Abstract
The gut microbiome is emerging as an important contributor to both cardiovascular disease risk and metabolism of xenobiotics. Alterations in the intestinal microbiota are associated with atherosclerosis, dyslipidemia, hypertension and heart failure. The microbiota have the ability to metabolize medications which can results in altered drug pharmacokinetics and pharmacodynamics or formation of toxic metabolites which can interfere with drug response. Early evidence suggests that the gut microbiome modulates response to statins and antihypertensive medications. In this review we will highlight mechanisms by which the gut microbiome facilitates the biotransformation of drugs and impacts pharmacological efficacy. A better understanding of the complex interactions of the gut microbiome, host factors and response to medications will be important for the development of novel precision therapeutics for targeting CVD.
Keywords: metabolomics, pharmacogenetics, drug, statin, gut microbiome, drug response, Cardiovascular Disease, Lipids and Cholesterol, Genetics, Hypertension, Pharmacology
Introduction
Cardiovascular disease (CVD) remains the leading cause of death worldwide. While evidence-based strategies such as high-dose statins are available, treatment response is individual and varied. Many individuals remain at high risk of developing CVD, despite adherence to recommended therapies. Genetic contributions have an important role in CVD pathogenesis, yet genetic variation alone may only account for 20% of the risk of developing CVD.1 Lifestyle factors such as poor diet and physical inactivity are well established contributors to the development of CVD,2, 3 however the mechanisms remain incompletely understood. Together, these observations suggest that an improved understanding of the environmental factors contributing to both CVD pathogenesis and pharmacological variability to cardiovascular drug treatment will enhance our efforts in decreasing CVD related morbidity and mortality.
Increasing evidence has linked the activity and composition of the gut microbiome to health and disease. The human body is colonized by trillions of microorganisms, collectively known as the microbiome. Together, these microbiota possess hundreds of times as many genes as coded in the human genome.4 The gut microbiome is a complex community that contributes to important host metabolic functions such as processing nutrients and regulation of the immune system which are necessary to maintain healthy host physiology.4 Advances in high throughput sequencing technology such as 16S amplicon sequencing or shotgun metagenomics has allowed for the identification of human-associated microorganisms without the need for culturing.5 Alterations in gut microbiome composition are associated with pathogenesis of several human diseases such as obesity6, diabetes,7 and cardiovascular disease including atherosclerosis,8, 9 dyslipidemia,10, 11 hypertension12 and heart failure.13 Importantly, intestinal microbiota impacts the metabolism of xenobiotics14, 15 and have been shown to be a contributing factor in drug variability.16
The gut microbiota can impact host function through the generation of bioactive metabolites such as peptides, antibiotics, amino acids, and bile acids that can mediate host receptor activation, signaling and immunomodulatory effects.17, 18 Several categories of gut derived metabolites have been linked with CVD such as short chain fatty acids (SCFA) and bile acids.19, 20 Most recently, trimethylamine-N-oxide (TMAO), a metabolite derived from dietary choline and carnitine by the actions of the gut microbiome, was discovered to be a causal contributor to CVD risk.21–24 Microbiome derived metabolites may also affect drug response by interfering with drug pharmacokinetics and pharmacodynamics.25, 26
In this review, we provide a brief overview of the contribution of the gut microbiome to the development of CVD. We outline how gut microbiome composition influences drug metabolism, with specific evidence for cardiovascular drugs, and discuss mechanisms by which the microbiome exerts its effects on CVD and variation in drug response.
Contribution of the gut microbiome to cardiovascular disease
There are several lines of evidence suggesting that the gut microbiome contributes to cardiovascular disease. Atherosclerotic plaques from patients with coronary artery disease contain DNA from a wide range of bacterial species,27 which correlate with bacterial abundance in the oral cavity and intestine.8 In mice, gut microbial transplants have demonstrated a causal role for microbiota in inducing inflammation and accelerating atherosclerosis 28, likely through modulation of SCFA production and inflammatory signaling.
Trimethylamine-N-oxide (TMAO), a metabolite derived from dietary choline and carnitine by the actions of the gut microbiome associates with incident and prevalent major cardiovascular events21, 24 as described in detail in recent reviews29–31 TMAO has also been shown to be elevated in patients with heart failure and associated with greater all-cause mortality.32, 33TMAO is pro-atherogenic, pro-thrombotic and a causal contributor to CAD risk.22–24 In animal models, TMAO decreases reverse cholesterol transport, 22 the mechanism by which macrophages remove cholesterol from peripheral tissue for excretion through the bile.
The intestinal microbiota also regulate host lipid metabolism10, 11 with effects that are similar in magnitude to that explained by host genetics and independent of body mass index.10 The mechanism of how the gut microbiome regulates plasma lipid levels is unknown; however the role of gut bacteria in bile acid metabolism has been well described.34–37 Since microbial enzymes have the potential to create dozens of bile acid metabolites, each with differing potential to activate host receptors, further investigation in humans into how gut-derived metabolites may interface with lipid metabolism is warranted.
Alterations in the gut microbiome have been observed in hypertensive animals12, 38 and patients.39 While mechanisms remain unclear, both SCFAs and sodium-dependent inflammation have been implicated as microbiome-mediated contributors to hypertension.12, 40, 41 While the evidence supports a causal role for microbiota in cardiovascular disease, many mechanistic questions remain.
Mechanisms by which the gut microbiome impact drug response
The United States spent $333.4 billion dollars on prescription drugs in 2017, accounting for 10% of total national health spending.42 Individual patients display significant variation in response to medication and drug related adverse events result in considerable morbidity and mortality.43–45 Therefore there is a strong interest in understanding the host and environmental factors underlying the variation in drug response and the occurrence of adverse events. Age, sex, nutritional status, disease states, along with genetic and environmental exposures can explain how individuals will respond to drug therapies.43, 45, 46 However, genetic factors associated with drug response or pharmacogenomics has only explained variability for a small proportion of drugs. For example, in the most recent pharmacogenomic meta-analysis of LDL-C response to statins in 40,000 individuals, only four loci reached genome-wide significance (APOE, LPA, SORT1 SLCO1B1), and together they explained only ~5% of the variation in LDL-C response to statin treatment.47 The microbiome is increasingly recognized as an under-explored contributor to variation in drug metabolism and pharmacological efficacy. Greater than 50 drugs display evidence for metabolism by the gut microbiome.14 Gut microbiota can directly and indirectly influence drug response either by interfering with drug pharmacokinetics or pharmacodynamics.
Pharmacokinetic interactions
Drug absorption is a complex process that is dependent on multiple factors including: drug solubility in gastrointestinal (GI) fluids, stability of the drug in the pH of the GI lumen, GI transit time, permeability across epithelial membranes and pre-systemic metabolism by host and microbial enzyme systems.48 The pH changes along the GI lumen affect not only drug stability but provide distinct microenvironments that are amenable to growth of certain microbiota. For example, areas with low pH create a harsh environment for bacterial growth and limit the diversity of bacteria residing there.48
Most xenobiotics are metabolized in the liver to more hydrophilic compounds in order to facilitate excretion from the body.49 Phase I drug metabolizing enzymes, encoded by the cytochrome P450 (CYP P450) system, are involved in oxidation, reduction and hydrolysis reactions while phase II enzymes perform conjugation reactions.45 The intestine itself is an important drug metabolizing organ as it also expresses many drug metabolizing enzymes and drug transporters and contributes to pre-systemic metabolism and drug transport from the intestinal lumen. The gut microbiota also possess the genetic machinery necessary to produce enzymes that metabolize orally administered drugs which are focused on two main reaction types- hydrolysis and reduction. Microbial metabolism transforms hydrophilic drugs into more hydrophobic compounds, which enhance their absorption across the gut lumen.48
Microbial activity can thus result in altered drug pharmacokinetics, activation of prodrugs, unwanted formation of toxic metabolites or inactivation of drugs.50 Therefore inter-individual differences in intestinal bacterial species also contribute to the variation in drug response.14, 16 Examples of microbial biotransformations are listed in the Figure. Since there is a paucity of examples within the cardiovascular field, many well-known microbial reactions from other areas that impact drug pharmacokinetics and pharmacodynamics are described.
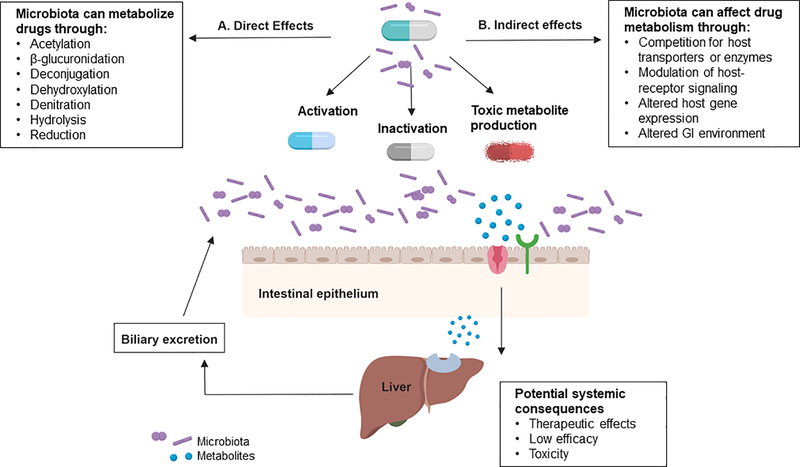
Figure 1.
Mechanisms by which the gut microbiome influence drug response. A. The gut microbiota produce enzymes that can directly metabolize drugs via biochemical reactions (acetylation, β-glucuronidation, deconjugation, dihydroxylation, denitration, hydrolysis, reduction, etc) interfering with drug pharmacokinetics. These reactions can biotransform prodrugs to activate metabolites, inactive compounds or lead to the formation of toxic metabolites. B. The gut microbiota can indirectly influence response to medication by the generation of microbial metabolites that can compete with host transport or detoxification systems, impact host receptor signaling pathways and alter host gene expression. Additional opportunities for microbiome-drug interactions exist for medications that undergo biliary excretion.
Absorption
A classic example in which the gut microbiome can interfere with drug bioavailability is with digoxin, a drug used for the treatment of heart failure. In 10% of patients, digoxin is converted to the inactive microbial metabolite dihydrodigoxin by, eggerthella lenta, limiting the amount of active drug that is absorbed into the systemic circulation. 51 Recent studies have identified a cytochrome-encoding operon, termed the cardiac glycoside reductase (cgr) that is activated by digoxin and is present in some strains of E. lenta but absent from nonreducing strains.52 The presence of the cgr operon but not the abundance of E. lenta itself predicts digoxin inactivation by the gut microbiome. Clinically coadminstration of digoxin with antibiotics53 or arginine rich diet52 will impair this microbial reaction and result in increased systemic digoxin levels and clinically relevant fluctuations in drug levels.
Distribution
In the treatment of inflammatory bowel disease, prodrugs are formulated with azo-bonds to overcome the acidic environment of the stomach and degradative enzymes of the small intestine in order to deliver drug to the colon. Microbiota of the colon produces azoreductases capable of reducing azo-bonds, and thereby releases the active drug at the desired site of action. Sulfasalazine, a prodrug, is converted to its pharmacologically active form, 5-amino 5-salicylic acid by the action of microbial azoreductases.54 The introduction of azo bonds has been applied to enhance drug distribution of additional drugs used to treat inflammatory bowel disease including olsalazine and balsalazide.50
Metabolism
Nitrazepam, a benzodiazepine, used for the treatment of anxiety and insomnia, undergoes nitro reduction by gut microbiota to 7-amino-nitrazepam, a teratogenic metabolite.55 Similarly, another sedative clonazepam is metabolized to 7-aminoclonazapam by the gut microbiome.48 Both of these reduction reactions are inhibited in the presence of antibiotics. 48 Other types of microbial reactions are listed in the Figure.
Excretion
The intravenous chemotherapeutic prodrug irinotecan, used to treat colorectal cancer is activated by hydrolysis by host enzymes to SN-38. In the liver SN-38 is inactivated by phase II enzymes by the glucuronidation pathway and the glucuronide metabolite is excreted to the intestine via the biliary route where it is exposed to gut bacteria56 Bacteria expressing β-glucuronidases remove the glucuronide group and reactivates the drug in the intestine resulting in severe diarrhea. Oral administration of a specific inhibitor of the β-glucuronidase enzyme in mice was shown to reduce irinotecan induced toxicity.57
Indirect effects
The gut microbiome can produce small molecules that can be metabolized by the same host drug metabolizing enzymes used for xenobiotics.15 The analgesic drug acetaminophen is primarily metabolized in the liver by phase II enzymes to the inactive acetaminophen sulfate and acetaminophen glucuronide. A minor metabolite of acetaminophen, N-acetyl p-benzoquinone imine (NAPQI) results in liver toxicity.58 A metabolomics study in 90 human subjects identified a microbial derived metabolite p-cresol that is systemically absorbed and undergoes sulfation in the liver.15 Both acetaminophen and p-cresol are substrates for the human cytosolic sulfotransferases 1A1 (SULT1A1). This competitive binding can impede the ability for the liver to detoxify acetaminophen, leading to NAPQI accumulation.
In addition to their direct effects on drug metabolism, the gut microbiota can regulate the expression of several CYP P450 enzymes and nuclear receptors. An RNA sequencing study comparing the transcript levels of phase I and phase II drug metabolizing enzymes and transporters in the liver of germ-free mice with conventional mice showed significant differential expression in many of these genes.59 Most notably, cyp3a11 the mouse ortholog of CYP3A4 in the human, which metabolizes more than 40% of the drugs on the market,45 was significantly decreased in the livers of germ-free mice. This means that any environmental factors known to perturb the gut microbiota such as antibiotics, prebiotics or diet may alter not only the bacteria themselves, but can have consequences on the host ability to metabolize many xenobiotics.
Pharmacodynamic Interactions
The microbiome can also modify the host response to drug efficacy and toxicity. Although not as well described for cardiovascular drugs, the mechanism by which the microbiome modifies both the toxicity and efficacy of chemotherapeutic drugs has recently described using a mechanistic framework called TIMER-- Translocation, Immunomodulation, Metabolism, Enzymatic degradation and Reduced diversity.60 For example in mice, cyclophosphamide causes intestinal villi shortening and translocation of commensal bacteria. This translocation resulted in a reduction in the bacterial species required to mediate the beneficial type 17 T helper (TH17) anti-tumor response.61 Furthermore, long-term treatment with vancomycin, commonly used for the treatment of gram-positive infections in the hospital, inhibited the antitumor effects of cyclophosphamide.
The gut microbiome has been shown to influence the antitumor immune responses in the treatment of melanoma with checkpoint inhibitors which target programmed cell death protein (PD-1).62 In 112 patients with melanoma undergoing treatment with anti-PD-1 therapy, the fecal microbiome of responders were enriched with Faecalibacterium and those with a high abundance had a significantly prolonged progression free survival compared with those with a low abundance. Additionally, fecal overabundance of the Bacteroidetes phylum was correlated with decreased occurrence of immune-mediated colitis, a common side effect seen with checkpoint inhibitor therapy.63 A favorable microbiome profile may help predict response and minimize toxicity with anti-PD-1 immunotherapy and further evaluation of modulating the gut microbiome in patients receiving checkpoint blockade is warranted.
The impact of the gut microbiome on cardiovascular therapeutics
The knowledge of how the microbiome influences drug response is still in its infancy and interaction of the gut microbiome with cardiovascular medications is only now being uncovered. Metagenomic sequencing of stool samples in 1135 participants from a cohort study in the Netherlands, revealed that the use of several pharmaceutical agents had a significant impact on the gut microbiome, including statins, beta-blockers, angiotensin-converting enzyme inhibitors, and platelet aggregation inhibitors.64 Some of these associations have been replicated in another cross-sectional study of 2700 individuals from the British TwinsUK cohort, confirming the association of several bacterial taxa with medications use, including beta-blockers and alpha blockers.65 At this time the specific effects and mechanisms are unknown (Table) and interventional studies will be required to clarify the potential bi-directional effects of drug-microbiota interactions.
Table.
Known and proposed mechanisms by which the gut microbiota may influence cardiovascular drug outcomes
| Drug | Bacteria | Mechanism(s) | Outcome |
|---|---|---|---|
| Known microbiome-drug interactions | |||
| Digoxin52 | Eggerthella lenta | Inactivation by reduction | Bacterial reductase activity decreases amount of active drug reaching target tissues |
| Proposed microbiome-drug interactions | |||
| Simvastatin66 | Not known | Microbial derived bile acids competing for host uptake transporters Alteration in bacterial communities with bile salt hydrolase (bsh) activity |
Decreased amount of drug reaching target tissues Variability in FXR receptor signaling |
| Rosuvastatin67, 68 | Not known | Alteration in host gene expression of bile acid metabolism pathways Alteration in bacterial communities with bile salt hydrolase (bsh) activity |
Variability in FXR receptor signaling |
| Atorvastatin69–71 | Not known | Decreased amount of secondary bile acids | Variability in FXR receptor signaling |
| Amlodipine72 | Not known | Pre-systemic metabolism by dehydrogenation | Decreased amount of active drug reaching target tissues |
| Captopril73 | Not known | Not known | Decreased intestinal permeability and improved villi length |
FXR- farnesoid X receptor
Gut microbiome and statin response
Interindividual variation to statin response is well known. High dose statins are expected to reduce LDL-C by at least 50%, however treatment response is individual and varied, with a high proportion of individuals exhibiting a suboptimal outcomes (<30% reduction in LDL-C).74 A meta-analysis of statin interventional trials encompassing 32,258 patients from 37 trials, showed the standard deviation of LDL-C reduction for all statins and doses ranged from 12.8 to 17.9% and the percentage of patients experiencing suboptimal responses ranged from 5.3 to 53.3%.74 This variability was not related to specific statin or dose. In the Jupiter study75, where healthy subjects with a median baseline LDL-C of 108 mg/dL (IQR 94–119 mg/dL) received rosuvastatin 20 mg, 46% experienced a LDL-C reduction of ≥50%, 43% experienced a LDL-C reduction between 0% and 50%, and 11% experienced no reduction or an increase in LDL-C compared with baseline. Genome-wide association (GWA) analysis has been applied to understanding the genetic basis of the variation in LDL-C lowering response, but has identified only a handful of variants.47, 76–78
Simvastatin, rosuvastatin and atorvastatin, three of the most commonly prescribed statin medications, display evidence for modulation by the gut microbiome. In a study involving plasma metabolomic profiling in 100 human subjects, baseline concentrations of three secondary, bacterial-derived bile acids, lithocholic acid, taurolithocholic acid and glycolithocholic acid, positively predicted the magnitude of simvastatin induced LDL-C lowering.66 In addition pre-treatment concentrations of chenodeoxycholic acid and deoxycholic acid were correlated with on-treatment plasma simvastatin concentrations. The authors showed that plasma levels of seven bile acids, including the previously mentioned lithocholic acid and taurolithocholic acid, were higher in minor allele carriers of the SNP rs4149056 in the gene encoding the organic anion transporter SLCO1B1. This is the same nonsynonymous variant identified in genome-wide association analysis associated with increased risk of simvastatin induced myothpathy.51 This SNP has been shown to reduce the activity of the transporter in vitro52 and has a strong impact on simvastatin pharmacokinetics.53 The mechanism(s) of the association of bile acids with LDL-C response to statin medications remains unknown. Gut derived bile acids may directly influence drug pharmacokinetics, and therefore drug response, by competing with drug transport mechanisms across the gut lumen, or by influencing uptake in the liver. Statins may also directly alter the abundance of specific bacterial species, modulating the abundance of bacteria encoding enzymes involved in bile acid metabolism; however these hypotheses remain to be tested.
A proof-of-concept study in human subjects with hyperlipidemia showed that 4–8 weeks of rosuvastatin treatment significantly altered the gut microbiome and the abundance of specific bacterial taxa which was correlated to the LDL-C lowering response of the drug.67 The phyla Firmicutes and Fusobacteria were negatively associated with LDL-C levels, but Cyanobacteria and Lentisphaerae were positively associated. However, this observational study lacked a control group, and the bacterial sequencing was performed only after treatment, thus the investigators were unable to assess the change in the microbiome. It has been hypothesized that the LDL-C response to statins may be due to the activity of bacteria containing bile salt hydrolases (bsh). A randomized placebo controlled clinical trial of 127 participants, treatment with Lactobacillus reuteri, a bacteria with elevated bile salt hydrolase (bsh) activity, was shown to significantly reduced LDL-C levels.79 In addition, individual changes in LDL-C were inversely correlated with circulating bile acids. Bile salt hydrolase activity has been characterized in Firmicutes,80 a species associated with LDL-C in response to rosuvastatin identified in the previous study.67 Treatment of mice with rosuvastatin modulated the composition of the bacterial species, and reduced hepatic expression of the CYP27a1,68 which encodes an enzyme regulating the conversion of 7α-hydroxycholesterol to chenodeoxycholic acid, a primary bile acid.35, 81, 82 Rosuvastatin, by inducing gene expression changes, altered the concentrations of bile acids which contribute to the composition of gut microbiota.
In an animal model, atorvastatin decreased the amounts of secondary bile acids in the small and large intestine.69 Treatment of rats fed a high-fat diet with atorvastatin showed significant alterations in gut microbial communities towards species similar to healthy control rats fed a normal diet.70 A cross-sectional study compared the gut microbiome in 15 untreated hypercholesterolemia patients, 27 atorvastatin treated hypercholesterolemia patients, with 19 healthy subjects.71 The untreated hypercholesterolemia patients showed a distinct bacterial signature with species associated with inflammation (e.g. Collinsella, Streptococcus) while atorvastatin treated patients had an increased abundance of putative anti-inflammatory species (Akermansia muciniphila, Faecalibacterium prausnitzii).
Rosuvastatin has also been associated with an uncommon side effect of fish odor syndrome which is mediated by the gut microbiome.83 Rosuvastatin contains a tertiary amine, which competed with TMA for metabolism by FMO3 in the liver, elevating levels of TMA and subsequent excretion in the urine, resulting in fish odor syndrome.
Gut microbiome and antihypertensive drugs
The data on the effects of the gut microbiome and specific antihypertensive medications is limited and mainly based on animal studies. Amlodipine is a calcium channel blocker that is relatively well absorbed across the GI tract with a bioavailability of 60%. The drug is primarily oxidized in the liver by CYP3A4 enzymes to inactive metabolites. Amlodipine, when incubated ex vivo with human and rat fecal suspensions over a 72 hour period, decreased in concentrations while its major metabolite increased.72 Pharmacokinetic analysis of orally administered amlodipine in rats showed a significantly increased amlodipine area under the curve when pre-treated with ampicillin, indicating enhanced absorption across the GI tract as a result of suppression of metabolic activity of the gut microbiota. A three day separation between the administration of ampicillin and amlodipine was employed to avoid a direct drug-drug interaction with the two medications. Therefore any alterations in the gut flora, such as seen with antibiotics, may result in variation in blood pressure control
Hypertensive rats treated with captopril reversed the dysbiotic state associated with hypertension by decreasing intestinal permeability and fibrosis and improving villi length.73 Further work is needed to determine which specific bacterial genera are associated with inflammation and hypertension and how these are impacted by antihypertensive drugs.
Anticoagulant, anti-platelet and anti-inflammatory drugs
Warfarin is a commonly prescribed anticoagulant used for the treatment of thromboembolic disorders and is subject to numerous drug and food interactions.84 Warfarin produces its effects by inhibiting the vitamin K dependent activation of clotting factors II, VII, IX and Z. Concomitant administration of warfarin and antibiotics has been shown to increase bleeding events with warfarin.85 Antibiotics may elevate the bleeding risk by interfering with warfarin metabolism through inhibition or induction of CYP enzymes.84 Alternatively, antibiotics may disrupt the intestinal flora resulting in an elimination of vitamin K producing bacteria, such as bacteriodes, leading to alteration in coagulation status.86
Aspirin is a commonly prescribed antiplatelet drug used to treat and prevent CVD by inhibition of cyclooxygenase (COX)-1 enzymes and prostaglandin synthesis. Non-steroidal anti-inflammatory drugs (NSAIDs) are known to damage the mucosa of the upper GI tract87 and gut microbiome composition is a contributing factor.88, 89 The gut microbiome composition was associated with use of the NSAIDs including aspirin.90 Four species were able to discriminate aspirin users from non-users (Prevotella species, Bacteroides species, a member of the Ruminococaceae family and Barnesiella species) with an area under the curve of the receiver operating curve 0.96 (95% CI 0.84–1.00). Mechanistic studies in mice reveals a bidirectional effect of another NSAID, indomethacin, on the gut microbiome- where indomethacin altered the composition of the microbiome and the microbiome altered indomethacin metabolism.89 Gut derived β-glucuronidases deconjugate indomethacin metabolites allowing for enterohepatic recycling of the drug, prolonged drug exposure and enhanced pharmacological effects. Aspirin resistance has been well-documented in the population and occurs in 6–60% of individuals, depending on how it is defined.91 While genetic studies have identified several genes associated with variation in response to aspirin,92 the contribution of the gut microbiome on variation in aspirin response had not been fully explored.93
Targeting of the gut microbiome and implications for drug development
While still in the early stages, efforts are ongoing to develop therapeutics which improve cardiovascular health through modulation of the microbiome, thus directly targeting microbes and microbial metabolism rather than the host. This approach has considerable appeal, as a precision medicine strategy that would potentially have far smaller chances of side effects compared with drugs that target host metabolism. For example small molecules inhibiting the microbial production of the TMAO precursor, trimethylamine, are currently being pursued.94, 95 Further, efforts to increase production of anti-inflammatory N-acyl phosphatidylethanolamines (NAPEs) have been successful in treating obesity and cardiometabolic disease in animal models. 96, 97
Knowledge of microbial reactions may be used in rational drug design as chemical groups that are known to be metabolized by the microbial species could be removed or modified to control drug delivery. Inhibitors of known microbial enzymes are also being developed. Targeting microbial reactions may be beneficial in limiting drug toxicity. For example, inhibition of bacterial β-glucuronidase may prove useful for irinotecan associated diarrhea.57, 60 A systems approach to drug development that incorporates host and microbial genetics, environmental inputs (e.g. diet, exercise, alcohol use) along with clinical and disease data (e.g. critical illness98) will need to be considered.
Challenges and Future Directions
There is a considerable challenge in understanding how to utilize knowledge of microbiome composition and function in clinical practice. Beyond descriptive studies which show differences in microbiome composition between groups of patients, we need longitudinal and interventional studies to understand the dynamic nature of the microbiome over time, particularly during development of disease, and in response to dietary, lifestyle, or pharmacological treatment of disease. Consideration of the microbiome as a factor in drug non-response, and in variability of responses will be important in understanding how much the microbiome plays a role in drug efficacy. At the same time, effective strategies to alter microbiome composition must be tested. Phytochemicals and other dietary substances are also metabolized by the gut microbiome, which independently influence disease risk, and potentially interact with medications.99 Simultaneous profiling of the gut microbiome along with metabolomics and other omics profiling, in conjunction with functional interrogation, will allow us to pinpoint the mechanisms by which the gut microbiome interacts with the host to modulate drug metabolism and response.25
Conclusion
Several drugs used for the treatment of CVD show evidence for an interaction with the gut microbiome. Our current understanding of microbial-host interactions and their impact on CVD and drug response is still superficial. Comprehensive deep phenotyping studies are required to understand the directionality and complex relationship between the host, the microbiome, and medications. Well-controlled clinical studies to investigate the impact of differences in microbiome composition on outcomes in response to treatment of disease are still needed. A deeper understanding of the molecular mechanisms by which the gut microbiome contributes to CVD risk and drug response will enable us to improve outcomes for patients with cardiovascular disease, and move towards microbiome-informed precision medicine.
Acknowledgments
Sources Funding: National Heart Lung Blood Institute Mentored Patient-Oriented Research Career Development Award (K23HL143161) to ST. American Heart Association Scientist Development Grant (15SDG24890015) to JFF.
Abbreviations
- bsh
bile salt hydrolase
- cgr
cardiac glycoside reductase
- COX
cyclooxygenase
- CVD
cardiovascular disease
- CYP P450
cytochrome P450
- FXR
farnesoid X receptor
- GI
gastrointestinal
- NAPEs
N-acyl phosphatidylethanolamines
- NAPQI
N-acetyl p-benzoquinone imine
- NSAIDs
Non-steroidal anti-inflammatory drugs
- PD-1
programmed cell death protein
- SCFA
short chain fatty acids
- SULT1A1
sulfotransferases 1A1
- TH17
type 17 T helper
- TMAO
trimethylamine-N-oxide
Footnotes
Disclosures: None
References
- 1.Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, Guiducci C, Perola M, Jula A, Sinisalo J, et al. A multilocus genetic risk score for coronary heart disease: Case-control and prospective cohort analyses. Lancet. 2010;376:1393–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young DR, Hivert MF, Alhassan S, Camhi SM, Ferguson JF, Katzmarzyk PT, Lewis CE, Owen N, Perry CK, Siddique J, et al. Sedentary behavior and cardiovascular morbidity and mortality: A science advisory from the american heart association. Circulation. 2016;134:e262–279. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson JF, Allayee H, Gerszten RE, Ideraabdullah F, Kris-Etherton PM, Ordovás JM, Rimm EB, Wang TJ, Bennett BJ. Nutrigenomics, the microbiome, and gene-environment interactions: New directions in cardiovascular disease research, prevention, and treatment: A scientific statement from the american heart association. Circ Cardiovasc Genet. 2016;9:291–313. doi: 10.1161/HCG.0000000000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: An integrative view. Cell. 2012;148:1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL. Analysis of the microbiome: Advantages of whole genome shotgun versus 16s amplicon sequencing. Biochem Biophys Res Commun. 2016;469:967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. [DOI] [PubMed] [Google Scholar]
- 7.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455:1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koren O, Spor A, Felin J, Fåk F, Stombaugh J, Tremaroli V, Behre CJ, Knight R, Fagerberg B, Ley RE, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A. 2011;108:4592–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlsson FH, Fak F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, Backhed F, Nielsen J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012;3:1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, Dekens JA, Brandsma E, Marczynska J, Imhann F, Weersma RK, et al. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ Res. 2015;117:817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caesar R, Nygren H, Oresic M, Backhed F. Interaction between dietary lipids and gut microbiota regulates hepatic cholesterol metabolism. J Lipid Res. 2016;57:474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marques FZ, Mackay CR, Kaye DM. Beyond gut feelings: How the gut microbiota regulates blood pressure. Nat Rev Cardiol. 2018;15:20–32. [DOI] [PubMed] [Google Scholar]
- 13.Tang WHW, Li DY, Hazen SL. Dietary metabolism, the gut microbiome, and heart failure. Nat Rev Cardiol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haiser HJ, Turnbaugh PJ. Developing a metagenomic view of xenobiotic metabolism. Pharmacol Res. 2013;69:21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A. 2009;106:14728–14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spanogiannopoulos P, Bess EN, Carmody RN, Turnbaugh PJ. The microbial pharmacists within us: A metagenomic view of xenobiotic metabolism. Nat Rev Microbiol. 2016;14:273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donia MS, Fischbach MA. Human microbiota. Small molecules from the human microbiota. Science. 2015;349:1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartolomaeus H, Balogh A, Yakoub M, Homann S, Marko L, Hoges S, Tsvetkov D, Krannich A, Wundersitz S, Avery EG, et al. The short-chain fatty acid propionate protects from hypertensive cardiovascular damage. Circulation. 2019;139:1407–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porez G, Prawitt J, Gross B, Staels B. Bile acid receptors as targets for the treatment of dyslipidemia and cardiovascular disease. J Lipid Res. 2012;53:1723–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, Fu X, Wu Y, Mehrabian M, et al. Gut microbial metabolite tmao enhances platelet hyperreactivity and thrombosis risk. Cell. 2016;165:111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma A, Buschmann MM, Gilbert JA. Pharmacomicrobiomics: The holy grail to variability in drug response? Clin Pharmacol Ther. 2019;106:317–328. [DOI] [PubMed] [Google Scholar]
- 26.ElRakaiby M, Dutilh BE, Rizkallah MR, Boleij A, Cole JN, Aziz RK. Pharmacomicrobiomics: The impact of human microbiome variations on systems pharmacology and personalized therapeutics. OMICS. 2014;18:402–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ott SJ, El Mokhtari NE, Musfeldt M, Hellmig S, Freitag S, Rehman A, Kuhbacher T, Nikolaus S, Namsolleck P, Blaut M, et al. Detection of diverse bacterial signatures in atherosclerotic lesions of patients with coronary heart disease. Circulation. 2006;113:929–937. [DOI] [PubMed] [Google Scholar]
- 28.Brandsma E, Kloosterhuis NJ, Koster M, Dekker DC, Gijbels MJJ, van der Velden S, Rios-Morales M, van Faassen MJR, Loreti MG, de Bruin A, et al. A proinflammatory gut microbiota increases systemic inflammation and accelerates atherosclerosis. Circ Res. 2019;124:94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang WH, Kitai T, Hazen SL. Gut microbiota in cardiovascular health and disease. Circ Res. 2017;120:1183–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown JM, Hazen SL. Microbial modulation of cardiovascular disease. Nat Rev Microbiol. 2018;16:171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown JM, Hazen SL. The gut microbial endocrine organ: Bacterially derived signals driving cardiometabolic diseases. Annu Rev Med. 2015;66:343–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang WH, Wang Z, Fan Y, Levison B, Hazen JE, Donahue LM, Wu Y, Hazen SL. Prognostic value of elevated levels of intestinal microbe-generated metabolite trimethylamine-n-oxide in patients with heart failure: Refining the gut hypothesis. J Am Coll Cardiol. 2014;64:1908–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuett K, Kleber ME, Scharnagl H, Lorkowski S, Marz W, Niessner A, Marx N, Meinitzer A. Trimethylamine-n-oxide and heart failure with reduced versus preserved ejection fraction. J Am Coll Cardiol. 2017;70:3202–3204. [DOI] [PubMed] [Google Scholar]
- 34.Gérard P Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens. 2013;3:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, Hill C, Gahan CGM. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci U S A. 2014;111:7421–7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ridlon JM, Kang D-J, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. [DOI] [PubMed] [Google Scholar]
- 37.Ryan KK, Tremaroli V, Clemmensen C, Kovatcheva-Datchary P, Myronovych A, Karns R, Wilson-Perez HE, Sandoval DA, Kohli R, Backhed F, et al. Fxr is a molecular target for the effects of vertical sleeve gastrectomy. Nature. 2014;509:183–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adnan S, Nelson JW, Ajami NJ, Venna VR, Petrosino JF, Bryan RM Jr., Durgan DJ. Alterations in the gut microbiota can elicit hypertension in rats. Physiol Genomics. 2017;49:96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G, Wu S, Liu W, Cui Q, Geng B, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017;5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, Haase S, Mahler A, Balogh A, Marko L, et al. Salt-responsive gut commensal modulates th17 axis and disease. Nature. 2017;551:585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, Brunet I, Wan LX, Rey F, Wang T, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci U S A. 2013;110:4410–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.National health care spending in 2017: Growth slows to post–great recession rates; share of gdp stabilizes. Health Affairs. 2019;38: 10.1377/hlthaff.2018.05085 [DOI] [PubMed] [Google Scholar]
- 43.Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364:1144–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shamliyan TA, Kane RL. Drug-related harms in hospitalized medicare beneficiaries: Results from the healthcare cost and utilization project, 2000–2008. J Patient Saf. 2016;12:89–107. [DOI] [PubMed] [Google Scholar]
- 45.Wilkinson GR. Drug metabolism and variability among patients in drug response. N Engl J Med. 2005;352:2211–2221. [DOI] [PubMed] [Google Scholar]
- 46.Roden DM, Wilke RA, Kroemer HK, Stein CM. Pharmacogenomics: The genetics of variable drug responses. Circulation. 2011;123:1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Postmus I, Trompet S, Deshmukh HA, Barnes MR, Li X, Warren HR, Chasman DI, Zhou K, Arsenault BJ, Donnelly LA, et al. Pharmacogenetic meta-analysis of genome-wide association studies of ldl cholesterol response to statins. Nat Commun. 2014;5:5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim D-H. Gut microbiota-mediated drug-antibiotic interactions. Drug Metab Dispos. 2015;43:1581–1589. [DOI] [PubMed] [Google Scholar]
- 49.Burton ME. Applied pharmacokinetics and pharmacodynamics : Principles of therapeutic drug monitoring. Baltimore: Lippincott Williams & Wilkins; 2006. [Google Scholar]
- 50.Sousa T, Paterson R, Moore V, Carlsson A, Abrahamsson B, Basit AW. The gastrointestinal microbiota as a site for the biotransformation of drugs. Int J Pharm. 2008;363:1–25. [DOI] [PubMed] [Google Scholar]
- 51.Saha JR, Butler VP Jr., Neu HC, Lindenbaum J. Digoxin-inactivating bacteria: Identification in human gut flora. Science. 1983;220:325–327. [DOI] [PubMed] [Google Scholar]
- 52.Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. Predicting and manipulating cardiac drug inactivation by the human gut bacterium eggerthella lenta. Science. 2013;341:295–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindenbaum J, Rund DG, Butler VP Jr., Tse-Eng D, Saha JR. Inactivation of digoxin by the gut flora: Reversal by antibiotic therapy. N Engl J Med. 1981;305:789–794. [DOI] [PubMed] [Google Scholar]
- 54.Oz HS, Ebersole JL. Application of prodrugs to inflammatory diseases of the gut. Molecules (Basel, Switzerland). 2008;13:452–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rafii F, Sutherland JB, Hansen EB Jr., Cerniglia CE. Reduction of nitrazepam by clostridium leptum, a nitroreductase-producing bacterium isolated from the human intestinal tract. Clin Infect Dis. 1997;25 Suppl 2:S121–122. [DOI] [PubMed] [Google Scholar]
- 56.Gupta E, Lestingi TM, Mick R, Ramirez J, Vokes EE, Ratain MJ. Metabolic fate of irinotecan in humans: Correlation of glucuronidation with diarrhea. Cancer Res. 1994;54:3723–3725. [PubMed] [Google Scholar]
- 57.Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, Venkatesh M, Jobin C, Yeh LA, Mani S, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoon E, Babar A, Choudhary M, Kutner M, Pyrsopoulos N. Acetaminophen-induced hepatotoxicity: A comprehensive update. J Clin Transl Hepatol. 2016;4:131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Selwyn FP, Cui JY, Klaassen CD. Rna-seq quantification of hepatic drug processing genes in germ-free mice. Drug Metab Dispos. 2015;43:1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol. 2017;14:356–365. [DOI] [PubMed] [Google Scholar]
- 61.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. Gut microbiome modulates response to anti-pd-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dubin K, Callahan MK, Ren B, Khanin R, Viale A, Ling L, No D, Gobourne A, Littmann E, Huttenhower C, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun. 2016;7:10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jackson MA, Verdi S, Maxan ME, Shin CM, Zierer J, Bowyer RCE, Martin T, Williams FMK, Menni C, Bell JT, et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat Commun. 2018;9:2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaddurah-Daouk R, Baillie RA, Zhu H, Zeng ZB, Wiest MM, Nguyen UT, Wojnoonski K, Watkins SM, Trupp M, Krauss RM. Enteric microbiome metabolites correlate with response to simvastatin treatment. PLoS One. 2011;6:e25482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Y, Song X, Zhou H, Zhou X, Xia Y, Dong X, Zhong W, Tang S, Wang L, Wen S, et al. Gut microbiome associates with lipid-lowering effect of rosuvastatin in vivo. Front Microbiol. 2018;9:530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nolan JA, Skuse P, Govindarajan K, Patterson E, Konstantinidou N, Casey PG, MacSharry J, Shanahan F, Stanton C, Hill C, et al. The influence of rosuvastatin on the gastrointestinal microbiota and host gene expression profiles. Am J Physiol Gastrointest Liver Physiol. 2017;312:G488–G497. [DOI] [PubMed] [Google Scholar]
- 69.Fu ZD, Cui JY, Klaassen CD. Atorvastatin induces bile acid-synthetic enzyme cyp7a1 by suppressing fxr signaling in both liver and intestine in mice. J Lipid Res. 2014;55:2576–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khan TJ, Ahmed YM, Zamzami MA, Mohamed SA, Khan I, Baothman OAS, Mehanna MG, Yasir M. Effect of atorvastatin on the gut microbiota of high fat diet-induced hypercholesterolemic rats. Sci Rep. 2018;8:662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khan TJ, Ahmed YM, Zamzami MA, Siddiqui AM, Khan I, Baothman OAS, Mehanna MG, Kuerban A, Kaleemuddin M, Yasir M. Atorvastatin treatment modulates the gut microbiota of the hypercholesterolemic patients. OMICS. 2018;22:154–163. [DOI] [PubMed] [Google Scholar]
- 72.Yoo HH, Kim IS, Yoo DH, Kim DH. Effects of orally administered antibiotics on the bioavailability of amlodipine: Gut microbiota-mediated drug interaction. J Hypertens. 2016;34:156–162. [DOI] [PubMed] [Google Scholar]
- 73.Santisteban MM, Qi Y, Zubcevic J, Kim S, Yang T, Shenoy V, Cole-Jeffrey CT, Lobaton GO, Stewart DC, Rubiano A, et al. Hypertension-linked pathophysiological alterations in the gut. Circ Res. 2017;120:312–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karlson BW, Wiklund O, Palmer MK, Nicholls SJ, Lundman P, Barter PJ. Variability of low-density lipoprotein cholesterol response with different doses of atorvastatin, rosuvastatin, and simvastatin: Results from voyager. Eur Heart J Cardiovasc Pharmacother. 2016;2:212–217 [DOI] [PubMed] [Google Scholar]
- 75.Ridker PM, Mora S, Rose L. Percent reduction in ldl cholesterol following high-intensity statin therapy: Potential implications for guidelines and for the prescription of emerging lipid-lowering agents. Eur Heart J. 2016;37:1373–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barber MJ, Mangravite LM, Hyde CL, Chasman DI, Smith JD, McCarty CA, Li X, Wilke RA, Rieder MJ, Williams PT, et al. Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS ONE. 2010;5:e9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chasman DI, Giulianini F, MacFadyen J, Barratt BJ, Nyberg F, Ridker PM. Genetic determinants of statin-induced low-density lipoprotein cholesterol reduction: The justification for the use of statins in prevention: An intervention trial evaluating rosuvastatin (jupiter) trial. Circ Cardiovasc Genet. 2012;5:257–264. [DOI] [PubMed] [Google Scholar]
- 78.Hopewell JC, Parish S, Offer A, Link E, Clarke R, Lathrop M, Armitage J, Collins R. Impact of common genetic variation on response to simvastatin therapy among 18 705 participants in the heart protection study. Eur Heart J. 2013;34:982–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jones ML, Martoni CJ, Prakash S. Cholesterol lowering and inhibition of sterol absorption by lactobacillus reuteri ncimb 30242: A randomized controlled trial. Eur J Clin Nutr. 2012;66:1234–1241. [DOI] [PubMed] [Google Scholar]
- 80.Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A. 2008;105:13580–13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Islam KBMS, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, Yokota A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–1781. [DOI] [PubMed] [Google Scholar]
- 82.Yokota A, Fukiya S, Islam KBMS, Ooka T, Ogura Y, Hayashi T, Hagio M, Ishizuka S. Is bile acid a determinant of the gut microbiota on a high-fat diet? Gut Microbes. 2012;3:455–459. [DOI] [PubMed] [Google Scholar]
- 83.Li M, Al-Sarraf A, Sinclair G, Frohlich J. Fish odour syndrome. CMAJ. 2011;183:929–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Holbrook AM, Pereira JA, Labiris R, McDonald H, Douketis JD, Crowther M, Wells PS. Systematic overview of warfarin and its drug and food interactions. Arch Intern Med. 2005;165:1095–1106. [DOI] [PubMed] [Google Scholar]
- 85.Lane MA, Zeringue A, McDonald JR. Serious bleeding events due to warfarin and antibiotic co-prescription in a cohort of veterans. Am J Med. 2014;127:657–663 e652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shearer MJ, Newman P. Metabolism and cell biology of vitamin k. Thromb Haemost. 2008;100:530–547. [PubMed] [Google Scholar]
- 87.Watanabe T, Tanigawa T, Nadatani Y, Nagami Y, Sugimori S, Okazaki H, Yamagami H, Watanabe K, Tominaga K, Fujiwara Y, et al. Risk factors for severe nonsteroidal anti-inflammatory drug-induced small intestinal damage. Dig Liver Dis. 2013;45:390–395. [DOI] [PubMed] [Google Scholar]
- 88.Takeuchi K, Satoh H. Nsaid-induced small intestinal damage--roles of various pathogenic factors. Digestion. 2015;91:218–232. [DOI] [PubMed] [Google Scholar]
- 89.Liang X, Bittinger K, Li X, Abernethy DR, Bushman FD, FitzGerald GA. Bidirectional interactions between indomethacin and the murine intestinal microbiota. Elife. 2015;4:e08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rogers MAM, Aronoff DM. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin Microbiol Infect. 2016;22:178 e171–178 e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mansour K, Taher AT, Musallam KM, Alam S. Aspirin resistance. Adv Hematol. 2009;2009:937352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beitelshees AL, Voora D, Lewis JP. Personalized antiplatelet and anticoagulation therapy: Applications and significance of pharmacogenomics. Pharmgenomics Pers Med. 2015;8:43–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim IS, Yoo DH, Jung IH, Lim S, Jeong JJ, Kim KA, Bae ON, Yoo HH, Kim DH. Reduced metabolic activity of gut microbiota by antibiotics can potentiate the antithrombotic effect of aspirin. Biochem Pharmacol. 2016;122:72–79. [DOI] [PubMed] [Google Scholar]
- 94.Wang Z, Roberts AB, Buffa JA, Levison BS, Zhu W, Org E, Gu X, Huang Y, Zamanian-Daryoush M, Culley MK, et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell. 2015;163:1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roberts AB, Gu X, Buffa JA, Hurd AG, Wang Z, Zhu W, Gupta N, Skye SM, Cody DB, Levison BS, et al. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat Med. 2018;24:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen Z, Guo L, Zhang Y, Walzem RL, Pendergast JS, Printz RL, Morris LC, Matafonova E, Stien X, Kang L, et al. Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity. J Clin Invest. 2014;124:3391–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.May-Zhang LS, Chen Z, Dosoky NS, Yancey PG, Boyd KL, Hasty AH, Linton MF, Davies SS. Administration of n-acyl-phosphatidylethanolamine expressing bacteria to low density lipoprotein receptor(−/−) mice improves indices of cardiometabolic disease. Sci Rep. 2019;9:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Otani S, Chihade DB, Coopersmith CM. Critical illness and the role of the microbiome. Acute Med Surg. 2019;6:91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Carmody RN, Turnbaugh PJ. Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J Clin Invest. 2014;124:4173–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]