Abstract
Human perivascular progenitor cells, including pericytes, are well-described multipotent mesenchymal cells giving rise to mesenchymal stem cells in culture. Despite the unique location of pericytes, specific antigens to distinguish human pericytes from other cell types are few. Here, we employed a human tissue microarray (Human Protein Atlas) to identify proteins that are strongly and specifically expressed in a pericytic location within human adipose tissue. Next, these results were cross-referenced with RNA sequencing data from human adipose tissue pericytes, as defined as a fluorescence activated cell sorting (FACS) purified CD146+CD34−CD31−CD45− cell population. Results showed that from 105,532 core biopsies of soft tissue, 229 proteins showed strong and specific perivascular immunoreactivity, the majority of which (155) were present in the tunica intima. Next, cross-referencing with the transcriptome of FACS-derived CD146+ pericytes yielded 25 consistently expressed genes/proteins, including 18 novel antigens. A majority of these transcripts showed maintained expression after culture propagation (56% of genes). Interestingly, many novel antigens within pericytes are regulators of osteogenic differentiation. In sum, our study demonstrates the existence of novel pericyte markers, some of which are conserved in culture that may be useful for future efforts to typify, isolate, and characterize human pericytes.
Keywords: pericyte, mesenchymal stem cell, pericyte markers
Introduction
Pericytes are mesenchymal progenitor cells that express mesenchymal stem cell (MSC) surface markers in situ and in culture, and they display multipotentiality toward osteogenic, adipogenic, and chondrogenic cell lineages. Pericytes are ubiquitously present around arterioles, capillaries, and venules in human organs [1–3]. Current data support the notion that pericytes have organ-specific functions in tissue repair while maintaining progenitor cell properties [4]. Traditional pericyte markers include NG2 [1], CD146 [5], platelet-derived growth factor receptor-beta (PDGFRβ) [5], and α-smooth muscle actin (αSMA) [6], with absence of endothelial cell (EC) specific antigens such as CD31, CD34, or von Willebrand factor (vWF). However, there exists a paucity of specific markers for the identification of human pericytes.
Here, we used the Human Protein Atlas (HPA) to help define such markers by identifying proteins expressed in a pericytic location within human adipose tissue. The HPA is a large tissue microarray (TMA) dataset used to provide protein expression information at the tissue, cellular, and, occasionally, subcellular level [7]. The HPA was initiated in 2003 and a first version of the public database www.proteinatlas.org was launched in 2005 [8], which contains >25,000 antibodies that have passed rigorous quality tests for antigen specificity and validation, leading to a collection of more than 10 million immunohistochemistry (IHC) images and 82,000 high-resolution immunofluorescence images. Within the HPA, the HPA subcellular classification (SubC) software package consists of a suite of tools that allows users to download all images of a particular organ, and then rapidly review them sequentially for staining patterns of interest. HPASubC uses a Playstation-style gamepad controller to rapidly evaluate images at a user-dependent rate of up to ∼1 image/s [9]. The HPASubC software has been used successfully to identify EC and/or smooth muscle cells (SMCs), specific proteins discovered within heart TMA images in the HPA [9]. This software was used in our study to reveal defining patterns of protein expression in adipose tissue-derived pericytes.
To identify novel antigen expression within pericytes, candidate surface markers of pericytes were identified via HPASubC analysis. Therefore, cell protein surface markers found from TMA were cross-referenced with RNA-Seq data from freshly sorted or cultured pericytes to validate protein expression. Pericytes from different tissue origins might express differential surface markers. Thus, these proteins were further investigated by TMA for expression in tissues other than adipose tissue. Our study demonstrates the existence of novel pericyte markers, some of which are conserved in culture that may be useful for future efforts to typify or isolate human pericytes.
Materials and Methods
HPASubC software
The HPASubC software has been validated, published [9,10], and can be downloaded from https://github.com/cornish/HPASubC. The HPASubC software consists of a suite of tools implemented as Python scripts. The scripts target Python 2.7 and depend on the standard library and several third-party modules. Mechanize-0.2.5 (https://pypi.python.org/pypi/mechanize) is used for a stateful interaction with the HPA website. Pygame (www.pygame.org) is used to manage image display and game controller input. RESTful API is used to parse HTML pages retrieved from the HPA website. Pyexiv2 (http://tilloy. net/dev/pyexiv2) is used to read and write the Exif metadata in the downloaded images. HPASubC uses the standard Exif UserComment tag to store JSON-encoded metadata, such as Ensembl gene (ENSG ID), tissue type, and antibody, directly in the downloaded HPA image files.
General HPASubC usage
A list of all proteins evaluated at HPA is first accessed by using normal_tissue.csv.zip, available at www.proteinatlas.org/about/download Next, a list of unique ENSGs IDs is selected and used with the download_images_from_gene_list.py script to download images from adult human normal tissues and generate a table of image identifiers. The download script writes the ENSG tissue and antibody data to the JPEG's Exif UserComment tag to maintain the context of the downloaded images. A Sony PlayStation style USB controller (Logitech Precision) is then used with the image_ viewer.py script to scroll through the images. Images are zoomed in on as needed, and images with the appropriate staining patterns are selected. The selected images are then scored with the image_scorer.py script by using score values of 0–3 that can be based on user parameters such as stain specificity. Additional HPA metadata such as protein names, expression across different tissues, and Entrez data about a protein of interest can be obtained by using the download_protein_data_from_gene_list.py script. Detailed instructions for running the scripts are included in the package's README.TXT file [9].
HPASubC usage to identify novel markers of pericytes
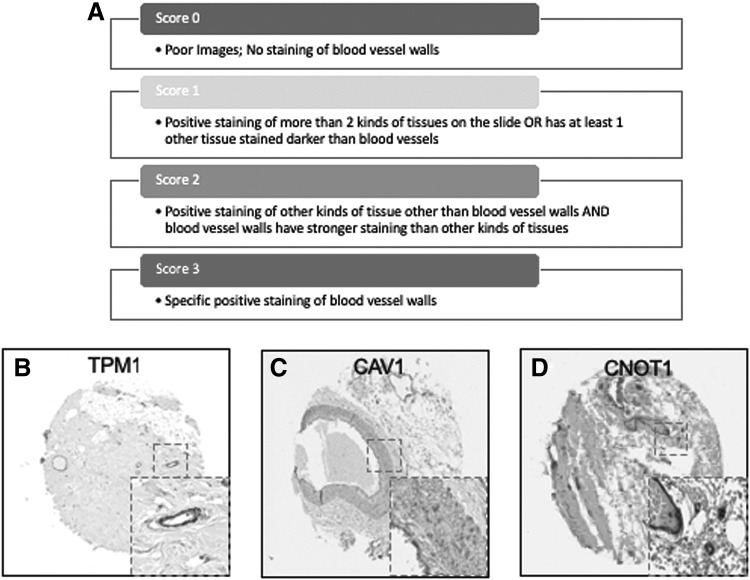
For identification of novel pericyte markers, all images with weak, moderate, or strong staining of blood vessels and moderate nonspecific staining of other tissues were excluded. Conversely, samples with strong, nonspecific staining were included. Images were scored on a scale of 0–3 based on the following parameters: 3: Specific positive staining of blood vessels; 2: Positive staining of tissues other than blood vessels AND blood vessels have stronger staining than other kinds of tissues; 1: Positive staining of more than two kinds of tissues on the slide OR has at least one other tissue stained more intensely than blood vessels; 0: Poor images; very little staining of blood vessels. However, it is challenging to distinguish whether pericytes or ECs are stained from the IHC images on the TMA. In this study, proteins that were repeat scored 3 were cross-referenced with fresh and cultured pericyte RNA-seq datasets.
Isolation of human pericytes from adipose tissue
Human adipose lipoaspirate specimens used for fluorescence activated cell sorting (FACS), culture, and RNA-seq analysis were obtained from six healthy female donors (age range: 35–63 years; body mass index range: 21.1–32.0) undergoing elective liposuction from lateral axillary areas at local plastic surgery clinics under IRB approval. Tissue dissociation and pericyte isolation were performed as previously described [11]. The cells were collected by centrifugation and incubated in Aldefluor assay buffer containing fluorescently labeled, mouse antihuman monoclonal antibodies (PE- or APC-Cy7-conjugated CD45, and PE- or PerCP-Cy5.5-conjugated CD146; BD BioSciences, San Jose, CA) for 20 min on ice to allow immunophenotypic staining for FACS. Cells collected by FACS were either lysed immediately for RNA extraction or seeded for culture expansion. To adjust for gene expression changes brought about in culture, after three passages, cultured pericytes were stained with CD31, CD45, and CD146 fluorescent antibodies and sorted by FACS a second time. Pericytes that had retained CD146 expression in culture were isolated for RNA-seq analysis.
RNA extraction and sequencing
Subpopulations of freshly isolated pericytes and their cultured counterparts were sorted into a cell lysis buffer solution, and RNA was extracted by using the Qiagen RNeasy Micro kit (Qiagen). Total RNA was quantified and measured for RNA integrity numbers of 7.7 or greater (2100 Agilent Bioanalyzer). For each subpopulation, complementary DNA (cDNA) was then prepared by using the NuGen's Ovation RNA-Seq System V2 kit from ∼100 ng of RNA pooled from two or three donors and used as input for Ovation's Ultralow DR Multiplex System 1–8 (Nugen, San Carlos, CA). High-throughput sequencing was conducted on an Illumina Hiseq 2000 platform to generate 100-bp paired-end reads at a depth of coverage of ∼79 million reads. Cluster formation achieved ∼86%, whereas the quality of reads exceeded 95%. Fragment sequence identification and alignment were performed by using TopHat and Bowtie, respectively. Samples were normalized by RPKM (reads per kilobase of exon per million mapped reads), which takes into account both library size and gene length in within-sample comparisons. Cross-library, individual gene comparisons were made possible by using a similar number of reads between libraries (ie, depths of coverage).
Results
Identification of membrane proteins specific to and consistently expressed on blood vessels
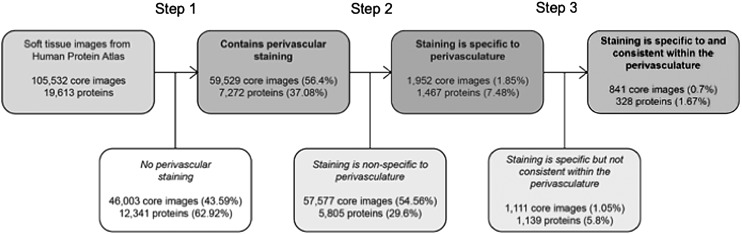
To identify new perivascular antigens, we analyzed proteins from HPA adult human normal soft tissue images. A total of 105,532 soft tissue core images with 19,613 proteins were obtained from the HPA to be analyzed (Fig. 1). In step 1, we parsed out the ENSG IDs, and we obtained 59,529 images (56.4% of total soft tissue images), which contained 7,272 proteins (37.08% of total proteins) with perivascular immunoreactivity. In step 2, images containing proteins with a high specificity for perivascular expression (qualitative score of 3) were selected for further analysis (Fig. 2A). This yielded 1,952 images (1.85% of total soft tissue images) with 1,467 different proteins (7.48% of total proteins) that were highly specific for blood vessels.
FIG. 1.
Schematic of the selection process of HPA images. All HPA records are obtained and used to identify Ensemble gene IDs of immunostained proteins. HPA “Soft tissue” images are parsed out to capture and download TMA images. Images were selected and scored by specificity and consistency to identify novel markers for pericyte cell sorting. In step 1, images were divided into those with or without clear perivascular staining. In step 2, those remaining images were divided into those in which immunostaining was either specific or nonspecific to the perivasculature. In step 3, those remaining images were divided based on whether or not perivascular immunostaining was consistent across images. In the end, 328 proteins were found to be both specific and consistent within the perivascular space within the normal adult human “soft tissue” TMA dataset. HPA, Human Protein Atlas; TMA, tissue microarray.
FIG. 2.
Scoring system of TMA imaging selection, and examples of specificity staining Score 3. Images with blood vessels were scored on a scale of 0–3 based on the following parameters: 3: specific positive staining of blood vessel walls; 2: positive staining of other kinds of tissue other than blood vessels AND blood vessel walls have stronger staining than other kinds of tissues; 1: positive staining of more than two kinds of tissues on the slide OR has at least one other tissue stained darker than blood vessel walls; 0: poor images; very little staining of blood vessel walls (A). Examples of specific and consistent staining within blood vessel walls (B–D). Magnified of the positive staining image was shown in gray dotted squares on lower right of each image.
In the TMA database, each protein of interest was probed by two or more antibodies. In some situations, one antibody might be more or less specific than the others. Thus, these images were further investigated to determine whether staining with different antibodies produced staining of the 1,467 proteins in the same perivascular location. Proteins with only 1 image with a score of 3 were excluded in Step 3, leaving 841 images (0.07%) of 328 proteins (1.67%) that were consistently found in the perivasculature (examples shown in Fig. 2B–D).
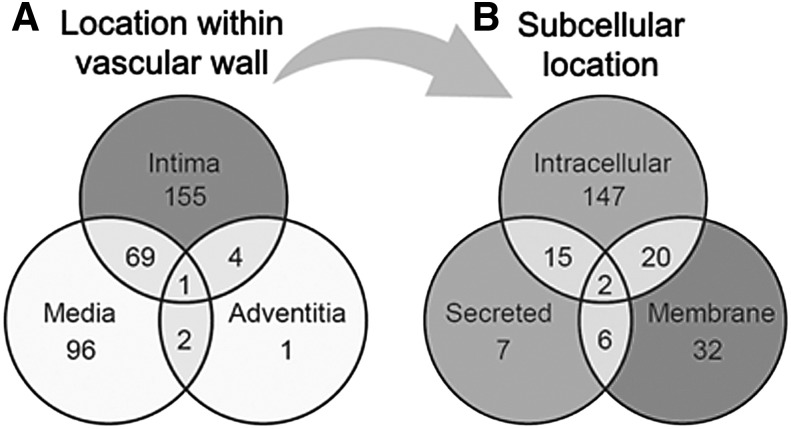
Pericytes reside within the tunica intima in a subendothelial location. Next, these 841 images of 328 proteins were analyzed to categorize their location within either the (1) tunica intima, (2) media, or (3) adventitia. Of the 328 remaining proteins, 155 are specific to the intima (eg, ENG), one (CDCP1) is specific to the adventitia, and 92 are specific to the media (eg, VWA9). Four were shared between the intima and adventitia, two (RDH16 and TNR) were shared between the adventitia and the media, 69 were shared between the intima and the media (eg, TPM1), and one (CAAP1) was shared among all three locations (Fig. 3A and Supplementary Table S1). Ninety-five media-specific proteins were excluded from further analysis, as medial SMCs are not considered to represent a multipotent cell type. Twenty-seven proteins were additionally excluded due to inconsistencies related to blood vessel staining pattern between samples (eg, DES). This left 229 proteins (Supplementary Table S2), which were consistently and specifically expressed within the tunica intima.
FIG. 3.
Histologic and subcellular location of perivascular antigens. (Left) Venn diagram to demonstrate the location of n = 328 perivascular antigens within different layers of the vessel wall, including the tunica intima, media, and adventitia. The majority of antigens were found within the tunica intima only (n = 155) (A). Next most common, n = 96 antigens were specific to the tunica media. n = 69 antigens showed shared immunostaining between both the tunica intima and the media. (Right) All antigens expressed in the tunica intima (n = 229) were next segregated by known subcellular location. The majority of proteins were restricted to the intracellular compartment (n = 147) (B) In total, n = 60 proteins with known membranous location were identified, including n = 32 proteins with expression restricted to the membrane. These 60 membranous proteins within the tunica intima were next used for cross-referencing with the transcriptome of human pericytes.
Among these 229 proteins (Supplementary Table S2), well-known pericyte markers ENG, Myl9 [12] have been reported to be associated with pericytes. Moreover, ABI3 [13], BECN1 [14], and ADPIOQ [15] have been reported to be highly expressed within pericytes. Endothelial markers, such as vWF [16], were included in this list, as it is difficult to distinguish from the TMA images whether ECs or perictytes are stained. In this study, these 229 proteins were further categorized by their subcellular location due to the nature of instability of secreted proteins and the inability to locate intracellular proteins.
The 229 proteins were then categorized by location within the cell to identify membrane proteins to serve as potential pericyte markers for FACS. One hundred and forty-seven proteins were intracellular only (eg, MYL3), 32 were restricted to the membrane (eg, GFRA3), and 7 were secreted proteins (such as COL15A1). Twenty proteins were both intracellular and membrane proteins (such as VWDE), 6 were both membrane and secreted proteins, 15 were both intracellular and secreted proteins, and 2 were present in all three locations (eg, C12orf76) (Fig. 3B and Supplementary Table S2). In sum, 60 out of the 229 proteins found to be specific to the tunica intima were also membrane-bound proteins, serving as possible novel surface marker proteins for pericyte isolation (shown in Table 1).
Table 1.
Sixty Membrane Proteins Specifically Expressed in the Tunica Intima of Vessel Wall in Normal Adult Human Adipose Tissue
| ABC7-42404400C24.1 | GFRA3 | RPRML |
| ABCB10 | GJD2 | SHISA7 |
| ABCB7 | GPR4 | SLC22A13 |
| ACKR1 | HLA-DRA | SLC22A15 |
| ADTRP | HTR5A | SLC25A48 |
| AKAP1 | IGSF23 | SLC30A7 |
| ALG12 | INTS1 | SPG11 |
| ATP2B4 | ITGA2 | ST3GAL5 |
| ATP2C2 | KCNH5 | ST3GAL6 |
| C12orf76 | KCNK18 | TBC1D2B |
| C1orf27 | LRRC4B | TIMM23 |
| CAV1 | LRRC8C | TIMMDC1 |
| CAV2 | LRRN4CL | TMC7 |
| CD93 | MMP16 | TMEM135 |
| CDCP1 | OTOP2 | TMEM70 |
| CNOT1 | PLA2G15 | TMPRSS9 |
| CYP2U1 | PSMD1 | TPM1 |
| ELOVL7 | RANBP17 | VSTM4 |
| ENG | RDH16 | WNT10A |
| FAM171A1 | RNF112 | XCR1 |
Confirmation of protein expression within an RNA-Seq data base of human CD146+ pericytes
To confirm whether these 60 membrane proteins expressed within the tunica intima were significantly expressed in purified human adult adipose-derived pericytes, a cross-comparison was next performed by using RNA Sequencing data from freshly sorted and cultured human adipose-derived pericytes [17] (Fig. 4). Adipose-derived pericytes were isolated by incubation in Aldefluor assay buffer containing fluorescently labeled, mouse antihuman monoclonal antibodies to CD146, as well as negative selection for CD31 and CD45.
FIG. 4.
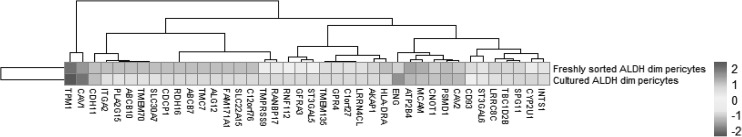
Heatmap of gene expression in adipose tissue-derived fresh sorted and cultured pericytes. CD146+Aldhdim human pericytes were derived from n = 6 lipoaspirate samples from healthy female donors. Total RNA-Seq was performed on purified human pericytes, either immediately after FACS (freshly sorted) or after culture propagation (cultured). Samples were normalized by RPKM. The culture time point (y-axis) and level of gene expression (x-axis) is color-coded from dark gray to light gray. CD146 (MCAM) expression is included as a positive control. FACS, fluorescence activated cell sorting; RPKM, reads per kilobase of exon per million mapped reads.
RNA-Seq data showed that 38 out of the 60 TMA-identified proteins were expressed in adipose-derived human pericytes (Tables 2 and 3). As a positive control, the pericyte marker CD146 (MCAM) used to isolate pericytes was also examined, which showed consistent expression among purified cells, both freshly after sorting and after culture expansion. Eight out of 38 have only weak expression in pericytes (RPKM = 1 freshly sorted and after culture). Five out of 38 had low expression (RPKM = 1) in freshly sorted pericytes, but increased expression after culture. Fourteen out of the 25 membrane proteins were consistently expressed both on freshly sorted pericytes and after culture. Several of these transcripts increased in expression after culture, such as TPM, CNOT1, CYP2U1, and INTS1 (Table 3). Many transcripts were detectable at lower levels after culture, including CAV1, ENG, ATP2B4, CAV2, PSMD1, SPG11, TBC1D2B, LRRC8C, ST3GAL6, and CD93. The remaining 11 genes were expressed on the membranes of freshly sorted pericytes, but they were lost after culture (HLA-DRA, AKAP1, TMEM135, GPR4, C1orf27, LRRN4CL, GFRA3, ST3GAL5, RNF112, TMPRSS9, and RANBP17) (Table 3).
Table 2.
Perivascular RNA Transcripts Expressed Among Both Freshly Sorted and Culture Expanded CD146+ Human Pericytes
| Gene | Freshly sorted pericytes (RPKM) | Cultured pericytes (RPKM) |
|---|---|---|
| TPM1 | 5654.46 | 21302.99 |
| CAV1 | 1406.75 | 5801.03 |
| ENG | 231.67 | 2120.38 |
| ATP2B4 | 1027.87 | 499.82 |
| CAV2 | 329.2 | 404.2 |
| PSMD1 | 432 | 396.5 |
| CNOT1 | 268.67 | 296.11 |
| SPG11 | 203.67 | 164 |
| CYP2U1 | 43.5 | 156.75 |
| TBC1D2B | 188 | 110.5 |
| INTS1 | 71 | 94 |
| LRRC8C | 233 | 79.6 |
| ST3GAL6 | 76 | 40 |
| CD93 | 126 | 11 |
Of the 25 genes/proteins expressed present in human pericytes by both TMA and RNA-Seq, 14 genes were expressed among both freshly sorted and cultured pericytes. Data were normalized by per million mapped reads (RPKM) dataset. Corresponding heatmap is shown in Fig. 4.
RPKM, reads per kilobase of exon per million mapped reads; TMA, tissue microarray.
Table 3.
Perivascular RNA Transcripts Expressed Among Freshly Sorted but Not Culture Expanded CD146+ Human Pericytes
| Gene | Freshly sorted pericytes (RPKM) | Cultured pericytes (RPKM) |
|---|---|---|
| HLA-DRA | 401 | 1 |
| AKAP1 | 211.25 | 1 |
| CD93 | 126 | 11 |
| TMEM135 | 115 | 1 |
| GPR4 | 104 | 1 |
| C1orf27 | 95.5 | 1 |
| LRRN4CL | 95 | 1 |
| GFRA3 | 45 | 1 |
| ST3GAL5 | 30 | 1 |
| RNF112 | 17 | 1 |
| TMPRSS9 | 8 | 1 |
| RANBP17 | 5 | 1 |
| SLC22A15 | 1 | 1 |
| C12orf76 | 1 | 1 |
| FAM171A1 | 1 | 1 |
| ALG12 | 1 | 1 |
| TMC7 | 1 | 1 |
| ABCB7 | 1 | 1 |
| RDH16 | 1 | 1 |
| CDCP1 | 1 | 1 |
Of the 25 genes/proteins expressed present in human pericytes by both TMA and RNA-Seq, 11 genes were expressed among freshly sorted pericytes, but they were lost after culture propagation. Data were normalized by per Million mapped reads (RPKM) dataset. Corresponding heatmap is shown in Fig. 4.
Confirmation of novel pericyte antigen expression in other tissues
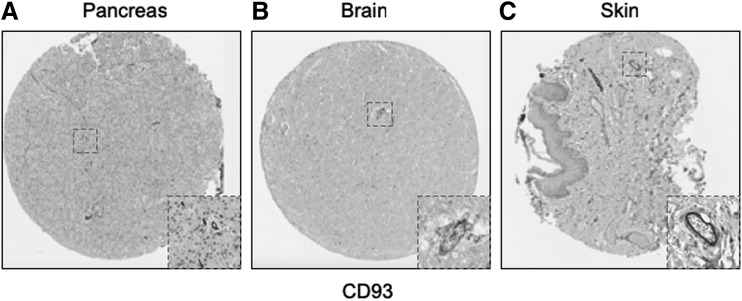
Emerging concepts suggest that pericytes may have tissue-specific properties [18–20], including tissue-specific antigen expression [21]. For this reason, we next examined immunolocalization of these 25 novel “pericyte” antigens in all other tissues available within the HPA (including 11 additional tissue depots from visceral and mesenchymal locations). Each image was again scored based on perivascular specificity (see again Fig. 2). A wide range of perivascular specificity was observed (Table 4), and most antigens showed significant variation across tissues in terms of relative specificity for blood vessels. Several markers were highly specific for a pericytic location across tissues, including, for example, CD93 (Fig. 5A–C, mean specificity score: 2.9) and ST3GAL5 (mean score: 2.58).
Table 4.
Twenty-Five Pericyte Antigens and Their Expression Across Diverse Human Tissues Within the Human Protein Atlas
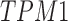 |
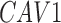 |
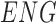 |
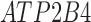 |
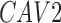 |
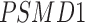 |
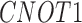 |
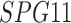 |
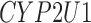 |
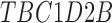 |
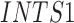 |
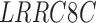 |
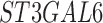 |
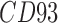 |
 |
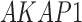 |
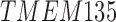 |
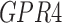 |
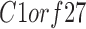 |
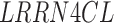 |
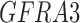 |
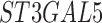 |
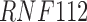 |
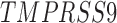 |
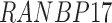 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Brain | 3 | 3 | 3 | 2 | 3 | 0 | 0 | 1 | 0 | 2 | 2 | 2 | 0 | 3 | 3 | 1 | 0 | 2 | 3 | 1 | 2 | 3 | 0 | 0 | 2 |
| Endocrine tissues | 2 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 3 | 3 | 0 | 0 | 1 | 2 | 2 | 1 | 0 | 0 | 3 | 0 |
| Skin | 2 | 1 | 3 | 1 | 2 | 0 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 3 | 3 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 1 | 3 |
| Muscle tissue | 0 | 2 | 0 | 2 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 3 | 3 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 3 | 1 |
| Lung | 3 | 1 | 1 | 1 | 0 | 0 | 0 | 2 | 2 | 0 | 1 | 0 | 1 | 3 | 2 | 0 | 0 | 2 | 1 | 2 | 2 | 3 | 1 | 2 | 1 |
| Liver and gallbladder | 0 | 2 | 2 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 3 | 3 | 0 | 0 | 2 | 1 | 2 | 2 | 3 | 0 | 0 | 1 |
| Pancreas | 3 | 3 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 1 | 0 | 1 | 0 | 3 | 3 | 0 | 0 | 2 | 0 | 2 | 3 | 3 | 2 | 3 | 2 |
| GI tract | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 0 | 1 | 1 | 1 | 2 | 1 | 3 | 2 | 1 | 2 | 3 | 2 | 2 | 3 | 3 | 1 | 3 | 3 |
| Kidney and urinary bladder | 2 | 2 | 0 | 0 | 2 | 0 | 0 | 2 | 1 | 1 | 1 | 1 | 0 | 3 | 2 | 1 | 1 | 2 | 2 | 2 | 1 | 3 | 0 | 0 | 1 |
| Male tissues | 1 | 1 | 1 | 0 | 2 | 0 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 2 | 2 | 1 | 2 | 2 | 1 | 1 | 0 | 3 | 0 | 3 | 1 |
| Female tissues | 2 | 0 | 3 | 0 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 3 | 3 | 1 | 2 | 2 | 1 | 1 | 2 | 3 | 1 | 3 | 2 |
| Bone marrow and immune system | 2 | 2 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 3 | 2 | 2 | 0 | 2 | 2 | 0 | 1 | 3 | 0 | 3 | 2 |
| Mean score | 1.8 | 1.6 | 1.3 | 0.6 | 1.6 | 0.2 | 0.4 | 1.3 | 0.6 | 1.1 | 0.8 | 1.2 | 0.8 | 2.9 | 2.6 | 0.8 | 0.8 | 1.9 | 1.6 | 1.6 | 1.7 | 2.6 | 0.8 | 2 | 1.9 |
Twenty-five novel “pericyte” antigens in all other tissues available within the HPA (including 11 additional tissue depots from visceral and mesenchymal locations). Each image was again scored based on perivascular specificity (see again Fig. 2).
HPA, Human Protein Atlas.
FIG. 5.
Representative examples of conserved pericytic CD93 immunoreactivity across tissues. Of all identified pericyte antigens, CD93 showed the highest specificity for a pericytic location across human tissues. Examples shown within the HPA TMA, including (A) pancreas, (B) brain, and (C) skin.
Discussion
In summary, using the HPA TMA, 60 novel membrane proteins were found to be consistently and specifically expressed in the tunica intima. Twenty-five of these proteins were confirmed by RNA-Seq data to be expressed in purified human adipose-derived pericytes. Of these, several pericyte antigens were conserved across organ systems. These 25 proteins serve as potential pericyte cell surface markers for future efforts in the FACS isolation of human adipose tissue pericytes. Interestingly, a large minority (7) of these 25 antigens expressed at the gene and protein level in human pericytes have known roles in regulating osteogenic differentiation. Specifically, GPR4, CAV1, TMEM135, ENG, SLC30A7, CDCP1, and ITGA2 have all been reported to have roles in MSC differentiation and osteogenesis [22–33]. Among the seven membrane proteins that have been reported to have functions in MSC differentiation and osteogenesis, ITGA2 and TMEM135 have positive regulatory effects on osteogenic differentiation [21,25,27,34]; whereas ENG, GPR4, CAV1, SLC30A7, and CDCP1 have negative regulatory effects [23,26,28,29,32,33].
As mentioned, and of these new pericyte antigens, a large minority have been previously reported to have functions in the regulation of osteogenic differentiation of cells. For example, both Integrin alpha-2 ITGA2 and TMEM-135 have been reported to positively regulate MSC osteogenic differentiation. The membrane glycoprotein ITGA2 regulates pericyte osteogenic differentiation and osteoblast maturation [25,27,32]. TMEM135 has also been shown to have effects on osteogenesis in bone marrow stromal cells [34]. Conversely, several pericyte antigens negatively regulate osteogenic differentiation. Endoglin (ENG, CD105) is a co-receptor for transforming growth factor beta (TGF-β) and canonical “MSC” marker, which has been previously observed to be expressed on human pericytes across tissues [3,35]. Most studies suggest that the absence of CD105 on mouse and human MSC results in increased osteogenic differentiation [33,36]. G protein-coupled receptor 4 (GPR4) is expressed in multiple organs, including the vasculature [23], and has been observed to inhibit the osteogenesis of bone marrow stem cells [24]. Caveolin-1 (CAV1) is a scaffolding protein of cholesterol-rich caveolae lipid rafts in the plasma membrane [37]. The caveolin-1 null mouse has increased bone mass and formation, and its bone marrow-derived MSCs have enhanced osteogenic potential [32]. SLC30A7(ZnT7) inhibits osteogenesis through Wnt/β-catenin signaling. CUB domain-containing protein 1 (CDCP1) is almost exclusively expressed in a subset of CD34(+) stem/progenitor cells in the bone marrow and is downregulated during MSC differentiation down osteogenic, adipogenic, and chondrogenic cell lineages [38]. Given the high number of negative regulators of ossification endogenously present within human pericytes, it is interesting to speculate whether these have functional relevance during pathophysiologic calcification and ossification of vessel walls.
The remaining 18 proteins have no reported function in osteogenesis on MSCs and can be divided into 5 categories based on current knowledge of their location or function: (1) highly expressed on ECs; (2) predominately expressed in neurons and the brain; (3) function in immune cells; (4) functions in stem cell differentiation, cancer progression, or cell metabolism; and (5) unknown.
Three proteins found in our study are markers that are shared with the endothelium or neurons. Chromosome 1 open reading frame 27 (C1orf27), SLC22A15 (FLIPT1), and CD93 are shared with the endothelium. C1orf27 exhibits cytoplasmic expression in epidermal tissue and plays a role in cell fate specification and development [39]. SLC22A15 (FLIPT1) is expressed mainly in ECs [26] and mediates the uptake of pharmaceutical drugs to different tissues [40]. CD93 is a highly glycosylated transmembrane protein expressed in ECs and stem cells [41], and it has been previously studied in EC adhesion and migration and tumor angiogenesis.
In our study, 13 proteins have been reported to function in cell metabolism (CYP2U1, TBC1D2B, AKAP1, PSMD1, TMEM70, TMPRSS956) [42], tumor progression (CAV-2, TPM1), and stem cell differentiation (ST3GAL5, ST3GAL6 ATP2B4, CNOT1, RANBP17) [22,29,42–50]. The functions of both C12orf76 and GFRA3 are currently unknown. However, the artemin-GFRA3 signaling system has a role in regulating early embryonic development and apoptosis [51]. The diverse roles of pericyte cell surface marker proteins demonstrate the importance of pericytes to several different systems in the body.
Recently, pericytes have emerged as tissue-specific progenitor cells, and traits of pericytes are different depending on their tissue of origin [52]. For example, previous studies have shown that in certain regions of the brain, pericytes are neural crest derivatives [53], whereas pericytes in other organs are mesoderm derivatives [54]. Further, neural versus non-neural pericytes have been suggested to have differences in abilities of differentiation. For example, recent studies identified that human cardiac pericytes express mesenchymal stem/stromal cell markers, including CD44, CD73, CD90, and CD105. These pericytes have the capacity to differentiate into osteo-, chondro-, and adipogenesis, but there is no potential for skeletal myogenesis in vitro [55]. On the other hand, in the central nervous system, pericytes play an essential role in the maturation and maintenance of the blood–brain barrier [56,57]. Our study demonstrates the tissue-intrinsic differences in human pericyte antigen expression. Comparing results from soft tissue and brain tissue in this study, some proteins such as CAV1, TPM1, and ENG (Fig. 4) showed specific staining in both brain and soft tissue. Other protein markers, such as CYP2U1, RNF112, and TMPRSS9, only showed specific staining in soft tissue but not in the brain.
Notably, the transcriptomic analysis of this study demonstrates large changes in pericyte gene expression before and after culture expansion. This is remarkable, since much of what we know about pericytes is based on in vitro culture experiments [58,59]. Gene expression profile changes after culture may be caused by a variety of reasons, including cell culture medium, stiffness and composition of the cell culture substrate, confluency, and cell cycle stage. Further investigation of these surface markers and their functions may aid in profiling the different conditions of stromal cells.
Several limitations to the study just mentioned exist. Some well-studied pericyte markers were not available for review within the currently available TMA, such as RGS5 and TBX18. Along these lines, the quality of each individual IHC antibody is another limitation. In some situations, when more than two antibodies were sometimes used to stain the same protein of interest, clear discrepancies were identified. However, this issue was mitigated by excluding any inconsistent or nonspecific staining. Unfortunately, some common pericyte antigens also were noted to have nonspecific staining within the TMA, including, for example, CD29 and NG2. To further mitigate any spurious results from TMA analysis, our cross-referencing with the transcriptome of FACS-derived pericytes identified a large number of novel antigens expressed on the cell membrane which could be used in the future to isolate or further partition human pericytes.
In sum, combined use of a human TMA dataset with the transcriptome of adipose tissue-derived human pericytes yielded 25 cell surface antigens, including 18 novel antigens expressed within human pericytes. Our study demonstrates the existence of novel pericyte markers, some of which are conserved in culture and that may be useful for future efforts to typify or isolate human pericytes.
Supplementary Material
Acknowledgments
A.W.J. was funded by NIH/NIAMS (R01 AR070773, K08 AR068316), NIH/NIDCR (R21 DE027922), USAMRAA (W81XWH-18-1-0121, W81XWH-18-1-0336, W81XWH-18-1-0613), the American Cancer Society (RSG-18-027-01-CSM), the Orthopedic Research and Education Foundation with funding provided by the Musculoskeletal Transplant Foundation, the Maryland Stem Cell Research Foundation, and the Musculoskeletal Transplant Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health, Department of Defense, or US Army. The authors thank Dr. Marc Halushka for his assistance in HPASubC analysis.
Author Disclosure Statement
None of the authors state that they have conflicts of interest with this article.
Supplementary Material
References
- 1. Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, Andriolo G, Sun B, Zheng B, et al. (2008). A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 3:301–313 [DOI] [PubMed] [Google Scholar]
- 2. Armulik A, Genove G. and Betsholtz C. (2011). Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev Cell 21:193–215 [DOI] [PubMed] [Google Scholar]
- 3. Crisan M, Corselli M, Chen WC. and Peault B. (2012). Perivascular cells for regenerative medicine. J Cell Mol Med 16:2851–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mills SJ,. Cowin AJ. and Kaur P. (2013). Pericytes, mesenchymal stem cells and the wound healing process. Cells 2:621–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zimmerlin L, Donnenberg VS, Rubin JP. and Donnenberg AD. (2013). Mesenchymal markers on human adipose stem/progenitor cells. Cytometry A 83:134–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Psaltis PJ,. Harbuzariu A, Delacroix S, Holroyd EW. and Simari RD. (2011). Resident vascular progenitor cells—diverse origins, phenotype, and function. J Cardiovasc Transl Res 4:161–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thul PJ. and Lindskog C. (2018). The Human Protein Atlas: a spatial map of the human proteome. Protein Sci 27:233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, et al. (2005). A Human Protein Atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics 4:1920–1932 [DOI] [PubMed] [Google Scholar]
- 9. Cornish TC,. Chakravarti A, Kapoor A. and Halushka MK. (2015). HPASubC: a suite of tools for user subclassification of Human Protein Atlas tissue images. J Pathol Inform 6:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anene DF,. Rosenberg AZ, Kleiner DE, Cornish TC. and Halushka MK. (2016). Utilization of HPASubC for the identification of sinusoid-specific proteins in the liver. J Proteome Res 15:1623–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kara M, Axton RA, Jackson M, Ghaffari S, Buerger K, Watt AJ, Taylor AH, Orr B, Hardy WR, Peault B. and Forrester LM. (2015). A role for MOSPD1 in mesenchymal stem cell proliferation and differentiation. Stem Cells 33:3077–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vanlandewijck M, He L, Mae MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Lavina B, et al. (2018). A molecular atlas of cell types and zonation in the brain vasculature. Nature 554:475–480 [DOI] [PubMed] [Google Scholar]
- 13. Giannoni P, Badaut J, Dargazanli C, De Maudave AF, Klement W, Costalat V. and Marchi N. (2018). The pericyte-glia interface at the blood-brain barrier. Clin Sci (Lond) 132:361–374 [DOI] [PubMed] [Google Scholar]
- 14. Nah J, Yoo SM, Jung S, Jeong EI, Park M, Kaang BK. and Jung YK. (2017). Phosphorylated CAV1 activates autophagy through an interaction with BECN1 under oxidative stress. Cell Death Dis 8:e2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Supakul S, Yao K, Ochi H, Shimada T, Hashimoto K, Sunamura S, Mabuchi Y, Tanaka M, Akazawa C, et al. (2019). Pericytes as a source of osteogenic cells in bone fracture healing. Int J Mol Sci 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zanetta L, Marcus SG, Vasile J, Dobryansky M, Cohen H, Eng K, Shamamian P. and Mignatti P. (2000). Expression of Von Willebrand factor, an endothelial cell marker, is up-regulated by angiogenesis factors: a potential method for objective assessment of tumor angiogenesis. Int J Cancer 85:281–288 [DOI] [PubMed] [Google Scholar]
- 17. Hardy WR,. Moldovan NI, Moldovan L, Livak KJ, Datta K, Goswami C, Corselli M, Traktuev DO, Murray IR, Peault B. and March K. (2017). Transcriptional networks in single perivascular cells sorted from human adipose tissue reveal a hierarchy of mesenchymal stem cells. Stem Cells 35:1273–1289 [DOI] [PubMed] [Google Scholar]
- 18. Hindle P, Khan N, Biant L. and Peault B. (2017). The infrapatellar fat pad as a source of perivascular stem cells with increased chondrogenic potential for regenerative medicine. Stem Cells Transl Med 6:77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tavazoie M, Van der Veken L, Silva-Vargas V, Louissaint M, Colonna L, Zaidi B, Garcia-Verdugo JM. and Doetsch F. (2008). A specialized vascular niche for adult neural stem cells. Cell Stem Cell 3:279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi S. and Gronthos S. (2003). Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J Bone Miner Res 18:696–704 [DOI] [PubMed] [Google Scholar]
- 21. Guimaraes-Camboa N, Cattaneo P, Sun Y, Moore-Morris T, Gu Y, Dalton ND, Rockenstein E, Masliah E, Peterson KL, et al. (2017). Pericytes of multiple organs do not behave as mesenchymal stem cells in vivo. Cell Stem Cell 20:345..e5–359.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoshikawa M, Go S, Suzuki S, Suzuki A, Katori Y, Morlet T, Gottlieb SM, Fujiwara M, Iwasaki K, Strauss KA. and Inokuchi J. (2015). Ganglioside GM3 is essential for the structural integrity and function of cochlear hair cells. Hum Mol Genet 24:2796–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang LV,. Radu CG, Roy M, Lee S, McLaughlin J, Teitell MA, Iruela-Arispe ML. and Witte ON. (2007). Vascular abnormalities in mice deficient for the G protein-coupled receptor GPR4 that functions as a pH sensor. Mol Cell Biol 27:1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tao SC,. Gao YS, Zhu HY, Yin JH, Chen YX, Zhang YL, Guo SC. and Zhang CQ. (2016). Decreased extracellular pH inhibits osteogenesis through proton-sensing GPR4-mediated suppression of yes-associated protein. Sci Rep 6:26835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Olivares-Navarrete R, Hyzy SL, Park JH, Dunn GR, Haithcock DA, Wasilewski CE, Boyan BD. and Schwartz Z. (2011). Mediation of osteogenic differentiation of human mesenchymal stem cells on titanium surfaces by a Wnt-integrin feedback loop. Biomaterials 32:6399–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nigam SK. (2018). The SLC22 transporter family: a paradigm for the impact of drug transporters on metabolic pathways, signaling, and disease. Annu Rev Pharmacol Toxicol 58:663–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Logan N. and Brett P. (2013). The control of mesenchymal stromal cell osteogenic differentiation through modified surfaces. Stem Cells Int 2013:361637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Levi B, Wan DC, Glotzbach JP, Hyun J, Januszyk M, Montoro D, Sorkin M, James AW, Nelson ER, et al. (2011). CD105 protein depletion enhances human adipose-derived stromal cell osteogenesis through reduction of transforming growth factor beta1 (TGF-beta1) signaling. J Biol Chem 286:39497–39509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Iwata M, Torok-Storb B, Wayner EA. and Carter WG. (2014). CDCP1 identifies a CD146 negative subset of marrow fibroblasts involved with cytokine production. PLoS One 9:e109304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Elsheikh SE,. Green AR, Rakha EA, Samaka RM, Ammar AA, Powe D, Reis-Filho JS. and Ellis IO. (2008). Caveolin 1 and Caveolin 2 are associated with breast cancer basal-like and triple-negative immunophenotype. Br J Cancer 99:327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Conze T, Lammers R, Kuci S, Scherl-Mostageer M, Schweifer N, Kanz L. and Buhring HJ. (2003). CDCP1 is a novel marker for hematopoietic stem cells. Ann N Y Acad Sci 996:222–226 [DOI] [PubMed] [Google Scholar]
- 32. Baker N, Zhang G, You Y. and Tuan RS. (2012). Caveolin-1 regulates proliferation and osteogenic differentiation of human mesenchymal stem cells. J Cell Biochem 113:3773–3787 [DOI] [PubMed] [Google Scholar]
- 33. Anderson P, Carrillo-Galvez AB, Garcia-Perez A, Cobo M. and Martin F. (2013). CD105 (endoglin)-negative murine mesenchymal stromal cells define a new multipotent subpopulation with distinct differentiation and immunomodulatory capacities. PLoS One 8:e76979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scheideler M, Elabd C, Zaragosi LE, Chiellini C, Hackl H, Sanchez-Cabo F, Yadav S, Duszka K, Friedl G, et al. (2008). Comparative transcriptomics of human multipotent stem cells during adipogenesis and osteoblastogenesis. BMC Genomics 9:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nassiri F, Cusimano MD, Scheithauer BW, Rotondo F, Fazio A, Yousef GM, Syro LV, Kovacs K. and Lloyd RV. (2011). Endoglin (CD105): a review of its role in angiogenesis and tumor diagnosis, progression and therapy. Anticancer Res 31:2283–2290 [PubMed] [Google Scholar]
- 36. Ishiy FAA, Fanganiello RD, Kobayashi GS, Kague E, Kuriki PS. and Passos-Bueno MR. (2018). CD105 is regulated by hsa-miR-1287 and its expression is inversely correlated with osteopotential in SHED. Bone 106:112–120 [DOI] [PubMed] [Google Scholar]
- 37. Zheng YZ,. Boscher C, Inder KL, Fairbank M, Loo D, Hill MM, Nabi IR. and Foster LJ. (2011). Differential impact of caveolae and caveolin-1 scaffolds on the membrane raft proteome. Mol Cell Proteomics 10:M110..007146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ren T, Piperdi S, Koirala P, Park A, Zhang W, Ivenitsky D, Zhang Y, Villanueva-Siles E, Hawkins DS, Roth M. and Gorlick R. (2017). CD49b inhibits osteogenic differentiation and plays an important role in osteosarcoma progression. Oncotarget 8:87848–87859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Makar AB,. McMartin KE, Palese M. and Tephly TR. (1975). Formate assay in body fluids: application in methanol poisoning. Biochem Med 13:117–126 [DOI] [PubMed] [Google Scholar]
- 40. Liang Y, Li S. and Chen L. (2015). The physiological role of drug transporters. Protein Cell 6:334–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bohlson SS,. Silva R, Fonseca MI. and Tenner AJ. (2005). CD93 is rapidly shed from the surface of human myeloid cells and the soluble form is detected in human plasma. J Immunol 175:1239–1247 [DOI] [PubMed] [Google Scholar]
- 42. Joshi M, Patil P B, He Z, Holgersson J, Olausson M. and Sumitran-Holgersson S. (2012). Fetal liver-derived mesenchymal stromal cells augment engraftment of transplanted hepatocytes. Cytotherapy 14:657–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boccuto L, Aoki K, Flanagan-Steet H, Chen CF, Fan X, Bartel F, Petukh M, Pittman A, Saul R, et al. (2014). A mutation in a ganglioside biosynthetic enzyme, ST3GAL5, results in salt & pepper syndrome, a neurocutaneous disorder with altered glycolipid and glycoprotein glycosylation. Hum Mol Genet 23:418–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chuang SS,. Helvig C, Taimi M, Ramshaw HA, Collop AH, Amad M, White JA, Petkovich M, Jones G. and Korczak B. (2004). CYP2U1, a novel human thymus- and brain-specific cytochrome P450, catalyzes omega- and (omega-1)-hydroxylation of fatty acids. J Biol Chem 279:6305–6314 [DOI] [PubMed] [Google Scholar]
- 45. Czachor A, Failla A, Lockey R. and Kolliputi N. (2016). Pivotal role of AKAP121 in mitochondrial physiology. Am J Physiol Cell Physiol 310:C625–C628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. England J, Granados-Riveron J, Polo-Parada L, Kuriakose D, Moore C, Brook JD, Rutland CS, Setchfield K, Gell C, et al. (2017). Tropomyosin 1: multiple roles in the developing heart and in the formation of congenital heart defects. J Mol Cell Cardiol 106:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kutay U, Hartmann E, Treichel N, Calado A, Carmo-Fonseca M, Prehn S, Kraft R, Gorlich D. and Bischoff FR. (2000). Identification of two novel RanGTP-binding proteins belonging to the importin beta superfamily. J Biol Chem 275:40163–40168 [DOI] [PubMed] [Google Scholar]
- 48. Martinu L, Masuda-Robens JM, Robertson SE, Santy LC, Casanova JE. and Chou MM. (2004). The TBC (Tre-2/Bub2/Cdc16) domain protein TRE17 regulates plasma membrane-endosomal trafficking through activation of Arf6. Mol Cell Biol 24:9752–9762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sangel P, Oka M. and Yoneda Y. (2014). The role of Importin-betas in the maintenance and lineage commitment of mouse embryonic stem cells. FEBS Open Bio 4:112–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vrbacky M, Kovalcikova J, Chawengsaksophak K, Beck IM, Mracek T, Nuskova H, Sedmera D, Papousek F, Kolar F, et al. (2016). Knockout of Tmem70 alters biogenesis of ATP synthase and leads to embryonal lethality in mice. Hum Mol Genet 25:4674–4685 [DOI] [PubMed] [Google Scholar]
- 51. Li J, Klein C, Liang C, Rauch R, Kawamura K. and Hsueh AJ. (2009). Autocrine regulation of early embryonic development by the artemin-GFRA3 (GDNF family receptor-alpha 3) signaling system in mice. FEBS Lett 583:2479–2485 [DOI] [PubMed] [Google Scholar]
- 52. Nakagomi T, Nakano-Doi A, Kawamura M. and Matsuyama T. (2015). Do vascular pericytes contribute to neurovasculogenesis in the central nervous system as multipotent vascular stem cells? Stem Cells Dev 24:1730–1739 [DOI] [PubMed] [Google Scholar]
- 53. Etchevers HC,. Vincent C, Le Douarin NM. and Couly GF. (2001). The cephalic neural crest provides pericytes and smooth muscle cells to all blood vessels of the face and forebrain. Development 128:1059–1068 [DOI] [PubMed] [Google Scholar]
- 54. Feng J, Mantesso A, De Bari C, Nishiyama A. and Sharpe PT. (2011). Dual origin of mesenchymal stem cells contributing to organ growth and repair. Proc Natl Acad Sci U S A 108:6503–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen WC,. Baily JE, Corselli M, Diaz ME, Sun B, Xiang G, Gray GA, Huard J. and Peault B. (2015). Human myocardial pericytes: multipotent mesodermal precursors exhibiting cardiac specificity. Stem Cells 33:557–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Armulik A, Genove G, Mae M, Nisancioglu MH, Wallgard E, Niaudet C, He L, Norlin J, Lindblom P, et al. (2010). Pericytes regulate the blood-brain barrier. Nature 468:557–561 [DOI] [PubMed] [Google Scholar]
- 57. Daneman R, Zhou L, Kebede AA. and Barres BA. (2010). Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature 468:562–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jones EA,. English A, Kinsey SE, Straszynski L, Emery P, Ponchel F. and McGonagle D. (2006). Optimization of a flow cytometry-based protocol for detection and phenotypic characterization of multipotent mesenchymal stromal cells from human bone marrow. Cytometry B Clin Cytom 70:391–399 [DOI] [PubMed] [Google Scholar]
- 59. Fekete N, Rojewski MT, Furst D, Kreja L, Ignatius A, Dausend J. and Schrezenmeier H. (2012). GMP-compliant isolation and large-scale expansion of bone marrow-derived MSC. PLoS One 7:e43255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.