Introductory
The maintenance of proteostasis is crucial for any organism to survive and reproduce in an ever-changing environment, but its efficiency declines with age1. Posttranscriptional regulators such as microRNAs control protein translation of target mRNAs with major consequences for development, physiology, and longevity2,3. Here we show that food odor stimulates organismal proteostasis and promotes longevity in Caenorhabditis elegans through mir-71-mediated inhibition of tir-1 mRNA stability in olfactory AWC neurons. Screening a collection of microRNAs that control aging3 we find that miRNA mir-71 regulates lifespan and promotes ubiquitin-dependent protein turnover, particularly in the intestine. We show that mir-71 directly inhibits the toll receptor domain protein TIR-1 in AWC olfactory neurons and that disruption of mir-71/tir-1 or loss of AWC olfactory neurons eliminates the influence of food source on proteostasis. mir-71-mediated regulation of TIR-1 controls chemotactic behavior and is regulated by odor. Thus, odor perception influences cell-type specific miRNA-target interaction to regulate organismal proteostasis and longevity. We anticipate that the proposed mechanism of food perception will stimulate further research on neuroendocrine brain-to-gut communication and may open the possibility for therapeutic interventions to improve proteostasis and organismal health via the sense of smell, with potential implication for obesity, diabetes and aging.
MicroRNA biogenesis is associated with improved stress tolerance and longevity in C. elegans4, and specific microRNAs have been linked to stress resistance and aging3. Changes in stress resistance and longevity are often associated with alterations in protein homeostasis (proteostasis)1. To identify microRNAs that control longevity via effects on organismal proteostasis, we used established in vivo assays to monitor ubiquitin-mediated turnover of fluorescently labeled model substrates in C. elegans5,6. The UbV-GFP protein is a ubiquitously expressed, ubiquitin fusion degradation (UFD) substrate. In wild-type worms, polyubiquitylation of the N-terminal ubiquitin moiety triggers degradation of UbV-GFP by the 26S proteasome. Disruption of ubiquitylation or of proteasomal turnover results in UbV-GFP stabilization in transgenic worms (Fig. 1a)5. We also monitored endoplasmic reticulum (ER)-associated protein degradation (ERAD) by following the turnover of an unstable form of the cathepsin L-like cysteine protease CPL-1 (CPL-1*-YFP) expressed in intestinal cells6. CPL-1*-YFP is normally retro-translocated out of the ER lumen for ubiquitin-mediated proteasomal degradation in the cytosol; loss of the ERAD-associated E3 ubiquitin-protein ligase SEL-1 triggers substrate stabilization (Supplementary Fig. 1f, 3d)7.
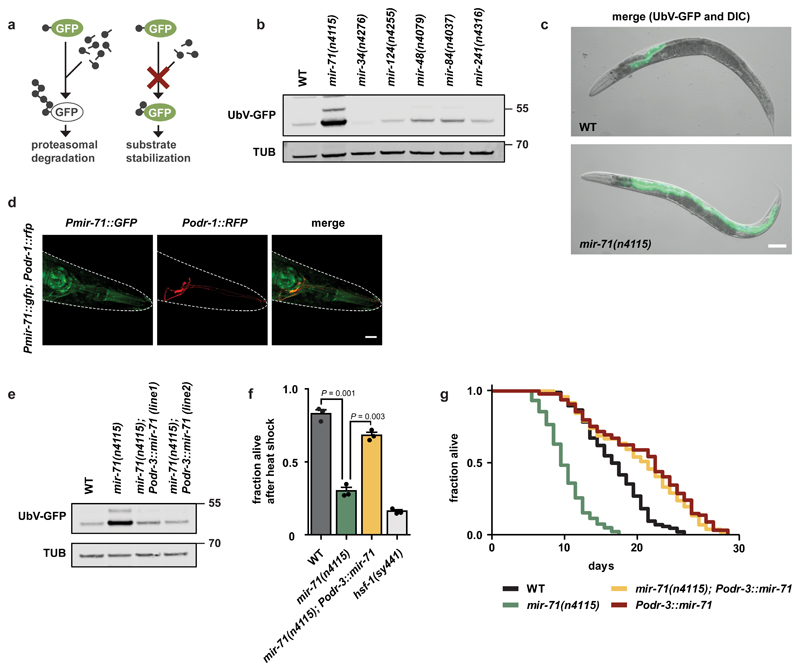
Figure 1. Expression of mir-71 in olfactory neurons supports proteostasis and longevity.
a, The ubiquitin fusion degradation (UFD) model substrate for monitoring ubiquitin-dependent degradation. b, The mir-71(n4115) deletion allele exhibits stabilization of the UFD substrate. Detection of the GFP signals via western blot showing UbV-GFP and tubulin (TUB) level. c, The UFD substrate accumulates mainly in the intestine upon mir-71(n4115) deletion. Representative fluorescent images of day 1 adult worms with indicated genotypes. Scale bar: 250 μm. d, mir-71 is expressed in olfactory (AWC) neurons. Confocal microscopy images showing localization of Pmir-71::GFP in green and AWC neurons in red. Scale bar: 15 µm. e, AWC-selective rescue of the mir-71(n4115) deletion mutant restores protein degradation. Western blot from day 1 adult worm lysates with indicated genotypes showing UbV-GFP and tubulin (TUB) level. f, AWC-selective expression of mir-71 increases survival upon heat stress. The hsf-1(sy441) mutant served as control. Bars show mean values ± SEM obtained from n=3 biological replicates using at least 50 worms (mean values represented by dots); statistics were determined by one-way ANOVA with post-hoc test. g, AWC-selective expression of mir-71 extends lifespan. For statistics details see Supplementary Table 1. b-e, Representative data derived from at least 3 independent experiments with similar results. b, e: Molecular weights are shown in kilodalton (kDa).
Of the microRNA loss-of-function mutants we tested, we found that mir-71(n4115) showed a substantial increase in both UbV-GFP and CPL-1*-YFP levels, particularly within the intestine, relative to wild-type worms (Fig. 1b, c, Supplementary Fig. 1a, g, h). Importantly, the levels of ubiquitylation-resistant K29/48RUbV-GFP and GFP were unaltered in mir-71(n4115) (Supplementary Fig. 1b)5, and wild-type and mir-71(n4115) animals showed comparable levels of UbV-GFP mRNA (Supplementary Fig. 1c). Overexpression of the proteasomal subunit RPN-6.1, which triggers degradation of ubiquitylated proteins8, suppressed the stabilization of UbV-GFP in mir-71(n4115) (Supplementary Fig. 1d). In contrast, loss of the E3 ligase HECD-1, which acts upstream of the 26S proteasome in substrate ubiquitylation, cannot be compensated by elevated RPN-6.1 level (Supplementary Fig. 1d)5. RNAi-mediated knockdown of hecd-1, as well as rpn-8, led to an additive defect when combined with mir-71(n4115) (Supplementary Fig. 1e). These data suggest that mir-71 regulates ubiquitin-dependent protein degradation via the 26S proteasome.
Consistent with ERAD defects, mir-71(n4115) worms are sensitive to ER stress induced by tunicamycin (TM), which blocks N-linked glycosylation of ER proteins9. Interestingly, the previously reported lifespan reduction of mir-71(n4115)3 was further decreased by TM treatment (Supplementary Fig. 1i). To further characterize the role of mir-71 in intestinal proteostasis, we performed tissue-specific rescue experiments10. Ubiquitous and pan-neuronal, but not hypodermal, expression of mir-71 in mir-71(n4115) rescued defective turnover of UbV-GFP (Supplementary Fig. 2a, b). These data suggest that loss of mir-71 expression in neurons stabilizes UbV-GFP in intestinal cells. Previous work found that mir-71 is expressed in olfactory neurons11. To identify the type of neuron required for mir-71-dependent regulation of proteostasis, we genetically abolished the specification of AWA, AWB/ADF, AWC, or PQR/PHA/PHB olfactory neurons by depleting the transcription factors required for their cell-fate determination, ODR-7, LIM-4, CEH-36, and CEH-14, respectively12. Abrogation of either AWB/lim-4 or AWC/ceh-36 development partially suppressed UbV-GFP accumulation in mir-71(n4115) (Supplementary Fig. 2c), suggesting that these olfactory neurons are required for mir-71-mediated regulation of intestinal homeostasis. Intriguingly, the AWCs belong to a class of ciliated olfactory neurons previously implicated in the regulation of longevity13. This led us to hypothesize a prominent role for AWC neurons in organismal proteostasis via mir-71.
To test this hypothesis, we first confirmed that mir-71 is expressed in AWC neurons. Indeed, we observed colocalization of Pmir-71::GFP and the AWC-specific marker Podr-1::RFP (Fig. 1d)11. Further, Podr-1::RFP expression revealed comparable AWC neuronal integrity in mir-71(n4115) and wild-type worms (Supplementary Fig. 2d). Intriguingly, selective expression of mir-71 in AWC (Podr-3) but not AWB neurons (Pstr-1), fully restored the ability of mir-71(n4115) animals to degrade UbV-GFP (Fig. 1e, Supplementary Fig. 2e, f)14. AWC-selective expression of mir-71 in mir-71(n4115) alleviated mortality induced by the proteasome inhibitor bortezomib (BTZ) (Supplementary Fig. 2g), as well as heat sensitivity and reduced lifespan (Fig. 1f, g). Further, Podr-3::mir-71 expression prolonged lifespan of wild-type worms (Fig. 1g), suggesting that mir-71 improves organismal physiology. Thus, mir-71 expression in AWC olfactory neurons is necessary and sufficient to coordinate organismal proteostasis, particularly in the intestine.
MicroRNAs regulate gene expression via complementary base pairing with target mRNAs. To identify potential targets of mir-71 important for proteostasis regulation, we compiled a list of genes that were differentially expressed in olfactory neurons (i) upon proteasomal inhibition of wild-type worms, and (ii) in the mir-71(n4115) deletion mutant, relative to wild-type (Supplementary Fig. 3a, Supplementary Table 2). We further reduced the list to AWC-specific mRNAs regulated in response to proteotoxic stress, based on RNA sequencing data and bioinformatic microRNA target prediction (Fig. 2a, Supplementary Table 3) (TargetScanWorm Release 6.2;15).
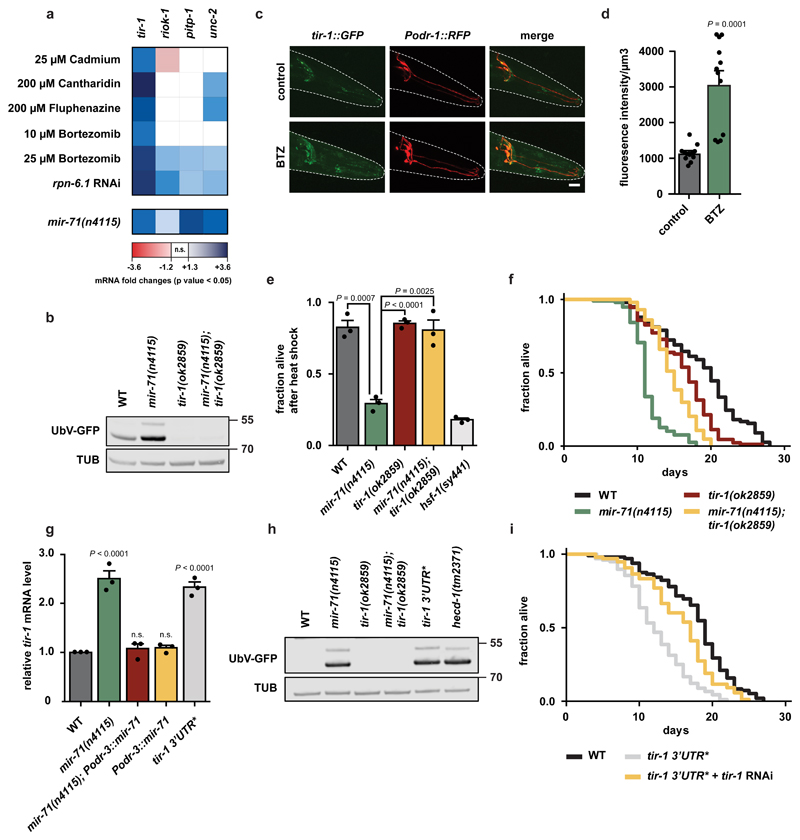
Figure 2. Negative regulation of tir-1 by mir-71 important for proteostasis and longevity.
a, mRNA level of predicted mir-71 targets (TargetScanWorm Release 6.2). Color-coded transcriptional responses of mRNAs expressed in AWC neurons under multiple proteotoxic stress conditions or in mir-71(n4115) deletion mutant. Expression levels are relative to untreated wild-type (WT) worms and were determined via RNA sequencing (upper panel) (n=3 biological replicates) or microarray (lower panel) (n=4 biological replicates), n.s.=not significant. b, h, Representative western blots from day 1 adult worms of indicated genotypes show UbV-GFP and tubulin (TUB) level. Data derived from at least 3 independent experiments with similar results. Molecular weights are shown in kDa. b, The tir-1(ok2859) deletion mutant suppresses mir-71(n4115)-induced proteolytic defects. c, Proteotoxic stress increases TIR-1 levels in AWC neurons. Confocal microscopy images showing tir-1::GFP in green and AWC neurons in red. Scale bar: 15 µm. d, Quantification of TIR-1::GFP fluorescence intensity shown in (c). DMSO was used as a solvent control but did not show a significant difference compared to the control condition (1322.24 ± 96.05, data not shown). Bars show mean values ± SEM obtained from n=12 individual animals (represented by dots). Statistics were determined by one-way ANOVA with post-hoc test. e, Heat stress sensitivity of mir-71(n4115) mutant is suppressed by tir-1(ok2859) deletion. The hsf-1(sy441) mutant served as control. Bars show mean values ± SEM obtained from n=3 biological replicates using at least 50 worms (mean values represented by dots); statistics were determined by one-way ANOVA with post-hoc test. f, tir-1(ok2859) deletion extends the lifespan of mir-71(n4115) worms. For statistics details see Supplementary Table 1. g, mir-71 negatively regulates tir-1 transcript level. Relative tir-1 mRNA levels measured by qRT-PCR in day 1 adult worms. Data normalized to wild-type (WT). Bars show mean values ± SEM obtained from n=3 biological replicates with 3 technical replicates each (mean values represented by dots); statistics were determined by one-way ANOVA with post-hoc test; ns=not significant). h, AWC-selective expression of mir-71 does not suppress proteostasis defects caused by the tir-1 3’UTR* mutation. i, The tir-1 3’UTR* mutation shortens lifespan. For statistics details see Supplementary Table 1.
We identified tir-1, riok-1, pitp-1, and unc-2 as potential targets of mir-71. In particular, tir-1 mRNA was strongly elevated in response to proteotoxic reagents and contains three predicted mir-71 binding sites within the 3’UTR (Fig. 2a, Supplementary Fig. 3a, Supplementary Table 2, 3). To test whether up-regulation of the identified genes might account for the defects in proteostasis of mir-71-deficient animals, we depleted the candidate mRNAs by RNAi. Only tir-1 depletion restored UbV-GFP levels in mir-71(n4115) mutants (Supplementary Fig. 3b). Further, degradation of both the UFD and ERAD substrates was restored in mir-71(n4115); tir-1(ok2859) double mutants (Fig. 2b, Supplementary Fig. 3c, d). Finally, ubiquitylation-resistant K29/48RUbV-GFP or GFP proteins, as well as UbV-GFP mRNA levels, were unaltered in tir-1(ok2859) (Supplementary Fig. 3e, f), revealing that the decrease in substrate protein levels did not result from transcriptional or translational changes.
TIR-1 is a toll receptor domain adapter protein that is important for innate immune responses and neuronal development11,16,17. To determine where tir-1 is expressed, we used CRISPR-Cas9-based gene editing to generate a tir-1::GFP reporter. We observed TIR-1::GFP expression in AWC olfactory neurons11,16. Further, proteotoxic stress via BTZ treatment increased the expression of TIR-1::GFP as well as tir-1 mRNA levels (Fig. 2a, c, d). Importantly, loss of tir-1 suppressed the heat stress sensitivity and the shortened lifespan of mir-71(n4115) mutants (Fig. 2e, f). We also evaluated a gain-of-function mutant tir-1(yz68), which mediates constitutive activation of TIR-118. In contrast to loss of TIR-1, the tir-1(yz68) mutant caused a slight increase in UbV-GFP levels, which further increased in the context of mir-71(n4115) (Supplementary Fig. 3g, h). These findings suggest that mir-71 negatively regulates tir-1.
To test whether tir-1 mRNA is a direct target of mir-71, we used a CRISPR/Cas9 approach to disrupt all three predicted mir-71 binding sites in an endogenous allele of tir-1 in wild-type worms (tir-1 3’UTR*) (Supplementary Fig. 4a). Indeed, tir-1 mRNA levels increased in tir-1 3’UTR* worms relative to wild-type (Fig. 2g, grey bar), indicating a direct repressive effect of the microRNA. Further, the mir-71(n4115) deletion mutant showed increased levels of tir-1 mRNA (Fig. 2g, green bar), in line with our microarray analysis (Fig. 2a). Expression of mir-71 in AWCs restored tir-1 mRNA levels back to wild-type in mir-71(n4115), demonstrating that the tir-1 de-regulation observed in mir-71 mutant animals occurs primarily in AWC neurons (Fig. 2g, green versus red bar).
Importantly, animals carrying the tir-1 3’UTR* allele accumulated both UFD and ERAD substrates, fully phenocopying mir-71(n4115)-related defects in protein degradation (Fig. 2h, Supplementary Fig. 4b-d). RNAi-mediated depletion of tir-1 or overexpression of RPN-6.1 suppressed the protein turnover defect in tir-1 3’UTR* animals (Supplementary Fig. 4e, f). Thus, the substrate stabilization caused by tir-1 3’UTR* depends on increased tir-1 levels and results from ubiquitin proteasome system (UPS) dysfunction. Overexpression of mir-71 in AWC neurons did not re-establish UPS function in tir-1 3’UTR* animals, highlighting the unique importance of tir-1 mRNA regulation for proteostasis (Supplementary Fig. 4g). Similar to loss of mir-71, the tir-1 3’UTR* mutation also reduced lifespan, which was rescued by RNAi-mediated depletion of tir-1 (Fig. 2i). These data suggest that mir-71/tir-1 represents a one-to-one microRNA/mRNA interaction that regulates proteome stability and longevity.
We previously showed that feeding worms with different bacterial food sources modifies protein degradation in the intestine5. As expected, UbV-GFP was efficiently degraded in wild-type worms grown on standard food, Escherichia coli (E. coli) strain OP50, but accumulated in worms fed E. coli C600, HT115, or HB101 bacteria (Fig. 3a). In contrast, mir-71(n4115) mutants did not display food-related changes in ubiquitin-dependent proteolysis, and this was restored by AWC-selective mir-71 expression (Fig. 3a, b, Supplementary Fig. 5a).
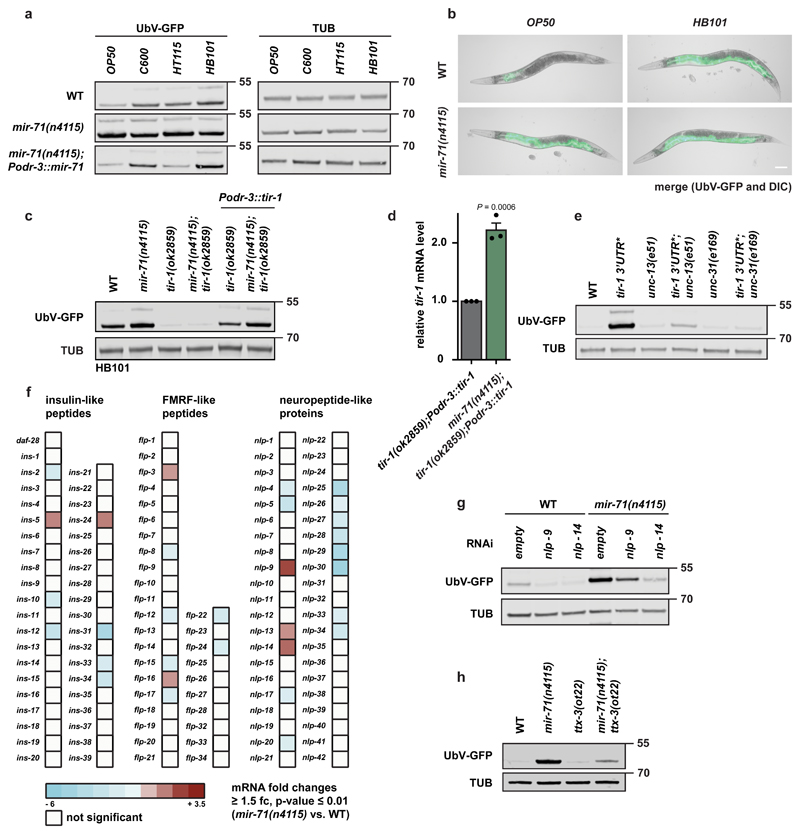
Figure 3. Food-dependent coordination of proteostasis is triggered by mir-71/tir-1 dynamics in AWC neurons.
a, c, e, g, h, Western blots of worm lysates with the indicated genotypes show UbV-GFP and tubulin (TUB) level. Data derived from at least 3 independent experiments with similar results. Molecular weights are shown in kDa. a, AWC-selective mir-71 expression is important for food perception and proteostasis. Animals were grown on OP50, C600, HT115, or HB101 bacteria prior to worm lysis. b, Fluorescent images of day 1 adult worms grown on OP50 or HB101 with indicated genotypes. Scale bar: 250 μm. Data derived from at least 3 independent experiments with similar results. c, AWC-selective tir-1 expression rescues food-dependent changes in protein degradation. Animals were grown on HB101 bacteria prior to worm lysis for western blotting. d, mir-71 negatively regulates tir-1 transcript level in AWC neurons. Relative tir-1 mRNA levels measured by quantitative real-time PCR (qRT-PCR) in day 1 adult worms. Data normalized to tir-1(ok2859); Podr-3::tir-1. Bars show mean values ± SEM obtained from n=3 biological replicates with 3 technical replicates each (mean values represented by dots); statistics were determined by two-tailed paired Student’s t-test. e, Neuronal signaling is required for changes in UPS activity caused by the tir-1 3’UTR* mutation. f, Microarray data of mir-71(n4115) compared to wild-type (WT) reveals misregulation of neuropeptide expression. n=4 biological replicates g, NLP-9 and NLP-14 neuropeptides are important for proteostasis signaling of the mir-71(n4115) mutant. Representative western blot of worms treated with RNAi against the indicated factors detect UbV-GFP and tubulin (TUB) levels. h, Reduced AIY interneuron activity caused by ttx-3(ot22) suppresses protein degradation defects of mir-71 deletion.
Similar to mir-71(n4115), tir-1 3’UTR* animals exhibited high levels of UbV-GFP, irrespective of different bacterial strains (Supplementary Fig. 5b). In contrast, tir-1(ok2859) loss-of-function mutants did not accumulate UbV-GFP when grown on HB101, which was reversed upon AWC-selective tir-1 expression (Fig. 3c, Supplementary Fig. 5b, c). We found that UbV-GFP and tir-1 mRNA levels were higher when Podr-3::tir-1 was expressed in mir-71(n4115) mutants compared to wild-type worms, consistent with loss of miRNA-mediated tir-1 suppression in mir-71(n4115) (Fig. 3d). These data suggest that food-related changes in proteostasis are mediated at least in part by mir-71-dependent, cell type-specific regulation of tir-1 in AWC olfactory neurons.
Cell-nonautonomous, neuroendocrine signaling pathways have been shown to trigger organismal regulation of proteotoxic stress and quality control19,20. Since UbV-GFP accumulated mainly in the intestine upon exposure to different bacteria or mir-71 deletion, we tested whether AWC-selective tir-1 regulation is communicated to peripheral tissues by neuronal signaling. We used an UNC-13 mutant to evaluate the consequence of blocking neurotransmitter release from small clear vesicles (SCV), and an UNC-31 mutant to block neuropeptide release from dense core vesicles (DCV)21,22. We found that both unc-13(e51) and unc-31(e168) suppressed the substrate stabilization caused by feeding HB101 bacteria (Supplementary Fig. 5a), by mir-71 loss-of-function (Supplementary Fig. 5d), and by tir-1 3’UTR* (Fig. 3e), suggesting that neuronal signal transduction is important for AWC-dependent coordination of proteostasis in peripheral tissues.
To determine the signaling molecules that trigger the cell-nonautonomous communication from the olfactory neurons to the intestine, we evaluated changes in the expression of neuropeptide genes in mir-71(n4115) compared to wild-type worms (Fig. 3f and Supplementary Table 4)23. We observed high upregulation of NLP-9 and NLP-14 in mir-71(n4115) worms (Fig. 3f). Further, RNAi-mediated depletion of NLP-9 and NLP-14 partially or fully suppressed mir-71(n4115)-dependent degradation defects, respectively (Fig. 3g). Thus, these data suggest that AWC neurons regulate intestinal homeostasis via secretion of the neuropeptides NLP-9 and NLP-14.
AWC neurons communicate with AIB and AIY interneurons, among others, to initiate food- and odor-derived behavior24. To validate the role of mir-71 for communication via the olfactory system, we analyzed a loss-of-function mutant for ttx-3, which encodes a LIM homeodomain protein necessary for AIY function25. Indeed, the mir-71(n4115)-dependent defects in protein degradation were suppressed in the ttx-3(ot22) mutant background, supporting the idea that signal transduction via AIY interneurons connects food-derived olfactory inputs and proteostasis (Fig. 3h).
To define the cause of food-related changes in proteostasis, we monitored degradation of UbV-GFP in wild-type worms exposed to mixtures of OP50 and HB101 bacteria. In contrast to HB101 alone, worms raised on HB101/OP50 mixtures (100:1, 10:1, and 1:1) showed significantly decreased substrate stabilization almost indistinguishable from worms exclusively exposed to OP50 (Fig. 4a). However, mir-71(n4115) and tir-1 3’UTR* worms did not distinguish the different mixtures and showed UbV-GFP substrate stabilization, which did not exceed effects with HB101 alone in all cases (Fig. 4a). The very low detection threshold for the proteostasis response to OP50 is consistent with C. elegans’ ability to detect low concentrations of volatile chemicals26. To acutely silence AWC activity specifically after development, we expressed a histamine-gated chloride channel (HisCl1) under control of the ceh-36 promoter and treated worms with histamine (Supplementary Fig. 5e)27. Histamine (HA) treatment of worms grown on OP50 or HB101 bacteria completely abolished the food-evoked differences in UbV-GFP stabilization (Fig. 4b). Strikingly, HA-treatment similarly suppressed degradation defects of mir-71(n4115) worms on both bacterial strains, which suggests a direct link between neuronal activity and mir-71 function in food-dependent regulation of proteostasis (Fig. 4c).
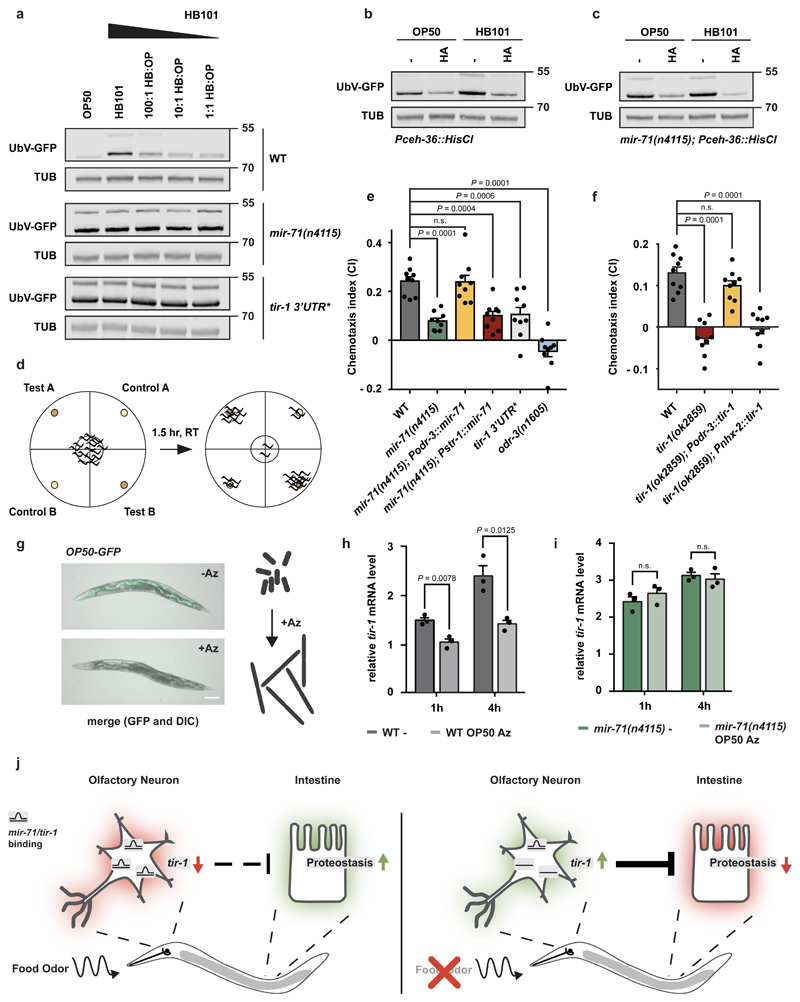
Figure 4. Coordination of food perception and organismal proteostasis.
a-c, g, Western blots showing UbV-GFP and tubulin (TUB). Data derived from at least 3 independent experiments with similar results. Molecular weights are shown in kDa. a, Low concentrations of OP50 alleviate HB101-dependent changes in proteostasis. Worms of indicated genotypes were grown on depicted mixture of HB101 and OP50. b-c, AWC silencing by histamine-gated chloride channel expression (Pceh-36::HisCl). Food-dependent proteostasis defects in WT (b) and mir-71(n4115) (c) are inhibited upon AWC silencing. Age-synchronized L4 larvae were grown on control (-) or histamine (HA) containing plates (10 mM HA) until day 1 of adulthood. d, Schematic overview of the chemotaxis assay: Plates are separated into test and control quadrants. Worms were transferred to the center of the plate. Animals in each quadrant were determined after 1.5 hr. Calculation of the chemotaxis index (CI) is shown below. e, f, AWC-selective expression of tir-1 or mir-71 is important for chemotaxis behavior (E. coli OP50). Chemotaxis assays were performed using worms of the indicated genotypes. Bars show mean values ± SEM obtained from n=9 independent experiments (mean values represented by dots) with at least 66 (e) or 80 animals (f). Statistics were determined by one-way ANOVA with post-hoc test; ns=not significant. g, E. coli treated with Aztreonam (Az) show abnormal cell growth and cannot be ingested by C. elegans. For ingestion control E.coli OP50 expressing GFP (OP50-GFP) were either treated with Az or left untreated before seeding them to NGM plates. Worms were grown on respective bacteria for 6 hr prior to imaging. Fluorescent images of day 1 adult worms. Scale bar: 250 μm. h, i, Food odor affects tir-1 mRNA level. Relative tir-1 mRNA levels measured by qRT-PCR in day 1 adult worms. Age-synchronized worms were grown on OP50 until day 1 of adulthood and transferred to plates without food (-) or Az-treated OP50 (OP50 Az) for 1 h or 4 hr. Data normalized to wild-type (WT) tir-1 level on food (control condition, data not shown). Bars show mean values ± SEM obtained from n=3 biological replicates with 3 technical replicates each (mean values represented by dots); statistics were determined by one-way ANOVA with post-hoc test; ns=not significant). j, Model: Odors related to bacterial food are sensed via ciliated AWC olfactory neurons. The olfactory stimulation of AWCs provokes cell-nonautonomous regulation of ubiquitin-dependent protein degradation and stress resistance in peripheral tissues including the intestine. A key regulator of this food response is the toll receptor domain protein TIR-1, which is dynamically regulated by direct binding between tir-1 mRNA and mir-71 (mir-71/tir-1) in AWC neurons. Absence of olfactory information (food odor) or abrogation of posttranscriptional regulation via mir-71 results in elevated tir-1 level. Chronic upregulation of tir-1 in AWC neurons in mir-71(n4115) alters diet-evoked communication and blocks adjustments of intestinal proteostasis, which correlates with decreased stress tolerance and lifespan.
To discern the role of AWC neurons in smelling volatile odors, we used standard chemotaxis assays to test whether tir-1 regulation is necessary for sensing a bacterial food source (Fig. 4d)28. Indeed, both mir-71(n4115) and tir-1 3’UTR* animals showed reduced food detection. The compromised chemosensation of mir-71(n4115) could be rescued by mir-71 expression in AWC but not AWB neurons (Fig. 4e, Supplementary Table 5). Similarly, chemotaxis defects of tir-1(ok2859) deletion animals were rescued by AWC-selective tir-1 but not intestinal expression (Fig. 4f, Supplementary Table 5), suggesting that cell type-specific tir-1 regulation is required both for olfactory food perception and proteostasis. To test this hypothesis, we measured the level of tir-1 mRNA in wild-type worms exposed to the smell of bacteria. We incubated OP50 bacteria with aztreonam (Az), an antibiotic that affects bacterial cell division, resulting in inedible bacterial filaments (Fig. 4g)29. We found that tir-1 mRNA levels increased in wild-type worms transferred to a plate without food, and the smell of inedible bacteria suppressed tir-1 elevation in this context (Fig. 4h). Interestingly, mir-71(n4115) mutants, which have already upregulated tir-1 mRNA, did not further increase in tir-1 mRNA levels upon transfer to a plate lacking food (Fig. 4i). These data suggest that the absence of an odor relieves mir-71 mediated repression of tir-1 in wild-type worms. Our findings support a direct link between olfactory food perception and mir-71-mediated inhibition of TIR-1 to control proteostasis and aging (Fig. 4j).
Previous reports indicated that neuroendocrine signaling communicates proteotoxic stress between neurons and the intestine19,20. However, little information has been gathered on the physiological role of these systems. Given that proteostasis is linked to aging and longevity1, the olfactory system appears of particular importance since food-derived volatile cues in D. melanogaster and C. elegans affect lifespan30,31. Here, we provide new mechanistic insights showing that a neuronal olfactory circuit rewires proteolytic networks in intestinal cells, thus establishing an underlying concept for the regulation of food adaption (Fig. 4j).
In this context, the primary AWC olfactory neurons are central in sensing and transducing food-derived information. Interestingly, AWCs are OFF-neurons which exhibit high activity (intracellular Ca2+ level) in the absence of food cues24, a condition that results in gradual upregulation of tir-1 mRNA (Fig. 4h) and might suggest a functional relationship between neuronal activity and tir-1 abundance. tir-1 is also chronically upregulated in mir-71(n4115) (Fig. 2a, 3d, 4i, Supplementary Fig. 3a), which indicates an aberrant neuroendocrine signaling that interferes with stress tolerance, aging, and organismal proteostasis. Notably, tir-1 is a Ca2+ signaling scaffold protein located near synapse structures important for proper AWC development16 and adaptive chemotactic behavior in neighboring AWA neurons32. Together with our findings that tir-1 levels are affected by food odor (Fig. 4h), this might suggest far-reaching consequences for the underlying chemosensory circuits originating from AWCs. However, in-depth activity studies in AWCs, connected primary sensory neurons, as well as interneurons will be needed to elucidate the precise role of TIR-1 within neuronal networks. Since we identified NLP-9 and NLP-14 as part of the mir-71/tir-1-dependent neuronal signaling towards the intestine (Fig. 3f, 3g), determining their exact origin and function, as well as respective receptors will further help to understand the functionality of this cell-nonautonomous pathway.
The TIR-1 ortholog sterile alpha and armadillo repeats containing protein 1 (SARM1) is predominantly expressed in the mammalian brain and facilitates clearance of damaged axons in response to trauma or disease, a process also known as Wallerian degeneration. Mechanistically, the SARM1-TIR domain provides NADase activity, which triggers depletion of the essential metabolite NAD+ and neuronal destruction33. Since changes in NAD+ levels are linked to proteasomal function, the NADase activity of SARM1 might directly affect ubiquitin-dependent protein degradation34. Here, the TIR domain from C. elegans TIR-1 provides a similar activity in axonal degeneration35. Interestingly, recent studies further associates SARM1 with the development of NAFLD (non-alcoholic fatty liver disease) induced by high fat diet36 as well as social interaction and cognitive flexibility in mice37. Together with our provided data on diet-dependent proteostasis and chemotactic behaviour this might indicate evolutionarily conserved concepts of TIR-1/SARM1.
The perception of food-related odors might coordinate food appreciation and selection with physiological and metabolic demands of the digestive tract. In this context, it is interesting to see that E. coli OP50 and HB101 differ in their nutrient composition; with higher carbohydrate level in HB101. Consequently, worms fed with HB101 show lower fat storage in comparison to OP50, indicative for diet-evoked metabolic changes38. Along this line, the sense of smell impacts the metabolic status especially in the context of obesity and fat storage in mice39 and regulates the production of digestive enzymes in humans40. Our observations show that the food source affects intestinal proteostasis5 (Fig. 3a, 4a, Supplementary Fig. 5a, b) which further suggests a direct link to the metabolic status of the organism. Together, the mechanistic insight on the organismal control via olfaction might open a new entry point to control and manipulate diet-related changes in proteostasis and metabolism with potential implication for associated diseases like obesity and diabetes.
Materials and Methods
Caenorhabditis elegans maintenance and transgenic lines
Nematodes were grown at 20°C (unless stated otherwise) on nematode growth medium (NGM) plates seeded with the bacterial Escherichia coli strain OP50 as a food source according to standard protocols and methods41,42. The N2 Bristol strain served as wild-type. All strains that were used in this study are listed in Supplementary Table 6. Transgenic strains expressing Pnhx-2::tir-1, Podr-3::tir-1 and Pstr-1::mir-71 were constructed via PCR amplification of N2 genomic DNA or cDNA (nhx-2 promoter 602 bp genomic DNA43, tir-1 2793 bp cDNA, tir-1 3’UTR 587 bp cDNA, odr-3 promoter 2755 bp genomic DNA44, str-1 promoter 4000 bp genomic DNA14, premature mir-71 94 bp genomic DNA). Oligonucleotides used in this study are listed in Supplementary Table 7. PCR fragments were cloned into the IR101 (pDEST R4-R3) vector containing a Prps0::hygR::mCherry cassette for selection of transgenic animals. The constructs were particle bombarded into N2 hermaphrodites and selected as described previously45. Microparticle bombardment was done with the Bio-Rad Biolistic PDS-1000/HE with ¼” gap distance, 9 mm macrocarrier to screen distance, 28 inches of Hg vacuum and a 1350 p.s.i. rupture disc. Per bombardment, about 1 mg of 1 µm microcarrier gold beads were coated with 7 µg linearized DNA. Animals were allowed to recover for 1 h at room temperature and were then transferred to 90 mm enriched peptone plates seeded with E. coli C600 bacteria. After 3-4 days 100 µl hygromycin B (100 mg/ml), 600 µl Antibiotic-Antimycotic 1006 (GIBCO Life technologies) and kanamycin (final concentration 50 mg/ml) were added to each plate to select transgenic animals by hygromycin B resistance. About 10 days after bombardment viable animals were screened for mCherry fluorescence. Individuals positive for mCherry were singled and screened for homozygosity. Strains expressing the UFD substrate (UbV-GFP) were described previously5.
CRISPR/Cas9 genome editing
CRISPR/Cas9 mutant alleles were generated as described46. Briefly, around 35-40 worms were injected with a mix containing 50 ng/μl Peft-3:Cas9sv40:nls:tbb-247, 50 ng/μl plk111 (U6prom:sgSRNA)48, 50 ng/μl repair templates (see Supplementary Table 8) for homology directed repair, 3 ng/μl Pmyo-2:mCherry and 100 ng/μl of a plasmid expressing a neomycin resistance cassette (pBCN44 from49). Injected F0 were singled out and stable lines were selected on OP50-seeded NGM plates, containing (400 μg/ml) of neomycin (G-418). All surviving F1 hermaphrodites within each plate were pooled and tested by PCR for the desired event. In case of the tir-1 3’UTR* mutant the introduced mutations were spotted by restriction digestion of the PCR product with KpnI. Non-transgenic animals were cloned out to obtain homozygous isolates and the absence of the extrachromosomal array was verified by PCR. All alleles were validated by Sanger sequencing of the respective loci.
Lifespan and survival assays
For lifespan assays 100 age-synchronized L4 larvae per strain were transferred to fresh NGM agar plates. Day 0 of the lifespan experiment refers to the first day after reaching adulthood. Within the first seven days animals were placed on new plates every day to separate them from F1s and to prevent starvation. After entering the post-reproductive phase, worms were only transferred when necessary. Survival was examined daily by checking pharyngeal pumping or avoidance behaviour in response to mechanical stimuli. All lifespan experiments were carried out in two biological replicates at 20°C. If indicated, NGM agar plates were supplemented with 25 μM fluorodeoxyuridine (FUdR) to prevent embryonic development and egg hatching50. Statistical details for all life span experiments are presented in Supplementary Table 1. To dissect sensitivity to endoplasmic reticulum (ER) stress conditions NGM plates were supplemented with 50 µg/µl tunicamycin (an equal amount of DMSO served as solvent control) followed by monitoring survival as described above. For proteasome inhibition NGM agar plates were supplemented with 10 μM bortezomib (BTZ) and L4 larvae were placed on the respective plates. Survival was determined for three consecutive days. Experiments were performed with 50 worms for three biological replicates. Heat stress was induced by incubation of age-synchronized L4 larvae at 32.5°C for 15 hr. Survival was analyzed in three biological replicates using 50-100 worms each.
Western blotting
To prepare whole worm lysates, 100 animals were collected in 80 µl 1x SDS loading buffer. Subsequently, samples were boiled at 95°C for 5 min, sonicated (30 sec at 60% amplitude) and again boiled at 95°C for 5 min. Following, samples were centrifuged at 15,000 rpm for 5 min. For western blotting, protein lysates were resolved by SDS-PAGE using NuPAGE® 4-12% Bis-Tris SDS-gels with the respective NuPAGE® MES SDS running buffer (Thermo Fisher Scientific; settings according to manufacturer’s instructions). A semi-dry blotting system (Bio-Rad, Trans-Blot Turbo) was applied with NuPAGE® transfer buffer. Antibodies were diluted in 1x Roti®-Block (Carl Roth). The following antibodies were used in this study: Mouse Monoclonal anti-alpha Tubulin (Clone B-5-1-2) (Sigma-Aldrich; Cat# T6074 RRID:AB_477582), Living Colors® A.v. Monoclonal Antibody (JL-8) (Mouse anti-GFP) (Clontech; Cat# 632380), Donkey anti-mouse IRDye® 800CW/680 (LI-COR; Cat# 926-32212 RRID:AB_621847). Visualization of fluorescent signals was achieved using the Odyssey scanner (LI-COR) and the Image Studio Lite v4.0 software. Images of all uncropped blots are shown in Supplementary Fig. 6.
Microscopy
For fluorescent and DIC images the M165 FC stereomicroscope with a DFC 340 FX camera (Leica Camera) or the Axio Zoom V16 microscope with an Axiocam 506 mono camera (Carl Zeiss Microscopy GmbH) was used and images were processed with the Zen 2.3 pro software (Carl Zeiss Microscopy GmbH). For close-up images of the C. elegans head region, the Confocal Laser Scanning Microscope Zeiss Meta 710 (Carl Zeiss Microscopy GmbH) was used. To paralyze worms for imaging, animals were treated with 0.1 µg/ml levamisole (Sigma Aldrich) or 60 mM sodium azide (Carl Roth). For analyzing TIR-1::GFP levels in AWC neurons age-synchronized L4 larvae of tir-1::gfp worms, also expressing RFP under an AWC-specific promotor (Podr-1::rfp), were grown until reaching day 1 of adulthood on OP50-seeded NGM plates supplemented with 25 µM BTZ or on plates supplemented with an equal volume of DMSO as solvent control, respectively. Worms were prepared as described above for confocal microscopy and obtained images were processed using ImageJ 1.52b. GFP fluorescence intensity was analyzed with the Imaris 9.l.2 software. Therefore, the volume of AWC neurons was determined in µm3 by creating a region of interest (ROI) based on RFP fluorescence with a threshold of 0.9 for background substraction. Within this ROI the GFP intensity was calculated as fluorescence intensity/µm3.
RNA interference
RNAi was performed via the feeding method established for C. elegans51,52. Age-synchronized worms were transferred to RNAi plates seeded with E. coli HT115 bacteria expressing the respective double-stranded RNA (dsRNA). Clones carrying the vector against a gene of interest were either taken from the RNAi Collection (Ahringer) (Source BioScience, Cat# 3318_Cel_RNAi_complete) or the ORF-RNAi Resource (Vidal)53 (Source BioScience, Cat# 3320_Cel_ORF_RNAi). As control condition, bacteria transformed with the empty pPD129.36 vector were used for feeding. To achieve an enhanced RNAi efficiency in neuronal tissues, additional knockdown of the RNaseT enzyme ERI-1 was performed as described54. Here, age-synchronized worms were fed HT115 bacteria expressing eri-1 dsRNA for 36 hours first. Followed by transfer to RNAi plates with HT115 bacteria expressing both eri-1 dsRNA and dsRNA of interest for additional 36 hours. Assessment of phenotypic changes and/or analysis via western blot was performed at day 1 adult stage (ca. 72 hours after placing L1 synchronized worms on RNAi plates).
RNA purification and qRT-PCR
Isolation of total RNA was performed using TRIzol (Invitrogen) and the QIAGEN RNeasy® kit. Age-synchronized worms were washed two times with M9 buffer, resuspended in 1 ml TRIzol reagent and frozen at -80°C for at least 1 hr. Worms were homogenized by adding silica beads, followed by tissue disruption with the Precellys 24-Dual cell homogenizer (Peq-Lab). Next, 1-bromo-3-chloropropane was added to the samples followed by vigorous mixing and phase separation via centrifugation. The aqueous phase was used to isolate total RNA with the RNeasy® Mini Kit (Qiagen) according to manufacturer’s instructions. cDNA synthesis was performed with 200 ng total RNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Gene expression levels were measured via quantitative real time PCR (qRT-PCR) with the Brilliant III Ultra-Fast SYBR® Green QPCR Master Mix (Agilent Technologies) and the Bio-Rad CFX96 Real-Time PCR Detection System. For each sample three technical replicates were analyzed and cdc-42 and pmp-3 were used for normalization55.
Microarray analysis and RNA sequencing
Expression profiling of mir-71 mutants versus WT was done using the Affymetrix™ C. elegans Gene Array 1.0. To this end, total RNA of around 200-300 age-synchronized day 1 adult hermaphrodites was extracted as described above. Microarray analysis was performed using four replicates per strain and was processed by the Cologne Center for Genomics (CCG, Cologne, Germany). Further quality control (analysis of RNA integrity number (RIN)) and gene expression profiling was performed. The obtained data were analyzed in detail with Partek® Genomic Suite® 6.6. After robust multi-array average (RMA) normalization, one-way ANOVA was performed to identify significant changes. Genome-wide transcriptional responses to proteotoxic stress were analyzed by RNA sequencing. Mid L4-staged N2 worms were put on OP50-seeded NGM plates containing 25 µM cadmium, 200 µM fluphenazine, 200 µM cantharidin or 10 µM and 25 µM BTZ, and then collected for total RNA extraction using the mirVana™ microRNA isolation kit (Ambion). Worms treated with rpn-6.1 RNAi were put on RNAi plates from early L3 stage on and harvested as day 1 adults. For each condition triplicates of ~2000 worms each were used for total RNA extraction. RNA quality control, library preparation of polyA-selected transcripts and subsequent Illumina® sequencing was conducted by the CCG. Raw data were analyzed by the in-house Bioinformatics facility, calculating Fragment Per Kilobase of exon per Million (FPKM) using Cufflinks and compute fold changes relative to untreated N2 worms. Significant changes were identified by analysis of variance (ANOVA). Filtering of the data and heatmap generation was done using Microsoft Excel. During establishment of the conditions, the various treatments were analyzed for their capacity to block the proteasome and to induce autophagy. Proteotoxic stress was applied by either, fluphenazine and cantharidin treatment-induced autophagy, or by directly blocking the proteasome with 10 or 25 µM BTZ or rpn-6.1 RNAi.
Bacterial feeding and chemotaxis assays
To study the impact of different food sources on proteostasis, worms were grown on different bacterial strains and UFD substrate (UbV-GFP) or tir-1 mRNA levels were monitored via western blotting and qRT-PCR, respectively. Here, age-synchronized L1 larvae were transferred to plates seeded with the respective E. coli strain. For inedible food experiments E. coli were treated with the antibiotic aztreonam (Az) that causes cell division stop and morphological changes inhibiting consequent ingestion by C. elegans29,56. OP50 were grown to log phase at 37°C, followed by Az treatment (final concentration 10 µg/ml) for additional 3 hr at 37°C and seeded to standard NGM plates 1 day before the experiment. OP50 expressing GFP (OP50-GFP) was used to control blocked ingestion after Az treatment (Fig. 4g). Age-synchronized worms were grown until day 1 of adulthood on E. coli OP50, transferred to Az containing plates, control plates or plates without food and cultivated for 6 hr. Following, animals were harvested for qRT-PCR as described above.
For studying chemotaxis of C. elegans, behavioural assays were adapted from S. Ward28. Chemotaxis studies were performed on freshly prepared Agar plates (2 % Agar; 5 mM KPO4, pH 6.0; 1 mM CaCl2; 1 mM MgSO4). Plates were separated in four equal quadrants (two test and two control quadrants). In each of the test quadrants 20 µl of OP50 bacteria containing 1 µM sodium azide were placed near the rim of the plate. LB medium was used as control condition. Age-synchronized worms were grown on NGM plates seeded with OP50 bacteria until day 1 of adulthood, rinsed from plates with S-Basal medium and washed three times. Animals were then transferred to the centre of the chemotaxis plate. After the residual S-Basal medium was soaked in, the plates were sealed with parafilm and after a 1.5 hr incubation at room temperature the number of worms in each quadrant was counted. Individuals that did not cross the inner circle were excluded. The chemotaxis index (CI) was determined with the following equation:
Silencing of AWC olfactory neurons
For functional silencing of AWC activity27, synchronized worms carrying Ex[ceh-36::HisCl; myo-2::GFP] (gift from T. Ishihara)57 were grown until L4 larval stage either on E. coli OP50 or HB101, respectively. L4s are finally transferred to histamine (HA) containing NGM plates (10 mM final concentration) or control plates and allowed to grow until day 1 of adulthood, followed by assessment of proteostasis capability using the UFD substrate (UbV-GFP) and according western blotting.
Statistical analysis
For statistical analysis the GraphPad Prism 5 software was used. A two-tailed paired Student’s t-test or one-way ANOVA with post-hoc test were used to analyze statistical significance of mRNA levels, fluorescence intensity in AWC neurons, chemotaxis, and proteotoxic stress assays. Results are given as mean and standard error of the mean (SEM). For survival assays, significance was determined using the Log-rank (Mantel-Cox) test.
Supplementary Material
Editorial summary.
Olfactory food perception is known to extend lifespan in C. elegans. Here, the authors demonstrate food-odor-dependent brain-to-gut communication that extends lifespan in worms. Food odor downregulates tir-1 mRNA in AWC neurons, in a manner dependent on the miRNA mir-71, which triggers downstream effects in the gut, owing to neuropeptide secretion, which promotes proteostasis and longevity.
Acknowledgments
We thank Y. Kohara and the Caenorhabditis Genetics Center (funded by the NIH National Center for Research Resources), the Dana-Farber Cancer Institute, Addgene and Geneservice Ltd for plasmids, cDNAs, and strains. We thank A. Segref for sharing unpublished strains and data on brain-to-gut regulation mechanisms. We thank the CECAD Imaging facility for support with confocal microscopy and the Cologne Center for Genomics for microarray analysis and RNA sequencing.
Funding:
This work is supported by grants of the Deutsche Forschungsgemeinschaft (DFG) (CECAD, FKZ: ZUK81/1, and SFB1218) and the European Research Council (consolidator grant 616499) to T.H. and grants of the Austrian Science Fund (FWF) (W-1207-B09 and SFB-F43-23) and European Research Council (ERC-StG-337161) to L.C.
Footnotes
Reporting Summary
Further information on research design is available in the Reporting Summary provided with this work.
Data availability
The authors declare that the main data supporting the findings of this study are available within the article and its Supplementary Information files. RNA sequencing and MicroArray data are deposited in the Gene Expression Omibus (GEO) database with the identifier [GSE124178] (RNA-seq) and [GSE124300] (MicroArray).
Author contributions:
F.F., F.O., and A.S. designed, performed and analyzed the experiments. A.S. and L.P. performed and analyzed RNA-sequencing experiments; T.D. and L.C. established gene-edited strains. S.M. performed stress assays. T.H. supervised the design and data interpretation; F.F., F.O., and T.H. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Competing interests:
The authors declare no competing interests.
References
- 1.Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol. 2011 doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 3.de Lencastre A, et al. MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol. 2010;20:2159–68. doi: 10.1016/j.cub.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mori Ma, et al. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab. 2012;16:336–47. doi: 10.1016/j.cmet.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segref A, Torres S, Hoppe T. A screenable in vivo assay to study proteostasis networks in Caenorhabditis elegans. Genetics. 2011;187:1235–40. doi: 10.1534/genetics.111.126797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denzel MS, et al. Hexosamine pathway metabolites enhance protein quality control and prolong life. Cell. 2014;156:1167–1178. doi: 10.1016/j.cell.2014.01.061. [DOI] [PubMed] [Google Scholar]
- 7.Ruggiano A, Foresti O, Carvalho P. Quality control: ER-associated degradation: protein quality control and beyond. J Cell Biol. 2014;204:869–79. doi: 10.1083/jcb.201312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vilchez D, et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–8. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 9.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 10.Boulias K, Horvitz HR. The C. elegans MicroRNA mir-71 Acts in Neurons to Promote Germline-Mediated Longevity through Regulation of DAF-16/FOXO. Cell Metab. 2012;15:439–50. doi: 10.1016/j.cmet.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsieh Y-W, Chang C, Chuang C-F. The microRNA mir-71 inhibits calcium signaling by targeting the TIR-1/Sarm1 adaptor protein to control stochastic L/R neuronal asymmetry in C. elegans. PLoS Genet. 2012;8:e1002864. doi: 10.1371/journal.pgen.1002864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hobert O. Terminal Selectors of Neuronal Identity. Curr Top Dev Biol. 2016;116:455–75. doi: 10.1016/bs.ctdb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Alcedo J, Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004;41:45–55. doi: 10.1016/s0896-6273(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 14.Troemel ER, Kimmel BE, Bargmann CI. Reprogramming chemotaxis responses: Sensory neurons define olfactory preferences in C. elegans. Cell. 1997;91:161–169. doi: 10.1016/s0092-8674(00)80399-2. [DOI] [PubMed] [Google Scholar]
- 15.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3’UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuang C-F, Bargmann CI. A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev. 2005;19:270–81. doi: 10.1101/gad.1276505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liberati NT, et al. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc Natl Acad Sci U S A. 2004;101:6593–8. doi: 10.1073/pnas.0308625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Y, Moussaif M, Choi S, Xu L, Sze JY. RFX transcription factor DAF-19 regulates 5-HT and innate immune responses to pathogenic bacteria in Caenorhabditis elegans. PLoS Genet. 2013;9:e1003324. doi: 10.1371/journal.pgen.1003324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153:1435–47. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prahlad V, Cornelius T, Morimoto RI. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–4. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madison JM, Nurrish S, Kaplan JM. UNC-13 interaction with syntaxin is required for synaptic transmission. Curr Biol. 2005;15:2236–2242. doi: 10.1016/j.cub.2005.10.049. [DOI] [PubMed] [Google Scholar]
- 22.Speese S, et al. UNC-31 (CAPS) Is Required for Dense-Core Vesicle But Not Synaptic Vesicle Exocytosis in Caenorhabditis elegans. J Neurosci. 2007;27:6150–6162. doi: 10.1523/JNEUROSCI.1466-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C. The ever-expanding neuropeptide gene families in the nematode Caenorhabditis elegans. Parasitology. 2005;131(Suppl):S109–27. doi: 10.1017/S0031182005009376. [DOI] [PubMed] [Google Scholar]
- 24.Chalasani SH, et al. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature. 2007;450:63–70. doi: 10.1038/nature06292. [DOI] [PubMed] [Google Scholar]
- 25.Hobert O, et al. Regulation of interneuron function in the C. elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron. 1997;19:345–57. doi: 10.1016/s0896-6273(00)80944-7. [DOI] [PubMed] [Google Scholar]
- 26.Bargmann CI. Chemosensation in C. elegans. WormBook. 2006:1–29. doi: 10.1895/wormbook.1.123.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pokala N, Liu Q, Gordus A, Bargmann CI. Inducible and titratable silencing of Caenorhabditis elegans neurons in vivo with histamine-gated chloride channels. Proc Natl Acad Sci U S A. 2014;111:2770–5. doi: 10.1073/pnas.1400615111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward S. Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proc Natl Acad Sci U S A. 1973;70:817–21. doi: 10.1073/pnas.70.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben Arous J, Laffont S, Chatenay D. Molecular and sensory basis of a food related two-state behavior in C. elegans. PLoS One. 2009;4:e7584. doi: 10.1371/journal.pone.0007584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Libert S, et al. Regulation of Drosophila life span by olfaction and food-derived odors. Science (80-. ) 2007 doi: 10.1126/science.1136610. [DOI] [PubMed] [Google Scholar]
- 31.Maier W, Adilov B, Regenass M, Alcedo J. A neuromedin u receptor acts with the sensory system to modulate food type-dependent effects on C. elegans lifespan. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue A, et al. Forgetting in C. elegans is accelerated by neuronal communication via the TIR-1/JNK-1 pathway. Cell Rep. 2013;3:808–19. doi: 10.1016/j.celrep.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 33.Essuman K, et al. The SARM1 Toll/Interleukin-1 Receptor Domain Possesses Intrinsic NAD+Cleavage Activity that Promotes Pathological Axonal Degeneration. Neuron. 2017;93:1334–1343.e5. doi: 10.1016/j.neuron.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsvetkov P, et al. NADH binds and stabilizes the 26S proteasomes independent of ATP. J Biol Chem. 2014;289:11272–81. doi: 10.1074/jbc.M113.537175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Summers DW, Gibson DA, DiAntonio A, Milbrandt J. SARM1-specific motifs in the TIR domain enable NAD+ loss and regulate injury-induced SARM1 activation. Proc Natl Acad Sci U S A. 2016;113:E6271–E6280. doi: 10.1073/pnas.1601506113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan Z-G, An X-S. SARM1 deletion restrains NAFLD induced by high fat diet (HFD) through reducing inflammation, oxidative stress and lipid accumulation. Biochem Biophys Res Commun. 2018;498:416–423. doi: 10.1016/j.bbrc.2018.02.115. [DOI] [PubMed] [Google Scholar]
- 37.Lin C-W, Hsueh Y-P. Sarm1, a neuronal inflammatory regulator, controls social interaction, associative memory and cognitive flexibility in mice. Brain Behav Immun. 2014;37:142–151. doi: 10.1016/j.bbi.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Brooks KK, Liang B, Watts JL. The Influence of Bacterial Diet on Fat Storage in C. elegans. PLoS One. 2009;4:e7545. doi: 10.1371/journal.pone.0007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riera CE, et al. The Sense of Smell Impacts Metabolic Health and Obesity. Cell Metab. 2017;26:198–211.e5. doi: 10.1016/j.cmet.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 40.Teff K. Nutritional implications of the cephalic-phase reflexes: endocrine responses. Appetite. 2000;34:206–213. doi: 10.1006/appe.1999.0282. [DOI] [PubMed] [Google Scholar]
- 41.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miedel MT, et al. A Pro-Cathepsin L Mutant Is a Luminal Substrate for Endoplasmic-Reticulum-Associated Degradation in C. elegans. PLoS One. 2012;7:e40145. doi: 10.1371/journal.pone.0040145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roayaie K, Crump JG, Sagasti A, Bargmann CI. The G alpha protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron. 1998;20:55–67. doi: 10.1016/s0896-6273(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 45.Radman I, Greiss S, Chin JW. Efficient and Rapid C. elegans Transgenesis by Bombardment and Hygromycin B Selection. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drexel T, Mahofsky K, Latham R, Zimmer M, Cochella L. Neuron type-specific miRNA represses two broadly expressed genes to modulate an avoidance behavior in C. elegans. Genes Dev. 2016;30:2042–2047. doi: 10.1101/gad.287904.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedland AE, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741–3. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katic I, Großhans H. Targeted Heritable Mutation and Gene Conversion by Cas9-CRISPR in Caenorhabditis elegans. Genetics. 2013 doi: 10.1534/genetics.113.155754. genetics.113.155754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Semple JI, Biondini L, Lehner B. Generating transgenic nematodes by bombardment and antibiotic selection. Nat Methods. 2012;9:118–119. doi: 10.1038/nmeth.1864. [DOI] [PubMed] [Google Scholar]
- 50.Mitchell DH, Stiles JW, Santelli J, Sanadi DR. Synchronous Growth and Aging of Caenorhabditis elegans in the Presence of Fluorodeoxyuridine 1. 1979;34:28–36. doi: 10.1093/geronj/34.1.28. [DOI] [PubMed] [Google Scholar]
- 51.Timmons L, Fire a. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 52.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rual JF, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jadiya P, Nazir A. A pre- and co-knockdown of RNAseT enzyme, Eri-1, enhances the efficiency of RNAi induced gene silencing in Caenorhabditis elegans. PLoS One. 2014;9:e87635. doi: 10.1371/journal.pone.0087635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol. 2008;9:9. doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Potluri L, et al. Septal and lateral wall localization of PBP5, the major D,D-carboxypeptidase of Escherichia coli, requires substrate recognition and membrane attachment. Mol Microbiol. 2010;77:300–23. doi: 10.1111/j.1365-2958.2010.07205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitazono T, et al. Multiple Signaling Pathways Coordinately Regulate Forgetting of Olfactory Adaptation through Control of Sensory Responses in Caenorhabditis elegans. J Neurosci. 2017;37:10240–10251. doi: 10.1523/JNEUROSCI.0031-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.