RRC1 interacts with SFPS and phyB in nuclear speckles and coordinately controls pre-mRNA splicing of a subset of genes involved in light signaling and circadian clock pathways.
Abstract
Light signals perceived by the phytochrome (phy) family of photoreceptors control gene expression at both transcriptional and posttranscriptional levels to promote photomorphogenesis. Recently, we identified a factor called SPLICING FACTOR FOR PHYTOCHROME SIGNALING (SFPS) that directly interacts with the photoreceptor phyB and regulates pre-mRNA splicing in Arabidopsis (Arabidopsis thaliana). To identify SFPS-interacting proteins, we performed an immunoprecipitation followed by a mass spectrometry and identified the Ser/Arg-like protein REDUCED RED-LIGHT RESPONSES IN CRY1CRY2 BACKGROUND1 (RRC1). Genetic analyses revealed that the sfps-2 rrc1-3 phenotypes are similar to those of the single mutants, suggesting that RRC1 and SFPS might function together. RNA sequence analyses of rrc1-3 identified a large number of genes whose pre-mRNA splicing is altered under dark and light conditions. Comparison of the sequence data revealed a subset of common genes coregulated by SFPS and RRC1 under dark and light conditions. Similar to SFPS, RRC1 also interacts with phyB, colocalizes in nuclear photobodies, and regulates light-dependent pre-mRNA splicing of a subset of genes. Taken together, these data suggest that although SFPS and RRC1 can regulate distinct subsets of genes, they also form a complex and coordinately control pre-mRNA splicing of a subset of genes involved in light signaling and circadian clock pathways to promote photomorphogenesis.
INTRODUCTION
The majority of the eukaryotic genes contain intervening noncoding sequences termed introns, which are absent from the mature mRNA. Removal of these introns is performed by the action of ribonuclear protein mega-particles termed spliceosomes that consist of five small nuclear ribonucleoproteins (snRNPs: U1, U2, U4, U5, and U6) and ∼200 proteins (Will and Lürhmann, 2006; Matera and Wang, 2014; Lee and Rio, 2015). Spliceosome assembly on the pre-mRNA is initiated by the recognition of 5ʹ and 3ʹ splicing sites. However, the splicing site consensus sequences are generally not sufficient to direct the assembly of a functional spliceosome. The auxiliary elements, known as exonic splicing enhancers (ESEs)/intronic splicing enhancers and silencers (ISSs), also contribute to the splicing site recognition (Fu and Ares, 2014; Lee and Rio, 2015). To discern the information coded in these cis-regulatory elements, in addition to the core spliceosome proteins, a large number of RNA binding proteins (RBPs) such as Ser/Arg (SR)-rich proteins and heterogeneous nuclear ribonucleoproteins, binding to different RNA sequences are involved. As these RBPs function as splicing regulators, the final outcome, including or skipping a particular exon, is mostly dependent on the delicate protein–protein and protein–RNA associations. In addition, both endogenous and environmental factors regulate the expression and/or activity of RBPs to precisely control pre-mRNA splicing (Staiger and Brown, 2013; Lee and Rio, 2015).
The SR proteins are a class of RBPs that usually contain N-terminal RNA binding domains or RNA recognition motifs, and the C-terminal Arg/Ser-rich (RS) region (Barta et al., 2010). The RS domain of the SR proteins, enriched in Arg and Ser dipeptides, was originally found in three splicing regulators, SUPPRESSOR OF WHITE APRICOT (SWAP), TRANSFORMER (Tra), and TRANSFORMER-2 (Tra-2), in Drosophila melanogaster (Boggs et al., 1987; Chou et al., 1987; Amrein et al., 1988). Numerous studies over the years have revealed the molecular mechanisms of SR proteins in regulating pre-mRNA splicing in multicellular eukaryotes (Fu, 1995; Long and Caceres, 2009; Reddy and Shad Ali, 2011; Richardson et al., 2011; Reddy et al., 2013). SR proteins essentially recognize the purine-rich sequences and typically function in the complex with other splicing regulators to promote splice site recognition (Erkelenz et al., 2013). Furthermore, it has been well documented that the pre-mRNA splicing often occurs co-transcriptionally. SR proteins are concentrated in diverse nuclear granules and can be recruited to the RNA polymerase II complex through a direct interaction with its carboxy-terminal domain (Roth et al., 1991). Thus, SR proteins might also play an important role during this co-transcriptional splicing regulation. Finally, recent studies have shown that the alternative splicing (AS) and phosphorylation status of SR proteins are modulated by diverse signaling events, leading to changes in the expression pattern and functional dynamics of SR proteins (de la Fuente van Bentem et al., 2006; Schindler et al., 2008; Carvalho et al., 2016). Therefore, splicing regulatory networks might also be subject to outside stimuli (Long and Caceres, 2009). Detailed molecular mechanisms of SR protein-mediated pre-mRNA splicing have been unraveled in animal system; however, it is still poorly understood in plants. Plants have three SR protein families that are not conserved in animals (Barta et al., 2010; Reddy and Shad Ali, 2011; Richardson et al., 2011; Reddy et al., 2013). This pattern suggests that the molecular mechanisms of some SR proteins in plants might be unique during splicing regulation.
The red/far-red light signals perceived by the phytochrome (phy) family of photoreceptors regulate photomorphogenic development in plants (Bae and Choi, 2008; Pham et al., 2018). There are five phytochromes (phyA to phyE) in Arabidopsis (Arabidopsis thaliana) that exist in the inactive red light–absorbing (Pr) form in the dark. Perception of red light induces photoconversion to the biologically active far-red-light–absorbing (Pfr) form, which migrates into the nucleus and triggers large-scale gene expression changes that drive photomorphogenesis (Quail, 2007; Klose et al., 2015). Recent evidence suggests that light signals perceived by phytochromes induce not only transcriptional reprogramming but also pre-mRNA splicing of a large number of genes (Shikata et al., 2014; Hartmann et al., 2016; Zhang et al., 2017; Cheng and Tu, 2018). Phytochromes might regulate pre-mRNA splicing directly and/or indirectly through regulating the circadian clock, chloroplast development, and photosynthesis as well as energy availability (Hong et al., 2010; Sanchez et al., 2010; Jones et al., 2012; Wang et al., 2012; Petrillo et al., 2014; Schlaen et al., 2015; Hartmann et al., 2016). Among these, SPLICING FACTOR FOR PHYTOCHROME SIGNALING (SFPS) and REDUCED RED-LIGHT RESPONSES IN CRY1CRY2 BACKGROUND1 (RRC1) are the only two genes that have been identified so far to be involved in early phytochrome-mediated pre-mRNA splicing events in Arabidopsis. RRC1 encodes an ortholog of the human potential splicing factor SR140, a SR-like protein (Will et al., 2002; Shikata et al., 2012). The loss-of-function rrc1 mutation caused pleiotropic developmental abnormalities, including seedling photomorphogenic phenotypes, early flowering, and death. RRC1 undergoes AS in response to light signals, implying the presence of a self-reinforcing circuitry (Hartmann et al., 2016). Recently, we have shown that the human SPF45-related splicing factor SFPS is involved in fine-tuning light responses via controlling pre-mRNA splicing (Xin et al., 2017). SFPS directly interacts with phyB in a red light–dependent manner. However, the regulatory mechanisms of SFPS activity remain to be understood. Here, we identified RRC1 as one of the SFPS-interacting proteins using immunoprecipitation followed by mass spectrometry (IP-MS). The sfps-2 rrc1-3 double mutant shows a similar phenotype as the sfps single mutant, suggesting they might function, in part, in the same pathway. The RNA deep sequencing data have identified a subset of genes involved in light signaling and the circadian clock whose pre-mRNA splicing is altered in these mutants under dark and light conditions. These data suggest that these two proteins function, in part, in the same complex and regulate pre-mRNA splicing of a subset of genes to promote photomorphogenesis.
RESULTS
SFPS Interacts with RRC1 in Vitro and in Vivo
SFPS is involved in light-regulated pre-mRNA splicing to promote photomorphogenesis (Xin et al., 2017). To identify SFPS-interacting proteins, we performed immunoprecipitation of SFPS-green fluorescent protein (GFP) from crude extracts of SFPSpro:SFPS-GFP/sfps-2 transgenic seedlings followed by mass spectrometry (IP-MS). One of the SFPS-interacting proteins identified was RRC1 (Supplemental Figures 1A and 1B). RRC1 was previously reported to be a functional splicing factor required for phyB signaling (Shikata et al., 2012). RRC1 contains an RNA recognition motif and a Suppressor-of-White Apricot domain at its N terminus and an RPR domain at its C terminus (Supplemental Figure 1C).
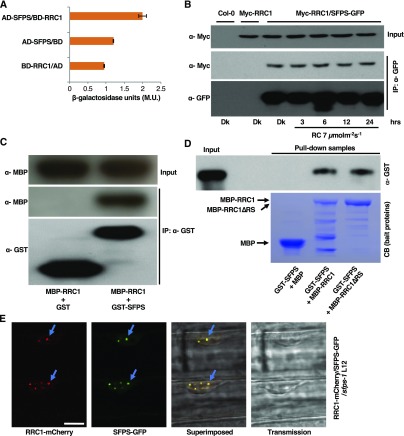
To further validate the IP-MS data, yeast two-hybrid and in vivo coimmunoprecipitation (Co-IP) assays were performed. Both assays show that SFPS strongly interacts with RRC1 under in vivo conditions (Figures 1A and 1B). Moreover, the SFPS-RRC1 interaction is not regulated by light, as up to 24 h of constant red-light treatment did not significantly alter the strength of interaction between SFPS and RRC1. Next, we sought to confirm whether SFPS directly interacts with RRC1 without any auxiliary proteins by performing an in vitro pull-down assay. Bacterially expressed glutathione-agarose–bound glutathione S-transferase (GST) and GST-SFPS were used as bait proteins to pull down maltose binding protein (MBP)-RRC1 prey protein. GST-SFPS, but not GST, interacts with bacterially expressed MBP-RRC1 (Figure 1C). Previously, it was shown that the RS domain of RRC1 is necessary for its biological function under red light (Shikata et al., 2012). To examine whether the RS domain is required for interaction with SFPS, we performed in vitro pull-down assays using the full-length RRC1 and a truncated version of RRC1 lacking the RS domain. The RS domain of RRC1 was not necessary for interaction with SFPS (Figure 1D).
Figure 1.
SFPS Interacts with RRC1 Both In Vivo and In Vitro.
(A) SFPS interacts with RRC1 in yeast two-hybrid assay. Full-length RRC1 ORF was fused to the GAL4 DNA binding domain (BD); full-length SFPS ORF was fused to the GAL4 activation domain (AD). β-Galactosidase activity was measured to quantify the strength of interaction between RRC1 and SFPS. Error bars represent sem (n > 3). M.U., Miller Unit.
(B) SFPS-GFP and Myc-RRC1 proteins co-immunoprecipitate in Arabidopsis seedlings. Total protein was extracted from double transgenic Arabidopsis seedlings expressing SFPS-GFP and Myc-RRC1. SFPS-GFP was immunoprecipitated with anti-GFP antibody. Myc-RRC1 and SFPS-GFP were detected using anti-Myc and anti-GFP antibodies, respectively.
(C) GST-SFPS precipitated MBP-RRC1 in vitro. Tagged proteins were expressed and purified from E. coli BL21 strain. Glutathione agarose–bound GST or GST-SFPS were used to pull down equal amount of purified MBP-RRC1 protein. Pull-down samples were loaded into the 8% SDS polyacrylamide gel and followed by immunoblotting. The precipitated MBP-RRC1 was detected by the anti-MBP antibody, and the control blot was probed with anti-GST antibody.
(D) The RS domain of RRC1 is not required for its interaction with SFPS. Both MBP-RRC1 and MBP-RRC1∆RS interact with GST-SFPS. GST-SFPS, MBP, MBP-RRC1, and MBP-RRC1∆RS were expressed in bacteria. Equal amounts of affinity-purified GST-SFPS were added to three separate tubes containing purified MBP, MBP-RRC1, or MBP-RRC1∆RS bound to amylose beads. Pull-down samples were loaded onto 8% SDS polyacrylamide gel followed by immunoblotting. The precipitated GST-SFPS was detected by the anti–GST-horseradish peroxidase antibody (top image) and the bait proteins were stained with Coomassie blue (CB; bottom image). The additional bands for the MBP-RRC1 and MBP-RRC1∆RS samples on the Coomassie-stained gel are degradation products of RRC1 in these lanes.
(E) RRC1-mCherry colocalizes with SFPS-GFP in nuclear speckles. Double transgenic seedlings expressing RRC1-mCherry and SFPS-GFP were grown under dark for 4 d and then exposed to red light for 24 h. Images were taken from the elongation zone of root tissue using confocal microscopy. Bar = 10 μm.
As reported previously (Shikata et al., 2012), constitutively expressed RRC1-mCherry localizes into the nucleus, forming nuclear speckles (Figure 1E; Supplemental Figure 2A). Since SFPS-GFP also localizes to the nuclear speckles (Xin et al., 2017), we examined whether SFPS-GFP colocalizes with RRC1-mCherry in nuclear speckles. Fluorescence confocal imaging of the nuclei of RRC1-mCherry/SFPS-GFP transgenic seedlings revealed that these two proteins colocalize with each other in nuclear speckles (Figure 1E; Supplemental Figure 2A). RRC1-mCherry also formed nuclear speckles in the sfps-2 background, suggesting that the nuclear speckle formation of RRC1 does not depend on SFPS (Supplemental Figure 2B). Overall, these data suggest that SFPS interacts with RRC1 in vitro and in vivo and might function together in a complex to regulate photomorphogenesis by controlling pre-mRNA splicing.
RRC1 Interacts with PhyB through the RS Domain
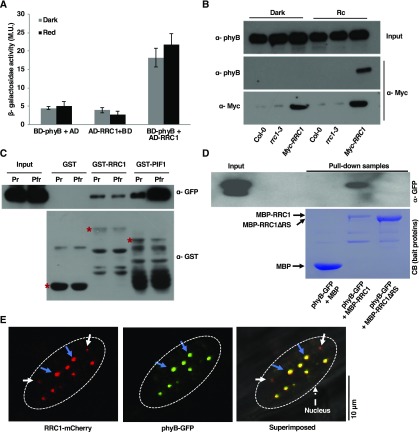
Because RRC1 is an essential component in phyB signaling and rrc1-3 showed altered red-light phenotypes (Shikata et al., 2012), the ability of RRC1 to physically interact with phyB was examined. Yeast two-hybrid analysis using full-length phyB and RRC1 indicated that these two proteins interact with each other (Figure 2A). To further validate the yeast two-hybrid result, in vivo Co-IP assay was performed using 35Spro:Myc-RRC1/rrc1-3 transgenic seedlings. To determine whether ectopically expressed Myc-RRC1 is biologically functional, seedlings of two independent transgenic lines were grown under continuous red light along with the wild-type and rrc1-3 mutant seedlings. Both independent transgenic lines complemented the long hypocotyl phenotype of the rrc1-3 under red light (Supplemental Figure 3), suggesting that the Myc-RRC1 fusion protein is biologically functional.
Figure 2.
RRC1 Interacts with phyB and Colocalizes in Nuclear Photobodies.
(A) RRC1 interacts with phyB in the yeast two-hybrid assay. Full-length RRC1 ORF was fused to the GAL4 activation domain (AD); full-length phyB ORF was fused to the GAL4 DNA binding domain (BD). β-Galactosidase activity was measured to quantify the strength of the interaction between RRC1 and phyB. Error bars represent sd (n ≥ 3). M.U., Miller Unit.
(B) Myc-RRC1 and phyB proteins co-immunoprecipitate in Arabidopsis seedling extracts. Total protein was extracted from 35Spro:Myc-RRC1 transgenic Arabidopsis seedlings expressing Myc-RRC1. Myc-RRC1 was immunoprecipitated with anti-Myc antibody. Myc-RRC1 and phyB were detected using anti-Myc and anti-phyB antibodies, respectively.
(C) GST-RRC1 interacts with phyB-GFP in vitro. GST, GST-RRC1, and GST-PIF1 were expressed and purified from E. coli. phyB-GFP was expressed in yeast cells. Glutathione agarose–bound GST, GST-RRC1, or GST-PIF1 were used to pull down equal amount of crude extracts of phyB-GFP (Pr or Pfr form) protein. Pull-down samples were loaded into the 8% (w/v) SDS polyacrylamide gel followed by immunoblotting. The precipitated phyB-GFP was detected by the anti-GFP antibody, and the control blot was probed with anti-GST antibody. Red stars indicate GST or GST-tagged proteins. The presence of additional bands for the GST-RRC1 and GST-PIF1 samples on the anti-GST blot are degradation products of these proteins in each lane.
(D) The RS domain of RRC1 is essential for its interaction with phyB. MBP-RRC1, but not MBP-RRC1∆RS, interacts with phyB-GFP in vitro. MBP, MBP-RRC1, and MBP-RRC1∆RS were expressed in bacteria, while phyB-GFP was expressed in yeast cells. Equal amounts of crude extracts of phyB-GFP (pfr form) was added to three separate tubes containing purified MBP, MBP-RRC1, or MBP-RRC1∆RS bound to amylose beads. Pull-down samples were loaded onto the 8% SDS polyacrylamide gel followed by immunoblotting. The precipitated phyB-GFP was detected by the anti-GFP antibody (top image), and the bait proteins were stained with Coomassie stain (CB; bottom image). The presence of additional bands for the MBP-RRC1 and MBP-RRC1∆RS samples on the Coomassie-stained gel is degradation products of RRC1 in these lanes.
(E) RRC1-mCherry colocalizes with phyB-GFP in nuclear speckles. Images were taken from the elongation zone of root tissue from double transgenic seedlings expressing RRC1-mCherry and phyB-GFP grown in the dark for 4 d and then exposed to red light for 6 h using confocal microscopy. Blue arrow indicates nuclear speckles in which RRC1-mCherry colocalizes with phyB-GFP; while white arrows indicate nuclear speckles in which RRC1-mCherry does not colocalize with phyB-GFP. Dotted ovals indicate nuclei. Bar = 10 μm.
Myc-RRC1 was then immunoprecipitated using anti-Myc antibodies, and the presence or absence of native phyB in the complex was detected using anti-phyB antibodies. RRC1 interacted with phyB in a light-dependent manner (Figure 2B). Additionally, in vitro pull-down assays were performed using bacterially expressed GST-RRC1 as a bait protein and phyB-GFP protein expressed in yeast (Saccharomyces cerevisiae) as a prey protein. GST-phytochrome-interacting factor1 (PIF1) and GST were used as positive and negative controls, respectively, for this assay. Prior to the assay, crude phyB-GFP protein was treated with phycocyanobilin and either kept in the dark (phyB-Pr) or exposed to the red light (phyB-Pfr). GST-RRC1, but not GST alone, interacted with both the Pr and Pfr forms of phyB-GFP. GST-PIF1 was used as a control and interacted more strongly with the Pfr form than the Pr form of phyB under the same conditions (Figure 2C). The light-independent interaction between GST-RRC1 and phyB-GFP was still weaker than the interaction between GST-PIF1 and the Pr form phyB-GFP. The RS domain of RRC1 is required for phyB signaling under red light (Shikata et al., 2012). To examine whether the RS domain is required for interaction with phyB, we performed in an vitro pull-down assay using the full-length and a truncated version of RRC1 lacking the RS domain. The RS domain of RRC1 was necessary for interaction with phyB (Figure 2D). These data suggest that phyB might regulate pre-mRNA splicing by direct physical interaction with RRC1 in response to light.
As both RRC1 and phyB formed nuclear speckles (Shikata et al., 2012; Klose et al., 2015), we examined whether these two proteins also colocalize in nuclear speckles. Confocal imaging of RRC1-mCherry/phyB-GFP nuclei clearly indicated that these proteins do form nuclear speckle in vivo. In addition, while some of the RRC1-mCherry speckles were distinct, the majority of the RRC1-mCherry speckles colocalized with phyB-GFP photobodies (Figure 2E). These results demonstrate that RRC1 and phyB not only colocalize in the nuclear speckles but also physically interact with each other directly.
The sfps-2rrc1-3 Double Mutant and sfps-2 Single Mutant Display Similar Phenotypes
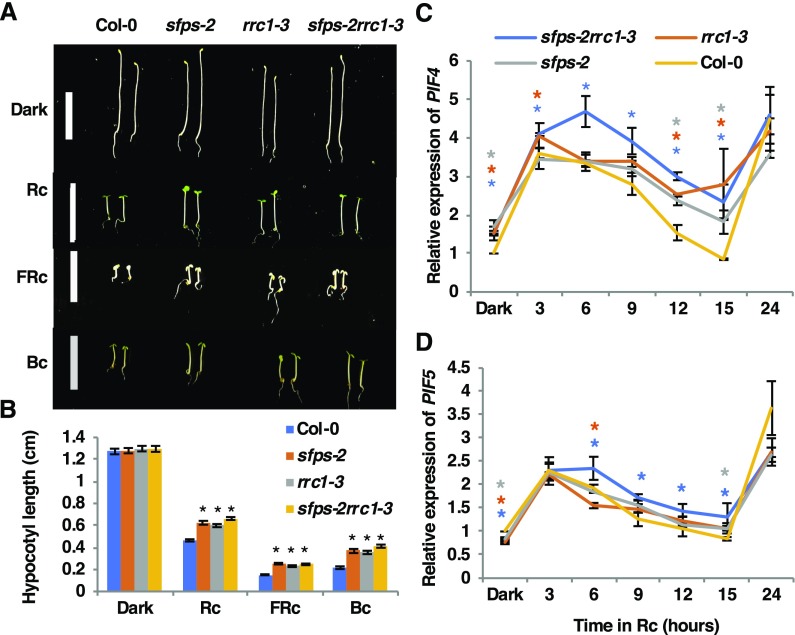
Previously, a genetic study showed that a null allele of rrc1 mutant called rrc1-4 is lethal, complicating genetic analysis. However, a weak allele of rrc1 mutant (rrc1-3; SALK_011832) displays significantly longer hypocotyl and flowers early compared with the wild type (Shikata et al., 2012), which is similar to the phenotypes displayed by sfps-2 mutant (Xin et al., 2017). To investigate the genetic relationships between SFPS and RRC1, the sfps-2 rrc1-3 double homozygous line was created and its phenotypes were compared with those of the parental single mutants and the wild type. The results showed that the average length of the hypocotyl of sfps-2 rrc1-3 double mutant was similar to that of the sfps-2 or rrc1-3 single mutant, under continuous red (Rc), far-red (FRc), and blue light (Bc) conditions (Figures 3A and 3B). Previously, rrc1-3 did not display a long hypocotyl phenotype under FRc and Bc conditions (Shikata et al., 2012). However, in our study a weak phenotype was observed under FRc and Bc conditions, possibly due to the use of a different fluence rate of light. In addition, in response to Rc over time, the expression of two basic-helix-loop-helix transcription factor genes, PIF4 and PIF5, which play pivotal roles in regulating hypocotyl elongation, was significantly higher in the double mutant compared with the parental genotypes at multiple time points (Figures 3C and 3D).
Figure 3.
SFPS and RRC1 Function Positively in Phytochrome Signaling.
(A) Photographs of Col-0, sfps-2, rrc1-3, and sfps-2 rrc1-3 mutant seedlings grown for 4 d in the dark and under continuous red- (at 7 μmol m−2 s−1), far-red (at 0.56 μmol m−2 s−1), and blue (0.73 μmol m−2 s−1)-light conditions. Bar, 1 = cm.
(B) Quantification of the hypocotyl lengths for 4-d-old seedlings grown under the conditions described in (A). Error bars indicate sem (n > 30), and asterisks indicate significant difference compared with Col-0 (P < 0.05) based on Student’s t test.
(C) and (D) Relative expression of PIF4 and PIF5 in different genotypes as indicated, respectively. PIF4 (C) and PIF5 (D) were upregulated in the sfps-2, rrc1-3, and sfps-2rrc1-3 mutant compared with Col-0 over 24 h of red-light irradiation (at 7 μmol m−2 s−1). Total RNA for RT-qPCR was extracted from 4-d-old Col-0, sfps-2, rrc1-3, and sfps-2 rrc1-3 seedlings grown in the dark and then irradiated with red light for 3, 6, 9, 12, 15, and 24 h. PP2A was used as an internal control. Each bar is the mean ± sem (n = 3 independent biological repeats, and each biological repeat include three technical repeats). The asterisks indicate significant difference (P < 0.05, based on Student’s t test) when comparing with Col-0 at each time point (blue, sfps-2 rrc1-3; orange, rrc1-3; gray, sfps-2).
RRC1 Regulates Gene Expression in Arabidopsis
To characterize the genes regulated by RRC1, RNA deep sequencing was performed to identify genome-wide changes in gene expression and pre-mRNA splicing from 4-d-old dark-grown rrc1-3 seedlings with or without an exposure to a 3-h red-light treatment. These data were compared with a previously published data set for the wild-type seedlings grown under identical conditions and sequenced using identical methods (Xin et al., 2017). To examine batch variation, the Surrogate Variable Analysis tool available as part of the Bioconductor R package (Mancini et al., 2017) was used. This analysis did not show any batch variation between the two data sets from independent experiments. With a false discovery rate (FDR) of 0.05 and a fold change of 1.5, a total of 5630 and 4031 genes were identified as differentially expressed under dark and light, respectively, in the rrc1-3 background compared with the wild type (Supplemental Figure 4A; Supplemental Data Set 1, I and III).
The influence of light on gene expression was also investigated in rrc1-3 mutant by comparing expression of genes in the dark versus exposure to a pulse of 3-h red light. These data show a total of 5711 genes that were differentially expressed in rrc1-3 mutant in response to light (Supplemental Data Set 1, V). By contrast, a total of 4291 genes were differentially expressed in the wild type in response to light (Xin et al., 2017). Volcano plots indicate that hundreds of genes were significantly regulated by RRC1 both in the dark and light (Supplemental Figures 4B and 4C). The expression patterns of these genes are shown in the heatmaps under dark conditions (Supplemental Figure 4D) and light conditions (Supplemental Figure 4E), displaying the opposite expression patterns of these genes in the wild type and the rrc1-3 mutants.
Detailed analyses of gene ontology (GO)-terms revealed the enrichment of 131 GO-terms in the dark, while in response to red-light treatment 230 GO-terms were found to be enriched (Supplemental Data Set 1, II and IV). Moreover, GO-terms such as photosynthesis, response to light-stimulus, and response to the red and far-red light were significantly enriched in the differentially expressed gene list. Many genes involved in light signaling and circadian clock pathways were differentially expressed in rrc1-3 mutant under both dark and light conditions (Supplemental Data Set 1, VI and VII). These data highlight the significance of RRC1 in regulating the expression of a large number of genes to promote photomorphogenesis.
RRC1 Controls Pre-mRNA Splicing of a Large Number of Genes in Arabidopsis
Because SFPS-GFP colocalizes with components of the U2 snRNP-associated splicing factors in nuclear speckles (Xin et al., 2017), a colocalization test for RRC1 with components of the U2 snRNP-associated splicing factors was also performed by transiently expressing RRC1-GFP with U2Aʹ-mCherry, U2AF35A, and U2AF65B in epidermal cells of Nicotiana benthamiana. Fluorescence confocal imaging of the epidermal cells clearly indicated that RRC1-GFP colocalizes with all three components (Supplemental Figure 5). These data suggest that RRC1 might be stored in nuclear speckles with the U2 snRNP-associated components and/or may function with these components in mediating pre-mRNA splicing in Arabidopsis.
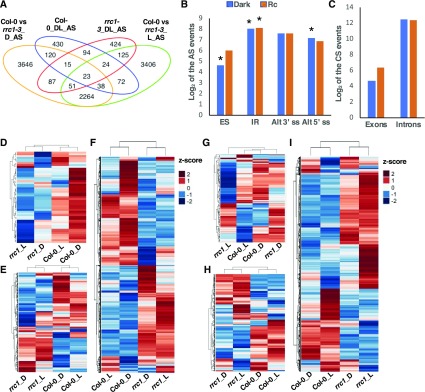
To investigate the role of RRC1 in regulating the genome-wide pre-mRNA splicing, RNA deep sequencing data were analyzed to identify altered splicing patterns in rrc1-3 mutant compared with the wild type, as described previously (Xin et al., 2017). The splicing pattern changes resulting from rrc1-3 mutation were evaluated by comparing the percentage of inclusion of exons (percent spliced-in [PSI]) and introns (percent intron retention [PIR]) in four different comparisons: the wild type versus rrc1-3 dark, the wild type versus rrc1-3 3 h Rc, the wild type dark versus 3 h Rc, and rrc1 dark versus 3 h Rc. A total of 6244 (FDR < 0.05 and Delta PSI or Delta PIR > 3%) differentially splicing events mapping to 4283 gene loci were identified between wild-type and rrc1-3 seedlings grown in the dark (Figure 4A; Supplemental Data Set 2 I). By contrast, a total of 6003 (FDR < 0.05 and Delta PSI or PIR > 3%) differentially splicing events covering 4159 genes were identified under light conditions (Figure 4A; Supplemental Data Set 2, III).
Figure 4.
RRC1 Controls Pre-mRNA Splicing of a Large Number of Genes in Arabidopsis.
(A) Venn diagram shows the number and overlap of the differentially splicing events (FDR < 0.05 and absolute value of Delta_PSI/PIR > 3%) in Col-0 and rrc1-3 mutant grown in the dark and light-treated samples. AS, differentially splicing events; D, dark condition; L, light condition.
(B) and (C) Bar graphs show the categories of the differentially spliced events in rrc1-3 compared with Col-0 wild type under dark (blue) and light (orange) conditions, respectively. (B) AS: ES, IR, Alt3ʹss, and Alt5ʹss. (C) CS events: exon or intron events. The asterisks indicate that differentially splicing events were significantly enriched in that category in the rrc1-3 mutant background.
(D) to (I) Heatmaps of pre-mRNA splicing profiles, based on Z-scores of PSI/PIR values of Col-0 and rrc1-3 in the dark (see [D] to [F]) and light conditions (see [G] to [I]). The Z-scores of PSI/PIR values for the corresponding events under light and dark have been added for comparison purpose only in the dark and light heatmaps. Hierarchical clustering was performed on genes with altered pre-mRNA splicing pattern between Col-0 and rrc1-3. Blue color indicates low Z-scores, whereas red color indicates high Z-scores. Red light intensity used was at 7 μmol m−2 s−1.
(D) and (G) Heatmaps of the quantified splicing profiles of ES events and CS exon changes.
(E) and (H) Heatmaps of the quantified splicing profiles of 5ʹ or 3ʹ AS events.
(F) and (I) Heatmaps of the quantified splicing profiles of IR events and CS intron changes.
Among these differentially splicing events, 2376 were found to be overlapping between light and dark conditions (Figure 4A), which is equivalent to ∼38 and ∼40% of the total differentially splicing events observed in the rrc1-3 mutant under dark and light conditions, respectively. These numbers were much smaller for the sfps-2 mutant (972 differentially splicing events covering 787 genes in darkness and 992 differentially splicing events covering 827 genes under light; Xin et al., 2017), indicating that RRC1 plays a more prominent role in regulating pre-mRNA splicing both in the dark and light conditions.
The altered splicing events were categorized into two broad classes: constitutive splicing (CS) events and AS events. CS covers those introns that were mostly spliced out and those exons that were mostly included in the mRNAs present in the wild-type plants. By contrast, AS events were defined as those in which 5′ and/or 3′ splice sites (ss) are in competition, and those events in which an intron in the wild-type plant was recognized in some cases but retained in others (Fu and Ares, 2014). AS events were further classified as exon skipping (ES), intron retention (IR), and altered splicing donor or acceptor sites (3ʹ- or 5ʹ-alt). The splicing efficiency changes in rrc1-3 mutant consisted of both CS and AS events based on the current genome annotation (Figure 4B) and could be considered as mis-splicing in the rrc1 background compared with the wild type. Strikingly, the CS events were much more pronounced, with a large number of splicing efficiency changes in the introns but only a few changes in the exons (Figure 4C). Because RRC1 is an ortholog of SR140, which is part of the U2-snRNP complex in human (Will et al., 2002), these data suggest that RRC1 might participate as a general splicing factor controlling a large amount of pre-mRNA splicing.
Heatmaps showed that the splicing patterns of both CS and AS events in the rrc1-3 mutant were in general opposite compared with the wild type under both dark and light conditions (Figures 4D to 4I). To identify the biological processes that are regulated by RRC1, GO analysis was performed for the genes displaying altered splicing patterns between the wild type and rrc1-3. A total of 220 and 259 GO categories (FDR < 0.05) were over-represented in the dark and light conditions, respectively (Supplemental Figure 6; Supplemental Data Set 2, II and IV). Several GO-terms related to light stimulus, red or far-red light signaling pathway, photosynthesis, and circadian rhythm were significantly enriched (P < 0.05) in the dark or light conditions (Supplemental Figures 6A and 6B; Supplemental Data Set 2, VIII and IX). An in-depth analysis of the enriched GO-terms revealed that the splicing patterns of the transcription factor genes (e.g., ELF3, REV8, PIF3, and others) that directly regulate photomorphogenesis and the hypocotyl elongation were also disrupted. Moreover, the Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis indicated that spliceosome, basal transcriptional factors, and mRNA surveillance pathways were significantly enriched in the differentially spliced gene lists (Supplemental Figures 6C and6D; Supplemental Data Set 2, VI and VII). These include the splicing regulators that are involved in the pre-mRNA processing pathways (e.g., SR30, RS31, SR45, and SKIP; Supplemental Data Set 2, I and III).
Interestingly, some of these splicing factor mutants also display photomorphogenic phenotypes (Wang et al., 2012). These data suggest that the splicing cascade might also directly or indirectly fine-tune the expression of light response genes. The altered splicing cascade coupled with the transcriptional regulation in the rrc1-3 compared with the wild type resulted in a small overlap between the differentially expressed genes and the differentially spliced genes under both dark and light conditions (Supplemental Figures 7A and 7B). Taken together, these data indicate that the transcriptional and the posttranscriptional regulation might coordinate with each other to control the downstream gene expression. Thus, the gene expression and the splicing changes were consistent with the hypo-sensitive phenotypes of rrc1-3 mutant in response to light.
RRC1 Is Involved in Light Regulation of Pre-mRNA Splicing
To determine whether RRC1 is indeed necessary for the light-regulated pre-mRNA splicing, we examined the Delta_PSI/PIR (PSI/PIR_D-PSI/PIR_Rc) to evaluate the light-induced splicing changes in rrc1-3 mutant seedlings. A total of 843 splicing events corresponding to 643 gene loci were found to undergo splicing pattern changes (Delta_PSI/PIR > 0.03, FDR < 0.05) in response to a 3-h red-light pulse in rrc1-3 mutant (Supplemental Data Set 2, V). As reported previously, 816 pre-mRNA splicing events covering 610 genes are light regulated in the wild-type background (Xin et al., 2017). To further compare the splicing efficiency changes of these 816 events in different genetic backgrounds, we examined the Delta_PSI/PIR (PSI/PIR_D-PSI/PIR_Rc) of these events in the wild-type Columbia (Col-0) and rrc1-3, as well as sfps-2, a splicing mutant we characterized previously (Xin et al., 2017).
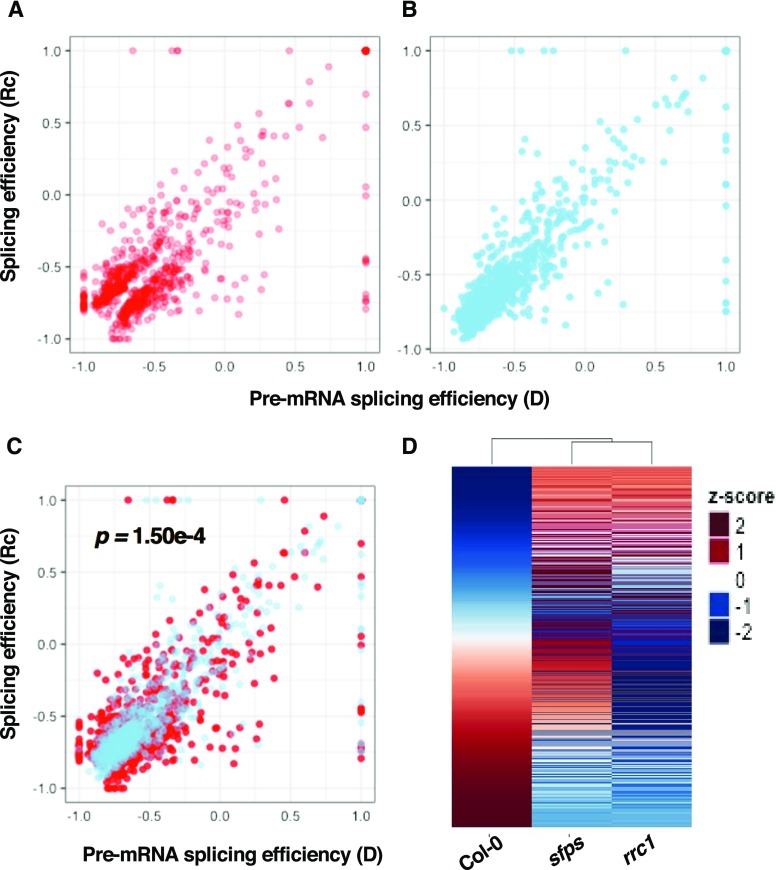
The folded roots of PSI/PIR in the dark and light conditions were graphed in the scatterplots, which showed that the splicing efficiency of light-regulated splicing events (774 of 816 events that are common among all three genotypes) was either upregulated or downregulated in the wild type upon light exposure (Figures 5A and 5B). However, the splicing efficiency changes of these events became opposite or much less dramatic in the rrc1-3 mutant compared with the wild type (Figures 5A to 5C). This pattern also appeared in the heatmaps, where the light-regulated splicing patterns were altered in both rrc1-3 and sfps-2 mutants compared with the wild type (Figure 5D; Supplemental Figures 8D and 8E). Notably, the splicing efficiency changes (Delta_PSI/PIR= PSI/PIR_D-PSI/PIR_Rc) of the 843 splicing events characterized in the rrc1-3 background were also significantly different from the Delta_PSI/PIR of the corresponding events in the wild type (Supplemental Figures 8A to 8C).
Figure 5.
RRC1 Is Involved in Light Regulation of Pre-mRNA Splicing.
(A) and (B) Scatterplots indicate the splicing efficiency changes for 774 light-regulated splicing events (the splicing events with the Delta_PSI/PIR > 3%, FDR < 0.5 while comparing Col-0 dark and Rc-irradiated samples) in Col-0 wild-type (A) and rrc1-3 (B) mutant backgrounds. The x axis is the splicing efficiency (folded root of PSI/PIR) in the dark, and the y axis is the splicing efficiency (folded root of PSI/PIR) after light treatment.
(C) Overlay of (A) and (B). The pair-wise t test indicates the Delta_PSI/PIR of the Col-0 wild type (Delta_PSI/PIR= Col-0_PSI/PIR_D – Col-0_PSI/PIR_Rc) is significantly different from the Delta_PSI/PIR of rrc1 mutant (Delta_PSI/PIR= rrc1-3_PSI/PIR_D – rrc1-3_PSI/PIR_Rc), P < 0.01.
(D) Heatmaps of the quantified splicing profiles of 774 light-regulated splicing events. Delta_PSI/PIR (Delta_PSI/PIR= PSI/PIR_D − PSI/PIR_Rc) values were used to quantify the splicing efficiency changes in response to light. The events are ranked by the value of Delta_PSI/PIR in Col-0 background. Blue color indicates low Z-scores, whereas red color indicates high Z-scores. Red light intensity used was at 7 μmol m−2 s−1.
To independently verify that the light-regulated pre-mRNA splicing events were altered in rrc1-3 compared with the wild type, RT-quantitative (q)PCR assays were performed for selected genes using RNA isolated under the same conditions used for RNA-seq experiment. The light-regulated splicing changes in rrc1-3 captured by RNA sequencing (RNA-seq) analysis were largely reproduced by RT-qPCR assays (Supplemental Figure 9). Overall, these data support our hypothesis that, similar to SFPS, RRC1 might function during light regulation of pre-mRNA splicing and fine-tune the transcript abundance of the light signaling and circadian clock–regulated genes in addition to the transcriptional regulation in Arabidopsis. These data also suggest that the posttranscriptional regulation adds stringency of gene regulation in modulating downstream light responses during photomorphogenesis.
Phytochromes regulate AS in a light-dependent manner (Shikata et al., 2014). To examine whether the splicing changes identified in rrc1-3 and sfps-2 mutants overlap with those reported earlier in the phyAB mutant, a Venn diagram was constructed from the differentially spliced genes displaying changes under dark and light conditions. The differentially spliced genes regulated by RRC1, SFPS, and phyAB in response to light overlapped among all three regulators (Supplemental Figure 10; Supplemental Data Set 3). These data suggest that phytochromes might regulate AS through RRC1 and SFPS in response to light.
RRC1 and SFPS Coordinately Regulate Pre-mRNA Splicing of a Subset of Genes
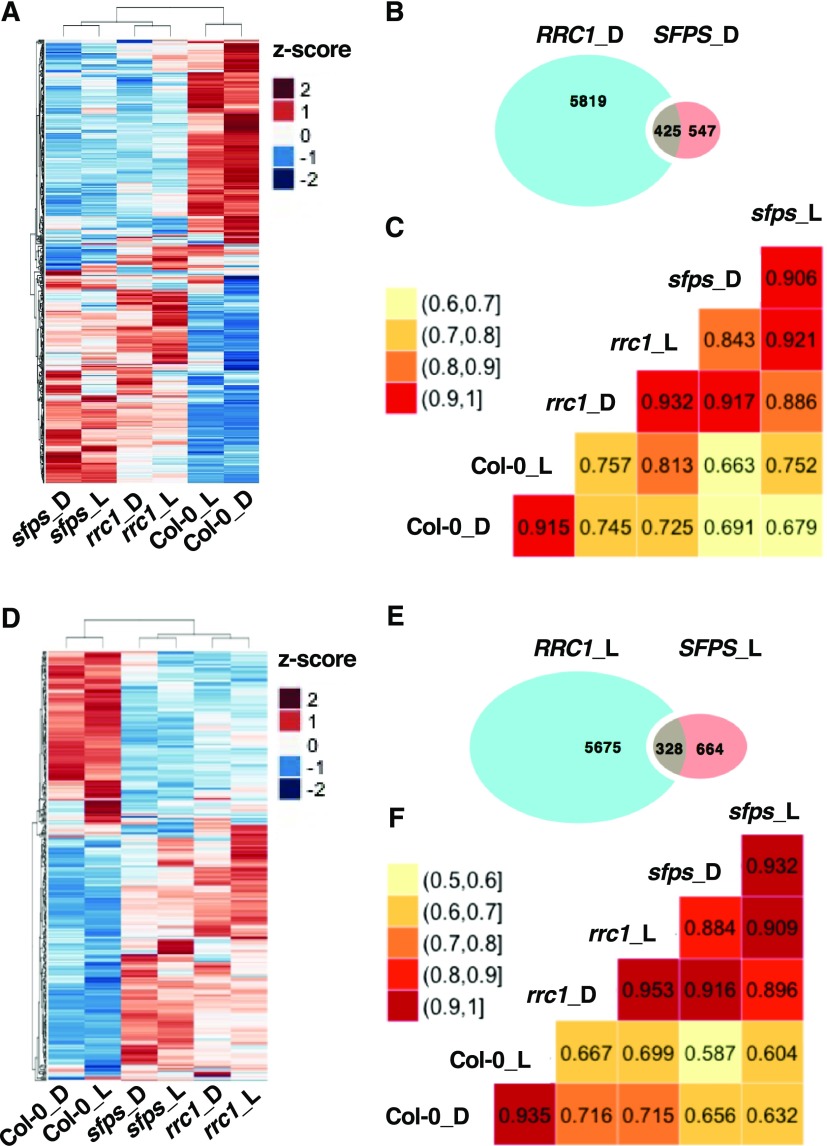
To investigate how RRC1 and SFPS proteins function in the same complex and affect the pre-mRNA splicing, a common subset of genes regulated by these two proteins was identified. The Venn diagrams revealed that 425 and 328 splicing events were coregulated by both SFPS and RRC1 under dark and light conditions, respectively. These events accounted for ∼43.7% (dark) and ∼33% (light) of SFPS-regulated splicing events but only ∼6.8% (dark) and 5.5% (light) RRC1-regulated events (Figures 6B and 6E). As shown in the heatmaps, the splicing efficiency changes of these coregulated splicing events were similar in the rrc1-3 and sfps-2 mutants but almost completely opposite to the patterns in the wild type (Figures 6A and 6D), which also indicates SFPS and RRC1 function in the same complex and coregulate pre-mRNA splicing of a subset of genes. Furthermore, the correlation coefficient matrices were computed to investigate the relationships among rrc1-3, sfps-2, and the wild-type samples, in regard to these coregulated targets under both dark and light conditions. The matrix plots also indicate that the splicing profiles for these coregulated targets in sfps-2 samples have higher correlation with rrc1-3 samples under both dark and light conditions (red color) compared with the wild type (Figures 6C and 6F, yellow color). Taken together, these data further indicate that RRC1 and SFPS function in the same complex collaboratively to regulate a large proportion of the SFPS targets but only a small fraction of the RRC1 targets. When one of these two proteins are missing, the whole complex is unable to function properly, resulting in the splicing site recognition deficiency.
Figure 6.
RRC1 and SFPS Coordinately Regulate Pre-mRNA Splicing of a Subset of Genes.
(A) and (D) Heatmaps of splicing efficiency of the differential spliced events that are coregulated by SFPS and RRC1, based on Z-scores of PSI/PIR values in the dark (A) and light (D) samples. Hierarchical clustering was performed on the differential spliced events that are coregulated by SFPS and RRC1. Blue color represents low Z-scores, whereas red color represents high Z-scores. Red light intensity used was at 7 μmol m−2 s−1.
(B) and (E) Venn graphs show the overlap of the splicing changes in sfps-2 and rrc1-3 mutant backgrounds compared with the wild type under dark (B) and light (E) conditions. In total, 425 and 328 overlapped targets were discovered under dark (B) and light (E) conditions, respectively.
(C) and (F) The correlogram to visualize the correlation of samples, based on the splicing efficiency (PSI/PIR) of the events that are coregulated by SFPS and RRC1 in dark (C) and light (F) conditions. The correlation coefficient of each pair of samples is labeled in the matrix plots.
To validate the RNA-seq results, we selected six common target genes of SFPS and RRC1 with altered splicing patterns for independent verification (Supplemental Figures 11A, 11D, 11G, 11J, 11M, and 11P, left column). Total RNA was isolated from three independent biological replicates from the wild-type, rrc1-3, sfps-2, and sfps-2 rrc1-3 double mutant plants, and RT-qPCR was performed using primers from the splicing junction regions of a particular isoform. Comparison of the RNA-seq (Supplemental Figures 11C, 11F, 11I, 11L, 11O, and 11R) and RT-qPCR (Supplemental Figures 11B, 11E, 11H, 11K, 11N, and 11Q) results confirmed that the RNA-seq data could be largely reproduced by an independent method.
RRC1 and SFPS Regulate Pre-mRNA Splicing of ELF3
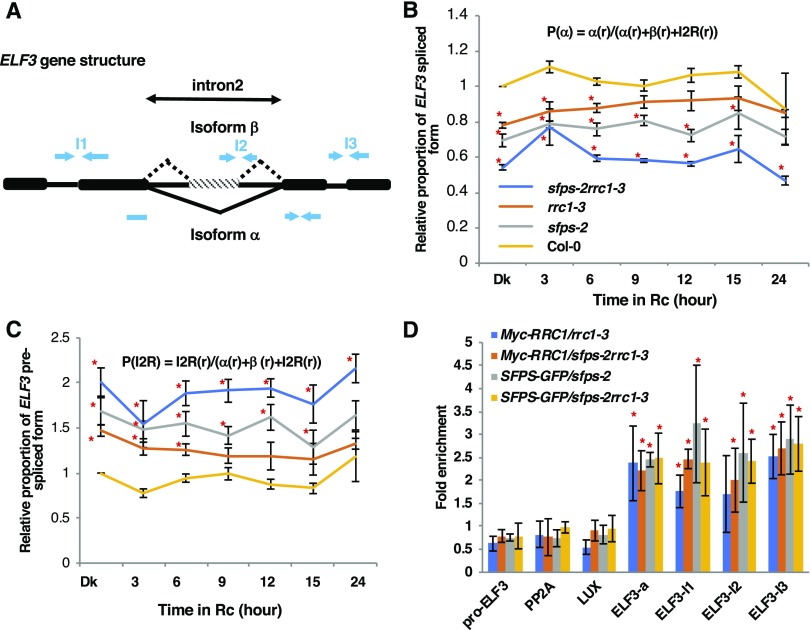
To further determine the mechanisms by which the SFPS-RRC1 complex regulates the pre-mRNA splicing of their common targets, we examined one of the target genes ELF3, for which splicing is defective in both sfps-2 and rrc1-3 (Supplemental Figures 11D to 11F; Kwon et al., 2014; Xin et al., 2017). The elf3 mutant displays a long hypocotyl and early flowering similar to sfps-2 and rrc1-3 mutants (Liu et al., 2001; Nusinow et al., 2011), although the elf3 phenotypes are much stronger than those of these mutants. To independently verify and quantify different isoforms of ELF3 over time under red light, RT-qPCR assays were performed on ELF3 transcripts. The mature spliced form of ELF3 was significantly reduced, while the unspliced form of ELF3 remained significantly higher in the rrc1-3, sfps-2, and sfps-2 rrc1-3 double mutants compared with the wild type (Supplemental Figures 12A and 12B). To eliminate the light-induced transcriptional regulation, the relative proportion of ELF3 transcripts was calculated (Figures 7B and 7C). These data also show that the spliced form of ELF3 is reduced, while the unspliced form of ELF3 is significantly enhanced in the rrc1-3 and sfps-2 mutants. Interestingly, the spliced and unspliced forms were additively downregulated and upregulated, respectively, in the sfps-2 rrc1-3 double mutant compared with sfps-2 and rrc1-3 single mutants (Figures 7B and 7C). These data suggest that both SFPS and RRC1 contribute to regulate splicing of ELF3.
Figure 7.
RRC1 and SFPS Regulate Pre-mRNA Splicing by Direct Association with ELF3 Transcript in Vivo.
(A) Schematic diagram indicates the exon–intron structure of ELF3 and the positions of primers used for the RIP assays.
(B) and (C) Relative proportion of the ELF3 spliced (B) and unspliced (C) forms is shown. Total RNA was extracted from Col-0, sfps-2, rrc1-3, and sfps-2 rrc1-3 double mutant seedlings grown under darkness and dark-grown seedling exposed to continuous red light (at 7 μmol m−2 s−1) for the times indicated. PP2A was used as an internal control. The error bar indicates sem (n = three biological repeats, each repeat includes three technical repeats). Red asterisks indicate significant difference (P < 0.05, based on Student’s t test) from Col-0.
(D) RIP assays show that RRC1 and SFPS associate with ELF3 pre-mRNA in vivo independent of each other. The RNA-protein complex was extracted from the genotypes indicated and immunoprecipitated by anti-GFP/anti-Myc antibodies. The abundance of each gene was quantified by RT-qPCR. The results were normalized to the input of each sample and then normalized to the wild type to calculate the fold-enrichment. Each bar is the mean ± sem (n = 3 independent biological repeats). Red asterisk indicates significant difference (P < 0.05) based on analysis of variance with post hoc Tukey’s honest significant difference test.
Previously, it was shown that SFPS associates with ELF3 pre-mRNA in vivo (Xin et al., 2017). To examine whether RRC1 also associated with ELF3 transcript, in vivo RNA immunoprecipitation (RIP) followed by RT-qPCR assays were performed in three biological replicates. Similar to SFPS, RRC1 associated with the ELF3 pre-mRNA in vivo, with similar strength (Figures 7A and 7D. To distinguish whether the binding of one factor depends on the other factor, RIP assay was performed using SFPS-GFP in sfps-2rrc1-3 and Myc-RRC1 in sfps-2rrc1-3 backgrounds and compared with the binding efficiency of SFPS-GFP in sfps-2 and Myc-RRC1 in rrc1-3 backgrounds, respectively. SFPS binding to ELF3 pre-mRNA did not depend on RRC1 and vice versa (Figure 7D). These data suggest that although these proteins interact with each other to form a complex, they can still bind to some of their common targets independently in vivo.
It is possible that both SFPS and RRC1 do not specifically bind to target RNA directly but rather indirectly by association with other RBPs. Alternatively, although rrc1-3 displayed phenotypes similar to those caused by the null allele rrc1-4 and a defect in ELF3 splicing (Figures 7B and 7C), rrc1-3 is a weak allele of rrc1 mutant and expresses a truncated protein (Shikata et al., 2012), which might interfere with SFPS binding to ELF3 pre-mRNA. However, sfps-2 is a null allele. Thus, RRC1 binding to ELF3 pre-mRNA may not require SFPS. In addition, the resolution of the in vivo RIP assay might not be sufficient to display small changes that might be present in their binding efficiency in each other mutant backgrounds.
RRC1 and SFPS Are Coexpressed and the Expression and Splicing of RRC1 Is Regulated by SFPS
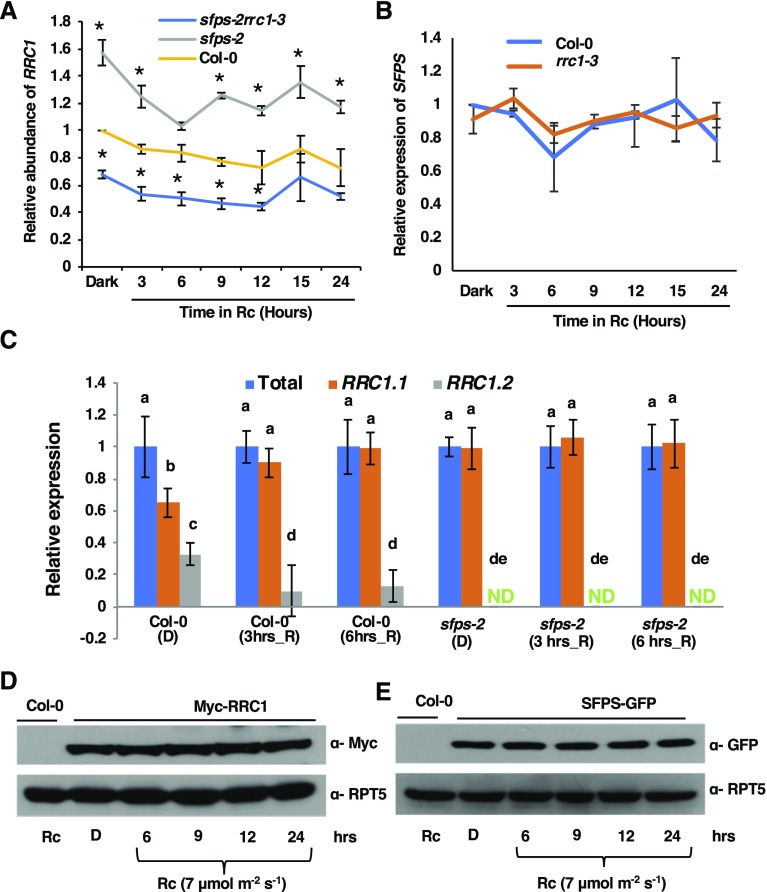
Since SFPS and RRC1 interact with each other and possibly function in a complex regulating similar pathways, coexpression of RRC1 and SFPS would be expected for such a complex to be functionally relevant in vivo. Analysis of the spatial expression patterns of RRC1 and SFPS on the publicly available Electronic Fluorescent Pictograph browser (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) showed that both genes were coexpressed throughout the plant at different developmental stages (Supplemental Figure 13). To examine whether the expression of RRC1 and SFPS was interdependent and/or regulated by light, RT-qPCR analysis was performed. The results indicate that no significant difference in expression was observed for either SFPS or RRC1 in response to light (Figures 8A and 8B). Interestingly, SFPS was found to be a negative regulator of RRC1 gene expression, as the mRNA abundance of RRC1 was significantly upregulated in sfps-2 mutant under both dark- and red-light-treated seedlings compared with the wild type. By contrast, the expression of SFPS was not dependent on RRC1 (Figures 8A and 8B).
Figure 8.
SFPS Regulates the Expression and Splicing of RRC1.
(A) RT-qPCR assays were performed to detect the transcript level of RRC1 in the wild type, sfps-2, and sfps-2rrc1-3 after long-term red-light irradiation (at 7 μmol m−2 s−1).
(B) RT-qPCR to detect the transcript level of SFPS in the wild type, and rrc1-3 after long-term red-light irradiation (at 7 μmol m−2 s−1). Total RNA was extracted from seedlings grown in darkness for 4 d and then transferred to red light for the period indicated. PP2A was used as an internal control. Each bar is the mean ± sem (n = 3 independent biological repeats, and each biological repeat includes three technical repeats).
(C) SFPS regulates the AS of RRC1. RT-qPCR assay was performed to detect total RRC1 transcript as well as two alternative splice variants RRC1.1 and RRC1.2 in the wild-type and sfps-2 mutant background under either dark or red-light (at 7 μmol m−2 s−1)–treated conditions. Total RNA was extracted from seedlings grown in darkness for 4 d and then transferred to red light for the period indicated. PP2A was used as an internal control. Each bar is the mean ± sem (n = 3 independent biological repeats, and each biological repeat includes three technical repeats). Statistical significance among different splice variants was determined using single factor analysis of variance and Tukey’s Tukey honest significant difference tests and is indicated by different letters. ND, not detected.
(D) Immunoblot shows the level of Myc-RRC1 after long-term red-light irradiation (at 7 μmol m−2 s−1).
(E) Immunoblot shows the level of SFPS-GFP after long-term red-light irradiation (at 7 μmol m−2 s−1). Total protein was extracted from the seedlings grown in the darkness for 4 d and then transferred to red light for the period as indicated. Anti-Myc and anti-GFP antibodies were used to detect Myc-RRC1 and SFPS-GFP, respectively. RPT5 was used as a control blot.
Recently, it was reported that the AS of RRC1 is regulated by light (Hartmann et al., 2016). The inclusion of exon 3 results in an RRC1.1 isoform that is translated into a functional protein, while the exclusion of exon 3 introduces a premature stop codon in the RRC1.2 isoform, resulting in a nonfunctional protein (Hartmann et al., 2016). RRC1 was one of the genes that displayed changes not only in AS but also in splicing efficiency of several introns in sfps-2 compared with the wild type under both dark and light conditions (Supplemental Figure 14; Xin et al., 2017). The AS of exon 3 in RRC1 was examined both in dark and light conditions over time in the wild type and sfps-2 mutants using RT-qPCR assays. The functional RRC1.1 isoform was slightly enriched in response to light in wild-type seedlings, as reported previously (Hartmann et al., 2016). However, in the sfps-2 mutant background, the RRC1.1 isoform was predominant under both dark and light conditions (Figure 8C). The altered splicing pattern of RRC1 might contribute to the increased abundance of RRC1 mRNA in the sfps-2 background. Overall, these data suggest that SFPS not only regulates the expression of RRC1 but also the AS and splicing efficiency of RRC1 in dark and light conditions.
To examine whether light affects the RRC1 and SFPS protein stability, dark-grown 35S:Myc-RRC1 and 35S:SFPS-GFP seedlings were exposed to continuous red light over the period of 24 h and the protein levels were tested by immunoblots. These data show that the light signal did not influence the posttranslational stability of either SFPS or RRC1, as SFPS-GFP and Myc-RRC1 protein abundances were similar in both dark- and light-treated seedlings (Figures 8D and 8E). However, because RRC1 undergoes AS in response to light, it is possible that the light signal regulates native RRC1 protein abundance through alternative splicing, which was not visible using the above 35S:Myc-RRC1 construct.
DISCUSSION
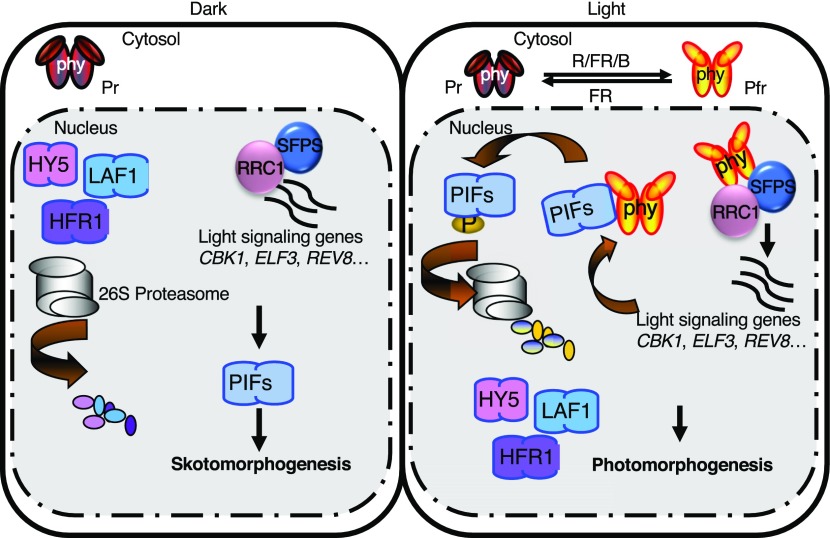
The data presented here provide strong evidence that SFPS and RRC1 form a complex and function as positive regulators in phytochrome signaling pathways. The association of SFPS and RRC1 detected by IP-MS was verified by multiple independent methods including yeast two-hybrid assays, in vivo and in vitro Co-IP, and colocalization assays in living plant cells. The rrc1 and sfps mutants display similar phenotypes as the phyB mutant, including long hypocotyls under light and early flowering. Further genetic analyses showed that the phenotypes of the sfps-2 rrc1-3 double mutant are similar to those of sfps-2, suggesting that these two proteins function, in part, in the same complex while modulating dark/light and vegetative/reproductive transitions. To date, of the many proteins ascribed roles in phytochrome signaling, SFPS and RRC1 are the only proteins regulating pre-mRNA splicing. Except for SFPS and RRC1, two of the five mutants impaired in the circadian clock and splicing (e.g., skip and prmt5) also display photomorphogenic phenotypes (Hong et al., 2010; Sanchez et al., 2010; Jones et al., 2012; Wang et al., 2012; Hernando et al., 2015; Schlaen et al., 2015). However, a role for these proteins in phytochrome signaling is still unknown. Thus, SFPS and RRC1 represent a distinct branch of phytochrome signaling components controlling pre-mRNA splicing to optimize photomorphogenesis (Figure 9).
Figure 9.
Model of RRC1-SFPS Function in Regulating Photomorphogenesis.
(Left) PIFs are constitutively nucleus localized, whereas phytochromes are in the cytosol in darkness. PIFs repress photomorphogenesis in the dark and light by promoting skotomorphogenesis. (Right) In response to light, phytochromes migrate into the nucleus, interact with PIFs, and induce rapid degradation of PIFs to promote photomorphogenesis. In addition, SFPS and RRC1 regulate pre-mRNA splicing of light signaling genes (e.g., CKB1, ELF3, REV8, and others) to fine-tune the light-regulated developmental processes in Arabidopsis. Phytochromes also interact with SFPS and RRC1 in vivo and regulate AS in response to light to promote photomorphogenesis.
Deep RNA-seq data indicate that RRC1 strongly regulates pre-mRNA splicing of a large number of genes under both dark and light conditions. While all four bona fide AS categories (IR, ES, and 3ʹ- or 5ʹ-alt) are affected in the mutant, the most prominent changes are the CS events. This is similar to the sfps mutant, which displayed a large number of CS events (Xin et al., 2017). Most of these CS events have not been categorized as AS events inThe Arabidopsis Information Resource 10 genome annotation. Thus, many of these may include either uncharacterized AS events and/or CS events in the wild-type plants that are altered in the rrc1 mutant. The RRC1 ortholog in human, SR140, is part of the U2-snRNP complex and participates in CS of pre-mRNAs (Will et al., 2002). Thus, RRC1 might be necessary for proper splicing of a large number of genes under dark and light conditions, such that mutation of RRC1 results in mis-splicing of these genes.
GO analysis revealed the enrichment of many biological processes including light stimulus, red or far-red light signaling pathway, photosynthesis, and circadian rhythm and others, suggesting that RRC1 might regulate many biological processes in addition to light signaling. Compared to the genes regulated by SFPS (∼950), RRC1 has a much broader influence (∼4200 genes) in regulating different developmental processes in Arabidopsis. Comparison of the RNA sequencing data also indicate that RRC1 and SFPS coregulate a subset of genes (∼400 splicing events), including several important regulators in the clock and light signaling pathways (e.g., ELF3, REV8, NPH3, and PAPP5). The coregulated genes account for only a small proportion of the differentially spliced loci in the rrc1 mutant (SFPS, ∼44% versus RRC1, ∼5% to 6%). At the same time, each protein regulates its own unique genes as well. These data suggest that RRC1 plays a more prominent role in the pre-mRNA splicing process compared to SFPS in Arabidopsis. Thus, these two proteins could function, in part, in the same complex regulating a small number of genes but also have distinct regulatory roles in plant development.
Although the detailed regulatory mechanisms have not been elucidated yet, our data and those of others suggest that the light signal affects both the splicing and transcription cascades as has recently been shown for cold responses (Calixto et al., 2018). Through RNA-seq, we have not only identified the potential targets of SFPS and RRC1 but also characterized many secondary effects that likely arise from the expression changes of the direct binding targets. For instance, when comparing the sequencing data between the wild type and mutants, one of the most significantly enriched categories is the RBPs, involved in mRNA processing (e.g., SR45, RS31, SR30, SKIP, and others) of which many changes have been experimentally verified by independent methods (Shikata et al., 2012; Xin et al., 2017). These splicing factors might undergo AS regulation and form distinct splicing networks. These particular splicing networks might adjust the whole-genome pre-mRNA splicing activities accordingly, when grown under different growth conditions or developmental stages (such as dark/light transitions or vegetative-to-reproduction growth). This phenomenon has also been observed in several independent sequencing studies, even across different species (Shikata et al., 2014; Wu et al., 2014). Thus, forming unique splicing networks through regulated pre-mRNA splicing of these splicing factors might be required to maintain the homeostasis of the cells, as well as giving immediate responses to outside stimuli.
In addition, SFPS and RRC1 control the pre-mRNA splicing of several vital transcription factors involved in the light signaling and circadian clock pathways (e.g., ELF3, REV8, PIF3, and others), which forms another layer of transcriptional regulation during the light responses. These transcription factors in turn regulate the transcription of PIFs, especially PIF4 and PIF5, further contributing to the secondary level of gene expression regulation. This indirect effect also explained why differentially expressed loci have little overlap with the differentially spliced loci in the sfps-2 and rrc1-3 mutants (Xin et al., 2017). The interconnected networks between transcription and splicing help the plants form a robust system (Kitano, 2004) that contributes to the establishment of the final transcriptome in plants.
Although a few studies have been done so far to understand how splicing regulators fine-tune light responses, the mechanisms by which these factors regulate light signaling are still unknown. It has been reported that the splicing regulatory proteins recognize distinct RNA sequences across the transcriptome (Fu and Ares, 2014; Lee and Rio, 2015), which suggests the existence of the splicing code. Deciphering the splicing code will require a comprehensive list of RBPs and their cis binding sites. For the RBPs functioning in the same complex, the target binding specificity and efficiency are usually influenced by each other. Our data suggest a collaborative role for SFPS and RRC1 in the same complex during fine-tuning of light responses; however, the detailed molecular mechanisms by which these two splicing factors regulate the target binding activities of each other have not been revealed yet.
Light signals perceived by phytochromes might regulate pre-mRNA splicing in multiple ways including regulation of the circadian clock, retrograde signaling pathway through chloroplast development, photosynthesis, and energy availability (Hong et al., 2010; Sanchez et al., 2010; Jones et al., 2012; Wang et al., 2012; Petrillo et al., 2014; Schlaen et al., 2015; Hartmann et al., 2016). Light also regulates AS by controlling the rate of transcriptional elongation by RNA polymerase II (Godoy Herz et al., 2019). Among these pathways, SFPS and RRC1 appear to function early and more directly, as these factors physically interact with photoactivated phyB. Based on the data presented here and elsewhere, we propose several potential hypotheses. First, the light signal perceived by phyB does not appear to regulate the abundance of SFPS and RRC1, despite phyB regulating the abundance of both positive- and negatively acting transcription factors posttranslationally (Pham et al., 2018). In addition, the light signal does not regulate the association of SFPS with RRC1 in vivo. Thus, phyB might regulate the activity of SFPS and RRC1. Current data show that the light signal affects the pre-mRNA splicing of RRC1 possibly through SFPS (Hartmann et al., 2016), whereas no obvious transcriptional regulation of RRC1 and SFPS has been detected by RT-qPCR upon light irradiation (Xin et al., 2017). Additionally, SR proteins usually undergo phosphorylation, which influences their target binding capacity (van Der Houven Van Oordt et al., 2000; de la Fuente van Bentem et al., 2006). Phytochromes have been shown to function as protein kinases (Yeh and Lagarias, 1998; Shin et al., 2016). Thus, RRC1 and/or SFPS might be phosphorylated by phytochromes in responses to light stimuli. Finally, light signals might affect spliceosome assembly, including affecting the association of the SFPS-RRC1 complex with the core spliceosome during transcription. The association of phyB with SFPS and/or RRC1 might play a vital regulatory role during this process. Further biochemical experiments are necessary to fully understand the detailed mechanisms by which phytochromes regulate pre-mRNA splicing.
In conclusion, phytochromes control the transcription of a large number of genes by regulating the activity and/or abundance of a diverse group of transcription factors. In darkness, light responses are suppressed by the negative components, such as PIFs, while positive regulators such as LONG HYPOCOTYL IN FAR-RED1 (HFR1), LONG HYPOCOTYL5 (HY5), and LONG AFTER FAR-RED LIGHT1 (LAF1) are degraded by the 26S proteasome. Upon light exposure, the negative components (PIFs) are degraded by the proteasome; however, the positive regulators are induced transcriptionally and/or stabilized posttranslationally, which initiates the light responses (Pham et al., 2018). Many efforts have focused on understanding the role of transcriptional programs in regulating photomorphogenesis; this study and others highlight the importance of posttranscriptional contributions (e.g., pre-mRNA splicing) in regulating photomorphogenesis (Xin et al., 2017; Zhang et al., 2017; Cheng and Tu, 2018). Thus, splicing networks composed of highly interconnected factors (e.g., SFPS and RRC1) facilitate the responses to external stimuli, such as light signals to fine tune photomorphogenesis (Figure 9). Identification and characterization of direct targets of SFPS and RRC1 will help further understand the mechanisms by which these proteins regulate photomorphogenesis.
METHODS
Plant Materials, Growth Conditions, and Measurements
The wild-type and mutant seeds used were in Col-0 background. Seeds were surface sterilized, plated on Murashige and Skoog medium, and stratified as described previously (Xin et al., 2017). Stratified seeds were exposed to white light for 3 h to induce germination before placing them back in the dark. To grow plants, 10- to 12-d-old seedlings were transplanted into pots containing moistened Metro-mix 200 soil (Sun Gro Horticulture) and grown under continuous light at 22°C.
To measure hypocotyl length, seedlings were grown either under continuous dark or different light (red, far-red, or blue) regimes. Images of 4-d-old seedlings were obtained, and absolute hypocotyl length of seedlings was measured using ImageJ tool. To document flowering time, plants were either grown under a short-day (8-h-light/16-h-dark) or a long-day (16-h-light/8-h-dark) regime. Number of days for flowering and the average number of rosette leaves at the time of flowering were counted. Experiments were performed in triplicates, and each set contained at least 30 seedlings/plants.
Construction of Vectors
To prepare 35Spro:Myc-RRC1, the full-length RRC1 (At5G25060) open reading frame (ORF) without a stop codon was amplified using the primers listed in Supplemental Data Set 4 and directionally cloned into pENTR following the protocol of pENTR/D-TOPO Cloning Kit (Life Technologies). The pENTR-RRC1 vector was recombined with the pEARLEYGATE203 (Earley et al., 2006) destination vector containing N-terminal Myc-tag using commercially available LR Clonase II enzyme mix (Life Technologies). To prepare 35Spro:RRC1-GFP, the pENTR-RRC1 vector was recombined with pB7FWG2 destination vector, containing C-terminal GFP-tag using LR Clonase II enzyme mix. To prepare 35Spro:RRC1-mCherry, the full-length RRC1 ORF was amplified using primers listed in Supplemental Data Set 4 and cloned into BamHI and XbaI restriction sites of vector. Preparation of 35Spro:SFPS-GFP was described previously by Xin et al. (2017). Binary vectors were transferred to Agrobacterium strain GV3101 and then transformed into plants through floral dip method as described previously by Clough and Bent (1998).
To construct pVP13-RRC1 bacterial expression vector, the pENTR-RRC1 vector was recombined with pVP13 destination vector, containing an N-terminal MBP-tag using LR Clonase II enzyme mix. For the pVP13-RRC1∆RS vector, truncated RRC1 without the C-terminal RS domain was amplified using the primers listed in Supplemental Data Set 4 and directionally cloned into pENTR following the protocol of pENTR/D-TOPO Cloning Kit (Life Technologies). The pENTR- RRC1∆RS was recombined with pVP13 destination vector using LR Clonase II enzyme mix. To prepare pDEST15-SFPS bacterial expression vector, full-length SFPS ORF without a stop codon was PCR amplified using primers listed in Supplemental Data Set 4 and directionally cloned into pENTR vector. The pENTR-SFPS vector was recombined with the pDEST15 destination vector (Thermo Fisher Scientific) containing N-terminal GST-tag using commercially available LR Clonase II enzyme mix. These vectors were transformed into Escherichia coli strain BL21 (DE3) for protein purification.
To prepare pGBKT7-RRC1 and pGBT9-RRC1 bait vector for yeast two-hybrid, full-length RRC1 ORF was amplified using primers mentioned in Supplemental Data Set 4 and cloned into BamHI/PstI restriction sites of pGBKT7 and pGBT9 vector. To prepare pGADT7-SFPS prey vector for yeast two-hybrid, full-length SFPS ORF was amplified with primers mentioned in the Supplemental Data Set 4 and cloned into BamHI/XhoI restriction sites of pGADT7 vector. To prepare pGAD424-RRC1 full-length RRC1 ORF was amplified using primers mentioned in Supplemental Data Set 4 and cloned into BamHI/PstI restriction sites of pGAD424. Preparation of BD-phyB constructs was described previously (Shimizu-Sato et al., 2002).
To prepare pYES2-PHYB-GFP vector, the full-length PHYB ORF was amplified using primers mentioned in Supplemental Data Set 4 and cloned into KpnI/SmaI restriction sites of pEZS-NL vector. Then the full-length PHYB-GFP was PCR amplified using primers mentioned in Supplemental Data Set 4 and cloned into KpnI/XbaI restriction sites of pYES2 vector (Life Technologies). The pYES2-PHYB-GFP construct was transformed into RKY1293 yeast (Saccharomyces cerevisiae) strain.
Yeast and Bacterial Protein Purification
To induce protein in bacterial culture, 2 mL of overnight-grown culture was transferred to a new flask containing 200 to 800 mL of Luria-Bertani medium and 100 μg/mL ampicillin. Flasks were incubated in the rotary shaker at 37°C and rotating at 200 rpm. Isopropyl β-d-thiogalactoside (final concentration, 0.1 mM) was added to the cultures reading OD = 0.5 and incubated on rotary shaker at 18°C and rotating at 250 rpm for 12 h for protein induction. Bacteria were pelleted at the end of the incubation period. Protein induction in yeast was performed following commercially available protocol (Invitrogen), with small modification. Phycocyanobilin (catalog no. P14137, Frontier Scientific) was included in the media during the protein induction.
In Vivo Co-IP and in Vitro Pull-Down Assays
To perform Co-IP assay, 4-d-old dark-grown seedlings that were either kept in the dark or exposed to constant red light for the indicated time period were frozen and ground in native extraction buffer. 35Spro:Myc-RRC1/SFPSpro:SFPS-GFP double transgenic seedlings were used to detect interaction between RRC1 and SFPS. SFPS-GFP was immunoprecipitated using anti-GFP antibody (catalog no. ab6556, Abcam), and the interacting partner Myc-RRC1 was detected using anti-Myc antibodies (catalog no. 2276S, Cell Signaling Technologies). 35Spro:Myc-RRC1 seedlings were used to detect interaction between RRC1 and phyB. Myc-RRC1 was immunoprecipitated using anti-Myc antibody (catalog no. 2276S, Cell Signaling Technologies), and the interacting partner was detected using anti-phyB antibody (catalog no. sc-9996, Santa Cruz Biotechnology). Composition of the native buffer and detailed Co-IP protocol were as described previously (Zhu et al., 2015).
To perform in vitro pull-down assays, bacterially expressed GST, GST-RRC1, GST-PIF1, GST-SFPS, MBP, MBP-RRC1, and MBP-RRC1∆RS were purified following the commercial kit protocol (catalog no. PI-16100, Thermo Fisher Scientific; catalog no. E8021S, New England Biolabs). To pull down MBP-RRC1, equal amounts of purified MBP-RRC1 were added to the glutathione beads containing attached GST or GST-SFPS. Tubes were incubated in rotatory mixer with gentle mixing for 3 h at 4°C. To pull down GST-SFPS, an equal amount of purified GST-SFPS was added to amylose beads containing attached MBP, MBP-RRC1, or MBP-RRC1∆RS. These tubes were gently mixed in a rotatory mixer for 3 h at 4°C. To pull down phyB-GFP, equal amounts of crude extracts of phyB-GFP were added to glutathione beads containing attached GST, GST-RRC1, or GST-PIF1. One set of tubes was kept in the dark (phyB-Pr), while another set was exposed to red light (at 7 μmol m−2 s−1; phyB-Pfr). After the incubation, glutathione or amylose beads were separated and washed thoroughly three times with 1× PBS containing 0.1% Tween 20. After final wash, beads were immersed in 1× SDS loading dye and boiled for 5 min at 65°C. Proteins were resolved on 6% SDS-PAGE gel. GST, GST-RRC1, GST-SFPS, and GST-PIF1 were detected using anti–GST-horseradish peroxidase antibodies (catalog no. RPN1236, GE Healthcare). MBP-RRC1 and phyB-GFP were detected using anti-MBP (catalog no. E8032S, New England Biolabs) and anti-GFP antibodies (catalog no. sc-9996, Santa Cruz Biotechnology).
Yeast Two-Hybrid Assays
pGBKT7-RRC1 and pGADT7-SFPS vectors were transformed into yeast strain AH109, and positive double transformants were selected on yeast drop-out medium (SD−Ade−His−Leu−Trp). pGAD424-RRC1, and D153-PHYB were cotransformed into yeast strain Y187 transformants were selected on yeast synthetic drop-out medium (SD−Leu−Trp). Liquid β-galactosidase assay to test the interaction between two proteins was performed following commercial protocol (Matchmaker Two-Hybrid System, Clontech Laboratories).
Transient Expression Assays
35Spro:RRC1-GFP was coinjected with 35Spro:U2Aʹ-mCherry, 35Spro:U2AF35A-mCherry, or 35Spro:U2AF65B-mCherry into Nicotiana benthamiana leaves using the infiltration method described previously (Xin et al., 2017). Colocalization of both proteins was observed between 24 and 36 h after infiltration under confocal microscope.
RNA-Seq Data Analysis
Four-day-old dark-grown seedlings of rrc1-3 with or without 3 h of red-light treatments were frozen. Total RNA isolation, preparation of samples for RNA-seq, and the data analysis were performed as described previously (Xin et al., 2017). Sequencing was performed on an Illumina Hiseq4000 instrument with 2 × 150-bp paired-ends reads. The sequencing yield is ∼120 million reads per sample. For each library, more than 90% of the reads were mapped to the unique loci of The Arabidopsis Information Resource 10 genome with the Tophat2 pipeline (Kim et al., 2013). To identify defects in rrc1-3 mutant, these data were compared with a previously published wild-type data set where the wild-type seedlings were grown under identical conditions and sequenced using identical methods as the rrc1-3 mutant (Xin et al., 2017). Differential gene expression was analyzed through Cufflink and Cuffdiff pipelines (Trapnell et al., 2012). Genes with FDR values lower than 0.05 and absolute log twofold change greater than 0.58 (1.5-fold) were considered as differentially expressed. AS was analyzed through ASpli_1.9 as part of the Bioconductor R package (Mancini et al., 2017), which makes use of junction reads information and quantifies the pre-mRNA splicing events through calculating PSI and PIR matrix {formulas: PSI(Altss) = #Jinclusion/(#Jinclusion + #Jexclusion); PSI(exon skipping) = (#Jstart + #Jend)/(#Jstart + #Jend + 2#Jexclusion); PIR(IR) = (#E1l + #IE2)/(#E1l + #IE2 + 2#jE1E2)}. The AS events with an absolute FDR < 5% and Delta PSI_PIR > 3% were deemed differentially spliced. The Z-scores of PSI_PIR values 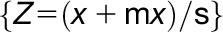 were calculated to generate heatmaps, where x is the value of the PSI_PIR, μ is the population mean, and σ is the sd. Square roots of PSI_PIR values
were calculated to generate heatmaps, where x is the value of the PSI_PIR, μ is the population mean, and σ is the sd. Square roots of PSI_PIR values 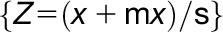 are calculated for scatterplots. Raw sequences (fastq files) and counts of genes, exons, introns, AS bins, and junctions used in this study have been deposited in the Gene Expression Omnibus database (accession number GSE114995). Primer sequences used for independent verification by RT-qPCR are listed in Supplemental Data Set 4.
are calculated for scatterplots. Raw sequences (fastq files) and counts of genes, exons, introns, AS bins, and junctions used in this study have been deposited in the Gene Expression Omnibus database (accession number GSE114995). Primer sequences used for independent verification by RT-qPCR are listed in Supplemental Data Set 4.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: PHYB (At2g18790), PIF4 (At2g43010), PIF5 (At3g59060), SFPS (AT1G30480), RRC1 (AT5G25060). Raw sequences (fastq files) and counts of genes, exons, introns, AS bins, and junctions used in this study have been deposited in the Gene Expression Omnibus database (accession no. GSE114995).
Supplemental Data
Supplemental Figure 1. IP-MS (Immunoprecipitation followed by mass spectrometry) to identify the interacting partner for SFPS.
Supplemental Figure 2. Nuclear speckle formation of RRC1-mCherry in sfps-2 background in Arabidopsis seedlings.
Supplemental Figure 3. Ectopically expressed 35Spro:Myc-RRC1 complements the hyposensitive phenotype of rrc1-3 mutant in response to red light.
Supplemental Figure 4. RRC1 regulates gene expression both in the dark and red light conditions.
Supplemental Figure 5. RRC1 co-localizes with U2 snRNP-associated component in discrete nuclear foci.
Supplemental Figure 6. GO-term and KEGG pathway enrichment analysis of genes that displayed AS pattern changes between wild type and rrc1-3 in the dark and light samples.
Supplemental Figure 7. Overlap between differentially expressed and differentially spliced genes regulated by RRC1 in the dark and light.
Supplemental Figure 8. RRC1 is involved in light regulation of pre-mRNA splicing.
Supplemental Figure 9. RRC1 is involved in light regulation of pre-mRNA splicing.
Supplemental Figure 10. Venn diagram of genes for which pre-mRNA splicing is regulated by phytochrome, SFPS, and RRC1.
Supplemental Figure 11. Validation of RNA-seq data by RT-qPCR for selected light signaling genes.
Supplemental Figure 12. RT-qPCR analyses of the mature and unspliced forms of ELF3 over time.
Supplemental Figure 13. Co-expression analyses of SFPS and RRC1 in Arabidopsis.
Supplemental Figure 14. The splicing changes of RRC1 in wild type and sfps-2 mutant under dark and light conditions, respectively.
Supplemental Data Set 1. List of genes differentially expressed in rrc1-3 under dark and light conditions and their GO analyses.
Supplemental Data Set 2. List of genes differentially spliced in rrc1-3 under dark and light conditions and their GO analyses.
Supplemental Data Set 3. List of genes differentially expressed and spliced in phyAB mutant under dark and light conditions and their GO analyses.
Supplemental Data Set 4. Primer sequences used in the experiments described in the text.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Marcelo J. Yanovsky for sharing ASpli_1.9 software and helping with data analyses, Inyup Paik for sharing pYES2-phyB-GFP expression vector, Mei-Chun Cheng for graphic design, Arlen Johnson for sharing RKY1293 yeast cells, and the members of the Huq laboratory for critical reading of the article. This work was supported by National Institutes of Health (grant GM-114297) and the National Science Foundation (MCB-1543813 to E.H.). We acknowledge the Texas Advanced Computing Center at The University of Texas at Austin for providing high performance computing, visualization, and database resources that have contributed to the research results reported in this aricle.
AUTHOR CONTRIBUTIONS
R.X., P.K.K., and E.H. designed experiments. R.X. and P.K.K. carried out experiments. R.X., P.K.K., and E.H. analyzed data. R.X., P.K.K., and E.H. wrote the article.
References
- Amrein H., Gorman M., Nöthiger R. (1988). The sex-determining gene tra-2 of Drosophila encodes a putative RNA binding protein. Cell 55: 1025–1035. [DOI] [PubMed] [Google Scholar]
- Bae G., Choi G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59: 281–311. [DOI] [PubMed] [Google Scholar]
- Barta A., Kalyna M., Reddy A.S.N. (2010). Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 22: 2926–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs R.T., Gregor P., Idriss S., Belote J.M., McKeown M. (1987). Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell 50: 739–747. [DOI] [PubMed] [Google Scholar]
- Calixto C.P.G., Guo W., James A.B., Tzioutziou N.A., Entizne J.C., Panter P.E., Knight H., Nimmo H.G., Zhang R., Brown J.W.S. (2018). Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. Plant Cell 30: 1424–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho R.F., Szakonyi D., Simpson C.G., Barbosa I.C.R., Brown J.W.S., Baena-González E., Duque P. (2016). The Arabidopsis SR45 splicing factor, a negative regulator of sugar signaling, modulates SNF1-related protein kinase 1 stability. Plant Cell 28: 1910–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.-L., Tu S.-L. (2018). Alternative splicing and cross-talk with light signaling. Plant Cell Physiol. 59: 1104–1110. [DOI] [PubMed] [Google Scholar]
- Chou T.B., Zachar Z., Bingham P.M. (1987). Developmental expression of a regulatory gene is programmed at the level of splicing. EMBO J. 6: 4095–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- de la Fuente van Bentem S., Anrather D., Roitinger E., Djamei A., Hufnagl T., Barta A., Csaszar E., Dohnal I., Lecourieux D., Hirt H. (2006). Phosphoproteomics reveals extensive in vivo phosphorylation of Arabidopsis proteins involved in RNA metabolism. Nucleic Acids Res. 34: 3267–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Erkelenz S., Mueller W.F., Evans M.S., Busch A., Schöneweis K., Hertel K.J., Schaal H. (2013). Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA 19: 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.D. (1995). The superfamily of arginine/serine-rich splicing factors. RNA 1: 663–680. [PMC free article] [PubMed] [Google Scholar]
- Fu X.-D., Ares M. Jr (2014). Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15: 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy Herz M.A., Kubaczka M.G., Brzyżek G., Servi L., Krzyszton M., Simpson C., Brown J., Swiezewski S., Petrillo E., Kornblihtt A.R. (2019). Light regulates plant alternative splicing through the control of transcriptional elongation. Mol. Cell 73: 1066–1074.e3. [DOI] [PubMed] [Google Scholar]
- Hartmann L., Drewe-Boß P., Wießner T., Wagner G., Geue S., Lee H.-C., Obermüller D.M., Kahles A., Behr J., Sinz F.H., Rätsch G., Wachter A. (2016). Alternative splicing substantially diversifies the transcriptome during early photomorphogenesis and correlates with the energy availability in Arabidopsis. Plant Cell 28: 2715–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando C.E., Sanchez S.E., Mancini E., Yanovsky M.J. (2015). Genome wide comparative analysis of the effects of PRMT5 and PRMT4/CARM1 arginine methyltransferases on the Arabidopsis thaliana transcriptome. BMC Genomics 16: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Song H.R., Lutz K., Kerstetter R.A., Michael T.P., McClung C.R. (2010). Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 21211–21216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.A., Williams B.A., McNicol J., Simpson C.G., Brown J.W., Harmer S.L. (2012). Mutation of Arabidopsis spliceosomal timekeeper locus1 causes circadian clock defects. Plant Cell 24: 4066–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. (2004). Biological robustness. Nat. Rev. Genet. 5: 826–837. [DOI] [PubMed] [Google Scholar]
- Klose C., Viczián A., Kircher S., Schäfer E., Nagy F. (2015). Molecular mechanisms for mediating light-dependent nucleo/cytoplasmic partitioning of phytochrome photoreceptors. New Phytol. 206: 965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y.-J., Park M.-J., Kim S.-G., Baldwin I.T., Park C.-M. (2014). Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. BMC Plant Biol. 14: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Rio D.C. (2015). Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84: 291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.L., Covington M.F., Fankhauser C., Chory J., Wagner D.R. (2001). ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.C., Caceres J.F. (2009). The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 417: 15–27. [DOI] [PubMed] [Google Scholar]
- Mancini E., Iserte J., Yanovsky M.J., Chemomoretz A. (2017). ASpli: Analysis of alternative splicing using RNA-Seq. R package version 1.6.0. https://rdrr.io/bioc/ASpli/man/ASpli-package.html#heading-1
- Matera A.G., Wang Z. (2014). A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15: 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrillo E., Godoy Herz M.A., Fuchs A., Reifer D., Fuller J., Yanovsky M.J., Simpson C., Brown J.W.S., Barta A., Kalyna M., Kornblihtt A.R. (2014). A chloroplast retrograde signal regulates nuclear alternative splicing. Science 344: 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham V.N., Kathare P.K., Huq E. (2018). Phytochromes and phytochrome interacting factors. Plant Physiol. 176: 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P.H. (2007). Phytochrome-regulated gene expression. J. Integr. Plant Biol. 49: 11–20. [Google Scholar]
- Reddy A.S.N., Shad Ali G. (2011). Plant serine/arginine-rich proteins: Roles in precursor messenger RNA splicing, plant development, and stress responses. Wiley Interdiscip. Rev. RNA 2: 875–889. [DOI] [PubMed] [Google Scholar]
- Reddy A.S.N., Marquez Y., Kalyna M., Barta A. (2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson D.N., Rogers M.F., Labadorf A., Ben-Hur A., Guo H., Paterson A.H., Reddy A.S.N. (2011). Comparative analysis of serine/arginine-rich proteins across 27 eukaryotes: Insights into sub-family classification and extent of alternative splicing. PLoS One 6: e24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M.B., Zahler A.M., Stolk J.A. (1991). A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J. Cell Biol. 115: 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S.E., et al. (2010). A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468: 112–116. [DOI] [PubMed] [Google Scholar]
- Schindler S., Szafranski K., Hiller M., Ali G.S., Palusa S.G., Backofen R., Platzer M., Reddy A.S. (2008). Alternative splicing at NAGNAG acceptors in Arabidopsis thaliana SR and SR-related protein-coding genes. BMC Genomics 9: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaen R.G., Mancini E., Sanchez S.E., Perez-Santángelo S., Rugnone M.L., Simpson C.G., Brown J.W., Zhang X., Chernomoretz A., Yanovsky M.J. (2015). The spliceosome assembly factor GEMIN2 attenuates the effects of temperature on alternative splicing and circadian rhythms. Proc. Natl. Acad. Sci. USA 112: 9382–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata H., Shibata M., Ushijima T., Nakashima M., Kong S.-G., Matsuoka K., Lin C., Matsushita T. (2012). The RS domain of Arabidopsis splicing factor RRC1 is required for phytochrome B signal transduction. Plant J. 70: 727–738. [DOI] [PubMed] [Google Scholar]
- Shikata H., Hanada K., Ushijima T., Nakashima M., Suzuki Y., Matsushita T. (2014). Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 18781–18786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S., Huq E., Tepperman J.M., Quail P.H. (2002). A light-switchable gene promoter system. Nat. Biotechnol. 20: 1041–1044. [DOI] [PubMed] [Google Scholar]
- Shin A.-Y., Han Y.-J., Baek A., Ahn T., Kim S.Y., Nguyen T.S., Son M., Lee K.W., Shen Y., Song P.-S., Kim J.-I. (2016). Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat. Commun. 7: 11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Brown J.W.S. (2013). Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25: 3640–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Der Houven Van Oordt W., Newton K., Screaton G.R., Cáceres J.F. (2000). Role of SR protein modular domains in alternative splicing specificity in vivo. Nucleic Acids Res. 28: 4822–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. (2012). SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 24: 3278–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L., Lürhmann R. (2006). Spliceosome structure and function. In The RNA World, 3rd ed., Gesteland R.F., Cech T.R., Atkins J.F., eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ), pp. 369–400. [Google Scholar]
- Will C.L., Urlaub H., Achsel T., Gentzel M., Wilm M., Lührmann R. (2002). Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 21: 4978–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.-P., Su Y.S., Chen H.-C., Chen Y.-R., Wu C.-C., Lin W.-D., Tu S.-L. (2014). Genome-wide analysis of light-regulated alternative splicing mediated by photoreceptors in Physcomitrella patens. Genome Biol. 15: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin R., Zhu L., Salomé P.A., Mancini E., Marshall C.M., Harmon F.G., Yanovsky M.J., Weigel D., Huq E. (2017). SPF45-related splicing factor for phytochrome signaling promotes photomorphogenesis by regulating pre-mRNA splicing in Arabidopsis. Proc. Natl. Acad. Sci. USA 114: E7018–E7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K.C., Lagarias J.C. (1998). Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95: 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Lin C., Gu L. (2017). Light regulation of alternative pre-mRNA splicing in plants. Photochem. Photobiol. 93: 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Bu Q., Xu X., Paik I., Huang X., Hoecker U., Deng X.W., Huq E. (2015). CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat. Commun. 6: 7245. [DOI] [PubMed] [Google Scholar]