Microorganisms produce membrane vesicles. One synthesis pathway seems to be pearling that describes the physical formation of vesicle chains from phospholipid vesicles via extended tubes. Bacteria with vesicle chains had been observed as well as bacteria with tubes, but pearling was so far not observed. Here, we report the observation of, initially, tubes and then vesicle chains during the growth of a flavobacterium, suggesting biopearling of vesicle chains. The flavobacterium is abundant during spring bacterioplankton blooms developing after algal blooms and has a special set of enzymes for laminarin, the major storage polysaccharide of microalgae. We demonstrated with fluorescently labeled laminarin that the vesicle chains bind laminarin or contain laminarin-derived compounds. Proteomic analyses revealed surface-attached degradative enzymes on the outer membrane vesicles. We conclude that the large surface area and the lumen of vesicle chains may contribute to the ecological success of this marine bacterium.
KEYWORDS: Flavobacteriia, outer membrane, outer membrane proteins, vesicle
ABSTRACT
Large surface-to-volume ratios provide optimal nutrient uptake conditions for small microorganisms in oligotrophic habitats. The surface area can be increased with appendages. Here, we describe chains of interconnecting vesicles protruding from cells of strain Hel3_A1_48, affiliating with Formosa spp. within the Flavobacteriia and originating from coastal free-living bacterioplankton. The chains were up to 10 μm long and had vesicles emanating from the outer membrane with a single membrane and a size of 80 to 100 nm by 50 to 80 nm. Cells extruded membrane tubes in the exponential phase, whereas vesicle chains dominated on cells in the stationary growth phase. This formation is known as pearling, a physical morphogenic process in which membrane tubes protrude from liposomes and transform into chains of interconnected vesicles. Proteomes of whole-cell membranes and of detached vesicles were dominated by outer membrane proteins, including the type IX secretion system and surface-attached peptidases, glycoside hydrolases, and endonucleases. Fluorescein-labeled laminarin stained the cells and the vesicle chains. Thus, the appendages provide binding domains and degradative enzymes on their surfaces and probably storage volume in the vesicle lumen. Both may contribute to the high abundance of these Formosa-affiliated bacteria during laminarin utilization shortly after spring algal blooms.
IMPORTANCE Microorganisms produce membrane vesicles. One synthesis pathway seems to be pearling that describes the physical formation of vesicle chains from phospholipid vesicles via extended tubes. Bacteria with vesicle chains had been observed as well as bacteria with tubes, but pearling was so far not observed. Here, we report the observation of, initially, tubes and then vesicle chains during the growth of a flavobacterium, suggesting biopearling of vesicle chains. The flavobacterium is abundant during spring bacterioplankton blooms developing after algal blooms and has a special set of enzymes for laminarin, the major storage polysaccharide of microalgae. We demonstrated with fluorescently labeled laminarin that the vesicle chains bind laminarin or contain laminarin-derived compounds. Proteomic analyses revealed surface-attached degradative enzymes on the outer membrane vesicles. We conclude that the large surface area and the lumen of vesicle chains may contribute to the ecological success of this marine bacterium.
INTRODUCTION
Open oceans and coastal seas are oligotrophic habitats for heterotrophic microorganisms. Key factors for their ecological success in the photic zone are the usage of light with proteorhodopsin or photosystem I for energy conservation (mixotrophic growth) and the efficient uptake of nutrients. Active transport occurs by high-affinity uptake systems in the membranes, most efficiently via energy-driven uptake across the outer and inner membranes. Limiting factors are the diffusive fluxes of nutrients toward the cell (1) and the surface area of the cells (2). A larger surface-to-volume ratio implies that more surface area feeds a given cytoplasmic volume. Consequently, small microorganisms thrive favorably in oligotrophic habitats (3, 4). The abundant bacterioplankton species are small, have reduced genomes, and are immobile, as seen for “Candidatus Pelagibacter ubique” (5). Cell surface extensions are an alternative evolutionary solution for a larger surface-to-volume ratio. Most microorganisms have low intracellular water activity and high turgor pressure that forces the inner membrane tightly against the murein sacculus or the outer cell membrane and forms the cell morphology. Therefore, cell surface extensions have to be well structured to be stable. For example, prosthecae are buds or stalks formed by an outgrowth of the cell containing cytosol, outer and inner membranes, and peptidoglycan (6). In oligotrophic environments, prosthecate bacteria are more abundant (7), for example, the genus Prosthecobacter that has a cytoskeleton of tubulins (8). In contrast, copiotrophic bacteria have appendages, such as fimbriae, pili, and flagella, to interact with nutrient-rich environments. These extracellular structures contribute indirectly to an increased nutrient uptake via attachment to surfaces and motility.
Marine bacterioplankton cells that were characterized by atomic force microscopy have cell surface structures on the nanometer scale; five out of six free-living coastal bacteria possessed distinct cell surface architectures (9, 10). These surface structures extend up to 6 μm from the cell. Approximately 30% of all free-living bacteria were conjointed by cell surface structures (9). The implications of these associations for the ecological functioning of free-living bacteria await exploration, including their relevance for nutrient uptake.
The present study was initiated following the detection of unusual extracellular structures on cells of a flavobacterial strain in electron micrographs. Strain Hel3_A1_48 was inactivated by blending cultures on a vortex mixer. This mechanosensitivity hampered the isolation. To get the abundant marine bacterium from a coastal bacterioplankton spring bloom in culture, we used dilution-to-extinction cultivation in an oligotrophic, defined liquid medium (11). Strain Hel3_A1_48 is putatively affiliated with the genus Formosa (11). The strain possesses a special set of enzymes for laminarin degradation, the storage polysaccharide released from lysed algae (12). The 16S rRNA gene phylotype of the strain was recurrent within a succession of microbial populations that thrive on the remains of annual spring algal blooms in the North Sea near Helgoland (13–15). In addition, Formosa-affiliated phylotypes are abundant species in bacterioplankton blooms following an algal bloom in the annual local climate cycle, as is evident from 16S rRNA gene amplicon sequences from the North Sea and the Atlantic coast of North America (16). The 16S rRNA gene phylotypes with a high sequence identity (> 97.5%) to strain Hel3_A1_48 are ubiquitous in marine bacterioplankton. They were also detected in the Mediterranean Sea (17), the Atlantic Ocean (18, 19), and the Pacific Ocean (20, 21).
To understand the nature of the large cellular protrusions of strain Hel3_A1_48, we characterized the appendages by electron microscopy and proteomics. Our observations indicated a protrusion of the outer membrane in the form of a tube that is unstable and transforms into a chain of interconnected vesicles in a process called pearling (22, 23).
Pearling is an abiotic morphogenic process in which metastable phospholipid vesicles protrude tubes from the membrane to stabilize the vesicle by shrinking in diameter. The tube subsequently transforms, starting at the distal end of the tube, into a chain of interconnected vesicles (“pearls on a string”) (22). Inducers of pearling affect the tension of the membrane either by forces (22, 24–26) or by modification of the membrane curvature and, thus, finally yield a membrane asymmetry (27, 28). The conformation as a chain of vesicles is supported by the energetic minimum of the mean curvature, described as a Delaunay shape (28). Vesicle chains have been reported for Shewanella oneidensis (29), Francisella novicida (30), Myxococcus xanthus (31), Flavobacterium columnare (32), and Flavobacterium psychrophilum (33). The chains of interconnected outer membrane vesicles (OMVs), as characterized by cryo-electron tomography, differ clearly in size and shape from pili and flagella, as seen for the mechanosensitive OMVs of surface-attached Shewanella oneidensis cells (29). For this bacterium, the biosynthesis of the vesicle chains was suggested to occur by budding from the outer membrane and immediate vesicle formation (29). Here, we report evidence for a variation of outer membrane vesicle synthesis, the intermediate formation of membrane tubes that pearl into chains of interconnected vesicles.
RESULTS
Morphology and fine structure of appendages.
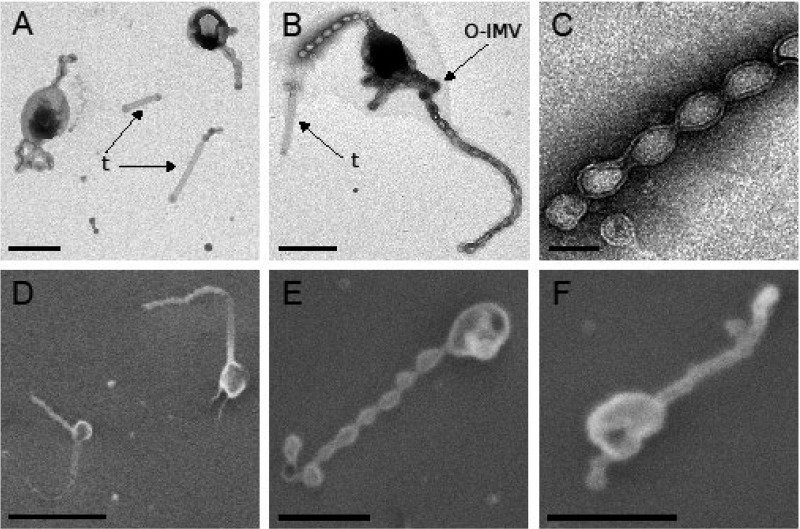
Strain Hel3_A1_48 cells appeared as oval-shaped cells with a length of 500 to 1,100 nm and a width of 300 to 600 nm in transmission electron microscopy (TEM) and scanning electron microscopy (SEM) micrographs (Fig. 1; see Fig. S1 and S2 in the supplemental material). The cells were directly stained and air-dried to avoid mechanical stress during centrifugation. Drying seemed to collapse the cytosol (Fig. 1; Fig. S1 and S2). Appending chains of at least two vesicles were observed on 69% of cells (n = 354). We observed up to five chains per cell (Fig. S1A), with a length variation between 1 and 10 μm (Fig. S1). Individual vesicles in the chains were consistently 80 to 100 nm in length and 50 to 80 nm in diameter. TEM images also revealed straight tubes (Fig. 1 A and B; Fig. S1B and D). They had lengths of 0.5 to 2 μm and diameters of 50 to 80 nm. Some appendages appeared as branching vesicle chains that may be an artifact from the preparation (Fig. S1E and I). Thicker vesicles were also detected, with a size of 160 to 250 nm in length and 130 to 200 nm in diameter (Fig. 1B; Fig. S1A, D, and G). They appeared more frequently at the end of a chain. This minor fraction was identified as vesicles with two membranes based on micrographs obtained by thin-section TEM (see below). The SEM images of cells dehydrated with ethanol and dried at the critical point confirmed the TEM observations (Fig. 1D to F; Fig. S2). Cell protuberances were chains of vesicles up to 2 μm long. Chains contained up to 20 vesicles, with lengths of 100 to 140 nm and diameters of 70 to 100 nm. Detached single vesicles, tubes, and chains of vesicles were also observed, with increasing quantities in samples that had undergone mechanical stress (centrifugation and resuspension) (data not shown), indicating a fragility of the cellular protrusions.
FIG 1.
Micrographs of strain Hel3_A1_48 cells in stationary growth phase. Membrane tubes (t) and thicker vesicles (assigned as O-IMVs) are indicated. Cells grown in HaHa_100V medium at 21°C to stationary phase were negatively stained with 1% uranyl acetate for transmission electron microscopy (TEM) (A to C). The cells were passively settled on a silica wafer, dehydrated by an ethanol series, and preserved using critical point drying for scanning electron microscopy (SEM) (D to F). Bar corresponds to 100 nm (C), 500 nm (A, B, E, and F), or 1 μm (D).
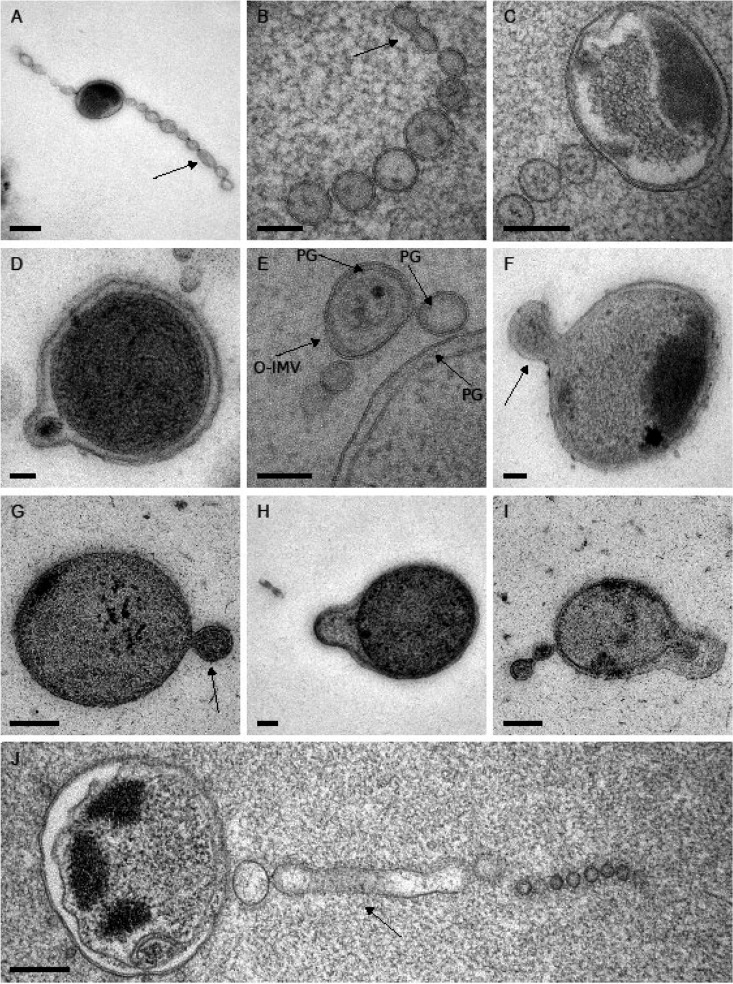
For high-resolution analysis of the vesicle systems, stationary cells were high-pressure frozen, freeze-substituted, and cut into 70-nm thin sections. The extracellular structures were predominantly vesicle chains with one membrane and a length of up to 1.8 μm. The chains were connected to cells or in close proximity (Fig. 2A, C, and J; see Fig. S3D in the supplemental material). Detached chains were rarely observed (Fig. 2B; Fig. S3C) and membrane tubes were also scarce (Fig. 2J). The presence of one membrane in the abundant vesicles suggested an origin from the outer membrane (Fig. 2A to C and J). These outer membrane vesicles (OMVs) had a length of 80 to 120 nm and a diameter in the range of 50 to 80 nm. Larger vesicles with two membranes were present as a minor fraction, with lengths of 120 to 200 nm and diameters of 90 to 160 nm (Fig. 2D to F; Fig. S3A, B, D, and E and Fig. S6A). They were annotated as outer and inner membrane vesicles (O-IMVs) (according to reference 34). The periplasm of many cells appeared as a large space with a span of 15 to 20 nm and was enlarged in the area of cellular protrusions (Fig. 2F to I; Fig. S3A). Several cells showed an onset of a protrusion of the outer membrane or of both membranes (Fig. 2G to I). This suggested that the vesicles are extrusions of the outer membrane of strain Hel3_A1_48 cells. Peptidoglycan is covalently linked to the outer membrane by lipoprotein(s) (35, 36) and may be extruded together with the outer membrane. This hypothesis was supported by peptidoglycan-like structures (37) that were detected in the periplasm close to the outer membrane (Fig. 2E; see Fig. S4 in the supplemental material) as well as within OMVs (Fig. 2E; Fig. S3C) and O-IMVs (Fig. 2E). Cells showed an internal structure that was heavily stained (Fig. 2H; Fig. S3A and B). Element analyses of the cellular content by energy-dispersive X-ray analysis (EDX) identified elevated concentrations of magnesium, potassium, calcium, uranium (originating from the stain), oxygen, and phosphorus for the heavily stained structure (see Fig. S5 in the supplemental material). This composition was consistent with the composition of polyphosphate granules (38, 39).
FIG 2.
Thin sections of strain Hel3_A1_48 in transmission electron micrographs. Highlighted is a small part of tube that has not pearled (A), a detached chain of vesicles with an incomplete pearling at one end of the chain (B), structures assigned as peptidoglycan (PG) (E), O-IMVs with two membranes (E to G), protrusions as onset of the synthesis of chains (F to I), and an attached tube that has incompletely pearled (J). Cells grown in HaHa_100V medium at 21°C to stationary phase were pelleted and prepared by high-pressure freezing, freeze-substitution, and sectioning into 70-nm thin sections for TEM. Bar corresponds to 50 nm (G, H), 100 nm (B, D, I), 200 nm (C, E, F), or 250 nm (A, J).
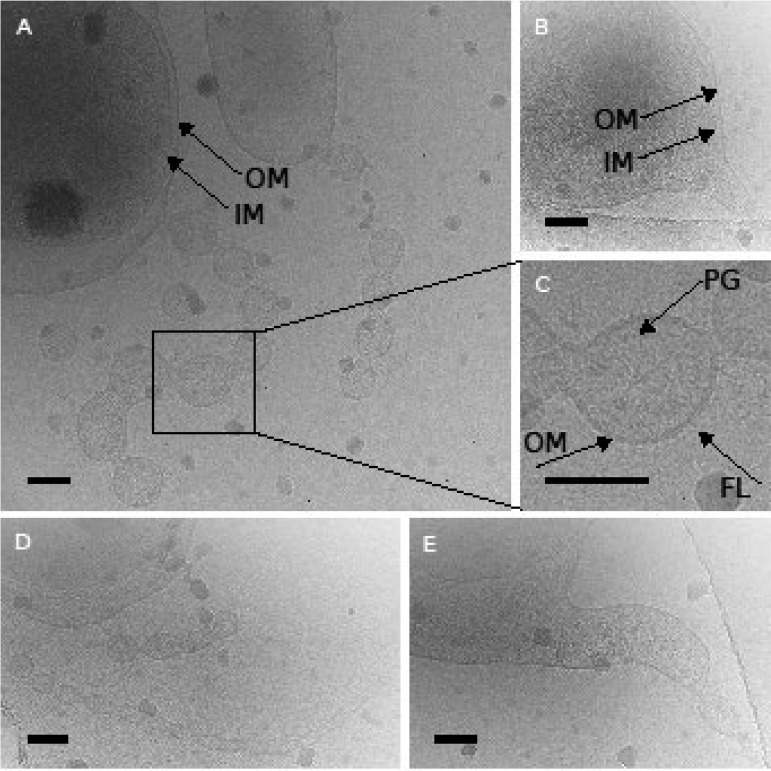
Cryo-electron micrographs showed a large periplasm (Fig. 3A and E; Fig. S6B), chains of vesicles (Fig. 3A; Fig. S6B), and membrane tubes (Fig. 3D and E). These structures had one membrane (Fig. 3A, C, D, and E; Fig. S6B). An electron-dense layer was visible on the inner side of the membrane (Fig. 3C), possibly representing peptidoglycan (40). The outer surface had a fuzzy layer (Fig. 3C), similar to the lipid-anchored protein layer on the outer membrane of other members of the phylum Bacteroidetes (41–43). A transition from a tubular appendage to a vesicle chain was observed at the end of an appendage structure (Fig. 3D). The cells appeared polymorphic, with protrusions of the cytoplasm (Fig. 3B) or even forming an inner membrane vesicle, both still enclosed by the outer membrane of the cell (Fig. S6A).
FIG 3.
Cryo-electron micrographs of stationary-phase cells of strain Hel3_A1_48 show a large periplasm (A, E), a protrusion of the cytoplasm (B), peptidoglycan and a fuzzy OM on the outside (C), and tubular appendages (D, E). Inner and outer membranes (IM and OM) as well as peptidoglycan (PG) and the fuzzy OM surface layer (FL) are indicated. Cells grown in HaHa_100V medium were directly frozen on the grid in liquid ethane and observed in a frozen state. Bars correspond to 100 nm.
The micrographs revealed a differential staining behavior of the cytoplasm and the periplasm where the cytoplasm stained more intensely. The lumen of the vesicles showed a staining behavior identical to that of the periplasm, an indication that the outer membrane is source of the vesicle membrane.
Biosynthesis of vesicle chains.
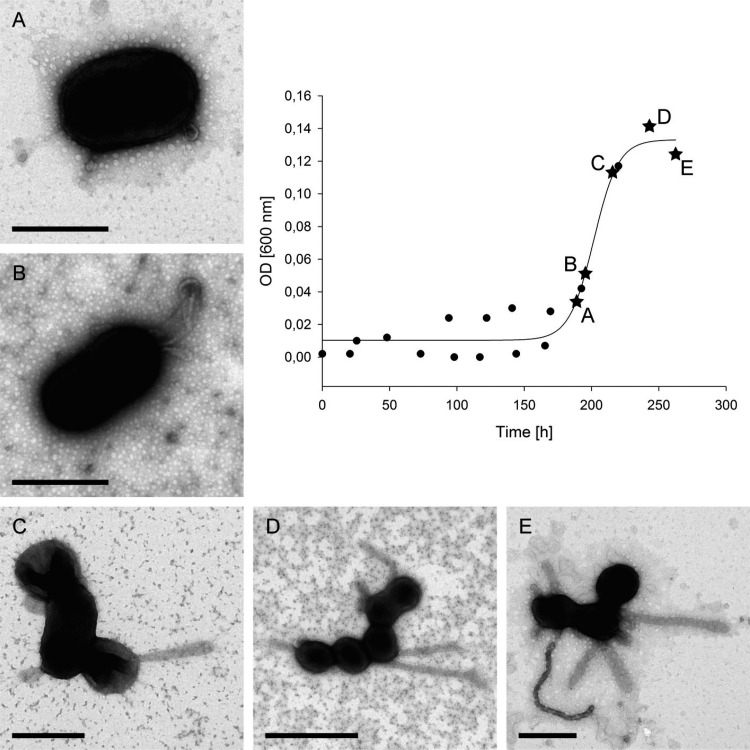
Formation of the extracellular structures was studied over the growth curve. Strain Hel3_A1_48 grew within 10 days to an optical density at 600 nm (OD600) of 0.14 (Fig. 4). The medium provided nutrition balanced in carbon and nitrogen with a small surplus of phosphate. The latter was used by the cells to store polyphosphate granules, as shown by SEM-EDX (Fig. S5). Cells possessed no or very short appendages in the early exponential phase (Fig. 4A and B). Tubes were observed at the end of the exponential growth phase (Fig. 4C). The early stationary phase revealed cells with tubes (Fig. 4D) and some tube chains of vesicles, likely formed by pearling (Fig. 4E). Cells sampled later in the stationary phase had more and longer vesicle chains (Fig. 1). Thus, the TEM micrographs of different growth phases suggested a protrusion of a membrane tube that in a later stage was transformed into a chain of vesicles.
FIG 4.
Appendage formation during growth of strain Hel3_A1_48 cells. The cells were grown in HaHa_100V medium at 21°C and sampled at different time points for TEM analyses. Cells were fixed and negatively stained with 1% uranyl acetate. Bar corresponds to 500 nm (A–C, E) and 1 μm (D).
The influence of growth conditions on the formation of appendages was tested by changing the carbon-to-nitrogen ratio in the medium. The strain grew on media with a carbon surplus (glucose) or a nitrogen surplus (Casamino Acids or peptone) (see Fig. S7 and S8 in the supplemental material). We also tested several organic sulfur compounds, without better growth of the strain. Electron micrographs revealed that the appendages were formed under all tested conditions (see Fig. S9 in the supplemental material).
Proteins in membranes and vesicles.
The strain Hel3_A1_48 is mechanosensitive. Shewanella oneidensis OMVs are fragile and can be observed only after glutaraldehyde fixation (29). These and our electron microscopy (EM) observations suggest that the nonattached vesicles detected in electron micrographs of cultures likely originated from vesicle chains that were originally on cell surfaces and broke off. We tested several methods for the preparation of outer membranes and of detached vesicles (see the methods section of the supplemental material). Analysis by SDS-PAGE and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF-MS) of outer membrane preparations (Fig. S10A) and of cell-free vesicle fractions (Fig. S10B) revealed a range of outer membrane proteins, mainly porins and outer membrane transporters, known as TonB-dependent receptors (see Table S1 in the supplemental material). Most abundant was a putative OmpL-like beta-barrel porin (GenBank accession number AOR25254) with a calculated mass of 34 kDa. Proteins of the flavobacterial type IX secretion system (T9SS, Por) (44) were detected in the vesicle fraction, namely, the gliding-motility-associated protein GldN and lipoproteins GldJ and GldK and a lamin tail domain (LTD)-containing protein (GenBank accession number AOR25680) with the C-terminal sorting domain of the T9SS. The predicted size of 61 kDa contrasted with sizes of 220 and 240 kDa on SDS gels, suggesting a covalent modification. These initial studies suggested an origin of the vesicles from the outer membrane and prompted an in-depth comparison of proteins present in a total membrane preparation from cells and in a cell-free vesicle fraction obtained as a 43-Svedberg (43S) pellet of a 0.2-μm filtrate.
Cells and vesicles used for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses of tryptic oligopeptides were harvested from cultures at exponential and stationary growth phases (OD600s of 0.108 to 0.134 [exponential phase], 0.141 to 0.145 [maximal OD], and 0.098 to 0.112 [stationary phase]) (see Fig. S11 and S12 in the supplemental material). Of 1,866 protein-coding sequences present in the genome, we detected 1,310 proteins in the membrane fraction and 374 proteins in the vesicle fraction originating from cultures in the exponential phase. Totals of 1,075 and 285 proteins were detected in membranes and vesicles representing stationary-phase cultures, respectively.
A PSORTb 3.0.2 (45) prediction annotated the cellular location of proteins. Outer membrane proteins were well represented in all four samples, with 78% to 89% of all predicted outer membrane proteins detected in the fractions. Proteins of the inner membrane were present in the membrane fraction (43% and 60% detection coverage), but this coverage was not observed in the vesicle samples (4% and 11% detection coverage) (see Table S2 in the supplemental material). This result supported an origin of the vesicles from the outer membrane. Cytosolic, periplasmic, and extracellular proteins had detection coverages of 8% to 84%, 47% to 82%, and 44% to 72%, respectively (see Table S4 in the supplemental material). The membrane fractions contained many cytosolic proteins due to a technical reason. To avoid losses of chains of vesicles, we did not wash the membrane fraction. Consequently, proteins loosely attached to the membranes were also harvested. In quantitative terms, outer membrane proteins represented 22% and 27% of the total detected proteins (quantified as normalized spectral abundance factors) in the vesicle preparation but only 6 to 7% in the membranes. Cytoplasmic proteins accounted for 7 and 8% of the vesicle preparation and 12 and 7% of the membrane fraction. Based on the PSORTb 3.0.2 definition of membrane proteins, this suggested an enrichment of outer membrane proteins in the vesicles (see Table S3 in the supplemental material).
In a second analysis, a set of well-characterized marker proteins (Table S4) and their relative abundance in samples (NSAF%) was used to compare the portion of the cellular compartments in the preparations. Periplasmic and outer membrane proteins increased in relative abundance from the exponential to the stationary phase (see Table S5 in the supplemental material), coinciding with the observed increase in appendage numbers and sizes on the cells, but also with the expected consumption of cytosolic proteins in the stationary phase. The relative abundance of outer membrane proteins was larger in the vesicle fraction, whereas proteins from the cytosol and the inner membrane were less abundant in vesicles in comparison to the membrane preparation. Thus, the marker protein analyses indicated an enrichment of outer membrane proteins in the vesicles.
Among the >1,000 detected proteins, 88 proteins had normalized spectral abundance factors (NSAF%) of >0.4 in one of the samples and were manually annotated, in addition to the automatic annotation (Table 1). Of these, 29 proteins were outer membrane proteins or associated with the OM according to PSORTb or a manual annotation, which focused on the type IX secretion system of flavobacteria, the T9SS complex. PSORTb analysis relies on the detection of beta barrels and transmembrane alpha helices for the identification of outer and inner membrane proteins, respectively. In this study, proteins of the T9SS complex and proteins possessing a peptidoglycan binding domain similar to that of OmpA were often not identified by PSORTb as outer membrane proteins and required manual annotation. GldB, -J, -K, -L, -M, and -N; SprA, -E, -F, and -T; and PorV and -U of the T9SS were detected in membranes and in vesicles (see Table S6 in the supplemental material), together with 20 proteins (five with NSAF% > 0.4) which possessed the T9SS C-terminal target domain (TIGRfam04131 and -04183) directing an export via T9SS to the outer membrane surface and a covalent modification of the exported protein with an anionic lipopolysaccharide (43) (see Table S7 in the supplemental material). Catalytic activities of these enzymes, which are expected to be located as lipoproteins on the surface of the outer membrane, include endonucleases, peptidases, and glycoside hydrolases for the degradation of biogenic polymers. Transport of the degradation products through the outer membrane may occur by porins for passive transport as well as by active transport systems with ExbD/TolR- and TonB-dependent outer membrane proteins. Both T9SS and TonB-dependent proteins were abundant in the membrane and vesicle fractions. The list of abundantly detected proteins also revealed experimental limitations. The 43S pellet included the vesicle fraction but also remains of lysed cells, namely, membranes, proteins, and protein complexes smaller than 0.2 μm but larger than 1.3-MDa molecular mass, such as chaperone or glutamine synthetase complexes (Table 1). Also, inner membrane proteins of large enzyme complexes spanning inner and outer membranes, such as ExbBD-TonB, may be exported as part of the whole complex in the formation of cellular appendages from outer membranes.
TABLE 1.
Most abundant proteins from the vesicle and the membrane fraction of the exponential and stationary growth phase cells of strain Hel3_A1_48a
| GenBank accession no. | Annotation | Mol wt (kDa) | NSAF% of: |
Localization according to: |
||||
|---|---|---|---|---|---|---|---|---|
| VE | VS | ME | MS | PSORTb 3.0.2 | Manually annotated | |||
| AOR25254 | OmpL-like OM-channel, beta-barrel porin-2 | 35 | 8.330 | 6.719 | 2.611 | 4.150 | Unknown | Outer membrane |
| AOR25288 | Protein with peptidoglycan binding domain similar to OmpA | 31 | 5.689 | 4.695 | 1.144 | 2.003 | Cytoplasmic membrane | Periplasm |
| AOR25736 | Gliding motility lipoprotein GldJ | 64 | 3.687 | 3.168 | 0.182 | 0.196 | Outer membrane | OM + periplasm |
| AOR26642 | Gliding motility lipoprotein GldK | 52 | 3.347 | 2.605 | 0.198 | 0.189 | Cytoplasmic | OM + periplasm |
| AOR26723 | OmpL-like OM-channel, beta-barrel porin-2 | 35 | 3.186 | 1.464 | 1.408 | 1.581 | Outer membrane | |
| AOR26645 | Gliding motility protein GldN | 41 | 2.615 | 2.859 | 0.156 | 0.145 | Cytoplasmic | OM + periplasm |
| AOR26575 | Protein with domain of unknown function (DUF5017) | 52 | 2.503 | 0.870 | 0.712 | 0.658 | Outer membrane | |
| AOR25738 | PorV (T9SS) | 47 | 2.179 | 3.286 | 0.288 | 0.212 | Outer Membrane | |
| AOR25966 | TonB-dependent outer membrane protein of SusC/RagA family | 112 | 1.852 | 3.622 | 1.712 | 1.805 | Outer membrane | |
| AOR25585 | Conserved protein (DUF4136) | 19 | 1.803 | 4.161 | 0.831 | 1.285 | Unknown | |
| AOR26486 | Protein with unknown function | 21 | 1.702 | 3.537 | 0.379 | 0.299 | Outer membrane | |
| AOR26716 | DUF1573 domain-containing protein | 17 | 1.324 | 2.463 | 0.553 | 0.955 | Unknown | |
| AOR25720 | Protein with unknown function | 35 | 1.217 | 2.147 | 0.099 | 0.056 | Unknown | |
| AOR26448 | TonB-dependent outer membrane protein of SusC/RagA family | 105 | 1.001 | 2.390 | 0.641 | 1.386 | Outer membrane | |
| AOR25874 | PorT family protein in outer membrane | 28 | 0.975 | 0.432 | 0.051 | 0.060 | Unknown | Outer membrane |
| AOR26302 | OmpA family protein, with peptidoglycan binding domain | 50 | 0.972 | 2.576 | 1.343 | 1.389 | Outer membrane | |
| AOR25767 | FKBP-type peptidyl-prolyl cis-trans isomerase | 38 | 0.893 | 0.324 | 0.127 | 0.074 | Unknown | |
| AOR26643 | Gliding motility protein GldL | 22 | 0.874 | 0.256 | 1.545 | 2.432 | Unknown | CM + periplasm |
| AOR26876 | C-terminal processing peptidase_S41_CPP | 53 | 0.794 | 1.065 | 0.145 | 0.144 | Outer membrane | |
| AOR26784 | DUF541 domain-containing protein (YggE) with kinase-interacting SIMPL domain | 26 | 0.760 | 0.564 | 0.279 | 0.505 | Unknown | |
| AOR25680 | T9SS C-terminal target domain-containing protein with lamin tail domain | 61 | 0.693 | 1.875 | 0.406 | 0.465 | Unknown | OM + extracellular |
| AOR25530 | Elongation factor Tu | 43 | 0.692 | 0.109 | 0.648 | 1.197 | Cytoplasmic | |
| AOR26574 | TonB-dependent receptor | 103 | 0.622 | 0.656 | 0.297 | 0.393 | Outer membrane | |
| AOR26993 | T9SS C-terminal target domain-containing protein | 40 | 0.620 | 0.317 | 0.117 | 0 | Extracellular | |
| AOR26505 | Protein with unknown function | 42 | 0.606 | 3.895 | 0.037 | 0.410 | Unknown | |
| AOR26632 | Chaperonin GroEL | 57 | 0.603 | 0.090 | 0.999 | 1.073 | Cytoplasmic | |
| AOR26184 | T9SS membrane protein PorP/SprF | 38 | 0.592 | 1.131 | 0.110 | 0.064 | Outer membrane | |
| AOR26178 | Protein with unknown function | 26 | 0.584 | 0.388 | 0.083 | 0.037 | Unknown | |
| AOR26182 | OmpH-like outer membrane protein | 19 | 0.550 | 1.089 | 0.202 | 0.256 | Unknown | Outer membrane |
| AOR26484 | T9SS C-terminal target domain-containing protein with sodium/calcium exchanger | 378 | 0.536 | 0.284 | 0.060 | 0.017 | Unknown | OM + extracellular |
| AOR26594 | Protein with unknown function | 48 | 0.535 | 0.389 | 0.014 | 0.016 | Outer membrane | |
| AOR25578 | Glutamine synthetase | 38 | 0.532 | 0.097 | 1.091 | 1.824 | Cytoplasmic | |
| AOR25822 | DUF1573 domain containing protein | 15 | 0.523 | 0.271 | 0.073 | 0.039 | Unknown | |
| AOR26504 | TonB-dependent outer membrane protein of SusC/RagA family | 105 | 0.521 | 0.648 | 0.277 | 0.310 | Outer membrane | |
| AOR25487 | T9SS C-terminal target domain-containing protein with fibronectin type III domain | 96 | 0.510 | 0.538 | 0.124 | 0.104 | Extracellular | |
| AOR26551 | MotA/TolQ/ExbB proton channel family protein for biopolymer transport | 28 | 0.500 | 0.102 | 1.095 | 1.220 | Cytoplasmic membrane | |
| AOR26233 | DUF2490 domain-containing protein, outer membrane beta-barrel protein | 28 | 0.499 | 1.263 | 0.034 | 0.027 | Outer membrane | |
| AOR25413 | DUF1566 domain-containing protein | 30 | 0.473 | 0.330 | 0.013 | 0.009 | Extracellular | |
| AOR26870 | Cytochrome c superfamily | 13 | 0.470 | 0.678 | 0.079 | 0.099 | Unknown | |
| AOR26555 | Biopolymer transporter ExbD/TolR | 18 | 0.464 | 0.061 | 0.469 | 0.683 | Unknown | Cytoplasmic membrane |
| AOR25345 | Protein with unknown function | 38 | 0.450 | 0.514 | 0.099 | 0.079 | Outer membrane | |
| AOR25794 | YceI family protein | 21 | 0.448 | 0.363 | 0.273 | 0.096 | Unknown | |
| AOR26879 | DUF4252 domain-containing protein | 21 | 0.438 | 0.570 | 0.107 | 0.138 | Cytoplasmic | |
| AOR26556 | Outer membrane protein with beta-barrel domain (PorT family protein) | 25 | 0.436 | 0.576 | 0.064 | 0.011 | Outer membrane | |
| AOR25309 | TPR 6 superfamily (Tol-Pal system protein YbgF) | 28 | 0.409 | 0.096 | 0.543 | 0.589 | Unknown | |
| AOR26266 | TonB-dependent outer membrane cobalamin receptor protein | 67 | 0.409 | 0.296 | 0.320 | 0.248 | Outer membrane | |
| AOR26066 | Protein with unknown function | 22 | 0.408 | 2.147 | 0.047 | 0.030 | Cytoplasmic membrane | |
| AOR25967 | RagB/SusD family nutrient uptake outer membrane protein | 52 | 0.398 | 0.710 | 0.348 | 0.637 | Extracellular | |
| AOR27028 | Prohibitin family in SPFH superfamily, regulator of protease activity HflC | 30 | 0.394 | 0.101 | 0.743 | 0.679 | Cytoplasmic | |
| AOR26810 | Transporter in putative MetA pathway of phenol degradation | 37 | 0.373 | 0.654 | 0.081 | 0.076 | Outer membrane | |
| AOR26085 | Zn-dependent peptidase of M16_C superfamily | 76 | 0.363 | 1.018 | 0.173 | 0.259 | Unknown | |
| AOR26098 | Protein with peptidoglycan binding domain similar to OmpA | 35 | 0.352 | 0.682 | 0.229 | 0.537 | Cytoplasmic membrane | |
| AOR25407 | T9SS C-terminal target domain-containing protein | 49 | 0.350 | 0.785 | 0.080 | 0.094 | Unknown | |
| AOR26761 | Periplasmic protein involved in polysaccharide biosynthesis/export | 28 | 0.336 | 0.237 | 0.198 | 0.445 | Unknown | Periplasmic |
| AOR25464 | Tetratricopeptide (TPR) repeat protein | 47 | 0.335 | 0.472 | 0.147 | 0.173 | Unknown | |
| AOR25606 | Tetratricopeptide (TPR) repeat protein | 55 | 0.326 | 0.426 | 0.178 | 0.304 | Unknown | |
| AOR25974 | Protein with unknown function | 31 | 0.306 | 0.631 | 0.080 | 0.089 | Unknown | |
| AOR25945 | 50S ribosomal protein L5 | 20 | 0.271 | 0 | 0.601 | 0.339 | Cytoplasmic | |
| AOR25710 | Protein with unknown function | 38 | 0.257 | 0.532 | 0.148 | 0.362 | Unknown | |
| AOR26447 | FoF1 ATP synthase subunit beta | 54 | 0.235 | 0.017 | 0.462 | 0.310 | Cytoplasmic | |
| AOR26084 | Zn-dependent peptidase of family M16 | 50 | 0.231 | 0.416 | 0.106 | 0.158 | Periplasmic | |
| AOR25320 | Amidohydrolase superfamily | 48 | 0.223 | 0.496 | 0.048 | 0.095 | Unknown | Cytoplasmic |
| AOR26257 | 30S ribosomal protein S2 | 29 | 0.219 | 0 | 0.568 | 0.323 | Cytoplasmic | |
| AOR26083 | 50S ribosomal protein L21 | 19 | 0.212 | 0 | 0.509 | 0.329 | Cytoplasmic | |
| AOR26932 | Molecular chaperone DnaK, HSP70 | 68 | 0.211 | 0 | 0.437 | 0.305 | Cytoplasmic | |
| AOR26975 | Peroxiredoxin (PRX) family | 24 | 0.208 | 0.092 | 0.338 | 0.416 | Cytoplasmic | |
| AOR26152 | Argininosuccinate synthase | 44 | 0.204 | 0.031 | 0.631 | 0.324 | Cytoplasmic | |
| AOR26449 | DUF4270 domain-containing protein | 61 | 0.202 | 0.428 | 0.139 | 0.137 | Unknown | |
| AOR26407 | ATP synthase Fo subunit B | 18 | 0.194 | 0.096 | 0.460 | 0.768 | Cytoplasmic membrane | |
| AOR25488 | Enoyl-ACP reductase | 30 | 0.182 | 0.084 | 0.243 | 0.440 | Cytoplasmic membrane | |
| AOR25526 | 50S ribosomal protein L1 | 24 | 0.181 | 0 | 0.456 | 0.223 | Cytoplasmic | |
| AOR26149 | Pyrroline-5-carboxylate reductase | 29 | 0.179 | 0.041 | 0.463 | 0.366 | Cytoplasmic | |
| AOR26577 | Protein with peptidoglycan binding domain similar to OmpA and TolB | 69 | 0.164 | 0.591 | 0.184 | 0.244 | Cytoplasmic membrane | |
| AOR25631 | DUF3347 domain-containing protein | 22 | 0.162 | 0.424 | 0.075 | 0.079 | Cytoplasmic | |
| AOR25933 | 30S ribosomal protein S4 | 23 | 0.147 | 0 | 0.684 | 0.421 | Cytoplasmic | |
| AOR26554 | Biopolymer transporter ExbD/TolR | 23 | 0.136 | 0 | 0.461 | 0.555 | Cytoplasmic | |
| AOR25718 | Bifunctional preprotein translocase subunit SecD/SecF | 107 | 0.130 | 0.026 | 0.483 | 0.368 | Cytoplasmic membrane | |
| AOR26774 | Type I glyceraldehyde-3-phosphate dehydrogenase | 36 | 0.111 | 0.042 | 0.175 | 0.471 | Cytoplasmic | |
| AOR26106 | 3-Deoxy-7-phosphoheptulonate synthase, chorismate mutase type II domain | 41 | 0.101 | 0 | 0.672 | 0.388 | Cytoplasmic | |
| AOR26764 | DUF3575 domain-containing protein | 23 | 0.086 | 0.478 | 0.036 | 0 | Unknown | |
| AOR26610 | O-Acetylhomoserine aminocarboxypropyl transferase | 46 | 0.085 | 0 | 1.382 | 0.040 | Cytoplasmic | |
| AOR26842 | Copper/zinc superoxide dismutase (SODC) family protein | 20 | 0.073 | 0.930 | 0.112 | 0.321 | Periplasmic | |
| AOR26578 | T9SS membrane protein PorP/SprF | 34 | 0 | 0.472 | 0.166 | 0.041 | Unknown | OM + periplasm |
| AOR25918 | Division/cell wall cluster transcriptional repressor MraZ | 17 | 0 | 0.067 | 0.204 | 0.612 | Cytoplasmic | |
| AOR25956 | 50S ribosomal protein L4 | 23 | 0 | 0 | 0.618 | 0.361 | Cytoplasmic | |
| AOR25649 | Ammonia channel protein AmtB | 43 | 0 | 0 | 0.521 | 0.040 | Cytoplasmic membrane | |
| AOR25940 | 30S ribosomal protein S5 | 18 | 0 | 0 | 0.448 | 0.305 | Cytoplasmic | |
| AOR26305 | Superoxide dismutase | 22 | 0 | 0 | 0.123 | 0.414 | Periplasmic | |
VE, vesicle fraction exponential phase; VS, vesicle fraction stationary phase; ME, membrane fraction exponential phase; MS, membrane fraction stationary phase; OM, outer membrane; CM, cytoplasmic membrane.
Besides enrichment or depletion of protein groups that are defined by their cellular location, we detected one protein with a highly varying relative abundance. A 34-fold-larger NSAF% in the membrane fraction of exponentially growing cells was observed for the O-acetylhomoserine aminocarboxypropyltransferase (EC 2.5.1.49), which captures methanethiol for the synthesis of methionine and, thus, provides a reduced sulfur source for biosynthesis. We tested dimethyl disulfide as a methanethiol precursor and did not observe improved growth of the strain (Fig. S8). However, in nature methanethiol may be the sulfur source for the species. In summary, our detailed proteomic analyses of vesicles and membranes indicated a dominant presence of outer membrane proteins in the vesicles.
Binding of laminarin.
Laminarin is a soluble storage polysaccharide of algae. It consists of a linear β-1,3-linked glucose chain with β-1,6-linked side chains. Strain Hel3_A1_48 has a small genome with three large operons coding for laminarin degradation (12). Fluorescein-labeled laminarin is expected to bind to surface-anchored domains to be hydrolyzed to oligosaccharides by a surface-attached endoglycosylhydrolase and to be imported into the periplasmic space of flavobacteria by an active transport mechanism, a TonB-dependent transporter system (46). We investigated whether the label also accumulated in the vesicle chains. Structured illumination micrographs detected the fluorescent label in the outside cells and also in the appendages (Fig. 5). The staining of the vesicle chains illustrated the binding of laminarin to a carbohydrate-binding module on the surface and/or a diffusion of a fluorescently labeled, laminarin-derived compound from the periplasm into the vesicle lumen because the uptake occurs in the outer membrane with a transporter system that is in physical contact with an energy-consuming protein complex located in the inner membrane. The latter would be indicative of the TonB-dependent “selfish” substrate uptake mechanism as described for Gramella forsetii (46, 47).
FIG 5.
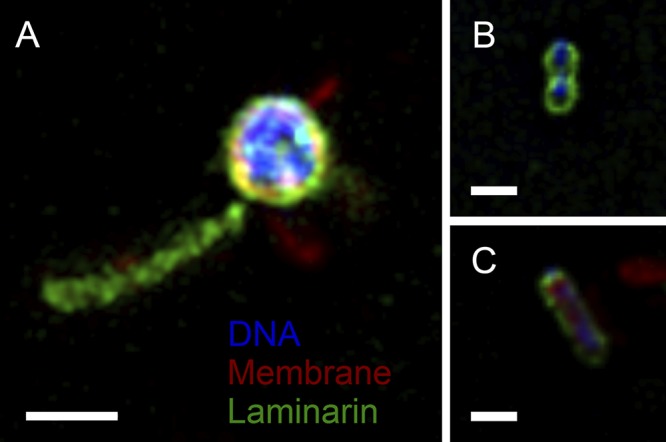
Superresolution structured illumination microscopy image of strain Hel3_A1_48 in stationary growth phase reveals the label of laminarin (green color) on the outside cells (Nile red membrane stain, red color; DAPI DNA stain, blue color) and on appendages (A) and on dividing cells (B, C). Bars correspond to 1 μm.
DISCUSSION
The flavobacterium strain Hel3_A1_48 has large surface appendages in the form of chains of vesicles. This study showed that the vesicles have one membrane; only a minor fraction of vesicles with two membranes were observed. In accordance, the vesicle proteome was dominated by proteins of the outer membrane. Growth phase-dependent observations showed the protrusion of membrane tubes. After suggested pearling into chains of vesicles, the vesicle lumina are expected to be interconnected.
The nanoscale size of the outer membrane vesicles, 80 to 100 nm long and 50 to 80 nm wide, coincides with vesicle observations in other bacterial cultures. Shewanella oneidensis forms chains of 100-nm vesicles (29), and Myxococcus xanthus forms chains of vesicles 30 to 60 nm in width (31). Flavobacteria—i.e., Flavobacterium columnare (32) and Flavobacterium psychrophilum (33)—form vesicles but some, such as Muricauda ruestringensis (48) and Flavobacterium columnare (32), also form appendages with one larger vesicle at the distal end. In all studies, thin-section TEM or cryo-EM images showed that the vesicles have only one membrane, and the outer membrane was suggested as a precursor of the vesicle membrane (29, 31–33; this study). Vesicles with two membranes (O-IMVs) were detected in this study as parts of outer membrane vesicle chains. Similar O-IMVs have also been observed for Shewanella vesiculosa (100 to 250 nm in size) (34), as well as Neisseria gonorrhoeae, Pseudomonas aeruginosa, and Acinetobacter baumannii (49).
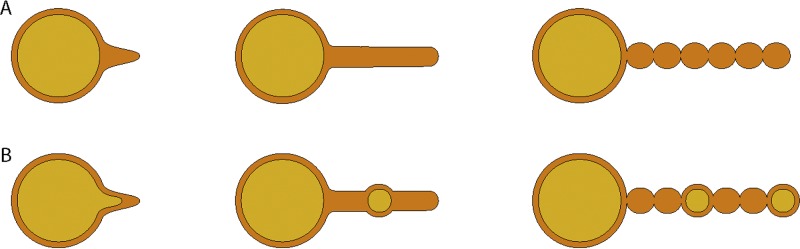
Membrane tubes are suggested to be an intermediate in the pearling process. Protrusion of the outer membrane can lead directly to the formation of interconnected vesicles, as observed by cryo-electron tomography on Shewanella oneidensis (29). In this case, membrane tubes are not a stable intermediate. We observed the appearance of membrane tubes mainly in exponentially growing cultures of strain Hel3_A1_48. The factors increasing the membrane tube stability and slowing down the pearling into vesicle chains are currently not understood. Membrane tubes with greater stability were observed on Francisella novicida, 40 nm in diameter and 0.3 to 1.5 μm in length. Tubes isolated from Francisella cultures pearled at 60°C within 5 minutes into vesicle chains and vesicles without cell contact (30). These observations suggest that extrusions of outer membranes form membrane tubes as intermediates that subsequently pearl into chains of vesicles (Fig. 6).
FIG 6.
Model of formation of the two types of vesicle chains by pearling. (A) Formation of an extension of the outer membrane, which elongates as a tube-like structure and then transforms into a vesicle chain by pearling. (B) Formation of an inner membrane vesicle which remains enclosed by the outer membrane and gets transported away from the cell due to the elongation of the outer membrane tube. This explains the different types of vesicles (O-IMVs and OMVs) in one vesicle chain and also the phenomenon that the end of the vesicle chain has a larger vesicle (Fig. S1A, D, and G).
Cells in the stationary growth phase consume endogenous carbon sources, with the consequence of a reduction of the cytosol volume and turgor pressure. In the absence of internal outer membrane degradation, the periplasmic space is broadened and the stability of the outer membrane morphology is weakened. For example, Pelagibacter ubique, a small mixotrophic photo- and chemotroph and the most abundant planktonic bacterium in the ocean, has an enlarged periplasm in the stationary phase (40) and loses its morphological shape in the absence of light due to collapsing outer membranes (50). Thus, nutrient limitation is one factor that contributes to vesicle formation. However, strain Hel3_A1_48 synthesizes outer membrane appendages already in exponential phase, suggesting a functional advantage conferred by these structures.
Electron micrographs stained with metal ions reveal the presence of negative charges, which includes the polar headgroups of lipids, acidic groups of peptidoglycan, and anionic groups of proteins and nucleic acids. These stains led to the annotation of appendages as chains of vesicles. However, due to the sample thicknesses, the absence of stains in the interconnection of vesicles was not detectable. Cryo-electron tomography revealed that Shewanella oneidensis chains had interconnected vesicles and that the lumina of neighboring vesicles were not separated by a membrane (29). In our study, we used fluorescently labeled laminarin to gain insight into the vesicles. The binding to the vesicles illustrated either binding to so-far-uncharacterized carbohydrate binding domains and/or a connection between periplasma and vesicle lumen. The latter may, thus, present additional storage space for oligosaccharides. Laminarin is hydrolyzed by an outer membrane-attached endoglucanase into oligosaccharides that are transported in an active manner, with energy consumption, into the periplasm. The active transport requires a proton gradient, a proton channel in the inner membrane (ExbBD), an energy transducer TonB protein anchored in the inner membrane and acting on the transporter in the outer membrane, and a SusCD homolog that imports the oligosaccharides (12). A TonB-dependent transporter in the outer membrane acts as a lock, with SusD serving as an outside cap and a motile plug domain on the periplasmic bottom of the transporter. Thus, imported oligosaccharides are caught in the periplasm. They can be further degraded and the monomers can be imported into the cell. The participation of the proton gradient across the inner membrane excludes the possibility of transport across a single outer membrane in vesicles.
The vesicle chains enlarge the surface of strain Hel3_A1_48 and, thus, allow a larger number of enzymes on the cell surface. In oligotrophic habitats, the location of hydrolytic depolymerases (glycoside hydrolases, peptidases, and endonucleases) on the outer membrane surface is advantageous over excretion into the surrounding sea. Functional active transport across the outer membrane provides another ecological advantage in oligotrophic habitats in comparison to organisms that have only porins as passive transporters in the outer membrane. In combination, such a “selfish” uptake (46, 47) limits the distance from the depolymerizing enzyme to the transporter in the outer membrane, thereby suppressing competition with other bacteria. Besides the advantage of having more surface enzymes per cell volume, the chains of vesicles may also provide an advantage by enlarging the surveillance space for polymeric nutrients. In contrast to monomers, polymers have large hydrodynamic radii and low diffusion coefficients. Recognition and binding of the substrate polymers by proteins on the appending vesicle chains is a first step for polymer utilization. This may just tie a rope between the cell and polymer and bring the polymer closer to the cell surface or, alternatively, may be a dedicated molecular machinery that transfers oligosaccharides onto membrane molecules and transports them to the active transporter.
MATERIALS AND METHODS
Cultivation.
Strain Hel3_A1_48 was cultivated in HaHa_100V medium (11) at 21°C with shaking at 55 rpm. A 1% (vol/vol) inoculum was used to inoculate triplicate cultures. Medium additions tested included (final concentration) the following: glucose (2 g/liter); Casamino Acids (0.5 g/liter); phosphate (200 μM); ammonium (0 to 1.6 mM); sodium carbonate (30 mM); and l-methionine, l-cysteine, and l-alanine (0.25 mM and 0.5 mM). Dimethyl disulfide (DMDS; 50 μM or 250 μM) was added to 100 ml HaHa_100V of medium in 1-liter glass bottles with a Teflon stopper. Growth was monitored at 600 nm.
Negative staining and TEM.
Cells were investigated unfixed unless otherwise stated. Fixation was performed using 1% (vol/vol) glutaraldehyde for 1 to 2 h at 21°C. A Formvar-coated 400-mesh copper grid (Quantifoil Micro Tools, Großlöbichau, Germany) was placed on 50 μl of a culture for 3 to 5 minutes. Then the grid was negatively stained with 1% (wt/vol) uranyl acetate for 1 min, washed three times in deionized water, and air-dried. The cells were imaged with a Zeiss EM 902A transmission electron microscope (Carl Zeiss, Oberkochen, Germany) with 80-kV acceleration voltage at calibrated magnifications. Cells were analyzed with the program MeasureIT (Olympus Soft Imaging System GmbH, Münster, Germany).
Thin-section TEM.
Nine milliliters of culture was centrifuged in 1.5-ml portions at 18,500 × g for 5 min. Cell pellets were suspended in 2 μl 20% (wt/vol) bovine serum albumin (BSA) in phosphate-buffered saline (PBS; for Epon embedding) or in 2 μl dextran (high molecular weight) in PBS (for Lowicryl HM20 embedding). After high-pressure freezing in an HPM 010 instrument (Abra Fluid AG, Widnau, Switzerland), freeze substitution was performed with a Leica EM AFS2 system (Leica Microsystems, Vienna, Austria) and cells were embedded into either Epon or Lowicryl HM20 (51). The Epon samples were stained with 1% (wt/vol) OsO4 in acetone and 2% (wt/vol) uranyl acetate in water. Lowicryl samples were stained in 0.1% (wt/vol) uranyl acetate in acetone. Thin sections of 70 nm were cut and floated onto 100-mesh Formvar-coated copper grids (G2410D; Plano GmbH, Wetzlar, Germany). Lowicryl sections were poststained with 2% (wt/vol) uranyl acetate in 70% methanol and Reynolds lead citrate. Sections were visualized with a transmission electron microscope (Biotwin CM 120; FEI, Eindhoven, The Netherlands). For EDX measurements, 300-nm sections were prepared on Formvar-coated slot grids (Quantifoil Micro Tools).
Cryogenic electron microscopy.
Carbon-coated mesh copper grids (C-flat; Protochips, Morrisville, NC, USA) were treated in a glow discharger (PELCO easiGlow; Ted Pella, Inc., Redding, CA, USA), and 3 μl of a freshly grown culture was applied. The sample was frozen in liquid ethane (52) using a Vitrobot (FEI, Hillsboro, OR, USA) (blot time, 2 s; plunge time, 4 s). The samples were kept in liquid nitrogen until visualization with 100 kV in a TF30 Polara microscope (FEI) equipped with an Ultrascan 4000 CCD camera (Gatan, Pleasanton, CA, USA).
Scanning electron microscopy.
After 50 μl of culture was spotted on a silica wafer (5 by 7 mm; Ted Pella, Inc., Redding, CA, USA), cells settled for up to 1 h at 21°C. Samples were dehydrated in an ethanol series (30%, 50%, 70%, 80%, 96%, and 99% [vol/vol], each for 10 min) and loaded into a critical point dryer (EM CPD 300; Leica, Vienna, Austria) (53). Then the wafer was mounted on an aluminum stub with sticky carbon tape (Plano GmbH) and the sample was imaged in a scanning electron microscope (Quanta 250 FEG; FEI) with 2 or 5 keV. Secondary electron images were recorded.
Energy dispersive X-ray analysis.
EDX was performed using a Quanta 250 FEG microscope equipped with a double detector system (XFlash 6/30 series; Bruker Nano GmbH, Berlin, Germany) with 10 and 20 kV on 300-nm thin sections. The EDX detector had an energy resolution of <123 eV at the Mn Kα line. Images were taken with the SEM using the scanning transmission electron microscopy (STEM) detector and processed using the Bruker software package Esprit 1.9.
Proteomic analyses.
Triplicate cultures grown in 250-ml cell culture bottles with slow shaking (10 rpm) on an orbital shaker were harvested in the exponential or stationary growth phase. Vesicles were prepared as described elsewhere (54). Cells for the membrane preparation were pelleted at 4,390 × g for 10 min. After removal of cell debris at 27,440 × g for 15 min (70.1 Ti rotor; Beckman Coulter, Brea, USA), the supernatant was filtered through a 0.2-μm-pore-size syringe filter. The vesicles in the filtrate were pelleted at 142,019 × g for 2 h (70.1 Ti rotor). The vesicle pellet was suspended in 100 mM KCl and 50 mM Tris (pH 7.5), frozen rapidly in liquid nitrogen, and kept at −80°C. The membrane preparation started with the lysis of cell pellets in 10 mM Tris-HCl (pH 7.5) and 10 mM EDTA (TE) by sonication on ice (3 times for 30 s at 30% intensity). Unbroken cells were removed at 8,000 × g for 10 min. Membranes in the supernatant were pelleted at 105,000 × g for 1 h (50.2 Ti rotor; Beckman Coulter). Pellets were frozen in liquid nitrogen and kept at −80°C. Pellets were resuspended in 500 μl TE buffer and homogenized using a pestle. After quantification with a Nanoquant assay (Roth, Karlsruhe, Germany), 25 μg protein per sample was separated on an SDS-PAGE lane. The Coomassie-stained separation lane was cut into 10 equal-sized pieces, which were destained by repeated washing (200 mM NH4HCO3 and 30% [vol/vol] acetonitrile at 37°C for 30 min). Proteins were in-gel digested for 16 h using trypsin (Promega, Madison, WI, USA). Peptides were eluted in an ultrasonic bath for 15 min and desalted using ZipTip columns (C18; Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. Peptide samples were separated by reversed-phase C18 column chromatography on a nanoAcquity ultraperformance liquid chromatography (UPLC) system (Waters Corporation, Milford, MA, USA) as described elsewhere (55). Mass spectrometry (MS) and MS/MS data were recorded using an online-coupled LTQ-Orbitrap classic mass spectrometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). We searched MS spectra against a target-decoy protein database, including sequences of strain Hel3_A1_48 (GenBank accession no CP017259) (12) and of common laboratory contaminants, using the Sorcerer-Sequest platform (v27.11; Thermo Fisher Scientific, San Jose, CA, USA) and Scaffold 4 (56). The false discovery rate (FDR) at the peptide and protein level was set to 0.01 (1%). Only proteins with a minimum of two peptide identifications were considered identified. We used only proteins that were detected in at least two out of three biological replicates. Contaminants were removed. To shift from weight abundance to molecule abundance, total spectral count (TSC) values were normalized to protein size by division by the molecular weight of the detected protein. The normalized spectral abundance factor (NSAF%) values were obtained by dividing the normalized TSCs by the sum of all TSCs for one replicate, giving the molecular incidence of each protein relative to all proteins in the sample (57). Then an average of the NSAF% values of the replicates was calculated (58, 59).
Superresolution structured illumination microscopy.
Cells were grown on 2 g/liter laminarin in HaHa_100V medium and then inoculated to 1% (vol/vol) into fresh HaHa_100V medium containing fluorescein-labeled laminarin (35 μM) (46). After fixation with 2% (vol/vol) formaldehyde, a 50-μl sample was heat fixed (50°C) on a glass microscope slide. The cells were counterstained with 4′6-diamidino-2-phenylindole (DAPI) for 7 min and Nile red for 30 min at final concentrations of 1 and 3 μg ml−1, respectively, and subsequently rinsed with ultrapure water. Cells were covered with Citifluor/VectaShield (4:1, vol/vol) and observed with superresolution structured illumination microscopy (SR-SIM) using a Zeiss ELYRA PS.1 microscope (Carl Zeiss, Oberkochen, Germany) with 561-, 488-, and 405-nm lasers and bandpass (BP) 573 to 613, BP 502 to 538, and BP 420 to 480 nm +LP 750 optical filters. Z-stack images were taken using a Plan-Apochromat 63×/1.4 oil objective and processed using the ZEN2011 software (Carl Zeiss).
Data accessibility.
All proteome raw data were uploaded to the ProteomeXchange Consortium (proteomecentral.proteomexchange.org) via the PRIDE partner repository (60) and are accessible with the data set identifier PXD012522.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daniela Tienken and Swantje Lilienthal for help during sample preparation for SEM, Carol Arnosti for providing the fluorescein-labeled laminarin, and Jan Brüwer for inhibition studies. We thank Jana Matulla, Frank Unfried, and Sebastian Grund for technical assistance with sample preparation and MS/MS analysis and Dirk Albrecht for MALDI measurements.
The project was financed by the Max Planck Society. The work was also financially supported by the DFG in the framework of the research unit FOR2406 “Proteogenomics of Marine Polysaccharide Utilization” (POMPU) via grants to R. Amann (AM 73/9-1), D. Becher (BE 3869/4-1), and T. Schweder (SCHW 595/10-1).
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00829-19.
REFERENCES
- 1.Schulz HN, Jørgensen BB. 2001. Big bacteria. Annu Rev Microbiol 55:105–137. doi: 10.1146/annurev.micro.55.1.105. [DOI] [PubMed] [Google Scholar]
- 2.Koch AL. 1996. What size should a bacterium be? A question of scale. Annu Rev Microbiol 50:317–348. doi: 10.1146/annurev.micro.50.1.317. [DOI] [PubMed] [Google Scholar]
- 3.Gasol JM, del Giorgio PA, Massana R, Duarte CM. 1995. Active versus inactive bacteria: size-dependence in a coastal marine plankton community. Mar Ecol Prog Ser 128:91–97. doi: 10.3354/meps128091. [DOI] [Google Scholar]
- 4.Young KD. 2006. The selective value of bacterial shape. Microbiol Mol Biol Rev 70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, Rappé MS. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 6.Staley JT. 1968. Prosthecomicrobium and Ancalomicrobium: new prosthecate freshwater bacteria. J Bacteriol 95:1921–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poindexter JS. 2006. Dimorphic prosthecate bacteria: the genera Caulobacter, Asticcacaulis, Hyphomicrobium, Pedomicrobium, Hyphomonas and Thiodendron, p 72–90. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (ed), The prokaryotes. Springer, New York, NY. [Google Scholar]
- 8.Jenkins C, Samudrala R, Anderson I, Hedlund BP, Petroni G, Michailova N, Pinel N, Overbeek R, Rosati G, Staley JT. 2002. Genes for the cytoskeletal protein tubulin in the bacterial genus Prosthecobacter. Proc Natl Acad Sci U S A 99:17049–17054. doi: 10.1073/pnas.012516899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malfatti F, Azam F. 2009. Atomic force microscopy reveals microscale networks and possible symbioses among pelagic marine bacteria. Aquat Microb Ecol 58:1–14. doi: 10.3354/ame01355. [DOI] [Google Scholar]
- 10.Malfatti F, Samo TJ, Azam F. 2010. High-resolution imaging of pelagic bacteria by atomic force microscopy and implications for carbon cycling. ISME J 4:427–439. doi: 10.1038/ismej.2009.116. [DOI] [PubMed] [Google Scholar]
- 11.Hahnke RL, Bennke CM, Fuchs BM, Mann AJ, Rhiel E, Teeling H, Amann R, Harder J. 2015. Dilution cultivation of marine heterotrophic bacteria abundant after a spring phytoplankton bloom in the North Sea. Environ Microbiol 17:3515–3526. doi: 10.1111/1462-2920.12479. [DOI] [PubMed] [Google Scholar]
- 12.Unfried F, Becker S, Robb CS, Hehemann J-H, Markert S, Heiden SE, Hinzke T, Becher D, Reintjes G, Krüger K, Avcı B, Kappelmann L, Hahnke RL, Fischer T, Harder J, Teeling H, Fuchs B, Barbeyron T, Amann RI, Schweder T. 2018. Adaptive mechanisms that provide competitive advantages to marine bacteroidetes during microalgal blooms. ISME J 12:2894–2906. doi: 10.1038/s41396-018-0243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, Kassabgy M, Huang S, Mann AJ, Waldmann J, Weber M, Klindworth A, Otto A, Lange J, Bernhardt J, Reinsch C, Hecker M, Peplies J, Bockelmann FD, Callies U, Gerdts G, Wichels A, Wiltshire KH, Glöckner FO, Schweder T, Amann R. 2012. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336:608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 14.Teeling H, Fuchs BM, Bennke CM, Krüger K, Chafee M, Kappelmann L, Reintjes G, Waldmann J, Quast C, Glöckner FO, Lucas J, Wichels A, Gerdts G, Wiltshire KH, Amann RI. 2016. Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. Elife 5:e11888. doi: 10.7554/eLife.11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chafee M, Fernàndez-Guerra A, Buttigieg PL, Gerdts G, Eren AM, Teeling H, Amann RI. 2018. Recurrent patterns of microdiversity in a temperate coastal marine environment. ISME J 12:237–252. doi: 10.1038/ismej.2017.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin-Platero AM, Cleary B, Kauffman K, Preheim SP, McGillicuddy DJ, Alm EJ, Polz MF. 2018. High resolution time series reveals cohesive but short-lived communities in coastal plankton. Nat Commun 9:266. doi: 10.1038/s41467-017-02571-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thiel V, Neulinger SC, Staufenberger T, Schmaljohann R, Imhoff JF. 2007. Spatial distribution of sponge-associated bacteria in the Mediterranean sponge Tethya aurantium. FEMS Microbiol Ecol 59:47–63. doi: 10.1111/j.1574-6941.2006.00217.x. [DOI] [PubMed] [Google Scholar]
- 18.Alonso C, Warnecke F, Amann R, Pernthaler J. 2007. High local and global diversity of Flavobacteria in marine plankton. Environ Microbiol 9:1253–1266. doi: 10.1111/j.1462-2920.2007.01244.x. [DOI] [PubMed] [Google Scholar]
- 19.D'ambrosio L, Ziervogel K, MacGregor B, Teske A, Arnosti C. 2014. Composition and enzymatic function of particle-associated and free-living bacteria: a coastal/offshore comparison. ISME J 8:2167–2179. doi: 10.1038/ismej.2014.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Preston CM, Marin IIR, Jensen SD, Feldman J, Birch JM, Massion EI, DeLong EF, Suzuki M, Wheeler K, Scholin CA. 2009. Near real‐time, autonomous detection of marine bacterioplankton on a coastal mooring in Monterey Bay, California, using rRNA‐targeted DNA probes. Environ Microbiol 11:1168–1180. doi: 10.1111/j.1462-2920.2009.01848.x. [DOI] [PubMed] [Google Scholar]
- 21.Choi CJ, Bachy C, Jaeger GS, Poirier C, Sudek L, Sarma VV, Mahadevan A, Giovannoni SJ, Worden AZ. 2017. Newly discovered deep-branching marine plastid lineages are numerically rare but globally distributed. Curr Biol 27:R15–16. doi: 10.1016/j.cub.2016.11.032. [DOI] [PubMed] [Google Scholar]
- 22.Bar-Ziv R, Moses E. 1994. Instability and “pearling” states produced in tubular membranes by competition of curvature and tension. Phys Rev Lett 73:1392. doi: 10.1103/PhysRevLett.73.1392. [DOI] [PubMed] [Google Scholar]
- 23.Yu Y, Granick S. 2009. Pearling of lipid vesicles induced by nanoparticles. J Am Chem Soc 13:14158–14159. doi: 10.1021/ja905900h. [DOI] [PubMed] [Google Scholar]
- 24.Fygenson DK, Marko JF, Libchaber A. 1997. Mechanics of microtubule-based membrane extension. Phys Rev Lett 79:4497–4500. doi: 10.1103/PhysRevLett.79.4497. [DOI] [Google Scholar]
- 25.Kantsler V, Segre E, Steinberg V. 2008. Critical dynamics of vesicle stretching transition in elongational flow. Phys Rev Lett 101:048101. doi: 10.1103/PhysRevLett.101.048101. [DOI] [PubMed] [Google Scholar]
- 26.Narsimhan V, Spann AP, Shaqfeh ES. 2014. The mechanism of shape instability for a vesicle in extensional flow. J Fluid Mech 750:144–190. doi: 10.1017/jfm.2014.248. [DOI] [Google Scholar]
- 27.Sanborn J, Oglecka K, Kraut RS, Parikh AN. 2013. Transient pearling and vesiculation of membrane tubes under osmotic gradients. Faraday Discuss 161:167–176. doi: 10.1039/C2FD20116J. [DOI] [PubMed] [Google Scholar]
- 28.Tsafrir I, Sagi D, Arzi T, Guedeau-Boudeville MA, Frette V, Kandel D, Stavans J. 2001. Pearling instabilities of membrane tubes with anchored polymers. Phys Rev Lett 86:1138–1141. doi: 10.1103/PhysRevLett.86.1138. [DOI] [PubMed] [Google Scholar]
- 29.Subramanian P, Pirbadian S, El-Naggar MY, Jensen GJ. 2018. Ultrastructure of Shewanella oneidensis MR-1 nanowires revealed by electron cryotomography. Proc Natl Acad Sci U S A 115:E3246–3255. doi: 10.1073/pnas.1718810115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCaig WD, Koller A, Thanassi DG. 2013. Production of outer membrane vesicles and outer membrane tubes by Francisella novicida. J Bacteriol 195:1120–1132. doi: 10.1128/JB.02007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Remis JP, Wei D, Gorur A, Zemla M, Haraga J, Allen S, Witkowska HE, Costerton JW, Berleman JE, Auer M. 2014. Bacterial social networks: structure and composition of Myxococcus xanthus outer membrane vesicle chains. Environ Microbiol 16:598–610. doi: 10.1111/1462-2920.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laanto E, Penttinen RK, Bamford JK, Sundberg LR. 2014. Comparing the different morphotypes of a fish pathogen—implications for key virulence factors in Flavobacterium columnare. BMC Microbiol 14:170. doi: 10.1186/1471-2180-14-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Møller JD, Barnes AC, Dalsgaard I, Ellis AE. 2005. Characterisation of surface blebbing and membrane vesicles produced by Flavobacterium psychrophilum. Dis Aquat Organ 64:201–209. doi: 10.3354/dao064201. [DOI] [PubMed] [Google Scholar]
- 34.Pérez-Cruz C, Carrión O, Delgado L, Martinez G, López-Iglesias C, Mercade E. 2013. A new type of outer membrane vesicles produced by the Gram-negative bacterium Shewanella vesiculosa M7T: implications for DNA content. Appl Environ Microbiol 79:1874–1881. doi: 10.1128/AEM.03657-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braun V, Rehn K. 1969. Chemical characterization, spatial distribution and function of a lipoprotein (murein‐lipoprotein) of the E. coli cell wall: the specific effect of trypsin on the membrane structure. Eur J Biochem 10:426–438. doi: 10.1111/j.1432-1033.1969.tb00707.x. [DOI] [PubMed] [Google Scholar]
- 36.Mizuno T. 1981. A novel peptidoglycan-associated lipoprotein (PAL) found in the outer membrane of Proteus mirabilis and other Gram-negative bacteria. J Biochem 89:1039–1049. [PubMed] [Google Scholar]
- 37.Beveridge TJ. 1999. Structures of Gram-negative cell walls and their derived membrane vesicles. J Bacteriol 181:4725–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hensgens CM, Santos H, Zhang C, Kruizinga WH, Hansen TA. 1996. Electron-dense granules in Desulfovibrio gigas do not consist of inorganic triphosphate but of a glucose pentakis(diphosphate). Eur J Biochem 242:327–331. doi: 10.1111/j.1432-1033.1996.0327r.x. [DOI] [PubMed] [Google Scholar]
- 39.Tocheva EI, Dekas AE, McGlynn SE, Morris D, Orphan VJ, Jensen GJ. 2013. Polyphosphate storage during sporulation in the gram-negative bacterium Acetonema longum. J Bacteriol 195:3940–3946. doi: 10.1128/JB.00712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao X, Schwartz CL, Pierson J, Giovannoni SJ, McIntosh JR, Nicastro D. 2017. Three-dimensional structure of the ultraoligotrophic marine bacterium “Candidatus Pelagibacter ubique.” Appl Environ Microbiol 83:e02807-16. doi: 10.1128/AEM.02807-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, McBride MJ, Subramaniam S. 2007. Cell surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J Bacteriol 189:7503–7506. doi: 10.1128/JB.00957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veith PD, Chen YY, Gorasia DG, Chen D, Glew MD, O’Brien-Simpson NM, Cecil JD, Holden JA, Reynolds EC. 2014. Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J Proteome Res 13:2420–2432. doi: 10.1021/pr401227e. [DOI] [PubMed] [Google Scholar]
- 43.Veith PD, Glew MD, Gorasia DG, Reynolds EC. 2017. Type IX secretion: the generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers. Mol Microbiol 106:35–53. doi: 10.1111/mmi.13752. [DOI] [PubMed] [Google Scholar]
- 44.McBride MJ, Nakane D. 2015. Flavobacterium gliding motility and the type IX secretion system. Curr Opin Microbiol 28:72–77. doi: 10.1016/j.mib.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 45.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reintjes G, Arnosti C, Fuchs BM, Amann R. 2017. An alternative polysaccharide uptake mechanism of marine bacteria. ISME J 11:1640–1650. doi: 10.1038/ismej.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, Rogowski A, Hamilton BS, Chen R, Tolbert TJ, Piens K, Bracke D, Vervecken W, Hakki Z, Speciale G, Munōz-Munōz JL, Day A, Peña MJ, McLean R, Suits MD, Boraston AB, Atherly T, Ziemer CJ, Williams SJ, Davies GJ, Abbott DW, Martens EC, Gilbert HJ. 2015. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517:165. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bruns A, Rohde M, Berthe-Corti L. 2001. Muricauda ruestringensis gen. nov., sp. nov., a facultatively anaerobic, appendaged bacterium from German North Sea intertidal sediment. Int J Syst Evol Microbiol 51:1997–2006. doi: 10.1099/00207713-51-6-1997. [DOI] [PubMed] [Google Scholar]
- 49.Pérez-Cruz C, Delgado L, López-Iglesias C, Mercade E. 2015. Outer-inner membrane vesicles naturally secreted by gram-negative pathogenic bacteria. PLoS One 10:e0116896. doi: 10.1371/journal.pone.0116896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steindler L, Schwalbach MS, Smith DP, Chan F, Giovannoni SJ. 2011. Energy starved Candidatus Pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PLoS One 6:e19725. doi: 10.1371/journal.pone.0019725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohen M, Santarella R, Wiesel N, Mattaj I, Gruenbaum Y. 2008. Chapter 21 Electron microscopy of lamin and the nuclear lamina in Caenorhabditis elegans. Methods Cell Biol 88:411–429. doi: 10.1016/S0091-679X(08)00421-4. [DOI] [PubMed] [Google Scholar]
- 52.Lepault J, Booy FP, Dubochet J. 1983. Electron microscopy of frozen biological suspensions. J Microsc 129:89–102. doi: 10.1111/j.1365-2818.1983.tb04163.x. [DOI] [PubMed] [Google Scholar]
- 53.Passey S, Pellegrin S, Mellor H. 2007. Scanning electron microscopy of cell surface morphology. Curr Protoc Cell Biol 37:4–17. doi: 10.1002/0471143030.cb0417s37. [DOI] [PubMed] [Google Scholar]
- 54.Kulkarni HM, Jagannadham MV. 2014. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology 160:2109–2121. doi: 10.1099/mic.0.079400-0. [DOI] [PubMed] [Google Scholar]
- 55.Otto A, Bernhardt J, Meyer H, Schaffer M, Herbst FA, Siebourg J, Mäder U, Lalk M, Hecker M, Becher D. 2010. Systems-wide temporal proteomic profiling in glucose-starved Bacillus subtilis. Nat Commun 1:137. doi: 10.1038/ncomms1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Searle BC. 2010. Scaffold: a bioinformatic tool for validating MS/MS‐based proteomic studies. Proteomics 10:1265–1269. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- 57.Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, Washburn MP. 2006. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40:303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kito K, Ito T. 2008. Mass spectrometry-based approaches toward absolute quantitative proteomics. Curr Genomics 9:263–274. doi: 10.2174/138920208784533647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang M, You J, Bemis KG, Tegeler TJ, Brown DP. 2008. Label-free mass spectrometry-based protein quantification technologies in proteomic analysis. Brief Funct Genomics 7:329–339. doi: 10.1093/bfgp/eln031. [DOI] [PubMed] [Google Scholar]
- 60.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All proteome raw data were uploaded to the ProteomeXchange Consortium (proteomecentral.proteomexchange.org) via the PRIDE partner repository (60) and are accessible with the data set identifier PXD012522.