The rubredoxin-like protein conserved in oxygenic phototrophs binds to the D1 protein and facilitates the formation of the D1/D2 heterodimeric reaction center complex of PSII.
Abstract
Oxygenic photosynthesis relies on accessory factors to promote the assembly and maintenance of the photosynthetic apparatus in the thylakoid membranes. The highly conserved membrane-bound rubredoxin-like protein RubA has previously been implicated in the accumulation of both PSI and PSII, but its mode of action remains unclear. Here, we show that RubA in the cyanobacterium Synechocystis sp PCC 6803 is required for photoautotrophic growth in fluctuating light and acts early in PSII biogenesis by promoting the formation of the heterodimeric D1/D2 reaction center complex, the site of primary photochemistry. We find that RubA, like the accessory factor Ycf48, is a component of the initial D1 assembly module as well as larger PSII assembly intermediates and that the redox-responsive rubredoxin-like domain is located on the cytoplasmic surface of PSII complexes. Fusion of RubA to Ycf48 still permits normal PSII assembly, suggesting a spatiotemporal proximity of both proteins during their action. RubA is also important for the accumulation of PSI, but this is an indirect effect stemming from the downregulation of light-dependent chlorophyll biosynthesis induced by PSII deficiency. Overall, our data support the involvement of RubA in the redox control of PSII biogenesis.
INTRODUCTION
Plants grow by converting solar energy into chemical energy through the operation of a photosynthetic electron transport chain located within the thylakoid membrane system found in chloroplasts. Two large multisubunit pigment-containing reaction center (RC) complexes, termed PSI and PSII, act in tandem to harvest light energy to drive electron flow through a series of electron carriers, ultimately leading to the synthesis of NADPH and ATP, which are used for carbon fixation and other key reactions necessary for plant survival (Flügge et al., 2016).
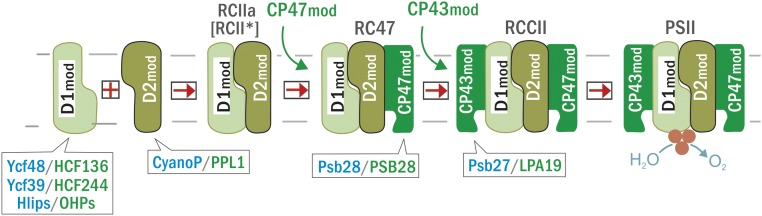
PSII is the protein complex of oxygenic photosynthesis that extracts electrons from water to release oxygen in one of the most demanding reactions in biology. PSII is found in the prokaryotic cyanobacteria as well as chloroplasts and is the most elaborate type of RC found in nature in terms of the number of protein subunits and cofactors (Umena et al., 2011). The biosynthesis of PSII has been intensively studied especially in cyanobacteria. The process involves the stepwise addition of four preassembled pigment/protein modules (D2mod, D1mod, CP47mod, and CP43mod; Figure 1), each containing one large chlorophyll (Chl) binding protein plus one or more neighboring, low-molecular-mass subunits within the PSII complex (Boehm et al., 2011; Komenda et al., 2012b). Efficient assembly of PSII is facilitated by accessory or auxiliary factors that bind to assembly complexes but are not present in the final holoenzyme (Komenda et al., 2012b).
Figure 1.
Simplified Scheme for the Modular Assembly of PSII.
Modules containing the D1 and D2 proteins combine to form the RCII complexes RCIIa and RCII*. They bind the CP47 module followed by the CP43 module to obtain a non-oxygen-evolving PSII core complex (RCCII), and finally, the monomeric and dimeric oxygen-evolving PSII complexes (PSII). Cyanobacterial accessory factors associating with the assembly complexes and their plant homologs are indicated in blue and green, respectively. For simplicity, the RCII complexes (RCIIa and RCII*) as well as PSII dimers and monomers are not distinguished in the scheme.
PSII assembly is initiated by the association of D1mod and D2mod to form the PSII reaction center (RCII) assembly complex (RCIIa; Figure 1). The D1/D2 heterodimer within PSII binds the cofactors needed for light-induced primary charge separation to drive water oxidation (Diner and Rappaport, 2002). RCIIa consists of D1/D2, PsbI, and the PsbE and PsbF subunits of cytochrome (Cyt) b559. In cyanobacteria, the association of D1mod with the lumenal Ycf48 accessory protein promotes the formation of RCIIa (Komenda et al., 2008). D1mod can further associate with a complex of Ycf39 and the high-light-inducible proteins (Hlips) that have a photoprotective role during the assembly of RCIIa (Knoppová et al., 2014; Staleva et al., 2015). During biogenesis, the Ycf39/Hlips complex remains associated with RCIIa, forming a larger assembly complex designated as RCII* (Knoppová et al., 2014). Assembly of RCII complexes (either RCII* or RCIIa) is also facilitated by CyanoP, which binds to the lumenal surface of D2mod (Knoppová et al., 2016). Homologs of CyanoP, Ycf48, Ycf39, and Hlips have been found in land plants (Figure 1; Meurer et al., 1998; Jansson et al., 2000; Ishihara et al., 2007; Li et al., 2019), and so it is thought that the process of PSII assembly including the early stages of RCII* formation is highly conserved in cyanobacteria and chloroplasts (Rühle and Leister, 2016; Li et al., 2019).
RCIIa (or RCII*) subsequently binds CP47mod (Boehm et al., 2011) to form the RC47 assembly intermediate (Boehm et al., 2012) and then CP43mod to give rise to the monomeric PSII core complex, RCCII (Boehm et al., 2011). Assembly of PSII is completed by the light-driven assembly of the Mn4O5Ca oxygen-evolving cluster that catalyzes water oxidation, attachment of lumenal extrinsic proteins, and dimerization to form the oxygen-evolving PSII dimer (Figure 1; Komenda et al., 2012b).
A number of other proteins have been implicated in the accumulation of functional PSII complexes, but their roles are currently unclear (Rühle and Leister, 2016). One of these is rubredoxin A (RubA), which is ubiquitously distributed in all types of oxygenic phototrophs (Calderon et al., 2013) and one of only two proteins possessing a rubredoxin-like domain within the photosynthetic membranes of Arabidopsis (Arabidopsis thaliana; Friso et al., 2004). RubA consists of a predicted C-terminal transmembrane helix connected via a linker to a soluble, redox-responsive rubredoxin-like domain (RD; Wastl et al., 2000; Shen et al., 2002b; Calderon et al., 2013; Guo et al., 2014). Rubredoxins are small, soluble proteins that typically contain one iron atom (although zinc, cobalt, or nickel can substitute) coordinated to four cysteinyl residues (Sieker et al., 1994). RDs are found in various (mainly archeal and bacterial) proteins, for instance flavorubredoxins, ruberythrins, nigerythrins, protein kinase G, or the essential LapB protein of Escherichia coli (Gomes et al., 2002; Iyer et al., 2005; Zhao et al., 2007; Prince and Jia, 2015; Wittwer et al., 2016; Prakash et al., 2018), and typically participate in electron transfer reactions or in redox regulation. RubA has been initially implicated in the redox control of the biogenesis of iron-sulfur (Fe-S) clusters of PSI in the cyanobacterium Synechococcus sp PCC 7002 (hereafter Synechococcus; Shen et al., 2002a, 2002b; Golbeck and Shen, 2006). More recently, the analysis of rubA knockout mutants in plants, algae, and cyanobacteria has suggested a conserved role in the accumulation of functional PSII (Calderon et al., 2013). Despite its vital importance for photosynthesis, the mechanism of RubA action in PSI and PSII biogenesis remains unknown.
In this study, we have discovered that RubA plays a role at an early stage in PSII biogenesis. We have found that the RubA protein binds to D1mod and that it is required for effective formation of the RCII complexes in Synechocystis sp PCC 6803 (hereafter Synechocystis) cells. We further show that the low levels of PSI seen in a photoautotrophically grown RubA-less mutant is an indirect effect of impaired PSII assembly on the light-dependent biosynthesis of Chl. Possible functional roles for RubA are discussed in light of our work.
RESULTS
RubA Associates with the D1 Assembly Module during the Biogenesis of PSII
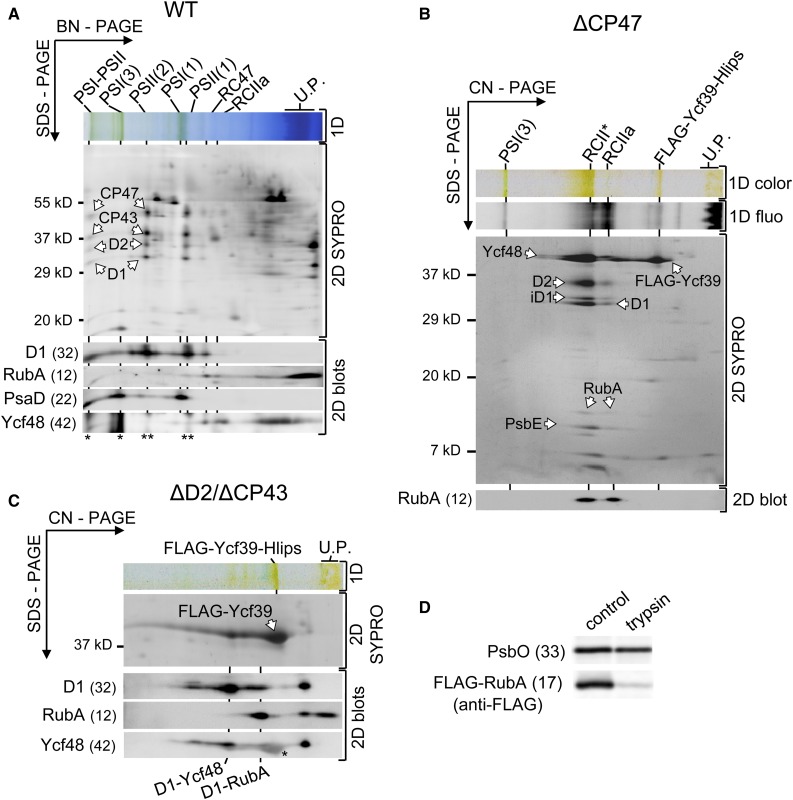
The RubA protein has previously been detected in thylakoid membranes (Shen et al., 2002b; Friso et al., 2004), but its precise location and function remain unknown. To gain more detailed information about the presence of RubA in various membrane complexes, we analyzed membranes isolated from the wild-type strain of Synechocystis using 2D blue native/SDS-PAGE in combination with immunoblotting using specific antibodies for RubA. Although the majority of RubA was detected in the region of small complexes and unassembled proteins, small amounts comigrated with the RC47 and RCIIa assembly intermediates of PSII (Figure 2A). Trace amounts also comigrated with the monomeric PSII core complex [PSII(1)] as well as with the previously detected PSII-PSI supercomplex (Bečková et al., 2017). Notably, RubA could not be detected in dimeric PSII [PSII(2)], suggesting that RubA is not a component of the fully assembled active PSII complex but rather associates with PSII assembly complexes.
Figure 2.
Localization and Topology of RubA in the Isolated Synechocystis Membranes.
(A) Membrane proteins were separated by blue native (BN) PAGE in the first dimension (1D) and further separated by SDS-PAGE in the second dimension. The 2D gel was stained with SYPRO Orange (2D SYPRO) and electroblotted, and the indicated proteins were immunodetected using specific antibodies (2D blots). The relative molecular masses in kD are shown on the side of the stained gel and in parentheses next to the names of the immunodetected proteins. The loaded sample contained 5 µg of Chl. Designation of complexes: PSI-PSII, supercomplex of PSI and PSII; PSI(3) and PSI(1), trimeric and monomeric PSI; PSII(2) and PSII(1), dimeric and monomeric PSII; RC47 and RCIIa are PSII assembly complexes consisting of the D1/D2 heterodimer with and without the CP47 antenna, respectively; U.P., unassembled proteins; WT, wild type. Asterisks indicate signals of PsaD (*) and D1 (**) present on the blot before the Ycf48 detection.
(B) Immunodetection of RubA in RCII complexes. The FLAG-Ycf39 protein with its interacting partners was purified from the CP47-less background that accumulates the RCII complexes. The preparation was analyzed using CN-PAGE. The gel was photographed (1D color), and Chl fluorescence (1D fluo) was detected. The subsequent 2D analysis was performed as described for (A). The complexes are designated as in (A); RCII*, PSII reaction center complex consisting of the D1/D2 heterodimer plus the Ycf39-Hlips complex. U.P., unassembled proteins.
(C) The FLAG-Ycf39 protein was immunopurified from a strain that lacks the D2 and CP43 subunits of PSII, so the complex assembly is arrested before the association of D1mod with D2mod. The preparation was analyzed using CN-PAGE in the first dimension (1D) and analyzed in the second dimension as described for (A). The complexes are designated as in (A) and (B). The asterisk on the blot developed after probing with the antibody against Ycf48 indicates a cross-reaction with the strong band of Ycf39. U.P., unassembled proteins.
(D) Trypsinization of membranes for the determination of the RubA membrane topology. Right-side-out membrane vesicles from the strain expressing an N-terminally FLAG-tagged RubA (FLAG-RubA) before and after 1 h of trypsinization were analyzed by SDS-PAGE, the gel was electroblotted, and FLAG-RubA as well as PsbO were detected using specific antibodies. Each loaded sample contained 3 µg of Chl.
To assess whether RubA is present in the RCII* assembly intermediate, we performed a RubA immunoblot analysis of FLAG-tagged Ycf39 complexes pulled down from a mutant lacking the CP47 antenna and therefore unable to assemble PSII beyond RCII* (Knoppová et al., 2014). The two-dimensional (2D) clear native electrophoresis (CN/SDS-PAGE) and the subsequent immunoblot analysis of the immunopurified FLAG-Ycf39 preparation clearly showed the presence of RubA in RCII* as well as in the dissociated RCIIa complex (Figure 2B). Comparing the SYPRO staining intensity of RubA and PsbE suggested a close to stoichiometric content of RubA in RCIIa but substoichiometric content in RCII*, indicating a possible steric constraint between RubA and the Ycf39-Hlip complex in RCII*.
To determine if RubA binds to D1mod before the formation of RCII*, we tested for the presence of RubA in FLAG-tagged Ycf39 complexes pulled down from a D2/CP43-less strain. In the absence of D2, the only assembly intermediate that can be coisolated with FLAG-Ycf39 is D1mod (Knoppová et al., 2014). In addition to the previously detected D1, PsbI, and Ycf48 components, the RubA protein also copurified with the FLAG-Ycf39-Hlips-D1mod complex (Figure 2C). Although the subunits of the purified complex partially dissociated during CN/SDS-PAGE, a fraction of RubA and D1 comigrated on the 2D gel, suggesting a physical interaction between these two proteins (Figure 2C).
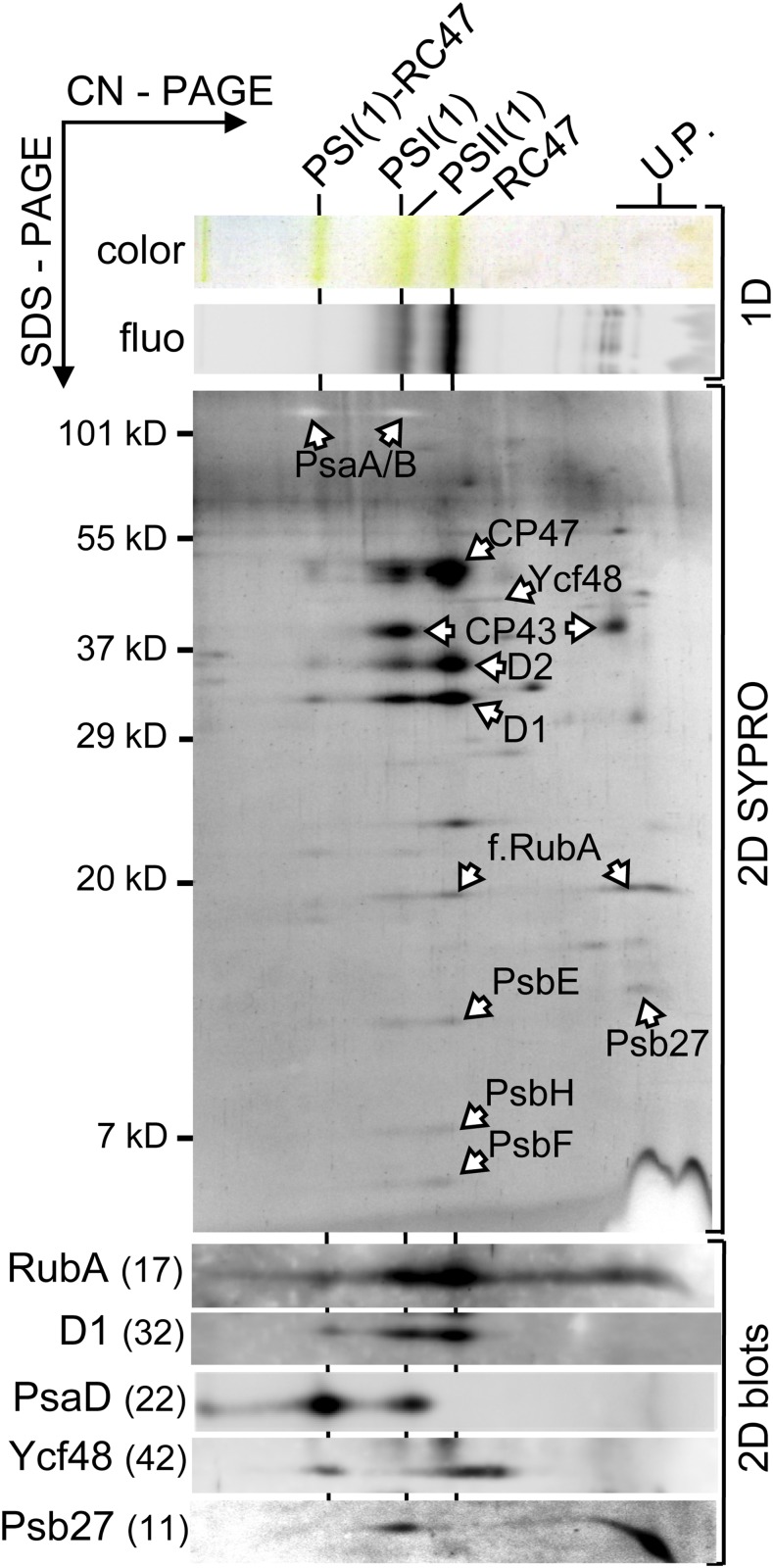
To further clarify the interacting partners of RubA, we constructed a strain expressing an N-terminal FLAG-tagged derivative of RubA under the control of the psbA2 promoter in the ∆rubA background (Supplemental Table 1). The resulting strain (∆rubA/psbA2pro:f.rubA) showed photoautotrophic growth and pigmentation comparable to the wild type, demonstrating that FLAG-RubA fulfills the physiological function of RubA (Supplemental Figure 1; Supplemental Table 2). The FLAG-RubA protein was isolated under native conditions using an anti-FLAG affinity gel. Analysis of the eluate by 2D CN/SDS-PAGE provided evidence that FLAG-RubA associates with the RC47 assembly complex and with the monomeric PSII core complex as well as with the recently described complex of RC47 and monomeric PSI (RC47-PSI(1); Figure 3; Bečková et al., 2017). The assembly factor Psb27, which is absent from active PSII complexes (Komenda et al., 2012a), was present in the FLAG-RubA eluate, signifying that the coisolated monomeric core of PSII did not originate from an active complex (Figure 3). Also, we could not detect any extrinsic subunits associated with oxygen-evolving PSII in the eluate, further supporting the conclusion that FLAG-RubA specifically interacts with PSII assembly complexes. Small amounts of PSI(1) were also detected in the eluate, but this most probably originated from the disintegration of the RC47-PSI(1) complex, as RubA cannot be detected in highly purified PSI-YFP complexes isolated using GFP-Trap (Supplemental Figure 2; Strašková et al., 2018).
Figure 3.
Protein Interacting Partners of RubA.
The FLAG-RubA preparation was analyzed by CN-PAGE in the first dimension. The native gel was photographed (1D color) and scanned for Chl fluorescence (1D fluo), and proteins were separated in the second dimension as described for Figure 2A. Designation of complexes is as in Figure 2A; f.RubA, FLAG-RubA; PSI(1)-RC47 is a complex containing monomeric PSI and RC47. U.P., unassembled proteins.
The 2D SDS-PAGE gel demonstrated the specific binding of the FLAG-RubA protein to PSII complexes, as the FLAG-RubA band comigrated with PSII subunits (Figure 3). The specific presence of the core PSII subunits was also confirmed by mass spectrometry (MS) analysis of the FLAG-RubA eluate (Supplemental Table 3). Unspecific binding of the identified proteins and complexes to the FLAG resin was excluded by a control isolation using wild-type cells. The resulting control preparation did not contain detectable amounts of PSII complexes and only trace amounts of trimeric PSI (Supplemental Figure 3). Overall, these biochemical data demonstrate that RubA can engage with D1mod before the formation of RCII complexes but can also be found in later PSII assembly intermediates.
RubA Is Exposed on the Cytoplasmic Side of the Membrane
Current structural models suggest that RubA consists of an N-terminal RD attached to a potential transmembrane α-helix via a 26-amino-acid-long flexible hinge (Wastl et al., 2000). To determine whether the N-terminal RD is orientated toward the lumenal or the cytoplasmic site of the membrane, we isolated right-side-out vesicles from the strain expressing FLAG-RubA and subsequently treated the vesicles with trypsin (Komenda, 2000). After 1 h of trypsinization, the control and treated samples were analyzed by immunoblot using FLAG-specific antibodies. We found that the vast majority of the FLAG signal disappeared after trypsinization (Figure 2D). Since RubA is FLAG-tagged at its N terminus, these data would place RD on the cytoplasmic side of the thylakoid membrane, as previously proposed (Wastl et al., 2000). By contrast, the lumenal PSII subunit PsbO (Umena et al., 2011) was protected from trypsin digestion, confirming the integrity of the right-side-out vesicles (Figure 2D).
RubA Is Required for the Accumulation of Both Photosystems
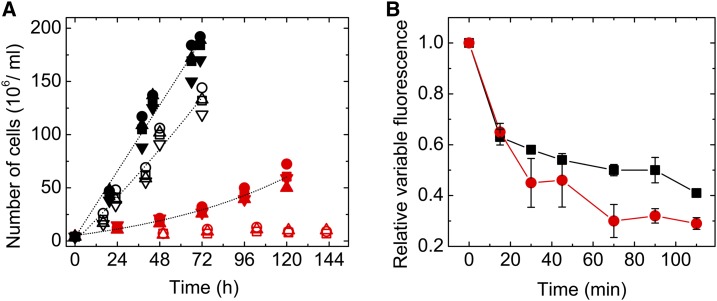
To clarify the physiological function of RubA, we constructed a Synechocystis rubA deletion mutant. The ∆rubA strain exhibited strongly impaired photoautotrophic growth in liquid cultures at standard illumination (40 µmol photons m−2 s−1), and the elimination of the rubA gene was lethal under alternating 15-min standard light/15-min dark conditions (Figure 4A). Furthermore, when wild-type and ∆rubA cells were exposed to high irradiance, the variable Chl fluorescence reflecting PSII function declined substantially faster in the mutant when compared with the wild type, indicating a high sensitivity of mutant cells to photoinhibition (Figure 4B).
Figure 4.
Photoautotrophic Growth and High Light-Induced PSII Photoinhibition of Wild-Type and ∆rubA Cells.
(A) The relative changes in cell number of the wild type (black symbols) and ∆rubA (red symbols) were assessed during exponential growth in liquid cultures under continuous (solid symbols) or fluctuating (15 min of light/15 min of dark; open symbols) light conditions. The circle, square, and triangle symbols represent three independent cultures.
(B) The same amount of wild-type (black) and ∆rubA (red) cells were exposed to 300 µmol photons m−2 s−1. Variable fluorescence was measured and normalized to the initial value (0 min). Mean values and the indicated se are derived from measurements of three independent cultures of each strain. The initial value measured for the ∆rubA mutant represented 20 ± 3% of the wild-type value.
The in vivo absorption spectra of the ∆rubA mutant showed a strong depletion of Chl as judged from the low absorption peak at ∼679 nm, which reflects reduced accumulation of Chl binding photosynthetic complexes, particularly PSI (Figure 5A). We isolated wild-type and mutant membranes and separated their complexes by CN-PAGE; and indeed, the color scan of the gel confirmed markedly lower amounts of trimeric and monomeric PSI [PSI(3)] and to a lesser degree PSI(1) in the ∆rubA strain compared with the wild type (Figure 5C, color). Moreover, substantially lower amounts of PSII(1) as well as dimeric PSII(2) were detected by Chl fluorescence in the membranes of ∆rubA (Figure 5C, fluo).
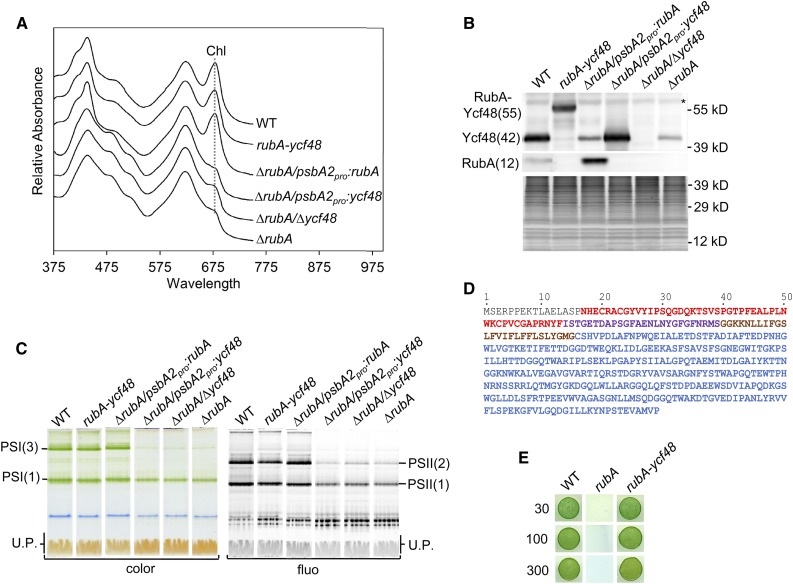
Figure 5.
Comparative Analysis of the Strains Lacking RubA and/or Ycf48 or Expressing a Fused Version of These Proteins.
(A) Whole-cell absorption spectra of the wild type, the strain lacking both RubA and Ycf48 but expressing a RubA-Ycf48 fusion protein (rubA-ycf48), the strain lacking RubA and expressing RubA from the psbA2 promotor (∆rubA/psbA2pro:rubA), the strain lacking RubA and expressing Ycf48 from the psbA2 promotor (∆rubA/psbA2pro:ycf48), the strain lacking both RubA and Ycf48 (∆rubA/∆ycf48), and the strain lacking RubA (∆rubA) were recorded when the photoautotrophic cultures reached an OD of 0.5 at 750 nm; they are shown after shifting for better visibility. The absorption peaks at 679 nm, which reflect Chl content, are designated by a dotted line.
(B) Membranes isolated from strains described in (A) were analyzed by SDS-PAGE. Proteins were subsequently blotted onto a PVDF membrane, and RubA as well as Ycf48 were detected using specific antibodies. For each strain, membranes corresponding to the same number of cells (1.4 × 108) were loaded into the lanes. The SYPRO-stained gel is shown to prove the equal loading of the lanes. The asterisk designates an unspecific cross-reaction. WT, wild type.
(C) Membranes isolated from strains described in (A) were analyzed by CN-PAGE, and the gel was photographed (color) and scanned for Chl fluorescence (fluo). Designation of complexes is as in Figure 2. For each strain, membranes corresponding to the same number of cells (2.5 × 108) were loaded into the lanes. U.P., unassembled proteins. WT, wild type.
(D) Amino acid sequence of the RubA-Ycf48 fusion protein. The RD (red), the rubredoxin linker peptide (purple), the transmembrane helix of RubA (brown), and the Ycf48 part (blue) are shown.
(E) Drops containing 1.25 × 104 cells of the wild type, ∆rubA, or the rubA-ycf48 strain were pipetted on a solid agar plate. The plate was photographed after 3 d of autotrophic growth under constant illumination at 30, 100, or 300 µmol photons m−2 s−1. WT, wild type.
Close Relationship between the RubA and Ycf48 Accessory Factors
In every known cyanobacterial genome, the rubA gene is upstream of the gene encoding Ycf48. Moreover, in Synechocystis, these two genes were shown to form one transcriptional unit (Kopf et al., 2014). In line with the transcriptomic data, we found that insertion of an antibiotic cassette into the coding region of the rubA gene led to a substantial decrease in the intracellular levels of Ycf48 (Figure 5B). To ensure that the low amounts of the photosystems in ∆rubA are not a consequence of the decreased accumulation of Ycf48, which is known to play a role in the assembly of PSII (Yu et al., 2018), we expressed an additional copy of ycf48 in the ∆rubA genetic background. The resulting ∆rubA/psbA2pro:ycf48 strain contained an even higher amount of Ycf48 than the wild type (Figure 5B). Nevertheless, the accumulation of PSII and PSI (Figure 5C) as well as photoautotrophic growth and pigmentation were not improved (Figure 5A; Supplemental Table 2). However, photoautotrophy of ∆rubA could be restored by expressing an extra copy of rubA, indicating that the phenotype of this mutant is specifically related to the absence of RubA, as the Ycf48 level remained low in the complemented ∆rubA/psbA2pro:rubA strain (Figures 5A to 5C; Supplemental Table 2; Calderon et al., 2013). Furthermore, deleting the ycf48 gene from ∆rubA did not change the growth and pigmentation of the strain (Figures 5A and 5C; Supplemental Table 2). Overall, these results confirm that RubA is important for photosystem accumulation and that the observed phenotype of ∆rubA is not due to decreased Ycf48 expression in the mutant.
The coupled expression of the genes coding for RubA and Ycf48 as well as the association of both of these proteins with D1mod during PSII biogenesis (Figure 2C) raised the possibility that they might act in concert and that perhaps they might once have functioned as a single protein. To test this, we constructed strain rubA-ycf48 that expressed RubA and Ycf48 as a fused protein (RubA-Ycf48; for the primary structure, see Figure 5D). The RubA-Ycf48 protein was detected in membranes using specific antibodies against both Ycf48 and RubA and had the expected predicted molecular mass (Figure 5B). Under standard growth conditions, the rubA-ycf48 strain contained somewhat lower amounts of PSII dimers than the wild type (Figure 5C), but otherwise its photoautotrophic growth and pigmentation were comparable to the wild type (Supplemental Table 2). Moreover, unlike ΔrubA, which could not proliferate on plates exposed to 30 µmol photons m−2 s−1, the rubA-ycf48 fusion strain could grow well even at 300 µmol photons m−2 s−1 (Figure 5E), which is an irradiance that is lethal for the single Δycf48 mutant (Yu et al., 2018). Overall, these data indicate that the RubA-Ycf48 fusion protein fulfills the physiological functions of RubA and Ycf48 almost as well as the individual proteins.
RubA Is Specifically Needed for the Formation of the RCII Complex
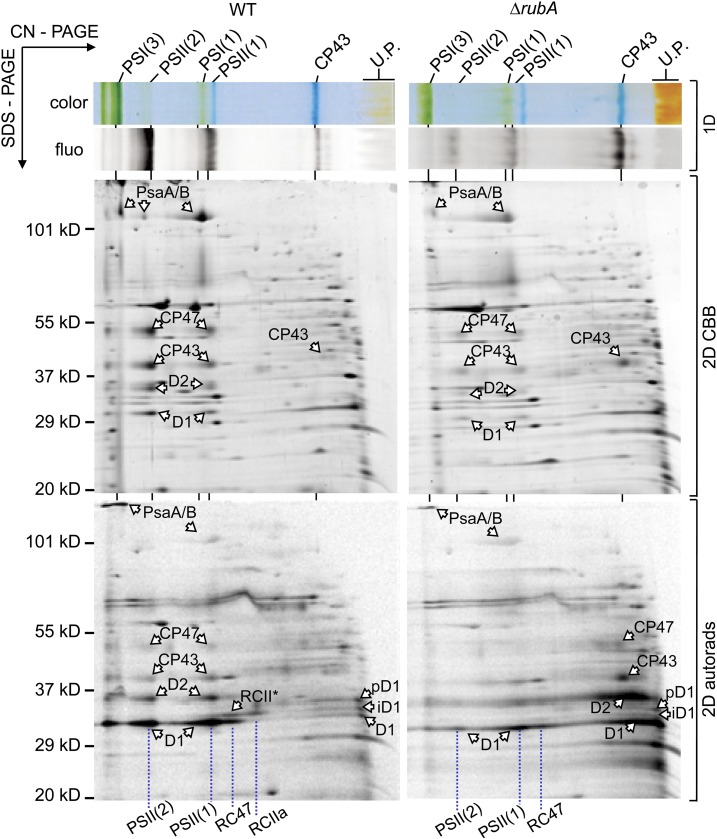
To gain detailed information about the synthesis and accumulation of the photosystems and their subunits, photoautotrophically grown cells of ∆rubA and the wild type were radioactively labeled with a mixture of [35S]Met/Cys. The membranes isolated from these cells were analyzed by 2D CN/SDS-PAGE, and the gel was stained by Coomassie Brilliant Blue (CBB), dried, and subjected to autoradiography. The CN gel and the stained 2D SDS-PAGE gel confirmed the low levels of PSI and PSII and revealed an excessive amount of unassembled CP43 in the mutant (Figure 6). The accumulation of this unassembled internal antenna suggests that the mutant membranes lack the RC47 assembly intermediate that binds CP43mod (Knoppová et al., 2014).
Figure 6.
2D Analysis of Radioactively Labeled Membrane Proteins of the Wild Type and ∆rubA.
Membranes isolated from radioactively labeled cells were analyzed by CN-PAGE in the first dimension (1D color and 1D fluo). After SDS-PAGE in the second dimension, the gel was stained (2D CBB), and the radiolabeled proteins were subsequently detected by autoradiography (2D autorads). Designation of complexes is as described in Figure 2; iD1, intermediate D1; pD1, precursor D1. Each loaded sample contained 5 µg of Chl. U.P., unassembled proteins. WT, wild type.
Importantly, combining the 2D gel with radiography revealed prominent differences in the distribution of newly synthesized D1 and D2 subunits in the wild-type and ∆rubA membranes. After a 30-min pulse, the majority of labeled D1 and D2 was assembled into monomeric and dimeric PSII complexes in the wild type, and minor amounts were as well detected in the RCII* and RCIIa complexes (Figure 6). In addition, small amounts of the three different forms of D1 (precursor D1 containing the C-terminal extension, a partially cleaved D1 intermediate, and mature D1) could be detected in the unassembled protein fraction of the wild type (Komenda et al., 2004).
In the ∆rubA strain, the D1 and D2 subunits as well as the CP47 and CP43 antennae were synthesized at close to wild-type levels (Figure 6, 2D autorads). However, in striking contrast to the wild type, the vast majority of labeled D1 and D2 remained in the unassembled protein fraction, consistent with a block in the formation of RCII complexes. Given that unassembled D1 and D2 are rapidly degraded in Synechocystis (Komenda et al., 2006, 2010), such a defect would explain the reduced accumulation of PSII seen in the absence of RubA.
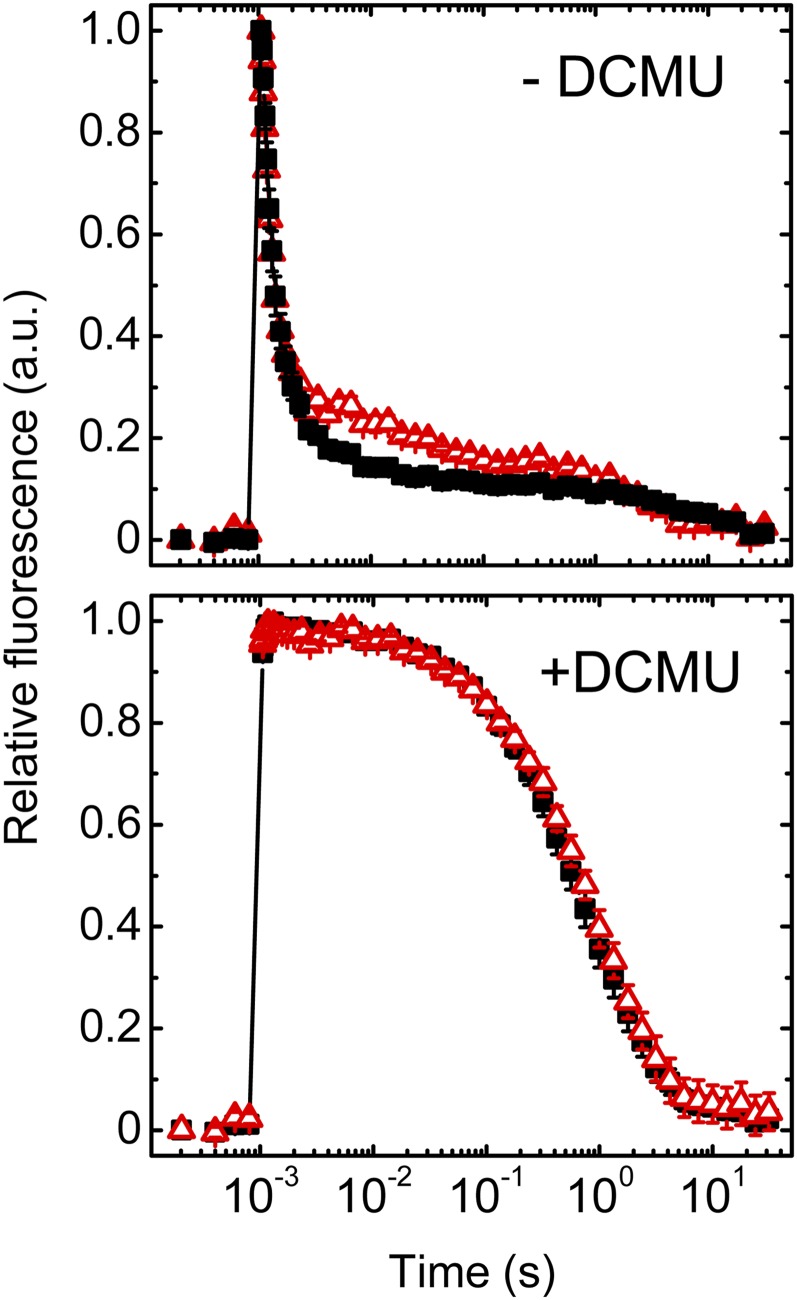
To characterize the functionality of PSII complexes that accumulate at low levels in the ∆rubA strain, we recorded the PSII variable fluorescence decay kinetics. This fluorescence parameter, gained after excitation of the dark-adapted samples by single turnover saturating flash, is proportional to the level of PSII and corresponds to the reduced QA electron acceptor of all active PSII. The subsequent relaxation of fluorescence in the dark arises from the reoxidation of QA−. The first, most rapid phase of the decay (half-time of ∼300–650 µs), reflecting forward electron flow to plastoquinone (PQ) QB acceptor (Vass et al., 1999), was similar in both strains, suggesting that the redox gap between the two quinone acceptors is largely unaffected in ∆rubA (Figure 7). The middle decay phase (half-time of 5–15 ms), reflecting QA− reoxidation when the QB site is empty and needs to bind oxidized PQ (Vass et al., 1999), showed a higher relative amplitude in the mutant (Figure 7). This phase is strongly influenced by the redox state of the PQ pool; therefore, its higher contribution to the decay kinetics might be explained by an enhanced accumulation of reduced PQ molecules (Deák et al., 2014). Finally, the slow phase (half-time of ∼1–20 s), which reflects charge recombination between QA−/QB− and the S2 state of the water-oxidizing complex, was comparable in the mutant and the wild type. The similarity in charge recombination between the wild type and ∆rubA was confirmed by the identical relaxation kinetics of flash-induced fluorescence monitored in the presence of DCMU that blocks electron transfer between QA and QB. Thus, we conclude that there are no substantial changes in the redox gaps within the PSII complex of the mutant, indicating that the assembled complexes are functionally intact and that the low PSII activity is the consequence of low PSII accumulation in the mutant cells (Figure 7; Table 1).
Figure 7.
Decay Kinetics of Flash-Induced Chl Fluorescence.
Equal numbers of wild-type (solid black squares) and ∆rubA (open red triangles) cells were dark adapted for 5 min in the absence (−DCMU) or presence (+DCMU) of DCMU and subsequently excited with single turnover flash at time = 1 ms. The relaxations of the flash-induced Chl fluorescence were recorded and are shown after normalization to the initial amplitude. Mean values (averages) and the indicated se are derived from five independent measurements. a.u., arbitrary units.
Table 1. Comparison of the Relative PSII and PSI Activities, ATP Levels, and Chl and Heme Contents in ΔrubA and in DCMU-Treated Wild-Type Cells.
| Parameter (%) | ΔrubA | Wild Type + DCMU |
|---|---|---|
| PSII activitya | 15 ± 5 | 8 ± 2 |
| PSI activityb | 33 ± 3 | 53 ± 3 |
| ATP contentc | 19 ± 2 | 21 ± 2 |
| Chl contentd | 35 ± 1 | 70 ± 4 |
| Heme contente | 144 ± 11 | 143 ± 9 |
Values represent percentages of the wild-type control levels that were taken as 100%. The PSII and PSI activities were determined from the oxygen-evolving capacities measured in the presence of artificial electron acceptors and from the Pm in the presence of artificial electron donors and acceptors, respectively. Values represent means ± sd for at least three independent cultures.
Oxygen evolution of the wild-type PSII complexes was 20 ± 4 pmol oxygen/h/cell.
The PSI activity of the wild type expressed as Pm was 71 ± 6 units/105 cells.
The ATP content of the wild type expressed as luminescence intensity was 24 ± 2 units/105 cells.
The Chl content of the wild type was 2.8 ± 0.2 µg Chl/108 cells.
The heme content of the wild type was 229 ± 8 pmol heme/108 cells.
Chl Biosynthesis Is Inhibited in the Photoautotrophically Grown ∆rubA Mutant
Although the accumulation of both photosystems is reduced in the ∆rubA strain, we could detect an association of RubA only with PSII and not with PSI (Figures 2 and 3; Supplemental Figure 2). The mutant was especially deficient in the content of trimeric PSI, whereas the monomeric PSI form was clearly less affected (Figure 5C). Such a preferential decrease in trimeric PSI has been previously observed in Synechocystis mutants with impaired Chl biosynthesis (Kopečná et al., 2013, 2015b).
Certain enzymatic steps in Chl biosynthesis need either plastoquinol, NADP(H), ATP, or oxygen (Willows, 2006; Steccanella et al., 2015). The production of these substances is dependent on PSII activity; therefore, we tested the potential effect of low PSII activity on Chl biosynthesis in wild-type cells. For this purpose, we grew the wild type autotrophically in the presence of 0.8 µM DCMU which suppressed oxygen-evolving PSII activity to a similar level to that found in ∆rubA (Table 1). Unlike in ∆rubA, PSII biogenesis in the DCMU-treated wild-type cells remained intact; assembly intermediates were converted into PSII(1) and PSII(2), and the accumulation of the PSII complexes was comparable to what was observed in the untreated wild-type cells (Supplemental Figures 4A and 4C). The reduction in PSII activity in both ∆rubA and the DCMU-treated wild-type cells led to similar low ATP levels and resulted in a reduced Chl content in comparison with the control wild-type cells. The lower Chl content was directly proportional to the reduction in the total amount of photooxidizable P700 (Pm), which reflects the number of active PSI complexes in the cell (Table 1).
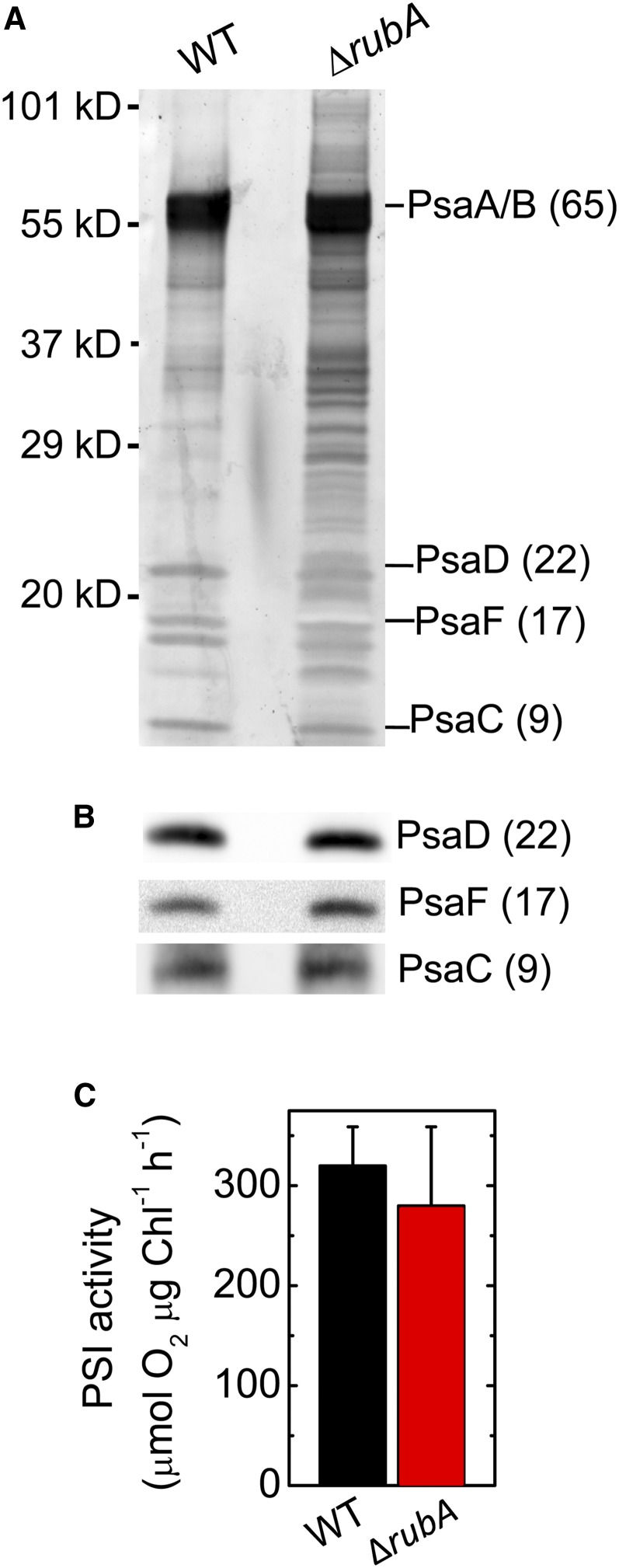
The ∆rubA mutant of the cyanobacterium Synechococcus has previously been reported to lack PSI-mediated electron transport activity due to the loss of the iron-sulfur clusters from the complex and their associated stromal subunits (Shen et al., 2002b). Therefore, we addressed whether the decreased Pm value in the Synechocystis ∆rubA mutant could be influenced by changes in the functional integrity of the complex. Using size-exclusion chromatography, we isolated fractions highly enriched in PSI(3) from detergent-solubilized membrane extracts of the Synechocystis wild-type and ∆rubA strains. SDS-PAGE and subsequent immunoblot analysis of the fractions indicated that the subunit composition of the PSI complexes isolated from the wild type and mutant was comparable (Figures 8A and 8B). Additionally, the rate of PSI-mediated electron transport in the isolated PSI-enriched membranes did not show significant differences (P = 0.197; Figure 8C). These data demonstrated that the low amount of P700+ in the ∆rubA mutant of Synechocystis is solely due to the reduced number of PSI complexes and that the functional integrity of the PSI complexes remained intact.
Figure 8.
Determination of the Structural and Functional Integrity of PSI in the ΔrubA Mutant.
(A) Electrophoretic analysis of PSI-enriched membranes. After separation of proteins by SDS-PAGE, the proteins were stained using SYPRO Orange. WT, wild type.
(B) Immunodetection of PsaD, PsaF, and PsaC subunits in the PSI-enriched membranes.
(C) PSI activities of the PSI-enriched membranes isolated from the wild type (black bar) and the ΔrubA mutant (red bar) measured by means of oxygen consumption in the presence of an artificial electron donor (dichlorophenolindophenol) and acceptor (methyl viologen). Each column and error bar represents a mean value (average) and the sd, respectively, for five independent measurements.
The same amounts of wild-type and mutant thylakoid particles (estimated according to Chl content) were used for all assays presented in (A) to (C). WT, wild type.
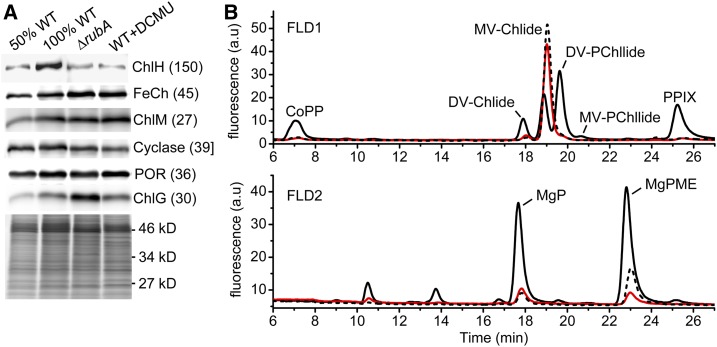
To estimate the effect of the low PSII activities on Chl biosynthesis, we assessed the relative abundances of particular enzymes as well as biosynthetic intermediates of the pathway. We found that the level of the catalytic H subunit of magnesium chelatase (MgCh), the enzyme responsible for the production of Mg-protoporphyrin IX (Mg-PPIX), was lower in ΔrubA as well as in DCMU-treated wild-type cells when compared with the control wild-type cells (Figure 9A). By contrast, the level of ferrochelatase (FeCh), the enzyme synthesizing heme and competing for the same substrate as MgCh, was relatively high in the mutant and in the DCMU-treated wild type. The reduced ratio of MgCh to FeCh suggests that Chl production was suppressed by channeling PPIX into the heme branch. This conclusion was supported by the relatively low Chl but higher heme content in ΔrubA and DCMU-treated wild-type cells compared with the control wild-type cells (Table 1). In addition, we detected a lower level of Mg-protoporphyrin IX monomethyl ester (MgPME) oxidative cyclase (cyclase) in DCMU-treated wild-type cells and overaccumulation of Chl synthase in the ΔrubA cells. HPLC analysis of pigments showed similarly low abundances of the tetrapyrrole biosynthetic intermediates in the ΔrubA and DCMU-treated wild-type cells (Figure 9B). Namely, the early precursor coproporphyrinogen III (CoPP) was hardly detectable, and the PPIX content was also substantially lower in ΔrubA and DCMU-treated wild-type cells in comparison with the control wild-type cells. These data indicate that low PSII activity led to the downregulation of Chl production at an early stage in biosynthesis and this resulted in decreased accumulation of PSI complexes (Table 1; Supplemental Figure 4).
Figure 9.
Analysis of the Tetrapyrrole Biosynthetic Pathway in ΔrubA and in the DCMU-Treated Wild Type.
(A) Levels of selected tetrapyrrole biosynthesis enzymes in the membranes of the wild type, ΔrubA, and the wild type grown in the presence of 0.8 µM DCMU. After 2 weeks of autotrophic cultivation, the cells were harvested. Membranes corresponding to the same amount of cells were analyzed by SDS-PAGE, and separated proteins were electroblotted and immunodetected. ChlG, chlorophyll synthase; ChlH, catalytic subunit of Mg-chelatase; ChlM, Mg-protoporphyrin IX methyltransferase; WT, wild type.
(B) Abundance of Chl precursors in the control wild-type (black solid line), ∆rubA (red line), and DCMU-treated wild-type (WT+DCMU; black dotted line) cells. Pigments were extracted by methanol from equal numbers of cells. The signals of the subsequent HPLC analysis were detected by a pair of fluorescence detectors (FLD1 and FLD2) set for different wavelengths to cover all Chl precursors. a.u., arbitrary units. DV, divinyl; MgP, Mg-protophorphyrin IX; MV, monovinyl.
Chl Biosynthesis Is Restored in the ∆rubA Mutant in the Dark
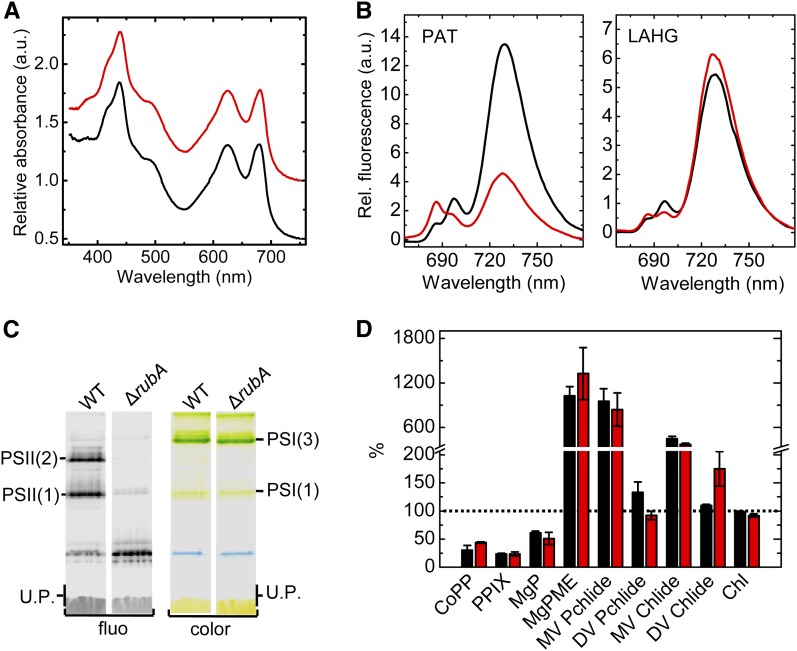
Synechocystis is a facultative photoautotroph that is able to proliferate in the dark by light-activated heterotrophic growth (LAHG). During LAHG, Synechocystis cells do not photosynthesize and instead drive their metabolism by energy and reductants derived solely from a carbohydrate supplement (Anderson and McIntosh, 1991). To verify that downregulated Chl biosynthesis and the consequent reduction in PSI accumulation are results of the low intracellular PSII activity, we grew the wild type and ΔrubA under LAHG conditions. We found that unlike during photoautotrophy, under LAHG conditions the in vivo absorption spectra of the wild-type and ΔrubA cells were very similar (Figure 10A). Nevertheless, CN-PAGE separation of the membrane complexes revealed that the accumulation of PSII remained impaired in the mutant, while the amount of the PSI complexes reached the wild-type control level (Figure 10B). The relative amounts of PSII and PSI in photoautotrophic and LAHG cultures of the wild type and ∆rubA were addressed by low-temperature Chl fluorescence spectroscopy on an equal cell basis. After excitation of Chl at 435 nm, the photoautotrophic ∆rubA cells exhibited remarkably lower fluorescence emission bands at 695 and 725 nm in comparison with the photoautotrophic wild-type cells (Figure 10C). This reflects lower amounts of active PSII and PSI complexes, respectively (Shen and Vermaas, 1994). The other PSII-related fluorescence peak at 685 nm was relatively high in the mutant and is likely to reflect the fluorescence emission from the CP43 antenna (Shen and Vermaas, 1994) that accumulates within the mutant (Figure 6). In the LAHG cells of ∆rubA, the PSII fluorescence peak at 695 nm remained relatively low. By contrast, the 725-nm fluorescence peak reflecting the PSI content was now comparable to that in LAHG cells of the wild type. Likewise, the Chl content of the mutant reached wild-type levels under LAHG conditions (Figure 10D). Moreover, the levels of the tetrapyrrole precursors reflecting the rate of Chl biosynthesis were similar in the LAHG cultures of ∆rubA and the wild type (Figure 10D). Therefore, these data confirmed that RubA in Synechocystis is specifically needed for the accumulation of PSII while the low PSI content of the photoautotrophic ∆rubA cells is an indirect effect of perturbed Chl biosynthesis in the light due to the lower PSII activity.
Figure 10.
Characterization of Photoautotrophic and LAHG-Grown Wild-Type and ΔrubA Cells.
(A) Whole cell absorption spectra of the wild type (black line) and ΔrubA (red line) grown for 6 d under LAHG conditions. The spectra are shown after normalization to the OD at 750 nm and are shifted for better visibility. a.u., arbitrary units.
(B) Separation of membrane protein complexes from LAHG-grown wild-type and ∆rubA cells using CN-PAGE. The gel was photographed (color) and scanned for Chl fluorescence (fluo). Designation of complexes is as in Figure 5C. a.u., arbitrary units.
(C) 77 K Chl fluorescence emission spectra from cells of the wild type (black line) and ∆rubA (red line) grown either photoautotrophically (PAT) or heterotrophically (LAHG). Equal amounts of cells were frozen in liquid nitrogen and excited at 435 nm. Spectra were normalized to the emission peak of the internal standard rhodamine at 570 nm. Curves represent mean values of three independent measurements. WT, wild type. U.P., unassembled proteins.
(D) Comparison of the relative cellular contents of Chl and its biosynthetic precursors in LAHG-grown wild-type (black bars) and ∆rubA (red bars) cultures. The Chl content was determined spectroscopically, while the relative abundances of Chl precursors were quantified by HPLC. Abbreviations are as in Figure 9B. The values were normalized to the autotrophically grown wild-type control levels that are taken as 100% and indicated by the dotted line. The columns and error bars represent means ± se, respectively, for at least three independent cultures.
DISCUSSION
In agreement with previous work (Calderon et al., 2013), we found that the abundance of PSII in Synechocystis is strongly reduced in the absence of RubA. We show here that the complexes that do accumulate (15%–20% of wild-type level) are functionally intact (Figure 7). These data indicate that RubA is involved in the accumulation but not the function of the oxygen-evolving PSII complex, in line with the absence of RubA from fully assembled PSII(2) complexes (Figure 2A).
A role for RubA in the assembly of PSII was clarified in pulse-labeling experiments that revealed a bottleneck in the formation of the RCII complex (Figure 6), previously shown to contain the D1/D2 heterodimer, Cyt b559, PsbI, and the Ycf48 accessory factor (Komenda et al., 2008). Pull-down experiments showed that RubA was present in the D1mod that combines with the D2mod to form RCII, and so RubA plays a role early in PSII assembly (Figures 2B and 2C). In contrast to the wild type, the RubA deletion strain showed a similar defect in PSII accumulation in the dark as well as in the light (Figures 5 and 10). These data therefore suggest that RubA plays an important role in the early stages of PSII assembly (Figure 11) even in the absence of photoinhibitory damage.
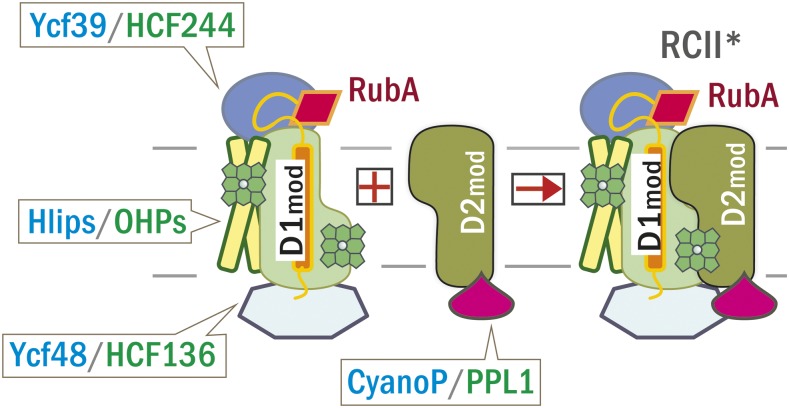
Figure 11.
Model for the Early Stages of PSII Assembly Showing the Locations of RubA and the Other Cyanobacterial/Plant Accessory Proteins.
Ycf48 (HCF136 in plants) and the chlorophyll-containing Ycf39/Hlip complex (HCF244/OHP in plants) assist in the insertion of Chl into D1mod, while CyanoP (PPL1 in plants) stabilizes D2mod. The transmembrane domain of RubA associates with the N-terminal region of D1, positioning the C-terminal tail of RubA in close proximity to the lumenal Ycf48 factor. The linker region of RubA allows the rubredoxin domain to bind at the interface of D1 and D2. All factors remain associated with the modules after they form the RCII* complex.
An additional role for RubA in protecting PSII assembly intermediates from photodamage is also possible given its presence in larger assembly complexes [RC47 and PSII(1); Figures 2 and 3] that are inactive for oxygen evolution and prone to photoinhibition (Komenda et al., 2010). Given that RubA is attached to D1mod containing the D1 subunit (Figure 2C), a role of RubA in replacing D1 during the repair of photodamaged PSII should also be considered. Both these features might explain the greater sensitivity of PSII in the ΔrubA strain to photoinhibition (Figure 4B).
The requirement of RubA for efficient formation of the D1/D2 heterodimer that holds all the redox-active components essential for PSII primary photochemistry (Diner and Rappaport, 2002) is rather intriguing. RubA possesses an RD that is known to participate in electron transfer reactions or redox regulation. Therefore, RubA might function to maintain PSII assembly complexes in an appropriate redox state during biogenesis to prevent light-induced irreversible damage and/or promote the insertion of the redox-active non-heme iron bound at the interface between D1 and D2. The other redox-active cofactor that is inserted during biogenesis and is located on the same cytoplasmic side of PSII as RD (Figure 2D; Wastl et al., 2000) is QA, the primary plastoquinone electron acceptor in PSII. The midpoint redox potential of RubA is +125 mV (Wastl et al., 2000), which is positive enough to allow RubA to keep the acceptor side of PSII oxidized and so prevent the accumulation of singly reduced QA (due to either inadvertent nonphotochemical reduction or lack of functional QB). Another possibility is that reduced RubA might reduce Cyt b559 within PSII and so act as part of a photoprotective cyclic electron transport pathway involving the oxidation and subsequent rereduction of P680+ via a pathway involving QA, RubA, Cyt b559, and the CarD2 carotenoid bound to D2 (Shinopoulos et al., 2014). A redox role for RubA is supported by our observation that the ΔrubA strain is able to proliferate in autotrophic media under continuous illumination but not when exposed to fluctuating light consisting of cycles of 15 min of light and 15 min of dark (Figure 4A). Together with the PSII deficiency of the ΔrubA strain, this would imply that RubA plays an especially important physiological role for the assembly of PSII under changeable redox conditions, such as dark-to-light transitions. It has also been reported that two Cys residues of RubA RD are reversibly oxidized upon light-to-dark transitions (Guo et al., 2014). So, the activity of RubA itself might be under redox control and a checkpoint for redox-regulated PSII assembly.
In all cyanobacteria, the rubA gene is transcribed together with the gene encoding Ycf48, which is a β-propeller protein located on the lumenal side of the membrane (Yu et al., 2018) and also is involved in promoting the formation of PSII RC (Komenda et al., 2008). RubA seems to play a more dominant role than Ycf48 in PSII assembly, as the phenotype of ΔrubA is more severe than that of the Δycf48 mutant and it was not altered by the overexpression of Ycf48 (Figures 5A and 5C). Consistent with this, the ΔrubA/Δycf48 double mutant has a similar phenotype to the ΔrubA single mutant. Nevertheless, a possible interplay between the action of RubA and Ycf48 is suggested by the effective complementation of the ΔrubA/Δycf48 mutant by the expression of a RubA-Ycf48 fusion protein in which the N terminus of Ycf48 is fused to the C terminus of RubA (Figures 5D and 5E). Additionally, both Ycf48 and RubA are detected in the RCII and RC47 assembly complexes (Figure 2A), and Ycf48 was detected in the pull-down assay using FLAG-tagged RubA as a bait (Figure 3). Based on these data, we speculate that Ycf48 and RubA might once have functioned as a single protein during PSII assembly and that in extant organisms Ycf48 still attaches to PSII in the lumen close to the C terminus of RubA (Figure 11).
Earlier studies showed that the rubA deletion mutant of Synechococcus was not able to grow photoautotrophically, and when grown using a carbohydrate supplement, PSII activity decreased to 77% of the wild-type levels (Shen et al., 2002b). No PSI activity was detected in the mutant, because of the loss of the Fe-S clusters of PSI and their associated stromal subunits (Shen et al., 2002a, 2002b). In Synechococcus, RubA is suspected to keep the strong reductant A1 oxidized during PSI biogenesis, so that A1− will not reduce the Fe-S cluster, which can then be inserted into the complex (Golbeck and Shen, 2006). However, unlike in higher plants and in the rest of cyanobacteria (including Synechocystis), where the PSI secondary electron acceptor A1 is a phylloquinone molecule, in Synechococcus, A1 is menaquinone, which is typical for bacterial type I reaction centers (Sakuragi et al., 2005). It is possible that the presence of different quinones in the proximity of the Fe-S clusters allows alternative ways of regulating the biogenesis of the complex. Nevertheless, our data show that, in contrast to Synechococcus, the absence of RubA in Synechocystis does not affect the subunit composition and the functional integrity of PSI (Figure 8).
Newly synthesized Chl has been previously shown to be preferentially channeled to trimeric PSI; therefore, accumulation of this complex is reliant on functional Chl biosynthesis (Kopečná et al., 2012). When PSII complexes in the wild type were inhibited by DCMU to give the same low activity displayed by ∆rubA (Table 1), the Chl biosynthetic pathway was similarly inhibited in both strains (Figure 9B). One possible step in Chl biosynthesis that could be regulated by PSII activity is the synthesis of Mg-PPIX catalyzed by MgCh. This enzyme consists of four subunits—the catalytic H subunit works together in a complex with Gun4 and the ChlI and ChlD subunits—and hydrolyzes ATP to drive the thermodynamically unfavorable insertion of Mg into PPIX (Adams et al., 2016). Therefore, it is likely that the reduced ATP content in cells with low PSII activity (Table 1) results in the feeble accumulation of Mg-PPIX, the first committed precursor of Chl (Figure 9B). Impaired production of Mg-PPIX was shown to downregulate the initial steps of the tetrapyrrole biosynthetic pathway (Hedtke et al., 2007; Kopečná et al., 2015a), which may explain the low levels of all precursors needed for Chl synthesis (Figure 9B). By contrast, the level of FeCh, the enzyme that competes for the same substrate as MgCh, was upregulated in ∆rubA (Figure 9A). FeCh is a key regulatory enzyme of the tetrapyrrole pathway in cyanobacteria as well as in plants (Sobotka et al., 2005, 2008; Woodson et al., 2011; Scharfenberg et al., 2015), and its elevated level in PSII mutants has been reported earlier (Sobotka et al., 2005). A combination of high FeCh and low MgCh activity could redirect most of the available PPIX into the heme branch away from Chl production. Furthermore, by a feedback loop, the increased amount of heme can inhibit the synthesis of 5-aminolevulinic acid, the first committed precursor of tetrapyrroles (Czarnecki and Grimm, 2012), further restricting the PPIX flow into the Chl biosynthetic pathway. How FeCh is connected to PSII is not known and needs further investigation.
Despite the comparable levels of Chl biosynthetic precursors in ∆rubA and in DCMU-treated wild-type cells, the latter still contains more Chl and more PSI complexes (Table 1; Supplemental Figure 4). We have previously shown that deletion of the PSII subunit CP47 also leads to PSI depletion, which is more extensive than in strains with inactivated PSII that are still able to accumulate CP47-containing PSII complexes (Bečková et al., 2017). These data suggest that the presence of RC47 is necessary for functional Chl biosynthesis in the light, and consequently, for the accumulation of PSI trimers. However, here we show that Chl biosynthesis, as well as PSI accumulation, was much more similar in the wild type and the PSII-deficient ∆rubA strains grown in the dark (Figure 10). These data indicate that assembled PSII complexes are not needed for Chl biosynthesis in the dark, which involves a different set of enzymes. In the dark, the light-independent protochlorophyllide oxidoreductase (DPOR), an enzyme producing chlorophyllide (Chlide), must replace its light-dependent version (Fang et al., 2017). This switch is particularly important under LAHG conditions (Kopečná et al., 2013), when the dissolved oxygen concentration is substantially lower and so more favorable for the nitrogenase-like, oxygen-sensitive DPOR (Yamazaki et al., 2006). Under such conditions, the oxygen-dependent cyclase enzyme preceding (D)POR is also downregulated and most probably partially replaced by the anoxygenic “bacterial-type” cyclase (BchE; Hasunuma et al., 2018). Indeed, the strong accumulation of both MgPME and protochlorophyllide (Pchlide), the substrates of cyclase and DPOR, was observed in the LAHG cells, implying a decline in the cyclase and POR activities (Figure 10D). Furthermore, the oxygen-dependent CoPP oxidase (HemF) responsible for the decarboxylation of CoPP is likely to be substituted by the oxygen-independent HemN under LAHG (Goto et al., 2010). These enzymes are structurally different from their “light” counterparts: they use different cofactors and electron donors/acceptors that might explain why in the dark Chl biosynthesis is not connected to PSII activity.
In summary, we have shown that RubA binds to D1mod during PSII assembly and plays a physiologically important role in the accumulation of the PSII reaction center assembly complex. The accumulation of PSI was also defective in the ∆rubA strain, but a specific interaction between RubA and PSI could not be confirmed (Supplemental Figure 2). Instead, we showed that the reduced PSI level is an indirect effect of low PSII activity on the light-dependent pathway of Chl biosynthesis needed for PSI accumulation. Thus, we conclude that the unique and ubiquitous presence of the rubA gene in all sequenced oxygenic phototrophs reflects the essential and universal role of RubA in the formation of the oxygen-evolving PSII complex.
METHODS
Construction and Cultivation of Strains
The Synechocystis sp PCC 6803 glucose-tolerant substrain GT-P was used as the wild type (Tichý et al., 2016). The ΔrubA strain was constructed by replacing the slr2033 gene (36–339 bp) by a zeocin antibiotic resistance cassette. The sequences upstream and downstream (350 bp) of the rubA gene were amplified using two sets of primers (Supplemental Table 4), and fusion PCR in conjunction with megaprimers (Ke and Madison, 1997) were used to anneal these on either side of the zeocin resistance cassette. The resulting PCR product was transformed into the GT-P Synechocystis substrain, and the transformed cells were fully segregated on BG-11 plates with increasing concentrations of zeocin. The Synechocystis strain expressing the RubA protein fused with 3xFLAG at the N terminus (FLAG-RubA) was prepared using the pPD-NFLAG plasmid essentially as described (Hollingshead et al., 2012). To construct the ΔrubA/Δycf48 mutant, the ΔrubA strain was transformed with chromosomal DNA isolated from the ycf48 disruption mutant (Komenda et al., 2008), and the transformed cells were fully segregated on increasing concentrations of chloramphenicol. The construction of ΔrubA/psbA2pro:rubA and ΔrubA/psbA2pro:ycf48 was performed by cloning rubA or ycf48, respectively, into the ΔrubA strain using the pPD-NFLAG plasmid. The in-frame fusion of the rubA and ycf48 genes was constructed by deleting the nucleotides between the codons specifying Gly-115 of RubA and Cys-29 of Ycf48 (Figure 5D) using inverse PCR (for primers, see Supplemental Table 4) and the template vector pYcf48WTgen (Yu et al., 2018). The religated PCR fragment was then cloned into Escherichia coli and sequenced prior to transformation of the Synechocystis mutant Δycf48 (Yu et al., 2018) to yield the rubA-ycf48 strain.
Unless stated otherwise, the Synechocystis strains were grown photoautotrophically in liquid BG-11 medium on a rotary shaker at 28°C under a moderate irradiance of 40 μmol photons m−2 s−1 given by white fluorescence tubes (standard light). The spectrum of the cultivation light (Supplemental Figure 5) was measured with a Spectrapen SP 110 spectrophotometer (PSI). The fluctuating light/dark conditions were achieved by alternating 15 min of standard light with 15-min dark intervals. The LAHG cultures were grown for 6 d in BG-11 medium supplemented with 5 mM glucose in darkness with 5-min daily illumination of standard light (Anderson and McIntosh, 1991). To mimic the low PSII activity measured in the ΔrubA mutant, a partial loss of PSII activity in the wild-type cells was artificially induced by the addition of 0.8 µM DCMU (Metz et al., 1986) to the growth medium. The DCMU-treated wild-type cell cultures were twice reinoculated to fresh DCMU-containing media and were harvested for characterization after 2 weeks of cultivation.
The absorption spectra of cells were measured by a UV-3000 spectrophotometer (Shimadzu). The number of cells was assessed by a Multisizer 4 (Beckman Coulter). For the determination of doubling times, five independent cultures of each strain were cultivated.
Assessment of Photosystem Activities and Integrities
The number of active PSII in the cell was assessed by measuring the oxygen evolution rate of the complexes in the presence of the artificial electron acceptors 0.5 mM K3Fe(CN)6 and 0.1 mM 2,5-dimethyl-p-benzoquinone (Sigma-Aldrich). The rates of oxygen evolution and consumption were measured using a Clark-type electrode. Variable fluorescence during high-light (300 μmol photons m−2 s−1) exposure was followed using an Aquapen-100 (PSI) after 5 min of dark incubation.
PSII electron transport kinetics were followed by the emission and subsequent relaxation kinetics of flash-induced Chl fluorescence in the absence or presence of 10 µM DCMU using a double-modulation fluorometer (PSI) as described (Vass et al., 1999).
The Pm to determine the amount of active PSI complexes was induced by a saturating pulse followed by a recovery period in the dark (Klughammer, 2008). Before the measurement, 1 mM 2,6-dichlorophenolindophenol, 10 mM L-ascorbate, and 1 mM methyl viologen (Sigma-Aldrich) were added as artificial electron donors and acceptor, respectively; 10 µM DCMU (Sigma-Aldrich) was added to exclude PSII-driven electron flow.
For the PSI and PSII assays with intact cells, five independent cultures were used.
The Chl fluorescence spectra was recorded at 77 K on equal amounts (108) of cells that were washed into fresh BG-11 medium, mixed with 2 nM rhodamine (Sigma-Aldrich), transferred into a sample holder, and frozen in liquid nitrogen. The frozen sample was excited at 435 nm, and the fluorescence emission was recorded between 550 and 800 nm by an Aminco Bowman Series 2 luminescence spectrometer (Spectronic Unicam). The assay was repeated on three independent cultures of each strain.
To assess the activity of isolated PSI, equal amounts of cellular membranes (normalized to Chl content) were isolated in MES buffer (25 mM MES/NaOH, pH 6.5, 10 mM MgCl2, and 5 mM CaCl2) and solubilized by n-dodecyl-β-D-maltoside (DDM; DDM/Chl = 20 [w/w]). The unsolubilized membranes were removed by Corning Costar Spin-X centrifuge tube filters (Sigma-Aldrich), and the solubilized membrane complexes were separated by a Superdex 200 (10/300 GL) size-exclusion column (Amersham Pharmacia Biotech) using MES buffer containing 0.06% (w/v) DDM at a flow rate of 1 mL/min. The minute fraction containing PSI(3) was collected and analyzed by protein immunoblot assay to determine the subunit composition of the PSI complexes. The electron transport rates mediated by PSI was measured by means of oxygen consumption in the presence of 1.5 mM l-ascorbate, 0.2 mM 2,6-dichlorophenolindophenol as electron donor, and 0.1 mM methyl viologen as electron acceptor. Ten micromolar DCMU was added to the sample to eliminate the effects of potential PSII contamination. Each isolate was measured five times. Since the wild-type data set gave a slightly higher average value than the mutant within the range of sd, we tested the significance of this difference using a one-tailed t test, with a significance level set to P < 0.05. The obtained t and P values were 0.9175 and 0.197, respectively.
Preparation of Cellular Membranes and Their Trypsinization
Cells from exponential growth phase (5 × 109) were pelleted, washed, and resuspended in buffer B (25 mM MES/NaOH, pH 6.5, 10 mM CaCl2, 10 mM MgCl2, and 25% (v/v) glycerol). They were broken mechanically in a Mini-Beadbeater (BioSpec) using balotina beads as described (Komenda and Barber, 1995). The membrane and soluble protein fractions were separated by centrifugation at high speed (65,000g, 20 min). The pelleted membranes were washed and resuspended in buffer B. The trypsinization of isolated membranes was performed as described (Dobáková et al., 2009).
Isolation of Protein Complexes
For the purification of protein complexes under native conditions, cells from 4-liter cultures (OD at 750 nm of ∼0.5) were centrifuged, washed, and broken as described above. The cellular membranes containing 1 mg/mL Chl were solubilized for 1 h with 1% (w/v) DDM at 10°C and centrifuged for 20 min at 65,000g to remove insolubilized membrane particles. The supernatant was loaded onto an anti-FLAG M2 affinity gel chromatography column (Sigma-Aldrich). Proteins bound to the column were washed six times with 2.5 resin volumes of buffer B containing 0.04% (w/v) DDM. FLAG-tagged protein complexes were eventually eluted with buffer B containing 0.04% (w/v) DDM and 150 μg/mL 3xFLAG peptide (Sigma-Aldrich).
The immunoprecipitation using green fluorescent protein-Trap (ChromoTek) was performed as described (Strašková et al., 2018).
Protein Electrophoresis, Immunoblotting, and Radiolabeling
For native electrophoresis, solubilized membrane proteins or isolated complexes were separated on 4 to 12% (w/v) CN (Wittig et al., 2007) or blue native (Schägger and von Jagow, 1991) gels in the first dimension. The protein gels were scanned and the Chl fluorescence image was taken by a LAS 4000 camera (Fuji). The individual components of the protein complexes were resolved by incubating the gel strip from the first dimension in 62.5 mM Tris/HCl, pH 6.8, containing 2% (w/v) SDS and 1% (w/v) DTT for 30 min at room temperature, and by subsequent separation in the second dimension by SDS-electrophoresis on a denaturing 12 to 20% (w/v) polyacrylamide gel containing 7 M urea (Dobáková et al., 2009). For standard single-dimension SDS-PAGE, membrane suspensions were solubilized at room temperature for 20 min after adding a one-fifth volume of 10% (w/v) SDS and 5% (w/v) DTT. Proteins were stained by CBB (Sigma-Aldrich); alternatively, in case of subsequent immunoblotting, they were stained by SYPRO Orange (Sigma-Aldrich). The SYPRO-stained gel was transferred onto a PVDF membrane that was subsequently incubated with specific primary antibody and then with secondary antibody conjugated with horseradish peroxidase (Sigma-Aldrich). The following primary antibodies were used in the study: anti-RubA (Shen et al., 2002b), anti-D1 (Komenda, 2005), anti-FLAG (Abgent, catalog number AP1013A; dilution 5000×), anti-PsaF (raised in rabbit against peptide 50-61 of the Synechocystis PsaF), anti-PsaD (Li et al., 1991), anti-Ycf48 (Yu et al., 2018), anti-PsaC (Agrisera, catalog number AS04 042P; dilution 5000×), anti-PsbO (Chapman et al., 1989), and anti-Psb27 (Komenda et al., 2012a). For protein labeling, cells containing 75 µg of total Chl were incubated for 30 min at an irradiance of 500 μmol photons m−2 s−1 in the presence of a mixture of [35S]Met and [35S]Cys (MP Biomedicals) as described previously (Dobáková et al., 2009). Membranes were isolated from the labeled cells and subjected to 2D protein separation described above. The 2D gel was stained by CBB, photographed, dried, exposed to a phosphorimager plate (GE Healthcare) overnight, and scanned with a Storm 860 (GE Healthcare).
Enzymatic Digestion and Protein Identification by MS
For identification of proteins by liquid chromatography-tandem mass spectrometry (MS/MS), the CBB-stained bands were cut from the gel, digested, and analyzed as described (Bučinská et al., 2018). For the matrix assisted laser desorption/ionization-time of flight (MALDI-TOF) MS analysis, proteins were digested for 8 h at 37°C in a cleavage buffer containing 25 mM 4-ethylmorpholine acetate, 5% (v/v) acetonitrile, and protease (100 ng of trypsin [Promega] or 20 ng of Lys-C or 10 ng of Asp-N [Roche]). Aliquots of Lys-C and Asp-N digests were further incubated for 4 h with Asp-N (10 ng) and trypsin (100 ng), respectively. Subsequently, 0.5 μL of each digest was deposited on the MALDI plate, air-dried at room temperature, and overlaid with 0.5 μL of the matrix solution (α-cyano-4-hydroxycinnamic acid in 50% (v/v) acetonitrile/0.1% (v/v) trifluoroacetic acid, 5 mg/mL [Sigma-Aldrich]). Peptide mass maps and MS/MS spectra were measured on an Ultraflex III MALDI-TOF instrument (Bruker Daltonics). For protein identification, MS/MS spectra were searched against the SwissProt 2019_02 database subset of Synechocystis proteins using an in-house MASCOT search engine.
Determination of Pigment Contents
The Chl content per cell was determined for five independent cultures after methanol extraction of pigments according to Ritchie (2006).
The detection of tetrapyrrole biosynthetic precursors was performed by an Agilent 1200 HPLC instrument (Agilent Technologies). The fluorescence detector FLD1 was set to excitation/emission maxima of 440/660 nm to detect the Chl precursors monovinyl-Chlide, divinyl-Chlide, monovinyl-PChlide, and divinyl-PChlide, while CoPP and PPIX were detected at 400/620 and 400/630 nm, respectively. Mg-protophorphyrin IX and MgPME were detected on the fluorescence detector FLD2 set to excitation/emission maxima of 416/595 nm (Pilný et al., 2015).
For determination of the heme content, the samples were extracted in acetone and injected into the HPLC instrument as described (Horáková et al., 2017). For the HPLC measurements of tetrapyrroles, samples from three independent cultures were taken.
Assessment of Cellular ATP Content
Three independent, exponentially growing cultures containing 2 × 109 cells were harvested by centrifugation, resuspended in 500 μL of deionized water, and broken mechanically for 20 s in Mini-Beadbeater (BioSpec) using 300 μL of balotina beads. The beads were removed by a quick spin at 3000g, and the phycobilisomes were precipitated with 2% (v/v) trichloroacetic acid. The precipitant was removed by centrifugation at maximum speed for 5 min, and the pH of the supernatant was adjusted to neutral with 90 mM Na3PO4 (pH 11.5). A 50-μL sample was mixed in wells with an equal amount of reaction mixture of a Cell Viability Kit (Promega). The ATP content of the cell lysate was reflected by a luminescence signal that was recorded by an Infinite F200 plate reader (Tecan) after a 1000-ms integration time.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: RubA (Slr2033), WP_010871719.1; Ycf48 (Slr2034), WP_010871720.1; D1 (Slr1311), WP_010871214.1; D2 (Sll0849), WP_010872429.1; CP43 (Sll0851), WP_014407100.1; CP47 (Slr0906), WP_010873685.1; Ycf39 (Slr0399), WP_010873152.1; Psb27 (Slr1645), WP_010873082.1; Psb28 (Sll1398), WP_010871246.1; HliC (Ss1633), WP_010872233.1; HliD (Ssr1789), WP_010871578.1; and CyanoP (Sll1418), WP_010872644.1.
Supplemental Data
Supplemental Figure 1. Comparison of whole-cell absorption spectra of WT and the strain expressing FLAG-RubA from the psbA2 promotor (∆rubA/psbA2pro:f.rubA).
Supplemental Figure 2. Immunodetection of RubA and Ycf48 in eluates prepared from solubilized membranes of the ΔCP47 strain containing (A) or lacking (B) YFP-tagged PSI complexes.
Supplemental Figure 3. Control co-immunopurification assay performed using WT membranes to exclude the unspecific binding of RubA and PSII subunits by resin.
Supplemental Figure 4. Effect of DCMU on the PSI and PSII accumulation in the autotrophic WT cells.
Supplemental Figure 5. The spectrum of the white light used for cultivation of strains in the study.
Supplemental Table 1. Synechocystis strains used in the study.
Supplemental Table 2. Growth and Chl content of WT and the strains constructed in this study.
Supplemental Table 3. Proteins identified in the affinity-purified FLAG-RubA preparation using MALDI-TOF MS.
Supplemental Table 4. Primers used in this study.
Acknowledgments
We thank John Golbeck for providing us the RubA-specific antibody. This work was supported by the Ministry of Education, Youth, and Sports of the Czech Republic (LO1416), by the Grant Agency of the Czech Republic (19-29225X), and by the Biotechnology and Biological Sciences Research Council (BB/L003260/1 and BB/P00931X/1).
AUTHOR CONTRIBUTIONS
É.K., J.Ko., P.J.N., and R.S. designed the study. R.S. and J.Y. constructed the strains used in this study. É.K., J.Kn., G.P., J.P., J.Y., P.H., J.Ko., and R.S. performed the research. É.K., J.K., P.J.N., and R.S. wrote the article. The whole study was supervised by J.Ko. and R.S. All authors discussed the results and commented on the article.
References
- Adams N.B.P., Brindley A.A., Hunter C.N., Reid J.D. (2016). The catalytic power of magnesium chelatase: A benchmark for the AAA(+) ATPases. FEBS Lett. 590: 1687–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S.L., McIntosh L. (1991). Light-activated heterotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803: A blue-light-requiring process. J. Bacteriol. 173: 2761–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bečková M., Gardian Z., Yu J., Koník P., Nixon P.J., Komenda J. (2017). Association of Psb28 and Psb27 proteins with PSII-PSI supercomplexes upon exposure of Synechocystis sp. PCC 6803 to high light. Mol. Plant 10: 62–72. [DOI] [PubMed] [Google Scholar]
- Boehm M., Romero E., Reisinger V., Yu J., Komenda J., Eichacker L.A., Dekker J.P., Nixon P.J. (2011). Investigating the early stages of photosystem II assembly in Synechocystis sp. PCC 6803: Isolation of CP47 and CP43 complexes. J. Biol. Chem. 286: 14812–14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M., Yu J., Reisinger V., Bečkova M., Eichacker L.A., Schlodder E., Komenda J., Nixon P.J. (2012). Subunit composition of CP43-less photosystem II complexes of Synechocystis sp. PCC 6803: Implications for the assembly and repair of photosystem II. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 3444–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bučinská L., Kiss É., Koník P., Knoppová J., Komenda J., Sobotka R. (2018). The ribosome-bound protein Pam68 promotes insertion of chlorophyll into the CP47 subunit of photosystem II. Plant Physiol. 176: 2931–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon R.H., García-Cerdán J.G., Malnoë A., Cook R., Russell J.J., Gaw C., Dent R.M., de Vitry C., Niyogi K.K. (2013). A conserved rubredoxin is necessary for photosystem II accumulation in diverse oxygenic photoautotrophs. J. Biol. Chem. 288: 26688–26696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman D.J., De Felice J., Davis K., Barber J. (1989). Effect of alkaline pH on photosynthetic water oxidation and the association of extrinsic proteins with Photosystem Two. Biochem. J. 258: 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecki O., Grimm B. (2012). Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J. Exp. Bot. 63: 1675–1687. [DOI] [PubMed] [Google Scholar]
- Deák Z., Sass L., Kiss E., Vass I. (2014). Characterization of wave phenomena in the relaxation of flash-induced chlorophyll fluorescence yield in cyanobacteria. Biochim. Biophys. Acta 1837: 1522–1532. [DOI] [PubMed] [Google Scholar]
- Diner B.A., Rappaport F. (2002). Structure, dynamics, and energetics of the primary photochemistry of photosystem II of oxygenic photosynthesis. Annu. Rev. Plant Biol. 53: 551–580. [DOI] [PubMed] [Google Scholar]
- Dobáková M., Sobotka R., Tichý M., Komenda J. (2009). Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 149: 1076–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Ge H., Huang X., Liu Y., Lu M., Wang J., Chen W., Xu W., Wang Y. (2017). Trophic mode-dependent proteomic analysis reveals functional significance of light-independent chlorophyll synthesis in Synechocystis sp. PCC 6803. Mol. Plant 10: 73–85. [DOI] [PubMed] [Google Scholar]
- Flügge U.-I., Westhoff P., Leister D. (2016). Recent advances in understanding photosynthesis. F1000 Res. 5: 2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G., Giacomelli L., Ytterberg A.J., Peltier J.B., Rudella A., Sun Q., Wijk K.J. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: New proteins, new functions, and a plastid proteome database. Plant Cell 16: 478–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golbeck J.H., Shen G. (2006). Photosystem I: The light-driven plastocyanin:ferredoxin oxidoreductase. Adv. Photosynth. Respir. 24: 529–547. [Google Scholar]
- Gomes C.M., Giuffrè A., Forte E., Vicente J.B., Saraiva L.M., Brunori M., Teixeira M. (2002). A novel type of nitric-oxide reductase: Escherichia coli flavorubredoxin. J. Biol. Chem. 277: 25273–25276. [DOI] [PubMed] [Google Scholar]
- Goto T., Aoki R., Minamizaki K., Fujita Y. (2010). Functional differentiation of two analogous coproporphyrinogen III oxidases for heme and chlorophyll biosynthesis pathways in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 51: 650–663. [DOI] [PubMed] [Google Scholar]
- Guo J., Nguyen A.Y., Dai Z., Su D., Gaffrey M.J., Moore R.J., Jacobs J.M., Monroe M.E., Smith R.D., Koppenaal D.W., Pakrasi H.B., Qian W.J. (2014). Proteome-wide light/dark modulation of thiol oxidation in cyanobacteria revealed by quantitative site-specific redox proteomics. Mol. Cell. Proteomics 13: 3270–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasunuma T., Matsuda M., Kato Y., Vavricka C.J., Kondo A. (2018). Temperature enhanced succinate production concurrent with increased central metabolism turnover in the cyanobacterium Synechocystis sp. PCC 6803. Metab. Eng. 48: 109–120. [DOI] [PubMed] [Google Scholar]
- Hedtke B., Alawady A., Chen S., Börnke F., Grimm B. (2007). HEMA RNAi silencing reveals a control mechanism of ALA biosynthesis on Mg chelatase and Fe chelatase. Plant Mol. Biol. 64: 733–742. [DOI] [PubMed] [Google Scholar]
- Hollingshead S., Kopecná J., Jackson P.J., Canniffe D.P., Davison P.A., Dickman M.J., Sobotka R., Hunter C.N. (2012). Conserved chloroplast open-reading frame ycf54 is required for activity of the magnesium protoporphyrin monomethylester oxidative cyclase in Synechocystis PCC 6803. J. Biol. Chem. 287: 27823–27833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horáková E., Changmai P., Vancová M., Sobotka R., Van Den Abbeele J., Vanhollebeke B., Lukeš J. (2017). The Trypanosoma brucei TbHrg protein is a heme transporter involved in the regulation of stage-specific morphological transitions. J. Biol. Chem. 292: 6998–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara S., Takabayashi A., Ido K., Endo T., Ifuku K., Sato F. (2007). Distinct functions for the two PsbP-like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis. Plant Physiol. 145: 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R.B., Silaghi-Dumitrescu R., Kurtz D.M. Jr., Lanzilotta W.N. (2005). High-resolution crystal structures of Desulfovibrio vulgaris (Hildenborough) nigerythrin: Facile, redox-dependent iron movement, domain interface variability, and peroxidase activity in the rubrerythrins. J. Biol. Inorg. Chem. 10: 407–416. [DOI] [PubMed] [Google Scholar]
- Jansson S., Andersson J., Kim S.J., Jackowski G. (2000). An Arabidopsis thaliana protein homologous to cyanobacterial high-light-inducible proteins. Plant Mol. Biol. 42: 345–351. [DOI] [PubMed] [Google Scholar]
- Ke S.H., Madison E.L. (1997). Rapid and efficient site-directed mutagenesis by single-tube ‘megaprimer’ PCR method. Nucleic Acids Res. 25: 3371–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klughammer C. (2008). Saturation pulse method for assessment of energy conversion in PS I. PAM Appl. Notes 1: 11–14. [Google Scholar]
- Knoppová J., Sobotka R., Tichý M., Yu J., Koník P., Halada P., Nixon P.J., Komenda J. (2014). Discovery of a chlorophyll binding protein complex involved in the early steps of photosystem II assembly in Synechocystis. Plant Cell 26: 1200–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoppová J., Yu J., Koník P., Nixon P.J., Komenda J. (2016). CyanoP is involved in the early steps of photosystem II assembly in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 57: 1921–1931. [DOI] [PubMed] [Google Scholar]
- Komenda J. (2000). Role of two forms of the D1 protein in the recovery from photoinhibition of photosystem II in the cyanobacterium Synechococcus PCC 7942. Biochim. Biophys. Acta 1457: 243–252. [DOI] [PubMed] [Google Scholar]
- Komenda J. (2005). Autotrophic cells of the Synechocystis psbH deletion mutant are deficient in synthesis of CP47 and accumulate inactive PS II core complexes. Photosynth. Res. 85: 161–167. [DOI] [PubMed] [Google Scholar]
- Komenda J., Barber J. (1995). Comparison of psbO and psbH deletion mutants of Synechocystis PCC 6803 indicates that degradation of D1 protein is regulated by the QB site and dependent on protein synthesis. Biochemistry 34: 9625–9631. [DOI] [PubMed] [Google Scholar]
- Komenda J., Reisinger V., Müller B.C., Dobáková M., Granvogl B., Eichacker L.A. (2004). Accumulation of the D2 protein is a key regulatory step for assembly of the photosystem II reaction center complex in Synechocystis PCC 6803. J. Biol. Chem. 279: 48620–48629. [DOI] [PubMed] [Google Scholar]
- Komenda J., Barker M., Kuviková S., de Vries R., Mullineaux C.W., Tichý M., Nixon P.J. (2006). The FtsH protease slr0228 is important for quality control of photosystem II in the thylakoid membrane of Synechocystis sp. PCC 6803. J. Biol. Chem. 281: 1145–1151. [DOI] [PubMed] [Google Scholar]
- Komenda J., Nickelsen J., Tichý M., Prásil O., Eichacker L.A., Nixon P.J. (2008). The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 283: 22390–22399. [DOI] [PubMed] [Google Scholar]
- Komenda J., Knoppová J., Krynická V., Nixon P.J., Tichý M. (2010). Role of FtsH2 in the repair of photosystem II in mutants of the cyanobacterium Synechocystis PCC 6803 with impaired assembly or stability of the CaMn4 cluster. Biochim. Biophys. Acta 1797: 566–575. [DOI] [PubMed] [Google Scholar]
- Komenda J., Knoppová J., Kopečná J., Sobotka R., Halada P., Yu J., Nickelsen J., Boehm M., Nixon P.J. (2012a). The Psb27 assembly factor binds to the CP43 complex of photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 158: 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J., Sobotka R., Nixon P.J. (2012b). Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr. Opin. Plant Biol. 15: 245–251. [DOI] [PubMed] [Google Scholar]
- Kopečná J., Komenda J., Bucinská L., Sobotka R. (2012). Long-term acclimation of the cyanobacterium Synechocystis sp. PCC 6803 to high light is accompanied by an enhanced production of chlorophyll that is preferentially channeled to trimeric photosystem I. Plant Physiol. 160: 2239–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopečná J., Sobotka R., Komenda J. (2013). Inhibition of chlorophyll biosynthesis at the protochlorophyllide reduction step results in the parallel depletion of photosystem I and photosystem II in the cyanobacterium Synechocystis PCC 6803. Planta 237: 497–508. [DOI] [PubMed] [Google Scholar]
- Kopečná J., Cabeza de Vaca I., Adams N.B., Davison P.A., Brindley A.A., Hunter C.N., Guallar V., Sobotka R. (2015a). Porphyrin binding to Gun4 protein, facilitated by a flexible loop, controls metabolite flow through the chlorophyll biosynthetic pathway. J. Biol. Chem. 290: 28477–28488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopečná J., Pilný J., Krynická V., Tomčala A., Kis M., Gombos Z., Komenda J., Sobotka R. (2015b). Lack of phosphatidylglycerol inhibits chlorophyll biosynthesis at multiple sites and limits chlorophyllide reutilization in Synechocystis sp. strain PCC 6803. Plant Physiol. 169: 1307–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M., Klähn S., Scholz I., Matthiessen J.K., Hess W.R., Voß B. (2014). Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res. 21: 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Zhao J.D., Warren P.V., Warden J.T., Bryant D.A., Golbeck J.H. (1991). PsaD is required for the stable binding of PsaC to the photosystem I core protein of Synechococcus sp. PCC 6301. Biochemistry 30: 7863–7872. [DOI] [PubMed] [Google Scholar]
- Li Y., Liu B., Zhang J., Kong F., Zhang L., Meng H., Li W., Rochaix J.-D., Li D., Peng L. (2019). OHP1, OHP2, and HCF244 form a transient functional complex with the photosystem II reaction center. Plant Physiol. 179: 195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz J.G., Pakrasi H.B., Seibert M., Arntzen C.J. (1986). Evidence for a dual function of the herbicide-binding D1 protein in photosystem II. FEBS Lett. 205: 269–274. [Google Scholar]
- Meurer J., Plücken H., Kowallik K.V., Westhoff P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17: 5286–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilný J., Kopečná J., Noda J., Sobotka R. (2015). Detection and quantification of heme and chlorophyll precursors using a high performance liquid chromatography (HPLC) system equipped with two fluorescence detectors. Bio Protoc. 5: e1390. [Google Scholar]
- Prakash D., Walters K.A., Martinie R.J., McCarver A.C., Kumar A.K., Lessner D.J., Krebs C., Golbeck J.H., Ferry J.G. (2018). Toward a mechanistic and physiological understanding of a ferredoxin:disulfide reductase from the domains Archaea and Bacteria. J. Biol. Chem. 293: 9198–9209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince C., Jia Z. (2015). An unexpected duo: Rubredoxin binds nine TPR motifs to form LapB, an essential regulator of lipopolysaccharide synthesis. Structure 23: 1500–1506. [DOI] [PubMed] [Google Scholar]
- Ritchie R.J. (2006). Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol and ethanol solvents. Photosynth. Res. 89: 27–41. [DOI] [PubMed] [Google Scholar]
- Rühle T., Leister D. (2016). Photosystem II assembly from scratch. Front. Plant Sci. 6: 1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuragi Y., Zybailov B., Shen G., Bryant D.A., Golbeck J.H., Diner B.A., Karygina I., Pushkar Y., Stehlik D. (2005). Recruitment of a foreign quinone into the A1 site of photosystem I: Characterization of a menB rubA double deletion mutant in Synechococcus sp. PCC 7002 devoid of FX, FA, and FB and containing plastoquinone or exchanged 9,10-anthraquinone. J. Biol. Chem. 280: 12371–12381. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. (1991). Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199: 223–231. [DOI] [PubMed] [Google Scholar]
- Scharfenberg M., Mittermayr L., von Roepenack-Lahaye E., Schlicke H., Grimm B., Leister D., Kleine T. (2015). Functional characterization of the two ferrochelatases in Arabidopsis thaliana. Plant Cell Environ. 38: 280–298. [DOI] [PubMed] [Google Scholar]
- Shen G., Vermaas W.F.J. (1994). Chlorophyll in a Synechocystis sp. PCC 6803 mutant without photosystem I and photosystem II core complexes: Evidence for peripheral antenna chlorophylls in cyanobacteria. J. Biol. Chem. 269: 13904–13910. [PubMed] [Google Scholar]
- Shen G., Antonkine M.L., van der Est A., Vassiliev I.R., Brettel K., Bittl R., Zech S.G., Zhao J., Stehlik D., Bryant D.A., Golbeck J.H. (2002a). Assembly of photosystem I. II. Rubredoxin is required for the in vivo assembly of FX in Synechococcus sp. PCC 7002 as shown by optical and EPR spectroscopy. J. Biol. Chem. 277: 20355–20366. [DOI] [PubMed] [Google Scholar]
- Shen G., Zhao J., Reimer S.K., Antonkine M.L., Cai Q., Weiland S.M., Golbeck J.H., Bryant D.A. (2002b). Assembly of photosystem I. I. Inactivation of the rubA gene encoding a membrane-associated rubredoxin in the cyanobacterium Synechococcus sp. PCC 7002 causes a loss of photosystem I activity. J. Biol. Chem. 277: 20343–20354. [DOI] [PubMed] [Google Scholar]
- Shinopoulos K.E., Yu J., Nixon P.J., Brudvig G.W. (2014). Using site-directed mutagenesis to probe the role of the D2 carotenoid in the secondary electron-transfer pathway of photosystem II. Photosynth. Res. 120: 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieker L.C., Stenkamp R.E., LeGall J. (1994). Inorganic microbial sulfur metabolism. Methods Enzymol. 243: 203–216. [DOI] [PubMed] [Google Scholar]
- Sobotka R., Komenda J., Bumba L., Tichý M. (2005). Photosystem II assembly in CP47 mutant of Synechocystis sp. PCC 6803 is dependent on the level of chlorophyll precursors regulated by ferrochelatase. J. Biol. Chem. 280: 31595–31602. [DOI] [PubMed] [Google Scholar]
- Sobotka R., McLean S., Žuberová M., Hunter C.N., Tichý M. (2008). The C-terminal extension of ferrochelatase is critical for enzyme activity and for functioning of the tetrapyrrole pathway in Synechocystis strain PCC 6803. J. Bacteriol. 190: 2086–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staleva H., Komenda J., Shukla M.K., Šlouf V., Kaňa R., Polívka T., Sobotka R. (2015). Mechanism of photoprotection in the cyanobacterial ancestor of plant antenna proteins. Nat. Chem. Biol. 11: 287–291. [DOI] [PubMed] [Google Scholar]
- Steccanella V., Hansson M., Jensen P.E. (2015). Linking chlorophyll biosynthesis to a dynamic plastoquinone pool. Plant Physiol. Biochem. 97: 207–216. [DOI] [PubMed] [Google Scholar]
- Strašková A., Knoppová J., Komenda J. (2018). Isolation of the cyanobacterial YFP-tagged photosystem I using GFP-Trap®. Photosynthetica 56: 300–305. [Google Scholar]
- Tichý M., Bečková M., Kopečná J., Noda J., Sobotka R., Komenda J. (2016). Strain of Synechocystis PCC 6803 with aberrant assembly of photosystem II contains tandem duplication of a large chromosomal region. Front. Plant Sci. 7: 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umena Y., Kawakami K., Shen J.R., Kamiya N. (2011). Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60. [DOI] [PubMed] [Google Scholar]
- Vass I., Kirilovsky D., Etienne A.-L. (1999). UV-B radiation-induced donor- and acceptor-side modifications of photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. Biochemistry 38: 12786–12794. [DOI] [PubMed] [Google Scholar]
- Wastl J., Duin E.C., Iuzzolino L., Dörner W., Link T., Hoffmann S., Sticht H., Dau H., Lingelbach K., Maier U.G. (2000). Eukaryotically encoded and chloroplast-located rubredoxin is associated with photosystem II. J. Biol. Chem. 275: 30058–30063. [DOI] [PubMed] [Google Scholar]
- Willows R.D. (2006). Chlorophyll synthesis. In The Structure and Function of Plastids, Wise R.R. and Hoober J.K., eds (Amsterodam, The Netherlands: Springer; ), pp. 295–313. [Google Scholar]
- Wittig I., Karas M., Schägger H. (2007). High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell. Proteomics 6: 1215–1225. [DOI] [PubMed] [Google Scholar]
- Wittwer M., Luo Q., Kaila V.R., Dames S.A. (2016). Oxidative unfolding of the rubredoxin domain and the natively disordered N-terminal region regulate the catalytic activity of Mycobacterium tuberculosis protein kinase G. J. Biol. Chem. 291: 27062–27072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson J.D., Perez-Ruiz J.M., Chory J. (2011). Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr. Biol. 21: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S., Nomata J., Fujita Y. (2006). Differential operation of dual protochlorophyllide reductases for chlorophyll biosynthesis in response to environmental oxygen levels in the cyanobacterium Leptolyngbya boryana. Plant Physiol. 142: 911–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Knoppová J., Michoux F., Bialek W., Cota E., Shukla M.K., Strašková A., Pascual G.A., Sobotka R., Komenda J., Murray J.W., Nixon P.J. (2018). Ycf48 involved in the biogenesis of the oxygen-evolving photosystem II complex is a seven-bladed beta-propeller protein. Proc. Natl. Acad. Sci. USA 115: E7824–E7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Ye Z., Zhao J. (2007). RbrA, a cyanobacterial rubrerythrin, functions as a FNR-dependent peroxidase in heterocysts in protection of nitrogenase from damage by hydrogen peroxide in Anabaena sp. PCC 7120. Mol. Microbiol. 66: 1219–1230. [DOI] [PubMed] [Google Scholar]