RST1, identified as a FREE1 suppressor, negatively regulates multivesicular body–mediated membrane sorting and degradation in plants.
Abstract
FYVE domain protein required for endosomal sorting1 (FREE1), a plant-specific endosomal sorting complex required for transport-I component, is essential for the biogenesis of multivesicular bodies (MVBs), vacuolar degradation of membrane protein, cargo vacuolar sorting, autophagic degradation, and vacuole biogenesis in Arabidopsis (Arabidopsis thaliana). Here, we report the characterization of RESURRECTION1 (RST1) as a suppressor of free1 that, when mutated as a null mutant, restores the normal MVB and vacuole formation of a FREE1-RNAi knockdown line and consequently allows survival. RST1 encodes an evolutionarily conserved multicellular organism–specific protein, which contains two Domain of Unknown Function 3730 domains, showing no similarity to known proteins, and predominantly localizes in the cytosol. The depletion of FREE1 causes substantial accumulation of RST1, and transgenic Arabidopsis plants overexpressing RST1 display retarded seedling growth with dilated MVBs, and inhibition of endocytosed FM4-64 dye to the tonoplast, suggesting that RST1 has a negative role in vacuolar transport. Consistently, enhanced endocytic degradation of membrane vacuolar cargoes occurs in the rst1 mutant. Further transcriptomic comparison of rst1 with free1 revealed a negative association between gene expression profiles, demonstrating that FREE1 and RST1 have antagonistic functions. Thus, RST1 is a negative regulator controlling membrane protein homeostasis and FREE1-mediated functions in plants.
INTRODUCTION
The endomembrane system is an evolutionarily conserved and essential cellular process for cell growth and responses to external signals that consists of several functionally distinct membrane-bound organelles, including the endoplasmic reticulum, Golgi apparatus, trans-Golgi network (TGN) or early endosome, prevacuolar compartment or multivesicular bodies (MVBs), and vacuole. The endomembrane system plays critical roles in membrane protein targeting, trafficking, and degradation (Surpin and Raikhel, 2004). To maintain proper protein homeostasis, membrane proteins destined for degradation are usually ubiquitinated and sorted into the intraluminal vesicles (ILVs) of prevacuolar compartments/MVBs for subsequent degradation in the vacuole/lysosome upon MVB-vacuole fusion (Cui et al., 2016, 2019). The sorting and packaging of ubiquitinated membrane proteins into MVBs is facilitated by a set of protein complexes named the endosomal sorting complex required for transport (ESCRT) machinery in eukaryotic cells (Gao et al., 2017; Isono and Kalinowska, 2017; Otegui, 2018).
In plants, unique ESCRT components have evolved for both the conserved ESCRT function and certain plant-specific functions (Gao et al., 2017; Li et al., 2019a). The plant unique ESCRT component called FYVE domain protein required for endosomal sorting1 (FREE1) is essential for ILV formation in MVBs (Gao et al., 2014). FREE1 may form different complexes that mediate multiple functions. FREE1 directly interacts with the ESCRT-I component VPS23 and binds to PI3P and ubiquitinated membrane cargoes to regulate MVB biogenesis and MVB-mediated sorting of membrane proteins (Barberon et al., 2014; Gao et al., 2014; Kolb et al., 2015; Belda-Palazon et al., 2016). FREE1 can also form a complex with PI3K regulatory subunit ATG6/VPS30 and autophagic regulator protein SH3P2 that regulates autophagic degradation (Zhuang et al., 2013; Gao et al., 2015). In addition, FREE1 is essential for vacuole biogenesis and seedling survival, while it remains unknown how FREE1 mediates vacuole biogenesis and coordinates multiple functions in plants.
We aimed to explore the multifaceted functions of FREE1 via a forward genetic screen for suppressor of free1 (sof) mutants (Zhao et al., 2015). We demonstrated the feasibility of this strategy and reported a novel plant-specific component of the ESCRT complex, Bro1-domain protein as FREE1 suppressor (BRAF; Shen et al., 2018).
Here, we report the identification of RESURRECTION1 (RST1) as sof negatively regulating the endomembrane trafficking pathway. We isolated four independent FREE1-related sof mutants, sof100, sof220, sof452, and sof453, having null mutations of the same gene RST1 (At3g27670), which encodes a protein with no similarity to any protein of known function. The RST1 protein is a multicellular organism–specific protein that contains two domain of unknown function 3730 (DUF3730) domains, mainly localizes in the cytosol, and accumulates substantially in free1 mutant cells. Ectopic overexpression of RST1 results in retarded seedling growth associated with enlarged MVBs and delayed vacuolar transport. Consistently, enhanced endocytic degradation of membrane vacuolar cargos occurred in the rst1 mutants. Further transcriptomic comparison of rst1 with free1 revealed an antagonistic relationship between FREE1 and RST1. Taken together, we propose that RST1 is a negative regulator controlling membrane protein homeostasis and FREE1-mediated functions in plants.
RESULTS
Isolation of RST1 as a FREE1 Suppressor
To elucidate the molecular regulation of FREE1, we used dexamethasone (DEX)-inducible DEX:FREE1-RNAi transgenic plants for a suppressor screen (Supplemental Figure 1A). Upon induction with DEX, the DEX:FREE1-RNAi transgenic plants showed barely detectable levels of FREE1 protein and displayed an almost identical seedling lethal phenotype and cellular defects as observed in the free1 T-DNA insertional mutant. Seeds from the DEX:FREE1-RNAi transgenic line were subjected to ethyl methanesulfonate (EMS) mutagenesis. To isolate sof mutants, the M2 seeds were screened for a survived growth phenotype following DEX induction. The sof mutants that only showed barely detectable levels of FREE1 protein with DEX induction were further isolated as FREE1-related sof mutants at the M3 generation. The FREE1-related sof mutants were outcrossed with Landsberg erecta wild type, and the F2 generation was used as the mapping population following a previously established mapping-by-sequencing workflow for detailed mutant gene identification (Supplemental Figure 1B; Zhao et al., 2015).
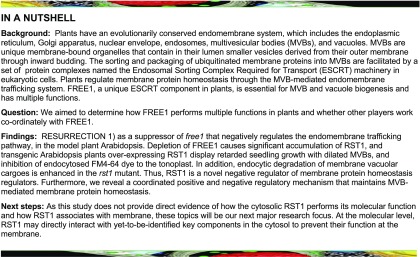
A subset of four FREE1-related sof mutants, sof100, sof220, sof452, and sof453, were further characterized to harbor EMS-induced missense mutations in RST1 (Figure 1A; Supplemental Figure 2). Next-generation sequencing–based mapping identified peaks in the same region on chromosome 3 (Supplemental Figure 2), suggesting that these four mutants (sof100, sof220, sof452, and sof453) may have meaningful mutations in the same gene. The fine mapping and annotation identified four premature stop codon mutations in RST1 (AT3G27670; Figure 1A). In Arabidopsis (Arabidopsis thaliana), RST1 is a single copy gene and genetically linked to stem cuticular wax distribution, embryo development, and defense response, with elusive underlying mechanisms (Chen et al., 2005; Mang et al., 2009). RST1 was also shown to precipitate with RRP4, a noncatalytic component of the RNA exosome complex with an unknown mechanism (Lange et al., 2014).
Figure 1.
Isolation of Four rst1 Alleles as sof Mutants Using FREE1-RNAi Transgenic Plants.
(A) Schematic illustration of RST1 showing T-DNA insertion sites and the sof mutations. *, stop.
(B) Seedling survival phenotype of sof100, sof452, and sof453 mutants. M3 seeds were plated on MS plates, and MS plates were supplied with 10 μM DEX.
(C) Detection of FREE1 protein extracted from sof100, sof452, and sof453 mutants. Immunoblot analysis of total protein with (+) or without (−) DEX using FREE1 antibody.
(D) Recovered ILV formation of MVBs in sof100, sof452, and sof453 mutants. The numbers of ILVs per MVB were quantified from 40 MVBs and are shown in the box and whisker plot for each genotype. On each box, the top, middle, and bottom of the box represent the 25th, 50th, and 75th percentiles, respectively. The bars are minimum and maximum. **P < 0.01; n.s., not significant (P > 0.05 in Student’s t test). Bar = 250 nm.
(E) Recovered large central vacuole in sof100, sof452, and sof453 mutants. The vacuoles were visualized using FM4-64 staining shown in magenta. Note that when grown on DEX medium, the sof mutants (sof100, sof452, and sof453) have a wild-type–like large vacuole, rather than the small fragmented vacuoles observed in FREE1-RNAi. Bar = 10 µm.
Loss of RST1 reverted the free1 lethal phenotype because (1) after 7 d of growth on DEX medium, sof100, sof220, sof452, and sof453 seedlings showed a wild-type–like viable normal growth phenotype, which is distinct from the lethal phenotype of FREE1-RNAi seedlings (Figure 1B and Supplemental Figure 3A); and (2) FREE1 protein was barely detected in these four sof lines with DEX induction (Figure 1C; Supplemental Figures 3B and 3C). These results indicate the involvement of RST1 in the FREE1-related pathway. Because FREE1-RNAi plants bypassed the embryo requirement of FREE1, our ability to identify mutants that suppress freel null mutants is limited.
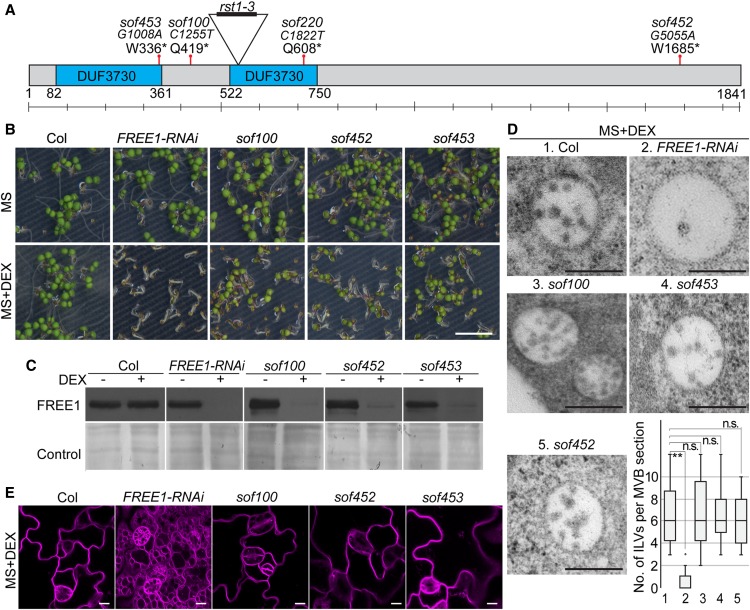
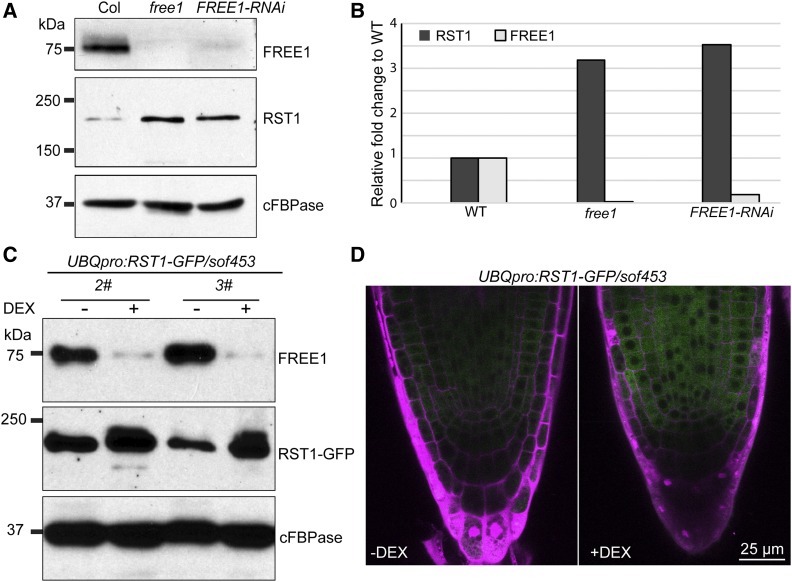
In a transmission electron microscopy (TEM) analysis, the ILVs in these sof mutants resembled those in the wild type. To determine whether the defective MVBs are restored to normal in the sof mutants, we performed a statistical analysis of the ILV number (Figure 1D) and MVB diameter (Supplemental Figure 3D). Statistical analyses showed that the number of ILVs per MVB section in the sof mutants was comparable to that in Columbia (Col), while the FREE1-RNAi plants displayed a significantly reduced number of ILVs per MVB section (Figure 1D). The differences in MVB diameter in Col, FREE1-RNAi, and the sof mutants were not significant (Supplemental Figure 3D). Subsequent analysis of the vacuole using FM4-64 staining of the tonoplast showed restored large central vacuoles in these sof mutants (sof100, sof452, and sof453; Figure 1E). Immunoblot analyses of the four mutants sof100, sof452, sof220, and sof453 detected no full-length RST1 protein, supporting the notion that these four mutants are rst1 null mutants (Figure 2A). RST1 antibody specifically recognized both endogenous RST1 protein and RST1-GFP protein (Figure 2B). Constitutive RST1-green fluorescent protein (GFP) expression in sof453 complemented the survived sof453 back to lethal phenotype, suggesting that RST1-GFP is a functional fusion protein and RST1 is the responsible gene for the rescued growth in these sof mutants (Figures 2C to 2E; Supplemental Figure 4). These results suggest that FREE1 may repress RST1, and RST1 may, in turn, repress the same MVB pathway. In this scenario, it is possible that a FREE1-independent MVB biogenesis pathway could have been activated without RST1.
Figure 2.
Complementation of sof453 with RST1-GFP Proteins.
(A) RST1 protein detected by a RST1-specific anti-peptide antibody. Anti-cFBPase antibodies were used as a loading control.
(B) RST1 antibody specifically recognizes endogenous RST1 and ectopically expressed RST1-GFP.
(C) Complementation of sof453 with UBQpro:RST1-GFP. The phenotype of 7-d-old sof453 seedlings expressing RST1-GFP grown on MS plates and MS plates supplied with (+) DEX.
(D) Seedling growth phenotypes and quantification of Col, FREE1-RNAi, rst1-3, sof453, and complemented lines on MS agar plates with or without DEX for 7 d. Bar graphs show means ± se (n = 11 to 15 plants). **P < 0.01; n.s., not significant (P > 0.05 in Student’s t test).
(E) Detection of RST1 protein and FREE1 protein in indicated lines grown on medium with (+) or without (−) DEX. Anti-cFBPase antibodies were used as loading control. Anti-RST1 antibodies and anti-FREE1 antibodies were used for RST1/RST1-GFP and FREE1 detection, respectively.
RST1 Encodes a Multicellular Organism–Specific Protein and Primarily Localizes in the Cytosol
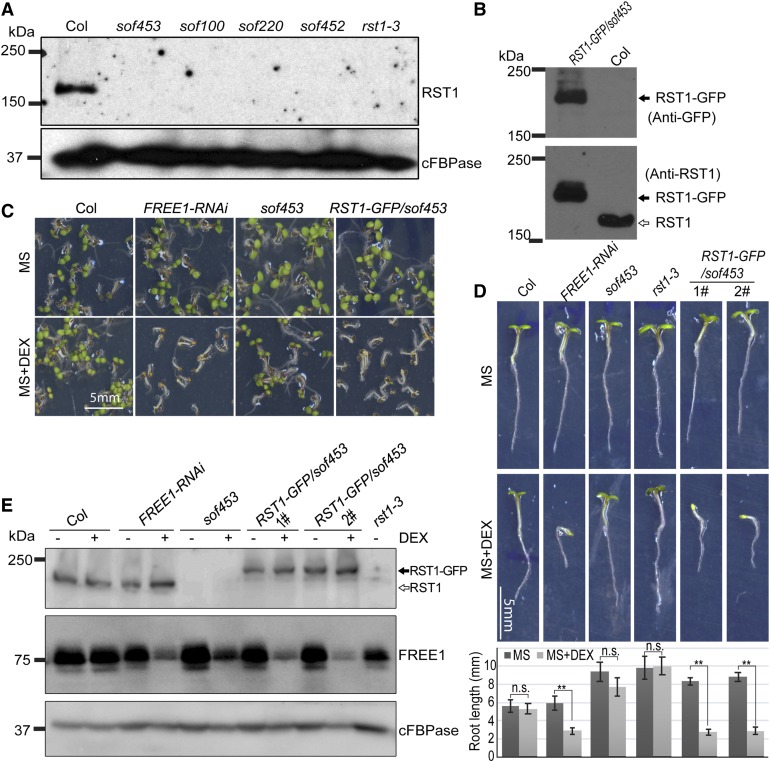
RST1 is a multicellular organism–specific protein; phylogenetic analysis identified RST1 homologs only in multicellular organisms and in Dictyostelium discoideum AX4, a unicellular eukaryote that can develop into a multicellular organism (Figure 3A; Supplemental Data Set 5), suggesting that RST1 may have specific functions in cell–cell communication. Little is known about the function of RST1 homologs in these organisms. The RST1 homolog in human (KIAA1797), which shows 30.2% identity at the amino acid level to Arabidopsis RST1, was identified as a brain cancer suppressor and may function as a component of the focal adhesion complex (Brockschmidt et al., 2012).
Figure 3.
RST1 Encodes a Conserved, Multicellular Organism–Specific Cytosolic Protein.
(A) Homologs of RST1 are found only in multicellular eukaryotic organisms. Molecular phylogenetic analysis of RST1 homologs by the maximum-likelihood method.
(B) Detection of RST1 protein and FREE1 protein in the indicated tissues. (1) Rosette leaf. (2) Stem leaf. (3) Stem. (4) Flower. (5) Young silique. (6) Old silique. (7) Dry seeds. (8) Seedlings. Anti-RST1 antibodies and anti-FREE1 antibodies were used for RST1 and FREE1 detection, respectively. LE, long exposure; SE, short exposure.
(C) Detection of RST1 and FREE1 expression level via qPCR analysis using RNA extracted from the indicated tissues. (1) Rosette leaf. (2) Stem leaf. (3) Stem. (4) Flower. (5) Young silique. (6) Old silique. (8) Seedlings. Error bars represent the sd from three technical replicates.
(D) RST1 is a cytosolic protein. Supernatant (S) and microsomal pellet fractions (P) were prepared from Arabidopsis seedlings. The membrane protein VSR serves as the membrane marker. The universally expressed cytoplasmic marker protein cFBPase was used as a cytosolic control. Anti-RST1 antibodies were used for RST1 detection.
At the transcriptional level, RST1 is constitutively expressed in all organs, with a relatively low basal expression level compared with FREE1 from Genevestigator data (Supplemental Figure 5). At the protein level, higher levels of RST1 protein were detected in stem and old green silique compared with in other tissues (Figure 3B). We also performed an RT-qPCR analysis to assess the expression levels of RST1 and FREE1 in different tissues (Figure 3C). Except for high expression in old green siliques, RST1 displayed a relatively low but constitutive expression pattern compared with FREE1 in all other tissues examined. Combined with our observations of the differed RST1 protein levels, it is possible that RST1 mainly function in the stem and silique, and regulation of RST1 protein level could mainly be at the posttranscriptional level.
In contrast to the membrane marker VSR protein (Tse et al., 2004), RST1 was mainly detected in both soluble fractions (S10 and S100), suggesting that RST1 is mainly a cytosolic protein. The weak RST1 band detected in pellet fraction P100 suggests that a small fraction of RST1 is able to associate with the intracellular membrane (Figure 3D). Consistently, the functional fusion RST1-GFP showed a diffused cytosolic pattern, without an obvious colocalization with the MVB-localized mCherry-FREE1 (Supplemental Figures 6A and 6B). We further detected the endogenous RST1 localization with RST1-specific antibody, and the result showed a consistent cytosolic pattern of RST1 (Supplemental Figure 6C). These results suggest that RST1 is mainly a cytosolic protein.
RST1 Accumulates in free1 and Functions as a FREE1-Specific Suppressor
When grown on DEX medium, the four sof mutants (sof100, sof220, sof452, and sof453) lacking both RST1 and FREE1 survived with normal MVBs and vacuoles, suggesting that FREE1 may repress RST1 that, in turn, represses the MVB function in the same pathway. In this hypothetic regulation mode, a certain FREE1-independent MVB pathway could have been relieved without both FREE1 and RST1. This hypothesis was supported because (1) we detected a significant over-accumulation of RST1 in the free1 mutant (Figures 4A and 4B), suggesting that over-accumulation of RST1 in free1 may block the FREE1-independent MVB pathway. Also, (2) a similar over-accumulation of RST1-GFP in FREE1-RNAi plants was observed in the UBQ:RST1-GFP complemented lines, suggesting that this over-accumulation of RST1-GFP in the free1 mutant is due to a posttranslational regulation (Figures 4C and 4D). However, neither a direct interaction in yeast cells nor an association between RST1 and FREE1 in plant cells was detected (Supplemental Figures 7A and 7B), suggesting that RST1 and FREE1 participate in different protein complexes and that RST1 accumulation is detrimental. Since the absence of yeast two-hybrid (Y2H) complementation is not suggestive of a lack of transient interactions, we cannot exclude the possible existence of transient interaction between RST1 and FREE1 during certain plant growth stages or under certain stress conditions.
Figure 4.
RST1 Accumulates in the free1 Mutant.
(A) Immunoblotting shows that RST1 accumulates in free1 mutant cells. Total proteins were extracted from wild type, free1 (T-DNA) mutants, and DEX-treated FREE1-RNAi plants. Anti-RST1 antibodies and anti-FREE1 antibodies were used for RST1 and FREE1 detection. Anti-cFBPase antibodies were used as the loading control.
(B) Quantification of the intensity of (A). WT, wild type.
(C) RST1-GFP accumulates in the free1 mutant in the complemented transgenic lines with constitutive expression of RST1. Anti-RST1 antibodies and anti-FREE1 antibodies were used for RST1/RST1-GFP and FREE1 detection, respectively. Anti-cFBPase antibodies were used as the loading control.
(D) Confocal image showing increased RST1-GFP signal in the free1 mutant in the complemented transgenic lines constitutively expressing RST1-GFP. PI staining (magenta) shows the cell morphology.
The FREE1-dependent MVB pathway relies on conserved ESCRT complexes (Gao et al., 2014). Among the conserved ESCRT complex components, the ESCRT-III component sucrose nonfermenting7 (SNF7) and the ESCRT-III–associated protein AAA ATPase Vps4/suppressor of K1 transport growth defect 1 (Vps4/SKD1) are employed using their dominant negative forms to block MVB biogenesis, and transgenic plants expressing either dominant negative mutants SNF7.1 (L22W) or SKD1 (E232Q) are seedling lethal with defective MVB biogenesis (Cai et al., 2014). To establish whether RST1 function is specific to free1 or applied similarly to other ESCRT mutants, we introduced DEX-inducible dominant negative mutations in either SNF7.1 (L22W) or SKD1 (E232Q) into rst1. The seedling lethal phenotype was observed for SNF7.1 (L22W) and SKD1 (E232Q) in rst1 (Supplemental Figure 8), suggesting that RST1 may function as a FREE1-specific suppressor and the undefined FREE1-independent MVB pathway may also require the conserved downstream ESCRT complexes.
The braf mutant had an increased FREE1 membrane association (Shen et al., 2018). We next investigated the subcellular localization of GFP-FREE1 and compared the membrane distribution of FREE1 and the ESCRT-I component VPS28 in the wild type versus rst1. GFP-FREE1 showed a similar distribution pattern in the wild type and rst1, whereas GFP-FREE1 in both the the wild type and rst1 formed enlarged structures when treated with wortmannin (Supplemental Figure 9A). In addition, the GFP-FREE1 level was slightly reduced in rst1 (Supplemental Figure 9B). Further ultrastructure analysis showed no alteration of MVB and Golgi structures in rst1 (Supplemental Figure 9C). FREE1 and VPS28 did not show increased membrane association in rst1 (Supplemental Figures 9D and 9F). These results suggest that, in contrast to BRAF (Shen et al., 2018), RST1 did not function through regulation of FREE1/ESCRT-I membrane distribution.
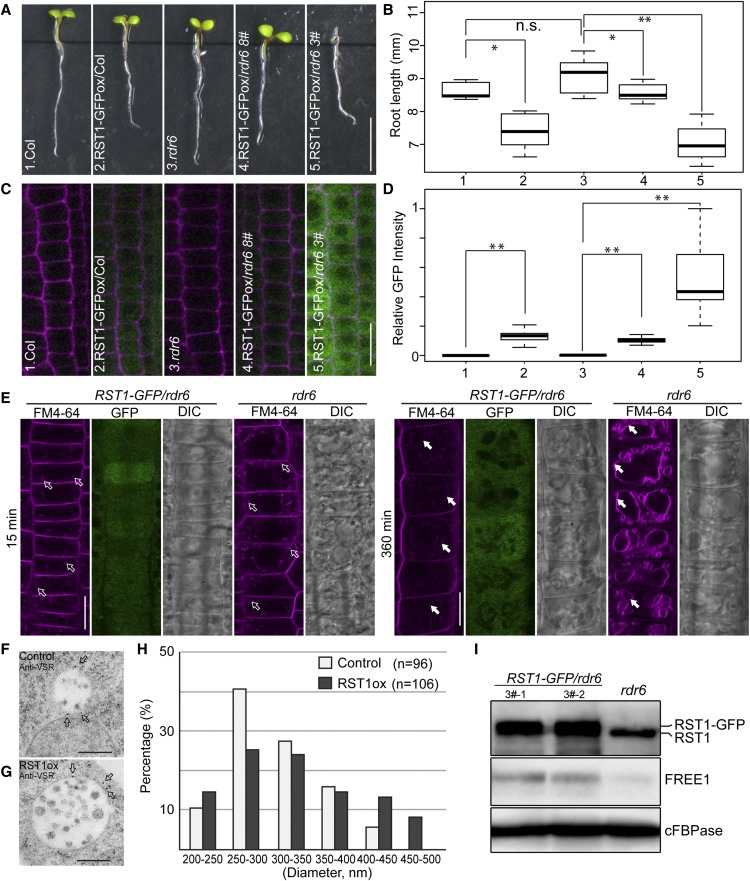
Increased RST1 Protein Levels Result in Retarded Growth, Enlarged MVBs, and Delayed Vacuolar Transport
Then RST1 may function as an inhibitor of the FREE1-independent MVB pathway, and we expect to see a free1-like phenotype in plants overexpressing RST1. We failed to isolate multiple transgenic lines highly expressing RST1-GFP in Col, suggesting that the level of RST1 may be under tight posttranscriptional silencing regulation. To avoid transgene silencing, we used rdr6-11 mutants with impaired gene silencing (Peragine et al., 2004; Luo and Chen, 2007). We then obtained several independent lines expressing RST1-GFP at different protein levels and observed a retarded seedling growth phenotype associated with increased RST1 protein level (Figures 5A to 5D; Supplemental Figure 10), suggesting that RST1 protein level is negatively associated with seedling growth. Therefore, the growth inhibition observed in the RST1 overexpression lines appears to be a relatively direct effect of increased RST1 protein level.
Figure 5.
RST1 Overexpression Arrests Seedling Growth, Inhibits FM4-64 Staining of the Tonoplast, and Results in Enlargement of the MVB.
(A) Root length analysis of 5-d-old transgenic Arabidopsis plants seedlings expressing RST1-GFP. Bar = 5 mm.
(B) Box and whisker plot show the root length in the indicated genotypes in (A) (n = 11 to 15 plants). On each box, the top, middle, and bottom of the box represent the 25th, 50th, and 75th percentiles, respectively. The bars are minimum and maximum. **P < 0.01; n.s., not significant (P > 0.05 in Student’s t test).
(C) Confocal images of RST1-GFP expression in independent lines. Bar = 20 µm.
(D) Quantitative analysis of GFP intensity of (C) in box and whisker plot (n = 5 cells per root from five seedlings). On each box, the top, middle, and bottom of the box represent the 25th, 50th, and 75th percentiles, respectively. The bars are minimum and maximum. **P < 0.01; n.s., not significant (P > 0.05 in Student’s t test).
(E) Inhibition of FM4-64 internalization through the tonoplast by RST1 overexpression. Representative images showing FM4-64 uptake at 15 and 360 min after staining. The unfilled white arrows (at 15 min) indicate the endosomes, while the filled arrows (at 360 min) indicate the vacuole membrane. DIC, differential interference contrast. Bar = 20 µm.
(F) to (H) Enlargement of MVB by RST1-GFP expression and increased proportion of enlarged MVBs in RST1ox lines in (G) compared with the control line (F). Diameters of MVBs were measured from the control (n = 96) and RST1-GFP expression line (n = 106) in (H).
(I) An increased RST1 protein level does not reduce the FREE1 protein level. Anti-RST1 antibodies and anti-FREE1 antibodies were used for RST1/RST1-GFP and FREE1 detection, respectively. Anti-cFBPase antibodies were used as loading control. Two independent lines were detected.
To further define whether increased RST1 protein level would disrupt the endomembrane trafficking system, we first performed time-lapse FM4-64 staining to track the endocytic pathway (Aniento and Robinson, 2005). FM4-64 is a lipophilic styryl membrane dye widely used to study endocytosis from the plasma membrane (PM) through the TGN to MVBs and finally reaching the tonoplast (Bolte et al., 2004; Gao et al., 2014). The time-course observation results showed that increased RST1 protein had no detectable effect on the early endocytic events (Figure 5E; Supplemental Figures 11A and 11B) at 15 and 30 min after staining, because FM4-64–labeled endosomes appeared similar in the RST1 overexpression lines and in the wild type. However, a significant decrease of tonoplast labeling was observed in the RST1 overexpression line compared with the rdr6 controls at 360 min after staining, supporting the conclusion that RST1 overexpression inhibits FM4-64 internalization to the tonoplast (Figure 5E; Supplemental Figures 11C, 11D, and 12).
Endocytic trafficking to vacuole has been well assayed using brefeldin A (BFA) treatments, as BFA treatments caused formation of the so-called “BFA bodies,” which indicated the redistribution of the early endosome/TGN into aggregates due to a block of recycling between the PM and early endosome/TGN (Geldner et al., 2003). The endocytic dye FM4-64 has been observed to localize to BFA bodies (Robinson et al., 2008; Chung et al., 2018). We did not detect a significant difference in BFA body accumulation of FM4-64 between the control and the RST1 overexpression lines (Supplemental Figure 11E), suggesting that RST1 does not play a role in these recycling endocytic events.
Next, we examined whether an increased RST1 protein level would lead to alterations in morphology or size of the MVBs. We repeatedly observed in RST1-GFP expression lines the morphological alterations of MVB in terms of size, which showed an increased proportion of large MVBs (Figures 5F to 5H). By contrast, we did not observe defects of ILV formation in MVBs. Enlarged MVBs have been observed in plants in which PI3K activity was inhibited by wortmannin treatment (Wang et al., 2009), a constitutively active form of RAB5 was overexpressed (Jia et al., 2013), or RAB7 activation was blocked through the mon1 mutation (Cui et al., 2014). We detected no reduction of FREE1 protein level in RST1 overexpression lines relative to the untransformed control (Figure 5I), suggesting that RST1 affects MVBs by bypassing FREE1. The increased MVB size in the RST1 overexpression lines suggested that RST1 participates in vacuolar trafficking as a molecular break that regulates either PI3P production, RAB5/RAB7 cycling between the GTP-bound active form and GDP-bound inactive form, or both.
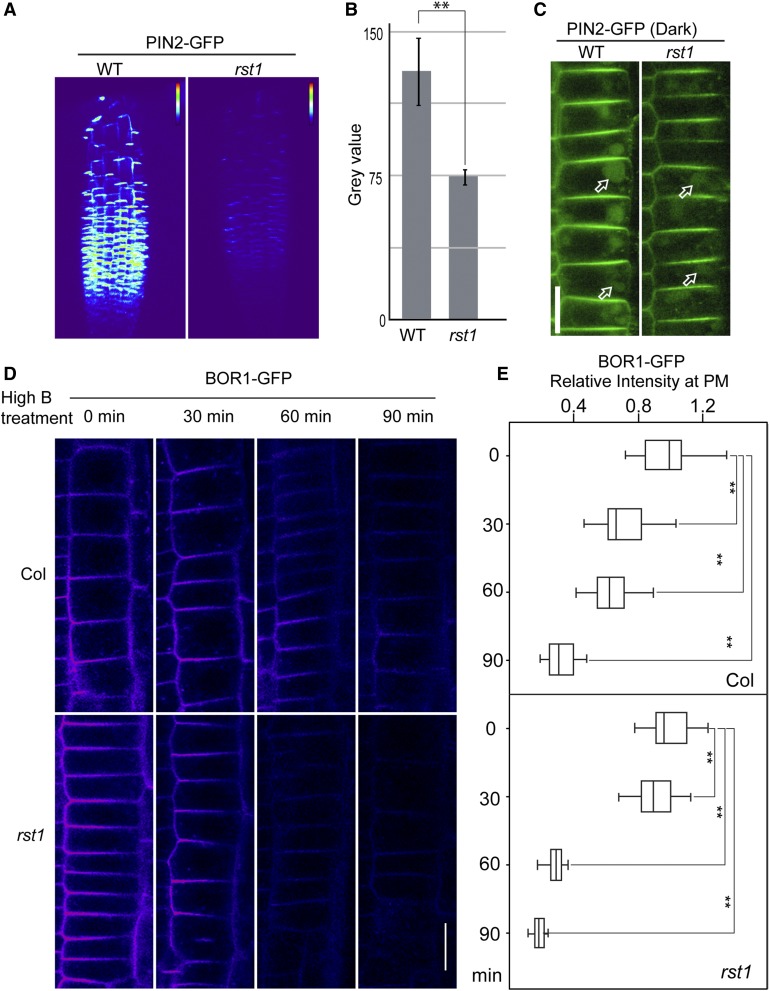
Enhanced Endocytic Degradation of Membrane Vacuolar Cargo in the rst1 Mutant
Our findings so far suggest that RST1 inhibits protein vacuolar trafficking. Thus, enhanced vacuolar trafficking would be expected in rst1. To test this possibility, we introduced two endocytic membrane cargo marker proteins PIN2-GFP and BOR1-GFP into the rst1 mutant (Takano et al., 2005; Leitner et al., 2012). A significant reduction in the basal level of PIN2-GFP was observed in rst1 (Figures 6A and 6B), suggesting that an enhanced endocytic degradation of PIN2-GFP may occur in rst1. Because GFP and GFP-related proteins are more stable in the lytic vacuoles under dark conditions, we next transferred the seedlings to dark conditions, and 6 h later observed GFP accumulation in the lytic vacuoles in both rst1 and the wild type, suggesting that vacuolar transport of PIN2-GFP was not affected in rst1 (Figure 6C).
Figure 6.
Enhanced Endocytic Degradation of Vacuolar Cargo in the rst1 Mutant.
(A) Reduced PIN2-GFP levels at the plasma membrane in the rst1 mutant. Three-dimensional animation of z-stacks was obtained. False color code was used for PIN2-GFP intensity visualization. WT, wild type.
(B) PIN2-GFP intensity quantification. The bar graph shows means ± se (n = 3). **P < 0.01; n.s., not significant (P > 0.05 in Student’s t test). WT, wild type.
(C) PIN2 accumulation in vacuole in the rst1 was detected. Bar = 20 µm. WT, wild type.
(D) High boron condition–induced endocytic BOR1-GFP degradation was enhanced in rst1. Representative epidermal cells showing BOR1-GFP in the wild type and rst1 at the indicated time points after transferring to high boron condition. Bar = 20 µm.
(E) Quantification of PM BOR1-GFP signal intensity compared with that at time 0 min in box and whisker plot (n = 5 cells per root from five seedlings.) On each box, the top, middle, and bottom of the box represent the 25th, 50th, and 75th percentiles, respectively. The bars are minimum and maximum. **P < 0.01; n.s., not significant (P > 0.05 in Student’s t test).
To further establish whether enhanced vacuolar trafficking occurs in the rst1 mutant for endocytic membrane cargo, we also compared the boron-induced endocytic degradation of BOR1-GFP in rst1 and the wild type (Takano et al., 2005). GFP signals of BOR1-GFP decreased faster in rst1 than in the wild type, especially in the epidermal cell layer by 60 min and 90 min after high boron treatment (Figures 6D and 6E; Supplemental Figure 13), supporting the conclusion that boron-induced endocytic degradation of BOR1-GFP is elevated in rst1.
The soluble vacuolar cargo marker protein spL-red fluorescent protein (RFP), which consists of an RFP fusion with the signal peptide and the sequence-specific vacuolar sorting signal of proricin for targeting to lytic vacuoles and has been widely used to analyze the vacuolar sorting of soluble vacuolar cargoes (Hunter et al., 2007; Gao et al., 2015), showed a similar vacuolar pattern in rst1 and the wild type (Supplemental Figure 14A), suggesting that vacuolar transport of soluble cargo was not affected in rst1. We further performed FM4-64 staining to compare the early endocytic process in rst1 and the wild type and found no significant difference, suggesting that RST1 is not involved in the early endocytic process (Supplemental Figure 14B)
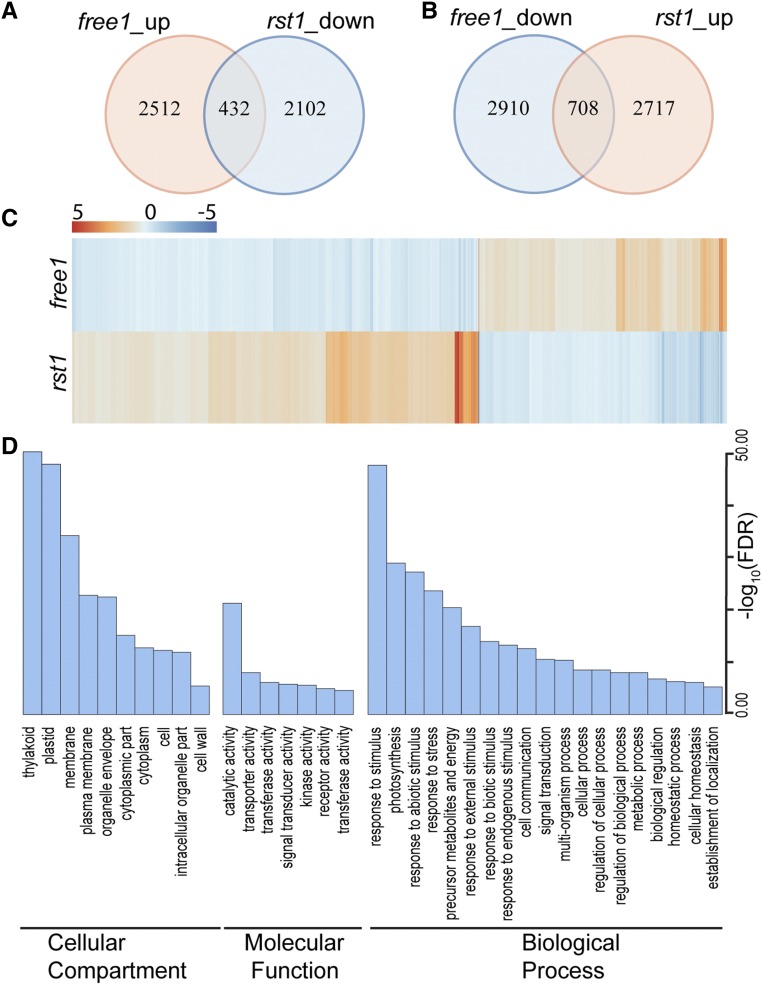
Transcriptome Analysis Reveals an Antagonistic Transcriptome Response in rst1 and free1
It is clear that RST1 exerts the opposite effect on vacuolar trafficking as FREE1 and seems to function by bypassing FREE1. This suggests that RST1 antagonizes FREE1 activity, perhaps by preventing FREE1 from forming a functional complex or acting independently to inhibit another undefined factor that works in parallel with FREE1. To determine whether such an antagonistic effect of RST1 and FREE1 can be reflected as a readout at the transcriptome level, we compared the gene expression profiles between free1 and rst1 using RNA sequencing (RNA-seq) analysis (Supplemental Data Sets 1 and 2). As illustrated in the heatmap, three biological replicates for each sample showed that these expression profiles are highly associated (Supplemental Figure 15), indicating the reproducibility of the RNA-seq data.
We then identified differentially expressed (DE) genes for each pairwise comparison using DESeq2 (Supplemental Data Sets 3 and 4; Love et al., 2014). Compared with the controls, 432 genes (17.2% of the free1-upregulated genes) from the free1-upregulated list are found in the rst1-downregulated (20.6% of the rst1-downregulated genes) list, while 708 genes (24.3% of the free1-upregulated list) from the free1-downregulated list are found in the rst1-upregulated (26.1% of the rst1-upregulated) list (Figures 7A and 7B), suggesting the opposite transcriptome response in rst1 and free1. By plotting the fold change of DE genes in rst1 and free1 using a heatmap (Figure 7C), we observed that the rst1 and free1 mutations had opposite effects on transcriptional readout.
Figure 7.
Antagonistic Effects of FREE1 and RST1 on Transcriptome by RNA-Seq.
(A) and (B) Venn diagram showing the overlap between the set of genes with altered expression in free1 (FREE1-RNAi line with DEX treatment compared with FREE1-RNAi line without DEX treatment) and rst1 (sof100 line compared with FREE1-RNAi line).
(C) Heatmap of the fold changes of mis-regulated genes in free1 and rst1. Changes in transcription abundance are indicated by intensities of colors expressed in log2 scales. Blue and red indicate down- and upregulation, respectively.
(D) Significantly enriched GO terms of mis-regulated genes in free1 and rst1. Bar chart showing the significance of GO terms expressed in –log10 (FDR) scales. FDR, false discovery rate.
To further analyze the genes that exhibited opposite transcriptomic responses in these two mutants, we analyzed the DE genes (Supplemental Data Sets 3 and 4) based on Gene Ontology (GO) annotation (Du et al., 2010; Tian et al., 2017). The genes involved in plastid-related processes, membrane-related processes, and stress-related responses were significantly enriched (Figure 7D), suggesting their functional relationship with RST1 and FREE1.
To determine whether the enhanced endocytic degradation of vacuolar cargo in rst1 mutant was caused by altered expression of certain ESCRT components (Gao et al., 2017), we compared the expression level of ESCRT components in free1 and rst1. Most of the ESCRT components showed no significant difference between the two mutants, except for TOL4, a putative ESCRT-0 functional homolog (Korbei et al., 2013). The significant alteration of TOL4 expression along with the increased expression in free1 and reduced expression in rst1 (Supplemental Figure 16) suggests that RST1 may inhibit MVB biogenesis and vacuolar trafficking through TOL4.
DISCUSSION
We used DEX-inducible DEX:FREE1-RNAi transgenic plants for a sof suppressor screen, and the identification of RST1 revealed a previously unexpected role for RST1 in the endomembrane trafficking pathway. The sof screening was a laborious but quite straightforward approach and could be applied to other seedling lethal genes or embryo defect mutants in other model systems. Our screen had several limitations that restricted the spectrum of genes identified. First, genes that have multiple homologs and function redundantly may represent FREE1 genetic regulators, while underrepresented in the screen. Second, genes that are essential for seedling survival may also represent FREE1 genetic regulators but be absent in our screen. Third, because FREE1-RNAi plants bypassed the embryo requirement of FREE1, our ability to identify mutants that suppress the free1 null mutants is limited.
By using this suppressor screening approach, we recently identified a plant-specific negative regulator, BRAF, that competitively binds to VPS23 to inhibit FREE1-mediated MVB biogenesis (Shen et al., 2018). The identification of RST1 as a suppressor of free1 negatively regulating the endomembrane trafficking pathway indicates that RST1 titrates out a certain component(s) of the FREE1-mediated pathway as a molecular switch. This study did not provide direct evidence on how the cytosolic RST1 performs its molecular function in MVB biogenesis and prepares membrane cargos for vacuolar transport. In the working model (Figure 8), we propose that RST1 may function as a molecular switch that provides quality surveillance of membrane cargos for vacuolar transport. When an interruption occurs in the whole cell trafficking system, such as in the free1 mutant, RST1 may function to switch off the whole trafficking pathway, that is, accumulation of RST1 in free1 is similar to a situation in which the traffic light was continuously switched back to red. It would be highly risky if cells grow without such a molecular switch to properly control the trafficking pathway in response to the changing environmental and growth signals. At the molecular level, RST1 may directly interact with yet-to-be-identified key components in the cytosol to prevent their function at the membrane. Indeed, our preliminary yeast screen results showed that RST1 interacted with VPS30/ATG6, which is a conserved essential component for vacuolar trafficking and plant growth (Fujiki et al., 2007; Qin et al., 2007).
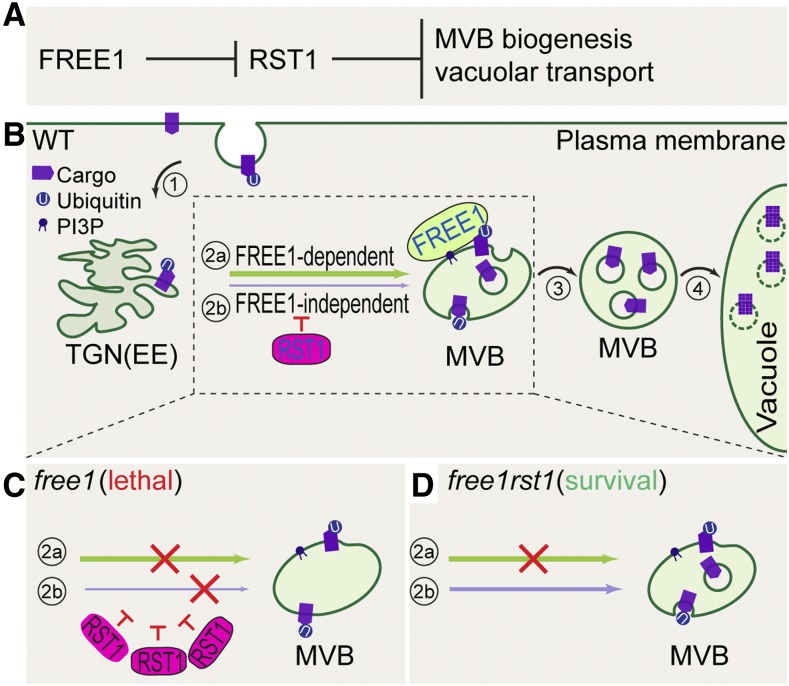
Figure 8.
Working Model for RST1 in the Negative Regulation of the MVB Biogenesis Pathway in Arabidopsis.
MVBs are endosomes containing membrane-bound ILVs, which are formed by budding into the lumen of the MVB. When marked for degradation with ubiquitination modification, surface cargo proteins are internalized to reach the TGN/EE compartments (route 1; in numbered circle) and are then sorted and packaged into ILVs in MVBs (route 2; in numbered circle). After maturation, MVBs become are filled with cargo-enriched ILVs (route 3; in numbered circle). Mature MVBs fuse with the vacuole to deliver cargo-enriched ILVs into the vacuolar lumen for degradation (route 4; in numbered circle).
(A) Genetic relationship between FREE1 and RST1. FREE1 represses RST1 and RST1 function downstream of FREE1 to negatively regulate MVB biogenesis and vacuolar transport.
(B) The FREE1 protein mediates most MVB biogenesis via incorporation into the ESCRT-I complex (route 2a; in numbered circle), while a FREE1-independent backup pathway may mediate MVB biogenesis when needed (route 2b; in numbered circle). In the wild type, RST1 inhibits the FREE1-independent MVB biogenesis pathway.
(C) In free1, substantial accumulation of RST1 leads to enhanced inhibition of FREE1-independent MVB biogenesis and trafficking, thus causing a total block of both the FREE1-dependent and -independent pathway, resulting in seedling lethality.
(D) In free1rst1, FREE1-independent MVB biogenesis and trafficking would be released from inhibition when RST1 is knocked out, thus reversing seedling lethality back to survival.
Solid arrows, secretory and endocytic trafficking routes toward vacuole; red T bar, inhibition effects of RST1; red cross, impediment of indicated pathways.
Genes functioning together tend to show coexpression patterns. We thus looked for gene networks associated with RST1 at string-db.org (Szklarczyk et al., 2017). Among the top-ten gene list, RST1 is coexpressed with At5g18525 (GFS12), At5g24350 (MIP2), and At3g08530 (CHC2), all of which are related to endomembrane trafficking (Kitakura et al., 2011; Li et al., 2013; Teh et al., 2015). RST1 is also coexpressed with At5g58410, which encodes an undefined E3 ubiquitin ligase (Supplemental Figures 17A and 17B). When comparing the expression level of these genes in free1 and rst1 from the RNA-seq data, both At5g58410 (E3) and At5g18525 (GFS12) showed increased expression in free1 but decreased expression in rst1 (Supplemental Figure 17 C), suggesting that these coexpressed genes may function together with RST1. Taken together, these data are in agreement with the hypothesis that RST1 preferentially functions antagonistically to FREE1, although future studies are needed to provide direct proof.
In summary, we demonstrate that RST1 functions as a negative regulator that inhibits the MVB biogenesis pathway. Based on our findings, we propose that at the genetic level, FREE1 represses RST1, and RST1 functions downstream of FREE1 to negatively regulate MVB biogenesis and vacuolar transport (Figure 8A). At the cellular level, FREE1 normally mediates the majority of housekeeping MVB biogenesis, while a FREE1-independent backup pathway might operate for MVB biogenesis and vacuolar transport under certain conditions when needed. Normally, RST1 inhibits this FREE1-independent pathway to restrict MVB biogenesis and vacuolar transport to a basal level (Figure 8B). In the free1 mutants, the significant RST1 accumulation exaggerated the inhibition of the FREE1-independent pathway, thus largely blocking the backup pathway, and resulting in seedling lethality (Figure 8C). In free1rst1 double mutants, as in the four sof mutants (sof100, sof220, sof452, and sof453) lacking RST1, the FREE1-independent pathway was relieved to mediate the backup pathway, thus rescuing the seedling lethality phenotype (Figure 8D). Much work remains to differentiate between the different possible mechanisms. We propose that RST1 encodes rate-limiting components integral to vacuolar trafficking pathways throughout development. RST1 is found throughout the multicellular eukaryotic organism, suggesting that RST1 inhibits conserved trafficking components rather than plant-specific factors, and likely a similar molecular breaks function of RST1 is applicable to other multicellular eukaryotic organisms. Recently, two other studies reported that RST1 also plays roles in RNA surveillance pathway, demonstrating that RST1 is a multifunctional protein in plants (Lange et al., 2019; Li et al., 2019b).
METHODS
Plant Material and Growth Conditions
All mutants and transgenic lines used in this study are in the Arabidopsis ecotype Col background. The FREE1-RNAi and rst1-3 were described previously (Mang et al., 2009; Gao et al., 2014). Arabidopsis seeds were sterilized and germinated on Murashige and Skoog (MS) plates (full-strength MS salts, 0.8% [w/v] phyto agar, and 3% [w/v] Suc, pH 5.7). Seedlings were then grown at 22°C under a 16-h light/8-h dark cycle for 5 d before being transferred to pots in a growth room at 22°C under a 16-h light/8-h dark cycle. The light intensity is ∼150 μmol/m2/s.
PI Staining
The roots of the 7-d-old seedlings were stained in liquid MS medium containing propidium iodide (PI; 10 µg/mL) for 2 min and then washed for 1 min with liquid MS medium. Confocal laser scanning microscopy imaging was then immediately performed using a TCS SP8 confocal microscope (Leica).
Chemical Treatment
For BFA treatments, Arabidopsis (Arabidopsis thaliana) seedlings were incubated in liquid MS medium containing BFA (Sigma-Aldrich) at a final concentration of 10 μg/mL for 30 min. For wortmannin treatment, wortmannin was prepared in DMSO and used at 33 μM for 30 min in the liquid MS medium. The solvent (DMSO) alone was added as the control. For boron treatment, BOR1-GFP seedlings were transferred into liquid medium containing 100 μM boric acid (+B) as previously used (Shen et al., 2018).
FM4-64 Staining
Seedlings were washed with liquid MS and then stained with FM4-64 (12 μM) in liquid MS overnight to examine vacuole morphology. Analysis of FM4-64 endocytosis was performed by incubation with FM4-64 (12 μM) for 5 min on ice, following by two washes with MS medium before time-course observation and image collection.
EMS Mutagenesis
The FREE1-RNAi transgenic line was mutagenized with EMS as described previously (Zhao et al., 2015). The screen for survived seedlings was performed on horizontally oriented MS plates with 3% (w/v) Suc supplemented with 10 µM DEX. Putative sof suppressors were transplanted into soil and genotyped to confirm homozygosis for the FREE1-RNAi transgenic insertion site.
Arabidopsis FREE1-RNAi Suppressor Screening
The mutant pool was established in a previous report (Zhao et al., 2015). Basically, seeds of the Arabidopsis (Col-0) line expressing a single insertion of pTA7002-FREE1-RNAi were mutagenized by EMS. Approximately 40,000 seeds corresponding to the progeny of 10,000 mutagenized M1 seeds were sown on MS plates supplemented with 10 μM DEX and grown for 5 d. Seedlings showing a survival phenotype were selected as sof. Selected M2 seedlings were planted into soil for individual M3 seed collection. Individual M3 seeds were screened on MS medium containing 10 μM DEX and hygromycin (50 mg/mL) again, and 7-d-old M3 seedlings were screened for the survival phenotype.
Immunoblot Analysis
Protein isolation and blotting were performed as described previously (Zhao et al., 2015; Shen et al., 2018). Total cell extracts from Arabidopsis seedlings were prepared in lysis buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 1% Triton X-100, and 1× Complete Protease Inhibitor Cocktail and then centrifuged at 14,000 rpm for 30 min at 4°C. The supernatant protein samples were boiled in 1× SDS sample loading dye at 95°C and then were subjected to gel electrophoresis on 10% SDS-PAGE gels. Proteins were transferred to nitrocellulose membranes (Bio-Rad) followed by blocking in PBS-0.05% Tween 20 with 5% milk powder and antibody incubation (4 µg/mL). Membranes were incubated with horseradish peroxidase–conjugated secondary antibodies, and Software Signal development was performed using the ECL Western Blotting system (GE Healthcare). Quantification of immunoblots was done using ImageJ.
Whole-Genome Sequencing–Based Mapping
Mapping was performed as described previously (Zhao et al., 2015). The sof lines sof453, sof100, sof220, and sof452 were outcrossed to Arabidopsis ecotype Landsberg erecta. For each mutant, the F2 population was grown on MS plates with 3% (w/v) Suc supplemented with 10 µM DEX and hygromycin (50 mg/mL), and 300 seedlings with the survival sof phenotype were pooled for a genomic DNA preparation. The genomic DNA was submitted to the DNA-sequencing facility for library preparation and sequencing on a HiSeq 2000 system (Illumina) to generate 100-bp pair-end reads, yielding >15-fold genome coverage. The reads were mapped to a Col-0 reference genome (The Arabidopsis Information Resource; https://www.arabidopsis.org) using BWA software (Li and Durbin, 2009), and putative single-nucleotide polymorphisms (SNPs) were identified as described using SAMTOOLS software (Li et al., 2009). The SNP frequency was plotted based on chromosomal location using SHOREmap software (Schneeberger et al., 2009; Sun and Schneeberger, 2015) based on 461,070 SNP markers using SHORE map outcross function. Relatively reliable loci were filtered as follows: consensus quality > 20 (error rate, 1%), total depth > 5. Only EMS-induced C/G-to-T/A transition SNP markers were further considered as candidates. The causative mutation within the mapping interval was annotated using the SHOREmap software annotate function.
Plasmid Construction
For the constructs used for genetic complementation in Arabidopsis, the cDNAs encoding the RST1 genes were synthesized in the plasmid pUC-Xba1-RST1 (without stop codon)-Kpn1 and then the plasmid pUC-Xba1-RST1 (without stop codon)-Kpn1 was double digested using Spe1 and Kpn1. Meanwhile, the pBI121-UBQ10-GFP vector plasmid was also double digested using Spe1 and Kpn1. The DNA fragment containing RST1 cDNA was gel recovered and was ligated into the digested pBI121 vectors. For DEX:VPS4 (E232Q) transgenic plants, the mutated coding regions were amplified from previous published plasmids (Cai et al., 2014), cloned into pDONR/Zeo, and then further cloned into the pTA7002-DEX-GW vector for plant transformation as described previously (Shen et al., 2018). For DEX:SNF7 (L22W) transgenic plants, the mutated coding regions were amplified from previous published plasmids (Cai et al., 2014), cloned into pDONR/Zeo, and then further cloned into the pTA7002-DEX-GW vector for plant transformation as described previously (Shen et al., 2018). For Y2H analysis, the cDNAs were cloned into the pGBKT7 and pGADT7 vectors. All constructs were confirmed by Sanger sequencing.
Ultrastructural Analysis and Immunogold Labeling using TEM
Ultrastructural analysis procedures for TEM sample preparation, thin sectioning, and immunogold labeling of high-pressure frozen, freeze-substituted tissues and HM20 embedding were performed as described previously (Cui et al., 2014; Gao et al., 2014; Zhuang et al., 2017; Shen et al., 2018). Briefly, the root tips of 4-d-old Arabidopsis seedlings subjected to the indicated treatments were cut and immediately frozen in a high-pressure freezer (EM PACT2, Leica), followed by subsequent freeze substitution in dry acetone containing 0.1% uranyl acetate at −85°C in an AFS freeze-substitution unit (Leica). Infiltration with Lowicryl HM20, embedding, and UV light polymerization were performed stepwise at −10°C. Immunogold labeling was performed as described previously with the homemade anti-VSR antibody and gold-coupled secondary antibody at 1:50 dilution. Sections were examined using a H-7650 transmission electron microscope with a charge-coupled device camera operating at 80 kV (Hitachi High-Technologies; www.hitachi-hightech.com/jp/).
Immunoprecipitation
Total cell extracts from Arabidopsis seedlings were prepared in lysis buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 0.4% Nonidet P-40, 5% glycerol, and 1× Complete Protease Inhibitor Cocktail, followed by brief centrifugation at 600g for 3 min at 4°C to remove intact cells and large cellular debris. The supernatant total cell extracts were then centrifuged at 14,000 rpm for 30 min at 4°C. The supernatant was incubated with GFP-Trap agarose beads (ChromoTek) for 4 h at 4°C in IP buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 0.2% Nonidet P-40, 5% glycerol, and 1× Complete Protease Inhibitor Cocktail) in a top to end rotator. After incubation, the beads were washed four times with ice-cold washing buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 0.05% Nonidet P-40, and 5% glycerol) and boiled in SDS sample buffer. Samples were analyzed by SDS-PAGE and immunoblot using the indicated antibodies.
Antibodies
Affinity-purified polyclonal antibody for RST1 was raised in rabbits at the Laboratory Animal Services Center of the Chinese University of Hong Kong against the synthetic peptide antigen acetyl-SASSDIDSDSYRNQEERLLC-amide (GeneScript), corresponding to 1049 to 1067 amino acids of RST1. The peptide was synthesised with an addition of a C-terminal cystein (underlined) for subsequent conjugation. Affinity purification was performed using CnBr-activated Sepharose 4B (catalog no. C9142, Sigma-Aldrich) column conjugated with the peptides. FREE1 and VSR antibodies were homemade. Antibodies including anti–fructose-1,6-bisphosphatase (cFBPase; AS04 043, Agrisera) and anti-GFP (633281, Clontech) were purchased from commercial sources.
Y2H Analysis
Y2H analysis was performed using pGBKT7 and pGADT7 vectors (Clontech) containing cDNAs. The paired plasmids (AD and BD) were co-introduced into the yeast strain AH109 by heat shock transformation. Positive transformants containing both AD and BD were selected on synthetic drop-out (SD) medium lacking Trp and Leu (SD−Trp−Leu). For determination of protein interactions, positive transformants were inoculated on SD medium lacking Ade, His, Trp, and Leu (SD−Ade−His−Trp−Leu). Each experiment was performed at least twice independently, and similar results were obtained.
RNA-Seq Analysis
For RNA-seq experiments, 11-d-old seedlings grown on 1/2 MS agar plates were treated with or without DEX in liquid 1/2 MS for 48 h. RNA-seq was performed using three biological replicates for FREE1-RNAi and sof100. Total RNA was extracted using TRI reagent (MRC). Total mRNA was prepared with 5 µg of total RNA using the NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB). RNA-seq libraries were constructed using the NEBNext mRNA Library Prep Reagent Set for Illumina (NEB) following the manufacturer’s protocols. Twelve libraries (three replicates for each sample) were pooled and sequenced on an Illumina HiSeq 2500 platform. Qualified unique reads were mapped to the Col genome using TOPHAT v.2.0.13 (Kim et al., 2013), with no mismatches permitted. Reads in gene regions were counted using an in-house Perl script (Huang et al., 2017). The expression fold change of each gene was calculated using the R package DESeq2 (Love et al., 2014) with the threshold for DE genes set to a fold change of 1.5 and a P-value < 0.05. Venn diagrams were generated using VENNY v.2.1 (http://bioinfogp.cnb.csic.es/tools/venny/index.html), and GO enrichment analysis of DE genes was performed on the agriGO website (Du et al., 2010; Tian et al., 2017).
Phylogenetic Analysis
The amino acid sequences of RST1 homologs were obtained from the National Center for Biotechnology Information by using BlastP using Arabidopsis RST1 as bait (Supplemental Data Set 5). Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016). The evolutionary history was inferred using the maximum likelihood method based on the JTT matrix–based model (Jones et al., 1992). The tree with the highest log likelihood (−31099.79) is shown. The initial tree(s) for the heuristic search was obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with a superior log likelihood value. The analysis involved 18 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 1060 positions in the final data set.
Confocal Microscopy
Samples were mounted in liquid MS medium and imaged with a Leica TCS SP8 confocal laser scanning microscope, using a 63× (numerical aperture 1.20) water immersion objective. Images with both GFP and RFP were collected using the sequential scanning mode to avoid possible crosstalk of fluorescent signals. The same microscope settings were used for both the control samples and the mutant/treated samples. For GFP observation, the excitation wavelength is 488 nm, while the emission wavelength is 500 to 550 nm. For PI, mCherry, and FM4-64 staining observation, the settings were adjusted to excitation wavelength 561 nm and emission wavelength 575 to 625 nm. Experiments were repeated at least three times using three different batches of seedlings.
Immunofluorescence Labeling in Arabidopsis Roots
Fixation and preparation of Arabidopsis roots for immunofluorescent labeling and confocal analysis were performed as described previously by Gao et al. (2014) and Shen et al. (2018). The roots of 5-d-old seedlings were fixed with 4% paraformaldehyde in PBS supplemented with 0.1% Triton X-100. Following cell wall digestion, permeabilization, and blocking with 3% BSA, the fixed roots were incubated with anti-VSR antibody or anti-RST1 antibody at 4 μg/mL diluted in 1% BSA at 4°C overnight. After a wash with PBS, the roots were probed with Alexa 568 goat anti-rabbit IgG (Invitrogen) secondary antibody for confocal observation.
RNA Extractions and Quantitative Real-Time RT-PCR Reaction
Total RNA was isolated from the indicated tissues by grinding the tissue in liquid nitrogen in the presence of TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 2 μg of total RNA using Moloney murine leukemia virus reverse transcriptase (Promega) according to the manufacturer’s instructions. For each qPCR, 1 μL of cDNA sample, 12.5 μL of iQ SYBR Green Supermix (Bio-Rad), and 0.5 μL of primer (from a 10 μM working solution) were used in a final volume of 25 μL supplemented with water. The RT-qPCRs were run on a MyiQ (Bio-Rad). Gene-specific primers for RST1 (5′-CGTGCTCCTCCAGCCAGTTC-3′; 5′-ATGTTGGAGCTGGCGATCCT-3′), FREE1 (5′-ACCGCAAGTCTTCGTCTGGT-3′; 5′-TGTGCGCTAACGAGGAAAGGG-3′), ACTIN (5′ GCACTTGCACCAAGCAGCAT-3′, 5′-ACGATTCCTGGACCTGCCTCA-3′), and UBQ (5′-AGTGGAAAGCTCCGACACCA-3′; 5′-CCACGAAGACGCAGGACCAA-3′) were used. The relative expression levels were compared after normalization to ACTIN transcript levels. Mean ± se results were plotted.
Microsome Analysis
Total cell extracts from Arabidopsis seedlings were prepared in lysis buffer containing 40 mM HEPES-KOH at pH 7.5, 1 mM EDTA, 10 mM KCl, 0.4 M Suc, 0.5 mM phenylmethylsulfonyl fluoride, 25 μg/mL leupeptin, and 1× Complete Protease Inhibitor Cocktail, followed by brief centrifugation at 600g for 3 min at 4°C. After discarding this low-speed pellet containing unbroken cells, nuclei, and debris, the cell extract was centrifuged at 10,000g for 10 min to separate soluble proteins (S10) from a fraction enriched in organelles (P10). The S10 fraction was further centrifuged at 100,000g for 30 min to separate membranes (P100) from soluble proteins (S100).
Quantification and Statistical Analysis
For immunoblot quantification, the relative intensities of each band were quantified using ImageJ (https://imagej.nih.gov/ij/); the control lane in each experiment was set as 1. For confocal image quantification, the relative fluorescence intensity was quantified using ImageJ, and the ratios of fluorescence intensities are shown. For each set of data, image quantification was performed in five cells per root in five individual seedlings. Statistically significant differences were determined using Student’s t test (*P < 0.05; **P < 0.01; n.s., P > 0.05). Sample numbers and the number of biological replicates for each experiment are indicated in the figure legends.
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are RST1 (AT3G27670), FREE1 (AT1G20110), SNF7A (AT2G19830), and Vps4 (AT2G27600).
Supplemental Data
Supplemental Figure 1. Flow chart of the sof suppressor screening and SOF gene identification.
Supplemental Figure 2. Four sof lines were mapped to the same region on chromosome 3.
Supplemental Figure 3. Isolation of four rst1 alleles as sof mutants using FREE1-RNAi transgenic plants.
Supplemental Figure 4. Characterization of UBQpro:RST1-GFP/sof453 complementation lines.
Supplemental Figure 5. Differential RST1 and FREE1 expression in different developmental stages of Arabidopsis based on Genevestigator microarray data.
Supplemental Figure 6. The cytosolic localization of the functional RST1-GFP fusion protein.
Supplemental Figure 7. RST1 does not interact with FREE1.
Supplemental Figure 8. Specific genetic interactions between rst1 mutation and the ESCRT pathway mutations.
Supplemental Figure 9. Mutation in RST1 did not alter the GFP-FREE1 MVB localization of GFP-FREE1 or the FREE1 and VPS28 membrane distribution.
Supplemental Figure 10. Characterization of UBQpro:RST1-GFP overexpression lines.
Supplemental Figure 11. Time-course images of FM4-64 uptake in Arabidopsis root cells.
Supplemental Figure 12. Inhibition of FM4-64 endocytic tracer staining of tonoplast by RST1 overexpression.
Supplemental Figure 13. Enhanced BOR1-GFP endocytic degradation in the rst1 mutant.
Supplemental Figure 14. Mutation in RST1 did not alter soluble cargo vacuolar transport and endocytosis.
Supplemental Figure 15. RNA-seq analysis of FREE1-RNAi and sof100 lines.
Supplemental Figure 16. Relative expression levels of ESCRT components and ESCRT-related proteins in free1 and rst1 by RNA-seq.
Supplemental Figure 17. RST1 Functional protein association network in Arabidopsis (action view) as visualized on the STRING website (http://string-db.org/).
Supplemental Data Set 1. RNA-seq RPKM data for FREE1-RNAi_DEX versus FREE1-RNAi_DEX.
Supplemental Data Set 2. RNA-seq RPKM data for sof100 versus FREE1-RNAi.
Supplemental Data Set 3. Shared genes upregulated in free1 but downregulated in rst1.
Supplemental Data Set 4. Shared genes downregulated in free1 but upregulated in rst1.
Supplemental Data Set 5. Alignment of sequences used for maximum-likelihood analysis.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
ACKNOWLEDGMENTS
This work was supported by grants from the Research Grants Council of Hong Kong (AoE/M-05/12, RIF R4005-18, G-CUHK 403/17, CUHK 14130716, 14102417, 14104716, 14100818, C4011-14R, C4012-16E, C4002-17G), the National Natural Science Foundation of China (31670179 and 91854201), and CUHK Research Committee to L.J., as well as by grants from the National Natural Science Foundation of China (31671467 and 31870171 to C.G.).
AUTHOR CONTRIBUTIONS
Q.Z., J.S., C.G., Y.C., and L.J. designed the research. Q.Z., J.S., C.G., Y.C., Y.W., J.C., L.C., W.C., Y.Z., S.H., Q.Z.Z., C.K.L., and K.P.L. conducted detailed studies on the function of the RST1 protein. Q.Z., Y.W., J.C., L.C., and X.C. performed RNA-seq analysis. Q.Z., J.S., C.G., Y.C., and L.J. analyzed the data. Q.Z. and L.J. wrote the article with comments from all authors. All authors discussed the results and approved the article.
References
- Aniento F., Robinson D.G. (2005). Testing for endocytosis in plants. Protoplasma 226: 3–11. [DOI] [PubMed] [Google Scholar]
- Barberon M., Dubeaux G., Kolb C., Isono E., Zelazny E., Vert G. (2014). Polarization of IRON-REGULATED TRANSPORTER 1 (IRT1) to the plant-soil interface plays crucial role in metal homeostasis. Proc. Natl. Acad. Sci. USA 111: 8293–8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Palazon B., et al. (2016). FYVE1/FREE1 interacts with the PYL4 ABA receptor and mediates its delivery to the vacuolar degradation pathway. Plant Cell 28: 2291–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., Talbot C., Boutte Y., Catrice O., Read N.D., Satiat-Jeunemaitre B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214: 159–173. [DOI] [PubMed] [Google Scholar]
- Brockschmidt A., et al. (2012). KIAA1797/FOCAD encodes a novel focal adhesion protein with tumour suppressor function in gliomas. Brain 135: 1027–1041. [DOI] [PubMed] [Google Scholar]
- Cai Y., Zhuang X., Gao C., Wang X., Jiang L. (2014). The Arabidopsis endosomal sorting complex required for transport III regulates internal vesicle formation of the prevacuolar compartment and is required for plant development. Plant Physiol. 165: 1328–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Goodwin S.M., Liu X., Chen X., Bressan R.A., Jenks M.A. (2005). Mutation of the RESURRECTION1 locus of Arabidopsis reveals an association of cuticular wax with embryo development. Plant Physiol. 139: 909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K.P., Zeng Y., Li Y., Ji C., Xia Y., Jiang L. (2018). Signal motif-dependent ER export of the Qc-SNARE BET12 interacts with MEMB12 and affects PR1 trafficking in Arabidopsis. J. Cell Sci. 131: jcs202838. [DOI] [PubMed] [Google Scholar]
- Cui Y., et al. (2019). A whole-cell electron tomography model of vacuole biogenesis in Arabidopsis root cells. Nat. Plants 5: 95–105. [DOI] [PubMed] [Google Scholar]
- Cui Y., Zhao Q., Gao C., Ding Y., Zeng Y., Ueda T., Nakano A., Jiang L. (2014). Activation of the Rab7 GTPase by the MON1-CCZ1 complex is essential for PVC-to-vacuole trafficking and plant growth in Arabidopsis. Plant Cell 26: 2080–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Shen J., Gao C., Zhuang X., Wang J., Jiang L. (2016). Biogenesis of plant prevacuolar multivesicular bodies. Mol. Plant 9: 774–786. [DOI] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y., Yoshimoto K., Ohsumi Y. (2007). An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 143: 1132–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Luo M., Zhao Q., Yang R., Cui Y., Zeng Y., Xia J., Jiang L. (2014). A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr. Biol. 24: 2556–2563. [DOI] [PubMed] [Google Scholar]
- Gao C., Zhuang X., Cui Y., Fu X., He Y., Zhao Q., Zeng Y., Shen J., Luo M., Jiang L. (2015). Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc. Natl. Acad. Sci. USA 112: 1886–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Zhuang X., Shen J., Jiang L. (2017). Plant ESCRT complexes: Moving beyond endosomal sorting. Trends Plant Sci. 22: 986–998. [DOI] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230. [DOI] [PubMed] [Google Scholar]
- Huang Z., Shi T., Zheng B., Yumul R.E., Liu X., You C., Gao Z., Xiao L., Chen X. (2017). APETALA2 antagonizes the transcriptional activity of AGAMOUS in regulating floral stem cells in Arabidopsis thaliana. New Phytol. 215: 1197–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter P.R., Craddock C.P., Di Benedetto S., Roberts L.M., Frigerio L. (2007). Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells. Plant Physiol. 145: 1371–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono E., Kalinowska K. (2017). ESCRT-dependent degradation of ubiquitylated plasma membrane proteins in plants. Curr. Opin. Plant Biol. 40: 49–55. [DOI] [PubMed] [Google Scholar]
- Jia T., Gao C., Cui Y., Wang J., Ding Y., Cai Y., Ueda T., Nakano A., Jiang L. (2013). ARA7(Q69L) expression in transgenic Arabidopsis cells induces the formation of enlarged multivesicular bodies. J. Exp. Bot. 64: 2817–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitakura S., Vanneste S., Robert S., Löfke C., Teichmann T., Tanaka H., Friml J. (2011). Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23: 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb C., Nagel M.K., Kalinowska K., Hagmann J., Ichikawa M., Anzenberger F., Alkofer A., Sato M.H., Braun P., Isono E. (2015). FYVE1 is essential for vacuole biogenesis and intracellular trafficking in Arabidopsis. Plant Physiol. 167: 1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbei B., Moulinier-Anzola J., De-Araujo L., Lucyshyn D., Retzer K., Khan M.A., Luschnig C. (2013). Arabidopsis TOL proteins act as gatekeepers for vacuolar sorting of PIN2 plasma membrane protein. Curr. Biol. 23: 2500–2505. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H., et al. (2014). The RNA helicases AtMTR4 and HEN2 target specific subsets of nuclear transcripts for degradation by the nuclear exosome in Arabidopsis thaliana. PLoS Genet. 10: e1004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H., et al. (2019). The RST1 and RIPR connect the cytosolic RNA exosome to the Ski complex in Arabidopsis. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner J., Petrášek J., Tomanov K., Retzer K., Pařezová M., Korbei B., Bachmair A., Zažímalová E., Luschnig C. (2012). Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc. Natl. Acad. Sci. USA 109: 8322–8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., et al. (2019a). The plant ESCRT component FREE1 shuttles to the nucleus to attenuate abscisic acid signalling. Nat. Plants 5: 512–524. [DOI] [PubMed] [Google Scholar]
- Li L., et al. (2013). MAG2 and three MAG2-INTERACTING PROTEINs form an ER-localized complex to facilitate storage protein transport in Arabidopsis thaliana. Plant J. 76: 781–791. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Natran A., Chen Y., Vercruysse J., Wang K., Gonzalez N., Dubois M., Inzé D. (2019b). A genetics screen highlights emerging roles for CPL3, RST1 and URT1 in RNA metabolism and silencing. Nat. Plants 5: 539–550. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Chen Z. (2007). Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell 19: 943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mang H.G., et al. (2009). The Arabidopsis RESURRECTION1 gene regulates a novel antagonistic interaction in plant defense to biotrophs and necrotrophs. Plant Physiol. 151: 290–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M.S. (2018). ESCRT-mediated sorting and intralumenal vesicle concatenation in plants. Biochem. Soc. Trans. 46: 537–545. [DOI] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18: 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Ma Z., Zhang L., Xing S., Hou X., Deng J., Liu J., Chen Z., Qu L.J., Gu H. (2007). Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 17: 249–263. [DOI] [PubMed] [Google Scholar]
- Robinson D.G., Jiang L., Schumacher K. (2008). The endosomal system of plants: Charting new and familiar territories. Plant Physiol. 147: 1482–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A.H., Nielsen K.L., Jørgensen J.E., Weigel D., Andersen S.U. (2009). SHOREmap: Simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6: 550–551. [DOI] [PubMed] [Google Scholar]
- Shen J., Zhao Q., Wang X., Gao C., Zhu Y., Zeng Y., Jiang L. (2018). A plant Bro1 domain protein BRAF regulates multivesicular body biogenesis and membrane protein homeostasis. Nat. Commun. 9: 3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Schneeberger K. (2015). SHOREmap v3.0: Fast and accurate identification of causal mutations from forward genetic screens. Methods Mol. Biol. 1284: 381–395. [DOI] [PubMed] [Google Scholar]
- Surpin M., Raikhel N. (2004). Traffic jams affect plant development and signal transduction. Nat. Rev. Mol. Cell Biol. 5: 100–109. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., Jensen L.J., von Mering C. (2017). The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45 (D1): D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J., Miwa K., Yuan L., von Wirén N., Fujiwara T. (2005). Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. USA 102: 12276–12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh O.K., Hatsugai N., Tamura K., Fuji K., Tabata R., Yamaguchi K., Shingenobu S., Yamada M., Hasebe M., Sawa S., Shimada T., Hara-Nishimura I. (2015). BEACH-domain proteins act together in a cascade to mediate vacuolar protein trafficking and disease resistance in Arabidopsis. Mol. Plant 8: 389–398. [DOI] [PubMed] [Google Scholar]
- Tian T., Liu Y., Yan H., You Q., Yi X., Du Z., Xu W., Su Z. (2017). agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45 (W1): W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse Y.C., Mo B., Hillmer S., Zhao M., Lo S.W., Robinson D.G., Jiang L. (2004). Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16: 672–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Cai Y., Miao Y., Lam S.K., Jiang L. (2009). Wortmannin induces homotypic fusion of plant prevacuolar compartments. J. Exp. Bot. 60: 3075–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., et al. (2015). Fast-suppressor screening for new components in protein trafficking, organelle biogenesis and silencing pathway in Arabidopsis thaliana using DEX-inducible FREE1-RNAi plants. J. Genet. Genomics 42: 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Wang H., Lam S.K., Gao C., Wang X., Cai Y., Jiang L. (2013). A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25: 4596–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Chung K.P., Cui Y., Lin W., Gao C., Kang B.H., Jiang L. (2017). ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc. Natl. Acad. Sci. USA 114: E426–E435. [DOI] [PMC free article] [PubMed] [Google Scholar]