Burial of organic carbon in marine and estuarine sediments represents a long-term sink for atmospheric carbon dioxide. Globally, ∼40% of organic carbon burial occurs in anoxic estuaries and deltaic systems. However, the ultimate controls on the amount of organic matter that is buried in sediments, versus oxidized into CO2, are poorly constrained. In this study, we used a combination of enzyme assays and metagenomic analysis to identify how subsurface microbial communities catalyze the first step of proteinaceous organic carbon degradation. Our results show that microbial communities in deeper sediments are adapted to access molecules characteristic of degraded organic matter, suggesting that those heterotrophs are adapted to life in the subsurface.
KEYWORDS: anaerobes, deep subsurface, extracellular enzymes, heterotrophs, peptidases, sediments
ABSTRACT
Anoxic subsurface sediments contain communities of heterotrophic microorganisms that metabolize organic carbon at extraordinarily low rates. In order to assess the mechanisms by which subsurface microorganisms access detrital sedimentary organic matter, we measured kinetics of a range of extracellular peptidases in anoxic sediments of the White Oak River Estuary, NC. Nine distinct peptidase substrates were enzymatically hydrolyzed at all depths. Potential peptidase activities (Vmax) decreased with increasing sediment depth, although Vmax expressed on a per-cell basis was approximately the same at all depths. Half-saturation constants (Km) decreased with depth, indicating peptidases that functioned more efficiently at low substrate concentrations. Potential activities of extracellular peptidases acting on molecules that are enriched in degraded organic matter (d-phenylalanine and l-ornithine) increased relative to enzymes that act on l-phenylalanine, further suggesting microbial community adaptation to access degraded organic matter. Nineteen classes of predicted, exported peptidases were identified in genomic data from the same site, of which genes for class C25 (gingipain-like) peptidases represented more than 40% at each depth. Methionine aminopeptidases, zinc carboxypeptidases, and class S24-like peptidases, which are involved in single-stranded-DNA repair, were also abundant. These results suggest a subsurface heterotrophic microbial community that primarily accesses low-quality detrital organic matter via a diverse suite of well-adapted extracellular enzymes.
IMPORTANCE Burial of organic carbon in marine and estuarine sediments represents a long-term sink for atmospheric carbon dioxide. Globally, ∼40% of organic carbon burial occurs in anoxic estuaries and deltaic systems. However, the ultimate controls on the amount of organic matter that is buried in sediments, versus oxidized into CO2, are poorly constrained. In this study, we used a combination of enzyme assays and metagenomic analysis to identify how subsurface microbial communities catalyze the first step of proteinaceous organic carbon degradation. Our results show that microbial communities in deeper sediments are adapted to access molecules characteristic of degraded organic matter, suggesting that those heterotrophs are adapted to life in the subsurface.
INTRODUCTION
A large fraction of the microorganisms in subsurface sediments are heterotrophs that metabolize aged, microbially altered organic matter (OM) (1–3). These communities’ metabolisms can be more than a million-fold slower than that of cells in culture (1, 4). A recent meta-analysis showed that only about 12% of cells in marine sediments belong to cultured species, while 27% belong to phyla that contain no cultured representatives (5). Consequently, the mechanisms by which these microorganisms access detrital organic matter are poorly understood (6).
In surface environments, where organic carbon (OC) metabolism is relatively rapid, heterotrophic microorganisms gain energy by metabolizing a combination of small molecules (<600 to 1,000 Da), which can be taken up directly via general uptake porins (7) and macromolecules, which must be broken down outside the cell by extracellular enzymes. Most freshly produced organic matter is macromolecular, and large molecules tend to be more bioavailable than small ones (8), so the nature and activity of extracellular enzymes present in surface environments are a major control on the rate of microbial carbon oxidation in such environments.
It is not clear whether microbial extracellular enzymes play the same role in subsurface sediments. It is conceivable that macromolecules are broken down primarily by nonenzymatic mechanisms in sediments. For instance, in soils, MnO4 catalyzes the depolymerization of proteins without requiring enzymes. Certain bacterial species can use TonB-dependent transporters to transport polysaccharides that are substantially larger than 600 Da into the periplasm (although enzymatic hydrolysis is still required prior to uptake into the cytoplasm [9]). Furthermore, some of the unique aspects of subsurface sediments suggest that extracellular enzymes might not be an effective strategy to obtain carbon or energy. In order for the production of extracellular enzymes to be part of a viable metabolic strategy, each enzyme must, over its lifetime, provide the cell with at least as much carbon or energy as was required to synthesize the enzyme (10–12). In subsurface sediments, where cell division times may be on the order of decades to millennia, enzyme lifetimes would need to be correspondingly longer to remain “profitable.” Since enzyme lifetimes are finite, there must exist a community metabolic rate below which extracellular enzyme lifetimes are too short to become profitable. That limit is difficult to quantify because enzyme lifetimes in any environment are poorly constrained (for an example, see reference 13). Thus, it is plausible that extracellular enzyme-mediated carbon acquisition is impractical in sediments in which metabolic rates are particularly slow.
While extracellular enzyme activity in surface sediments has frequently been reported, few reports exist of extracellular enzyme activity from deeper than 20 cm below the seafloor (cmbsf) (14). Enzyme activity has been reported for sapropels up to 389 cmbsf in the eastern Mediterranean Sea (15, 16) and in sediment from 600 to 630 cmbsf in Aarhus Bay sediments (17), as well as in a few other subsurface environments, such as the interior of seafloor basalts at the Loihi seamount (18). Furthermore, an analysis of transcriptomes from subsurface sediments of the Peru Margin revealed diverse exported peptidases and carbohydrate-active enzymes, which decreased in relative abundance with increasing depth (19).
In order to better understand how heterotrophic microorganisms in subsurface sediments access organic matter, we assayed a diverse set of peptidases (protein-degrading enzymes) in sediment cores from the White Oak River Estuary, NC. We paired these assays with analysis of the potential for extracellular peptidase production from existing metagenomic data sets. We chose this site because the porewater geochemistry and microbiology of these sediments have been well characterized (20–24) and because they contain abundant Bathyarchaeota and Thermoprofundales archaea, which appear to be capable of metabolizing detrital organic matter (17, 25–27). We focused on peptidases because protein degradation appears to be an important metabolism for some subsurface archaea (17) and because peptidases were more active than other enzymes in similar environments (15, 18). Because environmental samples contain a wide range of distinct peptidases at variable activities (28, 29), we measured the hydrolysis of 11 different substrates which may be hydrolyzed by structurally and genetically diverse extracellular peptidases.
RESULTS
Peptidase kinetics.
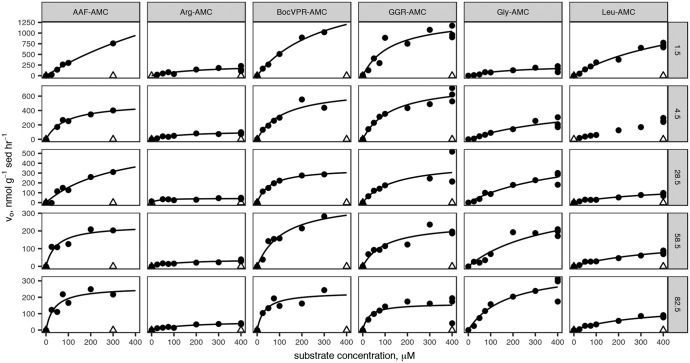
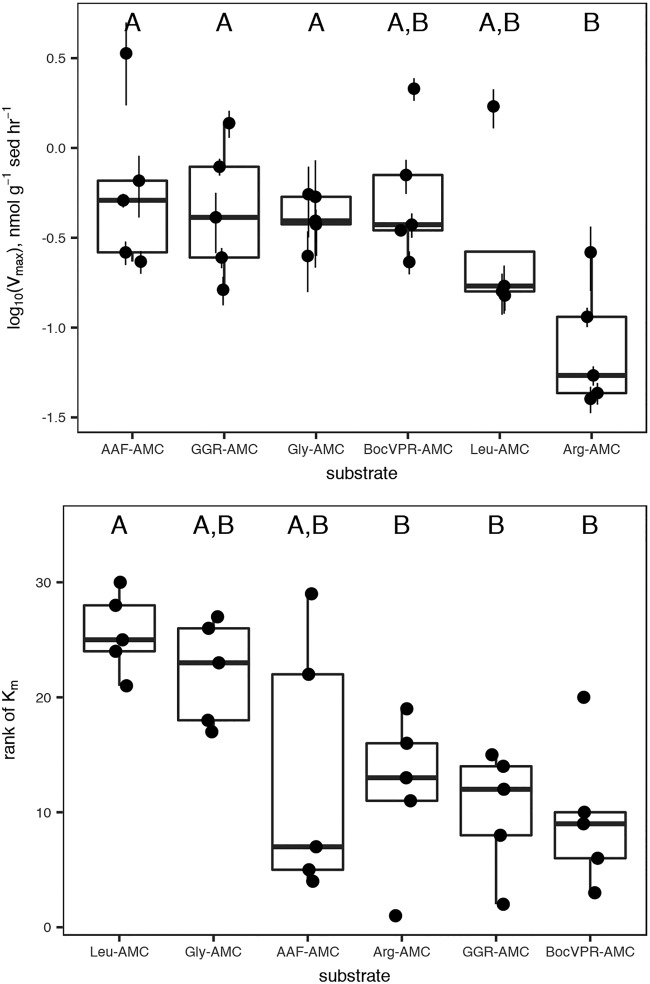
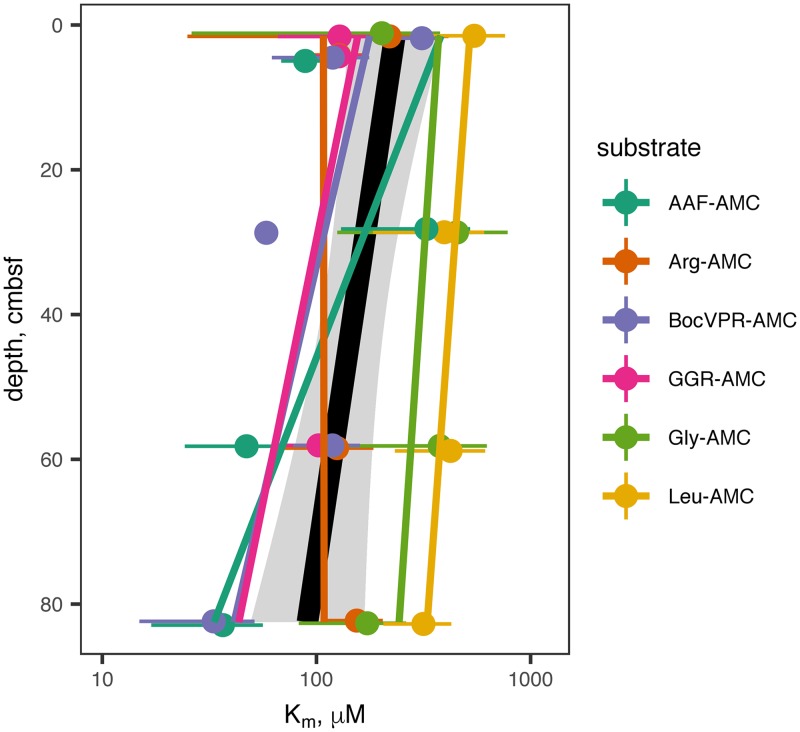
Sediment cores were sampled on two dates: first, to measure saturation curves for six structurally diverse peptidase substrates, and second, to measure a more targeted set of peptidases at high depth resolution in order to assess the relative ability of subsurface communities to access more degraded organic matter. Combining all samples, unambiguous hydrolysis of nine different peptidase substrates was observed. All peptidase substrates assayed with the more sensitive single-cuvette methodology were hydrolyzed much faster in untreated sediments than in autoclaved controls (Fig. 1). Kinetics of substrate hydrolysis were qualitatively consistent with the Michaelis-Menten rate law, v0 = (Vmax × [S])/Km + [S]), with estimated Vmax values ranging from 40 to 3,400 nmol g−1 sediment h−1 (median, 310 nmol g−1 sediment h−1; interquartile range, 190 to 560 nmol g−1 sediment h−1). Throughout the core, alanine-alanine-phenylalanine–7-amido-4-methylcoumarin (AAF-AMC), glycine-glycine-arginine-AMC (GGR-AMC), and Gly-AMC were hydrolyzed the fastest, and Arg-AMC was hydrolyzed the slowest (Fig. 2). Summed Vmax values for each substrate, a proxy for the total peptidolytic potential of the microbial community, decreased with depth from 9.09 μmol AMC g−1 sediment h−1 at the surface to 1.24 μmol AMC g−1 sediment h−1, or 13% of the surface value, at 82.5 cmbsf. Estimated Km values ranged from 36.1 μM to 1,310 μM (median, 138 μM; interquartile range, 102 to 326 μM) and trended downward (i.e., to greater substrate affinity) with increasing depth (Fig. 3). Km values for hydrolysis of Leu-AMC were the highest (i.e., lowest substrate affinity), while Km values for hydrolysis of Boc-valine-proline-arginine-AMC (Boc-VPR-AMC), GGR-AMC, and Arg-AMC were the lowest.
FIG 1.
Saturation curves for six substrates measured using the single-cuvette reader methodology at each of six depths. Dark circles indicate “live” samples; open triangles indicate autoclaved controls. Lines indicate nonlinear least-squares fits to the Michaelis-Menten rate law. Substrate abbreviations are given in the column headings and are defined in Table 1. Sediment depths are listed on row headings in centimeters below sediment-water interface.
FIG 2.
Vmax and Km values, shown individually with error bars indicating SEs of the nonlinear least-squares estimates, and collectively in a box-and-whisker plot. Substrates sharing a letter are not significantly different according to one-way analysis of variance (ANOVA) of log10-transformed data with Tukey honestly significant difference (HSD) post hoc analysis (Vmax) or Kruskal-Wallis test with Tukey HSD post hoc analysis (Km).
FIG 3.
Km values of extracellular peptidases as a function of depth. Details of substrates and the peptidases they correspond to are in Table 1. Error bars represent the SDs of replicate samples. Colored lines represent a linear least-squares regression for each substrate. The black line and gray shading represent linear regression and 95% confidence interval for all substrates taken together.
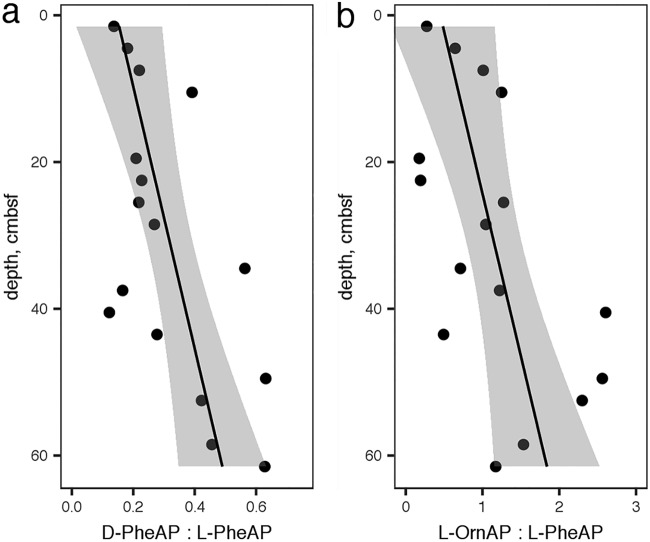
In a separate core, hydrolysis rates of d-Phe–AMC, l-Phe–AMC, and l-Orn–AMC were assessed. These were measured using a plate reader technique that proved insufficiently precise to accurately measure Vmax or Km, so we report only the observed hydrolysis rate v0, which was measured at a high substrate concentration (400 μM) and therefore approximates Vmax. Ratios of v0 for d-Phe–AMC/l-Phe–AMC hydrolysis and l-Orn–AMC/l-Phe–AMC hydrolysis rates increased approximately linearly downcore (Fig. 4).
FIG 4.
Ratios of v0 for d-phenylalanine aminopeptidase to l-phenylalanine aminopeptidase (a) and l-ornithine aminopeptidase to l-phenylalanine aminopeptidase (b). The linear regressions are given by the following: d-Phe–AP/l-Phe–AP = (5.60 ± 1.87) × 10−3 × depth + 0.146 (±0.067) and l-Orn–AP/l-Phe–AP = (2.26 ± 0.89) × 10−3 × depth + 0.451 (±0.326).
Microbial abundance, cell-specific peptidase activity, and organic carbon oxidation rates.
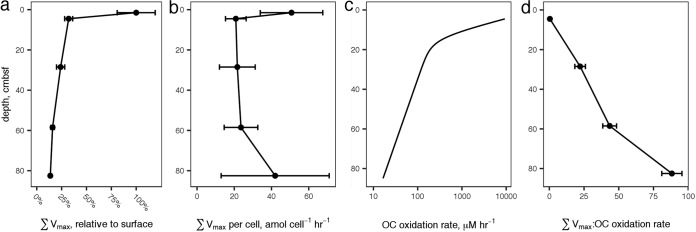
Concordantly with potential activities (Fig. 5a), cell counts decreased more or less steadily downcore from 4.5 × 108 cells ml−1 wet sediment at 1.5 cmbsf to 7.4 × 107 cells ml−1 wet sediment at 82.5 cmbsf. Consequently, cell-specific total potential peptidase activity was roughly constant at 32 ± 14 amol AMC cell−1 h−1 (Fig. 5b), with no significant trend as a function of depth. Most of the error in cell-specific peptidase activities results from variance in cell counts rather than in Vmax estimations.
FIG 5.
(a) The sum of all peptidase Vmax values, relative to the value at 4.5 cm, versus sediment depth. Error bars represent propagated errors of the Vmax estimates for the substrates. (b) Summed Vmax relative to cell count. Error bars represent propagated errors from summed Vmaxs and cell counts; errors are dominated by cell count uncertainty. (c) Organic carbon oxidation rates modeled from sulfate and methane profiles. (d) Summed Vmaxs relative to modeled carbon oxidation rates. Error bars represent propagated error from summed Vmaxs and cell counts. The propagated error is dominated by uncertainty in the cell counts.
Organic carbon oxidation rates were estimated using a 2-G model driven by porewater methane and sulfate concentrations. The total modeled organic carbon oxidation rate G at 82.5 cmbsf was approximately 0.17% relative to that at 4.5 cmbsf (the top of the model domain), a decrease of almost 3 orders of magnitude (Fig. 5c). Thus, summed Vmax relative to G, a proxy for the effort microbes exert compared to obtain complex organic carbon relative to the amount of carbon they metabolize, increased more than 200-fold in the deepest sediments relative to surface sediments (Fig. 5d).
Peptidase genes and microbial taxa.
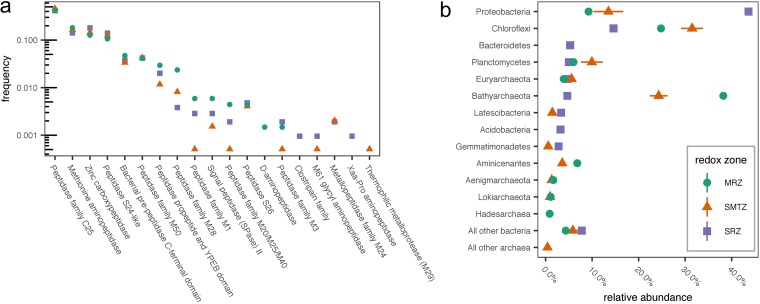
Samples for genomic analysis were taken from three broad sedimentary zones: the sulfate reduction zone (SRZ; 8 to 12 cmbsf), sulfate-methane transition zone (SMTZ; two distinct samples from nearby locations; 24 to 32 cmbsf and 26 to 30 cmbsf), and the methane-rich zone (MRZ; 52 to 54 cmbsf; data originally published by Baker et al. [22]). A total of 3,739 genes encoding extracellular peptidases were identified among metagenomes from the three depth zones examined, including 685 from the SRZ, 1,994 from the SMTZ, and 1,060 from the MRZ. Of the genes encoding peptidases, 0 to 71% (depending on the class of peptidase, algorithm, and sediment depth) contained a signal peptide (SP) and are likely secreted by the Sec-dependent transport system (see File S1 in the supplemental material). Among the genes associated with signal peptides, members of peptidase family C25, belonging to the gingipain family, were by far the most abundant at all depths, accounting for 41 to 45% of all SP-associated peptidases (Fig. 6a). Genes annotated as encoding extracellular methionine aminopeptidases and zinc carboxypeptidases were also abundant (13% to 19%). Together, these peptidase classes accounted for 73%, 76%, and 73% of exported peptidases in the SRZ, SMTZ, and MRZ, respectively. The composition of protein families was generally consistent with depth, particularly among the more abundant peptidases. Five peptidase annotations were an exception to this trend: peptidase family M1, peptidase family M20/M25/M40, peptidase family M3, M61 glycyl aminopeptidase, and thermophilic metalloprotease (M29) were found in much lower abundances at the SMTZ than the MRZ or SRZ. Given that those correspond to differences of one or a few total reads, these are well within the range of noise.
FIG 6.
(a) Frequency of reads for genes of various classes of extracellular peptidases that were associated with signal peptidases, relative to all genes for extracellular peptidases at that depth. SRZ, SMTZ, and MRZ, sulfate reduction zone (8 to 12 cmbsf), sulfate-methane transition zone (24 to 32 cmbsf), and methane-rich zone (24 to 28 cmbsf), respectively. (b) Relative abundances of phyla in bins at each depth. Only the 10 most abundant phyla at each depth are shown. The orange SMTZ points represent the average of two SMTZ samples, taken ∼500 m from each other, and error bars represent the ranges of the two sites.
Proteobacteria were by far the most abundant phylum in the SRZ sediments, with smaller contributions from Chloroflexi, Bacteroidetes, Planctomycetes, Euryarchaeota, and Bathyarchaeota (Fig. 6b). This community differed substantially from the communities in the SMTZ and MRZ, which were fairly similar to each other. In both sediment depths, Bathyarchaeota, Chloroflexi, and Proteobacteria were the dominant phyla.
DISCUSSION
Identities of extracellular peptidases present in White Oak River Estuary sediments.
The kinetics of fluorogenic substrate hydrolysis were consistent with the Michaelis-Menten rate law, and hydrolysis rates were dramatically lower in autoclaved controls than in “live” treatments, indicating that the substrates were hydrolyzed by enzymes rather than by abiotic factors. The enzyme substrates used in this study encompassed a diverse range of amino acid and peptide chemistries, including polar and nonpolar R groups at the P1 site (i.e., the amino acid N terminal to the scissile bond) and substrates with and without steric protecting groups, which must have been hydrolyzed by endopeptidases (which cleave proteins from within) and aminopeptidases (which cleave proteins from the N terminus), respectively. Peptide bonds adjacent to a diverse set of amino acid residues were cleaved, including glycine (the smallest amino acid), phenylalanine (among the largest amino acids), arginine (positively charged at porewater pH), and leucine (uncharged and hydrophobic), consistent with the presence of a diverse range of extracellular peptidases throughout the core.
The metagenomic results also indicated the potential for a diverse range of secreted peptidases, produced by a broad range of taxa, throughout the sediment column (Fig. 6). The metagenomic results represent a minimum estimate for the genomic potential for extracellular peptidase production, because they rely on the assumption that only those peptidases associated with signal peptides (SPs) are secreted. Non-SP-based enzyme secretion pathways may also contribute to the pool of extracellular enzymes, including Sec-independent secretion systems (30) and release of internal enzymes into the extracellular medium by viral lysis (31, 32).
The dominance of genes for exported gingipain-like endopeptidases (class C25) at all depths is consistent with rapid hydrolysis rates of fluorogenic substrates for endopeptidases. Gingipains are endopeptidases with preference for arginine at the P1 position (i.e., the N-terminal side of the hydrolyzed bond), which would be active toward the substrates GGR-AMC and Boc-VPR-AMC. Those were among the fastest-hydrolyzed substrates at each depth (Fig. 1 and 2), indicating that genes for C25 peptidases were likely expressed. Previously, gingipains have been identified in Thermoprofundales (formerly marine benthic group D) and in Bathyarchaeota, and they appear to be widespread in marine sediments (17, 19, 33). The M28 family, also among the most abundant annotations, contains a diverse range of aminopeptidases and carboxypeptidases, including leucine aminopeptidase, consistent with the observed hydrolysis of Leu-AMC (34). Genes for d-aminopeptidases were observed, consistent with hydrolysis of d-Phe–AMC.
Other abundant genes were annotated as methionine aminopeptidase, zinc carboxypeptidase, a C-terminal domain from bacterial prepeptidases, and peptidases from MEROPS families M24, S24, M50, and M28. Potential activities of these peptidases were not assayed. Zinc carboxypeptidases (M20) cleave enzymes from the carboxy terminus and have strong specificity for Gly at the P1 position (i.e., the position C terminal to the scissile bond) but little preference for the residue at the P1′ position (the position C terminal to the scissile bond; in a carboxypeptidase this would be the C terminus of the protein). Methionine aminopeptidases (M24) are metallopeptidases with preference for glycine at the P1 position.
S24 and M50 peptidases are less likely to be directly relevant to organic matter processing. S24 peptidases are involved in the SOS response for single-stranded-DNA repair (34). M50 peptidases are membrane-bound enzymes that act as sporulation factors in Bacillus subtilis, and possibly other bacteria (35, 36), and which are not secreted. However, DNA repair (37, 38) and spore formation (39, 40) both appear to be important survival mechanisms for microorganisms in subsurface sediments. The bacterial C-terminal prepeptidase domain is often found in secreted peptidases, but it is removed prior the peptidase becoming active and could be associated with a wide range of peptidases (41).
Each of these annotations is plausible in terms of what is known about peptidase activities in sediments, and the annotations were generally consistent with the observed activities. We did not assay for carboxypeptidases (e.g., MEROPS family M20) or methionine aminopeptidase, but carboxypeptidases have previously been observed to be active in estuarine sediments (42), and the generally broad substrate specificities of extracellular aminopeptidases suggest that methionine aminopeptidases could have contributed to the hydrolysis of the other aminopeptidase substrates (43). However, a note of caution is also warranted when interpreting peptidase annotations from deeply branching microorganisms: the high diversity of hydrolases makes precise annotations difficult, and the exact substrate specificities of the peptidases in these samples may differ somewhat from those inferred from the annotations (44). Thus, while these annotations are environmentally plausible and generally consistent with the fluorogenic enzyme assays, they should nevertheless be viewed with some skepticism.
Interestingly, the sets of peptidases identified in genomes, and the activities observed, varied among depths much less than the microbes present (compare the similar profiles in Fig. 6a to the notable differences among depths in Fig. 6b). It is possible that extracellular enzymes were produced only by a small subset of taxa that were present at all sediment depths, although this would be inconsistent with previous evidence that diverse taxa, including sulfate reducers and fermenters, produce extracellular enzymes in sediments (albeit deeper than those studied in this investigation [19]), with the widespread phylogenetic distribution of similar extracellular enzymes (45), and with previous observations of functional redundancy with respect to extracellular enzyme production in diverse systems (46–48).
Peptidase kinetics suggest adaptation of subsurface peptidases to degraded organic matter.
Heterotrophic microorganisms in subsurface sediments have little access to fresh organic matter. In the cores described here, which represented ∼275 years of sediment deposition, the organic matter oxidation rate decreased by at least 3 orders of magnitude between the surface and 82.5 cmbsf (Fig. 5c). It is challenging to determine what fraction of high- versus low-molecular-weight organic matter subsurface microorganisms metabolize. However, the fact that cell-specific Vmax was more or less constant downcore (Fig. 5b) suggests that the heterotrophic community relied on complex organic matter to similar degrees at all depths. The cell-specific Vmax values for Leu-AMC hydrolysis, 21 to 51 amol cell−1 h−1, are comparable to previous measurements in active environments such as surface sediments (2 to 100 amol cell−1 h−1) and seawater (mostly less than 100 amol cell−1 h−1 but with some measurements up to 10 nmol cell−1 h−1 [reference 49 and references therein]), consistent with communities that relied primarily on organic carbon derived from macromolecules.
The ratio of ΣVmax to OC oxidation rate is sensitive to the mix of enzymes included in the sum and to the substrate specificity of enzymes assayed (some enzymes are capable of hydrolyzing multiple substrates). The absolute value of that sum, therefore, is not particularly meaningful. The trend, however, is informative: as sediment depth increased, the potential activity of extracellular peptidases decreased much more slowly than the actual rate of organic carbon oxidation, so the ratio of ΣVmax to the OC oxidation rate increased dramatically (Fig. 5d). Vmax is a proxy for enzyme concentration, so the observed increase in the ratio of ΣVmax to OC oxidation rate combined with the trend in cell-specific ΣVmax suggests that deeper heterotrophic communities exhibited similar demand for detrital OM but that those enzymes returned bioavailable hydrolysate at a much lower rate because substrate concentrations were lower. The White Oak River subsurface communities were similar to their surface counterparts in terms of reliance on extracellular enzymes for bioavailable organic carbon, although subsurface metabolisms were considerably slower. However, enzyme kinetics and potential activities of d-phenylalanine aminopeptidase, l-phenylalanine aminopeptidase, and l-ornithine aminopeptidase all suggested microbial community adaptation to old, degraded organic matter in deeper sediments.
Most amino acids are biosynthesized as l-stereoisomers. As organic matter ages, the ratio of d-amino acids to l-amino acids (d/l ratio) increases with depth, due to abiotic racemization and increased abundance of d-amino acids derived from bacterial cell walls (1, 50). Accordingly, the potential activity of d-phenylalanyl aminopeptidase increased relative to that of l-phenylalanyl aminopeptidase, indicating an increased capacity to access degraded organic matter. Ornithine, which is a product of the release of urea from arginine, is another marker for degraded organic matter, while phenylalanine is more characteristic of fresher organic matter (51), and the potential activity of l-phenylalanine aminopeptidase relative to that of ornithine aminopeptidase followed the same increasing trend with depth. Finally, the decrease of Km values with increasing depth indicates peptidases that function more efficiently at lower substrate concentrations. It is intuitive that the concentration of enzyme-labile organic matter concentrations would decrease downcore, and the observed increase in ratio of ΣVmax to OC oxidation rate provides direct evidence of that. Taken together, these three observations provide strong evidence for a subsurface heterotrophic microbial community that is increasingly adapted to persist using degraded organic matter at increasing depth.
This evidence raises the ecological question of how selective pressure produces a heterotrophic community adapted to degraded organic matter. Modeling and genomic observations in older (thousands to millions of years), deeper sediments suggest that microbial growth rates are too slow for community adaptation by enhanced growth rates of more successful taxa; rather, communities in deeper sediments consist of taxa that were deposited at the sediment-water interface and died at the lowest rates (6, 52, 53).
If those findings can be generalized to the shallower environments investigated in this study, that poses a question: in which aquatic environments are microorganisms capable of gaining reproductive advantage by growing on recalcitrant organic carbon? The studies cited above addressed sites at which sedimentation rates, microbial respiration, and likely cell doubling times were considerably slower than in the sediments described here, so even if growth (as opposed to persistence) on recalcitrant organic carbon is not possible in those environments, it may have been in the White Oak River sediments. Alternately, microbial taxa may gain adaptations to metabolize recalcitrant organic matter in environments where labile organic matter is more abundant and growth rates are higher. This scenario would imply that organisms which primarily metabolize more labile organic matter would gain some selective advantage by also metabolizing recalcitrant organic matter. Finally, it is not entirely clear how the microorganisms in this study used the amino acids resulting from extracellular hydrolysis. In deeper sediments, heterotrophs appear to be energy limited rather than carbon limited (54), which would suggest that amino acids would likely be assimilated directly into proteins. However, if amino acids are catabolized, there may be an energetic advantage to incorporating d-amino acids: some catabolic pathways for l-amino acids involve conversion to the d-form prior to further processing, in which case uptake of d-amino acids could save energy (55). Further analysis of the mechanisms by which subsurface heterotrophs access degraded sedimentary organic matter may yield insights into how microorganisms survive in low-energy environments and into the processes that shape the pool of organic carbon that is preserved or oxidized over geological timescales.
MATERIALS AND METHODS
Study site.
Samples were collected from Station H in the White Oak River Estuary, 34°44.490′N, 77°07.44′W, first described by Gruebel and Martens (56). The White Oak River Estuary occupies a drowned river valley in the coastal plain of North Carolina. Station H is characterized by salinity in the range of 10 to 28 and water depth on the order of 2 m (21). The flux of ΣCO2 across the sediment-water interface was 0.46 ± 0.02 mmol m−2 h−1 (measured in May of 1987), primarily due to organic carbon oxidation via sulfate reduction. The sediment accumulation rate averages 0.3 cm year−1. The total organic carbon content is approximately 5% (21). For this study, push cores of 40 to 85 cm were collected from Station H by swimmers on 28 May 2013 and 22 October 2014. In 2013, cores were transported to the nearby Institute of Marine Sciences (University of North Carolina) at Morehead City, where they were sectioned and processed for enzyme activities, porewater geochemistry, and cell counts within 6 h of sample collection. Porewater sulfate in 2013 was depleted by 43.5 cm, and methane peaked at 79.5 cm (Fig. S1). In 2014, cores were transported on the day of sampling to the University of Tennessee, Knoxville, stored at 4°C, and processed for enzyme activities the following day. Samples for metagenomic analysis were collected separately in October 2010 from three sites (sites 1, 2, and 3, as previously described by Baker et al. [22]), all of which are within 550 m of Station H. Porewater geochemistry of those samples is described in Fig. S2 in the work of Lazar et al. (23). We note that although the samples for sequencing were taken at a different time and slightly different location than the samples for enzyme assays, the geochemistry and geomicrobiology of White Oak River sediments appear to be extremely stable and homogenous, possibly due to the fact that sediments are extremely fine and the estuarine water flow rates are low. In any case, the geochemistry of the sediments is stable over timescales of decades (21, 24, 57), and microbial abundances in the SMTZ were very similar even though they were collected at sites separated by ∼500 m (error bars in Fig. 6b are mostly smaller than the differences among depths).
Enzyme assays.
Enzyme assays were performed using different protocols in 2013 (data presented in Fig. 1 and 3) and 2014 (data presented in Fig. 4). In 2013, enzyme assays were performed according to a protocol similar to the one described by Lloyd et al. (17). Cores were sectioned into 3-cm intervals. The following intervals were selected for enzyme assays: 0 to 3 cm, 3 to 6 cm, 27 to 30 cm, 57 to 60 cm, and 81 to 83 cm. Each section was homogenized, and approximately 0.5 ml wet sediment was transferred into separate 5-ml amber glass serum vials, which had been preweighed and preloaded with 4 ml anoxic artificial seawater (Sigma Sea Salts; salinity = 15 and pH = 7.5) Samples were weighed again to determine the precise mass of wet sediment added, and then an appropriate quantity of 20 mM peptidase substrate stock dissolved in dimethyl sulfoxide (DMSO) was added, up to 90 μl, for final substrate concentrations of 0, 25, 50, 75, 100, 200, and 300 μM. Substrates are listed in Table 1. Triplicate incubations with 400 μM Arg-AMC, Gly-AMC, Leu-AMC, and Gly-Gly-Arg-AMC were also created, but these were omitted for Ala-Ala-Phe-AMC and Boc-Phe-Val-Arg-AMC because these two substrates are considerably more expensive. Each serum vial was vortexed and briefly gassed with N2 to remove oxygen introduced with the sample, and approximately 1.3 ml slurry was immediately removed, transferred to a microcentrifuge tube, and placed on ice to quench the reaction. The precise time of quenching was recorded. This was centrifuged at 10,000 × g within approximately 15 min. The supernatant was transferred to a methacrylate cuvette, and fluorescence was measured with a Turner Biosystems TBS-380 fluorescence detector set to UV mode (λex = 365 to 395 nm; λem = 465 to 485 nm). Samples were then incubated at 16°C, approximately the in situ temperature, and the sampling procedure was repeated after approximately 3 h. The rate of fluorescence production was calculated as the increase in fluorescence for each sample divided by the elapsed time between sample quenching. Killed controls were made using homogenized, autoclaved sediments from 35 to 45 cmbsf. However, we note that autoclaving does not completely destroy sediment enzymes because sorption to mineral surfaces stabilizes enzyme structure, vastly increasing their ability to maintain a functional conformation at high temperatures (58–60). We therefore used the autoclaved samples as a qualitative control for the null hypothesis that enzymes were responsible for none of the observed substrate hydrolysis, rather than as a quantitative method to distinguish enzymatic substrate hydrolysis from potential abiotic effects. In some sediments, a large fraction of fluorophore can sorb to particles, requiring a correction to observed fluorescence (15, 16). In order to test the extent of sorption, we incubated 120 nM AMC in White Oak River sediment slurry (12.5 g sediment/100 ml artificial seawater; pH = 7.5) over the course of 125 h. Fluorescence was stable over the entire incubation (Fig. S2). Further, we sometimes measured fluorescence calibration curves repeatedly over ∼3 h, and we observed no clear changes in the slopes of the curves. We thus concluded that sorption of the free fluorophore was negligible.
TABLE 1.
Substrates used in this study and the enzymes that hydrolyze thema
| Substrate | Abbreviation | Putative enzyme |
|---|---|---|
| l-Arginine–7-amido-4-methylcoumarin | Arg-AMC | Arginyl aminopeptidase |
| l-Glycine–7-amido-4-methylcoumarin | Gly-AMC | Glycyl aminopeptidase |
| l-Leucine–7-amido-4-methylcoumarin | Leu-AMC | Leucyl aminopeptidase |
| Carboxybenzoyl-glycine-glycine-arginine–7-amido-4-methylcoumarin | Z-GGR-AMC | Gingipain and other endopeptidases |
| Alanine-alanine-phenylalanine–7-amido-4-methylcoumarin | AAF-AMC | Clostripain and other endopeptidases |
| Boc-valine-proline-arginine-AMC | Boc-VPR-AMC | Gingipain and other endopeptidases |
| d-Phenylalanine–AMC | d-Phe–AMC | d-Phenyalanine aminopeptidase |
| l-Phenylalanine–AMC | l-Phe–AMC | l-Phenylalanine aminopeptidase |
| Ornithine-AMC | Orn-AMC | Ornithine aminopeptidase |
AMC, 7-amido-4-methylcoumarin, the moiety that becomes fluorescent after hydrolysis of the peptide bond. All amino acids are in the l-stereoconformation unless otherwise noted. Enzymes are described as “putative” because the substrate specificity of many environmental peptidases is fairly broad, so multiple peptidases may hydrolyze any given substrate.
In 2014, enzymes were assayed using a protocol based on the approach of Bell et al. (61), which was designed for soil enzyme assays. In this approach, peptidase substrates were mixed with sediment-buffer slurries in 2-ml wells of a deep-well plate. These plates were periodically centrifuged and 250-μl aliquots of supernatant were transferred into a black 96-well microplate. Fluorescence was read using a BioTek Cytation 3 microplate reader (λex = 360 nm; λem = 440 nm). Results from this method proved considerably noisier than those from the single-cuvette method used in 2013, so kinetic parameters (Vmax and Km) were not calculated for these data. Nevertheless, results were qualitatively similar to those from 2013, and we have reported Vmax from 2014 as v0 measured at a 400 μM substrate concentration, which was saturating. In October 2014, the following substrates were assayed: AAF-AMC, Arg-AMC, Boc-VPR-AMC, d-Phe–AMC, Gly-AMC, Leu-AMC, l-Phe–AMC, Orn-AMC, Z-Phe-Arg-AMC, and Z-Phe-Val-Arg-AMC. In October 2014, l-Phe–AMC, d-Phe–AMC, and Orn-AMC were assayed according to the same protocol in 3-cm core sections at 1.5, 4.5, 7.5, 10.5, 19.5, 22.5, 25.5, 28.5, 34.5, 37.5, 40.5, 43.5, 49.5, 52.5, 58.5, and 61.5 cmbsf.
Peptidase kinetic data were analyzed using R. All raw data and scripts related to enzyme analysis are posted at http://github.com/adsteen/WOR_enz_2013_2014. For samples taken using the more sensitive single-cuvette method, Michaelis-Menten parameters were estimated from nonlinear least-squares fits to kinetic data. In the case of Leu-AMC at 4.5 cm below seafloor, kinetic data could not successfully be fit to a Michaelis-Menten function, so no Km was reported and the value of v0 at the highest substrate concentration was substituted for Vmax. For analysis of correlations, data sets were qualitatively evaluated for homoskedasticity and normality of residuals using q-q plots and plots of residuals versus fitted values. When untransformed data met those criteria, the null hypothesis of no correlation was tested using linear least-squares regressions. When untransformed data failed to meet those criteria, data were log transformed. In cases in which either log-transformed data were heteroskedastic or residuals were nonnormally distributed, data were rank transformed and correlations were tested using Spearman’s ρ.
Geochemical and microbiological measurements.
Sediment porosity was measured by mass after drying at 80°C, according to the equation
Here mw represents mass lost after drying, ρw represents the density of pure water, md represents the mass of the dry sediment, S represents salinity in grams per kilogram, and ρds represents the density of dry sediment (assumed to be 2.5 g cm−3). Using an ion chromatograph (Dionex, Sunnyvale, CA), sulfate concentrations in porewater that was separated by centrifugation in 15-ml centrifuge tubes at 5,000 × g for 5 min, filtered at 0.2 μm, and acidified with 10% HCl were measured. Methane was measured using 3-ml sediment subsamples that were collected from a cutoff syringe, entering through the side of a core section, immediately after core extrusion. Subsamples were deposited immediately in 20-ml serum vials containing 1 ml 0.1 M KOH. These were immediately stoppered and shaken to mix sediment with KOH. Methane was later measured by injecting 500 μl bottle headspace into a gas chromatograph-flame ionization detector (GC-FID; Agilent, Santa Clara, CA) using a headspace equilibrium method (62).
Geochemical modeling.
Organic carbon remineralization rates as a function of depth were estimated by applying a multicomponent reaction-transport model to depth distributions of sulfate and methane concentration. The model is based on equations described by Boudreau (63) and includes only sulfate reduction and methane production due to lack of data regarding oxic and suboxic processes. Thus, the model is limited to depths greater than 4.5 cm, where sulfate reduction and methane production are the dominant processes and bioirrigation and bioturbation may be assumed to be negligible. The organic matter remineralization rate is parameterized using the multi-G model first proposed by Jørgensen (64); a two-component model was sufficient to accurately simulate the sulfate and methane data. For solutes, the upper boundary conditions were measured values at 4.5 cm, while the lower boundary conditions (200 cm) were set to zero gradient. The flux of reactive organic carbon to 4.5 cm was calculated from the sulfate flux across the 4.5-cm horizon and an estimate of methane burial below the lower boundary (the methane flux at the upper boundary was observed to be zero), with an assumed oxidation state of reactive carbon of −0.7. The model contains four adjustable parameters that are set to capture the major details of measured sulfate and methane data: first-order rate constants for both fractions of the reactive carbon pool, the partitioning factor for both fractions, and the rate constant for methane oxidation.
Cell enumeration.
Cells were enumerated by direct microscopic counts. One milliliter of sediment was placed in a 2-ml screw-cap tube with 500 μl 3% paraformaldehyde in phosphate-buffered saline (PBS), in which it was incubated overnight before being centrifuged for 5 min at 3,000 × g. The supernatant was removed and replaced with 500 μl PBS, vortexed briefly, and centrifuged again at 3,000 × g. The supernatant was subsequently removed and replaced with a 1:1 PBS-ethanol solution. Sediments were then sonicated using a Branson Ultrasonics SFX150 sonifier at 20% power for 40 s to disaggregate cells from sediments, diluted 40-fold into PBS prior to filtration onto a 0.2-μm polycarbonate filter (Fisher Scientific, Waltham, MA), and mounted onto a slide. Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) and enumerated by direct counts using a Leica epifluorescence microscope.
Metagenomic analysis.
To resolve the taxonomic distribution of extracellular peptidases, we searched a preexisting White Oak River de novo-assembled and -binned metagenomic data set (Table S2 in the work of Baker et al. [22]) for genes that were assigned extracellular peptidase functions. These assignments were based on best matches to extracellular peptidases in KEGG, pfam, and NCBI-nr (nonredundant) databases using the IMG annotation pipeline (70). Genes were additionally screened for signal peptidase motifs using the following programs: PrediSI setting the organism group to Gram-negative bacteria (65), PRED-Signal trained on archaea (66), the standalone version of PSORT v.3.0 trained against archaea (67), and SignalP 4.1 using Gram-negative bacteria as the organism group (68). All programs were used with default settings if not stated otherwise. Binned genomes from three different depth zones of White Oak River sediments were examined. The sulfate-rich zone (SRZ) genomes were obtained from sites 2 and 3, core sections 8 to 12 and 8 to 10 cm, respectively. The sulfate-methane transitions zone (SMTZ) genomes were recovered from sites 2 and 3 at depths of 30 to 32 cm and 24 to 28 cm. The methane-rich zone (MRZ) was from site 1 at 52 to 54 cm. To determine abundances of phyla, we used the 16S rRNA gene sequences that were automatically extracted from the Baker et al. [22] metagenomes by IMG/m. We used CLC Genomic Workbench 10.0 (CLC Bio, Aarhus, Denmark) to trim adaptors and make contigs from bidirectional sequences. We clustered operational taxonomic units (OTUs) at 97% similarity using Silva reference set 132 to identify the taxonomy of OTUs (69).
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael Piehler for access to his laboratory facilities at the University of North Carolina Institute of Marine Sciences, the captain of the R/V Capricorn for sampling assistance, and Terry Hazen for use of lab equipment at the University of Tennessee. We greatly appreciate two anonymous reviewers, whose constructive comments substantially improved the manuscript. Oliver Jeffers reminds A.D.S. that science is cool and worth doing.
Funding for K.H.M. was provided by NSF grant DBI-1156644 to Steven W. Wilhelm. Funding for A.D.S. was provided by NSF grant OCE-1431598, as well as a C-DEBI subaward (also to T.M.R.). This work is C-DEBI contribution number 485.
The authors declare no competing financial interests in relation to the work described.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00102-19.
REFERENCES
- 1.Lomstein BA, Niggemann J, Jørgensen BB, Langerhuus AT. 2009. Accumulation of prokaryotic remains during organic matter diagenesis in surface sediments off Peru. Limnol Oceanogr 54:1139–1151. doi: 10.4319/lo.2009.54.4.1139. [DOI] [Google Scholar]
- 2.Jørgensen BB, Marshall I. 2016. Slow microbial life in the seabed. Annu Rev Mar Sci 8:311–332. doi: 10.1146/annurev-marine-010814-015535. [DOI] [PubMed] [Google Scholar]
- 3.Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sørensen KB, Anderson R, Fredricks HF, Elvert M, Kelly TJ, Schrag DP, Sogin ML, Brenchley JE, Teske A, House CH, Hinrichs K-UK-U, Sorensen KB, Anderson R, Fredricks HF, Elvert M, Kelly TJ, Schrag DP, Sogin ML, Brenchley JE, Teske A, House CH, Hinrichs K-U. 2006. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci U S A 103:3846–3851. doi: 10.1073/pnas.0600035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoehler TM, Jørgensen BB. 2013. Microbial life under extreme energy limitation. Nat Rev Microbiol 11:83–94. doi: 10.1038/nrmicro2939. [DOI] [PubMed] [Google Scholar]
- 5.Lloyd KG, Steen AD, Ladau J, Yin J, Crosby L. 2018. Phylogenetically novel uncultured microbial cells dominate earth microbiomes. mSystems 3:e00055-18. doi: 10.1128/mSystems.00055-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orsi WD. 2018. Ecology and evolution of seafloor and subseafloor microbial communities. Nat Rev Microbiol 16:671–683. doi: 10.1038/s41579-018-0046-8. [DOI] [PubMed] [Google Scholar]
- 7.Benz R, Bauer K. 1988. Permeation of hydrophilic molecules through the outer membrane of gram-negative bacteria: review on bacterial porins. Eur J Biochem 176:1–19. doi: 10.1111/j.1432-1033.1988.tb14245.x. [DOI] [PubMed] [Google Scholar]
- 8.Benner R, Amon R. 2015. The size-reactivity continuum of major bioelements in the ocean. Annu Rev Mar Sci 7:185–205. doi: 10.1146/annurev-marine-010213-135126. [DOI] [PubMed] [Google Scholar]
- 9.Reintjes G, Arnosti C, Fuchs BM, Amann R. 2017. An alternative polysaccharide uptake mechanism of marine bacteria. ISME J 11:1640–1650. doi: 10.1038/ismej.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vetter YAY, Deming JJW, Jumars PPA, Krieger-Brockett BB. 1998. A predictive model of bacterial foraging by means of freely released extracellular enzymes. Microb Ecol 36:75–92. doi: 10.1007/s002489900095. [DOI] [PubMed] [Google Scholar]
- 11.Allison SD. 2005. Cheaters, diffusion and nutrients constrain decomposition by microbial enzymes in spatially structured environments. Ecol Lett 8:626–635. doi: 10.1111/j.1461-0248.2005.00756.x. [DOI] [Google Scholar]
- 12.Schimel JP, Weintraub MN. 2003. The implications of exoenzyme activity on microbial carbon and nitrogen limitation in soil: a theoretical model. Soil Biol Biochem 35:549–563. doi: 10.1016/S0038-0717(03)00015-4. [DOI] [Google Scholar]
- 13.Steen AD, Arnosti C. 2011. Long lifetimes of β-glucosidase, leucine aminopeptidase, and phosphatase in Arctic seawater. Mar Chem 123:127–132. doi: 10.1016/j.marchem.2010.10.006. [DOI] [Google Scholar]
- 14.Arnosti C, Bell C, Moorhead DLL, Sinsabaugh RLL, Steen ADD, Stromberger M, Wallenstein M, Weintraub M. 2014. Extracellular enzymes in terrestrial, freshwater, and marine environments: perspectives on system variability and common research needs. Biogeochemistry 117:5–21. doi: 10.1007/s10533-013-9906-5. [DOI] [Google Scholar]
- 15.Coolen MJL, Overmann J. 2000. Functional exoenzymes as indicators of metabolically active bacteria in 124,000-year-old sapropel layers of the eastern Mediterranean Sea. Appl Environ Microbiol 66:2589–2598. doi: 10.1128/AEM.66.6.2589-2598.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coolen MJL, Cypionka H, Sass AM, Sass H, Overmann J. 2002. Ongoing modification of Mediterranean Pleistocene sapropels mediated by prokaryotes. Science 296:2407–2410. doi: 10.1126/science.1071893. [DOI] [PubMed] [Google Scholar]
- 17.Lloyd K, Schreiber L, Petersen D, Kjeldsen K, Lever M, Steen AD, Stepanauskas R, Richter M, Kleindienst S, Lenk S, Schramm A, Jorgensen BB. 2013. Predominant archaea in marine sediments degrade detrital proteins. Nature 496:215–218. doi: 10.1038/nature12033. [DOI] [PubMed] [Google Scholar]
- 18.Meyers MEJ, Sylvan JB, Edwards KJ, Jacobson Meyers ME, Sylvan JB, Edwards KJ. 2014. Extracellular enzyme activity and microbial diversity measured on seafloor exposed basalts from Loihi seamount indicate the importance of basalts to global biogeochemical cycling. Appl Environ Microbiol 80:4854–4864. doi: 10.1128/AEM.01038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orsi WD, Richards TA, Francis WR. 2018. Predicted microbial secretomes and their target substrates in marine sediment. Nat Microbiol 3:32–37. doi: 10.1038/s41564-017-0047-9. [DOI] [PubMed] [Google Scholar]
- 20.Martens CS, Goldhaber MB. 1978. Early diagenesis in transitional sedimentary environments of the White Oak River Estuary, North Carolina. Limnol Oceanogr 23:428–441. doi: 10.4319/lo.1978.23.3.0428. [DOI] [Google Scholar]
- 21.Kelley CA, Martens CS, Chanton J. 1990. Variations in sedimentary carbon remineralization rates in the White Oak River estuary, North Carolina. Limnol Oceanogr 35:372–383. doi: 10.4319/lo.1990.35.2.0372. [DOI] [Google Scholar]
- 22.Baker BJ, Lazar CS, Teske AP, Dick GJ. 2015. Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome 3:14. doi: 10.1186/s40168-015-0077-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazar CS, Baker BJ, Seitz K, Hyde AS, Dick GJ, Hinrichs K-U, Teske AP. 2016. Genomic evidence for distinct carbon substrate preferences and ecological niches of Bathyarchaeota in estuarine sediments. Environ Microbiol 18:1200–1211. doi: 10.1111/1462-2920.13142. [DOI] [PubMed] [Google Scholar]
- 24.Lloyd KG, Alperin MJ, Teske A. 2011. Environmental evidence for net methane production and oxidation in putative ANaerobic MEthanotrophic (ANME) archaea. Environ Microbiol 13:2548–2564. doi: 10.1111/j.1462-2920.2011.02526.x. [DOI] [PubMed] [Google Scholar]
- 25.Kubo K, Lloyd KG, F Biddle J, Amann R, Teske A, Knittel K. 2012. Archaea of the miscellaneous crenarchaeotal group are abundant, diverse and widespread in marine sediments. ISME J 6:1949–1965. doi: 10.1038/ismej.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng J, Xu J, Qin D, He Y, Xiao X, Wang F. 2014. Genetic and functional properties of uncultivated MCG archaea assessed by metagenome and gene expression analyses. ISME J 8:650–659. doi: 10.1038/ismej.2013.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Z, Pan J, Wang F, Gu J-D, Li M. 2018. Bathyarchaeota: globally distributed metabolic generalists in anoxic environments. FEMS Microbiol Rev 42:639–655. doi: 10.1093/femsre/fuy023. [DOI] [PubMed] [Google Scholar]
- 28.Obayashi Y, Suzuki S. 2005. Proteolytic enzymes in coastal surface seawater: significant activity of endopeptidases and exopeptidases. Limnol Oceanogr 50:722–726. doi: 10.4319/lo.2005.50.2.0722. [DOI] [Google Scholar]
- 29.Steen AD, Arnosti C. 2013. Extracellular peptidase and carbohydrate hydrolase activities in an Arctic fjord (Smeerenburgfjord, Svalbard). Aquat Microb Ecol 69:93–99. doi: 10.3354/ame01625. [DOI] [Google Scholar]
- 30.Bendtsen JD, Kiemer L, Fausbøll A, Brunak S. 2005. Non-classical protein secretion in bacteria. BMC Microbiol 5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danovaro R, Dell’Anno A, Corinaldesi C, Magagnini M, Noble R, Tamburini C, Weinbauer M. 2008. Major viral impact on the functioning of benthic deep-sea ecosystems. Nature 454:1084–1087. doi: 10.1038/nature07268. [DOI] [PubMed] [Google Scholar]
- 32.Breitbart M, Felts B, Kelley S, Mahaffy JM, Nulton J, Salamon P, Rohwer F. 2004. Diversity and population structure of a near-shore marine-sediment viral community. Proc Biol Sci 271:565–574. doi: 10.1098/rspb.2003.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Z, Liu Y, Lloyd KG, Pan J, Yang Y, Gu J-D, Li M. 2018. Genomic and transcriptomic insights into the ecology and metabolism of benthic archaeal cosmopolitan, Thermoprofundales (MBG-D archaea). ISME J 13:885–901. doi: 10.1038/s41396-018-0321-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. 2018. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res 46:D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green DH, Cutting SM. 2000. Membrane topology of the Bacillus subtilis pro-ςK processing complex. J Bacteriol 182:278–285. doi: 10.1128/jb.182.2.278-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng L, Yan H, Wu Z, Yan N, Wang Z, Jeffrey PD, Shi Y. 2007. Structure of a site-2 protease family intramembrane metalloprotease. Science 318:1608–1612. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 37.Orsi WD, Edgcomb VP, Christman GD, Biddle JF. 2013. Gene expression in the deep biosphere. Nature 499:205–208. doi: 10.1038/nature12230. [DOI] [PubMed] [Google Scholar]
- 38.Bird JT, Tague E, Zinke L, Schmidt JM, Steen AD, Reese BK, Marshall I, Webster G, Weightman A, Castro H, Campagna SR, Lloyd KG. 2019. Uncultured microbial phyla suggest mechanisms for multi-thousand-year subsistence in Baltic Sea sediments. mBio 10:e02376-18. doi: 10.1128/mBio.02376-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Sullivan LA, Roussel EG, Weightman AJ, Webster G, Hubert CR, Bell E, Head I, Sass H, Parkes RJ. 2015. Survival of Desulfotomaculum spores from estuarine sediments after serial autoclaving and high-temperature exposure. ISME J 9:922–933. doi: 10.1038/ismej.2014.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lomstein BA, Langerhuus AT, D’Hondt S, Jørgensen BB, Spivack AJ, D’Hondt S, Jørgensen BB, Spivack AJ. 2012. Endospore abundance, microbial growth and necromass turnover in deep sub-seafloor sediment. Nature 484:101–104. doi: 10.1038/nature10905. [DOI] [PubMed] [Google Scholar]
- 41.Yeats C, Bentley S, Bateman A. 2003. New knowledge from old: in silico discovery of novel protein domains in Streptomyces coelicolor. BMC Microbiol 3:3. doi: 10.1186/1471-2180-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pantoja S, Lee C, Marecek JF. 1997. Hydrolysis of peptides in seawater and sediment. Mar Chem 57:25–40. doi: 10.1016/S0304-4203(97)00003-0. [DOI] [Google Scholar]
- 43.Steen AD, Vazin J, Hagen S, Mulligan K, Wilhelm S. 2015. Substrate specificity of aquatic extracellular peptidases assessed by competitive inhibition assays using synthetic substrates. Aquat Microb Ecol 75:271–281. doi: 10.3354/ame01755. [DOI] [Google Scholar]
- 44.Michalska K, Steen AD, Chhor G, Endres M, Webber AT, Bird J, Lloyd KG, Joachimiak A. 2015. New aminopeptidase from “microbial dark matter” archaeon. FASEB J 29:4071–4079. doi: 10.1096/fj.15-272906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimmerman AE, Martiny AC, Allison SD. 2013. Microdiversity of extracellular enzyme genes among sequenced prokaryotic genomes. ISME J 7:1187–1199. doi: 10.1038/ismej.2012.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wohl DL, Arora S, Gladstone JR. 2004. Functional redundancy supports biodiversity and ecosystem function in a closed and constant environment. Ecology 85:1534–1540. doi: 10.1890/03-3050. [DOI] [Google Scholar]
- 47.Reich PB, Walters MB, Ellsworth DS, Smith DP, Branco S, Glassman SI, Erlandson S, Vilgalys R, Liao H-L, Smith ME, Peay KG. 1997. From tropics to tundra: global convergence in plant functioning. Proc Natl Acad Sci U S A 94:13730–13734. doi: 10.1073/pnas.94.25.13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frossard A, Gerull L, Mutz M, Gessner MO. 2012. Disconnect of microbial structure and function: enzyme activities and bacterial communities in nascent stream corridors. ISME J 6:680–691. doi: 10.1038/ismej.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vetter YA, Deming JW. 1994. Extracellular enzyme activity in the Arctic Northeast Water polynya. Mar Ecol Prog Ser 114:23–34. doi: 10.3354/meps114023. [DOI] [Google Scholar]
- 50.Bada JL, Luyendyk BP, Maynard JB. 1970. Marine sediments: dating by the racemization of amino acids. Science 170:730–732. doi: 10.1126/science.170.3959.730. [DOI] [PubMed] [Google Scholar]
- 51.Dauwe B, Middelburg JJ, Herman PMJ, Heip C. 1999. Linking diagenetic alteration of amino acids and bulk organic matter reactivity. Limnol Oceanogr 44:1809–1814. doi: 10.4319/lo.1999.44.7.1809. [DOI] [Google Scholar]
- 52.Starnawski P, Bataillon T, Ettema TJG, Jochum LM, Schreiber L, Chen X, Lever MA, Polz MF, Jørgensen BB, Schramm A, Kjeldsen KU. 2017. Microbial community assembly and evolution in subseafloor sediment. Proc Natl Acad Sci U S A 114:2940–2945. doi: 10.1073/pnas.1614190114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bradley JA, Amend JP, LaRowe DE. 2019. Survival of the fewest: microbial dormancy and maintenance in marine sediments through deep time. Geobiology 17:43–59. doi: 10.1111/gbi.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morono Y, Terada T, Nishizawa M, Ito M, Hillion F, Takahata N, Sano Y, Inagaki F. 2011. Carbon and nitrogen assimilation in deep subseafloor microbial cells. Proc Natl Acad Sci U S A 108:18295–18300. doi: 10.1073/pnas.1107763108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Radkov AD, Moe LA. 2018. A broad spectrum racemase in Pseudomonas putida KT2440 plays a key role in amino acid catabolism. Front Microbiol 9:1343. doi: 10.3389/fmicb.2018.01343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gruebel KA, Martens CS. 1984. Radon-222 tracing of sediment-water chemical transport in an estuarine sediment. Limnol Oceanogr 29:587–597. doi: 10.4319/lo.1984.29.3.0587. [DOI] [Google Scholar]
- 57.Benninger LK, Martens CS. 1983. Sources and fates of sedimentary organic matter in the White Oak and Neuse River Estuaries. Water Resources Research Institute of the University of North Carolina, Raleigh, NC. [Google Scholar]
- 58.Stursova M, Sinsabaugh RL. 2008. Stabilization of oxidative enzymes in desert soil may limit organic matter accumulation. Soil Biol Biochem 40:550–553. doi: 10.1016/j.soilbio.2007.09.002. [DOI] [Google Scholar]
- 59.Carter DO, Yellowlees D, Tibbett M. 2007. Autoclaving kills soil microbes yet soil enzymes remain active. Pedobiologia (Jena) 51:295–299. doi: 10.1016/j.pedobi.2007.05.002. [DOI] [Google Scholar]
- 60.Schmidt J. 2016. Microbial extracellular enzymes in marine sediments: methods development and potential activities in the Baltic Sea deep biosphere. Master's thesis University of Tennessee, Knoxville, TN. [Google Scholar]
- 61.Bell CW, Fricks BE, Rocca JD, Steinweg JM, McMahon SK, Wallenstein MD. 2013. High-throughput fluorometric measurement of potential soil extracellular enzyme activities. J Vis Exp 2013:e50961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lapham LL, Chanton JP, Martens CS, Sleeper K, Woolsey JR. 2008. Microbial activity in surficial sediments overlying acoustic wipeout zones at a Gulf of Mexico cold seep. Geochem Geophys Geosyst 9:Q06001. [Google Scholar]
- 63.Boudreau BP. 1996. A method-of-lines code for carbon and nutrient diagenesis in aquatic sediments. Comput Geosci 22:479–496. doi: 10.1016/0098-3004(95)00115-8. [DOI] [Google Scholar]
- 64.Jørgensen BB. 1978. A comparison of methods for the quantification of bacterial sulfate reduction in coastal marine sediments: II. Calculation from mathematical models. Geomicrobiol J 1:29–47. doi: 10.1080/01490457809377722. [DOI] [Google Scholar]
- 65.Hiller K, Grote A, Scheer M, Münch R, Jahn D. 2004. PrediSi: prediction of signal peptides and their cleavage positions. Nucleic Acids Res 32:W375–W379. doi: 10.1093/nar/gkh378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bagos PG, Tsirigos KD, Plessas SK, Liakopoulos TD, Hamodrakas SJ. 2009. Prediction of signal peptides in archaea. Protein Eng Des Sel 22:27–35. doi: 10.1093/protein/gzn064. [DOI] [PubMed] [Google Scholar]
- 67.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman F. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 69.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Pillay M, Ratner A, Huang J, Woyke T, Huntemann M, Anderson I, Billis K, Varghese N, Mavromatis K, Pati A, Ivanova NN, Kyrpides NC. 2014. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res 42:D560–D567. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.