A systematic characterization of natural diversity, coexpression network analysis, and experimental validation for understanding the genetic architecture of senescence and stay-green in maize.
Abstract
Premature senescence in annual crops reduces yield, while delayed senescence, termed stay-green, imposes positive and negative impacts on yield and nutrition quality. Despite its importance, scant information is available on the genetic architecture of senescence in maize (Zea mays) and other cereals. We combined a systematic characterization of natural diversity for senescence in maize and coexpression networks derived from transcriptome analysis of normally senescing and stay-green lines. Sixty-four candidate genes were identified by genome-wide association study (GWAS), and 14 of these genes are supported by additional evidence for involvement in senescence-related processes including proteolysis, sugar transport and signaling, and sink activity. Eight of the GWAS candidates, independently supported by a coexpression network underlying stay-green, include a trehalose-6-phosphate synthase, a NAC transcription factor, and two xylan biosynthetic enzymes. Source–sink communication and the activity of cell walls as a secondary sink emerge as key determinants of stay-green. Mutant analysis supports the role of a candidate encoding Cys protease in stay-green in Arabidopsis (Arabidopsis thaliana), and analysis of natural alleles suggests a similar role in maize. This study provides a foundation for enhanced understanding and manipulation of senescence for increasing carbon yield, nutritional quality, and stress tolerance of maize and other cereals.
INTRODUCTION
Agricultural productivity is essentially the amount of carbohydrates generated by photosynthetic assimilation of CO2 by plant leaves and stored into heterotrophic organs harvested for human/industrial use. Improving the photosynthetic assimilation of crop plants therefore is a viable approach to increase agricultural productivity. However, the photosynthetic ability is inherently linked with the plant developmental program, and for monocarpic (annual) plants that are the major source of food, starts to decrease during the reproductive phase. For instance, during the grain-filling period in maize (Zea mays), one of the most important staple foods on earth, photosynthesis rates decline by as much as 50% within a 6-week period after anthesis (Ying et al., 2000). The decline of photosynthesis is triggered upon the onset of senescence, a highly regulated, well-coordinated, and biologically active process that marks the end of the life cycle of the leaf and ultimately the whole plant (Quirino et al., 2000; Buchanan-Wollaston et al., 2003; Lim et al., 2007; Wingler et al., 2009; Thomas, 2013).
Senescence is important for recycling resources, particularly nitrogen and carbon, from old organs to either the newly developing organs or storage sinks, thereby contributing to the fitness of a plant (Lim et al., 2007). However, most of the dry matter accumulating in grains of modern maize hybrids is fixed during the grain fill, with very little attributed to remobilization from organs developed during the pre-flowering period (Below et al., 1981; Cliquet et al., 1990; Ciampitti and Vyn, 2013). Thus, given the potential benefits of delaying senescence without adversely affecting grain yield, developing late senescing or stay-green cultivars with delayed physiological maturity and prolonged grain-filling period is an important crop breeding strategy (Duvick et al., 2010; Gregersen et al., 2013). In maize, selection for stay-green trait accounts for up to 63% of the total increase in dry matter accumulation in newer era hybrids released after the 2000s compared with older hybrids released in the 1930s (Lee and Tollenaar, 2007). Stay-green has been associated with increased drought tolerance in multiple crop species including wheat (Triticum aestivum), sorghum (Sorghum bicolor), barley (Hordeum vulgare), and maize (Tuinstra et al., 1997; Vijayalakshmi et al., 2010; Emebiri, 2013; Trachsel et al., 2016). Maize and sorghum stay-green lines possess increased resistance to diseases, especially those caused by stalk-rotting pathogens (King and Frederickson, 1976; Bertolini et al., 1983). Stay-green also improves produce quality such as higher Suc and protein content in maize and higher stalk carbohydrates in sorghum (McBee et al., 1983; Gentinetta et al., 1986). For certain production systems, the stay-green trait can also have serious negative consequences such as delayed dry down resulting in higher moisture in the stover (leaves, stalks, and husk), which interferes with timely mechanical harvesting with combine harvesters (Yang et al., 2010).
Senescence is a complex and dynamic process that, while developmentally programmed, can be initiated and/or accelerated by a number of internal and external factors and overlaps with other processes including autophagy, aging, and death. Genetic dissection of this quantitative trait in Arabidopsis (Arabidopsis thaliana) has implicated a wide range of processes and genes including those involved in the synthesis of hormones such as cytokinin and ethylene, sugar sensing and signaling, source–sink translocation, and autophagy and aging (Schippers, 2015; Wingler, 2018). Senescence is also promoted by several environmental factors including lack of light (darkness), excessive light intensity/dosage, drought, salinity, nutrient (e.g., nitrogen, sulfur, and iron) starvation, and biotic stresses (Quirino et al., 2000; Lim et al., 2007; Thomas, 2013; Schippers, 2015). The partitioning of sugars to various sinks and the associated signals emerging from the interplay of the photosynthetic output of the plant (source strength) and the need for the photoassimilate (sink strength) play a key role in the regulation of senescence (Lee and Tollenaar, 2007; Wingler et al., 2009; Thomas, 2013; Thomas and Ougham, 2014). A weaker source induces early senescence due to increased mobilization of dry matter from leaves and other organs to the developing sinks, thereby starving the source organs (Rajcan and Tollenaar, 1999; Thomas, 2013). Weaker sink demand also accelerates senescence (Lee and Tollenaar, 2007), likely due to the accumulation of sugars in leaves beyond a certain critical threshold followed by sugar-mediated signaling of leaf senescence (Wingler and Roitsch, 2008; Thomas, 2013). While the role of source–sink interaction in the regulation of senescence is well established, the underlying mechanisms and genetic elements are largely unknown.
Valuable insights into the genetic architecture of senescence have been provided by the forward genetic screens in multiple plant species; however, these mostly represent the major effect of genes producing visible mutant phenotypes. Biparental mapping populations have also been used to identify quantitative trait loci in Arabidopsis (Wingler et al., 2010; Chardon et al., 2014), maize (Zheng et al., 2009; Wang et al., 2012; Belícuas et al., 2014; Trachsel et al., 2016), rice (Oryza sativa; Jiang et al., 2004; Abdelkhalik et al., 2005; Yoo et al., 2007), sorghum (Tuinstra et al., 1997; Xu et al., 2000; Harris et al., 2007), and wheat (Vijayalakshmi et al., 2010; Bogard et al., 2011; Pinto et al., 2016). These studies have been useful in understanding the genetic architecture of senescence but often lack the resolution to identify the underlying genes. Transcriptomic analyses in Arabidopsis have identified several nuclear senescence-associated genes (SAGs) and transcription factors, and chloroplast genes (Breeze et al., 2011; Guo and Gan, 2012; Woo et al., 2016). We have performed transcriptomic analysis of leaf and internode tissues in the maize B73 inbred line undergoing premature senescence induced by lack of seed sink and identified genes involved in source–sink-regulated senescence (Sekhon et al., 2012). Differentially expressed (DE) genes during senescence have also been identified in sorghum (Johnson et al., 2015; Wu et al., 2016), wheat (Gregersen and Holm, 2007), and rice (Ma et al., 2005). While these studies provide general insights into global senescence-related transcriptome, identification of the underlying causal genes is difficult due to a large number of DE candidates.
Given the diversity of internal and external regulators of senescence, a systems genetics (SysGen) approach (Civelek and Lusis, 2014) is needed for a comprehensive understanding of the genes and genetic networks underlying this complex developmental process. We characterized a large proportion of natural variation captured in a diversity panel and constructed gene coexpression networks based on comparative transcriptomic analysis of stay-green and natural senescence. Investigation and integration of these system-level data revealed both known and novel genes/processes and provided several new and valuable insights into the regulation of senescence in maize and other grasses.
RESULTS
Characterization of the Natural Variation for Leaf Senescence in Maize
Maize diversity characterized in this study was captured in a panel that (1) includes maize lines from two major heterotic groups (Stiff Stalk Synthetic and Non-Stiff Stalk Synthetic), (2) includes only those inbred lines that flowered within a reasonable window in the midwestern United States, and (3) was genotyped by RNA sequencing (RNA-seq) of whole seedling mRNA (Hirsch et al., 2014). We evaluated natural variation for onset of leaf senescence in a subset of this panel in three environments by growing 328 inbred lines during summer 2011 in Verona, Wisconsin (W11); 364 inbred lines during summer 2013 in Arlington, Wisconsin (W13); and 223 inbred lines during summer 2017 in Pendleton, South Carolina (S17). The maximum quantum efficiency of PSII photochemistry (Fv/Fm), a reliable indicator of monocarpic senescence (Wingler et al., 2004, 2010), was recorded in dark-adapted, field-grown plants at 36 d after anthesis (DAA). We found substantial phenotypic variation ranging from complete senescence (Fv/Fm = 0) to lack of senescence (Fv/Fm = 0.82), with the latter signifying stay-green genotypes (Supplemental Figure 1; Supplemental Table 1). The genotypic variance was significant for all three environments and for the combined W11_13 analysis (Table 1). Consistent with the significant impact of environment on senescence, the genotype ☓ environment component was significant for the combined W11_13 analysis. The broad sense heritability of Fv/Fm observed in each environment was 37.3% (W11), 72.6% (W13), 41.2% (W11_13), and 43.7% (S17).
Table 1. SNPs and Candidate Genes Associated With Leaf Senescence in Maize.
| SNP no. | Environment(s)a | Chr | SNP Position (AGPv4) | P-value | LOSb | Maf | R2 (%) | Candidate Genec | Gene Descriptiond | DEe | Cox Netf | SAGf |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | W13 | 1 | 3806739 | 1.00E−06 | S | 0.04 | 5.8 | Zm00001d027361 | F-box protein PP2-A13 | Y** | SAG | |
| 2 | W11 | 1 | 27313091 | 4.05E−06 | S | 0.08 | 6.6 | Zm00001d028230 | ZmMST4/STP13 | Y** | SAG | |
| 3 | W13, W11_13 | 1 | 27701071 | 5.15E−07 | S | 0.03 | 6.2 | Zm00001d028241 | 3-Ketoacyl-CoA synthase | Y*** | ||
| 4 | W11, W11_13 | 1 | 159291867 | 5.76E−07 | S | 0.03 | 7.8 | Zm00001d030780 | Formin-like protein2 (FH2) | |||
| 5 | W11_13 | 1 | 169610725 | 3.02E−06 | S | 0.05 | 5.6 | Zm00001d030968 | Flowering locus K domain protein | |||
| 6 | W11_13 | 1 | 237181291 | 1.65E−06 | S | 0.10 | 5.9 | Zm00001d032750 | Zinc finger (C3HC4-type RING) | Y*** | ||
| 7 | W13 | 1 | 244123496 | 1.39E−06 | S | 0.06 | 5.7 | Zm00001d032937 | Mediator of ABA-regulated dormancy1 | SAG | ||
| 8 | W11 | 1 | 287023239 | 5.09E−06 | S | 0.04 | 6.4 | Zm00001d034224 | Glutathione S-transferase family protein | SAG | ||
| 9 | W13 | 1 | 295000279 | 1.78E−07 | SIG | 0.05 | 6.7 | Zm00001d034508 | Hydroxyproline O-galactosyltransferase | Y*** | SAG | |
| 10 | W11 | 2 | 3111873 | 1.04E−06 | S | 0.03 | 7.4 | Zm00001d001941 | Cell wall invertase (incw4) | Y*** | ||
| 11 | W11, W11_13 | 2 | 5236197 | 1.22E−09 | HSIG | 0.04 | 11.7 | Zm00001d002065 | mir3 (Pružinská et al., 2017) | Y*** | SAG | |
| 11a | W11, W11_13 | 2 | 5236197 | 1.22E−09 | HSIG | 0.04 | 11.7 | Zm00001d002066 | ZmIRX15-LIKE | Y*** | ||
| 12 | W13 | 2 | 28264867 | 2.22E−06 | S | 0.02 | 5.5 | Zm00001d002967 | AFG1-like ATPase family protein | Y*** | SAG | |
| 13 | S17 | 2 | 137366571 | 2.19E−06 | S | 0.09 | 10.8 | Zm00001d004762 | ABC transporter G family member25 | Y** | ||
| 14 | S17 | 2 | 137909220 | 2.36E−06 | S | 0.10 | 10.7 | Zm00001d004768 | Glucan endo-1,3-β-glucosidase | Y*** | SAG | |
| 15 | S17 | 2 | 194310598 | 1.42E−07 | SIG | 0.09 | 13.5 | Zm00001d005970 | Dof zinc finger protein DOF1.6 | Y** | ||
| 16 | W13 | 2 | 199698550 | 2.10E−08 | SIG | 0.03 | 7.8 | Zm00001d006148 | ZmPET191 | Y*** | Y | SAG |
| 17 | W11_13, W11 | 2 | 201398868 | 6.02E−07 | S | 0.04 | 6.4 | Zm00001d006198 | B-box zinc finger protein19 | Y*** | ||
| 18 | W11, W11_13 | 2 | 205075914 | 1.87E−10 | HSIG | 0.03 | 12.9 | Zm00001d006341 | Molybdenum cofactor sulfurase | Y*** | ||
| 19 | W13 | 2 | 230642666 | 2.74E−07 | S | 0.04 | 6.5 | Zm00001d007390 | Succinate dihydrogenase assembly factor2 | Y*** | SAG | |
| 20 | W13 | 2 | 239548398 | 3.97E−06 | S | 0.02 | 5.2 | Zm00001d007784 | C3H-transcription factor34 | Y*** | ||
| 21 | W13 | 2 | 242250761 | 4.93E−06 | S | 0.04 | 5.1 | Zm00001d007908 | Probable carboxylesterase 15 | Y*** | ||
| 22 | W11_13 | 3 | 3088968 | 1.78E−06 | S | 0.12 | 5.9 | Zm00001d039384 | Cytochrome P450 71A1 | Y*** | SAG | |
| 23 | W11 | 3 | 3980505 | 1.65E−06 | S | 0.09 | 7.1 | Zm00001d039426 | β-Glucuronosyltransferase GlcAT14B | Y** | SAG | |
| 24 | W13 | 3 | 33490908 | 3.48E−07 | S | 0.12 | 6.4 | Zm00001d040243 | Sugar transporter ERD6-like 3 | SAG | ||
| 25 | W13 | 3 | 33613659 | 3.48E−07 | S | 0.12 | 6.4 | Zm00001d040247 | Putative expansin-A17 | SAG | ||
| 26 | W11_13, W11 | 3 | 50460949 | 8.32E−09 | SIG | 0.02 | 8.7 | Zm00001d040569 | Programmed cell death2/zinc finger | |||
| 27 | W11_13, W13 | 3 | 126107238 | 2.11E−06 | S | 0.01 | 5.8 | Zm00001d041533 | Probable uridine nucleosidase2 | Y*** | SAG | |
| 28 | W13 | 3 | 183569800 | 8.18E−07 | S | 0.03 | 5.9 | Zm00001d042905 | Laccase-17 | Y*** | SAG | |
| 29 | W11_13, W11 | 3 | 197672135 | 1.12E−06 | S | 0.04 | 6.1 | Zm00001d043364 | Hypothetical protein | SAG | ||
| 30 | W11_13 | 3 | 207759321 | 3.87E−06 | S | 0.02 | 5.5 | Zm00001d043700 | Vesicle transport v-SNARE12 | SAG | ||
| 31 | W11 | 3 | 211449496 | 2.52E−06 | S | 0.07 | 6.9 | Zm00001d043838 | Transparent testa12 | SAG | ||
| 32 | W11_13 | 4 | 8260720 | 1.19E−06 | S | 0.02 | 6.1 | Zm00001d048928 | Di-glucose binding with kinesin motor | |||
| 33 | W11_13, W11 | 4 | 19863905 | 2.43E−07 | SIG | 0.02 | 6.9 | Zm00001d049191 | Methylthioribulose-1-phospate dehydratase | |||
| 34 | W13 | 4 | 172416278 | 9.34E−07 | S | 0.05 | 5.9 | Zm00001d051854 | Putative membrane protein (DUF2404) | |||
| 35 | W13 | 4 | 201215331 | 1.05E−06 | S | 0.05 | 5.8 | Zm00001d052798 | Trihelix-transcription factor 3 | Y*** | SAG | |
| 36 | W11_13 | 4 | 232855452 | 8.41E−07 | S | 0.08 | 6.3 | Zm00001d053518 | Putative leucine-rich repeat kinase | Y*** | SAG | |
| 37 | W13 | 4 | 241403663 | 3.29E−06 | S | 0.04 | 5.3 | Zm00001d053802 | dek10 | Y* | ||
| 38 | W13 | 4 | 244387849 | 1.31E−06 | S | 0.05 | 5.7 | Zm00001d053964 | Uncharacterized protein | Y*** | ||
| 39 | W13 | 5 | 10155281 | 4.56E−06 | S | 0.02 | 5.1 | Zm00001d013386 | Uncharacterized protein | Y*** | Y | |
| 40 | W11_13 | 5 | 211352713 | 2.13E−06 | S HSIG | 0.12 | 5.8 | Zm00001d017983 | Formin homolog4 | Y*** | SAG | |
| 41 | W13, W11_13 | 6 | 84476696 | 4.56E−10 | S | 0.02 | 9.7 | Zm00001d036328 | 2-Oxoglutarate dehydrogenase E1 | Y*** | SAG | |
| 42 | W11 | 6 | 151107104 | 4.05E−07 | S | 0.09 | 8.0 | Zm00001d038192 | Glutathione transferase41 | Y*** | ||
| 43 | W13 | 6 | 152051904 | 5.73E−07 | S | 0.07 | 6.1 | Zm00001d038229 | ZmGUX1 | Y*** | Y | |
| 43a | W13 | 6 | 152051904 | 5.73E−07 | SIG | 0.07 | 6.1 | Zm00001d038226 | ZmSWEET1b (Zhou et al., 2014) | Y** | ||
| 44 | W11_13, W13, W11 | 6 | 166156942 | 1.45E−08 | S | 0.05 | 8.4 | Zm00001d038870 | mlg3 | Y*** | Y | SAG |
| 45 | W11_13 | 7 | 32005835 | 4.80E−06 | S | 0.06 | 5.4 | Zm00001d019398 | ABC transporter G family member11 | Y*** | SAG | |
| 46 | W11_13 | 7 | 111244964 | 1.12E−06 | S | 0.17 | 6.1 | Zm00001d020396 | Trehalose-6-phospate synthase13(trps13) | Y*** | Y | |
| 47 | W13, W11_13 | 7 | 137480003 | 3.60E−06 | SIG | 0.04 | 5.2 | Zm00001d020951 | AT-hook motif nuclear-localized protein1 | Y*** | SAG | |
| 48 | W11_13, W13, W11 | 7 | 175969130 | 6.19E−08 | S | 0.02 | 7.6 | Zm00001d022353 | BSD domain-containing protein1 | Y*** | ||
| 49 | W11 | 7 | 181112708 | 5.40E−06 | SIG | 0.05 | 6.4 | Zm00001d022618 | SKP1-like protein 1A | Y*** | ||
| 50 | W13, W11_13 | 8 | 155438738 | 2.05E−08 | S | 0.06 | 7.8 | Zm00001d011596 | 30S ribosomal protein S20 chloroplastic | Y*** | ||
| 51 | W11 | 8 | 159901882 | 4.14E−06 | SIG | 0.13 | 6.6 | Zm00001d011721 | Brassinosteroid insensitive1a | SAG | ||
| 52 | W13 | 8 | 165321806 | 1.44E−07 | HSIG | 0.02 | 6.8 | Zm00001d011969 | NAC-transcription factor 9(nactf9) | Y*** | Y | |
| 53 | W13, W11_13 | 8 | 169165965 | 1.69E-09 | S | 0.03 | 9.0 | Zm00001d012159 | ZmBGLU42 | Y*** | ||
| 54 | W11_13, W11 | 8 | 179520170 | 1.75E−06 | S | 0.03 | 5.9 | Zm00001d012743 | Extensin-like receptor kinase(ZmPERK) | Y*** | Y | SAG |
| 55 | W11 | 9 | 1015574 | 4.19E−06 | S | 0.04 | 6.5 | Zm00001d044747 | B12D protein | Y*** | ||
| 56 | W13 | 9 | 5518351 | 1.13E−06 | SIG | 0.12 | 5.8 | Zm00001d044857 | AP2-EREBP-transcription factor 31 | Y*** | ||
| 57 | W11, W11_13 | 9 | 21687718 | 1.30E−07 | SIG | 0.06 | 8.7 | Zm00001d045427 | Barren stalk fastigiate1 | |||
| 58 | W13, W11_13 | 9 | 104674835 | 5.61E−08 | S | 0.01 | 7.3 | Zm00001d046761 | Gamma response1 | Y | ||
| 59 | W11_13 | 9 | 157775007 | 4.48E−06 | S | 0.09 | 5.4 | Zm00001d048524 | N-terminal amidase1 | Y*** | ||
| 60 | W11 | 10 | 90457182 | 2.98E−06 | HSIG | 0.12 | 6.8 | Zm00001d024839 | Glutathione S-transferase2 | Y*** | ||
| 61 | W13, W11_13 | 10 | 93455840 | 1.07E−09 | S | 0.02 | 9.3 | Zm00001d024894 | Putative thioredoxin superfamily protein | Y*** | ||
| 62 | S17 | 10 | 134987150 | 8.53E−07 | 0.03 | 11.7 | Zm00001d025964 | Homeobox-transcription factor 7 (AtHB7) | Y | SAG |
ABA, abscisic acid; ABC, ATP-binding cassette; AFG, ATPase family gene; AP2, APETALA2; AT, adenine-thymine; BSD, BTF2-like transcription factors, synapse-associated and DOS2-like proteins; Chr, chromosome; Cox Net, coexpression subnetwork; DOF, DNA-binding with one finger; Env, environment(s); ERD, Early-responsive to dehydration; EREBP, ethylene-responsive element binding proteins; Maf, minor allele frequency; SKP, S-Phase Kinase Associated Protein; v-SNARE, vesicle-soluble NSF attachment protein receptor.
The environment with lowest P-value appears first in the Environment(s) column and the P-value and R2 are provided for the first environment. Genes with bold and italicized font are those highlighted in Figure 3 with blue font.
LOS, level of significance indicated as HSIG, highly significant; SIG, significant; and S, suggestive (see text).
Bold and italicized font of gene ID indicate genes that highlighted in Figure 3 and Figure 4 with blue font.
Bold gene description indicates all genes that are included in Figure 4.
One, two, and three asterisks denote FDR corrected P ≤ 0.05, 0.01, and 0.001, respectively.
Blank cells indicate no data.
Identification of Loci Associated with Senescence by Exploiting Natural Allelic Diversity
To identify the genes and genetic elements underlying senescence, we tested association of 438,161 RNA-seq single-nucleotide polymorphism (SNP) markers reported for the maize diversity panel (Hirsch et al., 2014) with senescence, recorded as Fv/Fm, for individual environments (W11, W13, and S17) and for combined WI environments (W11_13) (Supplemental Figure 2). In total, 62 unique SNPs were identified to be associated with senescence in one or more environments, of which five were highly significant (P ≤ 5.45E−9), 11 were significant (P ≤ 2.73E−7), and 46 met a suggestive threshold (P ≤ 5.45E−6) based on a proposed criteria (see “Methods”; Table 1; Lander and Kruglyak, 1995; Li et al., 2012).
Based on the evaluation of a large collection of more than 2800 inbred lines, linkage disequilibrium in maize drops within ∼10 kb with an r2 of 0.2, but the extent of linkage disequilibrium is dependent on the germplasm included in the analysis and the region of the genome (Romay et al., 2013). Therefore, only the SNP with the lowest P-value within a 200-kb window (100 kb ± SNP position) is reported. Two SNPs are represented twice due to the presence of two candidate genes in the vicinities of each of these SNPs.
Total phenotypic variance (R2) explained by all SNPs ranged between 5.1 and 13.5%, with a median value of 6.4%. To investigate the role these SNPs in senescence, we defined the candidate regions represented by each SNP to span 100 kb on each flank of the SNP. The 200-kb window size is based on similar studies in maize (Li et al., 2013b, 2016; Diepenbrock et al., 2017) and aimed to facilitate the identification of the correct candidate gene associated with the SNP. A search for all annotated genes within these 200-kb windows surrounding the identified SNPs yielded 411 candidate genes, with a range of 1 to 17 candidate genes per window and a median of 6.5 (Supplemental Data Set 1). These genes were further scrutinized using a SysGen approach to identify the most likely candidates and their role in senescence.
Physiological and Metabolic Analysis of Senescence and Stay-Green
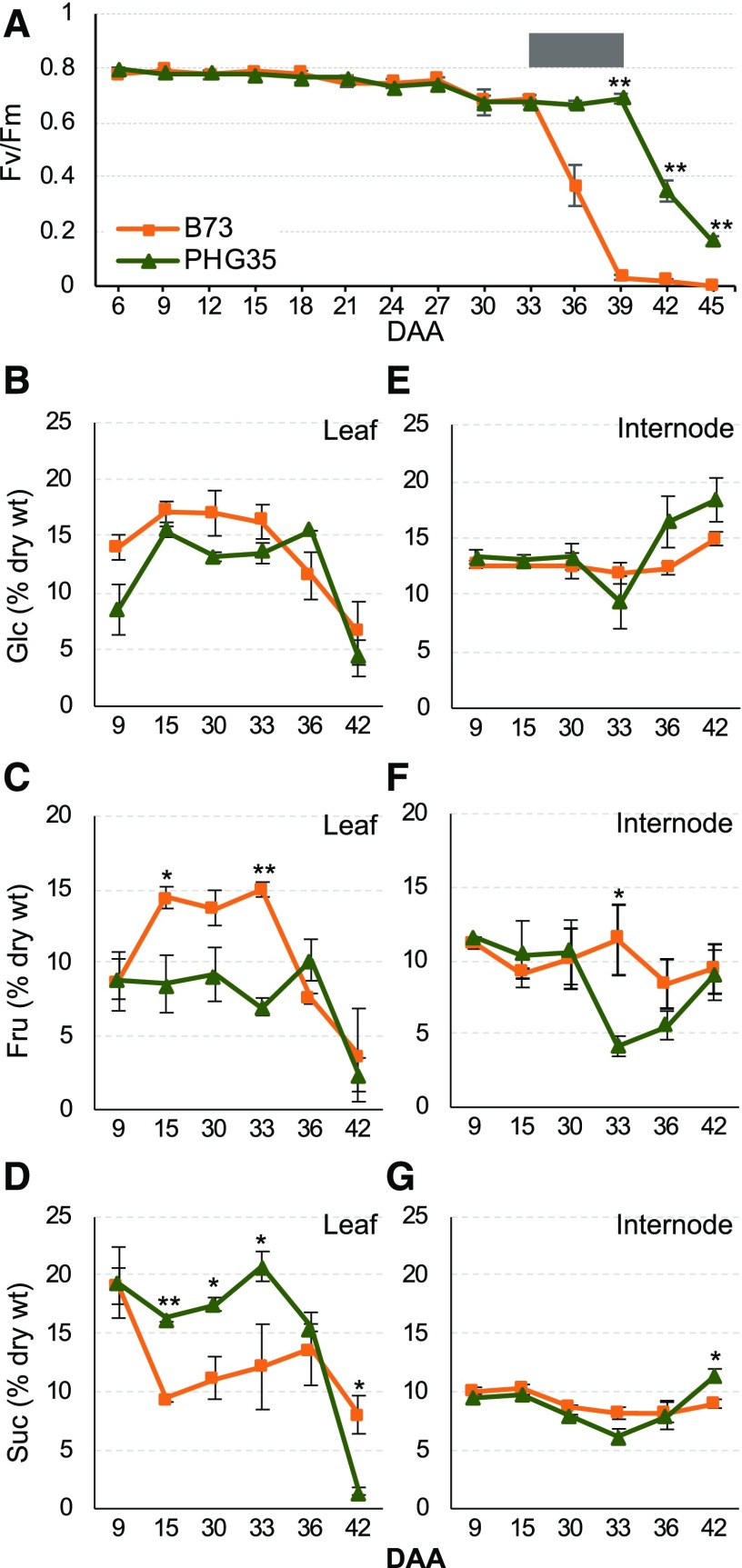
We chose two inbred lines for detailed analysis of senescence and stay-green: PHG35 and B73. PHG35, a Non-Stiff Stalk inbred line from Oh07-Midland/Iodent background, is a stay-green line (Popelka, 2012), while B73, a Stiff Stalk inbred line from Iowa Stiff Stalk Synthetic background, is a non-stay-green line. Photosynthetic activity, as indicated by Fv/Fm of dark-adapted ear leaf, started to decline in both inbred lines from 30 DAA, albeit the rate of decline was different (Figure 1A). B73 showed a sharper decline after 33 DAA, with complete loss of photosynthetic activity at 39 DAA, while PHG35 had very high activity up to 39 DAA but declined quickly thereafter. Both chlorophyll a (Chl a) and Chl b followed a similar trend, with a sharper decline in B73 after 33 DAA and a gradual loss in PHG35 (Supplemental Figure 3). Overall, the 33 to 39 DAA window represents a transition zone separating the non-stay-green and stay-green phenotype displayed by the two inbred lines (Figure 1A).
Figure 1.
Physiological and Metabolic Differences between a Non-Stay-Green (B73) and a Stay-Green (PHG35) Maize Inbred Line during Senescence.
(A) Onset and progression of senescence deduced from the maximum quantum efficiency of PSII (Fv/Fm). Gray box shows the transition zone separating non-stay-green and stay-green phenotype.
(B) to (D) Accumulation of Glc (B), Fru (C), and Suc (D) in leaf.
(E) to (G) Accumulation of Glc (E), Fru (F), and Suc (G) in internodes. Numbers on x axis denote DAA. Orange and dark green lines in all graphs represent B73 and PHG35, respectively. The error bars at each data point indicate se obtained from three biological replicates. One and two asterisks denote the significance of differences at P ≤ 0.05 and 0.01, respectively.
Since sugar status of the source and the sink tissues has a major role in senescence, we examined the accumulation of major nonstructural carbohydrates (NSCs) in leaf and internode tissues of B73 and PHG35 during post-flowering development. Up to 33 DAA, accumulation of hexoses (Glc and Fru) was higher in B73 leaves, but this trend reversed at 36 DAA with PHG35 accumulating more hexoses (Figures 1B and 1C). Accumulation of Suc showed an opposite trend, with higher accumulation in PHG35 up to 33 DAA, indicating active transport to sink (Figure 1D). The content of hexoses and Suc showed a sharp decline at 42 DAA, reflecting advanced senescence and death of the leaf tissue. The hexose-to-Suc ratio was higher in B73 during early stages but showed a reversed trend in PHG35 before the onset of senescence of this inbred line (Supplemental Figure 3). In the internodes, hexose and Suc concentration remained mostly unchanged in B73 except for a slight increase in Glc at 42 DAA (Figures 1E to 1G). PHG35 had lower amounts of hexose and Suc at 33 DAA, indicating remobilization of sugars to grain, but showed increased accumulation of all three sugars at later stages, eventually accumulating significantly higher Glc and Suc compared with B73. To summarize, the pattern of hexose accumulation in source tissue (i.e., leaf) is consistent with the onset of senescence in both inbreds. Glc and Fru hyperaccumulated in leaf of B73 before the observed decrease in Fv/Fm at 36 DAA, while such hyperaccumulation was later in PHG35, consistent with a drop in Fv/Fm at 42 DAA. Finally, PHG35 had higher soluble sugars in internodes at the end of the season compared with B73, indicating stronger alternative sink activity in the stay-green inbred.
Characterization of Transcriptome Underlying Natural Senescence
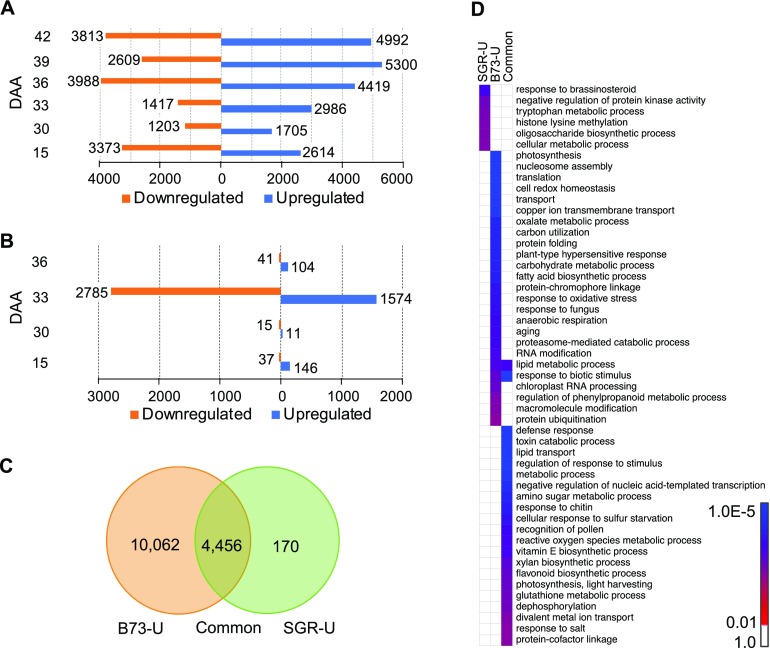
To understand the dynamics of the transcriptome during the onset and progression of senescence, we performed RNA-seq on the leaf at the ear-bearing node at key developmental stages selected based on the physiological and metabolic data (Sekhon et al., 2012). We focused on the normally senescing B73 inbred line and divided the leaf lifespan into the nonsenescent phase represented by 9 DAA and the senescent phase from 15 to 42 DAA (Supplemental Data Set 2). By comparing the expression at 9 DAA to each of the stages representing the senescence phase, we identified 14,518 (33.1%) genes showing differential expression at one or more stages during senescence (Supplemental Data Set 3). Distribution of up- and downregulated genes at different stages revealed that, except at 15 DAA, more genes were upregulated than down-regulated during senescence (Figure 2A). These DE genes were grouped into 22 clusters based on the kinetics of expression that included six clusters with 4005 (27.6%) upregulated genes, six clusters with 3043 (21.0%) downregulated genes, four clusters with 1346 (9.3%) genes upregulated during phenotypically visible senescence phase, and two clusters with 782 (5.37%) genes downregulated during phenotypically visible senescence phase (Supplemental Figure 4A). The other four clusters included remaining genes with other expression profiles.
Figure 2.
Transcriptome Dynamics Associated with Natural Senescence and Stay-Green Trait Deduced from Comparison of a Non-Stay-Green (B73) and a Stay-Green (PHG35) Inbred Line of Maize.
(A) Differentially expressed genes during the progression of natural senescence in B73.
(B) Differentially expressed genes underlying stay-green trait identified from comparison of B73 with PHG35.
(C) Genes unique to progression of natural senescence (B73_U), unique to stay-green (SGR_U), and common to both phenomena.
(D) Significantly overrepresented GOBPs for each of the categories described in (C).
Gene ontology (GO) enrichment analysis of the six upregulated and six downregulated clusters identified several were significantly enriched biological processes (GOBPs) and molecular functions (GOMFs; Supplemental Figure 4B). Notable upregulated GOBPs included autophagy, lipid metabolisms, carbohydrate metabolism, carbohydrate transport, and amino acid transport. Key downregulated GOBPs included chlorophyll biosynthesis chloroplast organization, multiple GOBPs related to photosynthesis and functioning of photosystem I (PSI) and PSII, and, surprisingly, abscisic acid biosynthesis. Notable upregulated GOMFs included amino acid transporters, kinases, glycosyltransferase activity, and transcription factor activity, while notable downregulated GOMFs included electron transport, chlorophyll binding, and carbon-nitrogen ligases. These results highlight the magnitude of functional transitions in the leaf during senescence and identify upregulated catabolic processes/activities and downregulated biogenesis processes.
Characterization of Transcriptome Underlying Stay-Green
To understand the regulation of stay-green, we compared the transcriptome of B73 (non-stay-green) and PHG35 (stay-green) at specific developmental stages relevant to the onset and progression of senescence. In total, 4713 (10.7%) of the genes were DE between these two genotypes at four selected stages (Figure 2B; Supplemental Data Set 4). The majority (70.7%) of the DE genes were observed at the 33 DAA stage, which precedes the appearance of visual symptoms of senescence in B73, but not in PHG35 (Figure 1A). Clustering of the DE genes based on expression kinetics produced one cluster (Supplemental Figure 5A), further supporting a synchronized transcriptional control of stay-green. Predominant enriched GOBPs included cell wall biogenesis, xylan biosynthesis, carbon fixation, regulation of autophagy, and lipid and amino acid metabolisms (Supplemental Figure 5B), while enriched GOMFs included abscisic acid binding, glycosyltransferase activity, carbohydrate and polysaccharide binding, and electron transport (Supplemental Figure 5C). Overall, this analysis suggests an important role for carbon fixation, transport, and incorporation of sugars into cell wall in the determination of stay-green trait in maize.
To understand the molecular activities specific to either progression of natural senescence or stay-green, and those shared between these two phenomena, we further examined the DE genes. Of 14,688 unique DE genes described in Supplemental Data Sets 3 and 4, 10,062 were unique to natural senescence, 170 were unique to stay-green, and 4456 were common (Figure 2C). Remarkably, response to brassinosteroids was one of the most significantly enriched GOBPs in stay-green set, and other notable terms included oligosaccharide synthesis, negative regulation of kinase activity, Trp metabolism, and histone Lys methylation (Figure 2D). The GOBPs enriched during natural senescence included photosynthesis, redox homeostasis, nucleosome assembly, carbon utilization, and aging. Finally, GOBPs enriched in the common group included defense response, lipid transport, glutathione metabolism, and xylan biosynthesis.
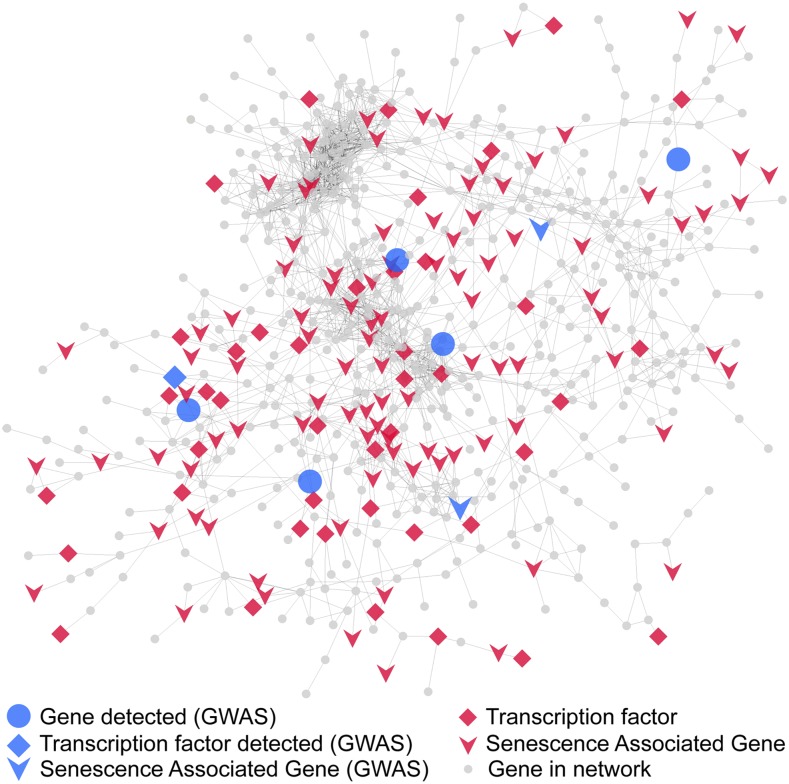
Coexpression network analysis of the 4713 DE genes underlying stay-green produced a predominant subnetwork with 663 unique genes (Figure 3; Supplemental Data Set 5). Highlighting a major role for transcriptional regulation in senescence, 45 genes (6.8%) in this subnetwork are putative transcription factors, with a majority (8 genes each) representing NAC and MADS families. Putative Arabidopsis orthologs of 109 of the genes (16.4%) in this subnetwork are DE during senescence and designated as SAGs (Li et al., 2017).
Figure 3.
A Major Coexpression Subnetwork Underlying Stay-Green.
Coexpression analysis of transcriptome deduced from comparison of B73 and PHG35 inbred lines produced a major subnetwork (n = 663 genes). Diamonds represent transcription factors, inverted arrows denote putative maize orthologs of SAGs in Arabidopsis, and the circles represent the rest of the annotated genes. Blue shapes are the genes that were also independently detected by GWAS.
Identification of Candidate Genes and Networks
The data generated from this study and previously published data on the mechanisms underlying senescence allowed us to implement a SysGen framework for discovery and prioritization of candidate genes. We used multiple criteria: (1) location of a SNP identified in the GWAS analysis, (2) annotated genes within a 100-kb window on each side of the SNP (200-kb window total), (3) relative expression of the 200-kb window genes in B73 and PHG35 inbred lines, (4) presence of a candidate gene in the stay-green coexpression network, and (5) deduced role of a candidate gene in senescence. For criterion 5, annotation of the 200-kb window genes, if available, was used as a term to search the literature and, based on role of a gene or the gene family in literature and/or the predicted orthology to SAGs in Arabidopsis (Li et al., 2014, 2017), the gene was considered to have a possible role in senescence. In the absence of any evidence based on criteria 3 to 5, the gene closest to the identified GWAS SNP was reported.
Using this strategy, the 62 unique GWAS associations identified in the study were first linked to 64 candidate genes (Table 1; Supplemental Data Set 1). Two SNPs, one each on chromosome 2 and chromosome 6, each had two candidate genes, one based on putative role in senescence based on the literature archive and the other indicated by the presence in the coexpression network. For SNP number 11 (Table 1), a Cys protease encoded by maize insect resistance3 (mir3) was considered a candidate due to reported role of these proteases in senescence in other species (Velasco-Arroyo et al., 2016; Pružinská et al., 2017). Likewise, for SNP number 43a, ZmSWEET1b was deemed a candidate based on reported role a SUGARS WILL EVENTUALLY BE EXPORTED TRANSPORTERS (SWEET) gene in senescence (Zhou et al., 2014).
Of the 64 candidate genes, 48 (75%) are DE during progression of senescence in B73, or in the comparison of B73 with PHG35 and therefore underlie stay-green, or during both processes (Supplemental Figure 6). Thirty (46.9%) of the candidate genes have been annotated as SAGs in Arabidopsis (Li et al., 2014, 2017), indicating both a substantial overlap and unique mechanisms used by cereals during senescence. Remarkably, 14 (21.9%) of the candidate genes are supported by more than one form of evidence and thus provide a very high-quality set of targets for further investigations (Figure 4; Table 1). Notable candidates in this set include a trehalose-6-phosphate (T6P) synthase (trps13), a NAC transcription factor (nactf9), an abscisic acid–responsive Group 3 late embryogenesis abundant protein (mlg3), a Cys protease (mir3), a β-glucosidase (ZmBGLU42), two cell wall–related genes (ZmIRX15-L and ZmGUX1), two sugar transporters (ZmSWEET1b and ZmMST4), a cell wall invertase (incw4), and a mitochondrial protein (dek10). Of these 14 candidates, 8 genes represent processes or functions relevant to senescence but lack conclusive experimental support, and the rest of the genes are novel. Eight genes of the 14 high-quality set are supported both by GWAS and coexpression network analysis and thus represent a core set of candidates (Figure 4).
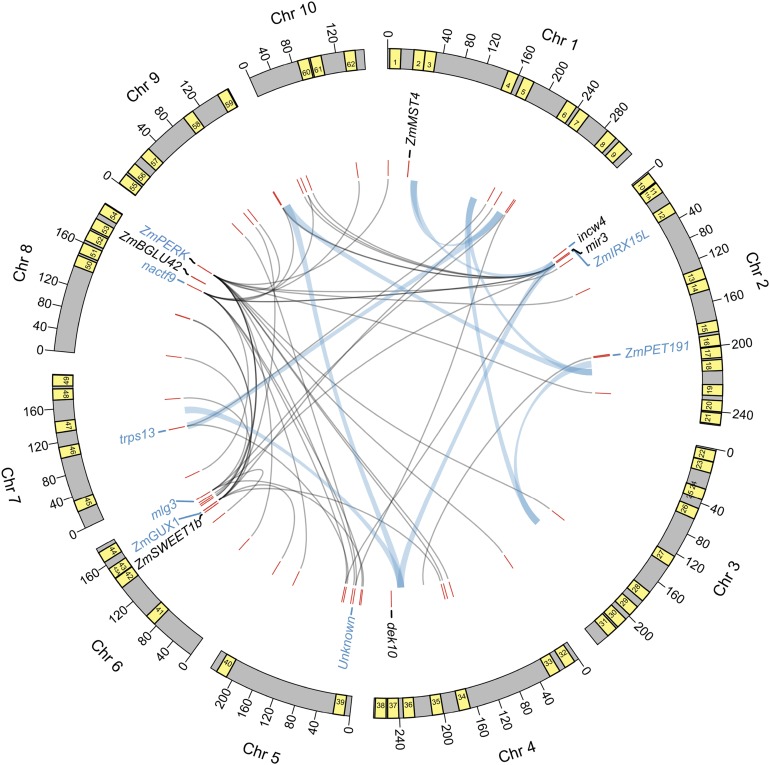
Figure 4.
Systems Genetic Analysis of Maize Leaf Senescence by Combining GWAS, Coexpression Networks, Pairwise Epistatic Interactions, and Prior Information.
Outer arcs denote 10 maize chromosomes along with 200-kb window around 62 GWAS SNPs shown as yellow boxes. Numbers in the boxes correspond to the serial number of the SNPs and candidate genes listed in Table 1. Selected candidate genes lying within the windows are shown underneath the arcs. Genes in blue font indicate those detected both by GWAS and coexpression network analysis (indicated by bold italicized font in Table 1) and in black font represent those detected only by GWAS but have a perceived role in senescence. Fine gray lines represent coexpression network connections of candidate genes with other genes in the genome. Thick light blue lines denote epistatic interaction between pairs of GWAS SNPs. Chr, chromosome.
The coexpression network and epistasis analysis revealed several putative interactions among the candidate genes and regions (Figure 4). One of the candidate SNPs (no. 36 in Table 1) was epistatic to three GWAS SNPs, while five others (nos. 3, 5, 18, 29, and 59) had two epistatic interactions, which suggests genetic interaction among the candidate regions or genes identified by GWAS. For instance, SNP number 5 harbors a candidate FLOWERING LOCUS K gene known to control flowering via suppression of FLOWERING LOCUS C transcription in Arabidopsis (Ripoll et al., 2009) and since FLOWERING LOCUS C also regulates senescence (Wingler, 2011), FLOWERING LOCUS K is expected to have a similar role. Interestingly, SNP number 5 is epistatic to SNP number 18 that lies near a candidate gene encoding for molybdenum cofactor sulfurase implicated in the regulation of senescence by controlling synthesis of xanthine dehydrogenase (Brychkova et al., 2008). Thus, while both candidates likely have a direct role in senescence independent of each other, our analysis suggests that these genes may be acting together. Likewise, several interactions were identified by coexpression networks underlying the stay-green trait (see “Discussion”). To summarize, a SysGen framework combining multiple pieces of evidence provides several highly valuable targets for future investigations to generate a novel mechanistic understanding of senescence and stay-green in maize and other cereals.
Functional Validation of Candidate Genes
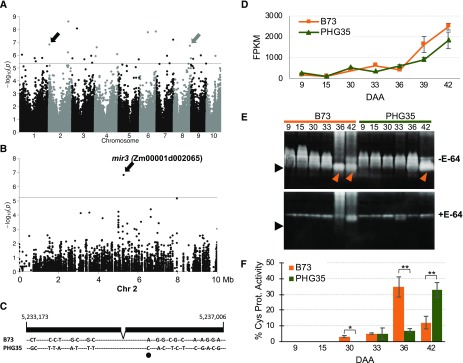
To examine the role of mir3 (Zm00001d002065), associated with a highly significant SNP (no. 11) on chromosome 2 (Figures 5A and 5B), we compared natural alleles of this gene in B73 and PHG35 that are polymorphic at 20 nucleotide positions throughout the coding region (Figure 5C). Five of these SNPs have a moderate-to-high predicted effect and one SNP has high predicted impact as this SNP creates a putative splice acceptor site (Figure 5C; Supplemental Table 2). For both inbred lines, expression of mir3 increased at 30 DAA, and B73 had higher transcript accumulation compared with PHG35 for most later developmental stages (Figure 5D). Cys protease activity of Mir3, which has a predicted molecular mass of 37.2 kD upon maturity and removal of signal peptide (Pechan et al., 1999), increased dramatically in B73 at 36 DAA as indicated by presence of an intense band of expected size in the presence of gelatin loss of the band upon addition of E-64 Cys protease inhibitor (Figure 5E). Importantly, the protease activity in PHG35 was delayed by 6 d as evident from significantly lower activity at 36 DAA but dramatic increase afterward such that the activity in PHG35 at 42 DAA was same as that in B73 at 36 DAA (Figures 5E and 5F). Overall, these data suggest that delayed Cys protease activity contributes to prolonged photosynthetic activity and stay-green in PHG35. Besides Mir3, several other Cys proteases are active in both inbred lines at multiple developmental stages as indicated by the presence of larger bands and disappearance of those bands upon addition of the inhibitor. A similar variety of Cys proteases have also been reported in senescing wheat leaves wherein proteases of 36 to 46 kD were associated with leaf senescence (Martínez et al., 2007). These data indicate that the onset of senescence is associated with natural allelic variation of mir3 and Cys protease activity in the leaves. Further studies will confirm the identity of Mir3 as the causal Cys protease in maize leaf senescence and stay-green.
Figure 5.
Functional Analysis of Natural Alleles of a Cys Protease Encoded by mir3.
(A) Manhattan plot of GWAS for natural senescence. The horizontal line indicates the suggested threshold (P ≤ 5.45E−6), and black and gray arrows indicate a Cys protease and a β-glucosidase gene discussed in the text, respectively.
(B) Regional association plot showing SNP used to identify mir3. Chr, chromosome.
(C) Sequence polymorphisms in alleles of mir3 in B73 and PHG35 inbred lines. One polymorphism with high predicted impact due to the generation of a putative splice acceptor site is marked by a black oval.
(D) Expression of mir3 in B73 and PHG35. Error bars are the se based on three biological replicates.
(E) Cys protease activity in leaves of B73 and PHG35 during the onset and progression of senescence. The protease activity was assayed in gels containing gelatin as a substrate in the absence (top) and presence (bottom) of E-64 protease inhibitor. The expected Mir3 band is marked by an arrow.
(F) Relative protease activity quantified based on signal intensity. Error bars are se obtained from biological replicates. One and two asterisks denote the significance of differences at P ≤ 0.05 and 0.01, respectively.
To further investigate the role of mir3, we annotated C1A papain-like Cys proteases (PLCPs) in four plant species based on the presence of the characteristic papain domain (Beers et al., 2004; van der Hoorn et al., 2004). Genome-wide search identified 63, 48, 36, and 30 PLCPs in maize, sorghum, Arabidopsis, and rice, respectively. Phylogenetic analysis of these protein sequences revealed that the maize Mir3 formed a close subgroup with Arabidopsis RD21A (AT1G47128), indicating a close evolutionary relationship between these two proteins (Supplemental Figure 7). Sequence analysis of the10 PLCPs in this subgroup revealed that RD21A, which encodes for a Cys protease (Koizumi et al., 1993), shares 66% amino acid sequence identity with maize Mir3 for complete protein sequence and 84% sequence identity for the papain domain (Supplemental Figure 8).
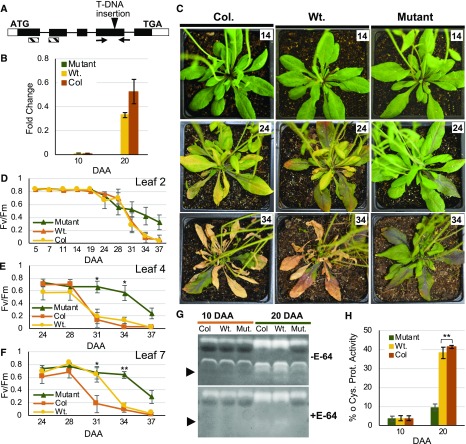
Based on this predicted orthology, we performed functional analysis of RD21A in Arabidopsis. A T-DNA insertion resulted in a complete loss of RD21A transcripts in Arabidopsis leaves (Figures 6A and 6B; Supplemental Figure 9). The mutant plants showed delayed onset of senescence at 24 DAA as evident from yellowing of the leaves in the wild-type Columbia (Col), and in the wild-type segregants derived from plants heterozygous for the insertional allele (Figure 6C). The Col and wild-type plants showed significantly advanced senescence in second and fourth rosette leaves at 28 DAA, in the seventh rosette leaf at 31 DAA, and complete senescence in all three leaves at 37 DAA (Figures 6D to 6F). However, the mutant plants maintained significantly higher photosynthetic activity at 37 DAA albeit at a lower level compared with earlier developmental stages, indicating delayed onset and a slower rate of senescence. The genotypic, transcriptional, and phenotypic characterization of RD21A was further confirmed with Cys protease zymography. The RD21A encoded protease, which has a predicted molecular mass of 33 kD upon maturity and removal of the signal peptide (Pružinská et al., 2017), was undetected in Col, the wild type, and the mutant plants at 10 DAA (Figures 6G and 6H). However, while RD21A abundance increased dramatically in the Col and wild-type plants, the protein was barely detectable in the mutants. Overall, these data further support the role of the mir3 ortholog in stay-green.
Figure 6.
T-DNA Insertional Mutation in the Putative mir3-Ortholog Leads to Loss of Protease Activity and Delayed Onset of Senescence in Arabidopsis.
(A) Gene structure of Arabidopsis Cys protease AT1G47128 (RD21A), showing exons (boxes), untranslated regions (open boxes), translated regions (dark shaded boxes), introns (lines connecting boxes), location of T-DNA insertion (vertical arrowhead), primers used for genotyping (horizontal arrows), and conserved peptidase C1A (papain) domain (hatched boxes).
(B) RT-qPCR data showing expression of RD21A in the mutant, wild-type segregants from the initial, heterozygous mutant in Col background (Wt.), and Col plants at two developmental stages in the combined tissue of leaf 4 and leaf 7. Error bars are se obtained from three biological replicates.
(C) Phenotypes of representative Col, wild-type segregant (Wt.), and mutant plants. Numbers in the white box in represent DAA.
(D) to (F) Fv/Fm of three individual rosette leaves with the number representing the location of the leaf in the rosette.
(G) Comparison of Cys protease activity in same tissue and genotypes used for RT-qPCR. The protease activity was assayed in gels containing gelatin as a substrate in the absence (top) and presence (bottom) of E-64 protease inhibitor with the expected RD21A band marked by an arrow.
(H) Relative protease activity quantified based on signal intensity. Error bars are se obtained from three biological replicates. One and two asterisks denote the significance of differences between and mutant and wild-type segregant (Wt.) at P ≤ 0.05 and 0.01, respectively.
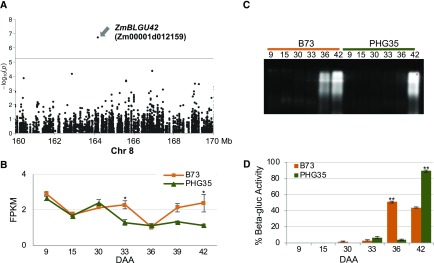
A highly significant SNP (no. 53) on chromosome 8 lies in gene ZmBLGU42 (Zm00001d012159; Figure 7A) which putatively encodes for a β-glucosidase orthologous to rice Os1bglu4 (Rouyi et al., 2014). We found that, compared with PHG35, transcript levels of ZmBLGU42 were higher in B73 at 33, 39, and 42 DAA but identical in both inbred lines at 36 DAA (Figure 7B). While the expression data were not conclusive, the assay of β-glucosidase activity in leaves of these inbred lines in zymograms by staining with 4-methylumbelliferyl-β-d-glucuronide revealed differential activity displayed by the B73 and PHG35 alleles. The onset of activity was earlier in B73 at 36 DAA, while PHG35 showed the activity at 42 DAA (Figures 7C and 7D). These data suggest that differential posttranscriptional activity of ZmBLGU42 alleles underlies differences in senescence phenotype.
Figure 7.
Functional Analysis of Natural Alleles of a GWAS Candidate Encoding for a β-Glucosidase.
(A) Regional association plot showing SNP used to identify the β-glucosidase gene ZmBLGU42 (Zm00001d012159).
(B) Expression of ZmBLGU42 in B73 and PHG35 based on RNA-seq data. Error bars are se obtained from biological replicates.
(C) Comparison of β-glucosidase activity in leaves of B73 and PHG35 at post-flowering stages covering the onset and progression of senescence. The β-glucosidase activity was assayed by staining with 4-methylumbelliferyl-β-d-glucuronide.
(D) Relative β-glucosidase activity quantified based on signal intensity. Error bars are se obtained from three biological replicates. One and two asterisks denote the significance of differences at P ≤ 0.05 and 0.01, respectively.
DISCUSSION
Through systematic genomic and transcriptomic analyses, implementation of SysGen framework, and experimental validations, we provide a broad model of senescence control and progression in a global staple crop. Sixty-two unique associations identified by the characterization of natural variation in a diversity panel of U.S. dent maize were linked to 64 candidate genes. These genes implicate diverse processes in senescence including proteolysis (a Cys protease), sugar signaling (a T6P synthase), sugar transport (two sugar transporters), unloading of sugars to sink (a cell wall invertase), and, remarkably, synthesis of cell wall polysaccharides. Most of the genes reported in this study are novel candidates that have not been experimentally validated despite their logical connection with senescence based on published literature. Half of the candidate genes have been designated as SAGs in Arabidopsis (Li et al., 2014, 2017), thus highlighting both mechanistic similarities and differences in monocot and eudicot senescence.
A small number of previous inheritance studies on maize senescence used biparental populations and identified valuable quantitative trait loci underlying the stay-green trait (Zheng et al., 2009; Wang et al., 2012; Belícuas et al., 2014). Our study expands this knowledge by evaluating the role of a larger number of alleles captured in a diversity panel encompassing a substantial portion of the genetic variation in U.S. midwestern dent maize.
The previous phenotyping has relied on leaf greenness produced by chlorophyll through green leaf area (Zheng et al., 2009; Wang et al., 2012), visual scoring of leaf greenness (Belícuas et al., 2014), or chlorophyll content (Trachsel et al., 2016). However, greenness can indicate either functional stay-green plants that maintain high levels of photosynthetic activity or visual stay-green plant that maintain high chlorophyll, but not the photosynthetic activity (Thomas and Howarth, 2000). For estimation of functional stay-green, we recorded maximum quantum efficiency of PSII photochemistry (Fv/Fm) in dark-adapted, field-grown plants. Aging and high chlorophyll content coupled with lower CO2 assimilation could cause oxidative damage to the leaf cells, and plants prevent such damage by systematic dismantling of photosynthetic apparatus and by nonphotochemical quenching (NPQ; Lu and Zhang, 1998; Wingler et al., 2004). The PSII centers are dismantled before PSI centers, resulting in a greater decrease in PSII activity in senescing leaves, while NPQ levels increase during this period (Ghosh et al., 2001; Wingler et al., 2004). Thus, NPQ, PSII activity of non-dark-adapted leaves (Fv′/Fm′), and PSII activity of dark-adapted leaves (Fv/Fm) are all reasonable parameters for measuring photosynthetic activity. Our approach to record Fv/Fm of dark-adapted plants, a widely used parameter to access photosynthetic activity and senescence (Maxwell and Johnson, 2000; Wingler et al., 2004, 2010; Bresson et al., 2018), provides a better estimation of photosynthetic activity and therefore senescence.
Three spatially and temporally distinct environments allowed us to capture a significant portion of the genetic architecture of senescence that may otherwise go unnoticed. Various environmental factors including heat, drought, nutrient status, and shading influence senescence (Woo et al., 2013; Schippers, 2015), and therefore lead to significant phenotypic variation across environments. Biotic stresses, mainly the prevalence of diseases that may also be accompanied by hypersensitive response and death of leaf tissue, may also affect senescence in a given environment. Variation in heritability across environments reflects the role of environment in regulation on senescence. Similar variation in heritability of Fv/Fm was observed in a rice diversity panel with the estimates in two distinct environments being 12% and 34% (Qu et al., 2017). Dramatic fluctuations in heritability across environment have been reported for various quantitative traits in maize (Asaro et al., 2016).
High levels of biological activity during the onset and progression of senescence are evident from a massive change in transcriptional activity in one-third of the annotated maize genes. By contrast, a much smaller fraction of genes (9.2%) underlie stay-green, indicating fewer and focused processes. The majority (92.4%) of these genes were DE at 33 DAA, indicating a swift and massive transcriptome changes that differentiate a stay-green and a normally senescing phenotype. Overrepresentation of transcripts related to cell wall biogenesis and, in particular, hemicellulose synthesis in the stay-green transcriptome indicates a role of plant cell walls as an alternative sink for storage of excess carbohydrates. Remarkably, identification of genes that are unique to the B73-PHG35 comparison, and hence likely determine presence or absence of stay-green in these two inbred lines, produced an even smaller number (170) of genes.
Response to brassinosteroids is a prominent GOBP enriched in this set, bringing a known but mechanistically less understood regulator of senescence to light. Impairment of brassinosteroids biosynthesis, as observed in det2 mutant, or signaling, as seen in bri1 and bes1 mutants, results in stay-green phenotype in Arabidopsis (Chory et al., 1994; Clouse, 1996; Yin et al., 2002). Interestingly, a putative maize ortholog of bri1 was also identified in our GWAS study (Table 1), providing additional support to the role of brassinosteroids in stay-green.
The role of epigenetic mechanisms, specifically histone Lys methylation, in stay-green is another important finding of this analysis. In Arabidopsis, histone demethylase activity of a JmjC domain-containing protein JMJ16 is responsible for suppression of age-dependent senescence (Liu et al., 2019). Coexpression network analysis of stay-green also provided some very intriguing insights. Substantial representation of transcription factors (6.8%) in the subnetwork demonstrates a major role of transcriptional regulation, particularly the NAC and MADS families, in specifying stay-green. Besides the genes that appeared in the SysGen analysis, this analysis provided other interesting candidates. For instance, a sorghum NAC ortholog of a coexpression subnetwork member nactf36 is a key determinant of pithy or juicy stems (Zhang et al., 2018) and hence likely plays a vital role in determining the sink strength and therefore senescence. These findings provide some very interesting and promising lines of investigation to be pursued in the future.
Identification of mir3, a putative Cys protease (Pechan et al., 1999) in our screen, highlights the role of proteolysis in senescence. Proteases are required for hydrolysis of ribulose-1,5-bisphosphate carboxylase/oxygenase and other chloroplastic proteins for recycling nitrogen and other nutrients (Guo et al., 2004; Breeze et al., 2011). A large number of proteases existing in plants respond to specific senescence-triggering factors (Martínez et al., 2007), and based on the activity of two natural alleles (B73 and PHG35) in zymograms, mir3 appears to have a key role in stay-green. T-DNA insertional knockout of mir3 ortholog RD21A in Arabidopsis also resulted in a modest, albeit significant, delay in leaf senescence in our study and another recent study (Pružinská et al., 2017). Suppression of a Cys protease HvPap-1 in barley also delayed abiotic stress-induced senescence, thus underscoring the importance of these proteases in exacerbating biotic stresses faced by crop plants (Velasco-Arroyo et al., 2016). Despite the well-established role of proteolysis in senescence, the extent to which proteases play a role in stay-green is poorly understood and, if anything, suppression of proteolytic activities is generally believed to accelerate senescence (Roberts et al., 2012; Thomas and Ougham, 2014). Could the observation that certain natural alleles of mir3 condition stay-green without any obvious adverse effects on plants fitness be due to the precise localization of this protein in the chloroplast? Future experiments involving Mir3-specific antibodies for identification of Mir3 targets and interacting partners, and transgenics in maize and other plant species will confirm the role of this protease in senescence and stay-green.
Key insights into the role of ZmBLGU42, a β-glucosidase orthologous to Arabidopsis BGLU42 (Zamioudis et al., 2014) and rice Os1bglu4 (Rouyi et al., 2014), in senescence comes from the early onset of glucosidase activity in the B73 compared with PHG35. The β-glucosidase activity also increases during Arabidopsis leaf senescence (Patro et al., 2014), although the exact role of this process remains to be determined. β-Glucosidase hydrolyzes cell wall oligosaccharides to release Glc and, during sugar starvation of senescing leaf cells, maintains energy homeostasis in these cells (Patro et al., 2014). Since cell walls remain intact until the late stages of senescence, hexoses generated by β-glucosidase may be important for energy-dependent senescence processes (Patro et al., 2014). Since hexoses act as signal molecules through the hexokinase1 (HXK1) pathway to control senescence (Nuccio et al., 2015; Wingler, 2018), β-glucosidase may have a role in sugar signaling. Indeed, leaf hexoses were increased in B73 compared with PHG35, while Suc levels remained stable, resulting in a higher hexose:Suc ratio during senescence, and this trend was delayed in PHG35 (Figure 1). Hexose levels also rise dramatically in senescing Arabidopsis leaves, while Suc levels remain stable (Quirino et al., 2001; Wingler et al., 2006). Plants have separate sensors for Suc and hexoses and any changes in Suc:hexose ratio initiate distinct signal transduction, leading to specific changes in transcription (Smeekens, 2000). Since the senescing leaves will produce less photoassimilate, the rise in hexose levels in senescing leaves instead of a decrease is inexplicable (Wingler and Roitsch, 2008). Cell wall hydrolysis by ZmBLGU42 encoded β-glucosidase may be at least one of the mechanism responsible for generating these hexoses.
A loss of carbon demand from the primary sink upon grain fill combined with the absence of a secondary sink(s) will also lead to accumulation of hexose in leaf cells as seen B73 compared with PHG35 at ∼33 DAA. Hexoses act as positive regulators of the hexose sensor HXK1, which promotes senescence either by suppressing cytokinin signaling or through the conversion of Glc to Glc 6-phosphate, which then inhibits Suc nonfermenting-1-related kinase1 (SnRK1), a suppressor of senescence (Zhang et al., 2009; Smeekens et al., 2010; Wingler, 2018). Our study supports a direct role of sugar-mediated signaling in stay-green by identification of trps13 that synthesizes T6P, a Glc disaccharide, from Glc 6-phosphate and UDP-Glc. T6P is a negative regulator of SnRK1 and required for the onset of senescence associated with high sugar accumulation in leaves (Wingler et al., 2012). T6P is crucial for maintaining optimum Suc levels in plants by controlling starch breakdown at the source in response to the sink demand, and by regulating Suc consumption and growth at the sink through SnRK1 and other unknown mechanisms (Figueroa and Lunn, 2016). Attenuation of T6P by expressing a rice T6P phosphatase in maize resulted in prolonged and higher photosynthetic activity (Oszvald et al., 2018). T6P is an important target for biotechnological manipulations to enhance crop productivity and stress tolerance, and our data linking this molecule to stay-green further highlight the need for future investigations to fully understand such roles.
Given the role of sugars in senescence signaling, transporting these sugars from leaf cells to an alternative sink, evident from higher sugars in PHG35 internodes, should weaken such signaling. Identification of two sugar transporters (ZmSWEET1b and ZmMST), a cell wall invertase (incw4), and a mitochondrial pentatricopeptide repeat protein (dek10) further supports a key role for sugar partitioning in senescence. Disruption of Suc transport from leaves often leads to senescence-like symptoms in maize as seen in sugar transport mutants tdy1, tdy2, and sut1 (Baker and Braun, 2007; Slewinski et al., 2009, 2012). Arabidopsis hexose transporter STP13, a putative ortholog of ZmMST4, is associated with senescence and nitrogen use efficiency (Nørholm et al., 2006; Schofield et al., 2009), and putative rice ortholog, OsMST4, transports hexoses generated by cell wall invertases (Wang et al., 2007).
Extracellular invertases hydrolyze and remove Suc from the vascular system into a sink tissue and therefore regulate senescence by determining sink strength, source–sink translocation of sugars, and intracellular hexose:Suc ratio (Balibrea Lara et al., 2004; Rolland et al., 2006; Ruan et al., 2010). Enhanced expression of a cell wall invertase encoded by incw2 (also know as miniature1) resulted in higher starch accumulation and grain yield in maize (Li et al., 2013a). While incw2 is expressed in the basal endosperm transfer layer connecting the maternal and the endosperm tissue, incw4 is expressed in both vegetative and reproductive organs (Kim et al., 2000), thus making this invertase a good candidate for unloading sugars to a vegetative sink. A coordinated induction of an extracellular invertase and a hexose transporter by cytokinin treatment in cell culture has been suggested to enhance the uptake of sugars by a sink (Ehness and Roitsch, 1997). dek10, a ubiquitously expressed protein in maize plants, controls sugar uptake by the grain sink by orchestrating the development of the basal endosperm transfer layer (Qi et al., 2017). Our study suggests a key role of the cell wall invertases, hexose transporters, cytokinin, and Dek10 nexus in regulating the sink strength of internode parenchyma or other vegetative tissues and thus enhancing the overall sink strength of plants, and further studies will explore such a role. Interestingly, dek10 also affects nucleosome assembly during senescence (Gal et al., 2015), thus warranting investigations into the role of epigenetic mechanisms in stay-green.
Support for a model that transport and storage of sugars in an alternative sink contribute to stay-green comes from the discovery of two hemicellulose synthesis genes (ZmIRX15-L and ZmGUX1) and a NAC transcription factor (nactf9) in our study. Putative Arabidopsis orthologs of ZmIRX15-L and ZmGUX1 are involved in the synthesis of the major hemicellulose xylan in the primary and secondary cell walls (Jensen et al., 2011; Bromley et al., 2013). Remarkably, Arabidopsis IRX15 (but not IRX15-L) has been shown to be coexpressed with a NAC transcription factor SECONDARY WALL-ASSOCIATED NAC DOMAIN2 that regulates genes involved in cellulose and hemicellulose synthesis (Hussey et al., 2011). The NAC transcription factors can also enhance or suppress senescence by regulating the synthesis of abscisic acid, an important regulator of senescence, and also contribute to transition from carbon-capture to the nitrogen-recycling phase (Liang et al., 2014; Thomas and Ougham, 2014). Such a role is also supported by GWAS candidate mlg3, a late embryogenesis abundant protein, which is regulated by abscisic acid (White and Rivin, 1995). Therefore, nactf9 appears to be one of the master regulators of stay-green, acting in conjunction with ZmIRX15-L, ZmGUX1, mlg3, and other candidate genes identified in our study, and future work will explore such a role.
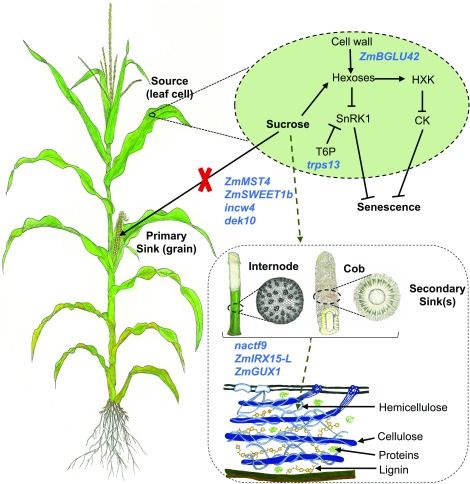
While both GWAS and transcriptome analyses produce false positives, our SysGen approach, which relies on combining GWAS, coexpression networks, and prior body of knowledge on senescence, provided a set of high-confidence candidate genes underlying stay-green and senescence (Figure 4). The SysGen framework identified source–sink communication as one of the key mechanisms underlying stay-green trait as 9 of the 14 high-confidence candidate genes play an important putative role in sugar transport and/or signaling (Figure 8). Cell wall appears to have an important role in senescence both as a source of sugars that can act in signaling at the source (leaf) and as a sink for excess sugars accumulating due to inactivity of the primary sink (in internode and other similar organs). In such a model, excess hexoses accumulating in the leaf cell due to loss of primary sink after completion of gain fill and the breakdown of cell wall polysaccharides by β-glucosidase (ZmBLU42), signal senescence through hexokinase (HXK1), and T6P (trps13).
Figure 8.
A Model for Regulation of Senescence by Source–Sink Communication.
Nine candidate genes (shown in blue font) identified from system genetic analysis are involved in sugar transport, signaling, and storage and thus likely regulate senescence. Suc generated via photosynthesis during the reproductive phase of the maize plant is destined for the primary sink (grain). Upon loss of sugar demand by the primary sink after completion of grain fill (indicated by a red cross mark), some of the candidate genes likely play role in transporting sugars to an alternative sink exemplified by stalk parenchyma or cob pith cells, while others are involved in the deposition of sugars into the secondary cell wall. Other candidate genes (shown within the light green oval) are likely important for regulation of senescence via sugar-mediated signaling. CK, cytokinin.
In stay-green genotypes, senescence signaling is attenuated by transport of Suc out of leaf cells by sugar transporters (ZmMST4, ZmSWEET1b) to alternative sinks. These sinks are activated by dek10, hydrolyzed by cell wall invertase (incw4), and incorporated as cell wall polysaccharides by the activity of a NAC (nactf9) and xylan synthesis genes ZmIRX15-L and ZmGUX1. Effective partitioning of sugars to alternative sinks achieved by the combined activity of these genes therefore is a major determinant of stay-green. The most significant finding of this study is however the novel candidate genes not placed in this model due to the lack of any experimental evidence. Further examination of the role of these genes in determining the sink strength and, in particular, the strength of alternative sink to allow diversion of photosynthate after grain fill will be valuable toward engineering stay-green genotypes with higher net energy content.
With ever-worsening climate for agriculture and decreasing availability of freshwater, the stay-green trait has proven to be effective in improving drought resistance. Another less explored but equally important advantage of stay-green is increased overall carbon yield in the form of sugars accumulating in multiple sinks. Extra carbon captured in stay-green could be diverted to stover for developing dual-purpose (grain/biomass) crops for food and biofuel and/or as animal feedstock. However, scant data are available on the molecular mechanisms that would enable such a diversion, and this study provides promising candidates. In certain agricultural production systems, stay-green has also been found to be an undesirable trait due to, for instance, high moisture in stover and grain (delayed dry down) at harvesting in stay-green maize hybrids grown in the midwestern United States. Identification of the genetic determinants of senescence holds the key to interventions such as genetically uncoupling stay-green from higher grain/stover moisture. Deeper insights into sources–sink aspects of senescence are also needed for understanding the determinants of annuality and perenniality of cereals. The detailed genetic architecture of senescence holds a great promise for both applied and fundamental aspects of senescence research. With an appreciable amount of conserved gene and genome functionality among cereals, the information presented here will potentially translate to other important grasses including sorghum and rice.
METHODS
Plant Materials and Phenotypic Data Collection on the Diversity Panel
A maize (Zea mays) diversity panel (Hirsch et al., 2014) was grown in three environments for phenotypic data collection on senescence (Supplemental Data Set 6). In summers 2011 and 2013, the panel was grown at the University of Wisconsin West Madison Agricultural Research Station in Verona and at the University of Wisconsin Arlington Research Station in Arlington, respectively. In summer 2017, the panel was grown at Clemson University Simpson Research Farm in Pendleton, South Carolina. At each location, the experiment consisted of two replications planted in a randomized complete block design. In each one-row plot, five to seven competitive plants were open pollinated and used for phenotypic data.
To record senescence at a uniform developmental stage, senescence data were recorded at 36 d after 50% of plants in a plot showed silk (elongated style) emergence. Judging by green color, chlorophyll content, and sugar accumulation in the plant, maize genotypes generally start to show the onset of senescence ∼30 DAA (Swank et al., 1982; Crafts-Brandner et al., 1984); thus, we recorded data at 36 DAA to ensure that a large portion of the variation is captured.
The maximum quantum efficiency of PSII photochemistry (Fv/Fm) was used as an indicator of senescence (Wingler et al., 2004, 2010). To ensure that the photosynthetic reaction centers are fully oxidized and available for photochemistry, data were recorded after allowing dark adaption for 2 h past sunset. Fv/Fm was recorded with a Pocket PEA fluorimeter (Hansatech Instruments) by exposing the leaves to a light intensity of 3500 µmol m−2 s−1 for 1 s. Data were recorded two nodes above the ear-bearing node on the leaf blade at the midpoint between the midrib and the leaf edge ∼12 inches from the collar (joint of leaf sheath and blade).
GWAS Analysis
Best linear unbiased predictions of the phenotypic values were calculated for each genotype in the individual environments (2011, 2013, and 2017) using the mixed linear model capability of JMP Pro v13.2.0 (SAS Institute). Best linear unbiased predictions were also calculated for the 2011 and 2013 environments combined since those were grown in similar geographical locations. The environments are named W11, W13, W11_13, and S17. GWAS was performed for all these environments using mixed linear model (Yu et al., 2006) implemented in Genome Association and Prediction Integrated Tool (Lipka et al., 2012).
A minor allele frequency cutoff of 0.01 was used. In total, 438,161 polymorphic markers were used for the GWAS analyses; however, since dense marker panels likely exhibit high levels of linkage disequilibrium, the effective number of markers for significance testing was determined by Genetic Type I Error Calculator (Li et al., 2012). This analysis showed that the effective number of markers is 183,449.
Three significance thresholds were calculated according to a Bonferroni correction based on the effective number of markers. Based on Lander and Kruglyak (1995), we classified SNPs according to three P-value thresholds. The three thresholds, referred to as suggestive, significant, and highly significant, are associated with a false positive result being expected to occur 1.0, 0.05, and 0.01 times, respectively, in a genome scan. The reason for the three thresholds is to attempt to control for both Type I (false positives) and Type II (false negatives) errors. The suggestive threshold allows an increased overall chance of false positives but greatly reduces the overall chance of false negatives, thereby reducing the chance of completely missing an important region. We follow the recommendation (Lander and Kruglyak, 1995) that markers with a P-value less than the suggestive threshold are only speculative and require further study. The highly significant threshold allows an increased overall chance of false negatives but greatly reduces the overall chance of false positives. The significant threshold attempts to balance both false positives and false negatives. The threshold P-values for this study were 5.45E−6 for suggestive, 2.73E−7 for significant, and 5.45E−9 for highly significant.
The SNP set used for GWAS is based on maize RefGen_v2, and the coordinates of the significant SNPs reported here were converted to RefGen_v4 using CrossMap v0.2.7 (Zhao et al., 2014). All SNP variants in the diversity panel were evaluated for their potential effect on gene function using snpEff (Cingolani et al., 2012).
Tissue and Phenotypic Data Collection for Transcriptomic Experiments
For transcriptomic analyses, inbred lines B73 and PHG35 lines were grown in a randomized complete block design with three replications (i.e., three blocks) at Clemson University Simpson Research Farm in Pendleton in summer 2017. In each block, a six-row plot of each inbred line was grown with row length and row-to-row distance being 4.57 and 0.762 m, respectively.
In each six-row plot, the primary and all subsequent ears of all plants were covered with shoot bags before silk emergence. Upon silk emergence in at least 80% of the plants in a plot, shoot bags were removed on the same day to allow open pollination. Supplemental hand pollinations were performed the same day to ensure maximum seed set, and this day was designated as 0 DAA. Only those plants that showed silk emergence at 0 DAA were included in the experiment.
Two tissues, leaf and internode, were collected at 3-d intervals from 0 to 45 DAA. At each stage and for each inbred line, three biological replicates of each tissue were collected. A biological replicate consisted of pooled tissue from two different, competitive (nonborder and well-spaced from neighbors) plants harvested from the same block and dissected to separate the leaf and internode tissue. The leaf samples consisted of a 30-cm section of the leaf blade at the primary ear-bearing node excluding ligule midrib and sheath. The internode samples consisted of a 10-cm section of the internode above the primary ear-bearing node excluding the nodes on either end. Samples were chopped into small pieces, flash frozen in liquid nitrogen, and stored at −80°C. All samplings were performed between 8:00 am and 12:00 pm.
Senescence data, recorded as Fv/Fm, were collected on 7 to 15 randomly chosen plants from the three blocks with the same approach as described above in the Methods section for the phenotypic data collection on the diversity panel.
RNA-Seq and Data Analysis
RNA extraction and sample preparation were performed as described previously (Sekhon et al., 2013). mRNA isolation, cDNA synthesis, and library preparation were performed by Novogene following the standard Illumina protocol. Sequencing on the Illumina HiSeq platform generated 150-bp paired-end reads. Reads were trimmed of low-quality bases and adapter sequences with the trimmomatic software package (Bolger et al., 2014). Reads were aligned to the reference maize genome (Zea_mays.AGPv4.40.gtf) (Jiao et al., 2017) with the Tophat2 v2.1.1 package (Trapnell et al., 2012) and Bowtie2 v.2.3.4.1 (Supplemental Table 3; Langmead and Salzberg, 2012).
One biological replicate (replicate 1) was dropped for both B73 and PHG35 at 42 DAA stage for all subsequent analyses due to a low number of reads resulting from the turnover of RNA at this late developmental stage. Fragments per kilobase million (FPKM) were determined with Cufflinks v.2.2.1 (Trapnell et al., 2012). The overlap of sequence reads with genes (exons) was determined with the HTSeq-count v.0.10.0 software package (Anders et al., 2015). Counts for each replicate and genotype were normalized to remove sample-specific effects with EdgeR (Robinson et al., 2010).
Relative pairwise changes in gene level expression were computed using the EdgeR package. Genes with a P ≤ 0.05 were corrected by the false discovery rate (FDR) ≤ 0.05 (Benjamini and Hochberg, 1995). To identify DE genes during natural senescence, log2 (fold change [FC]) was first calculated by subtracting the log2(FPKM) in B73 at 9 DAA from all other time points in B73 and, at each stage, genes with FDR-corrected P ≤ 0.05 and a log2(FC) of ≥ 1 were considered to be DE. The unique DE genes for each stage were combined to develop a list of all genes involved in natural senescence for all downstream analyses. To identify DE genes underlying stay-green, log2(FC) in B73 relative to PHG35 was calculated by subtracting log2(FPKM) in PHG35 at a stage from log2(FPKM) in B73 at that stage, and DE genes at each stage were those with P ≤ 0.05 and a log2(FC) of ≥ 1. All unique DE genes underlying stay-green were used for subsequent analyses.
For GO enrichment, candidate genes above the assigned thresholds of FC were clustered into functional pathways with the GOSeq software tool (Young et al., 2010) and filtered by P ≤ 0.05. JMP Pro v.13.0 was used to evaluate candidate genes via k-means clustering (Wang et al., 2011). Circular figures were prepared with the Circos plotting tool (Krzywinski et al., 2009). Genes within 200-kb region around the GWAS SNPs were identified with the BEDTOOLS package (https://bedtools.readthedocs.io/en/latest/).
Coexpression networks
Coexpression networks were constructed with DE genes underlying stay-green using our random matrix theory based approach (Luo et al., 2007). We filtered out genes that have FPKM value of zero in more than half of 40 samples as well as genes with maximum FPKM value > 1.0 across all. After filtering, we have 4474 DE genes for the coexpression network construction. The random matrix theory based method output a coexpression network with 856 genes represented in 48 subnetworks with size greater than two genes.
Estimation of Epistatic Interactions
All possible two-way interaction terms (additive by additive epistasis terms) were included in a model along with all the marker main effect terms (additive terms) chosen in the initial GWAS screening. The interaction terms to include in the final model were selected using the least absolute shrinkage and selection operator methodology (Tibshirani, 1996) implemented in JMP Pro v.13.2.0 (SAS Institute). The value of the Wald chi-square statistic for each included interaction term was used to determine the width of the connecting line in the network graph. Only those interactions with the Wald chi-square ≥ 1.0 are presented.
Annotation of Cys Proteases
A Pfam domain pepetidase_C1 (papain C1A, Pfam 00,112) was used as a query in EnsemblPlants (http://plants.ensembl.org/index.html) Biomart tool (Kinsella et al., 2011) to identify the PLCPs in maize, sorghum (Sorghum bicolor), rice (Oryza sativa), and Arabidopsis (Arabidopsis thaliana). The retrieved sequences were annotated by National Center for Biotechnology Information Conserved Domain Database (Marchler-Bauer and Bryant, 2004) to confirm the presence of papain_C1A. The protein alignments for a final set of 177 curated PLCPs were performed by ClustalW implemented in MEGA7 (Kumar et al., 2016). The phylogenetic tree was inferred using the neighbor-joining method (Saitou and Nei, 1987), and evolutionary distances were calculated using Poisson correction method (Zuckerkandl and Pauling, 1965). The Bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history (Felsenstein, 1985).
Cys Protease Zymography
Protease zymography was based on previously published protocols (Wagstaff et al., 2002; Martínez et al., 2007) and a soluble protein extraction buffer (Eason and De Vré, 1995). Briefly, each zymogram consisted of an SDS-PAGE gel with an 11% (v/v) separating gel containing 0.05% (v/v) gelatin (Type I from porcine skin, Sigma-Aldrich) and a 4% (v/v) stacking gel. Soluble protein (5 µg) was loaded in each lane in an equal volume representing 3 volumes of protein extract:1 volume of 4× protein loading buffer (100 mM Tris-HCl, pH 6.8, 20% [v/v] glycerol, 2% [w/v] SDS, and 0.04% [w/v] bromophenol blue) and electrophoresed at 100 V. Following electrophoresis, gels were washed for 1 h at room temperature in wash buffer (75 mM sodium-acetate, pH 4.5, 2% [v/v] Triton X-100, and 8 mM DTT) on an orbital shaker at 50 rpm to remove SDS. Gels were then developed for 16 h in incubation buffer (75 mM sodium-acetate, pH 4.5, and 8 mM DTT) at 37°C, washed briefly (3 × 5 min) in reverse osmosis water, stained for 1 h in LabSafe Gel Blue (G Biosciences), and imaged using a ChemiDoc MP Imaging System (Bio-Rad). To confirm Cys protease activity, gels were developed for 16 h in incubation buffer containing 20 µM E-64 Cys protease inhibitor and then processed in the same manner as done after the elecetrophoresis.
To quantify relative levels of Cys protease activity from the gel images, the peak area for the Cys protease was determined using ImageJ software (https://imagej.nih.gov/ij/docs/guide/index.html). The percentage of activity per sample was determined by calculating each time point and expressed that as a percentage of the total measured activity (peak area per time point/total measured peak area), allowing a comparison between the two genotypes. For both maize and Arabidopsis, three biological replicates were used for zymography, and each biological replicate consisted of pooled leaf tissues from two plants.
Arabidopsis Analysis
A mutant allele (stock: CS351853) of RD21A (AT1G47128) carrying a T-DNA insertion in the fourth exon of the gene in Col background was obtained from the Arabidopsis Information Resource (www.arabidopsis.org) and confirmed by sequencing and RT-PCR (Figures 6A and 6B; Supplemental Figure 9). Arabidopsis plants were subjected to 4 d of cold treatment and later transferred to a growth room at 22°C with 16-h day and 8-h night cycle. DNA from young leaves was isolated as described previously (Kim et al., 2016). Genotyping was performed using gene-specific forward (GSF, 5′-GGACCCTATGTTTCATTCAGTTTCA-3′), gene-specific reverse (GSR, 5′-TCAAAATTTTAGGCGGGATGACT-3′), and left border (LB, 5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′) primers with three combinations (GSF+GSR, LB+GSR, and GSF+GSR+LB) as described previously (O’Malley et al., 2015). PCR was performed in 20-μL reaction volume with reaction conditions as initial denaturation at 95°C for 3 min, 32 cycles of 30 s at 95°C, 30 s for 58°C and 1 min at 72°C, and the final extension at 72°C for 5 min.
For time-sensitive analyses, key plant developmental stages and specific leaves were identified using the published description of Arabidopsis ecotype Col development (Boyes et al., 2001). We used DAA, the number of days after the appearance of first flower (also known as stage 6), for comparative analysis. Total RNA was isolated from the second rosette leaf after 10 and 20 DAA in two biological replicates using TRIzol (Thermo Fisher Scientific) according to the manufacturer’s instructions. The first-strand cDNA synthesis was performed using SensiFAST cDNA synthesis kit (Bioline) using 1 µg of RNA. RT-qPCR was performed using SensiFAST kit (Bioline) according to manufacturer’s instructions using GSF and GSR primers. Normalization was based the Arabidopsis housekeeping gene ACTIN (AtACT-RT_F-GTTGGGATGAACCAGAAGGA; AtACT-RT_R-GAACCACCGATCCAGACACT). FCs were calculated using 2^(−ΔΔCT) method.
The onset of senescence was based on Fv/Fm data on one representative leaf from second, fourth, and seventh rosette using a light intensity of 3500 µmol m−2 s−1 for 1 s. DAA was used as a benchmark to ensure data collection at the same developmental stage in all plants. Data were collected on six plants belonging to each of the genotypic classes.
Annotation of Maize SAGs
A list of Arabidopsis SAGs was obtained from the leaf senescence database (Li et al., 2014), and the maize orthologs were identified as reported previously (Jiao et al., 2017) and accessed from MaizeGDB (Andorf et al., 2016).
Metabolic Analyses
NSCs were extracted from 50 mg of freeze-dried powdered leaf and internode tissue by adding 800 μL of 0.7 M perchloric acid (Tobias et al., 1992). Samples were incubated on ice with shaking for 20 to 30 min and centrifuged for 15 min at 12,000 rpm at 4°C. The supernatant was transferred to a new tube, neutralized to pH 7.0 by adding 240 μL of solution containing 2 M KOH and 0.4 M MES, and centrifuged for 10 min at 12,000 rpm at 4°C. The clear supernatant was transferred to a new tube and used for NSC analysis. Glc, Fru, and Suc were measured in the sequential reaction according to a published method (Zhao et al., 2010), with minor modifications. A Glc reagent containing 1 mM ATP, 1 mM NAD, 1 mM MgCl2, 2 U/mL hexokinase, and 4 U/mL Glc-6 phosphate dehydrogenase was added for Glc measurement, followed by addition of 2 U/reaction phosphoglucose isomerase for Fru measurement and, finally, the addition of 25 U/reaction of invertase for Suc measurements. The incubation time after adding enzymes was 30 min at 30°C for each reaction, and absorbance was taken at 340 nm. Purified Glc, Fru, and Suc (Sigma-Aldrich), each with at 0.5 mg/mL concentration, were used as standards with sequential dilutions. The amounts of NSC were estimated as Glc equivalents after comparison with the standards. Chl a and Chl b were measured and quantified following a previously published protocol (Lichtenthaler, 1987), with minor modifications as reported by Sekhon et al. (2012). For all metabolites, analyses were performed on three biological replicates.
Statistical Analysis
The metabolites, protease activity, and physiological data were analyzed using two-tailed Student’s t test using JMP Pro 14 (SAS Institute), and statistical significance was based on P ≤ 0.05. The values are presented as means ± se of the biological replicates.
Accession Numbers
Sequence data from this article can be found at the National Center for Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA505891.
Supplemental Data
Supplemental Figure 1. Distribution and correlation of Fv/Fm data across environments.
Supplemental Figure 2. Manhattan and quantile-quantile plots for GWAS analysis.
Supplemental Figure 3. The senescence-related decline in the content of chlorophyll a and chlorophyll b in leaves of B73 and PHG35 inbred lines.
Supplemental Figure 4. Dynamics of the transcriptome during the onset and progression of natural senescence in B73 inbred line of maize.
Supplemental Figure 5. Transcriptome dynamics associated with stay-green trait deduced from comparison of a stay-green (B73) and a stay-green (PHG35) inbred line of maize.
Supplemental Figure 6. Expression pattern of 48 differentially expressed GWAS candidate genes.
Supplemental Figure 7. Phylogeny of the papain-like Cys proteases (PLCP) in maize, sorghum, Arabidopsis, and rice.
Supplemental Figure 8. Multiple sequence alignment of predicted protein sequences belonging to a subgroup of 10 closely related PLCPs highlighted in Supplemental Figure 7.
Supplemental Figure 9. T-DNA insertion in Arabidopsis Cys protease AT1G47128 (RD21A) resulted in the loss of transcription.
Supplemental Table 1. Descriptive statistics and heritability of senescence measured as maximum quantum efficiency of photosystem II photochemistry (Fv/Fm) in the maize diversity panel.
Supplemental Table 2. SNP variants in maize Cys protease gene mir3 present in B73 and PHG35 and predicted consequences of these variants on gene activity.
Supplemental Table 3. Mapping summary of RNA-seq data.
Supplemental Data Set 1. List of candidate genes with 200-kb window around each selected GWAS SNP.
Supplemental Data Set 2. FPKM of biological replicates of B73 and PHG35 at various developmental stages included in the study.
Supplemental Data Set 3. Differentially expressed genes during at least one of the developmental stages covering natural senescence.
Supplemental Data Set 4. Differentially expressed underlying stay-green identified based on differential expression B73 compared with PHG35.
Supplemental Data Set 5. Genes represented in a major co-expression subnetwork underlying stay-green trait.
Supplemental Data Set 6. Inbred lines evaluated in the study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Jakyung Yi, Shannon Alford, Taylor DeHart, and many talented undergraduate student helpers for assistance in the collection of data and samples and Chris McMahan for helpful discussions. Special thanks to Yalan Huo for help in preparing graphics. We also thank anonymous reviewers for valuable suggestions to improve this article. This work was supported by Clemson University and by the U.S. Department of Energy Great Lakes Bioenergy Research Center, Department of Energy Office of Science (BER-DE-FC02-07ER64494).
AUTHOR CONTRIBUTIONS
R.S.S. and S.M.K. designed the research; R.S.S., R.K., B.S.F., A.J.A., and M.W.B. performed the research; R.S.S., C.S., F.L., T.M.B., W.C.B., and N.d.L. analyzed the data; and R.S.S., C.S., R.K., and S.M.K. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Abdelkhalik A.F., Shishido R., Nomura K., Ikehashi H. (2005). QTL-based analysis of leaf senescence in an indica/japonica hybrid in rice (Oryza sativa L.). Theor. Appl. Genet. 110: 1226–1235. [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. (2015). HTSeq--A Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andorf C.M., et al. (2016). MaizeGDB update: New tools, data and interface for the maize model organism database. Nucleic Acids Res. 44 (D1): D1195–D1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaro A., Ziegler G., Ziyomo C., Hoekenga O.A., Dilkes B.P., Baxter I. (2016). The interaction of genotype and environment determines variation in the maize kernel ionome. G3 (Bethesda) 6: 4175–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R.F., Braun D.M. (2007). tie-dyed1 Functions non-cell autonomously to control carbohydrate accumulation in maize leaves. Plant Physiol. 144: 867–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balibrea Lara M.E., Gonzalez Garcia M.C., Fatima T., Ehness R., Lee T.K., Proels R., Tanner W., Roitsch T. (2004). Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell 16: 1276–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers E.P., Jones A.M., Dickerman A.W. (2004). The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phytochemistry 65: 43–58. [DOI] [PubMed] [Google Scholar]
- Belícuas P.R., Aguiar A.M., Bento D.A.V., Câmara T.M.M., de Souza C.L. Junior (2014). Inheritance of the stay-green trait in tropical maize. Euphytica 198: 163–173. [Google Scholar]
- Below F.E., Christensen L.E., Reed A.J., Hageman R.H. (1981). Availability of reduced N and carbohydrates for ear development of maize. Plant Physiol. 68: 1186–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B Stat. Methodol. 57: 289–300. [Google Scholar]
- Bertolini M., Di Fonzo N., Gentinetta E., Motto M., Verdebio A., Salamini F., Fogher C., Lorenzoni C., Maggiore T., Bianchi A. (1983). Lo876o2. linea pura di mais “high lysine” sviluppata in Italia. Sementi Elette 19: 21–25. [Google Scholar]
- Bogard M., et al. (2011). Anthesis date mainly explained correlations between post-anthesis leaf senescence, grain yield, and grain protein concentration in a winter wheat population segregating for flowering time QTLs. J. Exp. Bot. 62: 3621–3636. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes D.C., Zayed A.M., Ascenzi R., McCaskill A.J., Hoffman N.E., Davis K.R., Görlach J. (2001). Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresson J., Bieker S., Riester L., Doll J., Zentgraf U. (2018). A guideline for leaf senescence analyses: From quantification to physiological and molecular investigations. J. Exp. Bot. 69: 769–786. [DOI] [PubMed] [Google Scholar]
- Bromley J.R., Busse-Wicher M., Tryfona T., Mortimer J.C., Zhang Z., Brown D.M., Dupree P. (2013). GUX1 and GUX2 glucuronyltransferases decorate distinct domains of glucuronoxylan with different substitution patterns. Plant J. 74: 423–434. [DOI] [PubMed] [Google Scholar]
- Brychkova G., Alikulov Z., Fluhr R., Sagi M. (2008). A critical role for ureides in dark and senescence-induced purine remobilization is unmasked in the Atxdh1 Arabidopsis mutant. Plant J. 54: 496–509. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Earl S., Harrison E., Mathas E., Navabpour S., Page T., Pink D. (2003). The molecular analysis of leaf senescence–A genomics approach. Plant Biotechnol. J. 1: 3–22. [DOI] [PubMed] [Google Scholar]
- Chardon F., Jasinski S., Durandet M., Lécureuil A., Soulay F., Bedu M., Guerche P., Masclaux-Daubresse C. (2014). QTL meta-analysis in Arabidopsis reveals an interaction between leaf senescence and resource allocation to seeds. J. Exp. Bot. 65: 3949–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J., Reinecke D., Sim S., Washburn T., Brenner M. (1994). A role for cytokinins in de-etiolation in Arabidopsis (det mutants have an altered response to cytokinins). Plant Physiol. 104: 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampitti I.A., Vyn T.J. (2013). Grain nitrogen source changes over time in maize: A review. Crop Sci. 53: 366–377. [Google Scholar]
- Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelek M., Lusis A.J. (2014). Systems genetics approaches to understand complex traits. Nat. Rev. Genet. 15: 34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliquet J.-B., Deléens E., Mariotti A. (1990). C and N mobilization from stalk and leaves during kernel filling by 13C and 15N tracing in Zea mays L. Plant Physiol. 94: 1547–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S.D. (1996). Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J. 10: 1–8. [DOI] [PubMed] [Google Scholar]
- Crafts-Brandner S.J., Below F.E., Wittenbach V.A., Harper J.E., Hageman R.H. (1984). Differential senescence of maize hybrids following ear removal: II. Selected leaf. Plant Physiol. 74: 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepenbrock C.H., et al. (2017). Novel loci underlie natural variation in vitamin E levels in maize grain. Plant Cell 29: 2374–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick D.N., Smith J.S.C., Cooper M. (2010). Long-term selection in a commercial hybrid maize breeding program. In Plant Breeding Reviews. (Oxford: John Wiley & Sons; ), pp. 109–151. [Google Scholar]
- Eason J.R., De Vré L. (1995). Ethylene‐insensitive floral senescence in Sandersonia aurantiaca (Hook.). N. Z. J. Crop Hortic. Sci. 23: 447–454. [Google Scholar]
- Ehness R., Roitsch T. (1997). Co-ordinated induction of mRNAs for extracellular invertase and a glucose transporter in Chenopodium rubrum by cytokinins. Plant J. 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Emebiri L.C. (2013). QTL dissection of the loss of green colour during post-anthesis grain maturation in two-rowed barley. Theor. Appl. Genet. 126: 1873–1884. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Figueroa C.M., Lunn J.E. (2016). A tale of two sugars: Trehalose 6-phosphate and sucrose. Plant Physiol. 172: 7–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal C., Moore K.M., Paszkiewicz K., Kent N.A., Whitehall S.K. (2015). The impact of the HIRA histone chaperone upon global nucleosome architecture. Cell Cycle 14: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentinetta E., Ceppi D., Lepori C., Perico G., Motto M., Salamini F. (1986). A major gene for delayed senescence in maize. Pattern of photosynthates accumulation and inheritance. Plant Breed. 97: 193–203. [Google Scholar]
- Ghosh S., Mahoney S.R., Penterman J.N., Peirson D., Dumbroff E.B. (2001). Ultrastructural and biochemical changes in chloroplasts during Brassica napus senescence. Plant Physiol. Biochem. 39: 777–784. [Google Scholar]
- Gregersen P.L., Holm P.B. (2007). Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biotechnol. J. 5: 192–206. [DOI] [PubMed] [Google Scholar]
- Gregersen P.L., Culetic A., Boschian L., Krupinska K. (2013). Plant senescence and crop productivity. Plant Mol. Biol. 82: 603–622. [DOI] [PubMed] [Google Scholar]
- Guo Y., Gan S.S. (2012). Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 35: 644–655. [DOI] [PubMed] [Google Scholar]
- Guo Y., Cai Z., Gan S. (2004). Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 27: 521–549. [Google Scholar]
- Harris K., Subudhi P.K., Borrell A., Jordan D., Rosenow D., Nguyen H., Klein P., Klein R., Mullet J. (2007). Sorghum stay-green QTL individually reduce post-flowering drought-induced leaf senescence. J. Exp. Bot. 58: 327–338. [DOI] [PubMed] [Google Scholar]
- Hirsch C.N., et al. (2014). Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26: 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey S.G., Mizrachi E., Spokevicius A.V., Bossinger G., Berger D.K., Myburg A.A. (2011). SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol. 11: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J.K., Kim H., Cocuron J.-C., Orler R., Ralph J., Wilkerson C.G. (2011). The DUF579 domain containing proteins IRX15 and IRX15-L affect xylan synthesis in Arabidopsis. Plant J. 66: 387–400. [DOI] [PubMed] [Google Scholar]
- Jiang G.H., He Y.Q., Xu C.G., Li X.H., Zhang Q. (2004). The genetic basis of stay-green in rice analyzed in a population of doubled haploid lines derived from an indica by japonica cross. Theor. Appl. Genet. 108: 688–698. [DOI] [PubMed] [Google Scholar]
- Jiao Y., et al. (2017). Improved maize reference genome with single-molecule technologies. Nature 546: 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S.M., Cummins I., Lim F.L., Slabas A.R., Knight M.R. (2015). Transcriptomic analysis comparing stay-green and senescent Sorghum bicolor lines identifies a role for proline biosynthesis in the stay-green trait. J. Exp. Bot. 66: 7061–7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-Y., Mahé A., Guy S., Brangeon J., Roche O., Chourey P.S., Prioul J.-L. (2000). Characterization of two members of the maize gene family, Incw3 and Incw4, encoding cell-wall invertases. Gene 245: 89–102. [DOI] [PubMed] [Google Scholar]
- Kim S.-R., Yang J., An G., Jena K. (2016). A simple DNA preparation method for high quality polymerase chain reaction in rice. Plant Breed. Biotechnol. 4: 99–106. [Google Scholar]
- King S.D., Frederickson R.A. (1976). Report on the international sorghum anthracnose-virulence nursery. Sorghum Newsletter 19: 105–106. [Google Scholar]
- Kinsella R.J., Kähäri A., Haider S., Zamora J., Proctor G., Spudich G., Almeida-King J., Staines D., Derwent P., Kerhornou A., Kersey P., Flicek P. (2011). Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database (Oxford) 2011: bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi M., Yamaguchi-Shinozaki K., Tsuji H., Shinozaki K. (1993). Structure and expression of two genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene 129: 175–182. [DOI] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E., Kruglyak L. (1995). Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat. Genet. 11: 241–247. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.A., Tollenaar M. (2007). Physiological basis of successful breeding strategies for maize grain yield. Crop Sci. 47: S-202–S-215. [Google Scholar]
- Li, et al. (2016). Identification of genetic variants associated with maize flowering time using an extremely large multi-genetic background population. Plant J. 86: 391–402. [DOI] [PubMed] [Google Scholar]
- Li H., et al. (2013b). Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 45: 43–50. [DOI] [PubMed] [Google Scholar]
- Li B., Liu H., Zhang Y., Kang T., Zhang L., Tong J., Xiao L., Zhang H. (2013a). Constitutive expression of cell wall invertase genes increases grain yield and starch content in maize. Plant Biotechnol. J. 11: 1080–1091. [DOI] [PubMed] [Google Scholar]
- Li M.-X., Yeung J.M.Y., Cherny S.S., Sham P.C. (2012). Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 131: 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhao Y., Liu X., Peng J., Guo H., Luo J. (2014). LSD 2.0: An update of the leaf senescence database. Nucleic Acids Res. 42: D1200–D1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhao Y., Liu X., Jiang Z., Peng J., Jin J., Guo H., Luo J. (2017). Construction of the leaf senescence database and functional assessment of senescence-associated genes. In Plant Genomics Databases: Methods and Protocols, van Dijk A.D.J., ed (New York, NY: Springer New York; ), pp. 315–333. [DOI] [PubMed] [Google Scholar]
- Liang C., Wang Y., Zhu Y., Tang J., Hu B., Liu L., Ou S., Wu H., Sun X., Chu J., Chu C. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 111: 10013–10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. In Methods in Enzymology, Lester Packer R.D., ed (New York: Academic Press; ), pp. 350–382. [Google Scholar]
- Lim P.O., Kim H.J., Nam H.G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58: 115–136. [DOI] [PubMed] [Google Scholar]
- Lipka A.E., Tian F., Wang Q., Peiffer J., Li M., Bradbury P.J., Gore M.A., Buckler E.S., Zhang Z. (2012). GAPIT: Genome association and prediction integrated tool. Bioinformatics 28: 2397–2399. [DOI] [PubMed] [Google Scholar]
- Liu P., Zhang S., Zhou B., Luo X., Zhou X., Cai B., Jin Y.H., Niu D., Lin J., Cao X., and Jin J.B. (2019). The histone H3K4 demethylase JMJ16 represses leaf senescence in Arabidopsis. Plant Cell 31: 430–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Zhang J. (1998). Modifications in photosystem II photochemistry in senescent leaves of maize plants. J. Exp. Bot. 49: 1671–1679. [Google Scholar]
- Luo F., Yang Y., Zhong J., Gao H., Khan L., Thompson D.K., Zhou J. (2007). Constructing gene co-expression networks and predicting functions of unknown genes by random matrix theory. BMC Bioinformatics 8: 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., et al. (2005). A microarray analysis of the rice transcriptome and its comparison to Arabidopsis. Genome Res. 15: 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bryant S.H. (2004). CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 32: W327–W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez D.E., Bartoli C.G., Grbic V., Guiamet J.J. (2007). Vacuolar cysteine proteases of wheat (Triticum aestivum L.) are common to leaf senescence induced by different factors. J. Exp. Bot. 58: 1099–1107. [DOI] [PubMed] [Google Scholar]
- Maxwell K., Johnson G.N. (2000). Chlorophyll fluorescence--A practical guide. J. Exp. Bot. 51: 659–668. [DOI] [PubMed] [Google Scholar]
- McBee G.G., Waskom R.M., Creelman R.A. (1983). Effect of senescence on carbohydrates in sorghum during late kernel maturity states. Crop Sci. 23: 372–376. [Google Scholar]
- Nørholm M.H.H., Nour-Eldin H.H., Brodersen P., Mundy J., Halkier B.A. (2006). Expression of the Arabidopsis high-affinity hexose transporter STP13 correlates with programmed cell death. FEBS Lett. 580: 2381–2387. [DOI] [PubMed] [Google Scholar]
- Nuccio M.L., Wu J., Mowers R., Zhou H.P., Meghji M., Primavesi L.F., Paul M.J., Chen X., Gao Y., Haque E., Basu S.S., Lagrimini L.M. (2015). Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat. Biotechnol. 33: 862–869. [DOI] [PubMed] [Google Scholar]
- O’Malley R.C., Barragan C.C., Ecker J.R. (2015). A user’s guide to the Arabidopsis T-DNA insertion mutant collections. Methods Mol. Biol. 1284: 323–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oszvald M., Primavesi L.F., Griffiths C.A., Cohn J., Basu S.S., Nuccio M.L., Paul M.J. (2018). Trehalose 6-phosphate regulates photosynthesis and assimilate partitioning in reproductive tissue. Plant Physiol. 176: 2623–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro L., Mohapatra P.K., Biswal U.C., Biswal B. (2014). Dehydration induced loss of photosynthesis in Arabidopsis leaves during senescence is accompanied by the reversible enhancement in the activity of cell wall β-glucosidase. J. Photochem. Photobiol. B 137: 49–54. [DOI] [PubMed] [Google Scholar]
- Pechan T., Jiang B., Steckler D., Ye L., Lin L., Luthe D.S., Williams W.P. (1999). Characterization of three distinct cDNA clones encoding cysteine proteinases from maize (Zea mays L.) callus. Plant Mol. Biol. 40: 111–119. [DOI] [PubMed] [Google Scholar]
- Pinto R.S., Lopes M.S., Collins N.C., Reynolds M.P. (2016). Modelling and genetic dissection of staygreen under heat stress. Theor. Appl. Genet. 129: 2055–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popelka M. (2012). Genetic Architecture of Stay-Green in Maize. (West Lafayette, IN: Purdue University; ). [Google Scholar]
- Pružinská A., Shindo T., Niessen S., Kaschani F., Tóth R., Millar A.H., van der Hoorn R.A.L. (2017). Major Cys protease activities are not essential for senescence in individually darkened Arabidopsis leaves. BMC Plant Biol. 17: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W., Tian Z., Lu L., Chen X., Chen X., Zhang W., Song R. (2017). Editing of mitochondrial transcripts nad3 and cox2 by Dek10 is essential for mitochondrial function and maize plant development. Genetics 205: 1489–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu M., Zheng G., Hamdani S., Essemine J., Song Q., Wang H., Chu C., Sirault X., Zhu X.-G. (2017). Leaf photosynthetic parameters related to biomass accumulation in a global rice diversity survey. Plant Physiol. 175: 248–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino B.F., Noh Y.S., Himelblau E., Amasino R.M. (2000). Molecular aspects of leaf senescence. Trends Plant Sci. 5: 278–282. [DOI] [PubMed] [Google Scholar]
- Quirino B.F., Reiter W.-D., Amasino R.D. (2001). One of two tandem Arabidopsis genes homologous to monosaccharide transporters is senescence-associated. Plant Mol. Biol. 46: 447–457. [DOI] [PubMed] [Google Scholar]
- Rajcan I., Tollenaar M. (1999). Source:sink ratio and leaf senescence in maize: II. Nitrogen metabolism during grain filling. Field Crops Res. 60: 255–265. [Google Scholar]
- Ripoll J.J., Rodríguez-Cazorla E., González-Reig S., Andújar A., Alonso-Cantabrana H., Perez-Amador M.A., Carbonell J., Martínez-Laborda A., Vera A. (2009). Antagonistic interactions between Arabidopsis K-homology domain genes uncover PEPPER as a positive regulator of the central floral repressor FLOWERING LOCUS C. Dev. Biol. 333: 251–262. [DOI] [PubMed] [Google Scholar]
- Roberts I.N., Caputo C., Criado M.V., Funk C. (2012). Senescence-associated proteases in plants. Physiol. Plant. 145: 130–139. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F., Baena-Gonzalez E., Sheen J. (2006). Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 57: 675–709. [DOI] [PubMed] [Google Scholar]
- Romay M.C., et al. (2013). Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biol. 14: R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouyi C., Baiya S., Lee S.-K., Mahong B., Jeon J.-S., Ketudat-Cairns J.R., Ketudat-Cairns M. (2014). Recombinant expression and characterization of the cytoplasmic rice β-glucosidase Os1BGlu4. PLoS One 9: e96712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y.L., Jin Y., Yang Y.J., Li G.J., Boyer J.S. (2010). Sugar input, metabolism, and signaling mediated by invertase: Roles in development, yield potential, and response to drought and heat. Mol. Plant 3: 942–955. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Schippers J.H.M. (2015). Transcriptional networks in leaf senescence. Curr. Opin. Plant Biol. 27: 77–83. [DOI] [PubMed] [Google Scholar]
- Schofield R.A., Bi Y.-M., Kant S., Rothstein S.J. (2009). Over-expression of STP13, a hexose transporter, improves plant growth and nitrogen use in Arabidopsis thaliana seedlings. Plant Cell Environ. 32: 271–285. [DOI] [PubMed] [Google Scholar]
- Sekhon R.S., Childs K.L., Santoro N., Foster C.E., Buell C.R., de Leon N., Kaeppler S.M. (2012). Transcriptional and metabolic analysis of senescence induced by preventing pollination in maize. Plant Physiol. 159: 1730–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon R.S., Briskine R., Hirsch C.N., Myers C.L., Springer N.M., Buell C.R., de Leon N., Kaeppler S.M. (2013). Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS One 8: e61005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slewinski T.L., Meeley R., Braun D.M. (2009). Sucrose transporter1 functions in phloem loading in maize leaves. J. Exp. Bot. 60: 881–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slewinski T.L., Baker R.F., Stubert A., Braun D.M. (2012). Tie-dyed2 encodes a callose synthase that functions in vein development and affects symplastic trafficking within the phloem of maize leaves. Plant Physiol. 160: 1540–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S. (2000). Sugar-induced signal transduction in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51: 49–81. [DOI] [PubMed] [Google Scholar]
- Smeekens S., Ma J., Hanson J., Rolland F. (2010). Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 13: 274–279. [DOI] [PubMed] [Google Scholar]
- Swank J.C., Below F.E., Lambert R.J., Hageman R.H. (1982). Interaction of carbon and nitrogen metabolism in the productivity of maize. Plant Physiol. 70: 1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H. (2013). Senescence, ageing and death of the whole plant. New Phytol. 197: 696–711. [DOI] [PubMed] [Google Scholar]
- Thomas H., Howarth C.J. (2000). Five ways to stay green. J. Exp. Bot. 51: 329–337. [DOI] [PubMed] [Google Scholar]
- Thomas H., Ougham H. (2014). The stay-green trait. J. Exp. Bot. 65: 3889–3900. [DOI] [PubMed] [Google Scholar]
- Tibshirani R. (1996). Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. B 58: 267–288. [Google Scholar]
- Tobias R.B., Boyer C.D., Shannon J.C. (1992). Alterations in carbohydrate intermediates in the endosperm of starch-deficient maize (Zea mays L.) genotypes. Plant Physiol. 99: 146–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel S., Sun D., SanVicente F.M., Zheng H., Atlin G.N., Suarez E.A., Babu R., Zhang X. (2016). Identification of qtl for early vigor and stay-green conferring tolerance to drought in two connected advanced backcross populations in tropical maize (Zea mays L.). PLoS One 11: e0149636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuinstra M.R., Grote E.M., Goldsbrough P.B., Ejeta G. (1997). Genetic analysis of post-flowering drought tolerance and components of grain development in Sorghum bicolor (L.) Moench. Mol. Breed. 3: 439–448. [Google Scholar]
- van der Hoorn R.A.L., Leeuwenburgh M.A., Bogyo M., Joosten M.H.A.J., Peck S.C. (2004). Activity profiling of papain-like cysteine proteases in plants. Plant Physiol. 135: 1170–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco-Arroyo B., Diaz-Mendoza M., Gandullo J., Gonzalez-Melendi P., Santamaria M.E., Dominguez-Figueroa J.D., Hensel G., Martinez M., Kumlehn J., Diaz I. (2016). HvPap-1 C1A protease actively participates in barley proteolysis mediated by abiotic stresses. J. Exp. Bot. 67: 4297–4310. [DOI] [PubMed] [Google Scholar]
- Vijayalakshmi K., Fritz A.K., Paulsen G.M., Bai G.H., Pandravada S., Gill B.S. (2010). Modeling and mapping QTL for senescence-related traits in winter wheat under high temperature. Mol. Breed. 26: 163–175. [Google Scholar]
- Wagstaff C., Leverentz M.K., Griffiths G., Thomas B., Chanasut U., Stead A.D., Rogers H.J. (2002). Cysteine protease gene expression and proteolytic activity during senescence of Alstroemeria petals. J. Exp. Bot. 53: 233–240. [DOI] [PubMed] [Google Scholar]
- Wang A.-Y., Li Y., Zhang C.-Q. (2012). QTL mapping for stay-green in maize (Zea mays). Can. J. Plant Sci. 92: 249–256. [Google Scholar]
- Wang W., Song W.G., Liu S.X., Zhang Y.M., Zheng H.Y., Tian W. (2011). A cloud detection algorithm for MODIS images combining Kmeans clustering and multi-spectral threshold method. Guangpuxue Yu Guangpu Fenxi 31: 1061–1064. [PubMed] [Google Scholar]
- Wang Y., Xu H., Wei X., Chai C., Xiao Y., Zhang Y., Chen B., Xiao G., Ouwerkerk P.B.F., Wang M., Zhu Z. (2007). Molecular cloning and expression analysis of a monosaccharide transporter gene OsMST4 from rice (Oryza sativa L.). Plant Mol. Biol. 65: 439–451. [DOI] [PubMed] [Google Scholar]
- White C.N., Rivin C.J. (1995). Sequence and regulation of a late embryogenesis abundant group 3 protein of maize. Plant Physiol. 108: 1337–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A. (2011). Interactions between flowering and senescence regulation and the influence of low temperature in Arabidopsis and crop plants. Ann. Appl. Biol. 159: 320–338. [Google Scholar]
- Wingler A. (2018). Transitioning to the next phase: The role of sugar signaling throughout the plant life cycle. Plant Physiol. 176: 1075–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A., Roitsch T. (2008). Metabolic regulation of leaf senescence: Interactions of sugar signalling with biotic and abiotic stress responses. Plant Biol. (Stuttg.) 10 (Suppl 1): 50–62. [DOI] [PubMed] [Google Scholar]
- Wingler A., Mares M., Pourtau N. (2004). Spatial patterns and metabolic regulation of photosynthetic parameters during leaf senescence. New Phytol. 161: 781–789. [DOI] [PubMed] [Google Scholar]
- Wingler A., Purdy S., MacLean J.A., Pourtau N. (2006). The role of sugars in integrating environmental signals during the regulation of leaf senescence. J. Exp. Bot. 57: 391–399. [DOI] [PubMed] [Google Scholar]
- Wingler A., Masclaux-Daubresse C., Fischer A.M. (2009). Sugars, senescence, and ageing in plants and heterotrophic organisms. J. Exp. Bot. 60: 1063–1066. [DOI] [PubMed] [Google Scholar]
- Wingler A., Purdy S.J., Edwards S.-A., Chardon F., Masclaux-Daubresse C. (2010). QTL analysis for sugar-regulated leaf senescence supports flowering-dependent and -independent senescence pathways. New Phytol. 185: 420–433. [DOI] [PubMed] [Google Scholar]
- Wingler A., Delatte T.L., O’Hara L.E., Primavesi L.F., Jhurreea D., Paul M.J., Schluepmann H. (2012). Trehalose 6-phosphate is required for the onset of leaf senescence associated with high carbon availability. Plant Physiol. 158: 1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, et al. (2016). Programming of plant leaf senescence with temporal and inter-organellar coordination of transcriptome in Arabidopsis. Plant Physiol. 171: 452–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H.R., Kim H.J., Nam H.G., Lim P.O. (2013). Plant leaf senescence and death - Regulation by multiple layers of control and implications for aging in general. J. Cell Sci. 126: 4823–4833. [DOI] [PubMed] [Google Scholar]
- Wu X.-Y., Hu W.-J., Luo H., Xia Y., Zhao Y., Wang L.-D., Zhang L.-M., Luo J.-C., Jing H.-C. (2016). Transcriptome profiling of developmental leaf senescence in sorghum (Sorghum bicolor). Plant Mol. Biol. 92: 555–580. [DOI] [PubMed] [Google Scholar]
- Xu W., Subudhi P.K., Crasta O.R., Rosenow D.T., Mullet J.E., Nguyen H.T. (2000). Molecular mapping of QTLs conferring stay-green in grain sorghum (Sorghum bicolor L. Moench). Genome 43: 461–469. [PubMed] [Google Scholar]
- Yang J., Carena M.J., Uphaus J. (2010). Area under the dry down curve (AUDDC): A method to evaluate rate of dry down in maize. Crop Sci. 50: 2347–2354. [Google Scholar]
- Yin Y., Wang Z.-Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191. [DOI] [PubMed] [Google Scholar]
- Ying J., Lee E.A., Tollenaar M. (2000). Response of maize leaf photosynthesis to low temperature during the grain-filling period. Field Crops Res. 68: 87–96. [Google Scholar]
- Yoo S.C., Cho S.H., Zhang H., Paik H.C., Lee C.H., Li J., Yoo J.H., Lee B.W., Koh H.J., Seo H.S., Paek N.C. (2007). Quantitative trait loci associated with functional stay-green SNU-SG1 in rice. Mol. Cells 24: 83–94. [PubMed] [Google Scholar]
- Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. (2010). Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Pressoir G., Briggs W.H., Vroh Bi I., Yamasaki M., Doebley J.F., McMullen M.D., Gaut B.S., Nielsen D.M., Holland J.B., Kresovich S., Buckler E.S. (2006). A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208. [DOI] [PubMed] [Google Scholar]
- Zamioudis C., Hanson J., Pieterse C.M. (2014). β-Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol. 204: 368–379. [DOI] [PubMed] [Google Scholar]
- Zhang, et al. (2018). Sweet sorghum originated through selection of Dry, a plant-specific NAC transcription factor gene. Plant Cell 30: 2286–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Primavesi L.F., Jhurreea D., Andralojc P.J., Mitchell R.A.C., Powers S.J., Schluepmann H., Delatte T., Wingler A., Paul M.J. (2009). Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 149: 1860–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., MacKown C.T., Starks P.J., Kindiger B.K. (2010). Rapid analysis of nonstructural carbohydrate components in grass forage using microplate enzymatic assays. Crop Sci. 50: 1537–1545. [Google Scholar]
- Zhao H., Sun Z., Wang J., Huang H., Kocher J.-P., Wang L. (2014). CrossMap: A versatile tool for coordinate conversion between genome assemblies. Bioinformatics 30: 1006–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H.J., Wu A.Z., Zheng C.C., Wang Y.F., Cai R., Shen X.F., Xu R.R., Liu P., Kong L.J., Dong S.T. (2009). QTL mapping of maize (Zea mays) stay‐green traits and their relationship to yield. Plant Breed. 128: 54–62. [Google Scholar]
- Zhou Y., Liu L., Huang W., Yuan M., Zhou F., Li X., Lin Y. (2014). Overexpression of OsSWEET5 in rice causes growth retardation and precocious senescence. PLoS One 9: e94210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L. (1965). Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins, Bryson V. and Vogel H.J., eds (New York: Academic Press; ), pp. 97–166. [Google Scholar]