The discovery of new plant growth-promoting bacteria is necessary for the continued development of biofertilizers, which are environmentally friendly and cost-efficient alternatives to conventional chemical fertilizers. Biofertilizer effects on plant growth can be inconsistent due to the complexity of plant-microbe interactions, as the same bacteria can be beneficial to the growth of some plant species and neutral or detrimental to others. We examined a set of bacterial endophytes isolated from plants growing in a unique petroleum-contaminated environment to discover plant growth-promoting bacteria. We show that strains of Plantibacter flavus exhibit strain-specific plant growth-promoting effects on four different plant species.
KEYWORDS: Arabidopsis, endophyte, host specificity, plant growth promotion, plant microbiology, Plantibacter flavus, basil, bok choy, lettuce
ABSTRACT
A collection of bacterial endophytes isolated from stem tissues of plants growing in soils highly contaminated with petroleum hydrocarbons were screened for plant growth-promoting capabilities. Twenty-seven endophytic isolates significantly improved the growth of Arabidopsis thaliana plants in comparison to that of uninoculated control plants. The five most beneficial isolates, one strain each of Curtobacterium herbarum, Paenibacillus taichungensis, and Rhizobium selenitireducens and two strains of Plantibacter flavus were further examined for growth promotion in Arabidopsis, lettuce, basil, and bok choy plants. Host-specific plant growth promotion was observed when plants were inoculated with the five bacterial strains. P. flavus strain M251 increased the total biomass and total root length of Arabidopsis plants by 4.7 and 5.8 times, respectively, over that of control plants and improved lettuce and basil root growth, while P. flavus strain M259 promoted Arabidopsis shoot and root growth, lettuce and basil root growth, and bok choy shoot growth. A genome comparison between P. flavus strains M251 and M259 showed that both genomes contain up to 70 actinobacterial putative plant-associated genes and genes involved in known plant-beneficial pathways, such as those for auxin and cytokinin biosynthesis and 1-aminocyclopropane-1-carboxylate deaminase production. This study provides evidence of direct plant growth promotion by Plantibacter flavus.
IMPORTANCE The discovery of new plant growth-promoting bacteria is necessary for the continued development of biofertilizers, which are environmentally friendly and cost-efficient alternatives to conventional chemical fertilizers. Biofertilizer effects on plant growth can be inconsistent due to the complexity of plant-microbe interactions, as the same bacteria can be beneficial to the growth of some plant species and neutral or detrimental to others. We examined a set of bacterial endophytes isolated from plants growing in a unique petroleum-contaminated environment to discover plant growth-promoting bacteria. We show that strains of Plantibacter flavus exhibit strain-specific plant growth-promoting effects on four different plant species.
INTRODUCTION
In recent years, associations between plants and their microbiomes have been increasingly scrutinized (1, 2). The discovery of numerous plant growth-promoting bacteria (PGPB) in the rhizosphere of plants has spawned more recent searches for such beneficial associations in the endosphere. PGPB have been isolated from a wide range of stressful environments, from the saline coast of the Yellow Sea (3) to acidic and metallic mine tailings (4) and the cold deserts of the Himalayas (5). PGPB can directly improve plant growth through various mechanisms, including improved nutrient acquisition, such as nitrogen and phosphorus uptake through nitrogen fixation (6) and phosphate solubilization (7), respectively, increasing the production of plant growth hormones like auxins, cytokinins, and gibberellins (8) and producing stress tolerance enzymes like 1-aminocyclopropane-1-carboxylate (ACC) deaminase (9). Some other benefits include the breakdown of toxic metals or pollutants and improved iron acquisition and pathogen protection through siderophore production (10).
PGPB have great potential for improving plant yield as natural biofertilizers (11–13). Traditionally, agriculture has relied on chemical fertilizers to provide plants with essential nutrients that may be limited in soil. The downsides to using chemical fertilizers are that they have long-term detrimental effects on the environment: they can leave residual salts in soils, which decrease long-term soil fertility, enter bodies of water through runoff, resulting in eutrophication (14), and decrease overall biodiversity (15). Biofertilizers, which contain beneficial microorganisms or natural compounds originating from microbes (16–18), are sustainable alternatives to stop or reduce the use of chemical fertilizers in organic and conventional agricultural systems. Biofertilizers are also advantageous in that they are more cost-efficient, provide a renewable source of nutrients, and can provide plants with a multitude of different benefits (19–23). Currently, biofertilizers are most commonly used as a means of improving crop nitrogen and phosphorus uptake through the application of PGPB like Azotobacter, Azospirillum, Bacillus, and Rhizobium species (11, 24–30).
The main limitation of using PGPB is that their benefits are dependent on the capability of plant-microbe associations. Plant benefits like nutrients, phytohormones, and siderophores are available only if they can be synthesized by microbes when associated with plants; however, plant-microbe associations can differ depending on plant species and environmental conditions (31). If the interaction is not favorable, then the growth of microbes may be limited, meaning that plant benefits from biofertilizers would be minimal. It is therefore important to discover a wide variety of PGPB that will improve the effectiveness and versatility of biofertilizers throughout different plants and environments.
The aim of our study was to examine a large collection of bacterial endophytes for plant growth promotion abilities. The bacteria in this collection were isolated from plants growing at a site where the atmosphere and soils were heavily contaminated by petroleum hydrocarbons (32). As stated by Lumactud et al. (32), soils sampled from the site had high levels of crude oil contamination (250,000 to 300,000 ppm). However, despite the toxicity of oil residues, plant growth appeared unimpeded. We hypothesized that these plants contain beneficial bacterial endophytes that are facilitating their survival through hydrocarbon degradation and/or direct improvement of shoot and root growth. Hydrocarbon degradation was previously examined and found to be present, although not dominant, in the collection (32), so the focus of this study was on direct plant growth promotion. To identify PGPB, the endophyte collection was screened for growth promotion using in planta rapid screens with Arabidopsis thaliana (herein referred to as Arabidopsis), and then the most promising beneficial bacteria were tested for growth promotion of lettuce, bok choy, and basil.
RESULTS
Microtiter plate growth promotion screens with Arabidopsis thaliana.
Inoculation with 18 of the 220 endophyte isolates (8%) resulted in Arabidopsis growth improvement, as measured by significant increases in number of buds and flowers, stem height, and/or total biomass in comparison to that of control plants for growth at a P of <0.05, while inoculation with 27 of the 220 endophyte isolates (12%) showed similar improvements at a P of <0.1 (Table 1). In total, 13 different genera were found to significantly improve Arabidopsis growth, with the majority of these improvements measured by increases in stem height, number of buds, and number of flowers.
TABLE 1.
Summary of plant growth promotion by different bacterial genera in the endophyte collection
| Endophyte genus | No. of isolates demonstrating significant plant growth promotion (P < 0.1) towards: |
% of growth-promoting isolates in this genus (present study) | |||
|---|---|---|---|---|---|
| Stem ht | No. of buds and/or flowers | Fresh biomass | Any trait | ||
| Bacillus | 3 | 4 | 1 | 5 | 20.8 |
| Curtobacterium | 1 | 2 | 1 | 3 | 18.8 |
| Microbacterium | 2 | 1 | 0 | 2 | 9.1 |
| Plantibacter | 2 | 2 | 0 | 2 | 25 |
| Arthrobacter | 0 | 1 | 1 | 2 | 25 |
| Brevundimonas | 1 | 1 | 0 | 1 | 20 |
| Pseudomonas | 1 | 1 | 0 | 1 | 8.3 |
| Rhizobium | 1 | 1 | 0 | 1 | 25 |
| Paenibacillus | 1 | 1 | 1 | 1 | 25 |
| Clavibacter | 0 | 1 | 0 | 1 | 16.7 |
| Micrococcus | 1 | 0 | 0 | 1 | 100 |
| Methylobacterium | 1 | 1 | 0 | 1 | 50 |
| Serratia | 1 | 0 | 0 | 1 | 50 |
| Unidentified genus | 2 | 4 | 1 | 6 | 10 |
| Total | 17 | 20 | 5 | 28 | 12.3 |
Five of the most promising plant growth-promoting bacteria (i.e., the “top five”) were selected for in-depth examinations based on their observed growth improvement of multiple Arabidopsis traits (Table 2). Isolate M132 was identified as Curtobacterium herbarum, M175 as Paenibacillus taichungensis, M259 as Plantibacter flavus, and M267 as Rhizobium selenitireducens. Isolate M251 had previously been identified as Plantibacter flavus strain 251 (33).
TABLE 2.
Species identities and growth benefits for Arabidopsis by the top five endophytes
| Isolate code | Arabidopsis trait improvements | Significance level | Species identity |
|---|---|---|---|
| M132 | Stem ht | 0.1 | Curtobacterium herbarum |
| No. of flowers | 0.05 | ||
| M175 | Stem ht | 0.05 | Paenibacillus taichungensis |
| No. of buds | 0.1 | ||
| Total biomass | 0.1 | ||
| M251 | Stem ht | 0.05 | Plantibacter flavus |
| No. of buds | 0.05 | ||
| No. of flowers | 0.1 | ||
| M259 | Stem ht | 0.05 | Plantibacter flavus |
| No. of buds | 0.05 | ||
| No. of flowers | 0.05 | ||
| M267 | Stem ht | 0.05 | Rhizobium selenitireducens |
| No. of buds | 0.1 | ||
| No. of flowers | 0.05 |
GA-7 box growth promotion tests with Arabidopsis thaliana.
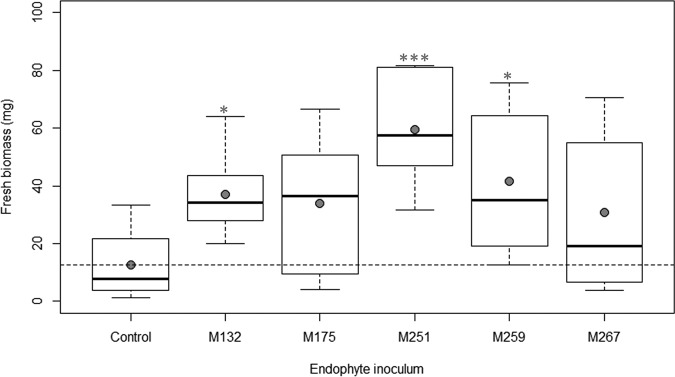
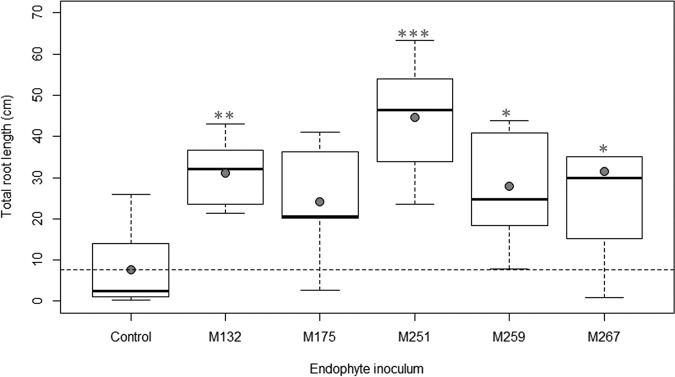
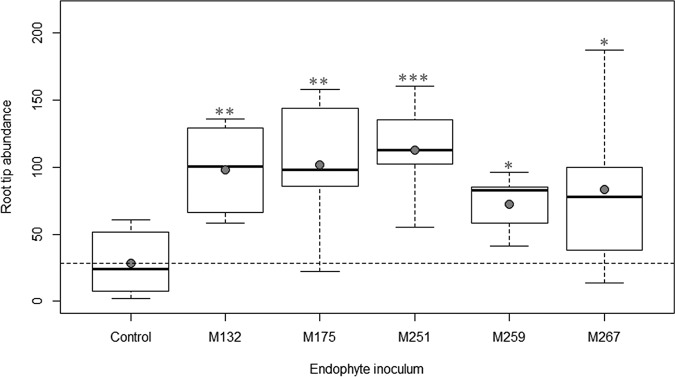
Inoculation of Arabidopsis with three of the top five endophytes (isolates M132, M251, and M259) resulted in significant improvement in total biomass at 21 days after inoculation in comparison to control plants (Fig. 1). Inoculation with four of the isolates (M132, M251, M259, and M267) increased Arabidopsis total root length, while all five isolates increased root tip abundance (Fig. 2 and 3). The greatest beneficial effects for Arabidopsis growth were seen from inoculation with M251, as these plants displayed a 4.7-fold increase in total biomass, a 5.8-fold increase in total root length, and a 3.9-fold increase in root tip abundance over uninoculated plants (all with a P of <0.001).
FIG 1.
Effects of the top five endophytes on total biomass of Arabidopsis plants at 21 days after inoculation. Filled circles represent means of results for each treatment. Horizontal dashed lines represent means of results for control plants. Statistical significance in comparison to control plants is noted with one (P < 0.05) or three (P < 0.001) asterisks.
FIG 2.
Effects of the top five endophytes on total root length of Arabidopsis plants at 21 days after inoculation. Filled circles represent means of results for each treatment. Horizontal dashed lines represent means of results for control plants. Statistical significance in comparison to control plants is noted with one (P < 0.05), two (P < 0.01), or three (P < 0.001) asterisks.
FIG 3.
Effects of the top five endophytes on root tip abundance of Arabidopsis plants. Filled circles represent means of results for each treatment. Horizontal dashed lines represent means of results for control plants. Statistical significance in comparison to control plants is noted with one (P < 0.05), two (P < 0.01), or three (P < 0.001) asterisks.
Aquaponic growth promotion tests with lettuce, basil, and bok choy.
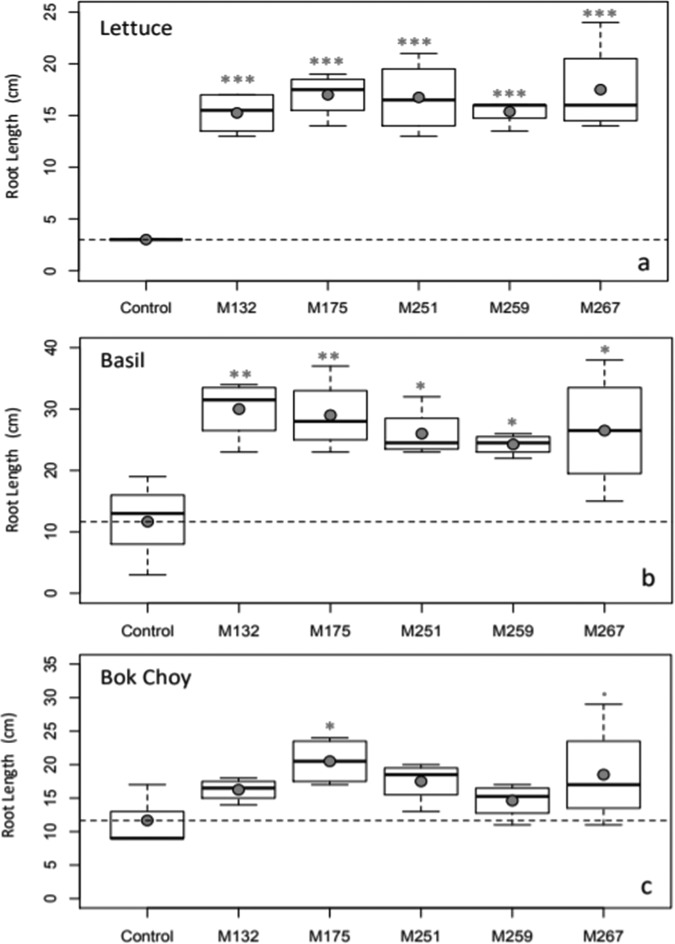
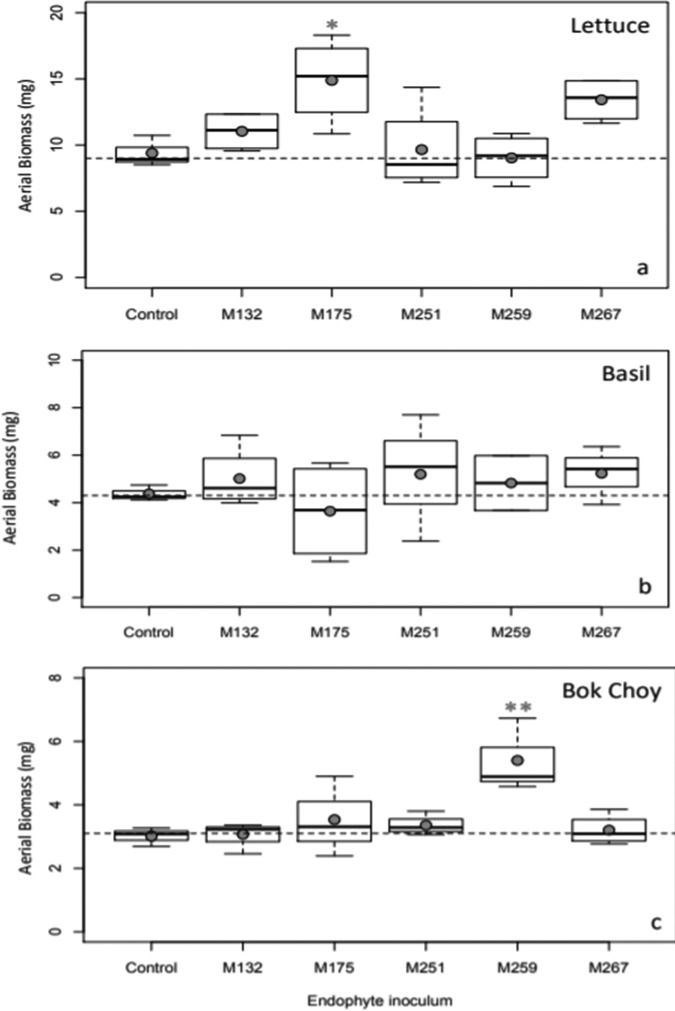
Plant inoculation with all of the top five isolates significantly improved the total root lengths of lettuce and basil, while inoculation only with isolates Paenibacillus taichungensis M175 and Rhizobium selenitireducens M267 significantly improved total root length of bok choy compared to uninoculated controls (Fig. 4). Isolates Paenibacillus taichungensis M175 and Plantibacter flavus M259 also improved aerial biomass for lettuce and bok choy, respectively (Fig. 5).
FIG 4.
Effects of the top five endophytes on root length of lettuce, basil, and bok choy plants. Filled circles represent means of results for each treatment. Horizontal dashed lines represent means of results for control plants. Statistical significance in comparison to control plants is noted with one (P < 0.05), two (P < 0.01), or three (P < 0.001) asterisks or with one dot (P < 0.1).
FIG 5.
Effects of the top five endophytes on aerial biomass of lettuce, basil, and bok choy plants. Filled circles represent means of results for each treatment. Horizontal dashed lines represent means of results for control plants. Statistical significance in comparison to control plants is noted with one (P < 0.05) or two (P < 0.01) asterisks.
Genomic comparison between Plantibacter flavus strain M251 and Plantibacter flavus strain M259 genomes.
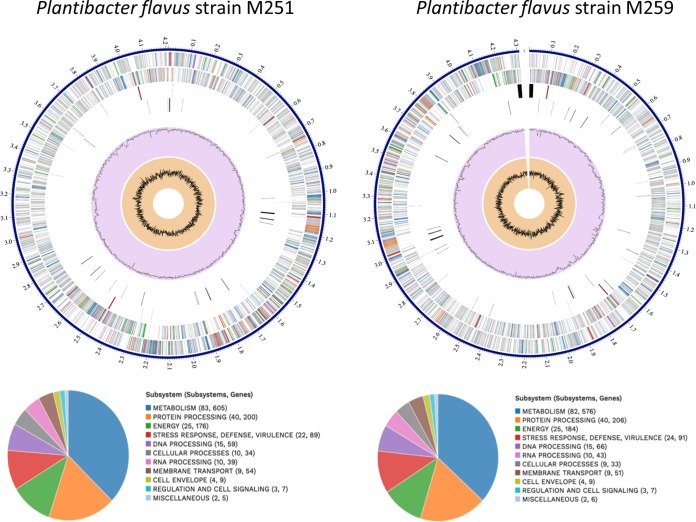
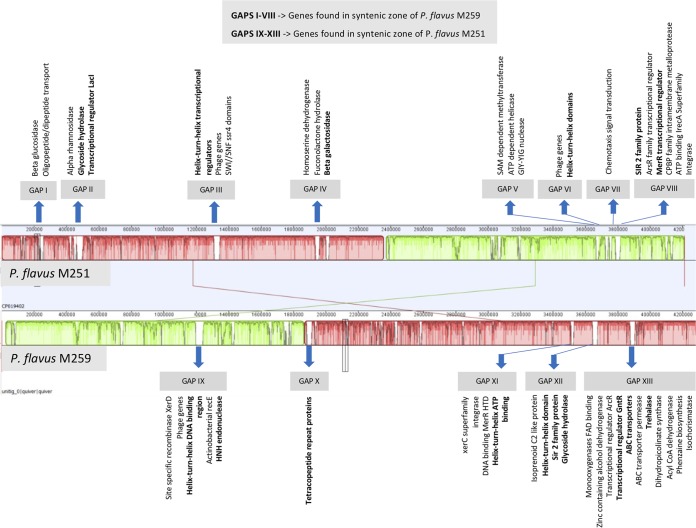
PacBio sequencing and assembly yielded one contig for M259, as it did previously for strain M251 (32). The full genomes of P. flavus strain M251 (4.2 Mbp) and P. flavus strain M259 (4.3 Mbp) and the summary of their metabolic features are depicted in Fig. 6. Both are specialized in carbohydrate and amino acid metabolism. Other features are listed in Tables 3 and 4. The two organisms are highly similar in size, nucleotides, and GC content. The Mauve alignment shows the genomes to be highly similar (Fig. 7), consisting of two large colinear blocks reflecting differing points of sequence origin. There are several regions missing from each of the genomes relative to each other. An examination of these islands proved that they consisted mostly of hypothetical proteins. Therefore, we conducted a PSI-BLAST search on the proteins. A number of these islands contained mobility-related genes (encoding phages, recombinases, and integrases) but also genes of other functions of interest (see below). Both P. flavus genomes contained genes for the synthesis of auxin, cytokinin, and ACC deaminase (1-aminocyclopropane-1-carboxylate deaminase). Only M251 contained genes for siderophore biosynthesis, and only P. flavus M259 contained genes for phosphate solubilization (Table 4). In addition, both contained numerous genes related to microbial antibiotic resistance: a comparison to DrugBank, CARD, and PATRIC AMR databases detected 27 genes related to antibiotic resistance in P. flavus strain M251 and 29 genes in P. flavus strain M259 (Table 4). Furthermore, using antiSMASH 5.0, we identified gene clusters in both genomes coding for production of secondary metabolites such as arginomycin (nonribosomal peptide synthetase), meilingmycin (T3PKS/type 3 polyketide synthase), carotenoid (terpene), and microansamycin (beta-lactone). All of these results attest to the high degree of similarity of the strains.
FIG 6.
Full genomes of Plantibacter flavus strains M251 and M259, showing general metabolic features (subsystems and number of annotated genes).
TABLE 3.
Comparison of general genomic characteristics between Plantibacter flavus strains M251 and M259
| Characteristic | Value for: |
|
|---|---|---|
| P. flavus strain M251 | P. flavus strain M259 | |
| No. of contigs | 1 | 1 |
| GC content (mol%) | 69.18 | 69.01 |
| No. of plasmids | 0 | 0 |
| Genome length (bp) | 4,211,682 | 4,311,036 |
| No. of conserved domains | 4,047 | 4,624 |
| No. of tRNA genes | 49 | 48 |
| No. of repeat regions | 7 | 24 |
| No. of rRNA genes | 6 | 6 |
| No. of hypothetical proteins | 1,601 | 1,786 |
| No. of proteins with functional assignments | 2,406 | 2,478 |
| No. of proteins with EC number assignments | 875 | 879 |
| No. of proteins with GO assignments | 758 | 767 |
| No. of proteins with pathway assignments | 674 | 683 |
| No. of proteins with PATRIC genus-specific family (PLfam) assignments | 3,972 | 3,791 |
| No. of proteins with PATRIC cross-genus family (PGfam) assignments | 3,973 | 3,875 |
| No. of genes related to antibiotic resistance in CARD database | 1 | 1 |
| No. of genes related to antibiotic resistance in NDARO database | 0 | 1 |
| No. of genes related to antibiotic resistance in PATRIC database | 27 | 29 |
| No. of genes with drug target (DrugBank) | 1 | 1 |
TABLE 4.
Genomic comparison between Plantibacter flavus strains M251 and M259
| Gene or protein and function | Presence of gene in indicated P. flavus strain |
|
|---|---|---|
| M251 | M259 | |
| Antibiotic resistance | ||
| Alr, Ddl, dxr, EF-G, EF-Tu, folA, Dfr, folP, gyrA, gyrB, iso-tRNA, kasA, MurA, rho, rpoB, rpoC, S10p, S12p | + | + |
| FabL-like | + | + |
| Cmx family | − | + |
| gidB | + | + |
| GdpD, PgsA | + | |
| MtrA, MtrB, VanO type | + | + |
| Plant growth promotion | ||
| ACC_deaminase (1-aminocyclopropane-1-carboxylate deaminase) | + | + |
| Siderophores | + | − |
| Auxin biosynthesis | + | + |
| Cytokinin biosynthesis | + | + |
| Phosphate solubilization | − | + |
| Biosynthesis of new products (antiSMASH 5.0.0rc1 search) | ||
| Arginomycin (nonribosomal peptide synthetase) | + | + |
| Meilingmycin (type 1 polyketide synthase [T1PKS]) | + | |
| Carotenoid (terpene) | + | |
| Microansamycin (beta-lactone) | + | |
| T3PKS-like product | ||
FIG 7.
Mauve alignment of Plantibacter flavus strain M251 and M259 genomes showing areas of missing genes in one relative to the other (gene islands). The sequence numbers of islands are given in Table S2 in the supplemental material. Gene family names in bold have functions equivalent to items in the putative plant association gene list of Levy et al. (34).
In a search for genes more relevant to plant association, we compared the proteomes of the two P. flavus strains to a list of 767 actinobacterial putative plant-associated protein families recently published by Levy et al. (34). From this search, we found that P. flavus strains M251 and M259 contained 53 and 70 putative plant-associated proteins, respectively (see Table S1 in the supplemental material). Several of the hypothetical proteins in the strain-specific islands proved to have functions similar to those present in the list of Levy et al. (34). These are depicted in Fig. 7 and listed in Table S2.
DISCUSSION
The largest and most developed Arabidopsis plants were generally those inoculated with the two strains of Plantibacter flavus. Similar results were seen with bok choy, which, in comparison to lettuce and basil, has the closest taxonomic relationship with Arabidopsis. These results provide evidence of direct plant growth by a Plantibacter species. Bacteria of the genus Plantibacter have been found in association with a variety of plants, including the phyllosphere of grasses (35), the endosphere of yarrow, goldenrod, dactylis, and clover (32), and the rhizosphere of wheat (36), maple sap (37), and rye flakes (38). Strains of Plantibacter have been seen to solubilize zinc in soil (35). The strains tested in this experiment are also known to degrade hydrocarbon contaminants (32).
Bacteria of the genus Plantibacter are members of the Microbacteriaceae, a family that contains many other genera that are commonly found living in association with plants. Within Microbacteriaceae, Plantibacter is most closely related to the genus Okibacterium, followed by Microbacterium (39). Strains of Okibacterium and Microbacterium have been discovered inside the plant endosphere (40, 41), while certain strains of Microbacterium have also been identified as plant growth promoters (42–44). Another closely related genus is Curtobacterium, a genus that includes known plant pathogens such as C. flaccumfaciens pv. flaccumfaciens (45, 46) in addition to plant growth promoters such as C. flaccumfaciens strain E108, C. albidum, and C. herbarum (47–49). The similarity of Plantibacter to other known plant growth-promoting genera suggests that other strains of Plantibacter, along with the two discovered in this study, are also likely to provide benefits for plant growth.
Inoculation with the top five endophytes had variable effects on different plants. For example, all five selected endophytes improved root growth in Arabidopsis, lettuce, and basil plants, while only two of these endophytes improved root growth in bok choy (Plantibacter flavus M259 and Paenibacillus taichungensis M175). Similarly, inoculation with Plantibacter flavus M259 significantly increased aerial biomass in Arabidopsis and bok choy plants but not in lettuce and basil. Differences were even seen between different strains of the same species, as a bok choy biomass increase was seen with inoculation with P. flavus M259 but not with P. flavus M251. These results emphasize that the benefits of endophyte inoculation are host specific. Host specificity has been reported in literature for legume colonization by Rhizobium strains (50–53) and nonlegume colonization by associative and free-living rhizospheric bacteria (54). Host specificity is known to be related to chemotaxis and microbial signaling (55–59), root exudation features (60–62), and plant defense responses (63). The beneficial plant-microbe interaction is facilitated by the production of specific bacterium-associated secondary metabolites (64), which may interact differently with certain plant species.
The genomic alignment and comparison of the two strains of P. flavus showed them to be highly similar to each other. Genomic annotation of P. flavus strains M251 and M259 revealed the presence of genes involved in various known growth promotion pathways. The most notable of these are pathways for phytohormone production, as microbial production of auxins and cytokinins are common contributors to plant growth promotion (65–68) since auxins and cytokinins play important roles in stimulating plant growth and development throughout all cell types (69–72). ACC deaminase benefits plant growth under stressful conditions, since ACC deaminase breaks down excess stress ethylene, which reduces general stress symptoms (73–75).
Despite the presence of these genes for known plant growth-promoting products, significant growth promotion was not seen from the inoculation of P. flavus strain M251 into bok choy. Even with a genome comparison, we can only speculate about what might cause this difference between the strains. Some notable genes are present in one strain but absent in the other: strain M259 produces acid phosphatase, which helps to solubilize phosphate in the environment, and M251 has genes related to siderophore production that are lacking in strain M259. This might be of particular importance in plants growing in aquaponic systems, where iron can become limiting. The extensive analysis of more than 3,000 bacteria recently published by Levy et al. (34) yields several candidate proteins of importance to plant-associated bacteria, albeit without any theoretical support. Some of these are lacking in strain M251 but present in strain M259 (flavodoxin 2, l-arabinose, molybdenum ABC transporter, some oxidoreductases, and transcriptional regulators from LysR and GntR families). Furthermore, both strains have unique islands containing hypothetical proteins that on deeper search prove to be functionally related to the proteins on this same list. Clearly, many mechanisms that affect host specificity in plant-bacterium interactions can be at play, and the mechanistic explanations may well be linked to the numerous genes of unknown function rather than to just those few for which mechanisms are well worked out.
We should also acknowledge that another factor may have affected our results, and that is colonization success. There is a good chance that our inoculate did not actually become established within the tissues of all of the replicates of our test species, leading to high variation in plant growth promotion effects. It was beyond the scope of this screening work to develop good inoculum detection methods or to mark our strains with antibiotic resistances. These should ideally be based on molecular detection, such as quantitative PCR, based on only bacterial genes (i.e., not genes also found in plant organelles).
The three non-Plantibacter select isolates (i.e., isolates M132, M175, and M267) also proved to be beneficial for plant growth, but these results are confirmatory rather than novel. Inoculation with M132 (Curtobacterium herbarum) improved both root and shoot growth for Arabidopsis plants as well as root growth for lettuce and basil. As previously mentioned, Curtobacterium strains have been noted as both plant pathogens and plant growth promoters. Examples of plant growth promotion by Curtobacterium include improving salinity tolerance in rice and barley (46, 47), increasing saffron yield (48), and protecting against the plant pathogen Pseudomonas syringae (76). Inoculation with M175 (Paenibacillus taichungensis) improved root growth for all plants and shoot growth for Arabidopsis, lettuce, and bok choy. Other examples of plant growth promotion by Paenibacillus species have been noted in laboratory (77) and field (78, 79) studies throughout the literature. Conserved plant-beneficial genes, including those for phosphate solubilization, auxin production, and nitrogen fixation, have also been seen throughout different strains of Paenibacillus (80). Inoculation with M267 (Rhizobium selenitireducens) benefited root growth of all plants. Bacteria of the genus Rhizobium are typically associated with the nitrogen fixation process in root nodules of leguminous plants. However, they have also been observed to benefit the growth of nonlegumes, such as peppers and tomatoes, and produce plant benefits like auxins and siderophores (81). Our study provides further evidence that strains of Curtobacterium, Paenibacillus, and Rhizobium can provide direct plant growth benefits.
To summarize, this study identified Plantibacter flavus as a novel plant growth-promoting endophyte and discovered interspecific microbial growth promotion for Arabidopsis thaliana, lettuce, basil, and bok choy plants. We also confirmed the effectiveness of Curtobacterium, Paenibacillus, and Rhizobium species as plant growth-promoting microbes. Considering the potential of endophytes to promote plant growth and health, the screening of successful matches between plants and microbes represents an important step toward agriculture sustainability.
MATERIALS AND METHODS
Plant sampling and endophyte isolation.
The 220 endophytes screened in this study were previously isolated in 2013 by Lumactud et al. (32) from stems of herbaceous plants (Achillea millefolium, Solidago canadensis, Trifolium aureum, and Dactylis glomerata) growing in oil-soaked soil in Oil Springs, Ontario, Canada. Briefly, stem samples were taken from five different plant species and then surface sterilized and macerated into a solution using a blender. The solution was spread onto tryptic soy agar (TSA) and Reasoner’s 2A agar (R2A) plates, which were incubated at 28°C. Distinct colonies were selected and streaked onto TSA to create pure bacterial cultures, which were then inoculated into tryptic soy broth (TSB) and incubated at 28°C and 128 rpm for 2 days. For more details on the sampling site and bacterial isolation process, see reference 32.
Seed sterilization.
Arabidopsis Col-0 seeds were surface sterilized in a microcentrifuge tube using the following protocol: wash with reverse osmosis water for 30 s, sterilization with 95% ethanol for 15 s, sterilization with 1% bleach for 2 min, inactivation of remaining bleach with 2% sodium thiosulfate for 10 min (82), and 6 washes with sterile water for 15 s each. For each step, 1 ml of the liquid was mixed with seeds via pipetting for the given length of time, after which seeds were allowed to settle to the bottom of the tube and surface liquid was removed and discarded. To confirm the effectiveness of the sterilization procedure, 200 μl of the final wash water was spread onto one TSA plate and one R2A plate. Plates were incubated at 28°C for 4 days and examined for growth. A lack of growth on either plate indicated that the sterilization procedure was successful.
Rapid screening tests (96-well microtiter plates) with Arabidopsis thaliana.
In planta screening tests were conducted with the 220 endophyte isolates to determine if direct plant growth promotion was present in the collection. Sterilized seeds were inoculated with bacteria by soaking them in 1 ml of TSB bacterial culture (treatments) or sterile TSB (controls) for 2 h. Individual seeds were then sown onto 1× Murashige and Skoog (MS) agar (pH adjusted to ∼6.0) in wells of a 96-well microtiter plate. The lids of the microtiter plates were elevated to allow for plant growth by manufacturing plastic spacers with a three-dimensional printer that allowed the lids to sit an additional 5 cm from the plate surface. Each microtiter plate contained 96 plants in total: 84 endophyte-treated plants (21 different endophyte treatments with four plant replicates each) and 12 uninoculated control plants. An image of the plants growing in a rapid screening test is shown in Fig. 8.
FIG 8.
Image of a microtiter plate used for a rapid screening test. Each microtiter plate contained 96 Arabidopsis plants consisting of 12 uninoculated control plants and 84 endophyte-treated plants. The plates were equipped with lid spacers to allow for taller growth.
Microtiter plates were stored at 4°C for 3 days to allow for seed stratification and then placed under a 68-W Floralight 16-h/8-h day-night light source (Lee Valley Tools) at room temperature to allow for growth. The number of leaves, stem height, number of buds, and number of flowers were recorded for each plant every 3 or 4 days. At the end of the growth period (29 or 30 days), plants were extracted from the medium and the total biomass was weighed using an analytical scale. Data for each plant growth trait were analyzed in R (see “Statistical analysis” below). Data from seeds that did not germinate were excluded from analysis.
Larger-scale tests (GA-7 boxes) with Arabidopsis thaliana.
To quantify plant growth promotion more accurately, in planta tests using GA-7 boxes (Magenta LLC, Lockport, IL) were conducted with five of the best-performing PGPB (“top five”), as selected from the screening test. Surface-sterilized seeds were sown onto petri plates containing 1× MS agar, stored at 4°C for 3 days to allow for seed stratification, and then transferred under a Floralight 16-h/8-h day-night light source at room temperature to grow for 4 days. Seedlings were aseptically removed using sterilized forceps and inoculated into 1 ml of TSB bacterial culture for each of the five endophytes selected from the screening tests (bacterial treatments) or sterile TSB (controls) for 30 min. To ensure relatively consistent concentrations, bacterial culture densities were adjusted to approximately 2.4 × 108 cells/ml before inoculation.
Inoculated seedlings were transferred to GA-7 boxes containing 150 ml of 1× MS agar. Two GA-7 boxes containing four seedlings each (eight replicates in total) were used for all treatments and controls. Lids of GA-7 boxes were left slightly open (i.e., not sealed tightly) to facilitate gas exchange. GA-7 boxes were transferred to the 68-W Floralight 16-h/8-h day-night light source to allow for plant growth. The number of leaves, buds, flowers, and stem height were recorded every 3 or 4 days. At 21 days after inoculation, the plants were extracted from the agar and weighed with an analytical scale to measure total fresh biomass. The roots were then separated from the shoots and scanned and analyzed using the WinRhizo software. Results for each plant growth characteristic were analyzed in R (see “Statistical analysis” below). Data from plants with fewer than six leaves at 21 days after inoculation were excluded from analysis, as they were thought to be damaged during the seedling transfer process.
Aquaponic growth tests with lettuce, basil, and bok choy.
Since the top five growth-promoting endophytes demonstrated growth promotion in Arabidopsis plants, they were subsequently tested for growth promotion in lettuce, bok choy, and basil. Plant seeds were germinated in water-soaked Rapid Rooter sponge plugs (International Horticultural Technologies, LLC, Hollister, CA). Five days after germination, seedlings were inoculated with bacteria by pipetting 50 μl of bacterial TSB culture containing approximately 106 cells/ml (treatments) or sterile broth (controls) and then transferred to plastic trays and placed under a 68-W Floralight 16-h/8-h day-night light source at room temperature. Six replicates per plant species were carried out for each treatment. After 10 days, sponge plugs containing the seedlings were transferred to Styrofoam floats in a tilapia-based aquaponics system, where plants were grown for 2 months at 23°C. At the end of the growth period, total root length, dried root weight, and aerial biomass were recorded for each plant.
Statistical analysis.
To evaluate the effects of each endophyte on plant growth, results for each category were compared between control plants and plants inoculated with each endophyte treatment. Before analysis, data were normalized using the Tukey ladder of power. Comparisons of transformed data were evaluated using a one-way analysis of variance (ANOVA), followed by Dunnett’s post hoc test. Statistical significance was noted at a probability level (P) of <0.05. All data transformations and statistical analyses were done in R version 3.4.2 using the rcompanion and multcomp packages, respectively. For data visualization, box plots were created in R using the boxplot function.
Isolate identification.
Bacterial isolates M132, M175, M259, and M267 were identified by Sanger sequencing of 16S rRNA gene regions. Genomic DNA was extracted from TSB cultures using the DNeasy blood and tissue kit (Qiagen). 16S rRNA gene regions were amplified through 20-μl PCR mixtures containing 10 μl HotStart Taq master mix (Qiagen), 1 μl of both forward and reverse 16S-specific primers (forward, AGAGTTTGATCCTGGCTCAG; reverse, TACCTTGTTACGACTT), and 1 μl genomic DNA template. The amplification protocol was as follows: initial denaturation at 95°C for 5 min, followed by 35 cycles of (denaturation at 94°C for 1 min, primer annealing at 55°C for 1 min, and extension at 72°C for 1 min 30 s), and finishing with a final extension at 72°C for 10 min. PCR products were visualized via gel electrophoresis with a 1% agarose gel, purified using the QIAquick PCR purification kit (Qiagen), and analyzed using the NanoDrop 1000 spectrophotometer (Thermo Scientific). Purified PCR products were sent to the Centre for Applied Genomics (SickKids Hospital, Toronto, Canada) for Sanger sequencing. Species were identified by comparing sequence results to GenBank 16S rRNA gene sequences using NCBI BLAST.
Genome sequencing.
Genomic DNA was isolated from Plantibacter flavus strain M259 using the QIAamp DNA kit (Qiagen). Using the genomic DNA, a PacBio whole-genome shotgun library of sheared long inserts was developed, sequenced using PacBio RS II (40× coverage), and assembled at the Génome Québec Innovation Centre). DNA contigs were annotated using PATRIC version 3.5.38 (83) and RASTtk.
Genome comparison and identification of plant-associated and plant growth promotion genes.
The genomes of P. flavus strain M251 (previously sequenced by Lumactud et al. [33]) and P. flavus strain M259 were compared using PATRIC version 3.5.17 and the RAST server (84) and antiSMASH version 5.0 (85). A search for plant-associated genes in the two strains was conducted using a list of 767 putative actinobacterial plant-associated proteins (34). Known plant growth-promoting genes were searched in the genomes using PATRIC and RAST, while possible new gene clusters for the biosynthesis of new products were identified using antiSMASH 5.0. Genomes were aligned using the Mauve package in Geneious 10.2.3 to identify areas of noncongruency. These regions were checked against genome comparisons in RASTtk and PATRIC. Where unique regions contained hypothetical proteins, we used PSI-BLAST to look for similar proteins with known functions.
Data availability.
GenBank accession numbers for the genomes used in this study are as follows: CP019402, Plantibacter flavus strain M251 (33); CP040750, Plantibacter flavus strain M259. GenBank accession numbers for the 16S rRNA gene sequences identified in this study are as follows: MH843493.1, Curtobacterium herbarum strain M132; MH843494.1, Paenibacillus taichungensis strain M175; MH843495.1, Plantibacter flavus strain M259; MH843496.1, Rhizobium selenitireducens strain M267.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rhea Lumactud (University of Toronto Scarborough) for isolating all endophytes in the collection, Keiko Yoshioka (University of Toronto) for providing us with Arabidopsis seeds, and Ripple Farms (Toronto, Ontario) for providing us with basil, lettuce, and bok choy seeds and growth facilities for the purposes of this study.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00383-19.
REFERENCES
- 1.Theis KR, Dheilly NM, Klassen JL, Brucker RM, Baines JF, Bosch TCG, Cryan JF, Gilbert SF, Goodnight CJ, Lloyd EA, Sapp J, Vandenkoornhuyse P, Zilber-Rosenberg I, Rosenberg E, Bordenstein SR. 2016. Getting the hologenome concept right: an eco-evolutionary framework for hosts and their microbiomes. mSystems 1:e00028-16. doi: 10.1128/mSystems.00028-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith DL, Gravel V, Yergeau E. 2017. Editorial: signaling in the phytomicrobiome. Front Plant Sci 8:611. doi: 10.3389/fpls.2017.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siddikee MA, Chauhan PS, Anandham R, Han GH, Sa T. 2010. Isolation, characterization, and use for plant growth promotion under salt stress, of ACC deaminase-producing halotolerant bacteria derived from coastal soil. J Microbiol Biotechnol 20:1577–1584. doi: 10.4014/jmb.1007.07011. [DOI] [PubMed] [Google Scholar]
- 4.Grandlic CJ, Mendez MO, Chorover J, Machado B, Maier RM. 2008. Plant growth-promoting bacteria for phytostabilization of mine tailings. Environ Sci Technol 42:2079–2084. doi: 10.1021/es072013j. [DOI] [PubMed] [Google Scholar]
- 5.Yadav AN, Sachan SG, Verma P, Saxena AK. 2015. Prospecting cold deserts of north western Himalayas for microbial diversity and plant growth promoting attributes. J Biosci Bioeng 119:683–693. doi: 10.1016/j.jbiosc.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Knoth JL, Kim SH, Ettl GJ, Doty SL. 2014. Biological nitrogen fixation and biomass accumulation within poplar clones as a result of inoculation with diazotrophic endophyte consortia. New Phytol 201:599–609. doi: 10.1111/nph.12536. [DOI] [PubMed] [Google Scholar]
- 7.Oteino N, Lally RD, Kiwanuka S, Lloyd A, Ryan D, Germaine KJ, Dowling DN. 2015. Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front Microbiol 6:745. doi: 10.3389/fmicb.2015.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sukumar P, Legué V, Vayssières A, Martin F, Tuskan GA, Kalluri UC. 2013. Involvement of auxin pathways in modulating root architecture during beneficial plant-microorganism interactions. Plant Cell Environ 36:909–919. doi: 10.1111/pce.12036. [DOI] [PubMed] [Google Scholar]
- 9.Khan AL, Halo BA, Elyassi A, Ali S, Al-Hosni K, Hussain J, Al-Harrasi A, Lee IJ. 2016. Indole acetic acid and ACC deaminase from endophytic bacteria improves the growth of Solanum lycopersicum. Electron J Biotechnol 21:58–64. doi: 10.1016/j.ejbt.2016.02.001. [DOI] [Google Scholar]
- 10.Loaces I, Ferrando L, Scavino AF. 2011. Dynamics, diversity, and function of endophytic siderophore-producing bacteria in rice. Microb Ecol 61:606–618. doi: 10.1007/s00248-010-9780-9. [DOI] [PubMed] [Google Scholar]
- 11.Sessitsch A, Mitter B. 2015. 21st century agriculture: integration of plant microbiomes for improved crop production and food security. Microb Biotechnol 8:32–33. doi: 10.1111/1751-7915.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margaritopoulou T, Roka L, Alexopoulou E, Christou M, Rigas S, Haralampidis K, Milioni D. 2016. Biotechnology towards energy crops. Mol Biotechnol 58:149–158. doi: 10.1007/s12033-016-9913-6. [DOI] [PubMed] [Google Scholar]
- 13.Shivlata L, Satyanarayana T. 2015. Thermophilic and alkaliphilic actinobacteria: biology and potential applications. Front Microbiol 6:1014. doi: 10.3389/fmicb.2015.01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhardwaj D, Ansari MW, Sahoo RK, Tuteja N. 2014. Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb Cell Fact 13:66–76. doi: 10.1186/1475-2859-13-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hallmann CA, Foppen RP, van Turnhout CA, de Kroon H, Jongejans E. 2014. Declines in insectivorous birds are associated with high neonicotinoid concentrations. Nature 511:341–343. doi: 10.1038/nature13531. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Gonzalez J, Sommerfeld M. 2016. Biofertilizer and biostimulant properties of the microalga Acutodesmus dimorphus. J Appl Phycol 28:1051–1061. doi: 10.1007/s10811-015-0625-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dakora F, Matiru V, Kanu A. 2015. Rhizosphere ecology of lumichrome and riboflavin, two bacterial signal molecules eliciting developmental changes in plants. Front Plant Sci 6:700. doi: 10.3389/fpls.2015.00700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delaplace P, Delory BM, Baudson C, Mendaluk-Saunier De Cazenave M, Spaepen S, Varin S, Brostaux Y, Du Jardin P. 2015. Influence of rhizobacterial volatiles on the root system architecture and the production and allocation of biomass in the model grass Brachypodium distachyon (L.) P. Beauv. BMC Plant Biol 15:195. doi: 10.1186/s12870-015-0585-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawalekar JS. 2013. Role of biofertilizers and biopesticides for sustainable agriculture. J Bio Innov 2:73–78. [Google Scholar]
- 20.Compant S, Duffy B, Nowak J, Clément C, Barka EA. 2005. Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl Environ Microbiol 71:4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuhegger R, Ihring A, Gantner S, Bahnweg G, Knappe C, Vogg G, Hutzler P, Schmid M, Van Breusegem F, Eberl L, Hartmann A, Langebartels C. 2006. Induction of systemic resistance in tomato by N-acyl-l-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ 29:909–918. doi: 10.1111/j.1365-3040.2005.01471.x. [DOI] [PubMed] [Google Scholar]
- 22.Pii Y, Mimmo T, Tomasi N, Terzano R, Cesco S, Crecchio C. 2015. Microbial interactions in the rhizosphere: beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process: a review. Biol Fertil Soils 51:403–415. doi: 10.1007/s00374-015-0996-1. [DOI] [Google Scholar]
- 23.Shakeel M, Rais A, Hassan MN, Hafeez FY. 2015. Root associated Bacillus sp. improves growth, yield and zinc translocation for basmati rice (Oryza sativa) varieties. Front Microbiol 6:1286. doi: 10.3389/fmicb.2015.01286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Timmusk S, Behers L, Muthoni J, Muraya M, Aronsson AC. 2017. Perspectives and challenges of microbial application for crop improvement. Front Plant Sci 8:49. doi: 10.3389/fpls.2017.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jayasinghearachchi HS, Seneviratne G. 2004. A bradyrhizobial-Penicillium spp. biofilm with nitrogenase activity improves N2 fixing symbiosis of soybean. Biol Fertil Soils 40:432–434. doi: 10.1007/s00374-004-0796-5. [DOI] [Google Scholar]
- 26.Rondon MA, Lehmann J, Ramirez J, Hurtado M. 2007. Biological nitrogen fixation by common beans (Phaseolus vulgaris L.) increases with bio-char additions. Biol Fertil Soils 43:699–708. doi: 10.1007/s00374-006-0152-z. [DOI] [Google Scholar]
- 27.Prudent M, Salon C, Souleimanov A, Emery RJN, Smith DL. 2015. Soybean is less impacted by water stress using Bradyrhizobium japonicum and thuricin-17 from Bacillus thuringiensis. Agron Sustain Dev 35:749–757. doi: 10.1007/s13593-014-0256-z. [DOI] [Google Scholar]
- 28.Desbrosses GJ, Stougaard J. 2011. Root nodulation: a paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 10:348–358. doi: 10.1016/j.chom.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Llorente BE, Alasia MA, Larraburu EE. 2016. Biofertilization with Azospirillum brasilense improves in vitro culture of Handroanthus ochraceus, a forestry, ornamental and medicinal plant. N Biotechnol 33:32–40. doi: 10.1016/j.nbt.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Saha M, Sarkar S, Sarkar B, Sharma BK, Bhattacharjee S, Tribedi P. 2016. Microbial siderophores and their potential applications: a review. Environ Sci Pollut Res Int 23:3984–3999. doi: 10.1007/s11356-015-4294-0. [DOI] [PubMed] [Google Scholar]
- 31.Talukdar D, Sharma R, Kumar R. 2017. Agriculture biotechnology, p 215–223. In Kumar R, Sharma AK, Ahluwalia SS (ed), Advances in environmental biotechnology. Springer Nature, Singapore. [Google Scholar]
- 32.Lumactud R, Shen SY, Lau M, Fulthorpe R. 2016. Bacterial endophytes isolated from plants in natural oil seep soils with chronic hydrocarbon contamination. Front Microbiol 7:755. doi: 10.3389/fmicb.2016.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lumactud R, Fulthorpe R, Sentchilo V, van der Meer JR. 2017. Draft genome sequence of Plantibacter flavus strain 251 isolated from a plant growing in a chronically hydrocarbon-contaminated site. Genome Announc 5:e00276-17. doi: 10.1128/genomeA.00276-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy A, Salas Gonzalez I, Mittelviefhaus M, Clingenpeel S, Herrera Paredes S, Miao J, Wang K, Devescovi G, Stillman K, Monteiro F, Alvarez BR, Lundberg D, Lu T, Lebeis S, Jin Z, McDonald M, Klein AP, Feltcher ME, Rio TG, Grant SR, Doty SL, Ley RE, Zhau B, Venturi V, Pelletier DA, Vorholt JA, Tringe SG, Woyke T, Dangl JL. 2017. Genomic features of bacterial adaptation to plants. Nat Genet 18:138–150. doi: 10.1038/s41588-017-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Behrendt U, Ulrich A, Schumann P, Naumann D, Suzuki K. 2002. Diversity of grass-associated Microbacteriaceae isolated from the phyllosphere and litter layer after mulching the sward; polyphasic characterization of Subtercola pratensis sp. nov., Curtobacterium herbarum sp. nov. and Plantibacter flavus gen. nov., sp. nov. Int J Syst Evol Microbiol 52:1441–1454. [DOI] [PubMed] [Google Scholar]
- 36.Costerousse B, Schonholzer-Mauclaire L, Frossard E, Thonar C. 2017. Identification of heterotrophic zinc mobilization processes among bacterial strains isolated from wheat rhizosphere (Triticum aestivum L.). Appl Environ Microbiol 84:e01715-17. doi: 10.1128/AEM.01715-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagacé L, Pitre M, Jacques M, Roy D. 2004. Identification of the microbial community of maple sap by using amplified ribosomal DNA (rDNA) restriction analysis and rDNA sequencing. Appl Environ Microbiol 70:2052–2060. doi: 10.1128/aem.70.4.2052-2060.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herranen M, Kariluoto S, Edelmann M, Piironen V, Ahvenniemi K, Iivonen V, Salovaara H, Korhola M. 2010. Isolation and characterization of folate-producing bacteria from oat bran and rye flakes. Int J Food Microbiol 142:277–285. doi: 10.1016/j.ijfoodmicro.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Evtushenko LI, Takeuchi M. 2006. The family Microbacteriaceae, p 1020–1098. In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (ed), The prokaryotes. Springer, New York, NY. [Google Scholar]
- 40.Wang H-F, Zhang Y-G, Li L, Liu W-H, Hozzein WN, Chen J-Y, Guo J-W, Zhang Y-M, Li W-J. 2015. Okibacterium endophyticum sp. nov., a novel endophytic actinobacterium isolated from roots of Salsola affinis C. A. Mey. Antonie Van Leeuwenhoek 107:835–843. doi: 10.1007/s10482-014-0376-0. [DOI] [PubMed] [Google Scholar]
- 41.Zinniel DK, Lambrecht P, Harris NB, Feng Z, Kuczmarski D, Higley P, Ishimaru CA, Arunakumari A, Barletta RG, Vidaver AK. 2002. Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl Environ Microbiol 68:2198–2208. doi: 10.1128/AEM.68.5.2198-2208.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karlidag H, Esitken A, Turan M, Sahin F. 2007. Effects of root inoculation of plant growth promoting rhizobacteria (PGPR) on yield, growth, and nutrient element contents of leaves of apple. Scientia Horticulturae 114:16–20. doi: 10.1016/j.scienta.2007.04.013. [DOI] [Google Scholar]
- 43.Sheng XF, Xia JJ, Jiang CY, He LY, Qian M. 2008. Characterization of heavy metal-resistant endophytic bacteria from rape (Brassica napus) roots and their potential in promoting the growth and lead accumulation in rape. Environ Pollution 156:1164–1170. doi: 10.1016/j.envpol.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 44.He LY, Zhang YF, Ma HY, Su LN, Chen ZJ, Wang QY, Qian M, Sheng XF. 2010. Characterization of copper-resistant bacteria and assessment of bacterial communities in rhizosphere soil of copper-tolerant plants. Appl Soil Ecol 44:49–55. doi: 10.1016/j.apsoil.2009.09.004. [DOI] [Google Scholar]
- 45.Agarkova IV, Lambrecht PA, Vidaver AK, Harveson RM. 2012. Genetic diversity among Curtobacterium flaccumfaciens pv. flaccumfaciens populations in the American high plains. Can J Microbiol 58:788–801. doi: 10.1139/w2012-052. [DOI] [PubMed] [Google Scholar]
- 46.Soares RM, Fantinato GGP, Darben LM, Marcelino-Guimarães FC, Seixas CDS, Carneiro C. 2013. First report of Curtobacterium flaccumfaciens pv. Flaccumfaciens on soybean in Brazil. Trop Plant Pathol 38:452–454. doi: 10.1590/S1982-56762013000500012. [DOI] [Google Scholar]
- 47.Cardinale M, Ratering S, Suarez C, Montoya AMZ, Geissler-Plaum R, Schnell S. 2015. Paradox of plant growth promotion potential of rhizobacteria and their actual promotion effect on growth of barley (Hordeum vulgare L.) under salt stress. Microbiol Res 181:22–32. doi: 10.1016/j.micres.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Vimal SR, Patel VK, Singh JS. 2019. Plant growth promoting Curtobacterium albidum strain SRV4: an agriculturally important microbe to alleviate salinity stress in paddy plants. Ecol Indic 105:553–562. doi: 10.1016/j.ecolind.2018.05.014. [DOI] [Google Scholar]
- 49.Díez-Méndez A, Rivas R. 2017. Improvement of saffron production using Curtobacterium herbarum as a bioinoculant under greenhouse conditions. AIMS Microbiol 3:354–364. doi: 10.3934/microbiol.2017.3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.William WC, Garya S. 1976. Chemotaxis of Rhizobium sp. to plant root exudates. Plant Physiol 57:820–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zuanazzi JAS, Clergeot PH, Quirion JC, Husson HP, Kondorosi A, Ratet P. 1998. Production of Sinorhizobium meliloti nod gene activator and repressor flavonoids from Medicago sativa roots. Mol Plant Microbe Interact 11:784–794. doi: 10.1094/MPMI.1998.11.8.784. [DOI] [Google Scholar]
- 52.Wang D, Yang S, Tang F, Zhu H. 2012. Symbiosis specificity in the legume: rhizobial mutualism. Cell Microbiol 14:334–342. doi: 10.1111/j.1462-5822.2011.01736.x. [DOI] [PubMed] [Google Scholar]
- 53.Iyer B, Rajkumar S. 2017. Host specificity and plant growth promotion by bacterial endophytes. Curr Res Microbiol Biotechnol 5:1018–1030. [Google Scholar]
- 54.Giri R, Dudeja SS. 2013. Root colonization of root and nodule endophytic bacteria in legume and non legume plants grown in liquid medium. J Microbiol Res Rev 1:75–82. [Google Scholar]
- 55.Geurts R, Bisseling T. 2002. Rhizobium Nod factor perception and signaling. Plant Cell 14:S239–S249. doi: 10.1105/tpc.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ortíz-Castro R, Contreras-Cornejo HA, Macías-Rodríguez L, López-Bucio J. 2009. The role of microbial signals in plant growth and development. Plant Signal Behav 4:701–712. doi: 10.4161/psb.4.8.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chauhan H, Bagyaraj D, Selvakumar G, Sundaram S. 2015. Novel plant growth promoting rhizobacteria-prospects and potential. Appl Soil Ecol 95:38–53. doi: 10.1016/j.apsoil.2015.05.011. [DOI] [Google Scholar]
- 58.Gutjahr C, Paszkowski U. 2009. Weights in the balance: jasmonic acid and salicylic acid signaling in root-biotroph interactions. Mol Plant Microbe Interact 22:763–772. doi: 10.1094/MPMI-22-7-0763. [DOI] [PubMed] [Google Scholar]
- 59.Zipfel C, Oldroyd GE. 2017. Plant signaling in symbiosis and immunity. Nature 543:328–336. doi: 10.1038/nature22009. [DOI] [PubMed] [Google Scholar]
- 60.Haichar FZ, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, Heulin T, Achouak W. 2008. Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2:1221–1230. doi: 10.1038/ismej.2008.80. [DOI] [PubMed] [Google Scholar]
- 61.Nelson MS, Sadowsky MJ. 2015. Secretion systems and signal exchange between nitrogen-fixing rhizobia and legumes. Front Plant Sci 6:491. doi: 10.3389/fpls.2015.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kawasaki A, Donn S, Ryan PR, Mathesius U, Devilla R, Jones A, Watt M. 2016. Microbiome and exudates of the root and rhizosphere of Brachypodium distachyon, a model for wheat. PLoS One 11:e0164533. doi: 10.1371/journal.pone.0164533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Samac DA, Graham MA. 2007. Recent advances in legume-microbe interactions: recognition, defense response, and symbiosis from a genomic perspective. Plant Physiol 144:582–587. doi: 10.1104/pp.107.096503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brader G, Compant S, Mitter B, Trognitz F, Sessitsch A. 2014. Metabolic potential of endophytic bacteria. Curr Opin Biotechnol 27:30–37. doi: 10.1016/j.copbio.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Asghar HM, Zahir ZA, Arshad M, Khaliq A. 2002. Relationship between in vitro production of auxins by rhizobacteria and their growth-promoting abilities in Brassica juncea L. Biol Fertil Soils 35:231–237. doi: 10.1007/s00374-002-0462-8. [DOI] [Google Scholar]
- 66.Khalid A, Arshad A, Zahir ZA. 2004. Screening plant growth-promoting rhizobacteria for improving growth and yield of wheat. J Appl Microbiol 96:473–480. doi: 10.1046/j.1365-2672.2003.02161.x. [DOI] [PubMed] [Google Scholar]
- 67.Dobbelaere S, Croonenborghs A, Thys A, Vande Broek A, Vanderleyden J. 1999. Phytostimulatory effect of Azospirillium brasilense wild type and mutant strains altered in IAA production on wheat. Plant Soil 212:153–162. doi: 10.1023/A:1004658000815. [DOI] [Google Scholar]
- 68.Fahad S, Hussain S, Bano A, Saud S, Hassan S, Shan D, Khan FA, Khan F, Chen Y, Wu C, Tabassum MA, Chun MX, Afzal M, Jan A, Jan MT, Huang J. 2015. Potential role of phytohormones and plant growth-promoting rhizobacteria in abiotic stresses: consequences for changing environment. Environ Sci Pollut Res Int 22:4907–4921. doi: 10.1007/s11356-014-3754-2. [DOI] [PubMed] [Google Scholar]
- 69.Su YH, Liu YB, Zhang XS. 2011. Auxin-cytokinin interaction regulates meristem development. Mol Plant 4:616–625. doi: 10.1093/mp/ssr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Omer ZS, Tombolini R, Broberg A, Gerhardson B. 2004. Indole-3-acetic acid production by pink-pigmented facultative methylotrophic bacteria. Plant Growth Regul 43:93–96. doi: 10.1023/B:GROW.0000038360.09079.ad. [DOI] [Google Scholar]
- 71.Kang SM, Joo GJ, Hamayun M, Na CI, Shin DH, Kim HY, Hong JK, Lee IJ. 2009. Gibberellin production and phosphate solubilization by newly isolated strain of Acinetobacter calcoaceticus and its effect on plant growth. Biotechnol Lett 31:277–281. doi: 10.1007/s10529-008-9867-2. [DOI] [PubMed] [Google Scholar]
- 72.Kang SM, Khan AL, Waqas M, You YH, Hamayun M, Joo GJ, Shahzad R, Choi KS, Lee IJ. 2015. Gibberellin-producing Serratia nematodiphila PEJ1011 ameliorates low temperature stress in Capsicum annuum L. Eur J Soil Biol 68:85–93. doi: 10.1016/j.ejsobi.2015.02.005. [DOI] [Google Scholar]
- 73.Glick BR. 2014. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res 169:30–39. doi: 10.1016/j.micres.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 74.Heydarian Z, Yu M, Gruber M, Glick BR, Zhou R, Hegedus DD. 2016. Inoculation of soil with plant growth promoting bacteria producing 1-aminocyclopropane-1-carboxylate deaminase or expression of the corresponding acdS gene in transgenic plants increases salinity tolerance in Camelina sativa. Front Microbiol 7:1966. doi: 10.3389/fmicb.2016.01966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang QY, Dodd IC, Belimov AA, Jiang F. 2016. Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase growth and photosynthesis of pea plants under salt stress by limiting Na+ accumulation. Funct Plant Biol 43:161–172. doi: 10.1071/FP15200. [DOI] [PubMed] [Google Scholar]
- 76.Barriuso J, Solano BR, Gutiérrez Mañero FJ. 2008. Protection against pathogen and salt stress by four plant growth-promoting rhizobacteria isolated from Pinus sp. on Arabidopsis thaliana. Phytopathology 98:666–672. doi: 10.1094/PHYTO-98-6-0666. [DOI] [PubMed] [Google Scholar]
- 77.De Souza R, Meyer J, Schoenfeld R, da Costa PB, Passaglia L. 2015. Characterization of plant growth-promoting bacteria associated with rice cropped in iron-stressed soils. Ann Microbiol 65:951–964. doi: 10.1007/s13213-014-0939-3. [DOI] [Google Scholar]
- 78.Furnkranz M, Eveline A, Muller H, Grube M, Huss H, Winkler J, Berg G. 2012. Promotion of growth, health, and stress tolerance of Styrian oil pumpkins by bacterial endophytes. Eur J Plant Pathol 134. doi: 10.1007/s10658-012-0033-2. [DOI] [Google Scholar]
- 79.Ker K, Seguin P, Driscoll BT, Fyles JW, Smith DL. 2012. Switchgrass establishment and seeding year production can be improved by inoculation with rhizosphere endophytes. Biomass Bioenergy 47:295–301. doi: 10.1016/j.biombioe.2012.09.031. [DOI] [Google Scholar]
- 80.Xie J, Shi H, Du Z, Wang T, Liu X, Chen S. 2016. Comparative genomic and functional analysis reveal conservation of plant growth-promoting traits in Paenibacillus polymyxa and its closely related species. Sci Rep 6:21239. doi: 10.1038/srep21329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.García-Fraile P, Carro L, Robledo M, Ramírez-Bahena M-H, Flores-Félix J-D, Fernández MT, Mateos PF, Rivas R, Igual JM, Martínez-Molina E, Peix Á, Velázquez E. 2012. Rhizobium promotes non-legumes growth and quality in several production steps: towards a biofertilization of edible raw vegetables healthy for humans. PLoS One 7:e38122. doi: 10.1371/journal.pone.0038122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miché L, Balandreau J. 2001. Effects of rice seed surface sterilization with hypochlorite on inoculated Burkholderia vietnamiensis. Appl Environ Microbiol 67:3046–3052. doi: 10.1128/AEM.67.7.3046-3052.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, Gerdes S, Henry CS, Kenyon RW, Machi D, Mao C, Nordberg EK, Olsen GJ, Murphy-Olson DE, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Vonstein V, Warren A, Xia F, Yoo H, Stevens RL. 2017. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 4:535–542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 8:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T. 2019. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GenBank accession numbers for the genomes used in this study are as follows: CP019402, Plantibacter flavus strain M251 (33); CP040750, Plantibacter flavus strain M259. GenBank accession numbers for the 16S rRNA gene sequences identified in this study are as follows: MH843493.1, Curtobacterium herbarum strain M132; MH843494.1, Paenibacillus taichungensis strain M175; MH843495.1, Plantibacter flavus strain M259; MH843496.1, Rhizobium selenitireducens strain M267.