Abstract
Phase separation of multivalent protein and RNA molecules enables cells the formation of reversible non-stoichiometric, membraneless assemblies. These assemblies, referred to as biomolecular condensates, help with the spatial organization and compartmentalization of cellular matter. Each biomolecular condensate is defined by a distinct macromolecular composition. Distinct condensates have distinct preferential locations within cells, and they are associated with distinct biological functions, including DNA replication, RNA metabolism, signal transduction, synaptic transmission, and stress response. Several proteins found in biomolecular condensates have also been implicated in disease, including Huntington’s disease, ALS, and several types of cancer. Disease-associated mutations in these proteins have been found to affect the material properties of condensates as well as the driving forces for phase separation. Understanding the intrinsic and extrinsic forces driving the formation and dissolution of biomolecular condensates via spontaneous and driven phase separation is an important step in understanding the processes associated with biological regulation in health and disease.
Keywords: phase separation, granules, biomolecular condensates, membraneless organelles, phase diagram, protein disorder
Introduction
Cells employ myriad ways to organize the enormous number of proteins, RNA, DNA, lipids, and other molecules within them into functional compartments, including membrane-bound organelles. Among the latter, the nucleus keeps DNA contained, lysosomes sequester destructive proteases and keep them from wreaking havoc on the cell, and the endoplasmic reticulum keeps the protein-producing machinery of ribosomes in one location. In addition to membrane-bound organelles, the cell also uses membrane-less organelles or bodies to maintain structure and organization. These structures often consist of networks of proteins and/or RNA that form by phase separation to create spatially separated, yet open, dynamic systems. Examples of membrane-less organelles that appear to form via phase separation include P granules, stress granules, promyelocytic leukemia (PML) bodies, nuclear speckles, and the centrosome.
Proteins involved in phase separation often contain dimerization or oligomerization domains, RNA-binding domains, and/or intrinsically disordered regions (IDR) that appear to mediate multivalent homo or hetero interactions. Another feature of phase separation is the existence of a concentration threshold known as a saturation concentration. Unlike protein-protein complexes that form as the concentration of one of the components increases, phase-separated structures only form once the concentration of the components has exceeded a given threshold, which depends on the system and its surroundings, such as the salt concentration, temperature, and other ions. Phase-separated structures can have different material properties. While the notion of liquid-like condensates has gained traction, there is growing awareness that condensates can be viscoelastic liquids, network fluids, liquid-crystalline, micellar, or semi/para-crystalline. Defining hallmarks of a liquid-like condensate are a spherical shape, the ability to fuse with other condensates, and the free diffusion of molecular components within each compartment. These properties allow liquid-like condensates to assemble and disassemble rapidly and concentrate components in a fluid, dynamic environment.
Dysregulation of material properties of condensates has been proposed to be a factor in several diseases, including amyotrophic lateral sclerosis (ALS), Huntington’s disease, and some types of cancer. Mutations implicated in these diseases often affect the driving forces for phase separation behavior, and this appears to also impact the normal functions, of the respective proteins and/or their interaction partners.
On February 20, 2019, several experts who work in the area of phase separation of protein and RNA molecules convened at the New York Academy of Sciences to discuss how phase separation contributes to biomolecular function and the onset of disease. The presenters discussed new tools for monitoring phase transitions within living cells, described atomic-level structural detail of phase-separated structures, and proposed new rationales for how phase transition plays a role in regulating protein function. This report describes the speakers’ presentations at the one-day symposium.
New tools to monitor phase separation
Clifford Brangwynne from Princeton University opened the meeting by describing his work on stress granule assembly. Stress granules are phase separated cytoplasmic condensates of protein and RNA that form in response to stress stimuli. In the lab, stress granules can be induced via heat shock or with arsenite treatment. Under these conditions, translating RNA/ribosome complexes disassemble, flooding the cytoplasm with exposed mRNA, which phase separate with stress granule proteins.
Brangwynne described the role of G3BP, a protein necessary for stress granule formation. G3BP knockout cells fail to form stress granules after arsenite treatment; adding back the wild type protein rescues this phenotype. The architecture of G3BP is similar to that of many proteins involved in phase transition, with an N-terminal oligomerization domain (NTF2), an IDR, and an RNA-binding domain. Unpublished work from Brangwynne’s lab used optogenetics to dissect the molecular features of G3BP in driving stress granule phase separation.
Brangwynne stressed the power of a tool like Corelets, which allows one to map phase transitions in living cells. In addition to Corelets, Brangwynne’s lab has developed other technologies to probe and quantify phase transitions within living cells, including Optodroplets1 and CasDrops.2
RNA localization in P granules
Geraldine Seydoux from Johns Hopkins School of Medicine discussed her work in delineating how mRNAs get into P granules. P granules are RNA/protein condensates that asymmetrically segregate toward one end of a dividing C. elegans embryo. After division, cells that contain P granules become germline cells.
Seydoux focused on two P granule proteins: MEG-3 and PGL-3. Typical of many proteins found in condensates, both contain oligomerization domains and an IDR. Ex vivo experiments of purified P granules reveal that the granules consist of at least two phases, a MEG-3-containing phase and a PGL-3 containing phase. Both ex vivo experiments of purified granules and experiments of granules assembled in vitro showed that the phases exhibit different physical properties. The PGL phase exhibits liquid-like behavior and dissolves quickly in buffer. In contrast, the MEG3 phase exhibits gel-like behavior and is more viscous and sturdier than the PGL phase.5 Seydoux showed that, in vitro, RNA localizes to the gel-like MEG3 phase and that, in vivo, MEG-3 is essential to recruit RNAs to P granules. Work in other RNA/protein condensates, including stress granules6 and balbiani bodies7 has also shown that RNA is found in a less dynamic gel phase. An intriguing possibility is that despite the liquid-like properties of RNA granules, RNAs in the granules are trapped in gel-like condensates.
The role of disorder in the nuclear pore complex
David Cowburn of the Albert Einstein College of Medicine presented his work on understanding how the nuclear pore complex (NPC) accomplishes its function of selectively allowing RNA and proteins to passively diffuse into and out of the nucleus. The NPC is an enormous multi-protein complex. The yeast complex, of which a high-resolution structure was published last year3, contains over 500 proteins. Structural analyses have been complicated due not only to the complex’s size, but also to the fact that the interior channel of the complex, which is responsible for the complex’s transport function, is intrinsically disordered.
The NPC channel consists of multiple intrinsically disordered phenylalanine/glycine (FG) repeat motifs that extend from nucleoporin folded segments. How these FG repeats provide both rapid and selective transport of proteins and RNA through the NPC is unknown and may provide new targets for therapeutics. Data from Cowburn’s and collaborators labs show that FG repeats specifically interact with transport factors with fast-binding kinetics, yet remain largely disordered.4 In addition, both isothermal titration calorimetry (ITC) and nuclear magnetic resonance (NMR) data demonstrate that the binding affinity for FG repeats for the transport factor NTF2 increases as the number of FG repeats increases. Because NMR experiments reveal that the binding sites on NTF2 do not change with increasing FG repeat number, Cowburn postulated that the dependency of binding affinity on FG repeat number is due to an increase in the local concentration of FG repeats, and thus an increase in the probability of rebinding, and not to an increase in the number of binding sites on NTF2.5
To better understand the mechanism of FG/NTF2 binding at an atomic level, Cowburn and collaborators conducted all-atom, long timescale molecular dynamics simulations. Consistent with the NMR data, the simulations show fast binding kinetics between FG repeats and NTF2, with most of the interactions within the FSFG repeat. The simulations provide additional insight on the binding mechanism, supporting a slide-and-exchange mechanism in which FG motifs reversibly slide in and out of NTF2 interaction sites, rapidly transitioning between strong and weak interactions site.6 Future work remains to be done on bridging these atomic-scale simulations with the experimental, biophysical data.7
Phase transitions driven by huntingtin
Rohit Pappu from the James McKelvey School of Engineering at Washington University in Saint Louis described his work on understanding two general mechanisms that drive phase separation: the use of block copolymeric architecture in proteins that drive phase separation and the modulation of phase separation by ligand binding.
Pappu uses exon 1 of huntingtin, the protein responsible for Huntington’s disease, as his model system. Huntingtin consists of an N-terminal amphipathic region, followed by a poly-glutamine (polyQ) region and a proline rich region. Expansion of the polyQ region correlates with Huntington’s disease severity and time of onset. Based on sequence, the entire length of the protein is predicted to be intrinsically disordered–a phrase that is often conflated with the protein being unstructured. Like huntingtin, many proteins involved in phase separation contain a block copolymeric architecture, i.e., blocks of repeating sequences that can either be folded or disordered.
Pappu showed how the length of the polyQ region affects the structure and phase separation behavior of huntingtin. Using intramolecular distances generated by single molecule fluorescence resonance energy transfer (smFRET) as constraints in atomic-level simulations, Pappu’s lab has generated a structural ensemble of huntingtin. The simulations revealed a “tadpole-like” architecture in which the amphipathic and polyQ regions form a globular head that becomes more compact as the length of the polyQ region increases. The proline-rich region extends as a semi-flexible tail. Huntingtin can undergo two phase transitions. In low concentrations, the protein exists primarily as monomers and oligomers. As the concentration increases past a given threshold, the protein shifts to form a micellar, or sphere-like, structure, in which the globular heads of monomers form an inner core surrounded by the proline-rich tail. Upon further concentration increases, a second threshold is reached in which the proteins form fibrils. The concentration thresholds between phase transitions depend on the length of the polyQ region: the longer the polyQ region, the lower the concentration needed to form the next phase.8
In addition to the polyQ region, ligand binding can also shift the phase concentration threshold. Profilin is known to bind to polyproline regions9 and has been shown to reduce huntingtin aggregation and cytotoxicity.10,11 Pappu showed that profilin shifts the concentration threshold for the monomer to micelle phase transition toward higher concentrations, thus suggesting that profilin preferentially binds huntingtin in the monomeric/oligomeric phase. Pappu also showed that profilin has weak, auxiliary interactions with the polyQ region, which may help explain why profilin preferentially binds the monomeric/oligomeric phase of huntingtin compared to the micellar phase, where the polyQ region is less accessible.8 Overall, the talk highlighted two major conceptual themes: the complexity of phase diagrams that often encompass more than just one type of phase boundary and the concept of polyphasic linkage, which refers to the ability of ligands to stabilize or destabilize phase separation via preferential interactions with a particular phase. This concept is of direct relevance to uncovering the role of a multitude of ligands in intracellular phase transitions.
Data blitz presentations
Carlos Castañeda of Syracuse University presented his results on how mutations in the protein ubiquilin-2 (UBQLN2) affect phase behavior. UBQLN2 is involved in protein quality control by shuttling poly-ubiquitinated proteins to the proteasome for degradation. Mutations in UBQLN2 have been associated with ALS.
Castañeda showed that UBQLN2 is recruited to stress granules in vivo and phase separates in vitro.18 Many of the ALS-associated mutations in UBQLN2 are found in its proline-rich region, which was also shown to be important for modulating phase separation behavior. Castañeda monitored the phase separation behavior of eleven ALS-linked mutations via turbidity assays and microscopy. While some mutations showed phase transition profiles similar to that of the wild-type protein, others significantly perturbed the phase transition behavior.12
Via NMR spectroscopy, Castañeda was able to identify a set of positions that show concentration-dependent chemical shift perturbations, suggesting that they are involved in UBQLN2 oligomerization.13 Since oligomerization is required for phase transition, Castañeda dubbed these positions “stickers” and proposed that they drive phase separation via self-association. Castañeda’s group created 95 point mutations in a set of sticker and spacer sites in a C-terminal construct of UBQLN2. Point substitutions in sticker regions disrupted the phase transition profile of UBQLN2 while substitutions in the spacer regions did not. He also observed qualitative trends in the type of substitutions. For example, hydrophobic substitutions only at sticker sites tended to promote phase transitions at lower temperatures while acidic substitutions at both sticker and spacer sites tended to change the shape of the phase transition profile, perhaps mimicking the effect of phosphorylation.14
Steven Metallo of Georgetown University discussed the importance of C-H-π interactions in promoting phase transitions. Metallo’s data is on the FSFG repeat found in the NPC; however, due to the prevalence of phenylalanine and glycine in phase separation motifs, he speculated that this could be a general theme among phase-separating proteins.
Metallo showed that an FSFG peptide forms a stiff hydrogel composed of micron-long branched fibers at high concentrations. Unlike gels that are driven by hydrophobic interactions, the FSFG gel becomes stiffer at lower temperatures. Metallo hypothesized that C-H-π interactions may be driving FSFG gel formation. C-H-π interactions exist when there is a highly polarized C-H bond that can interact with the pi orbital of an aromatic group. The interaction strength is not dependent on solvent hydrophobicity. Consistent with this hypothesis, FSFG gel strength was not dependent on solvent hydrophobicity. However, gel strength was significantly weaker in acetonitrile, which would be expected to compete with the C-H-π interaction due to its highly polarized C-H bond.
Metallo showed that a peptide containing a tri-FSFG repeat displayed similar behavior to the FSFG peptide, suggesting that C-H-π interactions may play a role in the full-length protein. Future work will focus on directly probing C-H-π interactions via NMR and infrared (IR) spectroscopy analyses.
Luke Berchowitz from Columbia University Irving School of Medicine demonstrated that some amyloid fibrils may play a functional role and can be broken down when they are no longer needed. This runs contrary to the popular belief that amyloid fibrils are considered terminal, irreversible, and pathological endpoints of phase-separated assemblies. Berchowitz focused on the yeast protein Rim4, an RNA-binding protein that is essential for meiosis. He used Semi-denaturing detergent agarose gel electrophoresis (SDD-AGE) to show that during meiosis I, Rim4 forms amyloid-like assemblies in vivo. These fibrils are dependent on the intrinsically disordered region of Rim4 and are cleared by the cells at the beginning of meiosis II.15
Berchowitz is interested in understanding how the cell breaks down and clears the large Rim4 amyloid fibrils in such a short period of time. Mass spectrometry revealed that Rim4 contains approximately 50 phosphorylated sites. Mutating 47 of these sites at once within the Rim4 IDR prevented Rim4 clearance, resulted in persistence of Rim4 into meiosis II, and caused cells to be stuck in meiosis II. Berchowitz showed that no single phosphorylation site governs Rim4 clearance. Rather, it is the combinatorial effect of several sites. Mutating 16 sites or fewer did not affect Rim4 clearance, while mutating 20 sites had an intermediate effect on clearance. He proposed that electrostatic repulsion between the negatively charged phosphorylation sites may destabilize the amyloid fibrils and allow them to be broken down by the cell.15
While Berchowitz’s data focused on Rim4 amyloid structures, he postulated that hyperphosphorylation may be a general mechanism by which the cell attempts to clear amyloid fibrils. Work by presenter Nicholas Fawzi has demonstrated, described below, has also shown that phosphorylation can prevent protein aggregation.16 Future work will investigate whether the hyperphosphorylated tau filaments seen in patients with Alzheimer’s disease may be an attempt by the cell to break the fibrils down instead of the pathological species of disease, as commonly thought.
Phase separation of a DNA sensor
Zhijuan “James” Chen of University of Texas Southwestern Medical Center described his work in understanding how DNA-induced phase separation of the protein cGAS contributes to its function. cGAS is a DNA sensor that binds to double stranded DNA in the cytoplasm, setting off a series of events that leads to activation of the innate immune system. Normally, self-DNA is relegated to the nucleus; therefore, if cGAS encounters double stranded DNA in the cytoplasm, it is likely to be from a pathogen. cGAS therefore can act as a sensor for many different viruses and bacteria.
Several lines of evidence suggest that cGAS is involved in liquid-liquid phase separation. First, cGAS and DNA form puncta structures in the cytoplasm.17 cGAS also contains an unstructured, highly positively charged N-terminal region, which is common among proteins that phase separate. DNA length positively correlates with cGAS enzymatic activity, indicating that multivalency plays a role. Finally, cGAS/DNA solutions turn turbid in vitro.
Work in Chen’s lab has revealed that cGAS and DNA separate into liquid droplets in vitro. Furthermore, both in vitro and in vivo experiments showed that cGAS activity is only present within the liquid droplets. No enzymatic activity is observed below concentrations required for phase separation, and separating droplets from cells via centrifugation revealed that cGAS activity is only present in the droplets and not the supernatant.18
Chen proposed that multivalent interactions between DNA and cGAS drive phase separation. Consistent with this, full-length cGAS phase separates more easily than a truncated form, suggesting a role for the unstructured N terminus. In addition, longer DNA drives phase separation more easily than short DNA sequences, suggesting the importance of multiple DNA/cGAS interaction sites. Chen also described different conditions that affect phase separation. Phase separation is inhibited by salt, suggesting that it is primarily driven by electrostatic interactions, and is difficult to achieve under physiological salt concentrations. However, phase separation is enhanced by zinc both in vitro and in vivo, which may be a mechanism by which the cell induces phase separation of cGAS.18
Chen proposed a model for how phase separation plays a key regulatory role for cGAS and may allow cells to strike a balance between immunity and autoimmunity.19 Because low levels of cytoplasmic DNA are not sufficient to induce phase separation and cGAS activation, Chen suggested that this allows cells to tolerate a small amount of self-DNA in the cytoplasm without activating an immune response and potentially creating an aberrant autoimmune reaction. When an infection is present, the large amount of foreign DNA in the cytoplasm induces phase separation and cGAS activity. This switch-like response, which is enabled by liquid phase separation, allows cGAS to elicit a robust and sensitive, yet general immune response to a variety of pathogens, independent of DNA sequence.
A phase-separated CO2 fixing organelle
Martin Jonikas from Princeton University discussed his lab’s work in uncovering the structure of the pyrenoid, an organelle found in many algae, which plays a key role in the global carbon cycle. In photosynthetic organisms, such as plants and algae, the enzyme Rubisco is responsible for fixing CO2 into sugar. In most land plants, the enzyme is distributed throughout the chloroplast; however, in algae, Rubisco localizes to a microcompartment known as the pyrenoid, into which CO2 is actively pumped. While Rubisco is inherently a very slow enzyme—its slow activity being thought to limit the growth of many land plants—the high concentration of substrate in the algal pyrenoid increases Rubisco activity. Jonikas hopes that understanding the pyrenoid may enable us to transfer its components into land plants, thus increasing Rubisco activity, and potentially improving crop growth and yield and reducing fertilizer needs.
Electron microscopy images of the pyrenoid reveal three compartments: a matrix, which is where Rubisco is found, a starch sheath that surrounds the matrix, and membrane tubules that traverse the matrix and are believed to bring CO2 into the matrix. Early electron micrographs of the pyrenoid suggested that it is a crystalline solid.20,21 However, Luke Mackinder and Elizabeth Freeman Rosenzweig in Jonikas’ lab obtained new data which indicate that the pyrenoid is a phase-separated liquid-like organelle; these data include fluorescence photobleaching showing that the pyrenoid matrix mixes like a liquid and imaging studies showing that the pyrenoid partially dissolves into the chloroplast during cell division.22
Jonikas’ lab has also worked to identify the components of the pyrenoid. Until recently, very little has been known about the proteins and molecules that make up the pyrenoid outside of Rubisco and Rubisco activase. Using transcriptomics to identify candidate proteins and fluorescence imaging to view protein localization, Luke Mackinder in Jonikas’ lab has identified approximately 90 proteins that localize to different compartments of the pyrenoid.23 In particular the study identified EPYC1, a protein that localizes to the matrix and is required for its formation. Mutant algae that lack EPYC1 are unable to grow under low-CO2 conditions, which would require a functioning pyrenoid. EM reveals that the cells have small pyrenoids that contain only the starch sheath and tubules, but no matrix. Although the total levels of Rubisco in mutants that lack EPYC1 are similar to wild type, the enzyme is found throughout the chloroplast, and not localized to the pyrenoid.24 Tobi Wünder in the laboratory of Jonikas’ collaborator, Oliver Mueller-Cajar, has shown that mixing purified EPYC1 and Rubisco yields phase-separated droplets, demonstrating that these proteins are both necessary and sufficient to form the pyrenoid matrix.25 Jonikas’ lab is now studying the molecular interactions between EPYC1 and Rubisco to understand the structural basis for their phase separation.
Jonikas and his collaborator Alistair McCormick from the University of Edinburgh have made preliminary attempts to transfer algal proteins into model land plants. They have transferred ten algal components into tobacco and Arabidopsis. Nearly all of the components localize correctly to the chloroplast without modification, an important first step towards ultimately transferring a pyrenoid into higher plants to improve crop yields.26
Phase-separated structures and biological patterns
Abby Dernburg from the University of California, Berkeley described her work on understanding the structure and function of the synaptonemal complex (SC). The SC is a structure consisting of several different proteins that forms in between homologous chromosome pairs during meiosis and holds them together during recombination and crossing over. Dernburg showed that the SC behaves like a phase-separated liquid crystal. It can be viewed via fluorescence in living C. elegans, in which it displays many liquid-like behaviors, e.g., rapid changes in shape and fusion of two or more particles. However, However, the molecules within this structure show a well-defined organization, making it different from many other phase-separated compartments..27
Dernburg believes that the phase-separating properties of the SC are important for its function. In addition to acting as a chromosomal glue, it has also been speculated that the SC may play a role in meiotic recombination. During recombination, many double-stranded breaks form; however, only a subset of these are selected to become a crossover event. In C. elegans, there is only one crossover per chromosome. In other animals, such as Drosophila, there can be multiple crossover events; however, they occur at widely-spaced intervals. Dernburg showed that the liquid-like property of the SC may determine the patterning of crossovers along the chromosome.27
Via FRAP experiments, Dernburg followed the dynamics and diffusion of proteins within the SC. In particular, ZHP-3 and −4, which localize to crossover sites, diffuse throughout the SC and slow down once they reach the crossover site.27,28 Dernburg proposed that the SC and crossover patterning is an example of a Turing pattern, in which biologic patterns can develop via interactions between components within a diffusive medium, such as the SC. She suggested that this Turing patterning may be a common theme among liquid-liquid phase separation systems in that it may create conditions for forming biological patterns that has yet to be fully appreciated.
Using phase separation to regulate protein degradation
Tanja Mittag from St. Jude Children’s Research Hospital discussed her work on how the cell’s protein quality control machinery targets proteins found in phase-separated particles. Mittag focused on the protein SPOP, a substrate adaptor of the cullin3-RING ubiquitin ligase. SPOP recruits substrate proteins to the ubiquitin ligase, which then ubiquitinates the protein, marking it for degradation. Mutations in SPOP have been linked to cancer, as many of its substrates play a role in oncogenesis.
Mittag proposed that SPOP targets its substrates, which are proteins that localize to phase-separated organelles, by undergoing phase separation with them. SPOP normally localizes to nuclear speckles29, dynamic structures that consist of pre-mRNA splicing factors found in the interchromatin regions of the nucleoplasm. However, in the presence of a substrate, such as the protein DAXX, SPOP leaves nuclear speckles and colocalizes with the substrate in liquid droplets.30
DAXX is a largely intrinsically disordered protein normally found in PML bodies31 that can phase separate and form liquid droplets on its own at high concentrations.30 Mittag showed that SPOP lowers the concentration threshold for phase transition, resulting in DAXX/SPOP liquid droplets at lower DAXX concentrations. Mittag identified residues in DAXX important for both SPOP binding and for DAXX/SPOP phase separation.30
Mittag also showed that phase separation of SPOP/DAXX mediates SPOP function in cells. Other components of the ubiquitin ligase complex colocalize to SPOP/DAXX droplets both in vitro and in vivo, and Mittag showed that ubiquitination can occur in the droplets. SPOP mutants implicated in cancer increase the concentration threshold for phase transition compared to wild type and abrogate SPOP/DAXX colocalization and SPOP activity, suggesting that phase transition is important for SPOP function 30. Whether phase separation is strictly necessary for ubiquitination is still unknown, however, because smaller complexes may be able to carry out similar functions.
Structural analyses of phase-separated proteins
Nicolas Fawzi from Brown University presented his work on visualizing the structure of and interactions between proteins in phase-separated materials at the atomic level. Fawzi discussed recent work on FUS and TDP-43, RNA-binding proteins that play roles in RNA processing and transcription.
Like many of the proteins discussed throughout the day, FUS contains an IDR and an RNA-binding domain. Mutations in the disordered domain of FUS have been associated with ALS and some forms of cancer, such as Ewing’s sarcoma. Both full-length FUS and the disordered domain phase separate in a salt- and RNA-dependent manner. In addition, FUS contains many serine and threonine residues, phosphorylation of which prevents phase separation.16
NMR spectra confirm that the N-terminal IDR of FUS is disordered at low concentrations when the protein is monomeric. NMR spectra of large FUS phase-separated assemblies largely resemble that of the monomeric species, suggesting that the protein remains disordered in phase-separated assemblies. These data imply that the disordered N-terminal region of FUS does not undergo any large-scale conformational changes upon phase separation and remains largely disordered.32
Fawzi also described ongoing efforts to investigate what specific contacts help to stabilize phase-separated assemblies using complementary NMR and molecular simulation approaches.
Fawzi also discussed structural features of phase separation for TDP-43, mutations of which have been implicated in ALS. The C-terminus of TDP-43 contains a conserved partially helical region that can phase separate. Fawzi showed that disrupting the helical region via mutation disrupted phase separation (33) and can lead to aggregation and co-aggregation with other RNA-binding proteins33 (see Figure 1). Fawzi’s data show the importance of the helical propensity of the C-terminus for TDP-43 phase separation, which they also demonstrated is correlated with cellular function.34,35
Figure 1:
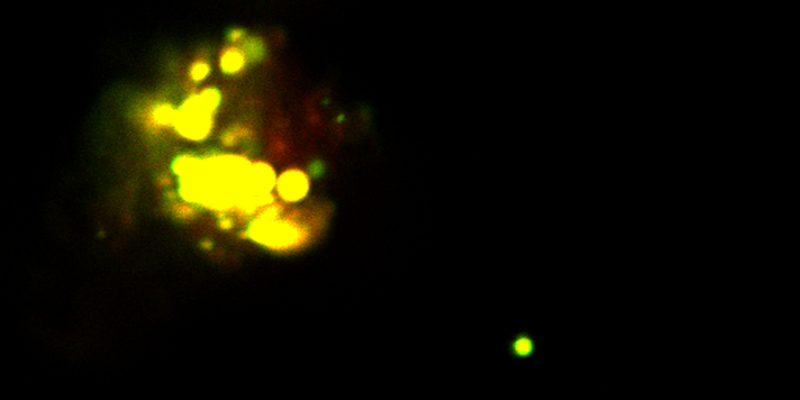
Liquid–liquid phase separation is the mechanism by which membrane-less organelles form. Some RNA-binding proteins with intrinsically disordered domains can undergo spontaneous LLPS in vitro. Fawzi and coworkers have shown that phase separation of the RNA-binding protein hnRNPA2 is altered by disease mutations, post-translational modifications, and protein–protein interactions. The image is a micrograph of hnRNPA2 low-complexity domain phase separation in the presence of salt and low temperature; hnRNPA2 and TDP-43 proteins aggregate together (in a test tube), providing a possible model for probing the structural details of their aggregation in neurodegenerative disease. Image credit: Veronica H. Ryan, Brown University.
Acknowledgments
The one-day symposium “Phase Separation in Biology and Medicine” was presented by the Chemical Biology Discussion Group of the New York Academy of Sciences. The scientific organizing committee for the symposium included Cliff Brangwynne (Princeton University), Jason Imbriglio (Merk), Neal Zondlo (University of Delaware), and Sara Donnelly (The New York Academy of Sciences).
Nicolas Fawzi thanks the members of his laboratory and was supported in part by Human Frontiers Science Program (RGP0045/2018), National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (R01GM118530), and the National Science Foundation (1845734) to N.L.F.
Footnotes
Competing interests
N.L.F. none.
References
- 1.Shin Y et al. Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell 168, 159–171.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shin Y et al. Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell 175, 1481–1491.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim SJ et al. Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hough LE et al. The molecular mechanism of nuclear transport revealed by atomic-scale measurements. eLife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayama R et al. Thermodynamic characterization of the multivalent interactions underlying rapid and selective translocation through the nuclear pore complex. J. Biol. Chem. 293, 4555–4563 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raveh B et al. Slide-and-exchange mechanism for rapid and selective transport through the nuclear pore complex. Proc. Natl. Acad. Sci. U. S. A. 113, E2489–2497 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sparks S, Temel DB, Rout MP & Cowburn D Deciphering the ‘Fuzzy’ Interaction of FG Nucleoporins and Transport Factors Using Small-Angle Neutron Scattering. Struct. Lond. Engl. 1993 26, 477–484.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Posey AE et al. Profilin reduces aggregation and phase separation of huntingtin N-terminal fragments by preferentially binding to soluble monomers and oligomers. J. Biol. Chem. 293, 3734–3746 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahoney NM, Rozwarski DA, Fedorov E, Fedorov AA & Almo SC Profilin binds proline-rich ligands in two distinct amide backbone orientations. Nat. Struct. Biol. 6, 666–671 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Shao J, Welch WJ, Diprospero NA & Diamond MI Phosphorylation of profilin by ROCK1 regulates polyglutamine aggregation. Mol. Cell. Biol. 28, 5196–5208 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao J, Welch WJ & Diamond MI ROCK and PRK-2 mediate the inhibitory effect of Y-27632 on polyglutamine aggregation. FEBS Lett. 582, 1637–1642 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dao TP et al. ALS-Linked Mutations Affect UBQLN2 Oligomerization and Phase Separation in a Position- and Amino Acid-Dependent Manner. Struct. Lond. Engl. 1993 (2019). doi: 10.1016/j.str.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dao TP et al. Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Mol. Cell 69, 965–978.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Jones HB, Dao TP & Castañeda CA Single Amino Acid Substitutions in Stickers, but Not Spacers, Substantially Alter UBQLN2 Phase Transitions and Dense Phase Material Properties. J. Phys. Chem. B (2019). doi: 10.1021/acs.jpcb.9b01024 [DOI] [PubMed] [Google Scholar]
- 15.Carpenter K, Bell RB, Yunus J, Amon A & Berchowitz LE Phosphorylation-Mediated Clearance of Amyloid-like Assemblies in Meiosis. Dev. Cell 45, 392–405.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monahan Z et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 36, 2951–2967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun L, Wu J, Du F, Chen X & Chen ZJ Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du M & Chen ZJ DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ablasser A & Chen ZJ cGAS in action: Expanding roles in immunity and inflammation. Science 363, (2019). [DOI] [PubMed] [Google Scholar]
- 20.Holdsworth RH The presence of a crystalline matrix in pyrenoids of the diatom, Achnanthes brevipes. J. Cell Biol. 37, 831–837 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kowallik K The crystal lattice of the pyrenoid matrix of Prorocentrum micans. J. Cell Sci. 5, 251–269 (1969). [DOI] [PubMed] [Google Scholar]
- 22.Freeman Rosenzweig ES et al. The Eukaryotic CO2-Concentrating Organelle Is Liquid-like and Exhibits Dynamic Reorganization. Cell 171, 148–162.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackinder LCM et al. A Spatial Interactome Reveals the Protein Organization of the Algal CO2-Concentrating Mechanism. Cell 171, 133–147.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackinder LCM et al. A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. Proc. Natl. Acad. Sci. U. S. A. 113, 5958–5963 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wunder T, Cheng SLH, Lai S-K, Li H-Y & Mueller-Cajar O The phase separation underlying the pyrenoid-based microalgal Rubisco supercharger. Nat. Commun. 9, 5076 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atkinson N et al. Introducing an algal carbon-concentrating mechanism into higher plants: location and incorporation of key components. Plant Biotechnol. J. 14, 1302–1315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rog O, Köhler S & Dernburg AF The synaptonemal complex has liquid crystalline properties and spatially regulates meiotic recombination factors. eLife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Köhler S, Rillo-Bohn R & Dernburg AF A compartmentalized signaling network mediates crossover control in meiosis. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagai Y et al. Identification of a novel nuclear speckle-type protein, SPOP. FEBS Lett. 418, 23–26 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Bouchard JJ et al. Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments. Mol. Cell 72, 19–36.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H et al. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell. Biol. 20, 1784–1796 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burke KA, Janke AM, Rhine CL & Fawzi NL Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol. Cell 60, 231–241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryan VH et al. Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation. Mol. Cell 69, 465–479.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conicella AE, Zerze GH, Mittal J & Fawzi NL ALS Mutations Disrupt Phase Separation Mediated by α-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain. Struct. Lond. Engl. 1993 24, 1537–1549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang A et al. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 37, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]


