Variation in centromere satellite sequence and copy number, and structural rearrangements produce asymmetries in polyploid wheat homeologs, highlighting the role in homolog pairing during meiosis.
Abstract
Centromeres mediate the pairing of homologous chromosomes during meiosis; this pairing is particularly challenging for polyploid plants such as hexaploid bread wheat (Triticum aestivum), as their meiotic machinery must differentiate homologs from similar homoeologs. However, the sequence compositions (especially functional centromeric satellites) and evolutionary history of wheat centromeres are largely unknown. Here, we mapped T. aestivum centromeres by chromatin immunoprecipitation sequencing using antibodies to the centromeric-specific histone H3 variant (CENH3); this identified two types of functional centromeric satellites that are abundant in two of the three subgenomes. These centromeric satellites had unit sizes greater than 500 bp and contained specific sites with highly phased binding to CENH3 nucleosomes. Phylogenetic analysis revealed that the satellites have diverged in the three T. aestivum subgenomes, and the more homogeneous satellite arrays are associated with CENH3. Satellite signals decreased and the degree of satellites variation increased from diploid to hexaploid wheat. Moreover, several T. aestivum centromeres lack satellite repeats. Rearrangements, including local expansion and satellite variations, inversions, and changes in gene expression, occurred during the evolution from diploid to tetraploid and hexaploid wheat. These results reveal the asymmetry in centromere organization among the wheat subgenomes, which may play a role in proper homolog pairing during meiosis.
INTRODUCTION
Allohexaploid bread wheat (Triticum aestivum) originated from interspecific hybridization and subsequent doubling of the A, B, and D genomes of closely related diploid progenitor species. The resulting 17-Gb wheat genome poses enormous challenges for genomic analysis (Marcussen et al., 2014). However, the abundant diploid, tetraploid, and hexaploid species in the wheat group and their wild relatives make wheat an excellent model for studying polyploidy.
Because of the presence of multiple similar chromosomes, polyploid wheat requires specific mechanisms to correctly pair homologous chromosomes and thus avoid the formation of unbalanced gametes during meiosis. Centromeres are essential for genome stability, and centromere coupling before the onset of homologous chromosome pairing during meiosis is critical for the faithful segregation of monocentric eukaryotic chromosomes to daughter cells (Pluta et al., 1995; Zhang et al., 2013b; Fukagawa and Earnshaw, 2014; Da Ines and White, 2015). These observations suggest that the centromeres in the subgenomes of polyploid species may function during meiosis to ensure genome stability via effects on chromosome pairing, in addition to mediating kinetochore organization and the attachment of spindle microtubules.
In most species, centromere identity and function are determined by highly conserved epigenetic mechanisms based on the presence of the centromeric-specific histone H3 variant CENH3, which is also known as CENP-A (Earnshaw and Rothfield, 1985; Palmer and Margolis, 1985; Zhong et al., 2002). The phosphorylation of histone H2A on Thr133 has also been associated with functional centromeres in plants (Dong and Han, 2012; Su et al., 2017). The DNA sequences of plant centromeres usually contain many copies of simple tandem repeats, interrupted by long-terminal repeat retrotransposons (Zhong et al., 2002; Hall et al., 2004; Comai et al., 2017). These tandem repeats, also called satellite repeats, occur in head-to-tail arrays and only those that are associated with CENH3 nucleosomes are considered to be part of the functional centromere (Sullivan et al., 2017). The unit length of most tandem repeat families in plants varies from 150 to 180 bp, for example, the 180-bp pAL1 in Arabidopsis (Arabidopsis thaliana), 155-bp CentO in rice (Oryza sativa), and 156-bp CentC in maize (Zea mays; Ananiev et al., 1998; Cheng et al., 2002; Melters et al., 2013), a length sufficient for wrapping around a single CENH3 nucleosome. These satellite repeats are highly phased with CENH3 nucleosomes (Gent et al., 2011; Zhang et al., 2013c; Iwata-Otsubo et al., 2017). By contrast, several types of satellite arrays of different unit sizes have been identified in potato (Solanum tuberosum), which possesses repeat-containing and repeat-free centromeres (Gong et al., 2012). Chromosome-specific tandem repeats and tandem-repeat-free centromeres were also identified in chicken DT40 cells (Shang et al., 2010).
Extensive variation within centromeric sequences has been observed among different species, on different chromosomes, and even in the same centromeres from different plant varieties (Cheng et al., 2002; Hall et al., 2003). Furthermore, variation in centromeric satellites during evolution has been associated with centromere function and location in several species (Henikoff et al., 2015; Aldrup-MacDonald et al., 2016; Maheshwari et al., 2017). For example, functional centromeres assemble on the youngest and most homogeneous arrays of pAL1 satellites in Arabidopsis (Maheshwari et al., 2017).
Several repeat sequences were identified in wheat centromeres by screening large DNA clones or bacterial artificial chromosomes (BACs) using the known cereal centromeric repeat CCS1 or CRW sequences (Jiang et al., 1996; Kishii et al., 2001; Cheng and Murata, 2003; Ito et al., 2004; Zhang et al., 2004; Liu et al., 2008; Li et al., 2013). Most of these sequences are centromeric Ty3/Gypsy retrotransposons that are associated with CENH3 nucleosomes in wheat (Li et al., 2013). One limitation of these studies is that clones with centromeric tandem repeats may be lost during screening. Indeed, the centromeric satellite repeats that are wrapped around CENH3 nucleosomes have not yet been described in wheat, although centromeric satellite-like sequences were detected, but did not associate with CENH3 (Li et al., 2013). Furthermore, little is known about the variation in centromeric sequences that occurred after polyploidization and whether each subgenome of polyploid wheat has its own unique centromeric satellites.
Polyploidization via interspecific hybridization followed by whole-genome duplication can lead to chromosome rearrangements and genome reorganization (Wendel et al., 2016). The genetic and epigenetic changes associated with this process have been extensively studied (Feldman and Levy, 2012; Song and Chen, 2015). For example, gain or loss and neo-functionalization or subfunctionalization of genes, as well as changes in gene expression after polyploidization, have been described in plants (Jackson and Chen, 2010). In wheat, changes in ploidy did not initially produce dramatic changes in gene expression and did not substantially alter epigenetic marks (Martín et al., 2018; Ramírez-González et al., 2018). However, asymmetric elimination of repetitive rDNA sequences and epigenetic modifications were observed in polyploid formation in wheat (Guo and Han, 2014). We recently detected centromeric repetitive sequence elimination, expansion, and multi-centromere formation induced by chromosome rearrangement in wheat aneuploids and in the offspring of their wide hybrids (Guo et al., 2016). Hybrid centromeres were detected in wheat and winter rye (Secale cereale) 1RS.1BL translocation lines, and wheat CENH3 was found at the fused centromeres (Wang et al., 2017). However, whether changes occur within the centromeres and whether such changes have distinct consequences is largely unknown.
With the availability of the reference sequences of distinct wheat chromosomes from species of different ploidy (Brenchley et al., 2012; Avni et al., 2017; Clavijo et al., 2017; Luo et al., 2017; Zhao et al., 2017; Zimin et al., 2017; International Wheat Genome Sequencing Consortium., 2018; Ling et al., 2018), it is now possible to perform a comprehensive investigation of the genetic composition, structure, and evolution of wheat centromeres. Here, we investigated functional centromeric regions of wheat via chromatin immunoprecipitation sequencing (ChIP-seq) using anti-CENH3 antibodies and compared the results with various wheat reference genomes: AABBDD, AABB, AA, and DD. We identified satellite repeats and analyzed the structure and organization of centromeres in hexaploid wheat and compared them with those of the diploid and tetraploid species that are extant relatives of its progenitors. Sequence variation within these long satellite sequences may influence their ability to associate with CENH3 nucleosomes, and the degree of variation in centromere satellites is different among polyploid wheats varieties. Our analysis revealed centromere features including structural rearrangements and satellite sequence variations that occurred in response to polyploidization during wheat evolution and may assist in chromosome pairing in hexaploid wheat.
RESULTS
Genome-Wide Mapping of DNA Sequences Associated with CENH3 Nucleosomes in Chinese Spring Wheat
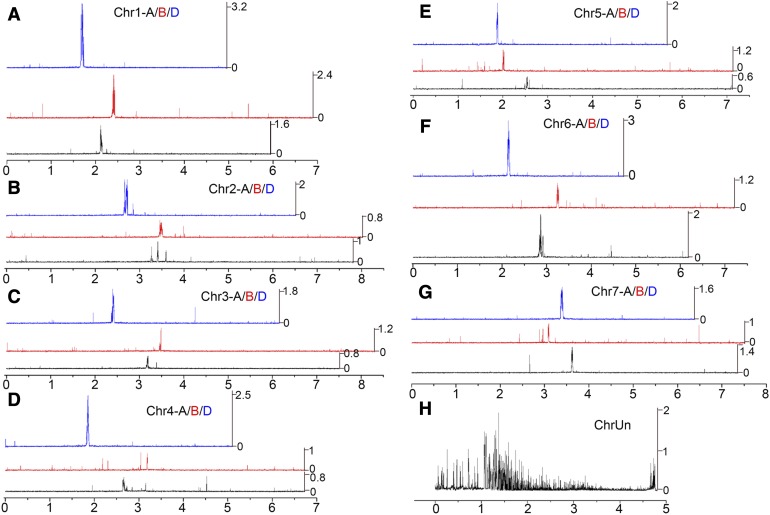
To identify functional centromeric regions in hexaploid wheat, we used data from ChIP-seq with antibodies specific to the centromeric histone variant CENH3. We remapped our previously reported anti-CENH3 ChIP-seq data set from the allohexaploid T. aestivum cv Chinese Spring (CS; GSE63752; Guo et al., 2016) to the International Wheat Genome Sequencing Consortium (IWGSC) RefSeq v1.0 reference genome for T. aestivum (IWGSC_v1.0) using Burrows-Wheeler Aligner (BWA)-MEM software (Li and Durbin, 2009). Approximately 57.4% of the 64 million 101-bp paired-end reads aligned to unique positions in the IWGSC_v1.0 genome. Figure 1 shows the genomic distribution of the unique ChIP-seq reads in 100-kb windows along the 21 wheat chromosomes and one group of unassigned scaffolds (ChrUn).
Figure 1.
Genome-Wide Mapping of CENH3 ChIP-Seq Reads from CS Wheat to the IWGSC Reference Genome (AABBDD).
(A) to (G) Homoeologous group Chr1 to Chr7.
(H) Unassigned scaffold. The mapped reads density was plotted in 100-kb windows along the 21 wheat chromosomes and one unassigned scaffold.
The x axis shows the positions (× 100 Mb) on each chromosome. The y axis represents the normalized density of uniquely mapped reads on each position, which was calculated based on the ratio of reads per million/100. The different colored lines represent the reads density in different wheat subgenomes (black, AA subgenome; red, BB subgenome; blue, DD subgenome).
In Burrows-Wheeler Aligner (BWA) general, enrichment of CENH3 peaks was observed in the centromere regions of almost all the 21 T. aestivum chromosomes (Figure 1). Compared with the AA and BB subgenomes, higher CENH3 enrichment was detected in the T. aestivum DD subgenome (Figure 1), suggesting that the DD centromeres contain more single and/or low copy sequences than the others. As expected, CENH3 binding subdomains (∼62 Mb) were also observed in the group of unassigned scaffolds (Figure 1H), indicating that many centromeric sequences have not yet been anchored in the assembled chromosomes.
Different centromeres showed different arrangements of subdomains that bind CENH3. For example, in Cen2A, three CENH3 subdomains are separated by two chromosomal domains (Figure 1B; Table 1). In Cen4B, two CENH3 subdomains are separated by a chromosomal domain (Figure 1D; Table 1). In Cen7B, three CENH3 subdomains are separated by two chromosomal domains (Figure 1G; Table 1). These results may be due to an assembly error in the reference genome, as fluorescence in situ hybridization (FISH) showed a single centromeric signal in each wheat metaphase chromosome (Guo et al., 2016).
Table 1. Positions of Functional CENH3-Binding Regions in the Wheat Centromeres.
| Chr | Chr Size (Mb) | Cen Location (Mb) | Chr | Chr Size (Mb) | Cen Location (Mb) | Chr | Chr Size (Mb) | Cen Location (Mb) |
|---|---|---|---|---|---|---|---|---|
| 1A | 592 | 210.0–216.3 | 1B | 688 | 237.5–244.0 | 1D | 494 | 166.0–174.0 |
| 2A | 779 | 326.2–327.8 | 2B | 799 | 344.0–351.6 | 2D | 650 | 264.4–272.5 |
| 339.2–342.2 | ||||||||
| 359.2–359.6 | ||||||||
| 3A | 749 | 317.0–320.0 | 3B | 829 | 345.8–347.0 | 3D | 641 | 237.0–243.5 |
| 348.4–349.1 | ||||||||
| 4A | 743 | 264.0–268.0 | 4B | 672 | 303.8–304.5 | 4D | 508 | 182.5–188.4 |
| 317.5–320.0 | ||||||||
| 5A | 708 | 252.5–255.5 | 5B | 711 | 199. 0–202.6 | 5D | 564 | 185.5–188.8 |
| 6A | 616 | 283.3–288.8 | 6B | 719 | 323. 0–327.7 | 6D | 472 | 211.9–217.5 |
| 7A | 735 | 360.2–363.9 | 7B | 749 | 294.3–294.7 | 7D | 637 | 336.0–342.0 |
| 296.4–296.6 | ||||||||
| 308.0–310.2 |
Cen, centromere. Chr, chromosome.
The locations and sizes of wheat centromeres were previously estimated based on the distribution of centromeric Ty3/Gypsy retrotransposon sequences along the T. aestivum 3B and Aegilops tauschii 3DS pseudochromosome (Choulet et al., 2014; Xie et al., 2017). The similar estimated sizes for centromere Ta3B and At3D obtained in the current study suggest that wheat chromosomes have a similar centromere composition. The size of the core region of CENH3 binding in wheat centromeres varies from 1.9 to 8.1 Mb (Table 1). Taking into account the core centromeric sequences located in the ChrUn scaffolds, the mean centromere size in bread wheat is ∼7.9 Mb, which is much larger than that of maize and rice centromeres (Yan et al., 2008; Wolfgruber et al., 2009).
Identification and Characterization of Centromeric Satellite Sequences in Wheat
Centromeric satellite repeats associated with CENH3 nucleosomes have not been reported in wheat and due to the highly repetitive nature of centromeres, the centromeric satellite sequences might not be fully assembled in the T. aestivum reference genome. To get around this problem, we first computationally identified repeats de novo in the T. aestivum genome and then examined whether these repeats were enriched in the anti-CENH3 ChIP data. To this end, we performed graph-based sequence clustering using RepeatExplorer software for de novo identification of repetitive sequences in wheat, as previously reported in other plant species (Macas et al., 2007; Novák et al., 2010). We randomly selected 1.14 million whole-genome shotgun 454 sequencing reads from T. aestivum CS (Brenchley et al., 2012) and analyzed them using the Web-based Galaxy RepeatExplorer software (https://repeatexplorer-elixir.cerit-sc.cz/galaxy/). All types of repeat clusters were reported, including putative satellite and dispersed repetitive sequences. Subsequently, ∼118 million 101-bp paired-end reads from Illumina sequencing of T. aestivum were used as the Input. BLAST analysis was performed to map the CENH3-ChIP-seq and Input reads to cluster repeats. The numbers of reads with significant BLAST hits to each cluster repeat for CENH3-ChIP and Input were calculated, and the CENH3-ChIP:Input ratio was taken as the CENH3-enrichment level.
We selected six clusters with CENH3-ChIP:Input enrichment >1.5 for further analysis (Table 2). Four of the clusters represent wheat centromeric-specific long terminal repeat retrotransposons. By contrast, clusters CL247 and CL6321 were identified as containing tandemly arranged satellite repeats with a unit size of 566 and 550 bp, respectively (Table 2). We named these satellites CentT566 and CentT550 for centromeric tandem repeat of Triticum. The alignment results indicate that they are two different types of satellites (Supplemental Figure 1A). The CentT550 repeat was a previously identified ∼550-bp centromeric satellite-like sequence, which was not associated with CENH3 (Li et al., 2013). The copy number of wheat centromeric satellites in the genome were much lower than that of centromeric retrotransposons (Table 2), as suggested previously (Li et al., 2013).
Table 2. Identification of Putative Centromere Repeat Sequences in Bread Wheat.
| Cluster | Genome Content (kb) | ChIP:WGS Ratio | Monomer (bp) | Repeat Type | Chromosomal Location |
|---|---|---|---|---|---|
| CL247 | 3,164 | 8.832 | 566 | Tandem repeat | Cen1B-7B, Cen5A, Cen1D and Cen3D |
| CL88 | 14,400 | 6.015 | 4791 | Ty3/Gypsy, CRW clade | All centromeres |
| CL18 | 58,000 | 3.237 | 7494 | Ty3/Gypsy, Quinta clade | All centromeres |
| CL99 | 12,500 | 2.019 | 3760 | Ty3/Gypsy, CRW clade | All centromeres |
| CL6321 | 679 | 1.915 | 580 | Tandem repeat | Cen1D, Cen2D, Cen4D, Cen6D and Cen3B |
| CL90 | 15,900 | 1.779 | 3379 | Ty3/Gypsy, CRW clade | All centromeres |
We performed FISH using the cloned probes from the clusters to confirm the centromere localization of the two satellite repeats. First, we checked the locations of the satellites in the diploid progenitors of bread wheat (Table 3). We found that the CentT566 signals were specifically located at all centromeres of the potential wheat B genome donor Aegilops speltoides (Ae92, Ae739, six chromosome pairs showed strong signals and one pair showed weak signals; Figures 2A and 2B). However, only two pairs of strong CentT566 signals and one pair of weak signals were detected in the wheat D genome donor Aegilops tauschii (accessions AL8/78 and TQ27; Figures 2D and 2E). Two to four pairs of CentT566 signals were observed in centromeric regions of accessions G1812, TMU06, and TMU38 of the wheat A genome donor Triticum urartu (Supplemental Figures 1B to 1D).
Table 3. Number of FISH Signals in Different Polyploid Wheat Varieties.
| Satellite | G1812 (AA) | TMU06 (AA) | TMU38 (AA) | Ae92 (BB) | Ae739 (BB) | AL8/78 (DD) | TQ27 (DD) | CS (AABBDD) |
|---|---|---|---|---|---|---|---|---|
| CentT566 | 3 pairs | 4 pairs | 3 pairs | 7 pairs | 7 pairs | 3 pairs | 3 pairs | 10 pairs |
| CentT550 | 2 pairs | 2 pairs | 0 pairs | 1 pairs | 1 pairs | 5 pairs | 5 pairs | 5 pairs |
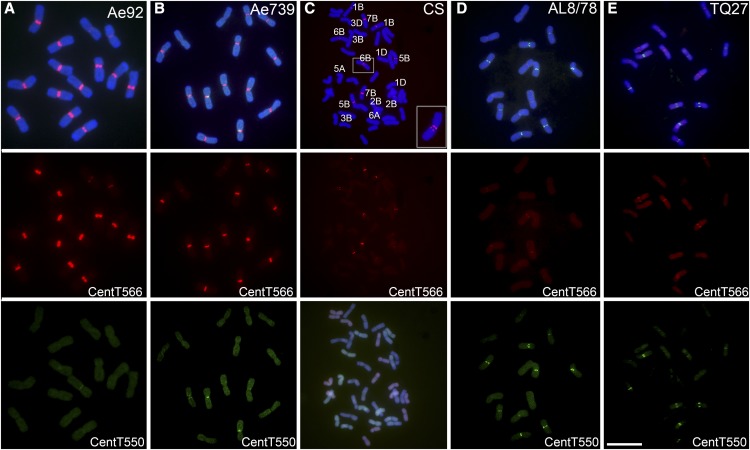
Figure 2.
FISH Confirms the Centromere Location of Satellite Repeats in Different Wheat Lines.
(A) and (B) FISH signals of repeat CentT566 (red) and CentT550 (green) in different lines of the potential wheat B genome donor Ae. speltoides Ae92 (A) and Ae739 (B).
(C) FISH signals of repeat CentT566 (red) and multi-color genomic in situ hybridization in bread wheat. The boxed insets show high-magnification images of chromosome 6B.
(D) and (E) FISH signals of repeat CentT566 (red) and CentT550 (green) in different accessions of the wheat D genome donor Ae. tauschii AL8/78 (D) and TQ27 (E).
For FISH, blue indicates chromosomes counterstained with 4′,6-diamidino-2-phenylindole. For multi-color FISH, the wheat AA genomic DNA is labeled in green, the DD genomic DNA is labeled in red, and the BB genomic DNA was used as a blocker (blue). Bar = 10 μm.
In contrast to the CentT566 repeats, the FISH signals of the CentT550 repeats were mainly abundant in the centromeric regions of the wheat D genome donor Ae. tauschii (Figures 2D and 2E), with four pairs of strong signals and one pair of weak signals. Only one pair of CentT550 signals was observed in the wheat B diploid progenitors (Figures 2A and 2B), and several weak signals were located in the pericentromeric regions of the wheat A genome donor (Supplemental Figures 1B to 1D). These results indicate that two types of centromeric satellites are abundant in the diploid wheat species that represent the extant relatives of the B and D genome donors.
In hexaploid wheat, CentT566 signals were observed in all centromeres of the BB subgenome, with strong signals detected in Cen2B, Cen3B, and Cen4B and weak signals detected in Cen1B, Cen5B, Cen6B, and Cen7B (Figure 2C). In addition, weak CentT566 signals were detected in Cen5A, Cen3D, and Cen1D (Figure 2C). Several weak CentT550 signals were detected in Cen1D, Cen2D, Cen4D, Cen6D, and Cen3B of bread wheat (Supplemental Figure 1E). These results are consistent with the proportions of the two satellites detected in the genome sequence (Table 2). The satellite shows different abundances in the subgenome in hexaploid wheat. No satellite signals were observed in centromeric regions of other chromosomes in bread wheat based on the FISH results and genome distribution (Figure 2C; Supplemental Figure 1E), which is consistent with the genomic distribution of the satellites (Supplemental Figures 2 and 3).
Furthermore, two separate CentT566 signals were observed in Cen6B of bread wheat (Figure 2C), while only one signal was present in all centromeres of the potential B genome donor Ae. speltoides (Figures 2A and 2B), suggesting that centromere expansion has occurred in hexaploid wheat. Taken together, our results show that in contrast to the canonical centromeric satellites with unit sizes ranging from 150 to 180 bp, the centromeres of diploid wheat varieties have two type of satellites with unit sizes >500 bp. The FISH observations suggest that the number of centromeric satellite repeats varied in the wheat varieties with different ploidies; perhaps these repeats expanded locally during evolution from the diploid progenitors to hexaploid wheat, yielding repeat-containing and repeat-free centromeres in bread wheat.
Detection of a Partial CentT566 Satellite Sequence in a Wheat Centromeric Retrotransposon
We also identified a 223-bp satellite-like sequence (TAIL5_TA#Sate) from a wheat centromeric region using RepeatMasker (Tempel, 2012). This sequence was found within the CL88 cluster sequence, belonging to the Ty3/Gypsy CRW clade. This satellite-like sequence could be aligned with the CentT566 sequence (Supplemental Figure 4A), suggesting that a part of the CentT566 satellite sequence became inserted into the centromeric retrotransposon during evolution. The corresponding FISH signals of TAIL5_TA#Sate were abundant in various B genome diploids species (Ae. speltoides and Aegilops longissima), with five to seven pairs of strong signals (Supplemental Figure 4C). By contrast, only a few weak signals were detected in the A- and D-genomes of diploids (Supplemental Figures 4B and 4D). In T. aestivum, the TAIL5_TA#Sate signals were located at the centromeres of all three subgenomes, with different signal strengths (Supplemental Figure 4E). The changes in centromeric sequence positions appear to reflect the dynamics of centromeric retrotransposons during the evolution of hexaploid wheat. The CentT566 satellite repeats in the B genomes might have invaded the two other subgenomes, indicating that transfer of centromeric sequences between subgenome has occurred.
CENH3 Nucleosomes Are Highly Phased with Specific Sites on the Wheat Centromeric Satellite DNA Monomers around WW Dinucleotides
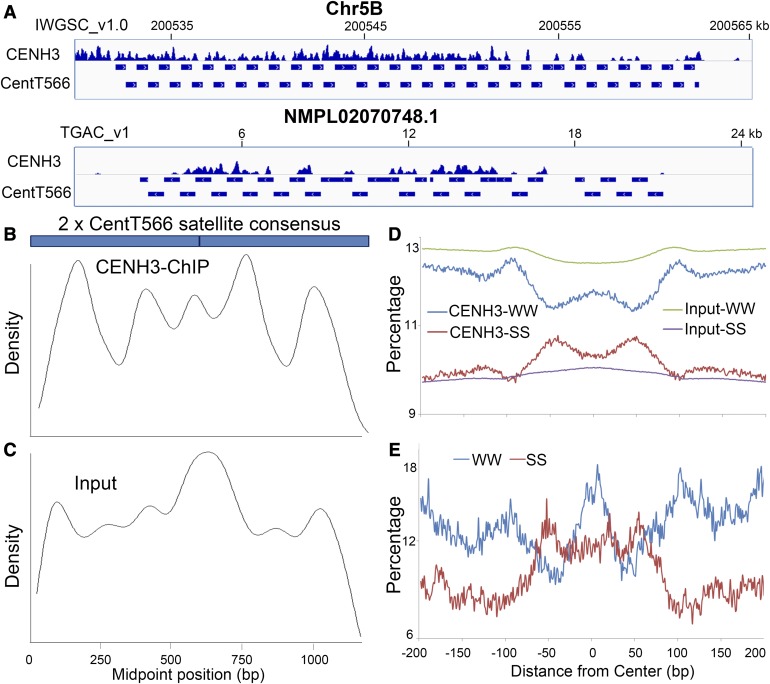
The TGACv1 version of the CS genome was assembled using PacBio sequence reads, and a recently study improved the assembly accuracy of the genomic scaffolds, especially in centromeric regions (Clavijo et al., 2017). Here, we discovered dozens of kilobase-sized tandem arrays of wheat satellite repeats CentT566 and CentT550 that were organized within CENH3 nucleosomes in the IWGSC_v1.0 and TGACv1 versions of the CS wheat genome (Figure 3A; Supplemental Figure 5A).
Figure 3.
CENH3 Nucleosomes Are Highly Phased with Specific Sites on Monomers of the Wheat Centromeric Satellite CentT566 around WW Dinucleotides.
(A) Snapshot of the distribution of the wheat centromeric satellite repeat CentT566 occupied by CENH3 nucleosomes in two versions of the bread wheat reference genome (IWGSC_v1.0 and TGACv1). The horizontal scale marks the centromere locus (in kilobases) on each chromosome or scaffold.
(B) and (C) Distribution of the midpoints of CENH3 ChIP (B) and input (C) reads (100 to 200 bp) along the dimerized CentT566 satellite consensus sequence. Five major sites occupied by the CENH3 nucleosome position were observed in dimerized CentT566. The same nucleosome binding position is indicated in the input sample.
(D) Sequence features of CENH3 and canonical nucleosomal DNA. The dinucleotide frequency of WW and SS is displayed within ±200 bp from the center. The green curve represents input-WW, the blue curve represents CENH3-WW, the red curve represents CENH3-SS, and the purple curve represents input-SS. The x axis represents the distance from the center (bp); the y axis represents the frequency of dinucleotides.
(E) Dinucleotides frequencies associated with CentT566-related CENH3 nucleosomes. Blue curve represents WW; red curve represents SS.
We wanted to know whether the CENH3 nucleosomes randomly bind to the sites of wheat centromeric satellites of large monomer size, or whether they bind to specific sites. We adopted a similar strategy to that used to analyze human and mouse centromeres (Hasson et al., 2013; Iwata-Otsubo et al., 2017). The CENH3-ChIPed and Input-seq reads, which represent the CENH3 and bulk nucleosome fragments, respectively, were merged and mapped to the dimer CentT566 and CentT550 consensus satellite sequences using BWA-MEM software (Li and Durbin, 2009). The distribution of the midpoint of the fragments was treated as the location of the sequence on CENH3 or canonical nucleosomes. Two major binding peaks were identified in the dimer CentT550 satellite consensus sequence for CENH3-ChIP-seq reads, and the locations were the same on each monomer satellite, indicating phasing of CENH3 nucleosomes on centromeric CentT550 satellite sequences (Supplemental Figure 5B, red line). The input nucleosomes are present at the same positions on the CentT550 monomer (Supplemental Figure 5B, green line), suggesting that most of CentT550 repeats were wrapped around CENH3 nucleosomes.
Five major binding positions were found on CentT566 dimer sequences in the CENH3-ChIP-seq reads. The first of these on each monomer was most frequently associated with CENH3 nucleosomes (Figure 3B), indicating the presence of strong and specific positions for wrapping around CENH3 nucleosomes. However, for input nucleosomes, broad peaks were present at multiple positions throughout the dimer repeat sequence (Figure 3C), indicating that some CentT566 repeats are not associated with CENH3 nucleosomes.
We used nucleR (Flores and Orozco, 2011) to align the nucleosome centers from 4,925,556 well-positioned canonical nucleosomes with the input data set and analyzed the profiles of SS (S = G or C) and WW (W = A or T) dinucleotides within a region ±200 bp from the center. The SS dinucleotides were abundant in the center and flanking regions, and the WW dinucleotides were enriched in regions ±100 bp from the center (Figure 3D, green and purple lines). This result is consistent with the nucleosome positioning signals in other species (Ioshikhes et al., 2011; Zhang et al., 2013c). We also aligned 840,542 well-positioned CENH3 nucleosome centers from the CENH3 ChIP-seq data. The WW dinucleotides were enriched in the center and regions ±100 bp from the center but decreased in flanking regions ±50 bp from the center (Figure 3D, red line). The distribution pattern of SS dinucleotides was opposite that of WW dinucleotides (Figure 3D, blue line). The distribution of SS/WW dinucleotides within ±200 bp from 4425 well-positioned CENH3 nucleosomes centers on the CentT566 satellite was similar to that of whole CENH3 nucleosomes (Figure 3E), showing strong preferences for WW dinucleotides at the centers of CENH3 nucleosomes containing the CentT566 satellite. This preference was also observed for CENH3 nucleosomes containing the CentT550 satellites (Supplemental Figure 5C). These results differ from the observations for humans, rice, and maize in which SS-enriched regions favor CENH3 nucleosome centering (Ioshikhes et al., 2011; Zhang et al., 2013c), suggesting that WW dinucleotides function in the positioning of CENH3 nucleosomes in wheat.
CentT566 Polymorphisms Related to CENH3 Binding Appeared during Hexaploid Wheat Evolution
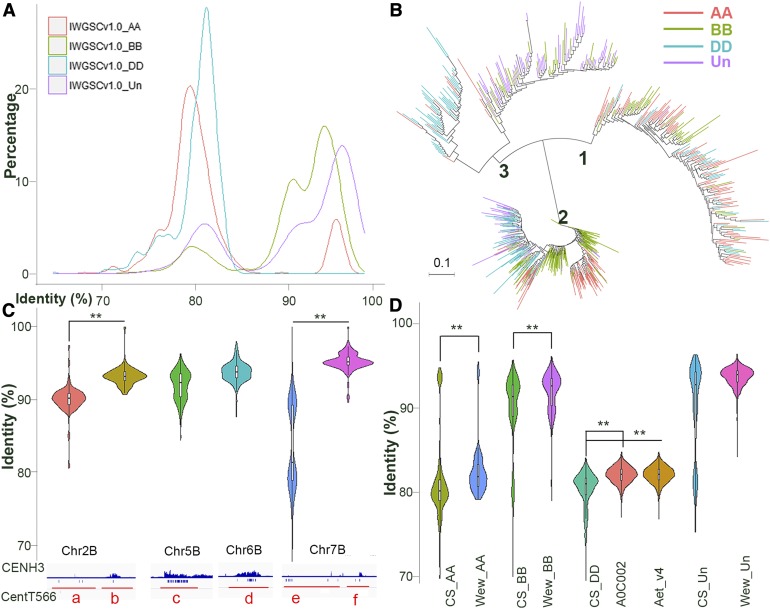
Centromeric satellites are associated with centromere function, and the variations in these highly repetitive sequences have been linked to centromere location or CENH3 binding (Henikoff et al., 2015; Maheshwari et al., 2017). These variations occur by mutation and unequal crossing over during evolution; however, little is known about how satellite sequences vary after polyploidization. To compare the sequence identity of wheat centromeric satellites after polyploidization, we identified all CentT566 satellite sequences in T. aestivum and calculated their sequence identity. Overall, the average pairwise identity between all CentT566 repeats identified in the T. aestivum genome was 87.99%, revealing a considerable level of genetic variation among the CentT566 tandem repeats. Two major sequence identity peaks with ∼80 and 95% identity were identified in T. aestivum genome assemblies IWGSC_v1.0 and TGACv1, and the 95% identity peak was the main peak (Supplemental Figure 6A).
We then investigated the patterns of sequence identity of CentT566 between different subgenomes. As expected, the distribution of CentT566 sequence identity in IWGSC_v1.0 unassigned scaffolds was similar to that of CentT566 distributed genome wide (Figure 4A, purple line), with the 95% as the main peak, revealing that most of the CentT566 from the unassigned scaffolds have the highest sequence identity. Only one major peak with 81% sequence identity was identified in the DD subgenome of bread wheat (Figure 4A, blue line). However, two levels of CentT566 identity were observed in the AA/BB subgenomes, with 79% as the main peak in the AA subgenome and 94% as the main peak in the BB subgenome (Figure 4A, red line for AA, green line for BB).
Figure 4.
CentT566 Satellite Sequence Variation Influences the Association with CENH3 Nucleosomes, and Sequence Polymorphisms in Wheat Genomes of Different Ploidy Levels.
(A) Sequence identity with the CentT566 satellite consensus sequence, as sampled in three subgenomes and one unassigned scaffold (Un) of CS IWGSC_v1.0 wheat. AA in red; BB, green; DD, cyan blue; and Un, purple. The x axis represents the sequence identity; the y axis represents the proportion of the CentT566 satellites.
(B) Phylogenetic tree of all CentT566 repeats with branches colored by subgenomes in bread wheat genome assemblies IWGSC_v1.0.
(C) Violin plot of the sequence identity in the CentT566 consensus sequence, as sampled in specific regions shown in Supplemental Figure 2A (a to f). The first track represents the enrichment of CENH3 reads in the corresponding regions. The second track represents the distribution of CentT566 satellites in the corresponding regions. **P < 0.001 (two-tailed Student’s t test)
(D) Violin plot of the sequence identity with the CentT566 consensus sequence, as sampled in specific wheat genomes and subgenomes of different ploidy levels. A0C002 and Aet_v4.0 are two versions of the wheat Ae. tauschii genomes; IWGSC_v1.0_AA, BB, DD, and Un represent the subgenomes of bread wheat, TRIDC_Wew_AA, BB and unassigned (Un) represent the subgenomes of wild emmer wheat T. turgidum. **P < 0.001 (two-tailed Student’s t test)
We constructed a phylogenetic tree of all CentT566 instances (Supplemental Files 1 and 3) colored by subgenomes in bread wheat genome assemblies IWGSC_v1.0 to describe the patterns of diversity (Figure 4B). Using the TGACv1 genome reference produced a similar tree (Supplemental Figure 6B, Supplemental Files 2 and 4). We partitioned the all the CentT566 repeats into three subpopulations (Figure 4B). Overwhelmingly, two subclades, 1 and 3, were mainly found in the AA and DD subgenomes, respectively, and some CentT566 repeats from unassigned scaffolds belonged to the DD subpopulations, suggesting that these repeats are from the DD subgenome. Most of the CentT566 repeats from the BB subgenome were classified into subclade 2. Some repeats from AA, DD, and ChrUn also belong to this subclade. Clusters 1 and 3 had the most divergence and the greatest branch lengths, whereas cluster 2 was more homogeneous. This phylogenetic relationship was consistent with the distribution of sequence identity and the divergent CentT566 satellite sequences within three subgenomes.
Several observations suggest that only some tandem arrays are associated with CENH3 nucleosomes. For example, Cen6B had two separate FISH signals for the CentT566 satellite but only one CENH3 signal (Figure 2C). Moreover, the genomic distribution of satellites in wheat genome shows that most of the CentT566 satellite arrays are associated with CENH3 nucleosomes in BB centromeres, whereas most CentT566 sequences in AA and DD centromeres were located in the pericentromeric or other genomic regions (Supplemental Figures 2 and 3). The phylogenetic tree and the distribution of sequence identity indicate that the CentT566 satellite arrays associated with CENH3 nucleosomes have fewer sequence polymorphisms than the CentT566 satellite arrays not associated with CENH3. The high sequence identity in different regions containing CentT566 repeats supports this hypothesis. Indeed, the sequence identity of CENH3 binding CentT566 satellite arrays was significantly higher than that of CentT566 satellites not bound to CENH3 (Figure 4C). This result suggests that the centromeric satellites with more homogeneous identity represent the centromere cores.
We then compared the sequence identities between the subdomains of the wheat genome and the corresponding progenitor genomes. Overall, a high level of sequence identity and few polymorphisms for the CentT566 satellites were detected between the BB subgenomes in T. aestivum and wild emmer wheat (Triticum turgidum; Figure 4D), as most CENH3 binding CentT566 repeats were observed in the BB subgenome (Supplemental Figure 2). Surprisingly, the identity with the consensus sequence was higher in the progenitors wild emmer wheat (AA/BB subgenomes) and Ae. tauschii (DD genome) compared with the corresponding subgenomes of bread wheat (Figure 4D), indicating that more polymorphisms in CentT566 satellites were generated during the evolution from the diploid and tetraploid to hexaploid wheat.
Gene Distribution and Expression in CENH3 and H3 Subdomains of Wheat Centromeres
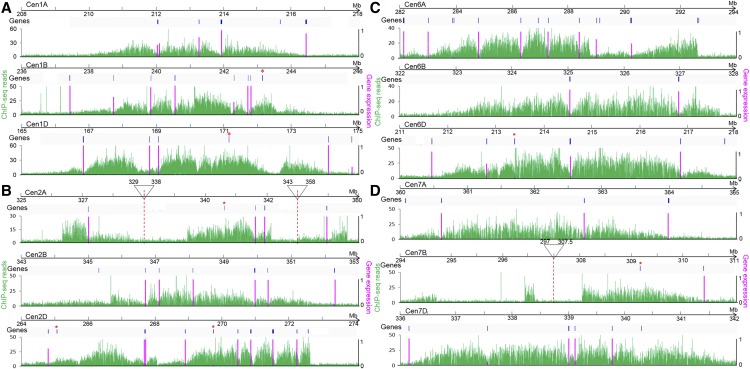
We observed an uneven distribution of CENH3 peaks within the 21 wheat centromeres (Figure 5; Supplemental Figure 7), representing CENH3-enriched and CENH3-depleted subdomains, as previously observed in the centromeres of rice, maize, and other species (Yan et al., 2008; Su et al., 2016). To distinguish the two subdomains, we examined the distribution of genes located in CENH3-enriched or CENH3-depleted subdomains in bread wheat.
Figure 5.
Gene Distribution and Expression in CENH3 and H3 Subdomains of Wheat Centromeres in Homoeologous Group Chromosomes.
(A) Cen1A, 1B, and 1D. 1A: 208,000,000 to 218,000,000; 1B: 236,000,000 to 246,000,000; 1D: 165,000,000 to 175,000,000.
(B) Cen2A, 2B, and 2D. 2A: 325,000,000 to 329,000,000, 338,000,000 to 343,000,000, 358,000,000 to 360,000,000; 2B: 343,000,000 to 353,000,000; 2D: 264,000,000 to 274,000,000.
(C) Cen6A, 6B, and 6D. 6A: 282,000,000 to 294,000,000; 6B: 322,000,000 to 328,000,000; 6D: 211,000,000 to 218,000,000.
(D) Cen7A, 7B, and 7D. 7A: 360,000,000 to 365,000,000; 7B: 294,000,000 to 297,000,000, 307,500,000 to 311,000,000; 7D: 336,000,000 to 342,000,000.
The horizontal scale marks a 10-Mb region including the core centromere for each chromosome. The first track shows the chromosomal locations of annotated genes. The asterisks represent the genes located within the CENH3 binding subdomain (Supplemental Data Set 1). The green line in the second track represents the enrichment of CENH3 ChIP-seq reads in each 100-bp window. The vertical purple bars show the percentage of gene expression in 18 different tissues and developmental stages from 56 public RNA-seq data sets.
To this end, IWGSC_v1.0 gene models were annotated in each wheat centromere. In total, 133 high-confidence genes were identified in wheat centromeric regions, but only seven genes located within CENH3-occupied subdomains were detected (Supplemental Data Set 1), indicating that most centromeric genes are located in the H3 subdomains. We detected 49, 34, and 50 genes in the centromeres of the AA, BB, and DD subgenomes, respectively.
We investigated the expression levels of the putative centromeric genes using the 56 publicly available RNA-seq data sets from 18 different tissues and developmental stages (Supplemental Data Set 2). Most genes had expression levels of Reads Per Kilobase per Million mapped reads (RPKM) >=0.5; only 30 of 133 genes were not expressed in any sample. However, the seven genes within CENH3-occupied subdomains were not expressed in most situations. Only three of the seven genes were expressed in a specific tissue and/or developmental stage (TraesCS1B01G152400 and TraesCS2D01G242300 in spikes during anthesis; TraesCS6D01G182500 in leaves/shoots; Figures 5A to 5C; Supplemental Data Set 3). By contrast, most genes (99 of the 126) located within H3 subdomains were expressed in at least one tissue (Figure 5; Supplemental Figure 7; Supplemental Data Set 2). These results indicate that genes within centromeric H3 subdomains are expressed in most cases, but genes associated with CENH3 nucleosomes are expressed only in a tissue- and/or stage-specific manner.
Rearrangement of Centromeres from the DD Subgenome of Ae. tauschii versus T. aestivum
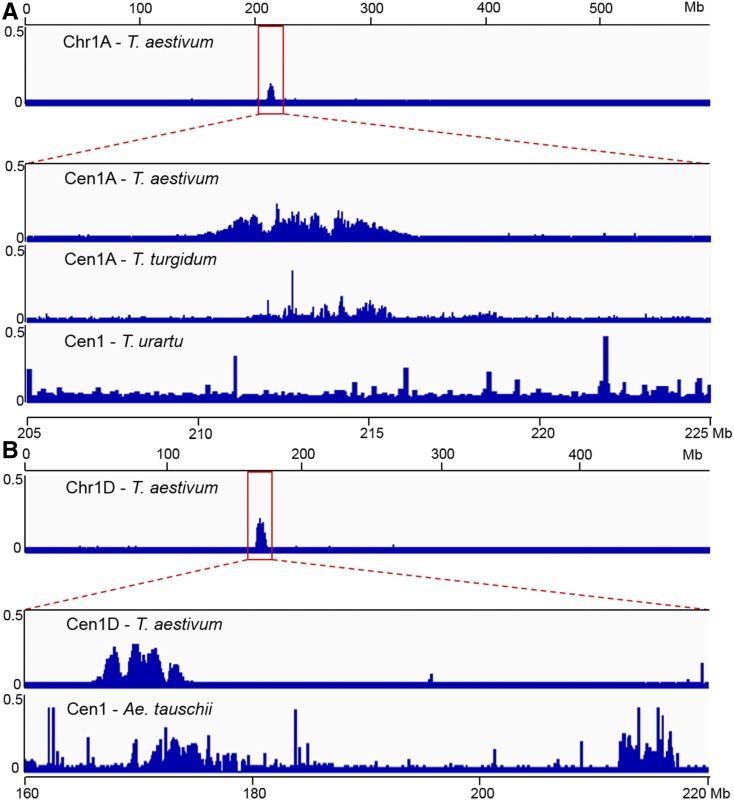
To further investigate the changes that occurred within the centromeric regions during wheat evolution, we remapped the CENH3-ChIP-seq reads from CS to the genome of wild emmer T. turgidum (AABB), the AA progenitor T. urartu, and the DD progenitor Ae. tauschii. Approximately 41.2, 52.8, and 33.8% of the reads were aligned to distinct positions in the AABB, AA, and DD chromosomes of the reference genomes, respectively. Some significant peaks coincided with the distribution of fam12 or CRW sequences in the genome-wide maps of AABB and DD (Figure 6; Supplemental Figures 8 and 9), indicating that these peaks indeed represent centromeric regions.
Figure 6.
Genome-Wide Mapping of CS CENH3 ChIP-Seq Reads to the AA, AABB, DD, and AABBDD References Genomes.
(A) Distribution of the density of CS CENH3 ChIP-seq reads along Chr1A of the AABBDD reference genome (first track) using the Integrative Genomic Viewer tool. The second track shows the enrichment of CENH3 reads in the enlarged view of Cen1A (205,000,000 to 225,000,000) in CS wheat. The third and fourth tracks represent the distribution of CS CENH3 ChIP-seq reads along the corresponding centromeric regions of chromosome 1A from the AABB and AA reference genomes, respectively. The x-coordinates of the zoomed-in-tracks of T. turgidum and T. uratu are the same as that of T. aestivum.
(B) Distribution of the density of CS CENH3 ChIP-seq reads along Chr1D of the AABBDD reference genome (first track). The second track shows the enrichment of CENH3 reads in the enlarged view of Cen1D (160,000,000 to 220,000,000) in CS wheat. The third track represents the distribution of CS CENH3 ChIP-seq reads along the corresponding centromeric region of Chr1D from the DD reference genome. The x-coordinate of the zoomed-in-tracks of Ae. tauschii is the same as that of T. aestivum.
However, in contrast to the AA, BB, and DD subgenomes of bread wheat, some narrow peaks were also observed on the chromosome arms of wild emmer and Ae. tauschii wheat (Supplemental Figures 8 to 10). Furthermore, no significant peaks and only a few traces of centromeric sequences were preserved in the centromeres of the AA diploid T. urartu (Figure 6A; Supplemental Figure 10). These results suggest that centromeric sequences were mainly retained, but some changes also occurred, from the progenitor of DD and wild emmer (AABB) to hexaploid wheat. The A genome donor T. urartu was involved in the initial hybridization event during hexaploid wheat evolution, suggesting intense divergence of the centromeric DNA sequences between the AA progenitor and AA subgenome in wild emmer and bread wheat during and/or after polyploidization.
The availability of assembled reference genomes with different ploidy levels allowed us to investigate the genetic consequences of polyploidization and adaptation in wheat. We performed BLAST and BLAT analyses to detect the orthologs of high-confidence centromeric genes between Ae. tauschii and the CS wheat DD subgenome. Most of the centromeric genes (40/50) in the DD subgenome of CS have an ortholog in the corresponding chromosomes of Ae. tauschii (Supplemental Data Set 3). Six genes in bread wheat have corresponding DNA sequences in the unassigned scaffolds of the Ae. tauschii genome (Supplemental Data Set 3), which likely belong to centromeric regions. The gene TraesCS7D01G290400 has two orthologs, including one on Chr4 and one in the unassigned scaffold NWVB01000004.1 of Ae. tauschii (Supplemental Data Set 3). The gene TraesCS3D01G201900.1 has two corresponding DNA sequences in the unassigned scaffolds NWVB01109126.1 and NWVB01000101.1 (Supplemental Data Set 3), likely due to the abundance of dispersed duplicated genes in Ae. tauschii (Luo et al., 2017).
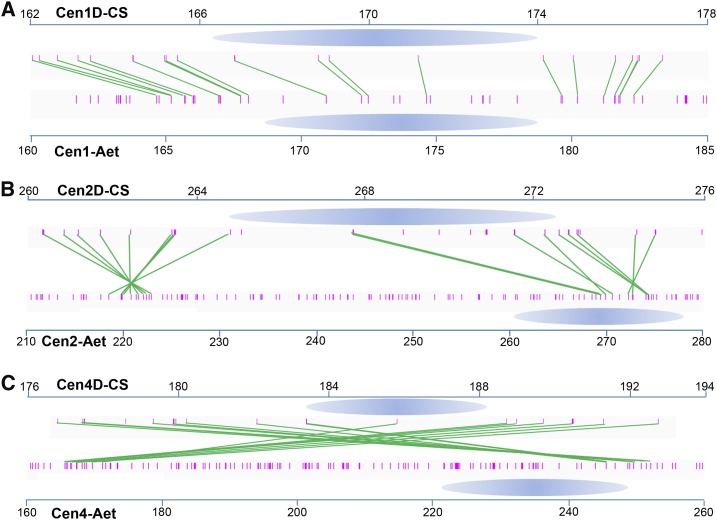
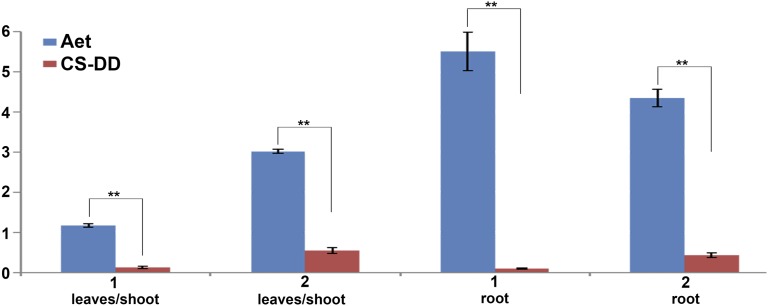
Collinearity analysis of genes within and surrounding the centromeric regions revealed no significant changes in centromeric positions from Ae. tauschii to the wheat DD subgenome (Figure 7; Supplemental Figure 11). The high confidence genes in Cen1D and Cen7D have orthologs with a consistent order on Cen1 and Cen7 of Ae. tauschii, respectively (Figure 7A; Supplemental Figure 11D). Interestingly, two separate domains on Chr2, Chr3, Chr4, Chr5, and Chr6 of Ae. tauschii were syntenic to the corresponding pericentromeres of the wheat DD subgenome (Figures 7B and 7C; Supplemental Figures 11A to 11C). Furthermore, several centromeric genes of Ae. tauschii lack orthologs at the corresponding positions in the centromeres of the DD subgenome of CS wheat (Figure 7; Supplemental Figure 11) and were therefore apparently lost from the centromeres of the wheat DD subgenome. These results suggest that large chromosomal deletions occurred from the progenitor of DD to bread wheat. Moreover, the gene orders were reversed within the centromeres of the two genomes, suggesting that inversions spanning the centromere occurred after polyploidization (Figure 7; Supplemental Figure 11). Orthologs of two highly expressed genes in Ae. tauschii, AET1Gv20340900 and AET6Gv20506100, showed reduced expression levels when associated with CENH3 nucleosomes in the DD centromeres of CS wheat (Figure 8). Taken together, the detection of gene loss, changes in gene expression levels, and inversions indicate that wheat centromeres have undergone rearrangement during and/or after polyploidization.
Figure 7.
Rearrangement of Centromeres between DD in Ae. tauschii and DD in T. aestivum after Polyploidization.
Syntenic relationship between the CS wheat DD and Ae. tauschii centromeres. The centromeres are marked as blue ovals. Short red bars represent the annotated genes in the CS wheat and Ae. tauschii reference genomes. Orthologous gene pairs are connected by green lines. Orthologous gene pairs between CS and Ae. tauschii were detected on both sides of the two centromeres. The horizontal scale marks the centromere locus (in megabases) on each chromosome.
(A) CS-Cen1D versus Aet-Cen1.
(B) CS-Cen2D versus Aet-Cen2.
(C) CS-Cen4D versus Aet-Cen4.
Figure 8.
Comparison of Centromeric Gene Expression between the CS Wheat DD Subgenome and Ae. tauschii.
The genes (1, TraesCS1D01G134500 and 2, TraesCS6D01G182500) associated with CENH3 nucleosomes in the DD centromere of CS wheat were significantly reduced compared with those of highly expressed orthologous genes in Ae. tauschii (AET1Gv20340900 and AET6Gv20506100). The expression levels from the RNA-seq data sets of leaves/shoot and root tissues were used for the two genes, respectively. The y axis represents the average gene expression level, calculated based on RPKM value. The error bars show the sd for four replicates per tissue. **P < 0.001 (two-tailed Student’s t test).
DISCUSSION
Classical plant centromeric sequences include interspersed satellite repeats and Ty3/Gypsy long terminal repeat retrotransposons, as demonstrated in barley (Hordeum vulgare), rice, maize, and Brachypodium distachyon (Hudakova et al., 2001; Yan et al., 2008; Wolfgruber et al., 2009; Li et al., 2018). However, wheat centromeric satellite repeats that are associated with CENH3 nucleosomes had not been identified. The methods used to detect centromeric sequence in previous studies were based on the screening of large DNA clones or BACs using known wheat centromeric sequences, such as the cereal centromeric repeat CCS1 and CRW sequences (Cheng and Murata, 2003; Liu et al., 2008; Li et al., 2013). However, BAC clones with centromeric satellites may be lost during the first step of screening satellite repeats. Here, we used CENH3-ChIP followed by high-throughput sequencing and performed graph-based clustering to identify centromeric sequences genome wide. This method is not dependent on the reference genome, which makes the results more comprehensive than those of previous studies. We identified two types of centromeric satellite repeats, which are associated with CENH3 nucleosomes. The CentT550 satellite is a satellite-like repeat, which was previously shown to not associate with CENH3 (Li et al., 2013). The CentT566 and CentT550 satellite sequences share no similarity (Supplemental Figure 1A), suggesting that they have different origins.
Our study uncovered several differences between wheat centromeres and the typical centromeres of other plant species. First, the unit size of the typical centromeric tandem repeats ranges from 150 to 180 bp, whereas the two subgenome-abundant satellite repeats in wheat centromeres are 566 and 550 bp long. Second, typical centromeres are mainly composed of large tracts of tandem repeats. However, the genomic portion of wheat centromeric satellites is very low, and the strength of both satellite signals in centromeres decreased from the diploid progenitors to hexaploid wheat. Several very weak CentT550 signals were detected in bread wheat (Supplemental Figure 1E). Lastly, most typical centromeres contain satellite repeats specific to a certain species. However, several wheat centromeres lacked satellite signals; the presence of the long unit-sized satellite may differentiate the centromeres of hexaploid wheat into repeat-containing and repeat-free centromeres. The presence of satellite-containing and satellite-free centromeres in the same species was also reported in potato and chicken cells. Several chromosome-specific centromeric satellites in potato have a unit size of up to several kilobases, whereas no satellite repeats were identified in some potato centromeres (Gong et al., 2012). The unit size of centromeric satellite repeats in chicken cells is dozens of base pairs (Shang et al., 2010). We therefore hypothesize that the presence of centromeric satellites with a very large unit size resulted in low copy numbers of these satellite due to the action transposable elements, resulting in the loss of satellite repeats in some cases.
Genomic variation is the basis of genetic diversity and is associated with differences in gene expression and function (Hamilton, 2002; Haraksingh and Snyder, 2013). The variation within centromeric satellites and its role in the association of satellites with CENH3 nucleosomes were largely unknown prior to recent reports in human and Arabidopsis (Henikoff et al., 2015; Maheshwari et al., 2017). These reports described significant genetic variation within centromeric satellites and identified young, homogeneous satellites as preferentially associated with CENH3 nucleosomes. However, the polymorphisms of satellites within wheat centromeres and their variation after polyploidization were previously not known. In the current study, FISH and analysis of the genomic distribution of the satellites indicated that only a portion of the satellites are associated with CENH3 (Figure 2; Supplemental Figures 2 to 3). Two peaks of sequence identity were present in all of the wheat reference genomes, and the phylogenetic tree of all the copies of CentT566 reveals the divergence and homogeneous CentT566 satellites in the three subgenomes (Figures 4A and 4B). Our results suggest that satellites with fewer polymorphisms preferentially associate with CENH3 nucleosomes in various wheat subgenomes (Figure 4C). The CentT566 satellites from the unassigned scaffolds have the highest sequence identity. They are likely to represent centromere cores and may be too homogeneous to assemble (Figure 4A). This finding is consistent with reports from other species (Henikoff et al., 2015; Maheshwari et al., 2017).
Moreover, we detected higher levels of polymorphism of the CentT566 satellite in the corresponding subgenomes of CS wheat than in the wheat progenitors Ae. tauschii and wild emmer wheat (Figure 4D), indicating that new polymorphisms of CentT566 satellite repeats arose during the evolution from diploid and tetraploid wheat to hexaploid wheat. Dynamic changes of centromeric retrotransposons were also observed in wheat (Li et al., 2013). However, the current diploid and tetraploid wheat species are, strictly speaking, not the progenitors of hexaploid wheat, but the offspring of the actual progenitors. With the very fast-evolving centromeric sequences (even lines within one species are different in the FISH analyses; Figure 2; Supplemental Figure 1), there is some uncertainty in deducing their composition at the time of polyploidization from their current genotypes.
Polyploids occur in most plant species due to interspecific hybridization and genome doubling. Rapid genomic and epigenomic changes have been documented in response to the genome shock that occurs during the early stage of plant polyploidy (Feldman and Levy, 2012; Song and Chen, 2015). In hybrid species, the transcriptional patterns of CENH3 genes from two different parents might change to fit the new genomic environment, and the new CENH3 expression patterns might inevitably lead to changes in centromeres. The identification of two types of CENH3 genes in wheat suggests that functional differentiation had occurred during evolution (Yuan et al., 2015). On the other hand, the centromere acts as the hub of chromosome movement during cell division, and the centromeres of allopolyploids must adopt the proper conformations for association during meiosis to avoid the formation of unbalanced gametes. The behavior of centromeres in wide wheat hybrids likely generates chromosomal biodiversity, with implications for speciation (Guo et al., 2016). The asymmetrical evolution of polyploid subgenomes has been reported in Brassica, cotton (Gossypium hirsutum), and wheat (Liu et al., 2014; Zhang et al., 2015; Pont and Salse, 2017). Such asymmetry, including gene loss, differences in gene expression, transposable element amplification, and structural rearrangements among different subgenomes might drive diploidization in polyploid plants (Thomas et al., 2006; Guo and Han, 2014; Renny-Byfield et al., 2014), which is essential for genome stability and ultimately for speciation. Two subgenome-abundant centromeric satellites in diploid wheat progenitors, and the dynamic changes of centromeres during the evolution of polyploid wheat, point to the asymmetric distribution of centromeric sequences within the three subgenomes in hexaploid wheat. This may reflect the role of centromeres in homologous chromosome pairing during the early phase of meiosis in polyploid species (Da Ines and White, 2015).
METHODS
Plant Materials
All wheat materials including diploid, tetraploid, and hexaploid species used in this study (Triticum aestivum and Triticum urartu; Aegilops speltoides, Aegilops longissima, and Aegilops tauschii) are listed in Table 4, and synthetic tetraploid wheat plants were generated in our laboratory. All wheat seeds were germinated at room temperature for several days until the root tips were 2 to 3 cm long, and the plants were then transplanted into soil and grown in the greenhouse at 20°C and under full natural light.
Table 4. Species and Lines of Diploid and Hexaploid Wheat Used in this Study.
| Species and Cultivars | Line Designation | Genome |
|---|---|---|
| T. urartua | G1812 | AA |
| TMU06 | AA | |
| TMU38 | AA | |
| Ae. speltoidesa | Ae92 | SS |
| Ae739 | SS | |
| Ae346 | SS | |
| Ae. longissimaa | TL05 | SlSl |
| Ae. tauschiia | AL8/78 | DD |
| TQ27 | DD | |
| T. aestivumb | CS | BBAADD |
Diploid (2n=2X=14)
Hexaploid (2n=6X=42)
Mapping of ChIP-Seq Reads to the Wheat Reference Genome
The ChIP-seq experiment with anti-CENH3 antibodies was performed according to a previously described method (Liu et al., 2015). The wheat-specific anti-αCENH3 antibodies were used for ChIP (Yuan et al., 2015). Quality control of raw 101-bp paired-end reads was performed with FastQC software, and adapters and low-quality reads were trimmed using the Trimmomatic (Bolger et al., 2014) with the parameters “ILLUMINACLIP:adapter.fa:2:30:10 LEADING:20 TRAILING:20 MINLEN:36 SLIDINGWINDOW:4:20”. The trimmed reads were mapped to the reference genomes of bread wheat (IWGSC_v1.0 and TGACv1; Clavijo et al., 2017; Zimin et al., 2017), wild emmer (TRIDC_Wew; Avni et al., 2017), and Ae. tauschii (Aet_v4.0 and AOC001; Luo et al., 2017; Zhao et al., 2017) using the BWA-MEM software with default parameters (Li and Durbin, 2009). The uniquely mapped reads with mapping quality values > 20 were used for further analysis. The alignment SAM files were converted to BAM files and sorted, and duplicates were removed using SAMtools (Li et al., 2009). Read coverage and enrichment were counted in each 100-kb window along the chromosomes, and the plots were produced using OriginPro software (Stevenson, 2011). The adjusted CENH3 ChIP-seq peaks calculated based on reads per million values were displayed with Integrative Genome Viewer (Thorvaldsdóttir et al., 2013). The centers of CENH3-containing and canonical nucleosomes were determined using nucleR (Flores and Orozco, 2011), and the CENH3 nucleosomes on the CentT566 satellite were identified based on the distribution of CentT566 sequences. The paired-end reads were joined using SeqPrep software with the parameters “-q 30 -L 25” (https://github.com/jstjohn/SeqPrep). Merged reads obtained from CENH3 ChIP-seq and Input-seq were aligned to a dimerized CentT566 satellite consensus sequence using BWA-MEM software (Li and Durbin, 2009), and the midpoints of the merged reads along the dimerized consensus sequence were used to generate nucleosome midpoint position plots. The anti-CENH3 ChIP-seq data were obtained from the Gene Expression Omnibus (GEO) database (accession number GSE63752), and the input data were obtained from Sequence Read Archive study PRJNA420988 (SRR6350669). Data processing and analysis were performed using Perl, and the figures were plotted with R.
Mapping of RNA-Seq Reads
The 56 public RNA-seq data sets from 18 different tissues of bread wheat cv CS and public RNA-seq data sets from two different tissues of Ae. tauschii (listed in Supplemental Data Set 2) were collected from the GEO database (Barrett et al., 2013). The reads were quality trimmed as described for the ChIP-seq reads. The Hisat2, StringTie, and Ballgown pipelines were used to analyze the RNA-seq data sets (Pertea et al., 2016). The trimmed reads from different lines were aligned to the bread wheat IWGSC_v1.0 and Ae. tauschii Aet_v4.0 reference genomes using HISAT2 software (Kim et al., 2015). Transcript assembly and quantification were performed for each sample using StringTie, including high confidence reference annotation (Pertea et al., 2015). The annotated centromeric genes were defined according to the centromere positions on the tested chromosomes. The genes associated with CENH3 nucleosomes were defined according to the CENH3-enrichment level. The reads per kilobase per million mapped reads (RPKM) value was treated as the gene expression level. The expression levels of centromeric genes in different tissues were calculated using the mean RPKM values for each tissue. Four replicate RNA-seq samples for leaves/shoot and root tissues were used for the comparison of expression levels of orthologous genes between CS and Ae. tauschii. Error bars show the sd. Double asterisks denote significant differences with P < 0.001 (two-tailed Student’s t test).
Identification of Orthologous Genes between Ae. tauschii and the D Subgenome of CS Wheat
The sequences of the annotated high-confidence class coding genes of CS wheat D subgenome centromeres (IWGSC_v1.0) were used as queries to identify the genes in Ae. tauschii. The protein and coding sequence nucleotide sequences were searched against the Ae. tauschii database using BLASTP and BLASTN with an E-value cut-off of 10−5 (Camacho et al., 2009). The bidirectional best BLAST hit for every gene was interrogated by bit score, E-value, and percentage identity. The coding sequence sequences of IWGSC_v1.0 centromeric genes were also used as queries for BLAST analysis (Kent, 2002) to the Ae. tauschii reference genome to confirm the results of bidirectional best BLAST hit analysis.
Identification and Characterization of Repeats
A set of 1.14 million randomly selected whole-genome shotgun 454 sequence reads from bread wheat cultivar CS (Brenchley et al., 2012) were analyzed using Web-based Galaxy RepeatExplorer software (https://repeatexplorer-elixir.cerit-sc.cz/galaxy/). The genome portion of each repeat cluster was determined using this software. The cluster repeat sequences were subsequently used as a database for BLAST searches. Approximately 118 million 101-bp paired-end reads from Illumina sequencing of bread wheat by IWGSC were treated as input. The CENH3-ChIP-seq and input reads were then subjected to BLAST analysis (Camacho et al., 2009) against the repeat clusters using the parameters “-evalue 1e-8 -num_alignment 1 -wordsize 9 -dust no -gapopen 5 -gapextend 2 -penalty -3 -reword 2”. The ratio between the numbers for each repeat cluster was calculated and represents the CENH3-enrichment level. The high-ratio clusters were amplified with specific PCR primers (Supplemental Table 1), and the clones were confirmed by sequencing. The clones were then used as probes for FISH.
Characterization of Genetic Diversity within Centromeric Satellites in Bread Wheat
All the individual CentT566 repeats were extracted based on BLAST output for each subgenome in wheat. The cd-hit-est software was used to cluster similar CentT566 sequences into clusters using parameters “-c 0.90 -n 10” within each subgenome (Fu et al., 2012). The clusters with length larger than 500 bp were used for further analysis. Multiple sequence alignment and phylogenetic analysis of all CentT566 clusters were performed with ClustalW and MEGA7 software using the neighbor-joining method (Kumar et al., 2016). The phylogenetic tree with branches colored by subgenomes was constructed with iTOL (Letunic and Bork, 2019).
FISH and Genomic in Situ Hybridization
Root tip cells from different plants were prepared for FISH and genomic in situ hybridization as described previously (Fu et al., 2013; Guo et al., 2016). The probes of repeats CentT566, CentT550, and other genomic DNA sequences were labeled with Alexa Fluor-594-5-dUTP (red) or Alexa Fluor-488-dUTP (green). The repetitive sequences pAs1 and pSc119.2 were used to karyotype the chromosomes of bread wheat (Zhang et al., 2013a). Chromosome samples from different wheat lines were exposed to the same conditions and to equal amounts of probes. The images were acquired by confocal microscopy (Cell Observer spinning disk confocal microscope, Zeiss) using the same exposure time and were processed with Photoshop CS 6.0 (Adobe).
Genome-Wide Identification of Centromeric Satellite Repeats and Calculation of Percent Identity among Repeats
The MegaBLAST tool (Morgulis et al., 2008) was used to identify all CentT566 and CentT550 repeats within the different assembled wheat reference genomes with an E-value cut-off of 10−5. These repeats were further filtered to exclude those <500 bp long. The sequence identity with the CentT566 and CentT550 consensus sequences was calculated using R software. The method to get the consensus sequences was performed as described by Jonathan et al. (Gent et al., 2017). The clusters of repeat sequences from Input-seq reads were produced by RepeatExplorer (Novák et al., 2010), and the circular consensus sequences of all satellite copies were made using the Geneious version 8.0.4 De Novo Assembly tool with default parameters. The CentT566 and CentT550 consensus sequences used for identity analyses among the different wheat reference genomes are submitted to GenBank with accession numbers MN161205 and MN161206.
Accession Numbers
The anti-CENH3 ChIP-seq data were obtained from the GEO database (accession number GSE63752), and the input data were from Sequence Read Archive study PRJNA420988 (SRR6350669). All the RNA-seq data sets analyzed during this study are included in Supplemental Data Set 1. The centromere satellite DNA sequences are submitted to the GenBank with accession numbers MN161205 and MN161206.
Supplemental Data
Supplemental Figure 1. FISH confirmation of predicted centromeric satellites in various wheat lines.
Supplemental Figure 2. Distribution of CentT566 and CentT550 satellite sequences in the T. aestivum chinese spring genome.
Supplemental Figure 3. Distribution of CentT566 and CentT550 satellite sequences in wheat genomes of different ploidy levels.
Supplemental Figure 4. The satellite-like sequences were inserted within the centromeric retrotransposon and changed during wheat polyploidization.
Supplemental Figure 5. CENH3 nucleosomes are highly phased with specific sites on the wheat centromeric satellite CentT550 monomer and centered on WW dinucleotides.
Supplemental Figure 6. Characterization of genetic diversity within CentT566 satellite repeats in the T. aestivum.
Supplemental Figure 7. Gene annotations and expression within intermingled CENH3 and H3 subdomains of T. aestivum centromeric regions of homoeologous group chromosomes 3–5.
Supplemental Figure 8. Genome-wide mapping of CENH3 ChIP-seq reads from chinese spring wheat to the wild emmer T. turgidum reference genome (AABB).
Supplemental Figure 9. Genome-wide mapping of CENH3 ChIP-seq reads from chinese spring wheat to the reference genome of wheat DD progenitor Ae. tauschii.
Supplemental Figure 10. Genome-wide mapping of CENH3 ChIP-seq reads from chinese spring wheat to the reference genome of wheat AA progenitor T. urartu.
Supplemental Figure 11. Syntenic relationship between the CS wheat DD-subgenome and Ae. tauschii centromeres.
Supplemental Table 1. Primers used in this study.
Supplemental File 1. Sequence alignments file of CentT566 repeats in T. aestivum genome assemblies IWGSC_v1.0.
Supplemental File 2. Sequence alignments file of CentT566 repeats in T. aestivum genome assemblies TGACv1.
Supplemental File 3. Phylogenetic tree file of CentT566 repeats in T. aestivum genome assemblies IWGSC_v1.0.
Supplemental File 4. Phylogenetic tree file of CentT566 repeats in T. aestivum genome assemblies TGACv1.
Supplemental Data Set 1. Expression levels of annotated centromeric genes in chinese spring from different tissues and developmental stages.
Supplemental Data Set 2. RNA-seq metadata from different tissues and developmental stages of chinese spring wheat and Ae. tauschii.
Supplemental Data Set 3. Identification of orthologous genes between Ae. tauschii and the DD-subgenome of CS.
Acknowledgments
We thank Ingo Schubert from the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) and James A. Birchler from the University of Missouri for critical reading of the article and helpful comments. This work was supported by the National Natural Science Foundation of China (31630049 and 31320103912) and the National Key Research and Development Program of China (2016YFD0102001).
AUTHOR CONTRIBUTIONS
F.H., H.S., and Y.L. designed the work and wrote the article. S.H.D., Y.L., C.L., Q.S., Y.H., and F.H. performed research, analyzed data, and edited the article. All authors read and approved the final article.
Footnotes
Articles can be viewed without a subscription.
References
- Aldrup-MacDonald M.E., Kuo M.E., Sullivan L.L., Chew K., Sullivan B.A. (2016). Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genome Res. 26: 1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev E.V., Phillips R.L., Rines H.W. (1998). Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95: 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni R., et al. (2017). Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357: 93–97. [DOI] [PubMed] [Google Scholar]
- Barrett T., et al. (2013). NCBI GEO: archive for functional genomics data sets--Update. Nucleic Acids Res. 41: D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley R., et al. (2012). Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491: 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. (2009). BLAST+: Architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z.J., Murata M. (2003). A centromeric tandem repeat family originating from a part of Ty3/gypsy-retroelement in wheat and its relatives. Genetics 164: 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Dong F., Langdon T., Ouyang S., Buell C.R., Gu M., Blattner F.R., Jiang J. (2002). Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14: 1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choulet F., et al. (2014). Structural and functional partitioning of bread wheat chromosome 3B. Science 345: 1249721. [DOI] [PubMed] [Google Scholar]
- Clavijo B.J., et al. (2017). An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 27: 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Maheshwari S., Marimuthu M.P.A. (2017). Plant centromeres. Curr. Opin. Plant Biol. 36: 158–167. [DOI] [PubMed] [Google Scholar]
- Da Ines O., White C.I. (2015). Centromere associations in meiotic chromosome pairing. Annu. Rev. Genet. 49: 95–114. [DOI] [PubMed] [Google Scholar]
- Dong Q., Han F. (2012). Phosphorylation of histone H2A is associated with centromere function and maintenance in meiosis. Plant J. 71: 800–809. [DOI] [PubMed] [Google Scholar]
- Earnshaw W.C., Rothfield N. (1985). Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91: 313–321. [DOI] [PubMed] [Google Scholar]
- Feldman M., Levy A.A. (2012). Genome evolution due to allopolyploidization in wheat. Genetics 192: 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores O., Orozco M. (2011). nucleR: A package for non-parametric nucleosome positioning. Bioinformatics 27: 2149–2150. [DOI] [PubMed] [Google Scholar]
- Fu L., Niu B., Zhu Z., Wu S., Li W. (2012). CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S., Lv Z., Guo X., Zhang X., Han F. (2013). Alteration of terminal heterochromatin and chromosome rearrangements in derivatives of wheat-rye hybrids. J. Genet. Genomics 40: 413–420. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Earnshaw W.C. (2014). The centromere: Chromatin foundation for the kinetochore machinery. Dev. Cell 30: 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J.I., Schneider K.L., Topp C.N., Rodriguez C., Presting G.G., Dawe R.K. (2011). Distinct influences of tandem repeats and retrotransposons on CENH3 nucleosome positioning. Epigenetics Chromatin 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J.I., Wang N., Dawe R.K. (2017). Stable centromere positioning in diverse sequence contexts of complex and satellite centromeres of maize and wild relatives. Genome Biol. 18: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Wu Y., Koblízková A., Torres G.A., Wang K., Iovene M., Neumann P., Zhang W., Novák P., Buell C.R., Macas J., Jiang J. (2012). Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell 24: 3559–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Han F. (2014). Asymmetric epigenetic modification and elimination of rDNA sequences by polyploidization in wheat. Plant Cell 26: 4311–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Su H., Shi Q., Fu S., Wang J., Zhang X., Hu Z., Han F. (2016). De novo centromere formation and centromeric sequence expansion in wheat and its wide hybrids. PLoS Genet. 12: e1005997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A.E., Keith K.C., Hall S.E., Copenhaver G.P., Preuss D. (2004). The rapidly evolving field of plant centromeres. Curr. Opin. Plant Biol. 7: 108–114. [DOI] [PubMed] [Google Scholar]
- Hall S.E., Kettler G., Preuss D. (2003). Centromere satellites from Arabidopsis populations: Maintenance of conserved and variable domains. Genome Res. 13: 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B.A. (2002). Variations in abundance: Genome-wide responses to genetic variation and vice versa. Genome Biol. 3: 1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraksingh R.R., Snyder M.P. (2013). Impacts of variation in the human genome on gene regulation. J. Mol. Biol. 425: 3970–3977. [DOI] [PubMed] [Google Scholar]
- Hasson D., Panchenko T., Salimian K.J., Salman M.U., Sekulic N., Alonso A., Warburton P.E., Black B.E. (2013). The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat. Struct. Mol. Biol. 20: 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff J.G., Thakur J., Kasinathan S., Henikoff S. (2015). A unique chromatin complex occupies young α-satellite arrays of human centromeres. Sci. Adv. 1: pii: e1400234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudakova S., Michalek W., Presting G.G., ten Hoopen R., dos Santos K., Jasencakova Z., Schubert I. (2001). Sequence organization of barley centromeres. Nucleic Acids Res. 29: 5029–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: 7191. [DOI] [PubMed] [Google Scholar]
- Ioshikhes I., Hosid S., Pugh B.F. (2011). Variety of genomic DNA patterns for nucleosome positioning. Genome Res. 21: 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Nasuda S., Endo T.R. (2004). A direct repeat sequence associated with the centromeric retrotransposons in wheat. Genome 47: 747–756. [DOI] [PubMed] [Google Scholar]
- Iwata-Otsubo A., Dawicki-McKenna J.M., Akera T., Falk S.J., Chmatal L., Yang K., Sullivan B.A., Schultz R.M., Lampson M.A., Black B.E. (2017). Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis. Curr. Biol. 27: 2365–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S., Chen Z.J. (2010). Genomic and expression plasticity of polyploidy. Curr. Opin. Plant Biol. 13: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Nasuda S., Dong F., Scherrer C.W., Woo S.S., Wing R.A., Gill B.S., Ward D.C. (1996). A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc. Natl. Acad. Sci. USA 93: 14210–14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. (2002). BLAT--The BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishii M., Nagaki K., Tsujimoto H. (2001). A tandem repetitive sequence located in the centromeric region of common wheat (Triticum aestivum) chromosomes. Chromosome Res. 9: 417–428. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2019). Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47 (W1): W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Choulet F., Heng Y., Hao W., Paux E., Liu Z., Yue W., Jin W., Feuillet C., Zhang X. (2013). Wheat centromeric retrotransposons: The new ones take a major role in centromeric structure. Plant J. 73: 952–965. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zuo S., Zhang Z., Li Z., Han J., Chu Z., Hasterok R., Wang K. (2018). Centromeric DNA characterization in the model grass Brachypodium distachyon provides insights on the evolution of the genus. Plant J. 93: 1088–1101. [DOI] [PubMed] [Google Scholar]
- Ling H.Q., et al. (2018). Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 557: 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., et al. (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5: 3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Su H., Pang J., Gao Z., Wang X.J., Birchler J.A., Han F. (2015). Sequential de novo centromere formation and inactivation on a chromosomal fragment in maize. Proc. Natl. Acad. Sci. USA 112: E1263–E1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Yue W., Li D., Wang R.R., Kong X., Lu K., Wang G., Dong Y., Jin W., Zhang X. (2008). Structure and dynamics of retrotransposons at wheat centromeres and pericentromeres. Chromosoma 117: 445–456. [DOI] [PubMed] [Google Scholar]
- Luo M.C., et al. (2017). Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551: 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macas J., Neumann P., Navrátilová A. (2007). Repetitive DNA in the pea (Pisum sativum L.) genome: Comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics 8: 427–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S., Ishii T., Brown C.T., Houben A., Comai L. (2017). Centromere location in Arabidopsis is unaltered by extreme divergence in CENH3 protein sequence. Genome Res. 27: 471–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcussen T., Sandve S.R., Heier L., Spannagl M., Pfeifer M., Jakobsen K.S., Wulff B.B., Steuernagel B., Mayer K.F., Olsen O.A.; International Wheat Genome Sequencing Consortium (2014). Ancient hybridizations among the ancestral genomes of bread wheat. Science 345: 1250092. [DOI] [PubMed] [Google Scholar]
- Martín A.C., Borrill P., Higgins J., Alabdullah A., Ramírez-González R.H., Swarbreck D., Uauy C., Shaw P., Moore G. (2018). Genome-wide transcription during early wheat meiosis is independent of synapsis, ploidy level, and the Ph1 locus. Front. Plant Sci. 9: 1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters D.P., et al. (2013). Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 14: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgulis A., Coulouris G., Raytselis Y., Madden T.L., Agarwala R., Schäffer A.A. (2008). Database indexing for production MegaBLAST searches. Bioinformatics 24: 1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P., Neumann P., Macas J. (2010). Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11: 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer D.K., Margolis R.L. (1985). Kinetochore components recognized by human autoantibodies are present on mononucleosomes. Mol. Cell. Biol. 5: 173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11: 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta A.F., Mackay A.M., Ainsztein A.M., Goldberg I.G., Earnshaw W.C. (1995). The centromere: Hub of chromosomal activities. Science 270: 1591–1594. [DOI] [PubMed] [Google Scholar]
- Pont C., Salse J. (2017). Wheat paleohistory created asymmetrical genomic evolution. Curr. Opin. Plant Biol. 36: 29–37. [DOI] [PubMed] [Google Scholar]
- Ramírez-González R.H., et al. (2018). The transcriptional landscape of polyploid wheat. Science 361: 361. [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S., Gallagher J.P., Grover C.E., Szadkowski E., Page J.T., Udall J.A., Wang X., Paterson A.H., Wendel J.F. (2014). Ancient gene duplicates in Gossypium (cotton) exhibit near-complete expression divergence. Genome Biol. Evol. 6: 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang W.H., Hori T., Toyoda A., Kato J., Popendorf K., Sakakibara Y., Fujiyama A., Fukagawa T. (2010). Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 20: 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q., Chen Z.J. (2015). Epigenetic and developmental regulation in plant polyploids. Curr. Opin. Plant Biol. 24: 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson K.J. (2011). Review of OriginPro 8.5. J. Am. Chem. Soc. 133: 5621. [Google Scholar]
- Su H., Liu Y., Liu Y.X., Lv Z., Li H., Xie S., Gao Z., Pang J., Wang X.J., Lai J., Birchler J.A., Han F. (2016). Dynamic chromatin changes associated with de novo centromere formation in maize euchromatin. Plant J. 88: 854–866. [DOI] [PubMed] [Google Scholar]
- Su H., Liu Y., Dong Q., Feng C., Zhang J., Liu Y., Birchler J.A., Han F. (2017). Dynamic location changes of Bub1-phosphorylated-H2AThr133 with CENH3 nucleosome in maize centromeric regions. New Phytol. 214: 682–694. [DOI] [PubMed] [Google Scholar]
- Sullivan L.L., Chew K., Sullivan B.A. (2017). α Satellite DNA variation and function of the human centromere. Nucleus 8: 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel S. (2012). Using and understanding RepeatMasker. Methods Mol. Biol. 859: 29–51. [DOI] [PubMed] [Google Scholar]
- Thomas B.C., Pedersen B., Freeling M. (2006). Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16: 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. (2013). Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Liu Y., Su H., Guo X., Han F. (2017). Centromere structure and function analysis in wheat-rye translocation lines. Plant J. 91: 199–207. [DOI] [PubMed] [Google Scholar]
- Wendel J.F., Jackson S.A., Meyers B.C., Wing R.A. (2016). Evolution of plant genome architecture. Genome Biol. 17: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgruber T.K., et al. (2009). Maize centromere structure and evolution: Sequence analysis of centromeres 2 and 5 reveals dynamic Loci shaped primarily by retrotransposons. PLoS Genet. 5: e1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., et al. (2017). Sequencing and comparative analyses of Aegilops tauschii chromosome arm 3DS reveal rapid evolution of Triticeae genomes. J. Genet. Genomics 44: 51–61. [DOI] [PubMed] [Google Scholar]
- Yan H., Talbert P.B., Lee H.R., Jett J., Henikoff S., Chen F., Jiang J. (2008). Intergenic locations of rice centromeric chromatin. PLoS Biol. 6: e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Guo X., Hu J., Lv Z., Han F. (2015). Characterization of two CENH3 genes and their roles in wheat evolution. New Phytol. 206: 839–851. [DOI] [PubMed] [Google Scholar]
- Zhang H., et al. (2013a). Persistent whole-chromosome aneuploidy is generally associated with nascent allohexaploid wheat. Proc. Natl. Acad. Sci. USA 110: 3447–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33: 531–537. [DOI] [PubMed] [Google Scholar]
- Zhang J., Pawlowski W.P., Han F. (2013b). Centromere pairing in early meiotic prophase requires active centromeres and precedes installation of the synaptonemal complex in maize. Plant Cell 25: 3900–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Li W., Fellers J., Friebe B., Gill B.S. (2004). BAC-FISH in wheat identifies chromosome landmarks consisting of different types of transposable elements. Chromosoma 112: 288–299. [DOI] [PubMed] [Google Scholar]
- Zhang T., Talbert P.B., Zhang W., Wu Y., Yang Z., Henikoff J.G., Henikoff S., Jiang J. (2013c). The CentO satellite confers translational and rotational phasing on cenH3 nucleosomes in rice centromeres. Proc. Natl. Acad. Sci. USA 110: E4875–E4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., et al. (2017). The Aegilops tauschii genome reveals multiple impacts of transposons. Nat. Plants 3: 946–955. [DOI] [PubMed] [Google Scholar]
- Zhong C.X., Marshall J.B., Topp C., Mroczek R., Kato A., Nagaki K., Birchler J.A., Jiang J., Dawe R.K. (2002). Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14: 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin A.V., Puiu D., Hall R., Kingan S., Clavijo B.J., Salzberg S.L. (2017). The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum. Gigascience 6: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]