Abstract
Arabidopsis VIRE2-INTERACTING PROTEIN2 (VIP2) was previously described as a protein with a NOT domain, and Arabidopsis vip2 mutants are recalcitrant to Agrobacterium-mediated root transformation. Here we show that VIP2 is a transcription regulator and the C-terminal NOT2 domain of VIP2 interacts with VirE2. Interestingly, AtVIP2 overexpressor lines in Arabidopsis did not show an improvement in Agrobacterium-mediated stable root transformation, but the transcriptome analysis identified 1,634 differentially expressed genes compared to wild-type. These differentially expressed genes belonged to various functional categories such as membrane proteins, circadian rhythm, signaling, response to stimulus, regulation of plant hypersensitive response, sequence-specific DNA binding transcription factor activity and transcription regulatory region binding. In addition to regulating genes involved in Agrobacterium-mediated plant transformation, AtVIP2 overexpressor line showed differential expression of genes involved in abiotic stresses. The majority of the genes involved in abscisic acid (ABA) response pathway, containing the Abscisic Acid Responsive Element (ABRE) element within their promoters, were down-regulated in AtVIP2 overexpressor lines. Consistent with this observation, AtVIP2 overexpressor lines were more susceptible to ABA and other abiotic stresses. Based on the above findings, we hypothesize that VIP2 not only plays a role in Agrobacterium-mediated plant transformation but also acts as a general transcriptional regulator in plants.
Subject terms: Molecular engineering in plants, Abiotic
Introduction
The plant pathogen Agrobacterium tumefaciens causes neoplastic growth called crown galls on plants by transferring genetic material coded on the transfer DNA (T-DNA) from the tumor-inducing (Ti plasmid) to plant cells, resulting in the genome modification following integration (see reviews1–4). The genetic transformation of a plant cell by A. tumefaciens involves the synthesis and translocation of the T-DNA mediated by the virulence (vir) gene products, interaction of the translocated virulence proteins with their cognate partners in the host, and expression of the genes on the T-DNA following integration (see reviews1,2,5,6). Some of the virulence proteins, namely VirD2, VirD5, VirE2, VirE3, and VirF, are A. tumefaciens effector proteins that are directly translocated into the plant cell via the type IV secretion system (T4SS)7,8.
One of the translocated virulence proteins, VirE2, binds to the single-stranded DNA (ssDNA), in vitro, to form a telephone-cord like structure protecting it from degradation by nucleases9–12. Plant proteins that interact with VirE2 were surveyed by the yeast two-hybrid system, resulting in the identification of two VirE2-interacting proteins (VIP), VIP1 and VIP213. VirE2 is suggested to piggy-back on VIP1 for nuclear import via an importin α-dependent pathway and this process is up-regulated by a host MAP kinase that phosphorylates VIP114. VIP2 is a negative on TATA-less (NOT)-domain containing protein which likely functions as a transcription factor and is required for plant stable transformation, but not for transient T-DNA expression15. An Arabidopsis vip2 mutant was shown to be deficient in T-DNA integration due to changes in the expression of many genes, including core histones, suggesting that VIP2 has a role in T-DNA integration15.
A NOT2 homolog in yeast acts as a general negative regulator of gene expression. Similarly, in Drosophila, a NOT2 homolog, the Rga protein, has probable function in mediating interaction between chromatin proteins and the transcriptional complex16,17. NOT2 is the core member of the CCR4-NOT complex that regulates mRNA metabolism at both transcriptional and posttranscriptional levels18 and has a role in promoting transcriptional elongation by RNA polymerase II19,20. The NOT proteins (VIP2/NOT2b and NOT2a) are general transcriptional regulators essential for plant development. NOT2s acts as a scaffold to interact with RNA polymerase II, and promotes transcription of both protein coding and miRNA genes, and facilitates efficient DICER-LIKE1 recruitment in miRNA biogenesis21. VIP2/NOT2b interact with miRNA processing factors such as cap binding proteins, CBP80 and CBP2021, and the interaction is modulated by VirD522.
Here, we demonstrate that VIP2 is a transcription regulator and overexpression of VIP2 in Arabidopsis modulates expression of genes not only involved in Agrobacterium-mediated plant transformation but also genes associated with several other pathways including abiotic stresses. This finding is consistent with its role as transcription regulator. Interestingly, overexpression of VIP2 in Arabidopsis did not significantly increase Agrobacterium-mediated root transformation efficiency.
Results
VIP2 is a transcriptional regulator
Based on the VIP2 sequence, transcript profiling data of Arabidopsis vip2 mutant15, and nuclear localization data, we hypothesized that VIP2 is a transcriptional regulator. To explore this hypothesis, we first tested whether VIP2 can activate transcription in yeast when bound to DNA as previously described for VirE323. We therefore tested the VIP2 protein for transcriptional activation through yeast one-hybrid assay24. For this purpose, the VIP2 (GenBank # AF295433) open reading frame was fused to the GAL4 DNA binding domain (DBD) and cloned into a pGBKT7 yeast vector. The human lamin C (LamC) and topoisomerase I (TopI) genes cloned into the pGBKT7 vector were used as controls. We performed single and double dropout assays to confirm the DNA binding ability of the VIP2 protein in yeast. Undiluted and a 1:10 dilution of all the four clones were able to grow well on solid synthetic dropout (SD) media lacking tryptophan. However, 1:100 and 1:1000 dilutions showed little to no growth (Fig. 1a). The VIP2 fusion construct grew on media lacking Trp and His and containing 10 mM 3-amino-1,2,4-triazole (3-AT), while the LamC and TopI did not (Fig. 1a). These results suggest that the VIP2 protein can activate and express the his3 gene, allowing yeast to grow on a histidine and tryptophan-deficient medium. The above findings suggest VIP2 can activate transcription in yeast, and therefore potentially is a transcription regulator.
Figure 1.
Yeast one-hybrid assays and in planta transactivation assays suggest VIP2 is a putative transcription regulator. (a) Single and double dropout assays were carried out on SD medium lacking Trp, or Trp and His respectively, and containing 10 mM 3-AT. Undiluted, and dilutions of 1:10, 1:100 and 1:1000 were plated. The experiments were repeated twice. (b) A constitutive CaMV35S promoter driving VIP2 was fused to the DNA binding domain (DBD) of a GAL4/UAS transactivation expression system in combination with the GUS reporter protein. GUS histochemical staining of tobacco leaves bombarded with different reporter constructs pSAT6-uasP-GUS only, pSAT6-uasP-GUS and pSAT6-mGAL4-DBD constructs, and pSAT6-uasP-GUS and pSAT6-mGAL4-DBD-VIP2 constructs were carried out. The experiments were repeated three times with similar results.
To validate the transcription activation property of VIP2 in planta, a promoter trans-activation assay, as described earlier25, was used. Tobacco leaves were bombarded with different combinations of reporter constructs such as pSAT6-uasP-GUS only, pSAT6-uasP-GUS and pSAT6-mGAL4-DBD constructs and, pSAT6-uasP-GUS and pSAT6-mGAL4-DBD-VIP2 constructs (Fig. 1b). GUS histochemical staining was performed 48–72 h post DNA bombardment, and the following observations were recorded. We observed few blue spots in the controls, pSAT6-uasP-GUS alone (32 ± 8 blue spots) or pSAT6-uasP-GUS vector expressing the unfused GAL4 DBD (pSAT6-mGAL4-DBD; 69 ± 12 blue spots) and weak expression of the GUS reporter (Fig. 1b). In contrast, co-bombardment of pSAT6-uasP-GUS with pSAT6-mGAL4-DBD-VIP2 that expresses mGAL4-VIP2 fusion from the CaMV35S promoter resulted in a significantly higher number of blue spots (685 ± 181 blue spots) and increased expression of the GUS reporter (Fig. 1b). Based on the above results we hypothesized that VIP2 is efficient in transactivation of the UAS promoter when fused to a DBD. These results further confirmed that VIP2 functions as a transcriptional regulator in planta, corroborating the findings obtained in the yeast-one hybrid assay (see Fig. 1a).
VirE2 interacts with the C- terminal NOT domain of VIP2 in yeast expression system
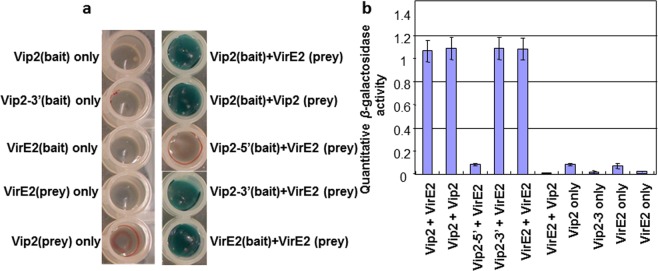
VIP2 was identified as a VIRE2 interacting protein using a yeast two hybrid system13,26. However, it was not clear which domain of VIP2 interacts with VirE2. It is especially important to know if the NOT domain is important for interaction. To determine this, the VIP2 gene was split into two regions; a C-terminal fragment containing the NOT2/NOT3/NOT5 plus 445 bp at the 3′ end and an N-terminal fragment minus the NOT domains (5′ end 1440 bp) as described15. Based on yeast growth, we concluded that VirE2 interacts with 3′ end of VIP2 that contain the NOT domain (Fig. 2a). In addition, both VIP2 and VirE2 proteins can form a dimer with themselves and activate the reporter genes (Fig. 2a). LacZ expression was quantified by β-galactosidase activity to further confirm protein-protein interactions (Fig. 2b).
Figure 2.
Yeast-two hybrid assay suggests the C-terminal of VIP2 protein interacts with VirE2. These various domains of VIP2 and full length VirE2 were cloned into yeast two-hybrid vectors pXDGAT-CY86 and pGADT7, respectively, and were independently transformed into AH109 and MaV204K yeast strains, followed by mating and selection in triple dropout media. (a) The LacZ expression in the various bait and bait/prey interactions containing either the full length proteins or the different VIP2 domains. (b) Quantitative LacZ activities between various haploid or diploid clones were further confirmed. The experiments were repeated twice with similar results.
Rescuing the tumorigenesis-deficiency phenotype in the Atvip2 mutant with constitutive expression of the AtVIP2 gene
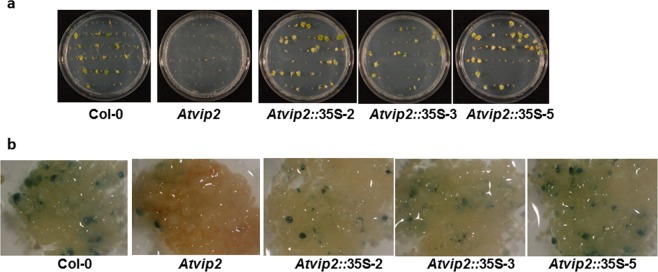
Previously, it was reported that the Atvip2 mutant is recalcitrant to stable transformation. However, complementation of the mutant was not shown15. Therefore, it is not definite if the transformation recalcitrance phenotype is due to the loss of function of VIP2. To address this, a construct containing the AtVIP2 gene (accession # AT5G59710.1) driven by the CaMV35S promoter was transformed into the Atvip2 mutant line. T1 plants of three independent Atvip2 lines expressing the AtVIP2 gene (Atvip2::35S-2, Atvip2::35S-3 and Atvip2::35S-5) were tested for restoration of the tumorigenesis-susceptibility phenotype. Root segments of all three transgenic lines along with the wild-type and the Atvip2 mutant were inoculated with a tumorigenic A. tumefaciens strain as described15. Four weeks after Agrobacterium inoculation, number of tumors produced in each plate was scored. As expected, the wild-type Col-0 was susceptible to transformation and Atvip2 was recalcitrant. The three transgenic Atvip2 lines expressing the AtVIP2 gene produced tumors at similar frequencies to that of Col-0 (Fig. 3a), indicating complementation of the tumorigenesis-deficiency phenotype of the Atvip2 mutant. Furthermore, another stable transformation assay with a non-tumorigenic A. tumefaciens strain with uidA gene within its T-DNA was performed on Atvip2 lines expressing the AtVIP2 gene as previously described27,28. GUS staining of calli indicated increased expression of the uidA gene in the complemented line compared to the Atvip2 mutant and the uidA expression was comparable to that of Col-0 (Fig. 3b). The data from this stable transformation assay are in accordance with the tumorigenesis data and further strengthen our conclusion that expression of the AtVIP2 gene in the Atvip2 mutant could restore the transformation-susceptibility phenotype. Taken together, these data suggest that mutation in the AtVIP2 gene in the Atvip2 mutant is responsible for the transformation recalcitrant phenotype.
Figure 3.
Complementation of the Atvip2 mutant by constitutive expression of AtVIP2. Three independent transgenic events were generated, and molecularly characterized before performing root transformation assays. (a) Roots of the wild-type, vip2 mutant and three transgenic Atvip2 lines expressing AtVIP2 cDNA were infected with a tumorigenic strain A. tumefaciens A208, at 1 × 107 CFU/ml concentration. Tumors incited on the roots were visualized and scored 4 weeks after infection. (b) Stable GUS expression. Roots of the wild-type, Atvip2 mutants and transgenic Atvip2 lines expressing AtVIP2 cDNA were inoculated with A. tumefaciens strain GV3101 carrying the uidA-intron gene within the T-DNA at 1 × 107 CFU/ml concentration. The inoculated roots were stained with X-Gluc 2-3 weeks post infection. All the experiments were repeated two times with similar results.
VIP2 is ubiquitously expressed in major plant organs in arabidopsis
Expression of the YFP-tagged AtVIP2 gene in Arabidopsis indicated the cell and tissue-specific expression of the AtVIP2 gene in the female gametophyte (megasporocytes and tapetum cells) organs, and not in other plant organs29. RT-PCR in the same samples showed expression of the AtVIP2 gene in mature flowers and flower buds, not in leaves, stem and roots29. Interestingly, we detected the expression of the AtVIP2 gene by semi-quantitative RT-PCR in many plant organs including young roots, leaves, stem, petioles and floral tissues (Fig. 4a). These findings contradict previously suggested expression of the AtVIP2 gene in specific Arabidopsis organelles29. To further confirm the AtVIP2 expression, Arabidopsis transgenic plants expressing an AtVIP2 Promoter:uidA fusion (Fig. 4b) were developed and 3-4 independent transgenic plants were analyzed by histochemical GUS staining (Fig. 4c). We observed the expression of the AtVIP2 promoter to be ubiquitous in most plant organs and specifically in the following tissues-: roots, root hairs, leaves, shoots, trichomes, and floral organs (Fig. 4c). Further, based on the GUS staining we concluded that the expression of AtVIP2 is significantly higher in vasculature and root tissues including lateral roots and root hairs. Additionally, we compared the expression of the AtVIP2 promoter with the CaMV35S promoter. Interestingly, GUS staining in AtVIP2 Promoter:uidA expressing plants was much stronger in many plant organs when compared to the transgenic Arabidopsis plants expressing a CaMV35S-uidA construct (Supplementary Fig. S1). Taken together, these data illustrate that the AtVIP2 gene is ubiquitously expressed in most plant cells and tissues. Our observations are consistent with the information provided at https://apps.araport.org/thalemine/portal.do?externalids=AT5G59710 (Supplementary Fig. S2). More interestingly, we report here the discovery of a plant specific promoter that is potentially stronger and ubiquitous in expression when compared to the CaMV35S promoter.
Figure 4.
AtVIP2 gene expression and the AtVIP2 promoter region. (a) The amplification of AtVIP2 transcripts from different plant organs (leaves, stem, root, petioles and flower) in Arabidopsis by semi-quantitative PCR. NC indicates negative control. (b) Schematic presentation of the different regulatory elements on the 1 kb promoter sequence identified upstream of the AtVIP2 coding sequence as shown in plant promoter database (http://ppdb.agr.gifu-u.ac.jp/ppdb/cgi-bin/index.cgi). (c) Analysis of Arabidopsis whole seedlings expressing GUS gene under the control of AtVIP2 promoter. Histochemical GUS staining of transgenic Arabidopsis whole seedlings; roots and root hairs; leaf; trichrome, stem, and floral organs. Pictures were taken 3-4 days after GUS staining.
Transgenic Arabidopsis plants overexpressing the AtVIP2 gene did not show enhanced transformation
Since the Atvip2 mutant is recalcitrant to Agrobacterium-mediated root transformation, we were interested to determine the effect of overexpressing AtVIP2 on root transformation. Transgenic Col-0 lines overexpressing AtVIP2 driven by the CaMV35S promoter were generated (Supplementary Fig. S3). A quantitative root tumor assay, as described above, was done using the tumorigenic A. tumefaciens strain A208. Tumor formation was monitored for a period of four weeks and data were recorded. Our results suggested that overexpression of AtVIP2 did not significantly improve transformation (Supplementary Figs. S4a, upper panel and S4b, left). The effect of AtVIP2 overexpression on another stable transformation assay was determined by calculating the frequency of PPT-resistant calli following infection with a disarmed A. tumefaciens strain GV3101 (pCAS1) as previously described28,30. There was no significant difference observed in the frequency of PPT-resistant calli formation between AtVIP2 overexpressing transgenic lines and vector control/or Col-0 wild-type (Supplementary Figs. S4a, lower panel and S4b, right). Taken together, these data suggest that overexpression of AtVIP2 in Arabidopsis does not improve root transformation.
VIP2 regulates transcription of defense genes, histones and galactolipid biosynthetic genes
To investigate the role of VIP2 in Agrobacterium-plant interaction, we carried out transcriptome profiling by comparing a homozygous Arabidopsis AtVIP2 overexpressor line with wild-type Col-0 plants following Agrobacterium infection. Soil grown Arabidopsis plants in the vegetative stage with fully formed rosette leaves were syringe infiltrated with the disarmed strain of A. tumefaciens, GV3101 harboring the uidA-intron gene within its T-DNA (OD600 = 0.215), and samples were collected at 0, 48 and 72 h after infection (HAI). RNA extracted from these samples were analyzed for gene expression using whole genome Affymetrix gene chip (ATH1) microarray. A total of 1,634 genes were differentially expressed (DE) between AtVIP2 overexpressor line and Col-0. To validate the microarray data, expression of 10 DE genes from AtVIP2 overexpressor plants were analyzed by RT-qPCR (Supplementary Fig. S5). The results obtained from RT-qPCR are in general agreement with the microarray results.
The Venn diagram for DE genes showed that many DE genes during Agrobacterium infection overlapped with DE genes of AtVIP2 overexpressor plants without Agrobacterium infection (Fig. 5a). To study the functional significance of all DE genes, we classified them based on Gene Ontology (GO) term enrichment (Fig. 5b). Various functional categories such as protein targeting to membrane, circadian rhythm, signaling, response to stimulus and regulation of hypersensitive response were enriched. Interestingly, GO terms associated with molecular functions such as sequence-specific DNA binding transcription factor activity and transcription regulatory region DNA binding were enriched. This shows the involvement of VIP2 in transcriptional regulation upon Agrobacteriuminfection with a T-DNA transfer competent strain. Many GO terms associated with stress or immune response were also observed. Interestingly, the majority of the DE genes in AtVIP2 overexpressor were from 0 HAI i.e., without Agrobacterium infection. Surprisingly, many of these genes were also DE in wild-type Col-0 upon Agrobacterium infection (Supplementary Fig. S6, Supplementary Table S1). GO term analysis of these commonly DE genes showed that they belonged to various functional categories such as ethylene-mediated signaling, response to ethylene, response to water, cellular response to hormone and defense response. Many genes in these GO terms are known to be involved in plant defense against various stress conditions31. Genes regulated by Agrobacterium infection and AtVIP2 overexpression include genes that encode WRKY70, receptor-like protein kinase THESEUS 1, Calmodulin-binding protein 60-like G (CBP60G), Calmodulin like 42 (CML42), Pathogenesis-related protein 5 (PR5), MATE efflux family protein, APS reductase 3, WRKY30 etc. These data suggest that at gene expression level, overexpression of AtVIP2 in Arabidopsis mimics gene regulation during Agrobacterium infection in wild-type Col-0. From the Mapman analysis, we additionally identified many genes involved in proteolysis that were down-regulated at 0 HAI in AtVIP2 overexpressor plants (Supplementary Fig. S7) compared to Col-0. The above data further strengthens the role for VIP2 as a transcriptional regulator.
Figure 5.
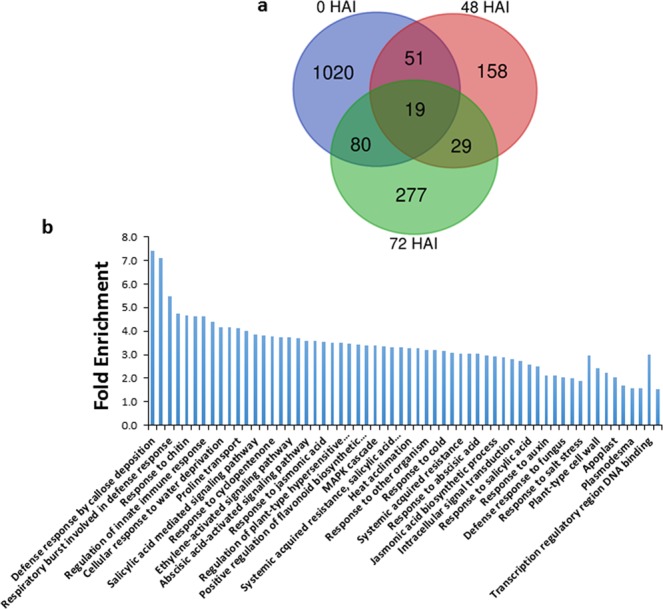
Transcript profiling of AtVIP2 overexpressor plants during different stages of Agrobacterium infection. (a) Venn-diagram showing overlap between differentially regulated (DE) genes, compared to Col-0, in AtVIP2 overexpressor plants during different time points after Agrobacterium infection. (b) GO term enrichment for up-regulated and down-regulated genes from all time points using Database for Annotation, Visualization and Integrated Discovery.
An elf-18 inducible gene, PP2-A5 (phloem protein 2 A5; At1g65390)32, was one the genes showing the highest level of induction in AtVIP2 overexpressor plants upon Agrobacterium infection. Some of the other genes induced are CAF1A (CCR4-associated factor 1a; At3g44260), β-glucosidase 18 (At1g52400), QQS (Qua-Quine Starch; At3g30720), and PCC1 (Pathogen and Circadian Controlled 1; At3g22231). Some of the genes with reduced expression during Agrobacterium infection in AtVIP2 overexpressor plants are TSA1 (TSK-associating protein 1; At3g15950), LEA25 (Late-embryogenesis abundant protein 25; At2g42560), LEA4-1 (At1g32560), CML41 (Calmodulin-like protein 41; At3g50770), and DEFL (Defensin-like; At3g05730).
Genes that were specifically regulated in AtVIP2 overexpressor plants during Agrobacterium infection process (AtVIP2 overexpressing plants at 48 and 72 HAI compared with AtVIP2 overexpressing plants at 0 HAI) belonged to GO terms such as galactolipid biosynthetic process, cellular response to phosphate starvation, response to fructose, response to sucrose and UDP-glycosyltransferase (UGT) activity (Supplementary Table S2). These GO categories were observed from the down-regulated data set. UGTs transfer glycosyl residues from activated nucleotide sugars to acceptor molecules (aglycones) containing an aromatic ring33. They were found to be involved in plant disease and defense responses. We also found that various histone genes that have been shown to be down-regulated in an Atvip2 mutant15 and a not2a Atvip2 mutant21, are up-regulated in our data set (Supplementary Fig. S8). Some up-regulated genes identified in our study encode proteins such as Arabinogalactan, CAF1, Nodulin-Like Protein and Protein Phosphatase 2 C which have been previously reported to be important for Agrobacterium-mediated plant transformation34–37.
ABRE motif containing genes are repressed in AtVIP2 overexpressors
Cis-acting elements are key regulators of gene expression. Therefore, in silico motif analysis to identify motifs that are specifically enriched in the promoters of DE genes was performed. Enrichment of mCACGTGk motif in the down-regulated gene set was identified (Supplementary Tables S3, S4). ACGTG is a core of the Abscisic Acid Responsive Element (ABRE) that is involved in the abscisic acid (ABA)-regulated gene expression38, and genes containing them responds to abiotic stress including drought stress and ABA treatment39. ABA, an abiotic stress hormone, has a major role in regulation of physiological processes during abiotic stress responses40. Some reports suggest that ABA is also involved in plant defense signaling against pathogens and is an essential component in integrating and fine-tuning abiotic and biotic stress-response signaling networks41,42. The above reports are in support of the GO term analysis, in that many of the signaling pathways enriched in our study (Fig. 5) have cross-talk with each other (reviewed in43). On the other hand, we did not find significantly enriched motifs in up-regulated gene set.
AtVIP2 overexpression causes low ABA content and sensitivity to abiotic stresses
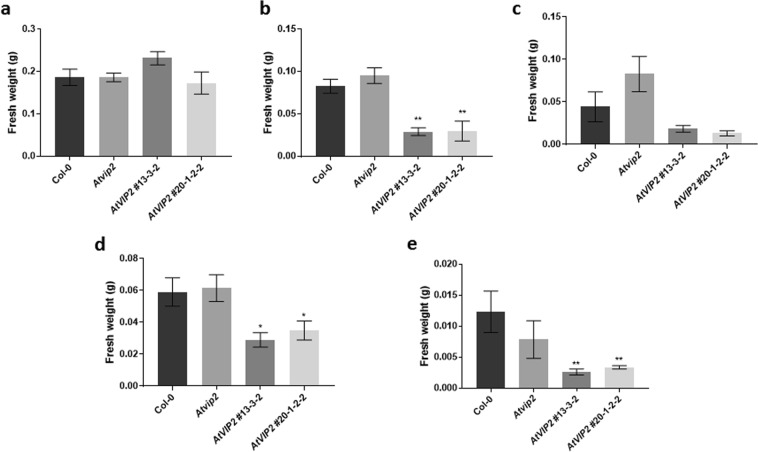
Since we observed enrichment of ABRE motifs in the promoters of down-regulated genes of AtVIP2 overexpressor plants, we further determined the ABA levels and the response of the AtVIP2 overexpressor lines to abiotic stress. We quantified the level of ABA in wild-type Col-0, AtVIP2 overexpressor, and Atvip2 knockout plants grown on half strength MS for three weeks. AtVIP2 overexpressor lines showed significantly less ABA content than Col-0 plants (Fig. 6). In addition, we studied response of Atvip2 and AtVIP2 overexpressor plants to various abiotic stresses by transplanting seedlings to half strength MS plates containing 100 mM NaCl, 5 µM ABA, 10 µM ABA or 75 mM mannitol. AtVIP2 overexpressor plants had lower fresh weights compared to wild-type plants when grown in 5 µM ABA, 100 mM NaCl or 75 mM mannitol (Fig. 7). The above data suggest that AtVIP2 regulates abiotic stress responses in plants, which is consistent with the speculated role as transcription regulator.
Figure 6.

ABA content of wild-type, Atvip2 mutant and AtVIP2 overexpression lines. Three weeks old seedlings grown in half MS media were used for ABA quantification. Error bars indicate SE of the mean (n ≥ 8). Asterisks indicate significant differences (*P ≤ 0.05 and **P ≤ 0.005) between Col-0 and other plants, as determined by two-way ANOVA, uncorrected Fisher’s LSD test.
Figure 7.
Effect of abiotic stress conditions on the growth of wild-type, Atvip2 and AtVIP2 overexpressor plants. Growth of plants under (a) normal (half strength MS), (b) 5 µM ABA, (c) 10 µM ABA, (d) 75 mM mannitol and (e) 100 mM NaCl. In all graphs, error bars indicate SE of the mean (n ≥ 8). Asterisks indicate significant differences (*P ≤ 0.05 and **P ≤ 0.005) between Col-0 and other plants, as determined by two-way ANOVA, uncorrected Fisher’s LSD test.
Discussion
Agrobacterium-mediated plant transformation is a complex process involving the functions of both bacterial virulence proteins and host proteins in various steps of the transformation process. Roles for plant genes in T-DNA transfer and integration during the transformation process have been shown or proposed44–46. VIP2 is one such plant gene required for Agrobacterium-mediated stable plant transformation15 and acts as a general transcriptional regulator controlling plant development21. In the present study, surprisingly, we showed that overexpression of AtVIP2 in Arabidopsis did not have a significant impact on Agrobacterium-mediated plant transformation. Our finding contradicts the data from Zhao and coworkers47 that illustrates up to 2.5-fold increase in Agrobacterium-mediated transformation efficiency in tobacco by the heterologous expression of the Triticum aestivum VIP2. It is not clear if endogenous VIP2 is expressed at saturated levels in tobacco. Nevertheless, these results show that over-expression of VIP2 can enhance transformation in some plant species, further confirming the role of VIP2 in Agrobacterium-mediated plant transformation. Another possible reason for the differences observed between our study, and Zhao et al.47 may be the use of homologous versus heterologous VIP2 gene sequences for overexpression. In a previous study, it has been shown that overexpression of AtVIP1 (a bZIP protein) in Arabidopsis did not alter either transient or stable transformation susceptibility48, whereas T-DNA transformation efficiency was improved when AtVIP1 expressing transgenic tobacco plants were retransformed with Agrobacterium49. Though the increased transformation efficiency was attributed to the lack of use of full-length VIP1 cDNA48,49, it is interesting to note that homologous expression of VIP1 did not alter transformation efficiency in Arabidopsis similar to our study. Based on the above findings, we speculate VIP2 is necessary but not a rate-limiting protein in Agrobacterium-mediated transformation in Arabidopsis.
A previous study22 showed that VirD5 interacts with AtVIP2 and competes with it for binding with cap binding proteins (CBPs; CBP20 and CBP80) that negatively regulates the Agrobacterium infection process. The interaction between AtVIP2 and VirD5 is very specific in that VirD5 interacts with AtVIP2 of Arabidopsis, but not with the homolog NOT2a of Arabidopsis or the ortholog OsNOT2 of rice22. It would be interesting to understand the role of AtVIP2 and VirD5 interaction in Agrobacterium-mediated plant transformation. In our previous publication, we demonstrated that VIP2 interacts with VIRE215. Wang et al.22 also showed that VIP2 and VIRE2 along with VIRD5 form a ternary complex in vivo. In this study, we specifically showed that VIRE2 interacts with C-terminal NOT domain of VIP2. We speculate that VIRE2 interacts with VIP2 to alter VIP2’s activity, which in turn will regulate expression of genes such as histones to enhance Agrobacterium-mediated plant transformation.
In this study, we also investigated AtVIP2 promoter activity in multiple tissues or organs and showed that the AtVIP2 promoter is highly active and expressed constitutively. Agrobacterium infection was previously shown to induce AtVIP2 expression in Arabidopsis15. We speculate that the VIP2 protein is present at saturated levels in Arabidopsis and therefore overproduction of this protein does not have an impact on plant transformation. However, overexpression of AtVIP2 resulted in differential expression of many genes that are important for Agrobacterium-mediated plant transformation (Fig. 5) including up-regulation of histone genes (Supplementary Fig. S8). In addition, expression of defense related genes were modulated in the AtVIP2 overexpressor plants. During Agrobacterium infection process, plant genes necessary for the transformation process are induced and host defense genes are repressed50,51. Surprisingly, we also found many genes involved in proteolysis were down-regulated in AtVIP2 overexpressor plants (Supplementary Fig. S7). Proteolysis of proteins coating the T-DNA is important for subsequent integration of T-DNA to the host genome52. It would be interesting to study how VIP2 affects proteolysis.
Genome-wide transcript profiling of Agrobacteriuminfected AtVIP2 overexpressor lines revealed the presence of many differentially regulated genes and GO terms. UGT is one of the GO terms specifically enriched in AtVIP2 overexpressor plants during Agrobacteriuminfection (Supplementary Table S2). Overexpression of barley UGT in wheat resulted in enhanced resistance to Fusarium graminearum53. Recently, it has been shown that expression of wheat UGT in tobacco and Arabidopsis reduced the efficiency of Agrobacterium-mediated plant transformation54. Though we did not find any relevant literature related to the galactolipid biosynthetic process or cellular response to phosphate starvation in relation to Agrobacterium-mediated plant transformation, it is important to note that alteration of membrane lipid composition is one of the adaptive mechanisms in higher plants to cope with phosphate starvation55. Highly induced genes in AtVIP2 overexpressor plants upon Agrobacterium infection include CAF1A, β-glucosidase 18, QQS and PCC1. CAF1A is involved in mRNA deadenylation and mediation of stress responses56. β-glucosidase 18 plays a role in the metabolism of abiotic stress hormone ABA57. An orphan gene, QQS, functions in modulation of carbon and nitrogen allocation58. RNAi silencing of PCC1, a regulator of defense against pathogens and stress-activated transition to flowering, resulted in plants being more susceptible to hemi-biotrophic oomycete pathogen, Phytophthora brassicae, and more resistant to the necrotrophic fungal pathogen Botrytis cinerea59.
GO terms related to abiotic stress responses such as response to salt stress, response to ABA, etc. are enriched among DE genes (Fig. 5). Motif analysis of DE genes dataset resulted in identification of ABRE containing motif (mCACGTGk) in the promoters of the down-regulated genes (Supplementary Table S3), and enrichment of these motifs in the 72 HAI down-regulated DE genes dataset (Supplementary Table S4). At 72 HAI, many abiotic stress responsive genes such as late embryogenesis abundant proteins (LEA) (At5g06760, At3g15670, At1g52690 and At2g35300), and calmodulin-like protein 41 (CML41) (At3g50770) were down-regulated. Endogenous ABA levels are an important factor in shaping plants tolerance to various abiotic stresses60–62. ABA induces expression of ABA-responsive genes via ABREs in their promoter regions60,63. ABRE containing elements are enriched in promoters of genes that are responsive to various abiotic stresses64. LEA proteins are intrinsically disordered proteins, have major role in abiotic stress tolerance in plants65,66. CMLs are major Ca2+ sensors, and they target many genes involved in various developmental processes and stress tolerance67,68. CML41 was induced by both plant cell wall derived oligosaccharides and bacterial flagellin peptide Flg22, and have been suggested to have a role in dampening plant immune responses69. In addition, recently, CML41 has been shown to have increased expression in response to elevated temperature70. Expression of TSA1 and DEFL was also reduced in AtVIP2 overexpressor plants upon Agrobacterium infection. TSA1 is involved in MeJA-induced ER body formation in plants71. Similar to our study, infection of a fungal pathogen Alternaria brassicicola represses the expression of DEFL72.
Results from our study along with previous reports15,21 showed that VIP2 is not only a regulator of plant genes involved in Agrobacterium-mediated plant transformation process but also a general transcriptional regulator that control multiple pathways. Our results showed that overexpression of AtVIP2 in Arabidopsis resulted in differential regulation of many abiotic stress related genes. This observation is consistent with the observation that the NOT2 domain of Fusarium oxysporum is suggested to regulate vegetative growth, conidiogenesis and virulence of the fungus by the transcriptional regulation of genes involved in multiple pathways controlling cell wall integrity, oxidative stress response, ROS production and fusaric acid production73 suggesting that VIP2 controls several physiological and metabolic pathways.
Modulating VIP2 expression might be a useful tool for enhancing plant transformation efficiency in plant species where endogenous VIP2 expression is at low level. However, susceptibility of VIP2 overexpressor lines to ABA and other abiotic stresses (Fig. 7) can be an issue. It requires further studies to understand how VIP2 regulates abiotic stress responses.
Methods
Yeast one-hybrid assay
The yeast one-hybrid assay was performed as described earlier24. VIP2 (GenBank # AF295433) open reading frame was fused to the GAL4 DNA binding domain (BD) and cloned into a pGBKT7 yeast vector containing the auxotrophic marker trp1 gene and the GAL4 upstream activating sequence (UAS promoter). The human lamin C and topoisomerase I genes cloned into the pGBKT7 vector were used as controls. These two genes are known to function as non-specific activators of the auxotrophic marker, but can be deactivated by the addition of 3-amino-1,2,4-triazole (3-AT). All clones were transformed into the MaV204K yeast strain. A single colony of each transformed MaV204K was picked and grown overnight in 100 µl liquid synthetic dropout (SD) medium lacking tryptophan in 96 well plates. All cultures were transferred into a new 96 well plate with three different dilutions of the liquid culture (1:10, 1:100 and 1:1000). Cultures were plated onto solid SD medium, lacking tryptophan and containing 3-AT, using 96-well pin replicator, and incubated at 30 °C overnight for the single-dropout assay. The three clones fused to the GAL4-BD were also replicated on SD medium lacking tryptophan and histidine, and containing 10 mM 3-AT for the double-dropout assay.
Transactivation analysis
A promoter containing three copies of the GAL4 upstream activating sequence (UAS), and VIP2 were fused to the DBD of GAL4 modified for optimal expression in plants (mGAL4) and expressed constitutively under the CaMV35S promoter from a pSAT6 vector (pSAT6-mGAL4-DBD-VIP2). Additionally, the pSAT6-uasP-GUS vector25 with GUS reporter gene fused to the minimal UAS promoter (uasP) was also included as a control. The biolistic approach for DNA delivery was performed using standard protocols and plasmids as described25.
Bait and Prey Construction
The full-length, C-terminal, and N-terminal of VIP2 and the VirE2 genes were PCR amplified using a high-fidelity platinum Pfx DNA polymerase (Invitrogen Inc.) and cloned into the pENTR/D Topo vector (Invitrogen, Carlsbad, CA). All these clones were then cloned into the yeast two-hybrid DNA BD vector pXDGAT-CY86 with the cyclohexamide sensitive gene (CYHS)74. The full-length VIP2 and VirE2 genes were also cloned into the DNA activation domain (AD) domain vector, pGADT7-Rec7G to be used as preys. The prey vector pGADT7-Rec7G was constructed from the pGADT7-Rec7 vector (Clontech, Mountain View, CA) by introducing the GATEWAY cassette at the SmaI site using standard cloning protocols and confirmed by sequencing. Recombination between pENTR/D Topo vectors and the destination vector was performed using the Clonase II enzyme mix according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Bait and prey constructs were confirmed to be in-frame with the DNA-BD or DNA-AD prior to transformations. Bait constructs were transformed into the MaV204K yeast strain (MAT α, leu2-3,112; trp1-901; his3 Δ200; ade2-101; cyh2R; can1R; gal4 Δ; gal80 Δ; GAL1::lacZ;HIS3UASGAL1::HIS3@LYS2; SPAL10::URA3)75. Prey constructs were transformed into AH109 yeast strain (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4 Δ, gal80 Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ, MEL1). All transformations were performed as described76. All bait strains were checked for auto activation of the reporter gene, his, by checking their growth on SD -His/-Trp supplemented with 0, 2.5, 5, 7.5, 10 and 15 mM 3-AT. The 3-AT concentration of 10 mM was found to inhibit auto activation of the both bait or prey clones, so it was used as media supplement in the rest of the study.
Mapping the interaction between VirE2 with VIP2
Pairs of various bait and prey constructs were co-transformed into AH109 strain to test the specificity and to map the interaction between VirE2 and VIP2 (Fig. 2a). Co-transformants were selected on solid SD medium lacking Ade, His, Trp, and Leu and supplemented with 10 mM 3-AT and X-gal. Positive clones were then re-grown in SD liquid medium under the same selection conditions. The selection was repeated four times by sub culturing positive clones and assayed for the activity of X-gal. Clones that maintained an ability to grow under selected conditions were then grown on liquid SD medium in a 96-well (Fig. 2a, left panel) plate along with yeast strains containing only bait or prey constructs as negative controls (Fig. 2a, right panel).
Validation and β-galactosidase assays
Co-transformed yeast strains with various VIP2 and VirE2 constructs, and only bait or prey, were grown on 96 well plates containing liquid SD medium that lacks Ade, His, Trp, and Leu supplemented with 10 mM 3-AT for 24 h. Absorbance at A600 was measured for three independent 100 µl cultures and then assayed for β-galactosidase activity using the yeast β-galactosidase assay kit (cat. 75768; Pierce Biotechnology, Inc.) following the manufacturer’s instructions. A600 and A420 were measured using multi-plate reader, Tecan infinite 200Pro. The β-galactosidase activity was calculated using the equation: β-galactosidase activity = 1000 × A420/T × V × A600 where T is time (in minutes) of incubation and V is volume of cells (ml) used in the assay.
Plant transformation experiments
We tested whether constitutive overexpression of AtVIP2 in the Atvip2 mutant and Col-0 plants could restore the wild-type phenotype and/or increase the transformation efficiency. Briefly, for over-expression studies, cDNA corresponding to the Arabidopsis VIP2 cDNA was amplified by RT-PCR along with GATEWAY adapter primers, sequence verified and cloned into the plant expression vector pMDC3277 driven by constitutive CaMV35S promoter. Wild-type Arabidopsis Col-0 and Atvip2 mutant plants were transformed by the floral dip method78. The transgenic plants were selected for resistance to hygromycin and also tested for the presence of the transgene using the GATEWAY adapter primers (attB1/B2), while the expression of the transgene was confirmed by semi-quantitative RT-PCR using gene specific primers. These transgenic plants were selfed and T1 homozygous seeds were collected.
Root tumor, GUS and callus assays
Arabidopsis root tumor assays were performed as described earlier15,28,30. For root tumor assays, axenic root segments from wild-type Col-0 and transgenic plants were infected with a tumor-inducing A. tumefaciens strain, native A208 containing the nopaline type Ti-plasmid pTiT3, co-cultivated for 48 h in the dark at room temperature and transferred to a hormone-free Murashige and Skoog media (MS) supplemented with cefotaxime (250 mg/l) and timentin (100 mg/l). Tumor numbers and phenotypes were recorded at 4 weeks after infection. Stable GUS transformation assays were performed as described earlier27,39. Root segments from Col-0 and transgenic plants were infected with disarmed strain of A. tumefaciens GV3101 carrying uidA-intron gene within the T-DNA and co-cultivated for 48 h at room temperature. The root segments were incubated on callus induction medium (CIM) for 2-3 weeks, and stained with X-gluc. Stable transformation callus assays were carried out as described earlier28,30. Root segments from wild-type Col-0 and transgenic plants were infected with a disarmed A. tumefaciens strain GV3101 containing pCAS128 and co-cultivated for 48 h in dark at room temperature. The root segments were incubated on CIM supplemented with phosphinothricin (PPT) at 10 mg/l, cefotaxime (250 mg/l) and timentin (100 mg/l). The number of root segments forming PPT-resistant calli was counted at 4 weeks after infection.
Tissue specific expression of VIP2
The promoter sequence upstream of the ORF (1 kb region, see ppdb: Plant promoter database, http://ppdb.agr.gifu-u.ac.jp/ppdb/cgi-bin/index.cgi, Fig. 4b)79 was PCR amplified, confirmed by sequencing, and fused to a reporter gene (uidA) in the pMDC162 vector, and mobilized into A. tumefaciens strain GV3101. Arabidopsis plants were transformed with the VIP2 Promoter:uidA fusion expression cassette via the floral dip transformation method and hygromycin-resistance transgenic T1 events were identified for promoter expression analysis. Three to four independent T1 lines were selected and screened by histochemical staining for GUS expression.
Microarray gene expression analysis
Three biological replicates were performed for each tissue sample. Total RNA was isolated as described previously15. RNA was quantified and evaluated for purity using a Nanodrop Spectrophotometer ND-100 (NanoDrop Technologies, Willington, DE) and Bioanalyzer 2100 (Agilent, Santa Clara, CA). Ten μg of total RNA was used for the expression analysis of each sample using the Arabidopsis ATH1 chip (Affymetrix, Santa Clara, CA). Probe labeling, chip hybridization and scanning were performed per the manufacturer’s instructions for one-cycle labeling (Affymetrix). Data normalization between chips was conducted using RMA (Robust Multichip Average)80. Presence/absence calls for each probe set were obtained using dCHIP81. Gene selections based on Associative T-test82 were made using Matlab (MathWorks, Natick, MA). In this method, the background noise present between replicates and technical noise during microarray experiments was measured by the residual present among a group of genes whose residuals are homoscedastic. Genes whose residuals between the compared sample pairs that are significantly higher than the measured background noise level were considered to be differentially expressed. A selection threshold of 2 for transcript ratios and a Bonferroni-corrected P value threshold of 2.19202E-06 were used. The Bonferroni-corrected P value threshold was derived from 0.05/N in these analyses, where N is the number of probes sets (22,810) on the chip.
Functional classification of genes was carried out using the Database for Annotation, Visualization and Integrated Discovery (DAVID) version 6.783 with the Benjamini correction and false discovery rate (FDR) <0.05, as well as MAPMAN (http://mapman.gabipd.org/) using log2 transformed value of transcript ratios.
In silico motif analysis
The 500 bp upstream region of one hundred genes from each class was downloaded from RSAT site (http://floresta.eead.csic.es/rsat/) with the option preventing overlap with neighbor genes (noorf). Promoters were analyzed using the online tool MotifSampler (http://bioinformatics.intec.ugent.be/MotifSuite/motifsampler.php). A precompiled background model of Arabidopsis with order set to 3 was used. The total number of runs was set to 50. MotifRanking (http://bioinformatics.intec.ugent.be/MotifSuite/motifranking.php) was used to rank highest scoring motifs based on LogLikelihood ratios.
ABA quantification
Freshly harvested plant tissues were frozen in liquid nitrogen and ground to a fine powder, and stored at −80 °C until ABA extraction. ABA quantification was carried out as described84. In brief, 1 ml of cold methanol:water (70:30, v-v) plus labeled ABA was added to 100 mg of powdered tissue. Samples were vortexed, sonicated, and extracted at 4 °C for 1 h. Samples were centrifuged at 16,000 × g for 5 min at 4 °C and the supernatants were dried using nitrogen gas. The residues were re-dissolved in 100% methanol, and the supernatant injected into an Agilent 1290 UHPLC connected to an Agilent 6430 Triple Quad mass spectrometer (Agilent Technologies). Separation was carried out using a Waters BEH C18 column (Waters Co., 1.76 µm, 2.1 × 150 mm). The following solvents were used at a flow rate of 0.4 ml min−1: (A) 0.05% formic acid/H2O and (B) acetonitrile/0.05% formic acid. The separation was achieved by starting with 5% solvent B, a gradient from 5% to 46% of solvent B over 19 min and a step to 90% B in 0.1 min, then a hold at 90% B for 2 min and a step to 5% solvent B in 0.1 min. The temperature of the UPLC column was set to 40 °C. The gas temperature was 300 °C, gas flow: 9 ml/min, nebulizer was 25 psi. Fragmentor and collision energy were optimized for each compound individually. The SRM analysis conditions for ABA and d6ABA (negative ion mode) were as follows: capillary = 4,000 V, fragmentor voltage = 100 V, collision energy = 4 V, dwell time = 200 ms and SRM transition (m/z) = 263/153 for unlabeled ABA and 269/159 for d6ABA. Relative amounts of ABA were based on comparison to the labeled hormone.
Abiotic stress experiments
Seeds of Arabidopsis ecotype Col-0 were surface sterilized in 75% ethanol for 2 min, and 30% bleach for 15 min, followed by four washes with sterile water. The seeds were sown onto half-strength MS medium containing 1% sucrose and 0.3% phytagel, stratified in the dark at 4 °C for 2 d and grown in growth chamber with 12.5 h day length at 24 °C. Seven days after stratification, seedlings were transferred to half-strength MS plates containing 5 µM ABA, 10 µM ABA, 75 mM mannitol, and 100 mM NaCl. Fifteen days after transfer, fresh weights of the plants were measured.
Supplementary information
Acknowledgements
This work was supported by Noble Research Institute, LLC and in part by National Science Foundation (grant # IOB 0445799). Authors thank Bonnie Watson (Noble Research Institute) for ABA quantification.
Author Contributions
A.A., V.R., B.V. and K.S.M. conceived and designed the experiments. A.A., V.R., B.V., M.R.M., B.D.P. and H.L. conducted the experiments. A.A., V.R. and Y.T. did data analyses. A.A., V.R., B.V. and K.S.M. wrote the manuscript.
Data Availability
All the data presented in the manuscript is publicly available. All transcriptome data is loaded into ArrayExpress with accession # E-MTAB-8326.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Vidhyavathi Raman and Ajith Anand contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-49590-3.
References
- 1.Gelvin SB. Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:223–256. doi: 10.1146/annurev.arplant.51.1.223. [DOI] [PubMed] [Google Scholar]
- 2.Gelvin SB. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev. 2003;67:16–37. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Escobar MA, Dandekar AM. Agrobacterium tumefaciens as an agent of disease. Trends Plant Sci. 2003;8:380–386. doi: 10.1016/S1360-1385(03)00162-6. [DOI] [PubMed] [Google Scholar]
- 4.Pitzschke A, Hirt H. New insights into an old story: Agrobacterium‐induced tumour formation in plants by plant transformation. The EMBO Journal. 2010;29:1021–1032. doi: 10.1038/emboj.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anand, A., Vaghchhipawala, Z. E. & Mysore, K. S. In Plant Transformation Technologies 31–49 (Wiley-Blackwell, 2010).
- 6.Hooykaas PJ, Shilperoort RA. Agrobacterium and plant genetic engineering. Plant Mol Biol. 1992;19:15–38. doi: 10.1007/BF00015604. [DOI] [PubMed] [Google Scholar]
- 7.Schrammeijer B, den Dulk-Ras A, Vergunst AC, Jurado Jacome E, Hooyakaas PJJ. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cervisiae as a model: evidence for transport of a novel effector protein VirE3. Nucl Acids Res. 2003;31:860–868. doi: 10.1093/nar/gkg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vergunst AC, et al. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- 9.Christie, P. J., Ward, J. E., Winans, S. C. & Nester, E. W. The Agrobacterium tumefaciens virE2 gene product is a single -stranded-DNA-binding protein that associates with T-DNa. J. Bacteriol170 (1988). [DOI] [PMC free article] [PubMed]
- 10.Citovsky V, Guralnick B, Simon MN, Wall JS. The molecular structure of Agrobacterium VirE2-single stranded DNA complexes involved in nuclear import. J Mol Biol. 1997;271:718–727. doi: 10.1006/jmbi.1997.1230. [DOI] [PubMed] [Google Scholar]
- 11.Citovsky V, Wong ML, Zambryski PC. Coooperative interaction of Agrobacterium VirE2 protein with single-stranded. DNA: Implications for the T-DNA transfer process. Pro Natl Acad Sci USA. 1989;86:1193–1197. doi: 10.1073/pnas.86.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Citovsky V, Zupan J, Warnick D, Zambryski PC. Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science. 1992;256:1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- 13.Tzfira T, Rhee Y, Chen M-H, Citovsky V. Nucleic acid transport in plant-microbe interactions: the molecules that walk through the walls. Annu. Rev. Microbiol. 2000;54:187–219. doi: 10.1146/annurev.micro.54.1.187. [DOI] [PubMed] [Google Scholar]
- 14.Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H. Trojan Horse Strategy in Agrobacterium Transformation: Abusing MAPK Defense Signaling. Science. 2007;318:453–456. doi: 10.1126/science.1148110. [DOI] [PubMed] [Google Scholar]
- 15.Anand A, et al. VIP2, a VIE2- plant-interactor protein is required for T-DNA integration and stable transformation by Agrobacterium. Plant Cell. 2007;19:1695–1708. doi: 10.1105/tpc.106.042903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collart MA, Struhl K. NOT1 (CDC39), NOT2 (CDC35), NOT3 and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 1994;8:525–537. doi: 10.1101/gad.8.5.525. [DOI] [PubMed] [Google Scholar]
- 17.Frolov MV, Benevolenskaya EV, Birchler JA. Regena (Rga), a Drosophila Homolog of the Global Negative Transcriptional Regulator CDC36 (NOT2) from Yeast, Modifies Gene Expression and Suppresses Position Effect Variegation. Genetics. 1998;148:317–330. doi: 10.1093/genetics/148.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denis CL, Chen J. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog Nucl Acids Res. 2003;73:221–250. doi: 10.1016/S0079-6603(03)01007-9. [DOI] [PubMed] [Google Scholar]
- 19.Kruk JA, Dutta A, Fu J, Gilmour DS, Reese JC. The multifunctional Ccr4–Not complex directly promotes transcription elongation. Genes & Development. 2011;25:581–593. doi: 10.1101/gad.2020911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reese JC. The control of elongation by the yeast Ccr4–Not complex. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2013;1829:127–133. doi: 10.1016/j.bbagrm.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, et al. NOT2 Proteins Promote Polymerase II–Dependent Transcription and Interact with Multiple MicroRNA Biogenesis Factors in Arabidopsis. The Plant Cell. 2013;25:715–727. doi: 10.1105/tpc.112.105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, et al. VirD5 is required for efficient Agrobacterium infection and interacts with Arabidopsis VIP2. New Phytologist. 2018;217:726–738. doi: 10.1111/nph.14854. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Rodriguez FM, Schrammeijer B, Hooykaas PJJ. The Agrobacterium VirE3 effector protein: a potential plant transcriptional activator. Nucl. Acids Res. 2006;34:6496–6504. doi: 10.1093/nar/gkl877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li JJ, Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262:1870. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 25.Tzfira T, et al. pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Molecular Biology. 2005;57:503–516. doi: 10.1007/s11103-005-0340-5. [DOI] [PubMed] [Google Scholar]
- 26.Tzfira T, Vaidya M, Citovsky V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001;20:3596–3607. doi: 10.1093/emboj/20.13.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narasimhulu SB, Deng X-B, Sarria R, Gelvin SB. Early transcription of Agrobacterium T-DNA genes in tobacco and maize. Plant Cell. 1996;8:873–886. doi: 10.1105/tpc.8.5.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nam J, et al. Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol Gen Genet. 1999;261:429–438. doi: 10.1007/s004380050985. [DOI] [PubMed] [Google Scholar]
- 29.Tian G-W, et al. High-Throughput Fluorescent Tagging of Full-Length Arabidopsis Gene Products in Planta. Plant Physiology. 2004;135:25–38. doi: 10.1104/pp.104.040139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaghchhipawala ZE, Vasudevan B, Lee S, Morsy MR, Mysore KS. Agrobacterium may delay plant nonhomologous end-joining DNA repair via XRCC4 to favor T-DNA integration. Plant Cell. 2012;24:4110–4123. doi: 10.1105/tpc.112.100495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang D, Wu W, Abrams SR, Cutler AJ. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. Journal of Experimental Botany. 2008;59:2991–3007. doi: 10.1093/jxb/ern155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Igarashi D, Tsuda K, Katagiri F. The peptide growth factor, phytosulfokine, attenuates pattern-triggered immunity. The Plant Journal. 2012;71:194–204. doi: 10.1111/j.1365-313X.2012.04950.x. [DOI] [PubMed] [Google Scholar]
- 33.Ross J, Li Y, Lim E, Bowles DJ. Higher plant glycosyltransferases. Genome biology. 2001;2:Reviews3004. doi: 10.1186/gb-2001-2-2-reviews3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anand A, et al. Identification of plant genes involved in Agrobacterium-mediated transformation by using virus-induced gene silencing as a functional genomics tool. Mol Plant Microbe Interact. 2007;20:41–52. doi: 10.1094/MPMI-20-0041. [DOI] [PubMed] [Google Scholar]
- 35.Gaspar YM, et al. Characterization of the Arabidopsis Lysine-Rich Arabinogalactan-Protein AtAGP17 Mutant (rat1) That Results in a Decreased Efficiency of Agrobacterium Transformation. Plant Physiology. 2004;135:2162–2171. doi: 10.1104/pp.104.045542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Endo M, et al. Increased frequency of homologous recombination and T‐DNA integration in Arabidopsis CAF‐1 mutants. The EMBO Journal. 2006;25:5579–5590. doi: 10.1038/sj.emboj.7601434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tao Y, Rao PK, Bhattacharjee S, Gelvin SB. Expression of plant protein phosphatase 2C interferes with nuclear import of the Agrobacterium T-complex protein VirD2. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:5164–5169. doi: 10.1073/pnas.0300084101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uno Y, et al. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proceedings of the National Academy of Sciences. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allu AD, Soja AM, Wu A, Szymanski J, Balazadeh S. Salt stress and senescence: identification of cross-talk regulatory components. Journal of Experimental Botany. 2014;65:3993–4008. doi: 10.1093/jxb/eru173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Derksen H, Rampitsch C, Daayf F. Signaling cross-talk in plant disease resistance. Plant Science. 2013;207:79–87. doi: 10.1016/j.plantsci.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Adie BAT, et al. ABA Is an Essential Signal for Plant Resistance to Pathogens Affecting JA Biosynthesis and the Activation of Defenses in Arabidopsis. The Plant Cell. 2007;19:1665–1681. doi: 10.1105/tpc.106.048041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asselbergh B, De Vleesschauwer D, Höfte M. Global Switches and Fine-Tuning—ABA Modulates Plant Pathogen Defense. Molecular Plant-Microbe Interactions. 2008;21:709–719. doi: 10.1094/MPMI-21-6-0709. [DOI] [PubMed] [Google Scholar]
- 43.Koornneef A, Pieterse CMJ. Cross Talk in Defense Signaling. Plant Physiology. 2008;146:839–844. doi: 10.1104/pp.107.112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bourras S, Rouxel T, Meyer M. Agrobacterium tumefaciens Gene Transfer: How a Plant Pathogen Hacks the Nuclei of Plant and Nonplant Organisms. Phytopathology. 2015;105:1288–1301. doi: 10.1094/PHYTO-12-14-0380-RVW. [DOI] [PubMed] [Google Scholar]
- 45.Gelvin SB. Integration of Agrobacterium T-DNA into the Plant Genome. Annual Review of Genetics. 2017;51:195–217. doi: 10.1146/annurev-genet-120215-035320. [DOI] [PubMed] [Google Scholar]
- 46.Nester EW. Agrobacterium: nature’s genetic engineer. Frontiers in Plant Science. 2014;5:730. doi: 10.3389/fpls.2014.00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao, P. et al. Cloning and characterization of TaVIP2 gene from Triticum aestivum and functional analysis in Nicotiana tabacum. Scientific Reports6, 37602, 10.1038/srep37602, https://www.nature.com/articles/srep37602#supplementary-information (2016). [DOI] [PMC free article] [PubMed]
- 48.Yong S, Lan-Ying L, Gelvin SB. Is VIP1 important for Agrobacterium-mediated transformation? The Plant Journal. 2014;79:848–860. doi: 10.1111/tpj.12596. [DOI] [PubMed] [Google Scholar]
- 49.Tzfira T, Vaidya M, Citovsky V. Increasing plant susceptibility to Agrobacterium infection by overexpression of the Arabidopsis nuclear protein VIP1. Proceedings of the National Academy of Sciences. 2002;99:10435–10440. doi: 10.1073/pnas.162304099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veena, Jiang H, Doerge RW, Gelvin SB. Transfer of T-DNA and Vir proteins to plant cells by Agrobacterium tumefaciens induces expression of host genes involved in mediating transformation and suppresses host defense gene expression. The Plant Journal. 2003;35:219–236. doi: 10.1046/j.1365-313X.2003.01796.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhou X, et al. Global analysis of differentially expressed genes and proteins in the wheat callus infected by Agrobacterium tumefaciens. PLoS One. 2013;8:e79390. doi: 10.1371/journal.pone.0079390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tzfira T, Vaidya M, Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature. 2004;431:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- 53.Li X, et al. Transgenic Wheat Expressing a Barley UDP-Glucosyltransferase Detoxifies Deoxynivalenol and Provides High Levels of Resistance to Fusarium graminearum. Molecular plant-microbe interactions: MPMI. 2015;28:1237–1246. doi: 10.1094/mpmi-03-15-0062-r. [DOI] [PubMed] [Google Scholar]
- 54.Zhou X, et al. Effects of the wheat UDP-glucosyltransferase gene TaUGT-B2 on Agrobacterium-mediated plant transformation. Acta Physiologiae Plantarum. 2017;39:15. doi: 10.1007/s11738-016-2317-1. [DOI] [Google Scholar]
- 55.Nicole G, Yuki N, Wolf-Rüdiger S, Hiroyuki O, Peter D. Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. The Plant Journal. 2008;56:28–39. doi: 10.1111/j.1365-313X.2008.03582.x. [DOI] [PubMed] [Google Scholar]
- 56.Walley JW, Kelley DR, Nestorova G, Hirschberg DL. & Dehesh, K. Arabidopsis Deadenylases AtCAF1a and AtCAF1b Play Overlapping and Distinct Roles in Mediating Environmental Stress Responses. Plant Physiology. 2010;152:866–875. doi: 10.1104/pp.109.149005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu Z-Y, et al. A Vacuolar β-Glucosidase Homolog That Possesses Glucose-Conjugated Abscisic Acid Hydrolyzing Activity Plays an Important Role in Osmotic Stress Responses in Arabidopsis. The Plant Cell. 2012;24:2184–2199. doi: 10.1105/tpc.112.095935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li L, et al. QQS orphan gene regulates carbon and nitrogen partitioning across species via NF-YC interactions. Proc Natl Acad Sci USA. 2015;112:14734–14739. doi: 10.1073/pnas.1514670112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mir R, et al. Pathogen and Circadian Controlled 1 (PCC1) regulates polar lipid content, ABA-related responses, and pathogen defence in Arabidopsis thaliana. J Exp Bot. 2013;64:3385–3395. doi: 10.1093/jxb/ert177. [DOI] [PubMed] [Google Scholar]
- 60.Fujita Y, Yoshida T, Yamaguchi-Shinozaki K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol Plant. 2013;147:15–27. doi: 10.1111/j.1399-3054.2012.01635.x. [DOI] [PubMed] [Google Scholar]
- 61.Lin PC, et al. Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiol. 2007;143:745–758. doi: 10.1104/pp.106.084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang ZY, Xiong L, Li W, Zhu JK, Zhu J. The plant cuticle is required for osmotic stress regulation of abscisic acid biosynthesis and osmotic stress tolerance in Arabidopsis. Plant Cell. 2011;23:1971–1984. doi: 10.1105/tpc.110.081943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Busk PK, Pagès M. Regulation of abscisic acid-induced transcription. Plant Molecular Biology. 1998;37:425–435. doi: 10.1023/a:1006058700720. [DOI] [PubMed] [Google Scholar]
- 64.Zhou J, et al. Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol Biol. 2007;63:591–608. doi: 10.1007/s11103-006-9111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Candat A, et al. The ubiquitous distribution of late embryogenesis abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress. Plant Cell. 2014;26:3148–3166. doi: 10.1105/tpc.114.127316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Banerjee A, Roychoudhury A. Group II late embryogenesis abundant (LEA) proteins: structural and functional aspects in plant abiotic stress. Plant Growth Regulation. 2015;79:1–17. doi: 10.1007/s10725-015-0113-3. [DOI] [Google Scholar]
- 67.Kim HS, et al. Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis. The Journal of biological chemistry. 2007;282:36292–36302. doi: 10.1074/jbc.M705217200. [DOI] [PubMed] [Google Scholar]
- 68.Zeng H, et al. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front Plant Sci. 2015;6:600. doi: 10.3389/fpls.2015.00600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Denoux C, et al. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Molecular plant. 2008;1:423–445. doi: 10.1093/mp/ssn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Naydenov M, et al. High-temperature effect on genes engaged in DNA methylation and affected by DNA methylation in Arabidopsis. Plant Physiol Biochem. 2015;87:102–108. doi: 10.1016/j.plaphy.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 71.Geem KR, et al. Jasmonic acid-inducible TSA1 facilitates ER body formation. Plant J. 2019;97:267–280. doi: 10.1111/tpj.14112. [DOI] [PubMed] [Google Scholar]
- 72.Tesfaye M, et al. Spatio-Temporal Expression Patterns of Arabidopsis thaliana and Medicago truncatula Defensin-Like Genes. PLOS ONE. 2013;8:e58992. doi: 10.1371/journal.pone.0058992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dai, Y. et al. CCR4-Not Complex Subunit Not2 Plays Critical Roles in Vegetative Growth, Conidiation and Virulence in Watermelon Fusarium Wilt Pathogen Fusarium oxysporum f. sp. niveum. Frontiers in Microbiology7, 10.3389/fmicb.2016.01449 (2016). [DOI] [PMC free article] [PubMed]
- 74.Ding X, Cory G, Song W-Y. A high-throughput system to verify candidate interactors from yeast two-hybrid screening using rolling circle amplification. Analytical Biochemistry. 2004;331:195–197. doi: 10.1016/S0003-2697(04)00304-5. [DOI] [PubMed] [Google Scholar]
- 75.Ito T, et al. Toward a protein–protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proceedings of the National Academy of Sciences. 2000;97:1143–1147. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gietz, R. D. & Woods, R. A. In Methods in Enzymology Vol. 350 87–96 (2002). [DOI] [PubMed]
- 77.Curtis MD, Grossniklaus U. A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 79.Yamamoto YY, Obokata J. ppdb: a plant promoter database. Nucl. Acids Res. 2008;36:D977–981. doi: 10.1093/nar/gkm785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 81.Li, C. & Wong, W. H. In The Analysis of Gene Expression Data: Methods and Software (eds Giovanni Parmigiani, Elizabeth S. Garrett, Rafael A. Irizarry, & Scott L. Zeger) 120–141 (Springer New York, 2003).
- 82.Dozmorov I, Centola M. An associative analysis of gene expression array data. Bioinformatics. 2003;19:204–211. doi: 10.1093/bioinformatics/19.2.204. [DOI] [PubMed] [Google Scholar]
- 83.Huang Da Wei, Sherman Brad T, Lempicki Richard A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2008;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 84.Almeida Trapp, M., De Souza, G. D., Rodrigues-Filho, E., Boland, W. & Mithöfer, A. Validated method for phytohormone quantification in plants. Front Plant Sci5, 10.3389/fpls.2014.00417 (2014). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data presented in the manuscript is publicly available. All transcriptome data is loaded into ArrayExpress with accession # E-MTAB-8326.