouse orthotopic mammary xenograft tumor model by mammary fat pad injection of LM2 ITGA9 KO vector control (LM2-ITGA9-KO-pLenti6.3) or LM2 ITGA9 KO with ITGA9 stable re-expression (LM2-ITGA9-KO-pLenti6.3-ITGA9) cells. It was found that the inhibitory effect of ITGA9 KO on mouse mammary xenograft tumor growth was reversed by stably re-expressing ITGA9 (Fig. 2c). Moreover, no mice injected with ITGA9 KO vector control cells developed lung metastasis (n = 5 mice); in contrast, all 5 mice injected with ITGA9 KO with ITGA9 stable re-expression cells developed lung metastasis as evidenced by the appearance of strong bioluminescent signals in the lungs detected by the IVIS imaging (Fig. 2d). These findings indicate that ITGA9 KO significantly reduces TNBC cell CSC-like property, tumor growth and metastasis.
Previous studies showed that ITGA9 plays important roles in normal angiogenesis.33,34 Since tumor angiogenesis is essential for tumor growth and metastasis, we then determined whether ITGA9 plays a role in TNBC tumor angiogenesis. IF staining of a vascular structure mark protein CD31 revealed extensive CD31 positive staining in mouse mammary tumors from injection of LM2 parental cells; in contrast, very limited CD31 positive staining was seen in mouse mammary tumors from injection of ITGA9 KO cells (Fig. S2, Fig. 2e). Strong CD31 positive staining was observed again in mouse mammary tumors from injection of ITGA9 KO with ITGA9 stable re-expression cells (Fig. 2e). These results suggest that ITGA9 plays a critical role in TNBC tumor angiogenesis.
ITGA9 down-regulation in mammary tumors by nanoparticle-mediated delivery of ITGA9 siRNA significantly reduces tumor angiogenesis, tumor growth and metastasis
We next determined whether down-regulating ITGA9 expression in TNBC tumor tissues using ITGA9 siRNA has an effect on tumor growth and metastasis. Knocking down a gene expression in tumor tissues by siRNA is challenging due to the intrinsic unstable nature of siRNA oligoes. Nanoparticle-mediated delivery of siRNA oligoes is one of the promising solutions. We used a hybrid nanoparticle (HNP) that was recently developed in our laboratory for efficient in vivo plasmid DNA delivery25 to deliver siRNA oligoes to tumor tissues. Western blot analysis showed that HNP-mediated delivery of ITGA9 siRNA oligoes is capable of reducing ITGA9 protein level in LM2 cells, which is comparable to the knockdown efficiency (60–70%) of a commonly-used commercial transfection reagent Lipofectamine-2000. No significant cytotoxicity was observed in HNP- or Lipofectamine-2000-transfected cells.
We then produced our third orthotopic mammary xenograft tumor model by injecting parental LM2 cells into nude mouse mammary fat pad. When tumors reached about 200 mm3, HNP-packaged control siRNA or ITGA9 siRNA oligoes were given to mice via intratumoral injection. After 3 weeks treatment, tumors treated with ITGA9 siRNA was significantly smaller than tumors treated with control siRNA oligoes (Fig. 3a). Western blot analysis showed that HNP-ITGA9 siRNA treatment significantly reduced tumor tissue ITGA9 level (Fig. 3b). Moreover, all 5 mice in control siRNA-treated group developed strong lung metastasis as evidenced by the appearance of strong bioluminescent signals in the lungs detected by IVIS imaging (Fig. 3c). In contrast, only very limited, focally and weakly bioluminescent signals were detected in the lungs of 5 mice in ITGA9 siRNA-treated group (Fig. 3c). These results indicate that ITGA9 siRNA treatment is capable of significantly reducing mammary tumor growth and suppressing lung metastasis, suggesting that ITGA9 could serve as a potential therapeutic target for treating triple negative breast cancer.
Fig. 3.
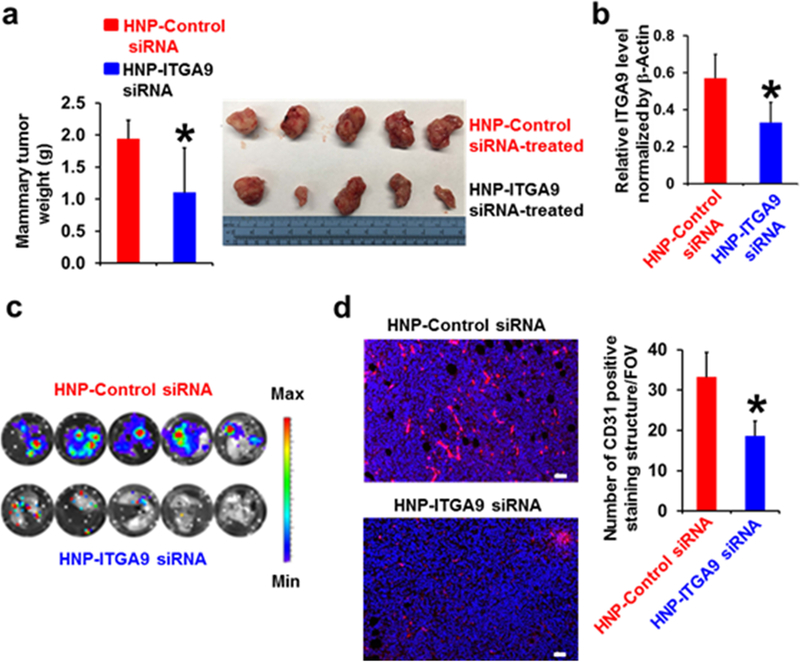
Down-regulation of ITGA9 expression in mouse mammary tumor tissues by nanoparticle-mediated delivery of ITGA9 siRNA oligoes significantly reduces tumor angiogenesis, tumor growth and metastasis. a The average mouse mammary tumor weight after 3-week siRNA treatment (mean ± SD, n=5). b Quantitation of Western blot analysis of ITGA9 level in mouse mammary tumors after 3-week siRNA treatment. The relative ITGA9 level is presented as the ratio of ITGA9 Western blot band intensity divided by the co-related β-Actin Western blot band intensity (mean ± SD, n=4–5). *p<0.05. c Images of mouse lung ex vivo IVIS bioluminescent imaging analysis. d Representative overlaid images of IF staining of CD31 (red) and nuclear DNA DAPI (blue) and quantifications of CD31 positive staining in mouse mammary tumors after 3-week siRNA treatment. The CD31 positive staining were counted and presented as numbers of CD31 positive staining structures per field of view (FOV) (mean ± SD, n=30). *p<0.05. Scale bar, 50 μm.
Consistent with above findings showing that mammary tumors resulting from injection of ITGA9 KO cells have very limited tumor angiogenesis (Fig. 2e), CD31 IF staining revealed that mammary tumors treated with ITGA9 siRNA oligoes display significantly less CD31 positive staining structures than tumors treated with control siRNA (Fig. 3d). These findings suggest that down-regulating tumor ITGA9 level significantly reduces TNBC tumor angiogenesis.
ITGA9 KO reduces β-catenin level through promoting β-catenin proteasome degradation and overexpressing β-catenin reverses the effect of ITGA9 KO on tumor growth and metastasis
We next determined the mechanism of ITGA9 down-regulation inhibiting TNBC cell CSC-like property, tumor angiogenesis, tumor growth and metastasis. One of the critical signaling pathways that play crucial roles in cancer stemness, tumor angiogenesis, tumor growth and metastasis is the canonical Wnt/β-catenin pathway; and β-catenin is the key signaling molecule in this pathway.15–17
The best-characterized mechanism of down-regulating β-catenin level is that β-catenin protein phosphorylation promotes its ubiquitylation and subsequent proteasome degradation.35 Non-phospho-β-catenin is considered as the active form of β-catenin. Western blot analysis revealed that ITGA9 KO caused a drastic decrease of total and non-phospho-β-catenin protein level, an effect that was significantly rescued by stably re-expressing ITGA9 in ITGA9 KO cells (Fig. 4a). Moreover, treatment with a proteasome inhibitor MG132 greatly recovered β-catenin protein level in ITGA9 KO cells (Fig. 4a). Particularly, MG132 treatment almost completely recovered the level of phospho-β-catenin (Ser33/37/Thr41), which is the main phospho-form of β-catenin that is ubiquitylated and degraded. The SCF β-TRCP complex is well-known to be responsible for the ubiquitination of phosphorylated-β-catenin, it is likely that ITGA9 KO causes β-catenin down-regulation mainly through the SCF β-TRCP complex-mediated ubiquitination of β-catenin and subsequently increasing its proteasome degradation. Moreover, this statement is further supported by the q-PCR analysis showing that ITGA9 KO has no significant effect on β-catenin mRNA level.
Fig. 4.
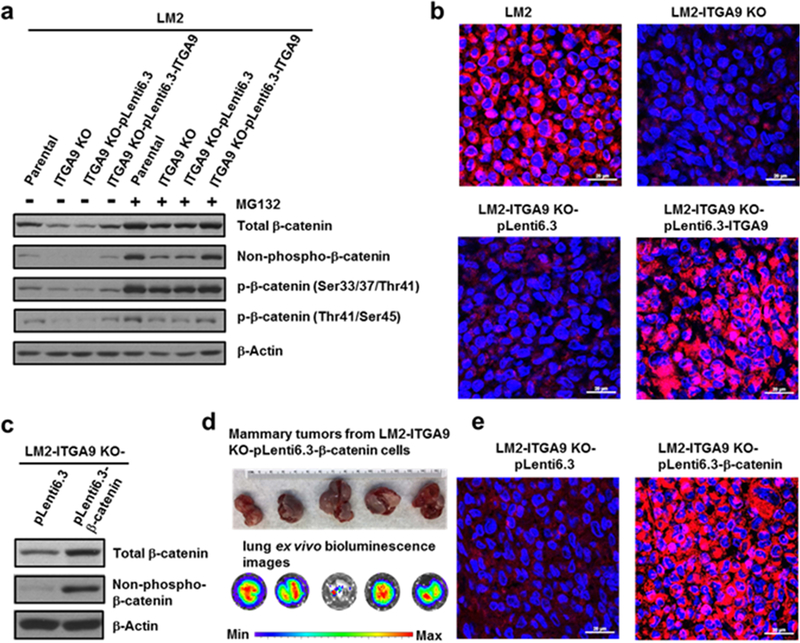
ITGA9 KO down-regulates β-catenin level and overexpressing β-catenin reverses the inhibitory effect of ITGA9 KO on tumor growth and metastasis. a Representative images of Western blot analysis of total β-catenin, non-phospho-β-catenin and phospho-β-catenin levels in ITGA9 KO and re-expressing cells with or without MG-132 treatment. Cells were treated with a vehicle control or MG-132 (10 μM) for 2 h and collected for Western blot analysis. b Representative overlaid confocal microscopy images of IF staining of β-catenin (red) and nuclear DNA DAPI (blue) in mouse mammary tumors from injection of LM2 parental, ITGA9 KO, ITGA9 KO vector control or ITGA9 re-expressing cells. Scale bar, 20 μm. c Representative Western blot showing β-catenin overexpression in ITGA9 KO cells. d Mouse mammary tumor images and lung ex vivo IVIS bioluminescence analysis images. e Representative overlaid confocal microscopy images of IF staining of β-catenin (red) and nuclear DNA DAPI (blue) staining in mouse mammary tumors from injection of vector control or β-catenin overexpressing ITGA9 KO LM2 cells. Scale bar, 20 μm.
β-catenin is a co-transcription factor and its nuclear localization indicates β-catenin activation increasing its target gene expression.35 β-catenin IF staining showed that β-catenin is mainly localized in nucleus in parental LM2 cells as evidenced by the merged pinkish color from β-catenin IF staining (red) and nuclear DNA DAPI fluorescent staining (blue) (Fig. S3a). Consistent with the results from Western blot analysis showing the diminished level of non-phospho-β-catenin in IGTA9 KO cells (Fig. 4a), very few and weakly pinkish color was viewed in the ITGA9 KO cells (Fig. S3a), which indicates significantly less β-catenin nuclear localization. In contrast, stably re-expressing ITGA9 in ITGA9 KO cells greatly recovered β-catenin nuclear localization (Fig. S3b). Confocal microscopy imaging of β-catenin IF staining revealed that significant β-catenin positive staining and nuclear localization are detected in mouse mammary tumors resulting from injection of LM2 parental cells; in contrast, diminished β-catenin positive staining and nuclear localization are observed in tumors from injection of LM2 ITGA9 KO cells (Fig. 4b). Increased β-catenin positive staining and nuclear localization were detected again in tumors from injection of ITGA9 KO with ITGA9 stable re-expression cells (Fig. 4b). These findings are supported by cell fractionation experiments showing that nuclear β-catenin protein level is dramatically decreased in ITGA9 KO cells; and ITGA9 re-expression in ITGA9 KO cells greatly recovered nuclear β-catenin protein level (Fig. S3c).
Nuclear-localized β-catenin interacts with TCF/LEF family of transcription factors and promotes its target gene expression.15 We further determined whether ITGA9 KO has an effect on β-catenin transcriptional activity using TOPflash (with wildtype TCF binding sites) and FOPflash (with TCF binding sites mutated) luciferase reporter assays. As shown in Fig. S4a, the TOPflash luciferase reporter activity was significantly lower in ITGA9 KO SUM-159 cells than the parental SUM-159 cells; and stably re-expressing ITGA9 significantly increased the TOPflash luciferase reporter activity. Due to LM2 cells stably expressing luciferase gene, we could not use this cell line for TOPflash luciferase reporter assay. However, Q-PCR analysis showed that the expression levels of three β-catenin target genes (VEGF-A: vascular endothelial growth factor-A; APC2: Adenomatous polyposis coli protein 2; and PDHK1: pyruvate dehydrogenase kinase 1) are significantly lower in ITGA9 KO LM2 cells than parental cells; and stably re-expressing ITGA9 greatly recover their expression levels (Fig. S4b). Together, these findings indicate that ITGA9 KO reduces β-catenin protein level in TNBC cells and tumors, resulting in decreased nuclear localization and transcriptional activity of β-catenin and causing down-regulation of its target gene expression.
To demonstrate the importance of β-catenin down-regulation in the inhibitory effect of ITGA9 KO on tumor growth and metastasis, we stably overexpressed β-catenin in ITGA9 KO LM2 cells to determine whether overexpressing β-catenin reverses the inhibitory effect of ITGA9 KO on tumor angiogenesis, tumor growth and metastasis. Overexpression of β-catenin in ITGA9 KO cells was confirmed by Western blot (Fig. 4c). Injection of ITGA9 KO with β-catenin overexpressing LM2 cells produced significantly larger mammary tumors (4.04 g ± 1.53, n=5) than injection of ITGA9 KO LM2 vector control cells (0.34 g ± 0.28, n=5) (Fig. 4d and Fig. 2c). Importantly, all 5 five mice injected with ITGA9 KO-β-catenin overexpressing LM2 cells developed strong lung metastasis (Fig. 4d). Confocal microscopy imaging of β-catenin IF staining revealed that mouse mammary tumors from injection of β-catenin overexpression cells display significant β-catenin positive staining and nuclear localization (Fig. 4e). Moreover, tumors from β-catenin overexpression cells also displayed significantly more CD31 positive staining (51.1 ± 7.98, n=30) than the tumors from vector control cells (8.4 ± 2.45, n=30) (Fig. S5). These results demonstrate a crucial role of β-catenin down-regulation in the inhibitory effect of ITGA9 KO on tumor growth and metastasis.
ITGA9 KO down-regulates β-catenin by increasing glycogen synthase kinase 3 (GSK3) activity through inhibiting the activity of protein kinase A (PKA)
We then determined the mechanism of how ITGA9 KO down-regulates β-catenin. β-Catenin ubiquitylation and subsequent proteasome degradation is preceded by its phosphorylation mediated mainly by the serine/threonine protein kinase glycogen synthase kinase 3α and 3β (GSK3α, 3β).15,35 The best-characterized mechanism that regulates GSK3 activity is its inhibitory phosphorylation of serine21 (Ser21) in GSK3α or of serine9 (Ser9) in GSK3β that reduces its kinase activity; and thus lower phospho-level of Ser21 in GSK3α or of Ser9 in GSK3β reflect higher activity of GSK3α and GSK3β, respectively.36 Western blot analysis showed that the phospho-level of Ser21 in GSK3α and of Ser9 in GSK3β are significantly lower in ITGA9 KO LM2 cells than the parental LM2 cells; and stably re-expressing ITGA9 reverses the observation (Fig. 5a). These results indicate that ITGA9 KO increases the activity of GSK3α and GSK3β leading to β-catenin down-regulation. This conclusion is further supported by the finding showing that inhibiting GSK3 activity with a GSK3 inhibitor lithium chloride (LiCl) greatly increased total β-catenin protein level and its target gene expression in ITGA9 KO LM2 cells (Fig. S6).
Fig. 5.
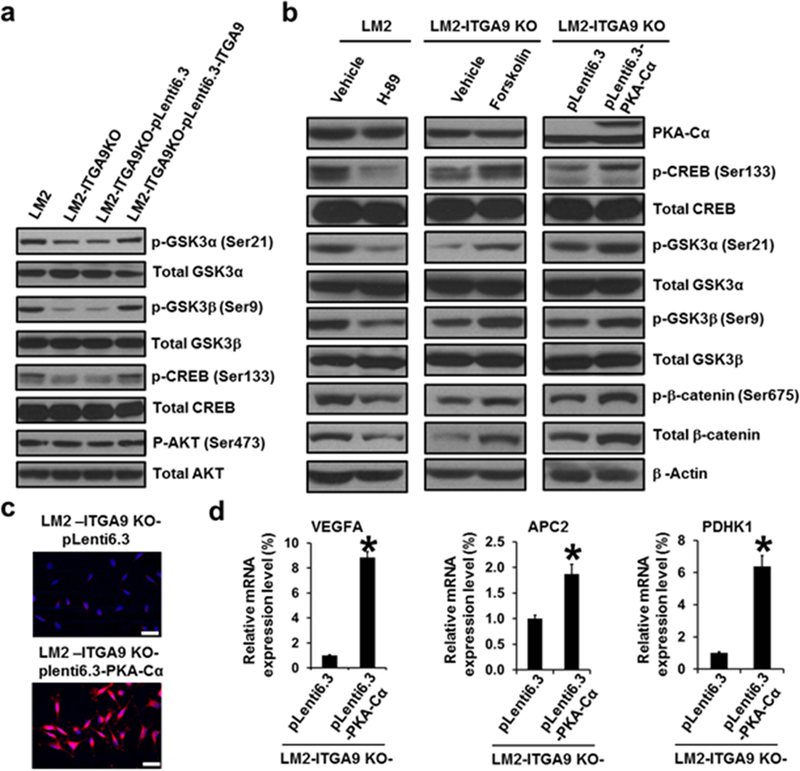
ITGA9 KO reduces β-catenin level by decreasing PKA activity and increasing GSK3 activity. a Representative images of Western blot analysis of phospho- and total levels of GSK3α, GSK3β, CREB and AKT in ITGA9 KO and re-expressing cells. b Representative images of Western blot analysis of PKA-Cα level, phospho- and total levels of CREB, GSK3α, GSK3β, and β-catenin in parental LM2 cells treated with a vehicle control or H-89 (10 μM, 24 h); and in ITGA9 KO LM2 cells treated with a vehicle control or Forskolin (10 μM, 24 h); and in vector control or PKA-Cα stably expressing ITGA9 KO LM2 cells. c Representative overlaid images of IF staining of β-catenin (red) and nuclear DNA DAPI (blue) in vector control and PKA-Cα stably expressing ITGA9 KO LM2 cells. Scale bar, 50 μm. d Q-PCR analysis of β-catenin target gene expression levels in vector control and PKA-Cα stably expressing ITGA9 KO LM2 cells. The mRNA level of each gene was determined by ABI specific gene expression assay and normalized by β-actin (mean ± SD, n=3). *p<0.05.
The inhibitory serine-phosphorylation of GSK3α (Ser21) and GSK3β (Ser9) are carried out mainly by several serine/threonine protein kinases such as PKA, AKT (also known as PKB), etc.36 Western blot analysis revealed no significant changes of the activation phosphorylation of AKT (Ser473) between the parental and ITGA9 KO LM2 cells (Fig. 5a), suggesting that ITGA9 KO causes no significant change of AKT activity. However, the phospho-level of CREB (p-CREB) (cAMP response element-binding protein), a substrate of PKA, was drastically lower in ITGA9 KO cells; and stably re-expressing ITGA9 recovered the level of p-CREB (Ser133) (Fig. 5a). These results suggest that ITGA9 KO may reduce PKA activity in TNBC cells, which leads to decreased inhibitory serine-phosphorylation of GSK3α (Ser21) and GSK3β (Ser9) increasing GSK3 activity. We then used following three approaches to demonstrate that ITGA9 KO decreases GSK3 phosphorylation via reducing PKA activity: (i) Parental LM2 cells treated with a PKA inhibitor H-89 showed reduced levels of p-CREB (Ser133), p-GSK3α (Ser21), p-GSK3β (Ser9) and total β-catenin (Fig. 5b). Moreover, the phosphorylation of β-catenin at Ser675 is carried out by PKA.37,38 It was found that LM2 cells treated with H-89 have significantly lower level of p-β-catenin (Ser675) (Fig. 5b). (ii) ITGA9 KO LM2 cells treated with a PKA agonist Forskolin showed increased levels of p-CREB (Ser133), p-GSK3α (Ser21), p-GSK3β (Ser9), p-β-catenin (Ser675) and total β-catenin (Fig. 5b). (iii) Stably expressing a constitutive active PKA catalytic subunit α (PKA-Cα) in ITGA9 KO cells also increased the levels of p-CREB (Ser133), p-GSK3α (Ser21), p-GSK3β (Ser9), p-β-catenin (Ser675) and total β-catenin (Fig. 5b). Moreover, β-catenin IF staining revealed that total and nuclear localized β-catenin staining is significantly reduced in H-89-treated LM2 cells compared to vehicle control-treated LM2 cells (Fig. S7a). In contrast, total and nuclear localized β-catenin staining was increased in Forskolin-treated or PKA-Cα stable expressing ITGA9 KO cells compared to vehicle control-treated ITGA9 KO cells or ITGA9 KO vector control cells (Fig. S7b, Fig. 5c). Further q-PCR analysis demonstrated that the expression levels of three β-catenin target genes are significantly increased in PKA-Cα stable expressing ITGA9 KO cells (Fig. 5d). Together, these results indicate that ITGA9 KO down-regulates β-catenin by increasing GSK3 activity through inhibiting the activity of PKA.
ITGA9 KO causes cytoplasmic translocation of integrin-linked kinase (ILK) that interacts with PKA regulatory subunit IIα (PKA-RIIα) leading to inhibition of PKA activity
We further determined the mechanism by which ITGA9 KO reduces PKA activity. The PKA holoenzyme is an inactive heterotetramer consisting of two regulatory subunits and two catalytic subunits. The binding of two cyclic adenosine monophosphates (cAMP) to each of the two regulatory subunits releases and activates the catalytic subunits, which phosphorylate serine and threonine residues on the substrate proteins. Proteins interacting with PKA regulatory or catalytic subunits may enhance or inhibit PKA activity.39–42 One of the important proteins that regulate integrin α9β1 signaling is the scaffold protein integrin-linked kinase (ILK), which was found to interact with ITGB1 cytoplasmic tail and other proteins at cellular membrane region.43,44 We hypothesize that ITGA9 KO disrupts the interaction between ITGA9 and ITGB1, which leads to ILK translocation from cellular membrane region to cytoplasm and interacts with PKA regulatory subunits and reduces PKA activity.
To test our hypothesis, we first performed ILK IF staining and found that strong ILK staining at cellular membrane region is observed in parental SUM-159 cells; in contrast, ILK staining is mostly seen at cytoplasm in ITGA9 KO SUM-159 cells (Fig. S8a). Moreover, re-expressing ITGA9 in ITGA9 KO cells greatly recovered ILK staining at cellular membrane region (Fig. S8b). Co-IF staining of ILK and PKA regulatory subunit IIα (PKA-IIα-reg) showed that PKA-IIα-reg is stained at cytoplasm in both parental and ITGA9 KO cells; and PKA-IIα-reg (green color) is co-localized with ILK (red color) in cytoplasm in IGTA9 KO cells as revealed by the merged yellowish color (Fig. 6a). In contrast, no significant co-localization between PKA-IIα-reg and ILK was detected in parental cells (Fig. 6a). We next performed two co-immunoprecipitation (Co-IP) assays to further demonstrate the interaction between ILK and PKA-IIα-reg in ITGA9 KO cells. First, after IP ILK, a significant more amount of PKA-IIα-reg was detected in ITGA9 KO cells, suggesting the increased interaction between ILK and PKA-IIα-reg in ITGA9 KO cells compared to the parental cells (Fig. 6b). The increase of ILK and PKA-IIα-reg interaction was reduced by stably re-expressing ITGA9 in ITGA9 KO cells (Fig. 6b). Second, after IP PKA-IIα-reg, a significantly more amount of ILK and PKA-Cα protein was detected in ITGA9 KO cells (Fig S9), indicating that more ILK and PKA-Cα are associated with PKA-IIα-reg in ITGA9 KO cells. Again, the increased interaction among ILK, PKA-IIα-reg and PKA-Cα in ITGA9 KO cells was decreased by re-expressing ITGA9 in ITGA9 KO cells (Fig. S9).
Fig. 6.
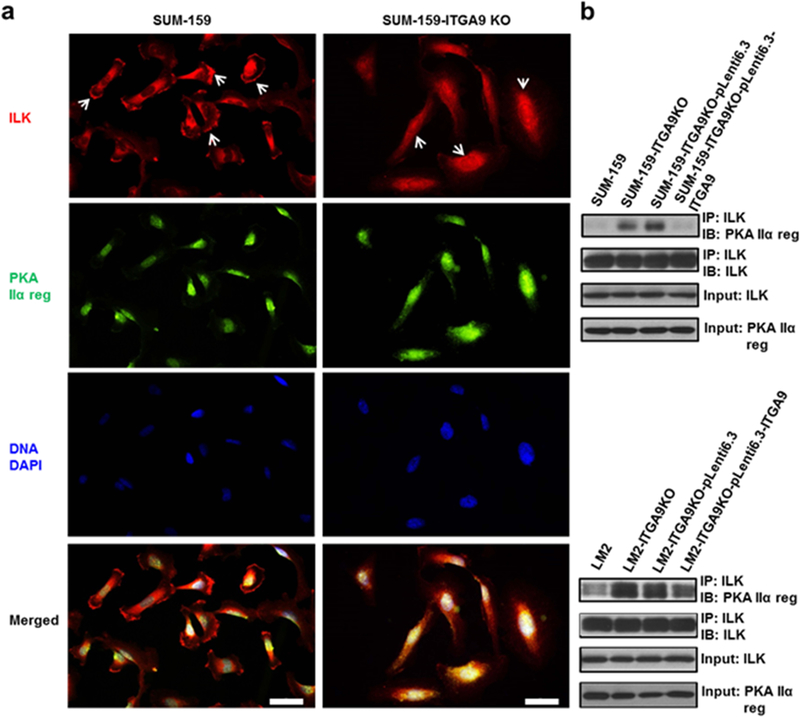
ITGA9 KO causes ILK relocation from cellular membrane region to cytoplasm and increases the interaction between ILK and PKA regulatory subunit IIα. a Representative images of IF staining of ILK (red), PKA regulatory subunit IIα (green) and nuclear DNA DAPI (blue) in parental and ITGA9 KO SUM-159 cells. White arrows point to representative ILK cellular membrane staining in parental cells and ILK cytoplasm staining in ITGA9 KO cells. Scale bar, 50 μm. b Representative Western blot images of Co-IP analysis determining the interaction between ILK and PKA regulatory subunit IIα in parental, ITGA9 KO and re-expressing LM2 and SUM-159 cells. IP: immunoprecipitating; IB: immunoblotting.
To demonstrate that increased interaction between ILK and PKA reduces PKA activity in ITGA9 KO cells, we used siRNA to knock down ILK level. It was found that knocking down ILK level in ITGA9 KO cells increased the phospho-levels of two PKA substrates: p-CREB (Ser133) and p-β-catenin (Ser675) (Fig. S10). Moreover, the up-regulation of p-CREB (Ser133) and p-β-catenin (Ser675) resulting from ILK knockdown in ITGA9 KO cells was reversed by treating cells with a PKA inhibitor H-89 (Fig. S10). These results indicate that ILK knockdown in ITGA9 KO cells increases PKA activity. As a result, ILK knockdown in ITGA9 KO cells increased the levels of p-GSK3α (Ser21) and p-GSK3β (Ser9), indicating that the activity of GSK3α and GSK3β is reduced leading to increased level of total β-catenin (Fig. S10), Again, this effect was reversed by treating cell with a PKA inhibitor H-89 (Fig. S10). Together, these results demonstrate that ITGA9 KO decreases PKA activity through ILK.
Discussion
Although ITGA9 has been shown to be involved in cell adhesion and motility,4 its expression pattern and its role in tumor growth and metastasis and the underlying mechanism remain largely unknown. By analyzing a large number of patients’ ITGA9 expression data, we found that ITGA9 expression level is significantly higher in TNBC than other breast cancer subtypes; and TNBC patients with higher ITGA9 level have significantly worse DMFS and RFS, implying an important role of ITGA9 in TNBC metastasis. Indeed, ITGA9 depletion in TNBC cells drastically reduced TNBC cell CSC-like property, tumor angiogenesis, tumor growth and tumor metastasis. Furthermore, ITGA9 down-regulation in TNBC tumor tissues by nanoparticle-mediated delivery of ITGA9 siRNA significantly decreased tumor growth and metastasis. Mechanistically, ITGA9 depletion caused ILK translocation from cellular membrane region to cytoplasm, where ILK interacted with PKA and reduced PKA activity leading to increased GSK3 activity and β-catenin down-regulation and inhibition of TNBC cancer stemness, tumor angiogenesis, tumor growth and metastasis. This study not only demonstrates a crucial role of ITGA9 in TNBC tumor growth and metastasis and the underlying mechanism, also identifies targets for developing new therapeutic strategies for treating metastatic TNBC, which currently lacks effective therapies representing a critical unmet clinic need.
One of important proteins that regulate integrin α9β1 function is ILK, a scaffold protein that interacts with the cytoplasmic tail of β1 integrin.43 ILK regulates integrin function by scaffolding β integrin cytoplasmic tail and other molecules critical for signal transduction initiated by integrin-mediated cell-matrix adhesion.44,45 In this study, we revealed a new mechanism for ILK regulation of integrin α9β1 function through the ITGA9/ILK/PKA/GSK3/β-catenin pathway.
Increased PKA activity is observed in cancer and PKA has been proposed as an important target for cancer treatment.40 Previous studies showed that cellular compartmentalized regulation of PKA activity is mainly achieved by A-kinase anchoring proteins, a group of proteins interacting with PKA regulatory subunits, recruiting PKA to cellular specific loci, and directing PKA kinase activity to specific substrates.39,46,47 By using Co-IF staining and Co-IP experiments, we demonstrated that ITGA9 KO significantly increases the interaction between ILK and PKA. Although it remains to be determined whether ILK directly interacts with PKA regulatory subunit or catalytic subunit, this interaction reduces PKA activity. It is likely that the interaction between ILK and PKA in ITGA9 KO cells causes PKA conformational change, which enhances the interaction between PKA regulatory subunits and its catalytic subunits and reduces PKA activity.
The finding that increased interaction between ILK and PKA leading to inhibition of PKA activity and down-regulation of β-catenin in ITGA9 KO TNBC cells is novel and significant. Although previous studies showed that the Wnt/β-catenin pathway is activated and enriched in TNBC, the underlying mechanism is poorly understood.18–20 Our findings showing that ITGA9 KO abolishes β-catenin pathway in TNBC provided new mechanistic insight for the strong activation of the Wnt/β-catenin pathway in TNBC.
Integrin α9β1 has been shown to interact with VEGF-A and thrombospondin-1 to promote angiogenesis,33,34 however, its role in tumor angiogenesis remains to be determined. In this study, we found that down-regulation of ITGA9 in TNBC cells or tumors drastically reduces tumor angiogenesis and re-expression of ITGA9 significantly recovers tumor angiogenesis. Mechanistically, ITGA9 down-regulation greatly decreased the angiogenic factor VEGF-A expression level via promoting β-catenin degradation. These findings identified a new mechanism for understanding the role of ITGA9 in angiogenesis.
The Wnt/β-catenin pathway plays pivotal roles in development, adult tissue homeostasis and cancer; and non-discriminatively targeting β-catenin for cancer therapy could be challenging due to the potential devastating effects on normal tissues.48 Alternatively, targeting abnormally expressed or activated molecules in cancer cells that enhance β-catenin signaling may achieve desired therapeutic effect with less unwanted side-effects. In this study, we found that ITGA9 down-regulation promotes β-catenin degradation, which may provide an alternative strategy for inhibiting β-catenin pathway through targeting ITGA9. Indeed, ours and other recent studies provided additional support to this idea of targeting abnormally expressed or activated molecules in cancer cells that enhance β-catenin signaling to reduce TNBC stemness, tumor growth and metastasis. 49,50
In summary, the findings from this study not only shed new light on TNBC biology and also reveal novel therapeutic opportunities with the potential to improve clinical outcomes of TNBC by targeting ITGA9.
Supplementary Material
Novelty & Impact Statements:
Integrin α9 (ITGA9) expression pattern in cancer and its role in tumor growth and metastasis remain largely unknown. This study demonstrates that ITGA9 is highly expressed in triple negative breast cancer (TNBC) and ITGA9 depletion suppresses TNBC stemness, tumor growth and metastasis through the ILK/PKA/GSK3/β-catenin pathway, which not only shed new light on TNBC biology also reveal novel therapeutic opportunities with the potential to improve clinical outcomes of TNBC by targeting ITGA9.
Acknowledgments
Funding: This study was supported in part by a Research Scholar Grant (RGS-15–026-01-CSM) from the American Cancer Society to C.Y., a research grant from Elsa U. Pardee Foundation to Z.W., and a NIH grant (R01CA133429) to G.T. This research was also supported by the Shared Animal Imaging and Histology Resources of the University of Kentucky Markey Cancer Center (P30CA177558) and The University of Kentucky Center for Appalachian Research in Environmental Sciences Center grant (1P30ES026529–01A1).
Abbreviations
- APC2
Adenomatous polyposis coli protein 2
- CREB
cAMP response element-binding protein
- CSC
cancer stem cell
- DMFS
distant metastasis free survival
- GSK3
glycogen synthase kinase 3
- HNP
hybrid nanoparticle
- ILK
integrin-linked kinase
- ITGA9
integrin α9
- KO
knockout
- PDHK1
pyruvate dehydrogenase kinase 1
- PKA
protein kinase A
- PKA-Cα
PKA catalytic subunit α
- PKA-RIIα
PKA regulatory subunit Iiα
- RFS
recurrence free survival
- TNBC
triple negative breast cancer
- VEGF-A
vascular endothelial growth factor-A
Footnotes
Conflict of interests: The authors declare that they have no conflict of interests.
References:
- 1.Seguin L, Desgrosellier JS, Weis SM, Cheresh DA. Integrins and cancer: regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 2015;25:234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–87. [DOI] [PubMed] [Google Scholar]
- 4.Høye AM, Couchman JR, Wewer UM, Fukami K, Yoneda A. The newcomer in the integrin family: integrin α9 in biology and cancer. Adv Biol Regul. 2012;52:326–39. [DOI] [PubMed] [Google Scholar]
- 5.Palmer EL, Rüegg C, Ferrando R, Pytela R, Sheppard D. Sequence and tissue distribution of the integrin alpha 9 subunit, a novel partner of beta 1 that is widely distributed in epithelia and muscle. J Cell Biol. 1993;123:1289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. [DOI] [PubMed] [Google Scholar]
- 7.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease, Nat Rev Clin Oncol. 2016;13:674–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar P, Aggarwal R. An overview of triple-negative breast cancer. Arch Gynecol Obstet. 2016;293:247–69. [DOI] [PubMed] [Google Scholar]
- 9.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. [DOI] [PubMed] [Google Scholar]
- 10.Thike AA, Cheok PY, Jara-Lazaro AR, Tan B, Tan P, Tan PH. Triple-negative breast cancer: clinicopathological characteristics and relationship with basal-like breast cancer. Mod Pathol. 2010;23:123–33. [DOI] [PubMed] [Google Scholar]
- 11.Kast K, Link T, Friedrich K, Petzold A, Niedostatek A, Schoffer O, et al. Impact of breast cancer subtypes and patterns of metastasis on outcome. Breast Cancer Res Treat. 2015;150:621–9. [DOI] [PubMed] [Google Scholar]
- 12.Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28:3271–7. [DOI] [PubMed] [Google Scholar]
- 13.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–34. [DOI] [PubMed] [Google Scholar]
- 14.Allen MD, Vaziri R, Green M, Chelala C, Brentnall AR, Dreger S, et al. Clinical and functional significance of α9β1 integrin expression in breast cancer: a novel cell-surface marker of the basal phenotype that promotes tumour cell invasion. J Pathol. 2011;223:646–58. [DOI] [PubMed] [Google Scholar]
- 15.Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. [DOI] [PubMed] [Google Scholar]
- 16.Holland JD, Klaus A, Garratt AN, Birchmeier W. Wnt signaling in stem and cancer stem cells. Curr Opin Cell Biol. 2013;25:254–64. [DOI] [PubMed] [Google Scholar]
- 17.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pohl S-G, Brook N, Agostino M, Arfuso F, Kumar AP, Dharmarajan A. Wnt signaling in triple-negative breast cancer. Oncogenesis. 2017;6:e310. doi: 10.1038/oncsis.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rangel MC, Bertolette D, Castro NP, Klauzinska M, Cuttitta F, Salomon DS. Developmental signaling pathways regulating mammary stem cells and contributing to the etiology of triple-negative breast cancer. Breast Cancer Res Treat. 2016;156:211–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH. Wnt/β-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol. 2010;176:2911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, V, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humphries B, Wang Z, Li Y, Jhan JR, Jiang Y, Yang C. ARHGAP18 Downregulation by miR-200b Suppresses Metastasis of Triple-Negative Breast Cancer by Enhancing Activation of RhoA. Cancer Res. 2017;77:4051–4064. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Humphries B, Xiao H, Jiang Y, Yang C. MicroRNA-200b suppresses arsenic-transformed cell migration by targeting protein kinase Cα and Wnt5b-protein kinase Cα positive feedback loop and inhibiting Rac1 activation. J Biol Chem. 2014;289:18373–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humphries B, Wang Z, Oom AL, Fisher T, Tan D, Cui Y, et al. MicroRNA-200b targets protein kinase Cα and suppresses triple-negative breast cancer metastasis. Carcinogenesis. 2014;35:2254–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Humphries B, Wang Z, Lang S, Huang X, Xiao H, et al. Complex Coacervation-Integrated Hybrid Nanoparticles Increasing Plasmid DNA Delivery Efficiency in Vivo. ACS Appl Mater Interfaces. 2016;8:30735–30746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Humphries B, Xiao H, Jiang Y, Yang C. Epithelial to mesenchymal transition in arsenic-transformed cells promotes angiogenesis through activating β-catenin-vascular endothelial growth factor pathway. Toxicol Appl Pharmacol. 2013;271:20–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao Y, Li Y, Tao H, Humphries B, Li A, Jiang Y, et al. Integrin α5 down-regulation by miR-205 suppresses triple negative breast cancer stemness and metastasis by inhibiting the Src/Vav2/Rac1 pathway. Cancer Lett. 2018;433:199–209. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Shu W, Lu MM, Morrisey EE. Wnt7b activates canonical signaling in epithelial and vascular smooth muscle cells through interactions with Fzd1, Fzd10, and LRP5. Mol Cell Biol. 2005;25:5022–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, Tan YS, Haslam SZ, Yang C. Perfluorooctanoic acid effects on steroid hormone and growth factor levels mediate stimulation of peripubertal mammary gland development in C57BL/6 mice. Toxicol Sci 2010;115:214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12:133–43. [DOI] [PubMed] [Google Scholar]
- 32.Shukla S, Meeran SM. Epigenetics of cancer stem cells: Pathways and therapeutics. Biochim Biophys Acta. 2014;1840:3494–3502. [DOI] [PubMed] [Google Scholar]
- 33.Vlahakis NE, Young BA, Atakilit A, Hawkridge AE, Issaka RB, Boudreau N, et al. Integrin alpha9beta1 directly binds to vascular endothelial growth factor (VEGF)-A and contributes to VEGF-A-induced angiogenesis. J Biol Chem. 2007;282:15187–96. [DOI] [PubMed] [Google Scholar]
- 34.Staniszewska I, Zaveri S, Del Valle L, Oliva I, Rothman VL, Croul SE, et al. Interaction of alpha9beta1 integrin with thrombospondin-1 promotes angiogenesis. Circ Res. 2007;100:1308–16. [DOI] [PubMed] [Google Scholar]
- 35.Stamos JL, Weis WI. The β-catenin destruction complex. Cold Spring Harbor perspectives in biology. 2013;5:a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beurel E, Grieco SF, Jope RS. Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol Ther. 2015;148:114–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taurin S, Sandbo N, Qin Y, Browning D, Dulin NO. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase. J Biol Chem. 2006;281:9971–6. [DOI] [PubMed] [Google Scholar]
- 38.Hino S, Tanji C, Nakayama KI, Kikuchi A. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase stabilizes beta-catenin through inhibition of its ubiquitination. Mol Cell Biol. 2005;25:9063–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torres-Quesada O, Mayrhofer JE, Stefan E. The many faces of compartmentalized PKA signalosomes. Cell Signal. 2017;37:1–11. [DOI] [PubMed] [Google Scholar]
- 40.Sapio L, Di Maiolo F, Illiano M, Esposito A, Chiosi E, Spina A, et al. Targeting protein kinase A in cancer therapy: an update. EXCLI J. 2014;13:843–55. [PMC free article] [PubMed] [Google Scholar]
- 41.Veglia G, Cembran A. Role of conformational entropy in the activity and regulation of the catalytic subunit of protein kinase A. FEBS J. 2013;280:5608–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor SS, Ilouz R, Zhang P, Kornev AP. Assembly of allosteric macromolecular switches: lessons from PKA. Nat Rev Mol Cell Biol. 2012;13:646–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hannigan GE, Leung-Hagesteijn C, Fitz-Gibbon L, Coppolino MG, Radeva G, Filmus J, et al. Regulation of cell adhesion and anchorage-dependent growth by a new beta 1-integrin-linked protein kinase. Nature. 1996;379:91–6. [DOI] [PubMed] [Google Scholar]
- 44.Qin J, Wu C. ILK: a pseudokinase in the center stage of cell-matrix adhesion and signaling. Curr Opin Cell Biol. 2012;24:607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghatak S, Morgner J, Wickström SA. ILK: a pseudokinase with a unique function in the integrin-actin linkage. BiochemSoc Trans. 2013;41:995–1001. [DOI] [PubMed] [Google Scholar]
- 46.Calejo AI, Taskén K. Targeting protein-protein interactions in complexes organized by A kinase anchoring proteins. Front Pharmacol. 2015;6:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skroblin P, Grossmann S, Schäfer G, Rosenthal W, Klussmann E. Mechanisms of protein kinase A anchoring. Int Rev Cell Mol Biol. 2010;283:235–330. [DOI] [PubMed] [Google Scholar]
- 48.Kahn M Can we safely target the WNT pathway? Nat Rev Drug Discov. 2014;13:513–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, Xiao Y, Lin HP, Reichel D, Bae Y, Lee EY, Jiang Y, Huang X, Yang C, Wang Z. In vivo β-catenin attenuation by the integrin α5-targeting nano-delivery strategy suppresses triple negative breast cancer stemness and metastasis. Biomaterials. 2019; 188:160–172. [DOI] [PubMed] [Google Scholar]
- 50.Lin CC, Lo MC, Moody R, Jiang H, Harouaka R, Stevers N, Tinsley S, Gasparyan M, Wicha M, Sun D. Targeting LRP8 inhibits breast cancer stem cells in triple-negative breast cancer. Cancer Lett. 2018; 438:165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


