Abstract
Activated nuclear factor-κB (NF-κB) plays an important role in the development of cardiovascular disease (CVD) through its regulated genes and microRNAs (miRNAs). However, the gene regulation profile remains unclear. In this study, primary mouse vascular endothelial cells (pMVECs) were employed to detect CVD-related NF-κB-regulated genes and miRNAs. Genechip assay identified 77 NF-κB-regulated genes, including 45 upregulated and 32 downregulated genes, in tumor necrosis factor α (TNFα)-treated pMVECs. Ten of these genes were also found to be regulated by NF-κB in TNFα-treated HeLa cells. Quantitative real-time PCR (qRT-PCR) assay confirmed the up-regulation of Egr1, Tnf, and Btg2 by NF-κB in the TNFα-treated pMVECs. The functional annotation revealed that many NF-κB-regulated genes identified in pMVECs were clustered into classical NF-κB-involved biological processes. Genechip assay also identified 26 NF-κB-regulated miRNAs, of which 21 were upregulated and 5 downregulated, in the TNFα-treated pMVECs. Further analysis showed that nine of the identified genes are regulated by seven of these miRNAs. Finally, among the identified NF-κB-regulated genes and miRNAs, 5 genes and 12 miRNAs were associated with CVD by miRWalk and genetic association database analysis. Taken together, these findings show an intricate gene regulation network raised by NF-κB in TNFα-treated pMVECs. The network provides new insights for understanding the molecular mechanism underlying the progression of CVD.
Keywords: Cardiovascular disease, Nuclear factor-κB (NF-κB), MicroRNA, Endothelial cell
1. Introduction
Cardiovascular disease (CVD) that involves heart and systemic blood vessels accounts for about half of global deaths and prevalence is increasing (Estruch, 2014; Mangge et al., 2014). As a barrier between the blood and surrounding tissue, endothelial cells (ECs) are extremely important for the maintenance of vascular function and homeostasis, and their dysfunction is mainly responsible for the progression of CVD (Widlansky et al., 2003; Tousoulis et al., 2010; Zhu et al., 2018). Endothelial dysfunction (ED) is characterized by changes in endothelial function and homeostatic control in response to extensive noxious stimuli (Madamanchi et al., 2005; Murdaca et al., 2012; Steyers and Miller, 2014). Endothelium undergoes a phenotypic modulation from normal to non-adaptive, which often manifests with impaired vascular reactivity, reduced nitric oxide (NO) availability, increased oxidative stress, and increased cellular adhesion molecules, such as vascular adhesion molecule-1 (VCAM-1) and intercellular adhesion molecule-1 (ICAM-1) (Madamanchi et al., 2005; Murdaca et al., 2012; Steyers and Miller, 2014). Investigation of altered gene expression in the response of ECs to external stimuli may provide clues for CVD research. The ED that is frequently found in atherosclerosis, hypertension, heart failure, and myocardial infarction leads to increased endothelial permeability and infiltration of inflammatory cells into the arterial wall (Boulanger, 1999; Lerman and Zeiher, 2005; Tousoulis et al., 2010). This process ultimately results in atherosclerosis (Zampetaki et al., 2013). Therefore, ED is a key step in the development of CVD events (Vanhoutte, 2009; Steyers and Miller, 2014).
Nuclear factor-κB (NF-κB) is a key inducible transcription factor family that contains five members including p65/RelA, p52, p50, c-Rel, and RelB (Zhou et al., 2017). NF-κB participates in physiological and pathological processes by regulating target genes (Zhou et al., 2017). Under stimulation by inducers (e.g. tumor necrosis factor α (TNFα)), the inhibitor-blocked NF-κB (e.g. IκBα) is liberated and subsequently moves into the cell nucleus to regulate its target genes (Zhou et al., 2017; Song Y et al., 2018). Many well-known NF-κB target genes such as TNF, E-selectin, ICAM-1, and VCAM-1 play pivotal roles in the progression of CVD (Vanhoutte, 2009; Murdaca et al., 2012; Steyers and Miller, 2014). Selective inhibition of the NF-κB upregulating endothelial-specific target genes, E-selectin and VCAM-1, attenuated chronic intermittent hypoxia-induced atherosclerosis in mice (Song DM et al., 2018). NF-κB-mediated upregulation of cell adhesion molecule 1 (CADM1), a recently identified target, has also been identified as a mechanism of TNFα-induced rat endothelial progenitor cell migration (Prisco et al., 2016).
MicroRNAs (miRNAs) are a class of short (approximately 22 nucleotides (nt)) endogenous noncoding RNAs that repress their target gene expression at the post transcriptional level by binding to 3' untranslated regions (3' UTRs) (Ma et al., 2011; Menghini et al., 2014). Reports about the crucial role of miRNA in the progression of CVD are increasing (Menghini et al., 2014). For example, the silence of miR-15 family members, including miR-497, miR-195, miR-16-2, miR-16-1, miR-15b, and miR-15a, reduces myocardial infarction size in mouse (Hullinger et al., 2012). Endogenous miR-221, increased by high-level glucose, ultimately results in ED including reduction of human umbilical vein endothelial cell (HUVEC) migration and c-kit expression, which are antagonized by antagomirs of miR-221 (Li et al., 2009). The five CVD-related miRNAs, miR-497 (Wei et al., 2016), miR-195 (Wei et al., 2016), miR-15a (Yang et al., 2015), miR-15b (Zhu et al., 2016), and miR-221 (Galardi et al., 2011), are all transcriptionally regulated by NF-κB. In this study, we have identified CVD-related NF-κB-regulated genes and miRNAs using primary mouse vascular endothelial cells (pMVECs) as approximate representations of the endothelial monolayer in blood vessels. The exposure of pMVECs to TNFα was used to cause the expression of potential NF-κB regulatory genes and miRNAs. RelA small interfering RNA (siRNA) was introduced to identify the genes and miRNAs specifically regulated by NF-κB.
2. Materials and methods
2.1. Mouse primary endothelial cell culture
The mouse experiments were approved by the Animal Care and Use Committee of the Southeast University (Nanjing, China). After sacrifice by cervical dislocation, the dead mice were soaked in 75% (v/v) ethanol for 1 min. Under sterile conditions, aorta was removed from the chest and immersed in sterile phosphate-buffered saline (PBS). Fats and fibrous tissues were removed from vascular adventitia by ophthalmic scissors and tweezers. The internal and external surfaces of blood vessels were washed with PBS containing antibiotics. Blood vessels were longitudinally cut open and cut into 1.5 mm×1.5 mm artery explants, and then placed into a petri dish coated with 1% (0.01 g/mL) gelatin at the density of 1 cm−2. The intima of the artery explant was in contact with the gelatin. The petri dish was then placed into a 5% (v/v) CO2 incubator (100% humidity) at 37 °C for 2 h. Primary endothelial cell special culture medium (PriCells, Wuhan, China) with 10% (v/v) fetal bovine serum (TransGen, Beijing, China) was then added to the dish without causing the artery explant to float. The medium was replaced after 72 h and thereafter every 48 h. The integrated pMVECs were dissociated into single cell suspensions using 0.25% (2.5 g/L) trypsin (Amresco, Boise, USA) containing 0.025% (0.25 g/L) ethylene diamine tetraacetic acid (EDTA). The pMVECs were used for experiments between the 4th and 7th passages.
2.2. Construction of the pMVEC model with different NF-κB activity
The NF-κB activator (TNFα) and NF-κB inhibitor (RelA siRNA) were used to construct pMVEC models as follows. pMVECs with about 50% confluency were first transfected with 30 nmol/L NF-κB RelA siRNA (Cell Signaling Technology, Danvers, MA, USA) by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the instructions. After 48 h, the transfected cells were stimulated with 30 ng/mL TNFα (Sigma, Shanghai, China) in the serum-free primary endothelial cell special culture medium at 37 °C for 30 min. Simultaneously, pMVECs were subjected to the same amount of Lipofectamine 2000 and treated with or without TNFα. From this, we created three pMVEC groups: RelA siRNA and TNFα co-treated pMVECs (thereafter depicted as siRNA-treated pMVECs), TNFα-treated pMVECs, and control pMVECs.
2.3. Genechip assay
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA from pMVECs. The extracted RNA samples underwent Genechip assay as in a previous study (Liu and Wang, 2013), to detect the global gene expression profile. Biotin-labeled complementary RNA (cRNA) generated by in vitro transcription reactions was fragmented and hybridized with Affymetrix, MOUSE 2.0T Genechip microarrays (Affymetrix, USA) at 45 °C for 16 h. The hybridized arrays were then reacted with streptavidin-phycoerythrin, and scanned by an Affymetrix Genechip Scanner 3000 7G (Thermo Fisher Scientific, Waltham, USA). Image analysis and quantification were performed with Genechip Operating Software (Thermo Fisher Scientific). After normalization and background filtration, the differentially expressed genes (DEGs) were analyzed in the siRNA-treated pMVECs, TNFα-treated pMVECs, and control pMVECs. The gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) analyses on these DEGs were performed by Database for Annotation, Visualization and Integrated Discovery (DAVID) online servers (http://david.abcc.ncifcrf.go).
Similarly, the miRNAs isolated from the pMVECs were hybridized with Affymetrix MICRORNA 3.0 Genechip (Affymetrix) to detect the global miRNA expression profile. After normalization and background filtration, the differentially expressed miRNAs (DEMs) were analyzed in the siRNA-treated pMVECs, TNFα-treated pMVECs, and control pMVECs.
2.4. qRT-PCR assay
Total RNA was extracted from pMVECs and mouse hepatoma Hepa1-6 cells and 1 µg of the extracted RNA was reversely transcribed into cDNA by PrimeScript™ RT Master Mix (TaKaRa, Dalian, China). The cDNAs were then used to detect gene expression by quantitative real-time PCR (qRT-PCR) on an ABI StepOne™ real-time PCR system (Applied Biosystems, Life Technologies, Grand Island, NY, USA) with SYBR Green Mix (Roche, Shanghai, China). The primers (5'→3') of the determined genes were as follows: RelA, AGACACAGA TGATCGCCACC and GGCTTGGGGACAGAAGT TGA; Cxcl10, CCACGTGTTGAGATCATTGCC and GAGGCTCTCTGCTGTCCATC; Btg2, GCACTGA CCGATCATTACAA and GGATCAACCCACAG GGTCAG; Egr1, CAATTGATGTCTCCGCTGC and AAAAGGACTCTGTGGTCAGGTG; Tnf, CGTCGT AGCAAACCACCAA and ATAGCAAATCGGCTG ACGGT; Icam1, CCTCCGGACTTTCGATCTT and CTGGTCCGCTAGCTCCAAAA; Vcam1, GGAGG TCTACTCATTCCCTGA and GGTGGGGATGAA GGTCGTTT; GADPH, TGGCAAAGTGGAGATTG TT and CTCGCTCCTGGAAGATGG. The expression of GADPH was used as an internal control. The melting curve was used to analyze the specificity of amplification. Relative expression was calculated by the comparative C T method. At least three biological replications for each gene were performed and the data were shown as mean±standard deviation (SD). Significant differences were assessed by one-way analysis of variance (ANOVA) using SPSS 13.0 (SPSS Inc., Chicago, IL, USA) for windows. P<0.05 was regarded as statistically significant.
2.5. Prediction of the target gene of NF-κB-regulated miRNA
The target genes of NF-κB-regulated miRNAs were identified according to the method used in our previous study (Zhou et al., 2014) with the online miRWalk program (Dweep et al., 2011) being used initially to predict the genes. The miRNA binding sites on 3' UTR of mRNA were predicted by setting the following parameters: the longest transcript, minimum seed length of 7 nt, and P-value of 0.05. The comparative platform of the miRWalk web was used to analyze the output result. An miRNA target that was predicted by at least six programs was regarded as a high-confidence nominated target. We then identified the targets of NF-κB-regulated miRNA from the nominated targets according to the previous criterion that miRNA downregulates its targeted mRNA (Zhou et al., 2014). In other words, the change in their expression level is inversely correlated to that of miRNA.
2.6. Analyses of CVD-associated NF-κB target genes and miRNAs
Five thousand and eleven disease-related mouse genes are collected in the genetic association database (GAD) (Becker et al., 2004). The collected genes were compared with the detected DEGs to find those related to CVD. The miRWalk database contains 324 disease-related mouse miRNAs (Dweep et al., 2011). The collected miRNAs were compared with the DEMs to find out the CVD-related miRNAs. Finally, these genes and miRNAs were visualized by the Cytoscape program (Shannon et al., 2003).
3. Results
3.1. Qualitative evaluation of pMVECs
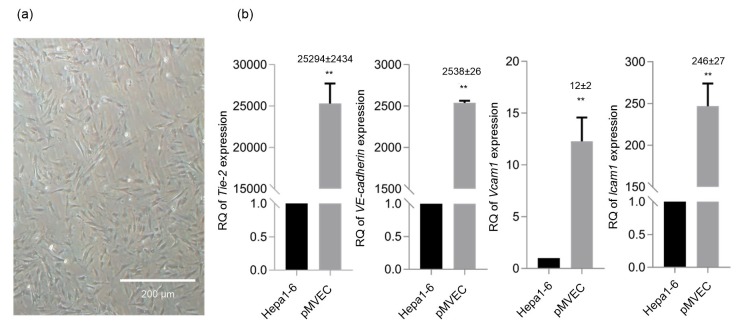
The pMVECs were isolated and cultured as described in Section 2. As shown in Fig. 1a, the cultured pMVECs presented adherent growth patterns and polygons. When pMVECs grew into a confluent monolayer, they showed a typical cobblestone-like arrangement. Tie-2, VE-cadherin, Vcam1, and Icam1 were endothelial-specific markers (Lim et al., 2003; Sun et al., 2014). Expression of these four genes was therefore detected by qRT-PCR to qualify isolated pMVECs. As a result, the expression of the four genes was significantly increased in pMVECs compared with Hepa1-6 cells (Fig. 1b). Of note, the expression change of Tie-2 increased 25 294-fold in pMVECs compared with Hepa1-6 cells (Fig. 1b).
Fig. 1.
Quality evaluation of cultured pMVECs
(a) Morphology of pMVECs. (b) Expression of Tie-2, VE-cadherin, Vcam1, and Icam1 in pMVECs compared with Hepa1-6. Data are shown as mean±standard deviation. Detection for Tie-2 and VE-cadherin expression, n=4; detection for Vcam1 and Icam1 expression, n=3. The significant differences were assessed by one-way analysis of variance (ANOVA). ** P<0.01. Numbers over the stars represent the fold change of gene expression in pMVECs compared with Hepa1-6. pMVECs, primary mouse vascular endothelial cells; RQ, relative quantitation
3.2. Qualitative evaluation of the pMVEC model with different NF-κB activity
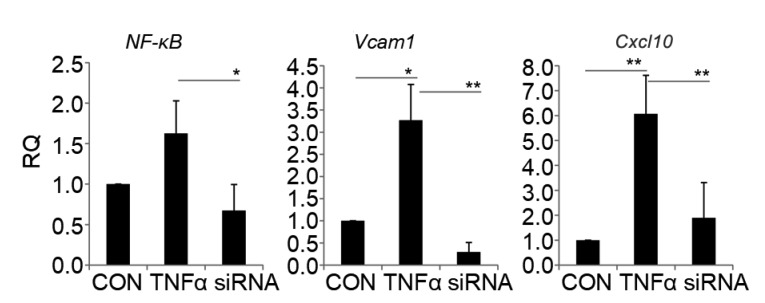
To identify CVD-related NF-κB-regulated genes and miRNAs, pMVEC models with different NF-κB activity were constructed by treating pMVECs with NF-κB activator (TNFα) and its inhibitor (RelA siRNA). The models were then evaluated by qRT-PCR assay. The result showed that the expression of RelA was significantly increased by TNFα and repressed by RelA siRNA (Fig. 2). In general, the activated NF-κB was transported into the nucleus to regulate expression of target genes (Zhou et al., 2017). To further evaluate the cell models, we used the qRT-PCR assay to detect the expression of two well-known NF-κB-regulated genes, Cxcl10 (Hein et al., 1997; Varley et al., 2003) and Vcam1 (Iademarco et al., 1992). We found that both were significantly increased by TNFα and repressed by RelA siRNA, which is consistent with the NF-κB levels (Fig. 2). These findings validate the pMVEC model for different NF-κB activity.
Fig. 2.
Expression of NF-κB (RelA), Cxcl10, and Vcam1 in pMVECs
RQ was calculated with the comparative C T method and normalized against GADPH. The data are shown as mean±standard deviation (n=4). The significant differences were assessed by one-way analysis of variance (ANOVA). * P<0.05, ** P<0.01. RQ, relative quantitation; pMVECs, primary mouse vascular endothelial cells; CON, control pMVECs; TNFα, TNFα-treated pMVECs; siRNA, siRNA-interfered pMVECs
3.3. Identification of NF-κB-regulated genes
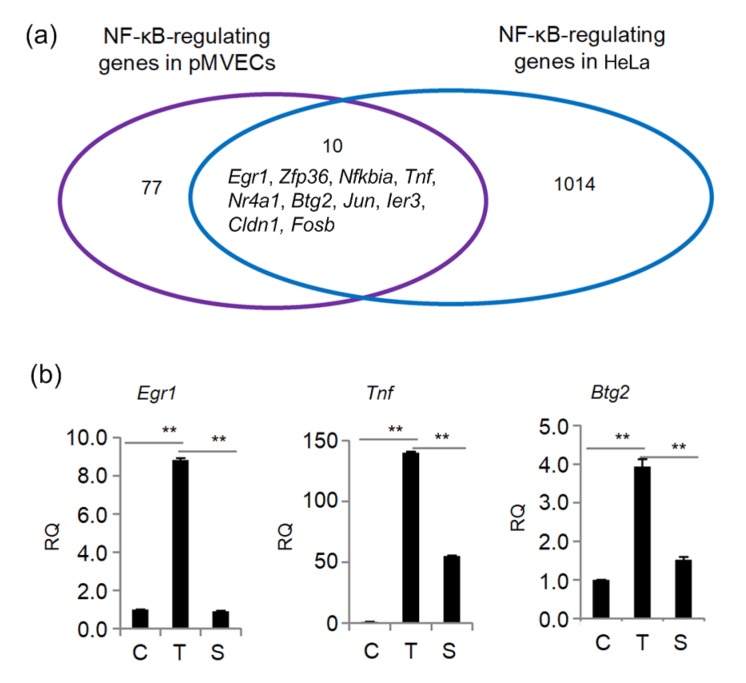
In order to identify NF-κB-regulated genes, the RNA samples from pMVECs with different NF-κB activity were analyzed by Genechip assay. Upon stimulation by TNFα, the expression of NF-κB potentially-regulated genes changed. Ninety were markedly upregulated and eighty-five downregulated compared with control cells (fold change ≥1.5) (Fig. 3a). To find reliable NF-κB-regulated genes, the gene expression profile of the RelA siRNA-interfered pMVECs was compared with that of the TNFα-treated pMVECs. As shown in Fig. 3, the expression of 45 TNFα-increased and 32 TNFα-inhibited genes was markedly antagonized by RelA siRNA (fold change ≥1.2). The fold changes of expression of the 77 genes that responded to the TNFα and RelA siRNA treatments are shown in Table S1. These 77 genes are regarded as NF-κB-regulated genes in TNFα-treated pMVECs. By comparing these with the NF-κB-regulated gene profile identified in similarly treated HeLa cells (Xing et al., 2013), we found ten NF-κB-regulated genes, including Egr1, Zfp36, Nfkbia, Tnf, Nr4a1, Btg2, Jun, Ier3, Cldn1, and Fosb, that are also regulated by this factor in the TNFα-treated HeLa cells (Fig. 4a). To further confirm these NF-κB-regulated genes, the expression of Egr1, Tnf, and Btg2 was detected by qRT-PCR assay. The results demonstrated that their expression was significantly enhanced by TNFα and then blocked by RelA siRNA (Fig. 4b), which is consistent with the Genechip assay. To explore the biological processes influenced by the identified NF-κB-regulated genes, 77 NF-κB-regulated genes were submitted to the DAVID database online for GO annotation. All the GO terms with P-value of <0.05 are presented in Table 1. The results indicate that the NF-κB-regulated genes are mainly clustered into GO terms of signal transduction, immunity and defense, stress response, intracellular signaling cascade, and mesoderm development, all of which are well-documented, classical biological processes involving NF-κB (Xing et al., 2013). It was also noted that four new NF-κB-regulated genes belonging to the Olfr gene family, Olfr480, Olfr490, Olfr1109, and Olfr1463, are clustered under the GO signal transduction term. To better understand the molecular function of these NF-κB-regulated genes, we analyzed the KEGG pathway influenced by the NF-κB-regulated genes and found ten NF-κB-regulated genes clustered into other important pathways such as the Toll-like receptor signaling pathway (Table 2).
Fig. 3.
NF-κB-regulated genes in TNFα-treated pMVECs
(a) Number of NF-κB-regulated genes. (b) Hierarchical cluster analysis of NF-κB-regulated genes. The horizontal axis shows comparison groups of gene expression change. T vs. C group represents the fold change of gene expression in the TNFα-treated pMVECs compared with control pMVECs. T vs. S group represents the fold change of gene expression in the TNFα-treated pMVECs compared with siRNA-interfered pMVECs. The left vertical axis shows clusters of NF-κB-regulated genes. Red indicates upregulated and green downregulated genes. pMVECs, primary mouse vascular endothelial cells (Note: for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article)
Fig. 4.
Verification of NF-κB-regulated genes in TNFα-treated pMVECs
(a) Comparison of the NF-κB-regulated gene profile in TNFα-treated pMVECs with TNFα-treated HeLa cells. (b) Expression of Egr1, Tnf, and Btg2 in TNFα-treated pMVECs. Data are shown as mean±standard deviation (n=3). The significant differences were analyzed by one-way analysis of variance (ANOVA). ** P<0.01. pMVECs, primary mouse vascular endothelial cells; RQ, relative quantitation. C, control pMVECs; T, TNFα-treated pMVECs; S, siRNA-interfered pMVECs
Table 1.
GO analysis of NF-κB-regulated genes by the DAVID online server
| GO term | Count | P-value | Fold enrichment | Clustered gene |
| BP00148: immunity and defense | 15 | 0.0005 | 2.7545 | Ier3, Ccl3, Tnf, Nfkbia, Nr4a1, Il11ra1, Itpr3, Fos, Gstp1, Antxr1, Dusp8, Cd200, Oas1E, H2-M10.5, H2-T10 |
| BP00178: stress response | 5 | 0.0042 | 7.4293 | Ier3, Nr4a1, Nfkbia, Itpr3, Dusp8 |
| BP00111: intracellular signaling cascade | 9 | 0.0060 | 3.1588 | Zfp36, Spry1, Btg2, Dgkg, Jun, Il11ra1, Nfkbia, Itpr3, Dusp8 |
| BP00248: mesoderm development | 6 | 0.0320 | 3.2951 | Bmp4, Fos, Spry1, Il11ra1, Myom1, Angpt2 |
| BP00102: signal transduction | 20 | 0.0426 | 1.4984 | Zfp36, Ccl3, Tnf, Il11ra1, Nfkbia, Nr4a1, Itpr3, Spry1, Wisp1, Vmn1R94, Btg2, Dgkg, Jun, Cd200, Dusp8, Angpt2, Olfr480, Olfr490, Olfr1109, Olfr1463 |
GO, gene ontology; NF-κB, nuclear factor-κB; DAVID, Database for Annotation, Visualization and Integrated Discovery
Table 2.
KEGG pathway analysis of NF-κB-regulated genes by the DAVID online server
| KEGG pathway | Count | P-value | Fold enrichment | Clustered gene |
| mmu04620: Toll-like receptor signaling pathway | 5 | 0.0012 | 9.9930 | Fos, Ccl3, Tnf, Jun, Nfkbia |
| mmu04660: T cell receptor signaling pathway | 4 | 0.0191 | 6.7072 | Fos, Tnf, Jun, Nfkbia |
| mmu05330: allograft rejection | 3 | 0.0321 | 10.2342 | Tnf, H2-M10.5, H2-T10 |
| mmu05332: graft-versus-host disease | 3 | 0.0321 | 10.2342 | Tnf, H2-M10.5, H2-T10 |
| mmu04940: type I diabetes mellitus | 3 | 0.0373 | 9.4220 | Tnf, H2-M10.5, H2-T10 |
| mmu04010: MAPK signaling pathway | 5 | 0.0380 | 3.7332 | Fos, Tnf, Jun, Nr4a1, Dusp8 |
KEGG, Kyoto encyclopedia of genes and genomes; NF-κB, nuclear factor-κB; DAVID, Database for Annotation, Visualization and Integrated Discovery; MAPK, mitogen-activated protein kinase
3.4. Identification of NF-κB-regulated miRNAs
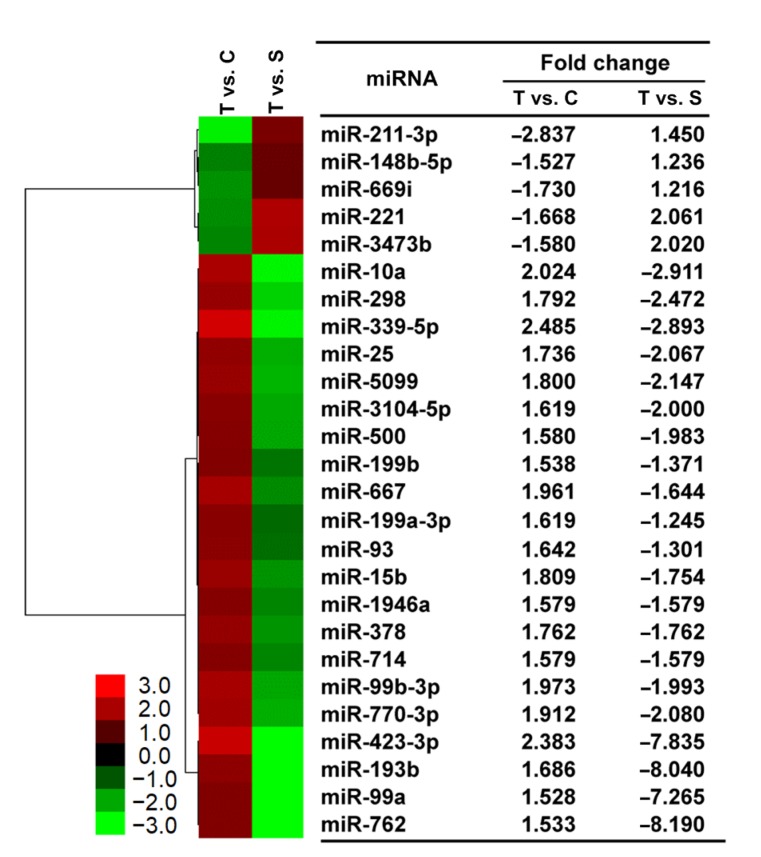
To explore NF-κB-controlled miRNAs in pMVECs, we analyzed miRNAs isolated from pMVECs with different NF-κB activity (Fig. 2) using the Genechip assay. The results show that expression of 41 and 15 miRNAs was markedly increased and inhibited in TNFα-treated pMVECs, respectively. To increase the reliability of our findings, we compared the miRNA expression profile of RelA siRNA-interfered pMVECs with that of TNFα-treated pMVECs and found that the expression of 21 TNFα-increased and 5 TNFα-inhibited miRNAs was antagonized by the siRNA with fold change more than 1.2 (Fig. 5). Therefore, 26 miRNAs are regarded NF-κB-regulated in TNFα-treated pMVECs.
Fig. 5.
NF-κB-regulated miRNAs in TNFα-treated pMVECs
Hierarchical cluster analysis of NF-κB-regulated miRNAs. The horizontal axis shows comparison groups of expression change of miRNAs. T vs. C group represents the fold changes of miRNA expression in TNFα-treated pMVECs compared with control pMVECs. T vs. S group represents the fold change of miRNA expression in the TNFα-treated pMVECs compared with siRNA-interfered pMVECs. The left vertical axis shows clusters of NF-κB-regulated miRNAs whose fold change is listed in the right-hand table. Red indicates upregulated and green downregulated miRNAs. pMVECs, primary mouse vascular endothelial cells (Note: for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article)
3.5. Analysis of target genes of NF-κB-regulated miRNAs
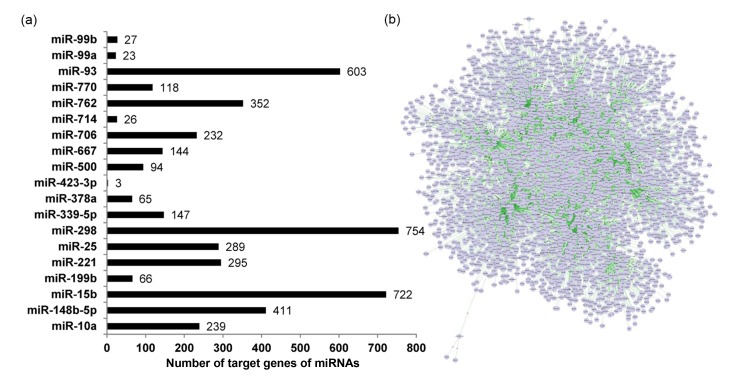
We submitted the identified NF-κB-regulated miRNAs to the miRWalk server in order to predict their target genes, according to the parameters and criteria in Section 2. Any miRNA target predicted by at least six miRWalk programs was regarded with high confidence as a nominated target of NF-κB-regulated miRNAs. From this computational approach, we found high-confidence nominated targets for a total of 19 NF-κB-regulated miRNAs. Of these, miR-423-3p had the fewest target genes (i.e. three), while miR-298 had the most (i.e. 754) (Fig. 6a). We also found that one miRNA had multiple nominated target genes and that several miRNAs had the same nominated target gene. Thus, a total of 3205 nominated target genes were hit by these NF-κB-regulated miRNAs. The Cytoscape program showed that these miRNAs and their target genes construct a complex regulatory network in the TNFα-treated pMVECs (Fig. 6b). The above nominated target profile was subsequently compared with the NF-κB-regulated gene profile to find expression changes for several of them. According to the criterion that changes in expression of the target gene are inversely correlated with that of miRNA (Zhou et al., 2014), we found that changes in expression of nine nominated target genes were inversely correlated to those of the seven corresponding NF-κB-regulated miRNAs in pMVECs treated with TNFα and RelA siRNA (Table 3). The nine genes were therefore considered as targets of the seven NF-κB-regulated miRNAs. For example, when pMVECs were subjected to TNFα stimulation and RelA siRNA-interference, the expression of Btg2 and Fos changed contrary to that of miR-221. Therefore, Btg2 and Fos were regarded as the target genes of miR-221 (Table 3). Accordingly, we identified the target genes of miR-15b, miR-25, miR-298, miR-339-5p, miR-500, and miR-93, as well as miR-221 (Table 3).
Fig. 6.
Nominated targets of NF-κB-regulated miRNAs and their constructed regulatory network
(a) Prediction of target genes of NF-κB-regulated miRNAs. (b) Regulatory network formed by NF-κB-regulated miRNAs and their predicted target genes
Table 3.
Expression changes of NF-κB-regulated miRNAs and their predicted target genes in pMVECs
| miRNA | Target gene (n=9) | Fold change |
|
| TNFα vs. control | TNFα vs. siRNA | ||
| miR-15b | 1.809 | −1.754 | |
| Wisp1 | −1.500 | 1.632 | |
| Myom1 | −1.719 | 1.261 | |
| miR-221 | −1.668 | 2.061 | |
| Fos | 1.959 | −2.302 | |
| Btg2 | 1.612 | −1.383 | |
| miR-25 | 1.736 | −2.067 | |
| Btg2 | 1.612 | −1.383 | |
| miR-298 | 1.792 | −2.472 | |
| Grhl2 | −1.745 | 1.335 | |
| Fem1a | −1.803 | 1.276 | |
| miR-339-5p | 2.485 | −2.893 | |
| Cadm4 | −1.586 | 1.692 | |
| miR-500 | 1.580 | −1.983 | |
| Myom1 | −1.719 | 1.261 | |
| miR-93 | 1.642 | −1.301 | |
| Grhl2 | −1.745 | 1.335 | |
NF-κB, nuclear factor-κB; pMVECs, primary mouse vascular endothelial cells; TNFα vs. control, TNFα-treated pMVECs compared with control pMVECs; TNFα vs. siRNA, TNFα-treated pMVECs compared with siRNA-interfered pMVECs
3.6. Analyses of CVD-related NF-κB-regulated genes and miRNAs
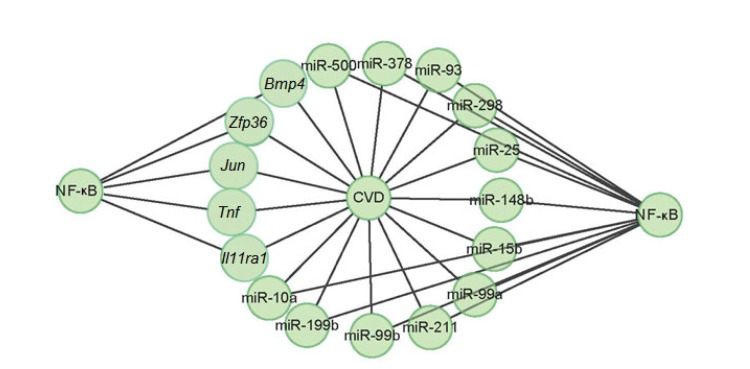
A series of disease-related mouse genes and miRNAs were collected in the GAD (Becker et al., 2004) and miRWalk (Dweep et al., 2011) databases. To fully understand the involvement of NF-κB in CVD, the identified NF-κB-regulated genes and miRNAs were analyzed by these databases. The GAD analysis found five NF-κB-regulated genes related to CVD (Fig. 7). For example, Zfp36, Tnf, Jun, Il11ra1, and Bmp4 were related to the cardiovascular system phenotype (Table 4). miRWalk analysis identified twelve NF-κB-regulated miRNAs related to CVD (Fig. 7). For example, miR-10a was related to atherosclerosis and plaque atherosclerotic (Table 5). Therefore, 5 genes and 12 miRNAs were identified as CVD-related NF-κB-regulated genes and miRNAs in the TNFα-treated pMVECs.
Fig. 7.
Involvement of NF-κB in the process of cardiovascular disease (CVD) via its regulated genes and miRNAs
Table 4.
CVDs-related NF-κB-regulated genes in TNFα-treated pMVECs
| Gene | Cardiovascular disease (CVD)-related phenotype |
| Zfp36 | Heart inflammation; cardiovascular system phenotype |
| Tnf | Cardiovascular system phenotype |
| Jun | Vasculature congestion; thin ventricular wall; cardiovascular system phenotype |
| Il11ra1 | Abnormal vascular development; cardiovascular system phenotype |
| Bmp4 | Abnormal heart development; abnormal aorta morphology; abnormal ventricular septum morphology; abnormal atrial septum morphology; double outlet right ventricle; abnormal endocardial cushion morphology; cardiovascular system phenotype |
NF-κB, nuclear factor-κB; TNFα, tumor necrosis factor α; pMVECs, primary mouse vascular endothelial cells
Table 5.
CVDs-related NF-κB-regulated miRNAs in TNFα-treated pMVECs
| miRNA | Cardiovascular disease (CVD) |
| miR-10a | Atherosclerosis; plaque atherosclerotic |
| miR-148b | Stroke |
| miR-15b | Hypertension; reperfusion injury; stroke |
| miR-199b | Cardiomegaly; cardiomyopathies; heart failure |
| miR-211 | Vascular diseases |
| miR-25 | Cardiomegaly; heart diseases; heart septal defects ventricular; stroke; inflammation artery; infarction middle cerebral |
| miR-298 | Brain injuries; hemorrhage; infarction middle cerebral artery; intracranial hemorrhages |
| miR-378 | Myocardial ischemia; heart diseases |
| miR-500 | Cardiomyopathies; cardiomyopathy dilated; heart failure |
| miR-93 | Heart diseases; heart septal defects ventricular |
| miR-99a | Heart defects congenital; infarction middle cerebral artery |
| miR-99b | Infarction middle cerebral artery |
NF-κB, nuclear factor-κB; TNFα, tumor necrosis factor α; pMVECs, primary mouse vascular endothelial cells
4. Discussion
NF-κB-regulated genes and miRNAs are involved in the pathogenesis of cardiovascular events. For example, upregulation of NF-κB-regulated genes, such as ICAM-1, VCAM-1, TNF, and IL6, contributes to the process of ED in CVD patients (Vanhoutte, 2009; Murdaca et al., 2012; Steyers and Miller, 2014). Overproduction of metalloproteinase-1 (MMP-1), MMP-3, and MMP-9, which are target genes of NF-κB (Bond et al., 1998; Vincenti et al., 1998; Borghaei et al., 2004), promotes atherosclerotic plaque rupture and myocardial infarction (Newby, 2008). The silence of NF-κB-regulated miRNAs, including miR-497, miR-195, miR-15b, and miR-15a (Yang et al., 2015; Wei et al., 2016; Zhu et al., 2016), reduces myocardial infarction size (Hullinger et al., 2012). Therefore, NF-κB plays a molecular role in the development of CVD. The NF-κB-regulated genes and miRNAs identified in this study provide new insights into cardiovascular events.
We used qualified pMVECs to represent the endothelial monolayer in blood vessels. The treatment of pMVECs with TNFα was used to detect a series of genes and miRNAs potentially regulated by NF-κB. RelA siRNA-interference was used to identify genes and miRNAs specifically regulated by NF-κB in the TNFα-treated pMVECs. A total of 77 NF-κB-regulated genes, of which 45 were upregulated and 32 downregulated, were identified. Among these, 10 NF-κB-regulated genes in the TNFα-treated pMVECs were also regulated in TNFα-treated HeLa cells (Xing et al., 2013). Among the genes we identified, some were well-known NF-κB target genes, such as TNF (Collart et al., 1990) and NFKBIA (Sun et al., 1993). The regulation of Egr1, Tnf, and Btg2 by NF-κB was also confirmed by qRT-PCR assay in TNFα-treated pMVECs. These results demonstrate the reliability of the NF-κB-regulated genes identified in TNFα-treated pMVECs.
We identified a total of 26 NF-κB-regulated miRNAs, 21 of them upregulated and 5 downregulated. It was noted that several miRNAs such as miR-199a (Chen et al., 2008) and miR-221 (Galardi et al., 2011; Santhekadur et al., 2012) have been documented as NF-κB-regulated miRNAs in human cell lines or tissues. This supports the reliability of the findings in our study. We also identified several new NF-κB-regulated miRNAs, such as miR-15b, a member of miR-15 family, and nine target genes of the seven identified NF-κB-regulated miRNAs. For example, both Btg2 and Fos were identified as target genes of miR-221, and Btg2 was identified as a target gene of miR-25.
As expected, we found 5 CVD-related NF-κB-regulated genes and 12 CVD-related NF-κB-regulated miRNAs by informatics analysis. For example, Zfp36, identified as a NF-κB target gene, was highly expressed in ECs overlying atherosclerotic lesions and exerted an anti-inflammatory effect by attenuating inflammatory cytokine/chemokine expression in ECs (Zhang et al., 2013). miR-15b, miR-221, and miR-199a, identified as NF-κB-target miRNAs, were also documented to take part in the CVD process. Silence of miR-15b reduced myocardial infarction size in mouse (Hullinger et al., 2012). Upregulation of the miR-221/222 cluster in initial atherogenic stages increased ED and EC apoptosis (Chistiakov et al., 2015). miR-221/222 also indirectly contributed to cardiovascular pathology by participating in fat and glucose metabolism in non-vascular tissues (Chistiakov et al., 2015). Overexpression of miR-199a suppressed cardiomyocyte autophagy (Chen et al., 2017; Li et al., 2017) and induced cardiac hypertrophy in miR-199a-transgenic mice (Li et al., 2017). The underlying mechanism of miR-199a-impaired cardiomyocyte autophagy was achieved by silencing its target genes Hspa5 and Gsk3b (Chen et al., 2017;
Li et al., 2017). As mentioned above, NF-κB-regulated miRNAs actually regulate their target genes. Therefore, NF-κB and its downstream molecular consequences give rise to a complex regulatory network, which may provide useful clues for uncovering the underlying mechanism in the development and progression of CVD.
In GO annotation, four new NF-κB-regulated genes, Olfr480, Olfr490, Olfr1109, and Olfr1463, were clustered into GO terms of signal transduction. Olfactory receptors encoded by the four genes interact with odorant molecules in the nose to initiate a neuronal response that triggers the perception of smell (Shiao et al., 2012). Therefore, NF-κB may play a role in the process of olfaction through its regulated Olfr family genes. This finding extends the understanding of the biological role of NF-κB. KEGG analysis indicates that NF-κB cross-talk with other CVD-related pathways, such as the Toll-like receptor signaling pathway, contributes to the development and progression of CVD (Frantz et al., 2007).
In conclusion, we have revealed a complex regulatory network constructed by NF-κB-regulated genes, NF-κB-regulated miRNAs, and their target genes in TNFα-stimulated pMVECs. This network suggests that NF-κB plays a variety of molecular roles in the development of CVD events. The finding provides new insights into understanding the molecular mechanisms underlying CVD events including CVD-related ED.
Acknowledgments
We thank Prof. Jin-ke WANG (State Key Laboratory of Bioelectronics, Southeast University, Nanjing, China) for his work on this study.
List of electronic supplementary materials
NF-κB-regulated genes in the TNFα-treated pMVECs
Footnotes
Project supported by the Natural Science Foundation of Guangdong Province (Nos. 2017A030310606 and 2016A030307039), the Science and Technology Planning Project of Guangdong Province (Nos. 2014A070713039 and 2016A030303063), the Science and Technology Planning Project of Chaozhou City (No. 2016GY18), and the National Natural Science Foundation of China (No. 31770584)
Contributors: Fei ZHOU designed the project, performed data analysis, and wrote the paper. Hui ZHU, Yun LI, and Mao-xian WANG conducted the experimental studies, evaluated the data, and partially wrote the manuscript. Wen-xin DU and Ju-hong WANG helped in the data analysis and revised the manuscript. All authors read and approved the final manuscript. Therefore, all authors have taken part in the study and take responsibility for the integrity and security of the data.
Electronic supplementary materials: The online version of this article (https://doi.org/10.1631/jzus.B1800631) contains supplementary materials, which are available to authorized users
Compliance with ethics guidelines: Hui ZHU, Yun LI, Mao-xian WANG, Ju-hong WANG, Wen-xin DU, and Fei ZHOU declare that they have no conflict of interest.
All institutional and national guidelines for the care and use of laboratory animals were followed.
References
- 1.Becker KG, Barnes KC, Bright TJ, et al. The genetic association database. Nat Genet. 2004;36(5):431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 2.Bond M, Fabunmi RP, Baker AH, et al. Synergistic upregulation of metalloproteinase-9 by growth factors and inflammatory cytokines: an absolute requirement for transcription factor NF-κB. FEBS Lett. 1998;435(1):29–34. doi: 10.1016/S0014-5793(98)01034-5. [DOI] [PubMed] [Google Scholar]
- 3.Borghaei RC, Rawlings PL, Jr, Javadi M, et al. NF-κB binds to a polymorphic repressor element in the MMP-3 promoter. Biochem Biophys Res Commun. 2004;316(1):182–188. doi: 10.1016/j.bbrc.2004.02.030. [DOI] [PubMed] [Google Scholar]
- 4.Boulanger CM. Secondary endothelial dysfunction: hypertension and heart failure. J Mol Cell Cardiol. 1999;31(1):39–49. doi: 10.1006/jmcc.1998.0842. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, Wang FY, Zeng ZY, et al. MicroRNA-199a acts as a potential suppressor of cardiomyocyte autophagy through targeting Hspa5 . Oncotarget. 2017;8(38):63825–63834. doi: 10.18632/oncotarget.19133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen R, Alvero AB, Silasi DA, et al. Regulation of IKKβ by miR-199a affects NF-κB activity in ovarian cancer cells. Oncogene. 2008;27(34):4712–4723. doi: 10.1038/onc.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chistiakov DA, Sobenin IA, Orekhov AN, et al. Human miR-221/222 in physiological and atherosclerotic vascular remodeling. Biomed Res Int, 2015:354517. 2015 doi: 10.1155/2015/354517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collart MA, Baeuerle P, Vassalli P. Regulation of tumor necrosis factor alpha transcription in macrophages: involvement of four κB-like motifs and of constitutive and inducible forms of NF-κB. Mol Cell Biol. 1990;10(4):1498–1506. doi: 10.1128/MCB.10.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dweep H, Sticht C, Pandey P, et al. miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44(5):839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Estruch R. Cardiovascular mortality: how can it be prevented? Nefrologia. 2014;34(5):561–569. doi: 10.3265/Nefrologia.pre2014.Apr.12481. [DOI] [PubMed] [Google Scholar]
- 11.Frantz S, Ertl G, Bauersachs J. Mechanisms of disease: Toll-like receptors in cardiovascular disease. Nat Clin Pract Cardiovasc Med. 2007;4(8):444–454. doi: 10.1038/ncpcardio0938. [DOI] [PubMed] [Google Scholar]
- 12.Galardi S, Mercatelli N, Farace MG, et al. NF-κB and c-Jun induce the expression of the oncogenic miR-221 and miR-222 in prostate carcinoma and glioblastoma cells. Nucleic Acids Res. 2011;39(9):3892–3902. doi: 10.1093/nar/gkr006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hein H, Schlüter C, Kulke R, et al. Genomic organization, sequence, and transcriptional regulation of the human eotaxin gene. Biochem Biophys Res Commun. 1997;237(3):537–542. doi: 10.1006/bbrc.1997.7169. [DOI] [PubMed] [Google Scholar]
- 14.Hullinger TG, Montgomery RL, Seto AG, et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res. 2012;110(1):71–81. doi: 10.1161/CIRCRESAHA.111.244442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iademarco MF, McQuillan JJ, Rosen GD, et al. Characterization of the promoter for vascular cell adhesion molecule-1 (VCAM-1) J Biol Chem. 1992;267(23):16323–16329. [PubMed] [Google Scholar]
- 16.Lerman A, Zeiher AM. Endothelial function: cardiac events. Circulation. 2005;111(3):363–368. doi: 10.1161/01.CIR.0000153339.27064.14. [DOI] [PubMed] [Google Scholar]
- 17.Li YX, Song YH, Li F, et al. MicroRNA-221 regulates high glucose-induced endothelial dysfunction. Biochem Biophys Res Commun. 2009;381(1):81–83. doi: 10.1016/j.bbrc.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Song Y, Liu L, et al. miR-199a impairs autophagy and induces cardiac hypertrophy through mTOR activation. Cell Death Differ. 2017;24(7):1205–1213. doi: 10.1038/cdd.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim YC, Garcia-Cardena G, Allport JR, et al. Heterogeneity of endothelial cells from different organ sites in T-cell subset recruitment. Am J Pathol. 2003;162(5):1591–1601. doi: 10.1016/S0002-9440(10)64293-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu YX, Wang JK. Effects of DMSA-coated Fe3O4 nanoparticles on the transcription of genes related to iron and osmosis homeostasis. Toxicol Sci. 2013;131(2):521–536. doi: 10.1093/toxsci/kfs300. [DOI] [PubMed] [Google Scholar]
- 21.Ma XD, Becker Buscaglia LE, Barker JR, et al. MicroRNAs in NF-κB signaling. J Mol Cell Biol. 2011;3(3):159–166. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madamanchi NR, Vendrov A, Runge MS. Oxidative stress and vascular disease. Arterioscler Thromb Vasc Biol. 2005;25(1):29–38. doi: 10.1161/01.ATV.0000150649.39934.13. [DOI] [PubMed] [Google Scholar]
- 23.Mangge H, Becker K, Fuchs D, et al. Antioxidants, inflammation and cardiovascular disease. World J Cardiol. 2014;6(6):462–477. doi: 10.4330/wjc.v6.i6.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menghini R, Stöhr R, Federici M. MicroRNAs in vascular aging and atherosclerosis. Ageing Res Rev. 2014;17:68–78. doi: 10.1016/j.arr.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 25.Murdaca G, Colombo BM, Cagnati P, et al. Endothelial dysfunction in rheumatic autoimmune diseases. Atherosclerosis. 2012;224(2):309–317. doi: 10.1016/j.atherosclerosis.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 26.Newby AC. Metalloproteinase expression in monocytes and macrophages and its relationship to atherosclerotic plaque instability. Arterioscler Thromb Vasc Biol. 2008;28(12):2108–2114. doi: 10.1161/ATVBAHA.108.173898. [DOI] [PubMed] [Google Scholar]
- 27.Prisco AR, Hoffmann BR, Kaczorowski CC, et al. Tumor necrosis factor α regulates endothelial progenitor cell migration via CADM1 and NF-κB. Stem Cells. 2016;34(7):1922–1933. doi: 10.1002/stem.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santhekadur PK, Das SK, Gredler R, et al. Multifunction protein staphylococcal nuclease domain containing 1 (SND1) promotes tumor angiogenesis in human hepatocellular carcinoma through novel pathway that involves nuclear factor κB and miR-221. J Biol Chem. 2012;287(17):13952–13958. doi: 10.1074/jbc.M111.321646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shiao MS, Chang AYF, Liao BY, et al. Transcriptomes of mouse olfactory epithelium reveal sexual differences in odorant detection. Genome Biol Evol. 2012;4(5):703–712. doi: 10.1093/gbe/evs039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song DM, Fang GQ, Mao SZ, et al. Selective inhibition of endothelial NF-κB signaling attenuates chronic intermittent hypoxia-induced atherosclerosis in mice. Atherosclerosis. 2018;270:68–75. doi: 10.1016/j.atherosclerosis.2018.01.027. [DOI] [PubMed] [Google Scholar]
- 32.Song Y, Wu ZC, Ding W, et al. NF-κB in mitochondria regulates PC12 cell apoptosis following lipopolysaccharide-induced injury. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2018;19(6):425–435. doi: 10.1631/jzus.B1700488. [DOI] [Google Scholar]
- 33.Steyers CM III, Miller FJ., Jr Endothelial dysfunction in chronic inflammatory diseases. Int J Mol Sci. 2014;15(7):11324–11349. doi: 10.3390/ijms150711324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun SC, Ganchi PA, Ballard DW, et al. NF-κB controls expression of inhibitor IκBα: evidence for an inducible autoregulatory pathway. Science. 1993;259(5103):1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 35.Sun XH, He SL, Wara AKW, et al. Systemic delivery of microRNA-181b inhibits nuclear factor-κB activation, vascular inflammation, and atherosclerosis in apolipoprotein E-deficient mice. Circ Res. 2014;114(1):32–40. doi: 10.1161/CIRCRESAHA.113.302089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tousoulis D, Koutsogiannis M, Papageorgiou N, et al. Endothelial dysfunction: potential clinical implications. Minerva Med. 2010;101(4):271–284. [PubMed] [Google Scholar]
- 37.Vanhoutte PM. Endothelial dysfunction: the first step toward coronary arteriosclerosis. Circ J. 2009;73(4):595–601. doi: 10.1253/circj.CJ-08-1169. [DOI] [PubMed] [Google Scholar]
- 38.Varley CL, Armitage S, Hassanshahiraviz G, et al. Regulation of the C-X-C chemokine, mob-1, gene expression in primary rat hepatocytes. Cytokine. 2003;23(3):64–75. doi: 10.1016/S1043-4666(03)00198-4. [DOI] [PubMed] [Google Scholar]
- 39.Vincenti MP, Coon CI, Brinckerhoff CE. Nuclear factor κB/p50 activates an element in the distal matrix metalloproteinase 1 promoter in interleukin-1β-stimulated synovial fibroblasts. Arthritis Rheum. 1998;41(11):1987–1994. doi: 10.1002/1529-0131(199811)41:11<1987::AID-ART14>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 40.Wei W, Zhang WY, Bai JB, et al. The NF-κB-modulated microRNAs miR-195 and miR-497 inhibit myoblast proliferation by targeting Igf1r, Insr and cyclin genes. J Cell Sci. 2016;129(1):39–50. doi: 10.1242/jcs.174235. [DOI] [PubMed] [Google Scholar]
- 41.Widlansky ME, Gokce N, Keaney JF, Jr, et al. The clinical implications of endothelial dysfunction. J Am Coll Cardiol. 2003;42(7):1149–1160. doi: 10.1016/S0735-1097(03)00994-X. [DOI] [PubMed] [Google Scholar]
- 42.Xing YJ, Zhou F, Wang JK. Subset of genes targeted by transcription factor NF-κB in TNFα-stimulated human HeLa cells. Funct Integr Genomics. 2013;13(1):143–154. doi: 10.1007/s10142-012-0305-0. [DOI] [PubMed] [Google Scholar]
- 43.Yang JCS, Wu SC, Rau CS, et al. TLR4/NF-κB-responsive microRNAs and their potential target genes: a mouse model of skeletal muscle ischemia-reperfusion injury. Biomed Res Int, 2015:410721. 2015 doi: 10.1155/2015/410721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zampetaki A, Dudek K, Mayr M. Oxidative stress in atherosclerosis: the role of microRNAs in arterial remodeling. Free Radic Biol Med. 2013;64:69–77. doi: 10.1016/j.freeradbiomed.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 45.Zhang HC, Taylor WR, Joseph G, et al. mRNA-binding protein ZFP36 is expressed in atherosclerotic lesions and reduces inflammation in aortic endothelial cells. Arterioscler Thromb Vasc Biol. 2013;33(6):1212–1220. doi: 10.1161/ATVBAHA.113.301496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou F, Wang W, Xing YJ, et al. NF-κB target microRNAs and their target genes in TNFα-stimulated HeLa cells. Biochim Biophys Acta. 2014;1839(4):344–354. doi: 10.1016/j.bbagrm.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 47.Zhou F, Xu XH, Wang DY, et al. Identification of novel NF-κB transcriptional targets in TNFα-treated HeLa and HepG2 cells. Cell Biol Int. 2017;41(5):555–569. doi: 10.1002/cbin.10762. [DOI] [PubMed] [Google Scholar]
- 48.Zhu BB, Ye J, Ashraf U, et al. Transcriptional regulation of miR-15b by c-Rel and CREB in Japanese encephalitis virus infection. Sci Rep, 6:22581. 2016 doi: 10.1038/srep22581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu J, Zhu LWS, Yang JH, et al. Proteomic analysis of human umbilical vein endothelial cells exposed to PM2.5 . J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2018;19(6):458–470. doi: 10.1631/jzus.B1700103. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NF-κB-regulated genes in the TNFα-treated pMVECs