The functional organization of ribosomal DNA and ribosome assembly in plants is described, along with recent findings concerning a plant-specific mechanism involved in ribosome stress responses.
Abstract
The transcription of 18S, 5.8S, and 18S rRNA genes (45S rDNA), cotranscriptional processing of pre-rRNA, and assembly of mature rRNA with ribosomal proteins are the linchpins of ribosome biogenesis. In yeast (Saccharomyces cerevisiae) and animal cells, hundreds of pre-rRNA processing factors have been identified and their involvement in ribosome assembly determined. These studies, together with structural analyses, have yielded comprehensive models of the pre-40S and pre-60S ribosome subunits as well as the largest cotranscriptionally assembled preribosome particle: the 90S/small subunit processome. Here, we present the current knowledge of the functional organization of 45S rDNA, pre-rRNA transcription, rRNA processing activities, and ribosome assembly factors in plants, focusing on data from Arabidopsis (Arabidopsis thaliana). Based on yeast and mammalian cell studies, we describe the ribonucleoprotein complexes and RNA-associated activities and discuss how they might specifically affect the production of 40S and 60S subunits. Finally, we review recent findings concerning pre-rRNA processing pathways and a novel mechanism involved in a ribosome stress response in plants
INTRODUCTION
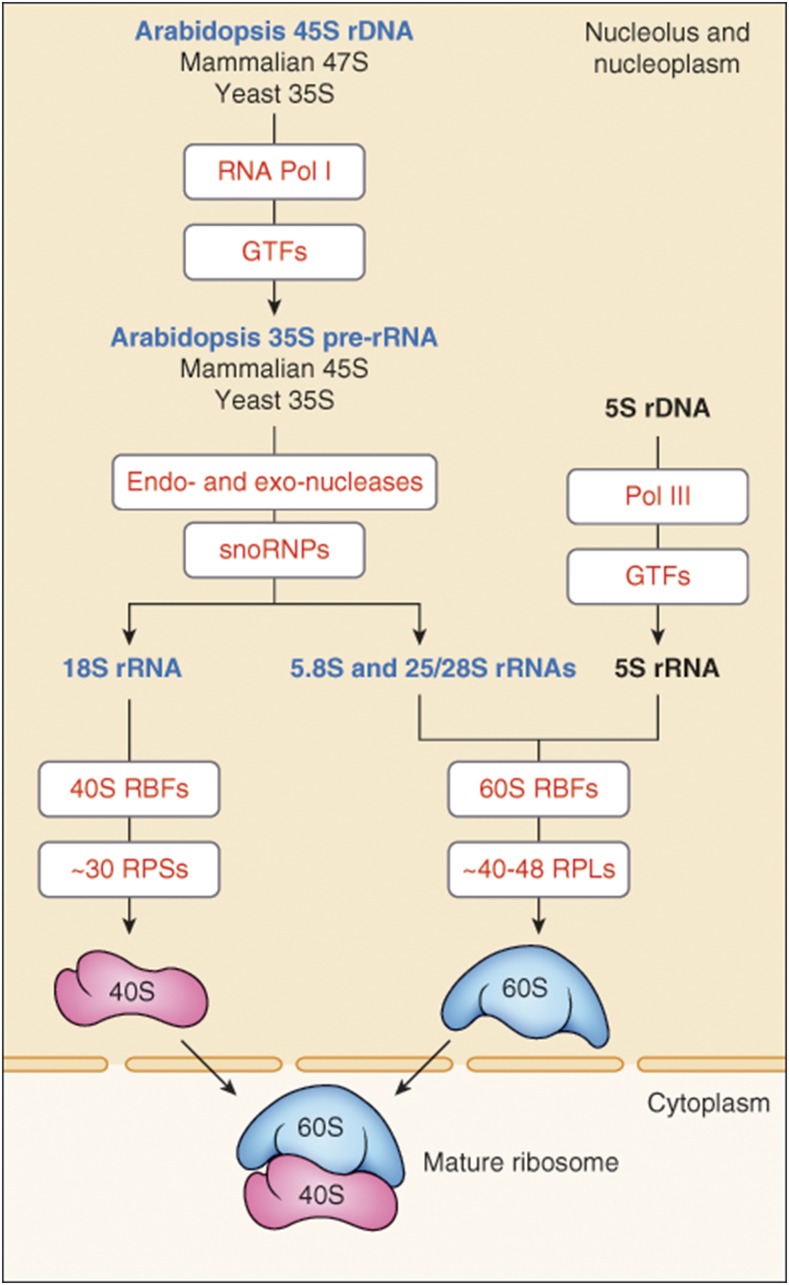
Eukaryotic ribosomes (80S) are made up of a large (60S) and a small (40S) ribosome subunit, with the S standing for the Svedberg unit sedimentation coefficient. 80S ribosome biogenesis involves the production and correct assembly of four rRNAs, 18S, 5.8S, 25S/28S, and 5S, and ∼80 ribosomal proteins (Figure 1).
Figure 1.
Graphic Representation of Ribosome Biogenesis in Eukaryotic Cells.
Transcription of rDNA (45S, 47S, and 35S in Arabidopsis, mammals, and yeast, respectively) requires RNA Pol I activity and a subset of GTFs. Transcribed transcript contains the 18S, 5.8S, and 25S (in Arabidopsis and yeast)/28S (in mammals) rRNAs and is first cotranscriptionally processed into pre-rRNA precursor (35S in Arabidopsis and yeast or 45S in mammals). Processing of pre-rRNAs into mature 18S, 5.8S, and 25S/28S rRNA involves multiple endonucleolytic and exonucleolytic cleavages and the modification of numerous rRNA residues, mainly pseudouridylation and 2′-O-ribose methylation, by snoRNP complexes (see Figure 3). 18S rRNAs assemble with ribosomal proteins RPSs (S for small) to form 40S ribosome subunits, while 5.8S and 25S/28S assemble with ribosomal proteins RPLs (L for large) and 5S rRNA transcribed by RNA Pol III. Assembly and transport of ribosomal particles from the nucleolus to the cytoplasm requires hundreds of specific 40S and 60S RBFs (Figures 4 and 5; Supplemental Data Sets 3 and 4). The 40S and 60S ribosomal subunits join to form translationally competent ribosomes.
The 18S, 5.8S, and 25S (or 28S in mammals) rRNAs are encoded by a single coding sequence or transcription unit known as 45S rDNA in plants, 47S rDNA in mammalian cells, and 35S rDNA in yeast. The 5S rRNA is encoded by transcriptionally separated 5S rDNA units. 45S, 47S, and 35S rDNA are transcribed by RNA polymerase I (Pol I) in a process requiring a large number of general transcription factors (GTFs; Russell and Zomerdijk, 2005; Saez-Vasquez and Echeverria, 2006; Schneider, 2012; Goodfellow and Zomerdijk, 2013). The resulting rRNA precursor transcripts (pre-rRNAs) are processed by numerous ribonucleoprotein (RNP) factors to obtain mature 18S, 5.8S, and 25S/28S rRNAs (Fromont-Racine et al., 2003; Dez and Tollervey, 2004; Chédin et al., 2007; Ebersberger et al., 2014; Weis et al., 2015a). 5S rDNA is transcribed by RNA Pol III via a process involving specific transcription factors TFIIIA, TFIIIB, and TFIIIC (reviewed in Ciganda and Williams, 2011; Layat et al., 2012; Orioli et al., 2012).
The 18S rRNAs assemble with ribosomal proteins of the small subunit (RPSs), while 5.8S and 25S/28S rRNAs assemble with ribosomal proteins of the large subunit (RPLs), as well as 5S rRNA, to form the small 40S and large 60S ribosome subunits, respectively. In mammalian, yeast, and plant cells, 40S ribosome subunits contain ∼30 RPSs, and 60S ribosome subunits contain ∼40 to 48 RPLs (Armache et al., 2010; Ben-Shem et al., 2011). Assembly of the ribosome subunits occurs cotranscriptionally and requires more than 200 accessory factors known as ribosome biogenesis factors (RBFs; Supplemental Data Sets 1to 4). Throughout transcription, the nascent pre-RNAs interact with RBFs, which are required for RNA processing and the assembly of RPSs and RPLs into preribosomes (Figure 1).
Nearly 50 years ago, it was demonstrated that transcribed amphibian rRNA genes have a characteristic Christmas tree appearance when chromatin is spread and visualized by electron microscopy (Miller and Beatty, 1969). In these observations, rDNA resembles the tree’s trunk and nascent rRNA transcripts its “branches,” whose tips are decorated with terminal “balls.” Since that time, it has been shown that the terminal balls are large U3 small nucleolar ribonucleoprotein (snoRNP)-associated complexes, named the 90S or SSU-processome (for small subunit), implicated in 45S pre-RNA processing and the assembly of pre-40S ribosome particles (Mougey et al., 1993). Similarly, pre-60S ribosome particles can be observed at the ends of nascent pre-RNAs after cotranscriptional cleavage events in pre-rRNAs that release pre-40S particles (Phipps et al., 2011; Henras et al., 2015).
The processing of pre-rRNAs and the assembly of preribosome subunit intermediates are relatively well understood in yeast and animals. In addition, the structures of several pre-40S complexes have been solved, including the 90S/SSU-processome and pre-60S complexes of yeast (Ben-Shem et al., 2011; Wu et al., 2016; Barandun et al., 2017; Chaker-Margot et al., 2017; Kater et al., 2017; Kressler et al., 2017; Sun et al., 2017; Ameismeier et al., 2018; Biedka et al., 2018; Sanghai et al., 2018; Zisser et al., 2018).
rDNA transcription, pre-rRNA processing, and ribosome biogenesis have been studied for many years in plants. Initially, most of these studies addressed questions concerning rDNA organization in different plant species, including maize (Zea mays), common wheat (Triticum aestivum), rice (Oryza sativa), radish (Raphanus sativus), Brassica oleracea and Brassica rapa, Arabidopsis (Arabidopsis thaliana), tobacco (Nicotiana tabacum), and potato (Solanum tuberosum; reviewed in Saez-Vasquez and Echeverria, 2006). Later, analysis of the Arabidopsis genome allowed the identification of all ribosomal proteins from the small and large ribosome subunits (Cooke et al., 1997; Barakat et al., 2001). In addition, biochemical approaches successfully identified and characterized RNA Pol I transcription and pre-rRNA processing complexes from Brassica species (Caparros-Ruiz et al., 1997; Saez-Vasquez and Pikaard, 1997, 2000; Saez-Vasquez et al., 2001; Sáez-Vasquez et al., 2004a). Since then, genetic approaches combined with proteomic analysis of the nucleolus have led to the identification and characterization of RBFs and the mechanisms implicated in the functional organization of rDNA and/or the processing of rRNA precursors in plants.
Here, we review these findings from plants and present an integrated picture of 45S rDNA organization and rRNA transcription in Arabidopsis. Based on yeast and animal models, we discuss the events tied to pre-rRNA transcription that lead to the formation of the largest U3 snoRNP and/or SSU-processome complex. We also review current knowledge of the protein complexes and biochemical activities required for the assembly of pre-60S particles in plants.
45S rDNA ORGANIZATION
Plant genomes contain hundreds to thousands of 45S rRNA genes, which are generally organized in a manner similar to the rDNA arrays of other organisms (Saez-Vasquez and Echeverria, 2006).
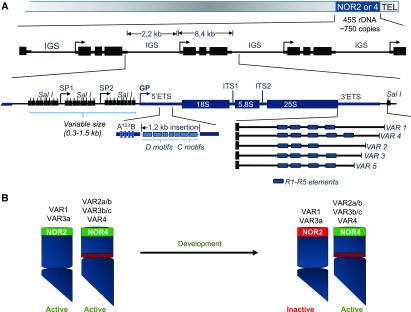
In the haploid Arabidopsis (ecotype Columbia-0 [Col-0]) genome, ∼750 copies of the 45S rDNA repeat unit are localized in tandem arrays at the tops of chromosomes 2 and 4, corresponding to Nucleolus Organizer Regions 2 and 4 (NOR2 and NOR4; Figure 2; Copenhaver and Pikaard, 1996a, 1996b). Under normal plant growth conditions, the rDNA of NOR4 is transcriptionally active, whereas the rDNA of NOR2 is transcriptionally silenced by repressive chromatin modifications (Chen and Pikaard, 1997; Fransz et al., 2002). Strikingly, the expression of rDNA within NOR2 is developmentally regulated (see section "Transcription of 45S rRNA Genes").
Figure 2.
45S rDNA Organization in Arabidopsis Accession Col-0.
(A) The figure illustrates the localization of 45S rDNA tandem repeat units in the NORs immediately adjacent to telomeres (TEL) on chromosomes 2 and 4. 45S rDNA units are separated by IGSs, which each contain repeated SalI box elements, SP1 and SP2, and the GP. The 45S rDNA transcribed sequence contains structural rRNAs (18S, 5.8S, and 25S) separated by external (5′ETS and 3′ETS) and internal (ITS1 and ITS2) transcribed spacers. Within the 5′ETS is the A123B motif and a 1.2-kb insertion, which contain D and C motifs. In the 3′ETS are polymorphic R1 to R5 elements that allow the 45S rDNA variants VAR1 through VAR5 to be discriminated.
(B) The image illustrates the distribution of 45S rDNA variants in NOR2 (VAR1 and VAR3a) and NOR4 (VAR2a/b, VAR3b/c, and VAR4; Chandrasekhara et al., 2016) and the repression of NOR2 during plant development.
As depicted in Figure 2, the 45S rDNA transcribed sequence is a total of ∼8.4 kb in length and contains the 18S (1800 bp), the 5.8S (161 bp), and 25S (3376 bp) rRNA sequences separated by internal transcribed spacers (ITS1 and ITS2) and flanked by external transcribed spacers (5′ETS and 3′ETS; Gruendler et al., 1989; Unfried et al., 1989; Unfried and Gruendler, 1990). Characterization of the 3′ETS revealed that 45S rDNA units are not all identical in Arabidopsis (Col-0). There are four major classes or variants of 45S rDNA (VAR1–VAR4), based on insertions/deletions in the 3′ETS (Figure 2A; Earley et al., 2010; Pontvianne et al., 2010). The 45S rDNA VAR1 class represents ∼50% of the copies and maps to the inactive NOR2, while rDNA VAR2 to VAR4 primarily map to the active NOR4 (Chandrasekhara et al., 2016). Nonetheless, some rDNA VAR3 copies map to NOR2 (Chandrasekhara et al., 2016), while the low-abundance rDNA variant VAR4 also maps near the centromere of chromosome 3 (Abou-Ellail et al., 2011). A fifth rRNA gene variant (VAR5) was reported by Havlová et al. (2016). The rDNA VAR5 was described as a VAR3 subtype based on the rDNA sequence length measured from the end of the 25S to the first SalI restriction site in the 3′ETS (Figure 2B). Remarkably, the translocation of rRNA genes from inactive NOR2 to active NOR4 results in the expression of normally silenced rDNA variants (Mohannath et al., 2016). This study demonstrated that the silencing of rRNA gene clusters in NOR2 depends on the chromosome on which they reside rather than on the sequences of these individual rRNA genes (Mohannath et al., 2016). In this context, the expression of rDNA VAR1 in nucleolin2 (nuc2) plants is thought to result from the relocation of rDNA VAR1 copies from NOR2 to NOR4 (Durut et al., 2014).
The 5′ETS is 1.8 kb and contains the U3 snoRNP binding site A123B and a 1.2-kb insertion found specifically in Arabidopsis but not in other cruciferous plant genomes (Sáez-Vasquez et al., 2004a). The 1.2-kb sequence contains five D (D1a, D1b, D2a, D12, and D2b) and two C (C1 and C2) motifs of unknown function (Figure 2A; Gruendler et al., 1991).
Each 45S rDNA coding sequence is separated from the adjacent ones by ∼2.2-kb intergenic spacer (IGS) regions, which contain the SalI boxes (also called A motifs), two spacer promoters (SP1 and SP2), and the gene promoter (GP; Figure 2A; Gruendler et al., 1991).
The minimal GP sequence is from −55/−33 upstream to +6 downstream relative to the transcription initiation site (+1; Doelling and Pikaard, 1995). Remarkably, the sequence surrounding the transcription initiation site (TATATAGGGGG, +1 is underlined) is highly similar among plant species (Delcasso-Tremousaygue et al., 1988; Cordesse et al., 1993; Fan et al., 1995; reviewed in Saez-Vasquez and Echeverria, 2006). Mutations in this consensus region abolish or inhibit rRNA transcription and perturb the position of transcription initiation (Doelling and Pikaard, 1995). The SP sequences share 90% similarity with the GP (from −92 to +6), but they are only ∼10% active compared with the GP (Doelling et al., 1993). The SP and GP also contain multiple sites homologous to small interfering RNA (siRNA) sequences, which are required for 45S rDNA silencing in hybrids (Preuss et al., 2008; Costa-Nunes et al., 2010).
Fewer studies have focused on the role of the IGS in rDNA organization or transcription. Two decades ago, the characterization of rRNA transgenes integrated into chromosomal regions outside NORs revealed that transgenic rDNA can associate with the nucleolus but that SalI boxes do not enhance RNA Pol I transcription in this context (Wanzenböck et al., 1997). By contrast, IGS might play a role in rDNA recombination (Urawa et al., 2001). Recently, major differences were reported in the length and/or organization of the IGS in A. thaliana. However, all the IGS rDNA sequences analyzed in this comprehensive study contain at least one SP and SalI box clusters of variable sizes ranging from 294 to 1505 bp (Havlová et al., 2016). Importantly, there is no obvious structural or functional relationship between the 3′ETS and IGS, as rDNA VAR1 to VAR5 display similar variability in terms of the number and/or length of SalI boxes and SP1/SP2 sequences (Havlová et al., 2016).
In animal cells, IGS transcripts are involved in rDNA repression (Bierhoff et al., 2010) and in nucleolar protein sequestering (Audas et al., 2012) in response to stress conditions. However, little is known about these mechanisms during plant development and/or in response to environmental stress.
TRANSCRIPTION OF 45S rRNA GENES
45S rDNA is transcribed by RNA Pol I in the nucleolus (Raska et al., 2004; Saez-Vasquez and Medina, 2008). RNA Pol I is a multisubunit protein enzyme that associates with additional GTFs to form larger RNA Pol I complexes or holoenzymes, which are competent to specifically initiate rDNA transcription (Saez-Vasquez and Pikaard, 1997; Seither et al., 1998; Albert et al., 1999).
In Arabidopsis, RNA Pol I consists of 14 protein subunits: 12 are subunits homologous or common to those of all nuclear RNA polymerases (RNA Pol I–Pol V), five of the RNA Pol I subunits are common to RNA Pol II and Pol III (subunits encoded by the same genes), and the two others are RNA Pol I-specific subunits (Ream et al., 2015). In plants, RNA Pol I holoenzyme purified from B. oleracea nuclear extracts was capable of binding specifically to the 45S rDNA promoter (GP) and initiating transcription in vitro (Saez-Vasquez and Pikaard, 1997, 2000). In addition, a CK2-like protein (CASEIN KINASE2) copurifies with RNA Pol I holoenzyme activity (Saez-Vasquez et al., 2001). In mammalian cells, CK2 phosphorylates TRANSCRIPTION INITIATOR FACTOR1A, thereby increasing rDNA transcription (Bierhoff et al., 2008). The role of CK2 in plant rRNA gene expression has not been reported; however, CK2 has been implicated in multiple developmental and stress response pathways in plants (Mulekar and Huq, 2014).
Little is known about the GTFs and/or accessory factors required for RNA Pol I transcription initiation, elongation, or termination in plants (reviewed in Saez-Vasquez and Echeverria, 2006). However, the TFIIB-related protein, pBrp, is a plant-specific GTF for RNA Pol I. pBrp does not interact with the rDNA promoter but forms a stable ternary complex with Arabidopsis TATA BINDING PROTEIN2 and the rDNA promoter (Imamura et al., 2008).
More is known about the functional organization of rDNA chromatin, mostly thanks to studies involving rRNA gene silencing in interspecific hybrids called nucleolar dominance (Tucker et al., 2010; Ge et al., 2013; Michalak et al., 2015). For example, in the hybrid Arabidopsis suecica, rDNA derived from one parent (Arabidopsis) is silenced by repressive chromatin modifications while the rDNA from the other parent (Arabidopsis arenosa) remains active (Chen et al., 1998; Pikaard, 2000; Lawrence et al., 2004). Studies of nucleolar dominance also demonstrate that in transcriptionally active rDNA chromatin, cytosines are hypomethylated and histone H3 is acetylated (H3Ac) and typically methylated at position Lys-4 (H3K4), whereas in inactive rDNA chromatin, cytosines are hypermethylated and histone H3 is typically methylated at position Lys-9 (H3K9; Lawrence et al., 2004; Lawrence and Pikaard, 2004).
More recently, studies of rDNA variants in Arabidopsis (Col-0) have allowed mechanisms controlling rDNA chromatin dynamics to be dissected in the context of nonhybrid plants. Together with the analysis of nucleolar dominance, this work addressed the question of how plants “select” one or another subset of 45S rDNA, because it is well known that not all rRNA gene copies are transcriptionally active in plants and other eukaryotic cells (Conconi et al., 1992; González-Melendi et al., 2001; Fransz et al., 2002; French et al., 2003).
As mentioned above, in Arabidopsis (Col-0), only a fraction of rRNA genes (those from NOR4) are transcribed throughout plant growth and development. NOR2 contains the rRNA VAR1 genes, which represent ∼50% of rDNA and are silenced ∼2 to 4 d after germination. Nevertheless, in globular embryos (Chen et al., 2016) and during the first few days after germination (Earley et al., 2006; Pontvianne et al., 2010), rRNA VAR1 transcripts can be detected, indicating that NOR2 is active during these stages of development (Figure 2B). The expression of rDNA VAR1 is correlated with the global decondensation of chromatin during germination (Layat et al., 2012).
Several protein factors have been described as transcriptional activators or repressors of NOR2 (Supplemental Data Set 1). For instance, rDNA VAR1 is expressed in mutant plants with deficiencies of NUC1 or NUC2, HISTONE DEACETYLASE6 (HDA6), SUVH5/6 SU(VAR)3-9HOMOLOG5 (SUVH5) and SUVH6 or ARABIDOPSIS TRITHORAX-RELATED PROTEINS5 (ATXR5) and ATXR6, FASCIATA1 (FAS1) or FAS2, Cell Division Cycle48A (CDC48A), MORPHOLOGY OF AGO1-52 SUPPRESSED2 (MAS2), and ALPHA THALASSEMIA-MENTAL RETARDATION X-LINKED (ATRX), which indicates that NOR2 fails to be silenced in the absence of these protein factors (Earley et al., 2006; Mozgová et al., 2010; Pontvianne et al., 2010, 2012, 2013; Durut et al., 2014; Mérai et al., 2014; Sánchez-García et al., 2015; Duc et al., 2017). By contrast, embryonic expression of rDNA VAR1 was repressed in plants with mutations in THALLO (THAL; Chen et al., 2016). Most of these proteins are histone chaperones (NUC1 and NUC2, FAS1 and FAS2, and ATRX), chromatin modifiers (SUVH5 and SUVH6, ATXR5 and ATXR6, and HDA6), and/or proteins involved in chromatin organization (CDC48 and MAS2). However, THAL is the homolog of Utp3, a protein that associates with the 90S/SSU-processome complex (Gallagher et al., 2004; Barandun et al., 2017). In Arabidopsis, NUC1 and NUC2 play antagonistic roles in rRNA gene expression. While NUC1 appears to facilitate rDNA transcription (Pontvianne et al., 2010), NUC2 induces and/or maintains a repressive rDNA chromatin state (Durut et al., 2014). Nonetheless, the expression of NUC2 appears to rescue the absence of NUC1 in nuc1 plants (Pontvianne et al., 2007; reviewed in Durut and Sáez-Vásquez, 2015).
Furthermore, rDNA VAR1 is expressed in mutant plants deficient for RNA RIBOSOMAL PROTEIN7 (RRP7) or SMALL ORGAN4/NUCLEOLAR PROTEIN53; SMO4/Nop53; Micol-Ponce et al., 2018), which are proteins that associate with the 90S/SSU-processome and 60S preribosome complexes.
Additional factors affect or have been linked to rDNA functional organization and expression. For instance, REGULATOR OF TELOMERE ELONGATION HELICASE1; RTEL1) is required for stabilizing 45S rDNA repeats, while simultaneous mutation of RTEL1 with RecQ-MEDIATED GENOME INSTABILITY PROTEIN2 (RMI2) provokes the increased loss of 45S rDNA repeats (Röhrig et al., 2016), similar to fas mutants (Pontvianne et al., 2013). Overexpression of TARGET OF RAPAMYCIN (TOR) induces the accumulation of 45S rRNA transcripts, whereas overexpression of RIBOSOMAL PROTEIN SMALL SUBUNIT6 (RPS6) represses this process (Ren et al., 2011; Kim et al., 2014). Moreover, 45S rRNA genes are overexpressed in plants with reduced FK506 BINDING PROTEIN (FKBP53) gene expression (Li and Luan, 2010). In addition, the 3xHMG-box1 (HIGH MOBILITY GROUP) protein binds to 45S rDNA loci via its basic N-terminal domain (Antosch et al., 2015). It is not known whether 3xHMG-box1 is related to Upstream Binding Protein (UBF) and/or High Mobility1(Hmo1), two HMG proteins involved in rDNA transcription in animal and yeast cells, respectively (Sanij et al., 2008; Albert et al., 2013). The precise roles of RTEL1, RMI2, FKBP53, TOR, RPS6, and 3xHMG-box1 in regulating rDNA VAR1 or, more generally, NOR gene expression have not been determined. Notably, plants deficient in these factors (or overexpressing 3xHMG-box1) typically show plant growth and developmental defects (Supplemental Data Set 1).
EARLY CLEAVAGE EVENTS OF 45S PRE-rRNA
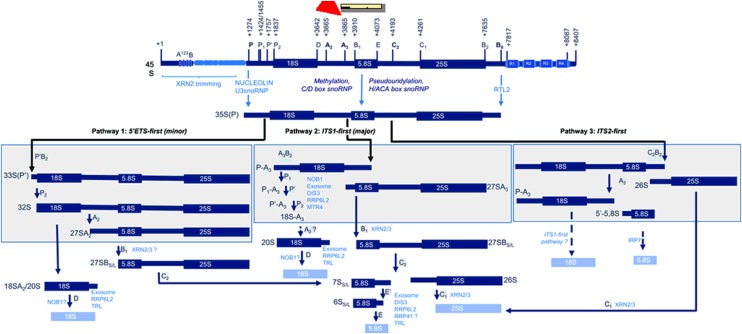
47S and 35S rRNAs are the longest pre-rRNA transcripts in animals and yeast, respectively (Venema and Tollervey, 1995; Fromont-Racine et al., 2003; Kressler et al., 2010; Henras et al., 2015; Tomecki et al., 2017). In Arabidopsis, the longest pre-rRNA detected to date is referred to as 45S pre-rRNA (Figure 3). Based on data from yeast and mammalian systems (Henras et al., 2015), Arabidopsis 45S pre-rRNA is likely subjected to similar cleavage events to those that generate 35S(P) pre-rRNA. This notion is supported by the experimental identification of cleavage sites and the characterization of plant RNP complexes and activities involved in the processing of the 5′ETS and 3′ETS (Sáez-Vasquez et al., 2004a; Comella et al., 2008; Zakrzewska-Placzek et al., 2010).
Figure 3.
35S Pre-rRNA Processing Pathways.
The figure illustrates the 45S pre-rRNA and cleavage sites in the ETS and ITS sequences. In the current models, the 5′ETS of 45S pre-rRNAs is trimmed by XRN2 before being cleaved at the P site by a nucleolin-U3 snoRNP complex. The 3′ETS is cleaved by the RNase III endonuclease, RTL2. Structural rRNAs are methylated by C/D snoRNP (2-O-ribose methylation) or pseudouridylated by H/ACA snoRNP complexes. The resulting 35S(P) pre-rRNA follows either pathway 1, 5′ETS-first (minor), pathway 2, ITS1-first (major), or pathway 3, ITS2-first. Specific processing events in pathway 1, 2, or 3 are shown in the boxes. The characterized protein factors required for RNA modification (C/D and H/ACA snoRNP and TRL), endonucleolytic cleavages (nucleolin-U3 snoRNP, RTL2, and NOB1), or exonucleolytic cleavages (XRN2, AtRRP6L2, and AtRRP41) are indicated.
The processing of pre-rRNA consists of exonucleolytic and endonucleolytic cleavages to remove external (5′ETS and 3′ETS) and internal (ITS1 and ITS2) transcribed sequences (Figure 3). The endonucleolytic cleavage sites in 45S pre-rRNA have been mapped in Arabidopsis or predicted based on yeast and human pre-rRNA processing maps (Tomecki et al., 2017). The initial cleavage site of 45S pre-rRNA is at the P site (+1275) in the 5′ETS, just after the 1.2-kp insertion present in Arabidopsis rRNA genes (Sáez-Vasquez et al., 2004a). Further cleavages occur at the P′ (+1757) and P2 (+1837) sites (Lange et al., 2008; Zakrzewska-Placzek et al., 2010). It appears that the reported P1 site is not an endonucleolytic cleavage site, because the expected P1-P′ fragments have not been detected (Figure 3). Instead, the accumulation of smaller and heterogeneous fragments mapping around P1 suggest that fragments between P and P′ are generated by exoribonucleolytic degradation (Lange et al., 2011; Sikorska et al., 2017).
The 268-bp ITS1 contains the D, A2, A3, and B1 cleavage sites, whereas the 190-bp ITS2 contains the E, C2, and C1 cleavage sites. A2, A3, and C2 are key cleavage sites that switch pre-rRNA processing to specific pathways: 5′ETS-first (minor) or ITS1-first (major; Figure 3; see below). The 3′ETS end and cleavage sites have not yet been precisely determined. However, two cleavage sites were detected upstream of the R repeat elements in nuc1 plants (Pontvianne et al., 2010), which might correspond to predicted B0 cleavage sites (Tomecki et al., 2017).
In yeast, cleavage of the 3′ETS is required for RNA Pol I transcription termination by a mechanism similar to the torpedo mechanism (West et al., 2004), which involves the RNase III activity of RNase three1(Rnt1) and 5′-3′ Exoribonuclease2 (Xrn2) proteins (Kawauchi et al., 2008). In the torpedo mechanism, cotranscriptional cleavage by Rnt1 releases nascent pre-rRNA from the RNA Pol I elongation complexes, allowing Xrn2 to degrade nascent rRNA still associated with the elongating complex, ultimately provoking the dissociation of RNA Pol I from rDNA. In Arabidopsis, genetic studies have shown that cleavage of the 3′ETS requires RNASE THREE LIKE2 (RTL2; Comella et al., 2008). The RTL2 cleavage sites in the 3′ETS have not yet been precisely mapped, and whether RTL2 is required for RNA Pol I termination remains unsettled. Nevertheless, RTL2 is required for 45S siRNA accumulation (Elvira-Matelot et al., 2016). Moreover, the transcription termination sequences reported in B. oleracea (Yang et al., 2015) were not detected in the Arabidopsis 3′ETS or IGS. This is in contrast to the evolutionary conservation among cruciferous plants of the rRNA gene promoter and 5′ETS A123B boxes (Doelling and Pikaard, 1996; Sáez-Vasquez et al., 2004a).
The 45S pre-rRNA of Arabidopsis has a much longer 5′ETS than pre-rRNAs of yeast, mammalian cells, or even other crucifers (Tomecki et al., 2017). In B. oleracea, biochemical studies have shown that a U3 snoRNP complex binds to the 5′ETS region of the rDNA and cleaves the pre-rRNA 5′ETS at the P site (Caparros-Ruiz et al., 1997; Sáez-Vasquez et al., 2004a, 2004b). In Brassica species and radish, the P site is located next to the A123B binding site of the nucleolin-U3 snoRNP complex, whereas in Arabidopsis, the P site is located ∼1.2 kb downstream of the A123B motif (Figure 3). In addition, the pre-rRNA 5′ETS in Arabidopsis is shortened by the exonuclease XRN2 before being cleaved at the P site (Zakrzewska-Placzek et al., 2010). The biological relevance of this XRN2-mediated trimming remains unknown.
PROCESSING OF PRE-rRNA: TWO ALTERNATIVE PATHWAYS
In all eukaryotic cells, mature 18S, 5.8S, and 25S/28S rRNAs are generated via distinct 40S (for 18S rRNA) and 60S (for 5.8S and 25S/28S rRNAs) pre-rRNA processing pathways (Eichler and Craig, 1994; Tomecki et al., 2017). Alternative processing steps have been described after cotranscriptional cleavages at A0, A1, and A2 sites in yeast (equivalent to P, P2, and A2 sites in Arabidopsis). In mammalian cells, cleavage of the 47S pre-rRNA can either start in the 5′ETS at A0 and 1(A1) sites (A0 and A1 in yeast) or in the ITS1 at site 2 (A3 in Arabidopsis and yeast), defining two distinct pathways: the minor 5′ETS-first pathway and the major ITS1-first pathway (Henras et al., 2015).
Based on the accumulation of specific pre-rRNA products and intermediates, two alternative pathways have also been described in Arabidopsis: the minor 5′ETS-first pathway and the major ITS1-first pathway (Figure 3). This preferential ITS1-first processing was first observed in xrn2 plants (Zakrzewska-Placzek et al., 2010), while the accumulation of 5′ETS-first transcripts was reported in biogenesis of ribosomes in xenopus1 (brx1) plants (Weis et al., 2015b). These two pathways also coexist in rice plants, with the ITS1-first mode being the major pathway (Hang et al., 2018). Environmental stimuli or stress conditions might prevent the accumulation of pre-rRNA from the ITS1-first pathway (Weis et al., 2015a).
In the 5′ETS-first pathway, the 35S(P) is cleaved at the P′, P2, and A2 sites and then at the B1 site, while in the ITS1-first pathway, the 35S(P) is cleaved at the A3 site and then at the P′ and P2 sites (Zakrzewska-Placzek et al., 2010; Weis et al., 2015a). Then, after the alternative A2 or A3 cleavage events, pre-rRNA precursors from both pathways are adenylated to be degraded and/or trimmed by the exosome. Uridylation of a small subset of intermediates might also occur, but the link between this modification and the exosome has not yet been established (Chekanova et al., 2000; Lange et al., 2008, 2011; Kumakura et al., 2013; Sikorski et al., 2015). Finally, 20S and 27SBS/L precursors from both alternative pathways are subjected to common processing events (at the C2, D, E, E′, and C1 sites) to obtain the mature 18S, 5.8S, and 25S rRNAs (reviewed in Weis et al., 2015a; Tomecki et al., 2017).
Alternative cleavages at either the A2 or A3 site were recently described in yeast (Choque et al., 2018). However, in contrast to Arabidopsis or mammalian cells, only cleavage at A2 is productive in yeast. Indeed, cleavage at A2 by Utp24 leads to the production of 20S pre-rRNAs and then mature 18S rRNAs (productive A2 pathway). Cleavage at A3 by RNase MRP results in the production of 23S rRNAs. These 23S rRNAs are targeted by Upt24, generating 11S and 17S rRNAs, which are subsequently degraded by TRAMP/Exosome (nonproductive A3 pathway; Choque et al., 2018).
In yeast, cleavages at the A0, A1, and A2 sites occur cotranscriptionally, and thus the larger 35S, 33S, and 32S pre-rRNAs are barely detectable. In Arabidopsis, these pre-rRNAs are easily detected (Zakrzewska-Placzek et al., 2010; Missbach et al., 2013; Weis et al., 2015b; Maekawa et al., 2018a), suggesting that cleavage at P, P′, and A2 occurs mostly posttranscriptionally, as suggested for the A0, 1(A1), and 2 sites in mammalian cells (Henras et al., 2015). Thus, the major ITS1-first processing pathway in Arabidopsis is analogous to the main processing pathway in mammalian cells, while the minor 5′ETS-first processing pathway is comparable to the pathway used in yeast.
A plant-specific ITS2 processing pathway was recently reported in Arabidopsis (Palm et al., 2019). The ITS2 pathway involves a first cleavage of 35S pre-rRNAs at the C2 site in the ITS2 and the accumulation of specific pre-rRNAs (Figure 3). Remarkably, the ITS2 pathway becomes visible in Arabidopsis irp7 plants (deficient for INVOLVED IN RNA PROCESSING7) after auxin treatment and in rapidly dividing cells. Moreover, IRP7, together with INVOLVED IN RNA PROCESSING8 (IRP8) and IRP9, are required for the normal processing of plant-specific 5′-5.8S pre-rRNAs. In irp9 plants, only the processing of 5′-5.8S pre-rRNAs is affected, while in irp7 and irp8, the accumulation of 27S-A3, P-A3, P′-A3, and 18S-A3 pre-rRNAs is affected as well. The IRP7 and IRP8 proteins are predicted to contain Acetylation Lower Binding Affinity (ALBA) and RNA Recognition Motif (RRM) protein domains, respectively, whereas IRP9 contains a nucleotide hydrolase domain and appears to be crucial for the processing of the plant-specific 5′-5.8S pre-rRNA (Palm et al., 2019).
The exoribonuclease activities involved in pre-rRNA processing are relatively well characterized in Arabidopsis. For instance, XRN2 is required for the 5′ trimming of 45S, 27S-A3, and 26S pre-rRNAs, while the exosome core complex Exo9 and the associated RRP44 and RRP6L2 proteins are necessary for the 3′ trimming of P-A3, P′-A3, 18S-A2/20S, and/or P-P′ and 5.8S pre-rRNAs. More precisely, the removal of 5.8S pre-rRNA, P-P′, and fragments cleaved at A3 requires the exosome core complex (Exo9), the RNA helicase MTR4, and either RRP6L2 and DEFECTIVE IN SISTER CHROMATID DISJOINING3 (DIS3) or both, while the elimination of 18S-A2 fragments depends mostly on RRP6L2 alone (Figure 3; Lange et al., 2008, 2011, 2014; Kumakura et al., 2013; Sikorski et al., 2015). Moreover, the nine-subunit exosome core has a distributive phosphorolytic activity involved in the degradation of P-P1 products (161, 168, 176, and 186 nucleotides) and the processing of 5.8S pre-rRNA. Therefore, the plant RNA exosome harbors a unique combination of hydrolytic and phosphorolytic ribonucleolytic activities, in contrast to exosome complexes in yeast and animal cells (Sikorska et al., 2017).
Pre-rRNA trimming and the degradation of rRNA processing by-products involve the terminal nucleotidyltransferase TRF4/5-LIKE (TRL). Poly(A) tails added to these by-products are up to 18 nucleotides long (Lange et al., 2008; Sikorski et al., 2015). Uridylation might also contribute to rRNA processing, because rRNA by-products can be uridylated in Arabidopsis (Sikorski et al., 2015).
Much less is known about the RNase activities responsible for the endonucleolytic cleavage of plant pre-rRNAs. On the one hand, Arabidopsis RTL2 is implicated in 3′ETS processing (Comella et al., 2008). Furthermore, Arabidopsis NOB1, a homolog of the yeast protein Nob1, is required for P-A3 pre-rRNA processing (Missbach et al., 2013). This finding is unexpected, because yeast Nob1 is required for the cleavage of 20S pre-rRNA (Fatica et al., 2003). On the other hand, genomic and proteomic analyses have identified an Arabidopsis homolog of the yeast protein Utp24 (Supplemental Data Set 2), which is likely involved in the cleavage of yeast A1 (equivalent to P2 in Arabidopsis) and A2 in the 5′ETS and ITS1, respectively. Indeed, it was recently shown that Utp24 is essential for early cleavages at three pre-rRNA sites in yeast (A0, A1, and A2) and humans [A0, 1(A1), and 2; Wells et al., 2016]. Similarly, Arabidopsis encodes homologs of the yeast proteins RMRP, Las1, and Rcl1, which are endoribonucleases required for cleavage at the A3, C2, and A1/A2 sites in yeast (Supplemental Data Sets 1 and 2; Kiss et al., 1992; Gasse et al., 2015; Wells et al., 2016; Goldfarb and Cech, 2017; Maekawa et al., 2018a). However, the predicted endonucleolytic activities of these Arabidopsis homologs remain to be tested.
A more detailed picture of the current knowledge on the endoribonuclease and exoribonuclease activities required for the processing of diverse pre-rRNAs and the degradation of rRNA by-products has been published by Tomecki et al. (2017).
rRNA PROCESSING FACTORS
Before the cleavage and removal of ETS and ITS segments from pre-rRNAs, the 18S, 5.8S, and 25S rRNA sequence regions of the precursor are subjected to a number of cotranscriptional sugar and base modifications (Sharma and Lafontaine, 2015). The most common rRNA modifications are 2′-O-ribose methylation and uridine isomerization (pseudouridylation). 2′-O-Methylation is guided by small nucleolar RNAs (snoRNAs) of the C/D box type and is accomplished by the protein FIBRILLARIN/Nop1, whereas pseudouridylation is guided by H/ACA box snoRNAs and is accomplished by the protein DYSKERIN/Cbf5p. The distinct snoRNA guides and FIBRILLARIN or DYSKERIN associate with additional proteins to form C/D box or H/ACA box snoRNPs (Massenet et al., 2017), respectively, which have also been identified in plants (Supplemental Data Sets 1 and 2).
In Arabidopsis, hundreds of C/D box snoRNAs have been identified, and the expected methylation sites have been mapped on the 18S, 5.8S, and 25S rRNA sequences (Barneche et al., 2001; Brown et al., 2001, 2003a, 2003b; Qu et al., 2001; Chen and Wu, 2009). Only a few snoRNAs have been associated with the cleavage of pre-rRNAs. For instance, the U3 and U14 C/D box and the E3 H/ACA box snoRNAs facilitate cleavages within the 5′ETS in yeast and animal cells (Hughes and Ares, 1991; Enright et al., 1996; Borovjagin and Gerbi, 1999). In plants, the U3 snoRNP is involved in processing at the P site in the 5′ETS (Sáez-Vasquez et al., 2004a), but a direct role for U3 snoRNA in pre-rRNA cleavage has not been demonstrated. By contrast, the base pairing of the HIDDEN TREASURE2 (HID2) C/D box snoRNA with rRNA is required for 25S pre-rRNA processing. Intriguingly, the predicted HID2 methylation target site in the 25S rRNA sequence is not affected in the hid2 mutant. HID2 is thought to play a role in promoting pre-rRNA processing and minimizing the production of aberrant rRNA species (Zhu et al., 2016).
The Arabidopsis genome encodes two functional FIBRILLARIN proteins, FIB1 and FIB2 (Supplemental Data Set 2; Barneche et al., 2000; Pih et al., 2000). Remarkably, it appears that the role of FIBRILLARIN is not limited to the methylation of rRNA. Indeed, FIBRILLARIN methylates histone H2A on rRNA gene promoters in both humans and Brassica species (Tessarz et al., 2014; Loza-Muller et al., 2015). Furthermore, a ribonuclease activity was demonstrated for the GAR domain of Arabidopsis FIB2 but not for that of FIB1 (Rodriguez-Corona et al., 2017). Because FIBRILLARIN copurifies with the NUCLEOLIN-U3 snoRNP complex, it was suggested that FIB2 might be involved in the cleavage of the P site in the 5′ETS (Rodriguez-Corona et al., 2017). More recently, analysis of fib2 knockout plants demonstrated that FIB2 binds to and negatively regulates the expression of innate immune response genes in Arabidopsis. Upon Pseudomonas syringae inoculation, FIB2 interacts with ELF18-INDUCED LONG-NONCODING RNA1 (ELENA1) and is then evicted from gene promoters to activate innate immune response genes (Seo et al., 2018). Taken together, these findings suggest that FIB1 and FIB2 play distinct roles in the modification of and interaction with RNA, DNA, and/or histone proteins in Arabidopsis.
In contrast to FIBRILLARIN, neither DYSKERIN/Cbf5p nor other H/ACA box snoRNP proteins appear to be associated with the processome. In yeast and mammalian cells, the functions of DYSKERIN/Cbf5p and H/ACA box snoRNPs are essential for ribosome biogenesis, pre-mRNA splicing, and telomere maintenance (Caton et al., 2018), but relatively little is known about these factors in plants. Several H/ACA box snoRNAs and a DYSKERIN/Cbf5p ortholog, NAP57, have been reported in Arabidopsis (Supplemental Data Set 1; Maceluch et al., 2001; Brown et al., 2003b). Characterization of a T-DNA insertion mutant in NAP57 showed that DYSKERIN/NAP57 is an essential protein in Arabidopsis (Lermontova et al., 2007), but the impact of rRNA pseudouridylation in plant growth and development remains unclear.
It was recently demonstrated that the histone deacetylase HD2A/C negatively regulates rRNA methylation and pre-rRNA processing in Arabidopsis (Supplemental Data Set 1; Chen et al., 2018). Remarkably, HD2C interacts with both pre-rRNAs and snoRNAs. Precisely how HD2A/C inhibits the methylation activity of C/D box snoRNPs remains an open question. HD2A/C likely does not affect rRNA pseudouridylation because most of the HD2A/C-interacting snoRNAs are C/D box snoRNAs.
The impact of rRNA modifications is not well understood in plants. However, rRNA modifications are thought to play roles in ribosome assembly and protein translation (Song and Nazar, 2002; Sloan et al., 2017). For example, ribose 2′-O-methylation and rRNA pseudouridylation can influence translational initiation, translational fidelity, and the ribosome’s affinity for specific (tRNA/IRES) ligands in yeast and mammalian cells (Jack et al., 2011; Marcel et al., 2013; Erales et al., 2017).
In yeast, several additional rRNA modifications have been described. These include the methylation of purine rings (m62A, m1A, and m7G) and pyrimidine rings [m5C, m3U, and 1-methyl-3-(3-amino-3-carboxypropyl)pseudouridine]. The latter, hypermodified uridine base contains an aminocarboxylation, and cytosine acetylation has also been detected (ac4C; Sharma and Lafontaine, 2015). However, the exact timing of these modifications remains unknown in most cases. In Arabidopsis, the RNA methyltransferase Nop2/Sun-like domain containing protein5/RNA (cytosine-5)-methyltransferase1 (NSUN5/Rcm1) is required for N5-methylation at position C2268 of the 25S rRNA (Burgess et al., 2015), while a ADENOSINE DIMETHYL TRANSFERASE1A (DIM1A/Dim1) homolog is required for N6-dimethylation at positions A1785 and A1786 of 18S rRNA (Wieckowski and Schiefelbein, 2012). In addition, the m5C and m7G RNA methyltransferase homologs OLIGOCELLULA2 (OLI2) and ROOT INITIATION DEFECTIVE2 (RID2), respectively, have been characterized in Arabidopsis (Fujikura et al., 2009; Ohbayashi et al., 2011). The roles of these proteins in rRNA methylation have not yet been demonstrated, but pre-rRNA accumulates in oli2 and rid2 plants (Ohbayashi et al., 2011; Kojima et al., 2018). The Arabidopsis genome also encodes homologs of yeast Essential for Mitotic Growth/Nucleolar Essential Protein1(Emg1/Nep1) and Killer toxin Resistant/RRNA cytidine Acetyltransferase1 (Kre33/Rra1; Supplemental Data Set 2). In yeast, these proteins are required for N1-methyltransferase and C-acetylation activities, respectively (Sharma and Lafontaine, 2015). Importantly, Emg1/Nep1 and Kre33/Rra1 associate with the processome in yeast (Thomas et al., 2011; Sondalle and Baserga, 2014; Sharma et al., 2015, 2017). Whether the plant homologs of Emg1/Nep1 and Kre33/Rra1 associate with 90S/SSU-processome complexes or have RNA methyltransferase or acetylation activity in Arabidopsis is currently unknown. Finally, the Arabidopsis enzyme PROTEIN ARGININE METHYLTRANSFERASE3 (PRMT3) is required for proper accumulation of the P-A3 fragment and promotes the major pre-rRNA processing pathway (Hang et al., 2014).
90S/SSU-PROCESSOME AND MATURATION OF PRE-40S PARTICLES
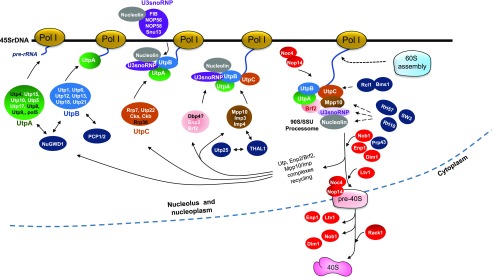
Ribosome assembly is initiated on nascent pre-rRNA transcripts, as was observed almost 50 years ago (Miller and Beatty, 1969). The largest preribosome particle identified to date in yeast and mammalian cells is the 90S or the SSU processome complex (Gallagher et al., 2004), a large U3 snoRNP complex from which the 40S ribosome particle is formed. The 90S/SSU processome is assembled cotranscriptionally in the nucleolus (reviewed in Phipps et al., 2011; Kressler et al., 2017). The genomes of Arabidopsis and rice contain orthologous genes for most yeast and human processome proteins (Supplemental Data Set 2), pointing to the existence of a 90S/SSU processome complex in plants and suggesting that animals, yeast, and plants share conserved mechanisms for 40S assembly (Figure 4).
Figure 4.
Model for Processome Assembly and the Maturation of Pre-40S Particles in Arabidopsis.
The picture illustrates UtpA (green), UtpB (blue), UtpC (yellow), U3 snoRNP (violet), Dbp4/Enp2/Brf2 (pink), and Mpp10/Imp3/Imp4 (brown) subcomplexes. Arabidopsis homologs (NuGWD1/Utp5, Nucleolin/Nrs1, Utp25, THAL1/Utp3, RH57/Rock1, RH10/Rrp3, and SW3/Dbp8) or specific (PCP1/2) factors that interact with processome factors or subunits are represented in dark blue. Arabidopsis homologs that interact with the processome and are required for pre-40S maturation and transport are represented in red. Known or predicted Arabidopsis homologs are labeled in white and those not yet identified in black. The anticipated assembly of pre-60S particles on nascent pre-RNA is shown. Assembly might initiate on full-length rRNA precursor or after cleavage at A2/A3 sites that release processome factor precursors and pre-40S particles. The figure was adapted and drawn from models of the assembly steps of the 90S/SSU-processome and pre-40S in yeast and mammalian cells (Soltanieh et al., 2014; Cerezo et al., 2019). Accession numbers for all the Arabidopsis factors are provided in Supplemental Data Set 2.
U3 is a C/D box small nucleolar RNA (U3 snoRNA) required for nucleolar processing (cleavage, not methylation) of pre-18S rRNA in all eukaryotes (Rothé et al., 2017). U3 snoRNA associates with proteins common to all C/D box snoRNAs, including Nop56, Nop58, Nop1/FIBRILLARIN, and Small nuclear protein13 (Snu13), and with the U3-specific proteins Rrp9/U3-55k/Sof1 (RRNA processing9/Suppressor of fibrillarin1), Mpp10p (M phase phosphoprotein10), Lcp5p (Lethal conditional pap1 [poly(A) polymerase] allele), Imp3 and Imp4 (Interacting with Mpp10), and Rrp9/U3-55k to form U3 snoRNP (Rothé et al., 2017). Protein-based affinity purification of the U3 snoRNP complex identified the Utp proteins (U three) and demonstrated that U3 and Utp proteins are the main components of the 90S/SSU processome (Supplemental Data Set 2; Gallagher et al., 2004; Phipps et al., 2011).
In yeast, the 90S/SSU processome contains the U3 snoRNP complex and more than 70 proteins, including 17 Utp proteins (Utp1–Utp17) and Rrp5 (for RRNA processing5; Dragon et al., 2002). The Utp proteins form the UtpA (Utp4, -8, -9, -10, and -15, Pol5, and Nan1), UtpB (Utp1, -6, -12, -13, -18, and -21), and UtpC complexes (Utp22, Rrp7, Rrp36, Cka, and Ckb). The Utp complexes associate with the U3 snoRNP, Mpp10 (Mpp10-Imp3-Imp4), and DEAD box protein4 (Dbp4)/ Essential nuclear protein2 (Enp2)/ TFIIB-related factor2 (Brf2) complexes in a sequential manner on the nascent pre-rRNA transcript to form 90S particles (Dragon et al., 2002; Grandi et al., 2002; Gallagher et al., 2004).
Although the 90S/SSU processome has not yet been directly observed in plants, some evidence indicates that a processome complex might exist (Figure 4). First, a complex of NUCLEOLIN and U3 snoRNP was isolated and characterized in B. oleracea (Sáez-Vasquez et al., 2004a; Samaha et al., 2010). Second, the Arabidopsis genome encodes most of the Utps (Matsumura et al., 2016) and U3 snoRNP (Kojima et al., 2007) homologs (Supplemental Data Set 2). Third, proteomic analysis identified most of the processome homolog proteins in purified Arabidopsis nucleolar fractions (Palm et al., 2016; Montacié et al., 2017).
Most genes encoding proteins in the U3 snoRNP and the UtpA, UtpB, and UtpC classes in yeast have corresponding gene homologs in the human, Arabidopsis, and rice genomes. Exceptions include Utp8, -9, and -16, which appear to be yeast-specific, and Pol5, Rrp36, Lcp5, Ltv1, Pfa1, Slx9, and Nop19, which are present in the yeast and human genomes but have not been detected in the Arabidopsis or rice genomes (Supplemental Data Set 2). These genes might have evolved to such an extent that they cannot be identified based on sequence homology. Among the Arabidopsis factors without a yeast or human RBF homolog, the following protein-protein interactions were demonstrated: PCP1 and PCP2 proteins (PLANT-SPECIFIC COMPONENT OF THE PRE-rRNA PROCESSING COMPLEX1 and -2) interact with UtpA and/or UtpB (Figure 4; Ishida et al., 2016).
Genes encoding components of U3 snoRNPs, which are single genes in yeast and human, are typically duplicated in Arabidopsis and rice, whereas genes in other processome categories are usually present in single copies in Arabidopsis, with some exceptions (Utp12, Utp3, Utp24, Kre33, Mrd1, Nsr1, Fal1, and Prp43). Interestingly, there are also differences in copy number between Arabidopsis and rice, and sometimes it is difficult to determine which rice gene is the homolog of a particular Arabidopsis gene (Supplemental Data Set 2).
In B. oleracea, the U3 snoRNP complex interacts with NUCLEOLIN and plays a key role coupling the transcription and processing of nascent pre-rRNAs. Indeed, the Brassica NUCLEOLIN-U3 snoRNP complex binds 5′ETS rDNA sequences and cleaves the pre-rRNA specifically at the primary cleavage site P in the 5′ETS rRNA (Sáez-Vasquez et al., 2004a). The NUCLEOLIN-U3 snoRNP complex from Brassica contains more than 80 proteins, including FIBRILLARIN, NOP56, NOP58, and ribosomal proteins from the small ribosome subunit (Samaha et al., 2010). Utp homologs have not been identified in the NUCLEOLIN-U3 snoRNP fraction. Importantly, the nucleolin-U3 snoRNP complex was purified based on these interactions with rDNA (Caparros-Ruiz et al., 1997; Sáez-Vasquez et al., 2004a; Samaha et al., 2010), in contrast to the yeast processome complex. It is likely that specific forms of NUCLEOLIN-U3 snoRNP bind rDNA while others associate with the SSU-processome. During transcription, the NUCLEOLIN-U3 snoRNP complex bound to rDNA might translocate to the pre-rRNA and/or processome (Sáez-Vasquez et al., 2004a, 2004b).
Only a few Arabidopsis homologs of yeast Utp-encoding genes have been functionally characterized (Supplemental Data Set 3). Several putative Utp homologs are essential for gametogenesis and/or embryogenesis, including SWA1, PWP2, NOC4, TORMOZ, and NOF1 (Shi et al., 2005; Griffith et al., 2007; Harscoët et al., 2010; Missbach et al., 2013; Ishida et al., 2016). Similarly, studies involving disrupting the genes encoding THAL1, RRP5, ENP1, SW2, or PUM24 (Li et al., 2009; Missbach et al., 2013; Chen et al., 2016; Shanmugam et al., 2017; Maekawa et al., 2018a) indicated that these factors are essential for gametogenesis and/or embryogenesis. By contrast, disrupting RRP7 affects Arabidopsisplant growth and development as well as abscisic acid sensitivity. Furthermore, Arabidopsis RRP7 is required for 18S rRNA maturation, and rrp7 deficiency mutants genetically interact with mas2, nuc1, and hda6 deficiency mutants (Micol-Ponce et al., 2018).
nuc1 plants show altered growth and leaf development (Pontvianne et al., 2007), while nuc2 plants display delayed flowering time (Durut et al., 2014). Plants with disrupted Arabidopsis PUMILIO23 (APUM23) or G-PATCH DOMAIN PROTEIN1 (GDP1) display growth and developmental defects reminiscent of nuc1 mutants (Abbasi et al., 2010; Zhang and Muench, 2015; Kojima et al., 2018). In addition, apum23/salt hypersensitive9 (sahy9) plants exhibit hypersensitivity to salt mediated by abscisic acid (Huang et al., 2018). By contrast, Arabidopsis glucose hypersensitive40 (ghs40) plants exhibit hypersensitivity to glucose and abscisic acid (Hsiao et al., 2016). Moreover, the GHS40 protein coimmunoprecipitates with SWA1 and other Utp protein homologs (Ishida et al., 2016).
The subcomplexes Mpp10/Imp3/Imp4 and Brf2/Dbp4/Enp2 are the last to assemble into the processome (Figure 4; Granneman et al., 2003; Soltanieh et al., 2014). Arabidopsis homologs of yeast Mpp10/Imp3/Imp4 and Dbp4 have been identified, but none have been functionally characterized. However, it has been demonstrated that an MPP10-like protein interacts with Arabidopsis homologs of THAL1 and NOF1 in a presumed processome complex (Chen et al., 2016).
Most enzymatic activities associated with the processome in yeast have functional homologs in Arabidopsis. However, only a few of these activities have been characterized (Supplemental Data Set 3). RNA helicases of the DEAE-, DEAH-, and DExH-box families play key roles in pre-RNA processing and ribosome biogenesis (Fatica and Tollervey, 2002; Kressler et al., 2010; Henras et al., 2015). Several plant helicases that are homologs of yeast and human proteins associated with the SSU-processome or involved in the 40S/60S pathways have been reported (Supplemental Data Sets 2 and 4). Among these are the RNA helicases SW3/RH36, RH57, and RH10, which are Arabidopsis homologs of yeast Dbp8, Rok1, and Rrp3, respectively (Figure 4; Liu et al., 2010; Hsu et al., 2014; Matsumura et al., 2016).
In Arabidopsis, SW3/RH36 gene expression is essential for gametogenesis, whereas the SW3/RH36 protein interacts with EIGHTEEN S FACTOR2 (ESF2). The yeast Esf2 protein is a component of the SSU processome required for pre-rRNA binding and enhancing Dbp8 ATPase activity (Liu et al., 2010). Moreover, rh57 plants are impaired in seedling growth and hypersensitive to glucose. In rh57 plants, 45S/35S pre-rRNA accumulates and 40S subunit levels decrease (Hsu et al., 2014). Finally, thermosensitive rh10 plants display a weak pointed leaf phenotype at 22°C, and their leaves become narrower at 26°C. The accumulation of 35/33S pre-rRNA is even more evident at 26°C (Matsumura et al., 2016).
The Arabidopsis protein RH7/PRH75 is required for processing of 35S and 18S pre-rRNAs and likely associates with the SSU-processome. RH7/PRH75 participates in cold tolerance, but the accumulation of 35S/18S pre-rRNAs is not further affected in rh7 plants under cold conditions (Huang et al., 2016; Liu et al., 2016). RH7/PRH75 is closely related to human DDX21, which might coordinate rRNA transcription and processing (Calo et al., 2015).
Homologs of the yeast proteins Dhr1 (DEAH-box RNA helicases), Dbp4, Fal1 (eukaryotic translation initiation factor Four A-Like), and Has1 (Helicase associated with Set1 helicases) have been identified in Arabidopsis and rice (Supplemental Data Set 2). These homologs were also detected in the proteome of the Arabidopsis nucleolus and thus might be associated with the plant processome (Pendle et al., 2005; Palm et al., 2016; Montacié et al., 2017). For example, STRS1 and STRS2 (STRESS RESPONSE SUPPRESSOR1 and -2) are two Arabidopsis homologs of yeast Has1 that show nucleolar localization. STRS proteins are involved in heat stress tolerance and epigenetic gene silencing (Khan et al., 2014). However, their role in pre-rRNA transcription or processing has not been determined.
Among other processome-associated factors are the Arabidopsis homologs of Utp11, Rrp20 (RRNA processing20), and the U3 snoRNP-specific protein U3-55K (Senapin et al., 2003; Pagnussat et al., 2005; Urbánek et al., 2005).
By combining the above information with current models of SSU processome organization and assembly in yeast (Phipps et al., 2011; Soltanieh et al., 2014; Barandun et al., 2017; Chaker-Margot et al., 2017), we developed a model for 90S/SSU-processome assembly in Arabidopsis (Figure 4). According to this model, the UtpA, UtpB, U3 snoRNP, and UtpC subcomplexes and related factors undergo stepwise assembly on the nascent 40S pre-rRNA while it is being transcribed by RNA Pol I. The Arabidopsis SSU-processome appears to exhibit some plant-specific attributes, including the interactions of PCP1/2 with UtpA and UtpB class proteins.
In yeast and animal cells, the first pre-40S particle is obtained following disassembly of the SSU processome. In addition to 20S pre-rRNA and 40S RPS, this pre-40S particle contains the assembly factor Enp1/Bystin, the endonuclease Nob1, its interacting partner Pno1/Dim2, the methyltransferase Dim1, and the RNA helicase Prp43/DHX15. In yeast, Dim1 dissociates from the pre-40S particle in the cytoplasm, while in mammals, DIM1 appears to dissociate in the nucleus. Subsequently, Ltv1, the kinase Hrr25/CKI, and the ATPase/kinase Rio2 are incorporated into the pre-40S particle in the nucleolus. The cytoplasmic export of the pre-40S particle requires the module Noc4/Utp19-Nop14/Utp2 and Crm1, which recognizes nuclear export signals (Schäfer et al., 2003; Kühn et al., 2009). In the cytoplasm, Enp1p/Bystin, Nob1, Ltv1, and other proteins dissociate from pre-40S particles while RACK1 and, later, RPS are incorporated to form mature 40S particles (Figure 4; reviewed in Cerezo et al., 2019).
Arabidopsis homologs of yeast Nob1, Noc4, and Enp1 are required pre-rRNA processing (Missbach et al., 2013), and Dim1 is required for N6-dimethylation of 18S rRNA (Wieckowski and Schiefelbein, 2012). Arabidopsis NOB1 and ENP1 are nuclear/nucleolar and nuclear/cytoplasmic proteins, respectively, like their yeast homologs, while Arabidopsis NOC4 and DIM1A strongly localize to the nucleolus (Wieckowski and Schiefelbein, 2012; Missbach et al., 2013). Finally, RACK1 copurifies with U3 snoRNP in Brassica species (Samaha et al., 2010) and with the 80S ribosome in Arabidopsis (Giavalisco et al., 2005). Plant genomes also encode homologs of Prp43/DHX15 and Nop14/Utp2, which remain uncharacterized (Figure 4; Supplemental Data Set 2).
Although the roles of many Arabidopsis homologs of the yeast and mammalian 90S/SSU processome and pre-40S particles remain unknown, current data indicate that they are evolutionarily conserved in plants and that their expression is required for plant growth and development. At the molecular level, disruption of these genes also affects pre-rRNA transcription and/or processing and, in some cases, provokes changes in the morphology and structure of the nucleolus (Supplemental Data Set 3).
MATURATION OF PRE-60S PARTICLES
The assembly of pre-60S particles begins on nascent pre-60S rRNA sequences after cotranscriptional cleavage of the pre-rRNA, leading to the release of pre-40S particles. A vast number of protein factors and complexes have been identified that are involved in 60S assembly (Phipps et al., 2011; Henras et al., 2015; Espinar-Marchena et al., 2017; Konikkat and Woolford, 2017; Bassler and Hurt, 2019; Klinge and Woolford, 2019).
The Arabidopsis genome contains homologs for most 60S RBFs, indicating that the mechanisms of 60S assembly are conserved in animals, yeast, and plants (Figure 5; Supplemental Data Set 4). With a few exceptions, most of these protein homologs of yeast and human 60S RBFs are encoded by single-copy genes in Arabidopsis. The disruption or inhibition of these orthologous genes thus causes pre-60S processing deficiencies along with plant growth and developmental defects, or lethality during gametogenesis or embryogenesis (Supplemental Data Set 4).
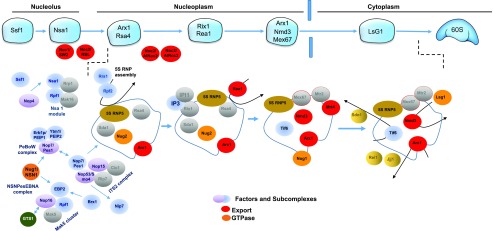
Figure 5.
Model for the Assembly and Maturation of Pre-60S Particles in Arabidopsis.
The picture illustrates the so-called yeast 60S particle in the nucleolus (Ssf1 and Nsa), nucleoplasm (Arx1/Rsa4, Rix1/Rea1, and Arx1/Nmd3/Mex67), and cytoplasm (Lsg1). Arabidopsis homologs of yeast processome factors and subcomplexes (blue and violet), protein export (red), and GTPase (orange) activities are represented. Those not yet identified are shown in gray. Plant GTS1, orthologous to human WDR39, is represented in dark green. The figure was adapted and drawn from simplified models of the assembly steps of the 60S particles in yeast (Shchepachev and Tollervey, 2016; Kressler et al., 2017). Accession numbers for all the Arabidopsis factors are provided in Supplemental Data Set 4.
GTPase homologs of the yeast and human proteins Nuclear GTPase1(Nug1)/NUCLEOSTEMIN1 (NS1), Nug2/Nucleolar GTPase2 (Nog2), Nog1, and Large Subunit GTPase1 (Lsg1/LSG1) are required for maturation of the pre-60S subunit in Arabidopsis (Im et al., 2011; Weis et al., 2014; Jeon et al., 2015; Zhao et al., 2015; Lee et al., 2017). However, no GTPase homologs have thus far been shown to mediate pre-40S subunit maturation.
Nucleostemin forms a large protein complex (>700 kD) that cofractionates with the pre-60S ribosomal subunit in yeast (Romanova et al., 2009). This appears to be the case in plants as well. Indeed, Arabidopsis NSN1 interacts with PESCADILLO (PES) and Epstein-Barr virus nuclear antigen binding protein2 (EBP2) orthologs (Jeon et al., 2015).
In yeast, Brx1 also interacts with Ebp2 and is required for the assembly of 60S ribosomal subunits (Shimoji et al., 2012). The Arabidopsis genome encodes two homologs of yeast Brx1: BRX1-1 and BRX1-2. brx1-1 and brx1-2 single mutants grow slowly and display defects in pre-rRNA processing. BRX1-1 and BRX1-2 likely play redundant roles in Arabidopsis, as genetic crosses between brx1-1 and brx1-2 did not yield any double homozygous plants (Weis et al., 2015b).
Arabidopsis PES interacts with PESCADILLO-INTERACTING PROTEIN1(PEIP1) and PEIP2, the homologs of Eukaryotic ribosome biogenesis1(Erb1)/BOP1Block of proliferation1 (BOP1), and Ytm1/ WD40 repeat2 protein (WDR2; Cho et al., 2013; Zografidis et al., 2014). Hence, plants might also express the trimeric PES-BOP1-WDR12 complex (Ahn et al., 2016), which is essential for the processing and maturation of mammalian 5.8S and 28S rRNAs. This complex is thought to associate with other proteins to form a much larger complex (Rohrmoser et al., 2007). Yeast Nug2 (NS2 in human) is a regulatory GTPase that monitors pre-60S maturation. Nug2 associates with two pre-60S remodeling factors, dynein-related AAA-type ATPase (Rea1) and its cosubstrate Ribosome assembly4 (Rsa4; Matsuo et al., 2014). In Arabidopsis and rice, the Nug2 homologs exhibit GTPase activity and are required for processing 27S pre-rRNA into 25S rRNA (Im et al., 2011).
No plant homologs of yeast Rsa4 have thus far been reported. However, characterization of the dwarf and short root1 (dsr1) mutant revealed that DSR1 is the Arabidopsishomolog of the yeast Rea1 and human MDN1 proteins (Li et al., 2016). Mutating a key amino acid position in ArabidopsisMDN1 (Glu to Lys at position 3838) induces pleiotropic developmental phenotypes, including slow germination, short roots, dwarf shoots, and reduced seed set (Li et al., 2016).
Moreover, homologs of the yeast Nog1 protein have been reported in Nicotiana benthamiana and Arabidopsisplants. The N. benthamiana homolog of Nog1 is required for processing of 35S, 33S, and 27S intermediates (Guo et al., 2018), while Arabidopsis NOG1-1 and NOG1-2 are involved in biotic and abiotic stress responses (Lee et al., 2017).
In Arabidopsis, the PES, NSN1, and NUG2/NOG2 homologs are encoded by single genes, and their expression is required for plant growth and/or development. Indeed, PES gene silencing causes growth arrest and cell death (Cho et al., 2013), NSN1 gene expression is required for embryogenesis and shoot apical meristem development (Wang et al., 2012; Jeon et al., 2015), while knockdown of NUG2/NOG2 leads to defective growth on medium containing cycloheximide (Im et al., 2011).
The Arabidopsis genome encodes two homologs of yeast Lsg1 and Rei1 (Required for isotropic bud growth1). Yeast Lsg1 and Rei1 proteins are involved in the recycling of the pre-60S subunit export adaptor Nmd3 (Nonsense-mediated mRNA decay3) and Arx1 (Associated with ribosomal export complex; Hedges et al., 2005; Hung and Johnson, 2006). In Arabidopsis, LSG1-2 physically associates with 60S/pre-60S particles, and in lsg1-2 plants, 18S pre-rRNA accumulates and monosome levels decrease, indicating that LSG1-2 is the functional homolog of yeast Lsg1 (Weis et al., 2014; Zhao et al., 2015). Arabidopsis LSG1-1 does not appear to be involved in ribosome biogenesis. The lsg1-2 mutants show severe defects in plant development and auxin responses, while lsg1-1 mutants grow more slowly than wild-type plants but do not show other defects (Weis et al., 2014; Zhao et al., 2015). Nevertheless, Arabidopsis plants in which both LSG1 genes are disrupted cannot be recovered, suggesting that LSG1-1 and LSG1-2 play partially redundant roles (Weis et al., 2014). By contrast, Arabidopsis plants with the simultaneous mutation of REIL1 and REIL2 are fully viable under optimal growth conditions. However, under cold conditions, the growth of reil1 reil2 double mutants is arrested, which is correlated with delayed accumulation of 60S particles (Schmidt et al., 2013). Arabidopsis REIL1, but not REIL2, complemented the temperature-dependent growth defect of the yeast ∆rei1 mutant. However, in contrast to yeast Rei1, Arabidopsis REIL1 is also present in translating ribosomes, pointing to the functional divergence of REIL proteins during plant evolution (Beine-Golovchuk et al., 2018).
The Arabidopsis genome also encodes homologs of the yeast export factors Nmd3 and Arx1 (Supplemental Data Set 4). Arabidopsis NMD3 plays a role in the nuclear export of 60S ribosomal subunits (Chen et al., 2012), but its functional interactions with LSG1-2, REIL1/2, or BRX1 have not been determined. In yeast, transport of 60S ribosome subunits from the nucleolus to the cytoplasm also requires the nucleolar proteins Noc1 to Noc3. The Noc1-Noc2 complex is likely required for the early transport of ribosome particles from the nucleolus to the nucleoplasm, while the Noc3-Noc2 complex appears to be involved in their subsequent transport from the nucleoplasm to the cytoplasm (Milkereit et al., 2001). In Arabidopsis, SLOW WALKER2 (SWA2) encodes a Noc1/Mak21 homolog that is essential for female gametophyte development, while REBELOTE (RBL) encodes a Noc2 homolog that is required for floral meristem termination (Prunet et al., 2008; Li et al., 2009). The Arabidopsis genome also contains uncharacterized second copies of genes encoding homologs of yeast Noc2 and Noc3p (Prunet et al., 2008).
The Arabidopsis proteins ARPF2 and ARRS1 are homologs of yeast Rpf2 and Rrs1 (Maekawa et al., 2018b). In yeast, Rpf2 and Rrs1 form a protein complex that binds to 5S rRNA and assembles 5S RNP into pre-60S particles (Madru et al., 2015). Arabidopsis ARPF2 and ARRS1 are encoded by single-copy genes. Mutating ARPF2 results in aborted embryos, while the overexpression of ARPF2 or ARRS1 results in shorter stems and abnormal leaf morphology.
The Arabidopsis genome also encodes homologs of the yeast/human exonucleases Rrp17 and Nop53/SMO4 as well as two Tif6 homologs (Oeffinger et al., 2009; Kato et al., 2010; Zhang et al., 2015). In Arabidopsis, smo4 plants exhibit reduced organ size (Zhang et al., 2015), while mutation of the Tif6 homolog gene At-eiF6;1 causes embryo lethality (Kato et al., 2010). The precise roles of these protein factors in 60S rRNA processing, assembly, and/or the export of ribosome particles remain unsettled.
Little is known about the RNA helicases involved in pre-60S ribosomal subunit processing and assembly in plants. For instance, Arabidopsis IRP6 (also named DRH1; Okanami et al., 1998) is involved in the processing of 27SB pre-rRNA at the C2 site (Palm et al., 2019). Remarkably, RNA helicases from Arabidopsis might affect the processing of different pre-rRNA substrates from their yeast counterparts. This is the case for mRNA transport regulator4 (MTR4), which is required for the processing of 18S pre-rRNA and degradation of rRNA processing by-products but not for 5.8S pre-rRNA processing, as observed in yeast (Lange et al., 2011).
Of the ∼70 yeast LSU-60S RBF proteins listed in Supplemental Data Set 4 (constructed from Schmidt et al., 2014; Biedka et al., 2018; Zisser et al., 2018), ∼50 have Arabidopsis homologs. The Arabidopsis genome also encodes homologs of human RBFs and proteins with homology to bacterial sequences. For instance, GIGANTUS1 (GTS1), the Arabidopsis homolog of human WD40-REPEAT protein (WDR39), interacts with L19e and NOP16 and regulates plant growth and development (Gachomo et al., 2014). Arabidopsis COLD SHOCK DOMAIN PROTEIN3 (CSP3), which has an RNA chaperone function like that of Escherichia coli cold shock proteins, interacts with RPL26A, RPL40A, GAR1, and NUC1 (Kim et al., 2013) and also with the cold tolerance-related RNA helicase RH7/PRH75 (Huang et al., 2016).
Taken together, the data indicate that the overall assembly of 60S ribosome particles occurs in a similar manner in plants, yeast, and mammalian cells (Figure 5). However, differences in the timing of events (transcriptional or posttranscriptional) and pre-rRNA cleavage patterns (5′ETS-first or ITS-first) suggest that there are differences in the assembly of 40S and 60S particles on nascent pre-rRNA transcripts. As observed for 40S-related RBF genes, it appears that some 60S homolog proteins are also encoded by two Arabidopsisgenes. In some cases, these duplicated genes have redundant ribosomal functions (REIL1/REIL2 and BRX1-1/BRX1-2), whereas in other cases, the two genes have diverged, leaving only one copy encoding the ancestral function in ribosome biogenesis (LSG1-1/LSG1-2). Plant RBFs might also have acquired some unique functions in ribosome biogenesis. For instance, ArabidopsisREIL1 is present in the translating ribosome, in contrast to its yeast homolog, which only interacts with pre-60S particles (Beine-Golovchuk et al., 2018). Likewise, while in yeast, a single Noc2 protein might interact with Noc1 and Noc3, in Arabidopsis, both NOC2 and RBL interact with SWA2/NOC1 and NOC3 (de Bossoreille et al., 2018). The functional redundancy and specificity of these protein-protein interactions in ribosome biogenesis have not been reported. Finally, studies about the nucleolar, nucleoplasmic, and cytosolic distribution of ribosomal proteins (Montacié et al., 2017; Palm et al., 2018) suggest that the assembly and maturation of 60S particles differ between plants and yeast.
RBFs IN CULTIVATED PLANT SPECIES
Most of our knowledge concerning the functional organization of plant rRNA genes, plant ribosome biogenesis, and related regulatory pathways comes from Arabidopsis plants of the Col-0 ecotype. However, RBFs have been described and characterized in several plant species of agronomic interest, providing important insights into rRNAs, ribosome biogenesis, and stress response pathways in plants.
In rice, genes for NUCLEOLIN and the DEAD-box helicase THERMOTOLERANT GROWTH REQUIRED1 (TOGR1) have been reported (Sripinyowanich et al., 2013; Wang et al., 2016). Interestingly, overexpression of NUCLEOLIN enhances salt adaptation, while TOGR1 is required for thermotolerance in rice. Furthermore, the characterization of SUPER APICAL DORMANT1 (SAD1) revealed an interaction between the Mediator complex and the RNA Pol I machinery in rice. SAD1 is an RNA Pol I subunit orthologous to yeast RPA34.5 and ArabidopsisNRPA14, and Mediator is a multifunctional complex involved in RNA transcription, elongation, termination, processing, and chromatin looping (Li et al., 2015; Ream et al., 2015). Other rice proteins showing homology to RBFs are listed in Supplemental Data Set 2. In some cases, however, there are several copy genes of particular RBF in rice, and it is therefore difficult to determine which copies are the homologs in Arabidopsis.
In maize, three protein-coding genes related to rRNA and ribosome biosynthesis have been characterized. The first, ZmDRH1, encodes a DEAE box RNA helicase homologous to RH14/DRH1 in Arabidopsis. The ZmDRH1 protein localizes to the nucleolus and interacts with the RNA binding protein MA6 and FIBRILLARIN (Gendra et al., 2004). The others, UNHEALTHY RIBOSOME BIOGENESIS2 (ZmURB2) and RIBOSOME EXPORT ASSOCIATED1(ZmREAS1) are required for kernel development and are involved in pre-rRNA processing and ribosome assembly (Qi et al., 2016; Wang et al., 2018).
In barley (Hordeum vulgare), a functional homolog of yeast rRNA PROCESSING PROTEIN46 (RRP46) was described. Yeast RRP46 is a critical component of the RNA exosome complex. Silencing of barley RRP46 provoked tip cell death in leaves and induced the accumulation of pre-rRNAs (Xi et al., 2009).
Finally, Simm et al. (2015) reported the identification of putative RBFs and their expression in tomato (Solanum lycopersicum). This analysis revealed that 25% of all predicted RBFs are encoded by more than one gene and that clusters of RBFs and RPs are coregulated.
RIBOSOME RESPONSE PATHWAY
In both animal and plant cells, rRNA and ribosome biogenesis can be disturbed by various stress conditions. These include nutrient starvation, changes in energy status, hypoxia, temperature changes, DNA damage, genotoxic stress, and other alterations to ribosome biogenesis factors. Disturbed ribosome biogenesis is typically accompanied by changes in nucleolar morphology and the activation of pathways leading to cell cycle arrest or apoptosis, which is commonly referred to as the ribosomal or nucleolar stress response (Boulon et al., 2010; Tsai and Pederson, 2014; Stepinski, 2016; Kalinina et al., 2018).
In animals, the ribosomal stress response involves the tumor protein53 (p53)/Murine Double Minute2 (MDM2) pathway (Boulon et al., 2010; van Sluis and McStay, 2017). MDM2 is an E3 ubiquitin ligase, and p53 is an antitumor transcriptional regulator that drives cell cycle arrest, senescence, and apoptosis. Under normal conditions, MDM2 targets, ubiquitinates, and programs the degradation of p53. By contrast, under stress conditions, MDM2 interacts with ribosomal proteins released from the nucleolus, which prevents p53 degradation (Gilkes and Chen, 2007; James et al., 2014). The ribosomal stress response also involves energy-dependent Nucleolar Silencing Complex (eNoSC) and Myb binding protein1A (MYBB1A). eNoSC is composed of the protein NUCLEOMETHYLIN (NML), the histone deacetylase Sirtuin-1 (SIRT1), and the histone methyltransferase Suppressor of Variegation39 Homolog1 (SUV39H1). In this pathway, eNoSC senses the energy status of the cell and represses rRNA transcription under glucose deprivation. As a result, MYBB1A translocates from the nucleolus to the nucleoplasm, inducing the activation of p53 by enhancing its acetylation by p300 (Murayama et al., 2008; Kumazawa et al., 2011).
Plants do not have homologs of the mammalian MDM2, p53, or NML proteins. However, the characterization of ANAC082, a NAC family transcriptional activator, suggests that plants have a specific ribosomal or nucleolar stress response pathway (Ohbayashi et al., 2017). ANAC082 was identified genetically in a mutagenized population of rid2 plants as the suppressor mutant suppressor of root initiation defective two1 (sriw1), which is able to rescue the impaired developmental abnormalities of rid2, including root length, leaf blade size, and venation patterns, but not the accumulation of rRNA precursors. Similarly, the sriw1 mutation also alleviates the developmental abnormalities of rid3, oligocellula5, and rh10 mutants, but not their impaired pre-rRNA processing. Also, ANAC082 expression is induced in temperature-sensitive RBF mutants, and ANAC082 appears to function downstream of ribosome biogenesis and nucleolar perturbations. Interestingly, the 5′ untranslated region of ANAC082 mRNA contains a uORF (upstream open reading frame) encoding a 37-amino acid peptide that might negatively regulate ANAC082 protein translation. Ribosomal stress or nucleolar perturbations might inhibit the expression of this uORF, allowing increased ANAC082 protein expression (Ohbayashi et al., 2017; reviewed in Ohbayashi and Sugiyama, 2018).
In Arabidopsis, the inhibition of RBF or RP gene expression induces a wide variety of plant growth and developmental phenotypes, including embryo lethality, delayed flowering time, and abnormal leaf and root phenotypes (Supplemental Data Sets 3 and 4). Cytological investigations have shown that the nucleolus becomes larger in tormoz (Griffith et al., 2007), nof1 (Harscoët et al., 2010), apum23 (Abbasi et al., 2010), rid2 (Ohbayashi et al., 2011; Shinohara et al., 2014), ren1 (Reňák et al., 2014), la1 (Fleurdepine et al., 2007), and domino1 (Lahmy et al., 2004) compared with wild-type plants. By contrast, the nucleolus is small in fem111 (Portereiko et al., 2006) and disorganized in nuc1 (Pontvianne et al., 2007) plants. The accumulation of pre-rRNA is affected in nof1, apum23, brx1, lsg1, and other mutants as well (Supplemental Data Sets 1 and 3).
Remarkably, some molecular phenotypes only become evident under specific growth conditions (Supplemental Data Sets 3 and 4). For instance, in response to high sugar concentrations, increased accumulation of pre-rRNA is observed in rh57 (Hsu et al., 2014) and apum24 (Maekawa et al., 2018a). At high temperatures, pre-rRNA accumulates in rh10 (Matsumura et al., 2016) and rid3 (Shinohara et al., 2014). At low temperatures, reil1 reil2 double mutants accumulate less ribosome subunits compared with wild-type plants (Beine-Golovchuk et al., 2018). Furthermore, rh10 nucleoli are enlarged at 28°C (Matsumura et al., 2016), whereas apum24 nucleoli become smaller in the presence of 50 mM Glc (Maekawa et al., 2018a).
Notably, APUM24 interacts with other pre-rRNA processing factors and recruits LAS1, the endonuclease involved in the processing of ITS2 (Figure 3). Perhaps Arabidopsis APUM24 links the nucleolar stress response pathway to altered sugar responses (Maekawa et al., 2018a). How APUM24 might sense the energy status of the cell remains unclear (reviewed in Maekawa and Yanagisawa, 2018).
We surveyed the expression of the genes listed in Supplemental Data Sets 3 and 4 using the Bio-Analytical Resource for Plant Biology (bar.utoronto.ca; Winter et al., 2007), which is primarily based on Affymetrix microarray experiments. We investigated developmental stage-specific effects and abiotic stress responses. In Supplemental Data Sets 3 and 4, we report the developmental stages at which the expression level of each gene is the highest. Several developmental stages are reported, such as dry seeds, seeds 24 h after imbibition, rosette leaves, and the apical meristem (including three situations: vegetative, transition, and inflorescence). For abiotic stress, the analyzed experiments deal with cold, osmotic stress, salt, drought, genotoxic stress, oxidative stress, UV-B light exposure, wounding, and heat treatment, as described by Winter et al. (2007).
The most striking result is that most of the genes surveyed show a remarkably homogeneous pattern of expression (Supplemental Data Sets 3 and 4). Not surprisingly, all these genes involved in the biosynthesis of rRNA and ribosomes are highly expressed in the most active tissues or organs, such as imbibed seeds, meristems, floral bud, and developing siliques and embryos. In some cases, there are small differences in the relative expression between the different stages of the apical meristem, suggesting that the transition from the vegetative to inflorescence meristem is associated with fine-tuning of the expression of genes involved in ribosome biogenesis, although this needs to be confirmed by studies at the protein level. An interesting observation is that many of the genes are expressed at unexpectedly high levels in dry seeds, which suggests that dry seeds contain all the mRNA necessary to immediately resume ribosome biogenesis as soon as the seeds are imbibed.
Finally, of the genes involved in stress responses, it is striking that most of the genes listed in Supplemental Data Sets 3 and 4 are induced by three major stresses: cold, heat, and UV-B light. This finding suggests that ribosome biogenesis can adapt to respond to these stresses by increasing the transcript levels of these key genes. Some of the genes are equally responsive to these various stresses, whereas others selectively respond to one or two of the stresses. The physiological significance of these differences is not yet clear, but the observed responses again suggest a fine-tuning of the regulation of rRNA and ribosome biogenesis. Surprisingly, very few of these genes respond to osmotic stress, drought, or wounding at the transcriptional level. An intriguing possibility is that the responses to these stresses might occur at the posttranscriptional level. In the future, it would be interesting to determine whether this is a general situation in plants or a situation restricted to Arabidopsis and several related species.
CONCLUDING REMARKS
Based on studies in Arabidopsisand other plant species, one can reasonably conclude that rRNA gene expression and ribosome assembly in plants resemble the corresponding processes in yeast and/or mammalian cells. However, these studies also revealed features of ribosome biogenesis particular to plants (i.e., features specific to plant growth and development and to the sessile lifestyle of plants, requiring them to adapt to local environmental conditions). With a few exceptions, all RBF genes detected in the yeast and human genomes are also present in Arabidopsis, rice, and presumably other plant species. However, there are significant differences in gene copy number. For instance, genes encoding the U3 snoRNP complex are duplicated in plants, whereas they are single-copy genes in yeast and animals. The possibility that duplicated genes for U3 snoRNP proteins could be subfunctionalized in Arabidopsisor rice is intriguing and should be investigated in future studies. This, together with the finding that the NUCLEOLIN-U3 snoRNP complex binds to rDNA in Brassica species (Sáez-Vasquez et al., 2004a, 2004b), suggests that the biosynthesis of rRNA and ribosomes might be more complex processes in plants than in yeast and animals. The combination of these processes could produce slightly different ribosomes depending on the developmental phase or stress conditions. Notably, in addition to the genes directly involved in transcription and assembly of the processome/pre-40S and pre-60S particles, all of the ribosomal protein genes belong to small multigene families (Barakat et al., 2001). Few situations are well documented concerning the respective functions of these duplicated genes. The case of the two nucleolin genes illustrates that duplicated genes can acquire distinct and sometimes antagonistic functions, allowing them to induce or silence distinct variants of rRNA genes.
It appears that some of the RBFs in yeast lack homologs in Arabidopsis, which suggests that either there is high sequence divergence between the species or that some of these factors simply do not exist in plant species. In turn, genetic, genomic, and proteomic analyses have revealed several nucleolar proteins that lack homology with yeast and/or mammalian RBFs and that might thus be considered plant-specific (such as PCP1 and PCP2). Functional characterization of these factors would provide novel insight into the impact of rRNA transcription and processing as well as ribosome assembly on plant growth, development, and adaptation to environmental conditions.
Studies concerning the functional organization of rDNA in Arabidopsis have also raised new questions. Rabanal et al. (2017) recently demonstrated that rRNA variants can be expressed from NOR2, from NOR4, or from both NORs in different Arabidopsis ecotypes. The functional or structural significance of sequence variation in the 3′ETS of Arabidopsis rRNA genes and in those of other plant species is also unclear. Variation in the 3′ETS likely does not result in ribosome heterogeneity, because these sequences are removed from mature rRNAs, although it cannot be completely excluded that these variants are genetically linked to discrete single-nucleotide polymorphisms that have yet to be detected in the mature RNAs. High-throughput DNA sequencing technologies that generate ultra-long reads (well over 10,000 bp) should begin to paint a better picture of 45S rDNA organization.
The modification of structural and catalytic rRNAs (18S, 5.8S, and 25S) might have a major impact on the fine-tuning of ribosome translational efficiency and capacity, as demonstrated in yeast and mammalian cells. How plant growth, development, or environmental conditions affect rRNA processing in plants is almost completely unknown. It would also be interesting to determine if plant IGS transcripts are involved in rDNA silencing and/or protein sequestering, as reported in mammalian cells in response to heat stress and acidosis.
The existence of two distinct pre-rRNA processing pathways remains controversial and raises many questions concerning the factors and conditions (developmental or environmental) that govern either the ITS1-first or 5′ETS-first processing pathways. The recent report of a third, plant-specific ITS2 processing pathway is intriguing and adds a new level of complexity to pre-rRNA processing in plants. Clearly, energy status and temperature affect RBF gene expression and pre-rRNA processing. However, the functional crosstalk between this processing and cellular and/or external signals remains unknown.
ANAC082 plays a role as a ribosomal stress response mediator in plants and is likely the plant functional homolog of mammalian p53. However, p53 is structurally different from ANAC082. Furthermore, other key players of the animal ribosomal stress response pathway are absent in plants. This suggests that the molecular mechanisms of ribosomal stress responses are different in plant cells compared with animal cells. The identification and characterization of ANAC082-related factors should help us to better understand ribosomal stress responses in plants.
Thus, although it is clear that plant growth and development are directly linked to ribosome synthesis and protein translation, many of the molecular mechanisms and signals that connect cellular and environmental conditions with rRNA synthesis and ribosome assembly remain to be characterized in plants. The question of the heterogeneity of ribosomes and their fine-tuned regulation is a challenge for the future. An associated issue is the protein synthesis capacity and specificity of slightly different ribosomes: can they preferentially translate different sets of mRNAs?
Accession Numbers
Sequence numbers from this article can be found in Supplemental Data Sets 1 to 4.
Supplemental Data
Supplemental Data Set 1. RBFs from S. cerevisiae and H. sapiens identified and/or characterized in Arabidopsis plants.
Supplemental Data Set 2. Components of the SSU processome and pre-40S Particles from S. cerevisiae and H. sapiens in Arabidopsis and O. sativa plants.
Supplemental Data Set 3. Characterization studies of the Arabidopsis SSU processome and pre-40S orthologs.
Supplemental Data Set 4. RBFs from S. cerevisiae and H. sapiens identified and/or characterized in Arabidopsis plants.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank D. Gagliardi, H. Lange, and T. Blevins for the detailed critical reading of the manuscript and C.S. Pikaard for comments and correction of the English. We also thank J. Salses and C. Pont for the identification of rice homologs of known RBFs and all the team members who have contributed to our research on rRNA dynamics and ribosome biogenesis in plants. We apologize for any articles we forgot to mention or that could not be included due to space restrictions. This work was supported by the CNRS and by the Agence Nationale de la Recherche (RiboStress (grant no. 17-CE12-0026-01 to J.S.-V.).
AUTHOR CONTRIBUTIONS
J.S.-V. and M.D. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Abbasi N., Kim H.B., Park N.I., Kim H.S., Kim Y.K., Park Y.I., Choi S.B. (2010). APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 64: 960–976. [DOI] [PubMed] [Google Scholar]
- Abou-Ellail M., Cooke R., Sáez-Vásquez J. (2011). Variations in a team: Major and minor variants of Arabidopsis thaliana rDNA genes. Nucleus 2: 294–299. [DOI] [PubMed] [Google Scholar]
- Ahn C.S., Cho H.K., Lee D.H., Sim H.J., Kim S.G., Pai H.S. (2016). Functional characterization of the ribosome biogenesis factors PES, BOP1, and WDR12 (PeBoW), and mechanisms of defective cell growth and proliferation caused by PeBoW deficiency in Arabidopsis. J. Exp. Bot. 67: 5217–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert A.C., Denton M., Kermekchiev M., Pikaard C.S. (1999). Histone acetyltransferase and protein kinase activities copurify with a putative Xenopus RNA polymerase I holoenzyme self-sufficient for promoter-dependent transcription. Mol. Cell. Biol. 19: 796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert B., Colleran C., Léger-Silvestre I., Berger A.B., Dez C., Normand C., Perez-Fernandez J., McStay B., Gadal O. (2013). Structure-function analysis of Hmo1 unveils an ancestral organization of HMG-box factors involved in ribosomal DNA transcription from yeast to human. Nucleic Acids Res. 41: 10135–10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameismeier M., Cheng J., Berninghausen O., Beckmann R. (2018). Visualizing late states of human 40S ribosomal subunit maturation. Nature 558: 249–253. [DOI] [PubMed] [Google Scholar]
- Antosch M., Schubert V., Holzinger P., Houben A., Grasser K.D. (2015). Mitotic lifecycle of chromosomal 3xHMG-box proteins and the role of their N-terminal domain in the association with rDNA loci and proteolysis. New Phytol. 208: 1067–1077. [DOI] [PubMed] [Google Scholar]
- Armache J.P., et al. (2010). Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc. Natl. Acad. Sci. USA 107: 19748–19753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audas T.E., Jacob M.D., Lee S. (2012). Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol. Cell 45: 147–157. [DOI] [PubMed] [Google Scholar]
- Barakat A., Szick-Miranda K., Chang I.F., Guyot R., Blanc G., Cooke R., Delseny M., Bailey-Serres J. (2001). The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 127: 398–415. [PMC free article] [PubMed] [Google Scholar]
- Barandun J., Chaker-Margot M., Hunziker M., Molloy K.R., Chait B.T., Klinge S. (2017). The complete structure of the small-subunit processome. Nat. Struct. Mol. Biol. 24: 944–953. [DOI] [PubMed] [Google Scholar]
- Barneche F., Steinmetz F., Echeverría M. (2000). Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem. 275: 27212–27220. [DOI] [PubMed] [Google Scholar]
- Barneche F., Gaspin C., Guyot R., Echeverría M. (2001). Identification of 66 box C/D snoRNAs in Arabidopsis thaliana: Extensive gene duplications generated multiple isoforms predicting new ribosomal RNA 2′-O-methylation sites. J. Mol. Biol. 311: 57–73. [DOI] [PubMed] [Google Scholar]
- Bassler J., Hurt E. (2019). Eukaryotic ribosome assembly. Annu. Rev. Biochem. 88: 281–306. [DOI] [PubMed] [Google Scholar]
- Beine-Golovchuk O., Firmino A.A.P., Dąbrowska A., Schmidt S., Erban A., Walther D., Zuther E., Hincha D.K., Kopka J. (2018). Plant temperature acclimation and growth rely on cytosolic ribosome biogenesis factor homologs. Plant Physiol. 176: 2251–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. (2011). The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529. [DOI] [PubMed] [Google Scholar]
- Biedka S., Micic J., Wilson D., Brown H., Diorio-Toth L., Woolford J.L. Jr (2018). Hierarchical recruitment of ribosomal proteins and assembly factors remodels nucleolar pre-60S ribosomes. J. Cell Biol. 217: 2503–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierhoff H., Dundr M., Michels A.A., Grummt I. (2008). Phosphorylation by casein kinase 2 facilitates rRNA gene transcription by promoting dissociation of TIF-IA from elongating RNA polymerase I. Mol. Cell. Biol. 28: 4988–4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierhoff H., Schmitz K., Maass F., Ye J., Grummt I. (2010). Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold Spring Harb. Symp. Quant. Biol. 75: 357–364. [DOI] [PubMed] [Google Scholar]
- Borovjagin A.V., Gerbi S.A. (1999). U3 small nucleolar RNA is essential for cleavage at sites 1, 2 and 3 in pre-rRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J. Mol. Biol. 286: 1347–1363. [DOI] [PubMed] [Google Scholar]
- Boulon S., Westman B.J., Hutten S., Boisvert F.M., Lamond A.I. (2010). The nucleolus under stress. Mol. Cell 40: 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.W., Clark G.P., Leader D.J., Simpson C.G., Lowe T. (2001). Multiple snoRNA gene clusters from Arabidopsis. RNA 7: 1817–1832. [PMC free article] [PubMed] [Google Scholar]
- Brown J.W., Echeverria M., Qu L.H. (2003a). Plant snoRNAs: Functional evolution and new modes of gene expression. Trends Plant Sci. 8: 42–49. [DOI] [PubMed] [Google Scholar]
- Brown J.W., Echeverria M., Qu L.H., Lowe T.M., Bachellerie J.P., Hüttenhofer A., Kastenmayer J.P., Green P.J., Shaw P., Marshall D.F. (2003b). Plant snoRNA database. Nucleic Acids Res. 31: 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess A.L., David R., Searle I.R. (2015). Conservation of tRNA and rRNA 5-methylcytosine in the kingdom Plantae. BMC Plant Biol. 15: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E., Flynn R.A., Martin L., Spitale R.C., Chang H.Y., Wysocka J. (2015). RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature 518: 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparros-Ruiz D., Lahmy S., Piersanti S., Echeverría M. (1997). Two ribosomal DNA-binding factors interact with a cluster of motifs on the 5′ external transcribed spacer, upstream from the primary pre-rRNA processing site in a higher plant. Eur. J. Biochem. 247: 981–989. [DOI] [PubMed] [Google Scholar]
- Caton E.A., Kelly E.K., Kamalampeta R., Kothe U. (2018). Efficient RNA pseudouridylation by eukaryotic H/ACA ribonucleoproteins requires high affinity binding and correct positioning of guide RNA. Nucleic Acids Res. 46: 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerezo E., Plisson-Chastang C., Henras A.K., Lebaron S., Gleizes P.E., O’Donohue M.F., Romeo Y., Henry Y. (2019). Maturation of pre-40S particles in yeast and humans. Wiley Interdiscip. Rev. RNA 10: e1516. [DOI] [PubMed] [Google Scholar]
- Chaker-Margot M., Barandun J., Hunziker M., Klinge S. (2017). Architecture of the yeast small subunit processome. Science 355: eaal1880. [DOI] [PubMed] [Google Scholar]
- Chandrasekhara C., Mohannath G., Blevins T., Pontvianne F., Pikaard C.S. (2016). Chromosome-specific NOR inactivation explains selective rRNA gene silencing and dosage control in Arabidopsis. Genes Dev. 30: 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chédin S., Laferté A., Hoang T., Lafontaine D.L., Riva M., Carles C. (2007). Is ribosome synthesis controlled by pol I transcription? Cell Cycle 6: 11–15. [DOI] [PubMed] [Google Scholar]
- Chekanova J.A., Shaw R.J., Wills M.A., Belostotsky D.A. (2000). Poly(A) tail-dependent exonuclease AtRrp41p from Arabidopsis thaliana rescues 5.8 S rRNA processing and mRNA decay defects of the yeast ski6 mutant and is found in an exosome-sized complex in plant and yeast cells. J. Biol. Chem. 275: 33158–33166. [DOI] [PubMed] [Google Scholar]
- Chen M.Q., et al. (2012). Arabidopsis NMD3 is required for nuclear export of 60S ribosomal subunits and affects secondary cell wall thickening. PLoS One 7: e35904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Wu S.H. (2009). Mining small RNA sequencing data: A new approach to identify small nucleolar RNAs in Arabidopsis. Nucleic Acids Res. 37: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J., Pikaard C.S. (1997). Epigenetic silencing of RNA polymerase I transcription: A role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 11: 2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Lu L., Qian S., Scalf M., Smith L.M., Zhong X. (2018). Canonical and non-canonical actions of Arabidopsis histone deacetylases in ribosomal RNA processing. Plant Cell 30: 134–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.C., Wang H.J., Jauh G.Y. (2016). Dual role of a SAS10/C1D family protein in ribosomal RNA gene expression and processing is essential for reproduction in Arabidopsis thaliana. PLoS Genet. 12: e1006408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J., Comai L., Pikaard C.S. (1998). Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc. Natl. Acad. Sci. USA 95: 14891–14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.K., Ahn C.S., Lee H.S., Kim J.K., Pai H.S. (2013). Pescadillo plays an essential role in plant cell growth and survival by modulating ribosome biogenesis. Plant J. 76: 393–405. [DOI] [PubMed] [Google Scholar]
- Choque E., Schneider C., Gadal O., Dez C. (2018). Turnover of aberrant pre-40S pre-ribosomal particles is initiated by a novel endonucleolytic decay pathway. Nucleic Acids Res. 46: 4699–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciganda M., Williams N. (2011). Eukaryotic 5S rRNA biogenesis. Wiley Interdiscip. Rev. RNA 2: 523–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comella P., Pontvianne F., Lahmy S., Vignols F., Barbezier N., Debures A., Jobet E., Brugidou E., Echeverria M., Sáez-Vásquez J. (2008). Characterization of a ribonuclease III-like protein required for cleavage of the pre-rRNA in the 3′ETS in Arabidopsis. Nucleic Acids Res. 36: 1163–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi A., Sogo J.M., Ryan C.A. (1992). Ribosomal gene clusters are uniquely proportioned between open and closed chromatin structures in both tomato leaf cells and exponentially growing suspension cultures. Proc. Natl. Acad. Sci. USA 89: 5256–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke R., Raynal M., Laudié M., Delseny M. (1997). Identification of members of gene families in Arabidopsis thaliana by contig construction from partial cDNA sequences: 106 genes encoding 50 cytoplasmic ribosomal proteins. Plant J. 11: 1127–1140. [DOI] [PubMed] [Google Scholar]
- Copenhaver G.P., Pikaard C.S. (1996a). RFLP and physical mapping with an rDNA-specific endonuclease reveals that nucleolus organizer regions of Arabidopsis thaliana adjoin the telomeres on chromosomes 2 and 4. Plant J. 9: 259–272. [DOI] [PubMed] [Google Scholar]
- Copenhaver G.P., Pikaard C.S. (1996b). Two-dimensional RFLP analyses reveal megabase-sized clusters of rRNA gene variants in Arabidopsis thaliana, suggesting local spreading of variants as the mode for gene homogenization during concerted evolution. Plant J. 9: 273–282. [DOI] [PubMed] [Google Scholar]
- Cordesse F., Cooke R., Tremousaygue D., Grellet F., Delseny M. (1993). Fine structure and evolution of the rDNA intergenic spacer in rice and other cereals. J. Mol. Evol. 36: 369–379. [DOI] [PubMed] [Google Scholar]
- Costa-Nunes P., Pontes O., Preuss S.B., Pikaard C.S. (2010). Extra views on RNA-dependent DNA methylation and MBD6-dependent heterochromatin formation in nucleolar dominance. Nucleus 1: 254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bossoreille S., Morel P., Trehin C., Negrutiu I. (2018). REBELOTE, a regulator of floral determinacy in Arabidopsis thaliana, interacts with both nucleolar and nucleoplasmic proteins. FEBS Open Bio 8: 1636–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcasso-Tremousaygue D., Grellet F., Panabieres F., Ananiev E.D., Delseny M. (1988). Structural and transcriptional characterization of the external spacer of a ribosomal RNA nuclear gene from a higher plant. Eur. J. Biochem. 172: 767–776. [DOI] [PubMed] [Google Scholar]
- Dez C., Tollervey D. (2004). Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 7: 631–637. [DOI] [PubMed] [Google Scholar]
- Doelling J.H., Pikaard C.S. (1995). The minimal ribosomal RNA gene promoter of Arabidopsis thaliana includes a critical element at the transcription initiation site. Plant J. 8: 683–692. [DOI] [PubMed] [Google Scholar]
- Doelling J.H., Pikaard C.S. (1996). Species-specificity of rRNA gene transcription in plants manifested as a switch in RNA polymerase specificity. Nucleic Acids Res. 24: 4725–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H., Gaudino R.J., Pikaard C.S. (1993). Functional analysis of Arabidopsis thaliana rRNA gene and spacer promoters in vivo and by transient expression. Proc. Natl. Acad. Sci. USA 90: 7528–7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon F., Gallagher J.E., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y., Beyer A.L., Hunt D.F., et al. (2002). A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duc C., et al. (2017). Arabidopsis ATRX modulates H3.3 occupancy and fine-tunes gene expression. Plant Cell 29: 1773–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durut N., et al. (2014). A duplicated NUCLEOLIN gene with antagonistic activity is required for chromatin organization of silent 45S rDNA in Arabidopsis. Plant Cell 26: 1330–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durut N., Sáez-Vásquez J. (2015). Nucleolin: Dual roles in rDNA chromatin transcription. Gene 556: 7–12. [DOI] [PubMed] [Google Scholar]
- Earley K., Lawrence R.J., Pontes O., Reuther R., Enciso A.J., Silva M., Neves N., Gross M., Viegas W., Pikaard C.S. (2006). Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 20: 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Pontvianne F., Wierzbicki A.T., Blevins T., Tucker S., Costa-Nunes P., Pontes O., Pikaard C.S. (2010). Mechanisms of HDA6-mediated rRNA gene silencing: Suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev. 24: 1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersberger I., Simm S., Leisegang M.S., Schmitzberger P., Mirus O., von Haeseler A., Bohnsack M.T., Schleiff E. (2014). The evolution of the ribosome biogenesis pathway from a yeast perspective. Nucleic Acids Res. 42: 1509–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler D.C., Craig N. (1994). Processing of eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 49: 197–239. [DOI] [PubMed] [Google Scholar]
- Elvira-Matelot E., Hachet M., Shamandi N., Comella P., Sáez-Vásquez J., Zytnicki M., Vaucheret H. (2016). Arabidopsis RNASE THREE LIKE2 modulates the expression of protein-coding genes via 24-nucleotide small interfering RNA-directed DNA methylation. Plant Cell 28: 406–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright C.A., Maxwell E.S., Eliceiri G.L., Sollner-Webb B. (1996). 5′ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3. RNA 2: 1094–1099. [PMC free article] [PubMed] [Google Scholar]
- Erales J., et al. (2017). Evidence for rRNA 2′-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. USA 114: 12934–12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinar-Marchena F.J., Babiano R., Cruz J. (2017). Placeholder factors in ribosome biogenesis: Please, pave my way. Microb. Cell 4: 144–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Yakura K., Miyanishi M., Sugita M., Sugiura M. (1995). In vitro transcription of plant RNA polymerase I-dependent rRNA genes is species-specific. Plant J. 8: 295–298. [DOI] [PubMed] [Google Scholar]
- Fatica A., Tollervey D. (2002). Making ribosomes. Curr. Opin. Cell Biol. 14: 313–318. [DOI] [PubMed] [Google Scholar]
- Fatica A., Oeffinger M., Dlakić M., Tollervey D. (2003). Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol. Cell. Biol. 23: 1798–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleurdepine S., Deragon J.M., Devic M., Guilleminot J., Bousquet-Antonelli C. (2007). A bona fide La protein is required for embryogenesis in Arabidopsis thaliana. Nucleic Acids Res. 35: 3306–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P., De Jong J.H., Lysak M., Castiglione M.R., Schubert I. (2002). Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl. Acad. Sci. USA 99: 14584–14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French S.L., Osheim Y.N., Cioci F., Nomura M., Beyer A.L. (2003). In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 23: 1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. (2003). Ribosome assembly in eukaryotes. Gene 313: 17–42. [DOI] [PubMed] [Google Scholar]
- Fujikura U., Horiguchi G., Ponce M.R., Micol J.L., Tsukaya H. (2009). Coordination of cell proliferation and cell expansion mediated by ribosome-related processes in the leaves of Arabidopsis thaliana. Plant J. 59: 499–508. [DOI] [PubMed] [Google Scholar]
- Gachomo E.W., Jimenez-Lopez J.C., Baptiste L.J., Kotchoni S.O. (2014). GIGANTUS1 (GTS1), a member of Transducin/WD40 protein superfamily, controls seed germination, growth and biomass accumulation through ribosome-biogenesis protein interactions in Arabidopsis thaliana. BMC Plant Biol. 14: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher J.E., Dunbar D.A., Granneman S., Mitchell B.M., Osheim Y., Beyer A.L., Baserga S.J. (2004). RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 18: 2506–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasse L., Flemming D., Hurt E. (2015). Coordinated ribosomal ITS2 RNA processing by the Las1 complex integrating endonuclease, polynucleotide kinase, and exonuclease activities. Mol. Cell 60: 808–815. [DOI] [PubMed] [Google Scholar]
- Ge X.H., Ding L., Li Z.Y. (2013). Nucleolar dominance and different genome behaviors in hybrids and allopolyploids. Plant Cell Rep. 32: 1661–1673. [DOI] [PubMed] [Google Scholar]
- Gendra E., Moreno A., Albà M.M., Pages M. (2004). Interaction of the plant glycine-rich RNA-binding protein MA16 with a novel nucleolar DEAD box RNA helicase protein from Zea mays. Plant J. 38: 875–886. [DOI] [PubMed] [Google Scholar]
- Giavalisco P., Wilson D., Kreitler T., Lehrach H., Klose J., Gobom J., Fucini P. (2005). High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Mol. Biol. 57: 577–591. [DOI] [PubMed] [Google Scholar]
- Gilkes D.M., Chen J. (2007). Distinct roles of MDMX in the regulation of p53 response to ribosomal stress. Cell Cycle 6: 151–155. [DOI] [PubMed] [Google Scholar]
- Goldfarb K.C., Cech T.R. (2017). Targeted CRISPR disruption reveals a role for RNase MRP RNA in human preribosomal RNA processing. Genes Dev. 31: 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Melendi P., Wells B., Beven A.F., Shaw P.J. (2001). Single ribosomal transcription units are linear, compacted Christmas trees in plant nucleoli. Plant J. 27: 223–233. [DOI] [PubMed] [Google Scholar]
- Goodfellow S.J., Zomerdijk J.C. (2013). Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes. Subcell. Biochem. 61: 211–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schäfer T., Kuster B., Tschochner H., Tollervey D., Gavin A.C., Hurt E. (2002). 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10: 105–115. [DOI] [PubMed] [Google Scholar]
- Granneman S., Gallagher J.E., Vogelzangs J., Horstman W., van Venrooij W.J., Baserga S.J., Pruijn G.J. (2003). The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60-80S ribonucleoprotein complexes. Nucleic Acids Res. 31: 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith M.E., Mayer U., Capron A., Ngo Q.A., Surendrarao A., McClinton R., Jürgens G., Sundaresan V. (2007). The TORMOZ gene encodes a nucleolar protein required for regulated division planes and embryo development in Arabidopsis. Plant Cell 19: 2246–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruendler P., Unfried I., Pointner R., Schweizer D. (1989). Nucleotide sequence of the 25S-18S ribosomal gene spacer from Arabidopsis thaliana. Nucleic Acids Res. 17: 6395–6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruendler P., Unfried I., Pascher K., Schweizer D. (1991). rDNA intergenic region from Arabidopsis thaliana: Structural analysis, intraspecific variation and functional implications. J. Mol. Biol. 221: 1209–1222. [DOI] [PubMed] [Google Scholar]
- Guo J., Han S., Zhao J., Xin C., Zheng X., Liu Y., Wang Y., Qu F. (2018). Essential role of NbNOG1 in ribosomal RNA processing. J. Integr. Plant Biol. 60: 1018–1022. [DOI] [PubMed] [Google Scholar]
- Hang R., Liu C., Ahmad A., Zhang Y., Lu F., Cao X. (2014). Arabidopsis protein arginine methyltransferase 3 is required for ribosome biogenesis by affecting precursor ribosomal RNA processing. Proc. Natl. Acad. Sci. USA 111: 16190–16195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang R., Wang Z., Deng X., Liu C., Yan B., Yang C., Song X., Mo B., Cao X. (2018). Ribosomal RNA biogenesis and its response to chilling stress in Oryza sativa. Plant Physiol. 177: 381–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harscoët E., Dubreucq B., Palauqui J.C., Lepiniec L. (2010). NOF1 encodes an Arabidopsis protein involved in the control of rRNA expression. PLoS One 5: e12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havlová K., Dvořáčková M., Peiro R., Abia D., Mozgová I., Vansáčová L., Gutierrez C., Fajkus J. (2016). Variation of 45S rDNA intergenic spacers in Arabidopsis thaliana. Plant Mol. Biol. 92: 457–471. [DOI] [PubMed] [Google Scholar]
- Hedges J., West M., Johnson A.W. (2005). Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24: 567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A.K., Plisson-Chastang C., O’Donohue M.F., Chakraborty A., Gleizes P.E. (2015). An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 6: 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao Y.C., Hsu Y.F., Chen Y.C., Chang Y.L., Wang C.S. (2016). A WD40 protein, AtGHS40, negatively modulates abscisic acid degrading and signaling genes during seedling growth under high glucose conditions. J. Plant Res. 129: 1127–1140. [DOI] [PubMed] [Google Scholar]
- Hsu Y.F., Chen Y.C., Hsiao Y.C., Wang B.J., Lin S.Y., Cheng W.H., Jauh G.Y., Harada J.J., Wang C.S. (2014). AtRH57, a DEAD-box RNA helicase, is involved in feedback inhibition of glucose-mediated abscisic acid accumulation during seedling development and additively affects pre-ribosomal RNA processing with high glucose. Plant J. 77: 119–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.K., Shen Y.L., Huang L.F., Wu S.J., Yeh C.H., Lu C.A. (2016). The DEAD-box RNA helicase AtRH7/PRH75 participates in pre-rRNA processing, plant development and cold tolerance in Arabidopsis. Plant Cell Physiol. 57: 174–191. [DOI] [PubMed] [Google Scholar]
- Huang K.C., Lin W.C., Cheng W.H. (2018). Salt hypersensitive mutant 9, a nucleolar APUM23 protein, is essential for salt sensitivity in association with the ABA signaling pathway in Arabidopsis. BMC Plant Biol. 18: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J.M., Ares M. Jr (1991). Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 10: 4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung N.J., Johnson A.W. (2006). Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 3718–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im C.H., Hwang S.M., Son Y.S., Heo J.B., Bang W.Y., Suwastika I.N., Shiina T., Bahk J.D. (2011). Nuclear/nucleolar GTPase 2 proteins as a subfamily of YlqF/YawG GTPases function in pre-60S ribosomal subunit maturation of mono- and dicotyledonous plants. J. Biol. Chem. 286: 8620–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura S., Hanaoka M., Tanaka K. (2008). The plant-specific TFIIB-related protein, pBrp, is a general transcription factor for RNA polymerase I. EMBO J. 27: 2317–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Maekawa S., Yanagisawa S. (2016). The pre-rRNA processing complex in Arabidopsis includes two WD40-domain-containing proteins encoded by glucose-inducible genes and plant-specific proteins. Mol. Plant 9: 312–315. [DOI] [PubMed] [Google Scholar]
- Jack K., Bellodi C., Landry D.M., Niederer R.O., Meskauskas A., Musalgaonkar S., Kopmar N., Krasnykh O., Dean A.M., Thompson S.R., Ruggero D., Dinman J.D. (2011). rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell 44: 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A., Wang Y., Raje H., Rosby R., DiMario P. (2014). Nucleolar stress with and without p53. Nucleus 5: 402–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y., Park Y.J., Cho H.K., Jung H.J., Ahn T.K., Kang H., Pai H.S. (2015). The nucleolar GTPase nucleostemin-like 1 plays a role in plant growth and senescence by modulating ribosome biogenesis. J. Exp. Bot. 66: 6297–6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina N.O., Makarova S., Makhotenko A., Love A.J., Taliansky M. (2018). The multiple functions of the nucleolus in plant development, disease and stress responses. Front. Plant Sci. 9: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater L., Thoms M., Barrio-Garcia C., Cheng J., Ismail S., Ahmed Y.L., Bange G., Kressler D., Berninghausen O., Sinning I., Hurt E., Beckmann R. (2017). Visualizing the assembly pathway of nucleolar pre-60S ribosomes. Cell 171: 1599–1610.e1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Konishi M., Shigyo M., Yoneyama T., Yanagisawa S. (2010). Characterization of plant eukaryotic translation initiation factor 6 (eIF6) genes: The essential role in embryogenesis and their differential expression in Arabidopsis and rice. Biochem. Biophys. Res. Commun. 397: 673–678. [DOI] [PubMed] [Google Scholar]
- Kawauchi J., Mischo H., Braglia P., Rondon A., Proudfoot N.J. (2008). Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev. 22: 1082–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., et al. (2014). The Arabidopsis STRESS RESPONSE SUPPRESSOR DEAD-box RNA helicases are nucleolar- and chromocenter-localized proteins that undergo stress-mediated relocalization and are involved in epigenetic gene silencing. Plant J. 79: 28–43. [DOI] [PubMed] [Google Scholar]
- Kim M.H., Sonoda Y., Sasaki K., Kaminaka H., Imai R. (2013). Interactome analysis reveals versatile functions of Arabidopsis COLD SHOCK DOMAIN PROTEIN 3 in RNA processing within the nucleus and cytoplasm. Cell Stress Chaperones 18: 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.K., Kim S., Shin Y.J., Hur Y.S., Kim W.Y., Lee M.S., Cheon C.I., Verma D.P. (2014). Ribosomal protein S6, a target of rapamycin, is involved in the regulation of rRNA genes by possible epigenetic changes in Arabidopsis. J. Biol. Chem. 289: 3901–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Marshallsay C., Filipowicz W. (1992). 7-2/MRP RNAs in plant and mammalian cells: Association with higher order structures in the nucleolus. EMBO J. 11: 3737–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S., Woolford J.L. Jr (2019). Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 20: 116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H., Suzuki T., Kato T., Enomoto K., Sato S., Kato T., Tabata S., Sáez-Vasquez J., Echeverría M., Nakagawa T., Ishiguro S., Nakamura K. (2007). Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 49: 1053–1063. [DOI] [PubMed] [Google Scholar]
- Kojima K., Tamura J., Chiba H., Fukada K., Tsukaya H., Horiguchi G. (2018). Two nucleolar proteins, GDP1 and OLI2, function as ribosome biogenesis factors and are preferentially involved in promotion of leaf cell proliferation without strongly affecting leaf adaxial-abaxial patterning in Arabidopsis thaliana. Front. Plant Sci. 8: 2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konikkat S., Woolford J.L. Jr (2017). Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast. Biochem. J. 474: 195–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D., Hurt E., Bassler J. (2010). Driving ribosome assembly. Biochim. Biophys. Acta 1803: 673–683. [DOI] [PubMed] [Google Scholar]
- Kressler D., Hurt E., Baßler J. (2017). A puzzle of life: Crafting ribosomal subunits. Trends Biochem. Sci. 42: 640–654. [DOI] [PubMed] [Google Scholar]
- Kühn H., Hierlmeier T., Merl J., Jakob S., Aguissa-Touré A.H., Milkereit P., Tschochner H. (2009). The Noc-domain containing C-terminus of Noc4p mediates both formation of the Noc4p-Nop14p submodule and its incorporation into the SSU processome. PLoS One 4: e8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumakura N., Otsuki H., Tsuzuki M., Takeda A., Watanabe Y. (2013). Arabidopsis AtRRP44A is the functional homolog of Rrp44/Dis3, an exosome component, is essential for viability and is required for RNA processing and degradation. PLoS One 8: e79219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazawa T., Nishimura K., Kuroda T., Ono W., Yamaguchi C., Katagiri N., Tsuchiya M., Masumoto H., Nakajima Y., Murayama A., Kimura K., Yanagisawa J. (2011). Novel nucleolar pathway connecting intracellular energy status with p53 activation. J. Biol. Chem. 286: 20861–20869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahmy S., Guilleminot J., Cheng C.M., Bechtold N., Albert S., Pelletier G., Delseny M., Devic M. (2004). DOMINO1, a member of a small plant-specific gene family, encodes a protein essential for nuclear and nucleolar functions. Plant J. 39: 809–820. [DOI] [PubMed] [Google Scholar]
- Lange H., et al. (2014). The RNA helicases AtMTR4 and HEN2 target specific subsets of nuclear transcripts for degradation by the nuclear exosome in Arabidopsis thaliana. PLoS Genet. 10: e1004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H., Holec S., Cognat V., Pieuchot L., Le Ret M., Canaday J., Gagliardi D. (2008). Degradation of a polyadenylated rRNA maturation by-product involves one of the three RRP6-like proteins in Arabidopsis thaliana. Mol. Cell. Biol. 28: 3038–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H., Sement F.M., Gagliardi D. (2011). MTR4, a putative RNA helicase and exosome co-factor, is required for proper rRNA biogenesis and development in Arabidopsis thaliana. Plant J. 68: 51–63. [DOI] [PubMed] [Google Scholar]
- Lawrence R.J., Pikaard C.S. (2004). Chromatin turn ons and turn offs of ribosomal RNA genes. Cell Cycle 3: 880–883. [PubMed] [Google Scholar]
- Lawrence R.J., Earley K., Pontes O., Silva M., Chen Z.J., Neves N., Viegas W., Pikaard C.S. (2004). A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 13: 599–609. [DOI] [PubMed] [Google Scholar]
- Layat E., Sáez-Vásquez J., Tourmente S. (2012). Regulation of Pol I-transcribed 45S rDNA and Pol III-transcribed 5S rDNA in Arabidopsis. Plant Cell Physiol. 53: 267–276. [DOI] [PubMed] [Google Scholar]
- Lee S., Senthil-Kumar M., Kang M., Rojas C.M., Tang Y., Oh S., Choudhury S.R., Lee H.K., Ishiga Y., Allen R.D., Pandey S., Mysore K.S. (2017). The small GTPase, nucleolar GTP-binding protein 1 (NOG1), has a novel role in plant innate immunity. Sci. Rep. 7: 9260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I., Schubert V., Börnke F., Macas J., Schubert I. (2007). Arabidopsis CBF5 interacts with the H/ACA snoRNP assembly factor NAF1. Plant Mol. Biol. 65: 615–626. [DOI] [PubMed] [Google Scholar]
- Li H., Luan S. (2010). AtFKBP53 is a histone chaperone required for repression of ribosomal RNA gene expression in Arabidopsis. Cell Res. 20: 357–366. [DOI] [PubMed] [Google Scholar]
- Li N., Yuan L., Liu N., Shi D., Li X., Tang Z., Liu J., Sundaresan V., Yang W.C. (2009). SLOW WALKER2, a NOC1/MAK21 homologue, is essential for coordinated cell cycle progression during female gametophyte development in Arabidopsis. Plant Physiol. 151: 1486–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P.C., Yu S.W., Li K., Huang J.G., Wang X.J., Zheng C.C. (2016). The mutation of Glu at amino acid 3838 of AtMDN1 provokes pleiotropic developmental phenotypes in Arabidopsis. Sci. Rep. 6: 36446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Yoshida A., Takahashi M., Maekawa M., Kojima M., Sakakibara H., Kyozuka J. (2015). SAD1, an RNA polymerase I subunit A34.5 of rice, interacts with Mediator and controls various aspects of plant development. Plant J. 81: 282–291. [DOI] [PubMed] [Google Scholar]
- Liu M., Shi D.Q., Yuan L., Liu J., Yang W.C. (2010). SLOW WALKER3, encoding a putative DEAD-box RNA helicase, is essential for female gametogenesis in Arabidopsis. J. Integr. Plant Biol. 52: 817–828. [DOI] [PubMed] [Google Scholar]
- Liu Y., Tabata D., Imai R. (2016). A cold-inducible DEAD-box RNA helicase from Arabidopsis thaliana regulates plant growth and development under low temperature. PLoS One 11: e0154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loza-Muller L., Rodríguez-Corona U., Sobol M., Rodríguez-Zapata L.C., Hozak P., Castano E. (2015). Fibrillarin methylates H2A in RNA polymerase I trans-active promoters in Brassica oleracea. Front. Plant Sci. 6: 976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maceluch J., Kmieciak M., Szweykowska-Kulińska Z., Jarmołowski A. (2001). Cloning and characterization of Arabidopsis thaliana AtNAP57: A homologue of yeast pseudouridine synthase Cbf5p. Acta Biochim. Pol. 48: 699–709. [PubMed] [Google Scholar]
- Madru C., Lebaron S., Blaud M., Delbos L., Pipoli J., Pasmant E., Réty S., Leulliot N. (2015). Chaperoning 5S RNA assembly. Genes Dev. 29: 1432–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S., Yanagisawa S. (2018). Nucleolar stress and sugar response in plants. Plant Signal. Behav. 13: e1442975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S., Ishida T., Yanagisawa S. (2018a). Reduced expression of APUM24, encoding a novel rRNA processing factor, induces sugar-dependent nucleolar stress and altered sugar responses in Arabidopsis thaliana. Plant Cell 30: 209–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S., Ueda Y., Yanagisawa S. (2018b). Overexpression of a Brix domain-containing ribosome biogenesis factor ARPF2 and its interactor ARRS1 causes morphological changes and lifespan extension in Arabidopsis thaliana. Front. Plant Sci. 9: 1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel V., et al. (2013). p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell 24: 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenet S., Bertrand E., Verheggen C. (2017). Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 14: 680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y., et al. (2016). A genetic link between epigenetic repressor AS1-AS2 and a putative small subunit processome in leaf polarity establishment of Arabidopsis. Biol. Open 5: 942–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y., Granneman S., Thoms M., Manikas R.G., Tollervey D., Hurt E. (2014). Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature 505: 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérai Z., et al. (2014). The AAA-ATPase molecular chaperone Cdc48/p97 disassembles sumoylated centromeres, decondenses heterochromatin, and activates ribosomal RNA genes. Proc. Natl. Acad. Sci. USA 111: 16166–16171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak K., Maciak S., Kim Y.B., Santopietro G., Oh J.H., Kang L., Garner H.R., Michalak P. (2015). Nucleolar dominance and maternal control of 45S rDNA expression. Proc. Biol. Sci. 282: 20152201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micol-Ponce R., Sarmiento-Mañús R., Ruiz-Bayón A., Montacié C., Sáez-Vasquez J., Ponce M.R. (2018). Arabidopsis RIBOSOMAL RNA PROCESSING7 is required for 18S rRNA maturation. Plant Cell 30: 2855–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P., Gadal O., Podtelejnikov A., Trumtel S., Gas N., Petfalski E., Tollervey D., Mann M., Hurt E., Tschochner H. (2001). Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105: 499–509. [DOI] [PubMed] [Google Scholar]
- Miller O.L. Jr., Beatty B.R. (1969). Visualization of nucleolar genes. Science 164: 955–957. [DOI] [PubMed] [Google Scholar]
- Missbach S., Weis B.L., Martin R., Simm S., Bohnsack M.T., Schleiff E. (2013). 40S ribosome biogenesis co-factors are essential for gametophyte and embryo development. PLoS One 8: e54084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohannath G., Pontvianne F., Pikaard C.S. (2016). Selective nucleolus organizer inactivation in Arabidopsis is a chromosome position-effect phenomenon. Proc. Natl. Acad. Sci. USA 113: 13426–13431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montacié C., Durut N., Opsomer A., Palm D., Comella P., Picart C., Carpentier M.C., Pontvianne F., Carapito C., Schleiff E., Sáez-Vásquez J. (2017). Nucleolar proteome analysis and proteasomal activity assays reveal a link between nucleolus and 26S proteasome in A. thaliana. Front. Plant Sci. 8: 1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougey E.B., O’Reilly M., Osheim Y., Miller O.L. Jr., Beyer A., Sollner-Webb B. (1993). The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 7: 1609–1619. [DOI] [PubMed] [Google Scholar]
- Mozgová I., Mokros P., Fajkus J. (2010). Dysfunction of chromatin assembly factor 1 induces shortening of telomeres and loss of 45S rDNA in Arabidopsis thaliana. Plant Cell 22: 2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulekar J.J., Huq E. (2014). Expanding roles of protein kinase CK2 in regulating plant growth and development. J. Exp. Bot. 65: 2883–2893. [DOI] [PubMed] [Google Scholar]
- Murayama A., et al. (2008). Epigenetic control of rDNA loci in response to intracellular energy status. Cell 133: 627–639. [DOI] [PubMed] [Google Scholar]
- Oeffinger M., Zenklusen D., Ferguson A., Wei K.E., El Hage A., Tollervey D., Chait B.T., Singer R.H., Rout M.P. (2009). Rrp17p is a eukaryotic exonuclease required for 5′ end processing of pre-60S ribosomal RNA. Mol. Cell 36: 768–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi I., Sugiyama M. (2018). Plant nucleolar stress response, a new face in the NAC-dependent cellular stress responses. Front. Plant Sci. 8: 2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi I., Konishi M., Ebine K., Sugiyama M. (2011). Genetic identification of Arabidopsis RID2 as an essential factor involved in pre-rRNA processing. Plant J. 67: 49–60. [DOI] [PubMed] [Google Scholar]
- Ohbayashi I., Lin C.Y., Shinohara N., Matsumura Y., Machida Y., Horiguchi G., Tsukaya H., Sugiyama M. (2017). Evidence for a role of ANAC082 as a ribosomal stress response mediator leading to growth defects and developmental alterations in Arabidopsis. Plant Cell 29: 2644–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okanami M., Meshi T., Iwabuchi M. (1998). Characterization of a DEAD box ATPase/RNA helicase protein of Arabidopsis thaliana. Nucleic Acids Res. 26: 2638–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orioli A., Pascali C., Pagano A., Teichmann M., Dieci G. (2012). RNA polymerase III transcription control elements: Themes and variations. Gene 493: 185–194. [DOI] [PubMed] [Google Scholar]
- Pagnussat G.C., Yu H.J., Ngo Q.A., Rajani S., Mayalagu S., Johnson C.S., Capron A., Xie L.F., Ye D., Sundaresan V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614. [DOI] [PubMed] [Google Scholar]
- Palm D., Simm S., Darm K., Weis B.L., Ruprecht M., Schleiff E., Scharf C. (2016). Proteome distribution between nucleoplasm and nucleolus and its relation to ribosome biogenesis in Arabidopsis thaliana. RNA Biol. 13: 441–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm D., Streit D., Ruprecht M., Simm S., Scharf C., Schleiff E. (2018). Late ribosomal protein localization in Arabidopsis thaliana differs to that in Saccharomyces cerevisiae. FEBS Open Bio 8: 1437–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm D., Streit D., Shanmugam T., Weis B.L., Ruprecht M., Simm S., Schleiff E. (2019). Plant-specific ribosome biogenesis factors in Arabidopsis thaliana with essential function in rRNA processing. Nucleic Acids Res. 47: 1880–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendle A.F., Clark G.P., Boon R., Lewandowska D., Lam Y.W., Andersen J., Mann M., Lamond A.I., Brown J.W., Shaw P.J. (2005). Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol. Biol. Cell 16: 260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps K.R., Charette J., Baserga S.J. (2011). The small subunit processome in ribosome biogenesis: progress and prospects. Wiley Interdiscip. Rev. RNA 2: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pih K.T., Yi M.J., Liang Y.S., Shin B.J., Cho M.J., Hwang I., Son D. (2000). Molecular cloning and targeting of a fibrillarin homolog from Arabidopsis. Plant Physiol. 123: 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard C.S. (2000). The epigenetics of nucleolar dominance. Trends Genet. 16: 495–500. [DOI] [PubMed] [Google Scholar]
- Pontvianne F., et al. (2010). Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet. 6: e1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., Matía I., Douet J., Tourmente S., Medina F.J., Echeverria M., Sáez-Vásquez J. (2007). Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol. Biol. Cell 18: 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., Blevins T., Chandrasekhara C., Feng W., Stroud H., Jacobsen S.E., Michaels S.D., Pikaard C.S. (2012). Histone methyltransferases regulating rRNA gene dose and dosage control in Arabidopsis. Genes Dev. 26: 945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., Blevins T., Chandrasekhara C., Mozgová I., Hassel C., Pontes O.M., Tucker S., Mokros P., Muchová V., Fajkus J., Pikaard C.S. (2013). Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev. 27: 1545–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portereiko M.F., Lloyd A., Steffen J.G., Punwani J.A., Otsuga D., Drews G.N. (2006). AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18: 1862–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss S.B., Costa-Nunes P., Tucker S., Pontes O., Lawrence R.J., Mosher R., Kasschau K.D., Carrington J.C., Baulcombe D.C., Viegas W., Pikaard C.S. (2008). Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol. Cell 32: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N., Morel P., Thierry A.M., Eshed Y., Bowman J.L., Negrutiu I., Trehin C. (2008). REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell 20: 901–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W., Zhu J., Wu Q., Wang Q., Li X., Yao D., Jin Y., Wang G., Wang G., Song R. (2016). Maize reas1 mutant stimulates ribosome use efficiency and triggers distinct transcriptional and translational responses. Plant Physiol. 170: 971–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L.H., Meng Q., Zhou H., Chen Y.Q. (2001). Identification of 10 novel snoRNA gene clusters from Arabidopsis thaliana. Nucleic Acids Res. 29: 1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabanal F.A., Nizhynska V., Mandakova T., Novikova P.Y., Lysak M.A., Mott R., Nordborg M. (2017). Unstable inheritance of 45S rRNA genes in Arabidopsis thaliana. G3 (Bethesda) 7: 1201–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I., Koberna K., Malínský J., Fidlerová H., Masata M. (2004). The nucleolus and transcription of ribosomal genes. Biol. Cell 96: 579–594. [DOI] [PubMed] [Google Scholar]
- Ream T.S., Haag J.R., Pontvianne F., Nicora C.D., Norbeck A.D., Paša-Tolić L., Pikaard C.S. (2015). Subunit compositions of Arabidopsis RNA polymerases I and III reveal Pol I- and Pol III-specific forms of the AC40 subunit and alternative forms of the C53 subunit. Nucleic Acids Res. 43: 4163–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M., Qiu S., Venglat P., Xiang D., Feng L., Selvaraj G., Datla R. (2011). Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol. 155: 1367–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reňák D., Gibalová A., Solcová K., Honys D. (2014). A new link between stress response and nucleolar function during pollen development in Arabidopsis mediated by AtREN1 protein. Plant Cell Environ. 37: 670–683. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Corona U., Pereira-Santana A., Sobol M., Rodriguez-Zapata L.C., Hozak P., Castano E. (2017). Novel ribonuclease activity differs between fibrillarins from Arabidopsis thaliana. Front. Plant Sci. 8: 1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhrig S., Schröpfer S., Knoll A., Puchta H. (2016). The RTR complex partner RMI2 and the DNA helicase RTEL1 are both independently involved in preserving the stability of 45S rDNA repeats in Arabidopsis thaliana. PLoS Genet. 12: e1006394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrmoser M., Hölzel M., Grimm T., Malamoussi A., Harasim T., Orban M., Pfisterer I., Gruber-Eber A., Kremmer E., Eick D. (2007). Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol. Cell. Biol. 27: 3682–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova L., Grand A., Zhang L., Rayner S., Katoku-Kikyo N., Kellner S., Kikyo N. (2009). Critical role of nucleostemin in pre-rRNA processing. J. Biol. Chem. 284: 4968–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothé B., Manival X., Rolland N., Charron C., Senty-Ségault V., Branlant C., Charpentier B. (2017). Implication of the box C/D snoRNP assembly factor Rsa1p in U3 snoRNP assembly. Nucleic Acids Res. 45: 7455–7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J., Zomerdijk J.C. (2005). RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem. Sci. 30: 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Vasquez J., Echeverria M. (2006). Polymerase I transcription. In Regulation of Transcription in Plants, Grasser K.D., ed (Oxford, UK: Blackwell; ), pp. 162–183. [Google Scholar]
- Saez-Vasquez J., Medina F.J. (2008). The plant nucleolus. In Botanical Research: Incorporating Advances in Plant Pathology, Vol. 47, Kader J.-C., Delseny M., eds (San Diego, CA: Elsevier Academic Press; ), pp. 1–46. [Google Scholar]
- Saez-Vasquez J., Pikaard C.S. (1997). Extensive purification of a putative RNA polymerase I holoenzyme from plants that accurately initiates rRNA gene transcription in vitro. Proc. Natl. Acad. Sci. USA 94: 11869–11874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Vasquez J., Pikaard C.S. (2000). RNA polymerase I holoenzyme-promoter interactions. J. Biol. Chem. 275: 37173–37180. [DOI] [PubMed] [Google Scholar]
- Saez-Vasquez J., Meissner M., Pikaard C.S. (2001). RNA polymerase I holoenzyme-promoter complexes include an associated CK2-like protein kinase. Plant Mol. Biol. 47: 449–459. [DOI] [PubMed] [Google Scholar]
- Sáez-Vasquez J., Caparros-Ruiz D., Barneche F., Echeverría M. (2004a). A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol. Cell. Biol. 24: 7284–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez-Vasquez J., Caparros-Ruiz D., Barneche F., Echeverría M. (2004b). Characterization of a crucifer plant pre-rRNA processing complex. Biochem. Soc. Trans. 32: 578–580. [DOI] [PubMed] [Google Scholar]
- Samaha H., Delorme V., Pontvianne F., Cooke R., Delalande F., Van Dorsselaer A., Echeverria M., Sáez-Vásquez J. (2010). Identification of protein factors and U3 snoRNAs from a Brassica oleracea RNP complex involved in the processing of pre-rRNA. Plant J. 61: 383–398. [DOI] [PubMed] [Google Scholar]
- Sánchez-García A.B., Aguilera V., Micol-Ponce R., Jover-Gil S., Ponce M.R. (2015). Arabidopsis MAS2, an essential gene that encodes a homolog of animal NF-κ B activating protein, is involved in 45S ribosomal DNA silencing. Plant Cell 27: 1999–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghai Z.A., Miller L., Molloy K.R., Barandun J., Hunziker M., Chaker-Margot M., Wang J., Chait B.T., Klinge S. (2018). Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature 556: 126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanij E., et al. (2008). UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 183: 1259–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer T., Strauss D., Petfalski E., Tollervey D., Hurt E. (2003). The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22: 1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S., Dethloff F., Beine-Golovchuk O., Kopka J. (2013). The REIL1 and REIL2 proteins of Arabidopsis thaliana are required for leaf growth in the cold. Plant Physiol. 163: 1623–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S., Dethloff F., Beine-Golovchuk O., Kopka J. (2014). REIL proteins of Arabidopsis thaliana interact in yeast-2-hybrid assays with homologs of the yeast Rlp24, Rpl24A, Rlp24B, Arx1, and Jjj1 proteins. Plant Signal. Behav. 9: e28224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D.A. (2012). RNA polymerase I activity is regulated at multiple steps in the transcription cycle: Recent insights into factors that influence transcription elongation. Gene 493: 176–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seither P., Iben S., Grummt I. (1998). Mammalian RNA polymerase I exists as a holoenzyme with associated basal transcription factors. J. Mol. Biol. 275: 43–53. [DOI] [PubMed] [Google Scholar]
- Senapin S., Clark-Walker G.D., Chen X.J., Séraphin B., Daugeron M.C. (2003). RRP20, a component of the 90S preribosome, is required for pre-18S rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 31: 2524–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J.S., Diloknawarit P., Park B.S., Chua N.H. (2018). ELF18-INDUCED LONG NONCODING RNA 1 evicts fibrillarin from mediator subunit to enhance PATHOGENESIS-RELATED GENE 1 (PR1) expression. New Phytol. 221: 2067–2079 [DOI] [PubMed] [Google Scholar]
- Shanmugam T., Abbasi N., Kim H.S., Kim H.B., Park N.I., Park G.T., Oh S.A., Park S.K., Muench D.G., Choi Y., Park Y.I., Choi S.B. (2017). An Arabidopsis divergent pumilio protein, APUM24, is essential for embryogenesis and required for faithful pre-rRNA processing. Plant J. 92: 1092–1105. [DOI] [PubMed] [Google Scholar]
- Sharma S., Lafontaine D.L.J. (2015). ‘View from a bridge’: A new perspective on eukaryotic rRNA base modification. Trends Biochem. Sci. 40: 560–575. [DOI] [PubMed] [Google Scholar]
- Sharma S., Langhendries J.L., Watzinger P., Kötter P., Entian K.D., Lafontaine D.L. (2015). Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 43: 2242–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Yang J., van Nues R., Watzinger P., Kötter P., Lafontaine D.L.J., Granneman S., Entian K.D. (2017). Specialized box C/D snoRNPs act as antisense guides to target RNA base acetylation. PLoS Genet. 13: e1006804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchepachev V., Tollervey D. (2016). Motoring toward pre-60S-ribosome export. Nat. Struct. Mol. Biol. 23: 3–4. [DOI] [PubMed] [Google Scholar]
- Shi D.Q., Liu J., Xiang Y.H., Ye D., Sundaresan V., Yang W.C. (2005). SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17: 2340–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoji K., Jakovljevic J., Tsuchihashi K., Umeki Y., Wan K., Kawasaki S., Talkish J., Woolford J.L. Jr., Mizuta K. (2012). Ebp2 and Brx1 function cooperatively in 60S ribosomal subunit assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 40: 4574–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara N., Ohbayashi I., Sugiyama M. (2014). Involvement of rRNA biosynthesis in the regulation of CUC1 gene expression and pre-meristematic cell mound formation during shoot regeneration. Front. Plant Sci. 5: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorska N., Zuber H., Gobert A., Lange H., Gagliardi D. (2017). RNA degradation by the plant RNA exosome involves both phosphorolytic and hydrolytic activities. Nat. Commun. 8: 2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski P.J., Zuber H., Philippe L., Sement F.M., Canaday J., Kufel J., Gagliardi D., Lange H. (2015). Distinct 18S rRNA precursors are targets of the exosome complex, the exoribonuclease RRP6L2 and the terminal nucleotidyltransferase TRL in Arabidopsis thaliana. Plant J. 83: 991–1004. [DOI] [PubMed] [Google Scholar]
- Simm S., Fragkostefanakis S., Paul P., Keller M., Einloft J., Scharf K.D., Schleiff E. (2015). Identification and expression analysis of ribosome biogenesis factor co-orthologs in Solanum lycopersicum. Bioinform. Biol. Insights 9: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan K.E., Warda A.S., Sharma S., Entian K.D., Lafontaine D.L.J., Bohnsack M.T. (2017). Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 14: 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltanieh S., Lapensée M., Dragon F. (2014). Nucleolar proteins Bfr2 and Enp2 interact with DEAD-box RNA helicase Dbp4 in two different complexes. Nucleic Acids Res. 42: 3194–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondalle S.B., Baserga S.J. (2014). Human diseases of the SSU processome. Biochim. Biophys. Acta 1842: 758–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Nazar R.N. (2002). Modification of rRNA as a ‘quality control mechanism’ in ribosome biogenesis. FEBS Lett. 523: 182–186. [DOI] [PubMed] [Google Scholar]
- Sripinyowanich S., Chamnanmanoontham N., Udomchalothorn T., Maneeprasopsuk S., Santawee P., Buaboocha T., Qu L.J., Gu H., Chadchawan S. (2013). Overexpression of a partial fragment of the salt-responsive gene OsNUC1 enhances salt adaptation in transgenic Arabidopsis thaliana and rice (Oryza sativa L.) during salt stress. Plant Sci. 213: 67–78. [DOI] [PubMed] [Google Scholar]
- Stepinski D. (2016). Nucleolus-derived mediators in oncogenic stress response and activation of p53-dependent pathways. Histochem. Cell Biol. 46: 119–139 [DOI] [PubMed] [Google Scholar]
- Sun Q., et al. (2017). Molecular architecture of the 90S small subunit pre-ribosome. eLife 6: e22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P., Santos-Rosa H., Robson S.C., Sylvestersen K.B., Nelson C.J., Nielsen M.L., Kouzarides T. (2014). Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 505: 564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S.R., Keller C.A., Szyk A., Cannon J.R., Laronde-Leblanc N.A. (2011). Structural insight into the functional mechanism of Nep1/Emg1 N1-specific pseudouridine methyltransferase in ribosome biogenesis. Nucleic Acids Res. 39: 2445–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R., Sikorski P.J., Zakrzewska-Placzek M. (2017). Comparison of preribosomal RNA processing pathways in yeast, plant and human cells: Focus on coordinated action of endo- and exoribonucleases. FEBS Lett. 591: 1801–1850. [DOI] [PubMed] [Google Scholar]
- Tsai R.Y., Pederson T. (2014). Connecting the nucleolus to the cell cycle and human disease. FASEB J. 28: 3290–3296. [DOI] [PubMed] [Google Scholar]
- Tucker S., Vitins A., Pikaard C.S. (2010). Nucleolar dominance and ribosomal RNA gene silencing. Curr. Opin. Cell Biol. 22: 351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unfried I., Gruendler P. (1990). Nucleotide sequence of the 5.8S and 25S rRNA genes and of the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Res. 18: 4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unfried I., Stocker U., Gruendler P. (1989). Nucleotide sequence of the 18S rRNA gene from Arabidopsis thaliana Co10. Nucleic Acids Res. 17: 7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urawa H., Hidaka M., Ishiguro S., Okada K., Horiuchi T. (2001). Enhanced homologous recombination caused by the non-transcribed spacer of the rDNA in Arabidopsis. Mol. Genet. Genomics 266: 546–555. [DOI] [PubMed] [Google Scholar]
- Urbánek P., Paces J., Paces V. (2005). An approach towards experimental cDNA sequence determination of predicted genes: An example from Arabidopsis U3-55k homologues. Gene 358: 67–72. [DOI] [PubMed] [Google Scholar]
- van Sluis M., McStay B. (2017). Nucleolar reorganization in response to rDNA damage. Curr. Opin. Cell Biol. 46: 81–86. [DOI] [PubMed] [Google Scholar]
- Venema J., Tollervey D. (1995). Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast 11: 1629–1650. [DOI] [PubMed] [Google Scholar]
- Wang D., Qin B., Li X., Tang D., Zhang Y., Cheng Z., Xue Y. (2016). Nucleolar DEAD-box RNA helicase TOGR1 regulates thermotolerant growth as a pre-rRNA chaperone in rice. PLoS Genet. 12: e1005844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang K., Du Q., Wang Y., Fu Z., Guo Z., Kang D., Li W.X., Tang J. (2018). Maize Urb2 protein is required for kernel development and vegetative growth by affecting pre-ribosomal RNA processing. New Phytol. 218: 1233–1246. [DOI] [PubMed] [Google Scholar]
- Wang X., Gingrich D.K., Deng Y., Hong Z. (2012). A nucleostemin-like GTPase required for normal apical and floral meristem development in Arabidopsis. Mol. Biol. Cell 23: 1446–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanzenböck E.M., Schöfer C., Schweizer D., Bachmair A. (1997). Ribosomal transcription units integrated via T-DNA transformation associate with the nucleolus and do not require upstream repeat sequences for activity in Arabidopsis thaliana. Plant J. 11: 1007–1016. [DOI] [PubMed] [Google Scholar]
- Weis B.L., Missbach S., Marzi J., Bohnsack M.T., Schleiff E. (2014). The 60S associated ribosome biogenesis factor LSG1-2 is required for 40S maturation in Arabidopsis thaliana. Plant J. 80: 1043–1056. [DOI] [PubMed] [Google Scholar]
- Weis B.L., Kovacevic J., Missbach S., Schleiff E. (2015a). Plant-specific features of ribosome biogenesis. Trends Plant Sci. 20: 729–740. [DOI] [PubMed] [Google Scholar]
- Weis B.L., Palm D., Missbach S., Bohnsack M.T., Schleiff E. (2015b). atBRX1-1 and atBRX1-2 are involved in an alternative rRNA processing pathway in Arabidopsis thaliana. RNA 21: 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells G.R., Weichmann F., Colvin D., Sloan K.E., Kudla G., Tollervey D., Watkins N.J., Schneider C. (2016). The PIN domain endonuclease Utp24 cleaves pre-ribosomal RNA at two coupled sites in yeast and humans. Nucleic Acids Res. 44: 5399–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S., Gromak N., Proudfoot N.J. (2004). Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432: 522–525. [DOI] [PubMed] [Google Scholar]
- Wieckowski Y., Schiefelbein J. (2012). Nuclear ribosome biogenesis mediated by the DIM1A rRNA dimethylase is required for organized root growth and epidermal patterning in Arabidopsis. Plant Cell 24: 2839–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., et al. (2016). Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature 534: 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi L., Moscou M.J., Meng Y., Xu W., Caldo R.A., Shaver M., Nettleton D., Wise R.P. (2009). Transcript-based cloning of RRP46, a regulator of rRNA processing and R gene-independent cell death in barley-powdery mildew interactions. Plant Cell 21: 3280–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K., Robin A.H., Yi G.E., Lee J., Chung M.Y., Yang T.J., Nou I.S. (2015). Diversity and inheritance of intergenic spacer sequences of 45S ribosomal DNA among accessions of Brassica oleracea L. var. capitata. Int. J. Mol. Sci. 16: 28783–28799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska-Placzek M., Souret F.F., Sobczyk G.J., Green P.J., Kufel J. (2010). Arabidopsis thaliana XRN2 is required for primary cleavage in the pre-ribosomal RNA. Nucleic Acids Res. 38: 4487–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Muench D.G. (2015). A nucleolar PUF RNA-binding protein with specificity for a unique RNA sequence. J. Biol. Chem. 290: 30108–30118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.R., Qin Z., Zhang X., Hu Y. (2015). Arabidopsis SMALL ORGAN 4, a homolog of yeast NOP53, regulates cell proliferation rate during organ growth. J. Integr. Plant Biol. 57: 810–818. [DOI] [PubMed] [Google Scholar]
- Zhao H., Lü S., Li R., Chen T., Zhang H., Cui P., Ding F., Liu P., Wang G., Xia Y., Running M.P., Xiong L. (2015). The Arabidopsis gene DIG6 encodes a large 60S subunit nuclear export GTPase 1 that is involved in ribosome biogenesis and affects multiple auxin-regulated development processes. J. Exp. Bot. 66: 6863–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P., Wang Y., Qin N., Wang F., Wang J., Deng X.W., Zhu D. (2016). Arabidopsis small nucleolar RNA monitors the efficient pre-rRNA processing during ribosome biogenesis. Proc. Natl. Acad. Sci. USA 113: 11967–11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisser G., Ohmayer U., Mauerhofer C., Mitterer V., Klein I., Rechberger G.N., Wolinski H., Prattes M., Pertschy B., Milkereit P., Bergler H. (2018). Viewing pre-60S maturation at a minute’s timescale. Nucleic Acids Res. 46: 3140–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zografidis A., Kapolas G., Podia V., Beri D., Papadopoulou K., Milioni D., Haralampidis K. (2014). Transcriptional regulation and functional involvement of the Arabidopsis pescadillo ortholog AtPES in root development. Plant Sci. 229: 53–65. [DOI] [PubMed] [Google Scholar]