Senescence results from two ineluctable laws of nature: every living entity will eventually die, and when it does, nothing will be lost, as everything will be recycled. At the organism level, senescence is an integral part of the plant life cycle, under strict age-dependent genetic control. This dismantlement of living tissues and recycling of essential elements aims to promote plant success (Schippers et al., 2015).
For a plant, the production of viable seeds requires a large investment of nutrients, all to increase the chances of survival of the next generation. To get this energy, the plant relocates necessary nutrients from sources to sinks (see figure). However, when a plant sacrifices leaves, and therefore photosynthesis units, for reproductive needs, it can ultimately cause the death of the whole plant. Delaying leaf senescence in “stay-green” crops has been a beneficial strategy for plant breeding, as crop productivity and senescence are associated (Thomas and Howarth, 2000). Although stay-green cultivars were developed decades ago, the genetic architecture of this trait remains poorly understood, as senescence is a complex developmental program dependent on both internal and external factors.
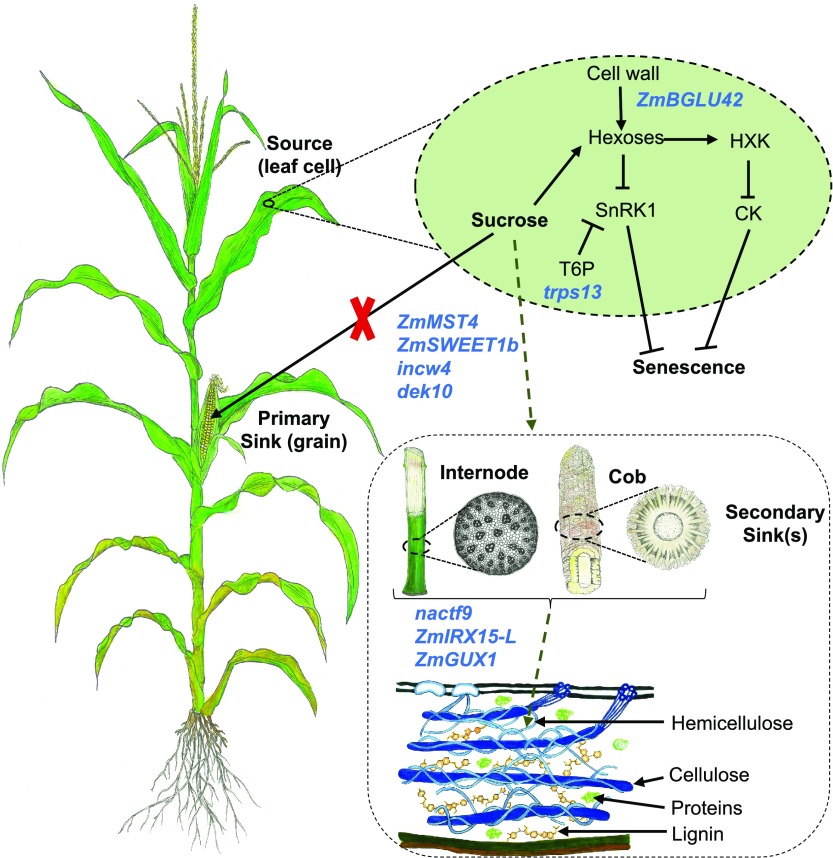
Model for the role of Sugar-Associated Candidate Genes (in Blue) in Maize Senescence.
Candidate genes are active in sugar signaling and transport between source and sink tissues. (Reprinted from Sekhon et al. [2019], Figure 8.)
Sekhon et al. (2019) revealed the genetic architecture of senescence in maize (Zea mays). They studied a stay-green line that maintained full photosynthetic capacity and grain filling for 6 critical additional days compared with a naturally senescent genotype. To reveal the genetic architecture of stay-green, the authors used a system genetics approach that integrated both gene identification by genome-wide association study in a U.S. maize diversity panel (Hirsch et al., 2014) and coexpression networks derived from transcriptomes at different developmental stages.
The system genetics framework revealed 64 candidates genes among which 48 were differentially expressed, and 30 were annotated as senescence-associated genes in Arabidopsis (Arabidopsis thaliana). Overall, gene expression profiles revealed the upregulation of autophagy as well as lipid, carbohydrate, and amino acid metabolism and/or transport but the downregulation of photosynthesis and abscisic acid synthesis.
The candidate genes revealed the role of cell wall and biological processes, such as proteolysis, and sugar signaling and transport in senescence in maize. The role of one of the candidate genes, mir3 (maize insect resistance3), which encodes a putative Cys protease, was validated in senescence. Knockout of mir3 in Arabidopsis delayed leaf senescence, suggesting the role of this gene in senescence in both monocots and dicots.
Sekhon et al. (2019) also explained the role of sugar partitioning and sugar signaling in senescence by placing nine candidate genes in a model (see figure). The candidate genes represent diverse processes, including sugar-mediated signaling (trps13), transport (ZmSWEET1b and ZmMST4), and control of sugar uptake (dek10). Cell walls also play a key role in stay-green lines, both as a source of sugars that potentially act in senescence signaling and as an alternative sink for excess sugars. The authors propose that the cell wall invertase (incw4) unloads sugars in the source, while they are incorporated in the sink by a NAC transcription factor (nacft9) or transformed into hemicellulose by ZmIRX15-L and ZmGUX1. nacft9 is proposed to be an important regulator of delayed senescence in the stay-green lines.
Epistatic interactions among the candidate genes were also detected. Other potentially valuable candidate genes require further investigation to reveal their mechanistic involvement in senescence.
Footnotes
Articles can be viewed without a subscription.
References
- Hirsch C.N., et al. (2014). Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26: 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schippers J.H., Schmidt R., Wagstaff C., Jing H.C. (2015). Living to die and dying to live: The survival strategy behind leaf senescence. Plant Physiol. 169: 914–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon RD, Saski C, Kumar R, Flinn BS, Luo F, Beissinger TM, Ackerman AJ, Breitzman MW, Bridges WC, de Leon N, Kaeppler SM (2019). Integrated genome-scale analysis identifies novel genes and networks underlying senescence in maize. Plant Cell 31: 1968–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H., Howarth C.J. (2000). Five ways to stay green. J. Exp. Bot. 51: 329–337. [DOI] [PubMed] [Google Scholar]