Plastics are widely used in the global economy, and each year, at least 350 to 400 million tons are being produced. Due to poor recycling and low circular use, millions of tons accumulate annually in terrestrial or marine environments. Today it has become clear that plastic causes adverse effects in all ecosystems and that microplastics are of particular concern to our health.
KEYWORDS: PET, cutinase, microbial plastic degradation, polyamides, polyethylene, polyethylene terephthalate, polypropylene, polystyrene, polyurethane, polyvinylchloride
ABSTRACT
Plastics are widely used in the global economy, and each year, at least 350 to 400 million tons are being produced. Due to poor recycling and low circular use, millions of tons accumulate annually in terrestrial or marine environments. Today it has become clear that plastic causes adverse effects in all ecosystems and that microplastics are of particular concern to our health. Therefore, recent microbial research has addressed the question of if and to what extent microorganisms can degrade plastics in the environment. This review summarizes current knowledge on microbial plastic degradation. Enzymes available act mainly on the high-molecular-weight polymers of polyethylene terephthalate (PET) and ester-based polyurethane (PUR). Unfortunately, the best PUR- and PET-active enzymes and microorganisms known still have moderate turnover rates. While many reports describing microbial communities degrading chemical additives have been published, no enzymes acting on the high-molecular-weight polymers polystyrene, polyamide, polyvinylchloride, polypropylene, ether-based polyurethane, and polyethylene are known. Together, these polymers comprise more than 80% of annual plastic production. Thus, further research is needed to significantly increase the diversity of enzymes and microorganisms acting on these polymers. This can be achieved by tapping into the global metagenomes of noncultivated microorganisms and dark matter proteins. Only then can novel biocatalysts and organisms be delivered that allow rapid degradation, recycling, or value-added use of the vast majority of most human-made polymers.
INTRODUCTION
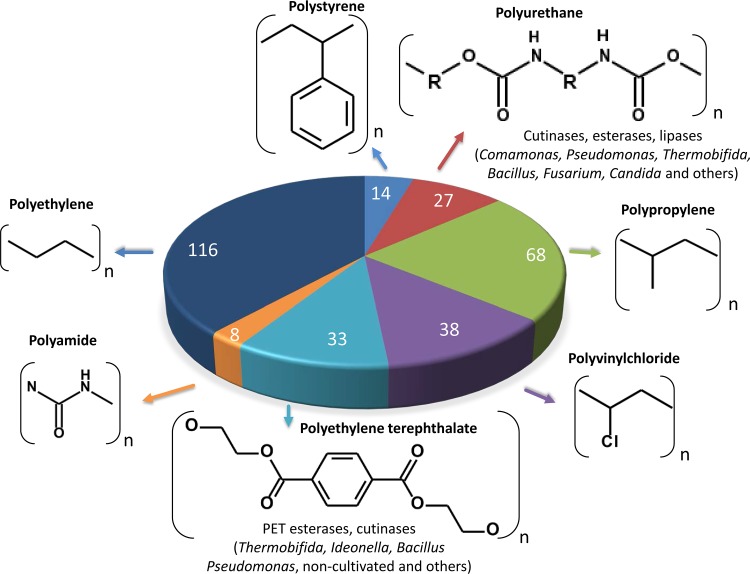
Altogether, synthetic polymers are produced worldwide at a scale of at least 350 to 400 million metric tons annually (1, 2; see also https://www.plasticsinsight.com/global-pet-resin-production-capacity, https://www.plasticsinsight.com/resin-intelligence/resin-prices/polyamide/, and https://www.plasticsinsight.com/world-plastics-production/). The main polymers that are produced and of importance to our economy are polyurethane (PUR), polyethylene (PE), polyamide (PA), polyethylene terephthalate (PET), polystyrene (PS), polyvinylchloride (PVC), and polypropylene (PP) (Fig. 1). With an increasing production and use of plastics, it is estimated that 5 to 13 million metric tons of plastic enter the ocean every year, with negative consequences for various ecosystems and for the health of humans and animals (1–3). Regarding only the Great Pacific Garbage Patch, more than 1.8 trillion pieces of plastic with an estimated weight of 80,000 tons have so far accumulated, with no end in sight (4–7). While a few reviews have recently been published focusing on the degradation of single types of plastic, only a few articles have addressed plastic degradation on a more global scale, addressing the degradation of several synthetic polymers (8). Therefore, the two main questions addressed in this review are as follows. (i) Which enzymes and microorganisms are currently known to be involved in high-molecular-weight polymer plastic degradation? (ii) What are the future challenges and technologies for identifying better enzymes acting on a highly diverse range of synthetic polymers?
FIG 1.
Main synthetic polymers globally produced in 2016. Numbers in the chart indicate the global annual production (millions of tons) of the specified synthetic polymer. Global annual plastic production was extracted from references 1–4, and https://www.plasticsinsight.com/global-pet-resin-production-capacity, https://www.plasticsinsight.com/resin-intelligence/resin-prices/polyamide/, and https://www.plasticsinsight.com/world-plastics-production/. Monomers are depicted above the chart. Indicated are the names of bacterial genera producing verified enzymes with available protein sequences that are known to be involved in the breakdown of the high-molecular-weight polymers (not the additives, plasticizers, etc.). For detailed references on the individual enzymes, refer to the main text. For PA, PE, PS, PVC, and PP, no defined enzymes that act on the polymer have been identified at the level of amino acid or DNA sequences. For enzymes acting on dimers or oligomers and feeding them into the different metabolic pathways, see the main text. For additional structural information on the polymers we refer to ChEBI (https://www.ebi.ac.uk/chebi/init.do).
Intriguingly, the currently best-known route of plastic destruction involves exposure to UV light together with mechanical disruption caused by waves and winds or grinding on marine rocks and sediments, which eventually breaks larger plastics into smaller pieces of micro- and nanoplastics (MP, with sizes of <5 mm, and NP, with sizes of <0.1 μm). So-called “weathering” and “photodegradation” are currently considered the main forces for initial depletion of plastics, and they mainly result in a modification of the chemical, physical, and mechanical properties of the plastics (9, 10). The resulting particles have a much larger surface area, which makes them amenable to further degradation (11). Notably, MPs and NPs are a concern to our health, as it is expected that they enter the food chain and end up in our intestines (12, 13). The fate of MPs or NPs in human or animal intestines has yet to be determined.
Therefore, removal of plastics from the environment using microbial enzymes has been a focus of recent research. The main challenge is that marine and terrestrial displaced plastics are highly stable and durable. Plastics have mainly been introduced since the 1960s and, given the relatively few decades since these human-made polymers became available, nature has only had a very short time to evolve highly active enzymes. Besides, many different types of plastics accumulate in the environment, and many of the frequently used plastics are mixtures containing additional solubilizers and other chemical agents to alter the mechanical and physical properties. These compounds are further targets for microbial biodegradation but may also interfere with degradative enzyme activities. It is assumed that the larger polymers are initially degraded by secreted exoenzymes into smaller subunits (multimers, dimers) that can be incorporated into the microbial cells. Once in the cells, either the oligomers or the degradation products of these are funneled through the classical degradation pathways to yield energy and/or serve as building blocks for catabolism or metabolism.
Within this framework, this review summarizes the main findings on microbial degradation of the polymers listed above. The chemical structures and some properties of these polymers are described in each of the following subsections. For a first overview on enzymes and microbes acting on the different plastics, see Fig. 1 and 2.
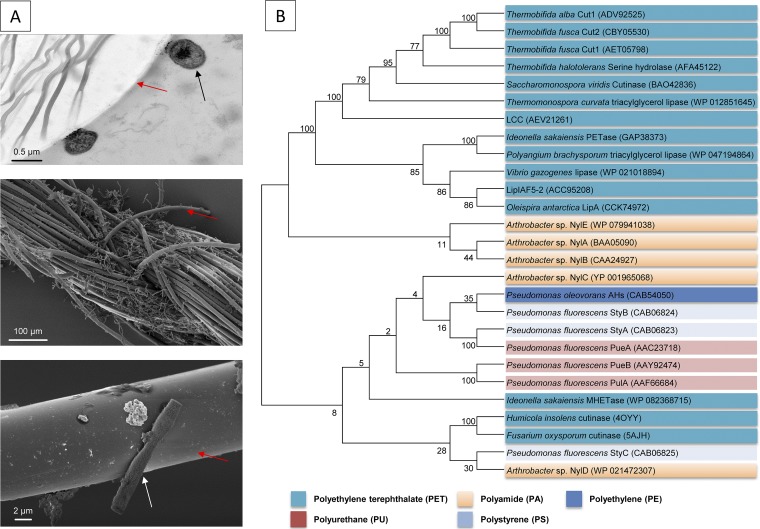
FIG 2.
(A) Electron microscopic images of Comamonas sp. strain DDHH 01 attached and hydrolyzing PET fibers. Comamonas sp. DDHH 01 was isolated from a sewage enrichment culture. Red arrows indicate PET fibers. Black and white arrows indicate bacterial cells. (Top) Transmission electron microscopy image of a PET fiber with attached Comamonas sp. cells. (Middle) Scanning electron microscope image of PET yarn with microcolonies. (Bottom) Closeup of a single cell on the surface of a single PET fiber. (B) Topology of a neighbor-joining tree containing representative sequences of most of the currently known synthetic polymer- or oligomer/monomer-degrading enzymes. The tree is based on amino acid sequence homologies. Overall, 27 known functional and verified enzymes were included in this alignment. This represents the majority of the currently known and biochemically characterized enzymes. PET hydrolases represent the largest fraction of known and studied enzymes. The alignment was calculated using T-Coffee in accurate mode (124). The tree was calculated with Molecular Evolutionary Genetics Analysis version 6 (MEGA6) (125) and is not rooted. A similarity and identity matrix for all included sequences, together with their accession numbers, is provided in the supplemental material (Table S1).
In general, it is believed that the microbial degradation of human-made polymers is a very slow process. This high resistance mainly stems from the high molecular weight of the fiber, the strong C-C bonds, and the extremely hydrophobic surface, which is very difficult to attack by enzymes. Notably, polymers are high-molecular-weight molecules, and they have amorphous and crystalline forms, which have different levels of degradability.
POLYMERS AND MICROBIAL DEGRADATION
Polyethylene terephthalate.
Polyethylene terephthalate (PET) is mainly used for production of PET bottles, PET foil, and fibers in the textile industry. PET is a polar, linear polymer of repeating units of the aromatic terephthalic acid and ethylene glycol. The PET monomer is designated bis(2-hydroxyethyl) terephthalate (BHET) (14). PET is a thermoplast and partly crystalline. The annual production of PET exceeded 30 million tons in 2017 (https://www.plasticsinsight.com/global-pet-resin-production-capacity).
Currently, only a few bacteria and fungi have been described for the partial degradation of PET to oligomers or monomers (8). All known PET hydrolases have relatively low turnover rates. Intriguingly, the trait for PET degradation appears to be limited to a few bacterial phyla, and most bacterial isolates with the potential for PET degradation are members of the Gram-positive phylum Actinobacteria (15). The best characterized examples originate from the genera Thermobifida and Thermomonospora (16–23). The enzymes involved in the degradation (e.g., PET hydrolase and tannase, MHETase) are typical serine hydrolases, e.g., cutinases (EC 3.1.1.74), lipases (EC 3.1.1.3), and carboxylesterases (EC 3.1.1.1). These enzymes possess a typical α/β-hydrolase fold, and the catalytic triad is composed of a serine, a histidine, and an aspartate residue (18, 24). They can also contain several disulfide bonds caused by cysteine residues, which promote thermal stability and specific binding to PET, as shown by the example of PETase from Ideonella sakaiensis 201-F6 (25).
Also, for the bacterium I. sakaiensis, usage of PET as a major energy and carbon source has been described (25). In addition to the PET hydrolase, the I. sakaiensis genome codes for a second enzyme that appears to be unique so far and which shares high similarity to the group of tannases, capable of degrading mono(2-hydroxyethyl) terephthalic acid. PET hydrolase as a secreted enzyme produces the intermediate mono(2-hydroxyethyl) terephthalic acid (MHET). MHET is internalized by the cell and hydrolyzed by MHETase. The resulting monomers are then used for bacterial metabolism. I. sakaiensis is affiliated with the phylum Betaproteobacteria and belongs to the order Burkholderiales.
The I. sakaiensis PETase three-dimensional (3D) structure was elucidated recently (26). The overall structure most resembles the structures of cutinases. Austin et al. showed that a double mutation (S238F/W159H), which narrows the active site of the enzyme and makes the protein even more like a cutinase resembling the enzyme from Thermobifida fusca, leads to an improved variant. The majority of the functionally verified PET hydrolases contain a C-terminal disulfide bond, promoting thermal and also kinetic stability (27–29). The only exception from this so far is a para-nitrobenzylesterase from Bacillus subtilis (30). An additional disulfide bond can be found in I. sakaiensis PETase, as well as in structural models of the functionally tested PET hydrolases described by Danso et al. (31). The structural data indicate that PETases bind the polymer with the hydrophobic surface and the substrate-binding cleft. In total, 4 MHET moieties are bound to the protein (one to subsite I and three to subsite II), whereby the ester bond to be cleaved is located between both subsites next to the catalytic serine. The MHETase from I. sakaiensis that further hydrolyzes MHET to ethylene glycol and terephthalic acid has been recently crystallized ligand free (2.05 Å) and with a nonhydrolyzable MHET analogue bound (2.1 Å). The enzyme possesses a lid domain that almost exclusively confers substrate specificity and activity toward MHET, with a kcat of 11.1 ± 1.4 s−1 (32).
While the I. sakaiensis enzymes are the best-studied models, other enzymes and organisms have been identified as potent PET degraders. Currently, four enzymes from Thermobifida species, one from Saccharomonospora, and one from the phylum Thermomonospora are known to act on PET. These actinobacterial enzymes are often Ca2+-dependent, especially in terms of their thermal stability (33), and they are partially inhibited by their released hydrolysis products MHET and BHET (33). Therefore, efforts have been made to overcome this limitation; one approach lies in the combination of polyester hydrolases with other enzymes to improve substrate binding and catalytic properties (26, 34, 35).
Besides the actinobacterial PET hydrolases, fungal cutinases showed activity on PET substrates as well. The most prominent examples are cutinases of the phyla Fusarium and Humicola. The latter was also used together with the lipase CalB from Candida antarctica in order to circumvent the previously mentioned product inhibition by BHET and MHET (34). While CalB completely converted to terephthalic acid, the Humicola-derived enzyme was limited in the last reaction step and accumulated the intermediate MHET.
Complementary to the above outlined activity-based approaches, a hidden Markov model (HMM) motif-based large-scale global search of existing genome and metagenome databases has been developed for the presence of potential PET hydrolases (31). Using this approach, >800 potential PET hydrolases were identified in bacterial and archaeal genomes and metagenomes, and several enzymes were functionally verified (e.g., PET2, PET4, PET6, and PET12). These findings imply that PET hydrolase-encoding genes are globally distributed in marine and terrestrial metagenomes (31).
Using an in silico genome mining approach, a cutinase from Pseudomonas pseudoalcaligenes (PpCutA) and a putative lipase from Pseudomonas pelagia (PpelaLip) were identified as potential enzymes acting on polyesters in general. Further experimental work using recombinant enzymes of PpCutA and PpelaLip verified the hydrolytic activities of both enzymes on different types of polyesters, including the hydrolysis of polyoxyethylene terephthalate (36). In their study, the authors used structurally different ionic phthalic acid-based polyesters with an average molecular weight ranging from 1,770 to 10,000 g/mol and semicrystalline polyesters with crystallinity below 1% to test and verify the microbial degradation. Notably, the identified organism belongs to a biotechnologically important novel species within the genus Pseudomonas, which was designated Pseudomonas pertucinogena (37).
In addition to the metagenome-derived PET esterases described above, colleagues recently reported on the functional screening of metagenomes and the characterization of selected enzymes. Among those were the metagenome-derived esterases MGS0156 and GEN0105, which hydrolyzed polylactic acid (PLA) and polycaprolactone, as well as bis(benzoyloxyethyl)-terephthalate. For MGS0156, 3D structural data at 1.95 Å indicate a modified α/β-hydrolase fold with a lid domain and a highly hydrophobic active site (38). The closest homologue to MGS0156 is an enzyme from Desulfovibrio fructosivorans with 70% sequence similarity.
In summary, PETases represent the best-explored and -studied class of enzymes with respect to the hydrolysis of synthetic polymers.
Polyurethanes.
Polyurethanes (PUR) can be synthesized by using different polyether or polyester polyols. PUR is a polymer of organic units connected by carbamate. The additional incorporation of aromatic ring structures has further impact on the physical and chemical properties of the polymer. PUR is a widely used synthetic polymer for the production of foams, insulation materials, textile coatings, and paint to prevent corrosion (39). With over 27 tons produced annually (2), it ranks fifth among the most often produced synthetic polymers.
To date, only bioactivities that act on the ester-based PUR have been reported (40, 41). Biodegradation was achieved by either bacteria or fungi. With respect to bacteria capable of degrading PUR, Gram-negative Betaproteobacteria from the genus Pseudomonas have been most frequently linked with PUR activities. One of the first enzymes identified to act on PUR was the PueB lipase from Pseudomonas chlororaphis (42, 43). This organism codes for at least one additional enzyme active on PUR, which was designated PueA (44). Both enzymes are lipases; PUR is degraded by the secreted hydrolases, and the degradation is tightly regulated. Their respective genes are part of a larger gene cluster encompassing seven open reading frames (ORFs) (45). Pseudomonas protegens strain Pf-5 uses a similar mechanism to degrade dispersions of the polyester PUR. In this strain, however, it was shown that PUR degradation is tightly regulated by mechanisms of carbon catabolite control and that both lipase genes, pueE and pueB, appear to be essential for growth on PUR dispersions (46). In a similar manner, Pseudomonas putida was reported to degrade PUR at relatively high rates (47). The bacterium needed 4 days to grow and consume the added colloidal PUR. Yet another example comes from Comamonas acidovorans TB-35. This strain produces a PUR-active enzyme that is an esterase and which was designated PudA (48, 49). PudA shows a hydrophobic PUR-surface-binding domain and a distinct catalytic domain, and its surface-binding domain is considered to be essential for PUR degradation. PudA acts as a 62-kDa monomer, and it releases diethylene glycol and adipic acid at an optimum temperature of 45°C and an optimum pH of 6.5.
Within this context, it is perhaps notable that often enzyme activities that are reported are based on clearing zones in agar plates. However, these assays are not fully reliable. For instance, different enzymes from Pseudomonas spp. and Bacillus spp. showed significant esterase activities and partially or even completely cleared plates containing colloidal PUR. However, only the Pseudomonas sp. lipase significantly degraded the added PUR based on nuclear magnetic resonance (NMR) and infrared (IR) data (50). Furthermore, there is strong evidence that some B. subtilis and Alicycliphilus sp. isolates are able to degrade PUR (51–53).
In a recent publication, Schmidt and colleagues reported on microbial degradation of PUR (i.e., Impranil DLN). The authors of this study employed the known polyester hydrolases LC-cutinase, TfCut2, Tcur1278, and Tcur0390 in their assays and observed significant weight loss of the tested foils when incubated for extended time periods (200 h) at a temperature of 70°C (54). The observation that cutinases, otherwise known to degrade polyethylene terephthalate, also act on PUR could be attributed to the promiscuous nature of the Thermobifida-derived cutinases. Recent research on promiscuity of enzymes implies that lipolytic enzymes such as cutinases are very often highly promiscuous and can convert up to 78 different substrates (55).
While the list of PUR-active bacteria is steadily increasing, a larger number of fungi have also been reported to degrade polyurethane (41). Notably, the authors of that study identified a 21-kDa metallo-hydrolase from Pestalotiopsis microspora as a responsible enzyme in PUR degradation.
Additional studies identified Fusarium solani, Candida ethanolica (56), and Candida rugosa (57) as PUR degraders. While for C. rugosa, a lipase has been identified as the key enzyme involved in PUR metabolism, no enzymes were yet identified for C. ethanolica and F. solani. Other fungi reported belong to the Cladosporium cladosporioides complex, including the species Cladosporium pseudocladosporioides, Cladosporium tenuissimum, Cladosporium asperulatum, and Cladosporium montecillanum, and three others were identified as Aspergillus fumigatus, Penicillium chrysogenum (58), and Aspergillus flavus (59). In the case of A. flavus, it is assumed that secreted esterases are responsible for the degradation. However, no defined enzyme has yet been linked to the observed activities. In a similar study, it was recently reported that Aspergillus tubingensis colonizes PUR and acts on the surface of films made of PUR. However, no enzyme was linked with the PUR activities (60).
It is noteworthy that the above-mentioned PUR-active enzymes and organisms were all acting on ester-linked PUR. However, to the best of our knowledge, no enzymes have yet been described acting on polyurethane ethers.
Polyethylene.
Polyethylene (PE) consists of long-chain polymers of ethylene, and it is produced as either high-density (HD-PE) or low-density (LD-PE) polyethylene. PE is chemically synthesized by polymerization of ethane and is highly variable, since side chains can be obtained depending on the manufacturing process. Such modifications mainly have influence on crystallinity and molecular weight. The polymer is most frequently used in the packaging industry as one of the main packaging materials, and more than 100 million tons of PE are produced globally per year (2, 61) (Fig. 2).
Possible PE degradation has been affiliated with a surprisingly large number of bacterial genera. Among those were Gram-negative species affiliated with the genera Pseudomonas, Ralstonia, and Stenotrophomonas but also many Gram-positive taxa (e.g., Rhodococcus, Staphylococcus, Streptomyces, Bacillus, and others) (see references in Sen and Raut [62] and Restrepo-Florez et al. [63]). In addition, fungal genera affiliated with assumed PE degradation were reported; these included Aspergillus, Cladosporium, Penicillium, and others (see references in references 62, 63, and 64–69). In addition, a few studies linked the PE-degrading microbes with the complex gut microbiomes of invertebrates (70, 71).
It is notable that in almost all the above-mentioned studies on PE-degrading microorganisms, the authors reported on degradation of the polymers using commercial polymers that possibly contained chemical additives, and degradation was determined by measuring weight loss and by Fourier transform infrared spectroscopy (FTIR). Since weight loss and surface structure changes are most likely attributed to the degradation of chemical additives, which often make up a significant fraction of the polymer, the results in these studies need to be verified using more advanced technologies. None of these studies reveled biochemical mechanisms and enzymes involved in PE breakdown. Within this framework, a more recent publication identified a Penicillium-derived laccase as potentially involved in PE breakdown (72). Unfortunately, no detailed biochemical characterization was performed, and no sequence of the protein or the corresponding gene was deposited.
Polyamide.
Polyamide (PA) is a polymer of repeating units of aliphatic, semiaromatic, or aromatic molecules linked via amide bonds. Since the monomers for making this polymer can be very versatile, there are many different types of synthetic polyamides, with the most popular being nylon and Kevlar. Synthetic polyamides are mainly used in textiles, automotive applications, carpets, and sportswear (73).
Remarkably, proteins as well as natural silk are polyamides per se. Based on this, it should be expected that nature has evolved enzymes that act on these nonnative polymers. However, to date, there is no microorganism known that is able to fully degrade the intact high-molecular-weight polymer. In contrast, several studies are available on bacteria acting on either linear or cyclic nylon oligomers with rather short chain lengths. In one of the first studies, different bacteria were described to grow on various oligomers derived from nylon production (74). In wastewater of nylon factories, 8-caprolactam, 6-aminohexanoic acid, 6-aminohexanoic acid cyclic dimer, and 6-aminohexanoic acid oligomers accumulate. These compounds can serve as the carbon and nitrogen source for specially adapted bacteria. One of the first bacteria described growing on these mixtures of oligomers was Flavobacterium sp. strain KI72, which was later renamed Achromobacter guttatus KI72 and then recently named Arthrobacter sp. strain KI72 (74, 75). Nylon oligomer-degrading Arthrobacter isolates code in their genomes for different hydrolases and several aminotransferases involved in the initial degradation of the oligomers and the subsequent metabolism. In the case of strain KI72, the respective genes are located on an accessory plasmid, pOAD2 (76–78).
Three main enzymes are essential for the initial hydrolysis of cyclic and linear 6-aminohexanoate oligomers. The first one is a cyclic-dimer hydrolase (NylA), the second a dimer hydrolase (NylB), and the third an endo-type oligomer hydrolase (NylC). NylC is a typical esterase, but its 3D structure also reveals motifs with β-lactamase folds (79–87). Once the oligomers are hydrolyzed, the monomers are metabolized by different aminotransferases. The draft genome of Arthrobacter sp. KI72 carries, among others, two genes, designated nylD1 and nylE1, that are responsible for the secondary 6-aminohexanoate metabolism. The 6-aminohexanoate aminotransferase (NylD1) catalyzes the reaction of 6-aminohexanoate to adipate semialdehyde. It uses α-ketoglutarate, pyruvate, and glyoxylate as amino acceptors and generates glutamate, alanine, and glycine, respectively. The reaction relies on pyridoxal phosphate as a cofactor. The second enzyme, the adipate semialdehyde dehydrogenase (NylE1), catalyzes the reaction, leading from adipate semialdehyde to adipate. This enzyme requires NADP+ as a cofactor and is an oxidoreductase (88, 89).
More recently, diverse marine bacteria were reported to act on nylon. The authors of this study reported a significant weight loss over a time period of 3 months. In their study, Bacillus cereus, Bacillus sphaericus, Vibrio furnissii, and Brevundimonas vesicularis were identified as potential nylon degraders (90). The genes and enzymes associated with the nylon degradation, however, remain to be identified, and the possibility cannot be excluded that the weight loss observed was primarily linked to the degradation of chemical additives, as outlined above.
Rather than using the synthetic polymer, Oppermann and colleagues reported on 12 bacterial species capable of degrading the natural polymer poly-γ-glutamic acid. The high-molecular-weight polymer is synthesized by many Gram-positive bacteria as a major component of capsules and slime. In contrast to the synthetic polymer, however, it is a water-soluble molecule and is thus more easily accessible to microbial degradation (91).
The only enzyme that has so far been reported to act on high-molecular-weight nylon fibers was classified as a manganese-dependent peroxidase and originated from a white rot fungus. The activity of the native and purified enzyme, however, differed from that of lignolytic enzymes. Nylon-degrading activity was quantified by measuring the structural disintegration of nylon-66 membranes. The enzyme had a molecular weight of 43 kDa and was dependent on the presence of lactate and other alpha-hydroxy acids. Unfortunately, no gene or protein sequence was determined (92).
While the first reports were published in 1965 stating that, among others, Pseudomonas aeruginosa is able to convert oligomeric nylon, further studies have confirmed that P. aeruginosa and evolved strain PAO1 are able to efficiently degrade 6-aminohexanoate linear dimers (74, 93). The main enzymatic activities were assigned to a 6-aminohexanoate cyclic-dimer hydrolase and a 6-aminohexanoate dimer hydrolase. Other Pseudomonas species have, however, also been reported to utilize 6-aminohexanoate-dimers as a sole carbon and nitrogen source (94).
Polystyrene.
Polystyrene (PS) [poly(1-phenylethene)] polymer consists of styrene monomers. PS is a widely used synthetic polymer for packaging industries but many daily use articles (CD cases, plastic cutlery, petri dishes, etc.) are also produced from this polymer (95). In 2016, about 14 million tons were produced (https://www.plasticsinsight.com/global-pet-resin-production-capacity).
Unfortunately, there is no enzyme known today that can degrade the high-molecular-weight polymer. However, a first report was published recently by Krueger and colleagues on the identification of brown rot fungi able to attack polystyrol by employing hydroquinone-driven Fenton reactions. In this preliminary study, Gloeophyllum striatum DSM 9592 and Gloeophyllum trabeum DSM 1398 caused substantial depolymerization after 20 days of incubation. The most active Gloeophyllum strains caused almost 50% reductions in molecular weight (96). In an earlier study, the white rot fungi Pleurotus ostreatus, Phanerochaete chrysosporium, and Trametes versicolor and the brown rot fungus Gloeophyllum trabeum were affiliated with the depolymerization of polystyrene when coincubation together with lignin was performed (97). While these are first and promising reports on the degradation of the high-molecular-weight polymer, the enzymes involved in the depolymerizing reaction remain to be elucidated. As already outlined above, weight loss may have been caused by the degradation of chemical additives.
Similarly, several bacteria have been reported to form either alone or as members of consortium biofilms on polystyrene films and particles, thereby degrading the polymer. In these studies, mainly weight loss has been assayed. Unfortunately, in none of these studies were enzymes linked to the assumed depolymerization (98, 99).
While not a single bacterium is known to degrade the polymer, a larger number of bacterial genera that are capable of metabolizing the monomer styrene as a sole source of carbon are known. The biochemistry of styrene metabolism is well understood, and for more detailed reviews, see references 98 and 100–103 and references therein. Styrene degradation in bacteria is well studied in Pseudomonas, Xanthobacter, Rhodococcus, Corynebacterium, and others. It appears to be a widespread metabolism. Under aerobic conditions, styrene is oxidized by two different pathways, namely, (i) attacking the vinyl side chain and (ii) a rather unspecific aromatic ring, thereby forming primarily the intermediates 3-vinylcatechol, phenylacetic acid, and 2-phenylethanol. These intermediates are channeled into the Krebs cycle after ring cleavage. The degradation of the vinyl side chain involves the action of three key enzymes, a styrene monooxygenase, a styrene oxide isomerase, and a phenylacetaldehyde dehydrogenase (104). The styrene monooxygenase attacks the vinyl side chain to release epoxystyrene, which is then subjected to isomerization to form phenylacetaldehyde. The latter is oxidized to phenylacetic acid though the involvement of a dehydrogenase. In P. putida, the phenylacetic acid is activated to phenylacetyl-coenzyme A (CoA) and then subjected to β-oxidation to yield acetyl-CoA, which is directly fed into the Krebs cycle. The respective genes for side-chain oxygenation are frequently located in a single conserved gene cluster, often designated styABC(D) (105). Thereby, the styA and styB genes code for the styrene monooxygenase complex. The styrene monooxygenase is a two-component flavoprotein that catalyzes the NADH- and FAD-dependent epoxidation of styrene to styrene oxide. StyA is the actual monooxygenase, and StyB functions as flavin adenine dinucleotide (FAD) reductase, which transfers the electrons from NADH to FAD+ to supply StyA with the required electrons (106). The styC gene codes for the styrene isomerase (107), and styD is a phenylacetaldehyde dehydrogenase gene (108). The expression of the conserved cluster is regulated through either a two-component regulatory system or LysR-type regulators (109–111).
The direct ring cleavage of styrene is initiated by a dihydroxylation of the aromatic ring. This reaction is catalyzed by a 2,3-dioxygenase and followed by a 2,3-dihydrodiol dehydrogenase. The two key products that are formed are styrene cis-glycol and 3-vinylcatechol. The latter can then be degraded by subsequent meta- or orthocleavage to form acrylic acid, acetaldehyde, and pyruvate. The pathway is rather unspecific for the general degradation of various aromatic compounds, such as phenol or toluene (100–102).
The produced phenylacetaldehydes are of interest to different industries, as they can be considered building blocks for the production of different fine chemicals or pharmaceutical compounds. They can serve as the starting material to synthesize fragrances, flavors, pharmaceuticals, insecticides, fungicides, or herbicides (112). Recent studies have also shown that Pseudomonas putida, Rhodococcus zopfii, and other Gram-negative species can convert polystyrene (i.e., styrene oil) into the biodegradable polymer polyhydroxyalkanoate or other valuable compounds. The approach involves as a first step the pyrolysis of polystyrene to styrene oil. The styrene oil is then converted in a second step to polyhydroxyalkanoate or other compounds. While the overall concept of this two-step process is intriguing, it may not be feasible on a large scale, as the pyrolysis is a process that runs at 520°C and this is energetically very demanding (113–115).
Polyvinylchloride and polypropylene.
Polyvinylchloride (PVC) and polypropylene (PP) are both important polymers produced at higher levels than the above-named polymers. PVC is the third most frequently produced polymer, and only PE and PP are produced at higher levels. PVC is composed of repeating chloroethyl units and PP of repeating units of propane-1,2-diyl units (116, 117). In sharp contrast to their huge global production rate, hardly any reliable information is available on microbial degradation of both of these important polymers. Only a very few reports that describe the degradation of the polymers based on weight loss and using mixed species microbial communities have been published (118, 119). However, it is likely that these reports were in part misled by the degradation of the chemical additives rather than the polymer. Consequently, no defined enzymes or pathways that are responsible for the degradation of either of these two high-molecular-weight polymers are known.
MICROBIOMES OF INVERTEBRATES AS POSSIBLE SOURCES OF PLASTIC-DEGRADING BACTERIA
Recently, it was reported that invertebrates can degrade different plastics (70, 71, 120–123). While these studies demonstrated that the insects perform a mechanical grinding and shredding of the plastics, it has been critically discussed if, and to which extent, the microbiomes associated with the different insects are capable of truly degrading the synthetic polymers. In one of those studies, Yang and colleagues provided convincing evidence that Tenebrio molitor L. (mealworms) digested Styrofoam. The larvae lived over a month when fed on the Styrofoam. Within a 16-day period, nearly 50% of the ingested Styrofoam carbon was converted into CO2, and the residual Styrofoam was found in the feces. Labeling studies using α-13C- or β-13C-labeled polystyrol implied that the carbon compound was preferentially used to build lipids (71). One of the earliest reports on insects digesting plastics came from caterpillars. In 2017, a Spanish team reported on the fast biodegradation of PE by larvae of the wax moth (Galleria mellonella). The authors of this study presented evidence that larvae of the wax moth produced holes in PE films with considerable speed (120). The findings of this study were critically discussed later on, as the occurrence of ethylene glycol as well as the correct usage of the FTIR method could not be immediately verified (121). Further work by a Chinese and United States-based research team identified Bacillus sp. strain YP1 as the polyethylene-degrading bacterium responsible for PE degradation in Indian mealworms (70, 122). A related study from the same group identified bacteria affiliated with the genera Citrobacter and Kosakonia as main degraders for PE and PS in the guts of Tenebrio molitor (123).
Thus, grinding of larger plastic pieces into smaller parts might offer a solution in that it increases the surface area and thereby allows microorganisms to better attach to the surfaces.
FUTURE CHALLENGES IN MICROBIAL PLASTIC DEGRADATION RESEARCH
The diversity of known enzymes and microbes acting on synthetic polymers is still rather limited. Therefore, future work has to address the identification of organisms acting on the most dominant polymers. The main bottleneck lies in the initial breakdown of high-molecular-weight and highly robust polymers and their crystalline structures. Furthermore, the implementation of enzymes in processes that would allow the degradation of plastic polluting environmental niches is a challenge for future generations of microbiologists. Since current cultivation technologies have not yet resulted in the identification of highly active enzymes for most plastics, the diversity of noncultivated microorganisms (i.e., global metagenomes) and the so-called dark matter proteins offer a promising source for the identification of such biocatalysts. Thus, the further development of smart search algorithms for mining metagenome data sets is certainly a rewarding task. In parallel, the setup of reliable function-based assays for the detection of high-molecular-weight-polymer-active enzymes is important as well.
Since commercially available polymers and films thereof are often used as substrates, they contain additives, plasticizers, and other biodegradable impurities (for example, phthalates), which are much more easily broken down than the actual backbone. This therefore interferes with the results and frequently leads to the identification of false positives. Thus, the overall methodology linked to the analysis of microbial plastic degradation needs to be standardized and optimized.
Similarly, the development of cellulosome-like structures (i.e., “plastosomes”) in microbes to attack intact and crystalline fibers would certainly be a worthwhile project. Along these lines, the simple development of highly active enzymes for textile industries could already significantly reduce annual plastic pollution and would perhaps be one of the more realistic short-term goals.
Furthermore, using synthetic biology to generate microorganisms that would produce high-value compounds from plastic waste is a future challenge and would contribute to an improved circular use of plastics. Monomers and oligomers formed after the degradation could be used to build value-added products or even new (biodegradable) polymers.
Lastly, obtaining plastic-active enzymes and implementing them in the production of true biopolymers is a highly rewarding research task and would significantly reduce our global plastic problem.
Supplementary Material
ACKNOWLEDGMENTS
This work was in part supported by the BMBF within the program MarBiotech (FKZ 031A565) and by the EU Horizon 2020 project INMARE and MetaGenLig (FKZ 031B0571B) at the University of Hamburg.
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01095-19.
REFERENCES
- 1.Geyer R, Jambeck JR, Law KL. 2017. Production, use, and fate of all plastics ever made. Sci Adv 3:e1700782. doi: 10.1126/sciadv.1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.PlasticsEurope. 2018. PlasticsEurope, plastics—the facts 2018: an analysis of European plastics production, demand and waste data. PlasticsEurope, Brussels, Belgium. [Google Scholar]
- 3.Ellen MacArthur Foundation. 2017. The new plastics economy: rethinking the future of plastics and catalysing action. Ellen MacArthur Foundation, Cowes, United Kingdom. [Google Scholar]
- 4.Jambeck JR, Geyer R, Wilcox C, Siegler TR, Perryman M, Andrady A, Narayan R, Law KL. 2015. Marine pollution. Plastic waste inputs from land into the ocean. Science 347:768–771. doi: 10.1126/science.1260352. [DOI] [PubMed] [Google Scholar]
- 5.Derraik J. 2002. The pollution of the marine environment by plastic debris: a review. Mar Pollut Bull 44:842–852. doi: 10.1016/S0025-326X(02)00220-5. [DOI] [PubMed] [Google Scholar]
- 6.Cózar A, Echevarría F, González-Gordillo JI, Irigoien X, Úbeda B, Hernández-León S, Palma ÁT, Navarro S, García-de-Lomas J, Ruiz A, Fernández-de-Puelles ML, Duarte CM. 2014. Plastic debris in the open ocean. Proc Natl Acad Sci U S A 111:10239–10244. doi: 10.1073/pnas.1314705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lebreton L, Slat B, Ferrari F, Sainte-Rose B, Aitken J, Marthouse R, Hajbane S, Cunsolo S, Schwarz A, Levivier A, Noble K, Debeljak P, Maral H, Schoeneich-Argent R, Brambini R, Reisser J. 2018. Evidence that the Great Pacific Garbage Patch is rapidly accumulating plastic. Sci Rep 8:4666. doi: 10.1038/s41598-018-22939-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei R, Zimmermann W. 2017. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: how far are we? Microb Biotechnol 10:1308. doi: 10.1111/1751-7915.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Day M, Wiles DM. 1972. Photochemical degradation of poly(ethylene terephthalate). II. Effect of wavelength and environment on the decomposition process. J Appl Polym Sci 16:191–202. doi: 10.1002/app.1972.070160117. [DOI] [Google Scholar]
- 10.Mohammadian M, Allen NS, Edge M, Jones K. 1991. Environmental degradation of poly (ethylene terephthalate). Textile Res J 61:690–696. doi: 10.1177/004051759106101109. [DOI] [Google Scholar]
- 11.Welzel K, Müller RJ, Deckwer WD. 2002. Enzymatischer Abbau von Polyester-Nanopartikeln. Chemie Ingenieur Technik 74:1496–1500. doi:. [DOI] [Google Scholar]
- 12.Smith M, Love DC, Rochman CM, Neff RA. 2018. Microplastics in seafood and the implications for human health. Curr Environ Health Rep 5:375–386. doi: 10.1007/s40572-018-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Souza Machado AA, Kloas W, Zarfl C, Hempel S, Rillig MC. 2018. Microplastics as an emerging threat to terrestrial ecosystems. Glob Chang Biol 24:1405–1416. doi: 10.1111/gcb.14020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gubbels E, Heitz T, Yamamoto M, Chilekar V, Zarbakhsh S, Gepraegs M, Köpnick H, Schmidt M, Brügging W, Rüter J, Kaminsky W. 2018. Polyesters In Ullmann’s encyclopedia of industrial chemistry. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 15.Acero EH, Ribitsch D, Steinkellner G, Gruber K, Greimel K, Eiteljoerg I, Trotscha E, Wei R, Zimmermann W, Zinn M, Cavaco-Paulo A, Freddi G, Schwab H, Guebitz G. 2011. Enzymatic surface hydrolysis of PET: effect of structural diversity on kinetic properties of cutinases from Thermobifida. Macromolecules 44:4632–4640. doi: 10.1021/ma200949p. [DOI] [Google Scholar]
- 16.Kleeberg I, Hetz C, Kroppenstedt RM, Muller RJ, Deckwer WD. 1998. Biodegradation of aliphatic-aromatic copolyesters by Thermomonospora fusca and other thermophilic compost isolates. Appl Environ Microbiol 64:1731–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu X, Thumarat U, Zhang X, Tang M, Kawai F. 2010. Diversity of polyester-degrading bacteria in compost and molecular analysis of a thermoactive esterase from Thermobifida alba AHK119. Appl Microbiol Biotechnol 87:771–779. doi: 10.1007/s00253-010-2555-x. [DOI] [PubMed] [Google Scholar]
- 18.Wei R, Oeser T, Then J, Kuhn N, Barth M, Schmidt J, Zimmermann W. 2014. Functional characterization and structural modeling of synthetic polyester-degrading hydrolases from Thermomonospora curvata. AMB Express 4:44. doi: 10.1186/s13568-014-0044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei R, Oeser T, Zimmermann W. 2014. Synthetic polyester-hydrolyzing enzymes from thermophilic actinomycetes. Adv Appl Microbiol 89:267–305. doi: 10.1016/B978-0-12-800259-9.00007-X. [DOI] [PubMed] [Google Scholar]
- 20.Chen S, Tong X, Woodard RW, Du GC, Wu J, Chen J. 2008. Identification and characterization of bacterial cutinase. J Biol Chem 283:25854–25862. doi: 10.1074/jbc.M800848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimmermann W, Billig S. 2011. Enzymes for the biofunctionalization of poly(ethylene terephthalate). Adv Biochem Eng Biotechnol 125:97–120. doi: 10.1007/10_2010_87. [DOI] [PubMed] [Google Scholar]
- 22.Ribitsch D, Acero EH, Greimel K, Dellacher A, Zitzenbacher S, Marold A, Rodriguez RD, Steinkellner G, Gruber K, Schwab H, Guebitz GM. 2012. A new esterase from Thermobifida halotolerans hydrolyses polyethylene terephthalate (PET) and polylactic acid (PLA). Polymers 4:617–629. doi: 10.3390/polym4010617. [DOI] [Google Scholar]
- 23.Kawai F, Oda M, Tamashiro T, Waku T, Tanaka N, Yamamoto M, Mizushima H, Miyakawa T, Tanokura M. 2014. A novel Ca2+-activated, thermostabilized polyesterase capable of hydrolyzing polyethylene terephthalate from Saccharomonospora viridis AHK190. Appl Microbiol Biotechnol 98:10053–10064. doi: 10.1007/s00253-014-5860-y. [DOI] [PubMed] [Google Scholar]
- 24.Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J. 1992. The alpha/beta hydrolase fold. Protein Eng 5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida S, Hiraga K, Takehana T, Taniguchi I, Yamaji H, Maeda Y, Toyohara K, Miyamoto K, Kimura Y, Oda K. 2016. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351:1196–1199. doi: 10.1126/science.aad6359. [DOI] [PubMed] [Google Scholar]
- 26.Austin HP, Allen MD, Donohoe BS, Rorrer NA, Kearns FL, Silveira RL, Pollard BC, Dominick G, Duman R, El Omari K, Mykhaylyk V, Wagner A, Michener WE, Amore A, Skaf MS, Crowley MF, Thorne AW, Johnson CW, Woodcock HL, McGeehan JE, Beckham GT. 2018. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc Natl Acad Sci U S A 115:E4350–E4357. doi: 10.1073/pnas.1718804115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roth C, Wei R, Oeser T, Then J, Follner C, Zimmermann W, Strater N. 2014. Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca. Appl Microbiol Biotechnol 98:7815–7823. doi: 10.1007/s00253-014-5672-0. [DOI] [PubMed] [Google Scholar]
- 28.Sulaiman S, You DJ, Kanaya E, Koga Y, Kanaya S. 2014. Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase. Biochemistry 53:1858–1869. doi: 10.1021/bi401561p. [DOI] [PubMed] [Google Scholar]
- 29.Then J, Wei R, Oeser T, Gerdts A, Schmidt J, Barth M, Zimmermann W. 2016. A disulfide bridge in the calcium binding site of a polyester hydrolase increases its thermal stability and activity against polyethylene terephthalate. FEBS Open Bio 6:425–432. doi: 10.1002/2211-5463.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ribitsch D, Heumann S, Trotscha E, Acero EH, Greimel K, Leber R, Birner-Gruenberger R, Deller S, Eiteljoerg I, Remler P, Weber T, Siegert P, Maurer KH, Donelli I, Freddi G, Schwab H, Guebitz GM. 2011. Hydrolysis of polyethyleneterephthalate by p-nitrobenzylesterase from Bacillus subtilis. Biotechnol Progress 27:951–960. doi: 10.1002/btpr.610. [DOI] [PubMed] [Google Scholar]
- 31.Danso D, Schmeisser C, Chow J, Zimmermann W, Wei R, Leggewie C, Li X, Hazen T, Streit WR. 2018. New insights into the function and global distribution of polyethylene terephthalate (PET)-degrading bacteria and enzymes in marine and terrestrial metagenomes. Appl Environ Microbiol 84:e02773-17. doi: 10.1128/AEM.02773-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palm GJ, Reisky L, Böttcher D, Müller H, Michels EAP, Walczak MC, Berndt L, Weiss MS, Bornscheuer UT, Weber G. 2019. Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate. Nat Commun 10:1717. doi: 10.1038/s41467-019-09326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barth M, Oeser T, Wei R, Then J, Schmidt J, Zimmermann W. 2015. Effect of hydrolysis products on the enzymatic degradation of polyethylene terephthalate nanoparticles by a polyester hydrolase from Thermobifida fusca. Biochem Eng J 93:222–228. doi: 10.1016/j.bej.2014.10.012. [DOI] [Google Scholar]
- 34.Carniel A, Valoni E, Nicomedes J, Gomes AD, de Castro AM. 2017. Lipase from Candida antarctica (CALB) and cutinase from Humicola insolens act synergistically for PET hydrolysis to terephthalic acid. Process Biochem 59:84–90. doi: 10.1016/j.procbio.2016.07.023. [DOI] [Google Scholar]
- 35.Wei R, Oeser T, Schmidt J, Meier R, Barth M, Then J, Zimmermann W. 2016. Engineered bacterial polyester hydrolases efficiently degrade polyethylene terephthalate due to relieved product inhibition. Biotechnol Bioeng 113:1658–1665. doi: 10.1002/bit.25941. [DOI] [PubMed] [Google Scholar]
- 36.Haernvall K, Zitzenbacher S, Wallig K, Yamamoto M, Schick MB, Ribitsch D, Guebitz GM. 2017. Hydrolysis of ionic phthalic acid based polyesters by wastewater microorganisms and their enzymes. Environ Sci Technol 51:4596–4605. doi: 10.1021/acs.est.7b00062. [DOI] [PubMed] [Google Scholar]
- 37.Bollinger A, Thies S, Katzke N, Jaeger KE. 25 June 2018. The biotechnological potential of marine bacteria in the novel lineage of Pseudomonas pertucinogena. Microb Biotechnol doi: 10.1111/1751-7915.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hajighasemi M, Tchigvintsev A, Nocek BP, Flick R, Popovic A, Hai T, Khusnutdinova AN, Brown G, Xu X, Cui H, Anstett J, Chernikova TN, Bruls T, Le Paslier D, Yakimov MM, Joachimiak A, Golyshina OV, Savchenko A, Golyshin PN, Edwards EA, Yakunin AF. 2018. Screening and characterization of novel polyesterases from environmental metagenomes with high hydrolytic activity against synthetic polyesters. Environ Sci Technol 52:12388–12401. doi: 10.1021/acs.est.8b04252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seymour RB, Kauffman GB. 1992. Polyurethanes: a class of modern versatile materials. J Chem Educ 69:909. doi: 10.1021/ed069p909. [DOI] [Google Scholar]
- 40.Darby RT, Kaplan AM. 1968. Fungal susceptibility of polyurethanes. Appl Microbiol 16:900–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell JR, Huang J, Anand P, Kucera K, Sandoval AG, Dantzler KW, Hickman D, Jee J, Kimovec FM, Koppstein D, Marks DH, Mittermiller PA, Núñez SJ, Santiago M, Townes MA, Vishnevetsky M, Williams NE, Vargas MPN, Boulanger L-A, Bascom-Slack C, Strobel SA. 2011. Biodegradation of polyester polyurethane by endophytic fungi. Appl Environ Microbiol 77:6076–6084. doi: 10.1128/AEM.00521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howard GT, Crother B, Vicknair J. 2001. Cloning, nucleotide sequencing and characterization of a polyurethanase gene (pueB) from Pseudomonas chlororaphis. Int Biodeterior Biodegrad 47:141–149. doi: 10.1016/S0964-8305(01)00042-7. [DOI] [Google Scholar]
- 43.Howard GT, Blake RC. 1998. Growth of Pseudomonas fluorescens on a polyester–polyurethane and the purification and characterization of a polyurethanase–protease enzyme. Int Biodeterior Biodegrad 42:213–220. doi: 10.1016/S0964-8305(98)00051-1. [DOI] [Google Scholar]
- 44.Stern RV, Howard GT. 2000. The polyester polyurethanase gene (pueA) from Pseudomonas chlororaphis encodes a lipase. FEMS Microbiol Lett 185:163–168. doi: 10.1111/j.1574-6968.2000.tb09056.x. [DOI] [PubMed] [Google Scholar]
- 45.Howard GT, Mackie RI, Cann IK, Ohene-Adjei S, Aboudehen KS, Duos BG, Childers GW. 2007. Effect of insertional mutations in the pueA and pueB genes encoding two polyurethanases in Pseudomonas chlororaphis contained within a gene cluster. J Appl Microbiol 103:2074–2083. doi: 10.1111/j.1365-2672.2007.03447.x. [DOI] [PubMed] [Google Scholar]
- 46.Hung CS, Zingarelli S, Nadeau LJ, Biffinger JC, Drake CA, Crouch AL, Barlow DE, Russell JN Jr, Crookes-Goodson WJ. 2016. Carbon catabolite repression and impranil polyurethane degradation in Pseudomonas protegens strain Pf-5. Appl Environ Microbiol 82:6080–6090. doi: 10.1128/AEM.01448-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng YH, Shih YH, Lai YC, Liu YZ, Liu YT, Lin NC. 2014. Degradation of polyurethane by bacterium isolated from soil and assessment of polyurethanolytic activity of a Pseudomonas putida strain. Environ Sci Pollut Res Int 21:9529–9537. doi: 10.1007/s11356-014-2647-8. [DOI] [PubMed] [Google Scholar]
- 48.Akutsu Y, Nakajima-Kambe T, Nomura N, Nakahara T. 1998. Purification and properties of a polyester polyurethane-degrading enzyme from Comamonas acidovorans TB-35. Appl Environ Microbiol 64:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shigeno-Akutsu Y, Nakajima-Kambe T, Nomura N, Nakahara T. 1999. Purification and properties of culture-broth-secreted esterase from the polyurethane degrader Comamonas acidovorans TB-35. J Biosci Bioeng 88:484–487. doi: 10.1016/S1389-1723(00)87663-X. [DOI] [PubMed] [Google Scholar]
- 50.Biffinger JC, Barlow DE, Cockrell AL, Cusick KD, Hervey WJ, Fitzgerald LA, Nadeau LJ, Hung CS, Crookes-Goodson WJ, Russell JN. 2015. The applicability of Impranil® DLN for gauging the biodegradation of polyurethanes. Polym Degradation Stab 120:178–185. doi: 10.1016/j.polymdegradstab.2015.06.020. [DOI] [Google Scholar]
- 51.Shah Z, Krumholz L, Aktas DF, Hasan F, Khattak M, Shah AA. 2013. Degradation of polyester polyurethane by a newly isolated soil bacterium, Bacillus subtilis strain MZA-75. Biodegradation 24:865–877. doi: 10.1007/s10532-013-9634-5. [DOI] [PubMed] [Google Scholar]
- 52.Rowe L, Howard GT. 2002. Growth of Bacillus subtilis on polyurethane and the purification and characterization of a polyurethanase-lipase enzyme. Int Biodeterior Biodegrad 50:33–40. doi: 10.1016/S0964-8305(02)00047-1. [DOI] [Google Scholar]
- 53.Oceguera-Cervantes A, Carrillo-García A, López N, Bolaños-Nuñez S, Cruz-Gómez MJ, Wacher C, Loza-Tavera H. 2007. Characterization of the polyurethanolytic activity of two Alicycliphilus sp. strains able to degrade polyurethane and n-methylpyrrolidone. Appl Environ Microbiol 73:6214–6223. doi: 10.1128/AEM.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt J, Wei R, Oeser T, Dedavid e Silva L, Breite D, Schulze A, Zimmermann W. 2017. Degradation of polyester polyurethane by bacterial polyester hydrolases. Polymers 9:65. doi: 10.3390/polym9020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez-Martinez M, Coscolin C, Santiago G, Chow J, Stogios PJ, Bargiela R, Gertler C, Navarro-Fernandez J, Bollinger A, Thies S, Mendez-Garcia C, Popovic A, Brown G, Chernikova TN, Garcia-Moyano A, Bjerga GEK, Perez-Garcia P, Hai T, Del Pozo MV, Stokke R, Steen IH, Cui H, Xu X, Nocek BP, Alcaide M, Distaso M, Mesa V, Pelaez AI, Sanchez J, Buchholz PCF, Pleiss J, Fernandez-Guerra A, Glockner FO, Golyshina OV, Yakimov MM, Savchenko A, Jaeger KE, Yakunin AF, Streit WR, Golyshin PN, Guallar V, Ferrer M, The Inmare Consortium. 2018. Determinants and prediction of esterase substrate promiscuity patterns. ACS Chem Biol 13:225–234. doi: 10.1021/acschembio.7b00996. [DOI] [PubMed] [Google Scholar]
- 56.Zafar U, Houlden A, Robson GD. 2013. Fungal communities associated with the biodegradation of polyester polyurethane buried under compost at different temperatures. Appl Environ Microbiol 79:7313–7324. doi: 10.1128/AEM.02536-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gautam R, Bassi AS, Yanful EK. 2007. Candida rugosa lipase-catalyzed polyurethane degradation in aqueous medium. Biotechnol Lett 29:1081–1086. doi: 10.1007/s10529-007-9354-1. [DOI] [PubMed] [Google Scholar]
- 58.Álvarez-Barragán J, Domínguez-Malfavón L, Vargas-Suárez M, González-Hernández R, Aguilar-Osorio G, Loza-Tavera H. 2016. Biodegradative activities of selected environmental fungi on a polyester polyurethane varnish and polyether polyurethane foams. Appl Environ Microbiol 82:5225–5235. doi: 10.1128/AEM.01344-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mathur G, Prasad R. 2012. Degradation of polyurethane by Aspergillus flavus (ITCC 6051) isolated from soil. Appl Biochem Biotechnol 167:1595–1602. doi: 10.1007/s12010-012-9572-4. [DOI] [PubMed] [Google Scholar]
- 60.Khan S, Nadir S, Shah ZU, Shah AA, Karunarathna SC, Xu J, Khan A, Munir S, Hasan F. 2017. Biodegradation of polyester polyurethane by Aspergillus tubingensis. Environ Pollut 225:469–480. doi: 10.1016/j.envpol.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 61.Nowlin T. 2014. Global polyethylene business overview In Nowlin TE. (ed), Business and technology of the global polyethylene industry. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 62.Sen SK, Raut S. 2015. Microbial degradation of low density polyethylene (LDPE): a review. J Environ Chem Eng 3:462–473. doi: 10.1016/j.jece.2015.01.003. [DOI] [Google Scholar]
- 63.Restrepo-Florez JM, Bassi A, Thompson MR. 2014. Microbial degradation and deterioration of polyethylene—a review. Int Biodeterior Biodegrad 88:83–90. doi: 10.1016/j.ibiod.2013.12.014. [DOI] [Google Scholar]
- 64.Pathak VM, Navneet. 2017. Review on the current status of polymer degradation: a microbial approach. Bioresource Bioprocess 4:15. doi: 10.1186/s40643-017-0145-9. [DOI] [Google Scholar]
- 65.Ojha N, Pradhan N, Singh S, Barla A, Shrivastava A, Khatua P, Rai V, Bose S. 2017. Evaluation of HDPE and LDPE degradation by fungus, implemented by statistical optimization. Sci Rep 7:39515. doi: 10.1038/srep39515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamada-Onodera K, Mukumoto H, Katsuyaya Y, Saiganji A, Tani Y. 2001. Degradation of polyethylene by a fungus, Penicillium simplicissimum YK. Polym Degradation Stab 72:323–327. doi: 10.1016/S0141-3910(01)00027-1. [DOI] [Google Scholar]
- 67.Bonhomme S, Cuer A, Delort A, Lemaire J, Sancelme M, Scott G. 2003. Environmental biodegradation of polyethylene. Polym Degradation Stab 81:441–452. doi: 10.1016/S0141-3910(03)00129-0. [DOI] [Google Scholar]
- 68.Veethahavya KS, Rajath BS, Noobia S, Kumar BM. 2016. Biodegradation of low density polyethylene in aqueous media. Procedia Environ Sci 35:709–713. doi: 10.1016/j.proenv.2016.07.072. [DOI] [Google Scholar]
- 69.Vimala PP, Mathew L. 2016. Biodegradation of polyethylene using Bacillus subtilis. Procedia Technol 24:232–239. doi: 10.1016/j.protcy.2016.05.031. [DOI] [Google Scholar]
- 70.Yang J, Yang Y, Wu W-M, Zhao J, Jiang L. 2014. Evidence of polyethylene biodegradation by bacterial strains from the guts of plastic-eating waxworms. Environ Sci Technol 48:13776–13784. doi: 10.1021/es504038a. [DOI] [PubMed] [Google Scholar]
- 71.Yang Y, Yang J, Wu W-M, Zhao J, Song Y, Gao L, Yang R, Jiang L. 2015. Biodegradation and mineralization of polystyrene by plastic-eating mealworms: Part 1. Chemical and physical characterization and isotopic tests. Environ Sci Technol 49:12080–12086. doi: 10.1021/acs.est.5b02661. [DOI] [PubMed] [Google Scholar]
- 72.Sowmya HV, Ramalingappa B, Krishnappa M, Thippeswamy B. 2015. Degradation of polyethylene by Penicillium simplicissimum isolated from local dumpsite of Shivamogga district. Environ Dev Sustain 17:731–745. doi: 10.1007/s10668-014-9571-4. [DOI] [Google Scholar]
- 73.Palmer R. 2001. Polyamides, plastics In Encyclopedia of polymer science and technology. Wiley, Hoboken, NJ. [Google Scholar]
- 74.Tosa T, Chibata I. 1965. Utilization of cyclic amides and formation of omega-amino acids by microorganisms. J Bacteriol 89:919–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takehara I, Kato DI, Takeo M, Negoro S. 2017. Draft genome sequence of the nylon oligomer-degrading bacterium Arthrobacter sp. strain KI72. Genome Announc 5:e00217-17. doi: 10.1128/genomeA.00217-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Negoro S, Taniguchi T, Kanaoka M, Kimura H, Okada H. 1983. Plasmid-determined enzymatic degradation of nylon oligomers. J Bacteriol 155:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Negoro S, Kakudo S, Urabe I, Okada H. 1992. A new nylon oligomer degradation gene (nylC) on plasmid pOAD2 from a Flavobacterium sp. J Bacteriol 174:7948–7953. doi: 10.1128/jb.174.24.7948-7953.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kakudo S, Negoro S, Urabe I, Okada H. 1993. Nylon oligomer degradation gene, nylC, on plasmid pOAD2 from a Flavobacterium strain encodes endo-type 6-aminohexanoate oligomer hydrolase: purification and characterization of the nylC gene product. Appl Environ Microbiol 59:3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Negoro S, Ohki T, Shibata N, Sasa K, Hayashi H, Nakano H, Yasuhira K, Kato D-i, Takeo M, Higuchi Y. 2007. Nylon-oligomer degrading enzyme/substrate complex: catalytic mechanism of 6-aminohexanoate-dimer hydrolase. J Mol Biol 370:142–156. doi: 10.1016/j.jmb.2007.04.043. [DOI] [PubMed] [Google Scholar]
- 80.Yasuhira K, Uedo Y, Shibata N, Negoro S, Takeo M, Higuchi Y. 2006. Crystallization and X-ray diffraction analysis of 6-aminohexanoate-cyclic-dimer hydrolase from Arthrobacter sp. KI72. Acta Crystallogr Sect F Struct Biol Cryst Commun 62:1209–1211. doi: 10.1107/S1744309106045076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ohki T, Mizuno N, Shibata N, Takeo M, Negoro S, Higuchi Y. 2005. Crystallization and X-ray diffraction analysis of 6-aminohexanoate-dimer hydrolase from Arthrobacter sp. KI72. Acta Crystallogr Sect F Struct Biol Cryst Commun 61:928–930. doi: 10.1107/S1744309105028812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nagai K, Yasuhira K, Tanaka Y, Kato D, Takeo M, Higuchi Y, Negoro S, Shibata N. 2013. Crystallization and X-ray diffraction analysis of nylon hydrolase (NylC) from Arthrobacter sp. KI72. Acta Crystallogr Sect F Struct Biol Cryst Commun 69:1151–1154. doi: 10.1107/S1744309113024263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kinoshita S, Terada T, Taniguchi T, Takene Y, Masuda S, Matsunaga N, Okada H. 1981. Purification and characterization of 6-aminohexanoic-acid-oligomer hydrolase of Flavobacterium sp. Ki72. Eur J Biochem 116:547–551. doi: 10.1111/j.1432-1033.1981.tb05371.x. [DOI] [PubMed] [Google Scholar]
- 84.Yasuhira K, Tanaka Y, Shibata H, Kawashima Y, Ohara A, Kato D, Takeo M, Negoro S. 2007. 6-Aminohexanoate oligomer hydrolases from the alkalophilic bacteria Agromyces sp. strain KY5R and Kocuria sp. strain KY2. Appl Environ Microbiol 73:7099–7102. doi: 10.1128/AEM.00777-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Negoro S, Ohki T, Shibata N, Mizuno N, Wakitani Y, Tsurukame J, Matsumoto K, Kawamoto I, Takeo M, Higuchi Y. 2005. X-ray crystallographic analysis of 6-aminohexanoate-dimer hydrolase: molecular basis for the birth of a nylon oligomer-degrading enzyme. J Biol Chem 280:39644–39652. doi: 10.1074/jbc.M505946200. [DOI] [PubMed] [Google Scholar]
- 86.Kinoshita S, Negoro S, Muramatsu M, Bisaria VS, Sawada S, Okada H. 1977. 6-Aminohexanoic acid cyclic dimer hydrolase. A new cyclic amide hydrolase produced by Achromobacter guttatus KI74. Eur J Biochem 80:489–495. doi: 10.1111/j.1432-1033.1977.tb11904.x. [DOI] [PubMed] [Google Scholar]
- 87.Kinoshita S, Kageyama S, Iba K, Yamada Y, Okada H. 1975. Utilization of a cyclic dimer and linear oligomers of ε-aminocaproic acid by Achromobacter guttatus KI 72. Agric Biol Chem 39:1219–1223. doi: 10.1271/bbb1961.39.1219. [DOI] [Google Scholar]
- 88.Takehara I, Fujii T, Tanimoto Y, Kato DI, Takeo M, Negoro S. 2018. Correction to: Metabolic pathway of 6-aminohexanoate in the nylon oligomer-degrading bacterium Arthrobacter sp. KI72: identification of the enzymes responsible for the conversion of 6-aminohexanoate to adipate. Appl Microbiol Biotechnol 102:815. doi: 10.1007/s00253-017-8682-x. [DOI] [PubMed] [Google Scholar]
- 89.Takehara I, Fujii T, Tanimoto Y, Kato DI, Takeo M, Negoro S. 2018. Metabolic pathway of 6-aminohexanoate in the nylon oligomer-degrading bacterium Arthrobacter sp. KI72: identification of the enzymes responsible for the conversion of 6-aminohexanoate to adipate. Appl Microbiol Biotechnol 102:801–814. doi: 10.1007/s00253-017-8657-y. [DOI] [PubMed] [Google Scholar]
- 90.Sudhakar M, Priyadarshini C, Doble M, Sriyutha Murthy P, Venkatesan R. 2007. Marine bacteria mediated degradation of nylon 66 and 6. Int Biodeterior Biodegrad 60:144–151. doi: 10.1016/j.ibiod.2007.02.002. [DOI] [Google Scholar]
- 91.Oppermann FB, Pickartz S, Steinbüchel A. 1998. Biodegradation of polyamides. Polym Degradation Stab 59:337–344. doi: 10.1016/S0141-3910(97)00175-4. [DOI] [Google Scholar]
- 92.Deguchi T, Kitaoka Y, Kakezawa M, Nishida T. 1998. Purification and characterization of a nylon-degrading enzyme. Appl Environ Microbiol 64:1366–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Prijambada ID, Negoro S, Yomo T, Urabe I. 1995. Emergence of nylon oligomer degradation enzymes in Pseudomonas aeruginosa PAO through experimental evolution. Appl Environ Microbiol 61:2020–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kanagawa K, Oishi M, Negoro S, Urabe I, Okada H. 1993. Characterization of the 6-aminohexanoate-dimer hydrolase from Pseudomonas sp. NK87. J Gen Microbiol 139:787–795. doi: 10.1099/00221287-139-4-787. [DOI] [PubMed] [Google Scholar]
- 95.Maul J, Frushour BG, Kontoff JR, Eichenauer H, Ott K-H, Schade C. 2007. Polystyrene and styrene copolymers In Ullmann’s encyclopedia of industrial chemistry. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 96.Krueger MC, Hofmann U, Moeder M, Schlosser D. 2015. Potential of wood-rotting fungi to attack polystyrene sulfonate and its depolymerisation by Gloeophyllum trabeum via hydroquinone-driven fenton chemistry. PLoS One 10:e0131773. doi: 10.1371/journal.pone.0131773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Milstein O, Gersonde R, Huttermann A, Chen MJ, Meister JJ. 1992. Fungal biodegradation of lignopolystyrene graft copolymers. Appl Environ Microbiol 58:3225–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ho BT, Roberts TK, Lucas S. 2018. An overview on biodegradation of polystyrene and modified polystyrene: the microbial approach. Crit Rev Biotechnol 38:308–320. doi: 10.1080/07388551.2017.1355293. [DOI] [PubMed] [Google Scholar]
- 99.Chauhan D, Agrawal G, Deshmukh S, Roy SS, Priyadarshini R. 2018. Biofilm formation by Exiguobacterium sp. DR11 and DR14 alter polystyrene surface properties and initiate biodegradation. RSC Adv 8:37590–37599. doi: 10.1039/C8RA06448B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mooney A, Ward PG, O’Connor KE. 2006. Microbial degradation of styrene: biochemistry, molecular genetics, and perspectives for biotechnological applications. Appl Microbiol Biotechnol 72:1. doi: 10.1007/s00253-006-0443-1. [DOI] [PubMed] [Google Scholar]
- 101.Dobson ADW, O’Leary ND, O'Connor KE. 2002. Biochemistry, genetics and physiology of microbial styrene degradation. FEMS Microbiol Rev 26:403–417. doi: 10.1111/j.1574-6976.2002.tb00622.x. [DOI] [PubMed] [Google Scholar]
- 102.Tischler D. 2015. Microbial styrene degradation, p 7–22. Springer International Publishing, Cham, Switzerland. doi: 10.1007/978-3-319-24862-2_2. [DOI] [Google Scholar]
- 103.Oelschlägel M, Zimmerling J, Tischler D. 2018. A review: the styrene metabolizing cascade of side-chain oxygenation as biotechnological basis to gain various valuable compounds. Front Microbiol 9:490. doi: 10.3389/fmicb.2018.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tischler D, Eulberg D, Lakner S, Kaschabek SR, van Berkel WJH, Schlomann M. 2009. Identification of a novel self-sufficient styrene monooxygenase from Rhodococcus opacus 1CP. J Bacteriol 191:4996–5009. doi: 10.1128/JB.00307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Velasco A, Alonso S, García JL, Perera J, Díaz E. 1998. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J Bacteriol 180:1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Morrison E, Kantz A, Gassner GT, Sazinsky MH. 2013. Structure and mechanism of styrene monooxygenase reductase: new insight into the FAD-transfer reaction. Biochemistry 52:6063–6075. doi: 10.1021/bi400763h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oelschlägel M, Gröning JAD, Tischler D, Kaschabek SR, Schlömann M. 2012. Styrene oxide isomerase of Rhodococcus opacus 1CP, a highly stable and considerably active enzyme. Appl Environ Microbiol 78:4330–4337. doi: 10.1128/AEM.07641-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Crabo AG, Singh B, Nguyen T, Emami S, Gassner GT, Sazinsky MH. 2017. Structure and biochemistry of phenylacetaldehyde dehydrogenase from the Pseudomonas putida S12 styrene catabolic pathway. Arch Biochem Biophys 616:47–58. doi: 10.1016/j.abb.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.O'Leary ND, O'Mahony MM, Dobson AD. 2011. Regulation of phenylacetic acid uptake is sigma54 dependent in Pseudomonas putida CA-3. BMC Microbiol 11:229. doi: 10.1186/1471-2180-11-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.O’Leary ND, Duetz WA, Dobson AD, O’Connor KE. 2002. Induction and repression of the sty operon in Pseudomonas putida CA-3 during growth on phenylacetic acid under organic and inorganic nutrient-limiting continuous culture conditions. FEMS Microbiol Lett 208:263–268. doi: 10.1016/S0378-1097(02)00473-1. [DOI] [PubMed] [Google Scholar]
- 111.O’Leary ND, Mooney A, O'Mahony M, Dobson AD. 2014. Functional characterization of a StyS sensor kinase reveals distinct domains associated with intracellular and extracellular sensing of styrene in P. putida CA-3. Bioengineered 5:114–122. doi: 10.4161/bioe.28354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sheldon RA, Van Bekkum H. 2008. Fine chemicals through heterogeneous catalysis. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- 113.O’Leary ND, O’Connor KE, Ward P, Goff M, Dobson AD. 2005. Genetic characterization of accumulation of polyhydroxyalkanoate from styrene in Pseudomonas putida CA-3. Appl Environ Microbiol 71:4380–4387. doi: 10.1128/AEM.71.8.4380-4387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ward PG, Goff M, Donner M, Kaminsky W, O’Connor KE. 2006. A two step chemo-biotechnological conversion of polystyrene to a biodegradable thermoplastic. Environ Sci Technol 40:2433–2437. doi: 10.1021/es0517668. [DOI] [PubMed] [Google Scholar]
- 115.Savoldelli J, Tomback D, Savoldelli H. 2017. Breaking down polystyrene through the application of a two-step thermal degradation and bacterial method to produce usable byproducts. Waste Manage 60:123–126. doi: 10.1016/j.wasman.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 116.Fischer I, Schmitt WF, Porth H, Allsopp MW, Vianello G. 2014. Poly(vinyl chloride). In Ullmann’s encyclopedia of industrial chemistry. Wiley‐VCH, Weinheim, Germany. [Google Scholar]
- 117.Karger-Kocsis J, Bárány T. 2019. Polypropylene handbook. Springer Nature Switzerland, Basel, Switzerland. [Google Scholar]
- 118.Cacciari I, Quatrini P, Zirletta G, Mincione E, Vinciguerra V, Lupattelli P, Giovannozzi Sermanni G. 1993. Isotactic polypropylene biodegradation by a microbial community: physicochemical characterization of metabolites produced. Appl Environ Microbiol 59:3695–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Iakovlev VV, Guelcher SA, Bendavid R. 2017. Degradation of polypropylene in vivo: a microscopic analysis of meshes explanted from patients. J Biomed Mater Res B Appl Biomater 105:237–248. doi: 10.1002/jbm.b.33502. [DOI] [PubMed] [Google Scholar]
- 120.Bombelli P, Howe CJ, Bertocchini F. 2017. Polyethylene bio-degradation by caterpillars of the wax moth Galleria mellonella. Curr Biol 27:R292–R293. doi: 10.1016/j.cub.2017.02.060. [DOI] [PubMed] [Google Scholar]
- 121.Weber C, Pusch S, Opatz T. 2017. Polyethylene bio-degradation by caterpillars? Curr Biol 27:R744–R745. doi: 10.1016/j.cub.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 122.Yang Y, Chen J, Wu W-M, Zhao J, Yang J. 2015. Complete genome sequence of Bacillus sp. YP1, a polyethylene-degrading bacterium from waxworm’s gut. J Biotechnol 200:77–78. doi: 10.1016/j.jbiotec.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 123.Brandon AM, Gao S-H, Tian R, Ning D, Yang S-S, Zhou J, Wu W-M, Criddle CS. 2018. Biodegradation of polyethylene and plastic mixtures in mealworms (larvae of Tenebrio molitor) and effects on the gut microbiome. Environ Sci Technol 52:6526–6533. doi: 10.1021/acs.est.8b02301. [DOI] [PubMed] [Google Scholar]
- 124.Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 125.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.