Abstract
Astrocytes are susceptible to HIV infection and potential latent HIV reservoirs. Tat is one of three abundantly expressed HIV early genes in HIV-infected astrocytes and has been shown to be a major pathogenic factor for HIV/neuroAIDS. In this study, we sought to determine if and how Tat expression would affect HIV infection and latency in astrocytes. Using the VSV-G-pseudotyped red-green HIV (RGH) reporter viruses, we showed that HIV infection was capable of establishing HIV latency in astrocytes. We also found that Tat expression decreased the generation of latent HIV-infected cells. Activation of latent HIV-infected astrocytes showed that treatment of GSK126, a selective inhibitor of methyltransferase Ezh2 that is specifically responsible for tri-methylation of histone 3 lysine 27 (H3K27me3), led to activation of significantly more latent HIV-infected Tat-expressing astrocytes. Molecular analysis showed that H3K27me3, Ezh2, MeCP2, and Tat all exhibited a similar bimodal expression kinetics in the course of HIV infection and latency in astrocytes, although H3K27me3, Ezh2 and MeCP2 were expressed higher in Tat-expressing astrocytes and their expression were peaked immediately preceding Tat expression. Subsequent studies showed that Tat expression alone was sufficient to induce H3K27me3 expression, likely through its regulation of Ezh2 and MeCP2 expression. Taken together, these results showed for the first time that Tat expression induced H3K27me3 expression and contributed to HIV latency in astrocytes and suggest a new role and novel mechanism for Tat in HIV latency.
Keywords: HIV-1, Tat, H3K27me3, MeCP2, Ezh2, latency
INTRODUCTION
Macrophages/microglia and astrocytes are major target cells for HIV infection in the central nervous system (CNS) (Burdo et al, 2013; Kaul et al, 2005). Astrocytes are the most abundant and long-lived cell type in CNS (Churchill and Nath, 2013; Kramer-Hammerle et al, 2005; Sofroniew and Vinters, 2010). They are susceptible to cell-free HIV infection as well as cell-cell contact HIV infection in vitro (Carroll-Anzinger et al, 2007; Dimitrov et al, 1993; Hubner et al, 2009; Luo and He, 2015). HIV-infected astrocytes have also been documented in vivo (Gorry et al, 2003; Saito et al, 1994; Tornatore et al, 1994). Up to 20% of perivascular astrocytes have been found to be HIV-infected and to be correlated with the severity of encephalitis and dementia (Luo and He, 2015), ascertaining the important roles of HIV infection of astrocytes in HIV/neuroAIDS. Astrocytes have been proposed to be HIV latent reservoirs in the CNS (Barat et al, 2018; Diaz et al, 2015; Huang and Nair, 2017; Thompson et al, 2011). However, the underlying molecular mechanisms remain largely unknown. Understanding these mechanisms is expected to help contribute to development of strategies for complete HIV eradication.
Tat is one of the three early-encoded HIV-1 proteins translated from the multiply spliced viral RNA transcript following HIV infection (Sabatier et al, 1991; Schwartz et al, 1990). It is required for HIV transcription and elongation to produce full-length viral transcripts through binding to TAR, cyclin T and CDK9 and phosphorylation of the C-terminal domain of RNA polymerase II (Wei et al, 1998). In addition, Tat has been shown to be a major pathogenic factor for HIV/neuroAIDS. Tat protein is detected in the HIV-infected brain (Hudson et al, 2000) and is secreted from HIV-infected-microglia/macrophages and astrocytes (He et al, 1997). It can cause direct neurotoxicity (Aprea et al, 2006; Brailoiu et al, 2006; Caporello et al, 2006; Kruman et al, 1998; Norman et al, 2007; Orsini et al, 1996), or indirect neurotoxicity through its chemokine-like activity and infiltration of monocytes/macrophages and lymphocytes into the CNS (Albini et al 1998; Benelli et al, 2000; de Paulis et al, 2000; Jones et al, 1998; Lafrenie et al, 1996; Park et al, 2001), or through its interaction with astrocytes (Fan and He, 2016a; Fan and He, 2016b). Nevertheless, it is not known whether and how abundant Tat expression following non-productive HIV infection of astrocytes would affect HIV latency in astrocytes.
In the current study, we first aimed to determine effects of Tat on establishment of HIV latency in astrocytes. We took advantage of astrocytoma cell line U373.MG and its derivative U373.MG.Tat that stably express Tat protein (Zhou et al, 2004), infected them with VSV-G-pseudotyped red-green reporter viruses, compared HIV replication and latency in these cells, and determined the molecular changes in the course of HIV infection and latency. We showed that Tat expression resulted in a lower level of HIV latency in astrocytes. Meanwhile, we demonstrated for the first time that Tat expression led to increased histone 3 tri-methylation at lysine 27 (H3K27me3) and subsequently facilitated HIV latency in astrocytes, likely through regulation of MeCP2 and Ezh2 expression.
MATERIALS AND METHODS
Cells
Human embryonic kidney 293T, human astrocytoma U373.MG, and human T lymphoblastoid cell line Jurkat and were obtained from American Tissue Culture Collection (Manassas, VA) and maintained in Dulbecco modified Eagle medium (DMEM, for 293T and U373.MG), or in Roswell Park Memorial Institute 1640 medium (RPMI-1640, for Jurkat), supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin-streptomycin-L-glutamine. U373.MG.Tat stably expressing Tat was established through transfection with pTat.myc, followed by selection of stable cell clones in the presence of G418 (Invitrogen, CA) (Zhou et al, 2004) and was maintained in DMEM containing 0.5 μg/ml G418. Human near-haploid cell line HAP1 and its MeCP2-knockout derivative Hap1-MeCP2−) were obtained from Horizon Discovery (Waterbeach Cambridge, United Kingdom) and maintained in Iscove modified Eagle medium (IMEM) supplemented with 10% heat-inactivated FBS and 1% penicillin-streptomycin-L-glutamine.
Antibodies and reagents
Tri-methyl-histone 3 (H3K27me3) antibody (G.299.10), histone 3 (H3) antibody (PA5-16183), and Ezh2 antibody (144CT2.1.5) were purchased from Life Technologies (Grand Island, NY). PCNA (PC10) antibody and MeCP2 (H300) antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). GSK126 (A34465) was purchased from Apexbio Technology LLC (Houston, TX).
Plasmids
The red green HIV (RGH) plasmid was obtained from NIH AIDS Reagent Program (donated by Dr. I. Sadowski and Dr. S. Viviana of University of British Columbia, Canada) (Dahabieh et al, 2013). pHCMV-G was a kind gift from Dr. J. Sodroski of Harvard Medical School, and it encodes the glycoprotein from vesicular stomatitis virus (VSV-G) under the control of the CMV promoter. pcDNA3 was purchased from Clontech (Mountain View, CA). pTat.myc was constructed as previously described (Liu et al, 2002). The MeCP2 promoter-driven luciferase reporter plasmid was purchased from Switchgear Genomics (Carlsbad, CA).
Cell-free HIV virus preparation and infection
293T were transfected with RGH/pHCMV-G plasmids (3:1) using a standard calcium phosphate precipitation method. The transfection medium was replaced with fresh medium 16 hr post transfection. The culture medium was collected 72 hr after the medium change, removed of cell debris, purified and concentrated by passing through a 20% sucrose cushion at 100,000 g for 2 hr, and then suspended in PBS, aliquoted and stored at liquid nitrogen. Viral titers were determined by the reverse transcriptase (RT) assay as previously described (Lopez-Herrera et al, 2005) and expressed as counts per minute (cpm). For cell-free HIV infection, U373.MG and U373.MG.Tat (1 × 106) were infected with HIV equivalent to 10,000 cpm RT in a final volume of 1 ml DMEM complete medium at 37°C, 5% CO2 4 hr. At the end of the infection, unbound viruses were removed, the cells were rinsed with multiple PBS washes and then cultured in fresh medium at a density of 0.3-1.0 × 106 cells/ml. The percentage of actively infected and latent cells was determined based on GFP and mCherry expression by flow cytometry.
Luciferase activity assay
The Renilla luciferase activity was measured using the luciferase assay system (Promega, Madison, WI) according to the manufacturer’s directions. Briefly, transfected cells were washed with PBS and collected in 1× Renilla luciferase lysis buffer at 72 hr post transfection. Lysates were centrifuged briefly to obtain clear cell lysates. The clear lysates were mixed with Renilla luciferase substrate, and the luciferase activity was measured by an Opticomp Luminometer (MGM Instruments, Hamden, CT).
Western blotting and protein quantitation
Cell lysates were prepared at 48 hr post-transfection unless stated otherwise using a modified RIPA buffer containing a higher concentration of NaCl (50 mM Tris.HCl, pH 8.0, 0.5% NP-40, 2 Mm EDTA, 500 mM NaCl, 10% glycerol) and separated on a 12% polyacrylamide-SDS gel. The proteins were transferred onto nitrocellulose membranes, probed with appropriate primary antibodies and HRP-conjugated secondary antibodies (0.5 μg/ml), and visualized using enhanced chemiluminescence reagents. ImageJ was used to quantitate protein expression levels on the blots.
RNA isolation and qRT-PCR
Total RNA was isolated from cells using the TRIzol (Invitrogen) according to the manufacturer's instructions. Total RNA (1 μg) was used to synthesize cDNA using an iScript cDNA synthesis kit (Bio-Rad) and used as the template for PCR using Sso Advanced SYBR green Supermix and the CFX96 real-time PCR detection system (Bio-Rad). The primers and their sequences are as follows: for β-actin, 5′-AAA CTG GAA CGG TGA AGG TG-3′ and 5′-AGA GAA GTG GGG TGG CTT TT-3′; for Tat from RGH, 5’-AGC CTT AGG CAT CTC CTA TGG-3’ and 5’′-CTA TTC CTT CGG GCC TGT CGG GT-3’; for Tat from U373.MG.Tat, 5′-AAA CTG GAA CGG TGA AGG TG-3′ and 5′-AGA AAT GAG CTT TTG CTC CTC TGC-3’. Threshold cycle (CT) values were calculated using Bio-Rad CFX manager software. The 2-ΔΔC T value was calculated to represent the fold change of the target gene mRNA using β-actin as the reference.
Histone methyltransferase activity assay
Transfected cells were trypsinized and washed in cold PBS. Nuclear pellets were obtained following first treatment with hypotonic buffer, then with an EpiQuik Nuclear Extraction Kit (#OP-0002, Epigentek, Farmingdale, NY). An equal amount of nuclear lysate was used to determine the histone methyltransferase activity using an EpiQuikTM histone methyltransferase activity/inhibition assay kit (H3K27) (#P-3005, Epigentek).
miRNA analysis
Total RNA (500 ng) was used to synthsize cDNA by using a qScript microRNA cDNA synthesis kit (Quanta PN 95107, QuantaBio, Beverly, MA). Then, synthesized cDNA at an equivalent of 10 ng initial total RNA was mixed with Perfecta SYBR Green SuperMix (Quanta PN 95054) and Universal PCR Primer (Quanta PN 95109) with miR-132 Perfecta microRNA Assay Primer (has-miR-132a-3p) in 10 μl qPCR reactions. The qPCR plates were run in a CFX96 real-time PCR detection system (Bio-Rad) using a two-step cycling protocol (95 °C for 2 min followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s) and followed by a melting curve. Threshold cycle (CT) values were calculated using Bio-Rad CFX manager software. 2(-ΔΔCT) value was calculated to represent the fold change of the target miRNA.
Data analysis.
Where appropriate, values were expressed as Mean ± SD of triplicate experiments. All statistical analyses were performed by one-way and two-way analyses of variances followed by Bonferroni correction or Dunnett's test. A p value of <0.05 was considered statistically significant and shown as *; a p value of <0.01 was considered highly significant and shown as **; and a p value of <0.001 was considered strongly significant and shown as ***. All data are representative of multiple repeated experiments.
RESULTS
Establishment of HIV latency in astrocytes and its inhibition by Tat
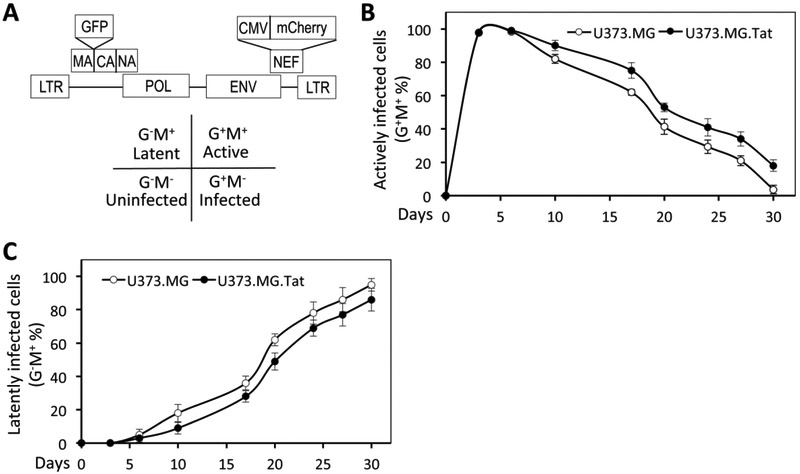
Astrocytes possess several attributes to be HIV reservoirs and several studies suggest that these cells can harbor integrated latent HIV (Carroll-Anzinger et al, 2007; Churchill and Nath, 2013; Gorry et al, 2003; Luo and He, 2015). To ascertain the notion that HIV is capable of establishing latency in astrocytes, we took advantage of a recently developed dual red-green fluorescent HIV reporter virus (RGH) (Dahabieh et al, 2013) (Fig. 1A), in which the green fluorescence protein gene (gfp) is inserted in-frame between matrix (MA) and capsid (CA) of gag gene, and a CMV-driven mCherry-expressing cassette (CMV-mCherry) is inserted in place of nef gene. In addition, env gene is disrupted by Kpn I digestion, blunted and re-ligation. RGH was designed in such a way that GFP expression alone (G+M−), mCherry expression alone (G−M+), expression of both GFP and mCherry (G+M+) in RGH-infected cells would be used to represent infected but not integrated, latent infection, and integrated/active infection, respectively (Dahabieh et al, 2013). Human astrocytoma cell line U373.MG were then infected with VSV-G-pseudotyped RGH using a standard HIV latency protocol, which have proven to generate the maximal number of HIV latent cells (Luo and He, 2015). The infected cells were then monitored for the percentage of G+M+ cells (active infection) and G−M+ cells (latent infection) every 3-5 days by flow cytometry. The percentage of G+M+ cells (active infection) showed an initial increase during the first three days (Fig. 1B), representative of de novo infection and replication (Dahabieh et al, 2013), then a gradual decrease to a background level at day 30 post infection. On the other hand, the percentage of G−M+ cells (latent infection) showed a gradual increase to about 95% at day 30 (Fig. 1C). These results confirm that HIV infection is capable of establishing latency in astrocytes.
Figure 1. HIV infection and latency in astrocytes.
A. Schematic of the red green dual HIV reporter virus genome (RGH). GFP was inserted in-frame between MA and NC gene, while a CMV-driven mCherry cassette was inserted in place of Nef gene. Env gene was disrupted by Kpn I digestion, blunted and re-ligation (filled-in). Expression of GFP (G+M−), mCherry (G−M+), or both (G+M+) was used to represent unintegrated infection, latent infection, and active replicating infection, respectively. B. U373.MG and U373.MG.Tat were infected with VSVG-pseudotyped RGH viruses and analyzed every 3-5 days by flow cytometry for active replicating cells (G+M+). C. U373.MG and U373.MG.Tat were infected with VSVG-pseudotyped RGH viruses and analyzed every 3-5 days by flow cytometry for latently infected cells (G−M+).
HIV-1 Tat is a major factor in HIV/neuroAIDS pathogenesis(Hudson et al, 2000). It is secreted from HIV-infected microglia/macrophage and astrocytes in the CNS (He et al, 1997; Hudson et al, 2000), it is expressed abundantly as one of the early genes in astrocytes (Bagashev and Sawaya, 2013; He et al, 1997), and it is present in the CNS of HIV-infected individuals with HIV suppression by antiretroviral therapy (Heaton et al, 2010; Yilmaz et al, 2012). Therefore, U373.MG.Tat, an U373.MG derivative that stably express HIV-1 Tat protein (Zhou et al, 2004) were in parallel infected with VSV-G-pseudotyped RGH to determine if Tat expression would affect establishment of HIV infection and latency. The infected cells were similarly monitored for the percentage of G+M+ cells (active infection) and G−M+ cells (latent infection). The percentage of G+M+ cells in VSV-G-pseudotyped RGH-infected U373.MG.Tat cells was slightly higher than that in VSV-G-pseudotyped RGH-infected U373.MG cells at each point of analysis beginning at day 10 post infection and lasting until day 30 post infection (Fig. 1B). In parallel, the percentage of G−M+ cells in VSV-G-pseudotyped RGH-infected U37.MG.Tat cells was about 9% lower than that in VSV-G-pseudotyped RGH-infected U37.MG cells (Fig. 1C). The differences of the percentage of latent HIV-infected cells between U373.MG and U373.MG.Tat appeared to be independent of the amount of input viruses and starting number of cells (data not shown). These results showed that ectopic Tat expression led to increased active HIV replication while decreased establishment of HIV latency in astrocytes.
Selective HIV activation in Tat-expressing astrocytes by GSK126
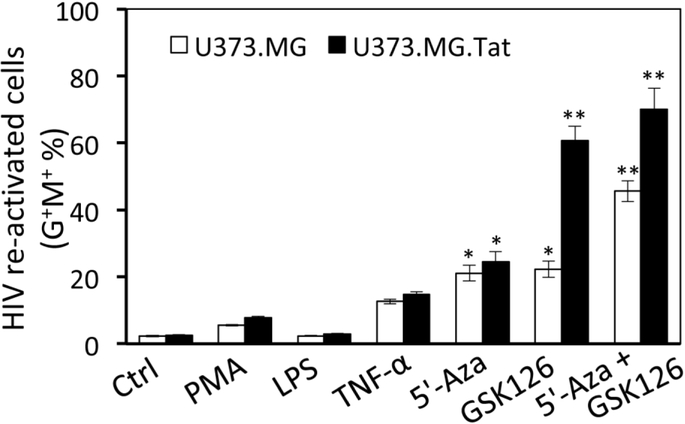
HIV latency involves multiple molecular mechanisms (Khan et al, 2018; Matsuda et al, 2015; Mbonye and Karn, 2017). To determine possible HIV latency mechanisms in astrocytes, the same number of HIV latent (G−M+) U373.MG and U373.MG.Tat at day 30 to 40 post-infection with VSV-G-pseudotyped RGH were treated with HIV activation reagents PMA, LPS, TNF-α, or 5’-azacytide (5’-Aza), one DNA methyltransferase inhibitor and analyzed for their effects on HIV activation, the percentage of G+M+ cells determined by flow cytometry. Compared to the control, LPS treatment led to little HIV activation, while PMA, TNF-α, 5’-Aza treatment showed about 9%, 16%, and 21% HIV activation, respectively (Fig. 2). There were no differences between U373.MG and U373.MG.Tat for all these four reagents. Then, these cells were treated with GSK126, a new and most potent, highly selective, S-adenosyl-methionine-competitive, small molecule inhibitor of histone methyltransferase enhancer of zeste homolog 2 (Ezh2), which is a subunit of polycomb repressive complex 2 (PRC2) (McCabe et al, 2012; Van Aller et al, 2014). Compared to the control and other four reagents, GSK126 treatment led to HIV activation in about 23% U373.MG and in 61% U373.MG.Tat. Co-treatment of 5’-Aza and GSK126 led to HIV activation in 46% U373.MG and 70% U373.MG.Tat, suggesting a likely additive effect of these two reagents on HIV activation. These results confirmed that HIV latency in astrocytes is susceptible to activation by external stimuli and provide the first evidence that Tat expression may regulate PRC2 through Ezh2 during HIV latency and subsequently affect establishment of HIV latency.
Figure 2. HIV activation in astrocytes.
The same number of HIV latent (G−M+) U373.MG and U373.MG.Tat at day 30 post infection of VSVG-RGH were treated with PMA (7.5 nM), LPS (10 ng/ml), TNF-α (10 ng/ml), 5’-Aza (5 μM), or GSK126 (7.5 μM) for 4 days and analyzed by flow cytometry for active replicating cells (G+M+). Untreated cells were included as a control (Ctrl). The data were Mean ± SD of triplicates and representative of three independent experiments.
Relationship among Ezh2, H3K27me3, MeCP2, and Tat expression during HIV-1 infection and latency
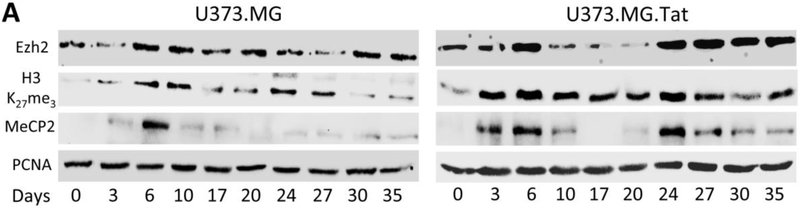
PRC2 is one of the two classes of polycom-group proteins and has four subunits: Suz12, Eed, Ezh1 or Ezh2, and RbAp48, it trimethylates and binds to histone 3 (H3) on lysine 27 (H3K27me3) through Ezh1 or Ezh2, and represses transcription including HIV transcription and latency (Friedman et al, 2011; Khan et al, 2018; Matsuda et al, 2015). MeCP2 has been shown to stimulate Ezh2 expression and H3k27 methylation (Hite et al, 2009; Mann et al, 2010). Therefore, we next determined the relationship among Ezh2, H3K27me3, MeCP2, and Tat expression in the course of HIV infection and latency in both U373.MG and U373.MG.Tat. VSV-G-pseudotyped RGH-infected U373.MG and U373.MG.Tat were harvested every 3-5 days post infection and analyzed for Ezh2, H3K27me3, and MeCP2 by Western blotting, followed by quantitation or Tat expression by qRT-PCR. In U373.MG (left panel, Fig. 3A), Ezh2 expression showed gradual increases, peaked at day 6 post-infection, decreased to and remained at the basal level, and began to increase again at day 30 (Fig. 3B). H3K27me3 expression showed gradual increases, peaked at day 6 post-infection and remained the level up to day 20 post-infection, began to increase again until day 27 and then decreased to the basal level (Fig. 3C). MeCP2 expression showed gradual increases, peaked at day 6 post-infection, then decreased to and remained at the basal level until day 35 post-infection (Fig. 3D). In comparison, Ezh2, H3K27me3, and MeCP2 in U373.MG.Tat (right panel, Fig. 3A) all exhibited a distinct synchronized bimodal expression kinetic with first peak at day 6 post-infection, and second peak at day 24 post-infection (Fig. 3B-D). In addition, of note was that Ezh2, H3K27me3 and MeCP2 all generally expressed at a higher level in U373.MG.Tat than that in U373.MG. To determine Tat mRNA expression, qRT-PCR was performed using primers that were specific for Tat mRNA from RGH infection or from U373.MG.Tat. Tat mRNA from RGH infection exhibited a bimodal kinetic with the first peak at day 3 post-infection and the second peak at day 20 post-infection in both U373.MG and U373.MG.Tat (Fig. 3E), both of which immediately preceded the peaked expression of all Ezh2, H3K27me3 and MeCP2. Interestingly, Tat mRNA expression in U373.MG.Tat also followed a similar bimodal kinetic (Fig. 3F), suggesting regulatory effects of RGH infection or RGH Tat expression on CMV-driven Tat expression in U373.MG.Tat.
Figure 3. Ezh2, H3K27me3, MeCP2, and Tat expression in relation to HIV infection and latency in astrocytes.
U373.MG and U373.MG.Tat were infected with VSVG-pseudotyped RGH viruses and analyzed every 3-5 days by Western blotting (A), followed by densitometry quantitation for Ezh2 (B), H3K27me3 (C), and MeCP2 (D), or by qRT-PCR for Tat mRNA expression from RGH (E) or in U373.MG.Tat (F) using RGH Tat mRNA-specific, or Tat.myc mRNA-specific primers. PCNA was included as a loading control and used as a quantitation reference for Western blotting. β-actin was used an internal control and used as used as a quantitation reference for qRT-PCR. All were expressed as relative levels (Rel.). The data were Mean ± SD of triplicates and representative of three independent experiments.
Tat expression led to increased global H3K27me3 level
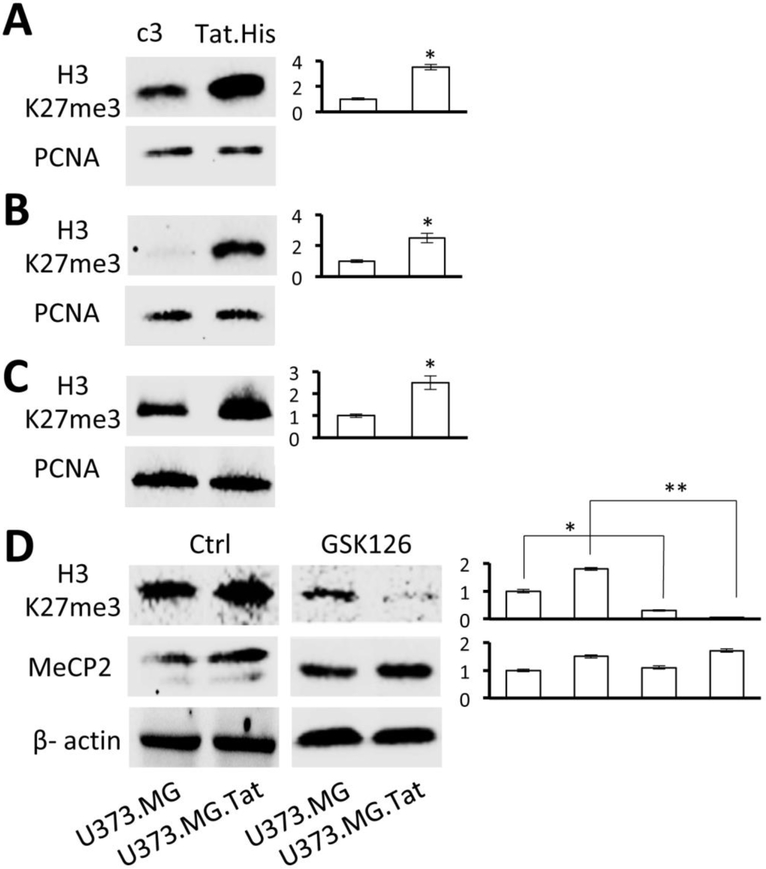
H3K27me3 have been directly linked to establishment of HIV latency(Friedman et al, 2011; Khan et al, 2018; Kim et al, 2011), but it is not known whether any HIV proteins are involved in H3K27me3 regulation. The findings above raised the possibility that HIV Tat might be involved. To address this possibility, three different cell lines (U373.MG, Jurkat, and 293T) were transiently transfected with Tat expression plasmid and analyzed for H3K27me3 expression by Western blotting. Tat expression alone showed to be sufficient to induce a significantly higher level of H3K27me3 expression in U373.MG (Fig. 4A), Jurkat (Fig. 4B), and 293T (Fig. 4C). Similarly, a significantly higher level of H3K27me3 and MeCP2 was detected in U373.MG.Tat than U373.MG, and GSK126 treatment of these cells led to significant decreases in H3K27me3 expression but little changes in MeCP2 expression in both U373.MG and U373.MG.Tat (Fig. 4D). These results demonstrated that HIV Tat expression directly induced H3K27me3 expression, which likely contributes to Tat regulatory roles in HIV latency.
Figure 4. Direct effects of Tat expression on H3K27me3 level.
U373.MG (A), Jurkat (B), and 293T (C) were transfected with cDNA3 or Tat.His expression plasmids, cultured for 48 hr, and analyzed by Western blotting for H3k27me3 expression. PCNA was included as a loading control and used as a quantitation reference. D. U373.MG and U373.MG.Tat at day 30 post-infection of RGH were treated with GSK126 (7.5 μM) for 4 days and analyzed by Western blotting for H3k27me3 and MeCP2 expression. β-actin was included as a loading control and used as a quantitation reference. The data were Mean ± SD of triplicates and representative of three independent experiments.
Requirement of MeCP2 for Tat-induced H3K27me3
We have recently shown that Tat induces miR-132 expression, which in turn down-modulates MeCP2 expression (Rahimian and He, 2016). MeCP2 has been shown to co-localize with nucleosomes and bind to CpG islands within the promoters and repress transcription (Rube et al, 2016). As discussed above, MeCP2 has been shown to be a positive regulator of EZH2 expression and H3K27me3 expression (Hite et al, 2009; Mann et al, 2010). Thus, we next sought to determine the roles of MeCP2 in Tat-induced H3K27me3 expression. We took advantage of a human haploid cell line Hap1-MeCP2−, in which the MeCP2 gene was knocked out using the CRISPR/Cas9 genome editing technology. MeCP2 expression was detected in Hap1 (Mock, right half, Fig. 5), but not in its derivate cell line of Hap1-MeCP2− (Mock, left half, Fig. 5). Then, both Hap1 and Hap1-MeCP2− were transfected with Tat, doubled amount of Tat (Tat*), RGH, or RGH and Tat (RGH+Tat) and analyzed for Ezh2 and H3K27me3 expression by Western blotting. Ezh2 expression showed slight increases with Tat, RGH, or both, but giving rise to little H3K27me3 expression in Hap1-MeCP2−.In comparison, Tat, RGH, or both led to significantly increased MeCP2, Ezh2, and H3K27me3 expression in Hap1. These results showed that Tat or RGH up-regulated MecP2 and subsequently Ezh2 expression and that MecP2 was involved in Tat-induced H3K27me3 expression.
Figure 5. Effects of MeCP2 knockout on the relationship among Tat, EZH2, and H3K27me3 expression.
Hap-1 and MeCP2-knockout Hap-1 (Hap1-MeCP2−) were transfected with Tat expression plasmid, a doubled amount of Tat expression plasmid (Tat*), RGH plasmid, or RGH plasmid plus Tat plasmid (RGH+Tat), cultured for 48 hr, and analyzed by Western blotting for MeCP2, Ezh2, and H3k27me3 expression. cDNA3 was used to normalize the total amount of DNA transfected, while untreated cells were included as a control (Mock). β-actin was included as a loading control and used as a quantitation reference. Protein expressions were normalized to β-actin and calculated as folds of change over Mock, which was set at 1.0. The data were Mean ± SD of triplicates and representative of three independent experiments.
Up-regulation of miR-132, MeCP2, H3K27me3 expression, and H3K27-specific methyltransferase activity by Tat expression
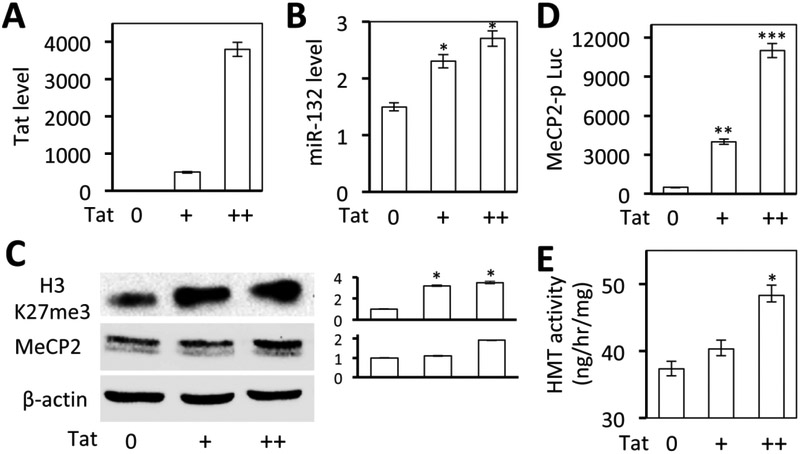
We have recently shown that Tat induces miR-132 expression, which in turn down-modulates MeCP2 expression (Rahimian and He, 2016). The findings above also suggest the possibility that Tat expression up-regulates MeCP2 expression. To ascertain the relationship between Tat, miR-132, and MeCP2 expression, U373.MG were transfected with increased amounts of Tat expression plasmid [0, 1X (+), 2X (++)], along with a MeCP2 promoter-driven luciferase reporter gene and analyzed for Tat and miR-132 expression by qRT-PCR, the luciferase gene expression, H3K27me3 and MeCP2 expression, and histone methyltransferase activity. Tat expression (Fig. 6A) led to increased miR-132 expression (Fig. 6B) and luciferase activity (Fig. 6C). Consistent with the findings above, Tat expression also led to increased H3K27me3 and MeCP2 expression (Fig. 6D). Importantly, Tat expression led to increased histone methyltransferase activity (Fig. 6E). These results demonstrated that Tat expression up-regulated MeCP2 expression, which may or may not be through miR-132, or through reciprocal regulation between miR-132 and MeCP2(Klein et al, 2007; Martinowich et al, 2003; Rahimian and He, 2016; Su et al, 2015). These results further confirm that Tat expression led to increased H3K27me3 and H3K27-specific methyltransferase (Ezh2) activity.
Figure 6. Effects of Tat expression on MeCP2 promoter activity and miR-132 expression.
A-C. 293T were transfected with an increasing amount of Tat expression plasmid, cultured for 48 hr, and harvested for RNA isolation, followed by qRT-PCR for Tat (A) miR132 expression (B), or harvested for cell lysates, followed by Western blotting for H3K27me3 and MecP2 expression (C). cDNA3 was used to normalize the total amount of DNA transfected. β-actin was included as a loading control and used as a quantitation reference. D. 293T were co-transfected with MecP2 promoter-driven luciferase reporter plasmid and an increasing amount of Tat expression plasmid, cultured for 48 hr, and harvested for the luciferase reporter gene assay. cDNA3 was used to normalize the total amount of DNA transfected. pCMV-β-gal was used to normalize the transfection efficiency variations among all transfections. E. 293T were transfected with increasing amounts of Tat expression plasmids, cultured for 48 hr, and harvested for nuclear lysates, followed by histone methyltransferase (HMT) activity assay. cDNA3 was used to normalize the total amount of DNA transfected. The data were Mean ± SD of triplicates and representative of three independent experiments.
DISCUSSION
In the study, we started with establishment of HIV latent astrocytes using VSV-G-pseudotyped RGH reporter viruses. We showed that RGH viruses were capable of establishing latency in both U373.MG and U373.MG.Tat (Fig. 1), which differed by about 10%, suggesting that Tat expression led to fewer HIV-infected cells entering the latency stage. We then attempted to activate the latent HIV-infected cells with several known activation reagents and found that GSK126, a selective inhibitor of histone 3 methyltransferase Ezh2 differentially activated HIV-infected U373.MG.Tat from HIV-infected U373.MG (Fig. 2), raising the possibility for the first time that Tat expression might promote histone 3 tri-methylation at lysine 27 (H3K27me3) and subsequently lead to HIV repression. Next, we analyzed expression kinetics of Ezh2, H3K27me3, and MeCP2 in relation to Tat expression in the course of HIV infection and latency in astrocytes. We showed that all exhibited similar bimodal expression kinetics with higher levels of expression in U373.MG.Tat than U373.MG (Fig. 3). Interestingly, we noticed that Tat expression oscillated immediately prior to Ezh2, H3K27me3, and MeCP2 expression. Subsequent studies showed that Tat expression alone was sufficient to induce H3K27me3 expression level (Fig. 4), that MeCP2 was required for Tat-induced H3K27me3 expression (Fig. 5) and H3K27 methyltransferase activity (Ezh2) (Fig. 6). Taken together, these results demonstrated that Tat expression induced H3K27me3 expression and HIV latency, likely through its regulation of MeCP2 and Ezh2 expression.
HIV-1 infection of astrocytes leads to abundant expression of HIV-1 early gene products such as Tat despite its restricted nature (Gorry et al, 2003; Messam and Major, 2000; Ranki et al, 1995). Since Tat is an essential transcription factor in HIV-1 replication, it is not workable to knock out or knock down Tat to determine the roles of Tat in HIV latency in astrocytes in this study. Instead, we took advantage of a stable Tat-expressing cell line U373.MG and used it in the study. It is conceivable that Tat inhibited latency development, as it functions to transactivate HIV-1 LTR promoter of provirus and HIV replication. However, the finding that GSK126 treatment led to activation of more latent Tat-expressing astrocyte indicated that Tat expression was involved in the regulation of the histone methyltransferase Ezh2 and H3K27me3 expression. Ezh2 is the only methyltransferase responsible for the tri-methylation of H3K27. Therefore, we studied the H3K27me3 expression along with Ezh2 during HIV infection and latency. The absence of new infection from viral gene expression in this RGH model would allow us to study the dynamic regulation of the LTR activity. There were two peaks of Ezh2, H3K27me3, MeCP2, and Tat expressions during this period. This could be a complicated process in which cellular/viral factors induced by HIV-1 transcription and exogenous Tat expression together promoted methylation of H3K27 in the early stage at day 6. Then followed by de-methylation of H3K27me3 process in the middle of the process. This could be induced from the inhibition of activity of methyltransferase Ezh2 from day 12 to day 20. Thus, it would be interesting to identify the inhibitory factors of methyltransferase Ezh2 compared with the first peak of induction through global gene expression analysis. The second peak of H3K27me3, EZH2, and MeCP2 expression occurred on day 24 prior to HIV latency. expression preceded Ezh2, H3K27me3 and MeCP2 expression in both peaks, supporting the notion that Tat expression alone or Tat expression from HIV infection is responsible for regulation of their expression.
Ezh2 expression and activity could be regulated by many different factors (Cao et al, 2002; Fujii et al, 2011; Mann et al, 2010; Margueron and Reinberg, 2011; Sander et al, 2008; Tang et al, 2004), and MecP2 is one of them (Mann et al, 2010). A major portion of MeCP2-bound nucleosomes contains H3K27me3 in brain fractions (Thambirajah et al, 2012). MecP2 are highly expressed in the brain and is recognized as epigenetic DNA methylation marks to repress gene transcription (Chahrour et al, 2008). Therefore, it is likely that Ezh2 activity induced by Tat is mediated by regulation of MeCP2 expression. Using human haploid cell model HAP1 in which MecP2 is knocked out through CRISPR/Cas9 genome editing technology, we found that Tat and/or RGH expression increased global H3K27me3 expression only when MeCP2 was expressed. Tat and/or RGH regulated Ezh2 expression regardless of MeCP2 expression. Ezh2 was dramatically down regulated without MeCP2, implying the importance of MeCP2 in maintaining Ezh2 at a certain level. However, up-regulation of Ezh2 by Tat and HIV-1 both in absence of MeCP2 and in the presence of MecP2 suggested the epigenetic changes leading HIV-1 to latency could be MeCP2-dependent and independent and that a threshold level of Ezh2 expression would be required for Tat- and HIV-induced H3K27me3 expression. All these results would support the notion that MeCP2 play a fundamental role in HIV latency.
Increased expression of miR-132, a brain-enriched microRNA specifically targeting MecP2 (Rahimian and He, 2016), was Tat expression dose-dependent. More interestingly, we showed that Tat transactivated MeCP2 promoter activity. There is a possibility that MiR-132 and MeCP2 are mutually regulated to maintain their proper function. The other alternative could be that Tat regulation of MeCP2 expression involves other mechanisms. MeCP2 is known for its gene repression function. MeCP2 has been shown to be a positive regulator of Ezh2 expression (Mann et al, 2010). Almost all cell types in adult mammalian tissues express MeCP2 (Song et al, 2014). However, it is for sure that Tat and RGH were not capable of increasing H3K27me3 expression without MeCP2 expression.
DNA methylation and histone modifications including histone methylation and deacetylation are two major epigenetic mechanisms involved in regulation and maintenance of HIV latency. Inhibitors of histone deacetylases (HDACi), inhibitors of DNA methylation, or combination of histone methyltransferase inhibitors and HADCi are often used to re-activate proviruses (Blazkova et al, 2009; Bullen et al, 2014; Colin and Van Lint, 2009). H3K27 tri-methylation (H3K27me3) and H2A ubiquitylation via PRC should be involved in HIV-1 latency and contribute to epigenetic gene silencing (Kim et al, 2011). Ezh2 is a core member of PRC2 and directly mediates H3K27me3 of the chromatin (Simon and Lange, 2008) and is required for the maintenance of HIV-1 latency in Jurkat (Friedman et al, 2011); it also directly serve as a recruitment platform to regulate DNA methylation through its association with and regulation of the activity of DNA methyltransferase (Kanduri et al, 2013; Vire et al, 2006). A recent study has shown that pretreatment with the Ezh2 inhibitor and subsequent with HDACi is a more effective approach to activate HIV latency in a primary resting T-cell model (Friedman et al, 2011; Tripathy et al, 2015). The findings from our study support a new role of Tat being a negative regulator to suppress HIV gene expression through induction of H3K27me3 expression and clearly provide the direct link between Tat expression and HIV infection, H3K27me expression, and HIV latency and could also provide a new strategy to activate HIV latency in astrocytes.
Acknowledgments
This work was supported in part by the grants NIH/NINDS R01NS090960 and NIH/NIDA R01DA043162 (to JJH).
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
REFERENCES
- Albini A, Ferrini S, Benelli R, Sforzini S, Giunciuglio D, Aluigi MG, Proudfoot AE, Alouani S, Wells TN, Mariani G, Rabin RL, Farber JM, Noonan DM (1998). HIV-1 Tat protein mimicry of chemokines. Proc Natl Acad Sci U S A 95: 13153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprea S, Del Valle L, Mameli G, Sawaya BE, Khalili K, Peruzzi F (2006). Tubulin-mediated binding of human immunodeficiency virus-1 Tat to the cytoskeleton causes proteasomal-dependent degradation of microtubule-associated protein 2 and neuronal damage. J Neurosci 26: 4054–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagashev A, Sawaya BE (2013). Roles and functions of HIV-1 Tat protein in the CNS: an overview. Virol J 10: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barat C, Proust A, Deshiere A, Leboeuf M, Drouin J, Tremblay MJ (2018). Astrocytes sustain long-term productive HIV-1 infection without establishment of reactivable viral latency. Glia 66: 1363–1381. [DOI] [PubMed] [Google Scholar]
- Benelli R, Barbero A, Ferrini S, Scapini P, Cassatella M, Bussolino F, Tacchetti C, Noonan DM, Albini A (2000). Human immunodeficiency virus transactivator protein (Tat) stimulates chemotaxis, calcium mobilization, and activation of human polymorphonuclear leukocytes: implications for Tat-mediated pathogenesis. J Infect Dis 182: 1643–51. [DOI] [PubMed] [Google Scholar]
- Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, Verdin E, Olive D, Van Lint C, Hejnar J, Hirsch I (2009). CpG methylation controls reactivation of HIV from latency. PLoS Pathog 5: e1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brailoiu E, Brailoiu GC, Mameli G, Dolei A, Sawaya BE, Dun NJ (2006). Acute exposure to ethanol potentiates human immunodeficiency virus type 1 Tat-induced Ca(2+) overload and neuronal death in cultured rat cortical neurons. J Neurovirol 12: 17–24. [DOI] [PubMed] [Google Scholar]
- Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF (2014). New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med 20: 425–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdo TH, Lackner A, Williams KC (2013). Monocyte/macrophages and their role in HIV neuropathogenesis. Immunol Rev 254: 102–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002). Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–43. [DOI] [PubMed] [Google Scholar]
- Caporello E, Nath A, Slevin J, Galey D, Hamilton G, Williams L, Steiner JP, Haughey NJ (2006). The immunophilin ligand GPI1046 protects neurons from the lethal effects of the HIV-1 proteins gp120 and Tat by modulating endoplasmic reticulum calcium load. J Neurochem 98: 146–55. [DOI] [PubMed] [Google Scholar]
- Carroll-Anzinger D, Kumar A, Adarichev V, Kashanchi F, Al-Harthi L (2007). Human immunodeficiency virus-restricted replication in astrocytes and the ability of gamma interferon to modulate this restriction are regulated by a downstream effector of the Wnt signaling pathway. J Virol 81: 5864–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY (2008). MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320: 1224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M, Nath A (2013). Where does HIV hide? A focus on the central nervous system. Curr Opin HIV AIDS 8: 165–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin L, Van Lint C (2009). Molecular control of HIV-1 postintegration latency: implications for the development of new therapeutic strategies. Retrovirology 6: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahabieh MS, Ooms M, Simon V, Sadowski I (2013). A doubly fluorescent HIV-1 reporter shows that the majority of integrated HIV-1 is latent shortly after infection. J Virol 87: 4716–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paulis A, De Palma R, Di Gioia L, Carfora M, Prevete N, Tosi G, Accolla RS, Marone G (2000). Tat protein is an HIV-1-encoded beta-chemokine homolog that promotes migration and up-regulates CCR3 expression on human Fc epsilon RI+ cells. J Immunol 165: 7171–9. [DOI] [PubMed] [Google Scholar]
- Diaz L, Martinez-Bonet M, Sanchez J, Fernandez-Pineda A, Jimenez JL, Munoz E, Moreno S, Alvarez S, Munoz-Fernandez MA (2015). Bryostatin activates HIV-1 latent expression in human astrocytes through a PKC and NF-kB-dependent mechanism. Sci Rep 5: 12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov DS, Willey RL, Sato H, Chang LJ, Blumenthal R, Martin MA (1993). Quantitation of human immunodeficiency virus type 1 infection kinetics. J Virol 67: 2182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, He JJ (2016a). HIV-1 Tat Induces Unfolded Protein Response and Endoplasmic Reticulum Stress in Astrocytes and Causes Neurotoxicity through Glial Fibrillary Acidic Protein (GFAP) Activation and Aggregation. J Biol Chem 291: 22819–22829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, He JJ (2016b). HIV-1 Tat Promotes Lysosomal Exocytosis in Astrocytes and Contributes to Astrocyte-mediated Tat Neurotoxicity. J Biol Chem 291: 22830–22840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Cho WK, Chu CK, Keedy KS, Archin NM, Margolis DM, Karn J (2011). Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J Virol 85: 9078–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Tokita K, Wada N, Ito K, Yamauchi C, Ito Y, Ochiai A (2011). MEK-ERK pathway regulates EZH2 overexpression in association with aggressive breast cancer subtypes. Oncogene 30: 4118–28. [DOI] [PubMed] [Google Scholar]
- Gorry PR, Ong C, Thorpe J, Bannwarth S, Thompson KA, Gatignol A, Vesselingh SL, Purcell DF (2003). Astrocyte infection by HIV-1: mechanisms of restricted virus replication, and role in the pathogenesis of HIV-1-associated dementia. Curr HIV Res 1: 463–73. [DOI] [PubMed] [Google Scholar]
- He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay CR, Sodroski J, Gabuzda D (1997). CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature 385: 645–9. [DOI] [PubMed] [Google Scholar]
- Heaton RK, Clifford DB, Franklin DR Jr., Woods SP, Ake C, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, Rivera-Mindt M, Vigil OR, Taylor MJ, Collier AC, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, McCutchan JA, Abramson I, Gamst A, Fennema-Notestine C, Jernigan TL, Wong J, Grant I, Group C (2010). HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology 75: 2087–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hite KC, Adams VH, Hansen JC (2009). Recent advances in MeCP2 structure and function. Biochem Cell Biol 87: 219–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Nair M (2017). A CRISPR/Cas9 guidance RNA screen platform for HIV provirus disruption and HIV/AIDS gene therapy in astrocytes. Sci Rep 7: 5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner W, McNerney GP, Chen P, Dale BM, Gordon RE, Chuang FY, Li XD, Asmuth DM, Huser T, Chen BK (2009). Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science 323: 1743–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson L, Liu J, Nath A, Jones M, Raghavan R, Narayan O, Male D, Everall I (2000). Detection of the human immunodeficiency virus regulatory protein tat in CNS tissues. J Neurovirol 6: 145–55. [DOI] [PubMed] [Google Scholar]
- Jones M, Olafson K, Del Bigio MR, Peeling J, Nath A (1998). Intraventricular injection of human immunodeficiency virus type 1 (HIV-1) tat protein causes inflammation, gliosis, apoptosis, and ventricular enlargement. J Neuropathol Exp Neurol 57: 563–70. [DOI] [PubMed] [Google Scholar]
- Kanduri M, Sander B, Ntoufa S, Papakonstantinou N, Sutton LA, Stamatopoulos K, Kanduri C, Rosenquist R (2013). A key role for EZH2 in epigenetic silencing of HOX genes in mantle cell lymphoma. Epigenetics 8: 1280–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul M, Zheng J, Okamoto S, Gendelman HE, Lipton SA (2005). HIV-1 infection and AIDS: consequences for the central nervous system. Cell Death Differ 12 Suppl 1: 878–92. [DOI] [PubMed] [Google Scholar]
- Khan S, Iqbal M, Tariq M, Baig SM, Abbas W (2018). Epigenetic regulation of HIV-1 latency: focus on polycomb group (PcG) proteins. Clin Epigenetics 10: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HG, Kim KC, Roh TY, Park J, Jung KM, Lee JS, Choi SY, Kim SS, Choi BS (2011). Gene silencing in HIV-1 latency by polycomb repressive group. Virol J 8: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH (2007). Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci 10: 1513–4. [DOI] [PubMed] [Google Scholar]
- Kramer-Hammerle S, Rothenaigner I, Wolff H, Bell JE, Brack-Werner R (2005). Cells of the central nervous system as targets and reservoirs of the human immunodeficiency virus. Virus Res 111: 194–213. [DOI] [PubMed] [Google Scholar]
- Kruman II, Nath A, Mattson MP (1998). HIV-1 protein Tat induces apoptosis of hippocampal neurons by a mechanism involving caspase activation, calcium overload, and oxidative stress. Exp Neurol 154: 276–88. [DOI] [PubMed] [Google Scholar]
- Lafrenie RM, Wahl LM, Epstein JS, Hewlett IK, Yamada KM, Dhawan S (1996). HIV-1-Tat protein promotes chemotaxis and invasive behavior by monocytes. J Immunol 157: 974–7. [PubMed] [Google Scholar]
- Liu Y, Li J, Kim BO, Pace BS, He JJ (2002). HIV-1 Tat protein-mediated transactivation of the HIV-1 long terminal repeat promoter is potentiated by a novel nuclear Tat-interacting protein of 110 kDa, Tip110. J Biol Chem 277: 23854–63. [DOI] [PubMed] [Google Scholar]
- Lopez-Herrera A, Liu Y, Rugeles MT, He JJ (2005). HIV-1 interaction with human mannose receptor (hMR) induces production of matrix metalloproteinase 2 (MMP-2) through hMR-mediated intracellular signaling in astrocytes. Biochim Biophys Acta 1741: 55–64. [DOI] [PubMed] [Google Scholar]
- Luo X, He JJ (2015). Cell-cell contact viral transfer contributes to HIV infection and persistence in astrocytes. J Neurovirol 21: 66–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann J, Chu DC, Maxwell A, Oakley F, Zhu NL, Tsukamoto H, Mann DA (2010). MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology 138: 705–14, 714 e1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D (2011). The Polycomb complex PRC2 and its mark in life. Nature 469: 343–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE (2003). DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302: 890–3. [DOI] [PubMed] [Google Scholar]
- Matsuda Y, Kobayashi-Ishihara M, Fujikawa D, Ishida T, Watanabe T, Yamagishi M (2015). Epigenetic heterogeneity in HIV-1 latency establishment. Sci Rep 5: 7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbonye U, Karn J (2017). The Molecular Basis for Human Immunodeficiency Virus Latency. Annu Rev Virol 4: 261–285. [DOI] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A 3rd, Diaz E, LaFrance LV, Mellinger M, Duquenne C, Tian X, Kruger RG, McHugh CF, Brandt M, Miller WH, Dhanak D, Verma SK, Tummino PJ, Creasy CL (2012). EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492: 108–12. [DOI] [PubMed] [Google Scholar]
- Messam CA, Major EO (2000). Stages of restricted HIV-1 infection in astrocyte cultures derived from human fetal brain tissue. J Neurovirol 6 Suppl 1: S90–4. [PubMed] [Google Scholar]
- Norman JP, Perry SW, Kasischke KA, Volsky DJ, Gelbard HA (2007). HIV-1 trans activator of transcription protein elicits mitochondrial hyperpolarization and respiratory deficit, with dysregulation of complex IV and nicotinamide adenine dinucleotide homeostasis in cortical neurons. J Immunol 178: 869–76. [DOI] [PubMed] [Google Scholar]
- Orsini MJ, Debouck CM, Webb CL, Lysko PG (1996). Extracellular human immunodeficiency virus type 1 Tat protein promotes aggregation and adhesion of cerebellar neurons. J Neurosci 16: 2546–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IW, Wang JF, Groopman JE (2001). HIV-1 Tat promotes monocyte chemoattractant protein-1 secretion followed by transmigration of monocytes. Blood 97: 352–8. [DOI] [PubMed] [Google Scholar]
- Rahimian P, He JJ (2016). HIV-1 Tat-shortened neurite outgrowth through regulation of microRNA-132 and its target gene expression. J Neuroinflammation 13: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranki A, Nyberg M, Ovod V, Haltia M, Elovaara I, Raininko R, Haapasalo H, Krohn K (1995). Abundant expression of HIV Nef and Rev proteins in brain astrocytes in vivo is associated with dementia. AIDS 9: 1001–8. [DOI] [PubMed] [Google Scholar]
- Rube HT, Lee W, Hejna M, Chen H, Yasui DH, Hess JF, LaSalle JM, Song JS, Gong Q (2016). Sequence features accurately predict genome-wide MeCP2 binding in vivo. Nat Commun 7: 11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatier JM, Vives E, Mabrouk K, Benjouad A, Rochat H, Duval A, Hue B, Bahraoui E (1991). Evidence for neurotoxic activity of tat from human immunodeficiency virus type 1. J Virol 65: 961–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Sharer LR, Epstein LG, Michaels J, Mintz M, Louder M, Golding K, Cvetkovich TA, Blumberg BM (1994). Overexpression of nef as a marker for restricted HIV-1 infection of astrocytes in postmortem pediatric central nervous tissues. Neurology 44: 474–81. [DOI] [PubMed] [Google Scholar]
- Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, Moller P, Stilgenbauer S, Pollack JR, Wirth T (2008). MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood 112: 4202–12. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Felber BK, Benko DM, Fenyo EM, Pavlakis GN (1990). Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J Virol 64: 2519–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Lange CA (2008). Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res 647: 21–9. [DOI] [PubMed] [Google Scholar]
- Sofroniew MV, Vinters HV (2010). Astrocytes: biology and pathology. Acta Neuropathol 119: 7–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Feodorova Y, Guy J, Peichl L, Jost KL, Kimura H, Cardoso MC, Bird A, Leonhardt H, Joffe B, Solovei I (2014). DNA methylation reader MECP2: cell type- and differentiation stage-specific protein distribution. Epigenetics Chromatin 7: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M, Hong J, Zhao Y, Liu S, Xue X (2015). MeCP2 controls hippocampal brain-derived neurotrophic factor expression via homeostatic interactions with microRNA132 in rats with depression. Mol Med Rep 12: 5399–406. [DOI] [PubMed] [Google Scholar]
- Tang X, Milyavsky M, Shats I, Erez N, Goldfinger N, Rotter V (2004). Activated p53 suppresses the histone methyltransferase EZH2 gene. Oncogene 23: 5759–69. [DOI] [PubMed] [Google Scholar]
- Thambirajah AA, Ng MK, Frehlick LJ, Li A, Serpa JJ, Petrotchenko EV, Silva-Moreno B, Missiaen KK, Borchers CH, Adam Hall J, Mackie R, Lutz F, Gowen BE, Hendzel M, Georgel PT, Ausio J (2012). MeCP2 binds to nucleosome free (linker DNA) regions and to H3K9/H3K27 methylated nucleosomes in the brain. Nucleic Acids Res 40: 2884–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson KA, Cherry CL, Bell JE, McLean CA (2011). Brain cell reservoirs of latent virus in presymptomatic HIV-infected individuals. Am J Pathol 179: 1623–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornatore C, Chandra R, Berger JR, Major EO (1994). HIV-1 infection of subcortical astrocytes in the pediatric central nervous system. Neurology 44: 481–7. [DOI] [PubMed] [Google Scholar]
- Tripathy MK, McManamy ME, Burch BD, Archin NM, Margolis DM (2015). H3K27 Demethylation at the Proviral Promoter Sensitizes Latent HIV to the Effects of Vorinostat in Ex Vivo Cultures of Resting CD4+ T Cells. J Virol 89: 8392–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aller GS, Pappalardi MB, Ott HM, Diaz E, Brandt M, Schwartz BJ, Miller WH, Dhanak D, McCabe MT, Verma SK, Creasy CL, Tummino PJ, Kruger RG (2014). Long residence time inhibition of EZH2 in activated polycomb repressive complex 2. ACS Chem Biol 9: 622–9. [DOI] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F (2006). The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439: 871–4. [DOI] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA (1998). A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92: 451–62. [DOI] [PubMed] [Google Scholar]
- Yilmaz A, Price RW, Gisslen M (2012). Antiretroviral drug treatment of CNS HIV-1 infection. J Antimicrob Chemother 67: 299–311. [DOI] [PubMed] [Google Scholar]
- Zhou BY, Liu Y, Kim B, Xiao Y, He JJ (2004). Astrocyte activation and dysfunction and neuron death by HIV-1 Tat expression in astrocytes. Mol Cell Neurosci 27: 296–305. [DOI] [PubMed] [Google Scholar]