Polyhydroxybutyrate (PHB) granules are a store of carbon and energy in bacteria and archaea and play an important role in stress adaptation. Recent studies have highlighted distinct roles of several granule-associated proteins (GAPs) in regulating the size, number, and localization of PHB granules in free-living bacteria, though our knowledge of the role of GAPs in bacteria associated with plants is still limited. Here we report distinct roles of core and accessory phasins associated with PHB granules of Sinorhizobium fredii NGR234, a broad-host-range microsymbiont of diverse legumes. Core phasins PhaP2 and PhaP1 are conserved major phasins in free-living cells. PhaP2 and accessory phasin PhaP3, encoded by an auxiliary gene on the symbiosis plasmid, are major phasins in nitrogen-fixing bacteroids in cowpea nodules. GAPs and metabolic profiles can vary in different phaP mutants. Contrasting symbiotic performances between mutants lacking PHB synthases, depolymerase, or phasins were revealed.
KEYWORDS: PHB, legume, phasin, rhizobium, symbiosis
ABSTRACT
The exact roles of various granule-associated proteins (GAPs) of polyhydroxybutyrate (PHB) are poorly investigated, particularly for bacteria associated with plants. In this study, four structural GAPs, named phasins PhaP1 to PhaP4, were identified and demonstrated as true phasins colocalized with PHB granules in Sinorhizobium fredii NGR234, a facultative microsymbiont of Vigna unguiculata and many other legumes. The conserved PhaP2 dominated in regulation of granule size under both free-living and symbiotic conditions. PhaP1, another conserved phasin, made a higher contribution than accessory phasins PhaP4 and PhaP3 to PHB biosynthesis at stationary phase. PhaP3, with limited phyletic distribution on the symbiosis plasmid of Sinorhizobium, was more important than PhaP1 in regulating PHB biosynthesis in V. unguiculata nodules. Under the test conditions, no significant symbiotic defects were observed for mutants lacking individual or multiple phaP genes. The mutant lacking two PHB synthases showed impaired symbiotic performance, while mutations in individual PHB synthases or a PHB depolymerase yielded no symbiotic defects. This phenomenon is not related to either the number or size of PHB granules in test mutants within nodules. Distinct metabolic profiles and cocktail pools of GAPs of different phaP mutants imply that core and accessory phasins can be differentially involved in regulating other cellular processes in the facultative microsymbiont S. fredii NGR234.
IMPORTANCE Polyhydroxybutyrate (PHB) granules are a store of carbon and energy in bacteria and archaea and play an important role in stress adaptation. Recent studies have highlighted distinct roles of several granule-associated proteins (GAPs) in regulating the size, number, and localization of PHB granules in free-living bacteria, though our knowledge of the role of GAPs in bacteria associated with plants is still limited. Here we report distinct roles of core and accessory phasins associated with PHB granules of Sinorhizobium fredii NGR234, a broad-host-range microsymbiont of diverse legumes. Core phasins PhaP2 and PhaP1 are conserved major phasins in free-living cells. PhaP2 and accessory phasin PhaP3, encoded by an auxiliary gene on the symbiosis plasmid, are major phasins in nitrogen-fixing bacteroids in cowpea nodules. GAPs and metabolic profiles can vary in different phaP mutants. Contrasting symbiotic performances between mutants lacking PHB synthases, depolymerase, or phasins were revealed.
INTRODUCTION
The metabolic ability to synthesize polyhydroxyalkanoates (PHAs), which are polyoxoesters of (R)-hydroxyalkanoic acid monomers, has been described for both bacteria and archaea (1). The most common PHA is poly(3-hydroxybutyrate) (PHB). High-molecular-weight PHB consisting of >103 3-hydroxybutyrate residues is present in the form of inclusion bodies (PHB granules) in prokaryote cells (2). The biosynthesis of PHB starts with the condensation of two molecules of acetyl coenzyme A (acetyl-CoA) by PhbA (3-ketothiolase) to give acetoacetyl-CoA, which is then reduced by PhbB (acetoacetyl-CoA reductase) to form 3-hydroxybutyryl-CoA. This compound is further polymerized to PHB by PhbC (PHB synthase). PHB degradation is initiated by PhaZ (PHB depolymerase) to release the 3-hydroxybutyrate monomer. PHB granules are accumulated under nutrient-limiting conditions (nitrogen, oxygen, phosphorus, etc.) but with an excess of carbon sources (1) and are considered a store of intracellular carbon and energy. Therefore, the PHB synthesis is tightly connected with the tricarboxylic acid (TCA) cycle, though the molecular mechanisms of acetyl-CoA partition between two processes remain elusive.
Beyond the storage function, the ability to accumulate PHB can enhance resistance of various bacteria, such as Aeromonas hydrophila, Azospirillum brasilense, Escherichia coli, Methylobacterium extorquens, and Ralstonia eutropha, to various stresses such as UV radiation, osmotic shock, hydroxyl radicals, and high or low temperatures (3–7). These protective roles of PHB have been considered a general phenomenon regardless of individual species or the type of stressing agent, but the underlying mechanisms are not well understood (8). The protective role of PHB is partially due to its hydrolysis product 3-hydroxybutyrate, which can act as a compatible solute protecting the activity of enzymes under osmotic and thermal stresses (9, 10). It was reported that methyl-esterified dimers and trimers of 3-hydroxybutyrate, produced by diverse bacteria during PHB degradation, have greater hydroxyl radical-scavenging activity than the monomer 3-hydroxybutyrate (6). Recent studies also show that a structural protein, PhaP, associated with PHB granules from Azotobacter sp. strain FA-8 has chaperone-like functions and stress-protecting effects in E. coli (11, 12). The phaP genes encode a group of low-molecular-weight proteins known as phasins, which are the major proteins surrounding the hydrophobic core of PHB granules. Other documented granule-associated proteins (GAPs) include PhbC, PhaZ, the transcriptional regulator of PHB synthesis PhaR, and some uncharacterized putative GAPs (13), while the suspected phospholipids in the surface layer are not found in vivo (14). The term carbonosome has been proposed for the complex organized subcellular structures of PHB granules surrounded by structural, biosynthetic, catabolic, and regulatory proteins (15). The importance of various GAPs has been highlighted by the distinct roles of several PhaPs and another GAP, PhaM, in the model strain R. eutropha H16. Among seven documented PhaPs, PhaP1Reu is the most abundant phasin in R. eutropha H16 and regulates the amount of PHB, as well as size and number of granules, while PhaM, which interacts with both PhbC and the nucleoid, is responsible for maximal activity of PHB synthase and subcellular localization of granules (16–18). These pioneering results suggest that the pool of GAPs and their putative roles during bacterial adaptations to fluctuating environmental conditions remain largely unexplored.
Rhizobia can form nitrogen-fixing nodules on legumes and play an important role in sustainable agriculture and the global nitrogen cycle. To reduce one molecule of N2, 16 ATPs are required in the oxygen-sensitive nitrogen fixation process (19). Within nodule cells, rhizobia differentiate into nongrowing and nitrogen-fixing bacteroids that receive carbon and all the essential nutrients from the legume host (20). Low levels of free oxygen may limit the TCA cycle of bacteroids, possibly leading to the biased partition of acetyl-CoA to the biosynthesis of PHB (21). However, the accumulation level of PHB in bacteroids varies between different rhizobium-legume systems, such as with PHB granules present in Glycine max (soybean) nodules but not in Medicago sativa (alfalfa) nodules (22, 23). This contrasting feature could be partially due to variations in microenvironments of nodules from two legumes. This view is supported by the fact that bacteroids within nodules of soybean and alfalfa, respectively, are phosphate limited and nonlimited (24, 25). However, PHB granules became visible in bacteroids of the phaZ mutant of Sinorhizobium meliloti Rm1021 in alfalfa nodules (26), indicating simultaneously active biosynthesis and depolymerization of PHB in S. meliloti bacteroids in alfalfa. The symbiotic performance of the phbC mutant of various rhizobia associated with corresponding legumes can be either defective (S. meliloti and Azorhizobium caulinodans) or enhanced (Rhizobium etli) (27–29). Our knowledge on GAPs of PHB in rhizobia is restricted to phasins of S. meliloti Rm1021 and Bradyrhizobium diazoefficiens USDA110 (27, 30, 31). Among the PhaPs differentially expressed in various culture media, PhaP4Bdi is the most abundant phasin in USDA110, and a phaP1Bdi phaP4Bdi double mutant formed a single, large cytoplasmic PHB granule (30, 31). PhaP1Sme and PhaP2Sme of Rm1021 may equally contribute to PHB production and symbiotic adaptation (27). Our previous transcriptomic analyses of Sinorhizobium fredii NGR234, characterized by its broad host range, revealed that three annotated phasin genes, phaP1, phaP2, and phaP3, exhibited distinct expression patterns within nodules of Vigna unguiculata and Leucaena leucocephala compared to those in free-living cultures (32), implying potential differentiated roles of these phasins under free-living and symbiotic conditions.
In this study, we aimed to characterize PhaP1, PhaP2, PhaP3, and other putative phasins of S. fredii NGR234 within nodules of V. unguiculata and under the nutrient-limiting conditions of the stationary phase. To this end, subcellular localizations of PhaPs and other representative GAPs, identified from proteomic analysis of purified PHB granules, were determined in wild-type NGR234 and phbC mutants. The number and size of PHB granules, and PHB amounts from individual and combined mutants of phaPs at stationary phase and within V. unguiculata nodules, were compared. Contrasting roles of different PhaPs in regulating granule size and number and PHB amounts under free-living and symbiotic conditions were revealed. These results were further discussed with the evidence of distinct metabolic profiles of phaP mutants, the pools of GAPs associated with representative mutants of phaP, and the contrasting phyletic distribution patterns of PhaPs in the genus Sinorhizobium.
RESULTS
Characterization of synthase and phasins of PHB in NGR234 at stationary phase.
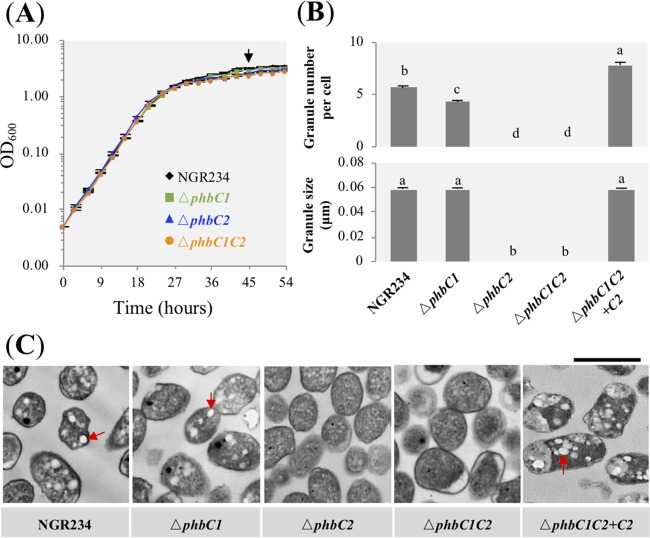
Within the genome of S. fredii NGR234, there are two genes, phbC1 (NGR_c34290) and phbC2 (NGR_c14000), encoding PHB synthase. In our exploratory experiments, a phbC1 phbC2 (here referred to as phbC1C2) insertion mutant was constructed, and it had no PHB granules under free-living conditions. Then the ΔphbC1, ΔphbC2, and ΔphbC1 ΔphbC2 (here referred to as ΔphbC1C2) in-frame deletion mutants were constructed; these mutants had a growth rate similar to that of wild-type NGR234 in TY medium (see Materials and Methods) (Fig. 1A). The number of PHB granules at stationary phase was slightly reduced in the ΔphbC1 mutant compared to that in the wild-type strain (Duncan’s test, alpha = 0.05), while no granule could be observed in the ΔphbC2 and ΔphbC1C2 mutants (Fig. 1B and C). A genetic complementation experiment showed that the wild-type phbC2 gene can restore the defects of the ΔphbC1C2 mutant in forming PHB granules (Fig. 1B and C). In line with these observations from transmission electronic microscopy (TEM) pictures, PHB quantification experiments showed that PhbC2 was the major PHB synthase, while no significant difference in PHB content was observed among the ΔphbC1 mutant, the ΔphbC1C2 mutant complemented with phbC2 (here referred to as ΔphbC1C2+C2), and NGR234 (see Table S1 in the supplemental material).
FIG 1.
Characteristics of PHB granules in free-living NGR234 and mutants lacking PHB synthase. (A) Growth curves of test strains. Bacterial cells were collected at stationary phase, as indicated by an arrow. (B) Number and size of PHB granules. A total of 150 cells were scored for each strain. Values represent means ± SEMs. Different letters indicate significant difference (alpha = 0.05, Duncan’s test). (C) Transmission electronic microscopy pictures. Red arrows indicate PHB granules. Bar, 1 μm.
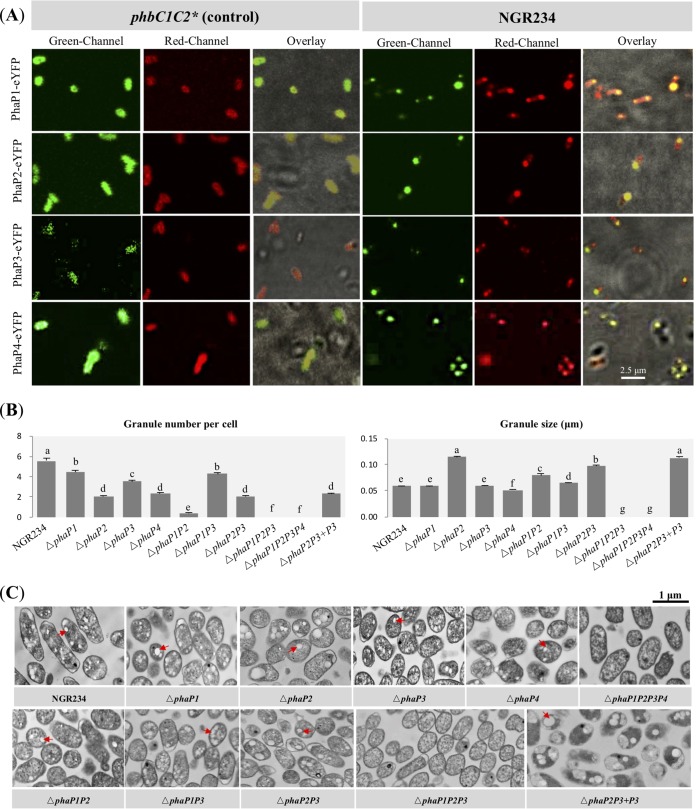
Three phasins were annotated in NGR234, i.e., PhaP1 (NGR_c03360), PhaP2 (NGR_c13240), and PhaP3 (NGR_a00900). Furthermore, the electrospray ionization mass spectrometry (ESI-MS) analyses described below led to the identification of a fourth putative phasin, PhaP4 (NGR_b11320). To test whether these putative phasins could be associated with PHB granules, PhaP1-enhanced yellow fluorescent protein (eYFP), PhaP2-eYFP, PhaP3-eYFP, and PhaP4-eYFP driven by a constitutively transcribed promoter were individually introduced into wild-type NGR234, the phbC1C2 mutant (for eYFP fusions of PhaP1, PhaP2, and PhaP3), or the ΔphbC1C2 mutant (for PhaP4-eYFP). The uniform fluorescence of the phbC1C2 or ΔphbC1C2 cells indicated soluble localization of free eYFP fused with PhaP1, PhaP2, PhaP3, or PhaP4 (Fig. 2A). In contrast, these phasin-eYFP fusions were colocalized with PHB granules, stained with Nile red in wild-type NGR234 (Fig. 2A).
FIG 2.
Characteristics of PHB granules in free-living NGR234 and mutant strains lacking genes encoding phasins. (A) Subcellular colocalization of predicted phasins with PHB granules. PhaP1-eYFP, PhaP2-eYFP, PhaP3-eYFP, and PhaP4-eYFP fusions were constitutively expressed in free-living NGR234 and its phbC1C2 mutant (*, ΔphbC1C2 used for expressing PhaP4-eYFP). The phbC1C2 (or ΔphbC1C2) mutant served as a control with no PHB accumulation. Fluorescence microscopic images were generated after staining with Nile red in the red channel (indicating the localization of PHB granules) or without staining in the green channel (showing the localization of phasins). (B) Number of PHB granules within each bacterial cell and size of PHB granules. A total of 150 cells were scored for each strain (58 cells were scored for the ΔphaP1P2 mutant; these values were obtained under the same conditions as for Fig. 1, and the same values for NGR234 were used in this case). Values represent means ± SEMs. Different letters indicate significant difference (alpha = 0.05, Duncan’s test). (C) Transmission electronic microscopy pictures of bacterial cells at stationary phase. Red arrows indicate PHB granules. Bar, 1 μm.
ΔphaP1, ΔphaP2, ΔphaP3, and ΔphaP4 in-frame deletion mutants and ΔphaP1P2, ΔphaP1P3, ΔphaP2P3, ΔphaP1P4, ΔphaP2P4, ΔphaP3P4, ΔphaP1P2P3, and ΔphaP1P2P3P4 mutants were constructed. At stationary phase, the relative contributions to PHB synthesis decreased in the order from PhaP2, PhaP1, PhaP4, to PhaP3 according to PHB content detected in these 12 mutants (Table S1). The dominant role of PhaP2 at stationary phase was further supported by the fact that the largest PHB granule, around two times larger than that of the wild-type NGR234, was found in the ΔphaP2 mutant (Duncan’s test, alpha = 0.05). Among the double mutants, the ΔphaP1P2 mutant synthesized the smallest amount of PHB (0.060 μg in 1 ml of bacterial culture with an optical density at 600 nm [OD600] of 1.0), 8.6% to 19.4% of the PHB content in the other five double mutants (Table S1). Although a few PHB granules can be observed in TEM pictures of the ΔphaP1P2 mutant, the amount of PHB in the ΔphaP1P2 mutant was not significantly different from that of the ΔphaP1P2P3 or ΔphaP1P2P3P4 mutant, for which no PHB granule was found in test TEM pictures (Fig. 2B and C and Table S1). Therefore, PhaP3 and PhaP4 had relatively minor roles in regulating the biosynthesis of PHB granules at stationary phase. The difference in the numbers of PHB granules among test mutants except the ΔphaP1P2P3 and ΔphaP1P2P3P4 mutants can be more than 10 times, with the average granule number below 0.40 in the ΔphaP1P2 mutant and 4.46 granules in the ΔphaP1 mutant (Fig. 2B). This was supported by PHB quantification results (Table S1). Notably, these numbers of granules and PHB amounts in test phasin mutants were lower than for NGR234 in different degrees (Fig. 2B and Table S1), suggesting cumulative contributions by the four phasins.
Role of phasins and PHB synthase in the symbiosis with V. unguiculata.
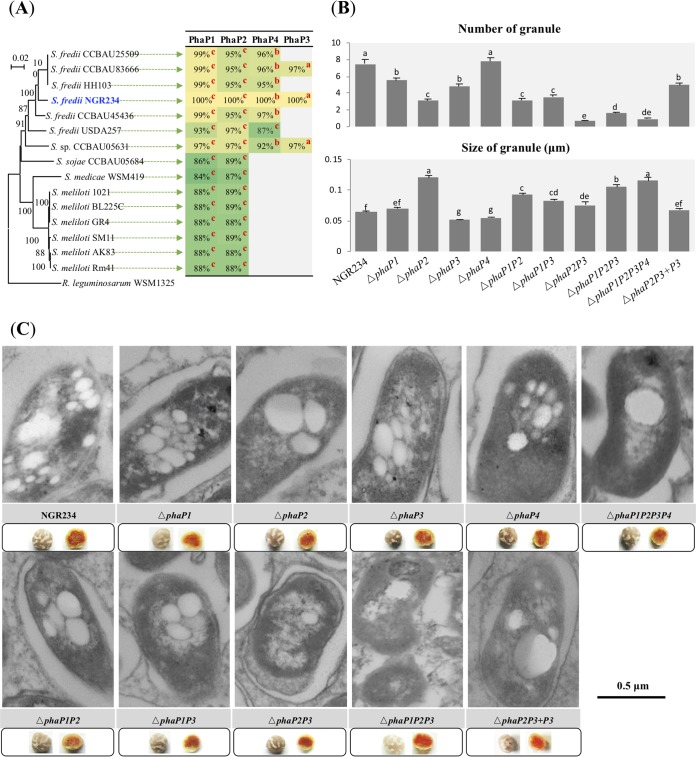
Sequence analysis demonstrated that PhaP1 and PhaP2 are highly conserved phasins encoded by core genes on chromosomes of Sinorhizobium, while PhaP4 and PhaP3 are restricted to a subset of species/strains, i.e., are encoded by accessory genes (Fig. 3A). The phaP4 homologs were found in chromids of Sinorhizobium sp. strain CCBAU05631 and all analyzed S. fredii strains except USDA257. A phaP4 homolog in USDA257 is located in a chromosomal region of 2 Mb displaying similarity to chromids of other S. fredii strains (33, 34). The phaP3 homologs were present in the symbiosis plasmid of Sinorhizobium sp. CCBAU05631 and S. fredii CCBAU83666 and NGR234. Mutants lacking individual or multiple phasin-coding genes were inoculated on V. unguiculata (Fig. 3B); little variation in leaf chlorophyll content and shoot dry weight was found among plants inoculated with these strains under the test conditions (Table S2). All of these phasin deletion mutants formed pink nodules (Fig. 3C). Similar to the case with free-living cells, PhaP2 played a dominant role in regulating PHB granule size in nodules (Fig. 3B and C). In contrast to its minor role in regulating PHB synthesis in stationary-phase cultures, PhaP3 seemed to have a greater role in PHB biosynthesis than PhaP1 in nodules, as the PHB content of the ΔphaP3 mutant was only half of that detected in the ΔphaP1 mutant or NGR234 (Table S1). Moreover, ΔphaP2P3 and ΔphaP3P4 bacteroids harbored 32.6% and 42.6% of the PHB content detected in the ΔphaP1P2 and ΔphaP1P4 mutants, respectively (Table S1). This was also supported by the number of PHB granules observed in representative TEM pictures of bacteroids (Fig. 3B and C). This is in line with the strong upregulation of the phaP3 gene in V. unguiculata nodules (32). A genetic complementation experiment showed that the wild-type phaP3 gene can significantly increase the granule number and PHB content in the ΔphaP2P3 mutant in nodules but not in stationary-phase culture (Fig. 2B and 3B and Table S1). Therefore, phaP3 located in symbiosis plasmid plays a greater role in regulating the number of PHB granules under symbiotic condition than in free-living culture. Although individual mutants of phaP1 and phaP4 had little defects in the number or size of PHB granules in bacteroids (Fig. 3B and C), PHB quantification results for various double mutants, as well as for the ΔphaP1P2P3 and ΔphaP1P2P3P4 mutants, suggest that PhaP1 and PhaP4 make a cumulative contribution to PHB biosynthesis in bacteroids (Table S1).
FIG 3.
Characteristics of PHB granules in bacteroids of NGR234 and mutant strains lacking genes encoding phasins. (A) Neighbor-joining phylogenetic tree of the rpoB gene and phyletic distribution of PhaP1 to PhaP4 in Sinorhizobium. Identity values of homologs of PhaPs (query coverage above 60%) in corresponding genomes are shown. Superscript c, b, and a represent genome locations of homologous phaP genes on the chromosome, chromid, and symbiosis plasmid, respectively. The background colors were scaled from the minimum similarity value of 84% (green) to 100% (yellow). (B) Number of PHB granules within each bacterial cell and size of PHB granules. Seventy-five bacterial cells (mutants) or 50 bacterial cells (wild-type NGR234) were scored. Values represent means ± SEMs. Significant difference between means is indicated by different letters based on Duncan’s test (alpha = 0.05). (C) Pictures of ultrathin sections of nodules obtained under transmission electronic microscopy. Pictures of more bacterial cells are shown in Fig. S1.
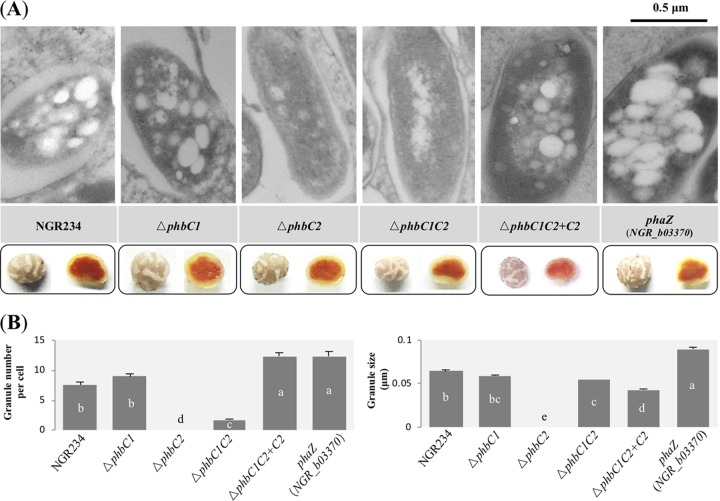
In bacteroids from V. unguiculata nodules induced by mutants lacking PHB synthase, PHB granules were barely found for the ΔphbC2 and ΔphbC1C2 mutants (Fig. 4A), and bacteroids of these two mutants harbored less than 17% of the PHB content detected in wild-type NGR234 (Table S1). A significant increase of granule number was observed for the NGR_b03370 mutant lacking a putative PHB depolymerase (PhaZ) compared to that for wild-type NGR234 (12.32 ± 0.80 versus 7.52 ± 0.50 per cell, average ± standard error of the mean [SEM]; Duncan’s test, alpha = 0.05 [Fig. 4B]). The average size of PHB granules was slightly but significantly larger in this phaZ (NGR_b03370) mutant than that of wild-type NGR234 (0.08 ± 0.002 versus 0.064 ± 0.002, average ± SEM; Duncan’s test, alpha = 0.05). All of these mutants formed pink nodules (Fig. 4A), and the inoculated plants were indistinguishable from each other regarding the chlorophyll content of leaf (Table S2), though a significant decrease of shoot dry weight was observed for the plants inoculated with the ΔphbC1C2 mutant (Table S2; Kruskal-Wallis test followed by Mann-Whitney test with Bonferroni correction). This symbiotic defect and PHB biosynthesis of the ΔphbC1C2 mutant can be restored by complementation using the wild-type phbC2 gene (Table S2 and Fig. 4). Moreover, the ΔphbC1C2 mutant occupied only 25.2% of V. unguiculata nodules in the presence of an equal amount of wild-type NGR234 cells (P < 0.05 [Fig. S2]).
FIG 4.
Characteristics of PHB granules in bacteroids of NGR234 and mutant strains lacking genes encoding PHB synthases or PHB depolymerase. (A) Pictures of ultrathin sections of nodules obtained under transmission electronic microscopy. Pictures of intact nodules and nodule halves are shown. Pictures of more bacterial cells are shown in Fig. S1. (B) Number of PHB granules within each bacterial cell and average size of PHB granules. Seventy-five bacterial cells (mutants) or 50 bacterial cells (wild-type NGR234) were scored. Values represent means ±SEMs. Significant difference between means is indicated by different letters based on Duncan’s test (alpha = 0.05).
Cocktail proteins associated with PHB granules in NGR234.
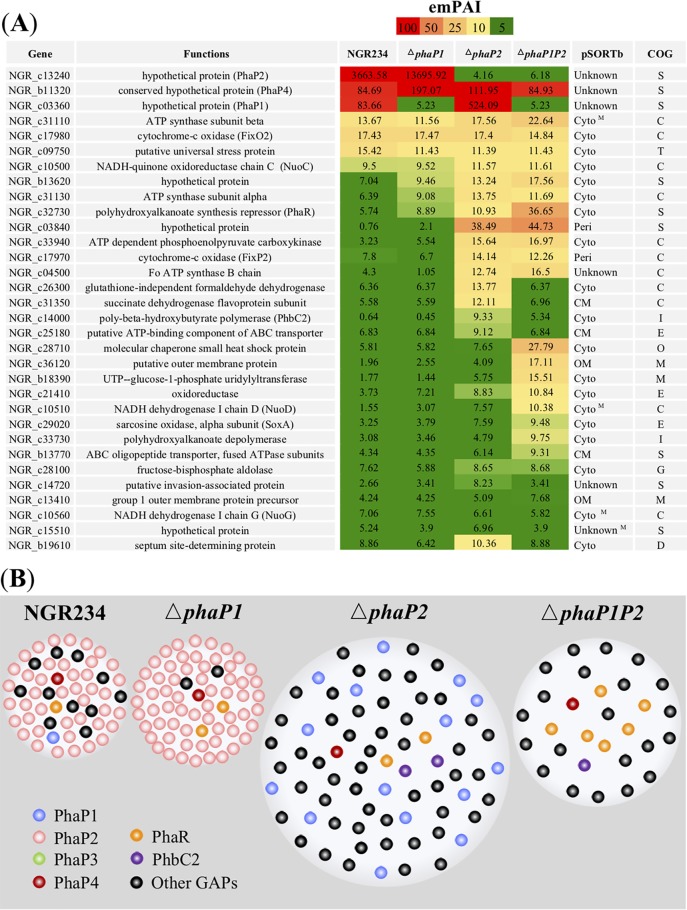
Among four phasins, the in-frame deletion of phaP2 caused the most dramatic change in the size of PHB granules (Fig. 2B). It has been reported on the basis of our previous transcriptome sequencing (RNA-seq) analysis (32) that phaP2 was constitutively transcribed under both free-living and symbiotic conditions, while phaP1 and phaP3 were down- and upregulated, respectively, in V. unguiculata nodules. The phaP4 gene was also upregulated in nodules, though to a lesser extent than phaP3 (32). In this study, we further investigated whether the change of granule size in the ΔphaP2 mutant would lead to a shift of the abundance and/or diversity of proteins associated with PHB granules. The ΔphaP1 and ΔphaP1P2 mutants were also included for comparison. PHB granules and associated proteins (Fig. S3A) were enriched as described in Materials and Methods for stationary-phase cultures. The enriched PHB-associated proteins compared to crude proteins of bacteria at stationary phase could be observed in SDS-PAGE for NGR234 (Fig. S3B). There was also notable variation in electrophoresis patterns for PHB granule-associated proteins (GAPs) from the ΔphaP1, ΔphaP2, and ΔphaP1P2 mutants (Fig. S3C). Bands corresponding to PhaP1, PhaP2, and PhaP4 were identified by MS analysis (Fig. S3B and S3C), confirming the reliability of test mutants. The nanospray ESI-MS analysis was used to identify proteins associated with PHB granules (Fig. 5A; a complete list of significant hits of P < 0.05 for each strain in three independent experiments is shown in Data Set S1). In wild-type NGR234 and the ΔphaP1 mutant, PhaP2 was the most abundant protein associated with PHB granules. PhaP1 was the most abundant GAP in the ΔphaP2 mutant. A conserved hypothetical protein, NGR_b11320, was found to be the most abundant GAP in the ΔphaP1P2 mutant and could be found in NGR234 and the ΔphaP1 and ΔphaP2 mutants. Further sequence analysis revealed that it contains a Phasin_2 domain (Pfam family PF09361), which is also present in PhaP1, PhaP2, and PhaP3. NGR_b11320 was named PhaP4 accordingly.
FIG 5.
PHB granule-associated proteins from NGR234 and strains lacking phaP1 and/or phaP2. (A) Exponentially modified protein abundance index (emPAI) for PHB granule-associated proteins (GAPs) identified using nanospray ESI-MS analysis. GAPs with a protein score above 1,500 (except NGR_b19610, serving as a control for a GAP with a protein score below 1,500 in the following colocalization experiment) in at least one test strain and identified from at least two out of three independent experiments (Data Set S1) are shown (emPAI values from the first experiment are shown). Subcellular localization predicted by pSORTb is shown as follows: OM, outer membrane; CM, cytoplasmic membrane; Cyto, cytoplasmic; Peri, periplasmic; superscript M, the protein may have multiple localization sites. The COG category of each protein is listed as follows: S (function unknown), C (energy production and conversion), T (signal transduction mechanism), I (lipid transport and metabolism), E (amino acid transport and metabolism), M (cell wall/membrane/envelope biogenesis), O (posttranslational modification, protein turnover, and chaperones), and G (carbohydrate transport and metabolism). (B) Schematic view of PHB granules and GAPs at stationary phase. The sizes of PHB granules in each strain are proportional to those average sizes determined for Fig. 2. The granule surface was largely occupied by PhaPs in NGR234 and the ΔphaP1 and ΔphaP2 mutants. The abundance of the regulator PhaR and the total pool of other GAPs excluding PhaPs increased in the ΔphaP1P2 mutant.
In contrast to typical phasins (PhaP1, PhaP2, and PhaP4), other putative GAPs identified in this study had lower abundances (Fig. 5B), such as cytochrome c oxidase FixO2 (NGR_c17980), ATP synthase subunit beta (NGR_c31110), NADH-quinone oxidoreductase chain C NuoC (NGR_c10500), and a putative universal stress protein (NGR_c09750) detected in all test strains. PHB granules from the ΔphaP1, ΔphaP2, and ΔphaP1P2 mutants seemed to have a relatively higher abundance than those from NGR234 in a hypothetical protein (NGR_b13620), ATP synthase subunit alpha (NGR_c31130), and polyhydroxyalkanoate synthesis repressor PhaR (NGR_c32730). Notably, the abundance of PhaR was particularly higher in the ΔphaP1P2 mutant than the other test strains. Similarly, PHB granules from the ΔphaP2 and ΔphaP1P2 mutants had a higher abundance in a hypothetical protein (NGR_c03840) and ATP-dependent phosphoenolpyruvate carboxykinase (NGR_c33940). Several proteins with a relatively high abundance in either the ΔphaP2 or ΔphaP1P2 mutant were also identified, such as a formaldehyde dehydrogenase (NGR_c26300), a succinate dehydrogenase flavoprotein subunit (NGR_c31350), and PhbC2 (NGR_c14000) in the ΔphaP2 mutant and NGR_c28710 (a small heat shock protein), NGR_c36120 (a putative outer membrane protein), and NGR_b18390 (a uridylyltransferase) in the ΔphaP1P2 mutant. Among these putative GAPs, the most abundant ones belong to the Clusters of Orthologous Groups (COG) categories S (function unknown), C (energy production and conversion) and T (signal transduction mechanism). A schematic overview of relative abundances of these GAPs in test strains at stationary phase is shown in Fig. 5B.
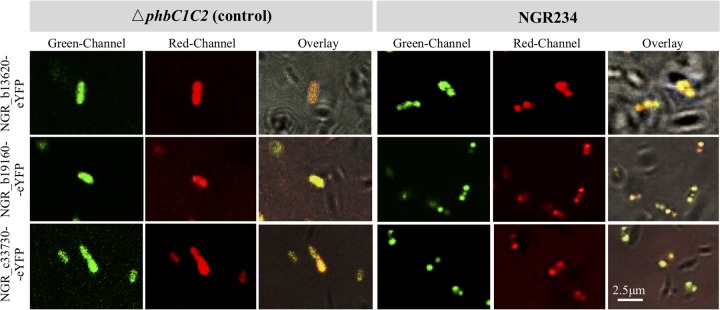
In addition to PhaP4 (NGR_b11320) (Fig. 2A), three representative GAPs of low abundances (NGR_b13620, NGR_c33730, NGR_b19610) (Fig. 5A and Data Set S1) were individually fused with the eYFP reporter and introduced into the ΔphbC1C2 mutant and NGR234. These constitutively expressed proteins were colocalized with PHB granules in NGR234 while having a uniform intracellular distribution in the ΔphbC1C2 mutant (Fig. 6).
FIG 6.
Subcellular colocalization of representative granule-associated proteins of low abundance. NGR_b13620-eYFP, NGR_b19160-eYFP, and NGR_c33730-eYFP fusions were constitutively expressed in free-living NGR234 and its ΔphbC1C2 mutant.
Metabolic characteristics of NGR234 and its derived mutants.
As described above, no direct relationship could be established between the number or size of PHB granules and the symbiotic performance of test mutants of genes involved in biosynthesis, depolymerization, and regulation of granule size and number. A Biolog GEN III plate was further used to compare the potential differences between these strains in their metabolic characteristics (Table S3). The ΔphaP1P2 mutant was impaired in utilizing l-alanine, l-lactic acid, propionic acid, d-fructose-6-phosphate, and glycyl-l-proline. The ΔphaP2P3 mutant exhibited reduced growth on acetoacetic acid, l-lactic acid, d-malic acid, and glycyl-l-proline. However, the ΔphaP1P2P3 grew better than the ΔphaP1P2 or ΔphaP2P3 mutant on l-alanine or acetoacetic acid. Similarly, the ΔphaP1P2P3P4 mutant and other mutants lacking individual or multiple phasins had their own characteristic metabolic profiles (Table S3). The ΔphbC1C2 mutant was distinct by its improved utilization of diverse nutrients compared to wild-type NGR234 and ΔphbC1, ΔphbC2, phaZ, and phasin mutants (Table S3).
DISCUSSION
PHB synthase is involved in effective symbiosis.
PHB granules could rarely be observed in the ΔphbC2 and the ΔphbC1C2 mutants of S. fredii NGR234 under both free-living and symbiotic conditions. This is consistent with the absence of PHB granules in the free-living phbC mutant of S. meliloti Rm1021 (27). Medicago truncatula or M. sativa plants inoculated with the phbC mutant of S. meliloti showed reduced shoot dry weight and competition in nodule occupancy (23, 27, 35). In this study, reduced shoot dry weight of V. unguiculata was observed when this plant was inoculated with the ΔphbC1C2 mutant rather than the mutant lacking a single PHB synthase. In contrast to the actively transcribed phbC2 of NGR234 under both free-living and symbiotic conditions, upregulation of phbC1 in nodules of V. unguiculata compared to free-living cultures was observed in our previous transcriptomic analyses (32). Moreover, the ΔphbC1C2 mutant exhibited a more drastic change from wild-type NGR234 in metabolic profiles than the ΔphbC2 or ΔphbC1 mutant did. All test strains in this study were able to use l-aspartate and l-malate, while the ΔphbC1C2 mutant showed enhanced ability to use d-aspartate and d-malate. Although less abundant in nature than l-amino acids, d-amino acids, including d-aspartate, have been detected and play diverse roles in plants, bacteria, and animals (36, 37). Transport and oxidization of d-malate into pyruvate by E. coli have been demonstrated (38). The ΔphbC1C2 mutant also exhibited increased metabolism of citrate. These metabolic profiles of the ΔphbC1C2 mutant indicated an upshift of the TCA cycle, which may be limited by the low-oxygen condition of infected nodule cells harboring nitrogen-fixing rhizobia (21). The downregulation of the TCA cycle in bacteroids is also supported by comparative transcriptomics of wild-type NGR234 in free-living culture and V. unguiculata nodules (32), where bacteroids exhibited reduced transcript levels of the genes encoding isocitrate dehydrogenase (NGR_c16430), α-ketoglutarate dehydrogenase E1 and E2 components (NGR_c31270 and NGR_c31260), and the catalytic subcomplex (SdhA/B, encoded by NGR_c31350 and NGR_c31340, respectively) of succinate dehydrogenase.
It is notable that there was a low number of PHB granules in the ΔphbC1C2 mutant in V. unguiculata nodules, whereas no PHB granules were found in a free-living culture of this mutant. Potential false positives could not be ruled out due to potential cytoplasmic changes in this mutant. This could be partially supported by PHB quantification results (Table S1) showing that no significant difference between the ΔphbC1C2 and ΔphbC2 mutants could be detected under both free-living and symbiotic conditions, though average PHB amounts were higher in nodules infected by the ΔphbC1C2 mutant than by the ΔphbC2 mutant. Alternatively, other proteins of similar function might exert a complementary effect to a limited extent. For example, NGR_b13620 shown as a putative GAP (Fig. 6) has a conserved domain (DUF3141), which shows a significant (E value, 2e−05) but low identity (23%) to poly(R)-hydroxyalkanoic acid synthase, class III PhaC subunit (WP_012267458.1). It is also annotated as 3-hydroxyalkanoate synthetase in many bacteria. Moreover, this gene is strongly induced in bacteroids in V. unguiculata nodules compared to free-living culture (log2 R = 7.16) (32). It has been reported that a phbC mutant of S. meliloti was unable to utilize acetoacetate, which is an intermediate in the degradation portion of the PHB cycle (39). The ΔphbC1 mutant rather than the ΔphbC1C2 or ΔphbC2 mutant of NGR234 showed impaired ability in acetoacetate utilization (Table S3). These findings imply that the differential regulation and diversity of enzymes involved in PHB cycle in different species may contribute to their distinct adaptation characteristics.
As shown by ESI-MS and colocalization experiments, there is another putative PHB depolymerase (NGR_c33730) that shows poor identity (9.16%) to NGR_b03370 in NGR234. Sequence alignment with a PHA depolymerase database (40) suggested that NGR_c33730 and NGR_b03370 might belong to intracellular (i-nPHAscl, no lipase box) and extracellular (e-dPHAscl, catalytic domain type 1, with lipase box GLSAG) depolymerase families, respectively. However, there was evidence that an extracellular PHAscl depolymerase from Paucimonas lemoignei only degrades native PHA granules (41). Although NGR_b03370 was transcribed at a relatively lower level than NGR_c33730 in free-living culture, it was more strongly upregulated in V. unguiculata nodules (32) and the phaZ (NGR_b03370) mutant in nodule cells harbored more PHB granules of larger size than the wild-type NGR234. Visible PHB granules in bacteroids were observed for the phaZ mutant but not wild-type Rm1021 of S. meliloti in Medicago nodules (26). However, these phaZ mutants of NGR234 and Rm1021 were indistinguishable from wild-type strains in symbiotic performance in this study and the earlier one (26). This is in line with the view that degradation of PHB may be not important in nitrogen-fixing nodules and the role of PHB during symbiosis could be a store of excess reductant rather than a carbon buffer (20). However, 3-hydroxybutyrate dehydrogenase (BdhA) acting downstream of PhaZ in the PHB degradation pathway is required for effective symbiosis of NGR234 on L. leucocephala but not Tephrosia vogelii, Macroptilium atropurpureum, or V. unguiculata plants (42). These findings suggest that the importance of PHB degradation can vary in different symbiosis systems and a buildup of 3-hydroxybutyrate could inhibit the symbiosis in certain cases.
It should be noted that the ΔphbC1C2 mutant of NGR234 still formed pink nodules and leaf chlorophyll content of inoculated V. unguiculata plants was similar to those inoculated with the wild-type NGR234. This is in line with results indicating that the phbC mutant of S. meliloti Rm1021 was able to fix nitrogen, though a host-dependent (Medi truncatula versus M. sativa) variation of nitrogenase activity was observed (23). However, host plants inoculated with these phbC mutants of Sinorhizobium exhibited a significant reduction in shoot dry weight and nodule occupancy as described above. The phbC mutant of A. caulinodans was Fix− in symbiosis with Sesbania rostrata (29), while an R. etli mutant lacking a functional PHB synthase exhibited a prolonged capacity to fix nitrogen in nodules of Phaseolus vulgaris (28). Therefore, PHB synthase can be involved in optimizing symbiosis in a rhizobium-host-dependent manner, or even in a way interacting with changing environments, as implied in a study of Rhizobium leguminosarum strains nodulating pea and bean (43).
Distinct roles of different phasins in regulating number and size of PHB granules.
Although PHB is thought as a source of carbon and energy for different cellular processes during adaptation and survival in the environment, a recent advance in this field has highlighted an active role of multifaceted phasins in reducing the deleterious effects of diverse stresses (13). Constitutively expressed fusion with eYFP has been developed as an effective tool to determine whether candidate proteins represent true GAPs of PHB (18, 44). In this study, four PhaPs harboring the Phasin_2 domain were demonstrated to colocalize with PHB granules under free-living condition using the same technique. In S. meliloti Rm1021, two putative phasins named as PhaP1 and PhaP2, showing 88.4% and 89.3% identity to PhaP1 and PhaP2 of NGR234, respectively, were redundant regarding their role in PHB production and symbiotic performance (27). However, four PhaPs in NGR234 seem to be differentially involved in PHB production under either free-living or symbiotic conditions.
At stationary phase, PhaP2 was the most abundant phasin on PHB granules of NGR234. Indeed, the ΔphaP2 mutant showed the most drastic change in PHB granule size among individual mutants of phasin genes. This is similar to the finding in R. eutropha H16 and B. diazoefficiens USDA110, in which PhaP1Reu and PhaP4Bdi, respectively, were the major phasin proteins (1, 30, 45). In contrast, PhaP1 and PhaP2 in S. meliloti Rm1021 made roughly equal contributions, as no obvious difference in either the size or number of PHB granules was found between individual phaP1 and phaP2 mutants (27). The phaP1P2 double mutant of Rm1021 had no detectable PHB granules (27), and this is in line with a significantly reduced number of PHB granules in the ΔphaP1P2 mutant compared to mutants lacking individual phasins and the other double mutants of NGR234 at stationary phase. Moreover, among diverse putative GAPs, PhaP1 was the most abundant one on PHB granules in the ΔphaP2 mutant of NGR234. These findings indicate that PhaP1 together with the major phasin PhaP2 is crucial for regulating PHB granule number in free-living cultures of NGR234. Although phaP3 and phaP4 had relatively low transcript levels under free-living conditions (32), a reduced number of PHB granules could also be observed in individual phaP3 and phaP4 mutants at stationary phase, indicating cumulative contributions by PhaP3 and PhaP4. The relatively lower importance of minority phasins was also reported for R. eutropha (18, 46), though PhaP5Reu, PhaP6Reu, and PhaP7Reu had distinct subcellular localizations (18).
Differential transcriptional profiles of different phasins have been reported for B. diazoefficiens USDA110 grown in different media (30). In R. eutropha H16, PhaP6Reu and PhaP7Reu could associate with PHB granules later than the other phasins (18). In contrast to the other three phasins, PhaP3 was not detected on PHB granules collected from free-living NGR234, possibly due to its lowest transcription level among these phasin genes in cultures (32). Instead, phaP3 located on the symbiosis plasmid was upregulated within V. unguiculata nodules (32). In line with these findings, the numbers of PHB granules in the ΔphaP1P3, ΔphaP2P3, and ΔphaP1P2P3 mutants compared to the ΔphaP1, ΔphaP2, and ΔphaP1P2 mutants, respectively, were similar at stationary phase but significantly lower within V. unguiculata nodules. Therefore, PhaP3 may play a more active role in regulating PHB granule number during symbiosis than in free-living cultures. The phaP4 gene was also upregulated in V. unguiculata nodules (to a lesser extent than phaP3) (32) and made a cumulative contribution to PHB biosynthesis.
It was reported that the phaP1P2 mutant of S. meliloti Rm1021 lacking PHB granules was able to fix nitrogen but that the shoot dry weight of inoculated M. truncatula plants was reduced compared to that of the wild-type strain (27). This Fix+ Eff− phenotype has also been reported for a glnD mutant of Rm1021 (47) and hemN1 mutants of S. fredii CCBAU45436 and Sinorhizobium sp. CCBAU05631 nodulating wild soybean (48). In this study, V. unguiculata plants inoculated with the ΔphaP1P2 or ΔphaP2P3 mutant of NGR234 exhibited slight but nonsignificant reduction in shoot dry weight compared to that of plants inoculated with the wild-type strain. Bacteroids of these two double mutants, as well as ΔphaP1P2P3 and ΔphaP1P2P3P4 bacteroids, had a reduced number of PHB granules, whereas no granules were observed in free-living cells of the ΔphaP1P2P3 or ΔphaP1P2P3P4 mutant.
Additional functions of phasins have been proposed, such as protective effect and chaperone activity (13). It was reported that a phasin from Azotobacter sp. FA8 can enhance the resistance of non-PHB-producing E. coli to both heat shock and superoxide stress, possibly by exerting chaperone-like activity (11). In the analysis of GAPs, PHB granules of the ΔphaP1P2 mutant at stationary phase had a lower abundance of phasin than the ΔphaP1 and ΔphaP2 mutants, while some other proteins of diverse functions were identified as putative GAPs, such as a small heat shock protein (NGR_c28710), oxidoreductase (NGR_c21410), sarcosine oxidase (NGR_c29020), and uridylyltransferase (NGR_b18390). This suggests that phasins may prevent PHB granules from associating with proteins of other cellular pathways by occupying the granule surface. For example, the most abundant GAPs identified in this study are those of unknown function and those involved in energy production/conversion and signal transduction. Although we could not exclude the probability of false-positive GAPs during the sample preparation, identification of GAPs of low abundance, multiple subcellular localizations, and those bound loosely to PHB granules is also possible, as discussed in a previous study of GAPs of PHB in R. eutropha H16 (49). For example, NGR_b13620, NGR_c33730, and NGR_b19160, of low abundance identified by the nanospray ESI-MS analysis in this study, were demonstrated as true GAPs in the subcellular colocalization experiments.
The low similarity levels among four PhaPs in NGR234 and their distinct transcription patterns under free-living and symbiotic conditions could lead to a complex interaction network, which remains unexplored. This hypothesis is supported by the distinct metabolic profiles of single or multiple mutants of PhaPs. It is noteworthy that in contrast to the conserved PhaP1 and PhaP2, accessory PhaP3 and PhaP4 have a restricted phyletic distribution in Sinorhizobium. This further supports the view that accessory functions have been extensively integrated into the core regulation network during environmental adaptation of rhizobia (34, 50–52).
Conclusions.
Among diverse GAPs of PHB, PhaP1, PhaP2, PhaP3, and PhaP4 were demonstrated as true phasins with distinct phyletic distribution patterns and transcriptional profiles. The conserved PhaP2 is the dominant phasin regulating PHB granule size under both free-living and symbiotic conditions. In free-living cells at stationary phase, the conserved PhaP1 and accessory PhaP3 and PhaP4 also contribute to the regulation of PHB granule number and size. The accessory phaP3 gene, located on the symbiosis plasmid and upregulated in nodules, and phaP2 are crucial in regulating PHB biosynthesis in nodules, and PhaP4 and PhaP1 made cumulative contributions. Under the test conditions, this study revealed the essential role of PHB synthases rather than PHB depolymerase or phasins of NGR234 in symbiotic interaction with V. unguiculata. The presence or absence of PHB granules could not account for the contrasting phenotypes of test mutants. Metabolic profiles of these mutants were strain specific under free-living conditions and should be further investigated in nodules. Variations in the pool of GAPs in representative phaP mutants imply that PHB is not just a store of carbon and reductant but may actively interact with other cellular pathways through direct binding of related proteins, particularly when the diversity and abundance of different phasins, the major components of the PHB granule cover, change under the fluctuating environmental conditions. These findings are also significant in the context of evolution, as core and accessory PhaPs play coordinated roles in these processes, highlighting the importance of integration between accessory and core functions during bacterial adaptations.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are shown in Table 1. E. coli was grown at 37°C in Luria broth (LB) medium supplemented with appropriate antibiotics. The broad-host-range strain S. fredii NGR234 (53) was grown at 28°C in TY medium (tryptone at 5 g/liter, yeast extract at 3 g/liter, and CaCl2 at 0.6 g/liter). S. fredii NGR234 is resistant to rifampin (Rif). The antibiotic concentrations used were 50 μg/ml of Rif, 50 μg/ml of kanamycin (Km), 30 μg/ml of gentamicin (Gen), and 10 μg/ml of tetracycline (Tet). The sucrose concentration for screening double-crossover mutant strains was 5% (wt/vol).
TABLE 1.
Plasmid and strains used in this study
| Plasmid or strain | Characteristicsa | Reference |
|---|---|---|
| Plasmids | ||
| pCM351 | Genr Tetr; broad-host-range cre-lox vector | 57 |
| pJQ200SK | Genr; P15A origin from pACYC184; lacZ sacB traJ | 54 |
| pVO155 | Kmr; pUC119 derivative for insertion into genome | 56 |
| pBBR1MCS-2 | Kmr; broad-host-range vector | 58 |
| pBBR1MCS-3 | Tetr; broad-host-range vector | 58 |
| pRK2013 | Kmr; ColE1 replicon, tra+ from RK2 | 55 |
| pBBR1MCS-2-PphaC-eyfp-n1 | Kmr; universal vector for construction of fusions N terminal to eyfp under the control of the phaCAB promoter | 44 |
| pBBR1MCS-3-PphaC-eyfp-n1 | Tetr; universal vector for construction of fusions N terminal to eyfp under the control of the phaCAB promoter | This work |
| pBBR1MCS-2-PphaC-phaP1-eyfp | Kmr; N-terminal fusion of PhaP1 to eYFP | This work |
| pBBR1MCS-2-PphaC-phaP2-eyfp | Kmr; N-terminal fusion of PhaP2 to eYFP | This work |
| pBBR1MCS-2-PphaC-phaP3-eyfp | Kmr; N-terminal fusion of PhaP3 to eYFP | This work |
| pBBR1MCS-3-PphaC-phaP1-eyfp | Tetr; N-terminal fusion of PhaP1 to eYFP | This work |
| pBBR1MCS-3-PphaC-phaP2-eyfp | Tetr; N-terminal fusion of PhaP2 to eYFP | This work |
| pBBR1MCS-3-PphaC-phaP3-eyfp | Tetr; N-terminal fusion of PhaP3 to eYFP | This work |
| pBBR1MCS-2-PphaC-b11320-eyfp | Kmr; N-terminal fusion of NGR_b11320 to eYFP | This work |
| pBBR1MCS-2-PphaC-b13620-eyfp | Kmr; N-terminal fusion of NGR_b13620 to eYFP | This work |
| pBBR1MCS-2-PphaC-c33730-eyfp | Kmr; N-terminal fusion of NGR_c33730 to eYFP | This work |
| pBBR1MCS-2-PphaC-b19160-eyfp | Kmr; N-terminal fusion of NGR_b19160 to eYFP | This work |
| pJQ200SK-phbC2 | Genr; pJQ200SK carrying phbC2 for in situ complementation | This work |
| pJQ200SK-phaP3 | Genr; pJQ200SK carrying phaP3 for in situ complementation | This work |
| Strains | ||
| Escherichia coli DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF) U169 deoR recA1 endA1 hsdR17(rk− mk+) phoA supE44 λ− thi-1 gyrA96 relA1 | |
| Sinorhizobium fredii | ||
| NGR234 | Rifr; wild type | 53 |
| phbC1 strain | Rifr Genr; NGR234 ΔphbC1::Gen | This work |
| phbC1C2 strain | Rifr Genr Kmr; NGR234 ΔphbC1::Gen with pVO155 inserted into phbC2 | This work |
| ΔphbC1 strain | Rifr; NGR234 with phbC1 precise deletion | This work |
| ΔphbC2 strain | Rifr; NGR234 with phbC2 precise deletion | This work |
| ΔphbC1C2 strain | Rifr; NGR234 with phbC1 and phbC2 precise deletion | This work |
| ΔphbC1C2+C2 strain | Rifr; ΔphbC1C2 strain complemented with phbC2 | This work |
| phaZ (NGR_b03370) strain | Rifr Kmr; NGR234 with pVO155 inserted into NGR_b03370 | This work |
| ΔphaP1 strain | Rifr; NGR234 with phaP1 precise deletion | This work |
| ΔphaP2 strain | Rifr; NGR234 with phaP2 precise deletion | This work |
| ΔphaP3 strain | Rifr; NGR234 with phaP3 precise deletion | This work |
| ΔphaP4 strain | Rifr; NGR234 with phaP4 precise deletion | This work |
| ΔphaP1P2 strain | Rifr; NGR234 with phaP1 and phaP2 precise deletions | This work |
| ΔphaP1P3 strain | Rifr; NGR234 with phaP1 and phaP3 precise deletions | This work |
| ΔphaP2P3 strain | Rifr; NGR234 with phaP2 and phaP3 precise deletions | This work |
| ΔphaP2P3+P3 strain | Rifr; ΔphaP2P3 strain complemented with phaP3 | This work |
| ΔphaP1P2P3 strain | Rifr; NGR234 with phaP1, phaP2, and phaP3 precise deletions | This work |
| ΔphaP1P2P3P4 strain | Rifr; NGR234 with phaP1, phaP2, phaP3, and phaP4 precise deletions | This work |
Genr, gentamicin resistance; Tetr, tetracycline resistance; Kmr, kanamycin resistance; Rifr, rifampin resistance.
Construction of mutants and complemented strains.
The primers used in this study are listed in Table 2. For precise deletion of coding sequences of phbC1, phbC2, phaP1, phaP2, phaP3, and phaP4 (NGR_b11320), a seamless assembly cloning kit (Taihe Biotechnology, Beijing, China) was used to construct knockout vectors derived from pJQ200SK (54). Briefly, 500- to 700-bp upstream and downstream fragments of target gene were amplified using primers (Table 2) carrying sequences corresponding to the ends of SmaI restriction sites in pJQ200SK. These two fragments were simultaneously mixed with linearized pJQ200SK (digested by SmaI) and incubated at 50°C for 15 min, and the positive E. coli clones were selected by antibiotic resistance and colony PCR after transformation. Then the integrative plasmid was conjugated into the wild-type NGR234 by triparental mating using the helper plasmid pRK2013 (55). Single-crossover clones were screened using a TY (Rif Gen) plate and by colony PCR. The resultant clones were cultured in liquid TY medium (Rif) for 24 to 36 h and screened on a TY (Rif) plate containing 5% sucrose. Double-crossover clones were verified by colony PCR and Sanger sequencing. The same method was used to construct double and multiple mutants. To generate the phaZ (NGR_b03370) mutant, an intragenic fragment of 388 bp, amplified using primers phaZF_BamHI and phaZR_XbaI, was digested by BamHI and XbaI and linked with the pVO155 (56) digested with the same endonucleases. The resultant vector was conjugated into NGR234. The NGR_b03370 mutant was selected on a TY (Rif Km) plate, followed by PCR verification. For the construction of the phbC1C2 double mutant, which served as a control in subcellular localization of PhaP1, PhaP2, and PhaP3, the cre-lox system was used (57). First, a 530-bp DNA fragment upstream and a 683-bp DNA fragment downstream from the coding sequence of phbC1 were amplified and ligated to pCM351 suicide plasmid using corresponding primers and endonucleases listed in Table 2. The resultant plasmid was conjugated into NGR234 through triparental mating with pRK2013 as the helper plasmid. The phbC1 mutant sensitive to Tet and resistant to Gen was selected. Then a pVO155 derivative carrying a 397-bp intragenic fragment of the phbC2 gene was constructed and conjugated into the phbC1 mutant using the same procedure as described above for the construction of the NGR_b03370 mutant. All constructed mutants were verified using PCR and Sanger sequencing. To perform a genetic complementation, the integrative pJQ200SK plasmid carrying corresponding gene (phaP3 or phbC2), upstream and downstream fragments of the gene was conjugated into mutant by triparental mating with pRK2013 helper plasmid. Primers used in this study are listed in Table 2. Single-crossover and subsequent double-crossover clones were screened and verified using the same method as the mutant construction using pJQ200SK plasmid. This allowed a complementation event with the cloned DNA being inserted into its original genome location by double crossover.
TABLE 2.
Primers used in this study
| Primer | DNA sequence (5′–3′)a | Description |
|---|---|---|
| phaP1F_NdeI | GGGAATTCCATATGGCTACCAAGAAGACCGAA | For construction of phaP1-eyfp fusion |
| phaP1R_BamHI | CGGGATCCCCGGCCTTCTTGAAGGTGGAG | |
| phaP2F_NdeI | GGGAATTCCATATGTTTAATTTCGACGACGCAAA | For construction of phaP2-eyfp fusion |
| phaP2R_BamHI | CGGGATCCCCGGCGGCGGCAGCAGACTTCA | |
| phaP3F_NdeI | GGGAATTCCATATGTCCAGGACCGCAGAAAAGC | For construction of phaP3-eyfp fusion |
| phaP3R_XmaI | CGCCCGGGCGGCCGTCTTGAGGTCTGCTA | |
| P123F_KpnI | CTAAAGGGAACAAAAGCTGGGTACCCAAAAATTCATCCTTCTCG | To transfer phaP1-, phaP2-, or phaP3-eyfp fusion into pBBR1MCS-3 |
| P123R_SpeI | CGGTGGCGGCCGCTCTAGAACTAGTTTACTTGTACAGCTCGTCC | |
| MCS-3F_SpeI | ACTAGTTCTAGAGCGGCCG | For generation of linear pBBR1MCS-3 plasmid |
| MCS-3R_KpnI | GGTACCCAGCTTTTGTTCC | |
| NGR_b11320F | AGTCGACGGTACCGCGGGCCATGTCGAAGAAAATATCCGAC | For construction of NGR_b11320-eyfp fusion |
| NGR_b11320R | GGTGGCGACCGGTGGATCCCGCATGACCATCTGCCGC | |
| NGR_b13620F | AGTCGACGGTACCGCGGGCCATGCCCAAGCAATCCACC | For construction of NGR_b13620-eyfp fusion |
| NGR_b13620R | GGTGGCGACCGGTGGATCCCGGGATGTCCTGCGCTCGC | |
| NGR_c33730F | AGTCGACGGTACCGCGGGCCATGTTCTACCAGCTTTACGAA | For construction of NGR_c33730-eyfp fusion |
| NGR_c33730R | GGTGGCGACCGGTGGATCCCGGGCCGATTTGCCGC | |
| NGR_b19160F | AGTCGACGGTACCGCGGGCCATGGCGAAAGTAATCGTTGT | For construction of NGR_b19160-eyfp fusion |
| NGR_b19160R | GGTGGCGACCGGTGGATCCCGTGCTGCCCTCCGTCC | |
| phbC1upF_EcoRI | GGAATTCACTGCTGCGCTACAAT | For phbC1 deletion |
| phbC1upR_NdeI | GGAATTCCATATGGTCGATCACCCGTCTT | |
| phbC1downF_AgeI | GACCGGTAAAGAGGCTATCCCCC | For phbC1 deletion |
| phbC1downR_SacI | GGCGAGCTCCGCAAACAGAAACAAA | |
| phbC2F_BamHI | CGGGATCCAGCAAGTTTGCCATCG | For phbC2 insertion |
| phbC2R_XbaI | GCTCTAGACATGGAGCGCCAGCGT | |
| phaZF_BamHI | CGGGATCCGCTGCGCGACAATAAC | For phaZ insertion |
| phaZR_XbaI | GCTCTAGAATCGTTCGTCCCGTGC | |
| phbC1upF | ATATCGAATTCCTGCAGCCCACTGCTGCGCTACAAT | For phbC1 precise deletion by pJQ200SK |
| phbC1upR | TAGCCTCTTTGTCGATCACCCGTCTT | |
| phbC1downF | GGTGATCGACAAAGAGGCTATCCCCC | For phbC1 precise deletion by pJQ200SK |
| phbC1downR | CTAGAACTAGTGGATCCCCCTCGCACGATACGAGAC | |
| phbC2upF | ATATCGAATTCCTGCAGCCCTTCGGCAATCGCTCAA | For phbC2 precise deletion and in situ complementation by pJQ200SK |
| phbC2upR | TTTGCCTTCTGTTCGACCGATTTGGG | For phbC2 precise deletion by pJQ200SK |
| phbC2downF | TCGGTCGAACAGAAGGCAAAGGAGAC | For phbC2 precise deletion by pJQ200SK |
| phbC2downR | CTAGAACTAGTGGATCCCCCAAATGGACGTTCTTCA | For phbC2 precise deletion and in situ complementation by pJQ200SK |
| phaP1upF | ATATCGAATTCCTGCAGCCCCAACTGGCGCAGGGAGA | For phaP1 precise deletion by pJQ200SK |
| phaP1upR | CGGCGGCTTCGGCCGGGTCAAAGGAAA | |
| phaP1downF | TGACCCGGCCGAAGCCGCCGAAAAGG | For phaP1 precise deletion by pJQ200SK |
| phaP1downR | CTAGAACTAGTGGATCCCCCCTGCACAACTACATCGCCC | |
| phaP2upF | ATATCGAATTCCTGCAGCCCGCGATAATCCAATGCGTTAC | For phaP2 precise deletion by pJQ200SK |
| phaP2upR | TCAGGCGGCGTGCTTTTCTTGTTTGCGTC | |
| phaP2downF | AAGAAAAGCACGCCGCCTGACGATAT | For phaP2 precise deletion by pJQ200SK |
| phaP2downR | CTAGAACTAGTGGATCCCCCGCCGCATTCTCCAACG | |
| phaP3upF | ATATCGAATTCCTGCAGCCCAACTTGATAGTCGCTTCGT | For phaP3 precise deletion and in situ complementation by pJQ200SK |
| phaP3upR | ACGGCAATCACGAAGCCAATCAAGGAAT | For phaP3 precise deletion by pJQ200SK |
| phaP3downF | ATTGGCTTCGTGATTGCCGTGGATTGAC | For phaP3 precise deletion by pJQ200SK |
| phaP3downR | CTAGAACTAGTGGATCCCCCTGATGGTAGTGCAGGCGA | For phaP3 precise deletion and in situ complementation by pJQ200SK |
| phaP4upF | CTTGATATCGAATTCCTGCAGCCCCCAGCTTGCCCAGACGA | For phaP4 precise deletion by pJQ200SK |
| phaP4upR | TGCACCCGCAACCAGCGAAGAACAGGAGGAAC | |
| phaP4downF | TGTTCTTCGCTGGTTGCGGGTGCAAGTG | For phaP4 precise deletion by pJQ200SK |
| phaP4downR | CGCTCTAGAACTAGTGGATCCCCCGCACGCTCTTCCACTGTT |
Restriction sites are in bold, the sequences at the ends of SmaI restriction sites in pJQ200SK are underlined, and the sequences at the end of the linear plasmid pBBR1MCS-2 or pBBR1MCS-3 are in italics.
Construction of strains carrying fusion proteins with eYFP.
The empty fusion vector pBBR1MCS-2-PphaC-eyfp-n1 carries the eyfp (enhanced yellow fluorescent protein) gene under the control of the constitutive phaCAB promoter of R. eutropha (44). Test genes (phaP1, phaP2, phaP3, NGR_b11320, NGR_b13620, NGR_c33730, and NGR_b19160) were amplified using primers listed in Table 2 and cloned in frame to the N terminus of the eyfp gene using corresponding endonucleases. The target_gene-eyfp fusion vector was conjugated into NGR234 by triparental mating using pRK2013 as the helper plasmid. The antibiotic resistance marker of pBBR1MCS-2-PphaC-eyfp-n1 (Kmr) is the same as that of the phbC1C2 mutant (Rifr Kmr). The PphaC-target_gene-eyfp fragment carrying phaP1, phaP2, or phaP3 was further amplified using primers P123F_KpnI and P123R_SpeI and cloned into pBBR1MCS-3 (58) using the seamless assembly cloning kit. The resultant pBBR1MCS-3-derived fusion vectors for phaP1, phaP2, and phaP3 were individually conjugated into the phbC1C2 mutant (Rifr Kmr). To simplify further experiments, a marker-free deletion mutant, the ΔphbC1C2 mutant, was constructed as described above using pJQ200SK. The pBBR1MCS-2-PphaC-eyfp-n1-derived fusion vectors for phaP4 (NGR_b11320), NGR_b13620, NGR_c33730, and NGR_b19160 were conjugated into this ΔphbC1C2 mutant as controls for subcellular colocalization experiments. All constructed vectors and strains carrying them were verified by PCR and Sanger sequencing.
Plant assay.
Seeds of V. unguiculata were surface sterilized in 17% (vol/vol) sodium hypochlorite solution for 5 min and washed five times using autoclaved deionized water. Then the treated seeds were germinated on 0.5% agar plates at 28°C in the dark for 48 h. Seedlings were inoculated with 1 ml of suspension (OD600 = 0.2) of rhizobia in 0.8% (wt/vol) NaCl solution and grown in vermiculite moistened with low-N nutrient solution [Ca(NO3)2·4H2O at 0.03 g, KCl at 0.075 g, MgSO4 at 0.06 g, K2HPO4 at 0.136 g, CaSO4·2H2O at 0.46 g, FeC6H5O7 at 0.075 g, H3BO3 at 2.86 mg, MnSO4 at 1.81 mg, CuSO4·5H2O at 0.8 mg, ZnSO4 at 0.22 mg, and H2MoO4 at 0.02 mg per liter of distilled water] in Leonard jars (59) at 25°C with a 12-h illumination period and harvested 30 days postinoculation (dpi). Leaf relative chlorophyll concentration was detected by a SPAD-502 meter (Konica Minolta) as described previously (22, 60). To minimize the sampling error, three leaflets of the third leaf were tested for each plant. The shoot dry weight was tested after drying in an oven at 60°C for 5 days. Nodules were used for preparing samples for transmission electron microscopy (TEM). For competitive nodulation, seedlings were inoculated with 1 ml of an inoculum combining equal volumes of bacterial suspension (OD600 = 0.2) of the wild type and mutant strains. Nodules of five plants for each treatment were collected 30 dpi and surface sterilized in 17% (vol/vol) sodium hypochlorite solution for 5 min. After washing five times with autoclaved deionized water, the nodules were crushed. Bacteria from each nodule were cultured on a TY plate continuously at 28°C for 3 days. Colony PCR was used to identify the wild type and mutants. At least two independent experiments were carried out.
TEM.
Bacterial cultures (1 ml) were harvested at stationary phase by centrifugation for 5 min at 8,000 × g. The pellets or nodules were fixed in 2.5% glutaraldehyde in 0.05 M cacodylate buffer (61). The resultant samples were washed with 0.1 M phosphate buffer and postfixed in the same buffer containing 1% (wt/vol) OsO4. The samples were washed again with 0.1 M phosphate buffer and dehydrated with increasing volumes of acetone (30%, 50%, 70%, 90%, and 100%). The samples embedded in Spurr epoxy were then cut into ultrathin sections (80 nm thick) using a Leica Ultracut C6i. The sections were stained with uranyl acetate and lead citrate and observed with a JEM-1230 transmission electron microscope. To determine the size and number of PHB granules by ImageJ software, 30 pictures (magnification, 25,000×) for cells from one free-living culture of each test strain and 15 pictures (mutants or complemented strains; magnification, 20,000×) or 10 pictures (wild-type NGR234; magnification, 20,000×) of 3 or 4 nodules from different plants were taken by the microscope. For each picture, five bacterial cells located on the diagonal line were selected for the determination of size and number of PHB granules.
Laser scanning confocal microscopy.
Stationary-phase cultures of strains carrying the eYFP-tagged fusions were collected and suspended in 0.8% sodium chloride and stained by addition of 0.01 volume of Nile red solution (100 μg of Nile red/ml of dimethyl sulfoxide) in the dark for 5 min. Nile red fluorescent dye was used for staining cellular PHB granules (62). The stained cells were washed once with 0.8% sodium chloride. Isometric stained cell culture and 1% agarose solution were mixed together to fix the cells, which were subsequently observed with an Olympus laser scanning confocal microscope using Olympus Fluoview version 4.0b software. The excitation light source was set to 488 nm (green fluorescent protein [GFP] channel) for detection of the eYFP signal and 546 nm (RFP-Channel) for Nile red signal.
Biolog metabolic assay.
A Biolog GEN III microplate (71 carbon source utilization assays and 23 chemical sensitivity assays) was used to compare metabolic profiles of test strains. The colonies of test strains on TY plates were individually suspended in the inoculating fluid IF-A (Biolog GEN III identification fluids, catalog no. 72401), and 100 μl of bacterial suspension was added into each well of the microplate. Then the microplates were incubated at 28°C for 72 h. Absorbance reading at 590 nm was carried out every 12 h using Biolog MicroStation system, and active (2), weak (1), or no (0) growth in each well was automatically defined by the integrated software of Biolog. Three independent experiments were performed for each strain.
Isolation and purification of PHB granules.
PHB granules were isolated by a method of two steps of sucrose density gradient centrifugation, modified from that described earlier (63). Briefly, 300-ml quantities of bacterial cultures grown in liquid TY medium were collected at stationary phase by centrifugation (5 min, 8000 × g, and 4°C) and washed once with 10 ml of potassium phosphate buffer (100 mM, pH 7.5). The pellets were ground into powder in the presence of liquid nitrogen. The bacterial powder was suspended and sonicated in 10 ml of Tris-HCl (10 mM, pH 8.0). After centrifugation, the insoluble fraction containing PHB granules was suspended with 5 ml Tris-HCl (10 mM, pH 8.0) and subject to ultracentrifugation (15 h, 28,000 × g, and 4°C, with a Beckman SW28 rotor) in a sucrose gradient. The gradient was prepared from 18 ml of 1.66 M, 8 ml of 1.33 M, and 8 ml of 1 M sucrose in Tris-HCl (10 mM, pH 8.0). The PHB granule layer (located between 1.66 M and 1.33 M sucrose solution) was separated and washed once with Tris-HCl (10 mM, pH 8.0). To improve the quality of purification, a second-round centrifugation in a sucrose gradient (15 h, 28,000 × g, and 4°C, with a Beckman SW28 rotor) was carried out. The gradient was prepared from 8 ml of 2 M, 14 ml of 1.66 M, 8 ml of 1.33 M, and 8 ml of 1 M sucrose in Tris-HCl (10 mM, pH 8.0). After ultracentrifugation, the PHB granules were washed twice with Tris-HCl (10 mM, pH 8.0) and then stored at –20°C.
SDS-PAGE
To obtain a global view of granule-associated proteins (GAPs) of PHB, granule pellets from representative strains were suspended with 1× loading buffer (0.6% [wt/vol] SDS, 1.25% [wt/vol] β-mercaptoethanol, 0.25 mM EDTA, 10% [vol/vol] glycerol, 0.001% [wt/vol] bromophenol blue, 12.5 mM Tris-HCl [pH 6.8]) and boiled at 100°C for 5 min. Then 20-μg quantities of proteins were separated by SDS-PAGE (polyacrylamide gel electrophoresis; 5% stacking gel, 15% separating gel, 0.1% SDS) and stained with Coomassie brilliant blue R-250.
Mass spectrometry analysis.
PHB granules were resuspended in a urea-sulfocarbamide solution (3.04 g of urea and 8.4 g of sulfocarbamide in 20 ml of double-distilled water [ddH2O]), in which GAPs from the surface of PHB granules were dissolved. After centrifugation (20 min and 14,000 × g), the supernatants were quantified by the Bradford method. Then 50 μg of GAPs from each strain was digested to peptide by trypsin. Nano-liquid chromatography (nano-LC) separation was achieved with a Waters (Milford, MA) nanoACQUITY nano-high-performance liquid chromatograph (nano-HPLC). Nanospray ESI-MS was performed on a Thermo Q-Exactive high-resolution mass spectrometer (Thermo Scientific, Waltham, MA) with a 70,000 MS scan resolution, 17,500 tandem MS (MS/MS) scan resolution, and top-10 MS/MS selection. Raw data from the mass spectrometer were preprocessed with Mascot Distiller 2.4 (Matrix Science, London, UK) for peak picking. The resultant peak lists were searched against an S. fredii NGR234 protein database using the Mascot 2.5 search engine (Matrix Science). The search parameters were as follows: carbamidomethyl as fixed modification of cysteine, oxidation of methionine and phosphorylation of serine, threonine, and tyrosine as variable modifications, two maximum missed cleavages for trypsin, MS mass tolerance of 10 ppm, and MS/MS mass tolerance of 0.02 Da. The exponentially modified protein abundance index (emPAI = 10PAI – 1, where PAI is the ratio of observed number of peptides per protein and observable number of peptides per protein), a label-free quantitative measure of protein abundance in proteomics (64), was used to determine the abundances of different GAPs.
Quantification of PHB.
Bacterial cultures (40 ml) in TY medium were collected at stationary phase by centrifugation (4 min and 13,000 × g). Then the cell pellets were homogenized with sodium hypochlorite at 37°C for 1 h. After centrifugation (20 min and 13,000 × g), the pellets were washed with 2 ml of ddH2O and precipitated with 2 ml of 1:1 alcohol-acetone. The pellets were resuspended in 2 ml of chloroform, and 100 μl of solution was used for determination of PHB levels after centrifugation (20 min and 14,000 × g). PHB was determined as chrotonic acid in 10 ml of H2SO4 (65, 66). For PHB quantification of bacteroids, 130- to 160-mg nodules (10 to 15 nodules randomly selected from five plants) were crushed with liquid nitrogen before homogenization with sodium hypochlorite at 37°C for 1 h. Three subsamples of stationary-phase cultures or nodules were used in PHB determination. Three independent experiments were performed.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dieter Jendrossek from Universität Stuttgart for providing the vector carrying eYFP.
This work was supported by the National Natural Science Foundation of China (31522003), the National Basic Research Program of China (973 program 2015CB158301), and the Innovative Project of State Key Laboratory of Agrobiotechnology (2018SKLAB1-2).
We declare that we do not have any commercial or associative interest connected with the work described here.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00717-19.
REFERENCES
- 1.Maestro B, Sanz JM. 2017. Polyhydroxyalkanoate-associated phasins as phylogenetically heterogeneous, multipurpose proteins. Microb Biotechnol 10:1323–1337. doi: 10.1111/1751-7915.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jendrossek D, Pfeiffer D. 2014. New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly (3-hydroxybutyrate). Environ Microbiol 16:2357–2373. doi: 10.1111/1462-2920.12356. [DOI] [PubMed] [Google Scholar]
- 3.Kadouri D, Jurkevitch E, Okon Y. 2003. Involvement of the reserve material poly-β-hydroxybutyrate in Azospirillum brasilense stress endurance and root colonization. Appl Environ Microbiol 69:3244–3250. doi: 10.1128/aem.69.6.3244-3250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q, Yu H, Xia Y, Kang Z, Qi Q. 2009. Complete PHB mobilization in Escherichia coli enhances the stress tolerance: a potential biotechnological application. Microb Cell Fact 8:47. doi: 10.1186/1475-2859-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao YH, Li HM, Qin LF, Wang HH, Chen GQ. 2007. Disruption of the polyhydroxyalkanoate synthase gene in Aeromonas hydrophila reduces its survival ability under stress conditions. FEMS Microbiol Lett 276:34–41. doi: 10.1111/j.1574-6968.2007.00904.x. [DOI] [PubMed] [Google Scholar]
- 6.Koskimäki JJ, Kajula M, Hokkanen J, Ihantola EL, Kim JH, Hautajärvi H, Hankala E, Suokas M, Pohjanen J, Podolich O, Kozyrovska N, Turpeinen A, Pääkkönen M, Mattila S, Campbell BC, Pirttilä AM. 2016. Methyl-esterified 3-hydroxybutyrate oligomers protect bacteria from hydroxyl radicals. Nat Chem Biol 12:332–338. doi: 10.1038/nchembio.2043. [DOI] [PubMed] [Google Scholar]
- 7.Nowroth V, Marquart L, Jendrossek D. 2016. Low temperature-induced viable but not culturable state of Ralstonia eutropha and its relationship to accumulated polyhydroxybutyrate. FEMS Microbiol Lett 363:fnw249. doi: 10.1093/femsle/fnw249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Obruca S, Sedlacek P, Koller M, Kucera D, Pernicova I. 2018. Involvement of polyhydroxyalkanoates in stress resistance of microbial cells: biotechnological consequences and applications. Biotechnol Adv 36:856–870. doi: 10.1016/j.biotechadv.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Soto G, Setten L, Lisi C, Maurelis C, Mozzicafreddo M, Cuccioloni M, Angeletti M, Ayub ND. 2012. Hydroxybutyrate prevents protein aggregation in the halotolerant bacterium Pseudomonas sp. CT13 under abiotic stress. Extremophiles 16:455–462. doi: 10.1007/s00792-012-0445-0. [DOI] [PubMed] [Google Scholar]
- 10.Obruca S, Sedlacek P, Mravec F, Samek O, Marova I. 2016. Evaluation of 3-hydroxybutyrate as an enzyme-protective agent against heating and oxidative damage and its potential role in stress response of poly(3-hydroxybutyrate) accumulating cells. Appl Microbiol Biotechnol 100:1365–1376. doi: 10.1007/s00253-015-7162-4. [DOI] [PubMed] [Google Scholar]
- 11.de Almeida A, Catone MV, Rhodius VA, Gross CA, Pettinari MJ. 2011. Unexpected stress-reducing effect of PhaP, a poly(3-hydroxybutyrate) granule-associated protein, in Escherichia coli. Appl Environ Microbiol 77:6622–6629. doi: 10.1128/AEM.05469-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mezzina MP, Wetzler DE, De Almeida A, Dinjaski N, Prieto MA, Pettinari MJ. 2015. A phasin with extra talents: a polyhydroxyalkanoate granule-associated protein has chaperone activity. Environ Microbiol 17:1765–1776. doi: 10.1111/1462-2920.12636. [DOI] [PubMed] [Google Scholar]
- 13.Mezzina MP, Pettinari MJ. 2016. Phasins, multifaceted polyhydroxyalkanoate granule-associated proteins. Appl Environ Microbiol 82:5060–5067. doi: 10.1128/AEM.01161-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bresan S, Sznajder A, Hauf W, Forchhammer K, Pfeiffer D, Jendrossek D. 2016. Polyhydroxyalkanoate (PHA) granules have no phospholipids. Sci Rep 6:26612. doi: 10.1038/srep26612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jendrossek D. 2009. Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J Bacteriol 191:3195–3202. doi: 10.1128/JB.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bresan S, Jendrossek D. 2017. New insights into PhaM-PhaC-mediated localization of polyhydroxybutyrate granules in Ralstonia eutropha H16. Appl Environ Microbiol 83:e00505-17. doi: 10.1128/AEM.00505-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfeiffer D, Jendrossek D. 2014. PhaM is the physiological activator of poly(3-hydroxybutyrate) (PHB) synthase (PhaC1) in Ralstonia eutropha. Appl Environ Microbiol 80:555–563. doi: 10.1128/AEM.02935-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeiffer D, Jendrossek D. 2012. Localization of poly(3-hydroxybutyrate) (PHB) granule-associated proteins during PHB granule formation and identification of two new phasins, PhaP6 and PhaP7, in Ralstonia eutropha H16. J Bacteriol 194:5909–5921. doi: 10.1128/JB.00779-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon R, Kahn D. 2004. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol 2:621–631. doi: 10.1038/nrmicro954. [DOI] [PubMed] [Google Scholar]
- 20.Udvardi M, Poole PS. 2013. Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol 64:781–805. doi: 10.1146/annurev-arplant-050312-120235. [DOI] [PubMed] [Google Scholar]
- 21.Poole P, Ramachandran V, Terpolilli J. 2018. Rhizobia: from saprophytes to endosymbionts. Nat Rev Microbiol 16:291–303. doi: 10.1038/nrmicro.2017.171. [DOI] [PubMed] [Google Scholar]
- 22.Li YZ, Wang D, Feng XY, Jiao J, Chen WX, Tian CF. 2016. Genetic analysis reveals the essential role of nitrogen phosphotransferase system components in Sinorhizobium fredii CCBAU 45436 symbioses with soybean and pigeonpea plants. Appl Environ Microbiol 82:1305–1315. doi: 10.1128/AEM.03454-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C, Saldanha M, Sheng X, Shelswell KJ, Walsh KT, Sobral BWS, Charles TC. 2007. Roles of poly-3-hydroxybutyrate (PHB) and glycogen in symbiosis of Sinorhizobium meliloti with Medicago sp. Microbiology 153:388–398. doi: 10.1099/mic.0.29214-0. [DOI] [PubMed] [Google Scholar]
- 24.Hu Y, Jiao J, Liu LX, Sun YW, Chen W, Sui X, Chen W, Tian CF. 2018. Evidence for phosphate starvation of rhizobia without terminal differentiation in legume nodules. Mol Plant Microbe Interact 31:1060–1068. doi: 10.1094/MPMI-02-18-0031-R. [DOI] [PubMed] [Google Scholar]
- 25.Yuan ZC, Zaheer R, Finan TM. 2006. Regulation and properties of PstSCAB, a high-affinity, high-velocity phosphate transport system of Sinorhizobium meliloti. J Bacteriol 188:1089–1102. doi: 10.1128/JB.188.3.1089-1102.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trainer MA, Capstick D, Zachertowska A, Lam KN, Clark SRD, Charles TC. 2010. Identification and characterization of the intracellular poly-3-hydroxybutyrate depolymerase enzyme PhaZ of Sinorhizobium meliloti. BMC Microbiol 10:92. doi: 10.1186/1471-2180-10-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, Sheng X, Equi RC, Trainer MA, Charles TC, Sobral BW. 2007. Influence of the poly-3-hydroxybutyrate (PHB) granule-associated proteins (PhaP1 and PhaP2) on PHB accumulation and symbiotic nitrogen fixation in Sinorhizobium meliloti Rm1021. J Bacteriol 189:9050–9056. doi: 10.1128/JB.01190-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cevallos MA, Encarnación S, Leija A, Mora Y, Mora J. 1996. Genetic and physiological characterization of a Rhizobium etli mutant strain unable to synthesize poly-beta-hydroxybutyrate. J Bacteriol 178:1646–1654. doi: 10.1128/jb.178.6.1646-1654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandon K, Michel-Reydellet N, Encarnacio S, Kaminski PA, Leija A, Cevallos MA, Elmerich C, Mora J. 1998. Poly-β-hydroxybutyrate turnover in Azorhizobium caulinodans is required for growth and affects nifA expression. J Bacteriol 180:5070–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida KI, Takemoto Y, Sotsuka T, Tanaka K, Takenaka S. 2013. PhaP phasins play a principal role in poly-β-hydroxybutyrate accumulation in free-living Bradyrhizobium japonicum. BMC Microbiol 13:290. doi: 10.1186/1471-2180-13-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quelas JI, Mesa S, Mongiardini EJ, Jendrossek D, Lodeiro AR. 2016. Regulation of polyhydroxybutyrate synthesis in the soil bacterium Bradyrhizobium diazoefficiens. Appl Environ Microbiol 82:4299–4308. doi: 10.1128/AEM.00757-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Tian CF, Chen WF, Wang L, Sui XH, Chen WX. 2013. High-resolution transcriptomic analyses of Sinorhizobium sp. NGR234 bacteroids in determinate nodules of Vigna unguiculata and indeterminate nodules of Leucaena leucocephala. PLoS One 8:e70531. doi: 10.1371/journal.pone.0070531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vinardell J-M, Acosta-Jurado S, Zehner S, Göttfert M, Becker A, Baena I, Blom J, Crespo-Rivas JC, Goesmann A, Jaenicke S, Krol E, McIntosh M, Margaret I, Pérez-Montaño F, Schneiker-Bekel S, Serranía J, Szczepanowski R, Buendía A-M, Lloret J, Bonilla I, Pühler A, Ruiz-Sainz J-E, Weidner S. 2015. The Sinorhizobium fredii HH103 genome: a comparative analysis with S. fredii strains differing in their symbiotic behavior with soybean. Mol Plant-Microbe Interact 28:811–824. doi: 10.1094/MPMI-12-14-0397-FI. [DOI] [PubMed] [Google Scholar]
- 34.Jiao J, Ni M, Zhang B, Zhang Z, Young JPW, Chan T-F, Chen WX, Lam H-M, Tian CF. 2018. Coordinated regulation of core and accessory genes in the multipartite genome of Sinorhizobium fredii. PLoS Genet 14:e1007428. doi: 10.1371/journal.pgen.1007428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aneja P, Zachertowska A, Charles T. 2005. Comparison of the symbiotic and competition phenotypes of Sinorhizobium meliloti PHB synthesis and degradation pathway mutants. Can J Microbiol 51:599–604. doi: 10.1139/w05-042. [DOI] [PubMed] [Google Scholar]
- 36.Vranova V, Zahradnickova H, Janous D, Skene KR, Matharu AS, Rejsek K, Formanek P. 2012. The significance of D-amino acids in soil, fate and utilization by microbes and plants: review and identification of knowledge gaps. Plant Soil 354:21–39. doi: 10.1007/s11104-011-1059-5. [DOI] [Google Scholar]
- 37.Aliashkevich A, Alvarez L, Cava F. 2018. New insights into the mechanisms and biological roles of D-amino acids in complex eco-systems. Front Microbiol 9:683. doi: 10.3389/fmicb.2018.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reed JL, Patel TR, Chen KH, Joyce AR, Applebee MK, Herring CD, Bui OT, Knight EM, Fong SS, Palsson BO. 2006. Systems approach to refining genome annotation. Proc Natl Acad Sci U S A 103:17480–17484. doi: 10.1073/pnas.0603364103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai G, Driscoll BT, Charles TC, Cai G, Driscoll BT. 2000. Requirement for the enzymes acetoacetyl coenzyme A synthetase and poly-3-hydroxybutyrate (PHB) synthase for growth of Sinorhizobium meliloti on PHB cycle intermediates. J Bacteriol 182:2113–2118. doi: 10.1128/JB.182.8.2113-2118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knoll M, Hamm TM, Wagner F, Martinez V, Pleiss J. 2009. The PHA Depolymerase Engineering Database: a systematic analysis tool for the diverse family of polyhydroxyalkanoate (PHA) depolymerases. BMC Bioinformatics 10:89. doi: 10.1186/1471-2105-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Handrick R, Reinhardt S, Focarete ML, Scandola M, Adamus G, Kowalczuk M, Jendrossek D. 2001. A new type of thermoalkalophilic hydrolase of Paucimonas lemoignei with high specificity for amorphous polyesters of short chain-length hydroxyalkanoic acids. J Biol Chem 276:36215–36224. doi: 10.1074/jbc.M101106200. [DOI] [PubMed] [Google Scholar]
- 42.Aneja P, Charles TC. 2005. Characterization of bdhA, encoding the enzyme D-3-hydroxybutyrate dehydrogenase, from Sinorhizobium sp strain NGR234. FEMS Microbiol Lett 242:87–94. doi: 10.1016/j.femsle.2004.10.043. [DOI] [PubMed] [Google Scholar]
- 43.Lodwig EM, Leonard M, Marroqui S, Wheeler TR, Findlay K, Downie JA, Poole PS. 2005. Role of polyhydroxybutyrate and glycogen as carbon storage compounds in pea and bean bacteroids. Mol Plant Microbe Interact 18:67–74. doi: 10.1094/MPMI-18-0067. [DOI] [PubMed] [Google Scholar]
- 44.Pfeiffer D, Wahl A, Jendrossek D. 2011. Identification of a multifunctional protein, PhaM, that determines number, surface to volume ratio, subcellular localization and distribution to daughter cells of poly (3-hydroxybutyrate), PHB, granules in Ralstonia eutropha H16. Mol Microbiol 82:936–951. doi: 10.1111/j.1365-2958.2011.07869.x. [DOI] [PubMed] [Google Scholar]
- 45.Mu H, Steinbu A. 2005. Influence of homologous phasins (PhaP) on PHA accumulation and regulation of their expression by the transcriptional repressor PhaR in Ralstonia eutropha H16. Microbiology 151:825–833. [DOI] [PubMed] [Google Scholar]
- 46.Kuchta K, Chi L, Fuchs H, Pötter M, Steinbüchel A. 2007. Studies on the influence of phasins on accumulation and degradation of PHB and nanostructure of PHB granules in Raistonia eutropha H16. Biomacromolecules 8:657–662. doi: 10.1021/bm060912e. [DOI] [PubMed] [Google Scholar]
- 47.Yurgel SN, Kahn ML. 2008. A mutant GlnD nitrogen sensor protein leads to a nitrogen-fixing but ineffective Sinorhizobium meliloti symbiosis with alfalfa. Proc Natl Acad Sci U S A 105:18958–18963. doi: 10.1073/pnas.0808048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu LX, Li QQ, Zhang YZ, Hu Y, Jiao J, Guo HJ, Zhang XX, Zhang B, Chen WX, Tian CF. 2017. The nitrate-reduction gene cluster components exert lineage-dependent contributions to optimization of Sinorhizobium symbiosis with soybeans. Environ Microbiol 19:4926–4938. doi: 10.1111/1462-2920.13948. [DOI] [PubMed] [Google Scholar]
- 49.Sznajder A, Pfeiffer D, Jendrossek D. 2015. Polyhydroxybutyrate (PHB) granule-associated proteins in Ralstonia eutropha H16. Appl Environ Microbiol 81:1847–1858. doi: 10.1128/AEM.03791-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian CF, Zhou YJ, Zhang YM, Li QQ, Zhang YZ, Li DF, Wang S, Wang J, Gilbert LB, Li YR, Chen WX. 2012. Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations. Proc Natl Acad Sci U S A 109:8629–8634. doi: 10.1073/pnas.1120436109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Remigi P, Zhu J, Young JPW, Masson-Boivin C. 2016. Symbiosis within symbiosis: evolving nitrogen-fixing legume symbionts. Trends Microbiol 24:63–75. doi: 10.1016/j.tim.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 52.Capela D, Marchetti M, Clérissi C, Perrier A, Guetta D, Gris C, Valls M, Jauneau A, Cruveiller S, Rocha EPC, Masson-Boivin C. 2017. Recruitment of a lineage-specific virulence regulatory pathway promotes intracellular infection by a plant pathogen experimentally evolved into a legume symbiont. Mol Biol Evol 34:2503–2521. doi: 10.1093/molbev/msx165. [DOI] [PubMed] [Google Scholar]
- 53.Pueppke SG, Broughton WJ. 1999. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol Plant Microbe Interact 12:293–318. doi: 10.1094/MPMI.1999.12.4.293. [DOI] [PubMed] [Google Scholar]
- 54.Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 55.Ditta G, Stanfield S, Corbin D, Helinski DR. 1980. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A 77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oke V, Long SR. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol Microbiol 32:837–849. doi: 10.1046/j.1365-2958.1999.01402.x. [DOI] [PubMed] [Google Scholar]
- 57.Marx CJ, Lidstrom ME. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33:1062–1067. doi: 10.2144/02335rr01. [DOI] [PubMed] [Google Scholar]
- 58.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 59.Vincent JM. 1970. A manual for the practical study of root nodule bacteria. Blackwell Scientific, London, United Kingdom. [Google Scholar]
- 60.Ling Q, Huang W, Jarvis P. 2011. Use of a SPAD-502 meter to measure leaf chlorophyll concentration in Arabidopsis thaliana. Photosynth Res 107:209–214. doi: 10.1007/s11120-010-9606-0. [DOI] [PubMed] [Google Scholar]
- 61.Van de Velde W, Guerra JCP, de Keyser A, de Rycke R, Rombauts S, Maunoury N, Mergaert P, Kondorosi E, Holsters M, Goormachtig S. 2006. Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula. Plant Physiol 141:711–720. doi: 10.1104/pp.106.078691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spiekermann P, Rehm BHA, Kalscheuer R, Baumeister D, Steinbüchel A. 1999. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch Microbiol 171:73–80. doi: 10.1007/s002030050681. [DOI] [PubMed] [Google Scholar]
- 63.Preusting H, Kingma J, Huisman G, Steinbüchel A, Witholt B. 1993. Formation of polyester blends by a recombinant strain of Pseudomonas oleovorans: different poly(3-hydroxyalkanoates) are stored in separate granules. J Environ Polym Degrad 1:11–21. doi: 10.1007/BF01457649. [DOI] [Google Scholar]
- 64.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. 2005. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Law JH, Slepecky RA, Law JH, Slepecky RA. 1961. Assay of poly-β-hydroxybutyric acid. J Bacteriol 82:33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quelas JI, Mongiardini EJ, Pérez-Giménez J, Parisi G, Lodeiro AR. 2013. Analysis of two polyhydroxyalkanoate synthases in Bradyrhizobium japonicum USDA 110. J Bacteriol 195:3145–3155. doi: 10.1128/JB.02203-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.