The jasmonic acid signaling repressor proteins JAZs physically interact with FHY3 and FAR1 and repress their transcriptional activity, thereby prioritizing growth over defense under shade.
Abstract
Increasing crop yield per unit of area can be achieved by increasing planting density. However, high-density planting could trigger shade avoidance responses, which cause exaggerated growth and increased susceptibility to various diseases. Previous studies have shown that the rapid elongation of plants under shade (i.e., reduced red to far-red ratios) is regulated by phytochromes and various phytohormones. However, the detailed molecular mechanisms governing the interaction among these signaling pathways are not well understood. Here, we report that loss-of-function mutants of FAR-RED ELONGATED HYPOCOTYL3 (FHY3) and FAR-RED-IMPAIRED RESPONSE1 (FAR1), which encode two homologous transcription factors essential for phytochrome signaling, exhibit an exaggerated shade avoidance phenotype. We show that FHY3 and FAR1 repress plant growth through directly activating the expression of two atypical basic helix-loop-helix transcriptional cofactors, PHYTOCHROME RAPIDLY REGULATED1 (PAR1) and PAR2, and that this process is antagonized by a group of JASMONATE ZIM-DOMAIN proteins, key repressors of the jasmonic acid (JA) signaling pathway, through physical interactions. Furthermore, we show that FHY3 interacts with MYC2, a key transcriptional regulator of JA responses, coordinately regulating JA-responsive defense gene expression. Our results unveil a previously unrecognized mechanism whereby plants balance their growth and defense responses through convergence of the phytochrome signaling pathway and JA signaling pathway under shade conditions.
INTRODUCTION
Light is a key environmental cue modulating plant growth and development; therefore, plants have evolved elaborate photoreceptor systems that sense the changes in their light environment. The model dicot plant Arabidopsis (Arabidopsis thaliana) possesses five phytochrome photoreceptors (phyA to phyE), which perceive red (R) and far-red (FR) light and modulate multifaceted growth and development processes, including seed germination, hypocotyl growth, chlorophyll synthesis, stomata opening, and flower initiation (Shin et al., 2009; Franklin and Quail, 2010; Li et al., 2011b; Wang and Wang, 2015). Phytochromes exist in two photoreversible forms: the inactive red light-absorbing (Pr) form and the active far-red light-absorbing (Pfr) form. Upon R light absorption, the Pr form is converted to the Pfr form, which can switch back to the Pr form upon FR irradiation (Whitelam et al., 1998). The activated Pfr form can be translocated to the nucleus, where it directly interacts with a group of transcription factors of the basic helix-loop-helix (bHLH) family, named phytochrome interacting factors (PIFs), triggering global gene expression changes and developmental programs (de Lucas and Prat, 2014; Leivar and Monte, 2014).
Among the phytochrome photoreceptors, phyA is the primary photoreceptor for perceiving FR light, whereas phyB to phyE are R light photoreceptors, with phyB playing a predominant role (Wang and Deng, 2003). Previous studies have shown that nuclear translocation of photoactivated phyA requires two chaperone proteins, FAR-RED ELONGATED HYPOCOTYL1 (FHY1) and FHY1-LIKE (FHL), and that two transposase-derived transcription factors, FHY3 and FAR-RED-IMPAIRED RESPONSE1 (FAR1), are required for transcriptional activation of FHY1 and FHL, phyA nuclear translocation, and subsequent FR responses (Lin et al., 2007). Numerous studies have subsequently shown that FHY3/FAR1 also function as key regulators of a diverse array of developmental and physiological responses, such as the UV-B response, circadian clock entrainment, flowering time control, chloroplast biogenesis, chlorophyll biosynthesis, homeostasis of reactive oxygen species, ABA signaling, branching, and plant architecture (Wang and Wang, 2015; Siddiqui et al., 2016).
When grown in the canopy density with decreased R/FR ratios of daylight, plants undergo multifaceted adjustments in growth and development, known as the shade avoidance syndrome (SAS), increasing a plant’s ability to compete for light and chance of reproductive success (Franklin, 2008; Casal, 2013). A major aspect of SAS is the allocation of more resources to elongation growth of plants, which may allow them to outcompete neighbors for light capture. Previous studies have identified numerous regulators of SAS acting downstream of the phytochrome photoreceptors in Arabidopsis, including the PIFs PIF1, PIF3, PIF4, PIF5, and PIF7, LONG HYPOCOTYL IN FR LIGHT (HFR1), PHYTOCHROME RAPIDLY REGULATED1 (PAR1), and PAR2. These factors act in concert to regulate the expression of downstream auxin biosynthetic or responsive genes (such as TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1, YUCCA, and INDOLE-3-ACETIC ACID INDUCIBLE29 [IAA29]) and cell wall-remodeling genes (such as XYLOGLUCAN ENDOTRANSGLYCOSYLASE15 [XTH15] and XTH33), and consequently elongation growth (Sessa et al., 2005; Bou-Torrent et al., 2008; Hornitschek et al., 2009; Leivar and Quail, 2011; Hao et al., 2012; Xie et al., 2017).
Another important aspect of the shade avoidance response is canopy shade-induced reduction in resistance to abiotic and biotic stresses, as a result of reduced resources allocated to defense (de Wit et al., 2013). The plant hormone jasmonic acid (JA) plays a key role in mediating many plant defense responses (Browse, 2009). Upon perception of JA by the F-box protein CORONATINE INSENSITIVE1 (COI1), a group of JASMONATE ZIM-DOMAIN (JAZ) proteins (key repressors of JA signaling) are targeted for rapid degradation by the 26S proteasome, which in turn unleashes and stabilizes several bHLH transcription factors (such as MYC2, MYC3, and MYC4), triggering downstream defense gene expression and JA-mediated responses (Fonseca et al., 2009; Fernández-Calvo et al., 2011). Previous studies have also established a genetic framework in which JA activates plant defense but represses plant growth through crosstalk with the phytochrome-mediated light signaling pathway, balancing growth and JA-mediated defense responses in the shade (Zhang and Turner, 2008; Hou et al., 2010; Robson et al., 2010). It was reported that both phyA and phyB are required for full JA responses (Moreno et al., 2009; Hou et al., 2010; Cerrudo et al., 2012). Inactivation of Arabidopsis phyB by FR light or simulated shade treatment reduces JA sensitivity by promoting the stability of PIFs and JAZ proteins while destabilizing MYC and DELLA proteins (a group of key repressors of gibberellic acid signaling and repressors of plant growth), thus relieving PIFs and JAZs from the inhibitory effect of DELLAs and allowing them to activate downstream genes and promote growth at the expense of compromised defense (de Lucas et al., 2008; Feng et al., 2008; Ballaré, 2009, 2014; Moreno et al., 2009; Hou et al., 2010; Yang et al., 2012; Chico et al., 2014; Leone et al., 2014). Moreover, previous studies showed that the accumulation of bioactive JA is reduced under shade environments in common milkweed (Asclepias syriaca) and lima bean (Phaseolus lunatus; Radhika et al., 2010; Agrawal et al., 2012). However, despite the strides made in this area of research, additional players and mechanisms likely exist and remain to be identified.
In this study, we report that the fhy3 and fhy3 far1 loss-of-function Arabidopsis mutants exhibit an exaggerated shade avoidance response and are insensitive to JA-mediated growth repression under shade. We found that, on the one hand, FHY3 and FAR1 directly upregulate the expression of PAR1 and PAR2, repressing shade-induced growth. On the other hand, FHY3 interacts with the JA signaling regulators MYC2/3/4, promoting JA-responsive defense gene expression and the defense response. Furthermore, we show that the transcriptional activities of FHY3 and FAR1 can be antagonized by JAZ proteins through physical interaction. Our collective data support the conclusion that the JAZ-FHY3 regulatory module plays a vital role in balancing the growth-defense tradeoff under shade conditions.
RESULTS
fhy3 and fhy3 far1 Mutants Exhibit Exaggerated Hypocotyl Elongation under Simulated Shade Conditions
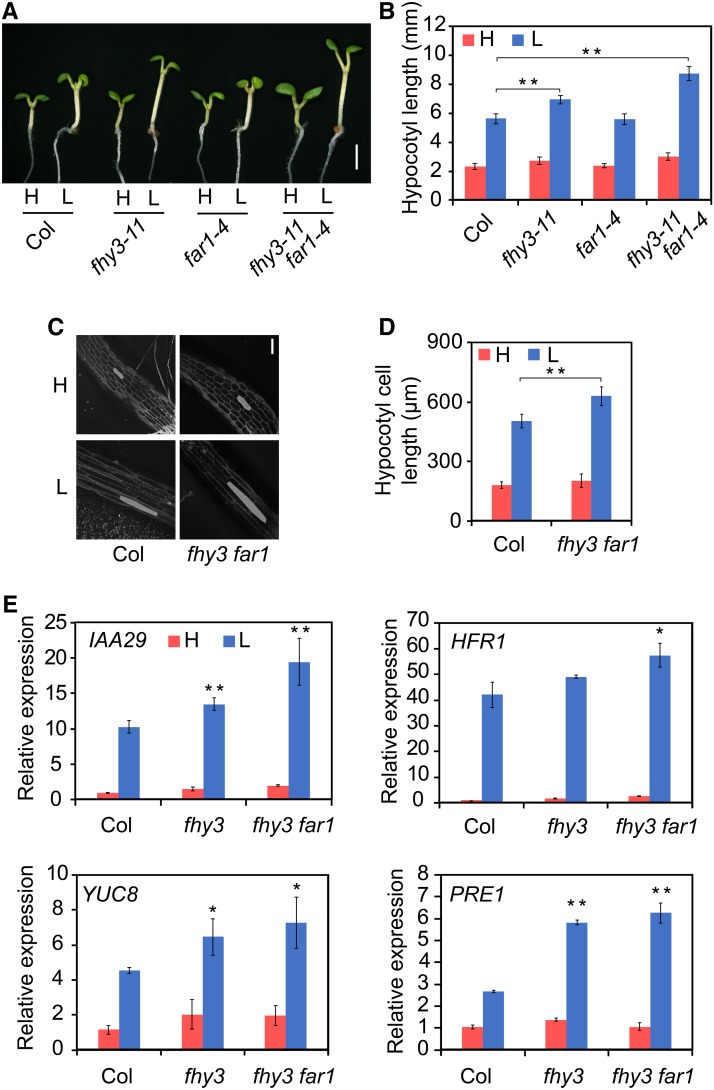
To determine whether FHY3 and FAR1 are involved in the regulation of shade avoidance responses, we examined the hypocotyl elongation phenotype of the wild type (Col-0 and No-0) and fhy3-11, far1-4, fhy3-11 far1-4, fhy3-4, far1-2, and fhy3-4 far1-2 mutants. Seedlings were grown for 3 d under white light (high R/FR) and then were either maintained under high R/FR or transferred to low R/FR (simulated shade) for another 3 d. Under white light, all mutant lines appeared similar to the wild type. By contrast, the fhy3 mutant, especially the fhy3 far1 double mutant, displayed a significantly longer hypocotyl than the wild type (Figures 1A and 1B; Supplemental Figure 1). Microscopy observation revealed that the more elongated hypocotyl length in the fhy3 mutant was primarily due to increased cell length (Figures 1C and 1D). Consistent with this, RT-qPCR analysis revealed that the expression of several representative shade-induced genes (IAA29, HFR1, YUC8, and PRE1) was more significantly upregulated by low R/FR treatment in the fhy3 single mutant and fhy3 far1 double mutant (Figure 1E). Thus, FHY3 and FAR1 act as repressors of elongation growth that prevent exaggerated growth under simulated shade.
Figure 1.
The fhy3 and fhy3 far1 Mutants Exhibit Exaggerated Hypocotyl Elongation under Simulated Shade.
(A) and (B) Hypocotyl length of the wild type and fhy3-11, far1-4, and fhy3-11 far1-4 mutants grown under white light and simulated shade conditions. Three-day-old seedlings were either retained in white light (H, high R/FR) or moved to simulated shade (L, low R/FR) for 3 d. Images of the representative seedlings are shown in (A). Bar = 2 mm. Quantification of hypocotyl length is shown in (B). Asterisks indicate significant differences between the indicated means with P < 0.01 by Student’s t test. Data are presented as means ± sd, n > 15.
(C) and (D) Hypocotyl cell length measurement in the wild type and fhy3-11 and fhy3-11 far1-4 mutants grown under white light and simulated shade conditions. Representative images of hypocotyl cells are shown in (C). Bar = 200 μm. Quantification of cell length is shown in (D). **, P < 0.01, Student’s t test. Data are means ± sd, including >50 cells from three to four independent seedlings.
(E) RT-qPCR of the expression of representative shade marker genes (IAA29, HFR1, YUC8, and PRE1) in wild-type, fhy3-11, and fhy3-11 far1-4 mutant seedlings grown under white light and simulated shade conditions. Significant differences between the wild type and mutants are indicated by asterisks: *, P < 0.05 and **, P < 0.01, Student’s t test. Data are means ± sd, n = 3.
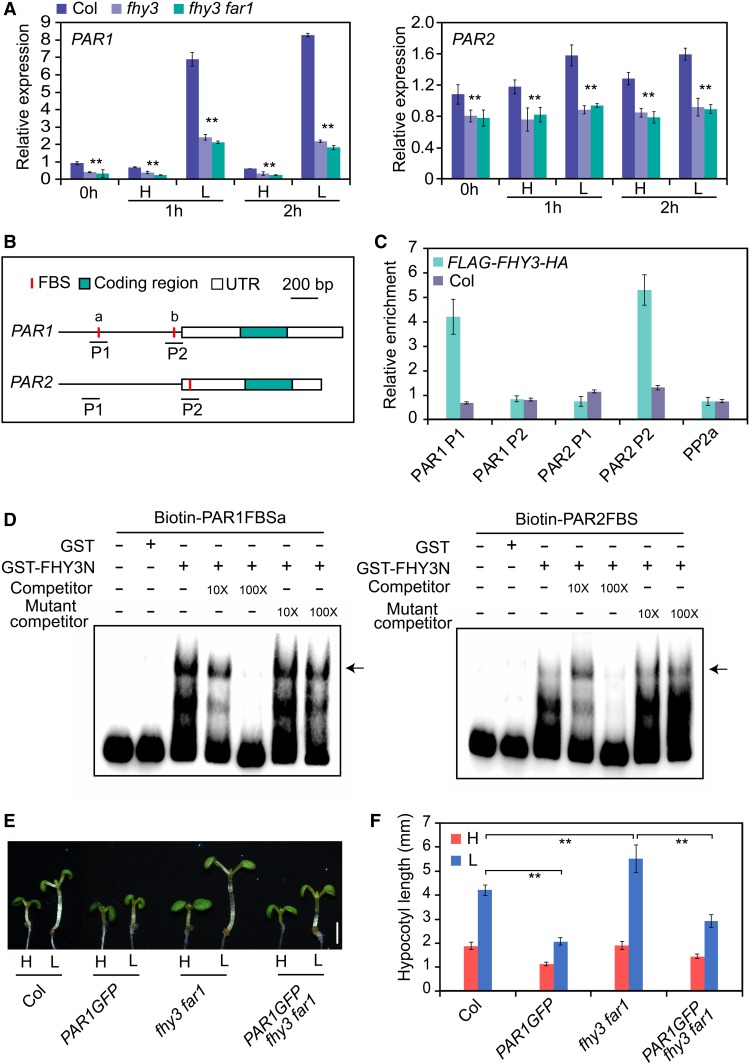
To investigate the molecular mechanisms by which FHY3 and FAR1 regulate elongation growth, we examined the previously published data of genome-wide binding sites of FHY3 (Ouyang et al., 2011) and found that PAR1 and PAR2 are potential direct targets of FHY3. To confirm this notion, we first examined the expression of PAR1 and PAR2 in the wild type and fhy3 and fhy3 far1 mutants under high and low R/FR ratios. The rapid induction of PAR1 and PAR2 by simulated shade was significantly reduced in the fhy3 and fhy3 far1 mutants compared with the wild type (Figure 2A), suggesting that FHY3 and FAR1 are required for shade-induced expression of PAR1 and PAR2. Consistent with this finding, immunoblot assay showed that the levels of FHY3 protein in Pro35S:FLAG-FHY3-HA transgenic seedlings rapidly increased after exposure to simulated shade (1, 4, and 10 h; Supplemental Figure 2A), despite the transcript levels of FHY3 and FAR1 in wild-type seedlings being mildly reduced after exposure to low R/FR (Supplemental Figure 2B). These observations suggest that the half-life of FHY3 protein increased in response to simulated shade treatment. Next, we examined the promoter sequences of PAR1 and PAR2 and found the presence of putative FHY3/FAR1 binding sites (FBS) in the promoters of both PAR1 and PAR2 (Figure 2B). Using chromatin immunoprecipitation (ChIP) and electrophoretic mobility shift assays (EMSAs), we verified that FHY3 could specifically recognize the FBS site in the PAR1 and PAR2 promoters. Notably, the region containing the FBSa site rather than the FBSb site in the PAR1 promoter was occupied by FHY3 and FAR1 (Figures 2C and 2D; Supplemental Figure 3). To further analyze the genetic relationship between FHY3/FAR1 and PAR1 in regulating SAS, we generated PAR1-GFP fhy3 far1 via genetic crosses and examined their responses to simulated shade treatment. Consistent with previous reports (Roig-Villanova et al., 2007; Galstyan et al., 2011), overexpressing PAR1 or PAR2 (PAR1-GFP or PAR2-GFP) displayed a reduced hypocotyl length phenotype, while the PAR-RNA interference (RNAi) lines with reduced expression of PAR1 and PAR2 exhibited an elongated hypocotyl length phenotype under simulated shade, in comparison with the wild type (Supplemental Figure 4). As expected, the PAR1-GFP fhy3 far1 plants showed a short hypocotyl phenotype compared with the PAR1-GFP plants under simulated shade conditions (Figures 2E and 2F), supporting the notion that PAR1 and PAR2 act downstream of FHY3 and FAR1 in regulating hypocotyl elongation in response to simulated shade.
Figure 2.
FHY3 and FAR1 Directly Activate PAR1 and PAR2 Expression under Simulated Shade.
(A) qRT-PCR analysis of PAR1 and PAR2 transcripts in wild-type, fhy3-11, and fhy3-11 far1-4 mutant seedlings. Five-day-old seedlings grown under high R/FR (H; 0 h) were exposed to low R/FR (L) for the indicated times. **, P < 0.01, Student’s t test. Data are means ± sd, n = 3.
(B) Schematic diagram of the genomic regions for PAR1 and PAR2. Letters a and b represent two FBS cis-elements in the PAR1 promoter. P1 and P2 indicate the fragments used for amplification in the ChIP-qPCR assay. UTR, untranslated region.
(C) ChIP-qPCR analysis of FHY3 binding to the PAR1 and PAR2 promoter regions. Ten-day-old seedlings of Pro35S:FLAG-FHY3-HA and the wild type were harvested and immunoprecipitated using anti-HA antibody. Values are means ± sd, n = 3.
(D) EMSA showing binding of the GST-FHY3N recombinant protein to biotin-labeled ProPAR1 (left panel) and ProPAR2 (right panel) probes. The arrows indicate the GST-FHY3N protein. Ten- and 100-fold molar excesses of unlabeled probes were used in the competition assay.
(E) and (F) Hypocotyl length of wild-type, PAR1GFP, fhy3-11 far1-4, and PAR1GFP/fhy3-11 far1-4 mutant seedlings grown under white light and simulated shade conditions. Three-day-old seedlings were either retained in white light or transferred to simulated shade for 3 d. Representative images of seedlings are shown in (E). Bar = 2 mm. Quantification of hypocotyl length is shown in (F). **, P < 0.01, Student’s t test. Data are presented as means ± sd, n > 15.
To test whether FHY3 also affects PAR1/PAR2 activity at the posttranscriptional level, we examined PAR1 protein levels in the Pro35S:PAR1-GFP and Pro35S:PAR1-GFP fhy3 far1 seedlings. We found that PAR1 protein displayed a similar accumulation pattern in both backgrounds, accumulating rapidly in response to low R/FR treatments (Supplemental Figures 5A and 5B). As the PAR1-GFP transgene is driven by the constitutive 35S promoter, the above results suggest that PAR1 is likely also regulated at the posttranscriptional level and that this regulation is independent of FHY3/FAR1. Consistent with this notion, no obvious interactions were observed between FHY3/FAR1 and PAR1/PAR2 protein in a yeast two-hybrid assay (Supplemental Figure 5C). Together, our results suggest that transcriptional regulation of PAR1 and PAR2 by FHY3 and FAR1 is a major mechanism repressing shade-induced hypocotyl elongation.
FHY3 and FAR1 Antagonize JA-Mediated Suppression of Hypocotyl Elongation under Simulated Shade Conditions
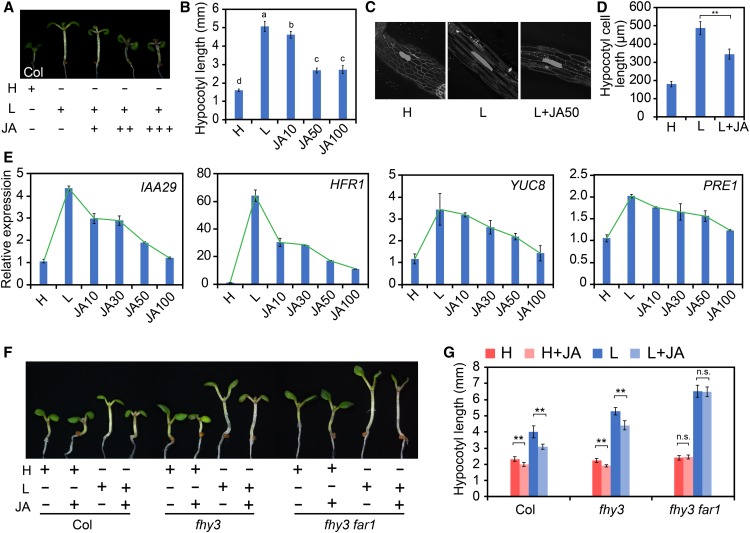
Previous studies have shown that JA can repress hypocotyl elongation and plays a crucial role in the regulation of shade-mediated growth-defense balance (Hou et al., 2010; Yang et al., 2012; Leone et al., 2014). To investigate whether FHY3 and FAR1 function in JA-mediated suppression of hypocotyl elongation, we compared the effect of JA on hypocotyl elongation in the wild type, coi1, JAZ1OE (Pro35S:JAZ1-GUS), fhy3, and fhy3 far1 grown under simulated shade conditions. As expected, the wild-type seedlings displayed a robust inhibition of hypocotyl elongation and shade-induced gene expression by JA treatment (Figures 3A to 3E), and this inhibition was not observed in the coi1 mutant and JAZ1OE lines (Supplemental Figures 6A and 6B). Strikingly, the fhy3 mutant displayed a much-reduced sensitivity, while the fhy3 far1 double mutant was nearly insensitive to the JA treatment under both high and low R/FR conditions (Figures 3F and 3G). Moreover, the PAR-RNAi plants also displayed a reduced response to JA (Supplemental Figure 4). These results support a role of FHY3 and FAR1, as well as PAR1 and PAR2, in JA-mediated suppression of hypocotyl growth.
Figure 3.
The fhy3 and fhy3 far1 Mutants Are Less Sensitive to JA-Mediated Repression of Hypocotyl Elongation.
(A) and (B) Visual image (A) and quantification (B) of the effect of JA on hypocotyl elongation of wild-type seedlings grown under simulated shade. Three-day-old seedlings were either kept in high R/FR or transferred to simulated shade with different concentrations of JA (0, 10, 50, or 100 μM) for 3 d. Different letters indicate significant differences by one-way ANOVA with SAS software (P < 0.05). Data are presented as means ± sd, n > 15.
(C) and (D) Phenotypic analysis of the effect of JA on hypocotyl cell elongation of wild-type seedlings grown under simulated shade. Representative images of hypocotyl cells are shown in (C). Quantification of cell length is shown in (D). Data are means ± sd, including >50 cells from three to four independent seedlings.
(E) qRT-PCR analysis of IAA29, HFR1, YUC8, and PRE1 expression in wild-type seedlings grown in white light or simulated shade with different concentrations of JA (0, 10, 50, or 100 μM). Data are presented as means ± sd, n = 3.
(F) and (G) Representative images (F) and quantification (G) of the effect of JA on wild-type, fhy3-11, and fhy3-11 far1-4 seedlings grown under simulated shade with or without 20 μM JA. **, P < 0.01, Student’s t test; n.s., no significance. Data are presented as means ± sd, n > 15. Bar = 2 mm.
FHY3/FAR1 Physically Interact with JAZ Proteins
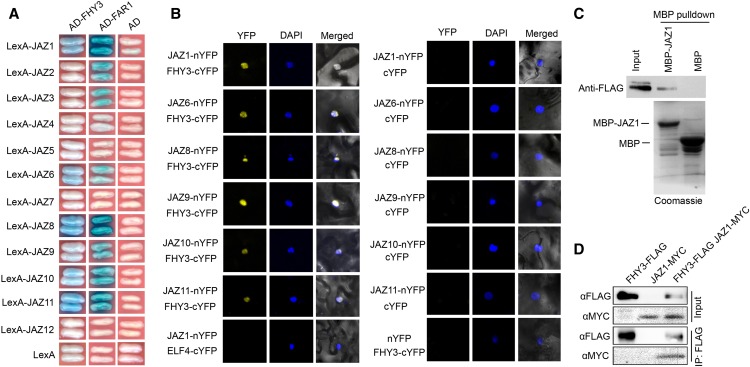
To investigate the possible mechanisms by which FHY3 and FAR1 regulate JA responses, we tested whether they physically interact with known signaling intermediates of the JA pathway using a yeast two-hybrid assay. FHY3 and FAR1 could physically interact with six Arabidopsis JAZ proteins (JAZ1, JAZ6, JAZ8, JAZ9, JAZ10, and JAZ11) but not with COI1 (Figure 4A; Supplemental Figure 7). The interactions between FHY3 and the JAZ proteins were supported by a bimolecular fluorescence complementation (BiFC) assay (Figure 4B). Additionally, the interaction between FHY3 and JAZ1 was further verified by pull-down and in vivo coimmunoprecipitation (Co-IP) assays (Figures 4C and 4D). To define the domains responsible for their interaction, we generated various deletion constructs for both JAZ1 and FHY3. The yeast two-hybrid results showed that the N-terminal domain of JAZ1 and the central transposase domain and the C-terminal SWIM domain of FHY3 are required for the interaction between JAZ1 and FHY3 (Supplemental Figure 8).
Figure 4.
FHY3 and FAR1 Interact with JAZs.
(A) Yeast two-hybrid assay showing interactions of JAZs with FHY3 and FAR1. The JAZ proteins were fused with the LexA DNA binding domain in pEG202. FHY3 and FAR1 were fused with the activation domain (AD) in pB42AD.
(B) BiFC assay confirming interactions between FHY3 and JAZs (JAZ1, JAZ6, JAZ8, JAZ9, JAZ10, and JAZ11) in N. benthamiana leaves. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI).
(C) In vitro pull-down assay verifying the interaction of FHY3 with JAZ1. The purified MBP and MBP-JAZ1 fusion proteins were incubated with the total protein extract from Pro35S:FLAG-FHY3-HA seedlings. The precipitates were subject to immunoblotting using anti-FLAG antibody. The purified MBP and MBP-JAZ1 proteins were separated by SDS-PAGE and stained by Coomassie blue.
(D) Co-IP assay showing that FHY3 associates with JAZ1 in N. benthamiana leaves in vivo. Protein extracts expressing FHY3-FLAG and JAZ1-MYC were immunoprecipitated using anti-FLAG antibody and detected using anti-FLAG (1:4000) or anti-MYC (1:1000) antibody, respectively.
JAZ Proteins Repress the Transcriptional Activity of FHY3 and FAR1 on PAR1
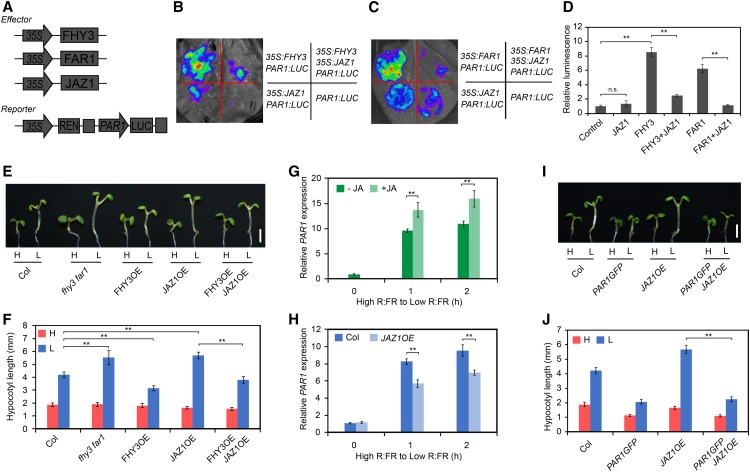
Previous studies have reported that overexpression of JAZ1 in Arabidopsis causes an exaggerated hypocotyl elongation phenotype (Robson et al., 2010; Supplemental Figure 9), similar to the fhy3 and fhy3 far1 mutants. Thus, we speculated that FHY3 and JAZ proteins might act antagonistically in regulating hypocotyl growth. To test this, we examined the effect of JAZ proteins on FHY3-mediated target gene expression in a transient transcriptional assay using Nicotiana benthamiana leaf. We used a dual-LUC reporter system that includes a firefly luciferase (LUC) gene driven by the PAR1 promoter and a Renilla luciferase (REN) gene driven by the constitutive 35S promoter. Both FHY3 and FAR1 could effectively activate the ProPAR1:LUC reporter gene expression, whereas coexpression of JAZ1 with FHY3 or FAR1 significantly repressed the expression of the ProPAR1:LUC reporter gene (Figures 5A to 5D). Similarly, several other JAZ proteins (JAZ3, JAZ6, JAZ8, and JAZ10) tested also repressed the activation of ProPAR1:LUC by FHY3 (Supplemental Figure 10). Moreover, genetic analysis revealed that the more elongated hypocotyl phenotype of the JAZ1OE transgenic plants under simulated shade conditions was fully rescued by overexpression of FHY3 (Figures 5E and 5F). Consistent with this, expression of shade-induced markers (IAA29 and PRE1) was also significantly reduced in the FHY3OE JAZ1OE plants, in comparison with the JAZ1OE plants (Supplemental Figure 11). In further support of this notion, we found that the expression levels of PAR1 and PAR2 were enhanced by JA treatment but repressed by JAZ1 (Figures 5G and 5H) and that overexpression of PAR1 could substantially suppress the hypocotyl elongation phenotype of the JAZ1OE plants (Figures 5I and 5J). Together, these data suggest that the exaggerated hypocotyl elongation phenotype of the JAZ1OE plants is mediated through FHY3, PAR1, and PAR2 (at least partially).
Figure 5.
JAZ1 Represses the Transcriptional Activity of FHY3 and FAR1.
(A) Schematic diagram of the PAR1 promoter-driven dual-LUC reporter and effector constructs.
(B) and (C) Transient expression assay in N. benthamiana leaves. Representative images of N. benthamiana leaves 72 h after infiltration are shown.
(D) Quantification of the transient expression assay. The relative LUC activities normalized to the REN activity are shown (LUC/REN). Significant differences are indicated: **, P < 0.01, Student’s t test; n.s., no significance. Values are means ± sd, n = 3.
(E) and (F) Hypocotyl length of wild-type, fhy3-11 far1-4, FHY3OE, JAZ1OE, and FHY3OE JAZ1OE seedlings grown under simulated shade conditions. Representative images of seedlings are shown in (E). Bar = 2 mm. Quantification of hypocotyl length is shown in (F). **, P < 0.01, Student’s t test. Data are means ± sd, n > 15.
(G) qRT-PCR analysis of PAR1 expression in wild-type seedlings treated with JA. Five-day-old seedlings were grown in high R/FR (0 h) and then transferred to low R/FR supplemented with 50 μM JA for 1 and 2 h. **, P < 0.01, Student’s t test. Data are means ± sd, n = 3.
(H) qRT-PCR analysis of PAR1 expression in wild-type and JAZ1OE plants. Five-day-old seedlings were grown in high R/FR (0 h) and then exposed to low R/FR for the indicated times. **, P < 0.01, Student’s t test. Data are means ± sd, n = 3.
(I) and (J) Hypocotyl length of wild-type, PAR1-GFP, JAZ1OE, and PAR1-GFP JAZ1OE seedlings grown under high and low R/FR ratios. Representative images of seedlings are shown in (I). Bar = 2 mm. Quantification of hypocotyl length is shown in (J). **, P < 0.01, Student’s t test. Data are means ± sd, n > 15.
Genome-Wide Effects of FHY3 and FAR1 on Shade-Responsive and JA-Responsive Gene Expression
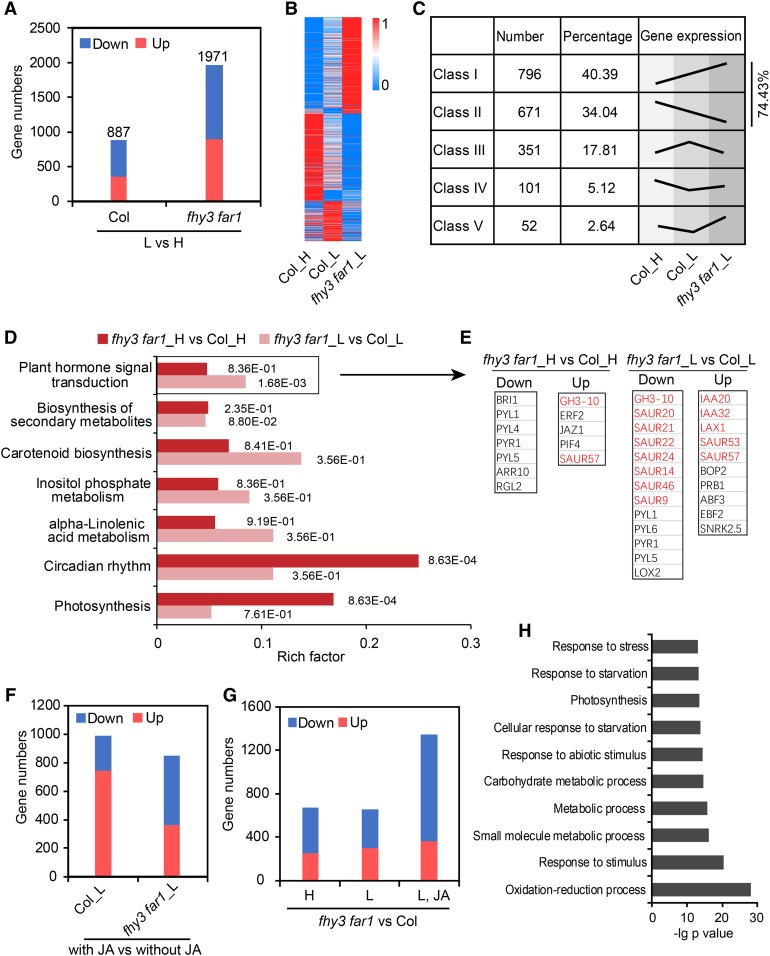
To further substantiate the role of FHY3/FAR1 in mediating the crosstalk between phytochrome-mediated signaling and JA-mediated signaling pathways under simulated shade, we performed RNA sequencing (RNA-seq) analysis using wild-type and fhy3 far1 mutant seedlings grown under high or low R/FR supplemented with or without JA (termed Col_H, Col_L, fhy3 far1_H, and fhy3 far1_L, respectively; Supplemental Data Set 1). The total number of differentially expressed genes (DEGs; both upregulated and downregulated combined) by shade were much higher in the fhy3 far1 mutant (1971 genes) than in the wild-type plants (887 genes; Figure 6A; Supplemental Data Set 2). These DEGs could be categorized into five classes (class I to V) in Col_H, Col_L, and fhy3 far1_L (Figures 6B and 6C). Notably, a large number of genes affected by shade (74.43%) displayed enhanced expression in the fhy3 far1 mutant (Figure 6C), suggesting that FHY3 and FAR1 may serve as a brake that prevents excessive expression of shade-responsive genes. In addition, the enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were substantially different for the DEGs in high and low R/FR conditions. In high R/FR conditions, FHY3/FAR1 primarily regulate genes involved in circadian rhythm and photosynthesis, whereas in low R/FR, the DEGs are more enriched in phytohormone signal transduction pathways, with particular enrichment for auxin-regulated genes (Figures 6D and 6E).
Figure 6.
RNA-Seq Analysis of the Wild Type and the fhy3 far1 Mutant in Response to Shade or JA Treatments.
(A) The number of DEGs (upregulated and downregulated) in Col and the fhy3-11 far1-4 mutant grown under simulated shade compared with that grown under white light (L vs H).
(B) Heatmap of DEGs in Col_H, Col_L, and fhy3 far1_L. The scale bar indicates the normalized FPKM value.
(C) These DEGs are clustered into five classes (I–V) based on their expression patterns in (B).
(D) KEGG analysis shows that diverse pathways are enriched among the DEGs regulated by FHY3/FAR1 in high and low R/FR conditions. Numbers next to the bars indicate corrected P values.
(E) Representative genes in the significantly enriched pathway of “plant hormone signal transduction” in (D). Auxin-related genes are shown in red.
(F) The number of DEGs (upregulated and downregulated) in Col and the fhy3 far1 mutant under shade supplemented with or without JA.
(G) DEGs between the wild type and the fhy3 far1 mutant grown under three conditions: high R/FR, low R/FR, and low R/FR plus JA treatment.
(H) Gene Ontology analysis of the genes downregulated in the fhy3 far1 mutant grown in simulated shade supplemented with JA.
On the other hand, we found that the number of genes upregulated by JA was substantially reduced in the fhy3 far1 mutant compared with the wild type grown in low R/FR (Figure 6F; Supplemental Data Set 3). Furthermore, comparison of the DEGs in three different growth conditions (high R/FR, low R/FR, with or without JA) showed that the number of downregulated genes was markedly increased in the sample grown under low R/FR with JA treatment, while the number of upregulated DEGs seemed not to be affected in plants grown under these three conditions (Figure 6G; Supplemental Data Set 4). These observations suggest that FHY3 and FAR1 play a more predominant role in regulating the expression of JA-induced downstream genes under low R/FR conditions. Moreover, Gene Ontology analysis showed that stress response-related genes were enriched in these downregulated genes in the fhy3 far1 mutant (Figure 6H). Together, these data suggest that FHY3 and FAR1 negatively regulate shade-responsive genes but positively regulate genes involved in JA signaling at the genome-wide level in response to simulated shade.
FHY3 and FAR1 Modulate JA-Dependent Defense Responses under Simulated Shade Conditions
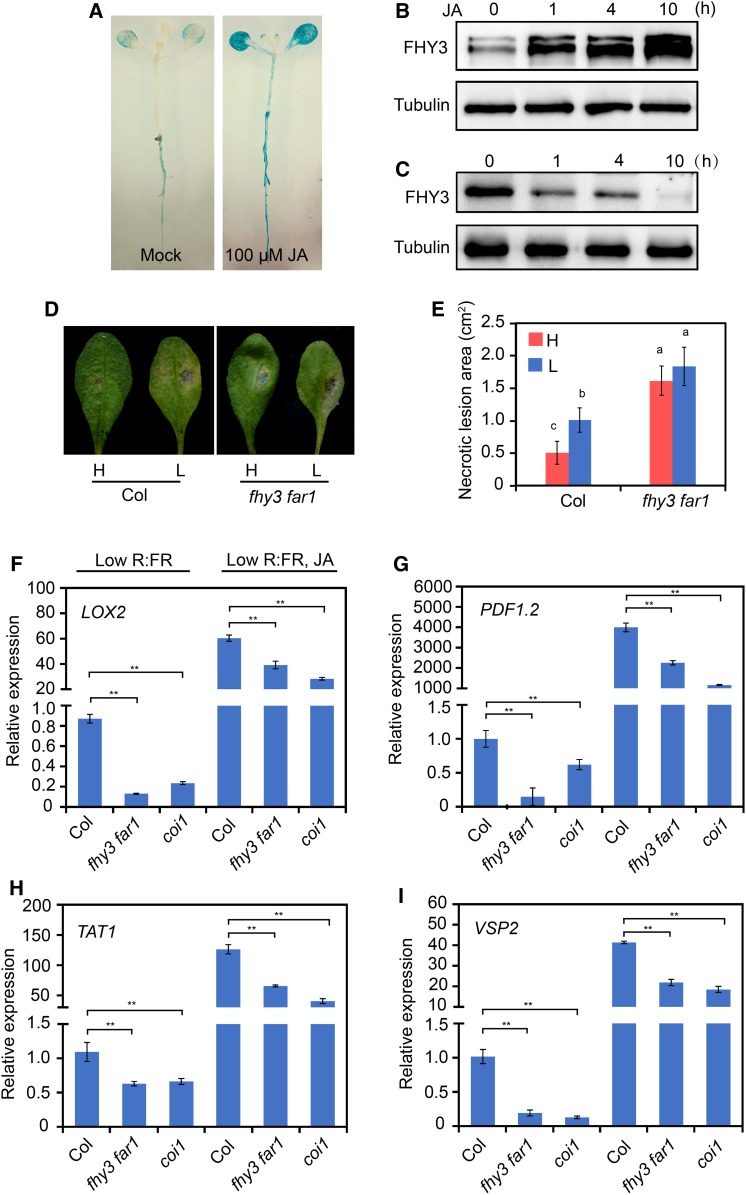
To further investigate whether FHY3 is involved in the JA signaling pathway, we examined the effects of JA on the transcript level and protein accumulation of FHY3. RT-qPCR analysis showed that JA treatment only caused a mild upregulation of the FHY3 transcript level (less than twofold; Supplemental Figure 12). However, histochemical staining of the Pro35S:GUS-FHY3 transgenic seedlings showed that in the presence of JA, accumulation of the FHY3-GUS fusion protein markedly increased upon JA treatment (Figure 7A). Consistent with this, immunoblot analysis also showed increased accumulation of FHY3 protein in 7-d-old Pro35S:FLAG-FHY3-HA transgenic seedlings treated with JA (Figure 7B). Consistent with this, JAZ1 overexpression inhibited FHY3 protein accumulation in the Pro35S:FHY3-FLAG JAZ1OE transgenic plants in response to simulated shade treatment (Figure 7C). These results suggest that FHY3 was predominantly regulated at a posttranscriptional level by JA.
Figure 7.
The fhy3 far1 Mutant Is More Susceptible to B. cinerea under Simulated Shade.
(A) Histochemical staining of 10-d-old Pro35S:GUS-FHY3 seedlings treated with 100 μM JA or an equal volume of ethanol (Mock) for 10 h.
(B) Immunoblot assay shows that JA promotes accumulation of FHY3 protein. Five-day-old Pro35S:FLAG-FHY3-HA seedlings were treated with 50 μM JA for the indicated times. FHY3 protein was detected with anti-FLAG antibody (1:4000; MBL). Tubulin was used as a loading control.
(C) Immunoblot assay shows that accumulation of FHY3 protein decreased in the Pro35S:FHY3-FLAG JAZ1OE seedlings treated with low R/FR for the indicated times.
(D) Disease symptoms of wild-type and fhy3-11 far1-4 plants incubated with B. cinerea. Four-week-old rosette leaves were incubated with 5 × 105 spores/mL and then placed under white light or simulated shade for 2 d before observation.
(E) Quantification of the lesion areas on rosette leaves after inoculation with B. cinerea spores. Different letters denote statistically significant differences by one-way ANOVA (P < 0.05). Data are means ± sd; three independent experiments were performed with similar results.
(F) to (I) qRT-PCR analysis of JA-responsive genes (LOX2, PDF1.2, TAT1, and VSP2) in wild-type, coi1-2, and fhy3-11 far1-4 plants grown under simulated shade conditions with or without JA application for 3 d. Asterisks indicate significant differences between the indicated means with P < 0.05 by Student’s t test. Data are means ± sd, n = 3.
Next, we examined whether FHY3 is involved in JA-mediated plant defense responses. We analyzed the defense responses of the wild type and the fhy3 far1 mutant against the necrotrophic fungus Botrytis cinerea under white light or simulated shade conditions. Two days after incubation with B. cinerea spores, the wild-type plants exhibited an increased susceptibility to B. cinerea under simulated shade. Intriguingly, the fhy3 far1 mutant was significantly more susceptible to this pathogen under both white light and shade conditions, suggesting that FHY3 and FAR1 play a role in defense against this pathogen (Figures 7D and 7E). Consistent with this notion, expression of several typical JA-responsive genes, including LOX2, PDF1.2, TAT1, and VSP2 (Lorenzo et al., 2004), was significantly reduced in the fhy3 far1 mutant as in the coi1 mutant, under simulated shade with or without JA treatment (Figures 7F to 7I). Taken together, these data suggest that FHY3 and FAR1 play a role in the JA-mediated defense response against necrotrophic pathogens.
FHY3 Interacts with MYC2 to Regulate JA-Responsive Defense Gene Expression
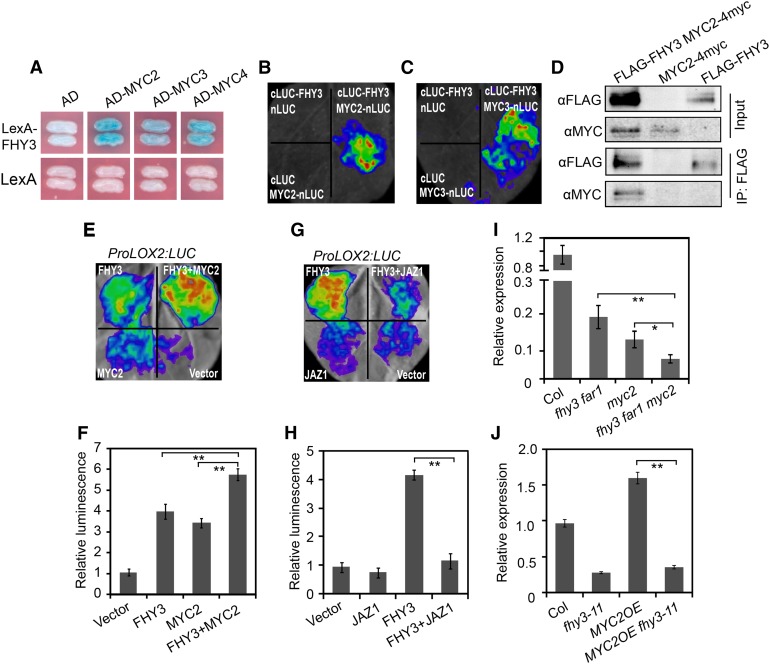
To explore the mechanism by which FHY3 and FAR1 participate in the JA-mediated plant defense response, we tested whether FHY3 could interact with MYC2, MYC3, and MYC4, three bHLH transcription factors that are master regulators of JA signaling. A yeast two-hybrid assay revealed that MYC2, MYC3, and MYC4 all directly interacted with FHY3 (Figure 8A). We confirmed the interaction between FHY3 with MYC2 and MYC3 using a firefly luciferase complementation imaging (LCI) assay and in vivo Co-IP assay (Figures 8B to 8D). To examine the effect of the FHY3-MYC2 interaction on the expression of downstream JA-responsive genes, we constructed a LUC reporter gene driven by the promoter of LOX2 (ProLOX2:LUC), which is a direct target of MYC2 (Hou et al., 2010). As expected, the expression level of ProLOX2:LUC was significantly induced by coexpression with either FHY3 or MYC2 (Figures 8E and 8F). In addition, similar to the reported repression of MYC2 activity by JAZ1 (Pauwels et al., 2010), the transcriptional activation activity of FHY3 on ProLOX2:LUC expression was also suppressed by JAZ1 (Figures 8G and 8H). Coexpression of FHY3 with MYC2 proteins activated the reporter gene expression to a higher level compared with the effect of FHY3 or MYC2 alone (Figures 8E and 8F). These results suggest that FHY3 could enhance the activity of MYC2 on JA-responsive genes. Consistent with this, RT-qPCR analysis showed that LOX2 expression was significantly reduced in the fhy3 far1 myc2 triple mutant compared with the fhy3 far1 and myc2 mutants (Figure 8I). In addition, the high expression level of LOX2 in the MYC2 overexpression lines (MYC2OE) disappeared when FHY3 was inactivated (Figure 8J). Moreover, by examining the publicly available target genes of FHY3 (Ouyang et al., 2011) and MYC2 (Dombrecht et al., 2007), we identified 66 putative common target genes potentially coregulated by FHY3 and MYC2. Among them, many are defense-related genes, such as JAZ8, ERF4, and ERF6 (Supplemental Data Set 5). Together, these results suggest that FHY3 acts together with MYC2 to regulate JA-responsive defense gene expression.
Figure 8.
FHY3 Interacts with MYC2 to Coordinately Regulate LOX2 Expression.
(A) Yeast two-hybrid assay shows that FHY3 interacts with MYC2, MYC3, and MYC4. FHY3 was fused with the LexA DNA binding domain in pEG202. MYC2, MYC3, and MYC4 were individually fused with the activation domain (AD) in pB42AD.
(B) Firefly LCI assay shows that FHY3 interacts with MYC2 in plant cells. N. benthamiana leaves were transformed with the construct pairs cLUC-FHY3/nLuc, cLUC-FHY3/nLuc-MYC2, and cLuc/nLuc-MYC2.
(C) Firefly LCI assay shows that FHY3 interacts with MYC3 in plant cells. N. benthamiana leaves were transformed with the construct pairs cLUC-FHY3/nLuc, cLUC-FHY3/nLuc-MYC3, and cLuc/nLuc-MYC3.
(D) Co-IP assay showing that FHY3 associates with MYC2 in planta. Protein extracts from 6-d-old seedlings expressing FLAG-FHY3-HA and MYC2-4myc were immunoprecipitated using anti-FLAG antibody and blotted with anti-FLAG or anti-MYC antibody, respectively.
(E) and (F) Transient expression assay shows that FHY3 and MYC2 coordinately regulate ProLOX2:LUC expression. The N. benthamiana leaves were infiltrated with A. tumefaciens transformed with the indicated reporter and effector constructs. (E) shows a representative image of luciferase activity in an N. benthamiana leaf. (F) shows quantification of the transient expression assay. **, P < 0.01, Student’s t test. Values are means ± sd, n = 3.
(G) and (H) Transient expression assay shows that JAZ1 represses the transcriptional activity of FHY3 on ProLOX2:LUC. (G) shows a representative image of luciferase activity in an N. benthamiana leaf. (H) shows quantification of the transient expression assay. **, P < 0.01, Student’s t test. Values are means ± sd, n = 3.
(I) qRT-PCR analysis of LOX2 expression in Col, fhy3-11 far1-4, myc2-2, and fhy3-11 far1-4 myc2-2 plants grown under simulated shade. *, P < 0.05 and **, P < 0.01, Student’s t test. Data are means ± sd, n = 3.
(J) qRT-PCR analysis of LOX2 expression in Col, fhy3-11, MYC2OE, and MYC2OE fhy3-11 seedlings grown under simulated shade. **, P < 0.01, Student’s t test. Data are means ± sd, n = 3.
DISCUSSION
As plants are sessile organisms, it is critical that the limited resources available are precisely allocated between growth and defense responses under shaded or other unfavorable environmental conditions. In this study, we unveiled a novel mechanism employed by plants to make such a decision: the Arabidopsis FHY3 and FAR1 proteins act at the nexus of the light and JA signaling pathways to coordinately regulate the balance between growth and defense. We show that under shade conditions, FHY3 and FAR1 repress plant growth through directly activating the expression of two negative regulators of elongation growth, PAR1 and PAR2, and that this process is antagonized by the JA signaling repressor JAZ proteins through physical interactions. We further show that FHY3 interacts with MYC2 to coordinately activate JA-responsive defense gene expression. Our results provide insight into the complex interplay between light and hormone signaling pathways that help plants to maximize growth and survival.
FHY3 and FAR1 Repress Elongation Growth by Activating PAR1 and PAR2
Previous studies have identified a group of atypical bHLH proteins, including HFR1, PAR1, and PAR2, that inhibit the shade avoidance response by forming non-DNA binding heterodimers with the PIF transcription factors, thereby influencing the expression of downstream auxin biosynthetic genes and cell wall-remodeling genes (Sessa et al., 2005; Roig-Villanova et al., 2007; Bou-Torrent et al., 2008; Hornitschek et al., 2009; Leivar and Quail, 2011; Hao et al., 2012). Furthermore, recent studies showed that shade reduces JA sensitivity by promoting the stability of PIFs and JAZ proteins (both of which promote plant growth) while destabilizing DELLA proteins (a group of key repressors of gibberellic acid signaling and repressors of plant growth), thus relieving PIFs and JAZs from the inhibitory effect of DELLAs and allowing them to activate downstream genes and promote growth at the expense of compromised defense (de Lucas et al., 2008; Feng et al., 2008; Ballaré, 2009, 2014; Hou et al., 2010; Yang et al., 2012; Chico et al., 2014; Leone et al., 2014; Xie et al., 2017).
In this study, we showed that the fhy3 and fhy3 far1 mutants display an exaggerated hypocotyl elongation phenotype under simulated shade. Thus, FHY3 and FAR1 define two novel repressors of the shade avoidance response. We demonstrate that FHY3 and FAR1 repress hypocotyl elongation by directly binding to the promoters of PAR1 and PAR2 and activating their expression in response to shade (Figures 2A to 2D and 5A to 5D). In support of this notion, we found that expression of PAR1 and PAR2 was significantly reduced in the fhy3 and fhy3 far1 mutants under simulated shade (Figure 2A) and that overexpression of PAR1 could largely suppress the long hypocotyl phenotype of the fhy3 far1 mutant under simulated shade conditions (Figures 2E and 2F). We also showed that, like other negative regulators of the shade response (HFR1, PIL1, and PAR1; Sessa et al., 2005; Roig-Villanova et al., 2006; Hornitschek et al., 2009), the accumulation of FHY3 protein was rapidly induced by simulated shade treatment (Supplemental Figure 2A). Together, these results suggest that, in response to simulated shade or vegetation proximity, increased FHY3 protein accumulation activates the expression of PAR1 and PAR2 to repress excessive elongation growth.
Interestingly, we found that JAZ proteins, which were stabilized under shade conditions (Robson et al., 2010; Leone et al., 2014), could repress the transcriptional activation activity of FHY3 and FAR1 on PAR1 through physical interactions, thus antagonizing the repressive function of FHY3 and FAR1 on elongation growth (Figure 5). We also showed that JA could promote the accumulation of FHY3 protein through a posttranscriptional mechanism (Figures 7A and 7B). These results suggest that JA signaling can enhance the activity of FHY3 and FAR1 via at least two distinct mechanisms: increasing its protein accumulation and unleashing them from the inhibition by JAZ proteins, and ultimately leading to repressed elongation growth. This notion is consistent with the earlier findings that the JAZ1 overexpression plants display an exaggerated hypocotyl elongation phenotype (Robson et al., 2010) and that the long hypocotyl phenotype of the JAZ1 overexpression plants can be substantially suppressed by overexpression of either FHY3 or PAR1 (Figures 5E and 5I). It is interesting that two JAZ genes, JAZ3 and JAZ8, were identified as two potential direct target genes of FHY3 in an earlier study (Ouyang et al., 2011), suggesting the existence of a possible feedback regulatory mechanism between FHY3 and JAZs.
It should be noted that previous studies reported that the shade avoidance response is attenuated in the Arabidopsis plants heterologously expressing oat (Avena sativa) phyA or overexpressing Arabidopsis phyB, concomitant with reduced PAR1 expression and other shade-induced genes (such as ATHB2, ATHB4, and HA2) that are proposed to be primary direct target genes of phytochrome signaling (Roig-Villanova et al., 2006). Although the detailed molecular mechanisms remain to be elucidated, the findings suggest that the rapid induction of PAR genes by shade is desensitized by overexpression of phytochromes; this may help the plants to fine-tune the levels of PAR gene expression and ensure a proper level of shade avoidance response. It will be interesting to investigate whether FHY3 and FAR1 are involved in this desensitizing mechanism. In addition, it has been shown that Arabidopsis phyA and phyB play antagonistic roles in regulating the SAS, which could help plants to distinguish vegetation proximity and canopy shade. Through the so-called FR-high irradiance response, phyA signaling could suppress excessive elongation growth under prolonged canopy shade conditions, thus conferring an important adaptive value for increasing plant fitness (Casal, 2013; Martínez-García et al., 2014). As FHY3 and FAR1 are essential for nuclear localization of photoactivated phyA and the phyA-mediated FR-high irradiance response (Lin et al., 2007), it will be interesting to further study the possible role of FHY3 and FAR1 in phyA-mediated attenuation of the shade avoidance response under deep canopy shade conditions.
FHY3 and FAR1 Promote the Defense Response by Interacting with MYC2 and Coordinately Regulate Downstream Defense Genes under Shade Conditions
Previous studies have shown that the MYC2, MYC3, and MYC4 transcription factors are required for JA-mediated defense against the necrotrophic pathogen B. cinerea and that these transcription factors are short-lived proteins subject to destabilization by FR light treatment or simulated shade that inactivates phyB (Chico et al., 2014). Moreover, it was found that in the shade, although production of JA could be triggered by pathogen attack, plants were still compromised in their ability to fight back, because of limited accumulation of these MYC proteins (de Wit et al., 2013; Chico et al., 2014). In this study, we showed that the fhy3 far1 mutant was significantly more susceptible to this pathogen under both white light and simulated shade conditions (Figures 7D and 7E), suggesting that FHY3 and FAR1 play a positive role in defense against this pathogen. Consistent with this notion, expression of several typical JA-responsive genes, including LOX2, PDF1.2, TAT1, and VSP2 (Lorenzo et al., 2004), was significantly reduced in the fhy3 far1 mutant as in the coi1 mutant under simulated shade with or without JA treatment (Figures 7F to 7I). Furthermore, we showed that FHY3 could physically interact with MYC2, MYC3, and MYC4 (Figures 8A to 8D) and that coexpression of FHY3 with MYC2 proteins together activated the ProLOX2:LUC reporter gene expression to a higher level than FHY3 or MYC2 alone (Figures 8E and 8F). These results suggest that FHY3 could enhance the activity of MYC proteins on JA-responsive genes. In support of this notion, we found that LOX2 expression was significantly reduced in the fhy3 far1 myc2 triple mutant compared with the fhy3 far1 and myc2 mutants (Figure 8I) and that the high expression level of LOX2 in the MYC2 overexpression lines (MYC2OE) disappeared when FHY3 was mutated (Figure 8J). Furthermore, our RNA-seq analyses revealed that FHY3 and FAR1 play a more predominant role in regulating the expression of JA-induced downstream genes, particularly stress response-related genes in Gene Ontology analysis, under low R/FR conditions (Figures 6G and 6H).
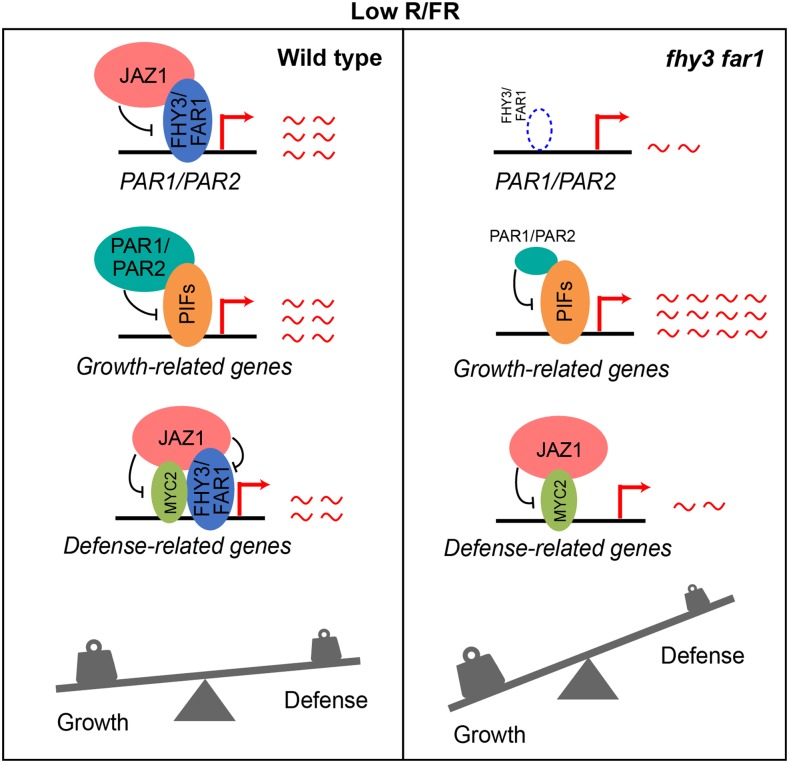
Based on these findings in conjunction with earlier reports, we propose a model for FHY3/FAR1 in mediating the balance between growth and defense in response to simulated shade. In wild-type plants, shade (low R/FR ratios) induces stabilization of PIFs, FHY3, and JAZ proteins while destabilizing MYC2 protein. On the one hand, FHY3 and FAR1 activate the expression of PAR1/PAR2, which inhibits the expression of growth-related genes by forming non-DNA binding heterodimers with PIFs, thus preventing an exaggerated elongation growth. On the other hand, FHY3/FAR1, together with MYC2, activate the expression of JA-responsive defense genes, while JAZ proteins inhibit the activity of FHY3 and FAR1 to maintain a proper level of defense gene expression and defense response. In the fhy3 far1 mutant, loss of FHY3 and FAR1 leads to reduced expression level of PAR1/PAR2, resulting in overexpression of growth-related genes and exaggerated elongation growth. Meanwhile, the expression of JA-responsive defense genes is also reduced due to the absence of FHY3 and FAR1 proteins. As a result, the balance between growth and defense is severely disrupted in the fhy3 far1 mutant plants under shade conditions (Figure 9).
Figure 9.
A Putative Model of FHY3 and FAR1 in Balancing Growth and Defense under Low R/FR Conditions.
In wild-type plants, shade (low R/FR ratios) induces stabilization of PIFs, FHY3, and JAZ proteins but destabilizes MYC2 protein. On the one hand, FHY3 and FAR1 activate the expression of PAR1/PAR2, which inhibits the expression of growth-related genes by forming non-DNA binding heterodimers with PIFs, thus preventing an exaggerated elongation growth. On the other hand, FHY3/FAR1, together with MYC2, activate the expression of JA-responsive defense genes, while JAZ proteins act to inhibit the activity of FHY3 and FAR1 to maintain a proper level of defense gene expression and defense response. In the fhy3 far1 mutant, loss of FHY3 and FAR1 leads to reduced expression level of PAR1/PAR2, resulting in overexpression of growth-related genes and exaggerated elongation growth. Meanwhile, the expression of JA-responsive defense genes is also reduced due to the absence of FHY3 and FAR1 proteins. As a result, the balance between growth and defense is severely disrupted in the fhy3 far1 mutant plants under shade conditions.
It should be noted that previous studies have also shown that FHY3 and FAR1 negatively modulate salicylic acid (SA) accumulation and antagonize SA-mediated resistance to the biotrophic pathogen Pseudomonas syringae. Thus, the fhy3 far1 double mutants exhibit high levels of SA and reactive oxygen species that trigger constitutive defense responses (Wang et al., 2016). In addition, mutual inhibition between the SA and JA signaling pathways has been well documented (Gupta et al., 2000; Kloek et al., 2001; Kunkel and Brooks, 2002). Like the fhy3 mutant (Wang et al., 2016), the coi1 mutant also displays enhanced expression of genes involved in SA-dependent responses (Kloek et al., 2001). Thus, enhanced SA signaling in the fhy3 far1 mutant may also contribute to the impairment of JA signaling. However, a recent study reported that low R/FR ratios could depress Arabidopsis immune responses against necrotrophic microorganisms via an SA-independent mechanism (Cerrudo et al., 2012). Therefore, the detailed crosstalk mechanism between SA and JA in response to shade remains to be further elucidated. In addition, recent studies have shown that DELLA proteins, a group of key repressors of the gibberellin signaling pathway, can suppress the activity of PIFs and JAZs (both of which are growth-promoting factors) and that shade (low R/FR ratios) can trigger the degradation of DELLAs, thereby unleashing the PIF and JAZ proteins to activate the expression of downstream growth-promoting genes, thus prioritizing growth over defense (Djakovic-Petrovic et al., 2007; de Lucas et al., 2008; Feng et al., 2008; Hou et al., 2010; Yang et al., 2012; Leone et al., 2014). Future work is required to further elucidate the complex regulatory interactions between FHY3 and FAR1 with various hormone signaling pathways in the coordinated regulation of plant growth and defense.
METHODS
Plant Materials and Growth Conditions
The wild-type and mutant Arabidopsis (Arabidopsis thaliana) plants used in this study are of the Col-0 genetic background unless otherwise indicated. The coi1-2 (Xu et al., 2002), fhy3-11 (SALK_002711; Stirnberg et al., 2012), far1-4 (SALK_031652; Stirnberg et al., 2012), and myc2-2 (Boter et al., 2004) mutants and the transgenic lines Pro35S:JAZ1-GUS (JAZ1OE; Thines et al., 2007), Pro35S:MYC2-4myc (MYC2OE; Chen et al., 2011), Pro35S:PAR1-GFP, Pro35S:PAR2-GFP, PAR-RNAi (Zhou et al., 2014), Pro35S:FLAG-FHY3-HA (Li et al., 2011a), and Pro35S:GUS-FHY3 (Wang and Deng, 2002) have been reported. The fhy3-4, far1-2, and fhy3-4 far1-2 mutants and the Pro35S:FLAG-FHY3-HA transgenic line are of the No-0 ecotype. The PAR1-GFP fhy3 far1, FHY3OE JAZ1OE, PAR1-GFP JAZ1OE, fhy3-11 far1-4 myc2-2, and MYC2OE fhy3-11 higher order mutant lines were generated by genetic crossing. Seeds were sterilized and germinated on half-strength Murashige and Skoog solid medium containing 1% (w/v) sucrose. After vernalization for 2 d at 4°C, plates were incubated in a growth chamber (Percival Scientific, cool white fluorescent bulb at 22°C) under continuous white light (LED light, PAR = 38 μmol m−2 s−1; red: 640–670 nm, 11 μmol m−2 s−1; FR: 720–750 nm, 1.2 μmol m−2 s−1) for 3 d. The plates were then either kept in white light or transferred to simulated shade (LED continuous white light plus FR, PAR = 30 μmol m−2 s−1; red: 640–670 nm, 22 μmol m−2 s−1; FR: 720–750 nm, 90 μmol m−2 s−1) for 3 d until hypocotyl measurement using ImageJ software.
Plasmid Construction
To generate yeast two-hybrid constructs for JAZs and COI1 and different deletion domains of JAZ1, the amplified coding regions were subcloned into the EcoRI and XhoI sites of pEG202 using the In-Fusion HD cloning kit (Clontech) to generate various LexA fusion constructs. The cDNAs of MYC2, MYC3, and MYC4 were amplified and subcloned into the EcoRI and XhoI sites of pB42AD to generate the AD-MYC2, AD-MYC3, and AD-MYC4 constructs. The AD-FHY3, AD-FAR1, and various deletion constructs of LexA-FHY3 were described previously (Liu et al., 2017). For the transient expression assay, promoters of PAR1 and LOX2 were amplified and cloned into pGreenII 0800-LUC to generate the ProPAR1:LUC and ProLOX2:LUC reporter genes. For the 35S promoter-driven effector constructs, the cDNAs of MYC2, JAZ1, JAZ3, JAZ6, JAZ8, JAZ9, JAZ10, and JAZ11 were amplified and cloned into the SPYNE vector at the BamHI and SalI sites. Pro35S:FHY3 SPYNE and Pro35S:FAR1 SPYNE have been described previously (Liu et al., 2017). To generate the Pro35S:FHY3-FLAG (FHY3OE) construct, the cDNA of FHY3 was amplified and subcloned into pCAMBIA1300-221-FLAG through the XbaI site. At least 10 independent T1 transgenic lines were selected on Murashige and Skoog medium containing hygromycin (50 mg/L) and confirmed using immunoblot analysis with anti-FLAG specific antibodies.
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were performed as described previously (Li et al., 2011a). Briefly, different combinations of activation domain and LexA fusion plasmids were cotransformed into the yeast strain EGY48, which already harbors the p8op:LacZ reporter. Transformants were grown on proper dropout plates (SD/-Trp/-Ura/-His) containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for blue color development.
EMSA
Preparation of the glutathione S-transferase (GST)-FHY3N fusion protein was described previously (Liu et al., 2017). For probe preparation, two complementary oligonucleotides (60 bp long) of the PAR1 and PAR2 promoter containing the FBS sites were synthesized, annealed, and labeled with biotin. The oligonucleotide sequences of the biotin-labeled probes are listed in Supplemental Data Set 6. EMSA was performed using a LightShift Chemiluminescent EMSA kit (Pierce) according to the manufacturer’s instructions. Briefly, biotin-labeled probes were incubated for 20 min with the purified proteins in a binding buffer at room temperature. The DNA-protein complexes were separated on 6% (w/v) native polyacrylamide gels, and the signal was detected using the Biostep Celvin S420 system (Biostep).
RNA Extraction and RT-qPCR
Total RNA was extracted from seedlings using Trizol (Invitrogen). The first-strand cDNA was synthesized from 1 μg of RNA using reverse transcriptase (Tiangen). The cDNA was diluted 1:10 and subjected to qPCR using SuperReal PreMix Plus (Tiangen) and a 7500 Real Time PCR System (Applied Biosystems) cycler according to the manufacturer’s manual. Gene expression levels were normalized to that of PP2A and were calculated relative to that of the wild type. Primers are listed in Supplemental Data Set 6. All experiments were replicated at least three times with similar results.
ChIP
ChIP experiments were performed using 10-d-old Pro35S:FLAG-FHY3-HA transgenic seedlings grown under long-day conditions, as previously described (Liu et al., 2017). In brief, seedlings (2 g) were homogenized and cross-linked for 10 min in 1% (v/v) formaldehyde solution under vacuum. Cross-linking was stopped by adding Gly to a final concentration of 0.125 M. The cross-linked chromatin complex was isolated using a nuclear lysis buffer (50 mM Tris-HCl at pH 8.0, 10 mM EDTA, and 1% [w/v] SDS, 1 mM PMSF, and cocktail), diluted fivefold with a ChIP dilution buffer (16.7 mM Tris-HCl at pH 8.0, 167 mM NaCl, 1.1% [v/v] Triton X-100, 1.2 mM EDTA, 1 mM PMSF, and one complete protease inhibitor cocktail [Roche]), and then sheared by sonication. The sonicated chromatin complex was then immunoprecipitated using anti-HA antibodies (2 μL; Cali-Bio). The beads were washed with a low-salt buffer (50 mM Tris-HCl at pH 8.0, 2 mM EDTA, 150 mM NaCl, and 1% Triton X-100), a high-salt buffer (50 mM Tris-HCl at pH 8.0, 2 mM EDTA, 500 mM NaCl, and 1% Triton X-100), LiCl buffer (10 mM Tris-HCl at pH 8.0, 1 mM EDTA, 0.25 M LiCl, 0.5% [v/v] Nonidet P-40, and 0.5% [w/v] deoxycholate), and TE buffer (10 mM Tris-HCl at pH 8.0 and 1 mM EDTA) and eluted with an elution buffer (1% [w/v] SDS and 0.1 M NaHCO3). After reverse cross-linking, the DNA was precipitated by phenol:chloroform:isoamyl alcohol (25:24:1) and analyzed by qPCR. The values were standardized to the input DNA to obtain the enrichment fold. Primers for the ChIP-qPCR are listed in Supplemental Data Set 6.
BiFC Assay
The N terminus and C terminus of the YFP were separately fused to JAZs, FHY3, or FAR1. Overnight cultured solutions of Agrobacterium tumefaciens strain EHA105 containing various combinations of nYFP and cYFP constructs were coincubated for 2 h and infiltrated into the leaves of 3-week-old Nicotiana benthamiana plants. After 3 d, YFP fluorescence was observed using a LSM 700 confocal microscope (Zeiss) with the following YFP filter setup: excitation at 515 nm and emission at 525 to 560 nm.
Immunoblotting
Seedlings were homogenized in extraction buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.1% (v/v) Tween 20, 1 mM PMSF, and 1 complete protease inhibitor cocktail (Roche). The extracts were centrifuged at 12,000g twice at 4°C for 10 min each. Proteins were separated by 10% (w/v) SDS-PAGE gels and blotted onto nitrocellulose filter membrane (GE). The proteins were then incubated with primary antibodies and subsequently the secondary antibody accordingly. The protein bands were visualized by the standard ECL method (Thermo Pierce, SuperSignal West Dura).
Pull-Down Assay
The JAZ1 coding region was cloned into pMAL-2c, resulting in maltose-binding protein (MBP)-JAZ1. The MBP and MBP-JAZ1 constructs were introduced into the Escherichia coli BL21 (DE3) strain. After protein purification, MBP and MBP-JAZ1 proteins were preincubated with 120 μL of amylose resin beads for 1 h at 4°C. The resin-bound MBP fusion protein was added to the total protein extract from Pro35S:FLAG-FHY3-HA seedlings and incubated for 3 h at 4°C. After washing, samples were loaded on 10% (w/v) SDS-PAGE gels, transferred to nitrocellulose membranes, and incubated with anti-FLAG antibodies (MEDICAL & BIOLOGICAL LABORATORIES CO., LTD., M185-7). The purified proteins were separated by SDS-PAGE on a 10% (w/v) acrylamide gel and stained with Coomassie Brilliant Blue.
Co-IP Assays
Pro35S:FHY3-FLAG and Pro35S:JAZ1-MYC constructs were introduced into A. tumefaciens strain EHA105 and cotransformed into the leaves of 6-week-old N. benthamiana plants simultaneously using the agroinfiltration-mediated infiltration method. Total protein was extracted using a homogenization buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.1% [v/v] Tween 20, 1 mM PMSF, and one complete protease inhibitor cocktail [Roche]), and the extract was mixed with anti-FLAG magnetic agarose beads (MBL, M185-10). After incubation overnight at 4°C, the beads were centrifuged (1000 rpm, 2 min, 4°C) and washed. The protein was eluted with 40 μL of loading buffer and analyzed by immunoblotting using anti-MYC antibody (MBL, 047-7). For Co-IP assay of FHY3 and MYC2, the FLAG-FHY3-HA MYC2OE line was generated by crossing FLAG-FHY3-HA with MYC2OE. F2 plants homozygous for both transgenes were selected based on antibiotic resistance and used for the Co-IP assay. Protein extracts were immunoprecipitated using anti-FLAG antibody (MBL, M185-11) and blotted with anti-FLAG (MBL, M185-7, 1:4000) or anti-MYC (MBL, M047-7, 1:1000) antibody.
LCI Assay
The firefly LCI assay was performed using N. benthamiana leaves. FHY3 cDNA was ligated into the KpnI/SalI sites of the 35S:cLUC vector. For the nLUC-MYC2 and nLUC-MYC3 constructs, cDNAs of MYC2 and MYC3 were ligated into the KpnI/SalI sites of the 35S:nLUC vector. Both the nLUC- and cLUC-fused constructs were coinfiltrated into N. benthamiana leaves via A. tumefaciens-mediated coinfiltration. The infiltrated plants were incubated for 3 d before examination using the NightSHADE LB985 Plant Imaging System (Berthold).
Transient Transcription Assay
The transient expression assay was performed as described previously (Li et al., 2011a). The A. tumefaciens (strain EHA105) solutions containing the reporter or effector constructs were coincubated for 2 h and infiltrated into the leaves of 3- to 4-week-old N. benthamiana plants. Plants were incubated under continuous white light (90 μmol m−2 s−1) for 3 d after infiltration. The firefly LUC activity was photographed after spraying with 1 mM luciferin (Goldbio). Firefly and the control Renilla LUC activities were assayed from leaf extracts collected 3 d after infiltration using the Dual-Glo Luciferase Assay System (Promega) and quantified using a Berthold LB942 luminometer.
GUS Staining
For histochemical GUS assays, 10-d-old Pro35S:JAZ1-GUS seedlings were treated with JA for the indicated time. Then the seedlings were immersed in the histochemical staining solution: 100 mM Na3PO4, pH 7.0, 1 mM EDTA, 1 mM potassium ferrocyanide, 1 mM potassium ferricyanide, 1% (v/v) Triton X-100, and 1 mg/mL X-Gal. Seedlings were incubated at 37°C for ∼10 h until chlorophylls were removed by incubation with 75% ethanol.
Pathogen Inoculation Assay
The pathogen inoculation assay was performed on 4-week-old plants. The fungal pathogen Botrytis cinerea (strain B0510) was incubated for 3 weeks on potato dextrose agar. Detached leaves were inoculated with 5-μL spores of B. cinerea (5 × 105 spores mL−1) suspended in potato dextrose broth, placed in Petri dishes with 0.8% (w/v) agar, and covered with lids. Five leaves were inoculated per plant. The lesion area of fungal infection was measured 2 d after inoculation.
RNA-Seq Analysis
The RNA-seq experiments included two genotypes (Col and fhy3 far1) and three treatments (white light, simulated shade without JA, and simulated shade with JA). Total mRNA was extracted from 6-d-old seedlings with three biological replicates. RNA-seq was conducted by Allwegene using HiSeq4000 (Illumina). Sequencing reads were mapped to the TAIR 10 Arabidopsis reference genome using TopHat with default parameters. The abundance of assembled transcripts was calculated in fragments per kilobase of exon model per million mapped fragments (FPKM). The TopHat and Cufflink software packages were used for the RNA-seq data analysis to identify DEGs. For the no-biological-repeat RNA-seq analysis, the read-count data needed to be standardized using TMM, and the threshold value of DEGs was | log2 (fold change)| > 1 and P adjusted < 0.005. The hierarchical clustering analysis was generated via the FPKM of DEGs. Gene Ontology enrichment was analyzed using the PANTHER Classification System (http://go.pantherdb.org/). The KEGG pathways were assigned using the KEGG software package (http://www.kegg.jp/) and considered significant at P < 0.05 (Fisher’s exact test).
Statistical Analysis
All statistics were calculated using the SPSS Statistics software. To determine statistical significance, we employed nonparametric t test and one-way ANOVA with Tukey’s posthoc test. A value of P < 0.05 was considered to indicate statistical significance. All sample sizes and significance thresholds are indicated in the figure legends. The results of the statistical report are included as Supplemental Data Set 7.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: FHY3 (At3g22170), FAR1 (At4g15090), PAR1 (At2g42870), PAR2 (At3g58850), IAA29 (At4g32280), HFR1 (At1g02340), YUC8 (At4g28720), PRE1 (At5g39860), JAZ1 (At1g19180), JAZ2 (At1g74950), JAZ3 (At3g17860), JAZ4 (At1g48500), JAZ5 (At1g17380), JAZ6 (At1g72450), JAZ7 (At2g34600), JAZ8 (At1g30135), JAZ9 (At1g70700), JAZ10 (At5g13220), JAZ11 (At3g43440), JAZ12 (At5g20900), MYC2 (At1g32640), MYC3 (At5g46760), MYC4 (At4g17880), LOX2 (At3g45140), PDF1.2 (At5g44420), TAT1 (At4g23600), and VSP2 (At5g24770).
Supplemental Data
Supplemental Figure 1. The fhy3 and fhy3 far1 mutants exhibit exaggerated hypocotyl elongation under simulated shade.
Supplemental Figure 2. FHY3 protein and mRNA levels in response to shade treatment.
Supplemental Figure 3. EMSA showing binding of FHY3 binding to FBS sites in the PAR1 and PAR2 promoters.
Supplemental Figure 4. PAR-RNAi seedlings are insensitive to JA treatment.
Supplemental Figure 5. FHY3 does not affect PAR1 protein expression pattern.
Supplemental Figure 6. coi1-2 and JAZ1OE displayed insensitive phenotype to JA treatment.
Supplemental Figure 7. Yeast two-hybrid assay shows that JAZ1 physically interacts with FHY3 and FAR1.
Supplemental Figure 8. Mapping the interactive domains of JAZ1 and FHY3.
Supplemental Figure 9. The coi1-2 mutant and JAZ1OE plants display exaggerated hypocotyl elongation under simulated shade.
Supplemental Figure 10. Transient expression assay shows the repressive effect of JAZs on the transcription activity of FHY3.
Supplemental Figure 11. Quantitative RT-PCR analysis of IAA29 and PRE1 expression.
Supplemental Figure 12. Quantitative RT-PCR analysis of FHY3 expression in response to JA treatment.
Supplemental Data Set 1. List of differentially expressed genes in six treatments.
Supplemental Data Set 2. List of genes regulated by shade in Col and fhy3 far1.
Supplemental Data Set 3. List of genes regulated by JA treatment in Col and fhy3 far1.
Supplemental Data Set 4. List of genes regulated by FHY3/FAR1 in high and low R/FR conditions.
Supplemental Data Set 5. Co-regulated genes of FHY3 and MYC2.
Supplemental Data Set 6. Primer sets used in this study.
Supplemental Data Set 7. Statistical report of t tests and ANOVA results for the data presented in each figure.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Daoxin Xie (Tsinghua University) for providing coi1-2, Chuanyou Li (Institute of Genetics and Developmental Biology, Chinese Academy of Science) for providing the JAZ1-GUS seeds, and Jianping Yang (Henan Agricultural University) for providing the PAR1-GFP, PAR2-GFP, and PAR-RNAi seeds. This work was supported by the National Natural Science Foundation of China (grants 31500239 and 31430008).
AUTHOR CONTRIBUTIONS
Y.L., H.W., M.M., Q.L., D.K., J.S., X.M., B.W., and Y.X. performed the research. Y.L., H.Y.W., and C.C. analyzed the data. H.Y.W. and Y.L. designed the research and wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Agrawal A.A., Kearney E.E., Hastings A.P., Ramsey T.E. (2012). Attenuation of the jasmonate burst, plant defensive traits, and resistance to specialist monarch caterpillars on shaded common milkweed (Asclepias syriaca). J. Chem. Ecol. 38: 893–901. [DOI] [PubMed] [Google Scholar]
- Ballaré C.L. (2009). Illuminated behaviour: Phytochrome as a key regulator of light foraging and plant anti-herbivore defence. Plant Cell Environ. 32: 713–725. [DOI] [PubMed] [Google Scholar]
- Ballaré C.L. (2014). Light regulation of plant defense. Annu. Rev. Plant Biol. 65: 335–363. [DOI] [PubMed] [Google Scholar]
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Torrent J., Roig-Villanova I., Galstyan A., Martínez-García J.F. (2008). PAR1 and PAR2 integrate shade and hormone transcriptional networks. Plant Signal. Behav. 3: 453–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205. [DOI] [PubMed] [Google Scholar]
- Casal J.J. (2013). Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64: 403–427. [DOI] [PubMed] [Google Scholar]
- Cerrudo I., Keller M.M., Cargnel M.D., Demkura P.V., de Wit M., Patitucci M.S., Pierik R., Pieterse C.M., Ballaré C.L. (2012). Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol. 158: 2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., et al. (2011). The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico J.M., Fernández-Barbero G., Chini A., Fernández-Calvo P., Díez-Díaz M., Solano R. (2014). Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. Plant Cell 26: 1967–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Prat S. (2014). PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol. 202: 1126–1141. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- de Wit M., Spoel S.H., Sanchez-Perez G.F., Gommers C.M.M., Pieterse C.M.J., Voesenek L.A.C.J., Pierik R. (2013). Perception of low red:far-red ratio compromises both salicylic acid- and jasmonic acid-dependent pathogen defences in Arabidopsis. Plant J. 75: 90–103. [DOI] [PubMed] [Google Scholar]
- Djakovic-Petrovic T., de Wit M., Voesenek L.A., Pierik R. (2007). DELLA protein function in growth responses to canopy signals. Plant J. 51: 117–126. [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chico J.M., Solano R. (2009). The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12: 539–547. [DOI] [PubMed] [Google Scholar]
- Franklin K.A. (2008). Shade avoidance. New Phytol. 179: 930–944. [DOI] [PubMed] [Google Scholar]
- Franklin K.A., Quail P.H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61: 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galstyan A., Cifuentes-Esquivel N., Bou-Torrent J., Martinez-Garcia J.F. (2011). The shade avoidance syndrome in Arabidopsis: A fundamental role for atypical basic helix-loop-helix proteins as transcriptional cofactors. Plant J. 66: 258–267. [DOI] [PubMed] [Google Scholar]
- Gupta V., Willits M.G., Glazebrook J. (2000). Arabidopsis thaliana EDS4 contributes to salicylic acid (SA)-dependent expression of defense responses: Evidence for inhibition of jasmonic acid signaling by SA. Mol. Plant Microbe Interact. 13: 503–511. [DOI] [PubMed] [Google Scholar]
- Hao Y., Oh E., Choi G., Liang Z., Wang Z.Y. (2012). Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 5: 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28: 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Lee L.Y.C., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894. [DOI] [PubMed] [Google Scholar]
- Kloek A.P., Verbsky M.L., Sharma S.B., Schoelz J.E., Vogel J., Klessig D.F., Kunkel B.N. (2001). Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26: 509–522. [DOI] [PubMed] [Google Scholar]
- Kunkel B.N., Brooks D.M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5: 325–331. [DOI] [PubMed] [Google Scholar]
- Leivar P., Monte E. (2014). PIFs: Systems integrators in plant development. Plant Cell 26: 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone M., Keller M.M., Cerrudo I., Ballaré C.L. (2014). To grow or defend? Low red:far-red ratios reduce jasmonate sensitivity in Arabidopsis seedlings by promoting DELLA degradation and increasing JAZ10 stability. New Phytol. 204: 355–367. [DOI] [PubMed] [Google Scholar]
- Li G., et al. (2011a). Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat. Cell Biol. 13: 616–622. [DOI] [PubMed] [Google Scholar]
- Li J., Li G., Wang H., Deng X.W. (2011b). Phytochrome signaling mechanisms. The Arabidopsis Book 9: e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Ding L., Casola C., Ripoll D.R., Feschotte C., Wang H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xie Y., Wang H., Ma X., Yao W., Wang H. (2017). Light and ethylene coordinately regulate the phosphate starvation response through transcriptional regulation of PHOSPHATE STARVATION RESPONSE1. Plant Cell 29: 2269–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García J.F., Gallemí M., Molina-Contreras M.J., Llorente B., Bevilaqua M.R., Quail P.H. (2014). The shade avoidance syndrome in Arabidopsis: The antagonistic role of phytochrome A and B differentiates vegetation proximity and canopy shade. PLoS One 9: e109275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno J.E., Tao Y., Chory J., Ballaré C.L. (2009). Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc. Natl. Acad. Sci. USA 106: 4935–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X., et al. (2011). Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23: 2514–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhika V., Kost C., Mithöfer A., Boland W. (2010). Regulation of extrafloral nectar secretion by jasmonates in lima bean is light dependent. Proc. Natl. Acad. Sci. USA 107: 17228–17233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F., Okamoto H., Patrick E., Harris S.R., Wasternack C., Brearley C., Turner J.G. (2010). Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22: 1143–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I., Bou J., Sorin C., Devlin P.F., Martínez-García J.F. (2006). Identification of primary target genes of phytochrome signaling: Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol. 141: 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I., Bou-Torrent J., Galstyan A., Carretero-Paulet L., Portolés S., Rodríguez-Concepción M., Martínez-García J.F. (2007). Interaction of shade avoidance and auxin responses: A role for two novel atypical bHLH proteins. EMBO J. 26: 4756–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G., Carabelli M., Sassi M., Ciolfi A., Possenti M., Mittempergher F., Becker J., Morelli G., Ruberti I. (2005). A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 19: 2811–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.-H., Lee D., Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui H., Khan S., Rhodes B.M., Devlin P.F. (2016). FHY3 and FAR1 act downstream of light stable phytochromes. Front. Plant Sci. 7: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P., Zhao S., Williamson L., Ward S., Leyser O. (2012). FHY3 promotes shoot branching and stress tolerance in Arabidopsis in an AXR1-dependent manner. Plant J. 71: 907–920. [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Wang H., Deng X.W. (2002). Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J. 21: 1339–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Deng X.W. (2003). Dissecting the phytochrome A-dependent signaling network in higher plants. Trends Plant Sci. 8: 172–178. [DOI] [PubMed] [Google Scholar]
- Wang H., Wang H. (2015). Multifaceted roles of FHY3 and FAR1 in light signaling and beyond. Trends Plant Sci. 20: 453–461. [DOI] [PubMed] [Google Scholar]
- Wang W., Tang W., Ma T., Niu D., Jin J.B., Wang H., Lin R. (2016). A pair of light signaling factors FHY3 and FAR1 regulates plant immunity by modulating chlorophyll biosynthesis. J. Integr. Plant Biol. 58: 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam G.C., Patel S., Devlin P.F. (1998). Phytochromes and photomorphogenesis in Arabidopsis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353: 1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Liu Y., Wang H., Ma X., Wang B., Wu G., Wang H. (2017). Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat. Commun. 8: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.L., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 109: E1192–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Turner J.G. (2008). Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS One 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Song M., Yang Q., Su L., Hou P., Guo L., Zheng X., Xi Y., Meng F., Xiao Y., Yang L., Yang J. (2014). Both PHYTOCHROME RAPIDLY REGULATED1 (PAR1) and PAR2 promote seedling photomorphogenesis in multiple light signaling pathways. Plant Physiol. 164: 841–852. [DOI] [PMC free article] [PubMed] [Google Scholar]