Abstract
Germline mutations in CDKN2A (p16) are commonly found in patients with family history of melanoma or personal history of multiple primary melanomas. The p16 tumor suppressor gene regulates cell cycle progression and senescence through binding of cyclin-dependent kinases (CDK) and also regulates cellular oxidative stress independently of cell cycle control. We identified a germline missense (c.350T>C, p.Leu117Pro) CDKN2A mutation in a patient who had history of four primary melanomas, numerous nevi, and self-reported family history of melanoma. This particular CDKN2A mutation has not been previously reported in prior large studies of melanoma kindreds or patients with multiple primary melanomas. Compared with wild-type p16, the p16L117P mutant largely retained binding capacity for CDK4 and CDK6 but exhibited impaired capacity for repressing cell cycle progression and inducing senescence, while retaining its ability to reduce mitochondrial reactive oxygen species. Structural modeling predicted that the Leu117Pro mutation disrupts a putative adenosine monophosphate (AMP) binding pocket involving residue 117 in the fourth ankyrin domain. Identification of this new likely pathogenic variant extends our understanding of CDKN2A in melanoma susceptibility and implicates AMP as a potential regulator of p16.
Keywords: AMP binding, CDKN2A, germline, melanoma, p16
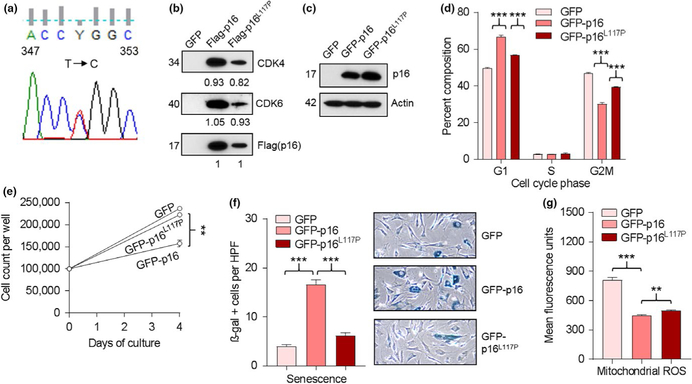
Germline mutations in CDKN2A (p16) have been reported in 20%–57% of melanoma patients with positive family history of melanoma (Goldstein et al., 2007) and in 11%–19% of those with multiple primary melanomas (Bruno et al., 2016; Hashemi et al., 2000). The p16 tumor suppressor gene regulates cell cycle progression and senescence through binding cyclin-dependent kinases (CDK) 4 and 6 and inhibition of the retinoblastoma pathway (Sharpless & DePinho, 1999). We have previously reported that p16 regulates cellular oxidative stress independently of cell cycle control (Jenkins et al., 2011) and that these activities were dissociated in various p16 mutants inherited in melanoma families (Jenkins et al., 2013). More recently, we showed that elevated reactive oxygen species (ROS) in p16-deficient cells are a consequence of aberrant mitochondrial biogenesis that is independent of CDK function (Tyagi et al., 2017). We encountered a male patient with history of four primary melanomas (first diagnosis at age 33), who had >50 nevi and self-reported family history of melanoma in his father and a sister. Germline determination of his melanocortin-1 receptor (MC1R) genotype (Cassidy et al., 2017) revealed homozygosity for a V60L mutation (not shown), considered to increase melanoma risk (Demenais et al., 2010). Sequencing of CDKN2A revealed a germline missense (c.350T>C, p.Leu117Pro) mutation (Figure 1a).
FIGURE 1.
Identification and characterization of the p16L117P mutant. Saliva was collected and genetic testing was performed under protocol 96762, approved by the University of Utah Institutional Review Board. Genomic DNA was isolated using an OGR-500 kit (DNA Genotek), and exons 1 and 2 of CDKN2A (Hashemi et al., 2000) were sequenced as described. (a) Chromatogram showing T → C substitution at residue 350 in CDKN2A. (b) A FLAG-tagged p16L117P construct was prepared by primer-directed site-directed mutagenesis (QuikChange II XL Kit, Agilent Technologies) using a wild-type human CDKN2A cDNA cloned into a Gateway (Thermo Fisher Scientific) vector with an N-terminal FLAG tag as a template. Both wild-type and p16L117P FLAG constructs were confirmed by DNA sequencing. Western blotting of anti-FLAG immunoprecipitates (1 mg eluates, Pierce Co-Immunoprecipitation Kit) from 293T cells (American Type Culture Collection) transfected with GFP (control vector), wild-type p16, or the p16L117P mutant using antibodies against CDK4 (Santa Cruz Biotechnology), CDK6 (Santa Cruz), or FLAG (Sigma). Numbers indicate densitometry values, normalized to FLAG signal for each construct. Representative of three experiments performed. (c) Western blotting of lysates of p16-deficient fibroblasts derived from p16−/−Arf+/+ mice (Jenkins et al., 2011) and obtained 48 h following lentiviral infection (Jenkins et al., 2011) with empty construct (GFP control) or constructs expressing wild-type p16 or the p16L117P mutant (site-directed mutagenesis as above, confirmed by sequencing). Representative of three experiments performed. (d) Cell cycle analysis by flow cytometry of propidium iodide-stained cells in (c). Error bars correspond to SEM of triplicate determinations. ***p < 0.001, 2-sided t tests. Representative of three experiments performed. (e) p16-deficient fibroblasts were infected with lentiviruses as in (c) for 2 days and then were plated into 6-well dishes (105 cells/well). Cell counts performed after 4 days. Error bars correspond to SEM of triplicate determinations. **p < 0.01, 2-sided t test. Representative of two experiments performed. (f) β-galactosidase staining of cells in (c). Error bars correspond to SEM of triplicate determinations. ***p < 0.001, 2-sided t tests. Representative photographs are shown. Representative of three experiments performed. (g) Detection of mitochondrial ROS by flow cytometry of cells in (c) stained with Mitosox Red (Life Technologies). Error bars correspond to SEM of triplicate determinations. ***p < 0.001, **p < 0.01, 2-sided t tests. Representative of three experiments performed
To assess the potential pathogenicity of this p16L117P variant, we initially characterized CDK-binding activity by immunoprecipitation. As shown in Figure 1b, p16L117P largely retained the capacity to bind CDK4 and CDK6 (as reflected by the ratios of their immunoblot intensities to the respective FLAG tag which served as an internal control) in 293T cells. Next, lentiviruses were used to express wild-type p16 and p16L117P in p16-deficient fibroblasts (Figure 1c) to assess potential differential effects on cell cycle and oxidative control (Jenkins et al., 2013). While wild-type p16 slowed cell cycle progression, as reflected by increase in the G1 fraction and decrease in the G2M fraction, this activity for the p16L117P mutant was significantly diminished (Figure 1d). Consistent with this result, the p16L117P mutant did not slow cell growth when expressed in p16-deficient fibroblasts compared to the wild-type p16 (Figure 1e). Similarly, the capacity of the wild-type p16 to induce senescence, as indicated by staining for β-galactosidase (Cotter, Florell, Leachman, & Grossman, 2007), was significantly reduced for the p16L117P mutant (Figure 1f). On the other hand, the ability of wild-type p16 to reduce mitochondrial ROS levels was preserved in the p16L117P mutant (Figure 1g). Thus, the Leu117Pro mutation in p16 impaired capacity for cell cycle control and senescence induction, but did not compromise oxidative regulation. We obtained similar results in these functional assays using human WM793 melanoma cells (Figure S1).
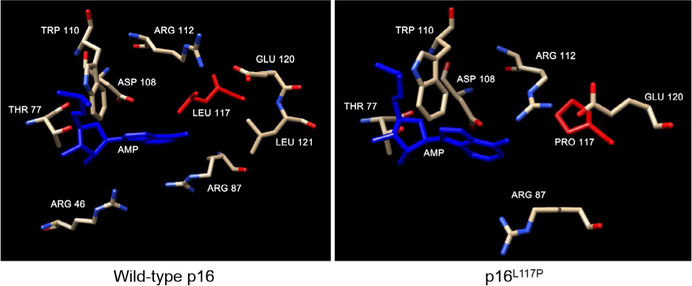
We performed structural modeling based on the published crystal structure of p16 (Russo, Tong, Lee, Jeffrey, & Pavletich, 1998). The 3D structures of p16 (Uniprot: P42771) and the Leu117Pro mutant were predicted using full homology modeling (Roy, Kucukural, & Zhang, 2010) and analyzed using Chimera visualization software (Pettersen et al., 2004). Analysis of the structures revealed overall agreement of predicted structure with the wild-type structure and that the overall structure of the Leu117Pro mutant was very similar to that of the wild type (Figure S2) with corresponding average root-mean-square deviation (RMSD) of atomic positions from the five best predicted superimposed structures of 1.12 and 1.06 and Å, respectively. The average RMSD between any two structures in the predicted sets was only 0.912 Å. Thus, any physiological effect of the mutation is likely related to changes in the local structure of a binding site in physical proximity to the mutation site. Modeling of potential binding pockets (Kallberg et al., 2012) revealed in both the wild-type and mutant proteins predicted an adenosine monophosphate (AMP) binding pocket involving residue 117 in the fourth ankyrin domain (Figure 2). This AMP binding pocket was found to be the most likely predicted binding pocket to exist in vivo through its multiplicity score. As depicted in Figure 2, there are significant structural differences between this pocket in the wild-type and Leu117Pro variant, showing that the orientation of the AMP ligand and the interacting residues in p16 are qualitatively different, possibly due to its loss of the two outlying binding residues. Because cyclic AMP (c-AMP) has been recognized as an important second messenger molecule (Berman et al., 2005) and c-AMP signaling is critical for differentiation and pigmentation in melanocytes (Rodriguez & Setaluri, 2014), we also investigated the probability of c-AMP binding to this site. However, negative results were obtained using the COACH (Yang, Roy, & Zhang, 2013) predictor tool (not shown). AMP is rapidly converted to adenosine, which binds to receptors that are highly conserved and broadly expressed (Holien et al., 2018), and is important in the regulation of cell proliferation via the AMP-activated protein kinase (Liu, Hu, Shan, Chen, & Tang, 2019). It is conceivable that AMP binding by p16 could contribute to the regulation of intracellular AMP levels and the maintenance of ATP balance during energy metabolism (Ke, Xu, Li, Luo, & Huang, 2018). Our findings thus provide impetus for further studies to elucidate the role of AMP binding to p16 in melanoma predisposition as well as treatment resistance. Our future studies will use molecular dynamics to assess the change in binding energies of this potential binding pocket attempting to further elucidate molecular mechanisms of pathogenicity of the mutation reported here as well as other p16 mutations we have previously characterized with loss of oxidative and/or cell cycle regulatory control (Jenkins et al., 2013).
FIGURE 2.
Structural modeling of the AMP binding site in the proximity of the L117P mutation in p16. Predicted structural elements of wild-type p16 (left panel) and p16L117P mutant (right panel). Structures of the wild-type and mutant were predicted using I-TASSER (Roy et al., 2010). Side chain of residue 117 is shown in red and AMP in blue. The other residues are depicted using a conventional color scheme (carbon in brown, nitrogen in blue, oxygen in red). The binding pocket for AMP was identified using RaptorX (Kallberg et al., 2012). The AMP binding site predictions were both found to be statistically significant (p < 0.05), and both the uSeqID/SeqID (unnormalized/normalized sequence identity) and uGDT/GDT (unnormalized/normalized global distance test) scores indicated high model and prediction quality, respectively (http://raptorx.uchicago.edu/documentation/#goto2)
We assessed the pathogenicity of the Leu117Pro mutation using common mutation effect predictor tools such as PolyPhen2 (Adzhubei, Jordan, & Sunyaev, 2013), FATHMM (Shihab et al., 2013), MutPred2 (Pejaver, Mooney, & Radivojac, 2017), and PANTHER (Mi, Muruganujan, & Thomas, 2013), which all predict that the mutation will have severe pathogenic effects. The results of PANTHER, which is an evolutionary/functional classification, are quite indicative of the seriousness of this variant. Because the mutation location is highly conserved in evolutionary times, it may be concluded that AMP binding in this location is very important for biological function and that any disruption of this binding may have serious biological impact that could be pathogenic. Moreover, MutPred2 predicts five statistically significant (p < 0.05) functional changes due to the Leu117Pro mutation, which are in order of significance: altered metal binding, loss of relative solvent accessibility, altered stability, altered transmembrane protein, and loss of helix (not shown). Germline mutations altering residues Arg47, Asp108, Trp110, Arg112, and Glu120 predicted to be adjacent to Leu117 (Figure 2) have been described in melanoma-prone families (McKenzie et al., 2010; Ruas & Peters, 1998). Mutations in these residues in cancers have been reported in both melanomas and squamous cell carcinomas in the Catalog of Somatic Mutations in Cancer (COSMIC, https://cancer.sanger.ac.uk/cosmic).
In summary, we have employed both experimental and modeling approaches to characterize a novel germline p16L117P mutation in a subject with both family history of melanoma and a personal history of multiple primary melanomas. To our knowledge, this mutation has not been previously reported in prior germline sequencing studies of patients with family history of melanoma or personal history of multiple primary melanomas (Hashemi et al., 2000; McKenzie et al., 2010; Orlow et al., 2007; Ruas & Peters, 1998). Although it was not possible to sequence CDKN2A in this subject’s family members to show co-segregation of the mutation with disease, we demonstrate that this p16L117P mutation compromises cell cycle regulatory function and is also likely to be pathogenic given structural modeling predictions. Given the conservation of oxidative regulatory function, this mutant appears to segregate with a group of previously characterized melanoma-predisposing p16 mutants (R99P, V126D, R24Q) in which we have demonstrated loss of cell cycle but retention of oxidative regulatory functions (Jenkins et al., 2013). Predicted effect on the AMP binding pocket by the Leu117Pro mutation suggests a role for p16 in AMP-mediated signaling in melanocytic cells, worthy of future investigation. Identification of this new pathogenic variant thus extends our understanding of CDKN2A in melanoma susceptibility.
Supplementary Material
Significance.
CDKN2A (p16) is a major melanoma predisposition gene. We identified a previously unreported germline mutation in p16 (p16L117P) in a patient with history of multiple primary melanomas and showed that this mutation impairs capacity for cell cycle control and senescence induction but does not compromise oxidative regulation. The Leu117Pro mutation is predicted to compromise an AMP binding site by structural modeling. Identification of this new pathogenic variant thus extends our understanding of CDKN2A in melanoma susceptibility.
ACKNOWLEDGEMENTS
D.G. was supported by NIH grant CA201757-01A1, the Department of Dermatology at the University of Utah, and the Huntsman Cancer Foundation. R.H. and J.F. have been partially supported by the Utah Center for Clinical and Translational Science funded by NCATS award 1ULTR001067 and a NLM Training grant T15-LM007124–18. Computer resources were provided by the University of Utah Center for High Performance Computing, which has been partially funded by the NIH Shared Instrumentation Grant 1S10OD021644-01A1.
Funding information
National Cancer Institute, Grant/Award Number: CA201757-01A1; U.S. National Library of Medicine, Grant/Award Number: T15-LM007124–18; National Center for Advancing Translational Sciences, Grant/Award Number: 1ULTR001067
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- Adzhubei I, Jordan DM, & Sunyaev SR (2013). Predicting functional effect of human missense mutations using PolyPhen-2. Current Protocols in Human Genetics. 10.1002/0471142905.hg0720s76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Ten Eyck LF, Goodsell DS, Haste NM, Kornev A, & Taylor SS (2005). The cAMP binding domain: An ancient signaling module. Proceedings of the National Academy of Sciences of the United States of America, 102, 45–50. 10.1073/pnas.0408579102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno W, Pastorino L, Ghiorzo P, Andreotti V, Martinuzzi C, Menin C, … Bianchi-Scarrà G (2016). Multiple primary melanomas (MPMs) and criteria for genetic assessment: MultiMEL, a multicenter study of the Italian Melanoma Intergroup. Journal of the American Academy of Dermatology, 74, 325–332. 10.1016/j.jaad.2015.09.053 [DOI] [PubMed] [Google Scholar]
- Cassidy PB, Liu T, Florell SR, Honeggar M, Leachman SA, Boucher KM, Grossman D (2017). A phase II randomized placebo-controlled trial of oral N-acetylcysteine for protection of melanocytic nevi against UV-induced oxidative stress in vivo. Cancer Prevention Research (Philadelphia, PA), 10, 36–44. 10.1158/1940-6207.CAPR-16-0162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter MA, Florell SR, Leachman SA, & Grossman D (2007). Absence of senescence-associated beta-galactosidase activity in human melanocytic nevi in vivo. Journal of Investigative Dermatology, 127, 2469–2471. 10.1038/sj.jid.5700903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demenais F, Mohamdi H, Chaudru V, Goldstein AM, Newton Bishop JA, Bishop DT, … Gruis N (2010). Association of MC1R variants and host phenotypes with melanoma risk in CDKN2A mutation carriers: A GenoMEL study. Journal of the National Cancer Institute, 102, 1568–1583. 10.1093/jnci/djq363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AM, Chan M, Harland M, Hayward NK, Demenais F, Bishop DT, … Yakobson E (2007). Features associated with germline CDKN2A mutations: A GenoMEL study of melanoma-prone families from three continents. Journal of Medical Genetics, 44, 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi J, Platz A, Ueno T, Stierner U, Ringborg U, & Hansson J (2000). CDKN2A germ-line mutations in individuals with multiple cutaneous melanomas. Cancer Research, 60, 6864–6867. [PubMed] [Google Scholar]
- Holien JK, Seibt B, Roberts V, Salvaris E, Parker MW, Cowan PJ, & Dwyer KM (2018). AMP and adenosine are both ligands for adenosine 2B receptor signaling. Bioorganic and Medicinal Chemistry Letters, 28, 202–206. 10.1016/j.bmcl.2017.11.019 [DOI] [PubMed] [Google Scholar]
- Jenkins NC, Jung J, Liu T, Wilde M, Holmen SL, & Grossman D (2013). Familial melanoma-associated mutations in p16 uncouple its tumor-suppressor functions. Journal of Investigative Dermatology, 133, 1043–1051. 10.1038/jid.2012.401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins NC, Liu T, Cassidy P, Leachman SA, Boucher KM, Goodson AG, … Grossman D (2011). The p16(INK4A) tumor suppressor regulates cellular oxidative stress. Oncogene, 30, 265–274. 10.1038/onc.2010.419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J (2012). Template-based protein structure modeling using the RaptorX web server. Nature Protocols, 7, 1511–1522. 10.1038/nprot.2012.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke R, Xu Q, Li C, Luo L, & Huang D (2018). Mechanisms of AMPK in the maintenance of ATP balance during energy metabolism. Cell Biology International, 42, 384–392. 10.1002/cbin.10915 [DOI] [PubMed] [Google Scholar]
- Liu Y, Hu X, Shan X, Chen K, & Tang H (2019). Rosiglitazone metformin adduct inhibits hepatocellular carcinoma proliferation via activation of AMPK/p21 pathway. Cancer Cell International, 19, 13 10.1186/s12935-019-0732-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie HA, Fung C, Becker TM, Irvine M, Mann GJ, Kefford RF, Rizos H (2010). Predicting functional significance of cancer-associated p16(INK4a) mutations in CDKN2A. Human Mutation, 31, 692–701. 10.1002/humu.21245 [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, & Thomas PD (2013). PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Research, 41, D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlow I, Begg CB, Cotignola J, Roy P, Hummer AJ, Clas BA, … Berwick M (2007). CDKN2A germline mutations in individuals with cutaneous malignant melanoma. Journal of Investigative Dermatology, 127, 1234–1243. 10.1038/sj.jid.5700689 [DOI] [PubMed] [Google Scholar]
- Pejaver V, Mooney SD, & Radivojac P (2017). Missense variant pathogenicity predictors generalize well across a range of function-specific prediction challenges. Human Mutation, 38, 1092–1108. 10.1002/humu.23258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004). UCSF Chimera – A visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25, 1605–1612. 10.1002/(ISSN)1096-987X [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, & Setaluri V (2014). Cyclic AMP (cAMP) signaling in melanocytes and melanoma. Archives of Biochemistry and Biophysics, 563, 22–27. 10.1016/j.abb.2014.07.003 [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, & Zhang Y (2010). I-TASSER: A unified platform for automated protein structure and function prediction. Nature Protocols, 5, 725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruas M, & Peters G (1998). The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochimica et Biophysica Acta, 1378, F115–F177. [DOI] [PubMed] [Google Scholar]
- Russo AA, Tong L, Lee JO, Jeffrey PD, & Pavletich NP (1998). Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature, 395, 237–243. 10.1038/26155 [DOI] [PubMed] [Google Scholar]
- Sharpless NE, & DePinho RA (1999). The INK4A/ARF locus and its two gene products. Current Opinion in Genetics and Development, 9, 22–30. 10.1016/S0959-437X(99)80004-5 [DOI] [PubMed] [Google Scholar]
- Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, … Gaunt TR (2013). Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Human Mutation, 34, 57–65. 10.1002/humu.22225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi E, Liu B, Li C, Liu T, Rutter J, & Grossman D (2017). Loss of p16(INK4A) stimulates aberrant mitochondrial biogenesis through a CDK4/Rb-independent pathway. Oncotarget, 8, 55848–55862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Roy A, & Zhang Y (2013). Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics, 29, 2588–2595. 10.1093/bioinformatics/btt447 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.