Viruses associated with mosquitoes have made a large impact on public and veterinary health. In the United States, several viruses, including WNV, DENV, and chikungunya virus (CHIKV), are responsible for human disease. From 2015 to 2018, imported Zika cases were reported in the United States, and in 2016 to 2017, local Zika transmission occurred in the states of Texas and Florida. With globalization and a changing climate, the frequency of outbreaks linked to arboviruses will increase, revealing a need to better detect viruses in vector populations. With the capacity of the LLMDA to detect viruses, bacteria, and fungi, this study highlights its ability to broadly screen field-collected mosquitoes and contribute to the surveillance and management of arboviral diseases.
KEYWORDS: Aedes aegypti, Aedes albopictus, Culex, Wolbachia, cell-fusing agent virus, Culex flavivirus, insect-specific virus, microarrays
ABSTRACT
Several mosquito-borne diseases affecting humans are emerging or reemerging in the United States. The early detection of pathogens in mosquito populations is essential to prevent and control the spread of these diseases. In this study, we tested the potential applicability of the Lawrence Livermore Microbial Detection Array (LLMDA) to enhance biosurveillance by detecting microbes present in Aedes aegypti, Aedes albopictus, and Culex mosquitoes, which are major vector species globally, including in Texas. The sensitivity and reproducibility of the LLMDA were tested in mosquito samples spiked with different concentrations of dengue virus (DENV), revealing a detection limit of >100 but <1,000 PFU/ml. Additionally, field-collected mosquitoes from Chicago, IL, and College Station, TX, of known infection status (West Nile virus [WNV] and Culex flavivirus [CxFLAV] positive) were tested on the LLMDA to confirm its efficiency. Mosquito field samples of unknown infection status, collected in San Antonio, TX, and the Lower Rio Grande Valley (LRGV), TX, were run on the LLMDA and further confirmed by PCR or quantitative PCR (qPCR). The analysis of the field samples with the LLMDA revealed the presence of cell-fusing agent virus (CFAV) in A. aegypti populations. Wolbachia was also detected in several of the field samples (A. albopictus and Culex spp.) by the LLMDA. Our findings demonstrated that the LLMDA can be used to detect multiple arboviruses of public health importance, including viruses that belong to the Flavivirus, Alphavirus, and Orthobunyavirus genera. Additionally, insect-specific viruses and bacteria were also detected in field-collected mosquitoes. Another strength of this array is its ability to detect multiple viruses in the same mosquito pool, allowing for the detection of cocirculating pathogens in an area and the identification of potential ecological associations between different viruses. This array can aid in the biosurveillance of mosquito-borne viruses circulating in specific geographical areas.
IMPORTANCE Viruses associated with mosquitoes have made a large impact on public and veterinary health. In the United States, several viruses, including WNV, DENV, and chikungunya virus (CHIKV), are responsible for human disease. From 2015 to 2018, imported Zika cases were reported in the United States, and in 2016 to 2017, local Zika transmission occurred in the states of Texas and Florida. With globalization and a changing climate, the frequency of outbreaks linked to arboviruses will increase, revealing a need to better detect viruses in vector populations. With the capacity of the LLMDA to detect viruses, bacteria, and fungi, this study highlights its ability to broadly screen field-collected mosquitoes and contribute to the surveillance and management of arboviral diseases.
INTRODUCTION
Mosquito-borne viruses emerge and reemerge at accelerating rates, causing significant morbidity and mortality in humans and animals (1). Due to globalization, mosquito vectors and associated arboviruses have been introduced into new geographic regions (2–5). One noteworthy example was the introduction of West Nile virus (WNV) into the New World. The virus was first detected in New York in 1999 and then spread throughout the United States (6) using several Culex species as vectors. The yellow fever mosquito, Aedes aegypti, and the Asian tiger mosquito, Aedes albopictus, are invasive mosquito species that are widespread in urban environments of tropical, subtropical, and temperate regions and are responsible for the emergence or reemergence of multiple mosquito-borne diseases caused by different viral agents, including dengue virus (DENV) (7–9), chikungunya virus (CHIKV), and, more recently, Zika virus (ZIKV). Since its introduction in Brazil in 2014, ZIKV has spread to the rest of South America and moved north to Central and North America, resulting in the local transmission of the virus in Florida and Texas in 2016 to 2017 (10–12).
These mosquito-borne viruses have proven to be difficult to manage and control despite considerable attention, and the ability to broadly screen mosquitoes for microbes has appeal on many fronts. Microarrays have the ability to detect multiple targets that would be missed by other more-specific or targeted assays and could reveal important components of the mosquito microbiome relevant to the transmission of viruses of public and veterinary health importance. Typically, microbial diversity associated with mosquitoes has been studied using both culture-dependent and -independent approaches (13–16). While culture-dependent approaches are time-consuming, molecular techniques such as reverse transcription PCR (RT-PCR) (17–19) and quantitative real-time PCR (qRT-PCR) (20–22) are typically designed to be specific at the species or family level. More recently, many new forms of next-generation sequencing (NGS) (23, 24) have proven to be effective to characterize the mosquito microbiome, but they require the depletion of host-derived nucleic acid in order to sensitively detect viruses (25, 26). For bacterial discovery, 16S rRNA sequencing is usually performed (27, 28), but it detects only conserved regions of the 16S rRNA gene of bacteria and does not allow for the detection of viruses and other microbes in the sample. Shotgun metagenomic sequencing provides the highest resolution to detect different kinds of microbes in a sample (29) but remains expensive and time-consuming and requires extensive bioinformatic expertise.
Accordingly, this study utilized the Lawrence Livermore Microbial Detection Array (LLMDA), which was designed to screen diverse samples for thousands of bacteria, viruses, fungi, and protozoa (30, 31). The LLMDA version used in this study detects 10,261 species of microbes, including 4,219 viruses, 5,367 bacteria, 293 archaebacteria, 265 fungi, and 117 protozoa (32). The LLMDA has been previously used to detect viral and bacterial pathogens from clinical and archeological samples (30, 33). We conducted a pilot study to evaluate the utility of the LLMDA to screen mosquito pools collected from multiple regions of Texas from 2016 to 2017 for mosquito-borne viruses. The LLMDA was able to detect and identify DENV serotype 2 (DENV-2), Rift Valley fever virus (RVFV), and Mayaro virus (MAYV) in spiked mosquito samples and WNV, Culex flavivirus (CxFLAV), and cell-fusing agent virus (CFAV) from field-collected mosquitoes. LLMDA results from field-collected mosquitoes were further confirmed using standard and/or quantitative PCR methods, and coinfection with multiple viruses was detected in spiked and field-collected mosquitoes. Viruses were detected in pools of mosquitoes of various size and in tissues, including midguts (MG) and salivary glands (SG). Additionally, Wolbachia was detected in field-collected Aedes aegypti and Culex mosquitoes.
RESULTS
In total, we analyzed 39 mosquito pools representing 512 individual mosquitoes (see Table S1 in the supplemental material). Ten pools were field-collected A. aegypti (n = 116), eight pools were colony-raised A. aegypti Liverpool (n = 80), eight pools were field-collected A. albopictus (n = 49), four pools were field-collected Culex spp. (n = 86), and six pools were field-collected Culex quinquefasciatus (n = 138). One pool was colony-raised C. quinquefasciatus (n = 10), and one pool was an equal mixture of colony-raised A. aegypti and C. quinquefasciatus (n = 20) to serve as a negative control. To understand the compartmentalization of bacteria within A. aegypti and Culex sp. mosquitoes, four additional pools were analyzed: one pool of 23 midguts (MG) and one pool of 23 salivary glands (SG) for each mosquito species (A. aegypti and Culex spp.).
LLMDA sensitivity and reproducibility.
In order to test the LLMDA sensitivity and reproducibility, we spiked known amounts of DENV serotype 2 (DENV-2) in A. aegypti Liverpool mosquito pools, each containing 10 female mosquitoes. Duplicate pools were spiked with 102 PFU/ml or 103 PFU/ml of virus, and two other pools were spiked with 104 PFU/ml or 105 PFU/ml (Table 1). According to our results, the limit of detection, or minimum amount of virus required to determine its presence or absence in the sample, is equal to or less than 103 PFU/ml and above 102 PFU/ml. The DENV-2 dilutions (103 PFU/ml, 104 PFU/ml, and 105 PFU/ml) were all detected using the array, with positive probes hybridizing to different regions of the DENV-2 genome (Fig. 1A). Because positive signals from more than 20% of the probes for DENV-2 were detected and were in several regions of the genome, these DENV-2-spiked samples are considered DENV positive. As seen in Table 1, the number of positive probes was close to matching the total number of probes present on the array for this target, especially for the samples spiked with the largest amount of virus. Additionally, the log CL ratio (ratio between the likelihood of the observed probe signal when assuming the target is present in the sample and the likelihood when assuming no target is present) was above 0, and therefore the samples were considered DENV positive. An increase in the log CL ratio, ranging from 56.7 to 224.6, was observed, correlating with the increase in amount of spiked virus. The reproducibility of the LLMDA was tested for two of the dilutions in duplicates (102 PFU/ml and 103 PFU/ml) and showed consistency. For the duplicates with 102 PFU/ml, no signal was recovered, and for the duplicates with 103 PFU/ml, the log CL ratios were similar, with respective values of 56.7 and 60.7.
TABLE 1.
LLMDA limit of detection and reproducibility in spiked mosquito pools
| Virus(es) | PFU/ml | LLMDA detection | Log CL ratio | No. of probes positive/total | Mosquito species |
|---|---|---|---|---|---|
| DENV-2 | 102 | Negative | A. aegypti | ||
| 102 | Negative | A. aegypti | |||
| 103 | Positive | 56.7 | 20/27 | A. aegypti | |
| 103 | Positive | 60.7 | 23/33 | A. aegypti | |
| DENV-2 + MAYV | 104, 104 | Positive | 197.1, 78.5 | 46/47, 20/25 | A. aegypti |
| 105, 104 | Positive | 224.6, 122.3 | 53/54, 25/25 | A. aegypti | |
| RVFV | 104 | Positive | 52.8 | 16/19 | C. quinquefasciatus |
| ZIKV | 104 | Negative | 0 | 3/27 | A. aegypti |
| 102 | Negative | 0 | 3/27 | A. aegypti |
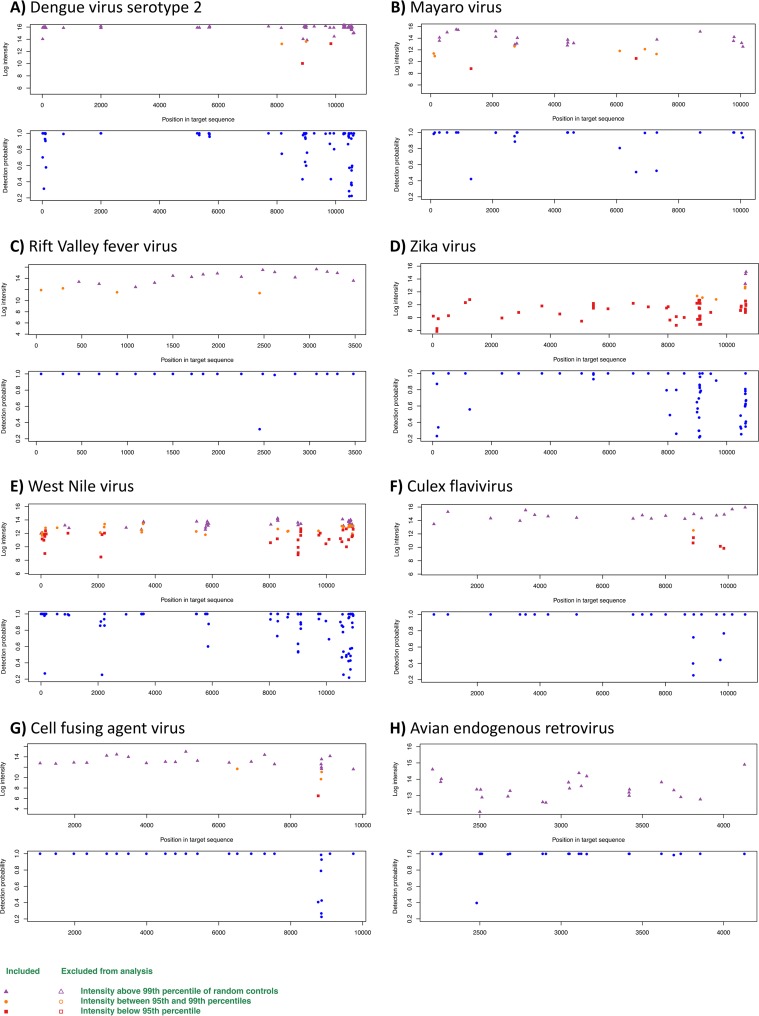
FIG 1.
LLMDA probe detection for DENV-2 (A), MAYV (B), RVFV (C), ZIKV (D), WNV (E), CxFLAV (F), CFAV (G), and avian endogenous retrovirus (H). For each virus, two graphs are shown; the upper panel represents the intensity of the probes according to the position of the target in the genome; the lower panel represents the probability of detection according to the genome region. Samples for which intensity was higher than the 99th percentile of that of the control probes are shown in purple, those for which intensity is between the 99th and 95th percentiles are shown in orange, and those for which the intensity was below the 95th percentile are in red.
Samples spiked with the largest amounts of DENV (105 PFU/ml and 104 PFU/ml) were coinfected with a known amount of Mayaro virus (MAYV) (104 PFU/ml). Both viruses were successfully detected by the LLMDA (Fig. 1A and B), demonstrating the ability of the LLMDA to detect viruses from different families if present in the same mosquito sample pool. Additionally, C. quinquefasciatus spiked with a known amount of Rift Valley fever virus (RVFV) (104 PFU/ml) also resulted in a positive signal, highlighting the ability of the LLMDA to detect other arboviruses of medical and veterinary importance (Fig. 1C). The A. aegypti homogenates spiked with ZIKV tested negative by the LLMDA. First, as seen in Fig. 1D, only 3 probes out of the 27 designed to detect ZIKV had a positive signal (the percentage of positive probes was therefore below the default threshold of 20). Second, the 3 high-intensity probes cover only a specific region of the genome instead of spanning across the genome. Third, the log CL ratio was equal to zero. These spiked samples were confirmed to be ZIKV positive using a quantitative PCR (qPCR) assay, with threshold cycle (CT) values of 20.63 and 28.96 for the samples spiked with 104 and 102 PFU/ml, respectively. In addition, densoviruses were detected in all of the DENV-2- and MAYV-spiked A. aegypti samples but were further tested by PCR for confirmation (see Table S4 in the supplemental material).
Application of LLMDA to detection of viruses from field-collected mosquitoes of known infection status.
In order to test the ability of the LLMDA to detect natural virus loads within mosquito pools, naturally WNV- and CxFLAV-infected mosquitoes previously collected in Chicago, IL, and College Station, TX, were used (Table 2). Of the two WNV-positive mosquito pools previously detected using qPCR (CT values, 15.16 and 19.95), only one was successfully identified as WNV by the LLMDA (Fig. 1E). In this particular case, 58 out of the 79 probes that characterized WNV were positive, and a log CL score of 115.3 was observed. Interestingly, of these two pools, one was found positive for Culex flavivirus (CxFLAV) by the microarray. In this sample, 19 out of 19 probes were positive (log CL ratio, 74.4), revealing the ability of the microarray to detect coinfections from naturally infected mosquito pools (Fig. 1F). The two CxFLAV-positive controls from College Station (CT values of 18.24 and 30.31) were not detected using the microarray (Table 2).
TABLE 2.
Comparison of LLMDA and qPCR results in naturally infected mosquito pools
| Virus | qPCR detection | Observed CT value | LLMDA detection | Log CL ratio | No. of probes positive/total | Mosquito species | Additional LLMDA virus detected | Log CL ratio | No. of probes positive/total |
|---|---|---|---|---|---|---|---|---|---|
| WNV | Positive | 15.16 | Positive | 115.3 | 58/79 | Culex spp. | CxFLAV | 74.4 | 19/19 |
| Positive | 19.95 | Negative | 0/79 | Culex spp. | CxFLAV | 0/19 | |||
| CxFLAV | Positive | 18.24 | Negative | 0/75 | C. quinquefasciatus | ||||
| Positive | 30.31 | Negative | 0/75 | C. quinquefasciatus |
Application of the LLMDA to detection of microbes from field-collected mosquitoes of unknown infection status.
(i) LLMDA viral analysis. Several viruses were detected in the field-collected mosquito pools (Fig. 1). A. aegypti from Lower Rio Grande Valley (LRGV) (n = 2) and San Antonio (n = 1) were found to be positive for cell-fusing agent virus (CFAV), an insect-specific flavivirus (Fig. 1G). All 21 probes designed for that virus on the array were positives (log CL ratio = 77). Aedes aegypti SG and MG pools were also positive for CFAV (log CL ratio = 77; positive probes/all target probes =21/21). Interestingly, one A. aegypti pool from the LRGV was found to be positive for the avian endogenous retrovirus (23 out of 23 expected probes; log CL ratio = 74.9) (Fig. 1H). None of the field-collected A. albopictus or Culex sp. samples tested positive for viruses, with the exception of the Culex population from Chicago (as described in the previous paragraph). To assess the accuracy of the LLMDA to detect the presence of insect-specific viruses, all samples were tested using conventional PCR methods with gene-specific primers designed for CFAV and CxFLAV (Tables 3 and 4). CFAV strain TX AR 11-1022 and CxFLAV strain M23873, obtained from the University of Texas Medical Branch (UTMB) World Reference Center for Emerging Viruses and Arboviruses (WRCEVA), were used as positive controls for the conventional PCR assay. Samples resulting in an amplicon were Sanger sequenced. The CFAV PCR assay confirmed the 5 microarray CFAV-positive pools and allowed the detection of 3 additional CFAV-positive pools. The CFAV strains detected in the A. aegypti pools from the LRGV showed 97.7% identity to CFAV strain from Puerto Rico (accession number GQ165810), while the CFAV strains from the A. aegypti population from San Antonio share 100% homology to a CFAV strain from Mexico (accession number KJ476731). Aedes aegypti SG and MG were both confirmed positive for CFAV (Table 4). For CxFLAV, only one of the two positive pools from Chicago identified by the microarray was confirmed positive by conventional PCR. While the microarray was not able to detect any CxFLAV as positive in the pools from College Station, these 2 pools were detected as CxFLAV positive by PCR (Table 3). CxFLAV strains from C. quinquefasciatus (College Station, TX) and Culex spp. (from Chicago) show 100% identity to a CxFLAV strain isolated from Culex pipiens in the United States (accession number KX512322).
TABLE 3.
LLMDA and conventional PCR detection of field-collected samples
| Locality | Mosquito species | Sample size | Virus | LLMDA detectiona | PCR detectiona | % identity by Sanger sequencing (accession no.) |
|---|---|---|---|---|---|---|
| LRGV | A. aegypti | 96 | CFAV | 2 (9) | 3 (9) | 97.7 (GQ165810) |
| San Antonio | A. aegypti | 33 | CFAV | 1 (2) | 2 (2) | 100 (KJ476731) |
| Colony | A. aegypti | 40 | CFAV | 0 (4) | 0 (4) | |
| LRGV | A. albopictus | 4 | CFAV | 0 (3) | 0 (3) | |
| San Antonio | A. albopictus | 36 | CFAV | 0 (2) | 0 (2) | |
| College Station | A. albopictus | 9 | CFAV | 0 (3) | 0 (3) | |
| LRGV | C. quinquefasciatus | 25 | CxFLAV | 0 (2) | 0 (2) | |
| San Antonio | C. quinquefasciatus | 13 | CxFLAV | 0 (2) | 0 (2) | |
| College Station | C. quinquefasciatus | 100 | CxFLAV | 0 (2) | 2 (2) | 100 (KX512322) |
| Chicago | Culex spp. | 70 | CxFLAV | 2 (2) | 1 (2) | 100 (KX512322) |
| LRGV | Culex spp. | 16 | CxFLAV | 0 (2) | 0 (2) |
Number of positive pools (total number of pools tested).
TABLE 4.
LLMDA and conventional PCR detection of insect-specific viruses in mosquito midguts and salivary glands in 23 samples from the LRGV
| Mosquito species | Tissue | n | Virus detected | LLMDA detectiona | PCR detectiona |
|---|---|---|---|---|---|
| A. aegypti | Midguts | 23 | CFAV | Positive (1/1) | Positive (1/1) |
| A. aegypti | Salivary glands | 23 | CFAV | Positive (1/1) | Positive (1/1) |
| Culex spp. | Midguts | 23 | CxFLAV | Negative (0/1) | Negative (0/1) |
| Culex spp. | Salivary glands | 23 | CxFLAV | Negative (0/1) | Negative (0/1) |
Values in parentheses are number of positive pools/total number of pools tested.
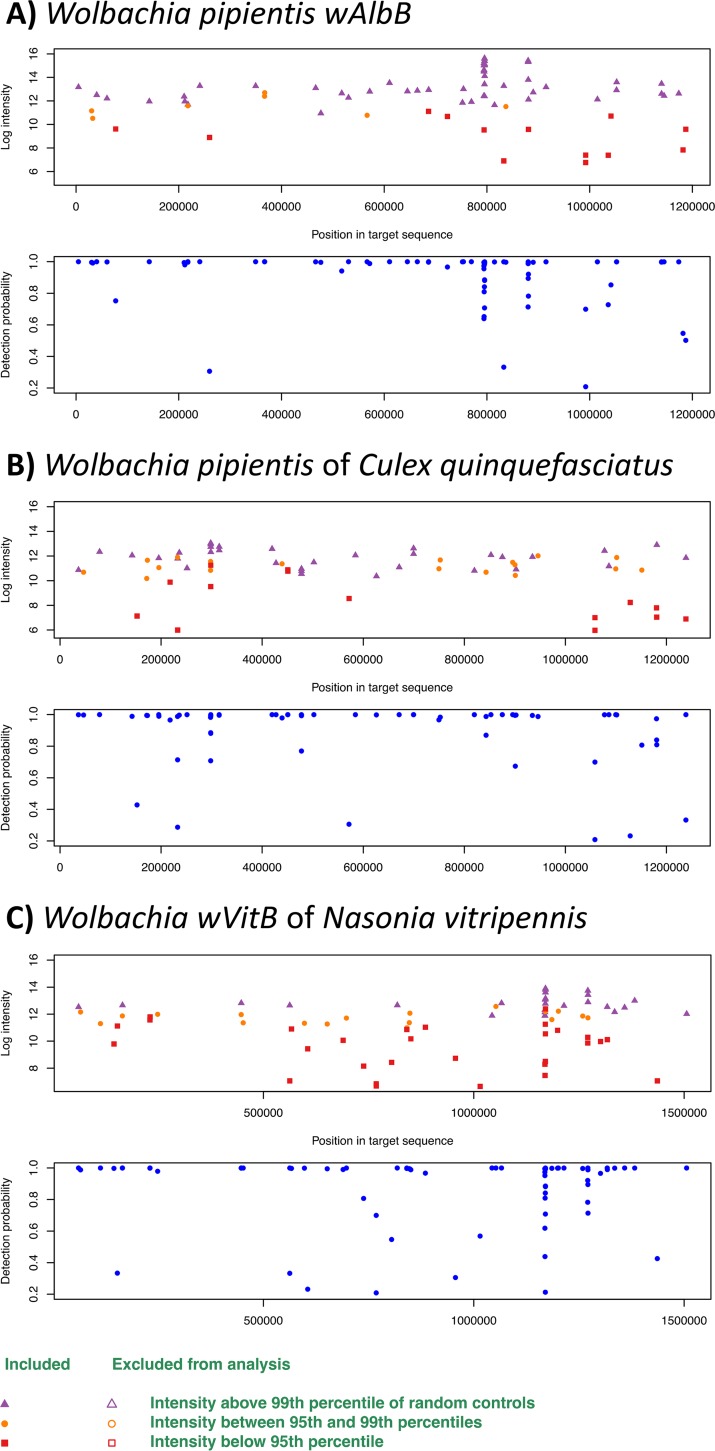
(ii) LLMDA bacterial analysis. Several A. albopictus and Culex sp. mosquito pools from Texas and Chicago were found to be naturally infected with Wolbachia (Fig. 2). A. albopictus mosquitoes from LRGV and San Antonio were infected with the Wolbachia pipientis symbiont of Aedes albopictus from the supergroup B (wAlbB) (log CL ratio = 199.7; positive probes/all target probes = 55/59) (Fig. 2A). Culex sp. mosquitoes from Chicago and Texas (LRGV) were infected with the Wolbachia pipientis symbiont of Culex pipiens from supergroup B (wPip) (log CL ratio = 95.5; positive probes/all target probes = 42/58) (Fig. 2B). In the San Antonio collection, one pool of Culex was found to be infected with wAlbB (log CL ratio = 199.7; probes detected/expected = 55/59), and one pool of A. albopictus was infected with the Wolbachia pipientis symbiont of Nasonia vitripennis from subgroup B (wVitB) (log CL ratio = 169.6; probes detected/expected = 50/56) (Fig. 2C). A few other bacteria, including Pseudomonas, Klebsiella, and Erwinia spp., were detected in various samples (Table S4). All mosquito pools identified as positive for Wolbachia using the microarray were subject to a Wolbachia surface protein gene (wsp) qPCR assay (Table 5). A. albopictus from the LRGV and San Antonio were confirmed to be harbor wspB. Additionally, these samples were found to be positive for the wspA gene. Whereas 2 A. albopictus pools from San Antonio were found to be positive with the LLDA, only one was confirmed using the wsp qPCR assay. The Culex spp. from San Antonio, TX, Chicago, IL, and the LRGV were all confirmed positive for the wspB gene, with CT values of 23.47, 29.77, and 19.99, respectively.
FIG 2.
LLMDA probe detection of Wolbachia strains. (A) Wolbachia pipientis wAlbB; (B) Wolbachia endosymbiont of Culex quinquefasciatus; (C) Wolbachia endosymbiont wVitB. For each bacterium, the upper panel represents the intensity of the probes according to the position of the target in the genome (>99th percentile of control in purple, 95th to 99th, in orange, and <95th in red). The lower panel represents the probability of detection according to the genome region.
TABLE 5.
Wolbachia detection in field mosquito sample from Texas and Chicago using LLMDA and qPCR with the wsp gene
| Locality | Mosquito species | n | LLMDA |
qPCR |
|||
|---|---|---|---|---|---|---|---|
| Detectiona | Strain | Detectiona | Wsp | CT value(s) | |||
| LRGV | A. albopictus | 4 | 1 (3) | wAlbB | 1 (3) | A + B | 25.0, 24.34 |
| San Antonio | A. albopictus | 36 | 2 (2) | wAlbB | 1 (1) | A + B | 19.37, 21.70 |
| wVitB | 1 (1) | B | 19.99 | ||||
| San Antonio | C. quinquefasciatus | 13 | 1 (2) | wAlbB | 1 (2) | B | 23.47 |
| Chicago | Culex spp. | 70 | 1 (2) | wPip | 1 (2) | B | 29.77 |
| LRGV | Culex spp. | 41 | 1 (4) | wPip | 1 (4) | B | 19.99 |
Number of positive pools (total number of pools tested).
DISCUSSION
Viruses.
The LLMDA version used in the study (v7) was developed in 2014 and can detect 4,219 viruses, 5,367 bacteria, 293 archaebacteria, 265 fungi, and 117 protozoa. We utilized this platform to evaluate its ability to screen mosquito pools for viruses and other microbes. Our study demonstrates that the LLMDA is a broad screening tool that can be used to detect introduced or emerging pathogens in mosquito populations as well as the presence of other insect-specific viruses and bacteria. The LLMDA is able to generate a comprehensive analysis of microbes circulating in mosquito populations of a specific area that could be used to implement future vector control programs. Because it is highly multiplexed and is based on random amplification, the LLMDA presents advantages over singleplex and multiplex PCR assays and cost and time advantages over next-generation sequencing. First, the sensitivity of the array was determined to be above 102 and below 103 PFU/ml using serial dilutions of DENV-2, a virus of major public health importance around the world. The array probes were designed to detect both conserved and unique regions of DENV using whole-genome sequences from 3,097 DENV genomes from all four serotypes, of which 403 were specific to DENV-2. The limit of detection of this virus in our array is within the range of viral detection from previous studies using the LLMDA (31, 34) and of other microarrays (35, 36). An interesting feature of the LLMDA is its ability to detect multiple infections from a single sample pool that would normally be missed if a gene-specific PCR approach is used. For example, the LLMDA detected both MAYV and DENV in mosquito pools coinfected with known amounts of both viruses. The LLMDA also successfully detected several viruses in field-collected mosquitoes of known (Table 2) and unknown (Table 3) infection status. For instance, in our study, one Culex sp. pool from Chicago, IL, was found to be dually infected with WNV and CxFLAV, which confirms prior studies documenting the cocirculation of these two viruses (37, 38). The presence of several viruses in a mosquito pool does not necessarily mean coinfection in a single mosquito, but coinfection of these two viruses has been previously reported (38, 39). Additionally, CxFLAV has been shown to interact with WNV transmission in Culex mosquitoes (40). This highlights the ability of the LLMDA to detect and identify two closely related viruses and viruses from different families within a sample if present.
LLMDA and PCR assays both detected the presence of CFAV and CxFLAV in several mosquito pools. When the LLMDA v7 array was designed in 2014, 22 CxFLAV sequences and one CFAV genome were publicly available. CxFLAV was detected in Culex sp. mosquito pools collected in Chicago, IL, but not in C. quinquefasciatus pools from College Station, TX. The inconsistency of the microarray to detect CxFLAV could be due to the variation in sequence between CxFLAV strains from different geographic origins or from different host species. Here the portion of the NS5 gene sequenced shows 100% homology to that of C. pipiens strain KX512322, but CxFLAV strains from different localities and different mosquito species have been shown by full genome analysis to cluster in two different clades (clade 1 and 2), with all the C. quinquefasciatus-related strains clustering together in clade 2 (41).
Additionally, the inconsistency of the results could be due to the difference in sensitivity between the two techniques and the fact that while the conventional PCR relies on the use of gene-specific primers, the microarray relies on the use of random primers during the amplification process. All Aedes sp. pools were found to be negative for CxFLAV.
CFAV was detected in A. aegypti from San Antonio, TX, and the LRGV, TX. Once again, the conventional PCR allowed the detection of CFAV in two additional samples, probably due to the difference in sensitivity between the two techniques. The tissue dissection revealed the presence of CFAV in both the MG and the SG, the two main barriers of arbovirus replication within the mosquito. This tropism suggests its potential for interaction with other viruses present within the mosquito. The ability of CFAV to transmit from one generation to the next (42), as well as its ability to interact with DENV in an A. aegypti cell line (43), makes it a promising candidate for paratransgenesis. Culex sp. pools were found to be negative for CFAV. The ability of the LLMDA to detect insect-specific viruses is of interest because it allows the characterization of ecological associations between insect-specific viruses and human pathogens that occur in nature. These could in turn be investigated for the impact of the insect-specific virus on the transmission of the human pathogen and serve as potential future vector control strategies.
The ZIKV strain PRVABC59 used in this study belongs to the Asian lineage and was not detected using the LLMDA. The LLMDA was designed in 2014, when the only ZIKV sequence available was that of the MR-766 African-lineage strain (accession number NC_012532.1). The two viral strains share only 87 to 90% homology (44, 45). Thus, it is likely that the genetic diversity of the PRVABC59 ZIKV strain compared to the MR-766 African strain did not allow for an efficient detection by the Zika probes present on the LLMDA. This result specifically highlights the need to design additional probes capable of recognizing the more-contemporary Asian lineage of ZIKV and, more broadly, the perpetual need to update the microarray as new viruses or viral strains are discovered or emerge.
Overall, this study was able to detect several viral symbionts. In the A. aegypti samples spiked with DENV-2 and/or MAYV, densoviruses were detected, but they were not detected in the nonspiked sample. This reflects the presence of the densoviruses in the C6/36 cells used to grow the different viruses (46–49). Surprisingly, endogenous avian retrovirus (EAV) was found in one pool of female A. aegypti collected from an autocidal gravid ovitrap (AGO) from the LRGV. EAVs are noninfectious ancient elements of virus that integrated into their host genome and are found in all species of the genus Gallus (50, 51). Many homeowners in the communities where mosquito trapping was done have chickens, and this result suggests that A. aegypti had previously fed on chickens or that chicken DNA had contaminated the mosquitoes. However, no human pathogen was detected using the LLMDA, presumably due to our limited set of field samples. In Texas, there was a total of 381 imported human Zika cases and 10 locally acquired ZIKV cases in the LRGV, with 6 cases in 2016 and 4 cases in 2017 (11, 12). In this context, the probability of detecting ZIKV-infected mosquitoes was low, especially because these mosquitoes were not being collected from or around the homes of human ZIKV cases. The use of the LLMDA for virus detection should be further tested using mosquitoes collected from regions with active arbovirus transmission areas and, if possible, from confirmed or probable human case households.
Although the number of viral species detected in our field samples is low, our results are comparable to those from other studies using microarrays to determine the virome of field-collected mosquitoes. For example, a study of 10 mosquito pools collected in Thailand revealed the presence of three different viruses: CxFLAV in Culex quinquefasciatus (n = 1), DENV-3 in Aedes aegypti (n = 1), and Japanese encephalitis virus (JEV) in two pools of Culex tritaeniorhynchus containing, respectively, 24 and 25 mosquitoes (35). Authors using pan-viral family primers coupled with conventional PCR also report low numbers of virus-positive pools. For example, in a study performed in Puerto Rico, 528 pools representing 1,584 mosquitoes lead to the identification of one insect-specific virus: CFAV in 67 pools (52). Other authors, using cell culture (observation of cytopathic effect [CPE]) followed by conventional PCR using pan-viral family primers to detect viruses in mosquito samples, have rarely detected extensive numbers of viral species. For example, in a study done in Brazil, researchers collected 950 adult female mosquitoes representing 16 species. From these, only two pools tested positive for flavivirus, and these were later identified as Nhumirim virus and Ilheus virus (53, 54).
The LLMDA is able to detect a wide variety of viruses, including mosquito-borne RNA viruses and insect-specific RNA viruses, and is able to detect coinfection in mosquito pools, making it an efficient tool for surveillance of known pathogens in understudied areas such as the LRGV. Given the recent interest in using bacteria or insect-specific viruses as a biocontrol tool and the role of coinfection in pathogen transmission, this tool can contribute to better understanding of disease dynamics in a particular region. However, periodic updates of probe sequences using genome data from more-contemporary strains is necessary to enable detection of emergent RNA virus genomes due to their high mutation rates.
Bacteria.
The LLMDA results show the presence of Wolbachia in several mosquito pools, which was confirmed with a qRT-PCR assay targeting the surface protein gene wsp. A. aegypti, the primary vector of dengue, Zika, and chikungunya viruses, was found to be negative for the presence of Wolbachia, which confirms previous observations (55). The secondary vector of these viruses, A. albopictus, was found to be infected with Wolbachia in 60% of the pools tested. The presence of Wolbachia in natural populations of A. albopictus has been previously reported (56), and A. albopictus is often found infected with group A (wAlbA) and B (wAlbB) strains, as suggested by our results. Additionally, a report of superinfection with the two strains has been published (55). Wolbachia has been shown to limit DENV transmission (57) and modulate CHIKV replication (58) in A. albopictus. The current study also detected Wolbachia in Culex populations from Chicago, San Antonio, and the LRGV, confirming previous studies in C. quinquefasciatus from Australia (83) and Brazil and Argentina (59) and in other Culex spp. in the United States, such as Culex pipiens (56, 60, 61). The presence of these endosymbionts in field populations in Texas is significant, since wPip (group B) has been reported to induce resistance to WNV in C. quinquefasciatus mosquitoes (62, 63). Because of its impact on transmission of human pathogens and on the mosquito reproduction, life span, and resistance to insecticides, knowledge of Wolbachia strains circulating in specific areas is needed if Wolbachia-based vector control strategies are to be implemented.
Overall, the number of bacterial hits in the mosquito pools was lower than expected, which might be explained by the lack of sufficient genomic sequences specific to insect-related bacterial species available during the array probe design, the low concentration of bacterial species in the samples, or the genetic divergence of the bacterial strains present in our samples compared to bacterial genomes used to develop the microarray. Additionally, the LLMDA was designed using only full genome sequences, and if at that time only partial bacterial sequences related to the mosquito microbiome were available, they would not have been included on the microarray. Since the development of this array, many studies have shown the importance of bacteria (64–66), viruses (67–69), and fungi (70, 71) in the epidemiology of mosquito-borne diseases, demonstrating the need to better characterize the mosquito microbiome. Updating the microarray with probes designed to detect the major components of insects’ microbiome could help alleviate the low number of bacterial hits detected in this study. In this study, we wanted to test the LLMDA’s ability to detect microbes present in mosquito samples without the need for a targeted enrichment. The LLMDA was successful at identifying viral pathogens without a baited approach, but it is not adequate to detect the whole bacterial community. Instead, the LLMDA seems to be efficient at detecting dominant bacterial species. Wolbachia has been reported to be the dominant member of A. albopictus and Culex mosquitoes (56) and has been successfully detected with the LLMDA. Other bacteria, including Pseudomonas, Klebsiella, and Erwinia spp., have been detected in Culex spp. and A. aegypti in our samples (see Table S4 in the supplemental material) and have already been reported in mosquitoes and their breeding sites (16, 29, 72–75). We encountered issues related to nonspecific probe binding in our samples, mostly to conserved regions of bacteria such as 23S or 16S rRNA genes, which might also explain the low number of bacterial species. Because we used a stringent threshold of determining a positive signal, i.e., at least 20% of probes being detected for a target sequence, and the criterion that probes should cover various regions of the genome, these nonspecific hits were not reported. In our case, after removal of nonspecific bacterial hits, Wolbachia was the most significant bacterial species confirmed to be present in the mosquito pools. Such challenges have been reported previously in low-biomass samples (76). Other approaches, such as shotgun metagenomic sequencing, would be alternative methods to characterize the microbiome.
In summary, to explore the potential usefulness of the LLMDA for biosurveillance, we took advantage of an ongoing mosquito surveillance program along the Texas-Mexico border in the LRGV where ZIKV circulated in 2016 to 2017, resulting in 10 cases of local transmission involving A. aegypti as the vector (12). A subset of the mosquito collections was tested using the LLMDA, and although no pools tested positive for ZIKV, the microarray was able to detect CFAV in A. aegypti populations from the LRGV and San Antonio, which could have an impact on the epidemiology of Aedes-vectored viral diseases. Similarly, CxFLAV was observed in several Culex populations. Wolbachia was detected at a high frequency in A. albopictus and Culex sp. mosquitoes but was not found in A. aegypti. Further characterization of the presence and strain types of locally occurring insect-specific viruses and Wolbachia is important (77, 78) for possible biologically based control interventions (66, 79, 80). This study presents the broad detection capability, sensitivity, and ease of use of the LLMDA approach for surveillance of mosquito-borne diseases of medical importance. This detection array could also aid in the surveillance of pathogens transmitted by other arthropod vectors, such as ticks. The study also demonstrated some limitations of the LLMDA and the need to develop an improved array including updated viral and bacterial full genomic sequences deposited in GenBank since 2014 for more-up-to-date biosurveillance studies.
MATERIALS AND METHODS
Mosquito samples.
Mosquitoes were collected in several locations in Texas (San Antonio and the LRGV) using three trapping methods. Autocidal gravid ovitraps (AGOs) (SpringStar Inc.), BG sentinel traps (Biogents), and Prokopack aspirators (John W. Hock Co.) were used (see Table S1 in the supplemental material). Whole female mosquitoes were pooled by trap and species, with a maximum size of 50 individuals per pool. Additionally, MG and SG of A. aegypti and Culex spp. were obtained by dissection of a subset of mosquitoes from the LRGV and pooled. These specimens were first surface sterilized (5 min in 70% ethanol) and rinsed twice in a sterile phosphate-buffered saline (PBS) solution, and then individual MG and SG were dissected under a dissecting microscope and rinsed in PBS.
LLMDA sensitivity and reproducibility.
Four different viruses were used in this assay: one alphavirus (Mayaro virus [MAYV] strain INHRR11a-10), two flaviviruses (DENV-2 strain INH125271 and ZIKV strain PRVABC59), and one bunyavirus (Rift Valley fever virus [RVFV] strain MP-12). For dengue virus, 100 μl of a 10-fold serial dilution (105 PFU/ml to 102 PFU/ml) of the virus was spiked into an A. aegypti Liverpool strain mosquito homogenate. The dilutions corresponding to 102 PFU/ml and 103 PFU/ml were done in duplicate to assess reproducibility. Additionally, 100 μl of MAYV virus at 104 PFU/ml was spiked into the mosquito homogenates containing 100 μl of DENV-2 at 104 PFU/ml and 100 μl of 105 PFU/ml. One hundred microliters of RVFV at 104 PFU/ml was spiked into a C. quinquefasciatus pool. One pool of A. aegypti and C. quinquefasciatus was used as a negative control. For ZIKV, two dilutions were tested, 104 PFU/ml and 102 PFU/ml. The ZIKV-spiked mosquito pools were tested by the ZIKV reverse transcription quantitative real-time PCR assay targeting the nonstructural protein 5 (NS5) gene (81, 82) to verify the presence/absence of infection (see Table S2 in the supplemental material).
LLMDA validation using field-collected sample of known status.
WNV-positive field-collected mosquitoes from Chicago, IL (2010), and CxFLAV-positive field-collected mosquitoes from College Station, TX (2013), were assessed on the LLMDA. These pools had previously tested positive in other studies using qRT-PCR targeting the envelope genes of WNV and CxFLAV (20, 39).
Mosquito sample preparation and nucleic acid extraction.
Three sample preparation methods were tested to evaluate different processing protocols that would optimize recovery of nucleic acid, retain the ability to isolate viruses, and remove surface exogenous nucleic acid. In method 1, mosquitoes were directly homogenized in TRIzol. In method 2, mosquitoes were homogenized in Hanks’ balanced salt solution (HBSS) (Thermo). In method 3, mosquitoes were washed in 70% ethanol for 5 min, followed by 2 PBS washes. Each mosquito pool was homogenized in a 2-ml microcentrifuge tube containing a single 2.8-mm stainless steel bead. Mosquitoes used for the MG and SG dissection were prepared following the procedure from method 3. Tubes were then centrifuged for 5 min at 15,000 × g. Nucleic acids were extracted from 100 μl of the homogenate supernatant using an RNA and DNA TRIzol extraction method.
LLMDA analysis.
The LLMDA v7 4x180K microarray consists of probes that targets both conserved and unique genomic regions of sequenced microbial species and has multiple probes per microbial genomic sequence to serve as an internal validation mechanism (34). All samples were analyzed using the LLMDA as described previously (30, 32). Briefly, RNA was reverse transcribed to cDNA using the phosphorylated random hexamer/SuperScriptIII (P-N6/SSIII) method, which uses the Superscript III reverse transcription kit (Invitrogen) and 5′-phosphorylated random hexamers (P-N6) (Eurofins MWG Operon) followed by the Qiagen QuantiTech whole transcriptome kit (30, 32). Each sample was loaded onto the LLMDA and allowed to hybridize for 40 h at 55°C in a rotator oven. After hybridization, the microarray was washed following standard manufacturer’s protocols with CGH wash buffers (Agilent) and further cleaned using a nitrogen gas stream to remove any particulates from the array surface. The microarray was then scanned and the data analyzed using a statistical method described previously (34). Briefly, the intensity of each probe is transformed into a positive or negative signal. A positive signal is obtained when the intensity of the probe exceeds an intensity threshold set to the 95th percentile of that for the negative controls (33). In other words, if the probe intensity is above the 95th percentile of the sum of the intensity of the random control probes on the array, then that probe is considered to have a positive signal. Given the different parameters used to validate our results, there is still a 5% chance for a false-positive probe signal (100% to 95%). A sample was assigned to a species when at least 20% of all the probes present for this particular species had a positive signal. Since we set a 20% threshold of all probes to assign a species as positive, there is still a certain probability that even with 20% of the probes lighting up, the sample would have a false-positive detection.
We then used a likelihood maximization algorithm to identify the target that explains the largest portion of the observed positive probe signals while minimizing the number of negative probe signals. The log likelihood for each of the possible targets was estimated from the BLAST similarity scores of the array feature and target sequences, together with the feature sequence complexity and other covariates derived from the BLAST results as described previously (34).
PCR assay to confirm microarray results.
Confirmation of the viral species detected in the field samples from San Antonio and the LRGV was performed by conventional PCR using gene-specific primers amplifying a 206-bp region of NS5 of CxFLAV (39) and a 340-bp fragment of the CFAV E gene (42). Additionally, the presence of Wolbachia in the mosquito samples was confirmed using quantitative PCR targeting the Wolbachia outer surface protein genes wspA and wspB (58) (see Table S3 in the supplemental material).
Supplementary Material
ACKNOWLEDGMENTS
We thank the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) at the University of Texas Medical Branch and the Centers for Disease Control and Prevention for providing the different positive controls used in this study. Field support for collecting mosquito samples was provided by Ester Carbajal, Edwin Valdez, Jose Juarez, Joel Obregon, Michelle Ximenez, Estefany Villalobos, and undergraduate researchers from Texas A&M University—San Antonio.
This work was performed, in part, under the auspices of the U.S. Department of Energy by the Lawrence Livermore National Laboratory under contract DE-AC52-07NA27344. This work was supported by an internal LLNL grant, by an NIH R21 grant (R21AI128953), and by Texas A&M AgriLife Research.
We have no conflict of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01202-19.
REFERENCES
- 1.Weaver SC, Reisen WK. 2010. Present and future arboviral threats. Antiviral Res 85:328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraemer MU, Sinka ME, Duda KA, Mylne A, Shearer FM, Brady OJ, Messina JP, Barker CM, Moore CG, Carvalho RG, Coelho GE, Van Bortel W, Hendrickx G, Schaffner F, Wint GR, Elyazar IR, Teng HJ, Hay SI. 2015. The global compendium of Aedes aegypti and Ae. albopictus occurrence. Sci Data 2:150035. doi: 10.1038/sdata.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powell JR, Tabachnick WJ. 2013. History of domestication and spread of Aedes aegypti—a review. Mem Inst Oswaldo Cruz 108(Suppl 1):11–17. doi: 10.1590/0074-0276130395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gratz NG. 2004. Critical review of the vector status of Aedes albopictus. Med Vet Entomol 18:215–227. doi: 10.1111/j.0269-283X.2004.00513.x. [DOI] [PubMed] [Google Scholar]
- 5.Lambrechts L, Scott TW, Gubler DJ. 2010. Consequences of the expanding global distribution of Aedes albopictus for dengue virus transmission. PLoS Negl Trop Dis 4:e646. doi: 10.1371/journal.pntd.0000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kilpatrick AM. 2011. Globalization, land use, and the invasion of West Nile virus. Science 334:323–327. doi: 10.1126/science.1201010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camargo S. 1967. History of Aedes aegypti eradication in the Americas. Bull World Health Organ 36:602–603. [PMC free article] [PubMed] [Google Scholar]
- 8.Hotez PJ. 2016. Zika in the United States of America and a fateful 1969 decision. PLoS Negl Trop Dis 10:e0004765. doi: 10.1371/journal.pntd.0004765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soper FL. 1963. The elimination of urban yellow fever in the Americas through the eradication of Aedes aegypti. Am J Public Health 53:7–16. doi: 10.2105/ajph.53.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Likos A, Griffin I, Bingham AM, Stanek D, Fischer M, White S, Hamilton J, Eisenstein L, Atrubin D, Mulay P, Scott B, Jenkins P, Fernandez D, Rico E, Gillis L, Jean R, Cone M, Blackmore C, McAllister J, Vasquez C, Rivera L, Philip C. 2016. Local mosquito-borne transmission of Zika virus—Miami-Dade and Broward counties, Florida, June–August 2016. MMWR Morb Mortal Wkly Rep 65:1032–1038. doi: 10.15585/mmwr.mm6538e1. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2018. Cumulative Zika virus disease case in the United States, 2015–2018. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 12.Martin E, Medeiros MCI, Carbajal E, Valdez E, Juarez JG, Garcia-Luna S, Salazar A, Qualls WA, Hinojosa S, Borucki MK, Manley HA, Badillo-Vargas IE, Frank M, Hamer GL. 2019. Surveillance of Aedes aegypti indoors and outdoors using autocidal gravid ovitraps in South Texas during local transmission of Zika virus, 2016 to 2018. Acta Trop 192:129–137. doi: 10.1016/j.actatropica.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Gusmao DS, Santos AV, Marini DC, Bacci M Jr, Berbert-Molina MA, Lemos FJ. 2010. Culture-dependent and culture-independent characterization of microorganisms associated with Aedes aegypti (Diptera: Culicidae) (L.) and dynamics of bacterial colonization in the midgut. Acta Trop 115:275–281. doi: 10.1016/j.actatropica.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 14.Kim CH, Lampman RL, Muturi EJ. 2015. Bacterial communities and midgut microbiota associated with mosquito populations from waste tires in East-Central Illinois. J Med Entomol 52:63–75. doi: 10.1093/jme/tju011. [DOI] [PubMed] [Google Scholar]
- 15.Pidiyar VJ, Jangid K, Patole MS, Shouche YS. 2004. Studies on cultured and uncultured microbiota of wild Culex quinquefasciatus mosquito midgut based on 16S ribosomal RNA gene analysis. Am J Trop Med Hyg 70:597–603. doi: 10.4269/ajtmh.2004.70.597. [DOI] [PubMed] [Google Scholar]
- 16.Zouache K, Raharimalala FN, Raquin V, Tran-Van V, Raveloson LH, Ravelonandro P, Mavingui P. 2011. Bacterial diversity of field-caught mosquitoes, Aedes albopictus and Aedes aegypti, from different geographic regions of Madagascar. FEMS Microbiol Ecol 75:377–389. doi: 10.1111/j.1574-6941.2010.01012.x. [DOI] [PubMed] [Google Scholar]
- 17.Kuno G. 1998. Universal diagnostic RT-PCR protocol for arboviruses. J Virol Methods 72:27–41. doi: 10.1016/S0166-0934(98)00003-2. [DOI] [PubMed] [Google Scholar]
- 18.Scaramozzino N, Crance JM, Jouan A, DeBriel DA, Stoll F, Garin D. 2001. Comparison of flavivirus universal primer pairs and development of a rapid, highly sensitive heminested reverse transcription-PCR assay for detection of flaviviruses targeted to a conserved region of the NS5 gene sequences. J Clin Microbiol 39:1922–1927. doi: 10.1128/JCM.39.5.1922-1927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eshoo MW, Whitehouse CA, Zoll ST, Massire C, Pennella TT, Blyn LB, Sampath R, Hall TA, Ecker JA, Desai A, Wasieloski LP, Li F, Turell MJ, Schink A, Rudnick K, Otero G, Weaver SC, Ludwig GV, Hofstadler SA, Ecker DJ. 2007. Direct broad-range detection of alphaviruses in mosquito extracts. Virology 368:286–295. doi: 10.1016/j.virol.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 20.Lanciotti RS, Kerst AJ, Nasci RS, Godsey MS, Mitchell CJ, Savage HM, Komar N, Panella NA, Allen BC, Volpe KE, Davis BS, Roehrig JT. 2000. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J Clin Microbiol 38:4066–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. 1992. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol 30:545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanciotti RS, Kerst AJ. 2001. Nucleic acid sequence-based amplification assays for rapid detection of West Nile and St. Louis encephalitis viruses. J Clin Microbiol 39:4506–4513. doi: 10.1128/JCM.39.12.4506-4513.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadeghi M, Popov V, Guzman H, Phan TG, Vasilakis N, Tesh R, Delwart E. 2017. Genomes of viral isolates derived from different mosquitos species. Virus Res 242:49–57. doi: 10.1016/j.virusres.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coffey LL, Page BL, Greninger AL, Herring BL, Russell RC, Doggett SL, Haniotis J, Wang C, Deng X, Delwart EL. 2014. Enhanced arbovirus surveillance with deep sequencing: identification of novel rhabdoviruses and bunyaviruses in Australian mosquitoes. Virology 448:146–158. doi: 10.1016/j.virol.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi M, Neville P, Nicholson J, Eden JS, Imrie A, Holmes EC. 2017. High-resolution metatranscriptomics reveals the ecological dynamics of mosquito-associated RNA viruses in Western Australia. J Virol 91:e00680-17. doi: 10.1128/JVI.00680-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fauver JR, Grubaugh ND, Krajacich BJ, Weger-Lucarelli J, Lakin SM, Fakoli LS, Bolay FK, Diclaro JW, Dabiré KR, Foy BD, Brackney DE, Ebel GD, Stenglein MD. 2016. West African Anopheles gambiae mosquitoes harbor a taxonomically diverse virome including new insect-specific flaviviruses, mononegaviruses, and totiviruses. Virology 498:288–299. doi: 10.1016/j.virol.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 27.Osei-Poku J, Mbogo CM, Palmer WJ, Jiggins FM. 2012. Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol Ecol 21:5138–5150. doi: 10.1111/j.1365-294X.2012.05759.x. [DOI] [PubMed] [Google Scholar]
- 28.Zink SD, Van Slyke GA, Palumbo MJ, Kramer LD, Ciota AT. 2015. Exposure to West Nile virus increases bacterial diversity and immune gene expression in Culex pipiens. Viruses 7:5619–5631. doi: 10.3390/v7102886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandler JA, Liu RM, Bennett SN. 2015. RNA shotgun metagenomic sequencing of northern California (USA) mosquitoes uncovers viruses, bacteria, and fungi. Front Microbiol 6:185. doi: 10.3389/fmicb.2015.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenstierne MW, McLoughlin KS, Olesen ML, Papa A, Gardner SN, Engler O, Plumet S, Mirazimi A, Weidmann M, Niedrig M, Fomsgaard A, Erlandsson L. 2014. The microbial detection array for detection of emerging viruses in clinical samples—a useful panmicrobial diagnostic tool. PLoS One 9:e100813. doi: 10.1371/journal.pone.0100813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thissen JB, McLoughlin K, Gardner S, Gu P, Mabery S, Slezak T, Jaing C. 2014. Analysis of sensitivity and rapid hybridization of a multiplexed microbial detection microarray. J Virol Methods 201:73–78. doi: 10.1016/j.jviromet.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 32.Jaing CJ, Thissen JB, Gardner SN, McLoughlin KS, Hullinger PJ, Monday NA, Niederwerder MC, Rowland RR. 2015. Application of a pathogen microarray for the analysis of viruses and bacteria in clinical diagnostic samples from pigs. J Vet Diagn Invest 27:313–325. doi: 10.1177/1040638715578484. [DOI] [PubMed] [Google Scholar]
- 33.Devault AM, McLoughlin K, Jaing C, Gardner S, Porter TM, Enk JM, Thissen J, Allen J, Borucki M, DeWitte SN, Dhody AN, Poinar HN. 2014. Ancient pathogen DNA in archaeological samples detected with a microbial detection array. Sci Rep 4:4245. doi: 10.1038/srep04245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardner SN, Jaing CJ, McLoughlin KS, Slezak TR. 2010. A microbial detection array (MDA) for viral and bacterial detection. BMC Genomics 11:668. doi: 10.1186/1471-2164-11-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kengluecha A, Lee JS, Pisarcik SE, Grubaugh ND, O'Guinn ML, Melanson VR, Jaichapor B, Petz LN, McMenamy SS, Long LS, Turell MJ. 2013. Evaluation of a field-portable DNA microarray platform and nucleic acid amplification strategies for the detection of arboviruses, arthropods, and bloodmeals. Am J Trop Med Hyg 88:245–253. doi: 10.4269/ajtmh.2012.12-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grubaugh ND, McMenamy SS, Turell MJ, Lee JS. 2013. Multi-gene detection and identification of mosquito-borne RNA viruses using an oligonucleotide microarray. PLoS Negl Trop Dis 7:e2349. doi: 10.1371/journal.pntd.0002349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Obara-Nagoya M, Yamauchi T, Watanabe M, Hasegawa S, Iwai-Itamochi M, Horimoto E, Takizawa T, Takashima I, Kariwa H. 2013. Ecological and genetic analyses of the complete genomes of Culex flavivirus strains isolated from Culex tritaeniorhynchus and Culex pipiens (Diptera: Culicidae) group mosquitoes. J Med Entomol 50:300–309. doi: 10.1603/me12159. [DOI] [PubMed] [Google Scholar]
- 38.Newman CM, Krebs BL, Anderson TK, Hamer GL, Ruiz MO, Brawn JD, Brown WM, Kitron UD, Goldberg TL. 2017. Culex flavivirus during West Nile virus epidemic and interepidemic years in Chicago, United States. Vector Borne Zoonotic Dis 17:567–575. doi: 10.1089/vbz.2017.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newman CM, Cerutti F, Anderson TK, Hamer GL, Walker ED, Kitron UD, Ruiz MO, Brawn JD, Goldberg TL. 2011. Culex flavivirus and West Nile virus mosquito coinfection and positive ecological association in Chicago, United States. Vector Borne Zoonotic Dis 11:1099–1105. doi: 10.1089/vbz.2010.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kent RJ, Crabtree MB, Miller BR. 2010. Transmission of West Nile virus by Culex quinquefasciatus say infected with Culex flavivirus Izabal. PLoS Negl Trop Dis 4:e671. doi: 10.1371/journal.pntd.0000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bittar C, Machado DC, Vedovello D, Ullmann LS, Rahal P, Araujo Junior JP, Nogueira ML. 2016. Genome sequencing and genetic characterization of Culex flavirirus (CxFV) provides new information about its genotypes. Virol J 13:158. doi: 10.1186/s12985-016-0614-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Contreras-Gutierrez MA, Guzman H, Thangamani S, Vasilakis N, Tesh RB. 2017. Experimental infection with and maintenance of cell fusing agent virus (Flavivirus) in Aedes aegypti. Am J Trop Med Hyg 97:299–304. doi: 10.4269/ajtmh.16-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang G, Asad S, Khromykh AA, Asgari S. 2017. Cell fusing agent virus and dengue virus mutually interact in Aedes aegypti cell lines. Sci Rep 7:6935. doi: 10.1038/s41598-017-07279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faria NR, Azevedo RDSDS, Kraemer MUG, Souza R, Cunha MS, Hill SC, Thézé J, Bonsall MB, Bowden TA, Rissanen I, Rocco IM, Nogueira JS, Maeda AY, Vasami FGDS, Macedo FLDL, Suzuki A, Rodrigues SG, Cruz ACR, Nunes BT, Medeiros DBDA, Rodrigues DSG, Queiroz ALN, da Silva EVP, Henriques DF, da Rosa EST, de Oliveira CS, Martins LC, Vasconcelos HB, Casseb LMN, Simith DDB, Messina JP, Abade L, Lourenço J, Alcantara LCJ, de Lima MM, Giovanetti M, Hay SI, de Oliveira RS, Lemos PDS, de Oliveira LF, de Lima CPS, da Silva SP, de Vasconcelos JM, Franco L, Cardoso JF, Vianez-Júnior JLDSG, Mir D, Bello G, Delatorre E, Khan K, et al. 2016. Zika virus in the Americas: early epidemiological and genetic findings. Science 352:345–349. doi: 10.1126/science.aaf5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haddow AD, Schuh AJ, Yasuda CY, Kasper MR, Heang V, Huy R, Guzman H, Tesh RB, Weaver SC. 2012. Genetic characterization of Zika virus strains: geographic expansion of the Asian lineage. PLoS Negl Trop Dis 6:e1477. doi: 10.1371/journal.pntd.0001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jousset FX, Barreau C, Boublik Y, Cornet M. 1993. A parvo-like virus persistently infecting a C6/36 clone of Aedes albopictus mosquito cell line and pathogenic for Aedes aegypti larvae. Virus Res 29:99–114. doi: 10.1016/0168-1702(93)90052-O. [DOI] [PubMed] [Google Scholar]
- 47.Chen S, Cheng L, Zhang Q, Lin W, Lu X, Brannan J, Zhou ZH, Zhang J. 2004. Genetic, biochemical, and structural characterization of a new densovirus isolated from a chronically infected Aedes albopictus C6/36 cell line. Virology 318:123–133. doi: 10.1016/j.virol.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 48.Paterson A, Robinson E, Suchman E, Afanasiev B, Carlson J. 2005. Mosquito densonucleosis viruses cause dramatically different infection phenotypes in the C6/36 Aedes albopictus cell line. Virology 337:253–261. doi: 10.1016/j.virol.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 49.Cataneo AHD, Kuczera D, Mosimann ALP, Silva EG, Ferreira AGA, Marques JT, Wowk PF, Santos C, Bordignon J. 2019. Detection and clearance of a mosquito densovirus contaminant from laboratory stocks of Zika virus. Mem Inst Oswaldo Cruz 114:e180432. doi: 10.1590/0074-02760180432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sacco MA, Nair VK. 2014. Prototype endogenous avian retroviruses of the genus Gallus. J Gen Virol 95:2060–2070. doi: 10.1099/vir.0.066852-0. [DOI] [PubMed] [Google Scholar]
- 51.Sacco MA, Flannery DM, Howes K, Venugopal K. 2000. Avian endogenous retrovirus EAV-HP shares regions of identity with avian leukosis virus subgroup J and the avian retrotransposon ART-CH. J Virol 74:1296–1306. doi: 10.1128/JVI.74.3.1296-1306.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cook S, Bennett SN, Holmes EC, De Chesse R, Moureau G, de Lamballerie X. 2006. Isolation of a new strain of the flavivirus cell fusing agent virus in a natural mosquito population from Puerto Rico. J Gen Virol 87:735–748. doi: 10.1099/vir.0.81475-0. [DOI] [PubMed] [Google Scholar]
- 53.Pauvolid-Correa A, Solberg O, Couto-Lima D, Kenney J, Serra-Freire N, Brault A, Nogueira R, Langevin S, Komar N. 2015. Nhumirim virus, a novel flavivirus isolated from mosquitoes from the Pantanal, Brazil. Arch Virol 160:21–27. doi: 10.1007/s00705-014-2219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pauvolid-Correa A, Kenney JL, Couto-Lima D, Campos ZM, Schatzmayr HG, Nogueira RM, Brault AC, Komar N. 2013. Ilheus virus isolation in the Pantanal, west-central Brazil. PLoS Negl Trop Dis 7:e2318. doi: 10.1371/journal.pntd.0002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kitrayapong P, Baimai V, O’Neill SL. 2002. Field prevalence of Wolbachia in the mosquito vector Aedes albopictus. Am J Trop Med Hyg 66:108–111. doi: 10.4269/ajtmh.2002.66.108. [DOI] [PubMed] [Google Scholar]
- 56.Muturi EJ, Ramirez JL, Rooney AP, Kim CH. 2017. Comparative analysis of gut microbiota of mosquito communities in central Illinois. PLoS Negl Trop Dis 11:e0005377. doi: 10.1371/journal.pntd.0005377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mousson L, Zouache K, Arias-Goeta C, Raquin V, Mavingui P, Failloux AB. 2012. The native Wolbachia symbionts limit transmission of dengue virus in Aedes albopictus. PLoS Negl Trop Dis 6:e1989. doi: 10.1371/journal.pntd.0001989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mousson L, Martin E, Zouache K, Madec Y, Mavingui P, Failloux AB. 2010. Wolbachia modulates Chikungunya replication in Aedes albopictus. Mol Ecol 19:1953–1964. doi: 10.1111/j.1365-294X.2010.04606.x. [DOI] [PubMed] [Google Scholar]
- 59.Morais SA, Almeida F, Suesdek L, Marrelli MT. 2012. Low genetic diversity in Wolbachia-infected Culex quinquefasciatus (Diptera: Culicidae) from Brazil and Argentina. Rev Inst Med Trop Sao Paulo 54:325–329. doi: 10.1590/s0036-46652012000600007. [DOI] [PubMed] [Google Scholar]
- 60.Rasgon JL, Scott TW. 2003. Wolbachia and cytoplasmic incompatibility in the California Culex pipiens mosquito species complex: parameter estimates and infection dynamics in natural populations. Genetics 165:2029–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morningstar RJ, Hamer GL, Goldberg TL, Huang S, Andreadis TG, Walker ED. 2012. Diversity of Wolbachia pipientis strain wPip in a genetically admixtured, above-ground Culex pipiens (Diptera: Culicidae) population: association with form molestus ancestry and host selection patterns. J Med Entomol 49:474–481. doi: 10.1603/ME11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Micieli MV, Glaser RL. 2014. Somatic Wolbachia (Rickettsiales: Rickettsiaceae) levels in Culex quinquefasciatus and Culex pipiens (Diptera: Culicidae) and resistance to West Nile virus infection. J Med Entomol 51:189–199. doi: 10.1603/me13152. [DOI] [PubMed] [Google Scholar]
- 63.Glaser RL, Meola MA. 2010. The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS One 5:e11977. doi: 10.1371/journal.pone.0011977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan CH, Wong PJ, Li MI, Yang H, Ng LC, O’Neill SL. 2017. wMel limits zika and chikungunya virus infection in a Singapore Wolbachia-introgressed Ae. aegypti strain, wMel-Sg. PLoS Negl Trop Dis 11:e0005496. doi: 10.1371/journal.pntd.0005496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, Rocha BC, Hall-Mendelin S, Day A, Riegler M, Hugo LE, Johnson KN, Kay BH, McGraw EA, van den Hurk AF, Ryan PA, O’Neill SL. 2009. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, chikungunya, and Plasmodium. Cell 139:1268–1278. doi: 10.1016/j.cell.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 66.Lambrechts L, Ferguson NM, Harris E, Holmes EC, McGraw EA, O’Neill SL, Ooi EE, Ritchie SA, Ryan PA, Scott TW, Simmons CP, Weaver SC. 2015. Assessing the epidemiological effect of Wolbachia for dengue control. Lancet Infect Dis 15:862–866. doi: 10.1016/S1473-3099(15)00091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goenaga S, Kenney JL, Duggal NK, Delorey M, Ebel GD, Zhang B, Levis SC, Enria DA, Brault AC. 2015. Potential for co-infection of a mosquito-specific flavivirus, Nhumirim virus, to block West Nile virus transmission in mosquitoes. Viruses 7:5801–5812. doi: 10.3390/v7112911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hall-Mendelin S, McLean BJ, Bielefeldt-Ohmann H, Hobson-Peters J, Hall RA, van den Hurk AF. 2016. The insect-specific Palm Creek virus modulates West Nile virus infection in and transmission by Australian mosquitoes. Parasit Vectors 9:414. doi: 10.1186/s13071-016-1683-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Romo H, Kenney JL, Blitvich BJ, Brault AC. 2018. Restriction of Zika virus infection and transmission in Aedes aegypti mediated by an insect-specific flavivirus. Emerg Microbes Infect 7:181. doi: 10.1038/s41426-018-0180-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dong Y, Morton JC Jr, Ramirez JL, Souza-Neto JA, Dimopoulos G. 2012. The entomopathogenic fungus Beauveria bassiana activate toll and JAK-STAT pathway-controlled effector genes and anti-dengue activity in Aedes aegypti. Insect Biochem Mol Biol 42:126–132. doi: 10.1016/j.ibmb.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramirez JL, Short SM, Bahia AC, Saraiva RG, Dong Y, Kang S, Tripathi A, Mlambo G, Dimopoulos G. 2014. Chromobacterium Csp_P reduces malaria and dengue infection in vector mosquitoes and has entomopathogenic and in vitro anti-pathogen activities. PLoS Pathog 10:e1004398. doi: 10.1371/journal.ppat.1004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Minard G, Tran FH, Dubost A, Tran-Van V, Mavingui P, Moro CV. 2014. Pyrosequencing 16S rRNA genes of bacteria associated with wild tiger mosquito Aedes albopictus: a pilot study. Front Cell Infect Microbiol 4:59. doi: 10.3389/fcimb.2014.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Minard G, Mavingui P, Moro CV. 2013. Diversity and function of bacterial microbiota in the mosquito holobiont. Parasit Vectors 6:146. doi: 10.1186/1756-3305-6-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thongsripong P, Chandler JA, Green AB, Kittayapong P, Wilcox BA, Kapan DD, Bennett SN. 2018. Mosquito vector-associated microbiota: metabarcoding bacteria and eukaryotic symbionts across habitat types in Thailand endemic for dengue and other arthropod-borne diseases. Ecol Evol 8:1352–1368. doi: 10.1002/ece3.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yadav KK, Datta S, Naglot A, Bora A, Hmuaka V, Bhagyawant S, Gogoi HK, Veer V, Raju PS. 2016. Diversity of cultivable midgut microbiota at different stages of the Asian tiger mosquito, Aedes albopictus from Tezpur, India. PLoS One 11:e0167409. doi: 10.1371/journal.pone.0167409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Altinli M, Gunay F, Alten B, Weill M, Sicard M. 2018. Wolbachia diversity and cytoplasmic incompatibility patterns in Culex pipiens populations in Turkey. Parasit Vectors 11:198. doi: 10.1186/s13071-018-2777-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mixão V, Mendes AM, Mauricio I, Calado M, Novo M, Belo S, Almeida A. 2016. Molecular detection of Wolbachia pipientis in natural populations of mosquito vectors of Dirofilaria immitis from continental Portugal: first detection in Culex theileri. Med Vet Entomol 30:301–309. doi: 10.1111/mve.12179. [DOI] [PubMed] [Google Scholar]
- 79.Kamtchum-Tatuene J, Makepeace BL, Benjamin L, Baylis M, Solomon T. 2017. The potential role of Wolbachia in controlling the transmission of emerging human arboviral infections. Curr Opin Infect Dis 30:108–116. doi: 10.1097/QCO.0000000000000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dutra HL, Caragata EP, Moreira LA. 2017. The re-emerging arboviral threat: hidden enemies: the emergence of obscure arboviral diseases, and the potential use of Wolbachia in their control. Bioessays 39:1600175. doi: 10.1002/bies.201600175. [DOI] [PubMed] [Google Scholar]
- 81.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. 2008. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liang W, He X, Liu G, Zhang S, Fu S, Wang M, Chen W, He Y, Tao X, Jiang H, Lin X, Gao X, Hu W, Liu Y, Feng L, Cao Y, Yang G, Jing C, Liang G, Wang H. 2015. Distribution and phylogenetic analysis of Culex flavivirus in mosquitoes in China. Arch Virol 160:2259–2268. doi: 10.1007/s00705-015-2492-1. [DOI] [PubMed] [Google Scholar]
- 83.O'Neill SL, Paterson HE. 1992. Crossing type variability associated with cytoplasmic incompatibility in Australian populations of the mosquito Culex quinquefasciatus Say. Med Vet Entomol 6:209–216. doi: 10.1111/j.1365-2915.1992.tb00608.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.