Abstract
Anorexia nervosa (AN) occurs nine times more often in females than in males. Although environmental factors likely play a role, the reasons for this imbalanced sex ratio remain unresolved. AN displays high genetic correlations with anthropometric and metabolic traits. Given sex differences in body composition, we investigated the possible metabolic underpinnings of female propensity for AN. We conducted sex‐specific GWAS in a healthy and medication‐free subsample of the UK Biobank (n = 155,961), identifying 77 genome‐wide significant loci associated with body fat percentage (BF%) and 174 with fat‐free mass (FFM). Partitioned heritability analysis showed an enrichment for central nervous tissue‐associated genes for BF%, which was more prominent in females than males. Genetic correlations of BF% and FFM with the largest GWAS of AN by the Psychiatric Genomics Consortium were estimated to explore shared genomics. The genetic correlations of BF%male and BF%female with AN differed significantly from each other (p < .0001, δ = −0.17), suggesting that the female preponderance in AN may, in part, be explained by sex‐specific anthropometric and metabolic genetic factors increasing liability to AN.
Keywords: eating disorder, fat‐free mass, female, genetic correlation, GWAS, shared genetics
1. INTRODUCTION
Anorexia nervosa (AN) is one of the most lethal psychiatric disorders and has established environmental and genetic risk factors (Chesney, Goodwin, & Fazel, 2014; Keshaviah et al., 2014). Female sex is the most robust and replicated risk factor, with nine females affected for each male case observed (Bulik et al., 2006; Micali, Hagberg, Petersen, & Treasure, 2013; Steinhausen & Jensen, 2015). Although historic diagnostic criteria for AN may have favored detection in females (e.g., presence of the amenorrhea criterion), most schemata did allow for the diagnosis of AN in males (American Psychiatric Association, 2013; World Health Organization, 1992). The focus of most work on gender differences in AN has been on sociocultural factors, such as personal evaluation of physical appearance and social pressures to be thin (Bakalar, Shank, Vannucci, Radin, & Tanofsky‐Kraff, 2015) although models based on biological and hormonal factors, such as growth, sex, and appetite‐regulating hormone abnormalities have also been posited (Culbert, Racine, & Klump, 2016; Schorr & Miller, 2017). However, collectively findings to date are not yet able to account for the widely disparate prevalences by sex.
The marked alterations in body composition, including fat mass (FM), fat‐free mass (FFM), and bone mineral density observed in AN are clinical characteristics of the illness, but have generally been considered to be sequelae of starvation (Westmoreland, Krantz, & Mehler, 2016). Females with AN show significantly greater FM deficits than affected males (Nagata et al., 2017) and, even after recovery, some individuals do not restore healthy body fat percentages (BF%; El Ghoch, Calugi, Lamburghini, & Dalle Grave, 2014). Moreover, lower BF% is a major risk factor for relapse (Bodell & Mayer, 2011). The causes of these particular sex differences have not yet been fully investigated.
Both AN and body composition as measured by bioelectrical impedance analysis are heritable (Schousboe et al., 2004; Tarnoki et al., 2014; Table S1). Significant negative single nucleotide polymorphism‐based autosomal genetic correlations (SNP‐r g) between AN and body mass index (BMI) and BF% were observed by the largest GWAS of AN conducted by the Eating Disorders Working Group of the Psychiatric Genomics Consortium (PGC‐ED; Duncan et al., 2017; Watson et al., 2018). This suggests shared etiology between those anthropometric traits and AN. Furthermore, AN shares common genetic variation with metabolic traits, such as insulin sensitivity and cholesterol. This revealed, for the first time, that a component of the genetic risk for AN is related to body composition and metabolism (Duncan et al., 2017; Hinney et al., 2017).
Phenotypic sex differences in body composition are also present in the general population; discernible as early as adolescence, females have on average higher BF% (Flegal et al., 2009), and less visceral adipose tissue and FFM than males (Paus, Wong, Syme, & Pausova, 2017), partially due to differences in adipocyte metabolism (Cheung & Cheng, 2016; Karastergiou & Fried, 2017; Link & Reue, 2017). Moreover, epidemiological findings indicate a female predominance at both tails of BMI, in extreme obesity (Kelly, Yang, Chen, Reynolds, & He, 2008; Lovre & Mauvais‐Jarvis, 2015) and in AN (Steinhausen & Jensen, 2015). Recent evidence shows clear biological sex differences in metabolism in rodent models (Arnold, 2017) and in humans (Mauvais‐Jarvis, 2015).
The observed phenotypic sex differences in body composition across the lifespan are partially due to genetic factors (Table S1 and Figure S1; Silventoinen et al., 2016, 2017). Heritability estimates from twin studies (twin‐h 2) of these epidemiological sex differences unveiled that twin‐h 2 estimates of BMI—a proxy of BF%—vary across the lifespan and show sex‐specific patterns, most apparent at the age of 13 years, from 20 to 30, and between ages 70 and 80 (Table S1 and Figure S1; Silventoinen et al., 2016, 2017). Although the twin‐h 2 varies somewhat, the specific genetic factors influencing BMI remain stable from decade to decade postadolescence, whereas environmental effects appear to change across time, especially in females (Haberstick et al., 2010). Additionally, several GWAS of proxy measures of BF% (Heid et al., 2010; Lindgren et al., 2009; Pulit et al., 2018; Randall et al., 2013; Winkler et al., 2017) and of BF% itself (Kilpeläinen et al., 2011; Lu et al., 2016) show clear sex differences in genome‐wide significant genomic loci and documented female‐specific heterogeneity in the genomic architecture extensively (for review, see Link & Reue, 2017; Pulit, Karaderi, & Lindgren, 2017; Small et al., 2018). Furthermore, studies have shown that BMI GWAS show tissue‐specific enrichment for the central nervous system (CNS; Finucane et al., 2015, 2018), whereas waist‐to‐hip ratio adjusted for BMI GWAS showed enrichment for adipose tissue (Finucane et al., 2018).
Convergent epidemiological and genetic findings show that the regulation of body composition varies between the sexes and is substantially influenced by both genetic and environmental factors. The primary goal of this study is to investigate whether a sex‐specific analysis of genetic determinants of body composition may partially explain the observed female preponderance in AN. We utilize new GWAS summary statistics from the PGC‐ED with about 16,000 cases, capitalizing on the availability of detailed and highly standardized body composition measurements and genetic data of 155,961 healthy and medication‐free individuals in the UK Biobank. Together, these provide a unique opportunity for a powerful investigation of the sex specificity of the genetic underpinnings of body composition and psychiatric traits and their relationship with AN.
2. METHODS
2.1. Genome‐wide association study of AN by the Eating Disorders Working Group of the Psychiatric Genomics Consortium
The meta‐analysis of GWAS on AN was a combined effort by the AN Genetics Initiative (Kirk et al., 2017; Thornton et al., 2018) and the PGC‐ED (www.med.unc.edu/pgc) and comprised 33 cohorts from 17 countries (Table S3) with 16,992 AN cases and 55,525 controls (Watson et al., 2018). The GWAS included 72,358 females (16,531 of whom are cases) and 24,454 males (460 of whom are cases; Table S2). The analysis includes additional samples from the Genetic Consortium for AN, the Wellcome Trust Case Control Consortium 3 (Boraska et al., 2014), and the UK Biobank (Sudlow et al., 2015). Case definitions established a lifetime diagnosis of AN via hospital or register records, structured clinical interviews, or online questionnaires based on standardized criteria—DSM‐III‐R, DSM‐IV, ICD‐8, ICD‐9, or ICD‐10—(American Psychiatric Association, 2013; World Health Organization, 1992), whereas in the UK Biobank cases self‐reported a diagnosis of AN (Davis et al., 2018). Quality control, imputation, GWAS, and meta‐analysis followed the standardized pipeline of the PGC, Rapid Imputation Consortium Pipeline (Ricopili; https://github.com/Nealelab/ricopili/tree/master/rp_bin). SNPs were excluded if they had a minor allele frequency (MAF) smaller than 1%, if no call was made in more than 2% of samples following imputation, if they were imputed with low confidence (INFO<0.7), or if they deviated substantially from Hardy–Weinberg equilibrium (controls p < 10−6, cases p < 10−10). Individuals were excluded if they showed inbreeding coefficients >0.2, or evidence of DNA contamination. Ancestry outliers were removed based on plotting of the first two principal components (PCs). The analysis was performed using imputed variant dosages and an additive model. The SNP‐based heritability (SNP‐h 2) of AN calculated using these data was 17% (SE = 1%), suggesting that a substantial fraction of the heritability of AN stems from common genetic variation across all autosomes (Watson et al., 2018).
2.2. GWASs of body composition: Study design and participants
Our study includes a cross‐sectional analysis of the baseline data from the epidemiological resource UK Biobank (www.ukbiobank.ac.uk; Allen, Sudlow, Peakman, Collins, & UK Biobank, 2014; Sudlow et al., 2015). To identify genetic variation associations with BF% and FFM that are not confounded by illnesses and their downstream effects or metabolism‐changing medication, we applied stringent exclusion criteria (Table S2). Due to this trait‐specific medication and illness filtering, the final analysis included 155,961 (45% female) healthy and drug‐free European ancestry participants comprising 32% of the genotyped UK Biobank participants. European ancestry was defined by 4‐means clustering of the first two PCs from the genetic data (Warren et al., 2017). Phenotypic characteristics separated by sex are presented in Table 1. All statistics were calculated in R 3.4.1 if not otherwise stated.
Table 1.
Phenotypic characteristics of individuals in the analyses
| Meta‐analyzed | Female | Male | |
|---|---|---|---|
| Number (%) | 155,961 | 70,700 (45%) | 85,261 (55%) |
| Age (years) | 54.9 ± 8.1 | 54.8 ± 8.0 | 55.0 ± 8.2 |
| Height (cm) | 170.4 ± 9.3 | 163.0 ± 6.2 | 176.4 ± 6.7 |
| Weight (kg) | 78.1 ± 15.1 | 69.6 ± 12.6 | 85.1 ± 13.2 |
| BMI (kg/m2) | 27.0 ± 4.2 | 26.2 ± 4.6 | 27.4 ± 3.8 |
| Waist circumference (cm) | 89.4 ± 12.6 | 82.3 ± 11.3 | 95.3 ± 10.3 |
| Hip circumference (cm) | 102.5 ± 8.1 | 102.0 ± 9.3 | 103.0 ± 6.9 |
| Waist‐to‐hip ratio | 0.9 ± 0.1 | 0.8 ± 0.1 | 0.9 ± 0.1 |
| Body fat (%) | 29.3 ± 8.2 | 35.3 ± 6.7 | 24.4 ± 5.5 |
| Fat mass (kg) | 23.0 ± 8.5 | 25.3 ± 9.1 | 21.2 ± 7.5 |
| FFM (kg) | 55.1 ± 11.6 | 44.4 ± 4.6 | 63.9 ± 7.4 |
| SES, Townsend deprivation index | −1.6 ± 2.9 | −1.7 ± 2.8 | −1.7 ± 2.9 |
BMI = body mass index; FFM = fat‐free mass; SES = socioeconomic status.
Data are n (%), or mean (SD).
2.3. Body composition assessment in healthy participants
Body composition was assessed with a rigorous and highly standardized protocol by UK Biobank using the same Tanita BC‐418 MA machines (Tanita Corporation, Arlington Heights, IL) for every participant. This body composition analyzer calculates FFM and FM from raw bioelectrical impedance data, using standard formulas including sex, age, height, and athleticism. Individuals whose hydration status might be compromised (e.g., suffering from diabetes mellitus or other endocrine diseases) were excluded (Table S3). Bioelectrical impedance technology has been extensively validated (Genton et al., 2003; Kyle et al., 2004; Lu et al., 2016), and results in more reliable estimates of body adiposity than BMI for healthy individuals (Mazzoccoli, 2016; Tanamas et al., 2016). Therefore, bioelectrical impedance analysis is the most feasible method in very large epidemiological samples, such as the UK Biobank, compared with proxy measures of adiposity, and does not expose participants to radiation unlike dual‐energy X‐ray absorptiometry.
2.4. GWASs on body composition
We calculated sex‐specific GWAS on residualized BF% and FFM, using BGENIE v1.2 (Bycroft et al., 2018). Our final analyses included 7,794,483 SNPs and insertion–deletion variants with an MAF >1%, imputation quality scores >0.8, and that were genotyped, or present in the Haplotype Reference Consortium (HRC) reference panel used for imputation by UK Biobank (McCarthy et al., 2016). We used an additive model on the imputed dosage data provided by UK Biobank, and residualized phenotypes prior to GWAS for factors related to assessment center, genotyping batch, smoking status, alcohol consumption, menopause, and for continuous measures of age, and socioeconomic status (SES) measured by the Townsend deprivation index (Townsend, 1987) as independent variables. We accounted for underlying population stratification by also including the first six PCs, calculated on the genotypes of the European subsample. We then meta‐analyzed these sex‐specific GWAS using METAL (http://csg.sph.umich.edu/abecasis/metal/; Willer, Li, & Abecasis, 2010) using an inverse variance weighted model with a fixed effect, to obtain sex‐combined results. Significantly associated SNPs (p < 5 × 10−8) were considered as potential index SNPs. SNPs in LD (r 2 > 0.2) with a more strongly associated SNP within 3,000 kb were assigned to the same locus using Functional Mapping and Annotation (FUMA; Watanabe, Taskesen, van Bochoven, & Posthuma, 2017). Overlapping clumps additionally were merged with a second clumping procedure in FUMA merging all lead SNPs with r 2 = 1 to genomic loci. After clumping, independent genome‐wide significant loci (5 × 10−8) were compared with entries in the NHGRI‐EBI GWAS catalog (MacArthur et al., 2017) using FUMA (Watanabe et al., 2017). Sex‐specific loci are defined as reaching genome‐wide significance (5 × 10−8) in either females or males while not showing at least suggestive significance in the opposite sex (5 × 10−6) with differences in beta estimates that remain significant after Bonferroni correction for the total number of significant genomic loci.
2.5. Genome‐wide SNP‐based heritability and partitioned heritability
Using BOLT‐LMM (Loh et al., 2015) on genotyped, genome‐wide, common genetic variants and linkage disequilibrium score regression (LDSC) implemented in LDSC v.1.0.0 (Bulik‐Sullivan et al., 2015) on genome‐wide summary statistics, we calculated the total phenotypic variance explained by common autosomal SNPs, SNP‐based heritability (SNP‐h 2). We included all genotyped and imputed autosomal variants for BF% and FFM and used the LD score reference files provided with the software. We tested for differences between the heritabilities by calculating SE using a block jackknife method implemented into the software. To identify tissue types associated with BF% and FFM, we performed a partitioned heritability analysis in LDSC v.1.0.0, ranking 10 cell type groups based on contribution to heritability after controlling for the effects of 53 functional annotations (Finucane et al., 2015).
2.6. Genetic correlations
Using an analytic extension of LDSC (Bulik‐Sullivan et al., 2015), we calculated SNP‐based bivariate genetic correlations (SNP‐r g) across the autosomes to examine the genetic overlap between AN and metabolic and psychiatric GWAS summary statistics. First, we calculated SNP‐r gs between anthropometric traits, namely our BF% and FFM GWASs with GWASs of childhood BMI (~8 years; Felix et al., 2016), childhood obesity (Bradfield et al., 2012), childhood FFM (Medina‐Gomez et al., 2017), adult FFM (Zillikens et al., 2017), and adolescence and young adulthood BMI (~15–25 years; Graff et al., 2013), to estimate the genomic overlap of body composition between different periods of life. Second, we calculated SNP‐r gs of these anthropometric traits across the lifespan with AN.
Additionally, we computed SNP‐r gs of AN (Supporting Information) with glycemic traits, such as insulin sensitivity assessed by the insulin resistance homeostatic model assessment (HOMA‐IR), fasting glucose, and insulin concentrations (Lagou, Mägi, & Hottenga, 2018; Manning et al., 2012; Scott et al., 2012), to investigate potential mediation of the relationship between body fat and AN. Physical activity is reported to be increased in AN patients (Achamrah, Coëffier, & Déchelotte, 2016; Shroff et al., 2006); therefore, we estimated the genetic overlap between physical activity (Hanscombe, 2018, Unpublished, Supporting Information) and AN. We explored the genomic contribution to the comorbidity of AN with psychiatric disorders and traits, including major depressive disorder (MDD; Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium et al., 2013), anxiety (Purves et al., 2017), schizophrenia (Schizophrenia Working Group of the Psychiatric Genomics Consortium et al., 2014), obsessive–compulsive disorder (OCD; International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF‐GC) and OCD Collaborative Genetics Association Studies (OCGAS), 2018), and neuroticism (Coleman, 2017, Unpublished, Supporting Information), as well as educational attainment (Okbay et al., 2016) by calculating SNP‐r gs. Information on all GWAS is presented in Table S4.
2.7. Sex‐specific analyses of genomic determinants
We investigated differences between sexes in heritability and genetic architecture to identify sex‐specific liability driven by genomic factors. We examined differences (δ) in the SNP‐h 2 estimates between males and females using a block jackknife approach (Supporting Information) and tested whether the SNP‐r gs between females and males were different from 1 to identify potential genetic differences related to sex. We calculated the SNP‐r g of the female and male GWASs with AN separately to investigate the differences in the relationship of these sex differences with the risk for AN. To test the statistical significance of all estimates, we calculated their SE and corresponding p value by applying a block jackknife method, as described and implemented in LDSC v1.0.0 by Bulik‐Sullivan et al. (2015) and in our Supporting Information.
As a sensitivity analysis, we repeated all SNP‐r g analysis with a female‐only GWAS of AN. However, due to the small number of male AN cases, it was impossible to perform a male‐only analysis. All methods are described in more detail in the Supporting Information. Stringent multiple testing correction was performed on each analysis, using matrix decomposition to detect the effective number of tests and subsequent Bonferroni correction of the p value thresholds.
3. RESULTS
3.1. GWAS of AN
The AN GWAS resulted in eight genome‐wide significant loci and showed enrichment for CNS cell types. It genetically correlated with a broad range of metabolic and psychiatric phenotypes, mirroring clinically observed comorbidity (for details, see Duncan et al., 2017 ; Watson et al., 2018).
3.2. GWAS of body composition measures in the UK Biobank
After quality control, we performed sex‐stratified association analyses on the continuous outcomes of BF% and FFM. Minimal inflation due to population stratification or other systematic biases was indicated by LDSC intercepts between 1.02 and 1.10 and lambda median statistic inflation values (λmedian) between 1.18 and 1.59 (Table S4 and Figure S3a,b). We identified 34 independent loci associated with meta‐analyzed BF% that are not reported to be associated with anthropometric traits in the GWAS catalog (MacArthur et al., 2017) and replicated 42 independent genome‐wide significant results (p < 5 × 10−8) after LD‐based and distance‐based clumping (Figure 1, Figure S4a, Table S5a,b). We identified one male‐specific locus in BF% (Table S5a). The meta‐analyzed GWAS of FFM yielded 83 novel loci and replicated 78 genomic risk loci previously associated with anthropometric traits (Figure 2, Figure S3b, Table S6a,b). We identified 13 male‐specific genomic loci in FFM (Table S6a). All genomic regions, region plots thereof, their annotations, including nearby protein coding genes (within 100 kb), and previous entries in the GWAS catalog are published on FUMA (http://fuma.ctglab.nl/browse) entries 20–25. Summary statistics are available for download www.topherhuebel.com/GWAS.
Figure 1.
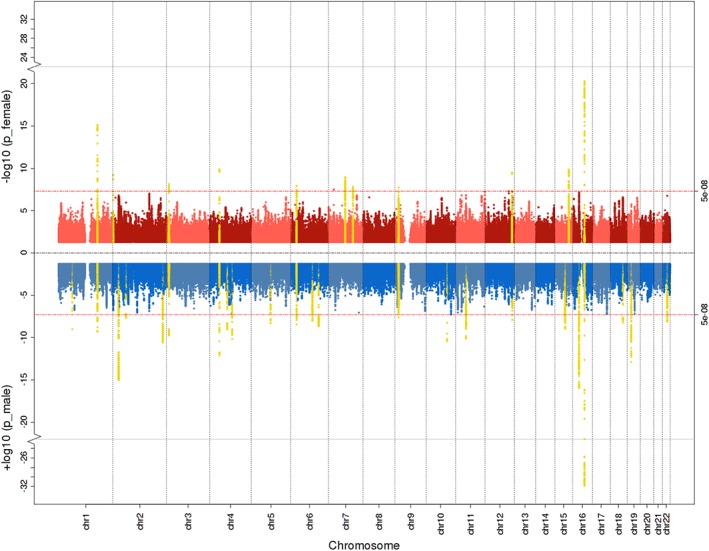
Miami plot for female (red), male (blue), and meta‐analyzed (yellow) genome‐wide body fat percentage (BF%) associations. Significant loci from the sex‐combined analyses are highlighted in yellow if they also reached genome‐wide significance in the sex‐specific genome‐wide association studies (GWASs). The genome‐wide significance threshold p < 5 × 10−8 is represented by the red horizontal lines. Chr = chromosome
Figure 2.
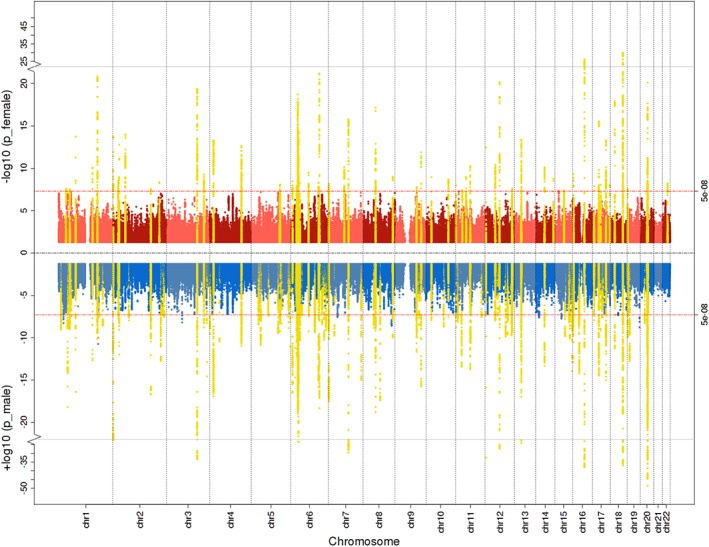
Miami plot for female (red), male (blue), and meta‐analyzed (yellow) genome‐wide fat‐free mass (FFM) associations. Significant loci from the sex‐combined analyses are highlighted in yellow if they also reached genome‐wide significance in the sex‐specific genome‐wide association studies (GWASs). The genome‐wide significance threshold p < 5 × 10−8 is represented by the red horizontal lines. Chr = chromosome
3.3. Genome‐wide SNP‐based and partitioned heritability
The SNP‐h 2 for BF% ranged between 29 and 33%, and for FFM between 43 and 51% (Figure 3), while that for AN is about 17–20% with an assumed population prevalence of 0.9% (Duncan et al., 2017; Watson et al., 2018). The SNP‐h 2 of FFMmale measured by LDSC was significantly higher than the SNP‐h 2 of FFMmeta (p < .001, δSNP‐h 2 = 5.6%). However, neither the SNP‐h 2 estimates for BF% nor for FFM measured by LDSC differed significantly between the sexes.
Figure 3.
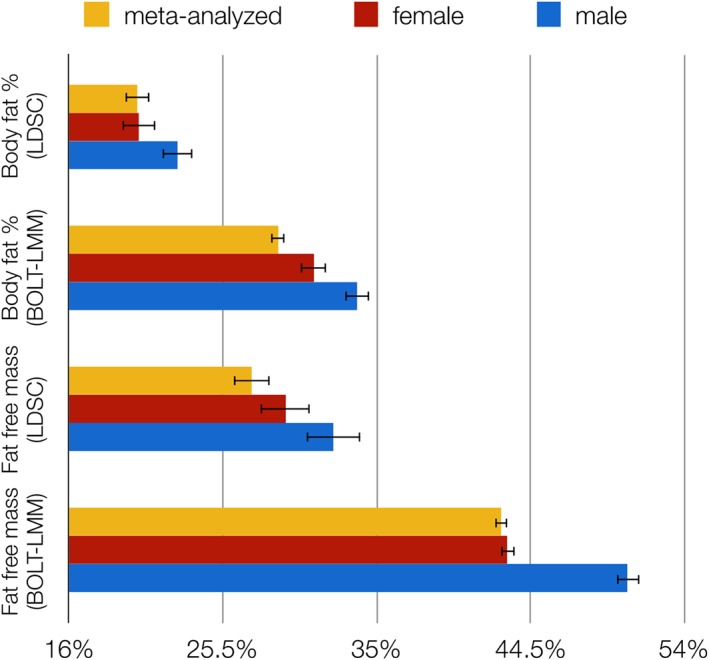
Sex‐specific single nucleotide polymorphism‐based heritability estimates (SNP‐h 2) for body fat percentage and fat‐free mass calculated by BOLT‐LMM (Loh et al., 2015) and linkage disequilibrium score regression (LDSC; Bulik‐Sullivan et al., 2015). Error bars represent SE. All estimated SNP‐h 2 were statistically significant
Partitioned heritabilities can estimate the proportion of the overall SNP‐h 2 that can be attributed to different cell type groups. BF%female showed an significant enrichment for the CNS cell type group with 14% of SNPs explaining an estimated 40% of the SNP‐h 2 (p = .004), whereas BF%male was significantly enriched for the “other” cell type group that contains adipose tissue with 20% of SNPs explaining an estimated 57% of the SNP‐h 2 (p = .004; Figure S4a,b). The FFMfemale and FFMmale were enriched for connective and bone tissue with 11% of SNPs explaining an estimated 47% of SNP‐h 2 in both sexes (p female = 6.65 × 10−6; p male = 2.29 × 10−7; Figure S5a,b). The meta‐analyzed FFMboth was also enriched for skeletal muscle with 10% of SNPs explaining an estimated 37% of SNP‐h 2 (p = .004, Figure S5c).
3.4. Genetic correlations of anthropometric traits across the lifespan
The significant SNP‐r g between BF%meta and BMIchildhood was 0.46 (SE = 0.04; p = 6.11 × 10−32) and between BF%meta and BMIadolescence/young adulthood was 0.48 (SE = 0.05; p = 9.24 × 10−25). Similarly, FFMchildhood and FFMadulthood showed a significant SNP‐r g of 0.69 (SE = 0.10; p = 2.70 × 10−12) and FFMchildhood also correlated genetically with FFMmeta in our UK Biobank sample (SNP‐r g = 0.30; SE = 0.04; p = 3.24 × 10−12).
BF%meta and FFMmeta correlated genetically (SNP‐r g = 0.26; SE = 0.02; p = 3.95 × 10−26). The SNP‐r g between BF%female and BF%male was significantly less than 1 (SNP‐r g = 0.89, SE = 0.03; p =1 = .0005), indicating heterogeneity in the genomic architecture between females and males (Figure 4).
Figure 4.
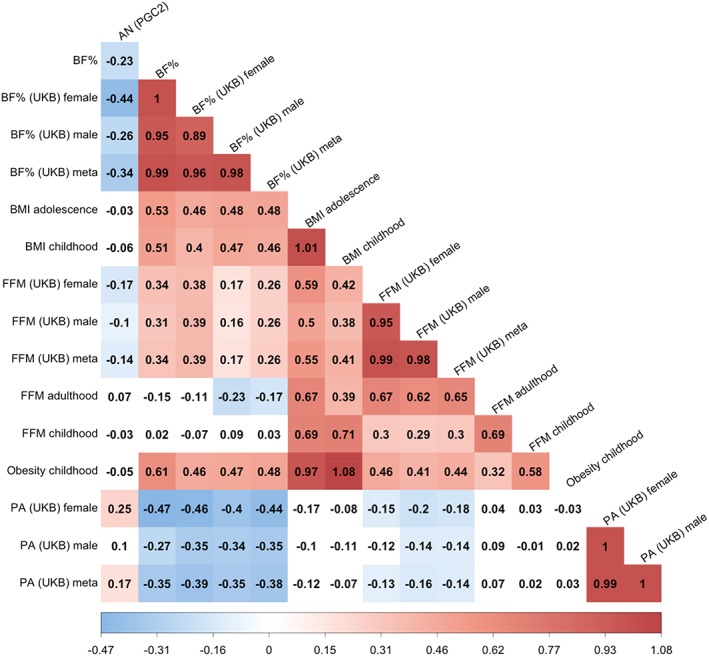
Heatmap of sex‐specific bivariate single nucleotide polymorphism‐based genetic correlations (SNP‐r2 g) of body fat percentage, BMI, fat‐free mass, physical activity, and obesity with AN. The strength of the correlation is reflected in the hue. Blue colors are negative SNP‐r gs, meaning that the same genetic variants influence both traits in opposite directions, and red are positive SNP‐r gs meaning that the same genetic variants influence traits in the same direction. Colored squares are significant after correction for multiple comparisons by matrix decomposition and Bonferroni correction (p Bonferroni = .05/10). The SNP‐r gs were calculated by linkage disequilibrium score regression (LDSC). AN = anorexia nervosa; BF% = body fat percentage; BMI = body mass index; FFM = fat‐free mass; PA = physical activity; PGC2 = 2nd freeze psychiatric genomics consortium; UKB = UK Biobank
3.5. Sex‐specific genetic correlations with AN
We calculated SNP‐r g between the sex‐specific and sex‐combined GWAS with AN to investigate sex differences. The genetic correlation between BF%female and AN was −0.44 (SE = 0.04; p = 8.28 × 10−27), whereas that between BF%male and AN was −0.26 (SE = 0.04; p = 1.04 × 10−13). These SNP‐r g were significantly different from each other (δSNP‐r g = −0.17; SE = 0.04; p = 4.23 × 10−5). AN showed a significant genetic correlation with FFMmeta (SNP‐r g = −0.14; SE = 0.03; p = 5.79 × 10−6) Physical activityfemale showed a significant SNP‐r g with AN (SNP‐r g = 0.25; SE = 0.06; p = 1.10 × 10−5), but physical activitymales did not (SNP‐r g = 0.10; SE = 0.06; p = .07). However, this difference was not statistically significant (δSNP‐r g = −0.13; SE = 0.07; p = .05; Figure 4) after multiple testing correction.
BMI‐adjusted fasting insulin concentrations and AN were genetically correlated (SNP‐r g = −0.24; SE = 0.06; p = 2.31 × 10−5). Fasting insulinfemale was genetically correlated with AN (SNP‐r g = −0.36; SE = 0.07; p = 5.29 × 10−7), but not fasting insulinmale (SNP‐r g = −0.16; SE = 0.05; p = .003). However, this difference in SNP‐r g between sexes did not reach significance (δSNP‐r g = −0.19; SE = 0.08; p = .02) after multiple testing correction. Sex‐ and age‐adjusted insulin resistance (HOMA‐IR) correlated significantly with AN (SNP‐r g = −0.29, SE = 0.07; p = 2.83 × 10−5; Figure 5), but no sex differences were observed.
Figure 5.
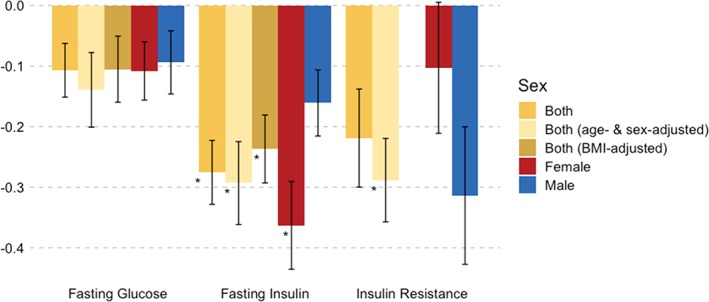
Sex‐specific bivariate single nucleotide polymorphism‐based genetic correlations (SNP‐r g) of fasting glucose, fasting insulin, and insulin resistance assessed by the HOMA‐IR with AN. The SNP‐r gs were calculated by linkage disequilibrium score regression (LDSC). Significant SNP‐r gs are marked with an asterisk (*) after correction for multiple comparisons by matrix decomposition and Bonferroni correction (p Bonferroni = .05/28). The error bars depict the SE. Summary statistics for BMI‐adjusted HOMA‐IR were not available. AN = anorexia nervosa; BMI = body mass index; HOMA‐IR = insulin resistance by homeostatic model assessment
AN was significantly correlated with MDDfemale (SNP‐r g = 0.26; SE = 0.07; p = 4.00 × 10−4) and anxietymeta (SNP‐r g = 0.25; SE = 0.05; p = 8.90 × 10−8). However, the difference between the male and female SNP‐r g with AN was not significant in MDD (δSNP‐r g = −0.004; SE = 0.16; p = .98). While the SNP‐r g between education years in females and males was significantly different from 1 (SNP‐r g = 0.91, SE = 0.02; p = 7.99 × 10−5), indicating sex differences, the SNP‐r g of education years with AN did not differ between females and males (δSNP‐r g = −0.02; SE = 0.03; p = .59; Figure 6). As sensitivity analysis, all SNP‐r gs were also calculated with a female only AN GWAS showing no meaningful differences (Table S8a).
Figure 6.
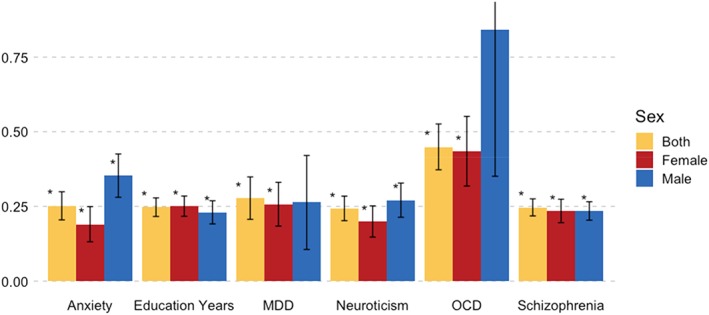
Sex‐specific bivariate single nucleotide polymorphism‐based genetic correlations (SNP‐r g) of probable anxiety disorder (anxiety), education years, MDD, neuroticism, OCD, and schizophrenia with anorexia nervosa. The SNP‐r gs were calculated by linkage disequilibrium score regression (LDSC). Significant SNP‐r gs are marked with an asterisk (*) after correction for multiple comparisons by matrix decomposition and Bonferroni correction (p Bonferroni = .05/28). The error bars depict the SE. The SE of the OCDmale reaches above 1 and has been cut off. MDD = major depressive disorder; OCD = obsessive–compulsive disorder
4. DISCUSSION
The latest GWAS on AN by the PGC‐ED presented evidence for a reconceptualization of AN as a metabo‐psychiatric disorder by identifying significant SNP‐r gs of AN with a variety of metabolic phenotypes, including body composition, lipid metabolism, and glycemic traits (Duncan et al., 2017; Watson et al., 2018). We extended the findings on the relationship between BF% and AN by replicating that genomic effects on BF% differ by sex (Heid et al., 2010; Lindgren et al., 2009; Pulit et al., 2018; Randall et al., 2013; Winkler et al., 2017) and showing that female‐specific effects on BF% have a significantly greater genetic correlation with AN (SNP‐r g = −0.44; SE = 0.04; p = 8.28 × 10−27) than male‐specific effects on BF% (SNP‐r g = −0.26; SE = 0.04; p = 1.04 × 10−13). This suggests that a specific set of genomic variation may be differentially active in females and may increase the liability for AN. The partitioned heritability analyses of SNP‐h 2 showed that BF%female was significantly enriched for CNS tissue while BF%male was enriched for adipose tissue, recapitulating prior findings in sex‐combined samples (Finucane et al., 2015, 2018; Willer et al., 2009). This indicates a sex‐specific enrichment for BF% and that BF% has associated genetic variation underlying its biology thereby validating the use of bioelectrical impedance analysis to measure body compartments. Moreover, our findings suggest that different tissues may be implicated in the regulation of BF% in females and males.
In our analysis of body composition across the lifespan, BF%childhood, BF%adolescence and young adulthood, and FFMchildhood were not genetically correlated with AN, whereas BF%adult and FFMadult was. However, GWASs of BF% and BMI as well as FFM were well correlated across the lifespan with SNP‐r g s of about ~0.60 across childhood, adolescence, young adulthood, and adulthood (Figure 4). This suggests that a proportion of BF%‐associated genomic variation may become operative at a later age and that this component may be correlated with risk for AN. This seems to overlap with the period—between 20 and 30 years of age—in which females and males show a significant difference in the twin‐h 2 of BMI (Figure S1; Silventoinen et al., 2016, 2017).
Additionally, we estimated SNP‐r g of AN with sex‐specific GWASs of physical activity and glycemic traits to investigate potential moderators and mediators of the relationship between body fat and AN. Only physical activityfemale and fasting insulinfemale were significantly genetically associated with AN. However, the differences between female and male SNP‐r gs were only nominally significant for both traits and did not survive correction for multiple testing emphasizing the need for larger sample sizes to examine sex differences.
In our sex‐specific investigation of the contribution of psychiatric disorders and behavioral traits to AN, genomic variation associated with MDD in females and OCD in males suggested a possible sex effect in their SNP‐r g with AN, but statistical tests did not confirm this. Power may be an issue; in particular, the current sample size of the OCD GWAS is relatively small. Consequently, some of our findings need to be interpreted cautiously, and this analysis should be repeated after much larger GWASs are available preferably with >10,000 cases of each sex. Some GWASs, however, are well powered and although the SNP‐r g of education years between males and females was significantly lower than 1—similar to BF%—we did not observe sex differences in the SNP‐r g of education years with AN, suggesting that metabolic traits may be more likely to contribute to the sex‐specific liability to AN than psychiatric or behavioral phenotypes.
Our investigation was limited by the small proportion of male AN cases in the primary AN GWAS (Table S2) not allowing for male‐only analyses. However, female‐only analyses did not show meaningful differences to the sex‐combined analyses (Table S8a). We were unable to include the X chromosome in the investigations as the genotype or summary level data for several GWASs in the PGC AN GWAS meta‐analysis were not available to us when the analyses were conducted. However, this should be incorporated in future studies. Most importantly, compared with prior BMI GWAS, our study benefited from arguably more homogeneously assessed body composition phenotypes, allowing us to differentiate between BF% and FFM more effectively (Kilpeläinen et al., 2011; Lu et al., 2016). Moreover, we adjusted for smoking behavior, alcohol consumption, and menopause and excluded participants taking weight altering medications and participants with somatic diseases or psychiatric disorders that affect body composition, such as cancers, diabetes, and MDD. This is a unique and important feature of our investigation and substantially reduced possible confounding of our GWAS.
Conclusion
Our results add further evidence that AN is both a psychiatric and metabolic disorder and suggest that an age‐dependent specific set of genomic variation may be differentially active in females that influences body composition, which may also contribute to liability for AN. Our work could have therapeutic implications, by considering exploring approaches to using body composition measures or genetic markers of body composition as predictors of clinical course or adverse outcome, and as a component of personalized treatment that considers an individual's propensity to lose therapeutically restored weight. Some individuals may be at greater risk of relapse, for example, when confronted with periods of negative energy balance, and this could be addressed in personalized treatment and relapse prevention (Bulik, 2016). Sex‐specific genetic and biological factors may partially underlie increased risk for AN in females which suggests that new and focused studies of body composition and metabolism in AN patients could increase our understanding of AN etiology and response to treatment.
Supporting information
Figure S1 Twin‐based heritabilities (twin‐h 2) of body mass index (BMI) across the lifespan as calculated by ACE models from Silventoinen et al. Blue values represent males and red females with error bars depicting 95% confidence intervals. After the age of 19, heritabilities are represented for a whole decade (Silventoinen et al., 2016, 2017)
Figure S2. Analysis workflow chart. AN = anorexia nervosa; BMI = body mass index; GWAS = genome‐wide association study; LDSC = linkage score disequilibrium regression; MAGIC = Meta‐Analyses of Glucose and Insulin‐related traits Consortium; MDD = major depressive disorder; OCD = obsessive–compulsive disorder; PGC = Psychiatric Genomics Consortium; SSGAC = Social Science Genetic Association Consortium; UKB = UK Biobank
Figure S3 (a) QQ plot for the body fat percentage (BF%) genome‐wide‐association study (GWAS). (b) QQ plot for the fat‐free mass (FFM) GWAS
Figure S4 (a) Manhattan plot of the meta‐analyzed genome‐wide association study (GWAS) of body fat percentage (BF%). The red line represents the genome‐wide significance threshold of 5 × 10−8. Chr = chromosome. (b) Manhattan plot of the meta‐analyzed genome‐wide association study (GWAS) of fat‐free mass (FFM). The red line represents the genome‐wide significance threshold of 5 × 10−8. Chr = chromosome
Figure S5 (a) Partitioned heritability by 10 cell type groups for body fat percentage in females. The black dashed lines at −log10(P) = 2.3 is the cutoff for Bonferroni significance. CNS = central nervous system SNP = single nucleotide polymorphism. (b) Partitioned heritability by 10 cell type groups for body fat percentage in males. The black dashed lines at −log10(P) = 2.3 is the cutoff for Bonferroni significance. CNS = central nervous system, SNP = single nucleotide polymorphism. (c) Partitioned heritability by 10 cell type groups for body fat percentage in the meta‐analyzed GWAS. The black dashed lines at −log10(P) = 2.3 is the cutoff for Bonferroni significance. CNS = central nervous system; SNP = single nucleotide polymorphism
Figure S6 (a) Partitioned heritability by 10 cell type groups for fat‐free mass (FFM) in females. The black dashed lines at −log10(P) = 2.3 is the cutoff for Bonferroni significance. CNS = central nervous system, SNP = single nucleotide polymorphism. (b) Partitioned heritability by 10 cell type groups for FFM in males. The black dashed lines at −log10(P) = 2.3 is the cutoff for Bonferroni significance. CNS = central nervous system, SNP = single nucleotide polymorphism. (c) Partitioned heritability by 10 cell type groups for fat‐free mass (FFM) in the meta‐analyzed GWAS. The black dashed lines at −log10(P) = 2.3 is the cutoff for Bonferroni significance. CNS = central nervous system; SNP = single nucleotide polymorphism
Table S1 Twin‐based (twin‐h 2) and single nucleotide polymorphism‐based heritability (SNP‐h 2) estimates derived from genome‐wide association studies (GWAS) for anorexia nervosa (AN) and anthropometric traits measured by bioelectrical impedance analysis of fat‐free mass (FFM), and body fat percentage (BF%)
Table S2. Sex data for the anorexia nervosa (AN) genome‐wide association study (GWAS) datasets
Table S3. Exclusion criteria by International Statistical Classification of Diseases (ICD‐10), British National Formulary (BNF), and UK Biobank variable
Table S4. Inflation statistics and heritability estimates on the observed scale of the genome‐wide association studies (GWAS). Attenuation ratio as calculated by Loh et al. (2017)
Table S5 (a) Genome‐wide significant loci of the body fat percentage genome‐wide association study (GWAS) including heterogeneity measures of the meta‐analysis and a z‐test to test for significant differences between the sexes. Sex‐specific loci are defined as reaching genome‐wide significance (5 × 10−8) in either females or males, not reaching suggestive significance in the opposite sex (5 × 10−6) and the differences in beta estimates is significant after Bonferroni correction for the total number of significant genomic loci. (b) Genomic loci associated with anthropometric traits in published genome‐wide association studies (GWAS). Previous associations have been retrieved from the GWAS catalog (https://www.ebi.ac.uk/gwas/). Data extracted from Functional Mapping and Annotation of Genome‐Wide Association Studies (FUMA, http://fuma.ctglab.nl/)
Table S6. (a) Genome‐wide significant loci of the fat‐free mass genome‐wide association study (GWAS) including heterogeneity measures of the meta‐analysis and a z‐test to test for significant differences between the sexes. Sex‐specific loci are defined as reaching genome‐wide significance (5 × 10−8) in either females or males, not reaching suggestive significance in the opposite sex (5 × 10−6) and the differences in beta estimates is significant after Bonferroni correction for the total number of significant genomic loci. (b) Genomic loci associated with anthropometric traits in published genome‐wide association studies (GWAS). Previous associations have been retrieved from the GWAS catalog (https://www.ebi.ac.uk/gwas/). Data extracted from Functional Mapping and Annotation of Genome‐Wide Association Studies (FUMA, http://fuma.ctglab.nl/)
Table S7. Heritability as estimated by BOLT‐LMM, v2.3.2, on genotyped single nucleotide polymorphisms (SNPs) on anthropometric traits in the UK Biobank. Variance explained by BOLT‐LMM's linear predictor—using the default mixture‐of‐Gaussians prior on SNP effect sizes, which accounts for larger‐effect SNPs—and variance theoretically explained by an optimal linear predictor, that is, SNP heritability (h 2g). Additionally, linkage disequilibrium score regression (LDSC) estimates are presented and the difference between BOLT‐LMM and LDSC estimates
Table S8. Genetic correlations for sex‐combined anorexia nervosa genome‐wide association study and genetic correlations across body composition, physical activity, and psychiatric and behavioral traits. (a) Genetic correlations for female‐only anorexia nervosa genome‐wide association study and genetic correlations across body composition, physical activity, and psychiatric and behavioral traits
Appendix S1 Supporting Information
Appendix S2 Supporting Information
Appendix S3 Supporting Information
ACKNOWLEDGMENTS
This study represents independent research part funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King's College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. High performance computing facilities were funded with capital equipment grants from the GSTT Charity (TR130505) and Maudsley Charity (980). Research reported in this publication was supported by the National Institute Of Mental Health of the National Institutes of Health under Award Number U01MH109514. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Dr. C.M.B. acknowledges funding from the Swedish Research Council (VR Dnr: 538‐2013‐8864) and the Klarman Family Foundation (the Anorexia Nervosa Genetics Initiative is an initiative of the Klarman Family Foundation). Dr. P.F.O. receives funding from the UK Medical Research Council (MR/N015746/1) and the Wellcome Trust (109863/Z/15/Z). Dr. M.G. acknowledges funding from the National Institutes of Health (R01HD057194). Dr. T.W. acknowledges funding by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health. Data on glycemic traits have been contributed by Meta‐Analyses of Glucose and Insulin‐related traits Consortium (MAGIC) investigators and have been downloaded from www.magicinvestigators.org. Data on the childhood BMI trait have been contributed by the EGG Consortium and has been downloaded from www.egg-consortium.org. This study was completed as part of approved UK Biobank study applications 16577 and 27546 to Dr. G.B. Dr. G.B. has received grant funding from and served as a consultant to Eli Lilly, and has received honoraria from Illumina and has served on advisory boards for Otsuka. Dr. C.M.B. is a grant recipient from and has served on advisory boards for Shire. She has received royalties from Pearson and Walker. All interests unrelated to this work. Dr. J.R.I.C., Dr. H.A.G., Dr. K.L.P., Dr. C.H., and Dr. P.F.O. have nothing to disclose.
Hübel C, Gaspar HA, Coleman JRI, et al. Genomics of body fat percentage may contribute to sex bias in anorexia nervosa. Am J Med Genet Part B. 2019;180B:428–438. 10.1002/ajmg.b.32709
Funding information GSTT Charity, Grant/Award Number: TR130505; Maudsley Charity, Grant/Award Numbers: 980, TR130505; NIH, Grant/Award Number: R01HD057194; UK Medical Research Council, Grant/Award Number: MR/N015746/1; Vetenskapsrådet, Grant/Award Number: Dnr: 538‐2013‐8864; Wellcome Trust, Grant/Award Number: 109863/Z/15/Z; National Institutes of Health, Grant/Award Number: R01HD057194; Eunice Kennedy Shriver National Institute of Child Health and Human Development; Wellcome Trust; Medical Research Council; Klarman Family Foundation; Swedish Research Council; South London and Maudsley NHS Foundation Trust; National Institute for Health Research
REFERENCES
- Achamrah, N. , Coëffier, M. , & Déchelotte, P. (2016). Physical activity in patients with anorexia nervosa. Nutrition Reviews, 74(5), 301–311. [DOI] [PubMed] [Google Scholar]
- Allen, N. E. , Sudlow, C. , Peakman, T. , Collins, R. , & UK Biobank . (2014). UK Biobank data: Come and get it. Science Translational Medicine, 6(224), 224ed4. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association . (2013). Diagnostic and statistical manual of mental disorders (5th ed). Washington, DC: American Psychiatric Association. [Google Scholar]
- Arnold, A. P. (2017). A general theory of sexual differentiation. Journal of Neuroscience Research, 95(1–2), 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakalar, J. L. , Shank, L. M. , Vannucci, A. , Radin, R. M. , & Tanofsky‐Kraff, M. (2015). Recent advances in developmental and risk factor research on eating disorders. Current Psychiatry Reports, 17(6), 42. [DOI] [PubMed] [Google Scholar]
- Bodell, L. P. , & Mayer, L. E. (2011). Percent body fat is a risk factor for relapse in anorexia nervosa: A replication study. International Journal of Eating Disorders, 44(2), 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boraska, V. , Franklin, C. S. , Floyd, J. A. B. , Thornton, L. M. , Huckins, L. M. , Southam, L. , … Bulik, C. M. (2014). A genome‐wide association study of anorexia nervosa. Molecular Psychiatry, 19(10), 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradfield, J. P. , Taal, H. R. , Timpson, N. J. , Scherag, A. , Lecoeur, C. , Warrington, N. M. , … Early Growth Genetics, Consortium . (2012). A genome‐wide association meta‐analysis identifies new childhood obesity loci. Nature Genetics, 44(5), 526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik, C. M. (2016). Towards a science of eating disorders: Replacing myths with realities: The fourth Birgit Olsson lecture. Nordic Journal of Psychiatry, 70(3), 224–230. [DOI] [PubMed] [Google Scholar]
- Bulik, C. M. , Sullivan, P. F. , Tozzi, F. , Furberg, H. , Lichtenstein, P. , & Pedersen, N. L. (2006). Prevalence, heritability, and prospective risk factors for anorexia nervosa. Archives of General Psychiatry, 63(3), 305–312. [DOI] [PubMed] [Google Scholar]
- Bulik‐Sullivan, B. K. , Loh, P. R. , Finucane, H. K. , Ripke, S. , Yang, J. , Schizophrenia Working Group of the Psychiatric Genomics Consortium , … Neale, B. M. (2015). LD score regression distinguishes confounding from polygenicity in genome‐wide association studies. Nature Genetics, 47(3), 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft, C. , Freeman, C. , Petkova, D. , Band, G. , Elliott, L. T. , Sharp, K. , … Marchini, J. (2018). The UK Biobank resource with deep phenotyping and genomic data. Nature, 562(7726), 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesney, E. , Goodwin, G. M. , & Fazel, S. (2014). Risks of all‐cause and suicide mortality in mental disorders: A meta‐review. World Psychiatry, 13(2), 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, O. K.‐W. , & Cheng, A. S.‐L. (2016). Gender differences in adipocyte metabolism and liver cancer progression. Frontiers in Genetics, 7, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbert, K. M. , Racine, S. E. , & Klump, K. L. (2016). Hormonal factors and disturbances in eating disorders. Current Psychiatry Reports, 18(7), 65. [DOI] [PubMed] [Google Scholar]
- Davis, K. A. S. , Coleman, J. R. I. , Adams, M. , Allen, N. , Breen, G. , Cullen, B. , … Hotopf, M. (2018). Mental health in UK Biobank: Development, implementation and results from an online questionnaire completed by 157 366 participants. BJPsych Open, 4(3), 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Duncan, L. , Yilmaz, Z. , Gaspar, H. , Walters, R. , Goldstein, J. , Anttila, V. , … Bulik, C. M. (2017). Significant locus and metabolic genetic correlations revealed in genome‐wide association study of anorexia nervosa. American Journal of Psychiatry, 174(9), 850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Ghoch, M. , Calugi, S. , Lamburghini, S. , & Dalle Grave, R. (2014). Anorexia nervosa and body fat distribution: A systematic review. Nutrients, 6(9), 3895–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix, J. F. , Bradfield, J. P. , Monnereau, C. , van der Valk, R. J. P. , Stergiakouli, E. , Chesi, A. , … Bone Mineral Density in Childhood Study BMDCS . (2016). Genome‐wide association analysis identifies three new susceptibility loci for childhood body mass index. Human Molecular Genetics, 25(2), 389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane, H. K. , Bulik‐Sullivan, B. , Gusev, A. , Trynka, G. , Reshef, Y. , Loh, P.‐R. , … Price, A. L. (2015). Partitioning heritability by functional annotation using genome‐wide association summary statistics. Nature Genetics, 47(11), 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane, H. K. , Reshef, Y. A. , Anttila, V. , Slowikowski, K. , Gusev, A. , Byrnes, A. , … Price, A. L. (2018). Heritability enrichment of specifically expressed genes identifies disease‐relevant tissues and cell types. Nature Genetics, 50(4), 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegal, K. M. , Shepherd, J. A. , Looker, A. C. , Graubard, B. I. , Borrud, L. G. , Ogden, C. L. , … Schenker, N. (2009). Comparisons of percentage body fat, body mass index, waist circumference, and waist‐stature ratio in adults. American Journal of Clinical Nutrition, 89(2), 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genton, L. , Hans, D. , Karsegard, V. L. , Kyle, U. G. , Slosman, D. O. , & Pichard, C. (2003). Comparison of dual energy X‐ray absorptiometry (DXA) with bioelectrical impedance analysis (BIA) in obese women. Clinical Nutrition, 22, S1.14558518 [Google Scholar]
- Graff, M. , Ngwa, J. S. , Workalemahu, T. , Homuth, G. , Schipf, S. , Teumer, A. , … Berndt, S. I. (2013). Genome‐wide analysis of BMI in adolescents and young adults reveals additional insight into the effects of genetic loci over the life course. Human Molecular Genetics, 22(17), 3597–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberstick, B. C. , Lessem, J. M. , McQueen, M. B. , Boardman, J. D. , Hopfer, C. J. , Smolen, A. , & Hewitt, J. K. (2010). Stable genes and changing environments: Body mass index across adolescence and young adulthood. Behavior Genetics, 40(4), 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid, I. M. , Jackson, A. U. , Randall, J. C. , Winkler, T. W. , Qi, L. , Steinthorsdottir, V. , … Lindgren, C. M. (2010). Meta‐analysis identifies 13 new loci associated with waist‐hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nature Genetics, 42(11), 949–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinney, A. , Kesselmeier, M. , Jall, S. , Volckmar, A.‐L. , Föcker, M. , Antel, J. , … Hebebrand, J. (2017). Evidence for three genetic loci involved in both anorexia nervosa risk and variation of body mass index. Molecular Psychiatry, 22(2), 192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF‐GC) and OCD Collaborative Genetics Association Studies (OCGAS) . (2018). Revealing the complex genetic architecture of obsessive‐compulsive disorder using meta‐analysis. Molecular Psychiatry, 23(5), 1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karastergiou, K. , & Fried, S. K. (2017). Cellular mechanisms driving sex differences in adipose tissue biology and body shape in humans and mouse models. Advances in Experimental Medicine and Biology, 1043, 29–51. [DOI] [PubMed] [Google Scholar]
- Kelly, T. , Yang, W. , Chen, C.‐S. , Reynolds, K. , & He, J. (2008). Global burden of obesity in 2005 and projections to 2030. International Journal of Obesity, 32(9), 1431–1437. [DOI] [PubMed] [Google Scholar]
- Keshaviah, A. , Edkins, K. , Hastings, E. R. , Krishna, M. , Franko, D. L. , Herzog, D. B. , … Eddy, K. T. (2014). Re‐examining premature mortality in anorexia nervosa: A meta‐analysis redux. Comprehensive Psychiatry, 55(8), 1773–1784. [DOI] [PubMed] [Google Scholar]
- Kilpeläinen, T. O. , Zillikens, M. C. , Stančákova, A. , Finucane, F. M. , Ried, J. S. , Langenberg, C. , … Loos, R. J. F. (2011). Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nature Genetics, 43(8), 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk, K. M. , Martin, F. C. , Mao, A. , Parker, R. , Maguire, S. , Thornton, L. M. , … Martin, N. G. (2017). The anorexia nervosa genetics initiative: Study description and sample characteristics of the Australian and New Zealand arm. The Australian and New Zealand Journal of Psychiatry, 51(6), 583–594. [DOI] [PubMed] [Google Scholar]
- Kyle, U. G. , Bosaeus, I. , De Lorenzo, A. D. , Deurenberg, P. , Elia, M. , Manuel Gómez, J. , … ESPEN . (2004). Bioelectrical impedance analysis‐part II: Utilization in clinical practice. Clinical Nutrition, 23(6), 1430–1145. [DOI] [PubMed] [Google Scholar]
- Lagou, V. , Mägi, R. , & Hottenga, J.‐J. J. (2018). Fasting glucose and insulin variability: Sex‐dimorphic genetic effects and novel loci. In Preparation. [DOI] [PMC free article] [PubMed]
- Lindgren, C. M. , Heid, I. M. , Randall, J. C. , Lamina, C. , Steinthorsdottir, V. , Qi, L. , … Giant Consortium . (2009). Genome‐wide association scan meta‐analysis identifies three loci influencing adiposity and fat distribution. PLoS Genetics, 5(6), e1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link, J. C. , & Reue, K. (2017). Genetic basis for sex differences in obesity and lipid metabolism. Annual Review of Nutrition, 37, 225–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh, P.‐R. , Kichaev, G. , Gazal, S. , Schoech, A. P. , & Price, A. L. (2018). Mixed‐model association for biobank‐scale datasets. Nature Genetics, 50(7), 906–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh, P.‐R. , Tucker, G. , Bulik‐Sullivan, B. K. , Vilhjálmsson, B. J. , Finucane, H. K. , Salem, R. M. , … Price, A. L. (2015). Efficient Bayesian mixed‐model analysis increases association power in large cohorts. Nature Genetics, 47(3), 284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovre, D. , & Mauvais‐Jarvis, F. (2015). Trends in prevalence of the metabolic syndrome. JAMA, 314(9), 950. [DOI] [PubMed] [Google Scholar]
- Lu, Y. , Day, F. R. , Gustafsson, S. , Buchkovich, M. L. , Na, J. , Bataille, V. , … Loos, R. J. F. (2016). New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nature Communications, 7, 10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur, J. , Bowler, E. , Cerezo, M. , Gil, L. , Hall, P. , Hastings, E. , … Parkinson, H. (2017). The new NHGRI‐EBI catalog of published genome‐wide association studies (GWAS catalog). Nucleic Acids Research, 45(D1), D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium , Ripke, S. , Wray, N. R. , Lewis, C. M. , Hamilton, S. P. , Weissman, M. M. , … Sullivan, P. F. (2013). A mega‐analysis of genome‐wide association studies for major depressive disorder. Molecular Psychiatry, 18(4), 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, A. K. , Hivert, M.‐F. , Scott, R. A. , Grimsby, J. L. , Bouatia‐Naji, N. , Chen, H. , … Langenberg, C. (2012). A genome‐wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nature Genetics, 44(6), 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvais‐Jarvis, F. (2015). Sex differences in metabolic homeostasis, diabetes, and obesity. Biology of Sex Differences, 6, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoccoli, G. (2016). Body composition: Where and when. European Journal of Radiology, 85(8), 1456–1460. [DOI] [PubMed] [Google Scholar]
- McCarthy, S. , Das, S. , Kretzschmar, W. , Delaneau, O. , Wood, A. R. , Teumer, A. , … Haplotype Reference Consortium . (2016). A reference panel of 64,976 haplotypes for genotype imputation. Nature Genetics, 48(10), 1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina‐Gomez, C. , Kemp, J. P. , Dimou, N. L. , Kreiner, E. , Chesi, A. , Zemel, B. S. , … Rivadeneira, F. (2017). Bivariate genome‐wide association meta‐analysis of pediatric musculoskeletal traits reveals pleiotropic effects at the SREBF1/TOM1L2 locus. Nature Communications, 8(1), 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micali, N. , Hagberg, K. W. , Petersen, I. , & Treasure, J. L. (2013). The incidence of eating disorders in the UK in 2000‐2009: Findings from the general practice research database. BMJ Open, 3(5), e002646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, J. M. , Golden, N. H. , Peebles, R. , Long, J. , Murray, S. B. , Leonard, M. B. , & Carlson, J. L. (2017). Assessment of sex differences in body composition among adolescents with anorexia nervosa. Journal of Adolescent Health, 60(4), 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay, A. , Beauchamp, J. P. , Fontana, M. A. , Lee, J. J. , Pers, T. H. , Rietveld, C. A. , … Benjamin, D. J. (2016). Genome‐wide association study identifies 74 loci associated with educational attainment. Nature, 533(7604), 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus, T. , Wong, A. P.‐Y. , Syme, C. , & Pausova, Z. (2017). Sex differences in the adolescent brain and body: Findings from the saguenay youth study. Journal of Neuroscience Research, 95(1–2), 362–370. [DOI] [PubMed] [Google Scholar]
- Pulit, S. L. , Karaderi, T. , & Lindgren, C. M. (2017). Sexual dimorphisms in genetic loci linked to body fat distribution. Bioscience Reports, 37(1), BSR20160184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit, S. L. , Stoneman, C. , Morris, A. P. , Wood, A. R. , Glastonbury, C. A. , Tyrrell, J. , … Lindgren, C. M. (2018). Meta‐analysis of genome‐wide association studies for body fat distribution in 694,649 individuals of European ancestry. Human Molecular Genetics. Advance online publication. 10.1093/hmg/ddy327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves, K. L. , Coleman, J. R. I. , Rayner, C. , Hettema, J. M. , Deckert, J. , McIntosh, A. M. , … Eley, T. C. (2017). The common genetic architecture of anxiety disorders. bioRxiv. 10.1101/203844 [DOI] [Google Scholar]
- Randall, J. C. , Winkler, T. W. , Kutalik, Z. , Berndt, S. I. , Jackson, A. U. , Monda, K. L. , … Heid, I. M. (2013). Sex‐stratified genome‐wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genetics, 9(6), e1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium , Ripke, S. , Neale, B. M. , Corvin, A. , Walters, J. T. R. , Farh, K.‐H. , … O'Donovan, M. C. (2014). Biological insights from 108 schizophrenia‐associated genetic loci. Nature, 511(7510), 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorr, M. , & Miller, K. K. (2017). The endocrine manifestations of anorexia nervosa: Mechanisms and management. Nature Reviews Endocrinology, 13(3), 174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schousboe, K. , Visscher, P. M. , Erbas, B. , Kyvik, K. O. , Hopper, J. L. , Henriksen, J. E. , … Sørensen, T. I. A. (2004). Twin study of genetic and environmental influences on adult body size, shape, and composition. International Journal of Obesity and Related Metabolic Disorders, 28(1), 39–48. [DOI] [PubMed] [Google Scholar]
- Scott, R. A. , Lagou, V. , Welch, R. P. , Wheeler, E. , Montasser, M. E. , Luan, J. ‘a. , … Barroso, I. (2012). Large‐scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nature Genetics, 44(9), 991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff, H. , Reba, L. , Thornton, L. M. , Tozzi, F. , Klump, K. L. , Berrettini, W. H. , … Bulik, C. M. (2006). Features associated with excessive exercise in women with eating disorders. International Journal of Eating Disorders, 39(6), 454–461. [DOI] [PubMed] [Google Scholar]
- Silventoinen, K. , Jelenkovic, A. , Sund, R. , Hur, Y.‐M. , Yokoyama, Y. , Honda, C. , … Kaprio, J. (2016). Genetic and environmental effects on body mass index from infancy to the onset of adulthood: An individual‐based pooled analysis of 45 twin cohorts participating in the COllaborative project of Development of Anthropometrical measures in Twins (CODATwins) study. American Journal of Clinical Nutrition, 104(2), 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silventoinen, K. , Jelenkovic, A. , Sund, R. , Yokoyama, Y. , Hur, Y.‐M. , Cozen, W. , … Kaprio, J. (2017). Differences in genetic and environmental variation in adult BMI by sex, age, time period, and region: An individual‐based pooled analysis of 40 twin cohorts. American Journal of Clinical Nutrition, 106(2), 457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, K. S. , Todorčević, M. , Civelek, M. , El‐Sayed Moustafa, J. S. , Wang, X. , Simon, M. M. , … McCarthy, M. I. (2018). Regulatory variants at KLF14 influence type 2 diabetes risk via a female‐specific effect on adipocyte size and body composition. Nature Genetics, 50(4), 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhausen, H.‐C. , & Jensen, C. M. (2015). Time trends in lifetime incidence rates of first‐time diagnosed anorexia nervosa and bulimia nervosa across 16 years in a Danish nationwide psychiatric registry study. International Journal of Eating Disorders, 48(7), 845–850. [DOI] [PubMed] [Google Scholar]
- Sudlow, C. , Gallacher, J. , Allen, N. , Beral, V. , Burton, P. , Danesh, J. , … Collins, R. (2015). UK Biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Medicine, 12(3), e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanamas, S. K. , Lean, M. E. J. , Combet, E. , Vlassopoulos, A. , Zimmet, P. Z. , & Peeters, A. (2016). Changing guards: Time to move beyond body mass index for population monitoring of excess adiposity. QJM, 109(7), 443–446. [DOI] [PubMed] [Google Scholar]
- Tarnoki, A. D. , Tarnoki, D. L. , Medda, E. , Cotichini, R. , Stazi, M. A. , Fagnani, C. , … Baffy, G. (2014). Bioimpedance analysis of body composition in an international twin cohort. Obesity Research & Clinical Practice, 8(3), e201–e298. [DOI] [PubMed] [Google Scholar]
- Thornton, L. M. , Munn‐Chernoff, M. A. , Baker, J. H. , Juréus, A. , Parker, R. , Henders, A. K. , … Bulik, C. M. (2018). The Anorexia Nervosa Genetics Initiative (ANGI): Overview and methods. Contemporary Clinical Trials, 74, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend, P. (1987). Deprivation. Journal of Social Policy, 16(02), 125. [Google Scholar]
- Warren, H. R. , Evangelou, E. , Cabrera, C. P. , Gao, H. , Ren, M. , Mifsud, B. , … UK Biobank CardioMetabolic Consortium BP working group . (2017). Genome‐wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nature Genetics, 49(3), 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, K. , Taskesen, E. , van Bochoven, A. , & Posthuma, D. (2017). Functional mapping and annotation of genetic associations with FUMA. Nature Communications, 8(1), 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, H. J. , Yilmaz, Z. , Thornton, L. M. , Hübel, C. , Coleman, J. R. I. , Gaspar, H. A. , … Bulik, C. M. (2018). Anorexia nervosa genome‐wide association study identifies eight loci and implicates metabo‐psychiatric origins. Manuscript submitted for publication. [DOI] [PMC free article] [PubMed]
- Westmoreland, P. , Krantz, M. J. , & Mehler, P. S. (2016). Medical complications of anorexia nervosa and bulimia. American Journal of Medicine, 129(1), 30–37. [DOI] [PubMed] [Google Scholar]
- Willer, C. J. , Li, Y. , & Abecasis, G. R. (2010). METAL: Fast and efficient meta‐analysis of genomewide association scans. Bioinformatics, 26(17), 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer, C. J. , Speliotes, E. K. , Loos, R. J. F. , Li, S. , Lindgren, C. M. , Heid, I. M. , … Genetic Investigation of ANthropometric Traits Consortium . (2009). Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nature Genetics, 41(1), 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, T. W. , Justice, A. E. , Cupples, L. A. , Kronenberg, F. , Kutalik, Z. , Heid, I. M. , & GIANT consortium . (2017). Approaches to detect genetic effects that differ between two strata in genome‐wide meta‐analyses: Recommendations based on a systematic evaluation. PLoS One, 12(7), e0181038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . (1992). ICD‐10: International statistical classification of diseases and related health problems: 10th revision. Geneva, Switzerland: World Health Organization. [Google Scholar]
- Zillikens, M. C. , Demissie, S. , Hsu, Y.‐H. , Yerges‐Armstrong, L. M. , Chou, W.‐C. , Stolk, L. , … Kiel, D. P. (2017). Large meta‐analysis of genome‐wide association studies identifies five loci for lean body mass. Nature Communications, 8(1), 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Twin‐based heritabilities (twin‐h 2) of body mass index (BMI) across the lifespan as calculated by ACE models from Silventoinen et al. Blue values represent males and red females with error bars depicting 95% confidence intervals. After the age of 19, heritabilities are represented for a whole decade (Silventoinen et al., 2016, 2017)
Figure S2. Analysis workflow chart. AN = anorexia nervosa; BMI = body mass index; GWAS = genome‐wide association study; LDSC = linkage score disequilibrium regression; MAGIC = Meta‐Analyses of Glucose and Insulin‐related traits Consortium; MDD = major depressive disorder; OCD = obsessive–compulsive disorder; PGC = Psychiatric Genomics Consortium; SSGAC = Social Science Genetic Association Consortium; UKB = UK Biobank
Figure S3 (a) QQ plot for the body fat percentage (BF%) genome‐wide‐association study (GWAS). (b) QQ plot for the fat‐free mass (FFM) GWAS
Figure S4 (a) Manhattan plot of the meta‐analyzed genome‐wide association study (GWAS) of body fat percentage (BF%). The red line represents the genome‐wide significance threshold of 5 × 10−8. Chr = chromosome. (b) Manhattan plot of the meta‐analyzed genome‐wide association study (GWAS) of fat‐free mass (FFM). The red line represents the genome‐wide significance threshold of 5 × 10−8. Chr = chromosome
Figure S5 (a) Partitioned heritability by 10 cell type groups for body fat percentage in females. The black dashed lines at −log10(P) = 2.3 is the cutoff for Bonferroni significance. CNS = central nervous system SNP = single nucleotide polymorphism. (b) Partitioned heritability by 10 cell type groups for body fat percentage in males. The black dashed lines at −log10(P) = 2.3 is the cutoff for Bonferroni significance. CNS = central nervous system, SNP = single nucleotide polymorphism. (c) Partitioned heritability by 10 cell type groups for body fat percentage in the meta‐analyzed GWAS. The black dashed lines at −log10(P) = 2.3 is the cutoff for Bonferroni significance. CNS = central nervous system; SNP = single nucleotide polymorphism
Figure S6 (a) Partitioned heritability by 10 cell type groups for fat‐free mass (FFM) in females. The black dashed lines at −log10(P) = 2.3 is the cutoff for Bonferroni significance. CNS = central nervous system, SNP = single nucleotide polymorphism. (b) Partitioned heritability by 10 cell type groups for FFM in males. The black dashed lines at −log10(P) = 2.3 is the cutoff for Bonferroni significance. CNS = central nervous system, SNP = single nucleotide polymorphism. (c) Partitioned heritability by 10 cell type groups for fat‐free mass (FFM) in the meta‐analyzed GWAS. The black dashed lines at −log10(P) = 2.3 is the cutoff for Bonferroni significance. CNS = central nervous system; SNP = single nucleotide polymorphism
Table S1 Twin‐based (twin‐h 2) and single nucleotide polymorphism‐based heritability (SNP‐h 2) estimates derived from genome‐wide association studies (GWAS) for anorexia nervosa (AN) and anthropometric traits measured by bioelectrical impedance analysis of fat‐free mass (FFM), and body fat percentage (BF%)
Table S2. Sex data for the anorexia nervosa (AN) genome‐wide association study (GWAS) datasets
Table S3. Exclusion criteria by International Statistical Classification of Diseases (ICD‐10), British National Formulary (BNF), and UK Biobank variable
Table S4. Inflation statistics and heritability estimates on the observed scale of the genome‐wide association studies (GWAS). Attenuation ratio as calculated by Loh et al. (2017)
Table S5 (a) Genome‐wide significant loci of the body fat percentage genome‐wide association study (GWAS) including heterogeneity measures of the meta‐analysis and a z‐test to test for significant differences between the sexes. Sex‐specific loci are defined as reaching genome‐wide significance (5 × 10−8) in either females or males, not reaching suggestive significance in the opposite sex (5 × 10−6) and the differences in beta estimates is significant after Bonferroni correction for the total number of significant genomic loci. (b) Genomic loci associated with anthropometric traits in published genome‐wide association studies (GWAS). Previous associations have been retrieved from the GWAS catalog (https://www.ebi.ac.uk/gwas/). Data extracted from Functional Mapping and Annotation of Genome‐Wide Association Studies (FUMA, http://fuma.ctglab.nl/)
Table S6. (a) Genome‐wide significant loci of the fat‐free mass genome‐wide association study (GWAS) including heterogeneity measures of the meta‐analysis and a z‐test to test for significant differences between the sexes. Sex‐specific loci are defined as reaching genome‐wide significance (5 × 10−8) in either females or males, not reaching suggestive significance in the opposite sex (5 × 10−6) and the differences in beta estimates is significant after Bonferroni correction for the total number of significant genomic loci. (b) Genomic loci associated with anthropometric traits in published genome‐wide association studies (GWAS). Previous associations have been retrieved from the GWAS catalog (https://www.ebi.ac.uk/gwas/). Data extracted from Functional Mapping and Annotation of Genome‐Wide Association Studies (FUMA, http://fuma.ctglab.nl/)
Table S7. Heritability as estimated by BOLT‐LMM, v2.3.2, on genotyped single nucleotide polymorphisms (SNPs) on anthropometric traits in the UK Biobank. Variance explained by BOLT‐LMM's linear predictor—using the default mixture‐of‐Gaussians prior on SNP effect sizes, which accounts for larger‐effect SNPs—and variance theoretically explained by an optimal linear predictor, that is, SNP heritability (h 2g). Additionally, linkage disequilibrium score regression (LDSC) estimates are presented and the difference between BOLT‐LMM and LDSC estimates
Table S8. Genetic correlations for sex‐combined anorexia nervosa genome‐wide association study and genetic correlations across body composition, physical activity, and psychiatric and behavioral traits. (a) Genetic correlations for female‐only anorexia nervosa genome‐wide association study and genetic correlations across body composition, physical activity, and psychiatric and behavioral traits
Appendix S1 Supporting Information
Appendix S2 Supporting Information
Appendix S3 Supporting Information


