Abstract
Background:
The turnover of cardiac ion channels underlying action potential duration (APD) is regulated by ubiquitination. Genome-wide association studies (GWAS) of QT interval identified several single-nucleotide polymorphisms located in or near genes involved in protein ubiquitination. A genetic variant upstream of LITAF (lipopolysaccharide-induced tumor necrosis factor) gene prompted us to determine its role in modulating cardiac excitation.
Methods:
Optical mapping was performed in zebrafish hearts to determine Ca2+ transients. Live cell confocal calcium imaging was performed on adult rabbit ventricular myocytes (ARbCM) to determine intracellular Ca2+handling. LTCC (L-type calcium channel) current was measured using whole cell recording. To study the effect of LITAF on Cav1.2 channel expression, surface biotinylation and westerns were performed. LITAF interactions were studied using co-immunoprecipitation and in situ proximity ligation assay (PLA).
Results:
LITAF knockdown in zebrafish resulted in a robust increase in calcium transients. Overexpressed LITAF in 3-week-old rabbit cardiomyocytes resulted in a decrease in ICa,L and Cavα1c abundance, whereas LITAF knockdown increased ICa,L and Cavα1c protein. LITAF-overexpressing decreases calcium transients in ARbCM, which was associated with lower Cavα1c levels. In tsA201 cells, overexpressed LITAF downregulated total and surface pools of Cavα1c via increased Cavα1c ubiquitination and its subsequent lysosomal degradation. We observed co-localization between LITAF and LTCC in tsA201 and cardiomyocytes. In tsA201, NEDD4-1, but not its catalytically inactive form NEDD4-1-C867A, increased Cavα1c ubiquitination. Cavα1c ubiquitination was further increased by co-expressed LITAF and NEDD4-1 but not NEDD4-1-C867A. NEDD4-1 knockdown abolished the negative effect of LITAF on ICa,L and Cavα1c levels in 3-week-old rabbit cardiomyocytes. Computer simulations demonstrated that a decrease of ICa,L current associated with LITAF overexpression simultaneously shortened APD and decreased calcium transients in rabbit cardiomyocytes.
Conclusions:
LITAF acts as an adaptor protein promoting NEDD4-1-mediated ubiquitination and subsequent degradation of LTCC, thereby controlling LTCC membrane levels and function and thus cardiac excitation.
Keywords: L-type calcium channels, L-type calcium current, ubiquitin, lipopolysaccharide-induced tumor necrosis factor, Ion Channels, Membrane Transport
Introduction
Ion channel proteins are regulated by various types of posttranslational modifications. In most cases, ubiquitination acts as a signal for endocytosis of the ion channels, which are subsequently degraded through lysosomal or proteasomal-dependent pathways. Ubiquitination also occurs at the exit of endoplasmic reticulum (ER) and the trans-Golgi-network (TGN).1–4 Several GWAS for loci that modify the QT interval and the risk for sudden cardiac death5, 6 have identified three single-nucleotide polymorphisms located in or near genes involved in protein ubiquitination: 1) The Ring finger protein 207 (RNF207) ubiquitin ligase;7, 8 2) Ring finger and FYVE-like domain containing E3 ubiquitin protein ligase (RFFL);9, 10 and 3) Lipopolysaccharide-induced tumor necrosis factor (LITAF), a regulator of endosomal trafficking11–13 and inflammatory cytokines,14, 15 and an adapter molecule for members of the NEDD4-like family of E3 ubiquitin ligases.16, 17 The genetic variant rs8049607 (Figure I in the Data Supplement) was associated with a modest QT-interval-prolongation effect (1.2 ms, p=5×10−15). 5, 6 It is approximately 41 kb upstream of the LITAF start codon18 and located within an intergenic enhancer region.19 Furthermore, data from expression quantitative trait loci (eQTL) analyses showed that rs8049607 was also associated with reduced LITAF mRNA transcript levels in the left ventricle (Figure I in the Data Supplement).20
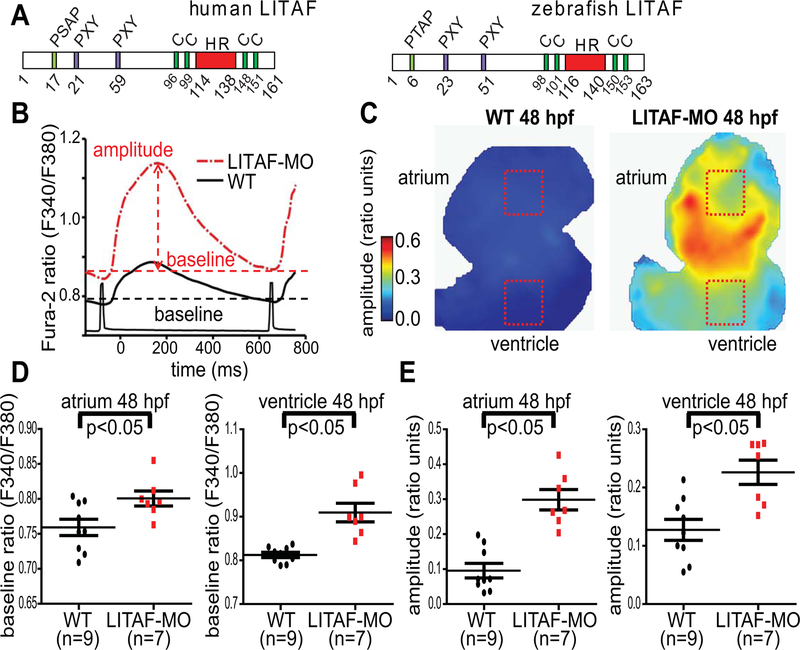
LITAF is a mediator of local and systemic inflammatory responses15 and is locally upregulated in a number of inflammatory diseases such as Crohn’s Disease and ulcerative colitis.21 Notably, whole-body LITAF deletion diminished experimental endotoxic shock and inflammatory arthritis in mice.15 Importantly, loss-of-function mutations in LITAF cause Charcot-Marie-Tooth (CMT) disease, an inherited peripheral neuropathy. These mutations22, 23 are clustered around the hydrophobic region required for membrane localization and cause mislocalization and impaired endosome-to-lysosome trafficking of membrane proteins11, 24 (Figure 1A).
Figure 1.
Genetic knockdown of LITAF increases calcium transients in the heart of zebrafish embryos. Zebrafish hearts from 48 hpf wild-type (WT) and LITAF morphants (MO) were stained with Fura-2 AM to measure Ca2+ transients. A, Structure of human and zebrafish LITAF with conserved PXY and P(S/T)AP motifs and the hydrophobic region (HR) required for membrane embedding.25 Also indicated are two conserved cysteine pairs for coordination of a zinc atom likely required for proper protein folding.25 B, Averaged ventricular Ca2+ transients (amplitudes and baselines) from regions of interest (ROIs) in panel C. C, Color maps of Ca2+ transient amplitudes from wild-type and LITAF morphant hearts. Color code depicts Ca2+ transient amplitudes in fluorescence ratio units (F340/F380). Squares indicate ROIs for measurements averaged in panel B. D, Mean baseline ratios. E, Mean Ca2+ transient amplitudes. One-way ANOVA, * p<0.05 for comparisons with wild-type (n=9 wild-type zebrafish embryos; n=7 LITAF morphants). Error bars depict SEM.
Although there has been some controversy as to the activity of LITAF as a transcription factor,14, 21, 26 a number of studies have established its functional role in endosomal trafficking and multivesicular body formation.11, 12, 27 Indeed, LITAF interacts with members of the endosomal sorting complex required for transport (ESCRT), including TSG101 [via LITAF’s tetrapeptide motif, P(S/T)AP (Figure 1A)] and STAM1 (physical interaction with LITAF); recruits them to the early endosomal membrane; and controls endosome-to-lysosome trafficking and exosome formation.11 The N-terminus of LITAF contains two PXY motifs (Figure 1A), which are important for interacting with members of the NEDD4 family of HECT domain ubiquitin ligases via their WW domains.12, 28 Based on the GWAS findings5, 6 and LITAF’s functional role in endosome-to-lysosome trafficking, we hypothesized that LITAF is a candidate for regulation of cardiac excitation, likely acting as an effector of ion channel complex trafficking or degradation through lysosomes. Therefore, we set out to investigate the possible role of LITAF in the regulation of ion channels in zebrafish heart and rabbit cardiomyocytes. In this study, we present data that support a role for LITAF in modulating membrane abundance and function of voltage-gated L-type calcium channels via the ubiquitin ligase NEDD4-1 in the heart.
Methods
We declare that all supporting data are available within the article (and its online supplementary files). All animal experiments and procedures were approved by the Rhode Island Hospital Institutional Animal Care and Use Committee. Experiments performed on zebrafish (D. rerio) are in in accordance with animal protocols approved by the Harvard Medical School Institutional Animal Care and Use Committee. An expanded Methods section is found in the Data Supplement.
Results
Genetic knockdown of LITAF in larval zebrafish hearts affects calcium transients
In an effort to delineate the effects of cardiac LITAF in vivo, we designed a morpholino oligomer to the zebrafish ortholog (LITAF) (Figure 1A) targeting the ATG. In initial dose-ranging studies, there was no evidence of cardiac or systemic toxicity (data not shown). Electrophysiological studies of the ventricular myocardium revealed that a LITAF knockdown in zebrafish larvae caused a robust increase in the amplitude of [Ca2+]i transients compared to control morphants at 48 hours post fertilization (Figure 1B–E). These data suggest that LITAF modulates Ca2+ handling in zebrafish. Since the phasic [Ca2+]i transient in zebrafish largely depends on Ca2+ influx through transmembrane Ca2+ channels,29 we reasoned that LITAF may be critical to regulating voltage-gated LTCC in the heart.
Effect of LITAF overexpression on Ca2+ cycling in adult rabbit cardiomyocytes (ARbCM)
Since our in vivo observations in zebrafish embryos showed an increase in Ca2+ transients in LITAF morphants (Figure 1B–E), we expected LITAF to also interfere with intracellular Ca2+ handling in rabbit cardiomyocytes. Therefore, we utilized cultured ARbCM, which in our hands and in agreement with Tian et al.30 largely preserve t-tubules in culture (data not shown), to study the effect of adenovirally expressed LITAF on cardiac Ca2+ cycling.
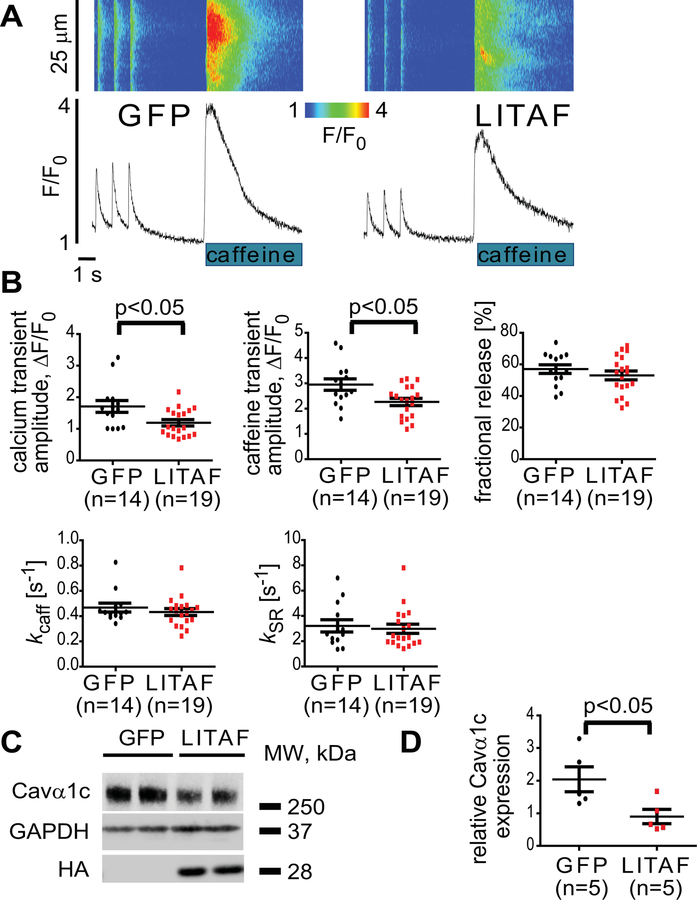
We performed live-cell confocal imaging and measured the amplitude of electrically evoked Ca2+ transients in cells overexpressing LITAF or GFP as control (Figure 2A–B). In order to determine the SR-Ca2+ load and assess NCX function, 20 mM caffeine was applied at the end of the experiments. We observed that LITAF overexpression in ARbCM significantly decreased the amplitude of Ca2+ transients (Figure 2A–B). This decrease was paralleled by a decrease in sarcoplasmic reticulum Ca2+ content assessed by rapid application of 20 mM caffeine. There were no changes in fractional release of Ca2+ (Figure 2B). NCX activity calculated by measuring the rate of decay of caffeine-induced Ca2+ transients was not different between the groups (Figure 2B). No change in SERCA2 activity was found as well (Figure 2B), which was calculated via a derived rate of decay, subtracting the rate of decay of caffeine-induced Ca2+ transients from the rate of decay of pacing-induced Ca2+ transients. This is corroborated by western blot data showing no significant changes in total levels of SERCA2, NCX, calsequestrin 2, or serine 16-phosphorylated phospholamban upon LITAF overexpression in cardiomyocytes (Figure II in the Data Supplement). Together, these data indicate that the reduction in Ca2+ transients is likely due to reduced Ca2+ influx, which is supported by a significant downregulation of the total pool of Cavα1c in LITAF-overexpressing cells (Figure 2C–D).
Figure 2.
Attenuation of Ca2+ transients and Cavα1c abundance by LITAF in adult rabbit cardiomyocytes. Cardiomyocytes were transduced with adenovirus expressing GFP or HA-LITAF. A, Representative confocal line scan images and corresponding Fluo-3 F/F0 time-dependent profiles at 1 Hz. B, Histograms depict mean data from Ca2+ transient amplitudes (GFP, 1.78±0.16 vs LITAF, 1.2±0.13 ΔF/F0), caffeine transient amplitudes (GFP, 2.8±0.15 vs LITAF, 2.1±0.19 ΔF/F0), fractional release and rates of Ca2+ removal by NCX (kcaff) and SERCA2 (kSR). Student’s t-test, p<0.05 (2–3 heart preparations). C, ARbCM lysates were probed with anti-Cavα1c, anti-HA and anti-GAPDH to indicate Cavα1c, exogenous LITAF and GAPDH (loading control) protein levels. D, Respective change in Cavα1c abundance, normalized to GAPDH (n=5 animals, performed in triplicate; mean±SEM). Student’s t-test, p<0.05.
LITAF modulates ICa,L and Cavα1c protein levels in 3-week-old cardiomyocytes (3wRbCM)
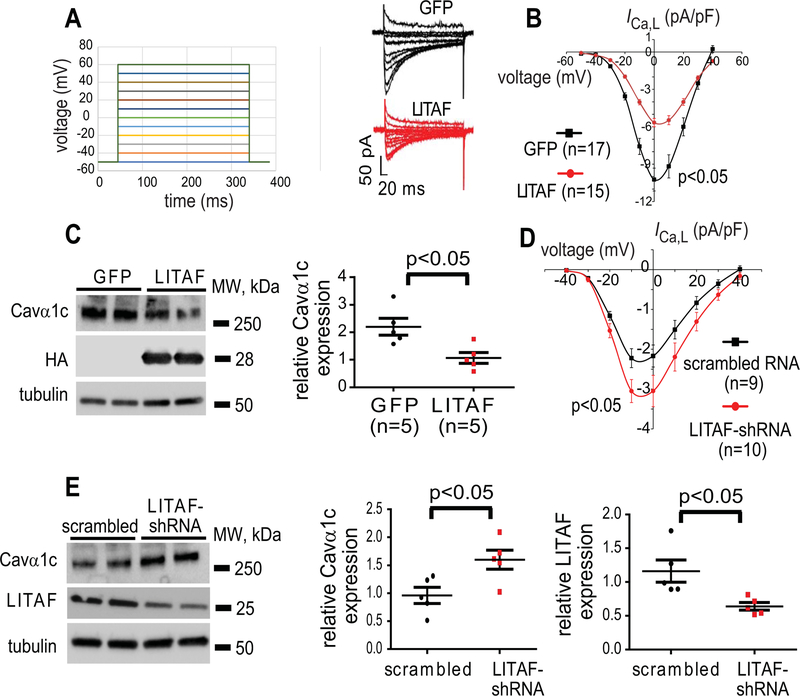
To study any effect of LITAF on Cavα1c and ICa,L in detail, we switched to cultured 3-week-old rabbit cardiomyocytes. We have recently developed this model system to investigate various ion channels underlying APD (data not shown).31 For example, these cells exhibit stable ICa,L current after 48 hours in culture (Figure 3A–B). The cells were transduced with adenovirus encoding GFP or HA-LITAF. LITAF overexpression caused a significant 45% decrease in maximal ICa,L density (from −10.2±0.9 pA/pF to −5.6±0.3 pA/pF; p<0.05) (Figure 3A–B) with no changes in voltage-dependent activation or inactivation kinetics (Figure III in the Data Supplement). Consistent with the electrophysiological data, Cavα1c protein levels were significantly downregulated by 45% in 3wRbCM overexpressing LITAF (p<0.05) (Figure 3C). By contrast, suppression of LITAF levels by shRNA resulted in a significant 40% upregulation in maximal ICa,L density (from −2.2±0.3 pA/pF to −3.1±0.4 pA/pF; p<0.05) (Figure 3D), which correlates with a significant 64% increase in Cavα1c abundance (Figure 3E) and a 30% downregulation of endogenous LITAF (Figure 3E). These data corroborate our findings in zebrafish and validate usage of both systems in the studies of LITAF effects.
Figure 3.
Control of ICa,L current and Cavα1c protein levels by LITAF in 3-week-old cardiomyocytes. Cardiomyocytes were transduced with adenovirus encoding GFP or HA-LITAF for 48 h. A, Left panel: voltage steps to measure I-V for ICa,L. Right panel: representative recordings of ICa,L in cardiomyocytes. B, Voltage dependence of ICa,L current density in cardiomyocytes transduced with GFP or LITAF (cells from 5 animals; mean±SEM; Student’s t-test, p<0.05). C, Protein levels of Cavα1c, HA-LITAF, and tubulin (left panel). Respective change in Cavα1c abundance, normalized to tubulin (n=5 animals, performed in triplicate; mean±SEM). Student’s t-test, p<0.05 (right panel). D, Mean current-voltage relationships of ICa,L peak currents for baseline conditions from cells expressing scrambled RNA or shRNA against endogenous LITAF (cells from 5 animals; mean±SEM; Student’s t-test, p<0.05). E, Protein levels of Cavα1c, total LITAF, and tubulin (left panel). Respective changes in Cavα1c and LITAF abundance, normalized to tubulin (n=5 animals, performed in triplicate) (right panel).
We also transduced neonatal rabbit cardiomyocytes with adenovirus encoding GFP (as a control) and HA-tagged LITAF and determined respective Cavα1c protein levels. Similar to ARbCM and 3wRbCM, we observed a significant downregulation of total Cavα1c protein levels by LITAF (Figure IVA in the Data Supplement). In addition, we used adenovirally expressed shRNA to knock down levels of endogenous LITAF in neonatal cardiomyocytes. After 48h, the knockdown efficiency of LITAF was approximately 60% (Figure IVB in the Data Supplement) compared to cells expressing scrambled control RNA. Lowered LITAF levels resulted in a significant, 3.5-fold, increase in total Cavα1c abundance (Figure IVB in the Data Supplement). Finally, quantitative PCR (qPCR) showed no changes in mRNA levels of LTCC in NRbCM overexpressing LITAF, ruling out transcriptional effects of LITAF on LTCC (Figure IVC in the Data Supplement).
LITAF has no significant effect on the major repolarizing K+ current, IKr in 3wRbCM.
The aforementioned GWAS5, 6 implied a role for LITAF in QT interval and therefore APD regulation. Interestingly, we have previously shown that two other genes identified in these studies, the RING finger ubiquitin ligases RNF207 and RFFL, affect a major repolarizing current in larger animals, (viz. IKr7, 9). Therefore, we set out to look for LITAF-dependent effects on this current. To this end, 3wRbCM were transduced with adenovirus encoding GFP or HA-LITAF and cultured for 48 hours. LITAF overexpression had no significant effect on IKr current (−30 mV: control: 0.72±0.13 pA/pF; LITAF: 0.77±0.14 pA/pF; p=0.87; 4 animals). Consistent with the electrophysiological data, we observed no change in the protein levels of HERG, which underlies IKr (Figure VA-B in the Data Supplement). Furthermore, our western blot data for other K+ channels, i.e. KvLQT1, Kir2.1, Kv4.2 and Kv1.4 showed no changes with LITAF overexpression (Figure VC-H in the Data Supplement). In summary, our data suggest a quite specific effect of LITAF on LTCC rather than potassium channels in cardiomyocytes.
Physical and functional interaction between LITAF and L-type Ca2+ channels in tsA201 cells
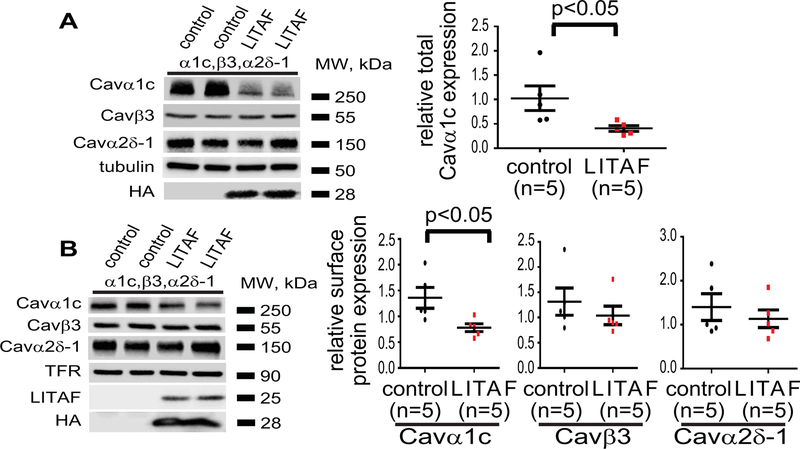
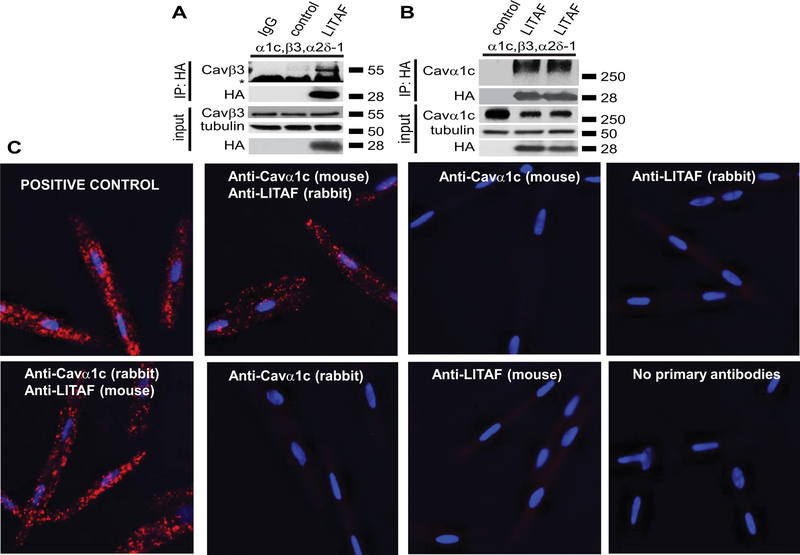
To explore the mechanisms underlying LITAF inhibition of LTCC, we used a heterologous expression system, viz. tsA201 cells, which are frequently used to study LTCC in vitro because they process the multi-subunit complex correctly and efficiently.32 We transfected tsA201 cells with expression plasmids for Cavα1c, Cavβ3, and Cavα2δ−1 to express functional LTCC and plasmid for HA-tagged LITAF or empty control plasmid. Cell-surface biotinylation was performed on transfected cells and protein levels of Cavα1c in total lysates and cell-surface fractions determined (Figure 4A–B). Overexpression of LITAF resulted in a significant downregulation of both total and surface-membrane levels of Cavα1c. In contrast, no changes in cell surface levels of Cavβ3 and Cavα2δ−1 were observed (Figure 4A–B). In order to see whether the functional interaction between LITAF and LTCC is based on a physical interaction, we performed co-immunoprecipitations with extracts from tsA201 cells co-transfected with expression plasmids for all three LTCC subunits and HA-tagged LITAF. Extracts were incubated with HA antiserum or isotype control and immunoprecipitates probed with anti-Cavβ3 (Figure 5A) or anti-Cavα1c (Figure 5B) antibody. Western blot analyses suggest that LITAF is found in a protein complex with Cavα1c as well as Cavβ3. Furthermore, to confirm co-localization between endogenous LITAF and LTCC in rabbit cardiomyocytes, we performed in situ proximity ligation assay (PLA). Representative images are shown in Figure 5. Specificity of the assay was shown by the lack of staining using mouse anti LITAF or rabbit anti-LTCC as negative controls (Figure 5C). Similarly, no signals were obtained omitting primary antibodies from the assay (Figure 5C). As a positive control for the assay, we used rabbit anti-Cavα2δ−1 and mouse anti-Cavα2δ−1 to detect endogenous Cavα2δ−1 (Figure 5C). Using the combination rabbit anti-LITAF and mouse anti-Cavα1c, the appearance of puncta suggests co-localization between LITAF and Cavα1c in cardiomyocytes (Figure 5C). Similar results were obtained using mouse anti-LITAF and rabbit anti-Cavα1c (Figure 5C).
Figure 4.
Functional interaction between LITAF and LTCC in tsA201 cells. Cells were transfected with plasmids for Cavα1c, Cavβ3, and Cavα2δ−1 to reconstitute functional LTCC, GFP, or HA-tagged LITAF. Cell-surface protein levels were determined by biotinylation: cell-surface proteins were biotinylated using sulfo-NHS-SS-biotin, purified with neutravidin beads from total cell lysates, subjected to SDS-PAGE and blotted onto a nitrocellulose membrane. A, A representative western blot shows total protein levels of Cavα1c, Cavβ3, Cavα2δ−1, HA-LITAF, and tubulin (left panel). Respective change in total Cavα1c abundance, normalized to tubulin levels (n=5, performed in triplicate; mean±SEM). Student’s t-test, p<0.05 (right panel). B, A representative western blot shows cell surface protein levels of Cavα1c, Cavβ3, Cavα2δ−1, transferrin receptor (TFR), total LITAF, and HA-LITAF (left panel). Respective changes in cell membrane protein levels of Cavα1c, Cavβ3, and Cavα2δ−1 normalized to transferrin receptor levels (n=5, performed in triplicate; mean±SEM). Student’s t-test, p<0.05 (right panel).
Figure 5.
Physical interaction between LITAF and LTCC in tsA201 cells and 3wRbCM. A, Immunoprecipitation (IP) of lysates from tsA201 cells transfected with plasmids for Cavα1c, Cavβ3, Cavα2δ−1, GFP, or HA-tagged LITAF using isotype control (lane 1) or HA antibody (lanes 2–3). A representative immunoblot against Cavβ3 shows an interaction between LITAF and the Cavβ3 subunit (IP panel) (the asterisk indicates the heavy chain of the IP capture antibody). Also shown is the immunopreciptated HA-LITAF protein. Input levels of Cavβ3, HA-LITAF, and tubulin are shown below. B, Immunoprecipitation (IP) of lysates from tsA201 cells transfected with plasmids for Cavα1c, Cavβ3, Cavα2δ−1, GFP, or HA-tagged LITAF using HA antibody. A representative immunoblot against Cavα1c shows an interaction between LITAF and the Cavα1c subunit (IP panel). Also shown is the immunopreciptated HA-LITAF protein. Input levels of Cavα1c, HA-LITAF, and tubulin are depicted below. C, Duo-link in situ PLA using rabbit anti-LITAF and mouse anti-Cavα1c antibodies (alternatively mouse anti-LITAF and rabbit anti-Cavα1c antibodies) in 3wRbCM, which are amenable to PLA and express detectable levels of LITAF and LTCC. Co-localization between molecules is indicated by red puncta. No puncta were detected in negative controls in which primary antibodies were omitted or only one antibody was used (rabbit anti-LITAF, mouse anti-Cavα1c, mouse anti-LITAF, or rabbit anti-Cavα1c antibodies). As positive control for the assay, a combination of rabbit polyclonal anti-Cavα2δ−1 and mouse monoclonal anti-Cavα2δ−1 was used to detect endogenous Cavα2δ−1. Nuclei were stained with DAPI (blue).
LITAF-dependent regulation of ubiquitination and degradation of Cavα1c in tsA201 cells
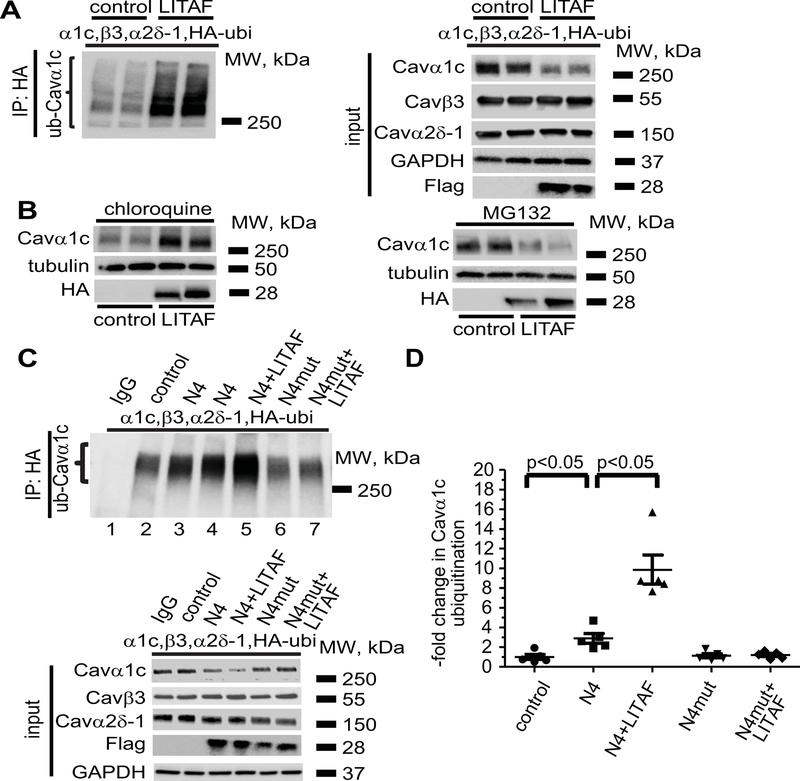
Since ubiquitination plays an important role in all three major protein degradation pathways,33 we examined whether Cavα1c was ubiquitinated in a LITAF-dependent manner. We reconstituted LTCC by transfecting tsA201 cells with expression plasmids encoding Cavα1c, Cavβ3, Cavα2δ−1, HA-tagged ubiquitin, and Flag-tagged LITAF, or empty control plasmid. Using anti-HA antibody to pull down ubiquitinated protein from cell lysates, followed by immunodetection of Cavα1c, we observed a LITAF-dependent increase in the ubiquitination level of Cavα1c (Figure 6A, left panel). Although total Cavα1c abundance was significantly reduced upon co-expression of LITAF, no changes in total Cavβ3, and Cavα2δ−1 levels were seen (Figure 6A, right panel). Next, we treated cells expressing functional LTCC and LITAF or control plasmid with chloroquine or MG132 to determine which protein degradation pathway is involved in LITAF-mediated Cavα1c downregulation. The lysosomal inhibitor chloroquine completely prevented the effect of LITAF on Cavα1c protein levels, whereas the proteasomal inhibitor MG132 did not impair downregulation of Cavα1c by LITAF (Figure 6B).
Figure 6.
LITAF-mediated ubiquitination and degradation of Cavα1c in tsA201 cells. Cells were transfected with plasmids for Cavα1c, Cavβ3, and Cavα2δ−1, HA-tagged ubiquitin, GFP as control, or Flag-tagged LITAF. Immunoprecipitation (IP) of lysates from transfected cells was performed with anti-HA antibody. A, A representative immunoblot shows levels of ubiquitinated Cavα1c (left panel) and input levels of Cavα1c, Cavβ3, Cavα2δ−1, Flag-tagged LITAF, and GAPDH (right panel). B, LITAF-mediated degradation of Cavα1c through lysosomes. Cells were transfected with plasmids for Cavα1c, Cavβ3, and Cavα2δ−1, GFP as control, or HA-tagged LITAF for 24 h and then treated with 10 μM chloroquine or 5 μM MG132 for 20 h. Representative western blots show total abundance of Cavα1c and tubulin of treated cells. C, Immunoprecipitation (IP) of lysates from cells transfected with plasmids for Cavα1c, Cavβ3, and Cavα2δ−1, HA-tagged ubiquitin, GFP as control, NEDD4-1, NEDD4-1-C867A, and/or Flag-tagged LITAF was performed with anti-HA antiserum. A representative immunoblot shows levels of ubiquitinated Cavα1c (top panel) and input levels of Cavα1c, Cavβ3, Cavα2δ−1, Flag-tagged LITAF, and GAPDH (bottom panel). D, Respective changes in the level of ubiquitinated Cavα1c, normalized to total Cavα1c (five experiments, performed in duplicate; mean±SEM). Student’s t-test, p<0.05.
Since LITAF binds to the ubiquitin ligase NEDD4-1,12 recently implicated in LTCC downregulation,34 we looked for a possible functional interaction between LITAF and NEDD4-1 with respect to ubiquitination of LTCC. To this end, we transfected tsA201 cells with expression plasmids for HA-tagged ubiquitin, Cavα1c, Cavβ3, and Cavα2δ−1, NEDD4-1, its catalytically inactive form NEDD4-1-C867A,35 Flag-tagged LITAF, or empty control plasmid. Immunoprecipitations were performed using anti-HA to pull down HA-ubiquitinated protein from cell extracts, followed by immunoblots against Cavα1c (Figure 6C–D). We found that NEDD4-1, but not NEDD4-1-C867A, increased Cavα1c ubiquitination compared to control (Figure 6C, cf. lanes 2, 3, 4, and 6). Cavα1c ubiquitination was further increased by co-expressed NEDD4-1 and LITAF (cf. lanes 3, 4, and 5). However, LITAF was not able to enhance Cavα1c ubiquitination when NEDD4-1-C867A was co-expressed (cf. lanes 6 and 7), suggesting that LITAF probably activates NEDD4-1, the ubiquitin ligase responsible for Cavα1c ubiquitination and degradation.
Functional interaction between LITAF, the ubiquitin ligase NEDD4-1 and L-type Ca2+ channels in 3-week-old cardiomyocytes
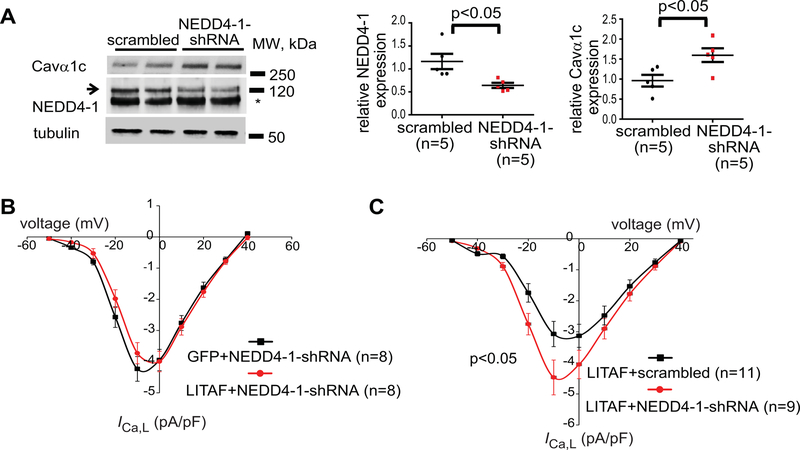
Since our data obtained from tsA201 cells suggest a possible functional interaction between LITAF and NEDD4-1 ubiquitin ligase in LTCC degradation, we designed adenovirus expressing shRNA against rabbit NEDD4-1. Transducing 3wRbCM with NEDD4-1 shRNA, endogenous NEDD4-1 protein was significantly downregulated and a concomitant increase in total Cavα1c levels observed (Figure 7A). To assess a possible requirement for NEDD4-1 in LITAF-mediated downregulation of LTCC in 3wRbCM, we transduced cells with adenovirus expressing GFP and NEDD4-1 shRNA (control) or LITAF and NEDD4-1 shRNA. ICa,L densities in cardiomyocytes expressing both LITAF and NEDD4-1 shRNA and cells expressing NEDD4-1 shRNA and GFP were virtually identical (Figure 7B), implying that NEDD4-1 is crucial for LITAF-mediated ICa,L regulation. To corroborate our findings in 3wRbCM, we also transduced cells with adenovirus expressing LITAF and scrambled RNA (control) or adenovirus expressing LITAF and NEDD4-1 shRNA (Figure 7C). In the presence of co-expressed NEDD4-1 shRNA, LITAF overexpression caused a significant 45% increase in peak ICa,L density compared to the control (from −3.1±0.4 pA/pF to −4.5±0.5 pA/pF; p<0.05), suggesting that the negative effect of LITAF on ICa,L densities requires the presence of NEDD4-1.
Figure 7.
NEDD4-1-dependent downregulation of LTCC by LITAF in 3-week-old rabbit cardiomyocytes. A, Protein levels of total Cavα1c, NEDD4-1, and tubulin in cells expressing scrambled control RNA or shRNA against endogenous NEDD4-1 (left panel; the asterisk indicates an unspecific band). Respective changes in NEDD4-1 and Cavα1c abundance, normalized to tubulin (n=5 animals, performed in triplicate; mean±SEM). Student’s t-test, p<0.05 (right panel). B, Mean current-voltage relationships of ICa,L peak currents for baseline conditions from cells expressing GFP and shRNA against endogenous NEDD4-1 (control) or LITAF and NEDD4-1 shRNA. C, 3wRbCM were transduced with adenovirus expressing scrambled RNA and LITAF (control) or LITAF and shRNA against endogenous NEDD4-1. Mean current-voltage relationships of ICa,L peak currents for baseline conditions from respective cells are depicted (cells from 5 animals; mean±SEM; Student’s t-test, p<0.05).
A decrease of ICa,L is responsible for shortening of APD and decreasing Ca2+ transient amplitude in myocytes with LITAF overexpression
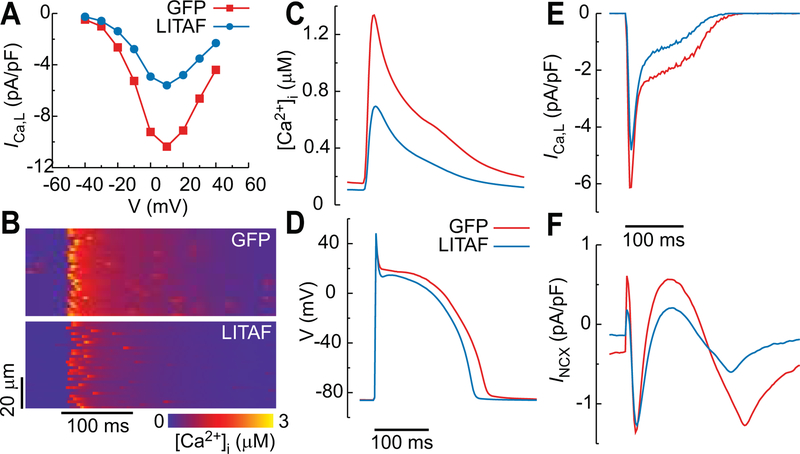
To study the causal relationship between LITAF overexpression and changes of electrophysiological phenotype, we used a physiologically detailed computational model of rabbit ventricular myocyte with membrane voltage coupled to spatially distributed subcellular Ca2+ dynamics. Details of the model are provided in the Data Supplement. We first determined the ICa,L current conductances in the model that reproduce the ICa,L peak current versus voltage curves measured in voltage-clamp mode (Figure 3B) for myocytes with GFP and LITAF. We found that reducing the number of functional sarcolemmal LTCC from 4 to 2 in each calcium release unit (CRU), where LTCCs are co-localized with RyR2 Ca2+ release channels, reproduced the approximately 50% reduction of whole-cell ICa,L current with LITAF compared to GFP. The whole-cell current is the summation of LTCCs from ~16,000 CRUs spatially distributed throughout the cell. Results in Figure 8A show that, with this 50% reduction in the total number of LTCCs, the model reproduces well the quantitative voltage-clamp measurements of Figure 3B. We then paced the myocytes at a 2.5 Hz in current clamp mode for ICa,L conductances (total number of LTCCs) corresponding to GFP and LITAF. The results show a 50% decrease of ICa,L conductance, resulting from LITAF overexpression, significantly reduces both local Ca2+ release (confocal linescan equivalent in Figure 8B) and whole cell Ca2+ transient amplitude (Figure 8C). It also shortens APD from 206 ms with GFP to 182 ms with LITAF (Figure 8D). The decrease of APD appears relatively small in view of the large decrease of ICa,L current during the action potential plateau phase (Figure 8E). Examination of other currents reveals that a shift of Na+/Ca2+ exchanger (NCX) current towards reverse mode during the AP plateau partly counterbalances the effect of decreased ICa,L current on APD. This shift is associated with a decrease of steady-state intracellular Na+ concentration [Na+]i (10.8 mM with GFP versus 9.4 mM with LITAF). Reduced [Na+]i with LITAF then promotes forward mode NCX current (Figure 8F), thereby prolonging APD and partly counterbalancing the effect of ICa,L reduction on APD shortening. We also note that, on the contrary, the reduced Ca2+ transient amplitude with LITAF promotes reverse mode NCX, but this effect is not as significant as the shift towards forward mode caused by [Na+]i reduction. LITAF overexpression in 3wRbCM showed a similar (insignificant) APD shortening (mean ventricular APD90 ± S.E.M.: LITAF 189±19 ms (n=21) vs. GFP 224±15 ms (n=25), p=0.29). Power analysis determined that we would need 122 cells from each group to achieve statistical significant results. Of note, morpholino-mediated downregulation of LITAF in zebrafish resulted in APD prolongation that did not reach statistical significance (mean ventricular APD80 ± S.E.M.: morphants 283±10 ms vs. WT 247±9 ms, p=0.11, n=7 and 9).
Figure 8.
Computer simulation of rabbit cardiomyocytes transduced with adenovirus encoding GFP and LITAF. A, ICa,L current versus clamped voltage. B-F, Current-clamp stimulation at 2.5 Hz: confocal linescan equivalent (B), cytosolic calcium concentration (C), action potential (D), ICa,L current (E), and NCX current (F).
Discussion
LITAF regulates endosomal trafficking11–13 and inflammatory cytokines,14, 15 and acts an adapter molecule for members of the NEDD4-like family of E3 ubiquitin ligases.16, 17 Sequence variation in LITAF (rs8049607) is associated with QT interval prolongation5, 6 and reduced LITAF mRNA expression in the left ventricle (Figure I in the Data Supplement).20 The present study provides the first empiric evidence that LITAF exerts an inhibitory effect on the abundance and function of cardiac LTCC, in part, by controlling NEDD4-1 ubiquitin ligases. Knockdown of LITAF in zebrafish larvae resulted in a robust increase in cardiomyocyte calcium transients on Fura-2 imaging. Overexpressed LITAF in 3-week-old rabbit cardiomyocytes resulted in a decrease in ICa,L and Cavα1c protein levels, whereas a LITAF knockdown increased ICa,L and Cavα1c protein levels. We observed a decrease in calcium transients in LITAF-overexpressing adult rabbit cardiomyocytes, which was accompanied by lower Cavα1c abundance. In tsA201 cells, overexpressed LITAF downregulated the total and surface pools of Cavα1c via increased Cavα1c ubiquitination and its subsequent lysosomal degradation. Co-immunoprecipitation showed that LITAF formed a complex with LTCC. Furthermore, in situ proximity ligation assay indicated co-localization between LITAF and LTCC in cardiomyocytes. In tsA201 cells, NEDD4-1, but not its catalytically inactive form NEDD4-1-C867A, increased Cavα1c ubiquitination compared to control. Cavα1c ubiquitination was further increased by co-expressed LITAF and NEDD4-1 but not NEDD4-1-C867A. Knockdown of NEDD4-1 using shRNA abolished the negative effect of LITAF on ICa,L and Cavα1c protein levels in 3-week-old rabbit cardiomyocytes. Computer simulations demonstrated that a decrease of ICa,L current associated with LITAF overexpression simultaneously shortened APD and decreased Ca2+ transient amplitude in rabbit ventricular myocytes.
LITAF: tissue and substrate specificity
Currently, we cannot rule out that LITAF/NEDD4-1-mediated downregulation of LTCC exists in other tissues, such as smooth muscle, somatodendritic neurons or endocrine cells, which also express Cavα1c at significant levels,36 as LITAF and NEDD4-1 are ubiquitously expressed proteins.37 Thus it is conceivable the reported SNP may also result in changes of ICa,L in non-cardiac tissue. Similarly, LITAF may also negatively affect forward trafficking of other α1-subunit isoforms that require a beta auxiliary subunit for surface expression, for example Cav1.3 and Cav2.3,38, 39 which are also expressed in the heart. In contrast, T-type calcium channels, which do not require a beta subunit for their function40 are likely not controlled by LITAF. Further studies are warranted to explore these possibilities.
NEDD4-1-mediated effects of LITAF on Cavα1c protein levels
The NEDD4 family of HECT ubiquitin ligases contains nine members,17 which are all expressed in the heart. Most studies have focused on NEDD4-1 and NEDD4-2, and a plethora of potential targets have been identified in vitro.17 Not surprisingly, various ion channels have been reported to be ubiquitinated by these ubiquitin ligases. For example, voltage-gated potassium (HERG41) or sodium channels (Nav1.542) are ubiquitinated by NEDD4-2, resulting in their lysosomal degradation. Interestingly, a recent report by Viard and colleagues34 suggests that NEDD4-1 promotes downregulation of newly synthesized Cavα1c at the endoplasmic reticulum/Golgi level in tsA201 cells. Prompted by these findings, we looked into the possibility that LITAF-dependent downregulation requires NEDD4-1. Here, we present data that clearly show that upon knockdown of endogenous NEDD4-1, the effect of LITAF on ICa,L is completely abolished in cardiomyocytes (Figure 7B), thus confirming an hypothesized requirement of a NEDD4-1-dependent polyubiquitination of Cavα1c. Prior authors did not observe any NEDD4-1-dependent increase in Cavα1c ubiquitination, in contrast to our findings of increased ubiquitination level of Cavα1c by NEDD4-1 and/or LITAF (Figure 6A, C & D). These differences likely reflect the discrete experimental conditions used. For example, in order to amplify the ubiquitination signal, we co-transfected tsA201 cells with an expression plasmid for HA-tagged ubiquitin. Additionally, we added 10 mM N-ethylmaleimide and iodocetamide to the lysis buffer to block reactive cysteines and thus prevent deubiquitination of proteins by deubiquitinases during sample processing.43 Notably, a significant LITAF-dependent increase in the overall ubiquitination level of total protein isolated from neonatal rabbit cardiomyocytes (NRbCM) was observed (data not shown). In agreement with the study by Rougier et al.34 who reported that Cavβ2 was essential for NEDD4-1-mediated regulation of Cavα1c in tsA201 cells, we observed an absolute requirement for the accessory subunit Cavβ3 (or Cavβ2; data not shown) in LITAF-mediated downregulation of Cavα1c in the same cell line (data not shown). This is corroborated by our co-IP findings in tsA201 cells, demonstrating that LITAF was found in a protein complex with Cavα1c as well as Cavβ3 (Figure 5A–B) or Cavβ2 (data not shown), and by our in situ proximity ligation assay, which demonstrates co-localization between LITAF and LTCC in cardiomyocytes (Figure 5C).
As shown in Figure 6B, the lysosomal inhibitor chloroquine but not the proteasomal inhibitor MG132 blocked LITAF-dependent Cavα1c downregulation. Experiments in tSA201 cells (Figure 6A) also show Cavα1c polyubiquitination, indicated by the high molecular smear of ubiquitinated Cavα1c, in the presence of LITAF. Polyubiquitination linked through Lys48 or Lys11 generally leads to proteasomal degradation. In contrast, Lys63 linkages perform non-degradative roles (e.g. cell signaling) but are also required for lysosomal degradation.44 Thus, we hypothesize that LITAF causes NEDD4-mediated Lys63 polyubiquitination of Cavα1c.
Contrary to the study by Rougier et al.,34 we did not notice a LITAF-dependent downregulation of the accessory subunits Cavβ3 (Cavβ2) and Cavα2δ−1. This scenario is reminiscent of the selective degradation of the B′β subunit of protein phosphatase 2A (PP2A) mediated via KLHL15, a ubiquitin ligase adapter.45 Although KLHL15 interacted with both the core AC dimer and the B′β monomer of PP2A, only the B′β subunit was ubiquitinated and degraded. It is conceivable that structural requirements mediated through LITAF could favor an exclusive ubiquitination of Cavα1c by NEDD4. Furthermore, it has been shown that both β and α2δ−1subunits can reach the membrane in the absence of alpha subunits.46–49 This could explain that no changes in respective subunits are seen on the membrane despite lower Cavα1c levels (Figure 4B).
LTCC levels on the membrane are determined by forward trafficking from the ER/Golgi apparatus as well as by channel internalization and recycling of endocytosed channel. One could entertain the possibility that LITAF provides NEDD4-mediated LTCC quantity control on the Golgi. Cardiomyocytes must maintain a sufficient number of LTCC channels on the membrane required for proper cardiac excitation. In contrast, limiting LTCC channels on the cell surface is required to prevent excessive calcium entry into the cell, a hallmark of cardiac dysfunction. We anticipate that regulatory pathways exist that allow tight control of LTCC forward trafficking by regulating stability and/or expression of LITAF. Such pathways could be adversely affected by certain cardiac diseases subsequently impacting ICa,L.
Regulation of NEDD4-1 ubiquitin ligase by LITAF
LITAF is a small zinc-binding monotopic membrane protein25 found on the Golgi apparatus and multivesicular bodies.12 It contains two N-terminal PXY motifs, which are required for physical interaction with WW-domains of the HECT ubiquitin ligases NEDD4-112 and ITCH.28 We demonstrated that LITAF interacts and co-localizes with LTCC subunits (Figure 5A–C) and forms a complex with LTCC. Previous studies have already observed co-localization of LITAF with NEDD4-1 on the Golgi apparatus12 and control of Cavα1c levels by NEDD4-1 at the ER/Golgi level.34 Therefore, it is very likely that the LITAF-mediated ubiquitination of Cavα1c is happening in the TGN (Figure VI in the Data Supplement). It is also conceivable that the physical adaptor LITAF promotes NEDD4-1 ubiquitination activity and recruits NEDD4-1 to its Cavα1c substrate on TGN membranes (Figure VI in the Data Supplement). Such a scenario is reminiscent of the small NEDD4 family-interacting proteins, NDFIP1 and NDFIP2.17 They contain cytoplasmic PXY motifs and are localized to the Golgi apparatus, endosomes and multivesicular bodies. Previously, NDFIP1 was shown to recruit various NEDD4 ubiquitin ligases to membranes, promote autoubiquitination of these ubiquitin ligases by relieving their inherent autoinhibition and induce substrate ubiquitination.50 Similarly, a recent study by Kang et al.51 revealed that NDFIP1 recruits NEDD4-2 to the Golgi apparatus to mediate polyubiquitination-dependent degradation of HERG. It is interesting to note that both LITAF and NDFIP1 have been reported to promote exosome secretion and both molecules could be detected in exosomes produced by cells overexpressing LITAF or NDFIP1, respectively.27, 52 Surprisingly, a LITAF double mutant harboring mutations in both PXY motifs (LITAF-Y23A-Y61A) behaved like wild-type LITAF regarding downregulation of ICa,L in cardiomyocytes (data not shown). We therefore believe that a direct physical interaction between LITAF and NEDD4 is not an absolute requirement for the observed LITAF-dependent downregulation of LTCC by NEDD4. One could envisage that both proteins are found in a larger multi-protein complex. Alternatively, despite the two PXY mutations, residual physical interaction between NEDD4 and LITAF could be sufficient for LTCC ubiquitination.
Transcriptional effects of LITAF
Although LITAF has recently been shown to target intracellular membranes,25 which would support its purported role in endosomal trafficking,11, 12 there have been numerous studies implicating LITAF, which is highly expressed in monocytes, macrophages, lymph nodes, and spleen,53 as an important mediator in systemic and chronic inflammation.54 Earlier studies identified LITAF as an LPS-induced transcription factor for TNF-α.55 Despite some controversy regarding these findings,14 recent studies using highly specific chromatin immunoprecipitation, confirmed LITAF’s role as a transcription factor. For example, LPS-induced LITAF acts as a transcriptional activator for TNF-α, which mediates a pro-inflammatory and pro-fibrogenic pattern in non-alcoholic fatty liver disease.26 We therefore entertained the possibility of LITAF-dependent transcriptional effects on Cavα1c expression. However, our qPCR data clearly indicated that Cavα1c transcript levels were not changed on LITAF overexpression in neonatal cardiomyocytes (Figure IVC in the Data Supplement), despite significantly decreased Cavα1c protein levels (Figure IVA in the Data Supplement). Currently, we cannot rule out any possible LITAF-mediated transcriptional effects on other ion channels or molecules underlying AP formation. Of note, we have not observed any significant changes in the total protein levels of HERG, KvLQT1, NCX, SERCA2 and PLN in LITAF-overexpressing cells (Figures IIA–B and VA-H in the Data Supplement).
LITAF, a potential candidate to alter QT interval
Based on the effects we have observed of LITAF on calcium transients in zebrafish (Figure 1), which largely depend on Ca2+ influx through transmembrane Ca2+ channels,29 and ICa,L in rabbit cardiomyocytes (Figure 3), we propose to add genetic variants of LITAF to the growing list of genetic risk factors for sporadic or drug-induced arrhythmias. Identification and expansion of molecular risk factors could help to identify ‘vulnerable’ patients and aid in the development of new strategies targeting individuals with a prolonged QT interval. The high degree of sequence conservation among known LITAF orthologs56 implies a conservation of function during vertebrate evolution from bony fish to mammals. Here, we provide strong evidence for a role of LITAF in regulation of calcium transients in zebrafish embryos (Figure 1), and in the control of LTCC levels on the membrane of rabbit cardiomyocytes (Figures 2–3). The association between LITAF and QT interval variation has been reported in various GWAS.5, 6 Indeed, computer simulation of rabbit ventricular myocytes predicted a modest APD shortening in the presence of overexpressed LITAF, primarily because of the compensatory response of NCX. We observed similar magnitude of shortening of the APD in 3wRbCM that did not reach statistical significance in the setting of large variance in the APD of cultured myocytes possibly due to cell-to-cell variability in remodeling of ion channels in culture. Complementing these experiments, LITAF knockdown in zebrafish resulted in prolongation APD that did not reach statistical significance. These observations may reflect a role for LITAF in damping dynamic changes in excitation similar to those reported for other regulators of membrane protein turnover. In this framework, changes in LITAF activity in either direction create vulnerabilities to other modulators of APD whether genetic or environmental.
Conclusions
Here, we provide both in vivo and in vitro data that imply a role for LITAF in the regulation of LTCC in the heart through modulation of the activity of the HECT ubiquitin ligase NEDD4-1. The end result is ubiquitination and subsequent lysosomal degradation of Cavα1c (Figure VI in the Data Supplement). This, in turn, results in lower LTCC levels on the cell surface and decreased ICa,L of cardiomyocytes. We conclude that LITAF controls membrane levels and function of LTCC and is a novel regulator of cardiac excitation.
Supplementary Material
Acknowledgments:
The authors are indebted to Dr. Diane Lipscombe (Department of Neuroscience, Brown University, Providence, RI, USA) for the Cav1.2, Cavb3, and CaVα2δ1 plasmids, Dr. Joan Massagué (Cancer Biology and Genetics Program, Memorial Sloan Kettering Cancer Center, New York, NY, USA) for NEDD4-1 and NEDD4-1-C867A plasmids and Dr. Ted Dawson (The Solomon H Snyder Department of Neuroscience, Johns Hopkins University, Baltimore, MD, USA) for the HA-tagged ubiquitin plasmid.
Sources of Funding: This work was supported by the National Institutes of Health [R01-HL-096669 to B.-R.C., T32HL094300 to G.K., R01HL110791 to G.K., R01HL134706 to G.K.]; and the Rhode Island Foundation [701-7131778 to K.S.M.].
Non-standard Abbreviations and Acronyms
- APD
Action potential duration
- GWAS
Genome-wide association studies
- LITAF
Lipopolysaccharide-induced tumor necrosis factor
- 3wRbCM
3-week-old rabbit cardiomyocytes
- ARbCM
Adult rabbit cardiomyocytes
- LTCC
L-type calcium channel
- ICa,L
LTCC current
- NEDD4
Neural precursor cell expressed developmentally down-regulated protein 4
- TGN
trans-Golgi-network
- ER
Endoplasmic Reticulum
- RNF207
Ring finger protein 207
- RFFL
Ring finger and FYVE-like domain containing E3 ubiquitin protein ligase
- eQTL
Expression quantitative trait loci
- CMT
Charcot-Marie-Tooth disease
- ESCRT
Endosomal sorting complex required for transport
- NCX
Sodium-calcium exchanger
- SERCA
Sarco/endoplasmic reticulum Ca2+-ATPase
- GFP
Green fluorescence protein
- HA
Hemagglutinin
- PLA
in situ proximity ligation assay
- [Na+]i
Intracellular Na+ concentration
- NRbCM
Neonatal rabbit cardiomyocytes
- NDFIP1
Nedd4 family interacting protein 1
- TNF-α
Tumor necrosis alpha factor
- qPCR
Quantitative polymerase chain reaction
- HERG
Human Ether-à-go-go-Related Gene
- HECT
Homologous to the E6-AP Carboxyl Terminus
Footnotes
Disclosures: None.
References
- 1.Scott PM, Bilodeau PS, Zhdankina O, Winistorfer SC, Hauglund MJ, Allaman MM, Kearney WR, Robertson AD, Boman AL, Piper RC. Gga proteins bind ubiquitin to facilitate sorting at the trans-golgi network. Nat Cell Biol 2004;6:252–259. [DOI] [PubMed] [Google Scholar]
- 2.Puertollano R, Bonifacino JS. Interactions of gga3 with the ubiquitin sorting machinery. Nat Cell Biol 2004;6:244–251. [DOI] [PubMed] [Google Scholar]
- 3.Abriel H, Staub O. Ubiquitylation of ion channels. Physiology (Bethesda) 2005;20:398–407. [DOI] [PubMed] [Google Scholar]
- 4.Gomez-Navarro N, Miller E. Protein sorting at the er-golgi interface. J Cell Biol 2016;215:769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newton-Cheh C, Eijgelsheim M, Rice K, de Bakker P, Yin X, Estrada K, Bis J, Marciante K, Rivadeneira F, Noseworthy P, et al. Common variants at ten loci influence qt interval duration in the qtgen study. Nat Genet 2009;41:399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeufer A, Sanna S, Arking DE, Müller M, Gateva V, Fuchsberger C, Ehret GB, Orrú M, Pattaro C, Köttgen A, et al. Common variants at ten loci modulate the qt interval duration in the qtscd study. Nat Genet 2009;41:407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roder K, Werdich AA, Li W, Liu M, Kim TY, Organ-Darling LE, Moshal KS, Hwang JM, Lu Y, Choi BR, et al. Ring finger protein rnf207, a novel regulator of cardiac excitation. J Biol Chem 2014;289:33730–33740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizushima W, Takahashi H, Watanabe M, Kinugawa S, Matsushima S, Takada S, Yokota T, Furihata T, Matsumoto J, Tsuda M, et al. The novel heart-specific ring finger protein 207 is involved in energy metabolism in cardiomyocytes. J Mol Cell Cardiol 2016;100:43–53. [DOI] [PubMed] [Google Scholar]
- 9.Roder K, Kabakov A, Moshal KS, Murphy KR, Xie A, Dudley S, Turan NN, Lu Y, MacRae CA, Koren G. Trafficking of the human ether-a-go-go-related gene (herg) potassium channel is regulated by the ubiquitin ligase rififylin (rffl). J Biol Chem 2019;294:351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gopalakrishnan K, Morgan EE, Yerga-Woolwine S, Farms P, Kumarasamy S, Kalinoski A, Liu X, Wu J, Liu L, Joe B. Augmented rififylin is a risk factor linked to aberrant cardiomyocyte function, short-qt interval and hypertension. Hypertension 2011;57:764–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SM, Chin LS, Li L. Charcot-marie-tooth disease-linked protein simple functions with the escrt machinery in endosomal trafficking. J Cell Biol 2012;199:799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shirk AJ, Anderson SK, Hashemi SH, Chance PF, Bennett CL. Simple interacts with nedd4 and tsg101: Evidence for a role in lysosomal sorting and implications for charcot-marie-tooth disease. J Neurosci Res 2005;82:43–50. [DOI] [PubMed] [Google Scholar]
- 13.Chin LS, Lee SM, Li L. Simple: A new regulator of endosomal trafficking and signaling in health and disease. Commun Integr Biol 2013;6:e24214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang X, Metzger D, Leeman S, Amar S. Lps-induced tnf-alpha factor (litaf)-deficient mice express reduced lps-induced cytokine: Evidence for litaf-dependent lps signaling pathways. Proc Natl Acad Sci U S A 2006;103:13777–13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merrill JC, You J, Constable C, Leeman SE, Amar S. Whole-body deletion of lps-induced tnf-α factor (litaf) markedly improves experimental endotoxic shock and inflammatory arthritis. Proc Natl Acad Sci U S A 2011;108:21247–21252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingham RJ, Gish G, Pawson T. The nedd4 family of e3 ubiquitin ligases: Functional diversity within a common modular architecture. Oncogene 2004;23:1972–1984. [DOI] [PubMed] [Google Scholar]
- 17.Yang B, Kumar S. Nedd4 and NEDD4-2: Closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death Differ 2010;17:68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, Fernandez Banet J, Billis K, García Girón C, Hourlier T, et al. The ensembl gene annotation system. Database (Oxford) 2016;2016. [DOI] [PMC free article] [PubMed]
- 19.Lek Wen TW, Anene G, Wong E, Tan HS, Pan B, Lee M, Autio M, Morley M, Margulies K, Cappola T, et al. Disease and phenotype relevant genetic variants identified from histone acetylomes in human hearts. bioRxiv 2019:536763.
- 20.Consortium G The genotype-tissue expression (gtex) project. Nat Genet 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, Bennett CL. Litaf/simple protein is increased in intestinal tissues from patients with cd and uc, but is unlikely to function as a transcription factor. Inflamm Bowel Dis 2007;13:120–121. [DOI] [PubMed] [Google Scholar]
- 22.Street VA, Bennett CL, Goldy JD, Shirk AJ, Kleopa KA, Tempel BL, Lipe HP, Scherer SS, Bird TD, Chance PF. Mutation of a putative protein degradation gene litaf/simple in charcot-marie-tooth disease 1c. Neurology 2003;60:22–26. [DOI] [PubMed] [Google Scholar]
- 23.Gerding WM, Koetting J, Epplen JT, Neusch C. Hereditary motor and sensory neuropathy caused by a novel mutation in litaf. Neuromuscul Disord 2009;19:701–703. [DOI] [PubMed] [Google Scholar]
- 24.Lacerda AF, Hartjes E, Brunetti CR. Litaf mutations associated with charcot-marie-tooth disease 1c show mislocalization from the late endosome/lysosome to the mitochondria. PLoS One 2014;9:e103454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho AK, Wagstaff JL, Manna PT, Wartosch L, Qamar S, Garman EF, Freund SM, Roberts RC. The topology, structure and pe interaction of litaf underpin a charcot-marie-tooth disease type 1c. BMC Biol 2016;14:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ceccarelli S, Panera N, Mina M, Gnani D, De Stefanis C, Crudele A, Rychlicki C, Petrini S, Bruscalupi G, Agostinelli L, et al. Lps-induced tnf-α factor mediates pro-inflammatory and pro-fibrogenic pattern in non-alcoholic fatty liver disease. Oncotarget 2015;6:41434–41452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu H, Guariglia S, Yu RY, Li W, Brancho D, Peinado H, Lyden D, Salzer J, Bennett C, Chow CW. Mutation of simple in charcot-marie-tooth 1c alters production of exosomes. Mol Biol Cell 2013;24:1619–1637, S1611–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eaton HE, Desrochers G, Drory SB, Metcalf J, Angers A, Brunetti CR. Simple/litaf expression induces the translocation of the ubiquitin ligase itch towards the lysosomal compartments. PLoS One 2011;6:e16873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bovo E, Dvornikov AV, Mazurek SR, de Tombe PP, Zima AV. Mechanisms of ca2+ handling in zebrafish ventricular myocytes. Pflugers Arch 2013;465:1775–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian Q, Pahlavan S, Oleinikow K, Jung J, Ruppenthal S, Scholz A, Schumann C, Kraegeloh A, Oberhofer M, Lipp P, et al. Functional and morphological preservation of adult ventricular myocytes in culture by sub-micromolar cytochalasin d supplement. J Mol Cell Cardiol 2012;52:113–124. [DOI] [PubMed] [Google Scholar]
- 31.Kabakov AY, Moshal K, Lu Y, Roder K, Nilufer T, Li W, Murphy K, Terentyev D, Koren G. 3-week-old rabbit cardiomyocytes (3wrbcm): A novel cellular model for studying cardiac excitation. Biophysical Journal 2019;116:230a. [Google Scholar]
- 32.McHugh D, Sharp EM, Scheuer T, Catterall WA. Inhibition of cardiac l-type calcium channels by protein kinase c phosphorylation of two sites in the n-terminal domain. Proc Natl Acad Sci U S A 2000;97:12334–12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clague MJ, Urbé S. Ubiquitin: Same molecule, different degradation pathways. Cell 2010;143:682–685. [DOI] [PubMed] [Google Scholar]
- 34.Rougier JS, Albesa M, Abriel H, Viard P. Neuronal precursor cell-expressed developmentally down-regulated 4–1 (NEDD4-1) controls the sorting of newly synthesized ca(v)1.2 calcium channels. J Biol Chem 2011;286:8829–8838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao S, Alarcón C, Sapkota G, Rahman S, Chen PY, Goerner N, Macias MJ, Erdjument-Bromage H, Tempst P, Massagué J. Ubiquitin ligase nedd4l targets activated smad2/3 to limit tgf-beta signaling. Mol Cell 2009;36:457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Striessnig J, Pinggera A, Kaur G, Bock G, Tuluc P. L-type ca. Wiley Interdiscip Rev Membr Transp Signal 2014;3:15–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 2014;13:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei SK, Colecraft HM, DeMaria CD, Peterson BZ, Zhang R, Kohout TA, Rogers TB, Yue DT. Ca(2+) channel modulation by recombinant auxiliary beta subunits expressed in young adult heart cells. Circ Res 2000;86:175–184. [DOI] [PubMed] [Google Scholar]
- 39.Berrow NS, Campbell V, Fitzgerald EM, Brickley K, Dolphin AC. Antisense depletion of beta-subunits modulates the biophysical and pharmacological properties of neuronal calcium channels. J Physiol 1995;482 ( Pt 3):481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leuranguer V, Bourinet E, Lory P, Nargeot J. Antisense depletion of beta-subunits fails to affect t-type calcium channels properties in a neuroblastoma cell line. Neuropharmacology 1998;37:701–708. [DOI] [PubMed] [Google Scholar]
- 41.Guo J, Wang T, Li X, Shallow H, Yang T, Li W, Xu J, Fridman MD, Yang X, Zhang S. Cell surface expression of human ether-a-go-go-related gene (herg) channels is regulated by caveolin-3 protein via the ubiquitin ligase NEDD4-2. J Biol Chem 2012;287:33132–33141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Bemmelen M, Rougier J, Gavillet B, Apothéloz F, Daidié D, Tateyama M, Rivolta I, Thomas M, Kass R, Staub O, et al. Cardiac voltage-gated sodium channel nav1.5 is regulated by NEDD4-2 mediated ubiquitination. Circ Res 2004;95:284–291. [DOI] [PubMed] [Google Scholar]
- 43.Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep 2009;10:1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swatek KN, Komander D. Ubiquitin modifications. Cell Res 2016;26:399–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oberg EA, Nifoussi SK, Gingras AC, Strack S. Selective proteasomal degradation of the b’β subunit of protein phosphatase 2a by the e3 ubiquitin ligase adaptor kelch-like 15. J Biol Chem 2012;287:43378–43389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bogdanov Y, Brice NL, Canti C, Page KM, Li M, Volsen SG, Dolphin AC. Acidic motif responsible for plasma membrane association of the voltage-dependent calcium channel beta1b subunit. Eur J Neurosci 2000;12:894–902. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi SX, Mittman S, Colecraft HM. Distinctive modulatory effects of five human auxiliary beta2 subunit splice variants on l-type calcium channel gating. Biophys J 2003;84:3007–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chien AJ, Carr KM, Shirokov RE, Rios E, Hosey MM. Identification of palmitoylation sites within the l-type calcium channel beta2a subunit and effects on channel function. J Biol Chem 1996;271:26465–26468. [DOI] [PubMed] [Google Scholar]
- 49.Davies A, Douglas L, Hendrich J, Wratten J, Tran Van Minh A, Foucault I, Koch D, Pratt WS, Saibil HR, Dolphin AC. The calcium channel alpha2delta-2 subunit partitions with cav2.1 into lipid rafts in cerebellum: Implications for localization and function. J Neurosci 2006;26:8748–8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mund T, Pelham HR. Control of the activity of ww-hect domain e3 ubiquitin ligases by ndfip proteins. EMBO Rep 2009;10:501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang Y, Guo J, Yang T, Li W, Zhang S. Regulation of the human ether-a-go-go-related gene (herg) potassium channel by nedd4 family interacting proteins (ndfips). Biochem J 2015;472:71–82. [DOI] [PubMed] [Google Scholar]
- 52.Putz U, Howitt J, Lackovic J, Foot N, Kumar S, Silke J, Tan SS. Nedd4 family-interacting protein 1 (ndfip1) is required for the exosomal secretion of nedd4 family proteins. J Biol Chem 2008;283:32621–32627. [DOI] [PubMed] [Google Scholar]
- 53.Myokai F, Takashiba S, Lebo R, Amar S. A novel lipopolysaccharide-induced transcription factor regulating tumor necrosis factor alpha gene expression: Molecular cloning, sequencing, characterization, and chromosomal assignment. Proc Natl Acad Sci U S A 1999;96:4518–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zou J, Guo P, Lv N, Huang D. Lipopolysaccharide-induced tumor necrosis factor-α factor enhances inflammation and is associated with cancer (review). Mol Med Rep 2015;12:6399–6404. [DOI] [PubMed] [Google Scholar]
- 55.Tang X, Marciano DL, Leeman SE, Amar S. Lps induces the interaction of a transcription factor, lps-induced tnf-alpha factor, and stat6(b) with effects on multiple cytokines. Proc Natl Acad Sci U S A 2005;102:5132–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin W, Wunderley L, Barrett AL, High S, Woodman PG. The charcot marie tooth disease protein litaf is a zinc-binding monotopic membrane protein. Biochem J 2016;473:3965–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.