Schizochytrium strains are usually studied as oil-producing strains, but they can also synthesize other secondary metabolites, such as astaxanthin. In this study, methanol was used as an inducer, and we explored its effects on the production of astaxanthin, a highly valuable substance in Schizochytrium. Methanol induced Schizochytrium to synthesize large amounts of astaxanthin. Transcriptomic analysis was used to investigate the regulation of signaling and metabolic pathways (mainly relative gene expression) in Schizochytrium grown in the presence of various concentrations of methanol. These results contribute to the understanding of the underlying molecular mechanisms and may aid in the future optimization of Schizochytrium for astaxanthin biosynthesis.
KEYWORDS: Schizochytrium limacinum, carotenoids, fatty acid, transcriptome
ABSTRACT
Schizochytrium is a promising source for the production of docosahexaenoic acid and astaxanthin. The effects of different methanol concentrations on astaxanthin, biomass, and production of the lipids, squalene, and total sterol in Schizochytrium limacinum B4D1 were investigated. Astaxanthin began to accumulate when the methanol concentration reached 3.2% and peaked at 5.6% methanol, with a 2,000-fold increase over that in the control. However, under cultivation with 5.6% methanol, the biomass, lipids, squalene, and total sterol decreased to various degrees. Transcriptomic analysis was performed to explore the effects of different methanol concentrations (0%, 3.2%, and 5.6%) on the expression profile of B4D1. Three key signaling pathways were found to play important roles in regulating cell growth and metabolism under cultivation with methanol. Five central carbon metabolism-associated genes were significantly downregulated in response to 5.6% methanol and thus were expected to result in less ATP and NADPH being available for cell growth and synthesis. High methanol conditions significantly downregulated three genes involved in fatty acid and squalene/sterol precursor biosynthesis but significantly upregulated geranylgeranyl diphosphate synthase, lycopene β-cyclase, and β-carotene 3-hydroxylase, which are involved in astaxanthin synthesis, thus resulting in an increase in the levels of precursors and the final production of astaxanthin. Additionally, the transcriptional levels of three stress response genes were upregulated. This study investigates gene expression profiles in the astaxanthin producer Schizochytrium when grown under various methanol concentrations. These results broaden current knowledge regarding genetic expression and provide important information for promoting astaxanthin biosynthesis in Schizochytrium.
IMPORTANCE Schizochytrium strains are usually studied as oil-producing strains, but they can also synthesize other secondary metabolites, such as astaxanthin. In this study, methanol was used as an inducer, and we explored its effects on the production of astaxanthin, a highly valuable substance in Schizochytrium. Methanol induced Schizochytrium to synthesize large amounts of astaxanthin. Transcriptomic analysis was used to investigate the regulation of signaling and metabolic pathways (mainly relative gene expression) in Schizochytrium grown in the presence of various concentrations of methanol. These results contribute to the understanding of the underlying molecular mechanisms and may aid in the future optimization of Schizochytrium for astaxanthin biosynthesis.
INTRODUCTION
Schizochytrium, which belongs to the thraustochytrids, is a unicellular, eukaryotic, heterotrophic marine microorganism. It is known for rapid growth and extensive accumulation of metabolites such as polyunsaturated fatty acids and carotenoids. Previous studies have focused mainly on the production of oils, especially docosahexaenoic acid (DHA), an important ω-3 fatty acid (1, 2). High-productivity strains can accumulate lipids to 50% to 70% of their dry cell weight (DCW), with DHA constituting 30% to 70% of the total fatty acids (3–5).
Thraustochytrids are often pigmented and have been found to contain carotenoids (5–8), including astaxanthin, canthaxanthin, β-carotene, zeaxanthin, and echinenone (6, 7, 9). Carotenoids, through their potent antioxidant activity and potential benefits to human health, may assist in the treatment of cancer and eye vision (10). The main application of carotenoids is as a colorant in food and feed, and the market for carotenoids in the nutrition, pharmaceutical, and cosmetic industries is continuously growing (3). The strong antioxidant characteristic of astaxanthin makes it the most important carotenoid in Schizochytrium (11, 12). Astaxanthin’s important role in cancer prevention (3) has attracted growing attention; researchers have studied its synthesis and have emphasized its value in the pharmaceutical and food industries (13). Natural sources of astaxanthin have been found in krill (14), microalgae (15), and the yeast Phaffia rhodozyma (16). However, the proportion of astaxanthin in those organisms is relatively low, ranging from 0.1% to 0.4% (17). In contrast, the cyst cells of Haematococcus pluvialis and its mutant strains contain astaxanthin to 4% of the DCW and have been successfully used in industrial applications (18, 19). The composition of carotenoids in Haematococcus, 80% of which comprises astaxanthin, may be advantageous for astaxanthin concentration and mass production. However, Haematococcus is an obligate photoautotroph and grows more slowly than heterotrophs, thus hindering increases in productivity. Because Schizochytrium strains are unicellular, eukaryotic, and heterotrophic, they are potential strains for astaxanthin production.
The astaxanthin synthetic pathway has been studied in some higher plants and microalgae (20–22). However, little is known regarding the pathway of Schizochytrium’s astaxanthin biosynthesis. In a previous study, Zhang et al. have proposed potential pathways for the production of fatty acids, squalene, sterol, and carotenoids in Schizochytrium and have also observed an interesting phenomenon in which the red color of Schizochytrium darkens with the induction of butanol (23), owing to astaxanthin accumulation.
Methanol, a short-chain alcohol, was used in this study. To better understand the molecular basis of the observed effects of methanol on the production of highly valuable substances (mainly astaxanthin), we conducted transcriptomic analysis of Schizochytrium limacinum B4D1 grown under different methanol concentrations. The results broaden the scientific understanding of astaxanthin biosynthesis in Schizochytrium and provide an important new functional genomics information resource. These results should facilitate future genetic studies on the molecular mechanisms and cell factory construction of Schizochytrium for astaxanthin biosynthesis.
RESULTS
Effects of methanol on biomass and metabolites in S. limacinum B4D1.
(i) Effects of methanol on biomass, lipid accumulation, and fatty acid composition. The effects of methanol on S. limacinum B4D1 at different concentrations (0%, 1.6%, 3.2%, 4.8%, 5.6%, and 6.4%) were examined. Methanol determination indicated that a small proportion of methanol was volatilized (∼23%) and utilized (∼10%) during cultivation, but most of the methanol (∼67%) remained in solution (Fig. 1), indicating that a small amount of methanol entered the S. limacinum B4D1 cells.
FIG 1.
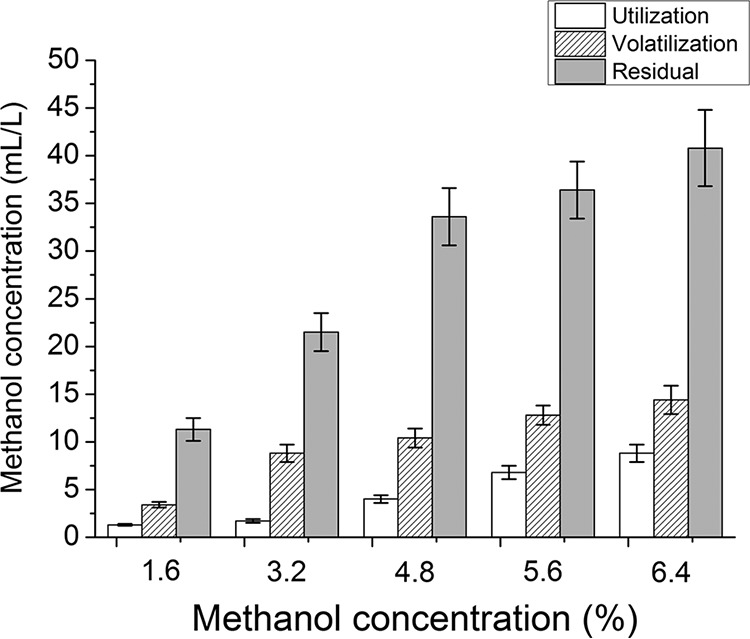
Determination of methanol utilization, methanol volatilization, and methanol residual in all groups. Shown are mean values and SEs of results from three separately grown cultures. Calculation formulas were as follows: methanol utilization = C3 – C2; methanol volatilization = C1 – C3. (C1, initial methanol concentration; C2, methanol concentration [with inoculation] at the end of fermentation; C3, methanol concentration [without inoculation] at the end of fermentation.)
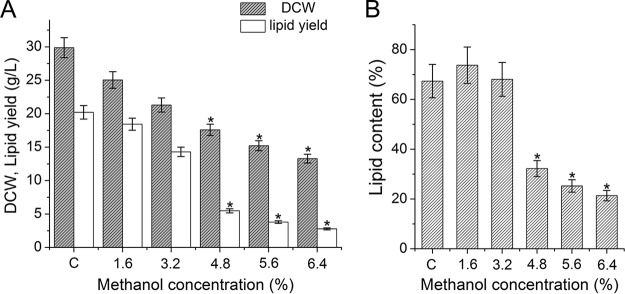
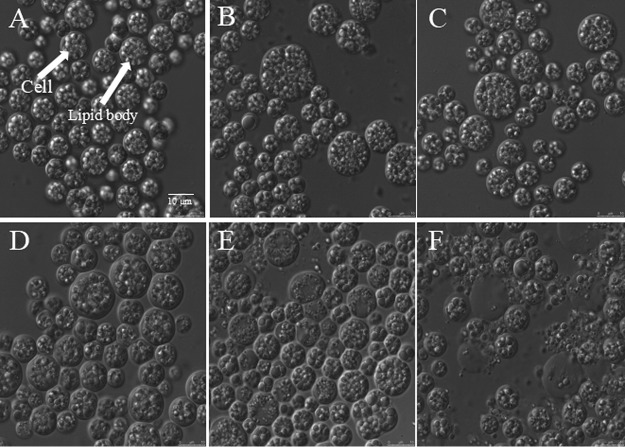
With increased methanol concentrations, both the DCW and lipid yield of S. limacinum B4D1 decreased (Fig. 2A). The proportion of lipid increased slightly or remained unchanged under culture with lower methanol concentrations (≤3.2%) but declined significantly (P ≤ 0.05) at higher methanol concentrations (≥4.8%) (Fig. 2B). These results were consistent with the microscopic images of S. limacinum B4D1. The cellular morphology changed slightly under lower methanol concentrations (≤3.2%) but changed significantly under higher methanol conditions (≥4.8%). Notably, when S. limacinum B4D1 was treated with 5.6% methanol, some cells began to lyse, which causes substances in the cell to flow out of the cell. In addition, the lipid body was significantly decreased when the methanol concentration increased from 4.8% to 6.4% (Fig. 3).
FIG 2.
Effects of methanol on dry cell weight (DCW) and lipid yield (A) and lipid content (B) of S. limacinum B4D1. Shown are mean values and SEs of results from three separately grown cultures. An asterisk indicates statistically significant differences in comparison to controls.
FIG 3.
Photomicrographs of S. limacinum B4D1 cells treated with methanol at concentrations of 0% (control) (A), 1.6% (B), 3.2% (C), 4.8% (D), 5.6% (E), and 6.4% (F). Scale bar, 10 μm.
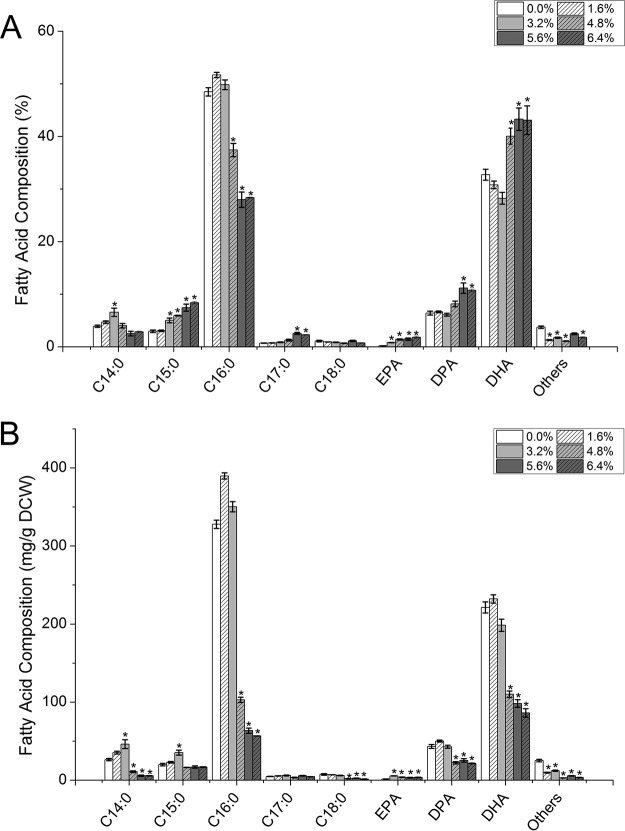
Methanol addition also significantly altered the fatty acid profiles of S. limacinum B4D1 and notably increased the proportions of DHA and eicosapentaenoic acid (EPA). C16:0 and DHA were the main saturated fatty acids and polyunsaturated fatty acids, respectively. Interestingly, the proportion of C16:0 was relatively stable at low methanol concentrations (≤3.2%) but significantly decreased (P ≤ 0.05) at high concentrations (≥4.8%) (Fig. 4A). Moreover, the DHA ratio also showed little variation at low methanol concentrations but markedly increased (P ≤ 0.05) at high concentrations. However, because of the sharp decline in biomass and lipid content, the DHA content (mg/g) decreased significantly (P ≤ 0.05) after treatment with high methanol concentrations (≥4.8%) (Fig. 4B). In addition, docosapentaenoic acid (DPA) showed a pattern similar to that of DHA, whereas EPA increased with increasing methanol concentrations.
FIG 4.
Effects of methanol on the fatty acid compositions of S. limacinum B4D1. (A) Percentage of total fatty acids; (B) percentage of dry cell weight. Shown are mean values and SEs of results from three separately grown cultures. An asterisk indicates statistically significant differences in comparison to controls.
(ii) Effects of methanol on squalene and sterol accumulation. Methanol also had significant effects on the accumulation of squalene and sterols in S. limacinum B4D1. When the methanol concentration increased from 1.6% to 3.2%, the squalene content increased significantly (P ≤ 0.05) but markedly decreased (P ≤ 0.05) under other methanol concentrations (Fig. 5A). In addition, the content of cholesterol, stigmasterol, and lanosterol decreased under methanol stress (Fig. 5B), and the inhibition increased with increasing methanol concentrations.
FIG 5.
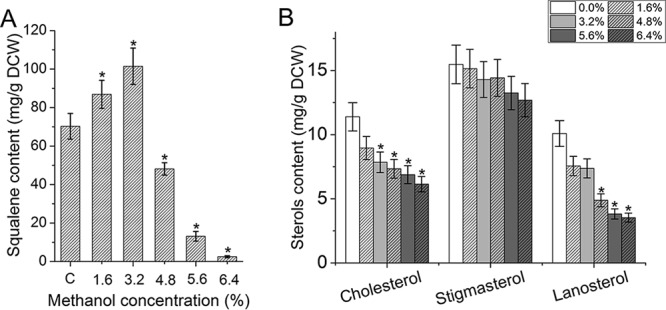
Effects of methanol on the squalene content (A) and sterol content (B) of S. limacinum B4D1. Shown are mean values and SEs of results from three separately grown cultures. An asterisk indicates statistically significant differences in comparison to controls.
(iii) Effects of methanol on carotenoid accumulation. The liquid culture of S. limacinum B4D1 with methanol stress was muddy orange or red, because of the accumulation of carotenoids. Among these carotenoids, astaxanthin was the main pigment of S. limacinum B4D1. The total astaxanthin content did not increase significantly under low methanol concentrations (≤3.2%); however, it increased sharply (P ≤ 0.05) under 4.8% methanol and peaked under 5.6% methanol, showing a >2,000-fold increase over the control levels. Furthermore, trans-astaxanthin was the most abundant isomer in all groups (more than 90% of total astaxanthin) (Fig. 6). In addition, the β-carotene, canthaxanthin, and lycopene content increased with increasing methanol concentration. Among the five groups with methanol added, the largest effect on β-carotene accumulation was observed under 5.6% methanol, whereas canthaxanthin and lycopene accumulated at 6.4% methanol.
FIG 6.
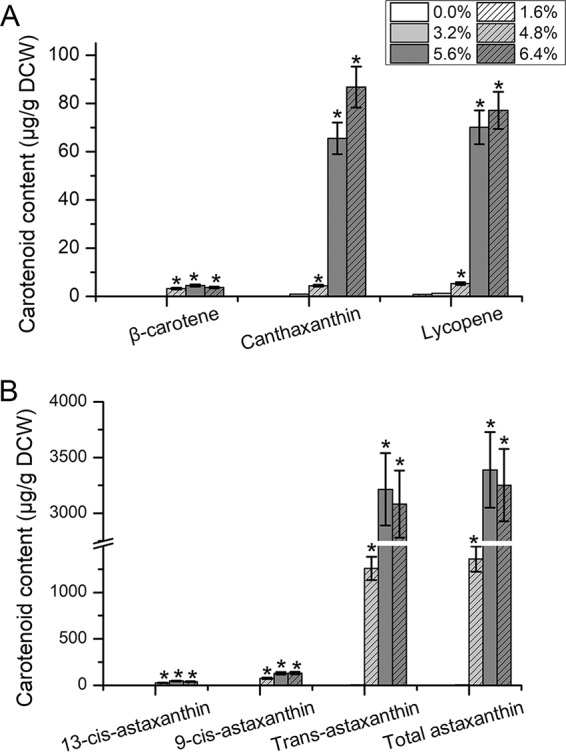
Effects of methanol on the composition of S. limacinum B4D1 carotenoids. (A) Content of carotenoids except astaxanthin; (B) astaxanthin content. Shown are mean values and SEs of results from three separately grown cultures. An asterisk indicates statistically significant differences in comparison to controls.
Transcriptomic analysis.
We performed gene expression profiling of S. limacinum B4D1 in fermentation culture under three different methanol concentrations (0% [control; C], 3.2% [low; L], and 5.6% [high; H]). After removing low-quality reads and trimming adapter sequences, we found that more than 2.07 × 106 ± 7.04 × 104, 4.23 × 106 ± 3.51 × 105, and 3.31 × 106 ± 6.60 × 105 read pairs mapped to the reference genome under each condition, reaching 14.08×, 28.69×, and 22.46× coverage, respectively (Table 1). A total of 1,918 genes showed differential expression patterns between L and C conditions: 927 and 991 genes were upregulated and downregulated, respectively. The number of differentially expressed genes (DEGs) between H and C conditions was 4,891, including 2,597 upregulated genes and 2,294 downregulated genes (Table 2). In total, 6,027 DEGs were identified by pairwise comparisons of the three different conditions. All genes mentioned herein are listed in Tables S1 to S5 in the supplemental material.
TABLE 1.
Summary of RNA-Seq data in three groups
| Groupa | No. of mapped reads | Total no. of bases (nt) | Coverage (×) |
|---|---|---|---|
| C | 2.07 × 106 | 2.80 × 108 | 14.08 |
| L | 4.23 × 106 | 5.71 × 108 | 28.69 |
| H | 3.31 × 106 | 4.47 × 108 | 22.46 |
C, control (0% methanol); L, 3.2% methanol concentration; H, 5.6% methanol concentration.
TABLE 2.
Analysis of DEGs in comparison of methanol conditionsa
| Comparison of conditions | No. of DEGs | No. of upregulated genes | No. of downregulated genes |
|---|---|---|---|
| L vs C | 1,918 | 927 | 991 |
| H vs C | 4,891 | 2,597 | 2,294 |
| H vs L | 4,451 | 2,362 | 2,089 |
C, control (0% methanol); L, 3.2% methanol concentration; H, 5.6% methanol concentration.
Transcriptomic analysis revealed that the mRNA levels of three genes for key signaling pathways that affect cell growth and metabolism changed with methanol stress. In the Ras signaling pathway, the gene encoding Ras (GTPase KRas) was significantly downregulated (1.60-fold), thus decreasing Ras activity and inhibiting cell growth. In the phospholipase D signaling pathway, the gene encoding phospholipase D1/2 (PLD) was significantly upregulated (nearly 2-fold), thus presumably enhancing phosphatidate production, activating the mTOR signaling pathway, and enhancing cell survival. Since gene expression may also depend on posttranscriptional regulatory mechanisms, this and other interpretations of the metabolic consequences should be considered tentative. Furthermore, in the AMPK signaling pathway, expression of the gene encoding 5′-AMP-activated protein kinase (AMPK) was significantly increased (1.56-fold), thereby increasing the activity of hydroxymethylglutaryl coenzyme A (CoA) reductase (HMGCR) to inhibit cholesterol synthesis; in addition, HMGCR also downregulated expression of the gene encoding sterol regulatory element-binding transcription factor 1 (SREBP1), thus further inhibiting the synthesis of fatty acids. In addition, genes encoding propionate CoA- transferase (pct), pyruvate dehydrogenase E2 component (DLAT), and phosphate acetyltransferase (pta), which are involved in pyruvate metabolism, were upregulated (1.42-, 1.59-, and 2.36-fold, respectively), thus enhancing acetyl-CoA production to some extent. However, most of the genes involved in fatty acid synthesis were significantly downregulated, thus resulting in lower fatty acid bioaccumulation.
(i) Differential gene expression in the central carbon metabolism pathway. According to gene annotation information, the expression of genes involved in the central carbon metabolism pathway among the three methanol concentration conditions (C, L, and H) were identified. Table S2 compares the transcript abundance for enzymes involved in central carbon metabolism pathways, including glycolysis (Embden-Meyerhof-Parnas pathway [EMP]), the tricarboxylic acid (TCA) cycle, and the pentose phosphate pathway (PPP). In EMP, in comparison with the C condition, 12 DEGs were identified under the H condition, with 2 genes upregulated and 10 genes downregulated. Notably, the hexokinase (HK), 6-phosphofructokinase (pfkA), and pyruvate kinase (pyk) genes, encoding three key enzymes, were all significantly downregulated (P ≤ 0.05), in comparison with the control (nearly 7-, 3-, and 13-fold decreases, respectively). There were 11 DEGs involved in the TCA cycle under the H condition, with 5 genes upregulated and 6 genes downregulated. The expression of genes encoding isocitrate dehydrogenase (IDH1, IDH3), 2-oxoglutarate dehydrogenase (sucA, sucB), and citrate synthase (gltA), which are rate-limiting enzymes, fluctuated with increasing methanol concentrations. In addition, genes encoding various enzymes involved in the PPP, including glucose-6-phosphate dehydrogenase (G6PD), 6-phosphogluconate dehydrogenase (6PGD), and ribose 5-phosphate epimerase (RPIA), were downregulated, whereas the ribulose-phosphate 3-epimerase (RPE) gene was upregulated by 1.21-fold (Table S2).
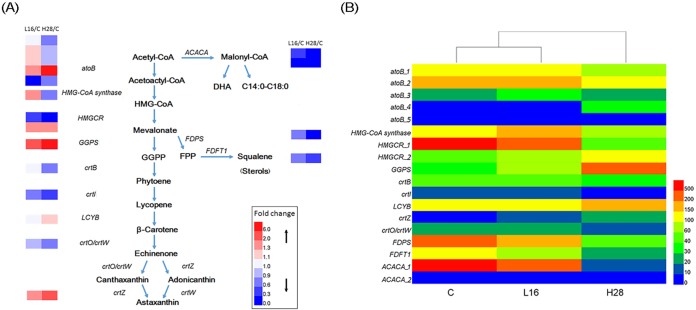
(ii) Analysis of differentially expressed genes associated with carotenoid biosynthesis. Through the analysis of the sample comparisons, we identified nine genes involved in carotenoid metabolism, and the biosynthetic pathway is presented in Fig. 7A. The transcriptomic analysis showed that these genes exhibited notably different levels in the carotenoid biosynthesis pathway. However, only the genes encoding geranylgeranyl diphosphate synthase (GGPS), lycopene β-cyclase (LCYB), and β-carotene 3-hydroxylase (crtZ) were upregulated to significantly different levels (P ≤ 0.05) under the H condition (Table S5; Fig. 7B).
FIG 7.
Differential expression of genes involved in the carotenoid biosynthesis pathway of S. limacinum B4D1. (A) Carotenoid biosynthesis pathway of S. limacinum B4D1 and relative expression of genes (fold change). (B) Relative gene fragments per kilobase per million (FPKM) in the carotenoid biosynthesis pathway. Genes encoding acetyl-CoA C-acetyltransferase (atoB), acetyl-CoA carboxylase (ACACA), farnesyl diphosphate synthase (FDPS), farnesyl-diphosphate farnesyltransferase (FDFT1), hydroxymethylglutaryl-CoA synthase (HMG-CoA synthase), hydroxymethylglutaryl-CoA reductase (HMGCR), geranylgeranyl diphosphate synthase, type III (GGPS), 15-cis-phytoene synthase (crtB), phytoene desaturase (crtI), lycopene β-cyclase (LCYB), β-carotene ketolase (crtO/crtW), and β-carotene 3-hydroxylase (crtZ) are shown.
(iii) Gene expression analysis of a competitive pathway of carotenoid synthesis. Starting with acetyl-CoA, there is a competitive pathway in carotenoid synthesis, i.e., the fatty acid synthesis pathway, and from mevalonate, there is another competitive pathway, i.e., the squalene/sterol synthesis pathway (Fig. 7A). In the fatty acid synthesis pathway, the acetyl-CoA carboxylase/biotin carboxylase gene (ACACA) encodes a key enzyme for the synthesis of fatty acid precursors, and its expression was markedly downregulated (2.11- and 101.80-fold from C to L and H conditions, respectively). Two genes encoding polyketide synthase (pks-1 and pks-2), a key enzyme for the synthesis of unsaturated fatty acids, were strongly downregulated (26.61- and 140.00-fold decreases from C to H conditions, respectively). In the squalene/sterol synthesis pathway, expression of the farnesyl diphosphate synthase (FDPS) and farnesyl-diphosphate farnesyltransferase (FDFT1) genes, which are key for the synthesis of precursors, was decreased by 5.01- and 3.41-fold from C to H conditions, respectively (Table S4).
(iv) Differential gene expression related to the antioxidant defense system. We also detected genes encoding three antioxidant enzymes: superoxide dismutase (SOD), l-ascorbate peroxidase (APX), and catalase (CAT). The transcription results indicated that under the H condition, four genes encoding SOD (SOD-1, SOD-2, SOD-3, and SOD-4) were significantly upregulated (P ≤ 0.05), compared with the transcription results under the C condition (3.25-, 5.79-, 3.75-, and 1.22-fold increases, respectively), whereas SOD-5 and SOD-6 showed no significant change. We also found that the H condition led to a significant upregulation (P ≤ 0.05) of CAT expression, which was mainly responsible for the disposal of free circulating H2O2. Furthermore, the expression of APX was also upregulated (Table S3). For glutathione peroxidase (GSH-Px), we detected three genes encoding GSH-Px which are part of a major protective system against endogenously and exogenously induced peroxidation. Two genes, GSH-Px-2 and GSH-Px-3, were significantly decreased by 3.20- and 2.50-fold, respectively (Table S3).
(v) Verification of gene expression through qRT-PCR. To validate the transcriptomic data, we used quantitative real-time PCR (qRT-PCR) to verify the transcriptome sequencing (RNA-seq) results for key genes involved in the central carbon metabolism pathways (HK, pfkA, pyk, IDH1, sucA, sucB, and gltA), fatty acid synthesis pathway (ACACA), squalene/sterol synthesis pathway (FDPS and FDFT1), carotenoid biosynthesis pathway (atoB-1, atoB-2, HMG-CoA synthase, HMGCR-1, HMGCR-2, crtB, GGPS, LCYB, crtI, crtO/crtW, crtZ), and antioxidant defense system (SOD-1, SOD-2, SOD-3, CAT, and APX). The results showed that the expression levels of most genes were consistent with the results from the transcriptomic analysis, thus indicating that the RNA-seq data were reliable (Fig. S1).
DISCUSSION
There is keen interest in the development of technologies for increasing the microbial production of carotenoids, especially astaxanthin. Schizochytrium has been found to synthesize large amounts of astaxanthin under methanol stress; this characteristic and its high growth rate make it a potential microorganism for astaxanthin production. To further understand and improve the capacity for astaxanthin production, we applied transcriptomic analysis to compare the expression of genes involved in important metabolic pathways under different methanol concentrations. We also investigated three key signaling pathways, thus providing new clues to understanding astaxanthin biosynthesis in S. limacinum B4D1 and laying a foundation for the future construction of cell factories for astaxanthin production.
The central carbon metabolism pathway is the main route for generating NADH and ATP, two important factors for cell growth (24). In most microorganisms, EMP and the TCA cycle are principally responsible for generating ATP and NADH. Our transcriptomic results showed that when S. limacinum B4D1 was cultured under the H condition, two phosphoglycerate kinase genes (pgk-1, pgk-2) and two pyk genes (pyk-1, pyk-2), encoding two key enzymes generating ATP, were downregulated by 1.66- and 2.33-fold and by 13.00- and 1.88-fold, respectively, compared with the C condition, whereas IDH1 and IDH3, encoding a key gene in NADH production, were upregulated (1.52- and 2.12-fold, respectively). Consequently, S. limacinum B4D1 was expected to generate less ATP and more NADH under the H condition. Likewise, the biomass under the 5.6% methanol (H) condition was significantly smaller (P ≤ 0.05) than that under the control (C) condition. Consequently, ATP, the most direct source of energy in organisms, plays a more important role in cell growth.
There are two known metabolic pathways for fatty acid biosynthesis in Schizochytrium: a fatty acid synthase pathway and a polyunsaturated fatty acid synthase pathway (23, 25, 26). Acetyl-CoA and NADPH are considered the same precursors for both pathways (11). Reducing power in the form of NADPH is assumed to be limiting for lipid accumulation (27). In most microorganisms, the PPP is principally responsible for generating NADPH, through the key enzymes G6PD and 6PGD. Various stress conditions, such as cold (28), oxidative (29), and high-salinity (30) conditions, have been found to improve the activity of G6PD and 6PGD and consequently maintain the abundant supply of NADPH. In addition, a study by Osada et al. has found that overexpressing G6PD and 6PGD to increase the production of NADPH promotes fatty acid biosynthesis in Fistulifera solaris (31). However, our transcriptomic data showed that when S. limacinum B4D1 was cultured under the H condition, G6PD and 6PGD were downregulated by 1.50-fold and 2.57-fold, respectively. Genes encoding other enzymes involved in the PPP, such as RPIA, were also downregulated, thus suggesting that the PPP decreased to some extent, so that less NADPH was generated than that under the C condition (see Table S2 in the supplemental material). Studies have also demonstrated that malic enzyme (ME) is also a main generator of NADPH for fatty acid synthesis in oleaginous microbes. This enzyme catalyzes the reversible oxidative decarboxylation of malate and links the glycolytic pathway and the citric acid cycle (32–34); however, ME gene responses vary in different environments. For example, the transcription of ME genes is not significantly affected by oxidative stress, whereas nitrogen starvation enhances ME gene expression in Drosophila melanogaster (35). Furthermore, Bi et al. have reported that the expression of ME genes under a high oxygen supply is significantly upregulated in Schizochytrium sp. strain HX-308 (24). However, our results showed that the expression of two genes encoding ME (ME-1, ME-2) was downregulated under the H condition. Downregulation of key genes in two pathways in microorganisms may lead to a decrease in NADPH used for biological metabolism.
In addition to NADPH, acetyl-CoA is a major topic in the lipid research field, because it is an indispensable precursor for fatty acid biosynthesis (36). Acetyl-CoA can be produced through various catabolic reactions, such as EMP, β-oxidation, and the catabolism of branched-chain amino acids, to supply energy for cells (37, 38). EMP plays a major role in shuttling carbon toward acetyl-CoA production. Our transcriptomic data revealed that many genes involved in EMP were significantly decreased (P ≤ 0.05) under the H condition (Table S2). HK, encoding a kinase that phosphorylates glucose to glucose-6-phosphate, was downregulated by 7.22-fold under the H condition. In contrast, previous studies have found that using glycerol as a carbon source or culturing under high oxygen supply enhances the expression of HK in Schizochytrium (24, 39). As a key enzyme in EMP, PfkA is responsible for catalyzing the ATP-dependent phosphorylation of fructose-6-phosphate to fructose 1,6-bisphosphate. The expression of pfkA transcripts decreased by 3.28-fold under the H condition. However, Cheng et al. (30) found that the expression of pfkA in a Nitzschia sp. strain increases under high-salinity stress conditions. In addition, we found that Pyk, which catalyzes phosphoenol pyruvate into pyruvate, was more sensitive to changes in methanol. The expression of two genes encoding Pyk was downregulated by 13.00- and 1.88-fold, respectively. Therefore, the expression of PPP- and EMP-related genes was downregulated to various degrees, thus suggesting that the NADPH and acetyl-CoA production of S. limacinum B4D1 decreased under the H condition.
Transcriptomic analysis also showed that ACACA, the rate-limiting enzyme in the first stage of fatty acid synthesis, which is responsible for catalyzing acetyl-CoA into malonyl-CoA, decreased over 100-fold under the H condition. Moreover, expression of two genes encoding Pks declined nearly 140- and 27-fold, respectively, compared with that under the C condition (Table S4). In conclusion, most key genes related to the synthesis of fatty acid and fatty acid precursors were significantly downregulated under the H condition and significantly decreased fatty acid bioaccumulation in S. limacinum B4D1, in agreement with the experimental results. Previous studies have reported that high concentrations of other short-chain alcohols, ethanol (40 ml/liter) and butanol (8 g/liter), lead to a decrease in both biomass (∼70% and ∼60%, respectively) and lipid accumulation (∼50% and ∼40%, respectively) (23, 40). In addition, ethanol and butanol also altered the fatty acid profiles of Schizochytrium. Its DHA percentage increased ∼20% in ethanol and decreased ∼30% in butanol. The EPA content in the presence of both ethanol and butanol increased by ∼70%. In this study, the results of methanol (5.6%) induction were similar to those of ethanol, decreasing biomass ∼50% and lipid ∼80% but increasing DHA ∼20%.
This study also found that higher methanol concentrations (≥4.8%) significantly decreased (P ≤ 0.05) squalene and sterol biosynthesis, and this decrease was more significant under increasing methanol concentrations (Fig. 5). Squalene and sterols are produced from farnesyl diphosphate via the mevalonate pathway (Fig. 7A). A study by Hong et al. has reported on the squalene synthase of Aurantiochytrium sp. strain KRS101 through recombinant methods and has shown it to catalyze the conversion of two molecules of farnesyl diphosphate into squalene in the presence of NADPH (41). Through transcriptomic analysis, we identified that two genes (FDPS and FDFT1) with important roles in synthesizing squalene/sterol precursors had lower expression under the H condition, thus suggesting that they are probably a major reason for the decrease in squalene/sterol synthesis (Table S4). The highest reported squalene levels have been achieved in Aurantiochytrium sp. strain18 W-13a (198 mg/g DCW) (42), Aurantiochytrium sp. strain Yonez 5-1 (318 mg/g DCW) (43), Aurantiochytrium mangrovei PQ6 (33 mg/g DCW) (44), and Schizochytrium sp. strain CCTCC M209059b (84 mg/g DCW) (45). This experiment also showed substantial accumulation of squalene (101 mg/g DCW) in S. limacinum B4D1 treated with 3.2% methanol. In conclusion, Schizochytrium may be a promising candidate microorganism for squalene accumulation.
The regulation of gene transcription is considered a critical mechanism to regulate astaxanthin biosynthesis and accumulation (20, 21). Two pathways have been identified as related to astaxanthin synthesis in Aurantiochytrium sp. strain SK4: the methylerythritol 4-phosphate (MEP) pathway and the mevalonate (MVA) pathway. However, most of the genes involved in the MEP pathway were not found in our transcriptomic data, a finding also consistent with other transcriptomic results (46). The MVA pathway is shown in Fig. 7A, and the related gene expression is shown in Fig. 7B and Table S5. Transcriptomic analysis revealed that three genes (GGPS, LCYB, and crtZ) in the MVA pathway were upregulated at significantly different levels (P ≤ 0.05) under the H condition than under the C condition. GGPS is the key gene for catalytic synthesis of GGPP, as catalyzed by phytoene synthase (crtB) into phytoene. As the first committed step in the pathway, phytoene biosynthesis has long been considered a bottleneck in carotenoid biosynthesis (47, 48). Previously, the expression of the crtB gene in Haematococcus cells has been shown to be upregulated by high light levels (49, 50). However, in this study, crtB was downregulated only 1.15-fold between the H and C conditions. LCYB encodes lycopene β-cyclase, which catalyzes the synthesis of β-carotene from lycopene, and β-carotene is an important precursor in astaxanthin synthesis. The results showed that LCYB was upregulated only 1.19-fold under the H condition. Kobayashi has found that astaxanthin is produced at the expense of β-carotene in two pathways: ketolase (crtO/crtW) and hydroxylation (crtZ) of β-carotene (51). Previous studies have also reported that crtO/crtW and crtZ are responsible for the rate-limiting steps in the catalysis of β-carotene to astaxanthin (49, 50, 52). We found that the transcripts of crtO/crtW were downregulated 1.49-fold, whereas crtZ was upregulated 2.43-fold under the H condition. Although the expression of crtB and crtO/crtW was lower under the H condition, it was consistent with that described in previous studies of astaxanthin synthetic strains (53–55). A study by Wen et al. has found that genes related to the astaxanthin synthesis pathway, such as ipi, lyc, and bkt, are upregulated when H. pluvialis is exposed to ethanol induction conditions, thus significantly increasing astaxanthin bioaccumulation (19). In addition, Vázquez et al. found that ethanol increases the astaxanthin content of Phaffia rhodozyma but is not conducive to its growth (56). Gu et al. reported that ethanol increases the rate of carotenoid formation in Phaffia rhodozyma, thus suggesting that ethanol improves the activity of ethanol dehydrogenase and carboxymethyl glutaric CoA reductase, which enhance the MVA pathway; consequently, a large number of precursors are synthesized, such as mevalonate, to promote the subsequent reaction, thus increasing the astaxanthin content (57). In the process of carotenoid synthesis, the concerted reactions of multiple enzymatic steps are considered key to enhancing astaxanthin biosynthesis (21). For instance, Miao et al. screened a Phaffia rhodozyma mutant strain and found that the carotenogenic genes responsible for astaxanthin biosynthesis from GGPP are significantly upregulated, thus resulting in high astaxanthin accumulation (58). In addition, the introduction of yeast astaxanthin biosynthesis genes in Saccharomyces cerevisiae significantly improves the bioaccumulation of astaxanthin as well as its intermediates in cells (59). Lu et al. successfully introduced β-carotene hydroxylase and ketolase into Escherichia coli cells and balanced the expression of the two enzymes to enhance astaxanthin production without the addition of an inducer (60). Many studies have reported that β-carotene hydroxylase and ketolase probably compete for their substrate, and only balancing the expression of these two enzymes might lead to the transformation from β-carotene to astaxanthin (61, 62). Therefore, further work is needed to explore the relationship between the expression of β-carotene hydroxylase and ketolase, as well as to balance their expression to increase Schizochytrium’s astaxanthin biosynthesis.
Microbes can produce excessive reactive oxygen species (ROS) under stress conditions (63, 64). For instance, methanol can be oxidized to generate formaldehyde and H2O2, both of which are highly toxic to living cells (65). In fact, for oleaginous microorganisms such as Schizochytrium, although most oxygen consumption is used to provide energy for cell growth and lipid accumulation, some oxygen will inevitably be converted into ROS under stress conditions (66). Gessler et al. found that an antioxidant defense system, consisting of enzymatic as well as nonenzymatic mechanisms, plays an important role in counteracting the various types of ROS (67). For the enzymatic defense system, SOD, which catalyzes the dismutation of the superoxide anion O2− into H2O and O2, is considered the first line of defense against ROS. Previous studies have found that SOD activity is enhanced in Ditylum brightwellii and Scenedesmus sp. strain in response to copper stress (68, 69). Likewise, Kumar et al. observed that SOD activity is significantly increased when microorganisms are exposed to high-salinity stress (70). In this study, six SOD genes were detected, four of which were upregulated to various degrees in S. limacinum B4D1 cultured under the H condition. However, additional studies are needed to explore the exact implication of each individual SOD gene in response to environmental change. Catalases (CAT) also play a central role in defense against oxidative stress, by catalyzing the breakdown of H2O2 into O2 and H2O. We observed a higher expression of CAT in S. limacinum B4D1 cultivated under the H condition, thus suggesting that more hydrogen peroxide was produced. In addition, the nonenzymatic defense system also counteracts ROS, such as low-molecular-weight metabolites (ascorbic acid, glutathione, thioredoxin, and astaxanthin) (21, 71). Schroeder et al. have shown that astaxanthin scavenges ROS and has antioxidant activity to protect cells from oxidative damage; in addition, the presence of substances causing oxidative damage alters carotenoid synthesis, thus resulting in high astaxanthin bioaccumulation (72). Likewise, in this study, although three glutathione peroxidases were downregulated under the H condition, astaxanthin bioaccumulation significantly increased (P ≤ 0.05) under treatment with 5.6% methanol (H) and probably enhanced the capability of the nonenzymatic defense system to protect cells from oxidative damage. Therefore, we speculated that under the H condition, the two antioxidant systems work in concert to decrease the damage of methanol stress in cells.
In conclusion, we examined a series of biological characteristics of S. limacinum B4D1, such as biomass and the content of fatty acids and astaxanthin, as induced by different concentrations of methanol. We additionally conducted a transcriptomic analysis to explore the changes in gene expression under methanol induction. The explorative analysis of signaling pathways, fatty acid, squalene/sterol, and astaxanthin synthesis pathways and the antioxidant defense system revealed possible reasons for the dramatic increase in astaxanthin content, all of which enhanced astaxanthin content directly or indirectly. Therefore, this work identified extensive DEGs involved in various signaling and metabolic pathways under different methanol conditions, thus providing important genetic information for engineering strategies in the future.
MATERIALS AND METHODS
Culture methods.
(i) Microorganism and medium composition. Schizochytrium limacinum B4D1 (CGMCC 8313) was preserved in 25% (vol/vol) glycerol at –80°C. The medium was divided into seed medium and fermentation medium. The seed medium and conditions were the same as those used in our previous study (23). The fermentation medium was composed of the following (per liter): artificial seawater salt, 15 g; glucose, 80 g; yeast extract, 4 g; peptone, 4 g; MgCl2·6H2O, 6.4 g; CaCl2·2H2O, 1 g; MgSO4·7H2O, 5 g; and KCl, 2 g. Both media were sterilized at 115°C for 20 min before inoculation.
(ii) Culture conditions. The culture of S. limacinum B4D1 was divided into two steps: (i) S. limacinum B4D1 was cultured in seed medium (250-ml flasks with 50 ml) at 26°C for 48 h on a shaker at 180 rpm; and (ii) 10% (vol/vol) seed culture was inoculated into fermentation medium (250-ml flasks with 50 ml) under the same culture conditions. The fermentation media were supplemented with six different concentrations of methanol (0%, 1.6%, 3.2%, 4.8%, 5.6%, and 6.4%). To prevent potential compositional shifts, we terminated fermentation when the glucose was completely consumed. The harvest times were as follows: 0% methanol, 3 days after inoculation; 1.6% methanol, 3 days after inoculation; 3.2% methanol, 4 days after inoculation; 4.8% methanol, 5 days after inoculation; 5.6% methanol, 6 days after inoculation; and 6.4% methanol, 7 days after inoculation.
(iii) Microscopy. When fermentation was stopped, the cells were observed with a TCS SP5 II microscope (Leica, Germany).
Determination methods.
(i) Methanol determination. Methanol concentrations were measured with an internal standard method with a gas chromatograph (GC) equipped with a flame ionization detector and capillary columns (Zebron ZB-WaxPlus; Phenomenex, USA). Nitrogen was used as the carrier gas. The injection volume was 1 μl. The program was as follows: oven temperature, 150°C; injector temperature, 180°C; and detector temperature, 180°C. Methanol was identified by comparison to methanol standards (73). The quantity of methanol was determined by using butanol (Sigma) as an internal standard. To determine whether methanol volatilized during shaking, a flask containing fermentation medium and methanol was also placed under the same conditions without inoculation. Methanol determination was divided into three indexes. First, we determined the initial methanol concentration in the medium (C1); second, we determined the medium methanol concentration (with inoculation) at the end of fermentation (C2); third, we determined the medium methanol concentration (without inoculation) at the end of fermentation (C3). The methanol utilization and methanol volatilization were calculated as follows: methanol utilization = C3 – C2; methanol volatilization = C1 – C3.
(ii) Glucose analysis. The glucose concentration was analyzed with a biosensor (SBA-40D; Shandong Academy of Science, China) (23).
(iii) Measurement of dry cell weight. The cells were harvested by centrifugation (14,972 × g, 5 min), washed with distilled water, and then dried in a freeze dryer (FD-1C-50; BJBYK Co., Beijing, China) until the weight was constant (23).
(iv) Lipid extraction. Freeze-dried biomass (0.1 to 0.2 g) was weighed to perform lipid extraction. The total lipids were extracted through an acid-heating procedure (74).
(v) Fatty acid analysis. Fatty acids were converted into fatty acid methyl esters (FAMEs) before determination, and the FAME samples were subsequently analyzed by GC (23). All FAMEs were identified by comparing the retention times with those of the Supelco 37 Component FAME mix (Sigma) or methyl all-cis-4,7,10,13,16-docosapentaenoate (Sigma).
(vi) Squalene and sterol determination. The squalene and sterol analysis was implemented as described previously (23). Squalene and sterol were isolated from total lipids by saponification and analyzed by GC/quadrupole time of flight mass spectrometry (GC/Q-TOF MS) (Agilent 7890A GC/7200 Q-TOF MS).
(vii) Carotenoid determination. Carotenoids were determined according to the method of Zhang et al. (23). Freeze-dried biomass was ground with liquid nitrogen, and 50 to 200 mg (according to its carotenoid content) of powder was weighed, placed in a tube, and mixed with an appropriate amount of acetone-methanol (ratio, 7:3 [vol/vol]). The tube was placed in an ultrasonic crusher (Sonics VibraCell VCX-130) in an ice bath (effective power, 200 W; cycle, 15 min; work, 3 s; interval, 3 s). The suspension was then centrifuged at 6,000 × g and 4°C for 10 min. The supernatant was evaporated to near-dryness under a stream of N2 gas, and the pigments were redissolved in 1 ml of acetone-methanol (ratio, 7:3 [vol/vol]).
Carotenoids were analyzed by high-performance liquid chromatography (LC20A; Shimadzu) with a C30 reversed-phase column (150 by 3.0 mm [inside diameter], 3-mm particle size; YMC). Mobile phase A was methanol-methyl tert-butyl ether (MTBE)-water (81/15/4), and mobile phase B was methanol-MTBE-water (8/90/2). The elution gradient was from 100% A and 0% B to 0% A and 100% B in 45 min. The flow rate was 1.0 ml/min, the injection volume was 3 μl, and the detection wavelength was 480 nm. All carotenoids were identified and determined using the carotenoid standards lycopene and β-carotene (Solarbio) and canthaxanthin and astaxanthin (Sigma).
Transcription studies.
(i) RNA isolation and library construction. We selected three methanol concentration groups (C, L, and H) for transcriptomic sequencing. For each group, three biological replicates were prepared. The samples were collected at a stage in which fermentation was terminated (for C, it was the 3rd day after inoculation, for L, it was the 4th day, and for H, it was the 6th day). The collected cells were immediately frozen in liquid nitrogen and stored at –80°C until RNA extraction. Total RNA was extracted with TRIzol (Invitrogen). Transcriptomic libraries were constructed according to the manufacturer’s instructions (Illumina, San Diego, CA), and 2 × 150 bp paired-end sequencing was performed on the Illumina MiSeq platform.
(ii) Transcriptomic analysis. The reference genome was Aurantiochytrium limacinum ATCC MYA-1381 (https://genome.jgi.doe.gov/portal/AurlimATCMYA1381_FD/AurlimATCMYA1381_FD.info.html).
Raw reads that failed the chastity filter and reads with an average quality of less than 20 were removed. The remaining reads were aligned to the annotated genome of A. limacinum ATCC MYA-1381 with TopHat v2.1.1 (75). The reads that mapped to the noncoding strands or to multiple loci were removed. Alignment files were then converted to BAM files in SAMtools v1.3.1 (76). Cufflinks v2.2.1 (77–80) was then used to quantify the expression of each transcript in each sample. Cuffdiff was used to identify DEGs. Genes with a P value of ≤0.05 and a fold change of ≥1.5 or ≤0.67 were considered differentially expressed.
(iii) Gene expression profiling: qRT-PCR.The quantitative real-time PCR (qRT-PCR) primers (Table 3) were designed in Primer Premier 5.0 software and synthesized by GENEWIZ (China). qRT-PCR analysis was performed on an ABI 7500 fast real-time PCR system (Applied Biosystems, USA) with SYBR Premix ExTaq (TaKaRa, China), according to the manufacturer’s instructions. The PCR process was 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, and 60°C for 34 s. The relative expression values were calculated with the 2–ΔΔCT method.
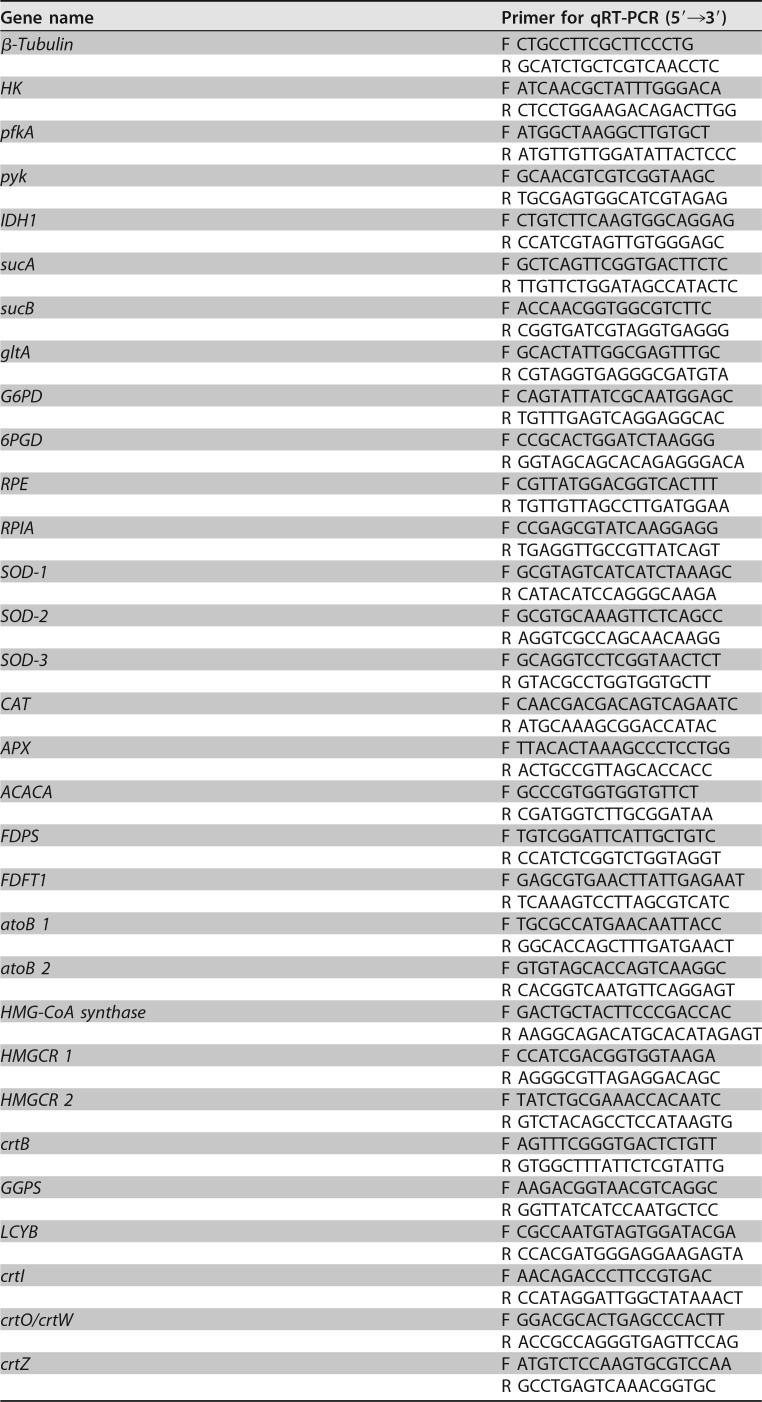
TABLE 3.
Primers for qRT-PCR in this study
Statistical analysis.
All experiments were performed in three biological replicates, and data were recorded as the mean and standard error (SE). Statistical analyses were performed with Student's t test (SPSS17.0). For all the data analysis, a P of ≤0.05 was considered statistically significant (indicated by an asterisk in the figures).
Accession number(s).
Raw sequencing reads have been deposited into the NCBI database under BioProject accession number PRJEB29668.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by The National Key Research and Development Program of China (2018YFA0902200, 2016YFD0501405), the Science and Technology Project of Tianjin (15PTCYSY00020), Youth Innovation Promotion Association CAS, the Key Research and Development Program of Shandong Province (2016GSF121030, 2017GSF21105, 2017CXGC0309), the Key Research and Development Program of Shandong Province (Food for Special Medical Purpose) (2018YYSP016), the National Natural Science Foundation of China (31170279, 41106124), the Natural Science Foundation of Shandong Province (ZR2011DM006, ZR2011CQ010), and the Project of Shandong Province Higher Educational Science and Technology Program (J17KA132).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01243-19.
REFERENCES
- 1.Gupta A, Barrow CJ, Puri M. 2012. Omega-3 biotechnology: thraustochytrids as a novel source of omega-3 oils. Biotechnol Adv 30:1733–1745. doi: 10.1016/j.biotechadv.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 2.Johnson MB, Wen ZY. 2009. Production of biodiesel fuel from the microalga Schizochytrium limacinum by direct transesterification of algal biomass. Energy Fuels 23:5179–5183. doi: 10.1021/ef900704h. [DOI] [Google Scholar]
- 3.Berman J, Zorrilla-Lopez U, Farre G, Zhu CF, Sandmann G, Twyman RM, Capell T, Christou P. 2015. Nutritionally important carotenoids as consumer products. Phytochem Rev 14:727–743. doi: 10.1007/s11101-014-9373-1. [DOI] [Google Scholar]
- 4.Chang GF, Gao NS, Tian GW, Wu QH, Chang M, Wang XG. 2013. Improvement of docosahexaenoic acid production on glycerol by Schizochytrium sp. S31 with constantly high oxygen transfer coefficient. Bioresour Technol 142:400–406. doi: 10.1016/j.biortech.2013.04.107. [DOI] [PubMed] [Google Scholar]
- 5.Raghukumar S. 2008. Thraustochytrid marine protists: production of PUFAs and other emerging technologies. Mar Biotechnol (NY) 10:631–640. doi: 10.1007/s10126-008-9135-4. [DOI] [PubMed] [Google Scholar]
- 6.Aki T, Hachida K, Yoshinaga M, Katai Y, Yamasaki T, Kawamoto S, Kakizono T, Maoka T, Shigeta S, Suzuki O, Ono K. 2003. Thraustochytrid as a potential source of carotenoids. J Am Oil Chem Soc 80:789–794. doi: 10.1007/s11746-003-0773-2. [DOI] [Google Scholar]
- 7.Armenta RE, Burja A, Radianingtyas H, Barrow CJ. 2006. Critical assessment of various techniques for the extraction of carotenoids and co-enzyme Q10 from the Thraustochytrid strain ONC-T18. J Agric Food Chem 54:9752–9758. doi: 10.1021/jf061260o. [DOI] [PubMed] [Google Scholar]
- 8.Singh P, Liu Y, Li LS, Wang GY. 2014. Ecological dynamics and biotechnological implications of thraustochytrids from marine habitats. Appl Microbiol Biotechnol 98:5789–5805. doi: 10.1007/s00253-014-5780-x. [DOI] [PubMed] [Google Scholar]
- 9.Quilodran B, Hinzpeter I, Hormazabal E, Quiroz A, Shene C. 2010. Docosahexaenoic acid (C22:6n-3, DHA) and astaxanthin production by Thraustochytriidae sp. AS4-A1 a native strain with high similitude to Ulkenia sp.: evaluation of liquid residues from food industry as nutrient sources. Enzyme Microb Technol 47:24–30. doi: 10.1016/j.enzmictec.2010.04.002. [DOI] [Google Scholar]
- 10.Johnson EJ. 2002. The role of carotenoids in human health. Nutr Clin Care 5:56–65. doi: 10.1046/j.1523-5408.2002.00004.x. [DOI] [PubMed] [Google Scholar]
- 11.Aasen IM, Ertesvag H, Heggeset TMB, Liu B, Brautaset T, Vadstein O, Ellingsen TE. 2016. Thraustochytrids as production organisms for docosahexaenoic acid (DHA), squalene, and carotenoids. Appl Microbiol Biotechnol 100:4309–4321. doi: 10.1007/s00253-016-7498-4. [DOI] [PubMed] [Google Scholar]
- 12.Chen LM, Liu XM, Li DM, Chen WX, Zhang K, Chen SL. 2016. Preparation of stable microcapsules from disrupted cell of Haematococcus pluvialis by spray drying. Int J Food Sci Technol 51:1834–1843. doi: 10.1111/ijfs.13155. [DOI] [Google Scholar]
- 13.Giovannetti R, Alibabaei L, Pucciarelli F. 2009. Kinetic model for astaxanthin aggregation in water-methanol mixtures. Spectrochim Acta A Mol Biomol Spectrosc 73:157–162. doi: 10.1016/j.saa.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 14.Xie D, Mu H, Tang T, Wang X, Wei W, Jin J, Wang X, Jin Q. 2018. Production of three types of krill oils from krill meal by a three-step solvent extraction procedure. Food Chem 248:279–286. doi: 10.1016/j.foodchem.2017.12.068. [DOI] [PubMed] [Google Scholar]
- 15.Prieto CVG, Ramos FD, Estrada V, Villar MA, Diaz MS. 2017. Optimization of an integrated algae-based biorefinery for the production of biodiesel, astaxanthin and PHB. Energy 139:1159–1172. doi: 10.1016/j.energy.2017.08.036. [DOI] [Google Scholar]
- 16.Feng ZZ, Li MY, Wang YT, Zhu MJ. 2018. Astaxanthin from Phaffia rhodozyma: microencapsulation with carboxymethyl cellulose sodium and microcrystalline cellulose and effects of microencapsulated astaxanthin on yogurt properties. LWT Food Sci Technol 96:152–160. doi: 10.1016/j.lwt.2018.04.084. [DOI] [Google Scholar]
- 17.Johnson EA, An GH. 1991. Astaxanthin from microbial sources. Crit Rev Biotechnol 11:297–326. doi: 10.3109/07388559109040622. [DOI] [Google Scholar]
- 18.Wang JX, Sommerfeld M, Hu Q. 2009. Occurrence and environmental stress responses of two plastid terminal oxidases in Haematococcus pluvialis (Chlorophyceae). Planta 230:191–203. doi: 10.1007/s00425-009-0932-4. [DOI] [PubMed] [Google Scholar]
- 19.Wen ZW, Liu ZY, Hou YY, Liu CF, Gao F, Zheng YB, Chen FJ. 2015. Ethanol induced astaxanthin accumulation and transcriptional expression of carotenogenic genes in Haematococcus pluvialis. Enzyme Microb Technol 78:10–17. doi: 10.1016/j.enzmictec.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 20.Gao ZQ, Li Y, Wu GX, Li GQ, Sun HF, Deng SZ, Shen YC, Chen GQ, Zhang RH, Meng CX, Zhang XW. 2015. Transcriptome analysis in Haematococcus pluvialis: astaxanthin induction by salicylic acid (SA) and jasmonic acid (JA). PLoS One 10:e0140609. doi: 10.1371/journal.pone.0140609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gwak Y, Hwang YS, Wang BB, Kim M, Jeong J, Lee CG, Hu Q, Han DX, Jin E. 2014. Comparative analyses of lipidomes and transcriptomes reveal a concerted action of multiple defensive systems against photooxidative stress in Haematococcus pluvialis. J Exp Bot 65:4317–4334. doi: 10.1093/jxb/eru206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng TT, Zhang ZK, Shahid MQ, Wei WL, Baloch FS, Wu JC, Lin SQ, Yang XH. 2017. RNA-Seq reveals differential expression patterns of genes associated with carotenoid accumulation in loquat. Acta Physiol Plant 39:168. doi: 10.1007/s11738-017-2463-0. [DOI] [Google Scholar]
- 23.Zhang K, Chen LM, Liu JM, Gao F, He RL, Chen WX, Guo W, Chen SL, Li DM. 2017. Effects of butanol on high value product production in Schizochytrium limacinum B4D1. Enzyme Microb Technol 102:9–15. doi: 10.1016/j.enzmictec.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Bi ZQ, Ren LJ, Hu XC, Sun XM, Zhu SY, Ji XJ, Huang H. 2018. Transcriptome and gene expression analysis of docosahexaenoic acid producer Schizochytrium sp. under different oxygen supply conditions. Biotechnol Biofuels 11:249. doi: 10.1186/s13068-018-1250-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo DS, Ji XJ, Ren LJ, Yin FW, Sun XM, Huang H, Zhen G. 2018. Development of a multi-stage continuous fermentation strategy for docosahexaenoic acid production by Schizochytrium sp. Bioresour Technol 269:32–39. doi: 10.1016/j.biortech.2018.08.066. [DOI] [PubMed] [Google Scholar]
- 26.Ren LJ, Sun XM, Ji XJ, Chen SL, Guo DS, Huang H. 2017. Enhancement of docosahexaenoic acid synthesis by manipulation of antioxidant capacity and prevention of oxidative damage in Schizochytrium sp. Bioresour Technol 223:141–148. doi: 10.1016/j.biortech.2016.10.040. [DOI] [PubMed] [Google Scholar]
- 27.Ratledge C. 2014. The role of malic enzyme as the provider of NADPH in oleaginous microorganisms: a reappraisal and unsolved problems. Biotechnol Lett 36:1557–1568. doi: 10.1007/s10529-014-1532-3. [DOI] [PubMed] [Google Scholar]
- 28.Valledor L, Furuhashi T, Hanak AM, Weckwerth W. 2013. Systemic cold stress adaptation of Chlamydomonas reinhardtii. Mol Cell Proteomics 12:2032–2047. doi: 10.1074/mcp.M112.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godon C, Lagniel G, Lee J, Buhler JM, Kieffer S, Perrot M, Boucherie H, Toledano MB, Labarre J. 1998. The H2O2 stimulon in Saccharomyces cerevisiae. J Biol Chem 273:22480–22489. doi: 10.1074/jbc.273.35.22480. [DOI] [PubMed] [Google Scholar]
- 30.Cheng RL, Feng J, Zhang BX, Huang Y, Cheng J, Zhang CX. 2014. Transcriptome and gene expression analysis of an oleaginous diatom under different salinity conditions. Bioenerg Res 7:192–205. doi: 10.1007/s12155-013-9360-1. [DOI] [Google Scholar]
- 31.Osada K, Maeda Y, Yoshino T, Nojima D, Bowler C, Tanaka T. 2017. Enhanced NADPH production in the pentose phosphate pathway accelerates lipid accumulation in the oleaginous diatom Fistulifera solaris. Algal Res 23:126–134. doi: 10.1016/j.algal.2017.01.015. [DOI] [Google Scholar]
- 32.Ratledge C. 2004. Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochimie 86:807–815. doi: 10.1016/j.biochi.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 33.Wynn JP, bin Abdul Hamid A, Ratledge C. 1999. The role of malic enzyme in the regulation of lipid accumulation in flamentous fungi. Microbiology 145:1911–1917. doi: 10.1099/13500872-145-8-1911. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Adams IP, Ratledge C. 2007. Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology 153:2013–2025. doi: 10.1099/mic.0.2006/002683-0. [DOI] [PubMed] [Google Scholar]
- 35.Rzezniczak TZ, Merritt TJS. 2012. Interactions of NADP-reducing enzymes across varying environmental conditions: a model of biological complexity. G3 (Bethesda) 2:1613–1623. doi: 10.1534/g3.112.003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan J, Cheng R, Lin X, You S, Li K, Rong H, Ma Y. 2013. Overexpression of acetyl-CoA synthetase increased the biomass and fatty acid proportion in microalga Schizochytrium. Appl Microbiol Biotechnol 97:1933–1939. doi: 10.1007/s00253-012-4481-6. [DOI] [PubMed] [Google Scholar]
- 37.Harris RA, Joshi M, Jeoung NH, Obayashi M. 2005. Overview of the molecular and biochemical basis of branched-chain amino acid catabolism. J Nutr 135:1527S–1530S. doi: 10.1093/jn/135.6.1527S. [DOI] [PubMed] [Google Scholar]
- 38.Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G. 2015. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab 21:805–821. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 39.Chen W, Zhou PP, Zhang M, Zhu YM, Wang XP, Luo XA, Bao ZD, Yu LJ. 2016. Transcriptome analysis reveals that up-regulation of the fatty acid synthase gene promotes the accumulation of docosahexaenoic acid in Schizochytrium sp. S056 when glycerol is used. Algal Res 15:83–92. doi: 10.1016/j.algal.2016.02.007. [DOI] [Google Scholar]
- 40.Sahin D, Tas E, Altindag UH. 2018. Enhancement of docosahexaenoic acid (DHA) production from Schizochytrium sp. S31 using different growth medium conditions. AMB Express 8:7. doi: 10.1186/s13568-018-0540-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong WK, Heo SY, Park HM, Kim CH, Sohn JH, Kondo A, Seo JW. 2013. Characterization of a squalene synthase from the thraustochytrid microalga Aurantiochytrium sp. KRS101. J Microbiol Biotechnol 23:759–765. doi: 10.4014/jmb.1212.12023. [DOI] [PubMed] [Google Scholar]
- 42.Kaya K, Nakazawa A, Matsuura H, Honda D, Inouye I, Watanabe MM. 2011. Thraustochytrid Aurantiochytrium sp. 18W-13a accumulates high amounts of squalene. Biosci Biotechnol Biochem 75:2246–2248. doi: 10.1271/bbb.110430. [DOI] [PubMed] [Google Scholar]
- 43.Nakazawa A, Kokubun Y, Matsuura H, Yonezawa N, Kose R, Yoshida M, Tanabe Y, Kusuda E, Van Thang D, Ueda M, Honda D, Mahakhant A, Kaya K, Watanabe MM. 2014. TLC screening of thraustochytrid strains for squalene production. J Appl Phycol 26:29–41. doi: 10.1007/s10811-013-0080-x. [DOI] [Google Scholar]
- 44.Hoang MH, Ha NC, Thom LT, Tam LT, Anh HTL, Thu NTH, Hong DD. 2014. Extraction of squalene as value-added product from the residual biomass of Schizochytrium mangrovei PQ6 during biodiesel producing process. J Biosci Bioeng 118:632–639. doi: 10.1016/j.jbiosc.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 45.Ren LJ, Sun GN, Ji XJ, Hu XC, Huang H. 2014. Compositional shift in lipid fractions during lipid accumulation and turnover in Schizochytrium sp. Bioresour Technol 157:107–113. doi: 10.1016/j.biortech.2014.01.078. [DOI] [PubMed] [Google Scholar]
- 46.Ye JR, Liu MM, He MX, Ye Y, Huang JC. 2019. Illustrating and enhancing the biosynthesis of astaxanthin and docosahexaenoic acid in Aurantiochytrium sp. SK4. Mar Drugs 17:E45. doi: 10.3390/md17010045.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cardozo KH, Guaratini T, Barros MP, Falcao VR, Tonon AP, Lopes NP, Campos S, Torres MA, Souza AO, Colepicolo P, Pinto E. 2007. Metabolites from algae with economical impact. Comp Biochem Physiol C Toxicol Pharmacol 146:60–78. doi: 10.1016/j.cbpc.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 48.Lorenz RT, Cysewski GR. 2000. Commercial potential for Haematococcus microalgae as a natural source of astaxanthin. Trends Biotechnol 18:160–167. doi: 10.1016/S0167-7799(00)01433-5. [DOI] [PubMed] [Google Scholar]
- 49.Steinbrenner J, Linden H. 2003. Light induction of carotenoid biosynthesis genes in the green alga Haematococcus pluvialis: regulation by photosynthetic redox control. Plant Mol Biol 52:343–356. doi: 10.1023/A:1023948929665. [DOI] [PubMed] [Google Scholar]
- 50.Vidhyavathi R, Venkatachalam L, Sarada R, Ravishankar GA. 2008. Regulation of carotenoid biosynthetic genes expression and carotenoid accumulation in the green alga Haematococcus pluvialis under nutrient stress conditions. J Exp Bot 59:1409–1418. doi: 10.1093/jxb/ern048. [DOI] [PubMed] [Google Scholar]
- 51.Kobayashi M. 2003. Astaxanthin biosynthesis enhanced by reactive oxygen species in the green alga Haematococcus pluvialis. Biotechnol Bioprocess Eng 8:322–330. doi: 10.1007/BF02949275. [DOI] [Google Scholar]
- 52.Steinbrenner J, Linden H. 2001. Regulation of two carotenoid biosynthesis genes coding for phytoene synthase and carotenoid hydroxylase during stress-induced astaxanthin formation in the green alga Haematococcus pluvialis. Plant Physiol 125:810–817. doi: 10.1104/pp.125.2.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Campisi L, Fambrini M, Michelotti V, Salvini M, Giuntini D, Pugliesi C. 2006. Phytoene accumulation in sunflower decreases the transcript levels of the phytoene synthase gene. Plant Growth Regul 48:79–87. doi: 10.1007/s10725-005-4831-9. [DOI] [Google Scholar]
- 54.Lotan T, Hirschberg J. 1995. Cloning and expression in Escherichia coli of the gene encoding β-C-4-oxygenase, that converts β-carotene to the ketocarotenoid canthaxanthin in Haematococcus pluvialis. FEBS Lett 364:125–128. doi: 10.1016/0014-5793(95)00368-j. [DOI] [PubMed] [Google Scholar]
- 55.Vidhyavathi R, Sarada R, Ravishankar GA. 2009. Expression of carotenogenic genes and carotenoid production in Haematococcus pluvialis under the influence of carotenoid and fatty acid synthesis inhibitors. Enzyme Microb Technol 45:88–93. doi: 10.1016/j.enzmictec.2009.05.005. [DOI] [Google Scholar]
- 56.Vázquez M, Santos V, Parajo JC. 1997. Effect of the carbon source on the carotenoid profiles of Phaffia rhodozyma strains. J Ind Microbiol Biotechnol 19:263–268. doi: 10.1038/sj.jim.2900376. [DOI] [Google Scholar]
- 57.Gu WL, An GH, Johnson EA. 1997. Ethanol increases carotenoid production in Phaffia rhodozyma. J Ind Microbiol Biotechnol 19:114–117. doi: 10.1038/sj.jim.2900425. [DOI] [PubMed] [Google Scholar]
- 58.Miao L, Chi S, Tang Y, Su Z, Yin T, Guan G, Li Y. 2011. Astaxanthin biosynthesis is enhanced by high carotenogenic gene expression and decrease of fatty acids and ergosterol in a Phaffa rhodozyma mutant strain. FEMS Yeast Res 11:192–201. doi: 10.1111/j.1567-1364.2010.00705.x. [DOI] [PubMed] [Google Scholar]
- 59.Ukibe K, Hashida K, Yoshida N, Takagi H. 2009. Metabolic engineering of Saccharomyces cerevisiae for astaxanthin production and oxidative stress tolerance. Appl Environ Microbiol 75:7205–7211. doi: 10.1128/AEM.01249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu Q, Bu YF, Liu JZ. 2017. Metabolic engineering of Escherichia coli for producing astaxanthin as the predominant carotenoid. Mar Drugs 15:E296. doi: 10.3390/md15100296.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lemuth K, Steuer K, Albermann C. 2011. Engineering of a plasmid-free Escherichia coli strain for improved in vivo biosynthesis of astaxanthin. Microb Cell Fact 10:29. doi: 10.1186/1475-2859-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scaife MA, Burja AM, Wright PC. 2009. Characterization of cyanobacterial beta-carotene ketolase and hydroxylase genes in Escherichia coli, and their application for astaxanthin biosynthesis. Biotechnol Bioeng 103:944–955. doi: 10.1002/bit.22330. [DOI] [PubMed] [Google Scholar]
- 63.Hama S, Tamalampudi S, Fukumizu T, Miura K, Yamaji H, Kondo A, Fukuda H. 2006. Lipase localization in Rhizopus oryzae cells immobilized within biomass support particles for use as whole-cell biocatalysts in biodiesel-fuel production. J Biosci Bioeng 101:328–333. doi: 10.1263/jbb.101.328. [DOI] [PubMed] [Google Scholar]
- 64.Tamalampudi S, Talukder MR, Hama S, Numata T, Kondo A, Fukuda H. 2008. Enzymatic production of biodiesel from Jatropha oil: a comparative study of immobilized-whole cell and commercial lipases as a biocatalyst. Biochem Eng J 39:185–189. doi: 10.1016/j.bej.2007.09.002. [DOI] [Google Scholar]
- 65.Yurimoto H, Sakai Y. 2009. Methanol-inducible gene expression and heterologous protein production in the methylotrophic yeast Candida boidinii. Biotechnol Appl Biochem 53:85–92. doi: 10.1042/BA20090030. [DOI] [PubMed] [Google Scholar]
- 66.Ruenwai R, Neiss A, Laoteng K, Vongsangnak W, Dalfard AB, Cheevadhanarak S, Petranovic D, Nielsen J. 2011. Heterologous production of polyunsaturated fatty acids in Saccharomyces cerevisiae causes a global transcriptional response resulting in reduced proteasomal activity and increased oxidative stress. Biotechnol J 6:343–356. doi: 10.1002/biot.201000316. [DOI] [PubMed] [Google Scholar]
- 67.Gessler NN, Aver'yanov AA, Belozerskaya TA. 2007. Reactive oxygen species in regulation of fungal development. Biochemistry (Mosc) 72:1091–1109. doi: 10.1134/S0006297907100070. [DOI] [PubMed] [Google Scholar]
- 68.Tripathi BN, Gaur JP. 2004. Relationship between copper- and zinc-induced oxidative stress and proline accumulation in Scenedesmus sp. Planta 219:397–404. doi: 10.1007/s00425-004-1237-2. [DOI] [PubMed] [Google Scholar]
- 69.Turrens JF. 1997. Superoxide production by the mitochondrial respiratory chain. Biosci Rep 17:3–8. doi: 10.1023/a:1027374931887. [DOI] [PubMed] [Google Scholar]
- 70.Kumar J, Singh VP, Prasad SM. 2015. NaCl-induced physiological and biochemical changes in two cyanobacteria Nostoc muscorum and Phormidium foveolarum acclimatized to different photosynthetically active radiation. J Photochem Photobiol B 151:221–232. doi: 10.1016/j.jphotobiol.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 71.Blokhina O, Virolainen E, Fagerstedt KV. 2003. Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann Bot 91:179–194. doi: 10.1093/aob/mcf118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schroeder WA, Johnson EA. 1995. Singlet oxygen and peroxyl radicals regulate carotenoid biosynthesis in Phaffia rhodozyma. J Biol Chem 270:18374–18379. doi: 10.1074/jbc.270.31.18374. [DOI] [PubMed] [Google Scholar]
- 73.Li D, Meng C, Wu G, Xie B, Han Y, Guo Y, Song C, Gao Z, Huang Z. 2018. Effects of zinc on the production of alcohol by Clostridium carboxidivorans P7 using model syngas. J Ind Microbiol Biotechnol 45:61–69. doi: 10.1007/s10295-017-1992-2. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y, Min QS, Xu J, Zhang K, Chen SL, Wang HJ, Li DM. 2016. Effect of malate on docosahexaenoic acid production from Schizochytrium sp B4D1. Electron J Biotechnol 19:56–60. doi: 10.1016/j.ejbt.2015.11.006. [DOI] [Google Scholar]
- 75.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roberts A, Pimentel H, Trapnell C, Pachter L. 2011. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 78.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. 2011. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol 12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.