Gut bacteria play a crucial role in the metabolism of dietary flavonoids, thereby contributing to their activation or inactivation after ingestion by the human host. Thus, bacterial activities in the intestine may influence the beneficial health effects of these polyphenolic plant compounds. While an increasing number of flavonoid-converting gut bacterial species have been identified, knowledge of the responsible enzymes is still limited. Here, we characterized Fcr as a key enzyme involved in the conversion of flavonoids of several subclasses by Eubacterium ramulus, a prevalent human gut bacterium. Sequence similarity of this enzyme to hypothetical proteins from other flavonoid-degrading intestinal bacteria in databases suggests a more widespread occurrence of this enzyme. Functional characterization of gene products of human intestinal microbiota enables the assignment of metagenomic sequences to specific bacteria and, more importantly, to certain activities, which is a prerequisite for targeted modulation of gut microbial functionality.
KEYWORDS: Eubacterium ramulus, enantiospecificity, flavanone, flavanonol, flavonoid, intestinal bacteria, naringenin, reductase
ABSTRACT
The human intestinal anaerobe Eubacterium ramulus is known for its ability to degrade various dietary flavonoids. In the present study, we demonstrate the cleavage of the heterocyclic C-ring of flavanones and flavanonols by an oxygen-sensitive NADH-dependent reductase, previously described as enoate reductase, from E. ramulus. This flavanone- and flavanonol-cleaving reductase (Fcr) was purified following its heterologous expression in Escherichia coli and further characterized. Fcr cleaved the flavanones naringenin, eriodictyol, liquiritigenin, and homoeriodictyol. Moreover, the flavanonols taxifolin and dihydrokaempferol served as substrates. The catalyzed reactions were stereospecific for the (2R)-enantiomers of the flavanone substrates and for the (2S,3S)-configured flavanonols. The enantioenrichment of the nonconverted stereoisomers allowed for the determination of hitherto unknown flavanone racemization rates. Fcr formed the corresponding dihydrochalcones and hydroxydihydrochalcones in the course of an unusual reductive cleavage of cyclic ether bonds. Fcr did not convert members of other flavonoid subclasses, including flavones, flavonols, and chalcones, the latter indicating that the reaction does not involve a chalcone intermediate. This view is strongly supported by the observed enantiospecificity of Fcr. Cinnamic acids, which are typical substrates of bacterial enoate reductases, were also not reduced by Fcr. Based on the presence of binding motifs for dinucleotide cofactors and a 4Fe-4S cluster in the amino acid sequence of Fcr, a cofactor-mediated hydride transfer from NADH onto C-2 of the respective substrate is proposed.
IMPORTANCE Gut bacteria play a crucial role in the metabolism of dietary flavonoids, thereby contributing to their activation or inactivation after ingestion by the human host. Thus, bacterial activities in the intestine may influence the beneficial health effects of these polyphenolic plant compounds. While an increasing number of flavonoid-converting gut bacterial species have been identified, knowledge of the responsible enzymes is still limited. Here, we characterized Fcr as a key enzyme involved in the conversion of flavonoids of several subclasses by Eubacterium ramulus, a prevalent human gut bacterium. Sequence similarity of this enzyme to hypothetical proteins from other flavonoid-degrading intestinal bacteria in databases suggests a more widespread occurrence of this enzyme. Functional characterization of gene products of human intestinal microbiota enables the assignment of metagenomic sequences to specific bacteria and, more importantly, to certain activities, which is a prerequisite for targeted modulation of gut microbial functionality.
INTRODUCTION
Flavonoids are secondary plant constituents that have attracted growing interest because of their health-promoting benefits (1). Such effects depend on their bioavailability, which may vary due to interindividual differences in absorption and metabolism (2). Following their ingestion, flavonoids are in general poorly absorbed and, thus, poorly metabolized by intestinal bacteria (3, 4). Identification of the bacterial species responsible for flavonoid conversion in the gut has therefore been expedited in recent years (5). However, knowledge of the involved bacterial enzymes is still limited.
The strict anaerobe Eubacterium ramulus is one of the most extensively studied flavonoid-degrading gut bacteria. It converts members of various flavonoid subclasses, such as flavonols, flavanonols, flavones, flavanones, and isoflavones (6–8). Moreover, E. ramulus is highly prevalent in the human intestine, reaching cell counts of 107 to 109 per g fecal dry weight (9, 10).
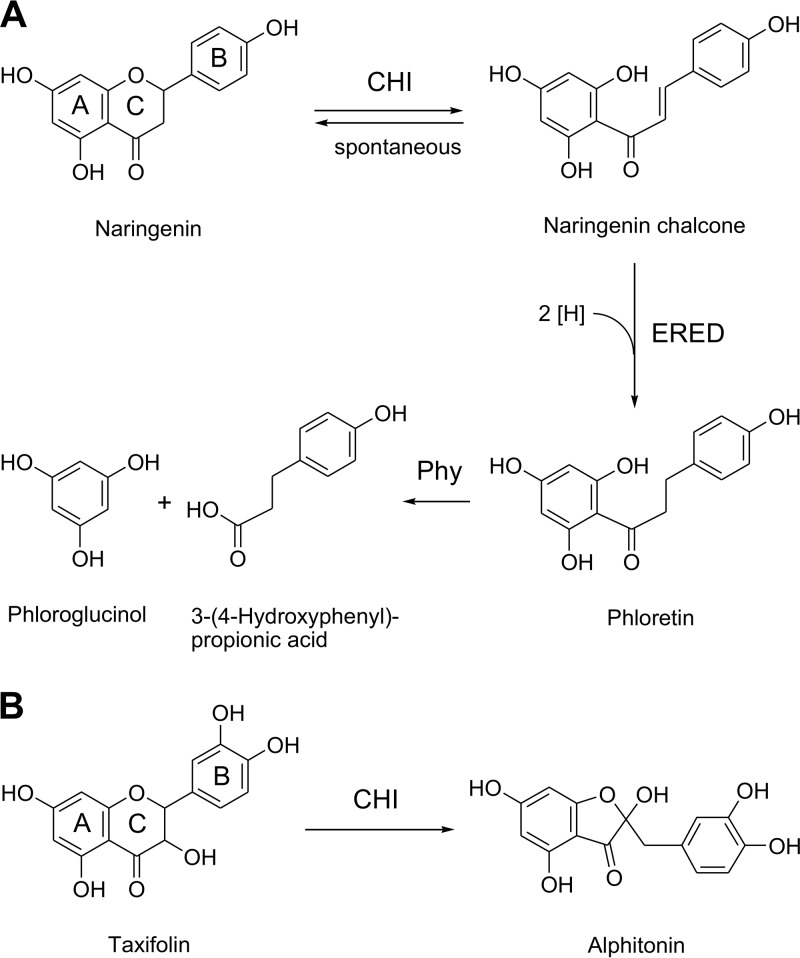
Three enzymes from E. ramulus involved in the degradation of flavonoids have been characterized so far, and the corresponding genes have been identified: chalcone isomerase (CHI), enoate reductase (ERED), and phloretin hydrolase (Phy) (11–13). CHI was reported to catalyze the C-ring fission of flavanones (e.g., naringenin) to chalcones (e.g., naringenin chalcone) followed by the ERED-catalyzed reduction of the chalcones to the corresponding dihydrochalcones (e.g., phloretin) (Fig. 1A) (11, 12). Phy cleaves the dihydrochalcone to monophenolic metabolites [e.g., 3-(4-hydroxyphenyl)propionic acid and phloroglucinol] (Fig. 1A) (13). Besides acting on flavanones, CHI was shown to catalyze the ring contraction of flavanonols (e.g., taxifolin) to isomeric auronols (e.g., alphitonin) (Fig. 1B) (14).
FIG 1.
Reported conversion of the flavanone naringenin (A) and the flavanonol taxifolin (B) by enzymes from Eubacterium ramulus. CHI, chalcone isomerase; ERED, enoate reductase; Phy, phloretin hydrolase.
To further characterize CHI, we aimed to compare the conversion rates for taxifolin and naringenin by adding ERED from E. ramulus as an auxiliary enzyme to monitor naringenin conversion as reported by Gall et al. (12). ERED was described to catalyze the NADH-dependent reduction of naringenin chalcone formed by CHI to give phloretin (12). During optimization of the CHI and ERED concentrations in the assay, we observed complete conversion of naringenin to phloretin even in the absence of CHI. Based on this finding, ERED from E. ramulus was characterized in the present study and found to be an NADH-dependent flavanone- and flavanonol-cleaving reductase (Fcr).
RESULTS
To characterize Fcr in detail, the enzyme was expressed in Escherichia coli and purified under anoxic conditions. The anaerobically grown recombinant E. coli cultures converted naringenin to phloretin (250 μM within 2 h of incubation), whereas under oxic conditions, naringenin conversion did not occur, although a much higher cell density was reached (see Fig. S1 in the supplemental material). Based on SDS-PAGE analysis, Fcr was expressed efficiently also in aerobically grown cells (data not shown) but was inactivated by oxygen. E. coli cells carrying the empty cloning vector did not convert naringenin either under anoxic or under oxic conditions.
The cell extracts prepared from Fcr-expressing E. coli grown anoxically converted naringenin at a maximal rate of 1.38 μmol min−1 (mg protein)−1. Fcr was purified from cell extracts of recombinant E. coli to apparent homogeneity (see Fig. S2A) with a yield of up to 7.3%. The observed size of 70 kDa is in agreement with the enzyme’s calculated molecular weight of 75 kDa. Native PAGE revealed a main band of 130 kDa, indicating a dimeric structure (Fig. S2B). In addition, minor bands occurred at 230 and 380 kDa. The purified enzyme catalyzed the cleavage of naringenin to phloretin in the presence of NADH at a specific activity of up to 15.6 μmol min−1 (mg protein)−1. No activity was observed in the absence of the coenzyme or when NADH was replaced by NADPH. The purified Fcr was sensitive to oxygen and freezing. Using anoxic conditions, freezing at −20°C resulted in a complete loss of activity. While bovine serum albumin (0.5 mg ml−1) and dithiothreitol (1 mM) did not protect the enzyme, the addition of glycerol (20% [vol/vol]) before freezing maintained 21% of the initial Fcr activity. Storage of the enzyme in an oxygen-free atmosphere at 4°C for 2 days preserved 93% of the activity.
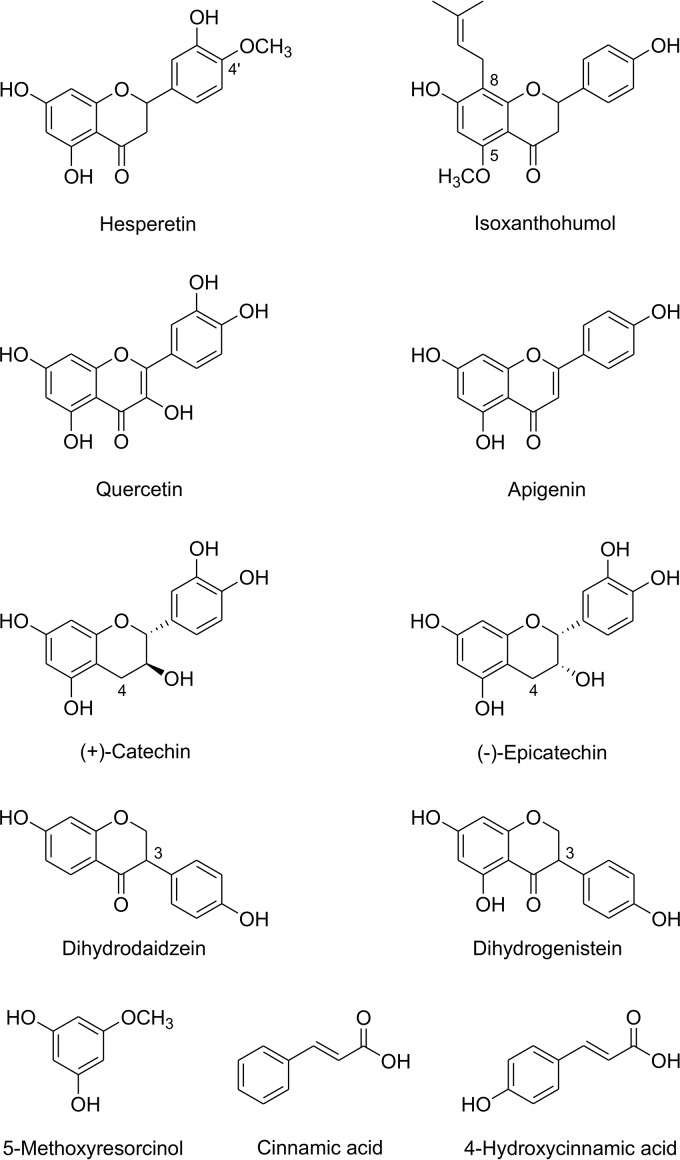
Besides cleaving naringenin, purified Fcr reductively cleaved the flavanones eriodictyol, liquiritigenin, and homoeriodictyol (structures in Fig. 2A) at product formation rates of 36%, 15%, and 10%, respectively, relative to that of naringenin (Fig. 3; see also Table S1). Eriodictyol was converted to 3-hydroxyphloretin and liquiritigenin to davidigenin. The product of homoeriodictyol conversion showed an absorption maximum of 288 nm, which is identical to that of the substrate. Due to the unavailability of a standard reference for the expected dihydrochalcone, this compound was isolated from enzymatic assay mixtures and its structure elucidated. Based on the exact mass of 303.0921 determined by liquid chromatography-mass spectrometry (LC-MS) in the negative ionization mode, the compound was identified as the expected 3-(4-hydroxy-3-methoxyphenyl)-1-(2,4,6-trihydroxyphenyl)propan-1-one (calculated exact mass of 303.0874) (structure in Fig. 2A). Two other flavanones, hesperetin and isoxanthohumol (structures in Fig. 4), were not cleaved by Fcr-containing cell extracts.
FIG 2.
Cleavage of flavanones (A) and flavanonols (B) catalyzed by Fcr.
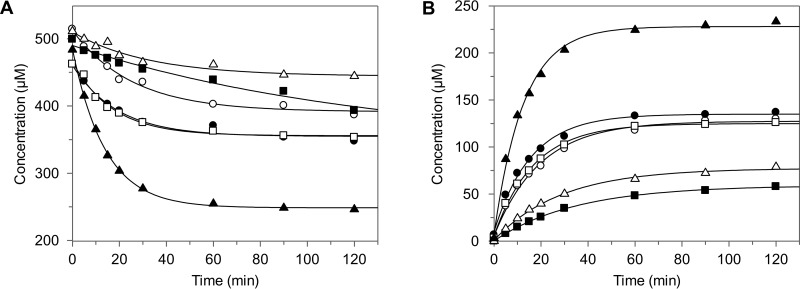
FIG 3.
Comparative kinetics of substrate conversion (A) and product formation (B) by purified Fcr. Racemic substrates were used at 500 μM. Each substrate and its corresponding product are depicted by the same symbol: naringenin and phloretin (▲), eriodictyol and 3-hydroxyphloretin (○), liquiritigenin and davidigenin (▵), homoeriodictyol and 3-(4-hydroxy-3-methoxyphenyl)-1-(2,4,6-trihydroxyphenyl)propan-1-one (■), dihydrokaempferol and 2-hydroxy-1-(2,4,6-trihydroxyphenyl)-3-(4-hydroxyphenyl)propan-1-one (●), taxifolin and 2-hydroxy-1-(2,4,6-trihydroxyphenyl)-3-(3,4-dihydroxyphenyl)propan-1-one (□).
FIG 4.
Compounds not converted by Fcr.
The purified Fcr also converted the flavanonols dihydrokaempferol and taxifolin (structures in Fig. 2B) at product formation rates of 46% and 40%, respectively, relative to that of naringenin (Fig. 3; Table S1). The absorption maxima of the products arising from dihydrokaempferol (289 nm) or taxifolin (290 nm) were similar or even identical to those of the substrates dihydrokaempferol (291 nm) and taxifolin (290 nm). Since authentic standards of the expected hydroxydihydrochalcones are not commercially available, both compounds were isolated from the enzymatic assay mixtures and subjected to LC-MS analysis in the negative ionization mode. The product of dihydrokaempferol conversion showed an exact mass of 289.0732, which indicates that it was 2-hydroxy-1-(2,4,6-trihydroxyphenyl)-3-(4-hydroxyphenyl)propan-1-one (calculated exact mass of 289.0718) (structure in Fig. 2B). Based on its determined exact mass of 305.0670, the taxifolin product was identified as 2-hydroxy-1-(2,4,6-trihydroxyphenyl)-3-(3,4-dihydroxyphenyl)propan-1-one (calculated exact mass, 305.0667) (structure in Fig. 2B).
Flavonoids of other subclasses, such as the flavonol quercetin, the flavone apigenin, the flavan-3-ols catechin and epicatechin, and the isoflavanones dihydrodaidzein and dihydrogenistein (structure in Fig. 4), were not cleaved by Fcr-containing cell extracts or the purified enzyme.
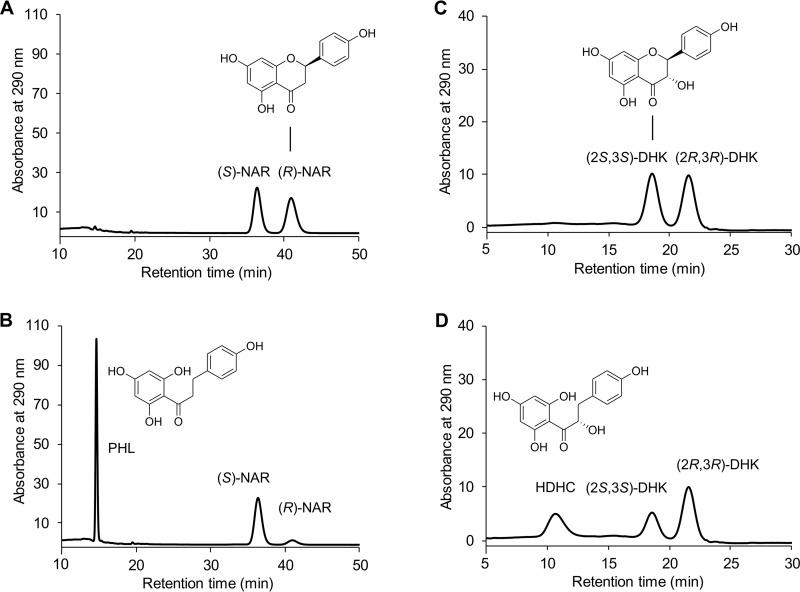
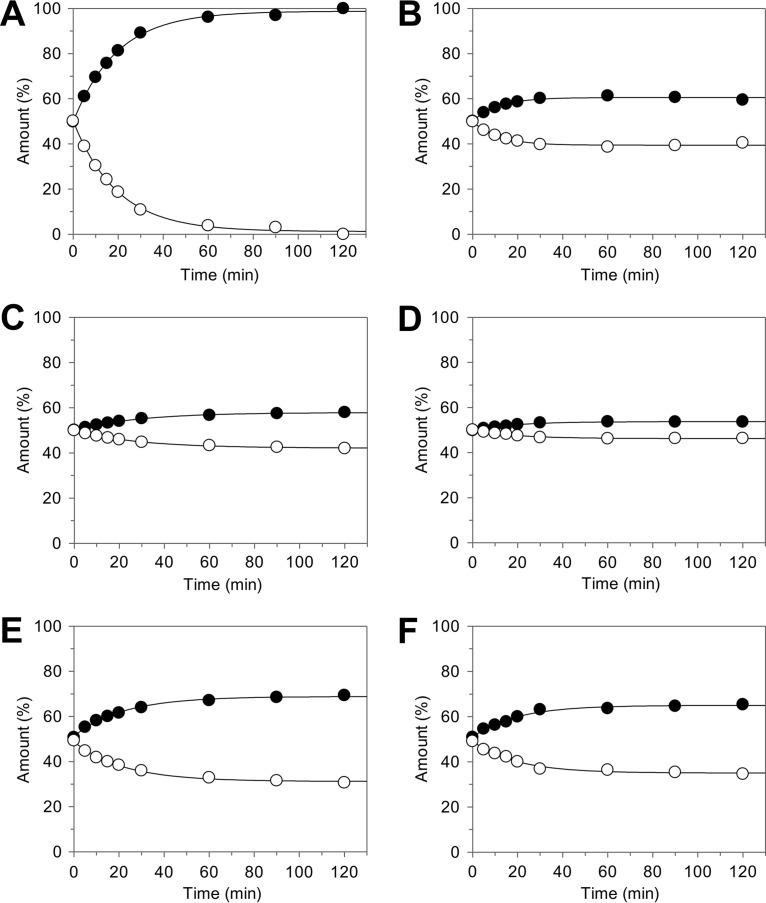
The transformation of only 50% of the racemic naringenin followed by its further conversion at much lower rates was observed with intact Fcr-expressing E. coli cells, resulting cell extracts, or the purified enzyme and indicated the stereospecificity of the catalyzed reaction. Chiral high-performance liquid chromatography (HPLC) analysis confirmed that exclusively (R)-naringenin was accepted as a substrate and converted to achiral phloretin (Fig. 5A and B). The preference of Fcr for the (R)-enantiomer was also observed for the other flavanone substrates, i.e., eriodictyol, liquiritigenin, and homoeriodictyol. In the case of the flavanonols dihydrokaempferol and taxifolin, which have two chiral centers, only the (2S,3S)-stereoisomer was converted (for dihydrokaempferol, see Fig. 5C and D). This was confirmed in control experiments with Fcr-containing cell extracts using the two (2R,3R)-stereoisomers, which were converted at rates of less than 1% compared to that of the (2S,3S)-analogues. The time-dependent ratios of flavanone enantiomers and flavanonol stereoisomers in the course of incubation with purified Fcr are depicted in Fig. 6A to F.
FIG 5.
Chiral HPLC analysis of flavonoid conversion by purified Fcr. Chromatograms of selected samples are depicted. From racemic naringenin (NAR) present at 0 h (A), phloretin (PHL) was formed only from the (R)-enantiomer after 0.5 h of incubation (B). From racemic dihydrokaempferol (DHK) present at 0 h (C), the corresponding hydroxydihydrochalcone (HDHC) was formed only from the (2S,3S)-stereoisomer after 1.5 h of incubation (D).
FIG 6.
Ratios of flavanone enantiomers and flavanonol stereoisomers in the course of substrate conversion by purified Fcr. Data are based on chiral HPLC analysis of samples that were analyzed by achiral HPLC as shown in Fig. 3. Flavanones naringenin (A), eriodictyol (B), liquiritigenin (C), and homoeriodictyol (D) comprised (S)-enantiomers (●) and (R)-enantiomers (○). Flavanonols dihydrokaempferol (E) and taxifolin (F) comprised (2R,3R)-stereoisomers (●) and (2S,3S)-stereoisomers (○).
Spontaneous flavanone racemization may interfere with the rate and the extent of enzymatic product formation. Therefore, we determined the racemization half-lives of the four flavanone substrates under assay conditions. Because of commercial unavailability of enantiomers or low stability, we used preparations enriched in the remaining (S)-enantiomer after enzymatic conversion of the racemate by Fcr. Enantiomers of naringenin and liquiritigenin were relatively stable, exhibiting half-lives of 11.8 h (k = [1.63 ± 0.26] 10−5 s−1) and 12.1 h (k = [1.59 ± 0.24] 10−5 s−1), respectively, while those of eriodictyol and homoeriodictyol rapidly racemized, with half-lives of 60 min (k = [19.4 ± 1.9] 10−5 s−1) and 44 min (k = [26.1 ± 2.4] 10−5 s−1), respectively (see Fig. S3).
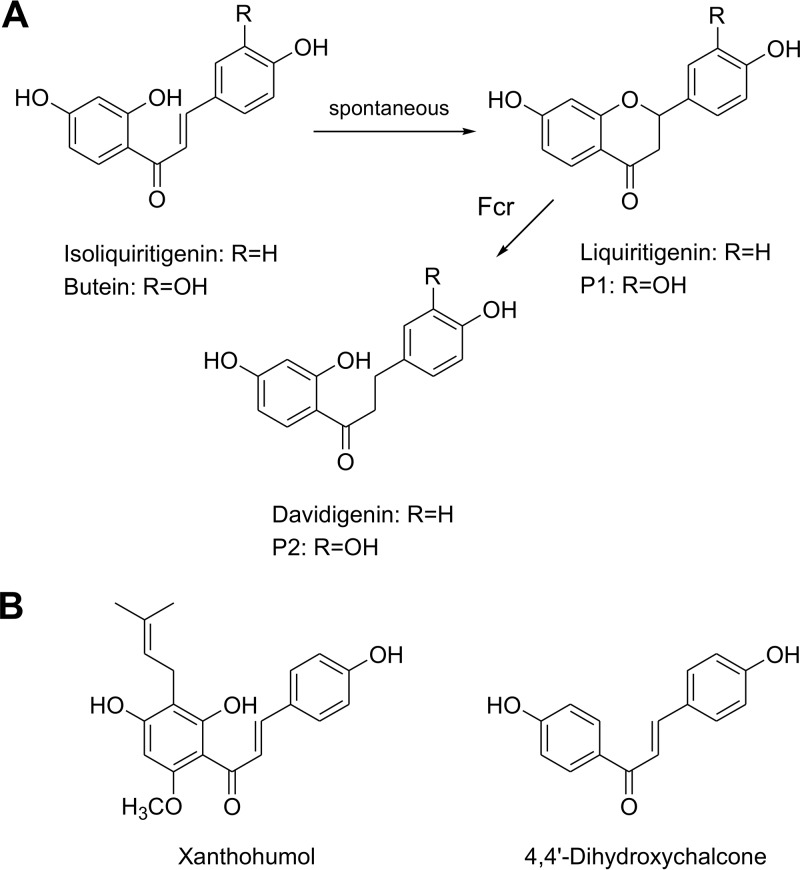
To clarify whether flavonoid conversion by Fcr involves the formation of chalcone intermediates, we tested several chalcones as substrates. Following incubation of isoliquiritigenin (300 μM) with Fcr-containing cell extract in the presence of NADH, liquiritigenin and davidigenin were formed within 5 h at low concentrations of 21 and 1.9 μM, respectively (structures in Fig. 7A). A similar liquiritigenin concentration of 23 μM was also observed after 5 h when the enzyme was absent, indicating spontaneous cyclization of isoliquiritigenin. Analogously, incubation of butein (300 μM) (structure in Fig. 7A) with Fcr-containing cell extract in the presence of NADH over 5 h resulted in the formation of two products, P1 (ca. 3.4 μM) and P2 (ca. 1.5 μM), while P1 (ca. 5.0 μM after 5 h) was also observed in the absence of Fcr. P1 and P2 are assumed to represent the cyclized flavanone and the corresponding dihydrochalcone, respectively (structures in Fig. 7A), but standard reference compounds were not available. Using identical assay conditions, the prenylated chalcone xanthohumol (structure in Fig. 7B) remained unchanged. The results obtained for isoliquiritigenin and butein suggested that, as reported previously (11), these chalcones spontaneously cyclize to the corresponding flavanones, which are subsequently cleaved by Fcr (Fig. 7A). To support this hypothesis, we applied the synthesized 4,4′-dihydroxychalcone (structure in Fig. 7B), which cannot cyclize due to missing hydroxy groups in the relevant positions. Incubation of this chalcone with Fcr-containing cell extract and NADH did not result in reduction to 4,4′-dihydroxydihydrochalcone, which was confirmed by applying the synthetized standard compound. In line with these results, the purified enzyme also did not convert any of the chalcones mentioned above. Taken together, the conversion of flavonoids by Fcr did not involve a chalcone structure. Instead, Fcr catalyzed the reductive ether cleavage of the heterocyclic C-ring of certain flavonoids. However, the ether bond present in the monocyclic 5-methoxyresorcinol (structure in Fig. 4) was not cleaved by Fcr-containing cell extract.
FIG 7.
(A) Spontaneous cyclization of chalcones to their corresponding flavanones and subsequent cleavage by Fcr. (B) Chalcones neither spontaneously cyclized nor converted by Fcr.
Since Fcr was originally described as ERED, we tested whether cinnamic acid or 4-hydroxycinnamic acid (structures in Fig. 4), which are typical substrates of bacterial EREDs, are reduced to 3-phenylpropionic acid and 3-(4-hydroxyphenyl)propionic acid, respectively. However, neither of these compounds was converted by Fcr-containing cell extracts.
DISCUSSION
Gut bacteria play a crucial role in the conversion of bioactive dietary compounds, including flavonoids (4). The number of bacterial species found to convert flavonoids has steadily increased, and their activities have been assessed (5). However, the majority of the underlying pathways and corresponding enzymes are still unknown or have not yet been characterized. E. ramulus has been identified as a prevalent human gut bacterium involved in the conversion of dietary flavonoids (6–8, 15).
Herein, the enzyme encoded by the ERED gene in E. ramulus DSM 16296 (KF154735, protein accession number AGS82961) was demonstrated to cleave several flavanones and flavanonols in an NADH-dependent manner. Previously, this enzyme was heterologously expressed in E. coli, purified as a protein with an apparent molecular weight of 70 kDa, and described as ERED, catalyzing the reduction of naringenin chalcone to phloretin (12) (Fig. 1). By combining ERED with CHI, Gall et al. observed the conversion of naringenin to phloretin; coexpression of ERED and CHI led to transformation of eriodictyol and homoeriodictyol in addition to naringenin (12). Based on this report, we intended to use ERED as an auxiliary enzyme in order to compare the rate of taxifolin conversion by CHI (14) to that of naringenin conversion. Unexpectedly, it turned out that phloretin formation from naringenin depended on ERED but not CHI concentration and occurred even in the absence of CHI.
Fcr under anoxic conditions NADH-dependently cleaved the following flavanones and flavanonols at decreasing rates: naringenin > dihydrokaempferol ∼ taxifolin ∼ eriodictyol > liquiritigenin ∼ homoeriodictyol (Fig. 3). A hydroxy or methoxy group at the C-3′ position of the B-ring (Fig. 2) attenuated the cleavage, as evidenced by comparing naringenin with eriodictyol or homoeriodictyol. The influence of the flavanonol-specific hydroxy substitution at C-3 of the C-ring was not really clear. Dihydrokaempferol was cleaved less efficiently than naringenin; however, similar rates were observed for taxifolin and eriodictyol. It is noteworthy that the hydroxy group at C-5 of the A-ring present in naringenin but not in liquiritigenin greatly favored enzymatic cleavage. Substitution of the hydroxy with a methoxy group at the B-ring C-4′ position prevented the corresponding flavanone, hesperetin, from being cleaved. Isoxanthohumol, which lacks the free C-5 hydroxy group and carries an additional prenyl moiety at C-8, also did not serve as a substrate. Members of flavonoid classes other than flavanones and flavanonols were not accepted as the substrates. These included flavan-3-ols without the carbonyl group at C-4 of the C-ring and isoflavanones carrying the aromatic substituent at C-3 of the C-ring.
While the compounds formed from naringenin, eriodictyol, and liquiritigenin by Fcr were identified by using reference standards, the products formed by C-ring cleavage from homoeriodictyol, dihydrokaempferol, and taxifolin were characterized by mass spectrometry as the corresponding dihydrochalcones (Fig. 2). Of these three metabolites, 2-hydroxy-1-(2,4,6-trihydroxyphenyl)-3-(3,4-dihydroxyphenyl)propan-1-one was previously described as a product of heterocyclic ring fission of taxifolin by Eggerthella sp. strain SDG-2 (16). As already demonstrated for neohesperidin dihydrochalcone and phloretin (17, 18), the novel dihydrochalcones described herein may have various interesting properties. For biological evaluation, enzymes such as Fcr may be utilized to produce dihydrochalcones that are difficult to obtain by chemical synthesis.
Our results show that Fcr does not reduce the C-C double bond of chalcones to form dihydrochalcones. This was clearly demonstrated for 4,4′-dihydroxychalcone (Fig. 7B). The small amounts of dihydrochalcones observed following the incubation of Fcr with cyclizable chalcones most likely resulted from the enzymatic cleavage of the spontaneously formed flavanones (Fig. 7A). For intact E. ramulus cells, however, reduction of xanthohumol rather than isoxanthohumol was recently reported (19). Fcr did not reduce C-C double bonds present in the C-ring of flavonols (e.g., quercetin) and flavones (e.g., apigenin) and also did not accept cinnamic acids as substrates (Fig. 4). Clostridial ERED enzymes have been shown to reduce cinnamic and 4-hydroxycinnamic acids to 3-phenylpropionic and 3-(4-hydroxyphenyl)propionic acids, respectively (20, 21).
Further evidence that Fcr accepts flavanones but not chalcones as substrates arises from the observed stereospecificity. Chalcones are spontaneously formed from both flavanone enantiomers in the course of racemization with identical rate constants. If the chalcones were bound and converted by the enzyme, preferred transformation of a single enantiomer would not occur. However, only one enantiomer of each of the investigated flavanone substrates was preferentially converted (Fig. 6). An enrichment of the nontransformed enantiomers was observed despite the competing spontaneous racemization. The racemization kinetics of flavanones has been little studied. The reported half-life for liquiritigenin of 6.3 h at 37°C (22) is in the same range as the value determined herein, namely, 12.1 h at 25°C. Our data indicate that the rate of racemization at pH 6.8 depends on the presence of a hydroxy or methoxy group at the C-3′ position of the B-ring. It can be concluded that, owing to the electron-withdrawing inductive effect of these meta-substituents, electron density at position 2 is reduced and ring opening accelerated. Accordingly, naringenin and liquiritigenin, which lack this meta-substituent, exhibited a 12- to 17-fold longer half-life than eriodictyol or homoeriodictyol. The enantioenrichment depends on the rates of both racemization and enzymatic conversion. This is obvious when comparing homoeriodictyol, a poor substrate with fast racemization (Fig. 6D), with naringenin, an excellent substrate with moderate racemization velocity (Fig. 6A). In the latter case, a complete enzymatic enantioseparation was achieved. In the case of flavanonols, racemization under assay conditions is negligible (23, 24), which is a further hint that a ring-opened derivative does not serve as a substrate for Fcr. Moreover, Fcr hardly converted the (2R,3R)-stereoisomers of taxifolin or dihydrokaempferol. This confirms the stereospecificity of this enzyme on the one hand and the negligible spontaneous racemization on the other hand.
The gene product assigned by Gall et al. was based on the amino acid motif NXRXDXXGG, which is highly conserved in several ERED sequences (12). However, the function of this motif is unknown. In addition, the primary sequence of Fcr exhibits a binding motif of a 4Fe-4S cluster [CXXCXXC(X)22C] and two dinucleotide binding motifs [GXGXXG(X)17E]. The presence of an Fe-S cluster may be the reason for the oxygen sensitivity of the enzyme. Fcr from E. ramulus and the hypothetical flavin adenine dinucleotide (FAD)-dependent oxidoreductase from E. oxidoreducens share the CXXC motif, employed by many redox proteins (e.g., thiol-disulfide oxidoreductases) for the formation, isomerization, and reduction of disulfide bonds and for other redox functions. The protein sequence of Fcr from E. ramulus DSM 16296 is identical to those of hypothetical proteins of other E. ramulus strains, including the strain E. ramulus DSM 15684T (ERK50666), present in databases and annotated as FAD-dependent oxidoreductases. Hypothetical gene products sharing lower levels of identity are also found in other flavonoid-degrading bacteria, such as Eubacterium oxidoreducens (SDB03427, 80% identity), Clostridium sp. strain SY8519 (BAK47992, 77% identity), and Flavonifractor plautii (CUQ13575, 42% identity) (10, 25–28).
In this study, we demonstrate that Fcr catalyzes the reductive cleavage of a cyclic ether bond, which represents an unusual reaction. However, bacteria can cleave ether bonds, and these conversions are catalyzed by a heterogeneous group of enzymes or enzyme systems exhibiting different mechanisms (29). The extensively studied lignin-depolymerizing β-etherases from sphingomonads use glutathione as a reducing cofactor (30–32). These enzymes stereospecifically cleave the β-O-4 aryl ether bonds, which account for 45% to 60% of linkages present in lignin. An NADPH-dependent benzyl-ether cleavage is catalyzed by pinoresinol reductase from the soil bacterium Sphingobium sp. strain SYK-6 (BAK65407) (33). This 36-kDa enzyme converted the lignan pinoresinol via lariciresinol to secoisolariciresinol. A benzyl-ether reductase (Ber) from Eggerthella lenta DSM 2243T (WP_015759938) with a much higher molecular weight of 61 kDa catalyzes the same sequential reduction of the furo[3,4-c]furan system present in pinoresinol (34). The primary sequence of the lignan-cleaving enzymes does not show any similarity to that of Fcr. Containing a central Fe/S binding motif and two C-terminal dinucleotide binding motifs and exhibiting a molecular weight of 74 kDa, Fcr considerably resembles the 2,4-dienoyl-coenzyme A (2,4-dienoyl-CoA) reductase from E. coli (KFB96642, 73 kDa), although the overall sequence identity of the two proteins is only 23%. The 2,4-dienoyl-CoA reductase is an NADPH-dependent iron-sulfur flavoenzyme that catalyzes the reduction of unsaturated fatty acids with double bonds at even-numbered carbon positions. Based on the crystal structure of the E. coli enzyme, the following reaction steps were proposed: (i) hydride transfer from NADPH to FAD, (ii) transfer of electrons, one at a time, to flavin mononucleotide (FMN) via the 4Fe-4S cluster, (iii) transfer of a hydride ion from the fully reduced FMN to the C-5 atom of the acyl-CoA substrate, and (iv) protonation of the C-4 atom of the substrate by a Tyr-His catalytic dyad (35). A similar electron transfer pathway may be postulated for catalysis by Fcr, namely a hydride transfer from NADH onto the C-2 of the substrate via FAD, 4Fe-4S, and FMN cofactors. The productive binding mode of the flavonoid substrate might enable this hydride transfer without steric hindrance at the chiral C-2 carbon. Our data on the enantiospecificity of the reaction provide evidence for a fine-tuned interplay of the hydride donor and the substrate. The orientation of the sterically demanding aryl substituent at position 2 affects the reaction course, and only compounds with a defined C-2 configuration serve as substrates. The (2S)-configured flavanones and the (2R)-configured flavanonols are homochiral and exhibit the same crucial molecular geometry around C-2. Hence, we assume a common binding mode involving interactions with substrate substituents, i.e., the 4′-OH and 5-OH groups, required for conversion. Consequently, in a productive binding mode, the C-2 substituent would be identically oriented.
The above-mentioned lignan reductases catalyze the enantio- or regiospecific sequential conversion of (+)- and (−)-pinoresinol via (+)- and (−)-lariciresinol to (−)- and (+)-secolariciresinol, respectively, through a direct hydride transfer from the NADPH cofactor to the substrate. Stacking interactions between the nicotinamide ring and the substrates were deduced from the crystal structures (36). Although known, SN2-type reactions are not common in enzymology because of the desolvated environment of the enzyme active site (37). Thus, a hybrid SN2/SN1 substitution for Fcr-catalyzed reactions is conceivable. The ring opening would be supported by protonation of the ring oxygen, irrespective of a pronounced SN2 or SN1 mechanism. Also, a base should be involved to abstract a proton from the nicotinamide. To clarify the general acid-base mechanism for Fcr-catalyzed reductions, further investigations are needed. These include the characterization of the involved enzyme-associated cofactors besides NADH.
With respect to the flavonoid metabolism by E. ramulus, it can be concluded that Fcr is responsible for the reductive ring cleavage of flavanones to dihydrochalcones. Thus, CHI does not appear to play an essential role in flavanone conversion as was previously assumed (11, 12). It is noteworthy that Fcr prefers (R)-configured flavanone substrates, although (S)-enantiomers predominate in plants and, thus, in plant-derived foods (38–40). However, besides spontaneous racemization, interconversion of enantiomers could be accelerated by ring-opening enzymes such as CHI. The conversion of flavanonols by enzymes from E. ramulus depends on the stereoisomeric form and the availability of NADH. While (2R,3R)-stereoisomers are isomerized by CHI to the corresponding auronols (14), Fcr NADH-dependently forms hydroxydihydrochalcones from (2S,3S)-flavanonols. However, the naturally occurring flavanonols exist in their (2R,3R)-form, which at least has been demonstrated for taxifolin (23, 41, 42). Nevertheless, the nonnatural stereoisomers of both flavanones and flavanonols may be formed as intermediates in the course of flavone and flavonol degradation by gut bacteria, including E. ramulus, but the corresponding enzymes catalyzing the preceding reduction of the C-2 to C-3 double bond of these flavonoids have not been identified yet.
MATERIALS AND METHODS
Chemicals.
Naringenin, eriodictyol, phloretin, (+)-taxifolin, hesperetin, (+)-catechin, (−)-epicatechin, apigenin, butein, and quercetin were purchased from Roth (Karlsruhe, Germany). Liquiritigenin and isoxanthohumol were from PhytoLab (Vestenbergsgreuth, Germany). Naringenin, eriodictyol, liquiritigenin, homoeriodictyol, and hesperetin were identified as racemic compounds by chiral HPLC analysis. (S)-Liquiritigenin, (±)-taxifolin, (±)-trans-dihydrokaempferol, (±)-catechin, 5-methoxyresorcinol, 4-hydroxycinnamic acid, 3-(4-hydroxyphenyl)propionic acid, and phloroglucinol were from Sigma-Aldrich (Munich, Germany). (S)-Homoeriodictyol and hesperetin dihydrochalcone were purchased from Extrasynthese (Genay, France). (+)-Dihydrokaempferol was from Arbonova (Turku, Finland), isoliquiritigenin from abcr (Karlsruhe, Germany), dihydrodaidzein from Toronto Research Chemicals (Toronto, Canada), 3-hydroxyphloretin from TransMIT PlantMetaChem (Giessen, Germany), cinnamic acid from Serva (Heidelberg, Germany), and 3-phenylpropionic acid from Acros (Geel, Belgium). Xanthohumol and dihydrogenistein were available from previous studies (43, 44).
Synthesis of 4,4′-dihydroxychalcone, 4,4′-dihydroxydihydrochalcone, and davidigenin.
For synthesis of 4,4′-dihydroxychalcone, 4-hydroxyacetophenone (1.36 g, 10.0 mmol) and 4-hydroxybenzaldehyde (1.22 g, 10.0 mmol) were dissolved in ethanol (7 ml). To the reaction mixture, 10 ml of a 60% aqueous KOH solution was added, stirred for 1 h, and kept overnight at room temperature. Under ice cooling, the resulting deep red solution was added to 2 M HCl (100 ml). The precipitate was filtered off, washed with water, and recrystallized from ethanol/water (4:1 [vol/vol]) to obtain a yellow solid (0.72 g, 30%): melting point (mp) = 198 to 200°C, literature value mp = 200°C (45); 1H NMR (500 MHz, dimethyl sulfoxide [DMSO]-d6) δ 6.82 (d, 2H, 3J = 8.5 Hz, 3-H, 5-H), 6.87 (d, 2H, 3J = 8.8 Hz, 3′-H, 5′-H), 7.59 (d, 1H, 3J = 15.4 Hz, COCH=CH), 7.66 (d, 1H, 3J = 15.4 Hz, COCH=CH), 7.68 (d, 2H, 3J = 8.5 Hz, 2-H, 6-H), 8.01 (d, 2H, 3J = 8.8 Hz, 2′-H, 6′-H), 10.13 (s, 2H, 4-OH, 4′-OH); 13C NMR (125 MHz, DMSO-d6) δ 115.43, 115.91 (C-3, C-5, C-3′, C-5′), 118.73 (COCH), 126.13, 129.60 (C-1, C-1′), 130.83, 131.04 (C-2, C-6, C-2′, C-6′), 143.26 (COCHCH), 159.95 (4-OH), 162.03 (4′-OH), 187.20 (CO); LC-MS (ESI) m/z 241 ([M+H]+), 95% purity.
For synthesis of 4,4′-dihydroxydihydrochalcone, 4,4′-dihydroxychalcone (0.17 g, 0.70 mmol) was dissolved in dry ethanol (10 ml) and 10% Pd/C (17 mg) was added. Under ice cooling, the reaction mixture was hydrogenated for 1 h under normal pressure. Pd/C was filtered off and the solvent was evaporated. The crude product was purified three times by column chromatography, using ethyl acetate/petroleum ether (1:1 [vol/vol]) twice and then dichloromethane/methanol (19:1 [vol/vol]), to obtain a colorless solid (59 mg, 35%): mp = 140 to 143°C; 1H NMR (500 MHz, DMSO-d6) δ 2.78 (t, 2H, 3J = 7.6 Hz, COCH2CH2), 3.13 to 3.16 (m, 2H, COCH2CH2), 6.63 to 6.65, 6.80 to 6.83 (each m, each 2H, 3-H, 5-H, 3′-H, 5′-H), 7.01 to 7.04 (m, 2H, 2-H, 6-H), 7.82 to 7.85 (m, 2H, 2′-H, 6′-H), 9.08, 10.25 (each s, each 1H, 4-OH, 4′-OH); 13C NMR (125 MHz, DMSO-d6) δ 29.17 (COCH2CH2), 115.14, 115.32 (C-3, C-5, C-3′, C-5′), 128.46 (C-1′), 129.30, 130.57 (C-2, C-6, C-2′, C-6′), 131.55 (C-1), 155.53 (C-4), 162.06 (C-4′), 197.57 (CO), the COCH2 signal is obscured by the DMSO signal; LC-MS (ESI) m/z 243 ([M+H]+), 95% purity.
For synthesis of davidigenin, isoliquiritigenin (256.3 mg, 1 mmol) was dissolved in dry ethanol (20 ml) and 15% Pd/C (40 mg) was added. The reaction mixture was hydrogenated for 60 min at 2.0 × 105 Pa. Pd/C was filtered off, and the solvent was evaporated to obtain a light yellow solid (232.0 mg, 90%): mp = 170 to 172°C; literature value mp = 205.9 to 206.4°C (46); 1H NMR (500 MHz, DMSO-d6) δ 2.80 (t, 2H, 3J = 7.6 Hz, COCH2CH2), 3.19 (t, 2H, 3J = 7.6 Hz, COCH2CH2), 6.23 (d, 1H, 4J = 2.2 Hz, 3′-H), 6.34 (dd, 1H, 3J = 8.8 Hz, 4J = 2.3 Hz, 5′-H), 6.65 (d, 2H, 3J = 8.6 Hz, 3-H, 5-H), 7.03 (d, 2H, 3J = 8.5 Hz, 2-H, 6-H), 7.78 (d, 1H, 3J = 8.8 Hz, 6′-H), 9.10, 10.56 (each s, each 1H, 4-OH, 4′-OH), 12.60 (s, 1H, 2′-OH); 13C NMR (125 MHz, DMSO-d6) δ 29.23 (COCH2CH2), 102.55 (C-3′), 108.29 (C-5′), 112.71 (C-1′), 115.19 (C-3, C-5), 129.34 (C-2, C-6), 131.14, 133.13 (C-1, C-6′), 155.63 (C-4), 164.38, 164.87 (C-2′, C-4′), 204.00 (CO), the COCH2 signal is obscured by the DMSO signal; LC-MS (ESI) m/z 259 ([M+H]+), 96% purity.
Melting points were determined on a Büchi 510 melting point apparatus. 1H NMR spectra (500 MHz) and 13C NMR spectra (125 MHz) were recorded on a Bruker Avance DRX 500 spectrometer (Cologne, Germany). LC-MS analyses were carried out on an API 2000 mass spectrometer (Applied Biosystems, Darmstadt, Germany) coupled to an Agilent 1100 LC system (Santa Clara, CA, USA) using a Luna C18 column (50 mm by 2.0 mm, 3 μm; Phenomenex, Aschaffenburg, Germany) in gradient mode (90% water to 100% methanol in 10 min, held for 10 min). Purity of the compounds was determined using the diode array detector (DAD) of the LC-MS instrument between 200 and 400 nm.
Gene cloning and heterologous expression in E. coli.
The ERED-encoding gene from Eubacterium ramulus DSM 16296 (accession number KF154735) was cloned by genetically fusing Strep-tag II to its C terminus using the StarGate cloning system (IBA, Göttingen, Germany). The PCR mixture (12.5 μl) for gene amplification contained 1× Q5 high-fidelity master mix (New England BioLabs, Frankfurt, Germany), 0.5 μM primers FP-CF (5′-AGC GGC TCT TCA ATG GCA GAA AAA AAT CAG TAT TTT CC-3′) and FP-CR (5′-AGC GGC TCT TCT CCC GAT AAT TTC CAT TGC TGC GGT-3′), and 300 ng of genomic DNA. Genomic DNA was isolated from E. ramulus using the InViSorb genomic DNA kit (InViTek, Berlin, Germany). The PCR program was as follows: 98°C for 30 s; 25 cycles of 98°C for 10 s, 64°C for 30 s, and 72°C for 2 min; and finally 72°C for 2 min. The gene was finally cloned in pPSG-IBA 3, and the resulting plasmid, pERED-1, was introduced into E. coli KRX (Promega). Successful cloning was verified by sequencing the integrated fragment using plasmid-directed standard primers T7 and PSG-rev.
E. coli KRX pERED-1 was grown either oxically in LB broth (Lennox; Roth) supplemented with carbenicillin (100 μg ml−1; Roth) with shaking (250 rpm) at 37°C or anoxically in LB broth supplemented with carbenicillin, cysteine HCl (0.5 mg ml−1; Roth), and resazurin (1 μg ml−1; Roth) using a gas phase of N2/CO2 (80:20 [vol/vol]) at 37°C. Gene expression was induced by rhamnose (0.1% [wt/vol]) at an optical density at 600 nm (OD600) of approximately 0.6 in aerobic cultures, followed by incubation at 25°C. For anaerobic cultures, rhamnose was added at an OD600 of approximately 0.2 and growth continued at 37°C. For testing naringenin conversion by intact recombinant E. coli cells, a stock solution of this flavanone dissolved in DMSO was added to the cells at a final concentration of 500 μM (final DMSO concentration, 1%) simultaneously with the induction of gene expression by rhamnose.
Preparation of cell extracts from E. coli and enzyme purification.
All procedures were performed under anoxic conditions using a MACS anaerobic workstation (Don Whitley Scientific Ltd., Shipley, UK) containing a gas atmosphere of N2/CO2/H2 (80:10:10 [vol/vol/vol]), airtight tubes, and gassed solutions (N2/CO2, 80:20 [vol/vol]). Bacteria were harvested by centrifugation (10,000 × g for 10 min at 4°C), washed with 50 mM potassium phosphate buffer (PPB; pH 6.8), and subsequently resuspended in the same buffer. Cells were disrupted by shaking with 0.1 mm zirconia-silica beads (Roth) for 20 s at 6.0 m s−1 in a FastPrep instrument (FP120 System; MP Biomedicals, Heidelberg, Germany). Cell debris and unbroken cells were sedimented by centrifugation (12,000 × g for 20 min at 4°C).
Purification of tagged proteins from cell extracts was performed using a Strep-Tactin Sepharose column (1 ml; IBA) as described previously (47). Protein concentrations were determined by the Bio-Rad Bradford assay using bovine serum albumin as a standard.
Enzyme assays.
Enzyme activities were assayed under anoxic conditions in the anaerobic workstation using airlock-equilibrated solutions and N2/CO2-gassed PPB. Assay mixtures contained 50 mM PPB, 300 to 750 μM flavonoid compound (stock solution in DMSO, final DMSO concentration of 5%), 1 mM NADH, and cell extract (20 to 200 μg protein ml−1) or purified Fcr (0.4 to 14 μg protein ml−1). Cinnamic acids were applied at 500 μM and 5-methoxyresorcinol at 2.5 mM final concentration. For chiral HPLC analyses, stock solutions of flavonoids (naringenin, eriodictyol, liquiritigenin, homoeriodictyol, taxifolin, and dihydrokaempferol) were prepared in 70% (vol/vol) aqueous methanol, resulting in a final concentration of 3.5% methanol. The assay mixture was incubated at 25°C. Samples were withdrawn at different time points and mixed with one volume of methanol/acetic acid (50:2.5 [vol/vol]) to stop the reaction. After centrifugation (12,000 × g for 5 min), an aliquot (5 to 10 μl) of the supernatant was analyzed by reversed-phase (RP) HPLC.
Enzyme activities were calculated based on the product formation using nonlinear regression of data points (GraFit 5; Erithacus Software, East Grinstead, UK). Initial rates, v, of product formation were determined by using the equation [P] = v (1 – e−kt) k−1, where [P] is the product concentration at time t and k is the corresponding first-order rate constant for the gradual deceleration of the reaction. Initial rates of substrate consumption were determined by using the equations [S] = [S0] e−kt and v = k [S0], where [S] is the substrate concentration at time t and [S0] is the initial substrate concentration.
HPLC analyses.
For RP-HPLC analysis, a Dionex UltiMate 3000 LC system (Thermo Scientific, Braunschweig, Germany) was used, which was equipped with a WPS-3000TSL autosampler, a LPG-3400SD pump, a DAD-3000 diode-array detector, a TCC-3000 thermostatted column compartment, and a Zorbax SB-C18 column (4.6 mm by 150 mm, 5 μm; Agilent, Santa Clara, CA, USA) and controlled by Chromeleon software. The column temperature was kept at 25°C. Aqueous 2% (vol/vol) acetic acid (solvent A) and acetonitrile (solvent B) served as the mobile phase in a gradient mode (B from 5% to 50% in 6 min, held for 4 min) at a flow rate of 1 ml min−1. Detection was at 280 or 290 nm; UV spectra were recorded in the range of 190 to 400 nm. For analysis of cinnamic and dihydrocinnamic acids, the gradient was modified (B from 18% to 44% in 16.5 min, from 44% to 100% in 0.5 min, held for 4 min), and besides UV, fluorescence detection (excitation at 260 nm, emission at 305 nm) was applied using a Dionex FLD-3400RS fluorescence detector (Thermo Scientific, Braunschweig, Germany).
For chiral HPLC analyses, a Daicel Chiralpak AD-RH column (150 mm by 4.6 mm, 5 μm; Chiral Technologies, Illkirch, France) was used. The column temperature was kept at 30°C. The mobile phase was a mixture of 0.1% (vol/vol) aqueous trifluoroacetic acid (solvent A) and methanol (solvent B). For analysis of samples from naringenin conversion, solvents were supplied in a gradient mode (B from 30% to 90% in 10 min, held for 40 min) at a flow rate of 0.4 ml min−1. Samples of conversion experiments with dihydrokaempferol, taxifolin, and eriodictyol were analyzed using a gradient (B from 30% to 90% in 20 min, held for 20 min) at a flow rate of 0.8 ml min−1. Liquiritigenin and homoeriodictyol samples were analyzed in isocratic mode utilizing 90% B and 75% B, respectively, for 22 min at a flow rate of 0.8 ml min−1. Modified gradients were used for the isolation of metabolites formed by Fcr from dihydrokaempferol (B, 40% for 20 min, from 40% to 90% in 10 min, from 90% to 100% in 20 min) and taxifolin (B, 30% for 10 min, 40% for 20 min, from 40% to 90% in 10 min, from 90% to 100% in 20 min), whereas the homoeriodictyol product was isolated according to the method described above. For analysis of samples from spontaneous racemization tests, the mobile phase was a mixture of 0.05% aqueous phosphoric acid and acetonitrile (70:30 [vol/vol]) at a flow rate of 0.4 ml min−1 for 40 min in the case of naringenin or a flow rate of 0.8 ml min−1 for 20 min for eriodictyol, liquiritigenin, and homoeriodictyol. Enantiomers of liquiritigenin, homoeriodictyol, taxifolin, and dihydrokaempferol were assigned by using the following standard reference compounds: (S)-liquiritigenin, (S)-homoeriodictyol, (2R,3R)-taxifolin, and (2R,3R)-dihydrokaempferol, respectively. Assignment of naringenin and eriodictyol enantiomers was performed according to Guo et al. (48) based on the sequence of elution resulting from corresponding chiral HPLC analysis.
Calibration curves of the corresponding standard compounds were used for quantification with the following exceptions owing to lacking reference compounds. The products of enzymatic cleavage of homoeriodictyol, dihydrokaempferol, and taxifolin were quantified based on calibration curves of their respective educts. Products formed from butein (P1 and P2) were quantified by using the butein calibration curve.
Determination of racemization rates of flavanones.
Preparations with excess amounts of (S)-naringenin, (S)-eriodictyol, and (S)-liquiritigenin, resulting from the preferred conversion of the (R)-enantiomer by Fcr, were used to determine the rates of spontaneous racemization. Therefore, 500 μM each racemic flavanone was incubated in 50 mM PPB containing 3.5% methanol and 1 mM NADH with Fcr-containing E. coli cell extract (final concentration, 152 μg protein ml−1) under anoxic conditions at 25°C. Samples withdrawn after 10 min (naringenin and eriodictyol) or 1 h (liquiritigenin) were lyophilized, and residues were dissolved in 70% (vol/vol) aqueous methanol. Solutions were diluted to final concentrations of ca. 80 μM flavanone and 3.5% methanol in 50 mM PPB and incubated at 25°C for 9 days. To determine homoeriodictyol racemization, a stock solution of the commercially available (S)-enantiomer was diluted to a final concentration of 150 μM. Samples (30 μl) were withdrawn and frozen at −20°C, thawed on ice, mixed with one volume of methanol/acetic acid (50:2.5 [vol/vol]), and centrifuged (14,000 × g for 5 min). Aliquots of supernatants (5 μl) were analyzed by chiral HPLC.
Similar to the kinetic analysis of enzymatic activities (see above), ratios of major and minor enantiomers were plotted versus time, and the first-order rate constants of racemization were determined by nonlinear regression of data points using the equations of exponential decay and exponential product formation, respectively, with offsets. Standard errors refer to the nonlinear regressions.
Isolation of dihydrochalcone metabolites and LC-MS analysis.
Racemic homoeriodictyol, dihydrokaempferol, or taxifolin (0.9 mg ml−1, approximately 3.3 mM) was incubated in 50 mM PPB containing 3.14% methanol and 27 mM NADH with cell extract from Fcr-expressing E. coli (final protein concentration, 50 to 100 μg ml−1) under anoxic conditions at 25°C for 45 h. Aliquots (700 μl) were lyophilized, and residues were extracted with 200 μl methanol by vortexing for 25 min. After centrifugation (12,000 × g for 5 min), 5-μl aliquots of the supernatant were applied to chiral HPLC as described above. Fractions containing the corresponding product were collected manually, pooled, and dried by vacuum centrifugation. Residues were dissolved in acetonitrile/water (1:1 [vol/vol]) and analyzed by high-resolution mass spectrometry (HRMS) on a Bruker micrOTOF-Q mass spectrometer (Cologne, Germany) connected to a Dionex UltiMate 3000 LC system (Thermo Scientific, Braunschweig, Germany) via an ESI interface as described previously (49). A Nucleodur C18 gravity column (50 mm by 2.0 mm, 3 μm; Macherey-Nagel, Düren, Germany) was used in gradient mode (90% water to 100% acetonitrile from 2 to 10 min, held for 15 min). Purity of the compounds was determined using the DAD between 195 and 360 nm.
Polyacrylamide gel electrophoresis.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed according to Laemmli (50) using 13% resolving gels combined with 5% stacking gels. Prestained marker proteins (Precision Plus Protein WesternC standards; Bio-Rad) were used as molecular weight standards. Protein bands were stained with Coomassie brilliant blue R-250 (0.2% [wt/vol] in methanol/acetic acid/water, 45:45:10 [vol/vol/vol]).
For native PAGE, precast gels with a linear gradient of 4% to 15% acrylamide (Mini-PROTEAN TGX; Bio-Rad) were used. Gels were run in Tris-glycine buffer (10 mM Tris, 77 mM glycine, pH 8.5) at 150 V. Marker proteins (NativeMark unstained protein standard; Life Technologies) were used to estimate molecular weights. Protein bands were stained as described above.
Sequence analyses.
DNA sequencing was carried out by Eurofins MWG Operon (Ebersberg, Germany). The Vector NTI Suite 9 software package (Invitrogen) was employed to process and assemble the sequenced DNA fragments. Sequence similarity searches were done with the Basic Local Alignment Search Tool (BLAST, www.ncbi.nlm.nih.gov/blast).
Supplementary Material
ACKNOWLEDGMENTS
We thank Anke Gühler for technical assistance and Janina Schmitz, Christian Breuer, and Jim Küppers for support in compound synthesis and analysis.
We have no conflict of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01233-19.
REFERENCES
- 1.Oteiza PI, Fraga CG, Mills DA, Taft DH. 2018. Flavonoids and the gastrointestinal tract: local and systemic effects. Mol Aspects Med 61:41–49. doi: 10.1016/j.mam.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Cassidy A, Minihane AM. 2017. The role of metabolism (and the microbiome) in defining the clinical efficacy of dietary flavonoids. Am J Clin Nutr 105:10–22. doi: 10.3945/ajcn.116.136051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonzales GB, Smagghe G, Grootaert C, Zotti M, Raes K, Van Camp J. 2015. Flavonoid interactions during digestion, absorption, distribution and metabolism: a sequential structure-activity/property relationship-based approach in the study of bioavailability and bioactivity. Drug Metab Rev 47:175–190. doi: 10.3109/03602532.2014.1003649. [DOI] [PubMed] [Google Scholar]
- 4.Kawabata K, Yoshioka Y, Terao J. 2019. Role of intestinal microbiota in the bioavailability and physiological functions of dietary polyphenols. Molecules 24:370. doi: 10.3390/molecules24020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braune A, Blaut M. 2016. Bacterial species involved in the conversion of dietary flavonoids in the human gut. Gut Microbes 7:216–234. doi: 10.1080/19490976.2016.1158395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider H, Schwiertz A, Collins MD, Blaut M. 1999. Anaerobic transformation of quercetin-3-glucoside by bacteria from the human intestinal tract. Arch Microbiol 171:81–91. doi: 10.1007/s002030050682. [DOI] [PubMed] [Google Scholar]
- 7.Schneider H, Blaut M. 2000. Anaerobic degradation of flavonoids by Eubacterium ramulus. Arch Microbiol 173:71–75. doi: 10.1007/s002030050010. [DOI] [PubMed] [Google Scholar]
- 8.Schoefer L, Mohan R, Braune A, Birringer M, Blaut M. 2002. Anaerobic C-ring cleavage of genistein and daidzein by Eubacterium ramulus. FEMS Microbiol Lett 208:197–202. doi: 10.1111/j.1574-6968.2002.tb11081.x. [DOI] [PubMed] [Google Scholar]
- 9.Simmering R, Kleessen B, Blaut M. 1999. Quantification of the flavonoid-degrading bacterium Eubacterium ramulus in human fecal samples with a species-specific oligonucleotide hybridization probe. Appl Environ Microbiol 65:3705–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoefer L, Mohan R, Schwiertz A, Braune A, Blaut M. 2003. Anaerobic degradation of flavonoids by Clostridium orbiscindens. Appl Environ Microbiol 69:5849–5854. doi: 10.1128/aem.69.10.5849-5854.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herles C, Braune A, Blaut M. 2004. First bacterial chalcone isomerase isolated from Eubacterium ramulus. Arch Microbiol 181:428–434. doi: 10.1007/s00203-004-0676-2. [DOI] [PubMed] [Google Scholar]
- 12.Gall M, Thomsen M, Peters C, Pavlidis IV, Jonczyk P, Grunert PP, Beutel S, Scheper T, Gross E, Backes M, Geissler T, Ley JP, Hilmer JM, Krammer G, Palm GJ, Hinrichs W, Bornscheuer UT. 2014. Enzymatic conversion of flavonoids using bacterial chalcone isomerase and enoate reductase. Angew Chem Int Ed Engl 53:1439–1442. doi: 10.1002/anie.201306952. [DOI] [PubMed] [Google Scholar]
- 13.Schoefer L, Braune A, Blaut M. 2004. Cloning and expression of a phloretin hydrolase gene from Eubacterium ramulus and characterization of the recombinant enzyme. Appl Environ Microbiol 70:6131–6137. doi: 10.1128/AEM.70.10.6131-6137.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braune A, Engst W, Elsinghorst PW, Furtmann N, Bajorath J, Gütschow M, Blaut M. 2016. Chalcone isomerase from Eubacterium ramulus catalyzes the ring contraction of flavanonols. J Bacteriol 198:2965–2974. doi: 10.1128/JB.00490-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braune A, Engst W, Blaut M. 2005. Degradation of neohesperidin dihydrochalcone by human intestinal bacteria. J Agric Food Chem 53:1782–1790. doi: 10.1021/jf0484982. [DOI] [PubMed] [Google Scholar]
- 16.Wang LQ, Meselhy MR, Li Y, Nakamura N, Min BS, Qin GW, Hattori M. 2001. The heterocyclic ring fission and dehydroxylation of catechins and related compounds by Eubacterium sp. strain SDG-2, a human intestinal bacterium. Chem Pharm Bull (Tokyo) 49:1640–1643. doi: 10.1248/cpb.49.1640. [DOI] [PubMed] [Google Scholar]
- 17.Choi BY. 2019. Biochemical basis of anti-cancer-effects of phloretin–a natural dihydrochalcone. Molecules 24:278. doi: 10.3390/molecules24020278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Chen B, Xie H, He Y, Zhong D, Chen D. 2018. Antioxidant structure-activity relationship analysis of five dihydrochalcones. Molecules 23:1162. doi: 10.3390/molecules23051162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paraiso IL, Plagmann LS, Yang L, Zielke R, Gombart AF, Maier CS, Sikora AE, Blakemore PR, Stevens JF. 2019. Reductive metabolism of xanthohumol and 8-prenylnaringenin by the intestinal bacterium Eubacterium ramulus. Mol Nutr Food Res 63:e1800923. doi: 10.1002/mnfr.201800923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun J, Lin Y, Shen X, Jain R, Sun X, Yuan Q, Yan Y. 2016. Aerobic biosynthesis of hydrocinnamic acids in Escherichia coli with a strictly oxygen-sensitive enoate reductase. Metab Eng 35:75–82. doi: 10.1016/j.ymben.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Mordaka PM, Hall SJ, Minton N, Stephens G. 2018. Recombinant expression and characterisation of the oxygen-sensitive 2-enoate reductase from Clostridium sporogenes. Microbiology 164:122–132. doi: 10.1099/mic.0.000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wistuba D, Trapp O, Gel-Moreto N, Galensa R, Schurig V. 2006. Stereoisomeric separation of flavanones and flavanone-7-O-glycosides by capillary electrophoresis and determination of interconversion barriers. Anal Chem 78:3424–3433. doi: 10.1021/ac0600499. [DOI] [PubMed] [Google Scholar]
- 23.Kiehlmann E, Li E. 1995. Isomerization of dihydroquercetin. J Nat Prod 58:450–455. doi: 10.1021/np50117a018. [DOI] [Google Scholar]
- 24.Elsinghorst PW, Cavlar T, Müller A, Braune A, Blaut M, Gütschow M. 2011. The thermal and enzymatic taxifolin-alphitonin rearrangement. J Nat Prod 74:2243–2249. doi: 10.1021/np200639s. [DOI] [PubMed] [Google Scholar]
- 25.Krumholz LR, Bryant MP. 1986. Eubacterium oxidoreducens sp. nov. requiring H2 or formate to degrade gallate, pyrogallol, phloroglucinol and quercetin. Arch Microbiol 144:8–14. doi: 10.1007/BF00454948. [DOI] [Google Scholar]
- 26.Winter J, Popoff MR, Grimont P, Bokkenheuser VD. 1991. Clostridium orbiscindens sp. nov., a human intestinal bacterium capable of cleaving the flavonoid C-ring. Int J Syst Bacteriol 41:355–357. doi: 10.1099/00207713-41-3-355. [DOI] [PubMed] [Google Scholar]
- 27.Yokoyama S, Niwa T, Osawa T, Suzuki T. 2010. Characterization of an O-desmethylangolensin-producing bacterium isolated from human feces. Arch Microbiol 192:15–22. doi: 10.1007/s00203-009-0524-5. [DOI] [PubMed] [Google Scholar]
- 28.Yokoyama S, Oshima K, Nomura I, Hattori M, Suzuki T. 2011. Complete genomic sequence of the O-desmethylangolensin-producing bacterium Clostridium rRNA cluster XIVa strain SY8519, isolated from adult human intestine. J Bacteriol 193:5568–5569. doi: 10.1128/JB.05637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White GF, Russell NJ, Tidswell EC. 1996. Bacterial scission of ether bonds. Microbiol Rev 60:216–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picart P, de Maria PD, Schallmey A. 2015. From gene to biorefinery: microbial β-etherases as promising biocatalysts for lignin valorization. Front Microbiol 6:916. doi: 10.3389/fmicb.2015.00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamimura N, Takahashi K, Mori K, Araki T, Fujita M, Higuchi Y, Masai E. 2017. Bacterial catabolism of lignin-derived aromatics: new findings in a recent decade: update on bacterial lignin catabolism. Environ Microbiol Rep 9:679–705. doi: 10.1111/1758-2229.12597. [DOI] [PubMed] [Google Scholar]
- 32.Husarcíková J, Voß H, Domínguez de María P, Schallmey A. 2018. Microbial β-etherases and glutathione lyases for lignin valorisation in biorefineries: current state and future perspectives. Appl Microbiol Biotechnol 102:5391–5401. doi: 10.1007/s00253-018-9040-3. [DOI] [PubMed] [Google Scholar]
- 33.Fukuhara Y, Kamimura N, Nakajima M, Hishiyama S, Hara H, Kasai D, Tsuji Y, Narita-Yamada S, Nakamura S, Katano Y, Fujita N, Katayama Y, Fukuda M, Kajita S, Masai E. 2013. Discovery of pinoresinol reductase genes in sphingomonads. Enzyme Microb Technol 52:38–43. doi: 10.1016/j.enzmictec.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Bess EN, Bisanz JE, Spanogiannopoulos P, Ang QY, Bustion A, Kitamura S, Alba DL, Wolan DW, Koliwad SK, Turnbaugh PJ. 2018. The genetic basis for the cooperative bioactivation of plant lignans by a human gut bacterial consortium. bioRxiv doi: 10.1101/357640. [DOI] [PMC free article] [PubMed]
- 35.Hubbard PA, Liang X, Schulz H, Kim JJ. 2003. The crystal structure and reaction mechanism of Escherichia coli 2,4-dienoyl-CoA reductase. J Biol Chem 278:37553–37560. doi: 10.1074/jbc.M304642200. [DOI] [PubMed] [Google Scholar]
- 36.Min T, Kasahara H, Bedgar DL, Youn B, Lawrence PK, Gang DR, Halls SC, Park H, Hilsenbeck JL, Davin LB, Lewis NG, Kang C. 2003. Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases and their relationship to isoflavone reductases. J Biol Chem 278:50714–50723. doi: 10.1074/jbc.M308493200. [DOI] [PubMed] [Google Scholar]
- 37.O'Hagan D, Schmidberger JW. 2010. Enzymes that catalyse SN2 reaction mechanisms. Nat Prod Rep 27:900–918. doi: 10.1039/b919371p. [DOI] [PubMed] [Google Scholar]
- 38.Krause M, Galensa R. 1991. Analysis of enantiomeric flavanones in plant extracts by high-performance liquid-chromatography on a cellulose triacetate based chiral stationary phase. Chromatographia 32:69–72. doi: 10.1007/BF02262470. [DOI] [Google Scholar]
- 39.Yanez JA, Remsberg CM, Miranda ND, Vega-Villa KR, Andrews PK, Davies NM. 2008. Pharmacokinetics of selected chiral flavonoids: hesperetin, naringenin and eriodictyol in rats and their content in fruit juices. Biopharm Drug Dispos 29:63–82. doi: 10.1002/bdd.588. [DOI] [PubMed] [Google Scholar]
- 40.Yanez JA, Andrews PK, Davies NM. 2007. Methods of analysis and separation of chiral flavonoids. J Chromatogr B Analyt Technol Biomed Life Sci 848:159–181. doi: 10.1016/j.jchromb.2006.10.052. [DOI] [PubMed] [Google Scholar]
- 41.Ohmura W, Ohara S, Hashida K, Aoyama M, Doi S. 2002. Hydrothermolysis of flavonoids in relation to steaming of Japanese larch wood. Holzforschung 56:493–497. doi: 10.1515/HF.2002.076. [DOI] [Google Scholar]
- 42.Vega-Villa KR, Remsberg CM, Takemoto JK, Ohgami Y, Yanez JA, Andrews PK, Davies NM. 2011. Stereospecific pharmacokinetics of racemic homoeriodictyol, isosakuranetin, and taxifolin in rats and their disposition in fruit. Chirality 23:339–348. doi: 10.1002/chir.20926. [DOI] [PubMed] [Google Scholar]
- 43.Hanske L, Loh G, Sczesny S, Blaut M, Braune A. 2010. Recovery and metabolism of xanthohumol in germ-free and human microbiota-associated rats. Mol Nutr Food Res 54:1405–1413. doi: 10.1002/mnfr.200900517. [DOI] [PubMed] [Google Scholar]
- 44.Matthies A, Clavel T, Gütschow M, Engst W, Haller D, Blaut M, Braune A. 2008. Conversion of daidzein and genistein by an anaerobic bacterium newly isolated from the mouse intestine. Appl Environ Microbiol 74:4847–4852. doi: 10.1128/AEM.00555-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klinke P, Gibian H. 1961. Über chalkone. Chem Ber 94:26–38. doi: 10.1002/cber.19610940105. [DOI] [Google Scholar]
- 46.Jesus AR, Marques AP, Rauter AP. 2016. An easy approach to dihydrochalcones via chalcone in situ hydrogenation. Pure Appl Chem 88:349–361. doi: 10.1515/pac-2016-0303. [DOI] [Google Scholar]
- 47.Schröder C, Matthies A, Engst W, Blaut M, Braune A. 2013. Identification and expression of genes involved in the conversion of daidzein and genistein by the equol-forming bacterium Slackia isoflavoniconvertens. Appl Environ Microbiol 79:3494–3502. doi: 10.1128/AEM.03693-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo X, Li C, Duan L, Zhao L, Lou H, Ren D. 2012. Separation of the enantiomers of naringenin and eriodictyol by amylose-based chiral reversed-phase high-performance liquid chromatography. Drug Discov Ther 6:321–326. doi: 10.5582/ddt.2012.v6.6.321. [DOI] [PubMed] [Google Scholar]
- 49.Schulz-Fincke AC, Tikhomirov AS, Braune A, Girbl T, Gilberg E, Bajorath J, Blaut M, Nourshargh S, Gütschow M. 2018. Design of an activity-based probe for human neutrophil elastase: implementation of the Lossen rearrangement to induce Förster resonance energy transfers. Biochemistry 57:742–752. doi: 10.1021/acs.biochem.7b00906. [DOI] [PubMed] [Google Scholar]
- 50.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.