Abstract
Vacuolar protein sorting-associated protein 13B (VPS13B), also known as COH1, is one of the VPS13 family members which is involved in transmembrane transport, Golgi integrity, and neuritogenesis. Mutations in the VPS13B gene are associated with Cohen syndrome and other cognitive disorders such as intellectual disabilities and autism spectrum disorder (ASD). However, the patho-physiology of VPS13B-associated cognitive deficits is unclear, in part, due to the lack of animal models. Here, we generated a Vps13b exon 2 deletion mutant mouse and analyzed the behavioral phenotypes. We found that Vps13b mutant mice showed reduced activity in open field test and significantly shorter latency to fall in the rotarod test, suggesting that the mutants have motor deficits. In addition, we found that Vps13b mutant mice showed deficits in spatial learning in the hidden platform version of the Morris water maze. The Vps13b mutant mice were normal in other behaviors such as anxiety-like behaviors, working memory and social behaviors. Our results suggest that Vps13b mutant mice may recapitulate key clinical symptoms in Cohen syndrome such as intellectual disability and hypotonia. Vps13b mutant mice may serve as a useful model to investigate the pathophysiology of VPS13B-associated disorders.
Keywords: Vps13b, Cohen syndrome, Learning disability
INTRODUCTION
VPS13B (also known as COH1) is one of the VPS13 protein family members which consist of four mammalian members; VPS13A, VPS13B, VPS13C, and VPS13D [1]. Each of VPS13 gene members except VPS13D has been shown to be associated with neurological disorders such as chorea-acanthocytosis and Parkinson’s disease [2–4]. VPS13B is located on chromosome 8q22, and spans 864 kb of genomic DNA sequences with 62 exons encoding a putative transmembrane protein of 4,022 amino acids [1, 5]. VPS13B was shown to be involved in the maintenance and function of Golgi complex, neuronal outgrowth, adipogenesis, and glycosylation [5–9]. Mutations in VPS13B gene have been identified in individuals with Cohen syndrome, a rare autosomal recessive disorder [10]. Clinical features of Cohen syndrome involve developmental delay, intellectual disability, microcephaly, hypotonia, and more [11–13]. In addition, VPS13B mutations have been found in individuals with ASD, which is a group of neurodevelopmental disorders characterized by impaired social-communication, and repetitive behaviors and restricted interests [14, 15]. Among VPS13 families, only Vps13a mutant mice are available as a model for chorea-acanthocytosis [16, 17].
In this study, we generated the first rodent model of Vps13b mutation by using CRISPR/Cas9 system. Vps13b exon 2 deletion mice showed impaired motor activity and spatial learning, suggesting that the Vps13b mutant mice can be a useful model to investigate the mechanism underlying neurological deficits associated with Cohen syndrome and other VPS13B-related diseases.
MATERIALS AND METHODS
Generation of Vps13b exon 2 deletion mutant mouse
Vps13b mutant mice were generated by using CRISPR/Cas9 system (Macrogen, Inc). Cas9 mRNA and single guide (sg) RNAs were microinjected to fertilized embryos of C57BL/6N mice. sgRNAs were designed to mark exon 2 of mouse Vps13b by using following sequences: 5′-ACGCTTAAATTTGAAGATGCTGG-3′ and 5′-CGAGTTAAAGTTGGACGTTCTGG-3′. Among 17 pups, deletion of vps13b was affirmed in 9 mice by mismatch sensitive nuclease (T7E1) assay. Sequences used for T7E1 assays were 5′-GCCAAGCTTCCTGAGAAGTG-3′ and 5′-ACGAG-GCAAAAAGCCTTGTA-3′. Mutations were confirmed by Sanger sequencing analyses. One of the male founder (#45) concealing 156bp deletion in exon2 was selected for breeding. The founder was crossed with wild type female mouse (C57BL/6N) to obtain heterozygous mutants. Male and female heterozygous mutants were crossed to obtain homozygous mutants. Each genotype was identified by PCR using primers used for T7E1 assay. Mice were bred up on a 12 h–12 h light-dark circadian cycle, and all behavioral experiments were conducted during the light phase. Food and water were always provided ad libitum. All experiments in this study were approved by the Institutional Animal Care and Use Committees at Seoul National University (SNU-130725-6).
RNA isolation, cDNA synthesis, RT-PCR, quantitative real-time PCR
RNA isolation method was previously described [18, 19]. Briefly, RNA was extracted from hippocampus of wild type mice, Vps13b heterozygous and homozygous mutant mice, respectively. Hippocampal tissue was homogenized with TRIZOL reagent (Invitrogen) and bead using Tissue lyser. Total RNA from tissue lysate was prepared using RNeasy MiniElute kit (Qiagen).
RT-PCR was performed as previously described [20]. Briefly, cDNA was synthesized by using SuperScriptIII and random hexamers following manufacturer’s instructions (Invitrogen). Quantitative real-time (qRT)-PCR was conducted with SYBR Premix Ex TaqII (Cat. # RR820A, Takara) and 7300 Real-Time PCR system (Applied Biosystems). Primers for Vps13b used in quantitative and conventional RT-PCR were shown in Table 1. Primers for Gapdh were 5′-GGAGCGAGATCCCTCCAAAAT-3′ and 5′-GGCT-GTTGTCATACTTCTCATGG-3′.
Table 1.
Primer sequences for RT-PCR
| Vps13b | Sense primer (5′ 3′) | Antisense primer (5′ 3′) | Product size (bp) |
|---|---|---|---|
| Exon 1–4 | 1:TCCAGCCTCTCTGCCTACTC | 5:CGGGGTTTTATGGATGATTTT | 424 bp (intact) 268 bp (del) |
| Exon 2–3 | 2:CGAGTTAAAGTTGGACGTTCTGG | 3:AGGTTCGGAGCCCAATTTTGT | 115 bp |
| Exon 3–4 | 4:AGTGGCCATATCCATGAACTG | 5:CGGGGTTTTATGGATGATTTT | 194 bp |
| Exon 22–23 | 6:CAGTAAAGAGTCTCACGCTACAG | 7:TGTTCCAGGGATGTCACCAGA | 110 bp |
Behavioral analysis
We used 2~5 month-old male (25~33 g) and female (18~22 g) mice for the behavioral experiments. Since we did not find any significant difference between male and female, we merged the results from both sexes. Behavioral analyses were performed by a test battery in a single cohort as the following order. Mice were tested in open field, elevated zero maze, light-dark box and Y-maze on consecutive days. After Y-maze, mice were handled for 5 days and trained in Morris water maze. Three days after finishing the water maze task, mice were trained for fear conditioning test. Three chamber test was performed 19 days after the fear conditioning test. Rotarod test was independently performed in a different cohort. The genotypes of the mice were blinded to the experimenters.
Open field test
Procedure for open field test was previously described [21]. Mice were placed in the center of open field box (40×40×40 cm) under dim light. Free movement of the mice were recorded for 30 min and analyzed by tracking software, EthoVision 9.0 (Noldus).
Elevated zero maze test
Elevated zero maze test was performed as described [22]. The maze (round-shaped, inner diameter: 50 cm, outer diameter: 60 cm, height of two walls: 20 cm) includes two close arms and two open arms, elevated to a height of 65 cm above the floor. Mice were placed in the center of the one closed arm, and allowed to move all arms freely for 10 min under bright light. The time spent in each arms was analyzed by EthoVision 9.0 (Noldus).
Light-dark box test
Specific experimental procedures were previously described [19, 23]. Briefly, light-dark box apparatus includes both light and dark chambers adhered to each other, and the mice could move freely from one another through the gate. The illumination of light chamber (20×30×20 cm, a large compartment) was 404 – 408 lux, while dark chamber (20×13×20 cm, a small apartment) was fully blocked from illuminator. Mice were first introduced to the dark chamber, and free movement between two chambers was recorded for 10 min. The time spent in each chamber was manually and blindly counted.
Y-maze test
We followed the protocols previously described [24]. Mice encountered to the center of the Y-maze with three identical arms at a 120o angle from each other under dim light. The movement was recorded for 8 min. Each arm was 30 cm long, 6 cm wide, and 15 cm height. The sequence of entries and the total number of each arm entries were recorded, and manually and blindly counted.
Morris water maze test
Test was performed as previously described [22, 24–27]. Briefly, mice were handled for 3 min for 5 successive days. Mice were trained in a swimming pool (140 cm diameter, 100 cm height) filled with a soluble white paint with multiple spatial cues on the walls of the test room. The temperature of the water was 21~22 degree of Celsius. Mice were released from four edges of the maze respectively, and allowed to find the hidden platform (round-shaped, 10 cm diameter), which was placed at the center of a target quadrant (TQ), within 60 s. Mice were trained with 4 trials (start swimming from four different edges of the maze) per day. Probe tests were conducted at day 7 and 11 with an absence of the platform, and training was not performed on the probe test days. The visual platform test was performed by placing a black flag on the platform after finishing hidden platform version of water maze test. The time spent in each quadrant and mean velocity were scored by EthoVision 9.0 (Noldus).
Contextual fear conditioning
Mice were trained and tested as formerly described [20, 25]. For conditioning, mice were located in the conditioning chamber (Coulbourn Instruments) for 3 min, and an electrical foot shock (3 s, 0.8 mA) was given through the floor grid. After the conditioning, mice went back to the home cages. After 24 h later, mice were disclosed to the same chamber for 3 min without a shock to recognize whether mice remember electric shock (fear memory) or not. Freezing level was automatically quantified by a software (Freeze Frame, Actimetrics).
Three chamber test
Sociability and social novelty recognition tests were conducted as previously described [19]. An apparatus we used had three sequential chambers and each chamber was connected with gates. For sociability test, a mouse (stranger 1) and an object were placed in a wired-cup sitting in two end chamber of the apparatus. A test mouse was placed into the center chamber and allowed to move whole apparatus for 10 min. For social novelty recognition test, the object was replaced to another mouse (stranger 2), with which test mouse had not met ever, and the test mouse was introduced and allowed to explore between stranger 1 and 2. The time spent exploring object, stranger 1, or stranger 2 was manually counted.
Rotarod test
Detailed experimental procedures were previously described [28]. Mice were habituated to the rotarod apparatus (LE8200, Harvard apparatus) slowly accelerated from 4 to 20 rpm over 11 min for two days. On the test day, the rod was accelerated from 4 to 40 rpm over 5 min and the latency to fall off the rod was recorded. The test was replayed for 3 days and the average time to fall was analyzed.
Statistical analysis
Data were analyzed followed procedures previously defined [29]. Statistical comparisons were made by using unpaired t-test for pairwise comparison and one-way or two-way ANOVA for multiple comparisons with appropriate post-hoc tests. In all cases, statistical significance was represented by *p<0.05, **p<0.01 and ***p<0.001. All data shown are mean±SEM. All the data were analyzed by using a software (GraphPad Prism 5).
RESULTS
Generation of Vps13b mutant mice
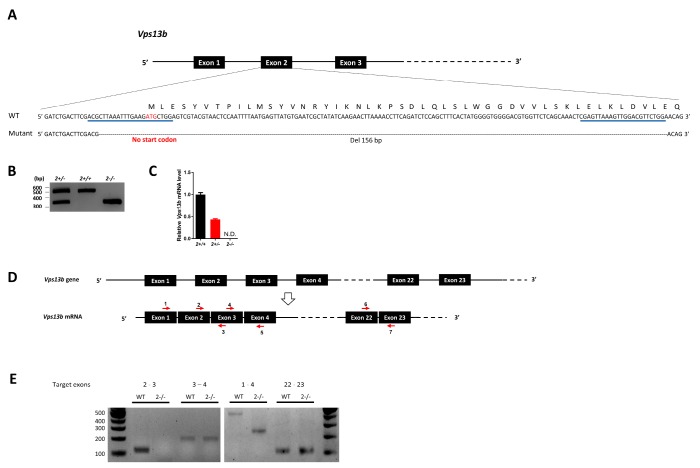
To model VPS13B-assoaicted disorders in rodent, we generated a Vps13b exon 2 deletion mutant mouse by using CRISPR/Cas9 system [30]. Single guide RNAs (sgRNAs) targeting the exon 2 of the Vps13b gene containing the start codon were injected into the mouse embryos (C57BL/6N background) together with Cas9 mRNA. Deletion mutants were screened by the mismatch sensitive nuclease assay and the deletions were found in 9 out of 17 mice. We selected a line harboring a 156 bp deletion around the start codon by performing Sanger sequencing (Fig. 1A). Genotypes were confirmed by PCR in which three genotypes were clearly distinguished (Fig. 1B). Heterozygous mice were crossed each other to obtain wild type (WT), heterozygous and homozygous mutant mice. We confirmed that the exon 2 is deleted in Vps13b mRNA in the hippocampus of Vps13b2−/− mice, while the transcript level is reduced by half in Vps13b2+/− mice (Fig. 1C). We found that other exons are normally expressed (Fig. 1D and 1E), suggesting that the deletion of exon 2 does not decrease the stability of the mutant mRNA.
Fig. 1.
Generation of Vps13b2−/− mice. (A) A schematic view of the CRISPR/Cas9-mediated generation of Vps13b mutant mice lacking exon 2. The nucleotide and amino acid sequences of wild type (WT) and mutant alleles are shown. The start codon (ATG) is indicated in red and the targeting sites for sgRNAs are underlined in blue. (B) A representative PCR genotyping result for Vps13b WT and mutants. (C) Relative Vps13b mRNA levels in the hippocampus of wild type, heterozygous and homozygous mutant mice measured by quantitative RT-PCR spanning exon 2–3. The mRNA levels were normalized by Gapdh mRNA levels. Vps13b2+/+, n=3 mice; Vps13b2+/−, n=2 mice Vps13b2−/−, n=4 mice. Experiments were triplicated. Data were represented as mean±SEM. (D) Schematics of genomic DNA and mRNA of Vps13b. The target sites of the primers used for RT-PCR are indicated by red arrows. (E) Conventional RT-PCRs were performed by using primers targeting different exons as indicated and cDNAs synthesized from total RNA extracted from the hippocampus. Note that primers targeting exons 2–3 did not produce any PCR product in Vps13b2−/− as shown in (C). Consistently, primers targeting exons 1–4 produce the shorter PCR product, confirming that the exon 2 is deleted in Vps13b2−/−.
Hypoactivity and impaired motor coordination in Vps13b2−/− mice
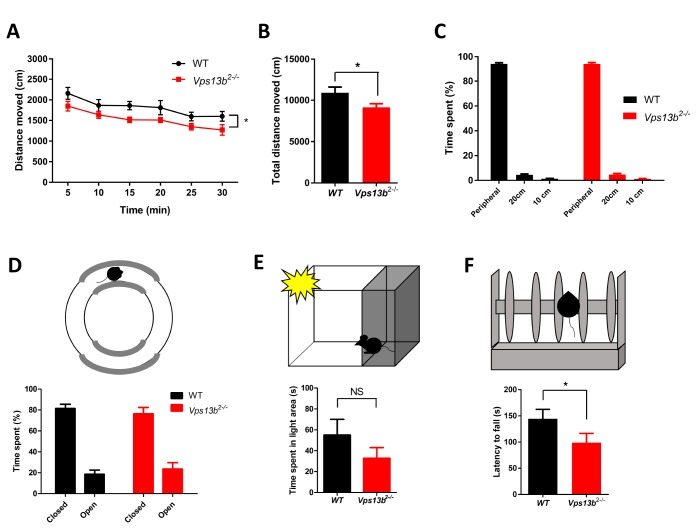
We investigated the behavioral phenotypes of the Vps13b2−/− mice in a series of analyses. Firstly, mice were subjected to open field test to examine the basal motor activity (Fig. 2A~C). Vps13b2−/− mice travelled the less distance during the test under dim light than WT littermates, suggesting that the Vps13b2−/− mice might be hypoactive (Fig. 2A and 2B). The time spent in the peripheral and the center of the arena were not different between genotypes, suggesting that the mutant mice show comparable anxiety level to WT littermates (Fig. 2C). Anxiety-like behavior was further examined in zero maze and light-dark box, where Vps13b2−/− and WT mice showed comparable performance, confirming that Vps13b2−/− mice show normal anxiety-related behaviors (Fig. 2D and 2E). To evaluate the motor coordination, the mice were tested on the accelerating rotarod. Vps13b2−/− mice fell off earlier than WT mice, suggesting that the motor coordination is impaired in Vps13b2−/− mice compared to WT littermates (Fig. 2F).
Fig. 2.
Reduced activity and impaired motor coordination in Vps13b2−/− mouse. (A) Distance moved measured in 5-min intervals across the 30-min test session in open field test (WT: n=9 mice; Vps13b2−/−: n=12 mice; two-way ANOVA, genotype x time, F 5,95=0.23, p=0.948; effect of genotype, F 1,95=5.04, p=0.037; effect of time, F 5,95=18.30, p<0.0001). (B) Total distance moved in open field test (WT: 10940±685.0 cm, n=9 mice; Vps13b2−/−: 9083±452.4 cm, n=12 mice; unpaired t-test, *p<0.05). (C) Time spent in peripheral (40 cm) and center (20 cm, 10 cm) zones in open field test (WT: n=9 mice; Vps13b2−/−: n=12 mice; two-way ANOVA, genotype x zone, F 2,38=0.01, p=0.990; effect of genotype, F 1,38=0.23, p=0.639). (D) Upper, Elevated zero maze apparatus. Lower, Time spent in closed and open compartment (WT: n=9 mice; Vps13b2−/−: n=12 mice; two-way ANOVA, genotype x sector, F 1,19=0.42, p=0.524; effect of genotype, F 1,19=1.86, p=0.189; effect of sector, F 1,19=54.49, p<0.0001). (E) Upper, Light dark box test apparatus. Lower, Time spent in lit area (WT: 54.98±15.04 s, n=9 mice; Vps13b2−/−: 32.79±10.29 s, n=12 mice; unpaired t-test, p=0.223). (F) Upper, Rotarod test apparatus. Lower, Latency to fall in accelerating (4 to 40 rpm across 5 min) rotarod test (WT: n=9 mice; Vps13b2−/−: n=9 mice; unpaired t-test, *p<0.05). Latency measured in three repeated trials were averaged. Data were represented as mean±SEM.
Impaired spatial learning in Vps13b2−/− mice
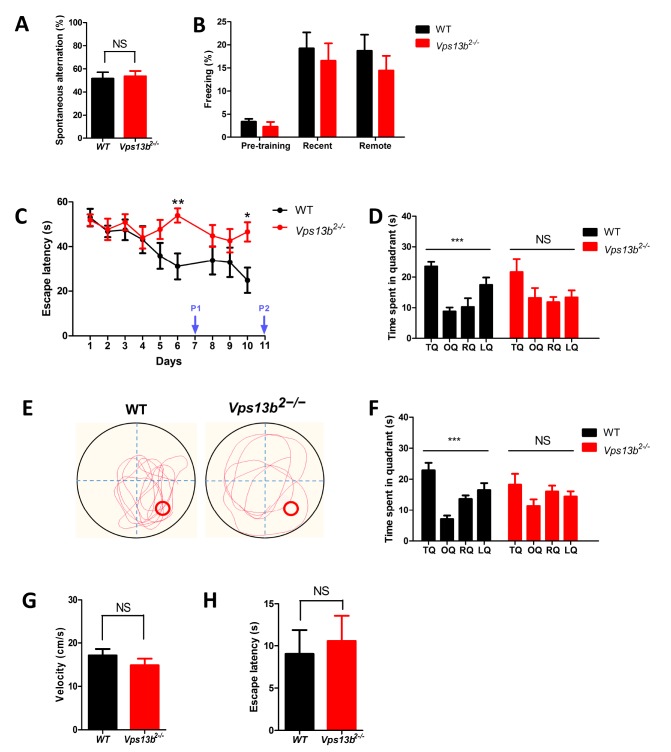
We examined the Vps13b2−/− mice in different types of learning and memory tests (Fig. 3). Working memory was assessed in Y-maze task, where the normal mice show spontaneous alterations among three arms (Fig. 3A). Vps13b2−/− mice showed the percentage of alteration comparable to that of WT littermates, suggesting that working memory assessed in Y maze is intact in the mutant mice (Fig. 3A). Hippocampus-dependent associative memory was assessed by using contextual fear conditioning paradigm (Fig. 3B). The freezing levels were comparable between genotypes when the memory was tested either 1 day or 4 weeks after training, showing the recent (1 day) and the remote (4 weeks) contextual fear memory are intact in Vps13b2−/− mice (Fig. 3B).
Fig. 3.
Spatial learning and memory deficit in Vps13b2−/− mouse. (A) Alternation percentage in Y maze test (WT: 51.67±5.40%, n=9 mice; Vps13b2−/−: 53.59±4.66%, n=12 mice; unpaired t-test, p=0.790). (B) Freezing levels in contextual fear conditioning. Freezing during pre-training test, retrieval 1 (recent), and retrieval 2 (remote) were tested (WT: n=9 mice, Vps13b2−/−: n=11 mice; two-way ANOVA, genotype x condition, F 2,54=0.15, p=0.862; effect of genotype, F 1,54 = 1.27, p=0.265; effect of condition, F 2,54=16.31, p <0.0001; Bonferroni posttests, NS: not significant). All data shown are mean±SEM. (C) The learning curve showing escape latency during training sessions (WT: n=9 mice, Vps13b2−/−: n=12 mice; two-way ANOVA, genotype x time, F 8,152=2.69, **p<0.001, effect of genotype, F 1,152=4.06, p=0.058, effect of time, F 8,152=4.100, ***p < 0.0001). (D) Time spent in each quadrant in probe test 1 at day 7 (WT: n=9 mice; Vps13b2−/−: n=12 mice; one-way ANOVA: WT, ***p<0.0001; Vps13b2−/−, p=0.106). (E) Representative swimming traces of WT and Vps13b2−/− mice during the probe test at day 11. (F) Time spent in each quadrant in probe test 2 at day 11 (WT: n=9 mice, one-way ANOVA: WT, ***p<0.0001; Vps13b2−/−: n=10 mice, p=0.242). (G) Mean velocity at probe test 1 (WT: 17.13±1.493 cm/s, n=9 mice; Vps13b2−/−: 14.86±1.522 cm/s, n=12 mice; unpaired t-test, p=0.311). (H) The escape latency in visible-platform version of the water maze (WT: 9.039±2.847 s; n=9 mice, Vps13b2−/−: 10.56±3.004 s, n=12 mice; unpaired t-test, p=0.725). All data shown are mean±SEM.
Next, we assessed hippocampus-dependent spatial learning and memory by testing the mice in the Morris water maze test. In the hidden platform version of this task, mice were trained to learn spatial cues around the maze to find a platform hidden under the water during training sessions (Fig. 3C). Hippocampus-dependent memory can be assessed in probe tests in which the platform is removed. We trained the mice for 10 days and preformed probe tests at day 7 and day 11 (Fig. 3D and 3E). Vps13b2−/− mice showed significantly longer latency to find the platform during training sessions, suggesting that the spatial learning is impaired in the mutants (Fig. 3C). In the first probe test (P1), Vps13b2−/− mice did not search selectively in the target quadrant where the platform was located during training sessions (Fig. 3D). Even with additional trainings, Vps13b2−/− mice did not spend significantly more time in the target quadrant in the second probe test (P2) [Fig. 3E and 3F]. Importantly, although the mutant mice were hypoactive in open field test (Fig. 2), swimming speeds during the probe tests were not different between genotypes (Fig. 3G). Furthermore, Vps13b2−/− mice showed comparable performance to WT littermates in the visible platform version of the water maze wherein a visible flag was placed on top of the platform, demonstrating that the mutant mice are not impaired in motivation, visuomotor function required to perform the water maze task (Fig. 3H).
Normal social behavior in Vps13b2−/− mice
Since VPS13B mutation was found in an autism patient [15], we examined whether Vps13b exon 2 deletion mutation affects social behaviors in mice by using three-chamber test. Social preference and social novelty recognition were examined (Fig. 4A). Both WT and the mutant mice showed significant preference to a novel mouse over a novel object and showed comparable sociability index (Fig. 4B). Also, the mutant mice were not impaired in social novelty recognition test (Fig. 4C). Taken together, Vps13b exon 2 deletion did not affect social behaviors in mice.
Fig. 4.
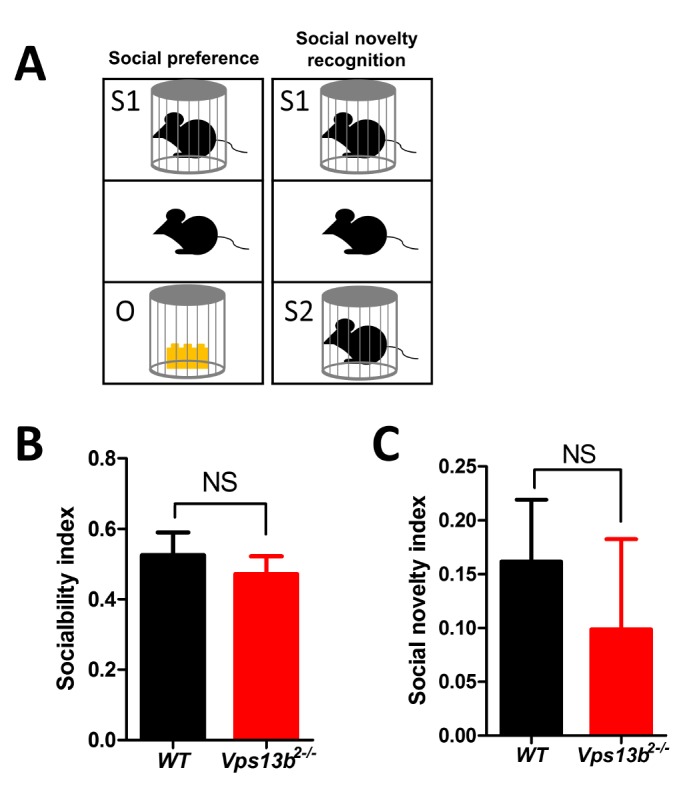
Normal social behavior in Vps13b2−/− mouse. (A) Schematics for social preference and social novelty recognition test in three-chamber test. (B) Sociability index was calculated by [Time spent interacting with object-stranger 1]/[Time spent interacting with object+stranger 1] (WT: 0.53±0.06, n=9 mice; Vps13b2−/−: 0.47±0.05, n=12 mice; unpaired t-test, p=0.516). (C) Sociability novelty index was calculated by [Time spent interacting with stranger 1-stranger 2]/[Time spent interacting with stranger 1+stranger 2] (WT: 0.16±0.06, n=9 mice; Vps13b2−/−: 0.10±0.08, n=12 mice; unpaired t-test, p=0.571). NS, not significant. All data shown are mean±SEM.
DISCUSSION
We have generated the first mouse model of Vps13b exon 2 deletion and examined behavioral phenotypes of the mutant mice. Major phenotypes are abnormalities in motor behaviors and spatial learning. Vps13b2−/− mice showed hypoactivity in the open field test. Motor coordination was further examined in the rotarod test, where the mutant mice showed clear deficits compared to WT littermates. These results suggest that Vps13b exon 2 deletion mutant may recapitulate motor deficits in Cohen syndrome patients such as hypotonia and motor clumsiness [13]. However, we could not find any morphological changes including microcephaly, retinal dystrophy and gross developmental delay in Vps13b2−/− mice compared to WT mice, which might be stemmed from great phenotypic variability even among human patients. In this study, we generated the Vps13b mutant by CRISPR/Cas9-mediated gene deletion. Mutant mice by knocking-in specific alleles identified in the human patients might recapitulate more features shown in Cohen syndrome. Vps13b is highly expressed in cerebellum (http://mouse.brain-map.org/gene/show/156104), a primary brain area controlling motor coordination and motor learning [31]. Interestingly, a recent study reported cerebellar hypoplasia in Cohen syndrome patients [32]. Further studies are required to examine if Vps13b exon 2 deletion affects cerebellar functions in mice.
Vps13a mutant mice have been generated as a chorea-acanthocytosis mouse model [17]. Chorea-acanthocytosis is a neurodegenerative disorder showing Huntington disease-like symptoms along with acanthocytosis in erythrocyte, which is associated with mutations in Vps13a [33, 34]. Interestingly, Vps13a mutant mice lacking exon 60 and 61 showed deficits in motor function in the open field and rotarod test, which are similar to behavioral phenotypes of Vps13b2−/− [17]. Vps13a mutant mice also showed significant deficits in social interaction [17], whereas Vps13b2−/− mice showed normal social behaviors. However, the behavioral and other phenotypes in Vps13a mutant were sensitive to the genetic background of the mice [16], suggesting that it would be worthwhile to examine phenotypes of Vps13b2−/− in another genetic backgrounds.
Although the Vps13b2−/− mice showed deficits in motor tests, the mutant mice performed normally in the visible platform version of the water maze, which enabled us to examine the spatial learning and memory of the mutant mice in the hidden platform version of the water maze. It is common that the motor phenotypes of a mutant mouse are different depending on the behavioral tests and environmental condition probably because each task requires different aspects of motor skills and motivation. We found that Vps13b exon 2 deletion significantly impaired learning in the water maze. Vps13b2−/− mice took significantly more time to find the hidden platform during training. Consistently, Vps13b2−/− mice did not show selective search in the target quadrant during the probe tests, demonstrating that Vps13b mutation significantly impairs spatial learning and memory. Interestingly, the mutant mice showed comparable performances to WT littermates in contextual fear conditioning test which is another hippocampus dependent task. The discrepancy may be derived from the differences between two tasks. Morris water maze is more complex and demanding behavioral task which requires multiple training sessions compared to the fear conditioning which is a simple one time associative learning [35, 36]. In addition, fear conditioning also involves emotional circuits including amygdala which may not be severely affected in the mutant mice [37]. Further electrophysiological studies such as examining synaptic plasticity in the hippocampus may reveal mechanisms for spatial learning and memory deficit in Vps13b mutant mice and shed light into understanding the pathophysiology of learning disability associated with Cohen syndrome.
ACKNOWLEDGEMENTS
Authors thank Macrogen for technical service generating Vps13b mutant mice. This work is supported by the National Honor Scientist Program (NRF-2012R1A3A1050385) through a grant to B.-K. K.; NRF-2016R1A1A3A04005452 and NRF-2017R1A3A1078210 to K.L.; NRF-2016R1D1A1B03931525 to C.-S.L.; NRF-2017M3C7A1026959 and NRF-2016R1E1A1A01941939 to Y.-S. L.
REFERENCES
- 1.Velayos-Baeza A, Vettori A, Copley RR, Dobson-Stone C, Monaco AP. Analysis of the human VPS13 gene family. Genomics. 2004;84:536–549. doi: 10.1016/j.ygeno.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 2.Lesage S, Drouet V, Majounie E, Deramecourt V, Jacoupy M, Nicolas A, Cormier-Dequaire F, Hassoun SM, Pujol C, Ciura S, Erpapazoglou Z, Usenko T, Maurage CA, Sahbatou M, Liebau S, Ding J, Bilgic B, Emre M, Erginel-Unaltuna N, Guven G, Tison F, Tranchant C, Vidailhet M, Corvol JC, Krack P, Leutenegger AL, Nalls MA, Hernandez DG, Heutink P, Gibbs JR, Hardy J, Wood NW, Gasser T, Durr A, Deleuze JF, Tazir M, Destée A, Lohmann E, Kabashi E, Singleton A, Corti O, Brice A. Loss of VPS13C function in autosomal-recessive parkinsonism causes mitochondrial dysfunction and increases PINK1/parkin-dependent mitophagy. Am J Hum Genet. 2016;98:500–513. doi: 10.1016/j.ajhg.2016.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rampoldi L, Dobson-Stone C, Rubio JP, Danek A, Chalmers RM, Wood NW, Verellen C, Ferrer X, Malandrini A, Fabrizi GM, Brown R, Vance J, Pericak-Vance M, Rudolf G, Carrè S, Alonso E, Manfredi M, Németh AH, Monaco AP. A conserved sorting-associated protein is mutant in chorea-acanthocytosis. Nat Genet. 2001;28:119–120. doi: 10.1038/88821. [DOI] [PubMed] [Google Scholar]
- 4.Ueno T, Kagawa T, Mizukawa N, Nakamura H, Sugahara T, Yamamoto T. Cellular origin of endochondral ossification from grafted periosteum. Anat Rec. 2001;264:348–357. doi: 10.1002/ar.10024. [DOI] [PubMed] [Google Scholar]
- 5.Kolehmainen J, Black GC, Saarinen A, Chandler K, Clayton-Smith J, Träskelin AL, Perveen R, Kivitie-Kallio S, Norio R, Warburg M, Fryns JP, de la Chapelle A, Lehesjoki AE. Cohen syndrome is caused by mutations in a novel gene, COH1, encoding a transmembrane protein with a presumed role in vesicle-mediated sorting and intracellular protein transport. Am J Hum Genet. 2003;72:1359–1369. doi: 10.1086/375454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duplomb L, Duvet S, Picot D, Jego G, El Chehadeh-Djebbar S, Marle N, Gigot N, Aral B, Carmignac V, Thevenon J, Lopez E, Rivière JB, Klein A, Philippe C, Droin N, Blair E, Girodon F, Donadieu J, Bellanné-Chantelot C, Delva L, Michalski JC, Solary E, Faivre L, Foulquier F, Thauvin-Robinet C. Cohen syndrome is associated with major glycosylation defects. Hum Mol Genet. 2014;23:2391–2399. doi: 10.1093/hmg/ddt630. [DOI] [PubMed] [Google Scholar]
- 7.Limoge F, Faivre L, Gautier T, Petit JM, Gautier E, Masson D, Jego G, El Chehadeh-Djebbar S, Marle N, Carmignac V, Deckert V, Brindisi MC, Edery P, Ghoumid J, Blair E, Lagrost L, Thauvin-Robinet C, Duplomb L. Insulin response dysregulation explains abnormal fat storage and increased risk of diabetes mellitus type 2 in Cohen Syndrome. Hum Mol Genet. 2015;24:6603–6613. doi: 10.1093/hmg/ddv366. [DOI] [PubMed] [Google Scholar]
- 8.Seifert W, Kühnisch J, Maritzen T, Lommatzsch S, Hennies HC, Bachmann S, Horn D, Haucke V. Cohen syndrome-associated protein COH1 physically and functionally interacts with the small GTPase RAB6 at the Golgi complex and directs neurite outgrowth. J Biol Chem. 2015;290:3349–3358. doi: 10.1074/jbc.M114.608174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seifert W, Kühnisch J, Maritzen T, Horn D, Haucke V, Hennies HC. Cohen syndrome-associated protein, COH1, is a novel, giant Golgi matrix protein required for Golgi integrity. J Biol Chem. 2011;286:37665–37675. doi: 10.1074/jbc.M111.267971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balikova I, Lehesjoki AE, de Ravel TJ, Thienpont B, Chandler KE, Clayton-Smith J, Träskelin AL, Fryns JP, Vermeesch JR. Deletions in the VPS13B (COH1) gene as a cause of Cohen syndrome. Hum Mutat. 2009;30:E845–E854. doi: 10.1002/humu.21065. [DOI] [PubMed] [Google Scholar]
- 11.El Chehadeh S, Aral B, Gigot N, Thauvin-Robinet C, Donzel A, Delrue MA, Lacombe D, David A, Burglen L, Philip N, Mon-cla A, Cormier-Daire V, Rio M, Edery P, Verloes A, Bonneau D, Afenjar A, Jacquette A, Heron D, Sarda P, Pinson L, Doray B, Vigneron J, Leheup B, Frances-Guidet AM, Dienne G, Holder M, Masurel-Paulet A, Huet F, Teyssier JR, Faivre L. Search for the best indicators for the presence of a VPS13B gene mutation and confirmation of diagnostic criteria in a series of 34 patients genotyped for suspected Cohen syndrome. J Med Genet. 2010;47:549–553. doi: 10.1136/jmg.2009.075028. [DOI] [PubMed] [Google Scholar]
- 12.El Chehadeh-Djebbar S, Blair E, Holder-Espinasse M, Moncla A, Frances AM, Rio M, Debray FG, Rump P, Masurel-Paulet A, Gigot N, Callier P, Duplomb L, Aral B, Huet F, Thauvin-Robinet C, Faivre L. Changing facial phenotype in Cohen syndrome: towards clues for an earlier diagnosis. Eur J Hum Genet. 2013;21:736–742. doi: 10.1038/ejhg.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen MM, Jr, Hall BD, Smith DW, Graham CB, Lampert KJ. A new syndrome with hypotonia, obesity, mental deficiency, and facial, oral, ocular, and limb anomalies. J Pediatr. 1973;83:280–284. doi: 10.1016/S0022-3476(73)80493-7. [DOI] [PubMed] [Google Scholar]
- 14.Murphy CH, Saddler NI, Devries MC, McGlory C, Baker SK, Phillips SM. Leucine supplementation enhances integrative myofibrillar protein synthesis in free-living older men consuming lower- and higher-protein diets: a parallel-group crossover study. Am J Clin Nutr. 2016;104:1594–1606. doi: 10.3945/ajcn.116.136424. [DOI] [PubMed] [Google Scholar]
- 15.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakimoto H, Nakamura M, Nagata O, Yokoyama I, Sano A. Phenotypic abnormalities in a chorea-acanthocytosis mouse model are modulated by strain background. Biochem Biophys Res Commun. 2016;472:118–124. doi: 10.1016/j.bbrc.2016.02.077. [DOI] [PubMed] [Google Scholar]
- 17.Tomemori Y, Ichiba M, Kusumoto A, Mizuno E, Sato D, Muroya S, Nakamura M, Kawaguchi H, Yoshida H, Ueno S, Nakao K, Nakamura K, Aiba A, Katsuki M, Sano A. A gene-targeted mouse model for chorea-acanthocytosis. J Neurochem. 2005;92:759–766. doi: 10.1111/j.1471-4159.2004.02924.x. [DOI] [PubMed] [Google Scholar]
- 18.Cho J, Yu NK, Choi JH, Sim SE, Kang SJ, Kwak C, Lee SW, Kim JI, Choi DI, Kim VN, Kaang BK. Multiple repressive mechanisms in the hippocampus during memory formation. Science. 2015;350:82–87. doi: 10.1126/science.aac7368. [DOI] [PubMed] [Google Scholar]
- 19.Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J, Ha S, Chung C, Jung ES, Cho YS, Park SG, Lee JS, Lee K, Kim D, Bae YC, Kaang BK, Lee MG, Kim E. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486:261–265. doi: 10.1038/nature11208. [DOI] [PubMed] [Google Scholar]
- 20.Isiegas C, McDonough C, Huang T, Havekes R, Fabian S, Wu LJ, Xu H, Zhao MG, Kim JI, Lee YS, Lee HR, Ko HG, Lee N, Choi SL, Lee JS, Son H, Zhuo M, Kaang BK, Abel T. A novel conditional genetic system reveals that increasing neuronal cAMP enhances memory and retrieval. J Neurosci. 2008;28:6220–6230. doi: 10.1523/JNEUROSCI.2935-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, Kim T, Lee HR, Kong YY, Kaang BK. Mind bomb-2 regulates hippocampus-dependent memory formation and synaptic plasticity. Korean J Physiol Pharmacol. 2015;19:515–522. doi: 10.4196/kjpp.2015.19.6.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim CS, Kim H, Yu NK, Kang SJ, Kim T, Ko HG, Lee J, Yang JE, Ryu HH, Park T, Gim J, Nam HJ, Baek SH, Wegener S, Schmitz D, Boeckers TM, Lee MG, Kim E, Lee JH, Lee YS, Kaang BK. Enhancing inhibitory synaptic function reverses spatial memory deficits in Shank2 mutant mice. Neuropharmacology. 2017;112(Pt A):104–112. doi: 10.1016/j.neuropharm.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 23.Shum FW, Ko SW, Lee YS, Kaang BK, Zhuo M. Genetic alteration of anxiety and stress-like behavior in mice lacking CaMKIV. Mol Pain. 2005;1:22–22. doi: 10.1186/1744-8069-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park SW, Ko HG, Lee N, Lee HR, Rim YS, Kim H, Lee KM, Kaang BK. Aged wild-type littermates and APPswe+PS1/ΔE9 mice present similar deficits in associative learning and spatial memory independent of amyloid load. Genes Genomics. 2010;32:63–70. doi: 10.1007/s13258-010-0833-8. [DOI] [Google Scholar]
- 25.Kim H, Kim HS, Kaang BK. Elevated contextual fear memory by SIRT6 depletion in excitatory neurons of mouse forebrain. Mol Brain. 2018;11:49. doi: 10.1186/s13041-018-0391-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S, Yu NK, Shim KW, Kim JI, Kim H, Han DH, Choi JE, Lee SW, Choi DI, Kim MW, Lee DS, Lee K, Galjart N, Lee YS, Lee JH, Kaang BK. Remote memory and cortical synaptic plasticity require neuronal CCCTC-binding factor (CTCF) J Neurosci. 2018;38:5042–5052. doi: 10.1523/JNEUROSCI.2738-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee YS, Ehninger D, Zhou M, Oh JY, Kang M, Kwak C, Ryu HH, Butz D, Araki T, Cai Y, Balaji J, Sano Y, Nam CI, Kim HK, Kaang BK, Burger C, Neel BG, Silva AJ. Mechanism and treatment for learning and memory deficits in mouse models of Noonan syndrome. Nat Neurosci. 2014;17:1736–1743. doi: 10.1038/nn.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Xu H, Wu LJ, Kim SS, Chen T, Koga K, Descalzi G, Gong B, Vadakkan KI, Zhang X, Kaang BK, Zhuo M. Identification of an adenylyl cyclase inhibitor for treating neuropathic and inflammatory pain. Sci Transl Med. 2011;3(65):65ra3. doi: 10.1126/scitranslmed.3001269. [DOI] [PubMed] [Google Scholar]
- 29.Qiu S, Chen T, Koga K, Guo YY, Xu H, Song Q, Wang JJ, Descalzi G, Kaang BK, Luo JH, Zhuo M, Zhao MG. An increase in synaptic NMDA receptors in the insular cortex contributes to neuropathic pain. Sci Signal. 2013;6:ra34–ra34. doi: 10.1126/scisignal.2003778. [DOI] [PubMed] [Google Scholar]
- 30.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dow RS, Moruzzi G. The physiology and pathology of the cerebellum. University of Minnesota Press; Minneapolis, MN: 1958. [Google Scholar]
- 32.Waite A, Somer M, O’Driscoll M, Millen K, Manson FD, Chandler KE. Cerebellar hypoplasia and Cohen syndrome: a confirmed association. Am J Med Genet A. 2010;152A:2390–2393. doi: 10.1002/ajmg.a.33569. [DOI] [PubMed] [Google Scholar]
- 33.Rampoldi L, Danek A, Monaco AP. Clinical features and molecular bases of neuroacanthocytosis. J Mol Med (Berl) 2002;80:475–491. doi: 10.1007/s00109-002-0349-z. [DOI] [PubMed] [Google Scholar]
- 34.Ueno S, Maruki Y, Nakamura M, Tomemori Y, Kamae K, Tanabe H, Yamashita Y, Matsuda S, Kaneko S, Sano A. The gene encoding a newly discovered protein, chorein, is mutated in chorea-acanthocytosis. Nat Genet. 2001;28:121–122. doi: 10.1038/88825. [DOI] [PubMed] [Google Scholar]
- 35.Frankland PW, Cestari V, Filipkowski RK, McDonald RJ, Silva AJ. The dorsal hippocampus is essential for context discrimination but not for contextual conditioning. Behav Neurosci. 1998;112:863–874. doi: 10.1037/0735-7044.112.4.863. [DOI] [PubMed] [Google Scholar]
- 36.Gerlai R. Behavioral tests of hippocampal function: simple paradigms complex problems. Behav Brain Res. 2001;125:269–277. doi: 10.1016/S0166-4328(01)00296-0. [DOI] [PubMed] [Google Scholar]
- 37.Johansen JP, Cain CK, Ostroff LE, LeDoux JE. Molecular mechanisms of fear learning and memory. Cell. 2011;147:509–524. doi: 10.1016/j.cell.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]